Abstract
Acetylcholinesterase (AChE) and beta-secretase (BACE-1) are two attractive targets in the discovery of novel substances that could control multiple aspects of Alzheimer’s disease (AD). Chalcones are the flavonoid derivatives with diverse bioactivities, including AChE and BACE-1 inhibition. In this study, a series of N-substituted-4-phenothiazine-chalcones was synthesized and tested for AChE and BACE-1 inhibitory activities. In silico models, including two-dimensional quantitative structure–activity relationship (2D-QSAR) for AChE and BACE-1 inhibitors, and molecular docking investigation, were developed to elucidate the experimental process. The results indicated that 13 chalcone derivatives were synthesized with relatively high yields (39–81%). The bioactivities of these substances were examined with pIC50 3.73–5.96 (AChE) and 5.20–6.81 (BACE-1). Eleven of synthesized chalcones had completely new structures. Two substances AC4 and AC12 exhibited the highest biological activities on both AChE and BACE-1. These substances could be employed for further researches. In addition to this, the present study results suggested that, by using a combination of two types of predictive models, 2D-QSAR and molecular docking, it was possible to estimate the biological activities of the prepared compounds with relatively high accuracy.
Keywords: in silico, QSAR, docking, chalcone, acetylcholiesterase inhibitor, BACE-1
1. Introduction
Alzheimer disease (AD) is an irreversible disorder resulting in dementia among the elderly [1]. This neurodegenerative ailment usually involves the impairment of the cerebral cortex through a complex process leading to the progressive cognitive decline and memory loss [2,3]. Currently, about 50 million people worldwide have been affected by the disease, thus creating a heavy burden on the health care system of many countries [4,5,6]. Therefore, the discovery of new drugs for AD is urgently needed.
The exact etiology of AD is currently not fully known which might be referred to as a neurological disorder caused by multiple factors, such as (i) the low concentrations of acetylcholine (ACh) in synaptic clefts; (ii) the accumulation of extracellular amyloid plaques and intracellular neurofibrillary tangles with hyperphosphorylated Tau protein; (iii) the homeostasis dysregulation of biometals; (iv) the oxidative stress [2,7,8,9]. The multifactorial nature of AD has transformed the paradigm of AD drug development from a single target to multiple targets, either with the multitarget-directed ligands or the cocktail therapy approach [10]. Acetylcholinesterase (AChE) and beta-secretase (BACE-1) are the two drug targets that attracted much attention among others. They are two critical enzymes in Alzheimer’s pathogenesis, responsible for the defects in the cholinergic signaling pathway [11] and the formation of beta-amyloid plaques [12].
For the last few years, chalcones, belonging to flavonoid derivatives, have attracted great interest due to their diverse bioactivities, encompassing antimalarial [13,14], antimicrobial [15,16,17,18,19,20,21,22,23,24,25,26,27], antioxidant [28], antitumor [29,30,31], anti-inflammatory [32,33,34], analgesic [34,35,36], to antiulcer [37,38]. Recent studies have also indicated the abilities of chalcones in inhibiting enzymes, including alpha-glucosidase [39,40], lipoxygenase [33,41], mammalian alpha-amylase [42,43], xanthine oxidase [44], monoamine oxidase (MAO) [45,46,47], especially acetylcholinesterase [48,49,50,51,52] and beta-secretase (BACE-1) [53,54]. Therefore, the research of bioactivities of chalcone derivatives on the function of human brain, especially AChE and BACE-1 provides great promise in the discovery of new therapeutic agents for a range of neurological diseases including AD.
In the current work, a series of N-substituted-4-phenothiazine-chalcones was synthesized and examined for AChE and BACE-1 inhibiting effects. These compounds served as an external validation set to evaluate two-dimensional quantitative structure–activity relationship (2D-QSAR) models developed for the AChE and BACE-1 inhibitors. Molecular docking investigation was carried out to provide an insight into the molecular binding abilities of these compounds with the enzymes, through which more detailed information on structure–activity relationship (SAR) were then revealed.
2. Results and Discussion
2.1. Chemistry
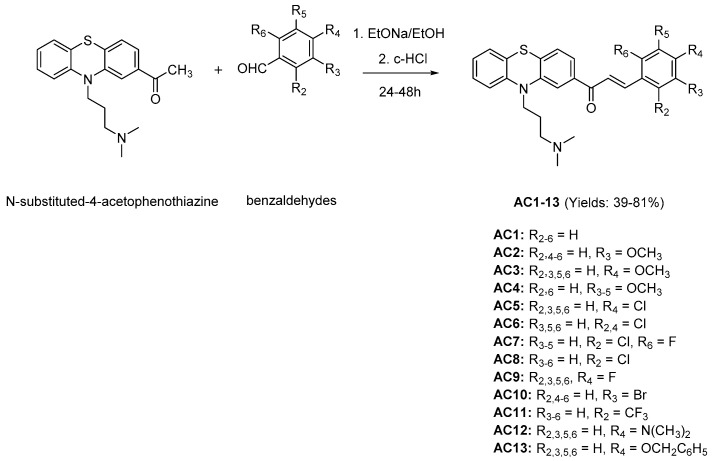
Claisen–Schmidt condensation reaction [55] was applied to synthesize 13 chalcones (Figure 1) with N-substituted-4-acetophenothiazine and benzaldehyde derivatives as reactive agents, ethanol as solvent and sodium ethanolate as catalyst. The synthetic yields were relatively high, ranging from 39% to 81%. The structures of these substances were elucidated via IR (infrared), HR-MS (high-resolution mass spectrometry), 1H-NMR (proton nuclear magnetic resonance), and 13C-NMR (carbon-13 nuclear magnetic resonance) spectra as described in the experimental section, and 11 of them were found as completely new compounds (AC2, AC4–13).
Figure 1.
Claisen–Schmidt condensation reaction in chalcones synthesis. EtONa/EtOH: Sodium ethanolate in ethanol, c-HCl: concentrated hydrochloric acid.
2.2. In Vitro Assays
The results of in vitro assays on AChE and BACE-1 inhibition are indicated in Table 1. From the biological investigation, the inhibitory activities of synthesized substances against the two enzymes were obtained with pIC50 values of 3.73–5.96 (IC50 186.21–1.10 µM) for AChE and 5.20–6.81 (IC50 6.34–0.16 µM) for BACE-1. Two compounds AC4 and AC12 exhibited the highest bioactivities in the synthesized substances with pIC50 values of 5.44 ± 0.08 (IC50 3.63 ± 0.61 µM, AChE) and 6.81 ± 0.09 (IC50 0.16 ± 0.03 µM, BACE-1), and 5.96 ± 0.10 (IC50 1.10 ± 0.24 µM, AChE) and 6.46 ± 0.05 (IC50 0.35 ± 0.04 µM, BACE-1), respectively. In the present work, galatamine, and quercetin were used as reference substances. Galantamine is a well-known AChE inhibitor used for the treatment of cognitive decline in mild to moderate AD. Quercetin is a flavonoid reported to have an inhibitory activity against BACE-1. These two compounds have been used as references in many studies on AChE and BACE-1 inhibition. The results show that the biological activities of the positive controls in this study are comparative to the previously published values [56,57,58]. On the other hand, the experimental assays also specified that AC12 had a higher activity than galantamine on AChE and all the synthesized chalcones exhibited the higher inhibiting effects than quercetin on BACE-1.
Table 1.
Acetylcholinesterase (AChE) and beta-secretase (BACE-1) inhibitory activities of synthesized N-substituted-4-phenothiazine-chalcones.
| AChE | BACE-1 | |||||
|---|---|---|---|---|---|---|
| Compound | IC50 (µM) * | pIC50 | IC50 (µM) * | pIC50 | ||
| Observed * | Predicted | Observed * | Predicted | |||
| AC1 | 33.88 ± 1.45 | 4.47 ± 0.02 | 4.80 | 6.34 ± 0.46 | 5.20 ± 0.03 | 6.52 |
| AC2 | 30.90 ± 2.10 | 4.51 ± 0.03 | 4.74 | 3.00 ± 0.00 | 5.52 ± 0.00 | 7.38 |
| AC3 | 11.48 ± 1.29 | 4.94 ± 0.05 | 4.74 | 4.48 ± 0.40 | 5.35 ± 0.04 | 7.37 |
| AC4 | 3.63 ± 0.61 | 5.44 ± 0.08 | 4.71 | 0.16 ± 0.03 | 6.81 ± 0.09 | 8.49 |
| AC5 | 60.26 ± 1.84 | 4.22 ± 0.01 | 4.82 | 1.24 ± 0.12 | 5.91 ± 0.04 | 7.25 |
| AC6 | 19.95 ± 1.57 | 4.70 ± 0.03 | 4.83 | 0.50 ± 0.00 | 6.30 ± 0.00 | 7.68 |
| AC7 | 40.74 ± 2.48 | 4.39 ± 0.03 | 4.76 | 0.45 ± 0.04 | 6.35 ± 0.04 | 7.63 |
| AC8 | 15.85 ± 1.08 | 4.80 ± 0.03 | 4.82 | 2.97 ± 0.05 | 5.53 ± 0.01 | 7.15 |
| AC9 | 25.12 ± 0.83 | 4.60 ± 0.01 | 4.76 | 3.72 ± 0.21 | 5.43 ± 0.02 | 7.12 |
| AC10 | 24.55 ± 1.09 | 4.61 ± 0.02 | 4.81 | 1.99 ± 0.21 | 5.70 ± 0.05 | 7.28 |
| AC11 | 186.21 ± 4.52 | 3.73 ± 0.01 | 4.59 | 0.40 ± 0.00 | 6.40 ± 0.00 | 7.99 |
| AC12 | 1.10 ± 0.24 | 5.96 ± 0.10 | 4.81 | 0.35 ± 0.04 | 6.46 ± 0.05 | 8.08 |
| AC13 | 11.75 ± 0.63 | 4.93 ± 0.02 | 4.63 | 3.03 ± 0.21 | 5.52 ± 0.03 | 7.20 |
| Galanthamine | 1.26 ± 0.12 | 5.90 ± 0.04 | 5.14 | - | - | - |
| Quercetin | - | - | - | 9.55 ± 0.37 | 5.02 ± 0.02 | 5.24 |
IC50: the half maximal inhibitory concentration, pIC50 = −logIC50. * Reported with standard deviation (SD).
2.3. Molecular Docking
The results of molecular docking study are expressed in Table 2 and in Supplementary Tables S1–S3 and Figures S1–S9. The interactions of 2 compounds with the highest pIC50 (AC4 and AC12) on both enzyme AChE and BACE-1 are indicated in Figure 2 and Figure 3. In this study, the co-crystallized complexes employed for AChE was 1DX6 (resolution: 2.30 Å) and for BACE-1 was 5HU1 (resolution: 1.50 Å). These were the complexes with high resolutions and the co-crystallized ligands were the drugs used in clinical (1DX6: galantamine) or in clinical development (5HU1: verubecestat). With this selection, the probability of docked compounds to be reached further optimization would be high. The results indicated that all studied substances were successfully docked into the binding pockets of AChE and BACE-1 with the docking scores of (−17.71)–(−27.80) kJ·mol−1 (AChE) and (−11.50)–(−22.51) kJ·mol−1 (BACE-1). A linear correlation between docking score and pIC50 on AChE of synthesized chalcone derivatives were revealed (Supplementary Figure S10).
Table 2.
