Abstract
The Mediator complex directs signals from DNA-binding transcription factors to RNA polymerase (Pol) II. Despite this pivotal position, mechanistic understanding of Mediator in human cells remains incomplete. Here, we quantified Mediator-controlled Pol II kinetics by coupling rapid subunit degradation with orthogonal experimental readouts. Consistent with a model of condensate-driven transcription initiation, large clusters of hypo-phosphorylated Pol II rapidly disassembled upon Mediator degradation. This was accompanied by a selective and pronounced disruption of cell type-specifying transcriptional circuits, whose constituent genes featured exceptionally high rates of Pol II turnover. Notably, transcriptional output of most other genes was largely unaffected by acute Mediator ablation. Maintenance of transcriptional activity at these genes was linked to an unexpected, CDK9-dependent compensatory feedback loop that elevated Pol II pause release rates genome-wide. Collectively, our work positions human Mediator as a globally acting coactivator that selectively safeguards the functionality of cell type-specifying transcriptional networks.
Introduction
DNA-binding transcription factors (TFs) guide the activity of RNA polymerase (Pol) II. Seminal in vitro studies established that eukaryotic TFs require intermediary factors to relay their regulatory information to Pol II core components1,2. Biochemical purification of this coactivator activity led to the identification of the large, multi-subunit Mediator complex in numerous eukaryotic model organisms2–9. Mediator is organized into biochemically separable, functional modules termed head, middle, tail and kinase module10–13. Initially described as an absolute requirement for activator-dependent transcription, Mediator also stimulates basal transcription in vitro 4. This posed the question whether Mediator is a general or a gene-specific factor14. Yeast in vivo studies yielded variable results, partly finding that Mediator mutants decrease global mRNA levels as much as Pol II mutants15, and partly describing mild effects upon nuclear depletion of individual Mediator subunits16. Strong transcriptional defects were only observed upon simultaneous depletion of head and tail Mediator subunits16. Compared to these and other studies in yeast17,18, mechanistic understanding of human Mediator is emerging slowly, owing to experimental challenges.
Human transcription regulation differs from yeast in several key aspects. Most importantly, human genomes allow the specification of different cell types from the same genomic potential. To this end, human genomes evolved distal cis-regulatory enhancer elements, which concentrate a plethora of TFs and coactivators, including Mediator19. High levels of coactivator binding were used to classify large stretches of clustered enhancers, termed super-enhancers (SEs), which predominantly drive the expression of cell type-specific genes20,21. A subset of SE-driven transcription factors interacts in interconnected gene regulatory networks (GRNs) that establish transcriptional circuits to control cell identity22–24. Recent models implicate the Mediator subunit MED1 with its intrinsically disordered region (IDR) in the formation of nuclear condensates at SE regions25,26. Molecular mechanisms of how Mediator subunits contribute to the organization of these subnuclear structures remain to be elucidated. Also, whether and how nuclear condensates influence Pol II transcription dynamics has not been addressed experimentally.
Enhancers are often megabases away from their target genes. Intricate mechanisms have evolved to ensure faithful communication with their target promoters, including tight regulation of chromatin architecture27,28. Mediator is located at the nexus of these processes, but whether it directly controls enhancer-promoter looping remains to be unambiguously resolved. Initial studies in mouse cells showed Mediator colocalization with cohesin and implicated both factors in the formation and maintenance of enhancer-promoter contacts at pluripotency genes29. A more recent study yielded opposite results, as overall chromatin architecture was largely unchanged after Mediator depletion30. Interestingly, the authors of the last study implicated Mediator as a globally required transcriptional regulator, as they observed 7-fold lower mRNA levels after long-term MED14 depletion for 60 hours. However, extended perturbation periods limit causal conclusions.
Almost three decades after the discovery of Mediator, these unresolved questions call for mechanistic studies of Mediator in human cells. Such studies have been hampered by the difficulty in rapidly perturbing essential Mediator components. Here we unravel direct Mediator functions using human cells with pharmacologically degradable Mediator subunit alleles. Acute Mediator loss strongly abrogated the transcription of SE-driven, cell type-specifying GRN genes, but only mildly impacted overall transcriptional output and genome architecture. Integrating nascent transcriptional readouts with mathemetical modeling and chromatin proteomics led us to propose a two-pronged mechanism for the observed GRN hyper-sensitivity. First, Mediator degradation disrupted large nuclear Pol II clusters. This was accompanied by a transcriptional collapse of associated GRN constituent genes, which were characterized by highly optimized Pol II dynamics. Second, we observed an unanticipated feedback loop that globally elevated Pol II pause release rates via increased activity of CDK9, a subunit of the P-TEFb complex that was previously shown to physically interact with Mediator31–34. This compensatory mechanism was sufficient to maintain transcriptional output at the vast majority of genes. However, it failed to rescue the transcription of GRN constituents, where Mediator optimized transcriptional output by enabling maximal rates of Pol II turnover. Our data position human Mediator as a general coactivator that facilitates transcription globally, but is strictly required for the functionality of cell type-specifying gene regulatory circuits.
Results
Acute Mediator subunit degradation in human cells
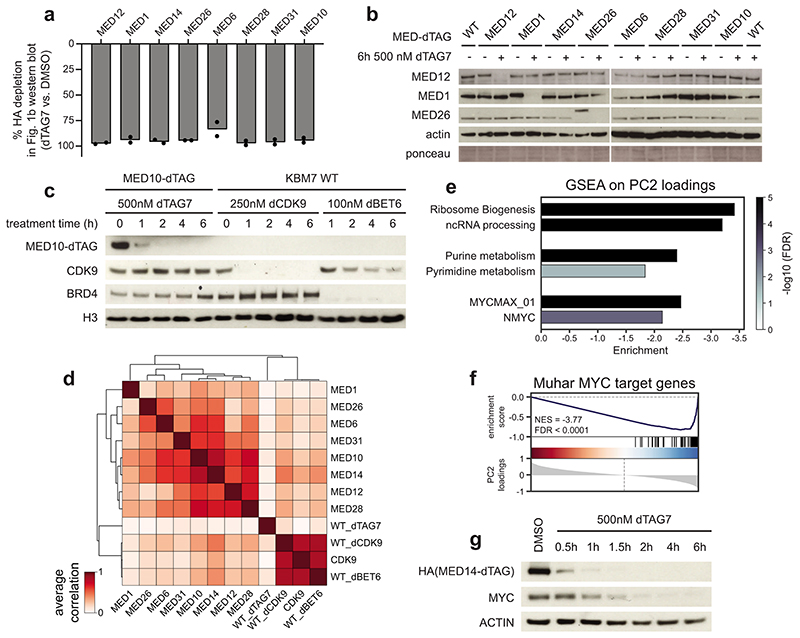
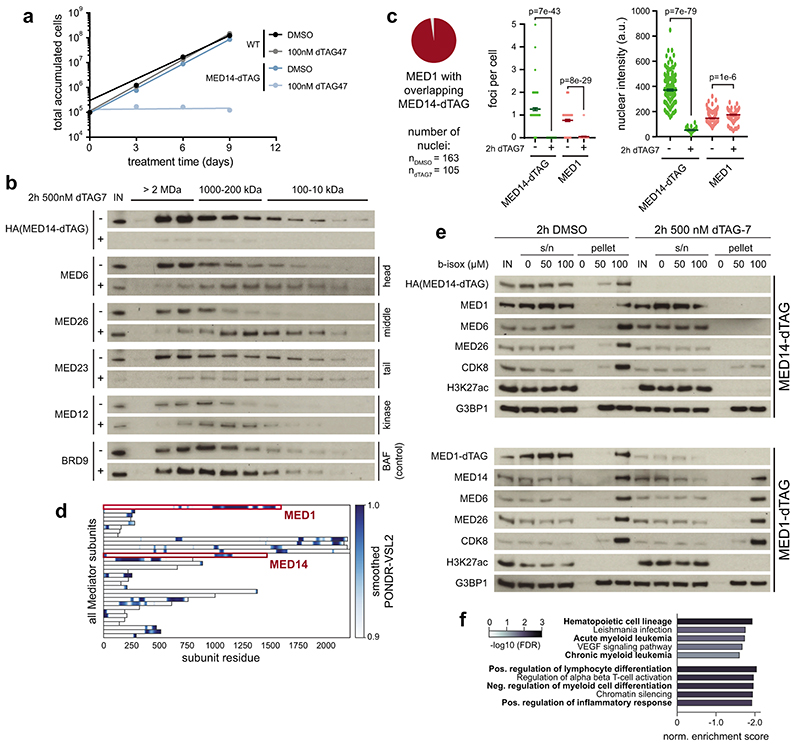
To uncover the direct function of human Mediator, we employed the degradation tag (dTAG) technology to generate eight endogenously tagged, degradation-sensitive Mediator alleles in near-haploid human KBM7 cells (Fig. 1a, Supplementary Table 1)35,36. This approach enables ligand-dependent recruitment of tagged subunits to the CRL4CRBN E3 ligase complex, leading to their fast and selective proteasomal degradation. Particular emphasis was put on core Mediator subunits, previously defined to support a minimal functional Mediator16,37. All tagged subunits were amenable to target-selective, ligand-dependent proteolysis with near-complete degradation evident after 6h of ligand exposure (Fig. 1b; Extended Data Fig. 1a,b). This allowed us to record transcriptional fingerprints of individual subunit degradation using spike-in normalized 3’-mRNA sequencing (Fig. 1c). We contextualized the results with BRD4/BET protein and CDK9 degradation, which are known to prompt global transcriptional arrest38,39. Mediator subunit depletion clearly differed from such global arrest and only mildly affected overall mRNA levels despite similar degradation kinetics and effectiveness (Fig. 1c,d; Extended Data Fig. 1c,d). Transcriptional profiles after subunit destabilization primarily differed in perturbation strength, i.e. quantitatively rather than qualitatively (Fig. 1e). Degradation of MED14 caused the most severe consequence on mRNA levels (Fig. 1c,e). Mediator subunit degradation segregated along principal component 2 (PC2), which distinguished impaired MYC-driven transcription (Fig. 1c, Extended Data Fig. 1e,f)40. Time-resolved immunoblots showed MED14 degradation already after 1h, followed by notable MYC destabilization as early as 2h (Extended Data Fig. 1g). Thus, MYC protein loss likely accounted for this secondary transcriptional phenotype, highlighting the need for readouts with higher kinetic resolution.
Fig. 1. Acute Mediator loss selectively abrogates the functionality of cell type-specifying transcriptional circuits.
a, MED-dTAG alleles and their location in the complex. H: head, M: middle, T: tail, K: kinase module. b, Destabilization of MED-dTAG fusion proteins. c, Principal component analysis of transcriptional fingerprints after 6h (n=3 independent drug treatments). Gene labels: dTAG-carrying cells treated with 500nM dTAG7. dCDK9, dBET6: direct degradation of CDK9 or BET proteins in wild-type cells with 250nM THAL-SNS-032 (dCDK9) or 100nM dBET6. d, Spike-in normalized mRNA log2 fold changes of degrader-treated cell lines (n=8,798 genes; median of 3 independent treatments). e, Genes deregulated by MED-dTAG degradation (left) converge on consensus targets (right). Red bars: genes deregulated by ≥3 perturbations. f, Immunofluorescence of MED1 foci (arrows) in MED14-dTAG cells. Maximum intensity projections of 3D images. Scale bars 1μm. g, Differences in TT-seq nascent transcript levels (n=2 independent treatments). Significantly deregulated (DESeq2 q < 0.01; dark grey), Super-enhancer (SE)-proximal (blue), and auto-regulatory transcription factor (TF) genes (red) are highlighted. Dark grey line: median log2 fold change of all n=19,559 transcription units. h, TT-seq signal of two auto-regulatory TFs, and an expression-matched control gene. H3K27ac and H3K4me3 ChIP-seq signals from KBM7 wild-type cells. i, Fold-change (color) and significance (size) of SE-driven cell identity and expression-matched control gene sets (data as in g). j, 24 auto-regulatory TFs constitute a KBM7 cell type-specifying gene regulatory network. Arrows: the given TF has binding motifs in the target TF’s SE region(s). Edge weight mirrors number of motifs. k, Cell type-specific impact of MED14 degradation on KBM7 and HCT-116 auto-regulatory TFs. Shaded areas: mean log2 fold changes (FC) of the underlying TF genes. Unprocessed western blot shown in Source Data. Violin plot elements: approximated density distribution with internal box plots showing medians with interquartile range and 1.5x whiskers.
