Abstract
Negative cofactor 2 (NC2), including two subunits NC2α and NC2β, is a conserved positive/negative regulator of class II gene transcription in eukaryotes. It is known that NC2 functions by regulating the assembly of the transcription preinitiation complex. However, the exact role of NC2 in transcriptional regulation is still unclear. Here, we reveal that, in Neurospora crassa, NC2 activates catalase-3 (cat-3) gene transcription in the form of heterodimer mediated by histone fold (HF) domains of two subunits. Deletion of HF domain in either of two subunits disrupts the NC2α–NC2β interaction and the binding of intact NC2 heterodimer to cat-3 locus. Loss of NC2 dramatically increases histone variant H2A.Z deposition at cat-3 locus. Further studies show that NC2 recruits chromatin remodeling complex INO80C to remove H2A.Z from the nucleosomes around cat-3 locus, resulting in transcriptional activation of cat-3. Besides HF domains of two subunits, interestingly, C-terminal repression domain of NC2β is required not only for NC2 binding to cat-3 locus, but also for the recruitment of INO80C to cat-3 locus and removal of H2A.Z from the nucleosomes. Collectively, our findings reveal a novel mechanism of NC2 in transcription activation through recruiting INO80C to remove H2A.Z from special H2A.Z-containing nucleosomes.
INTRODUCTION
In eukaryotic cells, transcription initiation is triggered by the assembly of a functional preinitiation complex (PIC) consisting of general transcription factors (GTFs) and RNA polymerase II (RNAP II) at class II gene promoters (1,2). The first step for sequential assembly of an active PIC at the class II gene promoter is either the binding of TBP of TFIID to the TATA box (1–3), or the recognizing and binding of TAFs of TFIID to TATA-less promoter elements, such as INR, MTE, and DPE (4,5). This is the rate-limiting step for PIC formation. The sequential assembly at the promoter in the following order: TFIIA, TFIIB, TFIIF-RNAP II, TFIIE, TFIIH and TFIIJ (3,6–8). The PIC assembly is initiated by a substantial number of transcriptional activators that either directly or indirectly associate with TBP to mediate the interaction between TFIIA and TFIIB with TBP (8,9).
In yeast, the PIC assembly is activated by TFIIA and TFIIB and inhibited by Negative cofactor 2 (NC2) (8). NC2 is initially identified as a transcriptional suppressor in HeLa cells, which represses basal TATA-dependent transcription through interacting with TBP in vitro (10,11). NC2 complex consists of two subunits, NC2α and NC2β, which are highly conserved proteins among eukaryotes (12). Previous studies have shown that in Saccharomyces cerevisiae, both NC2α/BUR6 and NC2β/YDR1 are critical for normal cell growth (13,14). Similarly, loss of NC2α/Drap1 leads to severe gastrulation defects in mice (15), indicating the earliest essential role for Drap1 in embryogenesis. NC2α and NC2β form a heterodimer through their N-terminal histone fold domains and bind to the TBP-TATA complex to repress the transcription of class II genes (16). Both TBP-binding domain and QA-rich domain of NC2β are required for the interaction between NC2 and TBP-TATA complex and its transcriptional repression (17). The association of DNA-bound TBP with NC2 complex obstructs its interaction with TFIIA and TFIIB, which inhibits PIC assembly for transcription initiation (10,18–20). The crystal structure of human NC2 recognizing TBP-DNA transcription complex showed that NC2 heterodimer binds to the bent DNA double helix on the underside of the preformed TBP-DNA complex, permitting the C terminus of NC2β to make specific contacts with the upper surface of TBP and block the recognition of TBP–DNA complex by TFIIB (21).
In addition to the well-established function as a transcriptional repressor, the NC2α also functions as transcriptional activator for GAL1 and GAL10 in vivo (13). The Drosophila homolog of NC2 purified from nuclear extracts is able to activate the transcription of genes containing TATA-less promoters with a downstream promoter element (DPE) in an in vitro transcription system (22). Consistent with this, yeast NC2 is required for the transcription of the HIS3 and HIS4 TATA-less core promoters in vivo (13,23). Interestingly, yeast NC2 also directly stimulates the activator-dependent transcription of genes with TATA-containing promoters both in vivo and in vitro; its stimulatory role requires the same surface of TBP that mediates the NC2 repression activity (18). Furthermore, recent studies showed that yeast NC2 activates the expression of some stress-induced genes in response to stress stimuli (e.g., heat or oxidative stress) through promoting the PIC assembly, indicating that NC2 plays an important role in regulating inducible genes (24–26).
In various environmental stresses, oxidative stress is a ubiquitous threat for aerobic organisms. It not only induces irreversible damage of cellular biomacromolecules, including lipids, proteins, and DNA, resulting in cellular dysfunction or cell death (27,28), but also triggers a series of physiological and pathological diseases such as pulmonary emphysema, adult respiratory distress syndrome (ARDS) (29) and colorectal cancer (30). Indeed, oxidative stress is caused by the imbalance that production of reactive oxygen species (ROS) exceeds the elimination capacity of ROS in cells. ROS is a family of unstable and highly reactive molecules that are products of intracellular oxidative metabolism (mitochondrial electron-transport chain) and extracellular ROS-inducing agents, such as radiation and heat shock (31,32). ROS mainly consists of superoxide anion (O2•−), hydroxyl radical (•OH), hydrogen peroxide (H2O2) and singlet oxygen (1O2) (33). In order to counteract the oxidative stress, aerobic organisms have evolved ROS scavengers such as catalases (CAT) that decomposes H2O2 into water and oxygen (31,34). Moreover, catalase is prevalent and highly conserved in aerobic organisms (35,36).
In N. crassa, there are three types of catalases, CAT-1, CAT-2 and CAT-3, which are differentially expressed during the asexual life cycle (37). CAT-1 is predominantly accumulated in conidia, CAT-2 is mainly found in aerial hyphae and conidia, and CAT-3 activity increases during exponential growth phase (37,38). Currently, biochemical study in N. crassa has demonstrated that CAT-3, present in growing mycelia, is the key catalase in mediating the resistance to oxidative stress, and its function could not be substituted by other catalases even under oxidative stress conditions (39). CAT-3 activity is also inducible under stress conditions, such as H2O2, heat shock, and menadione treatment (40), indicating that CAT-3 is critical for counteracting oxidative stress during the growth and development of mycelia in N. crassa. Given the exceedingly significant role of CAT-3 in resistance to oxidative stress, it is particularly important to explore the regulatory mechanism of cat-3 gene expression. Our previous studies showed that CPC1/GCN4 and GCN5 positively regulate the expression of cat-3 (41) and SWR complex-mediated H2A.Z deposition negatively regulates cat-3 transcription (42). However, the regulation of H2A.Z deposition at cat-3 locus is still unclear.
Here, we found that the histone fold domains-mediated NC2α/β heterodimer is essential for activating the cat-3 gene transcription through recruitment of the chromatin remodeling complex INO80C to remove histone variant H2A.Z from the nucleosomes around cat-3 promoter/TSS region. Interestingly, the C-terminal domain of NC2β is required not only for the binding of NC2 at cat-3 locus, but also for the INO80C-mediated H2A.Z removal from the nucleosomes around cat-3 locus. Taken together, these results provide a novel mechanism of NC2 in transcription activation of cat-3 which is important for resistance to oxidative stress.
MATERIALS AND METHODS
Strains and culture conditions
In this study, 87–3 (bd, a) (43) was used as the wild-type strain. The ku70RIP (bd, a) strain generated previously (44) was used as the host strain for creating the knockout strains (Nc2αKO, Nc2βKO, arp8KO, ies-4KO and ncu08417KO) and knock-in strains (Nc2αΔN, Nc2αΔHF, Nc2αΔC, Nc2βΔHF, Nc2βΔTBP, Nc2βΔR). The H2A.ZKO and cat-3KOstrains generated previously were also used in this study (42,45). The ino80KOstrain that has heterokaryotic and bd background in this study was generated previously (46). The heterokaryotic Nc2αKOH2A.ZKO strains were generated by replacing H2A.Z ORF region with basta-resistance bar gene in Nc2αKO background strain. The Nc2αKO, pqa-Myc-NC2α strain, Nc2αKO, pqa-Myc-NC2αΔN strain, Nc2αKO, pqa-Myc-NC2αΔHF strain, Nc2αKO, pqa-Myc-NC2αΔC strain and Nc2αKO, pqa-Myc-NC2β strain were generated by transferring pqa-Myc-NC2α, pqa-Myc-NC2αΔN, pqa-Myc-NC2αΔHF, pqa-Myc-NC2αΔC and pqa-Myc-NC2β constructs into the his-3 locus of Nc2αKO (his-3, a) host strain. Applying the same method, the Nc2βKO, pqa-Myc-NC2β strain, Nc2βKO, pqa-Myc-NC2βΔHF strain, Nc2βKO, pqa-Myc-NC2βΔTBP strain, Nc2βKO, pqa-Myc-NC2βΔR strain and Nc2βKO, pqa-Myc-NC2α strain were created. All strains used in this study possess the same bd background.
The medium for plate assays contained 1× Vogel's salts, 3% sucrose, and 1.5% (w/v) agar with or without H2O2 and in the absence or presence of 10−3 M QA. Liquid cultures were grown at 25°C with shaking in minimal medium (1× Vogel's and 2% glucose) for 18 h in constant light (LL). When QA was used, liquid cultures were grown in low-glucose medium (1× Vogel's, 0.1% glucose, 0.17% arginine) with 10−2 M QA.
Generation of antiserum against NC2α, NC2β, ARP8, TFIIB
The GST-NC2α (amino acids 95–401), GST-NC2β (amino acids 1–138), GST-ARP8 (amino acids 500–748) and GST-TFIIB (amino acids 19–348) fusion proteins were expressed in Escherichia coli BL21 cells, and the soluble recombinant proteins were purified and used as the antigens to immunize rabbits, which generated rabbit polyclonal antiserums, as described previously (47).
