Abstract
Anthropogenic activities are a key factor in the development of antibiotic resistance in bacteria, a growing problem worldwide. Nevertheless, antibiotics and resistances were being generated by bacterial communities long before their discovery by humankind, and might occur in areas without human influence. Bacteriophages are known to play a relevant role in the dissemination of antibiotic resistance genes (ARGs) in aquatic environments. In this study, five ARGs (blaTEM, blaCTX-M-1, blaCTX-M-9, sul1 and tetW) were monitored in phage particles isolated from seawater of two different locations: (i) the Mediterranean coast, subjected to high anthropogenic pressure, and (ii) the Antarctic coast, where the anthropogenic impact is low. Although found in lower quantities, ARG-containing phage particles were more prevalent among the Antarctic than the Mediterranean seawater samples and Antarctic bacterial communities were confirmed as their source. In the Mediterranean area, ARG-containing phages from anthropogenic fecal pollution might allow ARG transmission through the food chain. ARGs were detected in phage particles isolated from fish (Mediterranean, Atlantic, farmed, and frozen), the most abundant being β-lactamases. Some of these particles were infectious in cultures of the fecal bacteria Escherichia coli. By serving as ARG reservoirs in marine environments, including those with low human activity, such as the Antarctic, phages could contribute to ARG transmission between bacterial communities.
Keywords: bacteriophages, shellfish, fish, transduction, ARG, horizontal gene transfer
1. Introduction
Antibiotic resistance is one of the biggest threats to public health this century. The growing environmental presence of antibiotics and their derivative metabolites is mainly due to anthropogenic activities, such as medicine, agriculture, and industry. Antibiotics have been overused both in clinical settings and the livestock industry, including for prophylaxis, although their use as growth supplements has been prohibited in Europe since 2006 (EC Regulation No. 1831/2003) [1,2,3]. Epidemiological studies have verified that antibiotic consumption is directly connected with the emergence and dissemination of resistances [2].
Depending on their transport and persistence in soil or water, antibiotics can spread to surface water and groundwater, exerting a selective pressure on aquatic bacteria and the microbiota of aquatic animals, and leading to the emergence of resistant bacteria and the dissemination of antibiotic resistance genes (ARGs) [4,5].
A high demand for fish products has led to an increase in aquaculture in recent years [6]. However, these intensive fish production systems, with high concentrations of animals kept in limited areas of water, entail an increased risk of infectious diseases and consequently depend on the prophylactic and metaphylactic use of antimicrobials [6]. In addition to the antibiotic contamination of water bodies, which exerts a selective pressure on aquatic microbial communities, their use in the aquaculture industry causes the accumulation of antibiotic residues in fish and shellfish tissues, which in turn selects for resistances in the fish microbiota [6]. Although the presence of antibiotics, and consequently ARGs, in aquatic environments subjected to a high anthropogenic impact is well established [7], there is less information available about ARGs in areas with few human activities, such as polar aquatic environments, where ARGs have been detected [8,9].
Microorganisms have been producing natural antibiotics for billions of years to gain a selective advantage over competitors [2], and they simultaneously evolved mechanisms of antibiotic resistance [2]. As well as intrinsic resistance to antibiotics, innate to the species, bacteria may acquire resistance by mutation or by horizontal gene transfer (HGT) [3]. The most studied HGT mechanisms are conjugation and transformation, although in recent years, transduction, or gene transfer mediated by bacteriophages or phages, has attracted increasing attention as a means of ARG transmission in the environment [10,11].
Bacteriophages are the most abundant entities in the world (1031 phage particles have been estimated) and can be isolated from any habitat where bacteria exist [12]. The high persistence of phages in the environment enhances the efficiency of ARG dissemination in phage particles, which could, in turn, increase the chances of ARG transduction [13]. Phages can mobilize and spread ARGs by transduction, whether generalized, specialized, or lateral [14,15,16]. Phage particles carrying ARGs have been reported in different biomes, including sludge, soil, wastewater [10,11,15,17,18], marine viromes [19], and human and animal biomes [20,21,22]. Recent studies have reported the presence of ARG-carrying phage particles in vegetables and meat products classed as suitable for consumption, confirming that they can end up in food, and suggesting that their presence in feces is due to the fecal-oral route [23,24].
There is, however, no information about the occurrence of phage-associated ARGs in areas with very low human activity. The aim of this study was to assess ARG-carrying phage particles in two different environments: (i) Mediterranean coastal water, subjected to a high anthropogenic impact and (ii) Antarctic seawater, where the anthropogenic impact is low. Considering the negligible human influence in the Antarctic area, we also examined if phages induced from the bacterial fraction of marine communities could be confirmed as the source of the ARG-containing phage particles. Additionally, the impact of ARG-containing phage particles in Mediterranean seawater on the food chain was studied by analyzing their presence in marine fish, shellfish, and farmed fish.
2. Materials and Methods
2.1. Samples
Twenty seawater samples were collected from eleven different sampling sites (Med-1 to Med-11) off the Barcelona coast in February–October 2019 (Figure 1a). Ten seawater samples were collected at different sites off Livingston Island, the South Shetland Islands (LIV-1,2,3,4,5) near the Antarctic Spanish Base, and the Antarctic Peninsula (SW1, SW8, SW10, BISP and ROT) while on board the research vessel BIO Hespérides during the BLUEBIO-2 Antarctic Campaign (January–March 2019) within the Bluebio project (CTM2016-78901/ANT; Figure 1b). Antarctica seawater samples were not close to human bases or animal communities. Additionally, 50 samples of shellfish (n = 10 samples), Mediterranean fish (n = 10), Atlantic fish (n = 10), frozen fish (n = 10), and farmed fish (n = 10) were obtained in local supermarkets during 2018–2019. All the samples were collected in sterile containers, kept at 4 °C and analyzed within 24 h.
Figure 1.
Seawater sampling sites. (a) Barcelona coast (Mediterranean Sea). (b) Livingston Island (Antarctica). Stars represent the sampling sites in each area.
2.2. Phage Isolation from Seawater Samples
Five liters (L) of Antarctic seawater samples were filtered by 5 μm Swinnex® filter holders (Millipore, Merck, Darmstadt, Germany), followed by filtration through 3 μm mixed cellulose ester membranes (MCE) (Merck), and filtration through 0.2 μm Isopore™ polycarbonate membranes. Phage particles from the final filtrate were recovered by chemical flocculation [25]. An iron chloride solution (10 g/L) was used to flocculate the viral particles in the sample, which were then recovered by filtration through 1.2 µm Isopore™ membranes. Viral particles retained in the filters were resuspended using a 0.1M EDTA-0.2M MgCl2-0.2M ascorbic acid buffer to a final volume of 5 mL.
