Abstract
Background
Meningiomas are the most common primary intracranial tumours. They are classified as grade I, II, and III based on their histopathological features. While most meningiomas can be managed by surgery alone, adjuvant treatment may be required in case of recurrent, or high-grade tumours. To date, chemotherapy has proven ineffective in meningioma patients, reinforcing the need for novel therapeutic targets and molecular biomarkers.
Methods
Using meningioma tissues and in vitro models, we investigated microRNA levels in meningioma samples of different grades, as well as their regulation. Based on this, we also investigated candidate miRNAs expression in serum, and their potential as biomarkers.
Findings
We found that miR-497~195 cluster expression in meningioma decreases with increasing malignancy grade, and that Cyclin D1 overexpression correlated with downregulation of the miR-497~195 cluster. GATA binding protein 4, a transcription factor upregulated in malignant meningioma, caused increased cell viability by controlling the expression of the miR-497~195 cluster, resulting in increased Cyclin D1 expression. Accordingly, GATA-4 inhibition via the small-molecule inhibitor NSC140905 restored miR-497~195 cluster expression, resulting in decreased viability, and Cyclin D1 downregulation. Analysis of the miR-497~195 cluster expression in serum exosomes derived from high-grade meningioma patients, revealed lower levels of miR-497 compared to those of benign origin.
Interpretation
Our data suggest that GATA-4 could be a novel potential therapeutic target, and miR-497 could serve as a potential non-invasive biomarker for high-grade meningioma.
Keywords: MeningiomaGATA-4ExosomesmiRNAmiR-497Liquid biopsies
Research in context.
Evidence before this study
Meningioma is the most frequent primary intracranial tumour. Despite extensive genetic characterization of the mutational profile of these tumours, both treated and untreated meningiomas undergo long-term follow-up with MRI surveillance. There is no existing blood-based biomarker to guide when to treat, or how to best follow-up patients with meningioma, highlighting an unmet clinical need. Imaging is useful to detect a meningioma, but cannot define tumour grade. Thus, grading is based on histopathological characterization of tumour biopsies, which relies on tissue availability. Moreover, no effective pharmacological intervention for meningioma has been established to date, highlighting the need to find novel biomarkers and therapeutic targets for these tumours.
Added value of this study
In this study, we identified potential novel tissue-specific and circulating biomarkers, as well as a novel therapeutic target, for higher-grade meningioma tumours. We analysed tumour tissues and serum-derived exosomes covering all WHO grades, including the largest sample size to date of WHO III meningiomas. We also used a validation cohort to further support our findings in circulating exosomes. We described overexpression in malignant meningioma of a novel transcription factor, which acts as an oncoprotein in this tumour type, via downregulation of a miRNA cluster with tumour-suppressive characteristics.
Implications of all the available evidence
This study demonstrates the value of miRNAs as biomarkers for meningioma, especially in serum-derived exosomes. These can be used as liquid biopsies to facilitate or support meningioma diagnosis, which is currently solely dependent on tissue availability. Moreover, it proves that investigation of miRNAs regulation in meningioma can lead to the identification of new potential therapeutic targets. Further validation of our results may have clinical importance, leading to the establishment of novel biomarkers and treatment strategies.
Alt-text: Unlabelled box
1. Introduction
Meningiomas are the most common primary intracranial brain tumours, accounting for one third of all primary central nervous system (CNS) tumours [1]. According to the World Health Organization (WHO), meningiomas are classified as grade I, II, and III, with higher-grades associated with greater rates of morbidity and mortality [2]. Tumour recurrence depends on the histological grade of the tumour and the extent of resection; recurrence risks at 5 years for WHO I, II, and III meningioma are approximately 5–10%, 50%, and 80%, respectively [3,4]. Meningiomas can be asymptomatic, with tumours diagnosed as a result of imaging for other purposes [5]; when symptoms develop, they depend on the location of the tumour, and usually include seizure, headache, and focal neurological deficits [6]. The current standard treatment for meningioma is surgical excision, which can sometimes be challenging depending on tumour location, and presence of brain invasion [7]; adjuvant radiotherapy is used in higher-grade or recurrent meningiomas, especially when total resection is not possible. Available chemotherapies proved to be ineffective in meningioma patients, or met with significant toxicity, highlighting the need to find new therapeutic targets for meningioma treatment [8], [9], [10], [11].
The genetic landscape of meningioma has been well characterized over the years; these tumours commonly arise as a consequence of a genetic condition known as type 2 neurofibromatosis (NF2) [8]. Nearly all NF2-associated meningiomas, and the majority of sporadic tumours (~60%) carry the NF2 mutation, while the remaining non-NF2 related 40% of the tumours carry mutations in TRAF7, KLF4, SMO, PIK3CA, POLR2A, PRKAR1A, AKT3, and SUFU [3,12,13]. Despite this, prognostic tumour classification is based on histopathological characterization of the tumour [14], [15], [16].
Recently, some studies focussed on microRNAs (miRNAs) in order to identify specific molecular signatures for meningioma [17], [18], [19], [20], [21], [22], but investigations of their functions in this tumour type are lacking. miRNAs are small non-coding RNAs of around 20–22 nucleotides in length that function as negative gene regulators [23,24]. Interestingly, it has been shown that miRNAs can also be released from cells into the extracellular space via exosomes, vesicles of around 40–200 nm in size, which can be found in most body fluids, including the bloodstream [25], [26], [27], [28], [29].
This study focussed on the identification of a miRNA signature that could differentiate meningioma grades, its regulation, and the evaluation of its potential to be used as a biomarker, both in tissues and in serum-derived exosomes. We determined that Cyclin D1 overexpression in malignant meningioma[30], [31], [32] correlated with downregulation of the miR-497~195 cluster via upregulation of the GATA binding protein 4 (GATA-4) [33], a zinc finger transcription factor overexpressed in WHO III meningioma. Moreover, we investigated the synthetic lethality of NSC140905 (2-(1,3-benzodioxol-5-ylmethyl)butanedioic acid), a newly pioneered GATA-4 small-molecule inhibitor [34], in malignant meningioma cells. Interestingly, NSC140905 treatment led to a decrease in cell viability, and an increased miR-497~195 cluster expression, paralleled with a reduction of Cyclin D1, which is a predicted target of miR-497 and −195 [35].
Consistently with the analysis in tissues, isolation of circulating exosomes derived from high-grade meningioma patients showed lower levels of the miR-497~195 cluster when compared to those of benign origin. In particular, miR-497 showed a good diagnostic value in both the discovery and validation sets, suggesting its potential to be further investigated as a non-invasive biomarker for higher-grade meningioma tumours.
2. Material and methods
2.1. Clinical samples
Meningioma (MN) specimens were collected following the national ethical approvals (REC No: 14/SW/0119; IRAS project ID: 153,351) (Plymouth Hospitals NHS Trust: R&D No: 14/P/056 and North Bristol NHS Trust: R&D No: 3458), receiving a unique MN number. Blood was collected at the time point of surgery. ‘J’ specimens were collected via UK-Brain-Archive Information-Network (BRAIN UK; Ref no: 15/011; REC no: 14/SC/0098).
Clinical and histopathological data for all samples used in this study can be found in Tables S1–S3. All samples analysed here were untreated primary tumours, and were all tested for Epithelial Membrane Antigen (EMA), Vimentin, and Somatostatin Receptor 2 expression via immunohistochemistry to confirm that they consisted predominantly of viable meningioma.
Two frozen normal meninges were obtained from Analytical Biological Services Inc. and one human brain cerebral meninges was purchased from Novus Biologicals® (NB820–59183; lot B105014).
2.2. Cell culture
Human meningeal cells (HMC, Cat# 1400) were obtained from Sciencell™ and maintained following the manufacturer's protocol. The malignant meningioma cell line KT21-MG1-Luc5D (RRID:CVCL_JK00), the benign meningioma cell line Ben-Men-1 (RRID:CVCL_1959), and WHO I primary meningioma cells (PDMN) were maintained as previously described [36], [37], [38]. Briefly, tumour samples were washed twice with sterile PBS (Gibco, Life Technologies, Loughborough, UK), and transferred into a P100 sterile plate, where they were minced into small pieces and dissociated in DMEM (Gibco, Life Technologies, Loughborough, UK), with 10% FBS (Sigma Aldrich, Gillingham, UK), 100 U/mL penicillin/streptomycin, and 20 u/mL Collagenase III (Worthington Biomedical Corp., Lakewood, NJ) overnight at 37 °C. After incubation, cells were pelleted, re-suspended in complete medium (DMEM, 10% FBS, 1% d-(+)-glucose, 100 U/mL penicillin/streptomycin, 2 mM GlutaMAX™-I), and seeded in appropriate tissue culture flasks. WHO II meningioma primary cells were isolated from resected tumours following the same protocol as WHO I PDMN cells, but were maintained in Dulbecco's Modified Eagle Medium F-12 Nutrient Mixture (Ham) (DMEM/F-12 (1:1)(1X) + GlutaMAX™-I; Thermo Fisher Scientific, Loughborough, UK), supplemented with 20% FBS (Sigma Aldrich, Gillingham, UK), 1% d-(+)-glucose (Sigma Aldrich, Gillingham, UK), and 100 U/mL penicillin/streptomycin (Thermo Fisher Scientific, Loughborough, UK). All PDMN cells were used between passage 3 and 5, and were tested for Epithelial Membrane Antigen (EMA) and Vimentin expression via immunocytochemistry [37,38].
