Abstract
Attention deficit/hyperactivity disorder (ADHD) is a common neurodevelopmental disorder characterized by age-inappropriate symptoms of inattention, impulsivity, and hyperactivity that persist into adulthood in the majority of the diagnosed children. Despite several risk factors during childhood predicting the persistence of ADHD symptoms into adulthood, the genetic architecture underlying the trajectory of ADHD over time is still unclear. We set out to study the contribution of common genetic variants to the risk for ADHD across the lifespan by conducting meta-analyses of genome-wide association studies on persistent ADHD in adults and ADHD in childhood separately and jointly, and by comparing the genetic background between them in a total sample of 17,149 cases and 32,411 controls. Our results show nine new independent loci and support a shared contribution of common genetic variants to ADHD in children and adults. No subgroup heterogeneity was observed among children, while this group consists of future remitting and persistent individuals. We report similar patterns of genetic correlation of ADHD with other ADHD-related datasets and different traits and disorders among adults, children, and when combining both groups. These findings confirm that persistent ADHD in adults is a neurodevelopmental disorder and extend the existing hypothesis of a shared genetic architecture underlying ADHD and different traits to a lifespan perspective.
Subject terms: Genetic markers, ADHD
Introduction
Attention deficit/hyperactivity disorder (ADHD) is a common neurodevelopmental disorder that severely impairs the daily functioning of patients due to age-inappropriate levels of impulsivity and hyperactivity, and/or difficulties in focusing attention [1]. ADHD has a prevalence of 5–6% in childhood, and impairing symptoms persist into adulthood in around two-thirds of children with ADHD diagnosis, with an estimated adult prevalence around 3.4% [1, 2].
ADHD is a multifactorial disorder with heritability averaging 76% throughout the lifespan [3–5]. There is consistent evidence that both common and rare variants make an important contribution to the risk for the disorder [6–11]. Several genome-wide association studies (GWAS) and meta-analyses across those have been conducted [7], but only the largest GWAS meta-analysis (GWAS-MA) performed to date reported genome-wide significant loci [6]. This study concluded that common genetic variants (minor allele frequency, MAF > 0.01) account for 22% of the heritability of the disorder [6] and supported substantial genetic overlap between ADHD and other brain disorders and behavioral/cognitive traits [6, 12].
The presentation of ADHD symptoms changes from childhood to adulthood, with lower levels of hyperactivity in adulthood but a high risk for ongoing attention problems, disorganization, and emotional dysregulation [13, 14]. As in the general population, the pattern of psychiatric and somatic comorbid conditions in ADHD also changes substantially over time, with learning disabilities, oppositional defiant disorder, and conduct disorder being more prevalent in children, and substance use disorders, social phobia, insomnia, obesity, and mood disorders becoming more pronounced in adulthood [1, 15–18]. In addition, persistent ADHD in adults is, compared with the general population (and to cases with remitting ADHD), associated with higher risk for a wide range of functional and social impairments, including unemployment, accidents, and criminal behavior [7, 19–23].
Several risk factors measured in childhood predict the persistence of ADHD symptoms into adulthood, such as the presence of comorbid disorders, the severity of ADHD symptoms, being exposed to psychosocial adversity, as well as having a high polygenic risk score (PRS) for childhood ADHD [24–28]. Twin studies suggest that both stable and dynamic genetic influences affect the persistence of ADHD symptoms [4, 5, 29, 30]. However, specific genetic factors differentiating childhood and persistent ADHD into adulthood are not well understood due to the lack of longitudinal studies. Molecular studies, including the most recent GWAS-MA of ADHD [6], have been performed in children and adults either separately or jointly [6, 31–40], but large-scale analyses comparing their genetic basis are yet to be conducted.
Given this background, we set out to study the contribution of common genetic variants to the risk for ADHD from a lifespan perspective by conducting the largest GWAS-MAs performed so far on persistent ADHD in adults (diagnosed according to DSM-IV/ICD-10 criteria) and on ADHD in childhood (that may include remittent and persistent forms of the disorder) separately and jointly. For the first time, we estimated the genetic correlation between childhood and persistent ADHD, compared their patterns of genetic correlation with other traits and disorders, assessed the effect of childhood ADHD PRSs on persistent ADHD, and explored whether individuals in which ADHD symptoms may persist into adulthood could be distinguished already in childhood using genetic data.
Material and methods
Sample description
A total of 19 GWAS of ADHD comprising 49,560 individuals (17,149 cases and 32,411 controls), provided by the Psychiatric Genomics Consortium (PGC), the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), and the International Multi-centre persistent ADHD CollaboraTion (IMpACT), were analyzed. All participants were of European ancestry, had provided informed consent, and all sites had documented permission from local ethics committees.
The meta-analysis on persistent ADHD was conducted in 22,406 individuals (6,532 ADHD adult cases and 15,874 controls) using six datasets from the IMpACT consortium, two datasets from the PGC, and the adult subset from the iPSYCH cohort included in Demontis and Walters et al. [6]. The meta-analysis on ADHD in childhood included 27,154 individuals (10,617 cases and 16,537 controls), comprising two Brazilian and Spanish cohorts, seven datasets from the PGC, and the children subset from the iPSYCH cohort included in Demontis and Walters et al. [6]. All patients met DSM-IV/ICD-10 diagnostic criteria. In total, 7,086 new samples not included in Demontis and Walters et al. [6] were considered in the present study. Detailed information on each dataset is provided in Table S1 and in Supplementary Methods.
GWAS and meta-analyses
Genotyping platforms and quality control (QC) filters for each of the datasets are shown in Table S1. Pre-imputation QC at individual and SNP level were performed using the Rapid Imputation and COmputational PIpeLIne with the default settings (https://sites.google.com/a/broadinstitute.org/ricopili/). Non-European ancestry samples, related and duplicated individuals, and subjects with sex discrepancies were excluded. Phasing of genotype data was performed using the SHAPEIT2 algorithm, and imputation for unrelated samples and trios was performed with MaCH, IMPUTE2, or MINIMAC3 (http://genome.sph.umich.edu/wiki/Minimac3) depending on software availability at the time of imputation (Table S1) [41–43]. The European ancestry panel of the 1000 Genomes Project using genome build hg19 was considered as reference for genotype imputation (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/). After imputation, the association with ADHD of genotype dosages was tested using logistic regression in PLINK 1.9 [44], assuming an additive genetic model and including sex, the first ten principal components, and other relevant covariates for each case-control study (Table S1). GWAS summary statistics were filtered prior to meta-analysis, excluding variants with MAF < 0.01, and imputation quality scores (INFO) ≤ 0.8. Inverse-variance weighted fixed-effects meta-analyses were conducted using METAL [45] and results were filtered by effective sample size > 70% of the total, defined as [46]. The genome-wide significance threshold was set at P < 5.00E−08 to correct for multiple testing. Independent loci for variants exceeding this threshold were defined based on clumping using PLINK 1.9. Variants that were ±250 kb away from the index variant (variant with smallest P value in the region), with P value < 0.001, and with an estimated linkage disequilibrium (LD) of r2 > 0.2 with the index variant were assigned to a clump (p1 = 5.00E−08, p2 = 0.001, r2 = 0.2, kb = 250). Manhattan and Forest plots were generated using the “qqman” and “forestplot” R packages (3.4.4R version), respectively. The LocusZoom software [47] was used to generate regional association plots.
Details of downstream analyses for top signals identified are provided in the online supplement and include conditional analysis, Bayesian credible set analysis, and functional characterization of the significant variants.
SNP-based heritability (SNP-h2)
The SNP-h2 was estimated by single-trait LD score regression using summary statistics, HapMap 3 LD-scores, considering default SNP QC filters (INFO > 0.9 and MAF > 0.01) and assuming population prevalence of 3.4, 5.5, and 5% for persistent ADHD, ADHD on childhood, and ADHD across the lifespan, respectively, [48]. Data of 1,113,287, 1,072,558, and 1,092,418 SNPs from the GWAS-MA of persistent ADHD, ADHD on childhood, and ADHD across the lifespan, respectively, were considered to estimate the liability-scale SNP-h2. Partitioning and enrichment of the heritability by functional categories was analyzed using the 24 main annotations (no window around the functional categories) described by Finucane et al. [49]. Statistical significance was set using Bonferroni correction (P < 2.08E−03).
