Significance
Uncovering the role of defective metabolism in Parkinson’s disease (PD) may lead to the discovery of disease-modifying therapies. We recently linked branched-chain amino acid (BCAA) metabolism with PD, yet the underlying mechanisms were unknown. We now report the unexpected finding that BCAA metabolic dysfunction causes Parkinson’s-like motor deficits and neurodegeneration by inducing a state of hyperactive mitochondria. We found that the type 2 diabetes medication metformin is able to rescue neuronal viability by reducing mitochondrial respiration. These results offer mitochondrial hyperactivity as a new potential mechanism of mitochondrial dysfunction in Parkinson’s disease, and suggest that efforts to reduce mitochondrial respiration early in the disease—potentially by metformin treatment—may be efficacious.
Keywords: Parkinson’s disease, metformin, mitochondria, branched-chain amino acid metabolism, Caenorhabditis elegans
Abstract
Metabolic dysfunction occurs in many age-related neurodegenerative diseases, yet its role in disease etiology remains poorly understood. We recently discovered a potential causal link between the branched-chain amino acid transferase BCAT-1 and the neurodegenerative movement disorder Parkinson’s disease (PD). RNAi-mediated knockdown of Caenorhabditis elegans bcat-1 is known to recapitulate PD-like features, including progressive motor deficits and neurodegeneration with age, yet the underlying mechanisms have remained unknown. Using transcriptomic, metabolomic, and imaging approaches, we show here that bcat-1 knockdown increases mitochondrial respiration and induces oxidative damage in neurons through mammalian target of rapamycin-independent mechanisms. Increased mitochondrial respiration, or “mitochondrial hyperactivity,” is required for bcat-1(RNAi) neurotoxicity. Moreover, we show that post–disease-onset administration of the type 2 diabetes medication metformin reduces mitochondrial respiration to control levels and significantly improves both motor function and neuronal viability. Taken together, our findings suggest that mitochondrial hyperactivity may be an early event in the pathogenesis of PD, and that strategies aimed at reducing mitochondrial respiration may constitute a surprising new avenue for PD treatment.
Parkinson’s disease (PD) is the most common neurodegenerative movement disorder worldwide, and its prevalence is predicted to significantly increase as the population ages (1, 2). PD is characterized by a loss of dopaminergic neurons in the substantia nigra and the formation of pathological inclusions containing aggregated α-synuclein protein (3, 4). The mechanisms that induce neurodegeneration in PD are still poorly understood, and 90% of cases have no known cause (5).
In an effort to identify novel PD genes, we developed diseaseQUEST, a method that combines human genome-wide association studies with tissue-specific functional genetic networks and high-throughput behavioral screening in Caenorhabditis elegans (6). Using this computational-experimental framework, we discovered a link between PD and branched-chain amino acid transferase 1 (BCAT-1), which catalyzes the first step in branched-chain amino acid (BCAA) catabolism (7). We found that BCAT-1 expression is normally high in PD-susceptible regions of the healthy human brain, and that its expression is reduced in the substantia nigra of sporadic PD patients (6). While these findings suggest a correlation between defective BCAA metabolism and PD, animal models are needed to determine causality and underlying mechanisms.
The nematode C. elegans offers a unique system in which to study age-related neurologic disease due to its short lifespan and highly conserved nervous system signaling that gives rise to complex behaviors. Moreover, the worm is highly amenable to high-throughput screening approaches, allowing rapid testing of potential disease treatments (8). We previously showed that RNAi-mediated reduction of neuronal bcat-1 in C. elegans causes progressive, age-dependent motor dysfunction and accelerates dopamine neuron degeneration in worms expressing human α-synuclein (6), yet the underlying mechanisms have remained unknown. Here we used tissue-specific transcriptomics, high-resolution metabolomics, and functional imaging in aged worms and found that increased mitochondrial respiration leads to PD phenotypes. The type 2 diabetes medication metformin restores normal mitochondrial respiration levels, reduces neurodegeneration, and improves motor function, even with administration after disease onset. Our work offers mitochondrial hyperactivity as a potential mechanism underlying PD phenotypes, and supports the controlled reduction of mitochondrial respiration as a surprising potential treatment strategy for PD.
Results
Neuronal Transcriptional Analysis Reveals an Up-Regulation of Glycolysis and TCA Cycle Pathways in the bcat-1(RNAi) C. elegans Model of PD.
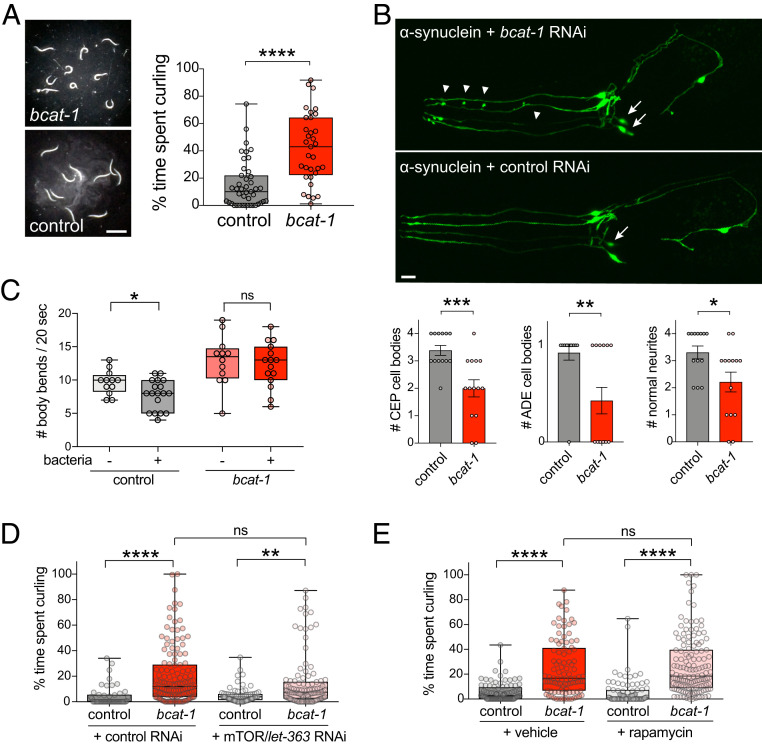
To model the reduction of BCAT-1 that was observed in the substantia nigra of patients with PD (6), we performed adult-specific RNAi-mediated knockdown of bcat-1 in C. elegans to avoid embryonic/larval lethality (9). Knockdown of bcat-1 exclusively in the nervous system (sid-1;Punc-119::sid-1) caused severe, spasm-like “curling” motor dysfunction in aged (day 8 adult) animals (Fig. 1A). By this timepoint, dopamine neurons were almost entirely lost in neuronal RNAi-sensitive worms expressing dopaminergic-specific α-synuclein (SI Appendix, Fig. S1A) consistent with previous reports (10); however, examination at the earlier timepoint of day 6 revealed that bcat-1 knockdown exacerbated the loss of cell bodies and the fragmentation and blebbing of neurites (Fig. 1B). Dopaminergic degeneration is associated with a loss of basal slowing response in C. elegans, in which worms normally slow down in the presence of a bacterial food source (11). Consistent with the observed structural damage (Fig. 1B), bcat-1 knockdown eliminated the worms’ basal slowing response (Fig. 1C).
