ABSTRACT
With the rapid increase in the frequency of azole-resistant species, combination therapy appears to be a promising tool to augment the antifungal activity of azole drugs against resistant Candida species. Here, we report the effect of aprepitant, an antiemetic agent, on the antifungal activities of azole drugs against the multidrug-resistant Candida auris. Aprepitant reduced the minimum inhibitory concentration (MIC) of itraconazole in vitro, by up to eight-folds. Additionally, the aprepitant/itraconazole combination interfered significantly with the biofilm-forming ability of C. auris by 95 ± 0.13%, and significantly disrupted mature biofilms by 52 ± 0.83%, relative to the untreated control. In a Caenorhabditis elegans infection model, the aprepitant/itraconazole combination significantly prolonged the survival of infected nematodes by ~90% (five days post-infection) and reduced the fungal burden by ~92% relative to the untreated control. Further, this novel drug combination displayed broad-spectrum synergistic interactions against other medically important Candida species such as C. albicans, C. krusei, C. tropicalis, and C. parapsilosis (ƩFICI ranged from 0.08 to 0.31). Comparative transcriptomic profiling and mechanistic studies indicated aprepitant/itraconazole interferes significantly with metal ion homeostasis and compromises the ROS detoxification ability of C. auris. This study presents aprepitant as a novel, potent, and broad-spectrum azole chemosensitizing agent that warrants further investigation.
KEYWORDS: Candida auris, fungal biofilms, azole resistance, Caenorhabditis elegans, metal ion homeostasis, reactive oxygen species (ROS)
Introduction
Candida auris is an emerging fungal pathogen that has been recently implicated in numerous global outbreaks of life-threatening candidemia, leading to significant morbidity and high rates of mortality [1,2]. Recently, the U.S. Centers for Disease Control and Prevention (CDC) has classified C. auris as an urgent threat that requires immediate intervention [3]. Unfortunately, resistance to major antifungal agents, including to the first-line therapeutics, was reported to be common among C. auris isolates [4]. According to the CDC, 90% of C. auris isolates are resistant to the antifungal activity of fluconazole and around 30% of isolates are resistant to amphotericin B [5]. The unique multidrug resistance nature of C. auris combined with its ability to be efficiently transmitted among patients are key challenges that hinder the development of effective control and containment of C. auris [1,6–8].
Currently, only three classes of systemic antifungal drugs (azoles, polyenes, and echinocandins) are available for the treatment of invasive Candida infections [9]. Azole drugs target ergosterol biosynthesis through the inhibition of lanosterol 14 alpha methyltransferase (Erg11p) resulting in depletion of the vital cell membrane component, ergosterol, and accumulation of cytotoxic metabolites [10]. Owing to safety, the spectrum of activity, and pharmacokinetic considerations such as oral bioavailability, azole antifungal drugs have gained preference as vital therapeutics to control various types of mycotic infections [11–14]. However, the extensive dependence on azole drugs has contributed to the increased rate of azole-resistance among fungal species [15,16]. Overexpression or mutation of ERG11, in addition to the upregulation of ABC and MFS efflux transporters, are known to be major triggers of azole resistance in Candida species [17].
Considering the limited therapeutic options and the increased rate of drug resistance in C. auris, there is a pressing need for novel antifungal agents and co-drugs capable of restoring or enhancing the antifungal activity of azole drugs. Historically, drug combinations have been utilized efficiently to treat various bacterial and viral infections and to fight cancer [18–21]. However, this approach has not been fully exploited in medical mycology despite the immediate need to discover new treatment strategies, especially with the current dearth in available therapeutics and the increased frequency of multidrug-resistant isolates.
In a whole-cell screening study aimed at identifying novel azole chemosensitizing agents (unpublished data), we noticed that the antiemetic agent aprepitant was able to restore the antifungal activity of fluconazole against the test C. auris isolate. Here, we investigated the interactions between aprepitant and different azole drugs against multiple C. auris isolates and other clinically important Candida species. Additionally, comparative transcriptomic profiling and mechanistic studies were performed to identify the potential mechanism by which aprepitant enhances the antifungal activities of azole drugs.
Materials and methods
Fungal strains, reagents, and chemicals
Sources and descriptions of the fungal strains used in this study are provided in Supplementary Table S1. Yeast-peptone dextrose (YPD) agar and broth were purchased from Becton, and Dickinson Company (Franklin Lakes, NJ). 3-(N-Morpholino) propanesulfonic acid (MOPS) was purchased from Sigma-Aldrich (St. Louis, MO). RPMI 1640 powder, supplemented with glutamine and lacking NaHCO3, was obtained from Thermo Fisher Scientific (Waltham, MA). Johns Hopkins Clinical Compounds Library (JHCCL) was obtained from the school of medicine at Johns Hopkins University and was used to identify the azole chemosensitizing activity of aprepitant. The library was screened at 16 µM against C. auris AR0390 in the presence or absence of a subinhibitory concentration of fluconazole (32 µg/ml). Fluconazole was purchased from Fisher Scientific (Pittsburgh, PA). Itraconazole, voriconazole, and aprepitant were purchased from TCI America (Portland, OR). Gentamicin sulfate was obtained from Chem-Impex International Inc. (Wood Dale, IL).
Microdilution checkerboard assays
Aprepitant and azole drugs (fluconazole, voriconazole, and itraconazole) were tested alone and in combinations against different Candida species using standard broth microdilution checkerboard assays, as previously described [22–24]. The ΣFICI (fractional inhibitory concentration index) was used to evaluate the interactions between the tested drugs. Interactions were deemed to be synergistic (SYN) when ΣFICI values were ≤0.50, additive (ADD) when ΣFICI values were >0.50 and ≤1, and indifferent (IND) when ΣFICI values were >1 and ≤4 [25].
Time-kill assay
To study the effect of aprepitant/itraconazole on the growth kinetics of C. auris, a time-kill assay was conducted, as previously described [26–29]. Briefly, exponential-phase C. auris AR0390 cells were diluted with RPMI 1640 medium to ~1 × 105 CFU/ml. Cells were incubated with aprepitant (10 µg/ml), itraconazole (1 µg/ml), or a combination of both agents for 24 h at 35°C. Following incubation, the number of viable cells was determined at specific time points (0, 6, 12, and 24 h). The time-kill curve was constructed by plotting viable colony forming units against time. The results are presented as the average values of triplicate measurements.
Effect of aprepitant/itraconazole combination against premature and mature C. auris biofilms
To assess the antibiofilm activity of aprepitant/itraconazole against C. auris, overnight cultures of C. auris AR0390 (~1 × 105 CFU/ml) in RPMI 1640 were transferred onto tissue culture-treated 96-well plates and incubated for 2 h at 35°C to allow cell adhesion. Then aprepitant (10 µg/ml), itraconazole (1 µg/ml), or a combination of both agents were added, and the plates were incubated for 24 h at 35°C. Cells treated with DMSO (1%), the drugs’ solvent, served as a negative control. Following incubation, RPMI 1640 medium was discarded and the formed biofilms were carefully washed twice with PBS before being quantified, using the XTT reduction assay [26,30].
To assess the activity of aprepitant/itraconazole against mature biofilms, C. auris cells (at ~1 × 105 CFU/ml) were seeded in tissue culture-treated 96-well plates and incubated for 24 h at 35°C to allow for the formation of mature biofilms [31–33]. Fresh RPMI 1640 media supplemented with aprepitant, itraconazole, or aprepitant/itraconazole, at the same concentrations indicated above, were added to the formed biofilms and incubated for another 24 h at 35°C. After incubation, biofilms were carefully washed and quantified by the XTT reduction method. All treatments were conducted in triplicates and data represent the average ± SD.
Effect of aprepitant/itraconazole against other Candida species
To investigate the effect of aprepitant on the antifungal activity of itraconazole against other medically important Candida species, checkerboard assays were utilized and ΣFIC indices were calculated as mentioned above. The aprepitant/itraconazole combination was assessed against C. albicans (n = 4), C. glabrata (n = 3), C. krusei (n = 3), C. tropicalis (n = 2), and C. parapsilosis (n = 2). Additionally, 5 µl samples from representative Candida cultures that displayed enhanced susceptibility to the aprepitant/itraconazole combination were spotted onto YPD agar plates and incubated for 24 h at 35°C before scanning the plates and determination of CFU counts.
RNA extraction
Exponential-phase cultures of C. auris AR0390 were treated (in duplicates) with DMSO (1%), aprepitant (10 µg/ml), itraconazole (1 µg/ml), or a combination of both drugs. All treated cultures were incubated for 3 h at 30°C. The cells were pelleted, washed twice with PBS, and RNA was isolated using an Ambion Ribopure yeast kit. The RNA quality was checked on a bioanalyzer Nano chip (Agilent) and all RNAs used for downstream experiments were determined to have RNA integrity numbers (RIN) of 9.5 and above. The SuperScript III First-Strand kit (Invitrogen) was used to prepare cDNA, following the manufacturer’s guidelines.
RNA sequencing and enrichment analysis of differentially expressed genes (DEGs)
Paired-end, 2 × 150 bp reads were sequenced on a NovaSeq6000. Reads were quality trimmed and Illumina TruSeq adapter sequences were removed using fastp [34]. HISAT2 (v2.1) was used to align reads to the Candida auris NCBI reference genome version B11221 [35]. FeatureCounts (v2.1) was used to count the read numbers mapped of each gene [36]. FPKM values of each gene were calculated based on the length of the gene in the associated annotation GTF file as well as reads count mapped to each gene. Differential expression analysis was performed on raw count data using the edgeR Bioconductor package (v3.16.5) [37,38]. The P values were adjusted using the Benjamini & Hochberg method [39]. A corrected P-value of 0.005 and an absolute foldchange of two was set as the threshold for significant differential expression. Differentially expressed genes were annotated by blasting the sequences of identified DEGs against the Candida Genome Database (CGD) and by using blasts against C. albicans SC5314 (taxid:237561) with an e-value cutoff of 0.01 [40]. Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented by the ClusterProfiler Bioconductor package (v2.4.3) [41]. GO terms with corrected P-values < 0.05 were considered significantly enriched within the differentially expressed genes lists.
