Abstract
Some species have a longer lifespan than others, but usually lifespan is correlated with typical body weight. Here, we study the lifetime evolution of the metabolic behaviour of Nothobranchius furzeri, a killifish with an extremely short lifespan with respect to other fishes, even when taking into account rescaling by body weight. Comparison of the gene expression patterns of N. furzeri with those of zebrafish Danio rerio and mouse (Mus musculus) shows that a broad set of metabolic genes and pathways are affected in N. furzeri during ageing in a way that is consistent with a global deregulation of chromatin. Computational analysis of the glycolysis pathway for the three species highlights a rapid increase in the metabolic activity during the lifetime of N. furzeri with respect to the other species. Our results highlight that the unusually short lifespan of N. furzeri is associated with peculiar patterns in the metabolic activities and in chromatin dynamics.
Keywords: metabolism, ageing, chromatin
1. Introduction
Nothobranchius furzeri is a species of killifish from the family Nothobranchiidae, which lives in pools in semi-arid areas with scarce and erratic precipitations in Africa. Because of its very short captive lifespan ranging from three to 12 months depending on the environment [1], it is considered by the scientific community to be an attractive live model for ageing [2]. A longitudinal study in N. furzeri showed quantitative correlation between gene expression variations during early adult life and lifespan [3]. Under the same conditions, it was also demonstrated that the mitochondrial respiratory chain complex I is a hub in a module of genes whose expression is negatively correlated with lifespan [3]. An interesting result was obtained treating both N. furzeri and Danio rerio with rotenone, an inhibitor of complex I, demonstrating a rejuvenating effect on the transcriptome [3].
Other studies in the literature reported on the mechanism of ageing in N. furzeri in comparison with mammals, showing some similar traits, including shortening of telomeres, mitochondrial dysfunctions, ageing-associated upregulation of translation and ribosomal processes and reduced regenerative capacity [4]. Herein, our aim was to understand in more depth the ageing behaviour of N. furzeri in comparison with other fishes, using as a model D. rerio, which has a longer lifespan and is a well-characterized animal model. To achieve this result, we compared transcriptomes, gene distances and metabolic genes and pathways of N. furzeri with respect to D. rerio in three different tissues (brain, liver and skin) at five different time points, ranging from sexual maturity up to average lifetime. We also compared under the same conditions our results with Mus musculus, as a mammalian example, to show if different mechanisms are displayed during ageing in different vertebrates (fishes versus mammals).
2. Material and methods
2.1. Lifespan analysis
Lifespan and weight of all available species were downloaded from the Fishbase online database [5]. For those species whose weight was not available, we estimated the weight using the formula a*Lb [6], where the length L and the parameters a, b were obtained from Fishbase. When more than one value was available for a single species, we considered the average value. As a whole, we were able to obtain data on 1002 species. Additionally, we considered the parameter K of dimension year−1, which essentially describes the growth speed of a species, being related to the exponential growth of its size [5]. Maximal longevity and weight of 332 teleosts were obtained from AnAge database [7]. Additionally, for five short-lived species lacking data in AnAge, we obtained the average weight W from the literature: Crystallogobius linearis (W = 0.2035 g) [8], Cyclothone braueri (W = 0.25 g) [9], Electrona risso (W = 2.7 g) [10], Eucyclogobius newberryi (W = 0.83 g) [11] and Galaxiella nigrostriata (W = 0.09 g) [12].
Lifetime and weight of D. rerio and eight different species of Nothobranchius (N. furzeri MZM and GRZ, Nothobranchius kilomberoensis, Nothobranchius rachovii, Nothobranchius korthausae, Nothobranchius kuhntae MT 03/04 and Beira, and Nothobranchius guentheri) were obtained from the literature when missing from the AnAge and Fishbase datasets [13–22], whereas the Eviota sigillata and N. furzeri weights were set to 0.09 g and 3 g, respectively, according to the Fishbase model. If the data were duplicated in both of the datasets, the data from the AnAge dataset were used and the data from the Fishbase dataset were discarded.
2.2. Transcriptomic data and gene annotation
We considered data for N. furzeri (strain MZM-04/10), D. rerio and M. musculus transcriptomes from three different tissues (brain, liver and skin) at five different time points, ranging from sexual maturity up to the average lifetime. In particular, N. furzeri data refer to animals 5, 12, 20, 27, 39 weeks old, while D. rerio and M. musculus samples were sequenced at 6, 12, 24, 36, 42 and 2, 9, 15, 24, 30 months old, respectively. Note that the last two time points refer to highly different probabilities of survival in each species (electronic supplementary material, figure S1). In particular the last time point corresponds to a survival rate of for D. rerio and M. musculus while only of N. furzeri individuals survive up to 39 weeks. For each species at each time point five replicates were available. All RNA-Seq data were published previously and are accessible at NCBI’s Gene Expression Omnibus (N. furzeri: GSE52462 and GSE66712, D. rerio: GSE74244, M. musculus: GSE75192) [23,24]. Nothobranchius furzeri raw sequences were aligned to a reference transcriptome ( [25]) using StringTie (v.1.3 [26]). For each transcriptome, genes with more than 10 counts in at least one time point were considered as expressed in a given tissue. This subset of genes was used as the background reference for enrichment analysis. Genes annotated as belonging to the histone deacetylase (HDAC) and histone acetyltransferase (HAT) classes and specific polycomb genes were selected for all the species using NCBI [27] annotation files, and additional hand-curated annotation from Uniprot was performed for D. rerio and M. musculus.
