Abstract
Major depressive disorder (MDD) is a psychiatric disorder characterized by abnormal resting state functional connectivity (rsFC) in various neural networks and especially in default-mode network (DMN). However, inconsistent findings, i.e., increased and decreased DMN rsFC, have been reported, which raise the question for the source of DMN changes in MDD. Testing whether the DMN abnormalities in MDD can be traced to either a local, i.e., intra-network, or a global, i.e., inter-network, source, we conducted a novel sequence of rsFC analyses, i.e., global FC, intra-network FC, and inter-network FC. Moreover, all analyses were conducted without global signal regression (non-GSR) and with GSR in order to identify the impact of specifically the global component of functional connectivity on within-network functional connectivity within specifically the DMN. In MDD our findings demonstrate (i) increased representation of global signal correlation (GSCORR) in DMN regions, as confirmed independently by degree of centrality (DC) and by an independent DMN template, (ii) increased within-network DMN rsFC, (iii) highly increased inter-network rsFC of both lower- and higher order non-DMN networks with DMN, (iv) high accuracy in classifying MDD vs. healthy subjects by using GSCORR as predictor. Further supporting the global, i.e., non-DMN source of within-network rsFC of the DMN, all results were obtained only when including the global signal, i.e., non-GSR, but not when conducting GSR. Together, we show for the first time increased global signal representation within rsFC of DMN as stemming from inter-network sources as distinguished from local sources, i.e., within- or intra-DMN.
Subject terms: Cognitive neuroscience, Depression
Introduction
Major depressive disorder (MDD) is a complex psychiatric syndrome characterized by pervasive disturbances, such as mood dysregulation, impaired cognitive control, suicidal thought and behavior [1]. MDD is ranked as one of the most burdensome disease and cause of disability world-wide [2] affecting more than 300 million of individuals [3]. This high prevalence of MDD makes brain-imaging investigation to obtain proper diagnosis and therapy urgent. However, major yet unresolved problems concern inconsistent findings in network abnormalities [4] and our lacking knowledge of their source, i.e., local vs. global sources.
The default-mode network (DMN) [5, 6] emerges as a critical circuit in MDD [4, 7–9]. However, findings are inconsistent as both resting state hyper- and hypo-functional connectivity (rsFC) within the DMN itself have been reported, i.e., intra-network rsFC: (1) some studies highlighted increased rsFC within DMN and specifically in anterior portions of the network [10–13]; while others (2) showed decreased rsFC within DMN [14–17].
Moreover, the DMN also showed aberrant connectivity patterns with other large-scale neural networks in MDD. Indeed, various researchers highlighted significant changes in DMN functional connectivity with other networks, i.e., inter-network rsFC, like salience network, central executive network, and visual, sensorimotor, and auditory networks [18–24]. The findings of alterations in both, intra- and inter-network DMN rsFC [4, 25], leaves open the source of abnormalities in DMN. Either DMN changes can be traced to a local source, i.e., related to changes within-network rsFC of DMN itself, or alternatively, to more global changes of networks outside the DMN that, based on inter-network rsFC, impact within-network rsFC of the DMN.
Global brain activity can be investigated with global signal correlation (GSCORR), where GS is defined as the spatial average of time varying BOLD signal across the brain [26–33]. The GS has been a matter of debate since it was hypothesized to reflect non-neuronal source of noise [34, 35] and thus it is often removed together with other “nuisance” signals, i.e., global signal regression (GSR) [36]. However, recent data show a physiological basis of the GS [31, 32]; these data suggest that removal of GS by GSR may eliminate one important source of neural activity, i.e., its global component that is also represented in local activity [26, 27, 30, 37]. This is further supported by studies showing relationship of GS to psychological features, e.g., trait personality [33, 38], as well as abnormalities in schizophrenia [26, 37, 39], autism [40, 41], and bipolar disorder [27]. While recent findings show changes in global rsFC in MDD [42–45], the impact of global rsFC on inter-network rsFC between DMN and non-DMN networks remains to be investigated—that is the focus of our study.
Our main aim is to investigate the global vs. local source of abnormal rsFC DMN changes in MDD. For this purpose, a novel sequence of rs-fMRI analyses was performed in four steps:
Our primary aim was to investigate the global brain topography of MDD as compared with a healthy control group. For that purpose, we applied two measures of global neuronal activity, that is, GSCORR, a measure based on global signal [27, 37], and, for independent confirmation, degree centrality (DC) [46, 47]. Moreover, we used an independent template of DMN [48–50] to validate the anatomical location of DMN. We hypothesized increased representation of global signal in MDD in specifically anterior and posterior midline regions of the DMN and in parahippocampal gyrus as suggested by previous GS investigation [27].
The second aim consisted in investigating intra-network rsFC within DMN. We hypothesized that those regions showing increased GS representation (particularly anterior and posterior DMN) would also be abnormally connected with each other in MDD, thus exhibiting increased intra-network rsFC. Moreover, we hypothesized that this would be manifest only in analysis without GSR while this difference would disappear when applying GSR.
The third specific aim was to explicitly test for the inter-network source of increased DMN rsFC in MDD. If DMN rsFC changes in MDD are globally based, one would expect increased rsFC from the various networks to the DMN. That was validated in various steps to demarcate the difference between the internal, i.e., within DMN, and external, i.e., non-DMN, components of within-network rsFC of the DMN.
A fourth aim was to test whether the hypothesized global changes could classify individual subjects (MDD vs. HC) implementing a multivariate pattern recognition analysis.
Importantly, we conducted all analyses with both inclusion (non-GSR) and exclusion (GSR) of the global signal that served the purpose to identify the specifically global, i.e., inter-network, component in local, i.e., intra-network, DMN activity. We hypothesized that, by eliminating the global component in the fMRI signal through the application of GSR in our analysis will abolish or reverse the differences between groups. However, this should hold only though if the DMN changes can be traced to a global (rather than local) source. Finally, we included as a proof of principle an additional MDD data set for replication where we conducted all of the above-mentioned analyses in the same way to further support our hypotheses and findings.
Methods
Subjects were recruited from the in-patients services of the Psychiatric Clinic of “IRCCS San Raffaele Turro” of Milano. After a complete description of the study, written informed consent was obtained. All research activities were approved by the local ethical committee. Clinical assessment, fMRI data acquisition, and preprocessing [51–63] are reported in Supplementary materials. Descriptive statistic and difference between MDD and HC are reported in Table S1. The local ethical committee approved all research activities.
fMRI data analysis
A graphical representation of the design of the study analysis can be found in Fig. S1 in Supplementary materials.
Calculation of global brain activity—GSCORR and DC
As a first step GSCORR was calculated as the Pearson correlation between the GS and all the other voxels in the whole brain gray matter with Fisher z transformation [57].
Second, to validate our results we calculated DC [64]. The DC analysis was performed for each subject by AFNI program 3dDegreeCentrality. As a further step a voxelwise group comparison in this study was carried out by AFNI’s function “3dttest+ +” for the two-sample t-test. The results were thresholded at p < 0.001 (FDR corrected; with a minimum cluster size of 40 voxels). The same procedure was controlled with a ROI-based approach applying GSR to the above-described analysis. Moreover we calculated the difference between Δ-(that is MDD-HC) non-GSR vs. Δ-GSR.
Third, for an independent confirmation, we compared the GSCORR values between MDD and HC extracted from a DMN template from existing literature [48, 50].
Fourth, the results of the GSCORR and DC were combined through an intersection analysis to establish the global topography specific for MDD (Global MDD network) and according to our hypothesis we tested for similarity between the Global MDD network and the DMN template [48, 50] (see Supplementary materials).
As a control analysis for measure specificity (see Supplementary materials), it was also calculated a similar analysis based on connectivity, e.g., global brain functional connectivity and a different analysis based on amplitude variance standard deviation of BOLD signal. Several control analysis for the impact of motion (Figs. S2 and S3), medication, number of depressive episodes, age, and sex are reported in Supplementary material.
Intra-DMN resting state functional connectivity
According to our hypothesis, for the intra-network FC analysis, the two key-nodes of the anterior and posterior DMN, i.e., the PACC and the PCC were expected to show a stronger FC in MDD when compared to HC. First we localized the two regions from the MNI provided with AFNI binaries since the two regions have been shown to be highly connected in the DMN [48, 53, 65–68]. Specifically, we used a seed region in the PACC (right and left are combined; x = ±8 y = 36 z = 3) and in PCC (right and left are combined; x = ±10 y = 56 z = 14).
