Abstract
Background
Microsatellite instability in colon cancer implies favorable therapeutic outcomes after checkpoint blockade immunotherapy. However, the molecular nature of microsatellite instability is not well elucidated.
Methods
We examined the immune microenvironment of colon cancer using assessments of the bulk transcriptome and the single-cell transcriptome focusing on molecular nature of microsatellite stability (MSS) and microsatellite instability (MSI) in colorectal cancer from a public database. The association of the mutation pattern and microsatellite status was analyzed by a random forest algorithm in The Cancer Genome Atlas (TCGA) and validated by our in-house dataset (39 tumor mutational burden (TMB)-low MSS colon cancer, 10 TMB-high MSS colon cancer, 15 MSI colon cancer). A prognostic model was constructed to predict the survival potential and stratify microsatellite status by a neural network.
Results
Despite the hostile CD8+ cytotoxic T lymphocyte (CTL)/Th1 microenvironment in MSI colon cancer, a high percentage of exhausted CD8+ T cells and upregulated expression of immune checkpoints were identified in MSI colon cancer at the single-cell level, indicating the potential neutralizing effect of cytotoxic T-cell activity by exhausted T-cell status. A more homogeneous highly expressed pattern of PD1 was observed in CD8+ T cells from MSI colon cancer; however, a small subgroup of CD8+ T cells with high expression of checkpoint molecules was identified in MSS patients. A random forest algorithm predicted important mutations that were associated with MSI status in the TCGA colon cancer cohort, and our in-house cohort validated higher frequencies of BRAF, ARID1A, RNF43, and KM2B mutations in MSI colon cancer. A robust microsatellite status–related gene signature was built to predict the prognosis and differentiate between MSI and MSS tumors. A neural network using the expression profile of the microsatellite status–related gene signature was constructed. A receiver operating characteristic curve was used to evaluate the accuracy rate of neural network, reaching 100%.
Conclusion
Our analysis unraveled the difference in the molecular nature and genomic variance in MSI and MSS colon cancer. The microsatellite status–related gene signature is better at predicting the prognosis of patients with colon cancer and response to the combination of immune checkpoint inhibitor–based immunotherapy and anti-VEGF therapy.
Keywords: immunotherapy, tumor microenvironment, tumor biomarkers, computational biology
Background
Colon cancer is a major cause of death worldwide. Approximately 25% of patients present with stage 4 tumors, and 25%–50% present with early-stage disease but develop metastatic disease.1 2 The prognosis has improved by early detection technologies, but the survival of patients with metastatic colon cancer remains poor.1 Thus, identifying an efficient therapy is urgently necessary for colon cancer. Colon cancer can be classified into mismatch-repair-deficient (dMMR) microsatellite instability (MSI-H) subtypes and mismatch-repair-proficient (pMMR) microsatellite stability (MSS) subtypes.3 dMMR-MSI colon tumors are characterized by a high tumor mutational burden (TMB) and high infiltration of activated CD8+ cytotoxic T lymphocytes (CTL) as well as activated Th1 cells with IFN-γ production.3 These features of MSI colon cancer make it a promising target for immunotherapy. A recent phase III study revealed pembrolizumab provided a clinically significant improvement in progression-free survival versus chemotherapy as first-line therapy for patients with MSI-H/dMMR metastatic colorectal cancer.4 Moreover, Keytruda (pembrolizumab) has been approved for the treatment of adult and pediatric patients with solid tumors with tissue TMB-high (≥10 mutations/megabase). Significant clinical benefit was obtained by anti-PD-1 therapy with pembrolizumab among patients with previously treated unresectable or metastatic MSI-H/dMMR non-colorectal cancer.5 These studies highlight the importance of microsatellite status and TMB value in immunotherapy. In contrast, MSS colon cancer has been long considered to have an inactive response to immune checkpoint inhibitors. The lack of immune infiltration and low TMB decrease the potential of obtaining benefits from immunotherapy and define MSI colon cancer as an “immune resistant” phenotype.6
The biological features and prognostic value of immune cell infiltration in tumors have been extensively reported in various tumor types.7–10 However, to date, most studies of colon cancer have been limited to bulk RNA-seq analysis.11 12 Considering the heterogeneous cell types in colon cancer, a single-cell transcriptome is urgently needed to analyze the difference in the molecular nature of immune cells in MSS and MSI colon cancer.
In the present study, we analyzed single-cell RNA-seq data, bulk RNA-seq data, and gene mutation data from public and in-house datasets. Through several computational biology methods, we depicted the molecular nature of immune cell populations from the bulk and single-cell levels in MSS and MSI colon cancer. The most important gene mutations associated with MSI tumors were identified. Then, we generated a differentially expressed gene profile from MSS and MSI colon cancer in order to identify a robust microsatellite status–related gene signature and thus predict the prognosis and differentiate between MSI and MSS tumors. A microsatellite status–related gene signature–based risk score (MSRS) was calculated for each patient. The association of the MSRS with immune cell populations, immune checkpoint inhibitors, and VEGF activities were further analyzed. A neural network using the expression profile of the microsatellite status–related gene signature was built and evaluated. We demonstrated that the microsatellite status–related gene signature is better at predicting the prognosis of patients with colon cancer and stratifying patients with a high risk of cancer-specific death, regardless of microsatellite instability.
Methods
Data source
Single cell-transcriptome files of GSE146771 and GSE98638 were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). GSE14333 is a colon cancer microarray dataset and was downloaded from GEO. The Cancer Genome Atlas (TCGA) RNA-seq transcriptome data and clinical information regarding colon cancer were downloaded via the UCSC Xena browser (https://xenabrowser.net/). The patient information is provided in online supplemental table 1 (online supplemental file 1). R software (V.3.5.3) and Python (V.3.6) were used for all the analyses in the manuscript. All bioinformatic methods are listed in online supplemental table 2 (Online supplemental file 1).
jitc-2020-001437supp001.pdf (22.9KB, pdf)
Immune cell abundance estimation
CIBERSORT R script was used to estimate the abundance of immune cell populations in the TCGA colon cancer cohorts by the bulk RNA-seq dataset.13 The cytotoxic T cell (CYT) score was obtained from cytolytic genes (calculated as the geometrical mean of PRF1 and GZMA).14 The observation of the T-cell infiltration score (TIS) was defined as the average of the standardized values for the subsets of T cells.15
ssGSEA analysis and GO analysis
Single-sample GSEA (ssGSEA) was applied to evaluate the enrichment scores for each sample. The “GSVA” package was used for ssGSEA analysis.16 The hallmark gene sets were downloaded from The Broad Institute. Tcm cell and Tem cell gene sets were from Bindea et al.17 The exhausted T score gene set was defined by Zheng et al.18 The correlation between the risk score and the enrichment scores was performed with Spearman’s coefficient. GO analysis was performed with the “clusterProfiler” package.19
Single-cell RNA-seq analysis
The single-cell transcriptome analysis for GSE146771 and GSE98638 was performed with “scanpy” in Python.20 The batch effect was removed by the BBKNN method. Nonlinear dimensional reduction was performed with the UMAP method. The gene features in each cluster were found by “scanpy”. The cell–cell interactions (CCIs) were calculated with the “SingleCellSignalR” package in R.21
Random forest algorithm for feature importance ranking
A random forest algorithm was applied on TCGA colon cancer NGS data to find the most important mutations associated with the microsatellite status in colon cancer. Briefly, the gene mutation dataset and microsatellite status were applied to find the most important gene mutations associated with microsatellite status in colon cancer. The “ranger” package was used to find the best hyperparameter in the regression process and build the model.22
DNA extraction, library preparation and targeted enrichment
NGS analyses were performed in a centralized clinical testing center (Nanjing Geneseeq Technology) according to protocols reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Zhejiang University. DNA extraction, library preparation, and targeted capture enrichment were performed with previously described methods with minor modifications.23 Briefly, genomic DNA from the white blood cells was extracted using the DNeasy Blood & Tissue Kit (Qiagen) and was used as the normal control to remove germline variations. After deparaffinizing of Formalin-fixed paraffin-embedded (FFPE) samples with xylene, genomic DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen). After quantification of DNA with Qubit 3.0 using the dsDNA HS Assay Kit (Life Technologies), a Nanodrop 2000 (Thermo Fisher) was used to evaluate DNA quality.
