Abstract
p53 (also known as TP53) mutation and amyloid formation are long associated with cancer pathogenesis; however, the direct demonstration of the link between p53 amyloid load and cancer progression is lacking. Using multi-disciplinary techniques and 59 tissues (53 oral and stomach cancer tumor tissue samples from Indian individuals with cancer and six non-cancer oral and stomach tissue samples), we showed that p53 amyloid load and cancer grades are highly correlated. Furthermore, next-generation sequencing (NGS) data suggest that not only mutant p53 (e.g. single-nucleotide variants, deletions, and insertions) but wild-type p53 also formed amyloids either in the nucleus (50%) and/or in the cytoplasm in most cancer tissues. Interestingly, in all these cancer tissues, p53 displays a loss of DNA-binding and transcriptional activities, suggesting that the level of amyloid load correlates with the degree of loss and an increase in cancer grades. The p53 amyloids also sequester higher amounts of the related p63 and p73 (also known as TP63 and TP73, respectively) protein in higher-grade tumor tissues. The data suggest p53 misfolding and/or aggregation, and subsequent amyloid formation, lead to loss of the tumor-suppressive function and the gain of oncogenic function, aggravation of which might determine the cancer grade.
Keywords: p53, Mutations, Misfolding, Aggregation, Amyloid, Cancer grades
Introduction
p53 (also known as TP53) plays a vital role as a tumor suppressor, and carries out different functions related to apoptosis, cell cycle arrest, DNA repair, senescence and metabolism (Aubrey et al., 2018; Lane and Crawford, 1979; Levine, 1997; Mello and Attardi,2018; Vousden and Lu, 2002). p53 functional loss was initially found to be associated with cancer-related mutations that resulting in either destabilization of the protein folding or its inability to bind to its consensus DNA sequence (Rivlin et al., 2011; Silva et al., 2018; Wang and Fersht, 2015a; Joerger et al., 2006). Mutant p53 not only displays its loss of tumor suppressor functions but also demonstrates a gain of oncogenic properties (Lang et al., 2004; Xu et al., 2011; Liu et al., 2010). Moreover, point mutations in any of the p53 domains such as the N-terminal, DNA-binding, or the tetramerization) lead to nuclear or cytoplasmic accumulation of p53 in cancer cells as well as in cancer tissues (Rivlin et al., 2011; Moll et al., 1992). The mutant p53 displays gain-of-function by interacting with wild-type (WT) p53, the p53-related proteins p63 and p73 (also known as TP63 and TP73, respectively), and other transcription factors (Xu et al., 2011). This leads to activation of gene transcription related to apoptosis, cellular growth, resistance and metabolic reprogramming (Mantovani et al., 2019; Moll et al., 1992). However, loss and gain of function of p53 might not always be associated with p53 gene alterations as WT p53 misfolding and accumulation might also contribute to cancer (Moll et al., 1992). Recent data suggest that p53 aggregation and amyloid formation are also correlated with the loss of p53 tumor-suppressive function and the gain of oncogenic function (Ghosh et al., 2017). Moreover, our group has recently demonstrated that p53 amyloids have prion-like properties in cells and can induce cancerous transformation in normal cells (Navalkar et al., 2021, 2022). Although previous data suggest that p53 amyloids might be associated with cancer, the relationship between the extent of p53 amyloid formation and cancer disease severity (such as cancer grade) is still not established.
In the present work, using 59 tissues (53 oral and stomach cancer tumor tissue samples of different cancer grades from Indian individuals with cancer, and six non-cancer oral and stomach tissue samples), we investigated the extent of p53 amyloid formation in different cancer grades. We found an increased level of p53 amyloids in biopsies from higher grade cancers for all individuals for both oral and stomach cancer types. Although most of these p53 accumulations were associated with p53 mutations, WT p53 accumulation and amyloid formation were also observed in certain cancer tissues. Both WT and mutant p53 amyloids were localized in the nucleus and/or as cytoplasmic aggregates consisting of transcriptionally inactive p53. However, with increasing grades of both cancer types, p53 amyloids sequestered more of their paralogs, such as p63 and p73, suggesting widespread deactivation of tumor-suppressive function due to p53 amyloid formation. Overall, the study reveals that p53 amyloid formation is not only plausible cause of cancer initiation but might also be a positive or essential factor for cancer severity and progression.
Results
Increased p53 amyloid load is seen in tissues from higher cancer grades
Several studies have previously suggested the formation of p53 aggregates in several tumor tissues (Ghosh et al., 2017; Moll et al., 1995; Ostermeyer et al., 1996). In this study, we examined whether p53 amyloids can be used as a prognosis factor for the tumor grade. For this, we used a small cohort of human cancer biopsies from Indian individuals with different grades of oral and stomach cancer, which are prevalent in the Indian population. Furthermore, among various cancers, the prevalence of p53 mutations is also high in both oral and stomach cancers (Olivier et al., 2010). We first confirmed the cancerous status of these tissues using hematoxylin and eosin (H&E) staining (Fig. S1). The data showed differential H&E staining as well as neoplastic cells with intense nuclear staining, indicating the presence of hyperproliferative cells. To examine the p53 status and its accumulation into amyloids in cancer tissues, a double immunofluorescence colocalization study was performed using the amyloid-specific antibody OC (Kayed et al., 2007) and a p53-specific antibody (DO-1, Santacruz Biotechnology, Dallas, TX, USA). The data suggest a high colocalization of p53 with amyloid (OC signal) in all of the cancer tissues of oral (Figs 1 and 2) and stomach cancers (Fig. 3). However, the corresponding non-cancerous tissues of oral and stomach origin (Fig. S2) either showed lower levels of p53 accumulation and/or negligible p53 colocalization with OC antibody. In the 59 biopsies (53 tumor tissues and six non-cancerous tissues) (Table S1), we found that >90% of both oral and stomach tumor tissues were positive for p53 in the amyloid state. Interestingly, when analyzed grade-wise, the p53 amyloid content (OC antibody staining) was significantly higher in samples of higher tumor grade for both oral and stomach cancers (Fig. 4A). Similar observations were also seen when fluorescence colocalization studies were performed with p53 antibody and amyloid-specific dye ThioS staining with selected cancer tissues (Fig. S2B–D). Previous reports have suggested that there is p53 oligomer formation in various tumor tissues by using oligomer-specific A11 antibody (Kayed et al., 2003). When we performed double immunofluorescence study using A11 antibody (red) and p53-specific antibody (green), the lower-grade cancer tissue of both oral and stomach origin showed a high degree of p53 colocalization with oligomer-specific antibody, whereas the higher-grade tissue did not. This suggests that p53 oligomers might be formed at the initial stage of cancer, the levels which subsequently go down in the tissues of higher grades (Fig. S3A).
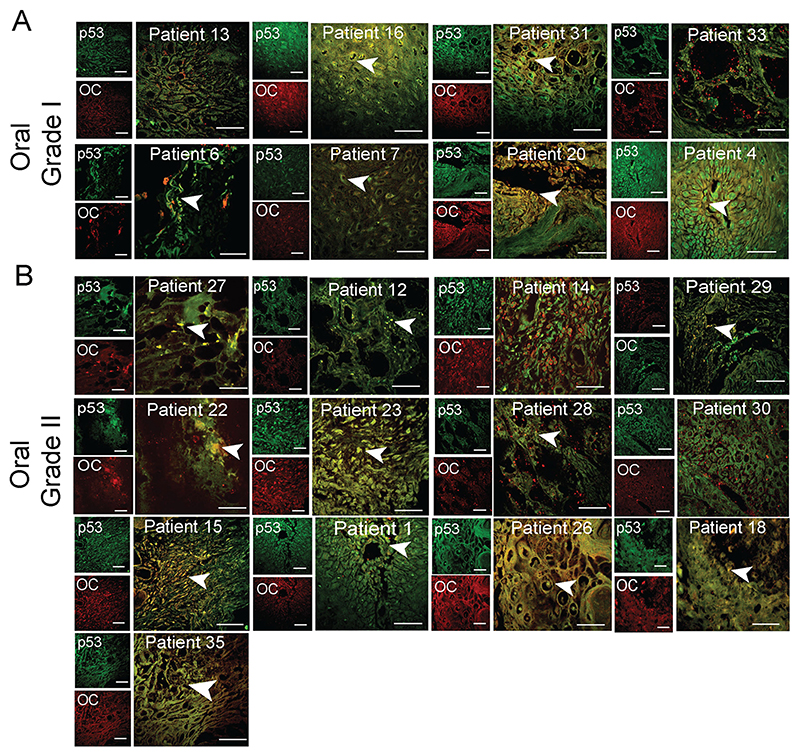
Fig. 1. p53 status in grade I and II of oral cancer tissues.
Immunohistochemistry using anti-p53 (DO-1) and anti-amyloid (OC) antibody showing p53 colocalization with amyloids. The yellow areas denote the colocalization of p53 and OC signals. Representative images of oral grade I (A) and oral grade II (B) are shown. The colocalized areas are shown with a white arrowhead. The patient number is denoted on all the images. Scale bars: 50 μm. Image representative of n=2 experiments.
