Abstract
Mosquitoes are insects of medical importance due their role as vectors of different pathogens to humans. There is a lack of information about the evolutionary history and phylogenetic positioning of the majority of mosquito species. Here we characterized the mitogenomes of mosquito species through low-coverage whole genome sequencing and data mining. A total of 37 draft mitogenomes of different species were assembled from which 16 are newly-sequenced species. We datamined additional 49 mosquito mitogenomes, and together with our 37 mitogenomes, we reconstructed the evolutionary history of 86 species including representatives from 15 genera and 7 tribes. Our results showed that most of the species clustered in clades with other members of their own genus with exception of Aedes genus which was paraphyletic. We confirmed the monophyletic status of the Mansoniini tribe including both Coquillettidia and Mansonia genus. The Aedeomyiini and Uranotaeniini were consistently recovered as basal to other tribes in the subfamily Culicinae, although the exact relationships among these tribes differed between analyses. These results demonstrate that low-coverage sequencing is effective to recover mitogenomes, establish phylogenetic knowledge and hence generate basic fundamental information that will help in the understanding of the role of these species as pathogen vectors.
Subject terms: Evolutionary genetics, Evolutionary biology
Introduction
Mosquitoes compose a large group of insects from the Culicidae family. There are around 3.567 valid species classified into two subfamilies (Anophelinae and Culicinae) and 41 genera (https://mosquito-taxonomic-inventory.info/ accessed on 21 Oct., 2019). The vast majority of mosquitoes species have anthropophilic behaviour towards reptiles and mammals including humans1. Because of that they can transmit many pathogens such as bacteria2, malaria protozoa3, filarial worms4 and arboviruses5 to the species they feed upon. Mosquitoes are responsible for the transmission of pathogens that cause outbreaks and epidemics annually in the tropical region, but the current globalization and land use change are increasing human-mosquito contact allowing the emergence of new mosquito-borne disease6–8. Several of the new emerging pathogens arose from forested environments where they circulate in a sylvatic cycle between wild animals and arthropod vector species such as mosquitoes9. Although there is abundant evidence that spillover occurs from sylvatic to urban environments, we know very little about the sylvatic cycle of these pathogens including the vector species that transmit them in their natural environment10. Therefore, basic knowledge about vector evolution and ecology is highly needed to better understand their role in the transmission cycle of pathogens11–13.
The huge improvement in nucleic acid sequencing platforms in the last decade has allowed an explosion of genomic information from a wide range of species. Mitogenomes, the entire mitochondrial genome, have been widely used as a target molecule to elucidate different aspects of metazoa species evolution such as population dynamics and phylogenetic relationships14. Complete mitogenomes are reliable tools to be used as a source of molecular markers in ecological and evolutionary studies because they provide genes with different evolutionary rates such as the most conserved rRNA genes (12S and 16S), the intermediate ND1-6 genes and the fast evolving cytochrome c oxidase subunit I (COI) gene, the most used molecular marker for species identification, allowing an accurate establishment of both ancient and recent speciation events15–17. In addition, mitogenomes have uniparental heritage, high copy number by cells and single-copy genes which facilitates DNA recovery and phylogenetic analysis18–20. Recently, some studies have sequenced a larger number of mitochondrial genomes from different mosquito species, but they are mostly focused on species from the Anopheles genus21–23. Mosquito mitogenomes are structurally conserved following the metazoa gene number and order, with few exceptions, showing 37 genes comprising 13 protein coding genes, 22 tRNAs and 2 rRNA genes24–26. Its size range varies from 14,820 bp for An. maculatus to 16,790 bp for Ae. aegypti (https://www.ncbi.nlm.nih.gov/genome/browse#!/organelles/culicidae accessed on 21 Oct, 2019).
Mitogenome sequencing has been a hard task using the first generation of sequence platforms based on the Sanger method. The first mosquito mitogenome was obtained after laborious steps such as mitochondria purification followed by DNA extraction, cloning and Sanger sequencing of several fragments27,28. Today there are a number of alternative approaches available to obtain mitogenomes which was only possible due to the improvement of the second and third generation sequencing platforms. Most of these strategies are based on PCR/Long Range PCR coupled with Next-generation sequencing (NGS), shotgun Whole Genome Sequencing or mitogenome sequencing through RNA-Seq data29,30. Other approaches available allow the recovery of mitogenomes by PCR amplification from environment samples and pooled DNA and mitogenome recovery from low-coverage sequencing31–33. Moreover, a number of bioinformatics tools were developed to specifically assembly and annotate mitogenomes34–38.
Most of the available mosquito mitogenomes belong to Anopheles species with fewer genomes for Culex, Aedes and other genera such as Haemagogus, Bironella, Sabethes, and Lutzia21–23,26,39, but there is no available molecular data for the large majority of the species. Aiming to contribute with this basic and fundamental knowledge we performed low-coverage whole genome sequencing and data mining on already published Culicidae SRA data to characterize the mitogenomes from different species and genera. Overall, we reconstructed and positioned 37 mitogenomes, 35 of them for the first time, representing 11 genera. Our evolutionary analysis covered a large diversity of the Culicinae subfamily evaluating 7 representatives from 11 existing tribes.
Results
Sequencing and mitogenome characterization
