Abstract
The gut microbiota guide the development of the host immune system by setting a systemic threshold for immune activation. Lipopolysaccharides (LPSs) from gut bacteria are able to trigger systemic and local proinflammatory and immunomodulatory responses, and this capability strongly relies on their fine structures. Up to now, only a few LPS structures from gut commensals have been elucidated; therefore, the molecular motifs that may be important for LPS–mammalian cell interactions at the gut level are still obscure. Here, we report on the full structure of the LPS isolated from one of the prominent species of the genus Bacteroides, Bacteroides vulgatus. The LPS turned out to consist of a particular chemical structure based on hypoacylated and mono-phosphorylated lipid A and with a galactofuranose-containing core oligosaccharide and an O-antigen built up of mannose and rhamnose. The evaluation of the immunological properties of this LPS on human in vitro models revealed a very interesting capability to produce anti-inflammatory cytokines and to induce a synergistic action of MD-2/TLR4- and TLR2-mediated signaling pathways.
Short abstract
Lipopolysaccharide of the gut commensal Bacteroides vulgatus shows novel structural features and a peculiar immunological behavior including a mild agonistic effect and combined TLR2/TLR4 activation.
Introduction
The gastrointestinal tract is in incessant contact with microorganisms, collectively defined as “microbiota”, which have profound impacts on the host immune system. The crosstalk between microbiota and the immune system at the level of the gut mucosa is crucial as it not only enables the tolerance of commensal microbes but also allows the immune system to recognize and fight pathogens thus preventing microbial invasion and infection.1,2 The nature of the microbial signals and the cellular mechanisms underlying such host–microbe interactions are still enigmatic and need further studies.
Innate immune recognition of microbes relies on the detection of conserved microbial structures, known as “microbe-associated molecular patterns” (MAMPs) such as lipopolysaccharides (LPSs), whose detection by the host immune system is at the basis of infection, inflammation, or symbiosis via an intracellular signaling.3,4 LPSs, the main constituents of the Gram-negative outer membrane, have a general structural architecture built up of three structural domains: a glycolipid portion termed lipid A, and a heteropolysaccharide composed of the core oligosaccharide (core OS) and the O-polysaccharide moiety (or O-chain).5,6 In mammals, the lipid A portion is recognized by the innate immunity receptor complex built up of myeloid differentiation protein-2 (MD-2) and Toll-like receptor 4 (TLR4).7 It is widely attested that the immunopotential of an LPS is correlated to its fine structure; namely, defined LPS molecules act as agonists of the MD-2/TLR4 complex, triggering the immune response culminating in production of proinflammatory cytokines. Nevertheless, some other LPSs act as antagonist agents by preventing the binding of toxic LPS to the receptor complex, therefore inhibiting or limiting the dangerous effects on infected cells.8
In this context, a still pending key question is how the molecular interactions between LPS and its receptors, and the subsequent signaling pathways, lead to disease vs commensalism or even mutualism. Since it is widely accepted that the innate immune responses, upon stimulation by LPS, are finely tuned by its chemical composition,3−8 one can envision that commensal gut microbiota express “modified” LPS structures well tolerated/not rejected by the host immune system.9 Therefore, a crucial point to be addressed is what commensal gut microbiota LPS looks like. To shed light on this, we have started a program aimed at looking for the structure to function relationship of LPS from gut microbiota. First we focused our attention on Bacteroides, the prominent Bacteroidetes representative in the gastrointestinal tract of the healthy American and Western European population, whose members can be considered as commensal, mutualist, or even pathobiont microorganisms.10Bacteroides surface structures have been shown to exert immunomodulatory effects. Examples are given by the capsular polysaccharide component from Bacteroides fragilis(11,12) but also the LPS from Bacteroides dorei that in a study by Vatanen et al. was shown to promote immunological tolerance.13 Recently, we demonstrated that Bacteroides vulgatus strain mpk, commensal of murine intestine, exerts strong immune-modulating properties leading to prevention of colitis-induction in several mouse models for experimental colitis.14−16 Furthermore, we showed that its isolated LPS (LPSBv) does not induce expression of proinflammatory cytokines but actively induces hyporesponsiveness toward subsequent LPS-stimuli in CD11c+ cells in murine in vitro systems,17 thereby fairly merging properties of TLR4 antagonists and agonists. Therefore, we proposed LPSBv acting as a “weak agonist” concerning its interaction with the murine MD-2/TLR4 receptor complex.17 Importantly, in the same work we also proved that the administration of purified LPSBv reestablishes intestinal immune homeostasis in a mouse model for experimental colitis and correlated these health-promoting effects to the weak agonistic properties of this LPS exploited through induction of a special type of LPS tolerance in intestinal lamina propria CD11c+ cells, via the MD-2/TLR4 receptor complex axis.17 Given that the administration of the sole LPSBv in mice caused the restoring of the homeostasis in intestine after severe inflammation, this phenomenon must of course be attributed to its chemical structure, and therefore, we have decided to establish the full structure of its LPS.
Here, we report on the full characterization of the LPSBv chemical structure achieved by the combined use of chemical, spectroscopic, and spectrometric techniques. Puzzled by the immunological properties of LPSBv observed in murine systems, we wondered whether the observed activity could be mirrored in human in vitro systems. Therefore, we also evaluated the impact of LPSBv on the human innate immune system by using HEK cell lines, human macrophages, and dendritic cells and discovered that to an intriguing chemical structure corresponded an equally intriguing immunological behavior.
Structural Characterization of B. vulgatus LPS: Purification and Chemical Analyses
In order to define the full structure of the LPSBv, a multidisciplinary approach was employed. In detail, a set of chemical analyses, on both intact and isolated LPS domains, was executed, furnishing the chemical composition of each LPS moiety.
LPS material was extracted from lyophilized bacterial cells by the hot phenol/water procedure.18 The extracted LPS underwent an enzymatic digestion by DNase, RNase, and proteases in order to remove cell contaminants. A further step of purification by size-exclusion chromatography and ultracentrifugation was also performed. The nature and purity of the LPSBv was determined by SDS-PAGE analysis after silver nitrate gel staining,19 disclosing the smooth nature of the extracted LPS, as proven by the ladderlike pattern in the upper part of the gel indicating the occurrence of high-molecular-weight species, i.e., the O-chain moiety (Figure S1a). In parallel, no bands were visible in an additional SDS-PAGE analysis followed by Coomassie Brilliant Blue gel staining (Figure S1b) which was indicative of the absence of any contaminating protein/lipoprotein in the isolated LPS material. Furthermore, the Micro BCA protein assay has been also executed disclosing a protein content of less than 1% (i.e., ≤0.8 μg/mL) in the presence of 2% SDS as requested by manufacturer protocol to remove most of the well-known lipid interferences (Figure S1c).20,21 However, it should be noted that the obtained value (≤0.8 μg/mL) (Figure S1c) was close to the lower limit of detection of the Micro BCA protein assay. For this reason (being close to the lower limit of detection) and due to the presence of lipids (i.e., the lipid A moiety), there are significant overestimations of the actual protein content (if any). In addition, as reported, the 2% SDS treatment remove only 70–80% of the lipid interferences.20,21
Chemical analyses22−24 executed on pure LPS revealed the presence of terminal, 4-substituted and 3,4-disubstituted l-rhamnopyranose (l-Rhap), terminal l-fucopyranose (l-Fucp), terminal galactofuranose (d-Galf), 3-substituted d-mannopyranose (d-Manp), 3-substituted d-glucopyranose (d-Glcp), 2,6-disubstituted d-galactopyranose (d-Galp), 6-substituted amino-d-glucopyranose (d-GlcpN), and 5-substituted Kdo. Fatty acid analysis25 showed the occurrence of tetradecanoic acid (C14:0), pentadecanoic acid (C15:0), hydroxypentadecanoid acid (C15:0(3-OH)), hydroxyhexadecanoid acid (C16:0(3-OH)), and hydroxyheptadecanoic acid (C17:0(3-OH)), in full agreement with previously reported data by Hashimoto et al. on the structure of lipid A from B. vulgatus IMCJ 1204.26
Structural Characterization of B. vulgatus LPS: NMR of the Saccharide Portion
An aliquot of the pure LPS isolated from B. vulgatus mpk underwent a full deacylation27 furnishing the complete LPS saccharide portion. The deacylated LPS fraction was then purified by gel permeation chromatography. Monosaccharide analysis of the isolated product confirmed the presence of the sugar residues detected in the intact LPS. The deacylated product was then analyzed by 1D and 2D NMR spectroscopy. A combination of homo- and heteronuclear 2D NMR experiments (DQF-COSY; TOCSY; NOESY; ROESY; 1H, 13C HSQC; 1H, 13C HMBC; and 31P and 1H, 31P HSQC) was performed in order to elucidate the complete saccharide sequence of LPSBv. In detail, each spin system was assigned on the basis of the spin connectivity observed in both the double-quantum-filtered correlation spectroscopy (DQF-COSY) and the total correlation spectroscopy (TOCSY) spectra; each carbon atom was identified through the analysis of the heteronuclear single-quantum coherence (HSQC) spectrum. The anomeric configuration of all sugar units was defined by intra-residual NOE contacts, detectable in the nuclear Overhauser effect spectroscopy (NOESY) spectrum and the 3JH-1,H-2 coupling constants from the DQF-COSY spectrum. The assignment of the relative configuration of each sugar residue was obtained through the observation of the vicinal 3JH,H coupling constant values. The combined study of rotating-frame NOE spectroscopy (ROESY), NOESY, and HMBC (heteronuclear multiple-bond correlation spectroscopy) spectra allowed the characterization of the entire primary structure of the LPS saccharide part. Finally, 31P and 31P, 1H HSQC experiments were pivotal to establish the location of the phosphate group decorating the LPSBv.
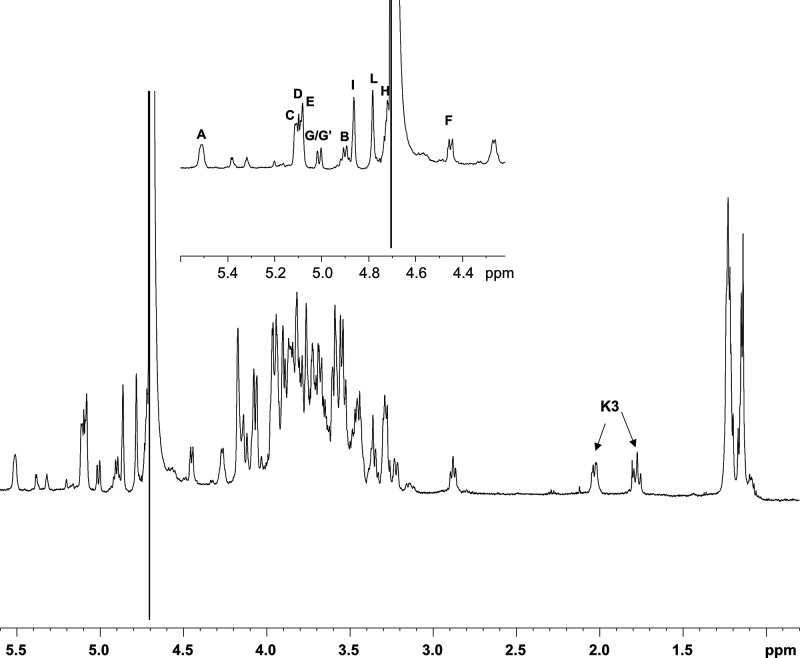
In the 1H NMR spectrum (Figure 1) 11 anomeric signals from 11 spin systems were clearly identified (A–L; Table 1); furthermore, the signals at δH 1.77 and 2.02 ppm were attributed to the H-3 methylene protons of the Kdo unit (K; Table 1). All the monosaccharide residues, except for D, were present as pyranose rings, according to both 13C chemical shift values and the presence of long-range correlations between C-1/H-1 and H-5/C-5 observable in the 1H, 13C HMBC spectrum (for the Kdo residue between C-2 and H-6).28 Conversely, residue D was present as a furanose ring as shown by the occurrence of low-field shifted ring carbon signals resonating around δC 82.0 ppm and anomeric carbon over δC 108 ppm and further confirmed by the intra-residual long-range H/C 1–4 correlations in the 1H, 13C HMBC spectrum.
