Abstract
The DNA polymerase Polκ plays a key role in translesion synthesis, an error-prone replication mechanism. Polκ is overexpressed in various tumor types. Here, we found that melanoma and lung and breast cancer cells experiencing stress from oncogene inhibition upregulated the expression of Polκ and shifted its localization from the cytoplasm to the nucleus. This effect was phenocopied by inhibition of the kinase mTOR, by induction of ER stress, or by glucose deprivation. In unstressed cells, Polκ is continually transported out of the nucleus by exportin-1. Inhibiting exportin-1 or overexpressing Polκ increased the abundance of nuclear-localized Polκ, particularly in response to the BRAF-targeted inhibitor vemurafenib, which decreased the cytotoxicity of the drug in BRAFV600E melanoma cells. These observations were analogous to how Escherichia coli encountering cell stress and nutrient deprivation can upregulate and activate DinB/pol IV, the bacterial orthologue of Polκ, to induce mutagenesis that enables stress tolerance or escape. However, we found that the increased expression of Polκ was not excessively mutagenic, indicating that non-catalytic or other functions of Polκ could mediate its role in stress responses in mammalian cells. Repressing the expression or nuclear localization of Polκ might prevent drug resistance in some cancer cells.
Introduction
Errors in DNA replication can lead to increased mutation rates, thereby contributing to cancer pathogenesis. For example, somatic or germline mutations in the proofreading domain of DNA polymerase delta (polδ) or epsilon (polε) can lead to tumors with markedly increased numbers of point mutations (1–3). Aside from these two main replicative polymerases, a number of other DNA polymerases have been identified that may contribute to cancer initiation or progression (4). For example, inactivation of DNA polymerase eta (polη) is associated with xeroderma pigmentosum variant (XP-V), which predisposes patients to UV-induced skin cancers (5). Additionally, DNA polymerase iota (polι) is upregulated in esophageal squamous cell cancer, and its expression levels positively correlate with lymph node metastasis/clinical stage (6). During the revision of this manuscript, a study identifying a role for multiple error-prone polymerases in resistance to targeted therapies, such as cetuximab, in colorectal cancer was published (7).
The roles of other DNA polymerases in this process are less well understood but likely could contribute to tumor progression. One such polymerase is DNA polymerase kappa (polκ), which is a member of the Y-family of DNA polymerases that plays an essential role in the DNA damage tolerance process of translesion synthesis (8, 9). Several previous studies have shown that overexpression of polκ can contribute to tumorigenesis and drug resistance in cancer (10–13). For example, overexpression of polκ in glioblastoma cells increases resistance to the DNA-damaging agent temozolomide (13), and it has also been found to be substantially overexpressed in lung cancer (10).
Polκ can replicate DNA in both an error-free and error-prone manner during translesion synthesis (14). It can bypass thymine glycols in a relatively error-free manner (15), whereas it bypasses N-2-acetylaminofluorene adducts in a more error-prone manner (16). When replicating on undamaged DNA, polκ has a markedly high error rate due to a relatively large active site and lack of a proofreading domain (17). Using in vitro assays, it has been shown to have error rates as high as 1 error per 200 base pairs when replicating on undamaged DNA (18). For this reason, it is considered an “error-prone” polymerase that can induce untargeted mutations while acting either directly at the replication fork or by filling in post-replication gaps (19). The range of errors introduced by polκ span virtually all substitutions, although to differing degrees (with a high rate of T→G substitutions), as well as a preponderance of deletions (17). These error rates are substantially higher than that found for the replicative polymerases polδ and polε.
In addition to these roles in DNA repair, recent data has also demonstrated that polκ may have a non-catalytic function (20). Human lympoblastic Nalm6 cells, which have intact p53 signaling and MSH2 activity, were engineered to express a catalytically dead (CD) D198A/E199A polκ mutant, which completely lost all polymerase activity yet maintained normal protein expression. The CD mutant was then compared to complete knockouts (KO) or wild-type (WT) cells for their ability to protect against a panel of genotoxic stressors. Remarkably, whereas the KO cells were highly sensitive to oxidizing agents such as hydrogen peroxide and menadione, the CD cells showed no such defects and were able to protect against these agents as well as WT cells. The mechanisms by which catalytically deficient polκ may protect against such damage remains unclear, but it is posited to be due to its interactions with other proteins such as the DNA repair protein REV1, and/or play a role in the repair of oxidized dNTPs.
Given the diverse role of polκ, it is important that cells regulate both its expression and access to DNA. How polκ is regulated can be informed by several decades of work studying DinB, the E. coli orthologue of polκ, which is regulated by the “SOS”/DNA damage response along with the RpoS/starvation stress response (21, 22). Under stress conditions, E. coli can temporarily increase their mutation rate using a process known as stress-induced mutagenesis (22–25). This hypermutation is enacted as part of double-strand DNA break repair, which becomes mutagenic due in part to the activity of DinB (26). This mutagenic process is regulated at three levels, as recently reviewed (24): (i) a double-strand break (27, 28), (ii) activation of the SOS DNA damage response (23), and (iii) activation of the generalized sigma S (RpoS) stress response (22). The SOS response, when coupled with the RpoS stress response, allows first for upregulation of DinB (29) and then subsequent usage of this error-prone polymerase for mutagenic repair, which results in the base substitutions and indels that are commonly observed (22). It is likely that deficiencies in mismatch repair contribute to this process since overexpression of MutL inhibits mutation in stationary phase but not during growth (30), whereas both MutS and MutH can be downregulated in part by the RpoS stress response pathway (31).
In contrast to those in E. coli, the mechanisms regulating the expression and localization of polκ in mammalian cells is less well understood. In normal human tissues, polκ is widely expressed at the mRNA level (32); whereas in the mouse, it is highly enriched in the adrenal cortex and testis (33). In the mouse, protein expression using a peptide-generated antibody was noted in adrenal cortex, pachytene cells in meiosis I, post-meiotic spermatids, and some epithelial cells in the lung and stomach (33). At the cellular level, numerous studies using overexpression of EGFP-polκ fusion proteins have demonstrated that polκ is strongly enriched in the nucleus (34, 35). However, antibody staining of endogenous polκ protein using antibodies generated with either peptide fragments or full-length proteins have shown variable expression in both the cytoplasm as well as the nucleus (33). Analysis of the polκ promoter has shown consensus binding sites for Sp1 and CREB, both of which have been reported to transcriptionally activate its expression (36, 37).
Although recent observations demonstrate that polκ can promote tumorigenesis and drug resistance (10–13), it is unknown what regulates its expression in cancer, and to what extent this depends on its mutagenic properties. Ectopic expression of polκ allows it to become part of the replication machinery, even in the absence of external stress, indicating that high levels of it could be sufficient to induce new mutations, likely in concert with other alterations in DNA repair machinery (34). This has important clinical implications since dysregulation of polκ expression could therefore contribute to tumorigenic phenotypes by affecting its normal subcellular localization. In this study, we demonstrate that deprivation of oncogenic signaling in melanoma, lung cancer, and breast cancer cell lines upregulates polκ and confines it to the nucleus. These pathways converge on PI3K/mTOR signaling, a central regulator of nutrient status in the cell (38) that may be analogous to the generalized stress factor sigma S (RpoS) in E. coli (24). When cells are nutrient replete and have intact mTOR signaling, polκ is primarily present in the cytoplasm; when cells are starved or mTOR is inhibited, polκ shifts primarily to the nucleus, suggesting that the cell can dynamically regulate polκ in response to cell stress. In line with this, we find that polκ can be rapidly exported back out of the nucleus by the nuclear export machinery, implying that cells may normally use nuclear export to prevent excess polκ in the nucleus. When this dynamic regulation is subverted by forced nuclear overexpression of polκ, we find that this increases resistance to the BRAFV600E inhibitor vemurafenib in melanoma cells. Surprisingly, we find little evidence that polκ is highly mutagenic in these melanoma cells, indicating it may play other currently unknown roles in inducing drug resistance. Our data suggest a mechanism by which mammalian cancer cells regulate the levels and localization of polκ, dysregulation of which can contribute to drug resistance.
Results
MAP kinase inhibition induces POLK mRNA upregulation and changes the subcellular localization of its protein
Given the role of stress in upregulating DinB/pol IV in E. coli, we first asked whether cell stress regulated polκ expression in cancer. We reasoned that drugs blocking oncogenic drivers would induce cell cycle arrest and ultimately apoptosis and would represent an extreme form of cell stress. In human melanoma, the most common activating mutation occurs at BRAFV600E, which activates downstream MAP kinase signaling via MEK/ERK signaling. This provides the melanoma cells with a substantial growth advantage. Small molecules targeting the BRAF/MEK/ERK pathway have been developed, some of which are being clinically used to treat melanoma patients (39–41). BRAF inhibitors such as vemurafenib can induce cell stress prior to overt death of the cancer cell, which can occur via the ER stress pathway (42) and is associated with stress-induced senescence (43). Based on this, we first asked whether inhibition of the MAP kinase pathway could induce polκ expression in melanoma.
To assess this, we treated the BRAFV600E mutant melanoma A375 cell line with the BRAFV600E kinase inhibitor PLX4032 (vemurafenib) (39) and measured the abundance of transcripts encoding Polκ (POLK mRNA) by qRT-PCR from 2 to 72 hours after exposure. This revealed an upregulation in expression of polκ at the transcript level that peaked at 24 hours and was sustained thereafter (Fig. 1A). We next examined expression of polκ at the protein level under similar conditions. We utilized an antibody raised against full-length human polκ protein and verified its specificity using shRNA knockdown and overexpression of endogenous polκ (fig. S1, A to C). Although polκ has been largely reported to be nuclear localized when overexpressed as an EGFP-tagged fusion protein (34, 35), we unexpectedly found that the Western blot using an antibody raised against the full-length protein showed that endogenous polκ was expressed in both cytoplasmic and nuclear fractions (Fig. 1B). This expression pattern is similar to that of the related family member pol eta (polη), where both cytoplasmic and nuclear expression is seen (Human Protein Atlas (44)). Whereas treatment with vemurafenib did not change overall protein levels of polκ, it instead induced a specific increase in nuclear polκ and corresponding decrease in cytoplasmic polκ (Fig. 1, B and C). We confirmed this redistribution using immunofluorescence, wherein acute exposure to PLX4032 induced a shift of polκ from the cytoplasm to the nucleus (Fig. 1, D and E). Cytoplasmic localization of a DNA polymerase that can shift from the cytoplasm to the nucleus has been reported previously (45–47), suggesting that this mode of regulation might be relevant for polκ under physiologic conditions. Because BRAF activates downstream MEK/ERK pathway signaling, we also examined whether downstream inhibitors would elicit similar effects. In two different BRAFV600E mutant melanoma cell lines (A375 and SK-MEL28), we found that both MEK and ERK inhibitors (40, 41) produced an upregulation of POLK mRNA and a nuclear shift of polκ that are similar to that seen after BRAF inhibition (Fig. 1, F to I and fig. S2, A to D).