2.3. RNA isolation and gene expression analysis
Total RNA was extracted using the Qiazol® reagent (Qiagen, Manchester, UK), following the manufacturer's protocol. RNA quality, integrity, and concentration were established using the NanoDrop ND-2000 (Thermo Fisher Scientific, Loughborough, UK).
RT-PCR was performed using 1 μg of total RNA with the TaqMan® MicroRNA Reverse Transcription Kit or the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Loughborough, UK), accordingly. Real Time PCR (qPCR) was conducted using the TaqMan® Fast Advanced Master Mix supplemented with TaqMan® assays (Thermo Fisher Scientific, Loghborough, UK) on a LightCycler® 480 II System (Roche Products Limited, Welwyn Garden City, UK), in three technical triplicates, employing the following assays (Thermo Fisher Scientific, Loughborough, UK): hsa-miR-195* (ID 002107), hsa-miR-15a* (ID 002419), hsa-miR-15b* (ID 002173), hsa-miR-16–1* (ID 002420) and hsa-miR-497* (ID 002368), CCND1 (Hs00765553_m1), BCL2 (Hs01048932_g1), and GATA4 (Hs00171403_m1). As internal controls, we used RNU6B (ID 001093) or GAPDH (Hs02786624_g1), accordingly. Gene expression levels were computed using the quantitative 2−(ΔΔCt) method, employing the HMC cells as calibrator [39].
2.4. Western blotting
Protein immunoblot was conducted as previously reported [40]. Briefly, tissues and cells were lysed using a RIPA buffer supplemented with protease (cOmplete™, EDTA free Protease inhibitor cocktail, Sigma Aldrich, Gillingham. UK) and phosphatase inhibitors (Santa Cruz Biotechnology Inc., Heidelberg, Germany). Protein concentration was estimated using the Coomassie Plus – The Better Bradford Assay™ Reagent (Thermo Fisher Scientific, Loughborough, UK), following the instructions of the supplier. Proteins were separated on a Laemmli SDS-PAGE, and transferred to a polyvinylidene difluoride membrane (Immun-Blot® PVDF Membrane, Bio-Rad). Membrane blocking, antibody incubation and washes were performed as previously described [37].
The following primary antibodies from Cell Signalling Technology (London, UK) were used: anti-GATA-4 (D3A3M, Cat# 36966S, RRID:AB_2799108), anti-Cyclin D1 (92G2, Cat# 2978T), anti-Bcl2 (124, Cat# 15071T, RRID:AB_2744528), anti-pRb (Ser807/811, D20B12, Cat# 8516S), and anti-CD9 (D8O1A, Cat# 13174S, RRID:AB_2798139). The anti-Rb (EPR17512, Cat# ab181616) antibody was purchased from Abcam (Cambridge, UK), while anti-CD63 (MX-49.129.5, Cat# sc-5275, RRID:AB_627877), and anti-Calnexin (H–70, Cat# sc-11397, RRID:AB_2243890) from Santa Cruz Biotechnology Inc. (Heidelberg, Germany), and anti-GM130 (35/GM130, Cat# 610823, RRID:AB_398142) from Becton Dickinson U.K. Ltd. (Swindon, UK). GAPDH (6C5, Cat# MAB374, RRID:AB_2107445, Merck Millipore, Watford, UK) was used as a loading control. Detection was achieved using the Pierce ECL or ECL Plus Western Blotting substrate (Thermo Fisher Scientific, Loughborough, UK). Membranes were exposed to Amersham Hyperfilm ECL (GE Healthcare Life Sciences, Chalfont Saint Giles, UK). Immunoreactive bands were acquired at a resolution of 600 dpi, quantified using the ImageJ software [41] and each band was normalized vs. the corresponding GAPDH value.
2.5. Immunohistochemistry
Paraffin sections (4 μm) were de-waxed, rehydrated, and incubated with primary antibody (GATA-4 25310, LOT A1218, Santa Cruz Biotechnology Inc, Heidelberg, Germany) at room temperature O/N, after antigen retrieval in Tris/EDTA for 30 min. Proteins were visualised with the Novolink Polymer detection system for Leica (Leica Biosystems, Newcastle, UK, RE 7140-K), according to the manufacturer's instructions. Slides were counterstained with haematoxylin (Sigma Aldrich, Gillingham, UK). The immunohistochemical results were reviewed ‘blind’ to the histological grade by a neuropathologist (DAH). Semi-quantitative assessment of staining intensity was assigned as follows: 0 (negative), 1 (low), 2 (moderate) and 3 (strong) [40]. Immunostaining was carried out in two batches, using internal positive control tissue to ensure consistency.
2.6. Lentiviral-mediated transduction
Ben-Men-1 and KT21-MG1 cells were plated in 6-well plates (1.5 × 105 cells/well) and left adhering O/N. Sub-confluent cells were washed once with PBS, and medium was replaced with complete medium supplemented with 8% protamine sulphate (Sigma Aldrich, Gillingham, UK). Lentiviral particles were added according to the manufacturer's protocol. ShRNA interference was conducted using the GATA4 shRNA (h) Lentiviral Particles (Cat# sc-35455-V, Santa Cruz Biotechnology Inc., Heidelberg, Germany), while GATA4 and hsa-miR-195–5p lentiviral-mediated transductions were conducted using the GATA4 pLenti-GIII-CMV-GFP-2A-Puro (Cat# LVP166711, Applied Biological Materials Inc., Richmond, BC, Canada) and the LentimiRa-GFP-hsa-miR-195–5p (Cat# mh15245, Applied Biological Materials Inc., Richmond, BC, Canada), respectively. As controls, we used the Control shRNA Lentiviral Particles (Cat# sc-108080, Santa Cruz Biotechnology Inc., Heidelberg, Germany), the pLenti-CMV-GFP-2A-Puro-Blank control virus (Cat# LVP690, Applied Biological Materials, Inc., Richmond, BC, Canada), and the Lenti-III-mir-GFP control virus (Cat# m002, Applied Biological Materials Inc., Richmond, BC, Canada), respectively. After 48 h of incubation, medium containing lentiviral particles was removed, cells were washed once with PBS, and transduced Ben-Men-1 and KT21-MG1 cells were selected for 4 days using 1 and 10 μg/mL puromycin (Thermo Fisher Scientific, Loughborough, UK), respectively.
2.7. The GATA-4 inhibitor NSC140905
The GATA-4 small-molecule inhibitor (2-(1,3-benzodioxol-5-ylmethyl)butanedioic acid), alias NSC140905, was synthesized and provided as a white powder by Angene International Ltd (Hong Kong, China) [34]. We enclosed the certificate of analysis including the material safety data sheet (MSDS), certificate of analysis (COA), LC-MS, and NMR spectrum in Supplemental Material 1. The NSC140905 was suspended in dimethyl sulfoxide (DMSO, Sigma Aldrich, Gillingham, UK) obtaining a stock solution of 4 mM, which was aliquoted and stored at −80 °C.
2.8. Cell viability assay
ATP levels were measured as an indicator of cell viability using the Cell Titer-Glo® Luminescent Cell Viability Assay Kit (G7570, Promega, Southampton, UK), according to the manufacturer's direction. Briefly, cells were plated in triplicate into a white, flat bottom 96-well plate at a density of 3500 cells/well, and allowed to adhere for 24 h. At the end of the desired time point, medium was replaced with fresh medium and the Cell Titer-Glo® reagent was added to each well. Plates were then shaken for 2 min at 450 rpm, and incubated in the dark for 10 min to allow signal stabilization. Luminescence was measured using the BMG Labtech FLUOstar® Omega plate reader (BMG Labtech, Aylesbury, UK).
2.9. Exosome isolation
Ben-Men-1, KT21-MG1, and WHO I PDMN cells were cultured in medium supplemented with Exo-FBS™ Exosome-depleted FBS (System Biosciences, Cambridge, UK). Briefly, cells were plated in 10-cm dishes (1 × 106 cells/dish) and left adhering O/N. Medium was replaced, and cells were cultured for three days. Supernatant was collected from confluent cells, and exosomes were harvested using the Total Exosome Isolation (from cell culture media) Reagent (Cat# 4478359, Gibco, Life Technologies, Loughborough, UK), following the manufacturer's recommendations. Immunoblot was performed using 40 µL of the exosomes resuspension, according to the Total Exosome RNA & Protein Isolation Kit (Cat# 4478545, Invitrogen, Thermo Fisher Scientific, Loughborough, UK) protocol.
After whole blood collection in gold Vacutainer® tubes (Becton Dickinson U.K. Ltd., Swindon, UK), specimens were allowed to clot undisturbed at room temperature for 30 min, and serum was obtained by centrifuging at 2400 x g for 10 min at 4 °C. Following centrifugation, serum was aliquoted (500 μL) into polypropylene tubes, and stored at −80 °C. Exosome isolation from patients’ serum samples was performed using the Total Exosome Isolation (from serum) Reagent (Cat# 4478360, Gibco, Life Technologies, Loughborough, UK), following the manufacturer's instructions.
2.10. Statistical analysis
Statistical analysis was performed using the unpaired Student's t-Test in experiments with two different groups, and the two-way or one-way ANOVA in experiments with three or more different groups, with the Tukey's Multiple Comparison as a post-test, using the MS Excel and GraphPad Prism softwares. Receiver operating characteristic (ROC) analysis was performed by GraphPad Prism. All cell lines were profiled at three different passages, to ensure data consistency. Data are expressed as mean ± SEM.
