Abstract
Objective
Physical exercise training is associated with increased glucose uptake in skeletal muscle and improved glycemic control. HDAC5, a class IIa histone deacetylase, has been shown to regulate transcription of the insulin-responsive glucose transporter GLUT4 in cultured muscle cells. In this study, we analyzed the contribution of HDAC5 to the transcriptional network in muscle and the beneficial effect of muscle contraction and regular exercise on glucose metabolism.
Methods
HDAC5 knockout mice (KO) and wild-type (WT) littermates were trained for 8 weeks on treadmills, metabolically phenotyped, and compared to sedentary controls. Hdac5-deficient skeletal muscle and cultured Hdac5-knockdown (KD) C2C12 myotubes were utilized for studies of gene expression and glucose metabolism. Chromatin immunoprecipitation (ChIP) studies were conducted to analyze Il6 promoter activity using H3K9ac and HDAC5 antibodies.
Results
Global transcriptome analysis of Hdac5 KO gastrocnemius muscle demonstrated activation of the IL-6 signaling pathway. Accordingly, knockdown of Hdac5 in C2C12 myotubes led to higher expression and secretion of IL-6 with enhanced insulin-stimulated activation of AKT that was reversed by Il6 knockdown. Moreover, Hdac5-deficient myotubes exhibited enhanced glucose uptake, glycogen synthesis, and elevated expression levels of the glucose transporter GLUT4. Transcription of Il6 was further enhanced by electrical pulse stimulation in Hdac5-deficient C2C12 myotubes. ChIP identified a ∼1 kb fragment of the Il6 promoter that interacts with HDAC5 and demonstrated increased activation-associated histone marker AcH3K9 in Hdac5-deficient muscle cells. Exercise intervention of HDAC5 KO mice resulted in improved systemic glucose tolerance as compared to WT controls.
Conclusions
We identified HDAC5 as a negative epigenetic regulator of IL-6 synthesis and release in skeletal muscle. HDAC5 may exert beneficial effects through two different mechanisms, transcriptional control of genes required for glucose disposal and utilization, and HDAC5-dependent IL-6 signaling cross-talk to improve glucose uptake in muscle in response to exercise.
Keywords: HDAC5, Interleukin 6, Diabetes, Insulin sensitivity, Exercise, Skeletal muscle
Abbreviations: AcH3K9, lysin 9 acetyl histone H3; ChIP, chromatin immunoprecipitation; EDL, extensor digitorum longus; EPS, electric pulse stimulation; GLUT4, glucose transporter isoform 4; HDACs, histone deacetylases; IL-6, interleukin 6; i.p. GTT, intraperitoneal glucose tolerance test; i.p. ITT, intraperitoneal insulin tolerance test; T2D, type 2 diabetes; WAT, white adipose tissue; WT, wild type
1. Introduction
Obesity and insulin resistance are strongly associated with the development of type 2 diabetes (T2D) [1]. Hyperglycemic conditions, if not treated in their early phase, can lead to severe micro- and macrovascular complications that cause comorbid diseases such as end-stage kidney damage or heart failure [[2], [3], [4]]. Skeletal muscle plays an important role in the disposal of glucose from the bloodstream that is initiated by both the activation of insulin signaling and muscle contraction [[5], [6], [7]]. Reduced glucose uptake in muscle, mainly through the insulin-sensitive glucose transporter GLUT4, and elevated hepatic gluconeogenesis have been attributed to the pathogenesis of insulin resistance and T2D [8].
Histone deacetylases (HDACs) are divided into three structurally defined subclasses and are critical for transcriptional regulation, cell cycle progression, and developmental events [9]. Several studies highlighted the role of histone deacetylase 5 (HDAC5), a class II HDAC, as a negative transcriptional regulator of GLUT4 in the skeletal muscle [[10], [11], [12]]. Pharmacological inhibition or deletion of HDAC5 in skeletal muscle and adipocytes was shown to increase the expression of GLUT4 and insulin-stimulated glucose uptake [12,13]. Mihaylova et al. reported amelioration of hyperglycemia in type 2 diabetes mouse models by the suppression of hepatic class IIa HDACs via adenoviral delivery of shRNAs directed against HDAC5 [14]. We recently demonstrated the role of hypothalamic HDAC5 in controlling food intake and body weight by modulating leptin signaling [15]. Interestingly, global HDAC5 KO animals demonstrated normal glucose and insulin tolerance despite elevated body adiposity and increased hepatic fat deposition due to leptin resistance and hyperphagia in mice [15]. These data indicate that the hypothalamic regulation of food intake and body weight may be disjointed from the peripheral regulation of glucose homeostasis by HDAC5. Collectively, these reports highlight the tissue-specific and context-dependent roles of HDAC5.
Physical activity can improve muscle glucose metabolism by facilitating insulin-dependent glucose uptake and increasing skeletal muscle insulin sensitivity [16]. Exercise has been associated with enhanced phosphorylation and nuclear exclusion of HDAC5, leading to increased transcription of GLUT4 genes [10,17]. Furthermore, exercise is linked to anti-inflammatory, metabolic and regenerative benefits, likely driven by the release of skeletal muscle-derived secretory factors such as interleukin 6 (IL-6) [[18], [19], [20], [21]]. IL-6 has insulin-like effects such as increasing glucose uptake and fatty acid oxidation in skeletal muscle [22,23]. However, the molecular signaling leading to IL-6 release from contracting muscle is not established to date [22,24].
In this study, we assessed the impact of HDAC5 on skeletal muscle energy metabolism under sedentary and exercise conditions. By combining in vivo studies of global HDAC5 KO mice with cell culture studies in HDAC5 knockdown muscle cells and ex vivo analyses of HDAC5 KO tissues, we explored the role of HDAC5 and IL-6 in mediating the beneficial effects of exercise on glucose metabolism.
2. Materials and methods
2.1. Animal experiments
HDAC5 knockout mice (KO) and corresponding wild-type (WT) littermate controls [15] were genotyped as previously described [25]. Male mice were used throughout the study. HDAC5 KO and WT mice 12–30 weeks old were group housed under a 12:12 h light–dark cycle (06:00 h lights on) with ad libitum access to low-fat chow (58% carbohydrates, 33% protein, 9% fat, R/M−H Extrudat, ssniff Spezialdiäten GmbH, Soest, Germany) and tap water. All of the experiments were approved by the Ethics Committee of the State Ministry of Agriculture, Nutrition, and Forestry, Az. 84-02.04.2017.A333 (North Rhine-Westphalia, Germany).
2.2. Measurements of body weight and body composition
Body weight was determined using an electronic scale (Sartorius, Göttingen, Germany). Fat mass and lean mass were measured via nuclear magnetic resonance technology (Whole Body Composition Analyzer, Echo MRI, Houston, TX, USA).
2.3. Exercise training protocol
In our chronic exercise intervention, male mice were trained on 4-lane treadmills (TSE Systems, Bad Homburg, Germany) for a maximum of 8 weeks, 1 h per day, 5 days per week followed by two days of recovery. During the 8 weeks, training parameters gradually increased from low intensities in the first week to higher intensities in the last week (a maximum speed up to 18 m/min, running time up to 45 min, and a treadmill slope of 5°). Tolerance tests were conducted in the last weeks of the training intervention. Running performance was measured before and at the end of the chronic treadmill training.
2.4. Running performance test
At the beginning and end of the chronic exercise intervention, mice underwent running performance tests on 1-lane calorimetric treadmills (TSE Systems, Bad Homburg, Germany) until exhaustion. At a 5° slope, the mice underwent a 15-min adaptation and warm-up phase until the subsequent measurement phase, which spanned a time course of ∼20 min maximum and gradually increased to speeds of up to 1.5 m/min. The mice were motivated by gentle tail tapping. Exhaustion was defined as the time point when an animal did not respond to motivation stimuli and interrupted the light barrier at the rear of the system three times within 15 s.
2.4. Intraperitoneal glucose and insulin tolerance tests (i.p. GTT and i.p. ITT)
HDAC5 KO and WT littermates were subjected to 6 h of fasting followed by an i.p. injection of 2 g/kg body weight (BW) glucose (glucose 20%, B. Braun Melsungen AG, Melsungen, Germany) during GTT or an i.p. injection of 0.75 U/kg BW of human recombinant insulin (Actrapid, Penfill, Denmark) during ITT. Tail blood glucose levels (mg/dL) were measured with a glucometer and standard glucose strips (Bayer Vital GmbH, Leverkusen, Germany) before (0 min) and 15, 30, 45, 60, and 120 min after injection.