For Mediterranean seawater samples, 100 mL was filtered through 0.22 μm low protein binding polyethersulfone (PES) membranes. Viral particles in filtered samples were then concentrated with PEG8000 following previously reported protocols [18,26].
Lastly, the suspensions of phages of both origins were dialyzed and concentrated with Amicon® Ultra Centrifugal Filters to volumes of 0.5 mL. The concentrated phage suspensions of both origins were used for the phage DNA extraction.
2.3. Phage Isolation from Induced Antarctic Bacterial Communities
Antarctic seawater samples were processed within 48 h after collection for phage induction from the bacterial communities. One liter of each sample was filtered by 0.2 μm Isopore™ polycarbonate membranes (Merck) to retain the marine bacterial communities. In order to enrich the bacterial communities, filters were incubated in 7 mL of Difco™ Marine Broth at 18 °C for 48–72 h without shaking. Phages were induced from the possible lysogens present in these bacterial communities by treatment with mitomycin C (0.5 µg/mL) and incubation overnight at 20 °C in the dark without shaking [27]. After induction, phages were purified by filtration through 0.22 μm low protein binding PES membranes and concentrated with Amicon® Ultra 15 mL centrifugal filters as described above. The suspensions were stored at −20 °C until processed for phage DNA extraction.
2.4. Phage DNA Extraction
As in previous studies [23,24], filtered and concentrated phage suspensions were treated with chloroform (1:10 (v/v)) to rule out the possible presence of vesicles containing DNA, vigorously vortexed for 5 min and centrifuged at 16,000× g for 5 min. The aqueous phase was incubated with DNase I (100 units/mL) to eliminate any free DNA in the samples outside the phage particles. DNase was inactivated by heating for 5 min at 75 °C.
These suspensions were then used to extract DNA from the phage capsids. Before breaking the capsids, aliquots were taken to confirm DNA removal by the DNase treatment and complete inactivation of DNase by heat treatment.
Viral capsids were broken using proteinase K (0.5 µg/mL) in 250 μL of proteinase K buffer and then incubated for 1 h at 55 °C [26]. The encapsidated DNA was extracted by phenol-chloroform (1:1) (v/v) and chloroform treatment. DNA was precipitated using 100% ethanol and 3 M sodium acetate and resuspended in 50 µL of ultrapure sterile water. DNA was quantified using a Nanodrop ND-1000 spectrophotometer.
2.5. Amplification of ARGs in the DNA Extracted from Phage Particles
Quantitative real-time PCR (qPCR) with TaqMan hydrolysis probes was performed using the StepOne Real Time PCR System in a 20 μL reaction mixture with the TaqMan® Environmental Master Mix 2.0 (Applied Biosystems, Foster City, CA, USA). The reaction contained 9 μL of the sample DNA or standards with known DNA concentration. The results were analyzed with the Applied Biosystems StepOne™ Instrument program.
A total of 11 ARGs with a range of mechanisms and clinical relevance were evaluated. Five genes conferred resistance to β-lactam antibiotics (blaTEM, blaCTX-M-1 group, blaCTX-M-9 group, blaOXA-48 and blaVIM), two quinolone resistance genes (qnrA and qnrS), mecA, a gene commonly found in Staphylococcus that confers resistance to methicillin [15], sul1, which confers resistance to sulfonamides and is frequently found in environmental and clinical bacterial populations [28], tetW, which confers resistance to tetracycline and is commonly found and used in aquaculture environments [29], and armA, which encodes aminoglycoside resistance and is widely distributed in Enterobacteriaceae [30]. Mediterranean and Antarctic seawater samples were tested for the presence of five ARGs only (blaTEM, blaCTX-M-1 group, blaCTX-M-9 group, tetW and sul1), and the fish samples were analyzed for the eleven ARGs.
For quantification, serial dilutions of gBlocks™ Gene Fragments (Integrated DNA Technologies, Coralville, IA, USA) of known concentration were used to generate the standard curves. All samples were run in triplicate (including the standards and negative controls). The number of gene copies (GC) was defined as the mean of the triplicate data obtained. To quantify the ARGs, we considered the results obtained within the threshold cycle and within the limit of quantification. This was determined by the last valid threshold cycle for each ARG assay (GC/μL are shown in Table S1) in the standard curve that is consistent in the diverse replicates.
To confirm that only the ARGs from the phage particles were evaluated, several controls were performed as previously described [24]. Briefly, an aliquot of each sample after DNase treatment and before desencapsidation was included in the qPCR in parallel with the DNA sample to confirm the absence of each ARG. Additionally, the controls were used to confirm the absence of bacterial 16S rDNA, which was verified by qPCR using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) and primers 338F/518R (Table S1).
2.6. Bacterial and Viral Indicators
Microbial indicators were evaluated in fish samples and Mediterranean seawater. To isolate bacteria and somatic phages from fish and shellfish samples, 20 g of each sample was homogenized for 2 min in 60 mL of phage buffer with the Stomacher homogenizer (IUL Instruments GmbH, Königswinter, Germany). Stomacher bags with filters (Afora, Barcelona, Spain) were used to enhance the separation of solid waste from the liquid fraction containing the microorganisms. For Mediterranean seawater, 100 mL of each sample was filtered through 0.45 µm mixed cellulose ester (MCE) EZ-Pak® filters (Merck), which were used to analyze the bacterial indicators. Somatic coliphages were analyzed as indicated below.
Total aerobic microorganisms resistant to ampicillin (AmpR) were evaluated on Tryptone Soy Agar (TSA) with Amp (100 μg/mL) at 37 °C. Total Escherichia coli and E. coli resistant to Amp were determined on Chromocult® Coliform Agar or Chromocult® Coliform Agar with Amp (100 μg/mL), respectively. Incubation was first performed for 2 h at 37 °C to accommodate potentially damaged microorganisms and then overnight at 44 °C. Presumed E. coli colonies were confirmed in MacConkey agar. Total Enterococcus were enumerated by membrane filtration, according to the ISO standard method [31]. Filters were incubated for 48 h at 37 °C in Difco™ Enterococcus agar (BD). Suspected Enterococcus colonies were confirmed by growing them in bile esculin agar for 3 h at 44 °C.