Structurally, MED14 acts as a central scaffold that anchors Mediator’s head and middle modules30,41–43. In line with this conserved, essential role of MED14, continuous degradation of MED14 abrogated cell growth (Extended Data Fig. 2a). Coupling acute, 2h MED14 degradation with size-exclusion chromatography showed major impacts on overall complex integrity (Extended Data Fig. 2b). Consistent with recently described coactivator condensates, we observed a highly focal distribution of both MED14 and MED1, with the majority of signal originating from large clusters of around 500nm diameter (Fig. 1f)25,26. These data are in good agreement with previously described stable Mediator clusters in mESC-derived epiblast-like cells26. Importantly, degradation of MED14 dispersed MED1 signal throughout the nucleus without affecting overall signal intensity (Fig. 1f, Extended Data Fig. 2c). This finding reflects MED14’s role as a central Mediator scaffold and suggests an inability of MED1 to maintain clusters, even though it contains an extended intrinsically disordered region (Extended Data Fig. 2d). In accordance, MED14 degradation, but not degradation of MED1, prevented co-precipitation of other Mediator members with biotinylated isoxazole hydrogels, either through direct effects on Mediator integrity, or indirectly by affecting other members of the transcription apparatus (Extended Data Fig. 2e)44. Together, acute MED14 degradation serves as a tool to rapidly disrupt overall Mediator integrity in human cells.
Nascent transcriptional profiling after MED14 degradation
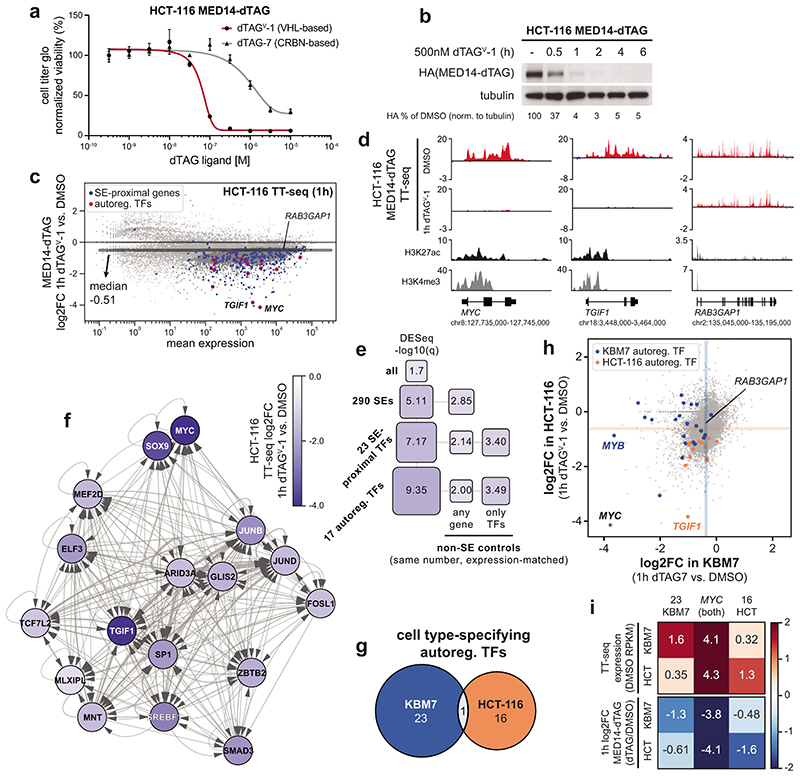
The high kinetic resolution afforded by MED14 degradation motivated us to record direct effects on transcription activity, rather than mRNA levels, using spike-in normalized transient transcriptome sequencing (TT-seq)45. Mediator disruption prompted a notably asymmetric response. Transcription of MYC or MYB had completely collapsed after 1h MED14 degradation, whereas most genes, including the expression-matched RAB3GAP1 control gene, were largely unaffected (Fig. 1g,h). Profound downregulation of additional cell identity genes supported a model where human Mediator safeguards the functionality of cell type-specifying transcriptional networks (Extended Data Fig. 2f). Cell type-specific genes are typically controlled by large, clustered enhancers (super-enhancers, SEs)20,21. A subset of SE-controlled transcription factors (TFs) is required to establish, maintain and alter gene regulatory network topology24. These TFs can be further refined to a core set of auto-regulatory TFs, which lock in cell identity by binding SE regions that drive their own expression46. Such auto-regulatory TFs were among the most Mediator-dependent genes independently of their steady state expression levels and formed a densely connected transcription regulatory circuit in KBM7 cells (Fig. 1i,j; Supplementary Table 2; see Supplementary Note).
To further evaluate if Mediator selectively drives the expression of cell type-specifying genes, we endogenously engineered MED14-dTAG also in HCT-116 colorectal carcinoma cells (Supplementary Table 1). These cells were sensitive to dTAGV-1 ligand hijacking the von-Hippel-Lindau CRL2VHL E3 ligase, but not dTAG7 (Extended Data Fig. 3a), likely due to previously observed lack of activity of CRBN-based degraders in HCT-116 cells38,47. MED14 degradation in HCT-116 cells was rapid, with near-complete degradation evident after 1h (Extended Data Fig. 3b). Also here, Mediator disruption strongly abrogated nascent transcription of SE-proximal genes and cell type-specifying auto-regulatory TFs. In line with data derived after MED14 degradation in KBM7 cells, global transcription was again much less affected (Extended Data Fig. 3c-f). As expected, auto-regulatory TFs in HCT-116 cells were distinct from those in KBM7, with MYC being the only TF in common (Extended Data Fig. 3g; Supplementary Table 2). Notably, the transcriptional sensitivity of a given TF gene to Mediator ablation was dependent on participation in a cell-type specific, auto-regulatory network (Fig. 1k; Extended Data Fig. 3h,i). These findings argue that lineage-specifying transcription is hyper-sensitive to Mediator disruption.
Impact of Mediator loss on chromatin conformation
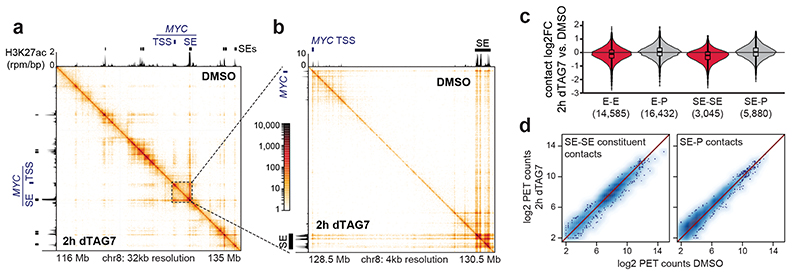
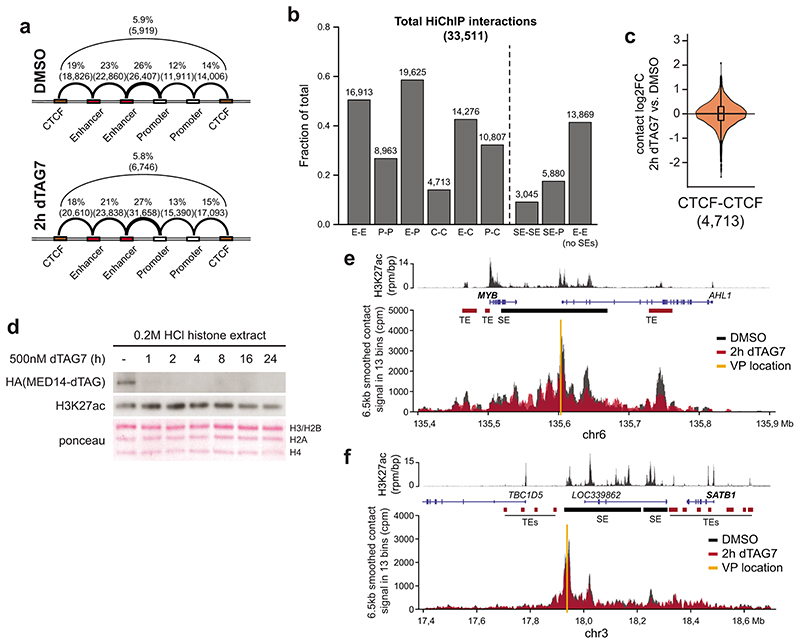
We next addressed if the observed selective shutdown in cell type-specifying transcription and Mediator cluster disassembly could be explained by a disruption of SE-promoter contacts. To that end, we coupled acute MED14 degradation in KBM7 cells with H3K27ac HiChIP analysis48. In agreement with a recent report30, Mediator degradation remained largely inconsequential for overall chromatin architecture, leaving the majority of enhancer-promoter contacts intact (Fig. 2a-c; Extended Data Fig. 4a-c). SE-involving contacts did display a subtle downward trend, albeit at a minor effect size (Fig. 2c,d). Global H3K27 acetylation was only mildly affected at early time points, although differential H3K27ac levels at SE loci might still contribute to the observed changes in contact frequency (Extended Data Fig. 4d). To rule out such immunoprecipitation-based biases of H3K27ac HiChIP, we confirmed these modest changes using in situ circularized chromosome conformation capture (4C-seq) on selected SE-proximal genes (Extended Data Fig. 4e,f)49. Nevertheless, based on their minor effect size, changes in chromatin architecture did not seem to account for the drastic defects in cell type-specifying transcription.
Fig. 2. Mediator is dispensible for maintenance of enhancer-promoter contacts.
a, H3K27ac HiChIP contact matrices of a ≈20Mb region around the MYC locus. Upper triangle shows 2h DMSO, lower triangle 2h MED14 degradation. Axis tracks show KBM7 H3K27ac ChIP-seq signal (capped at 2 rpm/bp) and super-enhancers (SEs). b, Zoom-in highlighting MYC SE–promoter and constituent–constituent contacts. c, Impact on H3K27ac HiChIP contact frequency (E: enhancer, P: promoter, SE: constituent). Brackets: number of quantified contacts. Violin plot elements: approximated density distribution with internal box plots showing medians with interquartile range and 1.5x whiskers. d, Paired-end tag (PET) counts of contacts involving SE constituents. TSS, transcription start site.