Fig. 1. Treatment of melanoma cells with BRAF or MAP kinase inhibitors modulates polκ expression and localization.
(A) qRT-PCR to detect the mRNA expression of polκ relative to the DMSO control was performed on A375 cells treated with DMSO or 5 μM PLX4032 for 2, 8, 24, 48, or 72 hours. Mean ± S.E.M, n=3 experiments; *P < 0.05, paired two-tailed t-test. (B) Western blot analysis for polκ was performed on A375 cells treated with DMSO (–) or 5 μM PLX4032 (+) for 48 hours. (C) Quantification of the Western blot data relative to the DMSO control. β-actin or Lamin B1 served as the loading control. Mean ± S.E.M from 5–8 independent experiments; ns = non-significant, **P < 0.01, paired two-tailed t-test. (D) Immunofluorescence staining of A375 cells treated with DMSO or 5 μM PLX4032 for 24 hours. scale bar, 10 μm. (E) Quantification of the percentage of cells with nuclear enrichment of polκ. Mean ± S.E.M; at least 2500 cells were counted for each sample across 22 fields of view. ***P < 0.001, paired two-tailed t-test. (F) Immunofluorescence staining of A375 cells treated with DMSO or MEK inhibitors (10 μM CI-1040, 10 μM U0126) for 24 hours. Scale bar, 10 μm. (G) Quantification of the percentage of cells with nuclear enrichment of polκ. Mean ± S.E.M; at least 1000 cells were counted for each sample across 18 fields of view. ***P < 0.001, paired two-tailed t-test. (H) Immunofluorescence staining of A375 cells treated with DMSO or ERK inhibitors (1 μM ulixertinib, 1 μM SCH772984) for 24 hours. Scale bar, 10 μm. (I) Quantification of the percentage of cells with nuclear enrichment of polκ. Mean ± S.E.M; at least 250 cells were counted for each sample across 6 fields of view. ***P < 0.001, paired two-tailed t-test.
DNA damage or cell cycle arrest is not sufficient for Polκ localization
The induction of DinB in bacteria relies upon two related systems: the SOS/DNA damage response and the RpoS-controlled general/starvation stress response. We reasoned that the nuclear localization of polκ in response to MAP kinase inhibition might act through one of these two mechanisms. To test this, we treated A375 melanoma cells with vemurafenib and then checked for markers of DNA damage (48), including gamma H2AX (γH2AX), 53BP1, phosphorylated Chk1 (p-Chk1), and phosphorylated Chk2 (p-Chk2) but saw no induction of any of these markers (fig. S3A). We also tested whether knockdown of p53, which is a regulator of the DNA damage response pathway (49), would prevent the observed subcellular shift but failed to see any change in the nuclear localization of polκ after treatment with PLX4032 (fig. S3, B to D). Another possibility was that cell cycle arrest was the signal, as would be expected during a starvation response or during MAP kinase inhibition. To test this, we treated A375 melanoma cells with either the CDK4/6 inhibitor PD0332991 (50) or the BRAFV600E inhibitor vemurafenib and then assessed the cell cycle using flow cytometry and polκ localization using immunofluorescence. As expected, both of these interventions led to a near complete G1 arrest (fig. S4A); however, treatment with the CDK4/6 inhibitor resulted in little to no nuclear polκ (fig. S4, B and C). Together, these data suggested to us that other mechanisms might be responsible for the induction of polκ.
mTOR inhibition rapidly induces Polκ nuclear accumulation
As we saw no effect of cell cycle inhibition or DNA damage, we next turned to mTOR signaling, a central sensor and effector of growth factor signaling, nutrient status, and stress (38). The mTOR pathway can be activated by RAF/MEK/ERK signaling either directly or by cross-pathway activation (51–54). In addition, BRAF inhibition with vemurafenib has recently been shown to induce the ER stress response (42, 55), which is associated with suppression of mTOR signaling (56). Based on this, we reasoned that MAP kinase inhibition might be acting to dampen downstream mTOR signaling to mediate the effect on polκ. To test this, we treated A375 melanoma cells with inhibitors of BRAF/MEK/ERK signaling for 24 hours and then measured mTOR pathway activation using levels of phosphorylated S6 (p-S6), a target of the kinase S6K (38, 57). We found that these MAP kinase inhibitors potently decreased levels of p-S6 (Fig. 2, A and B and fig. S5, A and B), and that this inversely correlated with the shift of polκ to the nucleus: Cells with the lowest level of p-S6 had the highest levels of nuclear polκ (Fig. 2C). These observations prompted us to then test whether inhibitors of the mTOR pathway itself would lead to an effect on polκ. We treated A375 melanoma cells with several inhibitors of the PI3K/mTOR pathway, including mTOR inhibitors (rapamycin or PP242) or a PI3K inhibitor (LY29002). We found that these inhibitors potently induced nuclear polκ to a level comparable to that seen with the BRAF/MEK/ERK inhibitors, but it occurred much more rapidly (Fig. 2, D and E). Whereas the MAP kinase inhibitors took ~24 hours for full induction of nuclear polκ, the mTOR pathway inhibitors could do this in as little as ~6 hours. In addition, drugs that activate the endoplasmic reticulum (ER) stress pathway, which can be activated by BRAF inhibitors (fig. S6, A and B) as well as dampened mTOR signaling, produced a similar effect (fig. S6, C and D).
Fig. 2. mTOR signaling regulates polκ subcellular localization.
(A) Immunofluorescence staining of A375 cells treated with DMSO or 5 μM PLX4032 for 24h hours. Scale bar, 10 μm. (B) Quantification of the percentage of cells with phosphorylated S6 (p-S6, S240/244). Mean ± S.E.M; at least 400 cells were counted for each sample across 8 fields of view. ***P < 0.001, paired two-tailed t-test. (C) Immunofluorescence staining of A375 cells treated with 5 μM PLX4032 for 3, 6, 12, or 24 hours. The bar graph shows the percentage of cells with nuclear enrichment of polκ (left axis, solid line) and the percentage of cells with phosphorylated S6 (p-S6, right axis, dashed line), Mean ± S.E.M; at least 150 cells were counted for each sample across 4–8 fields of view. The Pearson correlation coefficient (r) was calculated for the percentage of cells with nuclear enrichment of polκ vs. the percentage of cells with phosphorylated S6. (D) Immunofluorescence staining of A375 cells treated with DMSO, 0.5 μM rapamycin, 5 μM PP242, or 30 μM LY294002 for 6 hours. Scale bar, 10 μm. (E) Quantification of the percentage of cells with nuclear enrichment of polκ after 6 hours of treatment. Mean ± S.E.M; at least 200 cells were counted for each sample across 8–12 fields of view. **P < 0.01, ***P < 0.001, paired two-tailed t-test.
Polκ dysregulation occurs in other cancer types
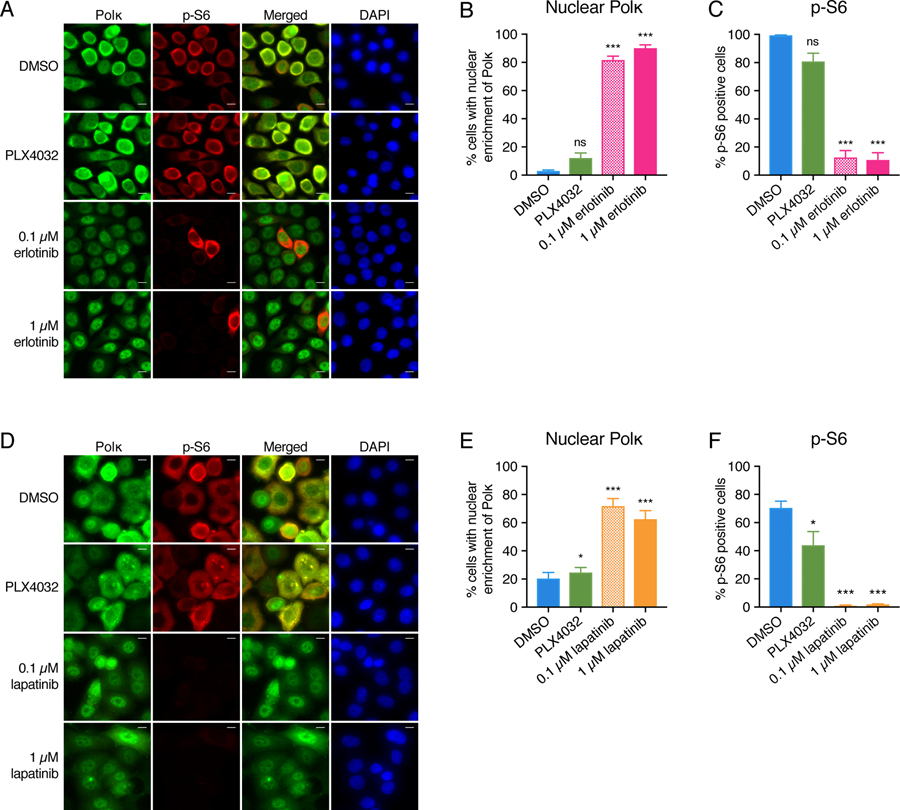
We next wished to determine if the observed effects on polκ were specific to melanoma/the BRAF pathway, or if they could be applied more broadly. To test this, we examined POLK mRNA levels and subcellular localization in breast and lung cancer cell lines, which harbor unique oncogenic dependencies that differ from melanoma. PC-9 lung cancer cells harbor activating mutations in EGFR and are critically dependent upon that growth factor signaling pathway. We treated PC-9 cells with either the EGFR inhibitor erlotinib (58) or the BRAFV600E inhibitor PLX4032 (which we would expect to have no effect as these cells contain wild-type BRAF) and then measured the expression of polκ and determined its localization. Both high and low doses of erlotinib caused induction of polκ mRNA and a shift of polκ to the nucleus in the PC-9 cells, but as expected, this effect was not seen with PLX4032 (Fig. 3, A and B and fig. S7A). Analogous to what we observed for melanoma, this shift of polκ to the nucleus was accompanied by a loss of p-S6 in the lung cancer cells (Fig. 3, A and C). A similar effect was seen in a breast cancer cell line. SK-BR3 cells, which overexpress the HER2 oncogene, were treated with the HER2 inhibitor lapatinib (59) or the BRAFV600E inhibitor PLX4032. Lapatinib caused a marked induction of polκ expression, nuclear accumulation of polκ, and suppression of p-S6 (Fig. 3, D to F and fig. S7B), an effect that was not seen with PLX4032 in this cell line as expected. These results demonstrate that the observed effects on polκ are specific to the driver oncogene, and that they can be generalizable to multiple cancer types.
Fig. 3. The effects on polκ and p-S6 are driver gene specific across tumor types.
