Highlights
-
•
Artificial Intelligence (AI) has revolutionized many aspects of the pharmaceuticals.
-
•
AI assistance to pharma industries helps to improve overall life cycle of product.
-
•
AI can be implemented in pharma ranging from drug discovery to product management.
-
•
Future challenges related to AI and their respective solutions have been expounded.
Artificial Intelligence (AI) has recently started to gear-up its application in various sectors of the society with the pharmaceutical industry as a front-runner beneficiary. This review highlights the impactful use of AI in diverse areas of the pharmaceutical sectors viz., drug discovery and development, drug repurposing, improving pharmaceutical productivity, clinical trials, etc. to name a few, thus reducing the human workload as well as achieving targets in a short period. Crosstalk on the tools and techniques utilized in enforcing AI, ongoing challenges, and ways to overcome them, along with the future of AI in the pharmaceutical industry, is also discussed.
Teaser
Artificial intelligence-integrated drug discovery and development has accelerated the growth of the pharmaceutical sector, leading to a revolutionary change in the pharma industry. Here, we discuss areas of integration, tools, and techniques utilized in enforcing AI, ongoing challenges, and ways to overcome them.
The use of artificial intelligence (AI) has been increasing in various sectors of society, particularly the pharmaceutical industry. In this review, we highlight the use of AI in diverse sectors of the pharmaceutical industry, including drug discovery and development, drug repurposing, improving pharmaceutical productivity, and clinical trials, among others; such use reduces the human workload as well as achieving targets in a short period of time. We also discuss crosstalk between the tools and techniques utilized in AI, ongoing challenges, and ways to overcome them, along with the future of AI in the pharmaceutical industry.
Artificial intelligence: things to know
Over the past few years, there has been a drastic increase in data digitalization in the pharmaceutical sector. However, this digitalization comes with the challenge of acquiring, scrutinizing, and applying that knowledge to solve complex clinical problems [1]. This motivates the use of AI, because it can handle large volumes of data with enhanced automation [2]. AI is a technology-based system involving various advanced tools and networks that can mimic human intelligence. At the same time, it does not threaten to replace human physical presence 3, 4 completely. AI utilizes systems and software that can interpret and learn from the input data to make independent decisions for accomplishing specific objectives. Its applications are continuously being extended in the pharmaceutical field, as described in this review. According to the McKinsey Global Institute, the rapid advances in AI-guided automation will be likely to completely change the work culture of society 5, 6.
AI: networks and tools
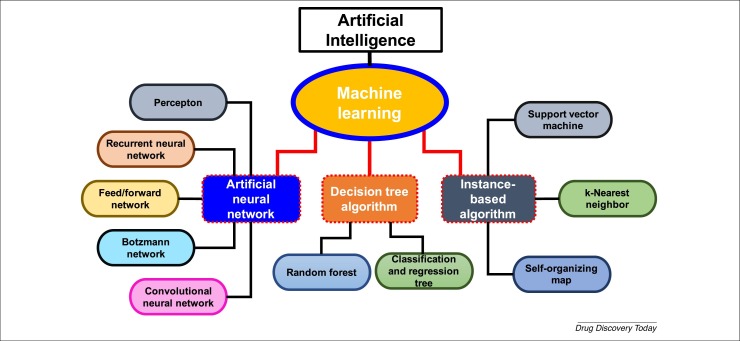
AI involves several method domains, such as reasoning, knowledge representation, solution search, and, among them, a fundamental paradigm of machine learning (ML). ML uses algorithms that can recognize patterns within a set of data that has been further classified. A subfield of the ML is deep learning (DL), which engages artificial neural networks (ANNs). These comprise a set of interconnected sophisticated computing elements involving ‘perceptons’ analogous to human biological neurons, mimicking the transmission of electrical impulses in the human brain [7]. ANNs constitute a set of nodes, each receiving a separate input, ultimately converting them to output, either singly or multi-linked using algorithms to solve problems [8]. ANNs involve various types, including multilayer perceptron (MLP) networks, recurrent neural networks (RNNs), and convolutional neural networks (CNNs), which utilize either supervised or unsupervised training procedures 9, 10.
The MLP network has applications including pattern recognition, optimization aids, process identification, and controls, are usually trained by supervised training procedures operating in a single direction only, and can be used as universal pattern classifiers [11]. RNNs are networks with a closed-loop, having the capability to memorize and store information, such as Boltzmann constants and Hopfield networks 11, 12. CNNs are a series of dynamic systems with local connections, characterized by its topology, and have use in image and video processing, biological system modeling, processing complex brain functions, pattern recognition, and sophisticated signal processing [13]. The more complex forms include Kohonen networks, RBF networks, LVQ networks, counter-propagation networks, and ADALINE networks 9, 11. Examples of method domains of AI are summarized in Figure 1 .
Figure 1.
Method domains of artificial intelligence (AI). This figure shows different AI method domains along with their subfields that can be implemented in different fields drug discovery and development.
Several tools have been developed based on the networks that form the core architecture of AI systems. One such tool developed using AI technology is the International Business Machine (IBM) Watson supercomputer (IBM, New York, USA). It was designed to assist in the analysis of a patient’s medical information and its correlation with a vast database, resulting in suggesting treatment strategies for cancer. This system can also be used for the rapid detection of diseases. This was demonstrated by its ability to detect breast cancer in only 60 s 14, 15.
AI in the lifecycle of pharmaceutical products
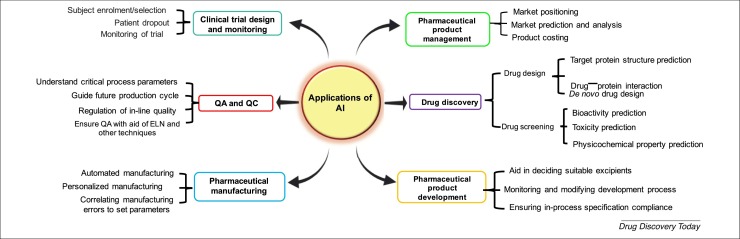
Involvement of AI in the development of a pharmaceutical product from the bench to the bedside can be imagined given that it can aid rational drug design [16]; assist in decision making; determine the right therapy for a patient, including personalized medicines; and manage the clinical data generated and use it for future drug development [17]. E-VAI is an analytical and decision-making AI platform developed by Eularis, which uses ML algorithms along with an easy-to-use user interface to create analytical roadmaps based on competitors, key stakeholders, and currently held market share to predict key drivers in sales of pharmaceuticals [18], thus helping marketing executives to allocate resources for maximum market share gain, reversing poor sales and enabled them to anticipate where to make investments. Different applications of AI in drug discovery and development are summarized in Figure 2 .
Figure 2.
Applications of artificial intelligence (AI) in different subfields of the pharmaceutical industry, from drug discovery to pharmaceutical product management.
AI in drug discovery
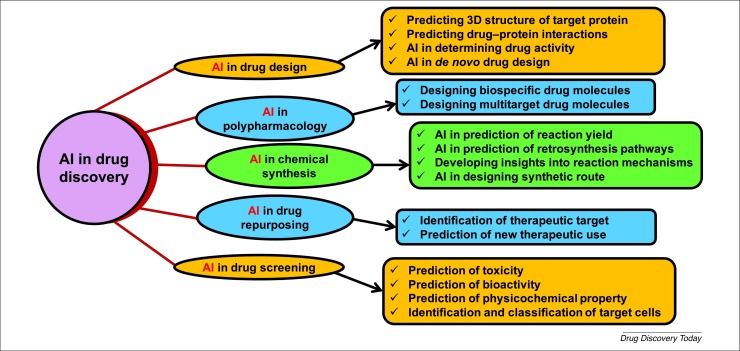
The vast chemical space, comprising >1060 molecules, fosters the development of a large number of drug molecules [19]. However, the lack of advanced technologies limits the drug development process, making it a time-consuming and expensive task, which can be addressed by using AI [15]. AI can recognize hit and lead compounds, and provide a quicker validation of the drug target and optimization of the drug structure design 19, 20. Different applications of AI in drug discovery are depicted in Figure 3 .
Figure 3.
Role of artificial intelligence (AI) in drug discovery. AI can be used effectively in different parts of drug discovery, including drug design, chemical synthesis, drug screening, polypharmacology, and drug repurposing.
Despite its advantages, AI faces some significant data challenges, such as the scale, growth, diversity, and uncertainty of the data. The data sets available for drug development in pharmaceutical companies can involve millions of compounds, and traditional ML tools might not be able to deal with these types of data. Quantitative structure-activity relationship (QSAR)-based computational model can quickly predict large numbers of compounds or simple physicochemical parameters, such as log P or log D. However, these models are some way from the predictions of complex biological properties, such as the efficacy and adverse effects of compounds. In addition, QSAR-based models also face problems such as small training sets, experimental data error in training sets, and lack of experimental validations. To overcome these challenges, recently developed AI approaches, such as DL and relevant modeling studies, can be implemented for safety and efficacy evaluations of drug molecules based on big data modeling and analysis. In 2012, Merck supported a QSAR ML challenge to observe the advantages of DL in the drug discovery process in the pharmaceutical industry. DL models showed significant predictivity compared with traditional ML approaches for 15 absorption, distribution, metabolism, excretion, and toxicity (ADMET) data sets of drug candidates 21, 22.