2.3. Differentially expressed genes
Differential expression analysis was performed using EdgeR ([28], R package version 3.12.1), considering only genes with more than 10 raw counts in at least one time point implementing the default Benjamini–Hochberg correction. For each time point, log2 expression of the fold change (log2FC) compared with the first time point was calculated and genes with corrected p-value <0.05 were considered as differentially expressed. Reactions containing at least one DE gene were considered as deregulated.
2.4. Pathways and gene ontology
Pathway and gene ontology analysis was performed using Panther [29] over all the genes differentially expressed in at least one time point on each tissue separately. For N. furzeri analysis in Panther we used D. rerio putative orthologous genes detected with OMA [30]. Kegg annotation of N. furzeri genes was used for further analysis.
2.5. Gene distance distribution
The distribution of genomic distances between expressed genes for each tissue in each species was obtained by calculating the distance between the end of one gene and the beginning of the next one. Random sampling (103 replica) was performed on the distributions in order to compare random inter-gene distances with the clustering occurring between either up- or downregulated genes. The sample size was chosen to be equal to the number of differentially expressed genes at each time point. The Kolmogorov–Smirnov test was used to compare the random distribution with the DE gene distribution using Python [31].
2.6. Metabolic genes and pathways
Metabolic pathways, reactions and related genes were downloaded from Kegg ([32]; last accession September 2018). A total of 1691 reactions belonging to 76 pathways and grouped into 11 superpathways involving genes expressed in at least one time point were considered. Reactions containing at least one DE gene were considered as deregulated. For D. rerio and M. musculus, KeggLinkDB [33] and BioMart [34] were used to map Kegg genes on RNA-seq data.
2.7. Model for metabolic deregulation
We use a constraint-based model where each reaction flux is a variable vi and each internal species imposes a mass constraint, as its overall production/consumption must be equal to zero. Some species are allowed to be out of balance, however, as they are inputs/outputs of the pathway. Considering only internal species, the mass balance condition can be expressed as A · v = 0, where A is the stoichiometric matrix of the pathway. We also take into account reaction reversibilities, imposing that vi > 0 for irreversible reactions, and add a minimal flux constraint |vi| > vmin for all reactions to avoid numerical issues during fold-change calculations; see the following section. Putting these three sets of constraints together, and denoting the set of irreversible reaction indices by , we define the set of feasible fluxes as
As the number of chemical reactions is larger than the number of internal species, the system is underdetermined and there are many possible solutions to the problem, represented by the set . Notice that we do not impose any further growth-related objective function—as is typical in flux balance analysis approaches. Instead, we propose a fold-change-based fitting mechanism for which no objective function is needed.
2.8. Model fitting
We assume that enzymatic abundances are proportional to reaction fluxes. That is,
| 2.1 |
where xi is the experimental enzymatic abundance associated with reaction i, vi is the corresponding model reaction flux and αi is an unknown, reaction-dependent proportionality constant. It is clear that without access to the value of αi we cannot compare absolute values of xi and vi. However, we can work at the level of fold changes without needing to determine the proportionality constants, as
| 2.2 |
In other words, we can associate a fold-change of factor λ in the model with a fold-change of the same factor in the experimental enzymatic abundances. Repeating this argument for all reactions, we find the pair of fluxes (u, v) that better approximate the experimental fold-changes. To do so, we define the squared total logarithmic error of a pair of model fluxes u, v given a pair of experimental values x, y as follows:
The best fit of the model to the data x, y is defined as the pair of feasible fluxes that minimize F(u, v|x, y) and are found numerically by using the minimize function from the scipy.optimize Python library.
2.9. Efficiency estimation
We define the efficiency of the glycolysis pathway as
where p, g are the pyruvate and glucose fluxes, respectively. The change in efficiency Δη between two time points can be defined as Δη = η1/η0. Is is easy to see that this quantity can be computed from the changes in glucose and pyruvate,
In turn, we can estimate Δp, Δg from the pair of solutions that better approximates the experimental data fold-changes; see above for details.
In summary, it is possible to estimate how the glycolysis pathway efficiency changes with time by fitting a constraint-based model to enzymatic abundance data.
3. Results
3.1. Allometric scaling of Nothobranchius furzeri in comparison with other fish
To better appreciate the exceptionally short lifespan of N. furzeri, we compared its properties with those of other fish species. Since a meaningful comparison could only be performed by also taking into account the relative body mass or body length of different species, we collected data on the total lifespan and weight for different fish species as detailed in the Material and methods section. Furthermore, we considered the growth speed of each fish species, encoded by the parameter K.
