Dogs and cats have gained a special position in human society by becoming our principal companion animals. In this context, efforts to ensure their health and well‐fare have increased exponentially, with in recent times a growing interest in assessing the impact of the gut microbiota on canine and feline health. Recent technological advances have generated new tools to not only examine the intestinal microbial composition of dogs and cats, but also to scrutinize the genetic repertoire and associated metabolic functions of this microbial community, revealing that the taxonomic composition and the metabolic repertoire of the intestinal microbial population of dogs and cats may be influenced by several factors, including diet, age and anthropogenic aspects, as well as intestinal dysbiosis.
Summary
Dogs and cats have gained a special position in human society by becoming our principal companion animals. In this context, efforts to ensure their health and welfare have increased exponentially, with in recent times a growing interest in assessing the impact of the gut microbiota on canine and feline health. Recent technological advances have generated new tools to not only examine the intestinal microbial composition of dogs and cats, but also to scrutinize the genetic repertoire and associated metabolic functions of this microbial community. The application of high‐throughput sequencing techniques to canine and feline faecal samples revealed similarities in their bacterial composition, with Fusobacteria, Firmicutes and Bacteroidetes as the most prevalent and abundant phyla, followed by Proteobacteria and Actinobacteria. Although key bacterial members were consistently present in their gut microbiota, the taxonomic composition and the metabolic repertoire of the intestinal microbial population may be influenced by several factors, including diet, age and anthropogenic aspects, as well as intestinal dysbiosis. The current review aims to provide a comprehensive overview of the multitude of factors which play a role in the modulation of the canine and feline gut microbiota and that of their human owners with whom they share the same environment.
Introduction
Being descendants of the Eurasian grey wolf and wild cat, domesticated dogs and cats, respectively, are among the first animals to have undergone profound anthropogenic changes (Sykes et al., 2020). Since their initial domestication, a process that commenced some 15 000 years ago, a large number of canine and feline breeds have been selected with an associated global dissemination of millions of these pets. Throughout recent millennia, dogs and cats have played an increasingly important role in human society. The ever‐closer relationship between these pets and their owners resulted in the former becoming the principal companion animals for humans. Furthermore, intense urbanization typical of the modern era has not only changed human habits, but has also completely altered the lifestyle of these pets (Dotson and Hyatt, 2008). In this context, concerns regarding pet health and well‐being are taken seriously and have consequently prompted a lot of research aimed at pet health promotion and disease prevention. In this context, the scientific community has in recent decades focused on gut health and the study of the intestinal bacterial population of dogs and cats, for the purpose of maintaining and promoting host health (Mondo et al., 2019).
The mammalian gastrointestinal tract (GIT) is inhabited by one of the most intricate and diverse communities of microorganisms in the biosphere, i.e. the gut microbiota, encompassing bacteria, archaea, viruses, fungi and protozoa (Suchodolski, 2011). Despite this diversity of microorganisms that may colonize the mammalian intestine, bacteria are by far the most abundant representatives of the mammalian intestinal population (Suchodolski, 2011, 2016). In this context, the long‐lasting mutualistic association (commensalism or symbiosis) of this indigenous microbial ecosystem with its mammalian host has laid the foundation for the establishment and subsequent consolidation of multiple trophic interactions. Specifically, the intestinal microbial community is involved in various metabolic and physiological activities, including degradation of otherwise non‐digestible complex carbohydrates, provision of energy sources to support intestinal epithelium integrity and to ensure its basal activity, education of the immune system, protection against pathogen colonization and production of metabolites, including short‐chain fatty acids (SCFAs), secondary bile acids, vitamins or other bacterially derived compounds such as trimethylamine‐N‐oxide (Moens and Veldhoen, 2012; Pickard et al., 2017; Moon et al., 2018; Mondo et al., 2019; Wernimont et al., 2020). Furthermore and similar to the human situation, the microbial community of the feline and canine intestine may be influenced by various factors, such as diet (Schmidt et al., 2018), age (Masuoka et al., 2017), metabolic disorders and inflammatory bowel diseases (Omori et al., 2017; Kalenyak et al., 2018). Despite the multiple functions exerted by the intestinal ecosystem and its involvement in supporting host health, our understanding of the gut microbiota of pet animals is rather scarce if at best incomplete when compared to the significant scientific progress achieved for similar studies of the human gut microbiota. In the present review, current knowledge on canine and feline intestinal community will be analysed. More specifically, we will discuss the composition of the gut microbiota of healthy dogs and cats, together with perturbations that this microbial community may undergo as a consequence of the onset of inflammatory bowel diseases, age or changes in dietary habits. Furthermore, we will discuss the impact of the close human–pet relationship on the microbiota of either party.
Overview of technical approaches for gut microbiota characterization
Originally, the study of canine and feline gut microbiota was based on culture‐dependent methods, involving cultivation and subsequent isolation of microorganisms by means of different growth media (Moon et al., 2018). Bacterial cultivation was typically used to detect specific enteropathogens, i.e. Salmonella spp. or Campylobacter spp., to determine active infection or to test for antibiotic sensitivity (Johnston et al., 2001; Pepin‐Puget et al., 2020). However, despite the advantages of these classical microbiological techniques, such as the possibility to perform physiological and biochemical analyses on isolated strains or to assess total viable bacterial load of a sample, culture‐dependent methods suffer from several disadvantages (Turroni et al., 2008). Indeed, samples require immediate processing and results are very much affected by cultivation media and laboratory conditions used, which are unlikely to faithfully reproduce the very complex intestinal environment. In this context, it has been stated that only a small portion of the intestinal biodiversity can be assessed by the application of culture‐dependent methods (Furrie, 2006; Suchodolski, 2016). However, in recent times these limitations have been overcome thanks to remarkable advances in sequencing technologies, opening up new research horizons to investigate ecological aspects of the gut microbiota and to perform culture‐independent approaches by means of cost‐effective, high‐throughput sequencing methods, such as metagenomics and metatranscriptomics. Furthermore, technological developments in metaproteomics and metabolomics areas have allowed researches to connect DNA sequences of the gut microbiota with their encoded functions.
Metagenomics allows the assessment of the composition, encoded activities and functional features of the entire intestinal microbial community, including bacterial taxa that have not yet been cultivated under in vitro, i.e. laboratory conditions, as well as the detection of non‐bacterial taxa that colonize the GIT such as protozoa, fungi, viruses and archaea (Hamady and Knight, 2009; Ventura et al., 2009; Blake and Suchodolski, 2016; Fig. 1). Historically, 16S rRNA gene‐based microbial profiling was the first culture‐independent method to be employed for the compositional profiling of the gut microbiota. This approach employs universal primers for PCR‐mediated amplification and subsequent sequencing of a single or multiple hypervariable regions of the 16S rRNA gene, which represents a conserved phylogenetic marker composed by highly conserved sequences interspersed with nine hypervariable regions (Neefs et al., 1993; Hamady and Knight, 2009; Milani et al., 2017a, 2017b,2017a, 2017b). However, despite the fact that the 16S rRNA microbial profiling method was and continues to be the most cost‐effective and popular technique to study gut microbiota composition, it has a number of serious limitations. There is currently no single standardized methodology for DNA extraction, nor is there agreement among the scientific community which primers are most appropriate for amplification or which hypervariable region of the 16S rRNA gene is to be targeted to achieve optimal sequencing efficiency of a DNA region with the highest level of taxonomic discriminatory power (Mancabelli et al., 2020). Furthermore, 16S rRNA gene‐based microbial profiling generates bacterial taxonomic composition data typically down to the genus level only, while it commonly fails in detecting underrepresented bacterial taxa, thereby causing biases in the interpretation and comparison of data from different studies (Chakravorty et al., 2007). More recently, the so‐called internal transcribed spacer (ITS) microbial profiling methods were introduced as a tool to offer a more refined taxonomic view of the gut microbiota (Milani et al., 2014, 2018). The ITS region is located between the 16S rRNA and 23S rRNA genes within the rRNA locus. This genomic portion is more variable at the interspecies level when compared to the 16S rRNA gene, and represents a more suitable phylogenetic marker to obtain an in‐depth view of the intestinal microbial population by providing an accurate species‐ or even subspecies‐level taxonomic resolution (Milani et al., 2017a, 2017b,2017a, 2017b). However, this method is currently only available for specific microbial genera such as Bifidobacterium and Lactobacillus (Milani et al., 2014, 2018).
Fig. 1.
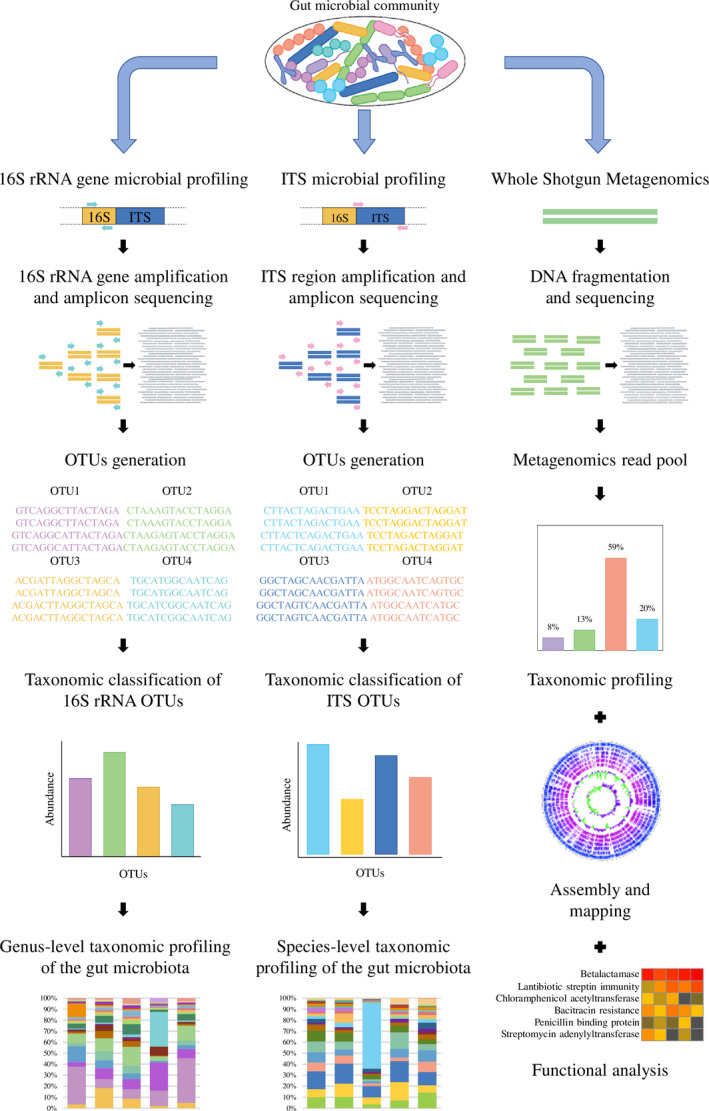
General overview of the metagenomic approaches available for gut microbiota characterization. Starting from DNA extraction of the intestinal microbial community, the subsequent high‐throughput sequencing provides taxonomic insight into the gut microbiota down to the genus and species levels for 16S rRNA gene and ITS microbial profiling respectively. In addition to the taxonomic composition, whole shotgun metagenomics allows the reconstruction of the microbial genomes and prediction of the bacterial functional features.
In‐depth whole metagenome shotgun sequencing (WMS), which involves high‐resolution sequencing of total microbial DNA extracted from a specific matrix, captures substantially more information when compared to gene or sequence‐specific amplification‐based approaches. Indeed, in addition to taxonomic composition, WMS offers in‐depth insights into the genomic content and compositional arrangement of bacterial consortia, also allowing predictions of their functional features (Lugli et al., 2019). Notably, functional classification of WMS data may be used to shed light on the microbial dark matter in order to identify the catabolic and anabolic activities of intestinal bacteria and to investigate the presence of genes involved in multiple processes, such as adhesion to the intestinal epithelium or antibiotic resistance (Rinke et al., 2013). Furthermore, WMS is not affected by the potential bias caused by the amplification step typical of the 16S rRNA or ITS microbial profiling methods. Despite these advantages, many of the large‐scale studies continue to involve 16S rRNA microbial profiling because of the high costs of high‐resolution WMS. In order to overcome the limitations of the 16S rRNA or ITS microbial profiling, shallow shotgun sequencing, which is a low‐depth WMS, has recently been proposed to combine species‐level taxonomic assignment with reduced cost while avoiding amplification bias of 16S rRNA‐based sequencing (Hillmann et al., 2018). However, regardless of the method used, metagenomics is a ‘relative approach’ (Vandeputte et al., 2017). Indeed, it provides a microbial profiling based on relative and not absolute abundance, thus hampering efforts to correlate microbiome features to quantitative data, including physiological parameters or metabolite levels (Vandeputte et al., 2017). Evaluation of the overall microbial abundance through quantitative approaches such as flow cytometry has recently been proposed to overcome this limitation so as to provide quantitative microbiome profiling (Vandeputte et al., 2017).
In addition to metagenomics, further ‘omics’ approaches, in particular metatranscriptomics and metabolomics, have been developed in order to move beyond mere taxonomic assignment or gene functional predictions. Indeed, metatranscriptomics identifies which genes, among the myriad of bacterial genes that constitute the gut microbiome, are actually transcribed through RNA sequencing of a given sample. In addition, metabolomics provides information on the production of microbial metabolites, thereby attempting to capture metabolic activity among and interactions between the gut microbiota and its host (Smirnov et al., 2016; Wang et al., 2020).
Insights into the gut microbiota of healthy dogs and cats
Combining culture‐dependent approaches with metagenomic methodologies for studying the canine and feline gut microbiota has shown that bacterial abundance and biodiversity gradually increase along the GIT (Ritchie et al., 2008; Suchodolski et al., 2008; Honneffer et al., 2017). Specifically, bacterial cultivation efforts revealed that the total microbial load of the stomach and small intestine of dogs and cats is lower than that found in the distal intestinal tract, with an overall bacterial abundance along the GIT that ranges from 102 up to 1014 colony‐forming units (CFU) per gram of luminal content (German et al., 2003; Mentula et al., 2005; Ritchie et al., 2008). Furthermore, the small intestine is inhabited by both aerobic and facultative anaerobic bacteria, while the large intestine is mostly colonized by anaerobic microbes, reflecting the microenvironment and oxygen availability of these different GIT compartments. Furthermore, cultivation‐independent molecular methods revealed variations of microbial richness along the length of the intestinal tract, with the greatest biodiversity recorded in the large intestine when compared to the stomach and small intestine. This observation is in accordance with the physiological functions of these different intestinal compartments, with the colon and caecum as the main fermentative sites in monogastric mammals (Suchodolski et al., 2008; Suchodolski, 2016; Honneffer et al., 2017). Despite the well‐accepted notion that variations in bacterial abundance, taxonomic composition and biodiversity occur along the GIT, much of the relevant published scientific literature bases its gut microbiota findings on the analysis of faecal samples due to practical difficulties and ethical constraints related to the collection of samples from each intestinal sector. However, although starting from the same environmental matrix (i.e. stool samples), comparison of the results obtained by different studies has revealed distinct differences between the bacterial populations present in the GIT of cats and dogs. In this context, multiple factors, including diet, environment, age, gender, genetics, diseases and relative therapies, have been shown to affect the intestinal microbial composition of an individual, promoting interindividual fluctuations among animals of the same species with each pet apparently possessing its own unique microbial gut ecosystem (David et al., 2014; Wernimont et al., 2020). Furthermore, non‐standard experimental procedures applied by different metagenomics‐based studies, due to different DNA extraction protocols, distinct primers employed for amplification of 16S rRNA gene‐associated hypervariable regions and/or varying bioinformatics methodologies for the interpretation and analysis of metagenomic data, have contributed to poor reproducibility across populations (Schloss, 2018). Despite this limitation in performing a straightforward comparison between findings of different studies, key bacterial players have transversely been found in feline and canine faeces in different studies, regardless of the metagenomic approaches used. Specifically, Fusobacteria, Bacteroidetes and Firmicutes were identified as the predominant and prevalent bacterial phyla characterizing the faecal microbiota of dogs and cats (Ritchie et al., 2010; Handl et al., 2011; Minamoto et al., 2012; Tun et al., 2012; Alessandri et al., 2019a, 2019b, 2019c). Within the Firmicutes phylum, Bacilli, Clostridia and Erysipelotrichi are the most representative bacterial classes for both canine and feline gut microbiota. Specifically, the Bacilli class is predominantly represented by Streptococcus and Lactobacillus genera in dogs and Enterococcus and Lactobacillus in cats (Handl et al., 2011). The Clostridia class is represented by Clostridium clusters IV (Ruminococcaceae family and Faecalibacterium prausnitzii), XI (Peptostreptococcaceae family) and XIVa (Lachnospiraceae family and Blautia), while the Erysipelotrichi is mainly represented by Turicibacter, Catenibacterium and Coprobacillus genera (Ritchie et al., 2010; Handl et al., 2011; Pilla and Suchodolski, 2019). Furthermore, Prevotella and Bacteroides, belonging to the Bacteroidetes phylum, are among the most abundant and prevalent bacterial genera of the faecal microbiota of both companion animals considered here (Alessandri et al., 2019a, 2019b, 2019c, 2020).
