Abstract
Noncoding RNA has a proven ability to direct and regulate chromatin modifications by acting as scaffolds between DNA and histone-modifying complexes. However, it is unknown if ncRNA plays any role in DNA replication and epigenome maintenance, including histone eviction and reinstallment of histone modifications after genome duplication. Isolation of nascent chromatin has identified a large number of RNA-binding proteins in addition to unknown components of the replication and epigenetic maintenance machinery. Here, we isolated and characterized long and short RNAs associated with nascent chromatin at active replication forks and track RNA composition during chromatin maturation across the cell cycle. Shortly after fork passage, GA-rich-, alpha- and TElomeric Repeat-containing RNAs (TERRA) are associated with replicated DNA. These repeat containing RNAs arise from loci undergoing replication, suggesting an interaction in cis. Post-replication during chromatin maturation, and even after mitosis in G1, the repeats remain enriched on DNA. This suggests that specific types of repeat RNAs are transcribed shortly after DNA replication and stably associate with their loci of origin throughout the cell cycle. The presented method and data enable studies of RNA interactions with replication forks and post-replicative chromatin and provide insights into how repeat RNAs and their engagement with chromatin are regulated with respect to DNA replication and across the cell cycle.
Keywords: RNA, DNA replication, chromatin, repeats
INTRODUCTION
Mammalian genome duplication relies on correct DNA replication and reestablishment of the local chromatin environment on the two newly synthesized sister chromatids. DNA replication is initiated by sequential activation of several origins of replication in S-phase (Bell and Dutta 2002; Mechali 2010). The replication program is carefully orchestrated according to genome organization, with a trend for transcriptionally active euchromatin to replicate early in S-phase (Marchal et al. 2019). The replication machinery disrupts chromatin ahead of the replication fork and must correctly restore the epigenetic landscape on both new daughter strands (Alabert and Groth 2012). Parental histones with their large variety of post-translational modifications (PTMs) are segregated onto the daughter strands and mixed with nucleosomes assembled from new histones (Alabert et al. 2015; Annunziato 2015). Parental histone recycling is remarkably accurate, allowing the histone PTM landscape to be reproduced after replication but with PTM levels twofold reduced due to dilution with new histones (Alabert et al. 2015). Post-replication, in a process termed chromatin restoration, new histones acquire modifications identical to those of the nearby parental histones in order to maintain epigenetic regulation and preserve cell identity. This is a highly heterogeneous process that can take minutes or hours depending on the nature and location of histone marks (Alabert et al. 2015; Reveron-Gomez et al. 2018), the regulation and mechanism of which is largely unknown. Generally, PTMs associated with active transcription are restored rapidly, with recent evidence in mouse embryonic stem cells (mESC) showing that RNA Polymerase II transcription recommences within 30 min after DNA replication (Stewart-Morgan et al. 2019).
The epigenetic landscape is largely shaped by histone modifications and DNA methylation, which are deposited and removed by histone and DNA modifiers (Rothbart and Strahl 2014). Recruitment of these factors to precise genomic sites is facilitated by transcription and recognition of chromatin features, including long noncoding RNAs (lncRNA) (Kouzarides 2007; Johnson and Straight 2017). Over the last decade, lncRNAs have broadened our understanding of the roles of RNA from encoding proteins to having important functions in almost all cellular pathways via a number of different functionalities, including binding to DNA, other RNAs and proteins (Quinn and Chang 2016). With the exception of Y-RNAs, which regulate replication initiation (Christov et al. 2006; Ge and Lin 2014), the role of RNA at ongoing replication forks and in chromatin maturation remains elusive.
Nascent chromatin capture (NCC) has been used to identify proteins associated with active replication forks and newly replicated DNA (Alabert et al. 2014). Surprisingly, a substantial number of these proteins have a described function in RNA binding and processing. Based on this and the many functional roles of RNA, such as the scaffolding of macromolecular complexes, we speculated that RNA could play a role in chromatin replication. To address this question, we developed an NCC-based method to detect RNA at active replication forks and maturing chromatin. The method utilizes pulse-labeling of DNA in replicating synchronized cells with biotin dUTP, enabling isolation of chromatin at specific time points post replication. We find a number of RNA transcripts arising from genes to be moderately enriched specifically at nascent chromatin. These genes are not replicated at the time of labeling, suggesting that the RNA associate with nascent chromatin in trans. In addition, we find considerable enrichment of specific groups of repeat containing RNAs at mid/late S-phase replicated DNA shortly after replication. These RNAs arise from DNA loci that are being replicated at the time of labeling, implying that these RNAs associate with nascent chromatin in cis. Notably, the repeat RNA species remain associated with the replicated chromatin loci for at least 10 h, suggesting that these RNAs are stably bound to chromatin. We propose that repeat RNAs are transcribed shortly after fork passage as a consequence of increased accessibility of DNA upon incorporation of new acetylated histones (Annunziato 2012) and that they remain associated with and influence chromatin structure at the loci across the cell cycle.
RESULTS
Development of NCC-RNA-seq method
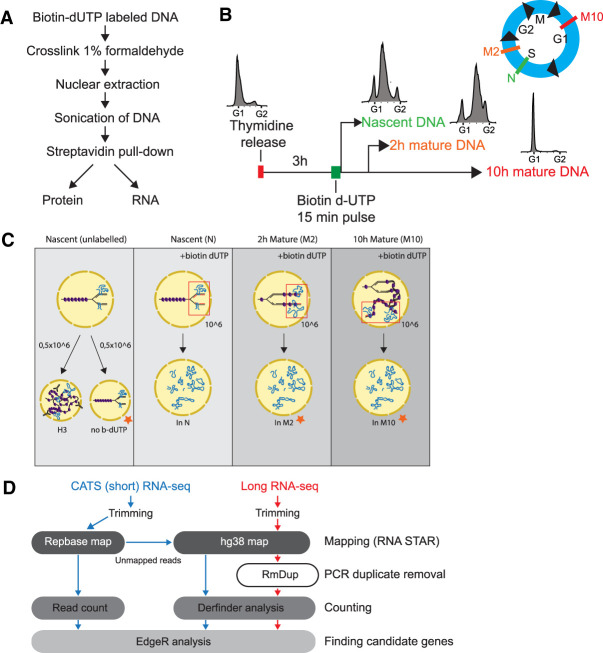
With the aim to identify specific RNAs associated with replication forks and maturing chromatin, we optimized the NCC technique for RNA isolation (Fig. 1A). This protocol allows for the isolation of active replication forks and nascent chromatin by incorporation of biotin-dUTP during DNA synthesis. Maturation of the pulse-labeled newly replicated chromatin can then be followed across the cell cycle (Alabert et al. 2014, 2015). To allow comparison of RNA-seq data with published NCC proteomics and DNA-seq data from HeLa cells (Alabert et al. 2014; Reveron-Gomez et al. 2018), we adapted the same synchronization strategy—that is, pulse labeling cells in mid-S phase 3 h after release from a G1/S block (Fig. 1B). We isolated RNA associated with nascent chromatin (N), and mature chromatin harvested 2 (M2), and 10 h (M10) after DNA replication (Fig. 1B,C; Alabert et al. 2015). We incorporated several controls, including a negative control without b-dUTP labeling (no b-dUTP) to account for binding of nonspecific RNAs and chromatin immunoprecipitation (ChIP) of histone H3, representing total chromatin-bound RNA. Input controls were included to account for bias between highly and lowly expressed RNAs. Importantly, we pull down PCNA, a key DNA polymerase processivity factor, in the nascent sample only, ensuring isolation of active replication forks (Supplemental Fig. S1B). We also observed an increase in the level of H3K9me3 with increasing chase time (Supplemental Fig. S1B), consistent with the restoration marks on new histones after chromatin replication.
FIGURE 1.
