Abstract
Identification of optimal schedules for combination drug administration relies on accurately estimating the correct pharmacokinetics, pharmacodynamics, and drug interaction effects. Misspecification of pharmacokinetics can lead to wrongly predicted timing or order of treatments, leading to schedules recommended based on incorrect assumptions about absorption and elimination of a drug and its effect on tumor growth. Here we developed a computational modeling platform and software package for combination treatment strategies with flexible pharmacokinetic profiles and multidrug interaction curves that are estimated from data. The software can be used to compare pre-specified schedules based on the number of resistant cells where drug interactions and pharmacokinetic curves can be estimated from user-provided data or models. We applied our approach to publicly available in vitro data of treatment with different tyrosine kinase inhibitors of BT-20 triple-negative breast cancer cells and of treatment with erlotinib of PC-9 non-small cell lung cancer cells. Our approach is publicly available in the form of an R package called ACESO (https://github.com/Michorlab/aceso) and can be used to investigate optimum dosing for any combination treatment.
Keywords: Mathematical Modeling, Cancer Evolution, Pharmacometrics, Drug Resistance
Introduction
A major obstacle in cancer research is the development of resistance to anticancer drugs (1,2). Although this phenomenon has long been studied, it has gained even more attention after the introduction of targeted therapies and technological advances such as next generation DNA/RNA sequencing (3–5). Targeted therapy differs from traditional cytotoxic chemotherapy in that it not only leads to more specific effects with reduced toxicity, but also promises a future of personally tailored treatment. Unfortunately, the initial clinical response to targeted therapies is almost always temporary, as acquired resistance to these drugs frequently develops (2,3).
Clonal evolution represents a key element for understanding and predicting the dynamics of resistance (5,6). In this context, cancer cells evolve specific (epi)genetic alterations during tumor growth that lead to the outgrowth of different subclones within a tumor cell population. This intratumor heterogeneity has important negative implications for targeted therapies because somatic mutations continue to occur even in the presence of drugs, leading to increased aggressiveness of the cancer as new drug-resistant cells with improved malignant potential emerge (1). Moreover, resistance might also be due to therapy-induced selection of a small resistant subpopulation of cells that was already present in the original tumor (7).
This realization has given rise to many stochastic mathematical models of genetic resistance, which consider the emergence of these alterations as random events during cell division (8–13). Goldie and Coldman presented some of the earliest work in the 1980s (14,15), proposing the use of branching process models to study pre-existing or acquired resistance to chemotherapy in tumor cell populations. Branching process models are a class of stochastic models that describe the growth and composition of populations made up of stochastically reproducing individuals (16,17). A subclass of these models is referred to as birth-death processes because each cell experiences a random birth or death event. Multi-type branching processes are convenient for modeling clonal evolution of tumor cells, as during the division process the emergence of random (epi)genetic alterations can give rise to tumor subclones with different fitness (i.e. proliferative capacity or resistance to apoptosis) than their ancestors (18). This approach has inspired other groups to mathematically characterize drug resistance dynamics and to investigate potential administration schedules that could delay the emergence of resistance (19–21). For instance, Hata et al. (7) used a branching process to model gefitinib-tolerant and gefitinib-resistant/persistent clones in epidermal growth factor receptor (EGFR)-mutant non-small-cell lung cancer and showed that acquired resistance can occur either by de novo acquisition of the EGFR-T790M mutation or by expansion of pre-existing EGFR-T790M-positive subclones under the selective pressure of gefitinib therapy. Branching process models have also been used to study multi-drug resistance (11,13) and the dynamics of metastasis formation (12,22).
We previously developed a comprehensive computational strategy to explore the evolutionary dynamics of heterogeneous tumor cell populations while taking pharmacokinetic and drug interaction effects into account (23–25). This approach consists of a cell-level description of the changes in sensitive and resistant cells over time and in response to treatment, using a birth-death-mutation process to model population dynamics. Our framework represents a departure from previously developed models where the birth and death rates of sensitive and resistant cells were not directly influenced by drug concentrations and were assumed to be constant over time. Additionally, the pharmacokinetic processes included in our previous work (20,24–26) were exponential declines of drug concentrations, a modeling choice that might not hold for all treatment and cell types. Furthermore, in vitro growth inhibition response of cells to a range of different drug concentrations is generally non-linear, which needs to be incorporated in a flexible framework (27,28).
In this paper, we developed a generalized modeling methodology that accounts for complex pharmacokinetic models and non-linear effects of drugs on the growth and death kinetics of tumor cells. We integrated this framework in an R package called ACESO (A Cancer Evolution Schedule Optimizer), providing users with an accessible tool to rationally identify optimum single-agent and combination treatment administration strategies for oncogene-driven cancers. We demonstrate the use of ACESO to explore optimum dosing strategies using publicly available data from the Harvard Medical School (HMS) Library of Integrated Network-based Cellular Signatures (LINCS) database (29). Additionally, we demonstrate how the new features and flexibility of ACESO can identify optimum administration schedules for erlotinib in non-small cell lung cancer (NSCLC) (20,21) by extending the previous model to account for Central Nervous System (CNS) metastases. Within these models, calculating the probability of developing resistance is a standard feature in ACESO. Our multi-scale framework represents a crucial step towards making clinically relevant predictions as it considers the most important aspects governing treatment response and cancer evolution.
Material and Methods
Model structure
The underlying model in ACESO is a multi-type branching process with time-dependent rates, which consists of multiple different types of cells; see the Quick Guide to Equations and Assumptions and Supplementary Information (SI) for a detailed description of the model and mathematical equations, respectively. The tumor-initiating cells with oncogene-activating mutations, denoted type-0 cells, are referred to as sensitive cells, since they contain all (epi)genetic alterations necessary for conferring the tumor phenotype, but are sensitive to anti-cancer therapies. These alterations can be a point mutation, amplification, deletion, inversion, methylation or other epigenetic change, or any other alteration that arises at a certain rate per cell division. Type-0 cells proliferate and die with rates b0(t) and d0(t), respectively. These rates can be modulated by the cytostatic and/or cytotoxic effect exerted by the drug(s) as a function of their concentrations over time.
Type-0 cells accumulate (epi)genetic alterations at probability ui per cell division, hence at rate uibi(t), to generate new clones harboring specific resistance mechanisms. This parameter can be a constant value or be modified by the effects of drugs. Newly emerging resistant cell types (denoted type-i cells, with i = 1,2,...) are again characterized by their birth and death rates, bi(t) and di(t), respectively, which may also exhibit a concentration-dependent profile. Each of these resistant cell types can then accumulate further (epi)genetic alterations and become resistant to additional drugs. Fig. 1A shows a schematic representation of a four-type branching process with cross-resistance. The current implementation of ACESO is limited to combinations of two drugs due to the difficulty of discerning the effect of each drug in combinations of more than two agents as well as interaction kinetics between them.
