Abstract
Cell competition is emerging as a quality control mechanism that eliminates unfit cells in a wide range of settings from development to the adult. However, the nature of the cells normally eliminated by cell competition and what triggers their elimination remains poorly understood. In mice, 35% of epiblast cells are eliminated prior to gastrulation. Here we show that cells with mitochondrial defects are eliminated by cell competition during early mouse development. Using single cell transcriptional profiling of eliminated mouse epiblast cells we identify hallmarks of cell competition and mitochondrial defects. We go on to demonstrate that mitochondrial defects are common to a range of different loser cell types and that manipulating mitochondrial function triggers cell competition. In the mouse embryo, cell competition eliminates cells with sequence changes in mt-Rnr1 and mt-Rnr2, and that even non-pathological changes in mitochondrial DNA sequence can induce cell competition. Our results suggest that cell competition is a purifying selection that optimises mitochondrial performance prior to gastrulation.
Introduction
Cell competition is a fitness sensing mechanism that eliminates cells that, although viable, are less fit than their neighbours. The cells that are eliminated are generically termed losers, while the fitter cells that survive are referred to as winners. Cell competition has been shown to act in a broad range of settings, from the developing embryo to the ageing organisms1–3. It has been primarily studied in Drosophila, where it was first described in the imaginal wing disc 4. Since then, it has also been found to be conserved in mammals. In the mouse embryo 35% of embryonic cells are eliminated between E5.5 and E6.5 and there is strong evidence that this elimination is through cell competition5–7. These and other studies identified a number of readouts of cell competition in the mouse embryo, such as relative low c-MYC expression, a loss of mTOR signalling, low TEAD activity, high P53 expression, or elevated levels of ERK phosphorylation 5–9. Importantly, there is a significant overlap with the markers of cell competition originally identified in Drosophila as well as those found in other cell competition models, such as Madin-Darby Canine Kidney (MDCK) cells – as reviewed1–3. In spite of the advance that having these cell competition markers signifies, given that they were primarily identified by using genetic models that rely on over-expression or mutation, we still have little insight into the over-arching features of the cells that are eliminated in the physiological context.
Mitochondria, with their diverse cellular functions ranging from determining the bioenergetic output of the cell to regulating its apoptotic response, are strong candidates for determining competitive cell fitness. During early mouse development mitochondria undergo profound changes in their shape and activity10. In the pre-implantation embryo mitochondria are rounded, fragmented and contain sparse cristae, but upon implantation they fuse to form complex networks with mature cristae11. The mode of replication of the mitochondrial genome (mtDNA), that encodes for vital components of the bioenergetic machinery, also changes during early mouse development. After fertilization, mtDNA replication ceases and its copy number per cell decreases with every division until post-implantation stages, when mtDNA replication resumes10. As the mutation rate of mtDNA is significantly higher than that of nuclear DNA12, 13, this increased replication most likely leads to an increased mutation load. In fact, inheritable mtDNA based diseases are reported with a prevalence of 5-15 cases per 100,000 individuals14, 15. A number of mechanisms have been proposed to reduce this mutation load, such as the bottleneck effect, purifying selection or biased segregation of mtDNA haplotypes16–21. However, how these mechanisms act at the molecular and cellular level is still poorly understood.
To understand the nature of the cells eliminated during early mouse post-implantation development, we have analysed their transcriptional profile by single-cell RNA sequencing and found that these cells share a cell competition signature. Analysis of the pathways mis-regulated identified mitochondrial dysfunction as a common feature. Importantly, our studies also found evidence of mtDNA mutations in the cells eliminated. Furthermore, we demonstrate that manipulating mitochondrial activity either by disrupting mitochondrial dynamics or by introducing non-pathological mtDNA changes is sufficient to trigger cell competition. These results therefore pinpoint mitochondrial performance as a key cellular feature that determines the competitive ability of embryonic cells and suggest that cell competition is acting as a purifying selection during early mammalian development.
Results
Loser cells have a distinct transcriptional profile
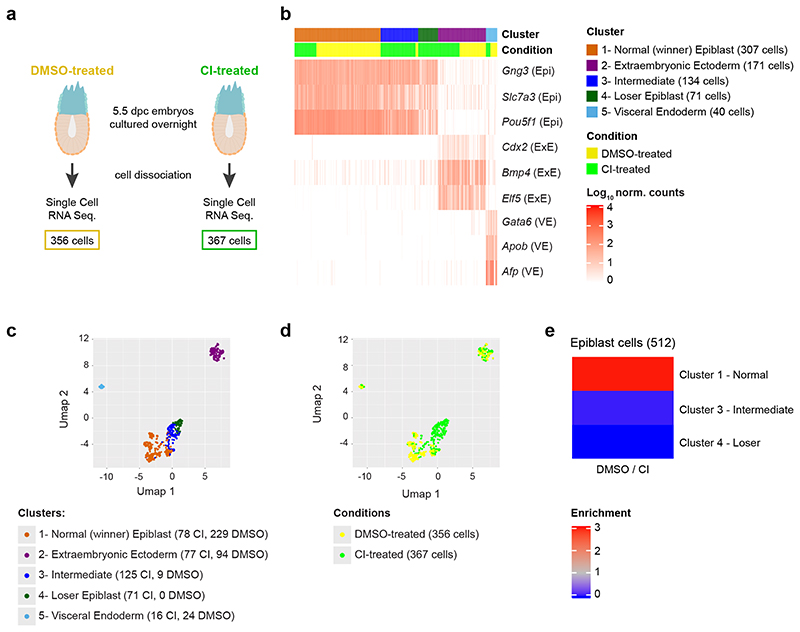
We have previously shown that in the early post-implantation mouse embryo about 35% of epiblast cells are eliminated and that these cells are marked by low mTOR signalling7. However, we currently do not understand the characteristics of these cells or what triggers their elimination. To answer these questions, we have analysed their transcriptional profile by single cell RNA sequencing (scRNA-seq). To ensure we can capture the eliminated cells, as we have done before7, we isolated embryos at E5.5 and cultured them for 16 hours in the presence of a caspase inhibitors (CI) or vehicle (DMSO) (Fig. 1a). Unsupervised clustering of the scRNA-seq data revealed five clusters: two corresponding to extra-embryonic tissues (visceral endoderm and extra-embryonic ectoderm) and three that expressed epiblast marker genes (Fig. 1b-c, Extended Data Fig. 1a-f and Methods). Interestingly, cells from CI- and DMSO-treated embryos are unequally distributed across the three epiblast clusters. In particular, one of these clusters (cluster 4) is only composed of cells from CI-treated embryos (Fig. 1d-e). It is worth noting that all epiblast clusters contained cells in G2/M and S phases of the cell cycle, suggesting they are all cycling (Extended Data Fig. 2a).
Fig. 1. Cells eliminated during early mouse embryogenesis have a distinct transcriptional profile.
a, Experimental design. The number of cells in the two conditions (DMSO-treated and CI-treated) refers to the number of cells that passed the quality control. b, Identification of the clusters according to known gene markers from the different embryonic regions56. Three clusters (clusters 1, 3 and 4) show marker genes of the epiblast (Epi), while the remaining clusters correspond to the extra-embryonic lineages visceral endoderm (VE; cluster 5) and extraembryonic ectoderm (ExE; cluster 2). The epiblast clusters are named “Winner”, “Intermediate” and “Loser” on the basis of the relative fraction of cells from CI-treated embryos they include (see panel e). c,d, UMAP visualization of the single-cell RNA-seq data, with cells coloured according to cluster (c) or condition (d). A region made up exclusively by cells from CI-treated embryos emerges. e, Ratio between the fraction of cells from DMSO-treated and CI-treated embryos in the three epiblast clusters. While the “winner” epiblast cluster shows an enrichment of cells from DMSO-treated embryos, the “intermediate” and the “loser” epiblast clusters are strongly enriched for cells from CI-treated embryos.
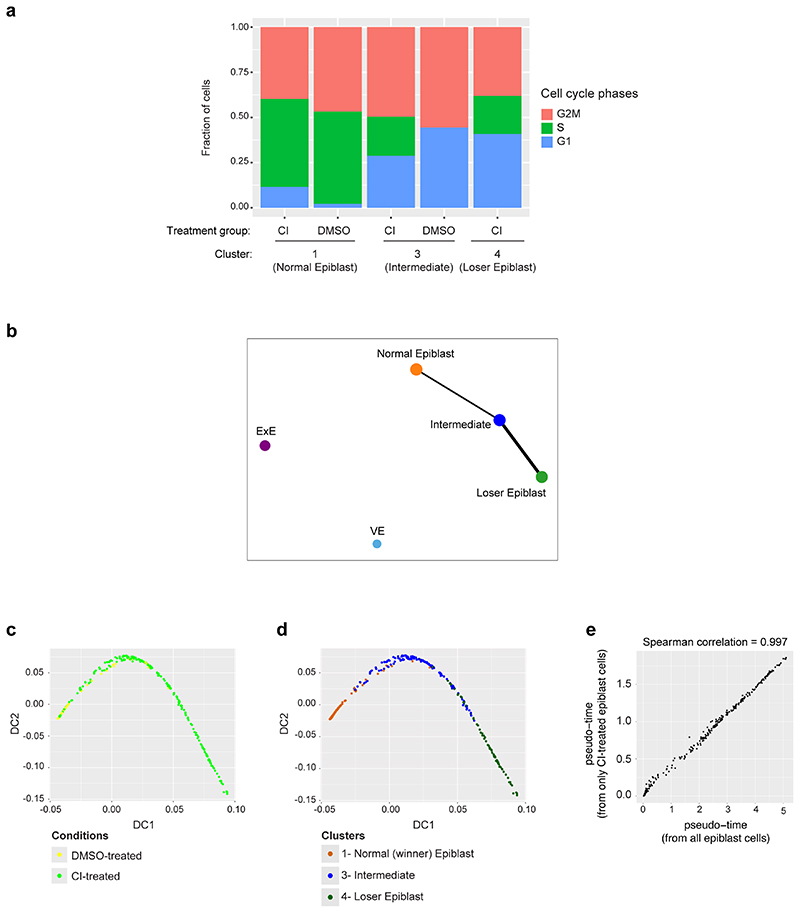
The three epiblast clusters are highly connected, as highlighted by a connectivity analysis carried out with PAGA22 (Extended Data Fig. 2b). Hence, to establish the relationship between these epiblast clusters we computed a diffusion map23. For this, we selected only cells captured from CI-treated embryos, to eliminate possible confounding effects due to the caspase inhibitor (Fig. 2a). However, when all epiblast cells are considered, the results remain unchanged (Extended Data Fig. 2c-e). This analysis identified a trajectory between the three epiblast clusters, with those cells unique to CI-treated embryos falling at one extreme end of the trajectory (corresponding to cluster 4; Fig. 2a) and with those cells present in both DMSO and CI-treated embryos at the other (corresponding to cluster 1; Fig. 2a and Extended Data Fig. 2d).
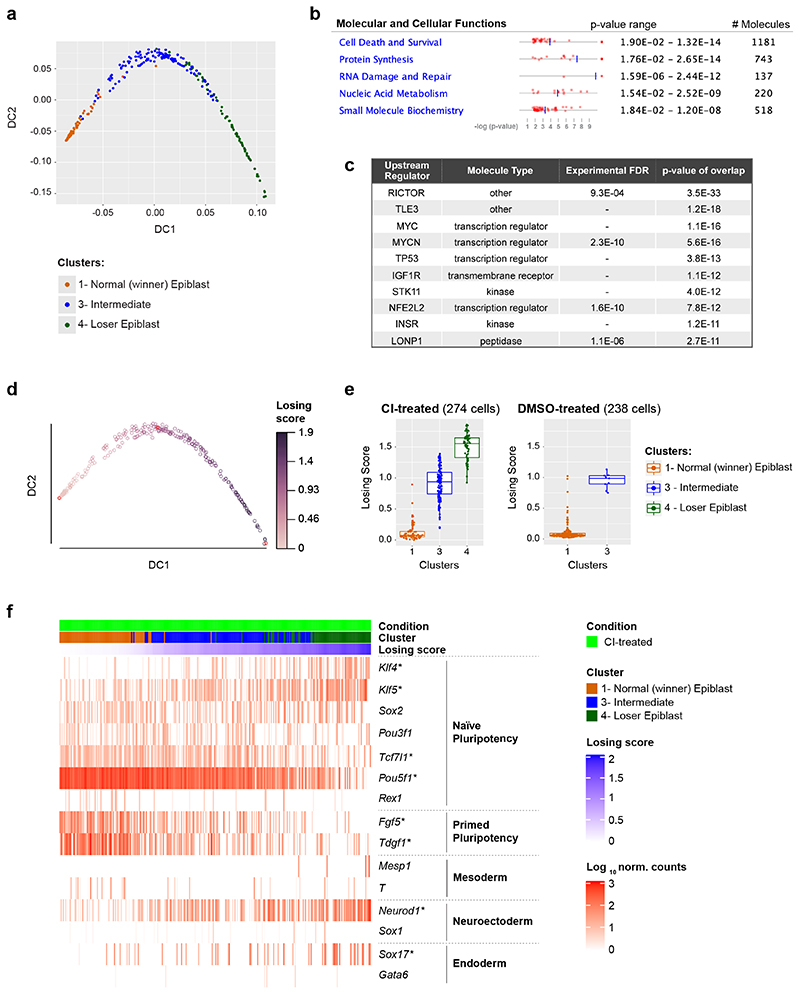
Fig. 2. A cell competition transcriptional signature is identified in cells eliminated during mouse embryonic development.
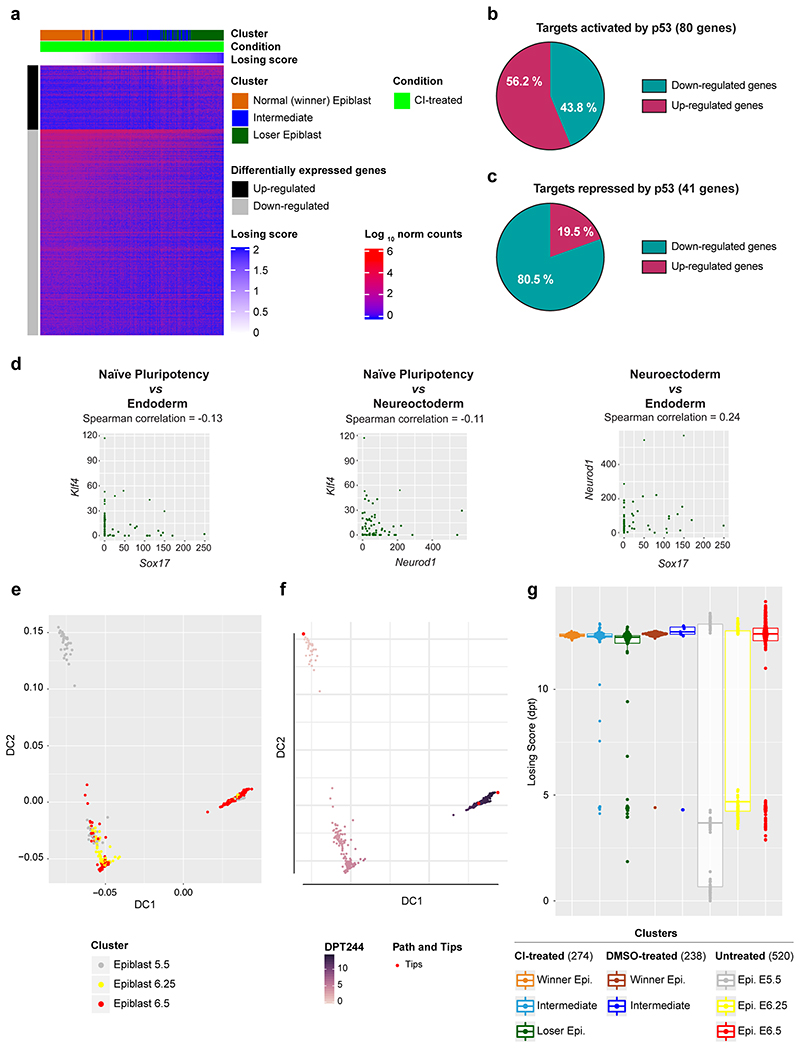
a, Diffusion map of epiblast cells (only from CI-treated embryos), coloured by cluster. b, c, IPA run on the list of genes differentially expressed along the diffusion trajectory (see Extended Data Fig. 2a) generated lists of top 5 molecular and cellular functions (b) and upstream regulators (c) found to be differentially activated in epiblast cells along the diffusion trajectory from winner (cluster 1) to loser status (cluster 4). d, Diffusion map of epiblast cells (only from CI-treated embryos) coloured by diffusion pseudotime coordinate (dpt). The winner and the loser clusters are found at the two extremities of the trajectory, hence the dpt can be interpreted as a “losing score”. e, Losing score of the cells in the three epiblast clusters in CI-treated (left) or DMSO-treated (right) embryos. The losing score of the cells from DMSO-treated embryos was obtained by projecting them on the diffusion map shown in panel d (see Methods). f, Expression levels in epiblast cells from CI-treated embryos of genes (in rows) that are markers for naïve pluripotency (Klf4, Klf5, Sox2, Pou3f1, Tcf7l1 and Pou5f1 and Rex1), primed pluripotency (Fgf5 and Tdgf1), mesoderm (Mesp1 and T), neuroectoderm (Neurod1 and Sox1) and endoderm (Sox17 and Gata6). Cells (in columns) are sorted by their losing scores. The genes marked with a * are differentially expressed along the trajectory. See methods for details on statistical analysis.
To further define the identity of the epiblast cells of CI-treated embryos we analysed the genes differentially expressed along the trajectory (see Methods and Extended Data Fig. 3a) using Ingenuity Pathway Analysis (IPA) to characterize gene signatures24. Importantly, we found that these differentially expressed genes fell under molecular and cellular function categories associated with cell death and survival, protein synthesis and nucleic acids (Fig. 2b). Analysis of the factors with enriched targets within the genes differentially expressed along the trajectory revealed RICTOR (an mTOR component), TLE3, MYC, MYCN, P53 and IGFR (that is upstream of mTOR) as the top upstream regulators (Fig. 2c). Breaking down the differentially expressed genes into those down-regulated or up-regulated along the winner-to loser trajectory revealed that the targets of RICTOR, MYC, MYCN and IGFR primarily fell within the down-regulated genes (Supplementary Tables 1 and 2). P53 activated targets were preferentially up-regulated and P53 repressed targets were preferentially down-regulated (Extended Data Fig. 3b-c). Moreover, genes related to protein synthesis were primarily found to be downregulated.
The observation that the genes differentially expressed along the trajectory fall into cell death categories, as well as being mTOR, MYC and P53 targets strongly suggests that cells at each end of the trajectory are the winners and losers of cell competition5–7. For this reason, we hereafter refer to those epiblast cells unique to CI-treated embryos as “loser” epiblast cells and to those at the opposite end of the trajectory as the “winner” epiblast cells. Those cells lying between these two populations on the trajectory are considered “intermediate”. Using this knowledge we can define a diffusion pseudotime (dpt) coordinate25 originating in the “winner” cluster that tracks the position of cells along the trajectory and that can be interpreted as a “losing score”, i.e., it quantifies how strong the signature of the “losing” state is in the transcriptome of a cell (see Fig. 2d-e).
