Abstract
Olfactory disorders have been increasingly reported in individuals infected with SARS‐CoV‐2, the virus causing the coronavirus disease 2019 (COVID‐19). Losing the sense of smell has a strong impact on the quality of life, since it may lead to malnutrition, weight loss, food poisoning, depression, and exposure to dangerous chemicals. Individuals who suffer from anosmia (inability to smell) also cannot sense the flavor of food, which is a combination of taste and smell. Interestingly, infected individuals have reported sudden loss of smell with no congested nose, as is frequently observed in common colds or other upper respiratory tract infections. These observations suggest that SARS‐CoV‐2 infection leads to olfactory loss through a distinct mechanism, which is still unclear. This article provides an overview of olfactory loss and the recent findings relating to COVID‐19. Possible mechanisms of SARS‐CoV‐2‐induced olfactory loss are also discussed.
Keywords: anosmia, coronavirus, olfaction, olfactory sensory neuron, SARS‐CoV‐2, smell loss
Loss of smell (anosmia) has been highly correlated with COVID‐19. In the nasal mucosa, the olfactory epithelium (OE) consists of olfactory sensory neurons (OSNs), progenitor cells (HBCs), and supporting (SUS) cells. SUS cells and HBCs are the only OE cell‐types equipped with the SARS‐CoV‐2 entry factors ACE2 and TMPRSS2. Infection of SUS cells by the virus would lead to robust inflammation and ultimately death of OSNs, resulting in anosmia.
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- COVID‐19
coronavirus disease 2019
- GBC
globose basal cell
- HBC
horizontal basal cell
- OB
olfactory bulb
- OE
olfactory epithelium
- OSN
olfactory sensory neuron
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
1. INTRODUCTION
A series of epidemiological studies have now confirmed the initial anecdotal observation that dysfunction of olfaction is a significant COVID‐19 symptom. In one study, the symptoms of more than 2 million individuals from Europe and USA were evaluated using a mobile application‐based symptom tracker (Menni et al., 2020). Of these, 65% of the subjects who tested positive for COVID‐19 reported loss of taste or smell, compared to 21.7% who tested negative for COVID‐19. A worldwide study conducted by the Global Consortium for Chemosensory Research (GCCR) included 4,039 participants from 41 different countries, who reported a COVID‐19 diagnosis (Parma et al., 2020). In this study, 89% of the participants reported loss of smell. Interestingly, nasal obstruction was not associated with smell loss, as commonly observed in other upper respiratory infections. In addition, 76% of the participants reported loss of taste and 46% had a reduction of chemesthesis (detection of chemicals that evoke tingling and burning sensations), indicating that the chemosensory impairment is not restricted to smell (Parma et al., 2020).
The incidence of anosmia in COVID‐19 patients however varies in different studies, ranging from 34% to 68% (Meng et al., 2020). This variability could be due to genetic factors, viral load, specificities of the different evaluated populations or methods used in the analysis. For example, in one study where 202 patients were analyzed 64.4% reported altered sense of smell or taste, and of these, only 34.6% also reported having a congested nose (Spinato et al., 2020). In another study where 417 mild to moderate COVID‐19 European patients were analyzed, 85.6% were anosmic or hyposmic (Lechien et al., 2020). In this case, only 76 patients did not suffer from nose obstruction or rhinorrhea, and among these, 66.2% suffered from anosmia and 13.5% from hyposmia (Lechien et al., 2020). In some cases, impairment of the sense of smell appeared before other clinical manifestations such as cough and fever, suggesting that it can serve as a clinical diagnosis for SARS‐CoV‐2 infection (Hopkins et al., 2020; Lechien et al., 2020). In most cases the sense of smell is recovered in average after two weeks or after the resolution of the other symptoms (Hopkins et al., 2020; Lechien et al., 2020). Also, only less than 10% of the participants in the GCCR study reported quality distortions in smell (parosmia) or smelling of an odorant that is not present (phantosmia) (Parma et al., 2020). Together, these observations suggest that in the majority of the cases the viral damage occurs peripherally rather than in the central nervous system (CNS). However, longer rates of recovery of the sense of smell and taste, that could be due to effects of the virus at the CNS, have also been observed, and should be further investigated (Hopkins et al., 2020).
Although there is now compelling clinical and epidemiological evidence that loss of the sense of smell is a marker of COVID‐19, there are still little data showing how SARS‐CoV‐2 can enter and efficiently replicate in cells of the olfactory tissues, causing smell loss. These points are addressed below, based both on published articles, and on a selected group of recent pre‐prints which have not yet passed through peer review. We will begin by describing how the olfactory system is anatomically organized. We will also describe some of the known examples of how the sense of smell can be perturbed or destroyed.
2. GENERAL ORGANIZATION OF THE OLFACTORY SYSTEM
Two different types of epithelia are found in the nasal cavity: the respiratory epithelium and the olfactory epithelium (OE). Most of the nasal cavity area is lined with the respiratory epithelium, a pseudostratified columnar epithelium composed of ciliated cells, secreting (goblet) cells and basal cells (Durante et al., 2020); (Reznik, 1990). The goblet cells secrete mucus that moistens the epithelium and the ciliated cells move the mucus (together with inhaled pathogens and irritants) up and away for expulsion from the body. The basal cells are small progenitor cells that can differentiate into all the cell types of the respiratory epithelium.
Odorant sensing initiates in the OE, which is located in the highest recesses of the nose (Morrison & Constanzo, 1990) (Figure 1a). The OE is also a pseudostratified columnar epithelium, but it contains highly specialized neuronal cells, the olfactory sensory neurons (OSNs), which are responsible for odorant detection. The odorants, small volatile molecules with varied chemical structures, are recognized by a large family of odorant receptors, expressed in the cilia of the OSNs (Buck & Axel, 1991). They enter the nasal cavity, reach the OE, and activate the OSNs. These neurons then transmit the sensory information to the olfactory bulb (OB), which relays it to the olfactory cortex and other higher brain centers, leading to odorant perception, emotions and behaviors.
FIGURE 1.
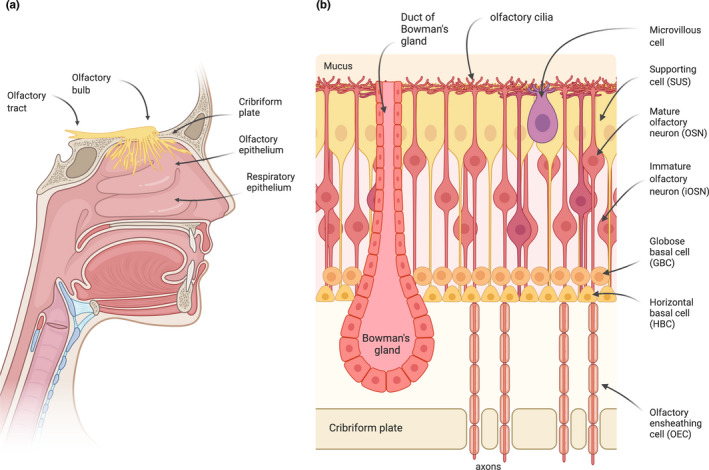
The olfactory system. (a) Odorants are detected by OSNs present in the olfactory epithelium (OE), a specialized neuroepithelium located in the highest recesses of the nose. The olfactory sensory neurons (OSNs) project a single unmyelinated axon to the olfactory bulb (OB). The axons of the OSNs and their associated olfactory ensheathing cells (OECs) form bundles that project through the perforations of the cribriform plate to the OB, where they synapse with the mitral and tufted cells, forming the glomeruli. The axons of these cells form the olfactory tract, which transmits the sensory information into the brain. (b) The different cell types that compose the olfactory epithelium are represented. Odorants are recognized by odorant receptors located in the cilia of the OSNs.
2.1. The olfactory epithelium
The OE is composed of different cell types: the supporting cells, the OSNs (mature and immature), the basal cells (globose and horizontal stem cells), microvillous cells, and Bowman's gland cells (Choi & Goldstein, 2018; Glezer & Malnic, 2019) (Figure 1b).
The supporting cells, also known as sustentacular cells, are attached to the basal lamina but their cell bodies are located more apically in the epithelium. They have a columnar shape and their apical region is covered with microvilli (Morrison & Costanzo, 1992). These cells provide support and insulation to the OSNs, in a similar manner to glial cells (Jafek, 1983; Liang, 2018; Morrison & Constanzo, 1990). The supporting cells express cytochrome P450 and other enzymes involved in metabolizing xenobiotic compounds, suggesting a role in detoxifying toxic inhaled substances to which the OE is exposed, and they also play a phagocytic role, to remove dead OSNs (Dahl et al., 1982; Suzuki et al., 1996; Thornton‐Manning et al., 1997).
The apical region of the OE also contains different types of microvillous cells. These non‐neuronal cells resemble the brush cells of the upper and lower airways, and like the supporting cells have a tuft of microvilli on their apical surface, extending into the nasal cavity (Moran et al., 1982). The microvillous cells are in general less numerous in the OE, when compared to the other cell types, and their function is still not well understood.
The OSNs are the predominant cell type in the OE. They are specialized bipolar cells whose cell bodies occupy a broad region in the middle of the epithelium (Figure 1b) (Glezer & Malnic, 2019). Each olfactory sensory neuron has one single dendrite from which cilia protrude into the mucus layer of the epithelium surface, and one unmyelinated axon that projects to the OB. The axons of the OSNs form the olfactory nerve bundles that cross through the openings of the cribriform plate to reach the OB, where they synapse with the mitral and tufted cells forming the glomeruli. The bundles of unmyelinated olfactory axons are surrounded by the olfactory ensheathing cells all the way from the OE to the OB (Li et al., 2005). These glial‐like cells provide protection and guidance to the olfactory axons during neuronal regeneration. A distinctive characteristic of the olfactory system is therefore that it has a direct access to the brain.
Another remarkable feature of the OE is its capability to generate new OSNs throughout life (Child et al., 2018; Fletcher et al., 2017; Schwob et al., 2017).The basal cells are small cells located in the basal region of the epithelium that can divide and differentiate to replace OSNs and all other cell types of the OE, during normal turnover or injury (Calof & Chikaraishi, 1989; Graziadei & Monti‐Graziadei, 1979). The basal cells can be subdivided into two different cell types: the horizontal basal cells (HBCs), which are located more basally in the OE, in direct contact with the basal lamina, and the globose basal cells (GBCs), which are located above the HBC layer (Holbrook et al., 2011). The HBCs proliferate at a low rate and show a multipotent progenitor phenotype, that can be massively recruited upon severe injury to regenerate all cell types in the OE (Carter et al., 2004; Herrick et al., 2017; Iwai et al., 2008; Leung et al., 2007).The GBCs are the actively proliferating stem cells that are responsible for the constant regeneration of the OSNs and of all the other cell types in the OE (Chen et al., 2004; Graziadei & Monti‐Graziadei, 1979; Huard et al., 1998).