(A) Immunofluorescence staining of PC-9 cells treated with DMSO, 5 μM PLX4032, or erlotinib (0.1 μM or 1 μM) for 24 hours. Scale bar, 10 μm. (B) Quantification of the percentage of cells with nuclear enrichment of polκ. Mean ± S.E.M; at least 100 cells were counted for each sample across 4 fields of view. ns = non-significant, ***P < 0.001, paired two-tailed t-test. (C) Quantification of the percentage of cells with phosphorylated S6 (p-S6, S240/244). Mean ± S.E.M; at least 100 cells were counted for each sample across 4 fields of view. ns = non-significant, ***P < 0.001, paired two-tailed t-test. (D) Immunofluorescence staining of SK-BR3 cells treated with DMSO, 5 μM PLX4032, or lapatinib (0.1 μM or 1 μM) for 24 hours. Scale bar, 10 μm. (E) Quantification of the percentage of cells with nuclear enrichment of polκ. Mean ± S.E.M; at least 200 cells were counted for each sample across 7 fields of view. *P < 0.05, ***P < 0.001, paired two-tailed t-test. (F) Quantification of the percentage of cells with phosphorylated S6 (p-S6, S240/244). Mean ± S.E.M; at least 200 cells were counted for each sample across 7 fields of view. *P < 0.05, ***P < 0.001, paired two-tailed t-test.
Glucose starvation phenocopies the effects on subcellular localization of Polκ
mTOR could elicit the observed effect on polκ through multiple downstream mechanisms, but because E. coli use the starvation response as part of DinB upregulation/activation, we wondered whether by inhibiting mTOR, we were mimicking a starvation response. To test this idea, we examined the effect of nutrient deprivation on polκ localization. We grew A375 melanoma cells in a variety of media conditions in which we selectively removed serum, glucose, or glutamine and measured polκ localization by immunofluorescence. Whereas we found that both glutamine and serum deprivation had small effects on nuclear polκ localization, glucose starvation resulted in a highly significant induction of nuclear polκ (fig. S8, A and B). Because glucose is a major carbon source for rapidly growing cancer cells, this may be analogous to the starvation response induced in E. coli by deprivation of lactose, which is an important carbon source for bacteria.
Exportin-1 plays a role in regulating the subcellular localization of Polκ
This data led us to ask what mechanisms the cells may use to control cytoplasmic versus nuclear localization of polκ. Two possibilities for the effects we observed was that they were mediated by either degradation of polκ or relocalization of polκ from the cytoplasm to the nucleus. A previous analysis of the protein structure of polκ demonstrated that the full-length protein contains a bipartite nuclear localization signal (NLS) towards the 3’ end of the gene (32, 35), and computational analysis (60) also indicated a likely nuclear export signal (NES). Deletion of the NLS region results in the protein completely localizing to the cytoplasm (35), which suggested to us that the localization results we saw above might be controlled by the nuclear import and/or export machinery. To test this, we utilized inhibitors of importin-β or exportin-1 (CRM1) and tested whether these affected the shift of polκ in response to BRAF inhibition in melanoma cells. We found that importazole (61), which inhibits importin-β, had little effect on polκ localization (fig. S9, A and B). In contrast, co-treatment with leptomycin B (62), which inhibits exportin-1, accelerated the rate at which polκ localized to the nucleus: whereas 3 hours of PLX4032 typically only leads to ~30% induction of nuclear polκ, in the presence of leptomycin B, this increased to 60% (Fig. 4, A and B). In addition, we noted that leptomycin B alone (even in the absence of PLX4032) induced a small but significant increase in nuclear polκ, indicating that polκ normally cycles between the cytoplasm and nucleus. To further test the contribution of the nuclear export machinery, we performed washout experiments. A375 cells were treated with PLX4032 (to induce nuclear polκ) and then the drug was washed away (Fig. 4C). After washout, polκ relocates to the cytoplasm within 24 hours. However, if leptomycin B is added after PLX4032 washout, polκ remains present in the nucleus (Fig. 4, D and E). In contrast, we found little evidence that PLX4032 directly affected polκ degradation. Treatment of A375 cells with the proteasome inhibitor MG132 (63) increased polκ protein levels to a similar extent in DMSO- or PLX4032-treated cells (fig. S10). Collectively, this data indicates that oncogenic signaling regulates the nuclear localization of polκ at least in part through the export machinery.
Fig. 4. Exportin-1 plays a role in the subcellular localization of polκ.
(A) Immunofluorescence staining of A375 cells treated with DMSO or 5 μM PLX4032 ± 20 μM leptomycin B (LMB) for 6 hours. Scale bar, 10 μm. (B) Quantification of the percentage of cells with nuclear enrichment of polκ after 3 or 6 hours. Mean ± S.E.M; at least 100 cells were counted for each sample across 4–8 fields of view. **P < 0.01, ***P < 0.001, paired two-tailed t-test. (C) Schema detailing the experiment design for the leptomycin B (LMB) recovery assay subsequently shown in (D and E). (D) Immunofluorescence staining of A375 cells treated with DMSO or 5 μM PLX4032 for 24 hours and then DMSO or 10 μM LMB for 24 hours. Scale bar, 10 μm. (E) Quantification of the percentage of cells with nuclear enrichment of polκ. Mean ± S.E.M; at least 100 cells were counted for each sample across 7 fields of view. *P < 0.05, **P < 0.01, ***P < 0.001, paired two-tailed t-test.
Polκ overexpression can lead to increased drug resistance but is minimally mutagenic on its own
These data suggested a model in which rapidly growing cancer cells generally keep polκ at relatively low levels and sequestered in the cytoplasm due to active nuclear export. Previous work has shown that forced overexpression of polκ, which is predominantly nuclear, can contribute to temozolomide resistance in glioblastoma, which occurs by activation of ATM-CHK1 signaling, which promotes homologous recombination-dependent DNA repair (13). This led us to ask whether nuclear polκ might contribute to drug resistance in melanoma as well either through its mutagenic activity or through non-mutagenic capabilities (20). To test this, we generated a doxycycline-inducible polκ overexpression construct for use in A375 melanoma cells (Fig. 5A). We then isolated multiple single cell clones from this population and expanded them in culture in the presence or absence of doxycyline for three months. As expected, these cells showed strong induction with increased nuclear localization of polκ after addition of doxycycline (fig. S1, B and C). We then tested both populations for sensitivity to either the BRAFV600E inhibitor PLX4032 or the CDK4/6 inhibitor PD0332991 (Fig. 5A). For both clones, the population that had experienced long-term polκ overexpression showed a modest increase in growth in PLX4032 across multiple different doses compared to their respective negative control cells (Fig. 5B), consistent with a mild increase in resistance to the drug. In contrast, there was no significant difference between the two populations with regards to their responses to PD0332991 (Fig. 5C).
Fig. 5. polκ overexpression can lead to increased drug resistance.
(A) Schema detailing how single cell clones of A375 cells containing the doxycyline (dox)-inducible polκ overexpression (OE) construct were created and how the dox- (-control) and dox+ (polκ OE) populations were generated and then used to measure drug resistance. (B) Drug resistance of the dox- and dox+ populations of two different clones of dox-inducible polκ overexpression cells to PLX4032 was determined by the CyQuant Direct assay. For each population, the relative cell viability at each dose, compared with the 0 µM dose control (DMSO), was calculated. Mean ± S.E.M, n=4–5 experiments; ns = non-significant, *P < 0.05, **P < 0.01, ***P < 0.001, Fisher’s Least Significant Difference (LSD) test. A comparison between the two populations in their response to PLX4032 by two-way ANOVA gave a p-value of 0.0130 for Clone #1 and of 0.0294 for Clone #2. (C) Drug resistance of the dox- and dox+ populations of two different clones of dox-inducible polκ overexpression cells to PD0332991 was determined by the CyQuant Direct assay. For each population, the relative cell viability at each dose, compared with the 0 µM dose control (DMSO), was calculated. Mean ± S.E.M, n=3–4 experiments; ns = non-significant, Fisher’s Least Significant Difference (LSD) test. A comparison between the two populations in their response to PD0332991 by two-way ANOVA gave a p-value of p = 0.3785 for Clone #1 and of p = 0.2278 for Clone #2.
To test whether this effect on drug resistance was likely mediated by new mutations, we used a mutation reporter assay (Fig. 6A), similar to (64). We stably overexpressed GFP in the dox-inducible polκ melanoma cells and FACS sorted them to yield a population that is 100% GFP+. In the presence of a factor that increases mutation rate, stochastic mutation of the GFP transgene will lead to a loss of GFP expression due to random frameshifts/indels/SNVs. As a positive control, we used CRISPR to knockout the canonical mismatch repair proteins MLH1, MSH2, MSH6 and PMS2. As expected, loss of the mismatch repair proteins led to a loss of GFP-positive cells as measured by FACS (Fig. 6, B and C). In contrast, we found that overexpression of polκ failed to increase mutation rate in this assay, even when combined with the defects in mismatch repair (Fig. 6, B and C). To confirm this, we isolated DNA from the A375 cells and performed MiSeq analysis of this transgenic insertion to a depth of ~10,000X. This analysis failed to demonstrate any new mutations in the polκ overexpressing clones, supporting the observation of the FACS readout. It is possible that polκ can only be fully mutagenic in concert with complete suppression of mismatch repair. Our studies with individual MMR CRISPRs did not show synergy with polκ, but we did not attempt to knockout all MMR genes at the same time. Thus, while we cannot fully exclude other low-level mutagenic activity induced by polκ in these cells, this data suggests polκ can contribute to BRAF inhibitor resistance in A375 melanoma cells but that mechanisms outside of its canonical ability to induce large numbers of new mutations may be contributory.
Fig. 6. Knockout of mismatch repair (MMR) genes but not overexpression of polκ causes increased mutagenesis.
(A) Schema detailing how single cell clones of A375 cells containing the SV40p-GFP and dox-inducible polκ overexpression (OE) constructs were created and how the dox- (-control) and dox+ (polκ OE) populations can be generated. Cas9 and sgRNAs against MMR genes (MLH1, MSH2, MSH6, or PMS2) or a negative control were added to the cells, and each knockout (KO) was validated using the Surveyor Mutation Detection Kit. Cells from each validated knockout were treated for 14 days with media −/+ dox, and then FACS was used to determine the percentage of GFP-negative vs. GFP-positive cells in each population. (B and C) GFP intensity was measured by flow cytometry after SV40p-GFP, dox-inducible polκ OE cells −/+ MMR KO were treated with media −/+ dox for 14 days. The bar graph shows the percentage of GFP-negative (gfp-) and GFP-positive (gfp+) cells for each sample from a representative experiment, performed using two different sets of sgRNAs against the MMR genes: sgRNAs #1 (B) and sgRNAs #2 (C).
