Abstract
B-lymphocyte development and selection is central to adaptive immunity and self-tolerance. These processes require B cell receptor (BCR)-signaling and occur in bone marrow, an environment with variable hypoxia, but whether Hypoxia-inducible factor (HIF) is involved is unknown. We show that HIF-activity is high in human and murine bone marrow Pro-B and Pre-B cells and decreases at the immature B cell stage. This stage-specific HIF-suppression is required for normal B cell development, since genetic-activation of HIF-1α in murine B cells led to reduced repertoire-diversity, decreased BCR-editing and developmental-arrest of immature B cells, resulting in reduced peripheral B cell numbers. HIF-1α activation lowered surface BCR, CD19 and BAFF-R and increased expression of pro-apoptotic BIM. BIM-deletion rescued the developmental block. Administration of a HIF-activator in clinical use markedly reduced bone marrow and transitional B cells, which has therapeutic implications. Together, our work demonstrates that dynamic regulation of HIF-1α is essential for normal B cell development.
Introduction
Immune cells operate in complex environments where oxygenation, nutrients, cytokines and antigens vary. Hypoxia-inducible factors (HIF) play a central role in integrating this information, enabling transcriptional responses to reduced environmental oxygen or receptor engagement, modulating many aspects of innate and adaptive immunity.
B cells develop in the bone marrow (BM) where the functional integrity and affinity of the B cell receptor (BCR) is tested. First, variable (V) diversity (D) and joining (J) genes recombine producing an immunoglobulin (Ig) μ heavy-chain, which binds surrogate light-chains forming the pre-BCR. Pre-BCR signaling initiate’s recombination of κ or λ light-chains, which together with the heavy-chain, yield a cell-surface IgM antibody, the BCR, defining an immature B cell. Immature B cells are selected based on stringent quality control (central tolerance), resulting in a diverse BCR repertoire with limited self-antigen recognition1.
These central tolerance mechanisms involve BCR-editing, whereby secondary rearrangements of Ig genes reprogram BCR specificity. BCR-editing promotes BCR diversity and salvages autoreactive or inefficient BCRs2. Immature B cells with inappropriate BCR signaling (high, indicating self-reactivity, or low, indicating functionally-impaired BCRs) undergo BCR-editing or clonal deletion, whilst sufficient signaling leads to positive selection3, 4. Following this checkpoint, surviving immature B cells upregulate the B cell-activating factor receptor (BAFF-R) and receive survival signals from BAFF, which sustains them as they enter the circulation as transitional B cells5, migrating to the spleen to complete maturation6.
In splenic mature and germinal-center B cells HIF regulates the generation and quality of humoral immune responses, being activated both by hypoxia and BCR signaling in these settings7, 8, 9. However, the role of HIF in early B cell development is unexplored. Since developing B cells may be exposed to hypoxia10, 11 and BCR signaling is central to assess antigen-receptor functionality, we hypothesized HIF would play a role in B cell development.
The HIF pathway consists of two alternative HIF-α transcription factors (HIF-1α/HIF-2α), three alternative prolyl hydroxylase domain enzymes (PHD1-3) acting as oxygen sensors, and a single E3 ubiquitin ligase (von Hippel Lindau, VHL) that mediates destruction of HIF-α’s following hydroxylation by PHDs in the presence of oxygen12, 13. We found variable HIF-activity at different developmental stages in BM B cells from humans and mice, suggesting that HIF-regulation may be important for B cell development. To investigate this, we generated a mouse with B cell-specific deficiency of VHL, a single non-redundant regulator of HIF-activation. Constitutive HIF-activation led to immature B cell developmental arrest and peripheral B cell loss. Dysregulated HIF-1α-activity was associated with limited B cell repertoire diversity and reduced BCR-editing and survival.
Mechanistically this involved reduced BCR expression and PI3K-signaling, failure to upregulate the BAFF-R and to suppress pro-apoptotic BIM. Deletion of one Hif1a allele resulted in graded effects implying a tunable response that may lead to different selection outcomes in a developing B cell. These data demonstrate that the dynamic regulation of HIF-1α is essential for normal B cell development and that HIF-1α contributes to the stringency of immature B cell selection by modulating the magnitude of key survival signals.
Results
Regulated HIF-lα-activity is required for human and murine B cell development
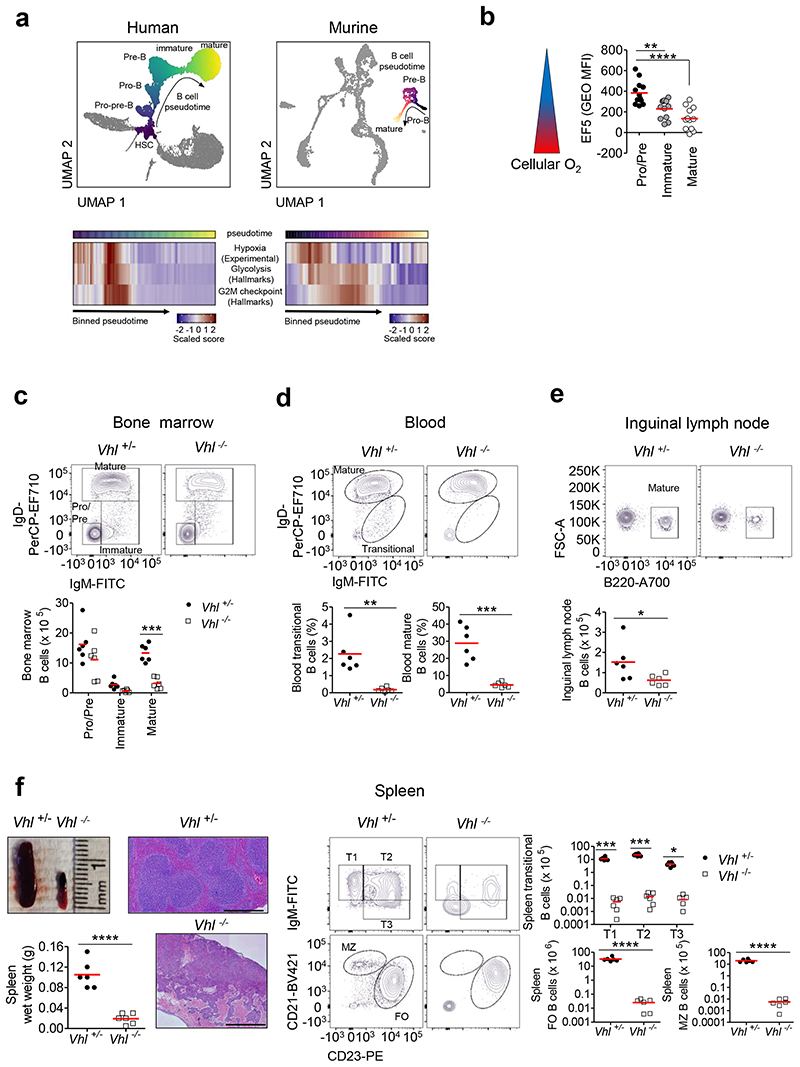
To investigate if HIF may be important in B cell development, we analyzed single cell RNA-sequencing data from human and murine BM cells, identified developing B cells based on canonical markers (Supplementary figures 1a-f,i-l) and determined hypoxia-dependent gene expression (Supplementary figures 1g-h) in B cells over developmental pseudotime. This demonstrated high expression of hypoxia-dependent genes in human Pre-pro-B, Pro-B and Pre-B cells, which decreased in immature B cells and remained low, showing that HIF-activity varies at different development stages within the BM. The expression of hypoxia-dependent genes during B cell development was temporally associated with increased expression of glycolysis and cell cycle genes, consistent with the conclusion that these processes may be HIF-regulated. A similar pattern of hypoxia-dependent gene expression was observed in murine BM B cells (Figure 1a). Furthermore, when we compared cellular O2 levels between different murine BM B cells using the hypoxia-probe EF5, we observed higher binding in Pro-B and/Pre-B cells, indicating that this subset has lower cellular O2. EF5-binding decreased in later developmental subsets (immature/mature) to levels similar to oxygenated blood B cells (Figure 1b, Supplementary figure 2a). Thus, dynamic regulation of HIF-activity coincides with cellular O2 levels in B cells.
Figure 1. Disruption of dynamic HIF regulation leads to severe peripheral B cell lymphopenia.
(a) Top: PAGA-initialized UMAP plot of 15101 cells (human cell atlas BM dataset) and 4075 cells (Tabula Muris BM dataset), illustrating cellular-differentiation trajectories in human and murine hematopoiesis. B cell-differentiation is colored according to pseudotime (dark-light). Other differentiation trajectories are de-colored. Bottom: Heatmaps of scaled geneset scores against pseudotime for human and murine BM B cells (averaged over 100 or 50 equal intervals respectively). Genesets used; Hypoxia (experimentally-derived), Glycolysis, G2M checkpoint (MSigDB Hallmarks). (b) Quantified EF5 levels (Geometric mean fluorescence-intensity (Geo MFI)) detected by anti-EF5 staining, in BM B cells Pro-B,Pre-B (B220+IgM-IgD-), immature (B220+IgM+IgD-) and mature (B220+IgM+IgD+). n=12 biologically independent C57BL/6J mice; data, pooled from four independent experiments. **P<0.01,****P<0.0001 one-way ANOVA Tukey post-test.
Flow cytometry showing the gating, frequency of live or absolute numbers of B cells from Vhl+/-Mb1-cre and Vhl-/-Mb1-cre mice in (c) BM, gated as in (b), (d) blood transitional (B220+IgM+IgD-) and mature (B220+IgM+IgD+), (e) Inguinal lymph node (ILN) mature (B220+) and (f) spleen transitional (T1-T3:B220+CD93+CD23 IgM), follicular (FO:B220+CD93-CD23+CD21+) and marginal zone (MZ:B220+CD93- CD23-CD21+). A single sample with no detectable T3 cells from a Vhl-/-Mb1-cre mouse is not depicted owing to the Log10-scale. Displayed, spleen wet weight and representative images of whole spleens and structure by hematoxylin and eosin staining. Scale-bar 500μm. (c-f) n=6 biologically independent mice per genotype. (c,f) *P<0.05,***P<0.001 two-way ANOVA Bonferroni post-test. (d,e,f) *P<0.05,**P<0.01,***P<0.001,****P<0.0001 unpaired two-sided t test. (b-f) Data, representative of nine independent experiments. Symbols represent individual mice, bars means ± S.D.
Collectively, these data suggested that variation in HIF-activity may be important for normal B cell development to proceed. To test this, we generated a mouse with B cell specific VHL-deficiency. Mice carrying loxP-flanked Vhl alleles14 were crossed with Cd79a (also known as Mb-1) cre mice15 (hereafter termed Vhl-/-Mb1-cre mice) to delete Vhl, activating HIF in the B cell lineage. The deletion efficiency for flow-sorted Vhl-/-Mb1-cre Pro-B and Pre-B cells was 84% (68-100%, 95%CI), for immature B cells 98% (97-98%, 95%CI) and for mature B cells 98% (97-99%, 95%CI). Mice heterozygous for wild-type (WT) and loxP-flanked Vhl (Vhl+/-Mb1-cre mice) were phenotypically identical to Vhl+/+Mb1-cre mice and used as controls.