In accordance with previous studies6, 8, 9, we also found evidence for miss-patterning in the eliminated epiblast cells, as a proportion of these cells co-expressed naïve pluripotency and differentiation markers (Fig. 2f and Extended Data Fig. 3d). To test if loser cells are developmentally delayed or advanced compared to control cells we projected our data onto a previously published diffusion map that includes epiblast cells from E5.5, E6.25 and E6.5 stage embryos26. We found that all epiblast cells, irrespective of the condition the embryos were cultured in (ie, DMSO or CI-treated) and of their losing state (ie, that they belonged to the winner, intermediate or loser cluster) mostly overlap with the E6.5 epiblast cells (Extended Data Fig. 3e-g). Cells from the loser cluster are slightly closer to the E6.25 stage than the winner and intermediate cells, as shown by their pseudo-time coordinate, but they remain far from the earlier E5.5 stage. This result combined with the higher expression of some differentiation markers observed in loser cells suggests that these cells are miss-patterned rather than developmentally delayed.
Loser cells have defects in mitochondrial function
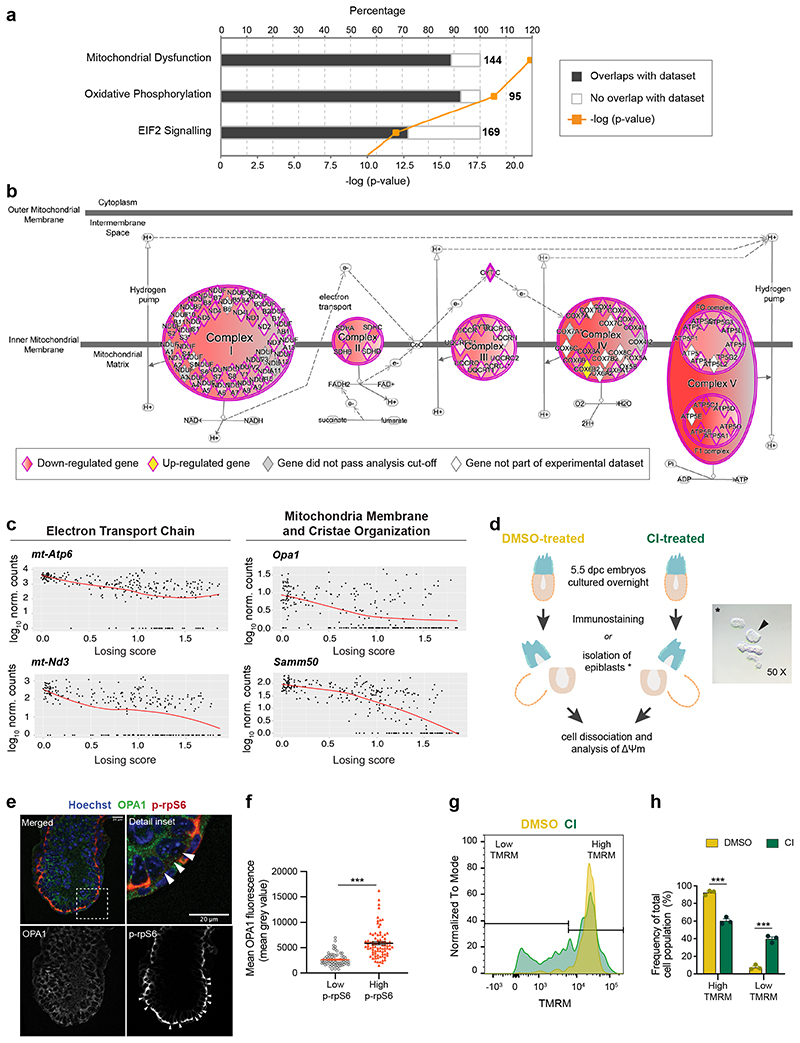
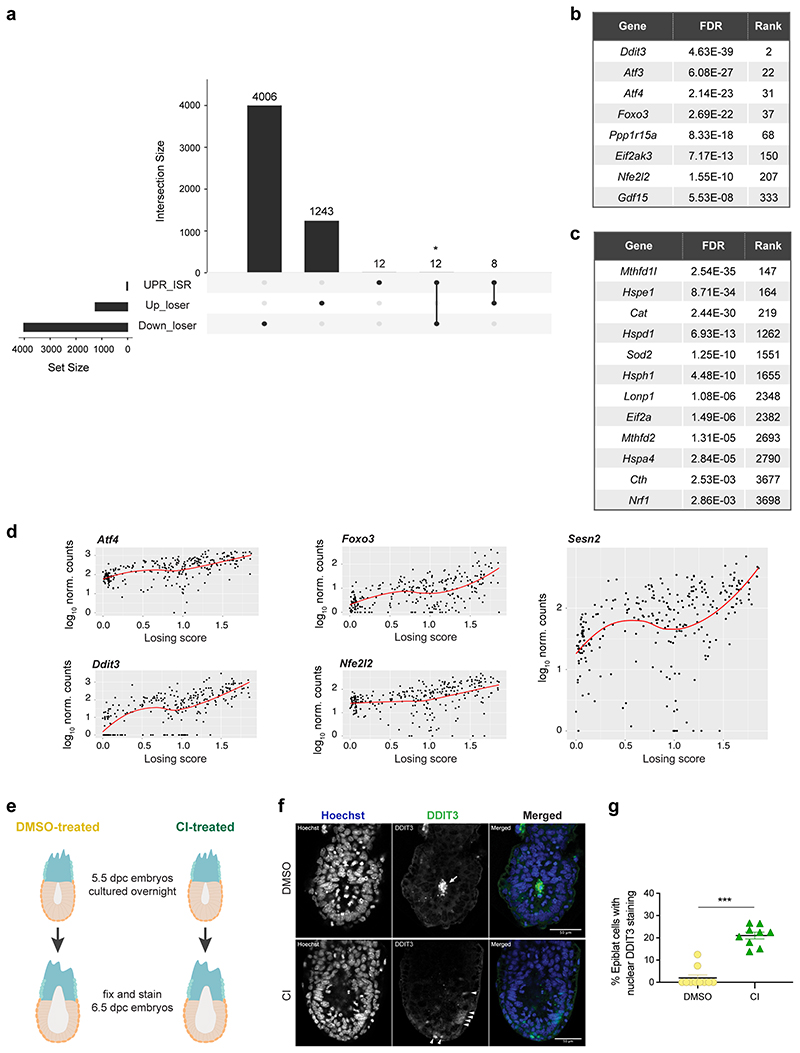
We next analysed using IPA the cellular pathways mis-regulated in loser epiblast cells and found that the top two pathways (mitochondrial dysfunction and oxidative phosphorylation) are related to mitochondrial function (Fig. 3a-b, Supplementary Table 1 and 2). For example, we found a down-regulation along the winner to loser trajectory of the mtDNA encoded mt-Nd3 and mt-Atp6, of regulators of mitochondrial dynamics such as Opa1 (optic atrophy 1), as well as of genes involved in mitochondrial membrane and cristae organisation such as Samm50 (Fig. 3c), suggesting that mitochondrial function is impaired in loser cells.
Fig. 3. Cells eliminated during early mouse embryogenesis have mitochondrial defects.
a, Top canonical pathways, identified by IPA, mis-regulated in loser cells in comparison to normal epiblast cells. The numbers at the end of each bar refer to total amount of genes involved in that pathway. The percentage refers to the number of genes found mis-regulated in loser cells relative to the number total genes within each pathway. b, Detail of changes in oxidative phosphorylation pathway identified in (a). Circular and oval shapes represent each of the ETC complexes (complexes I to V). Diamond shapes represent subunits of each ETC complex. Down-regulated genes in loser cells are coloured in shades of red. Darker shades correspond to lower FDR values. Cox6b2, coloured in yellow, was up-regulated in loser cells. Grey colour denotes genes that were not differentially expressed between loser and winner cells (FDR>0.01). White colour denotes genes from the Knowledge Base that were not tested (e.g., because they were not detected in our dataset). c, Expression levels of mitochondrial genes as a function of cells’ losing score. d, Experimental design adopted to assess mitochondria function in e-h. * Representative micrograph of one of the isolated epiblasts (arrow) used for Δψm analysis after embryo microdissection. e, Representative immunohistochemistry of OPA1 in E6.5 embryo where cell death was inhibited (CI-treated), quantified in (f). Loser cells are identified by low mTOR activation (low p-rpS6, arrowheads). Scale bar = 20 μm. f, Quantification of OPA1 fluorescence in normal epiblast cells and loser cells. N=6 embryos with a minimum of 8 cells analysed per condition. g, Representative histogram of flow cytometry analysis of TMRM probe, indicative of Δψm, in epiblast cells from embryos where cell death was allowed (DMSO-treated) or inhibited (CI-treated), quantified in (h). h, Frequency of epiblast cells with high or low TMRM fluorescence, according to range defined in (g) from embryos where cell competition was allowed (DMSO treated) or inhibited (CI-treated). Data obtained from 3 independent experiments and shown as mean ± SEM (g-h). 12 embryos per condition were pooled for each experiment. See methods for details on statistical analysis.
A recent body of evidence has revealed that stress responses, such as the integrated stress response (ISR) or the closely related unfolded protein response (UPR), when triggered in cells with impaired mitochondrial function prompt a transcriptional program to restore cellular homeostasis27–29. We observed that loser epiblast cells displayed a characteristic UPR-ISR signature30–33 and key regulators of this response, such as Atf4, Ddit3, Nrf2 and Foxo3 were all up-regulated in these cells (Extended Data Fig. 4a-d). Similarly, Sesn2, a target of p53 that controls mTOR activity34, was also up-regulated in loser cells (Extended Data Fig. 4d). These findings support that loser epiblast cells present mitochondrial defects, leading to the activation of a stress response in an attempt to restore cellular homeostasis35.
To validate the significance of the observed mitochondrial defects, we did two things. First, we asked if the changes of mitochondrial regulators at the mRNA level are also reflected at the protein level. We observed that in CI-treated embryos, loser cells that persist and are marked by low mTOR activity7, also show significantly lower OPA1 levels (Fig. 3d-f). We also found that DMSO-treated embryos showed strong DDIT3 staining (an UPR-ISR marker) in the dying cells that accumulate in the proamniotic cavity, and that in CI-treated embryos, DDIT3 expression was up-regulated in a proportion of epiblast cells (Extended Data Fig. 4e-g). The second thing we did to validate the importance of the mitochondrial defects was to study in loser epiblast cells their mitochondrial membrane potential (Δψm), an indication of mitochondrial health. We observed that while the cells of DMSO-treated embryos showed a high Δψm that fell within a narrow range, in CI-treated embryos the proportion of cells with a low Δψm significantly increased (Fig. 3d and 3g-h). Together, these results suggest that loser epiblast cells have impaired mitochondrial activity that triggers a stress response.
Mitochondrial dysfunction is common to different loser cells
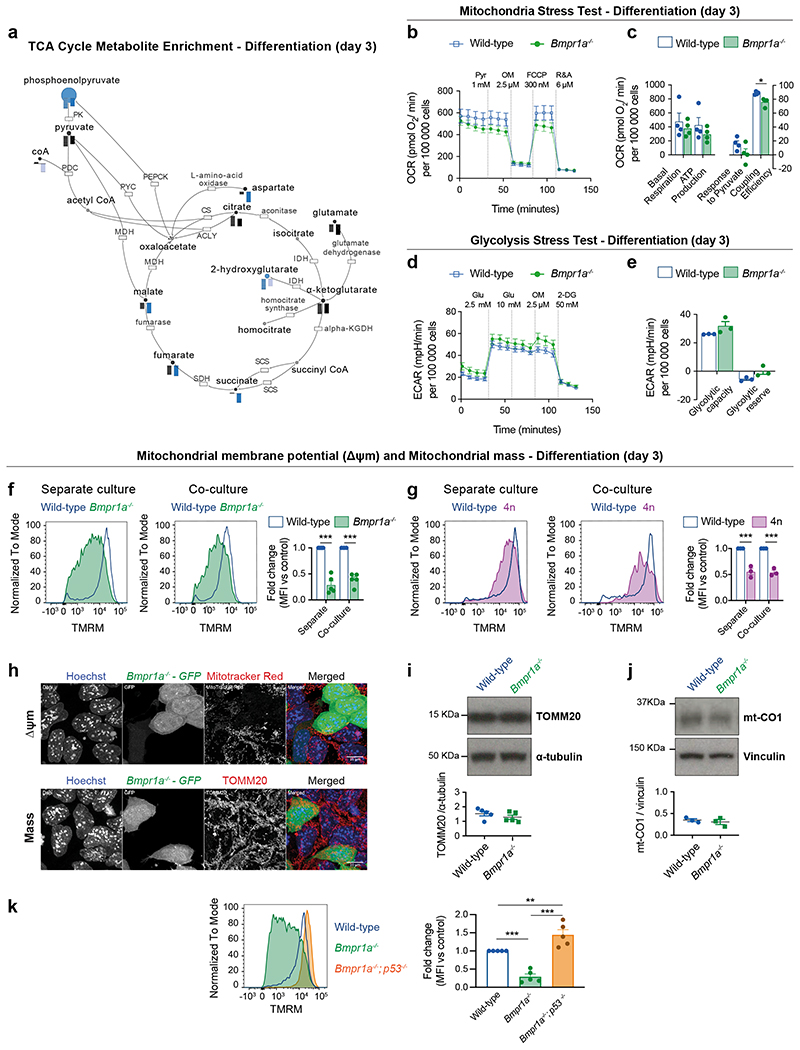
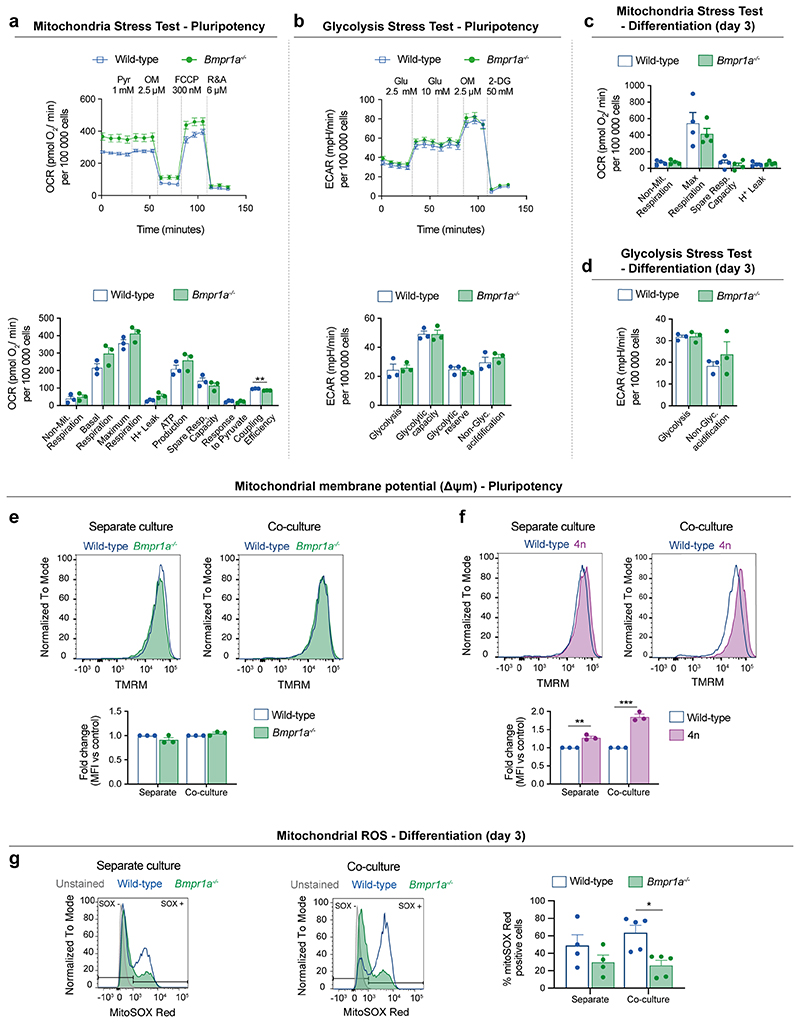
To address if mitochondrial defects are a common feature of loser cells eliminated by cell competition, we analysed ESCs that are defective for BMP signalling (Bmpr1a-/-) and tetraploid cells (4n)6. We first carried out a mass spectrometry analysis using the Metabolon platform and found that metabolites and intermediates of the TCA cycle, such as malate, fumarate, glutamate and α-ketoglutarate are depleted in both Bmpr1a-/- and 4n ESCs in differentiation culture conditions (Fig. 4a). Next, we performed an extracellular flux Seahorse analysis of Bmpr1a-/- ESCs to measure their glycolytic and oxidative phosphorylation (OXPHOS) rates. We observed that when these cells are maintained in pluripotency culture conditions that are not permissive for cell competition6, they but a higher OXPHOS rate than control cells (Extended Data Fig. 5a-b). In contrast, when Bmpr1a-/- cells are induced to differentiate, this phenotype is reversed, with mutant cells showing lower ATP generated through OXPHOS and a higher glycolytic capacity than controls (Fig. 4b-e and Extended Data Fig. 5c-d). This suggests that upon differentiation Bmpr1a-/- cells are unable to sustain proper OXPHOS activity.
Fig. 4. Mitochondrial defects are a common feature of cells eliminated by cell competition.
a, Metabolic enrichment analysis of the TCA cycle and intermediate metabolites obtained using Metabolon platform for defective cells (Bmpr1a-/-, left bar and 4n, right bar), in comparison to wild-type cells during differentiation. Bars indicate compound levels relative to wild-type cells. Blue bars indicate compounds that are significantly depleted (p<0.05) and light blue bars indicate compounds that are almost significantly depleted (0.05≤p≤0.1). Black bars indicate compounds that are depleted although not statistically significant in comparison to the levels found in wild-type cells. The enzymes on the pathway are represented as boxes and labelled by their canonical names. b-e, Metabolic flux analysis of wild-type and BMP-defective cells during differentiating conditions. Analysis of oxygen consumption rate (OCR) as a measure of mitochondria function (mitochondria stress test) (b). Detail of metabolic parameters found changed from the analysis of the mitochondria stress test (c). Analysis of extracellular acidification rate (ECAR) as a measure of glycolytic function (glycolysis stress test) (d). Detail of metabolic parameters found changed from the analysis of the glycolysis stress test (e). f-g, Δψm in defective mESCs undergoing differentiation in separate or co-culture conditions. Representative histograms of TMRM fluorescence and quantification for wild-type and Bmpr1a-/- (f) and wild-type and 4n (g). h, Representative micrographs of wild-type and Bmpr1a-/- cells co-cultured during differentiation and stained for a reporter of Δψm (MitoTracker Red, top panel) or mitochondria mass (TOMM20, bottom panel). Nuclei are stained with Hoechst. Scale bar = 10 μm. i-j, Western blot analysis of mitochondria mass markers TOMM20 (i) and mt-CO1 (j) for wild-type and Bmpr1a-/- cells during differentiation. k, Analysis of Δψm for wild-type, Bmpr1a-/- and Bmpr1a-/-;p53-/- cells during differentiation. Representative histogram of TMRM fluorescence and quantification. Data shown as mean ± SEM. Extracellular flux Seahorse data was obtained from 3 (d,e) or 4 independent experiments (b,c), with 5 replicates per cell type in each assay. Remaining was data obtained from 3 (g,j), or 5 (a,f,i,k) independent experiments. See methods for details on statistical analysis.