The KAPA Hyper Prep kit (KAPA Biosystems) was used to prepare libraries according to previously described methods.24 Briefly, a Covaris M220 instrument was used to shear 1–2 µg of genomic DNA into ~350 bp fragments. The KAPA Hyper DNA Library Prep kit (Roche Diagnostics) was used for end repair, A-tailing, and adaptor ligation of fragmented DNA, and Agencourt AMPure XP beads (Beckman Coulter) were used to perform size selection. PCR was performed to amplify DNA Libraries followed by purification using Agencourt AMPure XP beads.
Selective enrichment of 425 predefined cancer-related genes (Geneseeq Prime panel) was performed with a customized xGen lockdown probes panel (Integrated DNA Technologies). Blocking reagents were defined by Human cot-1 DNA (Life Technologies) and xGen Universal Blocking Oligos (Integrated DNA Technologies). Dynabeads M-270 (Life Technologies) and the xGen Lockdown Hybridization and Wash kit (Integrated DNA Technologies) were used to perform the capture reaction. PCR amplification was performed on captured libraries with KAPA HiFi HotStart ReadyMix (KAPA Biosystems). The KAPA Library Quantification kit (KAPA Biosystems) was used to quantify the purified library. Bioanalyzer 2100 was used to calculate the fragment size distribution.
Sequencing and bioinformatics analysis
Sequencing of the target enriched libraries was performed on the HiSeq4000 platform (Illumina) with 2×150 bp pair-end reads. bcl2fastq (v2.19) was used to demultiplex the sequencing data followed by analysis with Trimmomatic25 to remove low-quality (quality <15) or N bases. Then, alignment of the data to the hg19 reference human genome was performed with the Burrows-Wheeler Aligner (bwa-mem)26 followed by processing using the Picard suite (available at: https://broadinstitute.github.io/picard/) and the Genome Analysis Toolkit (GATK).27 VarScan228 and HaplotypeCaller/UnifiedGenotyper were used to call SNPs and indels in GATK, with the mutant allele frequency cut-off set as 0.5%. dbSNP and the 1000 Genome project were used to remove common variants. Germline mutations were filtered out by comparing with patient’s whole blood controls.
FACTERA29 and ADTEx30 were used to identify gene fusions and copy number variations, respectively. The log2 ratio cut-off for copy number gain and copy number loss was defined as 2.0 and 0.6, respectively, for tissue samples. TMB was defined as the number of somatic, coding, base substitution, and indel mutations per megabase of genome examined and was calculated as previously described.31 Briefly, all base substitutions, including nonsynonymous and synonymous alterations and indels in the coding region of targeted genes, were considered with the exception of known hotspot mutations in oncogenic driver genes and truncations in tumor suppressors.
A customized analysis algorithm was used to perform MSI testing. Briefly, the MSI sites within the panel (425-gene panel) coverage were defined with a sum of 52 mononucleotide repeats with a minimum of 15 bp repeats. The number of sequencing reads supporting each mononucleotide repeat length was tailed for each MSI site. The distribution of the length species within the tumor sample at the MSI site was compared with that summarized from a pool of normal samples. A significantly altered distribution of the length species in a site was considered unstable. A sample was defined as MSI if more than 40% of the evaluated site display instability.
Differentially expressed gene (DEG) analysis
The DEG analysis was applied with the “Limma” package on TCGA colon cancer transcriptome file.32 An empirical Bayesian method was applied to estimate the fold change between MSI-L and MSI-H or MSS and MSI-H using moderated t-tests. The adjusted p value for multiple testing was calculated using the Benjamini-Hochberg correction. The genes with an absolute log2 fold change greater than 1 and adjusted p value less than 0.05 were identified as DEGs.
Gene signature construction
The least absolute shrinkage and selection operator (LASSO) was applied to construct the microsatellite-related gene signature by TCGA colon cancer transcriptome data. L1-norm was used to penalize the weight of the features.33 A microsatellite-related gene signature–based risk score (MSRS) formula was established by including individual normalized gene expression values weighted by their LASSO Cox coefficients: .
Weighted correlation network analysis (WGCNA)
WGCNA was performed with the “WGCNA” package.34 A power of β=3 and a scale-free R2=0.95 were selected as soft-threshold parameters to ensure a signed scale-free co-expression gene network. A total of 12 non-gray modules were generated.
Survival analysis
Survival analysis was performed with the “survival” package.35 The HR was determined with univariate Cox regression analysis. The tROC analysis was performed for the evaluation of the AUC value in the follow-up period with the “survivalROC” package.36
Results
Tumor immune microenvironment in colon cancer with MSS or MSI status
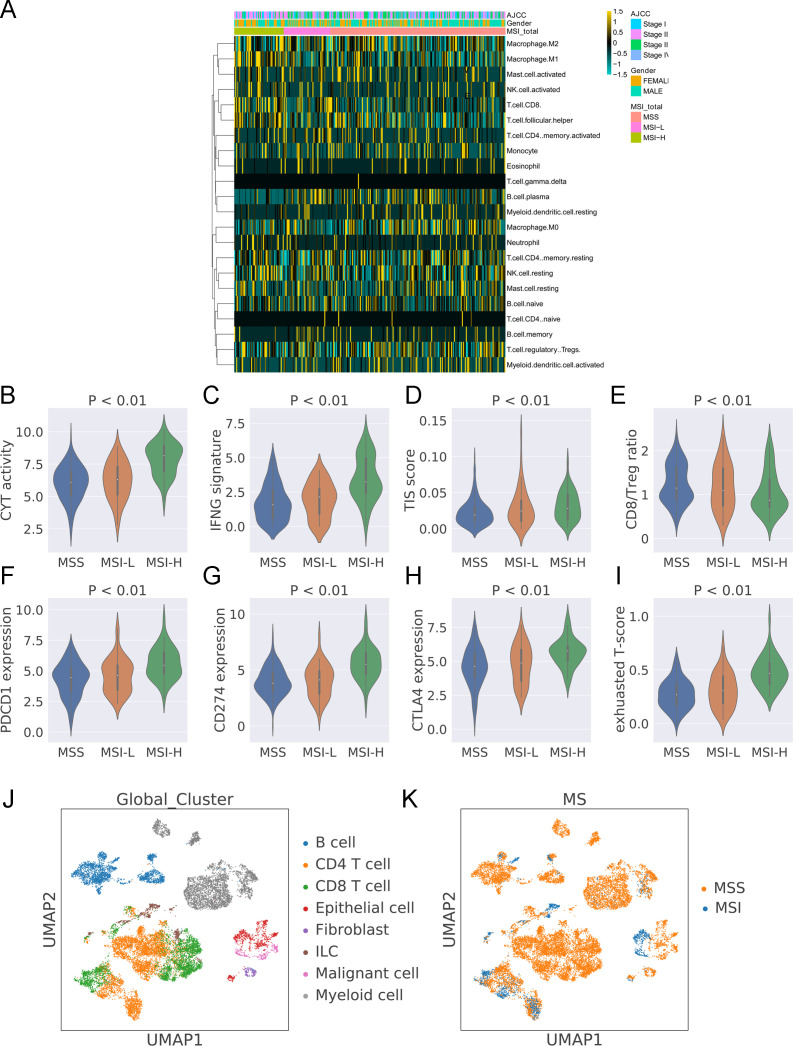
The tumor immune microenvironment of colon cancer is depicted with the 22 immune cell populations by the TCGA cohort (see online supplemental table S1 for detailed information). A higher abundance of CD8 T cells, activated NK cells, and M1 macrophages was identified in MSI colon cancer than in MSS colon cancer. Then, we estimated the CYT activity, CD8:Treg ratio, IFN-γ expression signature, and T-cell infiltration score (TIS) in MSI and MSS colon cancer. As expected, a higher CYT activity (figure 1B), IFN-γ expression signature (figure 1C), TIS (figure 1D), and CD8:Treg ratio (figure 1E) were found in MSI colon cancer. CTLA4, PD-L1 (CD274), and PD1 (PDCD1) are three important biomarkers for immune checkpoint inhibitor (ICI)–based immunotherapy. The expression levels of these three checkpoint molecules were greater in MSI-H colon than in MSS and MSI-L colon cancer (figure 1F–1H). Hence, we also evaluated the exhausted T-cell score with the ssGSEA algorithm. Despite greater CYT activity in MSI-H colon cancer, a higher exhausted T-cell score was observed in MSI-H colon cancer (figure 1I).
Figure 1.