Analysis of B cell development in control and Vhl-/-Mb1-cre mice showed no significant differences in the number of BM Pro-B and Pre-B cells (including each of the Hardy Fractions (Fr) A-D16; Supplementary figure 2b) and a minor reduction in Vhl-/-Mb1-cre immature B cells (Figure 1c). In contrast, we observed significant loss of recirculating BM mature B cells and severe peripheral B cell lymphopenia, including blood transitional and mature B cells, spleen transitional, follicular (FO) and marginal zone (MZ) B cells, inguinal lymph node (ILN) mature B cells and peritoneal B1 cells (Figures 1c-f, Supplementary figure 2c). Mice displayed additional clinical features of B cell lymphopenia17, 18 including low circulating IgM, elevated circulating levels of the B cell survival cytokine BAFF, and reduced BAFF-R expression in BM mature and peripheral B cells (Supplementary figures 2d-g). Spleen size was significantly reduced, with markedly disrupted architecture; Vhl-/-Mb1-cre spleens contained small collections of lymphocytes, fibrosis, a thickened capsule, cystic structures and multifocal-areas of subcapsular hemorrhage (Figure 1f) and few T cells. T cell development and numbers in other lymphoid compartments were unaffected (Supplementary figures 2h-k).
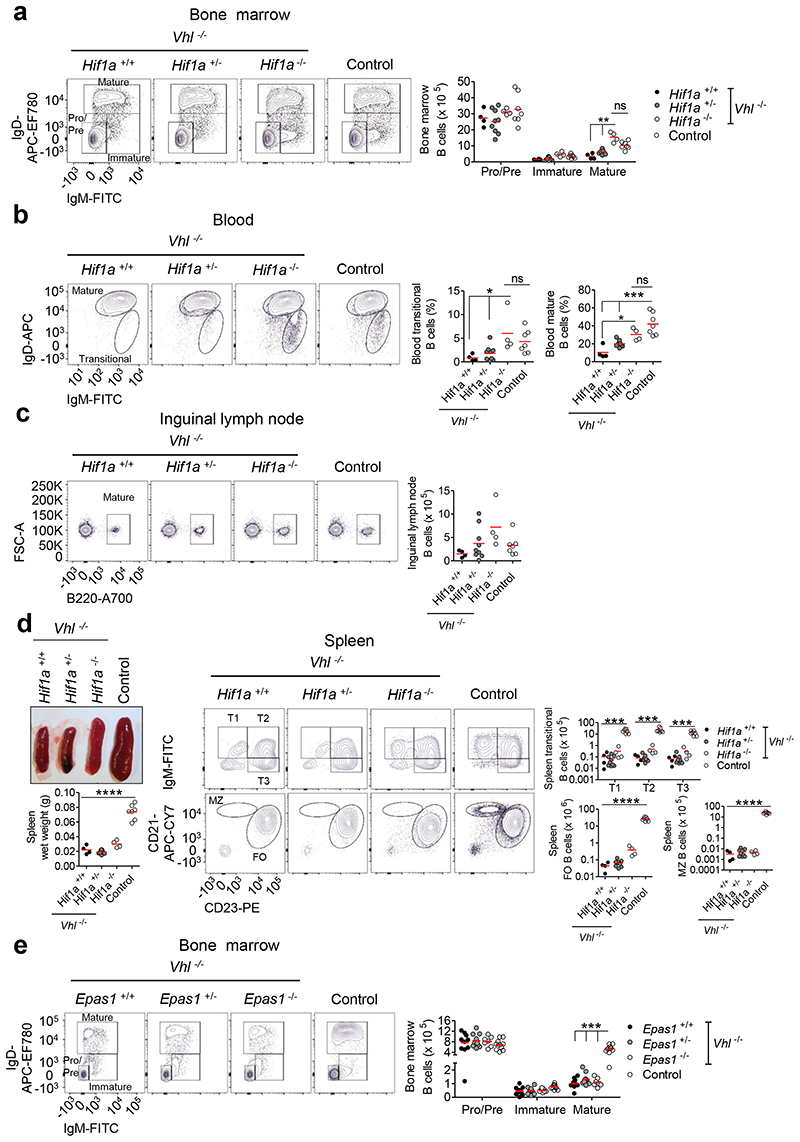
Effects of VHL loss-of-function could be via activation of HIF-1α, HIF-2α, or be HIF-independent. We assessed HIF-α expression in B cells lacking VHL and found high HIF-1α protein in all B cells, whereas HIF-2α was only detected in peripheral mature B cells (Supplementary figure 3a). To determine if HIF-1α mediated the effects of VHL loss, we crossed mice carrying loxP-flanked Hif1a alleles19 with Vhl-/-Mb1-cre mice (Supplementary figures 3b-c). Hif1a-deletion rescued BM mature B cells, blood transitional and mature B cells, ILN mature B cells and peritoneal B1 cells (Figures 2a-c, Supplementary figure 3d). Deletion of one Hif1a allele resulted in an intermediate phenotype indicating the effects of HIF-activity are tunable. However, spleen size and B cell numbers were not normalized (Figure 2d). To investigate the role of HIF-2α (encoded by Epas1) we crossed Vhl-/-Mb1-cre mice with mice carrying loxP-flanked Epas1 alleles20. HIF-2α-deletion did not rescue BM mature B cells (Figure 2e) indicating a HIF-1α-specific effect. These data demonstrate that uncontrolled HIF-1α-activity leads to marked defects in B cell development.
Figure 2. Hif1a-deletion in Vhl-/-Mb1-cre mice rescues peripheral B cell loss, indicating a HIF-1α-dependent effect.
Flow cytometry showing the gating, frequency of live or absolute numbers of B cells from Vhl-/- Mb1-cre mice carrying Hif1a+/+, Hif1a+/- and Hif1a-/- alleles, and from Vhl+/- Hif1a+/+Mb1-cre control mice in (a) BM, (b) blood, (c) ILN and (d) spleen. Displayed, spleen wet weight and a representative image of whole spleens from Vhl-/-Mb1-cre mice carrying Hif1a+/+, Hif1a+/- and Hif1a-/- alleles, and a Vhl+/- Hif1a+/+Mb1-cre control spleen. (e) Flow cytometry showing the gating and absolute numbers of B cells from Vhl-/-Mb1-cre mice carrying Epas1+/+, Epas1+/- and Epas1-/- alleles, and from Vhl+/-Epas1+/+Mb1-cre control mice in BM. (a-e) Gated as in Figure 1. (a,d,e) **P<0.01,***P<0.001 two-way ANOVA Bonferroni post-test. (b-d) *P<0.05,***P<0.001,****P<0.0001 one-way ANOVA Tukey post-test. (a-d) n=4 Vhl-/-Hif1a+/+Mb1-cre, 9 Vhl-/-Hif1a+/-Mb1-cre, 4 Vhl-/-Hif1a-/-Mb1-cre, 7 Vhl+/-Hif1a+/+Mb1-cre biologically independent mice. Data are pooled from two independent experiments. (e) n=10 Vhl-/- Epas1+/+Mb1-cre, 11 Vhl-/-Epas1+/-Mb1-cre, 7 Vhl-/-Epas1-/-Mb1-cre, 10 Vhl+/-Epas1+/+Mb1-cre biologically independent mice. Data are pooled from three independent experiments. (a-e) Symbols represent individual mice, bars means ± S.D.
We determined if the peripheral B cell lymphopenia observed in Vhl-/-Mb1-cre mice was B cell-intrinsic or due to aberrations in the microenvironment of secondary lymphoid organs. BM chimeras were generated by sub-lethal irradiation of muMt- mice, which lack B cells, followed by reconstitution with BM from Vhl+/-Mb1-cre or Vhl-/-Mb1-cre mice. Eight weeks after reconstitution, Vhl-/-Mb1-cre BM chimeras displayed severe peripheral B cell lymphopenia, similar to Vhl-/-Mb1-cre mice. With the caveat that muMt- spleens have reduced T cells21, a normalized spleen did not rescue peripheral B cell loss (Supplementary figure 4). Collectively, these data indicate a cell-intrinsic role of HIF-1α in B cell development.
Regulated HIF-1α-activity is necessary for the generation and diversification of the B cell repertoire
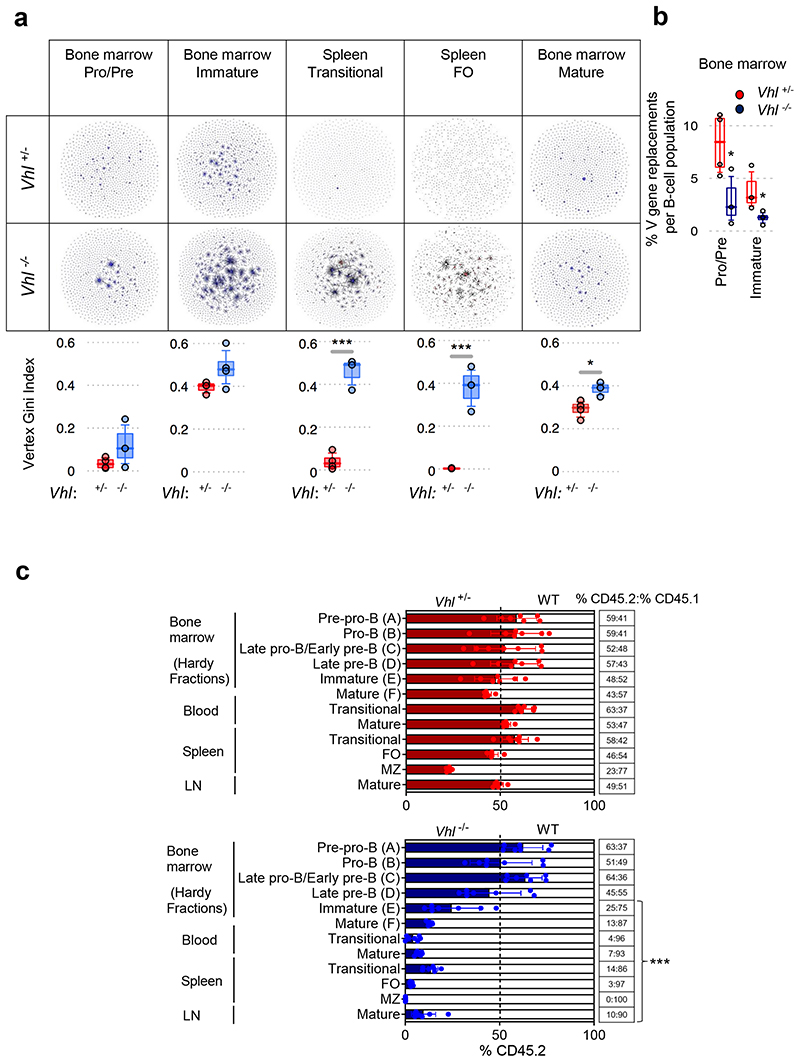
To determine the effect of HIF-1α on the formation of the pre-immune B cell repertoire, next-generation sequencing was employed to identify the rearranged IgHV-D-J sequences, and the relative clone sizes, providing a measure of BCR diversity22. Vhl-/-Mb1-cre mice demonstrated less IgHV-D-J diversity in Pro-B, Pre-B and immature B cells compared with Vhl+/-Mb1-cre controls. This restricted BCR repertoire persisted throughout subsequent development (in spleen transitional, FO and BM mature B cells; Figure 3a).
Figure 3. Regulated HIF-1α activity is required for the generation and diversification of the pre-immune B cell repertoire.
