Summary
Alzheimer disease (AD) is a devastating neurological disease associated with progressive loss of mental skills and cognitive and physical functions whose etiology is not completely understood. Here, our goal was to simultaneously uncover novel and known molecular targets in the structured layers of the hippocampus and olfactory bulbs that may contribute to early hippocampal synaptic deficits and olfactory dysfunction in AD mice. Spatially resolved transcriptomics was used to identify high-confidence genes that were differentially regulated in AD mice relative to controls. A diverse set of genes that modulate stress responses and transcription were predominant in both hippocampi and olfactory bulbs. Notably, we identify Bok, implicated in mitochondrial physiology and cell death, as a spatially downregulated gene in the hippocampus of mouse and human AD brains. In summary, we provide a rich resource of spatially differentially expressed genes, which may contribute to understanding AD pathology.
Subject Areas: Cellular Neuroscience, Omics, Transcriptomics
Graphical Abstract
Highlights
-
•
Spatial transcriptomics identifies differentially expressed genes with spatial patterns
-
•
Early application of spatial transcriptomics to olfactory bulbs from AD models
-
•
Bok gene is spatially differentially expressed in AD mouse and patient brains
-
•
Paip1 and Homer1 genes are regulated in a PolB-dependent manner
Cellular Neuroscience; Omics; Transcriptomics
Introduction
Alzheimer disease (AD) is a multifactorial and progressive neurodegenerative disorder that is the most common cause of dementia globally. Poor short-term memory and olfactory dysfunction are early signs in early stages of AD (Murphy, 2019; Rahayel et al., 2012). Emerging evidence suggests that olfactory dysfunction might predate the onset of cognitive decline. Often, olfactory dysfunction is perceived by the individual as a loss of taste, which worsens with the progression of the disease; however, the relationships between olfactory deficits, neurodegeneration, and dementia remain unclear (Murphy, 2019). Given the abundance of failed clinical trials for AD, it is apparent that neither the genes nor the cellular mechanisms that underlie this multifactorial disease are well understood. Hence, the molecular pathology of this devastating disorder is still in its infancy.
Genetically engineered mouse models are widely used to characterize molecular features of AD, including AD-associated differential gene expression. The triple transgenic AD model, 3xTg AD (3xAD), expresses three human gene variants: APP K670N/M671L, MAPT P301L, and PS1 M146V. Increased DNA damage occurs early in the course of AD disease pathology, and to test the importance of DNA repair, we created a modified version of the 3xAD strain by adding a deficiency in DNA polymerase beta (Polβ) (Sykora et al., 2015). Importantly, we had previously observed a loss of this protein and its polymerase activity in the brains of human patients with AD patient (Weissman et al., 2007). Behavioral and biochemical analyses of 3xTg AD/Polβ+/- (3xPB) mice revealed that these mice display more severe deficits in memory, learning, long-term potentiation (LTP), olfactory function, and mitochondrial homeostasis and higher levels of DNA damage and cell death than control mice (Hou et al., 2018; Misiak et al., 2017a; Sykora et al., 2015).
Gene expression changes contribute to AD disease, and defining a more detailed map of how gene expression changes in the neuronal structures of the brain, early in the disease process, may contribute to a better understanding of the disorder. Thus, the goal of the present study was to identify spatially expressed genes that were differentially expressed (DE) in the hippocampal and olfactory bulb (OFB) regions of 3xAD, 3xPB, and control mice. To accomplish this goal, gene expression was quantified using a powerful method known as spatial transcriptomics (ST), recently described by Stahl and colleagues (Stahl et al., 2016). In earlier studies, ST was used to analyze differential gene expression in neuronal tissue in neurodegenerative diseases (Chen et al., 2020; Maniatis et al., 2019). The key feature of ST is that it pairs quantitative transcriptomics with high-resolution tissue imaging; this allows gene expression profiles to be anchored to the physical map of the organ or tissue(s) of interest, which in this study are the hippocampal and OFB regions of the brain. The hippocampus and OFB play central roles in AD pathology, and these anatomical brain regions are each composed of layers of specialized cells. Thus, we focused our gene expression analyses on these brain regions.
ST is an attractive approach to resolve spatial differences in gene expression in AD brains. ST is unique, yielding a substantially different, richer dataset than more traditional transcriptome analysis using gene expression microarray technology. It combines high-resolution tissue imaging with unbiased spatially defined RNA sequencing (RNA-seq) using barcoded spots (100 um) on glass slides (Stahl et al., 2016). This affords the opportunity to characterize tissue morphology using unsupervised analyses at the molecular level. In comparison, previous RNA-seq of bulk brain tissue extracts is data rich but limited by the fact that it lacks spatial information. The alternative to bulk analysis is to perform single-cell/single-nuclei RNA-seq. However, this technique also does not yield spatial gene expression measures unless combined with fine dissection techniques. Furthermore, although the resolution of ST at the level of the spot is nominally lower than the resolution of single-cell transcriptomics, the ST dataset is again richer, because data points are anchored to a high-resolution image of the target tissue.
Our current AD research builds on previous strategies to identify gene expression changes that may identify novel markers or genes that can be targeted as biomarkers for interventions that halt or slow disease progression. We used male mice in an early phase of the disease and sought to identify key genes in either the hippocampus or OFBs to improve our understanding of the defects in synaptic transmission and odor sensing. We focused our analyses on three subsets of DE genes. First, we analyzed globally deregulated genes. Second, we identified significantly changed genes showing spatially restricted expression. Third, we defined genes that were significantly DE between 3xAD and 3xPB mice. Finally, we mined published data on these gene's expression in human AD brain tissue, to determine whether the results in mice translate to humans. One DE gene of interest is the gene encoding Bok. It was found to be specifically downregulated in mouse AD brains. In addition, we provide validation that it is also deregulated in human Alzheimer brains via immunohistochemistry and publicly available single-nucleus RNA-seq data. This is, to our knowledge, one of the largest studies performed using ST, which has integrated both behavioral and molecular characterizations of two brain regions.
Results
LTP and Olfaction in AD and Control Mice
In this study, we focused our molecular analyses on two regions of the mouse brain implicated in Alzheimer pathogenesis: the hippocampus and the OFBs from adult middle-aged male mice. We used male mice for a practical reason, as they perform better in smelling tests at this age. In addition, male mice show less pathology than females and we are interested in early gene expression changes before AD pathology is evident. Extensive ST analyses were performed with close to 50 tissue sections being analyzed with the transcriptome-wide in situ technology (three mice per genotype, and two sections per mouse). We used two inbred AD strains, namely, 3xAD (triple transgenic APP, PS1, and MAPT) and 3xPB (a quadrupole transgenic Polβ+/-, APP, PS1, and MAPT), and two control mouse strains, namely, WT (wild-type C57BL/6J) and PB (transgenic Polβ+/-). The AD models used here develop AD features much later than those of other AD mouse models and have been characterized previously (Hirata-Fukae et al., 2008; Hou et al., 2018; Liu et al., 2018; Misiak et al., 2017a; Sykora et al., 2015). In addition, behavioral changes are readily detected in our middle-aged mice (Hou et al., 2018; Misiak et al., 2017a; Sykora et al., 2015; www.alzforum.org/research-models).
We sought to evaluate mice in the early phase of the disease because we are interested in defining biomarkers of early disease. AD pathology was characterized by measures of inflammation (astrocytes or activated microglia) and patterns of Aβ deposition (intracellular or extracellular). The signal for astrocytes, marked by GFAP, was more abundant in 3xPB (Figure S1A). Astrocytes were mainly located in the hippocampus (Figure S1A), anterior olfactory nucleus, cortex, and cerebellum. In contrast, microglia, IBA-1 staining, were evenly distributed across the brains, but were like controls in morphology (Figure S1B). In both 3xPB mice, there were a few Aβ-positive extracellular deposits found (see arrows), but none were observed in the other mice (Figure S1C). Tau immunoreactivity was not assessed. Our results are consistent with similarly-aged mice described by Hirata-Fukae et al. (Hirata-Fukae et al., 2008), suggesting that the mice represent early aspects of AD pathology.
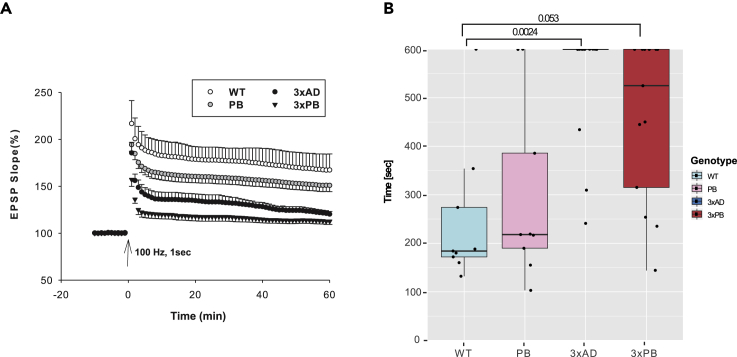
LTP measures synaptic transmission; is important for synaptic plasticity, memory, and learning; and is thought to be affected in AD. LTP specifically measures the ability of CA3 neurons to communicate with CA1 neurons via the Schaffer collateral synapses with modulation provided by the dentate gyrus (DG) (Caruana et al., 2012). Hippocampal LTP was significantly decreased in both AD strain mice (Figure 1A), down by 32.6% in 3xPB and 27.2% in 3xAD, relative to WT, as measured during the last 10 min of recording. PB mice also showed a significant decrease of 18.7% relative to WT mice. The significance of this finding is unknown but suggests that DNA repair is important for maintaining LTP.
Figure 1.
Application of ST to Understand Behavioral Changes in AD Mice
(A) Graphic representation of LTP results measured at the Schaffer collateral synapses. EPSP, excitatory postsynaptic potential. There were no differences in pre-synaptic transmissions. Six slices from a minimum of five mice per group were assessed, and the values represent the mean ± SE.
(B) Graphic representation of results of the buried food test. Mice n = 8–13 per genotype. Box plots hinges show 25th and 75 percentiles and sash mark shows median.
See also Figure S1.
As olfactory deficits are emerging as an early marker in AD neurodegeneration (Murphy, 2019; Rahayel et al., 2012), we also measured olfactory function using a buried food test. The results showed that 3xAD and 3xPB mice performed less well than WT and PB mice, whereas PB and WT mice perform similarly in this assay (Figure 1B). We note that the average body weight of 3xAD and 3xPB mice is higher than the average body weight of PB and WT mice (Figure S1D). Although PB mice have mild deficits in LTP, relative to the AD strains, they performed like WT mice in the buried food pellet test.