Real-time quantitative reverse transcription (RT-qPCR)
To validate the RNA-Seq results, we conducted quantitative real-time (RT) PCR assays to quantify the expression of six genes (ERG1, ERG2, ERG10, FRT1, ZRT2, and CTR1). Gene expression levels were calculated using the 2−ΔΔCT method [42]. Gene expression was internally normalized to ACT1 and compared to the untreated control (DMSO 1%). All primers used in this study were provided in Supplementary Table S2.
Effect on ROS levels in C. auris cells
The Image-iT™LIVE Green Reactive Oxygen Species (ROS) detection kit (Molecular Probes, Inc., Eugene, OR) was utilized to measure ROS levels, as previously described [27]. Briefly, exponential-phase C. auris AR0390 was adjusted to ~ 1 × 106 CFU/ml in PBS in the presence or absence of FeSO4 (50 µM). Cells were treated with either aprepitant (10 µg/ml), itraconazole (1 µg/ml), or a combination of both drugs, while DMSO (1%) treated culture was used as a negative control. All treatments were continued for 3 h at 35°C, then cells were pelleted, washed twice with PBS, and 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (carboxy-H2DCFDA) dye, at a final concentration of 50 µM, was added. Cells were then incubated for 1 h at 35°C. After incubation, cells were washed twice with PBS and the fluorescence intensity was measured spectrophotometrically at 495/529 nm. Data represent the average of three measurements ±SD.
Effect of iron supplementation on the antifungal activity of aprepitant/itraconazole
To investigate the effect of iron supplementation on the synergistic relationship between aprepitant and itraconazole against C. auris, a time-kill assay was performed as described above, and RPMI 1640 with/without ferrous sulfate (50 µM) was used as the assay medium. Cultures were incubated at 35°C for 24 h and then 5 µl samples of each treatment were spotted onto YPD agar plates and incubated at 35°C for another 24 h before scanning the plates.
Caenorhabditis elegans infection model
To assess the in vivo efficacy of the aprepitant/itraconazole combination, a C. elegans infection model was utilized as previously discribed [9,11,26]. Briefly, synchronized worms [strain AU37 genotype glp-4(bn2) I; sek-1(km4) X] at L4 phase were incubated in 5 ml of YPD broth containing C. auris AR0390 (adjusted at ~1 × 106 CFU/ml) for 3 h at room temperature. Following infection, worms were washed five times with PBS to remove non-ingested cells and then suspended in M9 buffer containing 20% RPMI 1640. Infected worms were distributed into groups (~20 worms/group) and then treated (in triplicates) with either DMSO (1%), aprepitant (10 µg/ml), itraconazole (1 µg/ml), or a combination of both drugs. Worms were then incubated with test agents at 25°C for 24 h before being washed twice with PBS and beaten vigorously with silicon carbide beads, for at least 2 min, to release C. auris cells. C. elegans lysates were serially diluted and plated over YPD agar plates containing gentamicin (100 µg/ml). Plates were incubated for 24 h at 35°C before viable CFU per worm was determined.
In another experiment, worms were incubated in 5 ml of YPD broth containing C. auris AR0390 (adjusted at ~1 × 106 CFU/ml) for 3 h at room temperature. Infected worms were treated with DMSO (1%), aprepitant (10 µg/ml), itraconazole (1 µg/ml), or a combination of both drugs. The survival of C. elegans was monitored and recorded for five days. Data are presented as percent survival of infected C. elegans using a Kaplan-Meier survival curve generated using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA).
Statistical analyses
Statistical analysis was performed using GraphPad Prism 6.0 (Graph Pad Software, La Jolla, CA, USA). One-way ANOVA and Dunnett’s test for multiple comparisons were used to assess the statistical significance (P < 0.05) between treated and untreated groups. Kaplan-Meier survival curves were assessed by the log-rank test for statistical significance.
Results
Azole chemosensitizing activity of aprepitant against C. auris isolates
To identify novel adjuvants to overcome azole resistance in C. auris, we explored the azole chemosensitizing activity of ~1600 FDA-approved drugs in the Johns Hopkins Chemical Library (unpublished data). Our initial screen identified aprepitant (an antiemetic agent) as a potent hit compound that restored the antifungal activity of fluconazole against the multidrug-resistant strain C. albicans AR0390. This observation encouraged us to explore the interactions between aprepitant and commonly used azole drugs in vitro against a panel of ten C. auris isolates obtained from the Antimicrobial Resistance Isolate Bank (CDC). To assess these interactions, we used standard microdilution checkerboard assays and calculated the fractional inhibitory concentration indices (ƩFICI). As shown in Table 1, aprepitant was able to enhance the antifungal activity of fluconazole against seven isolates with a synergistic relationship observed against four isolates (ƩFICI ranged from 0.09 to 0.50) and an additive relationship observed against three isolates (ƩFICI ranged from 0.51 to 0.63). Based on the current tentative breakpoints for azole resistance, aprepitant was able to restore the fluconazole susceptibility only in two fluconazole-resistant isolates, AR0388, and AR0390. When tested in combination with voriconazole, aprepitant was able to enhance the antifungal activity of voriconazole against eight isolates, with a synergistic relationship observed against two isolates (ƩFICI = 0.19) and an additive relationship observed against six isolates (ƩFICI ranged from 0.53 to 0.56). However, aprepitant failed to re-sensitize any of the tested voriconazole-resistant isolates (n = 3) that exhibited MIC ≥1 µg/ml. Interestingly, when combined with itraconazole, aprepitant was able to enhance the antifungal activity of itraconazole against all ten tested isolates, resulting in a synergistic relationship observed against eight isolates (ƩFICI ranged from 0.14 to 0.31) and an additive relationship observed against two isolates (ƩFICI = 0.52). Importantly, aprepitant was able to re-sensitize all tested isolates (n = 3) that displayed reduced susceptibility to itraconazole (MIC ≥1 µg/ml). Since aprepitant interacted more favorably with itraconazole against C. auris, the combination between aprepitant and itraconazole was selected for further investigation.
Table 1.
Effect of aprepitant on the antifungal activity of fluconazole (FLC), voriconazole (VRC), and itraconazole (ITC) against C. auris clinical isolates
|
C. auris Isolates |
MIC (µg/ml) |
Combination with FLC |
Combination with VRC |
Combination with ITC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| APR | FLC | VRC | ITC | APR/FLC (µg/ml) |
ƩFIC | Mode | APR/VRC (µg/ml) |
ƩFIC | Mode | APR/ITC (µg/ml) |
ƩFIC | Mode | |
| AR0381 | > 128 | 1 | 0.0078 | 0.25 | 0.5/0.5 | 0.50 | SYN | 8/0.007 | 1.06 | IND | 2/0.125 | 0.52 | ADD |
| AR0382 | 1 | 0.0625 | 0.25 | 1/0.5 | 0.51 | ADD | 8/0.031 | 0.56 | ADD | 8/0.0312 | 0.19 | SYN | |
| AR0383 | 256 | 0.5 | 0.5 | 2/256 | 1.02 | IND | 8/0.25 | 0.56 | ADD | 8/0.125 | 0.31 | SYN | |
| AR0384 | 128 | 0.5 | 0.25 | 2/64 | 0.52 | ADD | 8/0.25 | 0.56 | ADD | 8/0.0312 | 0.19 | SYN | |
| AR0385 | 256 | 4 | 0.5 | 2/256 | 1.02 | IND | 4/2 | 0.53 | ADD | 8/0.125 | 0.31 | SYN | |
| AR0386 | 256 | 2 | 0.5 | 16/128 | 0.63 | ADD | 4/1 | 0.53 | ADD | 8/0.125 | 0.31 | SYN | |
| AR0387 | 1 | 0.0312 | 0.125 | 0.5/0.5 | 0.50 | SYN | 4/0.015 | 0.53 | ADD | 8/0.0312 | 0.31 | SYN | |
| AR0388 | 256 | 0.5 | 1 | 8/8 | 0.09 | SYN | 8/0.062 | 0.19 | SYN | 2/0.125 | 0.14 | SYN | |
| AR0389 | 256 | 2 | 1 | 8/256 | 1.06 | IND | 8/2 | 1.06 | IND | 2/0.5 | 0.52 | ADD | |
| AR0390 | 256 | 0.5 | 1 | 8/8 | 0.09 | SYN | 8/0.062 | 0.19 | SYN | 4/0.125 | 0.16 | SYN | |
*ΣFICI (fractional inhibitory concentration index) was used to measure the interaction between the tested combinations. ΣFICI interpretation corresponded to the following definitions: synergism (SYN), ΣFICI ≤0.5; additivity (ADD), ΣFICI >0.5 and ≤1; and indifference (IND), ΣFICI >1 and ≤4.