(a) Clonal analysis of IgHV-D-J DNA rearrangements from BM B cells and IgHV-D-J RNA rearrangements from spleen B cells. Table contains sequencing-networks demonstrating that in Vhl-/-Mb1-cre B cells, IgHV diversity is reduced (number of dots) and the maximum clone size (relative size of dots) is increased. Below, quantification using the Vertex Gini Index. (b) High-throughput detection of IgHV secondary rearrangements in Vhl+/-Mb1-cre and Vhl-/-Mb1-cre B cells from BM (DNA). Plotted, frequency of unique BCR-sequences with the same stem region. (a-b) Gated on B cells Pro-B, Pre-B B220+IgM-, immature and transitional B220+IgM+IgD-, BM mature B220+IgM+IgD+ and FO B220+IgM+IgD+CD23+CD21+,*P<0.05,***P<0.005 one-sided Wilcoxon-tests. Graphs; box lines show the 25th, 50th and 75th percentiles; whiskers show the 10th and 90th percentiles. n=4 biologically independent mice per genotype from one experiment. (c) The relative proportion of B cell subsets in lethally-irradiated CD45.1 mice reconstituted for 8 weeks with 1:1 mixtures of CD45.1 WT and CD45.2 Vhl+/-Mb1-cre or Vhl-/-Mb1-cre BM. Gated on Pre-pro-B (FrA:B220+CD43+CD24-/lowBP-1-), Pro-B (FrB:B220+CD43+CD24+BP-1-), Late pro-B/Early pre-B (FrC:B220+CD43+/lowCD24+BP-1+), Late pre-B (FrD:B220+CD43-IgM-IgD-), immature (FrE:B220+CD43- IgM+IgD-), BM mature (FrF B220+CD43-IgM+IgD+), blood transitional (B220+IgM+IgD-), blood and ILN mature (B220+IgM+IgD+), spleen transitional (B220+CD93+), FO and MZ (gated as in Figure 1). Filled columns; mean %CD45.2, clear columns; mean %CD45.1, error bars S.D.; comparison between Vhl+/-Mb1-cre and Vhl-/-Mb1-cre B cells by two-way ANOVA Bonferroni post-test ***P<0.001, n=7 biologically independent mice per genotype; data, representative of two independent experiments. (a-c) Symbols represent individual mice.
During early development, repertoire diversity increases by IgHV-editing in Pre-B and immature B cells2. IgHV genes are edited by a form of secondary VDJ recombination known as IgHV gene replacement, so next-generation sequencing23 was used to determine the normalized percentage of IgHV gene replacements in Vhl-/-Mb1-cre and Vhl+/-Mb1-cre B cells. This revealed a significantly lower percentage of IgHV replacements per unique BCR in Vhl-/-Mb1-cre Pro-B, Pre-B and immature B cells (Figure 3b), which persisted in transitional B cells (Supplementary figure 5a). This narrowing of BCR diversity at early developmental stages (Pro-B, Pre-B, immature), and prior to B cell loss, was confirmed by testing the degree of clonal-overlap in early compared with late developmental subsets23. The probability of observing equal or greater BCR sequence overlap between early and late developmental B cells was significantly lower in Vhl-/-Mb1-cre mice (after accounting for differences in read-depth through subsampling, Supplementary figure 5b). V gene-usage differed distinctly between Vhl-/-Mb1-cre and control B cells (Supplementary figure 5c) further highlighting that activation of HIF-1α drives biased and restricted selection outcomes. Furthermore, using isotype-resolved BCR sequencing, we observed distinct differences in isotype-usage between Vhl-/-Mb1-cre and control plasma cells including increased IGHA-usage and reduced usage of IGHM, IGHG3 and IGH2B, whilst IGHE-usage was similar (Supplementary figure 5d). Thus, regulated HIF-1α-activity is required for diversification of the B cell repertoire, a key aspect of immunity.
Dysregulated HIF-1α leads to a cell-intrinsic failure in development at the immature B cell stage
To confirm the precise stage at which B cell development was disrupted by VHL loss, we generated mixed BM chimeras. Equal numbers of CD45.1 WT and CD45.2 control (Vhl+/-Mb1-cre) or Vhl-/-Mb1-cre BM cells were injected intravenously into lethally-irradiated CD45.1 WT recipient mice. Eight weeks after BM transfer, competitive reconstitution of developing B cells (Hardy FrA-F16) and peripheral B cells were analyzed. Our analysis confirmed a developmental block at the immature (FrE) B cell stage in Vhl-/-Mb1-cre mixed chimeras. This was accompanied by marked reductions in blood and spleen transitional B cells and blood, spleen, ILN and BM mature (FrF) B cells. Interestingly Vhl-/-Mb1-cre MZ B cells were absent from the spleen, with partial failure to reconstitute in Vhl+/-Mb1-cre heterozygote chimeras (Figure 3c, Supplementary figure 5e). In contrast to the parental and muMt- Vhl-/-Mb1-cre mice, splenic T cells were preserved in Vhl-/-Mb1-cre mixed chimeras, with similar numbers to Vhl+/-Mb1-cre or WT donors (Supplementary figure 5f), demonstrating that B cell loss is not due to low splenic T cell numbers. These findings confirmed that persistent HIF-1α-activation leads to a cell-intrinsic developmental block at the immature B cell stage.
Development defects linked to BCR signaling and survival
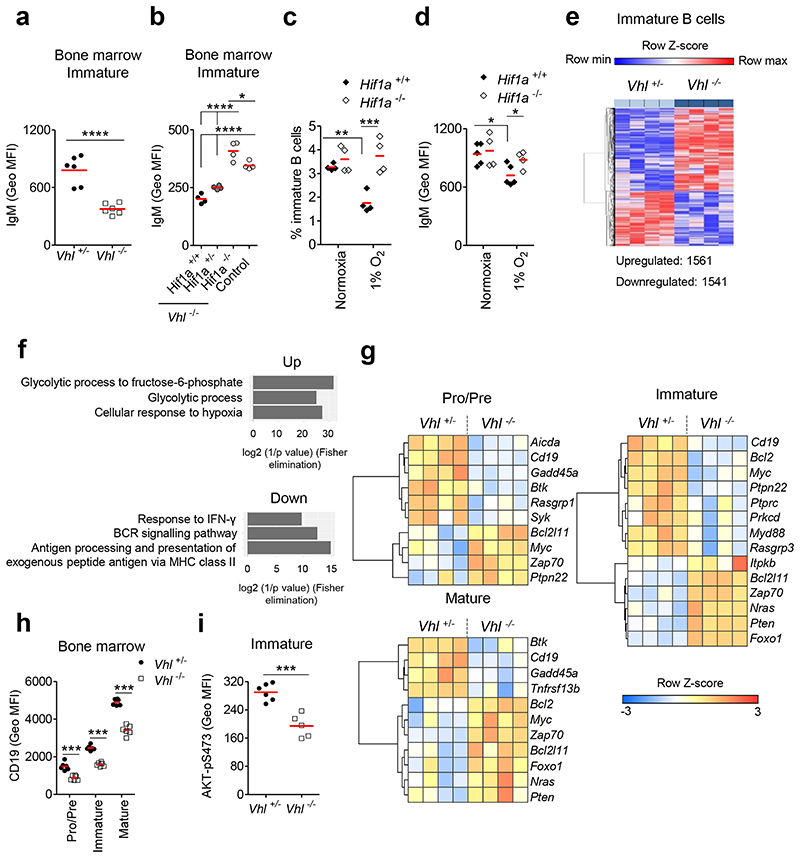
We observed reduced surface (s)IgM expression on Vhl-/-Mb1-cre immature B cells (Figure 4a), which was cell-intrinsic (Supplementary figure 6a) and normalized following Hif1a-deletion (Figure 4b). Sufficient sIgM expression is required for BCR-signaling and maturation3, suggesting one mechanism by which HIF-1α could affect B cell survival and selection. To directly test the effects of physiological HIF-1α-activation on this, we cultured Hif1a+/+Mb1-cre and Hif1a-/-Mb1-cre immature B cells under hypoxia (1% O2) for 3 days. The survival of Hif1a+/+Mb1-cre immature B cells was reduced under hypoxia and was unaffected in Hif1a-/-Mb1-cre immature B cells, demonstrating a HIF-lα-dependent effect (Figure 4c). Furthermore, hypoxic Hif1a+/+Mb1-cre immature B cells had significantly lower sIgM, which following Hif1a-deletion was restored to levels in oxygen-sufficient cells (Figures 4d). Collectively, these data demonstrate a role for HIF-1α and environmental oxygen in regulating BCR expression and the survival threshold of immature B cells.
Figure 4. Defects in development linked to BCR signaling.
Surface (s)IgM expression (Geo MFI) on immature B cells from (a) Vhl+/-Mb1-cre and Vhl-/-Mb1-cre mice, (b) Vhl-/-Mb1-cre mice carrying Hif1a+/+, Hif1a+/- and Hif1a-/- alleles, and control (Vhl+/-Hif1a+/+Mb1-cre) mice. (c) Frequency of live and (d) sIgM expression (Geo MFI) on Hif1a+/+Mb1-cre and Hif1a-/-Mb1-cre immature B cells cultured under normoxia or 1% O2 for 3d. (e) Heatmap of significantly differentially expressed genes in Vhl-/-Mb1-cre versus Vhl+/-Mb1-cre immature B cells (BH adjusted P<0.05). Columns represent individual mice. (f) Gene ontology (GO) analysis of significantly altered genes common to all Vhl-/-Mb1-cre BM B cells (Pro-B,Pre-B,immature,mature) when compared to Vhl+/-Mb1-cre controls. Top three GO terms (relevant to hypoxia and B cell biology) ranked by significance are plotted. (g) Heatmaps of row-scaled gene expression of selected differentially expressed transcripts in sorted BM B cells. Columns represent individual mice. Expression of (h) CD19 and (i) AKT-pS473 (Geo MFI) in BM B cells from Vhl+/-Mb1-cre and Vhl-/-Mb1-cre mice. (a-d,h-i) Gated as in Figure 1b. (e-g) Gated as in Figure 3a. (a,d,i) *P<0.05,***P<0.001,****P<0.0001 unpaired two-sided t test. (b) *P<0.05,****P<0.0001 one-way ANOVA Tukey post-test. (c,h) **P<0.01,***P<0.001 two-way ANOVA Bonferroni post-test. (a,h) n=6 per genotype, (b-c,e-g) n=4 per genotype, (d) n=5 Hif1a+/+Mb1-cre, 4 Hif1a-/-Mb1-cre and (i) n=6 Vhl+/-Mb1-cre, 5 Vhl-/-Mb1-cre, biologically independent mice. Data, (a) representative of nine independent experiments, (b-d) representative of at least two independent experiments, (e-g) from one experiment, (h) representative of six independent experiments, (i) representative of three independent experiments. (a-d,h-i) Symbols represent individual mice, bars means ± S.D.
To comprehensively investigate how HIF-1α shapes BCR signaling and survival, we performed global transcriptomic RNA-sequencing analysis on flow-sorted BM Pro-B, Pre-B, immature and mature B cells from Vhl-/-Mb1-cre and Vhl+/-Mb1-cre mice. We observed substantial differential gene expression between Vhl-/-Mb1-cre and Vhl+/-Mb1-cre B cells, particularly in immature B cells, with 3102 genes significantly differentially expressed relative to controls, of which 1561 genes were upregulated, and 1541 genes downregulated (Figure 4e, Supplementary figure 6b). Gene ontology terms represented in genes that were significantly differentially expressed in Vhl-/-Mb1-cre mice across all BM B cell subsets, included ‘cellular response to hypoxia’ indicative of HIF-activation. Consistently, genes involved in hypoxia-associated processes, including glycolysis, were increased. In line with our repertoire data, genes involved in pathways important for B cell selection and function were reduced, including BCR signaling, antigen processing and presentation of exogenous peptide antigen via MHC class II and response to IFN-γ (Figure 4f).
We focused on differences in the immature B cell transcriptome between Vhl-/-Mb1-cre and control mice, since this was where we observed the developmental block. Gene set enrichment analysis (GSEA) demonstrated significant negative enrichment for genes involved in pathways important for B cell development, including those relating to central tolerance; BCR-editing (DNA repair) and BCR signaling and survival (E2F targets, MYC targets). In contrast, apoptosis was positively enriched (Supplementary figure 6c).
