Abstract
Most antibodies produced in the body are of the IgA class. The dominant cell population producing them are plasma cells within the lamina propria of the gastrointestinal tract, but many IgA-producing cells are also found in the airways, within mammary tissues, the urogenital tract and inside the bone marrow. Most IgA antibodies are transported into the lumen by epithelial cells as part of the mucosal secretions, but they are also present in serum and other body fluids. A large part of the commensal microbiota in the gut is covered with IgA antibodies, and it has been demonstrated that this plays a role in maintaining a healthy balance between the host and the bacteria. However, IgA antibodies also play important roles in neutralizing pathogens in the gastrointestinal tract and the upper airways. The distinction between the two roles of IgA - protective and balance-maintaining - not only has implications on function but also on how the production is regulated. Here, we discuss these issues with a special focus on gut and airways.
Keywords: commensal microbiota, IgA, infection, mucosa
1. Introduction
More IgA is produced than any other antibody class. Mucosal surfaces are filled with IgA-producing plasma cells that in humans produce several grams of antibodies every day.1 Mucosally produced IgA is transported through the epithelial cell layer into the lumen.2 Most of the production is secreted into the gastrointestinal tract and significant amounts are also found in nasal and lung fluids, saliva, tears, and breast milk.3 Secretion of antibodies at mucosal surfaces is an evolutionary preserved mechanism that hinders pathogens from invading tissues.4,5 As pathogens may encounter IgA before they reach any tissues, IgA can potentially give rise to sterile immunity by hindering the initial infection.6 Several studies of disease-causing viruses, bacteria, toxins, or adjuvanted antigens applied at mucosal surfaces have demonstrated that antigen-specific IgA responses are triggered against them.7–10 Airway viruses can also trigger antigen-specific IgA responses, an example being the ongoing SARS-CoV-2 pandemic; in diseased individuals antigen-specific IgA antibodies are often detected in serum before either IgM or IgG.11,12 At the same time, IgA is often described as a non-inflammatory antibody class that hinders inflammation, and it has been suggested that a significant proportion of IgA is produced in a similar fashion as natural IgM in the absence of typical antigen activation.13 Why so much IgA is produced, where and how the IgA response is initiated, and what are its triggers for it to be initiated has been the focus for research for many years but there are still many unknowns, and no consensus view has been universally accepted.
There are more IgA-producing plasma cells at mucosal sites than there are other plasma cells in the body.1 At mucosal surfaces, a joining (J) chain is added to IgA when it is synthesized, creating multimers, primarily dimers.14,15 After being secreted from the plasma cell, the dimers bind to the poly-Ig receptor (pIgR) that is expressed at the basolateral side of mucosal epithelial cells before being transported to the apical side of the cells where it is released into the lumen by proteolytic cleavage.16 This leaves a part of the pIgR receptor, called the secretory component, binding to the molecule which protects the complex from proteolytic degradation.17 Significant numbers of IgA-producing plasma cells are also found at non-mucosal sites, and IgA antibodies are present in the circulation.18,19 Although steady-state serum levels of IgG are four to five times higher than for IgA in healthy adults, the amounts produced are actually quite similar as the half-life of the classes differs three to four times.20 In addition, IgA antibodies are present in other body fluids such as within the cerebrospinal fluid.21 Unlike IgA produced at mucosal surfaces, serum IgA is predominantly monomeric and not joined to the J chain or the secretory component.22 Adding to the complexity, humans, but not mice, possess two IgA isotypes, IgA1 and IgA2, transcribed from two distinct heavy chain constant regions, with the first one dominating in serum and most tissues and the second more often secreted in the lower gastrointestinal along with IgA1.23 The two classes are similar to each other, but may have different stability and effector functions due to differences in the hinge region and glycosylation patterns.24,25 In similar with the gastrointestinal tract, the airways contain epithelial cells expressing pIgR that transport IgA into the lumen.26,27 This mechanism is likely more important in the upper airways as pIgR expression is evident in mucous, serous and ciliated epithelial cells in the lung but is virtually absent in alveolar cells.28 In fact, transudation of IgG may play a more important role than IgA secretion in lung.29
In addition to the role of IgA at the mucosa, other locations where IgA may play a role in health and disease have been suggested. A relatively large proportion of plasma cells in the bone marrow are IgA producing, both in mice and humans.30,31 These produce antibodies reactive against intestinal antigens and appear to form in the gut as a consequence of recognizing commensal bacteria or pathogens.30,32,33 This would allow these antibodies to protect the host against sepsis if the mucosal barrier breaks down.30 More recently, IgA-producing plasma cells were described within brain tissue, in particular along meningeal venous sinuses.34–36 These have been suggested to play roles both in autoimmune diseases, where they then may hinder overt inflammation by production of IL10, but also in protecting the host against infection into brain tissues. Clonal links have been found between IgA produced in gut and other tissues, and IgA produced at non-mucosal sites has been found to be reactive toward microbiota present in the gut. Finally, plasma cells are often present in tumor tissues, and in this case IgA expression have been associated with either worse or, at least in the case of ovarian cancer, better outcomes.37,38
Most mucosal surfaces maintain a commensal microbiota.39 This is particularly true for the gut, which in humans has been estimated to contain a total of 1013-1014 bacteria belonging to around 1000 different species.40 In addition, the gut supports fungi and viruses at steady state that also influence the mucosal environment.41,42 Similarly, both the upper and lower airways maintain specific microbiotas.43 Many diseases are associated with changes in the composition of the microbiota, and in some cases, such alterations may contribute to the development of disease.44,45 This constant presence of stimuli plays an important role in the maturation of the epithelium as well as the mucosal immune system.46,47 At the same time, the constant presence of bacteria presents the immune system with a difficult challenge—how to differentiate non—self-molecular structures derived from beneficial microbiota (that should be tolerated) from those coming from pathogens (that should trigger immune responses against the infection).
Recent studies have found that a large part of the microbiota in the gut is covered with IgA antibodies, both in mice and humans.48–51 Two different mechanisms have been proposed. According to one model, bacteria are covered with IgA to protect the host from them.52 In other words, when bacteria with the ability to cause inflammation, colonize the gut, these will be detected by the immune system and a specific IgA response triggered. Subsequently, the bacteria are covered with IgA to avoid invasion and inflammatory responses, which enable the host to retain these beneficial, but pro-inflammatory, commensal strains. According to the other model, the IgA cover will hinder strong inflammatory antigen-specific immune response against bacteria, and thereby ensures that a regulatory environment forms that is associated with the formation of regulatory T cells (Treg).53 The two models are not mutually exclusive. Nevertheless, they would suggest different pathways for the generation of IgA. In the first case, inflammatory bacterial strains trigger specific protective immune responses that will subsequently maintain a healthy balance, while in the second an ongoing, presumably rather unspecific, IgA production is needed to maintain a non-inflammatory state that ensure that inflammatory responses are not triggered. Although it is possible that this represents two different pathways for IgA induction, for example, T independent or dependent induction, the proposed pathways nevertheless demonstrate that the functions associated with IgA production are closely linked to how it is induced—whether most IgA is derived from highly specific immune responses that are similar to antigen-specific IgG responses or if it is constantly produced in the absence of any specific trigger in similar with natural IgM.
2. Inductive and Effector Sites for Mucosal IgA Responses
While IgG is characteristic for specific systemic humoral responses, IgA is the archetypal mucosal antibody class. It is generally accepted that the production of these antibody classes represents the outcomes from separate systemic and mucosal immune systems that differ in inductive and effector sites.54,55 Although often described as entirely separate, there is some overlap between them, and antigen-specific IgG is often encountered in serum after oral immunization.7 Inductive sites for systemic responses are found in the spleen and in the lymph system, where antigen and antigen-loaded dendritic cells arrive via blood or afferent lymph for B and T cells to react to antigens.56 Proteinaceous antigens trigger both B and CD4+ T cells and following interactions between these, germinal centers (GC) form within the lymphoid follicles as a consequence of expansion of rapidly proliferating antigen-specific B cells.57,58 The production of antigen-specific serum antibodies is subsequently achived when the activated B cells differentiate into short-lived local plasma cells or migrate to bone marrow to become more long-lived.59–61
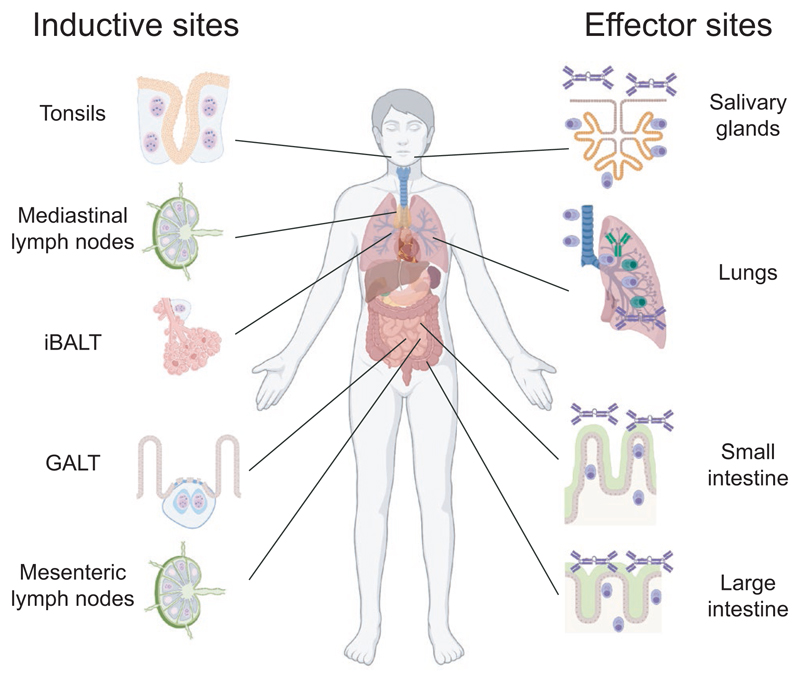
Inductive sites for the mucosal immune system, known as mucosal-associated lymphoid tissues (MALT), are non-encapsulated lymphoid follicles lacking afferent lymphatics that are embedded in the mucosa and submucosa (Figure 1).54 They can be further divided based on which areas they are found in, giving rise to inductive systems such as gut-associated lymphoid tissues (GALT), nasal-associated lymphoid tissues (NALT), or induced bronchial associated lymphoid tissues (iBALT).62 Distinct organ types make up each local system. Thus, GALT is made up of larger structures containing several lymphoid follicles (Peyer´s patches (PP) and colonic patches(CP)) and single follicles (isolated lymphoid follicles (ILF)), while NALT consist of several distinct tonsils (in humans) or structures along the nasopharyngeal wall (in mice).63,64 These structures are dependent on specific anlage formed during fetal life to develop, but differ between tissues and species with regard to whether they are dependent on microbiota to fully develop into mature tissues.63–65 In contrast, iBALT only develop as a consequence of inflammation and should possibly be considered a tertiary lymphoid organ.66–68 During inflammatory conditions, tertiary lymphoid tissues can also develop in the gut mucosa and these are hard to distinguish from ILF.69–71 To which extent tertiary tissues developing within the mucosa are distinct from those that form at non-mucosal inflammatory sites is not known.72,73 In addition to classical MALT, efferent lymph from the mucosa and MALT drain into lymph nodes, with, for example, mesenteric lymph nodes (MLN) receiving lymph from the gut and mediastinal lymph nodes (medLN) from the lungs.74,75 It is now appreciated that these lymph nodes should not be considered as one continuous organ; the MLN, for example, consist of several separate lymphoid follicles that handles antigens from different segments of the gut, and the arrival of an antigen to different follicles will influence whether it will trigger tolerogenic or inflammatory responses.76 Since lymph nodes are not situated within the mucosa and receive afferent lymph, they are not strictly part of the MALT but they still influence the induction of mucosal responses.
Figure 1.
Inductive and effector sites for gut and airway antibody mucosal responses. The mucosal systems for immune responses in the airways and the intestinal tract contain several sites involved in activation of adaptive immune response (inductive sites) and production of antibodies (effector sites). Inductive sites for airways include tonsils (considered the nasal-associated lymphoid tissue (NALT) equivalent in humans) in the upper airways, inducible bronchus-associated lymphoid tissues (iBALT) in the lower airways and mediastinal lymph nodes (medLN) that drain the lungs. Gut-associated lymphoid tissues (GALT), primarily made up of Peyer’s patches (PP) and isolated lymphoid follicles (ILF), function as inductive sites in the gut while mesenteric lymph nodes (MLN) drain the gut. Plasma cells producing IgA antibodies are found both in the upper airways, exemplified here by a salivary gland, and lower airways where both IgA and IgG are produced, and represent airway effector sites. In the gut, the lamina propria of the large and small intestine function as an effector site and is filled with plasma cells that produce IgA antibodies
Unlike systemic lymphoid tissues, some MALT maintain GC in their B cell follicles in the absence of any immunization. This is, for example, the case for PP in mice and tonsils in humans.63,77 GC are also commonly present in MLN during steady-state conditions.63,78 In some animal species, for example, birds, rabbits, and sheep, GALT play a role in the generation of primary diversity of B cell receptors as random mutations or parts of upstream pseudogenes are introduced into the V regions, but in human and mice they are mainly involved in shaping specific immune responses.79 However, even in mice and humans, the gut has been suggested to take part in early B cell development, and both VDJ recombination and selection of transitional B cells into specific B cell lineages have been described.80–83 Unexpectedly, some recent single-cell RNASeq studies have also found gene expression patterns suggestive for early B cell development in the meningeal tissues of the CNS,84,85 with two studies even reporting a brain associated lymphopoietic niche.86,87 This observation raises the possibility that not only bone marrow, but also other tissues able to maintain long-lived plasma cells are able to support primary B lymphopoiesis.
Although the formation of GC in GALT is influenced by antigen-specific interactions with the microbiota, it does not seem to be the only force promoting their development. Germ-free (GF) mice develop GC in PP although they are smaller than in individuals with a microbiota.69,78,88,89,90 The same is true for mice expressing just a single rearranged transgenic B cell receptor or even lacking B cell receptors altogether, suggesting that neither antigen-specific signals nor B cell receptor-mediated uptake of antigen and MHC II presentation are always required.91,92 These B cell receptor-independent models still require T cell signals for GC formation in PP, which is also true in normal mice.91–94 Thus, while T cells and certain signals derived from them are needed for PP GC formation, archetypical cognate B-T interactions through the MHC II and T cell receptors appear to be dispensable. Regardless, there is a strong tendency to form GC in MALT, even in the absence of signals required in peripheral organs´ GC formation.