In addition to these major cell types, olfactory glands, known as the Bowman's glands, are distributed throughout the mucosa (Getchell et al., 1984). These glands are located beneath the OE, and project narrow ducts onto the epithelial surface, through which they secrete the mucus that coats the epithelium. The mucus contains water, mucin glycoproteins, enzymes, antibodies, salts, and odorant‐binding proteins (OBPs). The OBPs carry the hydrophobic odorant molecules through the mucus to the cilia of the OSNs, the site of odorant detection (Bignetti et al., 1985; Heydel et al., 2013; Pelosi et al., 1982). The functions of the Bowman's glands are still not totally clear. Possible roles include transport of odorants, prevention of infection by microorganisms, protection against xenobiotic compounds through the secretion of biotransformation enzymes, and protection of the cilia from the OSNs (Heydel et al., 2001, 2013; Mellert et al., 1992; Solbu & Holen, 2012).
The OE is also populated with resident macrophages and dendritic cells, which surveil the neuroepithelium and sense pathogens and cell damage (Nan et al., 2001; Ruitenberg et al., 2008). Interestingly, these macrophages express receptors for the chemokine CX3CL1, also known as fractalkine, which is expressed by OSNs located in the intermediate neuronal layer of the epithelium (Ruitenberg et al., 2008). CX3CL1 modulates macrophage morphology and recruitment (Ruitenberg et al., 2008). Macrophages play important roles in the repair of the damaged OE, by removing pathogens, phagocytosing dead OSNs, and promoting neurogenesis (Borders et al., 2007).
Recent single‐cell transcriptome analysis of OE from healthy adult humans have identified, based on known olfactory marker genes, the presence of most of the cell types described above, including the basal stem cells and progenitors, immature and mature OSNs (Durante et al., 2020). These cells represent various stages of olfactory neuronal differentiation and confirm that adult olfactory neurogenesis occurs in the human OE. In this way, in circumstances where the OE is severely damaged, regeneration of the epithelium can occur and the sense of smell can be recovered.
3. OLFACTORY DISORDERS
Olfactory disorders can result from pathological processes at any point in the olfactory pathway, from the OE to the central brain regions. For example, anosmia is one of the first clinical signs of some neurodegenerative diseases, such as Parkinson's and Alzheimer's diseases (Doty & Hawkes, 2019). It is believed that neurodegeneration of central brain regions involved in olfactory processing may contribute to the disorder (Averback, 1983; Doty, 2007; Hyman et al., 1984). Nevertheless, the precise mechanisms that connect these diseases with olfactory loss are still unclear (Dibattista et al., 2020).
Smell loss can also be a consequence of genetic disorders, such as the ones that lead to ciliary defects, called olfactory ciliopathies (Jenkins et al., 2009; McEwen et al., 2008). These are human genetic disorders that affect ciliary assembly and/or protein transport to the cilia, which is the site of odorant signal transduction. Individuals with the Bardet–Biedl Syndrome, a pleiotropic ciliopathy which includes several different phenotypes such as retinal degeneration, obesity, renal and limb malformations, have partial or complete anosmia (Iannaccone et al., 2005; Kulaga et al., 2004; Reiter & Leroux, 2017). The Kallmann syndrome is another genetic disorder which causes olfactory deficits (Hardelin et al., 2000). In this case, individuals show defects in gonadal development associated with anosmia. The anosmia is a consequence of the absence or poor development of the OBs and olfactory tracts, and the gonadal defects are a consequence of hypothalamic gonadotropin‐releasing hormone (GnRH) deficiency (MacColl et al., 2002; Young et al., 2012).
More frequent causes of anosmia are however the presence of nasal polyps, allergies, head trauma, and other factors that lead to injury to the olfactory nerve. Viral infection of the upper respiratory tract is also one of the most common causes of olfactory dysfunction (denominated as post‐viral olfactory disorder) (Doty, 2019). Chronic inflammation of the nasal airways caused by the infection usually blocks the nasal passage and may also destroy the OSNs of the nose.
4. VIRAL INFECTION AND INFLAMMATION OF THE OLFACTORY EPITHELIUM
Different types of viruses were shown to infect the OE in animal models. In mice, the neurotropic vesicular stomatitis virus (VSV), a rhabdovirus, preferentially infects the OE compared to the nasal respiratory epithelium, and causes early inflammatory infiltration (Lundh et al., 1987). In ferrets and mice inoculated intranasally with the highly pathogenic avian influenza H5N1 virus, lesions were more severe in the olfactory than in the respiratory epithelium, and the virus antigen was found in OSNs, Bowman's glands and olfactory ensheathing cells (Iwasaki et al., 2004; Schrauwen et al., 2012). A recent study that modeled airborne transmission of the viruses in ferrets showed that while more cells of the OE were infected by the A/H5N1 virus than cells of the respiratory epithelium, the OE was less infected by the influenza A/H1N1, and A/H3N2 viruses, than the nasal respiratory epithelium (Richard et al., 2020). Mice infected with the Sendai virus, a murine counterpart of the human Parainfluenza virus, suffer from olfactory dysfunction (Tian et al., 2016). In this case, both apoptosis and proliferation of progenitor cells in the OE were decreased by viral infection (Tian et al., 2016). A different study showed that intranasal inoculation of the mouse hepatitis virus (MHV; a strain of murine coronavirus M‐CoV) that is destructive to the OB but not to the OE, promotes increase in the turnover of the epithelium cells, resulting in a higher proportion of immature OSNs compared to non‐infected mice (Schwob et al., 2001). These studies show that direct viral lesion to the OE is not an obligatory mechanism for post‐viral olfactory disorder development, however striking modifications can result due to viral infection in terms of neuronal renewal and function. Infection by the sialodacryoadenitis virus, a coronavirus, is associated with upper respiratory tract inflammation in rats and provokes lesions in the OE (Bihun & Percy, 1995). The Middle East respiratory syndrome coronavirus (MERS‐CoV) was shown to be able to infect the OE in inoculated dromedary camels (Adney et al., 2014). In humans, infection by coronaviruses was already known to cause mild upper respiratory tract infections, such as common cold (Zumla et al., 2016). Infections by the more pathogenic coronaviruses responsible for the severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), are however more severe.
The inflammatory responses in the OE in response to lesions caused by viral infection are still not well understood. It is interesting to note that the OE is highly damaged by intranasal infusion of pathogen components, or designed mimetic molecules, that trigger pro‐inflammatory gene expression through Toll‐like receptor (TLR) activation (Crisafulli et al., 2018; Hasegawa‐Ishii et al., 2017; Imamura & Hasegawa‐Ishii, 2016). It was shown that transgenic overexpression of the pro‐inflammatory cytokine TNF‐α promotes death of OSNs (Lane et al. 2010). One should also expect that viral infection in the OE would induce the innate immune system through the activation of receptors (such as endosome TLRs and cytoplasmatic viral receptors) that sense the presence of the foreign nucleic acid (ssRNA, in the case of coronaviruses), resulting in appropriate antiviral responses (Jensen & Thomsen, 2012; Schlee & Hartmann, 2016). Thus, viral infection may activate leukocytes and recruit immune cells to the OE, and possibly promote indirect cell death and complete disorganization of the epithelium architecture.
5. CELL INFECTION BY SARS‐COV‐2
The high incidence of smell loss in individuals infected by SARS‐CoV‐2 suggests that this virus may be able to infect the OE. One way to address this possibility is to analyze whether cells in the olfactory system are susceptible to SARS‐CoV‐2 entry and replication.
Since the original SARS outbreak in 2003, much has been learnt about the main mechanisms of coronaviruses (CoVs) entry and replication in the host cells. Like other CoVs, SARS‐CoV‐2 expresses in its membrane the spike protein (S protein), a glycosylated viral surface protein that anchors the virus to the host cell, mainly through binding to the human Angiotensin Converting Enzyme‐2 (ACE2) (Hoffmann, et al., 2020; Walls et al., 2020; Zhou et al., 2020). The S protein, which is assembled in a homotrimer, can be subdivided in two subunits, the S1 (head) subunit and S2 (stalk) subunit. The S1 subunit contains the binding site for the host cell receptor (receptor binding site, RBD). The S2 subunit harbors a transmembrane domain and is required for the fusion of the viral and cellular membranes. SARS‐CoV attaches to the surface of the host cell through the binding of the RBD in its S protein to ACE2. Entry into the cell depends on the priming of the S protein by cellular proteases, that cleave the S protein at the S1/S2 domain boundary and at the S2’ site, so that S1 dissociates and S2 undergoes a dramatic structural change causing the fusion of the virus and host cell membranes (Hoffmann, et al., 2020; Shang et al., 2020). An alternative mechanism for activation of the S protein after binding to ACE2, is through endocytosis and cleavage by the pH‐dependent cysteine protease cathepsin L (Simmons et al., 2005).
Therefore, for a cell to be infected by SARS‐CoV‐2 it would have to both, express a receptor for the S protein, and express at its surface (or in a cell in close proximity) proteases that are able to proteolytically activate the S protein. One of the best‐known SARS‐CoV and SARS‐CoV‐2 activating proteases is the cell surface protease TMPRSS2 (Hoffmann, et al., 2020). Single‐cell RNA‐sequencing data from several human organs have shown that cells in the nasal cavity respiratory epithelium (ciliated and goblet cells) express higher levels of both ACE2 and TMPRSS2 in comparison to lung and bronchiolar cells (Sungnak et al., 2020). Consistent with this, a more recent study demonstrated that SARS‐CoV‐2 displays an infectivity gradient in human primary cells from the upper to the lower respiratory tract (Hou et al., 2020). That was paralleled by the highest expression of ACE2 in the nasal cavity with decreasing expression throughout the lower respiratory tract, as measured by a very sensitive single‐cell RNA in situ analysis (Hou et al., 2020). TMPRSS2, however showed lower mRNA expression levels in the nasal mucosa than in all other respiratory tract regions (Hou et al., 2020). These results show not only that the nasal cavity is the main gate for SARS‐CoV‐2 entry to the lungs, but also, that it may serve as a viral reservoir enhancing virus dissemination.