Plate assay
Age-appropriate conidia were inoculated in petri dishes with 50 ml liquid medium containing Vogel's minimal medium (VM) and 2% glucose under static culture condition at 25°C in constant light (LL) until the exponential growth phase of mycelium. The disks of mycelium mat were cut with a cork borer for quantification. For each strain, individual mycelium disk was transferred into the centers of VM plates containing 3% sucrose and 1.5% (w/v) agar, and cultured at 25°C in constant light (LL). The response to oxidative stress was determined by analyzing disk diameters of strains on VM plates containing 3% sucrose and 1.5% (w/v) agar with or without H2O2 of indicated concentrations and/or in the absence or presence of 10−3 M QA. In order to exclude the effect of the growth rate of different strains on the H2O2 sensitivity, the calculation method used previously that the extent of relative growth rate to represent the extent of H2O2 sensitivity was also used in this study (41,42,45). In addition, in order to visually analyze the growth phenotype, all plates were photographed until the disk diameters of strains in medium without H2O2 and QA exactly extend to the edge of the plate. Then we directly analyzed the H2O2 sensitivity through visual observation of the disk diameters of strains in medium with oxidative stress.
Protein analyses
Cell extracts from the adhered mycelium mat incubated for 18 h were used for performing protein analyses. Protein extraction, quantification, and western blot analysis were performed as described previously (47). Equal amounts of total protein (40 μg) were loaded into each protein lane. After electrophoresis, proteins were transferred onto PVDF membrane by electroblot. Western blot analyses were performed by using antibodies against the proteins of interest.
In-gel assay for activity of catalases
Protein extraction and quantification prepared for the in-gel assay was same as previously depicted (37,45). Equal amounts of total protein (40 μg) were loaded into each protein lane of 7.5% native poly-acrylamide slab gel (45). After electrophoresis, the gel rinsed with ddH2O was soaked in 10 mM H2O2 with gently shaking for 10 min, and then straightway transferred into a mixture of freshly prepared 1% potassium hexacyanoferrate (III) and 1% iron (III) chloride hexahydrate. Catalase activity was visualized as a band where H2O2 was decomposed by catalases.
RNA analyses
For RT-qPCR assays, total RNA was extracted with TRIzol agent and treated with DNase I to digest genomic DNA according to the previous description (41,42,45). Each RNA sample (5 μg) was subjected to reverse transcription with M-MLV reverse transcriptase purchased from Pro-mega (M1705), and then amplified by real-time PCR (7500; ABI). The primers used for qPCR were shown in Supplementary Table S1. The relative value of gene expression was calculated using the 2−ΔΔCT method (48) by comparing the cycle numbers for each sample with that for the untreated control. The results were normalized to the expression level of β-tubulin gene.
ChIP analyses
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (49). Briefly, tissues were fixed with 1% formaldehyde for 15 min at 25°C with shaking, followed by stopped with glycine at a final concentration of 125 mM for 5 min. Cross-linked tissues were ground and resuspended at 0.5 g/5 ml in lysis buffer containing proteinase inhibitors (1 mM PMSF, 1 mg/ml pepstatin A and 1 mg/ml leupeptin). Chromatin was sheared by sonication to ∼500 bp fragments. 2 mg/ml protein was used as per immunoprecipitation and 10 μl was kept as the input DNA. ChIP was carried out with 8 μl of antibody to H2A.Z, 10 μl of antibody to NC2α, 10 μl of antibody to NC2β, 5 μl of antibody to INO80, 10 μl of antibody to ARP8 and 10 μl of antibody to TFIIB. Immunoprecipitated DNA was quantified using real-time PCR (7500; ABI) with primer pairs (see Supplementary Table S2). ChIP-quantitative PCR (qPCR) data were normalized by the input DNA and presented as a percentage of input DNA. Each experiment was independently performed at least three times.
Co-Immunoprecipitation (Co-IP)
Cell extracts from the adhered mycelium mat incubated for 18 h were used for performing co-immunoprecipitation analyses. Protein extraction, quantification, and co-immunoprecipitation assays were performed as described previously (47). Briefly, the 4 mg/ml protein extracts in extraction buffer were incubated with 5 μl of monoclonal antibody to c-Myc (HT101-02, TransGen Biotech), 10 μl of antibody to NC2α, 10 μl of antibody to NC2β, 10 μl of antibody to INO80, 10 μl of antibody to ARP8, 10 μl of antibody to IgG (HS101–01, TransGen Biotech) for 4 h at 4°C with rotation. Then the 40 μl precleaned protein G-Sepharose (17-0885-02, GE Healthcare) were added and incubated for 1 h at 4°C with rotation. The beads were washed three times with ice-cold extraction buffer, mixed with protein loading buffer, and boiled for 10 min, and the immunoprecipitated proteins were analyzed by Western blotting.
Expression and purification of recombinant proteins
The recombinant protein GST-NC2β, GST-GFP and His-ARP8 were expressed in E. coli BL21 cells, and the soluble recombinant proteins were purified as described previously (50). Briefly, the transformed bacteria were resuspended in lysis buffer containing 20 mM Tris–HCl (pH 7.3), 500 mM NaCl, 10% glycerol, 0.1% Triton X-100 and 1 mM PMSF, then disrupted by ultrasonication. The supernatant was collected and recombinant proteins were purified using an Ni-NTA agarose (Bio-Rad) or glutathione agarose 4B (Macherey-Nagel) affinity column. After washing with lysis buffer containing 0.3% GSH (m/v) or an increasing concentration gradient of imidazole (150, 200, 250, 300 and 400 mM), proteins were eluted and concentrated with Amicon-Ultra-10 filters (Millipore).
GST-pull down assay
The GST pull-down assay was performed as described previously (51). Briefly, approximately 10 μg of purified GST-NC2β or GST-GFP fusion proteins were incubated with His-ARP8 in 500 μl incubated buffer (50 mM Tris–HCl, pH 6.8, 250 mM NaCl, 1.5% glycerol, 0.6% Triton X-100, and 0.1% Tween) for 4 h at 4°C. The beads were washed three times with incubation buffer. The washed beads were boiled in 1× SDS loading buffer, and the precipitates were separated by SDS-PAGE, followed by western blot analysis with anti-GST (GenScript, catalog no. A00866) and anti-His (Abmart, catalog no. M30111) antibodies.
RESULTS
Nc2 mutants are extremely sensitive to H2O2-induced ROS stress
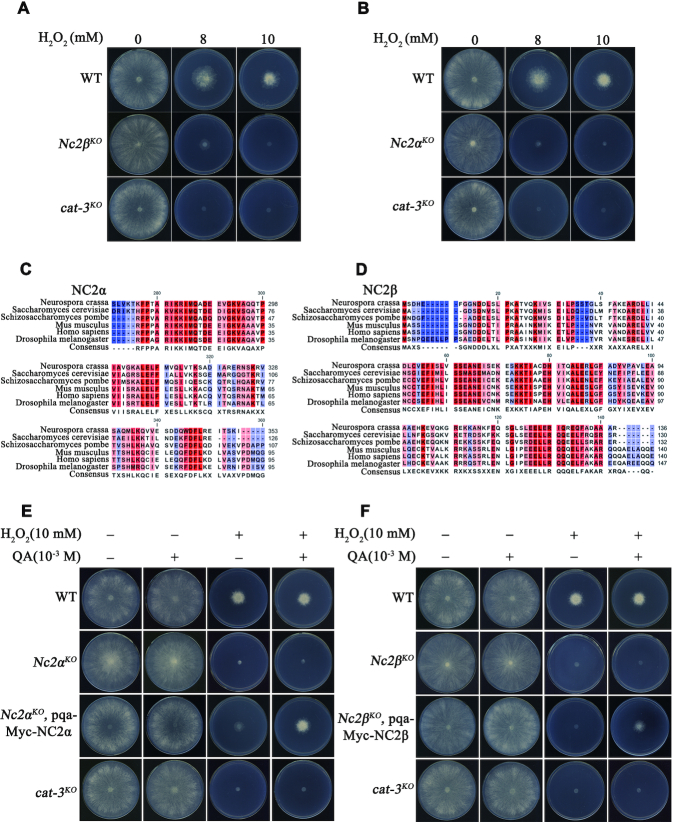
To characterize the transcription regulators that are involved in the regulation of cat-3 gene expression, we examined the H2O2 sensitivity of available knockout transcription regulator mutants with plate assays. As shown in Figure 1A, the strain with deletion of Nc2β gene (NCU02017) and cat-3KO strains showed similar H2O2-sensitive phenotypes compared to those of wild-type strains. Because the NC2 complex consists of NC2β and NC2α subunits in yeast and mammalian cells, we measured the H2O2 sensitivity of the deletion strain of Nc2α gene (NCU06405). As shown in Figure 1B, Nc2α deletion strain exhibited similar H2O2 sensitivity with Nc2βKO and cat-3KO strains, suggesting that the NC2 complex plays a critical role in the response to oxidative stress in N. crassa. Sequence alignment among NC2α and NC2β subunits in N. crassa with other eukaryotic counterparts revealed that the NC2 and its homologs are highly conserved among eukaryotes (Figure 1C and D, and Supplementary Figure S1A and B). Interestingly, the Nc2αKO and Nc2βKO strains exhibited hyphal formation and conidiation that was similar with the wild-type strains (Supplementary Figure S1C), indicating that both Nc2α and Nc2β genes are dispensable for N. crassa survival.
Figure 1.
Nc2 mutants are extremely sensitive to H2O2–induced ROS stress. (A, B) Plate assay analyzing mycelial growth on plates with 0, 8, or 10 mM H2O2 as indicated. Cultures were inoculated in plates at 25°C under constant light. (C, D) Amino acid sequence alignment of the conserved histone fold domain of NC2α (C) and NC2β (D) from Neurospora crassa, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Mus musculus, Homo sapiens, and Drosophila melanogaster. (E) Plate assay analysis showing mycelial growth of wild type (WT), Nc2αKO, Nc2αKO, pqa-Myc-NC2α and cat-3KO strains on plates with or without 10 mM H2O2 and in the absence or presence of 10−3 M QA. Cultures were inoculated in plates at 25°C under constant light. (F) The analysis of mycelial growth using plate assay in WT, Nc2βKO, Nc2βKO, pqa-Myc-NC2β and cat-3KO strains. The method tested in plates is same with (E).
To further confirm the H2O2 sensitivity phenotype of the Nc2αKO and Nc2βKO strains, two constructs carrying the sequences encoding Myc-tagged NC2α or NC2β driven by the quinic acid (QA)-inducible promoter, were transformed into Nc2αKO and Nc2βKO strains respectively. The ectopic expression of Myc-tagged NC2α or NC2β induced by QA rescued the H2O2 sensitivity phenotypes of Nc2αKO and Nc2βKO strains to those of WT on plate assays (Figure 1E and F), indicating that the observed H2O2-sensitive phenotype of each mutant was due to the deletion of respective NC2 subunits. Collectively, these results suggest that the NC2 complex plays a critical role in the response to oxidative stress in N. crassa.
