Abstract
Mitochondria play vital role in regulating the cellular energetics and metabolism. Further, it is a signaling hub for cell survival and apoptotic pathways. One of the key determinants that calibrate both cellular energetics and survival functions is mitochondrial calcium (Ca2+) dynamics. Mitochondrial Ca2+ regulates three Ca2+-sensitive dehydrogenase enzymes involved in tricarboxylic acid cycle (TCA) cycle thereby directly controlling ATP synthesis. On the other hand, excessive Ca2+ concentration within the mitochondrial matrix elevates mitochondrial reactive oxygen species (mROS) levels and causes mitochondrial membrane depolarization. This leads to opening of the mitochondrial permeability transition pore (mPTP) and release of cytochrome c into cytosol eventually triggering apoptosis. Therefore, it is critical for cell to maintain mitochondrial Ca2+ concentration. Since cells can neither synthesize nor metabolize Ca2+, it is the dynamic interplay of Ca2+ handling proteins involved in mitochondrial Ca2+ influx and efflux that take the center stage. In this review we would discuss the key molecular machinery regulating mitochondrial Ca2+ concentration. We would focus on the channel complex involved in bringing Ca2+ into mitochondrial matrix i.e. Mitochondrial Ca2+ Uniporter (MCU) and its key regulators Mitochondrial Ca2+ Uptake proteins (MICU1, 2 and 3), MCU regulatory subunit b (MCUb), Essential MCU Regulator (EMRE) and Mitochondrial Ca2+ Uniporter Regulator 1 (MCUR1). Further, we would deliberate on major mitochondrial Ca2+ efflux proteins i.e. Mitochondrial Na+/Ca2+/Li+ exchanger (NCLX) and Leucine zipper EF hand-containing transmembrane1 (Letm1). Moreover, we would highlight the physiological functions of these proteins and discuss their relevance in human pathophysiology. Finally, we would highlight key outstanding questions in the field.
Keywords: Mitochondrial calcium dynamics, MCU complex, MICU 1/2/3, MCUR1, EMRE, Letm1, NCLX
1. Introduction
Mitochondrial calcium (Ca2+) homeostasis regulates aerobic metabolism, cell survival, cellular Ca2+ signaling and mitochondrial reactive oxygen species (mROS) generation thereby driving several physiological and pathophysiological functions of the cell (De Stefani et al., 2016; Rizzuto et al., 2012). Mitochondria calibrate various cellular processes controlled by intracellular Ca2+ by buffering cytosolic Ca2+ and thereby maintaining cytosolic Ca2+ concentrations in range of 100 nM (Babcock et al., 1997; Carvalho et al., 2020). Within mitochondria, Ca2+ regulates three Ca2+-sensitive dehydrogenase enzymes (pyruvate dehydrogenase, iso-citrate dehydrogenase and α-ketoglutarate dehydrogenase) involved in tricarboxylic acid cycle (TCA) (Hajnoczky et al., 1995; Hansford, 1994; McCormack and Denton, 1979; McCormack et al., 1990). Therefore, mitochondrial Ca2+ directly regulates mitochondrial metabolism and controls adenosine triphosphate (ATP) generation (Hansford, 1994; McCormack and Denton, 1979; McCormack et al., 1990). However, sustained elevated levels of mitochondrial Ca2+ leads to elevated mROS, opening of mitochondrial permeability transition pore (mPTP) and release of cytochrome c (Boehning et al., 2003; Mammucari et al., 2018; Tanwar and Motiani, 2018). This leads to activation of pro-apoptotic factors eventually causing cell death (Rasola and Bernardi, 2011; Tanwar and Motiani, 2018). Mitochondrial Ca2+ further regulates several cell death pathways important for apoptosis as well as necrosis (La Rovere et al., 2016; Lemasters et al., 1998; Nakagawa et al., 2005; Rimessi et al., 2008). Deluca et al. reported mitochondrial Ca2+ uptake in isolated energized mitochondria (Deluca and Engstrom, 1961; Vasington and Murphy, 1962). This uptake was found to be dependent on mitochondrial membrane potential (Δψ ~ -180 mV) and was inhibited by the drug ruthenium red or its analog ruthenium 360 (Deluca and Engstrom, 1961; Ying et al., 1991). High electrochemical gradient of around —180 mV is responsible for Ca2+ entry into the mitochondria via a selective Ca2+ uniporter (Bernardi, 1999). Although there is high electrochemical gradient inside mitochondrial matrix for Ca2+, mitochondria have very low affinity for Ca2+ uptake. Mitochondrial Ca2+ uptake occurs at highly specialized regions of close contacts between mitochondria and endoplasmic reticulum (ER) called mitochondria associated membranes (MAMs) (Csordas et al., 2006; Rizzuto et al., 1998; Sharma et al., 2020; Vecellio Reane et al., 2020). ER resident Ca2+ release channel inositol-1,4,5-trisphosphate receptor (IP3R) releases Ca2+ in the MAMs region thereby elevating Ca2+ concentration (in μM range) in these nanodomains, which in turn activates mitochondrial Ca2+ uptake (Bartok et al., 2019; Rizzuto et al., 1993, 1998). Later on, the tools to measure free Ca2+ concentration of mitochondria were developed. Mitochondrial Ca2+ concentration ([Ca2+]m) was selectively measured with mitochondria targeting Ca2+-sensitive photoprotein aequorin (Rizzuto et al., 1992). Rizzuto et al. further reported rapid and transient increase of [Ca2+]m via agonist-stimulated elevations of cytosolic free Ca2+ concentrations (Rizzuto et al., 1992). In order to perform cellular functions a balanced regulation of mitochondrial Ca2+ influx and efflux is required, which is achieved through mitochondrial Ca2+ uptake and efflux players. Last few years have witnessed significant progress in the mitochondrial Ca2+ signaling field with the discovery of the several molecular components of the Mitochondrial Ca2+ Uniporter (MCU) complex that mediates mitochondrial Ca2+ uptake (Foskett and Philipson, 2015; Kamer and Mootha, 2015; Nemani et al., 2018; Pallafacchina et al., 2018; Pathak and Trebak, 2018). Moreover, the key players involved in mitochondrial Ca2+ efflux were also characterized recently (Giorgi et al., 2018; Kostic and Sekler, 2019; Lin and Stathopulos, 2019; Nita et al., 2015). In this review we will focus on critical components of mitochondrial Ca2+ influx and efflux 1. e. MCU complex and Mitochondrial Na+/Ca2+/Li+ exchanger (NCLX) & Leucine zipper EF hand-containing transmembrane1 (Letm1). We will discuss their activation mechanisms, associated regulatory components and their pathophysiological relevance.
2. Mitochondrial calcium (Ca2+) transport
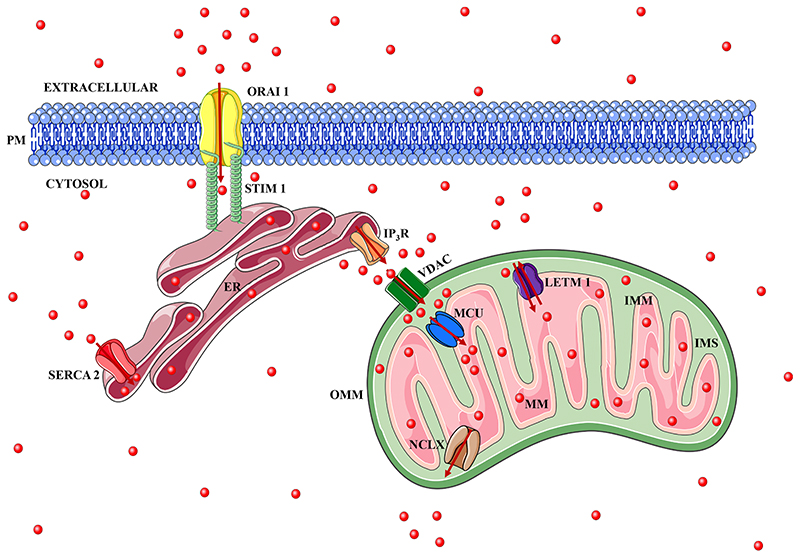
Ca2+ crosses the outer and inner mitochondrial membrane (IMM) in order to finally reach mitochondrial matrix. The outer mitochondrial membrane (OMM) is permeable to metabolites smaller than 5 kDa due to expression of Voltage-dependent anion channels (VDACs) that form pores in OMM (Madesh and Hajnoczky, 2001; Mertins et al., 2014; Rapizzi et al., 2002). VDACs allow Ca2+ to easily diffuse across the OMM. VDAC1 along with its two other isoforms VDAC2 and VDAC3 transport Ca2+ from cytosol to inter-membrane space (IMS) (Shoshan-Barmatz and Gincel, 2003). Although these three isoforms of VDACs are ubiquitously expressed, their ratio and sub-mitochondrial distribution varies among diverse tissues (Messina et al., 2012; Shoshan-Barmatz and Mizrachi, 2012). VDACs can mediate Ca2+ influx in both structurally open and closed conformations (Colombini, 2012; Shoshan-Barmatz et al., 2018; Tan and Colombini, 2007). Interestingly, VDAC in closed conformation shows increased selectivity and efficacy for Ca2+ transport (Colombini, 2012; Shoshan-Barmatz et al., 2018; Tan and Colombini, 2007). Rappizi et al. reported that the overexpression of VDAC in skeletal myotubes and HeLa cells leads to enhanced Ca2+ influx into mitochondria by virtue of enhanced mitochondrial Ca2+ uptake at MAMs (Rapizzi et al., 2002). In an exciting study, Rizzuto’s group demonstrated physical interaction of VDAC1 with the endoplasmic reticulum Ca2+-release channel inositol 1,4,5-trisphosphate receptors (IP3Rs) via a molecular chaperone glucose-regulated protein 75 (grp75) (Szabadkai et al., 2006). Unlike OMM, the IMM ion permeability is extremely stringent. Ca2+ uptake into the mitochondrial matrix is mediated via a highly Ca2+ selective channel called as Mitochondrial Ca2+ Uniporter (MCU) (Baughman et al., 2011; De Stefani et al., 2011). On the other hand, mitochondrial Ca2+ extrusion is mediated by a Na+/Ca2+ exchanger-like protein termed as Na+/Ca2+/Li+ exchanger (NCLX) (Palty et al., 2010) and Letm1 (2H+/Ca2+ exchanger) (Jiang et al., 2009). Mitochondrial Ca2+ extrusion can occur via two processes: either in a Na+ dependent manner or via a Na+ independent mechanism (Puskin et al., 1976). The concentration of Na+ in the mitochondrial matrix is lower than that in cytosol. This is due to functioning of Na+-H+ exchanger which is driven by pH gradient, that successfully transports Na+ out of the mitochondrial matrix (Murphy and Eisner, 2009). This Na+ gradient, in turn, is responsible for the activity of NCLX that extrudes Ca2+ from the mitochondrial matrix (Palty et al., 2010). The thermodynamic analysis of NCLX give rise to two school of thoughts, some investigators suggest that NCLX has 2:1 stoichiometry (Brand, 1985) showing an electro neutral nature of NCLX (2Na+/1 Ca2+) whereas according to others it is 3:1 (Baysal et al., 1994) exchanging 3Na+ ions for 1 Ca2+ ion, meaning it is electrogenic in nature. Further, NCLX can function in reverse mode and thereby it can transport Ca2+ into the mitochondrial matrix when the membrane potential of mitochondria dissipates (Boyman et al., 2013). For further details on NCLX thermodynamics and reverse mode functioning please refer to the detailed review focused exclusively on NCLX (Boyman et al., 2013). The NCLX and Letm1 are responsible for Ca2+ efflux from matrix to IMS and eventually Ca2+ is extruded to the cytosol via VDACs or other poorly characterized mechanisms (Giorgi et al., 2018). In the following sections, we will discuss the key molecular machinery regulating mitochondrial Ca2+ influx and efflux (please refer Fig. 1 for the diagrammatic illustration of intracellular Ca2+ handling machinery including molecular players involved in mitochondrial Ca2+ dynamics).
Fig. 1.
Intracellular Ca2+ handling machinery. Ligand binding to G protein-coupled receptors (GPCR) leads to phospholipase C (PLC) activation that further produces second messenger inositol-1,4,5 trisphosphate (IP3) which binds to its receptor, the IP3R, resulting in release of Ca2+ from ER to the cytosol. This depletion of ER Ca2+ stores activate canonical store-operated Ca2+ entry (SOCE) via plasma membrane localized Orai channels thus resulting in refilling of ER Ca2+ stores by sarcoplasmic/endoplasmic reticulum ATPase (SERCA) pumps or Ca2+ can be taken up by other organelles such as mitochondria. In Ca2+ enriched mitochondrial associated membranes (MAMs), Ca2+ released from IP3R crosses the OMM through VDACs and enters the intermembrane space (IMS), which is finally transported to mitochondrial matrix via MCU complex. While mitochondrial matrix Ca2+ is extruded via the Na+/Ca2+ exchange (NCLX) and Letm1.
3. MCU
The Mitochondrial Ca2+ Uniporter (MCU) is well conserved in metazoan and plants but absent in yeasts, few protozoan and fungal lineages (Bick et al., 2012; Cheng and Perocchi, 2015). Researchers have been extensively studying the Uniporter’s activity/biophysical properties for over 50 years even before the discovery of its molecular identity. In 2004, Kirichok et al. measured single-channel Ca2+ currents in mitoplasts isolated from COS-7 cells and confirmed existence of an inwardly rectifying, highly Ca2+ selective channel in the IMM (Kirichok et al., 2004). In 2010, using comparative physiology, evolutionary genomics, organelle proteomics and RNA interference studies first molecular component of the Uniporter complex named Mitochondrial Calcium Uptake 1 (MICU1) was discovered as a Ca2+ sensing regulator of the Uniporter (Perocchi et al., 2010). In 2011, the studies from two independent groups using different experimental approaches identified MCU (earlier known as CCDC109A) as a pore-forming subunit of mitochondrial Ca2+ uptake (Baughman et al., 2011; De Stefani et al., 2011). Baughman et al. utilized whole-genome phylogenetic profiling, genomewide RNA co-expression analysis and organelle-wide protein co‐expression analysis for discovering MCU (Baughman et al., 2011). They further reported that MCU forms oligomers in the mitochondrial inner membrane, and physically interacts with previously identified regulator MICU1 (Baughman et al., 2011). Silencing of MCU in cultured cells or in vivo in mouse liver led to decreased mitochondrial Ca2+ uptake without affecting mitochondrial respiration and membrane potential (Baughman et al., 2011). MCU has two coiled-coil (CC) domains and two transmembrane (TM) domains linked by a highly conserved acidic residue rich short loop facing IMS (Baughman et al., 2011). Both N- and C- termini of MCU are located in the mitochondrial matrix (Baughman et al., 2011; Martell et al., 2012). The linker sequence contains DIME motif, which acts as selectivity filter of MCU. The conserved negatively charged amino acids near and within the DIME motif are critical for MCU-mediated Ca2+ uptake (Baughman et al., 2011). Please refer Fig. 2 for diagrammatic illustration of domain architecture of MCU and other players involved in mitochondrial Ca2+ dynamics. Baughman et al further demonstrated that mutations with alanine at these conserved residues (E257A, S259A, D261A, E264A) render MCU non‐functional except S259A mutant, which confers resistance to most effective uniporter inhibitor Ru360 (Baughman et al., 2011). De Stefani et al. utilized in silico analysis to uncover the MCU, which was ubiquitously expressed along with MICU1 and is localized on the IMM (De Stefani et al., 2011). In HeLa cells siRNA mediated silencing of MCU led to substantial decrease in mitochondrial Ca2+ uptake while overexpression of MCU increased the mitochondrial Ca2+ uptake evoked upon histamine stimulation (De Stefani et al., 2011). Further, the authors purified MCU protein and recapitulated previously known electrophysiological properties as well as inhibitor sensitivity of the uniporter in planar lipid bilayers (De Stefani et al., 2011). In HeLa cells, the overexpression of a mutant MCU lacking two negatively charged residues (MCUD260Q,E263Q) of the putative pore-forming region exhibited no channel activity and decreased agonist-dependent mitochondrial Ca2+ transients (De Stefani et al., 2011). Further, the MICU1 over‐expression failed to rescue the decrease in the mitochondrial Ca2+ levels in MCU-silenced cells (De Stefani et al., 2011). Later on Chaudhuri et al utilized whole-mitoplast voltage-clamping technique and reported that RNAi-mediated knockdown of the MCU results in decreased mitochondrial Ca2+ current (IMiCa), whereas its overexpression leads to increase in IMiCa (Chaudhuri et al., 2013). Further, the authors demonstrated inhibition of MCU current by ruthenium red, which was abolished by a point mutation in the putative pore domain of MCU (S259A) thus establishing MCU as the pore-forming subunit of the uniporter complex (Chaudhuri et al., 2013).