2.5. ELISA analysis
Blood was collected in EDTA-coated microvettes and centrifuged at 4 °C and 2,000×g for 10 min to generate plasma. Plasma triglycerides, cholesterol, and non-esterified fatty acids were measured using commercial enzymatic assay kits (Wako Chemicals, Neuss, Germany). Plasma insulin was measured using a mouse ultrasensitive ELISA kit (DRG, Marburg, Germany). IL-6 was analyzed using a quantitative ELISA kit (R&D Systems, Minneapolis, MN, USA) in the supernatant of differentiated and electric pulse-stimulated cells as well as in blood plasma. All of the assays were conducted according to the manufacturer's instructions.
2.6. C2C12 cell culture
Mouse skeletal muscle C2C12 cells were cultured in high glucose (25 mM) DMEM medium (Life Technologies, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS) (Life Technologies) and 1% penicillin/streptomycin (Life Technologies) at 37 °C and 5% CO2. When the cells reached ∼80–90% confluence (day 0), differentiation was induced by adding high-glucose DMEM medium with 2% horse serum (Life Technologies) and 1% penicillin/streptomycin. The differentiation medium was changed every 24–48 h for up to 6 days. For protein expression analysis, the cells were serum starved for 5 h by changing the medium to DMEM medium with 0.01% horse serum. Then the cells were treated with 100 nM of insulin for 15 min and processed for Western blotting.
2.7. Electric pulse stimulation (EPS)
Electric pulse stimulation (EPS) experiments were conducted according to Lambernd et al. [26]. Briefly, myotubes were differentiated until day 6, followed by overnight starvation in DMEM without FBS. The starvation medium was changed to DMEM with 10% FBS directly before stimulation. EPS was performed using a C-Dish combined with a pulse generator (IonOptix, Milton, MA, USA) at the conditions of 1 Hz, 2 ms, and 11.5 V for 24 h.
2.8. SDS-PAGE and western blotting analysis
Tissues and cells were lysed in RIPA-buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% (v/v) Triton-X 100, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) including proteinase and phosphatase inhibitors (Complete and PhosSTOP, Roche Diagnostics, Mannheim, Germany) using a Bioruptor Pico (Diagnode, Seraing, Belgium). After centrifugation (12,000×g, 10 min, 4 °C), supernatants were collected and the protein content was determined using a BCA protein assay kit (Pierce, Rockford, IL, USA). Protein samples were reduced with Laemmli sample buffer containing DTT, separated by SDS-PAGE, and transferred to nitrocellulose membranes (Bio-Rad, München, Germany). Membranes were probed with primary antibodies (Supplementary Table 2) diluted in 1 × TTBS + 5% BSA and incubated overnight at 4 C under gentle agitation, followed by incubation with HRP-conjugated goat anti-rabbit IgG (Dianova, Hamburg, Germany) antibodies diluted 1:20,000 in 1 × TTBS + 5% BSA for 1 h at room temperature. Specific protein bands were visualized using enhanced chemiluminescence reagents for Western blotting analysis (Perkin Elmer, Waltham, MA, USA). Protein bands were quantified using Quantity One software (Bio-Rad).
2.9. Quantitative real-time polymerase chain reaction (qPCR)
Total RNA was extracted from frozen tissues or cells using a RNAeasy kit (Qiagen, Hilden, Germany) and quantified with a Nanodrop 2000 instrument (Thermo Fisher Scientific, Wilmington, DE, USA). cDNA templates for qPCR were synthesized from 1 μg of total RNA using a GoScript Reverse Transcription System (Promega, Madison, WI, USA). qPCR reactions were conducted using a Quantifast SYBR Green PCR Kit (Promega). Gene expression was evaluated using the ΔΔ-Ct method and Rpl32 was used as a housekeeping gene. The primer sequences are listed in Supplementary Table 3.
2.10. RNA extraction and gene expression analysis
Total RNA was isolated and purified from snap-frozen tissue using an RNeasy kit (Qiagen, Hilden, Germany). The quality of the isolated RNA was tested using an RNA 6000 nano kit (Agilent Technologies, Taufkirchen, Germany). Genome-wide expression analyses (n = 5 per genotype) were conducted with 150 ng of RNA according to an Ambion WT Expression kit and a WT Terminal Labeling kit (Thermo-Fischer Scientific, Darmstadt, Germany). All of the protocol steps were monitored using an RNA 6000 nano kit (Agilent Technologies, Taufkirchen, Germany). Mouse gene 1.0 ST arrays were hybridized with labeled complementary RNA samples. Data were collected with the GeneChip scanner 3000 7G and analyses of primary data were performed with the GDAS 1.4 package (Affymetrix), (Thermo-Fisher Scientific). The data were analyzed with Expression Console v1.1 and Transcriptome Analysis Console v2.0 (Affymetrix) as described [27]. Full datasets are available under accession number GSE150485 (available online at www.ncbi.nlm.nih.gov/geo/). Enrichment and canonical pathway analyses as well as potential upstream target analyses were conducted using Ingenuity pathway analysis software (Qiagen, Hilden, Germany) and ConsensusPathDB [28]. Cut-off values were set according to a p value < 0.05 for all analyses.
2.11. In vitro glucose uptake into C2C12 myotubes
For the glucose uptake assay, radioactively labeled [3H]2-deoxy-glucose (2.5 mCi/ml) (Analytic GmbH, Braunschweig, Germany) and [14C]mannitol (0.25 μCi/ml) (Perkin Elmer) were supplemented with Krebs-Henseleit (KRH) buffer (136 mM NaCl, 4.7 mM KCl, 41.25 mM MgSO4, 1.25 mM CaCl2, and 10 mM HEPES) and added to differentiated C2C12 myotubes. After 30 min, the cells were washed 3 times with ice-cold PBS and lysed in 100 μl of RIPA buffer. The radioactivity was measured in duplicate and normalized to the protein content.
2.12. Ex vivo glucose uptake in intact skeletal muscles and white adipose tissue (WAT)
Isolation of skeletal muscles and measurements of glucose uptake were conducted as previously described [29]. The extensor digitorum longus (EDL) and soleus muscles were removed from anesthetized HDAC5 KO and WT male mice. WAT was also removed and immediately dissected into 0.5 cm pieces. First, both muscles and WAT were incubated for 30 min at 30 °C in vials containing pre-oxygenated (95% O2/5% CO2) Krebs-Henseleit buffer (KHB) supplemented with 5 mM of HEPES, 5 mM of glucose, and 15 mM of mannitol. The muscles and WAT were then transferred to new vials and incubated for 30 min in KHB containing 5 mM of HEPES, 5 mM of glucose, and 15 mM of mannitol under basal conditions or in the presence of 120 nM of insulin (Actrapid, Novo Nordisk, Mainz, Germany). Muscles and WAT were then transferred to new vials with pre-oxygenized KHB supplemented with insulin and 15 mM of mannitol and incubated for 10 min. The muscles and WAT were then transferred to new vials containing pre-oxygenized KHB supplemented with 1 mM of [3H]2-deoxy-glucose (2.5 mCi/ml) (Analytic GmbH, Braunschweig, Germany) and 19 mM of [14C]mannitol (0.7 mCi/ml) (Perkin Elmer) for 20 min. All of the incubation steps were conducted under an atmosphere of 95% O2 and 5% CO2 at 30 °C with slight agitation. Muscles and WAT were immediately snap-frozen in liquid nitrogen and stored at −80 °C. Cleared muscle lysates were used to determine incorporated radioactivity by scintillation counting.
2.13. Glycogen synthesis
Differentiated C2C12 cells were starved for 90 min in serum- and glucose-free media with 12.5 mM of HEPES. After washing, media with 1 μCi/ml of 14C-glucose (Analytic GmbH) were added with or without 100 nM of insulin and the cells were incubated for 4 h at 37 °C while sealed with parafilm. The cells were then washed 2 times with ice-cold PBS and lysed by adding 100 μl of RIPA buffer (Sigma–Aldrich) with PMSF; 1 μl of the lysates was used for a BCA protein assay. Then 400 μl of 30% KOH and 35 μl of glycogen (60 mg/ml) were added to each sample. The samples were vortexed and incubated for 20 min at 80 °C. Then 500 μl of pure ethanol was added, and the lysates were vortexed and centrifuged at 4 °C and 10,000×g for 20 min. The supernatants were removed and the pellets washed once with 70% ethanol. After centrifugation (4 °C, 20 min, and 10,000×g), the supernatants were again removed and the pellets air-dried. The pellets were then dissolved in 500 μl of Milli-Q water and rotated at room temperature for 20 min. The samples were added to scintillation vials and the radioactivity was measured. The results were normalized to the protein content.