Aprepitant/itraconazole exerts a fungicidal effect against C. auris AR0390
To investigate the killing kinetics of the aprepitant/itraconazole combination against C. auris, a time-kill assay was conducted. As shown in Figure 1, aprepitant (at 10 µg/ml), by itself, did not produce any observable antifungal activity against the test isolate (AR0390). Itraconazole, at 1 × MIC (1 µg/ml), exhibited fungistatic activity for 12 h post-incubation; however, rapid fungal re-growth was observed thereafter. Interestingly, the combination of aprepitant and itraconazole (at the same concentrations each drug was tested alone) displayed fungicidal activity against the test isolate generating ~ 3-log10 reduction in the CFU count over 24 hours.
Figure 1.
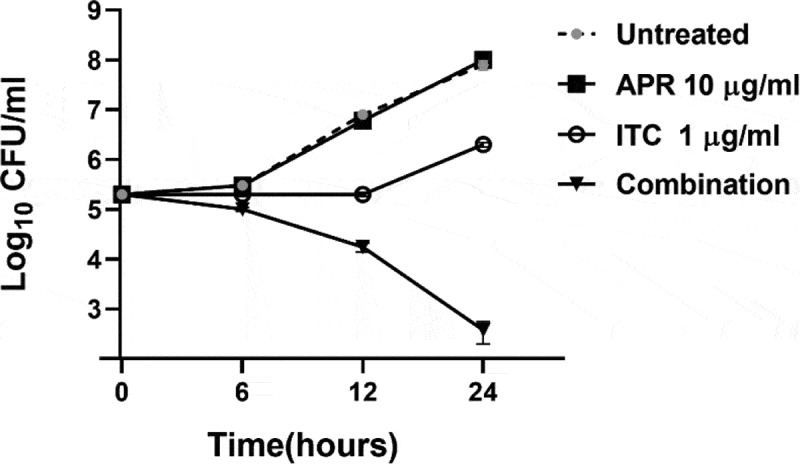
Time-kill assay of aprepitant (APR) at 10 µg/ml, itraconazole (ITC) at 1 µg/ml, or a combination of both agents at the same concentration as tested alone. Test agents were evaluated against C. auris AR0390 over a 48-hour incubation period at 35 °C. Cells treated with DMSO (1%) served as a negative untreated control. Error bars represent standard deviation values
Effect of aprepitant/itraconazole against C. auris biofilms
As shown above, the aprepitant/itraconazole combination exerted fungicidal activity against planktonic C. auris. To investigate whether a similar effect could be observed against C. auris biofilms, we utilized the XTT reduction assay to examine the effect of aprepitant/itraconazole against early-stage and mature biofilms. As shown in Figure 2(a), the combination of aprepitant (at 10 µg/ml) and itraconazole (at 1 µg/ml) significantly inhibited the biofilm-forming ability of the test isolate by ~95 ± 0.13% compared to the untreated control. However, single treatments with either aprepitant or itraconazole at the respective concentrations failed to interfere with the biofilm-forming ability of the test isolate.
Figure 2.
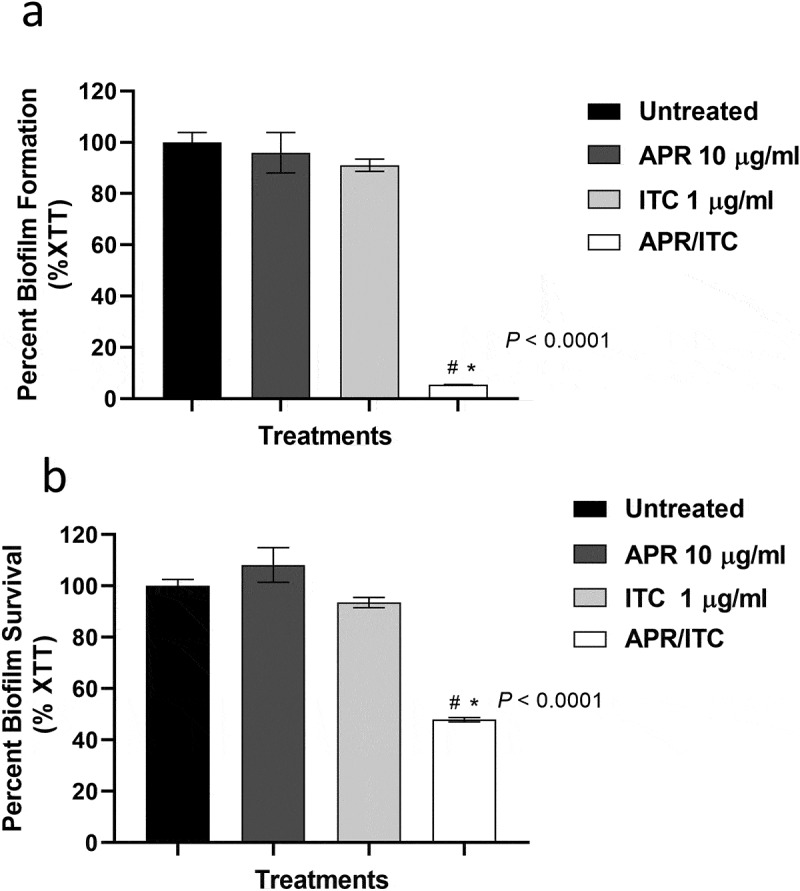
The anti-biofilm activity of aprepitant (APR)/itraconazole (ITC) against C. auris. (a) The inhibitory effect of the aprepitant (10 µg/mL)/itraconazole (1 µg/mL) combination on the formation of C. auris AR0390 biofilms as determined by the XTT reduction assay. (b) The ability of the aprepitant/itraconazole combination to diminish the metabolic activity of mature C. auris AR0390 biofilms. Data are shown as means ± SD. * indicates a statistical significance (P < 0.05) relative to the untreated control (P < 0.05), whereas #, indicates a statistical significance (P < 0.05) relative to the treatment with either aprepitant or itraconazole alone. Statistical significance was assessed by one-way ANOVA using Dunnett’s test for multiple comparisons
When tested against mature C. auris biofilms, the aprepitant/itraconazole combination, at the same concentrations, was able to significantly reduce the viability of C. auris AR0390 cells present in mature biofilms by 52 ± 0.83% (Figure 2(b)). As expected, neither aprepitant nor itraconazole alone, at the respective concentrations, was able to interfere with the viability of C. auris cells within mature biofilms. These data indicate the potent ability of aprepitant/itraconazole to interfere with both the early and late phases of C. auris biofilms.
Effect of aprepitant/itraconazole against other Candida species
The potent synergistic interaction observed between aprepitant and itraconazole against all C. auris isolates tested encouraged us to examine whether the aprepitant/itraconazole combination would exert a similar effect against other clinically important Candida species. Using standard microdilution checkerboard assays, we examined the interactions between aprepitant and itraconazole against a panel of 14 Candida species including C. albicans (n = 4), C. glabrata (n = 3), C. krusei (n = 3), C. tropicalis (n = 2), and C. parapsilosis (n = 2). As shown in Figure 3(a) and Supplementary Table S3, aprepitant exerted a synergistic relationship with itraconazole (ƩFICI ranged from 0.08 to 0.31) against all isolates, except for C. glabrata isolates where an indifferent effect was observed (ƩFICI = 1.02). These synergistic interactions were further demonstrated using spot assays. As shown in Figure 3(b), the aprepitant/itraconazole combination inhibited the visual growth of C. albicans (TWO7243), C. krusei (ATCC 14243), C. tropicalis (ATCC 1369), and C. parapsilosis (ATCC 22019), and reduced the CFU counts by ~ 2–4 orders of magnitude relative to the untreated control, as shown in Figure 3(c).
Figure 3.
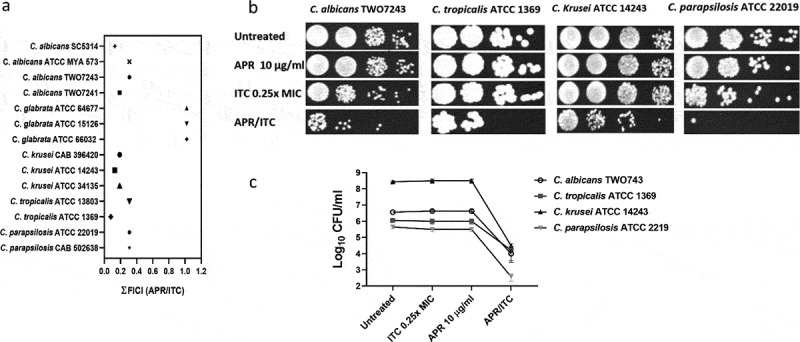
Interaction between aprepitant and itraconazole against different Candida species. (a) ƩFICI (fractional inhibitory concentration index) values were calculated from checkerboard assays. Cultures of C. albicans TWO7243, C. krusei ATCC 14243, C. tropicalis ATCC 1369, and C. parapsilosis ATCC 22019 were treated with APR (10 µg/ml) and ITC (0.25 × MIC), either alone or in combination. Treated cultures were incubated at 35°C for 24 h before being spotted onto YPD agar plates and reincubated for an additional 24 h. Plates were then scanned (b) and CFU count was performed (c)
Transcriptomic profiling of C. auris treated with aprepitant/itraconazole
To gain insight into the molecular mechanism(s) by which aprepitant interacts synergistically with azole drugs, we performed comparative transcriptomic analysis of C. auris AR0390 treated with DMSO (1%), aprepitant (10 µg/ml), itraconazole (1 µg/ml), or a combination of both drugs. A total of 81.9 million reads were obtained with over 95% mapping rates observed across all samples. The sequence reads of all samples were deposited in the NCBI sequence read archive (GEO) under the accession number of (GSE148749). The Bioconductor package edgeR was used to identify differentially expressed genes (DEGs) between groups. Genes with an adjusted P-value <0.05 and that showed greater than a two-fold (up or down) change in expression were considered differentially expressed. As shown in Figure 4(a), a total of 34 DEGs (19 upregulated + 15 downregulated) were identified in the aprepitant/itraconazole treated sample (Figure 4(a) and Table 2). Genes involved in ergosterol synthesis (ERG1, ERG2, ERG10, ERG24), stress responses (DDR48 and RTA3), and drug transporters (CDR1, MDR1, DUR3) were upregulated in the aprepitant/itraconazole treated sample. Additionally, the iron-regulated 1,3-beta glucan-linked cell wall gene (PIR1) and the vacuolar calcium P-type ATPase (PMC1) were also among the upregulated DEGs. On the other hand, the aprepitant/itraconazole combination resulted in the downregulation of genes responsible for metal ion transport such as FTR1 (iron transporter), ZTR2 (zinc transporter), and CTR1, which is important for both iron and copper transport. Also, several high-affinity glucose transporters (HGT6, HGT8, HGT12) were among the downregulated DEGs in the aprepitant/itraconazole treated sample.