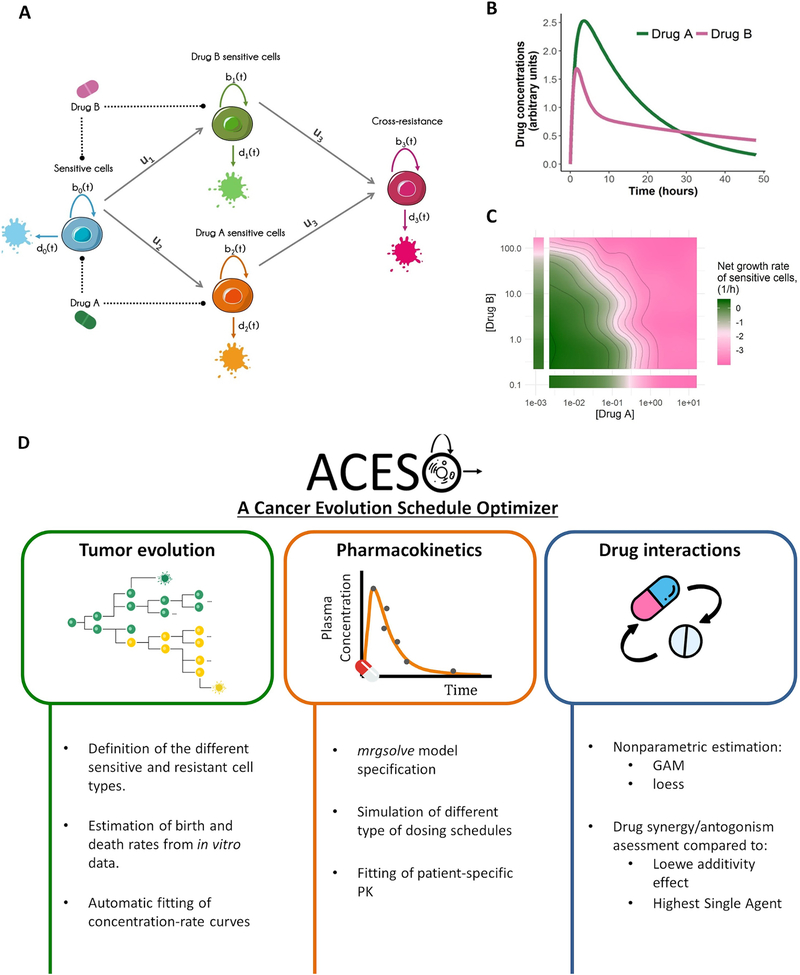
Figure 1.
Schematic representation of: A) a four-type branching process with cross-resistance. The type-0 cell (blue), which is sensitive to both drugs A and B, proliferates at rate b0(t), dies at rate d0(t) and can accumulate mutations at probability ui per cell division to give two different cell types, type-1 (green) and 2 (orange), which are sensitive to either drug A or drug B. Type-1 and 2 cells can mutate again and become resistant to both drug A and B (type 3 cells, magenta). Each resistant cell type has its own proliferation and death rates, bi(t) and di(t), with i = 1,2... B) Drug concentration-time profile governed by the pharmacokinetic parameters of drug A and B after a single-dose administration. C) Graphical evaluation of how the net growth rate of type-0 cells changes under the effects of different drug concentrations with a contour plot and isoboles (i.e. contour lines of equal drug effect). This representative example is based on the additive effect of the drugs in panel B with additional synergistic/antagonistic effects. D) Main functionalities of ACESO. PK refers to pharmacokinetics, GAM to Generalized Additive Models and loess to locally weighted scatterplot smoothing.
This quantitative approach considers situations in which there is preexisting resistance at the time of tumor diagnosis and treatment initiation as well as de novo resistance emerging during treatment. In previous work, we derived analytical solutions to calculate the expected number of cells of any type as a function of time and the probability of developing resistance after treatment initiation (24,25). These results are summarized in the Supplementary Information and allow for faster predictions without the need for stochastic simulations of the evolutionary process. This cell-level description of how heterogeneous cell populations evolve over time is then coupled to pharmacokinetic (PK) models, which describe how the drug concentrations change over time (Fig. 1B), and with information quantifying drug-drug interactions (Fig. 1C).
Estimation of birth and death rates
Data from in vitro cell viability and death assays are required to determine the growth and death rate parameters for each cell type under varying drug concentrations. Here, for each treatment condition (i.e. concentration level of one or more drugs) the corresponding rate constants of birth and death are estimated. One example of a viability assay that can be used to characterize cell proliferation is the MTS assay (30), where the number of viable cells is measured over time. This data is then used to identify a net growth rate constant assuming an exponentially growing cell population. The dynamics of tumor cell death can be characterized using apoptosis assays such as Annexin V/propidium iodide (PI) fluorescence-activated cell sorting (FACS) assays (31), where cells with positive Annexin V staining are considered dead. Our framework uses the fraction of dead cells resulting from these assays to determine the baseline death rate as well as the death rates in the presence of different drug concentrations (see SI for a detailed description). Finally, the net birth rate is calculated as the sum of the estimated cell growth and death rates for the different drug concentrations. Other assays for proliferation (e.g. KI67 protein assays (32)) and death (e.g. Caspase activation assays (33)) can also be used for rate estimation. The structure of an ideal database for ACESO is described in the SI.
Estimation of drug effect parameters
To estimate drug effects on birth and death rates, we perform the following steps: (i) In case of exposure to a single drug, ACESO includes several non-linear models (28) (see SI) that are fitted to the concentration-response data, which provides the values of the drug effect parameters. The selection of the best model is based on the lowest Akaike Information Criterion (AIC) value (34). (ii) The relationship between estimates of b or d and drug concentrations obtained from drug combination studies is established using nonparametric models (35,36). These methods do not assume any specific model structure a priori to fit the data, and thus, the birth/death rate vs. concentration profiles are described without assuming any mechanism of interaction, providing a large degree of flexibility.
Estimation of pharmacokinetic parameters
In order to implement more complex PK models and to simulate different routes of administration and dosing schedules, we integrated the mrgsolve R package (37) in ACESO, which allows for rapid simulations of ordinary differential equation-based PK models. When patient-specific PK data are available, user-defined or compartmental models available in ACESO using the mrgsolve model specification can be used to select the best model describing the drug concentrations over time. If data from different individuals are available, parameter estimates describing the median tendency of the data are reported, as estimation of inter-individual variability is not currently supported in ACESO. Selection between models is based on the minimum AIC value and inspection of the individual fits.
Assessing drug synergy/antagonism
ACESO helps to identify whether the combined action of two drugs is synergistic or antagonistic or whether there is no interaction. In order to quantify the degree of synergy/antagonism between two compounds, the most common approach is to compare the combination effect to a null reference model of no interaction. If the combination response is greater than what is expected from the reference model, the combination is classified as synergistic, while antagonism is defined as the combination producing less than the expected effect. There are several conventional approaches that define different null models to assess drug synergy/antagonism. The Loewe additivity model (38) quantifies a zero-interactive state for the combination of two drugs and defines synergy/antagonism as a combined inhibitory effect that is greater/lower than the sum of the individual effects of the drugs. Highest Single Agent (HSA), also known as Gaddum’s non-interaction model (39), is another popular model which defines “independent action” of drugs when the predicted effect of a combination is that of the most effective drug alone. According to this model, any combined effect stronger than the effect of a single drug is called synergism and a weaker effect antagonism. Additional information related to these models is provided in the SI. These two models are used in ACESO to define the pairwise drug combination effects. The Loewe/HSA scores reflect the difference between the measurement and the surface obtained under the no-interaction models, such that values less than zero represent antagonism and values greater than zero synergism.