Figure 1.
1H NMR of fully deacylated LPSBv. A zoom of the anomeric region is reported in the inset. Anomeric signals in the inset are as attributed in Table 1.
Table 1. 1H, 13C (italic) and 31P (bold) Chemical Shift Values of the Saccharide Region of the LPSBv.
| chemical shifts (δ) |
||||||||
|---|---|---|---|---|---|---|---|---|
| unit | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| A, 6-α-GlcpN | 5.5 | 3.22 | 3.81 | 3.27 | 4.07 | 4.12/3.80 | ||
| 90.4 | 54.3 | 69.9 | 70.1 | 73 | 68 | |||
| 2.99 | ||||||||
| B, 6-β-GlcpN | 4.9 | 2.86 | 3.52 | 3.43 | 3.55 | 3.69/3.47 | ||
| 99.6 | 55.6 | 72.3 | 69.8 | 74.5 | 61.3 | |||
| C, 3,4-α-Rhap (α-RhaI) | 5.11 | 4.16 | 3.96 | 3.85 | 3.85 | 1.23 | ||
| 101 | 70.2 | 78.9 | 73.6 | 68.6 | 16.9 | |||
| D, t-β-Galf | 5.07 | 4.06 | 3.96 | 3.92 | 3.72 | 3.56 | ||
| 108.6 | 81.8 | 77 | 82.8 | 70.3 | 62.7 | |||
| E, t-α-Fucp | 5.09 | 3.67 | 3.76 | 3.71 | 4.27 | 1.14 | ||
| 99.4 | 68.3 | 71.4 | 73.2 | 66.9 | 16.2 | |||
| F, 3-β-Glcp | 4.47 | 3.29 | 3.48 | 3.36 | 3.36 | 3.82/3.62 | ||
| 102.5 | 73.5 | 82.4 | 69.9 | 75.6 | 60.8 | |||
| G, t-α-Rhap (α-RhaII) | 5.01 | 3.93 | 3.81 | 3.34 | 3.96 | 1.22 | ||
| 100.8 | 70.4 | 69.7 | 71.9 | 67.4 | 17 | |||
| G′, 4-α-Rhap (α-Rha′II) | 5 | 3.93 | ND | 3.58 | 3.88 | 1.22 | ||
| 100.8 | 70.4 | ND | 79.4 | 67.5 | 17 | |||
| H, 2,6-β-Galp | 4.72 | 3.45 | 3.75 | 3.8 | 3.71 | 3.93 | ||
| 100.2 | 76.8 | 71.4 | 69.7 | 70.6 | 69 | |||
| I, 4-α-Rhap (α-RhaOC) | 4.86 | 3.9 | 3.94 | 3.58 | 3.89 | 1.22 | ||
| 99.7 | 70.3 | 70.3 | 79.4 | 67.5 | 17 | |||
| L, 3-β-Manp | 4.78 | 4.17 | 3.58 | 3.54 | 3.29 | 3.82/3.64 | ||
| 100.3 | 66.6 | 76.6 | 65 | 76.1 | 60.8 | |||
| K, 5-α-Kdop | 1.77/2.02 | 4.15 | 4.07 | 3.68 | 3.96 | 3.95/3.86 | ||
| 34.5 | 67.2 | 73.3 | 70.5 | 70.2 | 65.6 | |||
| 1.64 | ||||||||
Spin systems A (H-1 δ 5.50 ppm) and B (H-1 δ 4.90 ppm) were identified as the α-GlcpN and β-GlcpN of the lipid A moiety based on their H-2 proton signals, which correlated with two nitrogen-bearing carbon atoms at δC 54.3 and δC 55.6 ppm, respectively (Table 1, Figure S2). This hypothesis was also corroborated by the presence of an inter-residue contact between H-1 of B and H-6a,b of A observed in the NOESY spectrum (not shown). Furthermore, the occurrence of a correlation in the 31P, 1H HSQC spectrum (not shown), between the signal at δ 2.99 ppm and the anomeric proton signal of residue A (δH 5.50 ppm, Table 1, Figure S2), allowed the allocation of a phosphate group at such a position. Spin systems C, G, G′, and I were identified as α-rhamnopyranose residues as proven by the correlations, in the TOCSY spectrum, with the methyl group signals resonating at δH 1.23 and 1.24 ppm (δC 16.9 and 17.0 ppm; Table 1), respectively. As mentioned above, the α-anomeric configuration was determined on the basis of the 3JH-1,H-2 coupling constant values and the intra-residual NOE contact of H-1 with H-2, whereas the manno configuration was established by evaluation of 3JH,H coupling constant values.
Spin systems D, E, and H were attributed to galacto-configured sugar units. In detail, residue D (H-1 δ 5.07 ppm, Table 1), as stated above, was revealed to be a β-galactofuranose (anomeric carbon signal at δ 108.6 ppm, Table 1) based on its chemical shifts and intra-residual scalar and dipolar correlations. By contrast, spin system H (H-1 δ 4.72 ppm) was assigned to a β-galactopyranose, as proven by the chemical shift values of ring protons and the 3JH,H ring coupling constants; furthermore, the large 3JH-1,H-2 values and the NOE correlations of H-1 with H-3 and H-5 were indicative of the β-anomeric configuration. The galacto-configured sugar residue E (H-1 δ 5.09 ppm) was identified as an α-fucopyranose as shown by the correlations, in the TOCSY spectrum, with the methyl proton signal at δH 1.14 ppm (δC 16.2 ppm) and the 3JH-1,H-2 coupling constant values that confirmed the α-anomeric configuration.
Spin system F (H-1 δ 4.47 ppm) was assigned to a β-glucopyranose as indicated by the large 3JH,H ring coupling constants and the chemical shift values of ring protons, in agreement with the gluco-configuration of pyranose rings (Table 1). Moreover, the large 3JH-1,H-2 values, together with the NOE contacts of H-1 with H-3 and H-5, were diagnostic of the β-anomeric configuration. Finally, spin system L (H-1 δ 4.78 ppm) was attributed to a β-mannopyranose residue, as proven by the small 3JH-1,H-2 and 3JH-2,H-3 values, diagnostic of a H-2 equatorial orientation, whereas the intra-residual correlations between H-1 and H-5 in the NOESY and ROESY spectra allowed the β configuration assignment. The assignment of the Kdo unit (K) was achieved starting from the diastereotopic methylene signal, and its anomeric α-configuration was assigned on the basis of the chemical shift values of H-3 (δ 1.77/2.02 ppm, Table 1), and of the 3JH-7,H8a and 3JH-7,H-8b coupling constants.28,29 Moreover, the Kdo residue was phosphorylated at position O-4, due to the occurrence of a correlation with a signal at 1.64 ppm in the 31P, 1H HSQC spectrum (not shown).
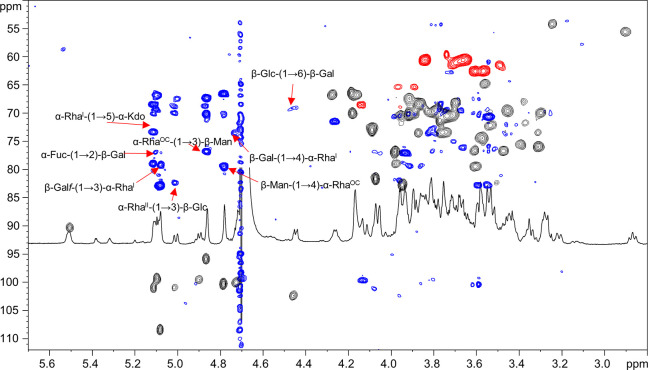
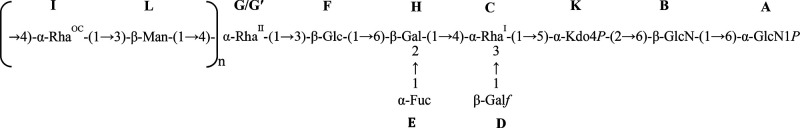
The downfield shifted carbon signals were indicative of glycosylation at position O-6 of A and B, O-5 of K, O-3 and O-4 of C, O-2 and O-6 of H, O-3 of F and L, and O-4 of I and G′, whereas D, E, and G were terminal sugar residues, in full accordance with the methylation data. The analysis of the inter-residual NOE contacts (observable in both the NOESY and ROESY spectra) and the long-range correlations in the HMBC spectrum (Figure 2) allowed the defining of the complete sugar sequence of the LPS from B. vulgatus mpk. Starting from the lipid A disaccharide backbone, made up of residue A and B, this latter sugar unit was found to be substituted at position O-6 by the Kdo residue (K); this was further confirmed by the weak downfield shift of signal of C-6 of residue B (δ 61.3 ppm, Table 1), which is consistent with the α-(2→6) ketosidic linkage of Kdo with the β-GlcN of the lipid A. The Kdo unit was, in turn, substituted at position O-5 by the rhamnose residue C, as suggested by the long-range correlation between the anomeric proton signal of residue C and the carbon atom at δ 73.3 ppm (C-5, Table 1) of residue K (Figure 2). Rhamnose C was revealed to be substituted at both positions O-3 and O-4 by the galacto-configured sugar units D and H, respectively, as showed by the occurrence of the strong NOE contacts between H-1 of D and H-3 of C and between H-1 of H and H-4 of C. These glycosydic linkages were also confirmed by the observation of the respective long-range correlations in the HMBC spectrum (Figure 2). Residue H, in turn, was substituted by the α-Fuc E at position O-2 and by the β-Glc F at position O-6, as proven by the long-range correlations between the H-1 of E and the C-2 signal of H, and H-1 of F and C-6 of H (Figure 2). This latter residue F turned out to be substituted at position O-3 by rhamnose G, as suggested by the long-range contact between the anomeric proton signal of G and the carbon atom signal at δ 82.4 ppm (C-3, Table 1) of β-glucose F. Interestingly, 2D NMR analysis allowed the identification of a second slight different glycoform of rhamnose G, referred to here as G′, differing for the presence of a glycosylated position, namely, position O-4 (Table 1). Sugar units I and L were attributed to the components of the disaccharide repeating unit of the O-chain moiety as demonstrated by the long-range correlation of the anomeric proton signal of β-Man L with the carbon atom signal C-4 of α-RhaOCI (Figure 2); further on, an HMBC correlation between the anomeric proton signal of I and C-3 of β-Man L was also clearly visible. These data were in full agreement with the LPS O-chain repeating unit previously elucidated by Hashimoto et al.26 Finally, we were also able to establish the biological repeating unit; in fact, residue G′, assigned to a 4-substituted α-Rha, was identified as the sugar likely connecting the O-chain moiety to the core region, as proven by the long-range correlation between H-1 of L and C-4 of G′ (Figure 2). This hypothesis was also confirmed by mass spectrometry (MS) and MS2 data (see below).
Figure 2.
Zoom of the overlapped 1H, 1H, 13C HMBC (blue) and 1H, 13C HSQC (black and red) NMR spectra. The key inter-residual long-range correlations involving sugar moieties (A–L) are indicated; letters are as in Table 1. Anomeric one-bond heteronuclear correlations were also reported in the spectrum.
