Abstract
Purpose
The last few decades have witnessed a rapid and global increase in multidrug-resistant bacteria (MDR) emergence.
Methods
The aim of the current study is to isolate the most common MDR bacteria from dairy farms and beef slaughterhouses followed by evaluation of their antimicrobial resistance pattern and assessment of the antibacterial activity of AgNPs-H2O2 as an alternative to conventional antibiotics. In this regard, 200 samples were collected from two dairy farms and one beef slaughterhouse located in Dakhliya Governorate, Egypt.
Results
Interestingly, out of 120 collected samples from dairy farms, the prevalence of the isolated strains was 26.7, 23.3, 21.7, 16.7, and 11.7% for S. typhimurium, E. coli O157:H7, L. monocytogenes, K. pneumoniae and P. aeruginosa, respectively. Meanwhile, the overall prevalence was 30, 25, 22.5, 17.5, and 5% for E. coli O157:H7, L. monocytogenes, S. typhimurium, P. aeruginosa, and K. pneumoniae, respectively, for the 80 samples collected from a beef slaughterhouse. The antimicrobial susceptibility pattern elucidated that all isolated strains exhibited resistance to at least four of the tested antimicrobials, with multiple-antibiotic resistance index values (MAR) ranging between 0.44 and 0.88. Furthermore, the commercial AgNPs-H2O2 product was characterized by transmission electron microscopy (TEM) and zeta potential that showed spherical particles with a surface charge of −0.192 mV. The antimicrobial activity of synergized nano-silver (AgNP) with H2O2 product toward MDR strains was assessed via measuring minimum inhibitory concentration (MIC), minimum bactericidal concentration (MBC), and time-kill curve.
Conclusion
The present data report high prevalence rates of MDR pathogens in dairy farms and abattoirs. More importantly, AgNPs-H2O2 exerted broad-spectrum bactericidal activity toward MDR bacterial strains, suggesting their promising usage as safe, ecofriendly, cost-effective antibacterial agents. To our knowledge, this study is a pioneer in investigating the potential alternative antimicrobial role of silver nanoparticles for control of multiple drug-resistant pathogens in Egypt.
Keywords: MDR pathogens, MIC, MBC, time-kill curve, AgNPs-H2O2
Introduction
Antimicrobial resistance (AMR) has been considered a serious global threat for animal and human health, food security, and development.1 This global threat primarily results from the indiscriminate use of antimicrobial agents, which in turn leads to a marked reduction or even losing their effectiveness.2,3 It should be stressed that the antimicrobial-resistant pathogens jeopardize the treatment capacity of infectious diseases within human and veterinary medicine.4,5 Clearly, the application of antimicrobials in food-producing animals, either for prophylactic and/or treatment purposes, generates a considerable selection pressure that contributes to the emergence, persistence, and transmission of antimicrobial resistance over the food supply.6,7 The food of animal origin could be contaminated with various zoonotic pathogens that might result from inappropriate production, processing methods in animal farms and/processing feed lines, which in turn results in transmission of these pathogens to the consumers.8–10 Among other diseases, mastitis is a multifactorial disease affecting milk production and quality in dairy farms.11 Taken into account, the major sources of milk contamination in dairy farms include handling, management type, and hygienic practices within the farm,12 and consequently numerous microorganisms can be derived from milk and the surrounding environment at farm level, representing vital sources of foodborne pathogens.13 This issue constitutes a major public health hazard, whereas the most frequently causing and isolated pathogens are Salmonella enterica, Escherichia coli O157:H7, Pseudomonas aeruginosa, Listeria monocytogenes, and Klebsiella pneumoniae.14–16 It is noteworthy to state that foodborne pathogens like Staphylococcus aureus, Streptococcus agalactiae, Escherichia coli, Pseudomonas aeruginosa, Corynebacterium bovis, and Bacillus cereus are considered primary causes of mastitis, which in turn leads to production losses and human illness due to the consumption of contaminated milk products.17 Additionally, L. monocytogenes is also considered a foodborne pathogen, which is transmitted via meat, poultry, dairy, and vegetable products,18 while E. coli O157:H7 is widely known as a main pathogen associated with food-borne illnesses observed in dairy products.19 Taken into consideration, beef cattle also harbor pathogenic E. coli.20 Moreover, Klebsiella pneumoniae is considered one of the environmental agents causing clinical and subclinical mastitis and reduces milk quality.21 This pathogen causes severe mastitis owing to its antibiotic resistance, rapid development of toxic shock and animal deaths.22 Furthermore, most human infections caused by Salmonella are foodborne origin acquired through foods of animal origin like milk and meat, or via animal contact and contaminated environments.23 Clearly, the inappropriate farm practices during the production, handling, and meat marketing facilitate transfer of foodborne pathogens to the meat and meat products. For combating this problem, the use of antibiotics has been considered the first choice of bacterial infection treatment in dairy cattle, particularly mastitis, resulting in dissemination of antibiotic residues in milk combined with potential risks of microbial resistance in the environment.24,25 Furthermore, cattle are exposed to a wide range of contaminants, including bacterial type through feces, feed, or the environment, resulting in transfer of these organisms during slaughtering on the carcass, which poses high risk to food safety.26,27 Moreover, the existence of commensal and environmental bacteria in the farm environment represents a major reservoir for transferring of antimicrobial resistance genes to pathogenic bacteria.28 In addition, some actions during slaughter at abattoirs, such as evisceration and splitting, could contribute to carcass contamination.29,30 This challenge is usually aggravated by the asymptomatic cattle carriers that represent a major public health hazard along the food chain.31,32 Hence, more strict hygienic measures combined with law enforcement at abattoirs and during slaughter procedures are the key elements to reduce meat contamination chances.33 Likewise, there is an urgent necessity to find environmentally ecofriendly alternatives to conventional antibiotics for combating the problem of widespread multidrug resistance (MDR) and its rapid emergence make.34
It is noteworthy to state that the recent years have witnessed a great progress about exploring the role played by nanotechnology in providing a great development and modification of nanoparticles (NPs) with exclusive physicochemical characters as a promising tool for use in medicine and farming to overcome the limitations caused by conventional antibiotics.35–38 The application of nanomaterials, principally Silver nanoparticles (AgNPs), boosted the attention in various aspects; academic side, industry, and nanomedicine field.39,40 AgNPs exhibited an amazing biocidal activity on a wide range of Gram-positive, Gram-negative bacteria, including foodborne pathogens such as Escherichia coli O157:H7, Salmonella, Listeria monocytogenes, Campylobacter jejuni, and Staphylococcus aureus.41–43 The suggested mechanism behind AgNPs’s actions includes their induction of cell death through generation of reactive oxygen species (ROS) in some bacteria including E. coli, K. pneumoniae, and Pseudomonas aeruginosa.36,44,45 Importantly, less reports of bacterial resistance have been documented towards AgNPs than against conventional antimicrobials.46 The antimicrobial activity of NPs is guarded by various factors, such as the size, shape, stability, and the used concentration of NPs.47–49 Taken into account, the most common and useful determinants of the relative biocidal activity of various synthetic nanomaterials are minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values.50
Given the above information, the present study was initially undertaken to display the prevalence rates and the existence of MDR foodborne pathogens, mainly five major species, that include Salmonella enterica, Escherichia coli O157:H7, Pseudomonas aeruginosa, Listeria monocytogenes, and Klebsiella pneumoniae isolated from various sources. These sources include milk, bulk tank milk, milking utensils, beef carcasses, walls, knives, and workers’ hands (swabs) from dairy farms and slaughterhouse in Dakahliya Governorate, Egypt. We also aimed to assess the antimicrobial activity of commercially synthesized AgNPs-H2O2 on representative MDR isolated pathogens combined with exploring the bactericidal mechanism of AgNPs using various cellular assays.
Materials and Methods
Ethical Considerations
The ethical approval of the present study was obtained from guidance of Research, Publication and Ethics of the Faculty of Veterinary Medicine, Mansoura University, Egypt, which complies with all relevant Egyptian legislations on research and publications. The dairy workers who participated provided informed consent during swab collection and the study was in accordance with the guidelines outlined in the Declaration of Helsinki.
Study Area, Samples Collection, and Preparation
In the present study, a total of 200 samples were collected from two dairy farms and one beef slaughterhouse located at El-Dakahlia Province, Egypt, during the period from September to November 2018. Regarding their distribution, the total number of samples collected from dairy farms was 120 at a rate of 20 samples from each source/farm, whereas these samples were collected and included samples from bulk tank milk (BTM) (100 mL), milking utensils, and dairy workers’ hand swabs. Regarding the beef slaughterhouse, the total number of collected samples was 80, at a rate of 20 samples from each source, and the samples included beef carcass samples (25 gm), wall, knives, and workers’ hand swabs that were collected under complete aseptic conditions. All samples were then transported into an icebox to the laboratory of Hygiene and Zoonoses Department, Mansoura University, and subjected for further processing. Later on, five major types of bacteria were isolated and characterized from collected samples; including E. coli O157: H7, Listeria monocytogenes, Salmonella typhimurium, Pseudomonas aeruginosa, and Klebsiella pneumonia, following the protocol described elsewhere.51 The isolation and characterization steps were carried out using appropriate selective culture media, and various incubation conditions then the colony characters, morphology, and biochemical profiles were done. Furthermore, the confirmation of the identified strains was carried out through serological identification and molecular characterization as described elsewhere.21,–52–55
Antimicrobial Susceptibility Testing and Isolation of MDR Bacteria
The antimicrobial susceptibility patterns for confirmed E. coli O157:H7, Listeria monocytogenes, Salmonella typhimurium, Pseudomonas aeruginosa, and Klebsiella pneumonia strains to selected antibiotics (Oxoid, Hampshire, UK) are mentioned in Table 1. These patterns were done by Kirby–Bauer disc diffusion test using Mueller-Hinton Agar (MHA) (BioMeriéux, Vienna, Austria) according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI).56 The isolates were classified as resistant, intermediate, or susceptible via measuring the inhibition zone diameter around each disk, as described by CLSI.56 The multiple drug resistance (MDR) index for each resistance pattern was then calculated from the number of resistances to antimicrobials of each strain, divided by total number of antimicrobials tested.57 The isolates that exhibited resistance to three or more antimicrobials were considered as multidrug-resistant strains.58
Table 1.
