Abstract
Nanopore technology holds remarkable promise for sequencing proteins and peptides. To achieve this, it is necessary to establish a characteristic profile for each individual amino acid through the statistical description of their translocation process. However, the subtle molecular differences among all twenty amino acids along with their unpredictable conformational changes at the nanopore sensing region result in very low distinguishability. Here we report the electrical sensing of individual amino acids using an α-hemolysin nanopore based on a derivatization strategy. Using derivatized amino acids as detection surrogates not only prolongs their interactions with the sensing region, but also improves their conformational variation. Furthermore, we show that distinct characteristics including current blockades and dwell time can be observed among all three classes of amino acids after 2,3-naphthalenedicarboxaldehyde (NDA)- and 2-naphthylisothiocyanate (NITC)-derivatization, respectively. These observable characteristics were applied towards the identification and differentiation of 9 of the 20 natural amino acids using their NITC derivatives. The method demonstrated herein will pave the way for the identification of all amino acids and further protein and peptide sequencing.
Keywords: nanopore, derivatization, amino acid, identification, proteomics
Graphical Abstract
A derivatization strategy was demonstrated for reliable identification of individual amino acids using an α-hemolysin nanopore.
The primary structure of proteins and peptides plays a significant role in their structural folding and functions. Very often subtle changes in a protein’s primary sequence can lead to debilitating pathologies.1-5 Traditional methods for proteome analysis and sequencing, such as mass spectrometry6-8 and Edman degradation,9 suffer from high cost, short reads, long turn-around time, and lack of sensitivity, so alternative approaches are sought. Nanopores made of either biological or inorganic materials with orifices of nanometer diameters and depth have been exploited as an exceptionally sensitive too for the analysis of individual biomolecules in real-time without the potential bias associated with signal amplification.10, 11 As a result, significant progress towards DNA and RNA sequencing has been realized through nanopore technology.12-17 Various other applications for nanopores in single molecular sensing have also been demonstrated.18-20
Recently, the focus of efforts has been directed towards amino acid identification and sequencing of proteins and peptides, which holds great promise for the advancement of proteomics.21-23 Research into proteins and peptides nanopore applications has been reported using the ionic current blockade signatures generated by their nanopore translocation.24 Various nanopore methods including ionic current blockade measurement in biological nanopores (i.e. bacteriophage T7 DNA packaging motor,25 FraC nanopores,26, 27 aerolysin,27-29 and α-hemolysin30) and inorganic perpendicular nanochannel,31 as well as recognition by tunneling current32 have been used to identify proteins and peptides. Nonetheless, nanopore sequencing of proteins and peptides still faces formidable open challenges,23 especially the feasibility of distinguishing individual amino acids. Recently, a small group of researchers have focused on nanopore sensing of individual amino acids.33-35 For example, a fingerprinting scheme has been reported in which only a subset of amino acids was labeled and detected.27 In another study, an elegant approach has been developed to detect 13 of 20 proteinogenic amino acids in an aerolysin nanopore with the help of a short peptide tag.36 However, all reported methods are unfeasible for the derivatization and differentiation of amino acids in situ.
Based on the Edman peptide degradation reaction,37,9, 38, 39 herein we demonstrate the efficient N-terminal derivatization of amino acids using aromatic tags can augment the distinguishability of different amino acids when they translocate through α-hemolysin (α-HL) nanopores. A panel of nine amino acids, including non-polar, polar and charged ones, could be discriminated individually. We envision this method can potentially be employed in the future development of nanopore sequencing of a protein or peptide analytes.
