Abstract
Survival of all living organisms requires the ability to detect attack and swiftly counter with protective immune responses. Despite considerable mechanistic advances, interconnectivity of signaling modules often remains unclear. A newly-characterized protein, IMMUNOREGULATORY RNA-BINDING PROTEIN (IRR), negatively regulates immune responses in both maize and Arabidopsis, with disrupted function resulting in enhanced disease resistance. IRR associates with, and promotes canonical splicing of, transcripts encoding defense signaling proteins, including the key negative regulator of pattern recognition receptor signaling complexes, CALCIUM-DEPENDENT PROTEIN KINASE 28 (CPK28). Upon immune activation by Plant Elicitor Peptides (Peps), IRR is dephosphorylated, disrupting interaction with CPK28 transcripts and resulting in accumulation of an alternative splice variant encoding a truncated CPK28 protein with impaired kinase activity and diminished function as a negative regulator. We demonstrate a novel mechanism linking Pep-induced post-translational modification of IRR with post-transcriptionally-mediated attenuation of CPK28 function to dynamically amplify Pep signaling and immune output.
One Sentence Summary
Plant innate immunity is promoted by post-translational modification of a novel RNA-binding protein that regulates alternative splicing of transcripts encoding defense signaling proteins to dynamically increase immune signaling capacity through deactivation of a key signal-buffering system.
Introduction
Innate immunity has been described as a double-edged sword, providing essential protection in the face of attack, but detrimental to the host if allowed to persist1. Accordingly, immune responses are tightly regulated by complex layers of dynamic checks and balances. Rapid and reversible modulation of the signaling components requisite for this dynamic regulation commonly occurs through posttranscriptional, translational and posttranslational mechanisms without requiring de novo transcription2–9. Collectively these mechanisms rapidly modulate signaling pathways through stochiometric changes in the relative quantity or functional state of key regulators. However, elucidation of how these regulatory layers assemble into defined modules, particularly with respect to temporal dynamics and cause-effect relationships, remains challenging.
Pattern-recognition receptors (PRRs) recognize microbe-, herbivore-, and parasitic plant-associated molecular patterns, as well as endogenous elicitors, to activate regulatory signaling networks that promote innate immune responses as a first line of defense10, 11. Following initiation of pattern-triggered immunity (PTI), endogenous plant hormones and messenger signals amplify initial inputs to coordinate immune outputs11. Among these, Plant Elicitor Peptides (Peps) have emerged as fundamental regulators of innate immunity across higher plants, with demonstrated ability to enhance resistance to a broad spectrum of insects, pathogens and nematodes in diverse species12–18. Peps are proteolytically released from PROPEP precursor proteins by the cysteine protease METACASPASE4 and activate PEP RECEPTORs (PEPRs) to coordinate downstream signals that trigger plant immune responses mediating resistance12, 19–24. Signaling by PEPRs requires SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) coreceptors and the plasma membrane-associated kinase BOTRYTIS-INDUCED KINASE 1 (BIK1)25–29. Acting as a positive regulator of both reactive oxygen species- and WRKY transcription factor-mediated downstream responses, BIK1 is rate-limiting for signaling through pattern recognition receptor complexes, including PEPRs, and is continuously turned-over to maintain signaling homeostasis30–32. The E3 ligases PLANT U-BOX PROTEINs PUB25 and PUB26 facilitate turnover by ubiquitylating BIK133. Phosphorylation of conserved residues in BIK1 and in PUB25/PUB26 by CALCIUM-DEPENDENT PROTEIN KINASE 28 (CPK28) enhances ubiquitin ligase activity and promotes BIK1 degradation. Through this mechanism, CPK28 negatively regulates immune receptor signaling by promoting BIK1 turnover32, 33.
Here we define a novel dynamic regulatory mechanism acting on the CPK28 buffering system mediated by a newly-described RNA-binding protein, termed IMMUNOREGULATORY RNA-BINDING PROTEIN (IRR). In the absence of immune challenge, IRR associates with CPK28 transcripts to promote canonical splicing into mRNA encoding full-length, functional proteins. Upon activation of PEPRs, IRR is transiently dephosphorylated, causing dissociation from CPK28 transcripts. Disruption of IRR interaction with CPK28 transcripts leads to increased levels of a retained-intron variant encoding a truncated protein that lacks EF-hand domains required for calcium-induced stimulation of kinase activity and exhibiting reduced functionality. Altered ratios of canonical versus retained-intron CPK28 transcripts modulate PEPR signaling sensitivity, with proportional increases in the retained intron variant resulting in amplified immune signaling and defense. These dynamic events directly link PEPR-induced dephosphorylation of IRR with post-transcriptionally-mediated attenuation of CPK28 function, providing a rapid mechanism to modulate PEPR signaling capacity and immune outputs.
Results
Pep signaling promotes IRR dephosphorylation.
To uncover post-translationally-modified regulators of immunity, we examined rapid Pep-induced protein phosphorylation changes in both Arabidopsis and maize. Suspension-cultured cells from each species were treated for 10 min with 100 nM Arabidopsis thaliana Pep1 (AtPep1) or Zea mays Pep3 (ZmPep3) respectively, and analyzed in comparison to water-treated controls using nano-liquid chromatography with tandem mass spectrometry (LC-MS/MS)34. In both species, a predicted RNA-binding protein containing an RNA Recognition Motif (RRM), IRR, was significantly dephosphorylated after Pep treatment (Supplementary Fig. 1), indicating a possible regulatory role in Pep-induced signal transduction across species. Encoded by At3g23900 and GRMZM2G132936 genes in Arabidopsis and maize respectively, IRR contains zinc finger and RRM domains with a carboxyl-terminal region highly enriched in serine/arginine (SR) dipeptides (Fig. 1a).
Figure 1. AtPep1 affects phosphorylation of IRR in a time- and concentration-dependent manner.
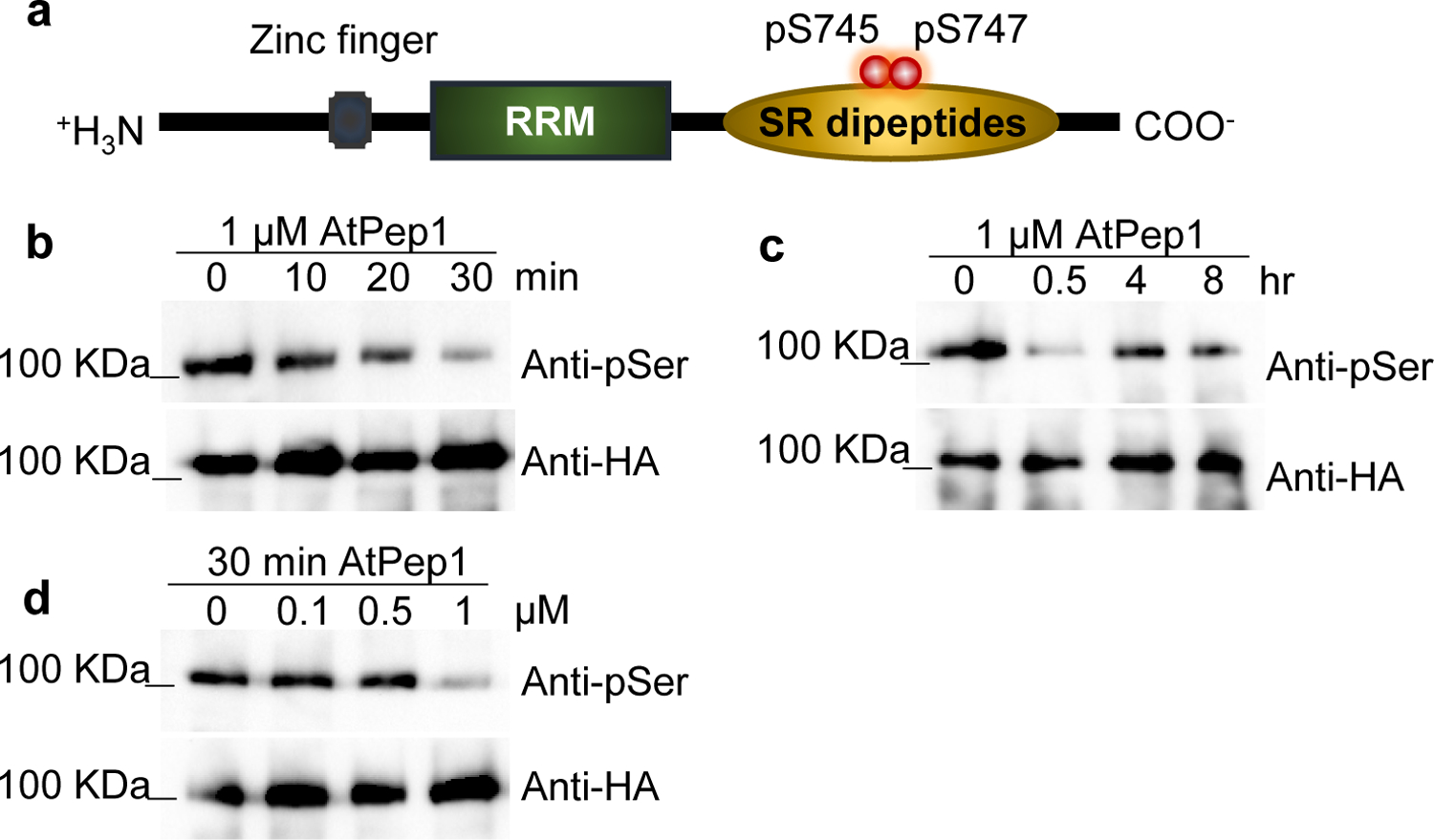
a, IRR phosphorylation sites affected by AtPep1 treatment. b, p35S:IRR-3xHA plants were treated with 1 μM solutions of AtPep1 for 0, 10, 20 and 30 min, or (c) 4 and 8 h. d, p35S:IRR-3xHA plants were treated with 0, 0.1, 0.5 and 1 μM solutions of AtPep1 for 30 min. IRR protein was subjected to immunoblot analysis with anti-phospho-Ser and anti-HA antibodies. Experiments in b–d were repeated three times independently, with similar results.
To confirm Pep-induced dephosphorylation of IRR in planta, Arabidopsis lines expressing an HA-tagged fusion of IRR (p35S:IRR-3xHA) were treated with AtPep1, followed by IRR immunoprecipitation and anti-phosphoserine antibody immunoblot analyses of phosphorylation state. Decreased IRR phosphorylation was observed within 30 min of AtPep1 treatment (Fig. 1b), while control anti-HA western blots demonstrated no change in total IRR protein levels. AtPep1-induced dephosphorylation was transient, with a recovery to resting levels of IRR phosphorylation within 4 h (Fig. 1c). Decreased IRR phosphorylation also occurred in a concentration-dependent manner (Fig. 1d). To confirm that phosphorylation detected by western blot corresponded to the serine residues observed in the initial phosphoproteomic study, S745 and S747, phosphoabolishing alanine substitutions at these positions were generated. Compared to the wild-type (Wt) IRR fusion protein, the phosphoabolishing variant (IRRS745A,S747A-YFP) showed constitutively decreased phosphorylation (Supplementary Fig. 2). To determine whether AtPep1-induced dephosphorylation was specific to IRR, phosphorylation of an unrelated serine/arginine (SR)-rich RRM protein previously reported as a phosphoprotein35, SR45, was examined after Pep treatment using plants expressing an SR45–3xHA fusion protein (Supplementary Fig. 3, 4). SR45 negatively regulates glucose and abscisic acid signaling during early seedling development, and recent transcriptional profiling studies have implicated it as a negative regulator of innate immunity36, 37. The phosphorylation levels of SR45 protein were unaltered by AtPep1 treatment (Supplementary Fig. 4), demonstrating that AtPep1-mediated dephosphorylation is not a general phenomenon in SR-rich RRM proteins.