Hypoxia and HIF have effects on DNA recombination and repair in other settings24. Taking this, together with the transcriptomic analysis, we hypothesized that HIF-1α might impair DNA repair and recombination of the Ig locus. To test this, we first examined the ability to repair X-ray induced DNA double-stranded breaks in control and Vhl-/-Mb1-cre B cells; this was equivalent in all BM B cells (Supplementary figure 6d). Next, we crossed Vhl-/-Mb1-cre mice with MD4 mice expressing a transgenic IgMaIgDa BCR specific for hen egg lysozyme25 (HEL; named IgHEL). IgHEL mice have a homogeneous B cell repertoire that does not undergo RAG1/2 DNA recombination and BCR-editing thus removing the requirement for DNA recombination and repair during these fundamental selection processes. The IgHEL transgene did not rescue B cell loss (Supplementary figure 6e). Importantly, we observed reduced sIgMa on IgHEL Vhl-/-Mb1-cre immature B cells (Supplementary figure 6f) indicating that the reduced BCR expression was not due to antigen-induced anergy, since these mice do not express HEL.
We next examined the expression of genes important in BCR signaling and selection1. Across all BM B cell subsets, we observed marked differences in gene expression between Vhl-/-Mb1-cre and Vhl+/-Mb1-cre B cells, which were most substantial in immature B cells. Vhl-/-Mb1-cre immature B cells expressed high levels of genes associated with Pre-B cells such as Foxo1 and Bcl2l11 (encoding pro-apoptotic BIM), and had altered expression of genes characteristic of immature B cells such as Cd19 and Myc, which were significantly lower, and Pten and Foxo1, which was significantly higher. This pattern of gene expression is consistent with a failure of developmental progression in immature B cells, when the newly formed immature BCR becomes strongly coupled to AKT via CD19 and PI3K, leading to suppression of nuclear FOXO1, downregulation of BIM and increased MYC that together promote survival and positive selection1, 26, 27, 28. Across all BM B cell subsets two genes were consistently differentially expressed in Vhl-/-Mb1-cre cells; Cd19 (reduced), and Bcl2l11 (increased) relative to controls (Figure 4g). CD19 is a component of the B cell co-receptor and deficiency causes a similar phenotype to that observed in Vhl-/-Mb1-cre mice and mixed chimeras29. We therefore assessed surface expression of CD19. Consistent with the transcriptional analysis, there was reduced surface expression of CD19 on Vhl-/-Mb1-cre BM B cells compared with controls (Figure 4h), which was HIF-1α-dependent and cell-intrinsic (Supplementary figures 7a-b). Collectively, reduced sIgM, repressed CD19 and Myc, and increased Pten and Foxo1 would be consistent with reduced BCR signaling through the PI3K pathway. This was confirmed by flow-cytometric analysis of AKT-pS473 expression, which was significantly lower in Vhl-/-Mb1-cre immature B cells versus Vhl+/-Mb1-cre controls (Figure 4i). These data suggest that HIF-1α reduces BCR signaling, thereby reducing survival and positive selection.
Developmental block linked to a BIM-dependent survival defect
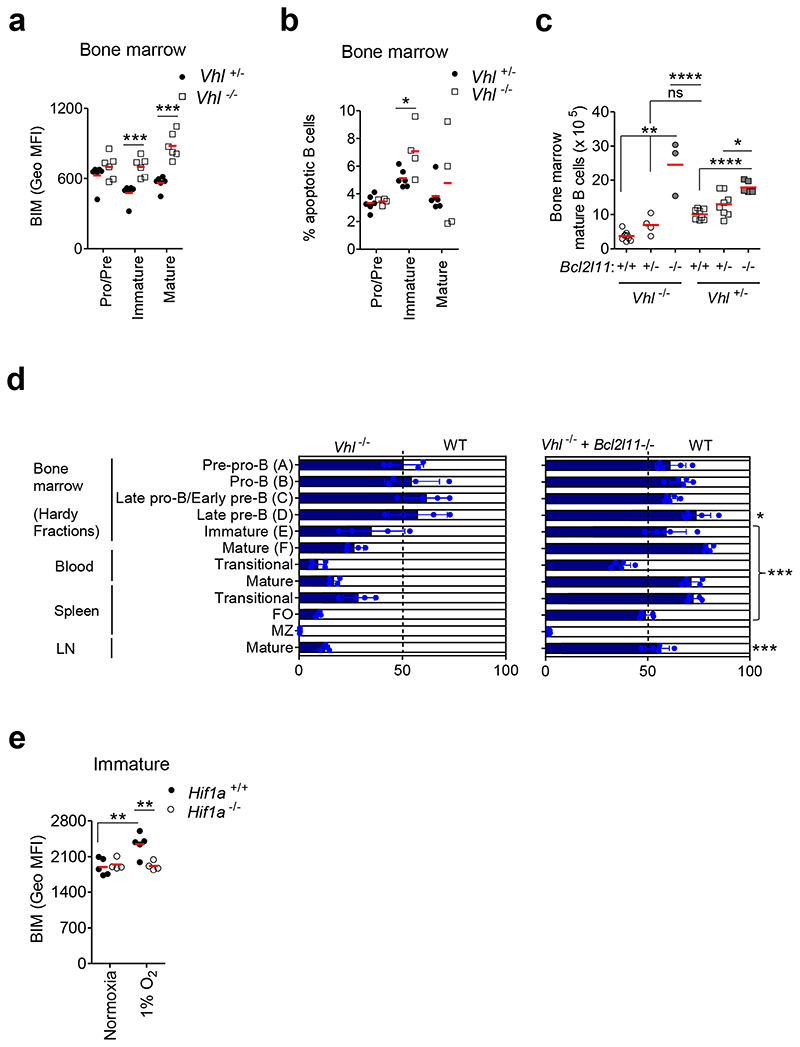
The pro-apoptotic factor BIM is highly expressed in editing immature B cells and is the principle driver of apoptosis at this developmental stage. Consistent with our transcriptomic data, we observed a failure to down-regulate BIM protein and increased apoptosis in Vhl-/-Mb1-cre BM B cells compared with Vhl+/-Mb1-cre controls (Figures 5a-b, Supplementary figure 7c). Deleting one Hif1a allele significantly reduced BIM expression, indicating a HIF-1α-dependent effect (Supplementary figure 7d). To determine if deleting BIM could alleviate BM mature B cell loss, Vhl+/-Mb1-cre and Vhl-/-Mb1-cre mice were crossed with Bcl2l11 knockout mice (Bcl2l11-/-)30. Deleting one or both Bcl2l11 alleles in Vhl-/-Mb1-cre mice increased BM mature B cells (Figure 5c). We then determined if BIM-deletion could reverse the developmental block in immature B cells. Equal numbers of CD45.1 WT and Vhl-/-Mb1-cre, Vhl-/-Mb1-cre + Bcl2l11-/- or control Vhl+/-Mb1-cre BM cells were injected intravenously into lethally-irradiated CD45.1 WT recipient mice. Eight weeks after BM transfer, competitive reconstitution of BM and peripheral B cells were analyzed. With the exception of MZ B cells, BIM-deletion reversed the developmental block observed in Vhl-/-Mb1-cre immature B cells and beyond (Figure 5d, Supplementary Figure 7e). To test the effect of environmental oxygen on BIM, Hif1a+/+Mb1-cre immature B cells were cultured in hypoxia (1% O2) for 24h. Hypoxic immature B cells had significantly higher BIM expression compared to their normoxic counterparts, linking environmental oxygen to BIM-dependent survival. Hypoxia-induced BIM expression was prevented by Hif1a-deletion, confirming a HIF-lα-dependent effect (Figure 5e). Collectively these data indicate that the hypoxic response limits the life-span of B cells, BIM-dependently.
Figure 5. Developmental block linked to BIM-dependent survival.
(a) BIM expression (Geo MFI) in BM B cells from Vhl+/-Mb1-cre and Vhl-/-Mb1-cre mice. (b) Percentage apoptotic BM B cells from Vhl+/-Mb1-cre and Vhl-/-Mb1-cre mice, measured by PO-PRO-1 and 7-AAD staining. (c) Enumeration of BM mature B cells, by flow cytometry, from Vhl+/-Mb1-cre and Vhl-/-Mb1-cre mice carrying Bcl2l11+/+, Bcl2l11+/- and Bcl2l11–/– alleles. (d) The relative proportion of B cell subsets in lethally irradiated CD45.1 mice reconstituted for 8 weeks with 1:1 mixtures of CD45.1 WT and CD45.2 Vhl-/-Mb1-cre or Vhl-/-Mb1-cre+Bcl2l11–/– BM. Gated as in Figure 3c. Filled columns; mean %CD45.2, clear columns; mean %CD45.1. Error bars S.D.; comparison between Vhl-/-Mb1-cre and Vhl-/-Mb1-cre+Bcl2l11–/– BM by two-way ANOVA Bonferroni post-test *P<0.05,***P<0.001. (e) BIM expression (Geo MFI) in Hif1a+/+Mb1-cre and Hif1a-/-Mb1-cre immature B cells cultured in 1%O2 for 24h. Gated on (a-b) Pro-B, Pre-B B220+IgM-IgD-, (a-b,e) immature B220+IgM+IgD- and (a-c) mature B220+IgM+IgD+. (a,e) **P<0.01,***P<0.001 two-way ANOVA Bonferroni post-test. (b) *P<0.05 unpaired two-sided t test, (c) *P<0.05,**P<0.01,****P<0.0001 one-way ANOVA Tukey post-test. (a) n=6 per genotype, (b) n=6 Vhl+/-Mb1-cre and 4 Vhl-/-Mb1-cre, (c) n=9 Vhl-/-Mb1-cre+Bcl2l11+/+, 4 Vhl-/-Mb1-cre+Bcl2l11+/-, 3 Vhl-/-Mb1-cre+Bcl2l11–/–, 8 Vhl+/-Mb1-cre+Bcl2l11+/+, 8 Vhl+/-Mb1-cre+Bcl2l11+/- and 6 Vhl+/-Mb1-cre+Bcl2l11–/–, (d) n=4 WT:Vhl-/-Mb1-cre and 5 WT:Vhl-/-Mb1-cre+Bcl2l11-/- and (e) n=5 Hif1a+/+Mb1-cre and 4 Hif1a-/-Mb1-cre, biologically independent mice. Data, (a) representative of three independent experiments, (b) representative of two independent experiments, (c) pooled from three independent experiments, (d) from one experiment and (e) representative of two independent experiments. (a-e) Symbols represent individual mice, bars means ± S.D.