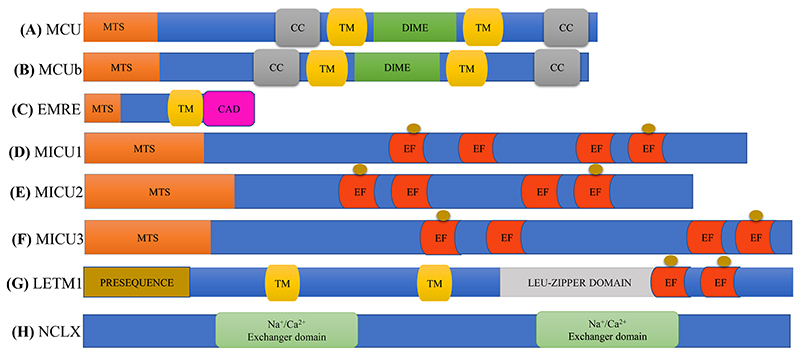
Fig. 2. Domain architecture of key molecular players involved in regulating mitochondrial Ca2+ dynamics.
(A-F): Linear overview of MCU, MCUb, EMRE and MICU1/2/3 showing the predicted domain structural components for each protein (Kamer and Mootha, 2015). (G): Linear overview of LETM1 structure, showing the specific sites and domains (Li et al., 2019). (H): Cartoon representing predicted domain architecture of NCLX protein (He and O’Halloran, 2014). Here, MTS stands for Mitochondrial targeting signal; CC stands for Coiled-coil domain; TM stands for transmembrane domain; DIME means conserved DIME motif; CAD stands for Carboxy-terminal acidic domain; EF means EF-hand domain; brown circles mean Ca2+ binding sites.
Subsequently, multiple groups have revealed X-ray and cryo-EM structure of MCU fungal orthologs (Baradaran et al., 2018; Fan et al., 2018; Nguyen et al., 2018; Yoo et al., 2018). More recently the structure of human MCU-EMRE subcomplex was revealed (Wang et al., 2019). For identifying pore architecture of the MCU, Oxenoid et al employed NMR and negative-stain EM techniques and suggested that MCU from Cae-norhabditis elegans forms a pentameric assembly (Oxenoid et al., 2016), which is still debatable. Subsequent study from the same group utilized paramagnetic nuclear magnetic resonance technique for demonstrating that the conserved DXXE signature sequence (Asp-X-X-Glu) of MCU forms pore gate and selectivity filter in Caenorhabditis elegans (Cao et al., 2017). They further reported that Ru360 directly blocks the selectivity filter by binding to Asp240 but not to Glu243 (Cao et al., 2017). Later on, Yoo et al. revealed high resolution cryo-EM structure (3.7 Å resolution) of full length MCU from Neurospora crassa (Yoo et al., 2018). The authors further demonstrated channel formation by four protomers (Yoo et al., 2018). A recent study by Fan et al. reported cryo‐EM structure of human MCU holocomplex (comprising MCU, EMRE, MICU1 and MICU2 subunits) in low and high Ca2+ conditions (Fan et al., 2020). Further, the authors reported well-resolved amino (N)-terminal domain (NTD) of MCU in uniplex dimer and demonstrated two main particle species-uniplex monomer and a V-shaped dimer in low Ca2+ while predominantly dimers in high Ca2+ concentrations (Fan et al., 2020). More recently, Zhuo et al. demonstrated intact structure of human MCU supercomplex consisting MCU-EMRE-MICU1-MICU2 (Zhuo et al., 2020). The authors reported MCU supercomplex as a 20- subunit-O shaped dimer of hetero-decamers along with MICU1 and MICU2 subunits. Furthermore, they demonstrated that EMRE plays an important role in the coupling of Ca2+ sensing MICUs with MCU (Zhuo et al., 2020). Similarly, Wang et al. reported cryo-EM structure of MCU-EMRE-MICU1-MICU2 holocomplex under resting Ca2+ conditions (Wang et al., 2020a). Another recent report by Wang et al. reported cryoEM structures of the human MCU holocomplex in the apo and Cambound states (Wang et al., 2020b). Collectively, these studies highlight a tetrameric configuration of MCU.
Using inflammatory and hypoxia models, Dong et al. recently reported that the conserved cysteine97 residue in the N-terminus of human MCU undergoes S-glutathionylation (Dong et al., 2017). The authors utilized biochemical assays and super-resolution imaging for demonstrating that the oxidation of this cysteine promotes oligomerization of MCU. This MCU oligomerization leads to an increase in the MCU channel activity resulting in enhanced mROS, mitochondrial Ca2+ overload, and cell death (Dong et al., 2017). Interestingly, Madesh’s group demonstrated that cytosolic Ca2+ concentration regulates MCU expression (Shanmughapriya et al., 2015). Shanmughapriya et al. demonstrated that MCU expression is dependent on Ca2+ signals generated by IP3 and SOCE signaling. The authors reported decrease in mitochondrial Ca2+ uptake and MCU expression in cells lacking either IP3R or STIM1/Orai1 (Shanmughapriya et al., 2015). In these cells, MCU was controlled by the Ca2+-dependent transcription factor, cyclic AMP response element-binding protein (CREB). Finally, it was revealed that CREB directly interacts with the MCU promoter to stimulate its expression (Shanmughapriya et al., 2015). Therefore, on one hand efficient MCU functioning helps in maintaining cellular Ca2+ levels and on the other hand cytosolic Ca2+ dynamics regulate MCU expression and activity. This clearly demonstrates an intricate crosstalk between cellular and mitochondrial Ca2+ signaling pathways. Recently, a study by Ghosh et al. revealed an important role of mitochondrial phospholipid, cardiolipin (CL) in MCU stability and activity in yeast model system (Ghosh et al., 2020). The authors heterologously expressed human MCU in the wild type yeast (lacks MCU) and in different yeast mutants defective in biosynthesis of phosphatidylethanolamine, phosphatidylcholine, or cardiolipin (CL) (Ghosh et al., 2020). They further showed that loss of CL specifically leads to a decrease in the MCU expression and Ca2+ uptake (Ghosh et al., 2020). Furthermore, they validated the physiological significance of this cardiolipin mediated stability and activity of MCU in pathophysiology of Barth syndrome (Ghosh et al., 2020). Please refer the “Pathophysiological relevance of MCU complex” section for further details on the role of MCU complex in human pathophysiology.
4. MCUb
The MCUb is a paralog of MCU and is characterized as dominant negative subunit of uniporter complex (Raffaello et al., 2013). MCUb forms homo as well as hetero-oligomers with MCU and acts as a negative regulator of MCU activity (Raffaello et al., 2013). Although MCUb does not form a Ca2+ permeable channel in planar lipid bilayers, co‐expression of MCUb with MCU leads to reduction in channel activity (Raffaello et al., 2013). It also contains two transmembrane domains just like MCU and shares 50% sequence similarity with MCU. Using RT-PCR analysis in HeLa cells and mouse tissues, the authors revealed lower as well as differential expression profile of MCUb in comparison to MCU (Raffaello et al., 2013). Recently, Lambert et al. revealed altered MCU stoichiometry and function in MCUb -/- HeLa cell lines (Lambert et al., 2019). The authors reported displacement of MCU from the functional MCU complex upon MCUb overexpression. This displacement decreased Ca2+ uptake by mitochondria (Lambert et al., 2019). Further during cardiac injury, MCUb incorporation into the MCU complex reduced mitochondrial Ca2+ overload (Lambert et al., 2019). Importantly, MCUb overexpression led to a decrease in myocardial infarct size post ischemia injury (Lambert et al., 2019). Moreover, the authors reported impaired mitochondrial energetics and contractile function upon MCUb incorporation (Lambert et al., 2019). Certainly, more studies from several independent groups are required to better understand physiological relevance of MCUb.
5. Mitochondrial Ca2+ uptake proteins (MICU1, MICU2 and MICU3)
The MICU proteins are known for their essential regulatory function on MCU activity. They are localized in IMS, contain Ca2+ binding EF hands and physically interact with MCU (Perocchi et al., 2010; Plovanich et al., 2013). Interestingly, MICU1 was discovered as a critical regulator of mitochondrial Ca2+ uptake well before the molecular identity of MCU (Perocchi et al., 2010). Mootha’s group utilized Mito-Carta inventory (compendium of the mammalian mitochondrial proteins) (Pagliarini et al., 2008) and integrative genomics approaches for the identification of MICU1 (Perocchi et al., 2010). The authors scanned MitoCarta for homologues of evolutionary conserved IMM proteins present in vertebrates, kinetoplastids (having evolutionary conserved MCU activity) (Carafoli and Lehninger, 1971; Docampo and Vercesi, 1989; Vercesi and Docampo, 1992) and absent in yeast S. cerevisiae (yeast doesn’t have MCU activity) (Balcavage et al., 1973; Carafoli and Lehninger, 1971; Uribe et al., 1992). This search was followed by RNAi of shortlisted genes to finally reach at MICU1 (Perocchi et al., 2010). Perocchi et al. reported that the silencing of MICU1 in the intact as well as permeabilized cells leads to reduction in mitochondrial Ca2+ entry but has no effect on mitochondrial respiration or membrane potential. The authors further demonstrated rescue of mitochondrial Ca2+ uptake in the sh-MICU1 knockdown cells with hairpin-insensitive MCU cDNA (Perocchi et al., 2010). Interestingly, by employing protein flux analysis Madesh’s group reported that MICU1 is localized to mitochondrial matrix and MICU1 N-terminal polybasic domain is essential for its interaction with CC domains of MCU (Hoffman et al., 2013). The work by the same group suggested that MICU1 is a gatekeeper of the MCU mediated Ca2+ uptake under basal conditions and knockdown of MICU1 in HeLa cells leads to basal accumulation of the mitochondrial Ca2+ thus generating excessive ROS (Mallilankaraman et al., 2012b). Later Csordàs et al. also proposed that MICU1 acts as a MCU gatekeeper by sensing cytosolic Ca2+ concentrations (Csordas et al., 2013). They also reported that MICU1 controls the threshold of the mitochondrial Ca2+ uptake (Csordas et al., 2013). Further, the authors showed mitochondrial Ca2+ accumulation upon MICU1 loss in mouse liver and cultured cells (Csordas et al., 2013). These studies collectively conclude that MCU is kept closed by MICUs at resting or low cytosolic Ca2+ concentrations thus preventing Ca2+ entry into mitochondrial matrix and an increase in the Ca2+ concentration is sensed by EF hands of MICUs eventually leading to opening of the MCU (Csordas et al., 2013; Hoffman et al., 2013; Mallilankaraman et al., 2012b). Similarly, overexpression of MICU1 in HeLa cells was shown to increase mitochondrial Ca2+ uptake upon histamine stimulation (Patron et al., 2014). Later on, the bioinformatics analysis led to discovery of MICU2 and MICU3 as two paralogs of MICU1 in humans (Plovanich et al., 2013). The authors demonstrated that the MCU, MICU1 and MICU2 reside within the same complex and stabilize protein expression of each other in HeLa and HEK293T cell lines (Plovanich et al., 2013). The RNAi mediated silencing of MICU1, MICU2 or both in mouse liver resulted in additive impairment of the mitochondrial Ca2+ signaling without affecting mitochondrial membrane potential and respiration (Plovanich et al., 2013). Both MICU2 and MICU3 have EF hand domains as well as have N-terminal mitochondrial targeting sequence (Plovanich et al., 2013). In an elegant study, using purified lipid bilayers and intact cells, it was reported that the MICU1-MICU2 heterodimers regulate activity of MCU (Patron et al., 2014). Patron et al. showed that the MCU is activated by MICU1 at higher Ca2+ concentrations and is inhibited by MICU2 at lower Ca2+ concentrations thus proposing MICU2 as a genuine negative regulator of MCU (Patron et al., 2014) (Please refer Fig. 3 for the diagrammatic illustration of Ca2+ mediated MCU activation). Recently, the same group reported differential expression of MICUs in diverse tissues; MICU3 is highly expressed in neuronal tissues and skeletal muscle while MICU2 in visceral organs (Patron et al., 2019). MICU3 physically interacts with MICU1 but not with MICU2 whereas MICU1 can form heterodimer with both MICU2 and MICU3 (Patron et al., 2019). The authors showed that MICU3 silencing results in impaired Ca2+ signaling in primary cortical neurons and therefore specifically regulating neuronal function (Patron et al., 2019). Interestingly, Raffaello and Rizzuto groups identified an alternative splice variant of MICU1 (MICU1.1) in skeletal muscle, which binds Ca2+ more efficiently than MICU1 (Vecellio Reane et al., 2016). The authors further reported that heterodimers of MICU1.1-MICU2 were able to activate MCU at lower Ca2+ concentrations as compared to MICU1-MICU2 heterodimers (Vecellio Reane et al., 2016). Recently, Mootha’s group reported stabilization of both MICU1 and MICU2 by Ca2+ and demonstrated higher binding affinity of MICU1 to Ca2+ as compared to MICU2 (Kamer et al., 2017). They further reported formation of MICU1-MICU2 heterodimers and their binding to Ca2+ with higher affinity in vitro (Kamer et al., 2017). In an exciting study Petrungaro et al. demonstrated that Mia40 (a mitochondrial chaperone) facilitates disulfide covalent bond that links MICU1-MICU2 Fheterodimers (Petrungaro et al., 2015). The presence of disulfide bond in the MICU1-MICU2 heterodimer enables its binding with MCU at low Ca2+ concentrations and dissociation at higher Ca2+ concentrations thereby regulating gatekeeping activity of the MICU1-MICU2 heterodimer (Petrungaro et al., 2015). In the absence of this bond, the receptor induced mitochondrial Ca2+ uptake was significantly increased (Petrungaro et al., 2015). Recently, Hajnóczky’s group reported that MICU1 regulates MCU inhibition by ruthenium red/Ru360 (Paillard et al., 2018). They identified a putative DIME interacting domain (DID) in MICU1, which is essential for the gatekeeping and cooperative activation of MCU (Paillard et al., 2018). The authors proposed that interaction of MICU1 with D ring of MCU pore is important for controlling the uniporter activity (Paillard et al., 2018). More recently, Madesh’s group demonstrated that MICU1 expression is increased upon inhibition of glycolysis, mitochondrial pyruvate transport and mitochondrial fatty acid transport (Nemani et al., 2020). Nemani et al. further reported that knockdown of mitochondrial pyruvate carrier (MPC) results in enhanced MICU1 expression via transcription factor early growth response 1 (EGR1). It implicates that mitochondrial pyruvate and fatty acid flux regulates MICU1-dependent activity of MCU (Nemani et al., 2020). Certainly, more studies are required for understanding the molecular mechanisms regulating expression and activity of MICUs.