Polκ regulates genes associated with drug resistance and immune surveillance
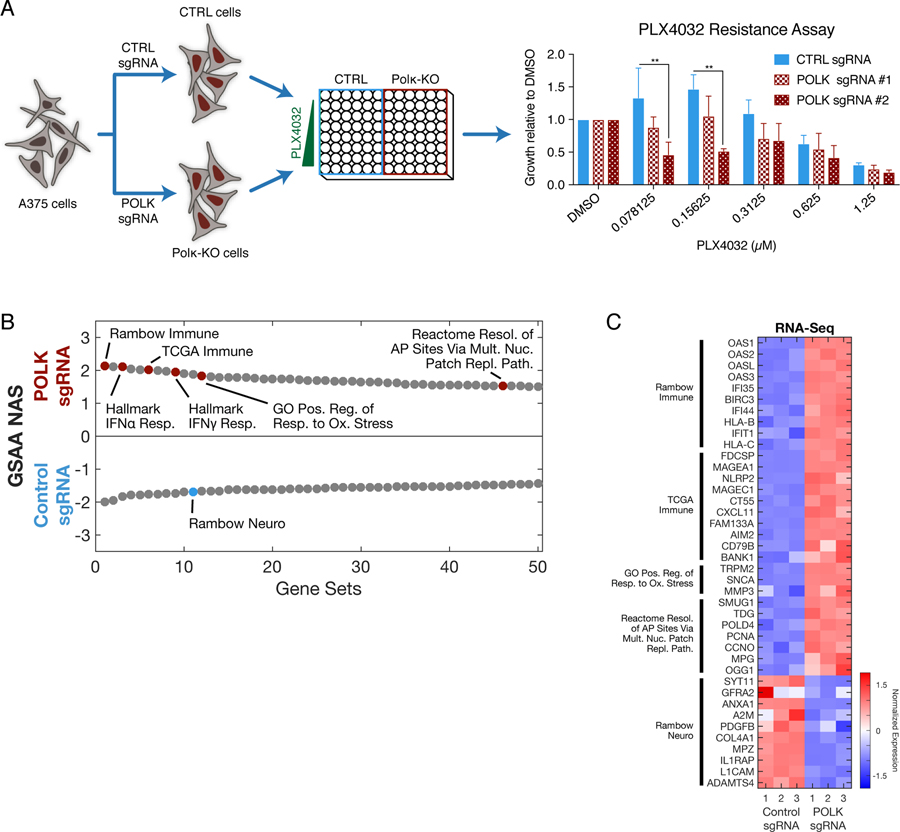
To gain further insight into the mechanisms by which polκ might regulate sensitivity to BRAF inhibitors, we performed CRISPR inactivation of polκ in A375 melanoma cells (fig. S1D) and measured their sensitivity to vemurafenib. This showed that at low doses of drug, knockdown of polκ enhanced sensitivity to vemurafenib, essentially the converse of what we observed with the overexpression of polκ (Fig. 7A). We then performed RNA-seq of the polκ CRISPR cells and compared their gene expression profile to cells that had been treated with a non-targeting control sgRNA. Overall, we found a total of 591 dysregulated genes (310 upregulated and 281 downregulated at a log2-fold change=2.0 and padj<0.05; table S3). We then performed Gene Set Enrichment Analysis (GSEA) on these genes to identify the most altered pathways (Fig. 7, B and C, and table S3). Amongst the most highly enriched pathways, we found that the polκ knockdown cells shared several gene signatures identified by Rambow et al. (65), as those associated with minimal residual disease after treatment with BRAF/MEK inhibitors, a state strongly associated with resistance to these therapies. For example, Rambow et al. identified both a “neuro” and “immune” subtype of cell associated with resistance, and we found that the polκ CRISPR cells strongly dysregulated several key genes expressed in those cells, such as ANGPTL4, LOXL2, ADAMTS4 and L1CAM. We also found that the polκ knockdown cells dysregulated pathways associated with the response to oxidative stress (TRPM2, SNCA, MMP3), which is consistent with the reports described above positing that the catalytically dead polκ may regulate drug resistance via repair of oxidized dNTPs (20). Further supporting this idea, we also identified numerous genes associated with the nucleotide patch patch pathway (SMUG1, TDG, POLD4). In addition to these pathways, we also unexpectedly found a strong enrichment for immune-related genes in the polκ knockdown cells. For example, we found an upregulation of genes associated with an immune signature in the TCGA cohort (FDSCP, MAGEA1, MAGEC1, CXCL11), and more specifically an increase in those associated with signaling mediated by IFN⍺/γ, such as OAS1 and OASL which are involved in the degradation of viral or cellular RNAs. This suggests a plausible idea that dysregulation of polκ (either by BRAF inhibition, starvation, or cellular stress more generally as we have shown) may influence the ability of tumor cells to respond to the immune system in vivo. Previous studies have suggested that error-prone polymerases such as polη or polι play a role in immune responses but this is thought to be primarily via their effects on somatic hypermutation (66–69). It is also possible that some of the effects we see are due to the CRISPR itself, but whether there is an additional effect of polκ on regulation of immune surveillance is an intriguing idea that will need further experimental validation in future studies.
Fig. 7. CRISPR of polκ demonstrates pathways involved in drug resistance and immune response.
(A) Schema detailing how A375 cells were treated with control or polκ sgRNA and Cas9, single cell clones were created, and the resulting cells were used to measure resistance to PLX4032. The graph shows the resistance of control and polκ KO cells to PLX4032 as determined by the CyQuant Direct assay. For each population, the relative cell viability at each dose, compared with the 0 µM dose control (DMSO), was calculated. Mean ± S.E.M, n=3 experiments; **P < 0.01, all other comparisons were non-significant, Fisher’s Least Significant Difference (LSD) test. (B) RNA-seq was performed on the control and polκ KO cells. Dual waterfall plot of top/bottom 50 gene sets from GSAA comparing POLK sgRNA versus control sgRNA ranked by Normalized Association Score (NAS). (C) Heatmap of top Leading Edge genes from selected gene sets (up to 10 genes per gene set).
Discussion
There have been multiple reports of how alterations to DNA replication and repair processes can contribute to tumor initiation and progression. For example, defects in mismatch repair proteins such as MLH1 (70, 71), MSH2 (72), MSH6 (73), or PMS2 (74) are associated with Lynch syndrome, which predisposes patients to a wide variety of tumors including colon and gynecologic cancers (75, 76). Large-scale genome sequencing efforts have identified point mutations in the replicative polymerases polδ or polε, which inactivate the proofreading capacity of these proteins and induce a large number of mutations in the tumors that emerge (1–3).
Several studies have now identified overexpression of polκ in human tumors such as lung cancer or glioblastoma (10, 11). Because polκ lacks a proofreading domain, its overexpression can be associated with increased mutation rates as well as drug resistance (12, 13). However, despite the potential importance of polκ in promoting tumor progression, little is known about the mechanisms by which it is regulated. In this study, we identified oncogenic signaling and nutrient status as one potential regulator of polκ expression and localization in cancer.
Polκ belongs to a family of related Y-family DNA polymerases which all function in translesion synthesis, a major DNA damage tolerance pathway (8, 9). In normal physiology, these polymerases play an important role in bypassing stalled replication forks that are induced by DNA damaging agents. Because they all lack proofreading domains, they have a propensity to introduce errors during replication, although depending on the specific lesion, they can also act in an error-free manner. For this reason, these Y-family polymerases can be a double-edged sword since they allow for replication past damaged regions of DNA and cell survival but may do so at a cost of new mutations (77). in vitro studies have demonstrated that these polymerases can act with extraordinarily low fidelity, with error rates ranging from 10−1 to 10−4, compared to ~10−6 for the normal replicative polymerases polδ or polε (17, 78, 79). For this reason, it is important that cells regulate the expression and localization of these polymerases such that they only act when the cell is under genomic stress.
Although DNA damage is likely a major endogenous inducer of polκ, our data would suggest that mammalian cells harbor the capacity to induce the expression of polκ under other forms of cell stress, namely loss of oncogenic signaling and/or nutrient starvation. This may be analogous to E. coli, whereby cells under starvation stress can upregulate DinB/pol IV (the bacterial orthologue of polκ) (21, 22). However, in E. coli, in addition to activating DinB, the RpoS and SOS pathways have other effects on the cells, including suppression of mismatch repair (31), and all of these processes are necessary to induce new mutations (24). Additionally, previous work has demonstrated that cancer cells harboring mutations in the proofreading domain of polε also experience loss of mismatch repair enzymes (80), and more recently it has been shown that suppression of mismatch repair is necessary for these polε mutants to dramatically increase their mutation rates (81). Notably, in mouse cells, overexpression of DINB1 (the mouse ortholog of polκ) has been shown to be highly mutagenic and causes a 10-fold increase in point mutation rate (82), although the status of mismatch repair was not directly assessed. In contrast, in our cultured A375 melanoma cells, we found little evidence for polκ being nearly this mutagenic, and no evidence that it cooperates with mismatch repair deficiency in generation of such mutations. In part, this could relate to the particularities of melanoma cell lines, which already have very high mutation burdens due to UV-induced DNA damage that occurs in the patients from whom these cell lines are derived (83). In future studies, it will be of great importance to elucidate the situation or cell types in which polκ may be more or less mutagenic.
Despite the lack of compelling evidence of mutagenesis in these melanoma cells, our studies do suggest that prolonged expression of polκ can be associated with modest resistance to BRAF inhibitors such as vemurafenib. Both genetic and non-genetic mechanisms of BRAF inhibitor resistance have been described (84–93), and many of the mutations that lead to drug resistance are pre-existing in the population when examined by deep sequencing. In our studies, it is possible that DNA mutation from polκ overexpression is mutagenic and that contributes to the drug resistance, but that our technologies were not sufficient to quantify this. However, it is important to note that we cannot exclude other, non-mutagenic functions of polκ that could account for the increased drug resistance. For example, non-catalytic functions of polκ (devoid of polymerase activity) are associated with the ability of cells to resist oxidative damage (20). In line with this, BRAF inhibitors such as vemurafenib are known to be potent inducers of ROS in melanoma cells (94), which leads to the activation of pyruvate dehydrogenase kinase (PDK). Our CRISPR and RNA-seq studies are consistent with these non-mutagenic effects of polκ, as we noted a significant enrichment for genes associated with BRAF/MEK-inhibitor resistance previously identified by Rambow, et al (65) as well as an increase in genes associated with oxidative stress response and nucleotide repair. We also unexpectedly noted that loss of polκ dysregulates a large number of genes associated with immune response, especially those in the IFN pathway. Whether polκ could have an unexpected interaction with the efficacy of immunotherapies is unexplored but will be important to functionally test in future studies. Moreover, given the availability of small molecule inhibitors of polκ (95, 96), it will be of future interest to determine whether the administration of such inhibitors could forestall the development of resistance to either targeted or immune-based therapies.
One unexpected finding in our study was the observation that polκ can exist in a cytoplasmic form, at least under certain conditions. Numerous prior studies have shown that polκ is primarily a nuclear protein, but these largely relied upon overexpression of an EGFP-polκ fusion protein (34, 35). In our hands, overexpression of polκ also strongly upregulated nuclear expression, which we believe likely reflects saturation of the export machinery. This would be consistent with our data using leptomycin B, which suggested that one important mechanism of regulation for polκ is nuclear-cytoplasmic shuttling. A few other studies have also observed scant cytoplasmic polκ in HeLa cells, including data from the Human Protein Atlas (44), which used antibodies raised against peptide antigens. One important difference in our studies is that we used a monoclonal antibody raised against the full-length protein. Examination of polκ transcript variants in Ensembl (97) reveals that humans transcribe at least two versions of polκ (polκ-201 and polκ-216), such that antibodies raised against short peptides versus full-length protein may recognize different transcripts with different localizations. The exact reasons for this discrepancy will await further studies of polκ structure regarding posttranslational modifications of nuclear localization or export signals or variations in transcript abundance in different cell types. Notably, another Y-family polymerase polη has also been reported to have nuclear localization when overexpressed as an EGFP fusion protein (98), yet data from the Human Protein Atlas (44) shows a predominantly cytoplasmic localization with nuclear enrichment in some cells, similar to what we saw with polκ.