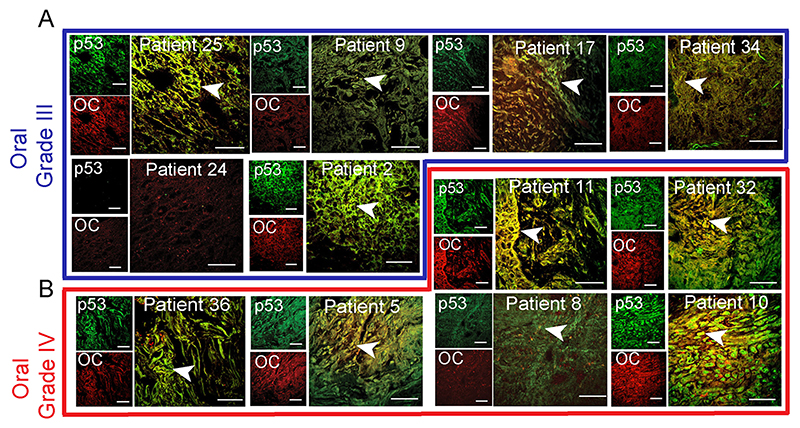
Fig. 2. p53 status in grade III and IV oral cancer tissues.
Immunohistochemistry using anti-p53 (DO-1) and anti-amyloid (OC) antibody showing p53 colocalization with amyloids. The yellow areas denote the colocalization of p53 and OC signals and marked with a white arrowhead. Representative images of oral grade III (A) and oral grade IV (B) were shown. The different oral cancer grades are highlighted in different color boxes or lines, oral grade III (blue) and oral grade IV (red). The patient number is denoted on all the images. Scale bars: 50 μm. Image representative of n=2 experiments.
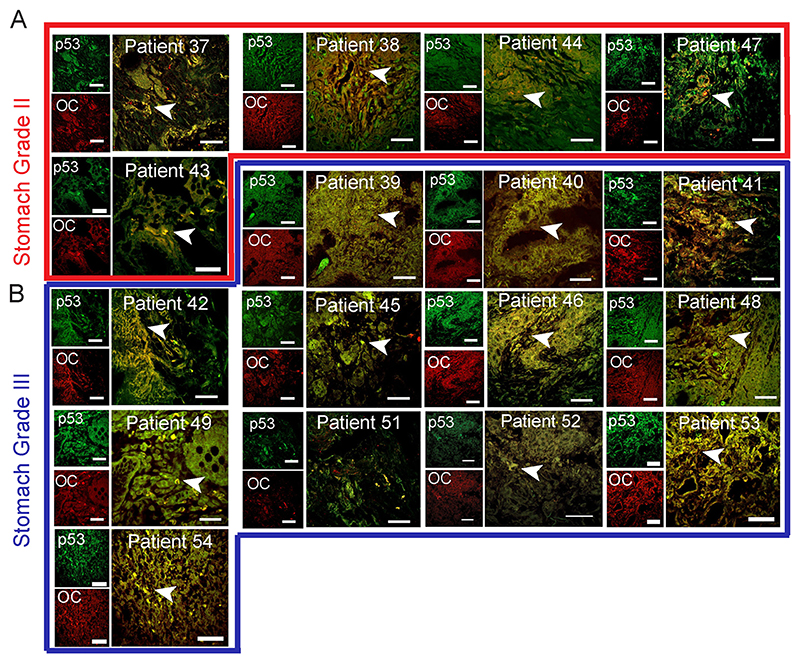
Fig. 3. p53 status in stomach cancer tissues.
Immunohistochemistry using anti-p53 (DO-1) and anti-amyloid (OC) antibody showing p53 colocalization with amyloids. The yellow areas denote the colocalization of p53 and OC signals and shown with a white arrowhead. Representative images of stomach grade II (A) and stomach grade III (B) are shown. The different stomach cancer grades are highlighted in different color boxes, stomach grade II (red) and stomach grade III (blue). The patient number is denoted on the images. Scale bar: 50 μm. Image representative of n=2 experiments.
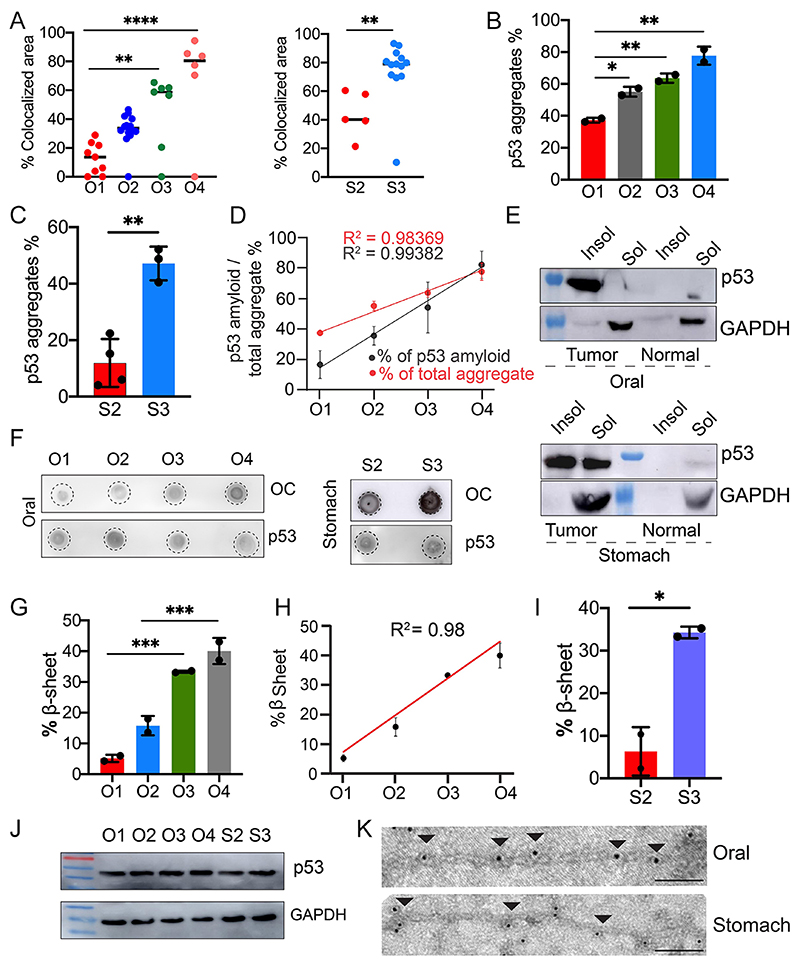
Fig. 4. p53 amyloid characterization in oral and stomach cancer tissues.
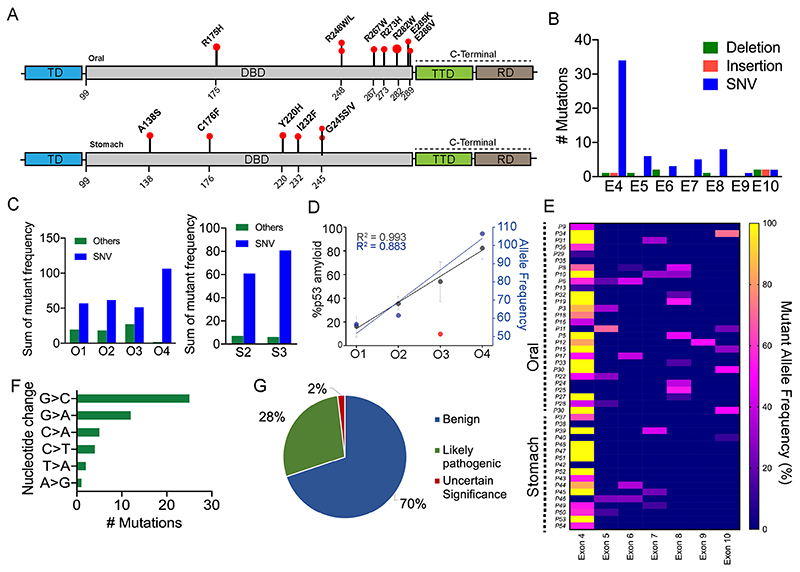
(A) The areas with colocalized of p53- and amyloid-specific OC antibody staining were quantified using ImageJ as the percentage of colocalized area for oral (left panel) and stomach (right panel), revealing that there is increased colocalization with higher cancer grades. The values were plotted as mean±s.e.m., n=3 independent experiments. (B,C) Total p53 aggregate quantification using a Seprion ligand-binding assay using anti-p53 antibody, showing the percentage of p53 aggregation for oral and stomach cancer tissues. The values are plotted as mean±s.d. (B) or mean±s.e.m. (C), for n=2 and n=3 individual data sets, respectively. (D) Correlation plot showing a strong positive correlation between the total p53 aggregation and p53 amyloid load in all the oral cancer grades. Results are mean±s.d. (n=2). (E) p53 expression as assessed by western blotting of the soluble (Sol) and insoluble (Insol) fraction from oral (top panel) and stomach (lower panel) tissues showing higher content of p53 in the insoluble fraction than in the soluble fraction. Normal, non-cancer tissues. GAPDH was used as a loading control. Image representative of n=2 experiments. (F) Dot blot showing amyloid content and p53 expression in different grades of oral (left panel) and stomach (right panel) cancer tissues. Image representative of n=2 experiments. (G–I) FTIR image analysis of oral tumor tissues with different grades, which was quantified for the amide I band in the 1620 cm−1 to 1640 cm−1 region showing an increased amount of β-sheet for the higher grades of cancer tissues. Percentage of β-sheet structure quantified from FTIR imaging of stomach cancer tissues showing the higher β-sheet structure in the higher grade of stomach cancer tissues. The values are plotted as mean±s.d., n=2 independent experiments. (J) p53 expression in tumor grades of both oral and stomach origin. The lower panel shows the expression of GAPDH for a loading control. Image representative of n=2 experiments. (K) Immunoelectron microscopy showing 10 nm gold particle decorations (arrowheads) on the fibrils due to the presence of p53 in fibrils isolated from oral and stomach cancer tissues. Scale bars: 200 nm. Images representative of n=3 independent experiments. O1–O4, oral grade I–IV; S2, S3, stomach grade II and III, respectively. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (one-way ANOVA followed by Tukey’s multiple comparison test).