3. Results
3.1. The miR-497~195 cluster is downregulated in malignant meningioma
Analysis of meningioma tumour specimens suggested that Cyclin D1 shows a trend to increase in WHO I, II, and III tumours compared to normal meningeal tissue (NMT), but no significant difference was observed among grades at the protein level, probably due to the presence of non-meningioma cells in the tumour tissue lysates and the small number of samples analysed (Fig. S1a). RT-qPCR analysis on a larger cohort of tumour tissues revealed that CCND1 is significantly overexpressed in WHO II and III tumours compared to WHO I (Fig. S1b). In vitro, we observed a significant Cyclin D1 overexpression in WHO I PDMN and KT21-MG1 cell line compared to HMC at the protein level (Fig. S1c), and a significant overexpression in the malignant (KT21-MG1) compared to the benign (Ben-Men-1) cell line, both at the protein and mRNA levels (Fig S1d). These observations are in agreement with previous reports stating that Cyclin D1 is upregulated in meningioma compared to normal tissue, correlating with tumour proliferation and recurrence [30], [31], [32].
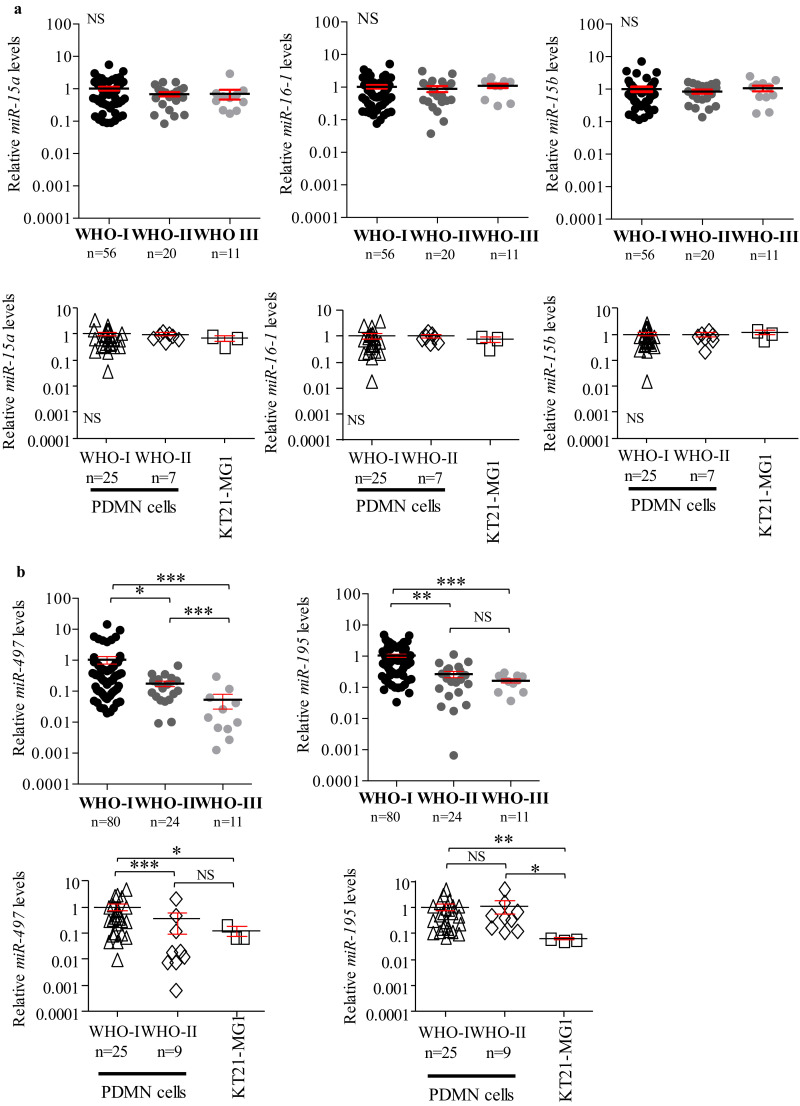
TargetScanHuman7.2 algorithm (Dataset S1), and previous reports in thyroid cancer cell lines [35], suggested that Cyclin D1 expression is regulated by the miR-15 family. Thus, we analysed the expression levels of this miRNA family, which consists of six members, grouped into three different clusters: miR-15a~16–1 on chromosome 13q14.3, miR-15b~16–2 on chromosome 3q26.1, and miR-497~195 on chromosome 17p13.1. Since miR-16–1 and miR-16–2 share the same sequence, they have both been profiled in this study as miR-16–1 [42]. Gene expression analysis demonstrated that miR-15a, −16–1, and −15b levels were unchanged between WHO I, II, and III meningioma (tissues and cells, Fig. 1a), whereas miR-497 and −195 were significantly lower in WHO II and III tissues compared to WHO I tumours (−0•55 and −1•045 Log10 folds for miR-497, and −0•85 and −0•98 Log10 fold for miR-195, respectively, Fig. 1b top). Normal meningeal tissue was not included in this analysis, as we aimed to find differences between tumour grades, not between normal and tumour tissue.
Fig. 1.
Gene expression analysis performed by RT-qPCR on the miR-15 family in meningioma tissues and cells. a MiR-15a, −16–1, and −15b expression levels in both tumours and cells. MiRNA levels were normalised to the mean of the WHO I meningioma samples (n = 56) and WHO I PDMN cells (n = 25), respectively. b MiR-497~195 cluster expression levels. MiRNA levels were normalised to the mean of the WHO I meningioma samples (n = 80) and WHO I PDMN cells (n = 25), respectively. Data are reported as mean ± SEM (One-way ANOVA; NS = not significant; * = p<0•05, ** = p<0•01, *** = p<0•005). KT21-MG1 cells were profiled at three different passages to ensure result consistency. N numbers for WHO I, II, and III tumours, and WHO I and II PDMN cells are reported in figure.
We also observed a significant decrease of the miR-497~195 cluster in the malignant KT21-MG1 cell line compared to WHO I PDMN cells (−0•92 and −1•19 Log10 folds for miR-497 and −195, respectively, Fig. 1b bottom). In WHO II PDMN cells we observed a significant decrease for miR-497 (−0•45 folds), whereas miR-195 levels were unchanged compared to WHO I (Fig. 1b). Our efforts to isolate and culture WHO III PDMN cells from tissues were unsuccessful, therefore in this study we used the KT21-MG1 cell line as an in vitro model of malignant meningioma.
3.2. GATA-4 is upregulated in malignant meningioma, controlling miR-497~195 cluster expression
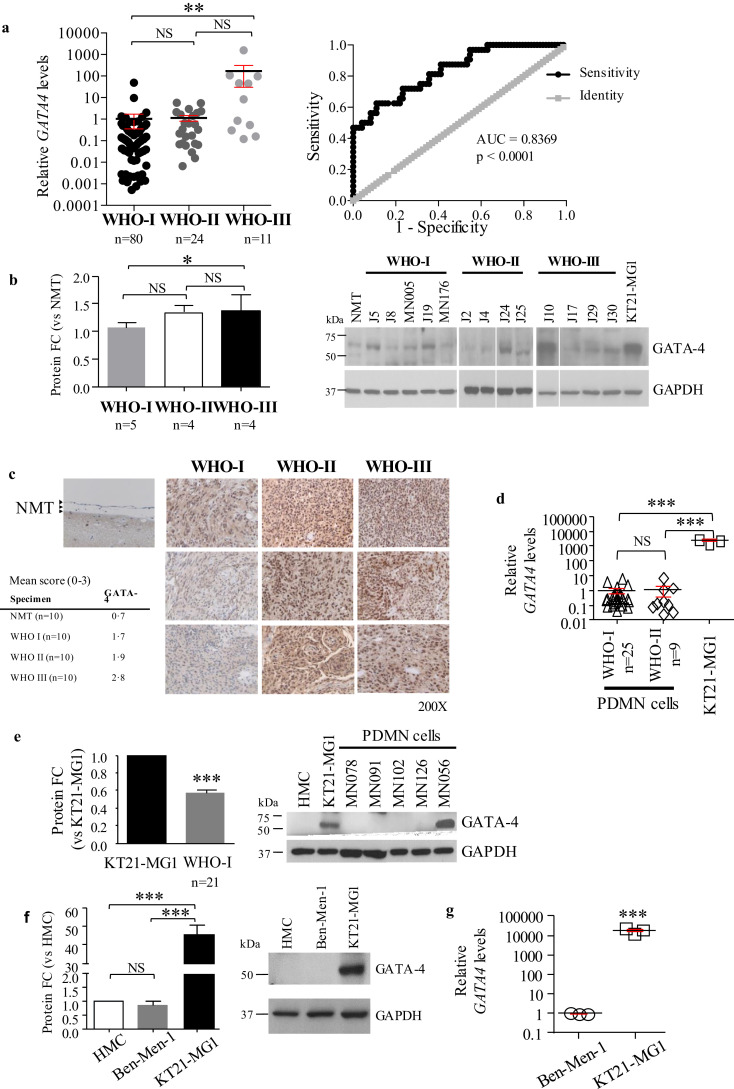
As GATA-4 has been suggested to be involved in the regulation of the miR-15 family in rat mesenchymal stem cells [33], we measured its expression levels in our meningioma tissues, and in vitro models. We showed that GATA-4 is significantly upregulated at the transcriptional level in WHO III meningioma tissues compared to benign tumours (2•63 Log10 folds), whereas no change was observed between WHO II and I tumours (Fig. 2a left). Receiver operating characteristic (ROC) analysis showed that GATA-4 has a good diagnostic value in discriminating between higher-grade (WHO II-III) and low-grade (WHO I) meningioma patients, at all sensitivity and specificity thresholds (AUC = 0•8369, p < 0•0001, Fig. 2a right).