The derivatization reagents, 2,3-naphthalenedicarboxaldehyde (NDA) and 2-naphthylisothiocyanate (NITC) were chosen due to their wide usage along with their high reaction rate and efficiency with most natural amino acids.9, 38-41 Nine amino acids were randomly selected from each of the three classes for derivatization with NDA and NITC, respectively (Fig. 1a), including polar uncharged amino acids: Serine (Ser, S), Tyrosine (Tyr, Y); charged amino acids: Histidine (His, H), Glutamic Acid (Glu, E), Aspartic Acid (Asp, D); and non-polar amino acids: Glycine (Gly, G), Alanine (Ala, A), Valine (Val, V); and Phenylalanine (Phe, F). The derivatization process is simple, efficient, and was performed under mild basic buffer condition as described in synthetic route I and II for NDA and NITC derivatives, respectively (Fig. S1, ESI†). In our nanopore analysis experiments, only purified amino acid derivatives were employed. Molecular structures of all derivatization products shown in Table S1 (ESI†) were characterized and confirmed by 1H and 13C nuclear magnetic resonance analysis (Fig. S4-S39, ESI†). However, due to the high efficiency of both chemistries used in our design, they can be readily employed for N-terminal modification of amino acids in aqueous solution for practical applications.
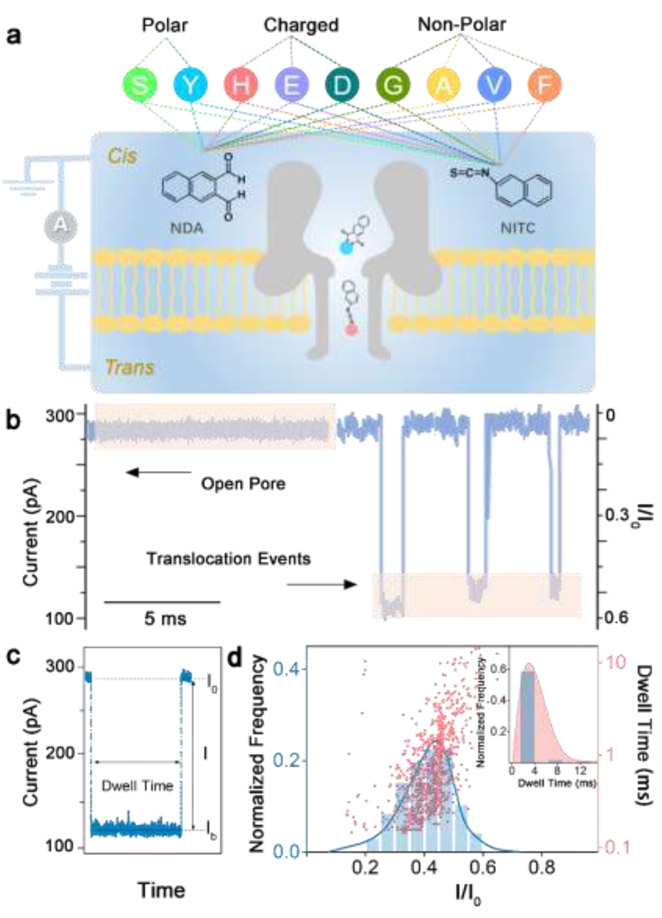
Fig. 1.
(a) Schematic of NDA and NITC derivatization of 9 amino acids in 3 classes and the experimental setup (not to scale). An external voltage is applied to the trans side of the lipid bilayer while the cis side is grounded. (b) Representative fragment of ionic current recording before (open pore) and after (translocation events) NITC-Tyr derivatives were added. (c) Illustration of typical signal events caused by translocation blockade. (d) Histogram of events per bin of current blockade I/I0 (left versus bottom axis) and scatter-plot of dwell time versus I/I0 (right versus bottom axis) produced by NITC-Tyr in an α-HL nanopore. Inset depicts the histogram of events per bin of dwell time for NITC-Tyr.
