Vertebrates synthesize a diverse set of steroids and bile acids that undergo bacterial biotransformations. The endocrine literature has principally focused on the biochemistry and molecular biology of host synthesis and tissue-specific metabolism of steroids. Host-associated microbiota possess a coevolved set of steroid and bile acid modifying enzymes that match the majority of host peripheral biotransformations in addition to unique capabilities. The set of host-associated microbial genes encoding enzymes involved in steroid transformations is known as the sterolbiome.
KEYWORDS: androgen; steroid-17,20-desmolase; Clostridium scindens; secondary bile acids; microbiome; cholesterol; sterolbiome; androgen
ABSTRACT
Vertebrates synthesize a diverse set of steroids and bile acids that undergo bacterial biotransformations. The endocrine literature has principally focused on the biochemistry and molecular biology of host synthesis and tissue-specific metabolism of steroids. Host-associated microbiota possess a coevolved set of steroid and bile acid modifying enzymes that match the majority of host peripheral biotransformations in addition to unique capabilities. The set of host-associated microbial genes encoding enzymes involved in steroid transformations is known as the sterolbiome. This review focuses on the current knowledge of the sterolbiome as well as its importance in medicine and agriculture.
INTRODUCTION
Host-associated bacteria are estimated to be roughly numerically equal to host cells (1) but possess ∼99% of the functional genes in the host (2). Microbiome compositions differ between health and disease states (3). Phenotypic transfer of a disease through fecal transplant establishes microbiomes as causal in the disease process (4). Further delving into mechanisms of causation requires determining relevant host-microbe interactions. In this regard, microbial metabolic contributions to the host metabolome have come sharply into focus (5, 6). However, we are still working toward identifying microbial genes associated with metabolites generated by host-associated microbiota. Much of the early mechanistic work on the human microbiome was with reference to the “glycobiome,” where complex pathways for polysaccharide degradation were determined (7–9). The glycobiome continues to present a “call to arms” in working out host-microbe interactions relating to the degradation of dietary and endogenous carbohydrates and the formation of short-chain fatty acids. It also provides a useful paradigm for organizing work focused on other areas of microbial metabolism.
The vertebrate host converts cholesterol to a diversity of sterols, neutral steroids, and bile acids, whose complexity is markedly enhanced by the metabolic activity of host-associated microbiota. My group previously introduced the term “sterolbiome” to describe “the genetic potential of the gut microbiome to produce endocrine molecules from endogenous and exogenous steroids in the mammalian gut” (10). The sterolbiome concept fits into the overarching and burgeoning field of microbial endocrinology (11), the notion of the bidirectional hormonal communication (interkingdom signaling) between host and microbe (Table 1). While much of the focus of microbial endocrinology has been on bacterial growth and virulence, this coevolved communication through small molecules has the potential to affect microbiome structure, as well as a variety of host functions, including appetite and digestion, immunity, endocrine function, cardiovascular health, mating and species recognition, and behavior. The original description of the sterolbiome focused solely on bile acids (10); however, here I expand upon this concept to include cholesterol and steroid hormones in both human and nonhuman vertebrates. The focus of this review is primarily on key biotransformations catalyzed by enzymes encoded by the vertebrate sterolbiome and the potential role of these pathways in medicine and agriculture.
TABLE 1.
Sterolbiome metabolites alter host physiology
| Host target (abbreviation) | Sterolbiome agonists | Disease relevance | Reference(s) |
|---|---|---|---|
| Nuclear receptor | |||
| Farnesoid X receptor (FXR) | DCA, LCA | Metabolic syndrome, colon cancer, liver cancer | 10, 57, 187 |
| Pregnane-activated receptor (PXR) | LCA, DCA | Metabolic syndrome, colon cancer, liver cancer | 10, 57, 187 |
| Vitamin D receptor (VDR) | 3-Oxo-LCA, LCA | Colon cancer, liver cancer, cholestasis | 10, 57, 187 |
| Androgen receptor (AR) | 11-Oxyandrogens | Colon cancer, prostate cancer, immune system function | 141, 188 |
| Retinoic acid receptor γ T (RORγT) | Allo-DCA, allo-LCA, isoalloLCA | Colon cancer, IBD,a liver regeneration, liver cancer | 61 |
| G protein-coupled receptor | |||
| TGR-5 | DCA, LCA | Metabolic syndrome, cancer, liver regeneration | 57 |
| M2,3-muscarinic receptor | DCA, LCA, TDCA, TLCA, GDCA, GLCA | Colon cancer, fetal heart arrhythmia | 57, 189–191 |
| Sphingosine-1-phosphate receptor 2 | TLCA, TDCA | Metabolic syndrome | 57 |
| Membrane androgen receptor | 11-Oxyandrogens | Prostate cancer, colon cancer | 192 |
| Host enzyme | |||
| 11β-Hydroxysteroid dehydrogenase1/2 | 7-Oxo-LCA, 11-oxyandrogens, 11-oxy-progesterone | Hypertension, metabolic syndrome | 97, 126 |
| Transcription/cell adhesion | |||
| β-Catenin | DCA, LCA | Colon cancer | 193 |
IBD, inflammatory bowel disease.
THE GUT STEROLBIOME
Cholesterol metabolism by gut bacteria.
Cholesterol is the precursor to all steroids, including sex steroids and bile acids. Roughly 1 g of cholesterol reaches the human gut each day, originating from diet, bile, and sloughed intestinal cells (12). Anaerobic bacteria have evolved enzymes to utilize cholesterol as an electron acceptor, resulting in the formation of coprostanol (12). In humans, coprostanol formation can be prevented through oral antibiotic treatment (13). Indeed, coprostanol is not observed in the gut contents of germfree (GF) animals (14). Moreover, conversion of cholesterol to coprostanol has been shown in vitro with fecal suspensions (12, 14). Unlike cholesterol, coprostanol is not absorbed efficiently in the gastrointestinal (GI) tract (15, 16). An inverse relationship between serum cholesterol levels and the fecal ratio of cholesterol to coprostanol has been reported. However, individual microbiomes differ in the capacity to metabolize cholesterol (16). Importantly, evidence in mice and rabbits suggests that coprostanoligenic gut bacteria may serve as probiotics that have potential to regulate serum cholesterol levels, although this was not observed in laying hens (17–19).
Unfortunately, work in this area has been hampered by the difficulty of isolating and culturing coprostanoligenic bacteria (20, 21). The first bacterium capable of converting cholesterol to coprostanol was isolated from rat cecal contents and displayed an absolute requirement for cholesterol or other unsaturated steroids (22), with limited growth in the absence of homogenized brain or brain lipid extracts (22). It was later shown that plasmenylethanolamine from brain lipid extraction is a growth factor for Eubacterium strain ATCC 21408 (20). In vitro studies also identified bacteria belonging to Lactobacillus, Bifidobacterium, and Clostridium as potentially capable of converting cholesterol to coprostanol (23).
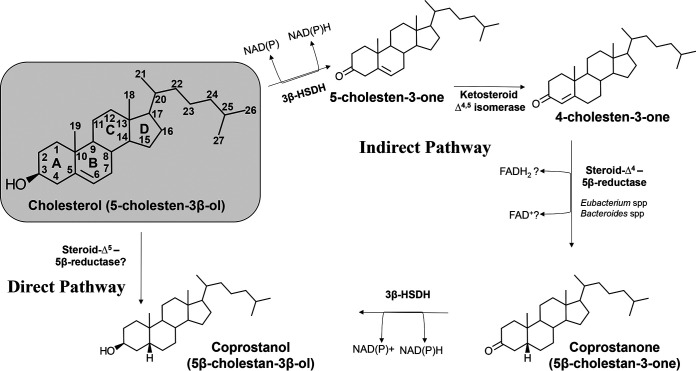
Two main biochemical pathways for conversion of cholesterol to coprostanol have been proposed, largely based on radiometric studies of whole cells or cecal contents (14, 24–26). The direct pathway (Fig. 1) involves the direct biohydrogenation of cholesterol (cholest-5-en-3β-ol) to coprostanol (5β-cholestan-3β-ol) (24, 25). Studies by Rosenfeld et al. showed that [3α-2H]- and [3α-3H]-cholesterol was converted to coprostanol in vivo and in vitro with retention of most of the label, suggesting that the 3β-hydroxyl group was not metabolized (24, 25). Thereafter, Björkhem et al. demonstrated removal and reinsertion of the 3α-3H label via enzyme-bound cofactors in sequential oxidative and reductive steps (14, 26). They showed using [4β-3H,4-14C]- and [3α-3H,4-14C]cholesterol extensive oxidoreduction at C-3, in addition to transfer of tritium from C-4 to C-6 indicating an isomerization reaction, which indicated an indirect pathway from cholesterol to coprostanol (26). Indeed, the greatest support is for the indirect pathway, which combines results from both radiometric studies in mixed rat cecal cultures as well as studies in whole cells of Eubacterium strain ATCC 21408, Eubacterium coprostanoligenes ATCC 51222, and Bacteroides sp. strain D8 (Table 2) (14, 22, 27–29). The indirect pathway involves oxidation of the 3β-hydroxy group (5-cholesten-3-one), followed by isomerization of the Δ5 bond to Δ4 (cholest-4-en-3-one), reduction of the Δ4 bond to coprostanone, and then reduction of the 3-oxo group to coprostanol (28) (Fig. 1). Addition of the above-mentioned intermediates to pure cultures resulted in formation of coprostanol (22, 27, 29). Identification of the genes responsible for coprostanol formation and determining substrate specificity for enzymes, particularly whether enzymes involved in isomerization and Δ4 reduction recognize both 3-keto and 3β-hydroxy group, will be important in testing the validity of the proposed direct and indirect pathway (Fig. 1).
FIG 1.
Microbial pathways for the conversion of cholesterol to coprostanol in the GI tract. Gray box, Cholesterol structure with numbering. There are three pathways proposed for the conversion of cholesterol to coprostanol. Refer to the text for a detailed description.
TABLE 2.
Bacterial species contributing to the sterolbiome
| Bacterial species | Source of isolation | Enzyme(s) or gene/operona | Substrate(s) | Product(s) | Reference(s) |
|---|---|---|---|---|---|
| Cholesterol metabolism | |||||
| Eubacterium ATCC 21408 | Large intestine | 3β-HSDH, Δ4-reductase, Δ4,5-isomerase | Cholesterol | Coprostanol | 22, 28 |
| E. coprostanoligenes ATCC 51222 | Hog sewage lagoon | 3β-HSDH, Δ4-reductase, Δ4,5-isomerase | Cholesterol, 4-cholesten-3-one, coprostanone | Coprostanol | 27 |
| Bacteroides sp. D8 | Large intestine | 3β-HSDH, Δ4-reductase, Δ4,5-isomerase | Cholesterol | Coprostanol | 29 |
| Eubacterium strain 403 | Baboon feces | 3β-HSDH, Δ4-reductase, Δ4,5-isomerase | Cholesterol, allocholesterol, 4-cholesten-3-one, coprostanone, androst-5-en-3β-ol-17-one | Coprostanol, 5β-androstane-3β-ol-17-one | 194 |
| Bile acid metabolism | |||||
| Clostridium perfringens | Large intestine | BSH, 3α-HSDH, 12α-HSDH | Conjugated bile acids | Unconjugated bile acids | 196 |
| Lactobacillus spp. | Large intestine | BSH | Conjugated bile acids | Unconjugated bile acids | 197, 198 |
| Bifidobacterium spp. | Large intestine | BSH | Conjugated bile acids | Unconjugated bile acids | 43, 207 |
| Enterococcus spp. | Large intestine | BSH | Conjugated bile acids | Unconjugated bile acids | 44 |
| Methanobrevibacter smithii | Large intestine | BSH | Conjugated bile acids | Unconjugated bile acids | 40 |
| Eggerthella lenta | Large intestine | 3α-HSDH, 3β-HSHD, 7α-HSDH, 12α-HSDH | Unconjugated bile acids | Oxo-epimers and bile acid epimers | 32, 83 |
| Ruminococcus gnavus | Large intestine | 3α-HSDH, 3β-HSHD | Unconjugated bile acids | Oxo-epimers and bile acid epimers | 31 |
| Escherichia coli | Large intestine | 7α-HSDH | Unconjugated bile acids | Oxo-epimers and bile acid epimers | 91 |
| Bacteroides spp. | Large intestine | BSH, 7α-HSDH | Conjugated bile acids; unconjugated primary bile acids | Unconjugated bile acids, oxo-bile acids | 52, 195, 199 |
| Clostridium paraputrificum | Large intestine | 12β-HSDH | Unconjugated bile acids | Oxo-epimers and bile acid epimers | 88, 89 |
| Blautia producta | Large intestine | 3α-HSDH, 3β-HSHD | Unconjugated bile acids | Oxo-epimers and bile acid epimers | 200 |
| Clostridium scindens | Large intestine | bai operon, 12α-HSDH, 7α-HSDH | Primary bile acids (CA, CDCA, UDCA) | Secondary bile acids (DCA, LCA), oxo-bile acids | 33, 38, 80, 201 |
| Clostridium hiranonis | Large intestine | bai operon, 12α-HSDH, 7α-HSDH | Primary bile acids (CA, CDCA, UDCA) | Secondary bile acids (DCA, LCA), oxo-bile acids | 202, 203 |
| Clostridium hylemonae | Large intestine | bai operon, 12α-HSDH, 7α-HSDH | Primary bile acids (CA, CDCA, UDCA) | Secondary bile acids (DCA, LCA), oxo-bile acids | 203, 204 |
| Clostridium bolteae | Large intestine | Bile acid conjugase | Primary bile acids (CA, CDCA, UDCA) | Tyro-, phenylalano-, leucho-bile acid conjugates | 95 |
| Clostridium limnosum | Large intestine | 7α-HSDH; 7β-HSDH | Primary bile acids (CA, CDCA, UDCA) | Oxo-epimers and bile acid epimers | 205 |
| Glucocorticoid metabolism | |||||
| Clostridium scindens ATCC 35704 | Large intestine | desABCD operon | Cortisol, cortisone, 11-desoxy-cortisol, allo-tetrahydrocortisol, 20α-dihydrocortisol | 11β-Hydroxyandrostenedione, 11-oxyandrogens | 76, 107–109, 207 |
| Butyricicoccus desmolans | Large intestine | desABE operon | Cortisol, cortisone, 11-desoxy-cortisol, allo-tetrahydrocortisol, 20β-dihydrocortisol | 11β-Hydroxyandrostenedione, 11-oxyandrogens | 110, 111 |
| Clostridium cadaveris | Large intestine | desABE operon | Cortisol, cortisone, 11-desoxy-cortisol, allo-tetrahydrocortisol, 20β-dihydrocortisol | 11β-Hydroxyandrostenedione, 11-oxyandrogens | 110 |
| Propionimicrobium lymphophilum | Urinary tract | desABE operon | Cortisol, cortisone, 11-desoxy-cortisol, allo-tetrahydrocortisol, 20β-dihydrocortisol | 11β-Hydroxyandrostenedione, 11-oxyandrogens | 109, 111 |
| Bifidobacterium adolescentis | Large intestine | desE | Cortisol, cortisone, 11-desoxy-cortisol, allo-tetrahydrocortisol, 20β-dihydrocortisol | 11β-Hydroxyandrostenedione, 11-oxyandrogens | 111, 112 |
| Eggerthella lenta | Large intestine | 21-Dehydroxylase | Corticosterone, DOC | 11β-Hydroxyprogesterone | 135 |
| Intestinibacillus sp. Marseille-P4005 | Large intestine | desABE operon | Cortisol, cortisone | 11β-Hydroxyandrostenedione, 11-oxyandrogens | 109 |
| Arcanobacterium urinimassiliense | Urinary tract | desABE operon | Cortisol, cortisone, 11-desoxy-cortisol, allo-tetrahydrocortisol, 20β-dihydrocortisol | 11β-Hydroxyandrostenedione, 11-oxyandrogens | 109 |
| Sex steroid metabolism | |||||
| C. innocuum | Large intestine | 17β-HSDH, 3β-HSDH, 5β-reductase | Estrone, progesterone, testosterone | 17β-Estradiol, 3β-hydroxy-5β-pregnan-20-one, 5β-androstan-3β,17β-diol | 138 |
| C. paraputrificum | Large intestine | 17β-HSDH, 3α-HSDH, 5β-reductase | Estrone, progesterone, testosterone | 17β-Estradiol, 3β-hydroxy-5β-pregnan-20-one, 5β-androstan-3α,17β-diol | 138 |
| Bacteroides fragilis | Large intestine | 17β-HSDH | (11β-Hydroxy)androstenedione | (11β-Hydroxy)testosterone | 137 |
| Eggerthella lenta 144 | Large intestine | 16-Dehydroxylase | 16α-Hydroxyprogesterone | 17α-Pregnanolone | 122, 123 |
| Large intestine | 17β-HSDH | Androstenedione | Testosterone | 82 | |
| Bacteroides melaninogenicus | Oral cavity | 5β-Reductase? | Estradiol, progesterone | 5β-Pregnane-3,20-dione? | 147 |
| Porphyromonas gingivalis | Oral cavity | 5β-Reductase? | Testosterone | DHT | 151 |
| Prevotella intermedia | Oral cavity | 5β-Reductase | Testosterone | DHT | 151 |
| Actinobacillus actinomycetemcomitans | Oral cavity | 5β-Reductase | Testosterone | DHT | 151 |
| Clostridium scindens VPI 12708 | Large intestine | 17α-HSDH | Androstenedione | Epitestosterone | 136 |
| 16-Ene-steroids | |||||
| Corynebacterium sp. | Axillae | 16-Reductase, 3α-HSDH, 3b-HSDH, Δ1-reductase, Δ4-reductase, 17β-HSDH | 5,16-Androstadien-3-ol | 1-Androstenedione, 4-androstenedione, pregna-1,4-dien-3,30-dione, pregna-4,6-dien-3,20-dione, 4,16-androstadien-3-one, 5β-androst-16-en-3-ol | 159, 160 |
BSH, bile salt hydrolase.