Gene-based and gene-set analyses
MAGMA software was undertaken for gene-based and gene-set association testing using summary data from our GWAS-MAs [50]. Variants were mapped to a gene if they were within 20 kb upstream or downstream from the gene according to dbSNP build 135 and NCBI 37.3 gene definitions. Genes in the MHC region (hg19:chr6:25-35M) were excluded from the analyses. LD patterns were estimated using the European ancestry reference panel of the 1000 Genomes Project. Gene sets denoting canonical pathways were downloaded from MSigDB (http://www.broadinstitute.org/gsea/msigdb), which integrates Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/), BioCarta (http://www.biocarta.com/), Reactome (https://reactome.org/), and Gene Ontology (GO) (http://www.geneontology.org/) resources. Bonferroni correction (P < 2.77E−06 for 18,038 genes in persistent ADHD; P < 2.75E−06 for 18,218 genes in childhood ADHD; P < 2.79E−06 for 17,948 genes in ADHD across the lifespan) and 10,000 permutations were used for multiple testing correction in the gene-based and gene-set analyses, respectively.
BUHMBOX analysis
The Breaking Up Heterogeneous Mixture Based On cross(X)-locus correlations (BUHMBOX) analysis [51] was used to test whether the genetic correlation between persistent ADHD and ADHD in childhood was driven by subgroup heterogeneity, found when there is a subset of children enriched for persistent ADHD-associated alleles. Subgroup heterogeneity was tested in each childhood dataset considering independent SNPs (r2 = 0.1, kb = 10,000) with MAF > 0.05 from the GWAS-MA of persistent ADHD using two different P value thresholds of P < 5.00E−05 (62 SNPs) and P < 1.00E−03 (710 SNPs). Results were meta-analyzed using the standard weighted sum of z-score approach, where z-scores are weighted by the square root of the effective sample size. The statistical power was calculated using 1,000 simulations, considering the ADHD children meta-analysis sample size, the odds ratios and risk allele frequencies from the GWAS-MA of persistent ADHD, and assuming 65% of heterogeneity proportion (π).
Sign test
The direction of the effect of variants associated with ADHD in childhood was tested in persistent ADHD and vice versa, using strict clumping (r2 = 0.05, kb = 500, p2 = 0.5) and different P value thresholds (1.00E−07, 5.00E−07, 1.00E−06, 5.00E−06, 1.00E−05, 5.00E−05, 1.00E−04, and 5.00E−04). The concordant direction of effect was evaluated using a one sample test of the proportion with Yates’ continuity correction against a null hypothesis of P = 0.50 with the “stats” R package.
Polygenic risk scoring
PRSs were constructed using different P value thresholds (P < 0.001, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, and 1) to select independent variants (p1 = 1, p2 = 1, r2 = 0.1, kb = 250) from the childhood GWAS-MA of ADHD and were then tested for association with persistent ADHD in each of the nine datasets, adjusting for the covariates included in the GWAS and using PRSice-2 (https://choishingwan.github.io/PRSice/). Best guess genotypes for nonambiguous strand variants present in all the persistent ADHD studies (missing rate < = 0.02) were included (NSNPs = 32,584 for P =1). Results from the nine PRS analyses at each P value threshold were combined using inverse-variance weighted meta-analysis.
Genetic correlation
Cross-trait LD score regression with unconstrained intercept was used to calculate genetic correlations (rg) between pairs of traits, considering HapMap3 LD-scores, markers with INFO ≥ 0.90, and excluding the MHC region (hg19:chr6:25-35M) [48]. Other ADHD datasets [6, 52] and phenotypes from the LD-hub centralized database [53] with heritability z-scores (observed heritability/observed standard error) >4 and with an observed heritability > 0.1 were considered (N = 139 out of 689 available traits). Statistical significance was set using Bonferroni correction (P < 3.60E−04). Pearson’s correlation coefficient (Pearson’s r) was calculated between the genetic correlations of persistent ADHD with the phenotypes from the LD-hub and the genetic correlations of ADHD in childhood with the phenotypes from the LD-hub.
Results
GWAS-MA of persistent ADHD in adults
The GWAS-MA of persistent ADHD in adults included 6,532 adult ADHD cases and 15,874 controls. Minimal population stratification or other systematic biases were detected (LD score regression intercept = 1.01, Fig. S1a). The proportion of heritability of persistent ADHD attributable to common single-nucleotide polymorphisms on the liability scale (SNP-h2) was 0.19 (SE = 0.024), with a nominally significant enrichment in the heritability of variants located in conserved genomic regions (P = 5.18E−03) and in the cell-specific histone mark H3K4me1 (P = 3.17E−02) (Fig. S2a). The gene-based analysis revealed six genes in four loci (ST3GAL3, FRAT1/FRAT2, CGB1, and RNF225/ZNF584) significantly associated with persistent ADHD, with ST3GAL3 being the most significant one (P = 8.72E−07) (Table S2a). The single-marker analysis showed no variants exceeding genome-wide significance, with the most significant signal being rs3923931 (P = 1.69E−07) (Fig. 1a and Table S3a). Similarly, no significant gene sets were identified in the pathway analysis after correction for multiple comparisons (Table S4a [excel file]).
Fig. 1. Manhattan plots of the three GWAS meta-analyses conducted.
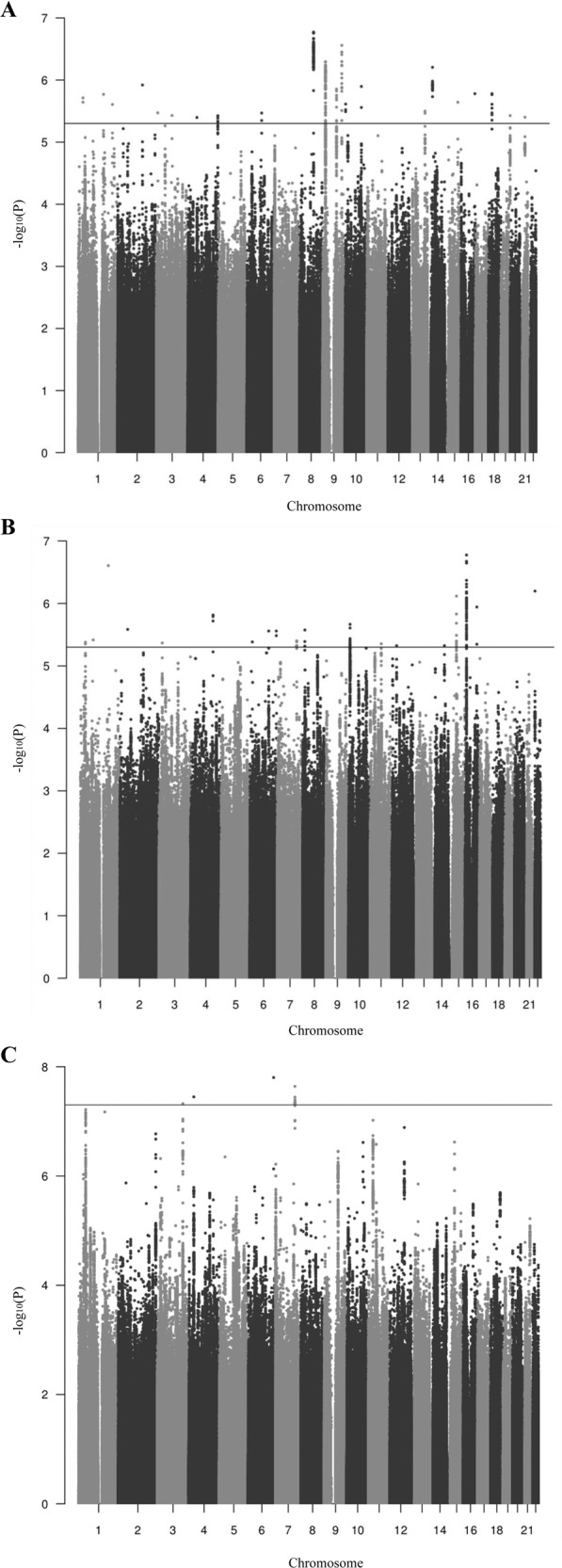
(a) GWAS-MA of nine cohorts of persistent ADHD in adults, (b) GWAS-MA of ten cohorts of ADHD in childhood, and (c) GWAS-MA of all datasets of ADHD across the lifespan (ADHD in childhood + persistent ADHD). Horizontal lines indicate suggestive (P value = 5.00E−06) and genome-wide significant (P = 5.00E−08) thresholds in a-b, and c, respectively.
GWAS-MA of ADHD in childhood
To compare the genetic background between persistent ADHD in adults and ADHD in childhood (that may include future remittent and persistent forms of the disorder), we conducted a GWAS-MA on children with ADHD in a total of 10,617 ADHD cases and 16,537 controls. We found no evidence of genomic inflation or population stratification (LD score regression intercept = 1.02, Fig. S1b). The liability-scale SNP-h2 for ADHD in childhood was 0.19 (SE = 0.021), with a significant enrichment in the heritability of variants located in conserved genomic regions after Bonferroni correction (P = 1.21E−06) (Fig. S2b). The gene-based analysis highlighted a significant association between FEZF1 and ADHD in childhood (P = 5.42E−07) (Table S2b). No single genetic variant exceeded genome-wide significance, with the top signal being in rs55686778 (P = 1.67E−07) (Fig. 1b and Table S3b), and no significant gene sets were identified in the pathway analysis after correction for multiple comparisons (Table S4b [excel file]).