The virtual chemical space is enormous and suggests a geographical map of molecules by illustrating the distributions of molecules and their properties. The idea behind the illustration of chemical space is to collect positional information about molecules within the space to search for bioactive compounds and, thus, virtual screening (VS) helps to select appropriate molecules for further testing. Several chemical spaces are open access, including PubChem, ChemBank, DrugBank, and ChemDB.
Numerous in silico methods to virtual screen compounds from virtual chemical spaces along with structure and ligand-based approaches, provide a better profile analysis, faster elimination of nonlead compounds and selection of drug molecules, with reduced expenditure [19]. Drug design algorithms, such as coulomb matrices and molecular fingerprint recognition, consider the physical, chemical, and toxicological profiles to select a lead compound [23].
Various parameters, such as predictive models, the similarity of molecules, the molecule generation process, and the application of in silico approaches can be used to predict the desired chemical structure of a compound 20, 24. Pereira et al. presented a new system, DeepVS, for the docking of 40 receptors and 2950 ligands, which showed exceptional performance when 95 000 decoys were tested against these receptors [25]. Another approach applied a multiobjective automated replacement algorithm to optimize the potency profile of a cyclin-dependent kinase-2 inhibitor by assessing its shape similarity, biochemical activity, and physicochemical properties [26].
QSAR modeling tools have been utilized for the identification of potential drug candidates and have evolved into AI-based QSAR approaches, such as linear discriminant analysis (LDA), support vector machines (SVMs), random forest (RF) and decision trees, which can be applied to speed up QSAR analysis 27, 28, 29. King et al. found a negligible statistical difference when the ability of six AI algorithms to rank anonymous compounds in terms of biological activity was compared with that of traditional approaches [30].
AI in drug screening
The process of discovering and developing a drug can take over a decade and costs US$2.8 billion on average. Even then, nine out of ten therapeutic molecules fail Phase II clinical trials and regulatory approval 31, 32. Algorithms, such as Nearest-Neighbour classifiers, RF, extreme learning machines, SVMs, and deep neural networks (DNNs), are used for VS based on synthesis feasibility and can also predict in vivo activity and toxicity 31, 33. Several biopharmaceutical companies, such as Bayer, Roche, and Pfizer, have teamed up with IT companies to develop a platform for the discovery of therapies in areas such as immuno-oncology and cardiovascular diseases [19]. The aspects of VS to which AI has been applied are discussed below.
Prediction of the physicochemical properties
Physicochemical properties, such as solubility, partition coefficient (logP), degree of ionization, and intrinsic permeability of the drug, indirectly affect its pharmacokinetics properties and its target receptor family and, hence, must be considered when designing a new drug [34]. Different AI-based tools can be used to predict physicochemical properties. For example, ML uses large data sets produced during compound optimization done previously to train the program [35]. Algorithms for drug design include molecular descriptors, such as SMILES strings, potential energy measurements, electron density around the molecule, and coordinates of atoms in 3D, to generate feasible molecules via DNN and thereby predict its properties [36].
Zang et al. created a quantitative structure–property relationship (QSPR) workflow to determine the six physicochemical properties of environmental chemicals obtained from the Environmental Protection Agency (EPA) called the Estimation Program Interface (EPI) Suite [35]. Neural networks based on the ADMET predictor and ALGOPS program have been used to predict the lipophilicity and solubility of various compounds [37]. DL methods, such as undirected graph recursive neural networks and graph-based convolutional neural networks (CVNN), have been used to predict the solubility of molecules [38].
In several instances, ANN-based models, graph kernels, and kernel ridge-based models were developed to predict the acid dissociation constant of compounds 35, 39. Similarly, cell lines, such as Madin-Darby canine kidney cells and human colon adenocarcinoma (Caco-2) cells have been utilized to generate cellular permeability data of a diverse class of molecules, which are subsequently fed to AI-assisted predictors [34].
Kumar et al. developed six predictive models [SVMs, ANNs, k-nearest neighbor algorithms, LDAs, probabilistic neural network algorithms, and partial least square (PLS)] utilizing 745 compounds for training; these were used later on 497 compounds to predict their intestinal absorptivity based on parameters including molecular surface area, molecular mass, total hydrogen count, molecular refractivity, molecular volume, logP, total polar surface area, the sum of E- states indices, solubility index (log S), and rotatable bonds [40]. On similar lines, RF and DNN-based in silico models were developed to determine human intestinal absorption of a variety of chemical compounds [41]. Thus, AI has a significant role in the development of a drug, to predict not only its desired physicochemical properties, but also the desired bioactivity.
Prediction of bioactivity
The efficacy of drug molecules depends on their affinity for the target protein or receptor. Drug molecules that do not show any interaction or affinity towards the targeted protein will not be able to deliver the therapeutic response. In some instances, it might also be possible that developed drug molecules interact with unintended proteins or receptors, leading to toxicity. Hence, drug target binding affinity (DTBA) is vital to predict drug–target interactions. AI-based methods can measure the binding affinity of a drug by considering either the features or similarities of the drug and its target. Feature-based interactions recognize the chemical moieties of the drug and that of the target to determine the feature vectors. By contrast, in similarity-based interaction, the similarity between drug and target is considered, and it is assumed that similar drugs will interact with the same targets [42].
Web applications, such as ChemMapper and the similarity ensemble approach (SEA), are available for predicting drug–target interactions [43]. Many strategies involving ML and DL have been used to determine DTBA, such as KronRLS, SimBoost, DeepDTA, and PADME. ML-based approaches, such as Kronecker-regularized least squares (KronRLS), evaluate the similarity between drugs and protein molecules to determine DTBA. Similarly, SimBoost utilized regression trees to predict DTBA, and considers both feature-based and similarity-based interactions. Drug features from SMILES, ligand maximum common substructure (LMCS), extended connectivity fingerprint, or a combination thereof can also be considered [42].
DL approaches have shown improved performance compared with ML because they apply network-based methods that do not depend on the availability of the 3D protein structure [43]. DeepDTA, PADME, WideDTA, and DeepAffinity are some DL methods used to measure DTBA. DeepDTA accepts drug data in the form of SMILES, whereby, the amino acid sequence is entered for protein input data and for the 1D representation of the drug structure [44]. WideDTA is CVNN DL method that incorporates ligand SMILES (LS), amino acid sequences, LMCS, and protein domains and motifs as input data for assessing the binding affinity [45].
DeepAffinity and Protein And Drug Molecule interaction prEdiction (PADME) are similar to the approaches described earlier [46]. DeepAffinity is an interpretable DL model that uses both RNN and CNN and both unlabeled and labeled data. It takes into account the compound in the SMILES format and protein sequences in the structural and physicochemical properties [47]. PADME is a DL-based platform that utilizes feed-forward neural networks for predicting drug target interactions (DTIs). It considers the combination of the features of the drug and target protein as input data and forecasts the interaction strength between the two. For the drug and the target, the SMILES representation and the protein sequence composition (PSC) are used for illustration, respectively [46]. Unsupervised ML techniques, such as MANTRA and PREDICT, can be used to forecast the therapeutic efficacy of drugs and target proteins of known and unknown pharmaceuticals, which can also be extrapolated to the application of drug repurposing and interpreting the molecular mechanism of the therapeutics. MANTRA groups compound based on similar gene expression profiles using a CMap data set and clusters those compounds predicted to have a common mechanism of action and common biological pathway [43]. The bioactivity of a drug also includes ADME data. AI-based tools, such as XenoSite, FAME, and SMARTCyp, are involved in determining the sites of metabolism of the drug. In addition, software such as CypRules, MetaSite, MetaPred, SMARTCyp, and WhichCyp were used to identify specific isoforms of CYP450 that mediate a particular drug metabolism. The clearance pathway of 141 approved drugs was done by SVM-based predictors with high accuracy [48].
Prediction of toxicity
The prediction of the toxicity of any drug molecule is vital to avoid toxic effects. Cell-based in vitro assays are often used as preliminary studies, followed by animal studies to identify the toxicity of a compound, increasing the expense of drug discovery. Several web-based tools, such as LimTox, pkCSM, admetSAR, and Toxtree, are available to help reduce the cost [35]. Advanced AI-based approaches look for similarities among compounds or project the toxicity of the compound based on input features. The Tox21 Data Challenge organized by the National Institutes of Health, Environmental Protection Agency (EPA), and US Food and Drug Administration (FDA) was an initiative to evaluate several computational techniques to forecast the toxicity of 12 707 environmental compounds and drugs [35]; an ML algorithm named DeepTox outperformed all methods by identifying static and dynamic features within the chemical descriptors of the molecules, such as molecular weight (MW) and Van der Waals volume, and could efficiently predict the toxicity of a molecule based on predefined 2500 toxicophore features [49]. The different AI tools used in drug discovery are listed in Table 1 .
Table 1.