Antibiotics Used to Test Antimicrobial Susceptibility of Five Types of Bacteria
| Bacteria | Antimicrobials |
|---|---|
| S. typhimurium | Sulfamethoxazole-trimethoprim (25 µg), Chloramphenicol (30 µg), Ciprofloxacin (5 µg), Amoxicillin (25 µg), Norfloxacin (10 µg), Nalidixic acid (30 µg), Cefotaxime (30 µg), Cephalexin (30 µg) |
| Listeria monocytogenes | Rifampicin (5 µg), Oxacillin (1 µg), Tetracycline (30 µg), Amoxicillin (10 µg), Gentamycin (10 µg), Cefotaxime (30 µg), Cefepime (30 µg), Sulfamethoxazole-trimethoprim (25 µg), Carbenicillin (30 µg), Ampicillin (10 µg), Chloramphenicol (30 µg) |
| Klebsiella pneumoniae | Neomycin (30 µg), Kanamycin (30 µg), Tetracycline (30 µg), Nalidixic acid (30 µg), Streptomycin (10 µg), Gentamycin (10 µg), Chloramphenicol (30 µg), Ampicillin (10 µg) |
| Pseudomonas aeruginosa | Ciprofloxacin (5 µg), Neomycin (30 µg), Norfloxacin (10 µg), Streptomycin (10 µg), Kanamycin (30 µg), Levofloxacin (5 µg), Carbapenem (100 µg), Cephalexin (30 µg), Cefepime (30 µg) |
| E. coli O157:H7 | Chloramphenicol (30 µg), Ampicillin (10 µg), Tetracycline (30 µg), Streptomycin (10 µg), Cefotaxime (30 µg), Carbenicillin (30 µg), Erythromycin (15 µg), Cephalexin (30 µg) |
Bacterial Preparation and Culture Conditions
The bacterial strains (N=10) which exhibited MDR to three or more antibiotics were prepared according to methods mentioned elsewhere.36 Briefly, all bacterial cultures were inoculated in Mueller Hinton broth (MHB) (BioMeriéux, Vienna, Austria) and grown aerobically at 37ºC/24 hours, then a loopful was streaked on MHA plates and sub-cultured for purification on the same medium. Pure colonies were subjected to harvesting and kept at −80ºC. Microbial inoculum of 0.5 McFarland was then prepared by the direct colony suspension method as recommended by the guidelines of the CLSI.56 The bacterial turbidity was adjusted spectrophotometrically using a 6715 UV/Visible scanning spectrophotometer (Jenway, Canada) to 0.08–0.12 at an optical density (OD) of 625 nm, giving a microbial suspension of (1–2)x108 colony forming unit (CFU)/mL.
Characterization of Commercial AgNPs-H2O2
AgNPs-H2O2 (Top Superpower-vision) was kindly purchased and provided as a commercial product by El-Delta center for nano silver technology company, Mansoura, Egypt. The stock solution of product composed of 45 nm silver nanoparticles (0.00004467 mL/liter) with Hydrogen Peroxide (50% liter) and natural herbs, peppermint (1 mL/liter) at a concentration of 5 mL/liter of water then the product was diluted in Mueller Hinton broth (MHB). The morphology and the average size of the commercial nanoproduct were characterized via TEM and zeta potential as described elsewhere,59 using the Malvern Instruments Ltd. Zeta Potential Ver. 2.3 at the Central Laboratory, Electron Microscope Unit, Faculty of Agriculture, Mansoura University, Mansoura, Egypt
In vitro Antimicrobial Activity of AgNPs-H2O2 Against MDR Strains
Determination of Minimum Inhibitory Concentration Value (MIC)
The inhibitory power of AgNPs-H2O2 product against MDR bacterial strains (N=5) was evaluated using MIC. The MIC was determined by broth microdilution method according to the guidelines of the CLSI.60 In detail, MIC was performed in 96-well microtiter plates by two-fold microdilution method. AgNPs-H2O2 mixture was prepared to a desired commercial concentration by dilution in sterile distilled H2O and then the mixture was diluted by 1/10 in sterile Mueller Hinton Broth (MHB). In this regard, a volume of 50 µL of MHB was seeded in each well, starting from the 2nd well to the 11th one followed by 2-fold microdilution of commercial nano-product in MHB. Later on, 100 µL of this mixture was inoculated in the first well, then 50 µL was transferred to the next wells except for the 12th one serving as a control negative (drug-free well). Then, 50 µL of bacterial suspension diluted to 1/150 in sterile MHB with microbial inoculum of 106 was dispensed in all wells followed by well mixing of the plates, and the plates were incubated at 35°C for 24 hours. The MIC is defined as the point at which bacterial growth was completely inhibited in microdilution wells that could be detected visually under transmitted light by the absence of turbidity. Hence, the first well showed no microbial growth was considered as the MIC, expressed in μg/mL. To determine the MICs of AgNPs of all tested strains, they were exposed to 0–100 μg/mL AgNPs. Furthermore, the MIC of each isolate was determined in triplicate for verification of the data. The experiment was included media alone and media which contain AgNPs as reference controls. As mentioned above, all samples were plated in triplicate to evaluate the reproducibility of the method and values were expressed as the average of three independent experiments.
Determination of Minimum Bactericidal Concentration (MBC)
The lowest concentration of AgNPs-H2O2 mixture required to kill 99.9% of final bacterial inoculum is known as MBC, which was determined after broth microdilution by sub-culturing 50 µL from all wells without visible turbidity on MHA plates and incubated at 35±1°C for 16–18 hours. The plates were then investigated for the presence or absence of bacterial colonies and the MBC was defined as the lowest concentration of NPs that completely inhibits the bacterial growth.61 The mode of activity of NPs product was assessed by MBC/MIC ratio, where scores of 1, 2, and 4 are considered bactericidal, and bacteriostatic if scores >4.62 Likewise, the lowest dilution without visible macroscopic bacterial growth was defined as MBC. This was done according to the CLSI method for antimicrobial drugs, described in the document M7-A9.63
Time-Kill Test (Time-Kill Curve)
The dynamic interaction between AgNPs-H2O2 and MDR bacterial strains was determined using time-kill test under AgNPs concentration equal to 0.25xMIC and 1xMIC as described elsewhere.56 Briefly, after reading the MIC for each AgNPs-H2O2 and MDR bacterial strains, three tubes with 10 mL MHB of 5×105 CFU/mL bacterial suspension were tested at 0.25×MIC and 1×MIC, and the third one was serving as growth control, while AgNPs-H2O2 was replaced for MHB as a control negative. All tubes were then incubated at 35±1°C for varied interval times (2, 4, 6, 8, 12, and 24 hours).64 The number of viable/dead bacterial cells (CFU/mL) of each tube was quantified on MHA plates using the agar plate method in relation to time intervals. These numbers were plotted on a graph describing the time-kill curve compared to positive and negative controls’ curves.48
Statistical Analysis
The statistical analysis was performed using statistical package SPSS, 23. The frequency of inhibited samples was compared between different concentrations of the drug. Moreover, kappa test was used to test the degree of agreement between MIC and MBC results. Results were considered significant at P<0.05, then the mean concentration of CFU were compared at different points of time using an analysis of variance (ANOVA) test to detect differences between means. Finally, Duncan multiple range test was used to make different comparisons between the means. Data were presented as means and standard errors and results were considered significant when P<0.05.
Results
The Prevalence of MDR Bacterial Species Among Examined Dairy Farms and Beef Slaughterhouse
The prevalence and frequency distribution of bacterial species from dairy farms and an abattoir are presented in Table 2. Among others, S. typhimurium was the most dominant bacterial species (26.7%) isolated from collected samples of examined dairy farms, where milking utensils harbored the highest prevalence of 35%, followed by workers’ hand swabs (25%), while the lowest prevalence was from BTM (20%). Following S. typhimurium, E. coli O157:H7 had the second prevalence level of 23.3%, whereas the frequency of isolation was 30%, 25%, and 15% for BTM, milking utensils, and workers’ hands swabs, respectively. The overall prevalence of L. monocytogenes was 21.7% among dairy farm samples while the recovery rate among the analyzed sources was 25%, 20%, and 20% for workers’ hands swabs, BTM, and milking utensils, respectively. Out of 120 collected samples from dairy farms, K. pneumoniae was recovered at a rate of 16.7%, whereas the BTM possessed the higher isolation rate of 25% followed by workers’ hands swabs (15%) and milking utensils (10%). On the other hand, P. aeruginosa has shown the lowest prevalence of the detected bacteria among all isolated species (11.7%), while the sequence of isolation rate from farm sources was 20%, 10%, and 5% from workers’ hands swabs, milking utensils, and BTM, respectively.
Table 2.