Developing nascent chromatin capture RNA-seq (NCC RNA-seq). (A) Workflow of NCC RNA-seq protocol. (B) Experimental setup illustrating synchronization, labeling and sample harvesting. FACS diagrams showing distribution of cells in cell cycle phases at harvest time points (also indicated in blue cell cycle illustration), N = nascent chromatin, M2 = 2 h mature chromatin, M10 = 10 h mature chromatin. (C) Schematic representation of sample setup for long RNA and short RNA sequencing. Orange stars indicate samples that were included in short but not long RNA-seq. Three out of four samples were labeled 3 h into S phase with a pulse of biotin dUTP. Nascent (N) and unlabeled samples were harvested 15 min after labeling, whereas mature samples were collected 2 h (M2) and 10 h (M10) later. The unlabeled sample in short RNA-seq was split in two and used for histone 3 (H3) immunoprecipitation and as a control for unspecific binding to Streptavidin T1 beads (no b-dUTP). Input controls were taken from indicated samples before cross-linking. (D) Bioinformatics pipeline for long (red) and short (blue) NCC RNA-seq.
Because isolated RNAs from the NCC samples were 20–4000 nt in length (Supplemental Fig. S1A), we performed sequencing of both long and short RNAs to obtain a comprehensive overview of RNAs associated with the replication complex and maturing chromatin. Long RNA-seq experiments were performed in quadruplicates with input and H3 controls taken at the time of labeling (Fig. 1C). Initial RNA-seq analysis using genome-guided reference transcriptome quantification revealed that more than 50% of mapped reads were lost due to reads mapping outside of known transcripts. However, mapping to the genome resulted in few unmapped reads. Instead of relying on annotated genes and transcripts, we performed ab initio transcriptome assembly using Trinity, which assembles transcripts based on the raw sequencing reads without a reference sequence (Grabherr et al. 2011). In addition, we mapped reads to the human genome version 38 (hg38) and used another annotation-free algorithm—DERfinder—to identify expressed regions based on clustering of reads in the genome, for example, unannotated exons (Frazee et al. 2014; Collado-Torres et al. 2017a,b). Both methods were analyzed with EdgeR for enrichment analysis (Jurka 2000; Jurka et al. 2005). Based on initial principal component analysis (PCA) and clustering of samples after EdgeR processing, one nascent replicate (N_112) was removed from the analysis since it clustered with the input samples instead of the other nascent samples (Supplemental Fig. S1C).
We prepared the short RNA-seq in triplicates using a capture and amplification by tailing and switching (CATS) strategy, which generates cDNA libraries ranging from 3 to 300 nt in length. In these experiments, we included cell cycle-matched input controls for all time points, H3 IP and a negative control without b-dUTP incorporation (Fig. 1C). No rRNA depletion or size selection was performed to reduce any potential enrichment bias (excluding the removal of primers during cDNA library preparation). This resulted in libraries that, in some instances, had more than 60% of reads mapping to repeat-containing RNAs. To mitigate the impact of multimapping repetitive reads and allow for meaningful quantification and statistical analysis of the data, we first mapped reads to human repeat sequences to uniquely map repeat-containing reads, and subsequently aligned the unmapped reads to the hg38 genome (Fig. 1D). Expressed regions were identified and quantified using DERfinder and the analysis of the repeat mapping and hg38 mapping was done with EdgeR (Supplemental Fig. S1D,E).
Long RNA sequencing identifies genes that are lowly yet specifically enriched at nascent chromatin, whereas GA-rich and telomeric RNAs are highly enriched at nascent, 2 and 10 h mature chromatin
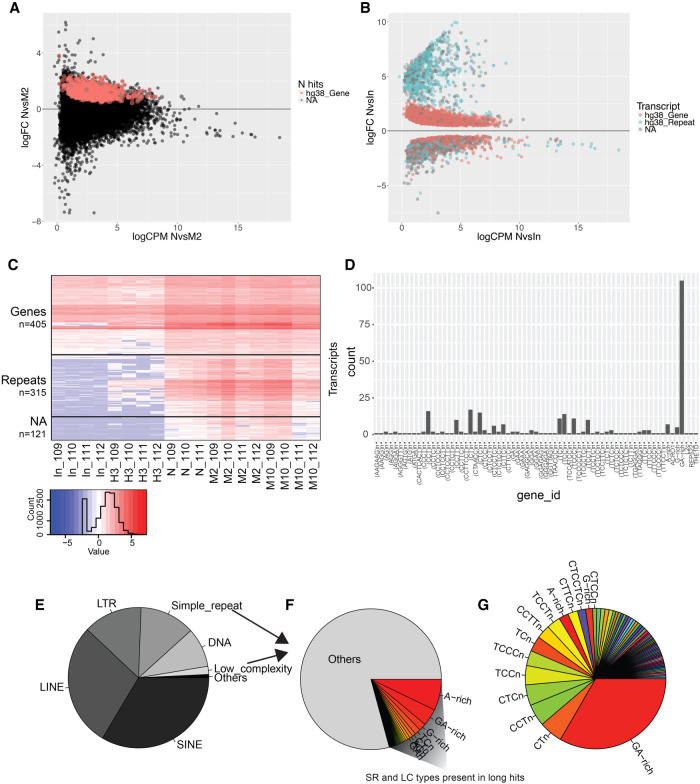
We first investigated the diversity of RNAs associated with replication forks. Interestingly, we found the nascent samples to cluster separately in the PCA plot, and when comparing data from the nascent samples (N) against all other samples (M2, M10, H3, and input), we found 737 RNAs specifically associated with the replication fork (nascent hits) (Fig. 2A; Supplemental Fig. S1C; Supplemental Table S1). Overlapping the RNAs with annotation files for repeats and genes showed that all nascent hits (except 15 unannotated regions) mapped to the exons of 309 different protein-coding genes, spread across all chromosomes. These RNAs displayed low enrichment between the nascent and the M2 samples with a log fold-change (logFC) < 2.5 (except one transcript with logFC 3.8). Based on this low enrichment, it is unlikely that the observed RNAs associate with all actively replicating forks, as this would likely have resulted in a much higher logFC between the nascent and mature chromatin. Similarly, we analyzed specific enrichment for transcripts in the M2 and M10 samples, respectively, but found only a subtle enrichment in terms of logFC. All three gene lists (N, M2, and M10 hits, Supplemental Table S1) were analyzed for characteristics via Enrichr, and although pathways, ontologies, etc. showed statistical enrichment, no consistent pattern of enrichment was found within each data set or in common (Chen et al. 2013; Kuleshov et al. 2016).
FIGURE 2.
GA-rich repeats are enriched in N, M, and M10 samples from long RNA-seq. (A) RNAs in long RNA-seq plotted according to log fold-change (logFC) versus average counts per million (CPM) from nascent (N) and 2 h mature (M2) sample comparison. Red and gray points indicate significantly enriched (FDR = <0.05) RNAs in N sample compared to M2. (B) Enriched RNAs (FDR = <0.05) from N versus input comparison plotted and colored according to RNA type (gene [red], repeat [blue], or not available [NA, gray]). (C) Heat map of long hits (n = 841) according to sample and repeat type. Colors represent relative difference of transcript levels in samples [log(CPM + 0.1)]. (D) Long hits repeats (n = 315) according to repeat type. (E) Relative proportion of repeat types in the human genome, n = 5,520,017 (UCSC repeat masker hg38 [Jurka 2000]). (F) Simple repeats (SR) and low complexity (LC) subtypes, n = 806,538 (14.6% of all repeats) with color coded proportions of repeats found in long hits. (G) Long hits repeats, n = 315 relative proportion and coloring according to F.
We also compared nascent, M2 and M10 samples against input and H3, which revealed a significant enrichment for a large number of RNAs (Fig. 2B). Based on this we created a hit list (long hits), composed of transcripts that were commonly enriched between N, M2 and M10 when individually compared to input and H3 (FDR ≤ 0.05 and logFC > 0) (Supplemental Table S1). Long hits consisted of 405 transcribed sequences annotated as being part of genes, 315 as repeats and 121 as unannotated regions (NA). In general, the RNAs that mapped to genes had a low logFC (typically below 2.5) whereas repeats and NAs had considerably higher enrichments (logFC 2.5–10) in the N, M2, and M10 samples over input (Fig. 2C; Supplemental Fig. S2A). These repeats are normally not detectable, or expressed at very low levels, but were enriched in the H3 control compared to input, indicating that the transcripts associate with chromatin and that their enrichment in the N, M2, and M10 samples does not reflect unspecific binding to the streptavidin beads.