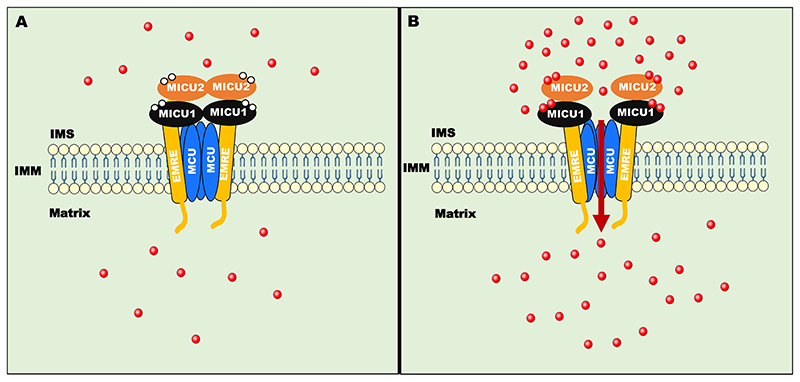
Fig. 3. MCU complex composition and its regulation by Ca2+.
A) The Uniporter complex consists of pore-forming subunit-MCU along with regulatory proteins-MICU1/2/3 and EMRE. At low mitochondrial intermembrane space (IMS) Ca2+ concentration, MICU1-MICU2 heterodimer inhibits MCU activity by virtue of inhibitory effect of MICU2. B) At high IMS Ca2+ concentration, Ca2+-dependent conformational change in MICU1-MICU2 dimer releases MICU2 dependent inhibition and MICU1 act as a positive regulator of MCU channel activity to cause Ca2+ uptake into the mitochondrial matrix. EMRE tether and stabilize MICU1/MICU2 to the MCU complex.
Importantly, the MICU1(-/-) mice showed ataxia and muscle weakness phenotype (Liu et al., 2016). Further, the MICU1(-/-) mice has increased resting mitochondrial Ca2+ levels, altered mitochondrial morphology, and reduced ATP thereby validating MICU1 as a molecular gatekeeper that prevents mitochondrial Ca2+ overload (Liu et al., 2016). Additionally, using MICU1(-/-) mice Hajnóczky’s group demonstrated critical role of MICU1 in adaptation to postnatal life and for tissue repair after injury (Antony et al., 2016). They reported failure of liver regeneration, impairment of cell cycle and necrosis upon MICU1 loss. In MICU1-deficient hepatocytes, the authors observed accelerated mPTP opening due to Ca2+ overload thus suggesting a role for MICU1 in liver regeneration (Antony et al., 2016). The individuals with MICU1 mutations show proximal myopathy, learning difficulties, elevated creatine kinase (CK) level, lethargy, fatigue, nystagmus, cataracts and a progressive extrapyramidal movement disorder (Lewis-Smith et al., 2016; Logan et al., 2014; Musa et al., 2019). The clinical MICU1 mutations result in increased agonist-induced mitochondrial Ca2+ uptake at low cytosolic Ca2+ concentrations and reduced cytosolic transients in fibroblasts (Logan et al., 2014). The loss of function mutations in MICU1 had no effect on mitochondrial membrane potential but caused mitochondrial fragmentation (Logan et al., 2014). A recent study reported impaired mitochondrial Ca2+ signaling upon loss of MICU1 in skeletal muscle of mice and patient derived fibroblast (homozygous MICU1 exon 1 deletion) (Debattisti et al., 2019). Taken together, these in vivo mice and human mutation studies clearly demonstrate the merit of targeting MICUs for calibrating mitochondrial Ca2+ signaling. A critical step in this direction was to determine the crystal structure. Excitingly, 1.9 Å crystal structure of MICU2 was recently reported, which shows its dimeric architecture (Wu et al., 2019). In both Ca2+-free and Ca2+- bound conditions, the authors characterized the interaction sites within the MICU2 homodimer, and MICU1-MICU2 heterodimer (Glu242 in MICU1 and Arg352 in MICU2 in apo heterodimer while Phe383 in MICU1 and Glu196 in MICU2 Ca2+-bound state) thereby proposing MICU1-MICU2 heterodimer model (Wu et al., 2019). More recently, the crystal structure of the apo form of the human MICU1-MICU2 heterodimer was reported wherein MICU1-MICU2 heterodimer acting as MCU gatekeepers was further corroborated (Park et al., 2020). Importantly, using high throughput screening MCU-i4 and MCU-i11 were identified as modulators of mitochondrial Ca2+ uptake (Di Marco et al., 2020). These compounds negatively regulate MCU activity resulting in decreased mitochondrial Ca2+ influx by specifically binding to MICU1. This inhibitory effect was not seen in MICU1 silenced or deleted cells (Di Marco et al., 2020). In skeletal muscle fibers MCU-i4 and MCU-i11 reduced mitochondrial Ca2+ uptake and impaired muscle growth (Di Marco et al., 2020). We are still in very early days of identification and characterization of the MICUs modulators. Definitely, significant efforts from multiple groups are required for addressing this highly relevant and outstanding need.
6. EMRE
The Essential MCU REgulator (EMRE) was identified using quantitative mass spectrometry of affinity-purified uniporter complex along with MICU1, MICU2, MCU and MCUb (Sancak et al., 2013). EMRE is an IMM localized protein with a single transmembrane domain and conserved carboxy terminal acidic domain (Sancak et al., 2013). Please refer Fig. 2 for diagrammatic illustration of domain architecture of MCU and other players involved in mitochondrial Ca2+ dynamics. The shRNA based knockdown or TALEN mediated knockout of EMRE in HEK-293 T cells resulted in impairment of mitochondrial Ca2+ uptake despite intact expression of the MCU protein and its oligomerization (Sancak et al., 2013). The EMRE is essential for MCU interaction with MICU1 and MICU2, which is critical for MCU channel activity (Sancak et al., 2013). In EMRE knockout cells, overexpression of MCU failed to restore mitochondrial Ca2+ uptake (Sancak et al., 2013). The EMRE protein expression was decreased in MCU deficient cells with no effect on mRNA levels suggesting post-transcriptional destabilization and degradation of EMRE (Sancak et al., 2013). EMRE regulates MCU channel activity by sensing mitochondrial matrix Ca2+ concentrations (Vais et al., 2016). Vais et al. utilized protease protection assays to demonstrate that the C terminus of EMRE is localized to matrix and acts as a sensor for matrix Ca2+ (Vais et al., 2016). The authors further proposed that the MCU activity is regulated by both cytoplasmic Ca2+ via MICU1/MICU2 as well as by matrix Ca2+ through EMRE thereby protecting mitochondria from Ca2+ depletion and Ca2+ overload (Vais et al., 2016). The deletion or charge neutralization of the acidic residues in EMRE C terminus abolished Ca2+ induced inhibition of MCU currents (Vais et al., 2016). In contrast to these studies, using a yeast expression system (reconstituting the mammalian MCU complex in yeast), Yamamoto et al. revealed opposing topology of EMRE (Yamamoto et al., 2016). The authors showed that the N-terminus of EMRE and not the C-terminus as reported earlier is located in the mitochondrial matrix (Yamamoto et al., 2016). Similarly, Tsai et al. demonstrated that the EMRE N-terminus is located in the mitochondrial matrix while its C-terminus is present in the intermembrane space (Tsai et al., 2016). The authors utilized mass‐tagging and MCU-EMRE fusion constructs for deriving the conclusions. They reported that EMRE physically interacts with MCU via its N-terminus and binds to MICU1 through C terminus (Tsai et al., 2016). Thus, the authors proposed a dual function of EMRE i.e. MCU activation and MICU1 regulation thereby protecting cells from mitochondrial Ca2+ overloaded and death (Tsai et al., 2016). Recently, same group reported a proteolytic pathway that maintains stoichiometrically suitable levels of MCU and EMRE thereby preventing dysfunctional assembly of uniporter subunits (Tsai et al., 2017). The authors further demonstrated that mitochondrial mAAA proteases AFG3L2 and SPG7 hydrolyze and degrade unassembled or excessive EMRE (Tsai et al., 2017). The protease resistant mutants of EMRE lead to induction of constitutive Ca2+ leakage into mitochondria (Tsai et al., 2017). Konig et al. studied interactome of mouse neuronal m-AAA proteases (IMM protein), mutations of which are associated with neurodegeneration in spinocerebellar ataxia (SCA28) and hereditary spastic paraplegia (HSP7) (Konig et al., 2016). The authors revealed a complex that regulates assembly of the MCU complex via EMRE biogenesis and m-AAA proteases that degrades non-assembled EMRE (Konig et al., 2016). The loss of the m-AAA protease led to augmented constitutively active MCU-EMRE channels that too without gatekeeping MICU subunits. This resulted in mitochondrial Ca2+ overload, mitochondrial permeability transition pore opening eventually causing neuronal death (Konig et al., 2016). A recent study reported cryo-EM structure of the human MCU-EMRE complex (Wang et al., 2019). Wang et al. reported that the juxta membrane loop of MCU and extended linker of EMRE are critical for EMRE-dependent MCU gating and EMRE stabilizes the juxta membrane loop of MCU (Wang et al., 2019). Further, the authors proposed dimerization of two MCU-EMRE complexes at the N-terminal domain (NTD) of human MCU (Wang et al., 2019). Another recent study reported cryo-EM structure of the intact human MCU-EMRE-MICU1-MICU2 supercomplex (MEMMS, 3.3-3.7 A) (Zhuo et al., 2020). Zhuo et al demonstrated that MCU forms hydrogen bonds with the N-terminal domain of EMRE. Further, the authors showed interactions between MICU1 G105, F106 and EMRE E93, E101 and no direct interactions of MICU1 with MCU indicating critical coupling function of EMRE (Zhuo et al., 2020). The authors further proposed that EMRE may act as lever for regulation of MCU matrix gate (Zhuo et al., 2020). Excitingly, the EMRE knockout mouse model was recently characterized (Liu et al., 2020). Liu et al. reported that although EMRE knockout mice are viable, they are smaller in size and are born less frequently. The authors further validated that EMRE is indeed critical for MCU function in vivo (Liu et al., 2020).
7. MCUR1
The Mitochondrial Ca2+ Uniporter regulator 1 (MCUR1) previously known as CCDC90A, was identified through a RNAi screening (Mallilankaraman et al., 2012a). It is a membrane protein essential for ruthenium red-sensitive MCU-mediated mitochondrial Ca2+ uptake in HEK293T, HeLa and primary human fibroblast cells (Mallilankaraman et al., 2012a). MCUR1 is ubiquitously expressed across mammalian tissues and is localized to IMM (Mallilankaraman et al., 2012a). Structurally, MCUR1 has two transmembrane domains and both its N- and C- termini face the inter-membrane space (Mallilankaraman et al., 2012a). The knockdown of MCUR1 had no effect on MCU localization and expression but led to decrease in Ca2+ uptake by energized mitochondria (Mallilankaraman et al., 2012a). Mallilankaraman et al. further demonstrated that MCUR1 overexpression enhances mitochondrial Ca2+ uptake. However, this increased Ca2+ influx was not observed upon MCU knockdown suggesting that both MCU and MCUR1 are critical for efficient mitochondrial Ca2+ uptake (Mallilankaraman et al., 2012a). Moreover, MCUR1 silencing led to disruption of oxidative phosphorylation and activation of AMP kinase-dependent pro-survival autophagy (Mallilankaraman et al., 2012a). Interestingly, the exact molecular mechanism by which MCUR1 regulates mitochondrial Ca2+ uptake is still not clear and its direct interaction with MCU is debatable. A rather contradictory study reported that MCUR1 is a cytochrome c oxidase assembly factor instead of MCU regulator (Paupe et al., 2015). Paupe et al. suggested that MCUR1 does not regulate MCU activity directly (Paupe et al., 2015). The decrease in MCU activity upon MCUR1 knockdown is due to change in mitochondrial membrane potential caused by defect in respiratory chain (Paupe et al., 2015). Later, Vais et al. employed patch clamp electrophysiology for directly measuring activity of MCU by recording MCU-mediated Ca2+ currents (IMCU) in mitoplasts isolated from wild-type HEK293 cells and MCUR1 knockdown HEK cells (Vais et al., 2015). The authors demonstrated significant decrease in ruthenium red-sensitive, inwardly rectifying, Ca2+ concentration-dependent Ca2+ currents upon knockdown of MCUR1. Further, the authors rescued the decrease in MCU-mediated Ca2+ currents by overexpressing shRNA-insensitive MCUR1 in MCUR1 stable knockdown cells (Vais et al., 2015). More recently, Madesh’s group suggested that MCUR1 functions as a scaffold protein for the MCU complex and it binds to conserved CC domains of both MCU and EMRE in COS-7 cells to form heterooligomeric complex (Tomar et al., 2016). Tomar et al. reported decrease in mitochondrial Ca2+ uptake and IMCU current in MCUR1 deficient mouse cardiomyocytes and endothelial cells (Tomar et al., 2016). The authors demonstrated that MCUR1 is essential for MCU complex assembly and loss of MCUR1 leads to disruption of MCU complex (Tomar et al., 2016). Interestingly, a recent report revealed that MCUR1 is a regulator of Ca2+ concentration required for induction of mitochondrial permeability transition (MPT) (Chaudhuri et al., 2016). Further, silencing of MCUR1 in HeLa cells resulted in increased Ca2+ threshold for MPT activation thus inhibiting cell death associated with mitochondrial Ca2+ overload (Chaudhuri et al., 2016). The role of MCUR1 in human pathophysiology is emerging in nature. Recently, it was reported that MCUR1 expression is increased in human hepatocellular carcinomas (HCC) (Ren et al., 2018). The upregulation of MCUR1 in HCC results in enhanced MCU-mediated mitochondrial Ca2+ uptake, increased cell proliferation and inhibition of apoptosis (Ren et al., 2018). The same group further reported that the higher MCUR1 expression is associated with HCC metastasis (Jin et al., 2019). It was shown that MCUR1 promotes epithelial-mesenchymal transition, invasion and metastasis of HCC cells (Jin et al., 2019). Certainly, more studies in different types of cancers are required for better understanding of MCUR1 role in tumorigenesis.