Frequency Distribution of Bacterial Species Recovered from Dairy Farms and Slaughterhouse
| Sampling Site | Sampling Source | Prevalence of Bacterial Isolates – Number (%) | ||||
|---|---|---|---|---|---|---|
| E. coli O157:H7 | L. monocytogenes | S. typhimurium | P. aeruginosa | K. pneumoniae | ||
| Dairy farms | BTM | 12 (30) | 8 (20) | 8 (20) | 2 (5) | 10 (25) |
| MU | 10 (25) | 8 (20) | 14 (35) | 4 (10) | 4 (10) | |
| WHSs | 6 (15) | 10 (25) | 10 (25) | 8 (20) | 6 (15) | |
| Total | 23.3 | 21.7 | 26.7 | 11.7 | 16.7 | |
| Beef slaughterhouse | Carcass | 6 (30) | 6 (30) | 6 (30) | 2 (10) | 0 |
| KSs | 8 (40) | 4 (20) | 2 (10) | 4 (20) | 2 (10) | |
| WSs | 2 (10) | 8 (40) | 6 (30) | 4 (20) | 0 | |
| WHSs | 8 (40) | 2 (10) | 4 (20) | 4 (20) | 2 (10) | |
| Total | 30 | 25 | 22.5 | 17.5 | 5 | |
Notes: Number of samples; 120/dairy farm samples, 80/slaughterhouse samples.
Abbreviations: BTM, bulk tank milk; MU, milking utensils; KSs, knives’ swabs; WSs, wall swabs; WHSs, workers’ hands swabs.
In accordance with samples collected from the beef slaughterhouse, out of the 80 collected samples, the overall prevalence was 30%, 25%, 22.5%, 17.5%, and 5% for E. coli O157:H7, L. monocytogenes, S. typhimurium, P. aeruginosa, and K. pneumoniae, respectively. Moreover, knife, workers’ hands swabs and carcasses were the major sources of E. coli O157:H7, with recovery rates of 40%, 40%, and 30%, respectively. Meanwhile, the hotspots of L. monocytogenes contamination in slaughterhouse were found to be high on wall swabs (40%), followed by carcasses (30%) and 20% on knife swabs. Most S. typhimurium in the slaughterhouse was isolated from wall swabs and carcasses (both were 30%), followed by workers’ hands swabs (20%), while the least was detected from knife swabs (10%). No major differences among different sources were found for P. aeruginosa isolation. K. pneumoniae was mostly isolated from knives’ and workers’ swabs at the rate of 10%, and could not be detected from the other sources.
Antimicrobial Resistance Patterns of Isolated Bacterial Species from Dairy Farms and Abattoir
Among all the isolated strains, 40 confirmed bacterial strains were isolated from dairy farms and the abattoir (five/each bacterial strain) that were subjected to antimicrobial sensitivity testing using disk-Bauer diffusion method to assess their resistance patterns (Table 3).
Table 3.
Multidrug Resistance Patterns (MDR) of Recovered Bacterial Species (N=40) from Dairy Farms and a Beef Slaughterhouse
| Bacterial Species | % of Resistant Dairy Farm Isolates | % of Resistant Beef Abattoir Isolates | MDR Pattern* | MAR Index |
|---|---|---|---|---|
| E. coli O157:H7 | 35.7 | 25 | E, AM, TE, S | 0.5 |
| L. monocytogenes | 46.2 | 20 | RA, CTX, TE, CN, C | 0.45 |
| S. typhimurium | 25 | 44.4 | SXT, C, NOR, AM, CIP | 0.63 |
| P. aeruginosa | 28.6 | 85.7 | CIP, NOR, S, LEV | 0.44 |
| K. pneumoniae | 70 | 50 | N, K, TE, C, NA, AM, CN | 0.88 |
Notes: *Erythromycin (E; 15 µg), Rifampicin (RA; 5 µg), Chloramphenicol (C; 30 µg), Norfloxacin (NOR; 10 µg), Levofloxacin (LEV; 5 µg), Neomycin (N; 30 µg), Nalidixic acid (NA; 30 µg), Ciprofloxacin (CIP; 5 µg), Tetracycline (TE; 5 µg), Amoxicillin (AM; 25 µg), Cefotaxime (CXT; 30 µg), Kanamycin (K; 30 µg), Trimethoprim/sulphamethoxazole (SXT; 25 µg), Gentamycin (CN; 10 µg), Streptomycin (S; 10 µg).
Abbreviation: MAR, multiple antibiotic resistance index.
As depicted in Table 3, the percentage of overall resistance patterns of the isolated strains from dairy farms with their corresponding bacteria was as follow: 37.5%, 46.2%, 25%, 28.6%, and 70%, for E. coli O157:H7, L. monocytogenes, S. typhimurium, P. aeruginosa, and K. pneumoniae, respectively. Clearly, these results indicate that K. pneumoniae showed the highest resistance level among the isolated bacterial species from dairy farms. Meanwhile, the percentage of the resistance level of isolated strains from beef abattoir samples vs their corresponding bacterial species was as follows: 85.7%, 50%, 44.4%, 25%, and 20% for P. aeruginosa, K. pneumoniae, S. typhimurium, E. coli O157:H7, and, L. monocytogenes, respectively. According to these results, a multiple antimicrobial resistances (MAR) index was determined and revealed that all isolated strains exhibited resistance to at least four antimicrobials.
Collectively, MAR index values ranged between 0.44–0.88, whereas K. pneumoniae had the highest MAR index of 0.88, with demonstrated resistance to six antimicrobials (neomycin, kanamycin, tetracycline, nalidixic acid, amoxicillin, and gentamycin). Recovered S. typhimurium strains had a MAR index of 0.63 with a resistance profile to five antimicrobials; norfloxacin, amoxicillin, ciprofloxacin, Chloramphenicol, and Trimethoprim/sulphamethoxazole. Furthermore, the confirmed E. coli O157:H7 strains exhibited resistance to erythromycin, amoxicillin, tetracycline, and streptomycin, with a MAR index of 0.5. Regarding L. monocytogenes, they demonstrated resistance to five antimicrobials; Rifampicin, Cefotaxime, tetracycline, gentamycin and chloramphenicol, with a MAR index value of 0.45. On the other hand, P. aeruginosa had the lowest MAR index value of 0.44 with a resistance pattern to ciprofloxacin, norfloxacin, streptomycin, and levofloxacin.
Characterization of AgNP-H2O2 Product
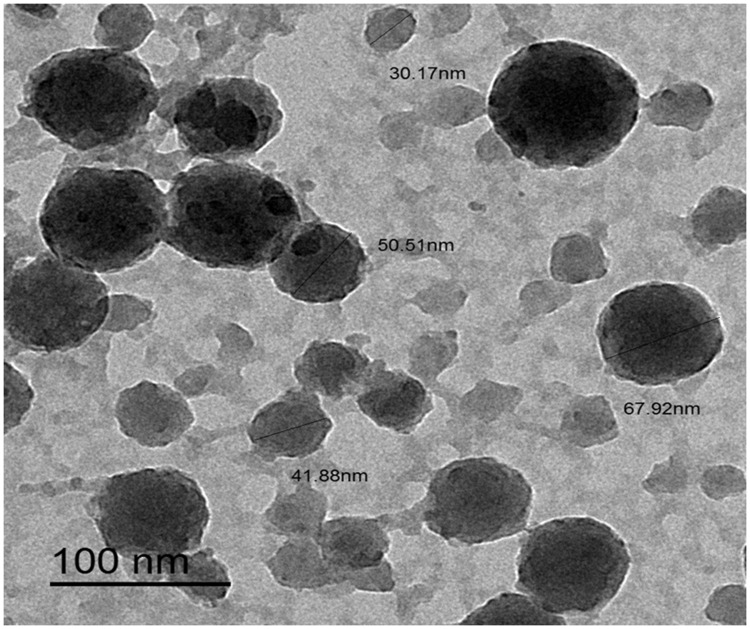
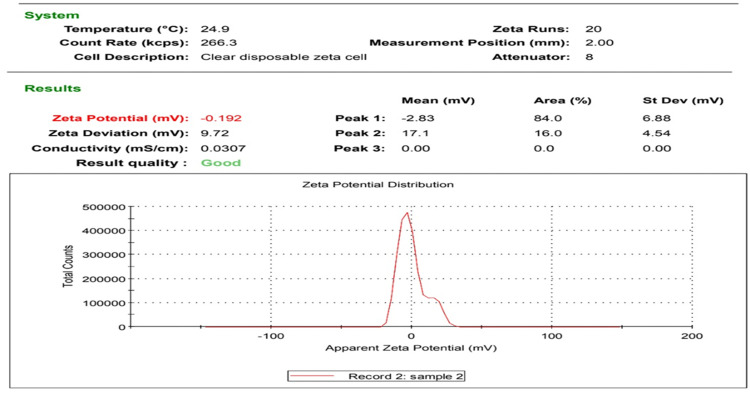
Figures 1 and 2 display the TEM images and zeta potential of the used AgNPs-H2O2 product. A stock solution of 100 nm silver nanoparticles product was made in culture media and further dilutions were made in Luria-Bertani broth. The morphology, shape, and size of NPs were measured by TEM and a Malvern Nano Zeta Sizer (Malvern Zetasize Nano-zs90). The nanoparticles exhibited spherical appearance with a well-defined particle size range (30.17–67.92 nm) that has a zeta potential estimation of −0.192 mV.
Figure 1.
Characterization of AgNPs-H2O2 product, TEM micrographs and size distribution for silver (scale bar: 100 nm), micrographs displays AgNPs-H2O2 with sizes ranging from (30.17–67.92 nm) based on TEM images.
Figure 2.
Zeta-potential of AgNPs-H2O2 product, −0.192 mV surface charge – good quality nanoparticles.