NC2 complex is a key factor for the activation of cat-3 gene transcription
The hyper-sensitivity to H2O2-induced ROS stress in Nc2αKO and Nc2βKO strains strongly suggested that NC2 complex, a well-known transcription regulator, may play a crucial role in the regulation of CAT-3 expression. We examined the CAT-3 activity using an in-gel experiment in both wild-type and Nc2KO strains cultured at the exponential growth phase of the mycelium. As displayed in Figure 2A and B, the bands corresponding to CAT-3 activity were extremely weak in the Nc2αKO and Nc2βKO strains compared to those in the WT strains. In addition, ectopic expression of Myc-tagged NC2α or NC2β in the Nc2αKO or Nc2βKO strains restored the activities of CAT-3 to levels similar to those of wild-type strains (Figure 2A and B). Since NC2 is an important transcription regulator, these genetic and biochemical results suggest that NC2 complex may participate in the regulation of cat-3 expression in N. crassa. As expected, levels of CAT-3 protein and cat-3 mRNA in Nc2αKO or Nc2βKO strains were dramatically reduced compared to those in the WT strains (Figure 2C–F). Ectopic expression of Myc-tagged NC2α or NC2β in Nc2 mutants restored the levels of CAT-3 expression to those of wild-type strains (Figure 2C and D). These results demonstrated that the NC2 complex is a key regulator for the transcription activation of cat-3 gene.
Figure 2.
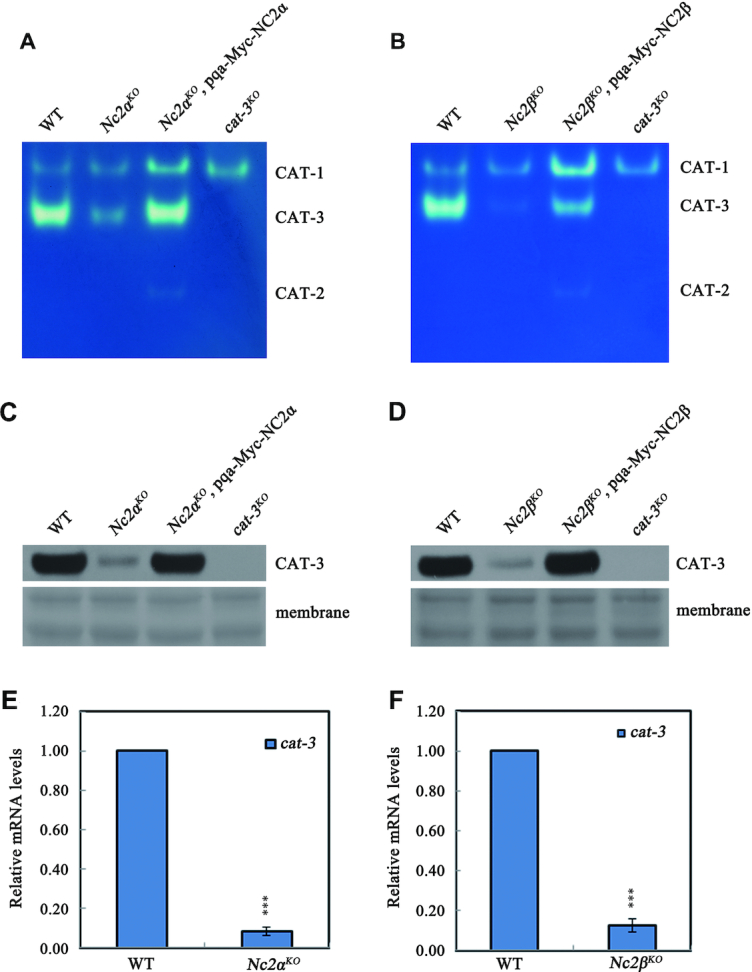
NC2 complex is a key factor for the activation of cat-3 gene transcription. (A, B) In-gel assay analysis of the CAT-3 activity levels in WT, Nc2αKO, Nc2βKO, Nc2αKO, pqa-Myc-NC2α and Nc2βKO, pqa-Myc-NC2β strains. (C, D) Western blot analyses showing the levels of CAT-3 protein in the WT, Nc2αKO, Nc2βKO, Nc2αKO, pqa-Myc-NC2α and Nc2βKO, pqa-Myc-NC2β strains. The membranes stained by coomassie blue represent the total protein in each sample and act as loading control for western blot. (E, F) RT-qPCR assays analyzing the levels of cat-3 mRNA in the WT, Nc2αKO and Nc2βKO strains. Error bars show s.d. (n = 3). Significance was evaluated by using a two-tailed t-test. ***P < 0.001 versus WT.
Conserved regions of NC2 subunits are required for transcriptional activation of cat-3
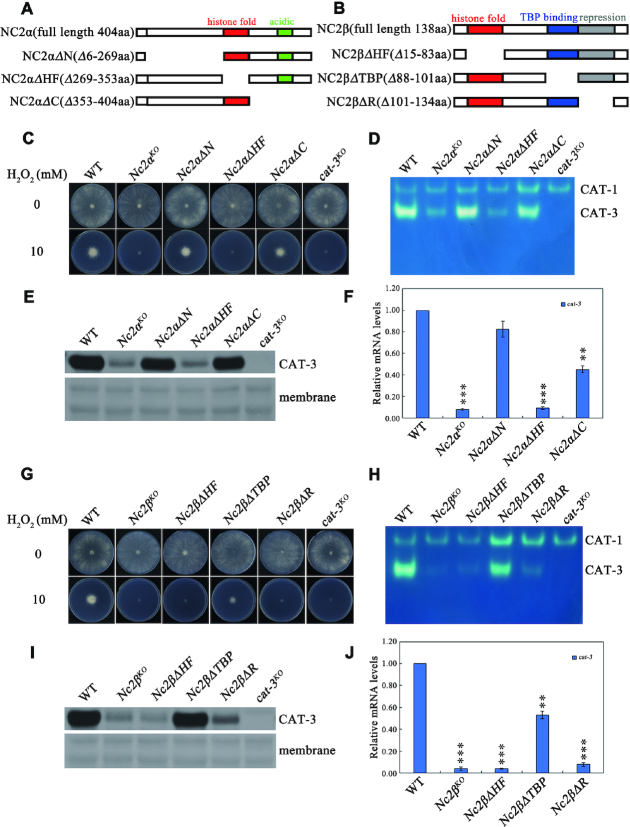
Alignment results revealed that N. crassa NC2α has an extended N-terminal region (from aa46 to aa268) compared to those of other eukaryotes (Supplementary Figure S1A), suggesting that NC2 subunits in N. crassa may function differently from homologues in other species. To characterize which region of NC2 subunits plays a role in the activation of cat-3 transcription, we generated a series of deletion strains across the NC2α or NC2β coding regions at the endogenous locus using knock-in methods (Figure 3A and B, and Supplementary Figure S2A and B). H2O2 sensitivity assays showed that Nc2αΔHF strain with a deletion of the histone fold domain of NC2α was extremely sensitive to H2O2, similar to the Nc2αKO strain (Figure 3C). However, Nc2αΔN and Nc2αΔC strains with a deletion either the N-terminal or the C-terminal region of NC2α exhibited the same H2O2 sensitivity as the wild-type strains (Figure 3C). Importantly, in-gel assays, western blot analyses and RT-qPCR assays revealed that the HF domain of NC2α, not the other parts, played a key role for activation of cat-3 expression (Figure 3D-F). Meanwhile, Nc2βΔHF and Nc2βΔR strains with a deletion of either the histone fold or repression domain of NC2β exhibited extreme H2O2 sensitivity similar to those of Nc2βKOstrain (Figure 3G), whereas Nc2βΔTBP strain with a deletion of TBP binding domain of NC2β was less sensitive to H2O2 than Nc2βΔHF and Nc2βΔR strains (Figure 3G). As expected, both HF domain and repression domain of NC2β, but not its TBP binding domain played a key role for maintenance of CAT-3 activity and the levels of CAT-3 protein and cat-3 mRNA (Figure 3H–J). Taken together, these data indicate that the activation of cat-3 gene transcription mainly requires the HF domains of both NC2 subunits and the C-terminal repression domain of NC2β.
Figure 3.
Conserved regions of NC2 subunits are required for transcriptional activation of cat-3. (A, B) Schematic drawing of N. crassa NC2α (A) and NC2β (B) subunits and their various deletion mutants. The position of the histone fold (red rectangle), acidic (green rectangle), TBP-binding (blue rectangle) and repression (gray rectangle) regions are indicated. (C) Plate assays analyzing mycelial growth of different deletion strains across NC2α coding region at endogenous locus on plates with 0 or 10 mM H2O2. Cultures were inoculated in plates at 25°C under constant light. (D) In-gel assay analysis of the CAT-3 activity levels in different deletion strains across NC2α coding region at endogenous locus. (E) Western blot analyses showing the levels of CAT-3 protein in different deletion strains across NC2α coding region at endogenous locus. The membrane stained by coomassie blue represents the total protein in each sample and acts as loading control for western blot. (F) RT-qPCR assays analyzing the levels of cat-3 mRNA in different deletion strains across NC2α coding region at endogenous locus. Error bars show s.d. (n = 3). Significance was evaluated by using a two-tailed t-test. **P < 0.01 and ***P < 0.001 versus WT. (G) Plate assays analyzing mycelial growth of different deletion strains across NC2β coding region at endogenous locus on plates with 0 or 10 mM H2O2. Cultures were inoculated in plates at 25°C under constant light. (H) In-gel assay analysis of the CAT-3 activity levels in different deletion strains across NC2β coding region at endogenous locus. (I) Western blot analyses showing the levels of CAT-3 protein in different deletion strains across NC2β coding region at endogenous locus. The membrane stained by coomassie blue represents the total protein in each sample and acts as loading control for western blot. (J) RT-qPCR assays analyzing the levels of cat-3 mRNA in different deletion strains across NC2β coding region at endogenous locus. Error bars show s.d. (n = 3). Significance was evaluated by using a two-tailed t-test. **P < 0.01 and ***P < 0.001 versus WT.