Figure 4.
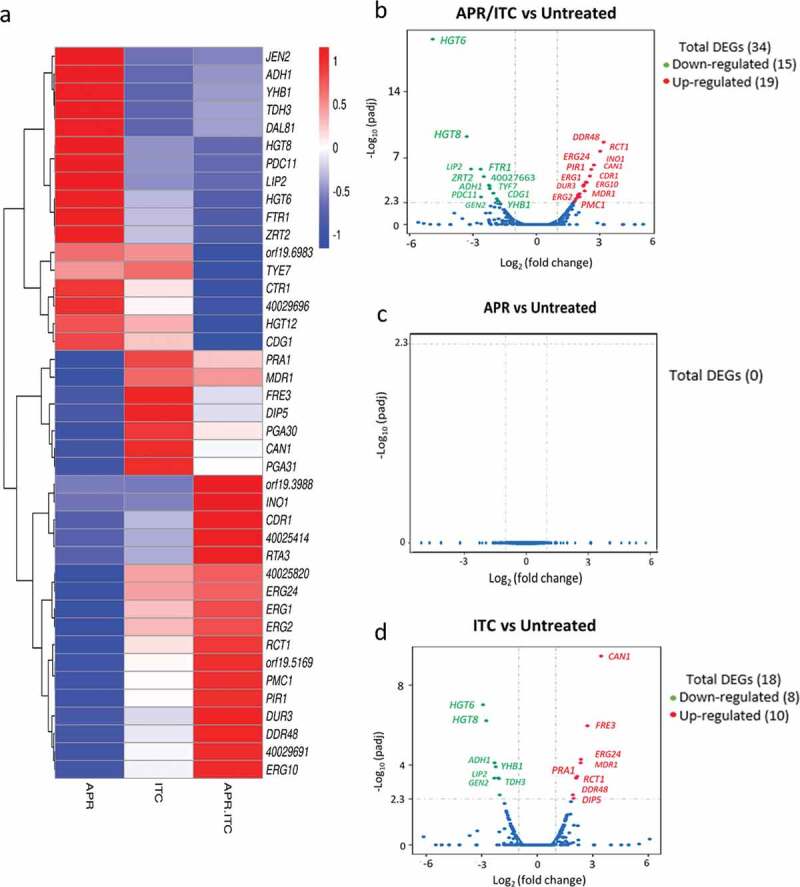
Transcriptional comparison of C. auris AR0390 treated with aprepitant (APR), itraconazole (ITC), or a combination of both versus the untreated control. (a) Heat map of FPKM values of DEGs of each treatment versus the untreated control, scaled by row. (b) Volcano blot of DEGs from C. auris AR0390 treated with aprepitant/itraconazole (APR/ITC) at 10/1 µg/ml. (c) Volcano blot of DEGs from C. auris AR0390 treated with aprepitant (APR) at 10 µg/ml. (d) Volcano blot of DEGs from C. auris AR0390 treated with itraconazole (ITC) at 1 µg/ml. Significant down-regulated DEGs are shown in green, whereas significant up-regulated DEGs are shown in red
Table 2.
List of differentially expressed genes (DEGs) for C. auris AR0390 treated with a combination of aprepitant (10 µg/ml) and itraconazole (1 µg/ml)
| ID | Name | Log2 Fold Change | P-value | CGD* Description |
|---|---|---|---|---|
| 40027169 | HGT6 | −4.90 | 3.1 x 10−20 | Putative high-affinity major facilitator superfamily (MFS) glucose transporter |
| 40027170 | HGT8 | −3.29 | 5.5 x 10−10 | High-affinity glucose transporter of the major facilitator superfamily |
| 40029385 | LIP2 | −3.09 | 1.5 x 10−6 | Secreted lipase, expressed in the alimentary tract, but not during oral infection |
| 40027445 | FTR1 | −2.63 | 1.5 x 10−6 | High-affinity iron permease, required for mouse virulence |
| 40029696 | - | −2.61 | 1.2 x 10−3 | Protein of unknown function |
| 40028877 | ZRT2 | −2.48 | 9.1 x 10−6 | Zinc transporter, essential for zinc uptake |
| 40027663 | CTR1 | −2.24 | 7.2 x 10−5 | Ortholog(s) have copper/iron transmembrane transporter activity |
| 40026070 | orf19.6983 | −2.21 | 8.5 x 10−5 | Protein of unknown function, repressed by nitric oxide |
| 40029380 | TYE7 | −2.20 | 1.7 x 10−4 | Transcription factor, control of glycolysis, required for biofilm formation |
| 40028922 | CDG1 | −2.04 | 5.2 x 10−4 | Putative cysteine dioxygenases, role in conversion of cysteine to sulfite |
| 40029539 | ADH1 | −2.03 | 5.2 x 10−4 | Alcohol dehydrogenase |
| 40026699 | PDC11 | −1.87 | 1.9 x 10−3 | Pyruvate decarboxylase, antigenic fluconazole, farnesol induced |
| 40026939 | HGT12 | −1.82 | 3.2 x 10−3 | Glucose, fructose, mannose transporter, major facilitator superfamily |
| 40029413 | JEN2 | −1.79 | 4.1 x 10−3 | Dicarboxylic acid transporter, regulated by glucose repression |
| 40027889 | YHB1 | −1.78 | 4.1 x 10−3 | Nitric oxide dioxygenase, acts in nitric oxide scavenging/detoxification |
| 40025324 | PMC1 | 1.78 | 2.3 x 10−5 | Vacuolar calcium P-type ATPase, mutants show increased resistance to fluconazole |
| 40027146 | DUR3 | 1.82 | 1.8 x 10−5 | High-affinity spermidine transporter |
| 40030172 | ERG2 | 1.88 | 9.2 x 10−6 | C-8 sterol isomerase, an enzyme important for ergosterol biosynthesis |
| 40029187 | MDR1 | 1.97 | 3.7 x 10−6 | Plasma membrane MDR/MFS multidrug efflux pump |
| 40026984 | orf19.3988 | 1.98 | 5.8 x 10−6 | Putative adhesin-like protein |
| 40025409 | ERG1 | 1.99 | 2.8 x 10−6 | Squalene epoxidase, ergosterol biosynthesis |
| 40030782 | ERG10 | 2.04 | 5.9 x 10−6 | Acetyl-CoA acetyltransferase, role in ergosterol biosynthesis |
| 40025820 | - | 2.04 | 2.3 x 10−6 | Protein of unknown function |
| 40029256 | PIR1 | 2.22 | 2.3 x 10−7 | 1,3-beta-glucan-linked cell wall protein, iron regulated |
| 40029691 | - | 2.23 | 2.7 x 10−7 | Protein of unknown function |
| 40025314 | CDR1 | 2.25 | 1.5 x 10−7 | Multidrug transporter of ABC superfamily |
| 40029715 | orf19.5169 | 2.29 | 9.7 x 10−7 | Has domain(s) with predicted amidase activity |
| 40025414 | - | 2.34 | 6.6 x 10−8 | Protein of unknown function |
| 40027963 | INO1 | 2.38 | 7.9 x 10−8 | nositol-1-phosphate synthase/glycosylation predicted |
| 40028953 | CAN1 | 2.53 | 1.2 x 10−8 | Basic amino acid permease |
| 40028243 | ERG24 | 2.60 | 2.2 x 10−9 | C-14 sterol reductase has a role in ergosterol biosynthesis |
| 40027842 | RCT1 | 2.72 | 4.8 x 10−10 | Fluconazole-induced protein, required for caspofungin tolerance |
| 40029690 | RTA3 | 3.02 | 1.4 x 10−11 | Putative lipid translocase influences the susceptibility of C. albicans to fluconazole |
| 40030199 | DDR48 | 3.19 | 1.1 x 10−12 | Immunogenic stress-associated protein |
*Descriptions of the identified DEGs obtained from the Candida Genome Database (CGD) combined with a tBLASTx search.
With regards to single treatments with either aprepitant or itraconazole, no significant change in C. auris’ transcriptome was observed in the aprepitant treated condition (Figure 4(c)). However, a total of 18 DEGs (10 upregulated + 8 downregulated) were detected in samples treated with itraconazole alone (Figure 4(d) and Table 3). Among the upregulated genes in the itraconazole treated condition were amino acid permeases (DIP5 and CAN1), stress-associated genes (DDR48 and PGA31), ergosterol biosynthesis (ERG24), and a redox related gene, ferric reductase (FRE3).
Table 3.