To further test the possibility that defective ESCs have impaired mitochondrial function, we assessed their Δψm. We found that whilst Bmpr1a-/- and 4n cells had a similar Δψm to control cells in pluripotency conditions (Extended Data Fig. 5e-f), upon differentiation both these cell types presented a loss of Δψm, irrespective of whether they were separate or co-cultured with wild-type cells (Fig. 4f-g). This reduction in Δψm is not due to excessive mitochondrial reactive oxygen species (ROS) production or to a lower mitochondrial mass within mutant cells since, as for example, Bmpr1a-/- cells have lower ROS levels and similar TOMM20 and mt-CO1 expression as control cells (Fig. 4h-j and Extended Data Fig. 5g). The fact that the loss of Δψm and lower OXPHOS activity can be observed even when loser cells are cultured separately, suggests that the mitochondrial dysfunction phenotype is an inherent property of loser cells and not a response to them being out-competed. These results also indicate that the mitochondrial defects are directly linked to the emergence of the loser status: in conditions that are not permissive for cell competition (pluripotency) mutant cells do not show defective mitochondrial function, but when they are switched to differentiation conditions that allow for cell competition, they display impaired mitochondrial function.
To further explore the relationship between mitochondrial activity and the competitive ability of the cell, we analysed the Δψm of BMP defective cells that are null for p53 (Bmpr1a-/-; p53-/- ESCs), as these are not eliminated by wild-type cells7. Remarkably, we observed that mutating p53 in Bmpr1a-/- cells not only rescues the loss of Δψm of these cells, but also causes hyperpolarisation of their mitochondria (Fig. 4k). These results not only suggest a role for P53 in regulating mitochondrial activity of ESCs, but also strongly support a pivotal role for mitochondrial activity in cell competition.
Impaired mitochondrial function triggers cell competition
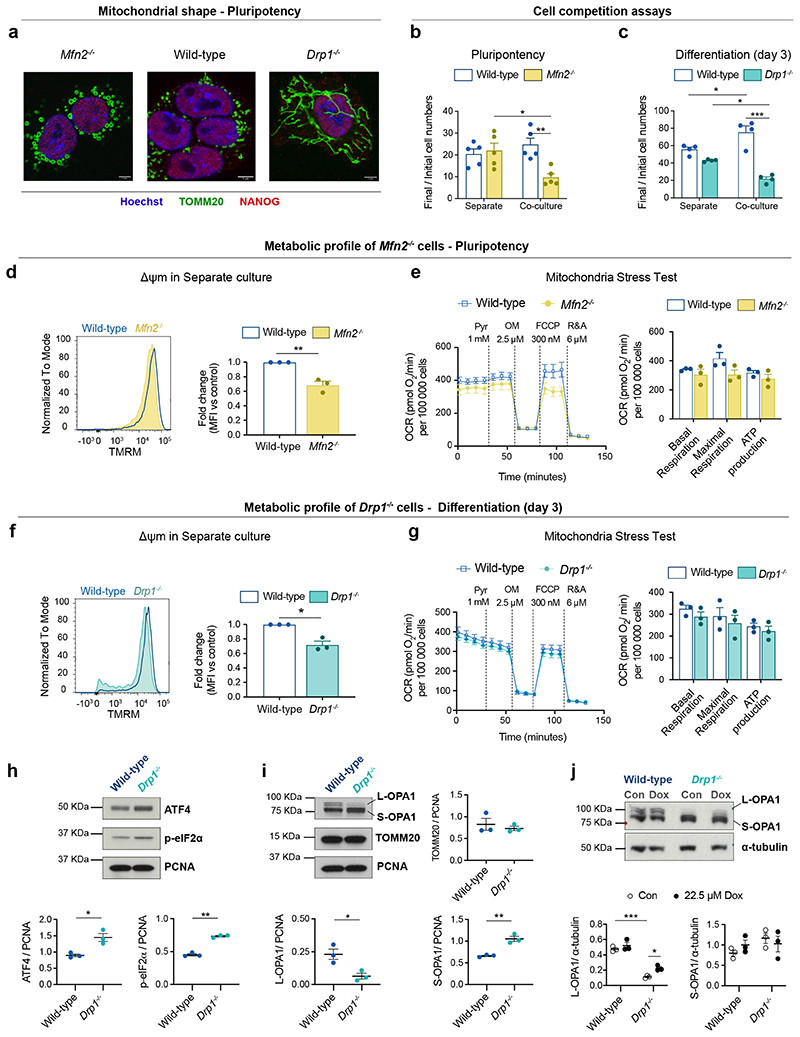
The mitochondrial defects observed in loser cells led us to ask if disrupting mitochondrial activity alone is sufficient to trigger cell competition. During the onset of differentiation, mitochondrial shape changes substantially. In pluripotent cells mitochondria have a round and fragmented shape, but upon differentiation they fuse and become elongated, forming complex networks10. Given that this change in shape correlates with when cell competition occurs, we tested if disrupting mitochondrial dynamics is sufficient to induce cell competition. MFN1 and MFN2 regulate mitochondrial fusion and DRP1 controls their fission36–38. We generated Mfn2-/- ESCs, that have enlarged globular mitochondria, and Drp1-/- ESCs, that show hyper-elongated mitochondria (Fig. 5a). We first tested the competitive ability of Mfn2-/- ESCs in pluripotency conditions, that we have previously found not to induce the out-competition of Bmpr1a-/- or 4n cells6. Interestingly, we found that although Mfn2-/- cells grow similarly to wild-type cells in separate cultures, they were out-competed in co-culture (Fig. 5b). Analysis of the Drp1 mutant cells showed that although they did not grow significantly slower than wild-type cells when cultured separately in differentiation inducing conditions, they were out-competed by wild-type cells in co-culture (Fig. 5c). The observation that disrupting mitochondrial dynamics can induce cell competition even in pluripotency culture conditions, suggests that mitochondrial activity is a dominant parameter determining the competitive ability of the cell.
Fig. 5. Manipulating mitochondria biology is sufficient to trigger cell competition.
a, Representative micrographs of wild-type, Mfn2-/- and Drp1-/- mESCs showing alterations in mitochondrial morphology in mutant cells. TOMM20 was used as a mitochondrial marker and NANOG as a pluripotency marker. Nuclei are stained with Hoechst. Scale bar = 5 μm. b,c, Cell competition assays between wild-type mESCs and cells with altered morphology, Mfn2-/- during pluripotency (b) and Drp1-/- during differentiation (c). The ratio of final/initial cell numbers in separate or co-culture is shown. d-j, Metabolic profile of Mfn2-/- and Drp1-/- mESCs. Analysis of mitochondrial membrane potential (Δψm) for wild-type and Mfn2-/- cultured separately during pluripotency (d) and for wild-type and Drp1-/- mESCs -/- after 3 days of differentiation in separate culture (f). Metabolic flux analysis of wild-type and Mfn2-/- mESCs cultured separately during pluripotency (e) and for wild-type and Drp1-/- after 3 days of differentiation in separate cultures (g). Data was collected from 3 independent experiments. h-j, Western blot analysis of markers of UPR and mitochondrial markers in wild-type and Drp1-/- after 3 days of differentiation in separate culture. p-eIF2α, phosphorylated eukaryotic initiation factor 2α. Cells were treated with doxycycline (Dox, 22.5 μM) or vehicle (Con) from day 1 of differentiation and samples were collected on day 3 (j). Data shown as mean ± SEM of 3 (d-j), 4 (c) or 5 (b) independent experiments. See methods for details on statistical analysis.
To establish how disruption of mitochondrial fusion and fission affects mitochondrial performance we compared the Δψm, respiration rates and mitochondrial ATP production of Mfn2-/- and Drp1-/- ESCs to that of wild-type cells (Fig. 5d-g). We found that whilst Mfn2-/- and Drp1-/- ESCs had lower Δψm than control cells (Fig. 5d,f), Mfn2-/- ESCs had lower maximal respiration rates but similar basal respiration and ATP production to controls and Drp1-/- ESCs showed similar respiration and ATP production to controls (Fig. 5e,g). This suggests that ATP production or respiration rates alone do not determine the relative competitive ability of ESCs.
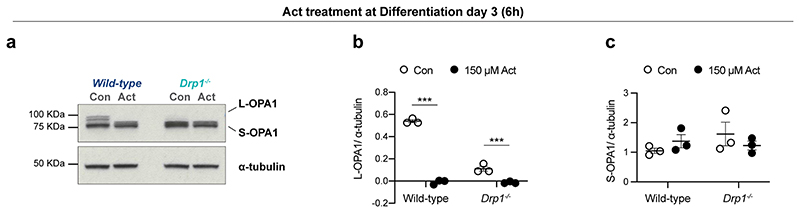
Besides mitochondrial dysfunction, another prominent signature of loser cells found in vivo was the UPR/ISR (Ext. Data Fig. 4). Since the loss of Drp1 has been associated with activation of the UPR39–41, we investigated if the Drp1-/- loser cells also showed evidence for the activation of the UPR/ISR. We observed that Drp1-/- cells show higher expression of ATF4 and p-eIF2α than wild-type counterparts, which is indicative of UPR/ISR activation (Fig. 5h)39–41. Another feature previously described upon loss of Drp1 is the proteolytic cleavage of OPA1, where short isoforms (S-OPA1) are accumulated in detriment of the long isoforms (L-OPA1)39. When we analysed the expression of OPA1 in wild-type and Drp1-/- cells we observed that while wild-type cells retain L-OPA1 expression, loser cells predominantly express the S-OPA1 isoforms and display almost no expression of L-OPA1 (Fig. 5i). This defect has been associated with mitoribosomal stalling, a phenotype that can be replicated by treating cells with actinonin (Extended Data Fig. 6)42. To test if the shift in isoform expression observed in Drp1-/- ESCs is due to aberrant mitochondrial translation we treated cells with doxycycline, that inhibits this translation43, and observed that this was sufficient to partially rescue L-OPA1 expression (Fig. 5j). This rescue together with the evidence for UPR/ISR expression suggest that Drp1-/- cells display defects in mitochondrial translation.
Loser epiblast cells accumulate mtDNA mutations
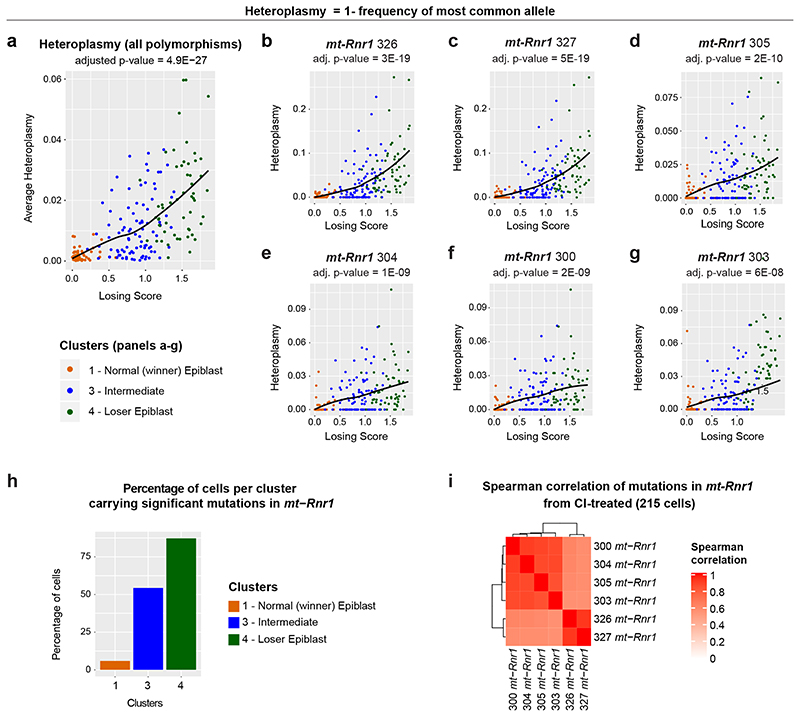
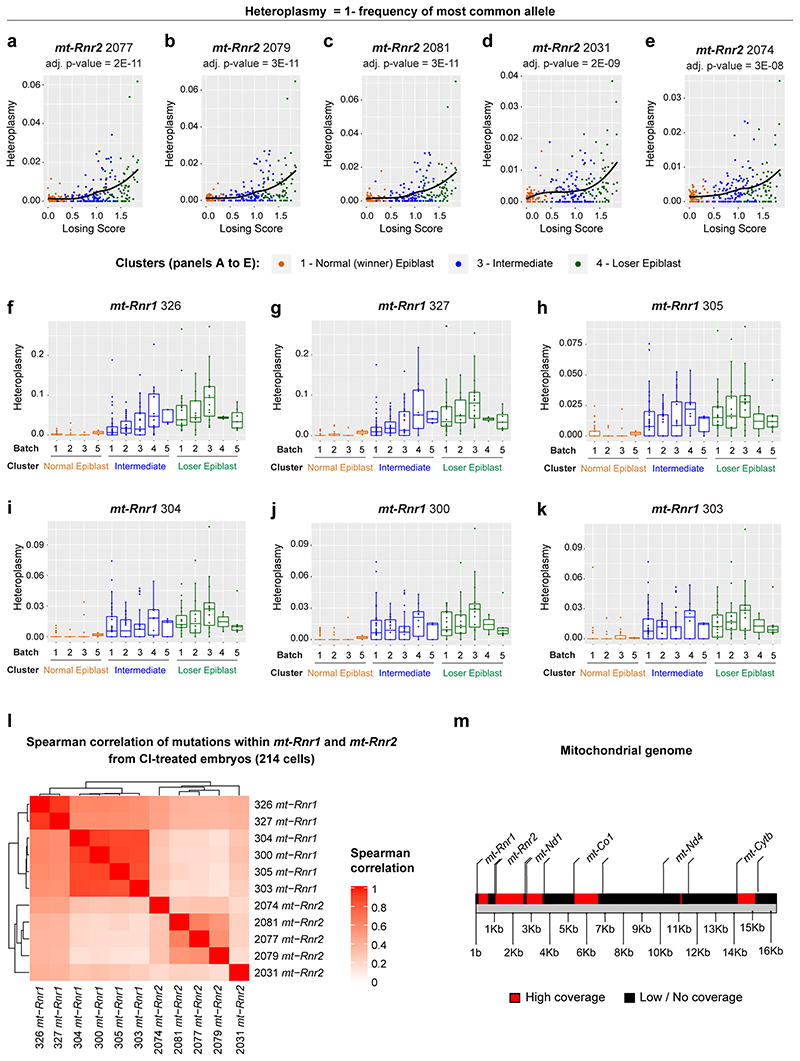
There is strong evidence for selection against aberrant mitochondrial function induced by deleterious mtDNA mutations in mammals21, 44–47. Given that we observe that cell competition selects against cells with impaired mitochondrial function, we asked if cell competition could be reducing mtDNA heteroplasmy (frequency of different mtDNA variants) during mouse development. It has been recently shown that scRNA-seq can be used to reliably identify mtDNA variants, although with a lower statistical power compared to more direct approaches, like mtDNA sequencing48. We therefore tested if mtDNA heteroplasmy is present in our scRNA-seq data and whether this correlates with the losing score of a cell. Our analysis revealed that the frequency of specific mtDNA polymorphisms increased with the losing score of epiblast cells (Fig. 6a), and such mtDNA changes occurred within mt-Rnr1 and mt-Rnr2 (Fig. 6b-h and Extended Data Fig. 7a-e). Moreover, these changes were not dependent on the litter from which the embryo came from (Extended Data Fig. 7f-k). As it was formally possible that these loser-specific sequence changes could originate from contaminating nuclear mitochondrial sequences (NUMTS) or from RNA editing, we performed several controls to confirm that mtDNA polymorphisms are the most likely source of these changes (see Methods section). For example, we considered only the RNA sequencing reads that are uniquely mapped to the mitochondrial genome and not to nuclear DNA, and we confirmed that the variants with highest heteroplasmy found in the “loser” cells were not present in any of the NUMTS that have previously been reported or could be identified using BLAST. Moreover, we verified that the observed sequence changes were not compatible with canonical RNA editing (see the Methods section for further details). It is worth noting that the sequence changes we detected in mt-Rnr1 and mt-Rnr2 strongly co-occurred in the same cell, with those closest together having the highest probability of co-existing (Fig. 6i and Extended Data Fig. 7l). This is suggestive of mtDNA replication errors that could be ‘scarring’ the mtDNA, disrupting the function of mt-Rnr1 (12S rRNA) and mt-Rnr2 (16S rRNA) and causing the loser phenotype. Importantly, the presence of these specific mtDNA mutations in the loser cells suggests that cell competition could be contributing to the elimination of deleterious mtDNA mutations during early mouse development. Of note, we only report mtDNA variants detected in regions of the genome with high sequencing coverage (Extended Data Fig. 7m), therefore the presence of other variations in mtDNA sequence between winners and loser cells cannot be excluded.