Comparison of the genetic background of persistent ADHD in adults and ADHD in childhood
We found a strong genetic correlation between persistent ADHD in adults and ADHD in childhood (rg = 0.81, 95% CI: 0.64–0.97), significantly different from 0 (P = 2.13E−21) and from 1 (P = 0.02). Sign test results provided evidence of a consistent direction of effect of genetic variants associated with ADHD in childhood in persistent ADHD and vice versa (P = 6.60E−04 and P = 4.47E−03, respectively, for variants with P < 5.00E−05 in each dataset) (Table S5). In addition, PRS analyses showed that childhood ADHD PRSs were associated with persistent ADHD at different predefined P value thresholds, with the P = 0.40 threshold (NSNPs = 20,398) explaining the most variance (r2 = 0.0041 and P = 1.20E−27) (Fig. 2a). The quintiles of the PRS built using this threshold showed the expected trend of higher ADHD risk for individuals in higher quintiles (Fig. 2b, Table S6).
Fig. 2. Polygenic risk scores for ADHD in childhood tested on persistent ADHD as target sample.
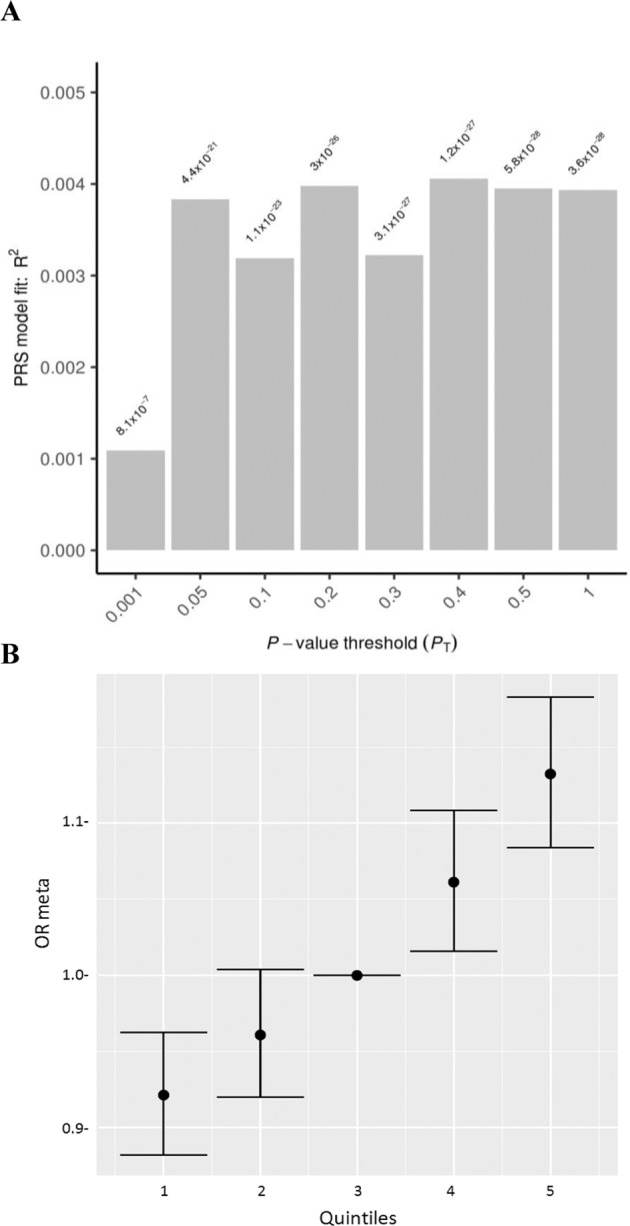
a Bar plot and b quintile plot of meta-analysis odds ratios (OR meta) with 95% confidence intervals for P value threshold = 0.4 using the third quintile as baseline.
We then tested whether the genetic correlation between persistent ADHD and ADHD in childhood was driven by a subset of children enriched for persistent ADHD-associated alleles using the Breaking Up Heterogeneous Mixture Based On Cross-locus correlations (BUHMBOX) analysis. We found no evidence of subgroup genetic heterogeneity in children, supporting that the sharing of persistent ADHD-associated alleles between children and adults was driven by the whole group of children, with a statistical power of 98.4 and 100% for thresholds of P < 5.00E−05 and P < 1.00E−03, respectively (Table S7).
GWAS-MA of ADHD across the lifespan
Given the strong genetic correlation between persistent ADHD in adults and in childhood, we performed a GWAS-MA of ADHD across the lifespan considering all datasets included in the GWAS-MAs. In total, 17,149 ADHD cases and 32,411 controls were included, and no evidence of genomic inflation or population stratification was found (LD score regression intercept = 1.03, Fig. S1c). The liability-scale SNP-h2 for ADHD across the lifespan was 0.17 (SE = 0.013), and a significant enrichment in the heritability of variants located in conserved genomic regions was observed after Bonferroni correction (P = 1.53E−06) (Fig. S2c). We identified four genome-wide significant variants (Figs. 1c and 3, Table 1a, and Fig. S3) and nine genes in seven loci (FEZF1, DUSP6, ST3GAL3/KDM4A, SEMA6D, C2orf82/GIGYF2, AMN, and FBXL17) significantly associated with ADHD across the lifespan (Table 1b). The most significantly associated locus was on chromosome 6 (index variant rs183882582-T, OR = 1.43 (95% CI: 1.26–1.60), P = 1.57E−08), followed by loci on chromosome 7 (index variant rs3958046), chromosome 4 (index variant rs200721207), and chromosome 3 (index variant rs1920644) (Table 1a, Fig. 3). The gene-set analysis showed a significant association of the “ribonucleoprotein complex” GO term with ADHD across the lifespan (P.adj = 0.021) (Table S4c [excel file]).
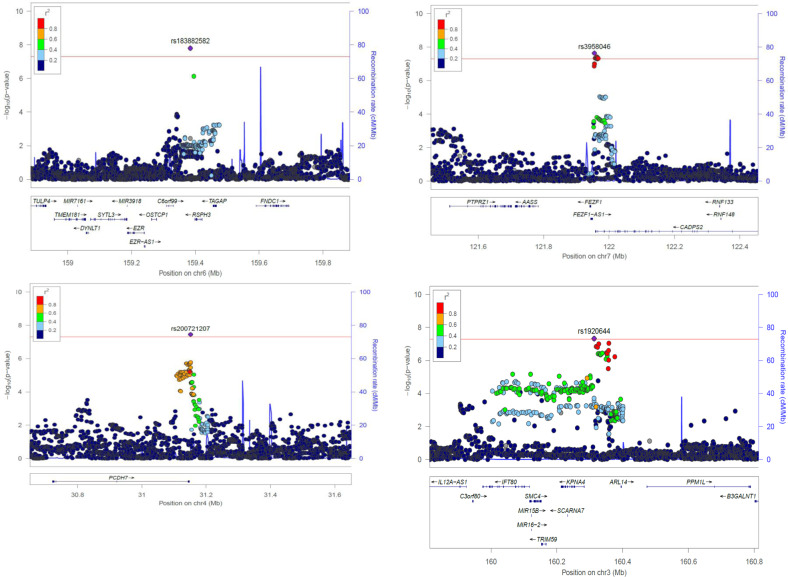
Fig. 3. Regional association plots for genome-wide significant loci identified in the GWAS meta-analysis of ADHD across the lifespan.
Each plot includes information about the locus, the location and orientation of the genes in the region, the local estimates of recombination rate (in the right corner), and the LD estimates of surrounding SNPs with the index SNP (r2 values are estimated based on 1000 Genomes European reference panel), which is indicated by color (in the upper left corner).
Table 1.