In a typical experiment, a single α-HL nanopore is inserted into a phosphate lipid bilayer that separates cis and trans compartments in an electrolyte solution. An external positive voltage (100 mV) is applied to the trans side of the bilayer, while the cis side is electrically grounded (Fig. 1a). The direction of protein insertion can be determined by the absolute value of open pore current under positive and negative voltages.42 In the present study, the tail of the mushroom shaped α-HL is inserted into the lipid bilayer, and all samples enter the nanochannel through the head of α-HL remaining on cis side. In the absence of amino acid derivatives, a steady ionic current I0 (~ 287 pA) flows through the nanopore (open pore, Fig. 1b). Each unmodified amino acid and derivatization reagent was first tested using an α-HL nanopore, and no obvious characteristic signal is observed for an extended recording time (Fig. S2, ESI†). In contrast, addition of any amino acid derivatives on the cis side of the bilayer induces transient events of the ionic current, with a representative translocation blockade profile for NITC-Tyr shown in Fig. 1b. Each blockade corresponding to the capture of an individual derivative in the pore is characterized by two parameters: current blockade I/I0 (I = I0 - Ib, Ib indicates the residual current induced by the analyte) and the blockade duration (dwell time) that represents the effective interaction time between the pore and the analyte (Fig. 1c). A typical histogram of the current blockades (I/I0) or the dwell time has a single narrow peak (Fig. 1d), which is characterized by the mean value of a Gaussian fit and its standard deviation (SD). In the case of NITC-Tyr in Fig. 1b, I/I0 was 0.439 ± 0.073 and the dwell time was 2.866 ± 0.540 ms. Similar experiments were performed separately for two series of NDA and NITC derivatives of 9 different amino acids. Corresponding superimposed histograms of the relative current blockade values are summarized in Fig. 2 and 3, where results are classified into polar uncharged, electrically charged, and non-polar uncharged amino acids. For the identification of individual amino acid, current blockade (I/I0) is used as the primary criterion as it can reflect the variation of the spatial structure of the molecule directly before and after modification, as confirmed previously.28 Dwell time is used as the secondary identification criterion when the current blockade is noneffective.
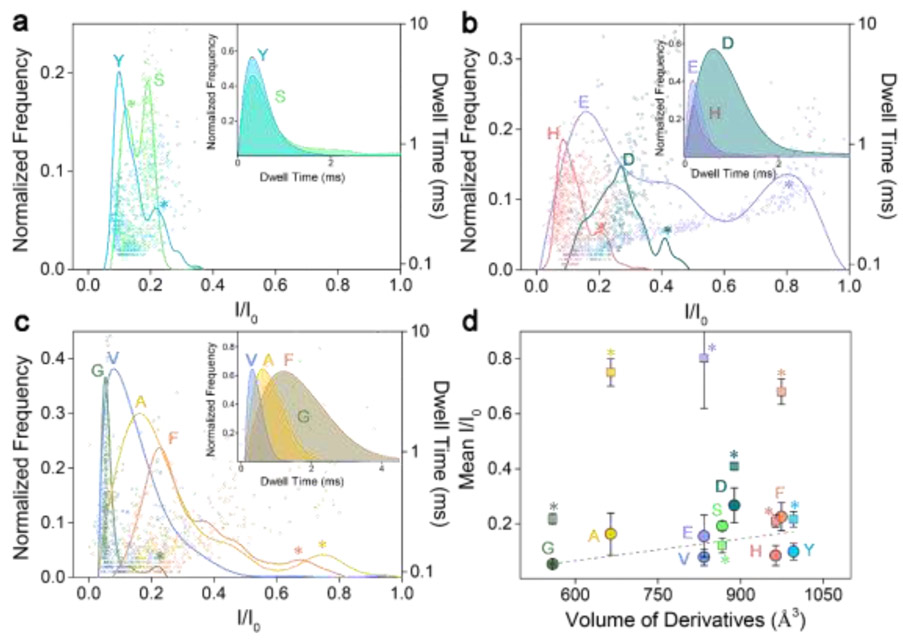
Fig. 2.
