Abstract
Purpose
The purpose of this study was to measure genomic changes that emerge with enzalutamide treatment using analyses of whole genome sequencing and RNA sequencing.
Experimental Design
One hundred and one tumors from men with mCRPC who had not been treated with enzalutamide (n=64) or who had enzalutamide-resistant mCRPC (n=37) underwent whole genome sequencing. Ninety-nine of these tumors also underwent RNA sequencing. We analyzed the genomes and transcriptomes of these mCRPC tumors.
Results
Copy number loss was more common than gain in enzalutamide-resistant tumors. Specially, we identified 124 protein-coding genes that were more commonly lost in enzalutamide-resistant samples. These 124 genes included eight putative tumor suppressors located at nine distinct genomic regions. We demonstrated that focal deletion of the 17q22 locus that includes RNF43 and SRSF1 was not present in any patient with enzalutamide-naïve mCRPC but was present in 16 percent (6/37) of patients with enzalutamide-resistant mCRPC. 17q22 loss was associated with lower RNF43 and SRSF1 expression and poor overall survival from time of biopsy (median overall survival of 19.3 months in 17q22 intact vs. 8.9 months in 17q22 loss, HR=3.44 [95% CI 1.338–8.867, log-rank p-value=0.006]). Finally, 17q22 loss was linked with activation of several targetable factors, including CDK1/2, Akt, and PLK1, demonstrating the potential therapeutic relevance of 17q22 loss in mCRPC.
Conclusion
Copy number loss is common in enzalutamide-resistant tumors. Focal deletion of chromosome 17q22 defines a previously unappreciated molecular subset of enzalutamide-resistant mCRPC associated with poor clinical outcome.
Keywords: Metastatic castration-resistant prostate cancer, 17q22 loss, Enzalutamide resistance, Druggable kinases, Wnt
INTRODUCTION
Metastatic castration-resistant prostate cancer (CRPC) is the lethal form of the disease, and the second leading cause of cancer-related mortality in men in the United States (1). Due to genomic sequencing efforts, we now know that recurrent genomic alterations in specific genes occur commonly in CRPC. These include: mutations in androgen receptor (AR), TP53, PTEN, and ETS fusions (2).
One common form of resistance to androgen deprivation therapy (ADT) is intracrine androgen production, leading to continued AR activation (3). The AR antagonist enzalutamide is one of the principal treatments for mCRPC patients and has demonstrated improved survival in men with metastatic CRPC in two phase III clinical trials (4–7). The majority of patients benefit from treatment with this agent (4,6). However, disease progression is inevitable, and little is known about mechanisms that contribute to clinical enzalutamide resistance (8,9).
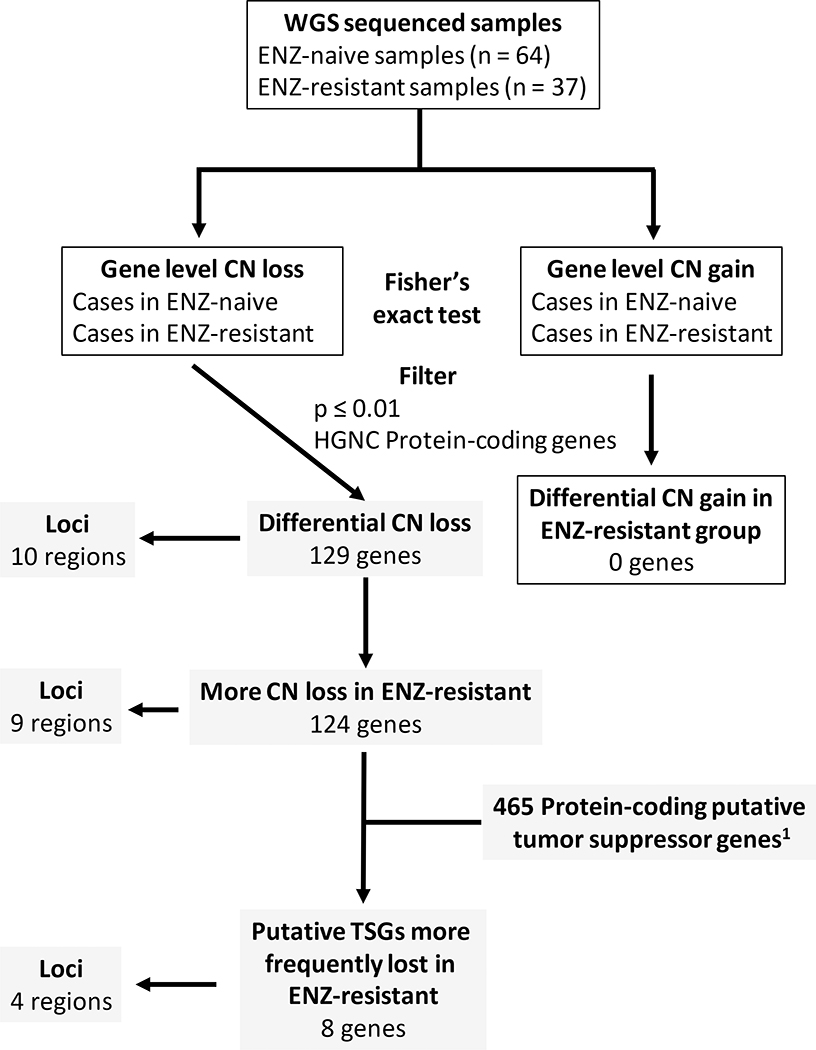
Seeking to clarify the molecular mechanisms that underlie enzalutamide resistance, we analyzed whole genome sequencing (WGS) and RNA sequencing (RNA-seq) of 101 CRPC metastases previously reported by Quigley et al (10), specifically comparing patients whose tumors were enzalutamide-naïve versus those whose tumors were enzalutamide-resistant. We hypothesized that there would be significant copy number differences between enzalutamide-resistant and -naïve samples and that specific copy number alterations would be linked to worse patient outcome. To that end, we examined copy number variation among these tumors, and compared the copy number loss events that were enriched in enzalutamide-resistant mCRPC tumors. Integrating these loci with putative tumor suppressor genes located there identified candidate genes whose loss may contribute to enzalutamide resistance.