To this end, Björkhem et al. partially purified NADH-dependent 3-oxo-Δ4-steroid 5β-reductase after generating cell extracts from rat cecal contents; however, the identity of the gene(s) is not known (14). The enzyme catalyzing the initial step of the indirect pathway has been proposed to be an oxygen-dependent enzyme, cholesterol oxidase (23). Presumably, this is because there is a well-characterized cholesterol oxidase expressed by aerobic soil bacteria. However, another plausible scenario under anaerobic conditions is oxidation of the 3β-hydroxyl group by an oxygen-independent NAD(P)H-dependent 3β-hydroxysteroid dehydrogenase (3β-HSDH) in the short-chain dehydrogenase, aldo/keto-reductase, or medium-chain reductase family (30). Examples of gut bacterial NAD(P)-dependent 3β-HSDH enzymes that recognize bile acids and neutral steroids have been reported (31, 32). NAD(P)H-dependent flavoproteins have been identified in gut bacteria capable of metabolizing bile acid 3-oxo-Δ4 intermediates (33) and progestins (34). Genes encoding 3-ketosteroid-Δ4,5-isomerase resulting in a 4-cholen-3-one intermediate have not been identified. The ketosteroid isomerase from Comamonas testosteroni (208) does not appear to have homologs in the gut microbiome based on a BLAST search (data not shown); however, the genomes of coprostanoligenic bacteria, such as Eubacterium VPI 21408, E. coprostanoligenes, and Bacteroides sp. D8, are reported to have been sequenced, but these sequences are not publicly available at present.
Bile salt biotransformations.
Bile acids are synthesized primarily in the liver from cholesterol through a multienzyme process (35). In aqueous solution, bile acids act as amphipathic detergents that self-associate into mixed micelles with cholesterol, lipid-soluble vitamins, and other lipids, including some pharmaceuticals. Chenodeoxycholic acid (CDCA; 3α,7α-dihydroxy-5β-cholan-24-oic acid) is the primary bile acid from which other bile acids are synthesized (35). In humans, cholic acid (CA; 3α,7α,12α-trihydroxy-5β-cholan-24-oic) is generated by the 12α-hydroxylation of CDCA, resulting in roughly equal proportions of the two primary bile acids in bile (35). In the liver, primary bile acids are then amidated (conjugated) to the amino groups of glycine or taurine, which lowers the pKa values by 2.4 to 5 units, respectively (36). The result is the formation of effective detergents involved in dietary lipid absorption in the small bowel. An initial step in microbial metabolism of bile salts in the intestines is the hydrolysis of the amide bond linking amino acid to bile acid.
Bile salt deconjugation.
Bile salt hydrolases (BSH; EC 3.5.1.24) are members of the choloylglycine hydrolase family, which includes penicillin V amidase (EC 3.5.1.11) (37) (Fig. 2). Bile salt hydrolysis is a substrate-limiting reaction and goes to completion in the GI tract, as evident in fecal bile acid profiles of healthy individuals who are not on antibiotics (38, 39). Bile salt hydrolase-encoding genes are generally constitutively expressed and intracellular, with some notable exceptions for both the former and the latter (38). Metagenomic surveys reveal that bsh genes are specific to vertebrate gut communities and widely distributed among bacterial genera from all major phyla (Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria) of the domain Bacteria and gut methanogens from the domain Archaea (Table 2) (40).
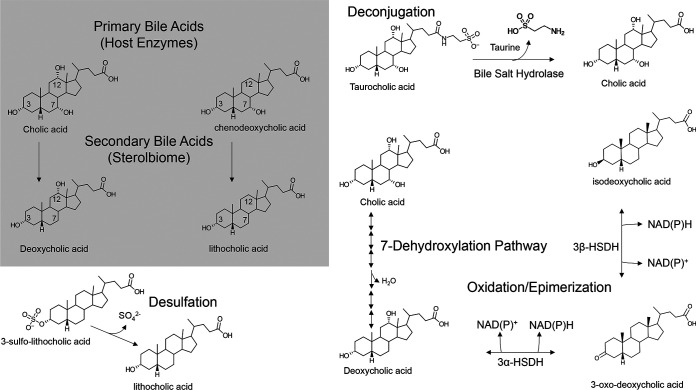
FIG 2.
Major bile salt biotransformation reactions in the GI tract. Gray box, structures of the two primary bile acids synthesized by the human liver and the major secondary bile acids generated from gut bacterial 7α-dehydroxylation. After conjugation to taurine or glycine in the liver, conjugated bile acids undergo hydrolysis in the large intestine catalyzed by bacterial bile salt hydrolase enzymes. Unconjugated primary bile acids can be reversibly oxidized and epimerized at each hydroxyl group by pyridine nucleotide-dependent hydroxysteroid dehydrogenase enzymes. Removal of the 7-hydroxyl group is catalyzed by a small population of clostridia that encode a bile acid-inducible (Bai) multienzyme pathway. Lithocholic acid, formed by bile acid 7-dehydroxylation of CDCA or UDCA, is highly toxic. When absorbed by colonocytes, LCA is sulfated at the 3-position. Bacteria have evolved desulfurases that hydrolyze 3-sulfo-LCA.
Despite differences in subunit composition, pH optimum, substrate specificity, and kinetic properties, BSH share conserved active-site amino acids (Cys2, Asp21, Asn175, and Arg228) in common with the BSH from Clostridium perfringens (38). Structures have been determined for some BSH, including those of Lactobacillus salivarius (41), C. perfringens (42), Bifidobacterium longum (43), and Enterococcus faecalis (44). Understanding the mechanisms for bile salt hydrolysis and the physiological effect of inhibition of BSH function in vivo may have important applications in both medicine and agriculture (45–48). The inhibition of BSH, particularly in lactobacilli, is predicted to improve weight gain in poultry (48, 49). Additional structural studies, particularly for BSH with diverse substrate specificity with bound substrates, would greatly aid in the development of inhibitors. Indeed, recent work identified a covalent BSH inhibitor that prevented deconjugation in conventional mice (209). Mechanisms for inverse relation between BSH activity and weight gain may include solubilization of dietary lipids (50), altered glucose and lipid metabolism due to changes in bile acid cellular signaling (51, 52), and reduction in the formation of lithocholic acid (LCA), which is hepatotoxic in poultry (53). Conversely, increased BSH activity functions to lower serum cholesterol in laboratory animals and humans (54). Thus, regulating the activity of certain sterolbiome enzymes may be important for production animals, as well as for cardiovascular health in humans.
Bile acid 7-dehydroxylation.
The fecal bile acid profile of healthy conventional vertebrates is predominated by hydrophobic secondary bile acids, particularly DCA and LCA (Fig. 2) (38). In contrast, the fecal bile acid profile of diseases such as inflammatory bowel disease and advanced cirrhosis (55) and patients with antibiotic-induced dysbiosis who acquire Clostridium difficile infection (56) are enriched in a mix of conjugated primary bile acids and unconjugated primary bile acids. Hydrophobic secondary bile acids have greater affinity for several host nuclear receptors and G-protein-coupled receptors than many primary bile acids (Table 1), indicating cross talk and coevolution between host and microbe (57). Among beneficial functions of hydrophobic secondary bile acids (DCA and LCA) are resistance against pathogens (56), regulation of serotonin synthesis (58), activation of cellular signaling that induces secretion of antimicrobial peptides (59), and regulation of immune function (57–61). The concentration of hydrophobic secondary bile acids may be key to the balance between normal physiological function, and the promotion of GI diseases because our diets and life span have changed drastically during the industrial and postindustrial era. Western diets, high in saturated fat and animal protein, select for increased bile acid secretion into the GI tract and expansion of Clostridium cluster XVIa, V, and IX organisms capable of converting host primary bile acids to hydrophobic secondary bile acids (Table 2) (62). Increased levels of DCA and LCA are associated mechanistically with chronic disease such as cholesterol gallstones (63), colon cancer (64–68), liver cancer (69, 70), and correlated with Alzheimer’s disease (71, 72).
In 1980, the first bacterium capable of converting CA to DCA, Eubacterium sp. strain VPI 12708 (73) (now C. scindens strain VPI 12708) (74), was isolated in pure culture, followed by the type strain Clostridium scindens ATCC 35704 (75). The addition of primary bile acids, such as CA and CDCA (but not ursodeoxycholic acid [UDCA]), to the culture medium of C. scindens strain VPI 12708 resulted in induction of multiple polypeptides on two-dimensional (2D) SDS-PAGE (76). During the late 1980s and early 1990s, a polycistronic bile acid-inducible (bai) regulon was cloned and sequenced (38). Radiolabeled cholic acid intermediates were separated after incubation with cell extracts of C. scindens strain VPI 12708 and identified by mass spectrometry (77). Chemical synthesis of each of these cholic acid intermediates, and addition of each to cell extracts of C. scindens strain VPI 12708, resulted in formation of DCA (78). The association of particular biochemical steps with purified native or recombinant Bai enzymes has expanded our understanding of this complex multistep process responsible for the formation of secondary bile acids such as DCA and LCA (reviewed previously [10, 38, 79]) (Fig. 2).
Recently, my group determined global differential gene expression (by transcriptome sequencing [RNA-Seq]) caused by the addition of CA or DCA (80). Several novel candidate genes involved in bile acid metabolism were identified, including several likely candidates for secondary bile acid efflux pumps. A recent report demonstrated that the bai gene cluster engineered in Clostridium sporogenes, or overexpressed in vitro, is sufficient to convert CA to DCA, indicating potential roles for BaiCD and BaiH in both the oxidative and reductive arms of the bile acid 7α-dehydroxylation pathway (210). Further work will be needed to determine the detailed substrate specificity of Bai enzymes and whether BaiCD and BaiH serve as the core reductive enzymes acting on the bile acid ring structure in bile acid 7α-dehydroxylating clostridia or serve an ancillary function as the bile acid concentration increases. As yet, genetic tools to manipulate the organisms encoding the bai pathway have yet to be reported.
Bile acid oxidation and epimerization.
Gut bacteria have evolved a suite of pyridine nucleotide-dependent hydroxysteroid dehydrogenases that are both regiospecific (C-3 versus C-7 hydroxy groups) and stereospecific (α- versus β-hydroxyl orientation) (79). For the bile acid chemist, these enzymes have practical application in the quantification of bile acids as well as the generation of bile acid standards (81). One of the more versatile bile acid oxidizers is Eggerthella lenta (formerly Eubacterium lentum). In rich growth medium under a nitrogen atmosphere, some Eggerthella lenta strains quantitatively convert cholic acid to trioxocholanoic acid, which is hypothesized to provide reducing equivalents for reductive carboxylation via the Wood-Ljundahl pathway (82, 83). E. lenta also epimerizes the 3α-hydroxy group, forming what are known as “iso”-bile acids (3β-hydroxy) (31, 82). Iso-bile acid derivatives of DCA and LCA make up a substantial quantity of bacterial fecal bile acid metabolites (39). Bacterial epimerization of bile acids results in partial hydrophilic character to both faces of the steroid ring system, which reduces the detergent strength of the bile acid and reduces its toxicity to both bacterial and host cells. The liver is capable of epimerization of iso-bile acids, loss of which results in an inability to solubilize lipids in the diet (84).