List of differentially expressed genes (DEGs) for C. auris AR0390 treated with itraconazole (1 µg/ml)
| ID | Name | Log2 Fold Change | P-value | CGD* Description |
|---|---|---|---|---|
| 40027169 | HGT6 | −2.9273 | 9.6 x 10−8 | Putative MFS glucose transporter |
| 40027170 | HGT8 | −2.7557 | 6.1 x 10−7 | High-affinity glucose transporter of the major facilitator superfamily |
| 40029385 | LIP2 | −2.3233 | 4.5 x 10−4 | Secreted lipase |
| 40029539 | ADH1 | −2.3056 | 7.7 x 10−5 | Alcohol dehydrogenase |
| 40027889 | YHB1 | −2.245 | 1.2 x 10−4 | Nitric oxide dioxygenase |
| 40029413 | JEN2 | −2.1117 | 4.4 x 10−4 | Dicarboxylic acid transporter |
| 40028527 | TDH3 | −2.0678 | 4.8 x 10−4 | NAD-linked glyceraldehyde-3-phosphate dehydrogenase |
| 40030546 | DAL81 | −2.0262 | 3.7 x 10−4 | RNA polymerase II repressing transcription factor binding activity |
| 40025268 | PGA30 | 1.9233 | 3.1 x 10−4 | Predicted GPI-anchored protein |
| 40026925 | DIP5 | 1.9692 | 4.7 x 10−4 | Dicarboxylic amino acid permease |
| 40028209 | PRA1 | 2.0941 | 4.5 x 10−4 | Cell surface protein that sequesters zinc from host tissue |
| 40027842 | RCT1 | 2.1026 | 4.5 x 10−4 | Fluconazole-induced protein, required for caspofungin tolerance |
| 40029187 | MDR1 | 2.1045 | 4.4 x 10−4 | Plasma membrane MDR/MFS multidrug efflux pump |
| 40030199 | DDR48 | 2.1539 | 3.7 x 10−4 | Stress-associated protein, induced by benomyl/caspofungin/ketoconazole |
| 40030120 | PGA31 | 2.3457 | 7.7 x 10−5 | Protein associated with cellular response to chemical stimulus |
| 40028243 | ERG24 | 2.3585 | 5.2 x 10−5 | C-14 sterol reductase has a role in ergosterol biosynthesis |
| 40025718 | FRE3 | 2.7106 | 1.09 x 10−6 | Protein with similarity to ferric reductase Fre10p |
| 40028953 | CAN1 | 3.4505 | 3.5 x 10−10 | Basic amino acid permease |
* Descriptions of the identified DEGs obtained from the Candida Genome Database (CGD) combined with a tBLASTx search.
In order to obtain an overall insight into the impact of aprepitant/itraconazole on the C. auris transcriptome, we performed a Gene Ontology (GO) analysis of the identified DEGs. Twenty-six GO terms were significantly overrepresented (enriched) in the aprepitant/itraconazole treated condition. However, no significant overrepresented GO terms were found in the itraconazole treated condition. Among the enriched GO terms, only one GO term related to membrane components was upregulated in the aprepitant/itraconazole sample (Figure 5(a) and Supplementary Table S4). However, a total of 25 GO terms were found to be downregulated in the aprepitant/itraconazole treated sample (Figure 5(b) and Supplementary Table S5). Notably, 18 downregulated GO terms were found to be associated with membrane transport processes, of which 11 GO terms were involved in metal ions and cation transport, indicating the negative impact that aprepitant/itraconazole combination has on the ions homeostasis in C. auris. Of note, since all genes that demonstrated differential expression in response to aprepitant/itraconazole treatments have orthologs in other Candida species, we expect this combination would have a similar mechanism of action in all susceptible species.
Figure 5.
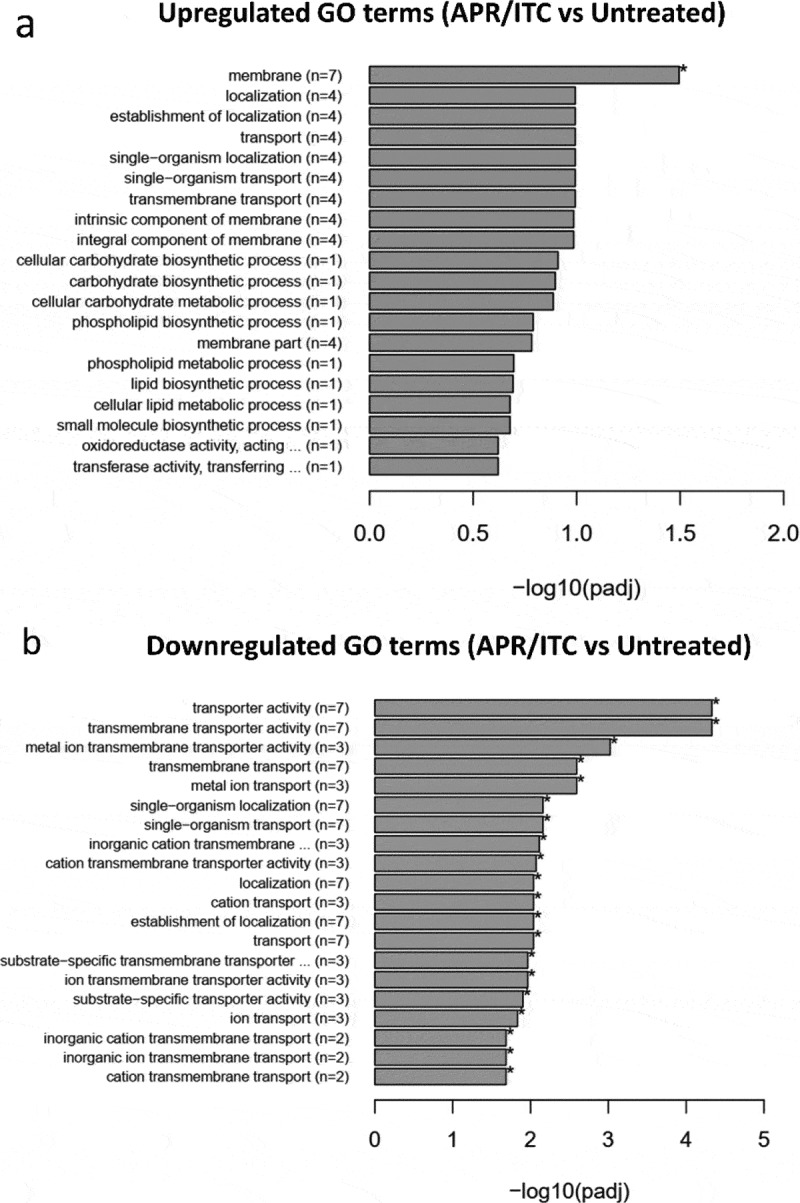
Enrichment analysis of all GO (gene ontology) terms of DEGs identified from the aprepitant/itraconazole combination (APR/ITC) versus the untreated control. GO analysis of DEGs was implemented by the ClusterProfiler R package and a P-value ≤ 0.05 was used as the cutoff parameter. Panel (a) displays up-regulated GO terms and panel (b) displays down-regulated DEGs
Validation of RNA-Seq data
Based on the RNA-Seq data, we selected six DEGs (three downregulated and three upregulated) exhibiting a broad range of differential expression for validation by RT-qPCR. Of note, three genes selected for analysis are involved in ergosterol biosynthesis and three genes are involved in metal ion homeostasis. Consistent with the RNA-Seq data, the qPCR analysis showed significant differential expression of the tested genes and confirmed the significant overexpression of ergosterol biosynthesis-related genes (ERG1, ERG2, ERG10) and the significant downregulation of metal transport-related genes (FTR1, ZRT2, and CTR1) only in the cells treated with the aprepitant/itraconazole combination (Figure 6).
Figure 6.
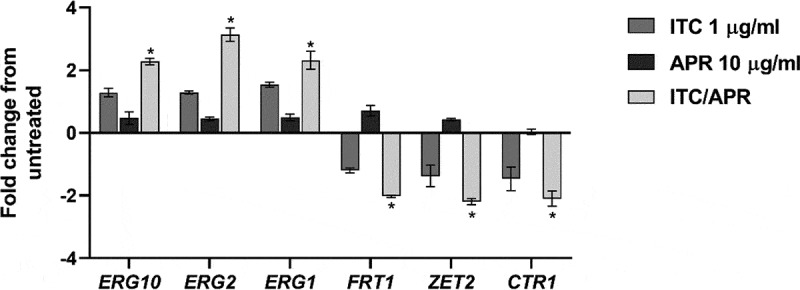
Quantification of C. auris selected genes that were differentially expressed only in the aprepitant/itraconazole combination (APR/ITC). Log phase C. auris AR0390 cells were exposed to APR (10 µg/ml), ITC (1 µg/ml), or a combination of both drugs for three hours. Following treatment, cells were harvested, lysed, and RNA was extracted. Gene expression was determined by quantitative RT-PCR using ACT1 as a housekeeping gene and the untreated sample as a reference control. An absolute two-fold change relative to the untreated control was set as a statistical cutoff value. The result is presented as mean ± SD
Effect of aprepitant on ROS production
Since metal ion homeostasis is critical for multiple biological processes, including redox regulatory mechanisms, we were curious to examine the effect of the aprepitant/itraconazole combination on the ability of C. auris to detoxify reactive oxygen species (ROS). As presented in Figure 7, aprepitant (at 10 µg/ml) significantly increased the intracellular ROS levels by ~ 300% relative to the untreated control, while itraconazole treatment (at 1 µg/ml) significantly increased the ROS levels by 270% relative to the untreated control. On the other hand, the aprepitant/itraconazole combination resulted in a more prominent effect by increasing the ROS production by ~ 600% relative to the untreated control. Interestingly, when the assay medium was supplemented with iron, we noticed that C. auris cells generated significantly lower levels of ROS as compared to iron-deficient media (Figure 7). This result indicates that both aprepitant and the aprepitant/itraconazole combination can interfere significantly with ROS detoxification in C. auris, apparently through the interference with metal ion transport.