The sequenced mosquito samples generated a total of 84.2 million paired-end reads representing the 17 species and eight genera (Aedeomyia, Aedes, Coquillettidia, Culex, Mansonia, Psorophora, Trichoprosopon and Uranotaenia). The amount of generated reads ranged from 1.1 million reads for Ur. pulcherrima to 11.3 million reads for Ae. taeniorhynchus (Table 1). Searching on the SRA database, we included raw sequencing datasets of additional 20 mosquito species for mitogenome characterization representing six genera (Aedes, Anopheles, Culex, Psorophora, Tripteroides and Toxorhynchites). Here, we characterized 35 mitogenomes for the first time and reassembled the An. aquasalis and Cx. nigripalpus mitogenomes that were recently published21,40. In summary, the newly characterized mitogenomes represent eight Culicidae genera that had no mitochondrial genome data available to the best of our knowledge (Aedeomyia, Coquillettidia, Mansonia, Psorophora, Trichoprosopon, Tripteroides, Toxorhynchites, and Uranotaenia).
Table 1.
General statistics for assembled draft mitogenomes.
| Species | Total of reads (Mi) | Final assembly (bp) | Coverage breadth* (%) | Mapped readsa | Final coverage depth | % of mito reads | Total of genes | PCG | rRNA | tRNA |
|---|---|---|---|---|---|---|---|---|---|---|
| Statistics for mitogenomes of species sampled and sequenced in this study | ||||||||||
| Ae. taeniorhynchus | 11.3 | 14,732 | 95.10 | 8174 | 37.73 | 0.07234 | 37 | 13 | 2 | 22 |
| Ma. wilsoni | 8.2 | 10,443 | 67.41 | 3522 | 23.27 | 0.04295 | 28 | 12 | 2 | 14 |
| Tr. digitatum | 7.6 | 8282 | 53.46 | 729 | 5.99 | 0.00959 | 28 | 10 | 2 | 16 |
| Ae. scapularis | 7 | 7795 | 50.32 | 218 | 1.99 | 0.00311 | 25 | 11 | 1 | 13 |
| Ma. titillans | 6.6 | 11,181 | 72.18 | 7338 | 46.60 | 0.11118 | 27 | 11 | 2 | 14 |
| Cq. chrysonotum | 6.2 | 12,032 | 77.67 | 1955 | 11.37 | 0.03153 | 35 | 13 | 2 | 20 |
| Cq. juxtamansonia | 5.5 | 7711 | 49.78 | 826 | 7.18 | 0.01502 | 18 | 8 | 2 | 8 |
| Ps. cingulata | 4.9 | 15,660 | 101.09 | 3551 | 16.33 | 0.07247 | 36 | 13 | 2 | 21 |
| Cx. nigripalpus | 4.8 | 14,492 | 93.55 | 3633 | 17.30 | 0.07569 | 37 | 13 | 2 | 22 |
| Cq. venezuelensis | 4.3 | 13,912 | 89.81 | 2706 | 13.42 | 0.06293 | 34 | 13 | 2 | 19 |
| Ad. squamipennis | 3.9 | 10,466 | 67.56 | 899 | 5.93 | 0.02305 | 27 | 9 | 2 | 16 |
| Cx. corniger | 3.9 | 5222 | 33.71 | 137 | 1.84 | 0.00351 | 20 | 9 | 2 | 9 |
| Cx. amazonensis | 2.5 | 15,265 | 98.54 | 3274 | 15.87 | 0.13096 | 36 | 13 | 2 | 21 |
| Cq. albicosta | 2.4 | 14,689 | 94.82 | 26,487 | 115.40 | 1.10363 | 36 | 13 | 2 | 21 |
| Ma. humeralis | 2.2 | 3699 | 23.88 | 193 | 3.91 | 0.00877 | 14 | 7 | 2 | 5 |
| Cq. hermanoi | 1.8 | 12,289 | 79.33 | 1958 | 11.79 | 0.10878 | 26 | 10 | 2 | 14 |
| Ur. pulcherrima | 1.1 | 13,845 | 89.37 | 781 | 4.23 | 0.07100 | 32 | 12 | 2 | 18 |
| Average | 4.95 | 11,277 | 72.8 | 66,381 | 20.01 | 0.11450 | – | – | – | – |
| Statistics for mitogenomes assembled from SRA data | ||||||||||
| Ae. alboannulatus | 45.6 | 14,314 | 92.40 | 2,645,908 | 22,749.53 | 5.80243 | 26 | 11 | 2 | 13 |
| Ae. camptorhynchus | 41 | 13,825 | 89.25 | 1,461,943 | 11,897.19 | 3.56571 | 30 | 12 | 2 | 16 |
| Ae. detritus | 83.6 | 14,707 | 94.94 | 2,228,562 | 19,597.21 | 2.66574 | 31 | 13 | 2 | 16 |
| Ae. fluviatilis | 6.2 | 14,360 | 92.70 | 3,22,138 | 3858.73 | 5.19577 | 17 | 12 | 1 | 4 |
| Ae. polynesiensis | 31.8 | 15,144 | 97.76 | 39,618 | 133.74 | 0.12458 | 34 | 12 | 2 | 20 |
| Ae. riversi | 17 | 5992 | 38.68 | 12,505 | 23.83 | 0.07356 | 17 | 10 | 1 | 6 |
| An. albimanus | 101.1 | 15,674 | 101.18 | 668,072 | 4304.92 | 0.66080 | 34 | 12 | 2 | 20 |
| An. aquasalis | 0.344 | 11,201 | 72.31 | 12,674 | 571.41 | 3.68002 | 14 | 9 | 2 | 3 |
| An. freeborni | 148.8 | 15,960 | 103.03 | 1,231,449 | 9200.00 | 0.82759 | 33 | 12 | 2 | 19 |
| An. nuneztovari | 76.9 | 12,741 | 82.25 | 42,462 | 446.26 | 0.05522 | 23 | 12 | 2 | 9 |
| An. quadriannulatus | 66.2 | 15,533 | 100.27 | 206,252 | 1341.11 | 0.31156 | 34 | 12 | 2 | 20 |
| Cx. australicus | 44.8 | 15,195 | 98.09 | 3,222,91 | 26,720.50 | 7.19395 | 32 | 13 | 2 | 17 |
| Cx. globocoxitus | 44.2 | 15,123 | 97.62 | 2,970,924 | 24,483.16 | 6.72155 | 31 | 12 | 2 | 17 |
| Cx. hortensis | 91.4 | 13,702 | 88.45 | 3,053,258 | 12,444.56 | 3.34054 | 25 | 11 | 2 | 12 |
| Cx. molestus | 27.2 | 11,440 | 73.85 | 558,460 | 4194.91 | 2.05316 | 21 | 13 | 2 | 6 |
| Cx. tarsalis | 37.8 | 15,831 | 102.19 | 3,037,171 | 28,602.31 | 8.03484 | 33 | 12 | 2 | 19 |
| Cx. torrentium | 47.8 | 12,652 | 81.67 | 558,460 | 4194.91 | 1.16833 | 26 | 13 | 2 | 11 |
| Ps.albipes | 135.6 | 15,791 | 101.94 | 1,087,575 | 8166.76 | 0.80205 | 33 | 12 | 2 | 19 |
| Tp. aranoides | 37.6 | 8990 | 58.03 | 287,439 | 4887.87 | 0.76447 | 9 | 7 | 2 | 0 |
| Tx. amboinensis | 100 | 15,775 | 101.83 | 2,220,264 | 14,215.32 | 2.22026 | 34 | 13 | 2 | 19 |
| Average | 59.24 | 13,698 | 88.2 | 1,293,401 | 5108 | 2.6311 | – | – | – | – |
PCG protein coding genes.
*Coverage breadth was calculated in relation to the average of culicidae mitochondrial genomes length (15,491 bp). Bp represents base pairs.
aMapped Reads against final assembly using MIRAbait tool.
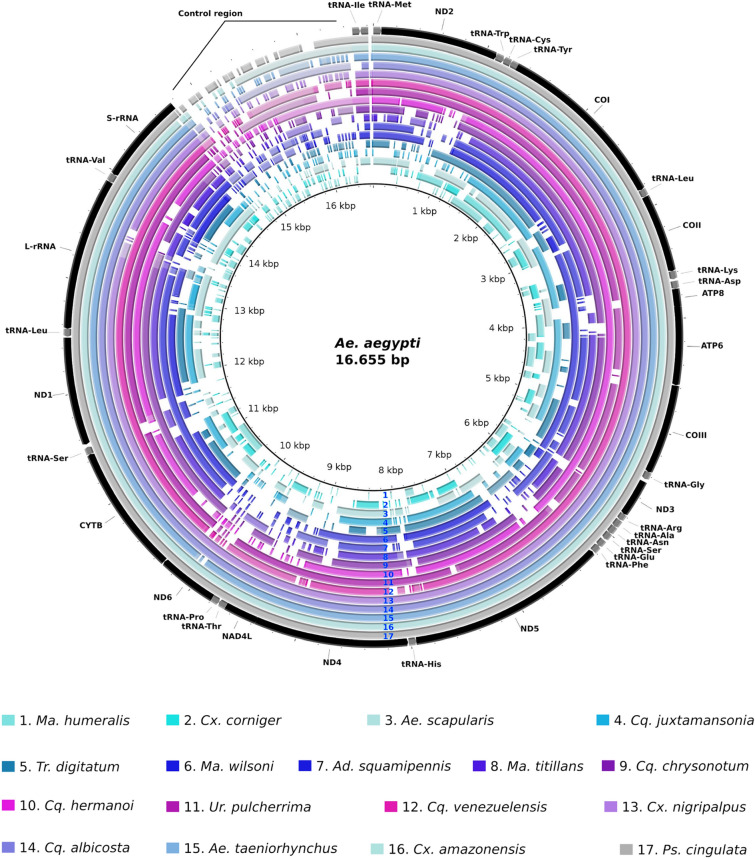
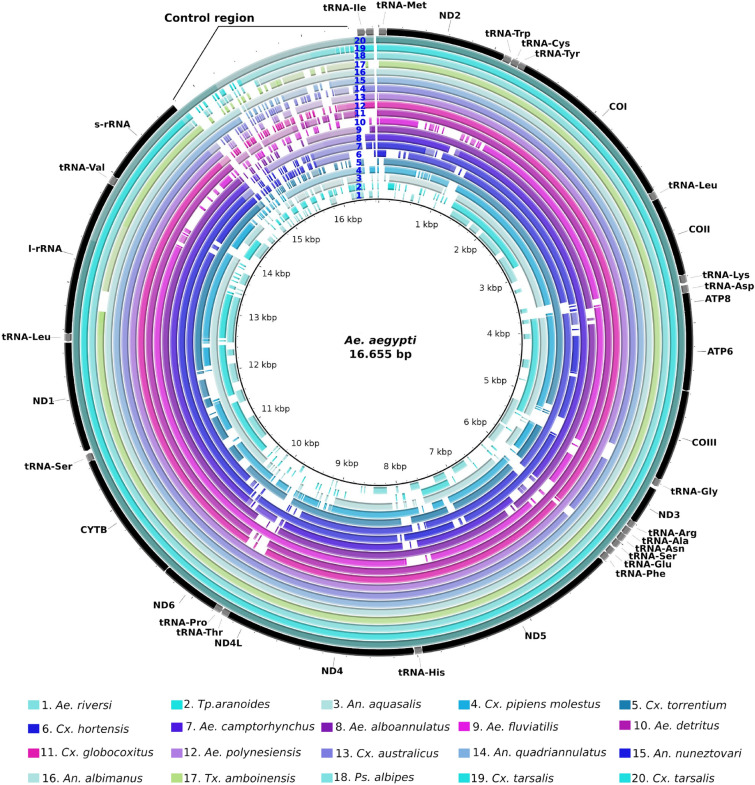
The coverage breadth of the sequenced draft mitogenomes ranged from 3699 to 15,660 bp for Ma. humeralis and Ps. cingulata, respectively (Table 1, Fig. 1) with an average coverage breadth of 72.80% and a coverage depth average of 20.01 fold (Table 1). Annotation of the protein coding genes (PCG) identified in the field-collected mosquitoes ranged from seven to 13. All 17 mitogenomes showed the two rRNA genes, except Ae. scapularis genome. In addition, tRNAs annotation ranged from five to 21 genes, except for Ae. taeniorhynchus and Cx. nigripalpus that showed all tRNAs genes (Table 1). Although some PCGs were not assembled, we could annotate the barcode COI in all 17 mitogenomes (Supplementary Table 1). The mitogenomes characterized from SRA data showed a coverage breadth ranging from 5992 to 15,960 bp for Ae. riversi and An. freeborni respectively (Fig. 2). In general those assemblies showed an average coverage breadth of 88.42% and from nine to 34 out of 37 mitochondrial genes were annotated with MITOS (Table 1). Although some of the SRA data came from RNA-Seq, we were able to identify almost all PCGs of these mosquito species. PCGs annotation ranged from seven for Tp. aranoides to 13 for other species (Supplementary Table 1, Fig. 2).
Figure 1.