Pol II clusters and enhancer transcription depend on MED14
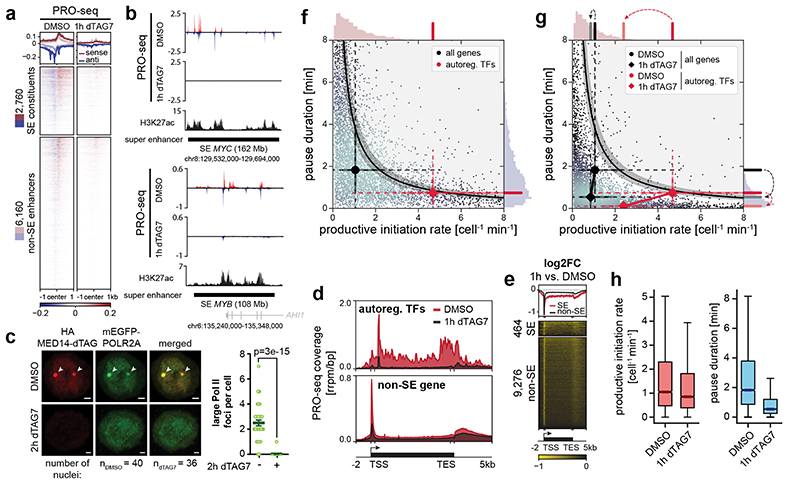
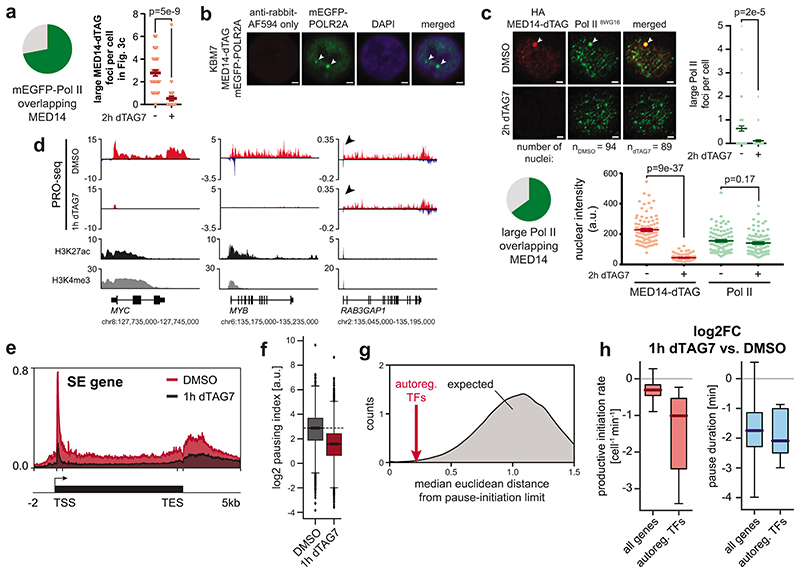
We next aimed to understand if the selectively abrogated transcription of cell type-specifying genes coincided with a loss of enhancer activity. Transcription of enhancer regions has emerged as a key proxy for their activity50. To record the genome-wide distribution of transcriptionally engaged polymerases, we employed spike-in normalized precision nuclear run-on sequencing (PRO-seq)51. Importantly, this assay does not provide information on Pol II recruitment in non-transcribing states, such as pre-initiation complexes52. Already 1h MED14 degradation eradicated transcribing polymerase at enhancers and SE constituents in particular, indicating lack of Pol II activity at these loci (Fig. 3a,b). To test emerging models of condensate-driven transcription initiation, we next wanted to address if Mediator directs the formation of Pol II clusters53,54. We hence engineered an N-terminal mEGFP fusion construct into the endogenous POLR2A locus of MED14-dTAG KBM7 cells (Supplementary Table 1). Indeed, large Pol II foci of around 500nm diameter26,55, which overlapped MED14 clusters at steady state, disassembled upon acute MED14 degradation (Fig. 3c; Extended Data Fig. 5a,b). These clusters likely represented non-transcribing Pol II, as evidenced by immunofluorescence experiments using an antibody directed towards hypo-phosphorylated Pol II (Extended Data Fig. 5c). Concomitantly, Mediator loss eradicated Pol II at SE target genes, and especially the subset of SE-controlled auto-regulatory TFs, in accordance with TT-seq results (Fig. 3d; Extended Data Fig. 5d,e). Pol II density in non-SE gene bodies was less affected, which again shows that the transcriptional output of the majority of genes is only subtly decreased after 1h Mediator ablation (Fig. 3d; Extended Data Fig. 5d). At human genes, Pol II pauses close to the promoter for several minutes before productively elongating a transcript. This pause might serve as a quality control checkpoint and has been shown to sterically limit upstream Pol II initiation56–58. We observed a profound and global decrease in promoter-proximal Pol II, indicative of a change in pause dynamics (Fig. 3e; Extended Data Fig. 5f). This unexpected finding prompted us to investigate how Mediator loss affects Pol II transcription kinetics.
Fig. 3. Mediator organizes Pol II clusters to optimize transcription dynamics of cell type-specifying gene regulatory networks.
a, Impact of 1h MED14 degradation on polymerase engagement at enhancers. b, PRO-seq signal of two super-enhancer (SE) regions. H3K27ac ChIP-seq signal from KBM7 wild-type cells. c, Fixed cell imaging of large Pol II foci (arrows) in MED14-dTAG mEGFP-POLR2A KBM7 cells. Maximum intensity projections of 3D images. Scale bars 1μm. Mean±s.e.m. with two-sided, unpaired t-test. Further quantification and controls in Extended Data Fig. 5a,b. d, Aggregate PRO-seq coverage of auto-regulatory transcription factor (TF) or non-SE metagenes. e, Enrichment heatmap of PRO-seq log2 fold changes (FC) over SE and non-SE metagenes. f, Steady-state productive initiation rates and pause durations for 6,791 transcription units in MED14-dTAG cells. The black central line denotes the theoretical pause-initiation limit with 15% uncertainty56. Markers indicate median gene set values with protruding whisker (dashed) and IQR (solid) proxies. Axis histograms highlight median values of auto-regulatory TFs. g, Pause-initiation values after 1h MED14 degradation. Histogram dashed arrows indicate median pause duration and productive initiation rate trajectories from DMSO to 1h MED14 degradation. h, Changes in productive initiation rates and pause durations for all n=6,791 transcription units. Box plot elements: medians with interquartile range, 1.5x whiskers and confidence region notches. TSS, transcription start site; TES, transcription end site.
Measuring Mediator-controlled Pol II turnover dynamics
To derive a quantitative kinetic model of Mediator-dependent human gene control, we performed integrative analysis of polymerase location (PRO-seq) and RNA synthesis activity (TT-seq). Combining information on the number of RNA molecules being synthesized (TT-seq) with genome-wide polymerase density maps (PRO-seq) informs on the number of transcribing polymerases and their turnover dynamics per unit time at near-nucleotide resolution56,59. This multi-omics modeling allowed us to calculate genome-wide rates of productive transcription initiation and pause duration56. Productive initiation events are defined as events that lead to successful Pol II escape not only from the promoter but also from the pause site into the gene body (see Supplementary Note). Transcriptional output increases with higher productive initiation rates and lower pause duration and is confined by steric considerations, manifesting as the pause-initiation limit56,59. Locating genes in the pause-initiation landscape revealed that SE-proximal auto-regulatory TFs operated closer to the theoretical pause-initiation limit than expected by chance (Fig. 3f; Extended Data Fig. 5g)56. This optimized location was predominantly governed by very high productive initiation rates (Fig. 3f). Acute loss of Mediator selectively diminished productive initiation rates of auto-regulatory TFs, whereas most genes maintained normal productive initiation rates (Fig. 3g,h; Extended Data Fig. 5h). Concomitantly, we observed an unexpected, global decrease in pause duration, indicating more efficient Pol II release into active elongation (Fig. 3g,h; Extended Data Fig. 5h). Together, Mediator organizes Pol II clusters and exerts highly selective control over productive initiation at SE-proximal cell type-specifying genes, while apparently having less influence on the bulk of genes. The global decrease in pause duration led us to hypothesize that defects in pre-initiation complex formation without Mediator are partially compensated by more efficient pause release of residually initiating Pol II.
MED14 loss triggers P-TEFb release from 7SK particles
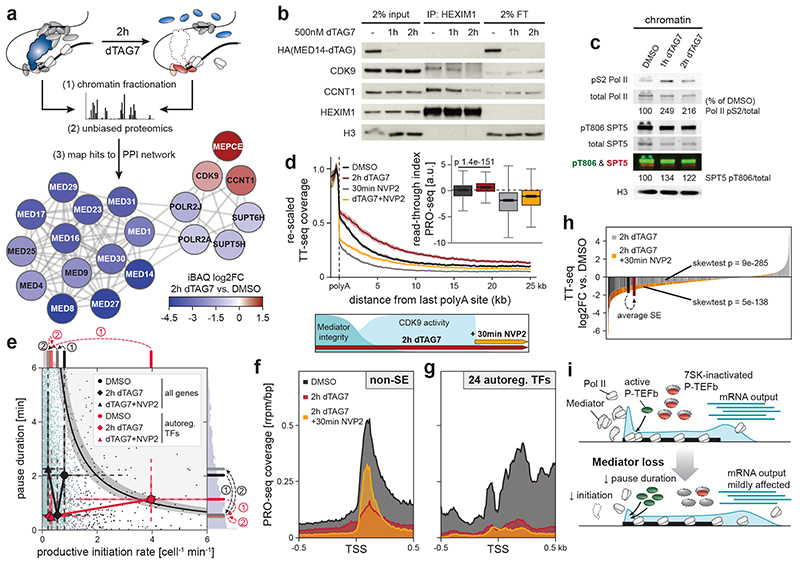
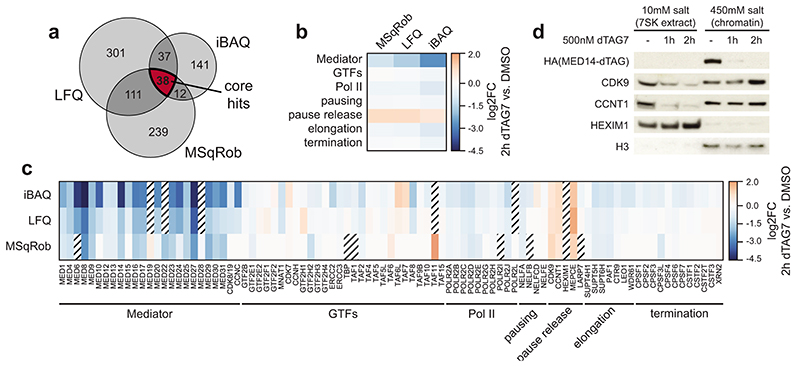
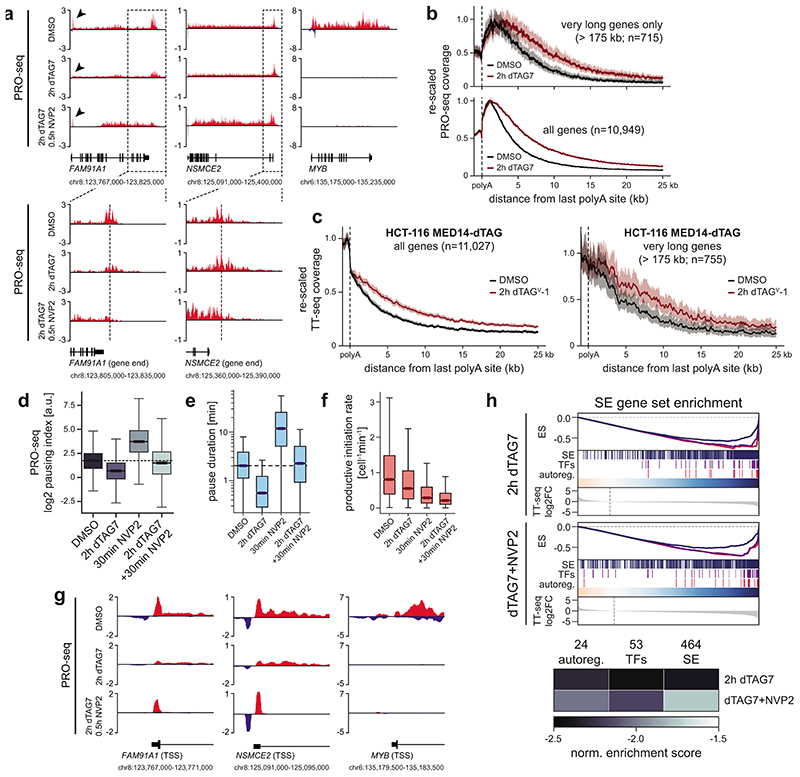
We next aimed to identify factors that might induce the observed changes in pause dynamics. To that end, we subjected purified chromatin fractions to unbiased, label-free mass spectrometry analysis. We intersected the results of three independent data analysis strategies to focus on high-confidence hits (Extended Data Fig. 6a; Supplementary Table 3; see Supplementary Note). Finally, we identified biochemically interacting hits by mapping differentially chromatin-bound proteins to publicly available protein-protein interaction data60. MED14 depletion was near-saturating at 95% and evicted other Mediator subunits off chromatin (Fig. 4a; Extended Data Fig. 6b,c). Binding of general transcription factors remained largely unchanged and Pol II components were only mildly decreased (Fig. 4a; Extended Data Fig. 6b,c). In contrast, three factors were more stably bound to chromatin with high confidence: the main pause-release factor CDK9, its cognate cyclin CCNT1, and MEPCE – all members of positive transcription elongation factor b (P-TEFb) and 7SK regulatory complexes (Fig. 4a; Extended Data Fig. 6c,d)61. As we elaborate in the discussion, this finding is in apparent disagreement with previous reports, which positioned Mediator as a positive regulator of pause release by directly recruiting P-TEFb via its MED23 or MED26 subunits31–33. Unexpectedly, our data imply the presence of a mechanism, which leads to a net-increase of P-TEFb binding to chromatin upon acute Mediator loss. At steady state, the majority of P-TEFb is inactivated by incorporation into 7SK RNPs, which consist of 7SK RNA, HEXIM1/2, LARP7, and MEPCE61. Although several transcriptional stresses, including CDK9 inhibition, are known to release P-TEFb from 7SK particles, functional impacts on Pol II dynamics remain elusive61. MED14 degradation also triggered 7SK disassembly, as indicated by decreased CDK9/CCNT1 binding to HEXIM1 (Fig. 4b). P-TEFb release led to increased phosphorylation of the chromatin-bound CDK9 targets pS2 Pol II and pT806 SPT5 (Fig. 4c)61,62. SPT5 phosphorylation is known to regulate Pol II velocity in the termination window at the 3’ end of genes63. Consistently, we observed vast CDK9-dependent read-through transcription, even at long genes where newly elongating Pol II has not yet reached the gene end (Fig. 4d; Extended Data Fig. 7a-c). The latter finding rules out that these phenotypes are due to differences in elongation complex formation close to the promoter. In summary, unbiased chromatin proteomics led us to identify P-TEFb hyper-activation by 7SK disassembly in response to Mediator loss.