Fig. 2.
GATA-4 is overexpressed in malignant meningioma. a Gene expression analysis by RT-qPCR of GATA-4 in meningioma tissues and ROC analysis is reported. GATA-4 levels were normalised to the mean of the WHO I specimens (n = 80). ROC analysis compares GATA-4 expression in WHO I vs. WHO II-III meningioma tumours (AUC and p value reported in figure, 95% confidence interval = 0•7552 to 0•9186). b Densitometry analysis and representative Western blot of GATA-4 in meningioma tumour lysates. WHO III tissues showed increased expression of GATA-4 compared to WHO I specimens (n = 5). c Representative images showing GATA-4 immunohistochemical staining in normal meninges (NMT, see black arrows), and WHO I, II, and III meningioma tumours. 200X magnification. Average score values for each cohort are reported. Single scores for the images reported in figure are as follows: NMT = 0; WHO I, top to bottom = 1, 1, 1; WHO II, top to bottom = 3, 3, 2; WHO III, top to bottom = 3, 3, 3. For single score values see Table S4 (samples represented in figure are highlighted in bold). d GATA-4 gene expression analysis by RT-qPCR in meningioma cells. Data have been normalised to the mean of WHO I PDMN cells (n = 25). e Densitometry analysis and representative Western blot of GATA-4 expression levels in meningioma cell lysates. f Densitometry analysis and representative Western blot of GATA-4 in normal human meningeal cells (HMC), the benign Ben-Men-1, and the malignant KT21-MG1 cell lines. g GATA-4 expression analysis by RT-qPCR in KT21-MG1 cells compared to Ben-Men-1. Data are reported as mean ± SEM (One-way ANOVA, Student's t-Test; NS = not significant; * = p<0•05, ** = p<0•01, *** = p<0•005). HMC, Ben-Men-1, and KT21-MG1 cell lines were profiled at three different passages to ensure result consistency. N numbers for WHO I, II, and III tumours, and PDMN cells are reported in figure.
Analysis of GATA-4 protein levels by Western blot analysis (including densitometry analysis, Fig. 2b) and immunohistochemical staining (Fig. 2c, and Table S4) confirmed that GATA-4 is significantly overexpressed in WHO III meningioma tissues when compared to WHO I (32% increase), but the increase in expression in WHO II tumours, compared with WHO I, did not reach statistical significance.
Analysis of GATA-4 expression in our in vitro meningioma model also showed that GATA-4 is significantly upregulated in malignant meningioma cells (KT21-MG1) compared to lower-grade primary meningioma cells (WHO I and II PDMN cells), and the benign Ben-Men-1 cell line, both at the transcriptional (3•37 and 4•25 Log10 folds, respectively, Fig. 2d and g) and protein levels (~1 and 48 fold increase, respectively, Fig. 2e and f). No difference was observed in GATA-4 expression between WHO I and II PDMN cells at the transcriptional level (Fig. 2d).
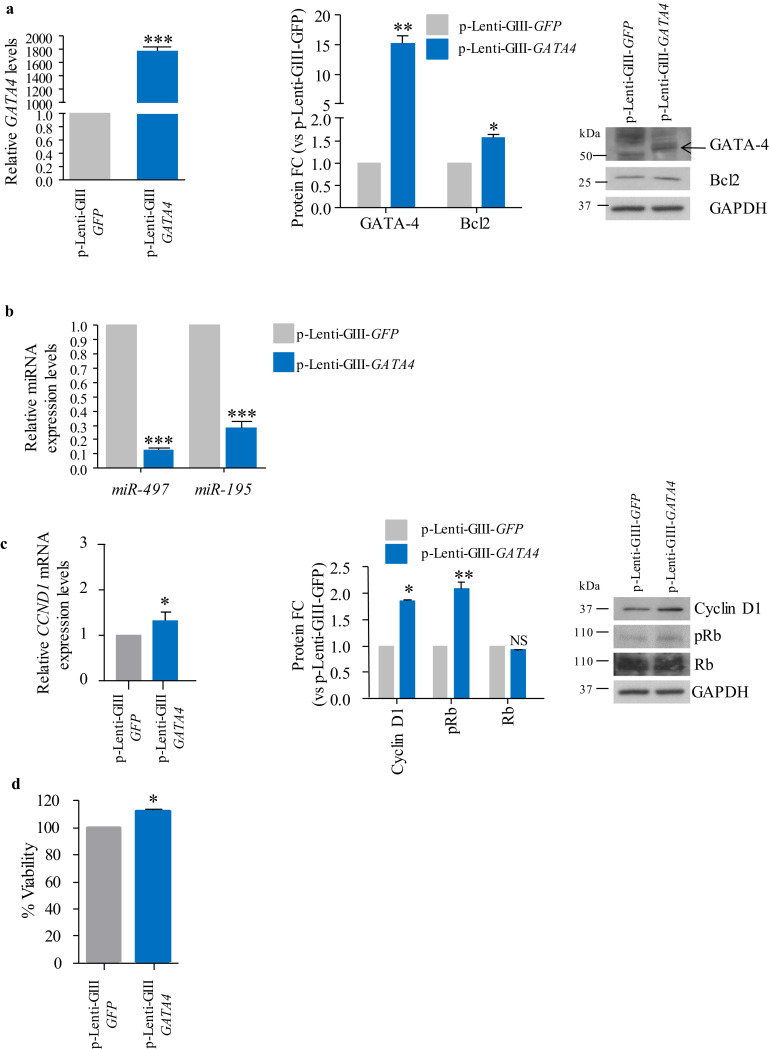
In order to determine whether GATA-4 overexpression leads to regulation of the miR-497~195 cluster in meningioma, we performed a lentiviral-mediated GATA4 overexpression in Ben-Men-1 cells, as this model expresses low levels of GATA4, and high levels of the miR-497~195 cluster compared to KT21-MG1 cells (Figs. 2f, g, and S2a, respectively). Following GATA4 transduction, we observed a significant increase of its expression levels (1792•77 and ~15 fold increase at the transcriptional and protein levels, respectively, Fig. 3a. See Fig. S2b for antibody specificity), and of BCL2 (1•52 and 1•13 fold increase at the protein and mRNA levels, respectively, as shown by Western blot densitometry analysis and RT-qPCR), a well-known GATA-4 transcriptional target [43], compared to p-Lenti-GIII-GFP-infected cells (Figs. 3a and S2c). Higher levels of GATA-4 in p-Lenti-GIII-GATA4-infected cells led to a significant miR-497 and −195 downregulation compared to scramble-infected cells (90% and 70% decrease, respectively, Fig. 3b), whereas no change was observed in the expression levels of other members of the miR-15 family (Fig. S2d). Moreover, following GATA4 overexpression in Ben-Men-1 cells, we observed an increase in Cyclin D1, both at the transcriptional and protein levels, a consequent increase in the phosphorylation levels of Rb (~2 fold increase, Fig. 3c right), and a significant increase in cell viability (~21% increase, Fig. 3d).
Fig. 3.
Ectopic expression of GATA-4 in the benign Ben-Men-1 cell line. a RT-qPCR, densitometry analysis and representative Western blot of GATA4 and BCL2 expression levels following lentiviral-mediated GATA4 transduction (p-Lenti-GIII-GATA4). b RT-qPCR analysis of the miR-497~195 cluster expression following GATA4 overexpression, compared to p-Lenti-GIII-GFP-infected cells. c RT-qPCR, densitometry analysis and representative Western blot showing increased Cyclin D1 expression levels and Rb phosphorylation following GATA-4 overexpression, compared to p-Lenti-GIII-GFP-infected cells. d Cell viability measured by ATP assay following p-Lenti-GIII-GATA4 transduction compared to control cells. Data are reported as mean ± SEM (Student's t-Test; NS = not significant; * = p<0•05, ** = p<0•01, *** = p<0•005). Transduction has been performed in three independent experiments, with one replica per repeat, to ensure data consistency.