My group’s in vitro work suggests that primary bile acids that are oxidized and epimerized at the 3-hydroxyl group are less rapidly converted to DCA or LCA by Clostridium scindens (82). Indeed, since C. scindens appears to lack appreciable 3β-HSDH activity (85), the conversion of CA or CDCA to iso-CA or iso-CDCA, respectively, would block removal of the 7α-hydroxyl group by C. scindens (38). This is because the removal of the 7α-hydroxyl group requires sequential oxidation of the 3α-hydroxy followed by ring-A oxidation. Urso-bile acids (7β-hydroxy) such as the therapeutic bile acid UDCA, are used to treat biliary disorders but are subject to 7β-dehydroxylation. The rate of bile acid 7β-dehydroxylation is lower than the rate of bile acid 7α-dehydroxylation (86). Urso-bile acids can be reversibly epimerized by gut bacteria and 7α-dehydroxylated by a few species of clostridia (87). In contrast, there is very little known about 12β-hydroxy bile acids (88, 89). Co-culture between E. lenta (12α-HSDH) and Clostridium paraputrificum (12β-HSDH) results in the formation of epicholic acid (88). The extent to which epicholic acid is 7α-dehydroxylated is not currently known. Structures have been determined for gut bacterial 3α-HSDH (90), 7α-HSDH (91), and 7β-HSDH (92), which are beginning to shed light on the regio- and stereospecificity of these enzymes.
Bile acid esterification, polymerization, and amidation.
There are reports in the literature that gut bacteria are capable of forming ethyl esters of bile acids as well as long-chain fatty acid esters of LCA (93). In addition, one report identified polyesters of DCA (94). These reactions very likely represent detoxification mechanisms by gut bacteria aimed at reducing the concentration of hydrophobic bile acids. The gut bacteria responsible for these reactions are not known, nor have the genes or gene products catalyzing these reactions been identified. A recent report indicates that Clostridium bolteae strains are capable of generating three new bile acid amides from phenylalanine (phenylalanocholic), tyrosine (tyrosocholic acid), and leucine (leuchocholic acid) (95). These conjugated bile acids were not deconjugated in mixed fecal suspension and were found to be potent farnesoid X receptor (FXR) agonists, suggesting that this may have physiological effects, potentially mediating host-microbe communication (95).
Corticosteroids.
Steroid hormones play critical roles in the regulation of salt and water balance, metabolism, and stress responses and in sexual differentiation and reproduction. The primary steroidogenic tissues include the adrenal cortex, the gonads, and the placenta, which are capable of de novo cholesterol and steroid biosynthesis from cholesterol (96). Vertebrates have evolved a suite of enzymes expressed in particular tissues in the body to modify steroid functional groups, altering their affinity to host receptors and thus their potency (96). The 11β-hydroxy group can be reversibly oxidized by NAD+-dependent 11β-HSD1 isoform, converting cortisol (active form) to cortisone (inactive form), the latter of which is not a ligand glucocorticoid receptor (97). The 17β-hydroxyl group is characteristic of potent androgens and estrogens, the oxidation of which by NADP-dependent 17β-HSD isoforms reversible (in)activates these steroids (HSD17B2/4) (96). 3-Keto/ring A reduction, primarily in the liver, to either 5α- or 5β-reduced forms inactivates steroids and is considered from the host point of view irreversible (96). Glucocorticoids and progesterone are also subject to 20α/20β-reduced through aldo-keto reductase (AKR1C1) (98). With the exception of metabolism at C-11, the human sterolbiome encodes enzymes capable of each reaction host tissues perform, as well as unique biotransformations. The human microbiome should thus be considered a potentially important component of the host endocrine system.
Steroid-17,20-desmolase pathway.
The adrenal product 11β-hydroxyandrostenedione (11β-OHAD) has been the subject of considerable recent controversy, after many years of marginal interest since its discovery in the 1950s. 11β-OHAD is formed primarily in the adrenal gland through 11β-hydroxylation of androstenedione, with some peripheral side chain cleavage of cortisol (99). Early clinical studies examining the treatment of ulcerative colitis by rectal infusion of cortisol provided the first evidence that human-associated bacteria also catalyze the side chain cleavage of cortisol (C21), resulting in 11β-OHAD (C19) (100, 101). Patients given doses of cortisol exhibited a spike in urinary excretion of 11β-OHAD derivatives (11-oxyandrogens). Oral antibiotic treatment prevented the spike in urinary 11-oxyandrogens (100, 101). This provided evidence that gut bacteria are capable of generating 11-oxyandrogens in the gut that are absorbed into the bloodstream. Formation of 11-oxyandrogens from cortisol in feces is common among those vertebrates that produce cortisol (102). Measurement of glucocorticoid levels in feces collected at a distance from an animal is a noninvasive measurement of stress in both the wild and captivity (103).
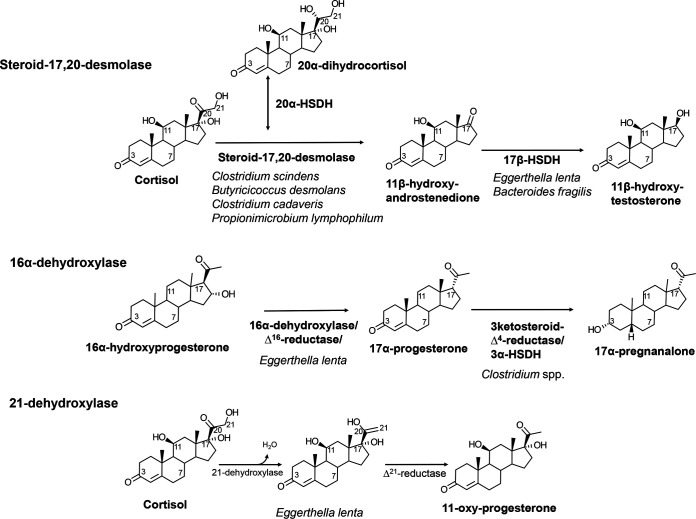
In 1981, Cerone-McLernon et al. definitively established that fecal microbes were capable of cortisol side chain cleavage by identifying radiolabeled products of cortisol incubated with human fecal suspensions (104). A few years later, Clostridium scindens ATCC 35704 was isolated and shown to generate two products from cortisol, 20α-dihydrocortisol and 11β-OHAD (Fig. 3) (75, 105). Both NADH-dependent cortisol 20α-HSDH and steroid-17,20-desmolase activities are induced in C. scindens ATCC 35704 by the addition of cortisol to the culture medium (106). The genes encoding NADH-dependent cortisol 20α-HSDH and steroid-17,20-desmolase were later identified by comparing mRNA profiles between cortisol-induced whole cells of C. scindens ATCC 35704 and uninduced control mRNA (107). A highly induced gene cluster was identified that was named desABCD for hypothesized desmolase activity (107).
FIG 3.
Metabolism of cortisol and 16α-hydroxyprogesterone by gut bacteria. The cortisol side chain is metabolized in two important pathways. The first is side chain cleavage, which converts a C21 glucocorticoid into a precursor to C19 11-oxyandrogens. Indeed, the conversion of 17-ketone to 17β-hydroxy by bacterial NAD(P)H-dependent 17β-HSDH along with 5α reduction by some clostridia produces a potent androgen. 16α-Hydroxyprogesterone is 16α dehydroxylated and Δ16 reduced, followed by reduction of the 3-oxo-Δ (123–125), resulting in 17α-pregnanalone.
desA and desB are annotated as “transketolase” genes, while desC is annotated as a zinc-dependent medium-chain dehydrogenase (MDR), and desD encodes a putative membrane transport protein (107). The desC gene from C. scindens ATCC 35704 was cloned and overexpressed in Escherichia coli and shown to encode a 40-kDa NADH-dependent 20α-HSDH with substrate specificity identical to that of the native enzyme (107). Phylogenetic analysis of DesC revealed that this protein appears to have evolved from MDR family proteins but has so far been found only in C. scindens (107). Our group recently reported the 2.0-Å apo-structure and proposed catalytic mechanism of DesC from C. scindens ATCC 35704 based on hybrid quantum mechanical molecular modeling (QM/MM) validated by site-directed mutagenesis and biophysical techniques (207).
The recombinant DesAB gene products from C. scindens were recently shown to catalyze the side chain cleavage of cortisol (108). Phylogenetic and sequence similarity network analysis revealed that desAB genes were found in Butyricicoccus desmolans and Clostridium cadaveris, as previously reported, as well as Intestinibacillus sp. strain Marseille-P4005, which remains uncharacterized with respect to cortisol metabolism (Table 2) (109, 110). Butyricicoccus desmolans and Clostridium cadaveris, as well as Bifidobacterium adolescentis, were also reported previously to express cortisol 20β-HSDH activity (110). Examination of the genomes of B. desmolans and C. cadavaris revealed desAB genes adjacent to a gene encoding a putative short-chain dehydrogenase family (SDR) our group hypothesized and demonstrated to encode cortisol 20β-HSDH, designated desE (111). The desC gene was not located in these genomes. We then solved the 2.0- and 2.2-Å structures of the DesE apo-complex and binary complex from B. adolescentis (112). The enzyme had strong directionality toward formation of 20β-dihydrocortisol. Interestingly, only some strains of B. adolescentis possess desE (111).
Steroid 16α-dehydroxylation and 3-ketosteroid-5β-reductase.
The host 16α-hydroxylates progesterone as a means to excrete it from the body (113). Intravenous administration of a human patient with 16α-hydroxyprogesterone resulted in urinary excretion of 17α-pregnanolone (3α-hydroxy-5β-17α-pregnan-20-one), which indicates that this excretion product is significantly modified in the gut and reabsorbed (114). In both humans and rodents, 16α-hydroxyprogesterone is reduced to 16α-hydroxypregnanolone by the liver, is secreted into bile, and undergoes enterohepatic circulation (EHC), encountering gut microbiota (114–117). It has been reported that 16α-dehydroxylation occurs in conventional rats but not in germfree rats (118). In fecal suspensions from rats (118) and humans (117), 16α-hydroxyprogesterone was converted to 17α-pregnanolone, implicating gut bacteria. This is significant because it indicates three important changes to 16α-hydroxyprogesterone: (i) removal of the 16α-hydroxy group, (ii) change in side chain stereochemistry, and (iii) reduction of 3-oxo-Δ4 to a 3α-hydroxy-5β derivative or 3α-hydroxy-5α derivative, both of which are absorbed from the GI tract and excreted via urine (Fig. 3). Thus, urinary metabolomics of pregnanolones capable of distinguishing side chain stereochemistry can differentiate the origin of the metabolite.
Later studies determined, after incubation of 16α-hydroxyprogesterone in the presence of human or rat fecal microbiota, the formation of 3α-hydroxy-5β-17α-pregnan-20-one or 3α-hydroxy-5α-17α-pregnan-20-one, respectively (119). Bokkenheuser et al. reported isolation of a strain identified as Bacteroides ruminicola subsp. brevis (strain 145) from rat feces with 3-ketosteroid reductase activity that resulted in the 5α-reduced derivative (120). However, steroid 16α-dehydroxylation appears so far limited to the Coriobacteriaceae, in particular strains of Eggerthella lenta or closely related taxa (121, 122). Bacteria expressing 16α-dehydroxylase activity were isolated and described as Gram-positive non-spore-forming obligate anaerobes that are nonmotile, with characteristics nearly identical to those of Eggerthella lenta, including in the case of strain 146 stimulation by addition of l-arginine, but did not reduce nitrate (121). Partial purification and characterization of 16α-dehydroxylase have been performed (122–125). Further work is needed to identify and characterize the gene(s) encoding 16α-dehydroxylase.
Steroid 21-dehydroxylation in the gut.
Corticosterone is synthesized at 1/10th the amount as cortisol; however, its metabolism by gut bacteria may have important consequences for the host (126). Work at the Karolinska Institute in the 1960s and 1970s sought to characterize corticosterone metabolism by the gut microbiota (114, 115, 117, 118). Corticosterone enters bile after ring A saturation (5α or 5β reduction) and glucuronidation in the liver (11). Comparison of steroid metabolism between germfree and conventional rodents indicates that gut bacteria catalyze the dehydroxylation of the side chain-terminal carbon (C-21) (Fig. 3) (127). Importantly, identification of radiolabeled metabolites of 3β,21-dihydroxy-5α-pregnane-20-one after incubation with rat cecal contents provided important clues to the substrate specificity of bacterial 21-dehydroxylase. Principally, derivatives with a C-20 hydroxyl group were not 21-dehydroxylated, but those with a C-20 ketone were 21-dehydroxylated (127). In addition, there were important sex differences observed, with male rats excreting increased 20-hydroxy derivatives, which underwent less 21-dehydroxylation than in female rats (127). Later studies by Honour and colleagues in the 1980s showed that corticosterone administration to rats led to increased blood pressure, which was ablated by neomycin treatment (128, 129).
A possible mechanistic link between the observation that gut bacteria convert corticosterone to 11β-hydroxyprogesterone, and that a relation exists between corticosterone, antibiotics, and hypertension, has been offered by the study of the inhibition of host 11β-hydroxysteroid dehydrogenase (11β-HSD) isoforms (97). Host 11β-HSD is expressed in mineralocorticoid target tissues such as colon, skin, artery smooth muscle, and renal tubules. Cortisol has affinity for the mineralocorticoid receptor (MR) on par with its physiological ligand, aldosterone, while the latter has low affinity (97, 126). Host 11β-HSD2 functions to convert cortisol to cortisone, which is not a ligand for MR. Patients deficient in 11β-HSD2 activity suffer from a severe form of hypertension that presents as aldosterone excess but in fact is due to excess cortisol binding to MR. Consumption of licorice is known to cause hypertension that presents in the same way as an apparent mineralocorticoid excess (130). Licorice contains the compound glycyrrhetinic acid, which causes hypertension through the inhibition of 11β-HSD2 (130). Endogenous 21-dehydroxylated derivatives of corticosterone were shown to inhibit 11β-HSD2 (126). From this, it has been hypothesized that gut microbial 21-dehydroxylation results in the formation of glycyrrhetinic acid-like factors (GALFs) that increase sodium and water retention through the inhibition of 11β-HSD1/2, resulting in hypertension observed in the Honour rat model (128, 129). Importantly, a consequence of bacterial metabolism of corticosterone derivatives is to affect the route of excretion of the microbial metabolites from fecal to renal (131). For example, 11-deoxycorticosterone (DOC) is a precursor of urinary 5β-pregnan-3α,20α-diol glucuronide (132), and corticosterone is a precursor to urinary 5β-pregnan-3α,20α-diol-11-one glucuronide (133). Additional work is needed to determine the role of microbial metabolism of corticosterone derivatives in hypertension.