Fig. 6. Intermediate and loser epiblast cells accumulate polymorphisms in mtDNA sequence.
a-g, mtDNA heteroplasmy (plotted as Heteroplasmy = 1- frequency of most common allele) in epiblast cells from CI-treated embryos. Average heteroplasmy (considering all eleven polymorphisms that have a statistically significant dependence on the losing score; see Methods) as a function of cells’ losing scores (a). mtDNA heteroplasmy for six positions within the mt-Rnr1 gene (b-g). The heteroplasmy at all these positions, as well as the average heteroplasmy, increase with the cells’ losing scores in a statistically significant way (the adjusted p-value estimated via a generalized linear model is indicated at the top of each plot). h, The bar plot indicates the fraction of epiblast cells in each of the cluster indicated on the x-axis (winner, intermediate, loser) that carry a mean heteroplasmy (computed on the six positions within the mt-Rnr1 indicated in the panels b-g) greater than 0.01. This shows that the level of mtDNA heteroplasmy in mt-Rnr1 is strongly associated with the loser status of the cells, since ~55% and ~87% of cells in the intermediate and the loser clusters, respectively, have heteroplasmic sequences in this gene compared to only ~5% of cells in the winner cluster. i, Spearman’s correlation coefficient between the mtDNA heteroplasmy at the six positions shown in panels (b-g). See methods for details on statistical analysis.
mtDNA sequence determines the competitive ability of a cell
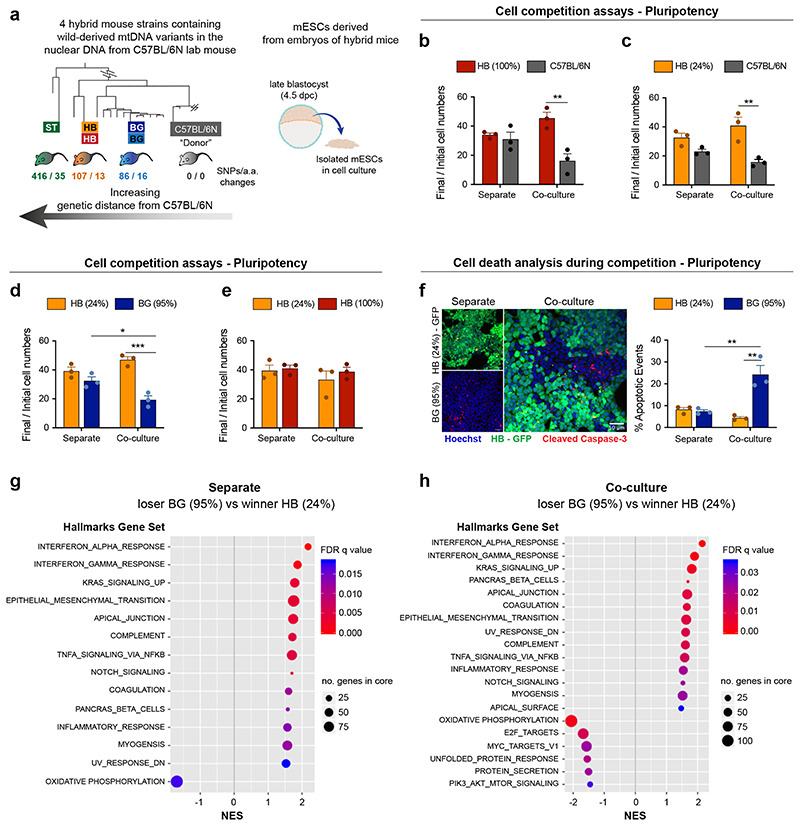
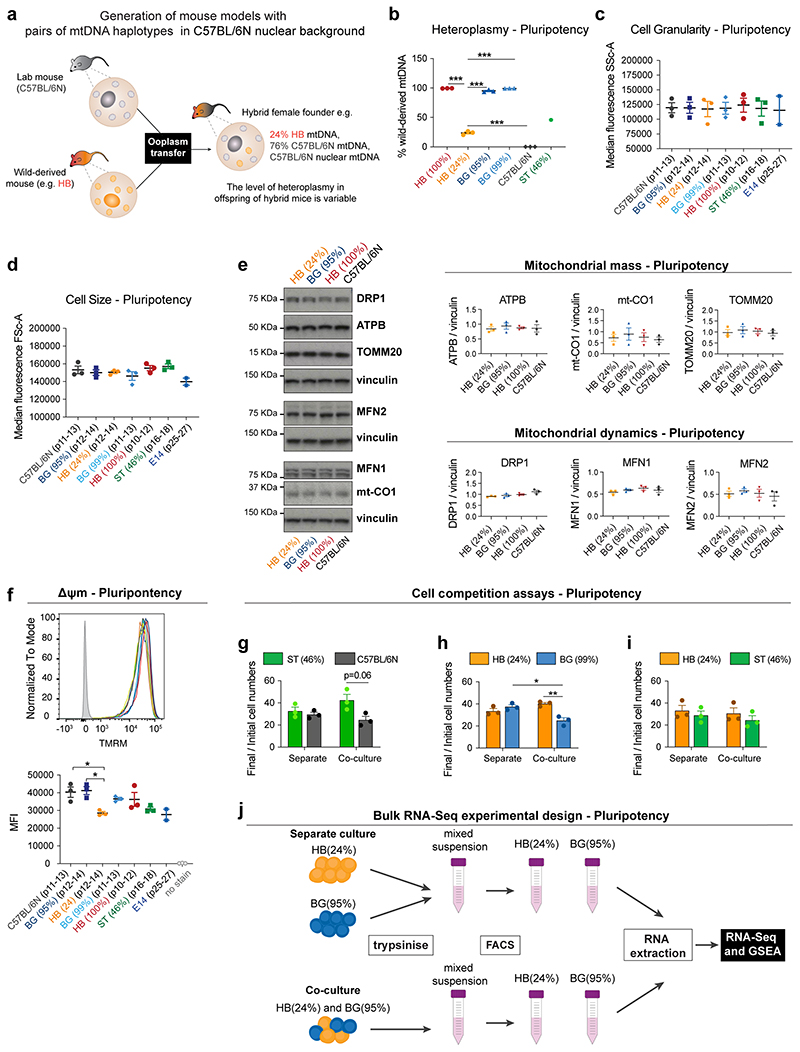
To explore this possibility further, we analysed if alterations in mtDNA can induce cell competition by testing the competitive ability of ESCs with non-pathological differences in mtDNA sequence. For this we compared the relative competitive ability of ESCs that shared the same nuclear genome background but differed in their mitochondrial genomes by a small number of non-pathological sequence changes. We derived ESCs from hybrid mouse strains that we had previously engineered to have a common nuclear C57BL/6N background, but mtDNAs from different wild-caught mice16. Each wild-derived mtDNA variant (or haplotype) contains a specific number of single nucleotide polymorphisms (SNPs) that lead to a small number of amino acid changes when compared to the C57BL/6N mtDNA haplotype. Furthermore, these haplotypes (BG, HB and ST) can be ranked according to their genetic distance from the C57BL/6N mtDNA (Fig. 7a and Extended Data Fig. 8a). Characterization of the isolated ESCs revealed that they have a range of heteroplasmy (mix of wild-derived and C57BL/6N mtDNAs) that is stable over several passages (Extended Data Fig. 8b). Importantly, these different mtDNA haplotypes and different levels of heteroplasmy do not alter cell size, cell granularity, mitochondrial mass or mitochondrial dynamics, nor do they substantially impact the cell’s Δψm (Extended Data Fig. 8c-f).
Fig. 7. Changes in mtDNA sequence can determine the competitive ability of a cell.
a, Derivation of mESCs from hybrid mouse strains, generated elsewhere by Burgstaller and colleagues. Neighbour-Joining Phylogenetic Analysis of mtDNA from wild and C57BL/6N mouse strains, that were used to generate hybrid mice (adapted from16), illustrates the genetic distance of the mtDNA from wild mouse strains to the C57BL/6N lab mouse. The number of single nucleotide polymorphisms and amino acid changes (SNPs/ a.a. changes) from wild to lab mouse strain is shown. mESCs were derived from embryos of hybrid mice, containing the nuclear background of a C57BL/6N lab mouse and mtDNA from three possible wild-derived strains (BG, HB or ST). b-e, Cell competition assays between cells derived from the embryos of hybrid mice performed in pluripotency maintenance conditions. The ratio of final/initial cell numbers in separate or co-culture is shown. f, Representative micrographs of cleaved caspase-3 staining and quantification of the percentage of apoptotic events in winners HB(24%) and loser BG(95%) mESCs maintained pluripotent and cultured in separate or co-culture conditions. g-h, Gene set enrichment analysis of differentially expressed genes from bulk RNA seq. in loser BG (95%) compared to winner HB (24%) mESCs maintained pluripotent and cultured in separate (g) or co-culture conditions (h). Gene sets that show positive normalized enrichment scores (NES) are enriched in loser cells, while gene sets that show negative NES are depleted in loser cells. Data obtained from 4 independent experiments (h). Remaining data shown as mean ± SEM of 3 independent experiments (b-f). See methods for details on statistical analysis.
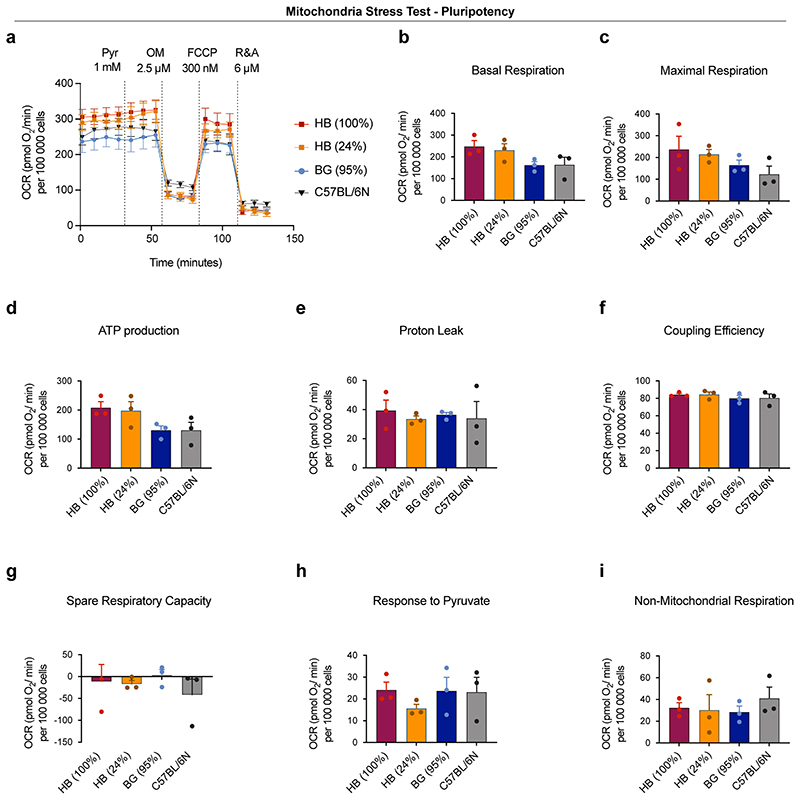
When we tested the competitive ability of these ESCs with different mtDNA content, in pluripotency culture conditions, we observed that cells carrying the mtDNAs that are most distant from the C57BL/6N mtDNA, such as the HB(100%), the HB(24%) and the ST(46%) ESCs could all out-compete the C57BL/6N line (Fig. 7b-c and Extended Data Fig. 8g). Similarly, when we tested the HB(24%) line against the BG(99%) or the BG(95%) lines (that have mtDNAs more closely related to the C57BL/6N mtDNA), we found that cells with the HB haplotype could also out-compete these ESCs (Fig. 7d and Extended Data Fig. 8h). In contrast, we observed that the HB(24%) ESCs were unable to out-compete either their homoplasmic counterparts, HB(100%), or the ST(46%) cells that carry the most distant mtDNA variant from C57BL/6N (Fig. 7e and Extended Data Fig. 8i). These results tell us three things. First, that non-pathological differences in mtDNA sequence can trigger cell competition. Second, that a competitive advantage can be conferred by only a small proportion of mtDNA content, as indicated by our finding that HB(24%) behave as winners. Finally, these findings suggest that the phylogenetic proximity between mtDNA variants can potentially determine their competitive cell fitness.
To characterise the mode of competition between cells with different mtDNA we focussed on the HB(24%) and the BG(95%) ESCs. Analysis of these cell lines revealed that specifically when co-cultured, the BG(95%) cells display high levels of apoptosis (Fig. 7f), indicating that their out-competition is through their elimination. To gain further insight we performed bulk RNA-seq of these cells in separate and co-culture conditions (Extended Data Fig. 8j) and analysed the differentially expressed genes by gene-set enrichment analysis (GSEA). We found that in separate culture the most notable features that distinguish BG(95%) from HB(24%) cells were a down-regulation of genes involved in oxidative phosphorylation and an up-regulation of those associated with cytokine activity (Fig. 8g). Interestingly, in the co-culture condition, in addition to these signatures, BG(95%) cells revealed a down-regulation in signature markers of MYC activity and mTOR signalling (Fig. 7h), whose downregulation are known read-outs of a loser status during cell competition in the embryo5–7 (Fig. 2c).
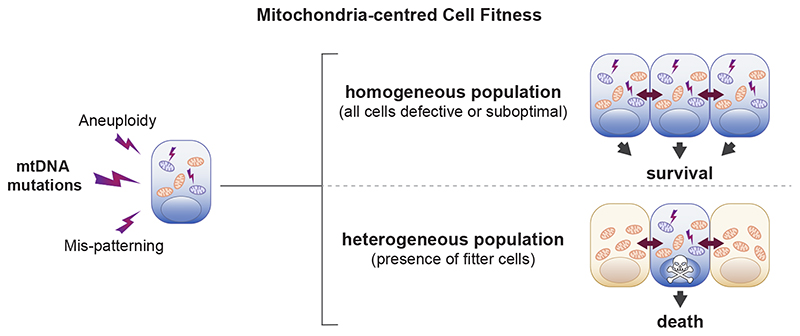
Fig. 8. Model of cell competition.
Summary of the main findings of the study. A range of cellular defects, such as aneuploidy, mis-patterning or mtDNA mutations cause alterations in mitochondria function, which affect the relative fitness of cells. The cells with suboptimal mitochondrial activity survive in a homogeneous population but are eliminated by cell competition in the presence of fitter cells.
To test if the down-regulation of genes involved in oxidative phosphorylation was also reflected at the functional level we compared oxygen consumption rates and mitochondrial ATP generation in HB(100%), HB(24%), BG(95%) and C57Bl/6N ESCs. We find that the winner cells HB(100%) and HB(24%) have higher basal respiration, higher maximal respiration and higher mitochondrial ATP production than the loser BG(95%) and C57BL/6N ESCs (Extended Data Fig. 9). These data indicate that the mtDNA differences that exist between winner and loser cells are sufficient to affect their mitochondrial performance and this ultimately determines their competitive ability. However, the observation that differentiating Drp1-/- ESCs are eliminated by cell competition but do not show differences in respiration rates or mitochondrial ATP production (Fig. 5b,e), suggests that respiration or ATP production rates alone are unlikely to be the mitochondrial parameters that control competitive cell fitness.
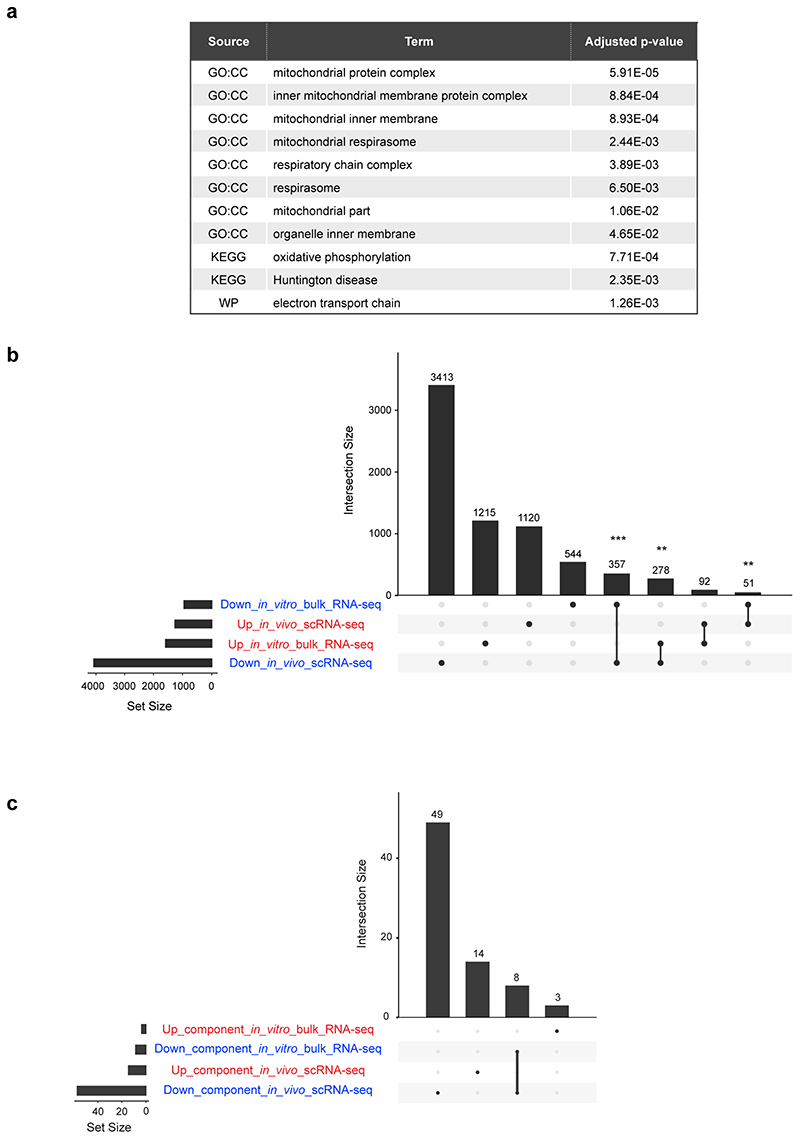
The finding that the genes down-regulated in BG(95%) cells when co-cultured with HB(24%) cells fell under functional categories relating to mitochondrial function (Extended Data Fig. 10a) led us to analyse the degree of overlap between these genes and the genes differentially expressed along the winner-to-loser trajectory in the embryo. We observed a significant overlap in mis-regulated genes (Extended Data Fig. 10b), as well as in the functional components that these genes can be categorised into (Extended Data Fig. 10c). This further highlights the importance of relative mitochondrial activity for determining the competitive ability of embryonic cells.
Discussion
The emerging role of cell competition as a regulator of cell fitness in a wide range of cellular contexts, from the developing embryo to the ageing tissue1–3, has highlighted the importance of understanding what cell types are normally eliminated by this process. With the aim of understanding this question, we have analysed the transcriptional identity of the cells eliminated in the early mouse embryo. We have found not only that they present a cell competition signature but also that they have impaired mitochondrial function and are marked by sequence changes in mt-Rnr1 and mt-Rnr2. Starting from these results, we leveraged in vitro models of cell competition to show that: (i) mitochondrial function is impaired in loser cells eliminated by cell competition, and (ii) differences in mitochondrial activity are sufficient to trigger cell competition in ESCs. Overall, this points to mitochondrial performance as a key determinant of the competitive ability of cells during early mammalian embryonic development. One implication of our findings is that a range of different types of defects, such as mis-patterning, karyotypic abnormalities or mtDNA mutations, all lead to dysfunctional mitochondria at the onset of differentiation and that ultimately it is their impaired mitochondrial function that triggers cell competition, inducing their elimination (Fig. 8).
Embryos are exposed to different microenvironments in vivo and when cultured ex-vivo. Similarly, ESCs also experience a different micro-environment to epiblast cells in the embryo. These different micro-environments could potentially affect the selective pressure and hence the transcriptional signature of loser cells. However, there are two reasons why we think that the loser cell signatures we identify here are conserved across systems. First, the transcriptional profile of our epiblast cells from cultured embryos is very similar to that of epiblast cells from freshly isolated embryos (see Extended Data Figure 3e-g). Second, the loser signature identified here is enriched for targets of P53 and depleted for mTOR and c-MYC targets. Given that these are regulators of cell competition identified by us and others in the embryo and in ESCs5–7, it suggests that the same pathways are inducing loser cell elimination in in vivo, ex-vivo and in ESC models of cell competition.
It is well known that the successful development of the embryo can be influenced by the quality of its mitochondrial pool10. Moreover, divergence from normal mitochondrial function during embryogenesis is either lethal or can lead to the development of mitochondrial disorders49. Deleterious mtDNA mutations are a common cause of mitochondrial diseases and during development selection against mutant mtDNA has been described to occur through at least two mechanisms: the bottleneck effect and the intra-cellular purifying selection. The bottleneck effect is associated specifically with the unequal segregation of mtDNAs during primordial germ cell specification, for example as seen in the human embryo50. In contrast to this, purifying selection, as the name implies, allows for selection against deleterious mtDNAs and has been proposed to take place both during development and post-natal life51. Importantly, purifying selection has been found to occur at the molecule and organelle level, as well as at the cellular level52. Our findings indicate that purifying selection can occur not only at the intra-cellular level but also inter-cellularly (cell non-autonomously). We show that epiblast cells are able to sense their relative mitochondrial activity and that those cells with mtDNA mutations, lower or aberrant mitochondrial function are eliminated. By selecting those cells with the most favourable mitochondrial performance, cell competition would not only prevent cells with mitochondrial defects from contributing to the germline or future embryo, but also ensure optimisation of the bioenergetic performance of the epiblast, therefore contributing to the synchronization of growth during early development.