Data availability
TNBC
Raw cell viability data used in the present analysis can be retrieved from the HMS LINCS Database (http://lincs.hms.harvard.edu/db/datasets/20259), where nine small molecule kinase inhibitors given in combination were chosen for analysis of combinatorial drug sensitivities in the BT-20 triple-negative breast cancer (TNBC) cell line (obtained from ATCC). Briefly, in these experiments cells were exposed during 72 hours to constant concentration levels of the drugs either as single agents or in combinations. Due to the lack of information regarding drug resistant cell lines in LINCS, we assumed that these cells had decreased birth and death rate parameters compared to their sensitive counterparts, and hence the proliferation and death rate parameters obtained for the parental BT-20 cell line were divided by different values depending on the drugs being analyzed and tested for sensitivity.
NSCLC
We used cell viability (MTS assay) and apoptosis (Annexin V/PI FACS assay) data from isogenic erlotinib-sensitive and erlotinib-resistant pairs of PC-9 NSCLC cell lines from a previous publication (40). In these experiments, the total numbers of viable and dead cells in culture were counted at 48, 60, and 72 hours after treatment initiation for different erlotinib concentrations and in the absence of the drug. In contrast to the TNBC data above, for the NSCLC data we used response kinetics to a single drug only and also did not have information on the growth patterns of resistant cell types.
Results
Using the building blocks outlined above, we created a multi-scale computational modeling platform that incorporates tumor evolution, pharmacokinetics and drug interaction dynamics (Fig. 1D). This computational platform is publicly available as an R package called A Cancer Evolution Schedule Optimizer (ACESO). This method can be applied to investigate best dose administration schedules for any combination treatment.
Optimum dosing strategies for TNBC
We analyzed the dynamics of heterogeneous TNBC cell lines during combination treatment. We obtained cell viability data resulting from the exposure of the BT-20 TNBC cell line to different concentrations of nine small molecule kinase inhibitors used as single-agents or in combination (see Data availability above). We focus our results section on the discussion of the combination of alpelisib and neratinib; results of analyses using all drug combinations in the database are shown in the SI. For the purpose of the model, BT-20 cells sensitive to both drugs are referred to as type-0 cells, whereas cells resistant to one of the drugs are defined as type-1 or type-2 cells.
Estimation of drug-sensitive cell growth and death rates
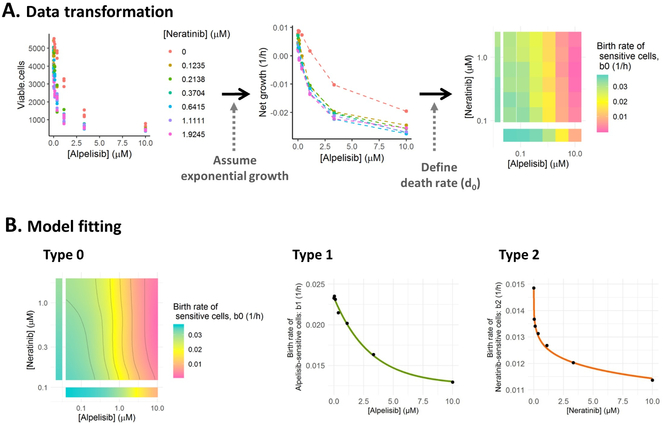
We first estimated tumor cell proliferation and death rates. To this end, we used an exponential model to characterize cell growth and estimated a net growth rate parameter for each treatment condition. Fig. 2A shows the relationship between viable cells measured 72h after exposure and the different concentration levels of the targeted therapies alpelisib and neratinib as well as the net growth rate estimates obtained from these measurements. Since no information regarding apoptosis assays was available to estimate the death rate of type-0 cells (d0) and assuming that these targeted therapies do not induce cell death per se, a different d0 value was defined for each concentration of drugs, which was equal to the minimum value needed to ensure that all birth rates of type-0 cells (b0) were positive regardless of the treatment condition. The values of b0 for the cells treated with alpelisib and neratinib are shown in Fig. 2A (right panel). These parameters were fitted using a Generalized Additive Model (GAM) to generate the predicted surface required for the simulation of the evolutionary process (Fig. 2B). GAMs flexibly model the relationship between the b0 values and each drug concentration by using a number of basis functions that can have a wide variety of shapes (36). The resulting isobolograms highlight points of equal drug contribution to the decrease of sensitive cell birth rates; Supplementary Fig. S1 shows the result of fitting GAMs to the b0 values obtained under additional drug combinations in our database.
Figure 2.
A) Estimation of the net growth rate of sensitive cells using drug combination data. 1) Viable cell counts under different drug concentrations of alpelisib and neratinib. 2) Net growth rate of sensitive cells as a function of the different drug concentrations. 3) Heatmap of type-0 birth rate parameters under different drug concentrations. B) Model fitting results for neratinib and alpelisib drug combination study, where the surface from type-0 (sensitive to both drugs) cell birth rates is characterized with a Generalized Additive Model (GAM) and the birth rates from resistant cell types (type-1 is neratinib-resistant and type-2 is alpelisib-resistant) are described with different non-linear models included in ACESO (exponential, Weibull, log-logistic…). The specific equation and parameter values for each curve are summarized in Table 1.
Estimation of drug-resistant cell growth and death rates
Once drug resistant cells have emerged from drug sensitive cells, they follow their own branching process model characterized by their corresponding birth and death rates. We considered situations in which drug concentrations did not have any influence on drug-resistant cell proliferation and therefore, these cells were only affected by the varying concentrations of the other drug in the combination. The birth rate-drug concentration profiles were then characterized using the non-linear models for single agents included in ACESO. Fig. 2B shows the fitting of neratinib-resistant (type-1) and alpelisib-resistant (type-2) cell birth rates, where an exponential decay model and a five parameter log-logistic function, respectively, best fit the data as indicated by the lowest AIC values (Table 1). The non-linear models used to characterize the other cell types in our database are also summarized in Table 1 along with the corresponding parameter estimates and standard errors. The best-fitting models based on AIC were exponential, log-logistic, and Weibull functions.
Table 1.