Superimposed histograms of I/I0 obtained from nanopore for NDA-modified amino acids, analyzed individually and grouped as: (a) polar, (b) charged, and (c) non-polar amino acids. Insets in a-c are superimposed histograms of dwell time for the derivatives. (d) Mean relative current blockade and standard deviation produced by each NDA amino acid derivative versus its spatial volume. The grey dashed line is obtained by a linear fit of the main peaks (represented by circles). All data was acquired in 3 M KCl, 10 mM Tris-HCl buffer, at 8.0 pH, 200 μM derivatives concentration, and under a 100 mV bias applied to the trans side. For each histogram, at least 600-1000 events were analyzed. Overlaps of current blockade subpopulations between each other are tagged with asterisks.
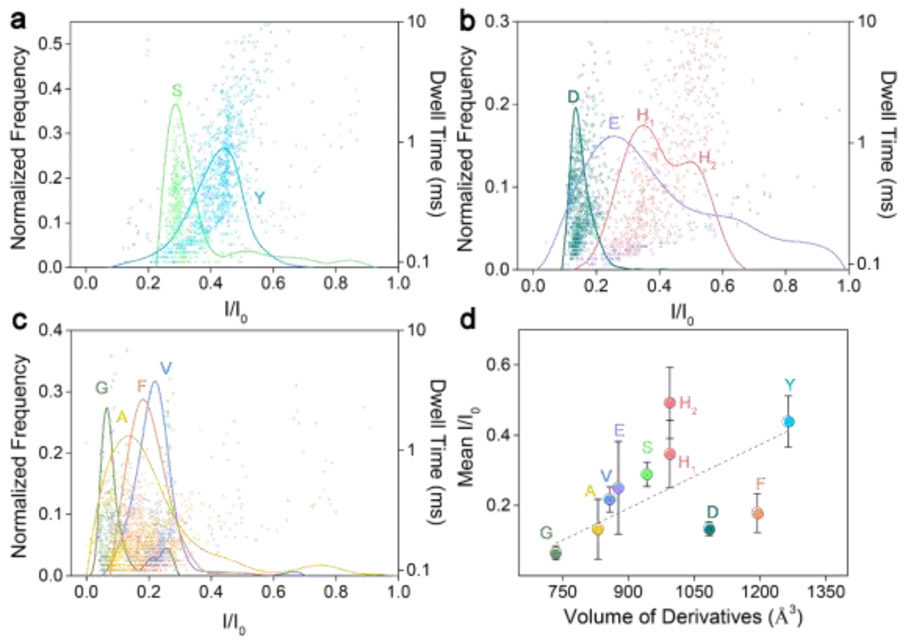
Fig. 3.
Superimposed histograms of I/I0 obtained from nanopore for NITC-modified amino acids, analyzed individually and grouped as: (a) polar, (b) charged, and (c) non-polar amino acids. (d) Mean relative current blockade and standard deviation produced by each NITC amino acid derivative versus its spatial volume. The grey dashed line is obtained by a linear fit of the main peaks (represented by the circle). All data was acquired in 3 M KCl, 10 mM Tris-HCl buffer, at 8.0 pH, 200 μM derivatives concentration, and under a 100-mV bias applied to the trans side. For each histogram, at least 600-1000 events were analyzed.
Fig. 2a-c show the superimposed histograms of each class of amino acids after modification with NDA, in which case three rigid benzoisoindolones are formed after linking with amino acids (Table S1, ESI†). This modification is expected to give a larger volume and various spatial structures to each individual amino acid, resulting in a fingerprint signal when translocated through the constriction region of the α-HL nanopore (width: 1.4 nm). The NDA derivatives all have effective interactions with the nanopore and each NDA derivative has its own characteristic distribution of I/I0 and dwell time (Fig. 2a-c, Table S2, ESI†). Furthermore, these characteristic signals from different amino acids in each class are clearly distinguishable. For instance, although two populations—corresponding to members (Y, S) of the polar family (Fig. 2a)—have similar dwell time distributions (inset in Fig. 2a), they can be clearly identified and distinguished from one another with mean I/I0 of 0.099 ± 0.029 and 0.192 ± 0.018, respectively.