Estimation of immune cell populations and immune signatures in TCGA-colon cancer. (A) Heatmap showing the estimation of immune cell populations in the TCGA-colon cancer cohort. (B) The ratio of immune cell populations and microsatellite status. The CYT activity (C), IFNG expression (D), TIS score (E), and CD8:Treg ratio (F) in colon cancer with MSS, MSI-L, and MSI-H. The CD274 (G), PDCD1 (H), and CTLA4 (I) expression and exhausted score (J) in colon cancer with MSS, MSI-L, and MSI-H. (K) UMAP clustering of tumor microenvironment cell populations with information regarding cell types from GSE146771. (L) UMAP clustering of tumor microenvironment cell populations with information regarding microsatellite status from GSE146771.
Single-cell RNA-seq analysis unravels the heterogeneous cell populations with differentially expressed genes in MSI and MSS colon cancer
The tumor tissues of colon cancer contain several cell populations such as malignant cells, immune cells (eg, CD8+ T cells, CD4+ T cells, B cells, and myeloid cells), and fibroblasts. To identify the molecular features in MSI and MSS colon cancer at the single-cell level, we analyzed the single-cell RNA-seq transcriptome data from GSE146771. The UMAP method was used to perform nonlinear dimensional reduction. The cells were clustered by their cell types rather than by the patients (figure 1J and online supplemental figure S1A). The microsatellite status was also shown by a UMAP plot (figure 1K). The Leiden method was applied to identify the clusters. The results revealed that the malignant cells, fibroblasts, CD8+ T cells, and CD4+ T cells from MSI and MSS colon cancer tissues were identified into different clusters (online supplemental figure S1B), indicating the molecular difference between colon cancer with MSS and MSI. Several important tumor microenvironment (TME) genes were selected to analyze the heterogeneity in colon cancer tissues (online supplemental figure S1C). MS4A1 was enriched in a subpopulation of B cells whereas FOXP3 was expressed in a subgroup of CD4+ T cells. MS4A6A, PLAUR, and PECAM1 are known to be featured genes expressed in macrophages. High expression of these genes was found in myeloid cells. EPCAM is overexpressed in epithelial cancers associated with enhanced malignant potential. EPCAM was nearly expressed in all epithelial cells and malignant cells in colon cancer.
jitc-2020-001437supp002.pdf (382.1KB, pdf)
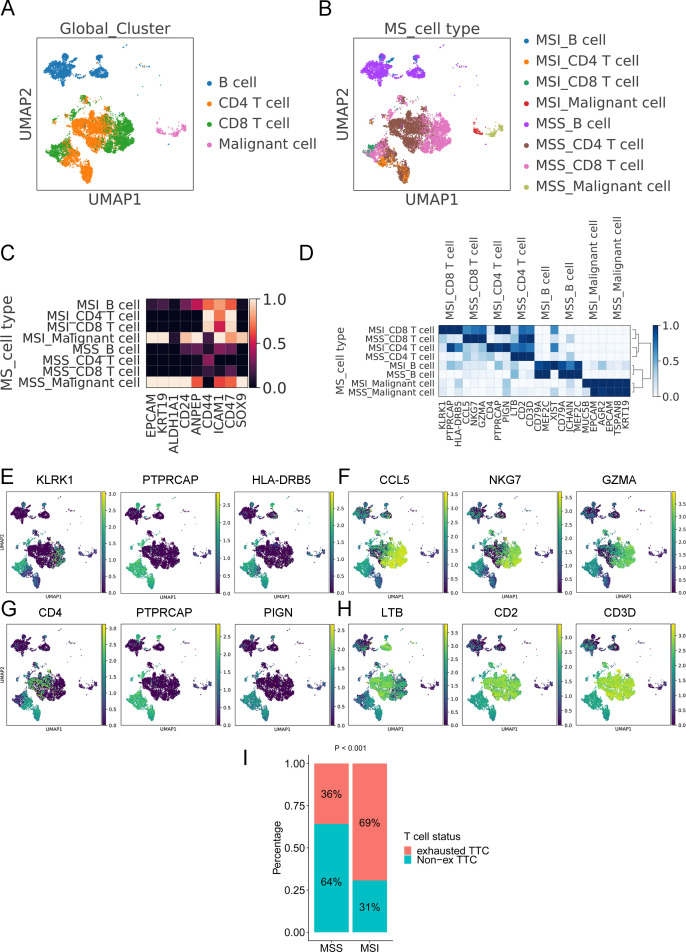
To further analyze the difference between MSS and MSI colon cancer at the single-cell level, CD8+ T cells, CD4+ T cells, B cells and malignant cells were extracted from the whole dataset. The UMAP method revealed the distribution of these cells and the microsatellite status in a two-dimensional plot (figure 2A and B). Several stem cell markers were selected to explore the heterogeneity in MSI and MSS tumors. ALDH1A1 was highly expressed in MSS tumors and lowly expressed in MSI tumors, and ICAM1 was highly expressed in MSI tumors and lowly expressed in MSS tumors (figure 2C). The featured gene expression in CD8+ T cells, CD4+ T cells, B cells and malignant cells from MSI and MSS colon cancer is shown in figure 2D. The CD8+ T cells and CD4+ T cells from MSI and MSS colon cancer had a significant difference in the most featured three genes. The CD8+ T cells from MSI colon cancer showed high KLRK1, PTPRCAP, and HLA-DRB5 expression (figure 2E); however, their counterparts from MSS colon cancer exhibited high expression of CCL5, NKG7, and GZMA (figure 2F). CD4, PTPRCAP, and PIGN were highly expressed in CD4+ T cells from MSI colon cancer (figure 2G), and LTB, CD2, and CD3D were highly expressed in CD4+ T cells from MSS colon cancer (figure 2H). The expression of three immune checkpoints (PD-L1, CTLA4, and PD1) are shown in online supplemental figure S2A–C. PD1 expression of single cells in MSI colon cancer was more homogeneous than its expression in MSS colon cancer. Most CD8+ T cells in MSI colon cancer had a high level of PD1 expression (indicated by the shape of the violin plot) whereas only a subgroup of CD8+ T cells in MSS colon cancer showed a high expression level of PD1. Using a reference with annotated T-cell status (exhausted and non-exhausted), we further analyzed the CD8+ T-cell status in MSS and MSS colon cancer. The results revealed a significantly higher percentage of exhausted CD8+ T cells in MSI colon cancer (69%) than in MSS colon cancer (36%) (p<0.001) (figure 2I).
Figure 2.
Molecular nature of colon cancer with MSS and MSI. (A) UMAP clustering of B cells, CD4+ T cells, CD8+ T cells, and malignant cells with information regarding cell types from GSE146771. (B) UMAP clustering of B cells, CD4+ T cells, CD8+ T cells, and malignant cells with information regarding microsatellite status and cell types from GSE146771. (C) The expression of stemness-related genes in each subgroup of cells. (D) The gene features in each subgroup of cells. (E) The expression of featured genes in CD8+ T cells from MSI colon cancer. (F) The expression of featured genes in CD8+ T cells from MSS colon cancer. (G) The expression of featured genes in CD4+ T cells from MSI colon cancer. (H) The expression of featured genes in CD4+ T cells from MSS colon cancer. (I) The ratio of exhausted CD8+ T cells and non-exhausted CD8+ T cells in colon cancer with MSS and MSI.
