Significance
In the present study we investigated the epigenetic pattern of genes involved in the regulation of glucocorticoid receptor signaling in two African populations of heavily traumatized individuals. The strongest link between regional methylation and posttraumatic stress disorder (PTSD) risk and symptoms was observed for NTRK2, which has been shown to play an important role in memory formation. NTRK2 methylation was not related to trauma load, suggesting that methylation differences preexisted the trauma. Furthermore, NTRK2 methylation was found to be related to memory and memory-related brain activity in healthy nontraumatized individuals. The present findings suggest that epigenetic modifications of NTRK2 are involved in memory modulation in health, and in influencing risk and symptoms of PTSD in case of traumatic experiences.
Keywords: PTSD, glucocorticoids, memory, epigenetics, NTRK2
Abstract
Extensive pharmacologic, genetic, and epigenetic research has linked the glucocorticoid receptor (GR) to memory processes, and to risk and symptoms of posttraumatic stress disorder (PTSD). In the present study we investigated the epigenetic pattern of 12 genes involved in the regulation of GR signaling in two African populations of heavily traumatized individuals: Survivors of the rebel war in northern Uganda (n = 463) and survivors of the Rwandan genocide (n = 350). The strongest link between regional methylation and PTSD risk and symptoms was observed for NTRK2, which encodes the transmembrane receptor tropomyosin-related kinase B, binds the brain-derived neurotrophic factor, and has been shown to play an important role in memory formation. NTRK2 methylation was not related to trauma load, suggesting that methylation differences preexisted the trauma. Because NTRK2 methylation differences were predominantly associated with memory-related PTSD symptoms, and because they seem to precede traumatic events, we next investigated the relationship between NTRK2 methylation and memory in a sample of nontraumatized individuals (n = 568). We found that NTRK2 methylation was negatively associated with recognition memory performance. Furthermore, fMRI analyses revealed NTRK2 methylation-dependent differences in brain network activity related to recognition memory. The present study demonstrates that NTRK2 is epigenetically linked to memory functions in nontraumatized subjects and to PTSD risk and symptoms in traumatized populations.
Posttraumatic stress disorder (PTSD) is a chronic pathological response to a traumatic event and characterized by reexperiencing of the traumatic event, avoidance of stimuli associated with the trauma, negative alterations in cognition and mood, and hyperarousal (1). Reexperiencing symptoms include intrusive daytime recollections, traumatic nightmares, and flashbacks in which components of the event are relived, even years after the events took place (2). The formation of an aversive memory trace after a traumatic experience is an important pathogenic mechanism for the development of PTSD (3–7). Consequently, factors that influence the formation and reactivation of the aversive memory trace might also influence risk and symptoms of PTSD.
Stress activates the hypothalamus–pituitary–adrenal (HPA) axis, which results in the release of glucocorticoid hormones from the adrenal cortex. It has long been recognized that glucocorticoids readily enter the brain and affect memory processes (8, 9). Studies investigating glucocorticoid signaling effects on distinct memory phases and studies discerning acute from chronic effects helped to disentangle the multifaceted actions of these stress hormones. For example, acute elevations of glucocorticoids at levels known to enhance the consolidation of memory of emotionally arousing information have been shown to impair the retrieval of already stored emotionally arousing information (10–14). Furthermore, growing evidence indicates that these acute glucocorticoid effects depend on emotional arousal-induced activation of noradrenergic transmission within the amygdala and on interactions of the amygdala with other brain regions (8, 15–19). The enhancement of memory for emotionally arousing events in most cases has an obvious adaptive value (20). However, in cases of extremely aversive experiences, this mechanism can lead to overly strong traumatic memories, which contribute to the development and symptoms of PTSD (3–7, 16).
When looking at changes in the glucocorticoid system in PTSD, it is important to note that this disorder is not characterized by persistently increased glucocorticoid levels (21), as in the case of chronic stress, but rather by enhanced HPA axis feedback (2, 22). Moreover, there is evidence suggesting that enhanced HPA axis feedback might represent a pretrauma risk factor for the disorder (23–25).
Interindividual differences in glucocorticoid receptor (GR) signaling might be partially driven by underlying epigenetic mechanisms. DNA hypermethylation of the GR gene (NR3C1) promoter in rodents (NR3C1-17) and humans (NR3C1-1F) (26) is associated with low maternal care, maternal depression, and perinatal stress (27, 28). Previously, we investigated whether epigenetic differences in the human NR3C1 promoter are related to aversive memory and the risk for PTSD. We found that increased DNA methylation at the NGFI-A binding site of the NR3C1 promoter was associated with less intrusive memory of the traumatic event and reduced PTSD risk in survivors of the Rwandan genocide (29). This association was independent of the trauma load, suggesting that the epigenetic changes preexisted the trauma. In healthy individuals, NR3C1 methylation was related to reduced picture recognition and differences in recognition memory-related brain activity (29).