MATERIALS AND METHODS
Tumor specimens and data processing
Tissue biopsy samples were obtained under an Institutional Review Board (IRB)-approved protocol at the participating West Coast Prostate Cancer Dream Team (WCDT) centers. All participating patients signed written, informed consent forms prior to study participation, and this protocol was conducted in accordance with recognized ethical guidelines under the supervision of the IRB at each center. Details for patient clinicopathologic features, tissue collection, sample pre-processing, WGS, and RNA-seq were previously described (10,11). WGS was performed on 101 mCRPC samples using the Illumina HiSeq platform. According to Prostate Cancer Clinical Trials Working Group-2 criteria (12), 37 samples were defined as enzalutamide-resistant based on disease progressed during treatment with enzalutamide; the other samples (n=64) were from patients not previously exposed to enzalutamide. Among these 101 samples with WGS data, 99 samples (enzalutamide-naive n=63; enzalutamide-resistant n=36) also underwent RNA-seq using Illumina NextSeq500 or Illumina HiSeq2500.
For WGS data analysis, reads were first aligned to hg38-decoy reference using the Isaac aligner (version 04.17.06.15) (13). We used Strelka (version 2.8.0) (14) for somatic mutations calling, and Annovar (15) and snpEff (16) for the annotation of the mutations. Copy numbers were derived from Canvas (version 1.28.0-O01073) (17) and CopyCat (https://github.com/chrisamiller/copyCat) with the copy number ratios being calculated by Circular Binary Segmentation (18). For RNA-seq data analysis, alignment to hg38-decoy reference was performed using STAR aligner (version 2.5.0b) with per-gene counts quantification on the basis of Illumina RNA-seq alignment app (version 1.1.0) (19). Expression of AR variant 7 (AR-V7) was measured from RNA-seq as described previously (20).
Mutation and copy number calling
Mutations were events that fulfilled the Strelka “PASS” designation and snpEff annotation of non-silent, splicing or exonic region. Copy number (CN) was defined as following: CN ≥ 3 (gain), 0.6 < CN ≤ 1.6 (shallow loss, i.e. mono-allelic), or CN ≤ 0.6 (deep loss, i.e. bi-allelic).
Curation of differential copy number alterations
To determine differential gene copy number variation between enzalutamide-naïve and -resistant samples, contingency table testing for each gene was performed using the Fisher’s exact test. More specifically, for each gene we built a contingency table accounting for copy number loss cases in enzalutamide-naïve and -resistant group and hypothesized that the proportion of gene loss was equal in the two groups. Applying multiple hypothesis correction did not yield differentially altered genes. As such, the genes meeting the uncorrected Fisher’s exact test p-value ≤ 0.01 were further analyzed for the enrichment of tumor suppressors. Differential copy number gains were tested and filtered in the same way. Cytobands were assigned to differentially altered copy number genes on the basis of hg38-decoy reference and hg38 cytoband annotation for standard chromosomes.
Clinical endpoints and survival analysis
Overall survival (OS) was measured from the time of biopsy. The median duration of follow-up for survival was approximately 14.5 months. The Kaplan-Meier method with log-rank testing and Cox proportional hazard (Cph) model was used to characterize the relationship between OS and cytobands containing putative tumor suppressors that showed differential loss between enzalutamide-naive and -resistant groups. Multivariable survival analysis with Cph model was performed to account for genomic and clinicopathologic factors. For genomic factors, RB1 2-hits, TP53 2-hits, PTEN 2-hits, and AR functional gain, as defined in the previous report (11), were selected. AR-V7 was recently shown as a prognostic factor in mCRPC and was also included (21). In addition, six of the eight clinicopathologic factors previously reported to be prognostic for OS were also examined (opioid analgesic use and serum albumin were not available in our cohort and thus excluded) (22). Genomic and clinicopathologic factors found to be prognostic in univariate Cph model at a significance level of < 0.05 were included in the subsequent multivariable survival analysis to evaluate the hazard ratio (HR) for each variable.
Gene set enrichment analysis
GSEAPreranked tool was used to perform gene set enrichment analysis. Candidate gene sets were defined in C2 canonical pathway reactome from the MSigDB database (version 6.2). We used DESeq2 to identify the genes that were differentially expressed in patients with 17q22-loss tumors (all enzalutamide resistant) relative to patients who had enzalutamide-resistant tumors but no 17q22 loss (23). Before DEseq2 analysis, low expression genes were excluded if they had fewer than 20 counts in all samples. We then ranked genes from highest confidence enrichment in patients with 17q22-loss tumors to highest confidence enrichment in patients who had enzalutamide-resistant tumors but no 17q22 loss. This ranked gene list was used as input for pathways analysis using GSEAPreranked tool with the number of permutation as 2,000.
Master regulator analysis
Kinase activity was inferred using the master regulator (MR) inference algorithm (MARINa) (24) compiled in the viper R package (25). Gene expression signatures and a regulatory network (regulome) are the two sources of data required as input for viper analysis. For each gene, we first used DESeq2 to calculate the Wald test statistics that quantified gene expression differences between samples with and without focal 17q22 deletion (23). The Wald test statistics from DESeq2 output was used as the gene expression signature input to MARINa. Each kinase regulator is comprised of positive (activated by the regulator) and negative (repressed by the regulator) targets. The kinase regulome used in this study was curated from several kinase databases as previously described (26).
Data visualization and statistical analysis
The OncoPrint plot was generated using the ComplexHeatmap R package (27). All statistical analyses were performed using R (v3.5.1). P-values were adjusted with a Benjamini-Hochberg (BH) correction to account for multiple comparisons.
RESULTS
Enzalutamide-resistance is associated with high copy number loss
Men with metastatic CRPC (mCRPC) were enrolled on the West Coast Prostate Cancer Dream Team (WCDT) biopsy protocol, and whole-genome sequencing (WGS) and RNA-sequencing were performed on samples with sufficient tumor present in the sample. Specifically, whole-genome sequences (WGS) were available for 101 samples (enzalutamide-resistant, n = 37; enzalutamide-naïve, n = 64) and RNA sequencing for 99 samples (enzalutamide-resistant, n = 36; enzalutamide-naïve, n = 63). The clinical characteristics and sequencing results have been previously reported (10,11). In this study, we sought to identify copy number alterations (CNAs) that were associated with enzalutamide resistance.