Strikingly, almost all the enriched long hit repeat-associated sequences were dinucleotide repeats (CT- or GA-rich) and distributed on all chromosomes, both in intergenic regions and gene bodies (Fig. 2D; Supplemental Fig. S2B,C). It is noteworthy to mention that G-rich telomere sequences (TTAGGG) were also observed to be significantly enriched (NvsIn FDR = 268 × 10−05). GA/CT-rich repeats account for <3% of the total number of repetitive sequences, composing ∼0.33% of the human genome, making it unlikely that the enrichment for this specific type of repeat RNA is by chance (Fig. 2E–G; Supplemental Fig. S2D,E). In order to establish if the transcripts were CT- or GA-rich, and to validate the enrichment independently of genome mapping (avoiding quantitative artifacts derived from multimappers), we performed transcriptome assembly, quantification and DE analysis. Mapping the enriched sequences from the transcriptome assembly analysis back to the genome provided parallel confirmation that the enriched repeats were exclusively GA- and not CT-rich RNAs.
Short RNA sequencing confirms GA enrichment and identifies centromeric and telomeric repeats enriched at nascent, 2 and 10 h mature chromatin
To get a comprehensive view of RNAs associated with replicated chromatin and to validate the GA-repeat enrichment observed in the long RNA-seq experiments, we additionally performed short RNA sequencing. Anticipating many repeat-containing RNAs, the resulting reads were analyzed by first mapping them to repeat sequences, then unmapped reads were subsequently mapped to hg38.
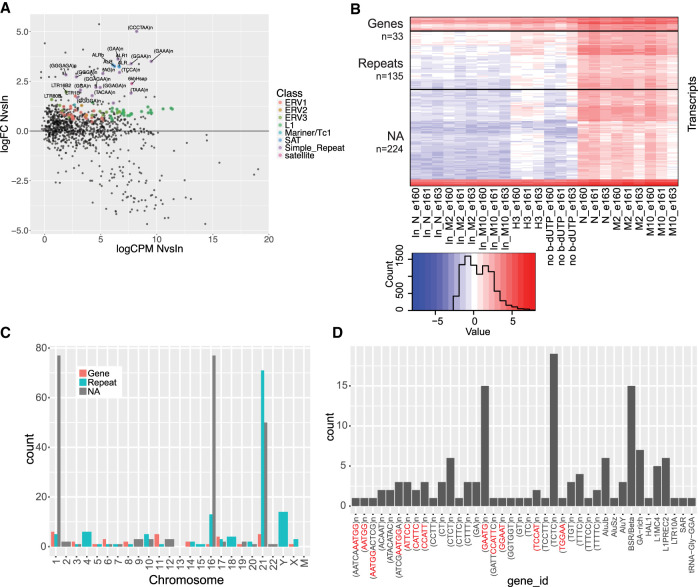
Similar to the long RNA-seq results, the independently analyzed repeat mapping showed no enriched RNAs at the N, M2, or M10 time points when compared to each other, but did present an enrichment of specific repeats in newly replicated and maturing chromatin samples when compared to controls (Fig. 3A; Supplemental Table S2). These repeats were mainly dinucleotide (GA/CT), alpha centromeric repeats and TERRA (telomere repeat) sequences, validating the GA repeat and TERRA enrichment observed in the long hits. The genome mapping analysis also did not show any specific hits at a single time-point, yet an enrichment was observed for repeats at nascent and maturing chromatin when compared to controls (short hits) (Fig. 3B; Supplemental Table S3). In contrast to the long RNA-seq, most of the enriched repeats mapped to a few chromosomes and were concentrated around heterochromatin regions, telomeres, centromeres and pericentromeres (Fig. 3C; Supplemental Fig. S3A). The enriched repeats are annotated mainly as GA/CT rich, centromeric and pericentromeric sequences, such as BSR/beta satellite repeats and CCATT repeats (centromeric repeats) (Fig. 3D; Catasti et al. 1994). CCATT repeats constitute most of HSATII repeats, which are mainly located on Chromosomes 1, 2, 10, and 16 (Tagarro et al. 1994). Repeat masking of transcripts showed that almost all of the unannotated hits (NAs) on Chr 1 and 16, including one hit on Chr 7 and one on Chr 22, were HSATII repeats (Supplemental Fig. S3B; Supplemental Table S4), whereas sequence analysis of the unmasked NA hits (n = 60) showed no strong enrichment of sequence motifs.
FIGURE 3.
Centromeric, telomeric, and GA-rich repeat RNAs are enriched in N, M2, and M10 samples from short RNA-seq. (A) Repbase mapped RNAs plotted according to log fold-change (logFC) versus average counts per million (CPM) in nascent (N) versus input (nascent) comparison (logCPM NvsIn). Colored points (according to repeat type) highlight significantly enriched repeat RNAs in N, M2, and M10 samples compared to all controls (inputs [N, M2, and M10], H3, and no b-dUTP). ALR = alpha repeat, CCCTAA = telomere repeat. (B) Heatmap of short hits (n = 392) according to sample and repeat type. Colors represent relative difference of transcript levels in samples [log(cpm + 0.1)]. (C) Short hits distribution on chromosomes and coloring according to RNA type. (D) Types of repeats present in short hit repeat RNAs (n = 135). Red nucleotides in repeats correspond to the same repeat sequence (CCATT)n or the reverse complementary (AATGG)n.
Taken together, the NCC short RNA sequencing revealed an enrichment of centromeric, pericentromeric (alpha, CCATT, HSATII, BSR) and telomeric repeats (TERRA) and GA-rich sequences shortly after genome replication (nascent chromatin), 2 h and 10 h later (mature chromatin), compared to controls. Although the short hits did not contain as many GA-rich RNA repeats as the long hits, we saw a tendency for most of the long hits to be enriched in the short RNA-seq N, M2, and M10 samples (Supplemental Fig. S3C). The NCC short RNA-seq thus substantiates the findings from long RNA-seq and identified a number of shorter RNA repeats located at telomeres and centromeres, exposing a similar enrichment pattern to GA-rich RNAs.
Repeat RNAs are replicated at the time of labeling
To further understand why RNA is associated with replicated chromatin, we wanted to investigate if the enriched transcripts in the NCC RNA-seq data were replicated at the time of labeling, an indicator of association in cis. To do so, we compared these data to our previously published NCC DNA-seq (Reveron-Gomez et al. 2018). This work sequenced NCC isolated DNA using the same HeLa S3 cell line and a similar NCC protocol (synchronization, labeling time, pulldown) as used in our study, allowing us to identify which genomic regions were replicated at the time of biotin labeling. Due to large differences between hg19 and hg38 in the centromeric regions where many of the short hits are located, we chose to reanalyze the data by first mapping to Repbase and subsequently to hg38.
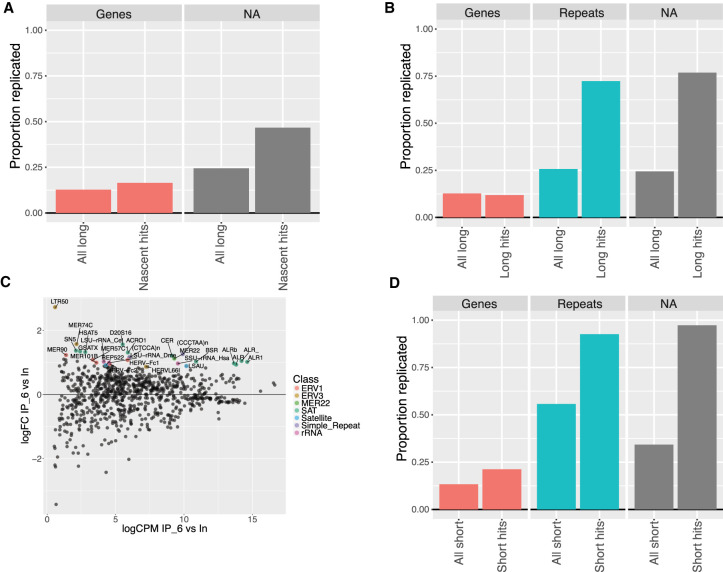
Of the 737 transcripts observed to be specifically enriched in the N samples, 127 (17%) overlapped with replicated loci identified by NCC DNA-seq (47 out of 309 genes) (Fig. 4A). These transcripts might thus be enriched due to concomitant transcription and replication of the loci. However, as this also roughly corresponds to the proportion expected based on overlap of the NCC DNA-seq data with all transcribed genes in the long RNA-seq data set (∼13%), the interactions are more likely to occur in trans.