Somatic coliphages were evaluated as fecal viral indicators in the samples [32]. The homogenates of fish and shellfish samples (10 mL each) were centrifuged for 15 min at 4000× g and the supernatant was filtered through 0.22 μm low protein binding PES membranes (Merck). Seawater samples (100 mL) were filtered through 0.22 μm low protein binding PES membranes (Merck) and concentrated with Amicon® Ultra 15 mL centrifugal filters (Merck). The filtrates were analyzed in duplicate for the presence of somatic coliphages according to the ISO standard method [33], which uses E. coli strain WG5 (ATCC 700078) as the bacterial host, and incubated at 37 °C for 18 h.
Each sample was analyzed for all the indicators in duplicate.
2.7. Propagation Cultures for Phages in Fish and Shellfish Samples
The ability of phage particles in the fish and shellfish samples to infect and propagate in enrichment cultures of the E. coli WG5 host strain was evaluated. This strain, used for the somatic coliphage count, was selected for its sensitivity to phage infection [23] and because it does not contain any of the ARGs targeted in this study or any prophage [34].
The propagation cultures were prepared with 1 mL of each phage suspension after the DNAse treatment and 1 mL of E. coli WG5 at the exponential phase (OD600 0.3) in 8 mL of Luria-Bertrani broth (LB) and incubated overnight at 37 °C with shaking. After incubation, phages were purified by filtration through 0.22 μm low protein binding PES membranes and treated with chloroform and DNase, as indicated above.
The infectivity of the phage particles carrying ARGs was evaluated using qPCR by comparing ARG abundance in packaged DNA before (direct quantification from the sample) and after phage propagation in the enrichment cultures. If the GC number of the ARGs in the phage particles increased after propagation, it was considered that the phage particles in this sample were able to infect and propagate in the host strain. Otherwise, if ARG levels did not vary or decreased, the particles in this sample were considered incapable of propagating.
3. Results
3.1. Comparison of ARGs in the Phage DNA Fraction from Mediterranean and Antarctic Seawater Samples
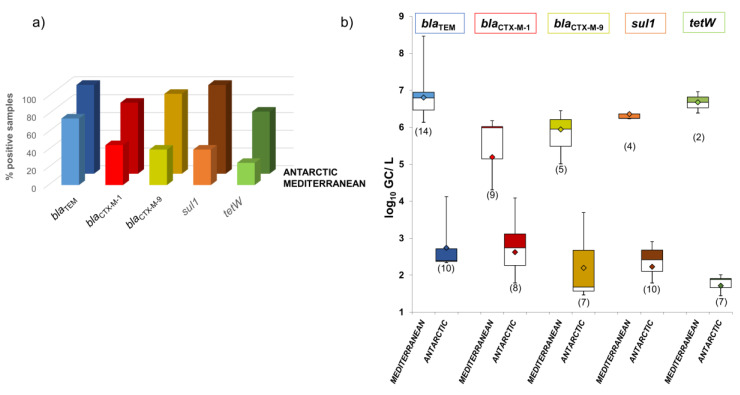
ARGs were detected in the phage DNA fraction isolated from Mediterranean and Antarctic seawater (Figure 2). For the five ARGs studied in both types of seawater, 25–60% fewer positive samples were obtained from the Mediterranean than the Antarctic (Figure 2a). blaTEM was the most prevalent ARG in both sites, found in 100% of Antarctic and 75% of Mediterranean samples (Figure 2a). sul1 was also found in 100% of Antarctic samples, but its prevalence in Mediterranean waters was much lower (40%). The second most prevalent ARG in the Mediterranean was blaCTX-M-1 (45%) but much less than in the Antarctic (80%) (Figure 2a).
Figure 2.
Antibiotic resistance genes (ARGs) in phage particles in Mediterranean and Antarctic seawater samples. (a) Percentage of positive samples for each ARG. (b) Abundance of each ARG in seawater (log10GC/L). Data are represented in boxplots; the diamond represents the average value of the positive samples, the upper squares include samples whose values are within the 75th percentile, and those in the lower white boxes are within the 25th percentile.
In contrast with the prevalence data, the abundance of ARGs (GC/L) in the Antarctic samples was 3–5 log10 units lower than in the Mediterranean samples (Figure 2b). blaTEM and tetW were the most abundant ARGs found in the Mediterranean, with average values close to 7 log10 GC/L, while blaTEM and blaCTX-M-1 were the most abundant in the Antarctic, with average values close to 3 log10 GC/L.
3.2. ARGs in Phage Particles Induced from Antarctic Bacterial Communities
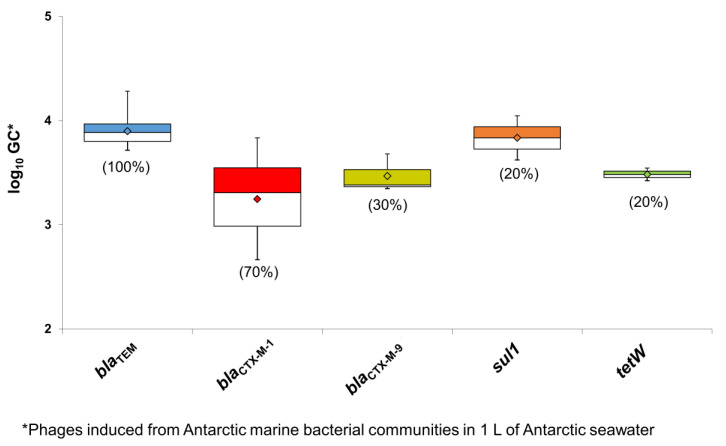
As mentioned, one of the aims of the study was to explore the origin of the ARG-containing phage particles found in the Antarctic area. We hypothesized that they could be produced by the Antarctic marine bacterial communities as part of an HGT mechanism. To test this hypothesis, the bacterial fraction of the samples was separated and treated with an inducing agent to force the production of phage particles, including those carrying ARGs. The results showed that particles carrying blaTEM were produced in 100% of the lysates, and particles carrying blaCTX-M-1 group were produced in 75% (Figure 3). Other ARG-containing particles were produced in a lower percentage but were still detectable.
Figure 3.