Comparative map of mitogenomes sequenced in relation to Ae. aegypti mitochondrial genome (NC_010241.1).
Figure 2.
Comparative map of mitogenomes characterized from SRA data in relation to Ae. aegypti mitochondrial genome (NC_010241.1).
Evolutionary analysis
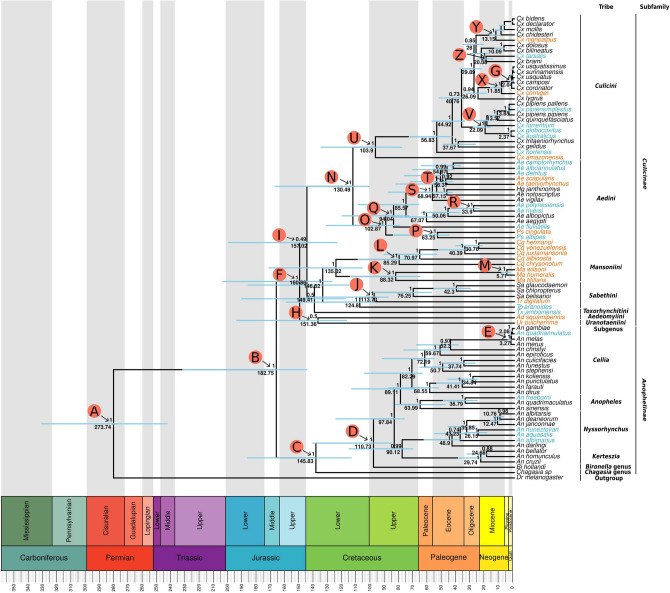
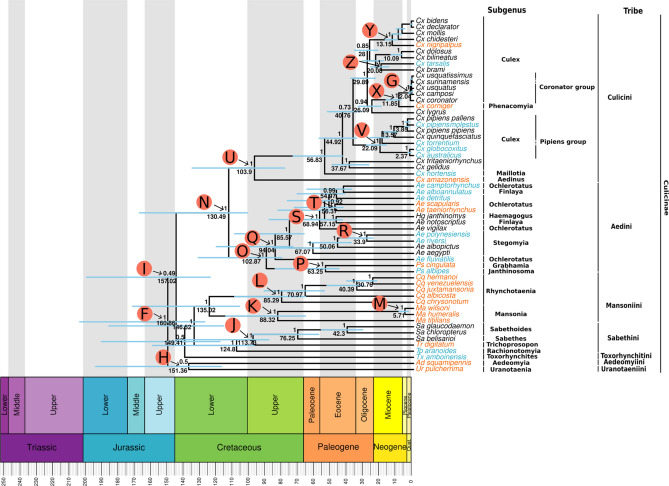
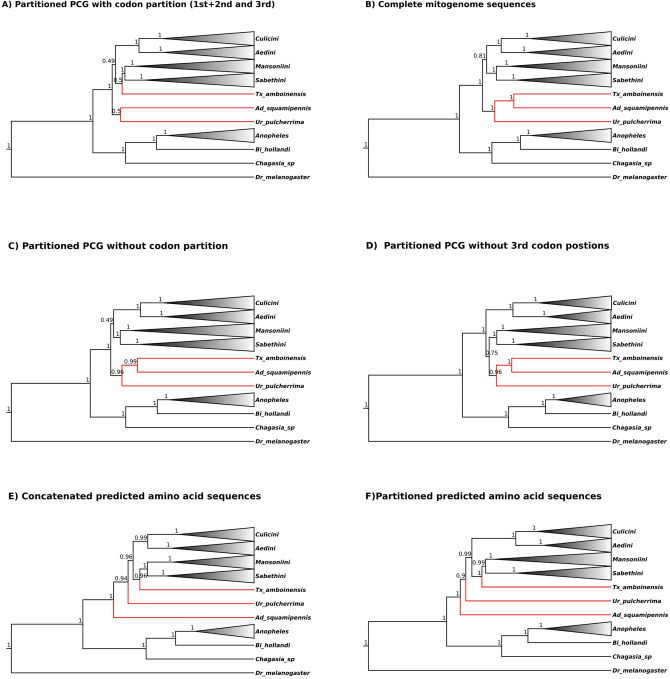
In order to establish the phylogenetic relationship of the Culicidae family we performed the phylogenetic analysis of 86 different mosquito mitogenomes representing 15 genera, including our 35 newly characterized mitogenomes. The analysis was based on nucleotide and amino acid datasets with or without partitioning. Since several genes showed nucleotide saturation at the third codon position (Supplementary file 1) we also performed phylogenetic analyses with codon partitions of each PCG and without the third codon positions (Supplementary Figs. 3–10). Topology of the phylogenetic trees built with those different alignments were mostly in agreement, but incongruences and variable positioning of some deep branches was observed for (Culicini + Aedini) + (Mansoniini + Sabethini) tribes and Aedeomyia, Uranotaenia and Toxorhynchites species (Figs. 3, 4 and 5 and Supplementary Fig. 1 and 2). In respect of the (Culicini + Aedini) + (Mansoniini + Sabethini) recovered relationship: using partitioned PCG taking or not into consideration the partitioning codon position showed a low posterior probability branch support of (0.49—Figs. 3, 4 and 5A,C), while complete mitochondrial genomes and concatenated and partitioned amino acid sequences supported this same clade with high branch support (0.81, 0.96, 0.99, respectively—Fig. 5B,E,F). Only the partitioned PCG without 3rd codon positions showed a different grouping with Mansoniini + Sabethini as a sister clade of (Tx. amboiensis + Ad. squamipennis) + Ur. pulcherrima, but with a relatively low posterior probability support (0.75). Regarding the variable positioning of Tx. amboinensis species, it was placed either as a basal clade of Mansoniini and Sabethini tribes in partitioned PCG taking into consideration (1st + 2nd and 3rd) codon positions but with a low posterior probability support value (0.5—Figs. 4, 5A) and concatenated and partitioned amino acid sequences with high branch support (0.96 and 1.0, Fig. 5E,F) or forming a clade with Ad. squamipennis and Ur. pulcherrima—(Tx. amboiensis + Ad. squamipennis) + Ur. pulcherrima—in complete mitochondrial genome tree (Fig. 5B, posterior probability of 1), partitioned PCG without codon partition (Fig. 5C, posterior probability of 0.99 and 0.96), partitioned PCG without 3rd codon position (Fig. 5D, posterior probability of 1 and 0.96). Moreover, a number of intra genus incongruences between the trees was observed in the Culex, Anopheles, and Aedes genera (Supplementary Fig. 1 and 2).
Figure 3.
Evolutionary timescale of Culicidae family. Tree was generated from BEAST using PCGs nucleotide sequences partitioned by gene and codon positions (1st + 2nd and 3rd separately). Blue bars represent the HPD95%. The numbers above and below the bars show the posterior probability and the predicted median dating respectively for each node. Specific words inside the circles represent the nodes discussed in the text. Light blue tip names represent mitogenomes characterized from SRA data. Orange tip names represent sequenced mitogenomes from this study.
Figure 4.
Zoom on the Culicinae branch of Fig. 3 showing in more detail the evolutionary timescale of Culicinae subfamily. Tree was generated from BEAST analysis of partitioned PCG taking into account the split of codon positions (1st + 2nd and 3rd codon position separately). Blue bars in the nodes represent the HPD95%. The numbers above and below the bars show the posterior probability and the predicted median dating respectively for each node. Specific words inside the circles represent the nodes discussed in the text. Light blue tip names represent mitogenomes characterized from SRA data. Orange tip names represent sequenced mitogenomes from this study.
Figure 5.
Bayesian inferred trees showing the incongruences in positioning of Aedeomyia, Uranotaenia and Toxorhynchites genera using different alignment datasets.
The evolutionary timescale of the Culicidae family showed the radiation of the last common ancestral between mosquitoes and Drosophila occurring during the Permian period around 273 million years ago (MYA) (Fig. 3, node A and Supplementary table 2). While the most recent common ancestor of the Culicidae family emerged in the Jurassic period around 182 MYA with the Anophelinae and Culicinae subfamilies origin (Fig. 3, node B). In the Anophelinae subfamily, the Chagasia genus was basal to Bironella and Anopheles genera with speciation in the Cretaceous period around 145 MYA (Fig. 3, node C). The last two genera showed speciation times from 110 to two MYA in the gambiae species complex (Fig. 3, nodes D and E, respectively).
The Culicinae subfamily formed a monophyletic group with the TMRCA (The most recent common ancestral) occurring around 160 MYA in the Jurassic period (Fig. 3, node F). Among Sabethini members, the Tripteroides genus was positioned as a basal lineage and the Trichoprosopon genus splited from other Sabethini species around 113 MYA (Fig. 4, node J). The Mansoniini tribe was placed as a sister clade to the Sabethini tribe with high posterior probability branch support (1.0) in all phylogenetic analysis performed (Supplementary Fig. 3–10). The Mansonia and Coquillettidia genera were both monophyletic with speciation processes starting around 88 and 85 MYA respectively (Fig. 4, nodes K and L, respectively).