jitc-2020-001437supp003.pdf (347.3KB, pdf)
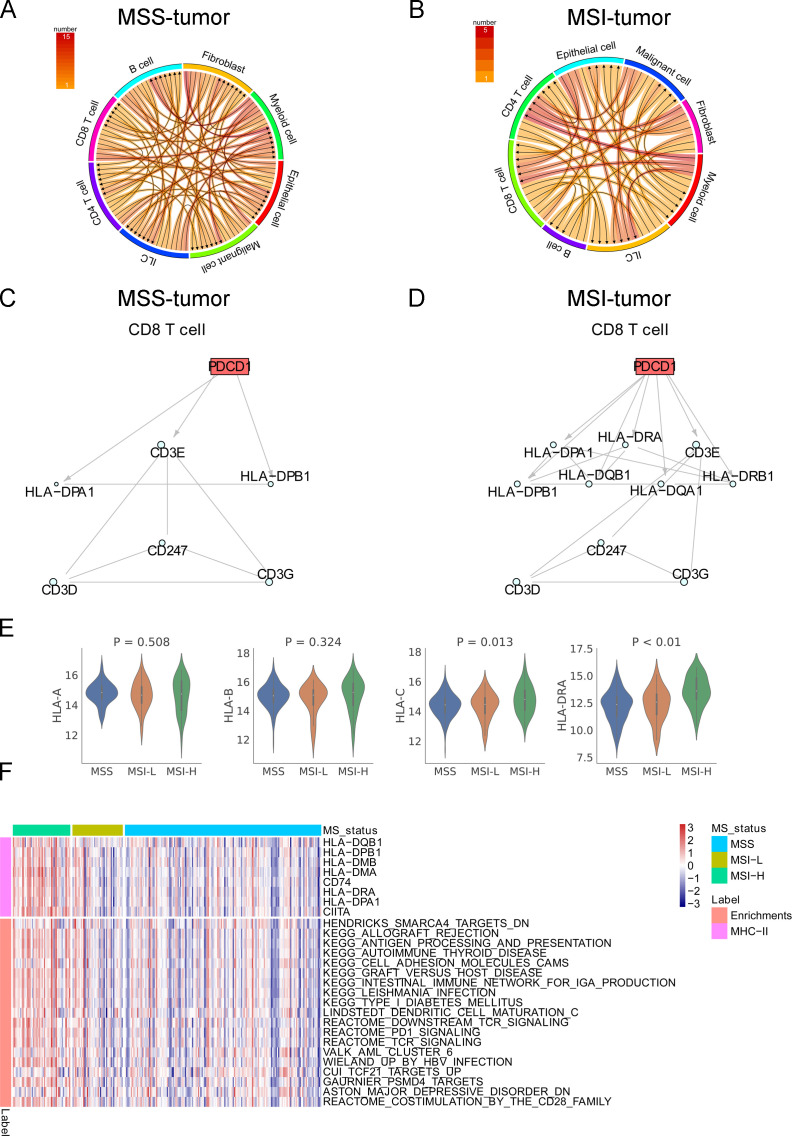
The CCIs in MSI and MSS tumors were explored with the single-cell transcriptome data. The directions and the number of ligand–receptor interactions are shown in figure 3A, B. The intracellular interactions of PD1 on CD8+ T cells from MSI and MSS tumors were therefore identified by the CCIs (figure 3C, D). A more complicated network with involvement of HLAs corresponding to MHC class II (DP, DQ, and DR) was found in MSI tumors. Greater expression of MHC class II molecules was found in MSI tumors than in MSS tumors, and ubiquitous expression of MHC class I molecules was observed between MSI and MSS tumors (figure 3E and online supplemental figure S3). ssGSEA suggested that MSI tumors harbored expression signatures of “PD-1 signaling”, “T-cell receptor signaling”, “graft-versus-host disease”, and “allograft rejection” (figure 3F). These findings indicated that there was high MHC-II expression consistent with a pro-immune/antitumor response in MSI tumors.
Figure 3.
CCIs in colon cancer with MSS and MSI. The directions and number of ligand–receptor interactions in MSS (A) and MSI (B) tumors in GSE146771. The intracellular network of PD1 in CD8+ T cells in MSS (C) and MSI (D) tumors. (E) The expression level of MHC-I molecules and MHC-II molecules (HLA-DRA) in MSI and MSS colon cancer in the TCGA colon cancer cohort. (F) The TCGA colon cancer transcriptome data were analyzed using the curated Molecular Signatures Database.
jitc-2020-001437supp004.pdf (846.5KB, pdf)
Association between microsatellite status and gene mutations in colon cancer
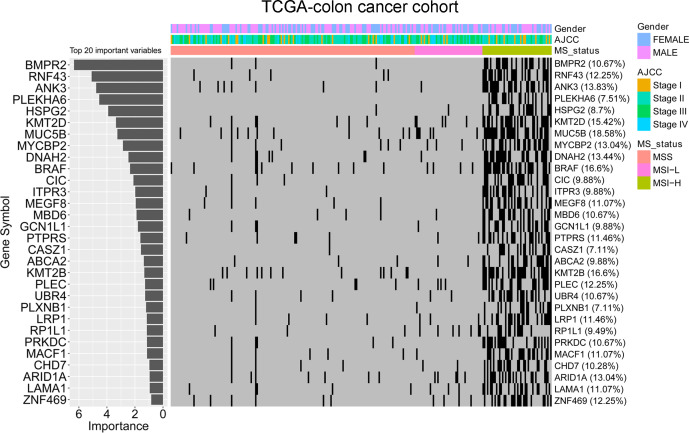
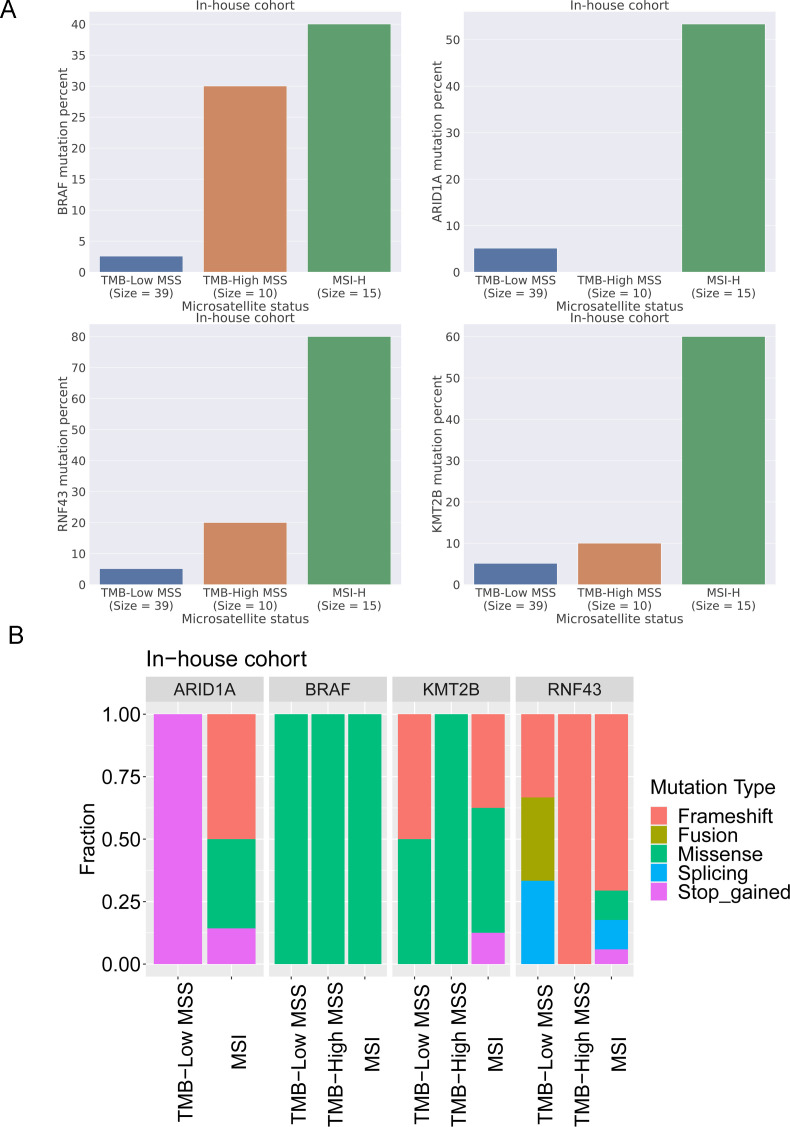
The random forest algorithm was applied to determine the importance of gene mutations associated with microsatellite status in colon cancer. Thirty of the most important gene mutations were identified in the TCGA cohort (figure 4). The results indicated that BMPR2, RNF43, and ANK3 were the most important gene mutations associated with the microsatellite status. MSI-H colon cancer tissues had a higher percentage of BMPR2 RNF43 and ANK3 mutations than MSI-L and MSS colon cancer tissues in the TCGA colon cancer cohort. To further validate the model mentioned previously, the mutation information of 39 TMB-low MSS colon cancer, 10 TMB-high MSS colon cancer, and 15 MSI colon cancer tissues were collected with the 425-panel as our in-house cohort (figure 5A). BRAF, ARID1A, RNF43, and KM2B were involved in the 425-panel. Our data revealed a significantly high frequency of BRAF, ARID1A, RNF43, and KM2B mutations in MSI colon cancer, which confirmed the random forest model. Moreover, the detailed mutation types of BRAF, ARID1A, RNF43, and KM2B are shown in figure 5B.
Figure 4.
Random forest algorithm on the TCGA colon cancer dataset identifying the most important genes associated with MSI status. The importance ranking of gene mutations and a heatmap showing the association of gene mutations and microsatellite status.
Figure 5.
In-house validation of the random forest model. (A) BRAF, ARID1A, RNF43, and KM2B mutation frequency in TMB-low MSS colon cancer, TMB-high MSS colon cancer, and MSI colon cancer. (B) BRAF, ARID1A, RNF43, and KM2B mutation types in TMB-low MSS colon cancer, TMB-high MSS colon cancer, and MSI colon cancer.