Furthermore, Fusobacterium, a genus belonging to the Fusobacteria phylum, was identified as one of the principal microbial players of the canine and feline gut microbiota, though with a lower average relative abundance in cats (Suchodolski et al., 2008; Alessandri et al., 2019a, 2019b, 2019c, 2020). Notably, in humans, Fusobacterium is associated with inflammatory bowel diseases, while the presence of a specific species of this genus in the human intestine, i.e. Fusobacterium nucleatum, is linked to the occurrence of colorectal cancer due to its potential carcinogenic features (Ng et al., 2019; Gethings‐Behncke et al., 2020). However, this negative association has not been observed for dogs and cats. Indeed, Fusobacterium has been found to be present at high abundance in the gut microbiota of healthy dogs and cats, and its common presence in the GIT of other carnivorous animals suggests that it is not harmful to its host (Ley et al., 2008; Swanson et al., 2011; Alessandri et al., 2019a, 2019b, 2019c, 2020). The ability of Fusobacterium species to degrade proteins to obtain their preferred growth substrates, i.e. amino acids and peptides, is probably the reason for its high abundance in carnivorous animals (Doron et al., 2014). Furthermore, not all members of the Fusobacterium genus should be considered as potentially harmful to the host. For example, it has been demonstrated that Fusobacterium varium not only acts as a potent antagonist of pathogen colonization, but is also able to exert anti‐inflammatory effects and sustain enterocytes by producing butyrate from protein fermentation (Potrykus et al., 2008). Indeed, while most of intestinal butyrate is obtained through bacterial fermentation of dietary fibres via two metabolic pathways, one involving the phosphorylation of the butyryl‐CoA to synthetize butyryl‐phosphate that is then transformed into butyrate via butyrate kinase and the other including the transfer of the CoA moiety of butyryl‐CoA to acetate leading to the production of butyrate and acetyl‐CoA, metagenomic data analyses revealed that butyrate can be also produced from proteins via the lysine pathways (Vital et al., 2014; Louis and Flint, 2017; Liu et al., 2018). In this context, specific ecological studies coupled with comparative genomic analyses are required to shed light on the significance of the high abundance of Fusobacterium and its role in the gut microbiota of dogs and cats.
With a reduced abundance but an equivalent prevalence of the co‐dominant Bacteroidetes, Firmicutes and Fusobacteria, the Proteobacteria and Actinobacteria phyla represent two other major taxa of the canine and feline gut microbiota (Tun et al., 2012; Alessandri et al., 2019a, 2019b, 2019c). Specifically, Proteobacteria are more abundant in dogs, while the relative abundance of Actinobacteria is higher in the feline faecal microbiota (Handl et al., 2011; Moon et al., 2018). Among the Proteobacteria, Sutterella, Succinivibrio, Anaerobiospirillum and Escherichia/Shigella are the most abundant genera in both canine and feline faeces (Handl et al., 2011). In this context, Escherichia/Shigella together with another genus of the Proteobacteria phylum, i.e. Salmonella, which is also a member of the Enterobacteriaceae, are generally considered gastrointestinal pathogens of public concern (Moon et al., 2018). However, most strains belonging to the abovementioned bacterial genera are non‐pathogenic and indeed contribute to the microbiome function of healthy hosts (Moon et al., 2018). Notably, a metagenomic study revealed that the Coriobacteriaceae family is the main representative of the Actinobacteria phylum, with Collinsella and Slackia as dominant genera in dogs, while Eggerthella and especially Olsenella being prevalent in cats (Handl et al., 2011). Furthermore, recently, the application of the bifidobacterial ITS microbial profiling method to multiple canine and feline faecal samples revealed the presence of several Bifidobacterium species in each processed sample (Alessandri et al., 2019a, 2019b, 2019c, 2020), thus strengthening the notion that bifidobacteria are ubiquitous commensal microorganisms of the intestinal microbial community of mammals, including dogs and cats (Milani et al., 2017a, 2017b,2017a, 2017b).
Although key bacterial members are commonly present in the faecal microbial community of healthy dogs and cats, a finding that is indicative of a core faecal microbiota, the taxonomic composition may be subject to changes and modulations due to the influence of several factors including diet (Bresciani et al., 2018; Schmidt et al., 2018; Alessandri et al., 2019a, 2019b, 2019c), age (Fahey et al., 2008; Masuoka et al., 2017), metabolic disorders (i.e. diabetes or obesity; Alexander et al., 2018; Salas‐Mani et al., 2018), intestinal dysbiosis (diarrhoea or inflammatory bowel diseases) or cancer (Honneffer et al., 2014; Omori et al., 2017; Kalenyak et al., 2018), as well as anthropogenic influences (Alessandri et al., 2019a, 2019b, 2019c; Fig. 2).
Fig. 2.
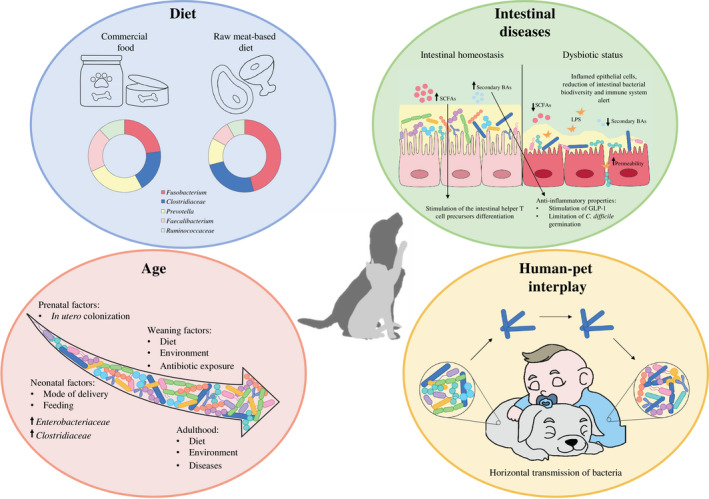
Schematic representation of the main factors influencing the canine and feline gut microbiota. Specifically, age, diet, perturbation of the gut microbiota homeostasis and human–pet interplay play a crucial role in the modulation of the intestinal microbial community of dogs and cats.
The effect of changes in dietary habits on canine and feline gut microbiota
Diet has been recognized as one of the main drivers influencing both biodiversity and functional features of the mammalian gut microbiota (Ley et al., 2008; David et al., 2014). Indeed, nutrients introduced through the diet not only act as sustenance for the host, but diet components that cannot be directly digested by the host may also represent nutrients for gut microorganisms.
Inclusion of prebiotics in canine and feline diet
Despite its original carnivorous classification, the domestic dog is currently considered as an omnivorous animal, since commercial pet foods are formulated to provide a balanced nutritional intake yet with the supplementation of high concentrations of fibre and carbohydrates (generally higher than 3% and 30%, respectively; Barry et al., 2012; Alessandri et al., 2019a, 2019b, 2019c), which is necessary to produce extruded dry diet (kibble) (Pilla and Suchodolski, 2019). The domestic cat, on the other hand, with its short colon represents an example of an evolutionary adaptation to a strict carnivorous diet and is still considered an obligate carnivore. Nonetheless, commercial feeds for domesticated felines are enriched in components of vegetable origin (Hooda et al., 2013; Rochus et al., 2014; Butowski et al., 2019). Furthermore, in recent years and based on a multitude of studies that have highlighted health‐promoting features of prebiotics, several companies have started to include prebiotic compounds in commercial pet foods (Deng and Swanson, 2015). Prebiotics are defined as selectively fermented ingredients that are able to induce specific changes in the composition and/or functional activities of the intestinal bacterial community, in order to confer benefits to the host (Gibson et al., 2004). Several non‐digestible carbohydrates are known to possess prebiotic features and range from the disaccharide lactulose to oligo‐ or polysaccharides such as fructo‐oligosaccharides (FOS), mannan‐oligosaccharides (MOS), xylo‐oligosaccharides (XOS) and galacto‐oligosaccharides (GOS) and inulin (Roberfroid et al., 2010; Slavin, 2013). To be effective, prebiotics have to withstand digestion by host enzymes and reach the distal part of the intestine where they favour the proliferation and metabolic activities of specific bacterial species able to promote evident beneficial effects to the host (Pinna and Biagi, 2016). By escaping upper intestinal hydrolysis and absorption and reaching the hindgut compartments, these complex carbohydrates will be selectively fermented by those microorganisms that possess genes encoding specific extracellular and/or intracellular glycosyl‐hydrolases (GH) and transport systems, required for the breakdown and uptake, respectively, of these carbohydrates (Vieira et al., 2013). The main products of this fermentative catabolic process are SCFAs, which include saturated aliphatic organic acids, mainly represented by butyrate, acetate and propionate. These particular organic acids are believed to improve host health by increasing mineral absorption, regulating bowel functions, decreasing the luminal pH thus limiting the proliferation of pH‐sensitive pathogenic bacteria, influencing the immune system and providing nutrients to enterocytes (Ratajczak et al., 2019). In this context, several studies have highlighted the beneficial effects of prebiotic inclusion in the diet of dogs and cats (Anderson et al., 1991; Barry et al., 2010; Nogueira et al., 2019; Ide et al., 2020).
A recent study assessed the impact of supplementation of 7.5% beet pulp, which is a commonly used dietary fibre for the formulation of commercial canine feed, consisting of a mixture of fermentable and non‐fermentable carbohydrates (Middelbos et al., 2010). This trial revealed a drastic shift of the Firmicutes/Fusobacteria ratio in favour of the former in the faecal microbiota of treated dogs, accompanied by a threefold increase of Faecalibacterium prausnitzii and proliferation of Eubacterium hallii (Middelbos et al., 2010). Another study was aimed at evaluating the impact of kestose (a type of FOS) supplementation on the canine gut microbiota and SCFA concentrations. While the Lactobacillus genus was shown to elicit a slight and statistically non‐significant increase in relative abundance, the Bifidobacterium taxa showed a marked trend of numerical enhancement during the eight weeks of treatment, thus supporting the bifidogenic effect of this specific FOS component. Furthermore, based on the observation that kestose was shown to cause a decreased relative abundance of Clostridium perfringens and a simultaneous increase of butyrate concentration, it was inferred that the increased concentration of butyrate lowers luminal pH, thus creating a less suitable environment for the proliferation of certain microbial species (Ide et al., 2020). Moreover, the administration of a FOS (4%)‐ or pectin (4%)‐based diet to cats was shown to cause an increase in the relative abundance of the Bifidobacterium genus and Lactobacillus genus, respectively, when compared to the control diet (supplemented with 4% cellulose). Furthermore, the total concentration of SCFAs was higher in faecal samples of treated cats when compared to the cellulose control (Barry et al., 2010). Taken together, these results highlight the important role of diet in modulating the intestinal bacterial community as well as its functional activities in both dogs and cats.
Raw meat‐based diet
Despite extensive efforts dedicated to the formulation of industrial pet food in order to guarantee a balanced diet as well as the inclusion of prebiotics to promote intestinal homeostasis, an apparently opposing trend has recently emerged. Indeed, it has become increasingly popular to feed dogs and cats with a raw meat‐based diet (RMBD), instead of the more conventional commercial dry food (van Bree et al., 2018). The choice of many pet owners to return to a ‘natural’ and ‘home‐made’ diet for their animals was driven by several factors. First, most industrial foods are obtained from the use of waste and/or by‐products from human‐oriented food processing industry, thus failing in formulating products of first choice. Furthermore, while the extruded and retort format, which is typical of commercial food, uses heat, moisture and pressure to process food, the RMB diet incorporates food that is essentially unprocessed (Algya et al., 2018). Second, pet owners can influence health and well‐being of their companion animals through the feed they provide to their pets (Schlesinger and Joffe, 2011). As the RMBD designation suggests, these formulations include raw meat, internal organs, cartilage, fat and fleshy bones from farm animals (ruminants, poultry and pigs), horses, game and fish (Fredriksson‐Ahomaa et al., 2017). Furthermore, though at a much reduced level when compared to the extruded diet, RMBD can also contain vegetables, eggs and dairy products (van Bree et al., 2018). The so‐called BARF (Bones and Raw Food or Biologically Appropriate Raw Food) diet is probably the most widespread and known example of RMBD. In parallel with the take‐up of these novel nutritional approaches, the advantages and disadvantages of feeding a natural diet were investigated. While it has been demonstrated that RMBD provides important health benefits to its animal consumer, including reduction in dental diseases coupled with fresher breath, alleviation of arthritis, increase of the immune response, healthier skin and a shiny coat, several issues of public concern were raised (Freeman et al., 2013). Indeed, some studies reported findings that discourage the administration of this natural diet since it does not guarantee a balanced nutritional intake and it increases the risk of exposure to zoonotic pathogens, encompassing Campylobacter spp., Salmonella spp., Yersinia spp. and pathogenic Escherichia coli strains, thus threatening the health of both animal companions and their owners (Schlesinger and Joffe, 2011; Kim et al., 2017; van Bree et al., 2018). However, these studies refer only to nutritional and pathogen contents, while they do not take into account any possible changes both in microbial composition and in functional activities of the gut microbiota of dogs and cats when they are fed on a RMB diet. Therefore, these investigations do not fully describe the advantages/disadvantages of this natural diet and care should be taken to draw far‐reaching conclusions from such findings.
Collectively, even if the studies investigating the impact of a RMBD on the gut microbiota of dogs and cats are based on different experimental design and sequencing protocols, they share common features (Table 1). In general, the BARF diet seems to favour the proliferation of the Fusobacterium genus and Clostridiaceae family. As already mentioned, members of the Fusobacterium genus were pointed out as potentially harmful microorganisms, being related to intestinal bowel diseases and colorectal carcinoma in humans (Castellarin et al., 2012; Kostic et al., 2013; Alhinai et al., 2019). In the same way, Clostridiaceae is a highly diverse family, including not only useful bacterial genera involved in nutrient digestibility, yet also pathogenic microorganisms, i.e. Clostridium difficile and Clostridium perfringens (Rajilic‐Stojanovic and de Vos, 2014). Based on these notions, it seems that the BARF diet undermines canine and feline health. However, by definition, these opportunistic pathogens are not always harmful to the host, but only in case of perturbations of the bowel homeostasis. Under normal circumstances, such bacteria behave like commensal microorganisms in the intricate microbial ecosystem of the host’s intestine. In this context, a positive correlation appears to exist between Clostridiaceae and protein digestibility and protein dietary content, suggesting that members of this microbial family are involved in protein digestion in the GIT of dogs or that they can thrive in a carbohydrate‐poor environment (Bermingham et al., 2017). Similarly, Clostridiaceae were shown to positively correlate with faecal health scores (measured through firmness of faecal material), while a negative correlation was observed with faecal output, indicating that the increased abundance of Clostridiaceae taxa in the faecal microbiota of dogs fed on a BARF diet is not detrimental for canine health, but, rather, is associated with protein degradation (Bermingham et al., 2017). To further confirm the involvement of this taxon in protein fermentation, various Clostridium species, in particular Clostridium ramosum, Clostridium rectum, Clostridium hiranonis and Clostridium perfringens, were detected at a significantly higher relative abundance level in faecal samples of healthy dogs fed on a natural diet when compared to the control group (Kim et al., 2017). In addition, it has recently been demonstrated that Clostridium perfringens is, in carnivores, responsible for the generation of butyrate from protein due to the presence, in its genetic repertoire, of a butyrate kinase‐encoding gene involved in the main butyrate‐synthesis pathway (Vital et al., 2015). Similarly, as mentioned in the previous paragraph, also some members of the Fusobacterium genus are able to produce butyrate via amino acid fermentation, starting from protein degradation (Potrykus et al., 2008). Therefore, the ability of proteolytic microorganisms to produce SCFAs may explain why only the faecal organic acid profiles were shown to be affected by diet, but not the total level of SCFAs, in either dogs or cats (Sandri et al., 2017; Butowski et al., 2019).
Table 1.