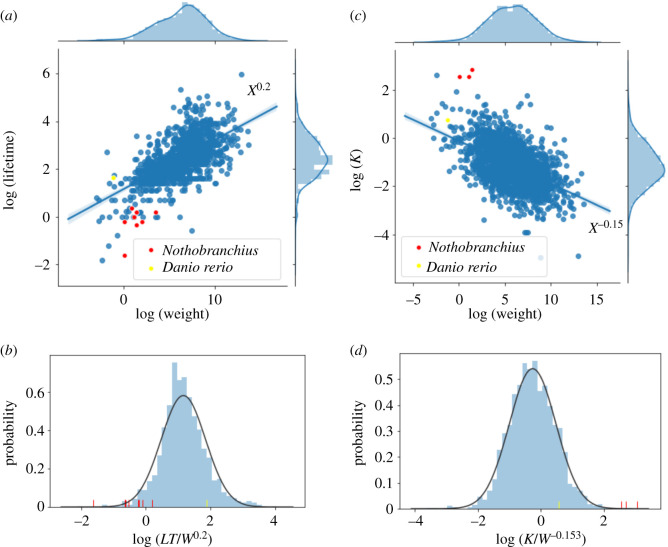
In figure 1a, we report a logarithmic plot of the lifetimes of 1441 fish species and 10 Nothobranchius species as a function of their weight. Regression analysis shows a relation between lifetimes LT and weight W scaling as (LT) ∝ (W)0.2. Nothobranchius species are reported in red and D. rerio in yellow. A plot of the lifetimes rescaled by their weight as LT/W0.20 is reported in figure 1b, showing that Nothobranchius species are in the left tail of the distribution while D. rerio lies in the middle. This means that Nothobranchius species live for a shorter time than would be expected for fishes of the same weight. Nothobranchius furzeri GRZ is the most extreme outlier since its value differs from the mean by more than four standard deviations (4σ), while Nothobranchius furzeri MZM differs by 2σ.
Figure 1.
Allometric scaling of fish species. (a) The lifetimes of 1441 fish species and 10 Nothobranchius species (in red) are plotted against their weight in a log–log plot. The fit yields a power law with exponent 0.20. Danio rerio is reported in yellow. (b) When lifetimes (LT) are rescaled by weights (W) as LT/W0.20 , Nothobranchius species all appear in the left-hand side of the distribution, particularly N. furzeri GRZ. (c) The growth rate K of 2109 fish species and three Nothobranchius species are plotted against their weight in a log–log plot. The fit yields a power law with exponent −0.15. (d) When growth rates (K) are rescaled by weights (W) as K/W−0.15, Nothobranchius species all appear in the right-hand side of the distribution, particularly N. furzeri GRZ. Danio rerio appears instead always close to the mean of the distribution.
Similar results were obtained considering the growth rate K, which we report in figure 1c for 2109 fish species and three Nothobranchius species as a function of their weight. Here, the fit yielded a power law with exponent −0.15. Again, when growth rates (K) were rescaled by weights (W) as K/W−0.15, both Nothobranchius furzeri GRZ and MZM are roughly 4σ above the mean (figure 1d), while D. rerio is very close to the mean.
3.2. Global deregulation of gene expression in Nothobranchius furzeri, Danio rerio and Mus musculus
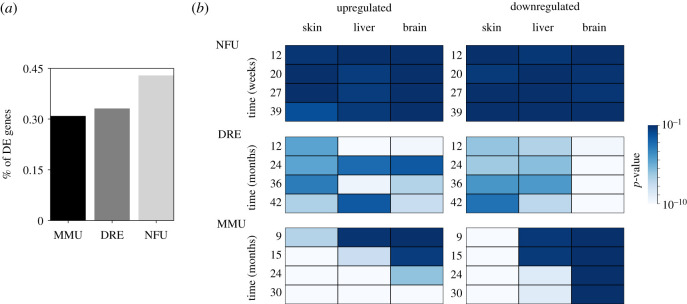
We evaluated differentially expressed (DE) genes from transcriptomes obtained from three different tissues (brain, liver and skin) of short-lived turquoise killifish N. furzeri as well as D. rerio and M. musculus under the same conditions (see Material and methods section). As a comparison, we also analysed M. musculus under the same conditions. Figure 2a shows that, while D. rerio and M. musculus have a similar percentage of DE genes, N. furzeri shows a larger fraction of DE genes during ageing. Moreover, the same trend was obtained when the comparison was performed separately for each tissue, for each time point or for a specific functional category (electronic supplementary material, figure S2).
Figure 2.
Deregulated genes during the ageing process. (a) Fraction of genes differentially expressed in the three species (MMU = M. musculus; DRE = D. rerio; NFU = N. furzeri) in at least one time point. (b) Probability that, at each time, genes differentially expressed are randomly distributed along the genome. The distribution of genomic distances between differentially regulated genes has been compared with the distribution obtained from random sampling of expressed genes for each species. The heatmap reports the average p-value obtained comparing the distributions of inter-gene distances using the Kolmogorov–Smirnov test (103 replica) Higher p-values indicate a distribution close to the null-model random distribution.
Gene ontology and analysis of differentially expressed pathways was performed on genes that were deregulated in at least one time point. We observed an enrichment of inflammatory pathways for both D. rerio and M. musculus while genes related to metabolic processes are in general depleted. Overall, the categories represented in both organisms are quite similar (see electronic supplementary material, dataset 1), while the cell cycle appears to be most affected in N. furzeri. This result could, however, be affected by the analysis performed only on D. rerio orthologues, which constitute about 80% of the genes. We calculated the enrichment of N. furzeri DE genes in five Kegg pathway classes, and we did not find significant results in most of the cases (see electronic supplementary material, dataset 1).