Results of molecular docking study of the synthesized N-substituted-4-phenothiazine-chalcones on AChE (1DX6) and BACE-1 (5HU1).
| Comp. | pIC50 (AChE) | pIC50 (BACE-1) | Docking Score (kJ·mol−1) (AChE: 1DX6) |
Docking Score (kJ·mol−1) (BACE-1: 5HU1 chain A, B) |
||
|---|---|---|---|---|---|---|
| Obs. * | Pred. | Obs. * | Pred. | |||
| AC1 | 4.47 ± 0.02 | 4.80 | 5.20 ± 0.03 | 6.52 | −25.83 | −17.82; −17.69 |
| AC2 | 4.51 ± 0.03 | 4.74 | 5.52 ± 0.00 | 7.38 | −26.03 | −17.22; −17.81 |
| AC3 | 4.94 ± 0.05 | 4.74 | 5.35 ± 0.04 | 7.37 | −27.67 | −16.28; −15.37 |
| AC4 | 5.44 ± 0.08 | 4.71 | 6.81 ± 0.09 | 8.49 | −24.49 | −16.71; −13.92 |
| AC5 | 4.22 ± 0.01 | 4.82 | 5.91 ± 0.04 | 7.25 | −17.71 | −20.77; −16.87 |
| AC6 | 4.70 ± 0.03 | 4.83 | 6.30 ± 0.00 | 7.68 | −25.45 | −18.79; −16.73 |
| AC7 | 4.39 ± 0.03 | 4.76 | 6.35 ± 0.04 | 7.63 | −17.94 | −19.51; −16.85 |
| AC8 | 4.80 ± 0.03 | 4.82 | 5.53 ± 0.01 | 7.15 | −26.27 | −20.36; −16.81 |
| AC9 | 4.60 ± 0.01 | 4.76 | 5.43 ± 0.02 | 7.12 | −27.80 | −20.97; −18.06 |
| AC10 | 4.61 ± 0.02 | 4.81 | 5.70 ± 0.05 | 7.28 | −27.30 | −22.51; −20.81 |
| AC11 | 3.73 ± 0.01 | 4.59 | 6.40 ± 0.00 | 7.99 | −25.33 | −19.35; −16.41 |
| AC12 | 5.96 ± 0.10 | 4.81 | 6.46 ± 0.05 | 8.08 | −22.15 | −18.25; −16.18 |
| AC13 | 4.93 ± 0.02 | 4.63 | 5.52 ± 0.03 | 7.20 | −23.30 | −11.50; −14.09 |
| Galantamine | 5.90 ± 0.04 | 5.14 | - | - | −28.53 | - |
| Verubecestat | - | - | - | 7.66 | - | −24.95; −22.43 |
| Quercetin | - | - | 5.02 ± 0.02 | 5.24 | - | −22.23; −23.95 |
Comp.: Compound, Obs.: Observed, Pred.: Predicted. * Reported with standard deviation (SD).
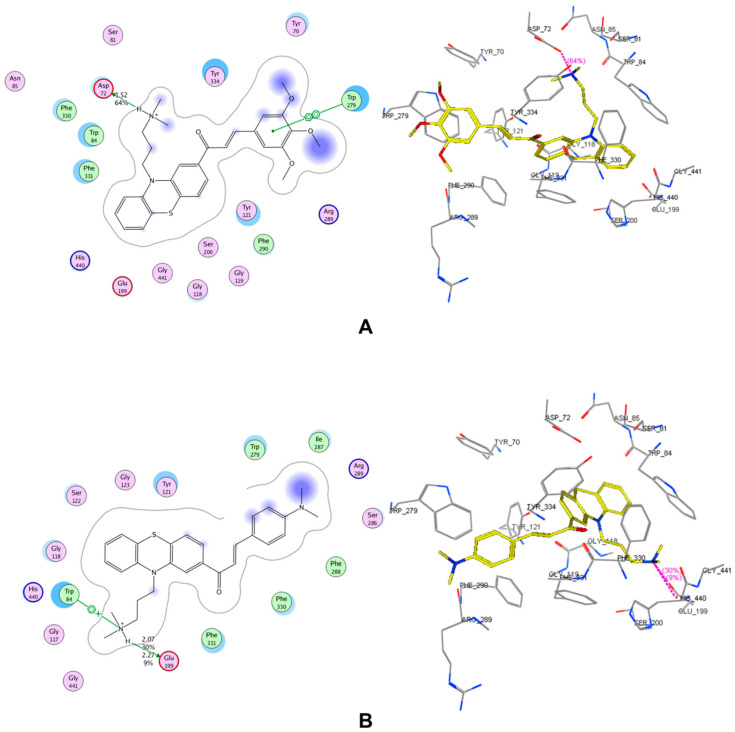
Figure 2.
Interactions in the binding pocket of acetylcholinesterase (AChE, complex 1DX6) made by (A) AC4 (observed pIC50: 5.44 ± 0.08), (B) AC12 (observed pIC50: 5.96 ± 0.10).
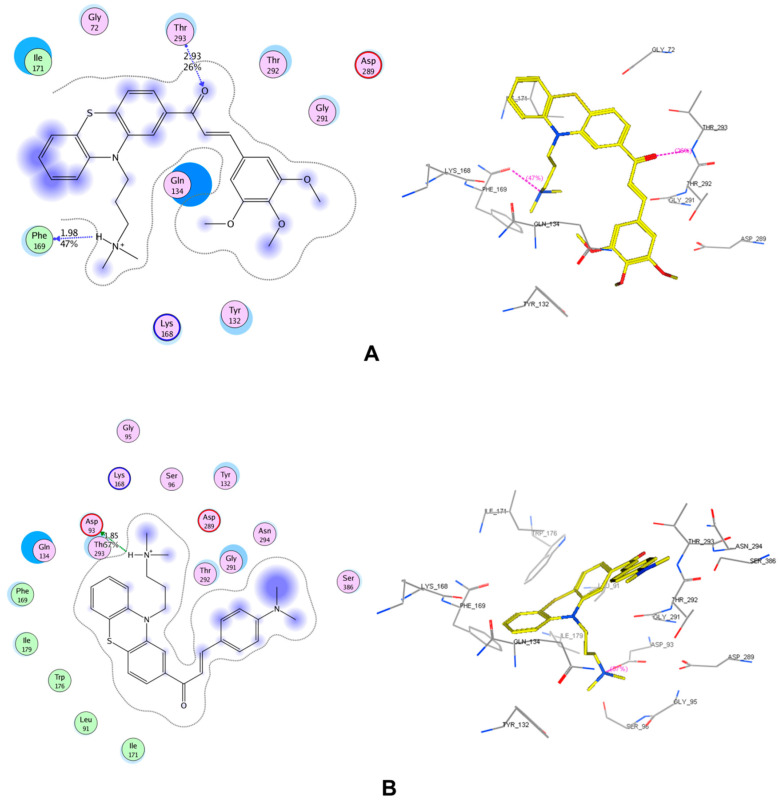
Figure 3.
Interactions in the binding pocket of beta-secretase (BACE-1, complex 5HU1-chain A) made by (A) AC4 (observed pIC50: 6.81 ± 0.09), (B) AC12 (observed pIC50: 6.46 ± 0.05).
The docking scores of most derivatives into AChE were not significantly different (except for AC5 and AC7). In addition, the differences in the observed pIC50 values could be partly explained by the analysis of the interactions between chalcone derivatives with AChE. These interactions include (i) arene–arene interactions with Trp84, Trp279, Tyr334; (ii) arene–cation interaction with Trp84; (iii) hydrogen bonding with Asp72, Glu199. These are residues of the active site-gorge of the enzyme where Trp84 and Tyr334 are exposed on the surface, particularly Asp72 on the top, Glu199 on the bottom, and Trp279 is on the entrance of the active-site gorge [59]. Eleven out of thirteen chalcone derivatives (except for AC5 and AC11) could make a hydrogen bond to Asp72. The substituents on the benzene rings of chalcone derivatives showed no strong interaction with the enzyme. However, the presences of different groups on this ring in the chalcone structures could lead to the change in scaffold’s shape as well as its interaction mode and intensity with the enzyme. There were sixsubstances with the same interaction modes (AC3, AC4, AC6, AC9, AC10, and AC11), in which AC4 was observed with the highest activity. This could be attributed to the strongest hydrogen bond between AC4 with Asp72 (score: 64%, length: 1.52 Å) compared to the other compounds.
Compared to galantamine, all the chalcone derivatives exhibited lower bioactivities (except for AC12). This could be explained by the fact that galantamine could create an arene-cation interaction with Phe330, a strong hydrogen bond to Glu199 (score: 36%, length: 1.85 Å), and a hydrogen bond (score: 22%, length: 3.07 Å) to Ser200 (a residue in catalytic triad). AC12 had the strongest observed bioactivity among the studied substances. This can be partly explained that AC12 could yield a strong hydrogen bond to Glu199. None of the rest compounds could interact with this residue. In addition, the arene–cation interaction between AC12 and Trp84 could also lead to the increased AChE inhibitory activity of this substance in relative to the others. The interaction with Trp84 (arene–arene interaction) also made the pIC50 value of AC1 equivalent to AC2 although AC1 only makes very weak hydrogen bond to Asp72 (score: 9%, length: 2.14 Å), whereas this interaction created by AC2 was very strong (score: 67%, length: 1.5 Å). Differences in the biological activity of the remaining substances can be explained by the strength of the interactions formed between those derivatives and the enzyme.
Molecular docking results on BACE-1 expressed that most compounds interacted with Asp93, Thr293 in the two chains of enzyme. AC1 and AC10 afforded the same interaction mode in both chains A and B of BACE-1 enzyme similar to verubecestate in this study. The others showed different interactions in the two chains. Two substances AC4 and AC12 (the most active compounds) both interacted with three residues: Asp93, Phe169 and Thr293. AC1 had a weak interaction with Asp93 in both chains of BACE-1 (chain A: hydrogen bond, score: 16%, length 2.22 Å; chain B: hydrogen bond, score: 30%, length 1.99 Å). Most of the remaining compounds formed strong hydrogen bonds with Asp93. Thus, there could be a correlation between the ability to create strong interactions with Asp93 with high BACE-1 inhibitory activity. AC13 exhibited many interactions with amino acid residues in both chains of the enzyme, except for the hydrogen bond with Asp93. The predicted and observed pIC50 value of this substance was as high as the others. In this study, quercetin exhibited the least bioactivity against BACE-1, but was the substance with the second highest docking score. Actually, it is not obvious that the most active compounds will always show the highest interaction energy and vice versa. Quercetin showed higher range of interaction energy due to some insignificant interactions with other elements or amino acids in the active site which have no contribution to the biological activity.
2.4. 2D-QSAR Models
The developed 2D-QSAR models for AChE and BACE-1 inhibitors were built upon the data curated from CheMBL database [60], and reported literatures [61,62,63,64,65,66,67,68,69,70,71,72]. The data was processed appropriately according to the guidelines of OECD (Organization for Economic Co-operation and Development) for an acceptable QSAR model [73] (further details are provided in the Materials and Methods section). The validation metrics for these models are showed in Table 3. The linear regressions between the observed bioactivities and those predicted from the QSAR models are expressed in Figure 4. The data sets used to build the models and the equations used for the calculations of the validation metrics are provided in Supplementary Tables S4–S6. Selected molecular descriptors used for building 2D-QSAR models are also described in detail in Supplementary Table S7. The values of selected molecular descriptors using in predicting pIC50 of the synthesized chalcone derivatives are provided in Supplementary Tables S8 and S9.
Table 3.
2D-QSAR models for AChE and BACE-1 inhibitors.
| AChE | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pIC50 = −0.928 + (2.348 × BCUT_SLOGP_3) − (0.150 × reactive) − (0.004 × PEOE_VSA + 1) − (0.005 × PEOE_VSA−3) − (0.002 × SlogP_VSA2) − (0.004 × SMR_VSA2) | ||||||||||||
| Internal Validation | External Validation | |||||||||||
| N | RMSE | R2 | RMSELOO | Q2LOO | N | RMSE | R2 | R2(PRED) | CCC | |||
| 50 | 0.18 | 0.70 | 0.22 | 0.57 | 22 | 0.16 | 0.78 | 0.78 | 0.64 | 0.69 | 0.11 | 0.88 |
| BACE-1 | ||||||||||||
| pIC50 = 1.268 + (0.870 × petitjean) + (6.370 × BCUT_PEOE_1) + (3.305 × a_ICM) − (0.478 × chiral_u) + (0.085 × rings) + (0.157 × a_Nn) + (0.006 × PEOE_VSA − 0) + (0.022 × PEOE_VSA − 6) − (0.260 × logS) + (0.009 × SlogP_VSA3) + (0.009 × SlogP_VSA5) | ||||||||||||
| Internal Validation | External Validation | |||||||||||
| N | RMSE | R2 | RMSELOO | Q2LOO | N | RMSE | R2 | R2(PRED) | CCC | |||
| 150 | 0.37 | 0.80 | 0.40 | 0.77 | 65 | 0.41 | 0.83 | 0.81 | 0.79 | 0.76 | 0.05 | 0.91 |
N: number of compounds; RMSE (root-mean-square error), R2 (squared correlation coefficient), RMSELOO (cross-validated root-mean-square error), Q2LOO (cross-validated squared correlation coefficient), CCC (concordance correlation coefficient), and (validation metrics suggested by Roy et al. [74]).