MALT are in general covered with a specialized follicle-associated epithelium (FAE) that allows for antigens to enter into the lymphoid structures below.95–97 This epithelium contains microfold (M) cells that, unlike other epithelial cells, can transport intact antigens from the mucosal lumen into the underlying tissues. Thus, instead of antigen entering through the efferent lymph as in lymph nodes, MALT will directly sample antigen from the lumen. This process is to some extent regulated, with molecular structures such as bacterial fimbriae, heat shock proteins, and IgA complexes promoting uptake, but other foreign antigens can also enter through unspecific processes.2,98,99 After entering, the antigens are not passively diffusing within the tissues. In GALT, for example, antigens enter into a specifically organized area named the subepithelial dome (SED) where DC, T, and B cells can directly interact with the M cells as well as with each other which ensures an optimal response.2,95,98,99,100,101 Given the rather non-specific function of M cells, non-proteinaceous macromolecules that contain microbe-associated molecular patterns (MAMP) will also enter. Thus, the MALT environment is distinct from that in lymph nodes or spleen; while these are thought to be essentially sterile and largely free from MAMP during steady-state conditions, MAMP are most likely ubiquitously present in MALT. Thus, it is unlikely that MAMP signals will function in a similar way as in systemic organs to determine whether there is an ongoing microbial invasion.102 In fact, even living bacteria may enter through the M cells into GALT, as has been shown for the commensal strain Alcaligenes, that colonize the PP after entering, and pathogens such as Brucella Abortus or Salmonella, that use this mechanism to get access to tissues.103,104
Major mucosal effector sites include the lamina propria along the gastrointestinal tract; lacrimal, nasal, and salivary glands in the upper airways; and mammary glands producing milk.105 In addition, cells producing secreted IgA are present within the lower airways and in the urogenital tract.106 Most IgA-producing mucosal plasma cells are thought to arrive to these effector tissues as plasmablasts that have left MALT after replicative expansion and class-switch recombination.107 The plasmablasts travel via the lymph system before they enter the bloodstream via ductus thoracicus and finally home to their effector sites. During steady state, IgA-producing plasmablasts dominate in blood compared to those expressing other classes, suggesting that they leave mucosal tissues continuously even when not reacting to novel pathogens or food antigen.31 The homing of these plasmablasts is controlled through expression integrins and chemokine receptors.59 In particular, the expression of integrin α4β1 and CCR10 has been associated with homing to mucosal surface with α4β7 and CCR10 specifically connected with gut responses.108–110
3. IgA Class-Switch Recombination
As for all antibody classes except IgM and IgD, class-switch recombination (CSR) is a prerequisite for IgA expression.63 This exchange of the heavy chain constant region from IgM to IgA occurs through a genomic deletion event that will still maintain the V region unaltered.111 Thus, antigen specificity is maintained while the functional characteristics of the antibody change. The process is controlled through regulated expression of activation-induced cytosine deaminase (AID), an enzyme that targets switch regions upstream of antibody heavy chain constant gene regions through deamination of cytosine into uracil within the DNA.112 Subsequently, the repair machinery that normally removes uracil residues introduced into DNA due to endogenous DNA damage will eliminate the AID-induced uracil; during this process, double-stranded breaks will develop in the switch regions that are specifically targeted by AID.113–115 An IgA expressing antibody gene is created when a break in the switch region upstream of IgM is joined to one occurring in the switch region upstream of IgA and the intervening part is deleted. This deletion process relies on the non-homologous end-joining machinery, likely as a consequence of cohesin meditated positioning of the switch regions with double-stranded breaks.116
The AID enzyme is also responsible for targeting V regions for mutations during GC proliferation, although in this case, the repair pathways used are slightly different than during CSR, resulting in point mutations rather than double-stranded DNA breaks. Importantly, both for CSR and mutation induction, DNA replication is required.112,117 Thus, it is unlikely that non-proliferating B cells will be able to undergo class-switch recombination or introduce mutations into their V regions.118,119
For CSR to occur, functional AID must be present in the cell nucleus, an even that is tightly regulated through several mechanisms to ensure that it only happens in proliferating B cells.120 The most critical signal for induction and activation of AID is transmitted through the surface receptor CD40, although other signals, such as MAMP recognition by TLR or BAFF receptor family members may substitute in some cases.120,121 These signals all activate NF-κB pathways that are likely critical in the activation process.122 Local cytokines act as cues that command CSR into different isotypes by activating sterile germline transcripts that open up the switch regions upstream of the antibody constant genes and make them targets for AID and position the constant regions.116,117 TGF-β has been closely linked to both induction of sterile IgA transcripts and for cells to undergo CSR to IgA,123 but signals from the two related cytokines a proliferation-inducing ligand (APRIL) or B-cell activating factor (BAFF) that activate the transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI) may also be able to promote the process.124,125 Other signals in the local milieu, such as interleukin 21 (IL21) and all-trans-retinoic acid (RA) play additional roles.126–128
As PP are surrounded by IgA-producing plasma cells in the mucosa, it has been assumed that CSR in GALT is efficiently skewed toward IgA. At the same time, IgA CSR appear to be rare in other organs. Recent observations have challenged the conclusion that IgA is the only or even major antibody class produced in MALT. In tonsils, for example, many IgG switched cells are present, although the mucosa in the upper airways, in similar with the gut, are dominated by IgA-producing plasma cells.77 In addition, in human GALT, as many as 10% of all B cells express IgG, and an evaluation of antibody heavy chain gene transcripts revealed that IgG transcripts were present in both sorted GC and memory B cells.77 Similarly, in mice relatively high numbers of IgG switched cells were detected in PP using single-cell RNASeq (Komban et al, manuscript in preparation). Recent studies have started to reveal signals needed for IgA CSR to occur in MALT. For example, in PP from mice with a normal microbiota, IgA dominated with some levels of IgG2b also being produced, whereas in germ-free animals or in animals with a limited bacterial microbiota, IgG1 was the dominant antibody class in PP with few IgA-switched cells detected.78 Skewing was also observed in experiments using microbiota transfers, where germ-free mice receiving microbiota from T cell-deficient mice reconstituted with Treg cells showed efficient induction of IgA, while in germ-free mice reconstituted with microbiota from T cell-deficient mice into which naïve CD4 cells had been transferred promoted IgG1.51 These observations are also in line with observations we made in transfer experiments with sorted T cells into nude mice. In this case, strong IgG responses were triggered in GALT after oral immunization in mice transferred with CD25-, presumably non-regulatory, T cells while if CD25+ T regulatory cells were co-transferred, the recipient mice could be immunized to produce IgA.129 In this model, the ability of T regulatory cells to produce active TGF-β was the factor needed to induce IgA production. However, other factors may also be involved, as both retinoic acid and IL21 have been shown to inhibit IgG switching when the cells are induced to IgA switching with TGF-β in vitro.130,131 Further investigations of the role for switch factors and cell types for efficient induction of IgA switching in MALT are clearly called for, as it seems that switching to other classes represents the default pathway in many cases.
4. T Dependent and Independent IgA Responses
An important division of systemic B responses is whether they are triggered by thymus (or T cell) dependent and independent antigens.132 Originally, this division was based on the observation that certain purified antigens, that is, proteins, required the thymus to give an optimal response, while others, that is, lipopolysaccharides (LPS) or highly repetitive antigens, did not.133,134 Eventually, the original observations led to the current view that it is whether the B cell interact with CD4 T cells through CD40-CD40L signals or not that determined the type of response—resulting in the term T cell-dependent or independent responses.135 Several studies have reported that B cells belonging to B1 and B2 marginal zone lineages are more often involved in T independent responses and B cells belonging to the follicular B2 lineage are more often involved in T dependent reactions.136 The classical view is that there are large differences in response triggered by the two types of antigens; in the case of a T dependent proteinaceous antigen, the B cell will proliferate in GC, will undergo somatic hypermutation of V regions and selection leading to affinity maturation of surviving cells, undergo class-switch recombination to other classes than IgM and will generate long-lived plasma and memory B cells, while in the case of a T independent antigen there will be proliferation of B cells but no GC formation, production of IgM of low affinity and that most plasma cells will be short-lived.132 Therefore, class switching, mutations of V regions, memory cell development, and long-lived antibody responses are all considered sign of T cell-dependent reactions, while a lack of these signs is associated with a T independent process. However, it should be pointed out that the division is a bit artificial in most infectious models, as essentially all pathogens will contain a mixture of T cell-dependent and independent antigens, often even covalently linked to each other, giving rise to a mixed responses where some T independent epitopes may give rise to T dependent responses. Nevertheless, one area where the distinction between the two types is important even clinically is during the development of subunit vaccines; in these polysaccharides are sometimes conjugated to proteins is to give more long-lived T dependent responses.137,138
Whereas the differences between T dependent and non-dependent responses to systemic antigens have been extensively characterized, it is much more problematic to define whether they should be seen as two separate arms in the mucosa.63 As IgA is a class-switched antibody, it must be assumed that cells expressing IgA have proliferated while they expressed AID at some time during their development. Hence, the most straight-forward assumption would be that all IgA-producing plasma cells in gut, expressing a switched antibody and being highly mutated, would have passed through GC in GALT. However, several mouse models and observations in humans have been made that in fact support the view that IgA production can be initiated in T independent systems (Table 1). Still, essentially all such observations come with caveats that suggest that T dependent reactions may non-the-less dominate in most cases.
Table 1. Arguments for and against that T cell-independent IgA responses play a major role in adult invidiuals.
| Argument for T cell-independent response | Argument for that it may not be relevant in healthy adults |
|---|---|
| LPS and polysaccharides, classical T independent antigens, trigger IgA responses at mucosal surfaces. | These are not encountered as single antigens at the mucosa but in combination with or even linked to proteins that could provide T cell help. When antibodies were cloned that were reactive against LPS in humans or polysaccharides in mice, these had gone through the process that introduce mutations in V regions—which is normally considered a tell-tale sign of having proliferated in GC. |
| B1 B cells transferred into immunocompromised hosts will start producing IgA in the gut. | The behavior of lymphocytes in hosts lacking competing cells is often unpredictable. When B1 and normal lymphocytes were transferred simultaneously, B1 cells were able to provide natural IgM in circulation but did not contribute to gut production. |
| Mice and humans that lack T cells or signals crucial for B-T interactions will still produce IgA in the gut, sometimes at close to normal levels, despite lacking GC that are typically associated with T dependent responses. | This may not be the case when competing with other pathways. When antibodies derived from gut IgA-producing plasma cells were cloned from such mice or memory cells from CD40L deficient humans, their V regions largely lacked mutations while essentially all V regions were mutated in normal individuals. |
| Studies have reported that IgA CSR may happen within the lamina propria outside of any organized lymphoid tissues. | Some of these studies were performed before ILF have been described and included these structures in the analysis. AID expression and cellular proliferation are closely linked to class- switch recombination, and several studies in mice and humans where the authors have carefully studied expression of AID, expression of proliferation markers, or molecular markers indicative of ongoing CSR to IgA have failed to detect any signs of it when ILF are carefully removed from the analysis |
Taken together, from these studies it can be concluded that in individuals lacking certain immunological pathways, IgA production in the gut can be T cell independent but that T dependent induction will totally dominate in immunoproficient individuals. Nevertheless, as will be discussed later, it is possible to imagine the existence of responses that are in between what is normally defined as T dependent and independent responses in MALT. The fact that high levels of mutations are found in antibody V regions from IgA secreting gut plasma cells in humans and mice demonstrate that the cells must have proliferated.139–141 Thus, the lack of data suggesting that extensive B cell proliferation occurs outside of organized lymphoid tissues make it unlikely that the vast amount of IgA plasmablasts continuously produced can be generated elsewhere.142 It should be noted that many early studies supporting that the lamina propria was an important site for IgA generation were performed before ILF had been defined as an entity separate from PP, and more recent studies have failed to identify molecular markers suggestive of ongoing CSR outside of organized lymphoid tissues such as ILF.140,142,143,144 Nevertheless, it is hard to exclude that T independent IgA CSR never occurs outside of GALT, although it must be concluded that in that case, it will only make a minor contribution to gut secreted IgA.
5. Antigen-Specific IgA Induction in Peyer’s Patches using NP-CT as a Model Antigen
We have developed a hapten-carrier system that allows us to track T cell-dependent antigen-specific B cell responses in PP following oral immunization.145 A traceable antigen was created by conjugating the well-known hapten 4-hydroxy-3-nitrophenyl (NP) to cholera toxin (CT) that act as a carrier to create an NP-CT conjugate. After oral immunization, CT triggers strong T cell-dependent responses in the gut,146 and we found that this was also the case for NP-CT.145 As possibly the most used hapten, NP has been extensively used to study antigen-specific antibody responses after systemic immunization, in particular, in C57 BL/6 mice.147 We adapted some of the well-established assays that exist for examining NP responses to be able to study gut responses. In a first study using NP-CT as an antigen, we were able to demonstrate that the response against a single antigen could be highly efficient, with 15% of all IgA plasma cells being reactive against NP and 30% against CT after repeated oral immunizations. Furthermore, we found that antigen-specific B cells invaded already existing GC in PP after the immunization, that affinity maturation was highly efficient, and that there was an exchange of cells not only between PP and effector sites but also between different inductive sites.145,148 Together these observations suggested that responses were coordinated by exchange of cells between tissues and that this helped to synchronize the response ensured that high-affinity clones generated in one PP could spread and "take over" GC in other PP lacking high-affinity clones.75
We had previously observed that CSR to IgA can occur before B cells enter GC in PP, and Reboldi et al149 from the laboratory of Jason Cyster later made the same observation.140,150 At first, this seemed to indicate that CSR to IgA in PP differed from that to IgG in systemic organs as the latter was thought to mainly occur in the GC, but it was in fact later demonstrated that CSR to IgG also often happens before B cells form GC and undergo somatic hypermutation.151 Reboldi et al also demonstrated that the area where pre-GC IgA CSR occurred was the SED region, where antigens enter PP through M cell-mediated transport and suggested that B cells will first migrate to the SED upon entering the PP and after that into GC after CSR. This observation, and that we found a subpopulation of activated B cells expressing CCR6, led us to study the SED region in some more detail using our antigen-specific system.101 Employing NP-CT as an oral antigen and transfer of Vh1-8hi expressing NP-reactive B cells that were labeled with GFP, we found that antigen-specific cells were not only present in SED at the start of the response, before cells entered the GC. Instead, all through the response, the GC and the SED regions harbored proliferating antigen-specific B cells.101,152 While B cell proliferation in SED was less dependent on antigen affinity than the subsequent entry into GC and proliferation within it, the presence of T cells supported SED proliferation.152 Nevertheless, some level of proliferation was maintained even when T cells were depleted, suggesting that CSR may be ongoing even in the absence of T cells. This was in line with the previous report from Reboldi et al that suggested that dendritic cells may play an important role in this process.150 Such proliferation may be important during the early response to antigen, since very few antigen-specific T cells have been activated at this time, while T cells are more important later during the response. However, pre-GC B cell proliferation in the SED region of the PP may be sufficient for IgA CSR even in the absence of T cells but will likely result in plasma cells with very limited numbers of mutations in their V regions because of limited replication and absence of specific T cell cues. Although it has not been directly tested yet, such a T independent pathway could be the one leading to IgA production in humans and mice lacking CD40 signals, which would explain the lack of V region mutations in IgA cells from such individuals.140,154,155
In mice lacking the SAP-SLAM pathway, CD40 signals are present, but traditional cognate interactions between B and T cells cannot occur154; in the laboratory of Ziv Shulman, Biram et al found that in these knock-out mice, PP GC will form but that in the absence of efficient interactions between B and T cells, antigen-specific cells will not enter into the GC after oral immunization and that the selection of B cells within the PP is disturbed.152,156 The situation in these mice is distinct to that in CD40 deficient mice that totally lack GC.93 Thus, while CD40 signals are strictly required for GC formation in GALT, traditional cognate B-T cell interactions involving the SAP-SLAM pathways are necessary for efficient responses as they facilitate entry into GC and/or selection of high-affinity clones. Nevertheless, even in the absence of SAP-SLAM interactions, some B cells will manage to form GC in PP.