2.14. Cloning, transfection, and stable and transient gene knockdown
DNA fragments of murine Il6 promotor were amplified from genomic DNA of mouse muscle tissue using S7 Fusion Polymerase (Biozym, Oldendorf, Germany) and then cloned into pGL4.14 luc2/hygro vector (Promega). Primer sequences are shown in Supplementary Table 4. Promoter fragments were transfected into C2C12 myoblasts using Lipofectamin 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Cells with a stable knockdown of Hdac5 were generated using pGIPZ vector containing the shRNA sequence (V3LMM_432050, Dharmacon, Lafayette, CO, USA). Transfection of the C2C12 cells with the isolated plasmids was performed with Lipofectamin 3000. Selection of stably transfected cells was conducted by adding 1.25 μg/μl of puromycin (Sigma–Aldrich, Steinheim, Germany) 24 h after transfection. Transfection of C2C12 myotubes with small interfering RNA (siRNA) was performed using ON-TARGET plus SMARTpool siRNA oligonucleotides specific for murine Il6 mRNA (L-043739-00-0005, Dharmacon) or ON-TARGET plus non-targeting siRNA (D-001810-10-05) as a negative control. Transfections were conducted according to the manufacturer's instructions. Briefly, DharmaFECT1 (T-2001-01, Dharmacon) and 50 nM of siRNA oligonucleotides were dissolved in antibiotic- and serum-free medium and incubated at room temperature for 20 min. The cells were then incubated with a mixture of siRNA oligonucleotides and DharmaFECTI in serum-containing medium.
2.15. Luciferase assays
C2C12 cells were cultivated in 12-well plates and transfected with 1 μg of a luciferase vector containing promoter fragments. Luciferase assays (Promega) were conducted according to the manufacturer's instructions. Briefly, 24 h after transfection, the cells were washed with ice-cold PBS and then reporter lysis buffer (Promega) was added. Lysis of the cells was facilitated by one cycle of freezing at −80 °C and thawing on ice. To measure the luciferase activity, 20 μl of clear extracts were used. Luciferase activity was measured by a luminometer (SpectraFluor Plus, Tecan), and the results of luciferase activity were normalized to the protein content.
2.16. ChIP analysis
Chromatin immunoprecipitation assays were conducted as previously described [30] with minor modifications. C2C12 myotubes were washed once with 1 × PBS and incubated at room temperature for 45 min with 2 mM of DSP (dithiobis-(succinimidyl propionate)) (Thermo-Fisher Scientific, Dreieich, Germany) for protein–protein crosslinking for 10 min with 1% formaldehyde for DNA-protein crosslinking. The reactions were stopped by adding 125 mM of glycine for 5 min. The cells were lysed for 10 min with lysis buffer (50 mM Tris–HCl, pH 8.0, 2 mM EDTA, 0.1% NP-40, 10% glycerol, and 2 mM dithiothreitol). To obtain the nuclear fraction, the cell lysates were centrifuged for 5 min at 4 °C at 1000×g. Nuclear fractions were then lysed in 200 μl of lysis buffer (50 mM EDTA, 1% SDS, and 2 mM dithiothreitol) and sonicated to obtain 200-500 bp chromatin fragments. Sonicated chromatin was diluted 1:9 with a diluting buffer (50 mM Tris–HCl, pH 8.0, 5 mM EDTA, 0.5% NP-40, and 200 mM NaCl) and then incubated with 1 μg of rabbit or mouse IgG (Cell Signaling 2729 and Santa Cruz 2025, respectively), anti-HDAC5 (Santa Cruz, Heidelberg, Germany, sc-133106), or anti-acetyl-histone H3 (Lys9) (Millipore, Darmstadt, Germany, 06-942) antibodies overnight, followed by incubation with 30 μl of SureBeads Protein A or G magnetic beads (Bio-Rad). The beads were washed in low-salt washing buffer (20 mM Tris–HCl, pH 8.0, 2 mM EDTA, 1% NP-40, 0.1% SDS, and 150 mM NaCl), high-salt washing buffer (20 mM Tris–HCl, pH 8.0, 2 mM EDTA, 1% NP-40, 0.1% SDS, and 500 mM NaCl), LiCl washing buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 1% NP-40, 1% Na-deoxycholate, and 250 mM LiCl), and TE buffer (10 mM Tris–HCl, pH 8.0, and 1 mM EDTA). Chromatin was eluted with DNA elution buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 100 mM NaHCO3, and 1% SDS) supplemented with 1 mg/ml of RNaseA (Qiagen) for 1 h at 37 °C with shaking at 900 rpm. The beads were then incubated in elution buffer containing 10 mg/ml of Proteinase K (Stratec, Germany) and agitated (900 rpm) for 2 h at 56 °C. Crosslinking was reversed overnight at 65 °C after the addition of 200 mM of NaCl. The DNA was then purified by phenol:chloroform:isoamyl alcohol (25:24:1) extraction and subjected to RT-PCR with primers directed against the Il6 promoter mapping to the region from 1 bp to 300 upstream (from −331 bp to −1 bp) of the transcription start site (forward 5′-AGAGTGCTCATGCTTCTTAGGG-3′ and reverse 5′-GTGCTGAGTCACTTTTAAAGAAAA-3′) and Slc2a4 (GLUT4) mapping to the region from −641 bp to −337 bp upstream of the transcription start site (forward 5′-ACATTTGGCGGAGCCTAAC-3′ and reverse 5′-AGACACGCCCCTAAGGTTC-3’) promoter.
2.17. Statistical analysis
All of the statistical analyses were conducted using GraphPad Prism V7 (GraphPad Software, San Diego, CA, USA). Groups were compared with two-tailed unpaired t-tests or one-way or two-way ANOVA followed by Bonferroni's post hoc analysis as stated in each figure legend. P values < 0.05 were considered significant. All of the results are presented as means ± SEM.
3. Results
3.1. HDAC5 regulates Il6 gene expression in skeletal muscle
To gain insight into the biological function of HDAC5 in skeletal muscle, we conducted microarray expression profiling with gastrocnemius muscle obtained from male 30-week-old global HDAC5 knockout mice (HDAC5 KO) and male wild-type littermates (WT) fed a chow diet. Consistent with our previous report [15], we found moderately elevated body weight due to an increase in fat mass in the HDAC5 KO mice compared to the WT mice (Supplementary Figs. S1A–C). In total, we identified 560 differentially regulated genes in the gastrocnemius muscle from the HDAC5 KO mice, and 289 and 271 transcripts were significantly up- or downregulated, respectively (Figure 1A). Interestingly, protein-encoding genes such as Klk3 or HtrA4 and genes involved in protein metabolism as well as histone modifications such as Mettl21, Klf10, or Prm1 were differentially expressed. We found that genes encoding members of the tyrosine phosphatase protein family (Dusp26 and Ptpn3) were upregulated. To further investigate the regulatory network of HDAC5 in skeletal muscle, we conducted Ingenuity pathway analyses. Interestingly, knockout of HDAC5 was associated with differential regulation of several key molecules involved in skeletal muscle growth, regeneration, and energy metabolism including TGFβ, VEGF, and insulin signaling (Supplementary Table 1) as well as other interconnected metabolic pathways (Supplementary Fig. S2). Most notably, we found substantial activation of IL-6 signaling, where 17 out of 22 genes of this pathway were transcriptionally upregulated in the HDAC5 KO mice compared to the WT controls (Figure 1B) (Supplementary Table 1). The impact of HDAC5 deficiency on IL-6 downstream genes was confirmed by qPCR analyses in the gastrocnemius muscle of the HDAC5 WT and KO mice (Figure 1C). We then generated a stable cell line of C2C12 cells with knockdown of Hdac5 (Supplementary Figs. S3A–B). Consistent with the data from knockout muscles, HDAC5 deficiency affected the same target genes in HDAC5 KD C2C12 myotubes (Figure 1D). Regression analyses further revealed a negative correlation between Il6 and Hdac5 mRNA levels in WT and HDAC5 KD myotubes (Figure 1E). Overall, these data strongly suggest a direct involvement of HDAC5 in Il6 transcription.
Figure 1.
