Abstract
Background -
DNA methylation patterns associated with habitual diet have not been well studied.
Methods -
Diet quality was characterized using a Mediterranean-style diet score (MDS) and the Alternative Healthy Eating Index score (AHEI). We conducted ethnicity-specific and trans-ethnic epigenome-wide association analyses for diet quality and leukocyte-derived DNA methylation at over 400,000 cytosine-guanine dinucleotides (CpGs) in five population-based cohorts including 6,662 European ancestry (EA), 2,702 African ancestry (AA), and 360 Hispanic ancestry (HA) participants. For diet-associated CpGs identified in epigenome-wide analyses, we conducted Mendelian randomization (MR) analysis to examine their relations to cardiovascular disease (CVD) risk factors and examined their longitudinal associations with all-cause mortality.
Results -
We identified 30 CpGs associated with either MDS or AHEI, or both, in EA participants. Among these CpGs, 12 CpGs were significantly associated with all-cause mortality (Bonferroni corrected p-value < 1.6×10−3). Hypermethylation of cg18181703 (SOCS3) was associated with higher scores of both MDS and AHEI and lower risk for all-cause mortality (p-value = 5.7×10−15). Ten additional diet-associated CpGs were nominally associated with all-cause mortality (p-value < 0.05). MR analysis revealed eight putatively causal associations for six CpGs with four CVD risk factors (BMI, triglycerides, high-density lipoprotein cholesterol concentrations, and type 2 diabetes; Bonferroni corrected MR p-value < 4.5×10−4). For example, hypermethylation of cg11250194 (FADS2) was associated with lower triglyceride concentrations (MR p-value = 1.5×10−14).and hypermethylation of cg02079413 (SNORA54; NAP1L4) was associated with BMI (corrected MR p-value = 1×10−6).
Conclusions -
Habitual diet quality was associated with differential peripheral leukocyte DNA methylation levels of 30 CpGs, most of which were also associated with multiple health outcomes, in EA individuals. These findings demonstrate that integrative genomic analysis of dietary information may reveal molecular targets for disease prevention and treatment.
Keywords: cardiovascular disease risk factors, all-cause death, Mendelian randomization, diet, DNA methylation, diet quality score, alternative healthy eating index
Journal Subject Terms: Cardiovascular Disease, Diet and Nutrition, Epidemiology, Epigenetics
Introduction
Epigenetic alterations are involved in the pathogenesis of many human diseases.1 DNA methylation, which commonly occurs at cytosine–guanine dinucleotide (CpG) sites, is a well-studied epigenetic modification that may affect gene expression and contribute to the development of chronic diseases, including cardiovascular disease.2–4
Several lines of evidence suggest that diet may be actively involved in epigenetic regulation, which impacts diet-related disease risk.5–8 Tremblay et al. measured genome-wide DNA methylation profiles before and after a six-week supplementation of daily dose of 3 grams of omega-3 polyunsaturated fatty acids (n-3 FAs) in 36 participants with BMI between 25 to 40 kg/m2.9 They found that n-3 FAs supplementation caused differential DNA methylation of 308 CpGs, which could be linked to 16 pathways related to cardiovascular disease (CVD) including inflammatory response and lipid metabolism.
While previous studies provide useful evidence that diet plays an important role in regulating the human epigenome, studies of DNA methylation signatures for overall diet quality, however, are few in number and limited by small sample sizes. Diet quality is crucial for chronic diseases prevention.10–12 In cohort studies, diet quality is often assessed using a variety of diet scores, including the Mediterranean-style diet score (MDS) and the Alternative Healthy Eating Index (AHEI) score.13–18 These studies showed that a higher diet score was associated with lower disease burden. A thorough insight into the biological mechanisms underlying diet-disease associations is important for disease prevention and treatment. To fill this knowledge gap, we conducted an epigenome-wide association study of diet quality, assessed by MDS and AHEI, with peripheral blood-derived DNA methylation in cohorts with representation of individuals of European as well as non-European ancestries.
Methods
The study design is presented in Figure 1. The datasets analyzed in the present study are available at the dbGAP repository phs000280.v5.p1 (ARIC), phs000007.v29.p10 (FHS), phs000741.v2.p1 (GOLDN), phs000209.v13.p3 (MESA), phs000853.v1.p1 (NAS), and phs000821.v1.p1 (LBC; phenotypic data). RS has a protocol for approving data requests (secretariat.epi@erasmusmc.nl). The informed consents given by KORA study participants do not cover data posting in public databases. However, data are available upon request from KORA Project Application Self-Service Tool (https://epi.helmholtz-muenchen.de/) Data requests can be submitted online and are subject to approval by the KORA Board. Methylation data of LBC have been submitted to the European Genome-phenome Archive under accession number EGAS00001000910. For ESTHER and InCHIANTI, the datasets used and/or analyzed during the current study are available from the corresponding author upon request. Data for WHI and CHS can be requested at https://www.whi.org/researchers/SitePages/Write%20a%20Paper.aspx and https://chs-nhlbi.org/node/6222, respectively. The study protocol was approved by each participating institutions’ Institutional Review Board. All participants provided written informed consent. Full descriptions of study populations, phenotypic definitions, DNA methylation profiling, and statistical analyses are available in the Supplemental Material.
Figure 1.