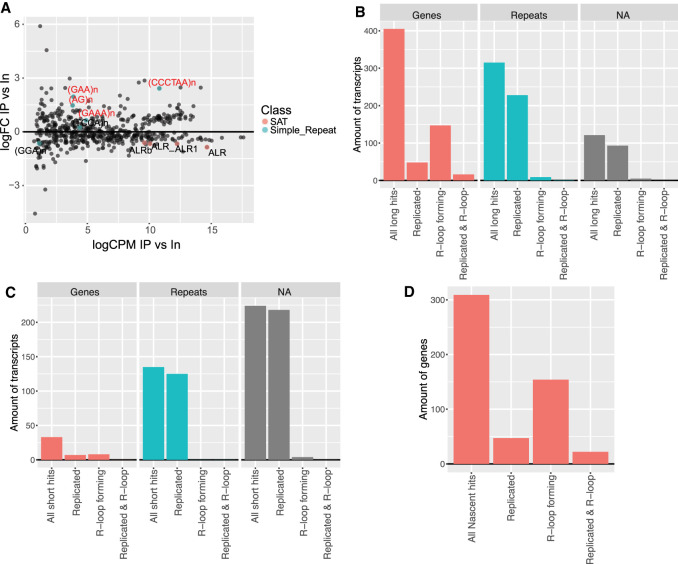
FIGURE 4.
NCC DNA-seq data show enriched repeat RNAs are replicated at time of labeling. (A) Proportion of replicated transcripts in entire long NCC RNA-seq transcriptome (all long) and in nascent hits, plotted according to transcript type (Genes = UCSC annotated gene in hg38 genome build or NA = not available). Transcripts are replicated if their mapped genomic locus overlaps with a replicated region in NCC DNA-seq. (B) Long hits compared to entire long NCC RNA-seq transcriptome as in A (repeats = UCSC genome browser RepeatMasker annotations for hg38). (C) Repbase analysis of NCC DNA-seq replicate (rep6). Repeat loci plotted according to log fold-change in streptavidin pull-downs (IP) versus input and significantly enriched repeats (P < 0.05) color coded according to repeat type. ALR = alpha repeat, CCCTAA = telomere repeat, BSR = beta satellite repeat. (D) Proportion of replicated transcripts in the entire short NCC RNA-seq transcriptome (all short) and in short hits.
On the other hand, long hit repeats and NAs showed a strong overlap with replicated regions compared to the overlap in the entire long NCC RNA-seq transcriptome (Fig. 4B). Most of the long hits were GA-rich. To find a common pattern in these sequences, we measured their C/G and GA/CT content, which showed that the vast majority of sequences had >40% C/G, >80% CT/GA content and that 99% were 199 nt or longer (Supplemental Fig. S4A–C). Based on this, we searched the genome for similar sequence compositions and identified ∼10,000 regions across all chromosomes (GA regions, Supplemental Table S5). Of these, 404 overlap with 435 long hit repeats and NAs and ∼37% are replicated in mid/late S-phase (Supplemental Fig. S4D). This high number of replicated regions, however, does not show evidence of GA regions being specifically replicated at the time of labeling since the replicated loci in the NCC DNA-seq data set covers ∼37% of the genome.
Mapping the NCC DNA-seq data to Repbase showed alpha, BSR centromeric and TERRA repeats were significantly enriched, indicating that these repeats were replicated at the time of labeling (Fig. 4C; Supplemental Table S6). This is in accordance with previous studies, which shows that pericentromeric and centromeric regions are replicated in mid/late S-phase (O'Keefe et al. 1992; Erliandri et al. 2014). Similarly, repeats and NA regions enriched in short hits overlap >90% with replicated loci identified in NCC DNA-seq, strongly suggesting that NCC-RNA enriched centromeric, pericentromeric and TERRA repeats, are replicated at the time of labeling (Fig. 4D). This suggests that repeat RNAs commonly enriched in N, M2, and M10 samples are associated with biotin-dUTP labeled chromatin in cis. It also suggests that the RNA is either being transcribed during locus replication, or reassociates with its locus after fork passage, and that this association persists at least 10 h after replication. The latter could be driven by the reassociation of RNA containing complexes with chromatin early in chromatin restoration.
NCC-enriched repeat RNAs do not form R-loops but coding genes prone to R-loop formation are enriched at replication forks
R-loops are DNA:RNA hybrids that typically form when a transcribed sequence invades and anneals to the complementary DNA strand in cis (Aguilera and Garcia-Muse 2012). G-rich RNA is prone to make R-loops, and alpha and TERRA sequences have been shown to facilitate specific functions via R-loop formation (Roy et al. 2008; Reddy et al. 2011; Balk et al. 2013; Arora et al. 2014; Groh et al. 2014; Kabeche et al. 2018; Lee et al. 2018). We thus wanted to investigate if the repeat RNAs enriched at nascent, 2 h and 10 h mature DNA could be associating with their parental locus in cis via R-loop formation. To do this, we reanalyzed published DRIP-seq data sets by applying the above-mentioned iterative repeat and reference RNA-seq mapping protocol (Hamperl et al. 2017).
The Repbase analysis confirmed TERRA as R-loop forming, in accordance with previous studies (Fig. 5A; Lee et al. 2018). The alpha repeats did not show any enrichment in the R-loop data, suggesting that alpha repeats are not associating with chromatin via R-loops in S- and G1-phase. Since mitotic cells are a small population in asynchronous cell cultures, our data do not dismiss alpha repeat R-loop formation in mitosis, which has previously been reported (Kabeche et al. 2018). Also, in accordance with previous studies, only “GAA” out of the simple dinucleotide repeats found in both the long and short hits showed statistical significance for R-loop formation, indicating that the GA-rich sequences in general do not form R-loops (Grabczyk et al. 2007; Reddy et al. 2011; Groh et al. 2014). These findings were further strengthened by the genome analysis that showed surprisingly little overlap of long and short hit repeats with R-loop forming regions (Fig. 5B,C). Of the 309 nascent hit genes, 154 were identified in the R-loop data, which included half of the few replicated genes (Fig. 5D). This proportion of R-loop forming genes was expected given that half of all expressed genes from the long RNA-seq overlapped with R-loop forming regions: 3863 of 6867 expressed genes were significantly enriched in the DRIP-seq data.
FIGURE 5.
DRIP-seq analysis shows that repeat RNAs enriched in N, M2, and M10 samples do not form R-loops. (A) R-loop analysis of DRIP-seq data mapping to Repbase sequences. Repeats enriched in NCC short RNA-seq Repbase mapping (N, M2, and M10 vs. controls) are highlighted with colors according to repeat type, whereas repeats that are enriched in the DRIP-seq data set are additionally labeled with red text. ALR = alpha repeat, CCCTAA = telomere repeat. (B) Long hits split and colored according to transcript type. Bars indicate number of transcripts in long hits (all long hits) overlapping with NCC DNA-seq (replicated), DRIP-seq (R-loop forming), or both (replicated and R-loop). (C) Same as B but for short hits. (D) Forty-seven genes out of 309 (nascent hits) overlap with replicated regions whereas 154 are prone to R-loop formation; 22 genes are present in all three data sets (nascent hits, replicated loci, and R-loops).