6. SARS‐COV2 AND THE OLFACTORY EPITHELIUM
The studies described above did however not include analysis of the OE. Do OSNs express the SARS‐CoV‐2 entry factors? Extensive transcriptome analysis indicated that while ACE2 and TMPRSS2 are expressed in the bulk mouse and human olfactory epithelia, analysis of the transcriptomes from single cells indicated that expression of these genes in OSNs is very low. Instead, they are expressed in non‐neuronal cell types, namely in the supporting cells, Bowman's glands and HBCs (Brann et al., 2020; Fodoulian et al., 2020). Co‐expression of ACE2 and TMPRSS2 was also detected in microvillous cells, though at lower levels (Fodoulian et al., 2020). Immunostaining experiments confirmed the transcriptomic analysis, and showed that the ACE2 protein is expressed in the supporting cells mostly localized in the dorsal region of the mouse OE (Brann et al., 2020; Fodoulian et al., 2020). Immunostaining of human nasal mucosa showed intense staining to ACE2 in supporting cells and Bowman's glands in the OE, and interestingly, while in the adjacent respiratory epithelium staining for ACE2 was also observed on the apical surface of the epithelium, the intensity of the staining was highly reduced compared to that of the OE (Chen et al. 2020). Increased expression of ACE2 in the OE relative to the respiratory epithelium was also observed in the mouse nasal mucosa (Bilinska et al., 2020). These results suggest the possibility that, within the nasal cavity, SARS‐Cov‐2 may show a higher tropism for cells in the OE when compared to cells in the respiratory epithelium.
Noteworthy, respiratory epithelium MUC5B secretory cells that express detectable levels of ACE2 and TMPRSS2 were not infected by SARS‐CoV‐2 in cell culture experiments (Hou et al., 2020), suggesting that co‐expression of ACE2 and TMPPRSS2 does not necessarily guarantee infection. Cells expressing ACE2 may be close to cells expressing the activating proteases so that the S protein can still be activated. Thus, it is possible that ACE2 and its activating protease do not necessarily need be expressed in the same cell to foster virus entry. Also, it is important to note that in some cases ACE2 expression can be induced by SARS‐CoV‐2 infection and inflammatory cytokines released in response to the virus (Codo et al., 2020; Ziegler et al., 2020).
6.1. Possible mechanisms of SARS‐CoV‐2 infection in the olfactory epithelium
The findings described above indicate that viral infection and replication might occur in the apical region of the OE, rather than in the layer containing the OSNs. An attractive hypothesis is that infection in non‐neuronal cells of the OE would indirectly impact the capacity of OSNs to sense odorants. A recent work showed that instillation of SARS‐CoV‐2 in the nasal cavity of golden Syrian hamsters, an animal model which has been successfully used for studying SARS‐CoV and SARS‐CoV‐2 infection, resulted in transient destruction of the OE (Bryche et al., 2020). The tissue injury was associated with infection of supporting cells but not of OSNs. Importantly, a major loss of the olfactory cilia was observed, indicating that even though the OSNs were not directly targeted, they were also seriously damaged. Furthermore, the authors observed massive infiltration of immune cells in the OE and lamina propria associated with the SARS‐CoV‐2 infection (Bryche et al., 2020). These results agree with the expression profiles of SARS‐CoV‐2 entry factors in the olfactory system and support a mechanism for SARS‐CoV‐2 induced anosmia where the virus would initially invade the supporting cells and other non‐neuronal cells that are essential for olfactory sensory neuron function, and also recruit inflammatory cells that would contribute to further damage to the OE (Figure 2). It is important to note though that more recent experiments have detected the presence of the virus in the OSNs from intranasally infected Syrian hamsters (Chan et al., 2020; Sia et al., 2020), suggesting the possibility that the OSNs could be directly infected by SARS‐CoV‐2.
FIGURE 2.
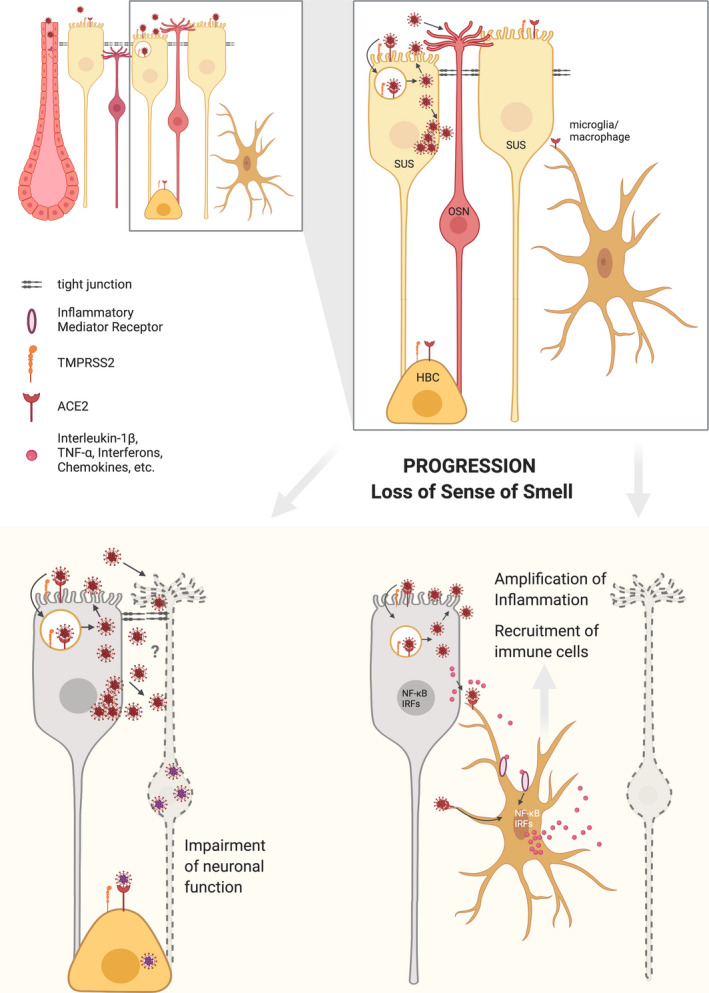
SARS‐CoV‐2 and olfactory neuron function. Supporting cells (SUS), horizontal basal cells (HBC) and Bowmans' gland cells (but not OSNs) express the SARS‐CoV‐2 entry factors ACE2 and TMPRSS2. It is therefore likely that SARS‐CoV‐2 primarily infects the supporting cells and gland cells which are located in the apical region of the epithelium. Damage to the supporting cells would indirectly lead to disruption of proper odorant signaling by OSNs. It remains to be determined whether the virus could infect the OSNs in an ACE2‐independent manner and directly interfere with their function. Importantly, infected supporting cells can release molecules that trigger innate immune signaling in resident microglia/macrophages, which are also responsive to the viral particles. In consequence, key pro‐inflammatory transcription factors (such as NF‐kappaB and IRFs) promote synthesis of interferons and inflammatory mediators that recruit and activate varied types of leukocytes. The olfactory neurons are vulnerable to inflammation, resulting in temporary olfactory loss until the viral infection is resolved and they are replenished by new neurons. Horizontal basal cells (HBCs), which are essential for regeneration of the epithelium after profound damage, express the SARS‐CoV‐2 entry proteins. Whether the virus can also infect these cells and perturb regeneration of the epithelium, remains unknown.
Supporting cells are crucial for proper odor sensing since they provide neurotrophic signaling and physical support to the OSNs. Contacts and cell junctions between the dendrites of the OSNs and the apical region of supporting cells have been observed in several vertebrate species (Breipohl et al., 1974; Menco, 1980; Steinke et al., 2008). In the rat OE, a large fraction of the OSN dendrites were actually shown to be enwrapped by the supporting cells (Liang, 2018), however the exact functional consequences of this wrapping are still not clear (Liang, 2020). One would expect therefore, that loss of the supporting cells would not only destroy OSN function, but also disorganize the whole epithelium. In fact, treatment of the OE with methimazole, an antithyroid drug that induces loss of smell in humans and preferentially targets supporting cells, is well known to cause severe damage to the whole epithelium (Bergstrom et al., 2003).
Interestingly, the toll receptor TLR3, which detects double stranded RNA and activates the pro‐inflammatory transcription factor NF‐κB, was shown to be preferentially expressed in the supporting cells of the mouse OE (Kanaya et al., 2014). Intranasal infusion of poly(I:C), a synthetic analog of virus double stranded RNA, lead to activation of NF‐κB in the supporting cells (Kanaya et al., 2014). These results indicate that these cells can trigger important events in innate immune antiviral responses. Supporting cells may also initiate leukocyte recruitment, either through activation of gene expression or by releasing danger signals upon viral infection and damage. In fact, such mechanisms have been proposed for ACE2‐positive respiratory epithelium cells infected by SARS‐CoV‐2 (Tay et al., 2020). Macrophages in the OE can also contribute to neuronal damage, since activation of these cells by SARS‐CoV‐2 could culminate in dysregulated expression and release of pro‐inflammatory molecules and chemotactic signals (Merad & Martin, 2020).
Only a few studies have so far analyzed the inflammatory responses to SARS‐CoV‐2 infection in the human OE. For example, post‐mortem analysis of COVID‐19 fatal cases revealed increased levels of TNF‐α in the OE (Torabi et al., 2020). Prominent leukocytic infiltrates were observed in damaged OE and olfactory nerve in two other lethal cases, one of them had reported anosmia (Kirschenbaum et al., 2020). Tomographic scan and magnetic resonance imaging of the nasal cavity of a patient infected by SARS‐CoV‐2 whose major symptom was a sudden and complete loss of smell without nasal obstruction, revealed a bilateral inflammatory obstruction of the olfactory clefts (Eliezer et al., 2020). Further analysis should reveal whether uncontrolled inflammation in the OE plays a key role in smell loss reported by COVID‐19 patients.
Nevertheless, the possibility that the virus could directly infect the OSNs by using alternative receptors cannot yet be excluded. Other cell surface proteins have been proposed to work as receptors for SARS‐CoV‐2, as for example CD147 (also known as bagisin). It has been previously shown that CD147 can facilitate invasion of host cells by SARS‐CoV (Chen et al., 2005), and one recent study suggests that SARS‐CoV‐2 may also invade cells through an interaction between the S protein and CD147 (Wang et al., 2020). CD147 is a highly glycosylated membrane protein that belongs to the immunoglobulin superfamily and is highly expressed in different mouse brain regions, including the OB (Fan et al., 1998.). Even though OSNs express very high levels of CD147 mRNA as indicated by transcriptome analysis of FACS sorted mature olfactory marker protein (OMP) positive OSNs (Magklara et al., 2011), and by transcriptomes from single human OSNs (Brann et al., 2020; Durante et al., 2020), there are no reports yet of CD147 protein detection in the olfactory organs.