Genome-wide significant loci in the GWAS meta-analysis of ADHD across the lifespan identified through (A) single-variant analysis and (B) gene-based analysis.
| Chr | BP | SNP | Effect allele | Freq effect allele | OR | CI 95% | P value | Gene |
|---|---|---|---|---|---|---|---|---|
| A | ||||||||
| 6 | 159384224 | rs183882582 | T | 0.98 | 1.43 | 1.26–1.60 | 1.57E−08 | RSPH3 (+14 kb) |
| 7 | 121955328 | rs3958046 | T | 0.40 | 1.09 | 1.06–1.10 | 2.28E−08 | CADPS2 (+3.2 kb)/FEZF1 (−13.9 kb)/FEZF1-AS1 (+5.2 kb) |
| 4 | 31151465 | rs200721207 | T | 0.66 | 1.10 | 1.06–1.13 | 3.56E−08 | PCDH7 (−3.0 kb) |
| 3 | 160313354 | rs1920644 | T | 0.52 | 1.09 | 1.05–1.12 | 4.74E−08 | BC125159 (+27.9 kb)/KPNA4 (−30 kb)/ARL14 (−81.6 kb) |
| Gene | Chr | Start | Stop | N SNPs* | N PARAM** | Z-STAT | P value | |
|---|---|---|---|---|---|---|---|---|
| B | ||||||||
| FEZF1 | 7 | 121921373 | 121971173 | 108 | 18 | 5.6 | 9.57E−09 | |
| DUSP6 | 12 | 89721837 | 89766296 | 103 | 12 | 5.4 | 3.51E−08 | |
| ST3GAL3 | 1 | 44153204 | 44416837 | 521 | 19 | 5.4 | 3.58E−08 | |
| SEMA6D | 15 | 47456403 | 48086420 | 1565 | 55 | 5.3 | 7.24E−08 | |
| KDM4A | 1 | 44095797 | 44191189 | 169 | 13 | 4.9 | 4.34E−07 | |
| C2orf82 | 2 | 233713724 | 233761111 | 138 | 17 | 4.8 | 7.74E−07 | |
| GIGYF2 | 2 | 233542015 | 233745287 | 511 | 19 | 4.8 | 8.36E−07 | |
| AMN | 14 | 103368993 | 103417179 | 101 | 21 | 4.6 | 2.56E−06 | |
| FBXL17 | 5 | 107174734 | 107738080 | 1273 | 35 | 4.6 | 2.59E−06 | |
The location (chromosome (Chr) and base position (BP)), effect allele and its frequency, odds ratio (OR) of the effect allele with 95% confidence interval (CI 95%) and association P values, along with genes in the locus are shown for each index variant ID (SNP). For the gene-based results, the number of single-nucleotide polymorphisms in the genes (*) and the number of relevant parameters used in the model by MAGMA software (**) are given.
One of the four loci identified in the single-variant analysis also reached genome-wide significance in the previous GWAS-MA on ADHD [6], and all of them showed consistent direction of the effect in that study (Table S8a). Significant loci reported by Demontis et al. [6] showed nominal association with ADHD across the lifespan in our study (Table S8b, c), with single variant hits showing the same direction of the effect (Table S8b).
Analyses conditioning on the index variant for the four ADHD-associated loci did not reveal new independent markers. These four significant loci were functionally characterized by obtaining Bayesian credible sets and searching for expression quantitative trait loci (eQTL) using available data in blood or brain [54, 55]. We found that credible sets for three of the four loci contained at least one eQTL within 1 Mb of the index variant. The credible set on chromosome 6 included the index variant (rs183882582) and rs12197454. This variant, in LD with the index variant (r2 = 0.56), was associated with the expression of RSPH3 in blood and brain (P.adj < 1.65E−05 and P.adj = 2.36E−07, respectively), and with the expression of VIL2 in blood (P.adj = 3.21E−03). The credible set for the second most associated locus on chromosome 7 included 24 variants. The index variant, rs3958046, and other variants in this set, were eQTLs for CADPS2 in brain (maximum P.adj = 2.91E−03). The credible set for the locus on chromosome 4 contained 50 variants, most of them located in or near PCDH7, but no eQTLs were identified. In the credible set for the locus on chromosome 3, which included 98 variants, the index variant, rs1920644, was associated with the expression of KPNA4, IFT80, and KRT8P12 in brain (P.adj = 1.16E−04, P.adj = 1.40E−03, and P.adj = 1.77E−03, respectively). Many other variants in this set were eQTLs for these genes and also for TRIM59, OTOL1, and/or C3orf80 in brain (P.adj < 0.05) (Table S9 [excel file]).
In a summary-data-based Mendelian randomization (SMR) analysis, we used summary data from the GWAS-MA of ADHD across the lifespan and the eQTL data in blood and brain from Westra et al. [54] and Qi et al. [55] to identify gene expression levels associated with ADHD. We found a significant association between ADHD across the lifespan and RMI1 expression in blood (PSMR = 5.36E−06) (Table S10 [in excel]), finding not likely to be an artifact due to LD between eQTL and other ADHD-associated variants given that the PHEIDI was 0.47.
Genetic correlation with other ADHD datasets and phenotypes
We found significant genetic correlations of ADHD in children and adults from the previous GWAS-MA [6] (N = 53,296) and persistent ADHD (rg = 0.85, SE = 0.04, P = 5.49E−99), ADHD in childhood (rg = 0.99, SE = 0.03, P = 5.02E−273), and ADHD across the lifespan (rg = 0.98, SE = 0.01, P < 2.23E−308) (Table S11). When removing sample overlap (LD score genetic covariance intercept = 0.75) and considering only the subset of new samples included in our GWAS-MA on ADHD across the lifespan (N = 7086), a significant genetic correlation was also obtained between their sample and ours (rg = 0.91, SE = 0.35, P = 8.70E−03).
We also observed significant genetic correlations between childhood ADHD symptom scores from a GWAS-MA in a population of children reported by the EAGLE consortium [52] (N = 17,666) and persistent ADHD (rg = 0.65, SE = 0.20, P = 1.10E−03), ADHD in childhood (rg = 0.98, SE = 0.21, P = 2.76E−06), and ADHD across the lifespan (rg = 0.87, SE = 0.19, P = 4.80E−06). Similarly, significant genetic correlations between GWAS of self-reported ADHD status from 23andMe (N = 952,652) and persistent ADHD (rg = 0.75, SE = 0.05, P = 2.49E−45), ADHD in childhood (rg = 0.63, SE = 0.05, P = 1.39E−42), and ADHD across the lifespan (rg = 0.72, SE = 0.04, P = 4.86E−88) were observed (Table S11).
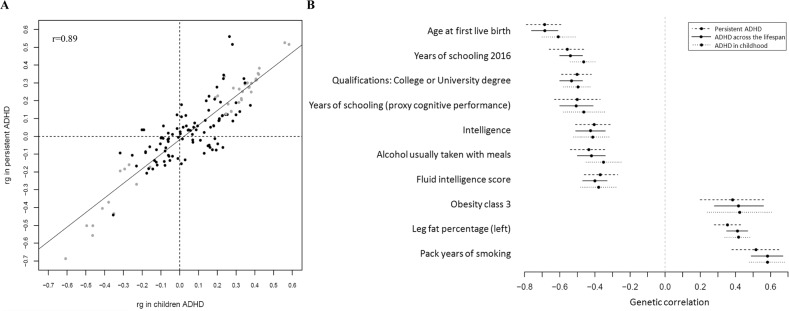
We also estimated the genetic correlation of persistent ADHD in adults, ADHD in childhood, and ADHD across the lifespan with all available phenotypes in LD-hub. Results for 139 phenotypes passed the QC parameters and 41 genetic correlations were significant after Bonferroni correction in both children and adults with persistent ADHD (Table S12 [excel file]). Again, the genetic correlations with ADHD were consistent across the lifespan, with similar patterns found in adulthood and childhood (Pearson’s r = 0.89) (Fig. 4a, Table S12 [excel file]). The strongest genetic correlations with ADHD were found for traits related to academic performance, intelligence, and risk-taking behaviors, including smoking and early pregnancy (Fig. 4b).
Fig. 4. Genetic correlation of ADHD and several traits.
a Dots represent genetic correlations (rg) for all traits considered (with h2 > 0.1 and z-score > 4) and those traits that met Bonferroni correction in both children and adult ADHD groups are presented in grey. r indicates Pearson’s correlation coefficient. b The ten strongest genetic correlations (with 95% confidence intervals) surpassing Bonferroni corrections in the children and persistent ADHD analysis are shown for each trait and ADHD.
Discussion
In the current study, we set out to explore the contribution of common genetic variants to the risk of ADHD across the lifespan by conducting GWAS-MAs separately for children and adults with persistent ADHD that meet DSM-IV/ICD-10 criteria. Using the largest GWAS datasets available from the PGC, the iPSYCH, and IMpACT consortia we found evidence for a common genetic basis for ADHD in childhood and persistent ADHD in adults and identified nine new loci associated with the disorder.