Our data indicates that mTOR and nutrient sensing regulate the localization of polκ, and this pathway more broadly is known to affect nuclear-cytoplasmic shuttling of multiple proteins (99–106). For example, PI3K-AKT-mTOR pathway signaling has been shown to prevent nuclear accumulation of glycogen synthase kinase 3β (GSK3β) such that inhibition of this pathway leads to a robust increase in nuclear GSK3β (104, 105). Furthermore, mTOR complex 1 (mTORC1)-dependent phosphorylation of the transcription factor TFEB promotes its association with members of the 14–3-3 family of proteins, thereby forcing its retention in the cytosol (103). However, the network connecting the mTOR pathway to the DNA damage response continues to be elucidated (107–110). One study (110) showed that mTOR-S6K signaling leads to phosphorylation and subsequent degradation of E3 ubiquitin ligase RNF168; hyperactivation of mTOR by the kinase LKB1 leads to decreased levels of RNF168 and increases DNA damage. One possible explanation for our observed results is that mTOR (or one of its downstream targets) could directly phosphorylate polκ and thereby affect its localization as has been shown for other proteins that undergo nuclear-cytoplasmic shuttling (111–114). An important area for future exploration is understanding the ways in which mTOR may interact with the maintenance of genome stability.
Materials and Methods
Cell culture
Cell culture was performed at 37°C in a humidified atmosphere containing 5% CO2. A375 and SK-MEL28 cells were obtained from ATCC. PC-9 cells were a gift from C. Rudin (MSKCC, NYC, USA). SK-BR3 cells were a gift from S. Chandarlapaty (MSKCC, NYC, USA). Cells were cultured in DMEM (A375 and SK-MEL28), RPMI1640 (PC-9), or DMEM/F12 (SK-BR3) supplemented with 2 mM glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated fetal bovine serum. Cell lines were regularly tested and verified to be mycoplasma negative by the MycoAlert™ Mycoplasma Detection Kit (Lonza).
Cell treatments
Inhibitors were maintained until collection unless otherwise noted: PLX4032 (Selleck), CI-1040 (Selleck), U0126 (Sigma Aldrich), Ulixertinib (Selleck), SCH772984 (Selleck), erlotinib (Selleck), rapamycin (Selleck), PP242 (Abcam), LY294002 (Sigma Aldrich), bleomycin (Sigma Aldrich), brefeldin A (Sigma Aldrich), thapsigargin (Sigma Aldrich), tunicamycin (Sigma Aldrich), importazole (Sigma Aldrich), leptomycin B (Sigma Aldrich), and MG132 (Sigma Aldrich). Lapatinib and PD0332991 were gifts from S. Chandarlapaty. Complete media consisted of DMEM (without glucose or glutamine) supplemented with 25 mM glucose, 6 mM glutamine, and 10% heat-inactivated fetal bovine serum. Media – glucose consisted of DMEM (without glucose or glutamine) supplemented with 6 mM glutamine and 10% heat-inactivated fetal bovine serum. Media – glutamine consisted of DMEM (without glucose or glutamine) supplemented with 25 mM glucose and 10% heat-inactivated fetal bovine serum. Media + 1% serum consisted of DMEM (without glucose or glutamine) supplemented with 25 mM glucose, 6 mM glutamine, and 1% heat-inactivated fetal bovine serum.
Generation of inducible polκ overexpression cells
For inducible overexpression of polκ, the human polκ open reading frame was amplified from a constitutive polκ overexpression plasmid that was a gift from J.S. Hoffmann (Toulouse, France) and cloned using the In-Fusion Cloning kit (Clontech) into the pSIN-TREtight-MCS-IRES-mCherry-PGK-Hygro vector, which has a doxycycline-inducible promoter and adds an IRES-mCherry to the C-terminus of polκ, to generate TetRE-POLK-IRES-mCherry. The pSIN-TREtight-MCS-IRES-mCherry-PGK-Hygro vector and its corresponding tet-activator (rtTA3) RIEP vector were gifts from S. Lowe (MSKCC, NYC, USA). The following primers were used for amplification:
TRE-hpolk F1 FW: CGGTACCCGGGGATCCCACCATGGATAGCACAAAG
TRE-hpolk F1 RV: TTAGTCTTCGCGGCCGCTTACTTAAAAAATATATCAAGGG
Lentivirus was produced by transfection of HEK-293T cells with TetRE-POLK-IRES-mCherry and the packaging plasmid psiAmpho (which was a gift from R. Levine, MSKCC, NYC, USA) at a 1:1 ratio. Transfection was performed using Fugene (Promega) reagent. The viral supernatant was collected 48 and 72 hours following transfection, filtered through a 0.45 μm filter (Thermo Fisher Scientific), and added to A375 cells with 8 μg/ml polybrene (Thermo Fisher Scientific). Infected cells were selected by treatment with 1 mg/ml hygromycin (Thermo Fisher Scientific). Lentivirus for RIEP (TetA) was produced the same way and then used to infect A375 cells already containing TetRE-POLK-IRES-mCherry. Doubly infected cells (TetRE-POLK-IRES-mCherry/TetA) were selected by treatment with 10 μg/ml puromycin (Thermo Fisher Scientific) and 1 mg/ml hygromycin. Single cell clones were then isolated, and their induction efficiency was tested using flow cytometry. Clones #1 and #2 showed the best induction after addition of 1 μg/ml doxycycline (Sigma Aldrich) compared to uninduced controls, so they were selected for future experiments.
To add constitutive GFP expression to these cells, we used pBABE-GFP, which was a gift from William Hahn (Addgene plasmid #10668). pBABE-GFP (SV40p-GFP) expresses humanized Renilla GFP (hrGFP) under the SV40 promoter in the pBABE backbone. Lentivirus for pBABE-GFP was produced in HEK-293T cells using the same protocol detailed above and then used to infect A375 cells already containing TetRE-POLK-IRES-mCherry/TetA (dox-inducible polκ overexpression cells). Infected cells were selected using flow cytometry for GFP-positive cells.
Inducible shRNA knockdown of polκ and p53
For inducible knockdown of polκ, two different shRNAs (115) targeting polκ were generated using PCR. The two polκ shRNAs (sh-polκ-1 and sh-polκ-2) were each cloned using restriction digestion and ligation into the LT3GEPIR vector (116), which allows doxycycline-inducible expression of a shRNA and GFP. The following primers were used for amplification:
miRE_XhoF: TACAATACTCGAGAAGGTATATTGCTGTTGACAGTGAGCG
miRE_EcoRI: TTAGATGAATTCTAGCCCCTTGAAGTCCGAGGCAGTAGGCA
The following oligos were used as the templates for amplification:
sh-polκ-1: TGCTGTTGACAGTGAGCGCAAGAAGAGTTTCTTTGATAAATAGTGAAGC
CACAGATGTATTTATCAAAGAAACTCTTCTTATGCCTACTGCCTCGGA
sh-polκ-2: TGCTGTTGACAGTGAGCGCAAGGATTTATGTAGTTGAATATAGTGAAGC
CACAGATGTATATTCAACTACATAAATCCTTATGCCTACTGCCTCGGA
Plasmids containing either a control hairpin (sh-Ctrl) or 2 different hairpins targeting p53 (sh-p53–1 and sh-p53–2) were gifts from S. Lowe. For all hairpins, lentivirus was produced by transfection of HEK-293T cells with each hairpin plasmid with the packaging plasmids psPAX2 and pMD2G at a 4:3:1 ratio. Transfection was performed using Lipofectamine 2000 (Life Technologies) reagent. The viral supernatant was collected at 48 and 72 hours following transfection and frozen at −80°C. Virus was later thawed and added to A375 cells, and infected cells were selected by treatment with 1 μg/ml puromycin. Induction after treatment with 1 μg/ml doxycycline was verified using flow cytometry.
Total RNA extraction, cDNA isolation and qRT-PCR analysis
Total RNA from treated cells was extracted using the Quick-RNA Mini-Prep kit (Zymo), and RNA concentration was determined using a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek). Reverse transcription of total RNA was performed using SuperScript III First-Strand Synthesis SuperMix (Thermo Fisher Scientific) according to the manufacturer’s guidelines. qRT-PCR reactions were detected on a CFX384 Touch machine (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). RNA expression levels were calculated using the comparative Ct method (2-ΔΔCT) normalized to β-actin. The qRT-PCR primer sets used in this study are listed in table S1.
Western blot analysis
For whole cell lysates, cells were washed 1x with phosphate buffered saline (PBS, Invitrogen) and then lysed in RIPA buffer (EMD Millipore) containing 1x HALT Combined Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific) for 20 minutes at 4°C. Cell lysates were clarified by centrifugation at 10,000 RPM for 10 minutes at 4°C. For cytoplasmic or nuclear fractions, the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Fisher Scientific) was used according to the manufacturer’s protocol. Protein concentration was measured with Bradford reagent (Sigma Aldrich), and samples were resolved by 4–15% or 12% SDS-PAGE gels (Bio-Rad) for polκ or p53, respectively. Proteins were transferred onto nitrocellulose membranes and subjected to standard immunoblotting. ECL Prime (Amersham) was used as the developing reagent.
Western blot antibodies:
polκ (ab57070, Abcam, 1:10,000), p53 (sc-126, Santa Cruz Biotechnology, 1:10,000), mcherry (ab167453, Abcam, 1:1000), lamin-B1 (ab133741, Abcam, 1:10,000), β-Actin (A5441, Sigma Aldrich, 1:20,000), anti-mouse (ab97046, Abcam, 1:10,000), anti-rabbit (ab97051, Abcam, 1:10,000)
Immunofluorescence
Cells were cultured on Millicell EZ SLIDE 4 or 8-well glass slides (EMD Millipore). Cells were fixed with 4% paraformaldehyde (Santa Cruz Biotechnology) at 37°C for 15 minutes, washed with PBS, and then blocked with 5% goat serum (Thermo Fisher Scientific) and 0.2% Triton X-100 (Thermo Fisher Scientific) in PBS for 1 hour. The cells were incubated with primary antibody in antibody dilution buffer (PBS with 1% bovine serum albumin (Sigma Aldrich) and 0.2% Triton X-100) at 4°C overnight, then washed 3 times with PBS, and incubated with Alexa-conjugated secondary antibody for 2 hours at RT. After washing 3 more times with PBS, cells were stained with 0.1 μg/ml DAPI (Thermo Fisher Scientific) and then mounted with Dako fluorescence mounting media (Agilent) and imaged on a Zeiss Axio Imager A2.