All the above information taken together allowed the establishing of the complete structure of the fully deacylated LPSBv as sketched below:
 |
1 |
Structural Characterization of B. vulgatus LPS: Mass Spectrometry Analysis
The fully deacylated LPSBv was also analyzed by electrospray ionization (ESI) mass spectrometry (MS). The corresponding spectrum is reported in Figure S3a. The detailed investigation of the double and triple charged ions in the spectrum in the mass range m/z 400–1130 allowed the determination of the fine structure of the LPS saccharide domain. In detail, the spectrum showed three main signal clusters (M1–M3) of double charged ions (m/z 821.3, 975.4, and 1129.4, Figure S3a and Table S1) originating from the monoisotopic molecules with a mass of 1644.47, 1952.58, and 2260.69 (calculated molecular masses 1644.6, 1952.8, and 2260.8 Da), respectively, differing by 308 u, which corresponded to the mass of an O-chain repeating unit (RU) built up of a hexose and a deoxyhexose, thus in full agreement with the NMR data. Briefly, M1 (the most abundant species) and M2 contained one and two O-chain RU, respectively, whereas M3 possessed three O-chain RU. Interestingly, the main peak m/z 667.3 was attributed to the double charged ion originating from the monoisotopic mass of 1336.36 (calculated molecular mass 1336.6 Da) which is consistent with an oligosaccharide composed of two hexosamines, one Kdo, two hexoses, two deoxyhexoses, and two phosphates. The structural sequence of the core OS was also corroborated by the ESI-MS2 investigation executed on precursor ion 1336.36 which is reported in Figure S3b. The spectrum showed the occurrence of a daughter ion at m/z 419.2 (Y2) originating from the monoisotopic mass 420.11 (calculated molecular mass 420.2 Da) assignable to the mono-phosphorylated diglucosamine backbone of the lipid A. Moreover, the daughter ion at m/z 915.4 (B3) relative to a mass of 916.24 (calculated mass 916.4 Da) was attributed to the oligosaccharide fragment made up of one Kdo, one phosphate, two hexoses, and two deoxy-hexoses. Finally, the ESI-MS spectrum revealed also the occurrence of the two double charged ions at m/z 627.3 and 748.3 relative to the monoisotopic masses 1256.39 and 1498.41 (calculated molecular masses 1256.6 and 1498.6 Da), respectively, which matched with the above-described core OS minus a phosphate group (1256.39) or carrying a further hexose unit (1498.41). Therefore, all above-mentioned MS and MS2 data were in full accordance with the NMR structural elucidation of the saccharide moiety of the LPSBv.
In order to define also the structure of the glycolipid portion of the LPSBv, an aliquot of pure LPS underwent a mild acid hydrolysis in order to selectively cleave the acid-labile linkage between the Kdo and the β-GlcN unit and to isolate the lipid A fraction. After purification, this latter was then analyzed by MALDI MS. The MALDI MS spectrum (Figure S4), acquired in negative polarity, matched perfectly with the MS analysis executed by Hashimoto et al.26 In detail, a high heterogeneity of lipid A species was immediately apparent as demonstrated by the occurrence of two clusters of signals differing in the number of the fatty acid chains (Figure S4). Moreover, each of these groups of ions was characterized by the presence of mass differences that can be explained with the presence of lipid A species differing in the fatty acid chain length. In detail, the most intense cluster of ions in the mass range m/z 1617–1717 Da (Figure S4) was consistent with mono-phosphorylated penta-acylated lipid A species whose fatty acid distribution and composition, for instance for the ion peak at m/z 1688.28, was determined as two C17:0(3-OH) as N-linked acyl chains, two C16:0(3-OH) as primary O-linked fatty acids, and C15:0 as a secondary O-acyl substitution. The less intense cluster of ions visible in the mass range m/z 1385–1479 Da (Figure S4) was attributed to mono-phosphorylated tetra-acylated lipid A species with the main ion peak at m/z 1436.02 assigned to a lipid A possessing the above-mentioned fatty acid distribution and composition but lacking one primary O-linked C16:0(3-OH).
Therefore, the overall data led to the delineation of the full structure of LPSBv as sketched in Figure 3. Moreover, a first indication of the 3D perspective of the dynamic behavior in water of both the tetra- and penta-acylated forms of LPSBv is reported in Figures S5–S7.
Figure 3.
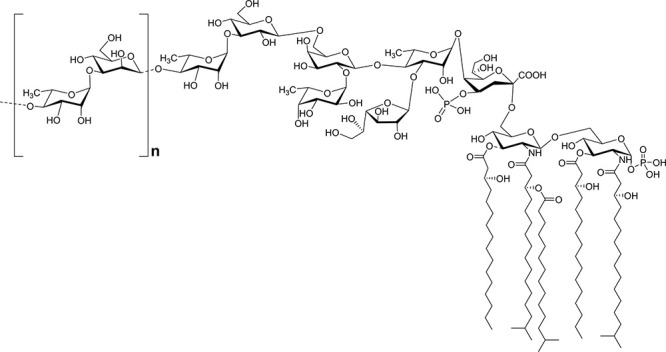
Complete structure of the LPS from B. vulgatus mpk. The lipid A is heterogeneous, being composed of a mixture of penta- and tetra-acylated mono-phosphorylated species. The dotted lines indicate the nonstoichiometric substitution. The O-chain repeating unit is indicated in square brackets.
Activation of Peripheral Blood-Monocyte-Derived Macrophages and of Peripheral Blood-Monocyte-Derived Dendritic Cells by B. vulgatus LPS and Lipid A
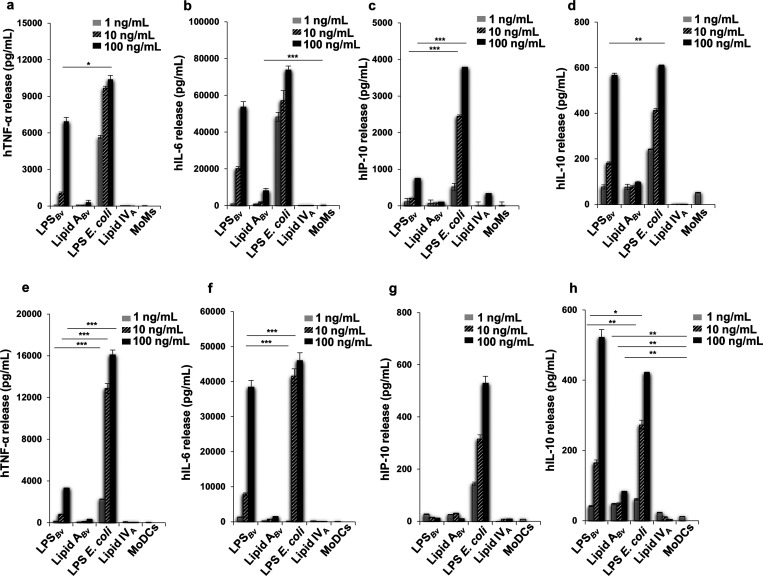
Once the structure was defined at a molecular level, we then passed to the immunological evaluation of LPSBv and its isolated lipid A portion (Lipid ABv) on human peripheral blood-monocyte-derived macrophages (MoMs) and on peripheral blood-monocyte-derived dendritic cells (MoDCs). LPSBv and lipid ABv were used to stimulate MoMs and MoDCs at the concentration of 1, 10, and 100 ng/mL. The agonistic hexa-acylated Escherichia coli O111:B4 LPS and the synthetic antagonist tetra-acylated lipid IVA at the same concentration as above were used as controls. Untreated cells were considered the negative control in all the experiments. The release of cytokines tumor necrosis factor-α (TNF-α) (Figure 4a,e), interleukin-6 (IL-6) (Figure 4b,f), the chemokine IFN-γ-inducible protein-10 (IP-10) (Figure 4c,g), and the interleukin-10 (IL-10) (Figure 4d,h) were quantified by ELISA after 12 h of stimulation. The choice has fallen on these cytokines/chemokines as they are the most representative to demonstrate the inflammatory (TNF-α and IL-6) and anti-inflammatory (IL-10) response following an LPS stimuli. Importantly, it is known that, following the recognition of LPS by TLR4, two signaling pathways mediated by the adaptor molecules MyD88 and TRIF, respectively, are triggered. Both pathways lead to the activation of the nuclear factor-κB (NF-κB) and the production of the proinflammatory mediators, while the only TRIF pathway promotes the production of type I interferon and the interferon-related cytokines/chemokines, such as the chemokine IP-10.30 Therefore, the measurement of this chemokine was chosen to assess whether LPSBv can activate the TRIF pathway in addition to the MyD88 pathway.
Figure 4.
Cytokine and chemokine release induced by LPSBv and lipid ABv in MoMs and MoDCs. Release of TNF-α (a, e), IL-6 (b, f), IP-10 (c, g), and IL-10 (d, h) in cell free supernatants of MoMs and MoDCs, respectively. Untreated cells are the negative control in these experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
In MoMs the amount of TNF-α triggered by LPSBv stimulation at 100 ng/mL was significantly reduced with respect to that of E. coli LPS at the same concentration (p < 0.5) but comparable to that of E. coli LPS at 10 ng/mL (Figure 4a). Likewise, a similar trend was observed for the cytokine IL-6 (Figure 4b) as at the concentration of 100 ng/mL no significant difference was detected with E. coli LPS. The chemokine IP-10 yield triggered by 100 ng/mL of LPSBv was appreciable (Figure 4c) but significantly lower (p < 0.001) than that of E. coli LPS at the same and minor concentration (10 ng/mL). Finally, upon stimulation with 100 ng/mL LPSBv or E. coli LPS, IL-10 production (Figure 4d) was comparable while at 10 ng/mL the difference between these values was highly significant (p < 0.01). Lipid ABv did not produce the release of the cytokines measured with the exception of IL-6 that was scantly but significantly produced (p < 0.001, for Lipid ABv vs untreated cells) at the concentration of 100 ng/mL (Figure 4b). As expected, no cytokine production was observed with lipid IVA.
In MoDCs, LPSBv triggered the production of TNF-α (Figure 4e) which was significantly lower than that of E. coli (p < 0.001 for all concentrations LPSBv vs E. coli LPS). IL-6 (Figure 4f) was produced in small amount at 1 and 10 ng/mL (p < 0.001 for LPSBv at 1 and 10 ng/mL vs E. coli LPS at the same concentrations) while at 100 ng/mL the two yields were comparable. No release of TNF-α and IL-6 was seen following lipid ABv stimulation at all the concentrations tested (Figure 4e,f). The chemokine IP-10 (Figure 4g) was poorly detectable upon stimulation with either LPSBv or lipid ABv. In line with the trend observed in MoMs, in MoDCs IL-10 (Figure 4h) release by LPSBv was lower than that elicited by E. coli LPS at 1 and 10 ng/mL (p < 0.01 and p < 0.05, respectively, for LPSBv vs E.coli LPS at the same concentrations) while at 100 ng/mL no statistical difference between the two values was found. Likewise, lipid ABv elicited an exiguous but still detectable production of IL-10 (p < 0.01 for lipid ABv vs untreated cells).
Furthermore, we also measured via ELISA the release of IL-6, IL-10 (Figure S8), and IFN-α by MoDCs after stimulation with LPSBv and E. coli LPS at different time points. This analysis confirmed the lower capacity of LPSBv in eliciting the production of IL-6 and IL-10 compared to E. coli LPS; finally, the absence of any detection of IFN-α, in addition to the poor detectability of the chemokine IP-10 release, further demonstrated that the LPSBv-induced IL-10 release is not regulated by IFN type I dependent pathways.
Therefore, the findings in MoDCs consolidated those achieved in MoMs and highlighted the peculiar ability of the LPSBv to stimulate selectively the release of IL-6 and the anti-inflammatory cytokine IL-10. It is noteworthy that the LPSBv stimulates the production of a low amount of TNF-α compared to IL-10 release, in contrast to what is observed with E. coli LPS which induced a high yield of TNF-α. This result defined a peculiar immunological property of LPSBv which is able to promote an anti-inflammatory response rather than the inflammation.