Furthermore, to quantify the total p53 aggregation in various grades of cancer tissues, we used an enzyme-linked-immunosorbent-assay (ELISA) that is based on the specific binding of polyionic and high-molecular-mass ligand to aggregated proteins (Maritschnegg et al., 2018). Consistent with the double immunofluorescence data (Figs 1, 2 and 3), the levels of p53 aggregates were increased many folds in the higher grades of oral and stomach cancer tissues, compared to the corresponding lower grades (Fig. 4B,C), exhibiting a positive correlation in oral cancer tissues (R2=0.98) (Fig. 4D). We found a high correlation between the total p53 aggregates (ELISA) and p53 amyloid (OC staining) (Fig. 4D). Interestingly, we further observed that in lower-grade samples, the p53 amyloid pool was much lower than total p53 aggregate; however, in higher cancer grade samples, p53 amyloid predominated, exhibiting a positive correlation (R2= 0.98 for total p53 aggregates and R2= 0.99 for total p53 amyloid) (Fig. 4D). The higher amount of p53 amyloids in higher cancer grades was also consistent with western blot analysis of soluble versus insoluble p53 fractions (Fig. 4E; Fig. S3B,C) and dot-blot analysis (Fig. 4F; Fig. S3D,E) from lysate isolated from various grades of cancer tissue extracts. It is important to note that, in the non-cancerous tissues, only faint band of p53 in soluble fraction was observed. It is a well-known fact that p53 is not detectable in normal tissue and cells as it readily degrades and is negatively regulated by MDM2 (Marine and Lozano, 2010; Francoz et al., 2006).
Overall, the data suggest an increased extent of p53 amyloid formation in higher grade oral and stomach cancers. This high extent of p53 amyloid formation at the higher grades of cancer could be due to widespread misfolding and amyloid amplification of p53. We next examined whether higher p53 amyloid loads could be correlated with the cancer grade for other cancers using bioinformatic analysis. For this, we hypothesized that if p53 amyloid load were correlated with cancer grade, it would be expected that there would be a greater extent of altered gene expression patterns associated with p53 amyloids. In this context, our previous study showed uniquely altered gene expressions associated with p53 amyloids in cells in contrast to the control cells and in cells with p53 cancer-associated mutations (Navalkar et al., 2021,2022). We first analyzed the correlation of these altered genes (i.e. those associated with p53 amyloids) in head and neck cancer tumor samples using the UALCAN database (https://ualcan.path.uab.edu/). We found that the enrichment of these unique genes was well correlated with cancer grades (Fig. S4A). Similar observations were also obtained with other cancers (Fig. S4B-G). The data, therefore, clearly showed that a higher amount of p53 amyloids is directly associated with higher grades of cancer and support the idea that p53 amyloids act as an oncogene for promoting cancer pathogenesis.
Characterization of p53 amyloid in cancer tissues
To characterize the amyloid content of the various cancer tissues, we performed Fourier-transform infrared (FTIR) imaging (Miller et al., 2013). The FTIR imaging with snap-frozen tumor biopsies showed a higher amount of β-sheet content in higher-grade oral cancers compared to the corresponding lower-grade and non-cancer tissues (Fig. 4G,H; Figs S5 and S6). Similar observations were also obtained for stomach cancer biopsies (Fig. 4I). It is important to note that the higher β-sheet content could be due to other protein amyloids along with p53 amyloids. However, our combined study of immunohistochemistry and label-free FTIR imaging on the identical tissue section (used adjacent sections for both the study) support that the presence of p53 amyloid might be mostly responsible for higher β-sheet-rich structure in these tissue sections (Fig. 4G–I; Figs S5 and S6). However, there is a possibility that amount of p53 expression and stabilization might be higher in higher grades of cancer compared to lower grades. To examine this, western blot analysis was performed for two individuals from each grade (I, II, III and IV) tissue for oral, and two individuals from each grade tissue for stomach (II and III) cancers. The data showed similar expression of p53 from grade I towards the higher-grade cancer (Fig. 4J; Fig. S7A). As our double immunofluorescence data showed that in the lower grade of cancer tissue, p53 oligomers are present but these are not present in the higher grade (Fig. S3A), therefore, the amount of total p53 might be same but its state or conformation might differ among grades with p53 amyloid dominating in the higher grade of cancer tissues.
We further isolated the total amyloid pool from representative oral and stomach cancer tissues, which showed a fibril-like morphology when observed under the electron microscope (EM) (Fig. 4K). A FTIR study of these p53 fibrils showed the presence of intense peaks at ~ 1627 for oral and ~ 1632 for stomach cancer in the amide I region (Figs S5 and S6) suggesting there is a β-sheet-rich amyloid structure (Jackson and Mantsch, 1995) in these p53 aggregates. Indeed, immunoelectron microscopy (immunoEM) using p53 antibody (primary) and 10 nm colloidal gold-conjugated secondary antibody confirmed p53 amyloid, as gold particles were aligned along the length of isolated fibrils from oral and stomach cancer (Fig. 4K; Fig. S5C).
WT and mutant p53 are responsible for amyloid formation in tumor tissues
p53 mutations are known to be associated with 50% of human cancers (Soussi et al., 2006). Most often, p53 mutations result in the accumulation of p53 as a punctate appearance in various cancer cells and tissues (Moll et al., 1995). Furthermore, it has been shown that mutant p53 preferentially accumulates in the nucleus, whereas WT p53 sequesters in the cytoplasm of the cancer cells (Moll et al., 1995). These p53 accumulations might be due to WT p53 destabilization, which could be further induced by p53 mutations (Kim et al., 2009; Soussi et al., 2006; Wang and Fersht, 2015a). Given that we observed a grade-wise increase in p53 amyloid formation (Fig. 4) in cancer tissues, we examined whether there is any link between p53 amyloids and p53 mutations. For p53 mutational status, we performed next-generation sequencing (NGS) for 48 cancer tissues (44 tumor and 4 non-cancerous tissues) (Fig. 5; Table S1), which showed TP53 mutations in ~93% of the tumor tissues (Table S1). Several SNVs, deletions, insertions and stop-gain mutations were observed in both of these tumor tissues. Although, the highest mutation type observed was single nucleotide variants (SNVs) in the p53-encoding gene in both oral and stomach cancer tissues. Interestingly, SNVs including various hotspot mutations (R175H, E286V, R267W, R248W, R282W, R248L, and E285K), stop-gain mutations (such as R306/*, R196/*, Q317/* and R213/*), and insertion and deletions were mostly detected in the DNA-binding domain (DBD) of the TP53 gene (the region encoding amino acids 94–312) (Fig. 5A; Table S1) in oral cancer tissues. In the tetramerization domain (amino acids 325–356), there was only one SNV (R337C), along with a stop-gain mutation R342/*, and no mutations were detected in the transactivation domain of the p53-encoding gene (Fig. 5A; Table S1). This is consistent with previous p53 mutation data, which suggests that p53 mutations mainly occur in the p53 DBD (Rivlin et al., 2011). We further found that the missense mutations in the DBD were highest at exon 4 followed by exon 8 (Fig. 5B,E). When we analyzed the extent of p53 mutations in various cancer grades, we observed an increased extent of SNV mutations in a higher grade of cancer tissues (Moll et al., 1995; Liu et al., 2010; Skinner et al., 2012). This suggests that p53 mutations could be one of the primary factors for the destabilization and amyloid aggregation of p53 (Fig. 5C–E). Our results provide support for the correlation between p53 SNVs and amyloid formation in different cancer grades, with the exception of oral cancer grade III, which exhibited a minimal number of SNVs (Fig. 5D). Previous data indicate that certain p53 mutations, such as R175H, R248W, and R282W, have a higher likelihood of causing misfolding and aggregation of the p53 protein (Ghosh et al., 2017; Palanikumar et al., 2021; Ferretti et al., 2022). A similar mutational status was also observed in the case of stomach cancer tissues where SNVs (G245V, I232F, Y220H, A138S, and C176F), deletions, and insertions were frequently observed in the DBD. We, however, observed neither stop-gain mutations nor SNVs in other domains of the p53-encoding gene. Furthermore, the transition of G>C was observed to be the highest in these tissue samples (Fig. 5F) compared to other transitions such as G> A, C>A, T>A, and A>G. It is important to note that many of these mutations that we observed are already known to be directly associated with human cancer (Mello and Attardi, 2013; Monti et al., 2020; Moll et al., 1995; Bauer et al., 2020; Manterola et al., 2018). However, for some mutations (e.g. P72R), further studies are required for their role in cancer pathogenesis (Fig. 5G). Although most of the cancer tissues containing p53 amyloids possess frequent mutations in p53 gene, we, however, observed p53 amyloids with WT protein in cancer tissues (patient numbers 35, 38 and 42). These data indicate that the misfolding, and aggregation followed by amyloid formation by p53, which is further promoted by cancer-associated mutations, might result in a higher amount of p53 amyloids in higher grades of cancer tissues.
Fig. 5. Detection of mutations in human oral and stomach biopsies by NGS.