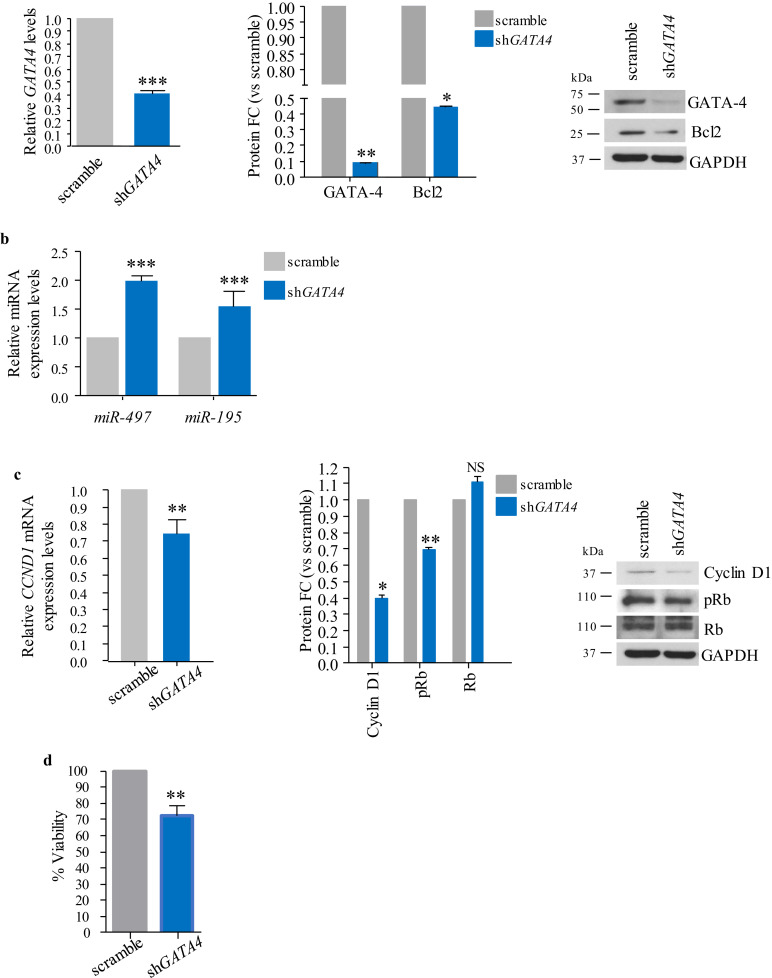
Complementary, lentiviral-mediated RNA interference of GATA4 in KT21-MG1 cells led to a significant reduction of GATA-4 and its target BCL2, both at the transcriptional (69% and 10%, respectively, Fig. 4a left and S2e) and protein levels (92% and ~56%, respectively, Fig. 4a right). Decreased GATA-4 levels led to a significant increase in the expression of the miR-497~195 cluster (2•05 and 1•16 for miR-497 and −195, respectively, Fig. 4b), whereas we did not observe a change in the expression levels of other members of the miR-15 family (Fig. S2f). Moreover, cells transduced with shGATA4 displayed significantly lower levels of Cyclin D1 compared to scramble-infected cells (35% and 60% reduction at the transcriptional and protein levels, respectively, Fig. 4c), which led to a small but significant decrease in Rb phosphorylation (~34% reduction, determined via densitometry analysis of Western blots, Fig. 4c right). Furthermore, shGATA4-transduced cells showed a significant decrease in cell viability compared to scramble-infected cells (~30%, Fig. 4d).
Fig. 4.
Lentiviral-mediated GATA4 RNA interference in KT21-MG1 cells. a RT-qPCR, densitometry analysis and representative Western blot showing levels of GATA4 and BCL2 following shGATA4 infection, compared to scramble-infected cells. b RT-qPCR analysis of the miR-497~195 cluster expression following shGATA4 infection. c RT-qPCR, densitometry analysis and representative Western blot showing Cyclin D1 and Rb phosphorylation levels following GATA4 knockdown, compared to scramble. d Cell viability measured by ATP assay following shGATA4 transduction compared to scramble-infected cells. Data are reported as mean ± SEM (Student's t-Test; NS = not significant; * = p<0•05, ** = p<0•01, *** = p<0•005). Transduction has been performed in three independent experiments, with one replica per repeat, to ensure data consistency.
Next, as GATA-4 has been predicted to be transcriptionally regulated by miR-195 (TargetScanHuman7.2, see Dataset S1), we performed a lentiviral-mediated hsa-miR-195–5p overexpression in KT21-MG1 cells. Interestingly, following hsa-miR-195–5p overexpression (Fig. S3a), we observed a significant downregulation of GATA-4, both at the transcriptional (16% decrease) and protein levels (~55% decrease, determined via densitometry analysis of Western blot), when compared to LentimiR-GFP–infected cells (Fig. S3b). Moreover, we also observed a significant decrease in Cyclin D1 (CCND1) expression (~40% decrease, Fig. S3d), as expected, and a significant increase in miR-497 expression (8•89 folds, Fig. S3c), supporting the hypothesis that GATA-4 plays a role in the regulation of miR-497 and −195 expression.
3.3. Synthetic GATA-4 blockade decreases cell viability in malignant meningioma cells
The observations outlined above suggested that GATA-4, which is upregulated in malignant meningioma (Fig. 2), plays a role in regulating the expression of the miR-497~195 cluster, and of Cyclin D1 (Figs. 3 and 4).
Therefore, we hypothesised that GATA-4 could represent a novel therapeutic target for WHO III meningioma, and investigated the effects of the NSC140905 compound, a newly pioneered GATA-4 small-molecule inhibitor [34].
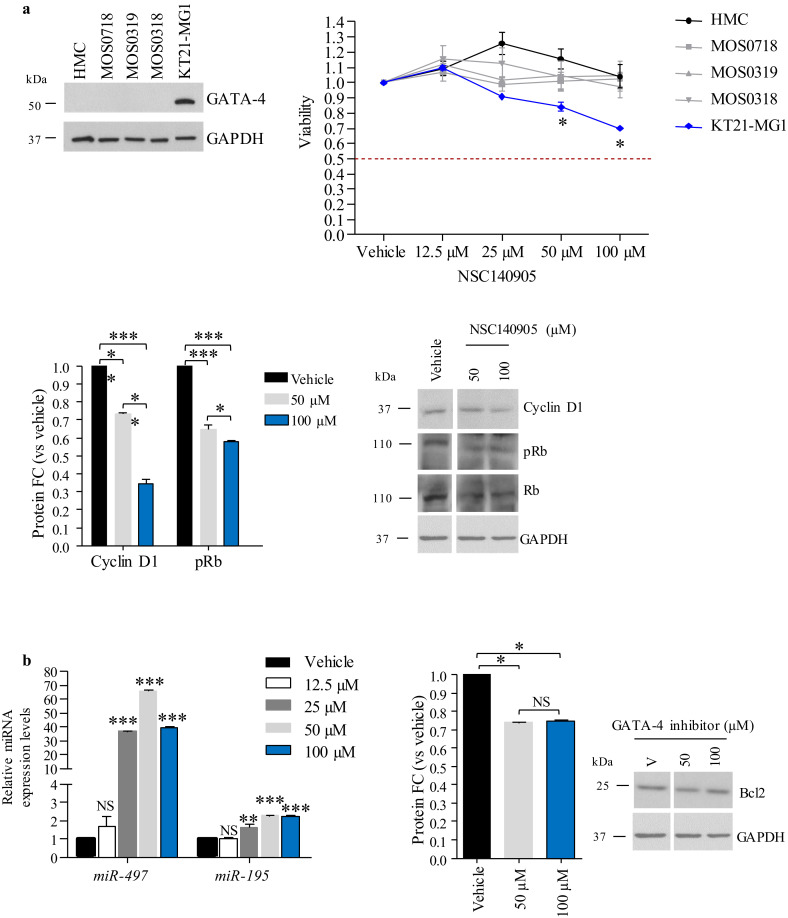
We evaluated the effects of the NSC140905 compound in KT21-MG1 cells at 24, 48, and 72 h, and assayed cell viability. As shown in Fig. S4, we observed a significant decrease in cell viability at 24 and 48 h, and at all three time points only at the highest doses tested (50 and 100 μM, respectively); therefore, we decided to focus all following studies on the 24 h time point with these two doses, which are in a comparable range as those used previously [34]. Vehicle-treated cells were exposed to 0•1% DMSO, which corresponds to the highest concentration used in this study. To understand whether the NSC140905 compound could have off target effects, we treated normal human meningeal cells (HMC) and primary Schwann cells (MOS), for which we could not detect GATA-4 expression via Western blot (Fig. 5a, top left), with the same range of concentrations used for KT21-MG1 cells, for 24 h. As shown in Fig. 5a (top right), treatment in HMC and MOS cells did not affect viability at any of the concentrations tested, whereas we observed a significant decrease in cell viability in KT21-MG1 cells at both 50 and 100 μM (~11% and ~31% decrease, respectively, Fig. 5a top right). Moreover, GATA-4 inhibition in KT21-MG1 cells was paralleled by a significant decrease in Cyclin D1 expression (~30% and ~70% decrease at 50 and 100 μM, respectively), and in the phosphorylation status of Rb (~40% and ~50% decreased phosphorylation at 50 and 100 μM, respectively, Fig. 5a bottom).
Fig. 5.
Inhibition of GATA-4 transcriptional activity following treatment with the small-molecule inhibitor NSC140905. a Left, representative Western blot of GATA-4 protein expression in HMC, primary Schwann cells (MOS0718, MOS0319, and MOS0318), and KT21-MG1 cells. Right, cell viability assessed by ATP assay after administration of the GATA-4 inhibitor for 24 h, compared to vehicle-treated cells (0•1% DMSO). Bottom, densitometry analysis and representative Western blot showing a decrease in Cyclin D1 expression and Rb phosphorylation levels following GATA-4 inhibition compared to control cells. b RT-qPCR analysis of the miR-497~195 cluster expression following GATA-4 inhibition, densitometry analysis and representative Western blot of Bcl2 following GATA-4 inhibition, compared to vehicle-treated cells (0•1% DMSO). Data are reported as mean ± SEM (Two-way and one-way ANOVA; NS = not significant; * = p<0•05, ** = p<0•01, *** = p<0•005). NSC140905 treatment has been performed in three independent experiments, with one replica per repeat, to ensure data consistency. We used the highest DMSO concentration (0•1% DMSO) as vehicle control in all experiments.