In the present study, we further examined the association between epigenetic modification of glucocorticoid signaling and PTSD by expanding our analysis to the genes involved in the regulation of GR signaling (RGRS) pathway in two independent samples of severely traumatized individuals. In a second-level analysis, after accounting for multiple comparisons within the RGRS pathway, we identified an epigenetic locus in neurotrophic tyrosine kinase receptor type 2 (NTRK2) to be significantly associated with traumatic memory, avoidance, and PTSD risk. Finally, to investigate if NTRK2 methylation might be related to memory processes prior to traumatic events, we investigated the association of this epigenetic locus with recognition memory performance and recognition memory-related brain activation in healthy, trauma-unexposed subjects.
Materials and Methods
African Samples, Conflict Survivors.
PTSD risk and symptomatology were assessed in two independent African samples. In the African sample 1, we included n = 463 survivors of the rebel war in northern Uganda (mean age 29 y, 18 to 55 y; 44.1% females) (Table 1). For the African sample 2, we investigated n = 350 survivors from the 1994 Rwandan genocide (mean age 34.8 y, 18 to 68 y; 49.1% females) (Table 1). This sample represents an extended cohort of individuals, previously investigated in a study linking epigenetic modifications of the NR3C1 gene with PTSD and traumatic memories (29).
Table 1.
PDS scores and percentage of lifetime PTSD diagnosis
| Avoidance | Intrusions | Hyperarousal | Sum | PTSD, % | Traumatic event types | |
| African sample 1 (n = 463) | 3.4 (0–7) | 3.5 (0–5) | 3.1 (0–5) | 10 (0–17) | 68.6 | 25.7 (3–59) |
| African sample 2 (n = 350) | 4.4 (0–7) | 4.2 (0–5) | 3.5 (0–5) | 11.7 (0–17) | 78.8 | 11.9 (0–24) |
A summary of PDS lifetime scores (mean, with range in the parentheses) and lifetime PTSD risk (percentage) according to DSM-IV criteria in survivors of the rebel war in Northern Uganda (African sample 1) and survivors of the 1994 Rwandan genocide (African sample 2). Please note that the calculation of traumatic event types was based on a checklist of 62 items in the African sample 1 and on a checklist of 36 items in the African sample 2.
All subjects had experienced traumatic situations and were examined according to Diagnostical and Statistical Manual of Mental Disorders IV (DSM-IV) criteria (1). Traumatic load was estimated by assessing the number of different traumatic event types experienced or witnessed (30). Taking into account known ceiling effects of trauma load on PTSD risk, individuals with extreme levels of trauma exposure were excluded for the current analyses (31).
Saliva samples were collected at the time-point of the main investigation for the DNA isolation. Study procedures were approved by the ethics committees of the University of Konstanz, Germany; the Mbarara University of Science and Technology, Mbarara, Uganda; the Gulu University, Uganda; Lacor Hospital, Gulu, Uganda; and the Ugandan National Council for Science and Technology, Uganda. Before the interview, all participants provided informed consent. For details see SI Appendix.
Swiss Sample, Healthy Young Adults.
Memory was assessed in a sample of healthy young adults from Basel, Switzerland (Swiss sample: n = 568; mean age 23.8 y, 18.3 to 36.8 y; 59% females). Subjects performed different consecutive tasks as described in detail previously (32, 33). For the purpose of the present study, we focused on a picture-based episodic memory recognition task that was tested 60 to 80 min after encoding.
For DNA isolation, subjects were reinvited for an additional blood sampling, which took place on average 360 d (median 341 d) after the main investigation. The investigation was carried out in accordance with the latest version of the Declaration of Helsinki. The ethics committee of the Cantons of Basel-Stadt– and Basel-Landschaft approved the experiments. Before the interview, all participants provided informed consent. For details, see SI Appendix.
fMRI Data Analyses.
Subject-level contrast estimates for correct recognition (correctly remembered pictures – pictures correctly identified as new) were used for association analyses with memory performance and DNA methylation (n = 498 subjects with complete dataset). More precisely, the dimension of the fMRI contrasts (participants × voxels) was first reduced using independent component analysis (ICA). ICA was used as an unbiased, data-driven method to reduce the dimensionality of those data to a lower number of statistically independent components (ICs). These components represent statistically independent latent sources that underlie the contrast estimates (voxel loadings). The decomposition also provides an estimation of each component’s activity strength per participant (participant scores). The subjects included in this analysis were part of a larger imaging genetics cohort of 1,576 subjects (34), providing a more robust estimation of the different components. Following a stability analysis, 13 ICs were identified and considered for further analyses. The IC’s participants scores relationship with correct recognition memory performance and NTRK2 methylation were examined by means of linear models. Correction for multiple comparisons across the 13 ICs and 2 independent variables was applied using the Bonferroni procedure, with a corrected threshold of PBonf = 0.0019. Age, sex, and two MR-related technical batches were included as covariates in all models. For details, see SI Appendix.
DNA Isolation from Human Samples.
Saliva samples were collected using an Oragene DNA Kit (DNA Genotek) and DNA was extracted using the precipitation protocol recommended by the manufacturer and then repurified.
Blood samples were collected using the 10.0-mL BD Vacutainer Plus plastic whole-blood tube, BD Hemogard closure with spray-coated K2EDTA (Becton Dickinson). DNA was isolated with QIAmp Blood Maxi Kit (Qiagen), using the recommended spin protocol (SI Appendix).