Examples of AI tools used in drug discovery
| Tools | Details | Website URL | Refs |
|---|---|---|---|
| DeepChem | MLP model that uses a python-based AI system to find a suitable candidate in drug discovery | https://github.com/deepchem/deepchem | [21] |
| DeepTox | Software that predicts the toxicity of total of 12 000 drugs | www.bioinf.jku.at/research/DeepTox | [22] |
| DeepNeuralNetQSAR | Python-based system driven by computational tools that aid detection of the molecular activity of compounds | https://github.com/Merck/DeepNeuralNet-QSAR | [23] |
| ORGANIC | A molecular generation tool that helps to create molecules with desired properties | https://github.com/aspuru-guzik-group/ORGANIC | [24] |
| PotentialNet | Uses NNs to predict binding affinity of ligands | https://pubs.acs.org/doi/full/10.1021/acscentsci.8b00507 | [25] |
| Hit Dexter | ML technique to predict molecules that might respond to biochemical assays | http://hitdexter2.zbh.uni-hamburg.de | |
| DeltaVina | A scoring function for rescoring drug–ligand binding affinity | https://github.com/chengwang88/deltavina | |
| Neural graph fingerprint | Helps to predict properties of novel molecules | https://github.com/HIPS/neural-fingerprint | |
| AlphaFold | Predicts 3D structures of proteins | https://deepmind.com/blog/alphafold | |
| Chemputer | Helps to report procedure for chemical synthesis in standardized format | https://zenodo.org/record/1481731 |
SEA was used to evaluate the safety target prediction of 656 marketed drugs against 73 unintended targets that might produce adverse effects [43]. Developed using an ML-based approach, eToxPred was applied to estimate the toxicity and synthesis feasibility of small organic molecules and showed accuracy as high as 72% [48]. Similarly, open-source tools, such as TargeTox and PrOCTOR, are also used in toxicity prediction [50]. TargeTox is biological network target-based drug toxicity risk prediction method that uses the guilt-by-association principle whereby entities that have similar functional properties share similarities in biological networks [51]. It can produce protein network data and unite pharmacological and functional properties in a ML classifier to predict drug toxicity [52]. PrOCTOR was trained using a RF model and took into account drug-likeliness properties, molecular features, target-based features, and properties of the protein targets to generate a ‘PrOCTOR score’, which forecasted whether a drug would fail in clinical trials owing to its toxicity. It also recognized FDA-approved drugs that later reported adverse drug events [53]. In another approach, Tox_(R)CNN involving a deep CVNN method evaluated the cytotoxicity of drugs that had been exposed to DAPI-stained cells [54].
AI in designing drug molecules
Prediction of the target protein structure
While developing a drug molecule, it is essential to assign the correct target for successful treatment. Numerous proteins are involved in the development of the disease and, in some cases, they are overexpressed. Hence, for selective targeting of disease, it is vital to predict the structure of the target protein to design the drug molecule. AI can assist in structure-based drug discovery by predicting the 3D protein structure because the design is in accordance with the chemical environment of the target protein site, thus helping to predict the effect of a compound on the target along with safety considerations before their synthesis or production [55]. The AI tool, AlphaFold, which is based on DNNs, was used to analyze the distance between the adjacent amino acids and the corresponding angles of the peptide bonds to predict the 3D target protein structure and demonstrated excellent results by correctly predicting 25 out of 43 structures.
In a study by AlQurashi, RNN was used to predict the protein structure. The author considered three stages (i.e., computation, geometry, and assessment) termed a recurrent geometric network (RGN). Here, the primary protein sequence was encoded, and the torsional angles for a given residue and a partially completed backbone obtained from the geometric unit upstream of this were then considered as input and provided a new backbone as output. The final unit produced the 3D structure as the output. Assessment of the deviation of predicted and experimental structures was done using the distance-based root mean square deviation (dRMSD) metric. The parameters in RGN were optimized to keep the dRMSD low between the experimental and predicted structures [56]. AlQurashi predicted that his AI method would be quicker than AlphaFold in terms of the time taken to predict the protein structure. However, AlphaFold is likely to have better accuracy in predicting protein structures with sequences similar to the reference structures [57].
A study was conducted to predict the 2D structure of a protein using MATLAB assisted by a nonlinear three-layered NN toolbox based on a feed-forward supervised learning and backpropagation error algorithm. MATLAB was used to train input and output data sets, and the NNs were learning algorithms and performance evaluators. The accuracy in predicting the 2D structure was 62.72% [58].
Predicting drug–protein interactions
Drug–protein interactions have a vital role in the success of a therapy. The prediction of the interaction of a drug with a receptor or protein is essential to understand its efficacy and effectiveness, allows the repurposing of drugs, and prevents polypharmacology [55]. Various AI methods have been useful in the accurate prediction of ligand–protein interactions, ensuring better therapeutic efficacy 55, 59. Wang et al. reported a model using the SVM approach, trained on 15 000 protein–ligand interactions, which were developed based on primary protein sequences and structural characteristics of small molecules to discover nine new compounds and their interaction with four crucial targets [60].
Yu et al. exploited two RF models to predict possible drug–protein interactions by the integration of pharmacological and chemical data and validating them against known platforms, such as SVM, with high sensitivity and specificity. Also, these modes were capable of predicting drug–target associations that could be further extended to target–disease and target–target associations, thereby speeding up the drug discovery process [61]. Xiao et al. adopted the Synthetic Minority Over-Sampling Technique and the Neighborhood Cleaning Rule to obtain optimized data for the subsequent development of iDrugTarget. This is a combination of four subpredictors (iDrug-GPCR, iDrug-Chl, iDrug-Enz, and iDrug-NR) for identifying interactions between a drug and G-protein-coupled receptors (GPCRs), ion channels, enzymes, and nuclear receptors (NR) respectively. When this predictor was compared with existing predictors through target-jackknife tests, the former surpassed the latter in terms of both prediction accuracy and consistency [62].
The ability of AI to predict drug–target interactions was also used to assist the repurposing of existing drugs and avoiding polypharmacology. Repurposing an existing drug qualifies it directly for Phase II clinical trials [19]. This also reduces expenditure because relaunching an existing drug costs ∼US$8.4 million compared with the launch of a new drug entity (∼US$41.3 million) [63]. The ‘Guilt by association’ approach can be utilized to forecast the innovative association of a drug and disease, which is either a knowledge-based or computationally driven network [64]. In a computationally driven network, the ML approach is widely used, which utilizes techniques such as SVM, NN, logistic regression, and DL. Logistic regression platforms, such as PREDICT, SPACE, and other ML approaches, consider drug–drug, disease–disease similarity, the similarity between target molecules, chemical structure, and gene expression profiles while repurposing a drug [65].
Cellular network-based deep learning technology (deepDTnet) has been explored to predict the therapeutic use of topotecan, currently used as a topoisomerase inhibitor. It can also be used for the therapy of multiple sclerosis by inhibiting human retinoic acid receptor-related orphan receptor-gamma t (ROR-γt) [66]. This platform is currently under a provisional US patent. Self-organizing maps (SOMs) are in the unsupervised category of ML and are used in drug repurposing. They use a ligand-based approach to search novel off-targets for a set of drug molecules by training the system on a defined number of compounds with recognized biological activities, which is later used for the analysis of different compounds [67]. In a recent study, DNN was used to repurpose existing drugs with proven activity against SARS-CoV, HIV, influenza virus, and drugs that are 3C-like protease inhibitors. In this, extended connectivity fingerprint (ECFP), functional-class fingerprints (FCFPs), and an octanol-water partition coefficient (ALogP_count) were considered to train the AI platform. From the results, it was concluded that 13 of the screened drugs could be carried toward further development based on their cytotoxicity and viral inhibition [68].
Drug–protein interactions can also predict the chances of polypharmacology, which is the tendency of a drug molecule to interact with multiple receptors producing off-target adverse effects [69]. AI can design a new molecule based on the rationale of polypharmacology and aid in the generation of safer drug molecules [70]. AI platforms such as SOM, along with the vast databases available, can be used to link several compounds to numerous targets and off-targets. Bayesian classifiers and SEA algorithms can be used to establish links between the pharmacological profiles of drugs and their possible targets [67].
Li et al. demonstrated the use of KinomeX, an AI-based online medium using DNNs for the detection of polypharmacology of kinases based on their chemical structures. This platform uses DNN trained with ∼14 000 bioactivity data points developed based on >300 kinases. Thus, it has practical application in studying the overall selectivity of a drug towards the kinase family and particular subfamilies of kinases, thus helping to design novel chemical modifiers. This study used NVP-BHG712 as a model compound to predict its primary targets and also its off-targets with reasonable accuracy [71]. One prominent instance is Cyclica’s cloud-based proteome-screening AI platform, Ligand Express, which is used to find receptors that can interact with a particular small molecule (the molecular description of which is in SMILE string) and produce on and off-target interactions. This helps in understanding the possible adverse effects of the drug [72].
AI in de novo drug design Over the past few years, the de novo drug design approach has been widely used to design drug molecules. The traditional method of de novo drug design is being replaced by evolving DL methods, the former having shortcomings of complicated synthesis routes and difficult prediction of the bioactivity of the novel molecule [36]. Computer-aided synthesis planning can also suggest millions of structures that can be synthesized and also predicts several different synthesis routes for them [73].