(A) Schematic of p53 domains with a lollipop plot showing the SNVs observed in the oral biopsies (upper panel) and stomach biopsies (lower panel). TD, transactivation domain; TTD, tetramerization domain; RD, regulatory domain. (B) Cohort samples showing the total mutations observed in different exons (denoted E4-E10) of the p53 gene. (C) Alternative allele frequency showing variation in mutation types for different grades of cancer tissues. (D) Correlation plot showing percent p53 amyloid and alternate allelic frequency of SNVs in different oral cancer grades. Results are mean±s.d. (n=2). (E) Heat map of all the cohort samples showing the mutation frequency in different p53 exonic regions. (F) Mutation frequency among different patients in the cohort showing nucleotide substitution. (G) Pie chart showing the clinical significance of the NGS-detected mutations as obtained from NCBI database search.
Cytoplasmic versus nuclear sequestration of misfolded p53 in tumor tissues
On the onset of stress (e.g. DNA damage), phosphorylated p53 enters the nucleus to carry out the transcriptional function by binding to its cognate DNA sequence (Sammons et al., 2020). However, p53 can be excluded from the nucleus (Lu et al., 2000) and shuttle between the nucleus and cytoplasm, with the nuclear localization signal (NLS) as well as nuclear export signal (NES) playing an essential role for this nucleocytoplasmic transportation (Liang and Clarke, 2001). Previous reports have suggested that mutant p53 accumulates preferentially in the nucleus, whereas WT p53 protein is sequestered and stabilized in the cytoplasm, thus rendering it non-functional (Moll et al., 1992).
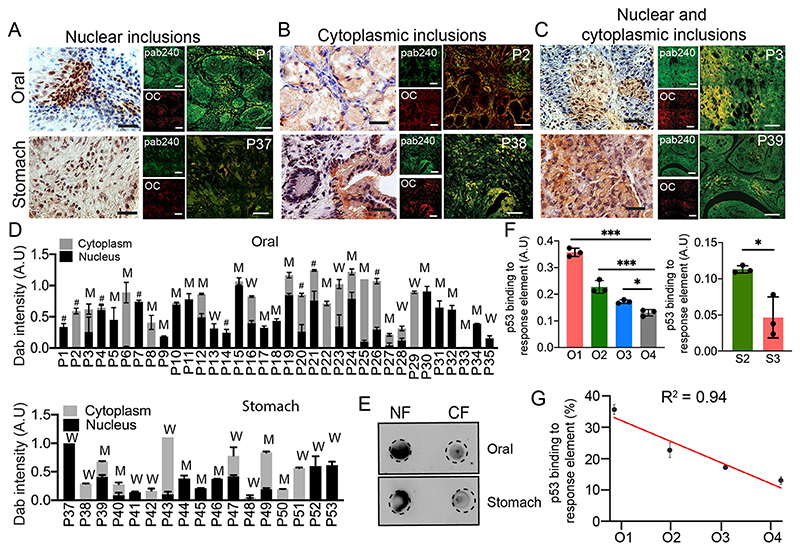
Given that we examined p53 amyloids in a relatively large number of cancer tissues with different grades and established the p53 mutational status, we examined the correlation between the mutational status of p53 and the location of p53 amyloids. For this, we used 3,3’-diaminobenzidine (DAB) staining for all tumor biopsies using a Pab240 monoclonal antibody (Santa Cruz Biotechnology), which recognizes misfolded p53 under non-denaturing conditions (Figs 6 and 7A–C). However, to examine whether DAB staining indeed recapitulates the p53 amyloids, we parallelly examined co-immunofluorescence with Pab240 and the amyloid-specific OC antibody or amyloid-specific dye ThioS (Fig. 7A–C, right panels; Fig. S7C–E) with selected tissues of different grades. The colocalization experiments with Pab240 antibody and OC antibody or ThioS dye (Fig. S7C–E) revealed colocalization of misfolded p53 with OC antibody in all cancer tissues (Fig. 7A–C) but not in the corresponding non-cancerous tissues (Fig. S7B). Thus, DAB staining along with p53 mutational status will allow us to understand the effect of p53 mutations on the localization of amyloid p53 in cancer tissues. The localization of p53 in selected biopsies (nuclear localization of p53 by immunofluorescence) was further confirmed by dot blot (Fig. 7E) analysis of nuclear and cytoplasmic extracts of these biopsies (from P1 and P39). The DAB staining of cancer tissues showed three distinct nucleo-cytoplasmic staining patterns (Fig. 7A–C). A total of 18 oral biopsies out of 36 (~50%) and seven stomach biopsies out of 17 (~41%) displayed only nuclear staining suggesting the accumulation of high levels of misfolded p53 protein in the nucleus (Fig. 7). Only cytoplasmic accumulation of p53 was observed in four oral biopsies out of 36 patients (~11%) and five stomach biopsies out of 15 (~33%). In all other biopsies [12 oral (~33%) and five stomach biopsies (~29%)], we observed p53 to localize in the nuclear as well as in the cytoplasm. When p53 localization is compared with mutational status, we observed that hot spot mutations either accumulate in the nucleus (such as R337C, R267W and R248W in oral cancer) or in both nucleus and cytoplasm (R175H and G245V). Next, we analyzed the extent of p53 localization in the nucleus versus cytoplasm in all the cancer tissues with pab240 antibody staining using ImageJ (Fig. 7D). Our data reveal that there is no correlation between p53 mutations and their sequestration either in the nucleus or in the cytoplasm as both WT and mutant p53 are seen to be sequestered in both nucleus and/or cytoplasm (Fig. 7D).
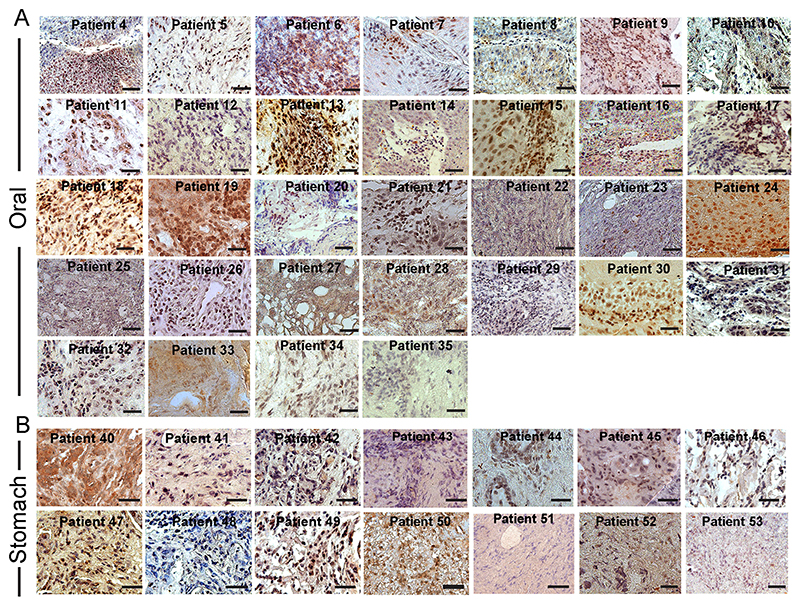
Fig. 6. Nuclear versus cytoplasmic inclusion of p53 in oral and stomach cancer tissue biopsies.
Immunohistochemical study showing misfolded p53 inclusion in (A) oral and (B) stomach cancer tissues using Pab240 antibody (Santa Cruz Biotechnology) and subsequently developed by DAB substrate (dark brown to light brown due to binding affinity). The tissues were counterstained with Harris Hematoxylin (blue and purple color). The data show nuclear versus cytoplasmic expression of p53 in various cancer tissues. Scale bars: 50 μm. Images representative of n=3 independent experiments.
Fig. 7. Nuclear and cytoplasmic localization of p53.
Immunohistochemical staining (DAB) for misfolded p53 using Pab240 antibody showing the representative image of nuclear inclusions (A), and cytoplasmic (B) and nucleo-cytoplasmic inclusion (C) for cancer tissues. Corresponding p53 amyloids are shown using colocalization of Pab240 antibody and amyloid-specific OC antibody staining. Representative images of both the oral (upper panel) and the stomach (lower panel) are shown. Images in A–C are representative of n=3 independent experiments. P denotes the patient number. Scale bars: 50 μm. (D) DAB intensity quantification (A.U, arbitrary units) showing the relative extent of misfolded p53 localization in nuclear, cytoplasmic and nucleo-cytoplasmic fractions as measured using ImageJ for individual oral (upper) and stomach (lower) cancer tissues. The data was plotted as a stacked bar plot with the average DAB intensity of one patient. The W denotes WT p53 in cancer tissues, M denotes mutated p53 (SNVs, deletions or insertions), and # denotes those tissue samples not analyzed by NGS. Results are given as mean±s.e.m. (n=3). (E) Dot blot using Pab240 showing higher load/inclusions of misfolded p53 in the nuclear fraction (NF) than in the cytoplasmic fraction (CF) for a nuclear-localized patient tissue. (F) p53 DNA-binding ability using tissue lysate of different grades of oral tissues (left panel) and stomach tissues (right panel) showing significant loss of p53 binding to DNA in higher grades of cancer. Results are given as mean±s.e.m. (n=3). (G) A correlation plot of p53 activity with oral cancer grades showing a decrease in p53 DNA-binding ability with an increase in cancer grades. O1–O4, oral grade I–IV; S2, S3, stomach grade II and III, respectively. Results are mean±s.d. (n=3). *P<0.05, ***P<0.001 (oneway ANOVA followed by Tukey’s multiple comparison test).