As observed following lentiviral-mediated GATA4 RNA interference (Fig. 4), decreased GATA-4 transcriptional activity, resulted from treatment with NSC140905, led to a significant increase in the expression levels of the miR-497~195 cluster (62•82 and 32•59 folds at 50 μM and 100 μM, respectively, for miR-497, and 2•29 and 2•24 folds at 50 and 100 µM, respectively, for miR-195), and a significant decrease of Bcl2 protein levels (27% and 26% decrease at 50 and 100 µM, respectively, Fig. 5b).
3.4. The miR-497~195 cluster is differentially expressed in circulating exosomes
The results outlined above showed that both miR-497 and −195 are downregulated in high-grade meningioma (WHO II and III) compared to benign tumours (WHO I, Fig. 1). Therefore, we analysed the miRNA cargo in circulating exosomes derived from meningioma patients, in order to understand whether these two candidates could be used as non-invasive biomarkers.
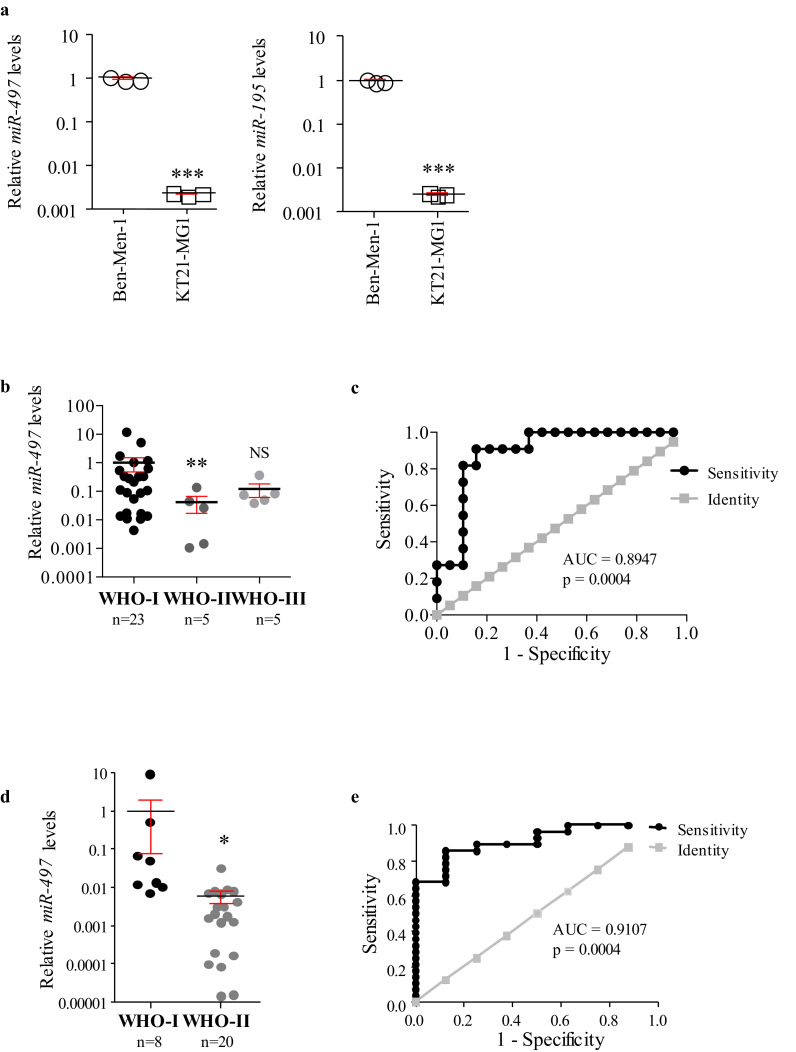
As separation of exosomes from other EVs is important [44], [45], [46], we monitored the quality of our preparations by investigating the expression of CD63 and CD9, well-known exosome biomarkers, and of Calnexin and GM130 as markers for the endoplasmic reticulum (ER) and Golgi, respectively, as previously reported [47]. We also wanted to understand whether our preparations contained subpopulations derived from non-meningioma cells, and whether these could affect the results observed. To do so, we expanded our analysis including exosomes isolated from meningioma cells culture media. Western blot analysis not only confirmed that both preparations (from cell culture media and meningioma patients’ serum samples) were similarly enriched in exosomes, but also that they did not show contamination by cellular organelles (Fig. S5a and S5b, respectively). qPCR analysis revealed that both miR-497 and −195 were significantly lower in exosomes derived from KT21-MG1 cell culture media compared to those derived from Ben-Men-1 media (−2•65 and −2•60 Log10 folds, respectively, Fig. 6a), mirroring their expression levels in the cells of origin (Fig. S2a), and strengthening the hypothesis of a differential transcriptional regulation dependent on tumour grade.
Fig. 6.
Evaluation of miRNA cargo in cell culture media and serum exosomes. a miR-497 and −195 expression levels in exosomes isolated from KT21-MG1 and Ben-Men-1 cell culture media. The analysis was performed in biological triplicates to ensure data consistency. b, c miR-497 expression levels in circulating exosomes and ROC analysis (discovery cohort). ROC analysis compares low- (WHO I) and higher-grade (WHO II-III) meningioma patients (AUC and p value reported in figure, 95% confidence interval = 0•7775 to 1•012). d, e miR-497 expression levels in circulating exosomes and ROC analysis for the validation cohort. ROC analysis compares WHO I and WHO II meningioma patients (AUC and p value reported in figure, 95% confidence interval = 0•8120 to 1•009). Data are reported as mean ± SEM (One-way ANOVA, Student's t-Test; NS = not significant; * = p<0•05, ** = p<0•01, *** = p<0•005). N numbers for WHO I, II, and III serum-derived exosomes are reported in figure.
Importantly, investigation of miR-497 and-195 expression in exosomes isolated from the meningioma patients’ serum samples discovery cohort showed that both are significantly downregulated in WHO II compared to WHO I (−1•30 and −0•31 Log10 folds, respectively) whereas there is a trend to decrease in WHO III compared to WHO I samples (−0•98 and −0•29 Log10 folds, respectively, Figs. 6b and S5c). ROC analysis showed that miR-497 has better sensitivity and specificity than miR-195 in distinguishing between low-grade (WHO I) and higher-grade (WHO II-III) meningioma patients (AUC = 0•8943, p = 0•0004 and AUC = 0•7843, p = 0•0102, respectively, Figs. 6c and S5d). RNA extracted from exosomes isolated from healthy volunteers’ serum samples as been used as a calibrator.
Although we could not collect a sufficient amount of WHO III samples for our validation cohort, we were able to confirm the results observed for both miR-497 and −195 in independent WHO I and II samples. In fact, both candidates showed a significant decrease in exosomes derived from WHO II meningioma patients compared to WHO I (−2•10 and −0•20 Log10 folds, respectively, Figs. 6d and S5e). In line with the observations in the discovery cohort, ROC analysis showed that miR-497 had better sensitivity and specificity compared to miR-195 in distinguishing between WHO I and II meningioma patients (AUC = 0•9107, p = 0•0004 and AUC = 0•6490, p = 0•2082, respectively, Figs. 6e and S5f).
4. Discussion
Our data suggest, for the first time, that GATA-4, a transcription factor is overexpressed in higher-grade meningioma, representing a potential novel therapeutic target and is involved in the regulation of miR-497, a potential novel circulating biomarker for malignant meningioma.
GATA-4 has been previously reported to be involved in the protection of mesenchymal stem cells from ischemia by downregulating members of the miR-15 family [33], but its role in cancer is not completely understood, yet. The GATA4 coding gene is located on chromosome 8p, which is a site frequently deleted in multiple tumour types, such as colorectal and oesophageal cancer [48,49]. Alternatively, GATA4 can be downregulated via epigenetic silencing, as observed in lung, ovarian, and HPV-driven oropharyngeal cancer, in glioblastoma multiforme, and in diffuse large B-cell lymphoma [50], [51], [52], [53], [54]. All these observations suggest that GATA-4 may play tumour suppressive roles in these disease settings. However, GATA4 amplification has recently been described in certain gastric cancers, indicating a more oncogenic function [55]; moreover, it has been shown to promote the expression of the anti-apoptotic factor Bcl2, and of Cyclin D2 in ovarian granulosa cell tumours, further supporting its role as an oncoprotein [56,57].
As we observed a significant GATA-4 overexpression in WHO III meningioma, our data suggest that it could play an oncogenic function in this tumour setting, and it should be further investigated as a potential biomarker and therapeutic target for malignant meningioma.
We show that GATA-4 is involved in the transcriptional regulation of the miR-497~195 cluster, which in turn regulates Cyclin D1. However, the mechanism through which GATA-4 exerts its role needs to be further clarified. It is predicted that the promoter/enhancer region of the miR-497~195 cluster contains binding sites for two GATA-4 zinc finger subunits (GATAD2A and GATAD2B) [58], suggesting a direct involvement of this factor in the transcriptional regulation of the miRNA cluster.