Next, FC maps were computed as maps of temporal correlation coefficients between the BOLD time-course averaged across voxels in the seed region and all the other voxels of the mask of our resulting global analysis [69–71]. The correlation coefficients were then transformed into z-values by means of the Fisher r-to-z transformation, to improve normality for group-level analysis. This procedure produced spatial maps in which the values of voxels represented the strength of the correlation with the ROIs.
As a second step a group comparison was carried out by AFNI’s function “3dttest+ +” for the two-sample t-test. The results were thresholded at p < 0.001 (FDR corrected; with a minimum cluster size of 40 voxels). The same procedure was controlled with a ROI-based approach applying GSR. Moreover we calculated the difference between Δ-non-GSR vs. Δ-GSR. As a further control a set of correlation matrices between ROIs defined from previous step is reported for MDD and HC for both conditions (non-GSR and GSR; see also Supplementary material and Tables S1 and S2)
Between-network functional connectivity
For the inter-network FC analysis, we used as seed various defined functional networks from a previous study [52, 72]. The original template consisted of 11 functional networks [52, 73, 74]. For the aim of this study, we included the DMN and (1) three functional networks corresponding to “primary sensory functions”, namely sensory/somatomotor (SS), auditory (Audi), and visual networks (Vis), (2) three functional areas corresponding to “attentional functions”, namely salience (Sal), ventral attention (VA) and dorsal attention (DA), (3) two “higher order cognitive functions” such as frontoparietal task control (FPTC), cingulo-opercular task control (COTC). The functional mask of the selected networks’ union was defined as the global mask, which was used to extract the global value for each of the following measurements.
First we generated a functional connectivity matrix [48] for each group (MDD and HC) and for each condition (non-GSR and GSR) to calculate the degree of connectivity between DMN and non-DMN networks and the difference between MDD and HC.
Second, a voxelwise FC map for each non-DMN network was computed as a map of temporal correlation coefficients between BOLD time course in each voxel of the brain and BOLD time course averaged across voxels in the seed regions. The correlation coefficients were then transformed into z-values by means of the Fisher r-to-z transformation, to improve normality for group-level analysis. This procedure produced spatial maps in which the values of voxels represented the strength of the correlation with the networks. A group comparison was carried out by AFNI’s function “3dttest+ +” for the two-sample t-test. The results were thresholded at p < 0.001 (FDR corrected; with a minimum cluster size of 40 voxels).
The same procedure was controlled with a ROI-based approach applying GSR. In addition, we calculated the difference between Δ-non-GSR vs. Δ-GSR.
Third, we tested for the connectivity within DMN (Intra-DMN FC), the connectivity between all other non-DMN network (non-DMN FC) and, finally, the correlation between DMN and non-DMN networks. We thus calculated for all three, (1) Intra-DMN FC, (2) nonDMN FC, (3) DMN–non-DMN corr: (a) a group comparison between MDD and HC and between non-GSR and GSR; (b) a group comparison for the Δ of non-GSR–GSR between MDD and HC; (c) a comparison between the three groups for the Δ of MDD-HC for non-GSR and GSR separately. Finally, (d) we correlated the DMN with the non-DMN signal for each group and each condition.
Individual classification of subjects—machine learning as support vector machine
To test whether the global origin of increased global connectivity in DMN allows to predict single subject data including differentiation of MDD vs. HC subjects, we conducted machine learning performing a multivariate pattern analyses on GSCORR by using Support Vector Machine for binary classification of MDD and HC as implemented in PRoNTo software version 2.1 (http://www.mlnl.cs.ucl.ac.uk/pronto). Two separate SVM were performed on GSCORR measures with non-GSR and GSR to test whether the hypothesized group difference between MDD and HC is lost or attenuated in the case of GSR.
In supervised pattern recognition analysis, the predictive function is usually defined during a training phase where the algorithm learns patterns from the provided data in order to predict a label or a target. Then, during a test phase, the algorithm is used to predict outcome in an independent data set. Predictions obtained are then compared to the true labels, providing measures of algorithm accuracy. In our study we performed a fivefold nested cross validation on subjects per group in order to optimize the model’s hyperparameter (i.e. soft-margin C ranged 0.01, 0.1, 1, 10, 100, 1000) and to compute model performance. Nested cross-validation scheme has been shown to provide more reliable estimate than other validation procedures such as leave-one-out [75].
Predictions obtained in the test set allow defining balanced accuracy (BA) value, computed as the average of the class accuracies (corresponding to the sensitivity for MDD and specificity for HC), positive and negative predictive values and area under the receiver operator curve (AUC). Each class accuracy was calculated averaging classification results across the fivefold of cross validation [76]. We also estimated the statistical significance (p < 0.05) of accuracies by using 1000 permutations of the labels during training phase.
Results
Increased representation of global activity in DMN in MDD
To investigate the specific alteration in GS topography of MDD, we first investigated GSCORR (non-GSR) topographical distribution in MDD by comparing it with healthy controls.
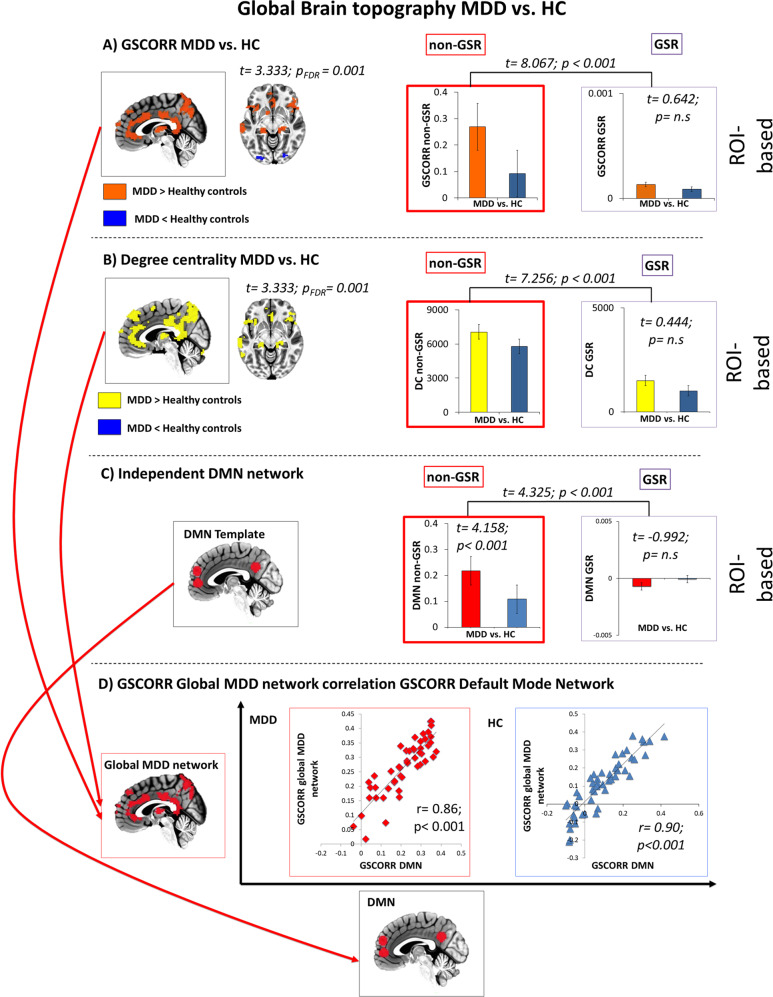
The two-sample t-test with GSCORR maps yielded a very significant difference (t = 3.333; p = 0.001; FDR corrected; cluster size voxels > 40) indicating a strongest increase of a distributed cluster (Fig. 1) comprising typical DMN regions, e.g., PACC and PCC.
Fig. 1. Global Level.
Global brain topography of MDD. a Global signal correlation (GSCORR) topography of MDD vs. HC. b Degree centrality (DC) topography of MDD vs. HC c Independent DMN template MDD vs. HC d correlation between GSCORR Global MDD Network and the GSCORR of the DMN template.
To further independently validate the global topographical patterns yielded by GSCORR findings, we conducted analysis using DC. A very similar topography was further confirmed by the DC distribution in MDD showing a prominent role of DMN regions in differentiating MDD from HC (t = 3.333; p = 0.001; FDR corrected; cluster size voxels >40).
We replicated the same results yielded by extracting GSCORR values from a DMN template [48], which confirmed that there is a significant difference in MDD vs. HC for the non-GSR condition (t = 4.158; p < 0.001, 95% CI: [0.153 0.257]; SE = 0.22).