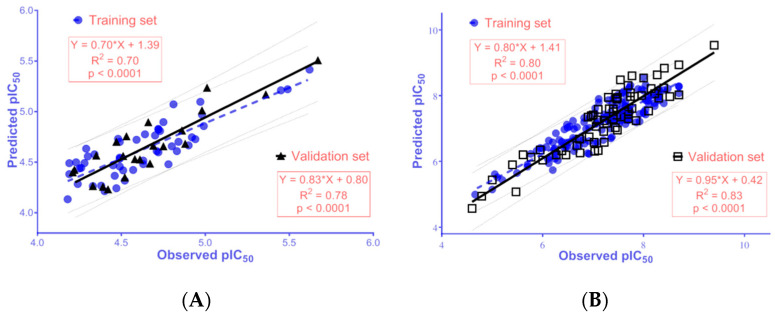
Figure 4.
The linear regression between observed pIC50 and those predicted from the 2D-QSAR models for inhibitors of (A) AChE and (B) BACE-1.
The results exhibited that the QSAR models meet the criteria thresholds for a good model, with R2 = 0.70–0.83 (>0.50), Q2LOO = 0.57–0.77 (>0.50), RMSE = 0.16–0.41 (<0.50), RMSELOO = 0.22–0.40 (<0.50), R2PRED = 0.78–0.81(>0.50), ≥ 0.65; CCC ≥ 0.85; ≥ 0.5; and ≤ 0.2 [75,76].
A comparison of the statistical results obtained from the present QSAR models and the previously published works is indicated in Table 4 and Table 5. Based on the statistical quality in the context of both internal and external validation criteria, theFn-sub models reported in this study is statistically significant and sufficiently robust to predict the biological activities of new ligands.
Table 4.
Comparison of this study with previous published works on 2D-QSAR model for AChE.
| Source | Model | Training Set | Validation Set | |||
|---|---|---|---|---|---|---|
| N | R2 | Q2 | N | R2PRED | ||
| This study | PLS | 55 | 0.70 | 0.57 | 22 | 0.78 |
| Roy et al. 2018 [77] | MLR | 284 | 0.52–0.74 | 0.50–0.71 | 142 | 0.50–0.63 |
| Niraj et al. 2015 [78] | PLS | 24 | 0.78 | 0.70 | 11 | 0.66 |
PLS: Partial least squares; MLR: Multiple linear regression; GFA: Genetic Function Approximation; N: number of compounds.
Table 5.
Comparison of this study with previous published works on 2D-QSAR model for BACE1.
| Source | Model | Training Set | Validation Set | |||
|---|---|---|---|---|---|---|
| N | R2 | Q2 | n | R2PRED | ||
| This study | PLS | 150 | 0.80 | 0.77 | 65 | 0.81 |
| Ambure et al. 2016 [79] | PLS | 52 | 0.83 | 0.76 | 22 | 0.81 |
| Ambure et al. 2016 [79] | MLR | 51 | 0.83 | 0.76 | 22 | 0.80 |
| Hossain et al. 2013 [80] | CoMFA | 71 | 1.00 | 0.77 | 35 | 0.77 |
| Hossain et al. 2013 [80] | CoMSIA | 71 | 1.00 | 0.73 | 35 | 0.71 |
| Hossain et al. 2013 [80] | PLS | 71 | 0.94 | 0.79 | 35 | 0.71 |
| Roy et al. 2018 [77] | MLR | 51 | 0.76–0.83 | 0.71–0.76 | 23 | 0.75–0.91 |
PLS: Partial least squares; MLR: Multiple linear regression; CoMFA: Comparative molecular field analysis; CoMSIA: Comparative similarity indices analysis; LHM: Linear heuristic method; N: number of compounds.
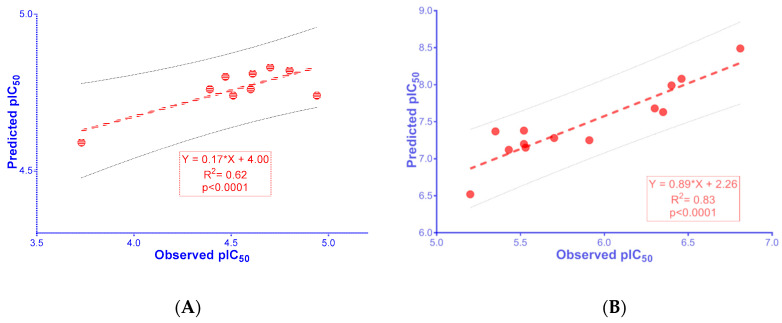
2D-QSAR model for AChE inhibitors was developed with optimal molecular descriptors, including BCUT_SLOGP_3 (the adjacency and distance matrix), reactive (physical property), PEOE_VSA+1 and PEOE_VSA–3 (partial charge), and SlogP_VSA2 and SMR_VSA2 (subdivided surface areas). The 2D-QSAR model showed a positive correlation with BCUT_SLOGP_3, and a negative correlation with reactive, PEOE_VSA+1, PEOE_VSA–3, SlogP_VSA2, and SMR_VSA2, indicating that new ligands with high BCUT_SLOGP_3, and low reactive, PEOE_VSA+1, PEOE_VSA–3, SlogP_VSA2, SMR_VSA2 values should have higher acetylcholinesterase inhibitory activities. However, as indicated in Table 3, in the developed 2D-QSAR for AChE inhibitors, BCUT_SLOGP_3 had the highest relative importance and it played a decisive role in the predicted pIC50 value. As shown in Supplementary Table S8, the studied compounds exhibited a common scaffold structure as N-substituted-4-phenothiazine; therefore, the BCUT_SLOGP_3 values were very close to each other, resulting in the similarity among the predicted pIC50 of the chalcone derivatives. When the structure changed from chalcone to galantamine, the BCUT_SLOGP_3 value displayed a small increase whilst the predicted biological activity changed significantly, suggesting that the developed QSAR model could provide some prediction on the difference in bioactivity of substances with different scaffolds. It can be observed from Table 1 and Figure 5, the 2D-QSAR for AChE in the present work could predict quite accurately the biological activity most of synthetic chalcone derivatives, with nine out of 13 compounds displaying correlations in the predicted and observed pIC50 values (R2 = 0.62).
Figure 5.
The linear regression between observed and predicted activities of synthesized chalcone derivatives against (A) AChE (values from AC1–3, AC6–11) and (B) BACE-1 (values from AC1–13).
Additionally, it can be seen that the 2D-QSAR model for BACE-1 inhibitors was developed with 11 molecular descriptors, namely petitjean and BCUT_PEOE_1 (adjacency and distance matrixes), a_ICM, chiral_u, rings, and a_Nn (atom counts and bond counts), PEOE_VSA–0 and PEOE_VSA–6 (partial charges), logS (physical properties), and SlogP_VSA3 and SlogP_VSA5 (subdivided surface areas). The 2D-QSAR model reported a positive correlation between pIC50 for BACE-1 inhibition with the descriptors of petitjean, BCUT_PEOE_1, a_ICM, rings, a_Nn, PEOE_VSA–0, PEOE_VSA–6, SlogP_VSA3, SlogP_VSA5, and a negative correlation with chiral_u, logS. This thus suggested that new ligands with high petitjean, BCUT_PEOE_1, a_ICM, rings, a_Nn, PEOE_VSA–0, PEOE_VSA–6, SlogP_VSA3, SlogP_VSA5, and low chiral_u, or logS values should have higher BACE-1 inhibiting effect. The descriptor BCUT_PEOE_1 was positively correlated with biological activity; it had the highest relative importance with a decisive role to the predicted pIC50 value (Table 3). The results shown in Table 1 indicate that the two substances, AC4 and AC12, had the highest observed and predicted pIC50 values. This could be due to these two substances having higher BCUT_PEOE_1 values than the others (high value of BCUT_PEOE_1 was also observed in AC11, which also exhibited a high bioactivity on BACE-1). The second factor contributing to the increased pIC50 values of AC4 and AC12 compared to other substances is the a_ICM descriptor which was the second most important parameter in the studied model and positively correlated with the pIC50 value (Table 3). In addition, high values at other descriptors such as PEOE_VSA–0, SlogP_VSA5 (Supplementary Table S9) also contributed to the increase of pIC50 of AC4 and AC12, though not much. The correlation between the QSAR predicted and observed pIC50 values for BACE-1 was obtained with R2 value of 0.83 (Figure 5).
3. Materials and Methods
3.1. Material and Instruments
All chemicals were obtained from commercial suppliers, and used without further purification. Melting points were determined on open capillary tubes and are uncorrected (using Gallenkamp apparatus). IR spectra were recorded on a Bruker FTIR Tensor 27 instrument. MS spectra were recorded on an Agilent 6200 Series TOF and 6500 Series Q-TOF LC/MS system. 1H-NMR and 13C-NMR spectra were recorded on an AV500 Bruker (500 MHz) spectrometer. Chemical shifts were reported in parts per million (ppm) downfield relative to tetramethylsilane as an internal standard. Peak splitting patterns were abbreviated as m (multiplet), s (singlet), bs (broad singlet), d (doublet), bd (broad doublet), t (triplet), and dd (doublet of doublets).
All computation processes were performed on a computer system with the processsor of Intel® CoreTM i&-7700 CPU @ 3.60 Hz, 16.0 GB of RAM, the Visual Graphic Card of NVIDIA GeForce GT 1030 2GB, and the operating system of 64 bit Windows 10 (Microsoft, Redmond, WA, USA).
3.2. Chemistry
The chemical synthesis of the chalcone compounds AC1–AC13 is indicated in the Figure 1 and as follows. N-substituted-4-acetophenothiazine and benzaldehyde derivatives in equimolar amounts were dissolved in ethanol and cooled in ice. Sodium ethanolate was then added. The resulting solution was stirred using an utrasonic probe. The chemical reaction was monitored by thin layer chromatography. Final mixture was cooled and acidified by a solution of concentrated HCl to pH ≈ 5–7 and left on for 24–48 h. The crude product appeared in solid or liquid. The solid was filtered, washed with cold water, and recrystallized from appropriate solvents to give the final product. With the liquid, post-reaction mixture was evaporated and purified by column chromatography with appropriate solvent systems to obtain the final product. The structures of the target compounds were elucidated by IR, MS, 1H-NMR, and 13C-NMR spectra.
(E)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-phenylprop-2-en-1-one (AC1), Yield: 68%. Mp: Liquid at room temperature. IR (ν cm–1, KBr): 2939 (νC-H sp3), 1658 (νC=O), 1599 (νC=C Ar), 1491 (νC=C Ar), 1061 (νC-N). MS: [M + H]+ m/z = 415.1814. 1H-NMR (500 MHz, MeOD, δ ppm): 7.76 (d, J = 16 Hz, 1H, CH=C), 7.71 (m, 2H, Ar-H), 7.66 (d, J = 16 Hz, 1H, CH=C), 7.61 (dd, J1 = 1 Hz, J2 = 8 Hz, 1H, Ar-H), 7.48 (d, 1H, Ar-H), 7.41 (m, 3H, Ar-H), 7.18 (m, 2H, Ar-H), 7.09 (dd, J1 =1 Hz, J2 = 7.5 Hz, 1H, Ar-H), 6.95 (m, 2H, Ar-H), 3.94 (t, J = 7 Hz, 2H, N-CH2), 2.51 (t, J = 7.5 Hz, 2H, N-CH2), 2.22 (s, 6H, N-CH3), 1.91 (m, 2H, CH2).13C-NMR (125 MHz, MeOD, δ ppm): 191.1, 146.7, 146.1, 145.5, 138.4, 136.1, 133.7, 131.8, 130.1, 129.7, 128.9, 128.3, 128.2, 125.2, 124.3, 124.1, 122.8, 117.2, 115.8, 57.6, 46.1, 45.2, 25.2.