Different nonlinear model and parameter estimates that best describe the data corresponding to only one drug sensitive cell types based on the data obtained from LINCS.
| Cells | Model | Parameters (Std. error*) | Death rate |
|---|---|---|---|
| Alpelisib-sensitive cells | Exponential decay model: E(C) = l + (u − l) · e−C/e | l=0.0126 h−1 (5 · 10−4) u=0.023 h−1 (2 · 10−4) e=3.047 μM (0.414) |
0.0146 h−1 |
| Neratinib-sensitive cells | 5-parameter log-logistic function: |
Emax = −0.02 h−1 (0.055) E0 = 0.0148 h−1 (1.3 · 10−4) h = 0.3 (0.055) EC50 = 0.63 μM (1.295) f = 0.086 (0.147) |
0.0093 h−1 |
| Trametinib-sensitive cells | 4-parameter log-logistic function: |
Emax = 0.0098 h−1(0.001) E0 = 0.0122 h−1 (2.5· 10−4) h = 0.43 (0.284) EC50 = 0.7 μM (1.56) |
0.0096 h−1 |
| Dactolisib-sensitive cells | Exponential decay model: E(C) = l + (u − l) · e−C/k | l = 0.00531 h−1 (4.8 · 10−4) u=0.01480 h−1 (4.9 · 10−4) k=0.03495 μM (0.007) |
0.0105 h−1 |
| Lapatinib-sensitive cells | 3-parameter Weibull function: | E0 = 0.0136 h−1 (4.2· 10−4) h = 2.32 (0.785) b= 30.38 μM (11.31) |
0.008 h−1 |
| Selumetinib-sensitive cells | 4-parameter log-logistic function: |
Emax = 0.0142 h−1 (4.6· 10−5) E0 = 0.01432 h−1 (2.6· 10−5) h = 4.4 (5.6) EC50 = 2.13 μM (1.45) |
0.0089 h−1 |
| Sacaratinib-sensitive cells | 4-parameter Weibull function: | u = 0.0118 h−1 (1.1· 10−4) l = 0.011 h−1 (1.9· 10−4) h = −3.37 (2.91) b= 0.91 μM (0.196) |
0.009 h−1 |
| NVP-TAE684-sensitive cells | 3-parameter log-normal model: E(C) = E0 · ϕ(h(log log C − log log b)) ϕ: cumulative distribution function of the standard normal distribution |
E0 = 0.0127 h−1 (6.7· 10−4) h = −2.09 (2.37) b= 3.94 μM (0.906) |
0.0092 h−1 |
Std. Error: Standard error
In addition, a value for the probability of emergence of (epi)genetic resistance mechanisms of 10−7 was defined for cells resistant to one drug, and of 10−8 for cross-resistance. These values were chosen because the baseline point mutation rate for genetically stable cells is around 10−10–10−11 per base per cell division (41) and we considered 1000 loci in the genome giving rise to resistance; similar rates may apply for the emergence of other resistance mechanisms such as amplifications, deletions, and methylation changes.
Identification of drug interaction dynamics
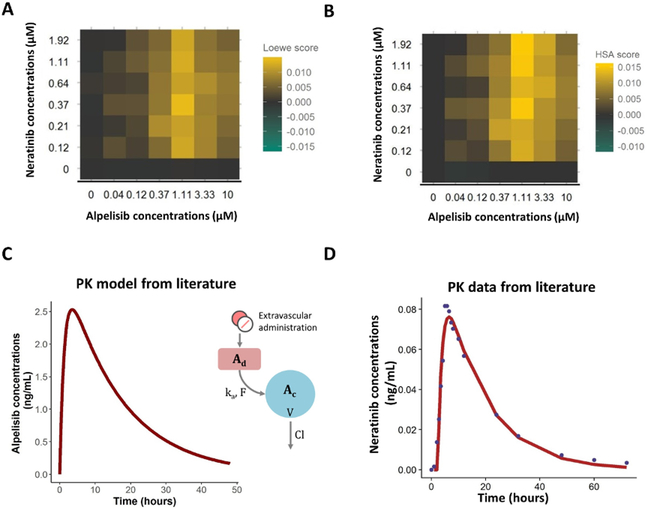
We next sought to understand if drug interactions played an important role in the combination studies. Specifically, we aimed to determine if the interaction of the drugs causes increased inhibition of the proliferation of drug-sensitive cells compared to an additive effect of the drugs (Loewe additivity principle) or the effect of each drug alone (HSA). The matrices in Fig. 3A–B show the values obtained by computing the differences between the measurements from the combination of alpelisib and neratinib and the surface obtained with the Loewe or HSA models, respectively, such that negative values represent antagonism and positive values synergism. In this case, both models revealed synergistic interactions between the drugs. Interaction matrices for the remaining drug combinations in this study are shown in Fig. S2–S3. The analysis using the Loewe model (Fig. S2) reported that the drugs with the greatest synergistic effect were the combination of alpelisib and neratinib, alpelisib with trametinib, and dactolisib with trametinib; whereas the highest antagonism values were found for the dactolisib and lapatinib combination. The matrices from the HSA model (Fig. S3) on the other hand, presented additional synergistic effects that were not found with the Loewe model (e.g. alpelisib with saracatinib or selumetinib) and less antagonistic interactions (although the highest antagonism was still found for the dactolisib and lapatinib combination). The discrepancy between the two figures is not surprising as each model has a different definition of synergism/antagonism.
Figure 3.
A-B) Concentration combination matrices that represent the difference between the measured effect on the birth rates of sensitive cells and the effect obtained under the Loewe additivity model (A) or Highest Single Agent model (B) for alpelisib and neratinib treatment. Yellow indicates synergy, green indicates antagonism and grey means no interaction. C) Pharmacokinetic model for alpelisib obtained from literature (see Supplementary Table S1) which consist on a one compartment model with first-order absorption and elimination (CL indicates total drug clearance, V is the apparent volume of distribution from the central compartment (Ac), Ka is the first-order absorption rate constant which drives the amount of drug from the depot compartment (Ad) to Ac and F is the bioavailability). D) Pharmacokinetic model for neratinib estimated using data from literature. Concentration data was best described with a one compartment model with first-order absorption and elimination. The solid line represents the prediction of the model and the solid points represent the observations. To see the parameter values go to Supplementary Table S1.
Pharmacokinetic models
We obtained relevant PK models and parameter estimates from previous studies (42,43) and used these estimates for the simulations of drug concentration profiles (Supplementary Table S1 and Fig. 3C). In the case of neratinib and dactolisib, pharmacokinetic parameters were estimated from the median concentration-time data scanned from the original publication using ACESO (Fig. 3D). For the examples outlined here, we repeated the PK dynamics for each cycle, but more complex models can also be simulated using ACESO.
Simulation of treatment effects
Once we defined the PK models, the birth/death/mutation rates of the different cell types, and the effect of different drugs concentrations and combinations on these rates, we used ACESO to identify optimum treatment combination schedules. The quality of a schedule was judged by its impact on the total number of different tumor cell types. For these simulations, we considered a population of 1 million sensitive cells and 1000 drug-resistant cells prior to treatment initiation. Simulations were implemented for each combination of drugs; the findings for alpelisib and neratinib combination are outlined below and the results for other drug combinations are summarized in the SI and Figures S4–S5.