HDAC5 regulates Il6 gene expression in muscle. (A) Volcano plot of differentially expressed genes analyzed by Affymetrix chip microarray from gastrocnemius muscle of the HDAC5 KO (KO) and wild-type (WT) male mice. (B) Representative heat map of IL-6 regulated genes analyzed by Affymetrix chip microarray from gastrocnemius muscle of the KO and WT mice. (C) Validation of differentially expressed genes in gastrocnemius muscle of the HDAC5 knockout (H5KO) and WT mice by qPCR (n = 5) (D) HDAC5 knockdown (H5KD) C2C12 myotubes (n = 6) by qPCR. (E) Correlation analysis with Pearson's coefficient between Il6 and HDAC5 mRNA obtained from WT and different batches of H5KD cells (n = 15). Results are represented as mean ± SEM. Statistical analyses were conducted using two-tailed unpaired Student's t-test. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
3.2. HDAC5 epigenetically regulates Il6 transcription in muscle
To explore the molecular mechanism of HDAC5-regulated Il6 transcription in muscle, we conducted chromatin immunoprecipitation (ChIP) for HDAC5 in the WT and HDAC5 KD myotubes. Il6 promoter analyses demonstrated strong amplification in the WT myotubes but no amplification in the HDAC5 KD myotubes, suggesting that HDAC5 was recruited to the Il6 promoter (Figure 2A). We next conducted ChIP-PCR analyses of the Il6 promoter with an antibody against lysine 9 acetyl histone H3 (AcH3K9) and found a marked increased signal in the HDAC5 KD myotubes compared to the WT controls (Figure 2B). H3K9 acetylation as a marker of active transcription [31] was further enriched in the Slc2a4 (GLUT4) promoter of the HDAC5 KD myotubes (Figure 2G,H). We next used luciferase reporter gene assays to assess Il6 promoter transactivation and found elevated transcriptional activity in the HDAC5 KD compared to the WT myotubes for the full-length construct and comparably higher activity for the construct with the first approximately 1,000 base pairs of the Il6 promoter (Figure 2C). Elevated reporter gene activity in the HDAC5 KD myotubes was also found for the promoter fragment with Il6 promoter base pairs −1000 to −2000, but not for −2000 to −3000 (Figure 2C). We then collected and analyzed cell culture media from the WT and HDAC5 KD myotubes following 5 h of serum starvation, respectively, and found substantially higher levels of IL-6 secretion in the HDAC5 KD compared to the WT cells (Figure 2D,E). Gene expression data revealed upregulation of Il6 in the HDAC5 KD myotubes (Figure 2F) to further prove that HDAC5 acts as a transcriptional regulator of IL-6.
Figure 2.
HDAC5 epigenetically regulates Il6 transcription and enhances Slc2a4 (GLUT4) expression in skeletal muscle in vitro. Mouse Il6 promoter ChIP-PCR analysis from the HDAC5 KD (H5KD) and wild-type (WT) C2C12 myotubes pulldown using HDAC5 antibody (A) and AcH3K9 antibody (B) (n = 3). (C) Luciferase assay for Il6 promoter fragments. The graph represents full length (Full), first 1000 bp (−1000), fragments from 1000 to 2000 bp (−2000), and fragments from 2000 to 3000 bp (−3000) of Il6 promoter cloned upstream of the luciferase gene. (D and E) Determination of IL-6 secretion in the media from the H5KD and WT C2C12 myotubes. Data were normalized to the total protein content and total volume, respectively (n = 10–12). (F) Il6 mRNA expression analysis from the H5KD and WT C2C12 myotubes (n = 7). Slc2a4 (GLUT4) promoter ChIP-PCR analysis from the HDAC5 KD (H5KD) and wild-type (WT) C2C12 myotubes using HDAC5 antibodies (G) and AcH3K9 antibodies (H) under basal conditions (n = 3). Results are represented as mean ± SEM. Statistical analyses were conducted using one-way ANOVA followed by Bonferroni's post-hoc test (A-B) or by two-tailed unpaired Student's t-test (C–F). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
3.3. HDAC5 deficiency augments skeletal muscle glucose uptake and glycogen synthesis
We next conducted in vitro glucose uptake assays in the HDAC5 WT and KD myotubes (Figure 3A) as well as ex vivo glucose uptake assays in oxidative soleus muscle, glycolytic extensor digitorum longus (EDL) muscle, and white adipose tissue (WAT) of the HDAC5 WT and KO mice (Figure 3G–I). The HDAC5 KD myotubes had significantly increased glucose uptake at basal level compared to the WT controls (Figure 3A), which coincided with both elevated Slc2a4 mRNA levels and GLUT4 protein abundance (Figure 3B,C). We further found significantly higher insulin-stimulated glycogen synthesis (Figure 3D), glycogen synthesis 2 (Gys2), protein phosphatase 1 regulatory subunit 3c (Ppp1r3c) expression (Figure 3E), and lower basal pGYSS641 (Figure 3F) protein levels in the HDAC5 KD cells. In line with our in vitro data, we found elevated insulin-stimulated glucose uptake in soleus muscle from the HDAC5 KO compared to WT mice, while basal glucose uptake was not significantly altered (Figure 3G, data not shown). Basal and insulin-stimulated glucose uptake was comparable between the WT and KO mice for the EDL and WAT of the KO mice (Figure 3H–I, data not shown).
Figure 3.
HDAC5 regulates glucose uptake in skeletal muscle.(A) In vitro glucose uptake under basal conditions in the HDAC5 KD (H5KD) and wild-type (WT) C2C12 myotubes (n = 15). (B) mRNA expression analysis of Slc2a4 (GLUT4) (n = 6). (C) Representative Western blotting and densitometry analysis of GLUT4 (n = 3). D) Glycogen synthesis under basal and insulin-stimulated conditions (n = 15). E) mRNA expression analysis of glycogen-related genes Gys2 and Ppp1r3c (n = 6). F) Representative Western blotting and densitometry analysis of pGYS following stimulation with insulin (n = 3) from the H5KD and WT C2C12 myotubes. Ex vivo glucose uptake under basal and insulin stimulated conditions from intact (G) soleus, (H) EDL muscle, and (I) WAT isolated from the WT and HDAC5 KO (H5KO) mice (n = 8). Results are represented as the mean ± SEM. Statistical analyses were conducted using two-way ANOVA followed by Bonferroni's post-hoc test (E and F) or two-tailed unpaired Student's t-test (A-D and G-I). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
3.4. HDAC5 deficiency is associated with elevated insulin-stimulated AKT activation independent of IL-6
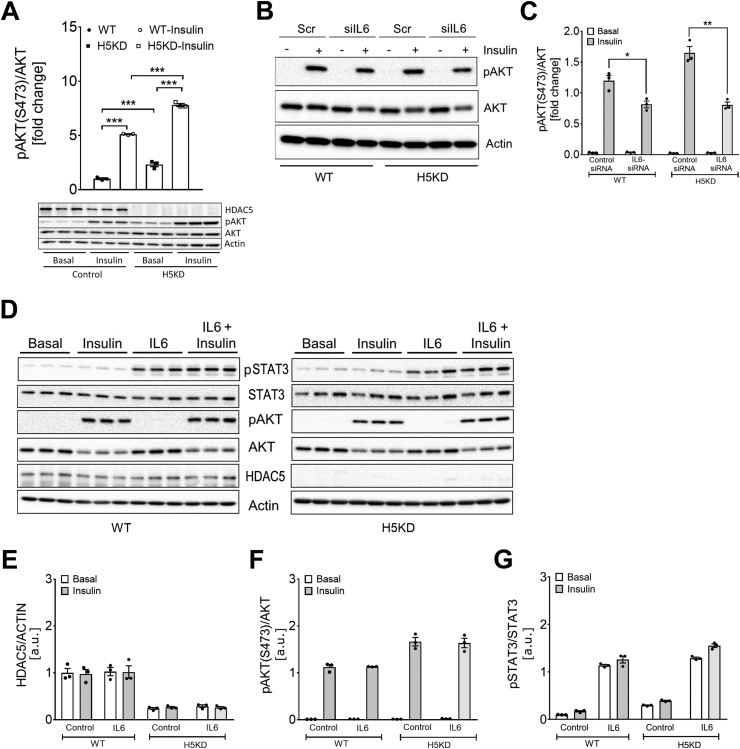
Prompted by the elevated insulin-stimulated muscle glucose uptake, we next assessed whether HDAC5 deficiency directly affects skeletal muscle insulin signaling. Western blotting analysis in the HDAC5 KD myotubes revealed enhanced levels of AKT phosphorylation both at baseline and after insulin stimulation compared to WT myotubes (Figure 4A). Knockdown of Il6 expression by siRNA technology led to substantially reduced Il6 mRNA levels (Supplementary Fig. S4) and a decrease in insulin-stimulated AKT phosphorylation in both the control and HDAC5 KD myotubes (Figure 4B,C). Treating the control and HDAC5 KD cells with recombinant IL-6 protein, however, did not affect the levels of HDAC5 protein expression (Figure 4D,E). Similarly, treating the control and HDAC5 KD myotubes with recombinant IL-6 protein had no effect on basal or insulin-stimulated AKT phosphorylation (Figure 4D,F). However, IL-6 treatment strongly induced STAT3 phosphorylation, consistent with previous reports [32] (Figure 4D,G).
Figure 4.