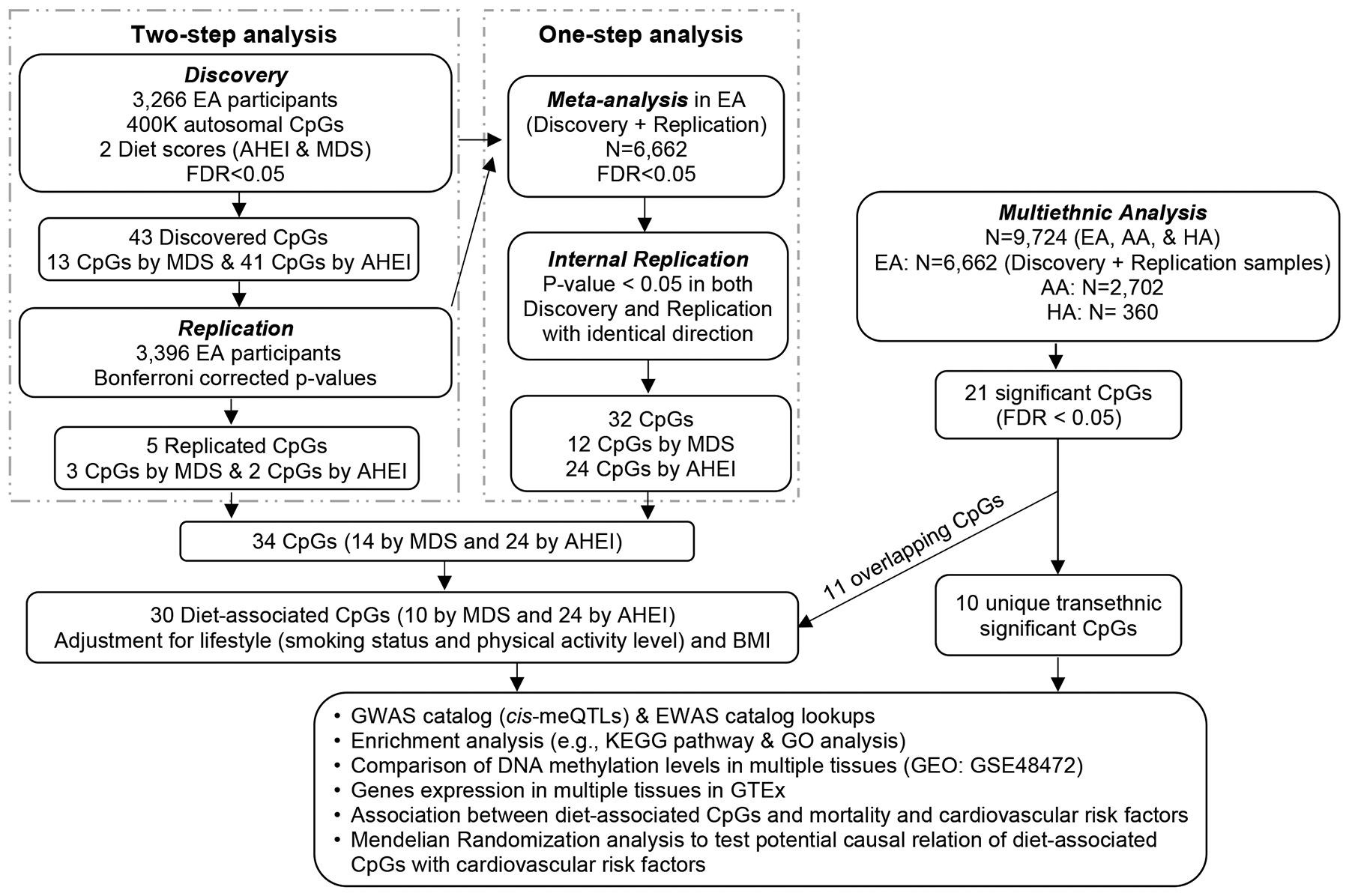
Study design flow chart. AA: African ancestry. EA: European ancestry. HA: Hispanic ancestry. CpGs: cytosine-guanine dinucleotides (DNA methylation sites). AHEI: Alternative Healthy Eating Index. MDS: Mediterranean-style diet score. FDR: false discovery rate. GWAS: genome-wide association study. cis-meQTLs: cis methylation quantitative loci. EWAS: epigenome-wide association study. GEO: Gene Expression Omnibus. GTEx: Genotype-Tissue Expression database. Discovery cohort: Framingham Heart Study (FHS). Replication cohorts: Atherosclerosis Risk in Communities (ARIC) Study, Genetics of Lipid Lowering Drugs and Diet Network (GOLDN), Multi-Ethnic Study of Atherosclerosis (MESA), and Rotterdam Study (RS). Cohorts for all-cause mortality includes: ARIC, FHS, ESTHER study, InChianti Study, Lothian Birth Cohort (LBC) Study 1921 and 1936, Cardiovascular Health Study (CHS), KORA F4 Study, Normative Aging Study (NAS), and Women’s Health Initiative (WHI).
Results
Epigenome-wide association analysis in European Ancestry (EA) participants.
We analyzed 403,087 autosomal CpGs. For each diet quality score, either MDS or AHEI, we conduct two analyses, a two-step analysis (i.e., discovery and replication) and an one-step analysis (i.e., meta-analysis of all cohorts with internal validation). For MDS, the discovery analysis identified 13 CpGs at false discovery rate (FDR) < 0.05 (corresponding p-value = 1.5×10−6; Supplemental Table 1; Supplemental Figure 1 [Manhattan plot] and Supplemental Figure 2 [Quantile-Quantile plot]). Of these CpGs, three replicated in the replication samples after Bonferroni correction (corresponding p-value < 0.004; Supplemental Table 1). The one-step analysis identified 12 CpGs associated with MDS at FDR < 0.05 (corresponding p-value = 1.2×10−6; Supplemental Table 2; Supplemental Figure 3 [Manhattan plot] and Supplemental Figure 2 [Quantile-Quantile plot]). Using models with adjustment for sex, age, and energy intake, the two analyses (two-step analysis and one-step analysis) identified 14 CpGs associated with MDS.
For AHEI, in the two-step analysis, the discovery step identified 41 CpGs at FDR < 0.05 (corresponding p-value = 6×10−6; Supplemental Table 3; Supplemental Figure 1 [Manhattan plot] and Supplemental Figure 2 [Quantile-Quantile plot]). Two CpGs replicated after Bonferroni correction (corresponding p-value < 0.001; Supplemental Table 3). The one-step analysis identified 24 CpGs at FDR < 0.05 (corresponding p-value = 3.1×10−6; Supplemental Table 4; Supplemental Figure 3 [Manhattan plot] and Supplemental Figure 2 [Quantile-Quantile plot]). The combination of the two-step analysis and the one-step identified 24 CpGs associated with AHEI using models adjusted for sex, age, and energy intake.
To reduce potential confounding effects by other lifestyle factors, we additionally adjusted for smoking status, physical activity, and BMI. Among the 14 CpGs associated with MDS, ten CpGs remained significant (p-value < 0.05/14; Figure 2), while all 24 CpGs associated with AHEI remained significant (p-value < 0.05/24; Figure 2). Overall, after adjustment for multiple confounders, we identified 30 CpGs associated with either MDS or AHEI, or both (Table 1). Pairwise correlations of the 30 CpGs were low to moderate, absolute Pearson r ranging from 0 to 0.66 (Supplemental Table 5). As shown in Supplemental Figure 4, regression coefficients in meta-analyses of all EA participants using MDS and AHEI were highly correlated, e.g., Pearson r was 0.97 for the regression coefficients of the top 500 CpGs in MDS versus AHEI. We therefore combined the CpGs identified using the two diet scores in the subsequent analyses.
Figure 2.