To identify CNA events—either amplification or deletion—that were enriched in enzalutamide-resistant samples, Fisher’s exact test was performed for each gene that was altered between the two groups (Fig. 1). From this analysis, we determined that copy number loss events in enzalutamide-resistant samples were more common than those in enzalutamide-naïve samples. Loss of loci with classical tumor suppressor genes, including TP53, RB1, and PTEN, were reported to be more common in mCRPC than hormone-naive cancers (2). However, only PTEN showed a trend toward greater frequency of loss in enzalutamide-resistant vs. -naïve samples (p = 0.04). (Fig. S1).
Figure 1. Identification of differential copy number alteration events between enzalutamide-naïve and -resistant samples.
Gene-level copy number (CN) gain and loss were analyzed separately. After Fisher’s exact test, differentially altered genes were defined through filtering according to unadjusted p-value and HGNC protein-coding genes. The list of genes was then intersected with previously annotated putative tumor suppressor genes by Davoli, et al (29). Locus was assigned on the basis of standard hg38 cytoband annotation. ENZ: enzalutamide; CN: copy number; HGNC: HUGO Gene Nomenclature Committee.
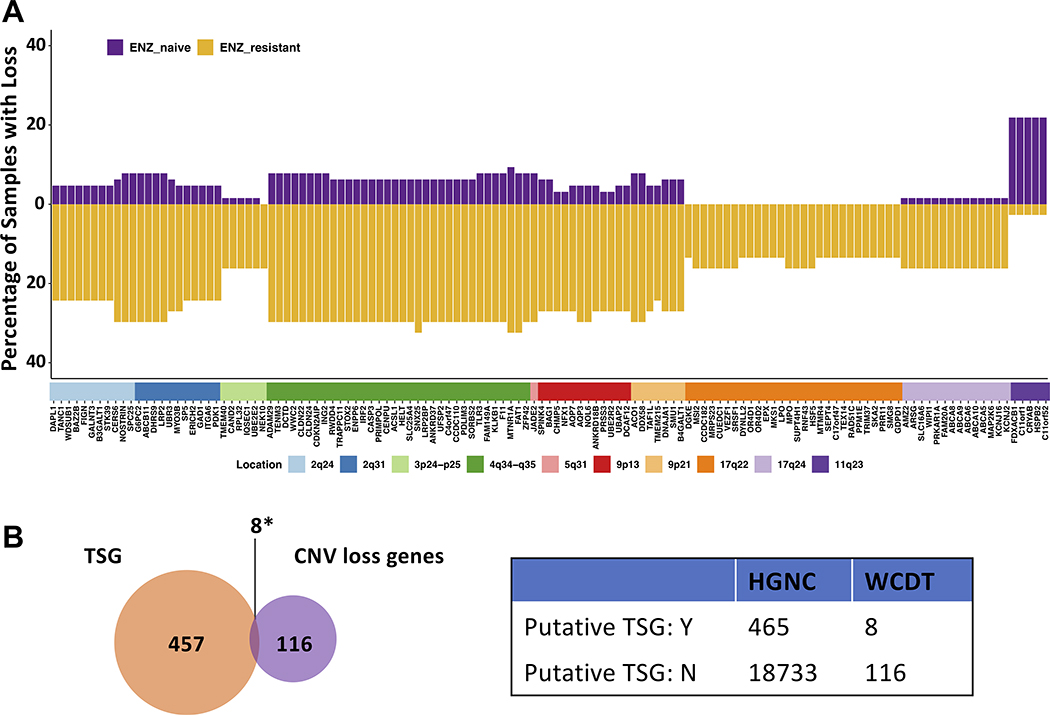
To identify other genomic loci that were different between enzalutamide-resistant vs. -naïve tumors, we examined copy number data and aimed to find changes in copy number linked with protein-coding genes that were statistically significant between these two groups (Fig. 1). No copy number gain events with significant differences were found between the two groups (Dataset S1). We hypothesized that copy number loss of specific genes, especially tumor suppressors, may contribute to enzalutamide resistance. Therefore, we focused on copy number loss events and identified 129 protein-coding genes that exhibited a differential copy number loss profile in enzalutamide-resistant tumors vs. enzalutamide-naïve tumors (unadjusted Fisher’s exact test p-value ≤ 0.01, Fig. 2A and Dataset S2). These differential loss genes were located on ten different chromosome regions (Fig. 2A). More specifically, 124 of these 129 protein-coding genes (located on nine regions) were more frequently lost in enzalutamide-resistant tumors vs. enzalutamide-naïve tumors (Fig. 2A). All of the nine loci that showed higher loss frequency in enzalutamide-resistant tumors were monoallelic (shadow loss, Fig. S2).
Figure 2. Enzalutamide-resistant samples contain more copy number losses than gains, and lost loci are enriched for putative tumor suppressor genes.
A, Waterfall plot showing the percentage of samples with copy number loss in enzalutamide-naïve and enzalutamide-resistant groups. The genes are ordered according to location on the genome. B, Left, Venn diagram showing the enrichment of putative tumor suppressor genes (TSG) from Davoli, et al (29). Right, contingency table for Fisher’s exact test. *p-value = 0.011. ENZ: Enzalutamide; TSG: Tumor suppressor gene; CNV: Copy number variation; Y: Yes; N: No.
Loss of tumor suppressor genes may confer enzalutamide resistance
We investigated our list of 124 protein-coding genes from loci that that were more frequently lost in enzalutamide-resistant versus -naïve samples and determined which of these genes were putative tumor suppressors by comparing with a list of previously curated putative tumor suppressors. The HUGO Gene Nomenclature Committee (HGNC) (28) examined a total of 19,198 protein-coding genes and 465 putative tumor suppressors were identified by Davoli et al (TUSON p-value ≤ 0.01) (Fig. 2B) (29). We compared our list of 124 protein-coding genes with this list of 465 putative tumor suppressor genes. Using these 465 putative tumor suppressor genes among all 19,198 protein-coding genes as the background for the prevalence of tumor suppressors, we determined that these putative tumor suppressor genes were over-represented in our list of protein-coding genes that had undergone copy number loss in enzalutamide-resistant tumors (Fisher’s exact test p = 0.011, odds ratio = 2.78, Fig. 2B).