Alpelisib and neratinib combination treatment
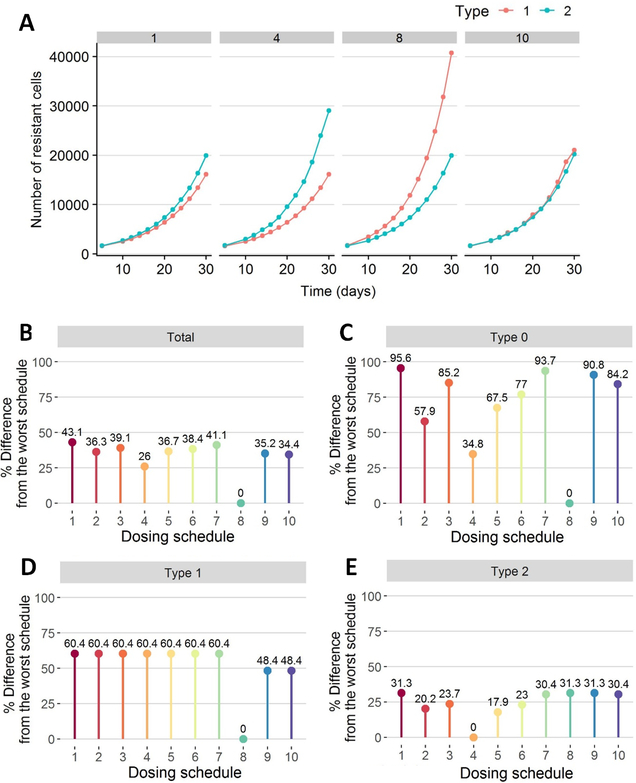
We tested different dosing schedules for alpelisib and neratinib combination therapy (Table 2). For these schedules, we then determined the dynamics of resistant cell populations over time (Fig. 4A) and compared the expected number of tumor cells 30 days after treatment obtained from each regimen (Fig. 4B:E). In the case of alpelisib, any modification from the maximally tolerated dose (MTD) administration of 300 mg once a day (QD) resulted in an increased number of alpelisib-sensitive cells after 30 days. This effect arises due to the large birth rate of alpelisib-sensitive cells in the absence of drug and the rapid clearance of alpelisib, both of which allow tumor cells to grow quickly during treatment breaks (Fig. 4A; regimen 8). In the case of neratinib, administering high dose pulses with low daily maintenance dosing gave similar results to the MTD schemas, since low concentrations of the drug had a considerable effect on the birth rate of type-2 cells (see Fig. 2) and therefore the schedule involving a high-dose pulse of 720 mg/week with an additional daily dose of 160 mg for the remainder of the week was comparable to the approved schedule of 240 mg QD (<1% difference in Fig. 4E). The synergistic effect between these two drugs, however, led to the best schedule of 300 mg QD alpelisib with 240 mg QD neratinib as it is the one that further decreases the number of type-0 cells and consequently, the total number of cells (43.1% improvement from the worst schedule in Fig. 4B), although the differences with respect to other regimens were small.
Table 2.
Different dosing schedules explored in the combination of alpelisib and neratinib to treat triple-negative breast cancer (TNBC) cell lines.
| Dosing regimen | Alpelisib | Neratinib |
|---|---|---|
| 1 | 300 mg QD | 240 mg QD |
| 2 | 300 mg QD | 120 mg QD |
| 3 | 300 mg QD | 360 mg QOD |
| 4 | 300 mg QD | 1200 mg QW |
| 5 | 300 mg QD | 720 mg QW + 40 mg QD r.m. |
| 6 | 300 mg QD | 720 mg QW + 80 mg QD r.m. |
| 7 | 300 mg QD | 720 mg QW + 160 mg QD r.m. |
| 8 | 525 mg QOD | 240 mg QD |
| 9 | 900 mg QW + 200 mg QD r.m. | 240 mg QD |
| 10 | 900 mg QW + 200 mg QD r.m. | 720 mg QW + 160 mg QD r.m. |
QD: once a day, QW: once a week, QOD: every other day, r.m.: remaining of the week.
Figure 4.
Effects of the treatment on the total number of the different cancer cell types (type-0: sensitive to both drugs, type-1: alpelisib-sensitive cells, type-2: neratinib-sensitive cells, total: the sum of type-0, type-1 and type-2 cells). The different dosing schedules explored are summarized in Table 2. A) Effect of four simulated dosing schedules on the number of resistant cells over time. B-E) Comparison of the different dosing schedules on the remaining fraction of cells after the treatment period (i.e. expected number of cancer cells after 30-day treatment divided by the initial number of cells before treatment administration): total number of cells (B), number of type-0 cells (C), number of type-1 cells (D) and number of type-2 cells (E). The bars represent the fraction of remaining cells whereas the numbers on top of the bars indicate the percent difference from the worst schedule; i.e. the highest bar represents the worst schedule while the shortest bar represents the best.
Optimum dosing strategies for NSCLC
We then sought to investigate optimum erlotinib dosing strategies for the treatment of NSCLC using the evolutionary cancer model from (20,40). NSCLCs that harbor mutations within EGFR gene are sensitive to the tyrosine kinase inhibitor (TKI) erlotinib (44). Unfortunately, all patients treated with this drug acquire resistance, most commonly as a result of a secondary mutation within EGFR (T790M mutation). In addition, approximately one third of the patients develop CNS metastases after initial response to EGFR TKIs (45).
Here, we extended the original two-type branching process to account for CNS metastases and to show the flexibility of ACESO to handle different scenarios of clonal evolution. As it has been previously suggested, the metastasized cells retain EGFR TKI sensitivity if sufficient drug concentration can be achieved in brain parenchyma or the cerebrospinal fluid (CSF) (45). We thus considered three cell types (Fig. 5A): primary tumor drug-sensitive cells, primary tumor drug-resistant cells and drug-sensitive metastasized cells – drug-sensitive cells that have disseminated to a new site and have the same proliferation and death rates as the original cells.
Figure 5.