(E)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-(3-methoxyphenyl)prop-2-en-1-one (AC2), Yield: 43%. Mp: Liquid at room temperature. IR (ν cm–1, KBr): 2970 (νC-H sp3), 1658 (νC=O), 1601 (νC=C Ar), 1462 (νC=C Ar), 1245, (νC-O), 1088 (νC-N). MS: [M + H]+ m/z = 445.1977. 1H-NMR (500 MHz, MeOD, δ ppm): 7.78 (d, J = 16 Hz, 1H, CH=C), 7.72 (d, J = 16 Hz, 1H, CH=C), 7.72 (dd, J1 = 1.5 Hz, J2 = 8.5 Hz, 1H, Ar-H), 7.58 (d, J = 1.5 Hz, 1H, Ar-H), 7.35 (m, 3H, Ar-H), 7.29 (d, J = 8 Hz, 1H, Ar-H), 7.24 (m, 1H, Ar-H), 7.15 (dd, J1 = 1.5 Hz, J2 = 7.5 Hz, 1H, Ar-H), 7.05 (dd, J1 = 1.5 Hz, J2 = 7.5 Hz, 1H, Ar-H), 7.03 (m, 1H, Ar-H), 7.00 (m, 1H, Ar-H), 4.06 (t, J = 7 Hz, 2H, N-CH2), 3.88 (s, 3H, OCH3), 2.56 (t, J = 7.5 Hz, 2H, N-CH2), 2.24 (s, 6H, N-CH3), 2.01 (m, 2H, CH2). 13C-NMR (125 MHz, MeOD, δ ppm): 191.5, 146.9, 146.1, 145.8, 138.2, 137.6, 133.9, 131.0, 129.0, 128.4, 128.2, 125.4, 124.4, 124.1, 123.2, 122.4, 117.7, 117.3, 115.9, 114.6, 57.8, 55.9, 46.2, 45.3, 25.4.
(E)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-(4-methoxyphenyl)prop-2-en-1-one (AC3), Yield: 41%. Mp: Liquid at room temperature. IR (ν cm–1, KBr): 2968 (νC-H sp3), 1655 (νC=O), 1593 (νC=C Ar), 1462 (νC=C Ar), 1254 (νC-O), 1036 (νC-N). MS: [M + H]+ m/z = 445.1923. 1H-NMR (500 MHz, MeOD, δ ppm): 7.76 (d, J = 15.5 Hz, 1H, CH=C), 7.69 (m, 2H, Ar-H), 7.64 (dd, J1 = 2 Hz, J2 = 8 Hz, 1H, Ar-H), 7.56 (d, J = 15.5 Hz, 1H, CH=C), 7.52 (d, J = 1.5 Hz, 1H, Ar-H), 7.23 (m, 2H, Ar-H), 7.13 (dd, J1 = 1.5 Hz, J2 = 7.5 Hz, 1H, Ar-H), 7.02 (d, J = 7.5 Hz, 1H, Ar-H), 6.97 (m, 3H, Ar-H), 4.03 (t, J = 7 Hz, 2H, N-CH2), 3.85 (s, 3H, OCH3), 2.70 (t, J = 7.5 Hz, 2H, N-CH2), 2.37 (s, 6H, N-CH3), 2.03 (m, 2H, CH2). 13C-NMR (125 MHz, MeOD, δ ppm): 191.4, 146.7, 146.3, 145.6, 138.9, 133.6, 131.7, 129.0, 128.8, 128.4, 128.2, 125.5, 124.3, 124.2, 120.3, 117.3, 115.8, 115.5, 57.4, 55.9, 45.9, 44.7, 24.7.
(E)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-(3,4,5-trimethoxyphenyl)prop-2-en-1-one (AC4), Yield: 51%. Mp: 167–168 °C. IR (ν cm–1, KBr): 2925 (νC-H sp3), 1657 (νC=O), 1582 (νC=C Ar), 1461 (νC=C Ar), 1280 (νC-O), 1124 (νC-N). MS: [M + H]+ m/z = 505.2132. 1H-NMR (500 MHz, MeOD, δ ppm): 7.75 (d, J = 15.5 Hz, 1H, CH=C), 7.74 (dd, J1 = 1.5 Hz, J2 = 7.5 Hz, 1H, Ar-H), 7.67 (d, J = 15.5 Hz, 1H, CH=C), 7.58 (d, J = 1.5 Hz, 1H, Ar-H), 7.28 (d, J = 8 Hz, 1H, Ar-H), 7.24 (td, J1 = 1.5 Hz, J2 = 8 Hz, 1H, Ar-H), 7.15 (dd, J1 = 1.5 Hz, J2 = 7.5 Hz, 1H, Ar-H), 7.09 (s, 2H, Ar-H), 7.05 (d, J = 7.5 Hz, 1H, Ar-H), 6.98 (td, J = 1Hz, J = 6.5 Hz, 1H, Ar-H), 4.06 (t, J =6.5 Hz, 2H, N-CH2), 3.92 (s, 6H, OCH3), 3.83 (s, 3H, OCH3), 2.28 (s, 6H, N-CH3), 2.02 (m, 2H, CH2). 13C-NMR (125 MHz, MeOD, δ ppm): 191.5, 154.9, 146.9, 146.4, 145.8, 141.7, 138.7, 133.9, 132.1, 129.0, 128.4, 128.2, 125.4, 124.5, 124.2, 122.3, 117.3, 115.8, 107.4, 61.2, 57.7, 56.8, 46.1, 45.2, 25.2.
(E)-3-(4-chlorophenyl)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-phenylprop-2-en-1-one (AC5), Yield: 78%. Mp: Liquid at room temperature. IR (ν cm–1, KBr): 2936 (νC-H sp3), 1657 (νC=O), 1590 (νC=C Ar), 1461 (νC=C Ar), 1089 (νC-N), 744 (νC-Cl). MS: [M + H]+ m/z = 449.1451. 1H-NMR (500 MHz, MeOD, δ ppm): 7.67 (d, J = 16 Hz, 1H, CH=C), 7.61 (m, 4H, Ar-H & CH=C), 7.44 (s, 1H, Ar-H), 7.35 (d, J = 7 Hz, 2H, Ar-H), 7.18 (t, J = 7.5 Hz, 1H, Ar-H), 7.12 (t, J = 7.5 Hz, 1H, Ar-H), 7.06 (d, J = 7.5 Hz, 1H, Ar-H), 6.93 (m, 2H, Ar-H), 3.91 (t, J = 5.5 Hz, 2H, N-CH2), 2.48 (t, J = 7 Hz, 2H, N-CH2), 2.20 (s, 6H, N-CH3), 1.92 (m, 2H, CH2). 13C-NMR (125 MHz, MeOD, δ ppm): 190.7, 146.7, 145.5, 144.3, 138.3, 137.4, 134.9, 133.7, 131.1, 130.2, 128.8, 128.3, 128.1, 125.1, 124.3, 124.1, 123.4, 117.2, 115.6, 57.6, 46.1, 45.2, 25.2.
(E)-3-(2,4-dichlorophenyl)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-phenylprop-2-en-1-one (AC6), Yield: 62%. Mp: Liquid at room temperature. IR (ν cm–1, KBr): 2940 (νC-H sp3), 1658 (νC=O), 1584 (νC=C Ar), 1463 (νC=C Ar), 1222 (νC-N). MS: [M + H]+ m/z = 483.1054. 1H-NMR (500 MHz, DMSO, δ ppm): 8.26 (d, J = 8.5 Hz, 1H, CH=C), 7.98 (m, 2H, Ar-H), 7.82 (dd, J1 = 1.5 Hz, J2 = 8 Hz, 1H), 7.78 (d, J = 2 Hz, 1H, Ar-H), 7.57 (d, J = 8 Hz, 1H, Ar-H), 7.56 (d, J = 8.5 Hz, 1H, CH=C), 7.35 (d, J = 8 Hz, 1H, Ar-H), 7.25 (td, J1 = 1.5 Hz, J2 = 8.5 Hz, 1H, Ar-H), 7.18 (dd, J = 1.5 Hz, J = 6.5 Hz, 1H, Ar-H), 7.08 (d, J = 7.5 Hz, 1H, Ar-H), 6.98 (td, J1 = 1 Hz, J2 = 6.5 Hz, 1H, Ar-H), 4.0 (t, J = 7 Hz, 2H, N-CH2), 2.35 (t, J = 7 Hz, 2H, N-CH2), 2.11 (s, 6H, N-(CH3)2), 1.83 (m, 2H, CH2). 13C-NMR (125 MHz, DMSO, δ ppm): 188.1, 137.2, 136.5, 135.6, 135.1, 131.3, 131.0, 129.8, 129.5, 128.0, 127.9, 127.2, 127.1, 125.3, 123.4, 122.8, 122.3, 116.2, 114.3, 56.2, 45.2, 44.7, 24.1.
(E)-3-(2-chloro-6-fluorophenyl)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-phenylprop-2-en-1-one (AC7), Yield: 43%. Mp: 152–155 °C. IR (ν cm–1, KBr): 2935 (νC-H sp3), 1660 (νC=O), 1596 (νC=C Ar), 1467 (νC=C Ar), 1062 (νC-N). MS: [M + H]+ m/z = 467.1368. 1H-NMR (500 MHz, DMSO, δ ppm): 7.81 (d, J = 16 Hz, 1H, CH=C), 7.74 (d, J = 16 Hz, 1H, CH=C), 7.63 (dd, J1 = 1.5 Hz, J2 = 8 Hz, 1H, Ar-H), 7.51 (m, 3H, Ar-H), 7.40 (m, 1H, Ar-H), 7.33 (d, J = 7.5 Hz, 1H, Ar-H), 7.23 (td, J1 = 1.5 Hz, J2 = 8.5 Hz, 1H, Ar-H), 7.16 (dd, J1 = 1.5 Hz, J2 = 7.5 Hz, 1H, Ar-H), 7.06 (d, J = 8 Hz, 1H, Ar-H), 6.97 (td, J1 = 1 Hz, J2 = 7.5 Hz, 1H, Ar-H), 3.97 (t, J = 7 Hz, 2H, N-CH2), 2.34 (t, J = 6.5 Hz, 2H, N-CH2), 2.10 (s, 6H, N-CH3), 1.82 (m, 2H, CH2). 13C-NMR (125 MHz, DMSO, δ ppm): 188.4, 144.8, 143.8, 136.4, 135.0, 133.1, 132.1, 130.9, 128.8, 128.0, 127.2, 126.2, 123.0, 122.8, 122.1, 121.4, 121.3, 116.1, 115.6, 114.2, 56.2, 45.2, 44.7, 24.0.
(E)-3-(2-chlorophenyl)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-phenylprop-2-en-1-one (AC8), Yield: 81%. Mp: 224–227 °C. IR (ν cm–1, KBr): 2957 (νC-H sp3), 1658 (νC=O), 1598 (νC=C Ar), 1468 (νC=C Ar), 1070 (νC-N). MS: [M + H]+ m/z = 449.1475. 1H-NMR (500 MHz, MeOD, δ ppm): 8.23 (d, J = 15.5 Hz, 1H, CH=C), 8.03 (dd, J1 = 1.5 Hz, J2 = 7 Hz, 1H, Ar-H), 7.80 (d, J = 1.5 Hz, 1H, Ar-H), 7.78 (d, J = 15.5 Hz, 1H, CH=C), 7.65 (s, 1H, Ar-H), 7.52 (dd, J1 = 1.5 Hz, J2 = 7.5 Hz, 1H, Ar-H), 7.44 (m, 2H, Ar-H), 7.37 (d, J = 8 Hz, 1H, Ar-H), 7.30 (t, J = 7.5 Hz, 1H, Ar-H), 7.22 (d, J = 8 Hz, 1H, Ar-H), 7.12 (d, J = 8 Hz, 1H, Ar-H), 7.04 (t, J = 7.5 Hz, 1H, Ar-H), 4.19 (t, J = 6.5 Hz, 2H, N-CH2), 3.24 (t, J = 8 Hz, 2H, N-CH2), 2.81 (s, 6H, N-CH3), 2.24 (m, 2H, CH2). 13C-NMR (125 MHz, MeOD, δ ppm): 190.9, 146.8, 145.5, 141.3, 138.5, 136.5, 134.6, 134.2, 132.8, 131.2, 129.3, 129.2, 128.7, 128.6, 128.5, 125.9, 125.4, 124.9, 124.6, 117.5, 115.9, 56.9, 45.2, 43.8, 23.4.