These results are consistent with prior analyses of our mice, which typically used older animals (Hou et al., 2018; Misiak et al., 2017a; Sykora et al., 2015). This motivated us to explore the transcriptional landscape using molecular tools to identify genes within the hippocampal and olfactory neuronal layers that might contribute to these behavioral changes seen in our AD strains.
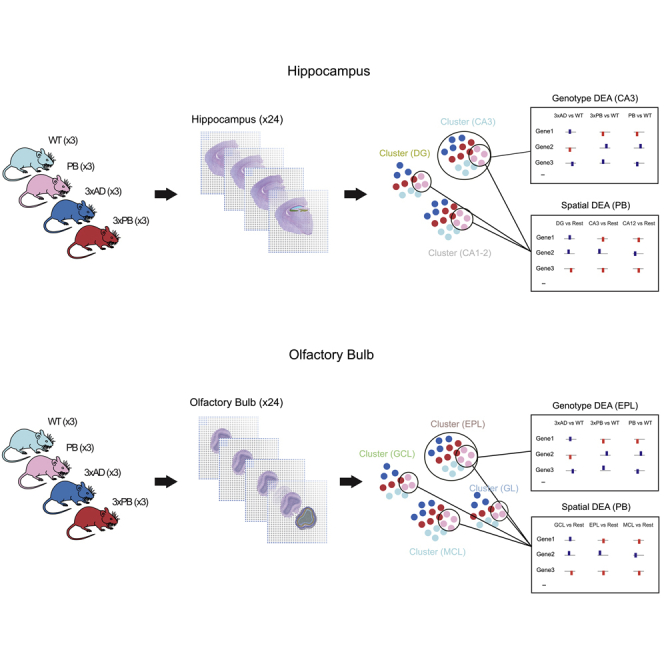
ST Identifies DE Genes and Molecular Clusters Corresponding to Anatomical Layers of the Mouse Hippocampus and OFB
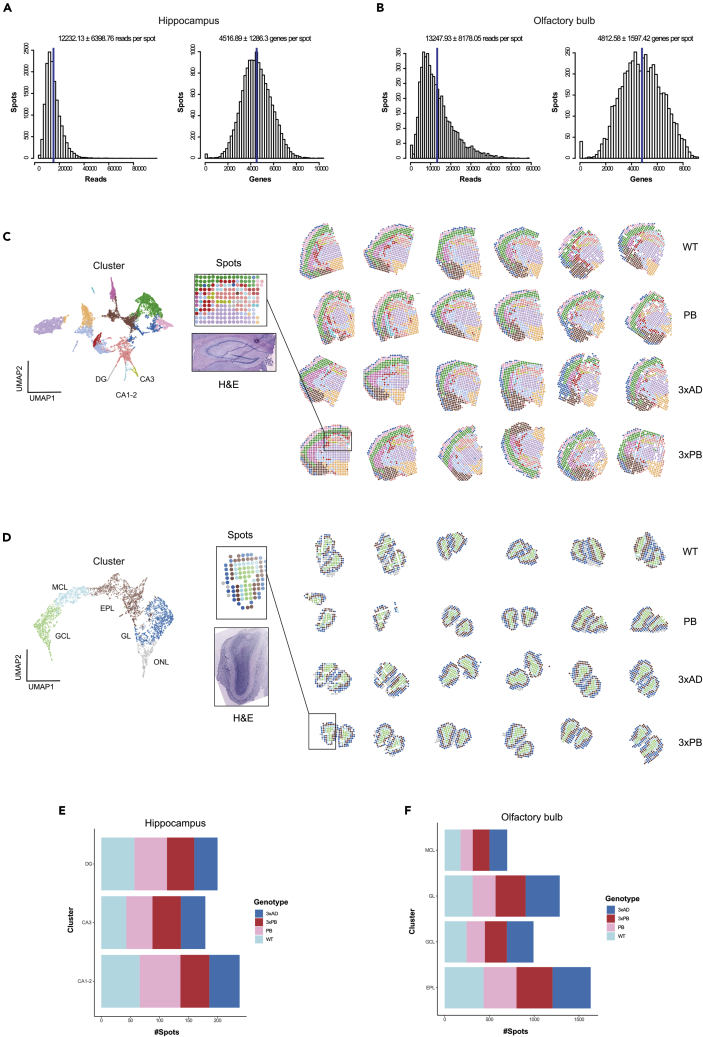
In brief, snap frozen tissue samples from three animals of each genotype, were cryo-sectioned, stained, and imaged on barcoded glass surface. The tissue sections, two sections per mouse, were then cryo-sectioned, stained, and imaged on a barcoded glass surface. The tissue sections were then permeabilized to allow for mRNA to hybridize to the polyT-barcoded probes. The resulting cDNA products were released from the surface, after an on-chip reverse transcription step, and prepared for Illumina sequencing. ST libraries were generated from coronal sections of the hippocampus and OFB, and a total of 48 tissue sections were analyzed. This resulted in a total of 15,062 spots and 22,701 unique genes for the hippocampus dataset with an average of 12,232 reads and 4,516 genes per spot (Figure 2A). Similarly, we obtained 5,059 spots and 20,471 unique genes for the OFB dataset with an average of 13,247 reads and 4,812 genes per spot (Figure 2B). Unsupervised clustering of the spots belonging to the hippocampus dataset was performed using factor analysis, an approach that is designed to account for batch effects and other sources of technical variation (Berglund et al., 2018; Maaskola et al., 2018). The annotation of the expression clusters was performed anatomically using the H&E images and the Allen Brain Atlas. This resulted in 14 well-defined gene expression clusters that correspond to the anatomical layers in the brain hemisphere (Figure 2C). We selected three clusters corresponding to hippocampal subfields CA1-2, CA3, and DG regions, well-known areas of pathological relevance for AD, for further analysis. Similarly, clustering analysis of the OFB dataset resulted in five well-defined clusters that clearly mapped the different layers of the OFB (Figure 2D). We selected the four clusters corresponding to the granular cell layer (GCL), mitral cell layers (MCL), the external plexiform layer (EPL), and glomerular layer (GL) for further analysis. The number of spots corresponding to the various clusters per genotype is shown in Figures 2E and 2F, while the breakdown of reads per spot is shown in Figures S2A and S2B. In addition, UMAP manifolds of the factors displaying spots colored by genotype, animal, and chip show the lack of batch effects and the power of the factor analysis to capture the biological differences (Figures S2C and S2D).
Figure 2.
Statistics and Unsupervised Clustering Analysis
(A) Reads and genes per spot distributions for the hippocampus dataset (median ± SD).
(B) Reads and genes per spot distributions for the OFB dataset (median ± SD).
(C) Factor analysis of the hippocampus dataset. UMAP manifold of the factor activities colored by cluster and genotype on the left and clusters overlaid onto the tissue sections on the right.
(D) Factor analysis of the OFB dataset. UMAP manifold of the factor activities colored by cluster and genotype on the left and clusters overlaid onto the tissue sections on the right.
(E) Graphic representation of the number of spots per cluster per mouse genotype (hippocampus). Three mice per genotype and two slices per mouse were used.
(F) Graphic representation of the number of spots per cluster per mouse genotype (olfactory bulb). Three mice per genotype and two slices per mouse were used.
See also Figure S2.
We next performed multiple differential expression analyses. To identify DE genes, we conducted a genotype-based analysis where every genotype was compared with WT. Via those comparisons, we aimed to detect genes that are DE between the strains and control within each investigated cluster (i.e., proxy for individual anatomic regions). In a separate analysis, we aimed to detect genes that were DE between the clusters for each genotype, i.e., CA1-2 versus CA3. By this analysis, we sought to detect genes that have distinct spatial patterns. This resulted, after applying thresholds of log2 fold-change 0.5 and adjusted p value 0.1, in 964 genes that are DE by genotype in the hippocampus (Figure S3A) and 993 genes that are DE by genotype in the OFB (Figure S3C). Venn diagrams depicting the overlap of the number of DE genes in the various anatomic regions are shown in Figures S3B–S3D. We did not find any genes changing in the opposite direction between the hippocampus and OFBs. We also conducted a direct comparison between 3xAD and 3xPB to identify gene expression changes induced by loss of the DNA repair protein, Polβ; a Venn diagram of that comparison is shown in Figures S3E and S3F. Multi-volcano plots of all the DE genes (Figures S4A and S5A) and genes in our selected lists are shown in Figures S4B and S5B.
Global Gene Expression Alterations in the Hippocampus and Olfactory Bulbs
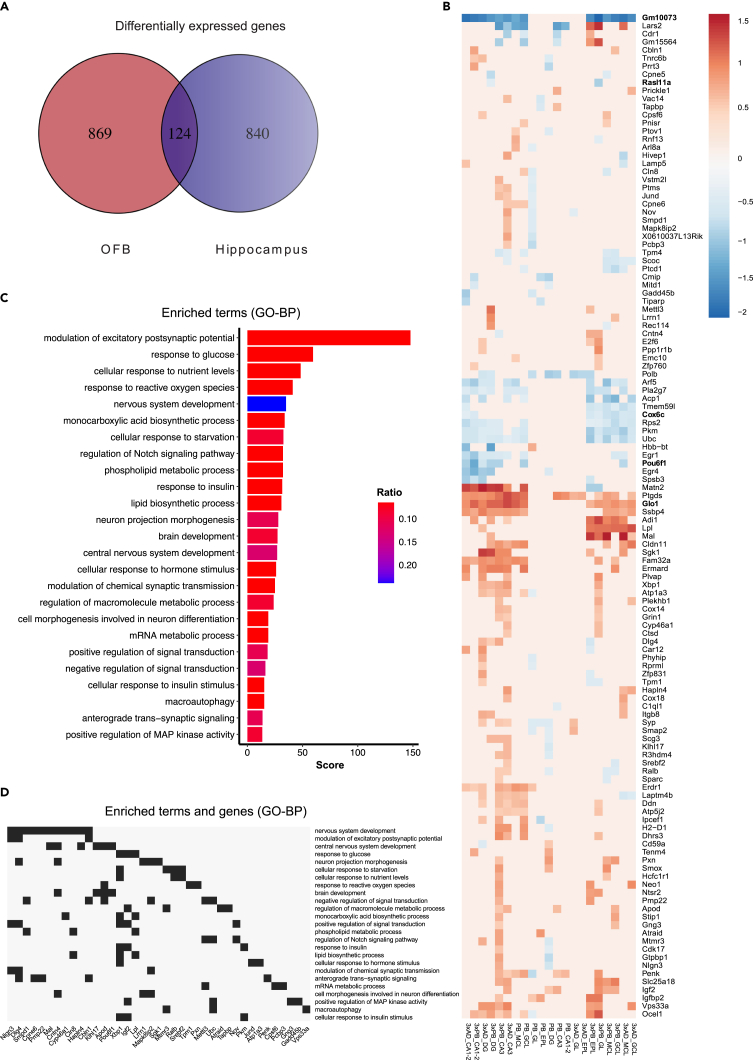
Using ST data from the hippocampal and OFB regions, the results were then examined for gene(s) DE in multiple layers/clusters of each brain region in each strain. These we called the globally deregulated genes and constituted the DE genes from both datasets that were DE in more than a single layer (i.e., clusters). Very few genes were DE across every layer within any given genotype (Figure S3 C-D). These findings exemplify the importance of defining the individual DE gene sets for each layer and analyzing them separately. A total of 124 genes were DE in one or more layers in the hippocampus and OFB (Figures 3A and 3B). The biological process gene ontology (GO) terms they fall into are shown in Figure 3C. The enriched genes within the terms are shown in Figure 3D. About one-third of the terms are related to metabolism, and another third are related to synaptic transmission and nervous system development. Response to reactive oxygen species, macroautophagy, and various kinase signaling terms were found within the remaining terms.