Fig. 1.
bcat-1 knockdown causes motor dysfunction and promotes neurodegeneration via mTOR-independent mechanisms. (A) Adult- and neuron-specific RNAi-mediated bcat-1 knockdown results in severe spasm-like “curling” motor dysfunction in aged (day 8 adult) animals. n = 43 for control, 33 for bcat-1(RNAi). Two-tailed t test. (Scale bar: 1 mm.) (B) In neuronal RNAi-sensitive worms expressing α-synuclein in dopaminergic neurons, bcat-1 knockdown accelerates degeneration of CEP and ADE dopaminergic cell bodies (arrows) and neurites (arrowheads) measured on day 6. n = 13 for control, 14 for bcat-1(RNAi). Two-tailed t test. Data are mean ± SEM. (Scale bar: 10 μm.) (C) While dopaminergic-dependent basal slowing behavior is still intact on day 5 in worms expressing α-synuclein in dopaminergic neurons, it is absent with additional knockdown of bcat-1. n = 12 each for (−)bacteria, 18 for control (+)bacteria, 15 for bcat-1 (+)bacteria. Two-tailed t tests. (D and E) Curling on day 8 is unaffected by mTOR/let-363 knockdown (D) or rapamycin treatment (E) in neuronal RNAi-sensitive worms (strain TU3311) with bcat-1 knockdown. In D, n = 48 for control, 134 for bcat-1 control, 72 for control let-363, 106 for bcat-1 let-363. Two-way ANOVA with Tukey’s post hoc test. In E, n = 92 for control vehicle, 97 for bcat-1 vehicle, 87 for control rapamycin, 139 for bcat-1 rapamycin. Two-way ANOVA with Tukey’s post hoc test. ns, not significant. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Boxplots show minimum, 25th percentile, median, 75th percentile, and maximum.
To understand how bcat-1 reduction causes neurodegeneration and motor deficits, we first investigated the mammalian target of rapamycin (mTOR) pathway, which was previously implicated in bcat-1 regulation of lifespan (12). However, we found that neither loss of mTOR/let-363 nor rapamycin treatment had any effect on curling behavior, suggesting that the TOR pathway does not interact with bcat-1 to regulate PD-related phenotypes (Fig. 1 D and E). mTOR suppression did reduce body length (SI Appendix, Fig. S1B), consistent with the known role of mTOR in growth (13).
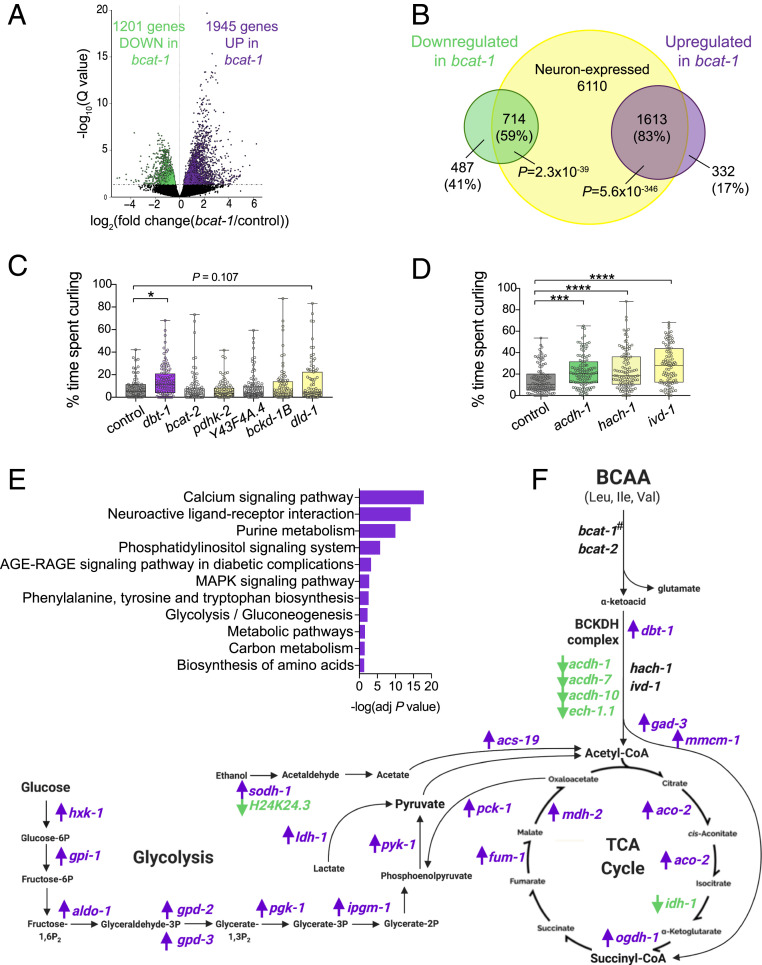
We next decided to take an unbiased and tissue-specific approach to uncover the mechanisms of bcat-1(RNAi) neurotoxicity, using our method for the isolation and RNA sequencing of adult C. elegans tissues (14). Neuronal RNAi-sensitive C. elegans were treated with bcat-1 or control RNAi, and neurons were collected on day 5 of adulthood, a timepoint that precedes severe motor dysfunction (6), to shed light on mechanisms that precede and drive disease rather than reporting transcriptional changes from dying neurons. bcat-1(RNAi)-treated samples and control RNAi-treated samples were distinct (SI Appendix, Fig. S2A), and the majority of the differentially expressed (false discovery rate < 0.05) genes (Fig. 2A and Dataset S1) (15) were previously identified as neuronally expressed in adult wild-type C. elegans (16) (Fig. 2B). Gene Ontology (GO) analysis also revealed that genes up-regulated in bcat-1(RNAi) neurons were largely neuronal, including such GO terms as chemical synaptic transmission, synapse organization, and regulation of neurotransmitter levels (SI Appendix, Fig. S2 B and C).
Fig. 2.
bcat-1-related PD phenotypes are associated with neuronal up-regulation of glycolysis and TCA cycle genes. Neurons were isolated on day 5 from neuronal RNAi-sensitive worms with adult-specific bcat-1 knockdown and were RNA-sequenced. (A) Volcano plot showing up-regulated (purple) and down-regulated (green) genes in bcat-1(RNAi) neurons (false discovery rate < 0.05). (B) Venn diagram confirming that the majority of differentially expressed genes are expressed in neurons. P values: hypergeometric distributions. (C and D) Adult-specific knockdown of BCAA pathway components in neuronal RNAi-sensitive worms (strain TU3311) shows that disruption of dbt-1 (C), acdh-1, hach-1, or ivd-1 (D) caused curling on day 8. n = 70 in C and 99 in D for controls, 100 for dbt-1, 95 for bcat-2, 91 for pdhk-2, 111 for Y43F4A.4, 85 for bckd-1B, 90 for dld-1, 111 for acdh-1, 107 for hach-1, and 96 for ivd-1. One-way ANOVA with Dunnett’s post hoc test. (E) KEGG analysis of genes up-regulated in bcat-1(RNAi) neurons. (F) BCAA, glycolysis, and TCA cycle pathway schematic with genes up-regulated (purple) and down-regulated (green) in bcat-1(RNAi) neurons. #bcat-1 expression could not be measured due to RNA-sequencing of the bcat-1 RNAi itself. n = 5 independent collections/group. *P < 0.05; ***P < 0.001; ****P < 0.0001. Boxplots show minimum, 25th percentile, median, 75th percentile, and maximum.
Several genes involved in BCAA metabolism were differentially expressed in response to bcat-1 knockdown (Fig. 2F and Dataset S1) (15). To test whether disrupting these BCAA pathway components produces motor dysfunction similar to that produced by bcat-1(RNAi), neuronal RNAi-sensitive worms were fed RNAi as adults and tested for curling on day 8. The acyl-CoA dehydrogenase acdh-1 was the most down-regulated neuronal gene (log2 fold change, −2.44; Padj = 2.87 × 10−5), while dbt-1, which encodes the catalytic core of the branched chain α-ketoacid dehydrogenase complex (BCKDHC), was increased in bcat-1(RNAi) neurons (log2 fold change, 0.55; Padj = 0.04). Disruption of either acdh-1 or dbt-1 alone was sufficient to induce curling (Fig. 2 C and D). Since acdh-1 functions in valine and isoleucine metabolism but not in leucine metabolism (17), we also tested hach-1, a hydrolase specific to valine, and ivd-1, a dehydrogenase specific to leucine. In each case, knockdown was sufficient to produce curling (Fig. 2D), indicating that defects in the metabolism of any of the three BCAAs can produce motor dysfunction. Therefore, normal BCAA metabolism appears to be critical to neuronal maintenance with age.