One particular difference of SARS‐CoV‐2 when compared to SARS‐CoV is the presence of a four amino acid insertion at the S1/S2 boundary constituting a cleavage site (RRAR) for furin, a ubiquitous protease expressed in most cells including lungs, liver, small intestine, and olfactory organs (Brann et al., 2020; Hoffmann et al., 2020; Ueha et al., 2020). Similar furin‐cleavage sites are found in S proteins of MERS‐CoV and many other pathogenic human viruses, including Ebola, HIV‐1 and highly aggressive strains of avian influenza. Experiments using human lung cells in culture showed that both MERS‐CoV and SARS‐CoV‐2 depend on furin‐mediated pre‐cleavage of their S proteins at the S1/S2 site for subsequent S protein activation by TMPRSS2 at the S’ cleavage site (Hoffmann et al., 2020). In addition, cleavage by furin and by TMPRSS2 together, enhance SARS‐CoV‐2 entry in Vero cells (Hou et al., 2020).
In the OE, immunostaining experiments detected the presence of furin in the supporting cells and Bowman's gland, but not in the OSNs (Ueha et al., 2020). Furin mRNA was however detected in transcriptomic data from sorted mature (OMP positive) OSNs (Magklara et al., 2011), and in transcriptomes from single human OSNs (Brann et al., 2020; Durante et al., 2020).
Two recent studies suggest that S protein furin‐cleavage products bind to NRP1, which is expressed in the OSNs, but also abundantly expressed in endothelial and epithelial cells in both the respiratory and olfactory epithelia, and that this interaction enhances SARS‐CoV‐2 entry in cultured cells (Cantuti‐Castelvetri et al., 2020; Daly et al., 2020). Further work is however needed to confirm whether alternative receptors such as CD147 and NRP1 and alternative proteases such as furin or cathepsins play a role in SARS‐CoV2 infection in the OE.
Thus, as a result of the infection by SARS‐CoV‐2, loss of OSNs would occur, resulting in anosmia. The observation that in most cases the sense of smell in infected individuals is rapidly recovered indicates that the damage occurs in the peripheral olfactory system, and that the OE is able to self‐regenerate in the course of the weeks following the infection. Therefore, even though ACE2 was found to be expressed in HBCs, renewal seems to continue.
7. INVASION OF THE CENTRAL NERVOUS SYSTEM THROUGH THE OLFACTORY ROUTE
Whether SARS‐CoV‐2 is able to invade the CNS through the olfactory route like some neurotropic viruses is still an open question. If as discussed above SARS‐CoV‐2 can really not infect the OSNs directly, the chances of such an invasion would be at least reduced. However, since neurological problems have been associated with COVID‐19 and anosmia in some cases (Aragao et al., 2020; Laurendon et al., 2020; Paterson et al., 2020; Politi et al., 2020), the possibility of invasion through the OB should be carefully examined.
7.1. Viral invasion of the CNS through the olfactory pathway
Early studies conducted in rodents showed that OSNs are able to uptake labeled proteins (tracers) and transport them to the OB glomerular region (Kristensson & Olsson, 1971), indicating a possible mechanism for central nervous system invasion by viruses that were known to use the olfactory route (Johnson, 1964; Nir et al., 1965; Sabin & Olitsky, 1937). Even so, different viral features such as coating/envelope, replication mechanisms, budding pathways and cell targets in the OE must determine specific viral abilities to invade the brain.
Many neurotropic viruses were shown to reach the central nervous system after interacting with the nasal mucosa in animal models, including MHV (Perlman et al., 1989), the Borna disease virus (BDV) (Shankar et al., 1992), the pseudorabies virus (PrV)(Babic et al., 1994), the herpes simplex virus type 1 and 2 (HSV‐1/HSV‐2)(Allavena et al., 2011; Barnett et al., 1993), the human coronavirus OC43 (HCoV‐OC43) (St‐Jean et al., 2004) and adenovirus (Lemiale et al., 2003). After infecting the OE, the vesicular stomatitis virus VSV can spread along the olfactory nerves into the glomeruli of the OBs, and afterwards, progressively spread to the brain (Lundh et al., 1987). The parainfluenza virus type 1 is also transported from the OE to the OB (Mori et al., 1995). In this case, supporting cells are not infected by the virus, while OSNs show persistent viral labeling, compatible with a non‐cytolytic infection.
Not all the neurotropic viruses are however found in the olfactory nerve fibers. Experiments with luciferase expressing HSV‐1 constructs, for example, showed that in intranasally infected mice this virus spreads from the OE to the trigeminal nerve, mostly excluding the olfactory nerve (Shivkumar et al., 2013).
7.2. Can SARS‐CoV‐2 invade the CNS through the olfactory pathway?
Experiments in mice showed that SARS‐CoV can enter the nervous system through the nasal route. In these experiments, transgenic mice expressing the human ACE2 (hACE2) under the control of the human cytokeratin 18 promoter (K18‐hACE2 mice) were infected intranasally with SARS‐CoV, and the virus was subsequently found in the brain (Netland et al., 2008). More recent experiments showed that intranasal inoculation of SARS‐CoV‐2 in mice that had their ACE2 gene replaced by the hACE2 gene leads to high levels of the virus RNA in the brain (Sun et al., 2020).
The mechanisms through which SARS‐CoV and SARS‐CoV‐2 invade the mouse brain are however unclear. As mentioned above, transcriptomic and immunostaining analysis showed that ACE2 is not expressed in the OSNs nor in neurons in the OB (Brann et al., 2020; Fodoulian et al., 2020). Therefore, if SARS‐CoV‐2 can be transported to the OB through the olfactory axons, it would have to do it in an ACE2‐independent manner. Interestingly, in the experiments with the SARS‐CoV inoculated K18‐hACE2 transgenic mice, a 60‐hr delay between the time of intranasal inoculation and detection of the virus in the OB was observed (Netland et al., 2008). After that, the virus rapidly spread from the OB to trans neuronally connected regions of the brain (Netland et al., 2008). These results suggest that the virus had first to replicate and accumulate in the OE, possibly in non‐neuronal cell types, before it was able to be transported to the OB. Thus, the ability of the OE to serve as an efficient reservoir for SARS‐CoV‐2 replication and amplification, could facilitate brain invasion by the virus (Butowt & Bilinska, 2020).
The ensheathing cells that surround the axons of the OSNs form a continuous channel throughout the olfactory nerve, from the basal region of the OE to the OB (Li et al., 2005). Diffusion through these channels has also been considered as an olfactory sensory neuron independent mechanism of viral entry to the brain (Butowt & Bilinska, 2020; van Riel et al., 2015). These ensheathing cell channels were shown to survive axonal degeneration, and stably persist and provide support for regeneration of the new axons (Li et al., 2005). As mentioned above, in ferrets subjected to intranasal inoculation of the avian influenza virus H5N1, the viral antigen was also found in the ensheathing cells (Schrauwen et al., 2012). This extracellular route could be used to transfer SARS‐Cov‐2 in an ACE2‐independent manner to the OB, once the virus crosses the basal lamina.
Interestingly, a mechanism that would prevent viral transport from the OE to the OB is the fast induction of apoptosis in infected OSNs (Mori et al., 2002, 2004). Viruses that block or delay apoptosis of the OSNs are therefore more likely to use the olfactory nerve as a route to enter the brain. Since OSNs are unlikely direct targets for SARS‐CoV‐2 (based on the lack of ACE2 expression), they could be able to bypass this preventive mechanism.
So far there is no experimental evidence showing that SARS‐CoV‐2 can invade the brain through the olfactory route. In the experiments mentioned above where golden Syrian hamsters were intranasally infected with SARS‐CoV‐2, the virus was not detected in the OBs, nor in the central nervous system (Bryche et al., 2020). These results suggest that brain invasion by SARS‐CoV‐2 through this pathway is not a common pathogenic mechanism for this virus. Nevertheless, as the authors note, the lack of detection of the virus in the brain could be due to the fact that a small number of animals were analyzed (Bryche et al., 2020).
Alterations in the OBs in COVID‐19 patients with anosmia have been recently reported, suggesting that in these cases SARS‐CoV‐2 could have reached the OB through the olfactory nerve (Aragao et al., 2020; Politi et al., 2020). The finding that ACE2 is highly expressed by pericytes in blood vessels in the OB (Brann et al., 2020; Fodoulian et al., 2020), raises the alternative possibility that the virus could use the hematogenous route to infect the OB, what would induce vascular inflammation and cause anosmia (Brann et al., 2020).
Importantly, not only the olfactory nerve, but also the trigeminal nerve, may serve as a route for entry of pathogens into the brain (Perlman et al., 1989). The trigeminal nerve can not only detect physical stimuli (mechanical and temperature) but also chemicals (denominated as chemesthesis) (Hummel & Frasnelli, 2019). The nose and oral trigeminal nerve endings are typically activated by irritant chemicals, such as air pollutants and other noxious stimuli. This activation triggers protective physiological responses, including decreased respiration rate, sweating and increased salivation. Many of these chemicals are recognized by transient receptor potential (TRP) channels and lead to diverse sensations, for example the burning (capsaicin), cooling (menthol), pungency (allicin) and spicy (thymol) sensations (Viana, 2011).
Trigeminal nerve endings that innervate the olfactory epithelium can branch to innervate the olfactory bulb (Schaefer et al., 2002). A trigeminal route of invasion by SARS‐CoV‐2 could explain some neurological symptoms shown by COVID‐19 patients, like for example loss of facial sensation and headaches.
8. CONCLUDING REMARKS
Mounting evidence indicates that SARS‐CoV‐2 infection can cause anosmia in a large percentage of the infected individuals. The cellular and molecular mechanisms underlying this effect remain largely unknown. OSNs are not likely candidates for SARS‐CoV‐2 infection since they lack expression of the bona‐fide viral entry factors. Loss of the sense of smell may be however attributed to viral damage to other epithelial cell types. The non‐neuronal supporting cells, HBCs and Bowmans’ gland cells are equipped with the viral entry proteins, and therefore, are likely to be targets for SARS‐CoV‐2 infection and replication. These cell types would serve as a reservoir of viral replication what would cause cell damage and inflammation and ultimately disruption of olfactory sensory neuron function. Future work is required to verify whether the virus is able to directly infect the OSNs using alternative receptors, and/or whether it is able to enter the brain through the olfactory or trigeminal route, even if in a small number of cases.