(E)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-(4-fluorophenyl)prop-2-en-1-one (AC9), Yield: 57%. Mp: 138–140 °C. IR (ν cm–1, KBr): 2931 (νC-H sp3), 1656 (νC=O), 1593 (νC=C Ar), 1463 (νC=C Ar), 1061 (νC-N). MS: [M + H]+ m/z = 433.1754. 1H-NMR (500 MHz, MeOD, δ ppm): 7.79 (m, 2H, Ar-H), 7.75 (d, J = 15.5 Hz, 1H, CH=C), 7.68 (d, J = 6 Hz, 1H, Ar-H), 7.64 (d, J = 15.5 Hz, 1H, CH=C), 7.55 (s, 1H, Ar-H), 7.24 (d, J = 8.5 Hz, 1H, Ar-H), 7.22 (d, J = 8.5 Hz, 1H, Ar-H), 7.18 (d, J = 8 Hz, 1H, Ar-H), 7.16 (d, J = 8 Hz, 1H, Ar-H), 7.14 (d, J = 8.5 Hz, 1H, Ar-H), 7.04 (m, 1H, Ar-H), 6.98 (m, 1H, Ar-H), 4.04 (t, J = 6.5, 2H, N-CH2), 2.73 (t, J = 7.5, 2H, N-CH2), 2.39 (s, 6H, N-CH3), 2.04 (m, 2H, CH2). 13C-NMR (125 MHz, MeOD, δ ppm): 191.1, 134.0, 132.7, 132.6, 132.0, 131.9, 129.0, 128.4, 128.2, 125.5, 124.4, 124.2, 122.7, 117.3, 117.1, 116.9, 115.8, 115.4, 115.2, 56.4, 45.8, 44.7, 24.7.
(E)-3-(3-bromophenyl)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-phenylprop-2-en-1-one (AC10), Yield: 51%. Mp: 178–180 °C. IR (ν cm–1, KBr): 2932 (νC-H sp3), 1659 (νC=O), 1598 (νC=C Ar), 1462 (νC=C Ar), 1059 (νC-N)). MS: [M + H]+ m/z = 493.0949 and 495.1410. 1H-NMR (500 MHz, MeOD, δ ppm): 7.94 (s, 1H, Ar-H), 7.70 (m, 4H, Ar-H & CH=C), 7.57 (m, 1H, Ar-H), 7.55 (d, J = 16 Hz, 1H, CH=C), 7.35 (m, 1H, Ar-H), 7.23 (m, 2H, Ar-H), 7.12 (dd, J1 = 1 Hz, J2 = 6.5 Hz, 1H), 7.02 (d, J = 8 Hz, 1H, Ar-H), 6.97 (t, J = 7.5 Hz, 1H, Ar-H), 4.02 (t, J = 6 Hz, 2H, N-CH2), 2.52 (t, J = 7.5 Hz, 2H, N-CH2), 2.22 (s, 6H, N-CH3), 1.99 (m, 2H, CH2). 13C-NMR (125 MHz, MeOD, δ ppm): 190.9, 146.9, 145.7, 144.1, 138.6, 138.3, 134.3, 134.0, 132.2, 131.7, 129.0, 128.6, 128.4, 128.2, 125.3, 124.5, 124.4, 124.1, 124.0, 117.3, 115.8, 57.8, 46.2, 45.3, 25.4.
(E)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-(2-trifluoromethyl)phenyl)prop-2-en-1-one (AC11), Yield: 58%. Mp: Liquid at room temperature. IR (ν cm–1, KBr): 2942 (νC-H sp3), 1662 (νC=O), 1592 (νC=C Ar), 1463 (νC=C Ar), 1123 (νC-N). MS: [M + H]+ m/z = 483.1721. 1H-NMR (500 MHz, MeOD, δ ppm): 8.13 (d, J = 15.5 Hz, 1H, CH=C), 8.10 (m, 1H, Ar-H), 7.77 (m, 1H, Ar-H), 7.73 (d, J = 15.5 Hz, 1H, CH=C), 7.70 (m, 2H, Ar-H), 7.59 (m, 1H, Ar-H), 7.54 (d, J = 10 Hz, 1H, Ar-H), 7.23 (m, 2H, Ar-H), 7.11 (m, 1H, Ar-H), 7.01 (t, J = 8.5 Hz, 1H, Ar-H), 6.96 (m, 1H, Ar-H), 4.00 (m, 2H, N-CH2), 2.51 (m, 2H, N-CH2), 2.22 (s, 3H, N-CH3), 2.21 (s, 3H, N-CH3), 1.96 (m, 2H, CH2). 13C-NMR (125 MHz, MeOD, δ ppm): 190.5, 146.9, 145.6, 140.5, 138.1, 134.9, 134.2, 133.7, 131.2, 129.9, 129.5, 129.0, 128.4, 128.2, 127.1, 127.0, 126.7, 125.2, 124.4, 124.1, 117.3, 115.8, 57.7, 46.2, 45.3, 25.3.
(E)-3-(4-(dimethylamino)phenyl)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-phenylprop-2-en-1-one (AC12), Yield: 39%. Mp: Liquid at room temperature. IR (ν cm–1, KBr): 2936 (νC-H sp3), 1647 (νC=O), 1572 (νC=C Ar), 1459 (νC=C Ar), 1168 (νC-N). MS: [M + H]+ m/z = 458.2298. 1H-NMR (500 MHz, MeOD, δ ppm): 7.76 (d, J = 15.5 Hz, 1H, CH=C), 7.60 (m, 3H, Ar-H), 7.51 (s, 1H, Ar-H), 7.43 (d, J = 15.5 Hz, 1H, CH=C), 7.22 (d, J = 8 Hz, 1H, Ar-H), 7.20 (d, J = 8 Hz, 1H, Ar-H), 7.11 (d, J = 7 Hz, 1H, Ar-H), 6.99 (d, J = 8 Hz, 1H, Ar-H), 6.95 (t, J = 7.5 Hz, 1H, Ar-H), 6.72 (d, J = 9 Hz, 2H, Ar-H), 3.98 (t, J = 6.5 Hz, 2H, N-CH2), 3.01 (s, 6H, N-CH3), 2.52 (t, J = 7.5 Hz, 2H, N-CH2), 2.22 (s, 6H, N-CH3), 1.96 (m, 2H, CH2). 13C-NMR (125 MHz, MeOD, δ ppm): 191.5, 153.9, 147.8, 146.7, 145.8, 139.4, 132.9, 131.9, 128.9, 128.4, 128.1, 125.4, 124.1, 123.7, 117.3, 117.0, 115.8, 113.0, 57.7, 46.1, 45.3, 40.2, 25.3.
(E)-3-(4-(benzyloxy)phenyl)-1-(10-(3-(dimethylamino)propyl)-10H-phenothiazin-2-yl)-3-phenylprop-2-en-1-one (AC13), Yield: 40%. Mp: 183-185 oC. IR (ν cm–1, KBr): 2936 (νC-H sp3), 1655 (νC=O), 1592 (νC=C Ar), 1459 (νC=C Ar), 1247 (νC-O), 1060 (νC-N). MS: [M + H]+ m/z = 521.2243. 1H-NMR (500 MHz, MeOD+DMSO, δ ppm): 7.80 (d, J = 15.5 Hz, 1H, CH=C), 7.79 (m, 2H, Ar-H), 7.74 (dd, J1 = 1.5 Hz, J2 = 8 Hz, 1H, Ar-H), 7.67 (d, J = 15.5 Hz, 1H, CH=C), 7.62 (d, J = 1.5 Hz, 1H, Ar-H), 7.51 (d, J = 7 Hz, 2H, Ar-H), 7.44 (m, 2H, Ar-H), 7.38 (m, 1H, Ar-H), 7.33 (d, J = 7.5 Hz, 1H, Ar-H), 7.28 (td, J1 = 1.5 Hz, J2 = 8.5 Hz, 1H, Ar-H), 7.20 (dd, J = 2 Hz, J = 7.5 Hz, 1H), 7.13 (m, 2H, Ar-H), 7.10 (d, J = 8 Hz, 1H, Ar-H), 7.00 (td, J1 = 1.5 Hz, J2 = 8 Hz, 1H, Ar-H), 5.22 (s, 2H, OCH2), 4.10 (t, J = 7 Hz, 2H, N-CH2), 2.55 (t, J = 7 Hz, 2H, N-CH2), 2.24 (s, 6H, N-CH3), 2.20 (m, 2H, CH2). 13C-NMR (125 MHz, MeOD + DMSO, δ ppm): 190.8, 162.4, 146.8, 145.9, 145.8, 138.9, 138.3, 133.1, 131.9, 129.2, 129.1, 128.8, 128.4, 128.2, 125.1, 124.3, 124.2, 120.7, 117.4, 116.6, 115.9, 71.1, 57.8, 46.2, 45.6, 25.5.
3.3. Acetylcholinesterase Inhibitory Activity Assay
AChE inhibitory activities of chalcones were determined by Ellman’s colourimetric method using purified acetylcolinesterase from electric eel (Type VI) and acetylthiocholine iodide (ATCI) as a substrate and galantamine as a reference. The assay was performed in 96-well microtiter plates in the same condition for both chalcones and control substance. 25 µL of 100 mM sodium phosphate buffer pH 8, 25 µL of sample and 25 µL acetylcholinesterase solutions containing 0.54 U/mL were mixed in each well of the plate and allowed to incubate for 15 min at 25 °C. Subsequently, 25 µL of a solution of ATCI (15 mM, dissolved in water) and 125 µL of 3 mM DTNB (5,5’-dithio-bis-nitro benzoic acid) were added. The absorbance at 405 nm was recorded during the first 5 min of the reaction. A control reaction, which was considered to have 100% activity, was carried out using the same volume of methanol/water instead of tested solutions. All samples and the positive control (galantamine) were assayed in triplicate. Percentages (%) of AChE inhibitions of tested compounds were calculated from the absorbance values as indicated in Equation (1):
| % I = [(A0E − A0) − (Ac − A0C)]/(A0E − A0) | (1) |
where I is the percent inhibition of acetylcholinesterase; A0E is the absorbance value of the control blank sample with enzyme; A0 is the absorbance value of blank sample; Ac is the absorbance value of the tested sample; A0C is the absorbance value of blank test sample.
The content of each sample was indicated in Table 6.
Table 6.
The content of each sample in AChE in vitro assay.
| Samples | ATCI | DTNB | Buffer | Chalcone | AChE |
|---|---|---|---|---|---|
| Control blank sample with enzyme (A0E) | + | + | + | − | + |
| Blank sample (A0) | + | + | + | − | − |
| Tested sample (AC) | + | + | + | + | + |
| Blank test sample (A0C) | + | + | + | + | − |
+: present; −: absent.
Linear recurrent equations indicating the correlation between common logarithm of the concentration of investigated compounds (µM) and their percentages of AChE inhibition (%) were built, from which the IC50 values (concentration that inhibits 50% AChE activity) of studied chalcones were extrapolated. The method was as described earlier [81].
3.4. β-secretase Inhibitory Activity Assay
β-secretase (BACE-1) Activity Detection Kit (Fluorescent) was purchased from Sigma–Aldrich and used to determine the effect of the synthesized chalcones on β-secretase activity. The assay was carried out according to the manufacturer’s protocol. The enzyme solution (0.3 units/uL, 2 μL) was reacted with the 50 μM of the substrate (7-methoxycumarin- 4-acetyl-(Asn670, Lue671)-amyloid β/A4 precursor protein 770 fragment 667–676-(2,4-dinitrophenyl))Lys-Arg-Arg amide trifluoroacetate salt and sulfated polysaccharide samples (2–5 mg/mL) in a fluorescence assay buffer in different wells. Baseline readings were measure immediately on a Hitachi F-7000 (excitation: 320 nm; emission: 405 nm) fluorescence spectrophotometer and repeated after 2 h incubation at 37 °C. Quercetin was used as the positive control. The method was as described earlier [82,83].
3.5. Building 2D-QSAR Model
3.5.1. Data Collection and Ligand Preparation
The dataset of AChE inhibitors was obtained from CheMBL database [60], and the BACE-1 inhibitor database was collected from reported literatures [61,62,63,64,65,66,67,68,69,70,71,72]. Initial processing of input data was then carried out, including rejecting substances with similar structures (use Cluster codes tool in MOE 2008.10 [84]), retaining compounds with the same bioassay method, and correcting IC50 appropriately. A total of 72 derivatives, with AChE inhibitory activities which were determined using Ellman’s method on the enzyme of Electrophorus electricus and galantamine as a reference, were finally obtained. The total number of BACE-1 inhibitors with the same FRET (Fluorescence Resonance Energy Transfer) test method was 215 (Supplementary Tables S4 and S5). IC50 values were then converted into pIC50 for convenient calculations. The structures were built directly in Sybyl X 2.0 [85]. These final datasets were used to build 2D-QSAR models with optimal molecular descriptors.
3.5.2. Molecular Descriptors Calculation and Processing
One hundred and eighty-four 2D molecular descriptors were calculated in MOE 2008.10, and then processed to eliminate redundant or irrelevant features for improving model quality and reducing computational time consumption [86]. Firstly, the molecular descriptors were filtered and removed useless or correlated attributes using RapidMiner 5.3.008 [87]. Subsequently, they were processed further by BestFirst search method of Weka 3.8 [88] to figure out parameters having the optimal values with 10-fold cross validation. Selected molecular descriptors used for building 2D-QSAR models in this study are described in detail in the Supplementary Table S7.