Kyoto Encyclopedia of Genes and Genomes (KEGG) functional analysis of genes up-regulated in bcat-1(RNAi) neurons suggested enrichment of several metabolic pathways, including glycolysis/gluconeogenesis, carbon metabolism, metabolic pathways, and purine metabolism (Fig. 2E and SI Appendix, Fig. S2D). We further examined carbon metabolism, in particular glycolysis and the tricarboxylic acid (TCA) cycle, since BCAA metabolism is closely linked with these bioenergetic pathways (7) (Fig. 2F) and BCAT-1 localizes to mitochondria in C. elegans (18). The majority (19 of 25) of the differentially expressed genes in these pathways were up-regulated in bcat-1(RNAi) neurons (Fig. 2F), suggesting that bcat-1 knockdown may promote a coordinated transcriptional response to alterations in BCAA metabolism.
Mitochondrial Hyperactivity Coincides with TCA Cycle Depletion and Is Required for bcat-1(RNAi) Neurotoxicity.
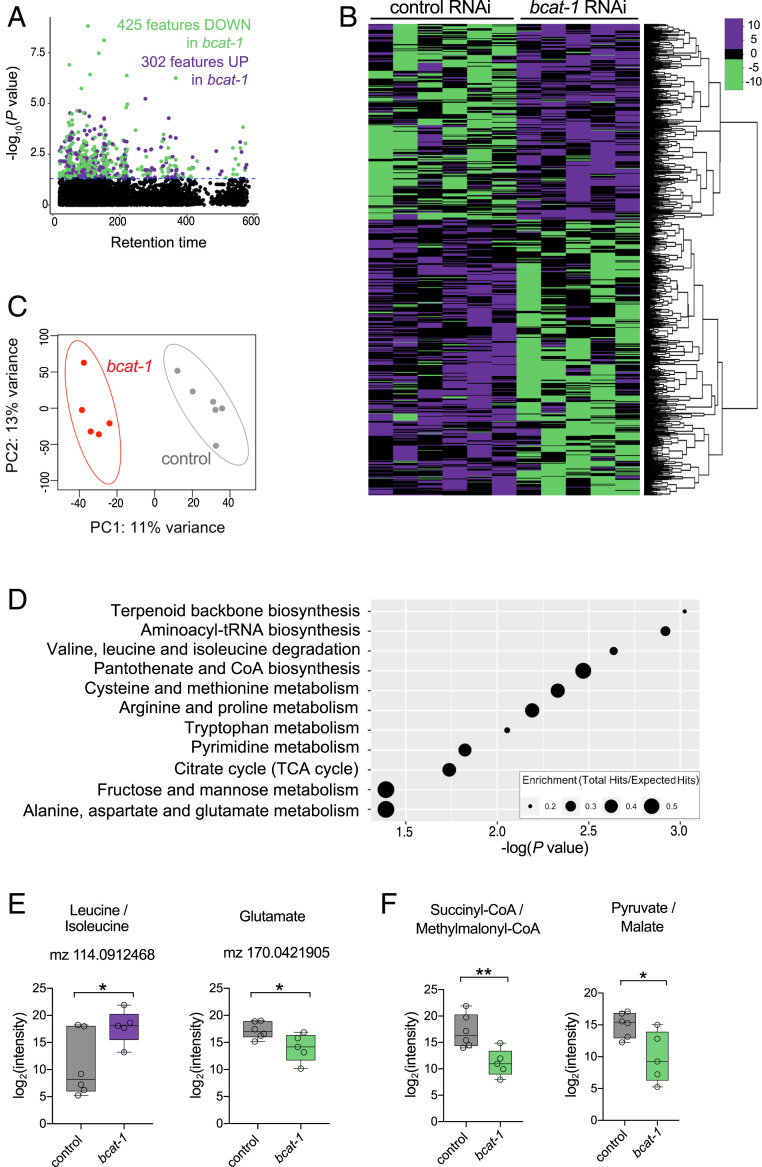
To better understand the effects of bcat-1 knockdown on metabolism, we next performed untargeted high-resolution metabolomics (19). Worms were fed bcat-1 or control RNAi as adults and collected on day 5, the same timepoint as our transcriptomics analysis. Significantly altered features (P < 0.05) were detected in bcat-1(RNAi) worms compared with control worms (Fig. 3 A and B and Dataset S2) (20), and partial least squares discriminant analysis showed distinct separation of the groups (Fig. 3C).
Fig. 3.
Metabolomics analysis reveals a depletion of TCA cycle metabolites in bcat-1(RNAi) worms. Neuronal RNAi-sensitive worms were collected for metabolomics on day 5 following adult-specific bcat-1 knockdown. (A and B) Manhattan plot (A) and heat map (B) showing features that were increased (purple) and decreased (green) (P < 0.05) in bcat-1(RNAi) worms. The color values in B represent relative intensities of features that have been log2-transformed and centered. (C) Partial least squares-discriminant analysis. (D) Pathway analysis using the mummichog algorithm. (E) As expected, features putatively annotated as leucine/isoleucine and glutamate were increased and decreased, respectively, in bcat-1(RNAi) worms. (F) TCA cycle metabolites/precursors were decreased in bcat-1(RNAi) worms. n = 6 independent collections for control, 5 for bcat-1. Two-tailed t tests. *P < 0.05; **P < 0.01. Boxplots show minimum, 25th percentile, median, 75th percentile, and maximum.
Several of the metabolic pathways that were significantly altered in bcat-1(RNAi) worms were related to BCAA metabolism, including valine, leucine, and isoleucine degradation and alanine, aspartate, and glutamate metabolism (Fig. 3D). As expected, features putatively annotated as leucine/isoleucine and glutamate were increased and decreased, respectively, in bcat-1(RNAi) worms (Fig. 3E and SI Appendix, Fig. S3). Consistent with our transcriptomics analysis, the citrate cycle (TCA cycle) was also a significantly altered metabolic pathway in bcat-1(RNAi) worms (Fig. 3D), suggesting that our transcriptomics and metabolomics analyses converge on the TCA cycle. However, while the expression of TCA cycle genes was largely increased (Fig. 2F), the levels of the annotated TCA cycle substrates and precursors pyruvate/malate and succinyl-CoA/methylmalonyl-CoA were significantly decreased (Fig. 3F). These findings suggest that bcat-1 knockdown reduces steady-state levels of TCA cycle metabolites, whereas expression of glycolysis and TCA cycle enzymes is increased.