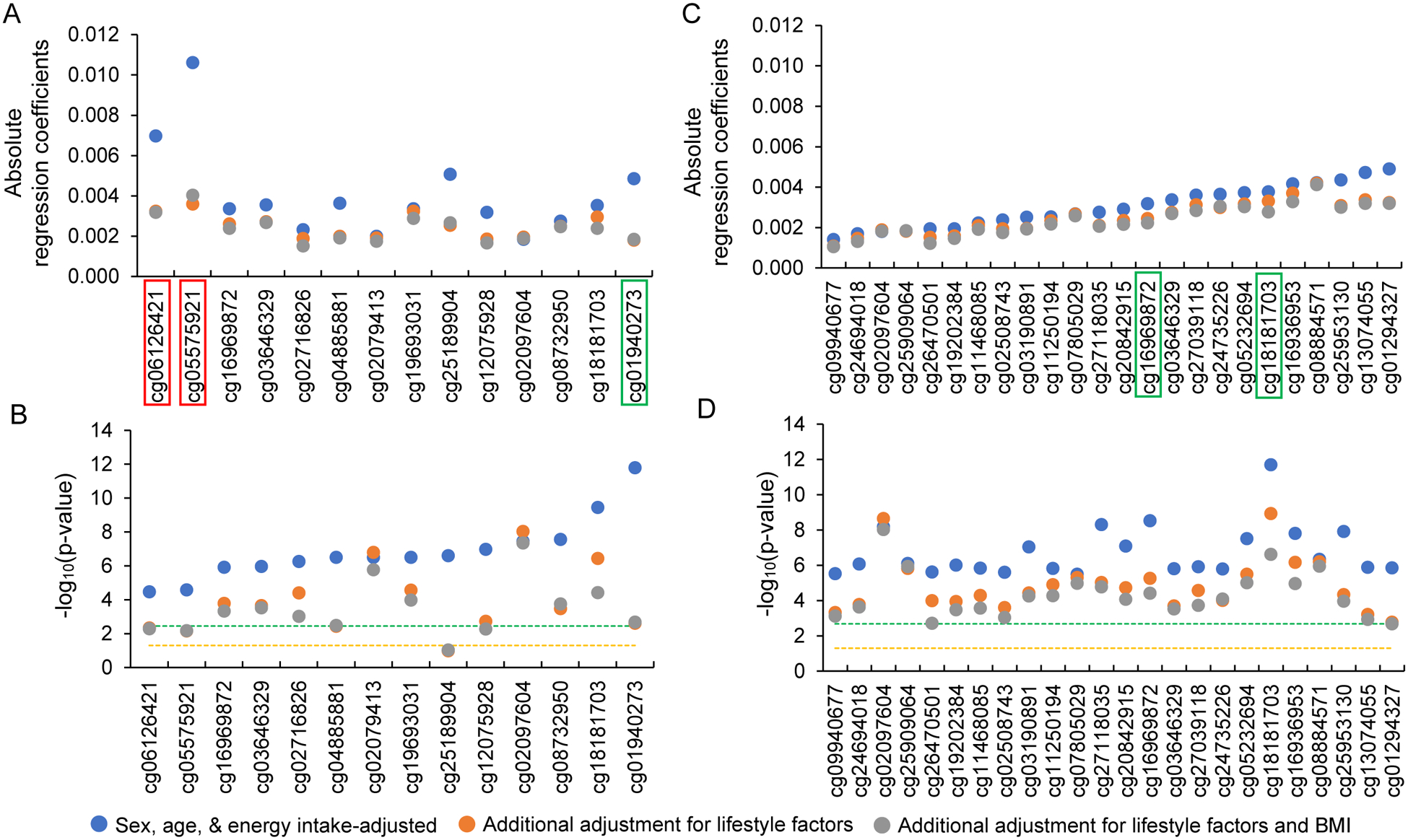
Effect of additional adjustment for lifestyle factors (smoking and physical activity) and BMI in European ancestry participants. A and B are 14 CpGs identified using the Mediterranean-style diet score (MDS). C and D are 24 CpGs identified using the Alternative Healthy Eating Index (AHEI). CpGs highlighted in red-colored rectangle are those identified in the two-step analysis alone and CpGs highlighted in green-colored rectangle are those identified in both one-step and two-step analyses. Orange colored dash line represents -log10 of 0.05 and green colored dash line represents -log10 of Bonferroni corrected p-value threshold, i.e., 0.05/14 for MDS and 0.05/24 for AHEI. Four CpGs (cg05575921, cg06126421, cg12075928, and cg25189904) in MDS analysis became non-significant after Bonferroni correction in models with adjustment for lifestyle factors and BMI, whereas all 24 CpGs in AHEI analysis remained significant.
Table 1.
Diet scores-associated CpGs in European Ancestry (EA) meta-analysis
| CpG | CHR | Position | Gene | Diet | Meta-analysis in all EA participants | ||||
|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P | Direction | I-squared | |||||
| cg04885881 | 1 | 11123118 | MDS | 0.004 | 0.001 | 3.2E-07 | +, +, +, +, + | 0.12 | |
| cg24735226 | 1 | 65096537 | CACHD1 | AHEI | −0.004 | 0.001 | 1.6E-06 | −, −, −, −, − | 0 |
| cg07805029 | 1 | 92953256 | GFI1 | AHEI | 0.003 | 0.001 | 3.1E-06 | +, +, +, +, + | 0 |
| cg19693031 | 1 | 145441552 | TXNIP | MDS | 0.003 | 0.001 | 3.1E-07 | +, +, +, +, + | 0.14 |
| cg24694018 | 1 | 145457621 | POLR3GL | AHEI | 0.002 | 0.0003 | 8.3E-07 | +, +, +, +, + | 0 |
| cg01940273 | 2 | 233284934 | MDS | 0.005 | 0.001 | 1.6E-12 | +, +, +, +, + | 0 | |
| cg20842915 | 7 | 39665132 | RALA | AHEI | 0.003 | 0.001 | 8.1E-08 | +, +, +, +, + | 0 |
| cg02508743 | 8 | 56903623 | LYN | AHEI | −0.002 | 0.001 | 2.5E-06 | −, −, −, −, − | 0 |
| cg27039118 | 8 | 116575902 | TRPS1 | AHEI | 0.004 | 0.001 | 1.2E-06 | +, +, +, +, + | 0 |
| cg02716826 | 9 | 33447032 | SUGT1P1;AQP3 | MDS | 0.002 | 0.0005 | 5.6E-07 | +, +, +, +, + | 0 |
| cg25953130 | 10 | 63753550 | ARID5B | AHEI | 0.004 | 0.001 | 1.2E-08 | +, +, +, +, + | 0 |
| cg03190891 | 10 | 97201172 | SORBS1 | AHEI | −0.003 | 0.0005 | 9.0E-08 | −, −, −, −, − | 0 |
| cg02079413 | 11 | 2986505 | SNORA54;NAP1L4 | MDS | −0.002 | 0.0004 | 3.1E-07 | −, −, −, −, − | 0.14 |
| cg11250194 | 11 | 61601937 | FADS2 | AHEI | 0.003 | 0.001 | 1.5E-06 | +, +, +, +, + | 0 |
| cg11468085 | 11 | 67435577 | ALDH3B2 | AHEI | −0.002 | 0.0005 | 1.4E-06 | −, −, −, −, − | 0.06 |
| cg25909064 | 11 | 120082805 | OAF | AHEI | 0.002 | 0.0004 | 8.0E-07 | +, +, +, +, + | 0 |
| cg03646329 | 13 | 48987165 | LPAR6;RB1 | AHEI | 0.003 | 0.001 | 1.5E-06 | +, +, +, +, + | 0 |
| MDS | 0.004 | 0.001 | 1.1E-06 | +, +, +, +, + | 0 | ||||
| cg16969872 | 13 | 79968324 | RBM26 | AHEI | 0.003 | 0.001 | 3.0E-09 | +, +, +, +, + | 0 |
| MDS | 0.003 | 0.001 | 1.2E-06 | +, +, +, +, + | 0.25 | ||||
| cg09940677 | 14 | 103415458 | CDC42BPB | AHEI | −0.001 | 0.0003 | 2.9E-06 | −, −, −, −, − | 0 |
| cg13074055 | 14 | 106329206 | AHEI | 0.005 | 0.001 | 1.3E-06 | +, +, +, +, + | 0 | |
| cg27118035 | 16 | 31891978 | ZNF267 | AHEI | −0.003 | 0.0005 | 4.9E-09 | −, −, −, −, − | 0 |
| cg08732950 | 16 | 89023389 | CBFA2T3 | MDS | −0.003 | 0.0005 | 2.8E-08 | −, −, −, −, − | 0 |
| cg02097604 | 17 | 17750910 | TOM1L2 | AHEI | 0.002 | 0.0003 | 6.6E-09 | +, +, +, +, + | 0 |
| MDS | 0.002 | 0.0003 | 3.6E-08 | +, +, +, +, + | 0 | ||||
| cg16936953 | 17 | 57915665 | VMP1 | AHEI | 0.004 | 0.001 | 1.5E-08 | +, +, +, +, + | 0 |
| cg18181703 | 17 | 76354621 | SOCS3 | AHEI | 0.004 | 0.001 | 2.0E-12 | +, +, +, +, + | 0 |
| MDS | 0.004 | 0.001 | 3.5E-10 | +, +, +, +, + | 0 | ||||
| cg19202384 | 17 | 79894511 | PYCR1 | AHEI | 0.002 | 0.0004 | 9.9E-07 | +, +, +, +, + | 0.04 |
| cg01294327 | 19 | 2291373 | LINGO3 | AHEI | 0.005 | 0.001 | 1.4E-06 | +, +, +, +, + | 0 |
| cg26470501 | 19 | 45252955 | BCL3 | AHEI | 0.002 | 0.0004 | 2.4E-06 | +, +, +, +, + | 0.03 |
| cg08884571 | 19 | 45901453 | PPP1R13L | AHEI | −0.004 | 0.001 | 4.6E-07 | −, −, −, −, − | 0 |
| cg05232694 | 20 | 48809539 | AHEI | 0.004 | 0.001 | 3.1E-08 | +, +, +, +, + | 0 | |