Strains identified as Eubacterium lentum (now Eggerthella lenta) capable of corticosteroid 21-dehydroxylation were isolated previously (134). Substrates for bacterial 21-dehydroxylation include aldosterone, corticosterone, DOC, and cortisol (11, 104). In vitro studies with E. lenta VPI 11122 indicate that 21-dehydroxylase is coupled with Δ21-reductase activity (NADH:flavin oxidoreductase) (135). The enzyme was reported to be rapidly and irreversibly inactivated by oxygen (135). When DOC was a substrate, only progesterone was detected as a product in the presence of crude cell extracts of E. lentum VPI 11122 (135). The genes encoding 21-dehydroxylase and Δ21-reductase have yet to be identified.
Reduction of ring A, 17-keto metabolism.
Gut bacteria are capable of desulfation and hydrolysis of glucuronides as well as ring A oxidation/reduction and reduction of 17-ketosteroids (14). Clostridium scindens strain VPI 12708 has been shown previously to express 17α-HSDH activity, which converts androstenedione to epitestosterone (136). E. lenta (82) and Bacteroides spp. (137) have been shown to express 17β-HSDH, which converts androstenedione to testosterone. C. paraputrificum is capable of converting 3-oxo-Δ4-steroids to 3α,5β-reduced derivatives, whereas Clostridium innocuum produces 3β,5β-reduced derivatives (14, 138). Incubation of radiolabeled cortisol and corticosterone results in a mixture of metabolites with these modifications in addition to side chain cleavage (steroid-17,20-desmolase) or 21-dehydroxylation (104, 139). Some modifications preclude side chain cleavage by C. scindens. Our group recently showed that C. scindens and recombinant DesAB recognizes allotetrahydrocortisol (3α,5α-reduced) (109) but not dihydro- or tetrahydrocortisol (5β-reduced) (108). The conversion of allodihydrocortisone to allodihydro-11-keto-testosterone through the combination of steroid-17,20-desmolase and bacterial 17β-HSDH generates a potent androgen (140, 141).
Conversion of estrone (17-keto) to estradiol (17β-hydroxy) by gut bacteria after β-glucuronidation allows resorption of a major estrogen (142, 143). The genes involved in estrogen metabolism have been termed the estrolbolome (144). Deconjugation of estrogens leads to increased levels of circulating estrogens associated with development and progression of breast cancer (144, 145). Similarly, recent evidence indicates that free and glucuronidated testosterone and dihydrotestosterone (DHT) are also detected in high relative concentrations in both male and female mouse GI tract as well as >20-fold higher concentrations than serum (146). This was also observed in the feces of young adult male humans with fecal levels >70-fold that of serum (146). Comparison of GF versus conventional mice indicates that β-glucuronidase is the major microbial activity, which releases a highly potent androgen into the large intestine (146). The host-associated microbiota should be recognized as a contributor to the metabolism of endocrine molecules whose function changes with diurnal rhythm, diet, and antibiotics and through other environmental and stochastic processes.
THE ORAL STEROLBIOME
Steroids are excreted in saliva, where they are metabolized by oral microbiota. Strains of Bacteroides melaninogenicus have been shown to increase in abundance in the oral cavity during pregnancy (147). Indeed, B. melaninogenicus was shown to import radiolabeled estradiol and progesterone, which could replace vitamin K as an essential growth factor (147). Metabolism of progesterone and estradiol was observed, and the product was speculated to be 5β-pregnane-3,20-dione (147). Measurement of salivary cortisol is an important diagnostic tool for monitoring disorders of steroid metabolism (148). The extent to which cortisol is metabolized by the oral microbiota, in particular whether side chain cleavage occurs, is not known. Approaches focusing on determining the cortisol-induced transcriptome of the oral microbiome (149) may provide a way to identify genes induced by cortisol and other steroids. Increased salivary cortisol has been shown to affect plaque formation in pregnant women and thus risk for dental caries and periodontal disease (150). Sex hormones are also known to affect oral health, and oral bacteria are likely to contribute to the metabolism of salivary steroids. Indeed, both mixed oral microbiota and strains of Porphyromonas gingivalis, Prevotella intermedia, and Actinobacillus actinomycetemcomitans were capable of converting [14C]testosterone to [14C]4-androstenedione and DHT (151). The study of the oral sterolbiome is only beginning but has the potential to impact oral health as well as interpretation of how well salivary metabolomes reflect steroid profiles in serum.
THE SKIN STEROLOBIOME
The skin represents an important barrier to the external environment, with a diverse biogeography ranging from “tropical” to “desert.” Cortisol has been measured in sweat at concentrations comparable to those in saliva. Indeed, liquid chromatography-tandem mass spectrometry (LC-MS/MS) approaches have been developed to measure cortisol levels in sweat (152). Gower and Ruparelia reported that pregnanolone is side chain cleaved by axillary bacteria (153). The metabolism of cortisol, particularly through steroid-17,20-desmolase, has not as yet been demonstrated by skin bacteria but may be relevant given that eccrine sweat glands in skin display colocalized expression of MR and 11β-HSD isoforms (154, 155). If skin microbiota are capable of steroid-17,20-desmolase, the presence of resulting “GALFs” that inhibit 11β-HSD isoforms could locally alter reductase/dehydrogenase activities of 11β-HSD isoforms, affecting the ratio of cortisol to cortisone and thus MR function. Such a scenario would be expected to alter bacterial growth due to alterations in glucocorticoid and sodium concentrations in sweat (156). Studies on cortisol metabolism by skin microbiota sampled across biogeographically distinct niches could be initiated to see if this hypothesis is worth pursuing.
The host enzyme cytochrome P450 17A1 (CYP17A1) catalyzes key reactions such as 17α-hydroxylation and 17,20-lyase activities involved in steroidogenesis. In addition, CYP17A1 catalyzes the formation of 16-androstenes in the testes, which are secreted in sweat (157). Studies starting in the 1950s revealed that malodor generated in the underarm (axilla) is caused by the biotransformation of odorless axillary secretions by resident microorganisms from the genera Micrococcus, Corynebacterium, Propionibacterium, and Staphylococcus (153). Indeed, washing of the axillae of men with a germicidal solution caused a reduction in formation of 5α-androstenone to undetectable levels (158). Studies on the biotransformations of 5,16-androstadien-3-ol by Corynebacterium isolates in culture revealed strain-dependent formation of products, including 5α-androstenol, 5α-androstenone, and 3α-androstenol (159). These 16-ene-steroids are reported to have a “urine-like” or “musk-like” odor (153). Testosterone is also metabolized to multiple metabolites, including DHT, by Corynebacterium spp., indicating the expression of Δ4-5α-reductase, Δ4-5β-reductase, 3α-HSDH, 3β-HSDH, and 17β-HSDH (160).
THE UROGENITAL STEROLBIOME
Recent work has begun to define the urinary microbiome in both health and disease, with particular focus on urinary tract infection (161, 162) and prostate cancer (163, 164). Urinary steroid profiles are used diagnostically for endocrine disorders and certain forms of steroid hypertension (96). Therefore, determining the extent to which the microbiota inhabiting the urinary tract is capable of metabolizing urinary steroids may be expected to be important for the interpretation of urinary steroids. Phylogenetic analysis of the 20β-HSDH (DesE) from Butyricicoccus desmolans identified several gut bacterial isolates, as well as urinary tract isolates of Propionimicrobium lymphophilum (111). Recent phylogenetic and sequence similarity network analysis of DesA revealed that P. lymphophilum and Arcanobacterium urinimassiliense possess the desEAB gene cluster that is found in a slightly different gene order in gut bacteria, including B. desmolans, Clostridium cadaveris, and Intestinibacillus sp. Marseille-P4005 (109). Incubation of P. lymphophilum ACS-093-V-SCH5 with cortisol or 20β-dihydrocortisol, but not 20α-dihydrocortisol, resulted in quantitative conversion to 11β-hydroxyandrostenedione, indicating expression of functional DesAB and DesE (111).
A recent metagenomic comparison between male patients with prostate cancer versus negative biopsy found a significant association with P. lymphophilum (164). The urinary tract runs through the prostate gland. Surgically removed prostate tissue has been reported to harbor a microbiome, although their spatial orientation within the tissue is not well delineated and biopsy through the GI tract prior to surgery may confound interpretation (165). It is possible that the urinary tract in some men is colonized by DesAB-expressing microbes, whereas others are DesAB negative. Since cortisol is excreted largely through the urine (∼200 nM), it is conceivable that side chain cleavage of cortisol may expose the prostate to 11-oxyandrogens and represent a risk factor for the development of prostate cancer (109). The significance of bacterial steroid-17,20-desmolase activity to host physiology and pathophysiology remains to be determined. However, work by Storbeck et al. has convincingly demonstrated that derivatives of the primary adrenal steroid 11β-hydroxyandrostenedione act as nuclear androgen receptor agonists as potent as dihydrotestosterone and play an important role in the development and progression of prostate cancer (140, 141). Indeed, my group’s recent work has shown that P. lymphophilum and purified recombinant DesAB are capable of the side chain cleavage of endogenous cortisol derivatives as well as pharmaceutical glucocorticoids (109). Indeed, initial cell culture work indicates that the side chain cleavage product of prednisone promotes the growth of prostate cancer cells to an even greater extent than the most potent endogenous androgen receptor ligand, DHT (109). Understanding the structure and catalytic mechanism of enzymes involved in cortisol metabolism is a key step to hasten the development of strategies that reduce the formation of disease-promoting bioactive steroids in individuals.
THE STEROLBIOME AND VERTEBRATE SOCIAL COMMUNICATION
Olfactory pathways have evolved in both invertebrates and vertebrates to facilitate responses to environmental clues that ensure survival and reproduction. In particular, chemosensory G protein-coupled receptors and 4-transmembrane domain receptors allow vertebrates to sense steroids and bile acids in the environment (166). The emission of complex steroid and bile acid profiles, phenotypes that result from the host genome and microbiome, provides olfactory “detectors” in the environment, with a readout of the internal state and identity of the emitter. Mapping out these patterns is the goal of metabolomics, which can be correlated with animal and/or neuronal behavior.
There is considerable diversity among animals in terms of the bile acid side chain structure and the degree and stereochemistry of bile acid hydroxyl groups (167, 168). Social communication, at least in rats and mice, may at least partially explain their unique formation of muricholic acids. Accessory olfactory system neuronal pattern activity was shown to be discriminable for unconjugated primary and secondary bile acids, including ω-muricholic acid, a secondary bile acid in high abundance in the feces of rats and mice (169). Thus, while many bile acids, such as CA, CDCA, DCA, and LCA, are common to most vertebrates, taxon-specific information may be conveyed by the presence of certain olfactory stimulating bile acids such as primary and secondary muricholic acids (169). The secondary bile acid DCA is detected by olfaction in a number of vertebrates (166). Because bile acid profiles offer a “molecular fingerprint” that changes with diet (64), health (62), and sex (170), the ability of vertebrates to sense such patterns may offer clues about potential mates, as well as health and vigor of emitters (169). Species-specific patterns of fecal bile acids, in combination with other chemical profiles, provide a means to either avoid predation or locate prey. In this way, complex steroid profiles, generated by host and microbiota, may serve as either pheromones (intraspecies) or kairomones (interspecies) (166).
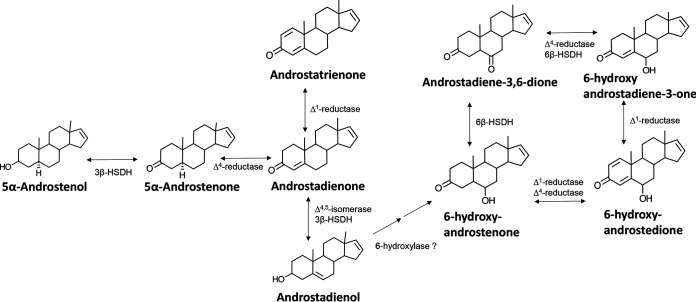
Pigs and humans respond to unconjugated, volatile androstanes (153). Male pigs secrete 16-ene-steroids such as androstenone, which induces attraction and mating behavior in estrous females (153). Humans are more genetically diverse in their responses to volatile androstanes, with some finding these odors pleasant while others find them repellant (171). The locations of 16-ene-steroids are reported to be the testes, adrenal gland, and ovaries (153). The source is suggested to be pregnenolone, rather than androgens (153). The axilla in humans is the major site for the secretion of 16-ene-steroids, which as described above undergo numerous metabolic biotransformations by axillary aerobic bacteria (159). 16-Ene-steroids are suggested to act as pheromones in humans (153). Indeed, 16-ene-steroids are generated in large abundance by males, but not females, and females respond to the olfactory effects of 16-ene-steroids, whereas males do not (153). Corynebacterium spp. isolated from the axilla have been reported to generate a complex mixture of 16-ene-steroids in vitro (159). Proposed biochemical pathways for microbial 16-ene-steroid metabolism remain speculative; however, initial identification of steroid metabolites suggests a potentially rich area of future research (Fig. 4; Table 2) (159). Indeed, because the microbiota biotransform 16-ene-steroids, there may be a role for bacteria in mate preference and other social interactions between humans (172). Such is the case for commensal bacteria in Drosophila, which generate pheromones (173). Bile alcohols also function as pheromones in sea lampreys and kairomones in invertebrates (174–176).
FIG 4.
Schematic of postulated pathways for Corynebacterium metabolism of 5,16-androstadien-3-ol. Modified from a model proposed by Austin and Ellis (159) to explain diverse metabolites identified after mixed or single culture of axilla isolates of Corynebacterium spp. Note that genes have yet to be identified and enzymes characterized for these proposed pathways; thus, these remain speculative.
THE ROLE OF BIOLOGICAL SEX IN THE STEROLBIOME
A complex interplay is being unraveled between sex-dependent circulating hormones, the immune system, the gut microbiome, and susceptibility to autoimmune diseases (177). Correlations have been found between serum testosterone or estradiol levels and gut microbial abundance and diversity (178). Altered expression of estrogen receptor (ER-β) affects the composition of the microbiome (179). Sex differences in microbiome composition have been demonstrated in 89 inbred strains of mice and were shown to be hormone dependent after gonadectomy and hormone treatment (180). Gonadectomy also resulted in sex-dependent changes in biliary bile acid profile (180). Sex differences in the adaptive and innate immune system are well known, with females having a more robust immunity than males (181). Circulating hormones differ between males and females, and most immune cells have receptors for estrogens, testosterone, secondary bile acids, and progesterone (182). Transfer of male rodent microbiota into young females conferred sustained testosterone elevation and metabolic changes that could be attenuated by the androgen receptor antagonist flutamide (183). This indicates that gut bacteria are capable of regulating sex hormone production, although the mechanisms are still poorly understood. Likewise, the structures of host-associated microbiomes are affected by steroid composition and the immune system. Distinct female and male pig gut microbiota appear to be driven by the testes, as castration of males resulted in a gut microbiome that overlapped that of females (184). The study of the role of the relationship between sex and the microbiota, termed the “microgenderome,” is still in its infancy (185, 186) and represents the bidirectional interactions between host physiology, steroids, and microbiota.