HIF-1α limits light-chain editing
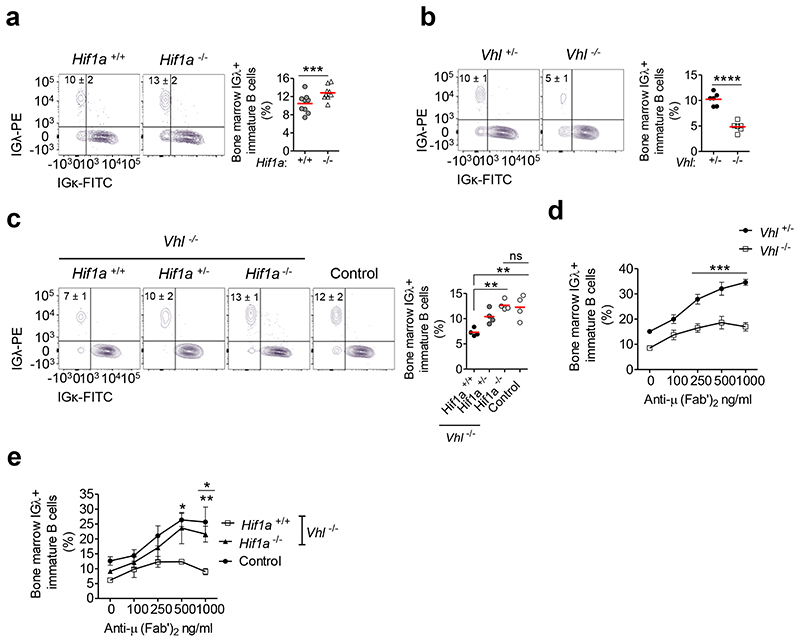
Our data demonstrate that down-regulation of HIF-1α-activity during B cell development is required for generating a normal complement of B cells. To further understand the physiological role of HIF-1α in B cell development, we deleted Hif1a in the B cell lineage (Hif1a-/-Mb1-cre mice). These mice demonstrated a normal complement of B cells and IgHV diversity (Supplementary figures 8a-e). These data are consistent with other studies using B cell-specific genetic manipulation of the HIF-VHL pathway9 but contrast with results in Hif1a-/- RAG-2-deficient chimeras, where reduced Pro-B and Pre-B cells were observed31; in the latter, Hif1a-deletion will have been in both B and T cells, which may account for the discrepancy. Since our studies revealed a role for HIF-1α in BCR signaling and BIM-dependent survival, which are both required for light-chain editing, a key central tolerance mechanism, we assessed the effect of Hif1a-deletion on this. Light-chain editing mainly occurs in immature B cells expressing autoreactive or inefficient BCRs2, 3, and in murine models, the frequency of IGλ+ B cells correlates with the extent of receptor-editing32. We observed increased IGλ-usage in Hif1a-/-Mb1-cre immature B cells compared with Hif1a+/+Mb1-cre controls (figure 6a). This was opposite to the reduced frequency of IGλ+ immature B cells observed in the presence of constitutive HIF-1α-activation in Vhl-/-Mb1-cre mice (Figure 6b). This was cell-intrinsic (Supplementary figure 8f). Furthermore, Hif1a-deletion in Vhl-/-Mb1-cre mice normalized the frequency of IGλ+ immature B cells (Figure 6c), establishing a HIF-1α-dependent effect. To interrogate the functional consequences of this, we assessed the ability of Vhl-/-Mb1-cre immature B cells to undergo BCR-editing on auto-antigen encounter. BM cultures were exposed to anti-μ (Fab’)2 as a self-antigen mimetic, for 3 days. Control Vhl+/-Mb1-cre immature B cells increasingly edited their BCRs in response to increasing anti-μ (Fab’)2, whilst Vhl-/-Mb1-cre immature BCRs edited significantly less (Figure 6d). This was HIF-1α-dependent (Figure 6e). These data highlight a cell-intrinsic role for HIF-1α in light-chain editing and central tolerance processing.
Figure 6. Regulated HIF-1α activity is required for BCR-editing.
Flow cytometry illustrating the frequency of lGλ+ and IGκ+ immature B cells from (a) Hif1a+/+Mb1-cre and Hif1a-/-Mb1-cre mice, (b) Vhl+/-Mb1-cre and Vhl-/-Mb1-cre mice and (c) from Vhl-/-Mb1-cre mice carrying Hif1a+/+, Hif1a+/- and Hif1a-/- alleles, and from control (Vhl+/- Hif1a+/+Mb1-cre) mice. Percentage of IGλ+ Vhl+/-Mb1-cre and Vhl-/-Mb1-cre immature B cells cultured with anti-μ (Fab’)2 for 3 days from (d) Vhl+/-Mb1-cre and Vhl-/-Mb1-cre mice, and (e) from Vhl-/-Mb1-cre mice carrying Hif1a+/+ and Hif1a-/- alleles, and from control (Vhl+/-Hif1a+/+Mb1-cre) mice. Gated on (a-c) immature B220+ IgM+ IgD, (d,e) immature B220+CD43-CD93+IgD-CD23- CD24++. (a) ***P<0.001 two-sided Wilcoxon Rank-Sum test, (b) ****P<0.0001 unpaired two-sided t test, (c) **P<0.01 one-way ANOVA Tukey post-test and (d,e) *P<0.05,**P<0.01,***P<0.001 two-way ANOVA Bonferroni post-test. In e, stars below the line are comparisons between control and Vhl-/-Hif1a+/+Mb1-cre and stars above the line are comparisons between Vhl-/-Hif1a-/-Mb1-cre and Vhl-/-Hif1a+/+Mb1-cre. (a) n=9 per genotype, (b) n=6 per genotype, (c) n= 4 per genotype, (d) n=5 per genotype and (e) n=3 Vhl-/-Hif1a+/+Mb1-cre, 4 Vhl-/-Hif1a-/-Mb1-cre and 4 Vhl+/-Hif1a+/+Mb1-cre, biologically independent mice. Data are, (a) pooled from two independent experiments, (b) representative of three independent experiments, (c) from one experiment, (d) representative of three independent experiments and (e) from one experiment. (a-c) Symbols represent individual mice, bars means ± S.D. (d,e) Error bars, S.E.M.
A small molecule HIF-activator blocks B cell development
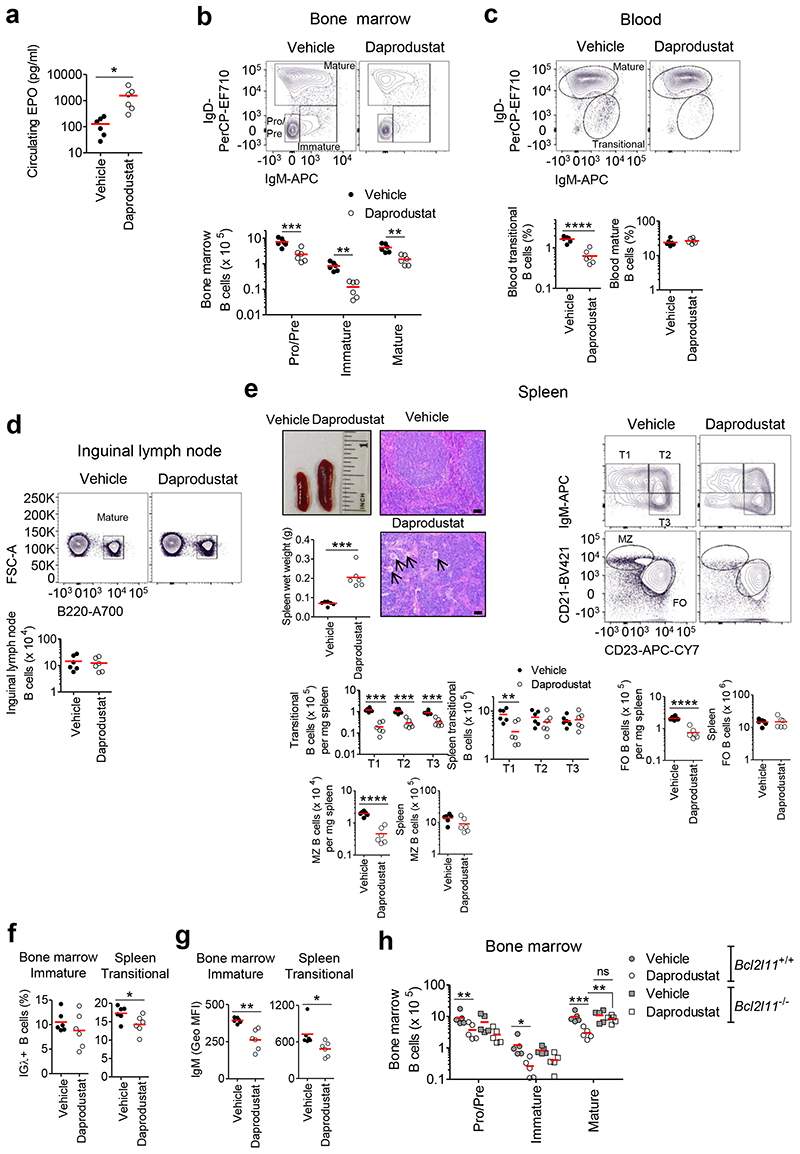
Small molecule inhibitors of PHDs activate HIF, increase hematocrit, and are under clinical evaluation for treating renal anemia33. To examine whether our observations may be of direct relevance to this therapeutic approach, C57BL/6J mice were dosed every 12h for 8 days with vehicle or 30 mg/kg of the small molecule pan-PHD inhibitor, daprodustat. HIF-activation in vivo was confirmed via erythropoietin (EPO) ELISA (Figure 7a). In daprodustat-treated mice, we observed marked reductions in BM Pro-B, Pre-B, immature and mature B cells, and blood transitional B cells. Blood and ILN mature B cells were unaffected (Figures 7b-d). Per mg spleen, B cell numbers were reduced in daprodustat-treated mice. However, the spleens were globally enlarged, with prominent extra-medullary hematopoiesis, and so total B cell numbers were similar to vehicle-treated controls (Figure 7e). Activation of HIF for 8 days by daprodustat resulted in a similar developmental defect in BM and transitional B cells as observed in Vhl-/-Mb1-cre mice, whereas in the periphery FO B cells, which normally turn over every eight weeks, were preserved, implying normal survival of the majority of peripheral B cells and a specific effect of HIF on the survival of BM B cells. In daprodustat-treated mice, the frequency of IGλ+ immature and transitional B cells was decreased, as was sIgM expression (Figures 7f-g). In the BM, daprodustat-treatment modestly reduced BAFF-R expression in immature and mature B cells but had no effect on expression in peripheral B cells. One potential reason (among several) for the effect of daprodustat being less than the effect of VHL-loss is that with the dosing schedule used, the level of HIF-activation was lower (Supplementary figures 8g-h). BM B cell loss in daprodustat-treated mice was in part BIM-dependent since in Bcl2l11-/- mice, immature, but particularly mature, B cells were less sensitive to the effects of daprodustat (Figure 7h), supporting the link between HIF-1 and BIM-dependent death. Of note, reduced Pro-B, Pre-B and immature B cells observed in both WT and Bcl2l11-/- mice treated with daprodustat could in part be due to indirect actions on the BM as a result of global HIF-activation (increased hematopoiesis, EPO, angiogenesis), which does not occur with B cell specific HIF-activation in Vhl-/-Mb1-cre mice. Collectively, these data highlight the potential of this approach to therapeutically target specific B cell subsets, in a reversible and titratable manner. This has important therapeutic implications in B cell-related diseases such as in autoimmunity and B cell malignancies.
Figure 7. Developing B cells are reduced in daprodustat treated mice.
C57BL/6J mice were treated for 8 days with vehicle (2% DMSO, PBS) or 30 mg/kg daprodustat every 12 h. (a) Erythropoietin (EPO) ELISA on serum from vehicle and daprodustat treated mice. Flow cytometry showing the gating, frequency of live or absolute numbers of B cells from vehicle or daprodustat treated mice, in (b) BM, (c) blood, (d) ILN and (e) spleen. Displayed, spleen wet weight and representative images of whole spleens and structure by hematoxylin and eosin staining from vehicle and daprodustat treated mice. Arrows identify megakaryocytes, characteristic of extra-medullary hematopoiesis. Scale-bar 100 μm. (f) Percentage of IGλ+ BM immature and spleen transitional B cells and (g) mean sIgM expression (GEO MFI) on BM immature and spleen transitional B cells, from vehicle and daprodustat treated mice. (h) Flow cytometry showing the enumeration of BM B cell subsets from WT (Bcl2l11+/+) or Bcl2l11–/– mice treated for 8 days with vehicle or 30 mg/kg daprodustat every 12 h. (b-h) Gated as in Figure 1. (a,c-g) *P<0.05,**P<0.01,****P<0.0001 unpaired two-sided t test, (b,e,h) *P<0.05,**P<0.01, ***P<0.001 two-way ANOVA Bonferroni post-test or one-way ANOVA Tukey post-test. (a-g) n=6 biologically independent mice per group. Data are representative of two independent experiments, (h) n=5 biologically independent mice per group, from one experiment. (a-h) Symbols represent individual mice, bars means ± S.D.