Abundance of ARGs in the phage DNA isolated from phage particles induced from Antarctic bacterial communities present in one liter of Antarctic seawater (log10 GC/L). Data are represented in boxplots; the diamond represents the average value of the positive samples, the upper squares include samples whose values are within the 75th percentile, and those in the lower white boxes are within the 25th percentile.
The Antarctic ARG counts were calculated from phage particles induced from marine bacteria isolated from 1 L of seawater. Nevertheless, this abundance should be treated with caution, as the cultivation of the bacterial communities using marine broth selected the bacteria able to grow in such conditions. In addition, different phage particles may be induced at different rates. Finally, the integrity of phage particles could have been compromised during their storage from the end of the sampling campaign until phage DNA extraction in the laboratory. Despite these considerations, all ARGs in the induced phages were detected with a similar abundance. These results point to the marine bacterial communities as a plausible source of the ARG-carrying phage particles found in the Antarctic seawater.
3.3. Consequences of Anthropogenic Pollution in Mediterranean Environments
The higher abundance of ARG-carrying phage particles in the Mediterranean could be attributed to the anthropogenic impact as well as production by the bacterial communities. These particles could be taken up by aquatic fauna and mediate the transfer of ARGs through the food chain. To evaluate the consequences of the ARG-carrying phage particles in Mediterranean coastal waters, a set of samples of marine fish, shellfish, and farmed fish from the area were analyzed.
The microbial indicators confirmed the influence of anthropogenic pollution in seawater and in fish samples. Aerobic bacteria resistant to ampicillin were detected in all the matrices analyzed (Table 1). Predictably, the highest positive values of E. coli were obtained in Mediterranean seawater samples, which contained ampicillin-resistant E. coli. The similarity of the average values of E. coli and ampicillin-resistant E. coli in the Mediterranean samples suggests that most of the E. coli isolates were ampicillin-resistant. Other fecal indicators, Enterococcus spp., were also present in all the samples studied (Table 1), their abundance in Mediterranean seawater and fish being similar to E. coli counts. Somatic coliphages, used as viral indicators of fecal pollution [35], were detected in all samples except in Mediterranean fish.
Table 1.
Bacterial (CFU/25 g) and viral (PFU/25 g) indicators in Mediterranean seawater, fish and shellfish samples. -; only one positive sample, no SD.
| Microorganism | Mediterranean Seawater * | Fish | |||||
|---|---|---|---|---|---|---|---|
| Shellfish | Mediterranean | Atlantic | Farm | Frozen | |||
| n | 20 | 10 | 10 | 10 | 10 | 10 | |
| Total aerobic bacteria AmpR | % positive samples | 100 | 90 | 80 | 70 | 100 | 40 |
| Average | 2.4 × 103 | 4.6 × 107 | 5.1 × 104 | 5.7 × 106 | 3.8 × 106 | 1.0 × 108 | |
| SD | 5.6 × 103 | 4.9 × 107 | 5.9 × 104 | 9.4 × 106 | 8.3 × 106 | 1.2 × 108 | |
| E. coli | % positive samples | 85 | 0 | 10 | 10 | 0 | 0 |
| Average | 6.9 × 102 | 0 | 3.7 × 101 | 7.5 × 101 | 0 | 0 | |
| SD | 2.1 × 103 | 0 | − | − | 0 | 0 | |
| E. coli AmpR | % positive samples | 60 | 0 | 0 | 0 | 0 | 0 |
| Average | 6.8 × 102 | 0 | 0 | 0 | 0 | 0 | |
| SD | 1.9 × 103 | 0 | 0 | 0 | 0 | 0 | |
| Enterococcus spp. | % positive samples | 90 | 30 | 10 | 40 | 20 | 10 |
| Average | 1.0 × 102 | 1.4 × 102 | 7.5 × 101 | 7.3 × 102 | 1.9 × 102 | 1.1 × 102 | |
| SD | 1.5 × 102 | 1.2 × 102 | − | 1.2 × 103 | 5.3 × 101 | − | |
| Somatic coliphages | % positive samples | 40 | 70 | 0 | 30 | 10 | 20 |
| Average | 1.0 × 103 | 1.1 × 103 | 0 | 9.2 × 101 | 7.5 × 101 | 6.6 × 102 | |
| SD | 1.3 × 103 | 9.9 × 102 | 0 | 5.2 × 101 | - | 8.2 × 102 | |
* CFU or PFU/L.
3.4. Prevalence of ARGs in Phage DNA Fraction Isolated from Shellfish and Fish Samples
ARGs in DNA extracted from phage particles isolated from shellfish and fish were quantified by qPCR. Samples were considered positive when the amplification values of the target gene were within the limit of quantification, previously defined with the standard curves. Those samples outside the limit of quantification but within the limit of detection and those that did not show any increase in fluorescence in the qPCR cycles (undetermined samples) were excluded from the ARG prevalence study.
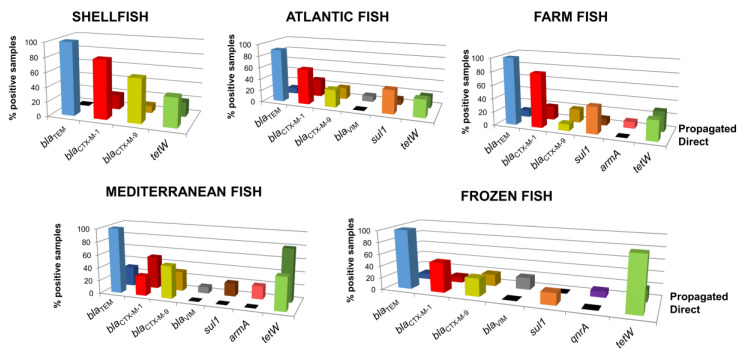
All samples were analyzed for ARGs, either directly or after propagation in E. coli cultures, with 10–100% of samples testing positive (Figure 4). An increase in ARG count in the DNA after the enrichment step revealed that the phage particles were able to infect a fecal strain and propagate, and consequently a fraction of the particles in these samples were infectious. On the contrary, in samples without an increase in GC numbers or no detection of ARGs (not shown in Figure 4), the isolated phage particles were likely non-infectious or the propagation strain was not a suitable host.
Figure 4.
Percentage of positive samples for each ARG in each fish matrix and shellfish analyzed directly from the samples (light color bars) and after propagation in the E. coli enrichment culture (dark color bars).