Antimicrobial Activity of AgNPs-H2O2 on MDR Bacteria
The microtiter broth dilution method was used to assess the bactericidal activity of AgNPs-H2O2 against MDR bacteria (Table 4). MIC values of the product to inhibit bacterial growth were 6.25, 12.5, 3.125, 6.25, and 25 µg/mL for E. coli O157:H7, L. monocytogenes, S. typhimurium, P. aeruginosa, and K. pneumoniae, respectively. The MBC of AgNPs-H2O2 ranged between 6.25 and 50 µg/mL for all tested strains. The MBC/MIC ratio is a measure that indicates the bactericidal capacity of the investigated compound. The association between MIC and MBC tests was measured by Kappa test. High association levels between both tests (83, 82, and 80%) were detected for S. typhimurium, E. coli O157:H7, and K. pneumoniae, respectively. However, a low association (30%) was recorded for both L. monocytogenes and P. aeruginosa (Table 4).
Table 4.
MIC, MBC Values, and MBC/MIC Ratio of Commercial AgNPs-H2O2 Product Against MDR Bacterial Species
| MDR Bacteria | Concentration (µg/mL) | MBC/MIC Ratio | P-value | Degree of Agreement Between MIC and MBC Tests %* | |
|---|---|---|---|---|---|
| MIC | MBC | ||||
| E. coli O157:H7 | 6.25 | 12.5 | 2 | <0.0001 | 82 |
| L. monocytogenes | 12.5 | 50 | 4 | <0.0001 | 30 |
| S. typhimurium | 3.125 | 6.25 | 2 | <0.0001 | 83 |
| P. aeruginosa | 6.25 | 50 | 8 | <0.0001 | 30 |
| K. pneumoniae | 25 | 50 | 2 | <0.0001 | 80 |
Note: *Kappa test association.
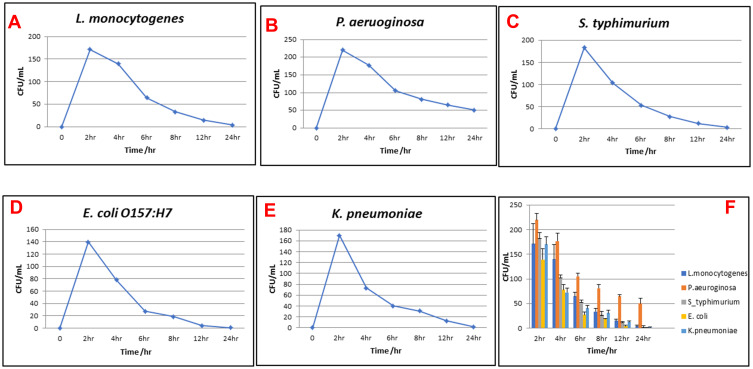
In the current study, AgNPs-H2O2 exerted a bactericidal effect toward all tested strains except for P. aeruginosa that expressed bacteriostatic action. As previously mentioned, the bactericidal effect of AgNPs-H2O2 on MDR bacteria, namely; E. coli O157:H7, L. monocytogenes, S. typhimurium, P. aeruginosa, and K. pneumoniae; isolated from dairy farms and a beef abattoir was assessed using various tests (MIC, MBC, and time-kill assay). The time-dependent bactericidal activity of AgNP product is shown in Figure 3 and this assay was done with respect to the MIC value of each selected strain. The bactericidal activity was evaluated by the actual reduction of cell viabilities (CFU/mL) at time intervals 2, 4, 6, 8, 12, and 24 hours for each isolate. All tested strains had high growth at 2 and 4 hours post-contact with AgNPs-H2O2 product. Importantly, the growth declined at 6 hours and reached complete inhibition at 24 hours, with the exception of P. aeruginosa, which showed instant bacterial growth even after 24 hours.
Figure 3.
Time-kill assays of AgNPs-H2O2 against multidrug-resistant bacteria. (A) L. monocytogenes, (B) P. aeruginosa, (C) S. typhimurium, (D) E. coli O157:H7, (E) K. pneumoniae, and (F) comparison of time-kill assay of AgNPs-H2O2 against all tested MDR bacteria. The measurements were made at 0, 2, 4, 6, 8, 12, and 24 hours post-treatment, viability of microorganisms expressed by CFU/mL, data points represented by mean±SEM for triplicates of each experiment.
Discussion
Cattle are considered a main reservoir for several zoonotic pathogens.65–67 The present study reported novel interesting data about the relatively high prevalence of various zoonotic MDR bacteria isolated from dairy farms and a beef slaughterhouse combined with an evaluation of their antimicrobial resistance patterns and assessment of the antibacterial activity of AgNPs-H2O2 as an alternative to conventional antibiotics. To our knowledge, the current work is the first study that involves exploring the role of silver nanoparticles for control of multiple drug-resistant pathogens isolated from dairy farms and beef slaughterhouses in Egypt.
Several previous studies revealed that several zoonotic pathogens, including E. coli O157:H7, colonize the intestine of cattle and excreted in the feces and are not being expelled in the milk;65–67 however, the fecal contamination of milk during its collection has been reported.68 Notably, as depicted in Table 2, our study reported a relative high prevalence rate of 23.3% and 22.5% for E. coli O157:H7 isolated from dairy farms and a beef abattoir, respectively. The prevalence rate was 30% in both BTM and carcasses from dairy farms and the slaughterhouse, respectively. The present prevalence is higher than reported in some previous studies.69,70 This variation might be attributed to poor hygienic measures adapted at both farms and abattoirs,69,70 besides the skin/fecal-carcass contamination at processing plants that play a vital role in carcass contamination.66 On the contrary, the lower prevalence rates of E. coli O157:H7 (2.7%, 3.2%, and 8%) from beef samples in abattoirs were reported in previous studies.71–73 Also, a low isolation rate of 10% from BTM was recorded elsewhere.74 In accordance with L. monocytogenes, it is classified as the third main pathogen that spread via food and its existence in milk and dairy products, besides having adverse effects on the dairy industry and public health.75–78 L. monocytogenes can be transmitted to humans through contaminated milk and meat.79,80 In the present study, the prevalence rate of L. monocytogenes was 21.7% and 25% from dairy farms and the slaughterhouse, respectively. Taking this into account, milking utensils had the highest isolation rate, at 25%, followed by 20% for both BTM and workers’ hands swabs. These results concurred with several previous findings which isolated L. monocytogenes by a percentage of 21–26% from raw milk.81,82 Additionally, another previous study reported that 20% of BTM samples harbored L. monocytogenes in a dairy farm.83 All these findings refer to infected animals, bad silage quality, and inadequate hygienic measures as possible sources of L. monocytogenes contamination in dairy farms.84 Our findings also reveal a high prevalence level of L. monocytogenes in the abattoir, which is supported by several previous studies which indicated that transportation stress of animals results in increasing shedding rates of the bacterium.85,86
In fact, S. typhimurium is one of the most frequent zoonotic pathogens that causes several human diseases due to consumption of contaminated foods, including meats and raw milk.87,88 Interestingly, our obtained data from dairy farms showed high prevalence rates of S. typhimurium (26.7%), where there was ascending order of frequency of isolation levels that were 35%, 25%, and 20% from milking utensils, workers’ hands swabs, and BTM, respectively. These results are closely correlated with those reported in a previous study where Salmonella spp. could be isolated from bulk tank milk and cull dairy cow fecal samples.89 On the other hand, lower isolation rates of 9% and 8% of Salmonella from milk and workers’ hands swabs were reported in a previous study in Sharkia, Egypt.90 Another previous study did not identify Salmonella in BTM from dairy farms.91 The variations of the current results vs the previous ones could be attributed to several factors including the differences in hygienic and sanitation practices applied during milking.87,92 Notably, high prevalence rates (26.7%) of S. typhimurium were found in the slaughterhouse, whereas the highest frequency level of isolation (30%) was reported from carcass and wall swabs’ samples. These data are in agreement with some previous reports.33,71 Carcasses contaminated during evisceration or hide removal, repeated use of the slaughtering devices for different animals, improper utensils’ sterilization, and sharpening knives on unclean objects all collectively contribute to high opportunities of Salmonella contamination in abattoirs.33,92
Regarding P. aeruginosa, it has been considered a pathogen of a wide host range. In cattle, P. aeruginosa has been linked to cause many diseases, particularly mastitis.93 Previous studies have shown that the presence of P. aeruginosa in the milk samples may be due to many unhygienic maintenance defects that include inadequate cleaning of refrigerators or bulk coolers and may be due to contamination of milk with polluted water at farm level.94 Remarkably, the data obtained from our current study shows that the distribution of P aeruginosa among the dairy farms and slaughterhouses was 11.7% and 17.5%, respectively. Nearly similar isolation rates of 9.4% of P. aeruginosa were reported in previous works.94,95 On the other hand, lower isolation rates of 3.6 and 5.4% were indicated in some previous reports.96,97 This difference could be attributed to the previous mentioned factors related to unhygienic practices that lead to contamination of milk with polluted water at farm level.94 In the same line, Klebsiella spp. are Gram-negative bacteria with considerable effects on milk production and animal survival.98 It has been involved in mastitis outbreaks.99 The overall prevalence of K. pneumoniae in our present results from dairy farms and an abattoir was 16.7% and 5%, respectively. These findings revealed the existence of this bacterium in the animal environment in various sources. A lower prevalence rate of 8.6% was reported in a previous report from dairy farms.100 Klebsiella spp. are frequently shed in feces of healthy cows that results in higher contamination levels in the bedding, and manure in holding pens and alleyways that together lead to higher prevalence of Klebsiella on the skin of the teat and consequently milk contamination.101