Infinium 450K and EPIC BeadChip Methylation Analyses.
DNA isolated from saliva (African samples) or peripheral blood (Swiss sample) was investigated with the 450K (African sample 2, Swiss sample) or EPIC (African sample 1) array (Illumina) (SI Appendix).
For preprocessing, data were extracted and analyzed using the R package RnBeads v0.99.9 (35). The background was subtracted using the “noob” method in the methylumi package (36), and the signal was further normalized using the SWAN algorithm (37). Postprocessing was further done for each of the three samples separately, combining the B values of the preprocessed data of all batches per sample (SI Appendix).
Finally, we used the genome-wide regional segmentation analysis and clustered the individual CpGs to regional elements, as previously described (35, 38). In short, the genome (GRCh37/hg19) was segmented in 5-kb sliding-window regional elements. Regional methylation was then calculated as mean value of DNA-methylation for CpGs clustered to every element, across all 12 genes represented in the RGRS (Gene ontology [GO]:2000322) (SI Appendix). For the extensive list of the regional methylation elements annotated to the RGRS pathway genes and examined in this study, please refer to SI Appendix, Table S1.
Statistical Analyses.
The association of the epigenetic regulation of the RGRS pathway with PTSD was assessed by first modeling PTSD lifetime risk against every regional methylation element annotated to the RGRS gene set (SI Appendix, Table S1), by logistic regression. To account for trauma load as a principal factor in the development of PTSD (31), sum of lifetime traumatic event types was used as a covariate. The relationship of DNA methylation at the NTRK2 regional element (chr9:87285001–87290000) with Posttraumatic Diagnostic Scale (PDS) sum- and subscores was assessed with linear regression using NTRK2 enhancer element methylation as a quantitative predictor and sum of life traumatic event types as a covariate.
In the Swiss sample, recognition performance as dependent variable was modeled against the DNA methylation of predefined NTRK2 regional element (chr9:87285001–87290000). The interaction of NTRK2 DNA methylation with the valence of pictures used in the emotional picture-encoding task was additionally tested in a linear mixed model.
Wherever appropriate, Bonferroni correction was implemented to account for multiple testing procedures. The significance threshold was set to P = 0.05. Statistical analyses were done in R (R v3.6.0; R Development Core Team 2017), using the cpg.assoc (39) and nlme (40) R packages.
All laboratory procedures were conducted in a blind, randomized order. For details, see SI Appendix.
Results
To investigate the epigenetic role of GR signaling in PTSD, we first modeled PTSD lifetime risk against DNA methylation at every regional element annotated to the predefined GO pathway “regulation of glucocorticoid receptor signaling” (GO:2000322). In the discovery cohort (African sample 1) results revealed that regional element methylation was nominally associated with PTSD risk at 4 of 12 genes within the RGRS pathway: NTRK2, CLOCK, NR3C1, and NCOA2 (Ps < 0.05) (SI Appendix, Table S1). After applying Bonferroni correction for all examined regional methylation elements (for 175 regional methylation elements annotated to the RGRS pathway) (SI Appendix, Table S1), the gene encoding NTRK2 (NTRK2; formerly known as TrkB-tyrosine receptor kinase B) remained significantly negatively associated with PTSD risk (Pnominal = 0.00019, PBonferroni < 0.05) (Table 2). The same epigenetic mark was also negatively associated with lifetime PTSD risk in the African sample 2 (Pnominal = 0.0023) (Table 2). The specific regional methylation element is located in the 5′ enhancer region of NTRK2 (SI Appendix, Table S2).
Table 2.
NTRK2 regional DNA methylation element (chr9:87285001–87290000) associations with lifetime PTSD risk and symptoms
| Sample | Phenotype | t-Statistic | P value | Effect size f2 |
| African sample 1 (n = 463) | PTSD risk | −3.76 | <0.001 | 0.17 (0.10, 0.25) |
| PDS Sum | −3.48 | <0.001 | 0.16 (0.08, 0.24) | |
| Avoidance | −3.50 | < 0.001 | 0.17 (0.09, 0.24) | |
| NTRK2 methylation (0.50 ± 0.08)* | Intrusions | −3.28 | <0.001 | 0.15 (0.07, 0.23) |
| Hyperarousal | −1.45 | n.s. | — | |
| African sample 2 (n = 350) | PTSD risk | −3.07 | 0.002 | 0.18 (0.08, 0.27) |
| PDS Sum | −3.04 | 0.003 | 0.16 (0.07, 0.25) | |
| Avoidance | −3.38 | <0.001 | 0.21 (0.11, 0.31) | |
| NTRK2 methylation (0.50 ± 0.11)* | Intrusions | −2.19 | 0.029 | 0.13 (0.03, 0.24) |
| Hyperarousal | −1.80 | n.s. | — |
Fixed-effects analysis t-statistics and nominal P values are shown. Sum of life traumatic event types was used as a covariate in the multiple regression model. Effect sizes of NTRK2 regional DNA methylation are provided as Cohen’s f2 (90% CI). n.s., not significant.
(Mean ± SD).