Although the different matrices gave heterogeneous results, β-lactamase genes and tetW were the most detected ARGs in all the samples. Among the β-lactamase genes, blaTEM was the most prevalent, found in 90–100% of samples, followed by blaCTX-M-1 and blaCTX-M-9, which were identified in 10–80% of the samples. blaVIM was only detected in Atlantic, Mediterranean and frozen fish after propagation. Using direct analysis, tetW was detected in 3–90% of the samples, with the highest prevalence in frozen fish. sul1 was determined in 20–40% of Atlantic, farmed and frozen fish samples by direct analysis and in Mediterranean fish after propagation.
The matrices with the widest range of ARGs were Mediterranean fish and frozen fish (seven in each). After the enrichment step, five ARGs were detected in an increased number of samples of Mediterranean fish, three of them (blaVIM, sul1 and armA) only after propagation (Figure 4). The only matrix in which qnrA was detected was frozen fish, with one positive sample after propagation, which also tested positive for blaTEM, blaCTX-M-1, blaCTX-M-9, blaVIM and tetW. armA was only detected after propagation in two Mediterranean fish samples and in one farmed fish sample. Finally, mecA, qnrS, and blaOXA-48 were not detected in any samples.
3.5. Abundance of ARGs in the Phage DNA Fraction Isolated from Shellfish and Fish Samples
ARG abundance in shellfish and fish was evaluated only by direct analysis of the samples (Figure 5). The abundance of ARGs in phage particles obtained after propagation studies with an enrichment step cannot be considered as an absolute value and was therefore not included in the abundance study.
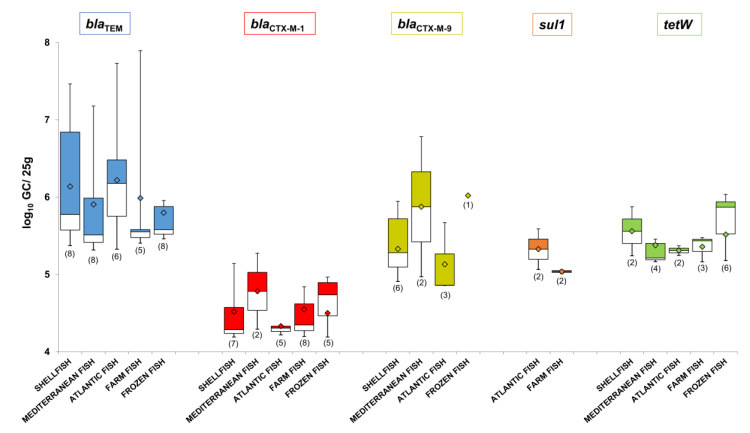
Figure 5.
Abundance of ARGs in the DNA isolated from the phage particles of each fish matrix and shellfish (log10 GC/25 g) grouped by ARG. Data is represented in boxplots; the diamond represents the average value of the positive samples, the upper squares include samples whose values are within the 75th percentile, and those in the lower white boxes are within the 25th percentile.
The most prevalent gene, blaTEM, was also the most abundant, with average values higher than 6 log10 GC/25 g of sample (Figure 5) and the highest value (7.9 log10 GC/25 g) observed in a farmed fish sample. In contrast, despite being one of the most prevalent ARGs, blaCTX-M-1, was the least abundant, detected at less than 5 log10 GC/25 g in most of the samples tested. The second most abundant ARG was blaCTX-M-9, particularly in Mediterranean and frozen fish, with average values close to 6 log10 GC/25 g. A similar level of abundance was found for the prevalent gene tetW, with the highest value of 6 log10 GC/25 g obtained in frozen fish samples, 90% of which were positive for this ARG. The sul1 gene was detected in both Atlantic and farmed fish at average values higher than 5 log10 GC/25 g.
Comparing the abundance of all ARGs in the phage fraction of each type of fish reveals higher values in shellfish, Mediterranean, and Atlantic fish than in farmed and frozen fish, with some exceptions (Figure S1).
4. Discussion
Antibiotic resistance is as ancient as some of the earliest bacterial communities, but little is known about the presence and persistence of antibiotics in the environment before they were harnessed for the benefit of humanity [36]. Seawater from the Antarctic, an area with a presumably limited anthropogenic influence, was expected to have a low prevalence of ARGs, but surprisingly, it was found to be higher than in Mediterranean samples. An explanation for these results could be the difference in sample volumes analyzed. Following reported protocols for environmental samples [18,25], larger volumes of seawater were collected and processed from the Antarctic to compensate for the expected limited ARG presence. We are aware that the volume affects the limit of detection, and the use of higher volumes of Mediterranean seawater could have led to more positive samples. Nevertheless, when a Mediterranean sample was positive, the abundance of quantified ARGs was much greater than in the Antarctic samples, being around 4 log units more with 50 times less volume, suggesting that ARGs in the Mediterranean samples are unlikely to have been underestimated. Independently of the number of positive samples, the presence of the same ARGs in both ecosystems is striking. In natural ecosystems, the production of antibiotics, innate in many bacterial communities, is a protective mechanism against competitors. In 2011, a study demonstrated the presence of ARGs in bacterial populations isolated from permafrost sediments, supporting that ARGs pre-date the human use of antibiotics and that antibiotic resistance occurs naturally in nature [37].
The most prevalent ARGs in Antarctic seawater were β-lactamase genes and sul1, in agreement with previous reports of β-lactamase genes (especially from CTX-M groups) in remote polar areas [38]. There was a notable presence of sul1, which expresses resistance to synthetic or semi-synthetic antibiotics such as sulphonamides. A recent study detected resistance against these antibiotics in bacteria recovered from freshwater samples collected close to Antarctic research stations [39], which was attributed to poor waste water treatment. Nevertheless, sul1 is not necessarily an indicator of human activity, as it is usually present in environmental bacteria from soils and waters, even in the Antarctic [40]. Furthermore, sul1 is associated with an integron, which could explain its wide dissemination even in the absence of selective pressure by antibiotics [41].
Despite the samples analyzed from Antarctica were not close to human bases, the growing human activity in the Antarctic area, including scientific expeditions, military stations, and tourism [42], could be responsible for disseminating human-origin antibiotic resistance [38]. Another contributing factor may be migrating wildlife, which by connecting anthropogenic and remote areas could act as biological vectors spreading antibiotic resistant bacteria and ARGs to the Antarctic [36].