Genome build 37. Regression coefficients are DNA methylation change for per standard deviation change in diet scores from analyses using sex, age, and energy intake adjusted models. Direction order (from left to right): FHS, ARIC, GOLDN, MESA, and RS. AHEI: Alternative Healthy Eating Index. MDS: Mediterranean-style diet score
Functional and regulatory annotation of diet-associated CpGs.
Relative to the whole set of CpGs analyzed, the 30 diet-associated CpGs were enriched in gene body regions (p-value = 9.3×10−4). The mean whole blood-derived DNA methylation levels of the 30 CpGs were moderately associated with those measured in muscle, omentum, and spleen (Supplemental Figure 5), with Spearman ranked r = 0.56 (n=6), 0.60 (n=6), and 0.62 (n=3); p-value = 1.5×10−3, 6.1×10−4, and 3.5×10−4, respectively.19
Among the 30 CpGs, 26 CpGs were annotated to 27 protein-coding genes (Supplemental Table 6). Based on the GTEx expression dataset,20 the annotated genes were differentially expressed in several tissues (Supplemental Figure 6 and Supplemental Table 7), e.g., differential expression was reported for 17 genes in muscle and 12 genes in small intestine (Bonferroni corrected p-value = 0.03 and 0.04, respectively). Gene set analyses did not reveal significant enrichment of pathways. Several genes, however, have important biological functions relevant to diet-associated diseases, e.g., SORBS1 (annotated to cg03190891) and FADS2 (annotated to cg11250194) play crucial roles in insulin signaling and fatty acids metabolism, respectively.
GWAS analysis
We identified 4,925 cis-meQTL variants for 23 of the 30 CpGs in the FHS (Supplemental Material). We found that 68 cis-meQTL variants for ten CpGs exactly matched a GWAS reported single nucleotide polymorphism (SNP) in the NHGRI-EBI GWAS Catalog21 (p-value < 5×10−8; Supplemental Table 8). For example, rs174550 for cg11250194 (FADS2) was associated with plasma omega-6 polyunsaturated fatty acid concentrations.22 Overall, these ten CpGs were linked to 35 unique traits, of which many are also diet-associated, such as lipid levels and chronic kidney disease.23,24
Associations of diet-associated CpGs with CVD risk factors.
In the EWAS catalog (Supplemental Table 9), we found that 26 (of 30) CpGs have been reported to be associated with one or more CVD risk factors, e.g., hypermethylation of cg18181703 (SOCS3) was associated with lower BMI and lower risk of type 2 diabetes.25–27 We conducted bidirectional Mendelian Randomization (MR) analysis to examine the potential causal relations between diet-associated CpGs and CVD risk factors, i.e., CpG → CVD trait and CVD trait → CpG. The MR analysis in direction of CpG to CVD trait was performed for 22 (of 30) CpGs that had cis-meQTL variants and summary results from the selected GWAS. We found significant putatively causal association for eight CpG-trait pairs after Bonferroni correction for 22 CpGs and five traits (corresponding MR p-value < 4.5×10−4) and nominally significant putatively causal association for 14 CpG-trait pairs (MR p-value < 0.05; Supplemental Table 10). For example, as shown in Figure 3, hypermethylation of cg11250194 (FADS2) was associated with lower triglyceride concentrations (MR p-value = 1.5×10−14) and hypermethylation of cg02079413 (SNORA54; NAP1L4) was associated with higher BMI (MR p-value = 1×10−6). We also observed unexpected associations in the MR analysis. For example, hypermethylation of cg26470501 (BCL3) was positively associated with BMI (MR p-value = 6.5×10−5; Supplemental Table 10; Figure 3), which was not consistent with the positive association that we observed between diet and this CpG and the inverse association between this CpG and BMI.25,28 In the opposite direction, MR analyses linking CVD traits to CpG, revealed no significant putative causal association after correction for multiple testing (p-value < 0.002; 0.05/30 diet-associated CpGs; Supplemental Table 11). Nevertheless, we observed two nominally significant associations: higher BMI was associated with hypomethylation of cg18181703 (p-value = 0.04) and higher waist-to-hip ratio adjusted for BMI (WHRadjBMI) was associated with hypomethylation of cg25953130 (p-value = 0.02).
Figure 3.