Microsatellite status–related gene signature predicts the prognosis and clinical outcome of patients with colon cancer
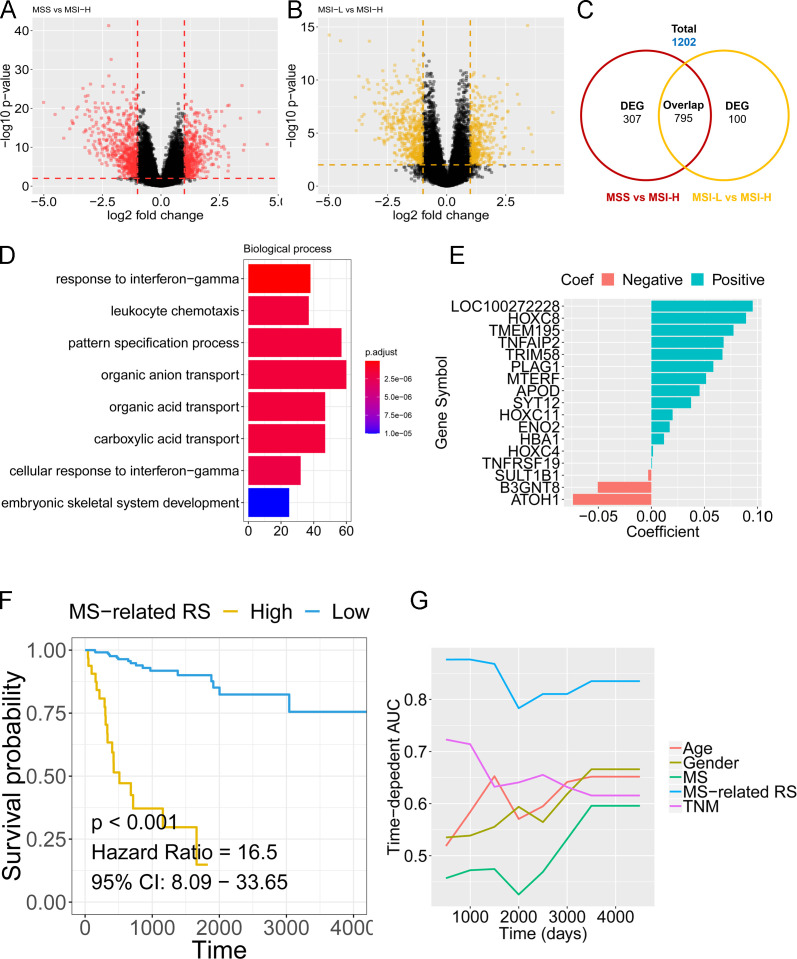
DEG analysis was performed for MSS and MSI colon cancer in the TCGA cohort. The volcano plot in figure 6A, B shows the DEGs among MSS and MSI-H, MSI-L, and MSI-H colon cancer. The Venn diagram in figure 6C shows that a total of 1202 genes were involved in the microsatellite status–related DEGs (online supplemental file 1). GO analysis revealed that the response to INF-γ, leukocyte chemotaxis, pattern specification process, organic anion transport, and other biological processes were enriched with the microsatellite status–related DEGs (figure 6D). The 1202 microsatellite status–related DEGs were used to build a prognostic model to predict the cancer-specific survival (CSS) of patients with colon cancer. LASSO Cox regression penalized the unimportant features in the regularization process, and 17 features (LOC100272228, HOXC8, TMEM195, TNFAIP2, TRIM58, PLAG1, MTERF, APOD, SYT12, HOXC11, ENO2, HBA1, HOXC4, TNFRSF19, SULT1B1, B3GNT8, and ATOH1) were finally selected as the microsatellite status–related gene signature (figure 6E, online supplemental table S3). The expression levels of the 17 genes in MSS and MSI colon cancer of different TNM stages are shown in (online supplemental figure 4-5). The MSRS for each patient was calculated based on the expression levels of the 17 genes with the coefficient in the model. The risk score formula was established as follows: . The patients with a higher MSRS had a worse CSS than patients with a lower MSRS (HR 16.5, p<0.001, best cut-off method with cut-off value 2.93) (figure 6F). Time-dependent ROC analysis further confirmed the higher accuracy of the MSRS (mean AUC values higher than 0.8 during the follow-up period) than other clinicopathological traits.
Figure 6.
Construction of a microsatellite status–related gene signature by TCGA colon cancer dataset. (A) The volcano plot shows the DEGs between MSS and MSI-H. (B) The volcano plot shows the DEGs between MSI-L and MSI-H. (C) The Venn plot shows the total genes and overlapping genes from the volcano plots. (D) Gene ontology analysis by the total genes from the Venn plot. (E) The coefficient calculated by LASSO Cox regression. (F) Kaplan-Meier analysis showed that patients with colon cancer with a higher MSRS exhibited an unfavorable CSS in the TCGA cohort. (G) tROC analysis indicated a higher prediction efficacy of the MSRS than other clinicopathological traits.
jitc-2020-001437supp010.pdf (183.1KB, pdf)
jitc-2020-001437supp011.pdf (202KB, pdf)
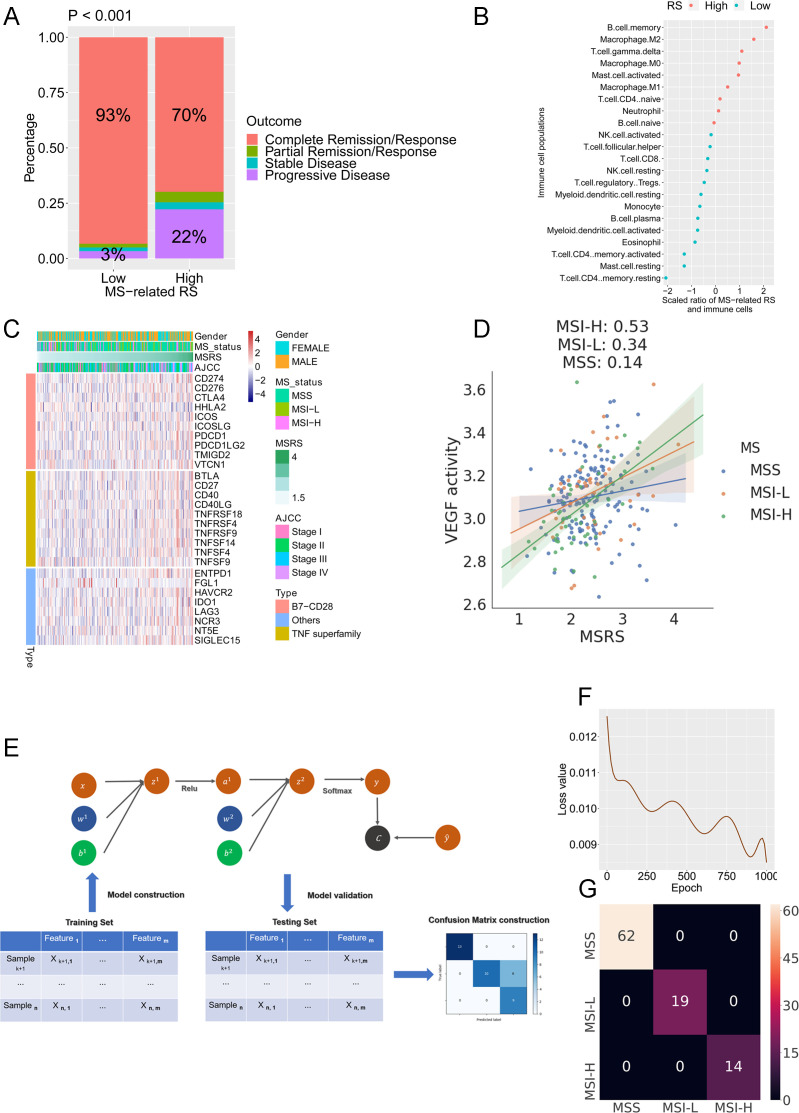
The relationship of treatment outcome and the MSRS indicated that patients with a lower MSRS had a higher fraction of complete remission (CR) (93% vs 70%) and a lower percentage of progressive disease (PD) (3% vs 22%) status (figure 7A). The association of the microsatellite status–related signature-based RS with the immune populations were calculated with Pearson’s coefficient (figure 7B), indicating a significant correlation with CD4+ T cells (negative) and memory B cells (positive). Next, we extended the analysis to include the immune checkpoint molecules, including the B7-CD28 family (CD274, CD276, CTLA4, HHLA2, ICOS, ICOSLG, PDCD1, PDCD1LG2, TMIGD2, VTCN1), TNF superfamily (BTLA, CD27, CD40, CD40LG, TNFRSF18, TNFRSF4, TNFRSF9, TNFSF4, TNFSF9) and others (ENTPD1, FGL1 HAVCR2, IDO1, LAG3, NCR3, NT5E, SIGLEC15) (figure 7C). In total, seven immune checkpoint molecules, including ENTPD1, SIGLEC15, TNFRSF4, TNFSF4, TNFSF14, PDCD1LG2, and HAVCR2, were upregulated in the high MSRS group (Cor >0.25). An external cohort (GSE14333) was used to confirm the results. The patients with a low MSRS had a more favorable survival outcome than patients with a high MSRS (online supplemental figure S6A). The association between the MSRS and expression pattern of immune checkpoint molecules in the GSE14333 showed the same tendency as it did in the TCGA cohort (online supplemental figure 6SB). For instance, the association of TNFSF4, HAVCR2, and ENTPD1 with the MSRS are shown in (online supplemental figure 6C–E). The counterparts in the TCGA cohort are shown in (online supplemental figure 6SF–H). As recently the combination of regorafenib plus nivolumab has shown a manageable safety profile and encouraging antitumor activity in colon cancer,37 we further investigated the association of the MSRS and VEGF activities in MSS, MSI-L, and MSI-H tumors. The results indicated a strikingly higher association of the MSRS and VEGF in MSI-H tumors (0.53) than in MSI-L tumors (0.34) and MSS tumors (0.14) (figure 7D). Several studies have discovered that PD-1 blockade therapy reactivates effector T cells and also promotes the proliferation of Tcm cells, improving antitumor immunity.38 39 Thus, the association between Tcm activity/Tem activity/Tscm activity and exhausted T-cell score/MSRS was analyzed. The results indicated more significant association between Tcm activity/Tem activity/Tscm activity and MSRS/exhausted score in MSI tumor compared with the association in MSS tumor (online supplemental figure S7A–F).