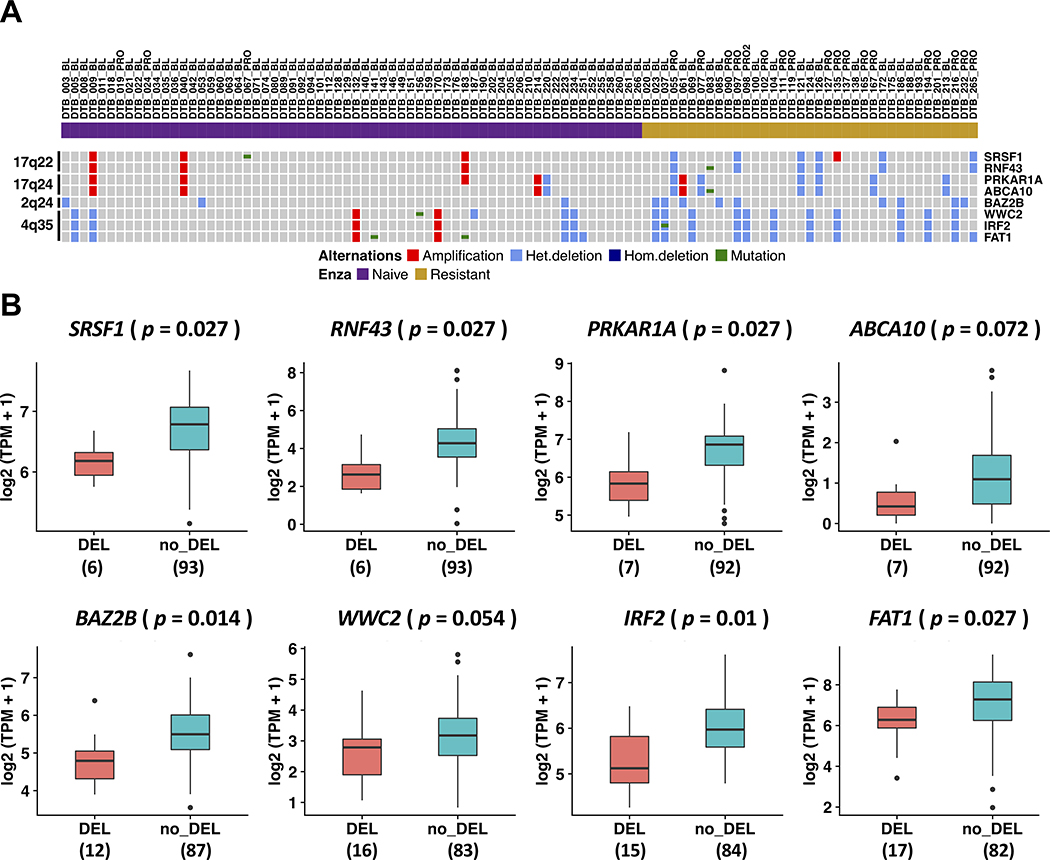
In total, we identified eight genes that were present in both our list of 124 protein-coding genes and the list of 465 putative tumor suppressor genes from Davoli, et al (29). These genes were located on four cytobands: 17q22 (SRSF1 and RNF43), 17q24 (PRKAR1A and ABCA10), 2q24 (BAZ2B), and 4q35 (WWC2, IRF2, and FAT1) (Figs. 2B and 3A). Among these eight genes, FAT1 loss was the most common in enzalutamide-resistant samples (twelve cases in enzalutamide-resistant samples [12 out of 37, 32.4%] vs. only five cases in enzalutamide-naïve samples [5 out of 64, 7.8%]). More intriguing was the loss of RNF43 and SRSF1 on 17q22 since this loss was only identified in patients resistant to enzalutamide (6 out of 37, Fig. 3A). Of our six patients whose tumors harbored 17q22 loss, 2 patients had received only enzalutamide while four others had received both enzalutamide and abiraterone. Importantly, none of the tumors from patients who had received abiraterone alone harbored 17q22 loss. In most cases, these genes along with other protein-coding genes on the same cytobands were lost concomitantly (Fig. S2).
Figure 3. Genetic alterations and gene expression of eight putative tumor suppressor genes from loci more frequently lost in enzalutamide-resistant mCRPC.
A, An OncoPrint indicating the copy number alterations and mutation status of tumor suppressor genes in each sample. B, Expression levels of tumor suppressor genes in samples with copy number loss versus samples without copy number loss reported as log2(TPM + 1). p-value was determined by two-tailed Student’s t-test and adjusted with Benjamini-Hochberg (BH) method.
With the transcriptomic data available for 99 samples, we next examined the expression of these eight putative tumor suppressor genes in samples with copy number loss versus samples without copy number loss. As shown in Fig. 3B, there was a statistically significant reduction in gene expression for six of the eight putative tumor suppressor genes whose loci were more frequently lost in enzalutamide-resistant vs. -naïve tumor samples (SRSF1, RNF43, PRKAR1A, BAZ2B, IRF2, and FAT1). A trend towards a statistically significant reduction in expression was observed for the other two genes—ABCA10 (adjusted p = 0.072) and WWC2 (adjusted p = 0.054).