The NTRK2 regional DNA methylation association with PTSD risk remained significant also after correcting for comorbidity of depression, as assessed by the Hopkins Symptom Checklist (HSCL-D) [z-value(459) = −3.09, P = 0.002 for African sample 1; z-value(291) = −3.14, P = 0.001 for African sample 2]. Furthermore, the association of NTRK2 with PTSD risk remained significant upon correcting for chronic somatic comorbidities (e.g., cancer, inflammatory disorders, or chronic pain), which have been assessed in the African sample 1 [z-value(458) = −3.95, P = 7.8e-05]. The data available in the current study, didn’t allow us to exclude potential further confounders (e.g., somatic medication, environmental hazards, or nutrition), which might have affected methylation of genes of RGRS pathway.
We further focused on the NTRK2 regional element and examined the associations between DNA methylation of this specific region and PDS sum and subscores. NTRK2 regional element DNA methylation was significantly negatively associated with PDS sum scores, and specifically with intrusion and avoidance symptoms in both African samples (Table 2). Hyperarousal was not significantly associated with NTRK2 methylation in either sample (Table 2). NTKR2 regional methylation was not related to the sum of traumatic life event types (African sample 1: P > 0.2, f2 = 0.06, 95% CI [0.00 to 0.15]; African sample 2: P > 0.5, f2 = 0.01, 95% CI [0.00, 0.05]), suggesting that traumatic life events did not induce methylation changes, but rather that the methylation differences preceded the traumatic events.
Since NTRK2 regional element DNA methylation differences were predominantly associated with memory-related PTSD symptoms (i.e., intrusions and avoidance), and the methylation differences seem to precede traumatic events, we next investigated the relationship between NTRK2 regional element methylation and memory in a sample of healthy, trauma-unexposed Swiss population. The interaction with valence was not significant (P > 0.05, mixed model), therefore we further examined the effects of NTRK2 methylation on recognition in a simple linear model. The analysis revealed a significant negative association of the NTRK2 regional element methylation with correct recognition memory performance, t(530) = −2.04, P = 0.042. Thus, individuals with higher methylation levels had poorer recognition memory performance. NTRK2 methylation was neither significantly associated with valence or arousal ratings in the picture task, nor with mood or trait anxiety (SI Appendix, Table S3).
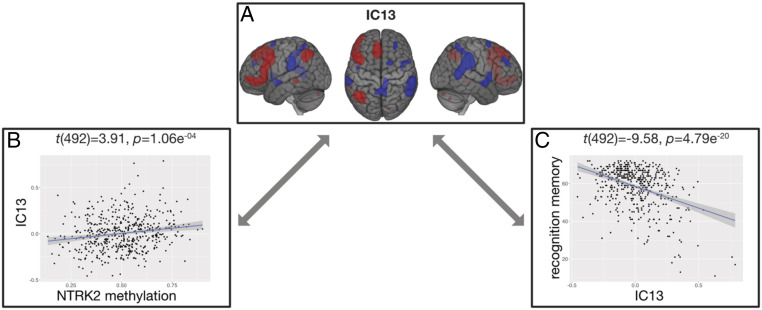
As we found a significant association of NTRK2 methylation with recognition memory performance, we next used functional MRI to capture brain activation patterns related to this cognitive task (41). ICA was applied to decompose brain activation into coactivation networks across participants (SI Appendix). Thirteen components were identified (SI Appendix, Fig. S1). After correction for multiple comparisons, 4 of the 13 ICs were significantly associated with memory performance (SI Appendix, Table S4). Correlation analyses revealed that one IC (IC13) was significantly positively associated with NTRK2 regional element methylation, t(492) = 3.91, P = 1.06e-04, (Fig. 1B). This component was negatively associated with correct recognition performance, t(492) = −9.58, P = 4.79e-20 (Fig. 1C). The main voxel loadings of this component included left frontal, left parietal, left temporal, and right cerebellar regions (positive loadings), as well as bilateral supramarginal, right frontal, right precuneus, and insula (negative loadings) (Fig. 1A and SI Appendix, Table S5).
Fig. 1.
Brain activation, recognition performance, and NTRK2 methylation in healthy subjects. (A) IC13, voxel loadings. Voxels with positive loadings (colored in red) corresponded to voxels that were more activated during correct recognition, whereas voxels with negative loadings (colored in blue) corresponded to voxels that were less activated during correct recognition. (B) Association between NTRK2 enhancer element DNA methylation and IC13 participants’ score. (C) Association between correct recognition performance and IC13 participants’ score. The dots displayed in B and C represent individual participants.
The amount of NTRK2 regional element methylation ranged from 0.3 to 0.8, with an average of 0.5 (β-value). Importantly, there were no significant differences in methylation distribution, variance, or median between the African cohorts and the Swiss sample used in this study (all P > 0.05).
Discussion
The present study investigated the methylation patterns of the RGRS pathway in two independent African samples of conflict survivors. Several members of this pathway were found to be epigenetically associated with PTSD. Previously, we and others reported that methylation of NR3C1 is linked to PTSD (29, 42, 43). The present study corroborates those findings by showing that NR3C1 was one of differentially methylated RGRS pathway member genes in an additional, independent cohort of severely traumatized individuals from Uganda.