The integrity of NC2 heterodimer is necessary for NC2 targeting to cat-3 locus to activate its expression
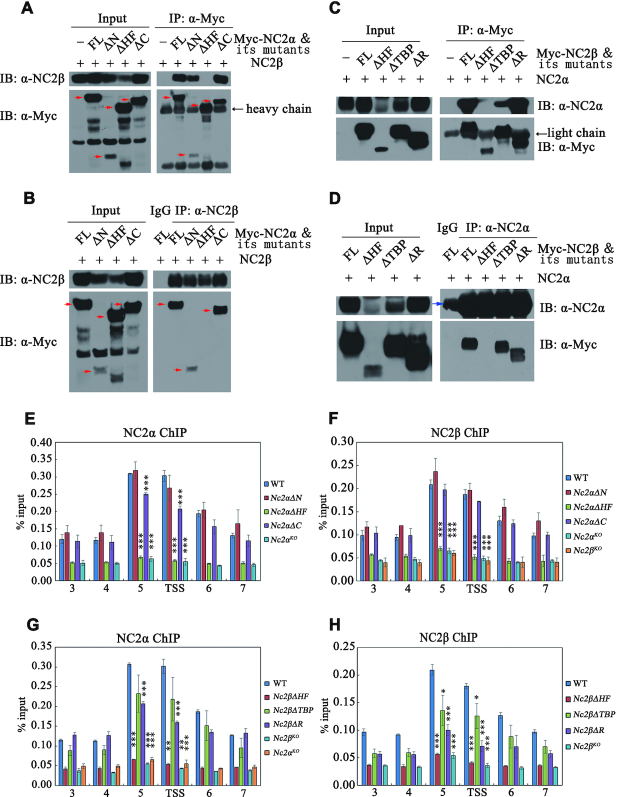
To further investigate whether NC2 complex directly regulates cat-3 transcription by binding to the cat-3 promoter, we generated NC2α- and NC2β-specific antibodies, which recognized a specific band at predicted molecular weight in the wild-type strain but not in the corresponding Nc2αKO and Nc2βKO strains (Supplementary Figure S3A and B). ChIP assays using respective antibodies showed that the enrichment of NC2α and NC2β is high at cat-3 locus, especially at its promoter (primer pairs 5) and TSS region in the wild-type strains but not in the Nc2αKO and Nc2βKO strains (Supplementary Figure S3C and D), indicating that NC2α and NC2β bind directly to the cat-3 locus to activate its transcription. In addition, ChIP assays showed that deletion of NC2α resulted in severely reduced enrichment of NC2β at cat-3 promoter/TSS region (Figure 4A). Similarly, NC2β is required for the binding of NC2α at cat-3 promoter/TSS region (Figure 4B). These results indicated that α and β subunits of NC2 are associated with cat-3 locus in an interdependent manner.
Figure 4.
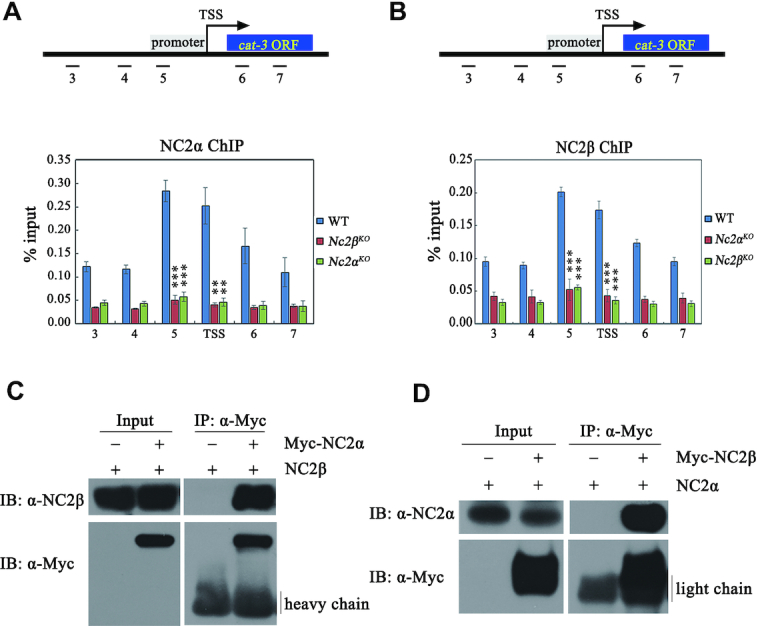
The integrity of NC2 heterodimer is a prerequisite for NC2 to bind to cat-3 locus and to activate its expression. (A, B) ChIP assays showing the binding levels of NC2α (A) and NC2β (B) at cat-3 locus in Nc2αKO, Nc2βKO and wild-type strains. Short black lines (primer pairs 3–7) under the schematic diagram of cat-3 gene represent the regions detected by ChIP-qPCR. TSS, transcription start site; ORF, open reading frame. Error bars show s.d. (n = 3). Significance was assessed by using a two-tailed t-test. **P < 0.01 and ***P < 0.001 versus WT. (C, D) Immunoprecipitation assays showing the interaction between Myc-NC2α or Myc-NC2β and endogenous NC2β or NC2α protein, respectively.
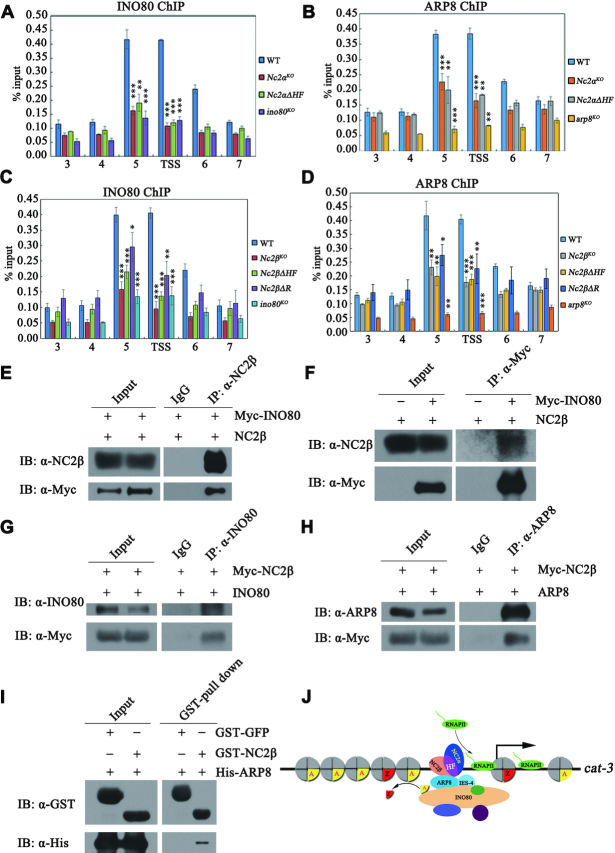
To further confirm the above results, we performed co-immunoprecipitation (Co-IP) experiments to assess the interaction between NC2α and NC2β in vivo. Co-IP assays showed that NC2α subunit strongly interacted with NC2β subunit in Nc2αKO, Myc-NC2α and Nc2βKO, Myc-NC2β transformants (Figure 4C and D). These results demonstrated that NC2α and NC2β function as a complex in N. crassa. To determine which domains of NC2 subunits are involved in NC2α-NC2β interaction, we performed Co-IP assays in Nc2αKO expressing Myc-NC2α or Myc-NC2α-deletion mutant strains and in Nc2βKO expressing Myc-NC2β or Myc-NC2β-deletion mutant strains. As shown in Figure 5A and B, the interaction between NC2α and NC2β was abolished in absence of the HF domain of NC2α, whereas both N- and C-terminal regions of NC2α were not required for the interaction with NC2β. Co-IP analyses also revealed that the HF domain of NC2β but not the other domains was required for its interaction with NC2α (Figure 5C and D). These results demonstrated that HF domains of both subunits are required for the formation of NC2 complex which may play a key role for the binding of NC2 at cat-3 locus. To confirm this possibility, we performed ChIP assays in WT and a series of deletion strains across the NC2α or NC2β coding regions at endogenous locus with NC2α- or NC2β-specific antibodies, respectively. As exhibited in Figure 5E and F, deletion of HF domain of NC2α but not its other domains led to severely reduced enrichment of NC2α and NC2β at cat-3 promoter/TSS region. Similarly, ChIP assays showed that the HF domain of NC2β subunit was essential for the binding of NC2α and NC2β at cat-3 promoter/TSS region (Figure 5G and H).
Figure 5.
HF domains in NC2 subunits are essential for its integrity and ability to bind at cat-3 locus. (A, B) Mapping of the NC2α region responsible for the interaction with NC2β in Nc2αKO strain with ectopically expressing wild-type Myc-NC2α or its various deletion mutants. Immunoprecipitation assays with anti-Myc antibody (A) or anti-NC2β antibody (B), the eluates were detected by western blot analysis using anti-Myc (α-Myc) and anti-NC2β (α-NC2β) antibodies. The red arrows denote specific bands, and the black arrow shows heavy chain. (C, D) Mapping of the NC2β region responsible for the interaction with NC2α in Nc2βKO strain with ectopically expressing Myc-NC2β or its various deletion mutants. Immunoprecipitation assays with anti-Myc antibody (C) or anti-NC2α antibody (D), the eluates were detected by western blot analysis using anti-Myc (α-Myc) and anti-NC2α (α-NC2α) antibodies. The blue arrow denotes heavy chain of IgG, the black arrow shows light chain. (E, F) ChIP assays showing the binding levels of NC2α (E) and NC2β (F) at cat-3 locus in different deletion strains across NC2α coding region at endogenous locus. (G, H) ChIP assays showing the binding levels of NC2α (G) and NC2β (H) at cat-3 locus in different deletion strains across NC2β coding region at endogenous locus. Error bars show s.d. (n = 3). Significance was assessed by using a two-tailed t-test. *P < 0.05, **P < 0.01 and ***P < 0.001 vs. WT.
The ectopic expression of Myc-tagged NC2β, but not NC2βΔHF, NC2βΔR, NC2βΔTBP fully restored the resistance to H2O2-induced ROS stress, CAT-3 activity, and CAT-3 expression of Nc2βKO strains (Supplementary Figure S4A–C), implying that the activation of cat-3 transcription by NC2β requires both the HF domain and the repression domain. The ectopic expression of Myc-tagged NC2α, NC2αΔN, NC2αΔC, but not NC2αΔHF restored the resistance to H2O2-induced ROS stress, CAT-3 activity, and CAT-3 expression of Nc2αKO strains (Supplementary Figure S4D–F), indicating that the function of NC2α in activating the expression of cat-3 gene is mediated through its HF domain. More importantly, the ectopic expression of Myc-tagged NC2α histone fold domain (NC2αHF) fully rescued the H2O2 resistance, the levels of CAT-3 protein and the CAT-3 activity in Nc2αKO strains (Supplementary Figure S5A–C). Taken together, these results demonstrated that the binding of the NC2 complex at the cat-3 locus is dependent on the HF domains of both NC2α and NC2β and the repression domain of NC2β.