Focal deletion of 17q22 containing SRSF1 and RNF43 is linked to poor overall survival
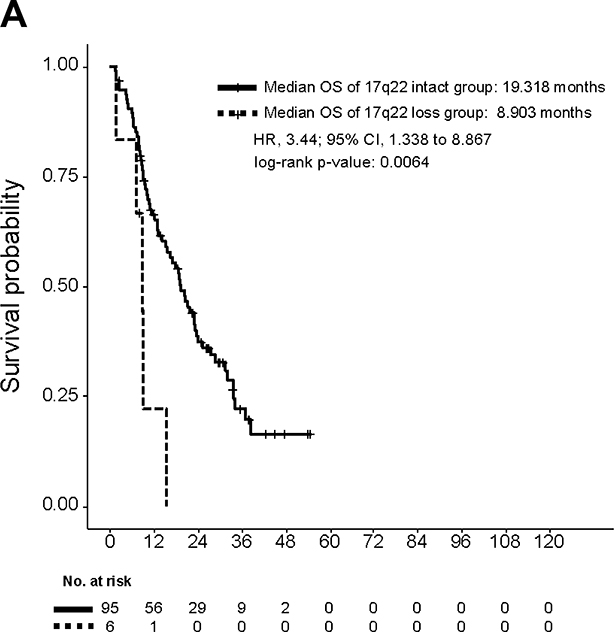
Next, we sought to determine if loss of any of the four cytobands harboring the eight putative tumor suppressor genes identified in our analysis was prognostic for overall survival (OS). To determine if loss of these loci was linked to poor outcomes, we performed survival analysis stratified by deletion of these four specific cytobands containing the putative tumor suppressors of interest. This analysis revealed that focal deletion of 17q22 that contains SRSF1 and RNF43 was associated with a significantly lower OS (Fig. 4; Fig. S3). Median OS of patients whose tumors did not harbor 17q22 loss was 19.3 months versus 8.9 months for patients whose tumors harbored 17q22 loss (log-rank p = 0.0064, Fig. 4).
Figure 4. Focal deletion of 17q22 containing RNF43 and SRSF1 is associated with poor overall survival.
Overall survival of patients harboring 17q22 loss compared with all other patients. Overall survival is calculated from the date when biopsy was taken.
Univariate survival analysis showed that clinicopathologic features including serum hemoglobin, serum lactate dehydrogenase, serum alkaline phosphatase, prostate-specific antigen (PSA), ECOG performance status and absence of visceral metastasis, and genomic factor such as RB1 2-hits, were associated with poor OS (Table S1). To determine whether 17q22 loss was predictive of worse OS after accounting for these clinicopathologic features and well-characterized genomic factor in mCRPC tumors, we performed multivariable survival analysis using Cox proportional hazard (Cph) model. 17q22 loss was prognostic (HR= 4.634 [95% CI 1.698 – 12.643], p =0.0028) after adjusting for these factors (Table 1).
Table 1.
Multivariable survival analysis
| HR | lower_0.95 | upper_0.95 | p_val | |
|---|---|---|---|---|
| Hemoglobin (g/dL) | 0.0326 | 0.0038 | 0.2772 | 0.0017 |
| Lactate dehydrogenase (IU/L) | 1.8081 | 0.8389 | 3.8971 | 0.1306 |
| Alkaline phosphatase (U/L) | 1.791 | 1.2043 | 2.6636 | 0.004 |
| Prostate-specific antigen (ng/mL) | 1.1044 | 0.923 | 1.3213 | 0.2781 |
| ECOG performance status >0 | 1.4309 | 0.8918 | 2.2959 | 0.1375 |
| Absence of visceral metastasis | 0.4272 | 0.2482 | 0.7354 | 0.0022 |
| RB1 2-hits | 1.4681 | 0.6826 | 3.1575 | 0.3257 |
| 17q22 focal loss | 4.634 | 1.6984 | 12.643 | 0.0028 |
Note: Laboratory values were measured at time of biopsy and modeled as log values for multivariable survival analysis. OS was measured from the time of biopsy.
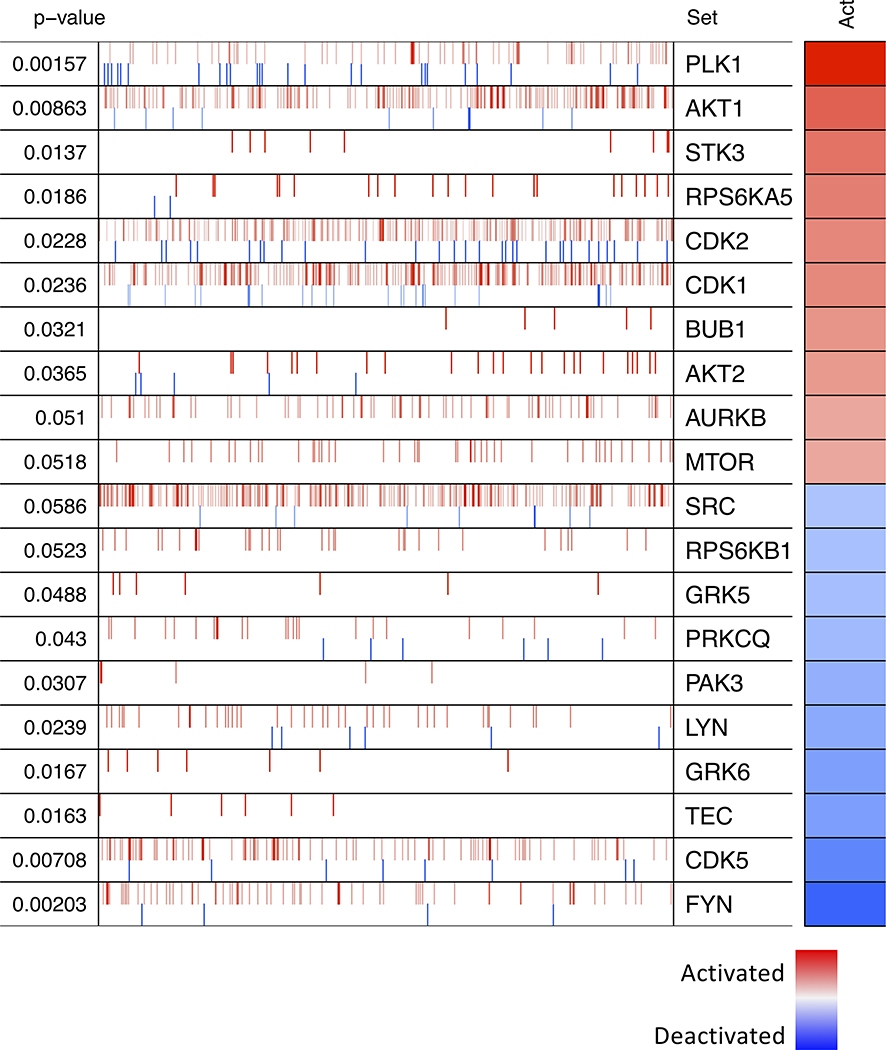
To identify potentially targetable factors that were differentially activated in tumors with 17q22 loss, we performed Master Regulator (MR) analysis—an algorithm that allows one to identify differentially activated regulators in a given comparison based on the enrichment of each regulator’s gene targets (24,25). This analysis predicted several kinases to be differentially activated in samples with 17q22 loss (Fig. 5). Among these were polo-like kinase 1 (PLK1), AKT1, and cyclin-dependent kinases (CDK1 and CDK2), all of which are targetable with small molecule inhibitors that are currently being investigated in clinical trials (Fig. 5).