The diversification between Culex and Aedini taxa occurred in the Cretaceous period around 130 MYA (Fig. 4, node N). While the split of Aedes and Psorophora genera occurred around 102 MYA, and the speciation of Ps. albipes and Ps. cingulata occurred in Paleogene around 63 MYA (Fig. 4, node O and P, respectively). Among the Aedes species, Ae. fluviatilis was recovered as the basal and early diverged species (94 MYA, node Q in Fig. 4) from the genus in eight out of nine phylogenetic reconstructions performed (Fig. 4 and Supplementary Fig. 3–10). Ae. polynesiensis and Ae. riversi were close to Ae. albopictus (Fig. 4, node R). Another clade formed closely to Ae. aegypti clade was composed by species from Ochlerotatus (Ae. vigilax, Ae. taeniorhynchus, Ae. scapularis, Ae. detritus, and Ae. camptorhynchus), Finlaya subgenera (Ae. notoscriptus and Ae. alboannulatus), and Haemagogus genus, in which Ae. vigilax was the basal species (Fig. 4, node S). The Finlaya subgenus has a paraphyletic status when the positioning of Ae. alboannulatus and Ae. notoscriptus is observed (Fig. 4). The neotropical species Ae. taeniorhynchus and Ae. scapularis formed a clade and diverged between themselves around 56 MYA (Fig. 4, node T). Among Culex species, Cx. amazonensis a member of the subgenus Aedinus, showed to be the basal and the earlier diverged species from the genus with the split from the other species occurring around 103 MYA (Fig. 4, node U). The pipiens group, from Culex subgenus, originated around 22 MYA in which the Australian species Cx. australicus and Cx. globocoxitus were placed in basal position in relationship to other Cx. pipiens species (Fig. 4, node V). Cx. torrentium grouped in the pipiens group. Cx. corniger, a member of Phenacomyia subgenus, was a basal species to the coronator group from Culex subgenus (node G), as well as, Cx. nigripalpus that speciated from Cx. chidesteri, Cx. mollis, Cx. declarator and Cx. bidens (Fig. 4, nodes X and Y, respectively). Moreover, Cx. tarsalis formed a clade with Cx. brami (Fig. 4, node Z).
Discussion
Mitogenomes have been widely used to elucidate the evolutionary history of several species of animals and plants and can also be used as barcode sequences for species identification16,41,42. The first mosquito mitogenome from An. gambiae was sequenced using the Sanger method on PCR amplified fragments27 and further mitochondrial genomes were slowly sequenced along with whole genome projects. Currently, most studies have been using whole genome sequencing or PCR amplification followed by high throughput sequencing to characterize several mitogenomes at once22,23,26 in a wide range of insect species showing promising results to reconstruct mitogenomes32,43. Here, we performed low-coverage whole genome sequencing to assemble and characterize the mitogenomes from seventeen mosquito species. We were able to generate 16 new draft mitogenomes from Culicidae species belonging to eight different genera. This approach has already been used in other studies demonstrating a cost-effective way to recover mitogenomes for evolutionary studies32,43–46. Richter et al. (2015)32 suggested that a minimum of 10 million reads are needed to recover mitogenomes with higher coverage breadth and datasets having around 1 million reads usually generate highly incomplete mitogenomes. Even though our study used different organisms and algorithm to obtain the mitogenomes in comparison to Richter et al. (2015)32, we were able to assemble nearly complete mitogenomes with as low as 1.1 million reads (Table 1). Besides, our draft mitogenomes assembled contained enough phylogenetic markers necessary for robust phylogenetic analysis.
Additionally, we reconstructed mitochondrial genomes from available RNA-Seq data. We were able to reconstruct 19 additional nearly complete draft mitogenomes for Culicidae species that had no mitogenomes available. No study has been able to reconstruct complete mitochondrial genomes from RNA-Seq data, mainly due to the endonuclease activity on transcripts or loss of mitochondrial transcripts due to the enrichment steps normally used during the sequencing library construction30. However, the remaining mitochondrial data available in different RNA-Seq datasets may still be used to retrieve mitogenomic sequences47–49. The datasets used for mitogenomes characterization contained around 0.073 to 8.034% of mitochondrial reads. In total, we were able to assemble 37 draft mitogenomes in this study representing 11 genera (Anopheles, Uranotaenia, Aedeomyia, Toxorhynchites, Tripteroides, Trichoprosopon, Mansonia, Coquillettidia, Psorophora, Aedes, and Culex).
Several efforts have been made to better understand the taxonomic status of groups inside of the Culicidae family, but most studies that included a substantial number of species employed only morphological data50 and the ones using molecular information suffer from limited sampling/taxonomic breadth12,21–23,51–53 and those with limited molecular markers54. Hence, there are still many non-studied species and unresolved phylogenetic relationships in genera such as Aedes, Armigeres, Coquillettidia, Culex, Mansonia, Mimomyia, Psorophora, Topomyia, Tripteroides, Toxorhynchites, Uranotaenia, and Wyeomyia50.
The phylogenetic analysis, including the 37 new mitogenomes assembled in this study comprising 11 Culicidae genera, is highly congruent regarding the monophyly of large species groups. Culicinae and Anophelinae subfamilies and Anopheles, Sabethes, Mansonia, Coquillettidia, Psorophora, and Culex genera were monophyletic. Moreover, we observed similar dating estimates as reported in the literature, for some key ancestors. For instance, our estimates of the common ancestor of Drosophila and Culicidae were around 273 MYA (HPD95%: 243.79–332.41), while other studies suggested that the common ancestor existed around 259 and 260 MYA using mitogenomes and phylogenomics analysis respectively13,53. The ancestral of Anophelinae and Culicinae subfamilies occurred in the Jurassic period around 182 MYA (HPD95%: 145.88–232.95). Similar estimates were obtained in other studies around 190–195 MYA55,56. Different evolutionary rates of molecular markers, limited species sampling and different algorithms used to reconstruct the species phylogeny could result in different time estimates13.
The evolutionary history of the Anophelinae subfamily has been more extensively studied considering the number of species analyzed, the morphological and molecular markers used including whole phylogenomic analysis52. A recent study using the mitochondrial genomes from several species, proposed a number of taxonomic status changes such as the elevation of some groups (Cellia, Anopheles, Kerteszia and Nyssorhynchus) from the subgenus to genus level21. Our results corroborate the monophyly of Cellia, Anopheles, Kerteszia and Nyssorhynchus subgenera although, our sampling of the Anopheles genus is insufficient to directly compare with Foster and collaborators21 on whether Anopheles should be split into multiple genera. Our phylogenetic analysis diverged from Foster’s study21 regarding the positioning of Kertezia subgenus that was sister group of Nyssorhynchus (Fig. 3) while Foster et al. (2017)21 recovered an uncertain grouping of Kertezia with other Anophelinae subgenera. In addition, Foster et al. (2017)21 and Neafsey et al. (2015)52 have not assessed the temporal diversification of some basal groups such as Chagasia and Bironella. In our analysis, these groups showed to be the early diverged lineages from Anophelinae subfamily, emerging in the Upper and Lower Cretaceous, respectively (Fig. 3) and Bironella genus showed to be an ancestral lineage in relation to Anopheles genus, including all Anopheles subgenera assessed in our study such as Kerteszia, Nyssorhynchus, Anopheles, and Cellia. Those results contrast with Foster et al. (2017)21 regarding Bironella positioning that suggest it grouped within Anopheles genus but with a low branch support. Previous studies, using both nuclear ribosomal sequences and fragments of mitochondrial genes COI and COII of Bi. gracilis57 and Bi. hollandi58, have already suggested the positioning of Bironella within the Anopheles genus. Although the number of the species analyzed, different molecular markers and phylogenetic approaches used in these studies, these contrasting results show that Bironella genus position and phyletic status are still open and a wide sampling of the genus and molecular markers are needed to uncover it. Regarding the Anopheles species, our analysis using mitogenomes showed a similar positioning as previously presented in other studies12,13,40,58–60.
The radiation in the Culicinae subfamily is older than Anophelinae around 160 MYA (HPD95%: 128.09–204.91) in the Jurassic period (Supplementary Fig. 11). In the Culicinae subfamily, we detected three low supported deep branch clades in the partitioned PCG taking into account the split of codon positions (Fig. 4), the Ad. squamipennis + Ur. pulcherrima (node H), the (Culicini + Aedini) + (Mansoniini + Sabethini) (node I) and the Tx. amboinensis basal positioning to Mansoniini + Sabethini clade. Regarding node I, there are substantial evidence in the literature corroborating the (Culicini + Aedini) + Sabethini grouping26,61 and all our complementary eight phylogenetic reconstructions using maximum likelihood and bayesian inference recovered this clade including Mansoniini as a sister group of Sabethini tribe (Fig. 5 and Supplementary Fig. 3–10). On the other hand, we found different results on the deep branching patterns and branch support for Ad. squamipennis, Ur. pulcherrima and Tx. amboinensis mainly depending on the molecule (nucleic acid or amino acid) used for phylogenetic reconstruction (Fig. 5). In short, amino acid based reconstruction placed Ad. squamipennis as the earliest divergent species followed by Ur. pulcherrima and Tx. amboinensis as an early divergent clade of Mansoniini + Sabethini group with all high branch support, while nucleic acid based reconstruction show a more variable branching pattern grouping these tree species in a highly supported basal clade to the remaining Culicinae species or with lower support as a sister clade of Mansoniini + Sabethini species (Fig. 5B–D). Other studies, based on morphological characters, suggested that Aedeomyia, Uranotaenia and Toxorhynchites genera are ancient and basal groups in the Culicinae subfamily62–65 suggesting that our phylogenetic reconstruction based on amino acid sequences may have recovered the true position of those genera. This is in line with the widespread knowledge that conserved amino acid sequences are more appropriate to recover deep branching patterns21,66,67, but due to the high branch support of a clade encompassing the three studied species from these genera in the nucleotide based trees (Fig. 5) and two studies based on six nuclear genes and 18S rDNA have shown the positioning of Ur. sapphirina more closely related to Culicini and Aedini tribes, respectively54,68, additional phylogenetic analysis including more species and nuclear molecular markers will be needed to test the hypothesis raised in our study.