Discussion
Hypoxic environments exist in BM where stringent selection and central tolerance processes occur as B cells develop. Our study demonstrates that B cell development is profoundly influenced by HIF-1α. We observed high HIF-activity in early B cell development and that subversion of the normal physiological down-regulation observed at later developmental stages led to developmental arrest, reduced BCR diversity, defects in BCR function and BCR-editing. Our findings infer that HIF-1α impedes survival signals generated by the nascent BCR, which is first expressed at this stage, via decreased sIgM, failure to suppress BIM, and attenuation of downstream signaling required for B cell selection and survival. We show that regulated HIF-1α-activity is necessary to maintain BCR expression and survival signals including appropriate PI3K-signaling and repression of BIM. We found that HIF-1α reduced expression of the pro-survival BAFF-R, suggesting that regulated HIF-1α-activity may be required for maintaining appropriate BAFF-R levels necessary for B cell selection and survival. An interesting notion is that reduced BCR expression may be due to increased antigen-independent signaling, leading to death34, linking a dependence on HIF-1 suppression for appropriate spontaneous BCR signaling.
Advances in non-invasive imaging have demonstrated dynamic heterogeneity in BM pO2, with steep gradients and variation with location, bone-type, injury and bone-remodeling10, 11, 35. Oxygenation levels in BM span a broad range, over which HIF is progressively responsive. Our results imply that temporal and spatial control of HIF-activity by these dynamic hypoxic gradients profoundly modulates B cell biology. Our observation that HIF-activity is dynamically regulated during human and murine B cell development in the BM supports this; with HIF being more active in early developmental Pro-B and Pre-B cells, supporting the proliferative burst by sustaining glycolysis36, and less active in immature and mature B cells. Thus, we propose a model in which oxygen gradients encountered by BM B cells as they develop provide a signal through HIF-1α-activation to coordinate location with B cell differentiation and survival, in a dynamic manner.
We show that HIF-1α restrains the diversity of the pre-immune B cell repertoire by limiting IgHV-editing and increasing apoptosis. Furthermore, a key central tolerance mechanism, light-chain editing was modulated by HIF-1α-activity, being increased when Hif1a was deleted and reduced when HIF-1α was activated. In our mouse models with activated HIF-1α, inactivation of a single Hif1a allele resulted in an intermediate phenotype and transient HIF-activation under hypoxia reduced BCR expression and survival suggesting that both the magnitude and duration of HIF-activation is tunable, leading to differing outcomes in the selection of immature B cells. Our data show that major selection signals are influenced by HIF-1α, expanding the aspects of B cell biology modulated by this pathway. It is known that constitutive HIF-activation in germinal center B cells reduces proliferation, isotype-switching and affinity maturation8, and that HIF-1α drives production of the anti-inflammatory cytokine IL-10 in B cells9. It is interesting to speculate that the suppressive effects of HIF-1α on B cell development may be advantageous in acute hypoxia-related pathology. For example, blood loss potently activates HIF-1α via iron-deficiency and hypoxemia, and the effect of this on B cell development would help redirect BM capacity to red cell generation in an emergency setting. Taken together, appropriate responses to oxygenation are crucial across the lifespan of a B cell, and manipulating HIF should have considerable therapeutic potential in B cell-mediated pathologies such as autoimmunity and malignant-transformation.
Methods
Human BM single cell RNA sequencing analysis
Raw count tables were downloaded from the human cell atlas data portal (https://preview.data.humancellatlas.org/) for n=1 human BM sample ‘MantonBM1’. Quality control metrics were calculated using the calculateQCmetrics function in the Scater R package (version 1.10.1). The expression matrix was subset to the “MantonBM1” sample, and cells with <20% mitochondrial counts and < 500 unique features were discarded. Doublets were classified using the Scrublet python package (version 0.2)37 and cells predicted to be doublets were discarded. Data were normalized and log-transformed using the Scanpy python package (version 1.4.1)38. Highly variable genes were detected using Scanpy, using settings min_mean = 0.01, max_mean = 4, min_disp =1. Principal components analysis was performed using the runPCA function in Scater, subsetting the expression matrix to highly variable genes, centering and scaling features. The first 10 principal components were used after examination of scree plots of variance explained. Uniform Manifold Approximation and Projection (UMAP) embeddings and Leiden clusters were calculated in Scanpy, setting n_neighbors = 15 and clustering resolution = 1. Partition-based graph abstraction (PAGA) initialized UMAP embeddings39 were calculated using Scanpy setting threshold = 0.03. This procedure was conducted for initial analysis of the whole dataset and for reanalysis of the data subset to the hematopoiesis compartment, however during the subcompartment analysis variable genes were calculated with min_mean = 0.01, max_mean = 8, min_disp =1, and UMAP was calculated using n_neighbors = 30.
Murine BM single cell RNA sequencing analysis
Raw count tables for Tabula Muris data40 were downloaded from https://github.com/czbiohub/tabula-muris using scripts provided therein for n=2 biologically independent mice (Data names10X_P7_2 and 10X_P7_3). Droplet based sequencing data were subset to data from BM, and quality control metrics were calculated using the calculateQCmetrics function in Scater. Cells with fewer than 250 unique features were discarded. Data were normalized and log transformed as described for human scRNAseq data, and highly variable genes calculated in Scanpy, using settings min_mean =0.005, max_mean = 8, min_disp = 1. Principal components analysis was performed as described above, retaining the first 20 principal components after examination of scree plots of variance explained. UMAP plots and Leiden clustering solutions were calculated in Scanpy using n_neighbors = 30, and resolution = 0.5. PAGA-initialized UMAP embeddings were calculated using Scanpy setting threshold = 0.1.
Single cell RNA sequencing pseudotime estimation
Pseudotime was calculated using the Slingshot R package (version 1.1.3)41. Principal curves were calculated using PAGA-initialized UMAP coordinates as input.
Single cell RNA sequencing marker gene calculation
Marker genes were calculated using the tf-idf metric42. Genes were ranked by the tf-idf metric, and p values calculated by a hypergeometric test, and corrected for multiple testing (Benjamini-Hochberg method).
Single cell geneset scoring
Aggregate geneset scores were calculated using the AddModuleScore function in the Seurat R package (version 3.0.0)43. Human genesets (Ensembl ID) were converted to murine orthologs using a conversion table downloaded from Ensembl BioMart (https://www.ensembl.org/index.htmlx).
Microarray analysis of human peripheral blood B cells exposed to hypoxia
Leukocyte cones, a waste product of the blood donation process, were obtained at random from unselected anonymous NHSBT blood donors. B cells were then isolated from peripheral blood mononuclear cells following ficoll density gradient centrifugation. Isolated B cells were cultured in normoxia or 1% O2 for 24 h and RNA was extracted using a RNeasy Mini kit (Qiagen). Leukocyte cones were obtained from anonymous NHSBT blood donors who provided specific blood donor consent for their donated materials to be used in appropriate ethically approved studies. The overarching study received ethical approval from the East of England – Cambridge Central Research Ethics Committee (REC reference 08/H0308/176).
Raw .CEL files were pre-processed using the oligo R package (version 1.46.0). Differential expression analysis was conducted using the limma R package (version 3.38.3). False Discovery Rate adjusted p values were calculated using the Benjamini-Hochberg method.
Mice
C57BL/6J Vhl-/-14, Hif1a-/-19 and Epas1-/-20 mice were crossed with Cd79a-cre (Mb1-cre) mice15 to delete the target gene. All mice with loxP-flanked alleles and their respective controls were hemizygous for mb-1 and Cre. To generate Vhl-/-Mb1-cre mice carrying Hif1a+/+, Hif1a+/- and Hif1a-/- alleles, Vhl-/-Hif1a+/-Mb1+/+ mice were crossed with Vhl-/-Hif1a+/-Mb1cre/cre mice. Control mice (Vhl+/- Hif1a+/+Mb1-cre) were age-matched to experimental littermates. To generate Vhl-/-Mb1-cre mice carrying Epas1+/+, Epas1+/- and Epas1-/- alleles, Vhl-/-Epas1+/-Mb1+/+ mice were crossed with Vhl-/- Epas1+/-Mb1cre/cre mice. Control mice (Vhl+/-Epas1+/+Mb1-cre) were age-matched to experimental littermates. The mice were backcrossed for at least 8 generations and maintained on a C57BL/6J background. These mice along with C57BL/6-Tg(IghelMD4)4Ccg/J (IgHEL; JAX stock #002595)25, muMt- mice (Jax stock #002288)44, CD45.1 (B6.SJL-Ptprca Pepcb/BoyCrl; JAX stock #002014)45 and Bcl2l11–/– mice30 (B6.129S1/Sv-Bcl2l11tm1.1Ast/Kieg, # EM: 00602, EMMA mouse repository) were housed in specific pathogen-free animal facilities (at 20-23°C, with 40–60% humidity, 12 hour light/12 hour dark cycle). All experiments included age and litter-matched mice that were not selected for gender. The Resource Equation was used to determine sample size for the majority of experiments. Randomization was genetic and where possible, investigators were blinded to the genetic status. When appropriate, a randomization algorithm was used (excel) to randomly allocate mice into experimental groups. For mice carrying loxP-flanked Vhl alleles, deletion efficiency was determined by real-time PCR of genomic DNA. The degree of excision was calculated by comparison of Vhl intact DNA relative to an unexcised gene Actb. The primers and Probes used were Vhl forward 5′-GCTTGCGAATCCGAGGG, Vhl reverse 5′-TCCTCTGGACTGGCTGCC, Vhl Probe 5′-E6-FAM-CCCGTTCCAATAATGCCCCGG (Life Technologies) and Actb (Mouse assay ID: Mm00607939_s1; Life Technologies). For analysis of EF5 levels C57BL/6J mice were injected intravenously with 10mM EF5 (dosed in 0.1ml/10g body weight), then culled 3 hours later for analysis. For single BM chimeras, muMT mice were irradiated with one dose of 6 Gy and injected intravenously with 2 x106 BM cells from Vhl+/-Mb1-cre or Vhl-/-Mb1-cre mice. For mixed chimeras, CD45.1 mice were irradiated with two doses of 4.5 Gy spaced 3 hours apart and were injected intravenously with 2x106 BM cells containing 1:1 mixtures of WT CD45.1 and CD45.2 Vhl+/-Mb1-cre, Vhl-/-Mb1-cre or Vhl-/-Mb1-cre + Bcl2l11-/- BM. Chimeric mice were left to reconstitute for 8 weeks prior to analysis. For PHD inhibitor studies, C57BL/6J or Bcl2l11-/- mice received vehicle (2% DMSO in phosphate buffered saline (PBS)) or 30 mg/kg daprodustat (Insight Biotechnology Ltd) via intraperitoneal injection dosed in 0.1ml/10g body weight, every 12 hours for 8 days. All procedures were ethically approved by the University of Cambridge Animal Welfare and Ethical Review Body (AWERB) and complied with the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012, under the authority of a UK Home Office License. The ARRIVE (Animal Research: Reporting In Vivo Experiments) guidelines https://www.nc3rs.org.uk/arriveguidelines were used for planning, conducting and reporting of experiments. For hematoxylin and eosin staining, wax-embedded or frozen spleen sections (4 or 7 μm) were rehydrated or fixed in acetone (10 min, -20°C), placed in hematoxylin for 25 seconds, rinsed, placed in eosin for 25 seconds, then dehydrated and mounted. Images were digitized on an Axioimager Z2 Upright Microscope (Zeiss) using Zen Blue Pro 2012 software (version 1.1.2.0; Zeiss).