Activation of Bone Marrow Monocyte-Derived Macrophages by B. vulgatus LPS and Lipid A
We then continued to analyze whether LPSBv and lipid ABv showed the same ability to induce the above profile of cytokine yield also in murine bone marrow monocyte-derived macrophages (BMDMs), which is the common model to analyze the immunopotential of LPS. The release of TNF-α, IL-6, IP-10, and IL-10 (Figure S9a–d) was quantified after 12 h of stimulation by ELISA. In contrast to human cells, in BMDMs stimulated with LPSBv at all concentrations the TNF-α production (Figure S9a) was scant and significantly lower (p < 0.001, for LPSBv vs E. coli LPS at all concentrations) than that induced by E. coli LPS at all concentrations tested. Likewise, IL-6 (Figure S9b), IP-10 (Figure S9c), and IL-10 (Figure S9d) values were undetectable or scantly detectable (p < 0.05 for IL-10: LPSBv at 100 ng/mL vs untreated cells). Conclusively, human but not murine cells seemed to be particularly sensitive to the immunopotential of LPSBv.
Activation by LPSBv and Lipid ABv in HEK293 Cell Models
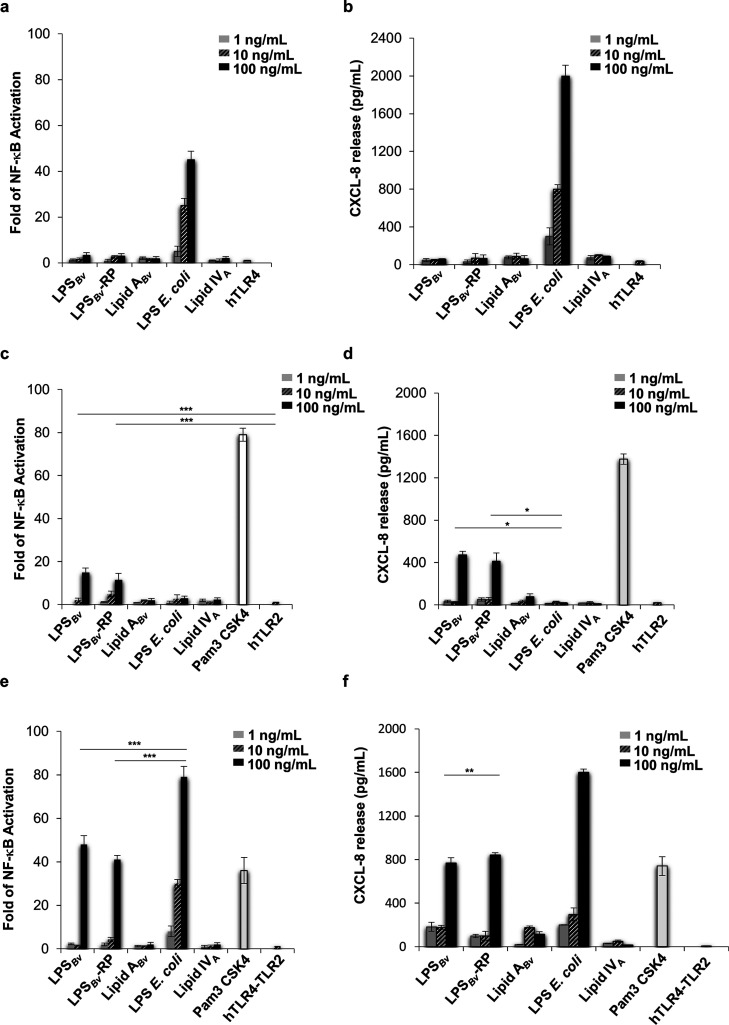
To acquire major insights on the biological properties of LPSBv and lipid ABv we tested them in the model of HEK293 cell line stably transfected with the human CD14/MD-2/TLR4 complex. As above, LPSBv and lipid ABv, E. coli LPS, and the lipid IVA were analyzed at the concentration of 1, 10, and 100 ng/mL. After 6 h, the activation of the nuclear factor-κB (NF-κB) (Figure 5a) and the production of chemokine (C-X-C motif) ligand 8 (CXCL-8 or IL-8) were measured (Figure S8b) as the most common read-outs used to evaluate the HEK293 cell activation upon LPS stimulation. As shown in Figure 5a, in HEK293 hTLR4 the level of NF-κB activation induced by LPSBv (100 ng/mL) roughly reached 7% compared to that of E. coli LPS at the same concentration while the cells were not responsive to lipid ABv. No CXCL-8 release was detected upon LPSBv and lipid ABv exposure (Figure 5b). At 10 and 100 ng/mL E. coli LPS stimulated high levels of NF-κB activation and CXCL-8 production, and no response was seen upon stimulation with lipid IVA, in accordance with the acylation degree.
Figure 5.
Analysis of the immunopotential of LPSBv, LPSBv-RP, and lipid ABv. HEK293 cells expressing hTLR4/MD2-CD14 (a, b) or hTLR2 (c, d) or hTLR4/MD2-CD14 and hTLR2 (e, f) were treated with LPSBv, LPSBv-RP, or lipid ABv, with E. coli LPS, with lipid IVA at the concentration of 1, 10, and 100 ng/mL, or with TLR2 agonist Pam3 CSK4 (500 ng/mL) (c–f). Untreated cells were the negative control in these experiments. Fold of NF-κB activation in HEK293 hTLR4/MD2-CD14 (a), HEK293 hTLR2 (c), and HEK293 hTLR4/MD2-CD14 hTLR2 (e). Release of CXCL-8 release supernatants of HEK293 hTLR4/MD2-CD14 (b), HEK293 hTLR2 (d), and HEK293 hTLR2 hTLR4 cells (f). *p < 0.05, **p < 0.01, ***p < 0.001.
These results did not clarify the immunopotential of LPSBv and its lipid A detected in MoMs and MoDCs. It can be argued that, in contrast to HEK293 hTLR4, which are cells that do not express the TLRs,31 macrophages and dendritic cells show the full panel of TLRs. Therefore, another TLR like TLR2 could cooperatively work with the TLR4. In this context, the issue of TLR2 activation by LPS is very controversial.32 However, very recently it has been reported that Helicobacter pylori LPS is able to activate TLR2,33 underlining that some atypical LPS could show a TLR2-activating capacity. Therefore, we tried to understand whether LPSBv or lipid ABv could induce a TLR2 response. The HEK293 expressing hTLR2 were used to assess the immunologic impact of LPSBv and lipid ABv at the concentration of 1, 10, and 100 ng/mL as above. E. coli LPS and lipid IVA were used in parallel. Pam3CSK4 (Pam3 CSK4) (500 ng/mL), a synthetic triacylated lipopeptide (LP), which mimics the acylated amino terminus of bacterial LP, represented the positive control for a TLR2 ligand. As expected, no activation of NF-κB (Figure 5c) and no CXCL-8 (Figure 5d) release were detected upon stimulation with E. coli LPS and lipid IVA. LPSBv (100 ng/mL), but not lipid ABv, elicited a visible degree of NF-κB activation (p < 0.001 vs untreated cells). With LPSBv CXCL-8 production was consistent with NF-κB results (p < 0.05 for LPSBv vs E. coli LPS, both at 100 ng/mL), and lipid ABv triggered only a scant yield. As expected, Pam3 determined high NF-κB activation and CXCL-8 production. These results matched with our previous observation of a slight activation of TLR2 in murine TLR2-HEK cells in response to LPSBv stimulation.17
Then, HEK293 hTLR4 cells were transfected with hTLR2 and analyzed under the same conditions as above (Figure 5e,f) to evaluate the mutual influence of both TLRs (TLR4 plus TLR2) upon stimulation with LPSBv and lipid ABv. In contrast to the results in HEK293 hTLR4, activation of NF-κB and CXCL-8 release induced a certain degree of activation following stimulation with LPSBv (100 ng/mL) although they were significantly lower (p < 0.001 and p < 0.01 for NF-κB and CXCL-8, respectively: LPSBv vs E. coli LPS, both at 100 ng/mL) than those of E. coli LPS at the same concentration. As controls, HEK293 null cells or carrying pcDNA3, stimulated as HEK293 hTLR2 and HEK293 hTLR4 hTLR2 cells, did not trigger any NF-κB activation and CXCL-8 production (Figure S10).
Importantly, LPS preparations containing highly immunoactive contaminants, i.e., lipoproteins and phospholipids, that could be responsible for the observed TLR2-mediated signaling, have been previously reported.34 Therefore, despite the extensive steps of purifications and the results of both the SDS-PAGE and the Micro BCA protein assay, we performed a further step of “repurification” of the LPSBv to eliminate possible lipoproteins present as previously described,35,36 preceded by several washes with a mixture of chloroform–methanol (1:2, v/v) and chloroform–methanol–water (3:2:0.25, v/v) to remove traces of phospholipids possibly present. No detectable proteins were observed in the repurified LPS (LPSBv-RP) analyzed by Micro BCA protein assay (Figure S1c), which was then employed for immunological studies on HEK293 cell lines. As shown in Figure 5, immunological tests executed on HEK293 hTLR4, HEK293 hTLR2, and HEK293 hTLR4 hTLR2 showed a similar behavior between the LPSBv and the repurified LPS (LPSBv-RP), thus further confirming that the LPSBv actually activates the TLR2-mediated signaling.
Competition Tests of LPSBv and Lipid ABv with Agonistic LPS in HEK293 Cell Model
Some underacylated lipid A and/or unusual LPS, containing, for example, long fatty acid chains, do show an inhibitory activity against endotoxically active LPS in human cells.37−40 In line with this issue, we tested the ability of LPSBv or Lipid ABv to compete with the hexa-acylated, fully immunocompetent E. coli LPS to engage the hTLR4 mediated signaling. With this aim, HEK 293 hTLR4 cells were preincubated for 1 h with LPSBv or lipid ABv and stimulated with 10 and 100 ng/mL of E. coli LPS for 4 h (Figure S11a–d). NF-κB activation and CXCL-8 production were quantified after this time. Lipid IVA was used in parallel under the same conditions as for LPSBv and lipid ABv as a control of the inhibitory effect. Under these experimental conditions, LPSBv and lipid ABv did not significantly inhibit the activity of NF-κB upon E. coli LPS stimulation at all the concentrations tested, albeit the trend of NF-κB was reduced with respect to the only stimulation with E. coli LPS (Figure S11a,b). This kinetics was confirmed and strengthened with the CXCL-8 yield as lipid ABv (1 and 10 ng/mL) plus E. coli LPS (10 ng/mL) reduced significantly the production of CXCL-8 of E. coli LPS (10 ng/mL) alone (p < 0.01) (Figure S11c). Likewise, LPSBv and Lipid ABv (1 and 10 ng/mL) plus E. coli LPS (100 ng/mL) induced a lower release of CXCL-8 than E. coli LPS (100 ng/mL) alone (p < 0.01) (Figure S11d). As expected, lipid IVA showed an inhibitory activity on E. coli LPS for NF-κB and CXCL-8 production at all the concentrations tested (p < 0.001). Therefore, LPSBv and lipid ABv show a weak inhibitory activity toward E. coli LPS. These data are in agreement with the observation that LPSBv is only a slight competitive inhibitor of E. coli LPS for binding to murine TLR4.17
Discussion
The involvement of the LPS in the host-gut microbiota cross-talk is receiving a growing interest as it is shown to play a crucial role, depending on its fine structure, triggering or preventing inflammation by modulating the host immune responses. Different LPS structures are at the basis of such divergent activities, with gut commensal LPS exerting only weak immune inflammatory reactions. Within this frame, the structural characterization of LPS from gut microbiota will potentially provide peculiar chemical characteristics which will be surely related to interesting immunological properties.