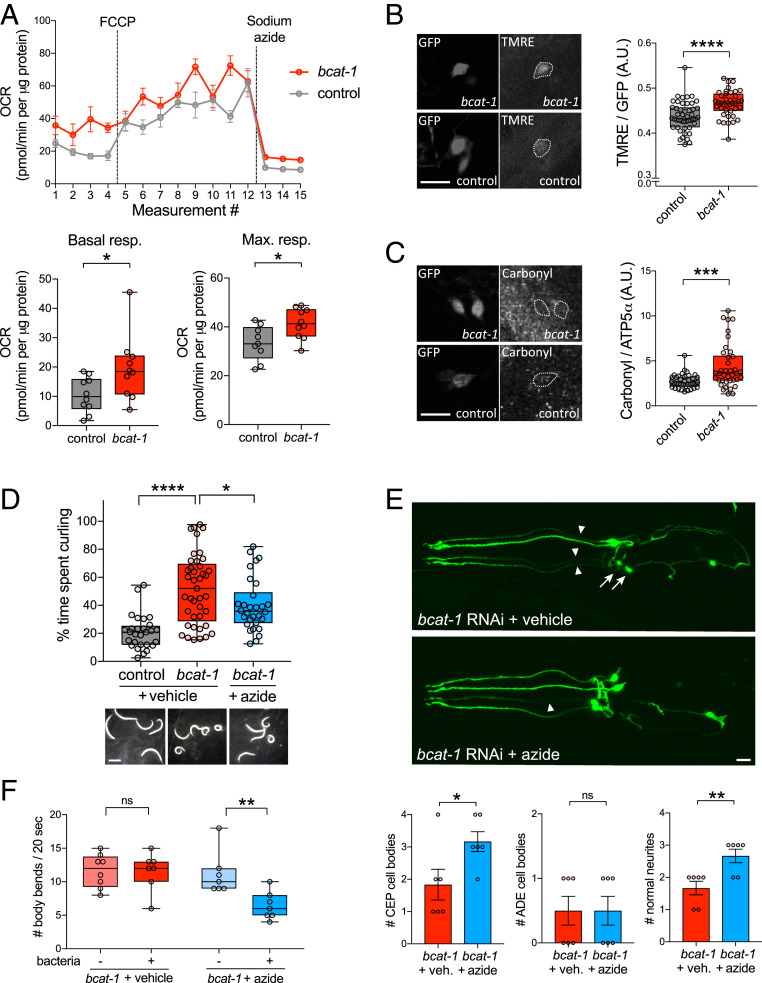
It is possible that low levels of TCA cycle metabolites leads to a reduction in mitochondrial respiration. Alternatively, low steady-state levels of metabolites may reflect greater TCA cycle turnover driven by more abundant components, which would result in higher levels of mitochondrial respiration. To distinguish between these possibilities, we measured mitochondrial activity in worms with bcat-1 knockdown. The oxygen consumption rate (OCR) was assayed on day 5, and both basal and maximal respiration rates were found to be significantly increased in bcat-1(RNAi) worms (Fig. 4A). Since OCR is necessarily measured in whole worms (21), and bcat-1 knockdown in nonneuronal tissues also increases OCR (SI Appendix, Fig. S4A), we next chose to specifically examine mitochondrial function in dopaminergic neurons. Using tetramethylrhodamine ethyl ester (TMRE) dye, which accumulates only in actively respiring mitochondria, we found that dopaminergic neurons expressing α-synuclein had increased TMRE fluorescence intensity with bcat-1 knockdown (Fig. 4B and SI Appendix, Fig. S4 B and C). These data support the conclusion that bcat-1 reduction leads to mitochondrial hyperactivity.
Fig. 4.
bcat-1(RNAi) neurotoxicity is dependent on increased mitochondrial respiration and is associated with oxidative damage. (A) Mitochondrial respiration was increased on day 5 in neuronal RNAi-sensitive bcat-1(RNAi) worms (strain CF512). n = 10 wells totaling 102 worms for control basal, 9 wells totaling 92 worms for control maximum, 10 wells totaling 100 worms per bcat-1 condition. Two-tailed t tests. max., maximal; resp., respiration. (B) Mitochondrial activity, as measured by TMRE fluorescence, was increased on day 5 in CEP α-synuclein–expressing dopaminergic neurons with bcat-1 knockdown. n = 15 worms totaling 47 CEPs for control, 11 worms totaling 39 CEPs for bcat-1. Two-tailed t test. (Scale bar: 10 μm.) (C) Protein carbonylation was increased on day 8 in CEP α-synuclein–expressing dopaminergic neurons with bcat-1 knockdown. (Scale bar, 10 μm.) n = 10 worms totaling 37 CEPs for control, 10 worms totaling 38 CEPs for bcat-1. Two-tailed t test. (D) bcat-1(RNAi) worms (strain CF512) treated with 100 μM sodium azide starting on day 4 had reduced curling motor dysfunction on day 8. n = 28 worms for control, 41 for bcat-1+vehicle, 30 for bcat-1+azide. One-way ANOVA with Tukey’s post hoc test. (Scale bar: 0.5 mm.) (E) Azide treatment starting on day 4 reduced neurodegeneration of α-synuclein–expressing CEP dopaminergic cell bodies (arrows) and neurites (arrowheads) with bcat-1 knockdown, as measured on day 8. n = 6 worms per group. Two-tailed t tests. Data are mean ± SEM. veh., vehicle. (Scale bar: 10 μm.) (F) Azide treatment starting on day 3 restored basal slowing on day 5 in bcat-1(RNAi) worms expressing α-synuclein in dopaminergic neurons. n = 7 per group except n = 8 for bcat-1 + vehicle (−)bacteria. Two-tailed t tests. A.U., arbitrary units. ns, not significant. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Boxplots show minimum, 25th percentile, median, 75th percentile, and maximum.
Since mitochondrial respiration produces reactive oxygen species, we next tested for evidence of oxidative damage. Protein carbonylation levels were measured at the day 8 endpoint in α-synuclein–expressing dopaminergic neurons with or without bcat-1 knockdown. Indeed, increased mitochondrial respiration was correlated with an eventual increase in levels of carbonylated proteins (Fig. 4C and SI Appendix, Fig. S4D), suggesting that disruption of bcat-1 dysregulates the cellular redox state such that neurons incur protein damage.
To further test whether increased mitochondrial respiration is required for bcat-1(RNAi) neurotoxicity, we exposed worms to 100 μM sodium azide, which inhibits complexes IV and V of the electron transport chain. This dose is 400-fold lower than the typical concentration used to immobilize worms (21), and indeed we found no difference in the total activity levels (activity index) of azide-treated bcat-1(RNAi) worms compared with controls (SI Appendix, Fig. S4E). However, azide treatment did significantly improve the motor function of bcat-1(RNAi) worms, in terms of both curling level and body wave number (Fig. 4D and SI Appendix, Fig. S4E). Moreover, azide treatment reduced the degeneration of CEP dopaminergic neurons and neurites (Fig. 4E) and restored the basal slowing response in bcat-1(RNAi) worms (Fig. 4F). These results strongly suggest that mitochondrial hyperactivity is a causative factor in bcat-1–associated PD phenotypes.
Metformin Reduces Neurodegeneration and Restores Normal Levels of Mitochondrial Respiration.
While disease mechanisms in PD are thought to involve decreased mitochondrial function (22), our findings instead suggest that elevated mitochondrial respiration may also contribute to PD pathogenesis, potentially preceding an ultimate loss of mitochondrial function. Thus, strategies to restore mitochondrial homeostasis early in the disease may be efficacious. We recently identified the type 2 diabetes medication metformin as one of the top-performing candidates in a high-throughput screen for drugs that improve motor function in bcat-1(RNAi) worms (23). Among several known targets, metformin can act as a complex I inhibitor (24), raising the possibility that reducing mitochondrial respiration may constitute a surprising new treatment avenue for PD.