Loss of function irr mutants display enhanced Pep-induced immune responses.
To examine effects of IRR loss-of-function on AtPep1 response output, two T-DNA insertional knockout mutants of IRR (SALK_015201, termed irr-1, and SALK_066572, termed irr-2) were analyzed for altered sensitivity to AtPep1 treatment in comparison to an SR45 insertional mutant line (SALK_123442) as a control. Presence of T-DNA insertions and absence of target gene expression was confirmed for all lines by PCR (Supplementary Fig. 5). Under normal growth conditions in soil or on tissue culture medium, both irr knockout lines were developmentally and morphologically indistinguishable from wild-type. However, both irr knockout lines demonstrated visible growth differences in the presence of AtPep1, which inhibits primary root elongation in Arabidopsis seedlings22, 24. Wild-type Col-0 (Wt) seedlings grown in media supplemented with 0.1 μM AtPep1 have primary roots approximately half the length of seedlings grown on water-supplemented medium (Fig. 2a), while the PEPR double knockout line, pepr1/pepr2, which is fully insensitive to AtPeps, demonstrates no root growth inhibition after AtPep1 treatment. In contrast, primary roots of irr1–1 and irr1–2 were significantly shorter than Wt roots when grown on medium supplemented with both 0.1 μM and 1 μM AtPep1, indicating hypersensitivity to the peptide (Fig. 2a, Supplementary Fig. 6a), whereas sr45 primary root growth was indistinguishable from Wt (Supplementary Fig. 6b). This result indicated a potential role for IRR as a negative regulator of AtPep1-induced responses. Additional layers of signaling and output downstream of AtPep1 were investigated, including production of secondary messengers, kinase activation and relative expression of AtPep1-associated marker genes. AtPep1 promotes generation of secondary-messenger reactive oxygen species (ROS) through stimulation of NADPH oxidase activity within minutes of PEPR activation22, 38. While the timing of AtPep1-induced ROS production was the same in both Wt and irr knockouts, the magnitude of ROS generated was greater in irr lines (Fig. 2b, Supplementary Fig. 7a). AtPep1 treatment also stimulates phosphorylation-mediated activation of MAP KINASES (MPK) 3, 6 and 4/11, which are integral to many plant signaling pathways responsive to biotic and abiotic stresses39, 40. MPK3/6/4/11 activation after AtPep1 treatment was probed through western blotting with anti-phospho-p44/42 MAPK antibody, revealing that phosphorylation of MPK3, MPK6 and MPK4/11 were more intense and prolonged in irr knockouts than in Wt, with detectable activity continuing 60 min after Pep treatment (Supplementary Fig. 7b). In correspondence with upregulation of second messenger and MAP kinase activities, expression of the AtPep1-responsive marker genes PLANT DEFENSIN 1.2 (PDF1.2) and PATHOGENESIS-RELATED PROTEIN 1 (PR-1) was also significantly increased 24 h after AtPep1 treatment in irr knockouts as compared to Wt (Fig. 2, c, d)12. Expression of the AtPep1-induced marker gene TYROSINE AMINOTRANSFERASE 3 (TAT3) was also observed as marginally increased in irr mutants, but was not statistically significant (Supplementary Fig. 7c). The irr-1 knockout phenotype was rescued through complementation with the IRR gene driven by its native promoter: two independent lines expressing pIRR:IRR-YFP in the irr-1 background behaved as Wt plants in assays of AtPep1-induced root growth inhibition and ROS production (Supplementary Fig. 8).
Figure 2. IRR mutants are hypersensitive to AtPep1 treatment.
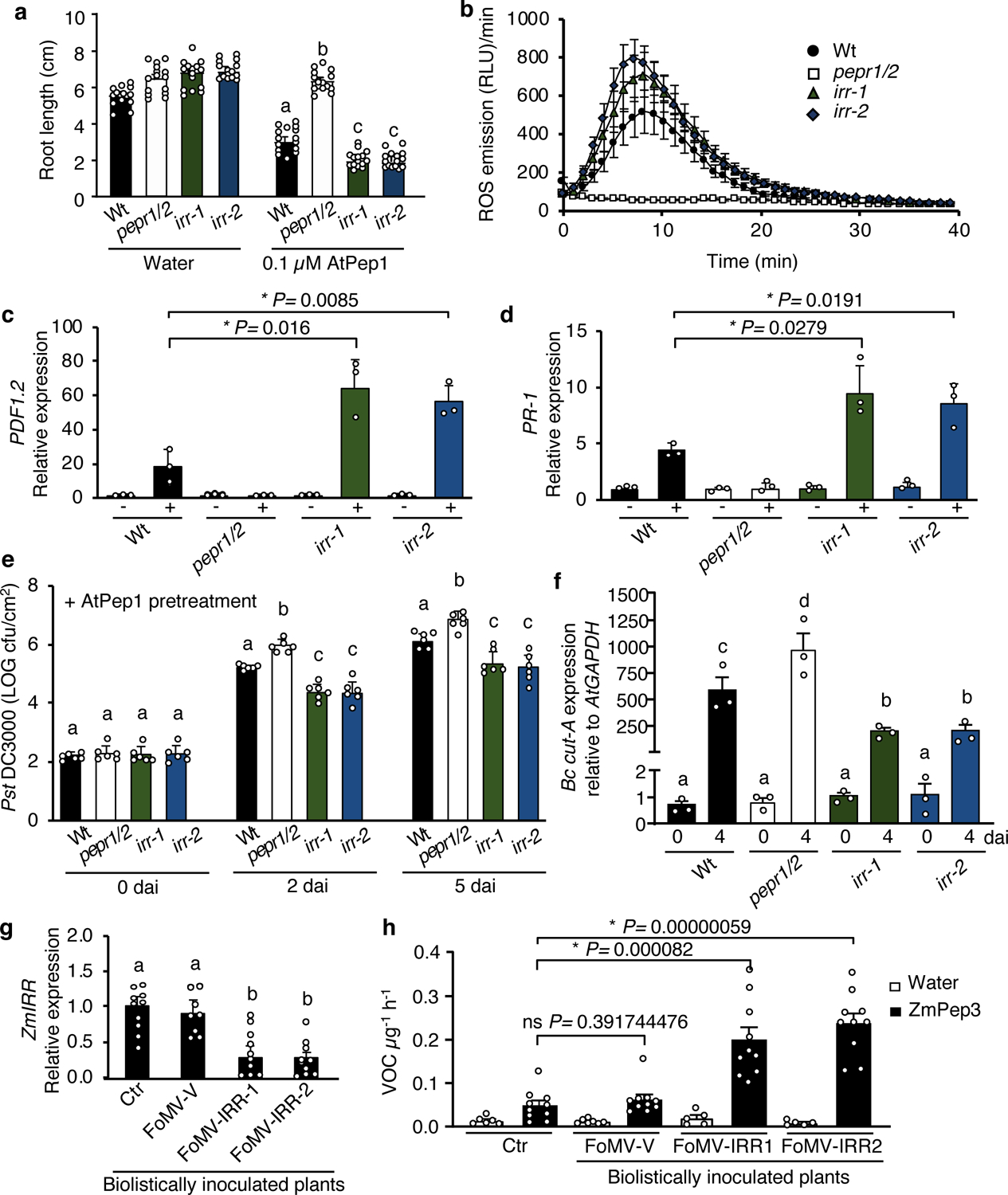
a, Arabidopsis seedlings were treated with water or a solution of 0.1 μM AtPep1, and root length measured after 15 days of treatment. Values represent the mean ± SD of 15 seedlings. b, Total ROS production was registered continuously using luminol fluorescence for 40 min after addition of 0 or 1 μM AtPep1, then summed. The values are the mean ± SEM. c, The relative expression level of AtPep1-inducible genes PDF1.2 and PR-1 (d) was determined by real-time qRT-PCR using mRNA from entire seedlings treated with water (−) and a solution of 1 μM AtPep1 (+) for 24 hours. Values represent the fold-change in expression versus the water-treated wild-type (Wt) samples after normalization against GAPDH expression. Error bars indicate the SD of three biological replicates. e, Pst DC3000 infection assay of wild-type (Wt), pepr1/pepr2 (pepr1/2), irr-1 and irr-2 plants after pretreatment via infiltration with a solution of 1 μM AtPep1 24 hours prior to infection. Bars indicate samples immediately following inoculation (0), or 2 and 5 days after inoculation (dai). Error bars indicate SD, n = 6. f, Botrytis cinerea infection in wild-type, pepr1/pepr2 and irr mutants. Quantification of in planta growth of B. cinerea. qPCR was used to analyze the relative level of B. cinerea Cutinase A genomic DNA (Bc cut-A) compared with Arabidopsis GAPDH (Bc cut-A/AtGAPDH). Error bars indicate SD, n = 15. Experiments in a–f were repeated at least three times independently, with similar results. g, Analysis of ZmIRR gene expression in maize plants carrying Foxtail Mosaic Virus-based viral-induced gene silencing constructs targeting ZmIRR (FoMV-IRR-1 and FoMV-IRR-2) relative to empty viral vectors (FoMV-V) or uninoculated control plants (Ctr). The bar graphs display the relative expression levels of ZmIRR mRNA in leaf 5 as determined by real-time qRT-PCR. Values represent the fold-change in expression versus the uninoculated control (Ctr) samples after normalization against ZmRPL17 expression. h, Total volatile organic compounds (VOCs) from maize leaves inoculated with viral-induced gene silencing constructs targeting ZmIRR (FoMV-IRR-1 and FoMV-IRR-2) relative to empty viral vectors (FoMV-V) or uninoculated control plants (Ctr). VOC were measured 16 hours posttreatment with water or with a 5 μM solution of ZmPep3. All error bars indicate the SD of 5–10 biological samples. Experiments in g and h were repeated two times independently, with similar results. For all graphs, different letters represent significant differences (one-way ANOVA followed by Tukey’s test corrections for multiple comparisons; P < 0.05). The asterisk indicates significant differences using Student t-tests (two-tailed distribution, unpaired), with P < 0.05; ns, not significant.
To assess whether enhanced AtPep1-induced immune responses in irr lines translated to increased disease resistance, plants were challenged with both the hemibiotrophic bacterial pathogen, Pseudomonas syringae pv. tomato DC3000 (Pst DC3000), and the necrotrophic fungal pathogen, Botrytis cinerea. When inoculated with Pst DC3000 by leaf infiltration, bacterial proliferation was similar in both wild-type and irr-1 plants after two and five days (Supplementary Fig. 9a), whereas the pepr1/pepr2 double mutant was more susceptible to Pst DC3000, as previously reported24. In contrast, after inducing immunity through pre-infiltration of leaves with AtPep1 24 h prior to Pst DC3000 inoculation, bacterial proliferation was significantly reduced in irr-1 and irr-2 knockout plants when compared to wild-type (Fig. 2e). Upon challenge with B. cinerea, four days post-inoculation, the average lesion area of irr leaves was smaller than for Wt (Supplementary Fig. 9b). Correspondingly, quantification of fungal proliferation through quantitative RT-PCR analysis of relative ratio of B. cinerea Cutinase A DNA versus A. thaliana GAPDH DNA revealed significantly lower pathogen levels in both irr mutant lines as compared to wild-type (Fig. 2f). To examine whether irr lines were hypersensitive to another peptide elicitor of immune responses, flg22-induced responses were evaluated (Supplementary Fig. 10). Interestingly, irr lines did not display increased flg22-sensitivity as measured by root growth inhibition and ROS accumulation. Furthermore, compared to Wt, irr lines did not have enhanced flg22-induced resistance to Pst DC3000 two days post-inoculation, and showed only marginal decreases in proliferation after five days. Together this indicates that IRR-based negative regulation more strongly affects AtPep1 signaling than flg22, and that IRR may have varying degrees of influence depending on immune input.