Fig. 4. Compensatory P-TEFb activation boosts non-super-enhancer (SE) output to shape the Mediator hyper-dependence of cell type-specifying transcription.
a, Chromatin proteomics strategy and results. Chromatin fractions were subjected to unbiased, label-free mass spectrometry. High-confidence hits were mapped to public protein-protein interaction (PPI) data60. The largest-connected-component network shows differential chromatin binding using isobaric absolute quantification (iBAQ). b, CDK9/CCNT1 co-immunoprecipitation with HEXIM1 inhibitory 7SK particles. c, CDK9 target phosphorylation on purified chromatin fractions. d, Aggregated TT-seq coverage past polyadenylation sites (n=10,949 transcription units; mean±bootstrapped confidence region). Treatment scheme: 2h MED14 degradation (500nM dTAG7) and/or CDK9 inhibition (500nM NVP2) in the last 30min. Boxplots: PRO-seq read-through indices (n=5,558 genes; Mann-Whitney U-test); medians with interquartile range, 1.5x whiskers and confidence region notches. e, Productive initiation rates and pause durations for 6,954 transcription units after combined MED14/CDK9 perturbation. Markers show median trajectories from DMSO control (circles) to 2h MED14 degradation (diamonds) and combined MED14/CDK9 perturbation (triangles). Axis histograms further illustrate these trajectories. f,g, Aggregated PRO-seq coverage around transcription start sites (TSS) of non-SE genes (f) or auto-regulatory transcription factors (TFs) (g). h, Rank-ordered TT-seq differential expression for 2h MED14 degradation (SE genes in black) or combined MED14/CDK9 perturbation (SE genes in red) compared to DMSO control (n=20,882 transcription units). Bold bars: mean SE ranks and log2 fold changes. P-values from all-gene distribution, two-sided skewness tests. i, Model how P-TEFb activation might compensate less efficient initiation in response to Mediator loss. This mechanism falls short of rescuing initiation defects at auto-regulatory TFs. Unprocessed western blots shown in Source Data.
Hyper-sensitive genes are refractory to P-TEFb compensation
We next addressed if P-TEFb hyper-activation shapes the transcription-regulatory consequences of acute Mediator depletion. To this end, we pharmacologically inhibited CDK9 in the last 30 min of 2h MED14 degradation. This allowed us to assess the role of CDK9 activity at a time of near-complete Mediator depletion (see scheme in Fig. 4d). Pause-initiation modeling confirmed that CDK9 activity was indeed responsible for decreased pause duration following Mediator loss (Fig. 4e; Extended Data Fig. 7d,e). Upon CDK9 inhibition, productive initiation rates decreased even further, implying the presence of residual Pol II initiation despite Mediator depletion (Fig. 4e; Extended Data Fig. 7f). We hence approximated promoter initiation activity by measuring if polymerases continued to enter the pause site. Indeed, blocking pause release after near-complete Mediator degradation substantially increased promoter-proximal Pol II occupancy (Fig. 4f; Extended Data Fig. 7g). We conclude that P-TEFb activation partially compensates Mediator loss by globally increasing pause release. The efficacy of this compensation is contingent on residual Pol II initiation.
Could this compensatory mechanism contribute to the apparent Mediator hyper-dependence of cell type-specifying transcription? In fact, additional CDK9 inhibition failed to re-accumulate promoter-proximal Pol II at auto-regulatory TFs and did not revert their initial drop in pause duration (Fig. 4e,g; Extended Data Fig. 7g). Hence, Mediator appeared absolutely required for successful Pol II initiation at these cell type-specifying genes. On a transcript level, preventing P-TEFb compensation rendered the Mediator phenotype less SE-selective (Fig. 4h; Extended Data Fig. 7h). Together, these experiments uncover a novel phenotypic buffering capacity of promoter-proximal pausing, where transient P-TEFb activation can partially compensate for defects in transcription initiation (Fig. 4i). Although this mechanism seemingly operates genome-wide, it falls short of compensating initiation defects at cell type-specifying genes, which are transcriptionally optimized for efficient Mediator cluster-driven Pol II recruitment. In summary, human Mediator contributes to global transcription, but is predominantly deployed to ensure the functionality of cell type-specifying transcriptional circuits.
Discussion
Here, we combined rapid pharmacologic degradation with multi-omics kinetic analyses to derive direct Mediator functions in human transcription regulation. We focused on MED14 degradation to disrupt overall Mediator integrity. MED14 was asymmetrically required to maintain Mediator and Pol II clusters and transcription of super-enhancer (SE-)driven cell type-specifying genes, but not overall genome architecture. Mediator did stimulate Pol II initiation globally, but appeared strictly required for the functionality of cell type-specifying transcriptional circuits. Two mechanisms contributed to the observed hyper-sensitivity of cell-type specifying gene regulatory networks (GRNs). First, Mediator sustained unusually high rates of Pol II turnover at GRN constituent genes. Second, a genome-wide increase in Pol II pause release unveiled a novel P-TEFb activating mechanism, which compensated for defective initiation at non-SE genes.
Mechanistic conclusions were enabled by our experimental setup, which coupled precise and fast perturbations with orthogonal nascent transcriptional readouts. Importantly, key TF proteins were undetectable within hours of MED14 degradation, likely due to their transcriptional shutdown, which highlights the need for time-resolved analyses. Previously reported global transcriptional collapse after long-term Mediator depletion30 might thus result from secondary effects that are due to a dysfunctional transcription factor machinery at the time of measurement. Our findings also reinforce the rationale and feasibility of pharmacologic inhibition of general transcriptional coactivators as a gene-selective therapeutic strategy64.
We surmised that globally decreased pause duration compensated for compromised Pol II initiation at most genes. We also showed that shorter pausing is most likely due to a CDK9 activity-dependent increase in pause release. However, less efficient establishment of paused Pol II complexes also might be a contributing factor. Of note, previous work has demonstrated a physical interaction between Mediator and P-TEFb and functionally implicated Mediator in contributing to P-TEFb recruitment31–34. Specifically, 48h knockdown of MED26 led to decreased P-TEFb binding at selected genes as assayed by ChIP-qPCR31. Another study described partially overlapping morphological defects in Drosophila embryos depleted of maternally-derived MED26 or Cdk9 mRNA33. In a third study, ChIP-qPCR in Med23 -/- mESCs showed decreased P-TEFb binding to selected genes32. Differences in kinetic resolution and cellular models render it difficult to unambiguously reconcile these findings with the apparently contradicting functional impact of acute MED14 degradation on Pol II pause dynamics. Among others, complex-independent roles of MED26 or MED23 may contribute and could be addressed in future studies. Overall, it remains to be addressed how acute Mediator disruption can prompt an increase in P-TEFb occupancy on chromatin and an ensuing increase in global pause release rates.
Our data are consistent with a model where increased CDK9 activity tips the balance to favor elongation licensing over putative early termination in the pause site56,57,59,65. These findings imply that pausing can serve as a phenotypic buffering mechanism to bridge periods of defective initiation. It might directly reflect the evolutionary advantage of why paused Pol II limits upstream transcription initiation, particularly in human cells56,57. Systematic biochemical analyses uncovered functional compensation that was driven by P-TEFb shedding from inhibitory 7SK particles. Although many stresses were known to trigger 7SK disassembly66, we here show a functional impact on pause duration. Importantly, our data do not provide direct evidence of the genomic regions that P-TEFb binds in response to Mediator disruption. Spatial information in this study is limited to the functional impacts on transcription dynamics, where we observe decreased pause duration at the vast majority of genes. It thus remains to be fully resolved how initiation defects are sensed in cells and how P-TEFb is recruited on a genome-wide scale66.
Mediator was predominantly required for productive Pol II initiation at cell type-specifying genes. We observed that Mediator organized clusters to recruit and supply Pol II, which is consistent with emerging models of condensate-driven transcription initiation53,54. Importantly, our fixed-cell imaging experiments do not allow any conclusions on whether these clusters are truly phase separated compartments67,68. Nonetheless, the fact that Mediator orchestrates the nuclear Pol II distribution and optimizes the transcriptional dynamics of SE-driven auto-regulatory TF genes argues for a strong mechanistic link between cluster formation and cell type-specifying transcription. This interpretation extends well-established biochemical principles of pre-initiation complex formation to human cells and positions human Mediator as a globally acting cofactor that selectively safeguards the functionality of cell type-specifying gene-regulatory networks69.