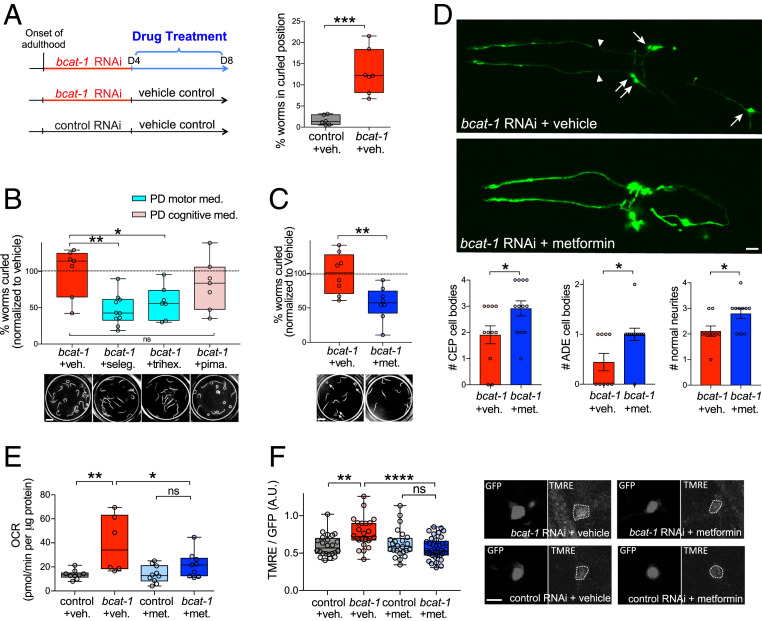
To investigate the neuroprotective action of metformin and its effects on mitochondria in the bcat-1(RNAi) PD worm model, we used the experimental paradigm established in our high-throughput drug screen (23). Neuronal RNAi-sensitive worms were fed bcat-1 or control RNAi as adults and on day 4 were transferred to plates seeded with heat-killed OP50 Escherichia coli and 50 μM drug or vehicle (0.05% DMSO) (Fig. 5A). The use of heat-killed bacteria eliminates any potential confounding effects of bacterial metabolism of the drugs, as has been documented for metformin (25). In addition, the day 4 timepoint for drug intervention is post-disease onset (6), mimicking the current clinical landscape in which PD diagnosis and treatment occurs well into the course of disease (26). On day 8, curling was quantified using our automated system (23). Exposure to bcat-1 RNAi until day 4, followed by vehicle treatment with heat-killed bacteria was sufficient to produce significant curling on day 8 (Fig. 5A). Using this paradigm, two PD medications currently prescribed for motor symptoms significantly reduced curling, whereas a PD drug indicated for cognitive symptoms had no effect (Fig. 5B). These data support the utility of this paradigm for new potential PD drug discoveries, as well as use of the curling phenotype as a model for motility defects in PD.
Fig. 5.
Metformin reduces neurodegeneration and restores normal levels of mitochondrial respiration in bcat-1(RNAi) worms. (A) Experimental design for drug treatments. Neuronal RNAi-sensitive worms (strain CF512) were fed bcat-1 or control RNAi as adults until day 4, then transferred to heat-killed OP50 E. coli and 50 μM drug or vehicle. Vehicle-treated bcat-1(RNAi) worms curled on day 8, as measured using our automated system. n = 6 wells totaling 91 worms for control, 7 wells totaling 57 worms for bcat-1. Two-tailed t test. (B) PD medications prescribed for motor symptoms (selegiline [seleg.] and trihexyphenidyl [trihex.]) reduced curling in bcat-1(RNAi) worms on day 8, whereas the antipsychotic pimavanserin (pima.) did not. n = 7 wells totaling 142 worms for bcat-1, 10 wells totaling 149 worms for selegiline, 7 wells totaling 86 worms for trihexyphenidyl, 7 wells totaling 103 worms for pimavanserin. One-way ANOVA with Dunnett’s post hoc test. (Scale bar: 1 mm.) (C) Metformin reduced curling (arrows) on day 8 in bcat-1(RNAi) worms expressing α-synuclein in dopaminergic neurons. n = 8 wells totaling 65 worms for vehicle, 8 wells totaling 121 worms for metformin. Two-tailed t test. (Scale bar: 1 mm.) (D) Metformin reduced neurodegeneration of α-synuclein–expressing dopaminergic cell bodies (arrows) and neurites (arrowheads) with bcat-1 knockdown, as measured on day 8. n = 11 for CEP vehicle, 12 each for CEP and ADE metformin, 9 each for ADE and neurites vehicle, 10 for neurites metformin. Two-tailed t tests. Data are mean ± SEM. (Scale bar: 10 μm.) (E) Metformin reduced basal mitochondrial respiration in bcat-1(RNAi) worms (strain CF512) on day 8. n = 9 wells totaling 144 worms for control vehicle, 6 wells totaling 50 worms for bcat-1 vehicle, 10 wells totaling 128 worms for control metformin, 9 wells totaling 113 worms for bcat-1 metformin. Two-way ANOVA with Tukey’s post hoc test. (F) Metformin reduced mitochondrial activity in α-synuclein–expressing CEP dopaminergic neurons with bcat-1 knockdown, as measured on day 6. n = 8 worms totaling 24 CEPs for control vehicle, 7 worms totaling 24 CEPs for bcat-1 vehicle, 9 worms totaling 24 CEPs for control metformin, 10 worms totaling 37 CEPs for bcat-1 metformin. Two-way ANOVA with Tukey’s post hoc. (Scale bar: 5 μm.) A.U., arbitrary units. met., metformin. ns, not significant. veh., vehicle. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Boxplots show minimum, 25th percentile, median, 75th percentile, and maximum.
To determine whether metformin rescues bcat-1–associated toxicity in worms with α-synuclein expression in dopaminergic neurons, we measured motor function and dopamine neuron degeneration. Worms treated with 50 μM metformin as described above (Fig. 5A) showed significantly reduced curling on day 8 (Fig. 5C). Remarkably, metformin also significantly rescued dopamine neuron viability. On day 8, the number of dopaminergic cell bodies and neurites with healthy morphologies was increased in metformin-treated bcat-1(RNAi) worms compared with vehicle-treated worms (Fig. 5D). Neuroprotection was detected as early as day 6, after only 2 d of metformin treatment (from day 4 to day 6), with dopaminergic neurons showing significantly improved neurite morphologies (SI Appendix, Fig. S5A).
We next asked whether metformin might exert its protective effects by suppressing bcat-1(RNAi)–driven mitochondrial hyperactivity. Indeed, we found that on day 8, metformin treatment had reduced both basal and maximal mitochondrial respiration down to levels of age-matched controls (Fig. 5E and SI Appendix, Fig. S5B). Similarly, using TMRE dye on day 6 (the latest timepoint for effective dye uptake), metformin treatment for only 2 d was already sufficient to restore normal mitochondrial activity levels in α-synuclein-expressing dopaminergic neurons of bcat-1(RNAi) worms (Fig. 5F and SI Appendix, Fig. S5 C and D). These data suggest that post-disease onset administration of metformin is able to correct aberrant mitochondrial respiration in the bcat-1(RNAi) worm model of PD.
Discussion
Here we demonstrate that defective BCAA metabolism recapitulates several major features of PD, including motor dysfunction and neurodegeneration. Consistent with this, the BCAA metabolic disorder maple syrup urine disease (MSUD), which arises from mutations in BCKDHC subunits, commonly presents with parkinsonian symptoms in adult patients (27). Neuronal loss in the substantia nigra has also been documented in MSUD (28). A meta-analysis of genome-wide association studies revealed associations between PD and the BCAA pathway genes methylcrotonoyl-CoA carboxylase 1 (MCCC1) and branched-chain ketoacid dehydrogenase kinase (BCKDK) (29), further implicating this pathway in disease risk. In addition, metabolomic profiling of PD patient urine revealed that BCAAs were elevated relative to controls, and that higher BCAA levels were correlated with greater disease severity (30). Nevertheless, how BCAA metabolic defects might induce PD-like symptoms, and how to abrogate these, remained unknown.
Our transcriptomics, metabolomics, respiration, and imaging approaches led to the surprising finding that bcat-1 reduction causes mitochondrial hyperactivity, consistent with reports from mice lacking the mitochondrial BCAT isoform (31). These findings appear to challenge the prevailing notion that mitochondrial function is solely decreased in PD, in particular due to complex I deficiency, which has been documented in peripheral tissues from PD patients (32) and postmortem patient brain samples (33). Critically, however, mitochondrial respiration levels in the brains of living PD patients remain unknown, and thus it is possible that an increase in respiration precedes the ultimate loss of function. Complex I inhibitors such as 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and rotenone are often used to model PD, and MPTP induces parkinsonism in humans (34). However, MPTP produces acute neurodegeneration that does not model the progressive nature of PD (35), and rotenone models suffer from issues of variability and subject mortality (36). While these compounds may model endpoint processes in the disease, our present study offers mitochondrial hyperactivity as a potential early pathological event in PD.