To investigate IRR function in maize, a Viral-Induced Gene Silencing (VIGS) system derived from Foxtail Mosaic Virus (FoMV) was deployed to silence ZmIRR (Supplementary Fig. 11a,b)41. Maize var. B73 seedlings were biolistically inoculated with FoMV-derived vectors carrying two different IRR sequence fragments, designated as FoMV-IRR-1 and FoMV-IRR-2. Control plants were inoculated with FoMV vector carrying no insert (FoMV-V). For each FoMV construct, 10 plants confirmed as infected were selected for further analysis. To evaluate relative ZmIRR silencing, ZmIRR transcript levels were compared among leaves of plants infected with FoMV-IRR-1/2, empty vector FoMV-V and uninoculated control plants using qRT-PCR. Expression levels of ZmIRR were similar in uninoculated and FoMV-V-infected maize plants, demonstrating that FoMV-V infection alone did not significantly affect ZmIRR gene expression (Fig. 2g). However, infection with either of the FoMV-IRR constructs significantly reduced ZmIRR expression (Fig. 2g). As irr knockout Arabidopsis mutants are hypersensitive to AtPep1, maize FoMV ZmIRR knockdown plants were tested for sensitivity to ZmPep3. In maize, ZmPep3 is a potent inducer of herbivore-associated volatile organic compounds (VOC) that serve as indirect defenses by recruiting parasitic wasps to attack Lepidopteran herbivore pests feeding on leaves14. VOC emission from FoMV-infected maize leaves after ZmPep3 treatment was measured by gas chromatography, and total VOCs emitted after peptide treatment were significantly higher from maize plants inoculated with FoMV-IRR-1/2 than from control plants (Fig. 2h). A direct comparison of relative ZmIRR expression versus total VOC emission levels in FoMV-IRR-1/2 confirmed that lower ZmIRR expression correlated with higher VOC emission (Supplementary Fig. 11c, d). Together these experiments suggest that IRR functions as a negative regulator of Pep-induced immune responses in both Arabidopsis and maize.
irr knockouts exhibit broad changes in defense gene expression and splicing patterns.
To better understand potential mechanisms underlying irr hypersensitivity to Pep treatment, global transcriptional patterns in irr-1 and wild-type plants were profiled by RNA-seq 24 h post-treatment with either water or 1 μM AtPep1. Water-treated irr-1 plants demonstrated extensive dysregulation of genes relating to the immune response, with over 600 genes differentially regulated in irr-1 compared to wild-type (Supplementary Fig. 12, Supplementary Table 1). Analysis of Gene Ontology (GO) for transcripts with increased basal expression levels in irr-1 revealed significant enrichment of immunity-related terms, with top categories including response to stimulus, defense response, immune system process and programmed cell death (Fig. 3a, Supplementary Table 2).
Figure 3. IRR is implicated in defense and alternative splicing.
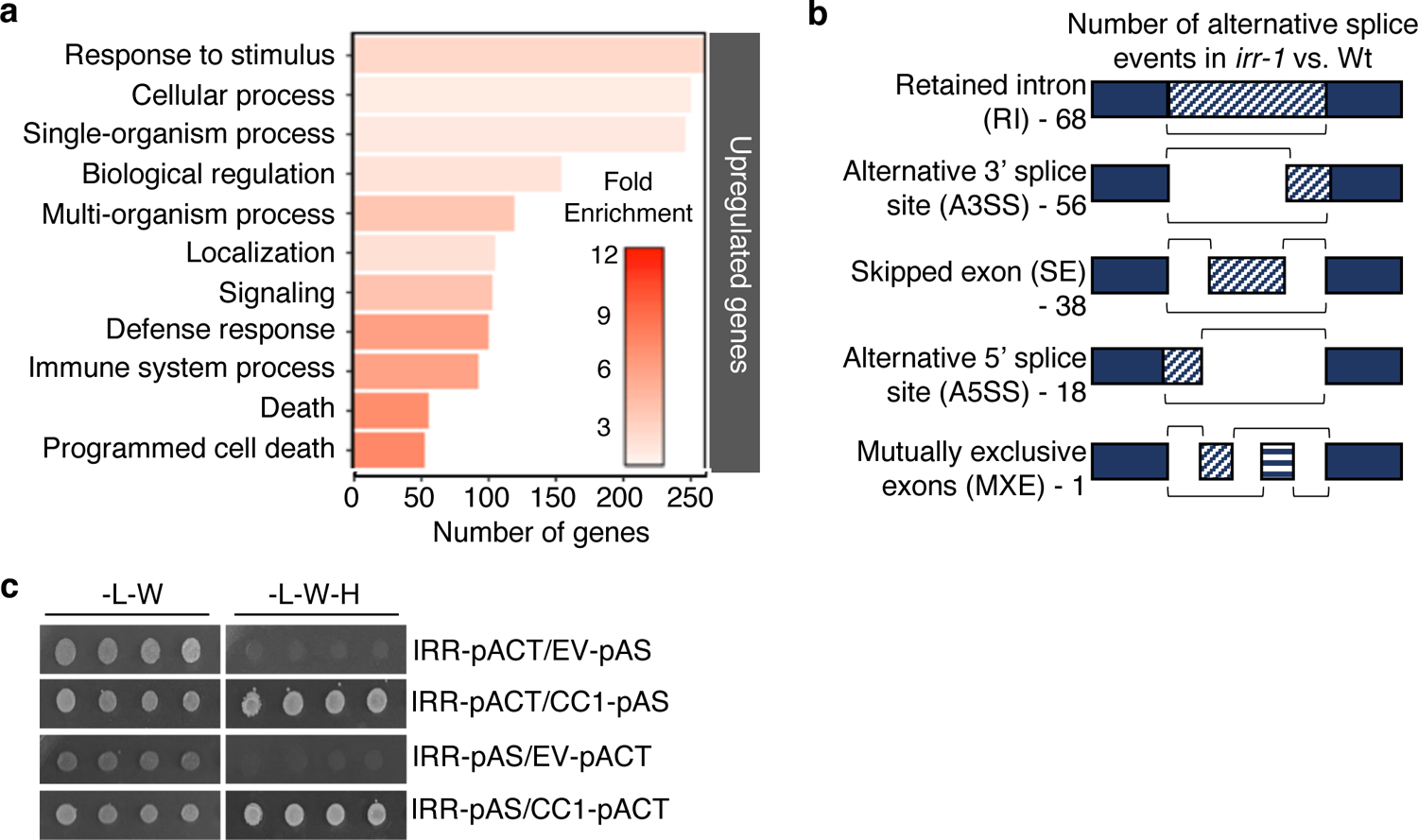
a, Gene Ontology (GO) term distribution of genes for which expression is upregulated in irr-1 versus wild-type (Wt) plants as analyzed through RNA-Seq. b, Number of alternative splice events in irr-1 versus Wt. Three biological replicates for each sample group were analyzed. c, IRR interacts with CC1-splicing factor in yeast. Yeast strain AH109 was co-transformed with IRR and CC1-splicing factor encoding proteins. The transformants were selected in media lacking leucine and tryptophan (-L-W), and interaction was tested in media lacking leucine, tryptophan and histidine (-L-W-H). This experiment was performed three times with similar results.
RNA-binding proteins with serine/arginine-enrichment and RRM domains are established as key to precursor-mRNA processing and alternative splicing in eukaryotes42, 43, thus we considered related roles for IRR. Notably, widespread changes in splicing patterns were observed in irr-1 as compared to wild-type, with differences in retained-intron, alternative 3′ splice site, skipped-exon, alternative 5′ splice site and mutually exclusive exon events (Fig. 3b, Supplementary Table 3). Among the alternative splicing patterns observed in the irr-1 knockout, retained-intron events were most abundant. Numerous transcripts encoding defense signaling proteins exhibited differing ratios of retained-intron variants encoding premature stop codons that would predictably result in truncated proteins and potentially modified functions (Supplementary Fig. 13). Negative regulators of plant immune signaling, such as CPK28, LESION-SIMULATING DISEASE 1 (LSD1), and JASMONATE-ZIM-DOMAIN PROTEIN 4 (JAZ4) all exhibited increased levels of retained-intron transcripts, whereas a positive regulator of immunity, CYSTEINE-RICH RECEPTOR-LIKE KINASE 13 (CRK13) had markedly decreased levels of a retained-intron isoform32, 44–47 (Supplementary Fig. 14a). To validate occurrence of retained-intron (RI) events observed through RNA-seq analysis, qRT-PCR was performed with primer sets unique to the intron of target genes that was retained at different ratios in irr-1 and wild-type plants. Amplification of the RI variant was performed in parallel with amplification using primers for the canonical splice form so that relative ratios could be compared. These analyses confirmed that in the irr-1 mutant, the ratio of CPK28, LSD1 and JAZ4 RI splice variants increased compared to wild-type, whereas the ratio of CRK13 RI splice variant decreased (Supplementary Fig. 14b).
In support of a role in mRNA splicing, IRR has been predicted to physically interact with the CC1-LIKE SPLICING FACTOR encoded by At2g1694048. Using a yeast two-hybrid system with co-expression of IRR and CC1 fused to bipartite transcription factor halves, IRR was found to physically interact with CC1 as predicted (Fig. 3c). Co-expression of CC1 with SR45, which has been demonstrated to mediate pre-mRNA splicing, also yielded a positive interaction49 (Supplementary Fig. 15). Because the sr45 knockout does not share the AtPep1-hypersensitive phenotype of irr knockouts, we conclude that while CC1 interaction with these proteins likely facilitates function, it is unlikely to contribute to target specificity.
The CPK28-RI splice variant underlies reduced negative regulation.
Retained-intron variants of transcripts encoding CPK28 were among the most significantly increased alternative splicing events observed in irr-1 knockouts, with a relative abundance of CPK28-RI approximately five-fold higher than in Wt plants, while levels of canonically-spliced CPK28 remained unchanged (Fig. 4a). Given the known function of CPK28 in negatively regulating AtPep1-PEPR complex activity by promoting turnover of the rate-limiting component BIK1, the role of IRR-mediated changes in CPK28 transcript splicing was examined32, 33. Calcium-dependent protein kinases are most commonly activated through exposure of the catalytic site as a result of conformational changes triggered by calcium ion-binding to the four regulatory EF hand domains50, 51. Partial losses of these EF hand domains can inhibit Ca2+-induced conformational changes, resulting in inactive CPKs with shielded catalytic domains50. The truncated CPK28 protein encoded by the CPK28-RI splice variant lacks two C-terminal EF hands (Fig. 4b), and was predicted to exhibit compromised catalytic activity as compared to full-length CPK28. Production of truncated CPK28 from CPK28-RI was confirmed through expression of cDNA encoding either canonical CPK28 or CPK28-RI as both a YFP-fusion in Arabidopsis and a GST-fusion in E. coli. In both cases, western blotting revealed that CPK28-RI was expressed to yield a truncated fusion protein of the expected size (Fig 4c,d). Using in vitro kinase activity assays, GST-CPK28 actively auto-phosphorylated and trans-phosphorylated both GST- and Histidine-tagged BIK1 protein, and the catalytically inactive variant GST- and His-BIK1K105A/K106A27, 32, whereas GST-CPK28-RI had greatly reduced phosphorylation activity (Fig. 4d, Supplementary Fig 16a). To deduce whether CPK28-RI was potentially active if exposed to higher levels of Ca2+, kinase activity was observed with a titration of Ca2+ concentrations up to 100 μM. Even as wild-type CPK28 showed increasing kinase activity as Ca2+ levels were increased, CPK28-RI activity remained attenuated (Supplementary Fig 16b). To ascertain whether CPK28-RI had reduced regulatory function in vivo, the cpk28 knockout mutant was complemented with either CPK28 or CPK28-RI expressed under control of the native promoter (Supplementary Fig. 16c). As determined by total ROS production, the AtPep1-hypersensitive phenotype of cpk28 was rescued through complementation with pCPK28:CPK28-YFP, but expression of pCPK28:CPK28-RI-YFP did not reduce sensitivity to AtPep1 (Supplementary Fig. 16d)32.