Flow Cytometry
Cell suspensions from BM (one femur), peripheral blood, spleens, inguinal lymph nodes and the peritoneal cavity were prepared by lysing red blood cells (RBCs) in 1ml RBC lysis buffer (0.155M NH4Cl, 12mM NaHCO3, 0.1M EDTA, distilled H2O) and quenching in 9ml RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin, 100 mg/ml streptomycin and 2 mM L-glutamine, then counted on a hemocytometer. Cells were blocked using FcR blocking reagent (1:100; Miltenyi Biotech) and stained with combinations of antibodies listed in Supplementary table 1, which were diluted in PBS supplemented with 0.5% bovine serum albumin (BSA). LIVE/DEAD Fixable Aqua or Blue Dead Cell Stain (diluted 1:200 in PBS; Invitrogen Thermo Fisher Scientific) was used for the detection and exclusion of dead cells. Intracellular staining was performed using a Cytofix/Cytoperm kit (BD Bioscience) with antibodies against antigens BIM (1 h incubation at room temperature), AKT-pS473 and EF5 (18 h incubation at 4°C; Supplementary table 1). For analysis of apoptotic B cells, cell suspensions were stained in PBS containing 7-aminoactinomycin D (7-AAD; 1:800) and PO-PRO-1 (1:300; Invitrogen Thermo Fisher Scientific) for 30 minutes (min) at 4°C. Fluorescence-minus-one (FMO) controls were used to set appropriate gates and as negative staining controls. FMO values were subtracted from expression data (EF5/IgM/CD19/AKT-pS473/BIM). EF5 GEO MFI values from vehicle treated mice were subtracted from GEO MFI values from EF5-treated mice. Total B cells were quantified both manually and using 123count eBeads Counting Beads (Invitrogen Thermo Fisher Scientific). Data were acquired on an LSR Fortessa cell analyzer or an Influx cell sorter (BD Biosciences) using FACSDiva 6.2 and FACSD 8.0.1 software, respectively (BD Biosciences) and analyzed with FlowJo software v 10 (Tree Star). A figure exemplifying the gating strategies for all figures generated from flow cytometry and cell sorting experiments is provided (Supplementary figure 9).
BCR amplification and sequencing
Total RNA was extracted from isolated BM B cells, Pro-B, Pre-B B220+IgM-, immature B220+IgM+IgD- and mature B220+IgM+IgD+ B cells and from spleen B cells, transitional (B220+IgM+IgD-) and follicular (B220+IgM+IgD+CD23+CD21+). Reverse transcription (RT) was performed in a 23 μL reaction: 14 μL of RT mix 1 (containing RNA template, 10 μM reverse primer mix, 1 μL dNTP (10mM), and nuclease-free water) was incubated for 5 min at 70°C. This mixture was immediately transferred to ice for 1 min, and the RT mix 2 (4 μL 5x FS buffer, 1 μL DTT (0.1M), 1 μL SuperScript®III (Thermo Fisher Scientific)) was added and incubated at 50°C for 60 min followed by 15 min inactivation at 70°C. cDNA was cleaned with Agencourt AMPure XP beads and PCR amplified with V-gene multiplex primer mix (10μM each forward primer) and 3’ universal reverse primer (10μM) using KAPA protocol and the thermal cycling conditions: 1 cycle (95°C - 5 min); 5 cycles (98°C - 5 sec; 72°C - 2 min); 5 cycles (65°C - 10 sec, 72°C - 2 min); 25 cycles (98°C - 20sec, 60°C - 1 min, 72°C - 2 min); 1 step (72°C - 10 min). Similarly, BCR amplification from DNA samples was performed with V-gene multiplex primer mix (10μM each forward primer) and 3’ universal reverse primer (10μM) using KAPA protocol and the thermal cycling conditions: 1 cycle (95°C - 5 min); 5 cycles (98°C - 5 sec; 72°C - 2 min); 5 cycles (65°C - 10 sec, 72°C - 2 min); 25 cycles (98°C – 20 sec, 60°C - 1 min, 72°C - 2 min); 1 step (72°C - 10 min). Primers are provided in Supplementary tables 2 and 3.
Sequencing libraries were prepared using Illumina protocols and sequenced using 300bp paired-ended MiSeq (Illumina). Raw reads were filtered for base quality (median Phred score >32) using QUASR version 6.X (http://sourceforge.net/projects/quasr/)46. Forward and reverse reads were merged together if they contained an identical overlapping region of >50bp, or otherwise discarded. Universal barcoded regions were identified in reads and orientated to read from V-primer to constant region primer. The barcoded region within each primer was identified and checked for conserved bases. Primers and constant regions were trimmed from each sequence, and sequences were retained only if there was >80% per base sequence similarity between all sequences obtained with the same barcode, otherwise discarded. The constant region allele with highest sequence similarity was identified by 10-mer matching to the reference constant region genes from the IMGT database47, and sequences were trimmed to give only the region of the sequence corresponding to the variable (VDJ) regions. Isotype usage information for each BCR was retained throughout the analysis hereafter. Sequences without complete reading frames and non-immunoglobulin sequences were removed and only reads with significant similarity to reference IgHV and J genes from the IMGT database using BLAST (version 2.7.1)48 were retained. Ig gene usages and sequence annotation were performed in IMGT V-QUEST (version 3.4.9), where repertoire differences were performed by custom scripts in python, and statistics were performed in R using one-sided Wilcoxon tests for significance (non-parametric test of differences between distributions).
The network generation algorithm and network properties were calculated as in22: each vertex represents a unique sequence, where relative vertex size is proportional to the number of identical reads. Edges join vertices that differ by single nucleotide non-indel differences and clusters are collections of related, connected vertices. A clone (cluster) refers to a group of clonally relate B cells, each containing BCRs with identical CDR3 regions and IgHV gene usage or differing by single point mutations. Each cluster should originate from the same pre-B cell. The vertex Gini index was calculated in a manner irrespective of sequencing depth through the subsampling of each sequencing sample to a specified depth. This is calculated from the distribution of the number of unique RNA or DNA molecules per vertex within subsampled BCR repertoires at a specified depth. The mean of 1000 repeats of resulting vertex Gini index was determined.
IgHV gene editing analyses were performed in a similar manner to23. For all BCRs, stem regions were identified (defined as N-IgHD-N-IgHJ regions starting 3bp downstream of the IgHV gene boundary). The number of unique BCR sequences sharing stem regions but with different IgHV gene usage (>95% difference in sequence identity in the IgHV region) and with different 5’ of the junctional region (defined as IgHV (last 3pb)-N-IgHD-N-IgHJ) was determined and compared to the total number of unique BCRs to give the percentage IgHV replacement. Sequences with joining regions (N-IgHD-N-IgHJ regions) shorter than 8 nucleotides were excluded from this percentage due to the potential of germline-encoded receptors. The BCR-seq data generated in this study is available at the SRA database (BioProject IDs: PRJNA574931, PRJNA574906, PRJNA574628).
RNA-Sequencing and analysis
Total RNA was extracted from isolated Pro-B, Pre-B (B220+ IgM-), immature (B220+ IgM+ IgD-) and mature (B220+IgM+IgD+) B cells from BM using a RNeasy Plus Micro kit (Qiagen). RNA quality and quantity was analyzed using an Agilent 2100 Bioanalyzer (Agilent). Library preparation was performed using a SMARTer® Universal Low Input RNA Kit (Takara Bio). Library quality was checked using the KappaQC qPCR kit and a bioanalyzer with 2100 Expert software (version B.02.10.SI764) and the samples were subjected to high-throughput paired-end sequencing (75 cycles) on a NextSeq500 sequencer (Illumina) at the Cambridge Genomic Services facility (https://www.cgs.path.cam.ac.uk/). Sequenced sample quality was checked using FastQC (version 0.11.7) and RNA-Seq reads were aligned to the mouse genome (UCSC mm10) using HISAT2 (version 2.1.0)49. The number of uniquely mapping reads was estimated using RSeQC (version 2.6.4)50 and genes counted using FeatureCounts from the Rsubread package51. Gene expression read counts were normalized and differentially expressed genes identified using DESeq2 (version 1.18.1)52. The Benjamini and Hochberg critical value was used for multiple testing correction. Genes with adjusted P value <0.05 were considered as having significantly altered expression. Gene set enrichment analysis (GSEA) was carried out by ranking changes in gene expression by the inverse of the adjusted P value with the sign of the log fold change. These lists were then analyzed using the GSEA package against public datasets (Hallmarks) and the MSigDB collection of gene pathways with the pre-ranked option using the classic enrichment statistic mode53. Gene ontology (GO) analysis was carried out on genesets, which were identified through filtering the differential expression results as described. Enrichment for GO terms within each geneset was performed using TopGo and enrichment was assessed against a gene universe, which was limited to all genes that were found to be expressed (count > 1) in at least one sample in the dataset. P values were calculated using the fisher slim method within the TopGo package. The RNA-seq data generated in this study is available at GEO: GSE129513.
Ex vivo exposures to hypoxia
RBC-lysed single-cell BM suspensions were cultured in RPMI-1640 supplemented with 15% FCS, 2 mM L-glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin, 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol. For hypoxic culture assays 4x106 BM cells were cultured in a humidified Whitley H35 Hypoxystation (Don Whitley Scientific) at 1% O2, 37°C, 5% CO2. For BCR-editing assays, 4x106 BM cells were stimulated with anti-IgM F(ab’)2 (Jackson Immunoresearch) and cultured for 3 days at 37°C, 5% CO2, 95% humidity.
Enzyme-linked immunosorbent assays
Total murine serum IgM was measured using an ELISA kit (Bethyl Laboratories) according the manufacturer’s instructions. Serum BAFF and EPO were measured using an ELISA kit (R&D Systems) according to the manufacturer’s instructions.
Western-blotting
Tissues were first homogenised then cells and tissues were sonicated in 50 mM TRIS-HCL pH7.4/120 mM NaCl/5 mM EDTA/0.5%NP40/1 mM DL-Dithiotheitol/1 mM Phenylmethylsulfonyl fluoride/2 mM sodium orthovanadate/2 mM NaF/20 mM β -glycerol phosphate/5 mM Sodium pyrophosphate/complete Mini EDTA-free Protease Inhibitor Cocktail Tablet, to generate lysates. Samples were centrifuged and supernatants collected for immuno-blotting. The following primary antibodies were used: HIF-1α (diluted 1:500, 14179; Cell Signaling), HIF-2α (diluted 1:200, AF2997; R&D Systems), β-actin (diluted 1:80000, A2228; Sigma-Aldrich). Secondary antibodies conjugated to horseradish peroxidase (HRP) were diluted 1:20000 (A90-337 and A120-208PP; Bethyl Laboratories), and 1:1000 (HAF109; R&D Systems). Proteins were detected using the SuperSignal West Femto kit (Thermo Scientific) and imaged on a Chemidoc MP Imaging system (Bio-rad). The uncropped imaged blots are provided as a Source Data file. Densitometric analysis was performed using ImageLab software version 6.0.1, which calculated background subtracted adjusted band volumes. Bands of interest were then multiplied by a normalization factor. This was generated by selecting a reference band in the protein loading controls and dividing all other loading control bands by the reference. The selected reference lane was either a normoxic control or one negative for HIF-α such that lanes negative for HIF-α have a value of 1.
Statistics
GraphPad Prism Software 5.01 (GraphPad Software, Inc.) was used for statistical analyses, and unpaired two-sided Student’s t tests, two-sided Mann-Whitney U tests, one-way ANOVA with Tukey post-tests or two-way ANOVA with Bonferroni post-tests were used for statistical comparisons between groups. For analysis of sequencing datasets, statistics used are described in the relevant methodology sections and figure legends.