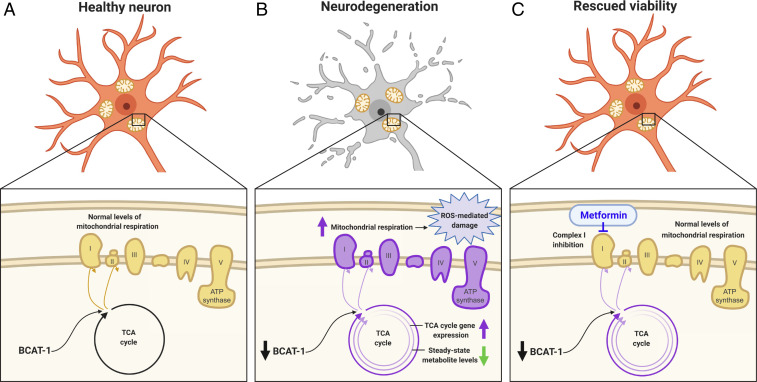
Our data support the model that loss of neuronal BCAT-1 causes neurodegeneration by increasing mitochondrial respiration and inducing oxidative damage (Fig. 6). BCAT-1 expression may naturally decrease with age, as has been shown in fish, worms, and mice (12), consistent with the age-dependence of PD symptom onset. The reduction in BCAT-1 causes an increase in TCA cycle gene expression and a depletion of TCA cycle metabolites (Fig. 6B), possibly due to greater TCA cycle turnover within individual mitochondria and/or increased mitochondrial biogenesis. Metformin treatment is able to restore normal levels of mitochondrial respiration and rescue neuronal viability, possibly through the inhibition of complex I (Fig. 6C). Collectively, our work supports a potential metabolic origin of PD and points to metformin as an existing Food and Drug Administration-approved drug with great promise for repurposing to PD.
Fig. 6.
Model. (A) In a healthy neuron, BCAT-1 normally provides metabolic input into the TCA cycle, giving rise to normal levels of mitochondrial respiration. (B) With a reduction of BCAT-1, TCA cycle gene expression is increased and steady-state levels of TCA cycle metabolites are decreased, possibly due to greater TCA cycle turnover. This leads to increased mitochondrial respiration and reactive oxygen species-mediated damage, ultimately causing neurodegeneration. (C) Metformin treatment restores mitochondrial homeostasis and rescues neuronal viability, potentially through complex I inhibition.
Materials and Methods
C. elegans Strains and Maintenance.
Worms were maintained at 20 °C on standard nematode growth medium (NGM) plates or high growth medium (HG) plates seeded with OP50 E. coli or HT115 RNAi E. coli, as indicated. The following strains were used in this study: wild-type worms of the N2 Bristol strain, neuron-only RNAi strain CQ511 [sid-1(pk3321)]; uIs69 [pCFJ90 (myo-2p::mCherry, unc-119p::sid-1)], and the following neuronal RNAi-sensitive strains: CF512 [fem-1(hc17); fer-15(b26)], CQ434 baIn11 [dat-1p::α-synuclein; dat-1p::gfp]; vIs69 [pCFJ90 (myo-2p::mCherry + unc-119p::sid-1)], and TU3311 uIs60 (unc-119p::sid-1, unc-119p::yfp).
RNAi and Drug Treatments.
Worms were synchronized from eggs by bleaching and plated onto HG plates seeded with OP50. At the L4 stage, worms were transferred to RNAi-seeded 100-mm NGM or HG plates containing carbenicillin and isopropyl β-d-1-thiogalactopyranoside (IPTG) that had been preinduced with 0.1 M IPTG at 1 h before transfer. Worms were transferred to fresh RNAi plates every 2 to 3 d. In all experiments, control RNAi refers to empty vector pL4440 in HT115 E. coli. For experiments with metformin or PD drugs, worms were transferred on day 4 to NGM plates seeded with 1 mL of heat-killed OP50 bacteria and 50 μM metformin (Sigma-Aldrich), 50 μM selegiline (MedChem Express), 50 μM trihexyphenidyl (MedChem Express), 50 μM pimavanserin (MedChem Express), or vehicle (0.5% DMSO). For sodium azide treatment, worms were transferred on day 3 or 4 (as indicated) to NGM plates seeded with 1 mL of heat-killed OP50 bacteria and 100 μM sodium azide or water. OP50 was killed by incubation at 65 °C for 30 min. For rapamycin experiments, rapamycin (LC Laboratories) was prepared in NGM medium at a final concentration of 100 μM, and worms were maintained on rapamycin or vehicle plates seeded with bcat-1 or control RNAi from L4 to day 8, with transfer to fresh plates every 2 to 3 d. As a positive control for let-363 RNAi phenotypic effects, worms were bleached onto NGM plates seeded with let-363 or control RNAi, and body length was measured on day 1 of adulthood using ImageJ software.
Curling and Other Motor Assays.
Manual quantification of individual worms’ curling level was performed as described previously (6). In brief, worms were picked into a drop of M9 buffer on a microscope slide, and 30-s videos were obtained. The percentage of time spent curling was quantified for each individual worm using a standard stopwatch. Automated curling analysis was performed as described previously (23). In brief, on the day of analysis, worms were rapidly washed twice with M9 buffer and dispensed into 96-well plates, and 30-s videos or a series of snapshots were obtained for individual wells containing 3 to 30 worms each. Our curling detection software quantified the number of worms in a curled position divided by the total number of worms detected, which was defined as the percentage of worms in a curled position. For body wave number and activity index, 30-s videos were obtained and analyzed by CeleST software as described previously (6, 37).
Neurodegeneration Assays.
Animals were mounted on 2% agarose pads in M9 and >40 mM sodium azide to immobilize worms. Images were obtained on a Nikon A1 confocal microscope at 40× magnification, and Z stacks were processed with Nikon NIS elements software. A region of interest was drawn around each CEP and ADE dopaminergic cell body. Before scoring, the threshold area (μm2) for defining a cell body as degenerated was defined, and all cell bodies were subsequently counted as present if their area exceeded this predetermined threshold. Neurites projecting anteriorly from CEP cell bodies were counted if they did not have degenerated morphologies, such as blebbing or fragmentation.
Basal Slowing Behavioral Assays.
On day 5, CQ434 baIn11 [dat-1p::α-synuclein; dat-1p::gfp]; vIs69 [pCFJ90 (myo-2p::mCherry + unc-119p::sid-1)] worms were individually picked onto NGM plates seeded with OP50. After 2 min, the number of body bends was manually counted for 20 s. Each worm was then picked onto an unseeded NGM plate. After 2 min, the number of body bends was manually counted for 20 s. If a worm curled during a 20-s measurement period, then the 20-s period was restarted when the worm resumed normal movement. The assay could not be performed on day 6 due to excessive curling. For azide experiments, treatment with 100 μM sodium azide or water was performed on day 3 to allow for the effect on basal slowing by day 5.
Neuron Isolation and Transcriptomics.