Comparing to the polar family, NDA derivatives of the charged family H, E, D exhibit wider distributions of I/I0, with the mean I/I0 of 0.084 ± 0.037, 0.154 ± 0.077, and 0.267 ± 0.063, respectively, as shown in Fig. 2b. A broad distribution of the current blockade was observed for NDA-Glu (E), possibly due to its negative charge and weak hydrophilicity, as well as the rigid benzoisoindolone structure resulting in multiple spatial orientations inside the nanopore. Although there is a serious current blockade overlap between E and D caused by the broad distribution, they can be further distinguished by their dwell time distribution (inset in Fig. 2b). In the non-polar family shown in Fig. 2c, G and V can be distinguished by using both I/I0 and the dwell time distribution, while A and F can be effectively distinguished by dwell time distribution regardless of their considerably overlapped I/I0 distribution (inset in Fig. 2c).
After geometrically optimizing the NDA amino acid derivatives to gain an accurate measurement of hydrodynamic volume (Fig. 2d), we plotted the mean relative current blockade against the hydrodynamic to probe the physical mechanism underlying the current blockade induced by different amino acid in a nanopore. Overall, the mean I/I0 increased as the excluded volume increases for each amino acid derivative with a fitting slope of 2.67, which agrees with the tendency found in previous reports.36, 43, 44 However, subpopulations located beside main peaks (tagged with asterisks in Fig. 2d) have random and irregular mean I/I0 distribution tendency. For most NDA derivatives, both criteria (current blockade and dwell time) are needed for differentiation among amino acids. This is likely due to the fact that the NDA modification cannot induce sufficient variation between amino acids, such as the narrow volume range, i.e., from 559 to 996 Å3 (Table S2, ESI†). In addition, the rigid benzoisoindolone structure formed in each NDA derivative causes complexity in its spatial conformation when translocating the nanopore, leading to a wide distribution on the histograms, which further dampens the discriminatory power.
To improve the discrimination between amino acids, we further modified these 9 amino acids with NITC (Table S1, ESI†). An increase in spatial structure complexity of the NITC-derivatives is confirmed by wider volume range, i.e., 734 – 1264 Å3 (Table S2, ESI†), which is expected to lead to higher discriminatory power and less uncertainty of the spatial orientation of derivatives inside the nanopore.
Results of all NITC derivatives confirm effective interactions with the nanopore by characteristic distribution of I/I0 and dwell time for each derivative. Although with some notable exceptions, the superimposed histograms of each family of derivatives exhibit well-separated populations with narrow distributions (Fig. 3). Two populations corresponding to S, Y of the non-polar amino acid family can be clearly identified and distinguished from one another by mean I/I0 of 0.289 ± 0.035 and 0.439 ± 0.073, respectively (Fig. 3a). Three populations corresponding to E, D, H of the charged amino acid family also can be clearly identified and distinguished by their characteristic distributions of I/I0, which are centered at 0.249 ± 0.132 (E), 0.134 ± 0.019 (D), 0.347 ± 0.096 (H1), 0.492 ± 0.201 (H2), respectively. Two equally distributed peaks observed for NITC-His (H) may be attributed to the rigidity of the molecular structure of His. The NITC modification strategy shows a more obvious advantage for identification of G, A, F, V in the non-polar family. As shown in Fig. 3c, these four amino acids can be discriminated through the mean I/I0 values: 0.065 ± 0.019 (G), 0.132 ± 0.085 (A), 0.178 ± 0.055 (F), and 0.217 ± 0.036 (V), regardless of minor overlaps between each other. NITC derivatization clearly affords a wider distribution of I/I0 (0.1–0.5) across the amino acids tested comparing to a previous report,36 which should improve identification sensitivity. While identification of all the 9 amino acids can be achieved by using the mean I/I0 (primary criterion) only, their dwell time distribution (secondary criterion) was also analyzed to further enhance the identification accuracy (Fig. S3 ESI†). Similar to NDA derivatives, NITC derivatives also exhibited an increasing trend of mean relative current blockade with increased hydrodynamic volume, but with a larger fitting slope of 6.04 that indicates enhanced discriminatory power by NITC derivatization (Fig. 3d).