Figure 3.
Identification of Genes Jointly Differentially Expressed in Hippocampi and Olfactory Bulb
(A) Venn diagram showing the numbers of statistically significant differentially expressed genes per region and the number of genes found in at least one region in both datasets.
(B) Heatmap showing the log2 fold-change for the set of shared genes, 124 genes, in (A) hierarchically clustered by anatomic region.
(C) GO biological process terms enriched from genes shown in (B). Score represents statistical confidence, and ratio represents the relative number of genes detected in the term divided by all the input genes.
(D) Gene clustergram showing the overrepresented genes from the GO terms shown in (C). Score represents statistical confidence, and ratio represents the relative number of genes detected in the term divided by all the inputted genes.
See also Figure S3.
Only three genes were differentially regulated across both AD datasets but absent from PB gene lists: Ubiquitin C (Ubc) and Gm10073 were downregulated in both AD strains, whereas Glyoxalase I (Glo1) was upregulated in both strains (Figure 3B). Ubc is a substrate for polyubiquitin reactions, which are integral to stress responses like DNA repair, innate immunity, proteome homeostasis, and the response to cellular stress. In this regard, UBC could promote dissolution of toxic protein aggregates, such as Aβ, a known issue in AD. In a recent large metadata analysis of human AD samples, UBC was identified as a key hub gene and downregulated in multiple brain regions (Patel et al., 2019), a result that is consistent with the present observations (e.g., downregulated in the hippocampus and OFB in AD but not PB mice). Gm10073 is a pseudogene of unknown function that is strongly downregulated in the AD mice. Glo1, the only universally upregulated gene in both hippocampus and OFBs of AD mice, promotes detoxification of methylglyoxal, a toxic by-product of high glucose in cells, via the glyoxalase system (Frandsen and Narayanasamy, 2018). This system converts methylglyoxal to lactate using glutathione as a cofactor, blocking production of advanced glycation end products. Methylglyoxal is neutralized by GLO1 and GLO2 enzymes. Thus, these proteins help prevent proteins, lipids, and nucleic acids from being derivatized by methylglyoxal. Using mass spectrometry on the OFB tissue, several GLO1-interacting partners were found to be downregulated at the protein level: GRHPR, glyoxylate and hydroxypyruvate reductase; NDUFB10, NADH dehydrogenase and oxidoreductase; TALDO1, transaldolase of the pentose phosphate pathway that provides ribose-5-phosphate for nucleic acid synthesis and NADPH for lipid biosynthesis; and TPI1, which catalyzes the transfer of a hydrogen atom from carbon 1 to carbon 2 in an intramolecular oxidation-reduction reaction (see Table S1 and Venn diagrams in Figure S3G for overlap between proteomics and transcriptomics).
To validate the increased expression of Glo1 in the mice, we ran western blots of GLO1 and GLO2 to evaluate the whole pathway. Western blot analysis confirmed that the abundance of GLO1 is higher in AD mice than control mice; however, the abundance of GLO2 protein was lower, consistent with the notion that the glyoxalase system is severely deregulated in our AD strains (Figure S6). Notably, in humans, GLO1 protein was found to be upregulated in the early stages of human AD but downregulated in middle and late stages (Kuhla et al., 2007).
Identification of Spatially DE Genes in Hippocampus
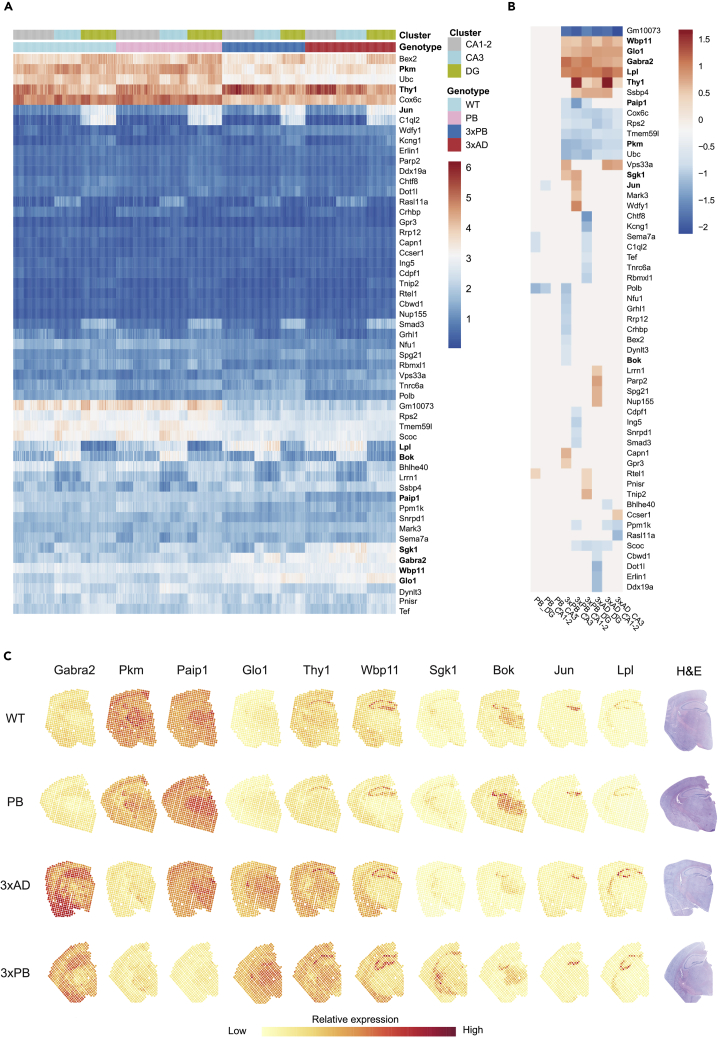
The hippocampus plays a critical role in memory and learning. All the highly confident DE genes were subjected to biological GO term analysis and the top 25 enriched terms are shown in Figure S7A, and a gene clustergram (Figure S7B) shows the overrepresented genes driving these term changes. There are multiple terms related to apoptosis and cell cycle, transcription, DNA metabolism (including DNA repair), and transforming growth factor β signaling. Protein degradation, deubiquitylation, and autophagy terms were also present. From the DE genes detected in the hippocampus dataset (964 genes, Figure S3A), we identified a list of 55 genes that showed significant differences in expression between genotypes and showed clear spatial patterns, a summary of these genes is provided in Table S2. They were subjected to hierarchically clustered (by rows) heatmaps of normalized expression (Figure 4A), log2-fold-change (Figure 4B), and individual gene plot (normalized gene expression plotted onto the tissue sections) analyses (Figure 4C).
Figure 4.
ST Analysis on Hippocampi Regions Reveals Differences among Stress Response Signaling Genes Globally and Regionally
(A) Heatmap of the spatially differentially expressed genes found in the hippocampi clustered by rows. Graphic colorized by normalized expression and separated by color-coded genotype and cluster region.
(B) Heatmap of log2-fold-change values of genes shown in (A) clustered by rows and columns.
(C) Graphic representation of normalized gene expression of selected genes overlaid onto brain sections. The genes represent the types of data seen. Shown are Gabra2, Pkm, Paip1, Glo1, Thy1, Wbp1, Sgk1, Bok, Jun, and Lpl. H&E-stained brain slices are shown on the right.
See also Figures S4 and S6–S9.
Several genes were DE across all hippocampal layers. Protein kinase muscle (Pkm), cytochrome c oxidase subunit 6c (Cox6c), ribosomal protein S2 (Rps2), and transmembrane 59 like (Tmem59l) were downregulated in the AD strains (Figures 4A and 4B). Two of these genes, Pkm and Cox6c, are important in glycolysis and respiration, respectively. Tmem59l plays a role in autophagy, which is a general stress response to starvation, growth factor deprivation, and endoplasmic reticulum (ER) stress. Downregulation of Tmem59l has been shown to prevent caspase-dependent neuronal cell death (Zheng et al., 2017). Based on its sequence similarity to Tmem59, it may directly regulate APP processing and Aβ localization (Ullrich et al., 2010). The upregulated genes were gamma-aminobutyric acid (GABA) receptor subunit alpha-2 (Gabra2), Lipoprotein lipase (Lpl) and WW domain-binding protein 11 (Wbp11). Gabra2 encodes a subunit of the chloride channel and receptor for the major neuroinhibitory transmitter GABA. Lpl protein is a triglyceride hydrolase that facilitates lipid uptake, including Aβ uptake, to promote its degradation (Nishitsuji et al., 2011). Importantly, it is also described as a microglia disease-associated marker (Keren-Shaul et al., 2017). Wbp11 encodes a splicing factor (Llorian et al., 2004), and its expression was significantly upregulated throughout the brain hemisphere. Representative gene expression plots for some of the genes can be seen in Figure 4C, and representative full gene plots for several of these genes are shown in Figure S8. As can be seen some genes show greater variability between mice than others, notably Gabra2 and Cox6c.