Regarding the Sabethini tribe, our results are in line with previous works that showed the monophyly of tribe, the basal positioning of Tripteroides (Tp. aranoides), and the sister positioning of Trichoprosopon genus (Tr. digitatum)22,26,54 (Fig. 4). Up to now, few studies have investigated the phylogenetic positioning and speciation time of the Mansoniini species. In our analysis, the eight species from the Mansoniini tribe formed a monophyletic group that is a sister group of the Sabethini tribe with a high posterior probability node support (1.0—Figs. 3, 4). Our results are in contrast with Reidenbach et al. (2009)54 analysis that positioned a single Coquillettidia species, Cq. pertubans, as a sister group of Aedini with a low posterior probability node support of 0.61. Our dataset covers a larger number of species from the Mansoniini tribe and more molecular markers than in Reidenbach’s study54, besides their study did not include any Mansonia species, which likely explain those differences.
Considering the Aedini tribe, our results showed the same basal positioning of Psorophora genus as observed by Reidenbach et al. (2009)54. Regarding Aedes genus, our results showed a paraphyletic group encompassing a single species from the Haemagogus genus which corroborates other findings with a larger number of Haemagogus species69. Besides, paraphyletic groups were observed for Ochlerotatus (Ae. fluviatilis, Ae. taeniorhynchus, Ae. scapularis, Ae. vigilax, Ae. detritus, and Ae. camptorhynchus) and Finlaya subgenus (Ae. notoscriptus, and Ae. alboannulatus), while Stegomyia subgenus (Ae. aegypti, Ae. albopictus, Ae. riversi, and Ae. polynesiensis) formed a monophyletic group (Fig. 4). A previous study, based on morphological cladistic analysis, suggested the monophyly of Ochlerotatus and Finlaya subgenera70. Inside of the Aedes genus, Ae. fluviatilis is the earliest branch in contrast to other studies that positioned it within Aedes branches69. Depending on the classification this species it is a member of Georgecraigius or Ochlerotatus taxa71–73, the basal positioning in our analysis renders Ochlerotatus group proposed by Reinert (2000)72, using morphological characters and supported by others74, non-monophyletic. In summary, the Aedes genus is a paraphyletic group showing several phylogenetic incongruences even considering studies that used different markers and species representatives. Hence, further reclassification is needed following the current knowledge of phylogenetic relationships of these species.
Regarding the Culex genus, our analysis showed that Cx. amazonensis and Cx. hortensis are the earliest diverged species from this genus. Our results are in agreement with Harbach’s. 201275 cladistic morphological analysis concerning the basal positioning of these species, however our mitogenomic data support Cx. amazonensis as the earliest divergent species instead of Cx. hortensis. Our analysis placed Cx. nigripalpus as a sister group of the clade composed by Cx. chidesteri, Cx. mollis, Cx. declarator, and Cx. bidens, while, Cx. corniger was placed as a sister lineage of the Coronator group. A previous study using a fragment of the COI gene, has already suggested this positioning76 and our mitogenomic analysis supported this placement. It has been discussed, if Cx. pipiens consist in a species or a group of sibling species of Pipiens group77. Some authors describe the Pipiens group harboring the following species: Cx. pipiens pipiens, Cx. quinquefasciatus, Cx. pipiens pallens, Cx. pipiens molestus, Cx. australicus, and Cx. globocoxitus77,78. Other similar species such as Cx. torrentium, has not been considered as a member of Pipiens group due to its genetic divergence to other species of the group79. A study based on analysis of ITS1 and ITS2 has already demonstrated the close relationship of Cx. torrentium with Pipiens group80. Our analysis have positioned Cx. torrentium within Pipiens group with Australian members Cx. globocoxitus and Cx. australicus as basal clade, suggesting that Cx. torrentium may be a true species from the Pipiens group. Although the lower divergence time among some members of the Pipiens group each “species” has specific ecological, physiological and behavioral characteristics79,81.
Conclusion
Overall, we characterized the phylogenetic position and speciation time of the main groups of the Culicidae family which emerged in the last 182 MYA between the Jurassic and Paleogene periods. Most of the different genera emerged in this range of time, but some recent speciation occurred in the Culex genus. Interestingly, a burst in mammals speciation also occurred in the Neogene period likely driving the speciation of these species at that time55,82. Furthermore, the new phylogenetic knowledge will allow us to propose new hypotheses about some mosquito traits emergence and maintenance related with vector competence. More in depth studies trying to tease apart different molecular mechanisms of vector competence considering the phylogeny of the Culicidae tree will benefit from the information generated in this work.
Material and methods
Mosquito sampling and taxonomic identification
Mosquito samples were collected in remnants of the Brazilian Atlantic forest and from the South border of the Brazilian Amazon forest. Three municipalities were sampled in the Brazilian Atlantic forest, state of Pernambuco: Recife, at the Parque Estadual Dois Irmãos (8°00′43.3″S 34°56′40.7″W); Moreno, at the Reserva Ecológica de Carnijó (8°08′20.7″S 35°04′47.3″W) and Camaragibe, at Aldeia (7°54′18.0″S 35°04′34.3″W). Three municipalities were sampled in the Brazilian Amazon forest, state of Mato Grosso: Sinop (− 12°04′73.9″S − 55°43′85.0″W); Sorriso (− 12°16′85.9″S − 55°70′68.3″W); and Ipiranga do Norte (− 11°61′08.2″S − 55°73′41.7″W). Different sampling methods were employed aiming to collect a large diversity of species. Diurnal sampling were performed with aspirators (HORST model) and entomological nets, larvae and pupae were collected on water pools and plant holes. Nocturnal sampling were performed using CDC-light traps and BG-Sentinel to sample mosquitoes attracted by light and odorants. The specimens were transported alive either to the Entomology department of Aggeu Magalhães Institute—Oswaldo Cruz Foundation (IAM/FIOCRUZ) or to the Molecular Biology and Immunology Laboratory—Federal University of Mato Grosso (LIBM/UFMT). Immature specimens were maintained in liquid water and fed with cat food (FRISKIES) until the emergence of adults. Adult mosquitoes were separated into morphological groups and dry stored in silica at room temperature until taxonomic identification. Taxonomic keys for neotropical Culicidae were used for species identification83,84. Besides the collection performed in this work, we included Ae. taeniorhynchus and Ae. scapularis samples provided by collaborators of the Entomology department of IAM, sampled respectively in the municipality of São Luis, state of Maranhão and in municipality of Juazeiro, state of Bahia. All collections were authorized by the regulatory organ—SISBIO under the license numbers: 58716-1 and 47284-2.
DNA extraction and sequencing
The specimens were macerated in ultrapure water using 40ul/specimen in single or pooled samples (Supplementary table 3) according to the number of specimens collected per species. Both male and female individuals from different collection points were included in the pools. Total DNA extractions were performed either by ethanol precipitation method85 or QIAprep Spin Miniprep extraction (QIAGEN) in order to improve mitochondrial DNA by enrichment as suggested by Quispe-Tintaya et al. (2013)86. All samples were assessed by quality and purity with NanoDrop 2000 (THERMO SCIENTIFIC) and quantified through Qubit dsDNA HS (High Sensitivity Assay) kit. The DNA library was prepared using the Nextera XT library preparation kit following the recommendations of the manufacturer (ILLUMINA, San Diego, CA, USA). DNA library was sequenced using a low-coverage whole genome sequencing strategy using the ILLUMINA Miseq platform. We employed a paired-end approach of 75 bases with Reagent Kit V3 of 150 cycles.
Dataset construction
A search on the National Center for Biotechnology Information (NCBI) was performed to recover previously characterized mitochondrial genomes from Culicinae and a subset of Anophelinae subfamily representing different genera comprising 50 mitogenomes (Supplementary table 4). Besides, we searched on the SRA database for mosquitoes raw sequence reads (Whole genome sequencing and RNA-Seq) available up to November, 2018, representing species that had no mitogenome available at that time (Supplementary table 5).
Quality control of sequences
The raw reads (sequenced in this study and recovered from SRA) were checked for quality using FastQC program (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ accessed on 21 Oct, 2019) and results were summarized on MultiQC tool87. Based on the excellent quality of our sequenced raw reads they were not trimmed (Supplementary Fig. 12) but, all SRA libraries were trimmed using the Trimmomatic tool v 0.3588 to remove adapters and ensure the quality of sequences (Phred score > 20).