Online Methods
Cell Culture, cellular growth and lentiviral transduction
Human, near-haploid chronic myeloid leukemia KBM7 cells were grown in IMDM supplemented with 10% FBS and 1x penicillin/streptomycin (Gibco). Human diploid HCT-116 colorectal carcinoma cells were grown in 15cm cell culture dishes in RPMI supplemented with 10% FBS and 1x penicillin/streptomycin (Gibco) and passaged every 3-4 days upon reaching ~70% confluency. Drosophila S2 cells were grown in Schneider’s Drosophila medium supplemented with 10% FBS (Gibco) and used as exogenous spike-in for all PRO-seq experiments.
For growth-over-time analysis, 105 cells were seeded in triplicates in 1mL medium +/- 100nM dTAG47 in 24-well plates, counted and split back to 105 cells every 3 days for a total of 9 days. Total accumulated cells were calculated by assuming exponential growth kinetics.
For CellTiter-Glo (CTG, Promega) viability-based drug dose-response assays, we seeded 6x103 MED14-dTAG HCT-116 cells in 50μL RPMI to 96-wells for 10 data points in triplicates. Serial drug dilutions were prepared in 2x concentration in RPMI, starting from 10μM final concentration, and 50μL were added to the 50μL cell suspension. Cells were incubated at 37°C for 3 days before performing CTG assay according to the manufacturer’s instructions. Briefly, CTG substrate was diluted 1:4 with water and added 1:1 to cells before measuring chemical luminescence on an M5 plate reader. Relative viability was calculated by normalizing each data point to the mean luminescence of the lowest drug concentration.
Stably Cas9-expressing KBM7 cells were generated with lentivirus produced from pLenti_Cas9_blast (Addgene #52962). Lentivirus was produced by seeding 4x106 HEK 293T cells to 10cm dishes 24h before transfection of 5μg transfer plasmid, 2.5μg pMD2.G (Addgene #12259) and 3.75μg psPAX2 (Addgene #12260) using PolyFect (Qiagen) following the manufacturer instructions. Medium was changed to 8mL fresh DMEM 8h after transfection. Viral supernatant was harvested after 72h and filtered through a.22μm syringe filter to remove cell debris. Viral supernatant was aliquoted and stored at -80°C until further use. 3x106 KBM7 cells in 3mL were transduced with different amounts of viral supernatant in 12-well plates using 8μg/mL polybrene and spin-infected for 45 min at 37°C and 2000rpm. Antibiotic selection with 10μg/mL blasticidin was started 24h after transduction.
Plasmids, cloning and chemicals used in this study
All plasmids and chemicals used in this study are summarized in Supplementary Table 4. To facilitate cloning of sgRNA cutting plasmids for endogenous dTAG knock-in, we generated a universal pX330A_sgX_sgPITCh cutting plasmid by slightly adapting published protocols70. Briefly, the U6-sgPITCh cassette from pX330S-2-PITCh (Addgene #63670) was cloned into pX330A_1-2x (Addgene #58766) with BsaI. Targeting sgRNA sequences for endogenous knock-in (see Supplementary Table 1) were then introduced under control of the first U6 cassette of pX330A_sgX_sgPITCh with standard BbsI-mediated oligo-anneal cloning. Targeting sgRNA sequences were selected based on location (as close as possible to the N-terminal start codon) and predicted sgRNA cleavage and off-target performance.
pCRIS-PITChv2 repair template plasmids were cloned following published procedures71. Briefly, primers were selected to contain 20-22bp microhomology sequences corresponding to genomic sequences immediately 5’ and 3’ of the sgRNA cleavage sites. Bases were added or removed to conserve the target reading frame, coding back the last cleaved amino acid whenever possible. The resulting Rep_X_F/R primers (see Supplementary Table 1) were used to PCR-amplify the dTAG-blast/puro cassettes from template plasmids (Addgene #91792 and #91793) and re-introduced into MluI-linearized pCRIS-PITChv2 backbone with NEBuilder 2x HiFi assembly (New England Biolabs).
For knock-in of mEGFP to the N-terminus of POLR2A, we modified pCRIS-PITChv2 repair plasmids in analogy to dTAG cassette carrying templates (as in Addgene #91792 and #91793). Coding sequence of mEGFP (A206K) was PCR amplified with primers adding N-terminal FLAG and C-terminal (GGGGS)x2 linker sequences. The resulting FLAG-mEGFP-SG fragment was introduced into MluI-linearized pCRIS-PITChv2 backbone with NEBuilder 2x HiFi assembly (New England Biolabs). 22 bp microhomology sequences immediately 5’ and 3’ of a POLR2A N-terminally targeting sgRNA cleavage site (see Supplementary Table 1) were then introduced using the same strategy as described for dTAG cassettes.
Genome editing for endogenous knock-in
For endogenous knock-in of dTAG cassettes, KBM7 cells were seeded to 2x105 cells/mL the day before transfection to ensure exponential cell growth. The next day, cells were transfected with PITCh sgRNA/Cas9 and repair template plasmids using Amaxa Nucleofector II with Reagent V (Lonza)70. Briefly, 2x106 cells/reaction were harvested by slow centrifugation at 90g and resuspended in 1mL warm, antibiotic-free IMDM per reaction. Cells were aliquoted to 2mL microcentrifuge tubes and again spun down at 100g in a tabletop centrifuge. After aspirating the supernatant, cells were resuspended in 100μL supplemented Amaxa reagent V mixed with 6μg DNA per plasmid (see Supplementary Table 1 for sgRNA and microhomology sequences). The cell/plasmid suspension was quickly transferred to the reaction cuvette and electroporated using the X-001 program. Immediately after the reaction, 700μL warm, antibiotic-free IMDM was added to the cuvette and the cell suspension was carefully transferred to 1.5mL pre-equilibrated, warm, antibiotic-free IMDM in 6-well plates using the provided single-use Pasteur pipettes.
Cells were allowed to recover for 5 days at 37°C before starting antibiotic selection of the pools in 10mL IMDM in T25 flasks (1μg/mL puromycin or 10μg/mL blasticidin, depending on the used dTAG cassette; see Supplementary Table 1). After 7 days of selection, surviving cells were seeded to 384-well plates at concentrations of 0.2-1 cells/well (50μL/well). Single-clone colonies were expanded to 24-well plates after 14 days and initially characterized using HA-immunoblotting. HA-positive clones were genotyped by gDNA PCR of the integration site followed by Sanger sequencing (Microsynth; see Supplementary Table 1 for genotyping primer sequences and editing outcomes). One working clone and up to two backup clones were selected and kept for further experiments.
Due to difficulties in obtaining MED12-dTAG clones, we here followed a slightly modified targeting strategy: stably Cas9-expressing (pLenti_Cas9_blast; Addgene #52962) KBM7_Cas9 cells were transfected with a truncated, Cas9-less version of pX330A_sgMED12_sgPITCh cutting plasmid. In this case, transfected DNA amounted to 2μg of Cas9-less cutting plasmid and 10μg of pCRIS-PITCh_v2_dTAG_puro repair plasmid. Clone selection and screening was performed as described above.
Endogenous knock-in of MED14-dTAG in HCT-116 colorectal carcinoma cells was performed following the same strategy as described above for KBM7 cells with minor adaptions. Briefly, 2x106 HCT-116 cells (~70% confluent) were trypsinized and transfected with 6μg per PITCh sgRNA/Cas9 and repair template plasmid using Amaxa Nucleofector II with Reagent V and electroporation program D-032 (Lonza)70. Cells were then selected using 10μg/mL blasticidin before single clone expansion and characterization as described above for KBM7 cells.
Secondary knock-in of mEGFP-POLR2A into MED14-dTAG KBM7 cells was achieved using the same strategy as for the parental MED14-dTAG knock-in cells with the following modifications: PITCh sgRNA/Cas9 and repair template plasmids were transiently transfected into MED14-dTAG KBM7 cells as described above, but using a FLAG-mEGFP-SG repair template with POLR2A N-terminal microhomologies (see Supplementary Table 1). The pool of transfected cells was allowed to recover for 5 days before fluorescence-activated cell sorting of the top 0.15% GFP-positive cells (Sony SH800 FACS cell sorter, Sony). Cells were expanded for 13 days before a second sort, followed by another 6 days of expansion and a third sort. Clones were isolated from GFP-positive pools by plating in limiting dilution to 96-well plates and subsequent clonal expansion. Successful mEGFP knock-in to the endogenous POLR2A locus was validated by PCR amplification and Sanger sequencing of the full-length genomic integration fragment (Microsynth; see Supplementary Table 1).
Whole cell lysis and immunoblotting
PBS-washed cell pellets were lysed in RIPA buffer (50mM Tris-HCl pH8.0, 150mM NaCl, 1% Triton-X-100, 0.5% Na-deoxycholate, 0.1% SDS, 1x Halt protease inhibitor cocktail, 25U/mL Benzonase) for 15 min on ice. Lysates were cleared by 15 min centrifugation at 4°C and 20,000g. Protein concentration was measured using BCA before adding 4x LDS sample buffer. 10-30μg protein were loaded on Bolt 4-12% Bis-Tris gels and blotted to nitrocellulose membranes. Membranes were blocked with 5% milk/TBST for 30 min at room temperature. Primary antibodies were added in milk or pure TBST for 1h at room temperature or 4°C overnight. Secondary antibodies were added for 1h at room temperature. Blots were developed using chemiluminescence films for HRP- or a ChemiDoc fluorescent imaging system for fluorescently labeled antibodies. Where applicable, western blots were quantified using ImageJ software (U.S. National Institutes of Health, Bethesda).
Used antibodies (see Supplementary Table 5): HA (1:1000, Cell Signaling Technology, #3724S and #2367S), GAPDH (1:1000, Santa Cruz Biotechnology, sc-365062), Histone H3 (1:10000 Abcam, ab1791), β-ACTIN (1:10000, Sigma Aldrich, A5441), MYC (1:1000, Santa Cruz Biotechnology, sc-764), MED10 (1:500, Abcam, ab110786), CDK9 (1:1000, Cell Signaling Technology, #2316S), BRD4 (1:5000, Bethyl Laboratories, A301-985A100), MED12 (1:2000, Bethyl Laboratories, A300-774A), MED1 (1:2000, Bethyl Laboratories, A300-793A), MED14 (1:1000, Bethyl Laboratories, A301-044A), MED26 (1:1000, Cell Signaling Technology, #14950S), MED6 (1:1000, Santa Cruz Biotechnology, sc-390474), MED23 (1:1000, Bethyl Laboratories, A300-425A), BRD9 (1:1000, Active Motif, #61537), CDK8 (1:1000, Cell Signaling Technology, #4101S), H3K27ac (1:1000, Abcam, ab4729), G3BP1 (1:500, Santa Cruz Biotechnology, sc-365338), α-TUBULIN (1:1000, Sigma Aldrich, T9026), CCNT1 (1:1000, Cell Signaling Technology, #81464S), HEXIM1 (1:1000, Abcam, ab25388), MED31 (1:500, Santa Cruz Biotechnology, sc-101189), pS2 Pol II (1:1000, Active Motif, #61083), Pol II (1:250, Santa Cruz Biotechnology, sc-899), pT806 SPT5 (1:1000, Robert Fisher lab, Icahn School of Medicine at Mount Sinai), SPT5 (1:1000, Santa Cruz Biotechnology, sc-133217), TUBULIN-Rhodamine (1:5000, Bio-Rad, #12004166).
Secondary antibodies: anti-mouse-HRP (1:5000, Jackson ImmunoResearch, 115-035-003), anti-rabbit-HRP (1:5000, Jackson ImmunoResearch, 111-035-003), anti-rat-HRP (1:5000, Dako, P0450), anti-rabbit-StarBright700 (1:5000, Bio-Rad, #12004162), anti-mouse-Dylight800 (1:5000, Bio-Rad, STAR117D800GA).