Within the RGRS pathway, NTRK2 was the gene with the most significant association of methylation differences with PTSD risk and symptoms. Specifically, NTRK2 methylation was negatively associated with intrusions and avoidance symptoms and with lifetime PTSD risk. Importantly, methylation of the NTRK2 regional element was independent of the trauma load, suggesting that methylation differences preexisted traumatic events. This idea is further supported by the finding that NTRK2 regional DNA methylation patterns in traumatized and healthy individuals were comparable. Indeed, epigenetic modifications can be set during a sensitive period around birth (44). In both rodents (45–47) and humans (27, 29, 48, 49), NR3C1 methylation is typically set perinatally.
In the African samples, NTRK2 methylation was related to traumatic memory-related symptoms (i.e., intrusions and avoidance) but not to hyperarousal. This pattern suggests that methylation might be related to PTSD through a regulation of memory processes. This idea is supported by the finding that NTRK2 methylation was associated with recognition memory performance in the healthy, nontraumatized Swiss sample. Moreover, the fMRI analysis in this sample revealed a relation of NTRK2 methylation with activation in brain regions previously shown to be involved in the regulation of recognition memory (50–52). Whereas glucocorticoid effects on memory consolidation processes have been predominantly found for emotionally arousing information (8), the methylation effects in the present study were independent of picture valence. However, it is possible that in the present study there was a heightened emotional arousal state even during the presentation of neutral pictures, as the memory task took place in an MRI scanner, a procedure that might induce arousal.
Furthermore, with the recognition test that took place 60 to 80 min after encoding, we were only able to assess epigenetic effects on molecular events during the early (but not late) phase of memory consolidation. Nevertheless, some of these early events, have been shown to be critical for the stabilization into long-term memory (53) and some depend on protein synthesis (54, 55). Furthermore, epigenetic changes may have affected recognition performance indirectly via influencing the storage of preexisting associative networks, given the known influence of such networks on the encoding of new information (56–58).
The observed DNA methylation differences in this study were associated with the 5′ enhancer region of the NTRK2 gene. A previous study has identified a regulatory element within that region that is required for cyclic AMP-responsive element binding (CREB)-dependent control of NTRK2 expression in neurons (59). Importantly, a genome-wide study on CREB occupancy suggests that CREB binding in the genome is restricted by means of a DNA methylation-dependent mechanism (60), resulting in dampened transcriptional activity from the affected locus. Indeed, DNA methylation of the 5′ regulatory region seems to be strongly negatively correlated with the expression of the NTRK2 gene across The Cancer Genome Atlas datasets (TGCA) (SI Appendix, Table S6) (61).
NTRK2 encodes the transmembrane receptor tropomyosin-related kinase B (TrkB) (62, 63). This receptor binds brain-derived neurotrophic factor (BDNF) as well as other neurotrophic factors (64, 65). Among its involvements in promoting neuronal survival and development, BDNF signaling has been shown to play an important role in memory consolidation through long-term potentiation induction (66–69). Importantly, BDNF-TrkB signaling through Erk1/2MAPK phosphorylation mediates the enhancement of fear memory induced by glucocorticoids (70). Down-regulation of TrkB signaling has been reported in neurodegenerative diseases and psychiatric disorders (71–73). Human genetics studies have also implicated NTRK2 with a variety of psychiatric conditions, such as suicide attempts in depressed patients (74), obsessive–compulsive disorder (75), anxiety (76), and mood disorder (77). However, to our knowledge there are no studies on epigenetic modifications of NTRK2 with regard to PTSD or learning and memory. In the present study we found that increased methylation of NTRK2 was associated with reduced memory in nontraumatized individuals. Thus, if a traumatic event is experienced by an individual with a high level of NTRK2 methylation, it is possible they may form a less traumatic memory and therefore have a reduced risk of developing PTSD.
In the present study, DNA methylation was assessed peripherally in saliva from the two African cohorts and in blood from the Swiss cohort. Several studies have demonstrated the use of peripheral blood (43, 78–81) or saliva (29) as a source of DNA to get insights to the phenotypes intrinsically linked to the CNS. Additionally, studies that have addressed brain to periphery variation of DNA methylation suggest that a part of the methylome signatures is significantly shared across the tissues (82, 83), especially in the genomic regions that contain a high density of cytosine guanine dinucleotides, as seen in the specific region of NTRK2 (84–88).
The present study may also have clinical implications, as the TrkB receptor is a druggable target. We found that DNA methylation at the NTRK2 regional element is negatively associated with memory, and with risk and symptoms of PTSD. Therefore, TrkB antagonists may be helpful in reducing initial consolidation or reconsolidation of aversive memories. Interestingly, a potent TrkB antagonist was found to have anxiolytic properties in animal studies (89, 90).
In conclusion, this study demonstrates that epigenetic differences in NTRK2 are related to memory functions in healthy nontraumatized subjects and to traumatic memory and PTSD risk in traumatized individuals. These results might contribute to a better understanding of the pathogenesis of PTSD, ultimately paving the way for the discovery of novel biomarkers and drug targets.