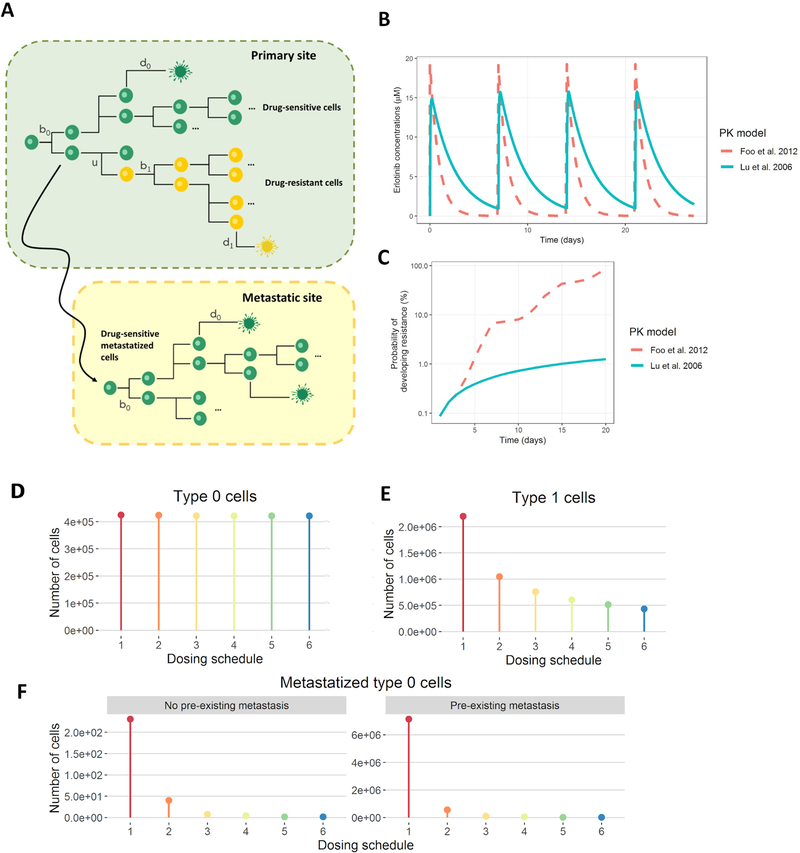
A) Schematic representation of the stochastic model proposed for NSCLC case study, where erlotinib-sensitive cell (green), proliferates at rate b0(t), dies at rate d0(t) and can accumulate mutations at rate u per cell division to give birth to the erlotinib-resistant cell type (yellow), which proliferates at rate b1(t) and dies at rate d1(t). Erlotinib-sensitive cell is also able to migrate to other sites (to CNS in this example) and continue growing at the same rates as the original cell type. B-C) Effect of two different pharmacokinetic models for erlotinib on the probability of developing resistance. The pharmacokinetic models described in 1) Lu et al. 2006 and 2) Foo et al. 2012 were coded in ACESO and a dosing schedule of 1600 mg once a week was simulated to explore the effects of the resulting erlotinib concentrations (B) on the risk of developing resistance for a period of 20 days (C). D-F) Effect of different dosing regimens in the number of cancer cells at 30 days. Erlotinib dosing schedules used in the simulations: 1) standard daily dosing of 150mg, 2) high-dose pulse of 1600mg once a week, 3) high-dose pulse of weekly 1600mg plus 50mg/day the rest of the week, 4) high-dose pulse of weekly 1800mg plus 50mg/day the rest of the week, 5) high-dose pulse of 1000mg day 1 and 2 followed by 100mg/day the rest of the week and 6) high-dose pulse of 1200mg day 1 and 2 followed by 50mg/day the rest of the week. Type-0 cells refer to primary tumor drug sensitive cells and type-1 cells to primary tumor drug resistant cells. For the case of pre-existing resistance, an initial number of 50 cells were assumed. The initial number of type-0 cells was set to 106 and we defined an erlotinib penetration to CNS of 5%.
Estimation of drug sensitive and resistant cell growth and death rates
We used publicly available data of erlotinib-sensitive and erlotinib-resistant cell birth and death rates (see Data availability section) (40). We then fit the concentration-response curves for the birth rates of each cell type using ACESO. The selected model for erlotinib-sensitive cells was a five parameter logistic function with a cell intrinsic birth rate of 0.04 h−1, a Emax of 0.003 h−1, a EC50 value of 0.76 μM, a Hill coefficient h of 1.5 and an asymmetry factor f of 8.26. A model was also fitted for the influence of erlotinib on the inhibition of drug-resistant cell growth. Although it was not the model with lower AIC, a linear equation with an intercept of 0.034 h−1 and a slope parameter of −0.0005 (μM·h)−1 was used to model the birth rates of the resistant PC-9 cell line as in the original article. The death rates obtained from the apoptosis assay counts for both cell types were almost identical for the different drug concentrations used in the experiments, and hence we defined a constant death rate of 0.12 day−1 and 0.06 day−1 for the drug sensitive and resistant cell populations, respectively.
Pharmacokinetic model
We then generated the erlotinib plasma drug concentration vs time profiles using the model. The corresponding parameters were obtained from a pharmacokinetic analysis of data from 1,047 patients with solid tumors (46) instead of using simpler equations for the PK obtained from scarce data points from the literature as in (20). Simulations represented in Fig. 5B were performed considering the administration of a 1600 mg dose of erlotinib once a week over one month of treatment. The simulations demonstrated differences in drug exposure obtained with each model, which had a large impact on the risk of developing resistance (Fig. 5C); depending on the drug clearance, drug concentrations drop faster and thereby cause cells to resume proliferation more quickly. This fact highlights the importance of properly characterizing the PK of drugs as they can have a large influence in selecting optimum dosing strategies. On the other hand, previous studies demonstrated that the concentration of erlotinib measured in the cerebrospinal fluid (CSF) during standard daily dosing of 150 mg is too low (5% of that in plasma according to (47)) to prevent EGFR-mutant NSCLC cell proliferation. Thus we assumed a linear pharmacokinetic process with a partition coefficient of 0.05 to simulate erlotinib concentrations in the metastatic site.
Simulation of treatment effects
The current analysis explores whether a higher dose of erlotinib provides an increased concentration in CNS that may prove to be more efficient in metastatic disease while preventing the development of resistance at the primary site. Based on the above considerations, the standard oral daily dosing schedule of 150 mg was compared to a high-dose pulse of 1600 mg once a week and four different combinations of low-dose continuous and high-dose pulsed strategies (Fig. 5D–F) assuming one month of treatment. Additional simulations increasing the daily dosing for erlotinib were not investigated, as daily doses above 200 mg are associated with unacceptable toxicity (48). In contrast, weekly high-dose pulses up to 2000 mg have been tolerated by NSCLC patients despite persistent nausea reported in patients receiving higher doses than 1200 mg (49).
Simulations were performed for cases of (i) an absence of pre-existing tumor cells at the metastatic site at the time of tumor diagnosis, and (ii) established CNS metastases at the time of the primary tumor diagnosis, assuming no pre-existing resistance in both cases. We used a mutation probability of 10−8 per erlotinib-sensitive cell division and a dissemination probability to CSF of 10−9.
The resulting numbers of the different cancer cell populations are shown in Fig. 5D–E. Variation in the number of sensitive cells in the primary tumor site from one schedule to the next was minimal. However, for erlotinib-resistant cells and especially for metastasized erlotinib-sensitive cells, using high-dose pulses considerably improved the results. We conclude that a combined low-dose continuous and high-dose pulsed erlotinib schedule was the most successful dosing strategy at preventing progression in patients with CNS metastases but did not show significantly delayed emergence of resistance due to the T790M EGFR mutation; however, once resistant cells arise, these schedules were also demonstrated to be more effective at killing the resistant cell population.
Discussion
In this work, we have built a multi-scale computational framework in R, called ACESO, which incorporates a continuous time multi-type branching process model to investigate the evolution of resistant clones. This model incorporates concentration-dependent birth and death rates as well as the probability per cell division that a resistance mechanism arises due to an (epi)genetic alteration. The use of mathematical models to search for optimized treatment schedules that delay the appearance of resistance in cancer cells is part of a growing effort to improve clinical trial design for cancer patients. Here we provide an open-source tool to contribute to this research by exploring the evolution of heterogeneous tumor cell populations while taking pharmacokinetic and drug interaction effects into account.