So why are there B cells in the SED region even when cells belonging to the same clone have already invaded the GC and started to mature there? Some of our observations give some clues. It is likely that antigen is a very limiting factor in most gut responses. A single antigen is diluted enormously in the sea of other antigens present in the gut, and the local level in PP is further limited by the M cell-mediated transport. Then, how can an antigen like CT give strong responses with affinity maturation, after an oral doses of only 10μg? As mentioned above, we found that activated antigen-specific B cells were present in the SED throughout the response, often in very close contact with M cells.101 However, they were not stationary there. By visualizing the tissue with 3D microscopy, we always observed some cells between the SED and GC, even several days after immunization.152 When we instead studied SED cells using live imaging, we also observed that the turnover of cells in the SED was relatively high and that cells in fact moved from the SED towards the GC.101 In addition, when migrating B cells had access to antigen, they would bind to this and transport it from the SED and toward the GC, possibly positioning it on their cell surface rather than degrading it. Careful sequencing of antibody genes from SED and GC cells also supported that such movements occurred.152 Thus, it appears that one important function of the cells in the SED is to bind to specific antigens to ensure that the GC is loaded with antigen.75 In fact, previous findings show that B cells in both LN and spleen can unspecifically transport antigen to GC using complement receptors, and transport of specific antigen recognized by the B cell receptor has been suggested to be important between lung and spleen.157,158 Thus, the SED region may act as a site that ensures that B cells transport antigens to FDC network of the GC when the antigen is present in the local gut environment, and attract newly arrived naive or activated B cells to enter the GC to compete for high-affinity binding.75
Based on these studies, a rather complex chain of the events during T dependent responses to oral antigens appear to take place (Figure 2). When entering the PP from blood, naive B cells will first enter the SED region, most likely in the absence of cognate T cell help. Here, they are tested for their ability to interact with antigens entering the SED through the M cells; only cells that bind antigen will start to proliferate and undergo IgA CSR. In the absence of cognate T cell help or if the cells are of low affinity, the SED response will wane over time, resulting in IgA-switched cells with low number of mutations in their V regions that subsequently become plasmablasts that can leave GALT to migrate to the mucosa. On the other hand, if cells have sufficient affinities and interact efficiently with T cells that recognize the same antigens, they will migrate toward the GC, where the cells will continue to proliferate and undergo affinity maturation. Some cells responding in the absence of cognate T cell interactions may still make it into the GC where additional proliferation will occur, a process that may still require non-cognate interactions with T cells (see below). In addition, some activated B cells will leave the GC, either to return to the SED, where they will reencounter antigens that can be transported back to the GC or to leave via the lymph, which will allow them to enter other PP where they are tested in the SED for their ability to interact with other local antigens. This process will allow for synchronization between spatially distinct GALT structures.
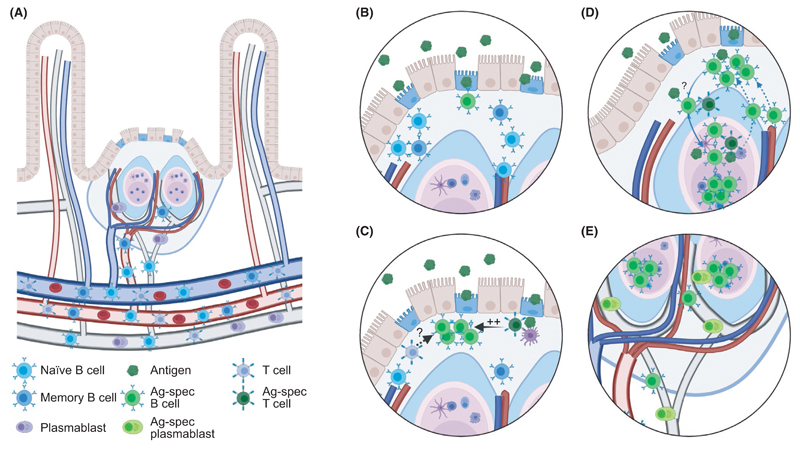
Figure 2. Antigen-specific B cell responses in Peyer´s patches.
(A) Peyer´s patches (PP) are supported by blood and lymph vessels that enables B and T cells to enter via high endothelial venules in areas between B cell follicles.237–239 These, as well as plasmablasts generated during the response, leave the organ via the lymph that subsequently pass the mesenteric lymph nodes before returning to blood when the thoracic ducts enters into the left subclavian vein. (B) Naive and memory B cells will enter the PP and migrate toward the subepithelial dome (SED) where they encounter antigens transported into the structure through M cells. When an antigen-specific naive or memory B cell encounters antigen, it will be activated in a process that is not dependent on antigen interactions but do not require high affinity. (C) Following activation, antigen-specific B cell will proliferate within the SED. The process is expedited if they interact with antigen-specific T cells and it may also benefit from non-cognate interactions between B and T cells. (D) Some activated B cells will enter into the preformed germinal centers (GC) where they will continue to proliferate, but SED proliferation will still be maintained. Entry and further proliferation within the GC will be dependent on antigen affinity as well as cognate interactions with T cells. B cells migrating from the SED may carry specific antigens using their B cell receptors that are loaded onto follicular dendritic cells in the GC. B cells showing signs of already having been in the GC are also present in the SED. It is not known if these migrate directly from the GC or leave as activated memory cells that subsequently reenter to the SED from blood. (E) Antigen-specific B cells and plasmablasts can leave the PP through the lymph. During the response, many of the B cells leaving appear to still have an activated phenotype
6. TI Antigens and GC Responses
When cognate B-T cell interactions occur during T dependent responses in PP, B cell proliferation increases in SED, and B cells with sufficiently high affinity are able to enter GC and give rise to highly mutated plasma and memory cells.152 Then, what about T independent antigens, how do they trigger PP responses? And why do IgA antibodies specific for them carry mutations in their V regions90,159,160? One possibility is that essentially all microbes are detected by the immune system as intact immunogenic particles that contain both antigens normally considered T independent and dependent. Hence, even when these antigens are not covalently linked, B cell receptors on the cell surface will trigger antigen uptake of the whole particle and present peptides derived from any protein within the microbe. Another possibility is that B cells activated against T independent antigens can actually enter GC in PP the absence of cognate B-T cell interactions.63,161 GALT is rather unique compared to other lymphoid organs; during systemic responses GC form following cognate T-B interactions but in GALT GC are already established when an antigen is encountered. Could it be that the cues that B cells need to enter an existing GC are different from those needed to initiate the formation of a GC? Several observations from systemic organs appear to support this hypothesis. For example, it was observed that activated B cells started to form rudimentary GC in the absence of cognate B-T interactions but that these collapsed if T cell help was not present.162 Possibly, if T cells reactive against other antigens maintain the GC, these newly activated B cells may survive longer. Furthermore, it has been demonstrated that B cells recognizing other antigens than those which triggered GC formation can enter GC after activation. While pre-existing T cells, able to provide cognate help to B cells, can facilitate this process, they are not indispensable for cells to enter.163,164 It is also supported also by the phenomenon of epitope spreading during autoimmune responses. Here, a response is started against a single epitope but is then spreading to non-related epitopes, as it has been demonstrated in an SLE mouse model.165 In this model, a knock-in of both a heavy and a light chain reactive against ribonuclear complexes made a proportion of the B cells auto-reactive, which resulted in spontaneous GC formation. However, the autoimmune response gradually spread to unrelated epitopes, presumably through invasion of these GC by B cells having other antigen specificities. Finally, non—antigen-specific non-activated B cells appear to be able to access the GC environment, possibly in order to evaluate their ability to recognize antigens within it, and recent experiments suggest that such bystander cells may sometimes even undergo diversification in GC.163,166 Thus, although not proven, it would seem feasible that T independent antigens can trigger GC entry of B cells in the PP and at least some proliferation within it, which could explain how mutations are introduced into their V regions.
The processes of somatic hypermutation and selecting high-affinity clones occur spatiotemporally distinct in the GC, occurring in the dark and light zones, respectively.58,167 However, even though typical T dependent antigens will give rise to efficient affinity maturation in PP, this process may be less efficient or even absent when T independent antigens are involved.145,161 One study that supported this idea found that antibodies that bound to the surface of microbiota, which to a large extent are made up of polysaccharides, did not show any notable decrease in binding when all mutations were removed from their V regions.168 Thus, in this study, germ-line antibodies, presumably present in naïve B cells, against T independent antigens have similar affinities as those produced in mutated post-GC B cells. However, other recent studies have instead found that mutations in IgA antibody genes do promote binding even to antigens classically defined as T independent. In these, binding of cloned antibodies to specific glycol-epitopes have been studied in humans and mice, and the conclusion was that high-affinity binding to specific epitopes did require the mutations to be present.90,160 Furthermore, mutation-dependent binding to the surface of microbiota, an area that is often heavily glycosylated, has been described both when antibodies from GALT B cells or LP plasma cells were cloned.78,169,170 Thus, although the role of affinity maturation to T independent antigens in the gut is still debated, most recent studies suggest that these antigens do induce affinity maturation when present on commensal microbiota.
7. T Cells Contributing to IgA Responses
As discussed above, the role of T dependent and independent antigens in gut responses cannot easily be answered by just comparing responses in mice lacking T cells with that in normal mice as redundancy will skew the results. Furthermore, to fully appreciate the role that T cell signals play in allowing GC to form versus in the activation of individual B cells and their entry into pre-existing GC, experiments must be done in mice where GC already exist but where activated B cells cannot access T cell help. However, what is clear, is that, in adult individuals, most IgA-producing antibodies plasma cells carry mutations in their V regions and for this reason, they must have proliferated extensively, most likely in a GC.
So, which T cells need to be present in PP or other MALT to form GC? In systemic organs, induction of T cells that enter into the T follicular helper (Tfh) lineage characterized by Bcl6 expression is critical for GC formation.171 But is that also true in MALT? In fact, there have been reports suggesting that Bcl6 expression may not even be needed for antigen-specific gut responses to T dependent antigens.172 And even if Tfh cells are needed—will they develop from naive T cells, or could other T-helper lineages develop into Tfh cells in MALT? Different types of Tfh cells that share phenotype with other lineages have been described, making them Tfh1, Tfh2, or Tfh17 cells; do these develop when Tfh cells differentiated toward other phenotypes or do Th1, Th2, and Th17 cells become Tfh cells.173,174 And what about T follicular regulatory (Tfr) cells—they seem to develop primarily from thymic T regulatory cells but can they also be derived from induced regulatory T cells or even naive cells.175
Some studies have suggested that antigen-specific IgA responses in PP may involve substantial cross-differentiation. Initially, Treg cells were implicated. In an antigen-specific mouse model recognizing bacterial fimbriae, depletion of Treg cells decreased IgA production and this could subsequently be rescued by transfer of CD25+ or Fox3 expressing cells.176 Furthermore, transfer or purified FoxP3-expressing Treg cells from spleen and lymph nodes into CD3ε-deficient mice lacking T cells led to a relative rapid induction of IgA production as well as GC formation with T cells within the PP.177 These transferred cells appeared to downregulate FoxP3 expression before becoming Tfh cells, making the pathway hard to track in the WT situation since lineage tracer mice were not used. Notably, later experiments revealed that the FoxP3-expressing cells did not only differentiate into Tfh cells but also directly regulated IgA response as Tfr cells in the transfer system, suggesting that Tfh and Tfr could belong to the same clones, making the two functions somewhat hard to distinguish from each other.51
Other experiments instead suggested that Th17 cells could be involved. It was first suggested that IL17 was needed for efficient mucosal responses and linked this to induction of Th17 cells.178 However, subsequent data suggested that this may be due to that IL17 supported pIgR-mediated transport of IgA rather than induction of B cells, but also that IL21 production from Th17 cells may play a role.127,179 Finally, using a tracer model it was shown that cells that had previously expressed IL17 may cross-differentiate into Bcl6 expressing Tfh phenotype that supported IgA CSR after transfer into mice lacking T cells.180
So how can these apparently contradictory results, that both Treg and Th17 cells develop into Tfh cells in PP, be integrated into a model for IgA responses in the normal situation. Is there competition between theses pathways? Or are they both minor pathways showing what can happen in artificial systems rather than indicators of what do happen in non-modified individuals? And if Th17 cells cross-differentiation is required, what would happen when a novel antigen is encountered? Would cells go through a short Th17 stage during the primary response or would we only be able to respond to previously encounted antigens? After all, T cells only become Th17 cells after they encounter antigens.
We recently started to address some of these questions by studying the roles of different T cell subsets in the response to specific antigens. A first study casted some light on the role of Treg and Tfh cells.129 This study was based on the fortuitous observation that in transgenic mice carrying a T cell receptor reactive against chicken ovalbumin, adjuvants were not needed for induction of specific IgA and IgG responses after an oral immunization with ovalbumin. This was also the case when T cells from the transgenic mice were transferred into T cell-deficient animals, and while CD25-non-regulatory T cells were sufficient for IgG responses, both non-regulatory and CD25+ Treg cells were required for IgA responses. In this transfer model, for the non-regulatory cells to support immunization in the absence of adjuvant, they had to be isolated from mice with a microbiota, and we found that the presence of a microbiota led to the occurrence of T cells in PP that expressed a recombined endogenous T cell receptor in addition to the transgenic T cell receptor. Thus, it seemed that the endogenous receptor detected antigens from the microbiota and that the transgene at the same time allowed for the cells to support the response against ovalbumin. A large fraction of these transferred non-regulatory cells expressed the markers PD-1, CXCR5, and Bcl6, suggesting that they were bona fide Tfh cells, and they also maintained this phenotype after transfer. At the same time, transferred CD25+ T regulatory cells did not have to express dual T cell receptors, and it was sufficient to transfer splenic, non-transgenic thymus-derived T regulatory cells to support IgA induction. Their role appeared to be to provide TGFβ and possibly other factors to support IgA CSR. Overall, this specific model suggested that when the ovalbumin-specific Tfh cells were at a non-limiting level in PP, due to constant activation through antigen-detection of the microbiota through expression of endogenous T cell receptors, adjuvants were not required for oral immunization. Furthermore, in the model thymic derived T regulatory cells were required to provide signals critical for the IgA CSR but this did not require antigen interactions.