DISCUSSION
In this work, we developed a method to investigate the interplay of RNAs with DNA replication and chromatin restoration. We used long- and short RNA sequencing and a dedicated bioinformatics pipeline to reveal that RNAs are associated with nascent and mature chromatin, followed by subsequent characterization of the nature of these RNAs (Fig. 1; Supplemental Fig. S1). The custom bioinformatics pipeline allowed us to investigate repeat reads normally discarded from analysis due to multimapping and their enrichment in different data sets such as the NCC RNA-seq, NCC DNA-seq, and DRIP-seq data.
In the long RNA sequencing we identified transcripts arising from genes that are significantly enriched at nascent chromatin, albeit at moderate abundance levels (Fig. 2A; Supplemental Table S1). A subtle enrichment of a transcribed gene at a replication fork could be explained by replication-transcription collisions, which can prevent release of the transcribed product. This is also concordant with the fact that only a small proportion of active replication forks would cover a single gene during 15 min labeling in mid S-phase, making a high enrichment unlikely. However, the majority of enriched genes are not replicated at the time of labeling, suggesting that they associate with replication forks in trans (Fig. 4A). It is imaginable that the enriched RNAs associate with replication forks due to specific RNA folds common between the otherwise diverse transcripts, but we are unable to address this hypothesis due to current limitations in computational tools. Thus, our study does not support a model where specific RNAs serve general functions during active DNA replication, but does not rule out site-specific functions such as epigenetic or transcriptional regulation. Interactions in trans could be envisioned through association with proteins, triple-helix formation or R-loop formation. There is, however, no overrepresentation of nascent hit genes in R-loop data compared to all transcribed genes (Fig. 5D).
We found three types of repeats associated with mid S-phase replicated DNA, associations that appear to be maintained into G1. GA-rich repeat RNAs were most strongly enriched in the long RNA-seq whereas alpha repeat RNAs were identified in the short RNA-seq and TERRA RNA in both (Figs. 2, 3; Supplemental Figs. S2, S3; Supplemental Tables S1–S4). We found the repeats and NA transcripts from the short RNA-seq to map to centromeric, pericentromeric and telomeric regions with most repeats mapping to Chr 16, 21, and Y. Repetitive and centromeric regions are difficult to sequence due to their size, repetitive nature, and polymorphism, and although hg38 does have an improved annotation of these areas compared to hg19, it is based on a “reference model” (Miga et al. 2014; Rosenbloom et al. 2015). This, and the fact that HeLa cells have a variable number of normal and abnormal chromosomes, may lead to a skewed mapping of repeats to chromosomes. The observed enrichment of repetitive RNAs could be explained by the sheer abundance of repetitive DNA in the genome. However, many other types of repeats are replicated at the time of labeling and transcribed, such as LINE and Alu repeats, which are not enriched in our NCC RNA-seq data (Supplemental Table S6). This suggests that the observed repeat RNAs are indeed retained in chromatin after being transcribed and copurifying with labeled DNA. This could be as a consequence of improper release of these specific types of repeat RNAs or interaction with chromatin either via protein or DNA binding.
Alpha repeat transcription and stalling of Pol II has been suggested to contribute to chromatin establishment and function of the centromeres via interaction and recruitment of histones and histone modifiers (Bergmann et al. 2011; Chan et al. 2012; Biscotti et al. 2015; Catania et al. 2015; Blower 2016; Molina et al. 2016). TERRA RNAs have likewise been proposed to facilitate chromatin compaction and play an important role in telomere maintenance during replication, but their transcription is believed to be initiated in less heterochromatic subtelomeric regions (Azzalin et al. 2007; Schoeftner and Blasco 2008; Deng et al. 2009; Porro et al. 2014; Rippe and Luke 2015; Montero et al. 2018). We found alpha and TERRA repeat RNAs to be associated with newly replicated DNA shortly after fork passage and the association to be maintained for at least 10 h after replication (Fig. 4). This suggests that alpha and TERRA RNAs are transcribed and inefficiently released, possibly due to Pol II stalling, on centromeres, pericentromeres and telomeres shortly after chromatin disruption by the replication fork. How transcription occurs or is even initiated in heterochromatin regions and whether stalling of Pol II and transcript retention is important for both repeat RNAs functions, is not known. Alpha RNA has been shown to induce chromatin changes via R-loop formation, which is also a well-known mechanism for TERRA RNA during alternative telomere lengthening (Balk et al. 2013; Castellano-Pozo et al. 2013; Pfeiffer et al. 2013; Arora et al. 2014; Velazquez Camacho et al. 2017; Kabeche et al. 2018; Lee et al. 2018). We found alpha repeat-containing RNAs to have little overlap with R-loop data, as opposed to TERRA RNAs, suggesting that alpha repeats do not form R-loops in general (Fig. 5A–C). It is also possible that some repeat RNAs are bound in complexes with chromatin regulators that reassociate with replicated DNA during chromatin maturation. Regardless, it is intriguing that repeat RNAs are present already at a very early time point after DNA replication, arguing that they can contribute to later steps in chromatin restoration.
We report a considerable enrichment for GA-rich RNA sequences in replicated DNA shortly (15 min) after replication fork passage and a maintained enrichment into G1 phase (10 h) (Fig. 4). Only a fraction of the genomic GA regions that match the sequence composition of long hit repeats and NA sequences, and overlap with replicated regions, are present in long hits (404/3802) (Supplemental Fig. S4). It is currently unknown if the remaining GA regions are transcribed or transcribed and then rapidly degraded. However, we cannot rule out that the actual number of genomic regions resembling the GA-rich hits is lower than 10,000 since our search was based on nucleic acid composition and not specific sequence motifs.
A particular property of GA-rich RNA sequences is their incapacity to form stable secondary structures. However, sequences with several (four or more) stretches of Gs (may also be interspersed with other nucleotides), have the ability to form tertiary structures known as G-quadruplexes (G4) (Gellert et al. 1962; Mukundan and Phan 2013). If loops and bulges are taken into consideration, all long hit repeat sequences are highly likely to produce G4 structures. By forming G4 structures the RNA would become more stable, which could explain why the GA-rich RNAs are not degraded compared to any other repeat type in the genome. Although GA-rich sequences have been reported to induce R-loop formation, we do not see R-loops form at the GA-rich RNA hits’ loci (Figure 5B,C; Roy et al. 2008). It is likely that the GA-rich regions form G4 structures both as RNA and DNA. If encountered by the replication fork, this could cause significant reduction in fork speed and potentially lead to replication stress (Mirkin and Mirkin 2007; Sabouri et al. 2014). A reduction in replication fork speed would increase the likelihood of the replicated locus to be labeled with biotin dUTP, which could explain the large number of GA-rich RNAs replicated at the time of labeling. Whether replication forks are stalling at GA-rich loci due to RNA or DNA G4 structure formation remains to be investigated.
Telomeres and centromeres are organized into heterochromatin, whereas GA-rich RNAs are often intergenic and lack active promoter signatures, raising the question of how these repeats are transcribed. Since we see repeat RNAs being associated with DNA shortly after fork passage (15 min), we hypothesize that DNA replication, due to incorporation of acetylated new histones and dilution of histone PTMs (Alabert and Groth 2012), may provide a window of opportunity for transcription in heterochromatin regions. These transcripts may be prone to degradation by the exosome as shown with PROMTs (Preker et al. 2008). However, if the RNA remains stably bound to chromatin (e.g., forming stable structures such as G4) it might be protected against degradation and thus accumulate. We speculate that centromeric, pericentromeric, TERRA and GA repeats, transcribed as a consequence of DNA replication and chromatin disruption, remain continuously present at their locus to perform vital functions such as in chromatin maintenance. It is unknown whether GA-rich repeat RNA serves a function at their locus in cis, and it will be interesting to investigate this in the future.