The HDAC5-IL-6 axis modulates pAKT signaling in skeletal muscle. Representative Western blotting and densitometry analysis of (A) AKT and pAKT obtained from the HDAC5 KD (H5KD) and wild-type (WT) C2C12 myotubes treated with 100 nM insulin for 15 min (n = 3). Western blotting (B) and densitometric analysis (C) of AKT and pAKT under basal and 100 nM insulin stimulated conditions from H5KD and WT following treatment with siIl6 or siScramble (Scr) siRNA oligonucleotides (n = 3). Representative Western blotting (D) and densitometric analysis (E-G) of pSTAT3, pAKT, and HDAC5 from the H5KD and WT C2C12 myotubes treated with recombinant IL-6 (IL6) following stimulation with 100 nM insulin for 15 min (n = 3). Results are represented as mean ± SEM. Statistical analyses were conducted using two-way ANOVA followed by Bonferroni's post-hoc test. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
3.5. HDAC5 deletion in mice enhances systemic glucose handling following exercise
We next investigated the HDAC5 WT and KO male mice to assess the role of HDAC5 in exercise-stimulated glucose uptake. At 30 weeks of age, the mice were subjected to an 8-week treadmill training protocol (Figure 5A; see methods). We measured the body composition and conducted running performance tests before and after the training; intraperitoneal glucose insulin tolerance tests and Slc2a4 mRNA expression measurements were conducted after the training intervention was complete.
Figure 5.
Endurance exercise training improves glucose tolerance in HDAC5 KO mice. Body composition, glucose, and insulin tolerance were tested after 8 weeks of chronic endurance exercise in the wild-type (WT) and HDAC5 KO (H5KO) male chow-fed mice and compared to respective sedentary littermate controls. (A) Experimental plan for the exercise intervention and in vivo measurements. The cohort of 30-week-old mice were subjected to treadmill training for 8 weeks. In vivo measurements included the determination of body composition by nuclear magnetic resonance (NMR) and intraperitoneal glucose or insulin tolerance tests (GTT and ITT), respectively. Running performance was tested before (Pre) and after chronic exercise (Post) as described in the methods section. Body composition in terms of (B) body weight, (C) lean mass, and (D) fat mass. (E) Running performance, (F) maximal oxygen uptake (VO2max), and (G) average respiratory exchange ratio (RER). Insulin tolerance test in (H) sedentary and (J) exercised mice and (I and K) area under the curve (AUC) for the sedentary and exercised mice, respectively. Glucose tolerance test from the (L) sedentary and (N) exercised mice and (M and O) AUC for the sedentary and exercised mice. (P) mRNA expression analysis of Slc2a4 (GLUT4) in gastrocnemius muscle (n = 4–5). Results are represented as mean ± SEM. Statistical analyses were conducted using two-way ANOVA followed by Bonferroni's post-hoc test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
In the WT mice, exercise intervention led to a mild reduction in body weight, whereas the HDAC5 KO mice were not affected by the training (Figure 5B–D). Both genotypes further displayed increased running performance after the training (Figure 5E). Moreover, in both genotypes, we observed a trend of increased VO2max (Figure 5F) and respiratory exchange ratio in response to the 8 weeks of training (Figure 5G). In the sedentary HDAC5 WT and KO mice, glucose tolerance was identical (Figure 5L and M). Notably, while insulin tolerance was only marginally affected by the exercise training in both genotypes (p = 0.1247) (Figure 5H–K), we observed marked improvements in glucose tolerance in the trained HDAC5 KO mice compared to the trained WT mice (Figure 5L–O). Slc2a4 mRNA expression was elevated in the gastrocnemius muscle from the HDAC5 KO mice in both sedentary and trained conditions (Figure 5P). Although the mean values of Slc2a4 mRNA were elevated in response to training in both genotypes, the increase did not reach statistical significance. Overall, these data indicate the important role of HDAC5 in glycemic regulation, specifically in the trained state.
3.6. HDAC5 deficiency augments EPS-induced Il6 transcription and secretion in C2C12 myotubes
To explore the molecular mechanism of the improved glucose tolerance of the exercised HDAC5 KO mice, we next adopted an in vitro exercise contraction model based on electrical pulse stimulation (EPS) [26] in the fully differentiated C2C12 WT and HDAC5 KD myotubes.
EPS treatment of the WT myotubes for 24 h tended to increase the phosphorylation of STAT3, AKT, and AMPK (Supplementary Figs. S5A–E). In the HDAC5 KD myotubes, AcSTAT3 and pAKT but not pSTAT3 or pAMPK levels were already elevated at baseline; EPS had no additional effect on Ac-STAT3, but amplified the pSTAT3, pAKT, and pAMPK levels (Supplementary Figs. S5A–E). EPS stimulation induced enhanced transcription and higher levels of IL-6 secretion in the HDAC5 KD compared to the WT myotubes (Figure 6A,B). ChIP-PCR-based promoter analyses of Il6 demonstrated a marked dissociation of HDAC5 from the Il6 promoter in the WT myotubes after EPS and the expected relative lack of HDAC5 binding to Il6 promoter in the HDAC5 KD myotubes (Figure 6C). Interestingly, ChIP-PCR analyses for histone H3K9 acetylation, a marker of activated chromatin, revealed a marked increase in AcH3K9 levels in the Il6 promoter of WT cells after EPS stimulation that was amplified at baseline and after EPS treatment as a result of the HDAC5 deficiency (Figure 6D). We further observed an increased expression of Slc2a4 (GLUT4) in response to EPS treatment in the WT myotubes (Figure 6E). Notably, in the HDAC5 KD myotubes, Slc2a4 expression was profoundly increased already at baseline and further elevated by EPS. ChIP-PCR signals of the Slc2a4 promoter using HDAC5 antibodies were reduced following EPS treatment of the WT myotubes, indicating decreased dissociation of HDAC5 from the Slc2a4 promoter (Figure 6F). Furthermore, ChIP-PCR signals of the Slc2a4 promoter using AcH3K9 antibodies were elevated after EPS treatment of C2C12 HDAC5 KD cells, consistent with a moderate increase in the acetylation of the Slc2a4 promoter (Figure 6G). Collectively, these data indicate that HDAC5 constitutes a negative epigenetic regulator of both Il6 and Slc2a4.
Figure 6.
HDAC5 regulates Il6 and Slc2a4 (GLUT4) expression in response to myotube contraction in vitro. (A) Cellular Il6 mRNA expression analysis and (B) IL-6 secretion in the supernatant media from the HDAC5 KD (H5KD) and wild-type (WT) C2C12 myotubes (n = 6). ChIP analysis from the H5KD and WT C2C12 myotubes following electro pulse stimulation (EPS) for 24 h. ChIP-PCR was conducted using (C) HDAC5 antibodies and (D) AcH3K9 antibodies, respectively, using primers directed against mouse Il6 promoter (n = 3). (E) Slc2a4 mRNA expression analysis in the EPS-treated H5KD and wild-type WT C2C12 myotubes (n = 6). Slc2a4 promoter ChIP-PCR analysis from the H5KD and WT C2C12 myotubes using (F) HDAC5 antibodies and (G) AcH3K9 antibodies, respectively, following EPS treatment for 24 h (n = 3). Results are represented as the mean ± SEM. Statistical analyses were conducted using one-way ANOVA followed by Bonferroni's post-hoc test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
4. Discussion
The present study provides a novel mechanism by which HDAC5 negatively regulates transcription of the Il6 gene in skeletal muscle through epigenetic regulation, contributing to the beneficial metabolic effects of physical exercise on glucose metabolism.
Previous studies provided evidence of an important role of class IIa HDACs in regulating energy metabolism in skeletal muscle [33]. Beneficial metabolic effects of exercise on energy metabolism were found to be associated with reduced class IIa HDAC activity in skeletal muscle, while inhibition of class IIa HDAC corepressor complex formation led to metabolic alterations in skeletal muscle that resemble an exercise response including increased whole-body energy expenditure, elevated fatty acid oxidation, and improved glycemia [34,35]. Activation of AMPK in vitro, which is associated with muscle contraction, has been shown to result in increased phosphorylation of HDAC5 that may cause nuclear exclusion of the protein and subsequently altered histone acetylation [11].
Transcriptional profiling of gastrocnemius muscle from the HDAC5-deficient mice (HDAC5 KO) and C2C12 myotubes after Hdac5 knockdown (HDAC5 KD) identified activation of genes annotated for the IL-6 pathway. Since the HDAC5 KD myotubes exhibited both increased transcription of Il6 and elevated levels of secreted IL-6 in the culture medium, we conclude that muscle-derived IL-6 production and release is regulated by HDAC5. In fact, HDAC5 chromatin immunoprecipitation assays revealed interactions of HDAC5 within the first 2 kb of the Il6 promoter and a substantially increased acetylation status of the Il6 promoter as assessed by AcH3K9-ChIP, indicating a role of HDAC5 in the epigenetic regulation of Il6 transcription.