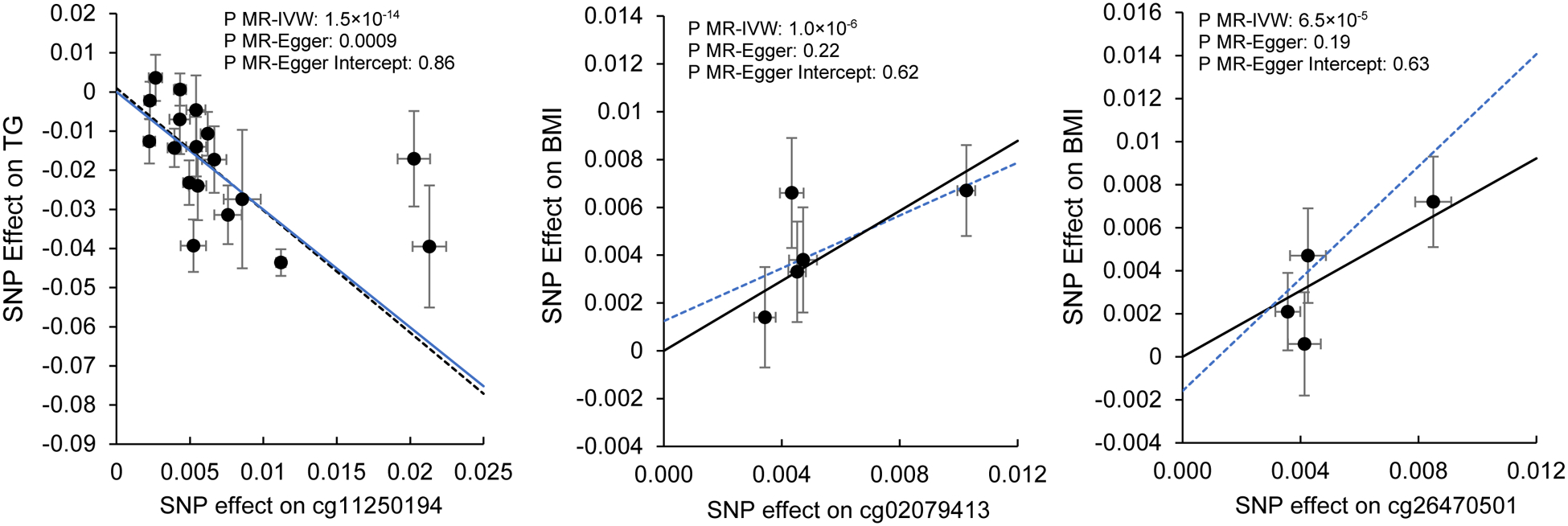
Mendelian Randomization (MR) analyses for associations between cg11250194 (FADS2) and triglycerides (TG), between cg02079413 (SNORA54; NAP1L4) and BMI, and between cg26470501 (BCL3) and BMI. IVW: inverse variance weighted. Symbols and bars represent effects size and standard errors of instruments variables (cis-meQTL variants) used in MR analysis. Solid line is for MR-IVW analysis and dashed line is for MR-Egger analysis. No horizontal pleiotropy effect was detected for all MR analyses.
Relations of diet-associated CpGs with mortality.
Of the 30 diet score-associated CpGs, the relations of 27 CpGs with all-cause mortality were examined in ten EA cohorts (N up to 10,083). Three CpGs were excluded because of missing data. After adjusting for multiple covariates (Figure 4), we found that 12 CpGs were significantly associated with all-cause mortality following Bonferroni correction (corresponding p-value < 1.6×10−3); ten additional CpGs were nominally associated with all-cause mortality (p-value < 0.05). The direction of the associations between CpGs and mortality was concordant with that for the diet-CpG associations, e.g., hypermethylation of cg18181703 (SOCS3), which was associated with higher scores of both AHEI and MDS, was associated with lower all-cause mortality (p-value = 5.7×10−15).
Figure 4.
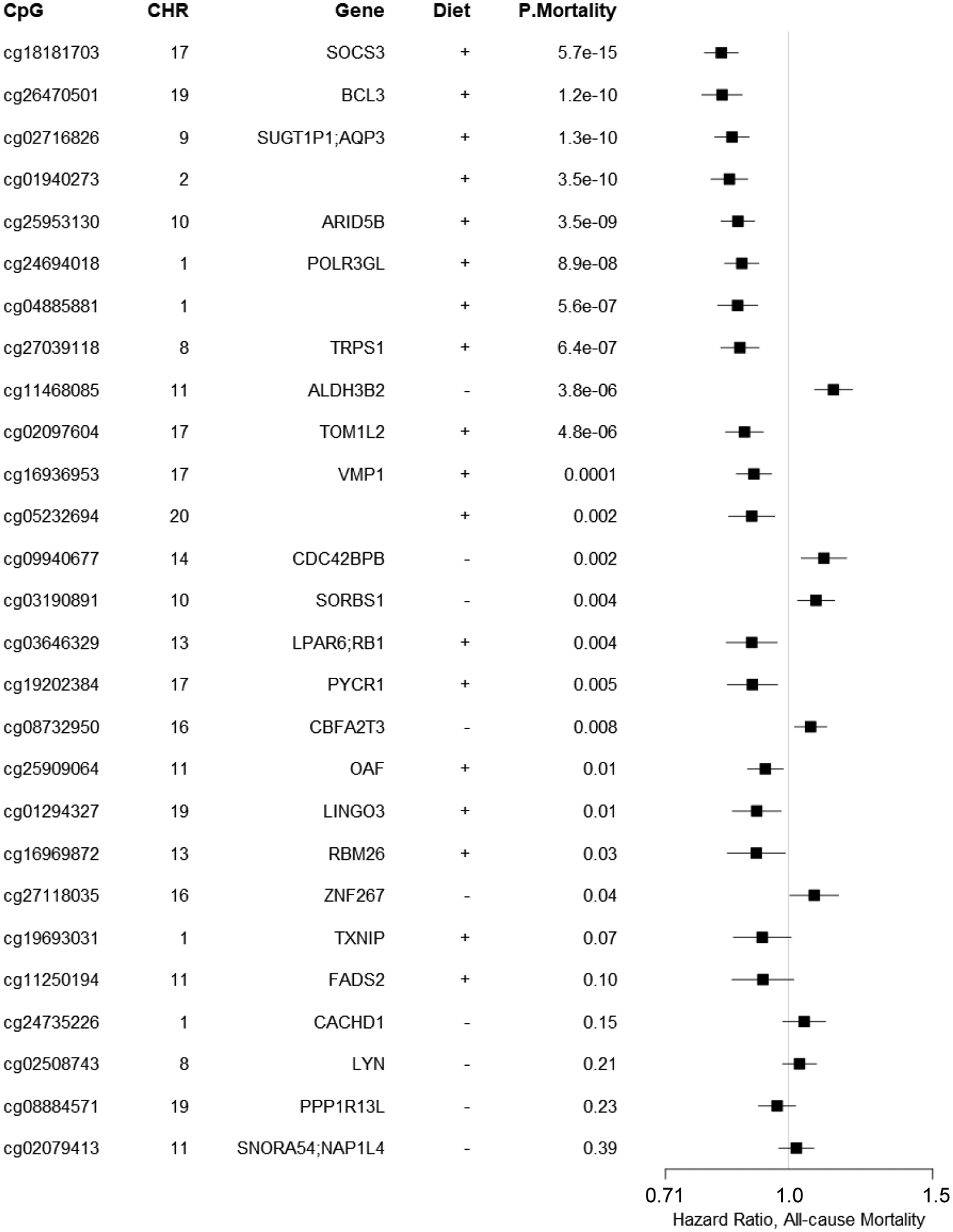
Meta-analysis of association between 30 diet-associated CpGs and all-cause mortality in 10 cohorts of European ancestry participants (n≈10,000). A positive sign for diet indicates that a higher dietary scores (MDS or AHEI, or both) were associated with DNA hypermethylation, whereas, a hazard ratio of over 1.0 indicates that DNA hypermethylation was associated with increased all-cause mortality. Models were adjusted for baseline covariates including sex, age, smoking status, physical activity level, alcohol intake, BMI, and prevalence disease status of hypertension, type 2 diabetes, cardiovascular disease, and cancer. Estimated leukocyte counts, technical variables, and kinship (for related study samples) were also considered. Hazard ratios and 95% confidence interval were estimated using Cox proportional hazard models and meta-analyzed using random effect models. X-axis is in logarithmic scale.