NC2 complex activates the transcription of cat-3 gene by antagonizing the deposition of H2A.Z at cat-3 locus
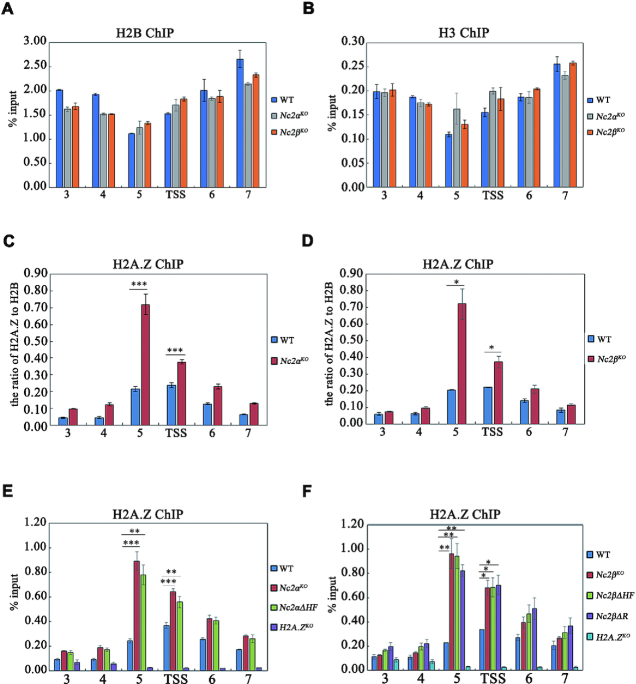
To test whether NC2 activates cat-3 gene transcription via altering the chromatin, we examined the occupancies of histone H2B and H3 at the cat-3 locus by ChIP assays in wild-type and Nc2 knockout strains. ChIP assays showed that the levels of H2B and H3 at cat-3 locus in Nc2KO strains remained the same as that of WT strain (Figure 6A and B), indicating that the nucleosome density is not affected at the cat-3 locus in Nc2 mutants. Our previous data showed that the occupancies of H2A.Z at cat-3 gene promoter/TSS region negatively regulate transcription of cat-3 (42), so we examined the occupancies of H2A.Z around cat-3 promoter/TSS region in Nc2KO and WT strains. ChIP assays using H2A.Z-specific antibody (42) showed that H2A.Z occupancies were dramatically increased at the cat-3 promoter/TSS region in the Nc2αKO and Nc2βKO strains compared to that of the WT strain (Figure 6C and D, and Supplementary Figure S6A and B), suggesting that NC2 complex activates cat-3 expression through regulating the deposition of H2A.Z at the nucleosomes around the cat-3 promoter/TSS region. ChIP assays further showed that the disruption of NC2 complex with HF deletion of NC2 subunits and C-terminus deletion of NC2β resulted in the increased occupancies of H2A.Z at the nucleosomes of cat-3 promoter/TSS region (Figure 6E and F). These results demonstrated that the HF domains of NC2 subunits and the C-terminal region of NC2β are required for the integrity of NC2 complex which activates cat-3 expression by antagonizing the deposition of H2A.Z at the nucleosomes around the cat-3 promoter/TSS region. Similar results were observed at other NC2 targeting genes such as heat shock protein genes (24). As shown in Supplementary Figure S6C and D, NC2β bound at promoter/TSS regions of heat shock protein genes hsp70/dnak and hsp90a. The deletion of NC2β resulted in increased occupancy of H2A.Z at hsp70/dnak and hsp90a (Supplementary Figure S6E and F), indicating that NC2 is involved in the removal of H2A.Z at these genes in Neurospora.
Figure 6.
NC2 complex activates the transcription of cat-3 gene by antagonizing the inhibition of H2A.Z at cat-3 locus. (A, B) ChIP assays showing the occupancy levels of H2B (A) and H3 (B) at cat-3 locus in Nc2αKO and Nc2βKO as well as wild type strains. (C, D) ChIP assays showing the relative occupancy levels of H2A.Z at cat-3 locus in WT, Nc2αKO (C) and Nc2βKO (D) strains. The relative occupancy levels of H2A.Z represent the ratio of H2A.Z to H2B. (E) ChIP assays analyzing the effects of deletion of HF domain of NC2α on the binding of H2A.Z at cat-3 locus. The H2A.ZKO strain was used as a negative control. (F) ChIP assays analyzing the effects of deletion of C-terminal repression or HF domain of NC2β on the binding of H2A.Z at cat-3 locus. The H2A.ZKO strain was used as a negative control. Error bars show s.d. (n = 3). Significance was assessed by using a two-tailed t-test. *P < 0.05, **P < 0.01 and ***P < 0.001.
To analyze the effect of H2A.Z deletion on cat-3 expression in Nc2 deletion strains, we tried to delete H2A.Z gene in Nc2αKO background strain by homologous recombination (Supplementary Figure S7A). Unfortunately, we only got the heterokaryotic Nc2αKOH2A.ZKO double mutant (Supplementary Figure S7B and C), suggesting that deletion of Nc2α and H2A.Z at the same time may be lethal in Neurospora. As shown in Supplementary Figure S7D and E, heterokaryotic Nc2αKOH2A.ZKO double mutant displayed reduced H2O2 sensitivity and derepressed cat-3 expression compared with those in Nc2αKO strain. Therefore, these results further indicate that NC2 activates cat-3 expression through the removal of H2A.Z at cat-3 locus.
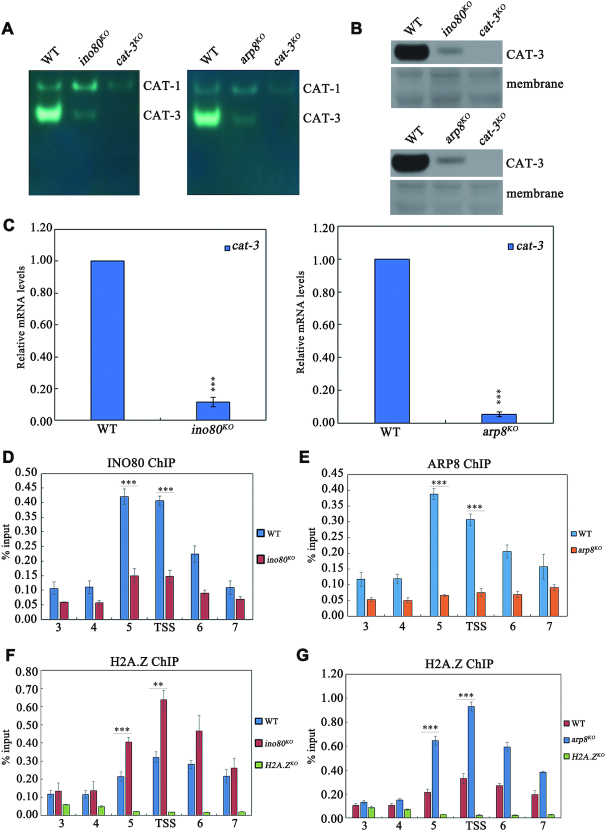
INO80C is involved in activating cat-3 transcription by mediating the removal of H2A.Z from nucleosomes around cat-3 locus
Previous studies suggested that the chromatin remodeling complex, INO80C, catalyzes the removal of histone variant H2A.Z from specific promoter-bound and H2A.Z-bearing nucleosomes to activate genes transcription (52–55). Therefore, we tested whether Neurospora INO80C is involved in the transcriptional activation of cat-3 by regulating the removal of H2A.Z from nucleosomes around the cat-3 locus. To address this question, we first investigated the effect of INO80C on cat-3 gene expression. As shown in Figure 7A–C and Supplementary Figure S8A–D, the deletion strains of INO80, ARP8 and IES-4 which are subunits of INO80 complex exhibited the obvious H2O2 sensitivity and the decreased levels of CAT-3 activity, CAT-3 protein and cat-3 mRNA compared to those of WT strain. Previous studies showed that the deletion of INO80 resulted in genome instability (56,57). To exclude the effect of genome instability on cat-3 expression, we treated WT strain for different time points with hydroxyurea (HU), which causes replication stress and genome instability (57). As shown in Supplementary Figure S8E, the increased cat-3 transcription was observed after treatment of wild-type strains with HU for indicated time points, indicating that HU-induced genome instability can stimulate the transcription of cat-3. This result is obviously different from the decreased cat-3 expression in INO80C deletion strains, indicating that the decreased cat-3 transcription in INO80C deletion strains is not caused by genome instability. Moreover, ChIP assays using INO80- (46) and ARP8-specific (Supplementary Figure S8F) antibodies showed that both INO80 and ARP8 predominantly occupied at cat-3 promoter/TSS region (Figure 7D and E). These results demonstrated that INO80C directly binds and functions at cat-3 locus for activating its transcription. Consistent with the low expression of cat-3 in ino80KO and arp8KO strains, ChIP assays using H2A.Z-specific antibody revealed that levels of H2A.Z enrichment at cat-3 promoter/TSS region was extremely increased compared to that in WT strain (Figure 7F and G). Taken together, these results indicated that INO80C activates the transcription of cat-3 gene by regulating the removal of H2A.Z from nucleosomes around cat-3 locus.
Figure 7.
INO80C is involved in activating cat-3 transcription by mediating the removal of H2A.Z from nucleosomes around cat-3 locus. (A) In-gel assay analysis of the CAT-3 activity levels in WT, ino80KO and arp8KO strains. (B) Western blot analyses showing the levels of CAT-3 protein in the WT, ino80KO and arp8KO strains. The membranes stained by coomassie blue represent the total protein in each sample and act as loading control for western blot. (C) RT-qPCR assays analyzing the levels of cat-3 mRNA in the WT, ino80KO and arp8KO strains. Error bars show s.d. (n = 3). Significance was evaluated by using a two-tailed t-test. ***P < 0.001 versus WT. (D, E) ChIP assays revealing the binding of INO80 (D) and ARP8 (E) at cat-3 locus. (F, G) ChIP assays analyzing the effects of deletion of INO80 (F) and ARP8 (G) on the binding of H2A.Z at cat-3 locus. The H2A.ZKO strain was used as a negative control. Error bars show s.d. (n = 3). Significance was evaluated by using a two-tailed t-test. **P < 0.01 and ***P < 0.001.