MATERIALS AND METHODS
Nascent chromatin capture
HeLa S3 cells were grown in spinner flasks with DMEM (Gibco-31966-047) including 10% FBS and 1% Pen/Strep. The NCC protocol was developed with minor changes from the original protocol (Alabert et al. 2014). Cells were grown in normal media and were cross-linked after 15 min, 2 or 10 h from the time of biotin-dUTP addition with 1% Formaldehyde for 15 min. All buffers had a pH of 7.5 and were prepared with DEPC treated water. Samples were snap-frozen before sonication of chromatin in a Diagenode Bioruptor (4°C, 30 s ON/90 sec OFF 30× cycles on high) and checked for correct size on an agarose gel. Sonicated samples were precleared before immunoprecipitation (IP) with Protein A dynabeads (Invitrogen 100-02D) by rotating 1 h at 4°C. Biotinylated chromatin was isolated with T1 streptavidin magnetic beads (Invitrogen 656-01) in the presence of RNase inhibitors (100 U/mL) (NEB M0314L) and decross-linked in TE buffer (incl. 0.5% SDS and Proteinase K) at 37°C for 10 h, followed by 6 h at 65°C (interval shaking) for RNA isolation or 40 min boiling in LSB buffer for protein isolation.
Cell sorting
Cells were fixed with 70% ethanol and left at 20°C for minimum 12 h. The cells were washed with six volumes of PBS-BSA (1%) before staining with a propidium iodide solution (10 μg/ml PI, 0.02 mg/ml RNaseA in PBS) for 15 min at RT. The samples were analyzed on a BD FACS Calibur.
Western blotting
Samples were loaded with equal volumes onto a gel (NuPAGE Novex 4%–12% Bis-Tris Protein Gels). Gels were blotted onto Amersham Hybond C-extra membranes (RPN303E) at 100V for 1.5 h. Membranes were checked for successful transfer with ponceau stain (Sigma-Aldrich 81462-1L), then blocked with 5% milk-PBS-tween for 30 min, before addition of antibody (PCNA: Abcam ab29, H3K9me3: Abcam ab8898) and overnight incubation at 4°C. Membranes were washed for 5 min in PBS-tween three times, incubated with secondary antibody for 1.5 h at room temperature and subsequently washed for 5 min in PBS-tween another three times. Blots were developed by addition of Chemiluminescent substrate (Thermo-Scientific 34078). For low abundance protein visualization, Femto (34095) was added in different proportions to the Chemiluminescent substrate mix before film exposure and development (GE Healthcare Amersham Hyperfilm ECL or AGFA HealthCare—CURIX Ortho HT-G Film).
RNA extraction
RNA was purified using a miRNeasy Purification Kit with on-column DNase digestion (QIAGEN, Cat No./ID: 217004) using manufacturer recommendations for low RNA amounts. RNA for the long RNA-seq was further purified and concentrated using a Zymo Clean and Concentrator-5 kit (Zymo Research) as per manufacturer recommendations.
RNA quality control prior to cDNA library construction
RNA quality was assessed on a Bioanalyzer 2100 instrument (Agilent Technologies) with an Agilent RNA Pico 6000 kit according to the manufacturer's recommendations. Samples were quantified on a Qubit 2.0 Fluorometer (Thermo Fisher Scientific) using an RNA HS assay. Where RNA concentrations were too low for the Qubit RNA HS assay, quantitation was determined directly from the Bioanalyzer assay.
Library preparation
Long RNA-seq: 10 ng of total RNA was used as input material for library preparation using the SMARTer Stranded Total RNA-Seq Kit—Pico Input Mammalian (Takara Bio, USA formerly Clontech Laboratories) according to manufacturer's instructions with the following modifications: Input and H3 samples were fractionated for 2 min while all other RNA samples were not fractionated given their shorter fragment size, reflected by RNA integrity numbers (RIN) of 2–3. Final library PCR amplification cycles were increased to 15 cycles. A negative control indicated no adverse effect to increased PCR cycles.
Short RNA-seq: 10 ng RNA was used for library preparation with the Diagenode CATS small RNA-seq according to manufacturer's protocol, using 10 PCR cycles. Library size selection was performed with Agencourt AMPure XP beads (Beckman Coulter) using a 1.4 volume ratio to remove primer-dimers.
Quantification and quality control of cDNA libraries
Indexed DNA libraries were analyzed individually using a Bioanalyzer 2100 instrument with a DNA High Sensitivity assay. Average library size was 392 bp (long RNA-seq) and 228 bp (short RNA-seq). Quantitation of DNA libraries was determined using a Qubit 2.0 Fluorometer (Thermo Fisher Scientific) using a DNA HS assay.
Long RNA-seq: Libraries were pooled into two groups at equimolar concentrations. Input and H3 samples were pooled into a single group and remaining samples into a separate group. Pooled libraries were analyzed using a LabChip GX instrument and DNA High Sensitivity Assay kit according to the manufacturer's instructions (PerkinElmer). PCR-competent library DNA concentration was verified using the universal KAPA Library Quantification Kit for Illumina Sequencing Platforms according to the manufacturer recommendations (KAPA Biosystems). An Applied Biosystems QuantStudio 7 Real-Time PCR machine (Life Technologies) was used for quantitative real-time PCR.
Short RNA-seq: Libraries were pooled according to experimental replicates with eight samples at equimolar concentrations including 2% PhiX (Illumina).
Sequencing
Long RNA-seq: Total RNA sequencing was performed using the Illumina HiSeq2500 platform in high output mode with version 4 chemistry for cluster generation and a paired-end 125 bp run configuration. Each library pool was run across a single lane.
Short RNA-seq: Multiplexed samples were run on an Illumina Nextseq500 with 75 single-end configuration.
Bioinformatics
Long RNA-seq: Reads were trimmed using trimmomatic version 0.32 with parameters “ILLUMINACLIP:/path_to_adapter.fa:2:30:5 SLIDINGWINDOW:8:25 MINLEN:50 HEADCROP:8”. Reads were mapped using RNA STAR version 2.6.0b and SAMtools version 1.8 “‐‐outFilterMultimapNmax 10 ‐‐outFilterMismatchNoverReadLmax 0,2 ‐‐outFilterScoreMinOverLread 0,8” to hg38 and PCR duplicates removed using RmDup version 0.1.19 (Li et al. 2009; Li 2011; Dobin et al. 2013; Bolger et al. 2014). BAM files were converted to BigWig, ucsc-bedgraphtobigwig (version 357) and BEDTools (version 2.27.1), and processed with DERfinder tool version 1.8.5 for annotation-free expressed region identification and counting (cutoff = 5, L = 126) (Quinlan and Hall 2010; Frazee et al. 2014; Quinlan 2014; Collado-Torres et al. 2017a,b). Differential expression statistics were done in EdgeR version 3.1 using an expressed region cutoff of >5 global counts in >1 sample (Robinson et al. 2010; McCarthy et al. 2012). One of the nascent sample libraries was removed because of high background, as evidenced from PCA analysis (Supplemental Fig. S1C). Overlap of expressed regions with gene and repeat annotations from UCSC “hg38_knownGene” and “hg38_rmsk” was performed using BEDTools version 2.24 intersect intervals and where regions overlapped with multiple annotations, the annotation with the longest overlap was chosen.
Transcriptome assembly: We ran Trinity transcriptome assembly on the long RNA-seq with parameters: Trinity ‐‐seqType fq ‐‐left reads_1.fq ‐‐right reads_2.fq ‐‐SS_lib_type FR ‐‐CPU 6 ‐‐quality_trimming_params “ILLUMINACLIP:/path_to_adapters.fa:2:30:5 SLIDINGWINDOW:8:25 MINLEN:50 HEADCROP:8 ‐‐min_kmer_cov 2 ‐‐bflyHeapSpaceMax 14G ‐‐bflyGCThreads 2 ‐‐bflyCPU 2” (Grabherr et al. 2011).