Immunofluorescence antibodies:
polκ (ab57070, Abcam, 1:500), p-S6 (5364, Cell Signaling, 1:1000), γH2AX (2577, Cell Signaling, 1:200), 53BP1 (ab175933, Abcam, 1:200), p-Chk1 (2348, Cell Signaling, 1:100), p-Chk2 (2661, Cell signaling, 1:500), anti-mouse Alexa-488 (4408, Cell signaling, 1:1000), anti-rabbit Alexa-488 (4412, Cell Signaling, 1:1000), anti-rabbit Alexa-594 (8889, Cell Signaling, 1:1000)
Quantification of nuclear vs. cytoplasmic enrichment of polκ
To quantify nuclear versus cytoplasmic enrichment, we created a nuclear mask from the DAPI image of each cell, which allowed us to quantify the number of cells per field. This mask was then overlaid onto the staining with polκ, and each cell was manually scored as to whether the staining was exclusively nuclear (i.e. perfectly overlapped with the DAPI mask) or not. The calculation of “% of cells with nuclear enrichment of polκ” was then calculated as the number of cells with strictly nuclear staining divided by the total number of cells. Because we found that this method correlated with our cell fractionation Western blots but was more scalable across many conditions and also allowed for additional stainings (i.e. phospho-S6), this method was applied to each figure.
Cell cycle analysis
Cells were treated with the indicated inhibitors for 24 hours and fixed in 70% ethanol (Thermo Fisher Scientific) at 4°C for at least 4 hours. Later, the cells were washed with PBS and stained with 20 μg of propidium iodide (Thermo Fisher Scientific), 200 μg of RNase A (Thermo Fisher Scientific), and 0.1% Triton X-100 in PBS for at least 30 minutes. The labeled cells were analyzed using a Fortessa flow cytometer, and the Dean-Jett Fox model in FlowJo was used to determine which cells were in G1, S, and G2/M.
Knockout of MMR genes and polκ using the Alt-R CRISPR-Cas9 system
Two sgRNAs for each MMR gene or polκ were designed using the CHOPCHOP online tool (117). Knockout was performed in A375 cells using the Alt-R CRISPR-Cas9 system (IDT), which utilizes sgRNAs against the gene of interest and labeled tracrRNAs to guide Cas9 protein to a specific genomic location. Proper cutting by Cas9 was verified using the Surveyor Mutation Detection Kit (IDT) after amplification of the targeted area using the associated forward and reverse primers. The sgRNA and validation primer sequences used in this study are listed in Table S2.
Drug resistance assays
Two different clones of doxycycline-inducible polκ overexpression cells (Clone #1 and Clone #2) were generated. We then divided each population equally and treated with or without 1 μg/ml doxycycline for ~3 months to generate polκ-overexpressing cells and their corresponding negative control population. All populations were then switched to media without doxycycline, plated in 96 well plates, and exposed to various doses of PLX4032 or PD0332991. After 4 (PLX4032) or 7 days (PD0332991), cell number was determined using the CyQUANT Direct Cell Proliferation Assay kit (Thermo Fisher Scientific). The technical replicates for each dose were averaged and then normalized against the 0 μM dose condition to determine the growth relative to DMSO. The PLX4032 resistance assay detailed above was also repeated for control and polκ KO (2 different sgRNAs) cells.
Mutagenesis assays
Dox-inducible polκ overexpression A375 cells (Clone #1 and Clone #2) constitutively expressing GFP under the SV40 promoter with control or MMR KO were generated. We then divided each population equally and treated with or without 1 μg/ml dox to generate polκ-overexpressing cells and their corresponding negative control population. After 14 days of treatment, both populations were tryspinized, stained with 1 μg/ml DAPI, and analyzed using a Fortessa flow cytometer. FlowJo was used to determine the percentage of GFP-negative and GFP-positive cells in each population.
RNA sequencing and analysis
Total RNA was extracted as described above. Purified RNA was delivered to GENEWIZ (South Plainfield, NJ) for mRNA preparation with the TruSeq RNA V2 kit (Illumina) and 150bp paired-end sequencing on the Illumina HiSeq2500. After quality control with FASTQC (Babraham Bioinformatics) and trimming with TRIMMOMATIC (118), reads were aligned to GRCh38 (Ensembl version 90) using STAR (119), with quality control via SeQC (120). Differential expression was calculated with DESeq2 (121) using the output of the --quantMode GeneCounts feature of STAR. Z-scores of log2 transformed normalized counts from the rlog function were used for heatmaps. Pathway analysis was performed with GSAA (122, 123) using the following parameters: gsametric Weighted_KS, demetric Signal2Noise, permute gene_set, rnd_type no_balance, scoring_scheme weighted, norm MeanDiv.
Statistical tests
The following statistical tests were used: paired 2-tailed t-test (all qRT-PCR, Western blot analysis, and immunofluorescence), chi-square tests (cell cycle analysis), Pearson correlation coefficient (comparison of rates of p-S6 loss vs. nuclear polκ enrichment), and two-way ANOVA and Fisher’s least significant difference (LSD) test (drug resistance assays).
Supplementary Material
Fig. S1. Validation of the Polκ antibody.
Fig. S2. MAPK inhibition in melanoma cell lines increases POLK mRNA levels and changes the subcellular localization of Polκ.
Fig. S3. DNA damage is not associated with the subcellular shift of Polκ.
Fig. S4. Cell cycle inhibition is not responsible for the shift in the subcellular localization of Polκ.
Fig. S5. MAPK inhibition decreases the abundance of phosphorylated S6.
Fig. S6. BRAF inhibition induces ER stress, and other inducers of ER stress also change the subcellular localization of Polκ.
Fig. S7. The effects on POLK mRNA levels are driver gene-specific across tumor types.
Fig. S8. Glucose starvation also modulates the subcellular localization of Polκ.
Fig. S9. Inhibiting importin-β does not prevent the nuclear shift of Polκ.
Fig. S10. Inhibition of the proteosome does not have a differential effect on the protein level of Polκ after treatment with DMSO or PLX4032.
Table S1. qRT-PCR primers used in the study.
Table S2. sgRNAs and validation primers used for MMR and Polκ knockout.
Table S3. RNA-seq and GSEA analysis on Polκ-CRISPR A375 cells.
Acknowledgements:
We thank S. Chandarlapaty (MSKCC, NYC) for providing lapatinib and PD0332991; J.S. Hoffmann (Toulouse, France) for the constitutive polκ overexpression plasmid; S. Lowe (MSKCC, NYC) for the pSIN-TREtight-MCS-IRES-mCherry-PGK-Hygro vector, its corresponding tet-activator (rtTA3) RIEP vector, and plasmids containing either a control hairpin (sh-Ctrl) or 2 different hairpins targeting p53 (sh-p53–1 and sh-p53–2); and R. Levine (MSKCC, NYC) for the packaging plasmid psiAmpho. We also thank Wenjing Wu for help with the figures.
Funding: This work was supported by the Ruth L. Kirschstein Predoctoral Individual (F31) National Research Service Award (5F31CA200341–03) (K.T.), the Ruth L. Kirschstein M.D/Ph.D. Predoctoral Individual (F30) National Research Service Award (F30CA220954) (N.R.C), a Medical Scientist Training Program Grant (T32GM007739) (N.R.C), the NIH Director’s New Innovator Award (DP2CA186572), Mentored Clinical Scientist Research Career Development Award (K08AR055368), the Melanoma Research Alliance, The Pershing Square Sohn Foundation, The Alan and Sandra Gerry Metastasis Research Initiative at the Memorial Sloan Kettering Cancer Center, and The Harry J. Lloyd Foundation and Consano (all to R.M.W.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The RNA-seq data have been deposited to the Gene Expression Omnibus (GEO), accession ID GSE145313. This and all other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. All materials, including plasmids used in this study, will be supplied upon request.