Moreover, as our results show that GATA-4 is involved in the regulation of key oncogenes, such as Cyclin D1 (directly, or indirectly through the regulation of the miR-497~195 cluster), we suggest it as a possible therapeutic target for malignant meningioma. To date, there are no FDA-approved drugs to specifically target GATA-4, but a study in 2011 identified four small molecular weight chemicals with lead-like properties able to bind to the DNA-binding domain of GATA-4, reducing its transcriptional activity [34]. One such compound, NSC140905, a derivative of succinic acid, proved to be more efficient than the others in inhibiting GATA-4 at the highest doses tested by the authors; however, they tested the efficiency of the compound on a cell line (HeLa) that does not express GATA-4, thus having to overexpress the protein before the treatment [34]. Therefore, we decided to investigate the effects of this compound on the malignant meningioma cell line KT21-MG1, which naturally expresses high levels of GATA-4. This molecule is a research compound, which has only been analysed for its ability to inhibit GATA-4 in vitro, and no in vivo data are available on its safety and effects.
In line with what we observed following lentiviral-mediated GATA4 RNA interference, treatment of KT21-MG1 cells with NSC140905 led to decreased Cyclin D1 expression and Rb phosphorylation, and ultimately to decreased cell viability, whilst not being effective in reducing normal cells viability (normal human meningeal cells and primary Schwann cells, MOS). Even though more experiments are needed to confirm these results, our observations provide proof of concepts that targeting GATA-4 is a potential valid strategy in malignant meningioma. To have a significant effect on cell viability, we had to use quite high concentrations of the GATA-4 small-molecule inhibitor (50 and 100 μM). These were the same concentrations used in the previous study [34], but they could lead to the manifestation of undesired effects; therefore, careful drug formulation and preclinical studies are warranted to determine any possible toxicity.
As meningioma surgery can be challenging depending on tumour location, and imaging cannot reveal tumour grade, it is important to find novel diagnostic biomarkers circulating in peripheral blood, which would allow a simple, non-invasive follow-up for patients. Identifying patients at higher risk of tumour progression will provide clinicians and patients additional prognostic information, as well as informing decision making on the frequency of follow-up imaging and the timing of surgery. Therefore, we measured the expression levels of the miR-497~195 cluster in meningioma patients’ serum-derived exosomes, randomizing the samples into a discovery and validation set. A previous study identified a 6-miRNA signature in meningioma patients’ serum that could be useful as a biomarker [18]. However, miRNAs in serum might not be associated with tumours, but could be a result of cell death and lysis, thus affecting their reliability as biomarkers. The advantage of our approach lays in the fact that miRNAs in serum primarily exist inside exosomes, and blood of cancer patients is twice as rich in exosomes compared to blood of healthy individuals [59]. Thus, we are more confident in the fact that the changes in circulating miRNAs observed are more likely due to tumour burden. Our approach is supported by a previous analysis in tissues, in which miR-497 has been suggested as a potential biomarker for atypical (WHO II) meningioma, when considered as part of a 4-miRNA signature (miR-222, −34a*, −136, and −497) [22], but its expression in blood or circulating exosomes has never been investigated in meningioma. Both miR-497 and −195 proved to be significantly downregulated in WHO II compared to WHO I samples in both cohorts, and showed a trend to decrease in WHO III compared to WHO I samples in the discovery set. Importantly, we were able to validate this result in an independent sample set.
This analysis has been limited by the scarce availability of WHO III meningioma serum samples; inclusion of more samples in our cohorts is warranted to strengthen the statistical power of our study. Nevertheless, our results suggest that miR-497 could be a novel potential circulating biomarker for higher-grade meningioma, as it showed a good diagnostic ability in discriminating between low- and higher-grade meningioma patients. In the future, access to serial samples from the same patient during follow-up would allow us to understand whether these two candidates can be used as biomarkers for clinical outcome, as well as diagnostic tools.
We recognise that our study is affected by a few limitations; I) we stratified our meningioma samples based on their WHO grades, which are assigned through histopathological assessment of the tumour, and are thus subjected to inter-observer variability. We took this decision as this is still the gold standard for diagnostic and prognostic meningioma stratification, and because we did not have a big enough sample size to stratify samples based on their mutational profile, which is mainly described for low-grade (WHO I) meningiomas. II) For all in vitro experiment, we compared WHO I and II PDMN cells to an established malignant cell line, KT21-MG1. Unfortunately, we did not receive enough samples to establish WHO III PDMN cells, and all the samples we could collect were clinically heterogeneous (either characterised by tumour progression, or by the presence of WHO III foci in lower-grade tumours). Therefore, the establishment of WHO III PDMN cells was not possible. Moreover, we decided to include WHO II PDMN cells only in RT-qPCR analysis as a lower amount of sample is needed compared to Western blot panels. Despite these drawbacks, our results are of clinical interest, as they could lead to the establishment of new, more reliable meningioma biomarkers, and a potential new therapeutic target.
In conclusion, this study shows that the transcription factor GATA-4 is overexpressed in malignant meningioma, where it negatively regulates miR-497~195 cluster expression and sustains cell viability, and suggests its potential to be used as a novel tissue-specific biomarker for higher-grade meningioma. Moreover, it proves its potential as a novel therapeutic target for malignant meningioma, and the ability of the small-molecule compound NSC140905 to efficiently inhibit GATA-4 activity in a cell line that endogenously expresses high levels of this transcription factor.
Furthermore, this is the first study to evaluate miRNA expression levels in circulating exosomes in meningioma. Our analysis in serum samples suggests that miR-497 could be a potential novel circulating biomarker.
Declaration of Interests
The authors declare no competing financial interests in relation to the work described.
Acknowledgments
Acknowledgements
We are grateful to Mr Agbolahan Sofela, MRCS, for reviewing the clinical data, and to the nurses involved in this study. Tissue samples were obtained from University Hospitals Plymouth NHS trust and Lancashire Teaching Hospitals NHS foundation Trust as part of the UK Brain Archive Information Network (BRAIN UK), which is funded by the Medical Research Council.
Funding sources
We thank Brain Tumour Research for supporting this work. Caterina Negroni was supported by a PUPSMD Postgraduate Research Award and Daniele Baiz was supported in part by the FP7 Marie Curie Actions (PCOFUND-GA-2012600181). The funders had no role in study design, data collection, data analysis and interpretation, writing of the manuscript, or decision to publish.
Author contributions
Daniele Baiz and C Oliver Hanemann equally conceived and designed the study; Kathreena Kurian and David Hilton provided and characterised clinical samples; Claire Adams conceived study ethics, sample collection and reviewed clinical data. Caterina Negroni performed most of the experiments; David Hilton generated IHC data; Caterina Negroni, Daniele Baiz, Emanuela Ercolano, and Claire Adams processed clinical samples and curated samples for the study biobank; Caterina Negroni and Daniele Baiz analysed the data and wrote the manuscript. C Oliver Hanemann participated in writing and revised the manuscript draft. All authors read, commented and approved the final version of the manuscript.
Footnotes
Funding. This study was funded by Brain Tumour Research, the University of Plymouth, and the FP7 Marie Curie Actions.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102941.