We then investigated if a particular pattern of deregulation occurred, comparing the distribution of DE genes on the genome with an appropriate null-model distribution where we assess its degree of randomness. In other words, our aim was to understand if the deregulated genes are randomly sampled throughout the genome or if they are localized into particular regions across the genome. Our analysis shows that the distribution of DE genes is dramatically different between the species and tissues (figure 2b). In fact, D. rerio shows a general non-random distribution of DE genes, while N. furzeri DE genes appear to be randomly distributed along the genome in all the cases considered. Mus musculus, in contrast, shows a completely different pattern with respect to both fishes, displaying a tissue- and time-specific regulation with an increasing clustering of upregulated DE genes during ageing (figure 2b).
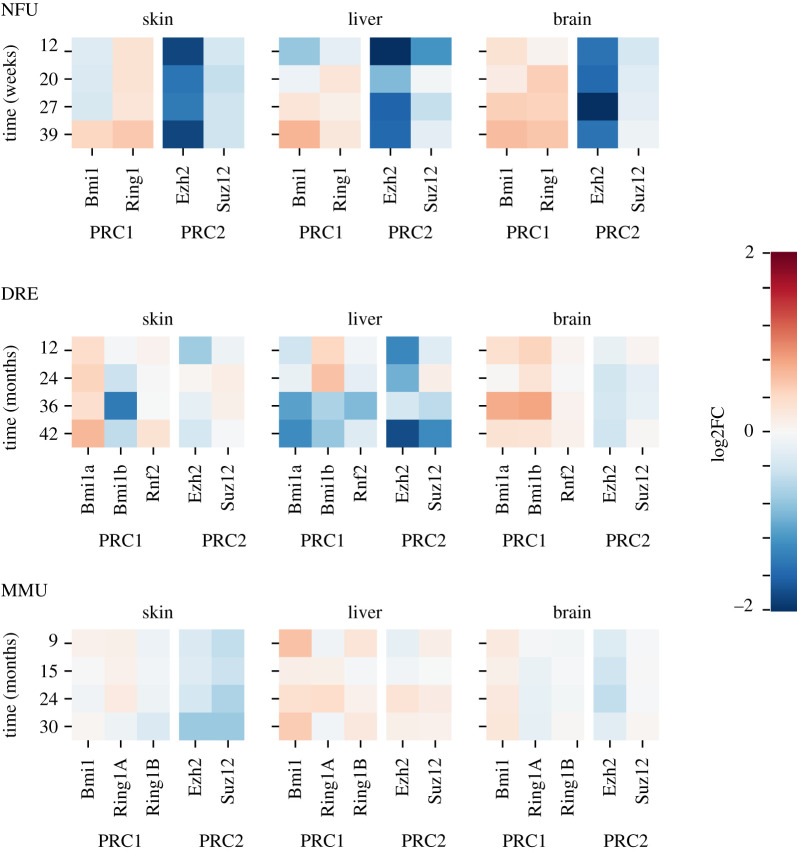
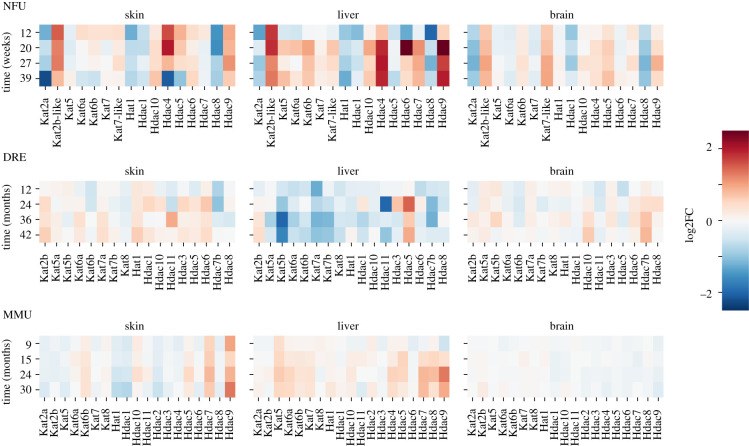
Since these data seem to suggest that the process of ageing in N. furzeri is related to a global, uniform deregulation of the chromatin state, we investigated the changes in expression of genes involved in chromatin remodelling, such as polycomb repressor complex 1 and 2 (PRC1 and PRC2) (BM1, RING, Ezh2 and Suz12), HDAC and HAT genes. As shown in figure 3, we found a specific and strong signal of deregulation in N. furzeri with a marked and constant decrease of PRC2 genes (Ezh2 and Suz12) and a slight increase of PRC1 (BM1 and RING) genes at late time points (figure 3). Ezh2 is closely related to the maintenance of heterochromatin state and its decrease suggests a shift towards open chromatin and a decrease in gene silencing [35]. In both mouse and D. rerio it is possible to observe a lower and time-dependent Ezh2 downregulation. Considering HDAC and HAT genes, N. furzeri shows a complex picture of DE genes in skin, liver and brain that is completely different with respect to D. rerio, where none of the genes is significantly deregulated (figure 4). Mus musculus also shows a different pattern in comparison with fishes and in particular brain does not show any DE HDAC or HAT genes (figure 4). The expression of most deregulated HAT and HDAC genes, including Kat2a, Kat2b, Hdac4, Hdac8 and Hdac9, shows a coherent expression through the different tissues and the opposite trend of genes belonging to different classes of deacetylase complexes can be attributed to their different roles (see electronic supplementary material, figure S3), coherently with observations reported in humans [36].