Supplementary Material
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Acknowledgements
This work was funded by the Wellcome Trust (19710), a Cambridge NIHR senior investigator award (NF-SI-0514-10122) and the Rosetrees Trust (G102721) to P.H. Maxwell, supporting N. Burrows, V.J. Bhute, A. Peñalver and J.E.G. Smith. R.J.M. Bashford-Rogers was supported by the Wellcome Trust (WT106068AIA). B.J. Stewart was supported by a Wellcome Clinical PhD fellowship (216366/Z/19/Z). T.M. Connor was supported by a MRC Clinic Research Training Fellowship (G0802266, ID 89800). S. Smith was supported by a summer studentship award from The Lister Institute of Preventive Medicine. M.G.B. Tran was supported by a NIHR CL-2006-14-006. M.R. Clatworthy was supported by a Chan-Zuckerberg Initiative Human Cell Atlas Technology Development Grant, a Versus Arthritis Cure Challenge Research Grant (21777), and an NIHR Research Professorship (RP-2017-08-ST2-002). The CIMR core facilities were supported by a Wellcome Trust/MRC center grant (097922/Z/11/Z) and a Wellcome Trust strategic award 100140. We thank R. Schulte, C. Cossetti, M. Maj and G. Grondys-Kotarba (CIMR Flow Cytometry Core Facility) and M. Gratian and M. Bowen (CIMR microscopy core facility) for their help and support, Y. Umrania for bioinformatics support, M. Reth (University of Freiburg, Germany) and M. Turner (Babraham Institute, Cambridge) for providing the Mb1-cre mice, and R. Johnson (Karolinska Institutet, Sweden) for providing Hif1a-/- mice. We thank J. Sale and M. Ashcroft for useful discussions and T. Wilson, K. Scott and D. Gale for experimental and technical support.
Footnotes
Author Contributions
N.B. led the research design, performed experiments, analyzed data, maintained the mouse lines and wrote the paper. R.J.M.B.-R. performed BCR amplification, sequencing and analysis and wrote the paper. V.J.B. performed experiments and bioinformatics analyses. A.P., J.E.G.S. and T.M.C. performed experiments and maintained the mouse lines. J.R.F., B.J.S., L.B., G.G., and E.P.performed bioinformatics analyses. M.D-L. helped with experimental and technical design. A.I., S.S. and M.G.B.T. performed experiments. P.A.L. helped with design of RNA-seq experiments. M.E., K.G.C.S. and B.J.P.H. contributed to research design. R.J.C. contributed to research and experimental design and provided guidance on manuscript preparation. M.R.C. contributed to research and experimental design and wrote the paper. P.H.M supervised the project and wrote the paper.
Competing Interests statement
P.H.M. is a scientific founder and equity holder of ReOx and has received speaker honoraria from Fibrogen, which both aim to develop PHD inhibitors as therapies. R.J.M.B-R. is a co-founder and consultant for Alchemab Therapeutics Ltd, which maps antibody repertoires to develop therapeutics for patients with hard-to-treat diseases, and is a consultant for Imperial College London and VHSquared, which develops Vorabodies (oral domain antibodies). R.J.C. is a scientific founder and equity holder of MIROBIO, which aims to develop novel therapies to treat a range of auto-immune diseases. All other authors declare no competing interests.
Data Availability
Data are available from the corresponding author upon request. Uncropped western blots for Supplementary figure 3a-c; 8h are provided as Source Data. The BCR-seq data generated in this study is available at the SRA database (https://www.ncbi.nlm.nih.gov/sra), BioProject Accession IDs: PRJNA574931, PRJNA574906, PRJNA574628. The RNA-seq and microarray datasets generated in this study are available at the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), GEO Accession IDs: GSE129513 and GSE152960.
Data in this study that was obtained from publically available sources can be accessed at:
-
1)
Tabula muris scRNAseq data https://tabula-muris.ds.czbiohub.org/ Data names: 10X_P7_2 and 10X_P7_3
-
2)
Human cell atlas scRNAseq data https://data.humancellatlas.org/explore/projects/cc95ff89-2e68-4a08-a234-480eca21ce79 Data name: MantonBM1
Code Availability
Custom scripts for the analysis of BCR repertoire differences are available at: Immune_receptor_NETWORK-GENERATION https://github.com/rbr1/Immune_receptor_NETWORK-GENERATION
References
- 1.Nemazee D. Mechanisms of central tolerance for B cells. Nat Rev Immunol. 2017;17:281–294. doi: 10.1038/nri.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun A, et al. VH replacement in primary immunoglobulin repertoire diversification. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E458–466. doi: 10.1073/pnas.1418001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kouskoff V, Lacaud G, Pape K, Retter M, Nemazee D. B cell receptor expression level determines the fate of developing B lymphocytes: Receptor editing versus selection. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7435–7439. doi: 10.1073/pnas.130182597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nemazee DA, Burki K. Clonal Deletion of Lymphocyte-B in a Transgenic Mouse Bearing Anti-Mhc Class-I Antibody Genes. Nature. 1989;337:562–566. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- 5.Tussiwand R, Rauch M, Fluck LA, Rolink AG. BAFF-R expression correlates with positive selection of immature B cells. European journal of immunology. 2012;42:206–216. doi: 10.1002/eji.201141957. [DOI] [PubMed] [Google Scholar]
- 6.Loder F, et al. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J Exp Med. 1999;190:75–89. doi: 10.1084/jem.190.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abbott RK, et al. Germinal Center Hypoxia Potentiates Immunoglobulin Class Switch Recombination. J Immunol. 2016;197:4014–4020. doi: 10.4049/jimmunol.1601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho SH, et al. Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature. 2016;537:234. doi: 10.1038/nature19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng XY, et al. Hypoxia-inducible factor-1 alpha is a critical transcription factor for IL-10-producing B cells in autoimmune disease. Nat Commun. 2018;9 doi: 10.1038/s41467-017-02683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruneboom A, et al. A network of trans-cortical capillaries as mainstay for blood circulation in long bones. Nature metabolism. 2019;1:236–250. doi: 10.1038/s42255-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spencer JA, et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 2014;508:269. doi: 10.1038/nature13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 13.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O-2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 14.Haase VH, Glickman JN, Socolovsky M, Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy RR, Hayakawa K. B cell development pathways. Annual review of immunology. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 17.Gaspar HB, Conley ME. Early B cell defects. Clin Exp Immunol. 2000;119:383–389. doi: 10.1046/j.1365-2249.2000.01192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreuzaler M, et al. Soluble BAFF Levels Inversely Correlate with Peripheral B Cell Numbers and the Expression of BAFF Receptors. J Immunol. 2012;188:497–503. doi: 10.4049/jimmunol.1102321. [DOI] [PubMed] [Google Scholar]
- 19.Ryan HE, et al. Hypoxia-inducible factor-1 alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- 20.Gruber M, et al. Acute postnatal ablation of Hif-2 alpha results in anemia. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2301–2306. doi: 10.1073/pnas.0608382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ngo VN, Cornall RJ, Cyster JG. Splenic T zone development is B cell dependent. J Exp Med. 2001;194:1649–1660. doi: 10.1084/jem.194.11.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bashford-Rogers RJM, et al. Network properties derived from deep sequencing of human B-cell receptor repertoires delineate B-cell populations. Genome Res. 2013;23:1874–1884. doi: 10.1101/gr.154815.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bashford-Rogers RJM, et al. Eye on the B-ALL: B-cell receptor repertoires reveal persistence of numerous B-lymphoblastic leukemia subclones from diagnosis to relapse. Leukemia. 2016;30:2312–2321. doi: 10.1038/leu.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bristow RG, Hill RP. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 25.Goodnow CC, et al. Altered Immunoglobulin Expression and Functional Silencing of Self-Reactive Lymphocytes-B in Transgenic Mice. Nature. 1988;334:676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 26.Aiba Y, Kameyama M, Yamazaki T, Tedder TF, Kurosaki T. Regulation of B-cell development by BCAP and CD19 through their binding to phosphoinositide 3-kinase. Blood. 2008;111:1497–1503. doi: 10.1182/blood-2007-08-109769. [DOI] [PubMed] [Google Scholar]
- 27.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, et al. Acetylation of FoxO1 Activates Bim Expression to Induce Apoptosis in Response to Histone Deacetylase Inhibitor Depsipeptide Treatment. Neoplasia. 2009;11:313–U315. doi: 10.1593/neo.81358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otero DC, Rickert RC. CD19 function in early and late B cell development. II. CD19 facilitates the pro-B/pre-B transition. J Immunol. 2003;171:5921–5930. doi: 10.4049/jimmunol.171.11.5921. [DOI] [PubMed] [Google Scholar]
- 30.Bouillet P, et al. Proapoptotic Bcl-2 relative bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 31.Kojima H, et al. Abnormal B lymphocyte development and autoimmunity in hypoxia-inducible factor 1 alpha-deficient chimeric mice. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2170–2174. doi: 10.1073/pnas.052706699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vela JL, Ait-Azzouzene D, Duong BH, Ota T, Nemazee D. Rearrangement of mouse immunoglobulin kappa deleting element recombining sequence promotes immune tolerance and lambda B cell production. Immunity. 2008;28:161–170. doi: 10.1016/j.immuni.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maxwell PH, Eckardt KU. HIF prolyl hydroxylase inhibitors for the treatment of renal anaemia and beyond. Nat Rev Nephrol. 2016;12:157–168. doi: 10.1038/nrneph.2015.193. [DOI] [PubMed] [Google Scholar]
- 34.Cornall RJ, et al. Polygenic autoimmune traits: Lyn, CD22, and SHP-1 are limiting elements of a biochemical pathway regulating BCR signaling and selection. Immunity. 1998;8:497–508. doi: 10.1016/s1074-7613(00)80554-3. [DOI] [PubMed] [Google Scholar]
- 35.Christodoulou C, et al. Live-animal imaging of native haematopoietic stem and progenitor cells. Nature. 2020;578:278. doi: 10.1038/s41586-020-1971-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kojima H, et al. Differentiation Stage-Specific Requirement in Hypoxia-Inducible Factor-1 alpha-Regulated Glycolytic Pathway during Murine B Cell Development in Bone Marrow. J Immunol. 2010;184:154–163. doi: 10.4049/jimmunol.0800167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 37.Wolock SL, Lopez R, Klein AM. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 2019;8:281. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19 doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf FA, et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20 doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaum N, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Street K, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. Bmc Genomics. 2018;19 doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young MD, et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018;361:594. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B-Cell-Deficient Mouse by Targeted Disruption of the Membrane Exon of the Immunoglobulin Mu-Chain Gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 45.Janowska-Wieczorek A, et al. Platelet-derived microparticles bind to hematopoietic stem/progenitor cells and enhance their engraftment. Blood. 2001;98:3143–3149. doi: 10.1182/blood.v98.10.3143. [DOI] [PubMed] [Google Scholar]
- 46.Watson SJ, et al. Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos T R Soc B. 2013;368 doi: 10.1098/rstb.2012.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lefranc MP. IMGT unique numbering for the variable (V), constant (C), and groove (G) domains of IG, TR, MH, IgSF, and MhSF. Cold Spring Harbor protocols. 2011;2011:633–642. doi: 10.1101/pdb.ip85. [DOI] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Kim D, Landmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357-U121. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 51.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic acids research. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the corresponding author upon request. Uncropped western blots for Supplementary figure 3a-c; 8h are provided as Source Data. The BCR-seq data generated in this study is available at the SRA database (https://www.ncbi.nlm.nih.gov/sra), BioProject Accession IDs: PRJNA574931, PRJNA574906, PRJNA574628. The RNA-seq and microarray datasets generated in this study are available at the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), GEO Accession IDs: GSE129513 and GSE152960.
Data in this study that was obtained from publically available sources can be accessed at:
-
1)
Tabula muris scRNAseq data https://tabula-muris.ds.czbiohub.org/ Data names: 10X_P7_2 and 10X_P7_3
-
2)
Human cell atlas scRNAseq data https://data.humancellatlas.org/explore/projects/cc95ff89-2e68-4a08-a234-480eca21ce79 Data name: MantonBM1
Custom scripts for the analysis of BCR repertoire differences are available at: Immune_receptor_NETWORK-GENERATION https://github.com/rbr1/Immune_receptor_NETWORK-GENERATION