Grzybowski et al. developed the Chematica program [74], now renamed Synthia, which has the ability to encode a set of rules into the machine and propose possible synthesizing routes for eight medicinally essential targets. This program has proven to be efficient both in terms of improving the yield and reducing expenses. It is also capable of providing alternate synthesizing strategies for patented products and is said to be helpful in the synthesis of compounds that have not yet been synthesized. Similarly, DNN focuses on rules of organic chemistry and retrosynthesis, which, with the aid of Monte-Carlo tree searches and symbolic AI, help in reaction prediction and the process of drug discovery and design, which is much faster than traditional methods 75, 76.
Coley et al. developed a framework in which a rigid forward reaction template was applied to a group of reactants to synthesize chemically feasible products with a significant rate of reaction. ML was used to determine the dominant product based on a score given by the NNs [23]. Putin et al. explored a DNN architecture called the reinforced adversarial neural computer (RANC) based on RL for de novo design of small organic molecules. This platform was trained with molecules represented as SMILES strings. It then generated molecules with predefined chemical descriptors in terms of MW, logP, and topological polar surface area (TPSA). RANC was compared with another platform, ORGANIC, where the former outperformed in generating unique structures without sufficient loss of their structure length [77].
Even RNN was based on the long short-term memory (LSTM) relating to molecules obtained from the ChEMBL database and fed as SMILES strings. This was used to generate a diverse library of molecules for VS. This approach was extended to procure novel molecules toward a particular target, such as targets for the 5-HT2A receptor, Staphylococcus aureus, and Plasmodium falciparum [78].
Popova et al. developed the Reinforcement Learning for Structural Evolution strategy for de novo drug synthesis, which involves generative and predictive DNNs to develop new compounds. In this, the generative model produces more unique molecules in terms of SMILE strings based on a stack memory, whereas the predictive models are used to forecast the properties of the developed compound [79]. Merk et al. also exploited the generative AI model to design retinoid X and PPAR agonist molecules, with desired therapeutic effects without requiring complex rules. The authors successfully designed five molecules, four out of which have shown good modulatory activity in cell assays, thereby emphasizing the use of generative AI in new molecule synthesis [80]. The involvement of AI in the de novo design of molecules can be beneficial to the pharmaceutical sector because of its various advantages, such as providing online learning and simultaneous optimization of the already-learned data as well as suggesting possible synthesis routes for compounds leading to swift lead design and development 78, 81.
AI in advancing pharmaceutical product development
The discovery of a novel drug molecule requires its subsequent incorporation in a suitable dosage form with desired delivery characteristics. In this area, AI can replace the older trial and error approach [82]. Various computational tools can resolve problems encountered in the formulation design area, such as stability issues, dissolution, porosity, and so on, with the help of QSPR [83]. Decision-support tools use rule-based systems to select the type, nature, and quantity of the excipients depending on the physicochemical attributes of the drug and operate through a feedback mechanism to monitor the entire process and intermittently modify it [84].
Guo et al. integrated Expert Systems (ES) and ANN to create a hybrid system for the development of direct-filling hard gelatin capsules of piroxicam in accordance with the specifications of its dissolution profile. The MODEL EXPERT SYSTEM (MES) makes decisions and recommendations for formulation development based on the input parameters. By contrast, ANN uses backpropagation learning to link formulation parameters to the desired response, jointly controlled by the control module, to ensure hassle-free formulation development [82].
Various mathematical tools, such as computational fluid dynamics (CFD), discrete element modeling (DEM), and the Finite Element Method have been used to examine the influence of the flow property of the powder on the die-filling and process of tablet compression 85, 86. CFD can also be utilized to study the impact of tablet geometry on its dissolution profile [87]. The combination of these mathematical models with AI could prove to be of immense help in the rapid production of pharmaceutical products.
AI in pharmaceutical manufacturing
With the increasing complexities of manufacturing processes along with increasing demand for efficiency and better product quality, modern manufacturing systems are trying to confer human knowledge to machines, continuously changing the manufacturing practice [88]. The incorporation of AI in manufacturing can prove to be a boost for the pharmaceutical industry. Tools, such as CFD, uses Reynolds-Averaged Navier-Stokes solvers technology that studies the impact of agitation and stress levels in different equipment (e.g., stirred tanks), exploiting the automation of many pharmaceutical operations. Similar systems, such as direct numerical simulations and large eddy simulations, involve advanced approaches to solve complicated flow problems in manufacturing [85].
The novel Chemputer platform helps digital automation for the synthesis and manufacturing of molecules, incorporating various chemical codes and operating by using a scripting language known as Chemical Assembly [23]. It has been successfully used for the synthesis and manufacture of sildenafil, diphenhydramine hydrochloride, and rufinamide, with the yield and purity significantly similar to manual synthesis [89]. The estimated completion of granulation in granulators of capacities ranging from 25 to 600 l can be done efficiently by AI technologies [90]. The technology and neuro-fuzzy logic correlated critical variables to their responses. They derived a polynomial equation for the prediction of the proportion of the granulation fluid to be added, required speed, and the diameter of the impeller in both geometrically similar and dissimilar granulators [91].
DEM has been widely utilized in the pharmaceutical industry, such as in studying the segregation of powders in a binary mixture, the effects of varying blade speed and shape, predicting the possible path of the tablets in the coating process, along with analysis of time spent by tablets under the spray zone [85]. ANNs, along with fuzzy models, studied the correlation between machine settings and the problem of capping to reduce tablet capping on the manufacturing line [92].
Meta-classifier and tablet-classifier are AI tools that help to govern the quality standard of the final product, indicating a possible error in the manufacturing of the tablet [93]. A patent has been filed, demonstrating a system capable of determining the most exquisite combination of drug and dosage regimen for each patient, using a processor receiving patient information, and designs the desired transdermal patch accordingly [94].
AI in quality control and quality assurance
Manufacturing of the desired product from the raw materials includes a balance of various parameters [93]. Quality control tests on the products, as well as maintenance of batch-to-batch consistency, require manual interference. This might not be the best approach in each case, showcasing the need for AI implementation at this stage [85]. The FDA amended the Current Good Manufacturing Practices (cGMP) by introducing a ‘Quality by Design’ approach to understand the critical operation and specific criteria that govern the final quality of the pharmaceutical product [95].
Gams et al. used a combination of human efforts and AI, wherein preliminary data from production batches were analyzed and decision trees developed. These were further translated into rules and analyzed by the operators to guide the production cycle in the future [93]. Goh et al. studied the dissolution profile, an indicator of batch-to-batch consistency of theophylline pellets with the aid of ANN, which correctly predicted the dissolution of the tested formulation with an error of <8% [96].
AI can also be implemented for the regulation of in-line manufacturing processes to achieve the desired standard of the product [95]. ANN-based monitoring of the freeze-drying process is used, which applies a combination of self-adaptive evolution along with local search and backpropagation algorithms. This can be used to predict the temperature and desiccated-cake thickness at a future time point (t + Δt) for a particular set of operating conditions, eventually helping to keep a check on the final product quality [97].
An automated data entry platform, such as an Electronic Lab Notebook, along with sophisticated, intelligent techniques, can ensure the quality assurance of the product [98]. Also, data mining and various knowledge discovery techniques in the Total Quality Management expert system can be used as valuable approaches in making complex decisions, creating new technologies for intelligent quality control [99].
AI in clinical trial design
Clinical trials are directed toward establishing the safety and efficacy of a drug product in humans for a particular disease condition and require 6–7 years along with a substantial financial investment. However, only one out of ten molecules entering these trials gain successful clearance, which is a massive loss for the industry [100]. These failures can result from inappropriate patient selection, shortage of technical requirements, and poor infrastructure. However, with the vast digital medical data available, these failures can be reduced with the implementation of AI [101].
The enrolment of patients takes one-third of the clinical trial timeline. The success of a clinical trial can be ensured by the recruitment of suitable patients, which otherwise leads to ∼86% of failure cases [102]. AI can assist in selecting only a specific diseased population for recruitment in Phase II and III of clinical trials by using patient-specific genome–exposome profile analysis, which can help in early prediction of the available drug targets in the patients selected 19, 101. Preclinical discovery of molecules as well as predicting lead compounds before the start of clinical trials by using other aspects of AI, such as predictive ML and other reasoning techniques, help in the early prediction of lead molecules that would pass clinical trials with consideration of the selected patient population [101].
Drop out of patients from clinical trials accounts for the failure of 30% of the clinical trials, creating additional recruiting requirements for the completion of the trial, leading to a wastage of time and money. This can be avoided by close monitoring of the patients and helping them follow the desired protocol of the clinical trial [102]. Mobile software was developed by AiCure that monitored regular medication intake by patients with schizophrenia in a Phase II trial, which increased the adherence rate of patients by 25%, ensuring successful completion of the clinical trial [19].
AI in pharmaceutical product management
AI in market positioning
Market positioning is the process of creating an identity of the product in the market to attract consumers to buy them, making it an essential element in almost all business strategies for companies to establish their own unique identity 103, 104. This approach was used in the marketing of pioneer brand Viagra, where the company targeted it not only for the treatment of men’s erectile dysfunction, but also for other problems affecting quality of life [105].