We found a highly similar proportion of the heritability of ADHD explained by common variants in children and in adults (SNP-h2 = 0.19), which is consistent with the SNP-h2 estimate reported in the recent GWAS-MA on ADHD [6] (SNP-h2 = 0.22), that included children and adults, and is in line with multiple studies supporting the stability of ADHD’s heritability from childhood to adulthood [3–5]. These results together with the 0.81 genetic correlation found between children and adults with persistent ADHD reinforce the hypothesis of the neurodevelopmental nature of persistent ADHD in adults. Consistently, the sign test and the PRS analysis confirmed the extensive overlap of common genetic risk variants for ADHD in childhood and adulthood.
In the view of the fact that children with ADHD may be an admixed group of individuals whose ADHD symptoms will persist or remit in adulthood, we ran a BUHMBOX analysis to elucidate if the potential “persistent” individuals could be distinguishable already in childhood. Our data supported genetic similarities in ADHD across the lifespan with no evidence of a subset of patients enriched for persistent ADHD-associated alleles within the group of children.
Despite not having identified specific genetic contributions for ADHD in children or persistent ADHD, our results are not inconsistent with evidence suggesting changes in the genetic contribution to ADHD symptoms from childhood into adulthood, as described in previous twin studies in the general population [4, 5, 29, 30]. Our study design and the still limited statistical power of the GWAS-MAs may have facilitated the identification of the shared genetic basis rather than specific genetic factors for persistence. Also, differences between the origin of the samples (population-based versus clinical) and/or discrepancies between self- and medical reports could explain why we found no group-specific genetic variants. In addition, given that Chen et al. [56] and Biederman et al. [57] reported that persistence of ADHD into adulthood indexed stronger familial aggregation of ADHD, we cannot yet discard influences of non-additive genetic effects, or other types of genetic variation, such as rare mutations or copy number variation, playing a role in the different ADHD trajectories across the lifespan.
We also found strong and significant positive genetic correlations of ADHD ascertained in clinical populations of adults, children, or both with other ADHD-related measures from general population samples, including the largest GWAS of self-reported ADHD status from 23andMe participants (N = 952,652) and the GWAS-MA of childhood rating scales of ADHD symptoms in the general population [52]. In agreement with previous reports, these data suggest that a clinical diagnosis of ADHD in adults is an extreme expression of continuous heritable traits [6] and that a single question about ever having received an ADHD diagnosis, as in the 23andMe sample, may be informative for molecular genetics studies.
Similar patterns of genetic correlation of ADHD with different somatic disorders and anthropometric, cognitive, and educational traits were identified for children and adults. These findings were highly similar to those observed in the recent GWAS-MA [6] and further extend the existing hypothesis of a shared genetic architecture underlying ADHD and these traits to a lifespan perspective.
We report 13 loci in gene- and SNP-based analyses for childhood ADHD, adult ADHD, and/or ADHD across the lifespan. Four ADHD-associated loci were previously identified by Demontis et al. [6], which was expected due to the sample overlap between the two datasets. The new loci identified in the present study mainly included genes involved in brain formation and function, such as FEZF1, a candidate for autism spectrum disorder implicated in the formation of the diencephalon [58, 59], RSPH3, which participates in neuronal migration in embryonic brain [60], CADPS2, which has been associated with psychiatric conditions due to its role in monoamine and neurotrophin neurotransmission [61–64], AMN, which is involved in the uptake of vitamin B12 [65, 66], essential for brain development, neural myelination, and cognitive function [67], and FBXL17, which has previously been related to intelligence [68].
The main limitation of this study is the sample overlap (85.7%) between the present GWAS-MAs and the previous one by Demontis et al. [6], which highlighted loci previously associated with ADHD. Although sample overlap may have inflated the genetic correlation found between these studies, the estimate remained strong and significant when excluding nonoverlapping datasets.
In summary, the present cross-sectional analyses identify new genetic loci associated with ADHD and, more importantly, support the hypothesis that persistent ADHD in adults is a neurodevelopmental disorder that shows a high and significant genetic overlap with ADHD in children. Future longitudinal studies will be required to disentangle the role of common genetic variants on ADHD remittance and/or persistence.
Funding and disclosure
VR has served on the speakers for Eli Lilly, Rubio, and Shire in the last 5 years. She has received travel awards from Eli Lilly and Co. and Shire for participating in psychiatric meetings. The ADHD Program has received unrestricted educational and research support from Eli Lilly and Co., Janssen-Cilag, Shire, Rovi, Psious, and Laboratorios Rubió in the past 2 years. MC received travel awards for taking part in psychiatric meetings from Shire. CF received travel awards for taking part in psychiatric meetings from Shire and Lundbeck. GEM received travel awards for taking part in psychiatric meetings from Shire. EJSS-B received speaker fees, consultancy, research funding, and conference support from Shire Pharma. Consultancy from Neurotech Solutions, Copenhagen University and Berhanderling, Skolerne & KU Leuven. Book royalties from OUP and Jessica Kingsley. Financial support—Arhus University and Ghent University for visiting Professorship. Editor-in-Chief JCPP—supported by a buy-out of time to University of Southampton and personal Honorarium. King’s College London received payments for work conducted by PA: consultancy for Shire, Eli Lilly, Novartis, and Lundbeck; educational and/or research awards from Shire, Eli Lilly, Novartis, Vifor Pharma, GW Pharma, and QbTech; speaker at events sponsored by Shire, Eli Lilly, Janssen-Cilag, and Novartis. JKB has been in the past 3 years a consultant to/member of advisory board of and/or speaker for Shire, Roche, Medice, and Servier. He is not an employee of any of these companies, and not a stock shareholder of any of these companies. He has no other financial or material support, including expert testimony, patents, royalties. In the past year, SVF received income, potential income, travel expenses continuing education support and/or research support from Tris, Otsuka, Arbor, Ironshore, Shire, Akili Interactive Labs, Enzymotec, Sunovion, Supernus, and Genomind. With his institution, he has US patent US20130217707 A1 for the use of sodium–hydrogen exchange inhibitors in the treatment of ADHD. He also receives royalties from books published by Guilford Press: Straight Talk about Your Child’s Mental Health, Oxford University Press: Schizophrenia: The Facts and Elsevier: ADHD: Non-Pharmacologic Interventions. He is principal investigator of www.adhdinadults.com. JK has given talks at educational events sponsored by Medice; all funds are received by King’s College London and used for studies of ADHD. HL has served as a speaker for Evolan Pharma and Shire and has received research grants from Shire; all outside the submitted work. K-PL served as a speaker for Eli Lilly and received research support from Medice, and travel support from Shire, all outside the submitted work. LAR reported receiving honoraria, serving on the speakers’ bureau/advisory board, and/or acting as a consultant for Eli Lilly, Janssen-Cilag, Novartis, and Shire in the last 3 years; receiving authorship royalties from Oxford Press and ArtMed; and receiving travel awards from Shire for his participation in the 2015 WFADHD meetings and from Novartis to take part of the 2016 AACAP meeting. The ADHD and juvenile bipolar disorder outpatient programs chaired by him received unrestricted educational and research support from the following pharmaceutical companies in the last 3 years: Janssen-Cilag, Novartis, and Shire. MC has received travel grants and research support from Eli Lilly and Co., Janssen-Cilag, Shire, and Lundbeck and served as consultant for Eli Lilly and Co., Janssen-Cilag, Shire, and Lundbeck. BF has received educational speaking fees from Medice and Shire. JAR-Q was on the speakers’ bureau and/or acted as consultant for Eli Lilly, Janssen-Cilag, Novartis, Shire, Lundbeck, Almirall, Braingaze, Sincrolab, Medice, and Rubió in the last 5 years. He also received travel awards (air tickets + hotel) for taking part in psychiatric meetings from Janssen-Cilag, Rubió, Shire, Medice, and Eli- Lilly. The Department of Psychiatry chaired by him received unrestricted educational and research support from the following companies in the last 5 years: Eli Lilly, Lundbeck, Janssen- Cilag, Actelion, Shire, Ferrer, Oryzon, Roche, Psious, and Rubió. The other authors have nothing to disclose. All members of the 23andMe Research Team are current or former employees of 23andMe, Inc. and hold stock or stock options in 23andMe. Authors of the ADHD Working group of the PGC that participated in this study: Catharina A. Hartman, Ziarih Hawi, Jennifer Crosbie, Sandra Loo, Josephine Elia, Russell Schachar, Christie Burton, Ted Reichborn-Kjennerud, Aribert Rothenberger, Søren Dalsgaard, Irwin Waldman, Mark Bellgrove, Hakon Hakonarson, Johannes Hebebrand, Anke Hinney, and Robert Oades have nothing to disclose. Joseph Biederman 2019–2020: he received research support from Genentech, Headspace Inc., Lundbeck AS, Neurocentria Inc, Pfizer Pharmaceuticals, Roche TCRC Inc., Shire Pharmaceuticals Inc., Sunovion Pharmaceuticals Inc., and Tris. He was a consultant for Akili, Avekshan, Jazz Pharma, and Shire/Takeda. Through MGH CTNI, he participated in a scientific advisory board for Supernus. Dr Biederman’s program has received departmental royalties from a copyrighted rating scale used for ADHD diagnoses, paid by Bracket Global, Ingenix, Prophase, Shire, Sunovion, and Theravance; these royalties were paid to the Department of Psychiatry at MGH. Tobias Banaschewski served in an advisory or consultancy role for Lundbeck, Medice, Neurim Pharmaceuticals, Oberberg GmbH, Shire, and Infectopharm. He received conference support or speaker’s fee by Lilly, Medice, and Shire. He received royalties from Hogrefe, Kohlhammer, CIP Medien, and Oxford University Press. James McGough is an expert testimony for Eli Lilly; DSMB for Sunovion. Benjamin Neale is a member of the scientific advisory board at Deep Genomics and consultant for Camp4 Therapeutics, Takeda Pharmaceutical, and Biogen. Ole Andreas Andreassen received speaker’s honorarium from Lundbeck, and is a consultant for HealthLytix.