Given the difference of the two measures (GSCORR is based on the degree of synchronization between time-series while DC is based on how many connections, i.e., edges, a node has), we first checked for their spatial similarity in representing the global brain topography. Our results confirm a similarity between GSCORR and DC topography (voxel based Pearson correlation: r = 0.88; p = 0.0001; FDR corrected). Secondly, we performed an intersection analysis (3dMean function in AFNI with the option –mask_inter) applying the same statistical threshold for the two-sample t-test (t = 3.333; p = 0.001; FDR corrected; cluster size voxels > 40). The result shows a network composed only by voxels shared by the two different analysis and differentiating MDD from HC. This specific “Global MDD network” (See Table S2 for more regional information) as the result of the intersection of the GSCORR and the DC maps allows establishing that the global abnormalities of MDD are independent from the calculation methodology, i.e., GSCORR and DC.
Finally, to check whether the regions obtained in the “Global MDD network”, as based on converging GSCORR and DC maps, really correspond to the DMN, we compared them with an independent DMN template [48]. We calculated the correlation between the GSCORR values of global MDD network and the DMN template, which was highly significant (for MDD r = 0.86 p < 0.001 95% CI: [0.772 0.921]; SE = 0.038 and for HC r = 0.90, p < 0.001 95% CI: [0.854 0.942]; SE = 0.023) (see also Supplementary Materials).
These analyses were conducted without regressing the GS (non-GSR). To investigate the impact of the global activity, i.e., GS on the topographical representation of DMN differences in MDD (vs. healthy), we conducted the same analyses with GSR. Applying GSR, no significant difference between the MDD and HC were found in the “Global MDD network” (for GSCORR; t = 0.64; p = 0.52; 95% CI: [0.031 0.247]; SE = 0.013; for DC t = 0.44; p = 0.65; 95% CI: [−0.001 0.001]; SE = 0.001; for GSCORR of the independent DMN template; t = 0.99; p = 0.32; 95% CI: [0.026 0.227]; SE = 0.003). This suggests global origin of DMN changes in MDD as also confirmed by our significant difference found between Δ-group non-GSR vs. Δ-group GSR both for GSCORR (t = 8.067; p < 0.001; 95% CI: [0.131 0.222]; SE = 0.22); for DC GSCORR (t = 7.256; p < 0.001; 95% CI: [0.11 0.247]; SE = 0.13) and for GSCORR of the independent DMN template (t = 3.325; p < 0.001; 95% CI: [0.191 0.268]; SE = 0.25). Moreover, we performed several control analysis to confirm our results on global brain connectivity (See Supplementary material).
Increased intra-DMN rsFC in MDD
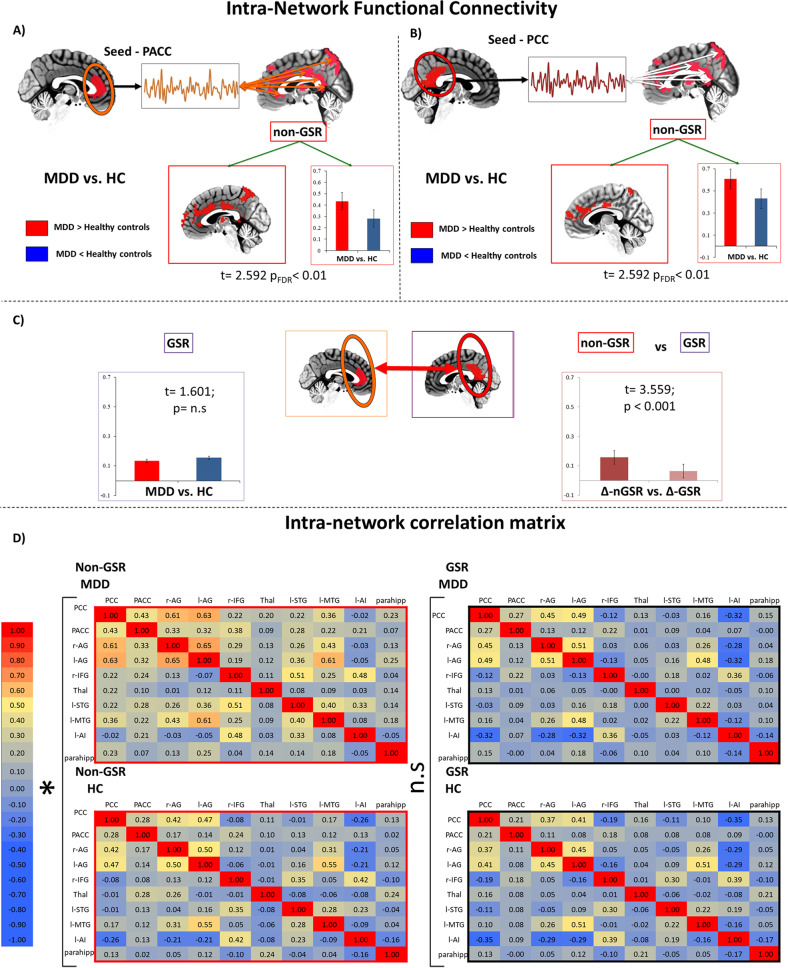
According to our hypothesis, we defined independently from our previous analysis PACC and PCC as ROIs and, in a second step, we used them consecutively as seed to calculate FC within the DMN network. We thus obtained PACC-FC within-DMN and PCC-FC within-DMN maps. The group comparison was carried out by AFNI’s function “3dttest+ +” for the two-sample t-test. The seed-based PACC rsFC connectivity within the DMN was significantly increased with all the other regions of the global MDD network, particularly posterior regions, in the case of MDD (t = 2.592, p < 0.01, FDR corrected). The same pattern was found for the seed-based PCC rsFC connectivity showing increased connectivity with anterior regions (t = 2.592; p < 0.01, FDR corrected).
We then repeated the same procedure with a ROI-based approach applying GSR. Interestingly, the two-sample t-test comparing the rsFC (GSR) between PACC and PCC in MDD vs. HC did not yield any significant difference (t = 1.601, p = 0.11; 95% CI: [−0.048 0.104]; SE = 0.037). Moreover, we found a significant difference between Δ-group non-GSR vs. Δ-group GSR (t = 3.559; p < 0.001; 95% CI: [0.041 0.146]; SE = 0.26).
We also calculated correlation coefficient matrices of the ROIs (resulting from global brain topography of step1) for both groups, i.e., MDD and HC and for both conditions, i.e., non-GSR and GSR. Thus, a group comparison between MDD and HC was carried (Fig. 2a-d, see Supplementary materials Step 2—Table S3a for non-GSR results and Table S3b for GSR results). These results indicate that in the case of non-GSR there is a significant difference between MDD and HC in almost of the ROI-dyads, while, on the opposite, this difference is lost or attenuated in the case of GSR.
Fig. 2. Intra-network level.
Intra network functional connectivity analysis in DMN. a Voxelwise PACC-FC of MDD vs. HC (non-GSR). b Voxelwise PCC-FC of MDD vs. HC (non-GSR). c Roi-based PACC-PCC-FC of MDD vs. HC (GSR) and a comparison of Δ-non-GSR vs. Δ-GSR. d Intra-network correlation matrices for both groups, i.e., MDD and HC, and for both conditions, non-GSR and GSR. *p < 0.01.
Altogether these findings suggest that the increased within-DMN rsFC is related to the global activity of the brain as the removal of the latter annulated the former. If, in contrast, increased within-DMN rsFC were of local source, removal of GSR should have no effect on MDD vs. HC within-network differences.
Increased rsFC between different networks and DMN in MDD
The findings so far show increased global activity presentation in DMN as well as increased intra-DMN rsFC in MDD. Both findings were only obtained in those analyses where the global signal was maintained. In contrast, both findings disappeared when regressing the GS. This strongly suggests that increased rsFC within the DMN is from global rather than local source, i.e, it stems from increased activity contribution between networks outside the DMN with the DMN itself. If so, one would expect that the networks other than the DMN should exhibit increased rsFC with the DMN. To test that hypothesis, we conducted three main lines of analyses.