Previous studies have demonstrated that an aerolysin nanopore with a narrower constriction of ∼1.0 nm is able to detect a bare cysteine,45 and differentiate certain peptides with one amino acid difference in length.46 A recent advancement demonstrates detection of more types of amino acids using a peptide as the carrier, resulting in various I/I0 distributions around 0.4 with only slight shifts for different modified amino acid.36 Whereas the NITC derivatization produced larger difference between I/I0 distributions of different amino acids (0.1–0.5), indicating improved sensitivity using small molecules as amino acid modifiers. In addition, it is overwhelmingly challenging to apply the peptide carrier method to practical protein sequencing due to various reaction conditions for modifying different amino acids. As demonstrated in the Edman degradation,9 N-terminal derivatization can efficiently increase the spatial size of all amino acids with similar reactivity within the same reaction, and thus can be readily applied to recognize all amino acids towards protein sequencing.
When an analyte translocates the nanopore under the applied voltage and the diffusion effect, a signal on the electrical current trace characterized by a current blockade and a dwell time can be generated as a result of the transient occupation of the nanopore lumen by the analyte and the interaction between analyte and nanopore. The low frequencies of translocation events for the selected amino acids demonstrate their weak interactions with the lumen of the α-HL nanopore, due to the smaller Van der Waals radii of the amino acids (~0.3–0.4 nm) comparing to the dimension of the α-HL nanopore constriction region (1.4 nm).47-49 As confirmed by the molecular structure modeling results, most NITC derivatives produced current blockades that positively correlate to their spatial size. Although exceptions (i.e. D, F) were observed, the general mechanism of electrical sensing of individual amino acids is to increase their spatial size by derivatization to promote interactions with the nanopore sensing region, thereby improving the signal-to-noise ratio. Further investigation using other types of biological nanopores is warranted to probe possible explanations for these exceptions, such as intra- and inter-molecular interactions between analytes.50
In conclusion, we have demonstrated a derivatization strategy for reliable identification of individual amino acids using α-HL nanopore. Compared to bare amino acids, both NDA-derived and NITC-derived amino acids can produce obvious fingerprint signals when translocating the nanopore. Furthermore, amino acids S, Y, D, E, H, G, A, F, V can be effectively identified with improved discriminatory power by NITC derivatization. While promising results were obtained in 9 amino acids, we do recognize the overall complexity of identifying all 20 amino acids. In particular, we need to develop more effective conjugation chemistry to derivatize proline, which does not have a primary amino group like others. Additionally, in-depth analysis is needed to better understand the interactions between amino acid derivatives with the lumen surface of biological nanopores. Novel characterizations of stochastic signals other than the traditional blockade and dwell time must be explored, and more advanced data analysis technology (e.g. machine learning, pattern recognition, etc.) should be applied to achieve even greater resolution. Nonetheless, compared to previous efforts on amino acid identification using nanopore, the presented method is readily applicable to future protein sequencing. We envision a potential “Sequencing-by-Hydrolysis” method, in which a nanopore will be used to identify the N-terminal amino acid of each peptide fragment in a peptide ladder generated from a peptide analyte, and then bioinformatics methods will be applied to reconstitute its full-length sequence.51
Supplementary Material
Acknowledgment
C. Liu acknowledges supports from the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under award number K22AI136686, and the South Carolina IDeA Networks of Biomedical Research Excellence Developmental Research Project funded by the National Institute of General Medical Sciences (NIGMS) of the NIH under award number P20RR016461. Q. Wang acknowledges the partial support from the BDSHC initiative of the University of South Carolina. D. Ma acknowledges the support (201806310084) from the State Scholarship Fund of the China Scholarship Council. L. Jing thanks the Youth Innovation Promotion Association CAS (2018042), National Natural Science Foundation of China (81671755).
Footnotes
The authors declare no competing financial interest.