With the help of technology and e-commerce as a platform, it has become easier for companies to get a natural recognition of their brand in the public domain. Companies exploit search engines as one of the technological platforms to occupy a prominent position in online marketing and help in the positioning of the product in the market, as also confirmed by the Internet Advertising Bureau. Companies continuously try to rank their websites higher than those of other companies, giving recognition to their brand in a short period [106].
Other techniques, such as statistical analysis methods, particle swarm optimization algorithms (proposed by Eberhart and Kennedy in 1995) in combination with NNs, provided a better idea about markets. They can help decide the marketing strategy for the product based on accurate consumer-demand prediction [107].
AI in market prediction and analysis
The success of a company lies in the continuous development and growth of its business. Even with access to substantial funds, R&D output in the pharmaceutical industry is falling because of the failure of companies to adopt new marketing technologies [108]. The advances in digital technologies, referred to as the ‘Fourth industrial revolution’, is helping innovative digitalized marketing via a multicriteria decision-making approach, which collects and analyzes statistical and mathematical data and implements human inferences to make AI-based decision-making models explore new marketing methodology [109].
AI also helped in a comprehensive analysis of the fundamental requirements of a product from the customer’s point of view as well as understanding the need of the market, which aid in decision-making using prediction tools. It can also forecast sales and analyze the market. AI-based software engages consumers and creates awareness among physicians by displaying advertisements directing them to the product site by just a click [110]. In addition, these methods use natural language-processing tools to analyze keywords entered by customers and relate them to the probability of purchasing the product 111, 112.
Several businesses to business (B2B) companies have announced self-service technologies that allow free browsing of health products, easily found by giving its specification, place orders, and track their shipping. Pharmaceutical companies are also introducing their online applications such as 1 mg, Medline, Netmeds, and Ask Apollo, to fulfill the unmet needs of the patients [109]. Prediction of the market is also essential for various pharmaceutical distribution companies, which can implement AI in the field, such as ‘Business intelligent Smart Sales Prediction Analysis’, which uses a combination of time series forecasting and real-time application. This helps pharmaceutical companies to predict the sale of products in advance to prevent costs of excess stock or prevent customer loss because of shortages [113].
AI in product cost
Based on the market analysis and cost incurred in the development of the pharmaceutical product, the company determines the final price of the product. The critical concept in applying AI to determine this price is harnessing its ability to mimic the thinking of a human expert to assess the factors that control the pricing of a product after its manufacture [114]. Factors, such as expenditure during research and development of the drug, strict price regulatory schemes in the concerned country, length of the exclusivity period, market share of the innovated drug after a year before are patent expiry, price of the reference product, and price-fixing policies determine the price of branded and generic drugs [115].
In ML, large sets of statistical data, such as product development cost, product demand in the market, inventory cost, manufacturing cost, and competitors’ product price, are analyzed by the software, subsequently developing algorithms for predicting the product price. AI platforms, such as In competitor, launched by Intelligence Node (founded in the year 2012), is a complete retail competitive intelligence platform that analyzes the competitor pricing data and helps retailers and brands to monitor the competition. Wise Athena and Navetti PricePoint enable the user to determine the pricing of their product, suggesting that pharmaceutical companies can adopt the same to assist product costing [116].
AI-based advanced applications
AI-based nanorobots for drug delivery
Nanorobots comprise mainly integrated circuits, sensors, power supply, and secure backup of data, which are maintained via computational technologies, such as AI 117, 118. They are programmed to avoid the collision, target identification, detect and attach, and finally excretion from the body. Advances in nano/microrobots give them the ability to navigate to the targeted site based on physiological conditions, such as pH, thus improving the efficacy and reducing systemic adverse effects [118]. Development of implantable nanorobots developed for controlled delivery of drugs and genes requires consideration of parameters such as dose adjustment, sustained release, and control release, and the release of the drugs requires automation controlled by AI tools, such as NNs, fuzzy logic, and integrators [119]. Microchip implants are used for programmed release as well as to detect the location of the implant in the body.
AI in combination drug delivery and synergism/antagonism prediction
Several combinations of drugs are approved and marketed to treat complex diseases, such as TB and cancer, because they can provide a synergistic effect for quick recovery 120, 121. The selection of precise and potential drugs for combination requires high-throughput screening of a considerable number of drugs, making the process tedious; for example, cancer therapy requires six or seven drugs as a combination therapy. ANNs, logistic regression, and network-based modeling can screen drug combinations and improve overall dose regimen 120, 122. Rashid et al. developed a quadratic phenotype optimization platform for the detection of optimal combination therapy for the treatment of bortezomib-resistant multiple myeloma using a collection of 114 FDA-approved drugs. This model recommended the combination of decitabine (Dec) and mitomycin C (MitoC) as the best two-drug combination and Dec, MitoC, and mechlorethamine as the superior three-drug combination [121].
Combination drug delivery can be more efficient if backed up by data on the synergism or antagonism of drugs administered together. The Master Regulator Inference Algorithm used ‘Mater regulator genes’ to efficiently predict 56% synergism. Other methods, such as Network-based Laplacian regularized least square synergistic drug combination, and RF, can also be used for the same [122].
Li et al. developed a synergistic drug combination model using RF for the prediction of synergistic anticancer drug combinations. This model was formed based on gene expression profiles and various networks, and the authors successfully predicted 28 synergistic anticancer combinations. They have reported three such combinations, although the remainder might also prove to be important [69]. Similarly, Mason et al. applied an ML approach, called the Combination Synergy Estimation, to predict potential synergistic antimalarial combinations based on a data set of 1540 antimalarial drug compounds [123].
AI emergence in nanomedicine
Nanomedicines use nanotechnology and medicines for the diagnosis, treatment, and monitoring of complex diseases, such as HIV, cancer, malaria, asthma, and various inflammatory diseases. In recent years, nanoparticle-modified drug delivery has become important in the field of therapeutics and diagnostics because they have enhanced efficacy and treatment 121, 124. A combination of nanotechnology and AI could provide solutions to many problems in formulation development [125].
A methotrexate nanosuspension was computationally formulated by studying the energy generated on the interaction between the drug molecules, monitoring the conditions that could lead to the aggregation of the formulation [83]. Coarse-grained simulation, along with chemical calculation, can aid the determination of drug–dendrimer interactions and evaluation of drug encapsulation within the dendrimer. In addition, software such as LAMMPS and GROMACS 4 can be used to examine the impact of surface chemistry on the internalization of nanoparticles into cells [83].
AI assisted the preparation of silicasomes, which is a combination of iRGD, a tumor-penetrating peptide, and irinotecan-loaded multifunctional mesoporous silica nanoparticles. This increased the uptake of silicasomes three–fourfold because iRGD improves the transcytosis of silicasomes, with improved treatment outcome and enhanced overall survival [124].
Pharmaceutical market of AI
To decrease the financial cost and chances of failures that accompany VS, pharmaceutical companies are shifting towards AI. There was an increase in the AI market from US$200 million in 2015 to US$700 million in 2018, and is expected to increase to $5 billion by 2024 [126]. A 40% projected growth from 2017 to 2024 indicates that AI will likely revolutionize the pharmaceutical and medical sectors. Various pharmaceutical companies have made and are continuing to invest in AI and have collaborated with AI companies to developed essential healthcare tools. The collaboration of DeepMind Technologies, a subsidiary of Google, with the Royal Free London NHS Foundation Trust for the assistance of acute kidney injury, is an example of this. Major pharmaceutical companies and AI players are detailed in Figure 4 [19].
Figure 4.
Leading pharmaceutical companies and their association with Artificial Intelligence (AI) organizations that are working in fields including oncology, cardiovascular diseases, and central nervous system disorders.
Ongoing challenges in adopting AI: leads on ways to overcome
The entire success of AI depends on the availability of a substantial amount of data because these data are used for the subsequent training provided to the system. Access to data from various database providers can incur extra costs to a company, and the data should also be reliable and high quality to ensure accurate result prediction. Other challenges that prevent full-fledged adoption of AI in the pharmaceutical industry include the lack of skilled personnel to operate AI-based platforms, limited budget for small organizations, apprehension of replacing humans leading to job loss, skepticism about the data generated by AI, and the black box phenomenon (i.e., how the conclusions are reached by the AI platform) [6].
Automation of certain tasks in drug development, manufacturing, and supply chains, clinical trials, and sales will take place with time, but these all fall under the category of ‘narrow AI’; where AI has to be trained using a large volume of data and, thus, makes it suitable for a particular task. Therefore, human intervention is mandatory for the successful implementation, development, and operation of the AI platform. However, the fear of unemployment could be a myth given that AI is currently is taking over repetitive jobs, while leaving scope for human intelligence to be used for developing more complicated insights and creativity.
Nevertheless, AI has been adopted by several pharmaceutical companies, and it is expected that a revenue of US$2.199 billion will be created by 2022 through AI-based solutions in the pharmaceutical sector, with an investment exceeding US$7.20 billion across 300+ deals between 2013 and 2018 by the pharmaceutical industry [127]. Pharmaceutical organizations need clarity about the potential of AI technology in finding solutions to problems once it has been implemented, along with understanding the reasonable goals that can be achieved. Skilled data scientists, software engineers with a sound knowledge of AI technology, and a clear understanding of the company business target and its R&D goal can be developed to utilize the full potential of the AI platform.