Figure 4. IRR affects the ratio of CPK28 retained-intron splice variants and CPK28 function.
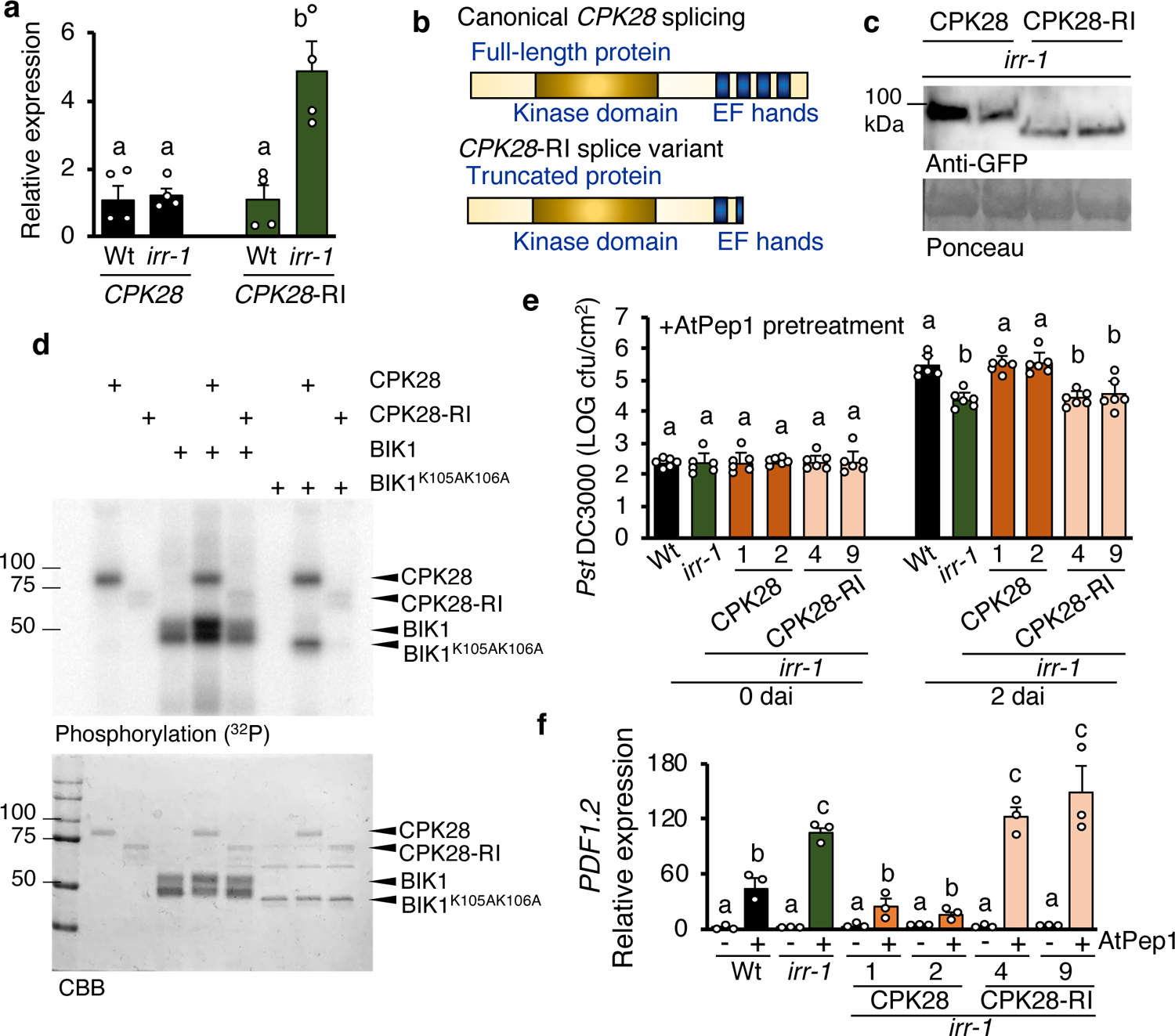
a, Relative expression of CPK28 and CPK28-RI splice variant in irr-1 plants. Values represent the fold change in expression versus the wild-type (Wt) control samples after normalization against ACTIN2 expression. Error bars indicate SEM, n = 4. b, Schema representing proteins translated from canonically spliced CPK28 transcript, which encodes a full-length protein containing four EF hands versus the retained intron-CPK28 splice variant, encoding a truncated protein missing two EF hands. c, Detection of CPK28-YFP and CPK28-RI-YFP fusion proteins as separated on a 7.5% SDS-PAGE gel using western blotting. Proteins were extracted from leaves of transgenic plants expressing pCPK28-CPK28-YFP and pCPK28-CPK28-RI-YFP in irr-1 background. Anti-GFP antibody was used to detect both proteins. Two independent events per line were analyzed. CPK28-YFP, 85 kDa; CPK28-RI-YFP, 75 kDa. Ponceau staining was used for verification of protein loading. d, Autoradiograph showing incorporation of P32 into GST-fused CPK28 and His-fused BIK1 recombinant proteins following in vitro kinase assays. Coomassie Brilliant Blue (CBB) stains are included as controls. e, Pst DC3000 infection assay of wild-type, irr-1, irr-1/pCPK28:CPK28-YFP and irr-1/pCPK28:CPK28-RI-YFP plants performed 24 h post-infiltration with a 1 μM solution of AtPep1. Bars indicate samples 0 and 2 days after inoculation (dai). Error bars indicate SD, n = 6. f, Relative expression levels of PDF1.2 as determined by real-time qRT-PCR for mRNA from entire seedlings treated for 24 h with water (−) or a solution 1 μM AtPep1 (+). Values represent the fold-change in expression versus the water-treated wild-type (Wt) control samples after normalization against ACTIN2 expression. Error bars indicate the SD of three biological replicates. Different letters represent significant differences (one-way ANOVA followed by Tukey’s test corrections for multiple comparisons; P < 0.05). Experiments were repeated three times independently, with similar results.
We hypothesized that one potential mechanism underlying irr-1 hypersensitivity to AtPep1 treatment is reduced negative regulation of PEPR signaling by CPK28 due to a loss of IRR function in promoting canonical splicing of CPK28 transcript to produce a functional full-length protein. In this case, increased expression of the canonical CPK28 transcript should fully or partially rescue the irr-1 phenotype. To test this hypothesis, Arabidopsis plants were transformed with cDNA constructs encoding both CPK28 transcript variants in the irr-1 mutant background under control of the native promoter and fused to a YFP tag. Two independent events for each transformation were selected, and protein expression confirmed by western blot, with plants expressing pCPK28:CPK28-RI-YFP producing a smaller protein than plants expressing pCPK28:CPK28-YFP (Fig. 4c). Expression of CPK28 transcript was approximately doubled in irr-1/pCPK28:CPK28-YFP plants as compared to Wt and irr-1, and expression of CPK28-RI transcript in irr-1/pCPK28:CPK28-RI-YFP plants was comparable to irr-1 (Supplementary Fig.17)
AtPep1-induced resistance to Pseudomonas syringae pv. tomato DC3000 (Pst) was examined in irr-1 plants expressing pCPK28:CPK28-YFP versus pCPK28:CPK28-RI-YFP. Without AtPep1 pretreatment, bacterial proliferation in irr-1 expressing YFP-tagged CPK28 or CPK28-RI was similar to Wt (Supplementary Fig.18). As previously observed, when pretreated with AtPep1 24 h prior to inoculation, irr-1 knockout plants were more resistant to Pst infection than Wt as measured by decreased bacterial proliferation after two days. In AtPep1-pretreated irr-1 plants expressing pCPK28:CPK28-YFP, Pst proliferation was similar to Wt, confirming that increased levels of canonical CPK28 transcript were able to rescue the irr-1 phenotype (Fig. 4e). In contrast, expression of pCPK28:CPK28-RI-YFP in irr-1 did not reduce the AtPep1-hypersensitive phenotype, as these lines exhibited the same enhanced AtPep1-induced restriction of bacterial proliferation as irr-1. AtPep1-sensitivity of irr-1 lines expressing pCPK28:CPK28-YFP was further examined through analysis of AtPep1-induced PDF1.2 marker gene expression. In irr-1 plants expressing pCPK28:CPK28-YFP, PDF1.2 expression was similar to Wt, whereas irr-1 lines expressing pCPK28:CPK28-RI-YFP maintained elevated PDF1.2 expression similar to irr-1 (Fig 4f). These experiments confirmed that increased levels of canonical CPK28 transcript was able to reduce the AtPep1-hypersensitive phenotype of irr-1. Together the evidence indicates that: (1) truncated CPK28-RI protein is not fully functional as a negative regulator, and (2) the inability of irr knockout plants to promote canonical CPK28 splicing for production of a functional full-length protein that negatively regulates PEPR signaling may be a significant contributor of irr hypersensitivity to AtPep1. Furthermore, relief of CPK28-mediated negative regulation of immunoregulatory receptor complex signaling appears integral to plant defense responses: Inoculation with Pst also triggers an increased proportion of CPK28-RI transcripts (Supplementary Fig. 19), demonstrating that CPK28 alternative splicing occurs during the course of a natural plant-microbe interaction.
Accumulation of CPK28-RI transcript is dependent on IRR phosphorylation state.
To ascertain whether levels of the CPK28-RI splice variant were affected by Pep signaling, plants were treated with AtPep1 and relative abundance of CPK28 and CPK28-RI transcripts analyzed by qRT-PCR. Notably, AtPep1 promoted significant increases in proportional CPK28-RI levels at 30 min (Fig. 5a), temporally coinciding with peak AtPep1-induced dephosphorylation of IRR (Fig. 1c). Elevated proportions of CPK28-RI returned to initial levels after 4 h (Fig. 5a), concurrent with recovery of IRR phosphorylation post-AtPep1 treatment. To determine whether IRR phosphorylation state contributed to CPK28-RI accumulation, irr-1 plants overexpressing an HA-tagged fusion of the phospho-abolishing mutant of IRR, IRRS745A,S747A, were examined. The irr-1:IRRS745A,S747A-3xHA plants exhibited higher levels of CPK28-RI than Wt or irr-1:IRR-3xHA plants, demonstrating a phenotype equivalent to irr-1 knockout plants (Fig. 5b). The inability of IRRS745A,S747A to reduce proportional CPK28-RI levels in the irr-1 background supports dephosphorylation of IRR as a contributing factor to the accumulation of CPK28-RI variant transcripts after AtPep1 treatment. Similarly, IRRS745A,S747A failed to rescue the AtPep1-hypersensitive phenotype of irr-1. Expression of the AtPep1-induced marker gene PDF1.2 remained elevated in irr-1:IRRS745A,S747A-3xHA lines, whereas irr-1 lines complemented with IRR phenocopied Wt (Fig. 5c). Furthermore, irr-1:IRRS745A,S747A-3xHA plants exhibited enhanced AtPep1-induced restriction of Pst proliferation comparable to irr-1, whereas Pst proliferation in irr-1:IRR-3xHA plants was similar to Wt (Fig. 5d). Like irr-1 lines, basal resistance to Pst of both irr-1:IRRS745A,S747A-3xHA and irr-1:IRR-3xHA was unchanged from Wt (Supplementary Fig. 20). Together these results suggest that phosphorylated IRR acts as a negative regulator of AtPep1-induced immune responses. In unchallenged wild-type plants, the population of IRR protein is predominantly phosphorylated on pS745 and pS747, and active as a negative regulator. However, upon perception of AtPep1, transient dephosphorylation of IRR temporarily attenuates negative regulatory function. When IRR is absent, as in irr knockouts, IRR-mediated negative regulation is fully relieved and results in enhanced immune responses.