Cell competition has been studied in a variety of organisms, from Drosophila to mammals, and it is likely that multiple different mechanisms fall under its broad umbrella1–3. In spite of this, there is considerable interest in understanding if there could be any common feature in at least some of the contexts where cell competition has been described. The first demonstration of cell competition in Drosophila was made by inducing clones carrying mutations in the ribosomal gene Minute 4 and this has become one of the primary models to study this process. Our finding that during normal early mouse development cell competition eliminates cells carrying mutations in mt-Rnr1 and mt-Rnr2 transcripts, demonstrates that in the physiological context mutations in ribosomal genes also trigger cell competition. While we identified 11 mutations specific to loser cells, we cannot exclude the presence of additional variants differentiating winners from losers in those positions that did not have sufficient coverage in our RNA-seq data. Our observation that mis-patterned and karyotypically abnormal cells show impaired mitochondrial activity indicates that during early mouse development different types of defects impair mitochondrial function and trigger cell competition. Interestingly, mtDNA genes are amongst the top mis-regulated factors identified during cell competition in the mouse skin53. In the Drosophila wing disc oxidative stress, a general consequence of dysfunctional mitochondria, underlies the out-competition of Minute and Mah-jong mutant cells54. Similarly, in MDCK cells a loss of Δψm occurs during the out-competition of RasV12 mutant cells and is key for their extrusion55. These observations raise the possibility that differences in mitochondrial activity may be a key determinant of competitive cell fitness in a wide range of systems. Unravelling what mitochondrial features can lead to cellular differences that can be sensed between cells during cell competition and if these are conserved in human systems will be key not only for understanding this process, but also to open up the possibility for future therapeutic avenues in the diagnosis or prevention of mitochondrial diseases.
Methods
Animals
Mice were maintained and treated in accordance with the Home Office’s Animals (Scientific Procedures) Act 1986 and covered by the Home Office project license PBBEBDCDA. All mice were housed on a 10 hr-14 hr light-dark cycle with access to water and food ad libitum. All mice were housed within Individually Ventilated cages (IVC). Temperature was maintained between 21-24C and humidity between 45-65%, although humidity. Mattings were generally set up in the afternoon. Noon of the day of finding a vaginal plug was designated embryonic day 0.5 (E0.5). Embryo dissection was performed at appropriate timepoints in M2 media (Sigma), using Dumont No.5 forceps (11251-10, FST). No distinction was made between male and female embryos during the analysis.
Cell lines, cell culture routine and drug treatments
E14 (RRID: CVCL_C320), kindly provided by Prof A. Smith, from Cambridge University, were used as wild-type control cells tdTomato-labelled or unlabelled. GFP-labelled or unlabelled cells defective for BMP signalling (Bmpr1a-/-), tetraploid cells (4n) and Bmp1a-/- null for p53 (Bmpr1a-/-;p53 -/ -) are described elsewhere 6, 7. Cells null for Dynamin-related protein 1 (Drp1-/-) or Mitofusin 2 (Mfn2-/-) were generated by CRISPR mutagenesis. Cells with different mitochondrial DNA (mtDNA) content in the same nuclear background were derived from embryos of hybrid mice, generated elsewhere 16.
Cells were maintained pluripotent and cultured at 37°C in 5% CO2 in 25 cm2 flasks (Nunc) coated with 0.1% gelatin (Sigma) in DPBS. Growth media (ES media) consisted of GMEM supplemented with 10% FCS, 1 mM sodium pyruvate, 2 mM L-glutamine, 1X minimum essential media non-essential amino-acids, 0.1 mM β-mercaptoethanol (all from Gibco) and 0.1% leukemia inhibitory factor (LIF, produced and tested in the lab). Cells derived from hybrid mice (C57BL/6N nuclear background) were maintained on 0.2% LIF. The growth media was changed daily, and cells were split every 3 days.
To manipulate mitochondrial translation during differentiation, wild-type and Drp1-/- mESCs were treated with doxycycline (Dox, 22.5 μM), from day 1 to day 3 of culture, or with actinonin (Act, 150 μM), for 6 hours on day 3 of culture in N2B27 media (see Differentiation and Cell competition assays). As control condition, cells were treated with vehicle (Con). Samples were collected on day 3 of differentiation for western blot analysis.
CRISPR mutagenesis
Drp1 and Mfn2 knockout ESCs were generated by CRISPR-Cas9 mediated deletion of Drp1 exon 2 and Mfn2 exon 3 respectively. sgRNA guides flanking Drp1 exon 2 or Mfn2 exon 3 were cloned into the PX459 vector (Addgene)57: Drp1 exon 2 upstream sgRNA: 5’ TGGAACGGTCACAGCTGCAC 3’; Drp1 exon 2 downstream sgRNA: 5’ TGGTCGCTGAGTTTGAGGCC 3’; Mfn2 upstream sgRNA: 5’ GTGGTATGACCAATCCCAGA 3’; Mfn2 downstream sgRNA: 5’ GGCCGGCCACTCTGCACCTT 3’. E14 ESCs were cotransfected with 1ug of each sgRNA expression using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. As control E14 ESCs were transfected in parallel with equal amount of empty PX459 plasmid. Following 6 days of Puromycin selection, single colonies were picked from both Drp1 sgRNA and empty vector transfected ESCs and screened for mutations. Drp1 exon 2 deletion was confirmed by PCR genotyping using the following primers: Drp1_genot F: 5’ GGATACCCCAAGATTTCTGGA 3’; Drp1_genot R: 5’ AGTCAGGTAATCGGGAGGAAA 3’, followed by Sanger Sequencing. Mfn2 exon 3 deletion was confirmed by PCR genotyping using the following primers: Mfn2_genot F: 5’ CAGCCCAGACATTGTTGCTTA 3’; Mfn2_genot R: 5’ AGCTGCCTCTCAGGAAATGAG 3’, followed by Sanger Sequencing.
Derivation of mESCs from hybrid mouse strains
The derivation of new mESC lines was adapted from 58. Cells were derived from embryos of hybrid mouse strains BG, HB and ST. These contain the mtDNA of C57BL/6N (Bl6) lab mouse and mtDNA variants from wild-caught mice 16.
Embryos were isolated at E2.5 (morula stage) and cultured in 4-well plates (Nunc, Thermo Scientific) containing KSOM media (Millipore) plus two inhibitors (KSOM+2i): 1 μM MEK inhibitor PDO325901 (Sigma-Aldrich) and 3 μM GSK-3 inhibitor CHIR9902 (Cayman Chemicals) for 2 days at 37°C in 5% CO2 incubator. To reduce evaporation, the area surrounding the wells was filled with DPBS. Embryos were further cultured in a fresh 4-well plates containing, N2B27+2i+LIF media: N2B27 media supplemented with 1 μM MEK inhibitor PDO325901 and 3 μM GSK-3 inhibitor and 0.1% LIF for up to 3 days until reaching the blastocyst stage. Each embryo was then transferred to a well of a 96-well plate coated with 0.1% gelatin in DPBS and containing 150 μL of N2B27+2i+LIF media per well. In these conditions, the embryos should attach to the wells allowing the epiblast to form an outgrowth. This plate was then incubated at 37°C in 5% CO2 incubator for 3 to 7 days until ES-like colonies start to develop from the epiblast outgrowth. Cells were passaged by dissociation with Accutase (Sigma) and seeded in gradual increasing surface area of growth (48-well, 24-well, 12-well plate, T12.5 and T25 flask), until new cell lines were established. At this stage cells were weaned from N2B27+2i+LIF media and then routinely cultured in ES media.
These new cell lines were then subjected to characterisation by flow cytometry (cell size, granularity and mitochondrial membrane potential) and Amplification Refractory Mutation System (ARMS)-qPCR assay16 (to determine heteroplasmy).
Heteroplasmy quantification by ARMS-qPCR assay
Every qPCR run consisted of the consensus and an ARMS assay. Consensus assay:
CO2-f: TCTTATATGGCCTACCCATTCCAA, CO2-R: GGAAAACAATTATTAGTGTGTGATCATG, CO2-FAM: 6FAMTTGGTCTACAAGACGCCACATCCCCT-BHQ-1®
(Amplicon length: 103bp)
ARMS-assays:
16SrRNA2340/Staudach-f: AAACCAACATATCTCATTGACCgAA (haplotype ST), 16SrRNA2340(3)G-f: AATCAACATATCTTATTGACCaAG (haplotype C57BL/6N), 16SrRNA2340(3)A-f: AATCAACATATCTTATTGACCgAA (haplotypes BG and HB), 16SrRNA2458-r: CAC CAT TGG GAT GTC CTG ATC, 16SrRNA-FAM: FAM-CAA TTA GGG TTT ACG ACC TCG ATG TT-BHQ-1®.
Lower-case letters indicate the intentional mismatch (ARMS), underlined letters indicate SNP-specific bases. (Amplicon length: 142bp BG, HB; 143bp ST).
Master-mixes for triplicate qPCR reactions contained 1 × buffer B2 (Solis BioDyne, Estonia); 4.5 mM MgCl2; 200 μM of the four deoxynucleotides (dNTPs, Solis BioDyne, Estonia), 0.7 units HOT FIREPol DNA polymerase (Solis BioDyne, Estonia), 300 nM of each primer and 100 nM hydroloysis probe. Per reaction 12 μL of master-mix and 3 μL DNA were transferred in triplicates to 384-well PCR plates (Life Technologies, Austria) using the automated pipetting system epMotion 5075TMX (Eppendorf, Germany). Amplification was performed on the ViiA 7 Real-Time PCR System using the ViiA™ 7 Software v1.1 (Life Technologies, USA). DNA denaturation and enzyme activation were performed for 15 min at 95°C. DNA was amplified over 40 cycles consisting of 95°C for 20 sec, 58°C for 20 sec and 72 °C for 40 sec for all assays.
The standard curve method was applied. Amplification efficiencies were determined for each run separately by DNA dilution series consisting of DNA from mice harbouring the respective analysed mtDNA. Typical results: slope = -3.462, -3.461, -3.576, -3.668; mean efficiency = 0.95, 0.94, 0.90, 0.87; and Y-intercept = 32.4, 33.8, 34.5, 31.9; for the consensus, C57BL/6N, HB and BG, and ST assays respectively (see Supplementary Fig. 1-4). Coefficient of correlation ≥ 0.99 in all assays in all runs. All target samples lay within the linear interval of the standard curves. To test for specificity, in each run a negative control sample, i.e. a DNA sample of a mouse harbouring the mtDNA of the non-analysed type in the heteroplasmic mouse (i.e. C57BL/6N or the respective wild-derived mtDNA) was measured. All assays could discriminate between C57BL/6N and wild-mouse mtDNA at a minimum level of > 1%. Target sample DNA was tested for inhibition by dilution in Tris-EDTA buffer (pH 8.0).
For the calculation of mtDNA heteroplasmy, always the assay detecting the minor allele (C57BL/6N or wild-derived <50%) was used. If both specific assays gave values >50% (what can happen around 50% heteroplasmy), the mean value of both assays was taken. All qPCR runs contained no template controls (NTCs) for all assays; these were negative in 100%.
ARMS-qPCR standard curves and detection limit
Mitochondrial DNA heteroplasmy was quantified by ARMS-qPCR, an established method in the field 16, 59–65. Calibration curves were created with a dilution series of DNA that matches 100% the respective assay. Therefore, for all assays necessarily divergent dilution series had to be used. The amount of DNA between the dilution series can diverge and are plotted as arbitrary units. Supplementary Fig. 1 shows standard curve produced for the consensus assay (detecting mtCo2 gene as measure of total mtDNA) and Supplementary Fig. 2-4 show standard curves produced for specific mtDNA variants (laboratory mouse mtDNA, C57BL/6N; wild-derived mice mtDNAs BG,HB and ST).
SNP-specific quantification of mtDNA
To test the SNP-specific quantification of mtDNA, mixtures of match and mismatch DNA were analysed in triplicates. All assays could discriminate between C57BL/6N and wild-mouse mtDNA (and vice versa) at a minimum level of 1%, as shown by the ARMS-qPCR typical false positive signal with the 100% mismatch DNA (detection limit, in all assays below 0.3%). The results and amplification plots for the specific quantification of HB and BG wild-mouse mtDNA from C57BL/6N mtDNA are shown in Supplementary Fig. 5 and Supplementary Table 9. The results and amplification plots for the specific quantification of ST wild-mouse mtDNA from C57BL/6N mtDNA are shown in Supplementary Fig. 6 and Supplementary Table 10. Average values of the triplicate values are shown.
Embryo experiments
Early mouse embryos were isolated at E5.5 (from pregnant CD1 females, purchased from Charles River, UK). Following dissection from the decidua, embryos were cultured overnight in N2B27 “poor” media (same formulation as N2B27 media but supplemented with 0.5xB27 supplement and 0.5xN2 supplement) with pan-caspase inhibitors (100 μM, Z-VAD-FMK, FMK001, R&D Systems, USA) or equal volume of vehicle (DMSO) as control. On the next morning, embryos were processed for single cell RNA-Seq (scRNA-seq) or functional validation (Δψm analysis and immunohistochemistry for markers of loser cells).
For the scRNA-seq and Δψm analysis embryos were dissociated into singe-cells. Briefly, up to 12 embryos were dissociated in 600 μL Acccutase (A6964, Sigma, UK) during 12 min at 37°C, tapping the tube every two minutes. Accutase was then neutralised with equal volume of FCS, cells span down and stained with TMRM, for Δψm analysis, or directly re-suspended in 300 μL DPBS with 1% FCS, for single cell sorting and RNA-seq. Sytox blue (1:1000, S34857, Thermo Fisher Scientific, UK), was used as viability staining.
Differentiation and Cell competition assays
Cell competition assays between wild-type and Bmpr1a-/-, 4n or Drp1-/- cells were performed in differentiating conditions. Cells were seeded onto fibronectin-coated plates (1:100, Merck) in DPBS during 1h at 37°C and grown in N2B27 media - to promote the differentiation of mESCs into a stage resembling the post-implantation epiblast, as cell competition was previously shown to occur in these conditions 6. N2B27 media consisted of 1:1 Dulbecco’s modified eagle medium nutrient mixture (DMEM/F12) and Neurobasal supplemented with N2 (1x) and B27 (1x) supplements, 2 mM L-glutamine and 0.1 mM β-mercaptoethanol - all from Gibco. Cell competition assays between wild-type and Mfn2-/- and between mESCs with different mtDNA content were performed in pluripotency maintenance conditions (ES media).
Cells were seeded either separately or mixed for co-cultures at a 50:50 ratio, onto 12 well plates, at a density of 8E04 cells per well, except for assays between wild-type and Mfn2-/- mESCs, where 3.2E05 cells were seeded per well. The growth of cells was followed daily and compared between separate or co-culture, to control for cell intrinsic growth differences, until the fourth day of culture. Viable cells were counted daily using Vi-CELL XR Analyser (Beckman Coulter, USA), and proportions of each cell type in co-cultures were determined using LSR II Flow Cytometer (BD Bioscience), based on the fluorescent tag of the ubiquitously expressed GFP or TdTomato in one of the cell populations.
Metabolomic analysis
The metabolic profile was obtained using the Metabolon Platform (Metabolon, Inc). Each sample consisted of 5 biological replicates. For each replicate, 1E07 cells were spun down and snap frozen in liquid nitrogen. Pellets from 5 independent experiments for each condition were analysed by Metabolon Inc by a combination of Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy (UPLC-MS/MS) and Gas Chromatography-Mass Spectroscopy (GC-MS). Compounds were identified by comparison to library entries of purified standards based on the retention time/index (RI), mass to charge ratio (m/z), and chromatographic data (including MS/MS spectral data) on all molecules present in the library. Samples were normalized to protein content measured by Bradford assay. Statistical analysis was done using Welch’s two-sample t-test and statistical significance defined as p ≤0.05.
Seahorse analysis
The metabolic function of cells was assessed by extracellular flux analysis using Seahorse XF24 (Agilent Technologies, UK). For assays ran during pluripotency, cells were seeded, on the day prior to the assay, onto 0.1% gelatin-coated (Sigma, UK) in 300 μL of ES media. All cell types were seeded at 5×104 cells per well, except for Bmpr1a-/- cells, that were seeded at 6E04 per well). For assays ran during differentiation, cells were seeded, the 3 days before the assay, onto fibronectin-coated fibronectin-coated plates (1:100, Merck, UK), in 300 μL of N2B27media. All cell types were seeded at 2.4E04 cells per well, except for Bmpr1a-/- cells, that were seeded at 3.2E04 cells per well.
On the day of the assay, cells were carefully washed twice with assay media and then left with a final volume of the 600 μL per well. The plate was then equilibrated on a non-CO2 incubator at 37°C for 30 min. The assay media consisted in unbuffered DMEM (D5030 – Sigma, UK), that was supplemented on the day of the assay according to the test performed. For the OCR measurements the assay media was supplemented with 0.5 g.L−1 of glucose (Sigma, UK) and 2 mM of L-glutamine (Life Technologies, UK), while for the ECAR measurements the media was supplemented with 1 mM of Sodium Pyruvate and 2 mM of L-glutamine (both from Life Technologies, UK), pH 7.4 at 37°C.
The protocol for the assay consisted of 4 baseline measurements and 3 measurements after each compound addition. Compounds (all from Sigma, UK) used in OCR and ECAR assays were prepared in the supplemented assay media. For the OCR assay, test the following compounds were added: 1 mM Pyruvate (Pyr), 2.5 μM oligomycin (OM), 300 nM Carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP) and a mixture of rotenone and antimycin A at 6 μM each (R&A). For the ECAR assay, the following compounds were added: 2.5 mM and 10 mM of glucose, 2.5 μM of oligomycin (OM), and a 50 mM of 2-deoxyglucose (2-DG). Each of the experiments was performed in 3 times, with 5 biological replicates of each cell type. For background correction measurements, 4 wells were left without cells (A1, B4, C3 and D6). Both ECAR and OCR measurements were performed on the same plate. The assay parameters for both tests were calculated following the Seahorse assay report generator (Agilent Technologies, UK).
At the end of the assay, cells were fixed and stained with Hoechst. Both OCR and ECAR were normalised to cell number, determined by manual cell counts using Fiji software. The normalisation of the data was processed on Wave Desktop software (Agilent Technologies, UK) and data exported to Prism 8 (GraphPad) for statistical analysis.