Figure 3.
Expression of polycomb repressive complex 1 and 2. Figure shows changes of expression in a set of genes belonging to polycomb repressive complex 1 (PRC1) and polycomb repressive complex 2 (PRC2) classes in the different species as a function of time. Expression is reported as log2FC compared with the initial time point.
Figure 4.
Differentially expressed HDAC and HAT genes. Figure shows the log2FC expression relative to the initial time point of genes belonging to the HDAC or HAT (Kat) classes. Expression is reported as log2FC compared with the initial time point. MMU = M. musculus; DRE = D. rerio; NFU = N. furzeri.
3.3. Modelling changes in metabolic pathways of N. furzeri, D. rerio and M. musculus during the ageing process
To investigate the possible origin of the shorter lifespan of N. furzeri with respect to D. rerio, we focused on possible differences in their metabolic activity. As a further comparison, M. musculus during ageing was also considered. Since information at the gene or pathway level does not appear to be significant (electronic supplementary material, dataset 1), we considered explicitly the contribution of DE genes to the deregulation of metabolic reactions. Metabolic networks for the three species were derived from the Kegg database [32]. In the three species, the global structure of the network is conserved, showing a good agreement in terms of compounds and reactions (see electronic supplementary material, figure S4).
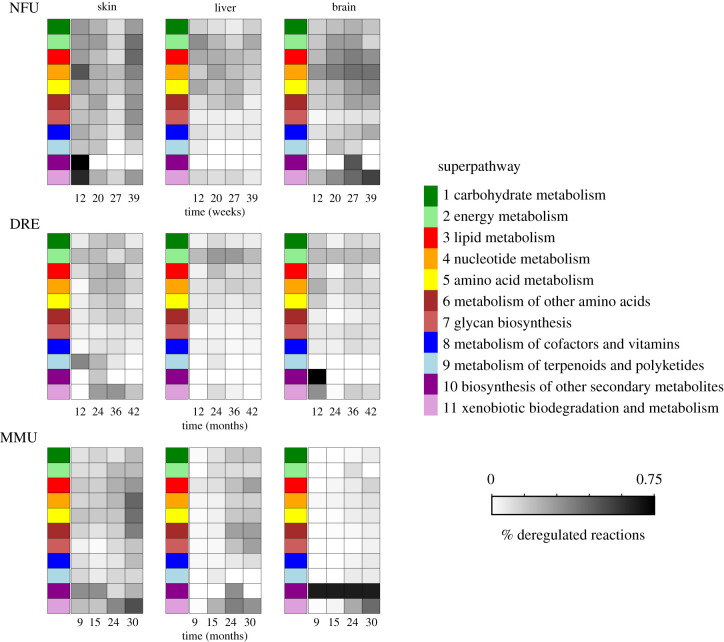
In order to highlight the possible deregulation of a metabolic network, DE genes annotated as enzymes were mapped on the metabolic network and reactions catalysed by at least one DE gene were considered as deregulated (for more details, see the Material and methods section). Figure 5 shows that, for all three organisms, the skin tissue has the highest amount of deregulated reactions for each category of metabolic pathway. When the three species are compared N. furzeri shows a higher number of deregulated superpathways; this is even more evident when we observe data at the single-pathway level (electronic supplementary material, figure S5).
Figure 5.
Deregulated metabolic reactions. Metabolic reactions containing at least one differentially expressed gene are considered as deregulated. Heatmaps show the percentage of deregulated metabolic reactions for each superpathway (see legend) as a function of time. MMU = M. musculus; DRE = D. rerio; NFU = N. furzeri.
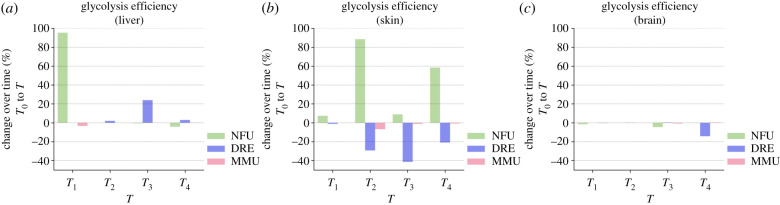
In order to verify whether deregulated reactions defined in terms of DE genes led to changes in terms of efficiency in the production and consumption of metabolites, we implemented a model based on flux balance analysis. We applied our model to a curated version of the fundamental glycolysis pathway, which is an ideal candidate to define metabolic efficiency. The model, described in detail in the Material and methods section, allows the changes in glycolysis efficiency (defined as the ratio between glucose income and pyruvate production) to be measured as a function of time based on gene expression changes. Figure 6 reports the relative change in efficiency at each time point in comparison with the initial one. Detectable changes in glycolysis were observed in liver and skin tissue, while in all three organisms brain efficiency is essentially constant (figure 6). Noticeably, N. furzeri shows an increase in pyruvate production, in accordance with an increase in metabolic rate during ageing, while D. rerio efficiency decreases.