Figure 7.
Microsatellite status–related gene signature serves as a predictor for clinical outcome and neural network-based machine learning model predicts microsatellite status in colon cancer. (A) Patients in the low-MSRS group had a higher ratio of complete remission. (B) The ratio of immune cell populations and MSRS. (C) The expression profile of costimulatory/coinhibitory immune checkpoints landscape in the TCGA cohort. (D) The association of MSRS and VEGF activities in MSS, MSI-L, and MSI-H tumors. (E) Schematic diagram of the neural network. (F) The loss value in the testing set decreased during the training process. (G) The confusion matrix in the testing cohort.
jitc-2020-001437supp006.pdf (216.4KB, pdf)
jitc-2020-001437supp007.pdf (467.5KB, pdf)
ssGSEA analysis was performed to evaluate the biological meaning underlying the MSRS (online supplemental figure S8A). Myogenesis, apical junctions, hedgehog signaling, TGF-β signaling, and epithelial mesenchymal transition were the hallmarks with most significantly correlated with the MSRS. To further confirm the results from the ssGSEA, WGCNA was performed. With a power of β=3 set as the optimal soft threshold to construct a scale-free signed network (online supplemental figure S8B, C), a total of 12 nongray modules were identified (online supplemental figure S8D, E). Among these nongray modules, two modules (turquoise and magenta) with the highest absolute correlation values with the MSRS were selected for further analysis. The genes from the two modules were applied to GO analysis. The results indicated that extracellular matrix reorganization–related enrichments (online supplemental figure S8E) and cell division–related enrichments (online supplemental figure S8F) were related to the genes involved in the turquoise and magenta modules, which confirmed the results from the ssGSEA analysis.
jitc-2020-001437supp008.pdf (498.3KB, pdf)
Subgroup survival analysis
The MSRS serves as a promising marker to predict DSS in different subgroups of patients with colon cancer in the TCGA cohort, including TNM stage I–II (HR 6.37, p<0.001) (online supplemental figure S9A), TNM stage III–IV (HR 9.36, p<0.001) (online supplemental figure S9B), age <66 years (HR 10.19, p<0.001) (online supplemental figure 9C), age >66 years (HR 9.76, p<0.001) (online supplemental figure 9D), MSI (HR 18.39, p<0.001) (online supplemental figure S9E), MSS (HR 6.24, p<0.001) (online supplemental figure S9F), female gender (HR 11.15, p<0.001) (online supplemental figure S9G), and male gender (HR 11.37, p<0.001) (online supplemental figure S9H).
jitc-2020-001437supp009.pdf (675.3KB, pdf)
Neural network construction to identify MSS and MSI colon cancer
To further use the microsatellite status–related gene signature, we built a neural network with the gene expression pattern of the microsatellite status–related gene signature to stratify MSS and MSI colon cancer. Figure 7E shows a schematic diagram of the neural network. The expression profile of the microsatellite status–related gene signature from the TCGA colon cancer cohort was split into the training dataset (2/3) and testing dataset (1/3). With increasing epochs in the training set, the loss value decreased in the testing set (figure 7F). The confusion matrix shows that all tumors were classified accurately in the testing set (figure 7G).
Discussion
Immunotherapy has achieved long-term durable responses for multiple types of previously difficult-to-treat solid cancers, such as lung cancer and melanoma.40 MSI-H tumors cancer have been approved for the application of ICI such as pembrolizumab.3 Nivolumab (anti-PD1) plus ipilimumab (anti-CTLA-4) has demonstrated high response rates, improving progression-free survival and overall survival at 12 months, and is a promising treatment option for patients with dMMR MSI-H metastatic colorectal cancer.41 In contrast, MSS colon cancer seems to obtain limited benefits from immunotherapy. Chalabi et al42 tested ipilimumab plus nivolumab in early stage dMMR and pMMR colon cancers. Major pathological responses were observed in 7/7 (100%) dMMR tumors, with 4/7 (57%) complete responses, and no major pathological responses were found in pMMR tumors. Interestingly, significant increases in T-cell infiltration, particularly in CD8+ T cells, were seen post-treatment in both pMMR and dMMR tumors, indicating the partial activation of CD8+ T cell–mediated immunity in pMMR tumors after immunotherapy without successful pathological responses.42 In our analysis on public single-cell transcriptome data from GEO database, we found a small subset of CD8+ T cells with high expression of ICI (eg, CTLA4 and PD1) in MSS tumors and generously high PD1-expressing CD8+ T cells in MSI tumors, which may explain the increased T-cell infiltration after ipilimumab plus nivolumab treatment in both MSI and MSS tumors. dMMR-MSI colon cancer is characterized by the abundant infiltration of CD8+ tumor-infiltrating lymphocytes, CD4+ T helper cells and activation of type I interferon signaling.43 In our analysis, we confirmed the enrichment of CD8+ T cells, CD4+ T cells, and M1-macrophages in MSI-H colon cancer. A high CYT activity, ratio of CD8+:Treg, IFNG expression, and TIS were observed in MSI-H colon cancer. CD8+ T-cell infiltration into the tumor microenvironment has been long considered as a favorable factor for clinical outcomes due to its role in tumor-killing ability. Thus, we analyzed the T-cell status in MSI colon cancer. T-cell receptors (TCRs) on the surface of T cells can bind with complexes of peptides with major histocompatibility complex (MHC) class I molecules presented on the surface of all cells.44 Nevertheless, the activation of T cells does not rely on recognition of peptide–MHC class I complexes by the TCR alone. A range of coinhibitory or costimulatory signals tune the response of T cell, which tumor cells exploit to escape destruction.45 Thus, coinhibitory receptors such as CTLA4 and PD1 on T cells and the ligand of PD1 (PD-L1) on tumor cells are the normal targets of ICI.46 We found that in MSI colon cancer, CTL4, CD274, and PD-L1 were all highly expressed, which neutralized the high CYT activity in MSI colon cancer. ssGSEA analysis with exhausted T-cell hallmarks further confirmed the conclusion.