Effects of feeding a raw meat‐based diet on canine and feline gut microbiota.
| Animal species | Effect of the raw meat‐based diet | References |
|---|---|---|
| Dog | Increase of the Fusobacteria and Proteobacteria phyla. Decrease of Firmicutes, especially of Ruminococcaceae and Erysipelotrichaceae family and the Faecalibacterium genus | Schmidt et al. (2018) |
| Dog | Diet affected 27 microbial families and 53 genera. Bacteroides, Prevotella, Peptostreptococcus and Faecalibacterium were reduced while Fusobacterium, Collinsella, Slakia Lactobacillus and Clostridium increased. | Bermingham et al. (2017) |
| Dog | Decreased proportion of Lactobacillus, Paralactobacillus and Prevotella with a parallel higher relative abundance of Clostridium, Bacteroides and Fusobacterium. | Sandri et al. (2017) |
| Dog | Decreased abundance of Prevotella, Faecalibacterium and Sutterella coupled with reduced GH families involved in the degradation of complex plant‐derived polysaccharides. Higher abundance of Fusobacteria and Actinobacteria and increased proportion of genes related to amino acid and fatty acid/lipid degradation pathways. | Alessandri et al. (2019) |
| Cat | Higher abundance of Fusobacterium, Eubacterium and Clostridium together with a reduced abundance of Prevotella | Butowski et al. (2019) |
| Cat | Greater proportions of Peptococcus, Pseudobutyrivibrio and significantly reduced abundance of Faecalibacterium and Succinivibrio. Higher abundance, yet not statistically significant of Fusobacterium and Clostridium. | Kerr et al. (2014) |
At the same time and in parallel with the numerical enhancement of proteolytic bacteria, a significant reduction was observed in the abundance of saccharolytic microorganisms, such as Prevotella and Faecalibacterium genera together with members of the Ruminococcaceae family (Herstad et al., 2017; Alessandri et al., 2019a, 2019b, 2019c). Notably, these taxa are generally associated with high‐fibre and carbohydrate intake since they are able to metabolize a wide variety of dietary glycans (David et al., 2014). In addition, the observation that differences in the proportion of macronutrients in the canine and feline diet cause gut microbiota compositional changes was confirmed from a functional point of view. Indeed, a shotgun metagenomics analysis involving a faecal sample per group of diet (commercial diet vs. BARF) revealed a higher abundance of GH families involved in the breakdown of complex plant‐derived polysaccharides being associated with a commercial food diet, while a greater proportion of genes involved in amino acid degradation and fatty acid/lipid degradation pathways was observed in the faecal samples from dog fed on a diet of raw meat (Alessandri et al., 2019a, 2019b, 2019c). In parallel, the RMBD was shown to play a role in modulating the intestinal metabolome as a BARF‐based diet causes an increase in faecal cholesterol and other metabolites, such as isomaltose, the inhibitory neurotransmitter gamma‐aminobutyric acid (GABA) and its precursor gamma‐hydroxybutyric acid (GHB; Schmidt et al., 2018).
Taken together, these changes emphasize the plasticity of the gut microbiota in adapting to different dietary components.
Evolution of the canine and feline intestinal community during their life span
For a long time, it was thought that development of the mammalian gut microbiota occurs immediately after birth. However, in recent years, the dogma of a sterile in utero environment has been challenged by different studies that have highlighted the existence, in humans and rat, of an intrauterine and placenta microbial community in healthy full‐term pregnancies (Aagaard et al., 2014; Perez‐Munoz et al., 2017; Mancino et al., 2019). Despite these publications, the assumption that the very first bacterial contamination of the gut microbiota may occur before birth is still highly controversial (Jimenez et al., 2008; Walker et al., 2017; Kuperman et al., 2019; Rackaityte et al., 2020). Instead, it is well accepted that the earliest colonizers of the mammalian GIT, including dogs and cats, are generally represented by facultative anaerobes, especially members of the Enterobacteriaceae family, which are believed to be responsible for the elimination of oxygen present in the gastrointestinal tract immediately following birth (Matamoros et al., 2013; Moon et al., 2018). This oxygen depletion event transforms the intestinal tract into a suitable environment for strictly anaerobic microorganisms (Moon et al., 2018). Substantial variations in the total bacterial load as well as in the taxonomic composition of the gut microbiota of newborns may occur depending on multiple factors, including mode of delivery (natural vs. C‐section delivery), type of feeding (breast or formula milk) and antibiotic exposure (Fugelli and Laerum, 1989; Del Chierico et al., 2015; Imoto et al., 2018). For example, it has been demonstrated that the gut microbiota of vaginally delivered infants, which have been in contact with the maternal vaginal and faecal microbiota, is generally colonized by mother‐to‐infant vertically transmitted microorganisms. Conversely, Caesarean section delivered infants are not directly exposed to maternal bacteria, and their intestinal tract is for this reason mainly colonized by skin‐ and environment‐associated microorganisms (Dominguez‐Bello et al., 2010; Backhed et al., 2015). Nevertheless, to date, no studies have attempted to evaluate the impact of delivery mode and/or feeding type on the canine and feline gut microbiota due to the difficulties in collecting faecal samples from pups and milk form dams. However, an ecological survey investigating the bifidobacterial population harboured by the intestinal tract of multiple mammals revealed the presence of shared bifidobacterial phylotypes among the faecal samples of mother–offspring dyads, including canine and feline dams with their respective puppies and kittens (Milani et al., 2017a, 2017b,2017a, 2017b). Furthermore, siblings from the same litter were shown to harbour a more similar bifidobacterial population when compared to pups from different litters (Milani et al., 2017a, 2017b,2017a, 2017b). These findings therefore support the notion that, from humans to other mammals, and including companion animals, vertical transmission events from mother to her offspring play a key role in defining the biodiversity and composition of the newborn gut microbiota. Furthermore, these results corroborate the current view that members of the Bifidobacterium genus are among the first colonizers of the intestinal tract of various animals that provide parental care to their offspring (Bunesova et al., 2014). However, in humans the relative abundance of this genus gradually declines during ageing, declining from the first years of life, adolescence and adulthood, to old age (Arboleya et al., 2016). A similar trend was observed for dogs and cats. Indeed, several studies reported a higher abundance of the Bifidobacterium taxa in puppies and kittens during the lactation and post‐weaning phases when compared to adult cats and dogs (Jia et al., 2011; Guard et al., 2017; Alessandri et al., 2019a, 2019b, 2019c).
Beyond the mode of delivery, feeding type represents another major factor that influences the early microbial colonization, thus playing a role in the modulation of the neonatal intestinal microbial population and relative functions. Indeed, mammalian milk not only acts as a vector for certain bacteria to transfer from mother to offspring, but it also provides a mix of nutrients as well as antimicrobial agents and immunoglobulins (Martin et al., 2003; O'Sullivan et al., 2015; Chastant‐Maillard et al., 2017; Milani et al., 2017a, 2017b,2017a, 2017b). In this context, it has been reported that canine milk is a natural source of Lactobacillus species that can be transmitted to suckling puppies (Martin et al., 2010). In parallel, a high abundance of lactobacilli was observed in the faeces and luminal content of puppies (Buddington, 2003; Vilson et al., 2018). Therefore, it is possible that canine milk, similarly to human milk, acts as a carrier of microorganisms that are then able to colonize the intestinal tract of pups. Certainly, studies aimed at evaluating the presence of shared Lactobacillus strains in milk and faecal samples of puppy or kitten and their corresponding canine or feline parents are required to validate this hypothesis.
Weaning marks another important step in the establishment and development of the host’s intestinal microbial community. As already observed in humans, the transition from a liquid to a solid and more diverse diet (around the fourth/fifth week of life in dogs and cats) is, indeed, generally accompanied by an increase in the biodiversity of the pups' gut microbiota (Backhed et al., 2015; Guard et al., 2017). In this context, a longitudinally study demonstrated that while Fusobacteria and Proteobacteria maintain similar percentages at all assessed time points (2, 21, 42 and 56 days of life), the Firmicutes phylum dominated the intestinal microbial community at 2 days of age, while Bacteroidetes was underrepresented to reach a higher relative abundance typical of an adult intestinal gut community starting from day 21 (Guard et al., 2017). Specifically, Lactobacillus, Clostridium spp. and Escherichia coli were prominent in the first days after birth to gradually decrease over time to give way to other multiple bacteria for intestinal colonization (Buddington, 2003; Guard et al., 2017). In a similar way, a culture‐based analysis revealed that Enterobacteriaceae and Enterococcus spp. dominated the gut microbiota of pre‐weanling kittens, while their abundance decreased throughout life (Masuoka et al., 2017). In addition, as observed in dogs, also the gut microbiota of lactating kittens is characterized by high numbers of Clostridium spp. but a low relative abundance of Bacteroides which, instead, will increase during adult life (Fahey et al., 2008). Although weaning causes profound changes in the gut microbiota of puppies and kittens with a bacterial community that begins to acquire the stability typical of the adult life, further changes may occur. Indeed, it has been demonstrated that eight weeks after birth the faecal microbial population of puppies is still significantly different from that of their mother, and by extension from that of adult dogs (Guard et al., 2017). Similarly, a shotgun metagenomic‐based longitudinal study of feline faecal samples revealed significant differences in both functional activities and taxonomic composition at 18 weeks of age when compared to the 30 week age point (Deusch et al., 2015). Despite the lack of long‐term studies focusing on the gut microbiota of dogs and cats during life, it is well accepted that following the evolutions of the first months of life, as it has been shown for humans, the canine and feline intestinal microbial community remains stable throughout adulthood to further changes during old age (O'Toole and Jeffery, 2015; Pilla and Suchodolski, 2019). In this context, human senescence is generally associated with consistent alterations in nutrition, lifestyle and physiology accompanied by a decline of the immune system (immunosenescence) resulting in a chronic low grade of inflammation with consequences on the intestinal microbial community (Candela et al., 2014; O'Toole and Jeffery, 2015). Indeed, ageing usually correlates with higher levels of gut oxygen concentration and reactive oxygen microbial species, thus inactivating strict anaerobes and compromising the overall functionality of the intestinal population, ultimately resulting in a reduced microbial biodiversity and pauperization of genes involved in SCFA production (Candela et al., 2014). In this context, age is, next to diet, an important factor affecting the gut microbiota composition of dogs and cats, which is consistent with similar observations in humans.
Metabolic disorders and alterations of the canine and feline gut microbiota
During recent decades, obesity and overweight have rapidly increased in pet populations, which has caused a major concern for companion animal health (Grzeskowiak et al., 2015). An imbalance between energy intake and energy expenditure is the main cause of obesity. However, additional factors contribute to the onset of this metabolic disorder. Indeed, as a consequence of domestication, dogs and cats have started to experience a sedentary life with reduced physical activity and ad libitum access to high‐caloric diet which, coupled with genetic predisposition and neutering, represent the basis of the multifactorial aetiology of obesity (Grzeskowiak et al., 2015; Osto and Lutz, 2015; Tal et al., 2020). In addition, advances in microbiome studies have demonstrated that gut microbiota plays a role in extra‐intestinal disorders, including obesity and diabetes (Park et al., 2015; Blake and Suchodolski, 2016; Forster et al., 2018, Chun et al., 2020). In this context, the relative abundance of the Actinobacteria and Fusobacteria phyla and Roseburia genus increased in obese dogs when compared to the lean group (Kieler et al., 2017; Chun et al., 2020). Specifically, among the Fusobacteria phylum, Fusobacterium mortiferum and Fusobacterium perfoetens were positively correlated with an overweight status, thus implying a contribution of these two bacterial species to canine obesity (Chun et al., 2020). Conversely, a general decrease of bacterial taxa involved in SCFA production was observed in obese dogs, including the Blautia and Eubacterium genera and Lachnospiraceae family (Handl et al., 2013; Forster et al., 2018). Furthermore, a parallel reduced abundance of carboxylic acids, encompassing linoleic acid, ferulic acid and colnelenic acid was registered in obese dogs (Forster et al., 2018). Since these metabolites act as antioxidants as well as anti‐inflammatory compounds, their decreased abundance may reflect the subclinical chronic inflammation and oxidative stress typical of an overweight condition (Forster et al., 2018). In obese cats, instead, Fusobacteria as well as Bacteroidetes and Clostridium cluster XIVa showed reduced abundance when compared to the lean group, while Enterobacteriaceae displayed an opposite trend (Kieler et al., 2016). A diet intervention based on monitoring calorie intake is the elective therapy for treating canine and feline obesity. Furthermore, inclusion of dietary prebiotics seems to aid in promoting health in overweight or obese dogs and cats (Li et al., 2017; Alexander et al., 2018; Apper et al., 2020). In this context, the administration of short‐chain fructo‐oligosaccharides to obese dogs was shown to impact on gut microbiota composition and fermentative activity coupled with an increased microbial biodiversity, which is considered a positive effect (Apper et al., 2020). Specifically, an increase in SCFA bacterial producers, i.e. Roseburia and Blautia, was observed in parallel to an increment in faecal butyrate concentration that not only acts as an anti‐inflammatory molecule decreasing intestinal permeability, but also regulates levels of anti‐inflammatory gut hormones such as the glucagon‐like peptide 1 (GLP‐1) responsible for enhancing glucose‐dependent insulin secretion by pancreatic β‐cells (Apper et al., 2020). Similarly, another study reported the increased abundance of butyrate in the faeces of obese dogs supplemented with inulin‐type fructans, as well as an increase in the relative abundance of Eubacterium and Turibicater, while the Proteobacteria phylum showed an opposite trend (Alexander et al., 2018).
Furthermore, an overweight condition is generally associated with several other comorbidities such as diabetes mellitus (DM), osteoarthritis or cardiovascular diseases that undermine canine and feline health (Grzeskowiak et al., 2015; Phillips et al., 2017). In this context, while dogs generally display type 1 DM, cats are more likely to be affected by type 2 DM (Wernimont et al., 2020). Similarly to obesity, altered gut microbiota composition has been related to diabetes (Blake and Suchodolski, 2016). Specifically, dogs with DM are characterized by both intestinal dysbiosis with higher proportion of Enterobacteriaceae and alterations of the faecal concentration of unconjugated bile acids (Jergens et al., 2019). In the same way, a decreased gut microbial biodiversity and a drastic reduction of butyrate‐producing bacteria occur in cats affected by type 2 DM (Kieler et al., 2019). However, data related to DM in cats and dogs are currently limited and therefore further analyses are required to better understand the possible role played by the gut microbiota to cause these metabolic disorders.
The canine and feline gut microbiota and GI diseases
The lifelong interaction between the complex microbial ecosystem that resides in the GIT and its host plays an essential role in influencing the host health status. Indeed, it has been convincingly demonstrated that a balanced microbial ecosystem is involved in multiple physiological and metabolic functions. Beyond its metabolic role in providing nutritional support to the host by degrading otherwise non‐digestible dietary compounds coupled with the production of various SCFAs and other metabolites that provide nutrients to colonocytes, the gut microbiota is believed to be engaged in a continuous dialogue with the host’s immune system (Honneffer et al., 2014). Notably, the intestinal microbial community modulates local and/or systemic immune responses, promotes intestinal barrier integrity, influences bowel homeostasis and functionality, and provides protection against enteropathogen colonization (Pickard et al., 2017; Alessandri et al., 2019a, 2019b, 2019c; Goto, 2019). The interactions between intestinal bacteria and the host immune system may be regulated either through direct contact between intestinal microorganisms and the innate immune system or indirectly by the release of microbial metabolites. The latter, in turn, can be represented by bacterial products such as SCFAs or vitamins as well as host primary metabolites that can be enzymatically converted into secondary metabolites by (elements of) the intestinal consortia (Suchodolski, 2016). In parallel, in order to ensure intestinal homeostasis, the gastrointestinal mucosa acts as a semipermeable barrier for nutrient absorption and immune sensing on the one hand, while preventing passage of potentially harmful microorganisms, compounds and antigens on the other (Salvo Romero et al., 2015). However, when functional or physiological impairments of the intestinal epithelial barrier occur, aberrant consequences such as alteration of intestinal permeability may affect intestinal homeostasis. Similarly, alterations in the abundance or in gut microbiota composition as well as drastic changes in its functional activities may cause dysregulation of the adaptive immune response, and activate an inflammatory process, which in turn may be associated with increased susceptibility to infections. One or a combination of these intestinal perturbations are generally related to the onset of different diseases such as chronic enteropathy (CE), acute haemorrhagic or non‐haemorrhagic diarrhoea syndrome, or even intestinal cancer in dogs and cats (Kieler et al., 2017; Gavazza et al., 2018; Minamoto et al., 2012; Redfern et al., 2017). In this context, several markers are considered as clear and common signs of intestinal dysbiosis (Heilmann and Steiner, 2018). Particularly, it seems that an increase of bioavailable oxygen in the intestinal lumen as a consequence of inflammation‐related enhanced intestinal barrier permeability plays a central role in the alteration of the gut microbiota composition in favour of facultative anaerobe proliferation, especially members of the Enterobacteriaceae family (Vazquez‐Baeza et al., 2016; Zeng et al., 2017). Furthermore, intestinal inflammation is commonly associated with bile acid dysmetabolism, i.e. altered bile acid transformation (Duboc et al., 2013). Primary bile acids are essential for dietary lipid digestion and absorption, but they also play an important anti‐inflammatory role. Indeed, once primary bile acids reach the hindgut compartments, they may be transformed into secondary bile acids by means of deconjugation and dehydroxylation through metabolic activities elicited by the intestinal microbial community with an accumulation of these secondary metabolites in the intestine where they can exert their anti‐inflammatory properties (Dawson and Karpen, 2015; Suchodolski, 2016). Specifically, while they counteract germination of C. difficile spores, they are also able to stimulate induction of GLP‐1, which is involved in increasing the amount of circulating insulin, or to activate the G protein‐coupled receptor TGR5 which, in turn, suppresses the pro‐inflammatory status induced by circulating bacterial cell wall‐associated lipopolysaccharides (LPS) typical of an inflammatory status (Dawson and Karpen, 2015; Blake and Suchodolski, 2016). Therefore, an imbalance in the primary to secondary bile acid ratio is an indication of intestinal dysbiosis with potential negative consequences for host metabolism (Guard et al., 2019). In addition, gut dysbiosis is commonly characterized by a significant reduction in the abundance of certain bacterial genera or species involved in the fermentation of complex carbohydrates, resulting in a decreased intestinal concentration of SCFAs, which play an important anti‐inflammatory role and which stimulate differentiation of intestinal helper T (Th) cell precursors into immune regulatory T cells (Treg) (Arpaia et al., 2013; Suchodolski, 2016). However, despite numerous studies in this field, it is not yet clear whether the functional and microbial taxonomic changes that occur in the intestine are the cause or effect of the inflammatory bowel diseases. This is made even more difficult for dogs and cats since in most cases the diagnosis of inflammatory bowel disease is established after a non‐responsive antibiotic treatment, which in itself will have altered the microbiota composition (Pilla and Suchodolski, 2019). However, investigating the microbial and metabolic alterations will be pivotal to identify potential diagnostic biomarkers in order to facilitate early detection of bowel diseases and to develop therapeutic interventions (Blake and Suchodolski, 2016). In the following sections, the main features of intestinal of both acute and chronic intestinal diseases as well as intestinal cancer of dogs and cats will be briefly covered.