To further analyze whether these aggregated and misfolded p53 have DNA-binding ability, we performed an ELISA assay using tissue lysates from nine oral (Fig. 7F, left panel) and three stomach cancer biopsies (Fig. 7F, right panel). The three tumor biopsies were chosen from each cancer grade. In all these cases and irrespective of p53 localization, the sequestered p53 was observed to be transcriptionally inactive due to its inability to bind to DNA. We also observed a much lesser DNA-binding capacity (higher p53 inactivation) in higher grades of cancer (Fig. 7F,G). It is important to note that one of the stomach cancer tissue samples harboring WT p53 amyloid also displayed no p53 DNA-binding capacity, suggesting its transcriptional inactivation. Therefore, the data suggest that irrespective of localization or mutation, the higher amount of p53 accumulation into amyloids resulted in the inability of p53 to bind to cognate DNA sequences in the higher grades of cancer.
p53 loss of function and higher sequestration of p63 and p73 with p53 amyloids in higher grades of cancer
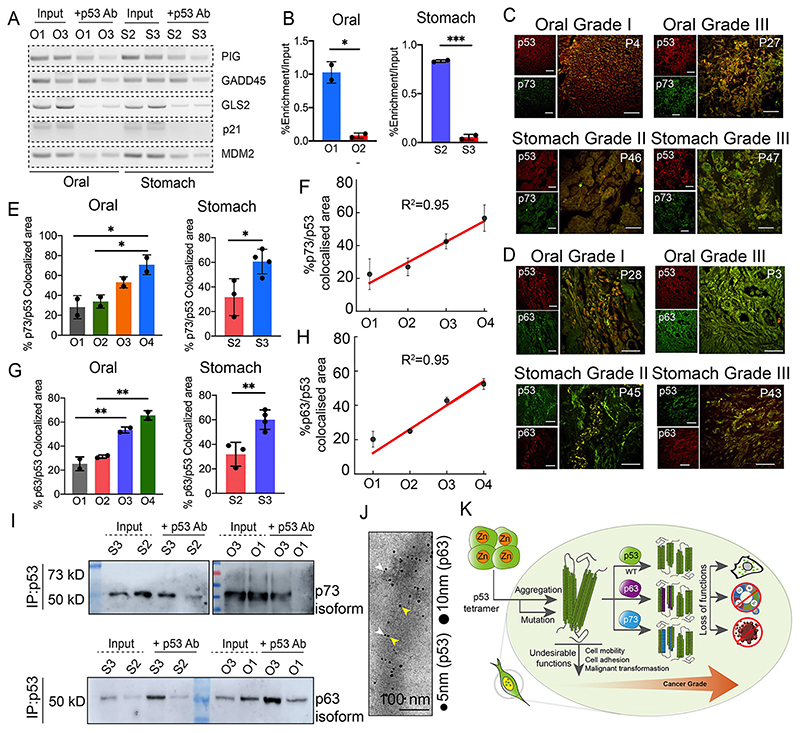
p53 amyloid formation has been shown to cause a loss of its tumor-suppressive abilities as well as a gain of oncogenic functions (Ghosh et al., 2017; Navalkar et al., 2021, 2022, 2020; Sengupta et al., 2022). As p53 is a transcription factor, it can recognize its target genes by binding to a consensus binding element that is located at the gene promoter (Rivlin et al., 2011). To examine the loss of function due to the gradual increase of p53 amyloids in higher grades of tumor tissues, we analyzed six oral and six stomach cancer tumor biopsies (three individuals each with cancer grade I and grade III of oral cancer, and three individuals each from cancer grade II and III of stomach cancer) by chromatin immunoprecipitation (ChIP) assay using an anti-p53-DO1 antibody (primer sequence listed in Table S2). With increased p53 accumulation (amyloids) from grade I to III, we observed a reduction in the extent of p53 bound to response elements (responsible for apoptosis or cell cycle arrest, such as in p21 (also known as CDKN1A), PIG (also known as TP53I3) and Gadd45 (GADD45A) (Fig. 8A,B). In contrast, lower-grade tumors with low p53 accumulation (amyloids) showed a high amount of p53 bound to response elements (Fig. 8A,B). The observation is also further confirmed by quantitative (q)PCR analysis using the p21 gene (Fig. 8B). The data suggests that more p53 accumulation as amyloids results in a lower amount of p53 being bound to its response element in higher cancer grades. We further hypothesized that widespread p53 inactivation, preventing it acting as a tumor suppressor, and a gain of p53 oncogenic functions in higher grades of cancer might happen not only due to the gradual increase of p53 amyloids but also due to the sequestration of other tumor suppressor proteins (e.g. p63 and p73) by p53 amyloids. In this context, it has been shown that p53 aggregates sequestered p53 paralogs, such as p63 and p73, in cells (Xu et al., 2011). Furthermore, p63 and p73 are known to be rarely mutated in tumors; however, their tumor suppressor functions are frequently inhibited by mutant p53 (Inoue and Fry, 2014). To examine the sequestration of p63 and p73 in p53 amyloids in these cancer tissues, co-immunofluorescence experiments were performed using p53 with p73 or p63 antibodies. We analyzed 12 oral and six stomach cancer samples (three individuals from each cancer grade). p53 was observed to colocalize with p73 (Fig. 8C) and p63 (Fig. 8D) in all the cancer grades for both oral and stomach cancer biopsies. The ImageJ analysis suggests that the percentage of colocalization was significantly greater in the higher cancer grades for both oral and stomach cancers (Fig. 8E–H). In oral cancer, the colocalization of p53 and p73 was ~30% in grade I, which increased up to 70% in grade IV, suggesting that p53 and p73 colocalization is highly correlated with oral cancer grade (Fig. 8F). Similar observations were also seen between stomach grade II (20% colocalization) and grade III (60% colocalization). Similar to p53 and p73 colocalization, we also observed a higher degree of colocalization of p53 and p63 in higher cancer grades for both oral and stomach cancer tissue. The colocalization of p53 and p63 was ~20–25% in oral and stomach grade I tissues, which increased to ~60% for both oral grade IV and stomach grade III tissues (Fig. 8G,H).
Fig. 8. Loss of tumor-suppressive function and gain of oncogenic function by p53 due to p53 amyloids in cancer tissues.
(A) Chromatin immunoprecipitation (ChIP) assay showing increased loss of p53 binding to its response element in higher grades when compared with lower grades. p53, is unable to bind to the response elements of PIG, GADD45A, GLS2 (also known as glutaminase 2), p21 and MDM2. Input represents 5% of the whole-cell extract. Image representative of n=2 experiments. (B) Quantitative real-time PCR verifying the functional loss of p53 from oral (left panel) and stomach (right panel) tissues showing its inability to bind to p21 promoter in a higher grade of cancer. The values were plotted as mean±s.e.m., n=2 independent experiments. (C) Double immunohistochemistry [using anti-p53 antibody (DO-1) and anti-p73 antibody] showing colocalization of p53 and p73 signals in oral cancers of different grades (O1, O3, upper panel) and stomach cancer (S2, S3, lower panel). The colocalization is higher in higher cancer-grade tissue. Images shown are from n=3 independent experiments. (D) Double immunohistochemistry showing colocalization of p53 and p63 signals in oral cancers (O1, O3, upper panel) and stomach cancer (S2, S3, lower panel). Image representative of n=3 experiments. P denotes the patient number. Scale bars: 50 μm. (E) Quantification of p53-p73 colocalization using ImageJ showing a significant increase in the extent of colocalization with increased cancer grades for both oral (left) and stomach (right panel). The values were plotted as mean±s.e.m., n=2 independent experiments. (F) Plot showing a high correlation between the percentage of p53-p73 colocalization and higher oral cancer grades. (G) Quantification of p53-p63 colocalization using ImageJ showing a significant increase in the co-aggregation with an increase with higher cancer grades for both oral (left) and stomach (right panel). The values were plotted as mean±s.d., n=2 and n=3, respectively, independent experiments. (H) Correlation plot showing a high positive correlation between the percentage of p53/p63 colocalization with higher oral cancer grades. (I) Western blot analysis of immunoprecipitated p53 from the stomach and oral cancer tissues using p73 and p63 antibodies showing that the p73 and p63 isoforms are co-immunoprecipitated (IP) with p53. The amount of p73 and p63 isoforms was higher in the higher cancer grades than in the corresponding lower grades. Input represents 5% of the whole-cell extract. Blots shown representative of n=2 experiments. (J) Co-immunoelectron microscopy showing 5 nm (gold-labeled secondary antibody against primary antibody for p53, white arrowheads) and 10 nm gold (gold-labeled secondary antibody against primary antibody for p63, yellow arrowheads) particle decorations on the fibrils due to the presence of p53 and p63 co-aggregation in fibrils isolated from grade III oral cancer tissues. Scale bars: 200 nm. Image representative of n=2 experiments. (K) Schematic showing p53 amyloid load is corelated with the increase in cancer grades. p53 amyloid can sequester other family members, p63 and p73 which are higher in higher cancer grades. p53 amyloid results in loss of function and gain of oncogenic properties of p53. O1-O4, oral grade I–IV; S2, S3, stomach grade II and III, respectively.