References and Notes:
- 1.Palles C, Cazier J-B, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Salguero I, Sherborne A, Chubb D, Carvajal-Carmona LG, Ma Y, Kaur K, Dobbins S, Barclay E, Gorman M, Martin L, Kovac MB, Tomlinson I, Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet 45, 136–144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Church DN, Briggs SEW, Palles C, Domingo E, Kearsey SJ, Grimes JM, Gorman M, Martin L, Howarth KM, Hodgson SV, NSECG Collaborators, Kaur K, Taylor J, Tomlinson IPM, DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum. Mol. Genet 22, 2820–2828 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erson-Omay EZ, Çağlayan AO, Schultz N, Weinhold N, Omay SB, Özduman K, Köksal Y, Li J, Serin Harmancı A, Clark V, Carrión-Grant G, Baranoski J, Çağlar C, Barak T, Coşkun S, Baran B, Köse D, Sun J, Bakırcıoğlu M, Moliterno Günel J, Günel M, Somatic POLE mutations cause an ultramutated giant cell high-grade glioma subtype with better prognosis. Neuro Oncol 17, 1356–1364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons JL, Nicolay NH, Sharma RA, Biological and therapeutic relevance of nonreplicative DNA polymerases to cancer. Antioxid. Redox Signal 18, 851–873 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F, The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 399, 700–704 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Sun H, Zou S, Zhang S, Liu B, Meng X, Li X, Yu J, Wu J, Zhou J, Elevated DNA polymerase iota (Poli) is involved in the acquisition of aggressive phenotypes of human esophageal squamous cell cancer. Int. J. Clin. Exp. Pathol 8, 3591–3601 (2015). [PMC free article] [PubMed] [Google Scholar]
- 7.Russo M, Crisafulli G, Sogari A, Reilly NM, Arena S, Lamba S, Bartolini A, Amodio V, Magrì A, Novara L, Sarotto I, Nagel ZD, Piett CG, Amatu A, Sartore-Bianchi A, Siena S, Bertotti A, Trusolino L, Corigliano M, Gherardi M, Bardelli A, Adaptive mutability of colorectal cancers in response to targeted therapies. Science. 366, 1473–1480 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R, The Y-family of DNA polymerases. Mol. Cell. 8, 7–8 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Sale JE, Lehmann AR, Woodgate R, Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol 13, 141–152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O-Wang J, Kawamura K, Tada Y, Ohmori H, Kimura H, Sakiyama S, Tagawa M, DNA polymerase kappa, implicated in spontaneous and DNA damage-induced mutagenesis, is overexpressed in lung cancer. Cancer Res 61, 5366–5369 (2001). [PubMed] [Google Scholar]
- 11.Wang H, Wu W, Wang H-W, Wang S, Chen Y, Zhang X, Yang J, Zhao S, Ding H-F, Lu D, Analysis of specialized DNA polymerases expression in human gliomas: association with prognostic significance. Neuro Oncol 12, 679–686 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bavoux C, Leopoldino AM, Bergoglio V, O-Wang J, Ogi T, Bieth A, Judde J-G, Pena SDJ, Poupon M-F, Helleday T, Tagawa M, Machado C, Hoffmann J-S, Cazaux C, Up-regulation of the error-prone DNA polymerase {kappa} promotes pleiotropic genetic alterations and tumorigenesis. Cancer Res 65, 325–330 (2005). [PubMed] [Google Scholar]
- 13.Peng C, Chen Z, Wang S, Wang H-W, Qiu W, Zhao L, Xu R, Luo H, Chen Y, Chen D, You Y, Liu N, Wang H, The Error-Prone DNA Polymerase κ Promotes Temozolomide Resistance in Glioblastoma through Rad17-Dependent Activation of ATR-Chk1 Signaling. Cancer Res 76, 2340–2353 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z, Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res 28, 4138–4146 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC, Human DNA polymerase kappa bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J. Biol. Chem 277, 37604–37611 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H, Error-prone bypass of certain DNA lesions by the human DNA polymerase kappa. Genes Dev 14, 1589–1594 (2000). [PMC free article] [PubMed] [Google Scholar]
- 17.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA, Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J. Biol. Chem 275, 39678–39684 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Yuan F, Xin H, Wu X, Rajpal DK, Yang D, Wang Z, Human DNA polymerase kappa synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res 28, 4147–4156 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinet A, Vessoni AT, Rocha CRR, Gottifredi V, Biard D, Sarasin A, Menck CFM, Stary A, Gap-filling and bypass at the replication fork are both active mechanisms for tolerance of low-dose ultraviolet-induced DNA damage in the human genome. DNA Repair (Amst) 14, 27–38 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Kanemaru Y, Suzuki T, Niimi N, Grúz P, Matsumoto K, Adachi N, Honma M, Nohmi T, Catalytic and non-catalytic roles of DNA polymerase κ in the protection of human cells against genotoxic stresses. Environ. Mol. Mutagen 56, 650–662 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Galhardo RS, Hastings PJ, Rosenberg SM, Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol 42, 399–435 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponder RG, Fonville NC, Rosenberg SM, A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell. 19, 791–804 (2005). [DOI] [PubMed] [Google Scholar]
- 23.McKenzie GJ, Harris RS, Lee PL, Rosenberg SM, The SOS response regulates adaptive mutation. Proc Natl Acad Sci USA 97, 6646–6651 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzgerald DM, Hastings PJ, Rosenberg SM, Stress-Induced Mutagenesis: Implications in Cancer and Drug Resistance. Annu. Rev. Cancer Biol 1, 119–140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosche WA, Foster PL, The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc Natl Acad Sci USA 96, 6862–6867 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM, SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell. 7, 571–579 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Pennington JM, Rosenberg SM, Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet 39, 797–802 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shee C, Cox BD, Gu F, Luengas EM, Joshi MC, Chiu L-Y, Magnan D, Halliday JA, Frisch RL, Gibson JL, Nehring RB, Do HG, Hernandez M, Li L, Herman C, Hastings PJ, Bates D, Harris RS, Miller KM, Rosenberg SM, Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. elife 2, e01222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galhardo RS, Do R, Yamada M, Friedberg EC, Hastings PJ, Nohmi T, Rosenberg SM, DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics. 182, 55–68 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris RS, Feng G, Ross KJ, Sidhu R, Thulin C, Longerich S, Szigety SK, Winkler ME, Rosenberg SM, Mismatch repair protein MutL becomes limiting during stationary-phase mutation. Genes Dev 11, 2426–2437 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsui HC, Feng G, Winkler ME, Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol 179, 7476–7487 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerlach VL, Aravind L, Gotway G, Schultz RA, Koonin EV, Friedberg EC, Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc Natl Acad Sci USA 96, 11922–11927 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velasco-Miguel S, Richardson JA, Gerlach VL, Lai WC, Gao T, Russell LD, Hladik CL, White CL, Friedberg EC, Constitutive and regulated expression of the mouse Dinb (Polkappa) gene encoding DNA polymerase kappa. DNA Repair (Amst) 2, 91–106 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Bergoglio V, Bavoux C, Verbiest V, Hoffmann J-S, Cazaux C, Localisation of human DNA polymerase kappa to replication foci. J. Cell Sci 115, 4413–4418 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Ogi T, Kannouche P, Lehmann AR, Localisation of human Y-family DNA polymerase kappa: relationship to PCNA foci. J. Cell Sci 118, 129–136 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Lemée F, Bavoux C, Pillaire MJ, Bieth A, Machado CR, Pena SD, Guimbaud R, Selves J, Hoffmann JS, Cazaux C, Characterization of promoter regulatory elements involved in downexpression of the DNA polymerase kappa in colorectal cancer. Oncogene 26, 3387–3394 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Zhu H, Fan Y, Shen J, Qi H, Shao J, Characterization of human DNA polymerase κ promoter in response to benzo[a]pyrene diol epoxide. Environ. Toxicol. Pharmacol 33, 205–211 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Laplante M, Sabatini DM, mTOR signaling in growth control and disease. Cell. 149, 274–293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O’Day SJ, Sosman JA, Kirkwood JM, Eggermont AMM, BRIM-3 Study Group, Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med 364, 2507–2516 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, Demidov LV, Hassel JC, Rutkowski P, Mohr P, Dummer R, Trefzer U, Larkin JMG, Utikal J, Dreno B, Nyakas M, Middleton MR, Becker JC, Casey M, Sherman LJ, METRIC Study Group, Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med 367, 107–114 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Germann UA, Furey BF, Markland W, Hoover RR, Aronov AM, Roix JJ, Hale M, Boucher DM, Sorrell DA, Martinez-Botella G, Fitzgibbon M, Shapiro P, Wick MJ, Samadani R, Meshaw K, Groover A, DeCrescenzo G, Namchuk M, Emery CM, Saha S, Welsch DJ, Targeting the MAPK Signaling Pathway in Cancer: Promising Preclinical Activity with the Novel Selective ERK1/2 Inhibitor BVD-523 (Ulixertinib). Mol. Cancer Ther 16, 2351–2363 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Beck D, Niessner H, Smalley KSM, Flaherty K, Paraiso KHT, Busch C, Sinnberg T, Vasseur S, Iovanna JL, Drießen S, Stork B, Wesselborg S, Schaller M, Biedermann T, Bauer J, Lasithiotakis K, Weide B, Eberle J, Schittek B, Schadendorf D, Meier F, Vemurafenib potently induces endoplasmic reticulum stress-mediated apoptosis in BRAFV600E melanoma cells. Sci. Signal 6, ra7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haferkamp S, Borst A, Adam C, Becker TM, Motschenbacher S, Windhövel S, Hufnagel AL, Houben R, Meierjohann S, Vemurafenib induces senescence features in melanoma cells. J. Invest. Dermatol 133, 1601–1609 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Uhlén M, Björling E, Agaton C, Szigyarto CA-K, Amini B, Andersen E, Andersson A-C, Angelidou P, Asplund A, Asplund C, Berglund L, Bergström K, Brumer H, Cerjan D, Ekström M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Pontén F, A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics 4, 1920–1932 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Bollum FJ, Potter VR, Incorporation of thymidine into deoxyribonucleic acid by enzymes from rat tissues. J. Biol. Chem 233, 478–482 (1958). [PubMed] [Google Scholar]
- 46.Prescott DM, Bollum FJ, Kluss BC, Is DNA polymerase a cytoplasmic enzyme? J. Cell Biol 13, 172–174 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Littlefield JW, McGovern AP, Margeson KB, Changes in the distribution of polymerase activity during DNA synthesis in mouse fibroblasts. Proc Natl Acad Sci USA 49, 102–107 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harper JW, Elledge SJ, The DNA damage response: ten years after. Mol. Cell. 28, 739–745 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Lakin ND, Jackson SP, Regulation of p53 in response to DNA damage. Oncogene 18, 7644–7655 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, Los G, Slamon DJ, PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res 11, R77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendoza MC, Er EE, Blenis J, The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem. Sci 36, 320–328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J, Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 370, 527–532 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J, Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci USA 101, 13489–13494 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carriere A, Romeo Y, Acosta-Jaquez HA, Moreau J, Bonneil E, Thibault P, Fingar DC, Roux PP, ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1). J. Biol. Chem 286, 567–577 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oslowski CM, Urano F, Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Meth. Enzymol 490, 71–92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin L, Wang Z, Tao L, Wang Y, ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 6, 239–247 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM, RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA 95, 1432–1437 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, van Kooten M, Dediu M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabárbara P, Seymour L, National Cancer Institute of Canada Clinical Trials Group, Erlotinib in previously treated non-small-cell lung cancer. N. Engl. J. Med 353, 123–132 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Kaufman B, Trudeau M, Awada A, Blackwell K, Bachelot T, Salazar V, DeSilvio M, Westlund R, Zaks T, Spector N, Johnston S, Lapatinib monotherapy in patients with HER2-overexpressing relapsed or refractory inflammatory breast cancer: final results and survival of the expanded HER2+ cohort in EGF103009, a phase II study. Lancet Oncol 10, 581–588 (2009). [DOI] [PubMed] [Google Scholar]
- 60.NES Finder 0.2, (available at http://research.nki.nl/fornerodlab/NES-Finder.htm).