To identify genes that display spatially restricted expression, we evaluated the heatmaps (Figures 4A and 4B) and the gene plots, which shows a gene's normalized expression values overlaid onto the tissue sections (Figure 4C). Several genes stood out by this analysis. Although Lpl was upregulated across all AD-associated clusters of the hippocampus (Figures 4A and 4B), it was visibly upregulated in the CA1-2 and CA3 sublayers of the hippocampus (Figures 4C and S8). We have previously demonstrated Lpl protein upregulation by western blotting and suggested that it may contribute to altered fat metabolism in our mice (Demarest et al., 2020). Thy1 was also upregulated in the AD strains' CA1 regions and throughout the brain hemisphere. Here, it should be noted that the Thy1 promoter drives expression of the AD transgenes. Two genes, Bcl-2-related ovarian killer (Bok) and Ras-like family 11 member A (Rasl11a), displayed decreased expression in the CA3 and thalamus regions (via inspection of images) (Figures 4C and S8). BOK is a multifunctional BCL2 family member protein that participates in the intrinsic mitochondrial pathway of apoptosis, and there are reports of BOK as a pro-apoptotic (Llambi et al., 2016) agent or not (D'Orsi et al., 2016). Interestingly, BOK may also be important for calcium homeostasis, mitochondrial dynamics, and bioenergetics (D'Orsi et al., 2016; Schulman et al., 2019). Rasl11a protein has been reported to enhance rDNA transcription via RNA polymerase I, and its downregulation here may impinge upon the assembly of rRNA components and translation specifically in the CA3 and thalamus regions (Figure S8). RNAscope was used to validate the localized expression changes of Gabra2, Bok, and Lpl (Figure S9).
The AP-1 transcription factor subunit and immediate-early gene, Jun, is an illustrative example of the power of spatially resolved gene expression analysis (Figures 4 and S8). Jun is a DE gene; however, Jun was visibly expressed in the DG of the AD strains (Figures 4C and S8) but was only DE in the CA1 of 3xPB mice (Figure 4B). Thus, we chose not to focus further attention on genes that display this type of expression pattern.
Genes Important in Translation and Stress Response Signaling Are Revealed in the Comparison of 3xAD and 3xPB Mice
At the outset of this project, we sought to gain insights into the differences between 3xAD and 3xPB mice because disease progression is faster in the DNA repair-deficient 3xPB mice (Hou et al., 2018; Misiak et al., 2017a; Sykora et al., 2015). A DE analysis was performed to directly compare 3xAD and 3xPB mice using all layers; Venn diagrams of the genes that overlap are shown in Figures S3E and S3F. Fewer genes were DE between these two strains than between either of them compared with WT mice. This is to be expected, as they are highly inbred. There were 144 highly confident DE genes in the hippocampus and 198 genes in the OFBs 3xPB (Figures S3E and S3F). The hippocampal genes that stood out by this analysis included Paip1, Eif3f, Sgk1, Rpa2, Gadd45a, Cdkn1a, and of course Polβ. In the hippocampus, the Poly(A) binding protein interacting protein 1, Paip1, was globally downregulated in 3xPB mice, but not in 3xAD mice. The protein encoded by this gene participates in translation initiation, and elements of LTP require nascent translation (Johnstone and Raymond, 2013). The gene Eif3f is also involved in translation regulation (Shi et al., 2006). The serum glucocorticoid-regulated kinase 1, Sgk1, was uniquely upregulated in 3xPB mice, visibly in the fornix, which is the major output track from the hippocampus. Sgk1 is a major stress response kinase and one of a few genes deregulated in both the hippocampus and OFBs in the 3xPB mice (Figure 3B). It participates in several stress-associated signaling cascades including NF-κβ and GSK3β-PPARα signaling (Lang et al., 2018). RPA2, GADD45A, CDKN1a (p21) and POLβ proteins are all important in the response to DNA damage, and this finding is consistent with our previous report that our 3xPB mice accumulate more DNA damage (Sykora et al., 2015).
Identification of Spatially DE Genes in Olfactory Bulbs
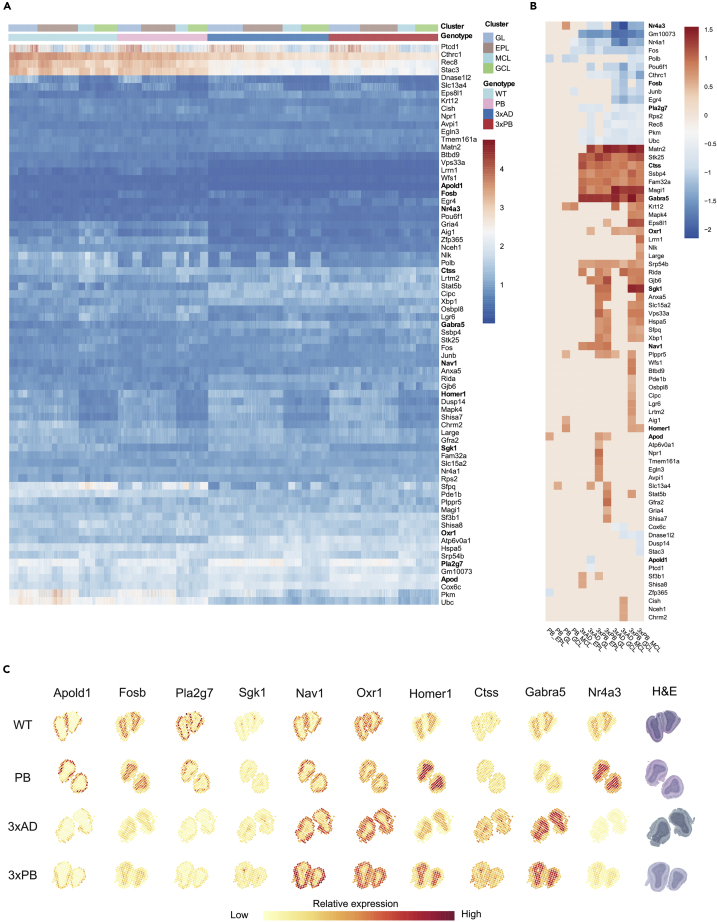
Olfactory function is emerging as an important early marker for neurodegeneration and AD (Murphy, 2019; Rahayel et al., 2012). AD mice suffer from a variety of progressive olfactory deficits; however, we do not have good biomarkers for AD olfactory dysfunction. All the OFB DE genes were subjected to biological GO analysis, and the top 25 enriched terms are shown in Figure S9A with the genes overrepresented in these terms shown in Figure S9B. Notably, there were multiple terms for transcription, ion transport including chemical synaptic transmission, TGFβ signaling, and programmed cell death. Using the same analysis strategy as discussed above for the hippocampus, we identified 73 DE genes that displayed spatial expression in the OFBs (Figures 5A and 5B). A summary of the genes discussed below is shown in Table S3.
Figure 5.
ST Analysis on Olfactory Bulbs Reveals Differences among Synaptic and Stress Response Signaling Genes both Globally and Regionally
(A) Heatmap of the differentially expressed genes found in the OFBs clustered by rows. Graphic colorized by normalized expression and separated by color-coded genotype and cluster region.
(B) Heatmap of log2-fold-change values of genes shown in (A) clustered by rows and columns.
(C) Graphic representation of normalized gene expression of selected genes overlaid onto olfactory bulb tissue sections. Genes changed by region include Apold1, Fosb, Pla2g7, Sgk1, Nav1, Oxr1, Homer1, Ctss, Gabra5, and Nr4a3. H&E-stained brain slices are shown on the right.
See also Figures S5, S10, and S11.
Most of the globally downregulated genes were shared with the hippocampus, except for phospholipase A2 group 7, Pla2g7. This protein is involved in the breakdown of oxidized phospholipids and is known to suppress mitochondrial apoptosis (Chen et al., 2007). In WT and PB mice, Pla2g7 showed marked expression in the outer layers of the OFBs, and this was lost in the AD strains (Figure 5C). Validation of some of the globally deregulated OFB genes was confirmed by mass spectrometric analysis (Table S1).
One POU family transcription factor, Pou6f1, was found to be significantly downregulated throughout the OFBs of the AD strains. OFB interneurons secrete the neuropeptide corticotropin-releasing hormone (CRH), which promotes synaptic plasticity in the OFB. The receptor for CRH, CRHR1, activates CREB-dependent transcription. Pou6f1 has a CREB-binding site in its promoter and loss of POU6f\F1 protein in CRHR1+ neurons causes reduced synaptic connectivity and dendritic complexity, whereas overexpression of POU6F1 induces dendrite outgrowth, branching, and synapse plasticity (McClard et al., 2018). Our results suggest that diminished expression of Pou6f1 in both the OFBs and hippocampus may substantially alter synaptic functionality and contribute to loss of the sense of smelling and LTP, in the AD strains. Interestingly, Blalock et al. demonstrated that Pou6f1 was downregulated in patients with incipient AD via microarray of hippocampal tissue (Blalock et al., 2004).
There are clear indications of stress across the OFBs. We detect upregulation of Glo1, discussed earlier, oxidant-stress response kinase (Stk25), cathepsin S (Ctss), and Fam32a globally (Figure 5B). STK25 is a kinase and Golgi protein that regulates Golgi morphology and cell death, in part by its translocation from the Golgi to the nucleus in response to stress. It plays roles in lipid metabolism, glycolysis, and glucose and insulin homeostasis. CTSS is a cysteine protease thought to contribute to autophagy, the degradation of proteins and organelles for self-protection. Fam32a and Srp54b are globally upregulated splicing factors. A recent proteomic study using brains of patients with AD patient deduced that many AD proteins are subjected to RNA splicing and that the alternatively spliced proteins correlate with AD pathology and cognition (Johnson et al., 2018). Another gene globally deregulated in the OFBs was Gabra5 (Figures 5B, 5C, and S10). It is a subunit of the GABA A receptor and was significantly upregulated across the AD OFBs but with greater expression in the GCL layer. Representative examples of the OFB globally deregulated gene expression patterns we saw are exemplified by Cox6C and Fam32a (Figure S10).
Proteomic analysis on the OFBs revealed 859 DE proteins, of which 20 were changed in opposite directions between the AD strains. None of the genes encoding these proteins were represented in our ST dataset. All DE proteins were subjected to GO-BP analysis; see Table S4 for the significant terms. Among the top 25 terms, RNA splicing (six terms) was overrepresented. The top term, plus two additional from the list of top 25 terms, was associated with mitochondrial electron transport. The single neuronal item, among these top terms, was associated with neuron projection development. See Table S4 for the full list of significant GO-BP terms. There were 92 proteins/genes that were shared between the proteomics and transcriptomics analyses. Up- and downregulated proteins were similar between the AD strains. Of the genes we discuss here, the direction of change for PKM, COX6C, and MAGI1 were consistent between the two types of analysis, whereas Nav1 and HSPA5 were not. The joint list of proteins/genes (Table S1) was subjected to GO-BP analysis. Among the top 25 terms, terms relating to cytoskeleton and actin organization (six terms), glycolysis (three terms), and antigen processing (three terms) were overrepresented. Four terms were related to neuronal function including generation and differentiation of neurons, synaptic transmission, and long-term synaptic potentiation. See Table S5 for the full list of GO-BP terms.