Next, to directly confirm the p53–p73 and p53–p63 co-aggregation, p53 was immunoprecipitated using anti-p53 antibody from oral biopsies of grade I (P4), III (P27)) and stomach biopsies of grade II (P46), III (P47) followed by western blot analysis with anti-p73 and p63 antibodies. The western blot signal confirms the presence of p73 and p63 isoforms along with immunoprecipitated p53 in higher-grade cancer samples (grade III) for both oral and stomach tissues (Fig. 8I; Fig. S8A–D). Based on the molecular mass of these proteins, we conclude that these are isoforms of p63 and p73 that could possibly be co-aggregating with the p53 fibrils in both the cancer tissue types owing to a gain-of-function property displayed by the p53 amyloids (Fig. 8I,K). However, in some of the cancer samples, we also observed faint expression of full-length p73 along with immunoprecipitated p53 in both oral and stomach samples (Fig. S8C). Furthermore, to examine the co-immunoprecipitation of p53 and p63, and p53 and p73, we also immunoprecipitated using antibody of p63 and p73 and then performed western blots with anti-p53 antibody (DO1). We indeed found a similar observation that, along with both p63 and p73, we found the presence of p53 (Fig. S8D). This suggests that p63 and p73 co-aggregate with p53. We further confirmed this using double immunoelectron microscopy of an oral cancer tissue (grade III) with an anti-p53 antibody (DOI) and anti-p63 antibody. We found both species in single fibrils (Fig. 8J; Fig. S8E). Moreover, it is important to note that due to the higher colocalization, at this moment, we were not able to examine whether p63 or p73 is also in the amyloid state using OC or ThioS staining in cancer tissues. A future study is required to determine whether p63 or p73 alone can form amyloid independently of p53, which can be associated with cancer.
Discussion
Amyloid formation is usually associated with neurodegenerative diseases, such as Alzheimer’s, Parkinson’s and prion disease (Chiti and Dobson, 2017; Dobson, 2001). Owing to amyloid formation, there is a loss of particular protein functions, and a gain of toxic functions, which leads to cell death and neurodegeneration (Eisenberg and Jucker, 2012). In contrast to disease-associated amyloid, amyloids that are associated with the normal function of the host organism have also been discovered, which are termed ‘functional amyloids’ (Maji et al., 2009; Liebman and Chernoff,2012; Fowler et al., 2005; Otzen, 2010). For example, yeast prions show prion-like transmissive properties similar to human prion proteins but provide a survival advantage to the host organism against harsh environmental conditions (Liebman and Chernoff,2012, 2012; Halfmann et al., 2012). Several mammalian functional amyloids have also been discovered, where these amyloid support normal function rather than causing cell death (Barnhart and Chapman, 2006; Maji et al., 2009; Fowler and Kelly, 2012; Chatterjee et al., 2022). Recently, it was suggested that p53 aggregation and amyloid formation are associated with loss of the normal tumor-suppressive function of p53 and gain of an oncogenic function (Ghosh et al., 2017; Navalkar et al., 2020, 2021, 2022; Silva et al., 2018; Marques et al., 2022). These studies suggest that p53 amyloids could serve as an oncogene and might cause cancer initiation in cells (Ghosh et al., 2017; Navalkar et al., 2021, 2022; Sengupta et al., 2022). Moreover, p53 amyloid formation not only leads to its loss of tumor-suppressive properties but might also cause the acquisition of tumorigenic function in cells by sequestering other tumor suppressor proteins (Xu et al., 2011) and/or by prion-like p53 amyloid amplification (Forget et al., 2013; Ghosh et al., 2017; Navalkar et al., 2021) similar to what is seen for the prion protein (Prusiner, 1998; Aguzzi and Heppner, 2000).
In contrast to most of the neurodegenerative amyloid diseases (Chiti and Dobson, 2017; Wolfe and Cyr, 2011; Crews and Masliah,2010), the p53 amyloid load in relation to the prognosis of cancer severity/grade had not been previously examined. To examine the relationship between p53 amyloid and cancer grades, we studied various grades of biopsies of the stomach and oral cancers from Indian individuals. We found that all the stomach and oral cancer tissues under study contained p53 amyloids (Figs. 1, 2 and 3). Interestingly, we observed an increase in p53 amyloid with increased cancer grades for both cancer types (Fig. 4). These data support the idea that widespread p53 deactivation and oncogenic gain of function of p53 could be associated with p53 amyloids. This is further supported by the fact that these p53 aggregates are neither functional nor able to bind the cognate DNA sequences necessary for their apoptotic activity, which in turn supports more cell survival in cancer, similar to action of the functional amyloid for yeast prion (Bradley et al., 2002; Edskes et al., 2014; Ness et al., 2002). Indeed, tumors containing p53 amyloids showed a striking reduction in the amount of p53 bound to response elements (genes responsible for apoptosis or cell cycle arrest, such as p21, PIG and Gadd45A) (Fig. 8A) in higher cancer grades than in the lower grades, which is expected to be due to greater accumulation of p53 amyloid. Hence, p53 amyloid formation probably gives more advantage to the cells leading to cancer progression (Fig. 8K). Previous data have suggested that cancer-associated mutations destabilize the p53 functional fold, which might induce aggregation and amyloid formation (Wang and Fersht, 2015b; Moll et al., 1992, 1996; Wang and Fersht, 2012; Levy et al., 2011; Rangel et al., 2014). Aggregation of wild-type p53 is also known in cancer cells and tissues (Moll et al., 1992, 1996; Ostermeyer et al., 1996; Wang and Fersht, 2015b). Consistent with this, the sequence information of the p53 gene in our cohort revealed p53 mutations in most of the cancer tissues, whereas a small number of tissues had WT p53 (Fig. 5). Interestingly, in contrast to a previous study (Moll et al., 1995), our sequence data and p53 amyloid localization (using DAB staining) study showed that both wild-type and mutant p53 form nuclear, cytoplasmic and nucleo-cytoplasmic p53 amyloids (Figs 6 and 7). Irrespective of nuclear or cytoplasmic localization and/or mutation status (WT or mutant protein), p53 showed severe functionality loss in these cancer tissues suggesting that misfolding of WT or misfolding initiated by p53 mutation can produce p53 amyloids, with subsequent loss of normal p53 function and a gain of tumorigenic function (Fig. 8). Consistent with this observation, it has been shown previously that many cancer tissues contain p53 aggregates both in the cytoplasm and in the nucleus and this occurs for both mutant p53 and WT p53 (Bosari et al., 1995, 1994; Moll et al., 1992; De Smet et al., 2017; Navalkar et al., 2020). Moreover, neuroblastoma and other cancer cells show cytoplasmic WT p53 segregation leading to p53 inactivation (Moll et al., 1996, 1992, 1995; Bosari et al., 1994, 1995). A previous study has demonstrated a correlation between cytoplasmic p53 deposits and poor prognosis for high-grade serous ovarian carcinoma patients (Iwahashi et al., 2022). The study indicated that oncogenic cytoplasmic p53 aggregates can contribute to disease progression. In contrast, our study shows that the presence of p53 amyloid is correlated with cancer grades irrespective of the localization of these amyloids in the nucleus and/or in cytoplasm, suggesting that extent of p53 aggregation and amyloid formation could be more predictive for tumor prognosis in these cancers.
Although, at this point, it is unclear how WT p53 would misfold, aggregate and form amyloid; it is, however, known that Zn2+ depletion (Wilcken et al., 2012; Butler and Loh, 2003) or the presence of various cellular factors (such as cholesterol secosterol aldehydes in inflammation-associated cancer; Nieva et al., 2011) might contribute to WT p53 aggregation and amyloid formation. It is interesting to note that individual domains of p53 are indeed amyloidogenic in nature (Ishimaru et al., 2003; Lee et al., 2003; Rigacci et al., 2008; Higashimoto et al., 2006); therefore, unless regulated and stabilized by p53-stabilizing chaperons such as CCT (also known as TRiC) (Trinidad et al., 2013), there is a possibility that a certain amount of p53 after synthesis could attain a misfolded state (unless degraded by proteasomal degradation), which might transform into the amyloid form. The minute amount of p53 amyloid also could be further amplified, similar to prion-like amplification. Furthermore, previous studies have suggested that truncated p53 isoforms, which are more aggregation-prone proteins, unless regulated by chaperon or other tumor suppressors, might lead to aggregation and cause oncogenicity of cells (Arsic et al., 2021). For example, aggregates of Δ133p53β are present in various cancer cells and tumor tissue biopsies. The aggregation and activity of Δ133p53β is further controlled by p63 family proteins or CCT chaperon complex (Arsic et al., 2021). Similarly, another truncated p53 isoform, Δ40p53, has been found to form cytoplasmic amyloid-like aggregates in cells (Melo Dos Santos et al., 2019), suggesting that, unless tightly regulated, both truncated p53 isoforms and full-length p53 might aggregate and cause oncogenicity to the cells.
In addition, there is the possibility that p53 aggregation and/or amyloid formation, either exclusively in the nucleus or cytoplasm or both compartments, could be prognostic factors, depending on the cancer type. The higher amount of p53 amyloid with increasing cancer grade suggests that, similar to prion-like amplification and spread (Ness et al., 2002; Chiti and Dobson, 2006; Bradley et al., 2002; Iwahashi et al., 2022), p53 amyloid amplification might result in widespread p53 inactivity and a gain of tumorigenic function in higher grade cancers. We further asked whether, apart from self-amplification, the p53 amyloid might also sequester other p53 paralogs such as p63 and p73 (as shown for p53 aggregates in cells; Xu et al., 2011), leading to a dominant-negative effect as proposed for p53 mutations (Mantovani et al., 2019). Both p63 and p73 are known to display transcriptional activity (Dötsch et al., 2010; Blandino and Dobbelstein, 2004) and have tumor-suppressor functions (McKeon, 2004). Therefore, p63 and p73 sequestration by p53 amyloids might render them non-functional resulting in disease progression and an increase in cancer grade. Indeed, we found increased colocalization of p63 and p73 isoforms with p53 amyloid in higher grades of both cancers (Fig. 6). The data suggest that p53 amyloid formation, its prion-like amplification and sequestration of other p53 paralogs might provide conducive environments for higher cancer grades. The present data, therefore, demonstrate that increased p53 amyloid formation can be a prognosis factor for cancer grade, and p53 aggregation inhibitors (Soragni et al., 2016; Palanikumar et al., 2021; Ferretti et al., 2022) might be a valuable target against cancer.