Figure 5. IRR associates with CPK28 transcript in a phosphorylation-dependent manner.
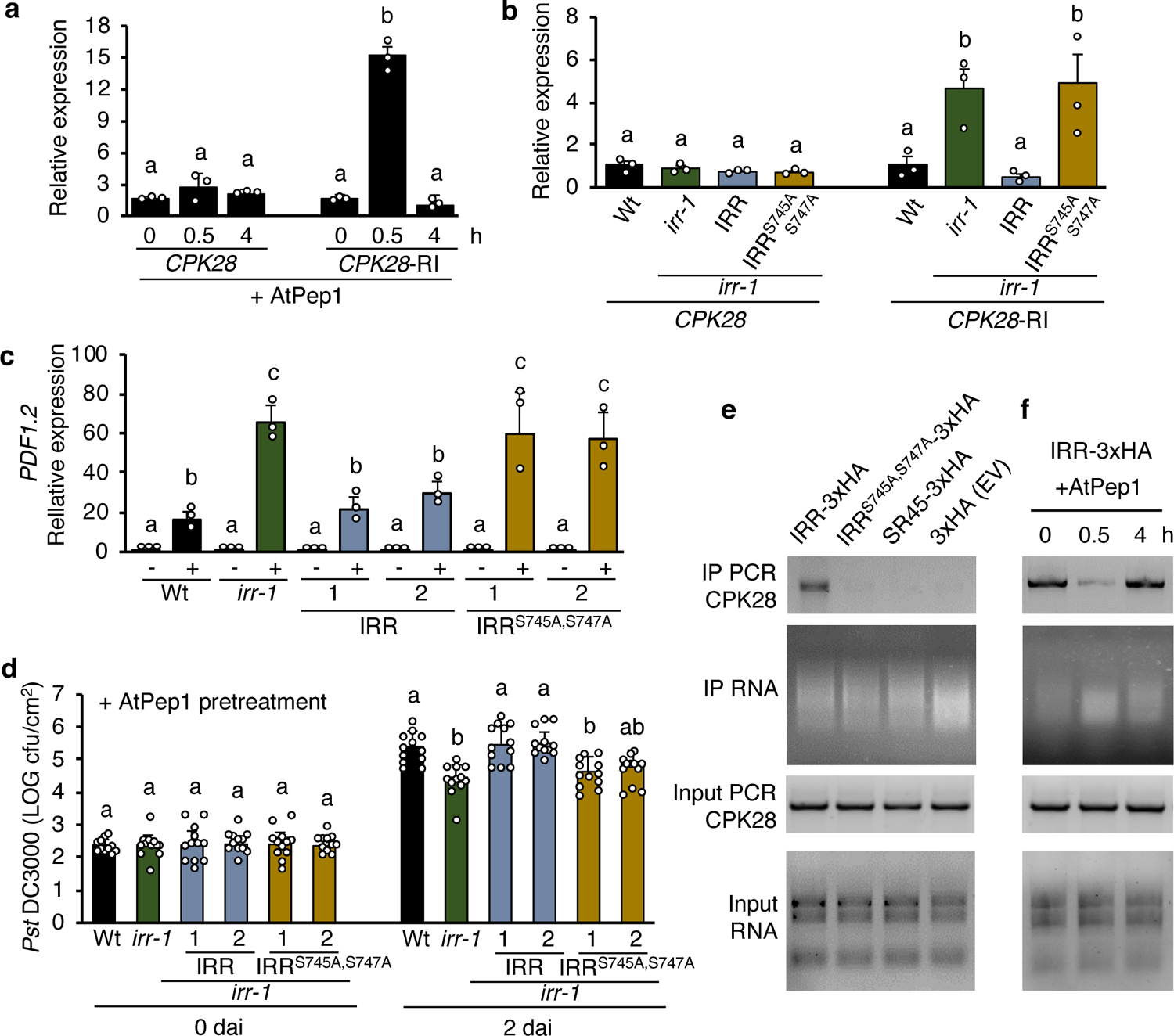
a, Relative expression of CPK28 and CPK28-RI splice variant in irr-1/IRR-3xHA lines analyzed by qRT-PCR. Plants were treated with solutions of 1 μM AtPep1 for 0, 0.5 or 4 h. Values represent the fold-change in expression versus 0 h samples after normalization against ACTIN2 expression. Error bars indicate SEM, n = 3. b, Relative expression of CPK28 and CPK28-RI in wild-type (Wt), irr-1 and transgenic lines overexpressing triple HA-tagged IRR (irr-1:IRR) and IRRS745A,S747A (irr-1:IRRS745A,S747A). Values represent the fold-change in expression versus the Wt control samples after normalization against ACTIN2 expression. c, The relative expression of AtPep1-inducible gene PDF1.2 was determined by real-time qRT-PCR using mRNA from whole seedlings treated for 24 h with wither water (−) or a 1 μM solution of AtPep1 (+). Values represent the fold-change in expression versus the water-treated Wt control samples after normalization against ACTIN2 expression. Two independent events per transgenic line overexpressing triple HA-tagged IRR (irr-1:IRR) and IRRS745A,S747A (irr-1:IRRS745A,S747A) were used. Error bars indicate the SD of three biological replicates. d, Pst DC3000 infection assay of wild-type, irr-1 and irr-1 overexpressing triple HA-tagged IRR (irr-1:IRR) and IRRS745A,S747A (irr-1:IRRS745A,S747A) plants 24 h after pretreatment by infiltration with a 1 μM solution of AtPep1. Bars indicate samples 0 and 2 days after inoculation (dai). Error bars indicate SD, n = 11–12. e, RNA immunoprecipitated (RIP) from transgenic plants overexpressing IRR-3xHA, IRRS745A,S747A-3xHA, SR45-3xHA and 3xHA (Empty vector, EV). RIP-PCR was performed to detect CPK28 transcripts. f, RIP from transgenic plants overexpressing IRR-3xHA after treatment with a 1 μM solution of AtPep1 for 0, 0.5 or 4 h. RIP-PCR was performed to detect CPK28 transcripts. The protein-RNA complex was immunoprecipitated using anti-HA magnetic beads. Input RNA was extracted from all samples as control. PCR reactions with primers to detect CPK28 transcript used cDNA templates reverse transcribed from input RNA samples. Different letters represent significant differences (one-way ANOVA followed by Tukey’s test corrections for multiple comparisons; P < 0.05). Experiments were repeated three times independently, with similar results.
IRR associates with CPK28 transcript in a phosphorylation-dependent manner.
To better understand how dephosphorylation of IRR might contribute to function, behavior of the IRRS745A,S747A mutant protein relative to wild-type IRR was assessed. Subcellular localization of YFP-tagged IRRS745A,S747A was examined as compared to IRR-YFP, and found to be unaffected (Supplementary Fig. 21). For many serine/arginine-rich RNA-binding proteins that function in precursor-mRNA processing, phosphorylation promotes interaction with other proteins in the splicing complex52, so the effect of mutant IRRS745A,S747A on physical interactions with the CC1 splicing factor was also examined. As determined by plate-based yeast two-hybrid and monitoring of yeast growth dynamic in liquid medium, mutation of the phosphorylation sites to alanine does not impair interaction with the CC1-splicing factor, nor alter interaction affinity (Supplementary Fig. 22a–c). To investigate whether AtPep1-induced dephosphorylation affects IRR stability and turnover, irr-1 plants overexpressing triple HA-tagged IRR were treated with AtPep1. No change in total IRR protein levels was observed by western blot after AtPep1 treatment (Supplementary Fig. 23a). However, cotreatment with AtPep1 and the proteasome inhibitor MG132, or the protein translation inhibitor cycloheximide (CHX), revealed that phosphorylation state may affect turnover rate. Treatment with MG132 or CHX alone did not affect IRR levels, but with cotreatments, IRR accumulated 30 and 120 min after MG132/AtPep1 treatment, and was depleted after CHX/AtPep1 treatment (Supplementary Fig. 23a, b). Mutant IRRS745A,S747A protein levels were significantly decreased by CHX treatment in both the absence and presence of AtPep1, suggesting that dephosphorylation of IRR promotes protein turnover (Supplementary Fig. 23c).
To ascertain whether IRR regulation of CPK28 transcript splicing might occur through association of the two in complex, RNA immunoprecipitation (RIP) coupled with PCR analysis was used to test for IRR-CPK28 transcript interactions. RIP-PCR demonstrated that IRR associates with CPK28 mRNA, with a CPK28 fragment amplified from RNA co-immunoprecipitating with IRR (Fig. 5e). Whether this association is direct or occurs through complex with additional factors is not known. The RNA-binding protein SR45 was used as a negative control since SR45-interacting transcripts have previously been identified by RIP-Seq, and CPK28 is not among them53. As expected, amplification of RNA co-immunoprecipitated with SR45 did not yield a CPK28 fragment, nor did amplification of RNA co-immunoprecipitated with an unfused triple-HA tag (Fig. 5e). Because mutant IRRS745A,S747A fails to promote canonical splicing of CPK28, in contrast to wild-type IRR, dependence of the IRR-CPK28 interaction on pSer745 and pSer747 was probed. No CPK28 fragment was amplified from RNA co-immunoprecipitation with IRRS745A,S747A (Fig. 5e), indicating that abolished phosphorylation at these sites disrupts association of IRR with CPK28 transcript in addition to eliminating IRR-stimulated canonical splicing of CPK28. All proteins were detected after protein-RNA complex immunoprecipitation (Supplementary Fig. 24).
Because phospho-abolishing mutations of IRR also abolished association of IRR with CPK28 transcript, the effects of AtPep1-mediated transient dephosphorylation of IRR on CPK28 transcript interactions were investigated. Coimmunoprecipitation of triple HA-tagged IRR with associated RNA was performed 0, 0.5, or 4 h post-treatment with AtPep1 to compare with IRR dephosphorylation dynamics (Fig. 1a–b). CPK28 transcript was abundant in IRR-interacting RNA at 0 h, but declined in RNA coimmunoprecipitated with IRR 30 min after AtPep1 treatment (Fig. 5f). Thus AtPep1-induced dephosphorylation of IRR parallels a disrupted association with CPK28 transcript. Predictably, 4 h after AtPep1 treatment, when IRR phosphorylation levels have recovered (Fig. 1b), increased amplification of CPK28 transcript from coimmunoprecipitated RNA was observed, indicating a reestablishment of IRR/CPK28 association (Fig. 5f). For all samples, input and coimmunoprecipitated RNA was determined to be of similar quantities and CPK28 transcript amplified similarly from input RNA collected from samples prior to coimmunoprecipitation (Fig. 5e,f).
Discussion
Alternative splicing is a fundamental layer of regulation in Eukaryotes, allowing for rapid adaptation to stress in the absence of de novo transcription. For both plants and animals, immune challenge is associated with widespread changes in splicing patterns, with pathogen infection, caterpillar infestation and elicitor treatments all resulting in large-scale reprogramming of plant mRNA splicing54–58. Serine/arginine-rich RNA Recognition Motif-containing (SR-RRM) proteins in particular have been implicated as potential regulators of immune-induced variation in splicing37, 57, 59, 60. Disruption of function or of proper trafficking of SR-RRM proteins can result in dysregulation of both stress-induced splicing patterns and disease resistance37, 59, 60. The critical role for SR-RRM proteins in regulating plant immunity is underscored by the finding that pathogens such as Phytophthora sojae have evolved effector proteins that physically interact with SR-RRMs to alter immune-induced splicing patterns and promote susceptibility61. Transcripts for many defense signaling proteins have been identified as targets of alternative splicing during immunity, including pattern recognition receptors, kinases, transcription factors, and Resistance proteins2, 58, 60, 62–64. Retained-intron events are frequently observed, with retention of early introns often associated with changes in transcript stability or translation, and retention of later introns generally resulting in production of truncated protein isoforms2, 55, 62. For truncated signaling protein variants that have been characterized, function is generally abolished or altered, supporting a role for alternative splicing as one mechanism to quickly remodel signaling pathways and resultant cellular output2, 55. Interestingly, altered levels of several CPK28 splice variants, including those with retained-introns have recently been reported after flg22 treatment in an MPK4-dependent manner58. While this indicates that CPK28 splicing is a common regulatory target for multiple inputs, our finding that flg22-induced responses are only modestly affected in irr knockouts along with similar findings for cpk28 knockouts indicate that flg22-induced CPK28 splicing may regulate sensitivity to signals other than flg2232. Examination of the degree of intersection between these and other MAMP/DAMP signaling pathways will be an interesting line of research for the future.