Concluding remarks and prospects
The advancement of AI, along with its remarkable tools, continuously aims to reduce challenges faced by pharmaceutical companies, impacting the drug development process along with the overall lifecycle of the product, which could explain the increase in the number of start-ups in this sector [23]. The current healthcare sector is facing several complex challenges, such as the increased cost of drugs and therapies, and society needs specific significant changes in this area. With the inclusion of AI in the manufacturing of pharmaceutical products, personalized medications with the desired dose, release parameters, and other required aspects can be manufactured according to individual patient need [85]. Using the latest AI-based technologies will not only speed up the time needed for the products to come to the market, but will also improve the quality of products and the overall safety of the production process, and provide better utilization of available resources along with being cost-effective, thereby increasing the importance of automation [128].
The most significant worry regarding the incorporation of these technologies is the job losses that would follow and the strict regulations needed for the implementation of AI. However, these systems are intended only to make work easier and not to completely replace humans [129]. AI can not only aid quick and hassle-free hit compound identification, but also contribute to suggestions of synthesis routes of these molecules along with the prediction of the desired chemical structure and an understanding of drug–target interactions and its SAR.
AI can also make major contributions to the further incorporation of the developed drug in its correct dosage form as well as its optimization, in addition to aiding quick decision-making, leading to faster manufacturing of better-quality products along with assurance of batch-to-batch consistency. AI can also contribute to establishing the safety and efficacy of the product in clinical trials, as well as ensuring proper positioning and costing in the market through comprehensive market analysis and prediction. Although there are no drugs currently on the market developed with AI-based approaches and specific challenges remain with regards to the implementation of this technology, it is likely that AI will become an invaluable tool in the pharmaceutical industry in the near future.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in the paper.
Acknowledgments
The authors acknowledge the Department of Pharmaceuticals, Ministry of Chemicals and Fertilizers, Government of India for financial support. R.K.T. would like to acknowledge the Science and Engineering Research Board (Statutory Body Established through an Act of Parliament: SERB Act 2008), Department of Science and Technology, Government of India for a grant (Grant #ECR/2016/001964) and N-PDF for funding (PDF/2016/003329) research in his laboratory.
Biographies

Rakesh K. Tekade, currently an associate professor in NIPER, Ahmedabad, is an academic-researcher with >10 years of teaching and research experience. Dr Tekade’s research group investigates the design, development, and characterization of targeted nanotechnology-based products for the site-specific delivery of therapeutic drugs, siRNA, miRNA, and so on, for the treatment of cancer, diabetes, arthritis, and neurological disorders. He has coauthored >100 peer-reviewed publications in international journals, contributed >50 international reference book chapters, five invited editorial articles, and four patent applications. Dr Tekade is an editor in chief of a Book Series entitled Advances in Pharmaceutical Product Development and Research Series.

Debleena Paul received a BSc in pharmacy from Maulana Abul Kalam Azad University of Technology and is currently pursuing her MS Pharm at NIPER-Ahmedabad under the guidance Rakesh K. Tekade. Her research focuses on the development of in situ gelling dusting powder for wound dressing applications.

Kiran Kalia is a professor of pharmacology and Director of NIPER, Ahmedabad; she is also a professor in lien in the Department of Biosciences, Sardar Patel University. She has research experience spanning over 35 years and has mentored several MSc and PhD students. She was an awardee of the Indian National Science Academy (INSA) Research fellowship and has received several CSIR Fellowships. Professor Kiran is an editorial board and review committee member of several international journals. Her research interests encompass proteomic markers for diabetic nephropathy from urine, genomic markers of the susceptibility of diabetic retinopathy, genomic alterations in oral cancer, and environmental biotechnology & toxicology studies.
References
- 1.Ramesh A. Artificial intelligence in medicine. Ann. R. Coll. Surg. Engl. 2004;86:334–338. doi: 10.1308/147870804290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miles J., Walker A. The potential application of artificial intelligence in transport. IEE Proc.-Intell. Transport Syst. 2006;153:183–198. [Google Scholar]
- 3.Yang Y., Siau K. MWAIS; 2018. A Qualitative Research on Marketing and Sales in the Artificial Intelligence Age. [Google Scholar]
- 4.Wirtz B.W. Artificial intelligence and the public sector—applications and challenges. Int. J. Public Adm. 2019;42:596–615. [Google Scholar]
- 5.Smith R.G., Farquhar A. The road ahead for knowledge management: an AI perspective. AI Mag. 2000;21 17–17. [Google Scholar]
- 6.Lamberti M.J. A study on the application and use of artificial intelligence to support drug development. Clin. Ther. 2019;41:1414–1426. doi: 10.1016/j.clinthera.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 7.Beneke F., Mackenrodt M.-O. Artificial intelligence and collusion. IIC Int. Rev. Intellectual Property Competition Law. 2019;50:109–134. [Google Scholar]
- 8.Steels L., Brooks R. Routledge; 2018. The Artificial Life Route to Artificial Intelligence: Building Embodied, Situated Agents. [Google Scholar]
- 9.Bielecki A., Bielecki A. Foundations of artificial neural networks. In: Kacprzyk Janusz., editor. Models of Neurons and Perceptrons: Selected Problems and Challenges. Springer International Publishing; 2019. pp. 15–28. Polish academy of sciences, Warsaw, Poland. [Google Scholar]
- 10.Kalyane D. Artificial intelligence in the pharmaceutical sector: current scene and future prospect. In: Tekade Rakesh K., editor. The Future of Pharmaceutical Product Development and Research. Elsevier; 2020. pp. 73–107. [Google Scholar]
- 11.Da Silva I.N. Springer; 2017. Artificial Neural Networks. [Google Scholar]
- 12.Medsker L., Jain L.C. CRC Press; 1999. Recurrent Neural Networks: Design and Applications. [Google Scholar]
- 13.Hänggi M., Moschytz G.S. Springer Science & Business Media; 2000. Cellular Neural Networks: Analysis, Design and Optimization. [Google Scholar]
- 14.Rouse M. 2017. IBM Watson Supercomputer.https://searchenterpriseai.techtarget.com/definition/IBM-Watson-supercomputer . Accessed 13 October 2020. [Google Scholar]
- 15.Vyas M. Artificial intelligence: the beginning of a new era in pharmacy profession. Asian J. Pharm. 2018;12:72–76. [Google Scholar]
- 16.Duch W. Artificial intelligence approaches for rational drug design and discovery. Curr. Pharm. Des. 2007;13:1497–1508. doi: 10.2174/138161207780765954. [DOI] [PubMed] [Google Scholar]
- 17.Blasiak A. CURATE. AI: optimizing personalized medicine with artificial intelligence. SLAS Technol. 2020;25:95–105. doi: 10.1177/2472630319890316. [DOI] [PubMed] [Google Scholar]
- 18.Baronzio G. Overview of methods for overcoming hindrance to drug delivery to tumors, with special attention to tumor interstitial fluid. Front. Oncol. 2015;5:165. doi: 10.3389/fonc.2015.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mak K.-K., Pichika M.R. Artificial intelligence in drug development: present status and future prospects. Drug Discovery Today. 2019;24:773–780. doi: 10.1016/j.drudis.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Sellwood M.A. Artificial intelligence in drug discovery. Fut. Sci. 2018;10:2025–2028. doi: 10.4155/fmc-2018-0212. [DOI] [PubMed] [Google Scholar]
- 21.Zhu H. Big data and artificial intelligence modeling for drug discovery. Annu. Rev. Pharmacol. Toxicol. 2020;60:573–589. doi: 10.1146/annurev-pharmtox-010919-023324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciallella H.L., Zhu H. Advancing computational toxicology in the big data era by artificial intelligence: data-driven and mechanism-driven modeling for chemical toxicity. Chem. Res. Toxicol. 2019;32:536–547. doi: 10.1021/acs.chemrestox.8b00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan H.S. Advancing drug discovery via artificial intelligence. Trends Pharmacol. Sci. 2019;40(8):592–604. doi: 10.1016/j.tips.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Brown N. Royal Society of Chemistry; 2015. Silico Medicinal Chemistry: Computational Methods to Support Drug Design. [Google Scholar]
- 25.Pereira J.C. Boosting docking-based virtual screening with deep learning. J. Chem. Inf. Model. 2016;56:2495–2506. doi: 10.1021/acs.jcim.6b00355. [DOI] [PubMed] [Google Scholar]