Mitogenome assembly and annotation
The mitogenomes were assembled using a baiting and iterative mapping approach implemented in MITObim 1.936. Different mosquito mitogenomes were used as reference genome such as Ae. vigilax, Ae. aegypti, Sa. belisarioi, Cx. quinquefasciatus (accession numbers can be found in Supplementary table 4) for the first capturing of reads considering the closest mitogenome available to each species analyzed. SRA reads were assembled using MITObim default parameters (-kbait parameter = 31). Also, we used a combination of parameters to generate a consensus sequence for the sequenced species. A first assembly was performed using -kbait = 15 followed by a second assembly step using -kbait = 31. The final consensus assembly was composed by the consensus of the two assemblies, which was then checked with well characterized mitogenomes to correct any potential assembly errors (e.g. the assembly of non alignable regions between mitogenomes). To assess the average coverage depth of each mitogenome, the reads were mapped against the assembled mitogenomes through the MIRAbait module from MIRA sequence assembler software89.
Complete nucleotide sequences of the mitogenomes characterized in this study, were aligned by MAFFT v 7.0 tool90 with previously characterized complete mitogenomes recovered from databases (Supplementary table 4 and Supplementary file 2). The non-aligned sites were removed using GBLOCKS tool v. 0.91b—default parameters, with exception for the allowed gap positions that was set with the “half” option91, to generate the final version of the mitochondrial genomes. Automatic gene annotation of the mitochondrial genomes were performed on MITOS2 web server (https://mitos2.bioinf.uni-leipzig.de/index.py accessed on 5 Dec., 2018)37 based on invertebrate genetic code against the metazoan Refseq 81. Comparative genomic maps were built using Ae. aegypti mitogenome (Accession number: NC_010241.1) as reference in BRIG (BLAST Ring Image Generator)92.
Evolutionary analysis
Evolutionary analysis were performed based on five possible alignment approaches: (I) complete nucleotide mitogenome alignment sequences, (II) partitioned nucleotide sequence of protein coding genes derived from complete and draft mitogenomes with partitioned codon positions (1st + 2nd and 3rd), (III) partitioned nucleotide sequence of protein coding genes without 3rd codon positions without codon partitioning, (IV) partitioned predicted amino acid sequences from coding regions and (V) concatenated alignment of amino acid sequences. Final alignment was visualized and checked on Aliview93. Nucleotide substitution saturation analysis was performed for each nucleotide gene alignment in DAMBE software94 evaluating 1st + 2nd and 3rd codon position separately through the Xia et al. test95. Nucleotide substitution models for I, II and III alignments were obtained with Smart model selection (SMS) implemented on PhyML webserver96. Protein evolutionary models were assessed for IV and V alignments using Prottest 3.4.297. All divergence dating analysis were based on a Bayesian Markov Monte Carlo approach (MCMC) performed on BEAST 1.8.4 package98 to infer the topology of Culicidae family and the speciation time of the common ancestor of clades in million years. A previous literature search was performed to obtain fossil dates representing the different Culicidae clades and calibrate the molecular clock analysis. Although there are several potential calibration points to the Culicidae tree we only kept the ones supported by fossil evidence. We used four calibration points representing the Diptera order, Culicidae family and Anophelinae and Culicinae subfamily (Supplementary table 6 and Supplementary Fig. 13).
Bayesian analysis was performed with at least three independent runs of 150 million generations sampling at each 1000 trees, for each alignment dataset. The effective sample size of each parameter (ESS) was evaluated by Tracer 1.7.199 and reached 200 for most of the important parameters for dating and tree likelihood. The analysis was performed under an uncorrelated relaxed molecular clock using a lognormal distribution and a Birth–Death model process of speciation as Tree Prior. For the complete mitochondrial genome alignment (alignment I) the GTR + G + I evolutionary model was used. For the partitioned gene analysis (alignment II) and partitioned predicted amino acids (alignment IV) each partition was set with a specific evolutionary model as previously described (Supplementary table 7). Besides, the partitioned gene analysis we also performed a more robust analysis based on the nucleotide saturation of each gene taking into account the codon position partitioning where the 1st and 2nd codon positions were split from the 3rd codon position. The concatenated protein analysis was performed under the mtREV + G + I evolutionary model. The posterior probability tree for each alignment dataset was built combining the three independent runs of each analysis with the LogCombiner program applying a burn-in of 25% and the consensus credible tree was obtained through the TreeAnnotator program. The timescale trees were plotted with Phyloch package version 1.5–3 (available on https://www.christophheibl.de/Rpackages.html accessed on 21 Oct, 2019) from R programming language. Tree topologies comparison were performed by plotting tanglegrams using the Dendextend R package100 based on trees obtained from BEAST analysis. Besides the bayesian analysis we also performed maximum likelihood phylogenetic analysis based on alignment I, III (concatenating individual alignments). The best evolutionary model was selected by the ModelFinder101 followed by the tree reconstruction using the IQ-TREE version 1.6.12102 performing the ultrafast bootstrapping103 with 1000 replicates. The consensus trees were visualized and edited on FigTree version 1.4.2 (available on https://tree.bio.ed.ac.uk/software/figtree/ accessed on 3 Jun, 2020).
Supplementary information
Acknowledgements
This research was supported by Fundação Oswaldo Cruz and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) under the project number PROEP/IAM (400742/2019-5) and CNPq for the research grant PQ-2 of Wallau, GL (303902/2019-1). da Silva, A.F received a masters scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). We thank Cláudio Júlio da Silva and Edivaldo José Apolinário from Núcleo de Vigilância à Saúde e do Meio Ambiente for the support in mosquito sampling from Moreno municipality. We thank the Núcleo de Bioinformática and Núcleo de Plataformas Tecnológicas (NPT) of IAM for the computational infrastructure and sequencing platform, respectively.
Author contributions
G.L.W. conceived, designed the research, performed mosquito sampling and wrote the paper. A.F.S. performed the mosquito sampling, molecular experiments, bioinformatics analysis and drafted the paper. L.C.M. performed the mosquito sampling, molecular experiments and contributed to drafting the manuscript. M.B.P. and C.J.S.P.V. performed the taxonomic identification and contributed to write the paper. R.V.M.B. and M.A.V.M.S. performed mosquito sampling and contributed to the paper. All authors read and approved the final version of the manuscript.
Data availability
Raw mitochondrial reads were submitted to European Bioinformatic Institute under the project number: PRJEB36702. The final mitogenome assemblies and raw phylogenetic tree files generated are available on Supplementary file 2 and Supplementary file 3, respectively (https://doi.org/10.6084/m9.figshare.12114129).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-74883-3.