Analysis of Δψm and mitochondrial ROS
For TMRM staining in single cells from early mouse epiblasts, embryos were dissected at E5.5 and cultured overnight in the presence or absence of caspase inhibitors. On the following morning, to avoid misleading readings, epiblasts were isolated initially by an enzymatic treatment with of 2.5% pancreatin, 0.5% trypsin and 0.5% polyvinylpyrrolidone (PVP40) - all from Sigma-Aldrich - to remove the visceral endoderm (VE). Embryos were treated during 8 min at 4°C, followed by 2 min at RT. The VE was then pealed with the forceps and the extraembryonic ectoderm removed to isolate the epiblasts. Twelve epiblasts were pooled per 600μL of Accutase (Sigma-Aldrich) for dissociation into single cells prior to staining. Reaction was stopped with equal volume of FCS and cells subjected to TMRM staining. Cells were incubated in 200 μL of 10 nM Nernstian probe tetramethylrhodamin methyl ester perchlorate (TMRM, T5428, Sigma, UK), prepared in N2B27 media. After incubating for 15 min at 37°C, cells were pelleted again and re-suspended in flow cytometry (FC) buffer (3% FCS in DPBS). Sytox blue (1:1000, S34857, ThermoFisher Scientific, UK), was used as viability staining.
Quantitative analysis of Δψm and mitochondrial ROS was performed by flow cytometry. Cells were grown in pluripotency or differentiating conditions. Cells were dissociated and pelleted to obtain 2E05 cells per sample for the staining procedure. For TMRM staining in mESCs, 2E05 cells of each cell line were resuspended in 200 μL of 10 nM TMRM (T5428, Sigma, UK), prepared in N2B27 media. Cells were incubated at 37°C for 15 min, and then resuspended in FC buffer (3% FCS in DPBS). For the analysis of mitochondrial ROS, cells were grown in differentiating conditions and stained on the third day of culture. Briefly, 2E05 cells of each cell line were resuspended in 200 μL of 5 μM solution of MitoSOX (M36008, Invitrogen, UK) prepared in N2B27 media. Cells were incubated at 37°C for 15 min, and then resuspended in FC buffer. Sytox blue was used as viability staining.
Stained cell suspensions with TMRM or MitoSOX were analysed in BD LSRII flow cytometer operated through FACSDiva software (Becton Dickinson Biosciences, UK). For TMRM fluorescence detection the yellow laser was adjusted for excitation at λ=562 nm, capturing the emission light at λ=585 nm for TMRM. MitoSOX fluorescence was analysed with the violet laser adjusted for excitation at λ=405 nm, capturing the emission light at λ=610 nm. In the case of GFP-labelled cell lines, for GFP fluorescence detection the blue laser was adjusted for excitation at λ=488 nm, capturing the emission light at λ=525 nm. Results were analysed in FlowJo Software v9 or v10.0.7r2. See FACS gating strategy in Supplementary Fig. 7.
Qualitative analysis of Δψm was performed by confocal microscopy. Wild-type and Bmpr1a-/- cells were grown in fibronectin-coated glass coverslips. On the third day of differentiation, cells were loaded with 200 nM MitoTracker Red probe (Life Technologies), prepared in N2B27 media, for 15 min at 37°C. Cells were then washed with DPBS and fixed with 3.7% formaldehyde for subsequent immunocytochemical staining of total mitochondria mass, with TOMM20 antibody.
Immunofluorescence
Cells were washed with DPBS and fixed with 3.7% formaldehyde (Sigma, UK) in N2B27, for 15 min at 37°C. Permeabilization of the cell membranes was done with 0.4% Triton X-100 in DPBS (DPBS-Tx), at RT with agitation. Blocking step with 5% BSA in DPBS-Tx 0.1% was performed for 30 min, at RT with agitation. Mitochondria were labelled with TOMM20 antibody (1:100, Santa Cruz Biotechnologies). Dead cells were labelled with cleaved caspase-3 antibody (1:400, CST-9664) and NANOG antibody was used to mark pluripotent cells (1:100, eBioscience). Secondary antibodies were Alexa Fluor 488 and 568 (1:600, Invitrogen). Primary antibody incubation was performed overnight at 4°C and secondary antibody incubation during 45 min, together with Hoechst to stain nuclei (1:1000, Thermo Scientific), at RT and protected from light. In both cases antibodies were diluted in blocking solution. Three 10 min washes with DPBS-Tx 0.1% were performed between each critical step and before mounting with Vectashield medium (Vector Laboratories).
Samples were imaged with a Zeiss LSM780 confocal microscope (Zeiss, UK) and processed with Fiji software 68. Mitochondria stainings were imaged with a 63x/1.4 Oil objective. For samples stained with TOMM20 antibody and MitoTracker Red, Z-stacks were acquired and processed for deconvolution using Huygens software (Scientific Volume Imaging, https://svi.nl/). Samples stained with cleaved caspase-3 were imaged with 20x/0.8 air objective. Imaging and deconvolution analysis were performed with the support and advice from Mr. Stephen Rothery from the Facility for Imaging by Light Microscopy (FILM) at Imperial College London.
Embryo immunofluorescent staining for p-rpS6, OPA1 and DDIT3 (CHOP) markers was performed as follows. Cultured embryos were fixed in 4% PFA in DPBS containing 0.01% Triton and 0.1% Tween 20 during 20 min at RT. Permeabilization of the membranes was done during 10 min in DPBS with 0.5% Triton. Embryos were blocked in 5% BSA in DPBS with 0.25% Triton during 45 min. Incubation with primary antibodies - CHOP (1:500, CST-2895), OPA1 (1:100, BD Biosciences - 612606) and p-rpS6 (1:200, CST - 5364) - was done overnight at 4°C in 2.5% BSA in DPBS with 0.125% Triton. On the following morning, hybridisation with secondary antibodies Alexa Fluor 568 and Alexa Fluor 488 (Invitrogen, diluted 1:600 in DPBS with 2.5% BSA and 0.125% Triton) was done next during 1h at RT. Hoechst was also added to this mixture to stain nuclei (1:1000, Invitrogen). Three 10 min washes with filtered DPBS-Tx 0.1% were performed between each critical step. All steps were done with gentle agitation.
Embryos were imaged in embryo dishes (Nunc) in a drop of Vectashield using Zeiss LSM780 confocal microscope at 40x/1.3 oil objective.
Further details about image acquisition and processing are specified in the Supplementary Table 8.
Western Blotting
Cells were washed in DPBS and lysed with Laemmli lysis buffer (0.05 M Tris- HCl at pH 6.8, 1% SDS, 10% glycerol, 0.1% β-mercaptoethanol in distilled water). Total protein quantification was done using BCA assay (Thermo Scientific, UK) and samples (15μg of protein per lane) were loaded into 12% Bis-Tris protein gels (BioRad). Resolved proteins were transferred into nitrocellulose membranes (GE Healthcare). The following primary antibodies, prepared in TBS-0.1% Tween containing 5% BSA were incubated overnight at 4°C with gentle agitation: rabbit anti-TOMM20 (1:1000, CST - 42406), mouse anti-ATPB (1:1000, Abcam - ab14730), rabbit anti-α-Tubulin (1:1000, CST- 2144), mouse anti-mt-CO1 (1:2000, Abcam - ab14705), rabbit anti-DRP1 (1:1000, CST- 8570), mouse anti-MFN1 (1:1000, Abcam - ab57602), mouse anti-MFN2 (1:500, Abcam - ab56889), mouse anti-Vinculin (1:1000, Sigma - V9131), mouse anti-OPA1 (1:1000, BD Biosciences - 612606), rabbit anti-ATF4 (1:1000, CST-11815), rabbit anti-PCNA (1:5000, Abcam - ab18197) and rabbit anti-p-eIF2α (Ser51, 1:1000, CST-9721). On the following morning, HRP-conjugated secondary antibodies (1:5000, sc-2004 and sc-2005, Santa Cruz), prepared in TBS-0.1% Tween containing 5% milk (Sigma) were incubated for 1h at RT under gentle agitation. Membranes were developed with ECL reagents (Promega) and mounted in cassette for time-time-controlled exposure to film (GE Healthcare).
Bulk RNA-Seq and Single cell RNA-Seq
For bulk RNA Seq in the competitive scenario between cells with different mtDNA, HB(24%) and BG(95%) mESCs were grown separately or in co-culture. On the third day of culture cells were dissociated and subjected to fluorescence activated cell sorting (FACS) to separate the cell populations in co-culture according to their GFP label. Propidium iodine (1:1000, 81845, Sigma, UK) were used as viability staining. See FACS gating strategy in Supplementary Fig. 8. To control for eventual transcriptional changes due to the FACS process, a mixture of the two separate populations was subjected to the same procedure as the co-cultured samples. Total RNA isolation was then carried out using RNA extraction Kit (RNeasy Mini Kit, Qiagen). PolyA selection/enrichment was the method adopted for library preparation, using the NEB Ultra II RNA Prep Kit. Single end 50bp libraries were sequenced on Illumina Hiseq 2500. Raw basecall files were converted to fastq files using Illumina’s bcl2fastq (version 2.1.7). Reads were aligned to the mouse genome (mm9) using Tophat2 version 2.0.11 69 with default parameters. Mapped reads that fell on genes were counted using featureCounts from Rsubread package 70. Generated count data were then used to identify differentially expressed genes using DESeq2 71. Genes with very low read counts were excluded. Finally, Gene Set Enrichment Analysis was performed using GSEA software 72, 73 on pre-ranked list generated by DESeq2.
To investigate the nature of cells eliminated by cell competition during early mouse embryogenesis by means of Single Cell RNA-Sequencing (scRNA-seq), early mouse embryos were dissected at E5.5 and cultured overnight in the presence or absence of caspase inhibitors. On the following morning, embryos were dissociated with Accutase and subjected to single-cell sorting into 384-well plates. Total RNA isolation was then carried out using a RNA extraction Kit (RNeasy Mini Kit, Qiagen). scRNA-seq was performed using the Smart-seq2 protocol74. PolyA selection/enrichment with Ultra II Kit (NEB) was the method adopted for library preparation.
Data processing, quality control and normalization
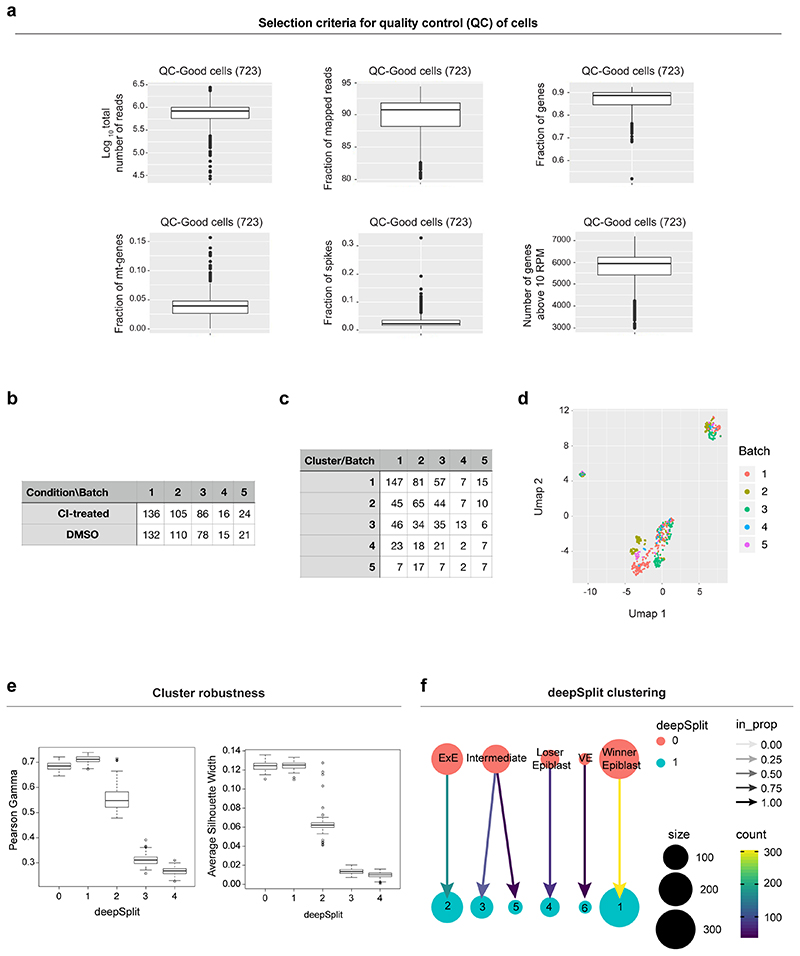
We performed transcript quantification in our scRNA-seq data by running Salmon v0.8.2 75 in the quasi-mapping-based mode. First, a transcriptome index was created from the mouse reference (version GRCm38.p4) and ERCC spike-in sequences. Then, the quantification step was carried out with the “quant” function, correcting for the sequence-specific biases (“--seqBias” flag) and the fragment-level GC biases (“--gcBias” flag). Finally, the transcript level abundances were aggregated to gene level counts. On the resulting raw count matrix including 1,495 cells, we apply a quality control to exclude poor quality cells from downstream analyses.
For the quality control we used the following criteria: we identified the cells that have a log10 total number of reads equal to or greater than 4, a fraction of mapped reads equal to or greater than 0.8, a number of genes with expression level above 10 reads per million equal to or greater than 3000 and a fraction of reads mapped to endogenous genes equal to or greater than 0.5. This resulted in the selection of 723 cells, which were kept for downstream analyses. Transcripts per million (TPM) normalization (as estimated by Salmon) was used.
Highly variable genes and dimensionality reduction
To identify highly variable genes (HVG), first we fitted a mean-total variance trend using the R function “trendVar” and then the variance was decomposed into biological and technical components with the R function “decomposeVar”; both functions are included in the package “scran” (version 1.6.9 76).
We considered HVGs those that have a biological component that is significantly greater than zero at a false discovery rate (Benjamini-Hochberg method) of 0.05. Then, we applied further filtering steps by keeping only genes that have an average expression greater to or equal than 10 TPM and are significantly correlated with one another (function “correlatePairs” in “scran” package, FDR<0.05). This yielded 1921 genes, which were used to calculate a distance matrix between cells defined as , where ρ is the Spearman’s correlation coefficient between cells. A 2D representation of the data was obtained with the UMAP package (version 0.2.0.0 https://cran.r-project.org/web/packages/umap/index.html) using the distance matrix as input.
Cell clustering and connectivity analysis
To classify cells into different clusters, we ran hierarchical clustering on the distance matrix (see above; “hclust” function in R with ward.D2 aggregation method) followed by the dynamic hybrid cut algorithm (“cutreeDynamic” function in R package “dynamicTreeCut” (https://CRAN.R-project.org/package=dynamicTreeCut) version 1.63.1, with the hybrid method, a minimum cluster size of 35 cells and a “deepSplit” parameter equal to 0), which identified five clusters. Cells from different batches were well mixed across these five clusters (see Extended Data Fig. 1), suggesting that the batch effect was negligible. The identity of the five clusters was established based on the expression of known marker genes of Epiblast, Visceral Endoderm and Extra-Embryonic Ectoderm, which were identified in a previous study56. The expression levels of some of the top markers is plotted in Figure 1b.
We performed a robustness analysis on the clustering by exploring in detail how the choices of genes, clustering parameters and algorithms affect the identity and the number of clusters. First, we quantified the cluster robustness by calculating Pearson’s gamma and the Average silhouette width obtained with 100 random subsets of 60% of the highly variable genes and different values of the deepSplit parameter. While the robustness at deepSplit=0 and 1 is similar, for greater values of deepSplit (corresponding to less conservative clustering) the robustness rapidly declines (Extended Data Figure 2a). The clustering with deepSplit = 0 and 1 (the more robust choices) yield very similar results, the only difference being the splitting of the intermediate cluster in two subclusters (Extended Data Figure 2b).
In addition to this, we also used Louvain clustering on the highly variable genes (resolution=0.3, k=20 with 20 principal components), which again produced very similar clusters.
We quantified the connectivity between the clusters (using only CI-treated cells) with PAGA22 implemented in the python library scanpy (version 1.4.7)77. The analysis revealed that the three epiblast clusters are connected with each other while the two extra embryonic tissues (Visceral Endoderm and Extra Embryonic Ectoderm) are isolated (Extended Data Figure 2c).
Identification of a single-cell trajectory in the epiblast
We calculated a diffusion map (“DiffusionMap” function in the R package “destiny” version 2.6.2 23 on the distance defined above on the epiblast cells from CI-treated embryos. The pseudotime coordinate was computed with the “DPT” function with the root cell in the winner epiblast cluster (identified by the function “tips” in the “destiny” package). Such pseudotime coordinate can be interpreted as a “losing score” for all the epiblast cells from the CI-treated embryos.
We estimated the losing scores of the epiblast cells from DMSO-treated embryos by projecting such data onto the diffusion map previously calculated (function “dm_predict” in the destiny package). Finally, for each of the projected cells, we assigned the losing score as the average of the losing scores of the 10 closest neighbours in the original diffusion map (detected with the function “projection-dist” in the destiny package).
While for the clustering and the trajectory analysis we used the highly variable genes computed from the whole dataset, we verified that all results concerning the separation between winner and loser epiblast cells (eg, clusters and losing score) remain unaffected if the highly variable genes are calculated using only the epiblast cells.
Mapping of data from epiblast cells onto published datasets
We compared the transcriptional profile of epiblast from embryos cultured in DMSO and CI with that of epiblast collected from freshly isolated embryos at different stages.
To do this, we considered the dataset published in26, which includes epiblast cells from embryos at the stages E5.5 (102 cells), E6.25 (130 cells) and E6.5 (288 cells). A diffusion map and a diffusion pseudotime coordinate were computed with these cells following the same procedure described in the section above (Extended Data Figure 2d-e). Then, we projected epiblast cells from CI and DMSO-treated embryos and we assigned to them a diffusion pseudotime coordinate as described above (Extended Data Figure 2f).
Differential gene expression analysis along the trajectory
To identify the genes that are differentially expressed along the trajectory, first we kept only genes that have more than 15 TPM in more than 10 cells (this list of genes is provided in Supplementary Table 4); then, we obtained the log-transformed expression levels of these genes (adding 1 as a pseudo-count to avoid infinities) as a function of the losing score and we fitted a generalized additive model to them (R function “gam” from “GAM” package version 1.16.). We used the ANOVA test for parametric effect provided by the gam function to estimate a p-value for each tested gene. This yielded a list of 5,311 differentially expressed genes (FDR < 0.01).
Next, we looked for groups of differentially expressed genes that share similar expression patterns along the trajectory. To this aim, similarly to what we did when clustering cells, we calculated a correlation-based distance matrix between genes, defined as , where ρ is the Spearman’s correlation coefficient between genes. Hierarchical clustering was then applied to this matrix (hclust function in R, with ward.D2 method) followed by the dynamic hybrid cut algorithm (dynamicTreeCut package) to define clusters (“cutreeDynamic” function in R with the hybrid method and a minimum cluster size of 100 genes and a deepSplit parameter equal to 0). This resulted in the definition of four clusters, three of genes that decrease along the trajectory (merged together for the GO enrichment and the IPA analysis) and one of increasing genes (Extended Data Fig. 2d). IPA (QIAGEN Inc., https://www.qiagenbio-informatics.com/products/ingenuity-pathway-analysis), was run on all genes differentially expressed (FDR < 0.01) along the trajectory from winner to loser cells (see Fig. 2a-d and Fig. 3a-c), using all the tested genes as a background (see Supplementary Table 4). This software generated networks, canonical pathways and functional analysis. The list of decreasing/increasing genes is provided in Supplementary Tables 1 and 2. The pathways found as mis-regulated in Figure 3 were: Mitochondrial Dysfunction, -log10(p-value) = 21.1; Oxidative Phosphorylation, - log10(p-value) = 18.6; EIF2 signalling, - log10(p-value) = 11.9. FDR for the genes shown in b ranges from 1.25E-51 (for Atp5b) to 5.42E-03 (for Ndufa11). Cox6b2, coloured in yellow, was found to be up-regulated in loser cells (FDR=2.69E-13).