References
- 1.Wei G, Su ZQ, Reynolds NP, Arosio P, Hamley IW, Gazit E and Mezzenga R, Chem. Soc. Rev, 2017, 46, 4661–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang WS, Yu XQ, Li Y, Su ZQ, Jandt KD and Wei G, Progress in Polymer Science, 2018, 80, 94–124. [Google Scholar]
- 3.Asandei A, Di Muccio G, Schiopu I, Mereuta L, Dragomir IS, Chinappi M and Luchian T, Small Methods, 2020, DOI: 10.1002/smtd.201900595. [DOI] [Google Scholar]
- 4.McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA and Spinner NB, American Journal of Human Genetics, 2006, 79, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katoh M and Katoh M, Clinical Cancer Research, 2007, 13, 4042–4045. [DOI] [PubMed] [Google Scholar]
- 6.Domon B and Aebersold R, Science, 2006, 312, 212–217. [DOI] [PubMed] [Google Scholar]
- 7.Angel TE, Aryal UK, Hengel SM, Baker ES, Kelly RT, Robinson EW and Smith RD, Chem. Soc. Rev, 2012, 41, 3912–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steen H and Mann M, Nature Reviews Molecular Cell Biology, 2004, 5, 699–711. [DOI] [PubMed] [Google Scholar]
- 9.Edman P, Acta Chemica Scandinavica, 1950, 4, 283–293. [Google Scholar]
- 10.Im J, Lindsay S, Wang X and Zhang P, Acs Nano, 2019, 13, 6308–6318. [DOI] [PubMed] [Google Scholar]
- 11.Waugh M, Briggs K, Gunn D, Gibeault M, King S, Ingram Q, Jimenez AM, Berryman S, Lomovtsev D, Andrzejewski L and Tabard-Cossa V, Nature Protocols, 2020, 15, 122–143. [DOI] [PubMed] [Google Scholar]
- 12.Deamer D, Akeson M and Branton D, Nature Biotechnology, 2016, 34, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, Jordan M, Ciccone J, Serra S, Keenan J, Martin S, McNeill L, Wallace EJ, Jayasinghe L, Wright C, Blasco J, Young S, Brocklebank D, Juul S, Clarke J, Heron AJ and Turner DJ, Nature Methods, 2018, 15, 201-+. [DOI] [PubMed] [Google Scholar]
- 14.Collins BC and Aebersold R, Nature Biotechnology, 2018, 36, 1051–1053. [DOI] [PubMed] [Google Scholar]
- 15.Cao C, Li M-Y, Cirauqui N, Wang Y-Q, Dal Peraro M, Tian H and Long Y-T, Nature Communications, 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loman NJ, Quick J and Simpson JT, Nature Methods, 2015, 12, 733–U751. [DOI] [PubMed] [Google Scholar]
- 17.Szalay T and Golovchenko JA, Nature Biotechnology, 2015, 33, 1087-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nivala J, Marks DB and Akeson M, Nature Biotechnology, 2013, 31, 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benner S, Chen RJA, Wilson NA, Abu-Shumays R, Hurt N, Lieberman KR, Deamer DW, Dunbar WB and Akeson M, Nature Nanotechnology, 2007, 2, 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallace EVB, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ and Bayley H, Chemical Communications, 2010, 46, 8195–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varongchayakul N, Song J, Meller A and Grinstaff MW, Chem. Soc. Rev, 2018, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson JWF and Reiner JE, Proteomics, 2018, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Restrepo-Perez L, Joo C and Dekker C, Nature Nanotechnology, 2018, 13, 786–796. [DOI] [PubMed] [Google Scholar]