Several regulatory response elements of the Il6 promoter are located in a ∼1000 bp segment of the 5’ flanking region in human and murine Il6 genes, including glucocorticoid response elements (GREs) and binding sites for activator protein 1 (AP-1), cyclic AMP (cAMP)-response element (CRE), CRE-binding protein (CREB), NF-IL-6, and NF-kB [36]. Most of the HDAC5-dependent promotor activity mapped to the -1kb region in our luciferase assays, indicating that this region is also subject to chromatin modulation through acetylation in muscle cells. Interestingly, in HL-60 promyelocytic cells, class I- and II-specific HDAC inhibitor trichostatin A had no effect on Il6 promoter accessibility, indicating tissue specificity in the regulation of chromatin in the Il6 gene [37]. IL-6 is also produced by immune cells such as monocytes and macrophages [38]. Consistent with our findings, a recent study reported increased AcH3K9 and H3K4me3 histone activation markers in the Il6 promoter region in macrophages after treatment with Scriptaid, an inhibitor of class IIa HDAC co–repressor complex formation [39]. However, while these data indicate epigenetic regulation of Il6 via histone acetylation in macrophages, the specific HDAC isoform in this cell type was not identified.
HDAC5 has been implicated in the regulation of Slc2a4 expression in skeletal muscle in particular in response to exercise, where nuclear localization of HDAC5 decreases the expression of GLUT4 [10,17]. Consistent with these findings, the HDAC5 KD myotubes exhibited elevated mRNA expression of Slc2a4, a higher amount of GLUT4 protein, and elevated insulin-stimulated glycogen synthesis. Moreover, intact isolated skeletal muscle from the HDAC5 KO mice demonstrated higher insulin-stimulated glucose uptake, indicating that HDAC5 negatively regulated Slc2a4 expression. Similar to IL-6, GLUT4 levels in muscle are upregulated in response to contraction and exercise [40,41]. As IL-6 has been found to induce translocation of GLUT4 via an AMPK-dependent pathway [22], HDAC5 may contribute to the beneficial effects of exercise on glycemia through two different mechanisms, transcriptional control in the nucleus and HDAC5-dependent IL-6 signaling to elevate glucose uptake in muscle in response to exercise. However, additional stimuli including electromyogenic, contractile, neuronal, and/or metabolic signals modulate the expression of GLUT4 through several transcription factors including MEF2, MyoD myogenic proteins, thyroid hormone receptors, Kruppel-like factor KLF15, NF1, and others [5,42].
Consistent with our previous report, the HDAC5 KO mice had higher fat mass and a trend of higher body weight without changes in lean mass than the WT littermates [15]. Both the WT animals and HDAC5 KO mice responded equally to the treadmill training with increased endurance capacity, whereas body weight was mildly reduced only in the WT mice. Interestingly, blockade of IL-6 receptor signaling by monoclonal antibodies was reported to abolish the beneficial effects of endurance exercise in reducing visceral fat mass in obese human subjects [43]. Since we did not observe significant alterations in fat mass after exercise training despite increased IL-6 signaling in the HDAC5KO mice, we presume that the fat mass-reducing effect of IL-6 is augmented more in the obese state with a higher proportion of visceral fat. Importantly, exercise training improved glucose tolerance and marginally affected insulin tolerance in the HDAC5 KO mice compared to the WT littermates, emphasizing the important function of HDAC5 on glycemic control in response to exercise.
We further investigated HDAC5- and contraction-dependent regulation of IL-6 and GLUT4 in the myotubes by applying EPS in vitro. In line with previous reports [[44], [45], [46]], EPS substantially increased transcription of Il6 and secretion of the hormone into the medium, thus validating the system. Consistent with previous studies, we also found increased phosphorylation of AKT in the C2C12 myotubes in response to EPS that has been attributed to activation of Rac1 [47,48]. However, further experiments are required to investigate the respective mechanism.
Compared to the WT cells, the HDAC5 KD myotubes had strongly elevated IL-6 expression and secretion in both the non-stimulated state and in response to EPS-induced contraction that was associated with reduced interaction of HDAC5 with Il6 promotor and increased AcH3K9 histone activation markers in the respective regions. Analogous effects regarding promotor association and AcH3K9 histone acetylation were observed for Slc2a4, strongly indicating that contraction-induced activation of IL-6 and GLUT4 production are mediated through the same epigenetic mechanism that alleviates inhibitory activity of HDAC5 in the respective promotor region.
Previous studies have shown that the exercise-dependent increase in GLUT4 expression requires binding of MEF2 to Slc2a4 promotor [49,50]. In the basal state, MEF2 is associated with class IIa HDAC5. Studies have shown that exercise may disrupt MEF/HDAC5 complexes and cause nuclear exclusion of the deacetylase, subsequently allowing activation of Slc2a4 transcription by MEF2 [10,51,52]. Phosphorylation of HDAC5, presumably by AMPK and CaMPKII, has been suggested to promote 14-3-3-protein-dependent nuclear exclusion of HDAC5 [11,53]. However, in skeletal muscle of exercise-trained AMPKα2 knockout mice, nuclear exclusion of HDAC5 was only partially reduced and training-induced increases in GLUT4 were normal in AMPKα2-KO muscles despite reduced AMPK signaling [54]. As AcH3K9 histone activation markers in Slc2a4 promotor are almost absent in both basal and contracted HDAC5 KD myotubes, it is unlikely that loss of HDAC5 is compensated by other class II deacetylases, suggesting a role of other pathways in regulating training-induced activation of Slc2a4 promoter activity. Similarly, additional activating factors conferring contraction-induced activation of Il6 promoter remain to be identified.
IL-6 can elicit either beneficial or deleterious responses to insulin signaling [55]. The resulting IL-6 phenotype may depend on several factors that include source, dose, duration of exposure, cell type, and potential interactions with soluble or cell surface IL-6 receptors. Induction of Il6 expression in skeletal muscle has been associated with enhanced glucose uptake, presumably due to an activation of the phosphatidylinositol 3-kinase (PI3K) pathway, as several reports identified enhanced PI3K/AKT signaling following stimulation with IL-6 in several cell types and reversal by PI3K inhibitors [[56], [57], [58], [59]]. Indeed, network analysis of overlapping canonical pathways indicated transcriptional activation of the PI3K pathway in the skeletal muscle of the HDAC5 KO mice (Supplementary Fig. S2). In the HDAC5 KD myotubes, we found elevated phosphorylation of AKT in both the basal and insulin-stimulated state, indicative of augmented insulin signaling and in line with increased glucose uptake and glycogen synthesis in the knockdown cells. Knockdown of Il6 reversed this effect in both the WT and HDAC5 KD myotubes, whereas stimulation of the cells with recombinant IL-6 resulted in STAT3 activation but not phosphorylation and activation of AKT. Thus, Hdac5 deficiency may increase AKT phosphorylation through pathways independent of IL-6 signaling. HDAC5 deficiency may lead to activation of further signaling pathways that could activate AKT (Supplementary Table 1).
HDAC inhibitors have been implicated to improve insulin resistance and type 2 diabetes in both preclinical and clinical settings [60], and isoform-specific compounds might be utilized to further study HDAC5-dependent pathways or even mimic the beneficial effects of exercise in skeletal muscle [61].
In conclusion, we identified HDAC5 as a negative epigenetic regulator of Il6 and Slc2a4 promoter transactivation and a gatekeeper for the systemic release of IL6 from skeletal muscle. Improved systemic benefits of exercise on glucose tolerance in HDAC5 KO mice were paralleled by diminished HDAC5 binding to Il6 and Slc2a4 promotor following muscle contraction in an ex vivo model. Overall, our data support a model whereby HDAC5 acts as an epigenetic suppressor of skeletal muscle glucose utilization that acts via the transcriptional blockage of Il6 and Slc2a4 expression. Exercise appears to diminish HDAC5 activity, thereby unleashing glucose-utilizing pathways controlled by insulin and/or IL-6.
Author contributions
Oleksiy Klymenko: Methodology, investigation, formal analysis, visualization, and writing original draft. Tim Brecklinghaus: Methodology and investigation. Matthias Dille: Investigation. Christian Springer: Investigation. Christian de Wendt: Investigation. Delsi Altenhofen: Investigation. Christian Binsch: Investigation. Birgit Knebel: Investigation and data analysis. Jürgen Scheller: Writing, review, and editing. Christopher Hardt: Investigation and data analysis. Ralf Herwig: Investigation and data analysis. Alexandra Chadt: Writing, review, editing, visualization, and project administration. Paul T. Pfluger: Conceptualization, funding, writing, review, and editing. Hadi Al-Hasani: Conceptualization, writing original draft, review, editing, visualization, supervision, and funding acquisition. Dhiraj G. Kabra: Conceptualization, supervision, formal analysis, funding and animal protocol acquisition, writing original draft, review, and editing.