Recently, Zulkifli et al. reported role of Saccharomyces cerevisiae homologs of MCUR1 (Put6 and Put7) in mitochondrial proline metabolism (Zulkifli et al., 2020). Further, the authors demonstrated that Put6 and Put7 are localized to inner mitochondrial membrane and form heterooligomeric complex. The authors showed that the loss of Put6 and Put7 leads to perturbed mitochondrial proline uptake and cellular redox balance (Zulkifli et al., 2020). While heterologous expression of human MCUR1 rescued proline-specific growth defects in put6Δ, put7Δ, and- put6Δput7Δ yeast cells (Zulkifli et al., 2020).
8. Pathophysiological relevance of MCU complex
The MCU complex plays an essential role in regulating several cellular functions including mitochondrial Ca2+ homeostasis, bioenergetics, autophagy, cytosolic Ca2+ buffering, secretory functions, cell survival, cell proliferation/migration and cell death among others (Giorgi et al., 2018; Mammucari et al., 2018; Pathak and Trebak, 2018; Paupe and Prudent, 2018). The MCU plays an important role in glucose sensing by pancreatic β cells (Tarasov et al., 2012). Literature advocates role of mitochondrial Ca2+ in macrophage polarization (Gu et al., 2017), embryonic development (Prudent et al., 2013) and inflammatory response during cystic fibrosis (Rimessi et al., 2015). In aging human skeletal muscle, Rizzuto’s group linked improved muscle function with exercise-induced increase in MCU expression (Zampieri et al., 2016). There have been numerous studies demonstrating MCU’s role in various pathophysiological conditions including ischemia, myocardial infarction, and neurodegenerative disorders (Mammucari et al., 2018). By utilizing gene trap strategy Pan et al. reported that MCU(-/-) mice present only a mild phenotype. The MCU(-/-) mice were slightly smaller in size as compared to wild type mice however organ weight was found proportional to body weight (Pan et al., 2013). This observation was rather surprising as MCU plays a critical role in the mitochondrial Ca2+ influx. Pan et al. reported that the mitochondria isolated from heart of MCU(-/-) mice were not able to uptake Ca2+ (Pan et al., 2013). Further, the authors reported altered phosphorylation and activity of pyruvate dehydrogenase enzyme in skeletal muscle of MCU(-/-) mice (Pan et al., 2013). Interestingly, the authors observed a significant decrease but not complete absence of mitochondrial matrix Ca2+ in the skeletal muscle cells of MCU(-/-) mice. The Ca2+ concentration in the mitochondrial matrix of MCU(-/-) mice was 25% of wild type littermate control mice. Pan and colleagues suggested that although the MCU mediated rapid mitochondrial Ca2+ influx was absent in MCU(-/-) mice, other alternative mechanisms must exist for slower Ca2+ influx. Further, the authors implicated that this physiological adaptation might be assisting MCU(-/-) mice in survival. However, it is important to note that NCLX is essential for cardiac function as conditional (cardiac specific) NCLX-/- mice is lethal with more than 87% lethality within 2 weeks of birth (Luongo et al., 2017). Please refer the NCLX section for the details.
Mammucari et al. reported that the MCU regulates skeletal muscle function and trophism in vivo (Mammucari et al., 2015). The authors further demonstrated that MCU overexpression protects from denervation-induced atrophy (Mammucari et al., 2015). Holmström et al. demonstrated impaired mitochondrial Ca2+ signaling in mitochondria isolated from MCU(-/-) mice without affecting basal cardiac function (Holmstrom et al., 2015). Another report revealed that Ca2+/ calmodulin-dependent protein kinase II (CaMKII) (activated in ischemia reperfusion, myocardial infarction and neurohumoral injury) regulates MCU current (IMCU) and opening of mPTP (Joiner et al., 2012). This eventually causes myocardial death thus implicating a critical role of MCU in heart failure (Joiner et al., 2012). The vital role of MCU in cardiac physiology was further demonstrated by using transgenic mice expressing a dominant negative form of MCU (Wu et al., 2015). These mice showed reduction in myocardial ATP generation and inhibition of fight or flight induced increase in heart rate (Wu et al., 2015). Independently, mice with cardiomyocyte-specific deletion of MCU showed abrogation of acute mitochondrial Ca2+ uptake without affecting basal mitochondrial Ca2+ levels (Kwong et al., 2015). The MCU deletion protected mice from acute ischemia-reperfusion injury (Kwong et al., 2015). Moreover, cardiomyocyte-specific expression of mutant MCU in adult mice leads to loss of mitochondrial Ca2+ uptake, reduced heart rate during fight or flight responses (Luongo et al., 2015). This imparts protection from ischemia-reperfusion mediated injury by reducing mPTP activation without affecting basal cardiac function (Luongo et al., 2015). In case of diabetic mouse hearts, the expression of MCU and EMRE is decreased but expression of MCUb is increased (Suarez et al., 2018). This leads to decrease in mitochondrial Ca2+ uptake thus affecting mitochondrial energetics and cardiac function during diabetic conditions (Suarez et al., 2018). Taken together, all these studies highlight a key role for MCU complex in cardiac physiology.
A recent study by Ghosh et al. demonstrated critical role of MCU in pathogenesis of Barth syndrome (rare genetic disorder) (Ghosh et al., 2020). Barth syndrome is characterized by skeletal muscle weakness, neutropenia and cardiomyopathy (Garlid et al., 2020). This disease is caused by mutations in Tafazzin (a transacylase required for acylating immature cardiolipin to mature cardiolipin) (Garlid et al., 2020). Ghosh et al. reported decrease in MCU stability and function in murine Tafazzin KO cells and Barth’s syndrome patient-derived B-lymphocytes and cardiac tissues (Ghosh et al., 2020). Therefore highlighting a crucial role of MCU stability and activity via cardiolipin in human physiology (Ghosh et al., 2020).The role of MCU complex in cancer biology is still emerging in nature. Early on, Pinton’s group identified miR-25 (overexpressed in human colon cancer) as a cancer-related MCU-targeting microRNA (Marchi et al., 2013). Marchi et al. reported decrease in MCU expression and mitochondrial Ca2+ uptake upon miR-25 overexpression thereby implicating role of MCU in tumorigenesis (Marchi et al., 2013). A recent study reported role of MCU in colorectal carcinogenesis via its crosstalk with receptor-interacting protein kinase 1 (RIPK1) (Zeng et al., 2018). RIPK1 is up-regulated in human colorectal cancer, which in turn results in increased mitochondrial Ca2+ uptake and bioenergetics (Zeng et al., 2018). A report from Mammucari’s group demonstrated role of MCU in triple-negative breast cancer (Tosatto et al., 2016). The authors observed a decrease in ROS production, reduction in hypoxia-inducible factor-1α (HIF-1α) expression and inhibition of breast cancer cell growth and metastasis in MCU-silenced cells (Tosatto et al., 2016). Interestingly, Tang et al. showed role of MCU in store-operated Ca2+ entry dependent cell migration in breast cancer (Tang et al., 2015). In MDA-MB-231 breast cancer cell line both pharmacological inhibition (with ruthenium Red) and siRNA mediated knockdown of MCU resulted in suppression of serum-induced migration (Tang et al., 2015). For further details regarding physiological and pathophysiological role of MCU complex please refer recent reviews on this topic (Arduino and Perocchi, 2018; De Stefani et al., 2016; Mammucari et al., 2017, 2018, 2016; Pathak and Trebak, 2018). Excitingly, Perocchi’s group identified mitoxantrone (an anti-cancer drug) as a highly specific human MCU inhibitor (Arduino et al., 2017). Future studies aimed at examining the effects of targeting MCU complex with mitoxantrone and other drugs (MICU inhibitors discussed earlier) would open up the doors for therapeutic modulation of MCU complex against a number of above discussed pathophysiological conditions.
9. NCLX
Mitochondrial Na+/Ca2+/Li+ exchanger (NCLX) proficiently mediates the exchange of Ca2+/Na+ and also the exchange of Li+/Ca2+. The action of NCLX was first reported in mitochondria isolated from cardiac cells loaded with Ca2+ (Carafoli et al., 1974). The fact was based on the observation that when Na+ was added to the solution containing isolated mitochondria there was a sudden efflux of Ca2+ from the mitochondria and in the absence of Na+ no Ca2+ extrusion was detected (Carafoli et al., 1974). Moreover, Li+ gave the similar results (Carafoli et al., 1974). The activity of Na+/Ca2+ exchange was later observed in the mitochondria isolated from several tissue types including liver (Gunter et al., 1988; Wingrove and Gunter, 1986), skeletal muscle, brain, parotid gland, adrenal cortex (Andrews et al., 2005; Crompton et al., 1978). Sekler’s group was first to discover the NCLX gene (Palty et al., 2004). Palty et al. cloned NCLX gene (earlier known as FLJ22233) from HEK293 cells and validated that NCLX is discrete from both Na+/ Ca2+/K+ exchanger (NCKX) and Na+/Ca2+ exchanger (NCX) in its capacity to catalyze Li+/Ca2+ exchange (Palty et al., 2004). The NCLX is highly specific for Ca2+ and doesn’t catalyze either Ba2+ or Zn2+ transport (Palty et al., 2004). Later on, the same group demonstrated that overexpression of NCLX increases the mitochondrial Ca2+ extrusion function while its silencing reduces this function (Palty et al., 2010). The authors also showed that NCLX is endogenously expressed on the mitochondrial cristae (Palty et al., 2010). The NCLX shares structural similarity with the Na+/Ca2+ exchangers (NCXs) that are members of Ca2+/cation antiporter (CaCA) gene family (Blaustein and Lederer, 1999; Khananshvili, 1991). It consists of two transmembrane domains known as α1 and α2 containing ion binding and ion exchange sites (Blaustein and Lederer, 1999; Khananshvili, 1991). Recently, Roy et al. used molecular modeling and fluorescence analysis approach to prove that α1 and α2 domains of NCLX play an important role in regulating cation selectivity (Roy et al., 2017). The authors further identified the distinctive residues critical for Li+ versus Na+ dependent Ca2+ efflux. Thereby, highlighting their functional significance under varying ionic concentration and mitochondrial membrane potential (Roy et al., 2017).
The NCLX influxes 3Na+ into mitochondria for extrusion of 1 Ca2+ out of mitochondria. Studies in primary astrocytes demonstrated that rise in cytosolic Na+ induced by glutamate is followed by activation of the mitochondrial Na+/Ca2+ exchanger, which in turn results in increased Na+ influx into the mitochondria (Bernardinelli et al., 2006). Parnis et al. corroborated NCLX’s critical role in mitochondrial Ca2+ efflux (Parnis et al., 2013). The silencing of NCLX dysregulated cytosolic Ca2+ levels in astrocytes because of perturbations in the Ca2+ entry through SOC pathway (Parnis et al., 2013). The authors further demonstrated that the coupling of Ca2+ influx via SOCE and mitochondria Ca2+ uptake is much robust than the ER Ca2+ release induced mitochondrial Ca2+ transients (Parnis et al., 2013). This data suggested that at least in astrocytes there is stronger functional crosstalk between SOCE and mitochondrial Ca2+ dynamics in comparison to Ca2+ release from ER (Parnis et al., 2013). However, in non-excitable cells it is demonstrated that in response to IP3, microdomains of high intracellular Ca2+ concentration are achieved at ER-Mitochondria contact sites. Riz-zuto et al. used mitochondria targeted recombinant aquorin to demonstrate that these microdomains were transiently generated and were sensed by mitochondria (Rizzuto et al., 1993). Therefore, it is suggested that in non-excitable cells the major route of mitochondrial Ca2+ influx is via Ca2+ microdomains established at ER-Mitochondria contact sites. An important factor that regulates source of Ca2+ (plasma membrane influx or ER release), which is taken up by mitochondria is the precise localization of these organelles within the cells. The organelle localization would in turn affect their relative positioning which is a critical factor regulating the mitochondrial Ca2+ uptake. For more details on the role of ER Ca2+ release in regulating mitochondrial Ca2+ influx, please refer to this relevant review (García-Sancho, 2014).
Interestingly, Sekler and Trebak groups recently delineated the molecular basis of the crosstalk between SOCE and NCLX mediated mitochondrial Ca2+ extrusion (Ben-Kasus Nissim et al., 2017). Nissim et al. demonstrated that exclusion of extracellular Na+ decreases cytosolic Na+ that in turn hinders NCLX function and inhibits Orai1 mediated SOCE (Ben-Kasus Nissim et al., 2017). Furthermore, the authors showed that silencing of NCLX neither affects the expression nor the interaction of STIM1 and Orai1 (Ben-Kasus Nissim et al., 2017). It was rather the ROS sensitivity of Orai1, which was modulated by NCLX functioning (Ben-Kasus Nissim et al., 2017).
The experiments performed on guinea pig myocytes for detecting the mitochondrial Ca2+ concentration showed that increased cytosolic Na+ levels activate NCLX that in turn depletes mitochondrial Ca2+ (Maack et al., 2006). Further, this leads to reduction in metabolic rate of mitochondria and decrease in ATP generation during cardiac ischemia (Maack et al., 2006). Moreover, the knockdown of NCLX causes substantial delay in glucose-dependent insulin secretion (Nita et al., 2012). The studies conducted on Parkinson’s disease have shown that the extrusion of mitochondrial Ca2+ via NCLX expression was neuro-protective. It was found that the MCU and MICU1 were transcriptionally up-regulated upon expression of mutant LRRK2 in primary mouse cortical neurons (Verma et al., 2017). Importantly, the knockdown of MCU and overexpression of NCLX were shown to be neuroprotective (Verma et al., 2017). The neuroprotective activity of NCLX was also observed in PTEN-Induced kinase 1(PINK1) deficient neurons, which are used as cellular model for Parkinson’s disease (Kostic et al., 2018). Purroy et al. used cellular models of Friedreich ataxia (neonatal rat cardiomyocytes and dorsal root ganglia neurons deficient in frataxin) and observed decrease in the NCLX expression (Purroy et al., 2018). Further, the studies using brain ischemia model have shown that the intra-axonal Ca2+ expulsion during ischemia mainly depends on Na+ inflow hence downregulation of NCLX plays a preventive role during such injuries (Nikolaeva et al., 2005). Recently, Luongo et al. have shown that tamoxifen-induced deletion of NCLX gene in adult mouse heart results in sudden death (Luongo et al., 2017). The lethality was associated with an extreme damage of myocardial tissue, resulting in cardiac failure. The mechanism behind the heart pathology is Ca2+ overload in mitochondria (Luongo et al., 2017). This leads to ROS generation and necrotic cell death, which was rescued by suppression of mPTP (Luongo et al., 2017). Conversely, overexpression of NCLX in mouse cardiac tissue by conditional transgenesis reduced ischemia-induced cardiac damage (Luongo et al., 2017). Recently, Trebak lab reported a dichotomous role of NCLX in colorectal cancer progression and metastasis (Pathak et al., 2020). Pathak et al. demonstrated that NCLX downregulation arrests tumor growth in xenografts and spontaneous colon cancer models by regulating cell-cycle progression. However, NCLX silencing led to an increase in metastatic potential of colon cancer cells through HIF1α signaling (Pathak et al., 2020). Taken together, these studies elegantly demonstrate a key role for NCLX in regulating pathophysiological conditions associated with perturbations in mitochondrial Ca2+ dynamics.