In this study, we fully established the LPS structure isolated from B. vulgatus mpk, a member of the genus Bacteroides which is predominant in mouse and human microbiota and whose health-promoting effects in murine model systems were previously described.17 Briefly, the lipid A resulted in a heterogeneous mixture of tetra- and penta-acylated species (Figure 3 and Figure S4) whose nature and distribution of the fatty acids were as previously reported.26 This lipid A blend resulted in being essentially phosphorylated only at the reducing glucosamine unit, and no lipid A species with a higher acylation and/or phosphorylation degree have been detected. As for the saccharide portion, the core region was built up of a hexa-saccharide bearing a phosphorylated Kdo unit, in turn, substituted at position O-5 by a Rhap residue. This latter was found to be a branched monosaccharide bearing a β-Galf unit at its position O-3 and, at position O-4, a β-Galp unit. The latter was, in turn, disubstituted by α-Fucp and, finally, by a β-Glcp which was established to be the residue substituted by the first rhamnose of the O-chain. The O-chain structure was built up of repeating units of β-Manp and α-Rhap, in agreement with what was previously published26 where we have further established the biological O-chain repeating unit which starts with α-Rhap. Notably, to the best of our knowledge, this is the first time that the full structure of an LPS from Bacteroides spp. is completely defined since so far only relatively little information and disjointed and partial structures are available concerning Bacteroides LPS saccharide domains.9
The investigation of the immunological properties of LPSBv in human macrophages showed a weaker capability to elicit proinflammatory cytokine production than E. coli LPS. Therefore, similar to what was observed in murine in vitro systems, it is tempting to translate in human immune cells the hypothesis of LPSBv as acting providing a basic anti-inflammatory intracellular transcription program, without exceeding a proinflammatory threshold, that could exert a “protective” role for the colonized host. In addition, our results showed that LPSBv was able to highly stimulate the release of IL-10, a potent anti-inflammatory cytokine that restrains pathogenic inflammation in the gut. Nevertheless, LPSBv promoted some production of the pleiotropic cytokine IL-6 whereas the proinflammatory cytokine TNF-α was only scantly stimulated. It might be speculated that LPSBv plays a key role in the host immune responses by balancing the excessive inflammation potentially induced by IL-6 with the anti-inflammatory activity of IL-10. The resulting immune balance is crucial to drive optimal immune responses without causing an overexuberant inflammation at the level of intestinal mucosa where microbiota proliferate.41
The use of HEK cell lines transfected with TLR4 confirmed a weaker agonistic effect of LPSBv compared to LPS from E. coli. In addition, HEK TLR2 cell lines showed a specific and significant interaction only with LPSBv. Interestingly, this interaction was upregulated by the coexpression of TLR4 and TLR2 on the HEK cell lines, showing a clear synergistic effect between the two TLRs in NF-κB activation and CXCL-8 production.
It is intriguing to speculate that the overall LPSBv particular architecture might have a role in the MD-2/TLR4 complexation with the LPS itself and the following downstream signaling. Within this frame, it has been recently demonstrated that total LPS from fecal samples of healthy adult humans, mainly composed of Bacteroidetes species, has the capability to facilitate host tolerance of gut microbes by potently antagonizing the host TLR4 pathway.42 Indeed, it is widely known that the immunogenicity, or the strength of the intracellular signaling, triggered by LPS via TLR4 is mostly mediated by the lipid A structure.43 Indeed, a reduction in the number of the lipid A acyl moieties typically correlates with a decreased immunopotency of the LPS. Contextually, hypo-acylated lipid A forms are conserved across Bacteroidetes species,44 as also confirmed by our data which are in agreement with previous reported information regarding the tendency of Bacteroides species to accumulate linear or branched fatty acids with 15–17 carbon atoms in their LPS lipid A moiety.42,44,45 Furthermore, both 1- and 4′- phosphates on the lipid A disaccharide backbone were demonstrated to be important moieties for MD-2/TLR4 receptor complex activation.4 Since LPSBv possesses only one phosphate at position 1 of the reducing glucosamine, this may also contribute to its weaker (compared to E. coli LPS) agonistic effects as a missing 4′-phosphate was demonstrated to result in a 100-fold reduction in endotoxic activity.46 Moreover, it has been recently demonstrated that the enzyme LpxF, which is responsible for removing a single phosphate from the lipid A, is conserved across commensal Bacteroidetes and serves to enhance bacterial resistance to antimicrobial peptides, thus favoring bacterial resilience in the intestine.47 Finally, recent studies revealed that not only do the the lipid A phosphorylation and acylation pattern determine LPS toxicity, but also the occurrence of either positive or negative charged groups, other than glucosamine backbone associated phosphates, can increase LPS immunopotency.48 In this case, besides Kdo and reducing glucosamine-phosphate, LPSBv contains no further charged groups either in the lipid A part or in the core OS.
Nevertheless, with this study we highlight the involvement also of TLR2 in the immunopotential of LPSBv. This is a fascinating aspect since it has been demonstrated that the TLR2 pathway establishes colonization for commensals of the human gut microbiota.49 In this frame, although it has been demonstrated that some atypical LPSs are able to activate the TLR2 signaling pathway,33 it is still unclear how TLR2 recognizes LPS. Given the controversial data about the presence of TLR2-stimulating contaminants in LPS preparations of Bacteroides spp., here we proved that the isolated LPSBv (as well as LPSBv-RP) was devoid of any immunoactive contaminant, thus further confirming the ability of this LPS to activate the TLR2 signaling in human cells, as also previously observed for murine TLR2-HEK cells in response to LPSBv stimulation.17 How to relate this crucial point to the structure of LPSBv will be a logical follow up of the present results. Within this context, under the structural point of view, the presence, in the inner part of the LPS core, of Galf, which is known to be immunogenic to mammals, is surely an innovative signature for bacterial LPS. Indeed, the occurrence of five membered sugar rings in the LPS core OS is very rare, and to the best of our knowledge, it has only been found in an environmental Shewanella(50) strain as well as in the thermophilic bacterium Thermomonas hydrothermalis.51 It is worth noting that particular bacterial hallmark monosaccharides, such as Galf, that can be recognized by lectins of the eukaryotic innate immunity system whose role is yet to be identified, have been recently reported.52−54 Indeed, we firmly believe that the combination of all the peculiar chemical features of the LPSBv molecule influences its net effect on human immune cells, and on this ground, the role of the Galf unit in LPSBv immunological properties is intriguing and requires a future in-depth study at the molecular level. In support of our hypothesis, a recent work by Erturk-Hasdemir et al. (2019)12 demonstrated that a covalently linked lipid anchor in polysaccharide A, the zwitterionic capsular polysaccharide of B. fragilis, was critical for the initiation of the anti-inflammatory immune response through the simultaneous activation of the TLR2 signaling (by the lipid anchor) and the Dectin-1 pathway (by the polysaccharide moiety). Similarly, we deem that the saccharide regions of LPSBv heavily contribute and collaborate with the lipid A portion in dictating the innate immune signaling mechanisms leading to the activation of an anti-inflammatory response.
By this study we intended to provide the complete structure of a commensal bacterium LPS and start the search for molecular motifs that might be important for LPS–eukaryotic cell interactions at the gut level. We are not aware of whether this new structure is a “chemical paradigm” of the intestinal microbiota LPS since, so far, only very few chemical structures of intestinal commensals have been established.9,55 Certainly, we can say that a thin border exists between the homeostatic balance maintained by commensal gut microbes and the war unleashed by bacterial pathogens invading gut mucosa. This thin line is, at least in part, chemically represented by the LPS structure which, in the case of commensals, responds to particular chemical requirements, such as peculiar fatty acids chemistry, location, and distribution and peculiar monosaccharide presence in the core region (i.e., Galf). In this specific case, this chemical structure also has a different immunological behavior, i.e., the mild activating effect and the combined TLR2/TLR4 activation.
Experimental Section
Extraction, Purification, and “Repurification” of LPS from B. vulgatus mpk
Bacterial dried cells (5 g) were extracted through the hot phenol/water procedure.18 The row LPS (300 mg) was checked by SDS-PAGE after gel silver staining19 revealing a smooth-type LPS only in the water phase. In order to remove cell contaminants, an intensive enzymatic treatment by RNase (Roth), DNase (Roth), and Proteinase K (Roth) at 37 and 56 °C was executed, followed by exhaustive dialysis. In order to further purify the LPS material, several ultracentrifugation steps (4 °C, 100 000g, 24 h) and a gel-filtration chromatography were also performed. The SDS-PAGE followed by silver staining was repeated to check the degree of purity of the isolated LPS confirming the previous analysis. Furthermore, an additional SDS-PAGE followed by Coomassie Brilliant Blue (Sigma-Aldrich) gel staining to evaluate the presence of protein/lipoprotein contaminants was also executed. In both SDS-PAGE analyses, the purified LPSBv was prepared at the desired concentration (1 mg/mL) and boiled for 10 min, and then, 0.5, 1, 2, 4, and 8 μL/well were loaded on a 13.5% SDS gel with a 5% stacking gel and separated using a mini-PROTEAN electrophoresis instrument (Bio-Rad Laboratories). Silver and Coomassie Brilliant Blue staining of the gels was executed according to the standard protocols.
In order to establish the protein content of the purified LPSBv, the Pierce Micro BCA protein assay (Thermo Scientific) has been performed following the manufacturer protocol, at an LPS concentration 100 times higher than the concentration used in the cellular experiments. Finally, the assay was repeated in the presence of 2% SDS as suggested by the manufacturer to remove the significant lipid interference.20,21
In order to further exclude the presence of any TLR2-stimulating contaminant in the LPSBv preparation, an aliquot of LPSBv, previously treated to remove nucleic acids and proteins, dialyzed, ultracentrifuged, and chromatographed as described above, underwent several washes with a mixture of chloroform–methanol (1:2, v/v) and chloroform–methanol–water (3:2:0.25, v/v) currently employed to remove phospholipids. Once the organic solvents were removed completely, the sample was subjected to the “repurification” protocol by Hirschfeld et al.36 to remove any traces of lipoproteins possibly contaminating the isolated LPSBv. The so-obtained LPS (LPSBv-RP) was analyzed through the Pierce Micro BCA protein assay (Thermo Scientific). Finally, LPSBv-RP and LPSBv were both employed for the immunological tests. No unexpected or unusually high safety hazards were encountered in this work.
Chemical Analyses of LPS from B. vulgatus mpk
LPSBv monosaccharide content was established by analysis of the acetylated O-methyl glycoside derivatives obtained by treatment with HCl/MeOH (1.25 M, 85 °C, 24 h) followed by an acetylation step with acetic anhydride in pyridine (85 °C, 30 min). The absolute configuration of each sugar unit was defined through the evaluation of the O-octylglycoside derivatives as previously described.22 The sugar linkage pattern was determined by the Ciucanu method:23,24 briefly, an aliquot of sample was suspended in DMSO to which NaOH in powder was added and then methylated with CH3I, hydrolyzed with trifluoroacetic acid (4 M, 100 °C, 4 h), carbonyl reduced with NaBD4, and acetylated with pyridine and acetic anhydride. The total fatty acid content was established on intact LPS by treating with HCl (4 M, 100 °C, 4 h) followed by NaOH (5 M, 100 °C, 30 min). The pH was adjusted to reach slight acidity. After extraction in chloroform, fatty acids were then methylated with diazomethane.25 All chemical analyses were performed by means of a gas–liquid chromatography (GLC-MS) Agilent Technologies 6850A instrument equipped with a mass selective detector 5973N and a Zebron ZB-5 capillary column (Phenomenex, 30 m × 0.25 mm i.d., flow rate 1 mL/min, He used as carrier gas) and using the following temperature program: 140 °C/240 °C at 3 °C/min.