Figure 6.
Change in glycolysis efficiency over time. Barplots displaying the change in glycolysis efficiency, defined as the ratio of glucose to pyruvate transformation, as estimated by the constraint-based metabolic model. The bars show the relative change in efficiency at a given time point T with respect to the initial time T0 for N. furzeri (green), zebrafish (blue), mouse (pink) and three different tissues: liver (a), skin (b) and brain (c). Overall, the figure shows how NFU increases its efficiency during its lifetime. MMU = M. musculus; DRE = D. rerio; NFU = N. furzeri.
4. Discussion
Every organism ages, but understanding the basic mechanisms of ageing is a longstanding open problem in evolutionary biology. Here we investigated this aspect considering N. furzeri, a species of killifish from the family Nothobranchiidae which has a significantly shorter lifespan with respect to other fishes, including D. rerio [1], even when the age is properly rescaled by the fish weight. We addressed two main questions: (i) Are differences in ageing behaviour due to a specific core of genes or is it a more global process involving a complex gene deregulation? (ii) Are ageing mechanisms distinct in different vertebrates?
To address the first question, our strategy was to compare the transcriptomes of N. furzeri in three different tissues (brain, liver and skin) at five different time points, ranging from sexual maturity up to average lifetime, with respect to D. rerio and quantify the percentage of deregulated genes. We found that N. furzeri has a larger fraction of deregulated genes (45%) with respect to D. rerio. We found a similar percentage of deregulated genes in D. rerio and in M. musculus related to similar pathways. We then computed the distribution of genomic distances between expressed genes in each species, comparing it with a null distribution obtained by picking random genes from each genome. Thus, we could measure, using the Kolmogorov–Smirnov test, how likely it is that DE genes are randomly distributed along the genome, introducing a new measure for positional enrichment analysis based only on differentially expressed genes [37,38]. Such an analysis could highlight clusters of genes and set the basis for higher order chromatin organization studies [39]. We thus used the Kolmogorov–Smirnov test to compare the DE gene cluster distribution with a random cluster distribution. We found that in N. furzeri the DE gene cluster distribution did not significantly deviate from the one obtained in a random cluster null model. The lack of localization of DE genes suggests the presence of global changes in the regulation of gene expression happening early during the lifetime of N. furzeri. On the other hand, in D. rerio and more clearly in M. musculus we found a clear non-random deregulation pattern, confirming that N. furzeri shows a peculiar pattern of gene deregulation. We should remember that this particular approach has some limitations, since it evaluates only the linear distance between genes without considering three-dimensional conformations or co-regulation [40]. However, the patterns we observed in the three organisms highlight the presence of a different organization in the distribution of DE genes.
In order to investigate in more depth the impact of ageing on the biology of these vertebrates, we focused our attention on genes involved in chromatin remodelling, such as polycomb genes. Polycomb functions are performed by multiprotein complexes that selectively occupy chromatin sites. They are usually subdivided into two main classes: polycomb repressive complex 1 and 2 (PRC1 and PRC2) [41]. Polycomb genes of both classes are almost constantly expressed in M. musculus, while there is a larger variability in their expression in D. rerio, where the different pattern of expression of bmi1a and bmi1b could indicate a slightly different role of these proteins during the zebrafish lifetime [42]. Our analysis shows instead a clear deregulation of polycomb genes in N. furzeri, suggesting that chromatin might be accessible in all the analysed tissues and at all the time points. Danio rerio and more clearly M. musculus show a tighter regulation of gene expression.
Since chromatin remodelling is due to the dynamic modification of chromatin architecture allowing for access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby controlling gene expression, we also investigated deregulation of HAT and HDAC genes. We found a deregulation of these genes in N. furzeri with respect to M. musculus and D. rerio in all three different tissues (brain, liver and skin) at all time points. All together these results show a global deregulation and suggest the presence of open chromatin in the short-lived N. furzeri without any specific pattern of organization of these genes. In D. rerio we found less deregulation and more organized patterns of deregulated genes. In M. musculus, the situation is similar but even more clearly organized into patterns and there are coordinated changes in gene expression.
To investigate whether we can observe an impact of gene deregulation at a higher level of complexity, we analysed the deregulation of metabolic pathways and reactions, using a reference Kegg database [32] for a total of 1691 reactions belonging to 76 pathways. We also grouped the genes into 11 superpathways and the reactions containing at least one DE gene were considered as deregulated. While the structure of the metabolic pathways was the same for all three species considered, we found a high deregulation of the reaction in N. furzeri considering the superpathways. To confirm these results, we introduced a network-based model to analyse the glycolysis efficiency, defined as the ratio of glucose to pyruvate transformation. Glycolysis was chosen because it is a highly conserved pathway and is fundamental in metabolism. Our analysis revealed changes in liver and skin tissue among the species, while in all three organisms brain efficiency was found to be essentially constant. Noticeably, N. furzeri showed an increase in pyruvate production as the metabolic rate increased during ageing, while in D. rerio the efficiency decreased with time. We decided to focus on metabolism because metabolic rate is usually associated with ageing, but it would also be possible to study anabolic reactions with the same network.