In addition, it is shown that ANP32E, a vertebrate-specific chaperon facilitates H2A.Z eviction (58,59), indicating that INO80 is not the only H2A.Z removal factor in vertebrate. Is there ANP32E homolog in Neurospora? Amino acid sequence alignment analysis showed that the C-terminal amino acid region of hypothetical protein NCU08417 in Neurospora is conserved with those of ANP32E, but the N-terminal region of NCU08417 is less conserved with those of ANP32E, suggesting that NCU08417 may be a homolog of ANP32E in Neurospora (Supplementary Figure S9A). Our results showed that the deletion strain of NCU08417 exhibited defects of H2O2 resistance, decreased cat-3 expression and increased H2A.Z occupancy at cat-3 promoter/TSS region (Supplementary Figure S9B-E), suggesting that NCU08417 is involved in the removal of H2A.Z around cat-3 locus in Neurospora. However, it needs further investigation of whether NCU08417 is the ANP32E homolog in Neurospora, because the protein sequence similarity is low (27%) between NCU08417 and vertebrate-specific ANP32E, and NCU08417 is lack of typical leucine-rich repeats domain conserved in ANP32E. Therefore, it will be an interesting question to explore whether NCU08417 is a H2A.Z-removal chaperone in Neurospora.
NC2 complex mediates the recruitment of INO80C to cat-3 locus to activate its expression
Given that the INO80 complex is a co-factor which is recruited to promoters in a specific transcription factor-dependent manner (60–62), we hypothesized that recruitment of INO80C to cat-3 locus was regulated in a NC2-dependent manner. To test this possibility, we examined the recruitment of INO80 and ARP8 at cat-3 locus through ChIP assays with INO80 and ARP8 antibodies in Nc2αKO, Nc2βKO and different domain deletion strains as well as wild-type strains. ChIP assays showed that the recruitments of INO80 and ARP8 to cat-3 promoter/TSS region were dependent on the binding of NC2 at these regions (Figure 8A–D), indicating that INO80C is recruited to cat-3 locus in a NC2-dependent manner. Since the INO80 complex acts as a co-factor for activating cat-3 expression in a NC2-dependent manner, we performed Co-IP assays to test the association of the INO80 complex with NC2 in N. crassa. As shown in Figure 8E, Myc-INO80 protein was immunoprecipitated with NC2β by the NC2β antibody, indicating that NC2β was able to interact with INO80C. Moreover, the interaction between INO80 and NC2β was confirmed in a reciprocal immunoprecipitation with anti-Myc antibody to precipitate NC2β protein (Figure 8F). Furthermore, Co-IP assays with anti-INO80 and ARP8 antibodies in the Nc2βKO, Myc-NC2β strains revealed that Myc-NC2β was immunoprecipitated by the INO80 antibody and ARP8 antibody, respectively (Figure 8G and H). In addition, GST-pull down assay showed that GST-NC2β interacted with His-ARP8 in vitro, indicating that NC2 directly interacts with INO80C in Neurospora (Figure 8I). Taken together, these results demonstrated that the recruitment of INO80C to the cat-3 locus is mediated through the association between the NC2 complex and the INO80 complex.
Figure 8.
NC2 complex mediates the recruitment of INO80C to cat-3 locus to activate its expression. (A, B) ChIP assays analyzing the effects of deletion of full-length NC2α or HF domain of NC2α on the binding of INO80 (A) and ARP8 (B) at cat-3 locus. (C, D) ChIP assays analyzing the effects of deletion of full length, C-terminal repression or HF domain of NC2β on the binding of INO80 (C) and ARP8 (D) at cat-3 locus. Error bars show s.d. (n = 3). Significance was assessed by using a two-tailed t-test. *P < 0.05, **P < 0.01 and ***P < 0.001 versus WT. (E, F) Immunoprecipitation assays showing the interaction between Myc-INO80 and endogenous NC2β. (G, H) Immunoprecipitation assays showing the interaction between Myc-NC2β and endogenous INO80 (G) or ARP8 (H). (I) GST pull-down assay showing the interaction between NC2β and ARP8 in vitro. Purified GST-NC2β or GST-GFP was incubated with His-ARP8. After being immunoprecipitated with glutathione-sepharose beads, the proteins were detected by western blot analysis with anti-His and anti-GST antibodies. (J) A model depicting the mechanism of NC2 regulating cat-3 expression. On the one hand, NC2 recruits INO80C to remove H2A.Z at cat-3 promoter and TSS, on the other hand, NC2 promotes the PIC assembly at cat-3 TSS region, ultimately resulting in the transcription activation of cat-3.
To further verify our results, we employed an inducible expression system where the qa-2 promoter driven Myc-NC2β in Nc2βKO strain is induced after treatment with QA. As shown in Supplementary Figure S10A–D, the increased QA concentrations induced the expression of Myc-NC2β, followed by the increased enrichment of NC2β and INO80 and reduced occupancy of H2A.Z at cat-3 promoter/TSS locus, resulting in cat-3 activation. Therefore, these results indicate that NC2 activates cat-3 expression through recruiting INO80C to remove H2A.Z around cat-3 locus.
DISCUSSION
Transcriptional control of catalase genes is an essential step in response to the environmental or intracellular oxidative stimuli. The precisely regulated expression of cat-3 provides a strong model to study the mechanisms of gene expression controlled by chromatin modifications (41,42,45). In this study, we found that the NC2 complex, a heterodimer of two histone-fold (HF) domain-containing transcription factors, is required for cat-3 gene activation and resistance to oxidative stress in N. crassa. The integrity of the NC2 heterodimer mediated by their HF domains is a prerequisite for activation of cat-3 expression, in which their HF domains are responsible for maintenance of the NC2α–NC2β interaction in vivo and targeting NC2 to cat-3 locus. ChIP assays revealed that the abundance of H2A.Z at cat-3 locus was dramatically increased in the absence of NC2. Loss of the INO80 and ARP8 subunits of INO80 complex resulted in a decreased cat-3 expression and an elevated deposition of H2A.Z at cat-3 locus similar to those in Nc2KO mutants. Strikingly, the interaction of NC2-INO80C and the binding activities of NC2 determined the recruitment of the INO80C to cat-3 gene promoter/TSS for removing H2A.Z from the nucleosomes, resulting in transcriptional activation of cat-3 expression. Collectively, we conclude that the NC2 complex recruits the INO80C complex to remove H2A.Z and activate cat-3 transcription to protect against oxidative stress in N. crassa.
The HF domains-mediated integrity of heterodimer is critical for NC2 to activate cat-3 gene expression to counteract oxidative stress
In S. cerevisiae, NC2 has been characterized as a general positive/negative regulator of class II gene transcription (12,14,25,63,64). However, the role of the NC2 heterodimer in transcription activation is still unclear. In the present study, we demonstrate that NC2α and NC2β regulate transcriptional activation of cat-3 in a heterodimer manner. Several lines of evidence presented here indicate that the HF domains of NC2α and NC2β governs NC2 heterodimer formation which is a prerequisite for the transcriptional activation of oxidative stress-responsive cat-3 gene. First, immunoprecipitation assays showed that NC2α and NC2β associate with each other through their HF domains in N. crassa (Figure 5A–D). Second, ectopic expression of Myc-NC2α in Nc2βKO strain or ectopic expression of Myc-NC2β in Nc2αKO strain cannot activate cat-3 expression (Supplementary Figure S11). Third, ChIP assays showed that deletion of each subunit or their HF domains of NC2α and NC2β proteins abolished the binding of NC2 subunits at cat-3 locus (Figure 5E–H). These results indicate that the HF domains is the key of NC2α/β heterodimerization and NC2α/β targeting to cat-3 locus. Similar to our results, NC2 lacking NC2β TBP domain or TBP domain and repression domain, could still activate the transcription of TATA-less DPE-containing genes in Drosophila (22). This suggests that the formation of HF-mediated NC2 heterodimer, but not the NC2β TBP domain or repression domain, is essential for NC2-mediated transcription activation of DPE-containing gene in vitro. Unlike Drosophila NC2β repression domain is dispensable for NC2-meiated transcription activation of DPE-containing gene, our results showed that Neurospora NC2β repression domain is involved in activating cat-3 expression in vivo (Figure 3H–J), and contributes to NC2 targeting to cat-3 locus (Figure 5G and H). Together, these results indicate that the HF domains-mediated integrity of heterodimer is critical for NC2 to activate cat-3 gene expression and counteract oxidative stress.
NC2 activates the transcription of cat-3 gene through removing histone variant H2A.Z from the nucleosomes around cat-3 locus
Previous studies showed that, in general, the function of NC2 in transcriptional repression/activation was associated with the assembly of transcription preinitiation complex (PIC) (16,25,64,65). It is also shown that blocking PIC assembly resulted in increased accumulation of H2A.Z at +1 nucleosome which covers the transcription start sites of most genes in yeast, indicating that the PIC is required to evict H2A.Z (66). Our results suggest that NC2 activates the transcription of cat-3 gene through removing histone variant H2A.Z from the nucleosomes around cat-3 promoter and TSS regions. Whether the increased H2A.Z occupancy at cat-3 promoter (locus 5) and TSS region in NC2 deletion strains is caused by the inhibition of PIC assembly in Neurospora? Our ChIP assays with TFIIB- (Supplementary Figure S12B) and RPB-1-specific (67) antibodies showed that deletion of NC2β resulted in reduced enrichment of TFIIB and RPB-1 at cat-3 TSS and ORF 5′ region (locus 6), but not cat-3 promoter under normal condition (Supplementary Figure S12C and D), indicating that the PIC assembly promoted by NC2 mainly occurs at cat-3 TSS and ORF 5′ region. However, NC2 was mainly occupied at cat-3 promoter and TSS region (Figure 4A and B, and Supplementary Figure S12A), and the localization of NC2 at cat-3 locus correlated well with those of INO80C and H2A.Z (Figures 6 and 7), but not that of TFIIB or RPB-1, suggesting that H2A.Z occupancy changes, at least at cat-3 promoter, are mediated by NC2 but not indirect effect of GTFs or RNAPII. In addition, the increased enrichment of TFIIB and RPB-1 at cat-3 locus is observed in WT strain (Supplementary Figure S12C and D), but not in Nc2βKO strain, after H2O2 treatment which can induce cat-3 expression in our previous studies (41,42,45). These results further indicate that NC2 activates cat-3 expression through promoting the recruitment of TFIIB and RPB-1 and the formation of PIC at cat-3 locus. Therefore, our results suggest that on the one hand, NC2 promotes the binding of TFIIB and RNAPII and the PIC assembly at cat-3 TSS region, on the other hand, NC2 recruits INO80C to remove H2A.Z at cat-3 promoter and TSS, ultimately resulting in the transcription activation of cat-3 (Figure 8J).