Acute bowel diseases
The main intestinal diseases for dogs and cats correspond to acute uncomplicated diarrhoea (AD) and acute haemorrhagic diarrhoea syndrome (AHDS; Suchodolski et al., 2012a, 2012b,2012a, 2012b). These two intestinal diseases differ in clinical outcomes. Indeed, while AD is characterized by mild symptoms, the clinical consequences of AHDS are more severe with haemorrhagic diarrhoea, dehydration, lethargy and anorexia associated with haemorrhagic lesions in the intestinal mucosa (Unterer et al., 2014; Ziese et al., 2018). Despite histological and symptomatic differences between the two diseases, AD and AHDS display similar shifts in the intestinal microbial composition (Suchodolski et al., 2012a, 2012b,2012a, 2012b; Guard et al., 2015). In this context, a study performed on dogs affected by AD or AHDS observed that in both cases the relative abundance of Blautia, Faecalibacterium, Ruminococcus and Turicibacter genera had significantly decreased, while the relative abundance of Clostridium spp. was shown to increase when compared to healthy controls (Suchodolski et al., 2012a, 2012b,2012a, 2012b). In accordance with these observations, another trial revealed a significant increase in the abundance of Clostridium spp., especially Clostridium perfringens, in AD or AHDS‐associated canine faecal samples with a simultaneous reduction of the Prevotella, Blautia, Faecalibacterium taxa, Lachnospiraceae and Ruminococcaceae families (Guard et al., 2015). In concert with changes in the taxonomic composition, AD and AHDS are generally associated with shifts in the metabolites produced. In this context, it was observed that acute episodes of diarrhoea not only affect the SCFA profiles, but also serum and urine metabolites, suggesting that acute bowel diseases elicit effects on the overall host‐associated metabolic profile (Guard et al., 2015). Specifically, a reduction in propionic acid production was observed in faecal samples of dogs affected by acute diarrhoea, while, although many members of the genera whose abundance decreases in AD or AHDS are known SCFA producers, the total level of butyrate increased. This inconsistency, as the authors suggested, may be the consequence of a reduced absorption or utilization of butyrate by the compromised enterocytes (Guard et al., 2015). On the other side, an increased abundance of Clostridium spp. was observed in both AD and AHDS diseases (Stoeckel et al., 2012; Guard et al., 2015; Leipig‐Rudolph et al., 2018). Although these species are commensal inhabitants of the GIT of healthy dogs and cats, the pathological role of clostridia in AHDS has recently been demonstrated since clostridia‐like bacteria were found on the necrotic mucosal surface of endoscopic biopsies of dogs with AHDS coupled with an abnormal proliferation of C. perfringens (Minamoto et al., 2014; Leipig‐Rudolph et al., 2018). Further investigations identified the netF‐positive type A C. perfringens bacterium is responsible for intestinal lesions typical of AHDS. Indeed, the netF gene, which encodes a pore‐forming toxin with cytotoxic activities, was shown to be present in the genome of C. perfringens strains isolated from different intestinal biopsies of dogs with AHDS (Leipig‐Rudolph et al., 2018). Moreover, the observation that recovery from AHDS is generally associated with a significant decrease in the abundance of netF‐positive type A C. perfringens, further supports the involvement of this toxin in the necrotizing mucosal lesions typical of dogs suffering from this disease (Ziese et al., 2018).
Similar to what was observed for dogs, faecal samples of cats with diarrhoea were shown to contain a high abundance of Proteobacteria, especially gamma‐ and beta‐Proteobacteria, Clostridium and E. coli. However, C. perfringens did not show a statistically significant increased relative abundance in cats with AD, thus indicating that this species does not play a significant role in feline GI diseases (Suchodolski et al., 2015). Instead, a role of E. coli in episodes of feline diarrhoea was proposed, given its statistically significant increase and its involvement in other GI diseases (Suchodolski et al., 2015). However, further studies are required to evaluate the precise role of this bacterial species as a microbial marker of dysbiosis or as an enteropathogen that contributes to intestinal diseases in cats. It should be noted that certain other taxa, such as Faecalibacterium and Roseburia, were shown to elicit decreased relative abundance in faecal samples of cats suffering from AD (Suchodolski et al., 2015).
Chronic intestinal diseases
The term ‘chronic enteropathy’ (CE) refers to a heterogeneous group of intestinal disorders that are generally classified in accordance to their response to treatment, encompassing diet‐responsive enteropathy (FRE), antibiotic‐responsive enteropathy (ARE) and immunosuppressant‐responsive enteropathy (IRE) better known as idiopathic inflammatory bowel diseases (IBD; Dandrieux, 2016). Despite having different aetiologies, CE disorders in dogs and cats are characterized by the overlap of persistent and recurrent clinical signs, including histological evidence of intestinal inflammation as well as vomiting, diarrhoea, hyporexia, abdominal pain and weight loss (Dandrieux, 2016). Furthermore, CEs occur spontaneously with similar multifactorial aetiology considering a combination of genetic susceptibility of the host, aberrant host immune system, dietary and/or environmental factors and altered intestinal microbial ecosystem (Minamoto et al., 2012; Vazquez‐Baeza et al., 2016). Although the involvement of the intestinal microbiota in the aetiology of CE is widely accepted, it is still difficult to define specific microbial biomarkers that can be directly associated with these intestinal disorders, due to the lack of a standard protocol for the study of the intestinal microbial community (Redfern et al., 2017). However, some common features have been identified. In dogs with CE, the relative abundance of Fusobacteria as well as some representatives of the Bacteroidetes phylum, i.e. Bacteroidaceae and Prevotellaceae decrease compared with healthy controls (Suchodolski et al., 2012a, 2012b,2012a, 2012b). Similarly, the relative abundance of several members of the Firmicutes phylum declines in dogs with CE, including Megamonas, Ruminococcus, Faecalibacterium, Blautia and Turicibacter and Lachnospiraceae family (Suchodolski et al., 2012a, 2012b,2012a, 2012b; Minamoto et al., 2015, 2019; Xu et al., 2016). Conversely, certain members of the Proteobacteria phylum and specifically the Enterobacteriaceae family, including in particular E. coli, were shown to elicit a significant relative abundance increase in dogs with CE (Suchodolski et al., 2012a, 2012b,2012a, 2012b; Minamoto et al., 2015; Xu et al., 2016).
In accordance to what has been observed in dogs, a study involving 16S rRNA microbial profiling revealed that bacterial taxa belonging to the Ruminococcaceae family and to the Turicibacter genus are significantly less abundant in cats with CE than in healthy controls (Marsilio et al., 2019). In the same way, some members of the Bacteroidetes, especially Bacteroides plebeius which has been associated with remission in humans with inflammatory bowel disease, and Actinobacteria phyla showed a trend towards a decrease abundance in case of CE (Marsilio et al., 2019). In contrast, representatives of the Enterobacteriaceae and Streptococcaceae families appear to increase in CE‐associated feline faeces coupled with Desulfovibrio spp., which are known to be toxic sulfide producers (Inness et al., 2007; Marsilio et al., 2019). Furthermore, various studies, employing different experimental procedures, including 16S rRNA sequencing and fluorescence in situ hybridization (FISH), highlighted that cats with CE generally show a decrease in the relative abundance of the Bifidobacterium genus, a trend that had not been documented for dogs suffering from CE (Inness et al., 2007; Marsilio et al., 2019).
As expected from changes in the taxonomic composition, a dysbiotic gut microbiota is also characterized by functional shifts with consequent impacts on the production of bacterial metabolites. Several SCFA‐producing bacteria such as Ruminococcus, Faecalibacterium, Turicibacter and Lachnospiraceae family significantly decreased in dogs and cats with CE. In this context, it has been demonstrated that faecal concentration of total SCFAs was significantly lower in dogs with CE than in the control group. Further scrutiny of the SCFA profile revealed that while butyrate showed only a decreasing trend, acetate and propionate levels were significantly reduced, implying an impact on the host immune system since SCFAs possess anti‐inflammatory features (Minamoto et al., 2019). Particularly, propionate is known to inhibit pro‐inflammatory cytokine production such as IL‐6, IL‐8 and TNF‐α (Moylan et al., 2020) and to simultaneously stimulate the expression of the anti‐inflammatory cytokine IL‐10 coupled with the Foxp3 transcriptional factor which is crucial for regulation of intestinal inflammation (Smith et al., 2013). In this context, a reduction of the number of Foxp3‐positive Treg cells was recorded in the duodenal mucosa of dogs with IBD, suggesting that a decrease in intestinal propionate levels plays a role in the pathogenesis of IBD in dogs (Maeda et al., 2016). In addition to alterations in faecal SCFA levels, CE in dogs is associated with decreased amino acid metabolism, indicating that CE‐associated gut microbiota is responsible for dysfunctional protein metabolism in the presence of intestinal inflammation (Minamoto et al., 2015). Furthermore, alteration in serum metabolite profile was pointed out as a typical sign of CE in both dogs and cats (Minamoto et al., 2015; Xu et al., 2016; Sakai et al., 2018). Notably, significantly lower levels of circulating tryptophan were detected in dogs and cats with CE, encompassing dogs with protein‐losing enteropathy, which is a particular form of CE characterized by hypoproteinaemia due to a drastic loss of protein in the intestinal tract (Kathrani et al., 2018; Sakai et al., 2018; Tamura et al., 2019). Tryptophan is an essential amino acid involved in protein synthesis and acts as a precursor for several bioactive compounds such as serotonin, melatonin and kynurenine (Richard et al., 2009). In addition, tryptophan may be used by intestinal bacteria in order to produce a range of indole compounds involved in activating anti‐inflammatory pathways (Lavelle and Sokol, 2020). Altogether, these findings highlight that tryptophan plays an important role in intestinal inflammatory diseases.
Bowel cancer
Besides acute or chronic gut diseases, alterations of the intestinal homeostasis are also believed to be involved in colonic carcinogenesis (Grzeskowiak et al., 2015; Gavazza et al., 2018). In fact, several studies have incriminated bacterial‐induced chronic bowel inflammation as promoter of a tumour‐permissive environment characterized by intestinal bacteria translocation into the circulatory system as well as mucosal infiltrations of tumour progression‐related cells (Garraway et al., 2018). Furthermore, in humans, a dysbiotic microbiota triggers a series of innate and adaptive immune responses involved in tumour genesis as well as the production of microbial metabolites such as lipoteichoic acids that through their binding to Toll‐like receptor 2 causes excessive secretion of pro‐inflammatory compounds or secondary bile acids that in this case can be detrimental by activating G protein‐coupled bile receptor 1, by promoting intestinal cell proliferation, DNA damage, cellular senescence and ultimately carcinogenesis (Meng et al., 2018). Although the relationship between intestinal dysbiosis and colorectal cancer has been widely debated in humans, only a small number of studies have been focused on the correlation between bowel cancer and gut microbiota in dogs and cats and, what is more, with discordant results (Omori et al., 2017; Garraway et al., 2018; Gavazza et al., 2018; Herstad et al., 2018). However, while the relative abundance of the Fusobacterium genus and the Enterobacteriaceae family increased in ileum and colon biopsies from cats with small cell GI lymphoma (Garraway et al., 2018), no differences of the abundance of these two bacterial taxa were observed in dogs affected by intestinal lymphoma when compared to the control (Omori et al., 2017; Gavazza et al., 2018; Herstad et al., 2018) and even one study reported a decreased abundance of Fusobacterium spp. in canine lymphoma biopsies (Gavazza et al., 2018). Conversely, while Faecalibacterium seems to decrease in case of canine lymphoma, the Streptococcus genus showed an opposite trend (Gavazza et al., 2018; Herstad et al., 2018). Despite these observations, further studies are required to comprehensively understand the implications of the gut microbiota in the onset of intestinal tumours in dogs and cats.
Therapeutic strategies for the treatment of inflammatory bowel diseases
In recent years, several treatments have been tested to try to restore the intestinal homeostasis in case of CE by manipulating the intestinal bacterial community. Antibiotics, prebiotics, probiotics, synbiotics, corticosteroids or even, though in exceptional cases, faecal microbiota transplantation (FMT) have been employed as therapeutic treatments (Manchester et al., 2019; Pilla et al., 2019; Sugita et al., 2019). However, there is no standard treatment that offers the best strategy to adopt for this scope. Indeed, typical treatments involve sequential trials starting with the less invasive diet, followed by antibiotics and ultimately immune‐suppressive drugs in non‐responsive dogs or cats (Dandrieux, 2016; Dandrieux et al., 2019).
Antibiotics have long been considered to represent the first port of call and indeed the gold standard for treatment of acute or chronic intestinal inflammation, and antibiotics are still considered one of the main components of sequential therapy for dogs and cats with CE. However, if the effectiveness of the use of antibiotics has been reported in case of infections, their real benefit to treat CE is uncertain since antibiotics are surrounded by many contradictions. Indeed, the employment of antibiotics can expose animals to risk factors such as a significant decline of the intestinal microbial biodiversity, reduction of beneficial bacteria and development of antibiotic‐resistant microorganisms (Suchodolski et al., 2009; Dandrieux et al., 2019; Werner et al., 2020). Metronidazole and/or tylosin are the most commonly prescribed antibiotics to treat GI diseases (Mondo et al., 2019). However, although several trials highlighted remission of dogs and cats after metronidazole or tylosin treatments, these antimicrobials are often administered in combination with dietary therapy or other drugs, thus preventing a complete understanding of the true impact of antibiotics in CE treatment (Munster et al., 2006; Makielski et al., 2019). In this context, the administration of prednisone (a corticosteroid) to dogs with IBD resulted as effective as the combined treatment with both prednisone and metronidazole (Jergens et al., 2010). In a similar way, the treatment with amoxicillin/clavulanic acid did not reduce mortality rate, duration of hospitalization or severity of clinical signs in dogs with AHDS, thus emphasizing the marginal ole of antibiotics in counteracting CE (Unterer et al., 2011). Furthermore, tylosin treatment induced a significant decrease in commensal taxa such as Fusobacterium, Faecalibacterium, Blautia and C. hiranonis, while Enterococcaceae and Peptostreptococcaceae increased probably due to their intrinsic or acquired antimicrobial resistance, thus arguing against the hypothesis that tylosin may elicit a beneficial effect in the restoration of a dysbiotic microbiota (Manchester et al., 2019). In addition, different studies reported that CE response to antibiotics is short‐lasting after cessation of the treatment with a high number of relapsing cases within a month (Dandrieux et al., 2019). Based on these findings, other solutions such as prebiotics, probiotics or synbiotics may be preferred for CE treatment.