The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under grant agreement n° 602805 (Aggressotype) as well as from the European Union H2020 Programme (H2020/2014–2020) under grant agreements n° 643051 (MiND), n° 667302 (CoCA), n° 728018 (Eat2beNICE), and n° 278948 (TACTICS). The work was also supported by the ECNP Network “ADHD across the Lifespan” (https://www.ecnp.eu/research-innovation/ECNP-networks/List-ECNP-Networks/ADHD.aspx). Over the course of this investigation, PR is a recipient of a pre-doctoral fellowship from the Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR), Generalitat de Catalunya, Spain (2016FI_B 00899). The iPSYCH project is funded by the Lundbeck Foundation (grant nos. R102-A9118 and R155-2014-1724) and the universities, and university hospitals of Aarhus and Copenhagen. ADB and NRM’s work is also supported by the EU’s Horizon 2020 programme (grant no. 667302, CoCA). Data handling and analysis was supported by NIMH (1U01MH109514-01 to Michael O’Donovan and ADB). CSM is a recipient of a Sara Borrell contract from the Instituto de Salud Carlos III, Ministerio de Economía, Industria y Competitividad, Spain (CD15/00199). K-PL and his team are supported by the Deutsche Forschungsgemeinschaft (DFG: CRU 125, CRC TRR 58 A1/A5, no. 44541416), ERA-Net NEURON/RESPOND, no. 01EW1602B, ERA-Net NEURON/DECODE, no. 01EW1902, and 5-100 Russian Academic Excellence Project. JH thanks Stiftelsen K.G. Jebsen, University of Bergen, the Western Norwegian Health Authorities (Helse Vest). HL thanks the Swedish research council. BC received financial support from the Spanish “Ministerio de Economía y Competitividad” (SAF2015-68341-R) and AGAUR (2017SGR738). MSA is a recipient of a contract from the Biomedical Network Research Center on Mental Health (CIBERSAM), Madrid, Spain. MR is a recipient of a Miguel de Servet contract from the Instituto de Salud Carlos III, Spain (CP09/00119 and CPII15/00023). The research leading to these results has received funding from the Instituto de Salud Carlos III (PI16/01505, PI17/00289, PI18/01788, PI19/00721 and P19/01224), and cofinanced by the European Regional Development Fund (ERDF), from the Pla estratègic de recerca i innovació en salut (PERIS); Generalitat de Catalunya (METAL-Cat; SLT006/17/287), from “la Marató de TV3” (092330/31) and from the Agència de Gestió d’Ajuts Universitaris i de Recerca-AGAUR, Generalitat de Catalunya (2017SGR1461). ES’s work is supported by a personal Hypatia grant from the Radboud University Medical Center. MH received a Veni grant from of the Netherlands Organization for Scientific Research (NWO, grant number 91619115). The NeuroIMAGE study was supported by NIH Grant R01MH62873 (to SVF), NWO Large Investment Grant 1750102007010 (to JKB), ZonMW grant 60-60600-97-193, NWO grants 056-13-015 and 433-09-242, and matching grants from Radboud University Nijmegen Medical Center, University Medical Center Groningen and Accare, and Vrije Universiteit Amsterdam. Organization for Scientific Research (NWO; grant 016-130-669). BF and MK’s work is supported by the Dutch National Science Agenda (NWA) for the NeurolabNL project (grant 400-17-602). This paper represents independent research part funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. BF received additional funding from a personal Vici grant of the Dutch. The authors declare no competing interests
Supplementary information
Acknowledgements
All the authors thank the patients and their families. High-performance computer capacity for handling and statistical analysis of iPSYCH data on the GenomeDK HPC facility was provided by the Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark, (grant to ADB) and Center for Genomics and Personalized Medicine, Aarhus, Denmark. This study is part of the International Multicentre persistent ADHD Collaboration (IMpACT; www.impactadhdgenomics.com). IMpACT unites major research centres working on the genetics of ADHD persistence across the lifespan and has participants in The Netherlands, Germany, Spain, Norway, the United Kingdom, the United States of America, Brazil, and Sweden. Principal investigators of IMpACT are: BF (chair), AR (co-chair), SVF, JH, BC, JAR-Q, PA, K-PL, JK, CHDB, JKB, SEJ, HL, AED, and EHG. This study uses data from the ADHD Working Group of the PGC, many of which contributed to this study and are named under the consortium author for this working group; additional members are at the time of submission of this manuscript: Anke R. Hammerschlag, Alexandra Philipsen, Alexandre Todorov, Alice Charach, Allison Ashley-Koch, Amaia Hervás, Ana Miranda, André Scherag, Anita Thapar, Anna Rommel, Anne Wheeler, Christine Cornforth, Danielle Posthuma, Elizabeth Corfield, Felecia Cerrato, Fernando Mulas, Franziska Degenhardt, Gláucia Chiyoko Akutagava Martins, Gun Peggy Strømstad Knudsen, Hans-Christoph Steinhausen, Herbert Roeyers, Hyo-Won Kim, Joanna Martin, Joel Gelernter, Joseph Sergeant, Juanita Gamble, Julia Pinsonneault, Jurgen Deckert, Kate Langley, Li Yang, Lindsey Kent, Manuel Mattheisen, Maria Jesús Arranz Calderón, Martin Steen Tesli, Meg Mariano, Michael Gill, Michael O’Donovan, Monica Bayes, Nick Martin, Niels Peter Ole Mors, Nigel Williams, Pak Sham, Patrick Sullivan, Patrick WL Leung, Paul Arnold, Paul Lichtenstein, Peter Holmans, Preben Bo Mortensen, Rachel Guerra, Raymond Walters, Richard Anney, Richard Ebstein, Ridha Joober, Sarah Anthony, Sarah Medland, Sarojini Sengupta, Søren Dinesen, Steve Nelson, Susan Smalley, Susann Scherag, Tammy Biondi, Tim Silk, Tinca Polderman, Tony Altar, Yanli Zhang-James, Yufeng Wang.
Author contributions
Conception or design of the work: PR, DD, CS-M, TZ, CPJ, OR, ES, AA-V, PA, BC, SVF, JH, SEJ, K-PL, MC, ADB, BF, JAR-Q, MSA, and MR. Acquisition, analysis or interpretation of the data: PR, DD, CS-M, TZ, MK, NRM, HW, IG-M, MP, LV, LA, VR, MC, CF, RB, GE, PA, AED, OG, MH, CPJ, SK-S, OR, ES, EJSS-B, PA, CHDB, JKB, BC, JH, SEJ, HL, K-PL, AR, LAR, MC, ADB, BF, JAR-Q, MSA, MR, Ole Andreas Andreassen, Tobias Banaschewski, Mark Bellgrove, Joseph Biederman, Christie Burton, Jennifer Crosbie, Søren Dalsgaard, Josephine Elia, Hakon Hakonarson, Catharina A. Hartman, Ziarih Hawi, Johannes Hebebrand, Anke Hinney, Sandra Loo, James McGough, Benjamin Neale, Robert Oades, Ted Reichborn-Kjennerud, Aribert Rothenberger, Russell Schachar, Irwin Waldman, Michelle Agee, Babak Alipanahi, Adam Auton, Robert K. Bell, Katarzyna Bryc, Sarah L. Elson, Pierre Fontanillas, Nicholas A. Furlotte, David A. Hinds, Karen E. Huber, Aaron Kleinman, Nadia K. Litterman, Jennifer C. McCreight, Matthew H. McIntyre, Joanna L. Mountain, Elizabeth S. Noblin, Carrie A.M. Northover, Steven J. Pitts, J. Fah Sathirapongsasuti, Olga V. Sazonova, Janie F. Shelton, Suyash Shringarpure, Chao Tian, Joyce Y. Tung, Vladimir Vacic, Xin Wang, and Catherine H. Wilson. Drafted the work or substantively revised it: PR, DD, CS-M, PA, EHG, AH, MH, PMK, AJL, OR, DLR, AS-O, BSdS, Ted Reichborn-Kjennerud, ES, TS, EJSS-B, Mark Bellgrove, Ziarih Hawi, PA, JK, CHDB, BC, Johannes Hebebrand, SVF, JKB, HL, AR, LAR, MC, ADB, BF, JAR-Q, MSA, and MR. Final approval of the version to be published: all authors. Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: PR, MSA, and MR.