Similar results were observed at other NC2 targeting genes such as heat shock protein genes (Supplementary Figure S6E and F) (24), indicating that NC2 is involved in the removal of H2A.Z at heat shock protein genes hsp70/dnak and hsp90a in Neurospora. Similar to our results, during embryonic stem (ES) cell differentiation into endoderm, transcription factor Foxa2 binds to H2A.Z nucleosomes occupied promoters to promote the recruitment of SWI/SNF and INO80 complexes, resulting in H2A.Z removal and nucleosome depletion, thus promoting ES cell differentiation (68). NC2 is involved in the removal of H2A.Z from nucleosomes around inducible genes such as cat-3, hsp70/dnak and hsp90a locus, indicating that the structure of H2A.Z-containing nucleosomes around these inducible genes is less stable than those of H2A-containing nucleosomes. Similar result is shown that Htz1/H2A.Z is more susceptible to dissociation from purified yeast chromatin than H2A or H3, suggesting that nucleosomes bearing Htz1/H2A.Z present in yeast chromatin are less stable than their canonical counterparts, and this property may serve to mark repressed/basal promoters with nucleosomes susceptible to histone loss during activation (53).
INO80C is involved in activating cat-3 transcription by mediating the removal of H2A.Z from nucleosomes around cat-3 locus
The deposition of H2A.Z into nucleosome is mediated by SWR1 complex (69,70), while the removal of H2A.Z from nucleosomes is catalyzed by INO80 complex (52–55). Previous studies showed that the removal of H2A.Z from nucleosomes is coupled with gene activation (53,71,72). As expected, deletion of the catalytic subunit INO80 and a structural subunit ARP8 resulted in extremely increased occupancy of H2A.Z at cat-3 promoter/TSS (Figure 7F and G), the result is consistent with the observation in Nc2KO strains. Similar to NC2, INO80C was also required for the transcriptional activation of cat-3 gene (Figure 7C). Deletion of either subunit of NC2 complex or their HF domains led to the dramatically decreased enrichment of INO80C at cat-3 promoter/TSS (Figure 8A–D). Furthermore, immunoprecipitation assays showed that NC2 associates with INO80C which is critical for recruitment of INO80C to cat-3 locus. Thus, the NC2-mediated removal of H2A.Z from the nucleosomes relies on chromatin remodeling complex INO80C. However, it is controversial about whether INO80C is involved in the removal of H2A.Z in yeast. It is shown that purified yeast INO80 complex can incorporate H2A into an H2A.Z nucleosome in vitro, indicating that it has a histone-exchange activity that replaces nucleosomal H2A.Z/H2B with free H2A/H2B dimers (54). In addition, INO80 translocates along DNA at the H2A–H2B dimer interface to displace DNA and promote H2A.Z exchange (73). During homologous recombination, the INO80 complex promotes presynaptic filament formation through removing H2A.Z (74). These studies indicate that INO80 complex participates in the removal of H2A.Z. However, some studies showed that deletion of yeast INO80 did not affect global H2A.Z occupancy (66,75). The reason for this discrepancy may be that different background strains or different experiment methods were used in different research groups. Consistent with former studies, our results indicate that the removal of H2A.Z from the nucleosomes around cat-3 locus is mediated by INO80 complex in N. crassa.
Here, we showed that the conserved transcription factor NC2 is essential for activation of cat-3 gene to counteract oxidative stress in N. crassa. Similar to our results, the increased yeast NC2α/BUR6 occupancy at heat shock-induced promoters of genes such as Cytosolic Catalase T (CTT1) occurs under the condition where its transcription is strongly induced upon a rapid heat shock, an inducer of oxidative stress (24). These findings suggested that in aerobic organisms, transcription factor NC2 is a conserved regulator of oxidative stress via controlling the expression of catalase gene. Moreover, NC2-INO80 complex is essential for transcriptional activation of cat-3 gene through removing H2A.Z from the nucleosomes around cat-3 locus, shedding light to a novel mechanism of NC2 in transcription regulation via changing the nucleosome composition. Similar mechanism has been observed in human, that is, purified in vitro recombinant human NC2 can facilitate chromatin remodeling complex ACF-mediated canonical nucleosome assembly, independently of a direct interaction with ACF (76). In addition, it has been shown that Mot1, Ino80C, and NC2 coordinate to suppress pervasive transcription in euchromatin and facultative heterochromatin in yeast and mammals (77). Therefore, our data demonstrated that NC2 is required for the removal of H2A.Z from the nucleosomes around cat-3 promoter/TSS, and this may be a conserved mechanism for other cat-3-like genes in eukaryotic organisms.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to Yubo He for the crucial revision of this manuscript and Prof. Xianbing Wang for pET-30a(+) and pGEX-KG-GFP plasmids.
Notes
Present address: Qing Dong, Department of Neurology, University of California, San Francisco, San Francisco, CA 94143, USA.
Contributor Information
Guofei Cui, State Key Laboratory of Agrobiotechnology and MOA Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Qing Dong, State Key Laboratory of Agrobiotechnology and MOA Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Jiabin Duan, State Key Laboratory of Agrobiotechnology and MOA Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Chengcheng Zhang, State Key Laboratory of Agrobiotechnology and MOA Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Xiao Liu, State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China; College of Life Sciences, University of the Chinese Academy of Sciences, Beijing 100049, China.
Qun He, State Key Laboratory of Agrobiotechnology and MOA Key Laboratory of Soil Microbiology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the grants from the National Key R&D Program of China [2018YFA0900500] and the National Natural Science Foundation of China [31771383] to Q.H.; National Natural Science Foundation of China [31970079], Beijing Natural Science Foundation [5202020] and CAS Pioneer Hundred Talents Program to X.L.
Conflict of interest statement. None declared.
REFERENCES
- 1. Orphanides G., Lagrange T., Reinberg D.. The general transcription factors of RNA polymerase II. Genes Dev. 1996; 10:2657–2683. [DOI] [PubMed] [Google Scholar]
- 2. Nikolov D.B., Burley S.K.. RNA polymerase II transcription initiation: a structural view. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nogales E., Louder R.K., He Y.. Structural insights into the eukaryotic transcription initiation machinery. Annu. Rev. Biophys. 2017; 46:59–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Juven-Gershon T., Kadonaga J.T.. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010; 339:225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Donczew R., Hahn S.. Mechanistic differences in transcription initiation at TATA-Less and TATA-Containing promoters. Mol. Cell. Biol. 2018; 38:e00448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goodrich J.A., Cutler G., Tjian R.. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996; 84:825–830. [DOI] [PubMed] [Google Scholar]
- 7. Roeder R.G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996; 21:327–335. [PubMed] [Google Scholar]
- 8. Thomas M.C., Chiang C.M.. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006; 41:105–178. [DOI] [PubMed] [Google Scholar]
- 9. Maldonado E., Hampsey M., Reinberg D.. Repression: targeting the heart of the matter. Cell. 1999; 99:455–458. [DOI] [PubMed] [Google Scholar]
- 10. Inostroza J.A., Mermelstein F.H., Ha I., Lane W.S., Reinberg D.. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992; 70:477–489. [DOI] [PubMed] [Google Scholar]
- 11. Meisterernst M., Roeder R.G.. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991; 67:557–567. [DOI] [PubMed] [Google Scholar]
- 12. Goppelt A., Meisterernst M.. Characterization of the basal inhibitor of class II transcription NC2 from Saccharomyces cerevisiae. Nucleic. Acids. Res. 1996; 24:4450–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2alpha homolog that has both positive and negative roles in transcription in vivo. Mol. Cell. Biol. 1997; 17:2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim S., Na J.G., Hampsey M., Reinberg D.. The Dr1/DRAP1 heterodimer is a global repressor of transcription in vivo. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:820–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iratni R., Yan Y.T., Chen C., Ding J., Zhang Y., Price S.M., Reinberg D., Shen M.M.. Inhibition of excess nodal signaling during mouse gastrulation by the transcriptional corepressor DRAP1. Science. 2002; 298:1996–1999. [DOI] [PubMed] [Google Scholar]
- 16. Goppelt A., Stelzer G., Lottspeich F., Meisterernst M.. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996; 15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 17. Yeung K.C., Inostroza J.A., Mermelstein F.H., Kannabiran C., Reinberg D.. Structure-function analysis of the TBP-binding protein Dr1 reveals a mechanism for repression of class II gene transcription. Genes Dev. 1994; 8:2097–2109. [DOI] [PubMed] [Google Scholar]
- 18. Cang Y., Prelich G.. Direct stimulation of transcription by negative cofactor 2 (NC2) through TATA-binding protein (TBP). Proc. Natl. Acad. Sci. U.S.A. 2002; 99:12727–12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie J., Collart M., Lemaire M., Stelzer G., Meisterernst M.. A single point mutation in TFIIA suppresses NC2 requirement in vivo. EMBO J. 2000; 19:672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim T.K., Zhao Y., Ge H., Bernstein R., Roeder R.G.. TATA-binding protein residues implicated in a functional interplay between negative cofactor NC2 (Dr1) and general factors TFIIA and TFIIB. J. Biol. Chem. 1995; 270:10976–10981. [DOI] [PubMed] [Google Scholar]
- 21. Kamada K., Shu F., Chen H., Malik S., Stelzer G., Roeder R.G., Meisterernst M., Burley S.K.. Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell. 2001; 106:71–81. [DOI] [PubMed] [Google Scholar]
- 22. Willy P.J., Kobayashi R., Kadonaga J.T.. A basal transcription factor that activates or represses transcription. Science. 2000; 290:982–985. [DOI] [PubMed] [Google Scholar]
- 23. Lemaire M., Xie J., Meisterernst M., Collart M.A.. The NC2 repressor is dispensable in yeast mutated for the Sin4p component of the holoenzyme and plays roles similar to Mot1p in vivo. Mol. Microbiol. 2000; 36:163–173. [DOI] [PubMed] [Google Scholar]