This model cast some light into the role of T cells but was limited in similar manners as previous studies by being based on non-natural circumstances that may skew the results. We have more recently been addressing the role of T cells play in GALT in a more physiological system that respond to a novel antigen in a more natural polyclonal situation (Gribonika et al, under revision). In this study, WT mice were immunized orally with CT, and an MHC II tetramer loaded with an immunodominant peptide from CT is used to identify antigen-specific cells that are subsequently investigated using multicolor flow cytometry and single-cell RNASeq sequencing coupled to cloning of T cell receptor genes.181,182 Using this approach, we did not find any support for the notion that antigen-specific Treg or Th17 cells were involved in CT-specific T cell responses or cross-differentiated to Tfh cells. Rather, cells with a Tfh phenotype dominated the antigen-specific response and did not share clonal relationships with Th17- or Treg-like cells, and a lineage tracer showed that the Tfh cells had not previously expressed IL17. Furthermore, we found that thymus-derived Treg populations shared transcriptional characteristic with Tfr cells, but had minimal clonal relationships and only minor transcriptional similarities with Tfh cells. The situation was very similar when steady-state non-selected T cells from PP were analyzed using single-cell RNASeq; Tfh-like cells made up more than 50% of all memory T cells and they showed very limited T cell receptor sharing with any other T cell lineages. Thus, although different transfer models have indicated that Th17 and Treg cells have the potential to cross-differentiate into the Tfh lineage in PP, we found little evidence for transcriptional or clonal relationships between the lineages in normal adult mice, neither for steady-state T cells nor cells responding to a novel antigen. Rather, the analysis suggested that direct differentiation of naïve CD4 T cells into the Tfh lineage totally dominated the response.
8. Do Model Antigen Responses in PP Reflect Responses to Pathogens and Commensal Microbiota?
With these observations using model antigens, an important question is how well they reflect responses to pathogens or other gut antigens. The responses that are triggered by CT and other related toxins represent a golden standard for mucosal vaccinology as they give strong protective responses made up of both secreted IgA and serum IgG, long-term production of antibodies, and development of long-lived memory B cells.146 However, although some mucosal vaccines are available, further development is needed to give responses of a similar magnitude, and these should ideally occur in the absence of the side effects associated with CT.7 Thus, in mice that tolerate CT better than humans, a CT-like response is a reasonable goal for mucosal vaccine response, both when it comes to levels of antibodies produced and types of cells induced. Similarly, certain mucosal infections give rise to long-term protection after infection, while others do not.8,10 To understand give long-term protection, it is likely that CT will represent a relatively good model.
When it comes to the IgA that covers the microbiota two schools of thought have developed (Figure 3). One favors the view that the IgA production represents a highly specific response to bacterial strains which may cause inflammation if they are not covered with IgA52; the other suggests that IgA binding to the microbiota is made in a rather unspecific manner in the absence of T cells help and thus hinders any inflammation, a role a bit akin to that of natural IgM.48,53 An argument for the latter view is that a large proportion of the gut bacterial microbiota is covered with IgA, and although some of these strains can cause inflammation, most will not. Another is that some cloned antibodies were of low affinity for the bacterial antigens.48 While these arguments are valid, it is important to consider the fact that antibacterial antibodies will mostly bind to polysaccharides on the bacterial surface.183 Although there is significant diversity among polysaccharides, many bacterial strains will carry identical or similar structures, and antibodies reactive to one strain will also be reactive against other strains.78,160 Furthermore, low-affinity binding to one structure cannot exclude high-affinity binding to another.184 Thus, the ability of the IgA antibodies to bind to many different bacterial strains may be a result of the fact that their production is induced by one specific strain, which can cause inflammation, but later antibodies from the same B cell clone will cross-react, possibly with lower affinity, with large number of other bacteria. When individual gut IgA antibodies were cloned from single mice or human donors, many of them were indeed able to bind to a relatively large proportion of gut microbiota representing many phyla.168,169 Thus, the pattern of binding is compatible with the hypothesis that a response against a specific structure will generate a cross-reactive response against similar structures on other bacterial strains. In line with this, some of these studies found that when Ig genes cloned from PP or plasmablasts were reversed to germ line by removing mutations binding was lost or diminished.78,90,160,169 This suggests affinity selection after introduction of mutations and would imply a role for GC in the process. One study did not see a correlation between mutations and binding, and also reported that both T cell-deficient and germ-free mice fed an antigen-free diet produced antibodies able to bind to bacterial antigens despite having never encountered them.168 How to interpret this was unclear until recently, when two studies have shed some light on the issue as they both found specific antibody clones enriched in PP GC of germ-free mice, possibly due to increased probability for recombination and reactivity against self-antigens.78,90 Thus, there was selection in GC of these clones even in germ-free mice, which can explain the presence of such clones in the gut. Taken together, current data would support a constant selection of B cell clones in PP GC. This will mostly be driven by antigens from the microbiota and will result in both affinity maturation but cross-reactivity to similar structures on other bacterial strains will still be maintained. The fact that essentially all IgA antibodies in gut are mutated certainly suggests that the cells producing them had passed through GC with PP being the most likely organ139,140,141,185; otherwise, a distinct, so far unidentified, area where IgA cells undergo rapid proliferation and acquisition of mutations must be posited.
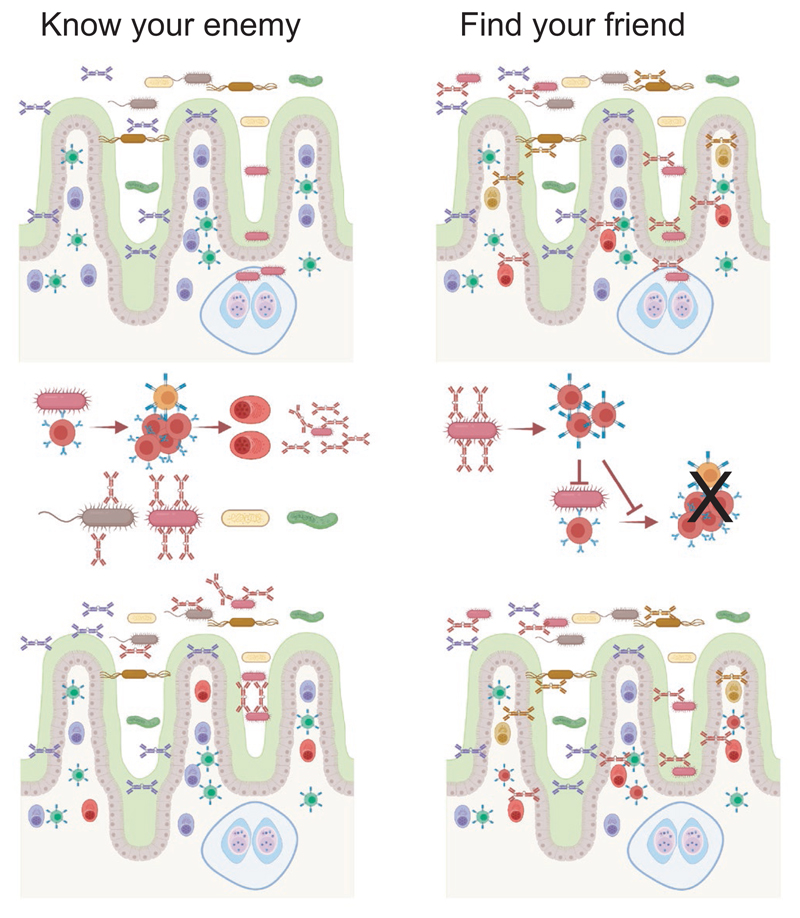
Figure 3.
Commensal binding of IgA antibodies and consequences of the induction pathways. Two mutually non-exclusive models for IgA binding to commensal microbiota has been proposed, and these have implications on the induction of antibodies. According to a "Know your enemy" model, bacterial strains able to trigger inflammation will activate antigen-specific responses in GALT, likely through T cell-dependent mechanisms, which ensures that this bacterial strain cannot invade into tissues again. IgA antibodies will cover other bacterial strains due to cross-reactivity between epitopes. According to the "Find your friend" model, broadly reactive IgA antibodies are produced in a T independent manner before the bacterial encounter, possibly even in the absence of antigen-specific interactions due to triggering of pattern recognition receptors on B cells. Bacteria covered with non-inflammatory IgA will trigger regulatory T cell responses if the bacteria manage to invade into tissue, which in turn ensures that specific responses or inflammation are not triggered
Overall, the outcome from reacting to commensal microbial antigens thus appears to be similar as the one we have observed using the NP-CT model antigen. Nevertheless, this does not necessary mean that the process of activation and selection is identical. After all, CT is an incredibly powerful antigen that also functions as an adjuvant.186 Maybe the toxin can directly influence B or T cells so that these are reacting in a different manner than those activated against other antigens? Recent experiments using single-cell RNASeq analysis of activated lymphocytes present in normal PP or antigen-specific cells reacting to CT antigens after oral immunization have been conducted in our laboratory. As discussed above, T cells were identified by a MHC II tetramer carrying an immunodominant peptide from CT181 whereas the B cells used in the analysis were GFP-expressing B1-8hi cells activated by NP-CT immunization (Gribonika et al, under revision; Komban et al, manuscript in preparation). The insights reached from these studies will be discussed in detail in upcoming publications, but what is clear from the studies is that the transcriptomic analysis does not identify extensive differences between B and T cells activated against CT antigens compared to other activated cells in the PP during steady state. Thus, although CT is without any doubt a powerful oral antigen, able to replace almost half of all IgA-producing plasma cells in the gut following repeated immunizations,145 there is no reason to believe that B or T cells would be activated through an non-physiological process in the response.
9. Mucosal Responses to Airway Infections
Much interest has focused on gut IgA production and responses during the last decades, both due to that the gut is the major IgA-producing organ and to an increased interest in the importance of the gut microbiota and immune responses against it. However, the still ongoing SARS-CoV-2 pandemic has put airborne pathogens and the antibody responses against them into the spotlight. The airways are arguably the most important entry sites for pandemic, epidemic, and endemic pathogens,187 and local secretion of airway antibodies has the potential to create truly sterilizing immunity by directly neutralizing pathogens before they are able to infect of epithelial cells or invade tissues.6 In the airways, IgA antibodies dominate the response in the upper airways with increasing proportions or IgG toward the lung.188 Studies in which purified IgG and secretory IgA antibodies have been transferred into naive recipients to test their role in protection has been performed in influenza mouse models, and while their functions are of course not separate in the normal situation, these studies have concluded that while IgG is most important for protection of the lower airways, secretion of IgA is the dominating humoral mechanism protecting the upper airways.189 This difference is also reflected in the localization of plasma cells—in the upper airways IgA production dominate but in the lower parts both IgA and IgG producing cells are present.190,191 A later study suggested that both IgG and IgA in the lower airways have equal neutralizing capacity, suggesting a safeguard mechanism to protect against infection.192
Despite their role in protecting the airways from contracting contagious diseases, many questions regarding how B cell system function in the airways are unanswered. Among these are how cells from different inductive sites collaborate and which cell movements are needed for efficient responses. Furthermore, similar to the gut, the airways maintain a commensal microbiota during healthy conditions; NALT tissues are in direct contact with a rather rich commensal microbiota in the nasal and mouth cavities, whereas fewer bacteria are present in the lower airways and probably even less in the medLN that drain them.193,194 A forthright interpretation of the fact that IgA plasma cells dominate in the upper airways and IgG in the lower airways, and that there are separate inductive sites for the upper (NALT) and lower (iBALT and medLN) airways would be that the immune responses of two parts are strictly separate. However, why would then so many IgG expressing cells be present in NALT/tonsils when they are not present in the mucosa or glands of the upper airways77,195,196,197? And, since it takes more than one week after infection for iBALT to be generated—do they play any role in a for viral clearance during primary response198? It appears that GC B cells in iBALT have different antigen specificity and B cell repertoire as compared to GC B cells in medLN after viral infection.199 This could be due to distinct T cell signals between mucosal and lymphoid tissues, different local antigen availability or to other, yet unidentified, stimuli. The same study also identifies that a large majority of GC B cells in iBALT are non-Influenza specific, even when mice are infected with influenza. This could be attributed to the characteristic of iBALT as a general priming site, where cells of many specificities can be activated.200 Future studies will need to address this and investigate whether already formed GC in iBALT are permissive for activated B cell entry, such as we have described for GC in PP.145
Another potential, but more restricted, role could be for the generation of tissue-resident memory cells.201 After the discovery that there are non-circulating effector memory-like CD8 T cells that do not leave the lungs, similar CD4 and, more recently, B cells were also identified.202–204 Recently, a newly identified subset of Tissue-resident helper (Trh) cells, which are independent of Tfh in lymphoid organs, has been shown to be crucial for the generation of iBALT, antigen-specific B cells, and local antibody production.205,206 Further, these cells are particularly important for B cell differentiation to plasmablasts upon reinfection.
To better understand de novo response in the lower airways to an actual infection, we recently conducted a single-cell RNASeq study using a mouse influenza model.207 At 7, 14, and 28 days after intranasal infection of mice with Influenza type A virus, hemagglutinin(HA)-specific B cells from lung (including iBALT), medLN, and spleen were isolated, and we performed single-cell RNASeq analysis paired with single-cell B cell receptor cloning on these cells. This relatively unbiased setup allowed us to determine how a typical response to influenza developed over space and time in the lower airways, and how B cells belonging to antibody clones mutated and distributed over the tissues. Although all cell subtypes were present in all organs, the distribution of cells was different between organs with GC B cells dominating in spleen and medLN and memory B cells within lung. Interestingly, there were no drastic differenced in the composition of cells within an organ between day 7 and 28. Memory cells from lung differed from those in other organs by expressing Cd69 and other genes suggestive for being tissue-resident memory cells. As judged by clonal relationships GC B cells, memory B cells, and plasmablasts within, but not between, organs shared origin. There was one notable exception from this—memory B cells in lung showed clonal relationships with GC and memory cells from both medLN and spleen. There was no apparent shift in the output from GC from memory B cells to plasmablasts; both cell types appeared to be generated all through the response. Neither did we see any the difference in affinity between the cells. However, plasmablasts more often belonged to large clones, whereas memory B cells showed a larger breath of antibody variants. Taken together, the data suggest that whereas GC responses are local in nature, those in iBALT are not sufficient to provide the lung with memory B cells and plasmablasts, but these also arrive from medLN and spleen. Thus, it appears that mucosal tissues in the airways, including lung, are a main site for memory B cell and plasmablast residency. These are maintained to protect the host from reinfection with the same or a variant pathogen.