- 26.Firth N.C. MOARF, an integrated workflow for multiobjective optimization: implementation, synthesis, and biological evaluation. J. Chem. Inf. Model. 2015;55:1169–1180. doi: 10.1021/acs.jcim.5b00073. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L. From machine learning to deep learning: progress in machine intelligence for rational drug discovery. Drug Discovery Today. 2017;22:1680–1685. doi: 10.1016/j.drudis.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Jain N. In silico de novo design of novel NNRTIs: a bio-molecular modelling approach. RSC Adv. 2015;5:14814–14827. [Google Scholar]
- 29.Wang Y. A comparative study of family-specific protein–ligand complex affinity prediction based on random forest approach. J. Comput.-Aided Mol. Des. 2015;29:349–360. doi: 10.1007/s10822-014-9827-y. [DOI] [PubMed] [Google Scholar]
- 30.King R.D. Comparison of artificial intelligence methods for modeling pharmaceutical QSARS. Appl. Artif. Intell. 1995;9:213–233. [Google Scholar]
- 31.Álvarez-Machancoses Ó, Fernández-Martínez J.L. Using artificial intelligence methods to speed up drug discovery. Expert Opin. Drug Discovery. 2019;14:769–777. doi: 10.1080/17460441.2019.1621284. [DOI] [PubMed] [Google Scholar]
- 32.Fleming N. How artificial intelligence is changing drug discovery. Nature. 2018;557 doi: 10.1038/d41586-018-05267-x. S55–S55. [DOI] [PubMed] [Google Scholar]
- 33.Dana D. Deep learning in drug discovery and medicine; scratching the surface. Molecules. 2018;23:2384. doi: 10.3390/molecules23092384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zang Q. In silico prediction of physicochemical properties of environmental chemicals using molecular fingerprints and machine learning. J. Chem. Inf. Model. 2017;57:36–49. doi: 10.1021/acs.jcim.6b00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X. Concepts of artificial intelligence for computer-assisted drug discovery. Chem. Rev. 2019;119:10520–10594. doi: 10.1021/acs.chemrev.8b00728. [DOI] [PubMed] [Google Scholar]
- 36.Hessler G., Baringhaus K.-H. Artificial intelligence in drug design. Molecules. 2018;23:2520. doi: 10.3390/molecules23102520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lusci A. Deep architectures and deep learning in chemoinformatics: the prediction of aqueous solubility for drug-like molecules. J. Chem. Inf. Model. 2013;53:1563–1575. doi: 10.1021/ci400187y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar R. Prediction of human intestinal absorption of compounds using artificial intelligence techniques. Curr. Drug Discovery Technol. 2017;14:244–254. doi: 10.2174/1570163814666170404160911. [DOI] [PubMed] [Google Scholar]
- 39.Rupp M. Estimation of acid dissociation constants using graph kernels. Mol. Inf. 2010;29:731–740. doi: 10.1002/minf.201000072. [DOI] [PubMed] [Google Scholar]
- 40.Chai S. A grand product design model for crystallization solvent design. Comput. Chem. Eng. 2020;135:106764. [Google Scholar]
- 41.Thafar M. Comparison study of computational prediction tools for drug–target binding affinities. Frontiers Chem. 2019;7:1–19. doi: 10.3389/fchem.2019.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Öztürk H. DeepDTA: deep drug–target binding affinity prediction. Bioinformatics. 2018;34:i821–i829. doi: 10.1093/bioinformatics/bty593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lounkine E. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahmud S.H. iDTi-CSsmoteB: identification of drug–target interaction based on drug chemical structure and protein sequence using XGBoost with over-sampling technique SMOTE. IEEE Access. 2019;7:48699–48714. [Google Scholar]
- 45.Gao K.Y. Interpretable drug target prediction using deep neural representation. In: Lang Jérôme., editor. Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence; IJCAI; 2018. pp. 3371–3377. [Google Scholar]
- 46.Feng Q. Padme: a deep learning-based framework for drug–target interaction prediction. arXiv. 2018 arXiv:1807.09741. [Google Scholar]
- 47.Karimi M. DeepAffinity: interpretable deep learning of compound–protein affinity through unified recurrent and convolutional neural networks. Bioinformatics. 2019;35(18):3329–3338. doi: 10.1093/bioinformatics/btz111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pu L. eToxPred: a machine learning-based approach to estimate the toxicity of drug candidates. BMC Pharmacol. Toxicol. 2019;20:2. doi: 10.1186/s40360-018-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayr A. DeepTox: toxicity prediction using deep learning. Frontiers Environ. Sci. 2016;3:80. [Google Scholar]
- 50.Basile A.O., Yahi A., Tatonetti N.P. Artificial intelligence for drug toxicity and safety. Trends Pharmacol. Sci. 2019;40(September (9)):624–635. doi: 10.1016/j.tips.2019.07.005. Epub 2019 Aug 2. PMID: 31383376; PMCID: PMC6710127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lysenko A. An integrative machine learning approach for prediction of toxicity-related drug safety. Life Sci. Alliance. 2018;1:YYY–ZZZ. doi: 10.26508/lsa.201800098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Basile A.O. Artificial intelligence for drug toxicity and safety. Trends Pharmacol. Sci. 2019;40:624–635. doi: 10.1016/j.tips.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gayvert K.M. A data-driven approach to predicting successes and failures of clinical trials. Cell Chem. Biolo. 2016;23:1294–1301. doi: 10.1016/j.chembiol.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jimenez-Carretero D. Tox_(R) CNN: deep learning-based nuclei profiling tool for drug toxicity screening. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan F., Zeng J. Deep learning with feature embedding for compound–protein interaction prediction. bioRxiv. 2016;2016 [Google Scholar]
- 56.AlQuraishi M. End-to-end differentiable learning of protein structure. Cell Syst. 2019;8:292–301. doi: 10.1016/j.cels.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hutson M. AI protein-folding algorithms solve structures faster than ever. Nature. 2019;XX:YYY–ZZZ. doi: 10.1038/d41586-019-01357-6. [DOI] [PubMed] [Google Scholar]
- 58.Avdagic Z. Artificial intelligence in prediction of secondary protein structure using CB513 database. Summit Transl. Bioinf. 2009;2009:1. [PMC free article] [PubMed] [Google Scholar]
- 59.Tian K. Boosting compound-protein interaction prediction by deep learning. Methods. 2016;110:64–72. doi: 10.1016/j.ymeth.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 60.Wang F. Computational screening for active compounds targeting protein sequences: methodology and experimental validation. J. Chem. Inf. Model. 2011;51:2821–2828. doi: 10.1021/ci200264h. [DOI] [PubMed] [Google Scholar]
- 61.Yu H. A systematic prediction of multiple drug–target interactions from chemical, genomic, and pharmacological data. PLoS One. 2012;7:e37608. doi: 10.1371/journal.pone.0037608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao X. iDrug-Target: predicting the interactions between drug compounds and target proteins in cellular networking via benchmark dataset optimization approach. J. Biomol. Struct. Dyn. 2015;33:2221–2233. doi: 10.1080/07391102.2014.998710. [DOI] [PubMed] [Google Scholar]
- 63.Persidis A. The benefits of drug repositioning. Drug Discov. World. 2011;12:9–12. [Google Scholar]
- 64.Koromina M. Rethinking drug repositioning and development with artificial intelligence, machine learning, and omics. Omics. 2019;23:539–548. doi: 10.1089/omi.2019.0151. [DOI] [PubMed] [Google Scholar]
- 65.Park K. A review of computational drug repurposing. Transl. Clin. Pharmacol. 2019;27:59–63. doi: 10.12793/tcp.2019.27.2.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeng X. Target identification among known drugs by deep learning from heterogeneous networks. Chem. Sci. 2020;11:1775–1797. doi: 10.1039/c9sc04336e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Achenbach J. Computational tools for polypharmacology and repurposing. Fut. Med. Chem. 2011;3:961–968. doi: 10.4155/fmc.11.62. [DOI] [PubMed] [Google Scholar]
- 68.Yi-Yu K. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed. J. 2020;43:355–362. doi: 10.1016/j.bj.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X. Prediction of synergistic anticancer drug combinations based on drug target network and drug induced gene expression profiles. Artif. Intell. Med. 2017;83:35–43. doi: 10.1016/j.artmed.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 70.Reddy A.S., Zhang S. Polypharmacology: drug discovery for the future. Expert Rev. Clin. Pharmacol. 2013;6:41–47. doi: 10.1586/ecp.12.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Z. KinomeX: a web application for predicting kinome-wide polypharmacology effect of small molecules. Bioinformatics. 2019;35:5354–5356. doi: 10.1093/bioinformatics/btz519. [DOI] [PubMed] [Google Scholar]
- 72.Cyclica . Cyclica; 2017. Cyclica Launches Ligand Express™, a Disruptive Cloud–Based Platform to Revolutionize Drug Discovery. [Google Scholar]
- 73.Corey E., Wipke W.T. Computer-assisted design of complex organic syntheses. Science. 1969;166:178–192. doi: 10.1126/science.166.3902.178. [DOI] [PubMed] [Google Scholar]
- 74.Grzybowski B.A. Chematica: a story of computer code that started to think like a chemist. Chem. 2018;4:390–398. [Google Scholar]
- 75.Klucznik T. Efficient syntheses of diverse, medicinally relevant targets planned by computer and executed in the laboratory. Chem. 2018;4:522–532. [Google Scholar]