ACKNOWLEDGEMENTS
B.M. is supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, 2016/24471‐0). I.G. is a member of the CEPID Redoxoma (FAPESP 2013/07937‐8) and is supported by FAPESP grant 2018/18633‐3. A.B‐C and D.S. are respectively supported by FAPESP grants 2019/26767‐2 and 2019/06982‐6. Figures were re‐drawn by Marco Bazelmans in BioRender (https://biorender.com/) on the basis of a draft provided by the author.
The authors have no conflict of interest to declare.
Glezer I, Bruni‐Cardoso A, Schechtman D, Malnic B. Viral infection and smell loss: The case of COVID‐19. J Neurochem 2021;157:930–943. 10.1111/jnc.15197
References
- Adney, D. R. , van Doremalen, N. , Brown, V. R. , Bushmaker, T. , Scott, D. , de Wit, E. , Bowen, R. A. , & Munster, V. J. (2014). Replication and shedding of MERS‐CoV in upper respiratory tract of inoculated dromedary camels. Emerging Infectious Diseases, 20, 1999–2005. 10.3201/eid2012.141280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allavena, R. E. , Desai, B. , Goodwin, D. , Khodai, T. , & Bright, H. (2011). Pathologic and virologic characterization of neuroinvasion by HSV‐2 in a mouse encephalitis model. Journal of Neuropathology and Experimental Neurology, 70, 724–734. 10.1097/NEN.0b013e3182275264 [DOI] [PubMed] [Google Scholar]
- Aragao, M. , Leal, M. C. , Cartaxo Filho, O. Q. , Fonseca, T. M. , & Valenca, M. M. (2020). Anosmia in COVID‐19 associated with injury to the olfactory bulbs evident on MRI. AJNR. American Journal of Neuroradiology, 41, 1703–1706. 10.3174/ajnr.A6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averback, P. (1983). Two new lesions in Alzheimer's disease. Lancet, 2, 1203. 10.1016/S0140-6736(83)91256-4 [DOI] [PubMed] [Google Scholar]
- Babic, N. , Mettenleiter, T. C. , Ugolini, G. , Flamand, A. , & Coulon, P. (1994). Propagation of pseudorabies virus in the nervous system of the mouse after intranasal inoculation. Virology, 204, 616–625. 10.1006/viro.1994.1576 [DOI] [PubMed] [Google Scholar]
- Barnett, E. M. , Cassell, M. D. , & Perlman, S. (1993). Two neurotropic viruses, herpes simplex virus type 1 and mouse hepatitis virus, spread along different neural pathways from the main olfactory bulb. Neuroscience, 57, 1007–1025. 10.1016/0306-4522(93)90045-H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom, U. , Giovanetti, A. , Piras, E. , & Brittebo, E. B. (2003). Methimazole‐induced damage in the olfactory mucosa: Effects on ultrastructure and glutathione levels. Toxicologic Pathology, 31, 379–387. 10.1080/01926230390201101 [DOI] [PubMed] [Google Scholar]
- Bignetti, E. , Cavaggioni, A. , Pelosi, P. , Persaud, K. , Sorbi, R. , & Tirindelli, R. (1985). Purification and characterisation of an odorant‐binding protein from cow nasal tissue. European Journal of Biochemistry, 149, 227–231. [DOI] [PubMed] [Google Scholar]
- Bihun, C. G. , & Percy, D. H. (1995). Morphologic changes in the nasal cavity associated with sialodacryoadenitis virus infection in the Wistar rat. Veterinary Pathology, 32, 1–10. 10.1177/030098589503200101 [DOI] [PubMed] [Google Scholar]
- Bilinska, K. , Jakubowska, P. , Von Bartheld, C. S. , & Butowt, R. (2020). Expression of the SARS‐CoV‐2 Entry Proteins, ACE2 and TMPRSS2, in Cells of the Olfactory Epithelium: Identification of Cell Types and Trends with Age. ACS Chemical Neuroscience, 11, 1555–1562. 10.1021/acschemneuro.0c00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borders, A. S. , Getchell, M. L. , Etscheidt, J. T. , van Rooijen, N. , Cohen, D. A. , & Getchell, T. V. (2007). Macrophage depletion in the murine olfactory epithelium leads to increased neuronal death and decreased neurogenesis. The Journal of Comparative Neurology, 501, 206–218. 10.1002/cne.21252 [DOI] [PubMed] [Google Scholar]
- Brann, D. H. , Tsukahara, T. , Weinreb, C. , Lipovsek, M. , Van den Berge, K. , Gong, B. , … Datta S. R. (2020). Non‐neuronal expression of SARS‐CoV‐2 entry genes in the olfactory system suggests mechanisms underlying COVID‐19‐associated anosmia. Science Advances, 6, (31), eabc5801. 10.1126/sciadv.abc5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breipohl, W. , Laugwitz, H. J. , & Bornfeld, N. (1974). Topological relations between the dendrites of olfactory sensory cells and sustentacular cells in different vertebrates. An Ultrastructural Study. Journal of Anatomy, 117, 89–94. [PMC free article] [PubMed] [Google Scholar]
- Bryche, B. , St Albin, A. , Murri, S. , Lacôte, S. , Pulido, C. , Ar Gouilh, M. … Meunier, N. (2020). Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS‐CoV‐2 in golden Syrian hamsters. Brain, Behavior, and Immunity. 10.1016/j.bbi.2020.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, L. , & Axel, R. (1991). A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell, 65, 175–187. 10.1016/0092-8674(91)90418-X [DOI] [PubMed] [Google Scholar]
- Butowt, R. , & Bilinska, K. (2020). SARS‐CoV‐2: Olfaction, brain infection, and the urgent need for clinical samples allowing earlier virus detection. ACS Chemical Neuroscience, 11, 1200–1203. 10.1021/acschemneuro.0c00172 [DOI] [PubMed] [Google Scholar]
- Calof, A. L. , & Chikaraishi, D. M. (1989). Analysis of neurogenesis in a mammalian neuroepithelium: Proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron, 3, 115–127. 10.1016/0896-6273(89)90120-7 [DOI] [PubMed] [Google Scholar]
- Cantuti‐Castelvetri, L. , Ojha, R. , Pedro, L. D. , Djannatian, M. , Franz, J. , Kuivanen, S. , Kallio, K. , Kaya, T. , Anastasina, M. , Smura, T. , Levanov, L. , Szirovicza, L. , Tobi, A. , Kallio‐Kokko, H. , Österlund, P. , Joensuu, M. , Meunier, F. A. , Butcher, S. , Winkler, M. S. , … Simons, M. (2020). Neuropilin‐1 facilitates SARS‐CoV‐2 cell entry and provides a possible pathway into the central nervous system. BioRxiv. 10.1101/2020.06.07.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, L. A. , MacDonald, J. L. , & Roskams, A. J. (2004). Olfactory horizontal basal cells demonstrate a conserved multipotent progenitor phenotype. Journal of Neuroscience, 24, 5670–5683. 10.1523/JNEUROSCI.0330-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F. , Zhang, A. J. , Yuan, S. , Poon, V. K.‐M. , Chan, C. C.‐S. , Lee, A. C.‐Y. , Chan, W.‐M. , Fan, Z. , Tsoi, H.‐W. , Wen, L. , Liang, R. , Cao, J. , Chen, Y. , Tang, K. , Luo, C. , Cai, J.‐P. , Kok, K.‐H. , Chu, H. , Chan, K.‐H. , Sridhar, S. , Chen, Z. , Honglin Chen, H. , To, K. K.‐W. , & Yuen, K.‐Y. (2020). Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID‐19) in golden Syrian hamster model: Implications for disease pathogenesis and transmissibility. Clinical Infectious Diseases, Mar 26, ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Shen W., Rowan N. R., Kulaga H., Hillel A., Ramanathan M., Lane A. P. (2020). Elevated ACE‐2 expression in the olfactory neuroepithelium: implications for anosmia and upper respiratory SARS‐CoV‐2 entry and replication. European Respiratory Journal, 56, 2001948. 10.1183/13993003.01948-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Fang, H. , & Schwob, J. (2004). Multipotency of purified, transplanted globose basal cells in olfactory epithelium. Journal of Comparative Neurology, 469, 457–474. 10.1002/cne.11031 [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Mi, L. I. , Xu, J. , Yu, J. , Wang, X. , Jiang, J. , Xing, J. , Shang, P. , Qian, A. , Li, Y. U. , Shaw, P. X. , Wang, J. , Duan, S. , Ding, J. , Fan, C. , Zhang, Y. , Yang, Y. , Yu, X. , Feng, Q. , … Zhu, P. (2005). Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. Journal of Infectious Diseases, 191, 755–760. 10.1086/427811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child, K. M. , Herrick, D. B. , Schwob, J. E. , Holbrook, E. H. , & Jang, W. (2018). The neuroregenerative capacity of olfactory stem cells is not limitless: Implications for aging. Journal of Neuroscience, 38(31), 6806–6824. 10.1523/JNEUROSCI.3261-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, R. , & Goldstein, B. J. (2018). Olfactory epithelium: Cells, clinical disorders, and insights from an adult stem cell niche. Laryngoscope Investig Otolaryngol, 3, 35–42. 10.1002/lio2.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codo, A. C. , Davanzo, G. G. , Monteiro, L. B. , de Souza, G. F. , Muraro, S. P. , Virgilio‐da‐Silva, J. V. , … Moraes‐Vieira, P. M. (2020). Elevated glucose levels favor SARS‐CoV‐2 infection and monocyte response through a HIF‐1α/Glycolysis‐dependent axis. Cell Metabolism, 32, 437–446.e5. 10.1016/j.cmet.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisafulli, U. , Xavier, A. M. , Dos Santos, F. B. , Cambiaghi, T. D. , Chang, S. Y. , Porcionatto, M. , Castilho, B. A. , Malnic, B. , & Glezer, I. (2018). Topical dexamethasone administration impairs protein synthesis and neuronal regeneration in the olfactory epithelium. Frontiers in Molecular Neuroscience, 11, 50. 10.3389/fnmol.2018.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, A. R. , Hadley, W. M. , Hahn, F. F. , Benson, J. M. , & McClellan, R. O. (1982). Cytochrome P‐450‐dependent monooxygenases in olfactory epithelium of dogs: Possible role in tumorigenicity. Science, 216, 57–59. 10.1126/science.7063870 [DOI] [PubMed] [Google Scholar]