As shown in Table 3, the antibiogram profile of the five isolated bacterial species from dairy farms and a beef abattoir revealed the emergence of multidrug resistance among all isolated strains. As illustrated in Table 3, the recorded percentage of resistance of isolated strains from dairy farms was 70%, 46.2%, 35.7%, 28.6%, and 25% for K. pneumoniae, L. monocytogenes, E. coli O157:H7, P. aeruginosa, and S. typhimurium, respectively. Meanwhile, among abattoir samples, the resistance pattern was 85.7% for P. aeruginosa strains, 50% for K. pneumoniae, then 44.4%, 25%, and 20% for S. typhimurium, E. coli O157:H7, and L. monocytogenes, respectively. Clearly, the present data reveals that MDR bacteria were predominant among isolated strains as foodborne pathogens that have been augmented through the last decades.102 The resistance pattern of the evaluated strains towards commonly used antibiotics in the veterinary field highlight that all bacterial strains had MAR to at least four antimicrobials. Our findings concurred with those reported elsewhere.103–107 The high resistance of E. coli O157:H7, L. monocytogenes, and K. pneumoniae to tetracycline correlated to the extensive use of tetracycline in dairy farms for treatment of infectious diseases.108 In the current study, all strains had a MAR index which ranged between 0.44–0.88, since it is well known that isolates which display a MAR index less than 0.2 are supposed to come from antibiotic-rare used sources.109 These data confirm that the usage of antimicrobial agents is highly linked to the evolution risk of MDR bacteria.1 It was formerly documented that resistant strains carefully chosen during an antimicrobial treatment extend for a long time in the intestinal tract when this treatment terminates. Moreover, these resistant strains could influence animal health and can be spread to other animals, particularly to their progeny and accompanying animals.110
Interestingly, examination of AgNPs-H2O2 product using TEM and zeta potential represent a major potential tool for determining the morphology, size, and surface charge of nanoparticles, and consequently in understanding the physical stability of the nanosuspensions.111 Nanosuspensions are known to have good physical activity when their nanocrystals have large positive or negative zeta potential values other than −30 mV to +30 mV because of the electrostatic repulsion of separate particles. The used product had a zeta potential value of −0.192 mV which refers to sufficient repulsive power inducing better physical colloidal stability with a high degree of dependability.112 Meanwhile, nanoparticles aggregation and flocculation are associated with small zeta potential values due to the attractive forces of van der Waals acting upon them, which targets physical instability.113
Over the past decades, global concern has been directed toward MDR bacteria as a major challenge for human and animal health.114 The control strategy of such bacteria is mainly based on the usage of conventional antibiotics. However, their inappropriate use can lead to a global disaster and push us to find alternatives for these drugs. Nanotechnology has been considered a biological nano weapon that helps in re-investigation of the biological characters of previously known antimicrobial compounds by controlling their size to modify their potential effects.115 In the present study, we aimed to assess the antimicrobial activity of AgNPs-H2O2 against MDR bacteria through several in vitro assays (MIC, MBC, and time-kill assay), and therefore, evaluating the efficacy of various concentrations and times on the cell viability of tested bacteria. MIC assay was used to determine the lowest concentration of the tested NPs necessary to completely inhibit the bacterial growth with increasing concentrations of the product. Our data revealed that all strains were sensitive to the tested NPs, where MIC values were as follows: 3.125 µg/mL for S. typhimurium, 6.25 µg/mL for both E. coli O157:H7 and P. aeruginosa, 12.5 µg/mL for L. monocytogenes, and 25 µg/mL for K. pneumoniae. MBC values of AgNPs-H2O2 indicating that S. typhimurium had the highest sensitivity toward NP product at 6.25 µg/mL, while E. coli O157:H7 needed a higher concentration to inhibit their growth at 12.5 µg/mL, whereas P. aeruginosa, L. monocytogenes, and K. pneumoniae growth totally declined at 50 µg/mL. These findings are consistent with those of Zarei et al,116 who reported antibacterial activity values of AgNPs against S. typhimurium were MIC of 3.12 µg/mL and MBC of 6.25 µg/mL. On the other hand, 2 µg/mL of AgNPs of 30 nm average size had a bacteriostatic effect on P. aeruginosa.117 For all MDR strains, except for P. aeruginosa, the MBC/MIC ratios indicated that AgNPs-H2O2 product has a bactericidal rather than bacteriostatic effect, which is favored clinically since bacterial death leads to a rapid infection termination, better clinical outcome, and less possibilities of resistance emergence and infection spread.115 Consequently, these actions in turn reduce the emergence of resistance mutations.118 It should be stressed that the silver nanoparticles have a broad-spectrum activity against Gram-negative bacteria including members of the genera Escherichia, Pseudomonas, and Salmonella, besides Gram-positive bacteria include Clostridium, Listeria, Staphylococcus, and Streptococcus,119 through attaching to the bacterial cell membrane that results in disruption of its function, invading bacteria, and releasing silver ions.120 The post-treatment bacterial survival can be evaluated by time-kill assays to describe the least time required to induce bacteriostatic or bactericidal actions. Rapid bacterial reproduction time is one of the main essentials of bacterial infectivity, a characteristic that could be a good target for impeding a viable infection. Analysis of association degree using Kappa test between MIC and MBC results might confirm the compatibility between the two methods of evaluation besides revealing the higher association degree and the more dependability of the assays. Hence, kappa test offers more information than a simple calculation of the raw amount of agreement.121 Assessing AgNPs-H2O2 product bactericidal power in relation to MIC values and time declared its bactericidal power in a dose- and time-dependent manner. Our results are consistent with that reported in a previous study,122 who found that E. coli count reduced from 107 to 101 CFU/mL at 14 hours post-treatment with silver ions,122 and, therefore, the activity of nanoparticles may be like that of silver ions.123 As observed in our study, the used nanoproduct is nanosilver in conjunction with H2O2. This combination has led to a rapid reduction in bacterial viability over time with complete bacterial death of MDR strains after 12–24 hours. This suggested actions might be attributed to higher bactericidal action by more than 100-folds attained by such a combination due to the Fenton-like reaction among the two agents produces a Hydroxy group (OH),124 that has been considered one of the most powerful biologically active ROS.42
Conclusions
Given the above information, our study highlights the worrying trend of higher MDR bacteria rates among dairy farms and beef abattoirs in Egypt, indicating the discriminative use of antimicrobials in treatment of infectious diseases. Clearly, there must an urgent call to find alternatives for these antimicrobials. More importantly, AgNPs-H2O2 proved a promising powerful bactericidal activity against MDR bacteria regardless of the resistance level against tested strains. Our present data also concludes that AgNPs-H2O2 can be recommended as an ecofriendly broad-spectrum bactericidal agent and our study suggests further future research in the same lines against other species of bacteria and similar nanoparticles. Furthermore, supervision of hygienic and biosecurity measures and antibiotics’ handling must be carefully checked.
Acknowledgments
The authors also thank the veterinarians and farm owners for their support and help in providing data and samples collection throughout the study.
Funding Statement
This research received no external funding.
Author Contributions
F.A.E., A.I.Z., R.R.S., and E.A.A.E. were involved in the conception of the idea, methodology design, laboratory work, performed data analysis, and interpretation. L.J.M.A., A.T., A.E.M., and E.K.E. participated in the design of the methodology, contributed their scientific advice during the sampling, the laboratory work, and data analysis. F.A.E. and E.K.E. drafted and prepared the manuscript for publication and were responsible for correspondence and revision. All authors contributed to data analysis, drafting, or revising the article, agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest for this work.
References
- 1.World Health Organization W. Global action plan on antimicrobial resistance; 2015. Available from: http://www.who.int/drugresistance/globalaction_plan/en/. Accessed June10, 2020.