CONCLUSIONS AND FUTURE DIRECTIONS
The sterolbiome concept encompasses host-associated microbiomes across vertebrates. Sterolbiome genes and pathways represent a modular aspect of the host endocrine system which may vary markedly between individuals and longitudinally within individuals through the course of life as part of the aging process or acutely after perturbations such as dietary changes or antibiotic use. The coevolution between host and microbiota suggests a normal physiological role of steroid metabolites unique to bacterial biotransformation. This interkingdom steroid signaling may become pathological in situations where the concentration of these metabolisms is altered by dysbiosis, exogenous intake of pharmaceutical steroids, or the Western diet. Mapping out sterolbiome genes and pathways in metagenomes, coupled with steroid metabolomics, is expected to allow researchers in the field to diagnose and treat pathological conditions. The vertebrate sterolbiome is predicted to have significant impact on microbiome structure, digestion, the gut-brain axis, innate and adaptive immune function, cancers, cardiovascular health, and behavior. The role of steroid metabolism both remote from tumors and in the tumor microenvironment may be one of many important contributing factors to cancer development and progression (206). Mapping out and manipulating the sterolbiome are expected to reveal mechanisms behind sex-dependent differences in microbiome composition and immune function. The identification of novel sterolbiome enzymes may be important for pharmaceutical production of steroids, and the development of inhibitors or the engineering of probiotics to modify bile acids and steroids may be important for growth of production animals.
ACKNOWLEDGMENTS
I thank Phillip Hylemon, Patricia Wolf, Lindsey Ly, and Heidi Doden for helpful comments and suggestions.
This work was supported by grants from the National Institutes of Health (R01GM134423 and R03AI147127).
Biography

Jason M. Ridlon, Ph.D., studied biology (B.Sc. 2002) at Bridgewater College in Virginia with Stephen F. Baron, a talented microbiologist trained by James G. Ferry and Phillip B. Hylemon. This led to an opportunity to completed doctoral and postdoctoral work at VCU School of Medicine with Phil Hylemon. There, he focused on the molecular biology and enzymology of the bile acid inducible (bai) regulon in Clostridium scindens and Clostridium hylemonae. In addition, he identified a gene cluster in C. scindens ATCC 35704 encoding the steroid-17,20-desmolase pathway involved in cortisol metabolism. In 2015, he joined the faculty in animal sciences at the University of Illinois at Urbana-Champaign, where his focus is maintained on the sterobiome. In the past several years, his lab has discovered and characterized genes encoding novel bile acid- and corticosteroid-metabolizing enzymes by both the gut and urinary tract microbiota. His long-term goals focus on modulating the sterolbiome to improve human health and animal production.
REFERENCES
- 1.Sender R, Fuchs S, Milo R, Lee T, Ahn H, Baek S. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch SV, Pedersen O. 2016. The human intestinal microbiome in health and disease. N Engl J Med 375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 4.Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, Knight R, Ley RE. 2014. Conducting a microbiome study. Cell 158:250–262. doi: 10.1016/j.cell.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koppel N, Balskus EP. 2016. Exploring and understanding the biochemical diversity of the human microbiota. Cell Chem Biol 23:18–30. doi: 10.1016/j.chembiol.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Fischbach MA. 2018. Microbiome: focus on causation and mechanism. Cell 174:785–790. doi: 10.1016/j.cell.2018.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouwerkerk JP, de Vos WM, Belzer C. 2013. Glycobiome: bacteria and mucus at the epithelial interface. Best Pract Res Clin Gastroenterol 27:25–38. doi: 10.1016/j.bpg.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Chiang HC, Bjursell MK, Gordon JI. 2004. Message from a human gut symbiont: sensitivity is a prerequisite for sharing. Trends Microbiol 12:21–28. doi: 10.1016/j.tim.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ridlon JM, Bajaj JS. 2015. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm Sin B 5:99–105. doi: 10.1016/j.apsb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyte M. 2014. Microbial endocrinology: host-microbiota neuroendocrine interactions influencing brain and behavior. Gut Microbes 5:381–389. doi: 10.4161/gmic.28682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macdonald IA, Bokkenheuser VD, Winter J, McLernon AM, Mosbach EH. 1983. Degradation of steroids in the human gut. J Lipid Res 24:675–700. [PubMed] [Google Scholar]
- 13.Midtvedt T, Frederichsen P. 1977. Influence of antibiotics on microbial intestinal transformation of cholesterol to coprostanol in man. Scand J Gastroenterol 12:669–672. doi: 10.3109/00365527709181701. [DOI] [PubMed] [Google Scholar]
- 14.Björkhem I, Gustafsson J, Wrange O. 1973. Microbial transformation of cholesterol into coprostanol: properties of a 3-oxo-Δ4-steroid-5β-reductase. Eur J Biochem 37:143–147. doi: 10.1111/j.1432-1033.1973.tb02968.x. [DOI] [PubMed] [Google Scholar]
- 15.Lichtenstein AH. 1990. Intestinal cholesterol metabolism. Ann Med 22:49–52. doi: 10.3109/07853899009147241. [DOI] [PubMed] [Google Scholar]
- 16.Sekimoto H, Shimada O, Makanishi M, Nakano T, Katayama O. 1983. Interrelationship between serum and fecal sterols. Jpn J Med 22:14–20. doi: 10.2169/internalmedicine1962.22.14. [DOI] [PubMed] [Google Scholar]
- 17.Li L, Buhman KK, Hartman PA, Beitz DC. 1995. Hypocholesterolemic effect of Eubacterium coprostanoligenes ATCC 51222 in rabbits. Lett Appl Microbiol 20:137–140. doi: 10.1111/j.1472-765X.1995.tb00410.x. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Batt SM, Wannemuehler M, Dispirito A, Beitz DC. 1998. Effect of feeding of a cholesterol-reducing bacterium, Eubacterium coprostanoligenes, to germ-free mice. Lab Anim Sci 48:253–255. [PubMed] [Google Scholar]
- 19.Li L, Baumann CA, Meling DD, Sell JL, Beitz DC. 1996. Effect of orally administered Eubacterium coprostanoligenes ATCC 51222 on plasma cholesterol concentration in laying hens. Poult Sci 75:743–745. doi: 10.3382/ps.0750743. [DOI] [PubMed] [Google Scholar]
- 20.Mott GE, Brinkley AW. 1979. Plasmenylethanolamine: growth factor for cholesterol-reducing Eubacterium. J Bacteriol 139:755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinkley AW, Gottesman AR, Mott GE. 1980. Growth of cholesterol-reducing Eubacterium on cholesterol-brain agar. Appl Environ Microbiol 40:1130–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eyssen HJ, Parmentier GG, Compernolle FC, De Pauw G, Piessens-Denef M. 1973. Biohydrogenation of sterols by Eubacterium ATCC 21408—nova species. Eur J Biochem 36:411–421. doi: 10.1111/j.1432-1033.1973.tb02926.x. [DOI] [PubMed] [Google Scholar]
- 23.Kriaa A, Bourgin M, Mkaouar H, Jablaoui A, Akermi N, Soussou S, Maguin E, Rhimi M. 2019. Microbial reduction of cholesterol to coprostanol: an old concept and new insights. Catalysts 9:167. doi: 10.3390/catal9020167. [DOI] [Google Scholar]
- 24.Rosenfeld RS, Fukushima DK, Hellman L, Gallagher TF. 1954. The transformation of cholesterol to coprostanol. J Biol Chem 211:301–311. [PubMed] [Google Scholar]
- 25.Rosenfeld RS, Gallagher TF. 1964. Further studies of the biotransformation of cholesterol to coprostanol. Steroids 4:515–520. doi: 10.1016/0039-128X(64)90098-4. [DOI] [Google Scholar]
- 26.Björkhem I, Gustafsson JA. 1971. Mechanism of microbial transformation of cholesterol into coprostanol. Eur J Biochem 21:428–432. doi: 10.1111/j.1432-1033.1971.tb01488.x. [DOI] [PubMed] [Google Scholar]
- 27.Ren D, Li L, Schwabacher AW, Young JW, Beitz DC. 1996. Mechanism of cholesterol reduction to coprostanol by Eubacterium coprostanoligenes ATCC 51222. Steroids 61:33–40. doi: 10.1016/0039-128x(95)00173-n. [DOI] [PubMed] [Google Scholar]
- 28.Parmentier G, Eyssen H. 1974. Mechanism of biohydrogenation of cholesterol to coprostanol by Eubacterium ATCC 21408. Biochim Biophys Acta 348:279–284. doi: 10.1016/0005-2760(74)90239-2. [DOI] [PubMed] [Google Scholar]
- 29.Gérard P, Lepercq P, Leclerc M, Gavini F, Raibaud R, Juste C. 2007. Bacteroides sp. strain D8, the first cholesterol-reducing bacterium isolated from human feces. Appl Environ Microbiol 73:5742–5749. doi: 10.1128/AEM.02806-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kisiela M, Skarka A, Ebert B, Maser E. 2012. Hydroxysteroid dehydrogenases (HSDs) in bacteria: a bioinformatic perspective. J Steroid Biochem Mol Biol 129:31–46. doi: 10.1016/j.jsbmb.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Devlin AS, Fischbach MA. 2015. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol 11:685–690. doi: 10.1038/nchembio.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mythen SM, Devendran S, Méndez-García C, Cann I, Ridlon JM. 2018. Targeted synthesis and characterization of a gene cluster encoding NAD(P)H-dependent 3α-, 3β-, and 12α-hydroxysteroid dehydrogenases from Eggerthella CAG:298, a gut metagenomic sequence. Appl Environ Microbiol 84:e02475-17. doi: 10.1128/AEM.02475-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang DJ, Ridlon JM, Moore DR II, Barnes S, Hylemon PB. 2008. Clostridium scindens baiCD and baiH genes encode stereo-specific 7alpha/7beta-hydroxy-3-oxo-delta4-cholenoic acid oxidoreductases. Biochim Biophys Acta 1781:16–25. doi: 10.1016/j.bbalip.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bokkenheuser VD, Winter J, Cohen BI, O’Rourke S, Mosbach EH. 1983. Inactivation of contraceptive steroid hormones by human intestinal clostridia. J Clin Microbiol 18:500–504. doi: 10.1128/JCM.18.3.500-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang JYL, Ferrell JM. 2019. Bile acids as metabolic regulators and nutrient sensors. Annu Rev Nutr 39:175–200. doi: 10.1146/annurev-nutr-082018-124344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofmann AF, Roda A. 1984. Physicochemical properties of bile acids and their relationship to biological properties: an overview of the problem. J Lipid Res 25:1477–1489. [PubMed] [Google Scholar]
- 37.Winston JA, Theriot CM. 2020. Diversification of host bile acids by members of the gut microbiota. Gut Microbes 11:158–171. doi: 10.1080/19490976.2019.1674124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ridlon JM, Kang DJ, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Kakiyama G, Muto A, Takei H, Nittono H, Murai T, Kurosawa T, Hofmann AF, Pandak WM, Bajaj JS. 2014. A simple and accurate HPLC method for fecal bile acid profile in healthy and cirrhotic subjects: validation by GC-MS and LC-MS. J Lipid Res 55:978–990. doi: 10.1194/jlr.D047506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. 2008. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu F, Hu XJ, Singh W, Geng W, Tikhonova IG, Lin J. 2019. The complex structure of bile salt hydrolase from Lactobacillus salivarius reveals the structural basis of substrate specificity. Sci Rep 9:12438. doi: 10.1038/s41598-019-48850-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossocha M, Schultz-Heienbrok R, von Moeller H, Coleman JP, Saenger W. 2005. Conjugated bile acid hydrolase is a tetrameric N-terminal thiol hydrolase with specific recognition of its cholyl but not of its tauryl product. Biochemistry 44:5739–5748. doi: 10.1021/bi0473206. [DOI] [PubMed] [Google Scholar]
- 43.Kumar RS, Brannigan JA, Prabhune AA, Pundle AV, Dodson GG, Dodson EJ, Suresh CG. 2006. Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J Biol Chem 281:32516–32525. doi: 10.1074/jbc.M604172200. [DOI] [PubMed] [Google Scholar]
- 44.Chand D, Panigrahi P, Varshney N, Ramasamy S, Suresh CG. 2018. Structure and function of a highly active bile salt hydrolase (BSH) from Enterococcus faecalis and post-translational processing of BSH enzymes. Biochim Biophys Acta Proteins Proteom 1866:507–518. doi: 10.1016/j.bbapap.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 45.Geng W, Lin J. 2016. Bacterial bile salt hydrolase: an intestinal microbiome target for enhanced animal health. Anim Health Res Rev 17:148–158. doi: 10.1017/S1466252316000153. [DOI] [PubMed] [Google Scholar]
- 46.Lin J. 2014. Antibiotic growth promoters enhance animal production by targeting intestinal bile salt hydrolase and its producers. Front Microbiol 5:33. doi: 10.3389/fmicb.2014.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones ML, Tomaro-Duchesneau C, Martoni CJ, Prakash S. 2013. Cholesterol lowering with bile salt hydrolase-active probiotic bacteria, mechanism of action, clinical evidence, and future direction for heart health applications. Expert Opin Biol Ther 13:631–642. doi: 10.1517/14712598.2013.758706. [DOI] [PubMed] [Google Scholar]
- 48.Feighner SD, Dashkevicz MP. 1987. Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl Environ Microbiol 53:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feighner SD, Dashkevicz MP. 1988. Effect of dietary carbohydrates on bacterial cholyltaurine hydrolase in poultry intestinal homogenates. Appl Environ Microbiol 54:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krogdahl A. 1985. Digestion and absorption of lipids in poultry. J Nutr 115:675–685. doi: 10.1093/jn/115.5.675. [DOI] [PubMed] [Google Scholar]