Notably, by mass spectrometric analysis, APOE was highly overexpressed, particularly in the OFB of 3xPB mice (Table S1). APOE is a lipoprotein, and one of its alleles, ApoE4, is found in 65% to 80% of patients with late-onset AD, reviewed in Misiak et al. (2017b). ApoE, by ST analysis, showed minor upregulation in the GL and EPL of AD OFBs. Apod, however, was upregulated in a few layers (Figure 5B). THY1 and MTOR (both downregulated) and MTOR-interacting proteins, AKT1S1 and LAMTOR2 (both upregulated), were deregulated in the mass spectrometric analysis (Table S1).
Olfaction is unaffected in PB mice. Therefore, we postulated that we might gain insight into the mechanism of olfactory dysfunction in AD, by identifying genes that are DE in the OFB of 3xAD and 3xPB mice, but not in PB mice. The AD strains had increased expression of oxidation resistance 1 (Oxr1), natriuetic peptide receptor 1 (Npr1), and neuron navigator 1 (Nav1). In contrast, expression of Apolipoprotein L domain containing 1 (Apold1) was lower in the outer layers of the OFBs (Figures 5B and 5C). Mice lacking Oxr1 display neurodegeneration, and point mutations in this protein contribute to epilepsy in humans (Oliver et al., 2011). In yeast and human cells, OXR1 is localized to mitochondria, where it protects them from oxidative stress. NAV1 associates with the growth cones and branch points of microtubes and is thought to be involved in axon guidance and synaptic maturation (Martinez-Lopez et al., 2005). Nav1 is upregulated in 3xPB mice EPL and GL. Overexpression of mouse NAV1 in cultured neurons leads to rearrangement of microtubules into bundles. NPR1 is a membrane bound guanylate cyclase that binds natriuretic peptides, which promote vasodilation (Pandey, 2018). Apold1 was downregulated in the GL, and this gene plays roles in endothelial cell signaling, vascular injury, and cellular permeability (Regard et al., 2004). Several of the genes discussed are shown in Figures 5A and 5B, their relative spatial expression patterns are shown in Figure 5C, and their full gene plot is shown in Figure S10.
Several immediate-early genes were downregulated in the AD strains: Fosb, Fos, Junb, Egr4, Nr4a1, and Nr4a3. The intermediate-early genes regulate transcription, synaptic plasticity, and intracellular signaling. Notably, Fosb and Nr4a3 showed clear expression in the MCL and GCL of WT and PB mice OFBs, whereas they were significantly downregulated in the AD strains' samples (Figures 5C and S10). Fosb is an alternatively spliced gene used as a marker for neuronal activity and is important for stress responses in the brain (Hiroi et al., 1997). NR4A3 functions in many pathways including neuronal differentiation, apoptosis, metabolism, and cell cycle progression (Paillasse and de Medina, 2015). In beta cells, knockdown of this gene reduced mitochondrial respiration in permeabilized cells (Reynolds et al., 2016). Some of these genes, Fos, Nr4a1, and Junb, were also downregulated in PB mice, so it is unclear if these genes contribute to the olfactory deficits in the AD strains because PB mice do not display smelling defects.
In WT mice, Polβ was found to be expressed throughout the OFBs (Figure S10) and Homer1 showed significant upregulation in the MCL and GCL in a Polβ-dependent manner. HOMER1 is a postsynaptic density scaffolding protein that regulates metabotropic glutamate receptor function and calcium release (Jardin et al., 2013). Homer1 displayed a clear spatial upregulation in the GCL and MCL (Figure 5C). Currently, it is unclear how or why loss of Polβ promotes increased expression of Homer1 or its functional consequences. We should note here that Paip1 and Homer1 genes are both located on mouse chromosome 13, whereas the Polβ gene is located on mouse chromosome 8. So, their change in expression is not due to a direct interaction with the inactivation of the Polβ gene. Obviously, we cannot rule out some off-target integration events or higher-order chromatin interactions.
As done earlier in the article, biological process GO terms linked to genes DE in the OFB of 3xAD and 3xPB mice were tabulated and analyzed. The most highly enriched GO terms were stress response signaling and lipid metabolism. Sgk1 and Xbp1 are among the few genes deregulated in both the OFBs and hippocampus (Figure 3B). Sgk1 was upregulated globally in 3xPB mice, but with a distinct pattern in the center of the OFB GCL (Figures 5C and S10). Xbp1 was globally upregulated across the OFBs of 3xPB mice (Figure S10). It is a multifunctional transcription factor that functions in the immune system, lipid metabolism, and ER stress response (Cisse et al., 2017).
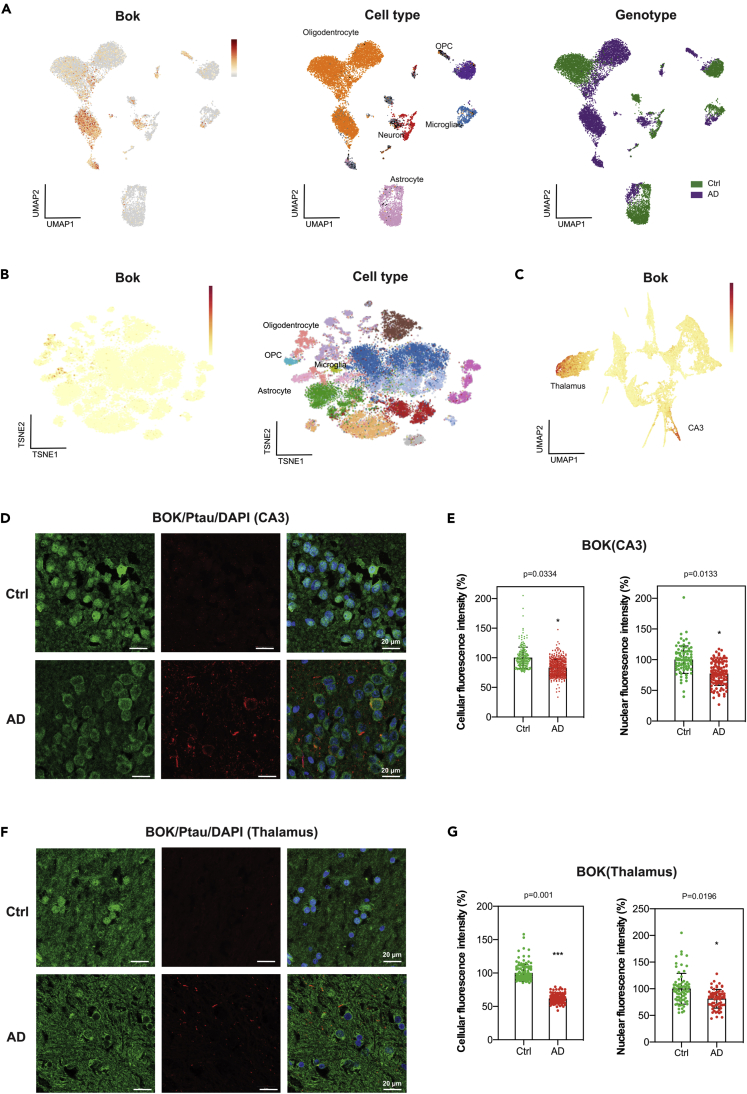
BOK, a Spatially Expressed Gene in Hippocampal CA3 Regions, Shows Loss of Expression in Mouse and Human AD Brains
Dysfunctional mitochondria contribute to AD pathology (Swerdlow, 2018). BOK is a mitochondrial protein that participates in mitochondrial apoptosis (D'Orsi et al., 2017; Einsele-Scholz et al., 2016) and mitochondrial dynamics (Schulman et al., 2019). Based on our ST data presented earlier, Bok is also downregulated in the hippocampus CA3 region and thalamus in 3xAD and 3xPB mice (Figures 4, 4C, and S8). This suggests the possibility that BOK could play a role in AD pathology. To determine if any of our DE genes, which also showed spatially restricted expression, were DE in human AD datasets, we obtained a human single-nuclei RNA-seq dataset composed of AD and healthy entorhinal cortex samples and probed for our genes (Grubman et al., 2019). UMAP plots of human AD BOK expression from this dataset are shown in Figure 6A. A subset of AD oligodendrocytes and cells of mixed origin appeared to upregulate BOK. BOK expression was also found altered in oligodendrocytes (upregulated) and neurons (downregulated) from parietal lobe tissue in another human single-nuclei RNA-seq AD dataset (Del-Aguila et al., 2019) (Figure 6B). In our datasets, we cannot distinguish the cell types of origin, but we find decreased expression of Bok in the CA3 region of the hippocampus and thalamus in 3xAD and 3xPB mice (Figures 4, S8, and 6C). Presumably, genes that display distinct layer-specific expression serve a specialized function in that tissue layer and thus, their DE in those layers, may impact cell biology to a greater extent. To evaluate BOK in human AD pathology, we performed BOK immunofluorescence labeling in hippocampal and thalamic brain sections from postmortem AD patients and non-demented controls. In both regions, there was a clear diminution of total cellular fluorescence signal for BOK, and this was largely driven by loss of nuclear BOK signal (Figures 6D–6G). These results are consistent with our observation of decreased hippocampal and thalamic BOK in our AD mouse models. An analysis of the biological implications of BOK loss is beyond the scope of this article.
Figure 6.
BOK Is Differentially Expressed in Human and Mouse AD Brains
(A) Single-nuclei RNA-seq generated from the entorhinal cortex of human AD and control postmortem brains (data from Grubman et al., 2019). UMAP manifold of single nuclei colored by BOK's normalized expression (red means higher expression level) together with genotype and cell type maps.
(B) Single-nuclei RNA-seq generated from the parietal lobe of human AD. t-SNE manifold of single nuclei colored by BOK (normalized expression) together with cell types. Data from Del-Aguila et al. (2019).
(C) UMAP manifold of factor activities colored by Bok's normalized expression. The manifold is the same as in Figure 2C. The regions corresponding to the thalamus and CA3 are labeled. Localization of Bok denoted with red dots.
(D) Alteration of BOK protein level and localization in AD hippocampi. Representative images of coronal CA3 sections from human control (Ctrl) and Alzheimer (AD, Braak VI) hippocampi. Scale bars, 20 μm.
(E) Quantification of images of BOK staining in hippocampi. The cellular and nuclear BOK fluorescence intensities were quantified within CA2/CA3 cells from Ctrl and AD hippocampi (cellular Ctrl, n = 237; AD, n = 493) (nuclear Ctrl, n = 86; AD, n = 115). Data are presented as mean ± SEM. Statistical significance was determined using per brain mean value; three brains per category were analyzed.
(F) Alteration of BOK protein levels and localization in AD thalami. Representative images of coronal sections from human control (Ctrl) and Alzheimer (AD, Braak VI) thalami. Scale bars, 20 μm.