Quant-seq 3’ mRNA transcriptional fingerprinting
Spike-in normalized Quant-seq (Lexogen) was performed in biological triplicates to record a transcriptional fingerprint of acute Mediator perturbation in the context of chemical (genetic) perturbations with known transcriptional consequences. Briefly, eight MED-dTAG (MED12, MED1, MED14, MED26, MED6, MED28, MED31, MED10), CDK9-dTAG and wild-type KBM7 cells were seeded to 5x105 cells/mL in 10mL T25 flasks two days before the experiment to avoid major differences in cell cycle and growth kinetics between different clones. On the day of the experiment, cells were counted and 8x105 cells/sample seeded to 24-well plates in 1mL for triplicate drug treatments. Drug treatments from 1000x DMSO stocks were performed for 6h with the following conditions: 500nM dTAG7 or DMSO for dTAG-carrying clones; 500nM dTAG7, 100nM dBET6, 250nM THAL-SNS-032 (dCDK9) or DMSO for wild-type KBM7 cells. Cell pellets were harvested by centrifugation, washed with cold PBS, snap-frozen and stored at -80°C.
The next day, cell pellets were thawed on ice and total RNA was extracted with RNeasy kits (Qiagen) after addition of SIRV-Set 3 exogenous spike-in mix (Lexogen). Briefly, 350μL buffer RLT were added to the pellets and the mix centrifuged through QIAshredder spin columns according to manufacturer instructions. Now, 10μL of 1:100 diluted (0.303ng/sample; according to manufacturer protocol) SIRV-Set 3 spike-in mix were added to the homogenized lysate before addition of 350μL ethanol and column purification according to manufacturer instructions. Total RNA was eluted in 30μL EB and stored at -80°C overnight.
The next morning, RNA was thawed on ice and concentration determined to 71-102ng/μL by NanoDrop. All samples were now diluted to 70ng/μL in 20μL and Quant-seq was performed in a 96-well PCR plate using 5μL (350ng RNA) as input. Samples were randomly distributed over the plate to avoid edge effects. Quant-seq libraries were now performed by carefully following the manufacturer’s instructions. Briefly, first strand (oligo-dT) cDNA synthesis was followed by RNA removal and second strand synthesis via random priming. The double-stranded library was bead-purified to remove reaction components before PCR amplification with i7 single-index primers for 13 cycles. Amplified libraries were again bead-purified according to the manufacturer’s protocol, yielding 0.67-3.16ng/μL DNA as measured by Qubit assay. A total of 11 representative low-, intermediate- and high-yield samples were checked for fragment size distribution on Bioanalyzer before pooling for 50bp single-read sequencing on a HiSeq 3000/4000 platform to obtain 2.4-9.6 million raw reads per sample (see Supplementary Table 6).
Quant-seq raw data were processed essentially as recommended by the library preparation kit manufacturer (Lexogen). Raw reads were trimmed using BBtools version 38.00 ‘bbduk.sh -ref “polyA.fa.gz”,”truseq.fa.gz” k=13 ktrim=r useshortkmers=t mink=5 qtrim=r trimq=10 minlength=20’ and aligned to a concatenated hg38_SIRV-Set-3 (sequences provided by Lexogen) index using STAR version 2.5.2b with ‘--outFilterType BySJout --outFilterMultimapNmax 20 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --outFilterMismatchNmax 999 --outFilterMismatchNoverLmax 0.6 --alignIntronMin 20 --alignIntronMax 1000000 --alignMatesGapMax 1000000 --outSAMattributes NH HI NM MD --outSAMtype BAM SortedByCoordinate’ 72. Aligned reads were then counted using htslib version 0.6.0 ‘htseq-count -m intersection-nonempty -s yes -f bam -r pos’ on a fused hg38_SIRV-Set-3.gtf ENSEMBL e87 geneID annotation73. Raw reads were normalized to counts per million (cpm).
Quant-seq data analysis (PCA, spike-in normalization, differential expression)
PCA analysis was performed on cpm values using the scikit-learn python package (v0.19.0)74. First, read counts were normalized using the StandardScaler.fit_transform function with standard settings. The first 10 principal components were calculated using the decomposition.PCA function. Per-gene PC loadings were computed by multiplication of the eigenvectors with the square root of the principal component eigenvalues. Gene set enrichment analysis (GSEA) of PC2 loadings was performed using GenePattern’s GSEAPreranked with collections KEGG_pathway, GO_process, and TF_Targets75. The top two gene sets containing at least 75 genes were shown. We additionally tested a published set of 100 high-confidence direct MYC target genes40.
Locally estimated scatterplot smoothing (LOESS) normalization was performed for spike-in normalization using the normalize.loess function of the affy v1.50.0 R package76. The LOESS distribution was fit on the SIRV-Set-3 spike-ins and then applied to the whole dataset. Average log2 fold changes were calculated from means of triplicate treatments, omitting genes with normalized log2(cpm) < 5.0 in average wild-type DMSO.
To identify the target gene spectrum of individual Mediator subunits, we performed DESeq2 v1.23.0 differential expression analysis on normalized counts relative to wild-type DMSO control77. The union of significantly deregulated genes (log2FC > +/-1.5; p < 0.05) was hierarchically clustered with correlation as distance metric and average as linkage method. The clustered list of deregulated genes was used as coordinates of a long “target genes scaffold” in the R BioCircos library78. Links connect dummy “MED-dTAG perturbation scaffolds” with the respective coordinate in the target gene scaffold, if the gene was deregulated by the given perturbation. A secondary heatmap outside the target gene scaffold indicates genes, which were deregulated by at least three MED-dTAG perturbations.
Size-exclusion chromatography to assess Mediator complex integrity
Size-exclusion chromatography of Mediator-containing nuclear extracts was performed similar to published procedures79. 100x106 MED14-dTAG KBM7 cells were washed with PBS and transferred to 2mL tubes. Cytoplasmic lysis was performed by resuspending with 500μL cold Buffer A (10mM Tris-HCl pH7.4, 10mM KCl, 1.5mM MgCl2, 0.5mM DTT, 0.1% Triton-X-100, 1x Halt protease inhibitor cocktail) and incubating for 10 min on ice. Nuclei were harvested by centrifugation at 4°C and 1000g for 5 min and the pellet was washed once with 1mL Buffer A. High-salt nuclear extracts were performed by adding 45μL Buffer C (20mM Tris-HCl pH7.4, 400mM KCl, 1.5mM MgCl2, 1mM DTT, 5 U/μL Benzonase, 1x Halt protease inhibitor cocktail) and incubating for 1h on ice with occasional vortexing. The lysate was clarified by centrifuging 15 min at 4°C and 20000g, yielding about 75μL nuclear extract at 5-10μg/mL total protein. Potential particles were removed by centrifugation through.22μm spin filter columns before injection into the HPLC system.
Nuclear extracts were loaded onto an Äkta FPLC system via a 50μL injection loop and separated with SEC running buffer (20mM Tris-HCl pH7.4, 200mM KCl) over a Superose 6 Increase 3.2/300mm gel filtration column (GE Lifesciences). Run parameters were set to 0.1 column volumes (CV) of initial equilibration, followed by sample injection and 1.5CV elution at a constant flow rate of 0.04mL/min (resulting in 0.7-0.8MPa run pressure). Elution fractions of 150μL were collected in 96-deep-well plates over the entire elution range. Eluates were directly diluted with 4x LDS sample buffer and 20μL per fraction (or 1-2μL input nuclear extract) loaded onto 4-12% Bis-Tris gels for western blot analysis.
Immunofluorescence microscopy of Mediator and RNA Pol II
1x106 MED14-dTAG KBM7 cells were spun onto poly-lysine pre-coated (30 min at 37°C; washed 3x with H2O) coverslips in 24-well plates for 15 min at 1800rpm. Cells were fixed by directly adding equal volume 4% methanol-free PFA (final 2%) and incubating 10 min at room temperature. Coverslips were washed 3x with PBS and then incubated for 5 min in 200μL/well permeabilization buffer (0.1% non-neutralized dibasic NaCitrate, 0.2% Triton-X-100). After again 3x washing with PBS, coverslips were incubated for 30 min in 200μL/well blocking buffer (2.5% BSA in PBS). Primary antibodies were added directly to the blocking solution at a concentration of 1:200 and incubated at 4°C overnight. The next morning, coverslips were washed with 3x PBS and incubated for 1-2h with 1:250 secondary antibody at room temperature. Coverslips were again washed with 3x PBS and labeled with DAPI for 5 min before mounting onto glass slides using Dako mounting medium.
The same procedure was followed for combined fixed-cell imaging of mEGFP-POLR2A (direct fluorescence) and MED14-dTAG (immunofluorescence; detected with anti-rabbit-AF594) KBM7 cells, including a negative control condition, where anti-HA primary antibody was omitted. For visualization of representative cells, AF594 signal was scaled to allow quantitative assessment of MED14-dTAG degradation, while mEGFP signal was scaled to resolve focal structures or lack thereof.
Images were acquired as 3D z-stacks on an LSM780 confocal fluorescence microscope in AiryScan mode (Zeiss) using a 63x oil objective. Maximum intensity projections were derived using the Zeiss ZEN Blue software and segmented for image quantification using a custom CellProfiler pipeline80. Large Mediator and Pol II foci of around 500nm diameter were segregated and classified using area- and intensity-based thresholds to quantify mutual overlap and number of large foci per cell.
Primary antibodies: HA (Cell Signaling Technology, #3724S and #2367S), MED1 (Abcam, ab64965), Pol II 8WG16 (Abcam, ab817). Secondary antibodies: anti-rabbit-AF594 (Invitrogen, A-21442), anti-mouse-AF488 (Invitrogen, A-11001).
Mediator subunit disorder prediction
Sequence-intrinsic disorder propensity was predicted for all human Mediator subunits. Canonical isoform FASTA sequences were obtained from Uniprot and subjected to PONDR-VSL2 disorder prediction software (http://www.pondr.com/)81. Results were compiled into a matrix by residue number and smoothed using a running mean window of 20 residues. Only high-confidence (VSL2 > 0.9) disordered regions were highlighted in a heatmap.
Biotinylated isoxazole precipitation of low complexity region-containing proteins
75x106 washed MED14-dTAG KBM7 cells were resuspended in 1.5mL EE-Buffer (50mM Tris-HCl pH7.4, 150mM NaCl, 1mM EDTA, 2.5mM EGTA, 10% glycerol, 0.1% Triton-X-100, 1x Halt protease inhibitor cocktail) and incubated 30 min at 4°C on a rotating wheel. Lysates were clarified by centrifugation for 15 min at 4°C and 20000g. 75μL lysate was removed as input and the rest was aliquoted into 3x400μL in fresh 1.5mL tubes. Biotinylated isoxazole from 100x DMSO stocks (Dalton Pharma Services) was added to the lysates to final concentrations of 0-100μM and the tubes were incubated for 2h at 4°C on a rotating wheel. Pellets were collected by 15 min centrifugation at 4°C and 20000g, supernatant was removed and kept and the pellet washed once with 500μL EE-Buffer. The resulting pellets were taken up in 75μL 1x LDS/EE-Buffer, heated 10 min at 95°C before loading 15μL per sample (input, supernatant, b-isox pellet) for western blot analysis.