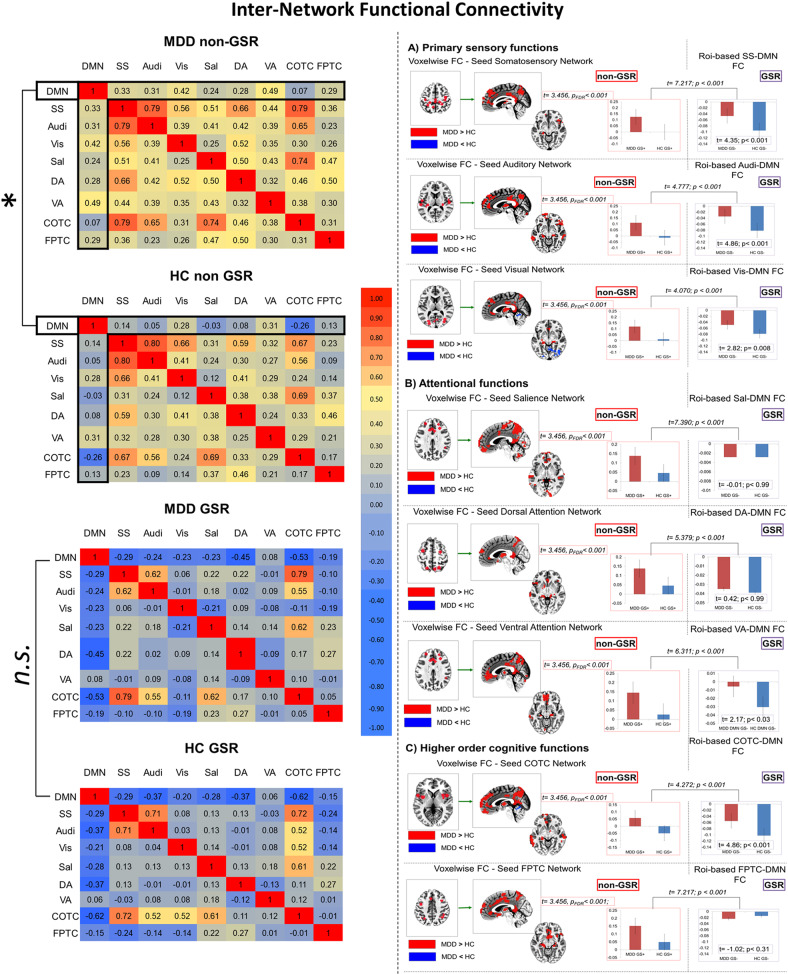
First, we correlated the rsFC of all networks with each other with both non-GSR and GSR. This yielded correlation maps on the left of Fig. 3. As can be seen in the first vertical column/row (marked), we obtained high correlation of DMN with almost all other networks in MDD during non-GSR, which differed significantly from HC (t = 3.456; p < 0.001, 95% CI: [0.195 0.293]; SE = 0.22). In contrast, that difference disappeared when using GSR as now the DMN–non-DMN network correlation no longer differed between MDD and HC (see also Fig. 4). These data suggest that within-network DMN rsFC is strongly connected with non-DMN rsFC.
Fig. 3. Inter-network level.
Left side of the figure: Correlation matrices for DMN and non-DMn networks in MDD and HC and in both conditions non-GSR and GSR. Right side of the figure: voxelwise (non-GSR) FC using as seed different networks (other than DMN) a priori defined. Roi/Networknetowork-based (GSR) FC of different networks with DMN. Δ-non-GSR vs. Δ-GSR at a roi/network-based level. a It shows the results for the analysis of primary sensory functions network, which are Somatosensory (SS), Auditory (Audi), and Visual (Vis) networknetowork. b It shows the results for the analysis of attentional functions networks, which are salience (Sal), dorsal attention (DA) and ventral attention (VA) network. c It shows the results for the analysis of higher order cognitive functions, which are CinguloCigulo-Opercular task control (COTC) and FrontoParietal task control (FPTC) networks. *p < 0.001.
Fig. 4. Comparison of i) Intra-DMN rsFC, ii) non-DMN rsFC and iii) DMN–non-DMN correlation.
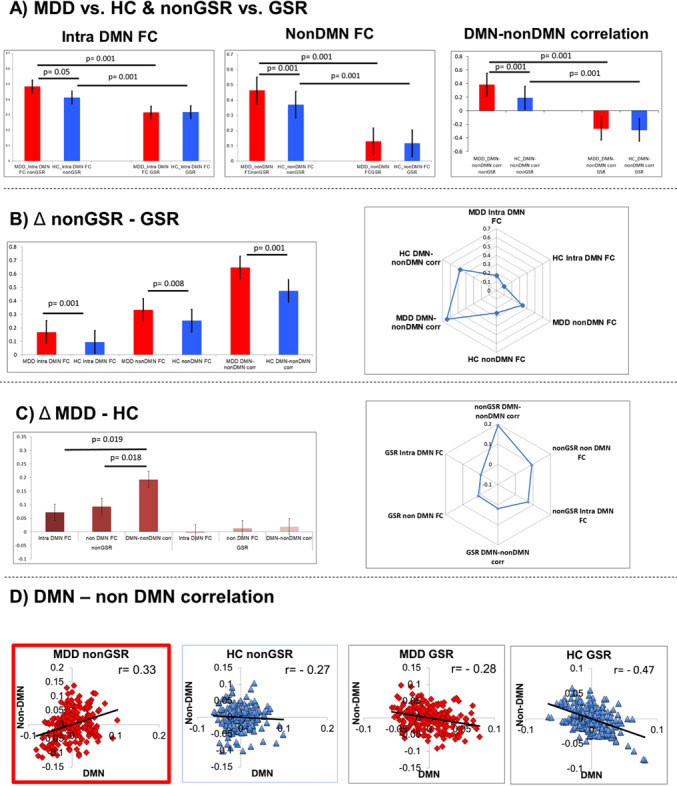
a For i) intra-DMN rsFC F (1, 98) = 5.94, p < 0.01; for ii) non-DMN rsFC F (1, 98) = 57,32, p < 0.001; for iii) DMN–non-DMN correlation F (1, 98) = 62.81, p < 0.01) Bonferroni post hoc test revealed that there was a significant difference between MDD and HC in non-GSR condition, this difference disappeared for the GSR condition. b For Δ non-GSR–GSR we found a significant difference between MDD and HC for all i) intra-DMN FC (t = 3.445, p < 0.001) for ii) non-DMN rsFC (t = 32,26, p < 0.001) and for iii) DMN–non-DMN rsFC (t = 3,56 p < 0.001). c For Δ MDD-HC we found a significant difference between DMN and non-DMN correlation with Intra-DMN FC (t = 2,43, p = 0.19) between DMN–non-DMN rsFC and non-DMN rsFC (t = 2,44, p = 0.18) while no differences were observed between Intra-DMN rsFC and non-DMN rsFC (t = −1399 p = 0.168). d scatter plot for DMN–non-DMN correlation for MDD non-GSR (r = 0.33, p = 0.19), for HC non-GSR (r = −27, p = 0.058); for MDD GSR (r = −29, p = 0.048); for HC GSR (r = −47, p = 0.0005).
To further support that assumption of the non-DMN source of increased within-DMN rsFC, we, in a second step, took various networks (other than DMN) as seed and investigated their rsFC to the rest of the brain including the DMN. A voxelwise FC map for each seed-networks was computed as a map of temporal correlation coefficients between BOLD time-course in each gray matter brain voxel and BOLD time-course averaged across voxels in the seed regions. We used as seed-networks different well established node template corresponding to SS, Audi, Vis (for primary sensory functions), Sal, VA, DA (for attentional functions), FPTC, COTC (for higher order cognitive functions).
In Fig. 3 (right part) it can be seen how, for almost all networks rsFC maps (non-GSR), there is a significant difference between MDD and HC (MDD > HC) in regions like PACC, PCC, and parahippocampal gyrus (t = 3.456, p < 0.001; FDR corrected; cluster size voxels >40).
We performed the same analysis with a ROI/network-based approach by excluding global signal, i.e., GSR; interestingly this difference in DMN is lost or attenuated and it shows a pattern indicating increased negative correlation of other networks with DMN in HC when GSR was applied. In addition, we calculated the difference between Δ-non-GSR vs. Δ-GSR (see Supplementary material Step 3).
Interestingly, in all cases of different networks-DMN FC (GSR) we see a pattern indicating an increased anticorrelation between different networks and DMN in HC when compared to MDD.
Third, to better distinguish the different sources of within-network rsFC of DMN, we compared three estimates including (1) intra-DMN rsFC, (2) non-DMN rsFC, and (3) DMN–non-DMN correlation with and without GSR for the two groups, i.e., MDD and HC. This yielded significant differences between MDD and HC in all three estimates in non-GSR, whereas these differences disappeared when applying GSR (upper row of Fig. 4).
To identify the global source of these differences, we compared the difference between non-GSR and GSR for all three estimates; that again yielded significant differences between MDD and HC for all three estimates—this suggests that the differences in MDD for all three estimates are really stemming from a global rather than local source (upper middle row of Fig. 4).