The greater abundance of ARGs in the Mediterranean samples can be explained by a high antibiotic selective pressure derived from human activity in the Mediterranean coast and the impact of human fecal pollution. These results are in line with the relative abundance of ARGs recently reported by Calero-Cáceres et al., [19] in the viromes of samples from diverse marine habitats. Specifically, the diversity and prevalence of phage ARGs in Indian Ocean samples, like the Mediterranean samples studied here, were relatively low, yet the abundance of certain genes was high, particularly those conferring resistance to β-lactams (especially blaTEM) and tetracyclines. Although a clear association has not yet been established, these findings shed light on the selective pressure generated by humans in natural environments, where bacteria can take advantage of phages to spread their genetic content in difficult conditions.
The ARGs detected in the phage DNA fraction isolated from lysogens of Antarctic bacteria allow us to suggest that these bacterial communities are the source of the ARGs in the phage particles. The particles would be formed after the packaging of bacterial DNA (including ARGs) by prophages inserted in the bacterial chromosome. Several studies reveal that temperate phages are widely distributed in Antarctic bacterial communities and prioritize lysogeny when bacterial concentration is low [43].
Our results therefore confirm the existence of a diversity of ARGs in an area as pristine as the Antarctic, and that bacteriophages in marine environments represent a potential vehicle for the horizontal transmission of ARGs between bacterial communities. The role of phages as HGT elements in marine environments has already been demonstrated, as they can disseminate key genes involved in oxygenic photosynthesis in cyanobacteria, which allows various bacterial communities to adapt to surface waters [44]. The demonstrated ability of phages to interact with their hosts has evolutionary and ecological implications.
On the other hand, the high abundance of phage-associated ARGs found in Mediterranean seawater may have important consequences for fish and shellfish, as well as consumers of fishery products. According to our results, fish microbiota showed a high percentage of resistance to β-lactam antibiotics, which represents both a direct health risk to consumers as well as a general health hazard if this resistance is transferred to human microbiota. These results are in accordance with a recent study in which ARGs were detected in wild fish, the most prevalent being blaTEM [45].
The most commonly used antibiotics in aquaculture are fluoroquinolones, sulfonamides, and tetracyclines [29]. The prevalence of tetW in our samples may reflect the overuse of tetracyclines in aquaculture and their spread in diverse environments [46]. Although the sulfonamide resistance gene sul1 has been reported as one of the most prevalent ARGs in phage DNA of environmental and food samples [7,23,24], in the current study it was detected only in a few samples of fish, being more prevalent in farmed fish, in accordance with the use of sulfonamides in aquaculture [47].
European food safety regulations establish that foodstuffs should not contain microorganisms in quantities representing a risk for human health [48], The samples analyzed in the current study were all suitable for consumption according to the microbiological parameters evaluated. In recreational coastal water, the European normative establishes that 500 CFU/100 mL of E. coli and 185 CFU/100 mL of intestinal enterococci in a sample indicate sufficient quality for consumption [49]. However, the detection of phages, including those capable of transferring ARGs to the natural microbiota and pathogens, is not contemplated in food safety practices. There is no legislation in place to control the spread of antibiotic resistance, ARGs, or the genetic elements that mobilize them.
5. Conclusions
This study confirms the presence of ARG-containing phage particles in aquatic environments subjected to a low or high anthropogenic influence, demonstrating that seawater is a reservoir of ARGs and that phages could play an important role in their transmission. In addition, human activity has a clear impact on the evolution of antibiotic resistance, the strongly anthropogenic Mediterranean area showing a higher abundance of ARGs than the relatively pristine Antarctic seawater. This high abundance of ARG-containing phage particles may have an important impact on fish and shellfish microbiota, contributing to the dissemination of those ARGs into microbial communities relevant to human health.
Acknowledgments
We thank the “Unidad de Tecnología Marina” (UTM) in the Antarctic Spanish Station in Livingston Island, and the ‘BIO-Hespérides’ crew for their logistic support.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/9/1293/s1, Figure S1: Abundance of ARGs in the DNA isolated from phage particles in fish and shellfish grouped by fish matrix. Table S1: Oligonucleotides used in this study [50,51,52,53,54].
Author Contributions
M.M., L.R.-R. and E.B. contributed to the conception and design of the study; P.B.-P., G.R., E.B., D.T.-A., C.G.-G. performed experiments and data collection; P.B.-P., G.R., E.B., M.M., L.R.-R., analyzed data; P.B.-P., M.M., L.R.-R. prepared the first draft; M.M., L.R.-R., E.B., C.A. reviewed and edited the final version. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Spanish Ministerio de Innovación y Ciencia (AGL2016-75536-P) and the Bluebio project (CTM2016-78901/ANT), the Agencia Estatal de Investigación (AEI), the European regional fund (ERF), the Generalitat de Catalunya (2017SGR170) and the Centre de Referència en Biotecnologia (XeRBa). P.B.-P. has a grant from the Spanish Ministry of Economy, Industry and Competitiveness (BES-2017-081296). L.R-R. is a Serra Húnter Fellow.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.World Health Organization Antibiotic Resistance. [(accessed on 8 January 2019)]; Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance.