As already mentioned in this review, prebiotics are non‐digestible compounds (frequently represented by fibres or carbohydrates) that promote proliferation of beneficial bacteria residing in the GIT of the host (Gibson et al., 2004). Probiotics, instead, are live microorganisms that, when consumed in adequate amounts, are able to confer health benefits to the host (Grzeskowiak et al., 2015). Among the various mechanisms of action, probiotics may exert their health benefits by inhibiting pathogenic bacteria by competition for nutrients or mucosal adhesion sites, by improving the intestinal barrier functions or by enhancing the immune responses (Sanchez et al., 2017). Synbiotics are formulated as a combination of synergistically acting probiotics and prebiotics aimed at promoting the survival and implantation of exogenous live microorganisms (i.e. the probiotic) through the supply of specific carbohydrates or fibres (representing the prebiotic; Gibson and Roberfroid, 1995; Markowiak and Slizewska, 2017). However, although several studies reported the beneficial effects of probiotics‐ and/or synbiotics‐based therapies to canine with CE (Table 2), no increase in the relative abundance of the administered microorganisms occur, thus suggesting that probiotic or synbiotic treatments have only negligible and transient effects on faecal microbial community (Garcia‐Mazcorro et al., 2011; Larsen et al., 2011; Rossi et al., 2014). For this reason, probiotic administration may be associated with standard immunosuppressive treatment.
Table 2.
Effects of probiotic or symbiotic administration to dogs with CE.
| Treatment | Type of disorder | Effect of the treatment | References |
|---|---|---|---|
| High‐fibre diet with probiotic blend | FRE | Resolution of clinical signs with improvement of faecal scores and Canine Chronic Enteropathy Clinical Activity Index (CCECAI) and histologic amelioration. | Rossi et al. (2020) |
| Enterococcus faecium NCIMB 10415 E1707 with FOS and gum Arabic | FRE | No differences in clinical efficacy, histologic scores or expression of specific cytokines emerged after the treatment. | Schmitz et al. (2015) |
| Enterococcus faecium NCIMB 10415 E1707 with FOS and gum Arabic | FRE | Increased intestinal biodiversity coupled with a slight increment of the Enterococcus genus relative abundance | Pilla et al. (2019) |
| Sour milk with three canine‐derived Lactobacillus strains | AD | Normalizing effects in stool consistency and improvement of the animal conditions with reduced vomiting and increased appetite. Remarkable reduction of C. perfringens alphatoxin‐ or enterotoxin‐producing strains | Gomez‐Gallego et al. (2016) |
| Probiotic VSL#3 | IBD | Increased relative abundance of the Faecalibacterium genus and improvement of histological scores with enhancement of regulatory T‐cell markers (FoxP3+ and TGF‐β) | Rossi et al. (2014) |
| Multi‐strain probiotic | IBD | Increased relative abundance of Lactobacillus spp., rapid clinical remission and increased expression of tight junction proteins | White et al. (2017) |
Although several studies have been carried out to understand the impact of probiotic/synbiotic‐based therapies on dogs with CE, currently available literature regarding such studies in cats is extremely scarce. Indeed, only a single study reported the use of a probiotic to treat intestinal disorders such as chronic constipation or idiopathic megacolon (Rossi et al., 2018). In this case, the administration of a probiotic blend resulted in significant clinical improvements coupled with reduced mucosal infiltration and enhanced histological parameters suggesting an anti‐inflammatory effect of the probiotic blend (Rossi et al., 2018). In light of this data and considering the benefits observed in dogs with CE, it is plausible to suggest that also in cats, treatments with probiotics or synbiotics may result in beneficial effects in case of intestinal bowel diseases. However, large‐scale trials are needed in order to support this hypothesis.
A newly emerging experimental frontier in the treatment of human intestinal diseases is represented by FMT, a procedure that aims to restore the dysbiotic gut microbiota of a patient affected by intestinal diseases by endoscopically or colonoscopically administering the faecal matter from a healthy donor (Chaitman et al., 2016). In human medicine, FMT has been successfully employed to treat recurrent and refractory C. difficile infections. In this situation, FMT is currently considered one of the most effective and safe solutions to eradicate C. difficile infection when compared to antibiotic therapies (Seekatz et al., 2014; Quraishi et al., 2017; Hui et al., 2019). Furthermore, the application of this procedure in other clinical settings, such as human IBD, has also resulted in beneficial outcomes for patients (Blanchaert et al., 2019). Based on these findings, FMT trials have been successfully performed in the veterinary field for the treatment of intestinal diseases in dogs and cats (Pereira et al., 2018; Niina et al., 2019; Sugita et al., 2019), resulting in successful recovery of a cat with IBD (Furmanski and Mor, 2017), faster resolution of diarrhoea and shorter hospitalization of puppies affected by parvovirus infection (Pereira et al., 2018), complete remission of both a dog affected by intermittent large bowel diarrhoea and C. difficile antigen and toxin A&B genes and proteins in its faecal sample and a dog with a prolonged history of vomiting and diarrhoea (Niina et al., 2019; Sugita et al., 2019).
Despite such promising outcomes, further clinical trials are required to develop a reliable protocol for a standardized FMT treatment in dogs and cats. Indeed, several factors that may influence the success of the FMT treatment should be taken into consideration, including the preservation of the donor faecal sample, timing of administration (single or multiple treatment) and the method of faecal supplementation (endoscopy, nasogastric tube, capsules, colonoscopy or retention enema).
The impact of human–pet interplay on gut microbiota composition of both humans and companion animals
Humans have been identified as major players in driving several evolutionary events, encompassing extinction or speciation through domestication, relocation and creation of novel ecosystems (Bull and Maron, 2016). Specifically, through domestication, humans have deliberately altered feeding, behaviour, habitat and genetic heritage of multiple animal species (Wang et al., 2013; Frantz et al., 2020). During the course of evolution, dogs and cats were among the first animals to have undergone a long and extensive domestication process starting from their direct ancestors, the grey wolf and wild cat, respectively, leading to the selection of a wide range of canine and feline breeds (Savolainen et al., 2002; Baca et al., 2018). However, apart from animal phenotypic and genotypic alterations, several studies have demonstrated that the anthropogenic influence has significantly modified the intestinal microbiota of domesticated animals when compared to their close, yet wild relatives (Metcalf et al., 2017; Alessandri et al., 2019a, 2019b, 2019c). In this milieu, dietary shifts, antibiotic exposure, reduced contact with nature and a concomitant close interaction with humans are only some of the multiple substantial changes imposed by domestication that may have impacted on the intestinal microbial community of animals (Ferrario et al., 2017; McKenzie et al., 2017). A recent study investigated the effects of artificial selection and close contact with humans on canine gut microbiota, comparing the 16S rRNA gene sequencing‐based core gut microbiota of dogs with that of humans and wolves (Alessandri et al., 2019a, 2019b, 2019c). This comparison highlighted that while six bacterial genera belonging to the core gut microbiota of wolves, i.e. Alistipes, Pseudomonas, Slackia, Subdoligranulum, Eubacterium coprostanoligenes and Barnesiella, have apparently been lost by the domesticated canine core gut microbiota, the latter have acquired five other microbial taxa, including Dorea, Parabacteroides, Streptococcus, U. m. of Bacteroidales order and U. m. of Clostridiales order, which are typical components of the human core intestinal microbiota (Alessandri et al., 2019a, 2019b, 2019c). These findings suggest that the shift from a natural and undomesticated lifestyle to cohabitation with humans has modulated the intestinal microbial community of domesticated dogs through horizontal transmission events. To support this observation, comparison of the gut microbiota of wolves with that of domestic dogs revealed that while the Streptococcus genus was absent in the intestinal community of wolves, it was detected in the canine one (Wu et al., 2017). Furthermore, representatives of the Cyanobacteria phylum were exclusively present in wolves, while members of the Verrucomicrobia taxa were found only in canine faeces (Wu et al., 2017). This strengthens the notion that human interference has played a role in the modulation of canine gut micro‐ecology. Moreover, it has been demonstrated that changes in feeding habits due to domestication has led to the differentiation in both composition and functional activities of the gut microbiota of dogs when compared to wolves (Lyu et al., 2018). Specifically, some bacterial taxa associated with a complex polysaccharide‐rich diet, including Ruminococcaceae, Desulfuromusa, Lactobacillus, Carnobacillus and Faecalibacterium, showed a significantly higher relative abundance in dogs, thus indicating that the composition of the canine gut microbiota has been influenced by dietary changes imposed by humans (Lyu et al., 2018). To further support this notion, it was observed that the canine gut microbiome is enriched in genes involved in carbohydrate metabolism pathways as well as genes encoding for GHs when compared to that present in wolves (Lyu et al., 2018).
However, cohabitation and the resulting close relationship between humans and their pets have not only significantly influenced the animal microbiota, but also the human‐associated microbial community (Song et al., 2013). In particular, it has been demonstrated that dog ownership influences the human skin microbiota. Indeed, the presence of a dog within a family generally leads to a higher microbial biodiversity of the adult skin, including hands and forehead, when compared to adults without dogs (Song et al., 2013). Furthermore, it seems that dogs may act as carriers of bacteria since not only dogs and their owners share various bacterial phylotypes between fur and skin, but it was also highlighted that adults that live together and simultaneously possess a pet share more skin bacteria than a couple without pets (Song et al., 2013). Pet ownership does not only impact on the adult microbial communities, but it also appears to play a role in modifying the infant gut microbiota. Indeed, a recent study showed that pre‐ and/or post‐natal exposure to household pets influences the infant gut microbiota (Tun et al., 2017). In general, regardless of birth variables, including type of delivery or intrapartum antibiotic prophylaxis, pet exposure induces a significant increase in species richness in the Firmicutes phylum, especially the Ruminococcus and Oscillospira genera, in the infant intestinal microbial community (Tun et al., 2017). In detail, while Oscillospira is associated with leanness and lower body mass index in both adults and infants, Ruminococcus is linked to the maintenance of the intestinal barrier integrity (Tun et al., 2017). Furthermore, both Ruminococcus and Oscillospira have been negatively related to the development of infant atopy and obesity, respectively, suggesting that pet‐associated microbiota may have a role in modulating the infant gut microbiota (Tun et al., 2017). In addition, it has been reported that perinatal pet exposure may impact on the composition of the infant intestinal microbiota and solicit the immune responses, thus inducing protection against infant wheezy bronchitis (Nermes et al., 2013). Despite the abovementioned positive influence of vertical transmission in dogs and cats, these animals may also act as a reservoir for opportunistic pathogens, such as Escherichia coli, Salmonella and Campylobacter that may be transmitted horizontally, and may thus cause zoonotic enteric diseases to humans (Morato et al., 2009; Lambertini et al., 2016; Moon et al., 2018). At the same time, the pet intestinal ecosystem may be a vector of antimicrobial resistance, thus representing a serious global health safety issue (De Graef et al., 2004). Overall, these results show that cohabitation and the close relationship between pet and their owners play an important, yet varied role in gut microbiota modulation with repercussions on the health of both parties.
Conclusions
During the course of evolution, dogs and cats have become the main companion animals for humans. Being an integral part of human life, interest for pet health and well‐being has rapidly increased during recent decades and has consequently promoted many studies concerning the canine and feline gut microbiota, which are known to influence the health status of the host. In this perspective, it has been demonstrated that diet, age, anthropogenic influences and several other environmental factors play a role in modulating both the taxonomical composition and the functional activities of the intestinal microbial community of dogs and cats. Furthermore, alterations of the intestinal homeostasis are generally associated with a dysbiotic gut microbiota and the onset of inflammatory bowel diseases that may be treated with different therapies, including prebiotics, probiotics, synbiotics, antibiotics or FMT as for humans IBD. Nevertheless, further research is necessary to deepen our understanding of the role of this microbial community and to better appreciate how it interacts with dogs and cats.
Conflict of interest
The authors declare that they have no competing interests.
Acknowledgments
This work was primarily funded by the EU Joint Programming Initiative – A Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) to DvS (in conjunction with Science Foundation Ireland [SFI], grant number 15/JP‐HDHL/3280). DvS is member of APC microbiome Ireland, funded by SFI through the Irish Government’s National Development Plan (grant numbers SFI/12/RC/2273‐P1 and SFI/12/RC/2273‐P2). The study is supported by Fondazione Cariparma, under TeachInParma Project (DvS and GA). We furthermore thank GenProbio srl for financial support of the Laboratory of Probiogenomics. This research benefited from the HPC (high‐performance computing) facility of the University of Parma, Italy.
Microbial Biotechnology (2020) 13(6), 1708–1732
Funding information
This work was primarily funded by the EU Joint Programming Initiative – A Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) to DvS (in conjunction with Science Foundation Ireland [SFI], grant number 15/JP‐HDHL/3280). DvS is member of APC microbiome Ireland, funded by SFI through the Irish Government’s National Development Plan (grant numbers SFI/12/RC/2273‐P1 and SFI/12/RC/2273‐P2). The study is supported by Fondazione Cariparma, under TeachInParma Project (DvS and GA).