- 51.Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CG. 2014. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A 111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao L, Seaton SC, Ndousse-Fetter S, Adhikari AA, DiBenedetto N, Mina AI, Banks AS, Bry L, Devlin AS. 2018. A selective gut bacterial bile salt hydrolase alters host metabolism. Elife 7:e37182. doi: 10.7554/eLife.37182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eyssen H, De Somer P. 1963. Toxicity of lithocholic acid for the chick. Poult Sci 42:1020–1022. doi: 10.3382/ps.0421020. [DOI] [Google Scholar]
- 54.Pavlović N, Stankov K, Mikov M. 2012. Probiotics—interactions with bile acids and impact on cholesterol metabolism. Appl Biochem Biotechnol 168:1880–1895. doi: 10.1007/s12010-012-9904-4. [DOI] [PubMed] [Google Scholar]
- 55.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. 2014. Bile acids and the gut microbiome. Curr Opin Gastroenterol 30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou H, Hylemon PB. 2014. Bile acids are nutrient signaling hormones. Steroids 86:62–68. doi: 10.1016/j.steroids.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. 2015. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, Mangelsdorf DJ, Kliewer SA. 2006. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A 103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. 2011. Induction of colonic regulatory T-cells by indigenous Clostridium species. Science 331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, Ha S, Nelson BN, Kelly SP, Wu L, Zheng Y, Longman RS, Rastinejad F, Devlin AS, Krout MR, Fischbach MA, Littman DR, Huh JR. 2019. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576:143–148. doi: 10.1038/s41586-019-1785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ridlon JM, Alves JM, Hylemon PB, Bajaj JS. 2013. Cirrhosis, bile acids and gut microbiota. Gut Microbes 4:382–387. doi: 10.4161/gmic.25723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berr F, Kullak-Ublick GA, Paumgartner G, Münzing W, Hylemon PB. 1996. 7 Alpha-dehydroxylating bacteria enhance deoxycholic acid input and cholesterol saturation of bile in patients with gallstones. Gastroenterology 111:1611–1620. doi: 10.1016/S0016-5085(96)70024-0. [DOI] [PubMed] [Google Scholar]
- 64.Ridlon JM, Wolf PG, Gaskins HR. 2016. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes 7:201–215. doi: 10.1080/19490976.2016.1150414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ocvirk S, O’Keefe SJ. 2017. Influence of bile acids on colorectal cancer risk: potential mechanisms mediated by diet-gut microbiota interactions. Curr Nutr Rep 6:315–322. doi: 10.1007/s13668-017-0219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. 2005. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res 589:47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 67.Bernstein C, Holubec H, Bhattacharyya AK, Nguyen H, Payne CM, Zaitlin B, Bernstein H. 2011. Carcinogenicity of deoxycholate, a secondary bile acid. Arch Toxicol 85:863–871. doi: 10.1007/s00204-011-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao H, Xu M, Dong W, Deng B, Wang S, Zhang Y, Wang S, Luo S, Wang W, Qi Y, Gao J, Cao X, Yan F, Wang B. 2017. Secondary bile acid-induced dysbiosis promotes intestinal carcinogenesis. Int J Cancer 140:2545–2556. doi: 10.1002/ijc.30643. [DOI] [PubMed] [Google Scholar]
- 69.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. 2013. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 70.Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, Ritz T, Longerich T, Theriot CM, McCulloch JA, Roy S, Yuan W, Thovarai V, Sen SK, Ruchirawat M, Korangy F, Wang XW, Trinchieri G, Greten TF. 2018. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360:eaan5931. doi: 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marksteiner J, Blasko I, Kemmler G, Koal T, Humpel C. 2018. Bile acid quantification of 20 plasma metabolites identifies lithocholic acid as a putative biomarker in Alzheimer’s disease. Metabolomics 14:1. doi: 10.1007/s11306-017-1297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.MahmoudianDehkordi S, Arnold M, Nho K, Ahmad S, Jia W, Xie G, Louie G, Kueider-Paisley A, Moseley MA, Thompson JW, St John Williams L, Tenenbaum JD, Blach C, Baillie R, Han X, Bhattacharyya S, Toledo JB, Schafferer S, Klein S, Koal T, Risacher SL, Kling MA, Motsinger-Reif A, Rotroff DM, Jack J, Hankemeier T, Bennett DA, De Jager PL, Trojanowski JQ, Shaw LM, Weiner MW, Doraiswamy PM, van Duijn CM, Saykin AJ, Kastenmüller G, Kaddurah-Daouk R, Alzheimer’s Disease Neuroimaging Initiative and the Alzheimer Disease Metabolomics Consortium . 2019. Altered bile acid profile associates with cognitive impairment in Alzheimer’s disease—an emerging role for gut microbiome. Alzheimers Dement 15:76–92. doi: 10.1016/j.jalz.2018.07.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.White BA, Lipsky RL, Fricke RJ, Hylemon PB. 1980. Bile acid induction specificity of 7alpha-dehydroxylase activity in an intestinal Eubacterium species. Steroids 35:103–109. doi: 10.1016/0039-128X(80)90115-4. [DOI] [PubMed] [Google Scholar]
- 74.Kitahara M, Takamine F, Imamura T, Benno Y. 2000. Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7alpha-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 50(Part 3):971–978. doi: 10.1099/00207713-50-3-971. [DOI] [PubMed] [Google Scholar]
- 75.Bokkenheuser VD, Morris GN, Ritchie AE, Holdeman LV, Winter J. 1984. Biosynthesis of androgen from cortisol by a species of Clostridium recovered from human fecal flora. J Infect Dis 149:489–494. doi: 10.1093/infdis/149.4.489. [DOI] [PubMed] [Google Scholar]
- 76.White BA, Cacciapuoti AF, Fricke RJ, Whitehead TR, Mosbach EH, Hylemon PB. 1981. Cofactor requirements for 7alpha-dehydroxylation of cholic and chenodeoxycholic acid in cell extracts of the intestinal anaerobic bacterium, Eubacterium species V.P.I. 12708. J Lipid Res 22:891–898. [PubMed] [Google Scholar]
- 77.Björkhem I, Einarsson K, Melone P, Hylemon PB. 1989. Mechanism of intestinal formation of deoxycholic acid from cholic acid in humans: evidence for a 3-oxo-delta4-steroid intermediate. J Lipid Res 30:1033–1039. [PubMed] [Google Scholar]
- 78.Hylemon PB, Melone PD, Franklund CV, Lund E, Björkhem I. 1991. Mechanism of intestinal 7 alpha-dehydroxylation of cholic acid: evidence that allo-deoxycholic acid is an inducible side-product. J Lipid Res 32:89–96. [PubMed] [Google Scholar]
- 79.Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. 2016. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7:22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Devendran S, Shrestha R, Alves JMP, Wolf PG, Ly L, Hernandez AG, Mendez C, Inboden A, Wiley J, Paul O, Allen A, Springer E, Wright CL, Fields CJ, Daniel SL, Ridlon JM. 2019. Clostridium scindens ATCC 35704: integration of nutritional requirements, the complete genome sequence, and global transcriptional responses to bile acids. Appl Environ Microbiol 85:e00052-19. doi: 10.1128/AEM.00052-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Campbell GR, Odling-Smee GW, Rowlands BJ, Irvine GB. 1989. Determination of bile acids and their conjugates in serum by HPLC using immobilized 3alpha-hydroxysteroid dehydrogenase for detection. Biomed Chromatogr 3:75–78. doi: 10.1002/bmc.1130030208. [DOI] [PubMed] [Google Scholar]
- 82.Harris SC, Devendran S, Méndez-García C, Mythen SM, Wright CL, Fields CJ, Hernandez AG, Cann I, Hylemon PB, Ridlon JM. 2018. Bile acid oxidation by Eggerthella lenta strains C592 and DSM 2243T. Gut Microbes 9:523–539. doi: 10.1080/19490976.2018.1458180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hylemon PB, Harris SC, Ridlon JM. 2018. Metabolism of hydrogen gases and bile acids in the gut microbiome. FEBS Lett 592:2070–2082. doi: 10.1002/1873-3468.13064. [DOI] [PubMed] [Google Scholar]
- 84.Shea HC, Head DD, Setchell KD, Russell DW. 2007. Analysis of HSD3B7 knockout mice reveals that a 3alpha-hydroxyl stereochemistry is required for bile acid function. Proc Natl Acad Sci U S A 104:11526–11533. doi: 10.1073/pnas.0705089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marion S, Studer N, Desharnais L, Menin L, Escrig S, Meibom A, Hapfelmeier S, Bernier-Latmani R. 2019. In vitro and in vivo characterization of Clostridium scindens bile acid transformations. Gut Microbes 10:481–503. doi: 10.1080/19490976.2018.1549420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.White BA, Fricke RJ, Hylemon PB. 1982. 7β-Dehydroxylation of ursodeoxycholic acid by whole cells and cell extracts of the intestinal anaerobic bacterium, Eubacterium species V.P.I. 12708. J Lipid Res 23:145–153. [PubMed] [Google Scholar]
- 87.Macdonald IA, Hutchison DM. 1982. Epimerization versus dehydroxylation of the 7alpha-hydroxyl- group of primary bile acids: competitive studies with Clostridium absonum and 7alpha-dehydroxylating bacteria (Eubacterium sp.). J Steroid Biochem 17:295–303. doi: 10.1016/0022-4731(82)90203-5. [DOI] [PubMed] [Google Scholar]
- 88.Edenharder R, Schneider J. 1985. 12 Beta-dehydrogenation of bile acids by Clostridium paraputrificum, C. tertium, and C. difficile and epimerization at carbon-12 of deoxycholic acid by cocultivation with 12 alpha-dehydrogenating Eubacterium lentum. Appl Environ Microbiol 49:964–968. doi: 10.1128/AEM.49.4.964-968.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Edenharder R, Pfützner A. 1988. Characterization of NADP-dependent 12beta-hydroxysteroid dehydrogenase from Clostridium paraputrificum. Biochim Biophys Acta 962:362–370. doi: 10.1016/0005-2760(88)90266-4. [DOI] [PubMed] [Google Scholar]
- 90.Bhowmik S, Jones DH, Chiu HP, Park IH, Chiu HJ, Axelrod HL, Farr CL, Tien HJ, Agarwalla S, Lesley SA. 2014. Structural and functional characterization of BaiA, an enzyme involved in secondary bile acid synthesis in human gut microbe. Proteins 82:216–229. doi: 10.1002/prot.24353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tanaka N, Nonaka T, Tanabe T, Yoshimoto T, Tsuru D, Mitsui Y. 1996. Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry 35:7715–7730. doi: 10.1021/bi951904d. [DOI] [PubMed] [Google Scholar]
- 92.Wang R, Wu J, Jin DK, Chen Y, Lv Z, Chen Q, Miao Q, Huo X, Wang F. 2017. Structure of NADP+-bound 7β-hydroxysteroid dehydrogenase reveals two cofactor-binding modes. Acta Crystallogr F Struct Biol Commun 73:246–252. doi: 10.1107/S2053230X17004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kelsey MI, Molina JE, Huang SK, Hwang KK. 1980. The identification of microbial metabolites of sulfolithocholic acid. J Lipid Res 21:751–759. [PubMed] [Google Scholar]
- 94.Benson GM, Haskins NJ, Eckers C, Moore PJ, Reid DG, Mitchell RC, Waghmare S, Suckling KE. 1993. Polydeoxycholic acid in human and hamster feces: a major product of cholate metabolism. J Lipid Res 34:2121–2134. [PubMed] [Google Scholar]
- 95.Quinn RA, Melnik AV, Vrbanac A, Fu T, Patras KA, Christy MP, Bodai Z, Belda-Ferre P, Tripathi A, Chung LK, Downes M, Welch RD, Quinn M, Humphrey G, Panitchpakdi M, Weldon KC, Aksenov A, da Silva R, Avila-Pacheco J, Clish C, Bae S, Mallick H, Franzosa EA, Lloyd-Price J, Bussell R, Thron T, Nelson AT, Wang M, Leszczynski E, Vargas F, Gauglitz JM, Meehan MJ, Gentry E, Arthur TD, Komor AC, Poulsen O, Boland BS, Chang JT, Sandborn WJ, Lim M, Garg N, Lumeng JC, Xavier RJ, Kazmierczak BI, Jain R, Egan M, Rhee KE, Ferguson D, Raffatellu M, Vlamakis H, Haddad GG, Siegel D, Huttenhower C, Mazmanian SK, Evans RM, Nizet V, Knight R, Dorrestein PC. 2020. Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579:123–129. doi: 10.1038/s41586-020-2047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schiffer L, Barnard L, Baranowski ES, Gilligan LC, Taylor AE, Arlt W, Shackleton CHL, Storbeck KH. 2019. Human steroid biosynthesis, metabolism and excretion are differentially reflected by serum and urine steroid metabolomes: a comprehensive review. J Steroid Biochem Mol Biol 194:105439. doi: 10.1016/j.jsbmb.2019.105439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Woods C, Tomlinson JW. 2015. The dehydrogenase hypothesis. Adv Exp Med Biol 872:353–380. doi: 10.1007/978-1-4939-2895-8_16. [DOI] [PubMed] [Google Scholar]
- 98.Jin Y. 2011. Activities of aldo-keto reductase 1 enzymes on two inhaled corticosteroids: implications for the pharmacological effects of inhaled corticosteroids. Chem Biol Interact 191:234–238. doi: 10.1016/j.cbi.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bloem LM, Storbeck KH, Schloms L, Swart AC. 2013. 11β-Hydroxyandrostenedione returns to the steroid arena: biosynthesis, metabolism and function. Molecules 18:13228–13244. doi: 10.3390/molecules181113228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nabarro JDN, Moxham A, Walker G, Slater J. 1957. Rectal hydrocortisone. Br Med J 2:272–274. doi: 10.1136/bmj.2.5039.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wade AP, Slater JDM, Kellie AE, Holliday ME. 1959. Urinary excretion of 17-ketosteroids following rectal infusion of cortisol. J Clin Endocrinol Metab 19:444–453. doi: 10.1210/jcem-19-4-444. [DOI] [PubMed] [Google Scholar]
- 102.Möstl E, Palme R. 2002. Hormones as indicators of stress. Domest Anim Endocrinol 23:67–74. doi: 10.1016/s0739-7240(02)00146-7. [DOI] [PubMed] [Google Scholar]
- 103.Palme R, Rettenbacher S, Touma C, El-Bahr SM, Möstl E. 2005. Stress hormones in mammals and birds: comparative aspects regarding metabolism, excretion, and noninvasive measurement in fecal samples. Ann N Y Acad Sci 1040:162–171. doi: 10.1196/annals.1327.021. [DOI] [PubMed] [Google Scholar]