Isolation of the LPSBv Saccharide Domain
An aliquot of pure LPS was treated with anhydrous hydrazine (2 mL), stirred at 37 °C for 90 min, cooled, poured into ice-cold acetone (20 mL), and allowed to precipitate. The precipitate was centrifuged (4000g, 30 min), washed with ice-cold acetone, dried, dissolved in water, and lyophilized. The O-deacylated product was then N-deacylated with 4 M KOH. The removal of salts was executed by gel-filtration chromatography on a Sephadex G-10 column (Pharmacia, 50 × 1.5 cm). The fully deacylated product was further purified on a Toyopearl TSK HW-50 instrument (Tosoh Bioscience).
NMR Spectroscopy
1D and 2D 1H NMR spectra were recorded on a Bruker 600 DRX instrument equipped with a cryoprobe. The solvent employed was D2O, and the temperature was 298 K and pD 7. Spectra calibration was performed with internal acetone (δH 2.225 ppm, δC 31.45 ppm). 31P NMR experiments were carried out with a Bruker DRX-400 spectrometer; aqueous 85% phosphoric acid was used as the external reference (δ = 0.00 ppm). The double-quantum filtered phase sensitive correlation spectroscopy (DQF-COSY) experiment was carried out by using data sets of 4096 × 256 points. Total correlation spectroscopy (TOCSY) experiments were executed with spinlock times of 100 ms, using data sets (t1 × t2) of 4096 × 256 points. Rotating frame Overhauser enhancement spectroscopy (ROESY) and nuclear Overhauser enhancement spectroscopy (NOESY) experiments were recorded by using data sets (t1 × t2) of 4096 × 256 points and by using mixing times between 100 and 400 ms. In all homonuclear experiments the data matrix was zero-filled in both dimensions to give a matrix of 4 K × 2 K points and was resolution enhanced in both dimensions by a cosine-bell function before Fourier transformation. The determination of coupling constants was obtained by 2D phase sensitive DQF-COSY.56,57 Heteronuclear single quantum coherence (1H, 13C HSQC) and heteronuclear multiple bound correlation (1H, 13C HMBC) experiments were recorded in 1H-detection mode by single-quantum coherence with proton decoupling in the 13C domain using data sets of 2048 × 256 points. 1H, 13C HSQC was executed using sensitivity improvement and in the phase-sensitive mode using Echo/Antiecho gradient selection, with multiplicity editing during the selection step.58 The 1H, 13C HMBC experiment was optimized on long-range coupling constants with the low-pass J filter to suppress one-bound connectivity, using gradient pulses for selection. A delay of 60 ms was employed for the evolution of long-range correlations. A long-range coupling constant value of 6 Hz was used. The data matrix in both heteronuclear experiments was extended to 2048 × 1024 points using forward linear prediction.59
Molecular Modeling
Computational studies and MD simulations of the following molecules were performed: LPSBv lipid A molecules (TetraLipA and PentaLipA) and tetra- and penta-acylated LPSBv (LPSBv-tetra and LPSBv-penta). The modeled 3D structures were in agreement with the NMR data as can be deduced from the monitoring of selected inter-residue distances (not shown).
Building of the Ligands
Compounds TetraLipA, PentaLipA, LPSBv-tetra, and LPSBv-penta were constructed using the Maestro interface.60 The 3D coordinates of E. coli LPS from PDB ID 3FXI were used as a template. The resulting structures were minimized using the OPLS3 force field, water as a solvent, and steepest descent as the method of minimization, with a maximum of 10 000 iterations.
Parameter Derivation
Parameterization of new units was performed using Gaussian0961 by optimizing the geometries with the 6-31G basis set at the Hartree–Fock level of theory. Charges were derived by applying the RESP methodology implemented in Antechamber, assigning the general Amber14 force field (GAFF) atom types.
Molecular Dynamics (MD) Simulations
All the MD simulations were carried out using Amber14.62 Each compound was solvated with water molecules (TIP3 force field), and Na+ counterions were added. Several steps of equilibration were performed before running the MD simulation. The first one consisted of 1000 steps of steepest descent minimization followed by 7000 steps of conjugate gradient minimization; a 100 kcal/(mol A2) harmonic potential constraint was applied to the solute. In the four subsequent steps, the harmonic potential was progressively lowered, respectively, to 10, 5, and 2.5 kcal/(mol A2)) for 600 steps of conjugate gradient minimization each time, and then, the whole system was minimized uniformly. In the following step, the system was heated from 0 to 100 K using the Langevin thermostat in the canonical ensemble (NVT) while applying a 20 kcal/(mol A2) harmonic potential restraint on the proteins and the ligand. The next step heated up the system from 100 to 300 K in the isothermal–isobaric ensemble (NPT) under the same restraint condition as in the previous step. In the last step, the same parameters were used to simulate the system for 100 ps, but no harmonic restraint was applied. At this point, the system was ready for the production run, which was performed using the Langevin thermostat under NPT ensemble, at a 2 fs time step. A 100 ns trajectory was generated and further analyzed.
ESI MS and MS/MS Analysis
MS spectra of the saccharide part of LPSBv were acquired on an amazon SL ion trap (IT) mass spectrometer with electrospray ionization (ESI) (Bruker Daltonics). The sample (75 μg) was dissolved in 50 μL of acetonitrile/water/formic acid solution (100:100:1, v/v/v) and analyzed in negative ion mode. The ion corresponding to core OS (m/z 667.3) was analyzed by MS/MS. Source parameters were as follows: sample flow, 3 μL/min; ion source temperature, 200 °C; nitrogen flow, 5 L/min at a pressure of 8 psi. Spectra were scanned in the 200–2000 m/z range. He was used as the collision gas in the IT. The system was calibrated using ESI-L tuning mix (Agilent Technologies).
Isolation of the Lipid A Fraction from LPSBv
Isolation of lipid A was achieved by mild acid hydrolysis by using acetate buffer (1 mL, pH = 4.4) promoted by SDS (1 mg/mL) on ca. 20 mg of isolated LPS at 100 °C. The solution was extracted three times with CHCl3/MeOH/H20 (100:30:30, v/v/v) and centrifuged (4 °C, 7000g, 15 min). The organic phase containing the lipid A was further purified by several washes with distilled water and then freeze-dried.
MALDI MS Analysis on Isolated A from LPSBv
Lipid A isolated after mild acid hydrolysis of LPSBv was analyzed with a MALDI-TOF ultrafleXtreme spectrometer (Bruker Daltonics). 100 μg of lipid A was dissolved in 100 μL of milli-Q water after 10 min of incubation at 40 °C in the ultrasonic bath. 9H-pyrido(3,4)-indole (10 mg) was mixed with 1 mL of CHCl3/MeOH (1:1, v/v), and 0.5 μL of the matrix was spotted onto the matrix. The sample was analyzed in reflectron, negative ion mode, scan range: m/z 700–3500. Instrument was calibrated within 1000–3150 Da mass range, using peptide mixture (Peptide Calibration Standard, Bruker Daltonics).
HEK 293 Cell Culture, Transfection, and Stimulation
The HEK293 cell line, stably transfected with human TLR4/MD2-CD14 (InvivoGen), was seeded into a 24-well plate at the concentration of 5 × 105 cells/mL. 48 h after seeding, the cells were transiently transfected through PolyFect transfection reagent (Qiagen) with a reaction mix containing 250 ng of Firefly luciferase reporter constructs, pGL3.ELAM.tk [harboring nuclear factor kappa B (NF-κB) promoter sequences], and 25 ng of Renilla luciferase reporter plasmid, pRLTK (as an internal control). For hTLR2, HEK293 cells or HEK293 cells hTLR4/MD2-CD14 were transfected through PolyFect transfection reagent with the plasmid pcDNA3-TLR2-YFP (pcDNA3-TLR2-YFP was a gift of Doug Bolenbock, Addgene plasmid 13016) and treated as above. The day after, the cells (HEK293 hTLR4/MD2-CD14, HEK293 hTLR2, or HEK293 hTLR4/MD2-CD14 hTLR2) were incubated with different concentrations of LPSBv, LPSBv-RP, or lipid ABv (1, 10, and 100 ng/mL) or with purified E. coli LPS (LPS-EB ultrapure; InvivoGen) or with synthetic lipid IVA used at the same concentrations as above, for 6 h to analyze NF-κB activity (dual luciferase reporter assay system, Promega) and to measure CXCL-8 release (DuoSet R&D system). In the experiments including the TLR2 in addition to LPSBv, LPSBv-RP, and lipid ABv, Pam3 CSK4 (Pam3 CSK4) (InvivoGen) (500 ng/mL) was used as a positive control. Where necessary, HEK293 cells only or HEK293 cells pcDNA3 were exposed to LPSs and lipid A or Pam3 CSK4, as above. For the competition assay, HEK 293 hTLR4/MD2-CD14 cells were primed with 1, 10, and 100 ng/mL of LPSBv or lipid ABv, or lipid IVA for 1 h and then exposed to E. coli LPS (10 and 100 ng/mL) for 4 h. After this time, NF-κB activity and CXCL-8 production were measured.
Human Macrophage and Dendritic Cell Isolation, Cultures, and Stimulation
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats obtained by the blood bank of Sapienza University or Sanquin Blood bank, Amsterdam, The Netherlands, from healthy adult volunteers (blood donors) following written informed consent in accordance with the Declaration of Helsinki. Each experiment included two buffy coats from two healthy adult volunteers and was carried out in triplicate.
For macrophages, PBMCs were obtained through a density gradient. CD14+ monocytes were isolated using the MACS microbead system (Miltenyi Biotec, Bergisch Gladbach, Germany). The monocytes were cultured for 6 days in RPMI 1640 (Lonza) supplemented with 10% heat-inactivated FBS (Euroclone Fetal Bovine Serum, GE Healthcare Life Sciences), 100 μM nonessential amino acids, 1 mM sodium pyruvate, 1000 U/mL penicillin, and 1000 U/mL streptomycin (all from Lonza) and 60 ng/mL GM-CSF (granulocyte-macrophage colony-stimulating factor) (Miltenyi Biotec) to obtain human macrophages (MoMs).
For dendritic cells, CD14+ monocytes were isolated using the MACS microbead system (Miltenyi Biotec, Bergisch Gladbach, Germany). The monocytes were cultured for 5 days in RPMI 1640 (Lonza) supplemented with 10% heat-inactivated FBS (HyClone Fetal Bovine Serum, GE Healthcare Life Sciences), 100 μM nonessential amino acids, 1000 U/mL penicillin, and 1000 U/mL streptomycin (all from Lonza), 20 ng/mL IL-4, and 50 ng/mL GM-CSF (both Miltenyi Biotec) to obtain immature human dendritic cells. MoDCs were characterized by immunostaining with CD11c, CD14, and CD80 (all from BD Pharmingen) through a flow cytometric analysis.
For MoMs and MoDCs stimulation, the cells were seeded in cell plates (2.5 × 105 cells/well for MoMs and 5 × 105 cells/well for MoDCs) and exposed to 1, 10, and 100 ng/mL of LPSBv or lipid ABv, or lipid IVA or E. coli LPS used at the same concentrations for 12 h. Cell supernatants were then collected and processed for ELISA to measure the levels of TNF-α, IL-10, IL-6, and IP-10.
As for the time-course of IL-6 and IL-10 release by MoDCs, cells were stimulated with LPSBv (10 ng/mL) after 1, 2, 4, 6, 8, and 24 h, and with 100 ng/mL after 1 and 24 h. Data are expressed as percentage of effect, and they are normalized over the MoDCs treated with LPS from E. coli (10 ng/mL at 24 h) set at 100%.