References
- Aagaard, K. , Ma, J. , Antony, K.M. , Ganu, R. , Petrosino, J. , and Versalovic, J. (2014) The placenta harbors a unique microbiome. Sci Transl Med 6: 237–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessandri, G. , Milani, C. , Mancabelli, L. , Mangifesta, M. , Lugli, G.A. , Viappiani, A. , et al (2019a) The impact of human‐facilitated selection on the gut microbiota of domesticated mammals. FEMS Microbiol Ecol 95: fiz121. [DOI] [PubMed] [Google Scholar]
- Alessandri, G. , Milani, C. , Mancabelli, L. , Mangifesta, M. , Lugli, G.A. , Viappiani, A. , et al (2019b) Metagenomic dissection of the canine gut microbiota: insights into taxonomic, metabolic and nutritional features. Environ Microbiol 21: 1331–1343. [DOI] [PubMed] [Google Scholar]
- Alessandri, G. , Ossiprandi, M.C. , MacSharry, J. , van Sinderen, D. , and Ventura, M. (2019c) Bifidobacterial dialogue with its human host and consequent modulation of the immune system. Front Immunol 10: 2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessandri, G. , Milani, C. , Mancabelli, L. , Longhi, G. , Anzalone, R. , Lugli, G.A. , et al (2020) Deciphering the bifidobacterial populations within the canine and feline gut microbiota. Appl Environ Microbiol 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, C. , Cross, T.L. , Devendran, S. , Neumer, F. , Theis, S. , Ridlon, J.M. , et al (2018) Effects of prebiotic inulin‐type fructans on blood metabolite and hormone concentrations and faecal microbiota and metabolites in overweight dogs. Br J Nutr 120: 711–720. [DOI] [PubMed] [Google Scholar]
- Algya, K.M. , Cross, T.L. , Leuck, K.N. , Kastner, M.E. , Baba, T. , Lye, L. , et al (2018) Apparent total‐tract macronutrient digestibility, serum chemistry, urinalysis, and fecal characteristics, metabolites and microbiota of adult dogs fed extruded, mildly cooked, and raw diets 1. J Anim Sci 96: 3670–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhinai, E.A. , Walton, G.E. , and Commane, D.M. (2019) The role of the gut microbiota in colorectal cancer causation. Int J Mol Sci 20: 5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.C. , Waldmeier, P.C. , and Tipton, K.F. (1991) The inhibition of monoamine oxidase by brofaromine. Biochem Pharmacol 41: 1871–1877. [DOI] [PubMed] [Google Scholar]
- Apper, E. , Privet, L. , Taminiau, B. , Le Bourgot, C. , Svilar, L. , Martin, J.C. , and Diez, M. (2020) Relationships between gut microbiota, metabolome, body weight, and glucose homeostasis of obese dogs fed with diets differing in prebiotic and protein content. Microorganisms 8: 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arboleya, S. , Watkins, C. , Stanton, C. , and Ross, R.P. (2016) Gut Bifidobacteria Populations In Human Health And Aging. Front Microbiol 7: 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia, N. , Campbell, C. , Fan, X. , Dikiy, S. , van der Veeken, J. , deRoos, P. , et al (2013) Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature 504: 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca, M. , Popovic, D. , Panagiotopoulou, H. , Marciszak, A. , Krajcarz, M. , Krajcarz, M.T. , et al (2018) Human‐mediated dispersal of cats in the Neolithic Central Europe. Heredity 121: 557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed, F. , Roswall, J. , Peng, Y. , Feng, Q. , Jia, H. , Kovatcheva‐Datchary, P. , et al (2015) Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17: 690–703. [DOI] [PubMed] [Google Scholar]
- Barry, K.A. , Wojcicki, B.J. , Middelbos, I.S. , Vester, B.M. , Swanson, K.S. , and Fahey, G.C. Jr (2010) Dietary cellulose, fructooligosaccharides, and pectin modify fecal protein catabolites and microbial populations in adult cats. J Anim Sci 88: 2978–2987. [DOI] [PubMed] [Google Scholar]
- Barry, K.A. , Middelbos, I.S. , Vester Boler, B.M. , Dowd, S.E. , Suchodolski, J.S. , Henrissat, B. , et al (2012) Effects of dietary fiber on the feline gastrointestinal metagenome. J Proteome Res 11: 5924–5933. [DOI] [PubMed] [Google Scholar]
- Bermingham, E.N. , Maclean, P. , Thomas, D.G. , Cave, N.J. , and Young, W. (2017) Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ 5: e3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, A.B. , and Suchodolski, J.S. (2016) Importance of gut microbiota for the health and disease of dogs and cats. Anim Front 6: 37–42. [Google Scholar]
- Blanchaert, C. , Strubbe, B. , and Peeters, H. (2019) Fecal microbiota transplantation in ulcerative colitis. Acta Gastroenterol Belg 82: 519–528. [PubMed] [Google Scholar]
- van Bree, F.P.J. , Bokken, G. , Mineur, R. , Franssen, F. , Opsteegh, M. , van der Giessen, J.W.B. , et al (2018) Zoonotic bacteria and parasites found in raw meat‐based diets for cats and dogs. Vet Rec 182: 50. [DOI] [PubMed] [Google Scholar]
- Bresciani, F. , Minamoto, Y. , Suchodolski, J.S. , Galiazzo, G. , Vecchiato, C.G. , Pinna, C. , et al (2018) Effect of an extruded animal protein‐free diet on fecal microbiota of dogs with food‐responsive enteropathy. J Vet Intern Med 32: 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddington, R.K. (2003) Postnatal changes in bacterial populations in the gastrointestinal tract of dogs. Am J Vet Res 64: 646–651. [DOI] [PubMed] [Google Scholar]
- Bull, J.W. , and Maron, M. (2016) How humans drive speciation as well as extinction. Proc Biol Sci 283: 20160600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunesova, V. , Vlkova, E. , Rada, V. , Killer, J. , and Musilova, S. (2014) Bifidobacteria from the gastrointestinal tract of animals: differences and similarities. Benef Microbes 5: 377–388. [DOI] [PubMed] [Google Scholar]
- Butowski, C.F. , Thomas, D.G. , Young, W. , Cave, N.J. , McKenzie, C.M. , Rosendale, D.I. , and Bermingham, E.N. (2019) Addition of plant dietary fibre to a raw red meat high protein, high fat diet, alters the faecal bacteriome and organic acid profiles of the domestic cat (Felis catus). PLoS One 14: e0216072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela, M. , Biagi, E. , Brigidi, P. , O'Toole, P.W. , and De Vos, W.M. (2014) Maintenance of a healthy trajectory of the intestinal microbiome during aging: a dietary approach. Mech Ageing Dev 136–137: 70–75. [DOI] [PubMed] [Google Scholar]
- Castellarin, M. , Warren, R.L. , Freeman, J.D. , Dreolini, L. , Krzywinski, M. , Strauss, J. , et al (2012) Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaitman, J. , Jergens, A.E. , Gaschen, F. , Garcia‐Mazcorro, J.F. , Marks, S.L. , Marroquin‐Cardona, A.G. , et al (2016) Commentary on key aspects of fecal microbiota transplantation in small animal practice. Vet Med 7: 71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty, S. , Helb, D. , Burday, M. , Connell, N. , and Alland, D. (2007) A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods 69: 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastant‐Maillard, S. , Aggouni, C. , Albaret, A. , Fournier, A. , and Mila, H. (2017) Canine and feline colostrum. Reprod Domest Anim 52(Suppl 2): 148–152. [DOI] [PubMed] [Google Scholar]
- Chun, J.L. , Ji, S.Y. , Lee, S.D. , Lee, Y.K. , Kim, B. , and Kim, K.H. (2020) Difference of gut microbiota composition based on the body condition scores in dogs. J Anim Sci Technol 62: 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandrieux, J.R. (2016) Inflammatory bowel disease versus chronic enteropathy in dogs: are they one and the same? J Small Anim Pract 57: 589–599. [DOI] [PubMed] [Google Scholar]
- Dandrieux, J. , Martinez Lopez, L.M. , Prakash, N. , and Mansfield, C.S. (2019) Treatment response and long term follow up in nineteen dogs diagnosed with chronic enteropathy in Australia. Aust Vet J 97: 301–307. [DOI] [PubMed] [Google Scholar]
- David, L.A. , Maurice, C.F. , Carmody, R.N. , Gootenberg, D.B. , Button, J.E. , Wolfe, B.E. , et al (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, P.A. , and Karpen, S.J. (2015) Intestinal transport and metabolism of bile acids. J Lipid Res 56: 1085–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Graef, E.M. , Decostere, A. , Devriese, L.A. , and Haesebrouck, F. (2004) Antibiotic resistance among fecal indicator bacteria from healthy individually owned and kennel dogs. Microb Drug Resist 10: 65–69. [DOI] [PubMed] [Google Scholar]
- Del Chierico, F. , Vernocchi, P. , Petrucca, A. , Paci, P. , Fuentes, S. , Pratico, G. , et al (2015) Phylogenetic and metabolic tracking of gut microbiota during perinatal development. PLoS One 10: e0137347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, P. , and Swanson, K.S. (2015) Gut microbiota of humans, dogs and cats: current knowledge and future opportunities and challenges. Br J Nutr 113(Suppl): S6–S17. [DOI] [PubMed] [Google Scholar]
- Deusch, O. , O'Flynn, C. , Colyer, A. , Swanson, K.S. , Allaway, D. , and Morris, P. (2015) A longitudinal study of the feline faecal microbiome identifies changes into early adulthood irrespective of sexual development. PLoS One 10: e0144881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez‐Bello, M.G. , Costello, E.K. , Contreras, M. , Magris, M. , Hidalgo, G. , Fierer, N. , and Knight, R. (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 107: 11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron, L. , Coppenhagen‐Glazer, S. , Ibrahim, Y. , Eini, A. , Naor, R. , Rosen, G. , and Bachrach, G. (2014) Identification and characterization of fusolisin, the Fusobacterium nucleatum autotransporter serine protease. PLoS One 9: e111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson, M.J. , and Hyatt, E.M. (2008) Understanding dog‐human companionship. J Business Res 61: 457–466. [Google Scholar]
- Duboc, H. , Rajca, S. , Rainteau, D. , Benarous, D. , Maubert, M.A. , Quervain, E. , et al (2013) Connecting dysbiosis, bile‐acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 62: 531–539. [DOI] [PubMed] [Google Scholar]
- Fahey, G.C. Jr , Barry, K.A. , and Swanson, K.S. (2008) Age‐related changes in nutrient utilization by companion animals. Annu Rev Nutr 28: 425–445. [DOI] [PubMed] [Google Scholar]
- Ferrario, C. , Alessandri, G. , Mancabelli, L. , Gering, E. , Mangifesta, M. , Milani, C. , et al (2017) Untangling the cecal microbiota of feral chickens by culturomic and metagenomic analyses. Environ Microbiol 19: 4771–4783. [DOI] [PubMed] [Google Scholar]
- Forster, G.M. , Stockman, J. , Noyes, N. , Heuberger, A.L. , Broeckling, C.D. , Bantle, C.M. , and Ryan, E.P. (2018) A comparative study of serum biochemistry. Metabolome and microbiome parameters of clinically healthy, normal weight, overweight, and obese companion dogs. Top Companion Anim Med 33: 126–135. [DOI] [PubMed] [Google Scholar]
- Frantz, L.A.F. , Bradley, D.G. , Larson, G. , and Orlando, L. (2020) Animal domestication in the era of ancient genomics. Nat Rev Genet 21: 449–460. [DOI] [PubMed] [Google Scholar]
- Fredriksson‐Ahomaa, M. , Heikkila, T. , Pernu, N. , Kovanen, S. , Hielm‐Bjorkman, A. , and Kivisto, R. (2017) Raw meat‐based diets in dogs and cats. Vet Sci 4: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, L.M. , Chandler, M.L. , Hamper, B.A. , and Weeth, L.P. (2013) Current knowledge about the risks and benefits of raw meat‐based diets for dogs and cats. J Am Vet Med Assoc 243: 1549–1558. [DOI] [PubMed] [Google Scholar]
- Fugelli, P. , and Laerum, E. (1989) Hospitals and primary health care. Tidsskr Nor Laegeforen 109: 17–18. [PubMed] [Google Scholar]
- Furmanski, S. , and Mor, T. (2017) First case report of fecal microbiota transplantation in a cat in Israel. Isr J Vet Med 72: 35–41. [Google Scholar]
- Furrie, E. (2006) A molecular revolution in the study of intestinal microflora. Gut 55: 141–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Mazcorro, J.F. , Lanerie, D.J. , Dowd, S.E. , Paddock, C.G. , Grutzner, N. , Steiner, J.M. , et al (2011) Effect of a multi‐species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS Microbiol Ecol 78: 542–554. [DOI] [PubMed] [Google Scholar]
- Garraway, K. , Johannes, C.M. , Bryan, A. , Peauroi, J. , Rossi, G. , Zhang, M. , et al (2018) Relationship of the mucosal microbiota to gastrointestinal inflammation and small cell intestinal lymphoma in cats. J Vet Intern Med 32: 1692–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavazza, A. , Rossi, G. , Lubas, G. , Cerquetella, M. , Minamoto, Y. , and Suchodolski, J.S. (2018) Faecal microbiota in dogs with multicentric lymphoma. Vet Comp Oncol 16: E169–E175. [DOI] [PubMed] [Google Scholar]
- German, A.J. , Day, M.J. , Ruaux, C.G. , Steiner, J.M. , Williams, D.A. , and Hall, E.J. (2003) Comparison of direct and indirect tests for small intestinal bacterial overgrowth and antibiotic‐responsive diarrhea in dogs. J Vet Intern Med 17: 33–43. [DOI] [PubMed] [Google Scholar]
- Gethings‐Behncke, C. , Coleman, H.G. , Jordao, H.W.T. , Longley, D.B. , Crawford, N. , Murray, L.J. , and Kunzmann, A.T. (2020) Fusobacterium nucleatum in the colorectum and its association with cancer risk and survival: a systematic review and meta‐analysis. Cancer Epidemiol Biomarkers Prev 29: 539–548. [DOI] [PubMed] [Google Scholar]
- Gibson, G.R. , and Roberfroid, M.B. (1995) Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr 125: 1401–1412. [DOI] [PubMed] [Google Scholar]
- Gibson, G.R. , Probert, H.M. , Loo, J.V. , Rastall, R.A. , and Roberfroid, M.B. (2004) Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev 17: 259–275. [DOI] [PubMed] [Google Scholar]
- Gomez‐Gallego, C. , Junnila, J. , Mannikko, S. , Hameenoja, P. , Valtonen, E. , Salminen, S. , and Beasley, S. (2016) A canine‐specific probiotic product in treating acute or intermittent diarrhea in dogs: A double‐blind placebo‐controlled efficacy study. Vet Microbiol 197: 122–128. [DOI] [PubMed] [Google Scholar]
- Goto, Y. (2019) Epithelial cells as a transmitter of signals from commensal bacteria and host immune cells. Front Immunol 10: 2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeskowiak, L. , Endo, A. , Beasley, S. , and Salminen, S. (2015) Microbiota and probiotics in canine and feline welfare. Anaerobe 34: 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guard, B.C. , Barr, J.W. , Reddivari, L. , Klemashevich, C. , Jayaraman, A. , Steiner, J.M. , et al (2015) Characterization of microbial dysbiosis and metabolomic changes in dogs with acute diarrhea. PLoS One 10: e0127259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guard, B.C. , Mila, H. , Steiner, J.M. , Mariani, C. , Suchodolski, J.S. , and Chastant‐Maillard, S. (2017) Characterization of the fecal microbiome during neonatal and early pediatric development in puppies. PLoS One 12: e0175718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guard, B.C. , Honneffer, J.B. , Jergens, A.E. , Jonika, M.M. , Toresson, L. , Lawrence, Y.A. , et al (2019) Longitudinal assessment of microbial dysbiosis, fecal unconjugated bile acid concentrations, and disease activity in dogs with steroid‐responsive chronic inflammatory enteropathy. J Vet Intern Med 33: 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady, M. , and Knight, R. (2009) Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res 19: 1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handl, S. , Dowd, S.E. , Garcia‐Mazcorro, J.F. , Steiner, J.M. , and Suchodolski, J.S. (2011) Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol 76: 301–310. [DOI] [PubMed] [Google Scholar]
- Handl, S. , German, A.J. , Holden, S.L. , Dowd, S.E. , Steiner, J.M. , Heilmann, R.M. , et al (2013) Faecal microbiota in lean and obese dogs. FEMS Microbiol Ecol 84: 332–343. [DOI] [PubMed] [Google Scholar]
- Heilmann, R.M. , and Steiner, J.M. (2018) Clinical utility of currently available biomarkers in inflammatory enteropathies of dogs. J Vet Intern Med 32: 1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herstad, K.M.V. , Gajardo, K. , Bakke, A.M. , Moe, L. , Ludvigsen, J. , Rudi, K. , et al (2017) A diet change from dry food to beef induces reversible changes on the faecal microbiota in healthy, adult client‐owned dogs. BMC Vet Res 13: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herstad, K.M.V. , Moen, A.E.F. , Gaby, J.C. , Moe, L. , and Skancke, E. (2018) Characterization of the fecal and mucosa‐associated microbiota in dogs with colorectal epithelial tumors. PLoS One 13: e0198342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmann, B. , Al‐Ghalith, G.A. , Shields‐Cutler, R.R. , Zhu, Q. , Gohl, D.M. , Beckman, K.B. , et al (2018) Evaluating the information content of shallow shotgun metagenomics. mSystems 3: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honneffer, J.B. , Minamoto, Y. , and Suchodolski, J.S. (2014) Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J Gastroenterol 20: 16489–16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honneffer, J.B. , Steiner, J.M. , Lidbury, J.A. , and Suchodolski, J.S. (2017) Variation of the microbiota and metabolome along the canine gastrointestinal tract. Metabolomics 13: 13–26. [Google Scholar]
- Hooda, S. , Vester Boler, B.M. , Kerr, K.R. , Dowd, S.E. , and Swanson, K.S. (2013) The gut microbiome of kittens is affected by dietary protein: carbohydrate ratio and associated with blood metabolite and hormone concentrations. Br J Nutr 109: 1637–1646. [DOI] [PubMed] [Google Scholar]