3.5.3. Database Division into Training Set and Validation Set
The database was divided into training set and validation set in a ratio of 70% to 30% using randomization method. The Rand function in MOE 2008.10 was used to split randomly the database of compounds, each of which was assigned a random number between 0 and 1.
3.5.4. Model Building and Validation
2D-QSAR model was built using the partial least square (PLS) method and then validated by the values of RMSE (root-mean-square error), R2 (squared correlation coefficient), RMSELOO (cross-validated root-mean-square error), Q2LOO (cross-validated squared correlation coefficient), and more widely used metrics , , , ; R2PRED, or CCC (concordance correlation coefficient) [89]. The equation used for calculation of these metrics are provided in the Supplementary Table S6.
3.6. Molecular Docking Procedure
Docking was firstly performed on 3 conformations of co-crystallized ligand to validate the procedure. The RMSD (Root-Mean-Square Deviation) value between re-docked conformations and the original bound ligand in the co-crystalized complex which was ≤ 1.5 Å would indicate the reliability of the binding ability prediction of new ligands [90]. Docking process was performed using complexes 1DX6 of AChE (resolution 2.3 Å) and 5HU1 (resolution 1.5 Å) of BACE-1 downloaded form Protein Data Bank [91] and FlexX program in BioSolveIT LeadIt [92] with default settings. This program applied the flexible-based docking methodology, in which the ligand was treated as a flexible component and the protein was kept rigid during docking process. FlexX used an incremental construction algorithm for the seach of ligand conformations. The base fragment was first placed into the active site by matching interaction geometries between the ligand and protein. Then, the remains were gradually built-up in conformity with a set of predefined rotatable torsion angles to account for ligand flexibility. The FlexX utilised empirical scoring functions to score and rank the docking poses [93,94].
The interactions between chalcone molecules and their target were rendered and analyzed in MOE 2008.10 program (hydrogen bonds, π–π interactions, cation–π interactions, ionic interactions). Moreover, van der Waals surface interactions were detected by the contact of hydrophilic and lipophilic surfaces of the ligands with those of binding points.
4. Conclusions
In this study, 13 N-substituted-4-phenothiazine-chalcone derivatives were synthesized and tested for AChE and BACE-1 inhibition with the pIC50 values of 3.73–5.96 (AChE) and 5.20–6.81 (BACE-1). Two 2D-QSAR models were built from curated data and validated through evaluation metrics. These models could be used to predict the bioactivities of chemical compounds with high reliabilities. The synthesized substances were considered as an external validation set to evaluate the developed 2D-QSAR model for AChE and BACE-1 inhibitors with relatively high correlations between the observed and estimated bioactivities (R2 = 0.62 for AChE and R2 = 0.83 for BACE-1). Eleven of the synthesized derivatives were newly discovered (AC2, AC4–13). The molecular docking model developed in the present work could be used to explain the difference in the observed biological activity compared to the predicted value as the 2D-QSAR models could not clearly interpreted. Thus, the combination of 2D and 3D models, between ligand- and structure-based drug designs, could allow for the prediction of the biological activities of chemical substances more accurately. Among the studied N-substituted-4-phenothiazine-chalcones, AC4 and AC12 were two derivatives with the strongest inhibitory activities against both AChE and BACE-1. These substances could be used in further studies. In addition, many substances need to be synthesized and tested for AChE and BACE-1 inhibitory activities to further validate the performance of the developed 2D-QSAR models.
Supplementary Materials
The following are available online. Table S1. Results of re-docking (RMSD in Å), Table S2: Docking results and ligand interaction (Co-crystallized 1DX6), Table S3. Docking results and ligand interaction (Co-crystallized 5HU1 chain A and B), Table S4. Dataset of 72 compounds used in the building of 2D-QSAR model for AChE inhibitors, Table S5. Dataset of 215 compounds used in the building of 2D-QSAR model for BACE-1 inhibitors, Table S6. Equations for calculation of 2D-QSAR validation metrics, Table S7. Selected descriptors used for building 2D-QSAR models, Table S8. Values of selected descriptors used in prediction of pIC50 of the synthesized chalcone derivatives (AChE), Table S9. Values of selected descriptors used in prediction of pIC50 of the synthesized chalcone derivatives (BACE-1, Table S10. Spectra of synthesized chalcone derivatives, Figure S1. Interactions of co-crystallized ligand in the protein complex 1DX6 (2D), Figure S2. Interactions of co-crystallized ligand in the protein complex 1DX6 (3D), Figure S3. Alignment of re-dock ligands with the native one in the binding pocket of of 1DX6, Figure S4. Interactions of co-crystallized ligand in the protein complex 5HU1-chain A (2D), Figure S5. Interactions of co-crystallized ligand in the protein complex 5HU1-chain A (3D), Figure S6. Alignment of re-dock ligands with the native one in the binding pocket of of 5HU1-chain A, Figure S7. Interactions of co-crystallized ligand in the protein complex 5HU1-chain B (2D), Figure S8. Interactions of co-crystallized ligand in the protein complex 5HU1-chain B (3D), Figure S9. Alignment of re-dock ligands with the native one in the binding pocket of of 5HU1-chain B, Figure S10. The linear regression between docking score and pIC50 on AChE of synthesized chalcone derivatives (A. observed values from AC1–3 and AC5–10, B. predicted values from AC1, AC4, AC6, AC8, AC10, AC13).
Author Contributions
Conceptualization, T.-D.T. and K.-M.T.; Data curation, T.-S.T. and K.-M.T.; Formal analysis, T.-S.T., M.-T.L., and K.-M.T.; Funding acquisition, K.-M.T.; Investigation, T.-S.T., M.-T.L., T.-C.-V.N., T.-H.T., T.-H.T., and K.-M.T.; Methodology, T.-S.T., M.-T.L., T.-H.T., and K.-M.T.; Resources, M.-T.L., T.-H.T., and K.-M.T.; Software, T.-S.T., T.-C.-V.N., T.-H.T., and K.-M.T.; Supervision, T.-D.T. and K.-M.T.; Validation, T.-S.T., M.-T.L., T.-H.T., T.-H.T., and K.-M.T.; Visualization, T.-S.T. and K.-M.T.; Writing—original draft, T.-S.T., M.-T.L., T.-H.T., T.-H.T., and K.-M.T.; Writing—review & editing, T.-S.T., M.-T.L., T.-C.-V.N., T.-H.T., T.-H.T., and K.-M.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research is funded by Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 106-YS.05-2015.31 to Khac-Minh Thai.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.Kim M., Park H.E., Lee S.-H., Han K., Lee J.H. Increased risk of Alzheimer’s disease in patients with psoriasis: A nationwide population-based cohort study. Sci. Rep. 2020;10:6454. doi: 10.1038/s41598-020-63550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ringman J.M. Update on Alzheimer’s and the Dementias: Introduction. Neurol. Clin. 2017;35:171–174. doi: 10.1016/j.ncl.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham W.V., Bonito-Oliva A., Sakmar T.P. Update on Alzheimer’s Disease Therapy and Prevention Strategies. Annu. Rev. Med. 2017;68:413–430. doi: 10.1146/annurev-med-042915-103753. [DOI] [PubMed] [Google Scholar]
- 4.Guo S., Getsios D., Revankar N., Xu P., Thompson G., Bobula J., Lacey L., Gaudig M. Evaluating disease-modifying agents: A simulation framework for Alzheimer’s disease. Pharmacoeconomics. 2014;32:1129–1139. doi: 10.1007/s40273-014-0203-5. [DOI] [PubMed] [Google Scholar]
- 5.Alzheimer’s Association 2015 Alzheimer’s disease facts and figures. Alzheimers Dement. 2015;11:332–384. doi: 10.1016/j.jalz.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Lalut J., Payan H., Davis A., Lecoutey C., Legay R., Santos J.S.-D.O., Claeysen S., Dallemagne P., Rochais C. Rational design of novel benzisoxazole derivatives with acetylcholinesterase inhibitory and serotoninergic 5-HT4 receptors activities for the treatment of Alzheimer’s disease. Sci. Rep. 2020;10:3014. doi: 10.1038/s41598-020-59805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blazer L.L., Neubig R.R. Small molecule protein-protein interaction inhibitors as CNS therapeutic agents: Current progress and future hurdles. Neuropsychopharmacology. 2009;34:126–141. doi: 10.1038/npp.2008.151. [DOI] [PubMed] [Google Scholar]
- 8.Querfurth H.W., LaFerla F.M. Alzheimer’s disease. N. Engl. J. Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 9.Tumiatti V., Minarini A., Bolognesi M.L., Milelli A., Rosini M., Melchiorre C. Tacrine derivatives and Alzheimer’s disease. Curr. Med. Chem. 2010;17:1825–1838. doi: 10.2174/092986710791111206. [DOI] [PubMed] [Google Scholar]
- 10.Gong C.X., Liu F., Iqbal K. Multifactorial Hypothesis and Multi-Targets for Alzheimer’s Disease. J. Alzheimers Dis. 2018;64:S107–S117. doi: 10.3233/JAD-179921. [DOI] [PubMed] [Google Scholar]
- 11.Rees T.M., Brimijoin S. The role of acetylcholinesterase in the pathogenesis of Alzheimer’s disease. Drugs Today. 2003;39:75–83. doi: 10.1358/dot.2003.39.1.740206. [DOI] [PubMed] [Google Scholar]
- 12.Vassar R. BACE1: The beta-secretase enzyme in Alzheimer’s disease. J. Mol. Neurosci. 2004;23:105–114. doi: 10.1385/JMN:23:1-2:105. [DOI] [PubMed] [Google Scholar]
- 13.Kumar D., Kumar M., Kumar A., Singh S.K. Chalcone and curcumin derivatives: A way ahead for malarial treatment. Mini Rev. Med. Chem. 2013;13:2116–2133. doi: 10.2174/13895575113136660101. [DOI] [PubMed] [Google Scholar]
- 14.Singh P., Anand A., Kumar V. Recent developments in biological activities of chalcones: A mini review. Eur. J. Med. Chem. 2014;85:758–777. doi: 10.1016/j.ejmech.2014.08.033. [DOI] [PubMed] [Google Scholar]
- 15.Lunardi F., Guzela M., Rodrigues A.T., Corrêa R., Eger-Mangrich I., Steindel M., Grisard E.C., Assreuy J., Calixto J.B., Santos A.R.S. Trypanocidal and Leishmanicidal Properties of Substitution-Containing Chalcones. Antimicrob. Agents Chemother. 2003;47:1449–1451. doi: 10.1128/AAC.47.4.1449-1451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Mello T.F., Cardoso B.M., Lopes S.N., Bitencourt H.R., Voltarelli E.M., Hernandes L., Aristides S.M., Lonardoni M.V., Silveira T.G. Activity of synthetic chalcones in hamsters experimentally infected with Leishmania (Viannia) braziliensis. Parasitol. Res. 2015;114:3587–3600. doi: 10.1007/s00436-015-4581-1. [DOI] [PubMed] [Google Scholar]