Then, we analyzed the molecular signature of MSI and MSS colon cancer at the single-cell level. The most significant featured genes were distinct in different immune cells and malignant cells from MSS and MSI colon cancer, which indicated the influence of microsatellite status on the tumor microenvironment. With single-cell transcriptome data with CD8+ T-cell annotation information (exhausted vs non-exhausted) as a reference, we further annotated the CD8+ T cells in MSI and MSS colon cancer. As a significantly higher percentage of exhausted CD8+ T cells were identified in MSI colon cancer than in MSS colon cancer, MSI tumors obtain a promising effect from ICI targeting PD1 (pembrolizumab and nivolumab), PDL1 (atezolizumab and durvalumab), and CTLA4 (ipilimumab). Characterizing cell–cell communication by analysis of ligand–receptor interactions in the tumor microenvironment can be performed by single-cell RNA-seq technology, which provides single-cell transcriptomes to define cell heterogeneity and also identifies profiles of ligand–receptor interactions and cell–cell signaling networks. In our single-cell transcriptome analysis on public single-cell transcriptome data, a complicated intracellular regulatory network that involved MHC-II molecules was identified in MSI tumors. As MHC-II expression is a functional antigen-presenting molecule that can promote CD4+ T-helper cell aid to the antitumor milieu,47 the MHC-II involvement in the PD1 network implies the importance of CD4+ T cells in antitumor immunity and ICI-based immunotherapy for MSI tumors.
Impaired DNA MMR facilitates the occurrence of frameshift mutations by insertions or deletions as well as single-base mismatches that can be point mutations in coding regions.10 Thus, dMMR MSI-H colon cancer is characterized by a high tumor mutation burden. The high mutational burden in MSI colon tumor creates many tumor-specific neoantigens, typically 10 to 50 times those of MSS colon cancer.43 The neoantigens may be processed and presented on MHC and recognized by T cells. This may be one potential reason for the high TIL infiltration in MSI colon cancer. Thus, it would be important to identify the important gene mutations that are associated with MSI tumors. BMPR2, RNF43, ANK3, and several other gene mutations were identified by a random forest algorithm in our analysis from the TCGA colon cancer dataset. One study revealed that a frameshift mutation of BMPR2 gene occurs in colon cancer with MSI-H.48 BMP signaling acts as a tumor suppressor in gastrointestinal tumorigenesis. This mutation of BMPR2 leads to the loss of BMPR2 expression and downregulation of BMP signaling in colon cancer.49 Another study demonstrated that truncating mutations of RNF43 were linked to the MSI-H phenotype in colon cancer.50 RNF43 acts as an E3 ubiquitin-protein ligase and can negatively regulate the Wnt signaling pathway.51 RNF43 mutation has been related to the sensitivity to small-molecule Wnt-specific acyltransferase porcupine inhibitors.52 Both our in-house dataset and the results from Giannakis et al50 confirmed the high frequency of RNF43 in MSI colon cancer. A BRAF mutation is involved in impaired mismatch repair, and the mutation frequency, as expected, is higher in TMB-high MSS colon cancer and MSI-H colon cancer than in TMB-low MSS colon cancer. ARID1A is a tumor suppressor that is involved in transcriptional regulation, proliferation, and chromatin remodeling.53 The high frequency of ARID1A in MSI colon cancer from our in-house cohort and the study by Cajuso et al53 further confirmed the accuracy of our random forest model.
In the next step, we analyzed the gene expression patterns in MSI tumors and MSS tumors from TCGA colon cancer dataset. A total of 1202 DEGs were found. The GO analysis by DEGs indicated enrichments in the IFN-γ response, which confirmed the previous results. As microsatellite status is not a good indicator of prognosis, we developed a gene signature–based risk score, the MSRS, to predict the cancer-specific survival of patients with colon cancer. The MSRS serves as an indicator of tumor-specific survival beyond MSI staging and TNM staging in total cohorts and subgroup analyses (eg, microsatellite status, TNM stage, age, and gender). Thus, we demonstrated that the MSRS is better at defining the prognosis of patients with colon cancer and identifying patients at a high risk of tumor recurrence, regardless of microsatellite instability. Furthermore, the MSRS also gives an indicator of tumor outcome and immunotherapy response. Patients with a lower MSRS have high probability of complete remission and active immunotherapy response. Pearson’s coefficients indicated a greater correlation between the MSRS and CD4+ T cells than between the MSRS and CD8+ T cells. One potential explanation is the “exhausted” status of CD8+ cytotoxic T cells in colon cancer. As we discussed previously, MSI colon cancer cells and a small subgroup of MSS colon cancer cells have a high expression level of coinhibitory factors such as CTLA4, PD1, and PD-L1, which impair the immune-killing activity of CD8+ T cells. The molecular feature underlying the MSRS implies the difference in the molecular nature in MSS and MSI colon cancer.
The benefits of CD8+ T cell–mediated antitumor responses might be blocked by high activity of the VEGF pathway, which mediates immune tolerance and also restricts T-cell infiltration into the tumor.54 In addition, one study revealed that VEGF-A induces the expression of the transcription factor TOX in T cells to drive an exhaustion-specific transcription program in T cells.55 Hence, it is logical to consider that combined blockade of PD-1 and VEGF-A may restore the antitumor functions of T cells and aid in better control of tumors. In our analysis, the MSRS showed a significantly higher correlation with VEGF pathway activity in MSI-H patients than in MSI-L patients and MSS patients. Thus, we conferred that the combination of a VEGF inhibitor (bevacizumab) and atezolizumab may be promising for patients with MSI-H tumors rather than for patients with MSS tumors, especially for patients with a high MSRS. Moreover, more significant association was found between Tcm activity/Tem activity/Tscm activity and MSRS/exhausted score in MSI tumor compared with the association in MSS tumor. Tcm/Tem presence in the tissue suggests that there is tumor antigen recognition. High tumor antigen recognition can lead to T-cell exhaustion. Therefore, a positive association between Tcm/Tem and exhaustion is plausible. Currently, Tscm is discussed as being those T cells that are precursor to exhausted T cells.56 57 They are thought to be the T cells that respond to checkpoint blockade. In our analysis, Tscm activity also significantly correlated with exhausted score as expected, especially in MSI-H tumors. The difference of association of the exhausted score and Tcm activity/Tem activity/Tscm activity in MSI and MSS tumor indicates the discrepancy of molecular pathways involved in the response to ICI-based immunotherapy. Furthermore, Tcm activity/Tem activity/Tscm activity showed a significant association with MSRS in MSI patients, implying the potential benefits of ICI-based immunotherapy on MSI tumor patients with high MSRS.
In summary, the featured gene expression in CD8+ T cells, CD4+ T cells, B cells, and malignant cells from MSI and MSS colon cancer showed a clear difference of the molecular nature and immune cell activities. A more homogeneous highly expressed pattern of PD1 was observed in CD8+ T cells from MSI colon cancer; however, a small subgroup of CD8+ T cells with high expression of checkpoint molecules was identified in MSS patients. BMPR2, RNF43, and ANK3 were the most important gene mutations associated with the microsatellite status in colon cancer. We also built a microsatellite status–related gene signature with which we can predict the microsatellite status with our neural network framework. The MSRS was calculated, which serves as an indicator of prognosis beyond MSI staging and the response to the combination of immunotherapy and anti-VEGF therapy.
jitc-2020-001437supp005.csv (26.9KB, csv)
Acknowledgments
We would like to thank Dr. Haowen Deng for the discussions and suggestions in building neural network.
Footnotes
XB and HZ contributed equally.
Contributors: WF, PZ, and FL supervised the project. XB and HZ designed the workflow and performed the bioinformatic analysis. WF, PZ, and FL led the study. WW, SC, XD, XZ, QF, ZT, LL, and YZ performed material preparation and data collection. XB and HZ wrote the first draft. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding: This research was supported by Major Scientific Project of Zhejiang Province (2017C03028) and National Natural Science Foundation of China Program (Grant No. 81602128).