Altogether, our results show that the short lifespan of N. furzeri, even when properly rescaled by its body weight, is due to a peculiar metabolic and chromatin dynamics that is very different not only from other fish species but also from mammals. It is only because of a comparative analysis of the time dependence of the metabolic and gene expression activity in fishes that one can fully appreciate the peculiarity of the ageing behaviour of N. furzeri. This can be seen as an example of exaptation in which the same gene and pathways common to all fishes are used in a different way by N. furzeri. On the hand, the ageing behaviour in mammals is closer to that observed in fish such as D. rerio rather than that in N. furzeri.
Supplementary Material
Supplementary Material
Acknowledgements
C.A.M.L.P. and S.Z. are grateful for hospitality at Ludwig-Maximilian University Munich.
Data accessibility
Accession numbers for the primary data analysed in this paper are reported in the methods section.
Authors' contributions
M.R.F., S.M. and S.Z. analysed the data. F.F.-C. performed numerical simulations. C.A.M.L.P. coordinated the project and wrote the paper.
Competing interests
We declare we have no competing interest.
Funding
S.Z. thanks the Alexander von Humboldt foundation for the Humboldt Research Award.
References
- 1.Dance A. 2016. Live fast, die young. Nature 535, 453–455. ( 10.1038/535453a) [DOI] [PubMed] [Google Scholar]
- 2.Harel I. et al. 2015. A platform for rapid exploration of aging and diseases in a naturally short-lived vertebrate. Cell 160, 1013–1026. ( 10.1016/j.cell.2015.01.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgart M. et al. 2016. Longitudinal RNA-seq analysis of vertebrate aging identifies mitochondrial complex I as a small-molecule-sensitive modifier of lifespan. Cell Syst. 2, 122–132. ( 10.1016/j.cels.2016.01.014) [DOI] [PubMed] [Google Scholar]
- 4.Platzer M, Englert C. 2016. Nothobranchius furzeri: a model for aging research and more. Trends Genet. 32, 543–552. ( 10.1016/j.tig.2016.06.006) [DOI] [PubMed] [Google Scholar]
- 5.Froese R, Pauly D. 2000. FishBase 2000: concepts, design and data sources. ICLARM, Los Ban os. Laguna, Philippines: ICLARM. [Google Scholar]
- 6.Cinco E. 1982. Length-weight relationships of fishes. ICLARM Tech. Rep. 7 Laguna, Philippines: ICLARM. [Google Scholar]
- 7.Tacutu R. et al. 2018. Human ageing genomic resources: new and updated databases. Nucleic Acids Res. 46, D1083–D1090. ( 10.1093/nar/gkx1042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.La Mesa M. 2001. Age and growth of Crystallogobius linearis (von Düben, 1845) (Teleostei: Gobiidae) from the Adriatic Sea. Sci. Mar. 65, 375–381. ( 10.3989/scimar.2001.65n4375) [DOI] [Google Scholar]
- 9.Badcock J, Merrett NR. 1976. Midwater fishes in the eastern North Atlantic—I. Vertical distribution and associated biology in 30°N, 23°W, with developmental notes on certain myctophids. Prog. Oceanogr. 7, 3–58. ( 10.1016/0079-6611(76)90003-3) [DOI] [Google Scholar]
- 10.Battaglia P, Andaloro F, Esposito V, Granata A, Guglielmo L, Guglielmo R, Musolino S, Romeo T, Zagami G. 2016. Diet and trophic ecology of the lanternfish Electrona risso (Cocco 1829) in the Strait of Messina (central Mediterranean Sea) and potential resource utilization from the Deep Scattering Layer (DSL). J. Mar. Syst. 159, 100–108. ( 10.1016/j.jmarsys.2016.03.011) [DOI] [Google Scholar]
- 11.Swenson RO. 1999. The ecology, behavior, and conservation of the tidewater goby, Eucyclogobius newberryi. Environ. Biol. Fishes 55, 99–114. ( 10.1023/A:1007478207892) [DOI] [Google Scholar]
- 12.Galeotti DM. 2013 Metapopulation theory explains black-stripe minnow (Pisces: Galaxiidae, Galaxiella nigrostriata) distribution in seasonal wetlands in south-west Western Australia. See https://ro.ecu.edu.au/theses/708 .