- 17.Passalacqua T.G., Dutra L.A., de Almeida L., Velasquez A.M., Torres F.A., Yamasaki P.R., dos Santos M.B., Regasini L.O., Michels P.A., Bolzani V.D.S., et al. Synthesis and evaluation of novel prenylated chalcone derivatives as anti-leishmanial and anti-trypanosomal compounds. Bioorg. Med. Chem. Lett. 2015;25:3342–3345. doi: 10.1016/j.bmcl.2015.05.072. [DOI] [PubMed] [Google Scholar]
- 18.Shakhatreh M.A., Al-Smadi M.L., Khabour O.F., Shuaibu F.A., Hussein E.I., Alzoubi K.H. Study of the antibacterial and antifungal activities of synthetic benzyl bromides, ketones, and corresponding chalcone derivatives. Drug Des. Dev. Ther. 2016;10:3653–3660. doi: 10.2147/DDDT.S116312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei Z.Y., Chi K.Q., Yu Z.K., Liu H.Y., Sun L.P., Zheng C.J., Piao H.R. Synthesis and biological evaluation of chalcone derivatives containing aminoguanidine or acylhydrazone moieties. Bioorg. Med. Chem. Lett. 2016;26:5920–5925. doi: 10.1016/j.bmcl.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Evranos-Aksoz B., Onurdag F.K., Ozgacar S.O. Antibacterial, antifungal and antimycobacterial activities of some pyrazoline, hydrazone and chalcone derivatives. Zeitschrift für Naturforschung C. 2015;70:183–189. doi: 10.1515/znc-2014-4195. [DOI] [PubMed] [Google Scholar]
- 21.Tran T.D., Do T.H., Tran N.C., Ngo T.D., Huynh T.N., Tran C.D., Thai K.M. Synthesis and anti Methicillin resistant Staphylococcus aureus activity of substituted chalcones alone and in combination with non-beta-lactam antibiotics. Bioorg. Med. Chem. Lett. 2012;22:4555–4560. doi: 10.1016/j.bmcl.2012.05.112. [DOI] [PubMed] [Google Scholar]
- 22.Yin B.T., Yan C.Y., Peng X.M., Zhang S.L., Rasheed S., Geng R.X., Zhou C.H. Synthesis and biological evaluation of alpha-triazolyl chalcones as a new type of potential antimicrobial agents and their interaction with calf thymus DNA and human serum albumin. Eur. J. Med. Chem. 2014;71:148–159. doi: 10.1016/j.ejmech.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y.H., Dong H.H., Zhao F., Wang J., Yan F., Jiang Y.Y., Jin Y.S. The synthesis and synergistic antifungal effects of chalcones against drug resistant Candida albicans. Bioorg. Med. Chem. Lett. 2016;26:3098–3102. doi: 10.1016/j.bmcl.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 24.Lahtchev K.L., Batovska D.I., Parushev S.P., Ubiyvovk V.M., Sibirny A.A. Antifungal activity of chalcones: A mechanistic study using various yeast strains. Eur. J. Med. Chem. 2008;43:2220–2228. doi: 10.1016/j.ejmech.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 25.Mateeva N., Eyunni S.V.K., Redda K.K., Ononuju U., Hansberry T.D., II, Aikens C., Nag A. Functional evaluation of synthetic flavonoids and chalcones for potential antiviral and anticancer properties. Bioorg. Med. Chem. Lett. 2017;27:2350–2356. doi: 10.1016/j.bmcl.2017.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patil S.A., Patil V., Patil R., Beaman K., Patil S.A. Identification of novel 5,6-dimethoxyindan-1-one derivatives as antiviral agents. Med. Chem. Med. Chem. 2017;13:787–795. doi: 10.2174/1573406413666170330094822. [DOI] [PubMed] [Google Scholar]
- 27.Lee J.S., Bukhari S.N., Fauzi N.M. Effects of chalcone derivatives on players of the immune system. Drug Des. Dev. Ther. 2015;9:4761–4778. doi: 10.2147/dddt.s86242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivakumar P.M., Prabhakar P.K., Doble M. Synthesis, antioxidant evaluation, and quantitative structure–activity relationship studies of chalcones. Med. Chem. Res. 2011;20:482–492. doi: 10.1007/s00044-010-9342-1. [DOI] [Google Scholar]
- 29.Yamali C., Gul H.I., Ozgun D.O., Hiroshi S.S., Umemura N., Kazaz C., Gul M. Synthesis and Cytotoxic Activities of Difluoro-Dimethoxy Chalcones. Anticancer Agents Med. Chem. 2017;17 doi: 10.2174/1871520617666170327123909. [DOI] [PubMed] [Google Scholar]
- 30.Sakagami H., Masuda Y., Tomomura M., Yokose S., Uesawa Y., Ikezoe N., Asahara D., Takao K., Kanamoto T., Terakubo S., et al. Quantitative Structure-Cytotoxicity Relationship of Chalcones. Anticancer Res. 2017;37:1091–1098. doi: 10.21873/anticanres.11421. [DOI] [PubMed] [Google Scholar]
- 31.Echeverria C., Santibanez J.F., Donoso-Tauda O., Escobar C.A., Ramirez-Tagle R. Structural antitumoral activity relationships of synthetic chalcones. Int. J. Mol. Sci. 2009;10:221–231. doi: 10.3390/ijms10010221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozdemir A., Altintop M.D., Turan-Zitouni G., Ciftci G.A., Ertorun I., Alatas O., Kaplancikli Z.A. Synthesis and evaluation of new indole-based chalcones as potential antiinflammatory agents. Eur. J. Med. Chem. 2015;89:304–309. doi: 10.1016/j.ejmech.2014.10.056. [DOI] [PubMed] [Google Scholar]
- 33.Jantan I., Bukhari S.N.A., Adekoya O.A., Sylte I. Studies of synthetic chalcone derivatives as potential inhibitors of secretory phospholipase A(2), cyclooxygenases, lipoxygenase and pro-inflammatory cytokines. Drug Des. Dev. Ther. 2014;8:1405–1418. doi: 10.2147/DDDT.S67370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdellatif K.R., Elshemy H.A., Salama S.A., Omar H.A. Synthesis, characterization and biological evaluation of novel 4’-fluoro-2’-hydroxy-chalcone derivatives as antioxidant, anti-inflammatory and analgesic agents. J. Enzym. Inhib. Med. Chem. 2015;30:484–491. doi: 10.3109/14756366.2014.949255. [DOI] [PubMed] [Google Scholar]
- 35.Heidari M.R., Foroumadi A., Noroozi H., Samzadeh-Kermani A., Azimzadeh B.S. Study of the anti-inflammatory and analgesic effects of novel rigid benzofuran-3, 4-dihydroxy chalcone by formalin, hot-plate and carrageenan tests in mice. Pak. J. Pharm Sci. 2009;22:395–401. [PubMed] [Google Scholar]
- 36.Ismail N.I., Ming-Tatt L., Lajis N., Akhtar M.N., Akira A., Perimal E.K., Israf D.A., Sulaiman M.R. Antinociceptive Effect of 3-(2,3-Dimethoxyphenyl)-1-(5-methylfuran-2-yl)prop-2-en-1-one in Mice Models of Induced Nociception. Molecules. 2016;21:1077. doi: 10.3390/molecules21081077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhiyaaldeen S.M., Amin Z.A., Darvish P.H., Mustafa I.F., Jamil M.M., Rouhollahi E., Abdulla M.A. Protective effects of (1-(4-hydroxy-phenyl)-3-m-tolyl-propenone chalcone in indomethacin-induced gastric erosive damage in rats. BMC Vet. Res. 2014;10:961. doi: 10.1186/s12917-014-0303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sashidhara K.V., Avula S.R., Mishra V., Palnati G.R., Singh L.R., Singh N., Chhonker Y.S., Swami P., Bhatta R.S., Palit G. Identification of quinoline-chalcone hybrids as potential antiulcer agents. Eur. J. Med. Chem. 2015;89:638–653. doi: 10.1016/j.ejmech.2014.10.068. [DOI] [PubMed] [Google Scholar]
- 39.Ansari F.L., Umbreen S., Hussain L., Makhmoor T., Nawaz S.A., Lodhi M.A., Khan S.N., Shaheen F., Choudhary M.I. Syntheses and biological activities of chalcone and 1,5-benzothiazepine derivatives: Promising new free-radical scavengers, and esterase, urease, and alpha-glucosidase inhibitors. Chem. Biodivers. 2005;2:487–496. doi: 10.1002/cbdv.200590029. [DOI] [PubMed] [Google Scholar]
- 40.Seo W.D., Kim J.H., Kang J.E., Ryu H.W., Curtis-Long M.J., Lee H.S., Yang M.S., Park K.H. Sulfonamide chalcone as a new class of alpha-glucosidase inhibitors. Bioorg. Med. Chem. Lett. 2005;15:5514–5516. doi: 10.1016/j.bmcl.2005.08.087. [DOI] [PubMed] [Google Scholar]
- 41.Hasan A., Khan K.M., Sher M., Maharvi G.M., Nawaz S.A., Choudhary M.I., Atta ur R., Supuran C.T. Synthesis and inhibitory potential towards acetylcholinesterase, butyrylcholinesterase and lipoxygenase of some variably substituted chalcones. J. Enzym. Inhib. Med. Chem. 2005;20:41–47. doi: 10.1080/14756360400015231. [DOI] [PubMed] [Google Scholar]
- 42.Najafian M., Ebrahim-Habibi A., Hezareh N., Yaghmaei P., Parivar K., Larijani B. Trans-chalcone: A novel small molecule inhibitor of mammalian alpha-amylase. Mol. Biol. Rep. 2011;38:1617–1620. doi: 10.1007/s11033-010-0271-3. [DOI] [PubMed] [Google Scholar]
- 43.Kashani-Amin E., Larijani B., Ebrahim-Habibi A. Neohesperidin dihydrochalcone: Presentation of a small molecule activator of mammalian alpha-amylase as an allosteric effector. FEBS Lett. 2013;587:652–658. doi: 10.1016/j.febslet.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 44.Hofmann E., Webster J., Do T., Kline R., Snider L., Hauser Q., Higginbottom G., Campbell A., Ma L., Paula S. Hydroxylated chalcones with dual properties: Xanthine oxidase inhibitors and radical scavengers. Bioorg. Med. Chem. 2016;24:578–587. doi: 10.1016/j.bmc.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathew B., Mathew G.E., Ucar G., Joy M., Nafna E.K., Suresh J. Monoamine oxidase inhibitory activity of methoxy-substituted chalcones. Int. J. Boil. Macromol. 2017;104:1321–1329. doi: 10.1016/j.ijbiomac.2017.05.162. [DOI] [PubMed] [Google Scholar]
- 46.Mathew B., Mathew G.E., Ucar G., Baysal I., Suresh J., Mathew S., Haridas A., Jayaprakash V. Potent and Selective Monoamine Oxidase-B Inhibitory Activity: Fluoro- vs. Trifluoromethyl-4-hydroxylated Chalcone Derivatives. Chem. Biodivers. 2016;13:1046–1052. doi: 10.1002/cbdv.201500367. [DOI] [PubMed] [Google Scholar]
- 47.Minders C., Petzer J.P., Petzer A., Lourens A.C. Monoamine oxidase inhibitory activities of heterocyclic chalcones. Bioorg. Med. Chem. Lett. 2015;25:5270–5276. doi: 10.1016/j.bmcl.2015.09.049. [DOI] [PubMed] [Google Scholar]
- 48.Liu H.R., Liu X.J., Fan H.Q., Tang J.J., Gao X.H., Liu W.K. Design, synthesis and pharmacological evaluation of chalcone derivatives as acetylcholinesterase inhibitors. Bioorg. Med. Chem. 2014;22:6124–6133. doi: 10.1016/j.bmc.2014.08.033. [DOI] [PubMed] [Google Scholar]
- 49.Liu H.R., Huang X.Q., Lou D.H., Liu X.J., Liu W.K., Wang Q.A. Synthesis and acetylcholinesterase inhibitory activity of Mannich base derivatives flavokawain B. Bioorg. Med. Chem. Lett. 2014;24:4749–4753. doi: 10.1016/j.bmcl.2014.07.087. [DOI] [PubMed] [Google Scholar]
- 50.Shah M.S., Khan S.U., Ejaz S.A., Afridi S., Rizvi S.U., Najam-Ul-Haq M., Iqbal J. Cholinesterases inhibition and molecular modeling studies of piperidyl-thienyl and 2-pyrazoline derivatives of chalcones. Biochem. Biophys. Res. Commun. 2017;482:615–624. doi: 10.1016/j.bbrc.2016.11.082. [DOI] [PubMed] [Google Scholar]
- 51.Bag S., Ghosh S., Tulsan R., Sood A., Zhou W., Schifone C., Foster M., LeVine H., III, Torok B., Torok M. Design, synthesis and biological activity of multifunctional alpha,beta-unsaturated carbonyl scaffolds for Alzheimer’s disease. Bioorg. Med. Chem. Lett. 2013;23:2614–2618. doi: 10.1016/j.bmcl.2013.02.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu H., Fan H., Gao X., Huang X., Liu X., Liu L., Zhou C., Tang J., Wang Q., Liu W. Design, synthesis and preliminary structure-activity relationship investigation of nitrogen-containing chalcone derivatives as acetylcholinesterase and butyrylcholinesterase inhibitors: A further study based on Flavokawain B Mannich base derivatives. J. Enzym. Inhib. Med. Chem. 2016;31:580–589. doi: 10.3109/14756366.2015.1050009. [DOI] [PubMed] [Google Scholar]