Appendix. Supplementary materials
References
- 1.Ostrom Q.T., Gittleman H., Truitt G., Boscia A., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro Oncol. 2018;20(suppl_4):iv1–iv86. doi: 10.1093/neuonc/noy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis D.N., Perry A., Reifenberger G. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Yuzawa S., Nishihara H., Tanaka S. Genetic landscape of meningioma. Brain Tumor Pathol. 2016;33(4):237–247. doi: 10.1007/s10014-016-0271-7. [DOI] [PubMed] [Google Scholar]
- 4.Brodbelt A.R., Barclay M.E., Greenberg D., Williams M., Jenkinson M.D., Karabatsou K. The Outcome of patients with surgically treated meningioma in England: 1999-2013. A cancer registry data analysis. Br J Neurosurg. 2019;33(6):641–647. doi: 10.1080/02688697.2019.1661965. [DOI] [PubMed] [Google Scholar]
- 5.Mawrin C., Chung C., Preusser M. Biology and clinical management challenges in meningioma. Am Soc Clin Oncol Educ Book. 2015:e106–e115. doi: 10.14694/EdBook_AM.2015.35.e106. [DOI] [PubMed] [Google Scholar]
- 6.Whittle I.R., Smith C., Navoo P., Collie D. Meningiomas. Lancet. 2004;363(9420):1535–1543. doi: 10.1016/S0140-6736(04)16153-9. [DOI] [PubMed] [Google Scholar]
- 7.Marosi C., Hassler M., Roessler K. Meningioma. Crit Rev Oncol Hematol. 2008;67(2):153–171. doi: 10.1016/j.critrevonc.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Rogers L., Barani I., Chamberlain M. Meningiomas: knowledge base, treatment outcomes, and uncertainties. A RANO review. J Neurosurg. 2015;122(1):4–23. doi: 10.3171/2014.7.JNS131644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamberlain M.C. Hydroxyurea for recurrent surgery and radiation refractory high-grade meningioma. J Neurooncol. 2012;107(2):315–321. doi: 10.1007/s11060-011-0741-z. [DOI] [PubMed] [Google Scholar]
- 10.Norden A.D., Raizer J.J., Abrey L.E. Phase II trials of erlotinib or gefitinib in patients with recurrent meningioma. J Neurooncol. 2010;96(2):211–217. doi: 10.1007/s11060-009-9948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaley T.J., Wen P., Schiff D. Phase II trial of sunitinib for recurrent and progressive atypical and anaplastic meningioma. Neuro Oncol. 2015;17(1):116–121. doi: 10.1093/neuonc/nou148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark V.E., Harmanci A.S., Bai H. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016;48(10):1253–1259. doi: 10.1038/ng.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark V.E., Erson-Omay E.Z., Serin A. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–1080. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers C.L., Perry A., Pugh S. Pathology concordance levels for meningioma classification and grading in NRG Oncology RTOG Trial 0539. Neuro Oncol. 2016;18(4):565–574. doi: 10.1093/neuonc/nov247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaubel R.A., Chen S.G., Raleigh D.R. Meningiomas with rhabdoid features lacking other histologic features of malignancy: a study of 44 cases and review of the literature. J Neuropathol Exp Neurol. 2016;75(1):44–52. doi: 10.1093/jnen/nlv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumgarten P., Gessler F., Schittenhelm J. Brain invasion in otherwise benign meningiomas does not predict tumor recurrence. Acta Neuropathol. 2016;132(3):479–481. doi: 10.1007/s00401-016-1598-1. [DOI] [PubMed] [Google Scholar]
- 17.Zhi F., Zhou G., Wang S. A microRNA expression signature predicts meningioma recurrence. Int J Cancer. 2013;132(1):128–136. doi: 10.1002/ijc.27658. [DOI] [PubMed] [Google Scholar]
- 18.Zhi F., Shao N., Li B. A serum 6-miRNA panel as a novel non-invasive biomarker for meningioma. Sci Rep. 2016;6:32067. doi: 10.1038/srep32067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Gewely M.R., Andreassen M., Walquist M. Differentially expressed micrornas in meningiomas grades I and II suggest shared biomarkers with malignant tumors. Cancers Basel. 2016;8(3) doi: 10.3390/cancers8030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M., Deng X., Ying Q., Jin T., Li M., Liang C. MicroRNA-224 targets ERG2 and contributes to malignant progressions of meningioma. Biochem Biophys Res Commun. 2015;460(2):354–361. doi: 10.1016/j.bbrc.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 21.Kliese N., Gobrecht P., Pachow D. MiRNA-145 is downregulated in atypical and anaplastic meningiomas and negatively regulates motility and proliferation of meningioma cells. Oncogene. 2013;32(39):4712–4720. doi: 10.1038/onc.2012.468. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig N., Kim Y., Mueller S. Posttranscriptional deregulation of signaling pathways in meningioma subtypes by differential expression of miRNAs. Neuro Oncol. 2015;17(9):1250–1260. doi: 10.1093/neuonc/nov014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esquela-Kerscher A., Slack F.J. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 24.Carthew R.W., Sontheimer E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., Li S., Li L. Exosome and exosomal microRNA: trafficking, sorting, and function. Genom Proteom Bioinform. 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lasser C., Alikhani V.S., Ekstrom K. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slomka A., Urban S.K., Lukacs-Kornek V., Zekanowska E., Kornek M. Large extracellular vesicles: have we found the holy grail of inflammation? Front Immunol. 2018;9:2723. doi: 10.3389/fimmu.2018.02723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawson C., Vicencio J.M., Yellon D.M., Davidson S.M. Microvesicles and exosomes: new players in metabolic and cardiovascular disease. J Endocrinol. 2016;228(2):R57–R71. doi: 10.1530/JOE-15-0201. [DOI] [PubMed] [Google Scholar]
- 29.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alama A., Barbieri F., Spaziante R. Significance of cyclin D1 expression in meningiomas: a preliminary study. J Clin Neurosci. 2007;14(4):355–358. doi: 10.1016/j.jocn.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Milenkovic S., Marinkovic T., Jovanovic M.B., Djuricic S. Berisavac, II, Berisavac I. Cyclin D1 immunoreactivity in meningiomas. Cell Mol Neurobiol. 2008;28(6):907–913. doi: 10.1007/s10571-008-9278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng G., Zhang L., Lv W., Dong C., Wang Y., Zhang J. Overexpression of cyclin D1 in meningioma is associated with malignancy grade and causes abnormalities in apoptosis, invasion and cell cycle progression. Med Oncol. 2015;32(1):439. doi: 10.1007/s12032-014-0439-0. [DOI] [PubMed] [Google Scholar]
- 33.Yu B., Gong M., He Z. Enhanced mesenchymal stem cell survival induced by GATA-4 overexpression is partially mediated by regulation of the miR-15 family. Int J Biochem Cell Biol. 2013;45(12):2724–2735. doi: 10.1016/j.biocel.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Hachem N., Nemer G. Identification of new GATA4-small molecule inhibitors by structure-based virtual screening. Bioorg Med Chem. 2011;19(5):1734–1742. doi: 10.1016/j.bmc.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 35.Yin Y., Hong S., Yu S. MiR-195 inhibits tumor growth and metastasis in papillary thyroid carcinoma cell lines by targeting CCND1 and FGF2. Int J Endocrinol. 2017;2017 doi: 10.1155/2017/6180425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow H.Y., Dong B., Duron S.G. Group I Paks as therapeutic targets in NF2-deficient meningioma. Oncotarget. 2015;6(4):1981–1994. doi: 10.18632/oncotarget.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bassiri K., Ferluga S., Sharma V. Global proteome and phospho-proteome analysis of merlin-deficient meningioma and schwannoma identifies PDLIM2 as a novel therapeutic target. EBioMedicine. 2017;16:76–86. doi: 10.1016/j.ebiom.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferluga S., Baiz D., Hilton D.A. Constitutive activation of the EGFR–STAT1 axis increases proliferation of meningioma tumor cells. Neuro Oncol Adv. 2020;2(1) doi: 10.1093/noajnl/vdaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Dunn J., Ferluga S., Sharma V. Proteomic analysis discovers the differential expression of novel proteins and phosphoproteins in meningioma including NEK9, HK2 and SET and deregulation of RNA metabolism. EBioMedicine. 2019;40:77–91. doi: 10.1016/j.ebiom.2018.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi S., Volden P., Timm D., Mao K., Xu X., Liang Q. Transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death. J Biol Chem. 2010;285(1):793–804. doi: 10.1074/jbc.M109.070037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thery C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3: Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 45.Gyorgy B., Szabo T.G., Pasztoi M. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68(16):2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor D.D., Shah S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods. 2015;87:3–10. doi: 10.1016/j.ymeth.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 47.Camacho L., Guerrero P., Marchetti D. MicroRNA and protein profiling of brain metastasis competent cell-derived exosomes. PLoS ONE. 2013;8(9):e73790. doi: 10.1371/journal.pone.0073790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Derks S., Postma C., Moerkerk P.T. Promoter methylation precedes chromosomal alterations in colorectal cancer development. Cell Oncol. 2006;28(5–6):247–257. doi: 10.1155/2006/846251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin L., Aggarwal S., Glover T.W., Orringer M.B., Hanash S., Beer D.G. A minimal critical region of the 8p22-23 amplicon in esophageal adenocarcinomas defined using sequence tagged site-amplification mapping and quantitative polymerase chain reaction includes the GATA-4 gene. Cancer Res. 2000;60(5):1341–1347. [PubMed] [Google Scholar]
- 50.Guo M., Akiyama Y., House M.G. Hypermethylation of the GATA genes in lung cancer. Clin Cancer Res. 2004;10(23):7917–7924. doi: 10.1158/1078-0432.CCR-04-1140. [DOI] [PubMed] [Google Scholar]
- 51.Wakana K., Akiyama Y., Aso T., Yuasa Y. Involvement of GATA-4/-5 transcription factors in ovarian carcinogenesis. Cancer Lett. 2006;241(2):281–288. doi: 10.1016/j.canlet.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 52.Kostareli E., Holzinger D., Bogatyrova O. HPV-related methylation signature predicts survival in oropharyngeal squamous cell carcinomas. J Clin Invest. 2013;123(6):2488–2501. doi: 10.1172/JCI67010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agnihotri S., Wolf A., Munoz D.M. A GATA4-regulated tumor suppressor network represses formation of malignant human astrocytomas. J Exp Med. 2011;208(4):689–702. doi: 10.1084/jem.20102099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pike B.L., Greiner T.C., Wang X. DNA methylation profiles in diffuse large B-cell lymphoma and their relationship to gene expression status. Leukemia. 2008;22(5):1035–1043. doi: 10.1038/leu.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chia N.Y., Deng N., Das K. Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut. 2015;64(5):707–719. doi: 10.1136/gutjnl-2013-306596. [DOI] [PubMed] [Google Scholar]
- 56.Kyronlahti A., Ramo M., Tamminen M. GATA-4 regulates Bcl-2 expression in ovarian granulosa cell tumors. Endocrinology. 2008;149(11):5635–5642. doi: 10.1210/en.2008-0148. [DOI] [PubMed] [Google Scholar]
- 57.Anttonen M., Pihlajoki M., Andersson N. FOXL2, GATA4, and SMAD3 co-operatively modulate gene expression, cell viability and apoptosis in ovarian granulosa cell tumor cells. PLoS ONE. 2014;9(1):e85545. doi: 10.1371/journal.pone.0085545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fishilevich S., Nudel R., Rappaport N. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database Oxf. 2017;2017 doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gallo A., Tandon M., Alevizos I., Illei G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE. 2012;7(3):e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.