Supplementary Material
Acknowledgments
This work was supported by Swiss National Science Foundation Grant 159740 (to D.J.-F.d.Q.) and by German Research Foundation grants (to I.-T.K. and T.E.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008415117/-/DCSupplemental.
Data Availability.
Deidentified data available on request from the authors: The data that support the findings of this study are available from the corresponding authors, upon reasonable request.
References
- 1.American Psychiatric Association , Diagnostical and Statistical Manual of Mental Disorders, (DSM-IV-TR), (American Psychiatric Association, Washington, DC, ed. 4, 2000). [Google Scholar]
- 2.Yehuda R., Post-traumatic stress disorder. N. Engl. J. Med. 346, 108–114 (2002). [DOI] [PubMed] [Google Scholar]
- 3.LeDoux J., The emotional brain, fear, and the amygdala. Cell. Mol. Neurobiol. 23, 727–738 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mineka S., Oehlberg K., The relevance of recent developments in classical conditioning to understanding the etiology and maintenance of anxiety disorders. Acta Psychol. 127, 567–580 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Phelps E. A., LeDoux J. E., Contributions of the amygdala to emotion processing: From animal models to human behavior. Neuron 48, 175–187 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Pitman R. K., Post-traumatic stress disorder, hormones, and memory. Biol. Psychiatry 26, 221–223 (1989). [DOI] [PubMed] [Google Scholar]
- 7.Yehuda R., Flory J. D., Differentiating biological correlates of risk, PTSD, and resilience following trauma exposure. J. Trauma. Stress 20, 435–447 (2007). [DOI] [PubMed] [Google Scholar]
- 8.de Quervain D., Schwabe L., Roozendaal B., Stress, glucocorticoids and memory: Implications for treating fear-related disorders. Nat. Rev. Neurosci. 18, 7–19 (2017). [DOI] [PubMed] [Google Scholar]
- 9.de Kloet E. R., Joëls M., Holsboer F., Stress and the brain: From adaptation to disease. Nat. Rev. Neurosci. 6, 463–475 (2005). [DOI] [PubMed] [Google Scholar]
- 10.de Quervain D. J., Roozendaal B., McGaugh J. L., Stress and glucocorticoids impair retrieval of long-term spatial memory. Nature 394, 787–790 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Roozendaal B., Carmi O., McGaugh J. L., Adrenocortical suppression blocks the memory-enhancing effects of amphetamine and epinephrine. Proc. Natl. Acad. Sci. U.S.A. 93, 1429–1433 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Quervain D. J., Roozendaal B., Nitsch R. M., McGaugh J. L., Hock C., Acute cortisone administration impairs retrieval of long-term declarative memory in humans. Nat. Neurosci. 3, 313–314 (2000). [DOI] [PubMed] [Google Scholar]
- 13.McGaugh J. L., Memory—A century of consolidation. Science 287, 248–251 (2000). [DOI] [PubMed] [Google Scholar]
- 14.McGaugh J. L., Consolidating memories. Annu. Rev. Psychol. 66, 1–24 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Roozendaal B., McGaugh J. L., Memory modulation. Behav. Neurosci. 125, 797–824 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Quervain D. J.-F., Aerni A., Schelling G., Roozendaal B., Glucocorticoids and the regulation of memory in health and disease. Front. Neuroendocrinol. 30, 358–370 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Roozendaal B., Barsegyan A., Lee S., Adrenal stress hormones, amygdala activation, and memory for emotionally arousing experiences. Prog. Brain Res. 167, 79–97 (2008). [DOI] [PubMed] [Google Scholar]
- 18.McGaugh J. L., Emotional arousal and enhanced amygdala activity: New evidence for the old perseveration-consolidation hypothesis. Learn. Mem. 12, 77–79 (2005). [DOI] [PubMed] [Google Scholar]
- 19.McGaugh J. L., The amygdala modulates the consolidation of memories of emotionally arousing experiences. Annu. Rev. Neurosci. 27, 1–28 (2004). [DOI] [PubMed] [Google Scholar]
- 20.McGaugh J. L., Making lasting memories: Remembering the significant. Proc. Natl. Acad. Sci. U.S.A. 110 (suppl. 2), 10402–10407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meewisse M.-L., Reitsma J. B., de Vries G.-J., Gersons B. P. R., Olff M., Cortisol and post-traumatic stress disorder in adults: Systematic review and meta-analysis. Br. J. Psychiatry 191, 387–392 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Pitman R. K. et al., Biological studies of post-traumatic stress disorder. Nat. Rev. Neurosci. 13, 769–787 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Zuiden M. et al., Pre-existing high glucocorticoid receptor number predicting development of posttraumatic stress symptoms after military deployment. Am. J. Psychiatry 168, 89–96 (2011). [DOI] [PubMed] [Google Scholar]
- 24.van Zuiden M., Kavelaars A., Geuze E., Olff M., Heijnen C. J., Predicting PTSD: Pre-existing vulnerabilities in glucocorticoid-signaling and implications for preventive interventions. Brain Behav. Immun. 30, 12–21 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Yehuda R. et al., Gene expression patterns associated with posttraumatic stress disorder following exposure to the World Trade Center attacks. Biol. Psychiatry 66, 708–711 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Turner J. D., Muller C. P., Structure of the glucocorticoid receptor (NR3C1) gene 5′ untranslated region: Identification, and tissue distribution of multiple new human exon 1. J. Mol. Endocrinol. 35, 283–292 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Oberlander T. F. et al., Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics 3, 97–106 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Weaver I. C. G. et al., The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: Altering epigenetic marks by immediate-early genes. J. Neurosci. 27, 1756–1768 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vukojevic V. et al., Epigenetic modification of the glucocorticoid receptor gene is linked to traumatic memory and post-traumatic stress disorder risk in genocide survivors. J. Neurosci. 34, 10274–10284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilker S. et al., How to quantify exposure to traumatic stress? Reliability and predictive validity of measures for cumulative trauma exposure in a post-conflict population. Eur. J. Psychotraumatol. 6, 28306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolassa I.-T. et al., Association study of trauma load and SLC6A4 promoter polymorphism in posttraumatic stress disorder: Evidence from survivors of the Rwandan genocide. J. Clin. Psychiatry 71, 543–547 (2010). [DOI] [PubMed] [Google Scholar]