Footnotes
A list of members of ADHD Working Group of the Psychiatric Genomics Consortium and 23andMe Research team and their affiliations are listed in Supplementary information.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Paula Rovira, Ditte Demontis, Anders D. Børglum, Barbara Franke, Josep Antoni Ramos-Quiroga, María Soler Artigas, Marta Ribasés.
Contributor Information
María Soler Artigas, Email: maria.soler@vhir.org.
Marta Ribasés, Email: marta.ribases@vhir.org.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41386-020-0664-5).
References
- 1.Franke B, Michelini G, Asherson P, Banaschewski T, Bilbow A, Buitelaar JK, et al. Live fast, die young? A review on the developmental trajectories of ADHD across the lifespan. Eur Neuropsychopharmacol. 2018;28:1059–88. doi: 10.1016/j.euroneuro.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polanczyk GV, Salum GA, Sugaya LS, Caye A, Rohde LA. Annual research review: a meta-analysis of the worldwide prevalence of mental disorders in children and adolescents. J Child Psychol Psychiatry. 2015;56:345–65. doi: 10.1111/jcpp.12381. [DOI] [PubMed] [Google Scholar]
- 3.Brikell I, Kuja-Halkola R, Larsson H. Heritability of attention-deficit hyperactivity disorder in adults. Am J Med Genet B Neuropsychiatr Genet. 2015;168:406–13. doi: 10.1002/ajmg.b.32335. [DOI] [PubMed] [Google Scholar]
- 4.Kan KJ, Dolan CV, Nivard MG, Middeldorp CM, van Beijsterveldt CE, Willemsen G, et al. Genetic and environmental stability in attention problems across the lifespan: evidence from the Netherlands twin register. J Am Acad Child Adolesc Psychiatry. 2013;52:12–25. doi: 10.1016/j.jaac.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Kuntsi J, Rijsdijk F, Ronald A, Asherson P, Plomin R. Genetic influences on the stability of attention-deficit/hyperactivity disorder symptoms from early to middle childhood. Biol Psychiatry. 2005;57:647–54. doi: 10.1016/j.biopsych.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 6.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faraone SV, Larsson H. Genetics of attention deficit hyperactivity disorder. Mol Psychiatry. 2019;24:562–75. doi: 10.1038/s41380-018-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganna A, Satterstrom FK, Zekavat SM, Das I, Kurki MI, Churchhouse C, et al. Quantifying the impact of rare and ultra-rare coding variation across the phenotypic spectrum. Am J Hum Genet. 2018;102:1204–11. doi: 10.1016/j.ajhg.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin J, O’Donovan MC, Thapar A, Langley K, Williams N. The relative contribution of common and rare genetic variants to ADHD. Transl Psychiatry. 2015;5:e506. doi: 10.1038/tp.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satterstrom FK, Walters RK, Singh T, Wigdor EM, Lescai F, Demontis D, et al. ASD and ADHD have a similar burden of rare protein-truncating variants. 10.1101/277707 (2018). [DOI] [PMC free article] [PubMed]
- 11.Hawi Z, Cummins TD, Tong J, Arcos-Burgos M, Zhao Q, Matthews N, et al. Rare DNA variants in the brain-derived neurotrophic factor gene increase risk for attention-deficit hyperactivity disorder: a next-generation sequencing study. Mol Psychiatry. 2017;22:580–4. doi: 10.1038/mp.2016.117. [DOI] [PubMed] [Google Scholar]
- 12.Brainstorm C, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaw P, Stringaris A, Nigg J, Leibenluft E. Emotion dysregulation in attention deficit hyperactivity disorder. Am J Psychiatry. 2014;171:276–93. doi: 10.1176/appi.ajp.2013.13070966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilens TE, Biederman J, Faraone SV, Martelon M, Westerberg D, Spencer TJ. Presenting ADHD symptoms, subtypes, and comorbid disorders in clinically referred adults with ADHD. J Clin Psychiatry. 2009;70:1557–62. doi: 10.4088/JCP.08m04785pur. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anker E, Bendiksen B, Heir T. Comorbid psychiatric disorders in a clinical sample of adults with ADHD, and associations with education, work and social characteristics: a cross-sectional study. BMJ Open. 2018;8:e019700. doi: 10.1136/bmjopen-2017-019700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biederman J, Faraone SV, Spencer T, Wilens T, Norman D, Lapey KA, et al. Patterns of psychiatric comorbidity, cognition, and psychosocial functioning in adults with attention deficit hyperactivity disorder. Am J Psychiatry. 1993;150:1792–8. doi: 10.1176/ajp.150.12.1792. [DOI] [PubMed] [Google Scholar]
- 17.Kessler RC, Adler LA, Barkley R, Biederman J, Conners CK, Faraone SV, et al. Patterns and predictors of attention-deficit/hyperactivity disorder persistence into adulthood: results from the national comorbidity survey replication. Biol Psychiatry. 2005;57:1442–51. doi: 10.1016/j.biopsych.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larson K, Russ SA, Kahn RS, Halfon N. Patterns of comorbidity, functioning, and service use for US children with ADHD, 2007. Pediatrics. 2011;127:462–70. doi: 10.1542/peds.2010-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalsgaard S, Østergaard SD, Leckman JF, Mortensen PB, Pedersen MG. Mortality in children, adolescents, and adults with attention deficit hyperactivity disorder: a nationwide cohort study. Lancet. 2015;385:2190–6. doi: 10.1016/S0140-6736(14)61684-6. [DOI] [PubMed] [Google Scholar]
- 20.Faraone SV, Asherson P, Banaschewski T, Biederman J, Buitelaar JK, Ramos-Quiroga JA, et al. Attention-deficit/hyperactivity disorder. Nat Rev Dis Prim. 2015;1:15020. doi: 10.1038/nrdp.2015.20. [DOI] [PubMed] [Google Scholar]
- 21.Kupper T, Haavik J, Drexler H, Ramos-Quiroga JA, Wermelskirchen D, Prutz C, et al. The negative impact of attention-deficit/hyperactivity disorder on occupational health in adults and adolescents. Int Arch Occup Environ Health. 2012;85:837–47. doi: 10.1007/s00420-012-0794-0. [DOI] [PubMed] [Google Scholar]
- 22.Voigt RG, Katusic SK, Colligan RC, Killian JM, Weaver AL, Barbaresi WJ. Academic achievement in adults with a history of childhood attention-deficit/hyperactivity disorder: a population-based prospective study. J Dev Behav Pediatr. 2017;38:1–11. doi: 10.1097/DBP.0000000000000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strine TW, Lesesne CA, Okoro CA, McGuire LC, Chapman DP, Balluz LS, et al. Emotional and behavioral difficulties and impairments in everyday functioning among children with a history of attention-deficit/hyperactivity disorder. Prev Chronic Dis. 2006;3:A52. [PMC free article] [PubMed] [Google Scholar]
- 24.Biederman J, Petty CR, Evans M, Small J, Faraone SV. How persistent is ADHD? A controlled 10-year follow-up study of boys with ADHD. Psychiatry Res. 2010;177:299–304. doi: 10.1016/j.psychres.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caye A, Spadini AV, Karam RG, Grevet EH, Rovaris DL, Bau CH, et al. Predictors of persistence of ADHD into adulthood: a systematic review of the literature and meta-analysis. Eur Child Adolesc Psychiatry. 2016;25:1151–9. doi: 10.1007/s00787-016-0831-8. [DOI] [PubMed] [Google Scholar]
- 26.Cheung CH, Rijdijk F, McLoughlin G, Faraone SV, Asherson P, Kuntsi J. Childhood predictors of adolescent and young adult outcome in ADHD. J Psychiatr Res. 2015;62:92–100. doi: 10.1016/j.jpsychires.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riglin L, Collishaw S, Thapar AK, Dalsgaard S, Langley K, Smith GD, et al. Association of genetic risk variants with attention-deficit/hyperactivity disorder trajectories in the general population. JAMA Psychiatry. 2016;73:1285–92. doi: 10.1001/jamapsychiatry.2016.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy A, Hechtman L, Arnold LE, Swanson JM, Molina BSG, Sibley MH, et al. Childhood predictors of adult functional outcomes in the multimodal treatment study of attention-deficit/hyperactivity disorder (MTA) J Am Acad Child Adolesc Psychiatry. 2017;56:687–95. doi: 10.1016/j.jaac.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pingault JB, Viding E, Galera C, Greven CU, Zheng Y, Plomin R, et al. Genetic and environmental influences on the developmental course of attention-deficit/hyperactivity disorder symptoms from childhood to adolescence. JAMA Psychiatry. 2015;72:651–8. doi: 10.1001/jamapsychiatry.2015.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rietveld MJ, Hudziak JJ, Bartels M, van Beijsterveldt CE, Boomsma DI. Heritability of attention problems in children: longitudinal results from a study of twins, age 3 to 12. J Child Psychol Psychiatry. 2004;45:577–88. doi: 10.1111/j.1469-7610.2004.00247.x. [DOI] [PubMed] [Google Scholar]