- 2.Ventola C.L. Antibiotic Resistance Crisis. Int. J. Med. Dev. Ctries. 2019;40:561–564. doi: 10.24911/ijmdc.51-1549060699. [DOI] [Google Scholar]
- 3.García J., García-Galán M.J., Day J.W., Boopathy R., White J.R., Wallace S., Hunter R.G. A review of emerging organic contaminants (EOCs), antibiotic resistant bacteria (ARB), and antibiotic resistance genes (ARGs) in the environment: Increasing removal with wetlands and reducing environmental impacts. Bioresour. Technol. 2020;307:123228. doi: 10.1016/j.biortech.2020.123228. [DOI] [PubMed] [Google Scholar]
- 4.Kraemer S.A., Ramachandran A., Perron G.G. Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms. 2019;7:180. doi: 10.3390/microorganisms7060180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho I.T., Santos L. Antibiotics in the aquatic environments: A review of the European scenario. Environ. Int. 2016;94:736–757. doi: 10.1016/j.envint.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 6.Santos L., Ramos F. Antimicrobial resistance in aquaculture: Current knowledge and alternatives to tackle the problem. Int. J. Antimicrob. Agents. 2018;52:135–143. doi: 10.1016/j.ijantimicag.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Calero-Cáceres W., Méndez J., Martín-Díaz J., Muniesa M. The occurrence of antibiotic resistance genes in a Mediterranean river and their persistence in the riverbed sediment. Environ. Pollut. 2017;223 doi: 10.1016/j.envpol.2017.01.035. [DOI] [PubMed] [Google Scholar]
- 8.Miller R.V., Gammon K., Day M.J. Antibiotic resistance among bacteria isolated from seawater and penguin fecal samples collected near Palmer Station, Antarctica. Can. J. Microbiol. 2009;55:37–45. doi: 10.1139/W08-119. [DOI] [PubMed] [Google Scholar]
- 9.Tan L., Li L., Ashbolt N., Wang X., Cui Y., Zhu X., Xu Y., Yang Y., Mao D., Luo Y. Arctic antibiotic resistance gene contamination, a result of anthropogenic activities and natural origin. Sci. Total Environ. 2018;621:1176–1184. doi: 10.1016/j.scitotenv.2017.10.110. [DOI] [PubMed] [Google Scholar]
- 10.Balcazar J.L. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog. 2014;10:e1004219. doi: 10.1371/journal.ppat.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lekunberri I., Subirats J., Borrego C.M., Balcázar J.L. Exploring the contribution of bacteriophages to antibiotic resistance. Environ. Pollut. 2017;220:981–984. doi: 10.1016/j.envpol.2016.11.059. [DOI] [PubMed] [Google Scholar]
- 12.Keen E.C. A century of phage research: Bacteriophages and the shaping of modern biology. BioEssays. 2015;37:6–9. doi: 10.1002/bies.201400152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calero-Cáceres W., Muniesa M. Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater. Water Res. 2016;95:11–18. doi: 10.1016/j.watres.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Chen J., Quiles-Puchalt N., Chiang Y.N., Bacigalupe R., Fillol-Salom A., Chee M.S.J., Fitzgerald J.R., Penadés J.R. Genome hypermobility by lateral transduction. Science. 2018;362:207–212. doi: 10.1126/science.aat5867. [DOI] [PubMed] [Google Scholar]
- 15.Colomer-Lluch M., Jofre J., Muniesa M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS ONE. 2011;6:e17549. doi: 10.1371/journal.pone.0017549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shousha A., Awaiwanont N., Sofka D., Smulders F.J.M., Paulsen P., Szostak M.P., Humphrey T., Hilbert F. Bacteriophages Isolated from Chicken Meat and the Horizontal Transfer of Antimicrobial Resistance Genes. Appl. Environ. Microbiol. 2015;81:4600–4606. doi: 10.1128/AEM.00872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calero-Cáceres W., Melgarejo A., Colomer-Lluch M., Stoll C., Lucena F., Jofre J., Muniesa M. Sludge as a potential important source of antibiotic resistance genes in both the bacterial and bacteriophage fractions. Environ. Sci. Technol. 2014;48 doi: 10.1021/es501851s. [DOI] [PubMed] [Google Scholar]
- 18.Colomer-Lluch M., Calero-Cáceres W., Jebri S., Hmaied F., Muniesa M., Jofre J. Antibiotic resistance genes in bacterial and bacteriophage fractions of Tunisian and Spanish wastewaters as markers to compare the antibiotic resistance patterns in each population. Environ. Int. 2014;73:167–175. doi: 10.1016/j.envint.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Calero-Cáceres W., Ye M., Balcázar J.L. Bacteriophages as Environmental Reservoirs of Antibiotic Resistance. Trends Microbiol. 2019;27:570–577. doi: 10.1016/j.tim.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Brown-Jaque M., Calero-Cáceres W., Espinal P., Rodríguez-Navarro J., Miró E., González-López J.J., Cornejo T., Hurtado J.C., Navarro F., Muniesa M. Antibiotic resistance genes in phage particles isolated from human feces and induced from clinical bacterial isolates. Int. J. Antimicrob. Agents. 2018;51:434–442. doi: 10.1016/j.ijantimicag.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Brown-Jaque M., Rodriguez Oyarzun L., Cornejo-Sánchez T., Martín-Gómez M.T., Gartner S., de Gracia J., Rovira S., Alvarez A., Jofre J., González-López J.J., et al. Detection of Bacteriophage Particles Containing Antibiotic Resistance Genes in the Sputum of Cystic Fibrosis Patients. Front. Microbiol. 2018;9:856. doi: 10.3389/fmicb.2018.00856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colomer-Lluch M., Imamovic L., Jofre J., Muniesa M. Bacteriophages carrying antibiotic resistance genes in fecal waste from cattle, pigs, and poultry. Antimicrob. Agents Chemother. 2011;55:4908–4911. doi: 10.1128/AAC.00535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larrañaga O., Brown-Jaque M., Quirós P., Gómez-Gómez C., Blanch A.R., Rodríguez-Rubio L., Muniesa M. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil. Environ. Int. 2018;115:133–141. doi: 10.1016/j.envint.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Gómez-Gómez C., Blanco-Picazo P., Brown-Jaque M., Quirós P., Rodríguez-Rubio L., Cerdà-Cuellar M., Muniesa M. Infectious phage particles packaging antibiotic resistance genes found in meat products and chicken feces. Sci. Rep. 2019;9:13281. doi: 10.1038/s41598-019-49898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poulos B.T., John S.G., Sullivan M.B. Iron chloride flocculation of bacteriophages from seawater. Methods Mol. Biol. 2018;1681:49–57. doi: 10.1007/978-1-4939-7343-9_4. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2001. p. 999. [Google Scholar]
- 27.Clokie M.R.J., Kropinski A.M. Bacteriophages Methods and Protocols Volume 2: Molecular and Applied Aspects. Volume 502. Springer; Berlin, Germany: 2009. [Google Scholar]