Short RNA-seq: Reads were trimmed according to the CATS protocol recommendations and mapped with RNA-STAR “ ‐‐outFilterMultimapNmax 10 ‐‐outFilterMismatchNoverReadLmax 0,2 ‐‐outFilterScoreMinOverLread 0,8 ‐‐outSAMunmapped Yes” to repeats (Repbase version 22.09) and quantified using IdxStats (SAMtools Version 1.2) (Jurka 1998, 2000; Jurka et al. 2005; Dobin et al. 2013; Bao et al. 2015). Unmapped reads were processed using BAM-toSAM, Filter SAM (SAMtools Version 1.2) “Type: The read is unmapped FLAG: Yes”, SAM to FASTQ picard Version 1.56.0 “Single or paired end: single, Rereverse bases and qualities of reads on negative strand: True”, before they were mapped to hg38 using RNA STAR Version 2.6.0b and SAMtools Version 1.8 “‐‐outSAMunmapped No ‐‐outFilterMultimapNmax 10 ‐‐outFilterMismatchNoverReadLmax 0,2 ‐‐outFilterScoreMinOverLread 0,8”. BAM files were converted to BigWig (ucsc-bedgraphtobigwig Version 357 and BEDTools Version 2.27.1) and processed in DERfinder tool Version 1.8.5 for annotation free Expressed region identification and counting (cutoff = 5, L = 76). Both data sets were analyzed in EdgeR Version 3.1 using an expressed region cutoff of >5 global counts in >1 samples. Repeat masking of NA hits was performed using CENSOR version and repeats with the highest score toward an NA region were selected (http://www.girinst.org/censor/index.php) (Jurka et al. 1996; Kohany et al. 2006).
NCC DNA-seq: Reads were first mapped to repeats (Repbase) using Bowtie 2 bowtie2 (version 2.2.6 and SAMtools version 1.2) with options “–L 20 –i S,1,0.5 ‐‐end-to-end –D 20 –R 10 ‐‐non-deterministic” and counted with SAMtools idxstats (Langmead and Salzberg 2012; Langmead et al. 2009). Subsequent analysis was performed in EdgeR with the two replicates analyzed separately against the input, setting the Biological Coefficient of Variation (BCV) manually to the calculated BCV for the NCC RNA-seq Repbase map (0.2119).
Unaligned reads were subsequently processed by duplication removal using RmDup and reads extended to 250 bp to better fit chromatin fragment size before being counted in genomic intervals of 250 bp (min overlap 0.5). Counts were adjusted to cpm in each replicate and replicate 6 and 7 summed before Input counts were subtracted in order to remove background. Peak calling was performed using MACS2 (macs2 version 2.1.0.20151222, numpy version 1.7.1, scipy version 0.12.0 and gnu_awk version 4.1.0) “bdgbroadcall ‐‐cutoff-peak 0.3 ‐‐cutoff-link 0.15 min_len 10000 max_gap 5000 max_link_gap 10000” (Zhang et al. 2008; Liu 2014).
DRIP-seq: Reads were mapped to Drosophila genome dm6 to filter out spike-in reads, unmapped reads were mapped to repeats and the rest of the reads mapped to hg38 reference sequence using Bowtie 2 “‐‐no-mixed True ‐‐no-discordant True ‐‐no-unal True ‐‐non-deterministic True”. Reads mapping to repeat sequences and hg38 were analyzed with the same pipeline as the short NCC RNA-seq, where repeats with less than two counts in three samples, and expressed regions (hg38 mapping) with less than 10 counts in any sample, were filtered out.
Enrichr analysis: Nascent hits (309 genes): http://amp.pharm.mssm.edu/Enrichr/enrich?dataset=f534544edac53e4b22daa24a89b86c66, M2 hits (96 genes): http://amp.pharm.mssm.edu/Enrichr/enrich?dataset=2925c372cc5a6a73a3f34d07df6d8ca4, M10 hits (218 genes): http://amp.pharm.mssm.edu/Enrichr/enrich?dataset=51559be99d0ddb399480a0cefda83380.
DATA DEPOSITION
Repeat data were obtained from the giri Repbase database where the repeat masking was also performed (https://www.girinst.org/Repbase/).
RNA STAR: https://github.com/alexdobin/STAR
SAMtools: https://github.com/samtools/samtools
Bowtie: http://bowtie-bio.sourceforge.net/bowtie2/index.shtml
MACS2: https://github.com/taoliu/MACS/
Trinity: https://github.com/trinityrnaseq/trinityrnaseq/wiki
DERfinder: https://www.bioconductor.org/packages/release/bioc/html/derfinder.html
Trimmomatic: http://www.usadellab.org/cms/?page=trimmomatic
UCSC: https://hgdownload.soe.ucsc.edu/gbdb/hg38/
EdgeR: https://bioconductor.org/packages/release/bioc/html/edgeR.html
All data sets are available on the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/): NCC RNA-seq: GSE139353 (Long RNA-seq: GSE139219, short RNA-seq: GSE139351), ChOR-seq (GSE110354), and DRIP-seq (GSE93368).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
COMPETING INTEREST STATEMENT
A.G. is co-founder and CSO of Ankrin Therapeutics.
Supplementary Material
ACKNOWLEDGMENTS
We thank Constance Alabert for establishing the NCC protocol and for help, together with Kyosuke Nakamura, to implement the protocol for RNA isolation. We furthermore thank Malte Thodborg, Sudeep Sahadevan, and Jens Vilstrup Johansen for help with valuable bioinformatics guidance and John Mattick for providing sequencing facilities and bioinformatic structure. Lastly, we thank the Karlene Cimprich laboratory for sharing the raw DRIP-seq data before publication and Stephan Hamperl and Michael Joseph Bocek for instrumental assistance during reanalysis of these data. Work in the Lund laboratory was supported by the Danish Council for Independent Research (Sapere Aude program) (4183-00179B), the Novo Nordisk Foundation (NNF18OC0030656, NNF17OC0028620), the Lundbeck Foundation, and the Danish Cancer Society (R124-A7493, R204-A12532). C.G-A. was supported by post.doc fellowships from the Lundbeck Foundation and research in the Groth laboratory was supported by the European Research Council (ERC2011StG, no. 281765), the Novo Nordisk Foundation (NNF14CC0001), the Danish Council for Independent Research (7016-00042B), and the Lundbeck Foundation (R198-2015-269).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.074757.120.