- 2.Beceiro A, Tomas M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev. 2013;26(2):185–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El Zowalaty ME, Al Thani AA, Webster TJ, et al. Pseudomonas aeruginosa: arsenal of resistance mechanisms, decades of changing resistance profiles, and future antimicrobial therapies. Future Microbiol. 2015;10(10):1683–1706. doi: 10.2217/fmb.15.48 [DOI] [PubMed] [Google Scholar]
- 4.Jasovsky D, Littmann J, Zorzet A, Cars O. Antimicrobial resistance-a threat to the world’s sustainable development. Ups J Med Sci. 2016;121(3):159–164. doi: 10.1080/03009734.2016.1195900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutchinson H, Finney S, Munoz-Vargas L, Feicht S, Masterson M, Habing G. Prevalence and transmission of antimicrobial resistance in a vertically integrated veal calf production system. Foodborne Pathog Dis. 2017;14(12):711–718. doi: 10.1089/fpd.2017.2310 [DOI] [PubMed] [Google Scholar]
- 6.Palma E, Tilocca B, Roncada P. Antimicrobial resistance in veterinary medicine: an overview. Int J Mol Sci. 2020;21(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McEwen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin Infect Dis. 2002;34(Suppl 3):S93–S106. doi: 10.1086/340246 [DOI] [PubMed] [Google Scholar]
- 8.Cook EAJ, de Glanville WA, Thomas LF, Kariuki S, de Clare Bronsvoort BM, Fèvre EM. Working conditions and public health risks in slaughterhouses in western Kenya. BMC Public Health. 2017;17(1):14. doi: 10.1186/s12889-016-3923-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fijałkowski K, Peitler D, Karakulska J. Staphylococci isolated from ready-to-eat meat–identification, antibiotic resistance and toxin gene profile. Int J Food Microbiol. 2016;238:113–120. doi: 10.1016/j.ijfoodmicro.2016.09.001 [DOI] [PubMed] [Google Scholar]
- 10.Soares Casaes Nunes R, Mere Del Aguila E, Paschoalin VMF. Safety evaluation of the coagulase-negative staphylococci microbiota of salami: superantigenic toxin production and antimicrobial resistance. Biomed Res Int. 2015;2015:1–17. doi: 10.1155/2015/483548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogeveen H, Van Der Voort M. Assessing the economic impact of an endemic disease: the case of mastitis. Rev Sci Tech. 2017;36(1):217–226. [DOI] [PubMed] [Google Scholar]
- 12.Mukasa D, Kankya C, Nakavuma JL. Listeria contamination of raw bovine milk and the factors influencing its occurrence in Greater Luweero District, Uganda. Microbiol Res J Int. 2016;1–10. [Google Scholar]
- 13.Manyi-Loh C, Mamphweli S, Meyer E, Okoh A. Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules. 2018;23(4):795. doi: 10.3390/molecules23040795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hudson CM, Bent ZW, Meagher RJ, Williams KP. Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS One. 2014;9(6):e99209. doi: 10.1371/journal.pone.0099209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krömker V, Leimbach S. Mastitis treatment—reduction in antibiotic usage in dairy cows. Reprod Domest Anim. 2017;52:21–29. [DOI] [PubMed] [Google Scholar]
- 16.Obaidat MM, Stringer AP. Prevalence, molecular characterization, and antimicrobial resistance profiles of Listeria monocytogenes, Salmonella enterica, and Escherichia coli O157: H7 on dairy cattle farms in Jordan. J Dairy Sci. 2019;102(10):8710–8720. doi: 10.3168/jds.2019-16461 [DOI] [PubMed] [Google Scholar]
- 17.Harmon RJ. Physiology of mastitis and factors affecting somatic cell counts. J Dairy Sci. 1994;77(7):2103–2112. [DOI] [PubMed] [Google Scholar]
- 18.Arslan S, Özdemir F. Prevalence and antimicrobial resistance of Listeria spp. in homemade white cheese. Food Control. 2008;19(4):360–363. doi: 10.1016/j.foodcont.2007.04.009 [DOI] [Google Scholar]
- 19.Carneiro LA, Lins MC, Garcia FR, et al. Phenotypic and genotypic characterisation of Escherichia coli strains serogrouped as enteropathogenic E. coli (EPEC) isolated from pasteurised milk. Int J Food Microbiol. 2006;108(1):15–21. doi: 10.1016/j.ijfoodmicro.2005.10.010 [DOI] [PubMed] [Google Scholar]
- 20.Varela-Hernandez JJ, Cabrera-Diaz E, Cardona-Lopez MA, et al. Isolation and characterization of Shiga toxin-producing Escherichia coli O157: h7and non-O157 from beef carcasses at a slaughter plant in Mexico. Int J Food Microbiol. 2007;113(2):237–241. doi: 10.1016/j.ijfoodmicro.2006.06.028 [DOI] [PubMed] [Google Scholar]
- 21.Langoni H, Guiduce MVS, Nóbrega DB, et al. Research of Klebsiella pneumoniae in dairy herds. Pesquisa Veterinária Brasileira. 2015;35(1):9–12. doi: 10.1590/S0100-736X2015000100003 [DOI] [Google Scholar]
- 22.Munoz M, Zadoks R. Patterns of fecal shedding of Klebsiella by dairy cows. J Dairy Sci. 2007;90(3):1220–1224. doi: 10.3168/jds.S0022-0302(07)71610-7 [DOI] [PubMed] [Google Scholar]
- 23.Doyle ME, Kaspar C, Archer J, Klos R White paper on human illness caused by salmonella from all food and non-food vectors. Madison, WI: Food Research Institute, University of Wisconsin—Madison; 2009. Available from: http://fri.wisc.edu/briefs/FRI_Brief_Salmonella_Human_Illness_6_09.pdf. Accessed Septimber 4, 2020. [Google Scholar]
- 24.Piepers S, de Vliegher S. Alternative approach to mastitis management–How to prevent and control mastitis without antibiotics? Braz J Vet Res Anim Sci. 2018;55(3):e137149. [Google Scholar]
- 25.Ruegg P, Tabone T. The relationship between antibiotic residue violations and somatic cell counts in Wisconsin dairy herds. J Dairy Sci. 2000;83(12):2805–2809. doi: 10.3168/jds.S0022-0302(00)75178-2 [DOI] [PubMed] [Google Scholar]
- 26.Arthur TM, Bosilevac JM, Brichta-Harhay DM, et al. Effects of a minimal hide wash cabinet on the levels and prevalence of Escherichia coli O157: H7 and Salmonella on the hides of beef cattle at slaughter. J Food Prot. 2007;70(5):1076–1079. doi: 10.4315/0362-028X-70.5.1076 [DOI] [PubMed] [Google Scholar]
- 27.Brichta-Harhay DM, Guerini MN, Arthur TM, et al. Salmonella and Escherichia coli O157: H7 contamination on hides and carcasses of cull cattle presented for slaughter in the United States: an evaluation of prevalence and bacterial loads by immunomagnetic separation and direct plating methods. Appl Environ Microbiol. 2008;74(20):6289–6297. doi: 10.1128/AEM.00700-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nandi S, Maurer JJ, Hofacre C, Summers AO. Gram-positive bacteria are a major reservoir of Class 1 antibiotic resistance integrons in poultry litter. Proc Natl Acad Sci. 2004;101(18):7118–7122. doi: 10.1073/pnas.0306466101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hui YH. Handbook of Meat and Meat Processing. CRC press; 2012. [Google Scholar]
- 30.Diyantoro WDK. Risk factors for bacterial contamination of bovine meat during slaughter in ten Indonesian abattoirs. Vet Med Int. 2019;2019:2707064. doi: 10.1155/2019/2707064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fegan N, Vanderlinde P, Higgs G, Desmarchelier P. A study of the prevalence and enumeration of Salmonella enterica in cattle and on carcasses during processing. J Food Prot. 2005;68(6):1147–1153. doi: 10.4315/0362-028X-68.6.1147 [DOI] [PubMed] [Google Scholar]
- 32.Tadesse G, Tessema TS. A meta-analysis of the prevalence of Salmonella in food animals in Ethiopia. BMC Microbiol. 2014;14(1):270. doi: 10.1186/s12866-014-0270-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madoroba E, Kapeta D, Gelaw AK. Salmonella contamination, serovars and antimicrobial resistance profiles of cattle slaughtered in South Africa. Onderstepoort J Vet Res. 2016;83(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Defoirdt T, Sorgeloos P, Bossier P. Alternatives to antibiotics for control of bacterial disease in aquaculture. Curr Opin Microbiol. 2011;14:251–258. [DOI] [PubMed] [Google Scholar]
- 35.Beyth N, Houri-Haddad Y, Domb A, Khan W, Hazan R. Alternative antimicrobial approach: nano-antimicrobial materials. Evid Based Complement Alternat Med. 2015;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurunathan S. Biologically synthesized silver nanoparticles enhances antibiotic activity against Gram-negative bacteria. J Ind Eng Chem. 2015;29:217–226. doi: 10.1016/j.jiec.2015.04.005 [DOI] [Google Scholar]
- 37.Yah CS, Simate GS. Nanoparticles as potential new generation broad spectrum antimicrobial agents. DARU J Pharm Sci. 2015;23(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patra JK, Das G, Fraceto LF, et al. Nano based drug delivery systems: recent developments and future prospects. J Nanobiotechnology. 2018;16(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vardanyan Z, Gevorkyan V, Ananyan M, Vardapetyan H, Trchounian A. Effects of various heavy metal nanoparticles on Enterococcus hirae and Escherichia coli growth and proton-coupled membrane transport. J Nanobiotechnology. 2015;13(1):69. doi: 10.1186/s12951-015-0131-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calderon-Jimenez B, Johnson ME, Montoro Bustos AR, Murphy KE, Winchester MR, Vega Baudrit JR. Silver nanoparticles: technological advances, societal impacts, and metrological challenges. Front Chem. 2017;5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones N, Ray B, Ranjit KT, Manna AC. Antibacterial activity of ZnO nanoparticle suspensions on a broad spectrum of microorganisms. FEMS Microbiol Lett. 2008;279(1):71–76. doi: 10.1111/j.1574-6968.2007.01012.x [DOI] [PubMed] [Google Scholar]
- 42.Xie Y, He Y, Irwin PL, Jin T, Shi X. Antibacterial activity and mechanism of action of zinc oxide nanoparticles against Campylobacter jejuni. Appl Environ Microbiol. 2011;77(7):2325–2331. doi: 10.1128/AEM.02149-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, He L, Mustapha A, Li H, Hu Z, Lin M. Antibacterial activities of zinc oxide nanoparticles against Escherichia coli O157: H7. J Appl Microbiol. 2009;107(4):1193–1201. doi: 10.1111/j.1365-2672.2009.04303.x [DOI] [PubMed] [Google Scholar]
- 44.Dakal TC, Kumar A, Majumdar RS, Yadav V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front Microbiol. 2016;7:1831. doi: 10.3389/fmicb.2016.01831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao S, Zhang Y, Pan X, et al. Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa. Int J Nanomedicine. 2019;14:1469–1487. doi: 10.2147/IJN.S191340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leid JG, Ditto AJ, Knapp A, et al. In vitro antimicrobial studies of silver carbene complexes: activity of free and nanoparticle carbene formulations against clinical isolates of pathogenic bacteria. J Antimicrob Chemother. 2012;67(1):138–148. doi: 10.1093/jac/dkr408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Hu C, Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int J Nanomedicine. 2017;12:1227–1249. doi: 10.2147/IJN.S121956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cavassin ED, de Figueiredo LFP, Otoch JP, et al. Comparison of methods to detect the in vitro activity of silver nanoparticles (AgNP) against multidrug resistant bacteria. J Nanobiotechnology. 2015;13(1):64. doi: 10.1186/s12951-015-0120-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rai M, Deshmukh S, Ingle A, Gade A. Silver nanoparticles: the powerful nanoweapon against multidrug‐resistant bacteria. J Appl Microbiol. 2012;112(5):841–852. doi: 10.1111/j.1365-2672.2012.05253.x [DOI] [PubMed] [Google Scholar]
- 50.Sullivan T, Chapman J, Regan F. Characterisation of Nano-antimicrobial Materials In: Nano-Antimicrobials. Springer, Berlin, Heidelberg; 2012:181–208. [Google Scholar]
- 51.Holt J, Krieg N, Sneath P, Staley J. Bergey’s manual of determinative bacteriology In: Bergey’s Manual of Determinative Bacteriology. 9th ed. Baltimore, MD: Williams & Wilkins Co.; 1994. [Google Scholar]
- 52.Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36(2):598–602. doi: 10.1128/JCM.36.2.598-602.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alvarez J, Sota M, Vivanco AB, et al. Development of a multiplex PCR technique for detection and epidemiological typing of salmonella in human clinical samples. J Clin Microbiol. 2004;42(4):1734–1738. doi: 10.1128/JCM.42.4.1734-1738.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Border PM, Howard JJ, Plastow GS, Siggens KW. Detection of Listeria species and Listeria monocytogenes using polymerase chain reaction. Lett Appl Microbiol. 1990;11(3):158–162. doi: 10.1111/j.1472-765X.1990.tb00149.x [DOI] [PubMed] [Google Scholar]
- 55.Ahmed AM, Motoi Y, Sato M, et al. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl Environ Microbiol. 2007;73(20):6686–6690. doi: 10.1128/AEM.01054-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clinical Laboratory Standards Institute C. Performance standards for antimicrobial disk susceptibility tests; approved standard. M02-A11 CLSI, Wayne; 2012.