- 28.Pruden A., Arabi M., Storteboom H.N. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 2012;46:11541–11549. doi: 10.1021/es302657r. [DOI] [PubMed] [Google Scholar]
- 29.Pereira A.M.P.T., Silva L.J.G., Meisel L.M., Pena A. Fluoroquinolones and Tetracycline Antibiotics in a Portuguese Aquaculture System and Aquatic Surroundings: Occurrence and Environmental Impact. J. Toxicol. Environ. Health Part A Curr. Issues. 2015;78:959–975. doi: 10.1080/15287394.2015.1036185. [DOI] [PubMed] [Google Scholar]
- 30.Galimand M., Sabtcheva S., Lambert T. Worldwide Disseminated armA Aminoglycoside Resistance Methylase Gene Is Borne by Composite Transposon Tn1548. Am. Soc. Microbiol. 2005;49:2949–2953. doi: 10.1128/AAC.49.7.2949-2953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ISO . Water Quality—Detection and Enumeration of Intestinal Enterococci—Part 2: Membrane Filtration Method. ISO; London, UK: 2000. ISO 7899-2:2000. [Google Scholar]
- 32.Jofre J. Perspectives in Medical Virology, Volume 17. Elsevier; Amsterdam, The Netherlands: 2007. Indicators of Waterborne Enteric Viruses; pp. 227–249. [Google Scholar]
- 33.ISO . Water Quality. Detection and Enumeration of Bacteriophages—Part 2: Enumeration of Somatic Coliphages. ISO; London, UK: 2000. ISO 10705-2. [Google Scholar]
- 34.Imamovic L., Misiakou M.-A., van der Helm E., Panagiotou G., Muniesa M., Sommer M.O.A. Complete Genome Sequence of Escherichia coli Strain WG5. Genome Announc. 2018;6 doi: 10.1128/genomeA.01403-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jofre J., Lucena F., Blanch A., Muniesa M. Coliphages as Model Organisms in the Characterization and Management of Water Resources. Water. 2016;8:199. doi: 10.3390/w8050199. [DOI] [Google Scholar]
- 36.Allen H.K., Donato J., Wang H.H., Cloud-Hansen K.A., Davies J., Handelsman J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 37.D’Costa V.M., King C.E., Kalan L., Morar M., Sung W.W.L., Schwarz C., Froese D., Zazula G., Calmels F., Debruyne R., et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 38.Hernández J., Stedt J., Bonnedahl J., Molin Y., Drobni M., Calisto-Ulloa N., Gomez-Fuentes C., Astorga-España M.S., González-Acuña D., Waldenström J., et al. Human-associated extended-spectrum β-lactamase in the Antarctic. Appl. Environ. Microbiol. 2012;78:2056–2058. doi: 10.1128/AEM.07320-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jara D., Bello-Toledo H., Domínguez M., Cigarroa C., Fernández P., Vergara L., Quezada-Aguiluz M., Opazo-Capurro A., Lima C.A., González-Rocha G. Antibiotic resistance in bacterial isolates from freshwater samples in Fildes Peninsula, King George Island, Antarctica. Sci. Rep. 2020;10:3145. doi: 10.1038/s41598-020-60035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Na G., Wang C., Gao H., Li R., Jin S., Zhang W., Zong H. The occurrence of sulfonamide and quinolone resistance genes at the Fildes Peninsula in Antarctica. Mar. Pollut. Bull. 2019;149 doi: 10.1016/j.marpolbul.2019.110503. [DOI] [PubMed] [Google Scholar]
- 41.Jiang H., Cheng H., Liang Y., Yu S., Yu T., Fang J., Zhu C. Diverse Mobile Genetic Elements and Conjugal Transferability of Sulfonamide Resistance Genes (sul1, sul2, and sul3) in Escherichia coli Isolates From Penaeus vannamei and Pork From Large Markets in Zhejiang, China. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tin T., Fleming Z.L., Hughes K.A., Ainley D.G., Convey P., Moreno C.A., Pfeiffer S., Scott J., Snape I. Impacts of local human activities on the Antarctic environment. Antarct. Sci. 2009;21:3–33. doi: 10.1017/S0954102009001722. [DOI] [Google Scholar]
- 43.Brum J.R., Hurwitz B.L., Schofield O., Ducklow H.W., Sullivan M.B. Seasonal time bombs: Dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 2016;10:437–449. doi: 10.1038/ismej.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindell D., Sullivan M.B., Johnson Z.I., Tolonen A.C., Rohwer F., Chisholm S.W. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. USA. 2004;101:11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marti E., Variatza E., Balcázar J.L. Bacteriophages as a reservoir of extended-spectrum β-lactamase and fluoroquinolone resistance genes in the environment. Clin. Microbiol. Infect. 2014;20:O456–O459. doi: 10.1111/1469-0691.12446. [DOI] [PubMed] [Google Scholar]
- 46.Tamminen M., Karkman A., Lõhmus A., Muziasari W.I., Takasu H., Wada S., Suzuki S., Virta M. Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ. Sci. Technol. 2011;45:386–391. doi: 10.1021/es102725n. [DOI] [PubMed] [Google Scholar]
- 47.Gao P., Mao D., Luo Y., Wang L., Xu B., Xu L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 2012;46:2355–2364. doi: 10.1016/j.watres.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Anonymous Commission Regulations (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. [(accessed on 23 August 2020)];Off. J. Eur. Union. 2005 :L338/1. Available online: https://eur-lex.europa.eu/eli/reg/2005/2073/oj. [Google Scholar]
- 49.Anonymous Directive 2006/7/EC of the European Parliament and of the Council of 15 February 2006 Concerning the Management of Bathing Water Quality. [(accessed on 23 August 2020)];Off. J. Eur. Union. 2006 L64/37 Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32006L0007&rid=1. [Google Scholar]
- 50.Lachmayr K.L., Kerkhof L.J., Dirienzo A.G., Cavanaugh C.M., Ford T.E. Quantifying nonspecific TEM beta-lactamase (blaTEM) genes in a wastewater stream. Appl. Environ. Microbiol. 2009;75:203–211. doi: 10.1128/AEM.01254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volkmann H., Schwartz T., Bischoff P., Kirchen S., Obst U. Detection of clinically relevant antibiotic-resistance genes in municipal wastewater using real-time PCR (TaqMan) J. Microbiol. Methods. 2004;56:277–286. doi: 10.1016/j.mimet.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 52.Colomer-Lluch M., Jofre J., Muniesa M. Quinolone resistance genes (qnrA and qnrS) in bacteriophage particles from wastewater samples and the effect of inducing agents on packaged antibiotic resistance genes. J. Antimicrob. Chemother. 2014;69:1265–1274. doi: 10.1093/jac/dkt528. [DOI] [PubMed] [Google Scholar]
- 53.Quirós P., Colomer-Lluch M., Martínez-Castillo A., Miró E., Argente M., Jofre J., Navarro F., Muniesa M. Antibiotic resistance genes in the bacteriophage DNA fraction of human fecal samples. Antimicrob. Agents Chemother. 2014;58:606–609. doi: 10.1128/AAC.01684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/JB.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.