- 104.Cerone-McLernon AM, Winter J, Mosbach EH, Bokkenheuser VD. 1981. Side-chain cleavage of cortisol by fecal flora. Biochim Biophys Acta 666:341–347. doi: 10.1016/0005-2760(81)90292-7. [DOI] [PubMed] [Google Scholar]
- 105.Winter J, Morris GN, O’Rourke-Locascio S, Bokkenheuser VD, Mosbach EH, Cohen BI, Hylemon PB. 1984. Mode of action of steroid desmolase and reductases synthesized by Clostridium “scindens” (formerly Clostridium strain 19). J Lipid Res 25:1124–1131. [PubMed] [Google Scholar]
- 106.Krafft AE, Hylemon PB. 1989. Purification and characterization of a novel form of 20 alpha-hydroxysteroid dehydrogenase from Clostridium scindens. J Bacteriol 171:2925–2932. doi: 10.1128/jb.171.6.2925-2932.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ridlon JM, Ikegawa S, Alves JM, Zhou B, Kobayashi A, Iida T, Mitamura K, Tanabe G, Serrano M, De Guzman A, Cooper P, Buck GA, Hylemon PB. 2013. Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J Lipid Res 54:2437–2449. doi: 10.1194/jlr.M038869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Devendran S, Mythen SM, Ridlon JM. 2018. The desA and desB genes from Clostridium scindens ATCC 35704 encode steroid-17,20-desmolase. J Lipid Res 59:1005–1014. doi: 10.1194/jlr.M083949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ly LK, Rowles JL, Paul HM, Alves JMP, Yemm C, Wolf PM, Devendran S, Hudson ME, Morris DJ, Erdman JW, Ridlon JM. 2020. Bacterial steroid-17,20-desmolase is a taxonomically rare enzymatic pathway that converts endogenous and pharmaceutical glucocorticoids into androgens. J Steroid Biochem Mol Biol 199:105567. doi: 10.1016/j.jsbmb.2019.105567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bokkenheuser VD, Winter J, Morris GN, Locascio S. 1986. Steroid desmolase synthesis by Eubacterium desmolans and Clostridium cadavaris. Appl Environ Microbiol 52:1153–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Devendran S, Méndez-García C, Ridlon JM. 2017. Identification and characterization of a 20β-HSDH from the anaerobic gut bacterium Butyricicoccus desmolans ATCC 43058. J Lipid Res 58:916–925. doi: 10.1194/jlr.M074914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Doden HL, Pollet RM, Mythen SM, Wawrzak Z, Devendran S, Cann I, Koropatkin NM, Ridlon JM. 2019. Structural and biochemical characterization of 20β-hydroxysteroid dehydrogenase from Bifidobacterium adolescentis strain L2-32. J Biol Chem 294:12040–12053. doi: 10.1074/jbc.RA119.009390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Calvin HI, Lieberman S. 1962. Studies on the metabolism of 16α-hydroxyprogesterone in humans. Conversion of urinary 17-isopregnanolone. Biochemistry 1:639–645. doi: 10.1021/bi00910a016. [DOI] [PubMed] [Google Scholar]
- 114.Laatikainen T, Vihko R. 1969. Identification of C19O2 and C21O2 steroids in the glucuronide fraction of human bile. Eur J Biochem 10:165–171. doi: 10.1111/j.1432-1033.1969.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 115.Laatikainen T. 1970. Identification of C1902 and C2102 steroids in the mono and disulphate fractions of human feces. Steroids 15:139–150. [DOI] [PubMed] [Google Scholar]
- 116.Shen H, Elliot WH, Doisy E, Doisy EA. 1954. Excretion of metabolites of progesterone-21-C14 after intragastric administration to rats. J Biol Chem 208:133–137. [PubMed] [Google Scholar]
- 117.Eriksson H, Gustafsson JA. 1971. Excretion of steroid hormones in adults. Steroid in feces from adults. Eur J Biochem 18:146–150. doi: 10.1111/j.1432-1033.1971.tb01225.x. [DOI] [PubMed] [Google Scholar]
- 118.Eriksson H, Gustafsson JA, Sjövall J. 1968. Steroids in germ-free and conventional rats. 4. Identification and bacterial formation of 17alpha-pregnane derivatives. Eur J Biochem 6:219–226. doi: 10.1111/j.1432-1033.1968.tb00441.x. [DOI] [PubMed] [Google Scholar]
- 119.Bokkenheuser VD, Winter J, Hylemon PB, Ayengar NK, Mosbach EH. 1981. Dehydroxylation of 16 alpha-hydroxyprogesterone by fecal flora of man and rat. J Lipid Res 22:95–102. [PubMed] [Google Scholar]
- 120.Bokkenheuser VD, Winter J, O’Rourke S, Ritchie AE. 1980. Isolation and characterization of fecal bacteria capable of 16 alpha-dehydroxylating corticoids. Appl Environ Microbiol 40:803–808. doi: 10.1128/AEM.40.4.803-808.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Winter J, O’Rourke S, Bokkenheuser VD, Hylemon PB, Glass TL. 1982. 16α-Dehydration of corticoids by bacteria isolated from rat fecal flora. J Steroid Biochem 16:231–237. doi: 10.1016/0022-4731(82)90171-6. [DOI] [PubMed] [Google Scholar]
- 122.Glass TL, Winter J, Bokkenheuser VD, Hylemon PB. 1982. Biotransformation of 16 alpha-hydroxyprogesterone by Eubacterium sp. 144: non-enzymatic addition of L-cysteine to delta 16-progesterone. J Lipid Res 23:352–356. [PubMed] [Google Scholar]
- 123.Glass TL, Lamppa RS. 1985. Purification and properties of 16 alpha-hydroxyprogesterone dehydroxylase from Eubacterium sp. strain 144. Biochim Biophys Acta 837:103–110. doi: 10.1016/0005-2760(85)90232-2. [DOI] [PubMed] [Google Scholar]
- 124.Glass TL, Saxerud MH, Casper HH. 1991. Properties of a 4-ene-3-ketosteroid-5 alpha-reductase in cell extracts of the intestinal anaerobe, Eubacterium sp. strain 144. J Steroid Biochem Mol Biol 39:367–374. doi: 10.1016/0960-0760(91)90048-a. [DOI] [PubMed] [Google Scholar]
- 125.Watkins WE, Glass TL. 1991. Characteristics of 16-dehydroprogesterone reductase in cell extracts of the intestinal anaerobe, Eubacterium sp. strain 144. J Steroid Biochem Mol Biol 38:257–263. doi: 10.1016/0960-0760(91)90134-q. [DOI] [PubMed] [Google Scholar]
- 126.Morris DJ, Ridlon JM. 2017. Glucocorticoids and gut bacteria: “The GALF Hypothesis” in the metagenomic era. Steroids 125:1–13. doi: 10.1016/j.steroids.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 127.Eriksson H, Gustafsson JA, Sjövall J. 1969. Steroids in germfree and conventional rats: 21-dehydroxylation by intestinal microorganisms. Eur J Biochem 9:550–554. doi: 10.1111/j.1432-1033.1969.tb00644.x. [DOI] [PubMed] [Google Scholar]
- 128.Honour J. 1982. The possible involvement of intestinal bacteria in steroidal hypertension. Endocrinology 110:285–287. doi: 10.1210/endo-110-1-285. [DOI] [PubMed] [Google Scholar]
- 129.Honour J. 2015. Historical perspective: gut dysbiosis and hypertension. Physiol Genomics 47:443–446. doi: 10.1152/physiolgenomics.00063.2015. [DOI] [PubMed] [Google Scholar]
- 130.Edwards CR, Stewart PM, Burt D, Brett L, McIntyre MA, Sutanto WS, de Kloet ER, Monder C. 1988. Localisation of 11beta-hydroxysteroid dehydrogenase—tissue specific protector of the mineralocorticoid receptor. Lancet 332:986–989. doi: 10.1016/S0140-6736(88)90742-8. [DOI] [PubMed] [Google Scholar]
- 131.Bokkenheuser VD, Suzuki JB, Polovsky SB, Winter J, Kelly WG. 1975. Metabolism of deoxycorticosterone by human fecal flora. Appl Microbiol 30:82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Horwitt BN, Dorfman RI, Shipley RA, Fish WR. 1944. Metabolism of steroid hormones: conversion of desoxycorticosterone to pregnanediol-3(alpha), 20(alpha), in man and in the chimpanzee. J Biol Chem 155:213–218. [Google Scholar]
- 133.Engel LL, Carter P, Fielding LL. 1955. Urinary metabolites of administered corticosterone. I. Steroids liberated by glucuronidase hydrolysis. J Biol Chem 213:99–106. [PubMed] [Google Scholar]
- 134.Bokkenheuser VD, Winter J, Dehazya P, Kelly WG. 1977. Isolation and characterization of human fecal bacteria capable of 21-dehydroxylating corticoids. Appl Environ Microbiol 34:571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Feighner SD, Bokkenheuser VD, Winter J, Hylemon PB. 1979. Characterization of a C21 neutral steroid hormone transforming enzyme, 21-dehydroxylase, in crude cell extracts of Eubacterium lentum. Biochim Biophys Acta 574:154–163. doi: 10.1016/0005-2760(79)90094-8. [DOI] [PubMed] [Google Scholar]
- 136.de Prada P, Setchell KD, Hylemon PB. 1994. Purification and characterization of a novel 17alpha-hydroxysteroid dehydrogenase from an intestinal Eubacterium sp. VPI 12708. J Lipid Res 35:922–929. [PubMed] [Google Scholar]
- 137.Winter J, O’Rourke-Locascio S, Bokkenheuser VD, Mosbach EH, Cohen BI. 1984. Reduction of 17-keto steroids by anaerobic microorganisms isolated from human fecal flora. Biochim Biophys Acta 795:208–211. doi: 10.1016/0005-2760(84)90067-5. [DOI] [PubMed] [Google Scholar]
- 138.Stokes NA, Hylemon PB. 1985. Characterization of delta 4-3-ketosteroid-5 beta-reductase and 3 beta-hydroxysteroid dehydrogenase in cell extracts of Clostridium innocuum. Biochim Biophys Acta 836:255–261. doi: 10.1016/0005-2760(85)90073-6. [DOI] [PubMed] [Google Scholar]
- 139.Winter J, Bokkenheuser VD. 1978. 21-dehydroxylation of corticoids by anaerobic bacteria isolated from human fecal flora. J Steroid Biochem 9:379–384. doi: 10.1016/0022-4731(78)90604-0. [DOI] [PubMed] [Google Scholar]
- 140.Storbeck KH, Bloem LM, Africander D, Schloms L, Swart P, Swart AC. 2013. 11β-Hydroxydihydrotestosterone and 11-ketodihydrotestosterone, novel C19 steroids with androgenic activity: a putative role in castration resistant prostate cancer? Mol Cell Endocrinol 377:135–146. doi: 10.1016/j.mce.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 141.Pretorius E, Africander DJ, Vlok M, Perkins MS, Quanson J, Storbeck KH. 2016. 11-Ketotestosterone and 11-ketodihydrotestosterone in castration resistant prostate cancer: potent androgens which can no longer be ignored. PLoS One 11:e0159867. doi: 10.1371/journal.pone.0159867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Järvenpää P, Kosunen T, Fotsis T, Adlercreutz H. 1980. In vitro metabolism of estrogens by isolated intestinal micro-organisms and by human faecal microflora. J Steroid Biochem 13:345–349. doi: 10.1016/0022-4731(80)90014-x. [DOI] [PubMed] [Google Scholar]
- 143.Adlercreutz H, Järvenpää P. 1982. Assay of estrogens in human feces. J Steroid Biochem 17:639–645. doi: 10.1016/0022-4731(82)90565-9. [DOI] [PubMed] [Google Scholar]
- 144.Plottel CS, Blaser MJ. 2011. Microbiome and malignancy. Cell Host Microbe 10:324–335. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kwa M, Plottel CS, Blaser MJ, Adams S. 2016. The intestinal microbiome and estrogen receptor-positive female breast cancer. J Natl Cancer Inst 108:djw029. doi: 10.1093/jnci/djw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Colldén H, Landin A, Wallenius V, Elebring E, Fändriks L, Nilsson ME, Ryberg H, Poutanen M, Sjögren K, Vandenput L, Ohlsson C. 2019. The gut microbiota is a major regulator of androgen metabolism in intestinal contents. Am J Physiol Endocrinol Metab 317:E1182–E1192. doi: 10.1152/ajpendo.00338.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kornman KS, Loesche WJ. 1982. Effects of estradiol and progesterone on Bacteroides melaninogenicus and Bacteroides gingivalis. Infect Immun 35:256–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Blair J, Adaway J, Keevil B, Ross R. 2017. Salivary cortisol and cortisone in the clinical setting. Curr Opin Endocrinol Diabetes Obes 24:161–168. doi: 10.1097/MED.0000000000000328. [DOI] [PubMed] [Google Scholar]
- 149.Duran-Pinedo AE, Solbiati J, Frias-Lopez J. 2018. The effect of the stress hormone cortisol on the metatranscriptome of the oral microbiome. NPJ Biofilms Microbiomes 4:25. doi: 10.1038/s41522-018-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tiznobaik A, Taheri S, Torkzaban P, Ghaleiha A, Soltanian AR, Omrani R, Shrininzad M. 2019. Relationship between dental plaque formation and salivary cortisol level in pregnant women. Eur Oral Res 53:62–66. doi: 10.26650/eor.20192484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Soory M. 1995. Bacterial steroidogenesis by periodontal pathogens and the effect of bacterial enzymes on steroid conversions by human gingival fibroblasts in culture. J Periodontal Res 30:124–131. doi: 10.1111/j.1600-0765.1995.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 152.Jia M, Chew WM, Feinstein Y, Skeath P, Sternberg EM. 2016. Quantification of cortisol in human eccrine sweat by liquid chromatography-tandem mass spectrometry. Analyst 141:2053–2060. doi: 10.1039/c5an02387d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gower DB, Ruparelia BA. 1993. Olfaction in humans with special reference to odorous 16-androstenes: their occurrence, perception and possible social, psychological and sexual impact. J Endocrinol 137:167–187. doi: 10.1677/joe.0.1370167. [DOI] [PubMed] [Google Scholar]
- 154.Kenouch S, Lombes M, Delahaye F, Eugene E, Bonvalet JP, Farman N. 1994. Human skin as target for aldosterone: coexpression of mineralocorticoid receptors and 11 beta-hydroxysteroid dehydrogenase. J Clin Endocrinol Metab 79:1334–1341. doi: 10.1210/jcem.79.5.7962326. [DOI] [PubMed] [Google Scholar]
- 155.Hirasawa G, Sasano H, Takahashi K, Fukushima K, Suzuki T, Hiwatashi N, Toyota T, Krozowski ZS, Nagura H. 1997. Colocalization of 11 beta-hydroxysteroid dehydrogenase type II and mineralocorticoid receptor in human epithelia. J Clin Endocrinol Metab 82:3859–3863. doi: 10.1210/jcem.82.11.4337. [DOI] [PubMed] [Google Scholar]
- 156.Hennebold JD, Daynes RA. 1998. Inhibition of skin 11beta-hydroxysteroid dehydrogenase activity in vivo potentiates the anti-inflammatory actions of glucocorticoids. Arch Dermatol Res 290:413–419. doi: 10.1007/s004030050328. [DOI] [PubMed] [Google Scholar]
- 157.Vasaitis TS, Bruno RD, Njar VC. 2011. CYP17 inhibitors for prostate cancer therapy. J Steroid Biochem Mol Biol 125:23–31. doi: 10.1016/j.jsbmb.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bird S, Gower DB. 1982. Axillary 5 alpha-androst-16-en-3-one, cholesterol and squalene in men; preliminary evidence for 5 alpha-androst-16-en-3-one being a product of bacterial action. J Steroid Biochem 17:517–522. doi: 10.1016/0022-4731(82)90010-3. [DOI] [PubMed] [Google Scholar]