There are several examples in the literature introducing different models to describe drug pharmacokinetics (Supplementary Fig. S6), tumor progression and drug effects (38,40,42,46). However, the implementation of our present goal previously required the use of different platforms for parameter estimation, model simulation or numerical/graphical diagnostics or programming skills, delaying the application of integrative quantitative analysis of cancer data. The objective of the current work was to integrate the tools required to characterize all necessary processes into a single computational framework. The application of our method to individual cancer types requires the availability of data on cell growth and death during different clinically tolerated concentrations of the drugs in order to parameterize the behavior of sensitive and resistant cancer cell populations. ACESO uses in vitro data because of the difficulty of obtaining such concentration-response curves from in vivo settings. This fact complicates the predictions of clinically relevant time scales for the emergence of resistance because growth kinetics in vitro occur on a time scale orders of magnitude faster than in vivo; however, as long as the relative differences in growth rates are constant between in vivo and in vitro situations or an in vitro-in vivo correlation model is included in the framework, the ranking of schedules based on in vitro rates is applicable to in vivo situations.
In this work we first applied ACESO to data from the HMS LINCS database. Due to the lack of data regarding apoptosis assays and drug resistant cell lines, our results serve as an example needing validation before clinical implementation. We also applied ACESO to an extension of a previously used model of EGFR-mutant cancer cells (20,40). The previous effort identified a low-dose/high-dose pulse erlotinib administration schedule as the predicted best intervention strategy for stage IV NSCLC, which was then tested in a prospective clinical trial (21) at the Memorial Sloan-Kettering Cancer Center (http://clinicaltrials.gov/show/NCT01967095). This trial established the maximally tolerated dose as 1200 mg for two days a week and 50 mg for the remainder of the week. Although the study was not powered to demonstrate a significant delay of the emergence of T790M-driven resistance due to larger than expected variability in patient PK, a significant reduction in the rate of progression due to CNS disease was observed (0% versus up to 33% given historical control). Using ACESO, we now expanded the original model (20) to include a population of drug-sensitive metastasized cells that are able to migrate to the CNS; we also used a population PK model (46). ACESO again predicted pulse dosing to be optimal since this strategy allows for increased drug concentrations in the CNS while the continued daily dosing controlled the progression of disease. This regimen is now being studied in a cohort of EGFR-mutant lung cancer patients with untreated CNS metastases.
The models represented in the current version of ACESO assume exponential cell growth, no spatial effects, and no competing interactions between cells. Therefore, microenvironmental phenomena such as tumor-immune cell interactions or a changing architecture of blood vessels are currently not considered but could, in the future, be incorporated by extending the methodology to more complex models. The model is able to describe any resistance mechanism that arises at a certain rate per cell division; such changes can include (epi)genetic alterations such as point mutations, amplifications, deletions, inversions, methylation of histone modification changes, or any other alterations. In order to parameterize such models, the kinetics of different cell types and interaction kinetics among them need to be measured in appropriate model systems. Apart from describing a growing population of tumor cells, our approach can describe any kind of cell population or bacterial infection where (epi)genetic changes cause drug resistance. Using ACESO, different strategies can also be tested for robustness due to variability in pharmacokinetic parameters among patients, variable growth and death rates of sensitive and resistant cells as well as different compositions of the tumor at the start of therapy. We propose use of our methodology for other cancer types and for therapeutic agents at the forefront of clinical development.
Supplementary Material
Significance: Findings introduce a computational modeling platform and software package for combination treatment strategies with flexible pharmacokinetic profiles and multidrug interaction curves that are estimated from data.
Acknowledgments
Financial Support: This work was supported by the Dana-Farber Cancer Institute Physical Science-Oncology Center; grant number NIH U54CA193461.
References
- 1.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies [Internet]. Nature Reviews Clinical Oncology. 2018. page 81–94. Available from: 10.1038/nrclinonc.2017.166 [DOI] [PubMed] [Google Scholar]
- 2.Groenendijk FH, Bernards R. Drug resistance to targeted therapies: déjà vu all over again. Mol Oncol. 2014;8:1067–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–81. [DOI] [PubMed] [Google Scholar]
- 4.Karaayvaz M, Cristea S, Gillespie SM, Patel AP, Mylvaganam R, Luo CC, et al. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat Commun. 2018;9:3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell. 2017;168:613–28. [DOI] [PubMed] [Google Scholar]
- 6.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foo J, Michor F. Evolution of acquired resistance to anti-cancer therapy. J Theor Biol. 2014;355:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beerenwinkel N, Schwarz RF, Gerstung M, Markowetz F. Cancer evolution: mathematical models and computational inference. Syst Biol. 2015;64:e1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwasa Y, Nowak MA, Michor F. Evolution of resistance during clonal expansion. Genetics. 2006;172:2557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komarova N Stochastic modeling of drug resistance in cancer. J Theor Biol. 2006;239:351–66. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto KN, Nakamura A, Haeno H. The evolution of tumor metastasis during clonal expansion with alterations in metastasis driver genes. Sci Rep. 2015;5:15886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson MD, Antal T. Competing evolutionary paths in growing populations with applications to multidrug resistance. PLoS Comput Biol. 2019;15:e1006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldie JH, Coldman AJ. Quantitative model for multiple levels of drug resistance in clinical tumors. Cancer Treat Rep. 1983;67:923–31. [PubMed] [Google Scholar]
- 15.Goldie JH, Coldman AJ. A mathematic model for relating the drug sensitivity of tumors to their spontaneous mutation rate. Cancer Treat Rep. 1979;63:1727–33. [PubMed] [Google Scholar]
- 16.Kimmel M, Axelrod DE. Branching Processes in Biology (Interdisciplinary Applied Mathematics). 2002; Available from: https://svsl46kph01.storage.googleapis.com/MTQ5MzkxNTU4NA==01.pdf
- 17.Haccou P, Haccou P, Jagers P, Vatutin VA,. Vatutin VA. Branching Processes: Variation, Growth, and Extinction of Populations. Cambridge University Press; 2005. [Google Scholar]
- 18.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. [DOI] [PubMed] [Google Scholar]