10. Human Mucosal Responses to SARS-CoV-2 Infection
It appears that in individuals infected with SARS-CoV-2, the major site for viral replication early during disease are the upper airways.208 Despite this, most studies of SARS-CoV-2 immune responses have focused on IgG production and systemic responses. In fact, a recent review argued that the role of the mucosal system in the response to SARS-CoV-2 was a neglected area that should be further addressed.209 While this is still true, some data have been published that look more into the roles that airway responses and IgA plays. Some reports found that IgA antibodies reactive to SARS-CoV-2 are often detectable before both IgM and IgG in serumou,11,12,210 and it was later reported that IgA and IgM levels in serum appear to decay faster than IgG although other studies did not find this.211–213 It is still unclear if this represents a more rapid decline in production or not; an alternative scenario is that short-lived plasmablasts expressing either class are gradually dying at a similar pace, but that the titers of IgG will show a slower decay due to differences in serum half-lives of antibody classes.20 It is also important to note that the early antibody decay in serum of any class will not give relevant information about long-term production.214 In addition, serum studies will not be able to determine whether IgA antibodies are secreted locally. Some studies have described the secreted IgA antibodies in saliva as well as in BAL and breast milk,210,215,216,217,218,219 and although the levels of antibodies in these body fluids appeared to decline as rapidly as in serum, further studies are needed to determine the result long term as some plasma cells at mucosal surfaces can be very long-lived.33,185 Interestingly, high background titers of IgA binding to SARS-CoV-2 antigens were noted in some study, which could potentially be linked to cross-reactive antibodies from previous infections with endemic coronaviruses.217–219 When it comes to virus neutralization it is still unclear what role IgA plays. Some studies have reported that SARS-CoV-2-specific IgA antibody levels in serum show a better correlation with the ability of the serum to neutralize SARS-CoV-2 than IgG levels do,210,220,221 at least early during the response but other studies have come to the opposite conclusion.155,222,223 Another found that IgA in nasal washes efficiently neutralized SARS-CoV-2.219 Regardless, one study demonstrated that the ability of secreted IgA to form dimers increased its ability to neutralize SARS-CoV-2 more than 10-fold, making secreted mucosal antibodies highly efficient.224
Although data are now emerging regarding IgA production and its potential role during SARS-CoV-2 infection, less has been done to characterize to which extent antigen-specific cells are part of a mucosal responses or not. As the major site of infection is the upper airways, it is important to determine to which extent they are primed in tonsils and if they will home to these areas to protect. Several studies have noted a strong plasmablast response, and some have also described that many plasmablasts express IgA with the majority of these being IgA1 expressing.210,225 In one of these, higher expression of CCR10, a chemokine receptor associated with mucosal homing, was found on plasmablast compared with naive and memory B cells, with IgA+ plasmablasts also expressing higher levels than IgG+ ones.210 Another recently published study showed ongoing development of more mutated memory B cells after recovery and demonstrated that some gut epithelial cells may still be infected at this time, which lead them to suggest that gut antigens may then drive the response.212
We will very soon submit a study that address some of the issues discussed above (Figure 4) (Lundgren et al, manuscript in preparation). In this, we studied the plasmablasts response in COVID-19 patients during disease and after recovery and compared it to healthy adults. Using a set of monoclonal antibodies detecting all human antibody classes,226 we found that IgA1, IgG1, and IgM plasmablasts dominated the early response, and that three months after recovery the number of plasmablasts in blood had normalized. As many as 70% of all plasmablasts expressed integrin β1, with approximately 30% of these also expressing integrin β1, and 80% CCR10. CCR9 expression was relatively rare. Interestingly, an increased number of plasmablasts expressed CD138 during disease compared to steady state, a marker potentially linked to GC origin and/or longevity.227,228 Taken together, it appears that a large proportion of plasmablasts during SARS-CoV-2 disease suggestive of a mucosal response with airway homing a likely outcome. In addition to these observations, we also detected increased levels of memory B cells expressing CD45RB and CD69, a phenotype that suggests that they may develop into resident memory B cells.229
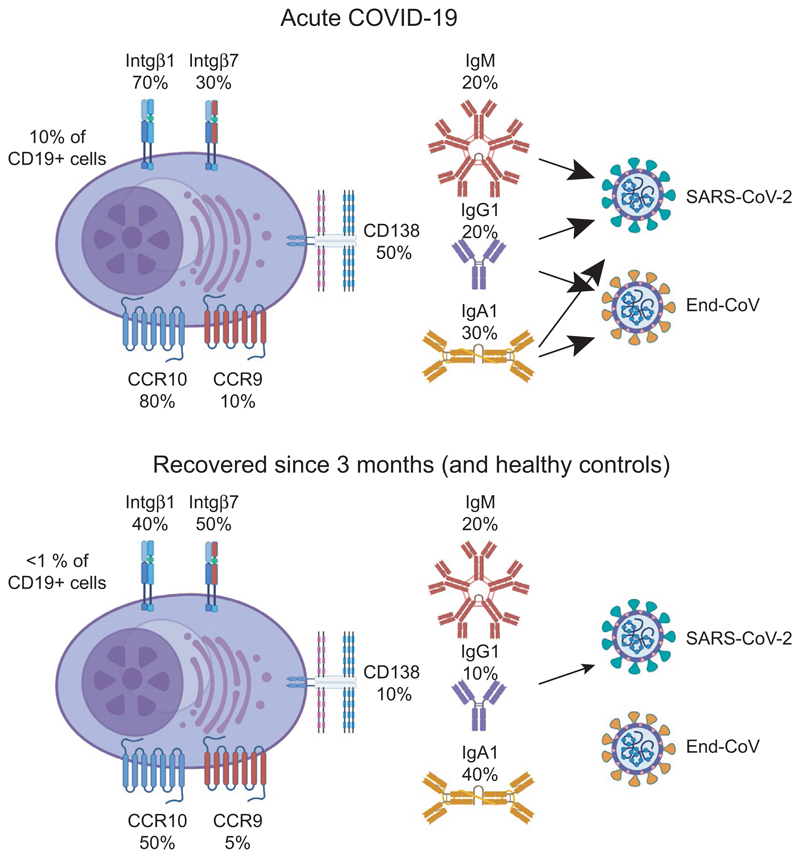
Figure 4.
Plasmablast responses during COVID-19 disease. Plasmablasts responses were studied in hospitalized COVID-19 patients during acute disease and three months after recovery. Plasmablast made up 10% of all CD19+ B cells in blood during active disease, but less than 1% 3 months after recovery or in healthy controls. Plasmablasts encountered in blood during active disease expressed higher levels of integrin β1, CCR10, and CD138 than those after recovery, and produced antibodies of IgA1, IgG1, and IgM class, which suggested that they many generated in a mucosal immune response. Antibodies of all classes produced by the plasmablasts during disease reacted with SARS-CoV-2, and both IgA and IgG antibodies showed cross-reactivity to endemic coronaviruses. After recovery, the plasmablasts in blood produced low levels of IgG antibodies against SARS-CoV-2 while antibodies of other classes could not be detected, and they did not show cross-reactivity
We also observed that antibodies produced by these early plasmablasts—both IgA and IgG—showed cross-reactivity to endemic coronaviruses. It has been suggested in previous studies that memory B cells generated during encounters with endemic coronavirus responses may be part of the response to SARS-CoV-2230,231 a situation that would be somewhat similar to the “original antigenic sin” observed during influenza.232,233 Interestingly, while cross-reactivity was readily detected during acute infection, neither the dwindling plasmablasts response against SARS-CoV-2 three months after infection or reactivity in memory B cells activated to antibody secretion showed cross-reactivity larger than background. Thus, whereas pre-existing cross-reactive memory may be recruited into the response early on, the response develops into higher specificity and less cross-reactivity over time, and the SARS-CoV-2 did not appear to replenish the memory against previously encountered coronaviruses to any large extent.
11. Conclusions
The development of the vertebrate immune system was influenced by the requirement to protect the semipermeable mucosal membranes that enables exchange of gases and nutrients.234 Here, in addition to protection from pathogens, the immune system needed to maintain a balance with commensal microbiota, which lead to the that specific secreted mucosal antibody classes developed already in bony fish.4,5 The duality that exists in the function of the mucosal immune system—maintenance of commensal strains and eradication of pathogenic ones—has fashioned its development all since. Here, we describe recent studies that show the mucosal system can respond to model antigens or novel pathogens, resulting in T cell-dependent IgA responses. We also discuss that the findings in these and previous studies indicate that most, if not all, IgA responses in adult individuals are generated through these pathways or at least are channeled into them. Thus, while truly T cell-independent IgA responses are certainly possible, they are unlikely to contribute much to overall IgA production in healthy adult individuals. Nevertheless, T cell-independent pathways may contribute during early life when the commensal microbiota is established.235 If future efforts in understanding IgA induction, production, and responses, it is important for researcher to be more careful in differentiating these scenarios.236 Germ-free mice reconstituted with a microbiota or immunodeficient mice into which purified cells are transferred are likely to be affected by homeostatic effects and/or redundancy and will be poor proxies of the situation in normal adult individuals already reacting to a complex microbiota. For researchers studying mucosal immunology to eventually reach a consensus view on how the IgA system protects against pathogens but at the same time maintains the commensal microbiota, it is important to perform studies in systems that resembles physiological conditions.
Acknowledgements
All figures were created using BioRender.com. MB is supported by grants from Swedish Research Council (2019-01708) and Swedish State Support for Clinical Research (ALFGBG-727081), and DA is supported by a European Research Council starting grant (B-DOMINANCE, 850638) and a Swedish Research Council starting grant (2017-01439). The work on COVID was supported by SciLifeLab/KAW national COVID-19 research program (2020-0182).
Funding information
FP7 Ideas: European Research Council, Grant/Award Number: B, DOMINANCE and 850638; Vetenskapsrådet, Grant/Award Number: 2017, 01439, 2019 and 01708; Science for Life Laboratory, Grant/Award Number: 2020-0182; Swedish State Support for Clinical Research, Grant/Award Number: ALFGBG and 727081
Footnotes
Conflict of Interest
The authors do not declare any conflicts of interest.
Data Availability Statement
Data sharing not applicable—no new data generated.
References
- 1.Pabst R, Russell MW, Brandtzaeg P. Tissue distribution of lymphocytes and plasma cells and the role of the gut. Trends Immunol. 2008;29:206–208. doi: 10.1016/j.it.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Corthésy B. Multi-faceted functions of secretory IgA at mucosal surfaces. Front Immunol. 2013;4:185. doi: 10.3389/fimmu.2013.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandtzaeg P. Secretory IgA: Designed for anti-microbial defense. Front Immunol. 2013;4:222. doi: 10.3389/fimmu.2013.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Z, Takizawa F, Casadei E, et al. Specialization of mucosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci Immunol. 2020;5:eaay3254. doi: 10.1126/sciimmunol.aay3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Y, Wang Q, Huang Z, Ding L, Xu Z. Immunoglobulins, mucosal immunity and vaccination in teleost fish. Front Immunol. 2020;11:567941. doi: 10.3389/fimmu.2020.567941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 7.Lycke N. Recent progress in mucosal vaccine development: potential and limitations. Nat Rev Immunol. 2012;12:592–605. doi: 10.1038/nri3251. [DOI] [PubMed] [Google Scholar]
- 8.Brandtzaeg P. Induction of secretory immunity and memory at mucosal surfaces. Vaccine. 2007;25:5467–5484. doi: 10.1016/j.vaccine.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Blutt SE, Conner ME. The gastrointestinal frontier: IgA and viruses. Front Immunol. 2013;4:402. doi: 10.3389/fimmu.2013.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amadou Amani S, Lang ML. Bacteria that cause enteric diseases stimulate distinct humoral immune responses. Front Immunol. 2020;11:565648. doi: 10.3389/fimmu.2020.565648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma H, Zeng W, He H, et al. Serum IgA: IgM, and IgG responses in COVID-19. Cell Mol Immunol. 2020;17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mkaddem SB, Christou I, Rossato E, Berthelot L, Lehuen A, Monteiro RC. IgA, IgA receptors, and their anti-inflammatory properties. Curr Top Microbiol Immunol. 2014;382:221–235. doi: 10.1007/978-3-319-07911-0_10. [DOI] [PubMed] [Google Scholar]
- 14.Halpern MS, Koshland ME. Noval subunit in secretory IgA. Nature. 1970;228:1276–1278. doi: 10.1038/2281276a0. [DOI] [PubMed] [Google Scholar]
- 15.Mestecky J, Zikan J, Butler WT. Immunoglobulin M and secretory immunoglobulin A: presence of a common polypeptide chain different from light chains. Science. 1971;171:1163–1165. doi: 10.1126/science.171.3976.1163. [DOI] [PubMed] [Google Scholar]
- 16.Brandtzaeg P. Mucosal and glandular distribution of immunoglobulin components: differential localization of free and bound SC in secretory epithelial cells. J Immunol. 1974;112:1553–1559. [PubMed] [Google Scholar]
- 17.Kaetzel CS. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol Rev. 2005;206:83–99. doi: 10.1111/j.0105-2896.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- 18.Isho B, Florescu A, Wang AA, Gommerman JL. Fantastic IgA plasma cells and where to find them. Immunol Rev. 2021 doi: 10.1111/imr.12980. [DOI] [PubMed] [Google Scholar]
- 19.Blanco E, Perez-Andres M, Arriba-Méndez S, et al. Age-associated distribution of normal B-cell and plasma cell subsets in peripheral blood. J Allergy Clin Immunol. 2018;141:2208–2219.:e16. doi: 10.1016/j.jaci.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 20.Turesson I. Distribution of immunoglobulin-containing cells in human bone marrow and lymphoid tissues. Acta Med Scand. 1976;199:293–304. doi: 10.1111/j.0954-6820.1976.tb06735.x. [DOI] [PubMed] [Google Scholar]
- 21.Sindic CJ, Delacroix DL, Vaerman JP, Laterre EC, Masson PL. Study of IgA in the cerebrospinal fluid of neurological patients with special reference to size, subclass and local production. J Neuroimmunol. 1984;7:65–75. doi: 10.1016/s0165-5728(84)80007-7. [DOI] [PubMed] [Google Scholar]
- 22.Leong KW, Ding JL. The unexplored roles of human serum IgA. DNA Cell Biol. 2014;33:823–829. doi: 10.1089/dna.2014.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandtzaeg P, Johansen F-E. Mucosal B cells: phenotypic characteristics, transcriptional regulation, and homing properties. Immunol Rev. 2005;206:32–63. doi: 10.1111/j.0105-2896.2005.00283.x. [DOI] [PubMed] [Google Scholar]
- 24.Steffen U, Koeleman CA, Sokolova MV, et al. IgA subclasses have different effector functions associated with distinct glycosylation profiles. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-019-13992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilian M, Reinholdt J, Lomholt H, Poulsen K, Frandsen EV. Biological significance of IgA1 proteases in bacterial colonization and pathogenesis: critical evaluation of experimental evidence. APMIS. 1996;104:321–338. doi: 10.1111/j.1699-0463.1996.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 26.Takemura T, Eishi Y. Distribution of secretory component and immunoglobulins in the developing lung. Am Rev Respir Dis. 1985;131:125–130. doi: 10.1164/arrd.1985.131.1.125. [DOI] [PubMed] [Google Scholar]