10. Letm1
Letm1 is shorthand for Leucine zipper EF hand-containing trans-membrane1. Letm1 is evolutionary conserved among eukaryotes and codes for mitochondrial inner membrane protein (Dimmer et al., 2008; Hasegawa and van der Bliek, 2007; Tamai et al., 2008). Letm2 is a paralog of Letm1, which is expressed in mammals, plants and yeast (Frazier et al., 2006; Tamai et al., 2008; Zhang et al., 2012). Endele et al identified Letm1 as a gene frequently deleted in Wolf-Hirschhorn syndrome (WHS) patients (Endele et al., 1999). Letm1 encodes an 83.4 kDa Ca2+ binding protein, containing two transmembrane domains, a leucine zipper domain and two EF hand motifs (Austin and Nowikovsky, 2019; Endele et al., 1999; Lee et al., 2017; Li et al., 2019). The initial report on role of Letm1 in regulating mitochondrial Ca2+ dynamics came from Clapham’s group (Jiang et al., 2009). Jiang et al. performed genome wide RNA interference screen for identifying genes responsible for the regulation of mitochondrial Ca2+ and H+ concentrations (Jiang et al., 2009). This screen in drosophila cells narrowed down to a mammalian homolog of Letm1, which regulates Ca2+/H+ exchange (Jiang et al., 2009). Further corroborating experiments like liposome reconstitution and overexpression of Letm1 exhibited that it is a mitochondrial Ca2+/H+ exchanger (Jiang et al., 2009). In subsequent studies Jiang et al. performed Letm1 knockdown in HEK293 cells and confirmed that Letm1 mediates Ca2+ influx into mitochondria at low mitochondrial Ca2+ levels (Jiang et al., 2013). They further generated Letm1 lacking mice and demonstrated lethality in homozygous and half of heterozygous knockout mice (Jiang et al., 2013). The surviving mice showed reduction in mitochondrial Ca2+ uptake at low cytosolic Ca2+ concentrations. Moreover, in Wolf-Hirschhorn syndrome (WHS) patients ATP generation was hindered and glucose metabolism was dysregulated due to loss of Letm1 function (Jiang et al., 2013). Nowikovsky et al. performed theoretical analysis on the direction of Ca2+ flux via mitochondrial membrane in energized mitochondria with different Ca2+/H+ stoichiometry (Nowikovsky et al., 2012). The authors proposed that Ca2+ extrudes mitochondria with 2H+: 1Ca2+/3H+: 1Ca2+ stoichiometry and enters mitochondria with 1H+: 1Ca2+ under physiological respiration conditions (Nowikovsky et al., 2012).
Interestingly, Clapham’s group has demonstrated role of Letm1 in mediating mitochondrial Ca2+ influx (Jiang et al., 2009; Tsai et al., 2014) while other groups in regulating mitochondrial Ca2+ extrusion (Austin et al., 2017; Shao et al., 2016). However, its role remains controversial (Austin and Nowikovsky, 2019; De Marchi et al., 2014). Importantly, Madesh’s group demonstrated role of Letm1 in both Ca2+ influx and efflux from mitochondria (Doonan et al., 2014). Doonan et al. reported that Letm1 silencing decreases mitochondrial Ca2+ influx without affecting expression and function of MCU (Doonan et al., 2014). Recently, Austin et al. reported that Letm1 silencing perturbs H+ mediated Na+ and K+ homeostasis which leads to K+ accumulation in the mitochondrial matrix (Austin et al., 2017). Further, Letm1 silencing resulted in specific reduction in Na+/Ca2+ exchange via NCLX (Austin et al., 2017). Therefore, the effects on mitochondrial Ca2+ efflux upon manipulation of Letm1 could be in part explained through its control over NCLX functioning (Austin et al., 2017).
Tamai et al. demonstrated importance of Letm1 in maintenance of tubular network of mitochondria and assembly of respiratory chain super complexes (Tamai et al., 2008). The Letm1 knockdown in HeLa cells led to decrease in number of cristae structures and induced mitochondrial swelling. Further, Letm1 silencing resulted in dismantling of complexes I, III and IV of respiratory chain (Tamai et al., 2008). Moreover, downregulation of BCS1L also resulted in insufficiency in the development of respiratory complexes I, III and IV (same as in the Letm1 knockdown), proposing that Letm1 and BCS1L coordinate in the biogenesis of respiratory chain complexes (Tamai et al., 2008). Similarly, Hasegawa et al. reported that the Letm1 silencing in human cells and in C. elegans causes mitochondrial swellings along the long edges (Hasegawa and van der Bliek, 2007). On the other hand, over expression of Letm1 resulted in swelling of cristae due to increased electron density of mitochondrial matrix (Hasegawa and van der Bliek, 2007). This study clearly demonstrates an essential role for Letm1 in regulating mitochondrial matrix volume (Hasegawa and van der Bliek, 2007). Recently, Nakamura et al. corroborated role of Letm1 in regulating mitochondrial shape (Nakamura et al., 2020). Letm1 altered the shape of in vitro reconstituted proteoliposomes leading to development of invaginations on artificial liposomes (Nakamura et al., 2020). However, mutant Letm1 failed to do so thereby implicating a key role for Letm1 in the organization of mitochondrial membrane morphology (Nakamura et al., 2020).
The role of Letm1 in maintaining osmotic balance, mitochondrial morphology, metabolism and dynamics is well established and these functions in part could be due to its role in maintaining mitochondrial ionic (K+, Na+ and Ca2+) homeostasis (Austin and Nowikovsky, 2019; Hashimi et al., 2013). Further, Letm1 was recently shown to couple mitochondrial translation to metabolic changes and cell survival in WHS derived fibroblasts (Durigon et al., 2018). Piao et al. demonstrated that overexpression of Letm1 decreases mitochondrial biogenesis and ATP synthesis thereby inducing cell death by necrosis (Piao et al., 2009). Moreover, the authors performed immuno-histochemical studies on human cancer tissues and found that Letm1 expression was higher in cancerous tissues in comparison to healthy tissues (Piao et al., 2009). Recently, overexpression of Letm1 was reported in esophageal squamous cell carcinoma (Yang et al., 2018). Further, Huang et al. showed that Letm1 silencing in bladder cancer cells (J82, EJ, T24 and 5637) inhibits their proliferation and invasion (Huang et al., 2017). Collectively, these studies point towards an evolving role of Letm1 in cancer progression. However, more studies are needed for clearly establishing its role in cancer biology and other diseases.
11. Concluding remarks and future perspectives
Although significance of mitochondrial Ca2+ dynamics and the electrophysiological characterization of the proteins involved in this process started emerging almost 50 years ago, the molecular identity of the key regulators of mitochondrial Ca2+ mobilization only started to evolve in last decade. Since identification of crucial players controlling mitochondrial Ca2+ influx and extrusion, this field has attained significant attention from the scientific community. This has led to several exciting discoveries in last few years. However, there are many more outstanding questions that need to be addressed in future for better understanding and possible targeting of these players in managing human pathophysiology. The emerging literature suggest that the perturbations in the expression and activity of MCU complex is associated with cardiovascular disorders, neuronal abnormalities and cancers. But the signaling cascades and molecular choreography that precisely regulate expression of these proteins in diverse disease conditions remain largely unexplored. Similarly, the role of post-translational modifications in modulating the activity of the mitochondrial Ca2+ handling toolkit under resting conditions and their alterations during pathophysiology is poorly understood. Further, so far the focus of the field has been on understanding the role of these proteins in mitochondrial physiology. In future, it would be interesting to examine if these players have any extra-mitochondrial function. Although most of these proteins have a mitochondrial targeting signal peptide, their extra-mitochondrial functions can’ t be completely ruled out. The association of mutations in mitochondrial Ca2+ handling toolkit with human diseases is emergent in nature and to this point only mutations in MICU1 and Letm1 are reported in human patients. Going forward, it would be fascinating to see if mutations in other components of MCU complex are associated with human diseases. The answers to above highlighted questions would certainly assist in better understanding of the mitochondrial Ca2+ dynamics and associated pathophysiological states. This knowledge would further augment the ongoing efforts on designing and characterization of small molecules for disease specific targeting of the mitochondrial Ca2+ handling machinery. Moreover, it would motivate both academia and pharma giants to investigate the possibilities of repurposing of FDA approved drugs for calibrating mitochondrial Ca2+ dynamics and thereby managing associated human disorders.
Acknowledgments
This work was supported by the DBT/Wellcome Trust India Alliance Fellowship (IA/I/19/2/504651) awarded to Rajender K Motiani. Authors also acknowledge RCB core funding and Science & Engineering Research Board (SERB) Start-up Research Grant (SRG/2019/000495) to RKM. JT acknowledges her Senior Research Fellowship from CSIR, India.
Abbreviations
- ATP
Adenosine Triphosphate
- CaCA
Ca2+/Cation Antiporter
- CK
Creatine Kinase
- CREB
Cyclic adenosine monophosphate Response ElementBinding protein
- DID
DIME Interacting Domain
- EGR1
Early Growth Response 1
- EMRE
Essential MCU Regulator
- EMT
Epithelial Mesenchymal Transition
- ER
Endoplasmic Reticulum
- HCC
Human Hepatocellular Carcinoma
- HIF-1α
Hypoxia Inducible Factor-1α
- HSP7
Hereditary Spastic Paraplegia 7
- IMM
Inner Mitochondrial Membrane
- IMS
Inter Membrane Space
- IP3R
Inositol-1,4,5-triphosphate Receptor
- LETM1
Leucine zipper EF-hand containing TransMembrane 1
- MAMs
Mitochondrial Associated Membranes
- MCU
Mitochondrial Calcium Uniporter
- MCUb
MCU Regulatory subunit b
- MCUR1
Mitochondrial Ca2+ Regulator 1
- MICU1
Mitochondrial Ca2+ Uptake protein 1
- Mptp
mitochondrial Permeability Transition Pore
- Mros
mitochondrial Reactive Oxygen Species
- NCLX
Na+/Ca2+/Li+ Exchanger
- NCKX
Na+/Ca2+/K+ Exchanger
- NCX
Na+/Ca2+ Exchanger
- OMM
Outer Mitochondrial Membrane
- PINK1
PTEN-Induced kinase 1
- PM
Plasma Membrane
- RIPK1
Receptor Interacting Protein Kinase 1
- Ru360
Ruthenium 360
- SERCA
Sarco/Endoplasmic Reticulum Ca2+ ATPase
- ShRNA
Short Hairpin RNA
- SiRNA
Small Interfering RNA
- SOCE
Store Operated Calcium Entry
- STIM1
Stromal Interaction Molecule 1
- TCA
Tricarboxylic Acid Cycle
- VDACs
Voltage Dependent Anion Channels
- WHS
Wolf-Hirschhorn Syndrome
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Andrews ZB, Diano S, Horvath TL. Mitochondrial uncoupling proteins in the CNS: In support of function and survival. Nat Rev Neurosci. 2005;6:829–840. doi: 10.1038/nrn1767. [DOI] [PubMed] [Google Scholar]
- Antony AN, Paillard M, Moffat C, Juskeviciute E, Correnti J, Bolon B, Rubin E, Csordas G, Seifert EL, Hoek JB, Hajnoczky G. MICU1 regulation of mitochondrial Ca(2+) uptake dictates survival and tissue regeneration. Nat Commun. 2016;7:10955. doi: 10.1038/ncomms10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduino DM, Perocchi F. Pharmacological modulation of mitochondrial calcium homeostasis. J Physiol. 2018;596:2717–2733. doi: 10.1113/JP274959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduino DM, Wettmarshausen J, Vais H, Navas-Navarro P, Cheng Y, Leimpek A, Ma Z, Delrio-Lorenzo A, Giordano A, Garcia-Perez C, Medard G, et al. Systematic identification of MCU modulators by orthogonal interspecies chemical screening. Mol Cell. 2017;67(711–723):e717. doi: 10.1016/j.molcel.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S, Nowikovsky K. LETM1: Essential for mitochondrial biology and cation homeostasis? Trends Biochem Sci. 2019;44:648–658. doi: 10.1016/j.tibs.2019.04.002. [DOI] [PubMed] [Google Scholar]
- Austin S, Tavakoli M, Pfeiffer C, Seifert J, Mattarei A, De Stefani D, Zoratti M, Nowikovsky K. LETM1-mediated K(+) and Na(+) homeostasis regulates mitochondrial Ca(2+) efflux. Front Physiol. 2017;8:839. doi: 10.3389/fphys.2017.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock DF, Herrington J, Goodwin PC, Park YB, Hille B. Mitochondrial participation in the intracellular Ca2+ network. J Cell Biol. 1997;136:833–844. doi: 10.1083/jcb.136.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcavage WX, Lloyd JL, Mattoon JR, Ohnishi T, Scarpa A. Cation movements and respiratory response in yeast mitochondria treated with high Ca2+ concentrations. Biochim Biophys Acta. 1973;305:41–51. doi: 10.1016/0005-2728(73)90229-6. [DOI] [PubMed] [Google Scholar]
- Baradaran R, Wang C, Siliciano AF, Long SB. Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature. 2018;559:580–584. doi: 10.1038/s41586-018-0331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartok A, Weaver D, Golenar T, Nichtova Z, Katona M, Bansaghi S, Alzayady KJ, Thomas VK, Ando H, Mikoshiba K, Joseph SK, et al. IP3 receptor isoforms differently regulate ER-mitochondrial contacts and local calcium transfer. Nat Commun. 2019;10:3726. doi: 10.1038/s41467-019-11646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, Koteliansky V, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal K, Jung D, Gunter K, Gunter T, Brierley G. Na (+)-dependent Ca2+ efflux mechanism of heart mitochondria is not a passive Ca2+/2Na+ exchanger. Am J Physiol Cell Physiol. 1994;266:C800–C808. doi: 10.1152/ajpcell.1994.266.3.C800. [DOI] [PubMed] [Google Scholar]
- Ben-Kasus Nissim T, Zhang X, Elazar A, Roy S, Stolwijk JA, Zhou Y, Motiani RK, Gueguinou M, Hempel N, Hershfinkel M. Mitochondria control store-operated Ca2+ entry through Na+ and redox signals. EMBO J. 2017;36:797–815. doi: 10.15252/embj.201592481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi P. Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol Rev. 1999;79:1127–1155. doi: 10.1152/physrev.1999.79.4.1127. [DOI] [PubMed] [Google Scholar]