Acknowledgments
The authors thank Andrea Cramer, Sylvia Jacob, Cornelia Köllmer, and Denise Schauer for expert technical assistance. We also thank Erik Olson for generously providing the Hdac5 knockout mice. This study was supported in part by the Federal Ministry of Education and Research, the Ministry of Science and Research of the State North Rhine-Westphalia (MIWF NRW), and a grant from the German Center for Diabetes Research (DZD) (to D.G.K. and P.T.P.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101062.
Conflict of interest
The authors declare no conflicts of interest associated with this manuscript.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.DeFronzo R.A., Ferrannini E., Groop L., Henry R.R., Herman W.H., Holst J.J. Type 2 diabetes mellitus. Nature Reviews Discovery Primers. 2015;1:15019. doi: 10.1038/nrdp.2015.19. [DOI] [PubMed] [Google Scholar]
- 2.Mattila T.K., de Boer A. Influence of intensive versus conventional glucose control on microvascular and macrovascular complications in type 1 and 2 diabetes mellitus. Drugs. 2010;70(17):2229–2245. doi: 10.2165/11585220-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 3.Kitasato L., Tojo T., Hatakeyama Y., Kameda R., Hashikata T., Yamaoka-Tojo M. Postprandial hyperglycemia and endothelial function in type 2 diabetes: focus on mitiglinide. Cardiovascular Diabetology. 2012;11:79. doi: 10.1186/1475-2840-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stehouwer C.D.A. Microvascular dysfunction and hyperglycemia: a vicious cycle with widespread consequences. Diabetes. 2018;67(9):1729–1741. doi: 10.2337/dbi17-0044. [DOI] [PubMed] [Google Scholar]
- 5.Richter E.A., Hargreaves M. Exercise, GLUT4, and skeletal muscle glucose uptake. Physiological Reviews. 2013;93(3):993–1017. doi: 10.1152/physrev.00038.2012. [DOI] [PubMed] [Google Scholar]
- 6.Cushman S.W., Goodyear L.J., Pilch P.F., Ralston E., Galbo H., Ploug T. Molecular mechanisms involved in GLUT4 translocation in muscle during insulin and contraction stimulation. Advances in Experimental Medicine & Biology. 1998;441:63–71. doi: 10.1007/978-1-4899-1928-1_6. [DOI] [PubMed] [Google Scholar]
- 7.Watson R.T., Pessin J.E. Bridging the GAP between insulin signaling and GLUT4 translocation. Trends in Biochemical Sciences. 2006;31(4):215–222. doi: 10.1016/j.tibs.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Petersen M.C., Shulman G.I. Mechanisms of insulin action and insulin resistance. Physiological Reviews. 2018;98(4):2133–2223. doi: 10.1152/physrev.00063.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park S.Y., Kim J.S. A short guide to histone deacetylases including recent progress on class II enzymes. Experimental & Molecular Medicine. 2020;52(2):204–212. doi: 10.1038/s12276-020-0382-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGee S.L., Hargreaves M. Exercise and myocyte enhancer factor 2 regulation in human skeletal muscle. Diabetes. 2004;53(5):1208–1214. doi: 10.2337/diabetes.53.5.1208. [DOI] [PubMed] [Google Scholar]
- 11.McGee S.L., van Denderen B.J., Howlett K.F., Mollica J., Schertzer J.D., Kemp B.E. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57(4):860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 12.Raichur S., Teh S.H., Ohwaki K., Gaur V., Long Y.C., Hargreaves M. Histone deacetylase 5 regulates glucose uptake and insulin action in muscle cells. Journal of Molecular Endocrinology. 2012;49(3):203–211. doi: 10.1530/JME-12-0095. [DOI] [PubMed] [Google Scholar]
- 13.Hu H., Li L., Wang C., He H., Mao K., Ma X. 4-Phenylbutyric acid increases GLUT4 gene expression through suppression of HDAC5 but not endoplasmic reticulum stress. Cellular Physiology and Biochemistry. 2014;33(6):1899–1910. doi: 10.1159/000362967. [DOI] [PubMed] [Google Scholar]
- 14.Mihaylova M.M., Vasquez D.S., Ravnskjaer K., Denechaud P.D., Yu R.T., Alvarez J.G. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145(4):607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabra D.G., Pfuhlmann K., Garcia-Caceres C., Schriever S.C., Casquero Garcia V., Kebede A.F. Hypothalamic leptin action is mediated by histone deacetylase 5. Nature Communications. 2016;7:10782. doi: 10.1038/ncomms10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whillier S. Exercise and insulin resistance. Advances in Experimental Medicine & Biology. 2020;1228:137–150. doi: 10.1007/978-981-15-1792-1_9. [DOI] [PubMed] [Google Scholar]
- 17.Niu Y., Wang T., Liu S., Yuan H., Li H., Fu L. Exercise-induced GLUT4 transcription via inactivation of HDAC4/5 in mouse skeletal muscle in an AMPKalpha2-dependent manner. Biochimica et Biophysica Acta - Molecular Basis of Disease. 2017;1863(9):2372–2381. doi: 10.1016/j.bbadis.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Belizario J.E., Fontes-Oliveira C.C., Borges J.P., Kashiabara J.A., Vannier E. Skeletal muscle wasting and renewal: a pivotal role of myokine IL-6. SpringerPlus. 2016;5:619. doi: 10.1186/s40064-016-2197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crescioli C. Targeting age-dependent functional and metabolic decline of human skeletal muscle: the geroprotective role of exercise, myokine IL-6, and vitamin D. International Journal of Molecular Sciences. 2020;21(3) doi: 10.3390/ijms21031010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leal L.G., Lopes M.A., Batista M.L., Jr. Physical exercise-induced myokines and muscle-adipose tissue crosstalk: a review of current knowledge and the implications for health and metabolic diseases. Frontiers in Physiology. 2018;9:1307. doi: 10.3389/fphys.2018.01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huh J.Y. The role of exercise-induced myokines in regulating metabolism. Archives of Pharmacal Research. 2018;41(1):14–29. doi: 10.1007/s12272-017-0994-y. [DOI] [PubMed] [Google Scholar]
- 22.Carey A.L., Steinberg G.R., Macaulay S.L., Thomas W.G., Holmes A.G., Ramm G. Interleukin-6 increases insulin-stimulated glucose disposal in humans and glucose uptake and fatty acid oxidation in vitro via AMP-activated protein kinase. Diabetes. 2006;55(10):2688–2697. doi: 10.2337/db05-1404. [DOI] [PubMed] [Google Scholar]
- 23.Jiang L.Q., Duque-Guimaraes D.E., Machado U.F., Zierath J.R., Krook A. Altered response of skeletal muscle to IL-6 in type 2 diabetic patients. Diabetes. 2013;62(2):355–361. doi: 10.2337/db11-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Khalili L., Bouzakri K., Glund S., Lonnqvist F., Koistinen H.A., Krook A. Signaling specificity of interleukin-6 action on glucose and lipid metabolism in skeletal muscle. Molecular Endocrinology. 2006;20(12):3364–3375. doi: 10.1210/me.2005-0490. [DOI] [PubMed] [Google Scholar]
- 25.Chang S., McKinsey T.A., Zhang C.L., Richardson J.A., Hill J.A., Olson E.N. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Molecular and Cellular Biology. 2004;24(19):8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambernd S., Taube A., Schober A., Platzbecker B., Gorgens S.W., Schlich R. Contractile activity of human skeletal muscle cells prevents insulin resistance by inhibiting pro-inflammatory signalling pathways. Diabetologia. 2012;55(4):1128–1139. doi: 10.1007/s00125-012-2454-z. [DOI] [PubMed] [Google Scholar]
- 27.Knebel B., Hartwig S., Jacob S., Kettel U., Schiller M., Passlack W. Inactivation of SREBP-1a phosphorylation prevents fatty liver disease in mice: identification of related signaling pathways by gene expression profiles in liver and proteomes of peroxisomes. International Journal of Molecular Sciences. 2018;19(4) doi: 10.3390/ijms19040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herwig R., Hardt C., Lienhard M., Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nature Protocols. 2016;11(10):1889–1907. doi: 10.1038/nprot.2016.117. [DOI] [PubMed] [Google Scholar]