Finally, we calculated the MDD-HC differences of all three estimates (for non-GSR and GSR) in order to compare which of the three estimates differs the most: this showed that the DMN–non-DMN correlation showed the strongest difference in MDD when compared to intra-DMN rsFC and non-DMN rsFC only for the non-GSR condition (but not for the GSR analysis; lower middle of Fig. 4). Finally, we plotted the DMN–non-DMN correlation for MDD and HC in both non-GSR and GSR; that yielded significantly positive correlation of DMN–non-DMN only in MDD but not in HC for non-GSR (lower Fig. 4). Whereas GSR introduced negative correlation or anticorrelation of DMN with non-DMN in both MDD and HC, which no longer showed any difference. This strongly suggests that the significantly higher DMN–non-DMN rsFC in MDD stems from a global, i.e., inter-network rather than local, i.e., within-DMN, source.
Machine learning: support vector machine (SVM)
To test whether the global origin of increased connectivity in DMN allows to predict single subject data including differentiation of MDD vs. HC subjects, through a machine learning approach we performed multivariate pattern analyses on GSCORR first with non-GSR expecting to find a robust differences between the two groups. Secondly, we performed the same analysis on GSCORR with GSR hypothesizing that the difference between groups is attenuated or even annulated.
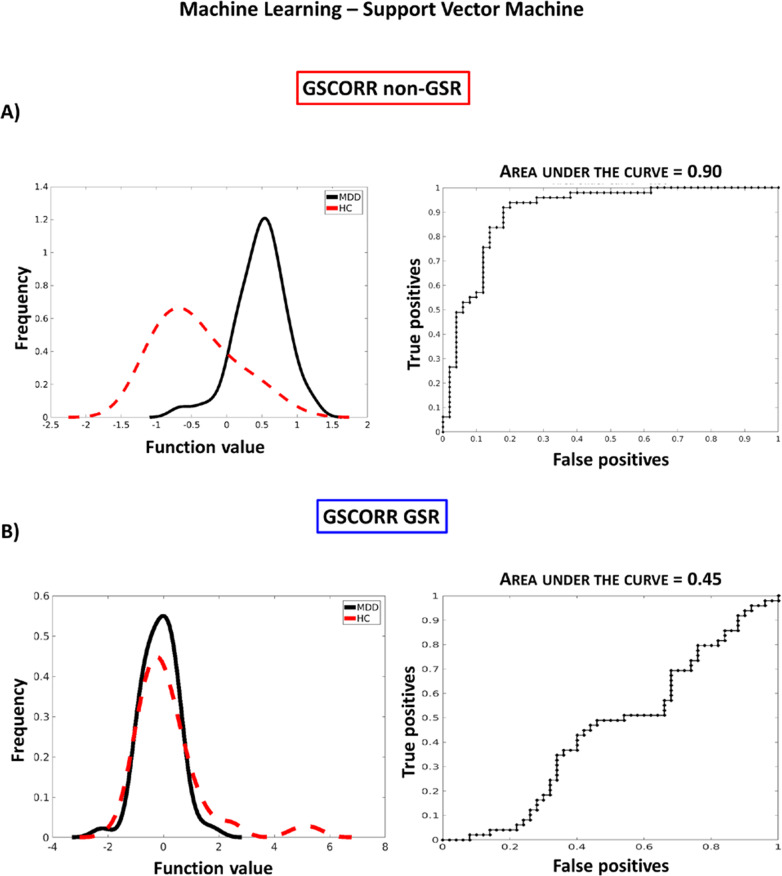
SVM for GSCORR non-GSR produced a BA of 85,94% (p = 0.001), MDD sensitivity of 93.88% (p = 0.001), HC specificity of 78% (p = 0.001) and the AUC was 90% (Fig. 5). Class predictive value for MDD was 80.70% and HC was 92.86%. Thus, model performance significantly exceeds the one expected by randomly guessing the labels, suggesting that the algorithm has successfully learned a predictive function [77]. Margin C = 1 obtained the best performance (Frequency of selection 100%).
Fig. 5. Machine learning diagnostic prediction.
a GSCORR non-GSR results showing a high diagnostic discrimination between MDD and HC, area under the curve-AUC = 0.90. b GSCORR GSR results showing no diagnostic discrimination between MDD and HC, area under the curve-AUC = 0.45.
SVM with GSCORR GSR produced a BA of 50.45% (p = 0.44), MDD sensitivity of 44.90% (p = 0.42), HC specificity of 56% (p = 0.56) and the AUC was 45% (Fig. 5). Class predictive value for MDD was 50% and HC was 50.91%. The model was not able to predict the differences between groups.
Altogether these findings demonstrated that the presumed global origin of increased connectivity in DMN allows a highly accurate classification of individual subjects. GSCORR non-GSR, differently from GSR, might not only characterize the functional organization of brain networks in MDD but might also contain a certain degree of information that seems to be clinically relevant in distinguishing MDD from HC. That further supports the importance and the relevance of our findings.
Replication data set
We applied the same sequence of analyses to another MDD data set for replicating the findings. As described in Supplementary material, all findings could be replicated showing also the discrepancy when either including or excluding the global signal in the data analyses. Hence, this further highlights the global source of increased within-DMN rsFC in MDD.
Discussion
We here investigated global, intra- and inter-networks rsFC in order to identify the local or global source of within-networks rsFC changes of DMN in MDD. Our results showed the following: (1) increased representation of resting state global signal in DMN and especially its cortical midline regions as further supported by DC and the use of an independent DMN template; (2) increased within-DMN rsFC, that is particularly between its anterior and posterior midline regions in MDD, which hold only in the analysis without GSR (non-GSR) but not with GSR; (3) increased inter-network rsFC connectivity of non-DMN networks with DMN that was obtained only in the analyses without GSR (non-GSR), while being abolished when applying GSR; and (iv) high individual classification of increased representation of global connectivity in DMN. Altogether these findings suggest a global rather than local source of within-network rsFC changes of DMN in MDD.
Global source of within-network DMN changes in MDD
Previous findings in psychiatric disorders like schizophrenia [26, 37, 39, 78], bipolar disorder [27], and MDD [42–45] demonstrated changes in global brain activity. Schizophrenia showed increased global activity [26, 39] as well as its abnormal representation in sensory and higher order association cortical networks [37]. While bipolar disorder was featured by abnormal, i.e., increased or decreased representation of global brain activity in specific regions likes motor cortex (mania), hippocampus (depression), or medial prefrontal cortex (euthymia) [27]. Studies in MDD, analyzing only GSR data, showed changes in global FC in medial prefrontal cortex [79] and other regions [42–45]. These findings in MDD leave open though the source of especially GS changes in DMN, are they due to within-network rsFC changes of DMN or, alternatively, do they stem from abnormally increased inter-network rsFC of DMN with non-DMN networks; that was the main focus in our study.
Changes in DMN activity have been most consistently observed in MDD [4, 25]. However, the direction of changes remains inconsistent [10–17, 76, 79]. Focusing on specific networks, these and other more globally oriented [42–45] studies mostly neglected the investigation of the global brain activity and specifically how it impacts within-network rsFC in DMN. Following the findings in schizophrenia and bipolar disorder, we here instead investigated the topography of global activity in MDD. For that purpose, we applied two measures for testing the topographic representation of global brain activity, i.e., GSCORR and DC. Both measures yielded similar results showing increased global connectivity within DMN in MDD. This concerned specifically the midline portion of the DMN, that is, anterior and posterior regions like PACC and PCC, that together shape a dynamic core within both the DMN and the brain as whole [48, 49, 80, 81] Our findings suggest that this dynamic core is abnormally strong in MDD as it is abnormally strong connected with the regions and networks outside this dynamic core, i.e., the non-DMN and their inter-network rsFC with DMN. That suggests a global, i.e., inter-network, rather than local, i.e., within-network, source of elevated DMN rsFC in MDD.
Our results extend previous data in various aspects. First, we show abnormal representation of global brain activity in MDD in specifically the DMN and its midline regions. Most interestingly, increased representation of global brain activity in DMN seems to hold specifically for MDD (as it was not reported in the other groups). This holds the promise that increased representation of global activity in DMN may serve as diagnostic marker, which is tentatively supported by our machine learning data where GSCORR allowed for high accuracy in distinguishing healthy vs. MDD subjects.
Second, our results show that intra-DMN rsFC does not stem from within DMN itself. Instead, our novel sequence of analyses strongly suggests that the source of increased DMN rsFC lies in the brain’s global activity and specifically, in the increased rsFC of the various networks (other than DMN) to the DMN. Thus, intra-DMN activity is strongly shaped and influenced by the brain’s global activity.