Previous studies have established alternative splicing as a contributor to immune regulation and revealed RNA-binding proteins and target transcripts involved in this process. In this study, we have characterized IRR as a new RNA-binding protein that regulates defense response strength, and have identified IRR-mediated changes in splicing, including targets encoding defense signaling proteins. Additionally, we have demonstrated how dynamic and site-specific posttranslational modification of IRR regulates association with, and alternative splicing of one of these transcripts, encoding the key defense regulator CPK28, to affect immune response outputs. It is likely that many of the other alternative splicing events observed in irr knockouts also impact immune regulation, and contribute to the overall AtPep1-hypersensitive phenotype of irr. For instance, the hyperactivation of MAPK phosphorylation observed in irr knockouts has been found to occur through PBLs other than BIK1, indicating that some IRR-mediated signaling is dependent on additional factors beyond CPK28-regulated stability of BIK165, 66. Future analysis of cpk28/irr double mutants will clarify which irr phenotypes are dependent on CPK28. This relationship and additional IRR targets are of interest for future study to increase overall understanding of both IRR and Pep-regulated immunity.
Based on our current findings, we propose one model for IRR function, in which IRR dynamically regulates the CPK28 immunomodulatory buffering system (Fig. 6). Prior to immune challenge, IRR is predominantly phosphorylated at S745 and S747, and associates with CPK28 transcripts, facilitating canonical splicing to produce a full-length, functional protein (Fig. 6a). Full-length CPK28 promotes BIK1 turnover to suppress immune receptor signaling by phosphorylating both BIK1 and the ubiquitin ligases PUB25 and PUB26 to promote BIK1 ubiquitylation and degradation32, 33. Upon Pep-induced activation of PEPRs, IRR is transiently dephosphorylated and dissociates from CPK28 transcripts, resulting in increased levels of the retained-intron CPK28-RI variant (Fig. 6b). The truncated protein encoded by CPK28-RI lacks EF hand motifs required for calcium-induced stimulation of kinase activity. Increased levels of this less-active CPK28-RI protein attenuate CPK28-mediated BIK1 degradation and temporarily enhance signaling capacity of PEPR complexes to amplify defense outputs. Re-phosphorylation of IRR facilitates equilibration back to immunoregulatory homeostasis, with CPK28 again buffering receptor complex function (Fig. 6c). Together this study defines a dynamic process that directly links PEPR-induced dephosphorylation of IRR with post-transcriptionally mediated attenuation of CPK28 function, revealing a new mechanism to modulate PEPR signaling capacity and immune response outputs. This regulatory module represents a strategy to temporarily derepress immune signaling during the acute response phase for promotion of a more robust protective response. Although this derepression of signal transmission is transient, on the order of hours, the downstream changes that are activated trigger a more lasting enhanced immunity, on the order of days. In sum, these findings reveal a new layer of complexity in the intricate regulatory programs used by plants to wield the double-edged sword of innate immunity for protection while minimizing detrimental effects of an inappropriately persistent response.
Figure 6. Proposed model by which IRR dynamically regulates CPK28 immunomodulatory buffering of PEPR-mediated immunity.
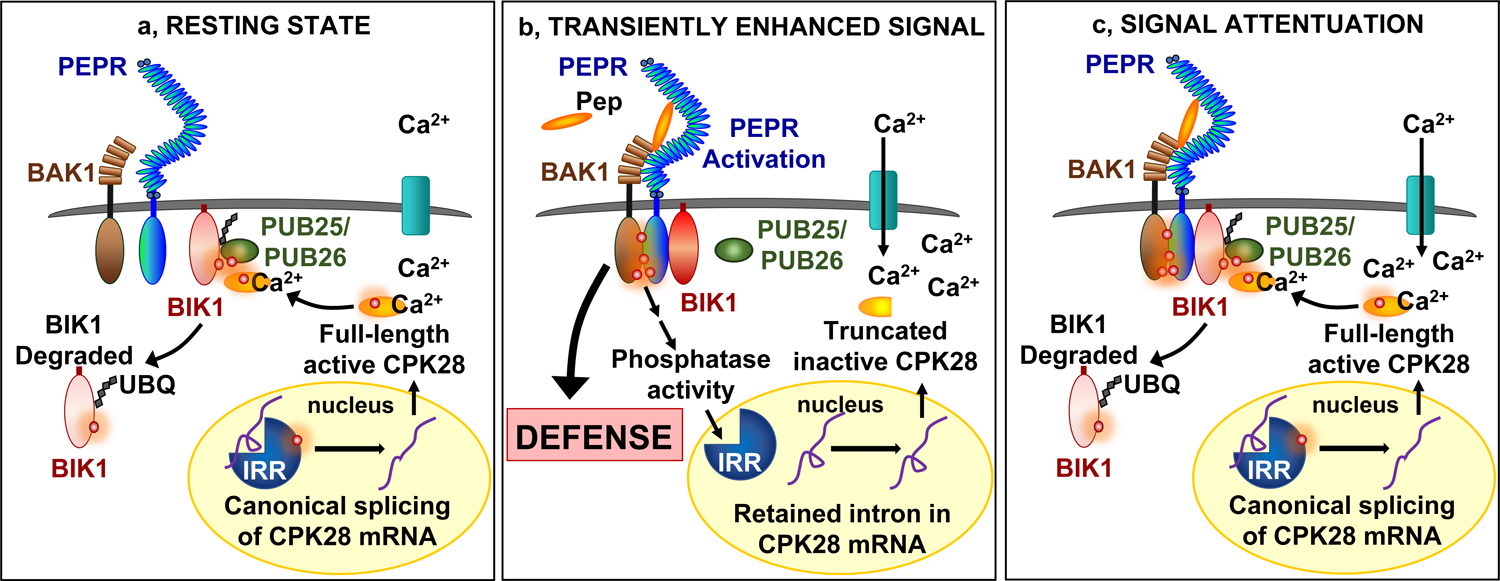
a, Resting state: phosphorylated IRR associates with transcripts encoding CPK28, facilitating canonical splicing into mRNA that produce a full-length protein which functions as a negative regulator of immune receptor complex signaling by phosphorylating PUB25/26 ubiquitin ligases to promote ubiquitylation and subsequent degradation of BIK1. b, Transiently enhanced signal: AtPep1 activates PEPR receptor complexes, leading to transient dephosphorylation of IRR and dissociation from CPK28 transcripts, resulting in temporary accumulation of a retained intron variant encoding a premature stop codon that yields a truncated and inactive CPK28 protein variant, reducing CPK28 buffering of the receptor complex and resulting in amplified PEPR-mediated immune signaling and defense output. c, Signal attenuation: recovery of IRR phosphorylation reestablishes association with CPK28 transcript to promote canonical splicing and resumed translation of a full-length protein to reinstitute negative regulation of PEPR signaling and immunity
Methods
Phosphoproteomic screen
Arabidopsis thaliana T87 suspension-cultured cells and maize suspension-cultured endosperm cells (var. Black Mexican Sweet) were treated with either water or 100 nM AtPep1 and ZmPep3, respectively. Cells were harvested after 10 min into liquid nitrogen. Protein extracted from these samples was subjected to a tryptic digest, labeled with iTRAQ mass tags, mixed and analyzed. Phosphopeptides were enriched using CeO2 affinity capture and analyzed separately. Peptides were separated by nano-LC using salt gradients on a three-phase capillary column with an LTQ Velos linear ion trap tandem MS in positive ion mode and data-dependent acquisitions67. Peptides were separated into three mass classes prior to scanning and each MS scan was followed by five MS/MS scans of the most intense parent ions. Data was extracted and searched using Spectrum Mill (Agilent). Peptide abundance and phosphorylation levels were quantified by spectral counting with counts for each protein representing the total number of peptides that matches to that protein.
Phosphorylation assay
Plants carrying p35S:IRR-3xHA and p35S:SR45–3xHA constructs were grown on plates containing half-strength MS media for 10–15 days, then transferred to half-strength MS liquid media and treated with varying concentrations of AtPep1 (0.1, 0.5 and 1 μM) for 30 min, or treated with 1 μM AtPep1 for different lengths of time (10, 20, 30 min and 4, 8 hours). Water treatment was used as a control. After treatment, approximately 1 g of whole seedlings was homogenized in extraction buffer [300 mM sucrose, 100 mM Tris-HCl pH 7.5, 25 mM EDTA, 25 mM NaF, 1 mM Na2MoO4, 1 mM PMSF, 0.5% Triton-X and protease inhibitor cocktail (Sigma), adjusted to pH 5.8–6.2] and centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was transferred to a new microcentrifuge tube, mixed with denaturing 1X loading buffer (NuPAGE LDS buffer) containing 10% β-mercaptoethanol and incubated at 95 °C for 8 min. Proteins were separated by SDS/PAGE and further analyzed by western blot. Anti-phosphoserine (1:1000, Sigma) antibody and anti-mouse-HRP-conjugated secondary antibody (1:2000, Sigma) were used with the Super Signal West Pico Maximum Chemiluminescent detection kit (Thermo Scientific) to detect proteins. The HA tagged proteins were detected with anti-HA (1:1000, Sigma) and anti-mouse HRP (1:1000, Sigma) antibodies.
Analysis of protein expression and stability
Plants carrying p35S:IRR-3xHA and p35S:IRRS745A,S747A-3xHA constructs were grown on plates containing half-strength MS media for 7 days, then transferred to half-strength MS liquid media and treated with 1 μM AtPep1 for varying lengths of time as indicated in the figures, or treated with peptide concomitantly with 10 μM cycloheximide (CHX), or with peptide concomitantly with 50 μM MG132. The solvent DMSO was used as control. After treatment, 500 mg of plant tissue was homogenized in extraction buffer and centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was homogenized with denaturing 1X loading buffer (NuPAGE SDS buffer) containing 10% β-mercaptoethanol and incubated at 95°C for 8 min. Proteins were separated by SDS/PAGE and further analyzed by western blot. Anti-HA (1:1000, Sigma) and anti-mouse HRP (1:1000, Sigma) antibodies were used with the Super Signal West Maximum Chemiluminescent detection kit (Thermo Scientific) to detect proteins.
RNA-seq
Sterile seeds were sown on half-strength MS media for 7–10 days and transferred to 24 well plates containing liquid half-strength MS. Approximately 15 plants were transferred to individual wells. Three hours after transfer, plants were treated with either water or 1 μM peptide for 24 hours at room temperature and constant light. The plant tissue was harvested in liquid nitrogen and total RNA was isolated with the Spectrum Plant Total RNA kit (Sigma) and treated with Turbo DNA-free kit (Ambion). RNA quality was checked using the Agilent2100 Bioanalyzer (Agilent Technologies). Three biological replicates per genotype per treatment were used. The preparation of RNA-Seq 250–300 bp insert cDNA library, Illumina HiSeq platform PE150 sequencing and bioinformatic data analysis were performed at the Novogene Corporation Inc. REViGO was used to create the summarized list of significantly enriched Gene Ontology terms68.