Figure 5. Kinase protein activities inferred using MARINa.
The top 10 kinases predicted to be most activated (red) or deactivated (blue) in the 17q22 loss group compared with all other samples are shown. The targets of each kinase are shown as tick marks with red vertical lines representing positive targets and blue vertical lines negative targets of a given kinase. Each row also illustrates the p-value and inferred differential activity (Act) for each kinase.
Discussion
Enzalutamide is commonly used as the first-line treatment for men with metastatic CRPC whose tumors are progressing despite ADT (4). Clinical benefit is seen in the majority of patients, but resistance is nearly universal (4). While several enzalutamide resistance mechanisms have been described in pre-clinical models, there are limited analyses examining acquired resistance mechanisms in tissue biopsies from patients treated with enzalutamide, and those reports were limited in scope (8,9). To our knowledge, our report represents one of the largest collection of patient metastases from those with known enzalutamide-naïve and -resistant mCRPC and sheds new light on alterations in genomic loci that may contribute to enzalutamide resistance and that are associated with poor outcomes.
When examining copy number gains in this cohort, we found no significant differences between patients who were enzalutamide-naïve compared to those who were enzalutamide-resistant, including for the AR locus on chromosome X and the MYC locus on chromosome 8 (Dataset S1). In contrast, when examining copy number loss in this cohort, we identified 124 protein-coding genes located on nine loci that that were more frequently lost in enzalutamide-resistant vs. -naïve tumors (unadjusted Fisher’s exact test p-value ≤ 0.01, Figs. 2A and S2). By comparing this list of 124 protein-coding genes with a collection of putative tumor suppressor genes (29), we determined that eight of these 124 genes were putative tumor suppressors: SRSF1, RNF43, PRKAR1A, ABCA10, BAZ2B, WWC2, IRF2, and FAT1 (Figs. 2B and 3A). Importantly, there was a significant association between copy number loss and reduced expression for six of these putative tumor suppressor genes (Fig. 3B), demonstrating that loss of these loci may contribute to functional changes in gene expression that may lead to enzalutamide resistance.
In accordance with the two-hit model proposed by Knudson, alterations in two alleles are required to cause a phenotypic change (30). In our cohort, monoallelic loss occurred in the four loci containing the eight putative tumor suppressor genes. In addition, inactivation of the second allele through mutation was uncommon for the eight putative tumor suppressors (Fig. 3A). We cannot rule out the possibility that post-transcriptional mechanisms may lead to loss of function of the remaining intact allele. Another possible explanation for our findings is haploinsufficiency, where loss of only a single copy is required for cancer development, as exemplified by p27KIP1 (31). Alternatively, hemizygous deletion of multiple genes, as we saw in several samples, may collectively contribute to the enzalutamide-resistant phenotype (Fig. S2).
It is noteworthy that our list of 124 protein-coding genes more commonly deleted from enzalutamide-resistant vs. -naïve tumors were located on nine recurrent loss regions (Figs. 2A and S2). Out of these nine regions, 17q22 and 4q34–35 contain a large number of protein-coding genes (Figs. 2A and S2). Importantly, previous genome-wide linkage analysis found an association between 17q22 and 4q35 and prostate cancer susceptibility genes among hereditary prostate cancer families (32). In our cohort, we found that focal deletion of 17q22 represented a previously underappreciated molecular subset of enzalutamide-resistant CRPC. This subset was only found in enzalutamide-resistant patients (6/37) and was associated with poor overall survival (HR 3.44 [95% CI 1.338–8.867], p = 0.0064) (Fig. 4). We hypothesized that poor prognosis of patients with tumors with 17q22 loss may be due to the putative tumor suppressors located there—RNF43 and SRSF1. RNF43 is an E3 ubiquitin ligase that is thought to negatively regulate the canonical Wnt signaling pathway, and loss of function in this gene has been well-described in multiple tumor types (33). In prostate cancer, comprehensive analysis of large datasets (680 primary tumors and 330 metastatic CRPC) revealed that RNF43 deep copy number loss is present in both primary and metastatic tumors (8/680 and 8/333, respectively) (34). Additional cases of monoallelic loss were found in both 48/680 primary tumors and 56/333 metastatic tumors. Importantly, the enzalutamide treatment status was not reported for patients included in that analysis (34). In our cohort, we did not find deep loss of RNF43, and monoallelic loss was present exclusively in enzalutamide-resistant tumors (6/37). This lack of RNF43 monoallelic loss in enzalutamide-naïve mCRPC appears to differ from the previous report by Armenia, et al (34), but the enzalutamide treatment status for patients included in that report was not reported. Differences in the copy number alteration analysis methods may also explain the differences seen between that report and our own. The splicing factor SRSF1 is also present on chromosome 17q22. The majority of the literature points to an oncogenic role for SRSF1 in numerous solid tumors (35–38). In patients with focal 17q22 deletion, SRSF1 was one of the 28 genes from that locus that was deleted (monoallelic loss) (Figs. 2A and S2). Thus, it is possible that loss of other genes from 17q22, including RNF43, may contribute to enzalutamide-resistance and poor survival in this subset (39). Alternatively, SRSF1 may play a different role in enzalutamide-resistant tumors than that previously described in prior studies. Further studies will be necessary to confirm this hypothesis.
A recent prospective study evaluated the genomic and transcriptomic features associated with primary resistance to abiraterone. The authors showed that non-responders had more frequent mutations/deletions in Wnt/β-catenin pathway and higher Wnt pathway activation scores (40). Another retrospective study found that Wnt-pathway activation, through mutations in CTNNB1 (activating mutations), APC and RNF43 (inactivating mutations), was prognostic of PSA progression-free survival on first-line abiraterone/enzalutamide and overall survival (41). Specially, low frequency (3 out of 137) of RNF43 mutations was found in that study. RNF43 is located at 17q22, and in our cohort, only enzalutamide-resistant patients had 17q22 loss (6 out of 37). Compared with the prior two studies, enzalutamide-resistant samples in our cohort were all taken post treatment. With the transcriptomic data available, we determined that enzalutamide-resistant patients with 17q22 loss had higher Wnt-pathway activation compared with those without 17q22 loss (Fig. S4). These data suggest that 17q22 loss could be a strong driver of enzalutamide resistance, possibly through Wnt-activation. This is of particular interest given that Wnt-pathway was shown to be more active in enzalutamide-resistant samples relative to enzalutamide-naïve samples (11).