Figure 7.
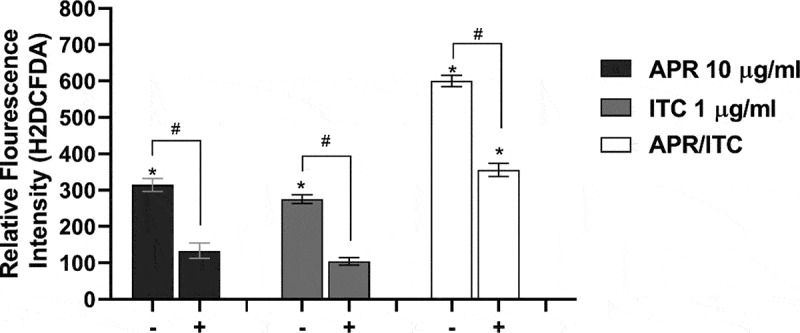
Effect of aprepitant/itraconazole on the ROS levels in the presence (+) or absence (-) of iron. Carboxy-H2DCFDA probe was used to analyze ROS levels within C. auris AR0390 in the presence (+) or absence of iron source (FeSO4 50 µM). PBS suspensions of C. auris cells, at 1× 106 CFU/ml, were treated with either aprepitant (APR, 10 µg/ml), itraconazole (ITC, 1 µg/ml), or a combination of both for 3 h, and then were treated with H2DCFDA for 1 h, before measuring the fluorescence intensity as an indication of ROS levels. Data are shown as means ± SD. An asterisk (*) indicates statistical significance (P < 0.05) between treated and untreated groups (P < 0.05), whereas #, indicates statistical significance (P < 0.05) between treatment groups in the presence or absence of iron. Statistical significance was assessed by one-way ANOVA using Dunnett’s test for multiple comparisons
Effect of metal ion supplementation on the synergistic relationship between aprepitant and itraconazole
Based on the data obtained from the transcriptomic analysis, several genes involved in metal ion transport were found to be downregulated in cells treated with the aprepitant/itraconazole combination. Thus, it was rational to investigate whether supplementation with exogenous metal ions would reverse the synergistic interaction observed between aprepitant and itraconazole. As shown in Figure 8(a), iron supplementation interfered significantly with the synergistic relationship of aprepitant and itraconazole, as demonstrated by a time-kill assay. Ferrous sulfate (at 50 µM) completely negated the fungicidal activity of the aprepitant/itraconazole combination against the test isolate (C. auris AR0390). This effect was further demonstrated by a spotting assay where iron supplementation restored the visible growth of C. auris colonies on YPD agar and eliminated the fungicidal effect of the aprepitant/itraconazole combination (Figure 8(b)). It should be noted that copper supplementation did not affect the antifungal activity of aprepitant/itraconazole (data not shown). Also, it was not possible to examine the effect of zinc supplementation on the relationship between aprepitant and itraconazole, as we noticed an immediate precipitation reaction when zinc sulfate was mixed with aprepitant. However, these results indicate that interference with iron homeostasis is a key mechanism that contributes to the synergistic relationship observed between aprepitant and itraconazole against C. auris.
Figure 8.
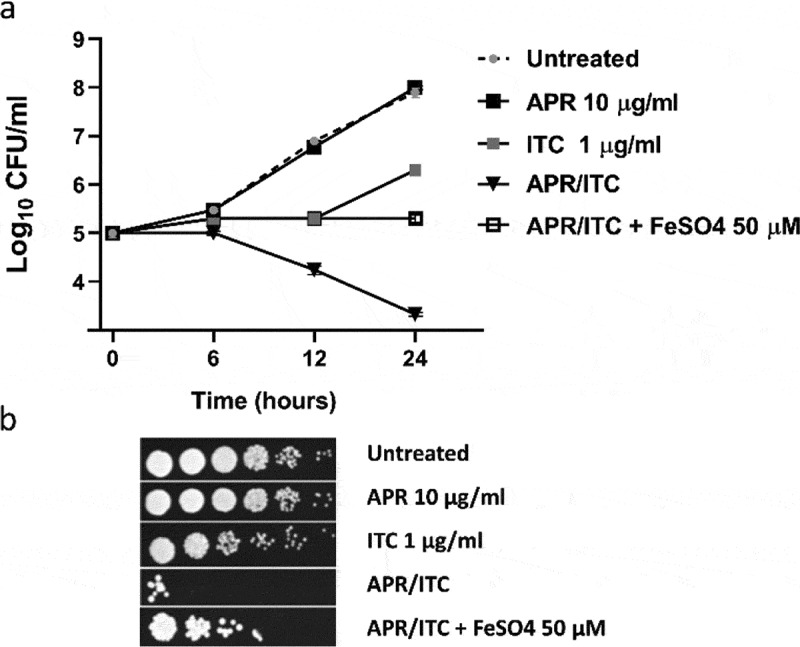
Effect of iron supplementation on the antifungal activity of aprepitant/itraconazole (APR/ITC). (a) Effect of iron supplement (FeSO4, at 50 µM) on the time-kill kinetics of aprepitant (10 µg/ml), itraconazole (1 µg/ml), or a combination of both drugs against C. auris AR0390. (b) Spot assays demonstrated the ability of iron supplementation to reverse the antifungal effect exerted by the aprepitant/itraconazole combination (10/1 µg/ml)
Efficacy of aprepitant/itraconazole in a Caenorhabditis elegans model of C. auris infection
To further corroborate the in vitro synergistic relationship observed between aprepitant and itraconazole, we tested the in vivo efficacy of aprepitant/itraconazole using C. elegans as an infection model. As shown in Figure 9(a), treatment with aprepitant alone at 10 µg/ml failed to reduce the fungal CFU burden in C. elegans nematodes infected with C. auris AR0390. However, itraconazole at 1 µg/ml (1 × MIC) was able to significantly reduce the fungal CFU burden by ~42 ± 3.6%, compared to the untreated control. Interestingly, the aprepitant/itraconazole combination was able to significantly reduce the C. auris CFU burden in the infected nematodes by ~92 ± 2.4%, compared to the untreated control.
Figure 9.
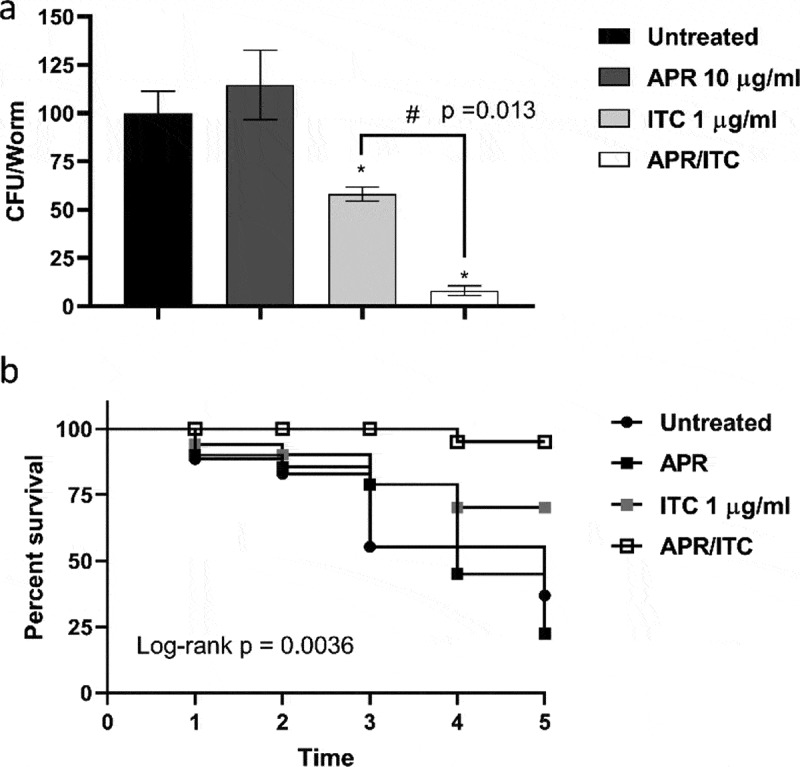
In vivo efficacy of aprepitant/itraconazole in Caenorhabditis elegans infected with C. auris. C. elegans nematodes were infected with 1 × 107 CFU C. auris AR0390 and then treated with aprepitant (APR), at 10 µg/ml, and itraconazole (ITC), at 1 µg/ml, either alone or in combination. Untreated worms served as a negative control. (a) C. auris AR0390 CFU burden/worm 24 h post-treatment. An asterisk (*) indicates statistical significance (P < 0.05) relative to the untreated control, while # indicates statistical significance (P < 0.05) compared to the ITC treatment. Statistical significance was assessed by one-way ANOVA using Dunnett’s test for multiple comparisons. (b) Kaplan-Meier survival curve, assessed by log-rank test for significance, of C. elegans nematodes infected by C. auris AR0390 and treated with APR (10 µg/ml), ITC (1 µg/ml), or a combination of both agents
Next, we assessed the effect of aprepitant/itraconazole on the survival of infected nematodes. As shown in Figure 9(b), only 20% of the untreated C. elegans nematodes survived C. auris infection for five days post-infection. As expected, aprepitant at 10 µg/ml did not improve the survival of infected nematodes with respect to the untreated control. However, itraconazole at 1 µg/ml (1 × MIC) enhanced the survival of the infected worms by ~ 40% (P < 0.05), compared to only 20% in the untreated group. On the other hand, the aprepitant/itraconazole combination significantly enhanced the survival of the infected nematodes as ~ 90% (P < 0.05) of C. elegans remained alive for five days. Altogether, these findings indicate that aprepitant/itraconazole has a potent in vivo efficacy of treating C. auris infection of C. elegans and warrants further investigation in higher animal models.