Supplementary Note
Further methods are described in detail in a Supplementary Note linked to this article.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Statistics and Reproducibility
Unpaired, two-tailed Student’s t-tests were performed for Fig. 3c and Extended Data Figs. 2c, 5a,c. Mann-Whitney U-test in Fig. 4c was performed using the SciPy v1.2.1 mannwhitneyu function with default settings (p = half of the two-sided p-value) (ref. 82). Skewness in Fig. 4h was tested using the SciPy v1.2.1 skewtest function with default settings (two-sided p-value), which tests the null hypothesis that the skewness of the given population is the same as that of a corresponding normal distribution82.
Box plots were generated using the Seaborn v.0.9.0 or Matplotlib v.2.2.3 boxplot function with standard settings: centre lines display median values and boxes display the Q3-Q1 interquartile range (IQR) with whiskers protruding as 1.5-times the IQR. When included, notches display the confidence region around the median, derived with standard settings83,84. Violin plots were generated using the Seaborn v0.9.0 violinplot function with standard settings, which displays an approximated density distribution of the underlying data points, as well as an internal boxplot with abovementioned elements83.
Fig. 1b and Extended Data Fig. 1b,g, 4d results are representative of two independently conducted cellular treatments and immunoblots. Fig. 4c results are representative of two independently conducted cellular treatments, immunoprecipitations and immunoblots. Fig. 4d results are representative of three independently loaded immunoblots from a single chromatin fractionation experiment. Extended Data Fig. 1c, 3b show results from a single cellular treatment and immunoblot experiment. Extended Data Fig. 2b shows results from a single size-exclusion chromatography experiment. Extended Data Fig. 2e results are representative of three independent biotinylated-isoxazole precipitation experiments. Extended Data Fig. 5b maximum intensity projection is representative of 9 nuclei acquired in one field of a single staining replicate and was performed in parallel to the Fig. 3c experiment. Extended Data Fig. 6d is representative of two independently loaded immunoblots from a single cellular treatment and nuclear fractionation experiment.
Extended Data
Extended Data Fig. 1. Extended characterization of chemically degradable MED-dTAG alleles.
a, MED-dTAG depletion mean of two independent image quantifications of the Fig. 1b immunoblot. b, Degrader treatment selectively destabilizes the tagged Mediator subunit without affecting other complex members. c, Time-resolved immunoblot of MED10-dTAG and direct pharmacologic degradation of CDK9 (dCDK9; THAL-SNS-032) or BRD4 (dBET6). d, Pearson correlation of average 3’ mRNA-seq log2 fold changes after 6h (n=3 independent drug treatments). For dTAG-carrying cell lines (only gene names shown), we compare dTAG7 vs. vehicle control in the same cell line. Other conditions represent drug vs. vehicle control in wild-type cells. e, Gene ontology (GO) terms enriched among negative PC2 loadings in Fig. 1c. Enrichment was calculated using the GSEAPreranked tool75. Negative enrichment indicates a strong influence of these terms on PC2 diversity and that the underlying genes are downregulated. f, Gene set enrichment analysis of top 100 core MYC target genes from (ref. 40) among PC2 loadings. g, Time-resolved immunoblot of MED14-dTAG degradation kinetics and its influence on MYC protein levels. Unprocessed western blots shown in Source Data.
Extended Data Fig. 2. MED14 degradation disrupts overall Mediator complex integrity.
a, Influence of long-term MED14 degradation on cell growth. b, Size-exclusion chromatography and western blotting of nuclear extracts after MED14 degradation. Mediator subunits of each submodule shifted to lower apparent molecular weight, indicating complex disassembly. BAF complex member BRD9 serves as negative control. c, Image quantification related to Fig. 1f. Pie chart: percent of n=125 MED1 foci with overlapping MED14-dTAG foci. Middle dot plot: mean±s.e.m number of foci per cell. Swarm plot: mean±s.e.m integrated nuclear fluorescence intensity. Unpaired, two-sided t-tests. 163 nuclei were quantified for DMSO and 105 for dTAG7. d, 20-residue running average-smoothed PONDR-VSL2 disorder prediction for human Mediator. Subunits are ordered by ascending index numbers from MED1 to MED31, followed by CDK8, CDK19, and CCNC. e, Influence of MED14 or MED1 degradation on co-precipitation of other Mediator subunits with biotinylated isoxazole pellets. MED14, but not MED1 degradation, prevents Mediator co-precipitation with IDR-enriching hydrogels. f, Cell identity gene sets enriched among downregulated transcription units in TT-seq after 1h MED14 degradation. Enrichment was calculated using the GSEAPreranked tool75. Unprocessed western blots shown in Source Data.
Extended Data Fig. 3. Acute transcriptional consequences of MED14 degradation in HCT-116 cells.
a, CellTiter-Glo viability-based 72h dose-response of dTAG7 and dTAGV-1 in HCT-116 MED14-dTAG cells. Mean±s.d. of n=3 drug treatments. b, Time-resolved immunoblot of MED14-dTAG degradation. c, Differences in TT-seq nascent transcript levels (n=2 independent treatments). Significantly deregulated (DESeq2 q < 0.01; dark grey), SE-proximal (blue), and auto-regulatory TF genes (red) are highlighted. Dark grey line: median log2 fold change of all n=21,629 transcription units. d, TT-seq signal of two auto-regulatory TFs, and an expression-matched control gene. H3K27ac and H3K4me3 ChIP-seq signals are from publically available data (GSE72622; see Supplementary Table 7)85. e, Fold-change (color) and significance (size) of SE-driven HCT-116 cell identity and expression-matched control gene sets (data as in c). f, Regulatory wiring of 17 auto-regulatory TFs in the HCT-116 cell type-specifying gene regulatory network. Arrows: the given TF has binding motifs in the target TF’s SE region(s). Edge weight mirrors number of motifs. g, Overlap of KBM7 and HCT-116 auto-regulatory TFs. h, Cell type-specific impact of 1h MED14 degradation. Auto-regulatory TFs in KBM7 (blue, e.g. MYB), HCT-116 (orange, e.g. TGIF1), or MYC (black) as the only shared TF are highlighted. Colored lines: median log2FC in the respective cell line. i, Mean steady state expression of auto-regulatory TFs in merged 1h and 2h DMSO TT-seq conditions and transcriptional defects after 1h MED14 degradation. Unprocessed western blot shown in Source Data.
Extended Data Fig. 4. Impact of MED14 degradation on overall chromatin architecture.
a, Genomic feature classes at H3K27ac HiChIP contact anchors. Only significant interactions called by hichipper/mango were used for anchor identification. Arcs indicate the percentage of anchor-anchor pairs annotated with the indicated feature in each of the samples. b, Total number of interactions common to DMSO and dTAG7 samples, which were used for quantification (E: enhancer, P: promoter, SE: constituent). c, Impact of Mediator loss on CTCF-CTCF contact strength as negative control. Bracket: number of quantified contacts. Violin plot elements: approximated density distribution with internal box plots showing medians with interquartile range and 1.5x whiskers. d, Impact of MED14 degradation on H3K27 acetylation. e, Pulldown-independent 4C-seq analysis of MYB SE constituent viewpoint (VP) after 2h MED14 degradation in triplicates. Top track shows KBM7 wild-type H3K27ac ChIP-seq. TE: typical enhancer, SE: super-enhancer f, Analogous to (e) with a SATB1 SE viewpoint. Unprocessed western blot shown in Source Data.
Extended Data Fig. 5. Impact of MED14 degradation on Pol II clusters and nascent transcription dynamics.
a, Image quantification related to Fig. 3c. Pie chart: percent of n=100 large Pol II foci, which overlap MED14-dTAG foci. Mean±s.e.m. with two-sided, unpaired t-test (n=40 nuclei in DMSO; n=36 nuclei in dTAG7 condition). b, Control imaging experiment related to Fig. 3c, omitting anti-HA primary antibody to rule out that Pol II foci are an HA channel bleed through artifact. c, Immunofluorescence of large hypo-phosphorylated Pol II foci (8WG16; arrows) in MED14-dTAG KBM7 cells. Maximum intensity projections of 3D images. Scale bars 1μm. Pie chart: percent of n=60 large Pol II foci, which overlap MED14-dTAG foci. Dot plots: changes in number of large Pol II foci per cell and integrated nuclear fluorescence intensity. Mean±s.e.m. with unpaired, two-sided t-tests (n=94 nuclei for DMSO; n=89 for dTAG7). d, PRO-seq signal of auto-regulatory TFs MYC and MYB, and the expression-matched control gene RAB3GAP1 after 1h MED14 degradation. Arrows highlight loss of promoter-proximal signal. H3K4me3 and H3K27ac ChIP-seq signal from KBM7 wild-type cells. e, Aggregated PRO-seq coverage over an SE-proximal metagene. TSS, transcription start site; TES, transcription end site. f, Changes in PRO-seq pausing index at n=7,643 genes after 1h MED14 degradation. g, Observed vs. expected median Euclidean distance of auto-regulatory TFs from the pause-initiation limit in Fig. 3f. The expected distribution was generated by randomly selecting the same number of genes from bulk. h, Changes in productive initiation rate and pause duration of all 6,791 genes vs. the 24 auto-regulatory TFs. Productive initiation rates selectively decrease for auto-regulatory TFs, while pause duration decreases globally. Box plot elements: medians with interquartile range and 1.5x whiskers.
Extended Data Fig. 6. Unbiased proteomics reveal increased P-TEFb levels on chromatin.
a, Overlap of three independent data analyses to detect high-confidence differentially chromatin-bound proteins (p < 0.1; see Supplementary Note). b,c, Differential chromatin binding of transcription regulators. Class averages are shown in b. Scratched boxes indicate missing values. GTFs: general transcription factors. d, Salt-based fractionation of 7SK- and chromatin-bound P-TEFb complexes. Unprocessed western blot shown in Source Data.
Extended Data Fig. 7. P-TEFb activation shapes the transcriptional response to Mediator loss.
a, PRO-seq read-through upon 2h MED14 degradation. Additionally inhibiting CDK9 with 500nM NVP2 in the last 30min reverses the read-through. Zoom-ins show 30kb windows around the polyadenylation site. Arrows highlight transcription start site (TSS) regions shown in g. b, Aggregated PRO-seq coverages show read-through even for long genes, where newly initiated Pol II has not yet reached the termination site. Mean±bootstrapped confidence region. c, Aggregated TT-seq coverages show read-through transcription after MED14 degradation also in HCT-116 cells. d,e, Changes in PRO-seq pausing index of all n=5,558 genes (d) and calculated pause duration at all n=6,954 transcription units (e) after MED14/CDK9 perturbation. f, Changes in productive initiation rates for all n=6,954 transcription units. Box plot elements: medians with interquartile range, 1.5x whiskers and confidence region notches. g, PRO-seq signal around transcription start sites (TSS) of two non-SE and one auto-regulatory TF gene. Paused polymerase does not re-accumulate at the MYB TSS upon combined MED14/CDK9 perturbation. h, TT-seq SE-gene set enrichment upon combined MED14/CDK9 perturbation. Less significant enrichment confirms that CDK9 activity aggravated the SE-selectivity of Mediator disruption.