- Daly, J.L. , Simonetti, B. , Antón‐Plágaro, C. , Williamson, M.K. , Shoemark, D.K. , Simón‐Gracia, L. … Yamauchi, Y. (2020). Neuropilin‐1 is a host factor for SARS‐CoV‐2 infection. BioRxiv. 10.1101/2020.06.05.134114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibattista, M. , Pifferi, S. , Menini, A. , & Reisert, J. (2020). Alzheimer's disease: What can we learn from the peripheral olfactory system? Frontiers in Neuroscience, 14, 440. 10.3389/fnins.2020.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty, R. L. (2007). Olfaction in Parkinson's disease. Parkinsonism & Related Disorders, 13(Suppl 3), S225–228. 10.1016/S1353-8020(08)70006-3 [DOI] [PubMed] [Google Scholar]
- Doty, R. L. (2019). Treatments for smell and taste disorders: A critical review. Handbook of Clinical Neurology, 164, 455–479. [DOI] [PubMed] [Google Scholar]
- Doty, R. L. , & Hawkes, C. H. (2019). Chemosensory dysfunction in neurodegenerative diseases. Handbook of Clinical Neurology, 164, 325–360. [DOI] [PubMed] [Google Scholar]
- Durante, M. A. , Kurtenbach, S. , Sargi, Z. B. , Harbour, J. W. , Choi, R. , Kurtenbach, S. , Goss, G. M. , Matsunami, H. , & Goldstein, B. J. (2020). Single‐cell analysis of olfactory neurogenesis and differentiation in adult humans. Nature Neuroscience, 23, 323–326. 10.1038/s41593-020-0587-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliezer M., Hautefort C., Hamel A‐L., Verillaud B., Herman P., Houdart E., & Eloit C. (2020). Sudden and complete olfactory loss of function as a possible symptom of COVID‐19. JAMA Otolaryngology–Head & Neck Surgery, 146, 674.. 10.1001/jamaoto.2020.0832. [DOI] [PubMed] [Google Scholar]
- Fan, Q. W. , Yuasa, S. , Kuno, N. , Senda, T. , Kobayashi, M. , Muramatsu, T. , & Kadomatsu, K. (1998). Expression of basigin, a member of the immunoglobulin superfamily, in the mouse central nervous system. Neuroscience Research, 30, 53–63. 10.1016/S0168-0102(97)00119-3 [DOI] [PubMed] [Google Scholar]
- Fletcher, R. B. , Das, D. , Gadye, L. , Street, K. N. , Baudhuin, A. , Wagner, A. , Cole, M. B. , Flores, Q. , Choi, Y. G. , Yosef, N. , Purdom, E. , Dudoit, S. , Risso, D. , & Ngai, J. (2017). Deconstructing olfactory stem cell trajectories at single‐cell resolution. Cell Stem Cell, 20(817–830), e818. 10.1016/j.stem.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodoulian, L. , Tuberosa, J. , Rossier, D. , Boillat, M. , Kan, C. , Pauli, V. , … Rodriguez, I . (2020). SARS‐CoV‐2 receptor and entry genes are expressed by sustentacular cells in the human olfactory neuroepithelium. BioRxiv. 10.1101/2020.03.31.013268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getchell, T. , Margolis, F. , & Getchell, M. (1984). Perireceptor and receptor events in vertebrage olfaction. Progress in Neurobiology, 23, 317–345. [DOI] [PubMed] [Google Scholar]
- Glezer, I. , & Malnic, B. (2019). Olfactory receptor function. Handbook of Clinical Neurology, 164, 67–78. [DOI] [PubMed] [Google Scholar]
- Graziadei, P. P. C. , & Monti‐Graziadei, G. A. (1979). Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. Journal of Neurocytology, 8, 1–18. 10.1007/BF01206454 [DOI] [PubMed] [Google Scholar]
- Hardelin, J. P. , Soussi‐Yanicostas, N. , Ardouin, O. , Levilliers, J. , & Petit, C. (2000). Kallmann syndrome. Advances in Oto‐Rhino‐Laryngology, 56, 268–274. [DOI] [PubMed] [Google Scholar]
- Hasegawa‐Ishii, S. , Shimada, A. , & Imamura, F. (2017). Lipopolysaccharide‐initiated persistent rhinitis causes gliosis and synaptic loss in the olfactory bulb. Scientific Reports, 7, 11605. 10.1038/s41598-017-10229-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick, D. B. , Lin, B. , Peterson, J. , Schnittke, N. , & Schwob, J. E. (2017). Notch1 maintains dormancy of olfactory horizontal basal cells, a reserve neural stem cell. Proceedings of the National Academy of Sciences, 114, E5589–E5598. 10.1073/pnas.1701333114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydel, J. M. , Coelho, A. , Thiebaud, N. et al (2013). Odorant‐binding proteins and xenobiotic metabolizing enzymes: Implications in olfactory perireceptor events. Anatomical Record (Hoboken), 296, 1333–1345. [DOI] [PubMed] [Google Scholar]
- Heydel, J. , Leclerc, S. , Bernard, P. , Pelczar, H. , Gradinaru, D. , Magdalou, J. , Minn, A. , Artur, Y. , & Goudonnet, H. (2001). Rat olfactory bulb and epithelium UDP‐glucuronosyltransferase 2A1 (UGT2A1) expression: In situ mRNA localization and quantitative analysis. Brain Research. Molecular Brain Research, 90, 83–92. 10.1016/S0169-328X(01)00080-8 [DOI] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , & Pohlmann, S. (2020). A multibasic cleavage site in the spike protein of SARS‐CoV‐2 is essential for infection of human lung cells. Molecular Cell, 78(779–784), e775. 10.1016/j.molcel.2020.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Kleine‐Weber, H. , Schroeder, S. , Krüger, N. , Herrler, T. , Erichsen, S. , Schiergens, T. S. , Herrler, G. , Wu, N.‐H. , Nitsche, A. , Müller, M. A. , Drosten, C. , & Pöhlmann, S. (2020). SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(271–280), e278. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook, E. H. , Wu, E. , Curry, W. T. , Lin, D. T. , & Schwob, J. E. (2011). Immunohistochemical characterization of human olfactory tissue. Laryngoscope, 121, 1687–1701. 10.1002/lary.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, C. , Surda, P. , Whitehead, E. , & Kumar, B. N. (2020). Early recovery following new onset anosmia during the COVID‐19 pandemic ‐ an observational cohort study. Journal of Otolaryngology ‐ Head & Neck Surgery, 49, 26. 10.1186/s40463-020-00423-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y.J. , Okuda, K. , Edwards, C. E. , Martinez, D. R. , Asakura, T. , Dinnon, K. H. , … Baric, R. S. (2020). SARS‐CoV‐2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell, 182, 429–446.e14. 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard, J. M. , Youngentob, S. L. , Goldstein, B. J. , Luskin, M. B. , & Schwob, J. E. (1998). Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non‐neural cells. The Journal of Comparative Neurology, 400, 469–486. [DOI] [PubMed] [Google Scholar]
- Hummel, T. , & Frasnelli, J. (2019). The intranasal trigeminal system. Handbook of Clinical Neurology, 164, 119–134. [DOI] [PubMed] [Google Scholar]
- Hyman, B. T. , Van Hoesen, G. W. , Damasio, A. R. , & Barnes, C. L. (1984). Alzheimer's disease: Cell‐specific pathology isolates the hippocampal formation. Science, 225, 1168–1170. 10.1126/science.6474172 [DOI] [PubMed] [Google Scholar]
- Iannaccone, A. , Mykytyn, K. , Persico, A. M. , Searby, C. C. , Baldi, A. , Jablonski, M. M. , & Sheffield, V. C. (2005). Clinical evidence of decreased olfaction in Bardet‐Biedl syndrome caused by a deletion in the BBS4 gene. American Journal of Medical Genetics. Part A, 132, 343–346. [DOI] [PubMed] [Google Scholar]
- Imamura, F. , & Hasegawa‐Ishii, S. (2016). Environmental toxicants‐induced immune responses in the olfactory mucosa. Frontiers in Immunology, 7, 475. 10.3389/fimmu.2016.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai, N. , Zhou, Z. , Roop, D. R. , & Behringer, R. R. (2008). Horizontal basal cells are multipotent progenitors in normal and injured adult olfactory epithelium. Stem Cells, 26, 1298–1306. 10.1634/stemcells.2007-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, T. , Itamura, S. , Nishimura, H. , Sato, Y. , Tashiro, M. , Hashikawa, T. , & Kurata, T. (2004). Productive infection in the murine central nervous system with avian influenza virus A (H5N1) after intranasal inoculation. Acta Neuropathologica, 108, 485–492. 10.1007/s00401-004-0909-0 [DOI] [PubMed] [Google Scholar]
- Jafek, B. W. (1983). Ultrastructure of human nasal mucosa. Laryngoscope, 93, 1576–1599. 10.1288/00005537-198312000-00011 [DOI] [PubMed] [Google Scholar]
- Jenkins, P. M. , McEwen, D. P. , & Martens, J. R. (2009). Olfactory cilia: Linking sensory cilia function and human disease. Chemical Senses, 34, 451–464. 10.1093/chemse/bjp020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S. , & Thomsen, A. R. (2012). Sensing of RNA viruses: A review of innate immune receptors involved in recognizing RNA virus invasion. Journal of Virology, 86, 2900–2910. 10.1128/JVI.05738-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R. T. (1964). The pathogenesis of herpes virus encephalitis. I. virus pathways to the nervous system of suckling mice demonstrated by fluorescent antibody staining. Journal of Experimental Medicine, 119, 343–356. 10.1084/jem.119.2.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya, K. , Kondo, K. , Suzukawa, K. , Sakamoto, T. , Kikuta, S. , Okada, K. , & Yamasoba, T. (2014). Innate immune responses and neuroepithelial degeneration and regeneration in the mouse olfactory mucosa induced by intranasal administration of Poly(I:C). Cell and Tissue Research, 357, 279–299. 10.1007/s00441-014-1848-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschenbaum, D. , Imbach, L. L , Ulrich, S. , Rushing, E. J. , Keller, E. , Reimann, R. R. , … Frontzek, K. (2020). Inflammatory olfactory neuropathy in two patients with COVID‐19. The Lancet, 396, 166. 10.1016/s0140-6736(20)31525-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensson, K. , & Olsson, Y. (1971). Uptake of exogenous proteins in mouse olfactory cells. Acta Neuropathologica, 19, 145–154. 10.1007/BF00688493 [DOI] [PubMed] [Google Scholar]
- Kulaga, H. M. , Leitch, C. C. , Eichers, E. R. , Badano, J. L. , Lesemann, A. , Hoskins, B. E. , Lupski, J. R. , Beales, P. L. , Reed, R. R. , & Katsanis, N. (2004). Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nature Genetics, 36, 994–998. 10.1038/ng1418 [DOI] [PubMed] [Google Scholar]