Figuratively put, the midline regions as dynamic core of DMN and the whole brain [48, 49] seems to abnormally related to the non-DMN regions and networks—hence, the title of our manuscript, all roads lead to the DMN. Extending this metaphor, the roads in the DMN seem to be abnormally strong in MDD. On the clinical side, this may explain why the various functions associated with non-DMN networks like executive function, movement/action, memory, reward/Sal, perception, and emotion are strongly tainted and shaped by those associated with the DMN as dynamic core, that is, internally oriented cognition like rumination and increased self-focus [82–84], in depressed subjects. The abnormally strong shaping of the various forms of externally oriented cognition (non-DMN) by internally oriented cognition (DMN) may thus be traced to spatio-temporal dysbalance of non-DMN and DMN, i.e., their abnormally positive inter-network rsFC.
The positive inter-network correlation of DMN with non-DMN in MDD stands in contrast to the usually observed negative correlation of DMN with other networks in the healthy brain. Our findings extend and differentiate previous observation of the loss of DMN-DAN (dorsal attention network) in various unconscious states [85] to the realm of mental disorder. Specifically, we observe that the whole network structure, that is, the anticorrelation of DMN with non-DMN networks including sensory and attentional networks is lost and replaced by their abnormally positive correlation. That suggests that the topography of the global brain activity, as measured by GS, is altered in MDD in a way that apparently is non-optimal for computational spatio-temporal processing considering the various symptoms. The impact of the abnormal spatio-temporal topography in MDD is supported by the fact that the positive DMN–non-DMN inter-network correlation is diminished when applying GSR like particularly in the case of DMN-SS, - Vis and -COTC.
We therefore propose that the topography of global brain activity may play a key role in MDD pathophysiology and other psychiatric disorders like schizophrenia [26] and bipolar disorder [27]. Their potential characterization by altered GS topography may distinguish psychiatric disorders from disorders of consciousness that show extreme reductions in the level of the overall GS corresponding to reduced levels of arousal [86]. Accordingly, albeit tentatively, we conceive psychiatric disorders as disorders of GS topography while disorders of consciousness may be disorders of the whole brain level of the overall GS.
Methodological implications and limitations
Methodologically, our results point out the need for including the global brain activity in the analyses of local vs global network changes in psychiatric disorders like MDD. Our results show that the comparison of the same data with and without GSR contains physiologically meaningful information results about the source of local within-network changes. That is supported by the fact that the removal of the global signal introduced some changes if not a complete reversal in some of our findings as well as by the various alternative ways of analyses we included.
Some limitations shall be mentioned. It can be possible that medications have a certain influence on the global signal and different connectivity patterns observed. However, medication effects are rather unlikely, as our results were not affected by their inclusion as co-variate. Future studies might want to extend these findings on psychiatric cohorts that are drug-free. Moreover, future studies may want to extend our results by investigating the representation of GS in task and, particularly, in rest-task relation [87–89]. These will shed new lights on the relevance of GS in understanding the physiological mechanism behind psychiatric disorders.
Conclusion
A major problem in brain imaging of psychiatric disorders like MDD is the inconsistency of findings. Conducting a novel sequence of global, intra- and inter-network analyses without (non-GSR) and with GSR, we demonstrate that increased rsFC within-DMN stems mainly from sources outside the DMN, i.e., non-DMN networks being abnormally strong connected to DMN. Such global-, rather than purely local source of increased within-network rsFC of DMN in MDD was further confirmed by the fact that all results only hold when including the global signal in the data (non-GSR) while they were abolished or reversed when removing the global signal, i.e., GSR. On the more methodological side, our results show the importance of including both type of analyses, with and without GSR as, otherwise, one important source of within-network rsFC, i.e., inter-network rsFC as global contribution to local within-network activity is excluded.
Together with our machine learning data, we here provide evidence for a predominantly global rather than local source of within-network rsFC changes of DMN in MDD. Our machine-learning results and the replication of these findings in an independent data set strongly suggests the utility of inter-network rsFC operating as global source of within-network DMN changes as potential biomarker for the diagnosis of MDD. Our data provide evidence that MDD may be a disorder of spatial topography of the brain’s global activity. More generally, this strongly supports the idea that psychopathological symptoms are driven by and based on abnormal spatio-temporal structure and organization with spatial dynamics, as in our case of GS topography in MDD, providing the “common currency” of neuronal and mental levels [90, 91]–this entails what recently has been introduced as “Spatio-temporal Psychopathology” [82, 92–98].
Funding and disclosure
The authors declare no potential conflicts of interest. This work was supported by the grant RF-2011-02349921 from the Italian Ministry of Health (to FB), by the, Michael Smith Foundation, EJLB-Canadian Institute of Health Research, Canada Research Chair (to GN), by the National Natural ScienceFoundation of China (No. 31271195), by the grant from the Ministry of Science and Technology of China, National Key R&D Program of China (2016YFC1306700) and from the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 785907 (Human Brain Project SGA2).
Supplementary information
Supplementary material - All roads lead to the default-mode network
Author contributions
AS and GN designed the framework and the logic of the analysis, AS analyzed the fMRI data; BV performed the machine learning analysis; SD supported for the fMRI analysis; SP and FB defined the fMRI protocol and scanned the patients; RZ and CC selected the patient and followed the ethical approval processing; AS and GN wrote the first draft of the manuscript, which was critically revised by FB, BV, SP, SD and CM. AS and GN reviewed the last version of the manuscript. Financial support was provided by FB (patients’ recruitment and scanning), CM and GN (data analysis). All authors approved the final version of the manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Andrea Scalabrini, Email: andrea.scalabrini@unich.it.
Georg Northoff, Email: georg.northoff@theroyal.ca.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41386-020-0785-x).
References
- 1.American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 5th ed. Washington, DC: American Psychiatric Publishing; 2013. [Google Scholar]
- 2.Murray CJ, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2197–223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. Depression. 2017. https://www.who.int/en/news-room/fact-sheets/detail/depression.
- 4.Kaiser RH, Andrews-Hanna JR, Wager TD, Pizzagalli DA. Large-scale network dysfunction in major depressive disorder: a meta-analysis of resting-state functional connectivity. JAMA Psychiatry. 2015;72:603–11. doi: 10.1001/jamapsychiatry.2015.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98:676–82. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckner RL, Andrews-Hanna JR, Schacter DL. The brain’s default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton JP, Etkin A, Furman DJ, Lemus MG, Johnson RF, Gotlib IH. Functional neuroimaging of major depressive disorder: a meta-analysis and new integration of baseline activation and neural response data. Am J Psychiatry. 2012;169:693–703. doi: 10.1176/appi.ajp.2012.11071105. [DOI] [PubMed] [Google Scholar]
- 8.Price JL, Drevets WC. Neurocircuitry of mood disorders. Neuropsychopharmacology. 2010;35:192. doi: 10.1038/npp.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegle GJ, Thompson W, Carter CS, Steinhauer SR, Thase ME. Increased amygdala and decreased dorsolateral prefrontal BOLD responses in unipolar depression: related and independent features. Biol Psychiatry. 2007;61:198–209. doi: 10.1016/j.biopsych.2006.05.048. [DOI] [PubMed] [Google Scholar]