- 27.Haimoto H, Nagura H, Imaizumi M, Watanabe K, Iijima S. Immunoelectronmicroscopic study on the transport of secretory IgA in the lower respiratory tract and alveoli. Virchows Arch A Pathol Anat Histopathol. 1984;404:369–380. doi: 10.1007/BF00695221. [DOI] [PubMed] [Google Scholar]
- 28.Ocak S, Pedchenko TV, Chen H, et al. Loss of polymeric immunoglobulin receptor expression is associated with lung tumourigenesis. Eur Respir J. 2012;39:1171–1180. doi: 10.1183/09031936.00184410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds HY. Immunoglobulin G and its function in the human respiratory tract. Mayo Clin Proc. 1988;63:161–174. doi: 10.1016/s0025-6196(12)64949-0. [DOI] [PubMed] [Google Scholar]
- 30.Wilmore JR, Gaudette BT, Gomez Atria D, et al. Commensal microbes induce serum IgA responses that protect against polymicrobial sepsis. Cell Host Microbe. 2018;23:302–311.:e3. doi: 10.1016/j.chom.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mei HE, Yoshida T, Sime W, et al. Blood-borne human plasma cells in steady state are derived from mucosal immune responses. Blood. 2009;113:2461–2469. doi: 10.1182/blood-2008-04-153544. [DOI] [PubMed] [Google Scholar]
- 32.Lemke A, Kraft M, Roth K, Riedel R, Lammerding D, Hauser AE. Long-lived plasma cells are generated in mucosal immune responses and contribute to the bone marrow plasma cell pool in mice. Mucosal Immunol. 2016;9:83–97. doi: 10.1038/mi.2015.38. [DOI] [PubMed] [Google Scholar]
- 33.Bemark M, Hazanov H, Strömberg A, et al. Limited clonal relatedness between gut IgA plasma cells and memory B cells after oral immunization. Nat Commun. 2016;7:12698. doi: 10.1038/ncomms12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fitzpatrick Z, Frazer G, Ferro A, et al. Gut-educated IgA plasma cells defend the meningeal venous sinuses. Nature. 2020;20:1–5. doi: 10.1038/s41586-020-2886-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rojas OL, Probstel A-K, Porfilio, et al. Recirculating Intestinal IgA-Producing Cells Regulate Neuroinflammation via IL-10. Cell. 2019;176:610–624.:e18. doi: 10.1016/j.cell.2018.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pröbstel A-K, Zhou X, Baumann R, et al. Gut microbiota-specific IgA+ B cells traffic to the CNS in active multiple sclerosis. Sci Immunol. 2020;5:eabc7191. doi: 10.1126/sciimmunol.abc7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharonov GV, Serebrovskaya EO, Yuzhakova DV, Britanova OV, Chudakov DM. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat Rev Immunol. 2020;348:1–14. doi: 10.1038/s41577-019-0257-x. [DOI] [PubMed] [Google Scholar]
- 38.Biswas S, Mandal G, Payne KK, et al. IgA transcytosis and antigen recognition govern ovarian cancer immunity. Nature. 2021;591:464–470. doi: 10.1038/s41586-020-03144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Chen D, Yu B, et al. Fungi in gastrointestinal tracts of human and mice: from community to functions. Microb Ecol. 2018;75:821–829. doi: 10.1007/s00248-017-1105-9. [DOI] [PubMed] [Google Scholar]
- 42.Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santacroce L, Charitos IA, Ballini A, et al. The human respiratory system and its microbiome at a glimpse. Biology (Basel) 2020;9:318. doi: 10.3390/biology9100318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol. 2019;20:1279–1290. doi: 10.1038/s41590-019-0451-9. [DOI] [PubMed] [Google Scholar]
- 45.Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med. 2019;216:20–40. doi: 10.1084/jem.20180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soderholm AT, Pedicord VA. Intestinal epithelial cells: at the interface of the microbiota and mucosal immunity. Immunology. 2019;158:267–280. doi: 10.1111/imm.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bunker JJ, Bendelac A. IgA Responses to microbiota. Immunity. 2018;49:211–224. doi: 10.1016/j.immuni.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macpherson AJ, Yilmaz B, Limenitakis JP, Ganal-Vonarburg SC. IgA function in relation to the intestinal microbiota. Annu Rev Immunol. 2018;36:359–381. doi: 10.1146/annurev-immunol-042617-053238. [DOI] [PubMed] [Google Scholar]
- 50.Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawamoto S, Maruya M, Kato LM, et al. Foxp3+ T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152–165. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 52.Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol. 2015;159:122–127. doi: 10.1016/j.clim.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato LM, Kawamoto S, Maruya M, Fagarasan S. The role of the adaptive immune system in regulation of gut microbiota. Immunol Rev. 2014;260:67–75. doi: 10.1111/imr.12185. [DOI] [PubMed] [Google Scholar]
- 54.Brandtzaeg P. Function of mucosa-associated lymphoid tissue in antibody formation. Immunol Invest. 2010;39:303–355. doi: 10.3109/08820131003680369. [DOI] [PubMed] [Google Scholar]
- 55.Holmgren J, Czerkinsky C. Mucosal immunity and vaccines. Nat Med. 2005;11:S45–53. doi: 10.1038/nm1213. [DOI] [PubMed] [Google Scholar]
- 56.Nicholson LB. The immune system. EssaysBiochem. 2016;60:275–301. doi: 10.1042/EBC20160017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. 2015;15:137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mesin L, Ersching J, Victora GD. Germinal center B cell dynamics. Immunity. 2016;45:471–482. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kunkel EJ, Butcher EC. Plasma-cell homing. Nat Rev Immunol. 2003;3:822–829. doi: 10.1038/nri1203. [DOI] [PubMed] [Google Scholar]
- 60.Tellier J, Nutt SL. Plasma cells: The programming of an antibody-secreting machine. Eur J Immunol. 2019;49:30–37. doi: 10.1002/eji.201847517. [DOI] [PubMed] [Google Scholar]
- 61.Brynjolfsson SF, Persson Berg L, Olsen Ekerhult T, et al. Long-lived plasma cells in mice and men. Front Immunol. 2018;9:2673. doi: 10.3389/fimmu.2018.02673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brandtzaeg P, Pabst R. Let’s go mucosal: communication on slippery ground. Trends Immunol. 2004;25:570–577. doi: 10.1016/j.it.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 63.Bemark M, Boysen P, Lycke NY. Induction of gut IgA production through T cell-dependent and T cell-independent pathways. Ann N Y Acad Sci. 2012;1247:97–116. doi: 10.1111/j.1749-6632.2011.06378.x. [DOI] [PubMed] [Google Scholar]
- 64.Kiyono H, Fukuyama S. NALT-versus Peyer’s-patch-mediated mucosal immunity. Nat Rev Immunol. 2004;4:699–710. doi: 10.1038/nri1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brandtzaeg P. Potential of nasopharynx-associated lymphoid tissue for vaccine responses in the airways. Am J Respir Crit Care Med. 2012;183:1595–1604. doi: 10.1164/rccm.201011-1783OC. [DOI] [PubMed] [Google Scholar]
- 66.Pabst R, Gehrke I. Is the bronchus-associated lymphoid tissue (BALT) an integral structure of the lung in normal mammals, including humans? Am J Respir Cell Mol Biol. 1990;3:131–135. doi: 10.1165/ajrcmb/3.2.131. [DOI] [PubMed] [Google Scholar]
- 67.Rangel-Moreno J, Carragher DM, de la Luz Garcia-Hernandez M, et al. The development of inducible bronchus-associated lymphoid tissue depends on IL-17. Nat Immunol. 2011;12:639–646. doi: 10.1038/ni.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moyron-Quiroz JE, Rangel-Moreno J, Kusser K, et al. Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nat Med. 2004;10:927–934. doi: 10.1038/nm1091. [DOI] [PubMed] [Google Scholar]
- 69.Lecuyer E, Rakotobe S, Lengliné-Garnier H, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 70.Lochner M, Ohnmacht C, Presley L, et al. Microbiota-induced tertiary lymphoid tissues aggravate inflammatory disease in the absence of RORgamma t and LTi cells. J Exp Med. 2011;208:125–134. doi: 10.1084/jem.20100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koscso B, Kurapati S, Rodrigues RR, et al. Gut-resident CX3CR1hi macrophages induce tertiary lymphoid structures and IgA response in situ. Sci Immunol. 2020;5:eaax0062. doi: 10.1126/sciimmunol.aax0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones GW, Jones SA. Ectopic lymphoid follicles: inducible centres for generating antigen-specific immune responses within tissues. Immunology. 2016;147:141–151. doi: 10.1111/imm.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruddle NH. Basics of inducible lymphoid organs. Curr Top Microbiol Immunol. 2020;426:1–19. doi: 10.1007/82_2020_218. [DOI] [PubMed] [Google Scholar]
- 74.Alon R, Sportiello M, Kozlovski S, et al. Leukocyte trafficking to the lungs and beyond: lessons from influenza for COVID-19. Nat Rev Immunol. 2021;21:49–64. doi: 10.1038/s41577-020-00470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lycke NY, Bemark M. The regulation of gut mucosal IgA B-cell responses: recent developments. Mucosal Immunol. 2017;10:1361–1374. doi: 10.1038/mi.2017.62. [DOI] [PubMed] [Google Scholar]
- 76.Esterhazy D, Canesso MCC, Mesin L, et al. Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature. 2019;569:126–130. doi: 10.1038/s41586-019-1125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao Y, Uduman M, Siu JHY, et al. Spatiotemporal segregation of human marginal zone and memory B cell populations in lymphoid tissue. Nat Commun. 2018;9:3857. doi: 10.1038/s41467-018-06089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nowosad CR, Mesin L, Castro TBR, et al. Tunable dynamics of B cell selection in gut germinal centres. Nature. 2020;588:321–326. doi: 10.1038/s41586-020-2865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tarlinton D. Sheepish B cells: evidence for antigen-independent antibody diversification in humans and mice. J Exp Med. 2008;205:1251–1254. doi: 10.1084/jem.20081057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wesemann DR, Portuguese AJ, Meyers RM, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501:112–115. doi: 10.1038/nature12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vossenkamper A, Blair PA, Safinia N, et al. A role for gut-associated lymphoid tissue in shaping the human B cell repertoire. J Exp Med. 2013;210:1665–1674. doi: 10.1084/jem.20122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tull TJ, Pitcher MJ, Guesdon W, et al. Human marginal zone B cell development from early T2 progenitors. J Exp Med. 2021;218:e20202001. doi: 10.1084/jem.20202001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Y, Chaudhary N, Yang N, et al. Microbial symbionts regulate the primary Ig repertoire. J Exp Med. 2018;215:1397–1415. doi: 10.1084/jem.20171761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cohen M, Giladi A, Raposo C. Meningeal lymphoid structures are activated under acute and chronic spinal cord pathologies. Life Sci Alliance. 2021;4:e202000907. doi: 10.26508/lsa.202000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zelco A, Borjesson V, de Kanter JK, et al. Single-cell atlas reveals meningeal leukocyte heterogeneity in the developing mouse brain. Genes & Dev. 2021 July 22; doi: 10.1101/gad.348190.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brioschi S, Wang W-L, Peng V, et al. Heterogeneity of meningeal B cells reveals a lymphopoietic niche at the CNS borders. Science. 2021;373:eabf9277. doi: 10.1126/science.abf9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schafflick D, Wolbert J, Heming M, et al. Single-cell profiling of CNS border compartment leukocytes reveals that B cells and their progenitors reside in non-diseased meninges. Nat Neurosci. 2021 July 12; doi: 10.1038/s41593-021-00880-y. [DOI] [PubMed] [Google Scholar]
- 88.Pollard M. Germfree animals and biological research. Science. 1964;145:247–251. doi: 10.1126/science.145.3629.247. [DOI] [PubMed] [Google Scholar]
- 89.Pollard M, Sharon N. Responses of the Peyer’s patches in germ-free mice to antigenic stimulation. Infect Immun. 1970;2:96–100. doi: 10.1128/iai.2.1.96-100.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen H, Zhang Y, Ye AY, et al. BCR selection and affinity maturation in Peyer’s patch germinal centres. Nature. 2020;582:421–425. doi: 10.1038/s41586-020-2262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bemark M, Sale JE, Kim HJ, Berek C, Cosgrove RA, Neuberger MS. Somatic hypermutation in the absence of DNA-dependent protein kinase catalytic subunit (DNA-PK(cs)) or recombination-activating gene (RAG)1 activity. J Exp Med. 2000;192:1509–1514. doi: 10.1084/jem.192.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Casola S, Otipoby KL, Alimzhanov M, et al. B cell receptor signal strength determines B cell fate. Nat Immunol. 2004;5:317–327. doi: 10.1038/ni1036. [DOI] [PubMed] [Google Scholar]
- 93.Bergqvist P, Gardby E, Stensson A, Bemark M, Lycke NY. Gut IgA class switch recombination in the absence of CD40 does not occur in the lamina propria and is independent of germinal centers. J Immunol. 2006;177:7772–7783. doi: 10.4049/jimmunol.177.11.7772. [DOI] [PubMed] [Google Scholar]
- 94.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 95.Mabbott NA, Donaldson DS, Ohno H, Williams IR, Mahajan A. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013;6:666–677. doi: 10.1038/mi.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kobayashi N, Takahashi D, Takano S, Kimura S, Hase K. The roles of Peyer’s patches and microfold cells in the gut immune system: relevance to autoimmune diseases. Front Immunol. 2019;10:337. doi: 10.3389/fimmu.2019.02345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kimura S, Mutoh M, Hisamoto M, et al. Airway M cells arise in the lower airway due to RANKL signaling and reside in the bronchiolar epithelium associated with iBALT in murine models of respiratory disease. Front Immunol. 2019;10:1150. doi: 10.3389/fimmu.2019.01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kanaya T, Williams IR, Ohno H. Intestinal M cells: Tireless samplers of enteric microbiota. Traffic. 2020;21:34–44. doi: 10.1111/tra.12707. [DOI] [PubMed] [Google Scholar]
- 99.Schulz O, Pabst O. Antigen sampling in the small intestine. Trends Immunol. 2013;34:155–161. doi: 10.1016/j.it.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 100.Lo DD. Vigilance or subversion? Constitutive and inducible M Cells in mucosal tissues. Trends Immunol. 2018;39:185–195. doi: 10.1016/j.it.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 101.Komban RJ, Strömberg A, Biram A, et al. Activated Peyer’s patch B cells sample antigen directly from M cells in the subepithelial dome. Nat Commun. 2019;10:2423. doi: 10.1038/s41467-019-10144-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bogoslowski A, Kubes P. Lymph Nodes: The unrecognized barrier against pathogens. ACS Infect Dis. 2018;4:1158–1161. doi: 10.1021/acsinfecdis.8b00111. [DOI] [PubMed] [Google Scholar]