Adult neuron isolation was performed as described previously (14). In brief, TU3311 uIs60 (unc-119p::sid-1, unc-119p::yfp) worms were synchronized by bleaching and transferred to fresh bcat-1 or control RNAi plates every 2 to 3 d starting at L4. Adult worms were separated from progeny every 2 d by sedimentation in M9 buffer. On day 5, worms were washed with M9 buffer and then incubated for 6.5 min with lysis buffer (200 mM dithiothreitol, 0.25% sodium dodecyl sulfate, 20 mM Hepes pH 8.0, and 3% sucrose). The worms were then washed with M9 buffer and incubated in 20 mg/mL pronase from Streptomyces griseus (Sigma-Aldrich) for 12 to 20 min, with vigorous pipetting every 2 to 3 min. When worm bodies were dissociated and the solution became cloudy, 2% fetal bovine serum (Gibco) was added to stop the reaction, and the mixture was filtered through 5-μm filters (EMD Millipore). GFP-labeled neurons were sorted using a FACSVantage SE with DiVa (BD Biosciences; 488-nm excitation for GFP detection), with age-matched N2 worms used as a negative control for setting gates. RNA was extracted, DNase-digested, and cleaned using Qiagen RNEasy MinElute columns as described previously (14). RNA sequencing libraries were prepared using the SMARTer Stranded Total RNA kit v2-Pico input mammalian according to the manufacturer’s instructions. Libraries were pooled and sequenced on the Illumina HiSeq 2000 platform, and the Galaxy Workflow System was used to analyze the RNA sequencing data. Reads were mapped to the C. elegans genome (WS245) using RNA STAR, and mapped reads that overlap with gene features were counted using htseq-count (mode = union). Per-gene counts were used as input for the DESeq2 R package for differential gene expression analysis. Five total independent collections were obtained for each condition (bcat-1 or control), and bcat-1 curling was confirmed for each replicate on day 8. Raw sequencing reads are available at National Center for Biotechnology Information BioProject PRJNA599166.
GO and KEGG Analyses.
Lists of up-regulated and down-regulated genes in bcat-1(RNAi) neurons were analyzed using gProfiler (38) with the following settings: C. elegans organism, only annotated genes, g:SCS threshold, user threshold 0.05, ENTREZGENE_ACC. REVIGO (39) was used to cluster and plot GO terms with a q value < 0.05.
High-Resolution Metabolomics.
CF512 [fem-1(hc17); fer-15(b26)] worms were synchonized by bleaching and sterilized by incubation at 25 °C from L2 to L4. Worms were transferred to fresh bcat-1 or control RNAi plates every 2 to 3 d starting at L4. On day 5, worms were washed with M9 buffer, and ∼500 worms per replicate were snap-frozen in liquid nitrogen. In parallel for each sample, ∼500 worms were incubated in M9 for 1 h to release gut bacteria, after which the starved worms were discarded and the bacteria were collected as a blank. Metabolites were extracted using acetonitrile (in a 2:1 ratio), which was added to all samples along with an internal standard. Bead beating was used to disrupt the worm cuticle and improve extraction. Each sample was placed in the bead beater at 6.5 m/s for 30 s, allowed to equilibrate on ice for 1 min, and then placed in the beater for another 30 s at the same speed. All processing was done either on ice or in a cold room. Nontargeted high-resolution mass spectrometry was run at the Clinical Biomarkers Laboratory at Emory University using a hydrophilic interaction liquid chromatography column (positive mode) and C18 column (negative mode) using chromatographic methods as described previously (40). Mass spectral data were generated on an orbitrap mass spectrometer in full scan mode scanning for mass range of 85 to 850 Da. Data were extracted using the R packages apLCMS (41) and xMSanalyzer (42). Features that were 1.5 times the intensity in the respective bacterial blank and were present in at least 9 of the 21 samples were subjected to further analysis. Features were imputed if missing with half the value of the minimum abundance, normalized to protein content of the sample, and log2-transformed.
All data processing, analysis, and visualization were done in R version 3.6.0, using the functions gplots::heatmap2() (43), ggplot2::ggplot() (44), and mixOmics::plsda() (45). Pathway analysis was done using the mummichog algorithm (46) hosted on the MetaboAnalyst (https://www.metaboanalyst.ca/) module “MS peaks to pathway” (47), using the C. elegans metabolic reference map available through KEGG. All feature annotations are of level 5 confidence (48). Five total independent collections were obtained for bcat-1(RNAi), and six independent collections were obtained for control RNAi. bcat-1 curling was confirmed for each replicate on day 8. Raw data are available at Dryad (https://doi.org/10.5061/dryad.5mkkwh72q).
Oxygen Consumption Measurements.
Whole-worm respiration was measured using a Seahorse XFe96 analyzer as described previously (21). In brief, worms were plated (5 to 25 worms per well) in 200 μL of M9 buffer containing dilute heat-killed OP50 to prevent starvation. The same dilute heat-killed OP50 solution was used as a blank. Baseline respiration was measured, followed by injection of carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP; final concentration 10 μM) to elicit maximal respiration, followed by injection of sodium azide (final concentration 40 mM) to reveal nonmitochondrial oxygen consumption. The first measurement after each injection was unstable and was censored. Basal mitochondrial respiration was defined as baseline respiration – azide measurement. Maximal mitochondrial respiration was defined as FCCP-induced respiration – azide measurement. Measurements were normalized to protein content per well, determined by bicinchoninic acid assay.
TMRE Staining.
NGM plates seeded with 1 mL of heat-killed OP50 were spotted with TMRE to a final concentration of 30 µM and allowed to dry. Worms were transferred to TMRE plates for 1 d and then mounted on 2% agarose pads with levamisole for imaging. Images of dopaminergic cell bodies were obtained with a Nikon A1 confocal microscope at 100× magnification, and Z stacks were processed with Nikon NIS elements software. For each cell, a region of interest was drawn around the cell body at the plane with the greatest GFP intensity. The corresponding TMRE intensity (tetramethylrhodamine-isothiocyanate [TRITC] channel) was normalized to the GFP intensity.
Protein Carbonylation Staining.
Staining was performed using Oxy-Blot (EMD Millipore; S7150) as described previously (49). In brief, worms were washed three times with M9 and then incubated in 1× 2,4-dinitrophenylhydrazine for 30 min at room temperature. The reaction was blocked by adding a 4× volume of neutralization buffer. Worms were washed twice with ice-cold water, sandwiched between two poly-l-lysine–coated slides, and incubated for 20 min in liquid nitrogen. The slide sandwiches were then freeze-cracked, and the worms were fixed with methanol followed by acetone and then blocked for 1 h at room temperature with 1× PBS, 0.2% gelatin, and 0.25% Triton X-100. The rabbit anti-DNP primary antibody was applied at 1:100 dilution in blocking solution, followed by incubation overnight at 4 °C. As an internal control, mouse anti-ATP5A antibody (Abcam; ab14748) was coincubated. After four 10-min washes with 0.25% Triton X-100 in 1× PBS, goat anti-mouse Cy5 (Invitrogen; A10524) and goat anti-rabbit 555 (Invitrogen; A32732) secondary antibodies were applied at 1:300 dilution in blocking solution and coincubated overnight at 4 °C. Images of dopaminergic cell bodies were obtained on a Nikon A1 confocal microscope at 60× magnification, and Z stacks were processed using Nikon NIS elements software. For each cell, a region of interest was drawn around the cell body at the plane with the greatest GFP intensity. The corresponding anti-DNP intensity (TRITC channel) was normalized to the anti-ATP5A intensity (Cy5 channel).
Statistical Analysis.
All comparisons between two groups were done using an unpaired two-tailed Student’s t test. For comparisons between multiple groups, one-way ANOVA or two-way ANOVA (for two variables (i.e., RNAi treatment with or without metformin) was performed with post hoc testing as indicated. Statistics were performed using GraphPad Prism 7.
Supplementary Material
Acknowledgments
We thank the C. elegans Genetics Center for strains (P40 OD010440), the Genomics Core Facility and Confocal Imaging Facility at Princeton University, Dr. Dean Jones and the Clinical Biomarkers Laboratory at Emory University, BioRender.com for model figure design software, and members of the C.T.M. lab for discussion. Strain UA44 was generously provided by G. Caldwell (University of Alabama). C.T.M. is the Director of the Glenn Center for Aging Research at Princeton and an HHMI-Simons Faculty Scholar. D.E.M. was supported by a Ruth L. Kirschstein National Research Service Award (NIA F32AG062036). Further support was provided by the NIH (NIEHS U2CES030163 and R01 ES023839, to G.W.M.; DP1 Pioneer Award NIGMS 5DP1GM119167, to C.T.M.) and the Glenn Foundation for Medical Research (CNV1001899, to C.T.M.).