- Lane, A. P. , Turner, J. , May, L. , & Reed, R. (2010). A genetic model of chronic Rhinosinusitis‐associated olfactory inflammation reveals reversible functional impairment and dramatic neuroepithelial reorganization. Journal of Neuroscience, 30(6), 2324–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurendon, T. , Radulesco, T. , Mugnier, J. , Gérault, M. , Chagnaud, C. , El Ahmadi, A‐A. , Varoquaux, A. (2020). Bilateral transient olfactory bulb edema during COVID‐19–related anosmia. Neurology, 95, 224–225. 10.1212/wnl.0000000000009850. [DOI] [PubMed] [Google Scholar]
- Lechien, J. R. , Chiesa‐Estomba, C. M. , De Siati, D. R. , Horoi, M. , Le Bon, S. D. , Rodriguez, A. , … Saussez, S. (2020). Olfactory and gustatory dysfunctions as a clinical presentation of mild‐to‐moderate forms of the coronavirus disease (COVID‐19): a multicenter European study. European Archives of Oto‐Rhino‐Laryngology, 277, (8), 2251–2261. 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemiale, F. , Kong, W. P. , Akyurek, L. M. , Ling, X. , Huang, Y. , Chakrabarti, B. K. , Eckhaus, M. , & Nabel, G. J. (2003). Enhanced mucosal immunoglobulin A response of intranasal adenoviral vector human immunodeficiency virus vaccine and localization in the central nervous system. Journal of Virology, 77, 10078–10087. 10.1128/JVI.77.18.10078-10087.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, C. T. , Coulombe, P. A. , & Reed, R. R. (2007). Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nature Neuroscience, 10, 720–726. 10.1038/nn1882 [DOI] [PubMed] [Google Scholar]
- Li, Y. , Field, P. , & Raisman, G. (2005). Olfactory ensheathing cells and olfactory nerve fibroblasts maintain continuous open channels for regrowth of olfactory nerve fibres. Glia, 52, 245–251. 10.1002/glia.20241 [DOI] [PubMed] [Google Scholar]
- Liang, F. (2018). Olfactory receptor neuronal dendrites become mostly intra‐sustentacularly enwrapped upon maturity. Journal of Anatomy, 232, 674–685. 10.1111/joa.12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F. (2020). Sustentacular cell enwrapment of olfactory receptor neuronal dendrites: An Update. Genes, 11, (5), 493. 10.3390/genes11050493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundh, B. , Kristensson, K. , & Norrby, E. (1987). Selective infections of olfactory and respiratory epithelium by vesicular stomatitis and Sendai viruses. Neuropathology and Applied Neurobiology, 13, 111–122. 10.1111/j.1365-2990.1987.tb00175.x [DOI] [PubMed] [Google Scholar]
- MacColl, G. , Bouloux, P. , & Quinton, R. (2002). Kallmann syndrome: Adhesion, afferents, and anosmia. Neuron, 34, 675–678. 10.1016/S0896-6273(02)00720-1 [DOI] [PubMed] [Google Scholar]
- Magklara, A. , Yen, A. , Colquitt, B. M. , Clowney, E. J. , Allen, W. , Markenscoff‐Papadimitriou, E. , Evans, Z. A. , Kheradpour, P. , Mountoufaris, G. , Carey, C. , Barnea, G. , Kellis, M. , & Lomvardas, S. (2011). An epigenetic signature for monoallelic olfactory receptor expression. Cell, 145, 555–570. 10.1016/j.cell.2011.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen, D. P. , Jenkins, P. M. , & Martens, J. R. (2008). Olfactory cilia: Our direct neuronal connection to the external world. Current Topics in Developmental Biology, 85, 333–370. [DOI] [PubMed] [Google Scholar]
- Mellert, T. K. , Getchell, M. L. , Sparks, L. , & Getchell, T. V. (1992). Characterization of the immune barrier in human olfactory mucosa. Otolaryngology ‐ Head and Neck Surgery, 106, 181–188. 10.1177/019459989210600221 [DOI] [PubMed] [Google Scholar]
- Menco, B. P. (1980). Qualitative and quantitative freeze‐fracture studies on olfactory and nasal respiratory epithelial surfaces of frog, ox, rat, and dog. Cell and Tissue Research, 211, 361–373. 10.1007/BF00234393 [DOI] [PubMed] [Google Scholar]
- Meng, X. , Deng, Y. , Dai, Z. , & Meng, Z. (2020). COVID‐19 and anosmia: A review based on up‐to‐date knowledge. American Journal of Otolaryngology, 41, 102581. 10.1016/j.amjoto.2020.102581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menni, C. , Valdes, A. M. , Freidin, M. B. , Sudre, C. H. , Nguyen, L. H. , Drew, D. A. , Ganesh, S. , Varsavsky, T. , Cardoso, M. J. , El‐Sayed Moustafa, J. S. , Visconti, A. , Hysi, P. , Bowyer, R. C. E. , Mangino, M. , Falchi, M. , Wolf, J. , Ourselin, S. , Chan, A. T. , Steves, C. J. , & Spector, T. D. (2020). Real‐time tracking of self‐reported symptoms to predict potential COVID‐19. Nature Medicine, 26(7), 1037–1040. 10.1038/s41591-020-0916-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad, M. , & Martin, J. C. (2020). Pathological inflammation in patients with COVID‐19: A key role for monocytes and macrophages. Nature Reviews Immunology, 20, 355–362. 10.1038/s41577-020-0331-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, D. T. , Rowley, J. C. 3rd , & Jafek, B. W. (1982). Electron microscopy of human olfactory epithelium reveals a new cell type: The microvillar cell. Brain Research, 253, 39–46. 10.1016/0006-8993(82)90671-0 [DOI] [PubMed] [Google Scholar]
- Mori, I. , Goshima, F. , Imai, Y. , Kohsaka, S. , Sugiyama, T. , Yoshida, T. , Yokochi, T. , Nishiyama, Y. , & Kimura, Y. (2002). Olfactory receptor neurons prevent dissemination of neurovirulent influenza A virus into the brain by undergoing virus‐induced apoptosis. Journal of General Virology, 83, 2109–2116. 10.1099/0022-1317-83-9-2109 [DOI] [PubMed] [Google Scholar]
- Mori, I. , Komatsu, T. , Takeuchi, K. , Nakakuki, K. , Sudo, M. , & Kimura, Y. (1995). Parainfluenza virus type 1 infects olfactory neurons and establishes long‐term persistence in the nerve tissue. Journal of General Virology, 76(Pt 5), 1251–1254. 10.1099/0022-1317-76-5-1251 [DOI] [PubMed] [Google Scholar]
- Mori, I. , Nishiyama, Y. , Yokochi, T. , & Kimura, Y. (2004). Virus‐induced neuronal apoptosis as pathological and protective responses of the host. Reviews in Medical Virology, 14, 209–216. 10.1002/rmv.426 [DOI] [PubMed] [Google Scholar]
- Morrison, E. , & Constanzo, R. (1990). Morphology of the human olfactory epithelium. The Journal of Comparative Neurology, 297, 1–13. [DOI] [PubMed] [Google Scholar]
- Morrison, E. E. , & Costanzo, R. M. (1992). Morphology of olfactory epithelium in humans and other vertebrates. Microscopy Research and Technique, 23, 49–61. 10.1002/jemt.1070230105 [DOI] [PubMed] [Google Scholar]
- Nan, B. , Getchell, M. L. , Partin, J. V. , & Getchell, T. V. (2001). Leukemia inhibitory factor, interleukin‐6, and their receptors are expressed transiently in the olfactory mucosa after target ablation. The Journal of Comparative Neurology, 435, 60–77. 10.1002/cne.1193 [DOI] [PubMed] [Google Scholar]
- Netland, J. , Meyerholz, D. K. , Moore, S. , Cassell, M. , & Perlman, S. (2008). Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. Journal of Virology, 82, 7264–7275. 10.1128/JVI.00737-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir, Y. , Beemer, A. , & Goldwasser, R. A. (1965). West Nile Virus infection in mice following exposure to a viral aerosol. British Journal of Experimental Pathology, 46, 443–449. [PMC free article] [PubMed] [Google Scholar]
- Parma, V. , Ohla, K. , Veldhuizen, M. G. , Niv, M. Y. , Kelly, C. E. , Bakke, A. J. , … Hayes, John E (2020). More than smell—COVID‐19 is associated with severe impairment of smell, taste, and chemesthesis. Chemical Senses, bjaa041.. 10.1093/chemse/bjaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, R. W. , Brown, R. L. , Benjamin, L. , Nortley, R. , Wiethoff, S. , Bharucha, T. , … Zandi, M. S. (2020). The emerging spectrum of COVID‐19 neurology: clinical, radiological and laboratory findings. Brain, awaa240. 10.1093/brain/awaa240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi, P. , Baldaccini, N. , & Pisanelli, A. (1982). Identification of a specific olfactory receptor for 2‐isobutyl‐3‐methoxypyrazine. The Biochemical Journal, 201, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman, S. , Jacobsen, G. , & Afifi, A. (1989). Spread of a neurotropic murine coronavirus into the CNS via the trigeminal and olfactory nerves. Virology, 170, 556–560. 10.1016/0042-6822(89)90446-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi, L. S. , Salsano, E. , & Grimaldi, M. (2020). Magnetic resonance imaging alteration of the brain in a patient with coronavirus disease 2019 (COVID‐19) and anosmia. JAMA Neurology, 77(8), 1028 10.1001/jamaneurol.2020.2125 [DOI] [PubMed] [Google Scholar]
- Reiter, J. F. , & Leroux, M. R. (2017). Genes and molecular pathways underpinning ciliopathies. Nature Reviews Molecular Cell Biology, 18, 533–547. 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik, G. K. (1990). Comparative anatomy, physiology, and function of the upper respiratory tract. Environmental Health Perspectives, 85, 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, M. , van den Brand, J. M. A. , Bestebroer, T. M. , Lexmond, P. , de Meulder, D. , Fouchier, R. A. M. , Lowen, A. C. , & Herfst, S. (2020). Influenza A viruses are transmitted via the air from the nasal respiratory epithelium of ferrets. Nature Communications, 11, 766. 10.1038/s41467-020-14626-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruitenberg, M. J. , Vukovic, J. , Blomster, L. , Hall, J. M. , Jung, S. , Filgueira, L. , McMenamin, P. G. , & Plant, G. W. (2008). CX3CL1/fractalkine regulates branching and migration of monocyte‐derived cells in the mouse olfactory epithelium. Journal of Neuroimmunology, 205, 80–85. 10.1016/j.jneuroim.2008.09.010 [DOI] [PubMed] [Google Scholar]