- 103.Kunisawa J, Kiyono H. Alcaligenes is commensal bacteria habituating in the gut-associated lymphoid tissue for the regulation of intestinal IgA responses. Front Immunol. 2012;3:65. doi: 10.3389/fimmu.2012.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang M, Gao Z, Zhang Z, Pan L, Zhang Y. Roles of M cells in infection and mucosal vaccines. Hum Vaccin Immunother. 2014;10:3544–3551. doi: 10.4161/hv.36174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Macpherson AJ, Mccoy KD, Johansen F-E, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal Immunol. 2008;1:11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- 106.Rudin A, Riise GC, Holmgren J. Antibody responses in the lower respiratory tract and male urogenital tract in humans after nasal and oral vaccination with cholera toxin B subunit. Infect Immun. 1999;67:2884–2890. doi: 10.1128/iai.67.6.2884-2890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brandtzaeg P, Kiyono H, Pabst R, Russell MW. Terminology: nomenclature of mucosa-associated lymphoid tissue. Mucosal Immunol. 2008;1:31–37. doi: 10.1038/mi.2007.9. [DOI] [PubMed] [Google Scholar]
- 108.Mora JR, Andrian, von Andrian UH. Differentiation and homing of IgA-secreting cells. Mucosal Immunol. 2008;1:96–109. doi: 10.1038/mi.2007.14. [DOI] [PubMed] [Google Scholar]
- 109.Habtezion A, Nguyen LP, Hadeiba H, Butcher EC. Leukocyte trafficking to the small intestine and colon. Gastroenterology. 2016;150:340–354. doi: 10.1053/j.gastro.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fenton TM, Jørgensen PB, Niss K, et al. Immune profiling of human gut-associated lymphoid tissue identifies a role for isolated lymphoid follicles in priming of region-specific immunity. Immunity. 2020;52:557–570.:e6. doi: 10.1016/j.immuni.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stavnezer J, Schrader CE. IgH chain class switch recombination: mechanism and regulation. J Immunol. 2014;193:5370–5378. doi: 10.4049/jimmunol.1401849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Feng Y, Seija N, Di Noia JM, Martin A. AID in antibody diversification: there and back again. Trends Immunol. 2020;41:586–600. doi: 10.1016/j.it.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krokan HE, Drabløs F, Slupphaug G. Uracil in DNA - occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 114.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 115.Maul RW, Gearhart PJ. Refining the Neuberger model: uracil processing by activated B cells. Eur J Immunol. 2014;44:1913–1916. doi: 10.1002/eji.201444813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang X, Zhang Y, Ba Z, Kyritsis N, Casellas R, Alt FW. Fundamental roles of chromatin loop extrusion in antibody class switching. Nature. 2019;575:385–389. doi: 10.1038/s41586-019-1723-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shen HM, Bozek G, Pinkert CA, et al. Expression of AID transgene is regulated in activated B cells but not in resting B cells and kidney. Mol Immunol. 2008;45:1883–1892. doi: 10.1016/j.molimm.2007.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tangye SG, Hodgkin PD. Divide and conquer: the importance of cell division in regulating B-cell responses. Immunology. 2004;112:509–520. doi: 10.1111/j.1365-2567.2004.01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity. 2013;46:83–101. doi: 10.3109/08916934.2012.749244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen Z, Ranganath S, Viboolsittiseri SS, et al. AID-initiated DNA lesions are differentially processed in distinct B cell populations. J Immunol. 2014;193:5545–5556. doi: 10.4049/jimmunol.1401549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yu H, Lin L, Zhang Z, Zhang H, Hu H. Targeting NF-κB pathway for the therapy of diseases: mechanism and clinical study. Signal Transduct Target Ther. 2020;5:209–223. doi: 10.1038/s41392-020-00312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stavnezer J, Kang J. The surprising discovery that TGF beta specifically induces the IgA class switch. J Immunol. 2009;182:5–7. doi: 10.4049/jimmunol.182.1.5. [DOI] [PubMed] [Google Scholar]
- 124.Castigli E, Wilson SA, Scott S, et al. TACI and BAFF-R mediate isotype switching in B cells. J Exp Med. 2005;201:35–39. doi: 10.1084/jem.20032000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cerutti A, Puga I, Cols M. Innate control of B cell responses. Trends Immunol. 2011;32:202–211. doi: 10.1016/j.it.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Park MH, Park SR, Lee MR, Kim YH, Kim PH. Retinoic acid induces expression of Ig germ line α transcript, an IgA isotype switching indicative, through retinoic acid receptor. Genes Genom. 2011;33:83–88. doi: 10.1007/s13258-010-0168-5. [DOI] [Google Scholar]
- 127.Cao AT, Yao S, Gong B, Nurieva RI, Elson CO, Cong Y. Interleukin (IL)-21 promotes intestinal IgA response to microbiota. Mucosal Immunol. 2015;8:1072–1082. doi: 10.1038/mi.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dullaers M, Li D, Xue R, et al. A T cell-dependent mechanism for the induction of human mucosal homing immunoglobulin A-secreting plasmablasts. Immunity. 2009;30:120–129. doi: 10.1016/j.immuni.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gribonika I, Eliasson DG, Chandode RK, et al. Class-switch recombination to IgA in the Peyer´s patches requires natural thymus-derived Tregs and appears to be antigen independent. Mucosal Immunol. 2019;12:1268–1279. doi: 10.1038/s41385-019-0202-0. [DOI] [PubMed] [Google Scholar]
- 130.Seo G-Y, Youn J, Kim P-H. IL-21 ensures TGF-beta 1-induced IgA isotype expression in mouse Peyer´s patches. J Leukoc Biol. 2009;85:744–750. doi: 10.1189/jlb.0708450. [DOI] [PubMed] [Google Scholar]
- 131.Seo G-Y, Jang Y-S, Kim H-A, et al. Retinoic acid, acting as a highly specific IgA isotype switch factor, cooperates with TGF-β1 to enhance the overall IgA response. J Leukoc Biol. 2013;94:325–335. doi: 10.1189/jlb.0313128. [DOI] [PubMed] [Google Scholar]
- 132.Allman D, Wilmore JR, Gaudette BT. The continuing story of T-cell independent antibodies. Immunol Rev. 2019;288:128–135. doi: 10.1111/imr.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mond JJ, Vos Q, Lees A, Snapper CM. T cell independent antigens. Curr Opin Immunol. 1995;7:349–354. doi: 10.1016/0952-7915(95)80109-x. [DOI] [PubMed] [Google Scholar]
- 134.Cooper MD. The early history of B cells. Nat Rev Immunol. 2015;15:191–197. doi: 10.1038/nri3801. [DOI] [PubMed] [Google Scholar]
- 135.Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. 2015;15:185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Allman D, Pillai S. Peripheral B cell subsets. Curr Opin Immunol. 2008;20:149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sell S. How vaccines work: immune effector mechanisms and designer vaccines. Expert Rev Vaccines. 2019;18:993–1015. doi: 10.1080/14760584.2019.1674144. [DOI] [PubMed] [Google Scholar]
- 138.Slifka MK, Amanna I. How advances in immunology provide insight into improving vaccine efficacy. Vaccine. 2014;32:2948–2957. doi: 10.1016/j.vaccine.2014.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barone F, Vossenkamper A, Boursier L, et al. IgA-producing plasma cells originate from germinal centers that are induced by B-cell receptor engagement in humans. Gastroenterology. 2011;140:947–956. doi: 10.1053/j.gastro.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bergqvist P, Stensson A, Lycke NY, Bemark M. T cell-independent IgA class switch recombination is restricted to the GALT and occurs prior to manifest germinal center formation. J Immunol. 2010;184:3545–3553. doi: 10.4049/jimmunol.0901895. [DOI] [PubMed] [Google Scholar]
- 141.Lindner C, Wahl B, Föhse L, et al. Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J Exp Med. 2012;209:365–377. doi: 10.1084/jem.20111980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Boursier L, Gordon JN, Thiagamoorthy S, Edgeworth JD, Spencer J. Human intestinal IgA response is generated in the organized gut-associated lymphoid tissue but not in the lamina propria. Gastroenterology. 2005;128:1879–1889. doi: 10.1053/j.gastro.2005.03.047. [DOI] [PubMed] [Google Scholar]
- 143.Barone F, Patel P, Sanderson JD, Spencer J. Gut-associated lymphoid tissue contains the molecular machinery to support T-cell-dependent and T-cell-independent class switch recombination. Mucosal Immunol. 2009;2:495–503. doi: 10.1038/mi.2009.106. [DOI] [PubMed] [Google Scholar]
- 144.Shikina T, Hiroi T, Iwatani K, et al. IgA class switch occurs in the organized nasopharynx- and gut-associated lymphoid tissue, but not in the diffuse lamina propria of airways and gut. J Immunol. 2004;172:6259–6264. doi: 10.4049/jimmunol.172.10.6259. [DOI] [PubMed] [Google Scholar]
- 145.Bergqvist P, Stensson A, Hazanov L, et al. Re-utilization of germinal centers in multiple Peyer´s patches results in highly synchronized, oligoclonal, and affinity-matured gut IgA responses. Mucosal Immunol. 2013;6:122–135. doi: 10.1038/mi.2012.56. [DOI] [PubMed] [Google Scholar]
- 146.Lycke N, Bemark M. Mucosal adjuvants and long-term memory development with special focus on CTA1-DD and other ADP-ribosylating toxins. Mucosal Immunol. 2010;3:556–566. doi: 10.1038/mi.2010.54. [DOI] [PubMed] [Google Scholar]
- 147.Heise N, Klein U. Somatic hypermutation and affinity maturation analysis using the 4-Hydroxy-3-Nitrophenyl-Acetyl (NP) System. Methods Mol Biol. 2017;1623:191–208. doi: 10.1007/978-1-4939-7095-7_16. [DOI] [PubMed] [Google Scholar]
- 148.Lycke NY, Bemark M. The role of Peyer´s patches in synchronizing gut IgA responses. Front Immunol. 2012;3:329. doi: 10.3389/fimmu.2012.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.He B, Xu W, Santini PA, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A2 class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26(6):812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 150.Reboldi A, Arnon TI, Rodda LB, Atakilit A, Sheppard D, Cyster JG. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer´s patches. Science. 2016;352:aaf4822. doi: 10.1126/science.aaf4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roco JA, Mesin L, Binder SC, et al. Class-switch recombination occurs infrequently in germinal centers. Immunity. 2019;51:337–350.:e7. doi: 10.1016/j.immuni.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Biram A, Strömberg A, Winter E, et al. BCR affinity differentially regulates colonization of the subepithelial dome and infiltration into germinal centers within Peyer’s patches. Nat Immunol. 2019;20:482–492. doi: 10.1038/s41590-019-0325-1. [DOI] [PubMed] [Google Scholar]
- 153.van Zelm MC, Bartol SJW, Driessen GJ, et al. Human CD19 and CD40L deficiencies impair antibody selection and differentially affect somatic hypermutation. J Allergy Clin Immunol. 2014;134:135–144. doi: 10.1016/j.jaci.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 154.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Marklund E, Leach S, Axelsson H, et al. Serum-IgG responses to SARS-CoV-2 after mild and severe COVID-19 infection and analysis of IgG non-responders. PLOS ONE. 2020;15(10):e0241104. doi: 10.1371/journal.pone.0241104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Biram A, Winter E, Denton AE, et al. B cell diversification is uncoupled from SAP-mediated selection forces in chronic germinal centers within Peyer’s patches. Cell Rep. 2020;30:1910–1922.:e5. doi: 10.1016/j.celrep.2020.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Batista FD, Harwood NE. The who, how and where of antigen presentation to B cells. Nat Rev Immunol. 2009;9:15–27. doi: 10.1038/nri2454. [DOI] [PubMed] [Google Scholar]
- 158.Bessa J, Zabel F, Link A, et al. Low-affinity B cells transport viral particles from the lung to the spleen to initiate antibody responses. Proc Natl Acad Sci USA. 2012;109:20566–20571. doi: 10.1073/pnas.1206970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kauffman RC, Bhuiyan TR, Nakajima R, et al. Single-cell analysis of the plasmablast response to Vibrio cholerae demonstrates expansion of cross-reactive memory B cells. MBio. 2016;7:e02021-e2116. doi: 10.1128/mBio.02021-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rollenske T, Szijarto V, Lukasiewicz J, et al. Cross-specificity of protective human antibodies against Klebsiella pneumoniae LPS O-antigen. Nat Immunol. 2018;19:617–624. doi: 10.1038/s41590-018-0106-2. [DOI] [PubMed] [Google Scholar]
- 161.Biram A, Shulman Z. T cell help to B cells: Cognate and atypical interactions in peripheral and intestinal lymphoid tissues. Immunol Rev. 2020;296:36–47. doi: 10.1111/imr.12890. [DOI] [PubMed] [Google Scholar]
- 162.de Vinuesa CG, Cook MC, Ball J, et al. Germinal centers without T cells. J Exp Med. 2000;191:485–494. doi: 10.1084/jem.191.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schwickert TA, Lindquist RL, Shakhar G, et al. In vivo imaging of germinal centres reveals a dynamic open structure. Nature. 2007;446:83–87. doi: 10.1038/nature05573. [DOI] [PubMed] [Google Scholar]
- 164.Schwickert TA, Alabyev B, Manser T, Nussenzweig MC. Germinal center reutilization by newly activated B cells. J Exp Med. 2009;206:2907–2914. doi: 10.1084/jem.20091225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Degn SE, van der Poel CE, Firl DJ, et al. Clonal evolution of autoreactive germinal centers. Cell. 2017;170:913–926.:e19. doi: 10.1016/j.cell.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Silver J, Zuo T, Chaudhary N, et al. Stochasticity enables BCR-independent germinal center initiation and antibody affinity maturation. J Exp Med. 2018;215:77–90. doi: 10.1084/jem.20171022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Biram A, Davidzohn N, Shulman Z. T cell interactions with B cells during germinal center formation, a three-step model. Immunol Rev. 2019;288:37–48. doi: 10.1111/imr.12737. [DOI] [PubMed] [Google Scholar]
- 168.Bunker JJ, Erickson SA, Flynn TM, et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science. 2017;358:eaan6619. doi: 10.1126/science.aan6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kabbert J, Benckert J, Rollenske T, et al. High microbiota reactivity of adult human intestinal IgA requires somatic mutations. J Exp Med. 2020;217:612. doi: 10.1084/jem.20200275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Benckert J, Schmolka N, Kreschel C, et al. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J Clin Invest. 2011;121:1946–1955. doi: 10.1172/JCI44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Choi J, Crotty S. Bcl6-Mediated transcriptional regulation of follicular helper T cells (TFH) Trends Immunol. 2021;42:336–349. doi: 10.1016/j.it.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang B, Liu E, Gertie JA, et al. Divergent T follicular helper cell requirement for IgA and IgE production to peanut during allergic sensitization. Sci Immunol. 2020;5:eaay2754. doi: 10.1126/sciimmunol.aay2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kurata I, Matsumoto I, Sumida T. T follicular helper cell subsets: a potential key player in autoimmunity. Immunol Med. 2021;44:1–9. doi: 10.1080/25785826.2020.1776079. [DOI] [PubMed] [Google Scholar]