- 53.Kang J.E., Cho J.K., Curtis-Long M.J., Ryu H.W., Kim J.H., Kim H.J., Yuk H.J., Kim D.W., Park K.H. Inhibitory evaluation of sulfonamide chalcones on β-secretase and acylcholinesterase. Molecules. 2012;18:140–153. doi: 10.3390/molecules18010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma L., Yang Z., Li C., Zhu Z., Shen X., Hu L. Design, synthesis and SAR study of hydroxychalcone inhibitors of human beta-secretase (BACE1) J. Enzym. Inhib. Med. Chem. 2011;26:643–648. doi: 10.3109/14756366.2010.543420. [DOI] [PubMed] [Google Scholar]
- 55.Wang Z. Comprehensive Organic Name Reactions and Reagents. John Wiley & Sons; New York, NY, USA: 2010. [Google Scholar]
- 56.Balkrishna A., Pokhrel S., Tomer M., Verma S., Kumar A., Nain P., Gupta A., Varshney A. Anti-Acetylcholinesterase Activities of Mono-Herbal Extracts and Exhibited Synergistic Effects of the Phytoconstituents: A Biochemical and Computational Study. Molecules. 2019;24:4175. doi: 10.3390/molecules24224175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paudel P., Seong S.H., Zhou Y., Ha M.T., Min B.S., Jung H.A., Choi J.S. Arylbenzofurans from the Root Bark of Morus alba as Triple Inhibitors of Cholinesterase, β-Site Amyloid Precursor Protein Cleaving Enzyme 1, and Glycogen Synthase Kinase-3β: Relevance to Alzheimer’s Disease. ACS Omega. 2019;4:6283–6294. doi: 10.1021/acsomega.9b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jannat S., Balupuri A., Hong S.S., Choi C., Choi Y.-H., Ku J.-M., Kim W., Leem J., Kim J., Shrestha A., et al. Inhibition of β-site amyloid precursor protein cleaving enzyme 1 and cholinesterases by pterosins via a specific structure−activity relationship with a strong BBB permeability. Exp. Mol. Med. 2019;51 doi: 10.1038/s12276-019-0205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sussman J.L., Silman I. Acetylcholinesterase: Structure and use as a model for specific cation-protein interactions. Curr. Opin. Struct. Biol. 1992;2:721–729. doi: 10.1016/0959-440X(92)90207-N. [DOI] [Google Scholar]
- 60.ChEMBL Database. [(accessed on 20 May 2019)]; Available online: https://www.ebi.ac.uk/chembl/
- 61.Beswick P., Charrier N., Clarke B., Demont E., Dingwall C., Dunsdon R., Faller A., Gleave R., Hawkins J., Hussain I., et al. BACE-1 inhibitors part 3: Identification of hydroxy ethylamines (HEAs) with nanomolar potency in cells. Bioorg. Med. Chem. Lett. 2008;18:1022–1026. doi: 10.1016/j.bmcl.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 62.Charrier N., Clarke B., Cutler L., Demont E., Dingwall C., Dunsdon R., Hawkins J., Howes C., Hubbard J., Hussain I., et al. Second generation of BACE-1 inhibitors. Part 1: The need for improved pharmacokinetics. Bioorg. Med. Chem. Lett. 2009;19:3664–3668. doi: 10.1016/j.bmcl.2009.03.165. [DOI] [PubMed] [Google Scholar]
- 63.Charrier N., Clarke B., Cutler L., Demont E., Dingwall C., Dunsdon R., Hawkins J., Howes C., Hubbard J., Hussain I., et al. Second generation of BACE-1 inhibitors part 3: Towards non hydroxyethylamine transition state mimetics. Bioorg. Med. Chem. Lett. 2009;19:3674–3678. doi: 10.1016/j.bmcl.2009.03.149. [DOI] [PubMed] [Google Scholar]
- 64.Chen S.H., Lamar J., Guo D., Kohn T., Yang H.C., McGee J., Timm D., Erickson J., Yip Y., May P., et al. P3 cap modified Phe*-Ala series BACE inhibitors. Bioorg. Med. Chem. Lett. 2004;14:245–250. doi: 10.1016/j.bmcl.2003.09.085. [DOI] [PubMed] [Google Scholar]
- 65.Clarke B., Demont E., Dingwall C., Dunsdon R., Faller A., Hawkins J., Hussain I., MacPherson D., Maile G., Matico R., et al. BACE-1 inhibitors part 2: Identification of hydroxy ethylamines (HEAs) with reduced peptidic character. Bioorg. Med. Chem. Lett. 2008;18:1017–1021. doi: 10.1016/j.bmcl.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 66.Ginman T., Viklund J., Malmstrom J., Blid J., Emond R., Forsblom R., Johansson A., Kers A., Lake F., Sehgelmeble F., et al. Core refinement toward permeable beta-secretase (BACE-1) inhibitors with low hERG activity. J. Med. Chem. 2013;56:4181–4205. doi: 10.1021/jm3011349. [DOI] [PubMed] [Google Scholar]
- 67.Hamada Y., Kiso Y. Advances in the identification of beta-secretase inhibitors. Expert Opin. Drug Discov. 2013;8:709–731. doi: 10.1517/17460441.2013.784267. [DOI] [PubMed] [Google Scholar]
- 68.Ng R.A., Sun M., Bowers S., Hom R.K., Probst G.D., John V., Fang L.Y., Maillard M., Gailunas A., Brogley L., et al. Design and synthesis of hydroxyethylamine (HEA) BACE-1 inhibitors: Prime side chromane-containing inhibitors. Bioorg. Med. Chem. Lett. 2013;23:4674–4679. doi: 10.1016/j.bmcl.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 69.Oehlrich D., Prokopcova H., Gijsen H.J. The evolution of amidine-based brain penetrant BACE1 inhibitors. Bioorg. Med. Chem. Lett. 2014;24:2033–2045. doi: 10.1016/j.bmcl.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 70.Weiss M.M., Williamson T., Babu-Khan S., Bartberger M.D., Brown J., Chen K., Cheng Y., Citron M., Croghan M.D., Dineen T.A., et al. Design and preparation of a potent series of hydroxyethylamine containing beta-secretase inhibitors that demonstrate robust reduction of central beta-amyloid. J. Med. Chem. 2012;55:9009–9024. doi: 10.1021/jm300119p. [DOI] [PubMed] [Google Scholar]
- 71.Woltering T.J., Wostl W., Hilpert H., Rogers-Evans M., Pinard E., Mayweg A., Gobel M., Banner D.W., Benz J., Travagli M., et al. BACE1 inhibitors: A head group scan on a series of amides. Bioorg. Med. Chem. Lett. 2013;23:4239–4243. doi: 10.1016/j.bmcl.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 72.Charrier N., Clarke B., Demont E., Dingwall C., Dunsdon R., Hawkins J., Hubbard J., Hussain I., Maile G., Matico R., et al. Second generation of BACE-1 inhibitors, Part 2: Optimisation of the non-prime side substituent. Bioorg. Med. Chem. Lett. 2009;19:3669–3673. doi: 10.1016/j.bmcl.2009.03.150. [DOI] [PubMed] [Google Scholar]
- 73.Gómez-Jiménez G., Gonzalez-Ponce K., Castillo-Pazos D.J., Madariaga-Mazon A., Barroso-Flores J., Cortes-Guzman F., Martinez-Mayorga K. The OECD Principles for (Q)SAR Models in the Context of Knowledge Discovery in Databases (KDD) Adv. Protein Chem. Struct. Biol. 2018;113:85–117. doi: 10.1016/bs.apcsb.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 74.Ojha P.K., Mitra I., Das R.N., Roy K. Further exploring rm2 metrics for validation of QSPR models. Chemom. Intell. Lab. Syst. 2011;107:194–205. doi: 10.1016/j.chemolab.2011.03.011. [DOI] [Google Scholar]
- 75.Thai K.-M., Bui Q.-H., Tran T.-D., Huynh T.-N.-P. QSAR modeling on benzo[c]phenanthridine analogues as topoisomerase I inhibitors and anti-cancer agents. Molecules. 2012;17:5690–5712. doi: 10.3390/molecules17055690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chirico N., Gramatica P. Real external predictivity of QSAR models: How to evaluate it? Comparison of different validation criteria and proposal of using the concordance correlation coefficient. J. Chem. Inf. Model. 2011;51:2320–2335. doi: 10.1021/ci200211n. [DOI] [PubMed] [Google Scholar]
- 77.Roy K., Ambure P., Kar S. How Precise Are Our Quantitative Structure–Activity Relationship Derived Predictions for New Query Chemicals? ACS Omega. 2018;3:11392–11406. doi: 10.1021/acsomega.8b01647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Niraj R.R., Saini V., Kumar A. QSAR analyses of organophosphates for insecticidal activity and its in-silico validation using molecular docking study. Environ. Toxicol. Pharmacol. 2015;40:886–894. doi: 10.1016/j.etap.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 79.Ambure P., Roy K. Understanding the structural requirements of cyclic sulfone hydroxyethylamines as hBACE1 inhibitors against Aβ plaques in Alzheimer’s disease: A predictive QSAR approach. RSC Adv. 2016;6:28171–28186. doi: 10.1039/C6RA04104C. [DOI] [Google Scholar]
- 80.Hossain T., Islam M.A., Pal R., Saha A. Exploring structural requirement and binding interactions of β-amyloid cleavage enzyme inhibitors using molecular modeling techniques. Med. Chem. Res. 2013;22:4766–4774. doi: 10.1007/s00044-013-0481-z. [DOI] [Google Scholar]
- 81.Tran T.-D.N., Nguyen V., Nguyen N.-S., Nguyen D.-M., Nguyen T.-T.-H., Le M.-T., Thai K.-M. Synthesis of Novel Chalcones as Acetylcholinesterase Inhibitors. Appl. Sci. 2016;6:198. doi: 10.3390/app6070198. [DOI] [Google Scholar]
- 82.Nuthakki V.K., Sharma A., Kumar A., Bharate S.B. Identification of embelin, a 3-undecyl-1,4-benzoquinone from Embelia ribes as a multitargeted anti-Alzheimer agent. Drug Dev. Res. 2019;80:655–665. doi: 10.1002/ddr.21544. [DOI] [PubMed] [Google Scholar]
- 83.Olasehinde T.A., Mabinya L.V., Olaniran A.O., Okoh A.I. Chemical characterization, antioxidant properties, cholinesterase inhibitory and anti-amyloidogenic activities of sulfated polysaccharides from some seaweeds. Bioact. Carbohydr. Diet. Fibre. 2019;18:100182. doi: 10.1016/j.bcdf.2019.100182. [DOI] [Google Scholar]
- 84.Chemical Computing Group MOE 2008.10 edition. [(accessed on 20 May 2019)]; Available online: https://www.chemcomp.com/
- 85.Sybyl X 2.0. [(accessed on 20 May 2019)]; Available online: https://sybyl-x.software.informer.com/2.0/
- 86.Demel M., Janecek A., Thai K.-M., Ecker G., Gansterer W. Predictive QSAR Models for Polyspecific Drug Targets: The Importance of Feature Selection. Curr. Comput. Aided Drug Des. 2008;4:91–110. doi: 10.2174/157340908784533256. [DOI] [Google Scholar]
- 87.RapidMiner 5.3.013. [(accessed on 20 May 2019)]; Available online: https://rapidminer.com/
- 88.Weka Software 3.8. [(accessed on 20 May 2019)]; Available online: https://waikato.github.io/weka-wiki/
- 89.Ngo T.D., Tran T.D., Le M.T., Thai K.M. Computational predictive models for P-glycoprotein inhibition of in-house chalcone derivatives and drug-bank compounds. Mol. Divers. 2016;20:945–961. doi: 10.1007/s11030-016-9688-5. [DOI] [PubMed] [Google Scholar]
- 90.Azam F., Madi A.M., Ali H.I. Molecular Docking and Prediction of Pharmacokinetic Properties of Dual Mechanism Drugs that Block MAO-B and Adenosine A(2A) Receptors for the Treatment of Parkinson’s Disease. J. Young Pharm. 2012;4:184–192. doi: 10.4103/0975-1483.100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Protein Data Bank. [(accessed on 20 May 2019)]; Available online: https://www.rcsb.org/
- 92.LeadIT 2.0.2. [(accessed on 20 May 2019)]; Available online: https://www.biosolveit.de/LeadIT/
- 93.Kitchen D.B., Decornez H., Furr J.R., Bajorath J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 94.Pagadala N.S., Syed K., Tuszynski J. Software for molecular docking: A review. Biophys. Rev. 2017;9:91–102. doi: 10.1007/s12551-016-0247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.