Material And Methods
Chemicals and reagents
All the chemicals and reagents used for the study were of the high purity and obtained from either Sigma-Aldrich (St. Louis, MO, USA) or Merck (Darmstadt, Germany). De-ionized and double-distilled water was obtained from a Milli-Q (MQ) system (Millipore Corp., Bedford, MA, USA). The DNA extraction kit was obtained from Qiagen. The seprion ELISA kit was obtained from Microsens Biotechnologies (Cambridge, UK). The p53 activity ELISA kit was obtained from Cayman Chemicals (George Town, USA).
Tumor biopsies from Indian individuals with cancer
A cohort of freshly frozen oral and stomach cancer and their corresponding non-cancer tissues of human origin were procured from the National Tumor Tissue Repository situated at the Tata Memorial Hospital, Mumbai, India. The entire study and the protocols were approved by the Institutional ethics committee (IITB-IEC/2019/046), Indian Institute of Technology Bombay, Mumbai, India (IIT Bombay). The National Tumor Tissue Repository situated at Tata Memorial Hospital, Mumbai India obtained informed consent from all tissue donors and all clinical investigation have been conducted according to the principles expressed in the Declaration of Helsinki. The study included 59 tissues (53 tumor tissues and six non-cancerous tissues). These tissues were segregated into different grades. For oral cancer tissues, four grades (grade I, II, III, and IV) were used, whereas, for stomach cancer tissues, grade II and grade III were used. Details of all the tissues are mentioned in Table S1. A minimum of five tissues per grade were selected for the study; however, in some grades, more than five tissues were used as per the prevalence. For the NGS study, a total of 48 tissues (44 tumors and four non-cancerous tissues) were taken to understand the p53 mutational status. For IHC studies, all 59 tissues were analyzed. The tissues were fixed and dehydrated, followed by clearing with xylene. The wax infiltration was carried out, and the tissues were sectioned (3–5 μm thickness) using a microtome and embedded onto glass slides.
H&E staining
The tissue sections were deparaffinized using the decreasing concentrations of xylene, starting with 100% xylene, followed by xylene and ethanol in a 1:1 ratio. This was further followed by rehydration in decreasing concentrations of ethanol from 100% to 50%, and finally washed with distilled water. Sections were stained for 2 min with 0.5% hematoxylin solution. Subsequently, 0.8% eosin, prepared in 95% ethanol was prepared to stain the sections for 1 min, and the slides were kept for 1 h in xylene. After that, the sections were mounted using dibutyl-phthalate polystyrene xylene (DPX) mounting medium and observed under a Leica DMi8 (Leica Microsystems, Germany) fluorescence microscope fitted with Andor Zyla cCMOS camera (Oxford Instruments, UK).
Immunohistochemistry of tissues
Paraffin-embedded fixed tumors and the corresponding non-cancerous tissue sections were used for the immunohistochemistry study. The entire work plan and protocols were approved in advance by the Institutional ethics committee (IITB-IEC/2019/046), IIT Bombay, Mumbai, India. The tissue sections were first deparaffinized and rehydrated, as mentioned above. The antigen retrieval was performed by incubating the sections for 2 min with TrypLE™ Express Enzyme (Thermo Fisher Scientific) at 37°C. The tissue sections were then washed with Tris-buffered saline with 0.1% Tween-20, pH 7.4 (TBST), and then incubated with 0.2% Triton X-100 in TBST for 10 min. The section blocking was performed with 2% BSA in TBST. The sections were then incubated overnight at 4°C with primary antibodies, such as mouse monoclonal anti-human p53 protein DO-1 (1:200; cat. no. sc-126, Santa Cruz Biotechnology, Dallas, TX, USA) or anti-human pab240 (1:500; cat. no. sc-99, Santa Cruz Biotechnology) and rabbit polyclonal oligomer-specific A11 (1:500; Kayed et al., 2003) or amyloid-specific (OC) antibody (1:500; Kayed et al., 2007). Tissues were washed with TBST followed by incubation with the secondary antibody, such as anti-mouse-IgG conjugated to FITC-488 (1:1000) or goat anti-rabbit-IgG conjugated to Alexa Fluor-647 (Life Technologies, Thermo Fisher Scientific) at room temperature for 2 h. To study the coaggregation of p53 with p63 or p73, rabbit monoclonal anti-human p53 protein SP5 (1:200; cat. no. MA5-16387, Invitrogen, USA) and mouse monoclonal anti-p63 antibody [1:500; cat. no. (D-9): sc-25268] or anti p73 antibody [1:500; cat. no. (E-4): sc-17823, Santa Cruz Biotechnology].
The Thioflavin S (Thio S) staining was performed after immunostaining with an anti-p53 primary antibody and subsequent incubation with an Alexa Fluor-555-conjugated anti-mouse-IgG secondary antibody (1:1000 dilution). The sections were then stained for 2 min with filtered 0.6% Thio S solution (Sigma-Aldrich) in the dark. The sections were washed twice with initially 50% ethanol and then with TBST buffer. The sections were then mounted using 1% 1,4-diazabicyclo-[2.2.2] octane (DABCO, Sigma-Aldrich) that was prepared in 90% glycerol and 10% phosphate-buffered saline (PBS) and left for drying. Imaging was performed using Zeiss Axio Observer.Z1 inverted confocal fluorescence microscope (Zeiss, Germany), which is fitted with a high-speed microlens-enhanced Nipkow spinning disc (CSU-X1, Yokogawa Electric Corporation, Tokyo, Japan).
Isolation of the tissue amyloid fibrils
The total pool of amyloid fibrils from oral and stomach tumor tissues was isolated using previously reported methods (Haltia et al., 1990) with certain modifications (Ghosh et al., 2017). Tumor tissues (150–200 mg) were homogenized for 20 min in 500 μl of 0.15 M NaCl and centrifuged at 9000 g for 1 h at 4°C. The supernatant was discarded, and the pellet was rehomogenized in the same buffer and centrifugation was undertaken at 9000 g for 1h at 4°C. After that, again the supernatant was removed, and the resulting pellet was suspended and homogenized in buffer containing 0.05 M Tris-HCl, 3mM NaN3 and 0.01 mM CaCl2, pH 7.5 (500 μl). Collagenase type I (Himedia) was then added to the total tissue at a ratio of 1:100 by weight (mg) and incubated at 37°C overnight. The next day, the homogenate was centrifuged at 28,000 g for 1 h at 4°C using an ultracentrifuge. The resulting pellet obtained was then homogenized in 0.15M NaCl (300 μl) and spun down at 28,000 g for 1 h at 4°C. This step was performed many times until the absorbance of the supernatant was recorded to be below 0.3. Next, the resulting pellet was suspended in 200 μl distilled water followed by homogenization and centrifugation for 1 h at 28,000 g at 4°C. All supernatants were then pooled together and NaCl (0.15M) was added to precipitate the fibrils. This mixture was centrifuged for 1 h at 28,000 g at 4°C. The final pellet containing the total amyloid fibrils was suspended in PBS buffer and stored at 4°C for future use.
Transmission electron microscopy
A 10 μl volume of the isolated amyloid fibril obtained as mentioned above was spotted on to a copper-coated formvar grids (Electron Microscopy Sciences, Hatfield, PA, USA). The grids were washed with MQ water and stained for 20 min using 10 μl of 0.1% uranyl formate solution (Electron Microscopy Sciences) prepared freshly and filtered with a 0.22 μm sterile syringe filter (Millipore, Billerica, MA, USA). For performing immunoelectron microscopy, 10 μl of anti-p53 DO-1 antibody as above (1:10) was spotted on the grid carrying the samples for 20 min. The grid was subsequently washed with MQ water followed by incubation for 20 min with 10 μl of 10 nm gold-labeled anti-mouse-IgG secondary antibody (1:10; Sigma-Aldrich). Next, the grid was washed and then stained with 0.1% uranyl formate solution. The images were acquired at 10,000× magnification at 200kV using JOEL FEG-TEM 200 (JEM-2100 F) (JEOL, Tokyo, Japan). The images were recorded using the Gatan Microscopy Suite® (Gatan, USA). For coimmunoprecipitation TEM, 10 μl anti-p53 (SP5, Invitrogen) antibody (1:10) and 10 μl anti-p63 antibody (Santa Cruz Biotechnology) was spotted on the grid carrying the samples for 20 min. The grid was subsequently washed with MQ water followed by incubation for 20 min with 10 μl of anti-mouse-IgG 10 nm gold-labeled secondary antibody (1:10) (Sigma-Aldrich) and 10 μl of anti-rabbit-IgG 5 nm gold-labeled secondary antibody (1:10) (Sigma-Aldrich). The grid was washed, stained with 0.1% uranyl formate solution and images acquired at 10,000× magnification at 200kV using JOEL FEG-TEM 200 (JEM-2100 F) (JEOL, Tokyo, Japan). The images were recorded digitally using the Gatan Microscopy Suite® (Gatan, USA).