- 174.Eisenbarth SC, Baumjohann D, Craft J, et al. CD4+ T cells that help B cells - a proposal for uniform nomenclature. Trends Immunol. 2021 July 6; doi: 10.1016/j.it.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Stebegg M, Kumar SD, Silva-Cayetano A, Fonseca VR, Linterman MA, Graca L. Regulation of the germinal center response. Front Immunol. 2018;9:117. doi: 10.3389/fimmu.2018.02469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tsuji M, Komatsu N, Kawamoto S, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer’s patches. Science. 2009;323:1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 178.Datta SK, Sabet M, Nguyen KPL, et al. Mucosal adjuvant activity of cholera toxin requires Th17 cells and protects against inhalation anthrax. Proc Natl Acad Sci USA. 2010;107:10638–10643. doi: 10.1073/pnas.1002348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Cao AT, Yao S, Gong B, Elson CO, Cong Y. Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J Immunol. 2012;189:4666–4673. doi: 10.4049/jimmunol.1200955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hirota K, Turner J-E, Villa M, et al. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cong Y, Bowdon HR, Elson CO. Identification of an immunodominant T cell epitope on cholera toxin. Eur J Immunol. 1996;26:2587–2594. doi: 10.1002/eji.1830261108. [DOI] [PubMed] [Google Scholar]
- 182.Nelson RW, Beisang D, Tubo, et al. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity. 2015;42:95–107. doi: 10.1016/j.immuni.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pabst O, Slack E. IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol. 2020;13:12–21. doi: 10.1038/s41385-019-0227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012;12:821–832. doi: 10.1038/nri3322. [DOI] [PubMed] [Google Scholar]
- 185.Landsverk OJB, Snir O, Casado RB, et al. Antibody-secreting plasma cells persist for decades in human intestine. J Exp Med. 2017;214:309–317. doi: 10.1084/jem.20161590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lycke N, Lebrero-Fernández C. ADP-ribosylating enterotoxins as vaccine adjuvants. Curr Opin Pharmacol. 2018;41:42–51. doi: 10.1016/j.coph.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 187.Ong CWM, Migliori GB, Raviglione M, et al. Epidemic and pandemic viral infections: impact on tuberculosis and the lung: a consensus by the World Association for Infectious Diseases and Immunological Disorders (WAidid), Global Tuberculosis Network (GTN), and members of the European Society of Clinical Microbiology and Infectious Diseases Study Group for Mycobacterial Infections (ESGMYC) Eur Resp J. 2020 October 1; doi: 10.1183/13993003.01727-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ito R, Ozaki YA, Yoshikawa T, et al. Roles of anti-hemagglutinin IgA and IgG antibodies in different sites of the respiratory tract of vaccinated mice in preventing lethal influenza pneumonia. Vaccine. 2003;21:2362–2371. doi: 10.1016/s0264-410x(03)00078-1. [DOI] [PubMed] [Google Scholar]
- 189.Renegar KB, Small PA, Boykins LG, Wright PF. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J Immunol. 2004;173:1978–1986. doi: 10.4049/jimmunol.173.3.1978. [DOI] [PubMed] [Google Scholar]
- 190.Soutar CA. Distribution of plasma cells and other cells containing immunoglobulin in the respiratory tract of normal man and class of immunoglobulin contained therein. Thorax. 1976;31:158–166. doi: 10.1136/thx.31.2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tourville DR, Adler RH, Bienenstock J, Tomasi TB. The human secretory immunoglobulin system: immunohistoligical localization of gamma A, secretory “piece”, and lactoferrin in normal human tissues. J Exp Med. 1969;129:411–429. doi: 10.1084/jem.129.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Onodera T, Takahashi Y, Yokoi Y, et al. Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc Natl Acad Sci USA. 2012;109:2485–2490. doi: 10.1073/pnas.1115369109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Thibeault C, Suttorp N, Opitz B. The microbiota in pneumonia: from protection to predisposition. Sci Transl Med. 2021;13:eaba0501. doi: 10.1126/scitranslmed.aba0501. [DOI] [PubMed] [Google Scholar]
- 194.Man WH, de Steenhuijsen Piters WAA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Micro. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.de Wilde PC, Kater L, Baak JP, van Houwelingen JC, Hené RJ, Slootweg PJ. A new and highly sensitive immunohistologic diagnostic criterion for Sjögren’s syndrome. Arthritis Rheum. 1989;32:1214–1220. doi: 10.1002/anr.1780321005. [DOI] [PubMed] [Google Scholar]
- 196.Quiding-Järbrink M, Granström G, Nordström I, Holmgren J, Czerkinsky C. Induction of compartmentalized B-cell responses in human tonsils. Infect Immun. 1995;63:853–857. doi: 10.1128/iai.63.3.853-857.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hirano T, Jiao X, Chen Z, Van Waes C, Gu X-X. Kinetics of mouse antibody and lymphocyte responses during intranasal vaccination with a lipooligosaccharide-based conjugate vaccine. Immunol Lett. 2006;107:131–139. doi: 10.1016/j.imlet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 198.GeurtsvanKessel CH, Willart MAM, Bergen IM, et al. Dendritic cells are crucial for maintenance of tertiary lymphoid structures in the lung of influenza virus-infected mice. J Exp Med. 2009;206:2339–2349. doi: 10.1084/jem.20090410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tan H-X, Esterbauer R, Vanderven HA, Juno JA, Kent SJ, Wheatley AK. Inducible bronchus-associated lymphoid tissues (iBALT) serve as sites of B cell selection and maturation following influenza infection in mice. Front Immunol. 2019;10:1999. doi: 10.3389/fimmu.2019.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Halle S, Dujardin HC, Bakocevic N, et al. Induced bronchus-associated lymphoid tissue serves as a general priming site for T cells and is maintained by dendritic cells. J Exp Med. 2009;206:2593–2601. doi: 10.1084/jem.20091472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Takamura S. Niches for the long-term maintenance of tissue-resident memory T cells. Front Immunol. 2018;9:1214. doi: 10.3389/fimmu.2018.01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Turner DL, Bickham KL, Thome JJ, et al. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 2014;7:501–510. doi: 10.1038/mi.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Graf BA, Lund FE, Randall TD. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat Immunol. 2019;20:97–108. doi: 10.1038/s41590-018-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Anderson KG, Sung H, Skon CN, et al. Cutting edge: intravascular staining redefines lung CD8 T cell responses. J Immunol. 2012;189:2702–2706. doi: 10.4049/jimmunol.1201682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Swarnalekha N, Schreiner D, Litzler LC, et al. T resident helper cells promote humoral responses in the lung. Sci Immunol. 2021;6:eabb6808. doi: 10.1126/sciimmunol.abb6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Son YM, Cheon IS, Wu Y, et al. Tissue-resident CD4+ T helper cells assist the development of protective respiratory B and CD8+ T cell memory responses. Sci Immunol. 2021;6:eabb6852. doi: 10.1126/sciimmunol.abb6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mathew NR, Jayanthan JK, Smirnov IV, et al. Single-cell BCR and transcriptome analysis after influenza infection reveals spatiotemporal dynamics of antigen-specific B cells. Cell Rep. 2021;35:109286. doi: 10.1016/j.celrep.2021.109286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wölfel R, Niemeyer D, Jones TC, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;382:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 209.Russell MW, Moldoveanu Z, Ogra PL, Mestecky J. Mucosal immunity in COVID-19: a neglected but critical aspect of SARS-CoV-2 infection. Front Immunol. 2020;11:611337. doi: 10.3389/fimmu.2020.611337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sterlin D, Mathian A, Miyara M, et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci Transl Med. 2020;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Iyer AS, Jones FK, Nodoushani A, et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol. 2020;5:eabe0367. doi: 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gaebler C, Wang Z, Lorenzi JCC, et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Anand SP, Prévost J, Nayrac M, et al. Longitudinal analysis of humoral immunity against SARS-CoV-2 Spike in convalescent individuals up to eight months post-symptom onset. CR Med. 2021;2:100290. doi: 10.1016/j.xcrm.2021.100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Baumgarth N, Nikolich-Zugich J, Lee FE-H, Bhattacharya D. Antibody responses to SARS-CoV-2: Let’s stick to known knowns. J Immunol. 2020;205:2342–2350. doi: 10.4049/jimmunol.2000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Fox A, Marino J, Amanat F, et al. Robust and specific secretory IgA against SARS-CoV-2 detected in human milk. iScience. 2020;23:101735. doi: 10.1016/j.isci.2020.101735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Cervia C, Nilsson J, Zurbuchen Y, et al. Systemic and mucosal antibody responses specific to SARS-CoV-2 during mild versus severe COVID-19. J Allergy Clin Immunol. 2021;147:545–557.:e9. doi: 10.1016/j.jaci.2020.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Isho B, Abe KT, Zuo M, et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Science Immunol. 2020;5:eabe5511. doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Pisanic N, Randad PR, Kruczynski K, et al. COVID-19 serology at population scale: SARS-CoV-2-specific antibody responses in saliva. J Clin Microbiol. 2020;59:e02204-e2220. doi: 10.1128/JCM.02204-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Butler SE, Crowley AR, Natarajan H, et al. Distinct features and functions of systemic and mucosal humoral immunity among SARS-CoV-2 convalescent individuals. Front Immunol. 2020;11:618685. doi: 10.3389/fimmu.2020.618685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Patil HP, Rane PS, Shrivastava S, et al. Antibody (IgA, IgG, and IgG Subtype) responses to SARS-CoV-2 in severe and nonsevere COVID-19 patients. Viral Immunol. 2021;34:201–209. doi: 10.1089/vim.2020.0321. [DOI] [PubMed] [Google Scholar]
- 221.Zeng W, Ma H, Ding C, et al. Characterization of SARS-CoV-2-specific antibodies in COVID-19 patients reveals highly potent neutralizing IgA. Signal Transduct Target Ther. 2021;6:35. doi: 10.1038/s41392-021-00478-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Klingler J, Weiss S, Itri V, et al. Role of Immunoglobulin M and A antibodies in the neutralization of severe acute respiratory syndrome coronavirus 2. J Infect Dis. 2021;223:957–970. doi: 10.1093/infdis/jiaa784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Gasser R, Cloutier M, Prévost J, et al. Major role of IgM in the neutralizing activity of convalescent plasma against SARS-CoV-2. Cell Rep. 2021;34:108790. doi: 10.1016/j.celrep.2021.108790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wang Z, Lorenzi JCC, Muecksch F, et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci Transl Med. 2021;13:eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Varnaitė R, García M, Glans H, et al. Expansion of SARS-CoV-2-specific antibody-secreting cells and generation of neutralizing antibodies in hospitalized COVID-19 patients. J Immunol. 2020;205:2437–2446. doi: 10.4049/jimmunol.2000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Blanco E, Perez-Andres M, Sanoja-Flores L, et al. Selection and validation of antibody clones against IgG and IgA subclasses in switched memory B-cells and plasma cells. J Immunol Methods. 2019;475:112372. doi: 10.1016/j.jim.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 227.Caraux A, Klein B, Paiva B, et al. Circulating human B and plasma cells. Age-associated changes in counts and detailed characterization of circulating normal CD138- and CD138+ plasma cells. Haematologica. 2010;95:1016–1020. doi: 10.3324/haematol.2009.018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Halliley JL, Tipton CM, Liesveld J, et al. Long-lived plasma cells are contained within the CD19-CD38hiCD138+ subset in human bone marrow. Immunity. 2015;43:132–145. doi: 10.1016/j.immuni.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Weisel NM, Weisel FJ, Farber DL, et al. Comprehensive analyses of B-cell compartments across the human body reveal novel subsets and a gut-resident memory phenotype. Blood. 2020;136:2774–2785. doi: 10.1182/blood.2019002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sokal A, Chappert P, Barba-Spaeth G, et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.:e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Yang F, Nielsen SCA, Hoh RA, et al. Shared B cell memory to coronaviruses and other pathogens varies in human age groups and tissues. Science. 2021;372:738–741. doi: 10.1126/science.abf6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yewdell JW, Santos JJS. Original antigenic sin: How original? How sinful? Cold Spring Harb Perspect Med. 2021;11:a038786. doi: 10.1101/cshperspect.a038786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Brown EL, Essigmann HT. Original antigenic sin: the downside of immunological memory and implications for COVID-19. mSphere. 2021;6:e00056-e121. doi: 10.1128/mSphere.00056-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sunyer JO. Fishing for mammalian paradigms in the teleost immune system. Nat Immunol. 2013;14:320–326. doi: 10.1038/ni.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li H, Limenitakis JP, Greiff V, et al. Mucosal or systemic microbiota exposures shape the B cell repertoire. Nature. 2020;584:274–278. doi: 10.1038/s41586-020-2564-6. [DOI] [PubMed] [Google Scholar]
- 236.Rollenske T, Macpherson AJ. Anti-commensal Ig—from enormous diversity to clear function. Mucosal Immunol. 2020;13:1–2. doi: 10.1038/s41385-019-0223-8. [DOI] [PubMed] [Google Scholar]
- 237.Azzali G. Structure, lymphatic vascularization and lymphocyte migration in mucosa-associated lymphoid tissue. Immunol Rev. 2003;195:178–189. doi: 10.1034/j.1600-065x.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 238.Ma B, Von Wasielewski R, Lindenmaier W, Dittmar KEJ. Immmunohistochemical study of the blood and lymphatic vasculature and the innervation of mouse gut and gut-associated lymphoid tissue. Anat Histol Embryol. 2007;36:62–74. doi: 10.1111/j.1439-0264.2006.00741.x. [DOI] [PubMed] [Google Scholar]
- 239.Yamaguchi K, Schoefl GI. Blood vessels of the Peyer’s patch in the mouse: I. Topographic studies. Anat Rec. 1983;206:391–401. doi: 10.1002/ar.1092060406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable—no new data generated.