There are limited treatment options for prostate cancer patients after progression on enzalutamide. Master regulator analysis of RNA-sequencing data identified several druggable kinases that were predicted to be differentially active in the subset of enzalutamide-resistant samples with focal 17q22 deletion, including polo-like kinase 1 (PLK1), Akt1, and cyclin-dependent kinases (CDK1 and CDK2) (Fig. 5). Preclinical experiments suggest that PLK1 inhibition can block CRPC tumor growth and that this may be explained in part by suppression of AR function (42,43). Given that PLK1 inhibitors have now entered clinical testing (44), our results suggest that PLK1 inhibitors warrant further examination in enzalutamide-resistant patients. Akt1 was also implicated in our master regulator analysis. Importantly, the combination of the AKT inhibitor AZD5363 plus enzalutamide was shown to delay the development of enzalutamide-resistant prostate cancer in pre-clinical models (45). Further, the Akt inhibitor ipatasertib plus abiraterone was recently shown to improve progression-free survival vs. abiraterone alone in a phase II study (46). A phase III study of this combination recently closed to accrual, and it will be important to determine if tumors harboring 17q22 loss may be particularly susceptible to this combination. Finally, proper cell division depends on CDKs, and these proteins are frequently activated in cancer. In the subset of samples with focal 17q22 deletion, we also found higher activities of CDK1 and CDK2. Prior work demonstrates that inhibition of CDK1 and CDK2 with NU2058 reduced cell proliferation in androgen-independent prostate cell lines (47), and CDK inhibitor trials are currently ongoing in metastatic CRPC.
In summary, our study demonstrates that focal deletion of 17q22, including the region with the genes RNF43 and SRSF1, defines a subset of mCRPC patients with poor prognosis. Our results also suggest that specific kinases may be activated in tumors with 17q22 loss. Importantly, drugs that block these kinases are in clinical testing in CRPC, and it will be important to correlate 17q22 loss with drug sensitivity. Finally, we did not identify any enzalutamide-naïve patients with 17q22 loss. However, because we did not have baseline tumor biopsies prior to enzalutamide for any of the patients whose enzalutamide-resistant tumor harbored 17q22 loss, our findings do not establish whether the loss of 17q22 emerges with enzalutamide treatment. Deeper sequencing, including single cell approaches, in tumor biopsies prior to enzalutamide will be necessary to answer that question. Finally, while RNF43 is a well-known tumor suppressor that negatively regulates the Wnt pathway, the role of SRSF1 remains less well defined. Further studies are thus necessary to establish the causal relationship between loss of the 17q22 locus—and all the genes that reside there—and enzalutamide resistance.
Supplementary Material
Translational Relevance.
Although enzalutamide prolongs survival of mCRPC patients, disease progression is nearly universal due to the development of drug resistance. Importantly, treatment options are limited for mCRPC patients who develop enzalutamide resistance. Thus, understanding mechanisms of resistance and identifying molecularly-defined patient subsets may lead to new approaches to overcome resistance. By analyzing the genomes and transcriptomes of enzalutamide-naïve and enzalutamide-resistant mCRPC tumors, we determined that focal deletion of chromosome 17q22 was present in 16 percent of patients with enzalutamide-resistant mCRPC—but none of the patients with enzalutamide-naïve CRPC—and was associated with poor overall survival from the time of biopsy. Master regulator analysis nominated several druggable targets including: CDK1/2, Akt, and PLK1 that may be activated in tumors with 17q22 loss.
ACKNOWLEDGEMENTS
This research was supported by: Stand Up To Cancer-Prostate Cancer Foundation - Prostate Cancer Dream Team Translational Cancer Research Grant (SU2C-AACR-DT0812: J.A.Urrutia, R.E.Reiter, M.Rettig, C.P.Evans, P.Lara, M.Gleave, T.M.Beer, G.V.Thomas, J.Huang, R.R.Aggarwal, D.A.Quigley, A.Foye, W.S.Chen, J.Youngren, A.S.Weinstein, J.M.Stuart, F.Y.Feng, E.J.Small, and J.J.Alumkal). Stand Up To Cancer (SU2C) is a division of the Entertainment Industry Foundation. This research grant was administered by the American Association for Cancer Research, the scientific partner of SU2C; Pacific Northwest Prostate Cancer SPORE/NCI (P50 CA097186: J.J.Alumkal); Department of Defense (DOD) Synergistic Idea Award (W81XWH-13–1-0420: J.J.Alumkal); National Institutes of Health (NIH) (K01LM012877: Z.Xia). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or DOD. Other support includes: Wayne D. Kuni and Joan E. Kuni Foundation (J.J.Alumkal) and a University of Michigan Rogel Scholar Award (J.J.Alumkal).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 2.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161(5):1215–28 doi 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montgomery RB, Mostaghel EA, Vessella R, Hess DL, Kalhorn TF, Higano CS, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res 2008;68(11):4447–54 doi 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 2014;371(5):424–33 doi 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scher HI, Beer TM, Higano CS, Anand A, Taplin ME, Efstathiou E, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1–2 study. Lancet 2010;375(9724):1437–46 doi 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367(13):1187–97 doi 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 7.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009;324(5928):787–90 doi 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efstathiou E, Titus M, Wen S, Hoang A, Karlou M, Ashe R, et al. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer. Eur Urol 2015;67(1):53–60 doi 10.1016/j.eururo.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han GC, Hwang J, Wankowicz SA, Zhang Z, Liu D, Cibulskis C, et al. Genomic Resistance Patterns to Second-Generation Androgen Blockade in Paired Tumor Biopsies of Metastatic Castration-Resistant Prostate Cancer. JCO Precision Oncology 2017;1:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 2018;175(3):889 doi 10.1016/j.cell.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 11.Chen WS, Aggarwal R, Zhang L, Zhao SG, Thomas GV, Beer TM, et al. Genomic Drivers of Poor Prognosis and Enzalutamide Resistance in Metastatic Castration-resistant Prostate Cancer. Eur Urol 2019. doi 10.1016/j.eururo.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scher HI, Halabi S, Tannock I, Morris M, Sternberg CN, Carducci MA, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol 2008;26(7):1148–59 doi 10.1200/JCO.2007.12.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raczy C, Petrovski R, Saunders CT, Chorny I, Kruglyak S, Margulies EH, et al. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 2013;29(16):2041–3 doi 10.1093/bioinformatics/btt314. [DOI] [PubMed] [Google Scholar]