(G) Quantification of images of BOK staining in thalami. The cellular and nuclear BOK fluorescence intensities were quantified within cells from Ctrl and AD thalami (cellular Ctrl, n = 127; AD, n = 133) (nuclear Ctrl, n = 74; AD, n = 67). Data are presented as mean ± SEM. Statistical significance was determined using per brain mean value; three brains per category were quantified.
Discussion
ST is an emerging technology that pairs quantitative transcriptomics with high-resolution tissue imaging and unbiased bioinformatic analyses, anchoring the expression data to the physical map of the organ or tissue(s) of interest. This type of analysis facilitates molecular characterization of tissue sections and cellular layers in an unbiased fashion. Here, we applied ST to chart gene expression in the hippocampus and OFBs of 3xAD, 3xPB, PB, and WT mice. We identify differential expression of novel and known genes in AD mouse brain and confirm spatially restricted differential expression of BOK in the brains of human patients with AD.
Genes that were globally deregulated may have profound and systemic implications, i.e., Ubc, Pkm, Cox6c, and Glo1, because all these genes can impact the metabolic state of the cell. As UBC protein is a substrate for polyubiquitin reactions, decreasing it may contribute to impaired proteasomal degradation and DNA repair while promoting inflammation and the accumulation of misfolded Aβ or tau. As mentioned earlier in the article, it has already been reported that UBC is a key hub gene and downregulated in multiple human AD brain regions (Patel et al., 2019). Downregulation of Pkm, which plays a role in glycolysis, and Cox6c, which functions in Complex IV of the electron transport chain, both have the potential to directly contribute to loss of ATP generation and to the glucose hypometabolism seen in patients with AD (Mosconi et al., 2008). There is a high demand for energy during synaptic transmissions (Liotta et al., 2012), thus any deviation from optimum energy production may impact neuronal function.
Glo1 is a part of the methylglyoxyal detoxification pathway, and increased expression here likely reflects the stress response to AD pathology. Interestingly, methylglyoxal, and the advanced glycation end products it produces, are elevated in patients with AD and may contribute to accumulation of insoluble Aβ- and APOE-dependent pathologies (Angeloni et al., 2014). With respect to LTP or smelling, derivatization of proteins by methylglyoxal could interfere with the dynamics of protein complex assemblies and thereby interfere with these functions. Furthermore, glutathione is used as co-substrate for methylglyoxal detoxification and deregulation of this pathway likely contributes to the depletion of glutathione, an important cellular antioxidant.
ST analysis also revealed that Gabra2 is upregulated across the hippocampus, whereas Gabra5 is upregulated in the OFBs in the AD strains. GABAergic synapses are critically important for the excitatory and inhibitory balance in the brain. There are 16 GABRA subunits, which form pentameric channels. Overexpression of Gabra2 modulates postsynaptic currents by its increased GABA affinity, which causes slower current deactivation (Dixon et al., 2014). Gabra5 is the main GABA A receptor responsible for tonic inhibition in the hippocampus (Glykys and Mody, 2006). It has been reported to be significantly downregulated in the CA1 area of human AD brain samples (age range of patient with AD: 75–85) (Rissman et al., 2007). GABA signaling also modulates neuronal homeostasis, adult neurogenesis, and neuroblast migration. Both proteins are known to regulate dendrite outgrowth and spine maturation. Overexpression of either subunit in their respective regions may modulate LTP in the hippocampus and synaptic transmission in the OFBs. Deregulation of these genes may contribute to AD pathology or be a compensatory response to AD pathology; further research is needed to clarify.
Our focus on genes that displayed spatially restricted expression patterns was an attempt to identify novel genes that may be linked to deficits in the specialized cellular layers of the hippocampus and OFBs in the AD strains. Among the spatially resolved genes that we identified, the pathways that these genes are associated with are highly cited as important in AD including lipid metabolism (Lpl, Pla2g7, Apold1), stress response genes (Glo1, Sgk1, Bok, Oxr1, Stk25, Ctss), and immediate-early genes (Fosb, Nr4a3). It is worth noting that deregulation of lipid metabolism genes in the outer layers of the OFB, GL, and EPL may alter odor information processing that is projected from the olfactory receptor neurons in the nasal cavity to the OFBs. ApoE protein is also expressed in these regions, and there is a clear association between ApoE4 and olfactory dysfunction in AD (Misiak et al., 2017b).
As mentioned earlier, the ST data presented showed spatially restricted DE of Bok in the hippocampus of AD mice. We propose that expression of Bok is relevant to AD, because of the strong association between mitochondrial dysfunction and AD and the established mitochondrial functions of BOK. Furthermore, our analyses confirm differential expression of BOK in brains of human patients with AD (Del-Aguila et al., 2019; Grubman et al., 2019). Our earlier analyses using microarray technology also suggested differential expression of Bok in AD mice (Hou et al., 2018), but without the added spatial context provided by the ST analysis it was not pursued. BOK protein has a variety of functions. It has been reported to regulate mitochondrial morphology (Schulman et al., 2019) and nucleotide metabolism (Srivastava et al., 2019) and suppress poly ADP-ribose polymerase-dependent cell death (D'Orsi et al., 2016). It can also function as a primary regulator of mitochondrial outer membrane permeabilization, an essential event in mitochondrial apoptosis (Llambi et al., 2016). BOK can trigger mitochondrial outer membrane permeabilization in the absence of other BCL-2 members, after inhibition of the proteasome (Zheng et al., 2018). Importantly, each of these factors may contribute to neuronal degeneration (Desler et al., 2017; Fang et al., 2019; Kam et al., 2018). In addition, hippocampal pyramidal neurons in the CA1 and CA3 display differential vulnerability to neuronal injury (Mattson et al., 1989), and Bok's preferential expression in CA3 may contribute to this phenomenon. Neuronal cell death in either the hippocampus or OFB could impede performance in function. In mouse and human AD brains, BOK showed decreased expression in the hippocampus and thalamus (Figures 4 and 6). Notably, in human AD, we saw that BOK protein was mainly decreased in the cell nuclei, and high levels of nuclear BOK have been associated with induced apoptosis (Bartholomeusz et al., 2006). Thus, we speculate that decreased nuclear BOK may be a compensatory response to prevent cell death in response to AD pathology. Importantly, the role of mitochondria in AD pathology is still emerging, and BOK may contribute to the mitochondrial AD stress response. Additional analyses will be necessary to determine more precisely BOK's role in responding to AD-associated pathology.
Our working hypothesis is that loss of DNA repair changes the rate of disease progression, and consistent with this, we typically find that 3xPB mice display more progressive AD features than 3xAD mice (Hou et al., 2018; Misiak et al., 2017a; Sykora et al., 2015). Thus, here we sought to compare the DE genes between 3xAD and 3xPB to identify candidate genes that might contribute to this phenomenon. In the hippocampus, we find genes important in translation, Paip1 and Eif3f, and stress and DNA damage response genes, Sgk1, Rpa2, Gadd45a, and Cdkn1a. LTP is one of the main mechanisms that facilitates acquisition, storage, and retrieval of information from neuronal circuits (Dringenberg, 2020). It is known that late LTP requires protein translation, and therefore loss of expression of translation genes in 3xPB may contribute to their diminished memory retention (Sykora et al., 2015). In the OFBs, Sgk1, Xbp1, Homer1, Mtor, and Apoe may all contribute to the differences between 3xPB and 3xAD. The alteration in the stress and DNA damage responsive genes are likely compensatory responses to loss of Polβ and expression of the AD transgenes. The energetic demands of responding to DNA damage may decrease the availability of ATP thereby slowing or delaying signal transduction cascades, increasing the presence of senescent cells, and contributing to the death of neurons. All these factors likely contribute to the behavioral changes between 3xPB and 3xAD mice. Moving forward, it will be important for us to evaluate the temporal deregulation of these genes and determine how their expression correlates with AD pathology and behavioral changes.
AD is a complex, multifactorial, progressive disorder. Here, ST was used to identify novel and known genes that may contribute to disease pathology or that contribute to the response to AD pathology. The DE genes may represent a pool of potential biomarkers for early AD. What is known about mitochondrial involvement in AD pathology is an expanding area of research, and this work identifies many genes that impinge upon mitochondrial functions. Although many of the genes we identified here were already known to be dysregulated in AD pathology, we identify BOK as a novel downregulated gene in both mice and human hippocampus and thalamus AD samples.
Limitations of the Study
Although ST is a powerful technology, we note the following limitations of the current study: (1) we are not able to ascribe any gene changes to a particular cell type in part because any given spot can contain multiple cells, (2) the tissue slices we analyzed did not encompass any anatomic region entirely, and (3) we have not identified the biological significance of any of the gene changes observed. In addition, as the OFB and hippocampus analyses were conducted on two separate cohorts of mice, of slightly different age, we cannot exclude that some of the gene changes are due to aging.
During the preparation of this manuscript, other techniques like Slide-seq (Rodriques et al., 2019) and 10X Visium and image-based techniques like SeqFISH, MERFISH, and STARmap have been developed that may yield similar results (Burgess, 2019). In addition, high-definition ST (Vickovic et al., 2019) is another technique, but with lower throughput.
Resource Availability
Lead Contact
Vilhelm A. Bohr (vbohr@nih.gov)
Materials Availability
No new materials were generated in this article.
Data and Code Availability
Processed and analyzed data and analysis scripts generated during this study are available at Mendeley https://doi.org/10.17632/6s959w2zyr.1 and GitHub https://github.com/jfnavarro/AD_POLB_ST. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (PubMed ID: 30395289) partner repositories with the dataset identifier PXD017766.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We would like to thank Jane Tian, and Drs. Magda Misiak, Yujun Hou, and Tyler Demarest for their help with tissue collection; Eduardo Zavala for help with western blotting; Dr. Yang Heng and Prof. Erik Boddeke for assistance with immunohistochemistry; and Reza Mirzazadeh for help. We thank Drs. Yujun Hou and Kala Puligilla for critically reading the manuscript and their comments. We thank Thomas Comptdaer for excellent immunofluorescence technical assistance. We are grateful to the IMPRT (Institut de Médecine Prédictive et de Recherche Thérapeutique, Lille, France) for access to the confocal microscopy platform and to M. Tardivel and A. Bongiovanni for their assistance with confocal microscopy analyses. We thank Vincent Deramecourt and Romain Perbet for providing human brain tissues from the Lille Neurobank (CHRU-Lille, France). We express gratitude to the patients with Alzheimer disease and their families who allowed us to perform brain autopsies. We thank Dr. Miriam Sander for editorial assistance.