Yeast two-hybrid
The IRR, SR45 and CC1-splicing factor coding regions were amplified from Arabidopsis cDNA using standard PCR and cloned into the pENTR/D-TOPO vector. To generate the IRR phosphoabolishing mutations, the QuickChange II XL site-directed mutagenesis kit (Agilent Technologies) was used to substitute serine to alanine (S745A, S747A). The fragments were transferred into pACT and pAS vectors by recombination. The primers used to amplify the fragments are listed in Supplementary Table 4. Yeast strain AH109 was transformed with the desired pairs of the pACT and pAS vectors as described69. The transformed yeast cells were grown on a synthetic complete medium lacking leucine and tryptophan (Clontech) and selected on a synthetic complete medium lacking leucine, tryptophan and histidine (Clontech). To check the strength of protein-protein interactions, four transformed yeast cells were grown individually in liquid synthetic complete medium lacking leucine, tryptophan and histidine for 2 days at 29°C, 200 rpm. Their OD was measured, adjusted to 0.05, and the cells were transferred to 96 well plate (250 μL per well). The plate was inserted into the microplate reader and agitated once every hour for 30 sec. The incubation temperature was 29°C. The OD was measured every hour until yeast growth was saturated.
MAP Kinase phosphorylation assay
Two-week-old seedlings were treated with 1 μM solutions of AtPep1 and then harvested in liquid nitrogen after 0, 5, 10, 30 and 60 min. Proteins were extracted with Lacus buffer (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 15 mM EGTA, 100 mM NaCl, 1 mM sodium fluoride, 1 mM sodium molybdate, 0.5 mM Na3VO4, 30 mM β-glycerol-phosphate, 0.1% Triton-X 100), as described previously70. The homogenized protein samples were centrifuged at 21.000 × g for 20 min at 4°C. 5x SDS loading buffer was added into each supernatant and 12 μl were subjected to immunoblot analysis. After transfer, protein was blocked for MAPK activation detection with 5% BSA for 1 hour. After washing three times in TBS-T, the membrane was incubated overnight at 4°C in TBS-T and α-p44/42 MAPK (Erk1/2) antibody (1:5000, CST). Following three washes with TBS-T, the membrane was incubated in α-rabbit-HRP (1:5000, Sigma) for 2 hours. Phosphorylated MAP kinases 3, 4/11, and 6 were detected using Pierce ECL western blotting substrate (Thermo Fisher).
RNA Immunoprecipitation (RIP)
Transgenic plants expressing p35S:IRR-3xHA, p35S:IRRS745A,S747A-3xHA or p35S:SR45–3xHA constructs were grown in plates containing half-strength MS media for 10–15 days. Transgenic plants expressing the empty vector p35S-3xHA were used as control. Plants were transferred to half-strength MS liquid media for approximately 16 hours prior to treatment with 1 μM AtPep1 for 30 min and 4 hours. Approximately 1 g of seedlings were carefully harvested and water-rinsed before cross-linking. The cross-linking was performed by immersing the seedlings in 0.5% formaldehyde and applying a vacuum four times (15 s/time). The seedlings were then kept at room temperature for 10 min, and glycine was added to a final concentration of 83 mM. Following this, the seedlings were again subjected to vacuum four times (15 s/time) and incubated for 5 min to quench the cross-linking. The seedlings were rinsed with water five times, wrapped in a few layers of Kimwipes to remove water and frozen in liquid nitrogen. The nuclear extract preparation was performed as described71. To immunoprecipitate the RNA-protein complex, the nuclear extract was resuspended in 900 μL of ChIP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl, and 160 units/mL RnaseOUT) and centrifuged at 16,000 × g for 10 min at 4°C. The supernatant was transferred to a clean microcentrifuge tube, and a small aliquot of the supernatant removed to probe as RNA inputs. Thirty microliters of anti-HA magnetic beads (Sigma) per sample was prepared by washing the beads five times with 1 mL of Binding/Washing buffer (150 mM NaCl, 20 mM Tris HCl, pH 8.0, 2 mM EDTA, 1% Triton X-100, and 0.1% SDS). The beads were resuspended in 50 μL of Binding/Washing buffer per sample and added to the diluted nuclear extraction. The samples were incubated on a rotator for 3 hours at 4°C. The anti-HA magnetic beads were washed six times with 1 mL Binding/Washing buffer containing 40 units/ml RnaseOUT and the magnetic separation rack. To elute the protein-RNA complexes, 60 μL of RIP Elution buffer (100 mM Tris HCl, pH 8.0, 10 mM EDTA, 1% SDS, and 800 units/mL RnaseOUT) was added to the beads, and the tubes were incubated on a rotator at room temperature for 10 min. The supernatant was saved. The elution was repeated with an additional 60 μL of RIP Elution buffer at 65°C for 10 min. The supernatants from both elution steps were combined. At the same time, 110 μL of RIP elution buffer was added to the 10 μL of RNA input sample. An aliquot of 1.2 μL Proteinase K (20 mg/mL, Invitrogen) was added to each IP or input sample. The tubes were incubated at 65°C for 1 hour. The RNA was isolated using Trizol reagent (Life Technologies). To facilitate RNA precipitation, 20 μg glycogen was added to the aqueous phase before the isopropanol precipitation. The RNA samples were further treated using a Turbo DNA-free kit (Ambion) and cleaned with the RNeasy MiniElute Kit (Qiagen). The RNA was eluted into 15 μL of RNase-free water. The cDNA synthesis and PCR reaction were performed as described in the Supplementary methods section.
In vitro kinase assay
Synthetic cDNA encoding either canonically spliced CPK28 or the CPK28-RI splice variant were made by Genscript. The BIK1 coding region was amplified from the cDNA of Arabidopsis. To generate the BIK1K105A/K106A kinase-dead mutant, the QuickChange II XL site-directed mutagenesis kit (Agilent Technologies) was used. All fragments were amplified by PCR and fused to GST in the N-terminal of pGEX6P-1 vector using the USER enzyme (New England Biolabs). BIK1 and BIK1K105A/K106A fragments were also fused to His in the N-terminal of pET vector using the In-Fusion® HD Cloning Kit (Takara). Primers used are listed in Supplementary Table 4. The GST-CPK28, GST-BIK1 and His-BIK1 variants were expressed and purified from Escherichia coli strain BL21. The in vitro kinase assay was performed in reaction buffer [50 mM Tris-HCl pH 7.5, 10 mM MgCl2, free Ca2+ (0, 0.5, 2 and 100 μM) buffered by 1 mM EGTA and CaCl2, 1 mM dithiothreitol, 0.1% (v/v) Triton X-100, 200 μM cold ATP, and 1 μCi (γ−32P)-ATP] at room temperature for 30 min.
Virus-Induced Genome Silencing (VIGS) in maize
A VIGS system derived from Foxtail mosaic virus (FoMV) was used to induce gene silencing in maize line B73. The 251 and 301 bp gene fragments of GRMZM2G132936 (ZmIRR) were amplified by primers listed in the Supplementary Table 4 and cloned into XhoI/ XbaI sites of the FoMV plasmid in an antisense orientation as described41, yielding plasmids pFoMV-IRR-1 and pFoMV-IRR-2. Seven-day old maize plants were inoculated with both plasmids by biolistic particle delivery system using gold particles (Seashell Technology). Control plants were biolistically inoculated with FoMV vector carrying no insert (FoMV-V). To confirm FoMV infection and gene silencing in plants, the total RNA was extracted from the fifth leaf of these plants using Trizol reagent (Life Technologies). cDNA was synthesized and PCR using primers designed to amplify a specific fragment from the FoMV genomic RNA was performed. Relative expression levels of ZmIRR were evaluated by qRT-PCR, using ZmRLP17 as reference gene. Primers used are listed in Supplementary Table 4.
Volatile emission assay
The fifth maize leaf was excised using a razor blade and supplied either 1 mL water or 5 μM ZmPep3 solution through the petiole overnight for 16 hours. Leaves were harvested and individually kept in closed glass tubes for one hour under the light while volatiles were collected on 50 mg Super Q (80/ 100 mesh; Alltech). Volatile compounds were eluted with methylene chloride containing nonyl acetate as an internal standard, and analyzed by GC as described72.
Supplementary Material
Acknowledgments
The authors thank Dr. Steven A. Whitham (Iowa State University Plant Sciences Institute) for providing the constructs for VIGS experiments, and Dr. Alexander Groisman (University of California San Diego Department of Physics) for use of his Biolistic inoculation apparatus. The authors also thank the Editor and Referees for their well-considered criticism and suggestions that guided the significant improvement of this work.
Funding
This work was funded by NSF CAREER Award #1943591, a Hellman Foundation Fellowship and UC San Diego Start-up funds to A.H. K.D. was additionally funded by Ciências sem Fronteiras/CNPq fellowship #200260/2015-4. E.P. was additionally funded by the Cell and Molecular Genetics (CMG) Training Program at the University of California, San Diego. Z.S. and S.P.B. were funded by NSF award #1546899.
Footnotes
Further information on research methods is available in the Supplementary Materials.