- 10.Greicius MD, Flores BH, Menon V, Glover GH, Solvason HB, Kenna H, et al. Resting-state functional connectivity in major depression: abnormally increased contributions from subgenual cingulate cortex and thalamus. Biol Psychiatry. 2007;62:429–37. doi: 10.1016/j.biopsych.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheline YI, Price JL, Yan Z, Mintun MA. Resting-state functional MRI in depression unmasks increased connectivity between networks via the dorsal nexus. Proc Natl Acad Sci USA. 2010;107:11020–25. doi: 10.1073/pnas.1000446107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Wang K, Liu Y, Song M, Song SW, Jiang T. Spontaneous brain activity observed with functional magnetic resonance imaging as a potential biomarker in neuropsychiatric disorders. Cogn Neurodyn. 2010;4:275–94. doi: 10.1007/s11571-010-9126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton JP, Furman DJ, Chang C, Thomason ME, Dennis E, Gotlib IH. Default-mode and task-positive network activity in major depressive disorder: implications for adaptive and maladaptive rumination. Biol Psychiatry. 2011;70:327–33. doi: 10.1016/j.biopsych.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bluhm R, Williamson P, Lanius R, Théberge J, Densmore M, Bartha R, et al. Resting state default‐mode network connectivity in early depression using a seed region‐of‐interest analysis: decreased connectivity with caudate nucleus. Psychiatry Clin Neurosci. 2009;63:754–61. doi: 10.1111/j.1440-1819.2009.02030.x. [DOI] [PubMed] [Google Scholar]
- 15.Anand A, Li Y, Wang Y, Wu J, Gao S, Bukhari L, et al. Activity and connectivity of brain mood regulating circuit in depression: a functional magnetic resonance study. Biol Psychiatry. 2005;57:1079–88. doi: 10.1016/j.biopsych.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 16.Veer IM, Beckmann CF, Van Tol MJ, Ferrarini L, Milles J, Veltman DJ, et al. Whole brain resting-state analysis reveals decreased functional connectivity in major depression. Front Syst Neurosci. 2010;4:41. doi: 10.3389/fnsys.2010.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan CG, Chen X, Li L, Castellanos FX, Bai TJ, Bo QJ, et al. Reduced default mode network functional connectivity in patients with recurrent major depressive disorder. Proc Natl Acad Sci USA. 2019;116:9078–83. doi: 10.1073/pnas.1900390116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheline YI, Barch DM, Price JL, Rundle MM, Vaishnavi SN, Snyder AZ, et al. The default mode network and self-referential processes in depression. Proc Natl Acad Sci USA. 2009;106:1942–94. doi: 10.1073/pnas.0812686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menon V. Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cogn Sci. 2011;15:483–506. doi: 10.1016/j.tics.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton JP, Chen MC, Gotlib IH. Neural systems approaches to understanding major depressive disorder: an intrinsic functional organization perspective. Neurobiol Dis. 2013;52:4–11. doi: 10.1016/j.nbd.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manoliu A, Riedl V, Zherdin A, Mühlau M, Schwerthöffer D, Scherr M, et al. Aberrant dependence of default mode/central executive network interactions on anterior insular salience network activity in schizophrenia. Schizophr Bull. 2014;40:428–37. doi: 10.1093/schbul/sbt037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambataro F, Wolf ND, Giusti P, Vasic N, Wolf RC. Default mode network in depression: a pathway to impaired affective cognition? Clin Neuropsychiatry. 2013;10:212–6. [Google Scholar]
- 24.Zeng LL, Shen H, Liu L, Wang L, Li B, Fang P, et al. Identifying major depression using whole-brain functional connectivity: a multivariate pattern analysis. Brain. 2012;135:1498–507. doi: 10.1093/brain/aws059. [DOI] [PubMed] [Google Scholar]
- 25.Zhong X, Pu W, Yao S. Functional alterations of fronto-limbic circuit and default mode network systems in first-episode, drug-naive patients with major depressive disorder: a meta-analysis of resting-state fMRI data. J Affect Disord. 2016;206:280–6. doi: 10.1016/j.jad.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Yang GJ, Murray JD, Repovs G, Cole MW, Savic A, Glasser MF, et al. Altered global brain signal in schizophrenia. Proc Natl Acad Sci USA. 2014;111:7438–43. doi: 10.1073/pnas.1405289111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Magioncalda P, Huang Z, Tan Z, Hu X, Hu Z, et al. Altered global signal topography and its different regional localization in motor cortex and hippocampus in mania and depression. Schizophr Bull. 2018;45:902–10. doi: 10.1093/schbul/sby138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu TT, Nalci A, Falahpour M. The global signal in fMRI: nuisance or information? Neuroimage. 2017;150:213–29. doi: 10.1016/j.neuroimage.2017.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, de Zwart JA, Schölvinck ML, Chang C, Ye FQ, Leopold DA, et al. Subcortical evidence for a contribution of arousal to fMRI studies of brain activity. Nat Commun. 2018;9:395. doi: 10.1038/s41467-017-02815-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy K, Fox MD. Towards a consensus regarding global signal regression for resting state functional connectivity MRI. Neuroimage. 2017;154:169–73. doi: 10.1016/j.neuroimage.2016.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schölvinck ML, Maier A, Ye FQ, Duyn JH, Leopold DA. Neural basis of global resting-state fMRI activity. Proc Natl Acad Sci USA. 2010;107:10238–43. doi: 10.1073/pnas.0913110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Kong R, Liegeois R, Orban C, Tan Y, Sun N, et al. Global signal regression strengthens association between resting-state functional connectivity and behavior. Neuroimage. 2019;196:126–41. doi: 10.1016/j.neuroimage.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Bolt T, Bzdok D, Nomi JS, Yeo BT, Spreng RN, et al. Topography and behavioral relevance of the global signal in the human brain. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-50750-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 2014;84:320–41. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Power JD, Plitt M, Laumann TO, Martin A. Sources and implications of whole-brain fMRI signals in humans. Neuroimage. 2017;146:609–25. doi: 10.1016/j.neuroimage.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. J Neurophysiol. 2009;101:3270–83. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang GJ, Murray JD, Glasser M, Pearlson GD, Krystal JH, Schleifer C, et al. Altered global signal topography in schizophrenia. Cereb Cortex. 2017;27:5156–69. doi: 10.1093/cercor/bhw297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scalabrini A, Mucci C, Esposito R, Damiani S, Northoff G. Dissociation as a disorder of integration - On the footsteps of Pierre Janet. Prog Neuropsychopharmacol Biol Psychiatry. 2020;101:109928. doi: 10.1016/j.pnpbp.2020.109928. [DOI] [PubMed] [Google Scholar]
- 39.Yang GJ, Murray JD, Wang XJ, Glahn DC, Pearlson GD, Repovs G, et al. Functional hierarchy underlies preferential connectivity disturbances in schizophrenia. Proc Natl Acad Sci USA. 2016;113:E219–28. doi: 10.1073/pnas.1508436113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gotts SJ, Simmons WK, Milbury LA, Wallace GL, Cox RW, Martin A. Fractionation of social brain circuits in autism spectrum disorders. Brain. 2012;135:2711–25. doi: 10.1093/brain/aws160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotts SJ, Saad ZS, Jo HJ, Wallace GL, Cox RW, Martin A. The perils of global signal regression for group comparisons: a case study of Autism Spectrum Disorders. Front Hum Neurosci. 2013;7:356. doi: 10.3389/fnhum.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han S, Wang X, He Z, Sheng W, Zou Q, Li L, et al. Decreased static and increased dynamic global signal topography in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2019;94:109665. doi: 10.1016/j.pnpbp.2019.109665. [DOI] [PubMed] [Google Scholar]
- 43.Abdallah C, Averill L, Collins K, et al. Ketamine treatment and global brain connectivity in major depression. Neuropsychopharmacology. 2017;42:1210–9. doi: 10.1038/npp.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheinost D, Holmes S, DellaGioia N, et al. Multimodal investigation of network level effects using intrinsic functional connectivity, anatomical covariance, and structure-to-function correlations in unmedicated major depressive disorder. Neuropsychopharmacology. 2018;43:1119–27. doi: 10.1038/npp.2017.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Wu H, Xu J, Shang J. Abnormal global functional connectivity patterns in medication-free major depressive disorder. Front. Neurosci. 2018;12:692. doi: 10.3389/fnins.2018.00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 47.Cole MW, Pathak S, Schneider W. Identifying the brain’s most globally connected regions. Neuroimage. 2010;49:3132–48. doi: 10.1016/j.neuroimage.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 48.de Pasquale F, Della Penna S, Snyder AZ, Marzetti L, Pizzella V, Romani GL, et al. A cortical core for dynamic integration of functional networks in the resting human brain. Neuron. 2012;74:753–64. doi: 10.1016/j.neuron.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Pasquale F, Corbetta M, Betti V, Della Penna S. Cortical cores in network dynamics. Neuroimage. 2018;180:370–82. doi: 10.1016/j.neuroimage.2017.09.063. [DOI] [PubMed] [Google Scholar]
- 50.Raichle ME. The brain’s default mode network. Annu Rev Neurosci. 2015;38:433–47. doi: 10.1146/annurev-neuro-071013-014030. [DOI] [PubMed] [Google Scholar]