Analysis of heteroplasmy in single-cell RNA seq dataset
We used STAR (version 2.7 78) to align the transcriptome of the epiblast cells from CI-treated embryos (274) to the mouse reference genome (mm10). Only reads that uniquely mapped to the mitochondrial DNA (mtDNA) were considered. From these, we obtained allele counts at each mtDNA position with a Phred Quality Score greater than 33 using the samtools mpileup function. Next, we applied filters to remove cells and mtDNA positions with a low coverage. First, we removed cells with fewer than 2,000 mtDNA positions covered by more than 50 reads. Second, we removed positions having less than 50 reads in more than 50% of cells in each of the three epiblast clusters (winner, intermediate and loser). These two filters resulted in 259 cells and 5,192 mtDNA positions (covered by ~700 reads per cell on average) being considered for further analyses.
Starting from these cells and positions, we applied an additional filter to keep only positions with a sufficiently high level of heteroplasmy. To this aim, for each position with more than 50 reads in a cell, we estimated the heteroplasmy as:
where fmax is the frequency of the most common allele. We kept only positions with H>0.01 in at least 10 cells.
Finally, using generalized additive models (see above), we identified the positions whose heteroplasmy H changes as a function of the cells’ losing score in a statistically significant way. We found a total of eleven significant positions (FDR < 0.001), six of them in the mt-Rnr1 gene and five in the mt-Rnr2 gene. All of these positions have a higher level of heteroplasmy in loser cells (see Fig. 6b-g and Extended Data Fig. 7f-k). The results remain substantially unaltered if the Spearman’s rank correlation test (in alternative to the generalized additive models) is used. The most common substitutions observed in each position are the following: mt-Rnr1 300 A-to-C; mt-Rnr1 303 T-to-G; mt-Rnr1 304 T-to-G; mt-Rnr1 305 C-to-G;mt-Rnr1 326 A-to-G; mt-Rnr1 327 C-to-G; mt-Rnr2 2031 T-to-G; mt-Rnr2 2074 C-to-G; mt-Rnr2 2077 A-to-C; mt-Rnr2 2079 C-to-T; mt-Rnr2 2081 A-to-G.
For the bar plot shown in Fig. 6h and the correlation heatmaps in Fig. 6i and Extended Data Fig. 7l, we took into account only cells that covered with more than 50 reads all the significant positions in the mt-Rnr1 gene (215 cells, Fig. 6h-6i) or in both the mt-Rnr1 and mt-Rnr2 genes (214 cells, Extended Data Fig. 7l).
As a negative control, we repeated the analysis described above using the ERCC spike-ins added to each cell. As expected, none of the positions was statistically significant, which suggested that our procedure is robust against sequence errors introduced during PCR amplification.
We also performed the mtDNA heteroplasmy analysis in cells from the Visceral Endoderm and the Extra-Embryonic Ectoderm in both DMSO and CI conditions: none of these cells have a mtDNA heteroplasmy higher than 0.01 in the 11 significant positions identified within mt-Rnr1 and mt-Rnr2 in loser epiblast cells, and the reference allele is always the most common. This reinforces the hypothesis that such variants are specific to loser epiblast cells and are not resulting from contamination.
To test the reliability of our heteroplasmy estimations, we used the RNA-seq data from two of the mtDNA cell lines (BG and HB, see Figure 7), for which the heteroplasmy was measured also by ARMS-qPCR. To do so, first we downloaded the fasta files of the two mtDNA cell lines from https://www.ncbi.nlm.nih.gov/nuccore/KC663619.1 and https://www.ncbi.nlm.nih.gov/nuccore/KC663620.1, then we identified the mtDNA positions that differ from the BL6 reference genome. Finally, on these different positions, the heteroplasmy H was computed as explained above. The values of heteroplasmy we found with our computational analysis were very close to those estimated by ARMS-qPCR: for HB(24%), ~17% from RNA-seq data vs ~24% measured by ARMS-qPCR; and for BG(95%), ~93% from RNA-seq data vs ~95% measured by ARMS-qPCR.
Since we are inferring mtDNA changes from RNA sequencing data, we have also considered additional potential sources for the sequence changes we observed. Specifically, one possible source is contamination from nuclear mitochondrial DNA segments (NUMTs). However, a NUMT contamination is very unlikely for the following reasons: (i) we considered only reads that uniquely mapped to the mitochondrial genome; (ii) the variants with highest heteroplasmy identified in “loser” cells (mt-Rnr1 326 and 327) were not present in any of the NUMTS previously reported79 or those that we identified using blastn (also taking into account the SNPs of the mouse strain we used); (iii) the variants detected are exclusively found in “loser” epiblast cells, and they are not detected in any other cell type from the same embryos, i.e., neither in “winner” epiblast cells nor in cells from extra-embryonic tissues; (iv) we estimated that if the variants with the strongest heteroplasmy (i.e., mtRnr-1 326 and 327) were present on a NUMT, in order for them to reach an heteroplasmy of ~20% (see Fig. 6B and 6C), the NUMT would have to be expressed at high levels, comparable or even higher than many mitochondrial genes. Another possible cause of the sequence changes is RNA editing. However, the majority of the changes that we found (see above) are not compatible with the canonical RNA editing in Metazoans, which consists of A-to-I (which would be read as A-to-G in RNA-sequencing) and Cto-U80.
Common features of scRNA-seq and bulk RNA-seq datasets
Differential expression analysis between the co-cultured winner HB(24%) and loser cell line BG(95%) was performed using the package EdgeR version 3.20.981.
Batches were specified in the argument of the function model.matrix. We fitted a quasi-likelihood negative binomial generalized log-linear model (with the function glmQLFit) to the genes that were filtered by the function filterByExpr (with default parameter). These genes were used as background for the gene enrichment analysis.
We set a FDR of 0.001 as a threshold for significance. The enrichment analysis for both the scRNA-seq and bulk RNA-seq datasets were performed using the tool g:Profiler 82. The list of up-regulated, down-regulated and background genes related to the DE analysis for the bulk RNA-seq dataset are provided in the Supplementary Tables 5, 6 and 7.
Quantification, Statistical Analysis and Reproducibility
The quantification of the DDIT3 and OPA1 expression in embryos was done by two distinct methods. DDIT3 expression was quantified by counting the number of epiblast cells with positive staining in the embryos of each group. The expression of OPA1 was quantified on Fiji software as the mean fluorescence across a 10 pixels width line drawn on the basal cytoplasm of each cell with high or low p-rpS6 fluorescence intensity, as specified in7. min of 8 cells were quantified per condition (high vs low mTOR activity) in each embryo. Six embryos treated with CI were analysed. Mean grey values of OPA1 fluorescence for each epiblast cell are pooled on the same graph.
Flow cytometry data was analysed with FlowJo Software v9 or v10.0.7r2. Western blot quantification was performed using Image Studio Lite v5.2.5 (LI-COR). Protein expression levels were normalised to loading controls vinculin, α-tubulin or PCNA. Normalisation of data from metabolic flux analysis with Seahorse was performed using Wave Desktop software (Agilent Technologies, UK) and data exported to Prism v8 (GraphPad) for statistical analysis.
All box plots show lower quartile (Q1, 25th percentile), median (Q2, 50th percentile) and upper quartile (Q3, 75th percentile). Box length refers to interquartile range (IQR, Q3-Q1). The upper whisker marks the minimum between the maximum value in the dataset and the IQR times 1.5 from Q3 (Q3+1.5 x IQR), while the lower whisker marks the maximum between the minimum value in the dataset and IQR times 1.5 from Q1 (Q1-1.5 x IQR). Outliers are shown outside the interval defined by box and whiskers as individual points.
The micrographs shown on Fig. 3d represents one of the micro dissected embryo epiblasts used for the experiment presented in Fig. 3g-h. The representative confocal microscopy images shown on Fig. 4h are from confocal imaging deconvolution performed from one experiment following reproducibility of observations from previous independent experiments. The representative confocal microscopy images were from one experiment following reproducibility of observations from previous independent experiments.
The statistical analysis of the results generated in wet lab was performed using GraphPad Prism version 8.0.0 for Mac (GraphPad Software, San Diego, California USA). Data was tested for normality using Shapiro-Wilk normality test. Two-tailed parametric or non-parametric statistical tests were applied accordingly. Statistical significance was considered with a confidence interval of 0.05%. * p<0.05; ** p<0.01;*** p<0.001.
Here we specify details about the statistical test and multiple comparisons test (when applicable) used for each experiment. The statistical significance of IPA analysis shown in Fig. 2b,c and Fig. 3a,b was calculated with right-tailed Fisher’s exact test (p<0.05). Data presented in Fig. 3f, Fig. 4i, Extended Data Fig. 4g were analysed by Mann-Whitney test. Data shown in Fig 4b-e and Extended Data Fig. 5a-d were analysed by unpaired t-test of Mann-Whitney test (see source data for details). Data shown in Fig 4j, Fig. 5e,g-i were analysed with unpaired t test. One sample t test was used to analysed data presented in Fig. 5d,f. Fig. 4k and Extended Data Fig. 8b-f shows data analysed by one-way ANOVA, followed by Holm-Sidak’s multiple comparisons test. Data presented in Fig. 3h, Fig. 4f-g, Fig. 5b-c,j, Fig. 7b-f, Extended Data Fig. 5e-g, 6b-c and 8g-i were analysed by two-way ANOVA, followed by Holm-Sidak’s multiple comparisons test. The statistical analysis on data from Extended Data Fig. 9 done with one-way ANOVA or Kruskal-Wallis test, followed by Holm-Sidak’s or Dunn’s multiple comparison test, respectively (see source data for details). ANOVA for parametric effects on GAM fit was used test statistical significance for data presented in Fig. 6a-g and Extended Data Fig. 7a-e, The adjusted p-values (indicated at the top of each plot) were computed using Benjamini & Hochberg method. The correlation coefficients shown in Fig. 6i, Extended Data Fig. 2e and 3d and 7l were calculated with Spearman’s rank correlation rho test (two-sided test, 0.95 confidence level). Data shown in Extended Data Fig. 3b-c, 4a and 10b-c was analysed with Fisher’s exact test (two-sided). Gene enrichment analysis shown in Extended Data Fig. 10 was tested for statistical significance with cumulative hypergeometric test. p-values were adjusted for multiple comparisons using the g:Profiler algorithm g:SCS (https://doi.org/10.1093/nar/gkm226). Finally, the statistical analysis on data presented in Supplementary Tables 5 and 6 was performed using empirical Bayes quasi-likelihood F-tests. p-values were adjusted for multiple comparisons using the Benjamini & Hochberg method.
Extended Data
Extended Data Fig. 1. Quality controls of scRNA-seq and clustering robustness analysis.
a, Selection criteria for quality control (QC) of all cells. A total of 723 passed the quality control (723 good quality cells) and were considered for downstream analysis. All these parameters were computed for each cell. Log10 total number of reads (top left): log10 of the sum of the number of reads that were processed in every cell; Fraction of mapped reads (top central): number of reads that are confidentially mapped to the reference genome divided by total number of reads that were processed for each cell. This number is automatically provided by Salmon v0.8.2; Fraction of genes (top right): number of reads mapped to endogenous genes divided by the total sum of reads that were processed; Fraction of mt-genes (bottom left): number of reads mapped to mitochondrial genes divided by the total sum of reads that were processed; Fraction of spikes (bottom central): number of reads mapped to ERCC spike-ins divided by the total sum of reads that were processed; Number of genes above 10 RPM (bottom right): number of genes with expression level above 10 reads per million. b, Number of good quality cells in each condition (rows) and batch (columns). c, Number of good quality cells per cluster (rows) and batch (columns). d, UMAP plot of the data with cells coloured by batch. In each batch there is a balanced distribution of cells in the two conditions and across the five clusters. e, The Pearson’s gamma (left panel) and the Average Silhouette Width (right panel) was calculated for each set of clusters obtained with 100 random subsamples of 60% of highly variable genes and different values of the deepSplit parameter (see Methods). The most robust clusters correspond to deepSplit values of 0 and 1. f, The changes in composition and number of clusters between the clustering obtained with deepSplit 0 (top) and 1 (bottom) are shown using the library “clustree”57. See methods for details on statistical analysis.
Extended Data Fig. 2. Cell cycle analysis and cluster connectivity.
a, Cell cycle analysis of epiblast cells from clusters 1, 3 and 4. Cell cycle phase was predicted with cyclone algorithm58 and shows that there are cells in S and G2M phase also in the loser and intermediate clusters. b, PAGA plot showing the connectivity of the five clusters of cells from CI-treated embryos. c-d, Diffusion map analysis in all epiblast cells (from DMSO and CI-treated embryos): cells are coloured according to the condition (c) and to the cluster (d). e, The pseudo-time coordinate of the CI-treated epiblast cells obtained from the diffusion map including all epiblast cells correlates extremely well (with the pseudo-time coordinate obtained in the diffusion map calculated only from CI-treated epiblast cells (Fig. 2a). See methods for details on statistical analysis.
Extended Data Fig. 3. Analysis on epiblast cells from DMSO and CI-treated embryos.
a, Heatmap showing the expression pattern of all genes differentially expressed along the trajectory from winning to losing cells in Fig. 2d. b-c, Overlap of genes differentially expressed along the trajectory joining winning and losing epiblast cells in CI-treated embryos (Fig. 2a and panel d) and genes targeted by p53. Pie charts show the percentage of genes up- or down-regulated in loser cells within the group of target genes that are activated (b) or repressed (c) by p53. There is an enrichment of activated/repressed targets among genes upregulated/downregulated in losing cells respectively (p-value=1E-4). The list of p53 targets is taken from59. d, Scatter plots of the expression levels of different marker genes plotted against each other in loser epiblast cells (cluster 4). Loser cells have higher expression of pluripotency markers as well as higher expression of some lineage-specific markers and the co-expression of these markers is only weakly correlated - the Spearman’s correlation coefficient is shown. e-g Our scRNA-seq data from epiblast cells is projected on top of previously published data from epiblast collected from freshly isolated embryos at different stages (E5.5, E6.25 and E6.5; data from26). First, a diffusion map (e) and a pseudotime coordinate (f) is computed for the epiblast cells from freshly isolated embryos. Then, a pseudotime coordinate is estimated for our data after projecting it onto the diffusion map. Panel g shows the pseudotime coordinates for both datasets, split by stage, treatment and cluster. See methods for details on statistical analysis.
Extended Data Fig. 4. Cells eliminated during early mouse embryogenesis have activated stress responses.
a, Overlap of genes differentially expressed along the trajectory joining winning and losing epiblast cells in CI-treated embryos (Fig. 2a and Extended Data Fig. 3a) and genes related to the unfolded protein response and integrated protein response pathways (UPR_ISR, see Supplementary Table 3). From the 32 genes related to the UPR & ISR pathways, 12 are down-regulated in loser cells, 8 genes are up-regulated in loser cells, and 12 genes are not differentially expressed between loser and winner cells. There is a statistically significant enrichment of UPR&ISR genes among the up-regulated genes in loser cells (odds ratio=3.0, p-value=0.012). The intersection between UPR-ISR genes and the down regulated genes is not significant (odds ratio=1.2, p value=0.69). b-c, List of genes from UPR-ISR pathways that are statistically significantly up-regulated (b) or down-regulated (c) in loser cells. d, Scatterplots with the expression levels of genes involved in stress responses in epiblast cells from CI-treated embryos as a function of cells’ losing score. e, Experimental design with the approach taken to validate the expression of the stress response marker DDIT3 in epiblast cells from DMSO or CI-treated embryos. f, Representative micrographs of DMSO (upper panel) or CI-treated embryos (100 μM, lower panel) stained for DDIT3, quantified in (g). Nuclei are labelled with Hoechst. In control embryos (DMSO-treated), dying cells in the cavity show very high DDIT3 expression (arrow), while live cells in the epiblast of the CI-treated embryos show more modest levels of DDIT3 expression (arrowheads). Scale bar = 20 μm. g, Quantification of the percentage of epiblast cells with nuclear DDIT3 expression. N=10 DMSO and N=9 CI-treated embryos. Data shown as mean ± SEM. See methods for details on statistical analysis.
Extended Data Fig. 5. Mitochondrial function in wild-type, Bmpr1a-/- and 4n mESCs.
a-d, Metabolic flux analysis of wild-type and Bmpr1a-/- mESCs. OCR profile and metabolic parameters assessed during the mitochondria stress test performed in pluripotency conditions (a). ECAR profile and metabolic parameters assessed during the glycolysis stress test performed in pluripotency conditions (b). Metabolic parameters from the mitochondria stress test found to be similar between wild-type and Bmpr1a-/- mESCs during differentiation – day 3 (c). Metabolic parameters from the glycolysis stress test found to be similar between wild-type and Bmpr1a-/- mESCs during differentiation – day 3 (d). Data obtained from 3 (a,b) or 5 (c,d) independent experiments, with 5 replicates per cell type in each assay. e-f, Analysis of mitochondrial membrane potential (Δψm) in defective mESCs maintained in pluripotency conditions, in separate or co-culture. Representative histograms of TMRM fluorescence and quantification for wild-type and Bmpr1a-/- (e) and wild-type and 4n (f). g, Analysis of mitochondrial ROS in wild-type and Bmpr1a-/- mESCs undergoing differentiation in separate or co-culture: representative histograms of mitoSOX Red fluorescence and quantification of the percentage of mitoSOX positive cells. Data shown as mean ± SEM from 3 (e-f) or 5 (g) independent experiments. See methods for details on statistical analysis.
Extended Data Fig. 6. Effect of actinonin in OPA1 expression in wild-type and Drp1-/- cells.
a, Western blot analysis of OPA1 expression in wild-type and Drp1-/- cells treated with actinonin (Act, 150 μM) during 6 hours on the third day of differentiation, quantified in (b-c). b-c, Expression levels of L-OPA1 (b) and S-OPA1 (c) relative to -tubulin. Data shown as mean ± SEM of 3 independent experiments.
Extended Data Fig. 7. Analysis of SNPs in mtDNA in epiblast cells.
a-e, mtDNA heteroplasmy (plotted as Heteroplasmy = 1- frequency of most common allele) in epiblast cells from CI-treated embryos for five positions within the mt-Rnr2 gene. All these positions have an heteroplasmy that increases with the cells’ losing scores in a statistically significant way - the adjusted p-values are indicated at the top of each plot. f-k, The variation in the heteroplasmy across the CI-treated cells is not due to a batch effect for the 6 significant positions within the mt-Rnr1 gene. The number of cells analysed per cluster (and batch) is as follows: number of cells in Normal Epiblast :42 (1),16 (2),18 (3),0 (4),2 (5); number of cells in Intermediate: 42 (1), 28 (2),28(3), 12 (4), 5 (5); number of cells in Loser Epiblast: 22 (1), 15(2), 20 (3), 2 (4), 7 (5). l, Correlation between the mtDNA heteroplasmy at all the statistically significant positions, six within the gene mt-Rnr1 and five within the gene mt-Rnr2. m, Schematic representation of the mitochondrial genome showing in red the positions that passed our filtering based on coverage and were considered for the heteroplasmy analysis. Only the genes that include these positions are indicated. See methods for details on statistical analysis.