- Hui, W. , Li, T. , Liu, W. , Zhou, C. , and Gao, F. (2019) Fecal microbiota transplantation for treatment of recurrent C. difficile infection: An updated randomized controlled trial meta‐analysis. PLoS One 14: e0210016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide, K. , Shinohara, M. , Yamagishi, S. , Endo, A. , Nishifuji, K. , and Tochio, T. (2020) Kestose supplementation exerts bifidogenic effect within fecal microbiota and increases fecal butyrate concentration in dogs. J Vet Med Sci 82: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto, N. , Morita, H. , Amanuma, F. , Maruyama, H. , Watanabe, S. , and Hashiguchi, N. (2018) Maternal antimicrobial use at delivery has a stronger impact than mode of delivery on bifidobacterial colonization in infants: a pilot study. J Perinatol 38: 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inness, V.L. , McCartney, A.L. , Khoo, C. , Gross, K.L. , and Gibson, G.R. (2007) Molecular characterisation of the gut microflora of healthy and inflammatory bowel disease cats using fluorescence in situ hybridisation with special reference to Desulfovibrio spp. J Anim Physiol Anim Nutr (Berl) 91: 48–53. [DOI] [PubMed] [Google Scholar]
- Jergens, A.E. , Crandell, J. , Morrison, J.A. , Deitz, K. , Pressel, M. , Ackermann, M. , et al (2010) Comparison of oral prednisone and prednisone combined with metronidazole for induction therapy of canine inflammatory bowel disease: a randomized‐controlled trial. J Vet Intern Med 24: 269–277. [DOI] [PubMed] [Google Scholar]
- Jergens, A.E. , Guard, B.C. , Redfern, A. , Rossi, G. , Mochel, J.P. , Pilla, R. , et al (2019) Microbiota‐related changes in unconjugated fecal bile acids are associated with naturally occurring, insulin‐dependent diabetes mellitus in dogs. Front Vet Sci 6: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, J. , Frantz, N. , Khoo, C. , Gibson, G.R. , Rastall, R.A. , and McCartney, A.L. (2011) Investigation of the faecal microbiota of kittens: monitoring bacterial succession and effect of diet. FEMS Microbiol Ecol 78: 395–404. [DOI] [PubMed] [Google Scholar]
- Jimenez, E. , Marin, M.L. , Martin, R. , Odriozola, J.M. , Olivares, M. , Xaus, J. , et al (2008) Is meconium from healthy newborns actually sterile? Res Microbiol 159: 187–193. [DOI] [PubMed] [Google Scholar]
- Johnston, K.L. , Swift, N.C. , Forster‐van Hijfte, M. , Rutgers, H.C. , Lamport, A. , Ballevre, O. , and Batt, R.M. (2001) Comparison of the bacterial flora of the duodenum in healthy cats and cats with signs of gastrointestinal tract disease. J Am Vet Med Assoc 218: 48–51. [DOI] [PubMed] [Google Scholar]
- Kalenyak, K. , Isaiah, A. , Heilmann, R.M. , Suchodolski, J.S. , and Burgener, I.A. (2018) Comparison of the intestinal mucosal microbiota in dogs diagnosed with idiopathic inflammatory bowel disease and dogs with food‐responsive diarrhea before and after treatment. FEMS Microbiol Ecol 94: fix173. [DOI] [PubMed] [Google Scholar]
- Kathrani, A. , Allenspach, K. , Fascetti, A.J. , Larsen, J.A. , and Hall, E.J. (2018) Alterations in serum amino acid concentrations in dogs with protein‐losing enteropathy. J Vet Intern Med 32: 1026–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr, K.R. , Dowd, S.E. , and Swanson, K.S. (2014) Faecal microbiota of domestic cats fed raw whole chicks v. an extruded chicken‐based diet. J Nutr Sci 3: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieler, I.N. , Molbak, L. , Hansen, L.L. , Hermann‐Bank, M.L. , and Bjornvad, C.R. (2016) Overweight and the feline gut microbiome ‐ a pilot study. J Anim Physiol Anim Nutr (Berl) 100: 478–484. [DOI] [PubMed] [Google Scholar]
- Kieler, I.N. , Shamzir Kamal, S. , Vitger, A.D. , Nielsen, D.S. , Lauridsen, C. , and Bjornvad, C.R. (2017) Gut microbiota composition may relate to weight loss rate in obese pet dogs. Vet Med Sci 3: 252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieler, I.N. , Osto, M. , Hugentobler, L. , Puetz, L. , Gilbert, M.T.P. , Hansen, T. , et al (2019) Diabetic cats have decreased gut microbial diversity and a lack of butyrate producing bacteria. Sci Rep 9: 4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. , An, J.U. , Kim, W. , Lee, S. , and Cho, S. (2017) Differences in the gut microbiota of dogs (Canis lupus familiaris) fed a natural diet or a commercial feed revealed by the Illumina MiSeq platform. Gut Pathog 9: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic, A.D. , Chun, E. , Robertson, L. , Glickman, J.N. , Gallini, C.A. , Michaud, M. , et al (2013) Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor‐immune microenvironment. Cell Host Microbe 14: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperman, A.A. , Zimmerman, A. , Hamadia, S. , Ziv, O. , Gurevich, V. , Fichtman, B. , et al (2019) Deep microbial analysis of multiple placentas shows no evidence for a placental microbiome. BJOG 127: 159–169. [DOI] [PubMed] [Google Scholar]
- Lambertini, E. , Buchanan, R.L. , Narrod, C. , and Pradhan, A.K. (2016) Transmission of bacterial zoonotic pathogens between pets and humans: the role of pet food. Crit Rev Food Sci Nutr 56: 364–418. [DOI] [PubMed] [Google Scholar]
- Larsen, N. , Vogensen, F.K. , Gobel, R. , Michaelsen, K.F. , Abu Al‐Soud, W. , Sorensen, S.J. , et al (2011) Predominant genera of fecal microbiota in children with atopic dermatitis are not altered by intake of probiotic bacteria Lactobacillus acidophilus NCFM and Bifidobacterium animalis subsp. lactis Bi‐07. FEMS Microbiol Ecol 75: 482–496. [DOI] [PubMed] [Google Scholar]
- Lavelle, A. , and Sokol, H. (2020) Gut microbiota‐derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 17: 223–237. [DOI] [PubMed] [Google Scholar]
- Leipig‐Rudolph, M. , Busch, K. , Prescott, J.F. , Mehdizadeh Gohari, I. , Leutenegger, C.M. , Hermanns, W. , et al (2018) Intestinal lesions in dogs with acute hemorrhagic diarrhea syndrome associated with netF‐positive Clostridium perfringens type A. J Vet Diagn Invest 30: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley, R.E. , Hamady, M. , Lozupone, C. , Turnbaugh, P.J. , Ramey, R.R. , Bircher, J.S. , et al (2008) Evolution of mammals and their gut microbes. Science 320: 1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Lauber, C.L. , Czarnecki‐Maulden, G. , Pan, Y. , and Hannah, S.S. (2017) Effects of the dietary protein and carbohydrate ratio on gut microbiomes in dogs of different body conditions. MBio 8: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Wang, J. , He, T. , Becker, S. , Zhang, G. , Li, D. , and Ma, X. (2018) Butyrate: a double‐edged sword for health? Adv Nutr 9: 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, P. , and Flint, H.J. (2017) Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19: 29–41. [DOI] [PubMed] [Google Scholar]
- Lugli, G.A. , Milani, C. , Duranti, S. , Alessandri, G. , Turroni, F. , Mancabelli, L. , et al (2019) Isolation of novel gut bifidobacteria using a combination of metagenomic and cultivation approaches. Genome Biol 20: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu, T. , Liu, G. , Zhang, H. , Wang, L. , Zhou, S. , Dou, H. , et al (2018) Changes in feeding habits promoted the differentiation of the composition and function of gut microbiotas between domestic dogs (Canis lupus familiaris) and gray wolves (Canis lupus). AMB Express 8: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, S. , Ohno, K. , Fujiwara‐Igarashi, A. , Uchida, K. , and Tsujimoto, H. (2016) Changes in Foxp3‐Positive Regulatory T Cell number in the intestine of dogs with idiopathic inflammatory bowel disease and intestinal lymphoma. Vet Pathol 53: 102–112. [DOI] [PubMed] [Google Scholar]
- Makielski, K. , Cullen, J. , O'Connor, A. , and Jergens, A.E. (2019) Narrative review of therapies for chronic enteropathies in dogs and cats. J Vet Intern Med 33: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancabelli, L. , Milani, C. , Lugli, G.A. , Fontana, F. , Turroni, F. , van Sinderen, D. , and Ventura, M. (2020) The impact of Primer Design on Amplicon‐based metagenomic profiling accuracy: detailed insights into bifidobacterial community structure. Microorganisms 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchester, A.C. , Webb, C.B. , Blake, A.B. , Sarwar, F. , Lidbury, J.A. , Steiner, J.M. , and Suchodolski, J.S. (2019) Long‐term impact of tylosin on fecal microbiota and fecal bile acids of healthy dogs. J Vet Intern Med 33: 2605–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancino, W. , Duranti, S. , Mancabelli, L. , Longhi, G. , Anzalone, R. , Milani, C. , et al (2019) Bifidobacterial transfer from mother to child as examined by an animal model. Microorganisms 7: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowiak, P. , and Slizewska, K. (2017) Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 9: 1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsilio, S. , Pilla, R. , Sarawichitr, B. , Chow, B. , Hill, S.L. , Ackermann, M.R. , et al (2019) Characterization of the fecal microbiome in cats with inflammatory bowel disease or alimentary small cell lymphoma. Sci Rep 9: 19208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, R. , Langa, S. , Reviriego, C. , Jiminez, E. , Marin, M.L. , Xaus, J. , et al (2003) Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr 143: 754–758. [DOI] [PubMed] [Google Scholar]
- Martin, R. , Olivares, M. , Perez, M. , Xaus, J. , Torre, C. , Fernandez, L. , and Rodriguez, J.M. (2010) Identification and evaluation of the probiotic potential of lactobacilli isolated from canine milk. Vet J 185: 193–198. [DOI] [PubMed] [Google Scholar]
- Masuoka, H. , Shimada, K. , Kiyosue‐Yasuda, T. , Kiyosue, M. , Oishi, Y. , Kimura, S. , et al (2017) Transition of the intestinal microbiota of cats with age. PLoS One 12: e0181739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros, S. , Gras‐Leguen, C. , Le Vacon, F. , Potel, G. , and de La Cochetiere, M.F. (2013) Development of intestinal microbiota in infants and its impact on health. Trends Microbiol 21: 167–173. [DOI] [PubMed] [Google Scholar]
- McKenzie, V.J. , Song, S.J. , Delsuc, F. , Prest, T.L. , Oliverio, A.M. , Korpita, T.M. , et al (2017) The effects of captivity on the mammalian gut microbiome. Integr Comp Biol 57: 690–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, C. , Bai, C. , Brown, T.D. , Hood, L.E. , and Tian, Q. (2018) Human gut microbiota and gastrointestinal cancer. Genomics Proteomics Bioinformatics 16: 33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentula, S. , Harmoinen, J. , Heikkila, M. , Westermarck, E. , Rautio, M. , Huovinen, P. , and Kononen, E. (2005) Comparison between cultured small‐intestinal and fecal microbiotas in beagle dogs. Appl Environ Microbiol 71: 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf, J.L. , Song, S.J. , Morton, J.T. , Weiss, S. , Seguin‐Orlando, A. , Joly, F. , et al (2017) Evaluating the impact of domestication and captivity on the horse gut microbiome. Sci Rep 7: 15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelbos, I.S. , Vester Boler, B.M. , Qu, A. , White, B.A. , Swanson, K.S. , and Fahey, G.C. Jr (2010) Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS One 5: e9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Lugli, G.A. , Turroni, F. , Mancabelli, L. , Duranti, S. , Viappiani, A. , et al (2014) Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol Ecol 90: 493–503. [DOI] [PubMed] [Google Scholar]
- Milani, C. , Duranti, S. , Bottacini, F. , Casey, E. , Turroni, F. , Mahony, J. , et al (2017a) The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Mangifesta, M. , Mancabelli, L. , Lugli, G.A. , James, K. , Duranti, S. , et al (2017b) Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J 11: 2834–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani, C. , Duranti, S. , Mangifesta, M. , Lugli, G.A. , Turroni, F. , Mancabelli, L. , et al (2018) Phylotype‐level profiling of lactobacilli in highly complex environments by means of an internal transcribed spacer‐based metagenomic approach. Appl Environ Microbiol 84: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamoto, Y. , Hooda, S. , Swanson, K.S. , and Suchodolski, J.S. (2012) Feline gastrointestinal microbiota. Anim Health Res Rev 13: 64–77. [DOI] [PubMed] [Google Scholar]
- Minamoto, Y. , Dhanani, N. , Markel, M.E. , Steiner, J.M. , and Suchodolski, J.S. (2014) Prevalence of Clostridium perfringens, Clostridium perfringens enterotoxin and dysbiosis in fecal samples of dogs with diarrhea. Vet Microbiol 174: 463–473. [DOI] [PubMed] [Google Scholar]
- Minamoto, Y. , Otoni, C.C. , Steelman, S.M. , Buyukleblebici, O. , Steiner, J.M. , Jergens, A.E. , and Suchodolski, J.S. (2015) Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes 6: 33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamoto, Y. , Minamoto, T. , Isaiah, A. , Sattasathuchana, P. , Buono, A. , Rangachari, V.R. , et al (2019) Fecal short‐chain fatty acid concentrations and dysbiosis in dogs with chronic enteropathy. J Vet Intern Med 33: 1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens, E. , and Veldhoen, M. (2012) Epithelial barrier biology: good fences make good neighbours. Immunology 135: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondo, E. , Marliani, G. , Accorsi, P.A. , Cocchi, M. , and Di Leone, A. (2019) Role of gut microbiota in dog and cat's health and diseases. Open Vet J 9: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, C.D. , Young, W. , Maclean, P.H. , Cookson, A.L. , and Bermingham, E.N. (2018) Metagenomic insights into the roles of Proteobacteria in the gastrointestinal microbiomes of healthy dogs and cats. Microbiologyopen 7: e00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morato, E.P. , Leomil, L. , Beutin, L. , Krause, G. , Moura, R.A. , and Pestana de Castro, A.F. (2009) Domestic cats constitute a natural reservoir of human enteropathogenic Escherichia coli types. Zoonoses Public Health 56: 229–237. [DOI] [PubMed] [Google Scholar]
- Moylan, H.E.C. , Nguyen‐Ngo, C. , Lim, R. , and Lappas, M. (2020) The short chain fatty acids butyrate and propionate protect against inflammation induced activation of mediators involved in active labor: implications for preterm birth. Mol Hum Reprod 26: 452–468. [DOI] [PubMed] [Google Scholar]
- Munster, M. , Horauf, A. , and Bilzer, T. (2006) Assessment of disease severity and outcome of dietary, antibiotic, and immunosuppressive interventions by use of the canine IBD activity index in 21 dogs with chronic inflammatory bowel disease. Berl Munch Tierarztl Wochenschr 119: 493–505. [PubMed] [Google Scholar]
- Neefs, J.M. , Van de Peer, Y. , De Rijk, P. , Chapelle, S. , and De Wachter, R. (1993) Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res 21: 3025–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nermes, M. , Niinivirta, K. , Nylund, L. , Laitinen, K. , Matomaki, J. , Salminen, S. , and Isolauri, E. (2013) Perinatal pet exposure, faecal microbiota, and wheezy bronchitis: is there a connection? ISRN Allergy 2013: 827934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, C. , Li, H. , Wu, W.K.K. , Wong, S.H. , and Yu, J. (2019) Genomics and metagenomics of colorectal cancer. J Gastrointest Oncol 10: 1164–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niina, A. , Kibe, R. , Suzuki, R. , Yuchi, Y. , Teshima, T. , Matsumoto, H. , et al (2019) Improvement in clinical symptoms and fecal microbiome after fecal microbiota transplantation in a dog with inflammatory bowel disease. Vet Med 10: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira, J.P.S. , He, F. , Mangian, H.F. , Oba, P.M. , and De Godoy, M.R.C. (2019) Dietary supplementation of a fiber‐prebiotic and saccharin‐eugenol blend in extruded diets fed to dogs. J Anim Sci 97: 4519–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori, M. , Maeda, S. , Igarashi, H. , Ohno, K. , Sakai, K. , Yonezawa, T. , et al (2017) Fecal microbiome in dogs with inflammatory bowel disease and intestinal lymphoma. J Vet Med Sci 79: 1840–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osto, M. , and Lutz, T.A. (2015) Translational value of animal models of obesity‐Focus on dogs and cats. Eur J Pharmacol 759: 240–252. [DOI] [PubMed] [Google Scholar]
- O'Sullivan, A. , Farver, M. , and Smilowitz, J.T. (2015) The influence of early infant‐feeding practices on the intestinal microbiome and body composition in infants. Nutr Metab Insights 8: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, P.W. , and Jeffery, I.B. (2015) Gut microbiota and aging. Science 350: 1214–1215. [DOI] [PubMed] [Google Scholar]
- Park, H.J. , Lee, S.E. , Kim, H.B. , Isaacson, R.E. , Seo, K.W. , and Song, K.H. (2015) Association of obesity with serum leptin, adiponectin, and serotonin and gut microflora in beagle dogs. J Vet Intern Med 29: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin‐Puget, L. , El Garch, F. , Bertrand, X. , Valot, B. , and Hocquet, D. (2020) Genome analysis of enterobacteriaceae with non‐wild type susceptibility to third‐generation cephalosporins recovered from diseased dogs and cats in Europe. Vet Microbiol 242: 108601. [DOI] [PubMed] [Google Scholar]