Data availability
Raw read sequences are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE146282. The data generated and analyzed in this study are included in the published article and Supplementary Information. All data are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
References
- 1.Kobayashi KS and Flavell RA, Shielding the double-edged sword: negative regulation of the innate immune system. J Leukoc Biol, 2004. 75(3): p. 428–33. [DOI] [PubMed] [Google Scholar]
- 2.Gassmann W, Alternative splicing in plant defense. Curr Top Microbiol Immunol, 2008. 326: p. 219–33. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Qian C, and Cao X, Post-Translational Modification Control of Innate Immunity. Immunity, 2016. 45(1): p. 15–30. [DOI] [PubMed] [Google Scholar]
- 4.Xu G, Greene GH, Yoo H, Liu L, Marques J, Motley J, and Dong X, Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature, 2017. 545(7655): p. 487–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nuhse TS, Bottrill AR, Jones AM, and Peck SC, Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J, 2007. 51(5): p. 931–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Withers J and Dong X, Post-translational regulation of plant immunity. Curr Opin Plant Biol, 2017. 38: p. 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tena G, Boudsocq M, and Sheen J, Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol, 2011. 14(5): p. 519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, and Shan L, Direct Ubiquitination of Pattern Recognition Receptor FLS2 Attenuates Plant Innate Immunity. 2011. 332(6036): p. 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng B, Liu C, de Oliveira MVV, Intorne AC, Li B, Babilonia K, de Souza Filho GA, Shan L, and He P, Protein Poly(ADP-ribosyl)ation Regulates Arabidopsis Immune Gene Expression and Defense Responses. PLOS Genetics, 2015. 11(1): p. e1004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macho AP and Zipfel C, Plant PRRs and the activation of innate immune signaling. Mol Cell, 2014. 54(2): p. 263–72. [DOI] [PubMed] [Google Scholar]
- 11.Yu X, Feng B, He P, and Shan L, From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu Rev Phytopathol, 2017. 55: p. 109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huffaker A, Pearce G, and Ryan CA, An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc Natl Acad Sci U S A, 2006. 103(26): p. 10098–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huffaker A, Dafoe NJ, and Schmelz EA, ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiol, 2011. 155(3): p. 1325–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huffaker A, Pearce G, Veyrat N, Erb M, Turlings TC, Sartor R, Shen Z, Briggs SP, Vaughan MM, Alborn HT, Teal PE, and Schmelz EA, Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc Natl Acad Sci U S A, 2013. 110(14): p. 5707–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trivilin AP, Hartke S, and Moraes MG, Components of different signalling pathways regulated by a new orthologue of AtPROPEP1 in tomato following infection by pathogens. 2014. 63(5): p. 1110–1118. [Google Scholar]
- 16.Lee MW, Huffaker A, Crippen D, Robbins RT, and Goggin FL, Plant elicitor peptides promote plant defences against nematodes in soybean. Mol Plant Pathol, 2018. 19(4): p. 858–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz C, Nadal A, Montesinos E, and Pla M, Novel Rosaceae plant elicitor peptides as sustainable tools to control Xanthomonas arboricola pv. pruni in Prunus spp. Mol Plant Pathol, 2018. 19(2): p. 418–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lori M, van Verk MC, Hander T, Schatowitz H, Klauser D, Flury P, Gehring CA, Boller T, and Bartels S, Evolutionary divergence of the plant elicitor peptides (Peps) and their receptors: interfamily incompatibility of perception but compatibility of downstream signalling. J Exp Bot, 2015. 66(17): p. 5315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hander T, Fernandez-Fernandez AD, Kumpf RP, Willems P, Schatowitz H, Rombaut D, Staes A, Nolf J, Pottie R, Yao P, Goncalves A, Pavie B, Boller T, Gevaert K, Van Breusegem F, Bartels S, and Stael S, Damage on plants activates Ca(2+)-dependent metacaspases for release of immunomodulatory peptides. Science, 2019. 363(6433). [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi Y, Pearce G, and Ryan CA, The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci U S A, 2006. 103(26): p. 10104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, and Ryan CA, PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell, 2010. 22(2): p. 508–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KA, Becker D, and Hedrich R, Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem, 2010. 285(18): p. 13471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tintor N, Ross A, Kanehara K, Yamada K, Fan L, Kemmerling B, Nurnberger T, Tsuda K, and Saijo Y, Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc Natl Acad Sci U S A, 2013. 110(15): p. 6211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross A, Yamada K, Hiruma K, Yamashita-Yamada M, Lu X, Takano Y, Tsuda K, and Saijo Y, The Arabidopsis PEPR pathway couples local and systemic plant immunity. Embo j, 2014. 33(1): p. 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Postel S, Kufner I, Beuter C, Mazzotta S, Schwedt A, Borlotti A, Halter T, Kemmerling B, and Nurnberger T, The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur J Cell Biol, 2010. 89(2–3): p. 169–74. [DOI] [PubMed] [Google Scholar]
- 26.Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, and Zhou JM, BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci U S A, 2013. 110(15): p. 6205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu D, Wu S, Gao X, Zhang Y, Shan L, and He P, A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci U S A, 2010. 107(1): p. 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T, Felix G, and Chinchilla D, Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem, 2010. 285(13): p. 9444–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tor M, de Vries S, and Zipfel C, The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell, 2011. 23(6): p. 2440–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, Chen S, and Zhou JM, The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe, 2014. 15(3): p. 329–38. [DOI] [PubMed] [Google Scholar]
- 31.Lal NK, Nagalakshmi U, Hurlburt NK, Flores R, Bak A, Sone P, Ma X, Song G, Walley J, Shan L, He P, Casteel C, Fisher AJ, and Dinesh-Kumar SP, The Receptor-like Cytoplasmic Kinase BIK1 Localizes to the Nucleus and Regulates Defense Hormone Expression during Plant Innate Immunity. Cell Host Microbe, 2018. 23(4): p. 485–497.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky FG, Rathjen JP, MacLean D, Romeis T, and Zipfel C, The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe, 2014. 16(5): p. 605–15. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Grubb LE, Wang J, Liang X, Li L, Gao C, Ma M, Feng F, Li M, Li L, Zhang X, Yu F, Xie Q, Chen S, Zipfel C, Monaghan J, and Zhou JM, A Regulatory Module Controlling Homeostasis of a Plant Immune Kinase. Mol Cell, 2018. 69(3): p. 493–504.e6. [DOI] [PubMed] [Google Scholar]
- 34.Walley JW, Sartor RC, Shen Z, Schmitz RJ, Wu KJ, Urich MA, Nery JR, Smith LG, Schnable JC, Ecker JR, and Briggs SP, Integration of omic networks in a developmental atlas of maize. Science, 2016. 353(6301): p. 814–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golovkin M and Reddy AS, An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1–70K protein. J Biol Chem, 1999. 274(51): p. 36428–38. [DOI] [PubMed] [Google Scholar]
- 36.Carvalho RF, Szakonyi D, Simpson CG, Barbosa IC, Brown JW, Baena-Gonzalez E, and Duque P, The Arabidopsis SR45 Splicing Factor, a Negative Regulator of Sugar Signaling, Modulates SNF1-Related Protein Kinase 1 Stability. Plant Cell, 2016. 28(8): p. 1910–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang XN, Shi Y, Powers JJ, Gowda NB, Zhang C, Ibrahim HMM, Ball HB, Chen SL, Lu H, and Mount SM, Transcriptome analyses reveal SR45 to be a neutral splicing regulator and a suppressor of innate immunity in Arabidopsis thaliana. BMC Genomics, 2017. 18(1): p. 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, and Zipfel C, Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell, 2014. 54(1): p. 43–55. [DOI] [PubMed] [Google Scholar]
- 39.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boiler T, Ausubel FM, and Sheen J, MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 2002. 415(6875): p. 977–83. [DOI] [PubMed] [Google Scholar]
- 40.Zhang M, Su J, Zhang Y, Xu J, and Zhang S, Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr Opin Plant Biol, 2018. 45(Pt A): p. 1–10. [DOI] [PubMed] [Google Scholar]
- 41.Mei Y, Zhang C, Kernodle BM, Hill JH, and Whitham SA, A Foxtail mosaic virus Vector for Virus-Induced Gene Silencing in Maize. Plant Physiol, 2016. 171(2): p. 760–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anko ML, Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin Cell Dev Biol, 2014. 32: p. 11–21. [DOI] [PubMed] [Google Scholar]
- 43.Jeong S, SR Proteins: Binders, Regulators, and Connectors of RNA. Mol Cells, 2017. 40(1): p. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, and Browse J, JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature, 2007. 448(7154): p. 661–5. [DOI] [PubMed] [Google Scholar]
- 45.Chung HS and Howe GA, A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell, 2009. 21(1): p. 131–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jabs T, Tschöpe M, Colling C, Hahlbrock K, and Scheel DJPotNAoS, Elicitor-stimulated ion fluxes and O2− from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. 1997. 94(9): p. 4800–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Acharya BR, Raina S, Maqbool SB, Jagadeeswaran G, Mosher SL, Appel HM, Schultz JC, Klessig DF, and Raina R, Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J, 2007. 50(3): p. 488–99. [DOI] [PubMed] [Google Scholar]
- 48.Waese J, Fan J, Pasha A, Yu H, Fucile G, Shi R, Cumming M, Kelley LA, Sternberg MJ, Krishnakumar V, Ferlanti E, Miller J, Town C, Stuerzlinger W, and Provart NJ, ePlant: Visualizing and Exploring Multiple Levels of Data for Hypothesis Generation in Plant Biology. Plant Cell, 2017. 29(8): p. 1806–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ali GS, Palusa SG, Golovkin M, Prasad J, Manley JL, and Reddy AS, Regulation of plant developmental processes by a novel splicing factor. PLoS One, 2007. 2(5): p. e471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liese A and Romeis T, Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim Biophys Acta, 2013. 1833(7): p. 1582–9. [DOI] [PubMed] [Google Scholar]
- 51.Klimecka M and Muszynska G, Structure and functions of plant calcium-dependent protein kinases. Acta Biochim Pol, 2007. 54(2): p. 219–33. [PubMed] [Google Scholar]
- 52.Manley JL and Tacke R, SR proteins and splicing control. Genes Dev, 1996. 10(13): p. 1569–79. [DOI] [PubMed] [Google Scholar]
- 53.Xing D, Wang Y, Hamilton M, Ben-Hur A, and Reddy AS, Transcriptome-Wide Identification of RNA Targets of Arabidopsis SERINE/ARGININE-RICH45 Uncovers the Unexpected Roles of This RNA Binding Protein in RNA Processing. Plant Cell, 2015. 27(12): p. 3294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carpenter S, Ricci EP, Mercier BC, Moore MJ, and Fitzgerald KA, Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol, 2014. 14(6): p. 361–76. [DOI] [PubMed] [Google Scholar]
- 55.Yang S, Tang F, and Zhu H, Alternative splicing in plant immunity. Int J Mol Sci, 2014. 15(6): p. 10424–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howard BE, Hu Q, Babaoglu AC, Chandra M, Borghi M, Tan X, He L, Winter-Sederoff H, Gassmann W, Veronese P, and Heber S, High-throughput RNA sequencing of pseudomonas-infected Arabidopsis reveals hidden transcriptome complexity and novel splice variants. PLoS One, 2013. 8(10): p. e74183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ling Z, Zhou W, Baldwin IT, and Xu S, Insect herbivory elicits genome-wide alternative splicing responses in Nicotiana attenuata. Plant J, 2015. 84(1): p. 228–43. [DOI] [PubMed] [Google Scholar]
- 58.Bazin J, Mariappan K, Jiang Y, Blein T, Voelz R, Crespi M, and Hirt H, Role of MPK4 in pathogen-associated molecular pattern-triggered alternative splicing in Arabidopsis. PLoS Pathog, 2020. 16(4): p. e1008401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu S, Zhang Z, Jing B, Gannon P, Ding J, Xu F, Li X, and Zhang Y, Transportin-SR is required for proper splicing of resistance genes and plant immunity. PLoS Genet, 2011. 7(6): p. e1002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z, Liu Y, Ding P, Li Y, Kong Q, and Zhang Y, Splicing of receptor-like kinase-encoding SNC4 and CERK1 is regulated by two conserved splicing factors that are required for plant immunity. Mol Plant, 2014. 7(12): p. 1766–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang J, Gu L, Zhang Y, Yan T, Kong G, Kong L, Guo B, Qiu M, Wang Y, Jing M, Xing W, Ye W, Wu Z, Zhang Z, Zheng X, Gijzen M, Wang Y, and Dong S, An oomycete plant pathogen reprograms host pre-mRNA splicing to subvert immunity. Nat Commun, 2017. 8(1): p. 2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dinesh-Kumar SP and Baker BJ, Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci U S A, 2000. 97(4): p. 1908–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang XC and Gassmann W, Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol, 2007. 145(4): p. 1577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J, Chen X, Liang X, Zhou X, Yang F, Liu J, He SY, and Guo Z, Alternative Splicing of Rice WRKY62 and WRKY76 Transcription Factor Genes in Pathogen Defense. Plant Physiol, 2016. 171(2): p. 1427–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, and Zhou J-M, A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature, 2012. 485(7396): p. 114–118. [DOI] [PubMed] [Google Scholar]
- 66.Rao S, Zhou Z, Miao P, Bi G, Hu M, Wu Y, Feng F, Zhang X, and Zhou J-M, Roles of Receptor-Like Cytoplasmic Kinase VII Members in Pattern-Triggered Immune Signaling. 2018. 177(4): p. 1679–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walley JW, Shen Z, Sartor R, Wu KJ, Osborn J, Smith LG, and Briggs SP, Reconstruction of protein networks from an atlas of maize seed proteotypes. Proc Natl Acad Sci U S A, 2013. 110(49): p. E4808–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Supek F, Bošnjak M, Škunca N, and Šmuc T, REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One, 2011. 6(7): p. e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gietz RD and Woods RA, Yeast Transformation by the LiAc/SS Carrier DNA/PEG Method, in Yeast Protocol, Xiao W, Editor. 2006, Humana Press: Totowa, NJ: p. 107–120. [Google Scholar]
- 70.Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, and Zipfel C, Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet, 2011. 7(4): p. e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu F and Copeland C, Nuclear Extraction from Arabidopsis thaliana. Bio-protocol, 2012. 2(24): p. e306. [Google Scholar]
- 72.Schmelz EA, Alborn HT, and Tumlinson JH, The influence of intact-plant and excised-leaf bioassay designs on volicitin- and jasmonic acid-induced sesquiterpene volatile release in Zea mays. Planta, 2001. 214(2): p. 171–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