Discussion
Drug repurposing is a valuable tool to identify novel antimicrobial agents and co-drugs capable of enhancing the activity of existing antimicrobials [27,43–46]. The risk posed by the recently identified fungal pathogen C. auris has prompted CDC to classify C. auris as an urgent threat that requires immediate action [3]. A hallmark trait of C. auris is its ability to resist multiple antifungal therapeutics, especially azoles. Here, we utilized a whole-cell screening assay to evaluate the azole chemosensitizing activity of ~1547 FDA-approved drugs and clinical molecules against the multidrug-resistant C. auris isolate AR0390. The screening data (unpublished) revealed aprepitant, an NK1 antagonist used for the treatment of chemotherapy-associated emesis, was able to restore the antifungal activity of fluconazole against the test isolate. This observation encouraged the exploration of the azole chemosensitizing activity of aprepitant against a panel of ten C. auris isolates, in addition to several clinically important Candida species. Checkerboard data indicate that aprepitant enhances the antifungal activity of azole drugs against various Candida species. More specifically, aprepitant interacted synergistically with itraconazole against the majority of Candida isolates tested. Additionally, combining aprepitant with itraconazole changed the fungistatic nature of itraconazole and produced a prominent fungicidal effect against C. auris AR0390 as demonstrated by the time-kill study. From a clinical point of view, this observation is of interest since fungicidal therapeutics are expected to improve the clinical outcome of invasive Candida infections, especially in patients with compromised immune systems [47,48].
A well-documented virulence factor of Candida species is the notable ability to form adherent structures, known as biofilms, on surfaces of medical devices [49–51]. Given the potent synergistic interaction and the fungicidal effect displayed by the aprepitant/itraconazole combination against planktonic C. auris cells, it was of interest to examine whether a similar effect could exist against the difficult-to-treat fungal biofilms. While neither aprepitant nor itraconazole alone was able to interfere with the biofilm-forming ability of C. auris, the aprepitant/itraconazole combination was effective at reducing the metabolic activity of fungal cells residing within premature and mature biofilms. These observations may be of clinical importance since biofilms are known to hinder the penetration of antifungal drugs and host immune effectors and also trigger the formation of Candida persister cells, leading to poor clinical outcomes [52,53].
To gain insight into the molecular mechanism by which aprepitant interacts synergistically with azole drugs against C. auris, we performed comparative transcriptomic profiling of C. auris cells treated with DMSO (1%), aprepitant (10 µg/ml), itraconazole (1 µg/ml), or the aprepitant/itraconazole combination (10/1 µg/ml). Our data indicate that aprepitant, at the tested concentration, has no significant effect on C. auris’ transcriptome. However, a total of 18 and 34 genes displayed significant changes in expression in groups treated with itraconazole and aprepitant/itraconazole respectively, compared to the untreated control. We focused our attention on the differentially expressed genes that were found to be specific to the aprepitant/itraconazole treated group. We noticed that several genes involved in metal ions transport were significantly downregulated in cells treated with aprepitant/itraconazole, relative to the untreated control. This observation suggests the aprepitant/itraconazole combination can interfere with metal ion homeostasis in C. auris. Since metal ions are key components of several essential genes, it is expected that disturbances in metal ion homeostasis would compromise multiple vital biological processes. One striking example is the critical role of metal ions, particularly iron, in the activity of many enzymes involved in the detoxification of reactive oxygen species (ROS) [54–56]. Thus, we sought to examine the effect of aprepitant/itraconazole on the ROS detoxification ability of C. auris. Interestingly, aprepitant by itself compromised the ROS detoxification ability of C. auris, resulting in elevated ROS levels (eight-times higher compared to the untreated control). This effect was even more pronounced with the aprepitant/itraconazole combination which generated ~ 15-times more ROS levels, compared to the untreated control. These increases in ROS levels could explain the observed fungicidal activity against planktonic and biofilms of C. auris as previously demonstrated in other studies [57–59]. Further supporting the critical role of iron homeostasis in controlling the intracellular ROS levels, we noticed that in iron supplemented media, C. auris cells treated with aprepitant/itraconazole produced significantly lower ROS levels, compared to iron-depleted media. Interestingly, similar observations have been recorded in plant and mammalian cells whereas high ROS levels were produced in iron depleting conditions [60,61], indicating that iron homeostasis has a conservative protective role against the ROS formation.
In Saccharomyces cerevisiae, iron homeostasis is known to be an important regulator of ergosterol biosynthesis [62]. In iron depleting conditions, yeast cells were reported to express higher mRNA levels of several ERG genes such as ERG1, ERG7, and ERG11 [62]. Consistent with this phenomenon, we noticed that several genes involved in ergosterol biosynthesis were significantly upregulated such as ERG1, ERG2, and ERG10. Our data also shows increased mRNA levels of ERG11, however, this upregulation didn’t meet our strict criteria for statistical significance. This upregulation of ERG genes could be viewed as a compensatory mechanism by which C. auris cells try to adapt to the reduced ergosterol content. Indeed, previous studies have shown that ergosterol depletion causes the upregulation of several genes involved in ergosterol biosynthesis [63,64].
Notably, iron homeostasis has been regarded as a potential target for other azole chemosensitizing agents such as doxycycline and lactoferrin [65,66]. Disturbance in iron transport was reported to affect the membrane permeability and enhance the activity of drugs that target cell membrane biosynthetic pathways such as azoles [67]. Additionally, iron depletion is thought to suppress the calcineurin pathway, leading to impairment of drug-induced stress responses in Candida [68]. Thus, we were interested to examine whether iron supplementation would weaken the synergistic interaction observed between aprepitant and itraconazole against Candida. Indeed, iron supplementation negated the fungicidal activity of aprepitant/itraconazole as shown in our time-kill kinetics. However, even in the presence of iron, the aprepitant/itraconazole combination was still able to exert a noticeable fungistatic activity against C. auris. This suggests that other additional mechanisms may contribute to the synergistic relationship between aprepitant and itraconazole. Of note, in contrast to iron, copper supplementation did not interfere with the antifungal activity of the aprepitant/itraconazole combination.
To conclude, our data indicate that aprepitant was able to enhance the antifungal activity of azole drugs, particularly itraconazole. The aprepitant/itraconazole combination displayed broad-spectrum antifungal activity and was fungicidal against planktonic and biofilms of C. auris, suggesting a promising clinical indication for the treatment of azole-recalcitrant infections. Furthermore, this novel drug combination demonstrated potent in vivo efficacy in a C. elegans infection model. The synergistic relationship between aprepitant and itraconazole appears to be mediated through potent interference with metal ion homeostasis, and the subsequent compromise in ROS detoxifying mechanisms and ergosterol biosynthesis, however, further molecular studies are needed to fully understand the mechanism of the azole chemosensitizing activity of aprepitant and its effect on the ergosterol pathway. Moreover, efficacy in higher animal models and further clinical studies are needed to fully assess the potential clinical use of aprepitant/itraconazole as a novel drug combination with promising antifungal activity against emergent multidrug-resistant C. auris.
Supplementary Material
Acknowledgments
The authors would like to thank BEI Resources and the US Centers for Disease Control and Prevention (CDC) for providing the clinical isolates used in this study. We also would like to thank Dr. Haroon Mohammad (Purdue University) for proofreading the manuscript.