- 24.Huang G, Voet A and Maglia G, Nature Communications, 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji Z, Kang X, Wang S and Guo P, Biomaterials, 2018, 182, 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang G, Willems K, Soskine M, Wloka C and Maglia G, Nature Communications, 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Restrepo-Perez L, Huang G, Bohlander PR, Worp N, Eelkema R, Maglia G, Joo C and Dekker C, Acs Nano, 2019, 13, 13668–13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piguet F, Ouldali H, Pastoriza-Gallego M, Manivet P, Pelta J and Oukhaled A, Nature Communications, 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao C, Cirauqui N, Marcaida MJ, Buglakova E, Duperrex A, Radenovic A and Dal Peraro M, Nature Communications, 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Muccio G, Rossini AE, Di Marino D, Zollo G and Chinappi M, Sci Rep, 2019, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boynton P and Di Ventra M, Sci Rep, 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao YA, Ashcroft B, Zhang PM, Liu H, Sen SM, Song W, Im J, Gyarfas B, Manna S, Biswas S, Borges C and Lindsay S, Nature Nanotechnology, 2014, 9, 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boersma AJ and Bayley H, Angewandte Chemie International Edition, 2012, 51, 9606–9609. [DOI] [PubMed] [Google Scholar]
- 34.Guo Y, Niu A, Jian F, Wang Y, Yao F, Wei Y, Tian L and Kang X, Analyst, 2017, 142, 1048–1053. [DOI] [PubMed] [Google Scholar]
- 35.Asandei A, Rossini AE, Chinappi M, Park Y and Luchian T, Langmuir, 2017, 33, 14451–14459. [DOI] [PubMed] [Google Scholar]
- 36.Ouldali H, Sarthak K, Ensslen T, Piguet F, Manivet P, Pelta J, Behrends JC, Aksimentiev A and Oukhaled A, Nature Biotechnology, 2020, 38, 176-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edman P, Archives of Biochemistry, 1949, 22, 475–476. [PubMed] [Google Scholar]
- 38.Molnar-Perl I, in Quantitation of Amino Acids and Amines by Chromatography: Methods and Protocols, ed. MolnarPerl I, 2005, vol. 70, pp. 163–198. [Google Scholar]
- 39.Checa-Moreno R, Manzano E, Miron G and Capitan-Vallvey LF, Journal of Separation Science, 2008, 31, 3817–3828. [DOI] [PubMed] [Google Scholar]
- 40.Fountoulakis M and Lahm HW, Journal of Chromatography A, 1998, 826, 109–134. [DOI] [PubMed] [Google Scholar]
- 41.Woo KL and Ahan YK, Journal of Chromatography A, 1996, 740, 41–50. [DOI] [PubMed] [Google Scholar]
- 42.Wei X, Zhang Z, Wang X, Lenhart B, Gambarini R, Gray J and Liu C, Nanotechnology and Precision Engineering, 2020, 3, 2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baaken G, Halimeh I, Bacri L, Pelta J, Oukhaled A and Behrends JC, Acs Nano, 2015, 9, 6443–6449. [DOI] [PubMed] [Google Scholar]
- 44.Chavis AE, Brady KT, Hatmaker GA, Angevine CE, Kothalawala N, Dass A, Robertson JWF and Reiner JE, Acs Sensors, 2017, 2, 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan B, Li S, Ying YL and Long YT, Analyst, 2020, 145, 1179–1183. [DOI] [PubMed] [Google Scholar]
- 46.Piguet F, Ouldali H, Pastoriza-Gallego M, Manivet P, Pelta J and Oukhaled A, Nature Communications, 2018, 9, 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasianowicz JJ, Brandin E, Branton D and Deamer DW, Proceedings of the National Academy of Sciences, 1996, 93, 13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson J, Sloman L, He Z and Aksimentiev A, Advanced Functional Materials, 2016, 26, 4830–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kennedy E, Dong Z, Tennant C and Timp G, Nature Nanotechnology, 2016, 11, 968. [DOI] [PubMed] [Google Scholar]
- 50.Zhang XY, Gong CC, Akakuru OU, Su ZQ, Wu AG and Wei G, Chem. Soc. Rev, 2019, 48, 5564–5595. [DOI] [PubMed] [Google Scholar]
- 51.Zhong HY, Zhang Y, Wen ZH and Li L, Nature Biotechnology, 2004, 22, 1291–1296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.