This work was, in part, supported by the National Institute on Aging, NIH (V.A.B., D.L.C., B.O., B.Y., and Y.W). This work was supported by an NIA AD Concept grant from the National Institute on Aging (V.A.B., AG000578). This work was supported by a grant from Olav Thon Stiftelsen to L.J.R., V.A.B., J.L., and T.T. L.J.R. and C.D. were supported by Nordea-fonden. T.R., M.S., and T.T. were supported by the Norwegian Research Council (NORBRAIN project 197467). This work was also supported by Labex (Excellence Laboratory) – Development of Innovative Strategies for a Transdisciplinary Approach to Alzheimer's disease [DISTALZ]. V.A.B. and M.-C.G. are also supported by INSTALZ, an EU Joint Program - Neurodegenerative Disease Research (JPND) project. The INSTALZ project is supported through the following funding organizations under the aegis of JPND - “http://www.jpnd.eu” www.jpnd.eu (Belgium, Research Foundation Flanders; Denmark, Innovation Fund Denmark; France, Agence Nationale de la Recherche; Sweden, Swedish Research Council; and United Kingdom, Medical Research Council). The project has received funding from the European Union’s Horizon 2020 Research and Innovation Program under grant agreement No 643417.
Author Contributions
Conceptualization and designed of the study done by J.L., D.L.C., V.A.B., T.T., J.F.N., and L.J.R. J.F.N. conducted the bioinformatics analysis with input from D.L.C. A.J. and Z.A. processed and imaged the tissues for S.T. B.Y. performed the smelling tests. Y.W. carried out the LTP analysis. B.O. assisted with the western blotting. T.T., T.R., and M.S. performed the mass spectrometry, and T.T., T.R., M.S., J.F.N., and D.L.C. performed data analysis. M.-C.G. performed immunohistochemistry on human AD brains. D.L.C. and J.F.N. wrote the original draft, and all other authors reviewed and edited the manuscript. Supervision was provided by V.A.B.., J.L., and T.T. Funding acquisition, V.A.B., J.L., T.T., L.J.R., and M.-C.G.
Declaration of Interests
J.L. and J.F.N are scientific advisors at 10x Genomics Inc, which provides commercial barcoded arrays. All other authors declare no competing interests.
Published: October 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101556.
Supplemental Information
DE proteins in italics are discussed in the text and only found in the proteomics dataset. Proteins in bold are discussed in the text. Fold-changes are shown
Terms were considered significant if they had a p value ≤0.05 and more than three proteins
Terms were considered significant if they had a p value ≤ 0.05 and more than three proteins/genes
References
- Angeloni C., Zambonin L., Hrelia S. Role of methylglyoxal in Alzheimer's disease. Biomed. Res. Int. 2014;2014:238485. doi: 10.1155/2014/238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomeusz G., Wu Y., Ali Seyed M., Xia W., Kwong K.Y., Hortobagyi G., Hung M.C. Nuclear translocation of the pro-apoptotic Bcl-2 family member Bok induces apoptosis. Mol. Carcinog. 2006;45:73–83. doi: 10.1002/mc.20156. [DOI] [PubMed] [Google Scholar]
- Berglund E., Maaskola J., Schultz N., Friedrich S., Marklund M., Bergenstrahle J., Tarish F., Tanoglidi A., Vickovic S., Larsson L. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018;9:2419. doi: 10.1038/s41467-018-04724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock E.M., Geddes J.W., Chen K.C., Porter N.M., Markesbery W.R., Landfield P.W. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc. Natl. Acad. Sci. U S A. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess D.J. Spatial transcriptomics coming of age. Nat. Rev. Genet. 2019;20:317. doi: 10.1038/s41576-019-0129-z. [DOI] [PubMed] [Google Scholar]
- Caruana D.A., Alexander G.M., Dudek S.M. New insights into the regulation of synaptic plasticity from an unexpected place: hippocampal area CA2. Learn Mem. 2012;19:391–400. doi: 10.1101/lm.025304.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Yang L., McIntyre T.M. Cytotoxic phospholipid oxidation products. Cell death from mitochondrial damage and the intrinsic caspase cascade. J. Biol. Chem. 2007;282:24842–24850. doi: 10.1074/jbc.M702865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.T., Lu A., Craessaerts K., Pavie B., Sala Frigerio C., Corthout N., Qian X., Lalakova J., Kuhnemund M., Voytyuk I. Spatial transcriptomics and in situ sequencing to study Alzheimer's disease. Cell. 2020;4:976–991.e19. doi: 10.1016/j.cell.2020.06.038. [DOI] [PubMed] [Google Scholar]
- Cisse M., Duplan E., Lorivel T., Dunys J., Bauer C., Meckler X., Gerakis Y., Lauritzen I., Checler F. The transcription factor XBP1s restores hippocampal synaptic plasticity and memory by control of the Kalirin-7 pathway in Alzheimer model. Mol. Psychiatry. 2017;22:1562–1575. doi: 10.1038/mp.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orsi B., Engel T., Pfeiffer S., Nandi S., Kaufmann T., Henshall D.C., Prehn J.H. Bok is not pro-apoptotic but suppresses poly ADP-ribose polymerase-dependent cell death pathways and protects against excitotoxic and seizure-induced neuronal injury. J. Neurosci. 2016;36:4564–4578. doi: 10.1523/JNEUROSCI.3780-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orsi B., Mateyka J., Prehn J.H.M. Control of mitochondrial physiology and cell death by the Bcl-2 family proteins Bax and Bok. Neurochem. Int. 2017;109:162–170. doi: 10.1016/j.neuint.2017.03.010. [DOI] [PubMed] [Google Scholar]
- Del-Aguila J.L., Li Z., Dube U., Mihindukulasuriya K.A., Budde J.P., Fernandez M.V., Ibanez L., Bradley J., Wang F., Bergmann K. A single-nuclei RNA sequencing study of Mendelian and sporadic AD in the human brain. Alzheimers Res. Ther. 2019;11:71. doi: 10.1186/s13195-019-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarest T.G., Varma V.R., Estrada D., Babbar M., Basu S., Mahajan U.V., Moaddel R., Croteau D.L., Thambisetty M., Mattson M.P. Biological sex and DNA repair deficiency drive Alzheimer's disease via systemic metabolic remodeling and brain mitochondrial dysfunction. Acta Neuropathol. 2020;140:25–47. doi: 10.1007/s00401-020-02152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desler C., Lillenes M.S., Tonjum T., Rasmussen L.J. The role of mitochondrial dysfunction in the progression of Alzheimer's disease. Curr. Med. Chem. 2017;40:5578–5587. doi: 10.2174/0929867324666170616110111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon C., Sah P., Lynch J.W., Keramidas A. GABAA receptor alpha and gamma subunits shape synaptic currents via different mechanisms. J. Biol. Chem. 2014;289:5399–5411. doi: 10.1074/jbc.M113.514695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einsele-Scholz S., Malmsheimer S., Bertram K., Stehle D., Johanning J., Manz M., Daniel P.T., Gillissen B.F., Schulze-Osthoff K., Essmann F. Bok is a genuine multi-BH-domain protein that triggers apoptosis in the absence of Bax and Bak. J. Cell Sci. 2016;129:2213–2223. doi: 10.1242/jcs.181727. [DOI] [PubMed] [Google Scholar]
- Fang E.F., Hou Y., Palikaras K., Adriaanse B.A., Kerr J.S., Yang B., Lautrup S., Hasan-Olive M.M., Caponio D., Dan X. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat. Neurosci. 2019;22:401–412. doi: 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen J.R., Narayanasamy P. Neuroprotection through flavonoid: enhancement of the glyoxalase pathway. Redox Biol. 2018;14:465–473. doi: 10.1016/j.redox.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glykys J., Mody I. Hippocampal network hyperactivity after selective reduction of tonic inhibition in GABA A receptor alpha5 subunit-deficient mice. J. Neurophysiol. 2006;95:2796–2807. doi: 10.1152/jn.01122.2005. [DOI] [PubMed] [Google Scholar]
- Grubman A., Chew G., Ouyang J.F., Sun G., Choo X.Y., McLean C., Simmons R.K., Buckberry S., Vargas-Landin D.B., Poppe D. A single-cell atlas of entorhinal cortex from individuals with Alzheimer's disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 2019;22:2087–2097. doi: 10.1038/s41593-019-0539-4. [DOI] [PubMed] [Google Scholar]
- Hirata-Fukae C., Li H.F., Hoe H.S., Gray A.J., Minami S.S., Hamada K., Niikura T., Hua F., Tsukagoshi-Nagai H., Horikoshi-Sakuraba Y. Females exhibit more extensive amyloid, but not tau, pathology in an Alzheimer transgenic model. Brain Res. 2008;1216:92–103. doi: 10.1016/j.brainres.2008.03.079. [DOI] [PubMed] [Google Scholar]
- Hiroi N., Brown J.R., Haile C.N., Ye H., Greenberg M.E., Nestler E.J. FosB mutant mice: loss of chronic cocaine induction of Fos-related proteins and heightened sensitivity to cocaine's psychomotor and rewarding effects. Proc. Natl. Acad. Sci. U S A. 1997;94:10397–10402. doi: 10.1073/pnas.94.19.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Lautrup S., Cordonnier S., Wang Y., Croteau D.L., Zavala E., Zhang Y., Moritoh K., O'Connell J.F., Baptiste B.A. NAD(+) supplementation normalizes key Alzheimer's features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc. Natl. Acad. Sci. U S A. 2018;115:E1876–E1885. doi: 10.1073/pnas.1718819115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardin I., Lopez J.J., Berna-Erro A., Salido G.M., Rosado J.A. Homer proteins in Ca(2)(+) entry. IUBMB Life. 2013;65:497–504. doi: 10.1002/iub.1162. [DOI] [PubMed] [Google Scholar]