Disclosure statement
The author(s) declare no competing interests.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Nett JE. Candida auris: an emerging pathogen “incognito”? PLoS Pathog. 2019;15:e1007638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sears D, Schwartz BS. Candida auris: an emerging multidrug-resistant pathogen. Inter J Infect Dis. 2017;63:95–98. [DOI] [PubMed] [Google Scholar]
- [3].CDC . Antibiotic resistance threats in the United States. Atlanta (GA): U.S. Department of Health and Human Services, Antibiotic Resistance Threats in the United States; 2019. (Accessed on 18 April 2020). Available from: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf [Google Scholar]
- [4].Eldesouky HE, Li X, Abutaleb NS, et al. Synergistic interactions of sulfamethoxazole and azole antifungal drugs against emerging multidrug-resistant Candida auris. Int J Antimicrob Agents. 2018;52:754–761. [DOI] [PubMed] [Google Scholar]
- [5].CDC . Antifungal susceptibility and resistance breakpoints in Candida auris; 2020. (Accessed on 18 April 2020). Available from: https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html
- [6].Jeffery-Smith A, Taori SK, Schelenz S, et al. Candida auris: a review of the literature. Clin Microbiol Rev. 2018;31. DOI: 10.1128/CMR.00029-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chowdhary A, Sharma C, Meis JF. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017;13:e1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Forsberg K, Woodworth K, Walters M, et al. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med Mycol. 2019;57:1–12. [DOI] [PubMed] [Google Scholar]
- [9].Mohammad H, Elghazawy NH, Eldesouky HE, et al. Discovery of a novel dibromoquinoline compound exhibiting potent antifungal and antivirulence activity that targets metal ion homeostasis. ACS Infect Dis. 2018;4:403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Haegler P, Joerin L, Krahenbuhl S, et al. Hepatocellular toxicity of imidazole and triazole antimycotic agents. Toxicol Sci. 2017;157:183–195. [DOI] [PubMed] [Google Scholar]
- [11].Eldesouky HE, Mayhoub A, Hazbun TR, et al. Reversal of azole resistance in Candida albicans by sulfa antibacterial drugs. Antimicrob Agents Chemother. 2018;62. DOI: 10.1128/AAC.00701-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sheehan DJ, Hitchcock CA, Sibley CM. Current and emerging azole antifungal agents. Clin Microbiol Rev. 1999;12:40–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lewis RE. Current concepts in antifungal pharmacology. Mayo Clin Proc. 2011;86:805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Allen D, Wilson D, Drew R, et al. Azole antifungals: 35 years of invasive fungal infection management. Expert Rev Anti Infect Ther. 2015;13:787–798. [DOI] [PubMed] [Google Scholar]
- [15].Zavrel M, Esquivel BD, White TC. The ins and outs of azole antifungal drug resistance: molecular mechanisms of transport. In: Gotte M, Berghuis A, Matlashewski G, et al., editors. Handbook of antimicrobial resistance. New York (NY): Springer; 2014:1. [Google Scholar]
- [16].Wiederhold NP. Antifungal resistance: current trends and future strategies to combat. Infect Drug Resist. 2017;10:249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Whaley SG, Berkow EL, Rybak JM, et al. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front Microbiol. 2016;7:2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Worthington RJ, Melander C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol. 2013;31:177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chiang CY, Uzoma I, Moore RT, et al. Mitigating the impact of antibacterial drug resistance through host-directed therapies: current progress, outlook, and challenges. mBio. 2018;9. DOI: 10.1128/mBio.01932-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pizzorno A, Padey B, Terrier O, et al. Drug repurposing approaches for the treatment of influenza viral infection: reviving old drugs to fight against a long-lived enemy. Front Immunol. 2019;10:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gurunathan S, Kang MH, Qasim M, et al. Nanoparticle-mediated combination therapy: two-in-one approach for cancer. Int J Mol Sci. 2018;19:3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gu WR, Guo DM, Zhang LP, et al. The synergistic effect of azoles and fluoxetine against resistant Candida albicans strains is attributed to attenuating fungal virulence. Antimicrob Agents Ch. 2016;60:6179–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sun LM, Liao K, Liang S, et al. Synergistic activity of magnolol with azoles and its possible antifungal mechanism against Candida albicans. J Appl Microbiol. 2015;118:826–838. [DOI] [PubMed] [Google Scholar]
- [24].Chen YL, Lehman VN, Averette AF, et al. Posaconazole exhibits in vitro and in vivo synergistic antifungal activity with caspofungin or FK506 against Candida albicans. Plos One. 2013;8:e57672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Silva LV, Sanguinetti M, Vandeputte P, et al. Milbemycins: more than efflux inhibitors for fungal pathogens. Antimicrob Agents Chemother. 2013;57(2):873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mohammad H, Eldesouky HE, Hazbun T, et al. Identification of a phenylthiazole small molecule with dual antifungal and antibiofilm activity against Candida albicans and Candida auris. Sci Rep. 2019;9:18941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Thangamani S, Eldesouky HE, Mohammad H, et al. Ebselen exerts antifungal activity by regulating glutathione (GSH) and reactive oxygen species (ROS) production in fungal cells. Biochim Biophys Acta Gen Subj. 2017;1861:3002–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mohammad H, Kyei-Baffour K, Younis W, et al. Investigation of aryl isonitrile compounds with potent, broad-spectrum antifungal activity. Bioorg Med Chem. 2017;25:2926–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Scorneaux B, Angulo D, Borroto-Esoda K, et al. SCY-078 is fungicidal against candida species in time-kill studies. Antimicrob Agents Chemother. 2017;61. DOI: 10.1128/AAC.01961-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chang W, Liu J, Zhang M, et al. Efflux pump-mediated resistance to antifungal compounds can be prevented by conjugation with triphenylphosphonium cation. Nat Commun. 2018;9:5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kubiczek D, Raber H, Gonzalez-García M, et al. Derivates of the antifungal peptide Cm-p5 inhibit development of Candida auris biofilms in vitro. Antibiotics (Basel). 2020;9. DOI: 10.3390/antibiotics9070363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lara HH, Ixtepan-Turrent L, Jose Yacaman M, et al. Inhibition of Candida auris biofilm formation on medical and environmental surfaces by silver nanoparticles. ACS Appl Mater Interfaces. 2020;12:21183–21191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dominguez EG, Zarnowski R, Choy HL, et al. Conserved role for biofilm matrix polysaccharides in Candida auris drug resistance. mSphere. 2019;4. DOI: 10.1128/mSphereDirect.00680-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chen S, Zhou Y, Chen Y, et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. [DOI] [PubMed] [Google Scholar]
- [37].Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Benjamini YH. Yosef Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- [40].Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chau AS, Mendrick CA, Sabatelli FJ, et al. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob Agents Chemother. 2004;48:2124–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Eldesouky HE, Salama EA, Hazbun TR, et al. Ospemifene displays broad-spectrum synergistic interactions with itraconazole through potent interference with fungal efflux activities. Sci Rep. 2020;10:6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Thangamani S, Maland M, Mohammad H, et al. Repurposing approach identifies auranofin with broad spectrum antifungal activity that targets Mia40-Erv1 pathway. Front Cell Infect Microbiol. 2017;7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Thangamani S, Mohammad H, Abushahba MFN, et al. Exploring simvastatin, an antihyperlipidemic drug, as a potential topical antibacterial agent. Sci Rep. 2015;5(1):16407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Eldesouky HE, Salama EA, Li X, et al. Repurposing approach identifies pitavastatin as a potent azole chemosensitizing agent effective against azole-resistant Candida species. Sci Rep. 2020;10(1):7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Andes D. Optimizing antifungal choice and administration. Curr Med Res Opin. 2013;29(Suppl 4):13–18. [DOI] [PubMed] [Google Scholar]
- [48].Graybill JR, Burgess DS, Hardin TC. Key issues concerning fungistatic versus fungicidal drugs. Eur J Clin Microbiol Infect Dis. 1997;16:42–50. [DOI] [PubMed] [Google Scholar]
- [49].Kojic EM, Darouiche RO. Candida infections of medical devices. Clin Microbiol Rev. 2004;17:255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nobile CJ, Johnson AD. Candida albicans biofilms and human disease. Annu Rev Microbiol. 2015;69:71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ramage G, Martinez JP, Lopez-Ribot JL. Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 2006;6:979–986. [DOI] [PubMed] [Google Scholar]
- [52].Cavalheiro M, Teixeira MC. Candida biofilms: threats, challenges, and promising strategies. Front Med (Lausanne). 2018;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wuyts J, Van Dijck P, Holtappels M. Fungal persister cells: the basis for recalcitrant infections? Plos Pathog. 2018;14:ARTN e1007301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Valasatava Y, Rosato A, Furnham N, et al. To what extent do structural changes in catalytic metal sites affect enzyme function? J Inorg Biochem. 2018;179:40–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Riordan JF. The role of metals in enzyme activity. Ann Clin Lab Sci. 1977;7:119–129. [PubMed] [Google Scholar]
- [56].Andreini C, Bertini I, Cavallaro G, et al. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. [DOI] [PubMed] [Google Scholar]
- [57].De Cremer K, De Brucker K, Staes I, et al. Stimulation of superoxide production increases fungicidal action of miconazole against Candida albicans biofilms. Sci Rep. 2016;6:27463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Delattin N, Cammue BP, Thevissen K. Reactive oxygen species-inducing antifungal agents and their activity against fungal biofilms. Future Med Chem. 2014;6:77–90. [DOI] [PubMed] [Google Scholar]
- [59].Tamura M, Cueno ME, Abe K, et al. Ions released from a S-PRG filler induces oxidative stress in Candida albicans inhibiting its growth and pathogenicity. Cell Stress Chaperones. 2018;23:1337–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tewari RK, Hadacek F, Sassmann S, et al. Iron deprivation-induced reactive oxygen species generation leads to non-autolytic PCD in Brassica napus leaves. Environ Exp Bot. 2013;91:74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bystrom LM, Guzman ML, Rivella S. Iron and reactive oxygen species: friends or foes of cancer cells? Antioxid Redox Signal. 2014;20:1917–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jorda T, Puig S. Regulation of ergosterol biosynthesis in saccharomyces cerevisiae. Genes (Basel). 2020;11. DOI: 10.3390/genes11070795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Henry KW, Nickels JT, Edlind TD. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob Agents Chemother. 2000;44:2693–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hu C, Zhou M, Wang W, et al. Abnormal Ergosterol biosynthesis activates transcriptional responses to antifungal azoles. Front Microbiol. 2018;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fiori A, Van Dijck P. Potent synergistic effect of doxycycline with fluconazole against Candida albicans is mediated by interference with iron homeostasis. Antimicrob Agents Chemother. 2012;56:3785–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kobayashi T, Kakeya H, Miyazaki T, et al. Synergistic antifungal effect of lactoferrin with azole antifungals against Candida albicans and a proposal for a new treatment method for invasive candidiasis. Jpn J Infect Dis. 2011;64:292–296. [PubMed] [Google Scholar]
- [67].Prasad T, Chandra A, Mukhopadhyay CK, et al. Unexpected link between iron and drug resistance of Candida spp.: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob Agents Chemother. 2006;50:3597–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hameed S, Dhamgaye S, Singh A, et al. Calcineurin signaling and membrane lipid homeostasis regulates iron mediated multidrug resistance mechanisms in Candida albicans. PLoS One. 2011;6:e18684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