- Pereira, G.Q. , Gomes, L.A. , Santos, I.S. , Alfieri, A.F. , Weese, J.S. , and Costa, M.C. (2018) Fecal microbiota transplantation in puppies with canine parvovirus infection. J Vet Intern Med 32: 707–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Munoz, M.E. , Arrieta, M.C. , Ramer‐Tait, A.E. , and Walter, J. (2017) A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome 5: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, A.M. , Coe, J.B. , Rock, M.J. , and Adams, C.L. (2017) Feline obesity in veterinary medicine: insights from a thematic analysis of communication in practice. Front Vet Sci 4: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard, J.M. , Zeng, M.Y. , Caruso, R. , and Nunez, G. (2017) Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev 279: 70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilla, R. , and Suchodolski, J.S. (2019) The Role of the canine gut microbiome and metabolome in health and gastrointestinal disease. Front Vet Sci 6: 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilla, R. , Guard, B.C. , Steiner, J.M. , Gaschen, F.P. , Olson, E. , Werling, D. , et al (2019) Administration of a synbiotic containing Enterococcus faecium does not significantly alter fecal microbiota richness or diversity in dogs with and without food‐responsive chronic enteropathy. Front Vet Sci 6: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna, C. , and Biagi, G. (2016) The utilisation of prebiotics and synbiotics in dogs. Ital J Anim Science 13: 3107. [Google Scholar]
- Potrykus, J. , White, R.L. , and Bearne, S.L. (2008) Proteomic investigation of amino acid catabolism in the indigenous gut anaerobe Fusobacterium varium. Proteomics 8: 2691–2703. [DOI] [PubMed] [Google Scholar]
- Quraishi, M.N. , Widlak, M. , Bhala, N. , Moore, D. , Price, M. , Sharma, N. , and Iqbal, T.H. (2017) Systematic review with meta‐analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Aliment Pharmacol Ther 46: 479–493. [DOI] [PubMed] [Google Scholar]
- Rackaityte, E. , Halkias, J. , Fukui, E.M. , Mendoza, V.F. , Hayzelden, C. , Crawford, E.D. , et al (2020) Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med 26: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic‐Stojanovic, M. , and de Vos, W.M. (2014) The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38: 996–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak, W. , Ryl, A. , Mizerski, A. , Walczakiewicz, K. , Sipak, O. , and Laszczynska, M. (2019) Immunomodulatory potential of gut microbiome‐derived short‐chain fatty acids (SCFAs). Acta Biochim Pol 66: 1–12. [DOI] [PubMed] [Google Scholar]
- Redfern, A. , Suchodolski, J. , and Jergens, A. (2017) Role of the gastrointestinal microbiota in small animal health and disease. Vet Rec 181: 370. [DOI] [PubMed] [Google Scholar]
- Richard, D.M. , Dawes, M.A. , Mathias, C.W. , Acheson, A. , Hill‐Kapturczak, N. , and Dougherty, D.M. (2009) L‐Tryptophan: basic metabolic functions, behavioral research and therapeutic indications. Int J Tryptophan Res 2: 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke, C. , Schwientek, P. , Sczyrba, A. , Ivanova, N.N. , Anderson, I.J. , Cheng, J.F. , et al (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Ritchie, L.E. , Steiner, J.M. , and Suchodolski, J.S. (2008) Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis. FEMS Microbiol Ecol 66: 590–598. [DOI] [PubMed] [Google Scholar]
- Ritchie, L.E. , Burke, K.F. , Garcia‐Mazcorro, J.F. , Steiner, J.M. , and Suchodolski, J.S. (2010) Characterization of fecal microbiota in cats using universal 16S rRNA gene and group‐specific primers for Lactobacillus and Bifidobacterium spp. Vet Microbiol 144: 140–146. [DOI] [PubMed] [Google Scholar]
- Roberfroid, M. , Gibson, G.R. , Hoyles, L. , McCartney, A.L. , Rastall, R. , Rowland, I. , et al (2010) Prebiotic effects: metabolic and health benefits. Br J Nutr 104(Suppl 2): S1–S63. [DOI] [PubMed] [Google Scholar]
- Rochus, K. , Janssens, G.P. , and Hesta, M. (2014) Dietary fibre and the importance of the gut microbiota in feline nutrition: a review. Nutr Res Rev 27: 295–307. [DOI] [PubMed] [Google Scholar]
- Rossi, G. , Pengo, G. , Caldin, M. , Palumbo Piccionello, A. , Steiner, J.M. , Cohen, N.D. , et al (2014) Comparison of microbiological, histological, and immunomodulatory parameters in response to treatment with either combination therapy with prednisone and metronidazole or probiotic VSL#3 strains in dogs with idiopathic inflammatory bowel disease. PLoS One 9: e94699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, G. , Jergens, A. , Cerquetella, M. , Berardi, S. , Di Cicco, E. , Bassotti, G. , et al (2018) Effects of a probiotic (SLAB51) on clinical and histologic variables and microbiota of cats with chronic constipation/megacolon: a pilot study. Benef Microbes 9: 101–110. [DOI] [PubMed] [Google Scholar]
- Rossi, G. , Cerquetella, M. , Gavazza, A. , Galosi, L. , Berardi, S. , Mangiaterra, S. , et al (2020) Rapid resolution of large bowel diarrhea after the administration of a combination of a high‐fiber diet and a probiotic mixture in 30 dogs. Vet Sci 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, K. , Maeda, S. , Yonezawa, T. , and Matsuki, N. (2018) Decreased plasma amino acid concentrations in cats with chronic gastrointestinal diseases and their possible contribution in the inflammatory response. Vet Immunol Immunopathol 195: 1–6. [DOI] [PubMed] [Google Scholar]
- Salas‐Mani, A. , Jeusette, I. , Castillo, I. , Manuelian, C.L. , Lionnet, C. , Iraculis, N. , et al (2018) Fecal microbiota composition changes after a BW loss diet in Beagle dogs. J Anim Sci 96: 3102–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvo Romero, E. , Alonso Cotoner, C. , Pardo Camacho, C. , Casado Bedmar, M. , and Vicario, M. (2015) The intestinal barrier function and its involvement in digestive disease. Rev Esp Enferm Dig 107: 686–696. [DOI] [PubMed] [Google Scholar]
- Sanchez, B. , Delgado, S. , Blanco‐Miguez, A. , Lourenco, A. , Gueimonde, M. , and Margolles, A. (2017) Probiotics, gut microbiota, and their influence on host health and disease. Mol Nutr Food Res 61. [DOI] [PubMed] [Google Scholar]
- Sandri, M. , Dal Monego, S. , Conte, G. , Sgorlon, S. , and Stefanon, B. (2017) Raw meat based diet influences faecal microbiome and end products of fermentation in healthy dogs. BMC Vet Res 13: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen, P. , Zhang, Y.P. , Luo, J. , Lundeberg, J. , and Leitner, T. (2002) Genetic evidence for an East Asian origin of domestic dogs. Science 298: 1610–1613. [DOI] [PubMed] [Google Scholar]
- Schlesinger, D.P. , and Joffe, D.J. (2011) Raw food diets in companion animals: a critical review. Can Vet J 52: 50–54. [PMC free article] [PubMed] [Google Scholar]
- Schloss, P.D. (2018) Identifying and overcoming threats to reproducibility, replicability, robustness, and generalizability in microbiome research. MBio 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, M. , Unterer, S. , Suchodolski, J.S. , Honneffer, J.B. , Guard, B.C. , Lidbury, J.A. , et al (2018) The fecal microbiome and metabolome differs between dogs fed Bones and Raw Food (BARF) diets and dogs fed commercial diets. PLoS One 13: e0201279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, S. , Glanemann, B. , Garden, O.A. , Brooks, H. , Chang, Y.M. , Werling, D. , and Allenspach, K. (2015) A prospective, randomized, blinded, placebo‐controlled pilot study on the effect of Enterococcus faecium on clinical activity and intestinal gene expression in canine food‐responsive chronic enteropathy. J Vet Intern Med 29: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seekatz, A.M. , Aas, J. , Gessert, C.E. , Rubin, T.A. , Saman, D.M. , Bakken, J.S. , and Young, V.B. (2014) Recovery of the gut microbiome following fecal microbiota transplantation. MBio 5: e00893‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavin, J. (2013) Fiber and prebiotics: mechanisms and health benefits. Nutrients 5: 1417–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov, K.S. , Maier, T.V. , Walker, A. , Heinzmann, S.S. , Forcisi, S. , Martinez, I. , et al (2016) Challenges of metabolomics in human gut microbiota research. Int J Med Microbiol 306: 266–279. [DOI] [PubMed] [Google Scholar]
- Smith, P.M. , Howitt, M.R. , Panikov, N. , Michaud, M. , Gallini, C.A. , Bohlooly, Y.M. , et al (2013) The microbial metabolites, short‐chain fatty acids, regulate colonic Treg cell homeostasis. Science 341: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S.J. , Lauber, C. , Costello, E.K. , Lozupone, C.A. , Humphrey, G. , Berg‐Lyons, D. , et al (2013) Cohabiting family members share microbiota with one another and with their dogs. eLife 2: e00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckel, M.C. , Morgenroth, F. , Buetefisch, C.M. , and Seitz, R.J. (2012) Differential grey matter changes in sensorimotor cortex related to exceptional fine motor skills. PLoS One 7: e51900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski, J.S. (2011) Intestinal microbiota of dogs and cats: a bigger world than we thought. Vet Clin North Am Small Anim Pract 41: 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski, J. (2016) Diagnosis and interpretation of intestinal dysbiosis in dogs and cats. Vet J 215: 30–37. [DOI] [PubMed] [Google Scholar]
- Suchodolski, J.S. , Camacho, J. , and Steiner, J.M. (2008) Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiol Ecol 66: 567–578. [DOI] [PubMed] [Google Scholar]
- Suchodolski, J.S. , Dowd, S.E. , Westermarck, E. , Steiner, J.M. , Wolcott, R.D. , Spillmann, T. , and Harmoinen, J.A. (2009) The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rRNA gene sequencing. BMC Microbiol 9: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski, J.S. , Dowd, S.E. , Wilke, V. , Steiner, J.M. , and Jergens, A.E. (2012a) 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS One 7: e39333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski, J.S. , Markel, M.E. , Garcia‐Mazcorro, J.F. , Unterer, S. , Heilmann, R.M. , Dowd, S.E. , et al (2012b) The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One 7: e51907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski, J.S. , Foster, M.L. , Sohail, M.U. , Leutenegger, C. , Queen, E.V. , Steiner, J.M. , and Marks, S.L. (2015) The fecal microbiome in cats with diarrhea. PLoS One 10: e0127378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita, K. , Yanuma, N. , Ohno, H. , Takahashi, K. , Kawano, K. , Morita, H. , and Ohmori, K. (2019) Oral faecal microbiota transplantation for the treatment of Clostridium difficile‐associated diarrhoea in a dog: a case report. BMC Vet Res 15: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, K.S. , Dowd, S.E. , Suchodolski, J.S. , Middelbos, I.S. , Vester, B.M. , Barry, K.A. , et al (2011) Phylogenetic and gene‐centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J 5: 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes, N. , Beirne, P. , Horowitz, A. , Jones, I. , Kalof, L. , Karlsson, E. , et al (2020) Humanity's best friend: a dog‐centric approach to addressing global challenges. Animals 10: 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal, M. , Weese, J.S. , Gomez, D.E. , Hesta, M. , Steiner, J.M. , and Verbrugghe, A. (2020) Bacterial fecal microbiota is only minimally affected by a standardized weight loss plan in obese cats. BMC Vet Res 16: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, Y. , Ohta, H. , Kagawa, Y. , Osuga, T. , Morishita, K. , Sasaki, N. , and Takiguchi, M. (2019) Plasma amino acid profiles in dogs with inflammatory bowel disease. J Vet Intern Med 33: 1602–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun, H.M. , Brar, M.S. , Khin, N. , Jun, L. , Hui, R.K. , Dowd, S.E. , and Leung, F.C. (2012) Gene‐centric metagenomics analysis of feline intestinal microbiome using 454 junior pyrosequencing. J Microbiol Methods 88: 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun, H.M. , Konya, T. , Takaro, T.K. , Brook, J.R. , Chari, R. , Field, C.J. , et al (2017) Exposure to household furry pets influences the gut microbiota of infant at 3–4 months following various birth scenarios. Microbiome 5: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni, F. , Ribbera, A. , Foroni, E. , van Sinderen, D. , and Ventura, M. (2008) Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek 94: 35–50. [DOI] [PubMed] [Google Scholar]
- Unterer, S. , Strohmeyer, K. , Kruse, B.D. , Sauter‐Louis, C. , and Hartmann, K. (2011) Treatment of aseptic dogs with hemorrhagic gastroenteritis with amoxicillin/clavulanic acid: a prospective blinded study. J Vet Intern Med 25: 973–979. [DOI] [PubMed] [Google Scholar]
- Unterer, S. , Busch, K. , Leipig, M. , Hermanns, W. , Wolf, G. , Straubinger, R.K. , et al (2014) Endoscopically visualized lesions, histologic findings, and bacterial invasion in the gastrointestinal mucosa of dogs with acute hemorrhagic diarrhea syndrome. J Vet Intern Med 28: 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte, D. , Kathagen, G. , D'Hoe, K. , Vieira‐Silva, S. , Valles‐Colomer, M. , Sabino, J. , et al (2017) Quantitative microbiome profiling links gut community variation to microbial load. Nature 551: 507–511. [DOI] [PubMed] [Google Scholar]
- Vazquez‐Baeza, Y. , Hyde, E.R. , Suchodolski, J.S. , and Knight, R. (2016) Dog and human inflammatory bowel disease rely on overlapping yet distinct dysbiosis networks. Nat Microbiol 1: 16177. [DOI] [PubMed] [Google Scholar]
- Ventura, M. , Turroni, F. , Canchaya, C. , Vaughan, E.E. , O'Toole, P.W. , and van Sinderen, D. (2009) Microbial diversity in the human intestine and novel insights from metagenomics. Front Biosci 14: 3214–3221. [DOI] [PubMed] [Google Scholar]
- Vieira, A.T. , Teixeira, M.M. , and Martins, F.S. (2013) The role of probiotics and prebiotics in inducing gut immunity. Front Immunol 4: 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilson, A. , Ramadan, Z. , Li, Q. , Hedhammar, A. , Reynolds, A. , Spears, J. , et al (2018) Disentangling factors that shape the gut microbiota in German Shepherd dogs. PLoS One 13: e0193507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital, M. , Howe, A.C. , and Tiedje, J.M. (2014) Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio 5: e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital, M. , Gao, J. , Rizzo, M. , Harrison, T. , and Tiedje, L.M. (2015) Diet is a major factor governing the fecal butyrate‐producing community structure across mammalia, aves and reptilia. ISME J 9: 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, R.W. , Clemente, J.C. , Peter, I. , and Loos, R.J.F. (2017) The prenatal gut microbiome: are we colonized with bacteria in utero? Pediatr Obes 12(Suppl 1): 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.D. , Zhai, W. , Yang, H.C. , Fan, R.X. , Cao, X. , Zhong, L. , et al (2013) The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun 4: 1860. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Zhou, Y. , Xiao, X. , Zheng, J. , and Zhou, H. (2020) Metaproteomics: a strategy to study the taxonomy and functionality of the gut microbiota. J Proteomics 219: 103737. [DOI] [PubMed] [Google Scholar]
- Werner, M. , Suchodolski, J.S. , Straubinger, R.K. , Wolf, G. , Steiner, J.M. , Lidbury, J.A. , et al (2020) Effect of amoxicillin‐clavulanic acid on clinical scores, intestinal microbiome, and amoxicillin‐resistant Escherichia coli in dogs with uncomplicated acute diarrhea. J Vet Intern Med 34: 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernimont, S.M. , Radosevich, J. , Jackson, M.I. , Ephraim, E. , Badri, D.V. , MacLeay, J.M. , et al (2020) The effects of nutrition on the gastrointestinal microbiome of cats and dogs: impact on health and disease. Front Microbiol 11: 1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R. , Atherly, T. , Guard, B. , Rossi, G. , Wang, C. , Mosher, C. , et al (2017) Randomized, controlled trial evaluating the effect of multi‐strain probiotic on the mucosal microbiota in canine idiopathic inflammatory bowel disease. Gut Microbes 8: 451–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Zhang, H. , Chen, J. , Shang, S. , Yan, J. , Chen, Y. , et al (2017) Analysis and comparison of the wolf microbiome under different environmental factors using three different data of Next Generation Sequencing. Sci Rep 7: 11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Verbrugghe, A. , Lourenco, M. , Janssens, G.P. , Liu, D.J. , Van de Wiele, T. , et al (2016) Does canine inflammatory bowel disease influence gut microbial profile and host metabolism? BMC Vet Res 12: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, M.Y. , Inohara, N. , and Nunez, G. (2017) Mechanisms of inflammation‐driven bacterial dysbiosis in the gut. Mucosal Immunol 10: 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziese, A.L. , Suchodolski, J.S. , Hartmann, K. , Busch, K. , Anderson, A. , Sarwar, F. , et al (2018) Effect of probiotic treatment on the clinical course, intestinal microbiome, and toxigenic Clostridium perfringens in dogs with acute hemorrhagic diarrhea. PLoS One 13: e0204691. [DOI] [PMC free article] [PubMed] [Google Scholar]