- 13.Genade T, Benedetti M, Terzibasi Tozzini E, Roncaglia P, Valenzano D, Cattaneo A, Cellerinoario A. 2005. Annual fishes of the genus Nothobranchius as a model system for aging research. Aging Cell 4, 223–233. ( 10.1111/j.1474-9726.2005.00165.x) [DOI] [PubMed] [Google Scholar]
- 14.Herrera M, Jagadeeswaran P. 2004. Annual fish as a genetic model for aging. J. Gerontol. A Biol. Sci. Med. Sci. 59, B101–B107. ( 10.1093/gerona/59.2.B101) [DOI] [PubMed] [Google Scholar]
- 15.Lucas Sánchez A, Almaida-Pagán P, Madrid JA, de Costa Ruiz J, Mendiola P. 2011. Age-related changes in fatty acid profile and locomotor activity rhythms in Nothobranchius korthausae. Exp. Gerontol. 46, 970–978. ( 10.1016/j.exger.2011.08.009) [DOI] [PubMed] [Google Scholar]
- 16.Markofsky A. et al. 1972. Age at sexual maturity and its relationship to longevity in the male annual cyprinodont fish, Nothobranchius guentheri. Exp. Gerontol. 7, 131–135. ( 10.1016/0531-5565(72)90007-1) [DOI] [PubMed] [Google Scholar]
- 17.Siccardi A, Garris H, Jones W, Moseley D, D’Abramo L, Watts S. 2009. Growth and survival of zebrafish (Danio rerio) fed different commercial and laboratory diets. Zebrafish 6, 275–280. ( 10.1089/zeb.2008.0553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tozzini ET. et al. 2013. Parallel evolution of senescence in annual fishes in response to extrinsic mortality. BMC Evol. Biol. 13, 77 ( 10.1186/1471-2148-13-77) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tozzini ET, Lefrancois C, Domenici P, Hartmann N, Graf M, Cellerino A. 2009. Effects of dietary restriction on mortality and age-related phenotypes in the short-lived fish Nothobranchius furzeri. Aging Cell 8, 88–99. ( 10.1111/j.1474-9726.2009.00455.x) [DOI] [PubMed] [Google Scholar]
- 20.Tozzini ET, Valenzano D, Benedetti M, Roncaglia P, Cattaneo A, Domenici L, Cellerino A. 2008. Large differences in aging phenotype between strains of the short-lived annual fish Nothobranchius furzeri. PLoS ONE 3, e3866 ( 10.1371/journal.pone.0003866) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valdesalici S, Cellerino A. 2003. Extremely short lifespan in the annual fish Nothobranchius furzeri. Proc. R. Soc. Lond. B 270(Suppl. 2), S189–S191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wendler S, Hartmann N, Hoppe B, Englert C. 2015. Age-dependent decline in fin regenerative capacity in the short-lived fish Nothobranchius furzeri. Aging Cell 14, 857–866. ( 10.1111/acel.12367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aramillo Irizar P. et al. 2018. Transcriptomic alterations during ageing reflect the shift from cancer to degenerative diseases in the elderly. Nat. Commun. 9, 327 ( 10.1038/s41467-017-02395-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF. 2012. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 39, D1005–-D1010. ( 10.1093/nar/gkq1184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reichwald K. et al. 2015. Insights into sex chromosome evolution and aging from the genome of a short-lived fish. Cell 163, 1527–1538. ( 10.1016/j.cell.2015.10.071) [DOI] [PubMed] [Google Scholar]
- 26.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. ( 10.1038/nprot.2016.095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Leary NA. et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. ( 10.1093/nar/gkv1189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. ( 10.1093/bioinformatics/btp616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. 2010. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38(Suppl 1), D204–D210. ( 10.1093/nar/gkp1019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altenhoff AM. et al. 2017. The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res. 46, D477–D485. ( 10.1093/nar/gkx1019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Virtanen P. et al. 2020. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272. ( 10.1038/s41592-019-0686-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2016. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. ( 10.1093/nar/gkw1092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujibuchi W, Goto S, Migimatsu H, Uchiyama I, Ogiwara A, Akiyama Y, Kanehisa M. 1998 DBGET/LinkDB: an integrated database retrieval system. Pacific Symposium on Biocomputing, Maui, HI, 4–9 January 1998, pp. 683–694. See http://psb.stanford.edu/psb-online/proceedings/psb98/fujibuchi.pdf . [PubMed]
- 34.Guberman JM. 2011. BioMart Central Portal: an open database network for the biological community. Database2011, bar041. ( 10.1093/database/bar041) [DOI]
- 35.Tan J-z, Yan Y, Wang X-x, Jiang Y, Xu HE. 2014. EZH2: biology, disease, and structure-based drug discovery. Acta Pharmacol. Sin. 35, 161–174. ( 10.1038/aps.2013.161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grozinger CM, Schreiberario SL. 2000. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl Acad. Sci. USA 97, 7835–7840. ( 10.1073/pnas.140199597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Preter K, Barriot R, Speleman F, Vandesompele J, Moreau Y. 2008. Positional gene enrichment analysis of gene sets for high-resolution identification of overrepresented chromosomal regions. Nucleic Acids Res. 36, e43 ( 10.1093/nar/gkn114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glaz J, Pozdnyakov V, Wallensteinario S. 2009. Scan statistics: methods and applications. Boston, MA: Birkhauser. [Google Scholar]
- 39.Fritz AJ. et al. 2018. Intranuclear and higher-order chromatin organization of the major histone gene cluster in breast cancer. J. Cell. Physiol. 233, 1278–1290. ( 10.1002/jcp.25996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szczepińska T, Pawłowski K. 2013. Genomic positions of co-expressed genes: echoes of chromosome organisation in gene expression data. BMC Res. Notes 6, 229 ( 10.1186/1756-0500-6-229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margueron R, Reinberg D. 2011. The polycomb complex PRC2 and its mark in life. Nature 469, 343–349. ( 10.1038/nature09784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dupret B, Völkel P, Bourhis XL, Angrand P-O. 2016. The polycomb group protein pcgf1 is dispensable in zebrafish but involved in early growth and aging. PLoS ONE 11, e0158700 ( 10.1371/journal.pone.0158700) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession numbers for the primary data analysed in this paper are reported in the methods section.