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: The studies involving human participants were reviewed and approved by the ethics committee of the First Affiliated Hospital, Zhejiang University School of Medicine.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: NGS data are available on reasonable request.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014;64:104–17. 10.3322/caac.21220 [DOI] [PubMed] [Google Scholar]
- 2.Garborg K, Holme Ø, Løberg M, et al. Current status of screening for colorectal cancer. Ann Oncol 2013;24:1963–72. 10.1093/annonc/mdt157 [DOI] [PubMed] [Google Scholar]
- 3.Ganesh K, Stadler ZK, Cercek A, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol 2019;16:361–75. 10.1038/s41575-019-0126-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andre T, Shiu K-K, Kim TW, et al. Pembrolizumab versus chemotherapy for microsatellite instability-high/mismatch repair deficient metastatic colorectal cancer: the phase 3 KEYNOTE-177 study. Am J Clin Oncol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marabelle A, Le DT, Ascierto PA, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair–deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol 2020;38:1–10. 10.1200/JCO.19.02105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DT L, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao X, Shi R, Zhang K, et al. Immune landscape of invasive ductal carcinoma tumor microenvironment identifies a prognostic and Immunotherapeutically relevant gene signature. Front Oncol 2019;9:903 10.3389/fonc.2019.00903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao X, Shi R, Zhao T, et al. Mast cell‐based molecular subtypes and signature associated with clinical outcome in early‐stage lung adenocarcinoma. Mol Oncol 2020;14:917–32. 10.1002/1878-0261.12670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao X, Shi R, Zhao T, et al. Immune landscape and a novel immunotherapy-related gene signature associated with clinical outcome in early-stage lung adenocarcinoma. J Mol Med 2020;98:805–18. 10.1007/s00109-020-01908-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Y, Freeman GJ. The microsatellite instable subset of colorectal cancer is a particularly good candidate for checkpoint blockade immunotherapy. Cancer Discov 2015;5:16–18. 10.1158/2159-8290.CD-14-1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mlecnik B, Bindea G, Angell HK, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 2016;44:698–711. 10.1016/j.immuni.2016.02.025 [DOI] [PubMed] [Google Scholar]
- 12.Lee SJ, Jun S-Y, Lee IH, et al. CD274, LAG3, and IDO1 expressions in tumor-infiltrating immune cells as prognostic biomarker for patients with MSI-high colon cancer. J Cancer Res Clin Oncol 2018;144:1005–14. 10.1007/s00432-018-2620-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen B, Khodadoust MS, Liu CL. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 2018;1711:243–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48–61. 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Zhao Y, Dai Y, et al. Immune landscape of colorectal cancer tumor microenvironment from different primary tumor location. Front Immunol 2018;9:1578 10.3389/fimmu.2018.01578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782–95. 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 18.Zheng C, Zheng L, Yoo J-K, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017;169:1342–56. 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]
- 19.Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabello-Aguilar S, Alame M, Kon-Sun-Tack F, et al. SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res 2020;48:e55–e. 10.1093/nar/gkaa183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright MN, Ziegler A. Ranger: a fast implementation of random forests for high dimensional data, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong L, Ding N, Tong X, et al. Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics 2019;9:5532–41. 10.7150/thno.34070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Yang N, Ou Q, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res 2018;24:3097–107. 10.1158/1078-0432.CCR-17-2310 [DOI] [PubMed] [Google Scholar]
- 25.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 2014;30:2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013. [Google Scholar]
- 27.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390–3. 10.1093/bioinformatics/btu549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amarasinghe KC, Li J, Halgamuge SK. Convex: copy number variation estimation in exome sequencing data using HMM. BMC Bioinformatics 2013;14:S2. 10.1186/1471-2105-14-S2-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang W, Ma Y, Yin JC, et al. Comprehensive genomic profiling identifies novel genetic predictors of response to anti–PD-(L)1 therapies in non–small cell lung cancer. Clin Cancer Res 2019;25:5015–26. 10.1158/1078-0432.CCR-19-0585 [DOI] [PubMed] [Google Scholar]
- 32.Smyth GK. Limma: linear models for microarray data. bioinformatics and computational biology solutions using R and Bioconductor. Springer, 2005: 397–420. [Google Scholar]
- 33.Tibshirani R. Regression shrinkage and selection via the LASSO. J R Stat Soc Series B 1996;58:267–88. 10.1111/j.2517-6161.1996.tb02080.x [DOI] [Google Scholar]
- 34.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Therneau TM, Lumley T. Package ‘survival’. Survival analysis. CRAN, 2014: 3. [Google Scholar]
- 36.Heagerty P, Saha PP. SurvivalROC: time-dependent ROC curve estimation from censored survival data. R package version 1.0, 2013. [Google Scholar]
- 37.Fukuoka S, Hara H, Takahashi N, et al. Regorafenib plus nivolumab in patients with advanced gastric or colorectal cancer: an open-label, dose-escalation, and dose-expansion phase Ib trial (REGONIVO, EPOC1603). JCO 2020;38:2053–61. 10.1200/JCO.19.03296 [DOI] [PubMed] [Google Scholar]
- 38.Enamorado M, Iborra S, Priego E, et al. Enhanced anti-tumour immunity requires the interplay between resident and circulating memory CD8+ T cells. Nat Commun 2017;8:1–11. 10.1038/ncomms16073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi K, Mishima K, Ohmura H, et al. Activation of central/effector memory T cells and T-helper 1 polarization in malignant melanoma patients treated with anti-programmed death-1 antibody. Cancer Sci 2018;109:3032–42. 10.1111/cas.13758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samstein RM, Lee C-H, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202–6. 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. JCO 2018;36:773–9. 10.1200/JCO.2017.76.9901 [DOI] [PubMed] [Google Scholar]
- 42.Chalabi M, Fanchi LF, Van den Berg JG, et al. Neoadjuvant ipilimumab plus nivolumab in early stage colon cancer. Annals of Oncology 2018;29:viii731 10.1093/annonc/mdy424.047 [DOI] [Google Scholar]
- 43.Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015;5:43–51. 10.1158/2159-8290.CD-14-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khalil DN, Smith EL, Brentjens RJ, et al. The future of cancer treatment: immunomodulation, cars and combination immunotherapy. Nat Rev Clin Oncol 2016;13:273–90. 10.1038/nrclinonc.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma P, Allison JP. The future of immune checkpoint therapy. Science 2015;348:56–61. 10.1126/science.aaa8172 [DOI] [PubMed] [Google Scholar]
- 46.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov 2018;8:1069–86. 10.1158/2159-8290.CD-18-0367 [DOI] [PubMed] [Google Scholar]
- 47.Johnson DB, Estrada MV, Salgado R, et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun 2016;7:1–10. 10.1038/ncomms10582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park SW, Hur SY, Yoo NJ, et al. Somatic frameshift mutations of bone morphogenic protein receptor 2 gene in gastric and colorectal cancers with microsatellite instability. APMIS 2010;118:824–9. 10.1111/j.1600-0463.2010.02670.x [DOI] [PubMed] [Google Scholar]
- 49.Beck SE, Jung BH, Fiorino A, et al. Bone morphogenetic protein signaling and growth suppression in colon cancer. Am J Physiol Gastrointest Liver Physiol 2006;291:G135–45. 10.1152/ajpgi.00482.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giannakis M, Hodis E, Jasmine Mu X, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet 2014;46:1264–6. 10.1038/ng.3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koo B-K, Spit M, Jordens I, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 2012;488:665–9. 10.1038/nature11308 [DOI] [PubMed] [Google Scholar]
- 52.Jiang X, Hao H-X, Growney JD, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A 2013;110:12649–54. 10.1073/pnas.1307218110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cajuso T, Hänninen UA, Kondelin J, et al. Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. Int J Cancer 2014;135:611–23. 10.1002/ijc.28705 [DOI] [PubMed] [Google Scholar]
- 54.Voron T, Colussi O, Marcheteau E, et al. Vegf-A modulates expression of inhibitory checkpoints on CD8+ T cells in tumors. J Exp Med 2015;212:139–48. 10.1084/jem.20140559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim CG, Jang M, Kim Y, et al. Vegf-A drives TOX-dependent T cell exhaustion in anti-PD-1-resistant microsatellite stable colorectal cancers. Sci Immunol 2019;4:eaay0555. 10.1126/sciimmunol.aay0555 [DOI] [PubMed] [Google Scholar]
- 56.Chu T, Zehn D. Charting the roadmap of T cell exhaustion. Immunity 2020;52:724–6. 10.1016/j.immuni.2020.04.019 [DOI] [PubMed] [Google Scholar]
- 57.Buchholz VR, Busch DH. Back to the future: effector fate during T cell exhaustion. Immunity 2019;51:970–2. 10.1016/j.immuni.2019.11.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001437supp001.pdf (22.9KB, pdf)
jitc-2020-001437supp002.pdf (382.1KB, pdf)
jitc-2020-001437supp003.pdf (347.3KB, pdf)
jitc-2020-001437supp004.pdf (846.5KB, pdf)
jitc-2020-001437supp010.pdf (183.1KB, pdf)
jitc-2020-001437supp011.pdf (202KB, pdf)
jitc-2020-001437supp006.pdf (216.4KB, pdf)
jitc-2020-001437supp007.pdf (467.5KB, pdf)
jitc-2020-001437supp008.pdf (498.3KB, pdf)
jitc-2020-001437supp009.pdf (675.3KB, pdf)
jitc-2020-001437supp005.csv (26.9KB, csv)