- 32.de Quervain D. J.-F. et al., A deletion variant of the alpha2b-adrenoceptor is related to emotional memory in Europeans and Africans. Nat. Neurosci. 10, 1137–1139 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Heck A. et al., Eexome sequencing of healthy phenotypic extremes links TROVE2 to emotional memory and PTSD. Nat. hum. behav. 1, 0081 (2017). [Google Scholar]
- 34.Heck A. et al., Genetic analysis of association between calcium signaling and hippocampal activation, memory performance in the young and old, and risk for sporadic Alzheimer disease. JAMA Psychiatry 72, 1029–1036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Assenov Y. et al., Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods 11, 1138–1140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis S., Du P., Bilke S., Triche J. T., Bootwalla M., methylumi: Handle Illumina methylation data. R Package Version 2.28.0. https://www.bioconductor.org/packages/release/bioc/html/methylumi.html. Accessed 5 May 2013.
- 37.Maksimovic J., Gordon L., Oshlack A., SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 13, R44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller F. et al., RnBeads 2.0: Comprehensive analysis of DNA methylation data. Genome Biol. 20, 55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barfield R. T., Kilaru V., Smith A. K., Conneely K. N., CpGassoc: An R function for analysis of DNA methylation microarray data. Bioinformatics 28, 1280–1281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinheiro J., Bates D., S. DebRoy, D. Sarkar; R Core Team, nlme: Linear and nonlinear mixed effects models. https://CRAN.R-project.org/package=nlme. Accessed 25 July 2013.
- 41.Axmacher N., Elger C. E., Fell J., The specific contribution of neuroimaging versus neurophysiological data to understanding cognition. Behav. Brain Res. 200, 1–6 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Yehuda R. et al., Lower methylation of glucocorticoid receptor gene promoter 1F in peripheral blood of veterans with posttraumatic stress disorder. Biol. Psychiatry 77, 356–364 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Labonté B., Azoulay N., Yerko V., Turecki G., Brunet A., Epigenetic modulation of glucocorticoid receptors in posttraumatic stress disorder. Transl. Psychiatry 4, e368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiele M. A., Domschke K., Epigenetics at the crossroads between genes, environment and resilience in anxiety disorders. Genes Brain Behav. 17, e12423 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Weaver I. C. G. et al., Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Mueller B. R., Bale T. L., Sex-specific programming of offspring emotionality after stress early in pregnancy. J. Neurosci. 28, 9055–9065 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suderman M. et al., Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc. Natl. Acad. Sci. U.S.A. 109 (suppl. 2), 17266–17272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGowan P. O. et al., Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 12, 342–348 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Radtke K. M. et al., Transgenerational impact of intimate partner violence on methylation in the promoter of the glucocorticoid receptor. Transl. Psychiatry 1, e21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aggleton J. P., Brown M. W., Interleaving brain systems for episodic and recognition memory. Trends Cogn. Sci. 10, 455–463 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Cabeza R., Ciaramelli E., Olson I. R., Moscovitch M., The parietal cortex and episodic memory: An attentional account. Nat. Rev. Neurosci. 9, 613–625 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ranganath C., Ritchey M., Two cortical systems for memory-guided behaviour. Nat. Rev. Neurosci. 13, 713–726 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Halder R. et al., DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat. Neurosci. 19, 102–110 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Izquierdo I. et al., Different molecular cascades in different sites of the brain control memory consolidation. Trends Neurosci. 29, 496–505 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Squire L. R., Barondes S. H., Variable decay of memory and its recovery in cycloheximide-treated mice. Proc. Natl. Acad. Sci. U.S.A. 69, 1416–1420 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuart G., Hulme C., The effects of word co-occurrence on short-term memory: Associative links in long-term memory affect short-term memory performance. J. Exp. Psychol. Learn. Mem. Cogn. 26, 796–802 (2000). [DOI] [PubMed] [Google Scholar]