Extended Data Fig. 8. Changes in mtDNA sequence are enough to trigger cell competition.
a, Illustration of the process of derivation of the mESCs lines from mice that are hybrid between the wild-caught strains (BG, HB or ST) and the lab mouse (C57BL/6N). These hybrid mice were generated elsewhere16 by ooplasmic transfer: the zygote of a C57BL/6N mouse was injected with ooplasm from a wild-caught mouse (orange, HB pictured). Therefore, these hybrid mice contain the nuclear background of the C57BL/6N strain and the mtDNA of wild-caught strain and potentially C57BL/6N mtDNA (heteroplasmic mice strains). mESCs lines were derived from the hybrid mice and characterised. b-f, Characterisation of the derived cell lines by flow cytometry, during pluripotency, in comparison to the wild-type cell line used in previous experiments (E14, 129/Ola background). Heteroplasmy analysis of the derived mESC lines from the hybrid mice, indicating the percentage of wild-derived mtDNA (b). Cell granularity (internal complexity) given as median fluorescence intensity of SSc-A laser (c). Cell size given as median fluorescence intensity of FSc-A laser (d). Analysis of the expression of mitochondrial markers: representative western blot and quantification of markers of mitochondrial mass (ATPB, mt-CO1 and TOMM20) and mitochondrial dynamics (DRP1, MFN1and MFN2), relative to vinculin, in cells derived from hybrid mice (e). f, Representative histograms and quantification of median TMRM fluorescence, indicative of Δψm, for the hybrid cell lines derived, in comparison to the wild-type cell line used in previous experiments (E14, 129/Ola background). g-i, Cell competition assays between hybrid cell lines maintained in pluripotency culture conditions. The ratio of final/initial cell numbers in separate or co-culture is shown. j, Experimental design for RNA-Seq and gene set enrichment analysis (GSEA). The isolation of RNA from winner HB(24%) and loser BG(95%) cells was performed after three days in separate or co-culture conditions, once cells have been subjected to FACS to isolate the two populations form mixed cultures. Data shown as mean ± SEM of 3 independent experiments. See methods for details on statistical analysis.
Extended Data Fig. 9. Metabolic flux analysis of the cells with different mtDNA variants: HB(100%), HB(24%), BG(95%) and C57BL/6N.
a, OCR profile during mitochondria stress test performed in pluripotency maintenance conditions. b-i, Metabolic parameters assessed during the during the mitochondria stress test performed in pluripotency conditions. Data obtained from 3 independent experiments, with 5 replicates per cell type in each assay. Error bars represent SEM. See methods for details on statistical analysis.
Extended Data Fig. 10. Common features of scRNA-seq and bulk RNA-seq datasets.
a, Terms significantly enriched among genes downregulated in BG(95%) (loser) ESCs in vitro when co-cultured with HB(24%) cells. The loss of mitochondrial activity emerges as a common feature between loser cells in vivo and in vitro. The gene enrichment analysis was performed using g-profiler tool (see Methods) and p-values were adjusted for multiple comparisons using the g:Profiler algorithm g:SCS (https://doi.org/10.1093/nar/gkm226). b, Intersection between differentially expressed genes along the trajectory from winning to losing epiblast cells (“in_vivo_scRNA-seq”; Fig. 2a and Extended Data Fig. 3a and genes differentially expressed between co-cultured HB(24%) (winner) and BG(95%) (loser) ESCs (“in_vitro_bulk_RNA-seq”). “Up” and “Down” here refer to genes up- or down-regulated in loser cells. For the intersection between down-regulated genes from scRNA-seq (in vivo) and down-regulated genes from bulk RNA-seq (in vitro): p-value, 1.71E-12; odds ratio 1.80. For the intersection between down-regulated genes from scRNA-seq (in vivo) and up-regulated genes from bulk RNA-seq (in vitro): p-value, 5.20E-3; odds ratio 0.67. For the intersection between up-regulated genes from scRNA-seq (in vivo) and down-regulated genes from bulk RNA-seq (in vitro): p-value, 4.87E-3; odds ratio 0.80. The intersection between up-regulated genes from sc-RNA-seq (in vivo) and up-regulated genes from bulk RNA-Seq (in vitro) is not statistically significant: p-value: 0.30, odds ratio 1.14. c, Intersection between the significantly enriched terms in genes upregulated or downregulated in loser cells in the epiblast of CI-treated embryos (“in_vivo_scRNA-Seq”) or in our in vitro model of competition between co-cultured HB(24%) (winner) and BG(95%) (loser) ESCs (“in_vitro_bulk_RNA-seq”). All the terms enriched among downregulated genes in vitro are also enriched in vivo. See methods for details on statistical analysis.
Supplementary Material
Acknowledgments
We would like to thank Stephen Rothery for guidance and advice with confocal microscopy. The Facility for Imaging by Light Microscopy (FILM) at Imperial College London is part-supported by funding from the Wellcome Trust (grant 104931/Z/14/Z) and BBSRC (grant BB/L015129/1). We thank James Elliot and Bhavik Patel from the LMS/NIHR Imperial Biomedical Research Centre Flow Cytometry Facility for support. We are thankful to George Chennell and Alessandro Sardini for guidance and support with Seahorse experiments. Research in Tristan Rodriguez lab was supported by the MRC project grant (MR/P018467/1) and the BBSRC project grant (BB/S008284/1) and by the British Heart Foundation (BHF) PhD studentships (FS/14/62/31288 and FS/17/64/33476). Work in the Scialdone lab is funded by the Helmholtz Association. Ana Lima was funded by a BHF centre of excellence PhD studentship. Shankar Srinivas was funded through Wellcome awards 103788/Z/14/Z and 108438/Z/15/Z.
Footnotes
Author Contributions
A.L. performed most of the experimental wet lab work. J.B. and A.L. derived heteroplasmic mESC lines. J.B. performed heteroplasmy measurements in heteroplasmic mESCs. B.P. generated Mfn2-/- and Drp1-/- mESCs and J.M.S did characterisation of mitochondria shape and pluripotency status. S.P.M participated in the metabolic characterisation of Drp1-/- cells. D.H. performed embryo dissections, treatments and cell dissociation prior to scRNA-seq experiments. G.L. did the bioinformatic analysis of scRNA-seq data. E.M., N.J. and A.G. participated in the analysis of mitochondrial DNA heteroplasmy. A.D.G. performed the metabolomic studies using Metabolon platform and participated in embryo dissections and immunohistochemistry stainings for validation of results obtained by scRNA-seq. T.K. collected the embryos for derivation of the mESCs with different mtDNA content. M.D, and M.K. performed the bioinformatic analysis of bulk RNA-seq experiments. N.J., S.S. and D.C. participated in the design of experimental work and analysis of results. A.L., G.L., A.S and T.R. interpreted results and wrote the paper. T.R. and A.S. directed and designed the research.
Competing Interests
The authors declare no competing interests.
Data Availability
Data were analysed with standard programs and packages, as detailed above. Authors can confirm that all relevant data are included in the paper and/ or its supplementary information files. Source data for Figures 2-5,7 and for Extended Data Figures 4-5, 7-8 are provided. RNA-seq raw as well as processed data are available through ArrayExpress, accession numbers EMTAB-8640, for scRNA-seq data, and E-MTAB-8692, for bulk RNA-seq data.
Code Availability
All the code used for generating the figures in the paper is available at https://github.com/ScialdoneLab/Cell-Competition-Paper-Figures.
References
- 1.Bowling S, Lawlor K, Rodriguez TA. Cell competition: the winners and losers of fitness selection. Development. 2019;146 doi: 10.1242/dev.167486. [DOI] [PubMed] [Google Scholar]
- 2.Diaz-Diaz C, Torres M. Insights into the quantitative and dynamic aspects of Cell Competition. Curr Opin Cell Biol. 2019;60:68–74. doi: 10.1016/j.ceb.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Madan E, Gogna R, Moreno E. Cell competition in development: information from flies and vertebrates. Curr Opin Cell Biol. 2018;55:150–157. doi: 10.1016/j.ceb.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Morata G, Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 5.Claveria C, Giovinazzo G, Sierra R, Torres M. Myc-driven endogenous cell competition in the early mammalian embryo. Nature. 2013;500:39–44. doi: 10.1038/nature12389. [DOI] [PubMed] [Google Scholar]
- 6.Sancho M, et al. Competitive interactions eliminate unfit embryonic stem cells at the onset of differentiation. Dev Cell. 2013;26:19–30. doi: 10.1016/j.devcel.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowling S, et al. P53 and mTOR signalling determine fitness selection through cell competition during early mouse embryonic development. Nat Commun. 2018;9:1763. doi: 10.1038/s41467-018-04167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz-Diaz C, et al. Pluripotency Surveillance by Myc-Driven Competitive Elimination of Differentiating Cells. Dev Cell. 2017;42:585–599.:e584. doi: 10.1016/j.devcel.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto M, Sasaki H. Epiblast Formation by TEAD-YAP-Dependent Expression of Pluripotency Factors and Competitive Elimination of Unspecified Cells. Dev Cell. 2019;50:139–154.:e135. doi: 10.1016/j.devcel.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 10.Lima A, Burgstaller J, Sanchez-Nieto JM, Rodriguez TA. The Mitochondria and the Regulation of Cell Fitness During Early Mammalian Development. Curr Top Dev Biol. 2018;128:339–363. doi: 10.1016/bs.ctdb.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Zhou W, et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khrapko K, et al. Mitochondrial mutational spectra in human cells and tissues. Proc Natl Acad Sci U S A. 1997;94:13798–13803. doi: 10.1073/pnas.94.25.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allio R, Donega S, Galtier N, Nabholz B. Large Variation in the Ratio of Mitochondrial to Nuclear Mutation Rate across Animals: Implications for Genetic Diversity and the Use of Mitochondrial DNA as a Molecular Marker. Mol Biol Evol. 2017;34:2762–2772. doi: 10.1093/molbev/msx197. [DOI] [PubMed] [Google Scholar]
- 14.Burgstaller JP, Johnston IG, Poulton J. Mitochondrial DNA disease and developmental implications for reproductive strategies. Mol Hum Reprod. 2015;21:11–22. doi: 10.1093/molehr/gau090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorman GS, et al. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- 16.Burgstaller JP, et al. MtDNA segregation in heteroplasmic tissues is common in vivo and modulated by haplotype differences and developmental stage. Cell Rep. 2014;7:2031–2041. doi: 10.1016/j.celrep.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnston IG, et al. Stochastic modelling, Bayesian inference, and new in vivo measurements elucidate the debated mtDNA bottleneck mechanism. Elife. 2015;4:e07464. doi: 10.7554/eLife.07464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latorre-Pellicer A, et al. Regulation of Mother-to-Offspring Transmission of mtDNA Heteroplasmy. Cell Metab. 2019 doi: 10.1016/j.cmet.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee HS, et al. Rapid mitochondrial DNA segregation in primate preimplantation embryos precedes somatic and germline bottleneck. Cell Rep. 2012;1:506–515. doi: 10.1016/j.celrep.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Burr SP, Chinnery PF. The mitochondrial DNA genetic bottleneck: inheritance and beyond. Essays Biochem. 2018;62:225–234. doi: 10.1042/EBC20170096. [DOI] [PubMed] [Google Scholar]
- 21.Sharpley MS, et al. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell. 2012;151:333–343. doi: 10.1016/j.cell.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf FA, et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20:59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angerer P, et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 24.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nature methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 26.Cheng S, et al. Single-Cell RNA-Seq Reveals Cellular Heterogeneity of Pluripotency Transition and X Chromosome Dynamics during Early Mouse Development. Cell Rep. 2019;26:2593–2607.:e2593. doi: 10.1016/j.celrep.2019.02.031. [DOI] [PubMed] [Google Scholar]
- 27.Topf U, Wrobel L, Chacinska A. Chatty Mitochondria: Keeping Balance in Cellular Protein Homeostasis. Trends Cell Biol. 2016;26:577–586. doi: 10.1016/j.tcb.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Melber A, Haynes CM. UPR(mt) regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res. 2018;28:281–295. doi: 10.1038/cr.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munch C. The different axes of the mammalian mitochondrial unfolded protein response. BMC Biol. 2018;16:81. doi: 10.1186/s12915-018-0548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Q, et al. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quiros PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 33.Mouchiroud L, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saveljeva S, et al. Endoplasmic reticulum stress-mediated induction of SESTRIN 2 potentiates cell survival. Oncotarget. 2016;7:12254–12266. doi: 10.18632/oncotarget.7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yun J, Finkel T. Mitohormesis. Cell Metab. 2014;19:757–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prudent J, McBride HM. The mitochondria-endoplasmic reticulum contact sites: a signalling platform for cell death. Curr Opin Cell Biol. 2017;47:52–63. doi: 10.1016/j.ceb.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Favaro G, et al. DRP1-mediated mitochondrial shape controls calcium homeostasis and muscle mass. Nat Commun. 2019;10:2576. doi: 10.1038/s41467-019-10226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quiros PM, et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol. 2017;216:2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Restelli LM, et al. Neuronal Mitochondrial Dysfunction Activates the Integrated Stress Response to Induce Fibroblast Growth Factor 21. Cell Rep. 2018;24:1407–1414. doi: 10.1016/j.celrep.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richter U, et al. A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr Biol. 2013;23:535–541. doi: 10.1016/j.cub.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 43.Moullan N, et al. Tetracyclines Disturb Mitochondrial Function across Eukaryotic Models: A Call for Caution in Biomedical Research. Cell Rep. 2015;10:1681–1691. doi: 10.1016/j.celrep.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kauppila JHK, et al. A Phenotype-Driven Approach to Generate Mouse Models with Pathogenic mtDNA Mutations Causing Mitochondrial Disease. Cell Rep. 2016;16:2980–2990. doi: 10.1016/j.celrep.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan W, et al. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart JB, et al. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freyer C, et al. Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat Genet. 2012;44:1282–1285. doi: 10.1038/ng.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ludwig LS, et al. Lineage Tracing in Humans Enabled by Mitochondrial Mutations and Single-Cell Genomics. Cell. 2019;176:1325–1339.:e1322. doi: 10.1016/j.cell.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chinnery PF, Hudson G. Mitochondrial genetics. Br Med Bull. 2013;106:135–159. doi: 10.1093/bmb/ldt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Floros VI, et al. Segregation of mitochondrial DNA heteroplasmy through a developmental genetic bottleneck in human embryos. Nat Cell Biol. 2018;20:144–151. doi: 10.1038/s41556-017-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burr SP, Pezet M, Chinnery PF. Mitochondrial DNA Heteroplasmy and Purifying Selection in the Mammalian Female Germ Line. Dev Growth Differ. 2018;60:21–32. doi: 10.1111/dgd.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajasimha HK, Chinnery PF, Samuels DC. Selection against pathogenic mtDNA mutations in a stem cell population leads to the loss of the 3243A-->G mutation in blood. Am J Hum Genet. 2008;82:333–343. doi: 10.1016/j.ajhg.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ellis SJ, et al. Distinct modes of cell competition shape mammalian tissue morphogenesis. Nature. 2019;569:497–502. doi: 10.1038/s41586-019-1199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kucinski I, Dinan M, Kolahgar G, Piddini E. Chronic activation of JNK JAK/STAT and oxidative stress signalling causes the loser cell status. Nat Commun. 2017;8:136. doi: 10.1038/s41467-017-00145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kon S, et al. Cell competition with normal epithelial cells promotes apical extrusion of transformed cells through metabolic changes. Nat Cell Biol. 2017;19:530–541. doi: 10.1038/ncb3509. [DOI] [PubMed] [Google Scholar]
- 56.Scialdone A, et al. Resolving early mesoderm diversification through single-cell expression profiling. Nature. 2016;535:289–293. doi: 10.1038/nature18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Czechanski A, et al. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat Protoc. 2014;9:559–574. doi: 10.1038/nprot.2014.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burgstaller JP, et al. Large-scale genetic analysis reveals mammalian mtDNA heteroplasmy dynamics and variance increase through lifetimes and generations. Nature communications. 2018;9:2488. doi: 10.1038/s41467-018-04797-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burgstaller JP, Schinogl P, Dinnyes A, Muller M, Steinborn R. Mitochondrial DNA heteroplasmy in ovine fetuses and sheep cloned by somatic cell nuclear transfer. BMC Dev Biol. 2007;7:141. doi: 10.1186/1471-213X-7-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang E, et al. Mitochondrial replacement in human oocytes carrying pathogenic mitochondrial DNA mutations. Nature. 2016;540:270–275. doi: 10.1038/nature20592. [DOI] [PubMed] [Google Scholar]
- 62.Yahata N, Boda H, Hata R. Elimination of mutant mtDNA by an optimized mpTALEN restores differentiation capacities of heteroplasmic MELAS-iPSCs. Molecular Therapy-Methods & Clinical Development. 2021;20:54–68. doi: 10.1016/j.omtm.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee H, et al. Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. Nat Commun. 2021;12:1190. doi: 10.1038/s41467-021-21464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venegas V, Halberg MC. Mitochondrial Disorders. Springer; 2012. Quantification of mtDNA mutation heteroplasmy (ARMS qPCR) pp. 313–326. [DOI] [PubMed] [Google Scholar]
- 65.Machado TS, et al. Real-time PCR quantification of heteroplasmy in a mouse model with mitochondrial DNA of C57BL/6 and NZB/BINJ strains. PLoS One. 2015;10:e0133650. doi: 10.1371/journal.pone.0133650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burgstaller JP, et al. MtDNA segregation in heteroplasmic tissues is common in vivo and modulated by haplotype differences and developmental stage. Cell Rep. 2014;7:2031–2041. doi: 10.1016/j.celrep.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee H-S, et al. Rapid mitochondrial DNA segregation in primate preimplantation embryos precedes somatic and germline bottleneck. Cell Rep. 2012;1:506–515. doi: 10.1016/j.celrep.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 73.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 75.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lun AT, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calabrese FM, Simone D, Attimonelli M. Primates and mouse NumtS in the UCSC Genome Browser. BMC Bioinformatics. 2012;13(Suppl 4):S15. doi: 10.1186/1471-2105-13-S4-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lukes J, Kaur B, Speijer D. RNA Editing in Mitochondria and Plastids: Weird and Widespread. Trends Genet. 2021;37:99–102. doi: 10.1016/j.tig.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 81.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reimand J, Arak T, Vilo J. g:Profiler--a web server for functional interpretation of gene lists (2011 update) Nucleic Acids Res. 2011;39:W307–315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were analysed with standard programs and packages, as detailed above. Authors can confirm that all relevant data are included in the paper and/ or its supplementary information files. Source data for Figures 2-5,7 and for Extended Data Figures 4-5, 7-8 are provided. RNA-seq raw as well as processed data are available through ArrayExpress, accession numbers EMTAB-8640, for scRNA-seq data, and E-MTAB-8692, for bulk RNA-seq data.
All the code used for generating the figures in the paper is available at https://github.com/ScialdoneLab/Cell-Competition-Paper-Figures.