- 24. Geisberg J.V., Holstege F.C., Young R.A., Struhl K.. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol. Cell. Biol. 2001; 21:2736–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Masson P., Leimgruber E., Creton S., Collart M.A.. The dual control of TFIIB recruitment by NC2 is gene specific. Nucleic Acids Res. 2008; 36:539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zanton S.J., Pugh B.F.. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 2006; 20:2250–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Valko M., Rhodes C.J., Moncol J., Izakovic M., Mazur M.. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006; 160:1–40. [DOI] [PubMed] [Google Scholar]
- 28. Valko M., Leibfritz D., Moncol J., Cronin M.T., Mazur M., Telser J.. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007; 39:44–84. [DOI] [PubMed] [Google Scholar]
- 29. Aggarwal S., Gross C.M., Sharma S., Fineman J.R., Black S.M.. Reactive oxygen species in pulmonary vascular remodeling. Compr Physiol. 2013; 3:1011–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu H., Liu X., Zhang C., Zhu H., Xu Q., Bu Y., Lei Y.. Redox imbalance in the development of colorectal cancer. J. Cancer. 2017; 8:1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abdal Dayem A., Hossain M.K., Lee S.B., Kim K., Saha S.K., Yang G.M., Choi H.Y., Cho S.G.. The role of reactive oxygen species (ROS) in the biological activities of metallic nanoparticles. Int. J. Mol. Sci. 2017; 18:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davidson J.F., Schiestl R.H.. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001; 21:8483–8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khan M.N., Mobin M., Abbas Z.K., AlMutairi K.A., Siddiqui Z.H.. Role of nanomaterials in plants under challenging environments. Plant Physiol. Biochem. 2017; 110:194–209. [DOI] [PubMed] [Google Scholar]
- 34. Feng C., Yang M., Lan M., Liu C., Zhang Y., Huang B., Liu H., Zhou Y.. ROS: crucial intermediators in the pathogenesis of intervertebral disc degeneration. Oxid Med Cell Longev. 2017; 2017:5601593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Glorieux C., Zamocky M., Sandoval J.M., Verrax J., Calderon P.B.. Regulation of catalase expression in healthy and cancerous cells. Free Radic. Biol. Med. 2015; 87:84–97. [DOI] [PubMed] [Google Scholar]
- 36. Hansberg W., Salas-Lizana R., Dominguez L.. Fungal catalases: function, phylogenetic origin and structure. Arch. Biochem. Biophys. 2012; 525:170–180. [DOI] [PubMed] [Google Scholar]
- 37. Chary P., Natvig D.O.. Evidence for three differentially regulated catalase genes in Neurospora crassa: effects of oxidative stress, heat shock, and development. J. Bacteriol. 1989; 171:2646–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peraza L., Hansberg W.. Neurospora crassa catalases, singlet oxygen and cell differentiation. Biol. Chem. 2002; 383:569–575. [DOI] [PubMed] [Google Scholar]
- 39. Michan S., Lledias F., Hansberg W.. Asexual development is increased in Neurospora crassa cat-3-null mutant strains. Eukaryot. Cell. 2003; 2:798–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Michan S., Lledias F., Baldwin J.D., Natvig D.O., Hansberg W.. Regulation and oxidation of two large monofunctional catalases. Free Radic. Biol. Med. 2002; 33:521–532. [DOI] [PubMed] [Google Scholar]
- 41. Qi S., He L., Zhang Q., Dong Q., Wang Y., Yang Q., Tian C., He Q., Wang Y.. Cross-pathway control gene CPC1/GCN4 coordinates with histone acetyltransferase GCN5 to regulate catalase-3 expression under oxidative stress in Neurospora crassa. Free Radic. Biol. Med. 2018; 117:218–227. [DOI] [PubMed] [Google Scholar]
- 42. Dong Q., Wang Y., Qi S., Gai K., He Q., Wang Y.. Histone variant H2A.Z antagonizes the positive effect of the transcriptional activator CPC1 to regulate catalase-3 expression under normal and oxidative stress conditions. Free Radic. Biol. Med. 2018; 121:136–148. [DOI] [PubMed] [Google Scholar]
- 43. Belden W.J., Larrondo L.F., Froehlich A.C., Shi M., Chen C.H., Loros J.J., Dunlap J.C.. The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev. 2007; 21:1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. He Q., Cha J., He Q., Lee H.C., Yang Y., Liu Y.. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 2006; 20:2552–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y., Dong Q., Ding Z., Gai K., Han X., Kaleri F.N., He Q., Wang Y.. Regulation of Neurospora Catalase-3 by global heterochromatin formation and its proximal heterochromatin region. Free Radic. Biol. Med. 2016; 99:139–152. [DOI] [PubMed] [Google Scholar]
- 46. Gai K., Cao X., Dong Q., Ding Z., Wei Y., Liu Y., Liu X., He Q.. Transcriptional repression of frequency by the IEC-1-INO80 complex is required for normal Neurospora circadian clock function. PLoS Genet. 2017; 13:e1006732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao Y., Shen Y., Yang S., Wang J., Hu Q., Wang Y., He Q.. Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J. Biol. Chem. 2010; 285:4355–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 49. Yang S., Li W., Qi S., Gai K., Chen Y., Suo J., Cao Y., He Y., Wang Y., He Q.. The highly expressed methionine synthase gene of Neurospora crassa is positively regulated by its proximal heterochromatic region. Nucleic Acids Res. 2014; 42:6183–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang X., Dong K., Xu K., Zhang K., Jin X., Yang M., Zhang Y., Wang X., Han C., Yu J. et al.. Barley stripe mosaic virus infection requires PKA-mediated phosphorylation of gammab for suppression of both RNA silencing and the host cell death response. New Phytol. 2018; 218:1570–1585. [DOI] [PubMed] [Google Scholar]
- 51. Yang M., Zhang Y., Xie X., Yue N., Li J., Wang X.B., Han C., Yu J., Liu Y., Li D.. Barley stripe mosaic virus gammab protein subverts autophagy to promote viral infection by disrupting the ATG7-ATG8 interaction. Plant Cell. 2018; 30:1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Poli J., Gasser S.M., Papamichos-Chronakis M.. The INO80 remodeller in transcription, replication and repair. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017; 372:20160290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang H., Roberts D.N., Cairns B.R.. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005; 123:219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Papamichos-Chronakis M., Watanabe S., Rando O.J., Peterson C.L.. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011; 144:200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Watanabe S., Radman-Livaja M., Rando O.J., Peterson C.L.. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science. 2013; 340:195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shen X., Mizuguchi G., Hamiche A., Wu C.. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000; 406:541–544. [DOI] [PubMed] [Google Scholar]
- 57. Papamichos-Chronakis M., Peterson C.L.. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat. Struct. Mol. Biol. 2008; 15:338–345. [DOI] [PubMed] [Google Scholar]
- 58. Mao Z., Pan L., Wang W., Sun J., Shan S., Dong Q., Liang X., Dai L., Ding X., Chen S. et al.. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res. 2014; 24:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Obri A., Ouararhni K., Papin C., Diebold M.L., Padmanabhan K., Marek M., Stoll I., Roy L., Reilly P.T., Mak T.W. et al.. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature. 2014; 505:648–653. [DOI] [PubMed] [Google Scholar]
- 60. Cai Y., Jin J., Yao T., Gottschalk A.J., Swanson S.K., Wu S., Shi Y., Washburn M.P., Florens L., Conaway R.C. et al.. YY1 functions with INO80 to activate transcription. Nat. Struct. Mol. Biol. 2007; 14:872–874. [DOI] [PubMed] [Google Scholar]
- 61. Ford J., Odeyale O., Shen C.H.. Activator-dependent recruitment of SWI/SNF and INO80 during INO1 activation. Biochem. Biophys. Res. Commun. 2008; 373:602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wimalarathna R.N., Pan P.Y., Shen C.H.. Co-dependent recruitment of Ino80p and Snf2p is required for yeast CUP1 activation. Biochem. Cell Biol. 2014; 92:69–75. [DOI] [PubMed] [Google Scholar]
- 63. Kim S., Cabane K., Hampsey M., Reinberg D.. Genetic analysis of the YDR1-BUR6 repressor complex reveals an intricate balance among transcriptional regulatory proteins in yeast. Mol. Cell. Biol. 2000; 20:2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yeung K., Kim S., Reinberg D.. Functional dissection of a human Dr1-DRAP1 repressor complex. Mol. Cell. Biol. 1997; 17:36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gilfillan S., Stelzer G., Piaia E., Hofmann M.G., Meisterernst M.. Efficient binding of NC2.TATA-binding protein to DNA in the absence of TATA. J. Biol. Chem. 2005; 280:6222–6230. [DOI] [PubMed] [Google Scholar]
- 66. Tramantano M., Sun L., Au C., Labuz D., Liu Z., Chou M., Shen C., Luk E.. Constitutive turnover of histone H2A.Z at yeast promoters requires the preinitiation complex. Elife. 2016; 5:e14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Duan J., Liu Q., Su S., Cha J., Zhou Y., Tang R., Liu X., Wang Y., Liu Y., He Q.. The Neurospora RNA polymerase II kinase CTK negatively regulates catalase expression in a chromatin context-dependent manner. Environ. Microbiol. 2020; 22:76–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li Z., Gadue P., Chen K., Jiao Y., Tuteja G., Schug J., Li W., Kaestner K.H.. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell. 2012; 151:1608–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C.. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004; 303:343–348. [DOI] [PubMed] [Google Scholar]
- 70. Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J.. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004; 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Adam M., Robert F., Larochelle M., Gaudreau L.. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 2001; 21:6270–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Santisteban M.S., Kalashnikova T., Smith M.M.. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000; 103:411–422. [DOI] [PubMed] [Google Scholar]
- 73. Brahma S., Udugama M.I., Kim J., Hada A., Bhardwaj S.K., Hailu S.G., Lee T.H., Bartholomew B.. INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat. Commun. 2017; 8:15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lademann C.A., Renkawitz J., Pfander B., Jentsch S.. The INO80 complex removes H2A.Z to promote presynaptic filament formation during homologous recombination. Cell Rep. 2017; 19:1294–1303. [DOI] [PubMed] [Google Scholar]
- 75. Jeronimo C., Watanabe S., Kaplan C.D., Peterson C.L., Robert F.. The histone chaperones FACT and Spt6 Restrict H2A.Z from intragenic locations. Mol. Cell. 2015; 58:1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kukimoto I., Elderkin S., Grimaldi M., Oelgeschlager T., Varga-Weisz P.D.. The histone-fold protein complex CHRAC-15/17 enhances nucleosome sliding and assembly mediated by ACF. Mol. Cell. 2004; 13:265–277. [DOI] [PubMed] [Google Scholar]
- 77. Xue Y., Pradhan S.K., Sun F., Chronis C., Tran N., Su T., Van C., Vashisht A., Wohlschlegel J., Peterson C.L. et al.. Mot1, Ino80C, and NC2 function coordinately to regulate pervasive transcription in yeast and mammals. Mol. Cell. 2017; 67:594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.