Multiethnic analysis
Although we observed largely consistent directions of effect in AA and HA participants for the 30 CpGs identified in EA participants, none of these CpGs was significant after Bonferroni correction (Supplemental Table 12). The transethnic meta-analysis identified 21 CpGs at FDR < 0.05 including 13 CpGs for AHEI with a corresponding p-value of 1.1×10−6 and 10 CpGs for MDS with a corresponding p-value of 7×10−7 (Supplemental Table 13). Of the 21 CpGs, ten CpGs were not among the 30 CpGs identified in EA participants and the correlations of the ten CpGs with the 30 CpGs were low to moderate, |r| ranging from 0 to 0.49 (Supplemental Table 14). The annotated genes for these ten CpGs (Supplemental Table 15) showed enrichment of lipid metabolism-related pathways (Supplemental Table 16). Nine of the ten CpGs were associated with nine unique traits in the EWAS catalog including serum triglyceride and HDL concentrations29 (Supplemental Table 17).
Discussion
In participants of EA ancestry, we identified 30 CpGs whose methylation in whole blood was associated with diet scores assessed, either MDS or AHEI, or both. Aligning cis-meQTL variants for these CpGs with GWAS catalog reported variants revealed that diet-associated differential DNA methylation can be linked to a series of metabolic and inflammatory disorders. Importantly, we also observed associations between these CpGs and all-cause mortality, which may reflect the importance of diet-induced epigenetic changes on health outcomes. Our study provides novel evidence that integrative genomic analysis of dietary information may be useful to highlight molecular targets for disease prevention and treatment.
Accumulating evidence has shown that epigenetic profiles may be regulated by dietary factors.6 A recent study found that women who had better adherence to the Mediterranean diet had greater DNA methylation levels at long interspersed nucleotide elements 1 (LINE-1), a surrogate marker of global genomic DNA methylation.8 In a small subgroup (n=36) of the Prevención con Dieta Mediterránea (PREDIMED) study, genome-wide methylation levels in peripheral blood derived DNA were assessed at baseline and again five years later.7 This study revealed that adherence to the Mediterranean diet may impact DNA methylation levels of several inflammation-related genes. None of the CpGs identified in this PREDIMED report, however, showed statistically significant differential DNA methylation in the meta-analysis in the present study.
Higher MDS and AHEI scores have been reported to be associated with lower body weight.17,18 Our observation that diet scores were positively associated with DNA methylation levels of cg18181703 (SOCS3) is therefore consistent with the inverse association of cg18181703 and BMI identified in multiple studies.25,28,30 Overall, by integrating association analysis and MR analysis, our data indicate that diet quality may affect BMI, alter DNA methylation of cg18181703, and impact long-term health. The association between cg18181703 and all-cause mortality also was consistent with observations in a small-scale epigenome-wide study.31 SOCS3 is a well characterized gene involved in immune system regulation, which suggests that the association of diet scores and cg18181703 may be relevant to inflammation and may partly explain the association of cg18181703 with all-cause mortality.
Several diet score-associated CpGs, such as cg19693031 (TXNIP) and cg02716826 (SUGT1P1; AQP3), have been reported to be associated with CVD risk factors.26,27 TXNIP, thioredoxin-interacting protein, is a key regulator of energy metabolism and a therapeutic candidate for type 2 diabetes.32 AQP3, aquaporin 3, is a member of water channel proteins that are associated with a number of diseases such as hypertension and congestive heart failure.33 Our MR analyses also support a causal link between methylation levels of diet-associated CpGs and CVD risk factors, e.g., hypermethylation of cg11250194 (FADS2) was associated with lower triglyceride concentrations. FADS2 is a key member of the fatty acid desaturase (FADS) family.34 This observation is consistent with the role of diet in the regulation of enzyme activity relevant to fatty acid desaturation.35 Therefore, the present study provides key evidence that diet may interact with the human genome via epigenetic mechanisms to impact health outcomes.
In a post-hoc analysis, we conducted lookup analysis for diet score-associated CpGs using data reported in a prior epigenome-wide association study of acute coronary syndrome.36 Although none of the 30 CpGs we report here was significant or correlated with the 47 CpGs identified in this prior study, we found that one of the 47 CpGs, cg14066471 was significantly associated with AHEI (p=0.0004) and nominally associated with MDS (p=0.004) after Bonferroni correction for 47 CpGs in our trans-ethnic analysis. This observation suggests that our data represent a good resource for other investigators to conduct lookup analyses. It also suggests that a candidate approach may identify additional CpGs associated with diet and diseases.
A major strength of the present study is its large sample size, which includes data from five US and European population-based cohorts, and the use of two common and well-studied diet scores. Several limitations warrant discussion. The diet scores were based on different versions of FFQs, which are prone to measurement errors due to self-reported diet data. In addition, although the associations remained significant for the majority of CpGs after adjustment for lifestyle factors, we cannot rule out the possibility of residual confounding. Although we showed a moderate correlation between peripheral blood-derived DNA methylation profiles and those from other tissues, we lacked data to analyze tissue-specific diet-associated DNA methylation changes which may be more directly related to the development of chronic diseases. Our study may lack power to detect diet-associated DNA methylation markers in AA and HA participants due to the smaller sample sizes (n=2,702 for AA and n=360 for HA) relative to EA participants (n=6,662).