- Sabin, A. B. , & Olitsky, P. K. (1937). Influence of host factors on neuroinvasiveness of vesicular stomatitis virus : Ii. effect of age on the invasion of the peripheral and central nervous systems by virus injected into the leg muscles or the eye. Journal of Experimental Medicine, 66, 35–57. 10.1084/jem.66.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, M. L. , Bottger, B. , Silver, W. L. , & Finger, T. E. (2002). Trigeminal collaterals in the nasal epithelium and olfactory bulb: A potential route for direct modulation of olfactory information by trigeminal stimuli. The Journal of Comparative Neurology, 444, 221–226. 10.1002/cne.10143 [DOI] [PubMed] [Google Scholar]
- Schlee, M. , & Hartmann, G. (2016). Discriminating self from non‐self in nucleic acid sensing. Nature Reviews Immunology, 16, 566–580. 10.1038/nri.2016.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen, E. J. A. , Herfst, S. , Leijten, L. M. , van Run, P. , Bestebroer, T. M. , Linster, M. , Bodewes, R. , Kreijtz, J. H. C. M. , Rimmelzwaan, G. F. , Osterhaus, A. D. M. E. , Fouchier, R. A. M. , Kuiken, T. , & van Riel, D. (2012). The multibasic cleavage site in H5N1 virus is critical for systemic spread along the olfactory and hematogenous routes in ferrets. Journal of Virology, 86, 3975–3984. 10.1128/JVI.06828-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob, J. E. , Jang, W. , Holbrook, E. H. , Lin, B. , Herrick, D. B. , Peterson, J. N. , & Hewitt Coleman, J. (2017). Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. The Journal of Comparative Neurology, 525, 1034–1054. 10.1002/cne.24105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob, J. E. , Saha, S. , Youngentob, S. L. , & Jubelt, B. (2001). Intranasal inoculation with the olfactory bulb line variant of mouse hepatitis virus causes extensive destruction of the olfactory bulb and accelerated turnover of neurons in the olfactory epithelium of mice. Chemical Senses, 26, 937–952. 10.1093/chemse/26.8.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J. , Wan, Y. , Luo, C. , Ye, G. , Geng, Q. , Auerbach, A. , & Li, F. (2020). Cell entry mechanisms of SARS‐CoV‐2. Proceedings of the National Academy of Sciences, 117, 11727–11734. 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar, V. , Kao, M. , Hamir, A. N. , Sheng, H. , Koprowski, H. , & Dietzschold, B. (1992). Kinetics of virus spread and changes in levels of several cytokine mRNAs in the brain after intranasal infection of rats with Borna disease virus. Journal of Virology, 66, 992–998. 10.1128/JVI.66.2.992-998.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivkumar, M. , Milho, R. , May, J. S. , Nicoll, M. P. , Efstathiou, S. , & Stevenson, P. G. (2013). Herpes simplex virus 1 targets the murine olfactory neuroepithelium for host entry. Journal of Virology, 87, 10477–10488. 10.1128/JVI.01748-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia, S. F. , Yan, L.‐M. , Chin, A. W. H. , Fung, K. , Choy, K.‐T. , Wong, A. Y. L. , Kaewpreedee, P. , Perera, R. A. P. M. , Poon, L. L. M. , Nicholls, J. M. , Peiris, M. , & Yen, H.‐L. (2020). Pathogenesis and transmission of SARS‐CoV‐2 in golden hamsters. Nature, 583, 834–838. 10.1038/s41586-020-2342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, G. , Gosalia, D. N. , Rennekamp, A. J. , Reeves, J. D. , Diamond, S. L. , & Bates, P. (2005). Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proceedings of the National Academy of Sciences, 102, 11876–11881. 10.1073/pnas.0505577102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solbu, T. T. , & Holen, T. (2012). Aquaporin pathways and mucin secretion of Bowman's glands might protect the olfactory mucosa. Chemical Senses, 37, 35–46. 10.1093/chemse/bjr063 [DOI] [PubMed] [Google Scholar]
- Spinato, G. , Fabbris, C. , Polesel, J. , Cazzador, D. , Borsetto, D. , Hopkins, C. , & Boscolo‐Rizzo, P. (2020). Alterations in smell or taste in mildly symptomatic outpatients with SARS‐CoV‐2 infection. JAMA, 323(20), 2089. 10.1001/jama.2020.6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinke, A. , Meier‐Stiegen, S. , Drenckhahn, D. , & Asan, E. (2008). Molecular composition of tight and adherens junctions in the rat olfactory epithelium and fila. Histochemistry and Cell Biology, 130, 339–361. 10.1007/s00418-008-0441-8 [DOI] [PubMed] [Google Scholar]
- St‐Jean, J. R. , Jacomy, H. , Desforges, M. , Vabret, A. , Freymuth, F. , & Talbot, P. J. (2004). Human respiratory coronavirus OC43: Genetic stability and neuroinvasion. Journal of Virology, 78, 8824–8834. 10.1128/JVI.78.16.8824-8834.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S‐H. , Chen, Q. , Gu, H‐J. , Yang, G. , Wang, Y‐X. , Huang, X‐Y. , … Wang, Y‐C. (2020). A mouse model of SARS‐CoV‐2 infection and pathogenesis. Cell Host & Microbe, 28, 124–133.e4. 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak, W. , Huang, N. , Becavin, C. et al (2020). SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nature Medicine, 26, 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y. , Takeda, M. , & Farbman, A. I. (1996). Supporting cells as phagocytes in the olfactory epithelium after bulbectomy. The Journal of Comparative Neurology, 376, 509–517. [DOI] [PubMed] [Google Scholar]
- Tay, M. Z. , Poh, C. M. , Renia, L. , MacAry, P. A. , & Ng, L. F. P. (2020). The trinity of COVID‐19: Immunity, inflammation and intervention. Nature Reviews Immunology, 20, 363–374. 10.1038/s41577-020-0311-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton‐Manning, J. R. , Nikula, K. J. , Hotchkiss, J. A. , Avila, K. J. , Rohrbacher, K. D. , Ding, X. , & Dahl, A. R. (1997). Nasal cytochrome P450 2A: Identification, regional localization, and metabolic activity toward hexamethylphosphoramide, a known nasal carcinogen. Toxicology and Applied Pharmacology, 142, 22–30. 10.1006/taap.1996.7975 [DOI] [PubMed] [Google Scholar]
- Tian, J. , Pinto, J. M. , Cui, X. , Zhang, H. , Li, L. , Liu, Y. , Wu, C. , & Wei, Y. (2016). Sendai virus induces persistent olfactory dysfunction in a murine model of PVOD via effects on apoptosis, cell proliferation, and response to odorants. PLoS One, 11, e0159033. 10.1371/journal.pone.0159033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi, A. , Mohammadbagheri, E. , Akbari Dilmaghani, N. , Bayat, A.‐H. , Fathi, M. , Vakili, K. , Alizadeh, R. , Rezaeimirghaed, O. , Hajiesmaeili, M. , Ramezani, M. , Simani, L. , & Aliaghaei, A. (2020). Proinflammatory cytokines in the olfactory mucosa result in COVID‐19 induced anosmia. ACS Chemical Neuroscience, 11, 1909–1913. 10.1021/acschemneuro.0c00249 [DOI] [PubMed] [Google Scholar]
- Ueha, R. , Kondo, K. , Kagoya, R. , Shichino, S. , Ueha, S. , & Yamasoba, T . (2020). Understanding olfactory dysfunction in COVID‐19: Expression of ACE2, TMPRSS2 and Furin in the nose and olfactory bulb in human and mice. BioRxiv. 10.1101/2020.05.15.097352 [DOI] [PubMed] [Google Scholar]
- van Riel, D. , Verdijk, R. , & Kuiken, T. (2015). The olfactory nerve: A shortcut for influenza and other viral diseases into the central nervous system. The Journal of Pathology, 235, 277–287. [DOI] [PubMed] [Google Scholar]
- Viana, F. (2011). Chemosensory properties of the trigeminal system. ACS Chemical Neuroscience, 2, 38–50. 10.1021/cn100102c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A. C. , Park, Y. J. , Tortorici, M. A. , Wall, A. , McGuire, A. T. , & Veesler, D. (2020). Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell, 181(281–292), e286. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Chen, W. , Zhou, Y‐S. , Lian, J‐Q. , Zhang, Z. , Du, P. … Chen, Z‐N. (2020). SARS‐CoV‐2 invades host cells via a novel route: CD147‐spike protein. BioRxiv. 10.1101/2020.03.14.988345 [DOI] [Google Scholar]
- Young, J. , Metay, C. , Bouligand, J. , Tou, B. , Francou, B. , Maione, L. , Tosca, L. , Sarfati, J. , Brioude, F. , Esteva, B. , Briand‐Suleau, A. , Brisset, S. , Goossens, M. , Tachdjian, G. , & Guiochon‐Mantel, A. (2012). SEMA3A deletion in a family with Kallmann syndrome validates the role of semaphorin 3A in human puberty and olfactory system development. Human Reproduction, 27, 1460–1465. 10.1093/humrep/des022 [DOI] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X.‐L. , Wang, X.‐G. , Hu, B. , Zhang, L. , Zhang, W. , Si, H.‐R. , Zhu, Y. , Li, B. , Huang, C.‐L. , Chen, H.‐D. , Chen, J. , Luo, Y. , Guo, H. , Jiang, R.‐D. , Liu, M.‐Q. , Chen, Y. , Shen, X.‐R. , Wang, X. I. , … Shi, Z.‐L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579, 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, C. G. K. , Allon, S. J. , Nyquist, S. K. , Mbano, I. M. , Miao, V. N. , Tzouanas, C. N. , Cao, Y. , Yousif, A. S. , Bals, J. , Hauser, B. M. , Feldman, J. , Muus, C. , Wadsworth, M. H. , Kazer, S. W. , Hughes, T. K. , Doran, B. , Gatter, G. J. , Vukovic, M. , Taliaferro, F. , … Zhang, K. (2020). SARS‐CoV‐2 Receptor ACE2 Is an Interferon‐Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell, 181(1016–1035), e1019. 10.1016/j.cell.2020.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla, A. , Chan, J. F. , Azhar, E. I. , Hui, D. S. , & Yuen, K. Y. (2016). Coronaviruses ‐ drug discovery and therapeutic options. Nature Reviews Drug Discovery, 15, 327–347. 10.1038/nrd.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]