- 57.Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46(1):165–170. doi: 10.1128/AEM.46.1.165-170.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarz S, Silley P, Simjee S, et al. Assessing the antimicrobial susceptibility of bacteria obtained from animals. J Antimicrob Chemother. 2010;65(4):601–604. doi: 10.1093/jac/dkq037 [DOI] [PubMed] [Google Scholar]
- 59.Jyoti K, Baunthiyal M, Singh A. Characterization of silver nanoparticles synthesized using Urtica dioica Linn. leaves and their synergistic effects with antibiotics. J Radiat Res Appl Sci. 2016;9(3):217–227. doi: 10.1016/j.jrras.2015.10.002 [DOI] [Google Scholar]
- 60.(CLSI) CLSI. Reference Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, Approved Standard. 2nd ed. 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087–1898, USA: NCCLS document M27- A2. CLSI; 2002. [Google Scholar]
- 61.Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: A review. J Pharm Anal. 2016;6(2):71–79. doi: 10.1016/j.jpha.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ayala-Núñez NV, Villegas HHL, Turrent L, Padilla CR. Silver nanoparticles toxicity and bactericidal effect against methicillin-resistant Staphylococcus aureus: nanoscale does matter. Nanobiotechnology. 2009;5(1–4):2–9. doi: 10.1007/s12030-009-9029-1 [DOI] [Google Scholar]
- 63.(CLSI) CLSI. M07: Methods for Dilution Antimicrobial Susceptibility for Bacteria That Grow Aerobically 11th Edition; 2015. [Google Scholar]
- 64.Konaté K, Mavoungou JF, Lepengué AN, et al. Antibacterial activity against β-lactamase producing Methicillin and Ampicillin-resistants Staphylococcus aureus: fractional Inhibitory Concentration Index (FICI) determination. Ann Clin Microbiol Antimicrob. 2012;11(1):18. doi: 10.1186/1476-0711-11-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McDaniel CJ, Cardwell DM, Moeller RB Jr, Gray GC. Humans and cattle: a review of bovine zoonoses. Vector Borne Zoonotic Dis. 2014;14(1):1–19. doi: 10.1089/vbz.2012.1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arthur TM, Brichta-Harhay DM, Bosilevac JM, et al. Super shedding of Escherichia coli O157: H7 by cattle and the impact on beef carcass contamination. Meat Sci. 2010;86(1):32–37. doi: 10.1016/j.meatsci.2010.04.019 [DOI] [PubMed] [Google Scholar]
- 67.Ferens WA, Hovde CJ. Escherichia coli O157: H7:animal reservoir and sources of human infection. Foodborne Pathog Dis. 2011;8(4):465–487. doi: 10.1089/fpd.2010.0673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heuvelink A, Van Den Biggelaar F, Zwartkruis-Nahuis J, et al. Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J Clin Microbiol. 1998;36(12):3480–3487. doi: 10.1128/JCM.36.12.3480-3487.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Njisane YZ, Muchenje V. Farm to abattoir conditions, animal factors and their subsequent effects on cattle behavioural responses and beef quality - A review. Asian-Australas J Anim Sci. 2017;30(6):755–764. doi: 10.5713/ajas.16.0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Founou LL, Founou RC, Essack SY. Antibiotic resistance in the food chain: a developing country-perspective. Front Microbiol. 2016;7:1881. doi: 10.3389/fmicb.2016.01881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McEvoy J, Doherty A, Sheridan J, et al. The prevalence and spread of Escherichia coli O157: H7 at a commercial beef abattoir. J Appl Microbiol. 2003;95(2):256–266. doi: 10.1046/j.1365-2672.2003.01981.x [DOI] [PubMed] [Google Scholar]
- 72.Taye M, Berhanu T, Berhanu Y, Tamiru F, Terefe D. Study on carcass contaminating Escherichia coli in apparently healthy slaughtered cattle in Haramaya University slaughter house with special emphasis on Escherichia coli O157: H7, Ethiopia. J Vet Sci Technol. 2013;4(1):132. [Google Scholar]
- 73.Abdissa R, Haile W, Fite AT, et al. Prevalence of Escherichia coli O157: h7in beef cattle at slaughter and beef carcasses at retail shops in Ethiopia. BMC Infect Dis. 2017;17(1):277. doi: 10.1186/s12879-017-2372-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Padhye NV, Doyle MP. Rapid procedure for detecting enterohemorrhagic Escherichia coli O157: H7 in food. Appl Environ Microbiol. 1991;57(9):2693–2698. doi: 10.1128/AEM.57.9.2693-2698.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shamloo E, Hosseini H, Abdi Moghadam Z, Halberg Larsen M, Haslberger A, Alebouyeh M. Importance of Listeria monocytogenes in food safety: a review of its prevalence, detection, and antibiotic resistance. Iran J Vet Res. 2019;20(4):241–254. [PMC free article] [PubMed] [Google Scholar]
- 76.Kasalica A, Vuković V, Vranješ A, Memiši N. Listeria monocytogenes in milk and dairy products. Biotechnol Anim Husbandry. 2011;27(3):1067–1082. doi: 10.2298/BAH1103067K [DOI] [Google Scholar]
- 77.Usman U, Kwaga J, Kabir J, Olonitola OS, Radu S, Bande F. Molecular characterization and phylogenetic analysis of listeria monocytogenes isolated from milk and milk products in Kaduna, Nigeria. Can J Infect Dis Med Microbiol. 2016;2016:1–7. doi: 10.1155/2016/4313827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States–major pathogens. Emerg Infect Dis. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Indrawattana N, Nibaddhasobon T, Sookrung N, et al. Prevalence of Listeria monocytogenes in raw meats marketed in Bangkok and characterization of the isolates by phenotypic and molecular methods. J Health Popul Nutr. 2011;29(1):26–38. doi: 10.3329/jhpn.v29i1.7565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ulusoy BH, Chirkena K. Two perspectives of Listeria monocytogenes hazards in dairy products: the prevalence and the antibiotic resistance. Food Qual Saf. 2019;3(4):233–241. [Google Scholar]
- 81.Vanegas MC, Vásquez E, Martinez AJ, Rueda AM. Detection of Listeria monocytogenes in raw whole milk for human consumption in Colombia by real-time PCR. Food Control. 2009;20(4):430–432. doi: 10.1016/j.foodcont.2008.07.007 [DOI] [Google Scholar]
- 82.Rahimi E, Ameri M, Momtaz H. Prevalence and antimicrobial resistance of Listeria species isolated from milk and dairy products in Iran. Food Control. 2010;21(11):1448–1452. doi: 10.1016/j.foodcont.2010.03.014 [DOI] [Google Scholar]
- 83.Latorre AA, Van Kessel JAS, Karns JS, et al. Molecular ecology of Listeria monocytogenes: evidence for a reservoir in milking equipment on a dairy farm. Appl Environ Microbiol. 2009;75(5):1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sağun E, Sancak Y, İşleyici Ö, Ekici K. The presence and prevalence of Listeria species in milk and herby cheese in and around Van. Turk J Vet Anim Sci. 2001;25(1):15–19. [Google Scholar]
- 85.Fenlon D, Wilson J, Donachie W. The incidence and level of Listeria monocytogenes contamination of food sources at primary production and initial processing. J Appl Bacteriol. 1996;81(6):641–650. doi: 10.1111/j.1365-2672.1996.tb03559.x [DOI] [PubMed] [Google Scholar]
- 86.Bailey GD, Vanselow BA, Hornitzky MA, et al. A study of the foodborne pathogens: campylobacter, Listeria and Yersinia, in faeces from slaughter-age cattle and sheep in Australia. Commun Dis Intell Q Rep. 2003;27(2):249–257. [PubMed] [Google Scholar]
- 87.El-Baz AH, El-Sherbini M, Abdelkhalek A, Al-Ashmawy MA. Prevalence and molecular characterization of Salmonella serovars in milk and cheese in Mansoura city, Egypt. J Adv Vet Anim Res. 2017;4(1):45–51. [Google Scholar]
- 88.Thung TY, Radu S, Mahyudin NA, et al. Prevalence, virulence genes and antimicrobial resistance profiles of Salmonella serovars from retail beef in Selangor, Malaysia. Front Microbiol. 2018;8:2697. doi: 10.3389/fmicb.2017.02697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Murinda SE, Nguyen LT, Ivey SJ, et al. Molecular characterization of Salmonella spp. isolated from bulk tank milk and cull dairy cow fecal samples. J Food Prot. 2002;65(7):1100–1105. doi: 10.4315/0362-028X-65.7.1100 [DOI] [PubMed] [Google Scholar]
- 90.El-Gedawy A, Ahmed H, Awadallah M. Occurrence and molecular characterization of some zoonotic bacteria in bovine milk, milking equipments and humans in dairy farms, Sharkia, Egypt. Int Food Res J. 2014;21(5). [Google Scholar]