In conclusion, the present study demonstrates that diet quality is associated with differential DNA methylation levels of 30 CpGs in leukocyte-derived DNA among EA participants. Our findings demonstrate that integration of dietary information and genomic data may reveal useful insights into the molecular effects at the intersection of diet, risk factors, and chronic diseases. Future studies with larger sample sizes, deeper coverage of DNA methylation, and more precise dietary measurement are needed to validate our findings and to investigate diet-associated DNA methylation patterns in larger ethnically diverse samples.
Supplementary Material
Acknowledgments:
We thank Mr. Michael Verbiest, Ms. Mila Jhamai, Ms. Sarah Higgins, Mr. Marijn Verkerk, and Lisette Stolk PhD for their help in creating the methylation database. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. CR reports receiving grant support from CONICYT PAI/INDUSTRIA 72170524. The authors thank the staff and participants of the ARIC study for their important contributions.
Sources and Funding: Infrastructure for the CHARGE Consortium is supported in part by the National Heart, Lung, and Blood Institute grant R01HL105756. This work was supported in part by the Intramural Research Program of the National Institutes of Health (NIH): National Heart Lung and Blood Institute, National Institute on Aging and the National Institute of Environmental Health Sciences.
The FHS (Framingham Heart Study) is funded by National Institutes of Health contract N01-HC-25195. The laboratory work for this investigation was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD. The analytical component of this project was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, and the Center for Information Technology, NIH, Bethesda, MD. JM is supported by the National Heart, Lung, and Blood Institute career transition award (1K22HL135075-01). MW is supported by NIH T32 (5T32HL125232).
The generation and management of the Illumina 450K methylation array data (EWAS data) for the Rotterdam Study (RS) was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Netherlands. The EWAS data was funded by the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, and by the Netherlands Organization for Scientific Research (NWO; project number 184021007) and made available as a Rainbow Project (RP3; BIOS) of the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL).
MESA (Multi-Ethnic Study of Atherosclerosis) and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, UL1-TR-000040, and DK063491. The MESA Epigenomics & Transcriptomics Study was funded by NIA grant 1R01HL101250-01 to Wake Forest University Health Sciences. Analysis of MESA data reported in this publication was also supported by NIA grant R03AG056959.
The Atherosclerosis Risk in Communities (ARIC) study has been funded by federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I, HHSN268201700005I).
Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) epigenetics data were generated with support from NIH NHLBI R01HL104135 (PI: Arnett) and analyzed with NIH NHLBI K01HL136700 (PI: Aslibekyan).
The ESTHER study was funded by grants from the Saarland state Ministry for Social Affairs, Health, Women and Family Affairs (Saarbrücken, Germany), the Baden-Württemberg state Ministry of Science, Research and Arts (Stuttgart, Germany), the Federal Ministry of Education and Research (Berlin, Germany) and the Federal Ministry of Family Affairs, Senior Citizens, Women and Youth (Berlin, Germany).
Cardiovascular Health Study (CHS) Infrastructure for the CHARGE Consortium is supported in part by the National Heart, Lung, and Blood Institute grant R01HL105756. The CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants U01HL080295, U01HL130114, K08HL116640, R01HL087652, R01HL092111, R01HL103612, R01HL105756, R01HL103612, R01HL111089, R01HL116747 and R01HL120393 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 from the National Institute on Aging (NIA), Merck Foundation / Society of Epidemiologic Research as well as Laughlin Family, Alpha Phi Foundation, and Locke Charitable Foundation. A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
The KORA study was initiated and financed by the Helmholtz Zentrum München – German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Furthermore, KORA research has been supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. The work was supported by the German Federal Ministry of Education and Research (BMBF) within the framework of the EU Joint Programming Initiative ‘A Healthy Diet for a Healthy Life’ (DIMENSION grant number 01EA1902A).
The Women’s Health Initiative (WHI) is funded by the National Heart, Lung and Blood Institute, U.S. Department of Health and Human Services, through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C. The Epigenetic Mechanisms of PM-Mediated CVD Risk (WHI-EMPC) was supported by National Institute of Environmental Health Science grant R01-ES020836. All contributors to WHI science are listed at https://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf.
Nonstandard Abbreviations and Acronyms
- CpG
cytosine–guanine dinucleotide
- CVD
cardiovascular disease
- n-3 FAs
omega-3 polyunsaturated fatty acids
- MDS
Mediterranean-style diet score
- AHEI
Alternative Healthy Eating Index
- EA
European Ancestry
- AA
African ancestry
- HA
Hispanic ancestry
- FDR
false discovery rate
- MR
Mendelian Randomization
Footnotes
Publisher's Disclaimer: Disclaimer: The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
Disclosure: None.
References:
- 1.Smith R, Mill J. Epigenetics and Chronic Diseases: An Overview. Epigenetic Aspects of Chronic Diseases. 2011:1–20. [Google Scholar]
- 2.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA Methylation in Cancer and Aging. Cancer Res. 2016;76:3446–3450. [DOI] [PubMed] [Google Scholar]
- 3.Zhong J, Agha G, Baccarelli AA. The Role of DNA Methylation in Cardiovascular Risk and Disease: Methodological Aspects, Study Design, and Data Analysis for Epidemiological Studies. Circ Res. 2016;118:119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agha G, Mendelson MM, Ward-Caviness CK, Joehanes R, Huan T, Gondalia R, Salfati E, Brody JA, Fiorito G, Bressler J, et al. Blood Leukocyte DNA Methylation Predicts Risk of Future Myocardial Infarction and Coronary Heart Disease. Circulation. 2019;140:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferguson JF, Allayee H, Gerszten RE, Ideraabdullah F, Kris-Etherton PM, Ordovás JM, Rimm EB, Wang TJ, Bennett BJ; American Heart Association Council on Functional Genomics and Translational Biology, Council on Epidemiology and Prevention, and Stroke Council. Nutrigenomics, the Microbiome, and Gene-Environment Interactions: New Directions in Cardiovascular Disease Research, Prevention, and Treatment: A Scientific Statement From the American Heart Association. Circ Cardiovasc Genet. 2016;9:291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Kutateladze TG. Diet and the epigenome. Nat Commun. 2018;9:3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arpon A, Riezu-Boj JI, Milagro FI, Marti A, Razquin C, Martínez-González MA, Corella D, Estruch R, Casas R, Fitó M, et al. Adherence to Mediterranean diet is associated with methylation changes in inflammation-related genes in peripheral blood cells. J Physiol Biochem. 2016;73:445–455. [DOI] [PubMed] [Google Scholar]