- 51.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–73. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 52.Huang Z, Liu X, Mashour GA, Hudetz AG. Timescales of intrinsic BOLD signal dynamics and functional connectivity in pharmacologic and neuropathologic states of unconsciousness. J Neurosci. 2018;38:2304–17. doi: 10.1523/JNEUROSCI.2545-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci USA. 2005;102:9673–8. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chai XJ, Castañón AN, Öngür D, Whitfield-Gabrieli S. Anticorrelations in resting state networks without global signal regression. Neuroimage. 2012;59:1420–8. doi: 10.1016/j.neuroimage.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He BJ. Scale-free properties of the functional magnetic resonance imaging signal during rest and task. J Neurosci. 2011;31:13786–95. doi: 10.1523/JNEUROSCI.2111-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephens GJ, Honey CJ, Hasson U. A place for time: the spatiotemporal structure of neural dynamics during natural audition. J Neurophysiol. 2013;110:2019–26. doi: 10.1152/jn.00268.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–54. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Dijk KR, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59:431–8. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnstone T, Ores Walsh KS, Greischar LL, Alexander AL, Fox AS, Davidson RJ, et al. Motion correction and the use of motion covariates in multiple‐subject fMRI analysis. Hum Brain Mapp. 2006;27:779–88. doi: 10.1002/hbm.20219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan CG, Craddock RC, Zuo XN, Zang YF, Milham MP. Standardizing the intrinsic brain: towards robust measurement of inner-individual variation in 1000 functional connectome. Neuroimage. 2013;80:246–62. doi: 10.1016/j.neuroimage.2013.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hassel S, Almeida JR, Kerr N, Nau S, Ladouceur CD, Fissell K, et al. Elevated striatal and decreased dorsolateral prefrontal cortical activity in response to emotional stimuli in euthymic bipolar disorder: no associations with psychotropic medication load. Bipolar Disord. 2008;10:916–27. doi: 10.1111/j.1399-5618.2008.00641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phillips ML, Travis MJ, Fagiolini A, Kupfer DJ. Medication effects in neuroimaging studies of bipolar disorder. Am J Psychiatry. 2008;165:313–20. doi: 10.1176/appi.ajp.2007.07071066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sackeim HA. The definition and meaning of treatment-resistant depression. J Clin Psychiatry. 2001;62:10–17. [PubMed] [Google Scholar]
- 64.Zuo XN, Ehmke R, Mennes M, Imperati D, Castellanos FX, Sporns O, et al. Network centrality in the human functional connectome. Cereb Cortex. 2012;22:1862–75. doi: 10.1093/cercor/bhr269. [DOI] [PubMed] [Google Scholar]
- 65.Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proc Natl Acad Sci USA. 2006;103:10046–51. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci USA. 2003;100:253–58. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 2009;19:72–78. doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laird AR, Eickhoff SB, Li K, Robin DA, Glahn DC, Fox PT. Investigating the functional heterogeneity of the default mode network using coordinate-based meta-analytic modeling. J Neurosci. 2009;29:14496–505. doi: 10.1523/JNEUROSCI.4004-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Biswal B, Zerrin Yetkin F, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo‐planar MRI. Magn Reson Med. 1995;34:537–41. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 70.Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8:700. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- 71.Zhang D, Raichle ME. Disease and the brain’s dark energy. Nat Rev Neurol. 2010;6:15. doi: 10.1038/nrneurol.2009.198. [DOI] [PubMed] [Google Scholar]
- 72.Power JD, Fair DA, Schlaggar BL, Petersen SE. The development of human functional brain networks. Neuron. 2010;67:735–48. doi: 10.1016/j.neuron.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. Functional network organization of the human brain. Neuron. 2011;72:665–78. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014;83:238–51. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Varoquaux G. Cross-validation failure: small sample sizes lead to large error bars. Neuroimage. 2018;180:68–77. doi: 10.1016/j.neuroimage.2017.06.061. [DOI] [PubMed] [Google Scholar]
- 76.Zhu X, Wang X, Xiao J, Liao J, Zhong M, Wang W, et al. Evidence of a dissociation pattern in resting-state default mode network connectivity in first-episode, treatment-naive major depression patients. Biol Psychiatry. 2012;71:611–7. doi: 10.1016/j.biopsych.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 77.Schrouff J, Mourão-Miranda J, Phillips C, Parvizi J. Decoding intracranial EEG data with multiple kernel learning method. J Neurosci Methods. 2016;261:19–28. doi: 10.1016/j.jneumeth.2015.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hahamy A, Calhoun V, Pearlson G, Harel M, Stern N, Attar F, et al. Save the global: global signal connectivity as a tool for studying clinical populations with functional magnetic resonance imaging. Brain Connect. 2014;4:395–403. doi: 10.1089/brain.2014.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murrough JW, Abdallah CG, Anticevic A, Collins KA, Geha P, Averill LA, et al. Reduced global functional connectivity of the medial prefrontal cortex in major depressive disorder. Hum Brain Mapp. 2016;37:3214–23. doi: 10.1002/hbm.23235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Margulies DS, Ghosh SS, Goulas A, Falkiewicz M, Huntenburg JM, Langs G, et al. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc Natl Acad Sci USA. 2016;113:12574–9. doi: 10.1073/pnas.1608282113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huntenburg JM, Bazin PL, Margulies DS. Large-scale gradients in human cortical organization. Trends Cogn Sci. 2018;22:21–31. doi: 10.1016/j.tics.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 82.Northoff G. Psychopathology and pathophysiology of the self in depression—neuropsychiatric hypothesis. J Affect Disord. 2007;104:1–14. doi: 10.1016/j.jad.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 83.Northoff G, Wiebking C, Feinberg T, Panksepp J. The ‘resting-state hypothesis’ of major depressive disorder-a translational subcortical-cortical framework for a system disorder. Neurosci Biobehav Rev. 2011;35:1929–45. doi: 10.1016/j.neubiorev.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 84.Northoff G. Is the self a higher-order or fundamental function of the brain? The “basis model of self-specificity” and its encoding by the brain’s spontaneous activity. Cogn Neurosci. 2016;7:203–22. doi: 10.1080/17588928.2015.1111868. [DOI] [PubMed] [Google Scholar]
- 85.Huang Z, Zhang J, Wu J, Mashour GA, Hudetz AG. Temporal circuit of macroscale dynamic brain activity supports human consciousness. Sci Adv. 2020;6:eaaz0087. doi: 10.1126/sciadv.aaz0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tanabe S, Huang Z, Zhang J, Chen Y, Fogel S, Doyon J, et al. Altered global brain signal during physiologic, pharmacologic, and pathologic states of unconsciousness in humans and rats. Anesthesiology. 2020;132:1392–1406. doi: 10.1097/ALN.0000000000003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scalabrini A, Ebisch SJH, Huang Z, Di Plinio S, Perrucci MG, Romani GL, et al. Spontaneous brain activity predicts task-evoked activity during animate versus inanimate touch. Cereb Cortex. 2019;29:4628–45. doi: 10.1093/cercor/bhy340. [DOI] [PubMed] [Google Scholar]
- 88.Scalabrini A, Huang Z, Mucci C, et al. How spontaneous brain activity and narcissistic features shape social interaction. Sci Rep. 2017;7:9986. doi: 10.1038/s41598-017-10389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scalabrini A, Mucci C, Northoff G. Is Our self related to personality? a neuropsychodynamic model. Front Hum Neurosci. 2018;12:346. doi: 10.3389/fnhum.2018.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Northoff G. Wainio-Theberge S. Evers K. Is temporo-spatial dynamics the “common currency” of brain and mind? In Quest of “Spatiotemporal Neuroscience.” Phys Life Rev. 2019. 10.1016/j.plrev.2019.05.002. [DOI] [PubMed]
- 91.Northoff G. Wainio-Theberge S. Evers K. Spatiotemporal neuroscience—what is it and why we need it. Phys Life Rev. 2020. 10.1016/j.plrev.2020.06.005. [DOI] [PubMed]
- 92.Northoff G. Spatiotemporal psychopathology I: no rest for the brain’s resting state activity in depression? Spatiotemporal psychopathology of depressive symptoms. J Affect Disord. 2016;190:854–66. doi: 10.1016/j.jad.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 93.Northoff G. Spatiotemporal Psychopathology II: how does a psychopathology of the brain’s resting state look like? Spatiotemporal approach and the history of psychopathology. J Affect Disord. 2016;190:867–79. doi: 10.1016/j.jad.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 94.Damiani S, Scalabrini A, Gomez-Pilar J, Brondino N, Northoff G. Increased scale-free dynamics in salience network in adult high-functioning autism. Neuroimage Clin. 2019;21:101634. doi: 10.1016/j.nicl.2018.101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Northoff G. Personal identity and cortical midline structure (CMS): do temporal features of CMS neural activity transform into “Self-Continuity”? Psychol Inq. 2017;28:122–31. [Google Scholar]
- 96.Northoff G. The brain’s spontaneous activity and its psychopathological symptoms–“Spatiotemporal binding and integration”. Prog Neuropsychopharmacol Biol Psychiatry. 2018;80:81–90. doi: 10.1016/j.pnpbp.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 97.Fingelkurts AA, Fingelkurts AA. Brain space and time in mental disorders: paradigm shift in biological psychiatry. Int J Psychiatry Med. 2019;54:53–63. doi: 10.1177/0091217418791438. [DOI] [PubMed] [Google Scholar]
- 98.Schrouff J, Monteiro JM, Portugal L, Rosa MJ, Phillips C, Mourão-Miranda J. Embedding anatomical or functional knowledge in whole-brain multiple kernel learning models. Neuroinformatics. 2018;16:117–43. doi: 10.1007/s12021-017-9347-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material - All roads lead to the default-mode network