- 91.Zeinhom MM, Abdel-Latef GK. Public health risk of some milk borne pathogens. Beni-Suef Univ J Basic Appl Sci. 2014;3(3):209–215. doi: 10.1016/j.bjbas.2014.10.006 [DOI] [Google Scholar]
- 92.Ledo J, Hettinga K, Luning P. A customized assessment tool to differentiate safety and hygiene control practices in emerging dairy chains. Food Control. 2019;111:107072. doi: 10.1016/j.foodcont.2019.107072 [DOI] [Google Scholar]
- 93.Daly M, Power E, Bjorkroth J, et al. Molecular analysis of Pseudomonas aeruginosa: epidemiological investigation of mastitis outbreaks in Irish dairy herds. Appl Environ Microbiol. 1999;65(6):2723–2729. doi: 10.1128/AEM.65.6.2723-2729.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Swetha CS, Babu A, Rao K, Sukumar B, Supriya R, Rao T. A study on the antimicrobial resistant patterns of Pseudomonas Aeruginosa isolated from raw milk samples in and around Tirupati, Andhra Pradesh. Asian J Dairy Food Res. 2017;36. [Google Scholar]
- 95.Patel JV. Study on prevalence of mastitis and antibiotic sensitivity of bacterial isolates recovered from crossbred cows of Anand district of Gujarat. Indian J Dairy Sci. 2012;65(6). [Google Scholar]
- 96.Banerjee S, Batabyal K, Joardar S, et al. Detection and characterization of pathogenic Pseudomonas aeruginosa from bovine subclinical mastitis in West Bengal, India. Vet World. 2017;10:738–742. doi: 10.14202/vetworld.2017.738-742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singh R, Sharma N, Soodan J, Sudhan N. Etiology and sensitivity of bacterial isolates from sub clinical mastitis in cattle from Jammu region. SKUAST J Res. 2005;4(2):223–224. [Google Scholar]
- 98.Munoz M, Ahlström C, Rauch B, Zadoks R. Fecal shedding of Klebsiella pneumoniae by dairy cows. J Dairy Sci. 2006;89(9):3425–3430. doi: 10.3168/jds.S0022-0302(06)72379-7 [DOI] [PubMed] [Google Scholar]
- 99.Langoni H, Corrêa C, Corrêa W, Barros J, Corrêa G. Mastites bovinas por Candida e Klebsiella. Rev Bras Med Vet. 1985;7(7):203–204. [Google Scholar]
- 100.Haftu R, Taddele H, Gugsa G, Kalayou S. Prevalence, bacterial causes, and antimicrobial susceptibility profile of mastitis isolates from cows in large-scale dairy farms of Northern Ethiopia. Trop Anim Health Prod. 2012;44(7):1765–1771. doi: 10.1007/s11250-012-0135-z [DOI] [PubMed] [Google Scholar]
- 101.Munoz M, Bennett G, Ahlström C, Griffiths H, Schukken Y, Zadoks R. Cleanliness scores as indicator of Klebsiella exposure in dairy cows. J Dairy Sci. 2008;91(10):3908–3916. doi: 10.3168/jds.2008-1090 [DOI] [PubMed] [Google Scholar]
- 102.Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect Drug Resist. 2015;8:49–61. doi: 10.2147/IDR.S55778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ranjbar R, Zeynali M, Sohrabi N, Ali A. Antibiotic resistance and prevalence of class 1 and 2 integrons in Escherichia coli isolated from hospital wastewater. Univ Med. 2018;37:209. doi: 10.18051/UnivMed.2018.v37.209-215 [DOI] [Google Scholar]
- 104.Oh H, Kim S, Lee S, et al. Prevalence, Serotype Diversity, Genotype and Antibiotic Resistance of Listeria monocytogenes Isolated from Carcasses and Human in Korea. Korean J Food Sci Anim Resour. 2018;38(5):851–865. doi: 10.5851/kosfa.2018.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sobur M, Haque Z, Sabuj A, et al. Molecular detection of multidrug and colistin-resistant Escherichia coli isolated from house flies in various environmental settings. Future Microbiol. 2019;14(10):847–858. doi: 10.2217/fmb-2019-0053 [DOI] [PubMed] [Google Scholar]
- 106.Benie C, Nathalie G, Adjéhi D, et al. Prevalence and antibiotic resistance of Pseudomonas aeruginosa isolated from bovine meat, fresh fish and smoked fish. Arch Clin Microbiol. 2017;08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Montso KP, Dlamini SB, Kumar A, Ateba CN. Antimicrobial resistance factors of extended-spectrum beta-lactamases producing Escherichia coli and Klebsiella pneumoniae isolated from Cattle Farms and Raw Beef in North-West Province, South Africa. Biomed Res Int. 2019;2019:4318306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tahoun AB, Elez RMA, Abdelfatah EN, Elsohaby I, El-Gedawy AA, Elmoslemany AM. Listeria monocytogenes in raw milk, milking equipment and dairy workers: molecular characterization and antimicrobial resistance patterns. J Glob Antimicrob Resist. 2017;10:264–270. doi: 10.1016/j.jgar.2017.07.008 [DOI] [PubMed] [Google Scholar]
- 109.Marian M, Aminah SS, Zuraini M, et al. MPN-PCR detection and antimicrobial resistance of Listeria monocytogenes isolated from raw and ready-to-eat foods in Malaysia. Food Control. 2012;28(2):309–314. doi: 10.1016/j.foodcont.2012.05.030 [DOI] [Google Scholar]
- 110.Roca-Saavedra P, Mendez-Vilabrille V, Miranda JM, et al. Food additives, contaminants and other minor components: effects on human gut microbiota—a review. J Physiol Biochem. 2018;74(1):69–83. [DOI] [PubMed] [Google Scholar]
- 111.Jiang J, Oberdörster G, Biswas P. Characterization of size, surface charge, and agglomeration state of nanoparticle dispersions for toxicological studies. J Nanopart Res. 2009;11(1):77–89. doi: 10.1007/s11051-008-9446-4 [DOI] [Google Scholar]
- 112.Joseph E, Singhvi G. Multifunctional nanocrystals for cancer therapy: a potential nanocarrier In: Nanomaterials for Drug Delivery and Therapy. William Andrew Publishing, Elsevier; 2019:91–116. [Google Scholar]
- 113.Hunter RJ. Zeta Potential in Colloid Science: Principles and Applications. Vol. 2 Academic press; 2013. [Google Scholar]
- 114.Webb GF, D’Agata EM, Magal P, Ruan S. A model of antibiotic-resistant bacterial epidemics in hospitals. Proc Natl Acad Sci. 2005;102(37):13343–13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lara HH, Ayala-Núñez NV, Turrent L, Padilla CR. Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World J Microbiol Biotechnol. 2010;26(4):615–621. doi: 10.1007/s11274-009-0211-3 [DOI] [Google Scholar]
- 116.Zarei M, Jamnejad A, Khajehali E. Antibacterial effect of silver nanoparticles against four foodborne pathogens. Jundishapur J Microbiol. 2014;7(1). doi: 10.5812/jjm.8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kora AJ, Arunachalam J. Assessment of antibacterial activity of silver nanoparticles on Pseudomonas aeruginosa and its mechanism of action. World J Microbiol Biotechnol. 2011;27(5):1209–1216. doi: 10.1007/s11274-010-0569-2 [DOI] [Google Scholar]
- 118.French G. Bactericidal agents in the treatment of MRSA infections—the potential role of daptomycin. J Antimicrob Chemother. 2006;58(6):1107–1117. doi: 10.1093/jac/dkl393 [DOI] [PubMed] [Google Scholar]
- 119.Percival SL, Bowler PG, Dolman J. Antimicrobial activity of silver-containing dressings on wound microorganisms using an in vitro biofilm model. Int Wound J. 2007;4(2):186–191. doi: 10.1111/j.1742-481X.2007.00296.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Morones JR, Elechiguerra JL, Camacho A, et al. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16(10):2346. doi: 10.1088/0957-4484/16/10/059 [DOI] [PubMed] [Google Scholar]
- 121.Viera AJ, Garrett JM. Understanding interobserver agreement: the kappa statistic. Fam Med. 2005;37(5):360–363. [PubMed] [Google Scholar]
- 122.Yamanaka M, Hara K, Kudo J. Bactericidal actions of a silver ion solution on Escherichia coli, studied by energy-filtering transmission electron microscopy and proteomic analysis. Appl Environ Microbiol. 2005;71(11):7589–7593. doi: 10.1128/AEM.71.11.7589-7593.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pal S, Tak YK, Song JM. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the Gram-negative bacterium Escherichia coli. Appl Environ Microbiol. 2007;73(6):1712–1720. doi: 10.1128/AEM.02218-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.He W, Zhou Y-T, Wamer WG, Boudreau MD, Yin -J-J. Mechanisms of the pH dependent generation of hydroxyl radicals and oxygen induced by Ag nanoparticles. Biomaterials. 2012;33(30):7547–7555. [DOI] [PubMed] [Google Scholar]