- 31.Akutagava-Martins GC, Salatino-Oliveira A, Kieling C, Genro JP, Polanczyk GV, Anselmi L, et al. COMT and DAT1 genes are associated with hyperactivity and inattention traits in the 1993 Pelotas Birth Cohort: evidence of sex-specific combined effect. J Psychiatry Neurosci. 2016;41:405–12. doi: 10.1503/jpn.150270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonvicini C, Faraone SV, Scassellati C. Attention-deficit hyperactivity disorder in adults: a systematic review and meta-analysis of genetic, pharmacogenetic and biochemical studies. Mol Psychiatry. 2016;21:1643. doi: 10.1038/mp.2016.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonvicini C, Faraone SV, Scassellati C. Common and specific genes and peripheral biomarkers in children and adults with attention-deficit/hyperactivity disorder. World J Biol Psychiatry. 2018;19:80–100. doi: 10.1080/15622975.2017.1282175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson S, Halleland H, Halmoy A, Jacobsen KK, Landaas ET, Dramsdahl M, et al. Genetic analyses of dopamine related genes in adult ADHD patients suggest an association with the DRD5-microsatellite repeat, but not with DRD4 or SLC6A3 VNTRs. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1470–5. doi: 10.1002/ajmg.b.30662. [DOI] [PubMed] [Google Scholar]
- 35.Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Roser C, Nguyen TT, et al. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm. 2008;115:1573–85. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- 36.Ribases M, Ramos-Quiroga JA, Hervas A, Bosch R, Bielsa A, Gastaminza X, et al. Exploration of 19 serotoninergic candidate genes in adults and children with attention-deficit/hyperactivity disorder identifies association for 5HT2A, DDC and MAOB. Mol Psychiatry. 2009;14:71–85. doi: 10.1038/sj.mp.4002100. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez-Mora C, Ramos-Quiroga JA, Bosch R, Corrales M, Garcia-Martinez I, Nogueira M, et al. Case-control genome-wide association study of persistent attention-deficit hyperactivity disorder identifies FBXO33 as a novel susceptibility gene for the disorder. Neuropsychopharmacology. 2015;40:915–26. doi: 10.1038/npp.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thissen AJ, Bralten J, Rommelse NN, Arias-Vasquez A, Greven CU, Heslenfeld D, et al. The role of age in association analyses of ADHD and related neurocognitive functioning: A proof of concept for dopaminergic and serotonergic genes. Am J Med Genet B Neuropsychiatr Genet. 2015;168:471–9. doi: 10.1002/ajmg.b.32290. [DOI] [PubMed] [Google Scholar]
- 39.Zayats T, Athanasiu L, Sonderby I, Djurovic S, Westlye LT, Tamnes CK, et al. Genome-wide analysis of attention deficit hyperactivity disorder in Norway. PLoS One. 2015;10:e0122501. doi: 10.1371/journal.pone.0122501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch KP, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49:884–97. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Magi R, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9:1192–212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–35. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han B, Pouget JG, Slowikowski K, Stahl E, Lee CH, Diogo D, et al. A method to decipher pleiotropy by detecting underlying heterogeneity driven by hidden subgroups applied to autoimmune and neuropsychiatric diseases. Nat Genet. 2016;48:803–10. doi: 10.1038/ng.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Middeldorp CM, Hammerschlag AR, Ouwens KG, Groen-Blokhuis MM, Pourcain BS, Greven CU, et al. A genome-wide association meta-analysis of attention-deficit/hyperactivity disorder symptoms in population-based pediatric cohorts. J Am Acad Child Adolesc Psychiatry. 2016;55:896–905. doi: 10.1016/j.jaac.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–9. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi T, Wu Y, Zeng J, Zhang F, Xue A, Jiang L, et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun. 2018;9:2282. doi: 10.1038/s41467-018-04558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Q, Brikell I, Lichtenstein P, Serlachius E, Kuja-Halkola R, Sandin S, et al. Familial aggregation of attention-deficit/hyperactivity disorder. J Child Psychol Psychiatry. 2017;58:231–9. doi: 10.1111/jcpp.12616. [DOI] [PubMed] [Google Scholar]
- 57.Biederman J, Faraone SV, Mick E, Spencer T, Wilens T, Kiely K, et al. High risk for attention deficit hyperactivity disorder among children of parents with childhood onset of the disorder: a pilot study. Am J Psychiatry. 1995;152:431–5. doi: 10.1176/ajp.152.3.431. [DOI] [PubMed] [Google Scholar]
- 58.Chapman NH, Nato AQ, Jr., Bernier R, Ankenman K, Sohi H, Munson J, et al. Whole exome sequencing in extended families with autism spectrum disorder implicates four candidate genes. Hum Genet. 2015;134:1055–68. doi: 10.1007/s00439-015-1585-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimizu T, Hibi M. Formation and patterning of the forebrain and olfactory system by zinc-finger genes Fezf1 and Fezf2. Dev Growth Differ. 2009;51:221–31. doi: 10.1111/j.1440-169X.2009.01088.x. [DOI] [PubMed] [Google Scholar]
- 60.Koukoulas I, Augustine C, Silkenbeumer N, Gunnersen JM, Scott HS, Tan SS. Genomic organisation and nervous system expression of radial spoke protein 3. Gene. 2004;336:15–23. doi: 10.1016/j.gene.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 61.Bonora E, Graziano C, Minopoli F, Bacchelli E, Magini P, Diquigiovanni C, et al. Maternally inherited genetic variants of CADPS2 are present in autism spectrum disorders and intellectual disability patients. EMBO Mol Med. 2014;6:795–809. doi: 10.1002/emmm.201303235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hattori K, Tanaka H, Wakabayashi C, Yamamoto N, Uchiyama H, Teraishi T, et al. Expression of Ca(2)(+)-dependent activator protein for secretion 2 is increased in the brains of schizophrenic patients. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:1738–43. doi: 10.1016/j.pnpbp.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 63.Sadakata T, Shinoda Y, Ishizaki Y, Furuichi T. Analysis of gene expression in Ca(2+)-dependent activator protein for secretion 2 (Cadps2) knockout cerebellum using GeneChip and KEGG pathways. Neurosci Lett. 2017;639:88–93. doi: 10.1016/j.neulet.2016.12.068. [DOI] [PubMed] [Google Scholar]
- 64.Velez JI, Chandrasekharappa SC, Henao E, Martinez AF, Harper U, Jones M, et al. Pooling/bootstrap-based GWAS (pbGWAS) identifies new loci modifying the age of onset in PSEN1 p.Glu280Ala Alzheimer’s disease. Mol Psychiatry. 2013;18:568–75. doi: 10.1038/mp.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fyfe JC, Madsen M, Hojrup P, Christensen EI, Tanner SM, de la Chapelle A, et al. The functional cobalamin (vitamin B12)-intrinsic factor receptor is a novel complex of cubilin and amnionless. Blood. 2004;103:1573–9. doi: 10.1182/blood-2003-08-2852. [DOI] [PubMed] [Google Scholar]
- 66.Tanner SM, Aminoff M, Wright FA, Liyanarachchi S, Kuronen M, Saarinen A, et al. Amnionless, essential for mouse gastrulation, is mutated in recessive hereditary megaloblastic anemia. Nat Genet. 2003;33:426–9. doi: 10.1038/ng1098. [DOI] [PubMed] [Google Scholar]
- 67.Venkatramanan S, Armata IE, Strupp BJ, Finkelstein JL. Vitamin B-12 and cognition in children. Adv Nutr. 2016;7:879–88. doi: 10.3945/an.115.012021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50:912–9. doi: 10.1038/s41588-018-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.