- 57.de Quervain D. J.-F., Papassotiropoulos A., Identification of a genetic cluster influencing memory performance and hippocampal activity in humans. Proc. Natl. Acad. Sci. U.S.A. 103, 4270–4274 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simons J. S., Spiers H. J., Prefrontal and medial temporal lobe interactions in long-term memory. Nat. Rev. Neurosci. 4, 637–648 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Kingsbury T. J., Krueger B. K., Ca2+, CREB and krüppel: A novel KLF7-binding element conserved in mouse and human TRKB promoters is required for CREB-dependent transcription. Mol. Cell. Neurosci. 35, 447–455 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X. et al., Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U.S.A. 102, 4459–4464 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding W. et al., DNMIVD: DNA methylation interactive visualization database. Nucleic Acids Res. 48, D856–D862 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakagawara A. et al., Cloning and chromosomal localization of the human TRK-B tyrosine kinase receptor gene (NTRK2). Genomics 25, 538–546 (1995). [DOI] [PubMed] [Google Scholar]
- 63.Valent A., Danglot G., Bernheim A., Mapping of the tyrosine kinase receptors trkA (NTRK1), trkB (NTRK2) and trkC(NTRK3) to human chromosomes 1q22, 9q22 and 15q25 by fluorescence in situ hybridization. Eur. J. Hum. Genet. 5, 102–104 (1997). [PubMed] [Google Scholar]
- 64.Barbacid M., The Trk family of neurotrophin receptors. J. Neurobiol. 25, 1386–1403 (1994). [DOI] [PubMed] [Google Scholar]
- 65.Klein R. et al., The trkB tyrosine protein kinase is a receptor for brain-derived neurotrophic factor and neurotrophin-3. Cell 66, 395–403 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reichardt L. F., Neurotrophin-regulated signalling pathways. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1545–1564 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skaper S. D., The biology of neurotrophins, signalling pathways, and functional peptide mimetics of neurotrophins and their receptors. CNS Neurol. Disord. Drug Targets 7, 46–62 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Minichiello L., TrkB signalling pathways in LTP and learning. Nat. Rev. Neurosci. 10, 850–860 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Gräff J., Tsai L.-H., Histone acetylation: Molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 14, 97–111 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Revest J.-M. et al., BDNF-TrkB signaling through Erk1/2 MAPK phosphorylation mediates the enhancement of fear memory induced by glucocorticoids. Mol. Psychiatry 19, 1001–1009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bernard R. et al., Altered expression of glutamate signaling, growth factor, and glia genes in the locus coeruleus of patients with major depression. Mol. Psychiatry 16, 634–646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pezet S., Malcangio M., Brain-derived neurotrophic factor as a drug target for CNS disorders. Expert Opin. Ther. Targets 8, 391–399 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Weickert C. S. et al., Reductions in neurotrophin receptor mRNAs in the prefrontal cortex of patients with schizophrenia. Mol. Psychiatry 10, 637–650 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Kohli M. A. et al., Association of genetic variants in the neurotrophic receptor-encoding gene NTRK2 and a lifetime history of suicide attempts in depressed patients. Arch. Gen. Psychiatry 67, 348–359 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alonso P. et al., Extensive genotyping of the BDNF and NTRK2 genes define protective haplotypes against obsessive-compulsive disorder. Biol. Psychiatry 63, 619–628 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Ernst C. et al., A deletion in tropomyosin-related kinase B and the development of human anxiety. Biol. Psychiatry 69, 604–607 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Deo A. J. et al., A large-scale candidate gene analysis of mood disorders: Evidence of neurotrophic tyrosine kinase receptor and opioid receptor signaling dysfunction. Psychiatr. Genet. 23, 47–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang S.-C. et al., Molecular variation at the SLC6A3 locus predicts lifetime risk of PTSD in the Detroit Neighborhood Health Study. PLoS One 7, e39184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klengel T. et al., Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 16, 33–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mehta D. et al., Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc. Natl. Acad. Sci. U.S.A. 110, 8302–8307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yehuda R. et al., Holocaust exposure induced intergenerational effects on FKBP5 methylation. Biol. Psychiatry 80, 372–380 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Hannon E., Lunnon K., Schalkwyk L., Mill J., Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics 10, 1024–1032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Braun P. R. et al., Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl. Psychiatry 9, 47-e1059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ladd-Acosta C. et al., DNA methylation signatures within the human brain. Am. J. Hum. Genet. 81, 1304–1315 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mill J. et al., Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 82, 696–711 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lister R. et al., Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dempster E. L. et al., Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum. Mol. Genet. 20, 4786–4796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davies M. N. et al., Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 13, R43 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cazorla M. et al., Identification of a low-molecular weight TrkB antagonist with anxiolytic and antidepressant activity in mice. J. Clin. Invest. 121, 1846–1857 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cazorla M. et al., Cyclotraxin-B, the first highly potent and selective TrkB inhibitor, has anxiolytic properties in mice. PLoS One 5, e9777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified data available on request from the authors: The data that support the findings of this study are available from the corresponding authors, upon reasonable request.