- 19.Bozic I, Reiter JG, Allen B, Antal T, Chatterjee K, Shah P, et al. Evolutionary dynamics of cancer in response to targeted combination therapy. Elife. 2013;2:e00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foo J, Chmielecki J, Pao W, Michor F. Effects of pharmacokinetic processes and varied dosing schedules on the dynamics of acquired resistance to erlotinib in EGFR-mutant lung cancer. J Thorac Oncol. 2012;7:1583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu HA, Sima C, Feldman D, Liu LL, Vaitheesvaran B, Cross J, et al. Phase 1 study of twice weekly pulse dose and daily low-dose erlotinib as initial treatment for patients with EGFR-mutant lung cancers. Ann Oncol. 2017;28:278–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haeno H, Gonen M, Davis MB, Herman JM, Iacobuzio-Donahue CA, Michor F. Computational modeling of pancreatic cancer reveals kinetics of metastasis suggesting optimum treatment strategies. Cell. 2012;148:362–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foo J, Michor F. Evolution of resistance to targeted anti-cancer therapies during continuous and pulsed administration strategies. PLoS Comput Biol. 2009;5:e1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foo J, Michor F. Evolution of resistance to anti-cancer therapy during general dosing schedules. J Theor Biol. 2010;263:179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakrabarti S, Michor F. Pharmacokinetics and Drug Interactions Determine Optimum Combination Strategies in Computational Models of Cancer Evolution. Cancer Res. 2017;77:3908–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu LL, Li F, Pao W, Michor F. Dose-Dependent Mutation Rates Determine Optimum Erlotinib Dosing Strategies for EGFR Mutant Non-Small Cell Lung Cancer Patients. PLoS One. 2015;10:e0141665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gadagkar SR, Call GB. Computational tools for fitting the Hill equation to dose-response curves. J Pharmacol Toxicol Methods. 2015;71:68–76. [DOI] [PubMed] [Google Scholar]
- 28.Ritz C, Baty F, Streibig JC, Gerhard D. Dose-Response Analysis Using R. PLoS One. 2015;10:e0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keenan AB, Jenkins SL, Jagodnik KM, Koplev S, He E, Torre D, et al. The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Syst. 2018;6:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cory AH, Owen TC, Barltrop JA, Cory JG. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991;3:207–12. [DOI] [PubMed] [Google Scholar]
- 31.Vermes I, Haanen C, Steffens-Nakken H, Reutellingsperger C. A novel assay for apoptosis Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184:39–51. [DOI] [PubMed] [Google Scholar]
- 32.Hogan T, Yates A, Seddon B. Analysing Temporal Dynamics of T Cell Division in vivo Using Ki67 and BrdU Co-labelling by Flow Cytometry. BIO-PROTOCOL [Internet]. 2017;7 Available from: https://bio-protocol.org/e2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufmann SH, Lee S-H, Meng XW, Loegering DA, Kottke TJ, Henzing AJ, et al. Apoptosis-associated caspase activation assays. Methods. 2008;44:262–72. [DOI] [PubMed] [Google Scholar]
- 34.Akaike H Factor analysis and AIC Selected Papers of Hirotugu Akaike. Springer; 1987. page 371–86. [Google Scholar]
- 35.Wood SN. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models [Internet]. Journal of the Royal Statistical Society: Series B (Statistical Methodology). 2011. page 3–36. Available from: 10.1111/j.1467-9868.2010.00749.x [DOI] [Google Scholar]
- 36.Cleveland WS, Grosse E, Shyu WM. Local regression models. Chapter 8 of Statistical Models in S (Edited by Chambers JM and Hastie TJ), 309–376. Chapman & Hall, London; 1991. [Google Scholar]
- 37.Baron KT, Hindmarsh AC, Petzold LR, Gillespie B, Margossian C. Pastoor D mrgsolve: Simulate from ODE-Based Population PK/PD and Systems Pharmacology Models Metrum Research Group; 2017. [Google Scholar]
- 38.Loewe S The problem of synergism and antagonism of combined drugs. Arzneimittelforschung. 1953;3:285–90. [PubMed] [Google Scholar]
- 39.Gaddum JH. Gaddum’s Pharmacology. Oxford University Press, USA; 1985. [Google Scholar]
- 40.Chmielecki J, Foo J, Oxnard GR, Hutchinson K, Ohashi K, Somwar R, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3:90ra59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhang H-EC, Ruddy DA, Krishnamurthy Radhakrishna V, Caushi JX, Zhao R, Hims MM, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med. 2015;21:440–8. [DOI] [PubMed] [Google Scholar]
- 42.De Buck SS, Jakab A, Boehm M, Bootle D, Juric D, Quadt C, et al. Population pharmacokinetics and pharmacodynamics of BYL719, a phosphoinositide 3-kinase antagonist, in adult patients with advanced solid malignancies. Br J Clin Pharmacol. 2014;78:543–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keyvanjah K, DiPrimeo D, Li A, Obaidi M, Swearingen D, Wong A. Pharmacokinetics of neratinib during coadministration with lansoprazole in healthy subjects. Br J Clin Pharmacol. 2017;83:554–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayat MA. Brain Metastases from Primary Tumors Volume 1: Epidemiology, Biology, and Therapy. Academic Press; 2014. [Google Scholar]
- 46.Lu J-F, Eppler SM, Wolf J, Hamilton M, Rakhit A, Bruno R, et al. Clinical pharmacokinetics of erlotinib in patients with solid tumors and exposure-safety relationship in patients with non-small cell lung cancer. Clinical Pharmacology & Therapeutics. Wiley Online Library; 2006;80:136–45. [DOI] [PubMed] [Google Scholar]
- 47.Togashi Y, Masago K, Fukudo M, Terada T, Fujita S, Irisa K, et al. Cerebrospinal fluid concentration of erlotinib and its active metabolite OSI-420 in patients with central nervous system metastases of non-small cell lung cancer. J Thorac Oncol. 2010;5:950–5. [DOI] [PubMed] [Google Scholar]
- 48.Milton DT, Azzoli CG, Heelan RT, Venkatraman E, Gomez JE, Kris MG, et al. A phase I/II study of weekly high-dose erlotinib in previously treated patients with nonsmall cell lung cancer. Cancer: Interdisciplinary International Journal of the American Cancer Society. Wiley Online Library; 2006;107:1034–41. [DOI] [PubMed] [Google Scholar]
- 49.Clarke JL, Pao W, Wu N, Miller VA, Lassman AB. High dose weekly erlotinib achieves therapeutic concentrations in CSF and is effective in leptomeningeal metastases from epidermal growth factor receptor mutant lung cancer. J Neurooncol. 2010;99:283–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TNBC
Raw cell viability data used in the present analysis can be retrieved from the HMS LINCS Database (http://lincs.hms.harvard.edu/db/datasets/20259), where nine small molecule kinase inhibitors given in combination were chosen for analysis of combinatorial drug sensitivities in the BT-20 triple-negative breast cancer (TNBC) cell line (obtained from ATCC). Briefly, in these experiments cells were exposed during 72 hours to constant concentration levels of the drugs either as single agents or in combinations. Due to the lack of information regarding drug resistant cell lines in LINCS, we assumed that these cells had decreased birth and death rate parameters compared to their sensitive counterparts, and hence the proliferation and death rate parameters obtained for the parental BT-20 cell line were divided by different values depending on the drugs being analyzed and tested for sensitivity.
NSCLC
We used cell viability (MTS assay) and apoptosis (Annexin V/PI FACS assay) data from isogenic erlotinib-sensitive and erlotinib-resistant pairs of PC-9 NSCLC cell lines from a previous publication (40). In these experiments, the total numbers of viable and dead cells in culture were counted at 48, 60, and 72 hours after treatment initiation for different erlotinib concentrations and in the absence of the drug. In contrast to the TNBC data above, for the NSCLC data we used response kinetics to a single drug only and also did not have information on the growth patterns of resistant cell types.