- Bernardinelli Y, Azarias G, Chatton JY. In situ fluorescence imaging of glutamate-evoked mitochondrial Na+ responses in astrocytes. Glia. 2006;54:460–470. doi: 10.1002/glia.20387. [DOI] [PubMed] [Google Scholar]
- Bick AG, Calvo SE, Mootha VK. Evolutionary diversity of the mitochondrial calcium uniporter. Science. 2012;336:886. doi: 10.1126/science.1214977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaustein MP, Lederer WJ. Sodium/calcium exchange: Its physiological implications. Physiol Rev. 1999;79:763–854. doi: 10.1152/physrev.1999.79.3.763. [DOI] [PubMed] [Google Scholar]
- Boehning D, Patterson RL, Sedaghat L, Glebova NO, Kurosaki T, Snyder SH. Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nat Cell Biol. 2003;5:1051–1061. doi: 10.1038/ncb1063. [DOI] [PubMed] [Google Scholar]
- Boyman L, Williams GS, Khananshvili D, Sekler I, Lederer W. NCLX: The mitochondrial sodium calcium exchanger. J Mol Cell Cardiol. 2013;59:205–213. doi: 10.1016/j.yjmcc.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD. Electroneutral efflux of Ca2+ from liver mitochondria. Biochem J. 1985;225:413–419. doi: 10.1042/bj2250413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Wang S, Cui T, Su XC, Chou JJ. Ion and inhibitor binding of the double-ring ion selectivity filter of the mitochondrial calcium uniporter. Proc Natl Acad Sci USA. 2017;114:E2846–E2851. doi: 10.1073/pnas.1620316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafoli E, Lehninger AL. A survey of the interaction of calcium ions with mitochondria from different tissues and species. Biochem J. 1971;122:681–690. doi: 10.1042/bj1220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafoli E, Tiozzo R, Lugli G, Crovetti F, Kratzing C. The release of calcium from heart mitochondria by sodium. J Mol Cell Cardiol. 1974;6:361–371. doi: 10.1016/0022-2828(74)90077-7. [DOI] [PubMed] [Google Scholar]
- Carvalho EJ, Stathopulos PB, Madesh M. Regulation of Ca2+ exchanges and signaling in mitochondria. Curr Opin Physiol. 2020;17:197–206. doi: 10.1016/j.cophys.2020.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri D, Artiga DJ, Abiria SA, Clapham DE. Mitochondrial calcium uniporter regulator 1 (MCUR) regulates the calcium threshold for the mitochondrial permeability transition. Proc Natl Acad Sci USA. 2016;113:E1872–1880. doi: 10.1073/pnas.1602264113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri D, Sancak Y, Mootha VK, Clapham DE. MCU encodes the pore conducting mitochondrial calcium currents. Elife. 2013;2:e00704. doi: 10.7554/eLife.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Perocchi F. ProtPhylo: identification of protein-phenotype and protein-protein functional associations via phylogenetic profiling. Nucl Acids Res. 2015;43:W160–168. doi: 10.1093/nar/gkv455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombini M. VDAC structure, selectivity, and dynamics. Biochim Biophys Acta. 2012;1818:1457–1465. doi: 10.1016/j.bbamem.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton M, Moser R, Lüdi H, Carafoli E. The interrelations between the transport of sodium and calcium in mitochondria of various mammalian tissues. Eur J Biochem. 1978;82:25–31. doi: 10.1111/j.1432-1033.1978.tb11993.x. [DOI] [PubMed] [Google Scholar]
- Csordas G, Golenar T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, Moffat C, Weaver D, de la Fuente Perez S, Bogorad R, Koteliansky V, et al. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca(2)(+) uniporter. Cell Metab. 2013;17:976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marchi U, Santo-Domingo J, Castelbou C, Sekler I, Wiederkehr A, Demaurex N. NCLX protein, but not LETM1, mediates mitochondrial Ca2+ extrusion, thereby limiting Ca2+-induced NAD (P) H production and modulating matrix redox state. J Biol Chem. 2014;289:20377–20385. doi: 10.1074/jbc.M113.540898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani D, Rizzuto R, Pozzan T. Enjoy the trip: Calcium in mitochondria back and forth. Annu Rev Biochem. 2016;85:161–192. doi: 10.1146/annurev-biochem-060614-034216. [DOI] [PubMed] [Google Scholar]
- Debattisti V, Horn A, Singh R, Seifert EL, Hogarth MW, Mazala DA, Huang KT, Horvath R, Jaiswal JK, Hajnoczky G. Dysregulation of mitochondrial Ca(2+) uptake and sarcolemma repair underlie muscle weakness and wasting in patients and mice lacking MICU1. Cell Rep. 2019;29(1274–1286):e1276. doi: 10.1016/j.celrep.2019.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca HF, Engstrom GW. Calcium uptake by rat kidney mitochondria. Proc Natl Acad Sci USA. 1961;47:1744–1750. doi: 10.1073/pnas.47.11.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marco G, Vallese F, Jourde B, Bergsdorf C, Sturlese M, De Mario A, Techer-Etienne V, Haasen D, Oberhauser B, Schleeger S, Minetti G, et al. A high-throughput screening identifies MICU1 targeting compounds. Cell Rep. 2020;30(2321–2331):e2326. doi: 10.1016/j.celrep.2020.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer KS, Navoni F, Casarin A, Trevisson E, Endele S, Winterpacht A, Salviati L, Scorrano L. LETM1, deleted in Wolf-Hirschhorn syndrome is required for normal mitochondrial morphology and cellular viability. Hum Mol Genet. 2008;17:201–214. doi: 10.1093/hmg/ddm297. [DOI] [PubMed] [Google Scholar]
- Docampo R, Vercesi AE. Ca2+ transport by coupled Trypanosoma cruzi mitochondria in situ. J Biol Chem. 1989;264:108–111. [PubMed] [Google Scholar]
- Dong Z, Shanmughapriya S, Tomar D, Siddiqui N, Lynch S, Nemani N, Breves SL, Zhang X, Tripathi A, Palaniappan P, Riitano MF, et al. Mitochondrial Ca(2+) uniporter is a mitochondrial luminal redox sensor that augments MCU channel activity. Mol Cell. 2017;65(1014–1028):e1017. doi: 10.1016/j.molcel.2017.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan PJ, Chandramoorthy HC, Hoffman NE, Zhang X, Cárdenas C, Shanmughapriya S, Rajan S, Vallem S, Chen X, Foskett JK, Cheung JY, et al. LETM1-dependent mitochondrial Ca2+ flux modulates cellular bioenergetics and proliferation. Faseb J. 2014;28:4936–4949. doi: 10.1096/fj.14-256453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durigon R, Mitchell AL, Jones AW, Manole A, Mennuni M, Hirst EM, Houlden H, Maragni G, Lattante S, Doronzio PN. LETM 1 couples mitochondrial DNA metabolism and nutrient preference. EMBO Mol Med. 2018;10:e8550. doi: 10.15252/emmm.201708550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endele S, Fuhry M, Pak SJ, Zabel BU, Winterpacht A. LETM1, a novel gene encoding a putative EF-hand Ca(2+)-binding protein, flanks the Wolf-Hirschhorn syndrome (WHS) critical region and is deleted in most WHS patients. Genomics. 1999;60:218–225. doi: 10.1006/geno.1999.5881. [DOI] [PubMed] [Google Scholar]
- Fan C, Fan M, Orlando BJ, Fastman NM, Zhang J, Xu Y, Chambers MG, Xu X, Perry K, Liao M, Feng L. X-ray and cryo-EM structures of the mitochondrial calcium uniporter. Nature. 2018;559:575–579. doi: 10.1038/s41586-018-0330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Zhang J, Tsai CW, Orlando BJ, Rodriguez M, Xu Y, Liao M, Tsai MF, Feng L. Structure and mechanism of the mitochondrial Ca(2+) uniporter holocomplex. Nature. 2020;582:129–133. doi: 10.1038/s41586-020-2309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foskett JK, Philipson B. The mitochondrial Ca(2+) uniporter complex. J Mol Cell Cardiol. 2015;78:3–8. doi: 10.1016/j.yjmcc.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AE, Taylor RD, Mick DU, Warscheid B, Stoepel N, Meyer HE, Ryan MT, Guiard B, Rehling P. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J Cell Biol. 2006;172:553–564. doi: 10.1083/jcb.200505060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sancho J. The coupling of plasma membrane calcium entry to calcium uptake by endoplasmic reticulum and mitochondria. J Physiol. 2014;592:261–268. doi: 10.1113/jphysiol.2013.255661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlid AO, Schaffer CT, Kim J, Bhatt H, Guevara-Gonzalez V, Ping P. TAZ encodes tafazzin, a transacylase essential for cardiolipin formation and central to the etiology of Barth syndrome. Gene. 2020;726:144–148. doi: 10.1016/j.gene.2019.144148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Basu Ball W, Madaris TR, Srikantan S, Madesh M, Mootha VK, Gohil VM. An essential role for cardiolipin in the stability and function of the mitochondrial calcium uniporter. Proc Natl Acad Sci. 2020;117:16383–16390. doi: 10.1073/pnas.2000640117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, Marchi S, Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol. 2018;19:713–730. doi: 10.1038/s41580-018-0052-8. [DOI] [PubMed] [Google Scholar]
- Gu L, Larson-Casey JL, Carter AB. Macrophages utilize the mitochondrial calcium uniporter for profibrotic polarization. FASEB J. 2017;31:3072–3083. doi: 10.1096/fj.201601371R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunter TE, Wingrove DE, Banerjee S, Gunter KK. Mechanisms of mitochondrial calcium transport, Cellular Ca2+ Regulation. Springer; 1988. pp. 1–14. [DOI] [PubMed] [Google Scholar]
- Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Decoding of cytosolic calcium oscillations in the mitochondria. Cell. 1995;82:415–424. doi: 10.1016/0092-8674(95)90430-1. [DOI] [PubMed] [Google Scholar]
- Hansford RG. Physiological role of mitochondrial Ca2+ transport. J Bioenerg Biomembr. 1994;26:495–508. doi: 10.1007/BF00762734. [DOI] [PubMed] [Google Scholar]
- Hasegawa A, vander Bliek AM. Inverse correlation between expression of the Wolfs Hirschhorn candidate gene Letm1 and mitochondrial volume in C. elegans and in mammalian cells. Hum Mol Genet. 2007;16:2061–2071. doi: 10.1093/hmg/ddm154. [DOI] [PubMed] [Google Scholar]
- Hashimi H, McDonald L, Stříbrná E, Lukeš J. Trypanosome Letm1 protein is essential for mitochondrial potassium homeostasis. J Biol Chem. 2013;288:26914–26925. doi: 10.1074/jbc.M113.495119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, O’Halloran DM. Analysis of the Na+/Ca2+ exchanger gene family within the phylum Nematoda. PLoS One. 2014;9:e112841. doi: 10.1371/journal.pone.0112841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman NE, Chandramoorthy HC, Shamugapriya S, Zhang X, Rajan S, Mallilankaraman K, Gandhirajan RK, Vagnozzi RJ, Ferrer LM, Sreekrishnanilayam K, Natarajaseenivasan K, et al. MICU1 motifs define mitochondrial calcium uniporter binding and activity. Cell Rep. 2013;5:1576–1588. doi: 10.1016/j.celrep.2013.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom KM, Pan X, Liu JC, Menazza S, Liu J, Nguyen TT, Pan H, Parks RJ, Anderson S, Noguchi A, Springer D, et al. Assessment of cardiac function in mice lacking the mitochondrial calcium uniporter. J Mol Cell Cardiol. 2015;85:178–182. doi: 10.1016/j.yjmcc.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Zhang J, Zhang X, Huang C, Hu G, Li S, Xie T, Liu M, Xu Y. Suppression of LETM1 by siRNA inhibits cell proliferation and invasion of bladder cancer cells. Oncol Rep. 2017;38:2935–2940. doi: 10.3892/or.2017.5959. [DOI] [PubMed] [Google Scholar]
- Jiang D, Zhao L, Clapham DE. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science. 2009;326:144–147. doi: 10.1126/science.1175145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Zhao L, Clish CB, Clapham DE. Letm1, the mitochondrial Ca2+/H+ antiporter, is essential for normal glucose metabolism and alters brain function in Wolf-Hirschhorn syndrome. Proc Natl Acad Sci USA. 2013;110:E2249–2254. doi: 10.1073/pnas.1308558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Wang J, Ji X, Cao H, Zhu J, Chen Y, Yang J, Zhao Z, Ren T, Xing J. MCUR facilitates epithelial-mesenchymal transition and metastasis via the mitochondrial calcium dependent ROS/Nrf2/Notch pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:136. doi: 10.1186/s13046-019-1135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner ML, Koval OM, Li J, He BJ, Allamargot C, Gao Z, Luczak ED, Hall DD, Fink BD, Chen B, Yang J, et al. CaMKII determines mitochondrial stress responses in heart. Nature. 2012;491:269–273. doi: 10.1038/nature11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer KJ, Grabarek Z, Mootha VK. High-affinity cooperative Ca(2+) binding by MICU1-MICU2 serves as an on-off switch for the uniporter. EMBO Rep. 2017;18:1397–1411. doi: 10.15252/embr.201643748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer KJ, Mootha VK. The molecular era of the mitochondrial calcium uniporter. Nat Rev Mol Cell Biol. 2015;16:545–553. doi: 10.1038/nrm4039. [DOI] [PubMed] [Google Scholar]
- Khananshvili D. Mechanism of partial reactions in the cardiac Na+-Ca2+ exchange systema A. Ann N Y Acad Sci. 1991;639:85–95. doi: 10.1111/j.1749-6632.1991.tb17291.x. [DOI] [PubMed] [Google Scholar]
- Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- Konig T, Troder SE, Bakka K, Korwitz A, Richter-Dennerlein R, Lampe PA, Patron M, Muhlmeister M, Guerrero-Castillo S, Brandt U, Decker T, et al. The m-AAA protease associated with neurodegeneration limits MCU activity in mitochondria. Mol Cell. 2016;64:148–162. doi: 10.1016/j.molcel.2016.08.020. [DOI] [PubMed] [Google Scholar]
- Kostic M, Katoshevski T, Sekler I. Allosteric regulation of NCLX by mitochondrial membrane potential links the metabolic state and Ca2+ signaling in mitochondria. Cell Rep. 2018;25(3465–3475):e3464. doi: 10.1016/j.celrep.2018.11.084. [DOI] [PubMed] [Google Scholar]
- Kostic M, Sekler I. Functional properties and mode of regulation of the mitochondrial Na(+)/Ca(2+) exchanger, NCLX. Semin Cell Dev Biol. 2019;94:59–65. doi: 10.1016/j.semcdb.2019.01.009. [DOI] [PubMed] [Google Scholar]
- Kwong JQ, Lu X, Correll RN, Schwanekamp JA, Vagnozzi RJ, Sargent MA, York AJ, Zhang J, Bers DM, Molkentin JD. The mitochondrial calcium uniporter selectively matches metabolic output to acute contractile stress in the heart. Cell Rep. 2015;12:15–22. doi: 10.1016/j.celrep.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rovere RM, Roest G, Bultynck G, Parys JB. Intracellular Ca2+ signaling and Ca2+ microdomains in the control of cell survival, apoptosis and autophagy. Cell Calcium. 2016;60:74–87. doi: 10.1016/j.ceca.2016.04.005. [DOI] [PubMed] [Google Scholar]