- 76.Segler M.H. Planning chemical syntheses with deep neural networks and symbolic AI. Nature. 2018;555:604–610. doi: 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- 77.Putin E. Reinforced adversarial neural computer for de novo molecular design. J. Chem. Inform. Modeling. 2018;58:1194–1204. doi: 10.1021/acs.jcim.7b00690. [DOI] [PubMed] [Google Scholar]
- 78.Segler M.H. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 2018;4:120–131. doi: 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Popova M. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018;4:eaap7885. doi: 10.1126/sciadv.aap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Merk D. De novo design of bioactive small molecules by artificial intelligence. Mol. Inf. 2018;37:1700153. doi: 10.1002/minf.201700153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schneider G., Clark D.E. Automated de novo drug design: are we nearly there yet? Angew. Chem. 2019;131:10906–10917. doi: 10.1002/anie.201814681. [DOI] [PubMed] [Google Scholar]
- 82.Guo M. A prototype intelligent hybrid system for hard gelatin capsule formulation development. Pharm. Technol. 2002;6:44–52. doi: 10.1208/pt060356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mehta C.H. Computational modeling for formulation design. Drug Discovery Today. 2019;24:781–788. doi: 10.1016/j.drudis.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 84.Zhao C. Toward intelligent decision support for pharmaceutical product development. J. Pharm. Innovation. 2006;1:23–35. [Google Scholar]
- 85.Rantanen J., Khinast J. The future of pharmaceutical manufacturing sciences. J. Pharm. Sci. 2015;104:3612–3638. doi: 10.1002/jps.24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ketterhagen W.R. Process modeling in the pharmaceutical industry using the discrete element method. J. Pharm. Sci. 2009;98:442–470. doi: 10.1002/jps.21466. [DOI] [PubMed] [Google Scholar]
- 87.Chen W. Mathematical model-based accelerated development of extended-release metformin hydrochloride tablet formulation. AAPS PharmSciTech. 2016;17:1007–1013. doi: 10.1208/s12249-015-0423-9. [DOI] [PubMed] [Google Scholar]
- 88.Meziane F. Intelligent systems in manufacturing: current developments and future prospects. Integr. Manuf. Syst. 2000;11:218–238. [Google Scholar]
- 89.Steiner S. Organic synthesis in a modular robotic system driven by a chemical programming language. Science. 2019;363:eaav2211. doi: 10.1126/science.aav2211. [DOI] [PubMed] [Google Scholar]
- 90.Faure A. Process control and scale-up of pharmaceutical wet granulation processes: a review. Eur. J. Pharm. Biopharm. 2001;52:269–277. doi: 10.1016/s0939-6411(01)00184-9. [DOI] [PubMed] [Google Scholar]
- 91.Landin M. Artificial intelligence tools for scaling up of high shear wet granulation process. J. Pharm. Sci. 2017;106:273–277. doi: 10.1016/j.xphs.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 92.Das M.K., Chakraborty T. ANN in pharmaceutical product and process development. In: Puri Munish., editor. Artificial Neural Network for Drug Design, Delivery and Disposition. Elsevier; 2016. pp. 277–293. [Google Scholar]
- 93.Gams M. Integrating artificial and human intelligence into tablet production process. AAPS PharmSciTech. 2014;15:1447–1453. doi: 10.1208/s12249-014-0174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kraft, D.L. System and methods for the production of personalized drug products. US20120041778A1.
- 95.Aksu B. A quality by design approach using artificial intelligence techniques to control the critical quality attributes of ramipril tablets manufactured by wet granulation. Pharm. Dev. Technol. 2013;18:236–245. doi: 10.3109/10837450.2012.705294. [DOI] [PubMed] [Google Scholar]
- 96.Goh W.Y. Application of a recurrent neural network to prediction of drug dissolution profiles. Neural Comput. Appl. 2002;10:311–317. [Google Scholar]
- 97.Drăgoi E.N. On the use of artificial neural networks to monitor a pharmaceutical freeze-drying process. Drying Technol. 2013;31:72–81. [Google Scholar]
- 98.Reklaitis R. PharmaHub; 2008. Towards Intelligent Decision Support for Pharmaceutical Product Development. [Google Scholar]
- 99.Wang X. 2009 International Conference on Computational Intelligence and Software Engineering. IEEE; 2009. Intelligent quality management using knowledge discovery in databases; pp. 1–4. [Google Scholar]
- 100.Hay M. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 101.Harrer S. Artificial intelligence for clinical trial design. Trends Pharmacol. Sci. 2019;40:577–591. doi: 10.1016/j.tips.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 102.Fogel D.B. Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: a review. Contemp. Clin. Trials Commun. 2018;11:156–164. doi: 10.1016/j.conctc.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kalafatis S.P. Positioning strategies in business markets. J. Bus. Ind. Marketing. 2000;15:416–437. [Google Scholar]
- 104.Jalkala A.M., Keränen J. Brand positioning strategies for industrial firms providing customer solutions. J. Bus. Ind. Marketing. 2014;29:253–264. [Google Scholar]
- 105.Ding M. Springer; 2016. Innovation and Marketing in the Pharmaceutical Industry. [Google Scholar]
- 106.Dou W. Brand positioning strategy using search engine marketing. Mis Quarterly. 2010:261–279. [Google Scholar]
- 107.Chiu C.-Y. An intelligent market segmentation system using k-means and particle swarm optimization. Expert Syst. Appl. 2009;36:4558–4565. [Google Scholar]
- 108.Toker D. A decision model for pharmaceutical marketing and a case study in Turkey. Ekonomska Istraživanja. 2013;26:101–114. [Google Scholar]
- 109.Singh J. Sales profession and professionals in the age of digitization and artificial intelligence technologies: concepts, priorities, and questions. J. Pers. Selling Sales Manage. 2019;39:2–22. [Google Scholar]
- 110.Milgrom P.R., Tadelis S. National Bureau of Economic Research; 2018. How Artificial Intelligence and Machine Learning Can Impact Market Design. [Google Scholar]
- 111.Davenport T. How artificial intelligence will change the future of marketing. J. Acad. Marketing Sci. 2020;48:24–42. [Google Scholar]
- 112.Syam N., Sharma A. Waiting for a sales renaissance in the fourth industrial revolution: machine learning and artificial intelligence in sales research and practice. Ind. Marketing Manage. 2018;69:135–146. [Google Scholar]
- 113.Mahajan K.N., Kumar A. Business intelligent smart sales prediction analysis for pharmaceutical distribution and proposed generic model. Int. J. Comput. Sci. Inform. Technol. 2017;8:407–412. [Google Scholar]
- 114.Duran O. Neural networks for cost estimation of shell and tube heat exchangers. Expert Syst. Appl. 2009;36:7435–7440. [Google Scholar]
- 115.Park Y. A literature review of factors affecting price and competition in the global pharmaceutical market. Value Health. 2016;19:A265. [Google Scholar]
- 116.de Jesus A. Emerj; 2019. AI for Pricing – Comparing 5 Current Applications. [Google Scholar]
- 117.Hassanzadeh P. The significance of artificial intelligence in drug delivery system design. Adv. Drug Delivery Rev. 2019;151:169–190. doi: 10.1016/j.addr.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 118.Luo M. Micro‐/nanorobots at work in active drug delivery. Adv. Funct. Mater. 2018;28:1706100. [Google Scholar]
- 119.Fu J., Yan H. Controlled drug release by a nanorobot. Nat. Biotechnol. 2012;30:407–408. doi: 10.1038/nbt.2206. [DOI] [PubMed] [Google Scholar]
- 120.Calzolari D. Search algorithms as a framework for the optimization of drug combinations. PLoS Comput. Biol. 2008;4:e1000249. doi: 10.1371/journal.pcbi.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wilson B., KM G. Artificial intelligence and related technologies enabled nanomedicine for advanced cancer treatment. Future Med. 2020;15:433–435. doi: 10.2217/nnm-2019-0366. [DOI] [PubMed] [Google Scholar]
- 122.Tsigelny I.F. Artificial intelligence in drug combination therapy. Brief. Bioinform. 2019;20:1434–1448. doi: 10.1093/bib/bby004. [DOI] [PubMed] [Google Scholar]
- 123.Mason D.J. Using machine learning to predict synergistic antimalarial compound combinations with novel structures. Front. Pharmacol. 2018;9:1096. doi: 10.3389/fphar.2018.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ho D. Artificial intelligence in nanomedicine. Nanoscale Horiz. 2019;4:365–377. doi: 10.1039/c8nh00233a. [DOI] [PubMed] [Google Scholar]
- 125.Sacha G.M., Varona P. Artificial intelligence in nanotechnology. Nanotechnology. 2013;24:452002. doi: 10.1088/0957-4484/24/45/452002. [DOI] [PubMed] [Google Scholar]
- 126.Pellat, G. and Anghelache, C. (Year) Governance in the EU Member States in the Era of Big Data, Publisher.
- 127.Research and Markets . Research and Markets; 2019. Global Growth Insight - Role of AI in the Pharmaceutical Industry 2018-2022: Exploring Key Investment Trends, Companies-to-Action, and Growth Opportunities for AI in the Pharmaceutical Industry. [Google Scholar]
- 128.Jämsä-Jounela S.-L. Future trends in process automation. Annu. Rev. Control. 2007;31:211–220. [Google Scholar]
- 129.Davenport T.H., Ronanki R. Artificial intelligence for the real world. Harvard Bus. Rev. 2018;96:108–116. [Google Scholar]