- Johnson E.C.B., Dammer E.B., Duong D.M., Yin L., Thambisetty M., Troncoso J.C., Lah J.J., Levey A.I., Seyfried N.T. Deep proteomic network analysis of Alzheimer's disease brain reveals alterations in RNA binding proteins and RNA splicing associated with disease. Mol. Neurodegener. 2018;13:52. doi: 10.1186/s13024-018-0282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone V.P., Raymond C.R. Postsynaptic protein synthesis is required for presynaptic enhancement in persistent forms of long-term potentiation. Front. Synaptic Neurosci. 2013;5:1. doi: 10.3389/fnsyn.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam T.I., Mao X., Park H., Chou S.C., Karuppagounder S.S., Umanah G.E., Yun S.P., Brahmachari S., Panicker N., Chen R. Poly(ADP-ribose) drives pathologic alpha-synuclein neurodegeneration in Parkinson's disease. Science. 2018;362:eaat8407. doi: 10.1126/science.aat8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B. A unique microglia type Associated with restricting development of Alzheimer's disease. Cell. 2017;169:1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Kuhla B., Boeck K., Schmidt A., Ogunlade V., Arendt T., Munch G., Luth H.J. Age- and stage-dependent glyoxalase I expression and its activity in normal and Alzheimer's disease brains. Neurobiol. Aging. 2007;28:29–41. doi: 10.1016/j.neurobiolaging.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Lang F., Stournaras C., Zacharopoulou N., Voelkl J., Alesutan I. Serum- and glucocorticoid-inducible kinase 1 and the response to cell stress. Cell Stress. 2018;3:1–8. doi: 10.15698/cst2019.01.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotta A., Rosner J., Huchzermeyer C., Wojtowicz A., Kann O., Schmitz D., Heinemann U., Kovacs R. Energy demand of synaptic transmission at the hippocampal Schaffer-collateral synapse. J. Cereb. Blood Flow Metab. 2012;32:2076–2083. doi: 10.1038/jcbfm.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Lu H., Stein E., Zhou Z., Yang Y., Mattson M.P. Brain regional synchronous activity predicts tauopathy in 3xTgAD mice. Neurobiol. Aging. 2018;70:160–169. doi: 10.1016/j.neurobiolaging.2018.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F., Wang Y.M., Victor B., Yang M., Schneider D.M., Gingras S., Parsons M.J., Zheng J.H., Brown S.A., Pelletier S. BOK is a non-canonical BCL-2 family effector of apoptosis regulated by ER-associated degradation. Cell. 2016;165:421–433. doi: 10.1016/j.cell.2016.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian M., Beullens M., Andres I., Ortiz J.M., Bollen M. SIPP1, a novel pre-mRNA splicing factor and interactor of protein phosphatase-1. Biochem. J. 2004;378:229–238. doi: 10.1042/BJ20030950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaskola J., Bergenstrahle L., Jurek A., Navarro J.F., Lagergren J., Lundeberg J. Charting tissue expression anatomy by spatial transcriptome decomposition. bioRxiv. 2018 doi: 10.1101/362624. [DOI] [Google Scholar]
- Maniatis S., Aijo T., Vickovic S., Braine C., Kang K., Mollbrink A., Fagegaltier D., Andrusivova Z., Saarenpaa S., Saiz-Castro G. Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science. 2019;364:89–93. doi: 10.1126/science.aav9776. [DOI] [PubMed] [Google Scholar]
- Martinez-Lopez M.J., Alcantara S., Mascaro C., Perez-Branguli F., Ruiz-Lozano P., Maes T., Soriano E., Buesa C. Mouse neuron navigator 1, a novel microtubule-associated protein involved in neuronal migration. Mol. Cell Neurosci. 2005;28:599–612. doi: 10.1016/j.mcn.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Mattson M.P., Guthrie P.B., Kater S.B. Intrinsic factors in the selective vulnerability of hippocampal pyramidal neurons. Prog. Clin. Biol. Res. 1989;317:333–351. [PubMed] [Google Scholar]
- McClard C.K., Kochukov M.Y., Herman I., Liu Z., Eblimit A., Moayedi Y., Ortiz-Guzman J., Colchado D., Pekarek B., Panneerselvam S. POU6f1 mediates neuropeptide-dependent plasticity in the adult brain. J. Neurosci. 2018;38:1443–1461. doi: 10.1523/JNEUROSCI.1641-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiak M., Vergara Greeno R., Baptiste B.A., Sykora P., Liu D., Cordonnier S., Fang E.F., Croteau D.L., Mattson M.P., Bohr V.A. DNA polymerase beta decrement triggers death of olfactory bulb cells and impairs olfaction in a mouse model of Alzheimer's disease. Aging Cell. 2017;16:162–172. doi: 10.1111/acel.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiak M.M., Hipolito M.S., Ressom H.W., Obisesan T.O., Manaye K.F., Nwulia E.A. Apo E4 alleles and impaired olfaction as predictors of Alzheimer's disease. Clin. Exp. Psychol. 2017;3:169. doi: 10.4172/2471-2701.1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L., Pupi A., De Leon M.J. Brain glucose hypometabolism and oxidative stress in preclinical Alzheimer's disease. Ann. N. Y Acad. Sci. 2008;1147:180–195. doi: 10.1196/annals.1427.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. Olfactory and other sensory impairments in Alzheimer disease. Nat. Rev. Neurol. 2019;15:11–24. doi: 10.1038/s41582-018-0097-5. [DOI] [PubMed] [Google Scholar]
- Nishitsuji K., Hosono T., Uchimura K., Michikawa M. Lipoprotein lipase is a novel amyloid beta (Abeta)-binding protein that promotes glycosaminoglycan-dependent cellular uptake of Abeta in astrocytes. J. Biol. Chem. 2011;286:6393–6401. doi: 10.1074/jbc.M110.172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver P.L., Finelli M.J., Edwards B., Bitoun E., Butts D.L., Becker E.B., Cheeseman M.T., Davies B., Davies K.E. Oxr1 is essential for protection against oxidative stress-induced neurodegeneration. Plos Genet. 2011;7:e1002338. doi: 10.1371/journal.pgen.1002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillasse M.R., de Medina P. The NR4A nuclear receptors as potential targets for anti-aging interventions. Med. Hypotheses. 2015;84:135–140. doi: 10.1016/j.mehy.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Pandey K.N. Molecular and genetic aspects of guanylyl cyclase natriuretic peptide receptor-A in regulation of blood pressure and renal function. Physiol. Genomics. 2018;50:913–928. doi: 10.1152/physiolgenomics.00083.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H., Dobson R.J.B., Newhouse S.J. A meta-analysis of Alzheimer's disease brain transcriptomic data. J. Alzheimers Dis. 2019;68:1635–1656. doi: 10.3233/JAD-181085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahayel S., Frasnelli J., Joubert S. The effect of Alzheimer's disease and Parkinson's disease on olfaction: a meta-analysis. Behav. Brain Res. 2012;231:60–74. doi: 10.1016/j.bbr.2012.02.047. [DOI] [PubMed] [Google Scholar]
- Regard J.B., Scheek S., Borbiev T., Lanahan A.A., Schneider A., Demetriades A.M., Hiemisch H., Barnes C.A., Verin A.D., Worley P.F. Verge: a novel vascular early response gene. J. Neurosci. 2004;24:4092–4103. doi: 10.1523/JNEUROSCI.4252-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds M.S., Hancock C.R., Ray J.D., Kener K.B., Draney C., Garland K., Hardman J., Bikman B.T., Tessem J.S. beta-Cell deletion of Nr4a1 and Nr4a3 nuclear receptors impedes mitochondrial respiration and insulin secretion. Am. J. Physiol. Endocrinol. Metab. 2016;311:E186–E201. doi: 10.1152/ajpendo.00022.2016. [DOI] [PubMed] [Google Scholar]
- Rissman R.A., De Blas A.L., Armstrong D.M. GABA(A) receptors in aging and Alzheimer's disease. J. Neurochem. 2007;103:1285–1292. doi: 10.1111/j.1471-4159.2007.04832.x. [DOI] [PubMed] [Google Scholar]
- Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., Welch J., Chen L.M., Chen F., Macosko E.Z. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman J.J., Szczesniak L.M., Bunker E.N., Nelson H.A., Roe M.W., Wagner L.E., 2nd, Yule D.I., Wojcikiewicz R.J.H. Bok regulates mitochondrial fusion and morphology. Cell Death Differ. 2019;I12I:2682–2694. doi: 10.1038/s41418-019-0327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Kahle A., Hershey J.W., Honchak B.M., Warneke J.A., Leong S.P., Nelson M.A. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene. 2006;25:4923–4936. doi: 10.1038/sj.onc.1209495. [DOI] [PubMed] [Google Scholar]
- Srivastava R., Cao Z., Nedeva C., Naim S., Bachmann D., Rabachini T., Gangoda L., Shahi S., Glab J., Menassa J. BCL-2 family protein BOK is a positive regulator of uridine metabolism in mammals. Proc. Natl. Acad. Sci. U S A. 2019;116:15469–15474. doi: 10.1073/pnas.1904523116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl P.L., Salmen F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., Giacomello S., Asp M., Westholm J.O., Huss M. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- Swerdlow R.H. Mitochondria and mitochondrial cascades in Alzheimer's disease. J. Alzheimers Dis. 2018:1403–1416. doi: 10.3233/JAD-170585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykora P., Misiak M., Wang Y., Ghosh S., Leandro G.S., Liu D., Tian J., Baptiste B.A., Cong W.N., Brenerman B.M. DNA polymerase beta deficiency leads to neurodegeneration and exacerbates Alzheimer disease phenotypes. Nucleic Acids Res. 2015;43:943–959. doi: 10.1093/nar/gku1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S., Munch A., Neumann S., Kremmer E., Tatzelt J., Lichtenthaler S.F. The novel membrane protein TMEM59 modulates complex glycosylation, cell surface expression, and secretion of the amyloid precursor protein. J. Biol. Chem. 2010;285:20664–20674. doi: 10.1074/jbc.M109.055608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickovic S., Eraslan G., Salmen F., Klughammer J., Stenbeck L., Schapiro D., Aijo T., Bonneau R., Bergenstrahle L., Navarro J.F. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman L., Jo D.G., Sorensen M.M., de Souza-Pinto N.C., Markesbery W.R., Mattson M.P., Bohr V.A. Defective DNA base excision repair in brain from individuals with Alzheimer's disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007;35:5545–5555. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.H., Grace C.R., Guibao C.D., McNamara D.E., Llambi F., Wang Y.M., Chen T., Moldoveanu T. Intrinsic instability of BOK enables membrane permeabilization in apoptosis. Cell Rep. 2018;23:2083–2094.e6. doi: 10.1016/j.celrep.2018.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Zheng X., Zhang L., Luo H., Qian L., Fu X., Liu Y., Gao Y., Niu M., Meng J. The neuron-specific protein TMEM59L mediates oxidative stress-induced cell death. Mol. Neurobiol. 2017;54:4189–4200. doi: 10.1007/s12035-016-9997-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DE proteins in italics are discussed in the text and only found in the proteomics dataset. Proteins in bold are discussed in the text. Fold-changes are shown
Terms were considered significant if they had a p value ≤0.05 and more than three proteins
Terms were considered significant if they had a p value ≤ 0.05 and more than three proteins/genes
Data Availability Statement
Processed and analyzed data and analysis scripts generated during this study are available at Mendeley https://doi.org/10.17632/6s959w2zyr.1 and GitHub https://github.com/jfnavarro/AD_POLB_ST. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (PubMed ID: 30395289) partner repositories with the dataset identifier PXD017766.