Supplementary Material
Acknowledgments
We thank R. Fisher (Icahn School of Medicine at Mount Sinai, New York, USA) for sharing pT806 SPT5 antibody. We thank the Biomedical Sequencing Facility at CeMM and the MPIMG Sequencing core for assistance with next generation sequencing. We thank the imaging core facility of the Medical University of Vienna for assistance with microscopy. We thank P. Lenart for critical review of image quantification procedures. We thank A. Mayer and M. Erb for feedback on this manuscript. M.G.J. was supported by a Boehringer Ingelheim Fonds PhD fellowship. T.V. was supported by the International Max Planck Research School for Genome Science, part of the Göttingen Graduate Center for Neurosciences, Biophysics, and Molecular Biosciences. B.A. is supported by the Austrian Science Fund (FWF) and Medical University of Vienna’s joint PhD program in Inflammation and Immunity (FWF1212). C.B. is supported by a New Frontiers Group award of the Austrian Academy of Sciences and by an ERC Starting Grant (European Union’s Horizon 2020 research and innovation programme, grant agreement n° 679146). B.N. was supported by an American Cancer Society Postdoctoral Fellowship PF-17-010-01-CDD. B.N. and N.S.G were supported by the Katherine L. and Steven C. Pinard Research Fund. D.H. is supported by the SPP2202 Priority Program Grant (HN 4/1-1) from the Deutsche Forschungsgemeinschaft (DFG). This project was further supported by a FWF Stand-Alone grant (P31690-B) awarded to the Winter lab.
Footnotes
Author contributions
M.G.J. and G.E.W. conceptualized this project. M.G.J., T.V., A.H., M.B., B.A., and D.H. designed and conducted experiments. M.G.J., B.S., S.D.M., A.H., H.I., and M.B. analyzed and interpreted original and published omics data. M.G.J., M.B., B.A., and S.C. generated cell lines. M.G.J., B.S., S.D.M., and M.B. visualized data. B.N. and F.M.F. synthesized the dTAGV-1 reagent. A.M., A.B., J.E.B, N.S.G, C.B., D.H., P.C., and G.E.W. supervised the work. M.G.J. and G.E.W. wrote the manuscript with input from all authors.
Competing interests
G.E.W., J.E.B., and B.N. are inventors on patent applications related to the dTAG system (WO/2017/024318, WO/2017/024319, WO/2018/148443, WO/2018/148440). The dTAGV-1 molecule is the subject of a patent application filed by Dana-Farber Cancer Institute. N.S.G. is a Scientific Founder, member of the Scientific Advisory Board (SAB) and equity holder in C4 Therapeutics, Syros, Soltego, B2S, Gatekeeper and Petra Pharmaceuticals. The Gray lab receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Janssen, Kinogen, Voroni, Her2llc, Deerfield and Sanofi. J.E.B. is now an executive and shareholder of Novartis AG, and has been a founder and shareholder of SHAPE (acquired by Medivir), Acetylon (acquired by Celgene), Tensha (acquired by Roche), Syros, Regenacy, and C4 Therapeutics.
Data availability
Next generation sequencing data are available through NCBI Gene Expression Omnibus under accession code GSE139468. Chromatin proteomics data have been deposited at PRIDE under dataset identifier PXD017611. Source data for Figs. 1, 4 and Extended Data Figs. 1–4, 6 are presented with the paper.
Code availability
Custom code used to analyze the data in this study is available at https://github.com/GWinterLab/Jaeger_Mediator_NatureGenetics_2020.
References
- 1.Kelleher RJ, 3rd, Flanagan PM, Kornberg RD. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–15. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–9. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–75. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 4.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 5.Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci U S A. 1996;93:8329–33. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang YW, et al. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci U S A. 1998;95:8538–43. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–63. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–5. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Kim YJ, Lis JT. Interactions between subunits of Drosophila Mediator and activator proteins. Trends Biochem Sci. 2005;30:245–9. doi: 10.1016/j.tibs.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soutourina J. Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol. 2018;19:262–274. doi: 10.1038/nrm.2017.115. [DOI] [PubMed] [Google Scholar]
- 12.Jeronimo C, Robert F. The Mediator Complex: At the Nexus of RNA Polymerase II Transcription. Trends Cell Biol. 2017;27:765–783. doi: 10.1016/j.tcb.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Eychenne T, Werner M, Soutourina J. Toward understanding of the mechanisms of Mediator function in vivo: Focus on the preinitiation complex assembly. Transcription. 2017;8:328–342. doi: 10.1080/21541264.2017.1329000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–72. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–28. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 16.Petrenko N, Jin Y, Wong KH, Struhl K. Evidence that Mediator is essential for Pol II transcription, but is not a required component of the preinitiation complex in vivo. eLife. 2017;6:e28447. doi: 10.7554/eLife.28447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeronimo C, et al. Tail and Kinase Modules Differently Regulate Core Mediator Recruitment and Function In Vivo. Mol Cell. 2016;64:455–466. doi: 10.1016/j.molcel.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrenko N, Jin Y, Wong KH, Struhl K. Mediator Undergoes a Compositional Change during Transcriptional Activation. Mol Cell. 2016;64:443–454. doi: 10.1016/j.molcel.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–86. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 20.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–26. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 23.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–51. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabari BR, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho W-K, et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Shendure J. Mechanisms of Interplay between Transcription Factors and the 3D Genome. Molecular Cell. 2019;76:306–319. doi: 10.1016/j.molcel.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Khattabi L, et al. A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers. Cell. 2019;178:1145–1158 e20. doi: 10.1016/j.cell.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi H, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, et al. Mediator MED23 regulates basal transcription in vivo via an interaction with P-TEFb. Transcription. 2013;4:39–51. doi: 10.4161/trns.22874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahlberg O, Shilkova O, Tang M, Holmqvist PH, Mannervik M. P-TEFb, the super elongation complex and mediator regulate a subset of non-paused genes during early Drosophila embryo development. PLoS Genet. 2015;11:e1004971. doi: 10.1371/journal.pgen.1004971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conaway RC, Conaway JW. The Mediator complex and transcription elongation. Biochim Biophys Acta. 2013;1829:69–75. doi: 10.1016/j.bbagrm.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erb MA, et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature. 2017;543:270–274. doi: 10.1038/nature21688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nabet B, et al. The dTAG system for immediate and target-specific protein degradation. Nature Chemical Biology. 2018;14:431–441. doi: 10.1038/s41589-018-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cevher MA, et al. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat Struct Mol Biol. 2014;21:1028–34. doi: 10.1038/nsmb.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winter GE, et al. BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Mol Cell. 2017;67:5–18 e19. doi: 10.1016/j.molcel.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olson CM, et al. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol. 2018;14:163–170. doi: 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhar M, et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science. 2018;360:800–805. doi: 10.1126/science.aao2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plaschka C, et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015;518:376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- 42.Nozawa K, Schneider TR, Cramer P. Core Mediator structure at 3.4 A extends model of transcription initiation complex. Nature. 2017;545:248–251. doi: 10.1038/nature22328. [DOI] [PubMed] [Google Scholar]
- 43.Tsai KL, et al. Mediator structure and rearrangements required for holoenzyme formation. Nature. 2017;544:196–201. doi: 10.1038/nature21393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–67. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwalb B, et al. TT-seq maps the human transient transcriptome. Science. 2016;352:1225–8. doi: 10.1126/science.aad9841. [DOI] [PubMed] [Google Scholar]
- 46.Saint-Andre V, et al. Models of human core transcriptional regulatory circuitries. Genome Res. 2016;26:385–96. doi: 10.1101/gr.197590.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nabet B, et al. Rapid and direct control of target protein levels with VHL-recruiting dTAG molecules. 2020 doi: 10.1101/2020.03.13.980946v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mumbach MR, et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13:919–922. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weintraub AS, et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell. 2017;171:1573–1588 e28. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 51.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise Maps of RNA Polymerase Reveal How Promoters Direct Initiation and Pausing. Science (New York, NY) 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wissink EM, Vihervaara A, Tippens ND, Lis JT. Nascent RNA analyses: tracking transcription and its regulation. Nature Reviews Genetics. 2019 doi: 10.1038/s41576-019-0159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cramer P. Organization and regulation of gene transcription. Nature. 2019;573:45–54. doi: 10.1038/s41586-019-1517-4. [DOI] [PubMed] [Google Scholar]
- 54.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo YE, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572:543–548. doi: 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gressel S, et al. CDK9-dependent RNA polymerase II pausing controls transcription initiation. Elife. 2017;6 doi: 10.7554/eLife.29736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shao W, Zeitlinger J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat Genet. 2017;49:1045–1051. doi: 10.1038/ng.3867. [DOI] [PubMed] [Google Scholar]
- 58.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ehrensberger Andreas H, Kelly Gavin P, Svejstrup Jesper Q. Mechanistic Interpretation of Promoter-Proximal Peaks and RNAPII Density Maps. Cell. 2013;154:713–715. doi: 10.1016/j.cell.2013.07.032. [DOI] [PubMed] [Google Scholar]
- 60.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–43. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanso M, et al. P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev. 2016;30:117–31. doi: 10.1101/gad.269589.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parua PK, et al. A Cdk9-PP1 switch regulates the elongation-termination transition of RNA polymerase II. Nature. 2018;558:460–464. doi: 10.1038/s41586-018-0214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bradner JE, Hnisz D, Young RA. Transcriptional Addiction in Cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krebs AR, et al. Genome-wide Single-Molecule Footprinting Reveals High RNA Polymerase II Turnover at Paused Promoters. Mol Cell. 2017;67:411–422 e4. doi: 10.1016/j.molcel.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y, Liu M, Chen LF, Chen R. P-TEFb: Finding its ways to release promoter-proximally paused RNA polymerase II. Transcription. 2018;9:88–94. doi: 10.1080/21541264.2017.1281864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mir M, Bickmore W, Furlong EEM, Narlikar G. Chromatin topology, condensates and gene regulation: shifting paradigms or just a phase? Development. 2019;146 doi: 10.1242/dev.182766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chong S, et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361:eaar2555. doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roeder RG. 50+ years of eukaryotic transcription: an expanding universe of factors and mechanisms. Nature Structural & Molecular Biology. 2019;26:783–791. doi: 10.1038/s41594-019-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sakuma T, Nakade S, Sakane Y, Suzuki K-IT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protocols. 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 71.Brand M, Winter GE. Locus-Specific Knock-In of a Degradable Tag for Target Validation Studies. Methods Mol Biol. 2019;1953:105–119. doi: 10.1007/978-1-4939-9145-7_7. [DOI] [PubMed] [Google Scholar]
- 72.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pedregosa F, et al. Scikit-learn: Machine learning in Python. Journal of machine learning research. 2011;12:2825–2830. [Google Scholar]
- 75.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 76.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 77.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui Y, et al. BioCircos.js: an interactive Circos JavaScript library for biological data visualization on web applications. Bioinformatics. 2016;32:1740–2. doi: 10.1093/bioinformatics/btw041. [DOI] [PubMed] [Google Scholar]
- 79.Aranda-Orgilles B, et al. MED12 Regulates HSC-Specific Enhancers Independently of Mediator Kinase Activity to Control Hematopoiesis. Cell Stem Cell. 2016;19:784–799. doi: 10.1016/j.stem.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Virtanen P, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nature Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Waskom M, et al. mwaskom/seaborn: v090 (July 2018) Zenodo; 2018. [DOI] [Google Scholar]
- 84.Hunter JD. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering. 2007;9:90–95. [Google Scholar]
- 85.McNamara RP, et al. KAP1 Recruitment of the 7SK snRNP Complex to Promoters Enables Transcription Elongation by RNA Polymerase II. Mol Cell. 2016;61:39–53. doi: 10.1016/j.molcel.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next generation sequencing data are available through NCBI Gene Expression Omnibus under accession code GSE139468. Chromatin proteomics data have been deposited at PRIDE under dataset identifier PXD017611. Source data for Figs. 1, 4 and Extended Data Figs. 1–4, 6 are presented with the paper.
Custom code used to analyze the data in this study is available at https://github.com/GWinterLab/Jaeger_Mediator_NatureGenetics_2020.