REFERENCES
- Aguilera A, Garcia-Muse T. 2012. R loops: from transcription byproducts to threats to genome stability. Mol Cell 46: 115–124. 10.1016/j.molcel.2012.04.009 [DOI] [PubMed] [Google Scholar]
- Alabert C, Groth A. 2012. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 13: 153–167. 10.1038/nrm3288 [DOI] [PubMed] [Google Scholar]
- Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. 2014. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 16: 281–293. 10.1038/ncb2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C, Barth TK, Reveron-Gomez N, Sidoli S, Schmidt A, Jensen ON, Imhof A, Groth A. 2015. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev 29: 585–590. 10.1101/gad.256354.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato AT. 2012. Assembling chromatin: the long and winding road. Biochim Biophys Acta 1819: 196–210. 10.1016/j.bbagrm.2011.07.005 [DOI] [PubMed] [Google Scholar]
- Annunziato AT. 2015. The fork in the road: histone partitioning during DNA replication. Genes (Basel) 6: 353–371. 10.3390/genes6020353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM. 2014. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun 5: 5220 10.1038/ncomms6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801. 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B. 2013. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol 20: 1199–1205. 10.1038/nsmb.2662 [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA 6: 11 10.1186/s13100-015-0041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A. 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374. 10.1146/annurev.biochem.71.110601.135425 [DOI] [PubMed] [Google Scholar]
- Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. 2011. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J 30: 328–340. 10.1038/emboj.2010.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscotti MA, Canapa A, Forconi M, Olmo E, Barucca M. 2015. Transcription of tandemly repetitive DNA: functional roles. Chromosome Res 23: 463–477. 10.1007/s10577-015-9494-4 [DOI] [PubMed] [Google Scholar]
- Blower MD. 2016. Centromeric transcription regulates Aurora-B localization and activation. Cell Rep 15: 1624–1633. 10.1016/j.celrep.2016.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M, Santos-Pereira JM, Rondon AG, Barroso S, Andujar E, Perez-Alegre M, Garcia-Muse T, Aguilera A. 2013. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell 52: 583–590. 10.1016/j.molcel.2013.10.006 [DOI] [PubMed] [Google Scholar]
- Catania S, Pidoux AL, Allshire RC. 2015. Sequence features and transcriptional stalling within centromere DNA promote establishment of CENP-A chromatin. PLoS Genet 11: e1004986 10.1371/journal.pgen.1004986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catasti P, Gupta G, Garcia AE, Ratliff R, Hong L, Yau P, Moyzis RK, Bradbury EM. 1994. Unusual structures of the tandem repetitive DNA-sequences located at human centromeres. Biochemistry 33: 3819–3830. 10.1021/bi00179a005 [DOI] [PubMed] [Google Scholar]
- Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Choo KHA, Wong LH. 2012. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl Acad. Sci 109: 1979–1984. 10.1073/pnas.1108705109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christov CP, Gardiner TJ, Szuts D, Krude T. 2006. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol Cell Biol 26: 6993–7004. 10.1128/MCB.01060-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Torres L, Jaffe AE, Leek JT. 2017a. derfinder: Annotation-agnostic differential expression analysis of RNA-seq data at base-pair resolution via the DER Finder approach. http://bioconductor.org/packages/release/bioc/html/derfinder.html 10.18129/B9.bioc.derfinder [DOI]
- Collado-Torres L, Nellore A, Frazee AC, Wilks C, Love MI, Langmead B, Irizarry RA, Leek JT, Jaffe AE. 2017b. Flexible expressed region analysis for RNA-seq with derfinder. Nucleic Acids Res 45: e9 10.1093/nar/gkw852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. 2009. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35: 403–413. 10.1016/j.molcel.2009.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erliandri I, Fu H, Nakano M, Kim JH, Miga KH, Liskovykh M, Earnshaw WC, Masumoto H, Kouprina N, Aladjem MI, et al. 2014. Replication of alpha-satellite DNA arrays in endogenous human centromeric regions and in human artificial chromosome. Nucleic Acids Res 42: 11502–11516. 10.1093/nar/gku835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazee AC, Sabunciyan S, Hansen KD, Irizarry RA, Leek JT. 2014. Differential expression analysis of RNA-seq data at single-base resolution. Biostatistics 15: 413–426. 10.1093/biostatistics/kxt053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Lin H. 2014. Noncoding RNAs in the regulation of DNA replication. Trends Biochem Sci 39: 341–343. 10.1016/j.tibs.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M, Lipsett MN, Davies DR. 1962. Helix formation by guanylic acid. Proc. Natl Acad. Sci. 48: 2013–2018. 10.1073/pnas.48.12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabczyk E, Mancuso M, Sammarco MC. 2007. A persistent RNA·DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res 35: 5351–5359. 10.1093/nar/gkm589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh M, Lufino MM, Wade-Martins R, Gromak N. 2014. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and Fragile X syndrome. PLoS Genet 10: e1004318 10.1371/journal.pgen.1004318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. 2017. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 170: 774–786 e719. 10.1016/j.cell.2017.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WL, Straight AF. 2017. RNA-mediated regulation of heterochromatin. Curr Opin Cell Biol 46: 102–109. 10.1016/j.ceb.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. 1998. Repeats in genomic DNA: mining and meaning. Curr Opin Struct Biol 8: 333–337. 10.1016/S0959-440X(98)80067-5 [DOI] [PubMed] [Google Scholar]
- Jurka J. 2000. Repbase Update: a database and an electronic journal of repetitive elements. Trends Genet 16: 418–420. 10.1016/S0168-9525(00)02093-X [DOI] [PubMed] [Google Scholar]
- Jurka J, Klonowski P, Dagman V, Pelton P. 1996. Censor—a program for identification and elimination of repetitive elements from DNA sequences. Comput Chemi 20: 119–121. 10.1016/S0097-8485(96)80013-1 [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467. 10.1159/000084979 [DOI] [PubMed] [Google Scholar]
- Kabeche L, Nguyen HD, Buisson R, Zou L. 2018. A mitosis-specific and R loop–driven ATR pathway promotes faithful chromosome segregation. Science 359: 108–114. 10.1126/science.aan6490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7: 474 10.1186/1471-2105-7-474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YW, Arora R, Wischnewski H, Azzalin CM. 2018. TRF1 participates in chromosome end protection by averting TRF2-dependent telomeric R loops. Nat Struct Mol Biol 25: 147–153. 10.1038/s41594-017-0021-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. 2014. Use Model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein–DNA interactions in embryonic stem cells. Methods Mol Biol 1150: 81–95. 10.1007/978-1-4939-0512-6_4 [DOI] [PubMed] [Google Scholar]
- Marchal C, Sima J, Gilbert DM. 2019. Control of DNA replication timing in the 3D genome. Nat Rev Mol Cell Biol 20: 721–737. 10.1038/s41580-019-0162-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali M. 2010. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol 11: 728–738. 10.1038/nrm2976 [DOI] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ. 2014. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res 24: 697–707. 10.1101/gr.159624.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. 2007. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 71: 13–35. 10.1128/MMBR.00030-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina O, Vargiu G, Abad MA, Zhiteneva A, Jeyaprakash AA, Masumoto H, Kouprina N, Larionov V, Earnshaw WC. 2016. Epigenetic engineering reveals a balance between histone modifications and transcription in kinetochore maintenance. Nat Commun 7: 13334 10.1038/ncomms13334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero JJ, Lopez-Silanes I, Megias D, Castells-Garcia FFMA, Blasco MA. 2018. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat Commun 9: 1548 10.1038/s41467-018-03916-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukundan VT, Phan AT. 2013. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J Am Chem Soc 135: 5017–5028. 10.1021/ja310251r [DOI] [PubMed] [Google Scholar]
- O'Keefe RT, Henderson SC, Spector DL. 1992. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol 116: 1095–1110. 10.1083/jcb.116.5.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer V, Crittin J, Grolimund L, Lingner J. 2013. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J 32: 2861–2871. 10.1038/emboj.2013.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J, Lingner J. 2014. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun 5: 5379 10.1038/ncomms6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854. 10.1126/science.1164096 [DOI] [PubMed] [Google Scholar]
- Quinlan AR. 2014. BEDTools: the Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics 47: 11.12.11–11.12.34. 10.1002/0471250953.bi1112s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. 2016. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17: 47–62. 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- Reddy K, Tam M, Bowater RP, Barber M, Tomlinson M, Edamura KN, Wang YH, Pearson CE. 2011. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res 39: 1749–1762. 10.1093/nar/gkq935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveron-Gomez N, Gonzalez-Aguilera C, Stewart-Morgan KR, Petryk N, Flury V, Graziano S, Johansen JV, Jakobsen JS, Alabert C, Groth A. 2018. Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication. Mol Cell 72: 239–249.e235. 10.1016/j.molcel.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K, Luke B. 2015. TERRA and the state of the telomere. Nat Struct Mol Biol 22: 853–858. 10.1038/nsmb.3078 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. 2015. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 43: D670–D681. 10.1093/nar/gku1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Strahl BD. 2014. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta 1839: 627–643. 10.1016/j.bbagrm.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Yu K, Lieber MR. 2008. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol 28: 50–60. 10.1128/MCB.01251-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouri N, Capra JA, Zakian VA. 2014. The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage. BMC Biol 12: 101 10.1186/s12915-014-0101-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236. 10.1038/ncb1685 [DOI] [PubMed] [Google Scholar]
- Stewart-Morgan KR, Reveron-Gomez N, Groth A. 2019. Transcription restart establishes chromatin accessibility after DNA replication. Mol Cell 75: 408–414. 10.1016/j.molcel.2019.06.035 [DOI] [PubMed] [Google Scholar]
- Tagarro I, Fernandezperalta AM, Gonzalezaguilera JJ. 1994. Chromosomal localization of human satellite-2 and satellite-3 by a FISH method using oligonucleotides as probes. Hum Genet 93: 383–388. 10.1007/BF00201662 [DOI] [PubMed] [Google Scholar]
- Velazquez Camacho O, Galan C, Swist-Rosowska K, Ching R, Gamalinda M, Karabiber F, De La Rosa-Velazquez I, Engist B, Koschorz B, Shukeir N, et al. 2017. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:dNA hybrid formation. eLife 6: e25293 10.7554/eLife.25293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.