- 8.Barchitta M, Maugeri A, Quattrocchi A, Barone G, Mazzoleni P, Catalfo A, De Guidi G, Iemmolo MG, Crimi N, Agodi A. Mediterranean Diet and Particulate Matter Exposure Are Associated With LINE-1 Methylation: Results From a Cross-Sectional Study in Women. Front Genet. 2018;9:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tremblay BL, Guénard F, Rudkowska I, Lemieux S, Couture P, Vohl MC. Epigenetic changes in blood leukocytes following an omega-3 fatty acid supplementation. Clin Epigenetics. 2017;9:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kris-Etherton P, Eckel RH, Howard BV, St Jeor S, Bazzarre TL; Nutrition Committee Population Science Committee and Clinical Science Committee of the American Heart Association. AHA Science Advisory: Lyon Diet Heart Study. Benefits of a Mediterranean-style, National Cholesterol Education Program/American Heart Association Step I Dietary Pattern on Cardiovascular Disease. Circulation. 2001;103:1823–1825. [DOI] [PubMed] [Google Scholar]
- 11.Appel LJ, Moore TJ, Obarzanek E, Vollmer WM, Svetkey LP, Sacks FM, Bray GA, Vogt TM, Cutler JA, Windhauser MM, et al. A clinical trial of the effects of dietary patterns on blood pressure. DASH Collaborative Research Group. N Engl J Med. 1997;336:1117–1124. [DOI] [PubMed] [Google Scholar]
- 12.Sacks FM, Svetkey LP, Vollmer WM, Appel LJ, Bray GA, Harsha D, Obarzanek E, Conlin PR, Miller ER 3rd, Simons-Morton DG, et al. Effects on blood pressure of reduced dietary sodium and the Dietary Approaches to Stop Hypertension (DASH) diet. DASH-Sodium Collaborative Research Group. N Engl J Med. 2001;344:3–10. [DOI] [PubMed] [Google Scholar]
- 13.Trichopoulou A, Costacou T, Bamia C, Trichopoulos D. Adherence to a Mediterranean diet and survival in a Greek population. N Engl J Med. 2003;348:2599–2608. [DOI] [PubMed] [Google Scholar]
- 14.Sotos-Prieto M, Bhupathiraju SN, Mattei J, Fung TT, Li Y, Pan A, Willett WC, Rimm EB, Hu FB. Association of Changes in Diet Quality with Total and Cause-Specific Mortality. N Engl J Med. 2017;377:143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung TT, Rexrode KM, Mantzoros CS, Manson JE, Willett WC, Hu FB. Mediterranean diet and incidence of and mortality from coronary heart disease and stroke in women. Circulation. 2009;119:1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiuve SE, Fung TT, Rimm EB, Hu FB, McCullough ML, Wang M, Stampfer MJ, Willett WC.. Alternative dietary indices both strongly predict risk of chronic disease. J Nutr. 2012;142:1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fung TT, Pan A, Hou T, Chiuve SE, Tobias DK, Mozaffarian D, Willett WC, Hu FB.. Long-Term Change in Diet Quality Is Associated with Body Weight Change in Men and Women. J Nutr. 2015;145:1850–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma J, Hennein R, Liu C, Long MT, Hoffmann U, Jacques PF, Lichtenstein AH, Hu FB, Levy D.. Improved Diet Quality Associates With Reduction in Liver Fat, Particularly in Individuals With High Genetic Risk Scores for Nonalcoholic Fatty Liver Disease. Gastroenterology. 2018;155:107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slieker RC, Bos SD, Goeman JJ, Bovée JV, Talens RP, van der Breggen R, Suchiman HE, Lameijer EW, Putter H, van den Akker EB, et al. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin. 2013;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galperin MY, Fernandez-Suarez XM, Rigden DJ. The 24th annual Nucleic Acids Research database issue: a look back and upcoming changes. Nucleic Acids Res. 2017;45:D1–D11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan W, Steffen BT, Lemaitre RN, , Wu JHY, Tanaka T, Manichaikul A, Foy M, Rich SS, Wang L, Nettleton JA, et al. Genome-wide association study of plasma N6 polyunsaturated fatty acids within the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet. 2014;7:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estruch R, Camafort M. The Mediterranean diet and plasma lipid profile. Rev Esp Cardiol (Engl Ed). 2015;68:279–281. [DOI] [PubMed] [Google Scholar]
- 24.Ma J, Jacques PF, Hwang SJ, Troy LM, McKeown NM, Chu AY, Fox CS. Dietary Guideline Adherence Index and Kidney Measures in the Framingham Heart Study. Am J Kidney Dis. 2016;68:703–715. [DOI] [PubMed] [Google Scholar]
- 25.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, Tsai PC, Ried JS, Zhang W, Yang Y, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V, Wahl S, Elliott HR, Rota F, Scott WR, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3:526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ligthart S, Marzi C, Aslibekyan S, Mendelson MM, Conneely KN, Tanaka T, Colicino E, Waite LL, Joehanes R, Guan W, et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 2016;17:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendelson MM, Marioni RE, Joehanes R, Liu C, Hedman ÅK, Aslibekyan S, Demerath EW, Guan W, Zhi D, Yao C, et al. Association of Body Mass Index with DNA Methylation and Gene Expression in Blood Cells and Relations to Cardiometabolic Disease: A Mendelian Randomization Approach. PLoS Med. 2017;14:e1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hedman AK, Mendelson MM, Marioni RE, Gustafsson S, Joehanes R, Irvin MR, Zhi D, Sandling JK, Yao C, Liu C, et al. Epigenetic Patterns in Blood Associated With Lipid Traits Predict Incident Coronary Heart Disease Events and Are Enriched for Results From Genome-Wide Association Studies. Circ Cardiovasc Genet. 2017;10:e001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu K, Zhang X, Wang Z, Hu Y, Sinha R. Epigenome-wide association analysis revealed that SOCS3 methylation influences the effect of cumulative stress on obesity. Biol Psychol. 2018;131:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Wilson R, Heiss J, Breitling LP, Saum KU, Schöttker B, Holleczek B, Waldenberger M, Peters A, Brenner H. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat Commun. 2017;8:14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alhawiti NM, Al Mahri S, Aziz MA, Malik SS, Mohammad S. TXNIP in Metabolic Regulation: Physiological Role and Therapeutic Outlook. Curr Drug Targets. 2017;18:1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeyaseelan K, Sepramaniam S, Armugam A, Wintour EM. Aquaporins: a promising target for drug development. Expert Opin Ther Targets. 2006;10:889–909. [DOI] [PubMed] [Google Scholar]
- 34.Lattka E, Illig T, Koletzko B, Heinrich J. Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr Opin Lipidol. 2010;21:64–69. [DOI] [PubMed] [Google Scholar]
- 35.Matthan NR, Ooi EM, Van Horn L, Neuhouser ML, Woodman R, Lichtenstein AH. Plasma phospholipid fatty acid biomarkers of dietary fat quality and endogenous metabolism predict coronary heart disease risk: a nested case-control study within the Women’s Health Initiative observational study. J Am Heart Assoc. 2014;3:e000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Zhu X, Yu K, Jiang H, Zhang Y, Deng S, Cheng L, Liu X, Zhong J, Zhang X, et al. Genome-Wide Analysis of DNA Methylation and Acute Coronary Syndrome. Circ Res. 2017;120:1754–1767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