- 159.Austin C, Ellis J. 2003. Microbial pathways leading to steroidal malodour in the axilla. J Steroid Biochem Mol Biol 87:105–110. doi: 10.1016/s0960-0760(03)00387-x. [DOI] [PubMed] [Google Scholar]
- 160.Nixon A, Mallet AI, Jackman PJ, Gower DB. 1986. Testosterone metabolism by isolated human axillary Corynebacterium spp.: a gas-chromatographic mass-spectrometric study. J Steroid Biochem 24:887–892. doi: 10.1016/0022-4731(86)90450-4. [DOI] [PubMed] [Google Scholar]
- 161.Imirzalioglu C, Hain T, Chakraborty T, Domann E. 2008. Hidden pathogens uncovered: metagenomic analysis of urinary tract infections. Andrologia 40:66–71. doi: 10.1111/j.1439-0272.2007.00830.x. [DOI] [PubMed] [Google Scholar]
- 162.Moustafa A, Li W, Singh H, Moncera KJ, Torralba MG, Yu Y, Manuel O, Biggs W, Venter JC, Nelson KE, Pieper R, Telenti A. 2018. Microbial metagenomics of urinary tract infections. Sci Rep 8:4333. doi: 10.1038/s41598-018-22660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Golombos DM, Ayangbesan A, O’Malley P, Lewicki P, Barlow L, Barbieri CE, Chan C, DuLong C, Abu-Ali G, Huttenhower C, Scherr DS. 2018. The role of gut microbiome in the pathogenesis of prostate cancer: a prospective, pilot study. Urology 111:122–128. doi: 10.1016/j.urology.2017.08.039. [DOI] [PubMed] [Google Scholar]
- 164.Shrestha E, White JR, Yu SH, Kulac I, Ertunc O, De Marzo AM, Yegnasubramanian S, Mangold LA, Partin AW, Sfanos KS. 2018. Profiling the urinary microbiome in men with positive versus negative biopsies for prostate cancer. J Urol 199:161–171. doi: 10.1016/j.juro.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cavarretta I, Ferrarese R, Cazzaniga W, Saita D, Lucianò R, Ceresola ER, Locatelli I, Visconti L, Lavorgna G, Briganti A, Nebuloni M, Doglioni C, Clementi M, Montorsi F, Canducci F, Salonia A. 2017. The microbiome of the prostate tumor microenvironment. Eur Urol 72:625–631. doi: 10.1016/j.eururo.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 166.Doyle WI, Meeks JP. 2018. Excreted steroids in vertebrate social communication. J Neurosci 38:3377–3387. doi: 10.1523/JNEUROSCI.2488-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hofmann AF, Hagey LR, Krasowski MD. 2010. Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res 51:226–246. doi: 10.1194/jlr.R000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang DQ, Carey MC. 2014. Therapeutic uses of animal biles in traditional Chinese medicine: an ethnopharmacological, biophysical chemical and medicinal review. World J Gastroenterol 20:9952–9975. doi: 10.3748/wjg.v20.i29.9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Doyle WI, Dinser JA, Cansler HL, Zhang X, Dinh DD, Browder NS, Riddington IM, Meeks JP. 2016. Faecal bile acids are natural ligands of the mouse accessory olfactory system. Nat Commun 7:11936. doi: 10.1038/ncomms11936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Phelps T, Snyder E, Rodriguez E, Child H, Harvey P. 2019. The influence of biological sex and sex hormones on bile acid synthesis and cholesterol homeostasis. Biol Sex Differ 10:52. doi: 10.1186/s13293-019-0265-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sergeant MJ. 2010. Female perception of male body odor. Vitam Horm 83:25–45. doi: 10.1016/S0083-6729(10)83002-X. [DOI] [PubMed] [Google Scholar]
- 172.Gower DB, Nixon A, Jackman PJ, Mallett AI. 1986. Transformation of steroids by axillary coryneform bacteria. Int J Cosmet Sci 8:149–158. doi: 10.1111/j.1467-2494.1986.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 173.Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. 2010. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci U S A 107:20051–20056. doi: 10.1073/pnas.1009906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hahn MA, Effertz C, Bigler L, von Elert E. 2019. 5α-Cyprinol sulfate, a bile salt from fish, induces diel vertical migration in Daphnia. Elife 8:e44791. doi: 10.7554/eLife.44791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Buchinger TJ, Li W, Johnson NS. 2014. Bile salts as semiochemicals in fish. Chem Senses 39:647–654. doi: 10.1093/chemse/bju039. [DOI] [PubMed] [Google Scholar]
- 176.Brant CO, Chung-Davidson YW, Li K, Scott AM, Li W. 2013. Biosynthesis and release of pheromonal bile salts in mature male sea lamprey. BMC Biochem 14:30. doi: 10.1186/1471-2091-14-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. 2013. Gender bias in autoimmunity is influenced by microbiota. Immunity 39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shin JH, Park YH, Sim M, Kim SA, Joung H, Shin DM. 2019. Serum level of sex steroid hormone is associated with diversity and profiles of human gut microbiome. Res Microbiol 170:192–201. doi: 10.1016/j.resmic.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 179.Menon R, Watson SE, Thomas LN, Allred CD, Dabney A, Azcarate-Peril MA, Sturino JM. 2013. Diet complexity and estrogen receptor β status affect the composition of the murine intestinal microbiota. Appl Environ Microbiol 79:5763–5773. doi: 10.1128/AEM.01182-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Org E, Mehrabian M, Parks BW, Shipkova P, Liu X, Drake TA, Lusis AJ. 2016. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 7:313–322. doi: 10.1080/19490976.2016.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Elderman M, de Vos P, Faas M. 2018. Role of microbiota in sexually dimorphic immunity. Front Immunol 9:1018. doi: 10.3389/fimmu.2018.01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Buskiewicz IA, Huber SA, Fairweather D. 2016. Sex hormone receptor expression in the immune system, p 45–60. In Neigh GN, Mitzelfelt MM (ed), Sex differences in physiology. Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 183.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. 2013. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 184.Xiao L, Estellé J, Kiilerich P, Ramayo-Caldas Y, Xia Z, Feng Q, Liang S, Pedersen AØ, Kjeldsen NJ, Liu C, Maguin E, Doré J, Pons N, Le Chatelier E, Prifti E, Li J, Jia H, Liu X, Xu X, Ehrlich SD, Madsen L, Kristiansen K, Rogel-Gaillard C, Wang J. 2016. A reference gene catalogue of the pig gut microbiome. Nat Microbiol 1:16161. doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- 185.Flak MB, Neves JF, Blumberg RS. 2013. Immunology. Welcome to the microgenderome. Science 339:1044–1045. doi: 10.1126/science.1236226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wallis A, Butt H, Ball M, Lewis DP, Bruck D. 2016. Support for the microgenderome: associations in a human clinical population. Sci Rep 6:19171. doi: 10.1038/srep19171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. 2009. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev 89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 188.Pretorius E, Arlt W, Storbeck KH. 2017. A new dawn for androgens: novel lessons from 11-oxygenated C19 steroids. Mol Cell Endocrinol 441:76–85. doi: 10.1016/j.mce.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 189.Sheikh Abdul Kadir SH, Miragoli M, Abu-Hayyeh S, Moshkov AV, Xie Q, Keitel V, Nikolaev VO, Williamson C, Gorelik J. 2010. Bile acid-induced arrhythmia is mediated by muscarinic M2 receptors in neonatal rat cardiomyocytes. PLoS One 5:e9689. doi: 10.1371/journal.pone.0009689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ibrahim E, Diakonov I, Arunthavarajah D, Swift T, Goodwin M, McIlvride S, Nikolova V, Williamson C, Gorelik J. 2018. Bile acids and their respective conjugates elicit different responses in neonatal cardiomyocytes: role of Gi protein, muscarinic receptors and TGR5. Sci Rep 8:7110. doi: 10.1038/s41598-018-25569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Vasavan T, Ferraro E, Ibrahim E, Dixon P, Gorelik J, Williamson C. 2018. Heart and bile acids—clinical consequences of altered bile acid metabolism. Biochim Biophys Acta Mol Basis Dis 1864:1345–1355. doi: 10.1016/j.bbadis.2017.12.039. [DOI] [PubMed] [Google Scholar]
- 192.Thomas P. 2019. Membrane androgen receptors unrelated to nuclear steroid receptors. Endocrinology 160:772–781. doi: 10.1210/en.2018-00987. [DOI] [PubMed] [Google Scholar]
- 193.Pai R, Tarnawski AS, Tran T. 2004. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell 15:2156–2163. doi: 10.1091/mbc.e03-12-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mott GE, Brinkley AW, Mersinger CL. 1980. Biochemical characterization of cholesterol-reducing Eubacterium. Appl Environ Microbiol 40:1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Stellwag EJ, Hylemon PB. 1976. Purification and characterization of bile salt hydrolase from Bacteroides fragilis subsp. fragilis. Biochim Biophys Acta 452:165–176. doi: 10.1016/0005-2744(76)90068-1. [DOI] [PubMed] [Google Scholar]
- 196.Gopal-Srivastava R, Hylemon PB. 1988. Purification and characterization of bile salt hydrolase from Clostridium perfringens. J Lipid Res 29:1079–1085. [PubMed] [Google Scholar]
- 197.O’Flaherty S, Briner Crawley A, Theriot CM, Barrangou R. 2018. The Lactobacillus bile salt hydrolase repertoire reveals niche-specific adaptation. mSphere 3:e00140-18. doi: 10.1128/mSphere.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Grill JP, Cayuela C, Antoine JM, Schneider F. 2000. Isolation and characterization of a Lactobacillus amylovorus mutant depleted in conjugated bile salt hydrolase activity: relation between activity and bile salt resistance. J Appl Microbiol 89:553–563. doi: 10.1046/j.1365-2672.2000.01147.x. [DOI] [PubMed] [Google Scholar]
- 199.Bennett MJ, McKnight SL, Coleman JP. 2003. Cloning and characterization of the NAD-dependent 7alpha-hydroxysteroid dehydrogenase from Bacteroides fragilis. Curr Microbiol 47:475–484. doi: 10.1007/s00284-003-4079-4. [DOI] [PubMed] [Google Scholar]
- 200.Edenharder R, Pfützner A, Hammann R. 1989. Characterization of NAD-dependent 3alpha- and 3beta-hydroxysteroid dehydrogenase and of NADP-dependent 7 beta-hydroxysteroid dehydrogenase from Peptostreptococcus productus. Biochim Biophys Acta 1004:230–238. doi: 10.1016/0005-2760(89)90272-5. [DOI] [PubMed] [Google Scholar]
- 201.Doden H, Sallam LA, Devendran S, Ly L, Doden G, Daniel SL, Alves JMP, Ridlon JM. 2018. Metabolism of oxo-bile acids and characterization of recombinant 12alpha-hydroxysteroid dehydrogenases from bile acid 7alpha-dehydroxylating human gut bacteria. Appl Environ Microbiol 84:e00235-18. doi: 10.1128/AEM.00235-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wells JE, Hylemon PB. 2000. Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces. Appl Environ Microbiol 66:1107–1113. doi: 10.1128/aem.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ridlon JM, Devendran S, Alves JM, Doden H, Wolf PG, Pereira GV, Ly L, Volland A, Takei H, Nittono H, Murai T, Kurosawa T, Chlipala GE, Green SJ, Hernandez AG, Fields CJ, Wright CL, Kakiyama G, Cann I, Kashyap P, McCracken V, Gaskins HR. 2020. The ‘in vivo lifestyle’ of bile acid 7α-dehydroxylating bacteria: comparative genomics, metatranscriptomic, and bile acid metabolomics analysis of a defined microbial community in gnotobiotic mice. Gut Microbes 11:381–404. doi: 10.1080/19490976.2019.1618173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ridlon JM, Kang DJ, Hylemon PB. 2010. Isolation and characterization of a bile acid inducible 7alpha-dehydroxylating operon in Clostridium hylemonae TN271. Anaerobe 16:137–146. doi: 10.1016/j.anaerobe.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sutherland JD, Williams CN. 1985. Bile acid induction of 7 alpha- and 7 beta-hydroxysteroid dehydrogenases in Clostridium limosum. J Lipid Res 26:344–350. [PubMed] [Google Scholar]
- 206.Xavier JB, Young VB, Skufca J, Ginty F, Testerman T, Pearson AT, Macklin P, Mitchell A, Shmulevich I, Xie L, Caporaso JG, Crandall KA, Simone NL, Godoy-Vitorino F, Griffin TJ, Whiteson KL, Gustafson HH, Slade DJ, Schmidt TM, Walther-Antonio MRS, Korem T, Webb-Robertson BM, Styczynski MP, Johnson WE, Jobin C, Ridlon JM, Koh AY, Yu M, Kelly L, Wargo JA. 2020. The cancer microbiome: distinguishing direct and indirect effects requires a systemic view. Trends Cancer 6:192–204. doi: 10.1016/j.trecan.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bernardi RC, Doden HL, Melo MCR, Devendran S, Pollet RM, Mythen SM, Bhowmik S, Lesley SA, Cann I, Luthey-Schulten Z, Koropatkin NM, Ridlon JM. 2020. Bacteria on steroids: the enzymatic mechanism of an NADH-dependent dehydrogenase that regulates the conversion of cortisol to androgen in the gut microbiome. bioRxiv doi: 10.1101/2020.06.12.149468. [DOI]
- 208.Cho HS, Choi G, Choi KY, Oh BH. 1998. Crystal structure and enzyme mechanism of delta 5-3-ketosteroid isomerase from Pseudomonas testosteroni. Biochemistry 37:8325–8330. doi: 10.1021/bi9801614. [DOI] [PubMed] [Google Scholar]
- 209.Adhikari AA, Seegar TCM, Ficarro SB, McCurry MD, Ramachandran D, Yao L, Chaudhari SN, Ndousse-Fetter S, Banks AS, Marto JA, Blacklow SC, Devlin AS. 2020. Development of a covalent inhibitor of gut bacterial bile salt hydrolases. Nat Chem Biol 16:318–326. doi: 10.1038/s41589-020-0467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Funabashi M, Grove TL, Wang M, Varma Y, McFadden ME, Brown LC, Guo C, Higginbottom S, Almo SC, Fischbach MA. 2020. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature 582:566–570. doi: 10.1038/s41586-020-2396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]