- Lambert JP, Luongo TS, Tomar D, Jadiya P, Gao E, Zhang X, Lucchese AM, Kolmetzky DW, Shah NS, Elrod JW. MCUB regulates the molecular composition of the mitochondrial calcium uniporter channel to limit mitochondrial calcium overload during stress. Circulation. 2019;140:1720–1733. doi: 10.1161/CIRCULATIONAHA.118.037968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Kang MG, Shin S, Kwak C, Kwon T, Seo JK, Kim JS, Rhee HW. Architecture mapping of the inner mitochondrial membrane proteome by chemical tools in live cells. J Am Chem Soc. 2017;139:3651–3662. doi: 10.1021/jacs.6b10418. [DOI] [PubMed] [Google Scholar]
- Lemasters JJ, Nieminen AL, Qian T, Trost LC, Elmore SP, Nishimura Y, Crowe RA, Cascio WE, Bradham CA, Brenner DA, Herman B. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim Biophys Acta. 1998;1366:177–196. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- Lewis-Smith D, Kamer KJ, Griffin H, Childs AM, Pysden K, Titov D, Duff J, Pyle A, Taylor RW, Yu-Wai-Man P, Ramesh V, et al. Homozygous deletion in MICU1 presenting with fatigue and lethargy in childhood. Neurol Genet. 2016;2:e59. doi: 10.1212/NXG.0000000000000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tran Q, Shrestha R, Piao L, Park S, Park J, Park J. LETM1 is required for mitochondrial homeostasis and cellular viability (Review) Mol Med Rep. 2019;19:3367–3375. doi: 10.3892/mmr.2019.10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin QT, Stathopulos PB. Molecular mechanisms of leucine zipper EF-hand containing transmembrane protein-1 function in health and disease. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JC, Liu J, Holmstrom KM, Menazza S, Parks RJ, Fergusson MM, Yu ZX, Springer DA, Halsey C, Liu C, Murphy E, et al. MICU1 serves as a molecular gatekeeper to prevent in vivo mitochondrial calcium overload. Cell Rep. 2016;16:1561–1573. doi: 10.1016/j.celrep.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JC, Syder NC, Ghorashi NS, Willingham TB, Parks RJ, Sun J, Fergusson MM, Liu J, Holmstrom KM, Menazza S, Springer DA, et al. EMRE is essential for mitochondrial calcium uniporter activity in a mouse model. JCI Insight. 2020;5 doi: 10.1172/jci.insight.134063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CV, Szabadkai G, Sharpe JA, Parry DA, Torelli S, Childs AM, Kriek M, Phadke R, Johnson CA, Roberts NY, Bonthron DT, et al. Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat Genet. 2014;46:188–193. doi: 10.1038/ng.2851. [DOI] [PubMed] [Google Scholar]
- Luongo TS, Lambert JP, Gross P, Nwokedi M, Lombardi AA, Shanmughapriya S, Carpenter AC, Kolmetzky D, Gao E, van Berlo JH, Tsai EJ, et al. The mitochondrial Na(+)/Ca (2+) exchanger is essential for Ca(2+) homeostasis and viability. Nature. 2017;545:93–97. doi: 10.1038/nature22082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luongo TS, Lambert JP, Yuan A, Zhang X, Gross P, Song J, Shanmughapriya S, Gao E, Jain M, Houser SR, Koch WJ, et al. the mitochondrial calcium uniporter matches energetic supply with cardiac workload during stress and modulates permeability transition. Cell Rep. 2015;12:23–34. doi: 10.1016/j.celrep.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maack C, Cortassa S, Aon MA, Ganesan AN, Liu T, O’Rourke B. Elevated cytosolic Na+ decreases mitochondrial Ca2+ uptake during excitation-contraction coupling and impairs energetic adaptation in cardiac myocytes. Circ Res. 2006;99:172–182. doi: 10.1161/01.RES.0000232546.92777.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madesh M, Hajnoczky G. VDAC-dependent permeabilization of the outer mitochondrial membrane by superoxide induces rapid and massive cytochrome c release. J Cell Biol. 2001;155:1003–1015. doi: 10.1083/jcb.200105057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Cardenas C, Doonan PJ, Chandramoorthy HC, Irrinki KM, Golenar T, Csordas G, Madireddi P, Yang J, Muller M, Miller R, et al. MCUR is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nat Cell Biol. 2012a;14:1336–1343. doi: 10.1038/ncb2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Doonan P, Cardenas C, Chandramoorthy HC, Muller M, Miller R, Hoffman NE, Gandhirajan RK, Molgo J, Birnbaum MJ, Rothberg BS, et al. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca(2+) uptake that regulates cell survival. Cell. 2012b;151:630–644. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Gherardi G, Rizzuto R. Structure, activity regulation, and role of the mitochondrial calcium uniporter in health and disease. Front Oncol. 2017;7:139. doi: 10.3389/fonc.2017.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Gherardi G, Zamparo I, Raffaello A, Boncompagni S, Chemello F, Cagnin S, Braga A, Zanin S, Pallafacchina G, Zentilin L, et al. The mitochondrial calcium uniporter controls skeletal muscle trophism in vivo. Cell Rep. 2015;10:1269–1279. doi: 10.1016/j.celrep.2015.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Raffaello A, Vecellio Reane D, Gherardi G, De Mario A, Rizzuto R. Mitochondrial calcium uptake in organ physiology: from molecular mechanism to animal models. Pflugers Arch. 2018;470:1165–1179. doi: 10.1007/s00424-018-2123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Raffaello A, Vecellio Reane D, Rizzuto R. Molecular structure and pathophysiological roles of the mitochondrial calcium uniporter. Biochim Biophys Acta. 2016;1863:2457–2464. doi: 10.1016/j.bbamcr.2016.03.006. [DOI] [PubMed] [Google Scholar]
- Marchi S, Lupini L, Patergnani S, Rimessi A, Missiroli S, Bonora M, Bononi A, Corra F, Giorgi C, De Marchi E, Poletti F, et al. Downregulation of the mitochondrial calcium uniporter by cancer-related miR-25. Curr Biol. 2013;23:58–63. doi: 10.1016/j.cub.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JG, Denton RM. The effects of calcium ions and adenine nucleotides on the activity of pig heart 2-oxoglutarate dehydrogenase complex. Biochem J. 1979;180:533–544. doi: 10.1042/bj1800533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JG, Halestrap AP, Denton RM. Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol Rev. 1990;70:391–425. doi: 10.1152/physrev.1990.70.2.391. [DOI] [PubMed] [Google Scholar]
- Mertins B, Psakis G, Essen LO. Voltage-dependent anion channels: the wizard of the mitochondrial outer membrane. Biol Chem. 2014;395:1435–1442. doi: 10.1515/hsz-2014-0203. [DOI] [PubMed] [Google Scholar]
- Messina A, Reina S, Guarino F, De Pinto V. VDAC isoforms in mammals. Biochim Biophys Acta. 2012;1818:1466–1476. doi: 10.1016/j.bbamem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Murphy E, Eisner DA. Regulation of intracellular and mitochondrial sodium in health and disease. Circ Res. 2009;104:292–303. doi: 10.1161/CIRCRESAHA.108.189050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa S, Eyaid W, Kamer K, Ali R, Al-Mureikhi M, Shahbeck N, Al Mesaifri F, Makhseed N, Mohamed Z, AlShehhi WA, Mootha VK, et al. A Middle Eastern Founder Mutation Expands the Genotypic and Phenotypic Spectrum of Mitochondrial MICU1 Deficiency: A Report of 13 Patients. JIMD Rep. 2019;43:79–83. doi: 10.1007/8904_2018_107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Matsui A, Akabane S, Tamura Y, Hatano A, Miyano Y, Omote H, Kajikawa M, Maenaka K, Moriyama Y. The mitochondrial inner membrane protein LETM1 modulates cristae organization through its LETM domain. Commun Biol. 2020;3:1–11. doi: 10.1038/s42003-020-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemani N, Dong Z, Daw CC, Madaris TR, Ramachandran K, Enslow BT, Rubannelsonkumar CS, Shanmughapriya S, Mallireddigari V, Maity S, SinghMalla P, et al. Mitochondrial pyruvate and fatty acid flux modulate MICU1-dependent control of MCU activity. Sci Signal. 2020;13 doi: 10.1126/scisignal.aaz6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemani N, Shanmughapriya S, Madesh M. Molecular regulation of MCU: Implications in physiology and disease. Cell Calcium. 2018;74:86–93. doi: 10.1016/j.ceca.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NX, Armache J-P, Lee C, Yang Y, Zeng W, Mootha VK, Cheng Y, Bai X-C, Jiang Y. Cryo-EM structure of a fungal mitochondrial calcium uniporter. Nature. 2018;559:570–574. doi: 10.1038/s41586-018-0333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaeva MA, Mukherjee B, Stys PK. Na+-dependent sources of intra-axonal Ca2+ release in rat optic nerve during in vitro chemical ischemia. J Neurosci. 2005;25:9960–9967. doi: 10.1523/JNEUROSCI.2003-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita II, Hershfinkel M, Fishman D, Ozeri E, Rutter GA, Sensi SL, Khananshvili D, Lewis EC, Sekler I. The mitochondrial Na+/Ca 2+ exchanger upregulates glucose dependent Ca 2+ signalling linked to insulin secretion. PLoS One. 2012;7:e46649. doi: 10.1371/journal.pone.0046649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita LI, Hershfinkel M, Sekler I. Life after the birth of the mitochondrial Na+/ Ca2+ exchanger, NCLX. Sci China Life Sci. 2015;58:59–65. doi: 10.1007/s11427-014-4789-9. [DOI] [PubMed] [Google Scholar]
- Nowikovsky K, Pozzan T, Rizzuto R, Scorrano L, Bernardi P. The pathophysiology of LETM1. J Gen Physiol. 2012;139:445–454. doi: 10.1085/jgp.201110757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenoid K, Dong Y, Cao C, Cui T, Sancak Y, Markhard AL, Grabarek Z, Kong L, Liu Z, Ouyang B, Cong Y, et al. Architecture of the mitochondrial calcium uniporter. Nature. 2016;533:269–273. doi: 10.1038/nature17656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard M, Csordas G, Huang KT, Varnai P, Joseph SK, Hajnoczky G. MICU1 interacts with the D-Ring of the MCU pore to control its Ca(2+) flux and sensitivity to Ru360. Mol Cell. 2018;72(778–785):e773. doi: 10.1016/j.molcel.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallafacchina G, Zanin S, Rizzuto R. Recent advances in the molecular mechanism of mitochondrial calcium uptake. F1000Res. 2018;7 doi: 10.12688/f1000research.15723.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palty R, Ohana E, Hershfinkel M, Volokita M, Elgazar V, Beharier O, Silverman WF, Argaman M, Sekler I. Lithium-calcium exchange is mediated by a distinct potassium-independent sodium-calcium exchanger. J Biol Chem. 2004;279:25234–25240. doi: 10.1074/jbc.M401229200. [DOI] [PubMed] [Google Scholar]
- Palty R, Silverman WF, Hershfinkel M, Caporale T, Sensi SL, Parnis J, Nolte C, Fishman D, Shoshan-Barmatz V, Herrmann S. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc Natl Acad Sci. 2010;107:436–441. doi: 10.1073/pnas.0908099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Liu J, Nguyen T, Liu C, Sun J, Teng Y, Fergusson MM, Rovira II, Allen M, Springer DA, Aponte AM, et al. The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat Cell Biol. 2013;15:1464–1472. doi: 10.1038/ncb2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee Y, Park T, Kang JY, Mun SA, Jin M, Yang J, Eom SH. Structure of the MICU1-MICU2 heterodimer provides insights into the gatekeeping threshold shift. IUCrJ. 2020;7:355–365. doi: 10.1107/S2052252520001840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnis J, Montana V, Delgado-Martinez I, Matyash V, Parpura V, Kettenmann H, Sekler I, Nolte C. Mitochondrial exchanger NCLX plays a major role in the intracellular Ca2+ signaling, gliotransmission, and proliferation of astrocytes. J Neurosci. 2013;33:7206–7219. doi: 10.1523/JNEUROSCI.5721-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak T, Gueguinou M, Walter V, Delierneux C, Johnson MT, Zhang X, Xin P, Yoast RE, Emrich SM, Yochum GS, Sekler I, et al. Dichotomous role of the human mitochondrial Na(+)/Ca2(+)/Li(+) exchanger NCLX in colorectal cancer growth and metastasis. eLife. 2020;9 doi: 10.7554/eLife.59686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak T, Trebak M. Mitochondrial Ca(2+) signaling. Pharmacol Ther. 2018;192:112–123. doi: 10.1016/j.pharmthera.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron M, Checchetto V, Raffaello A, Teardo E, Vecellio Reane D, Mantoan M, Granatiero V, Szabo I, De Stefani D, Rizzuto R. MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Mol Cell. 2014;53:726–737. doi: 10.1016/j.molcel.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron M, Granatiero V, Espino J, Rizzuto R, De Stefani D. MICU3 is a tissue-specific enhancer of mitochondrial calcium uptake. Cell Death Differ. 2019;26:179–195. doi: 10.1038/s41418-018-0113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paupe V, Prudent J. New insights into the role of mitochondrial calcium homeostasis in cell migration. Biochem Biophys Res Commun. 2018;500:75–86. doi: 10.1016/j.bbrc.2017.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paupe V, Prudent J, Dassa EP, Rendon OZ, Shoubridge EA. CCDC90A (MCUR) is a cytochrome c oxidase assembly factor and not a regulator of the mitochondrial calcium uniporter. Cell Metab. 2015;21:109–116. doi: 10.1016/j.cmet.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK. MICU1 encodes a mitochondrial EF hand protein required for Ca 2+ uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrungaro C, Zimmermann KM, Kuttner V, Fischer M, Dengjel J, Bogeski I, Riemer J. The Ca(2+)-dependent release of the Mia40-Induced MICU1-MICU2 dimer from MCU regulates mitochondrial Ca(2+) uptake. Cell Metab. 2015;22:721–733. doi: 10.1016/j.cmet.2015.08.019. [DOI] [PubMed] [Google Scholar]
- Piao L, Li Y, Kim SJ, Byun HS, Huang SM, Hwang SK, Yang KJ, Park KA, Won M, Hong J, Hur GM, et al. Association of LETM1 and MRPL36 contributes to the regulation of mitochondrial ATP production and necrotic cell death. Cancer Res. 2009;69:3397–3404. doi: 10.1158/0008-5472.CAN-08-3235. [DOI] [PubMed] [Google Scholar]
- Plovanich M, Bogorad RL, Sancak Y, Kamer KJ, Strittmatter L, Li AA, Girgis HS, Kuchimanchi S, De Groot J, Speciner L, Taneja N, et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One. 2013;8:e55785. doi: 10.1371/journal.pone.0055785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent J, Popgeorgiev N, Bonneau B, Thibaut J, Gadet R, Lopez J, Gonzalo P, Rimokh R, Manon S, Houart C, Herbomel P, et al. Bcl-wav and the mitochondrial calcium uniporter drive gastrula morphogenesis in zebrafish. Nat Commun. 2013;4:2330. doi: 10.1038/ncomms3330. [DOI] [PubMed] [Google Scholar]