- 14.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28(14):1811–7 doi 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 15.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38(16):e164 doi 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92 doi 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roller E, Ivakhno S, Lee S, Royce T, Tanner S. Canvas: versatile and scalable detection of copy number variants. Bioinformatics 2016;32(15):2375–7 doi 10.1093/bioinformatics/btw163. [DOI] [PubMed] [Google Scholar]
- 18.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004;5(4):557–72 doi 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21 doi 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohli M, Wang L, Xie F, Sicotte H, Yin P, Dehm SM, et al. Mutational landscapes of sequential prostate metastases and matched patient derived xenografts during enzalutamide therapy. PLoS One 2015;10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armstrong AJ, Halabi S, Luo J, Nanus DM, Giannakakou P, Szmulewitz RZ, et al. Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: the PROPHECY study. Journal of Clinical Oncology 2019;37(13):1120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halabi S, Lin C-Y, Kelly WK, Fizazi KS, Moul JW, Kaplan EB, et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. Journal of Clinical Oncology 2014;32(7):671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550 doi 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol 2010;6:377 doi 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Ye BH, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet 2016;48(8):838–47 doi 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017;32(2):204–20 e15 doi 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32(18):2847–9 doi 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 28.Braschi B, Denny P, Gray K, Jones T, Seal R, Tweedie S, et al. Genenames.org: the HGNC and VGNC resources in 2019. Nucleic Acids Res 2019;47(D1):D786–D92 doi 10.1093/nar/gky930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 2013;155(4):948–62 doi 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 1971;68(4):820–3 doi 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao H, Ouyang X, Banach-Petrosky W, Borowsky AD, Lin Y, Kim M, et al. A critical role for p27kip1 gene dosage in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci U S A 2004;101(49):17204–9 doi 10.1073/pnas.0407693101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillanders EM, Xu J, Chang BL, Lange EM, Wiklund F, Bailey-Wilson JE, et al. Combined genome-wide scan for prostate cancer susceptibility genes. J Natl Cancer Inst 2004;96(16):1240–7 doi 10.1093/jnci/djh228. [DOI] [PubMed] [Google Scholar]
- 33.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene 2017;36(11):1461–73 doi 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018;50(5):645–51 doi 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das S, Krainer AR. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res 2014;12(9):1195–204 doi 10.1158/1541-7786.MCR-14–0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer 2016;16(7):413–30 doi 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anczukow O, Akerman M, Clery A, Wu J, Shen C, Shirole NH, et al. SRSF1-Regulated Alternative Splicing in Breast Cancer. Mol Cell 2015;60(1):105–17 doi 10.1016/j.molcel.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang L, Huang J, Higgs BW, Hu Z, Xiao Z, Yao X, et al. Genomic Landscape Survey Identifies SRSF1 as a Key Oncodriver in Small Cell Lung Cancer. PLoS Genet 2016;12(4):e1005895 doi 10.1371/journal.pgen.1005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solimini NL, Xu Q, Mermel CH, Liang AC, Schlabach MR, Luo J, et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science 2012;337(6090):104–9 doi 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Dehm SM, Hillman D, Sicotte H, Tan W, Gormley M, et al. A prospective genome-wide study of prostate cancer metastases reveals association of wnt pathway activation and increased cell cycle proliferation with primary resistance to abiraterone acetate–prednisone. Annals of Oncology 2018;29(2):352–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velho PI, Fu W, Wang H, Mirkheshti N, Qazi F, Lima FA, et al. Wnt-pathway Activating Mutations Are Associated with Resistance to First-line Abiraterone and Enzalutamide in Castration-resistant Prostate Cancer. European urology 2020;77(1):14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Chen L, Wang H, Ahmad N, Liu X. Inhibition of Plk1 represses androgen signaling pathway in castration-resistant prostate cancer. Cell Cycle 2015;14(13):2142–8 doi 10.1080/15384101.2015.1041689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patterson JC, Varmeh S, Erlander MG, Yaffe MB. Combination of selective polo-like kinase 1 (PLK1) inhibitor PCM-075 with abiraterone in prostate cancer and non-androgen-driven cancer models. American Society of Clinical Oncology; 2018. [Google Scholar]
- 44.Gutteridge RE, Ndiaye MA, Liu X, Ahmad N. Plk1 Inhibitors in Cancer Therapy: From Laboratory to Clinics. Mol Cancer Ther 2016;15(7):1427–35 doi 10.1158/1535-7163.MCT-15–0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toren P, Kim S, Cordonnier T, Crafter C, Davies BR, Fazli L, et al. Combination AZD5363 with Enzalutamide Significantly Delays Enzalutamide-resistant Prostate Cancer in Preclinical Models. Eur Urol 2015;67(6):986–90 doi 10.1016/j.eururo.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 46.de Bono JS, De Giorgi U, Rodrigues DN, Massard C, Bracarda S, Font A, et al. Randomized Phase II Study Evaluating Akt Blockade with Ipatasertib, in Combination with Abiraterone, in Patients with Metastatic Prostate Cancer with and without PTEN Loss. Clin Cancer Res 2019;25(3):928–36 doi 10.1158/1078-0432.CCR-18–0981. [DOI] [PubMed] [Google Scholar]
- 47.Rigas AC, Robson CN, Curtin NJ. Therapeutic potential of CDK inhibitor NU2058 in androgen-independent prostate cancer. Oncogene 2007;26(55):7611–9 doi 10.1038/sj.onc.1210586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.