Bone Marrow-Monocyte-Derived Macrophage (BMDM) Isolation, Culture, and Stimulation
C57BL/6 mice were purchased from Charles River. BMDMs were derived from the bone marrow cells collected from 5 week old female mice, as already reported.39 Animal studies were conducted according to protocols approved by the University of Rome La Sapienza and adhered strictly to the Italian Ministry of Health guidelines for the use and care of experimental animals. BMDMs were differentiated during 7 days in RPMI 1640 (Lonza), supplemented with 10% of heat-inactivated FBS (HycloneTM, Euroclone), 1% of l-glutamine (Lonza), 1% sodium pyruvate (Lonza), 1% NEAA (Lonza), 0.5% 2-ME (Gibco), and 40 ng/mL macrophage colony-stimulating factor (M-CSF; Miltenyi Biotec). BMDMs were seeded into 24-well plate (5 × 105 cells per well) and were incubated with different concentrations of LPSBv or lipid ABv (1, 10, and 100 ng/mL), E. coli LPS, or with lipid IVA at the same concentrations for 12 h. After this, cell supernatants were then collected and processed for ELISA to measure the levels of TNF-α and IL-10, IL-6, and IP-10.
ELISA Assay
Cytokine and chemokine concentrations were determined by commercially available ELISA kits (Duo Set Kit R&D systems or ThermoFisher Scientific IFN alpha Human ELISA Kit). The absorbance was measured on a LT-4000 microplate reader (Labtech) (Hercules, CA).
Statistical Analysis
Data were presented as mean ± SD of at least 4 independent experiments (for human macrophages and dendritic cells, two donors for each experiment carried out in triplicate, as stated above). The statistical analysis has been carried out through GraphPad Prism software. For multiple group comparison the software used the multiple t test followed by Sidak–Bonferroni correction. Statistical significance was set at *p ≤ 0.05.
Acknowledgments
F.D.L. acknowledges Progetto STAR 2018 Linea 1 grant E66C18001330003. S.M.S. acknowledges Spanish Ministry of Science (ref. CTQ2017-88353-R). A.M., F.D.L., and A.S. acknowledge H2020 Marie Skłodowska-Curie ITN 2018 “SweetCrossTalk” grant 814102. A.M. acknowledges progetto POR SATIN POR-FESR 2014–2020 grant B61C17000070007 (OR3) and Progetto POR Campania Oncoterapia 2014–2020 grant B61G18000470007. A.S. acknowledges PRIN-MIUR 2017 Glytunes project. A.S. and F.C. acknowledge COST (European Cooperation in Science and Technology) Action CA18103 (INNOGLY). F.C. was financially supported by the NWO Spinoza award of Y.K.
Glossary
Abbreviations
- BCA
bicinchoninic acid
- CXCL-8
chemokine (C-X-C motif) ligand 8
- ESI MS
electrospray Ionization Mass spectrometry
- Fuc
fucose
- Gal
galactose
- Glc
glucose
- GLC-MS
gas–liquid chromatography mass spectrometry
- HEK cells
human embryonic kidney cells
- IL-6
interleukin-6
- IL-8
interleukin-8
- IL-10
interleukin-10
- IFN-α
interferon-α
- IP-10
IFN-γ inducible protein-10
- Kdo
keto-deoxy-d-manno-octanoic acid
- LPS
lipopolysaccharide
- LPSBv
B. vulgatus LPS
- MALDI MS
matrix-assisted laser desorption/ionization mass spectrometry
- MAMP
microbe-associated molecular pattern
- Man
mannose
- MD-2
myeloid differentiation protein-2
- MoDCs
peripheral blood-Monocyte-derived dendritic cells
- MoMs
peripheral blood-monocyte-derived macrophages
- NF-kB
nuclear factor-κB
- NMR
nuclear magnetic resonance
- OS
oligosaccharide
- Rha
rhamnose
- RU
repeating unit
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TLR2
toll-like receptor 2
- TLR4
toll-like receptor 4
- TNF-α
tumor necrosis factor α
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c00791.
Additional figures including staining images, assay results, NMR spectra, ESI-MS spectra, MALDI MS spectra, inter-residue distances, structures, and ELISA results (PDF)
Author Contributions
F.D.L., A.S., M.L.B., and A.M. conceived the study. F.D.L., A.S., S.M.S., F.C., W.J., M.L.B., and A.M. designed and executed the research and wrote the manuscript. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Dedication
This paper is dedicated to Prof. Jesús Jiménez-Barbero for his 60th birthday.
This paper was published ASAP on July 30, 2020. The abstract graphic and Figure 3 were updated. The revised paper was reposted on August 6, 2020.
Supplementary Material
References
- Sender R.; Fuchs S.; Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14 (8), e1002533 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler A. G.; Goodman A. L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2017, 2, 17026. 10.1038/nmicrobiol.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C.; Trent M. S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 2014, 83, 99–128. 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- Molinaro A.; Holst O.; Di Lorenzo F.; Callaghan M.; Nurisso A.; D’Errico G.; Zamyatina A.; Peri F.; Berisio R.; Jerala R.; Jiménez-Barbero J.; Silipo A.; Martín-Santamaría S. Chemistry of lipid A: at the heart of innate immunity. Chem. - Eur. J. 2015, 21 (2), 500–19. 10.1002/chem.201403923. [DOI] [PubMed] [Google Scholar]
- Raetz C. R.; Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo F.; De Castro C.; Lanzetta R.; Parrilli M.; Silipo A.; Molinaro A.. Lipopolysaccharides as microbe-associated molecular patterns: A structural perspective. In Carbohydrates in Drug Design and Discovery; Jiménez-Barbero J., Javier Canada F., Martín-Santamaría S., Eds.; Royal Society of Chemistry (RSC): London, UK, 2015; pp 38–63. [Google Scholar]
- Triantafilou M.; Triantafilou K. Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol. 2002, 23, 301–304. 10.1016/S1471-4906(02)02233-0. [DOI] [PubMed] [Google Scholar]
- Akira S.; Uematsu S.; Takeuchi O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Di Lorenzo F.; De Castro C.; Silipo A.; Molinaro A. Lipopolysaccharide structures of Gram-negative populations in the gut microbiota and effects on host interactions. FEMS Microbiol. Rev. 2019, 43 (3), 257–272. 10.1093/femsre/fuz002. [DOI] [PubMed] [Google Scholar]
- Davenport E. R.; Sanders J. G.; Song S. J.; Amato K. R.; Clark A. G.; Knight R. The human microbiome in evolution. BMC Biol. 2017, 15, 127. 10.1186/s12915-017-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff C. P.; Rhodes M. E.; Arnolds K. L.; Collins C. B.; Donnelly J.; Nusbacher N.; Jedlicka P.; Schneider J. M.; McCarter M. D.; Shaffer M.; Mazmanian S. K.; Palmer B. E.; Lozupone C. A. Diverse intestinal bacterial contain putative zwitterionic capsuar polysaccharides with anti-inflammatory properties. Cell Host Microbe 2016, 20, 535–547. 10.1016/j.chom.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erturk-Hasdemir D.; Oh S. F.; Okan N. A.; Stefanetti G.; Gazzaniga F. S.; Seeberger P. H.; Plevy S. E.; Kasper D. L. Symbionts exploit complex signaling to educate the immune system. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 26157. 10.1073/pnas.1915978116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanen T.; Kostic A. D.; d’Hennezel E.; Siljander H.; Franzosa E. A.; Yassour M.; Kolde R.; Vlamakis H.; Arthur T. D.; Hämäläinen A. M.; Peet A.; Tillmann V.; Uibo R.; Mokurov S.; Dorshakova N.; Ilonen J.; Virtanen S. M.; Szabo S. J.; Porter J. A.; Lähdesmäki H.; Huttenhower C.; Gevers D.; Cullen T. W.; Knip M.; Xavier R. J. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016, 165, 842–853. 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waidmann M.; Bechtold O.; Frick J. S.; Lehr H. A.; Schubert S.; Dobrindt U.; Loeffler J.; Bohn E.; Autenrieth I. B. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology 2003, 125, 162–177. 10.1016/S0016-5085(03)00672-3. [DOI] [PubMed] [Google Scholar]
- Müller M.; Fink K.; Geisel J.; Kahl F.; Jilge B.; Reimann J.; Mach N.; Autenrieth I. B.; Frick J. S. Intestinal colonization of IL-2 deficient mice with non-colitogenic B. vulgatus prevents DC maturation and T-cell polarization. PLoS One 2008, 3, e2376 10.1371/journal.pone.0002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimle A.; Gronbach K.; Beifuss B.; Schäfer A.; Harmening R.; Bender A.; Maerz J. K.; Lange A.; Michaelis L.; Maurer A.; Menz S.; McCoy K.; Autenrieth I. B.; Kalbacher H.; Frick J. S. Symbiotic gut commensal bacteria act as host cathepsin S activity regulators. J. Autoimmun. 2016, 75, 82–95. 10.1016/j.jaut.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Steimle A.; Michaelis L.; Di Lorenzo F.; Kliem T.; Münzner T.; Maerz J. K.; Schäfer A.; Lange A.; Parusel R.; Gronbach K.; Fuchs K.; Silipo A.; Öz H. H.; Pichler B. J.; Autenrieth I. B.; Molinaro A.; Frick J. S. Weak Agonistic LPS Restores Intestinal Immune Homeostasis. Mol. Ther. 2019, 27, 1974 10.1016/j.ymthe.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal O.; Jann K. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of procedure. Carbohydr. Chem. 1965, 5, 83–91. [Google Scholar]
- Kittelberger R.; Hilbink F. Sensitive silver-staining detection of bacterial lipopolysaccharides in polyacrylamide gels. J. Biochem. Biophys. Methods 1993, 26, 81–86. 10.1016/0165-022X(93)90024-I. [DOI] [PubMed] [Google Scholar]
- Kessler R. J.; Fanestil D. D. Interference by lipids in the determination of protein using bicinchoninic acid. Anal. Biochem. 1986, 159 (1), 138–42. 10.1016/0003-2697(86)90318-0. [DOI] [PubMed] [Google Scholar]
- Morton R. E.; Evans T. A. Modification of the bicinchoninic acid protein assay to eliminate lipid interference in determining lipoprotein protein content. Anal. Biochem. 1992, 204 (2), 332–4. 10.1016/0003-2697(92)90248-6. [DOI] [PubMed] [Google Scholar]
- Leontein K. Assignment of absolute configuration of sugars by glc of their acetylated glycosides formed from chiral alcohols. Carbohydr. Res. 1978, 62, 359–362. 10.1016/S0008-6215(00)80882-4. [DOI] [Google Scholar]
- Ciucanu I.; Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 1984, 131, 209–217. 10.1016/0008-6215(84)85242-8. [DOI] [Google Scholar]
- De Castro C.; Parrilli M.; Holst O.; Molinaro A. Microbe-associated molecular patterns in innate immunity: Extraction and chemical analysis of gram-negative bacterial lipopolysaccharides. Methods Enzymol. 2010, 480, 89–115. 10.1016/S0076-6879(10)80005-9. [DOI] [PubMed] [Google Scholar]
- Rietschel E. T. Absolute configuration of 3-hydroxy fatty acids present in lipopolysaccharides from various bacterial groups. Eur. J. Biochem. 1976, 64, 423–428. 10.1111/j.1432-1033.1976.tb10318.x. [DOI] [PubMed] [Google Scholar]