- 29.Jelenik T., Dille M., Muller-Luhlhoff S., Kabra D.G., Zhou Z., Binsch C. FGF21 regulates insulin sensitivity following long-term chronic stress. Molecular Metabolism. 2018;16:126–138. doi: 10.1016/j.molmet.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klymenko O., Huehn M., Wilhelm J., Wasnick R., Shalashova I., Ruppert C. Regulation and role of the ER stress transcription factor CHOP in alveolar epithelial type-II cells. Journal of Molecular Medicine (Berlin) 2019;97(7):973–990. doi: 10.1007/s00109-019-01787-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 32.Zhong Z., Wen Z., Darnell J.E., Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264(5155):95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 33.McGee S.L., Hargreaves M. Histone modifications and skeletal muscle metabolic gene expression. Clinical and Experimental Pharmacology and Physiology. 2010;37(3):392–396. doi: 10.1111/j.1440-1681.2009.05311.x. [DOI] [PubMed] [Google Scholar]
- 34.McGee S.L., Fairlie E., Garnham A.P., Hargreaves M. Exercise-induced histone modifications in human skeletal muscle. Journal of Physiology. 2009;587(Pt 24):5951–5958. doi: 10.1113/jphysiol.2009.181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaur V., Connor T., Sanigorski A., Martin S.D., Bruce C.R., Henstridge D.C. Disruption of the class IIa HDAC corepressor complex increases energy expenditure and lipid oxidation. Cell Reports. 2016;16(11):2802–2810. doi: 10.1016/j.celrep.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Luo Y., Zheng S.G. Hall of fame among pro-inflammatory cytokines: interleukin-6 gene and its transcriptional regulation mechanisms. Frontiers in Immunology. 2016;7:604. doi: 10.3389/fimmu.2016.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poplutz M.K., Wessels I., Rink L., Uciechowski P. Regulation of the Interleukin-6 gene expression during monocytic differentiation of HL-60 cells by chromatin remodeling and methylation. Immunobiology. 2014;219(8):619–626. doi: 10.1016/j.imbio.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka T., Narazaki M., Masuda K., Kishimoto T. Regulation of IL-6 in immunity and diseases. Advances in Experimental Medicine & Biology. 2016;941:79–88. doi: 10.1007/978-94-024-0921-5_4. [DOI] [PubMed] [Google Scholar]
- 39.Hu L., Yu Y., Huang H., Fan H., Hu L., Yin C. Epigenetic regulation of interleukin 6 by histone acetylation in macrophages and its role in paraquat-induced pulmonary fibrosis. Frontiers in Immunology. 2016;7:696. doi: 10.3389/fimmu.2016.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren J.M., Semenkovich C.F., Gulve E.A., Gao J., Holloszy J.O. Exercise induces rapid increases in GLUT4 expression, glucose transport capacity, and insulin-stimulated glycogen storage in muscle. Journal of Biological Chemistry. 1994;269(20):14396–14401. [PubMed] [Google Scholar]
- 41.Hiscock N., Chan M.H., Bisucci T., Darby I.A., Febbraio M.A. Skeletal myocytes are a source of interleukin-6 mRNA expression and protein release during contraction: evidence of fiber type specificity. The FASEB Journal. 2004;18(9):992–994. doi: 10.1096/fj.03-1259fje. [DOI] [PubMed] [Google Scholar]
- 42.Zorzano A., Palacin M., Guma A. Mechanisms regulating GLUT4 glucose transporter expression and glucose transport in skeletal muscle. Acta Physiologica Scandinavica. 2005;183(1):43–58. doi: 10.1111/j.1365-201X.2004.01380.x. [DOI] [PubMed] [Google Scholar]
- 43.Wedell-Neergaard A.S., Lang Lehrskov L., Christensen R.H., Legaard G.E., Dorph E., Larsen M.K. Exercise-induced changes in visceral adipose tissue mass are regulated by IL-6 signaling: a randomized controlled trial. Cell Metabolism. 2019;29(4):844–855. doi: 10.1016/j.cmet.2018.12.007. e843. [DOI] [PubMed] [Google Scholar]
- 44.Nedachi T., Fujita H., Kanzaki M. Contractile C2C12 myotube model for studying exercise-inducible responses in skeletal muscle. American Journal of Physiology. Endocrinology and Metabolism. 2008;295(5):E1191–E1204. doi: 10.1152/ajpendo.90280.2008. [DOI] [PubMed] [Google Scholar]
- 45.Farmawati A., Kitajima Y., Nedachi T., Sato M., Kanzaki M., Nagatomi R. Characterization of contraction-induced IL-6 up-regulation using contractile C2C12 myotubes. Endocrine Journal. 2013;60(2):137–147. doi: 10.1507/endocrj.ej12-0316. [DOI] [PubMed] [Google Scholar]
- 46.Chen W., Nyasha M.R., Koide M., Tsuchiya M., Suzuki N., Hagiwara Y. In vitro exercise model using contractile human and mouse hybrid myotubes. Scientific Reports. 2019;9(1):11914. doi: 10.1038/s41598-019-48316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nieuwoudt S., Mulya A., Fealy C.E., Martelli E., Dasarathy S., Naga Prasad S.V. In vitro contraction protects against palmitate-induced insulin resistance in C2C12 myotubes. American Journal of Physiology - Cell Physiology. 2017;313(5):C575–C583. doi: 10.1152/ajpcell.00123.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu F., Li N., Li Z., Zhang C., Yue Y., Liu Q. Electrical pulse stimulation induces GLUT4 translocation in a Rac-Akt-dependent manner in C2C12 myotubes. FEBS Letters. 2018;592(4):644–654. doi: 10.1002/1873-3468.12982. [DOI] [PubMed] [Google Scholar]
- 49.McGee S.L., Sparling D., Olson A.L., Hargreaves M. Exercise increases MEF2- and GEF DNA-binding activity in human skeletal muscle. The FASEB Journal. 2006;20(2):348–349. doi: 10.1096/fj.05-4671fje. [DOI] [PubMed] [Google Scholar]
- 50.Smith J.A., Collins M., Grobler L.A., Magee C.J., Ojuka E.O. Exercise and CaMK activation both increase the binding of MEF2A to the Glut4 promoter in skeletal muscle in vivo. American Journal of Physiology. Endocrinology and Metabolism. 2007;292(2):E413–E420. doi: 10.1152/ajpendo.00142.2006. [DOI] [PubMed] [Google Scholar]
- 51.Han A., He J., Wu Y., Liu J.O., Chen L. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. Journal of Molecular Biology. 2005;345(1):91–102. doi: 10.1016/j.jmb.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 52.McKinsey T.A., Zhang C.L., Lu J., Olson E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408(6808):106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grozinger C.M., Schreiber S.L. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proceedings of the National Academy of Sciences of the U S A. 2000;97(14):7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong H., Xie J., Zhang N., Yao L., Zhang Y. MEF2A binding to the Glut4 promoter occurs via an AMPKalpha2-dependent mechanism. Medicine & Science in Sports & Exercise. 2011;43(8):1441–1450. doi: 10.1249/MSS.0b013e31820f6093. [DOI] [PubMed] [Google Scholar]
- 55.Kim J.H., Bachmann R.A., Chen J. Interleukin-6 and insulin resistance. Vitamins & Hormones. 2009;80:613–633. doi: 10.1016/S0083-6729(08)00621-3. [DOI] [PubMed] [Google Scholar]
- 56.Chew G.S., Myers S., Shu-Chien A.C., Muhammad T.S. Interleukin-6 inhibition of peroxisome proliferator-activated receptor alpha expression is mediated by JAK2- and PI3K-induced STAT1/3 in HepG2 hepatocyte cells. Molecular and Cellular Biochemistry. 2014;388(1–2):25–37. doi: 10.1007/s11010-013-1896-z. [DOI] [PubMed] [Google Scholar]
- 57.Wegiel B., Bjartell A., Culig Z., Persson J.L. Interleukin-6 activates PI3K/Akt pathway and regulates cyclin A1 to promote prostate cancer cell survival. International Journal of Cancer. 2008;122(7):1521–1529. doi: 10.1002/ijc.23261. [DOI] [PubMed] [Google Scholar]
- 58.Nishikai-Yan Shen T., Kanazawa S., Kado M., Okada K., Luo L., Hayashi A. Interleukin-6 stimulates Akt and p38 MAPK phosphorylation and fibroblast migration in non-diabetic but not diabetic mice. PloS One. 2017;12(5) doi: 10.1371/journal.pone.0178232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J., Li Y., Shen B. PI3-K/Akt pathway contributes to IL-6-dependent growth of 7TD1 cells. Cancer Cell International. 2003;3(1):1. doi: 10.1186/1475-2867-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma S., Taliyan R. Histone deacetylase inhibitors: future therapeutics for insulin resistance and type 2 diabetes. Pharmacological Research. 2016;113(Pt A):320–326. doi: 10.1016/j.phrs.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 61.Marks P.A. The clinical development of histone deacetylase inhibitors as targeted anticancer drugs. Expert Opinion on Investigational Drugs. 2010;19(9):1049–1066. doi: 10.1517/13543784.2010.510514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.