References
- 1.Molaei G, Andreadis TG, Armstrong PM, Diuk-Wasser M. Host-feeding patterns of potential mosquito vectors in Connecticut, USA: molecular analysis of bloodmeals from 23 species of Aedes, Anopheles, Culex, Coquillettidia, Psorophora, and Uranotaenia. J. Med. Entomol. 2008;45:1143–1151. doi: 10.1093/jmedent/45.6.1143. [DOI] [PubMed] [Google Scholar]
- 2.Lundström JO, et al. Transstadial transmission of Francisella tularensis holarctica in mosquitoes, Sweden. Emerg. Infect. Dis. 2011;17:794–799. doi: 10.3201/eid1705.100426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox FE. History of the discovery of the malaria parasites and their vectors. Parasites Vectors. 2010;3:5. doi: 10.1186/1756-3305-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiawsirisup S, Nithiuthai S. Vector competence of Aedes aegypti (L.) and Culex quinquefasciatus (Say) for Dirofilaria immitis (Leidy) Southeast Asian J. Trop. Med. Public Health. 2006;37(Suppl 3):110–114. [PubMed] [Google Scholar]
- 5.Wilson ME, Schlagenhauf P. Aedes and the triple threat of DENV, CHIKV, ZIKV—arboviral risks and prevention at the 2016 Rio Olympic games. Travel Med. Infect. Dis. 2016;14:1–4. doi: 10.1016/j.tmaid.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Norris DE. Mosquito-borne diseases as a consequence of land use change. EcoHealth. 2004;1:19–24. doi: 10.1007/s10393-004-0008-7. [DOI] [Google Scholar]
- 7.Serra OP, Cardoso BF, Ribeiro AL, Santos FA, Slhessarenko RD. Mayaro virus and dengue virus 1 and 4 natural infection in culicids from Cuiabá, state of Mato Grosso, Brazil. Memorias do Inst. Oswaldo Cruz. 2016;111:20–29. doi: 10.1590/0074-02760150270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Vieira CJSP, et al. Detection of Ilheus virus in mosquitoes from southeast Amazon, Brazil. Trans. R. Soc. Trop. Med. Hyg. 2019;113:424–427. doi: 10.1093/trstmh/trz031. [DOI] [PubMed] [Google Scholar]
- 9.Weaver SC. Host range, amplification and arboviral disease emergence. Arch. Virol. Suppl. 2005;19:33–44. doi: 10.1007/3-211-29981-5_4. [DOI] [PubMed] [Google Scholar]
- 10.Weaver SC, Barrett AD. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2004;2:789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shragai T, Tesla B, Murdock C, Harrington LC. Zika and chikungunya: mosquito-borne viruses in a changing world. Ann. N. Y. Acad. Sci. 2017;1399:61–77. doi: 10.1111/nyas.13306. [DOI] [PubMed] [Google Scholar]
- 12.Chu H, et al. The phylogenetic relationships of known mosquito (Diptera: Culicidae) mitogenomes. Mitochondrial DNA Part A. 2018;29:31–35. doi: 10.1080/24701394.2016.1233533. [DOI] [PubMed] [Google Scholar]
- 13.Hao Y-J, et al. Complete mitochondrial genomes of Anophelesstephensi and An. dirus and comparative evolutionary mitochondriomics of 50 mosquitoes. Sci. Rep. 2017;7:7666. doi: 10.1038/s41598-017-07977-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behura SK. Molecular marker systems in insects: current trends and future avenues. Mol. Ecol. 2006;15:3087–3113. doi: 10.1111/j.1365-294X.2006.03014.x. [DOI] [PubMed] [Google Scholar]
- 15.Mandal SD, Chhakchhuak L, Gurusubramanian G, Kumar NS. Mitochondrial markers for identification and phylogenetic studies in insects—a review. DNA Barcodes. 2014;2:1–9. doi: 10.2478/dna-2014-0001. [DOI] [Google Scholar]
- 16.Hebert PDN, Cywinska A, Ball SL, de Waard JR. Biological identifications through DNA barcodes. Proc. Biol. Sci. R. Soc. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ratnasingham, S. & Hebert, P. D. N. BOLD: The Barcode of Life Data System (https://www.barcodinglife.org). Mol. Ecol. Notes.7, 355–364 (2007). [DOI] [PMC free article] [PubMed]
- 18.Curole JP, Kocher TD. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999;14:394–398. doi: 10.1016/S0169-5347(99)01660-2. [DOI] [PubMed] [Google Scholar]
- 19.Birky CW. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia, X. Rapid evolution of animal mitochondrial DNA. In Rapidly Evolving Genes and Genetic Systems 73–82 (Oxford University Press, Oxford, 2012). 10.1093/acprof:oso/9780199642274.003.0008
- 21.Foster PG, et al. Phylogeny of anophelinae using mitochondrial protein coding genes. R. Soc. Open Sci. 2017;4:170758. doi: 10.1098/rsos.170758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Aragão AO, et al. Description and phylogeny of the mitochondrial genome of Sabethes chloropterus, Sabethes glaucodaemon and Sabethes belisarioi (Diptera: Culicidae) Genomics. 2019;111:607–611. doi: 10.1016/j.ygeno.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Sun L, et al. The complete mt genomes of Lutzia halifaxia, Lt. fuscanus and Culex pallidothorax (Diptera: Culicidae) and comparative analysis of 16 Culex and Lutzia mt genome sequences. Parasites Vectors. 2019;12:368. doi: 10.1186/s13071-019-3625-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cameron SL. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu. Rev. Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 26.Lorenz C, et al. First record of translocation in Culicidae (Diptera) mitogenomes: evidence from the tribe Sabethini. BMC Genom. 2019;20:721. doi: 10.1186/s12864-019-6069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beard CB, Hamm DM, Collins FH. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol. Biol. 1993;2:103–124. doi: 10.1111/j.1365-2583.1993.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell SE, Cockburn AF, Seawright JA. The mitochondrial genome of Anopheles quadrimaculatus species A: complete nucleotide sequence and gene organization. Genome. 1993;36:1058–1073. doi: 10.1139/g93-141. [DOI] [PubMed] [Google Scholar]
- 29.Briscoe AG, Hopkins KP, Waeschenbach A. High-throughput sequencing of complete mitochondrial genomes. Methods Mol. Biol. 2016;1452:45–64. doi: 10.1007/978-1-4939-3774-5_3. [DOI] [PubMed] [Google Scholar]
- 30.Cameron SL. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst. Entomol. 2014;39:400–411. doi: 10.1111/syen.12071. [DOI] [Google Scholar]
- 31.Tang M, et al. Multiplex sequencing of pooled mitochondrial genomes—a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 2014;42:1–13. doi: 10.1093/nar/gkt1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richter S, Schwarz F, Hering L, Böggemann M, Bleidorn C. The utility of genome skimming for phylogenomic analyses as demonstrated for glycerid relationships (Annelida, Glyceridae) Genome Biol. Evol. 2015;7:3443–3462. doi: 10.1093/gbe/evv224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deiner K. Long-range PCR allows sequencing of mitochondrial genomes from environmental DNA. Methods Ecol. Evol. 2017;8:1888–1898. doi: 10.1111/2041-210X.12836. [DOI] [Google Scholar]
- 34.Plese B, et al. Trimitomics: An efficient pipeline for mitochondrial assembly from transcriptomic reads in non-model species. Mol. Ecol. Resour. 2019;19:1230–1239. doi: 10.1111/1755-0998.13033. [DOI] [PubMed] [Google Scholar]
- 35.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45:e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 2013;41:e129. doi: 10.1093/nar/gkt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernt M, et al. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 38.Al-Nakeeb K, Petersen TN, Sicheritz-Pontén T. Norgal: extraction and de novo assembly of mitochondrial DNA from whole-genome sequencing data. BMC Bioinform. 2017;18:510. doi: 10.1186/s12859-017-1927-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.da Lemos PS, et al. Characterization of mitochondrial genome of Haemagogus janthinomys (Diptera: Culicidae) Mitochondrial DNA Part A. 2017;28:50–51. doi: 10.3109/19401736.2015.1110793. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Villegas L, et al. Characterization of the complete mitogenome of Anopheles aquasalis, and phylogenetic divergences among Anopheles from diverse geographic zones. PLoS ONE. 2019;14:e0219523. doi: 10.1371/journal.pone.0219523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krzemińska U, et al. Population mitogenomics provides insights into evolutionary history, source of invasions and diversifying selection in the House Crow (Corvussplendens) Heredity. 2018;120:296–309. doi: 10.1038/s41437-017-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witt KE, et al. DNA analysis of ancient dogs of the Americas: identifying possible founding haplotypes and reconstructing population histories. J. Hum. Evol. 2015;79:105–118. doi: 10.1016/j.jhevol.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 43.Trevisan B, Alcantara DM, Machado DJ, Marques FP, Lahr DJ. Genome skimming is a low-cost and robust strategy to assemble complete mitochondrial genomes from ethanol preserved specimens in biodiversity studies. PeerJ. 2019;7:e7543. doi: 10.7717/peerj.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Therkildsen NO, Palumbi SR. Practical low-coverage genomewide sequencing of hundreds of individually barcoded samples for population and evolutionary genomics in nonmodel species. Mol. Ecol. Resour. 2017;17:194–208. doi: 10.1111/1755-0998.12593. [DOI] [PubMed] [Google Scholar]
- 45.Gan H, Schultz MB, Austin CM. Integrated shotgun sequencing and bioinformatics pipeline allows ultra-fast mitogenome recovery and confirms substantial gene rearrangements in Australian freshwater crayfishes. BMC Evol. Biol. 2014;14:19. doi: 10.1186/1471-2148-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doyle SR, Griffith IS, Murphy NP, Strugnell JM. Low-coverage MiSeq next generation sequencing reveals the mitochondrial genome of the Eastern Rock Lobster, Sagmariasus verreauxi. Mitochondrial DNA. 2015;26:844–845. doi: 10.3109/19401736.2013.855921. [DOI] [PubMed] [Google Scholar]