- 61.Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M, Weis K, Heald R, Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem. Biol 6, 700–708 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T, Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem 269, 6320–6324 (1994). [PubMed] [Google Scholar]
- 63.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL, Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 78, 761–771 (1994). [DOI] [PubMed] [Google Scholar]
- 64.Shor E, Schuyler J, Perlin DS, A Novel, Drug Resistance-Independent, Fluorescence-Based Approach To Measure Mutation Rates in Microbial Pathogens. MBio 10 (2019), doi: 10.1128/mBio.00120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rambow F, Rogiers A, Marin-Bejar O, Aibar S, Femel J, Dewaele M, Karras P, Brown D, Chang YH, Debiec-Rychter M, Adriaens C, Radaelli E, Wolter P, Bechter O, Dummer R, Levesque M, Piris A, Frederick DT, Boland G, Flaherty KT, Marine J-C, Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell. 174, 843–855.e19 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ, DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol 2, 537–541 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA, Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat. Immunol 2, 530–536 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Faili A, Aoufouchi S, Weller S, Vuillier F, Stary A, Sarasin A, Reynaud C-A, Weill J-C, DNA polymerase eta is involved in hypermutation occurring during immunoglobulin class switch recombination. J. Exp. Med 199, 265–270 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Faili A, Aoufouchi S, Flatter E, Guéranger Q, Reynaud C-A, Weill J-C, Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature. 419, 944–947 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 368, 258–261 (1994). [DOI] [PubMed] [Google Scholar]
- 71.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, Mutation of a mutL homolog in hereditary colon cancer. Science. 263, 1625–1629 (1994). [DOI] [PubMed] [Google Scholar]
- 72.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R, The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 75, 1027–1038 (1993). [DOI] [PubMed] [Google Scholar]
- 73.Miyaki M, Konishi M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Yasuno M, Igari T, Koike M, Chiba M, Mori T, Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat. Genet 17, 271–272 (1997). [DOI] [PubMed] [Google Scholar]
- 74.Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 371, 75–80 (1994). [DOI] [PubMed] [Google Scholar]
- 75.Lynch HT, de la Chapelle A, Genetic susceptibility to non-polyposis colorectal cancer. J. Med. Genet 36, 801–818 (1999). [PMC free article] [PubMed] [Google Scholar]
- 76.Mecklin JP, Järvinen HJ, Tumor spectrum in cancer family syndrome (hereditary nonpolyposis colorectal cancer). Cancer 68, 1109–1112 (1991). [DOI] [PubMed] [Google Scholar]
- 77.Bertolin AP, Mansilla SF, Gottifredi V, The identification of translesion DNA synthesis regulators: Inhibitors in the spotlight. DNA Repair (Amst) 32, 158–164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kunkel TA, DNA replication fidelity. J. Biol. Chem 279, 16895–16898 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Hoffmann J-S, Cazaux C, Aberrant expression of alternative DNA polymerases: a source of mutator phenotype as well as replicative stress in cancer. Semin. Cancer Biol 20, 312–319 (2010). [DOI] [PubMed] [Google Scholar]
- 80.Yoshida R, Miyashita K, Inoue M, Shimamoto A, Yan Z, Egashira A, Oki E, Kakeji Y, Oda S, Maehara Y, Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur. J. Hum. Genet 19, 320–325 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hodel KP, de Borja R, Henninger EE, Campbell BB, Ungerleider N, Light N, Wu T, LeCompte KG, Goksenin AY, Bunnell BA, Tabori U, Shlien A, Pursell ZF, Explosive mutation accumulation triggered by heterozygous human Pol ε proofreading-deficiency is driven by suppression of mismatch repair. elife 7 (2018), doi: 10.7554/eLife.32692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ogi T, Kato T, Kato T, Ohmori H, Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein dinB. Genes Cells. 4, 607–618 (1999). [DOI] [PubMed] [Google Scholar]
- 83.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, Kiezun A, Hammerman PS, McKenna A, Drier Y, Zou L, Ramos AH, Pugh TJ, Stransky N, Helman E, Kim J, Getz G, Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB, Kelley MC, Kefford RF, Chmielowski B, Glaspy JA, Sosman JA, van Baren N, Long GV, Ribas A, Lo RS, Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 4, 80–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lo RS, Shi H, Detecting mechanisms of acquired BRAF inhibitor resistance in melanoma. Methods Mol. Biol 1102, 163–174 (2014). [DOI] [PubMed] [Google Scholar]
- 86.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee M-K, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS, Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 468, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, Hatton C, Chopra R, Oberholzer PA, Karpova MB, MacConaill LE, Zhang J, Gray NS, Sellers WR, Dummer R, Garraway LA, MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci USA 106, 20411–20416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, Place CS, Taylor-Weiner A, Whittaker S, Kryukov GV, Hodis E, Rosenberg M, McKenna A, Cibulskis K, Farlow D, Zimmer L, Hillen U, Gutzmer R, Goldinger SM, Ugurel S, Dermatologic Cooperative Oncology Group of Germany (DeCOG), The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 4, 94–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goetz EM, Ghandi M, Treacy DJ, Wagle N, Garraway LA, ERK mutations confer resistance to mitogen-activated protein kinase pathway inhibitors. Cancer Res 74, 7079–7089 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson DB, Menzies AM, Zimmer L, Eroglu Z, Ye F, Zhao S, Rizos H, Sucker A, Scolyer RA, Gutzmer R, Gogas H, Kefford RF, Thompson JF, Becker JC, Berking C, Egberts F, Loquai C, Goldinger SM, Pupo GM, Hugo W, Schadendorf D, Acquired BRAF inhibitor resistance: A multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur. J. Cancer. 51, 2792–2799 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, Caponigro G, Hieronymus H, Murray RR, Salehi-Ashtiani K, Hill DE, Vidal M, Zhao JJ, Yang X, Alkan O, Kim S, Garraway LA, COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 468, 968–972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, Salton M, Dahlman KB, Tadi M, Wargo JA, Flaherty KT, Kelley MC, Misteli T, Chapman PB, Sosman JA, Graeber TG, Solit DB, RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 480, 387–390 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, Rosenberg M, Goetz EM, Sullivan RJ, Farlow DN, Friedrich DC, Anderka K, Perrin D, Johannessen CM, McKenna A, Cibulskis K, Kryukov G, Hodis E, Lawrence DP, Fisher S, Garraway LA, MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov 4, 61–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Corazao-Rozas P, Guerreschi P, Jendoubi M, André F, Jonneaux A, Scalbert C, Garçon G, Malet-Martino M, Balayssac S, Rocchi S, Savina A, Formstecher P, Mortier L, Kluza J, Marchetti P, Mitochondrial oxidative stress is the Achille’s heel of melanoma cells resistant to Braf-mutant inhibitor. Oncotarget 4, 1986–1998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mizushina Y, Motoshima H, Yamaguchi Y, Takeuchi T, Hirano K, Sugawara F, Yoshida H, 3-O-methylfunicone, a selective inhibitor of mammalian Y-family DNA polymerases from an Australian sea salt fungal strain. Mar. Drugs 7, 624–639 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yamanaka K, Dorjsuren D, Eoff RL, Egli M, Maloney DJ, Jadhav A, Simeonov A, Lloyd RS, A comprehensive strategy to discover inhibitors of the translesion synthesis DNA polymerase κ. PLoS ONE 7, e45032 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR, Flicek P, Ensembl 2018. Nucleic Acids Res 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR, Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells. Genes Dev 15, 158–172 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beck T, Hall MN, The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 402, 689–692 (1999). [DOI] [PubMed] [Google Scholar]
- 100.Kazyken D, Kaz Y, Kiyan V, Zhylkibayev AA, Chen C-H, Agarwal NK, Sarbassov DD, The nuclear import of ribosomal proteins is regulated by mTOR. Oncotarget 5, 9577–9593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Görner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schüller C, Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J 21, 135–144 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chughtai ZS, Rassadi R, Matusiewicz N, Stochaj U, Starvation promotes nuclear accumulation of the hsp70 Ssa4p in yeast cells. J. Biol. Chem 276, 20261–20266 (2001). [DOI] [PubMed] [Google Scholar]
- 103.Martina JA, Chen Y, Gucek M, Puertollano R, MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bautista SJ, Boras I, Vissa A, Mecica N, Yip CM, Kim PK, Antonescu CN, mTORC1 controls glycogen synthase kinase 3β nuclear localization and function. BioRxiv (2018), doi: 10.1101/277657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bechard M, Dalton S, Subcellular localization of glycogen synthase kinase 3beta controls embryonic stem cell self-renewal. Mol. Cell. Biol 29, 2092–2104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van der Vaart B, Fischböck J, Mieck C, Pichler P, Mechtler K, Medema RH, Westermann S, TORC1 signaling exerts spatial control over microtubule dynamics by promoting nuclear export of Stu2. J. Cell Biol 216, 3471–3484 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou X, Liu W, Hu X, Dorrance A, Garzon R, Houghton PJ, Shen C, Regulation of CHK1 by mTOR contributes to the evasion of DNA damage barrier of cancer cells. Sci. Rep 7, 1535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dominick G, Bowman J, Li X, Miller RA, Garcia GG, mTOR regulates the expression of DNA damage response enzymes in long-lived Snell dwarf, GHRKO, and PAPPA-KO mice. Aging Cell. 16, 52–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guo F, Li J, Du W, Zhang S, O’Connor M, Thomas G, Kozma S, Zingarelli B, Pang Q, Zheng Y, mTOR regulates DNA damage response through NF-κB-mediated FANCD2 pathway in hematopoietic cells. Leukemia 27, 2040–2046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xie X, Hu H, Tong X, Li L, Liu X, Chen M, Yuan H, Xie X, Li Q, Zhang Y, Ouyang H, Wei M, Huang J, Liu P, Gan W, Liu Y, Xie A, Kuai X, Chirn G-W, Zhou H, Gao D, The mTOR-S6K pathway links growth signalling to DNA damage response by targeting RNF168. Nat. Cell Biol 20, 320–331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nardozzi JD, Lott K, Cingolani G, Phosphorylation meets nuclear import: a review. Cell Commun. Signal 8, 32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tagawa T, Kuroki T, Vogt PK, Chida K, The cell cycle-dependent nuclear import of v-Jun is regulated by phosphorylation of a serine adjacent to the nuclear localization signal. J. Cell Biol 130, 255–263 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lam MH, House CM, Tiganis T, Mitchelhill KI, Sarcevic B, Cures A, Ramsay R, Kemp BE, Martin TJ, Gillespie MT, Phosphorylation at the cyclin-dependent kinases site (Thr85) of parathyroid hormone-related protein negatively regulates its nuclear localization. J. Biol. Chem 274, 18559–18566 (1999). [DOI] [PubMed] [Google Scholar]
- 114.Beals CR, Clipstone NA, Ho SN, Crabtree GR, Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev 11, 824–834 (1997). [DOI] [PubMed] [Google Scholar]
- 115.Pelossof R, Fairchild L, Huang C-H, Widmer C, Sreedharan VT, Sinha N, Lai D-Y, Guan Y, Premsrirut PK, Tschaharganeh DF, Hoffmann T, Thapar V, Xiang Q, Garippa RJ, Rätsch G, Zuber J, Lowe SW, Leslie CS, Fellmann C, Prediction of potent shRNAs with a sequential classification algorithm. Nat. Biotechnol 35, 350–353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IAM, Kwon JS, Guan Y, Sinha N, Zuber J, An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 5, 1704–1713 (2013). [DOI] [PubMed] [Google Scholar]
- 117.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E, CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42, W401–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire M-D, Williams C, Reich M, Winckler W, Getz G, RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xiong Q, Mukherjee S, Furey TS, GSAASeqSP: a toolset for gene set association analysis of RNA-Seq data. Sci. Rep 4, 6347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xiong Q, Ancona N, Hauser ER, Mukherjee S, Furey TS, Integrating genetic and gene expression evidence into genome-wide association analysis of gene sets. Genome Res 22, 386–397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Validation of the Polκ antibody.
Fig. S2. MAPK inhibition in melanoma cell lines increases POLK mRNA levels and changes the subcellular localization of Polκ.
Fig. S3. DNA damage is not associated with the subcellular shift of Polκ.
Fig. S4. Cell cycle inhibition is not responsible for the shift in the subcellular localization of Polκ.
Fig. S5. MAPK inhibition decreases the abundance of phosphorylated S6.
Fig. S6. BRAF inhibition induces ER stress, and other inducers of ER stress also change the subcellular localization of Polκ.
Fig. S7. The effects on POLK mRNA levels are driver gene-specific across tumor types.
Fig. S8. Glucose starvation also modulates the subcellular localization of Polκ.
Fig. S9. Inhibiting importin-β does not prevent the nuclear shift of Polκ.
Fig. S10. Inhibition of the proteosome does not have a differential effect on the protein level of Polκ after treatment with DMSO or PLX4032.
Table S1. qRT-PCR primers used in the study.
Table S2. sgRNAs and validation primers used for MMR and Polκ knockout.
Table S3. RNA-seq and GSEA analysis on Polκ-CRISPR A375 cells.