Footnotes
Competing interest statement: The method used for automated quantification of curling motor behavior was filed under Patent #62/989,317: C.T.M., S.S., D.E.M., R.K., W.K., “Novel High-Throughput Screening Method for Parkinson’s Phenotypes Using C. elegans.” This patent is specifically for the screening method, not for metformin treatment of Parkinson’s disease phenotypes.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009838117/-/DCSupplemental.
Data Availability.
Transcriptomics and metabolomics data have been deposited in National Center for Biotechnology Information BioProject PRJNA599166 and Dryad (https://doi.org/10.5061/dryad.5mkkwh72q), respectively.
References
- 1.Van Den Eeden S. K. et al., Incidence of Parkinson’s disease: Variation by age, gender, and race/ethnicity. Am. J. Epidemiol. 157, 1015–1022 (2003). [DOI] [PubMed] [Google Scholar]
- 2.de Lau L. M., Breteler M. M., Epidemiology of Parkinson’s disease. Lancet Neurol. 5, 525–535 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Forno L. S., Neuropathology of Parkinson’s disease. J. Neuropathol. Exp. Neurol. 55, 259–272 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Spillantini M. G. et al., Alpha-synuclein in Lewy bodies. Nature 388, 839–840 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Klein C., Westenberger A., Genetics of Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2, a008888 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao V. et al., An integrative tissue-network approach to identify and test human disease genes. Nat. Biotechnol. 36, 1091–1099 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valerio A., D’Antona G., Nisoli E., Branched-chain amino acids, mitochondrial biogenesis, and healthspan: An evolutionary perspective. Aging (Albany NY) 3, 464–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corsi A. K., Wightman B., Chalfie M., A transparent window into biology: A primer on Caenorhabditis elegans. Genetics 200, 387–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rual J. F. et al., Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao S., Gelwix C. C., Caldwell K. A., Caldwell G. A., Torsin-mediated protection from cellular stress in the dopaminergic neurons of Caenorhabditis elegans. J. Neurosci. 25, 3801–3812 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawin E. R., Ranganathan R., Horvitz H. R., C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26, 619–631 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Mansfeld J. et al., Branched-chain amino acid catabolism is a conserved regulator of physiological ageing. Nat. Commun. 6, 10043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaletsky R. et al., The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature 529, 92–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mor D. E., Sohrabi S., Kaletsky R., Keyes W., Tartici A., Kalia V., Miller G. W., Murphy C. T., Metformin rescues Parkinsons disease features induced by defective branched chain amino acid metabolism. NCBI. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA599166. Deposited 23 September 2020.
- 16.Kaletsky R. et al., Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS Genet. 14, e1007559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson E. et al., Metabolic network rewiring of propionate flux compensates vitamin B12 deficiency in C. elegans. eLife 5, e17670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meissner B. et al., Determining the sub-cellular localization of proteins within Caenorhabditis elegans body wall muscle. PLoS One 6, e19937 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Go Y. M. et al., Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicol. Sci. 148, 531–543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mor D. E., Sohrabi S., Kaletsky R., Keyes W., Tartici A., Kalia V., Miller G. W., Murphy C. T., Datasets associated with 'Metformin rescues Parkinson’s disease phenotypes caused by hyperactive mitochondria.' Dryad. 10.5061/dryad.5mkkwh72q, Deposited 22 September 2020. [DOI] [PMC free article] [PubMed]
- 21.Koopman M. et al., A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. Nat. Protoc. 11, 1798–1816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winklhofer K. F., Haass C., Mitochondrial dysfunction in Parkinson’s disease. Biochim. Biophys. Acta 1802, 29–44 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Sohrabi S., Mor D. E., Kaletsky R., Keyes W., Murphy C. T., High-throughput behavioral screen in C. elegans reveals novel Parkinson’s disease drug candidates. https://www.biorxiv.org/content/10.1101/2020.02.20.958751v1.full.pdf. Deposited 21 February 2020. [DOI] [PMC free article] [PubMed]
- 24.Vial G., Detaille D., Guigas B., Role of mitochondria in the mechanism(s) of action of metformin. Front. Endocrinol. (Lausanne) 10, 294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabreiro F. et al., Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153, 228–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng H. C., Ulane C. M., Burke R. E., Clinical progression in Parkinson disease and the neurobiology of axons. Ann. Neurol. 67, 715–725 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carecchio M. et al., Movement disorders in adult surviving patients with maple syrup urine disease. Mov. Disord. 26, 1324–1328 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiil R., Rokkones T., Late manifesting variant of branched-chain ketoaciduria (maple syrup urine disease). Acta Paediatr. (Stockh.) 53, 356–364 (1964). [DOI] [PubMed] [Google Scholar]
- 29.Nalls M. A. et al.; International Parkinson’s Disease Genomics Consortium (IPDGC); Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI); 23andMe; GenePD; NeuroGenetics Research Consortium (NGRC); Hussman Institute of Human Genomics (HIHG); Ashkenazi Jewish Dataset Investigator; Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC); Greek Parkinson’s Disease Consortium; Alzheimer Genetic Analysis Group , Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luan H. et al., Comprehensive urinary metabolomic profiling and identification of potential noninvasive marker for idiopathic Parkinson’s disease. Sci. Rep. 5, 13888 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.She P. et al., Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab. 6, 181–194 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker W. D. Jr., Boyson S. J., Parks J. K., Abnormalities of the electron transport chain in idiopathic Parkinson’s disease. Ann. Neurol. 26, 719–723 (1989). [DOI] [PubMed] [Google Scholar]
- 33.Schapira A. H. et al., Mitochondrial complex I deficiency in Parkinson’s disease. Lancet 1, 1269 (1989). [DOI] [PubMed] [Google Scholar]
- 34.Dauer W., Przedborski S., Parkinson’s disease: Mechanisms and models. Neuron 39, 889–909 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Tieu K., A guide to neurotoxic animal models of Parkinson’s disease. Cold Spring Harb. Perspect. Med. 1, a009316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cicchetti F., Drouin-Ouellet J., Gross R. E., Environmental toxins and Parkinson’s disease: What have we learned from pesticide-induced animal models? Trends Pharmacol. Sci. 30, 475–483 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Restif C. et al., CeleST: Computer vision software for quantitative analysis of C. elegans swim behavior reveals novel features of locomotion. PLOS Comput. Biol. 10, e1003702 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raudvere U. et al., g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu K. H. et al., High-resolution metabolomics assessment of military personnel: Evaluating analytical strategies for chemical detection. J. Occup. Environ. Med. 58, S53–S61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu T., Park Y., Johnson J. M., Jones D. P., apLCMS—Adaptive processing of high-resolution LC/MS data. Bioinformatics 25, 1930–1936 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uppal K. et al., xMSanalyzer: Automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics 14, 15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warnes G. R., et al. , gplots: Various R programming tools for plotting data. https://cran.r-project.org/web/packages/gplots/index.html. Accessed October 31, 2019.
- 44.Wickham H., ggplot2: Elegant Graphics for Data Analysis, (Springer, 2016). [Google Scholar]
- 45.Rohart F., Gautier B., Singh A., Lê Cao K. A., mixOmics: An R package for ’omics feature selection and multiple data integration. PLOS Comput. Biol. 13, e1005752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S. et al., Predicting network activity from high-throughput metabolomics. PLOS Comput. Biol. 9, e1003123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chong J., Wishart D. S., Xia J., Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinformatics 68, e86 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Schymanski E. L. et al., Identifying small molecules via high-resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 48, 2097–2098 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Goudeau J., Aguilaniu H., Carbonylated proteins are eliminated during reproduction in C. elegans. Aging Cell 9, 991–1003 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptomics and metabolomics data have been deposited in National Center for Biotechnology Information BioProject PRJNA599166 and Dryad (https://doi.org/10.5061/dryad.5mkkwh72q), respectively.