- 47.Raz T, et al. Protocol dependence of sequencing-based gene expression measurements. PLoS ONE. 2011;6:e19287. doi: 10.1371/journal.pone.0019287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian Y, Smith DR. Recovering complete mitochondrial genome sequences from RNA-Seq: a case study of Polytomella non-photosynthetic green algae. Mol. Phylogenet. Evol. 2016;98:57–62. doi: 10.1016/j.ympev.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 49.Nabholz B, Jarvis ED, Ellegren H. Obtaining mtDNA genomes from next-generation transcriptome sequencing: a case study on the basal Passerida (Aves: Passeriformes) phylogeny. Mol. Phylogenet. Evol. 2010;57:466–470. doi: 10.1016/j.ympev.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 50.Harbach RE. The Culicidae (Diptera): a review of taxonomy, classification and phylogeny. Zootaxa. 2007;1668:591–638. doi: 10.1017/CBO9781107415324.004. [DOI] [Google Scholar]
- 51.Demari-Silva B, et al. Mitochondrial genomes and comparative analyses of Culex camposi, Culex coronator, Culex usquatus and Culex usquatissimus (Diptera: Culicidae), members of the coronator group. BMC Genomics. 2015;16:831. doi: 10.1186/s12864-015-1951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neafsey D, et al. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science. 2015;347:1258522. doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X-G, et al. Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc. Natl. Acad. Sci. 2015;112:E5907–E5915. doi: 10.1073/pnas.1516410112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reidenbach KR, et al. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evol. Biol. 2009;9:298. doi: 10.1186/1471-2148-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang C, Davis KE, Delmer C, Yang D, Wills MA. Elevated atmospheric CO2 promoted speciation in mosquitoes (Diptera, Culicidae) Commun. Biol. 2018;1:182. doi: 10.1038/s42003-018-0191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moreno M, et al. Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malar. J. 2010;9:127. doi: 10.1186/1475-2875-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sallum MAM, et al. Phylogeny of Anophelinae (Diptera: Culicidae) based on nuclear ribosomal and mitochondrial DNA sequences: molecular phylogeny of Anophelinae. Syst. Entomol. 2002;27:361–382. doi: 10.1046/j.1365-3113.2002.00182.x. [DOI] [Google Scholar]
- 58.Freitas LA, et al. Diversification of the genus Anopheles and a neotropical clade from the Late Cretaceous. PLoS ONE. 2015;10:e0134462. doi: 10.1371/journal.pone.0134462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fontaine MC, et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347:1258524. doi: 10.1126/science.1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hua Y-Q, et al. The complete mitochondrial genome of Anopheles minimus (Diptera: Culicidae) and the phylogenetics of known Anopheles mitogenomes. Insect Sci. 2016;23:353–365. doi: 10.1111/1744-7917.12326. [DOI] [PubMed] [Google Scholar]
- 61.Besansky NJ, Fahey GT. Utility of the white gene in estimating phylogenetic relationships among mosquitoes (Diptera: Culicidae) Mol. Biol. Evol. 1997;14:442–454. doi: 10.1093/oxfordjournals.molbev.a025780. [DOI] [PubMed] [Google Scholar]
- 62.Harbach RE. Comparative and functional morphology of the mandibles of some fourth stage mosquito larvae (Diptera: Culicidae) Zoomorphologie. 1977;87:217–236. doi: 10.1007/BF00995821. [DOI] [Google Scholar]
- 63.Harbach RE, Kitching IJ. Phylogeny and classification of the Culicidae (Diptera) Syst. Entomol. 1998;23:327–370. doi: 10.1046/j.1365-3113.1998.00072.x. [DOI] [Google Scholar]
- 64.Krzywinski J, Wilkerson RC, Besansky NJ. Evolution of mitochondrial and ribosomal gene sequences in anophelinae (Diptera: Culicidae): implications for phylogeny reconstruction. Mol. Phylogenet. Evol. 2001;18:479–487. doi: 10.1006/mpev.2000.0894. [DOI] [PubMed] [Google Scholar]
- 65.Díaz-Nieto LM, et al. Distribution of mosquitoes in the south east of Argentina and first report on the analysis based on 18S rDNA and COI sequences. PLoS ONE. 2013;8:e75516. doi: 10.1371/journal.pone.0075516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rota-Stabelli O, Lartillot N, Philippe H, Pisani D. Serine codon-usage bias in deep phylogenomics: pancrustacean relationships as a case study. Syst. Biol. 2013;62:121–133. doi: 10.1093/sysbio/sys077. [DOI] [PubMed] [Google Scholar]
- 67.Inagaki Y, Simpson A, Dacks J, Roger A. Phylogenetic artifacts can be caused by leucine, serine, and arginine codon usage heterogeneity: dinoflagellate plastid origins as a case study. Syst. Biol. 2004;53:582–593. doi: 10.1080/10635150490468756. [DOI] [PubMed] [Google Scholar]
- 68.Shepard JJ, Andreadis TG, Vossbrinck CR. Molecular phylogeny and evolutionary relationships among mosquitoes (Diptera: Culicidae) from the northeastern United States based on small subunit ribosomal DNA (18S rDNA) sequences. J. Med. Entomol. 2006;43:443–454. doi: 10.1093/jmedent/43.3.443. [DOI] [PubMed] [Google Scholar]
- 69.Soghigian J, Andreadis TG, Livdahl TP. From ground pools to treeholes: convergent evolution of habitat and phenotype in Aedes mosquitoes. BMC Evol. Biol. 2017;17:262. doi: 10.1186/s12862-017-1092-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reinert JF, Harbach RE, Kitching IJ. Phylogeny and classification of tribe Aedini (Diptera: Culicidae) Zool. J. Linn. Soc. 2009;157:700–794. doi: 10.1111/j.1096-3642.2009.00570.x. [DOI] [Google Scholar]
- 71.Reinert JF, Harbach RE, Kitching IJ. Phylogeny and classification of Finlaya and allied taxa (Diptera: Culicidae: Aedini) based on morphological data from all life stages. Zool. J. Linn. Soc. 2006;148:1–101. doi: 10.1111/j.1096-3642.2006.00254.x. [DOI] [Google Scholar]
- 72.Reinert JF. New classification for the composite genus Aedes (Diptera: Culicidae: Aedini), elevation of subgenus Ochlerotatus to generic rank, reclassification of the other subgenera, and notes on certain subgenera and species. J. Am. Mosq. Control. Assoc. 2000;16:175–188. [PubMed] [Google Scholar]
- 73.Wilkerson RC, et al. Making mosquito taxonomy useful: a stable classification of tribe Aedini that balances utility with current knowledge of evolutionary relationships. PLoS ONE. 2015;10:e0133602. doi: 10.1371/journal.pone.0133602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reinert JF, Harbach RE, Kitching IJ. Phylogeny and classification of Aedini (Diptera: Culicidae), based on morphological characters of all life stages. Zool. J. Linn. Soc. 2004;142:289–368. doi: 10.1111/j.1096-3642.2004.00144.x. [DOI] [Google Scholar]
- 75.Harbach RE, Kitching IJ, Culverwell CL, Dubois J, Linton Y-M. Phylogeny of mosquitoes of tribe Culicini (Diptera: Culicidae) based on morphological diversity: phylogeny of Culicini. Zool. Scr. 2012;41:499–514. doi: 10.1111/j.1463-6409.2012.00546.x. [DOI] [Google Scholar]
- 76.Demari-Silva B, Vesgueiro FT, Sallum MAM, Marrelli MT. Taxonomic and phylogenetic relationships between species of the genus Culex (Diptera: culicidae) from Brazil inferred from the cytochrome c oxidase I mitochondrial gene. J. Med. Entomol. 2011;48:272–279. doi: 10.1603/ME09293. [DOI] [PubMed] [Google Scholar]
- 77.Harbach RE. Culex pipiens: species versus species complex taxonomic history and perspective. J. Am. Mosq. Control. Assoc. 2012;28:10–23. doi: 10.2987/8756-971X-28.4.10. [DOI] [PubMed] [Google Scholar]
- 78.Russell RC. A review of the status and significance of the species within the Culex pipiens group in Australia. J. Am. Mosq. Control. Assoc. 2012;28:24–27. doi: 10.2987/8756-971X-28.4s.24. [DOI] [PubMed] [Google Scholar]
- 79.Farajollahi A, Fonseca DM, Kramer LD, Marm Kilpatrick A. “Bird biting” mosquitoes and human disease: a review of the role of Culex pipiens complex mosquitoes in epidemiology. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2011;11:1577–1585. doi: 10.1016/j.meegid.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller BR, Crabtree MB, Savage HM. Phylogeny of fourteen Culex mosquito species, including the Culex pipiens complex, inferred from the internal transcribed spacers of ribosomal DNA. Insect Mol. Biol. 1996;5:93–107. doi: 10.1111/j.1365-2583.1996.tb00044.x. [DOI] [PubMed] [Google Scholar]
- 81.Zittra C, et al. Ecological characterization and molecular differentiation of Culexpipiens complex taxa and Culextorrentium in eastern Austria. Parasites Vectors. 2016;9:197. doi: 10.1186/s13071-016-1495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bininda-Emonds ORP, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 83.Lane J. Neotropical Culicidae. Sao Paulo: University of Sao Paulo; 1953. [Google Scholar]
- 84.Forattini, O. P. Culicidologia médica: identificaçäo, biologia e epidemiologia. 2 (Edusp, 2002).
- 85.Ayres CFJ, Melo-Santos MAV, Solé-Cava AM, Furtado AF. Genetic differentiation of Aedes aegypti (Diptera: Culicidae), the major dengue vector in Brazil. J. Med. Entomol. 2003;40:430–435. doi: 10.1603/0022-2585-40.4.430. [DOI] [PubMed] [Google Scholar]
- 86.Quispe-Tintaya W, White RR, Popov VN, Vijg J, Maslov AY. Fast mitochondrial DNA isolation from mammalian cells for next-generation sequencing. Biotechniques. 2013;55:133–136. doi: 10.2144/000114077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chevreux, B., Wetter, T. & Suhai, S. Genome sequence assembly using trace signals and additional sequence information. Ger. Conf. Bioinforma. 45–56 (1999).
- 90.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 92.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 95.Xia X, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 2003;26:1–7. doi: 10.1016/S1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- 96.Lefort V, Longueville J-E, Gascuel O. SMS: smart model selection in PhyML. Mol. Biol. Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Galili T. dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics. 2015;31:3718–3720. doi: 10.1093/bioinformatics/btv428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mitochondrial reads were submitted to European Bioinformatic Institute under the project number: PRJEB36702. The final mitogenome assemblies and raw phylogenetic tree files generated are available on Supplementary file 2 and Supplementary file 3, respectively (https://doi.org/10.6084/m9.figshare.12114129).