- Purroy R, Britti E, Delaspre F, Tamarit J, Ros J. Mitochondrial pore opening and loss of Ca2+ exchanger NCLX levels occur after frataxin depletion. Biochim Biophys Acta (BBA) Mol Basis Dis. 2018;1864:618–631. doi: 10.1016/j.bbadis.2017.12.005. [DOI] [PubMed] [Google Scholar]
- Puskin JS, Gunter TE, Gunter KK, Russell PR. Evidence for more than one C2+ ion transport mechanism in mitochondria. Biochemistry. 1976;15:3834–3842. doi: 10.1021/bi00662a029. [DOI] [PubMed] [Google Scholar]
- Raffaello A, De Stefani D, Sabbadin D, Teardo E, Merli G, Picard A, Checchetto V, Moro S, Szabo I, Rizzuto R. The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 2013;32:2362–2376. doi: 10.1038/emboj.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapizzi E, Pinton P, Szabadkai G, Wieckowski MR, Vandecasteele G, Baird G, Tuft RA, Fogarty KE, Rizzuto R. Recombinant expression of the voltagedependent anion channel enhances the transfer of Ca2+ microdomains to mitochondria. J Cell Biol. 2002;159:613–624. doi: 10.1083/jcb.200205091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasola A, Bernardi P. Mitochondrial permeability transition in Ca(2+)-dependent apoptosis and necrosis. Cell Calcium. 2011;50:222–233. doi: 10.1016/j.ceca.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Ren T, Wang J, Zhang H, Yuan P, Zhu J, Wu Y, Huang Q, Guo X, Zhang J, Ji L, Li J, et al. MCUR-Mediated mitochondrial calcium signaling facilitates cell survival of hepatocellular carcinoma via reactive oxygen species-dependent P53 degradation. Antioxid Redox Signal. 2018;28:1120–1136. doi: 10.1089/ars.2017.6990. [DOI] [PubMed] [Google Scholar]
- Rimessi A, Bezzerri V, Patergnani S, Marchi S, Cabrini G, Pinton P. Mitochondrial Ca2+-dependent NLRP3 activation exacerbates the Pseudomonas aeruginosa-driven inflammatory response in cystic fibrosis. Nat Commun. 2015;6:6201. doi: 10.1038/ncomms7201. [DOI] [PubMed] [Google Scholar]
- Rimessi A, Giorgi C, Pinton P, Rizzuto R. The versatility of mitochondrial calcium signals: from stimulation of cell metabolism to induction of cell death. Biochim Biophys Acta. 2008;1777:808–816. doi: 10.1016/j.bbabio.2008.05.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Murgia M, Pozzan T. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 1993a;262:744–747. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Simpson AW, Brini M, Pozzan T. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature. 1992;358:325–327. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- Roy S, Dey K, Hershfinkel M, Ohana E, Sekler I. Identification of residues that control Li+ versus Na+ dependent Ca2+ exchange at the transport site of the mitochondrial NCLX. Biochim Biophys Acta (BBA) Mol Cell Res. 2017;1864:997–1008. doi: 10.1016/j.bbamcr.2017.01.011. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Markhard AL, Kitami T, Kovacs-Bogdan E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA, Calvo SE, et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342:1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmughapriya S, Rajan S, Hoffman NE, Zhang X, Guo S, Kolesar JE, Hines KJ, Ragheb J, Jog NR, Caricchio R, Baba Y, et al. Ca2+ signals regulate mitochondrial metabolism by stimulating CREB-mediated expression of the mitochondrial Ca2+ uniporter gene MCU. SciSignal. 2015;8:ra23. doi: 10.1126/scisignal.2005673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Fu Z, Ji Y, Guan X, Guo S, Ding Z, Yang X, Cong Y, Shen Y. Leucine zipper-EF-hand containing transmembrane protein 1 (LETM1) forms a Ca 2 +/H+ antiporter. Sci Rep. 2016;6:34174. doi: 10.1038/srep34174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Arora S, Saurav S, Motiani RK. Pathophysiological Significance of Calcium Signaling at Mitochondria-Associated Endoplasmic Reticulum Membranes (MAMs) Curr Opin Physiol. 2020;17:234–242. [Google Scholar]
- Shoshan-Barmatz V, Gincel D. The voltage-dependent anion channel: characterization, modulation, and role in mitochondrial function in cell life and death. Cell Biochem Biophys. 2003;39:279–292. doi: 10.1385/CBB:39:3:279. [DOI] [PubMed] [Google Scholar]
- Shoshan-Barmatz V, Krelin Y, Shteinfer-Kuzmine A. VDAC1 functions in Ca(2 +) homeostasis and cell life and death in health and disease. Cell Calcium. 2018;69:81–100. doi: 10.1016/j.ceca.2017.06.007. [DOI] [PubMed] [Google Scholar]
- Shoshan-Barmatz V, Mizrachi D. VDAC1: from structure to cancer therapy. Front Oncol. 2012;2:164. doi: 10.3389/fonc.2012.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez J, Cividini F, Scott BT, Lehmann K, Diaz-Juarez J, Diemer T, Dai A, Suarez JA, Jain M, Dillmann WH. Restoring mitochondrial calcium uniporter expression in diabetic mouse heart improves mitochondrial calcium handling and cardiac function. J Biol Chem. 2018;293:8182–8195. doi: 10.1074/jbc.RA118.002066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Varnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175:901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai S, Iida H, Yokota S, Sayano T, Kiguchiya S, Ishihara N, Hayashi J-I, Mihara K, Oka T. Characterization of the mitochondrial protein LETM1, which maintains the mitochondrial tubular shapes and interacts with the AAA-ATPase BCS1L. J Cell Sci. 2008;121:2588–2600. doi: 10.1242/jcs.026625. [DOI] [PubMed] [Google Scholar]
- Tan W, Colombini M. VDAC closure increases calcium ion flux. Biochim Biophys Acta. 2007;1768:2510–2515. doi: 10.1016/j.bbamem.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Wang X, Shen Q, Yang X, Yu C, Cai C, Cai G, Meng X, Zou F. Mitochondrial Ca(2)(+) uniporter is critical for store-operated Ca(2)(+) entry-dependent breast cancer cell migration. Biochem Biophys Res Commun. 2015;458:186–193. doi: 10.1016/j.bbrc.2015.01.092. [DOI] [PubMed] [Google Scholar]
- Tanwar J, Motiani RK. Role of SOCE architects STIM and orai proteins in cell death. Cell Calcium. 2018;69:19–27. doi: 10.1016/j.ceca.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Tarasov AI, Semplici F, Ravier MA, Bellomo EA, Pullen TJ, Gilon P, Sekler I, Rizzuto R, Rutter GA. The mitochondrial Ca2+ uniporter MCU is essential for glucose-induced ATP increases in pancreatic beta-cells. PLoS One. 2012;7:e39722. doi: 10.1371/journal.pone.0039722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar D, Dong Z, Shanmughapriya S, Koch DA, Thomas T, Hoffman NE, Timbalia SA, Goldman SJ, Breves SL, Corbally DP, Nemani N, et al. MCUR is a Scaffold Factor for the MCU complex function and promotes mitochondrial bioenergetics. Cell Rep. 2016;15:1673–1685. doi: 10.1016/j.celrep.2016.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosatto A, Sommaggio R, Kummerow C, Bentham RB, Blacker TS, Berecz T, Duchen MR, Rosato A, Bogeski I, Szabadkai G, Rizzuto R, et al. The mitochondrial calcium uniporter regulates breast cancer progression via HIF-1alpha. EMBO Mol Med. 2016;8:569–585. doi: 10.15252/emmm.201606255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CW, Wu Y, Pao PC, Phillips CB, Williams C, Miller C, Ranaghan M, Tsai MF. Proteolytic control of the mitochondrial calcium uniporter complex. Proc Natl Acad Sci USA. 2017;114:4388–4393. doi: 10.1073/pnas.1702938114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MF, Jiang D, Zhao L, Clapham D, Miller C. Functional reconstitution of the mitochondrial Ca2+/H+ antiporter Letm1. J Gen Physiol. 2014;143:67–73. doi: 10.1085/jgp.201311096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MF, Phillips CB, Ranaghan M, Tsai CW, Wu Y, Willliams C, Miller C. Dual functions of a small regulatory subunit in the mitochondrial calcium uniporter complex. Elife. 2016;5 doi: 10.7554/eLife.15545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe S, Rangel P, Pardo J. Interactions of calcium with yeast mitochondria. Cell Calcium. 1992;13:211–217. doi: 10.1016/0143-4160(92)90009-h. [DOI] [PubMed] [Google Scholar]
- Vais H, Mallilankaraman K, Mak DD, Hoff H, Payne R, Tanis JE, Foskett JK. EMRE is a matrix Ca(2+) sensor that governs gatekeeping of the mitochondrial Ca(2+) uniporter. Cell Rep. 2016;14:403–410. doi: 10.1016/j.celrep.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vais H, Tanis JE, Muller M, Payne R, Mallilankaraman K, Foskett JK. MCUR, CCDC90A, is a regulator of the mitochondrial calcium uniporter. Cell Metab. 2015;22:533–535. doi: 10.1016/j.cmet.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasington FD, Murphy JV. Ca++ uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J Biol Chem. 1962;237:2670–2677. [PubMed] [Google Scholar]
- Vecellio Reane D, Rizzuto R, Raffaello A. The ER-mitochondria tether at the hub of Ca2+ signaling. Curr Opin Physiol. 2020;17:261–268. [Google Scholar]
- Vecellio Reane D, Vallese F, Checchetto V, Acquasaliente L, Butera G, De Filippis V, Szabo I, Zanotti G, Rizzuto R, Raffaello A. A MICU1 splice variant confers high sensitivity to the mitochondrial Ca(2+) uptake machinery of skeletal muscle. Mol Cell. 2016;64:760–773. doi: 10.1016/j.molcel.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Vercesi AE, Docampo R. Ca2+ transport by digitonin-permeabilized Leishmania donovani. Effects of Ca2+, pentamidine and WR-6026 on mitochondrial membrane potential in situ. Biochem J. 1992;284(Pt 2):463–467. doi: 10.1042/bj2840463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M, Callio J, Otero PA, Sekler I, Wills ZP, Chu CT. Mitochondrial calcium dysregulation contributes to dendrite degeneration mediated by PD/LBD-associated LRRK2 mutants. J Neurosci. 2017;37:11151–11165. doi: 10.1523/JNEUROSCI.3791-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Jacewicz A, Delgado BD, Baradaran R, Long SB. Structures reveal gatekeeping of the mitochondrial Ca(2+) uniporter by MICU1-MICU2. eLife. 2020a;9 doi: 10.7554/eLife.59991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Han Y, She J, Nguyen NX, Mootha VK, Bai XC, Jiang Y. Structural insights into the Ca(2+)-dependent gating of the human mitochondrial calcium uniporter. eLife. 2020b;9 doi: 10.7554/eLife.60513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nguyen NX, She J, Zeng W, Yang Y, Bai XC, Jiang Y. Structural mechanism of EMRE-dependent gating of the human mitochondrial calcium uniporter. Cell. 2019;177:1252–1261.:e1213. doi: 10.1016/j.cell.2019.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingrove DE, Gunter TE. Kinetics of mitochondrial calcium transport. II. A kinetic description of the sodium-dependent calcium efflux mechanism of liver mitochondria and inhibition by ruthenium red and by tetraphenylphosphonium. J Biol Chem. 1986;261:15166–15171. [PubMed] [Google Scholar]
- Wu W, Shen Q, Lei Z, Qiu Z, Li D, Pei H, Zheng J, Jia Z. The crystal structure of MICU2 provides insight into Ca(2+) binding and MICU1-MICU2 heterodimer formation. EMBO Rep. 2019;20:e47488. doi: 10.15252/embr.201847488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Rasmussen TP, Koval OM, Joiner ML, Hall DD, Chen B, Luczak ED, Wang Q, Rokita AG, Wehrens XH, Song LS, et al. The mitochondrial uniporter controls fight or flight heart rate increases. Nat Commun. 2015;6:6081. doi: 10.1038/ncomms7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Yamagoshi R, Harada K, Kawano M, Minami N, Ido Y, Kuwahara K, Fujita A, Ozono M, Watanabe A, Yamada A, et al. Analysis of the structure and function of EMRE in a yeast expression system. Biochim Biophys Acta. 2016;1857:831–839. doi: 10.1016/j.bbabio.2016.03.019. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ni W, Cui C, Qi W, Piao L, Xuan Y. Identification of LETM1 as a marker of cancer stem-like cells and predictor of poor prognosis in esophageal squamous cell carcinoma. Hum Pathol. 2018;81:148–156. doi: 10.1016/j.humpath.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Ying WL, Emerson J, Clarke MJ, Sanadi DR. Inhibition of mitochondrial calcium ion transport by an oxo-bridged dinuclear ruthenium ammine complex. Biochemistry. 1991;30:4949–4952. doi: 10.1021/bi00234a016. [DOI] [PubMed] [Google Scholar]
- Yoo J, Wu M, Yin Y, Herzik MA, Lander GC, Lee S-Y. Cryo-EM structure of a mitochondrial calcium uniporter. Science. 2018;361:506–511. doi: 10.1126/science.aar4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampieri S, Mammucari C, Romanello V, Barberi L, Pietrangelo L, Fusella A, Mosole S, Gherardi G, Hofer C, Lofler S, Sarabon N, et al. Physical exercise in aging human skeletal muscle increases mitochondrial calcium uniporter expression levels and affects mitochondria dynamics. Physiol Rep. 2016;4 doi: 10.14814/phy2.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Chen X, Cui W, Wen W, Lu F, Sun X, Ma D, Yuan Y, Li Z, Hou N, Zhao H, et al. RIPK1 Binds MCU to mediate induction of mitochondrial Ca(2+) uptake and promotes colorectal oncogenesis. Cancer Res. 2018;78:2876–2885. doi: 10.1158/0008-5472.CAN-17-3082. [DOI] [PubMed] [Google Scholar]
- Zhang B, Carrie C, Ivanova A, Narsai R, Murcha MW, Duncan O, Wang Y, Law SR, Albrecht V, Pogson B. LETM proteins play a role in the accumulation of mitochondrially encoded proteins in Arabidopsis thaliana and AtLETM2 displays parent of origin effects. J Biol Chem. 2012;287:41757–41773. doi: 10.1074/jbc.M112.383836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo W, Zhou H, Guo R, Yi J, Zhang L, Yu L, Sui Y, Zeng W, Wang P, Yang M. Structure of intact human MCU supercomplex with the auxiliary MICU subunits. Protein Cell. 2020 doi: 10.1007/s13238-020-00776-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulkifli M, Neff JK, Timbalia SA, Garza NM, Chen Y, Watrous JD, Murgia M, Trivedi PP, Anderson SK, Tomar D, Nilsson R, et al. Yeast homologs of human MCUR1 regulate mitochondrial proline metabolism. Nat Commun. 2020;11:4866. doi: 10.1038/s41467-020-18704-1. [DOI] [PMC free article] [PubMed] [Google Scholar]