- Hashimoto M.; Kirikae F.; Dohi T.; Adachi S.; Kusumoto S.; Suda Y.; Fujita T.; Naoki H.; Kirikae T. Structural study on lipid A and the O-specific polysaccharide of the lipopolysaccharide from a clinical isolate of Bacteroides vulgatus from a patient with Crohn’s disease. Eur. J. Biochem. 2002, 269, 3715–3721. 10.1046/j.1432-1033.2002.03062.x. [DOI] [PubMed] [Google Scholar]
- Holst O. Deacylation of lipopolysaccharides and isolation of oligosaccharide phosphates. Methods Mol. Biol. 2000, 145, 345–353. 10.1385/1-59259-052-7:345. [DOI] [PubMed] [Google Scholar]
- Birnbaum G. I. Conformations of ammonium. 3-deoxy-D-manno-2-octulosonate (KDO) and methyl -and -ketopyranosides of KDO: X-ray structure and 1H NMR analyses. J. Carbohydr. Chem. 1987, 6, 17–39. 10.1080/07328308708058858. [DOI] [Google Scholar]
- Silipo A.; Molinaro A.; Comegna C.; Sturiale L.; Cescutti P.; Garozzo D.; Lanzetta R.; Parrilli M. Full Structural Characterisation of the Lipooligosaccharide of a Burkholderia pyrrocinia Clinical Isolate. Eur. J. Org. Chem. 2006, 2006, 4874–4883. 10.1002/ejoc.200600520. [DOI] [Google Scholar]
- Yamamoto M.; Sato S.; Hemmi H.; Hoshino K.; Kaisho T.; Sanjo H.; Takeuchi O.; Sugiyama M.; Okabe M.; Takeda K.; Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 2003, 301 (5633), 640–3. 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- Hornung V.; Rothenfusser S.; Britsch S.; Krug A.; Jahrsdorfer B.; Giese T.; Endres S.; Hartmann G. Quantitative expression of toll-like receptor 1–10 mRNA cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 2002, 168, 4531–7. 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- Chandler C. E.; Ernst R. K. Bacterial lipids: powerful modifiers of the innate immune response. F1000Research 2017, 6, 1334. 10.12688/f1000research.11388.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarría-Velázquez C. O.; Torres-Martínez A. C.; Montaño L. F.; Rendón-Huerta E. P. TLR2 activation induced by H. pylori LPS promotes the differential expression of claudin-4, −6, −7 and −9 via either STAT3 and ERK1/2 in AGS cells. Immunobiology 2018, 223 (1), 38–48. 10.1016/j.imbio.2017.10.016. [DOI] [PubMed] [Google Scholar]
- Hashimoto M.; Waki J.; Nakayama-Imaohji H.; Ozono M.; Hashiguchi S.; Kuwahara T. TLR2-stimulating contaminants in glycoconjugate fractions prepared from Bacteroides fragilis. Innate Immun. 2017, 23 (5), 449–458. 10.1177/1753425917714313. [DOI] [PubMed] [Google Scholar]
- Manthey C. L.; Vogel S. N. Elimination of trace endotoxin protein from rough chemotype LPS. J. Endotoxin Res. 1994, 1, 84–91. 10.1177/096805199400100202. [DOI] [Google Scholar]
- Hirschfeld M.; Ma Y.; Weis J. H.; Vogel S. N.; Weis J. J. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J. Immunol. 2000, 165 (2), 618–22. 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- Miller S. I.; Ernst R. K.; Bader M. W. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 2005, 3 (1), 36–46. 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- Schromm A. B.; Brandenburg K.; Loppnow H.; Moran A. P.; Koch M. H.; Rietschel E. T.; Seydel U. Biological activities of lipopolysaccharides are determined by the shape of their lipid A portion. Eur. J. Biochem. 2000, 267 (7), 2008–13. 10.1046/j.1432-1327.2000.01204.x. [DOI] [PubMed] [Google Scholar]
- Lembo-Fazio L.; Billod J. M.; Di Lorenzo F.; Paciello I.; Pallach M.; Vaz-Francisco S.; Holgado A.; Beyaert R.; Fresno M.; Shimoyama A.; Lanzetta R.; Fukase K.; Gully D.; Giraud E.; Martín-Santamaría S.; Bernardini M. L.; Silipo A. Bradyrhizobium Lipid A: Immunological Properties and Molecular Basis of Its Binding to the Myeloid Differentiation Protein-2/Toll-Like Receptor 4 Complex. Front. Immunol. 2018, 9, 1888. 10.3389/fimmu.2018.01888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo F.; Palmigiano A.; Al Bitar-Nehme S.; Sturiale L.; Duda K. A.; Gully D.; Lanzetta R.; Giraud E.; Garozzo D.; Bernardini M. L.; Molinaro A.; Silipo A. The Lipid A from Rhodopseudomonas palustris Strain BisA53 LPS Possesses a Unique Structure and Low Immunostimulant Properties. Chem. - Eur. J. 2017, 23 (15), 3637–3647. 10.1002/chem.201604379. [DOI] [PubMed] [Google Scholar]
- Pandiyan P.; Bhaskaran N.; Zou M.; Schneider E.; Jayaraman S.; Huehn J. Microbiome Dependent Regulation of Tregs and Th17 Cells in Mucosa. Front. Immunol. 2019, 10, 426. 10.3389/fimmu.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Hennezel E.; Abubucker S.; Murphy L. O.; Cullen T. W. Total lipopolysaccharide from the human gut microbiome silences Toll-Like Receptor signaling. mSys 2017, 2 (6), e00046-17 10.1128/mSystems.00046-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg K.; Mayer H.; Koch M. H.; Weckesser J.; Rietschel E. T.; Seydel U. Influence of the supramolecular structure of free lipid A on its biological activity. Eur. J. Biochem. 1993, 218, 555–563. 10.1111/j.1432-1033.1993.tb18409.x. [DOI] [PubMed] [Google Scholar]
- Johne B.; Olsen J.; Bryan K. Fatty acids and sugars in lipopolysaccharides from Bacteroides intermedius Bacteroides gingivalis and Bacteroides loescheii.. Oral Microbiol. Immunol. 1988, 3, 22–27. 10.1111/j.1399-302X.1988.tb00600.x. [DOI] [PubMed] [Google Scholar]
- Rokosz A.; Meisel-Mikołajczyk F.; Kot K.; Mieszała M.; Szponar B.; Gamian A. Analysis of fatty acids from lipopolysaccharides of Bacteroides thetaiotaomicron and Bacteroides fragilis. Med. Dosw. Mikrobiol. 2001, 53 (2), 177–83. [PubMed] [Google Scholar]
- Rietschel E. T.; Kirikae T.; Schade F. U.; Mamat U.; Schmidt G.; Loppnow H.; Ulmer A. J.; Zähringer U.; Seydel U.; Di Padova F. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994, 8, 217–225. 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- Cullen T. W.; Schofield W. B.; Barry N. A.; Putnam E. E.; Rundell E. A.; Trent M. S.; Degnan P. H.; Booth C. J.; Yu H.; Goodman A. L. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 2015, 347, 170–5. 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo F.; Kubik Ł.; Oblak A.; Lorè N. I.; Cigana C.; Lanzetta R.; Parrilli M.; Hamad M. A.; De Soyza A.; Silipo A.; Jerala R.; Bragonzi A.; Valvano M. A.; Martín-Santamaría S.; Molinaro A. Activation of Human Toll-like Receptor 4 (TLR4)/Myeloid Differentiation Factor 2 (MD-2) by hypoacylated lipopolysaccharide from a clinical isolate of Burkholderia cenocepacia. J. Biol. Chem. 2015, 290 (35), 21305–19. 10.1074/jbc.M115.649087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J. L.; Lee S. M.; Li J.; Tran G.; Jabri B.; Chatila T. A.; Mazmanian S. K. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011, 332 (6032), 974–7. 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov E.; Korenevsky A.; Beveridge T. J. The structure of the carbohydrate backbone of the LPS from Shewanella putrefaciens CN32. Carbohydr. Res. 2002, 337, 1285–1289. 10.1016/S0008-6215(02)00160-X. [DOI] [PubMed] [Google Scholar]
- Di Lorenzo F.; Paciello I.; Fazio L. L.; Albuquerque L.; Sturiale L.; da Costa M. S.; Lanzetta R.; Parrilli M.; Garozzo D.; Bernardini M. L.; Silipo A.; Molinaro A. Thermophiles as potential source of novel endotoxin antagonists: the full structure and bioactivity of the lipo-oligosaccharide from Thermomonas hydrothermalis. ChemBioChem 2014, 15, 2146–2155. 10.1002/cbic.201402233. [DOI] [PubMed] [Google Scholar]
- Wesener D. A.; Dugan A.; Kiessling L. L. Recognition of microbial glycans by human intelectin-1. Curr. Opin. Struct. Biol. 2017, 44, 168–178. 10.1016/j.sbi.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesener D. A.; Dugan A.; Kiessling L. L. Recognition of microbial glycans by soluble human lectins. Curr. Opin. Struct. Biol. 2017, 44, 168–178. 10.1016/j.sbi.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodo F.; Marradi M.; Park J.; Ram A. F.; Penadés S.; van Die I.; Tefsen B. Galactofuranose-coated gold nanoparticles elicit a pro-inflammatory response in human monocyte-derived dendritic cells and are recognized by DC-SIGN. ACS Chem. Biol. 2014, 9 (2), 383–9. 10.1021/cb4008265. [DOI] [PubMed] [Google Scholar]
- Naito T.; Mulet C.; De Castro C.; Molinaro A.; Saffarian A.; Nigro G.; Bérard M.; Clerc M.; Pedersen A. B.; Sansonetti P. J.; Pédron T. Lipopolysaccharide from Crypt-Specific Core Microbiota Modulates the colonic epithelial proliferation-to-differentiation balance. mBio 2017, 8 (5), e01680-17 10.1128/mBio.01680-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piantini U.; Sorensen O. W.; Ernst R. R. Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 1982, 104, 6800–6801. 10.1021/ja00388a062. [DOI] [Google Scholar]
- Rance M.; Sørensen O. W.; Bodenhausen G.; Wagner G.; Ernst R. R.; Wüthrich K. Improved spectral resolution in COSY (1)H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 1983, 117, 479. 10.1016/0006-291X(83)91225-1. [DOI] [PubMed] [Google Scholar]
- States D. J.; Haberkorn R. A.; Ruben D. J. A two-dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J. Magn. Reson. 1982, 48, 286–292. 10.1016/0022-2364(82)90279-7. [DOI] [Google Scholar]
- Stern A. S.; Li K. B.; Hoch J. C. Modern spectrum analysis in multidimensional NMR spectroscopy: comparison of linear-prediction extrapolation and maximum-entropy reconstruction. J. Am. Chem. Soc. 2002, 124, 1982–1993. 10.1021/ja011669o. [DOI] [PubMed] [Google Scholar]
- Schrödinger Release 2019–1; Maestro: New York, NY, 2019.
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Petersson G. A.; Nakatsuji H.; Li X.; Caricato M.; Marenich A. V.; Bloino J.; Janesko B. G.; Gomperts R.; Mennucci B.; Hratchian H. P.; Ortiz J. V.; Izmaylov A. F.; Sonnenberg J. L.; Williams-Young D.; Ding F.; Lipparini F.; Egidi F.; Goings J.; Peng B.; Petrone A.; Henderson T.; Ranasinghe D.; Zakrzewski V. G.; Gao J.; Rega N.; Zheng G.; Liang W.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Throssell K.; Montgomery J. A. Jr.; Peralta J. E.; Ogliaro F.; Bearpark M.; Heyd J. J.; Brothers E. N.; Kudin K. N.; Staroverov V. N.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Millam J. M.; Klene M.; Adamo C.; Cammi R.; Ochterski J. W.; Martin R. L.; Morokuma K.; Farkas O.; Foresman J. B.; Fox D. J.. Gaussian 16, revision B.01; Gaussian, Inc.: Wallingford CT, 2016.
- Case D.; Berryman J.; Betz R.; Cerutti D.; Cheatham T. III; Darden T.; Duke R.; Giese T.; Gohlke H.; Goetz A.. AMBER 2015; University of California: San Francisco, 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.