FTIR imaging
Fixed tissue sections were used for the FTIR Imaging. Frozen tissue sections were deparaffinized and rehydrated as mentioned above. The tissue samples were scrapped and kept on the BaF2 window (38×19×4 nm) (Technosearch Instruments, India). BaF2 is known for its low absorbance in the whole UV-IR wavelength spectrum (200 nm to 12 μm) and also provides resistance in high-energy radiation. For background correction, a clean area of each BaF2 slide was measured and then subtracted from sample measurements. During FTIR spectra acquisition, a Vertex-80v vacuum optics bench (Bruker, Germany) was used, and for recording the FTIR spectra, a Vertex-80 FTIR machine (Bruker, Germany) was attached with a 3000 Hyperion microscope. The tissues were imaged at 15× magnification (2.7 μm resolution) in the focal plane array (FPA) mode in the wavenumber range of 1600–1700 cm−1 corresponding to the amide-I stretching (C=O) frequency of peptide bond. For the analysis of FTIR spectra, the OPUS-65 v6.5 software was used. Individual points on the tissue images were selected for the analysis. To eliminate any contribution from water in the FTIR spectrum near 1650 cm−1, background correction was done. The spectra were then subjected to baseline correction followed by Fourier Self Deconvolution (FSD) using the Lorentzian deconvolution function in the wavenumber range of 1700–1600 cm−1. Briefly, the deconvolution technique was used to get the single sharp peaks from the convoluted or broadened spectra. Notably, the deconvolution was performed by feeding noise and band deconvolution factors optimized for minimum noise and maximum sharpness in the spectra. After that, the peaks were assigned according to the protein secondary structures and were best fitted by the auto-fitting method in the OPUS 65 software with minimum root mean square (RMS) error. The area under each resulting peak, the fractional contribution of individual secondary structures, was integrated by the peak integration method in the OPUS-65 software. The integration value of the β-sheet is divided from the sum of all the secondary structures and multiplied by 100 to obtain the percentage abundance of β-sheet in every FTIR spectrum. FTIR imaging and double immunofluorescence with anti-p53 antibody along with the OC antibody were performed on the adjacent tissue sections of the same slides.
Dot blot assay
All the different grades of oral and stomach cancer tissues were lysed and homogenized using RIPA buffer, supplemented with the protease inhibitor cocktail (PIC; #5056489001, Merck Millipore). The lysate was centrifuged at 3000 g for 5 min at 4°C to remove the tissue debris. The tissue lysate or amyloid fraction containing p53 protein was used for dot blot assay. For this, 20 μg (total protein content) of lysate was used to detect p53 amyloid. 4 μl of the samples were spotted directly on the nitrocellulose membrane. After drying, the membrane was blocked using 5% non-fat skimmed milk powder (Himedia, India) prepared in TBST for 1 h. Next, the membrane was washed with TBST and incubated at 4°C overnight with anti-p53 antibody, 1:200 dilution or OC, 1:500 dilution. After incubation, the membranes were washed three times with TBST for 10 min, followed by a 2 h incubation at room temperature with HRP-tagged anti-mouse-IgG secondary antibody for p53 and anti-rabbit-IgG for OC (Thermo Fisher Scientific) at 1:10,000 dilution. Any nonspecific binding was removed by washing the blot with TBST for 10 min. The protein signals were developed with SuperSignal West Femto kit (Thermo Fisher Scientific).
Next-generation sequencing
A total of 48 oral and stomach tissues (44 tumor tissues and four normal tissues), as mentioned in Fig. S1, were taken for the NGS of the p53 gene. The genomic DNA was isolated from these tissues using QIA amp DNA Mini Kit (Qiagen) based on the manufacturer’s instructions. The genomic DNA quality and quantity were assessed using Nanodrop 2000 and Qubit (Thermo Fisher Scientific), respectively. The sequencing library was prepared using an Illumina-compatible Accel Amplicon library prep kit and TP53 Comprehensive Panel (Swift Biosciences) at Genotypic Technology Pvt. Ltd., Bangalore, India. Briefly, 20 ng of Qubit-quantified genomic DNA was taken as template for Multiplex PCR using Reagent G1 of the TP53 Comprehensive Panel, and PCR amplification was carried out for 22 cycles, following the manufacturer’s instructions. The amplicons were bead-purified in a 1:2 bead to sample ratio, followed by indexing whereby unique combinations of dual indices were added to the amplicons. Finally, the barcoded samples were bead-purified in a 0.8 bead to sample ratio. The purified libraries were quantified using Qubit fluorometer (Thermo Fisher Scientific) and qPCR assays. The libraries were paired-end sequenced using Illumina HiSeq X Ten sequencer (Illumina, San Diego, CA, USA) for 150 cycles as per the manufacturer’s instructions.
Raw reads obtained from Illumina HiSeq X Ten sequencing for 48 samples were processed using Trim Galore-v0.4.01to generate high-quality reads by removing adapters, reads with less than Q30 quality score, and reads with length less than 20. Next, these processed reads were mapped against chromosome 17 of the Grch37 genome using Bowtie2 v2.2.5 2 aligner to generate alignment files. The alignment files were used for the removal of PCR duplication and adding read group information using Picard v1.102 3 tool followed by GATK v4.1.4.1 4 for performing Realigner Target Creator, Indel Realigner and Haplotype Caller to generate variants. Variants were generated for 44 samples (out of 48 samples) in GATK Haplotype Caller by using the targeted regions bed file and reference (chromosome 17) sequence. Finally, the variant files were used for annotation using Variant Studio v3.0 5 tool. The sum of the mutant frequency was calculated by dividing the sum of total alternate frequency obtained for each individual by the number of tissues sequenced for that specific grade.
Seprion-ELISA amyloid quantification assay
The tumor tissues were weighed equally (~30 mg), and lysate was prepared using ice-cold NP-40 buffer, supplemented with protease inhibitor cocktail (Roche) to prepare a 2.5% (w/v) lysate. The tissues were minced, homogenized and incubated on ice for 30 min. The p53 aggregation was determined using the Seprion-ELISA kit (Microsens) as described previously (Haltia et al., 1990). The assay is based on a polyionic and high-molecular-mass ligand that is coated to the surface of the ELISA plate. Only the aggregated proteins can bind to the ligand in the presence of seprion capture buffer. Next, anti-p53 antibody was used for the sandwich ELISA to detect only the p53 aggregates and fibrils. The tissue lysate was added to the assay plate, and the absorbance was measured using a plate reader. The absorbance is proportional to the bound amount of aggregated or fibrillar p53.
DAB staining
The fixed tumor and non-cancerous tissues were deparaffinized and rehydrated, and enzymatic antigen retrieval was performed, as described above. The endogenous peroxidase was quenched by incubating slides with 3% H2O2 for 15 min. The tissue sections were subsequently washed with TBST, and were subsequently treated with 0.2% Triton X-100 in TBST for 10 min. We used a blocking buffer of TBST containing 5% BSA to block nonspecific antigenic sites. The sections were then incubated with monoclonal anti-mouse pab240 primary antibody (1:500; Santa Cruz Biotechnology) overnight at 4°C. Tissues were further incubated for 2 h at room temperature with HRP-tagged secondary antibody of goat anti-mouse-IgG (1:1000) (Life Technologies, Thermo Fisher Scientific). The sections were then washed with TBST three times. The sections were then incubated with DAB solution (Sigma-Aldrich) with 0.2% NiCl2 for 20 min and counterstained with hematoxylin. The sections were dehydrated again with increasing concentrations of ethanol and xylene. The slides were mounted with DPX mountant and observed using a Leica DMi8 (Leica Microsystems, Germany) fluorescence microscope fitted with Andor Zyla cCMOS camera (Oxford Instruments, UK), and images were analyzed using ImageJ2 software. The DAB intensity was quantified to determine the nuclear and cytoplasmic misfolded p53 in the sections. For that, the color deconvolution plugin of ImageJ was used. The built-in stain vector Hematoxylin and DAB (H DAB) was selected. Images stained only with DAB were selected, and total intensity was measured. Next, the threshold was adjusted so that only the nuclear intensity could be measured. The nuclear DAB intensity was then subtracted from the total intensity to obtain the cytoplasmic DAB intensity in the sections.
Chromatin immunoprecipitation
Chromatin immunoprecipitation assay was carried out to assess the functional status of p53 using the Magna ChIP™ G Tissue Kit (Millipore, USA). Briefly, 30 mg of fresh tissue biopsy was weighed and washed with PBS to remove any attached impurity. 1% formaldehyde was added for crosslinking, followed by glycine to stop the reaction. The tissue was manually disrupted, followed by sonication for 5 cycles with 1 min on and off, and centrifuged at 4°C to remove any cell debris. The supernatant was removed to fresh microfuge tubes in 125 μl aliquots. The supernatant was diluted in dilutant buffer and was incubated overnight in the presence of 20 μl of fully resuspended protein G magnetic beads and 10 μl anti-p53 antibody. The magnetic beads were pelleted with help of a magnetic separator rack, and the supernatant was discarded. The Protein G bead–antibody and bound chromatin complex was washed by resuspending the beads in 0.5 ml of cold buffers (in the order Low Salt Immune Complex Wash Buffer, High Salt Immune Complex Wash Buffer, LiCl Immune Complex Wash Buffer, one wash, TE Buffer, one wash) and then incubating for 5 min on a rotating platform. The supernatant fraction was carefully removed. Protein–DNA crosslink reversal was carried out, and DNA was eluted and purified using a QIAquick PCR purification kit (Qiagen) following the manufacturer’s instructions. The qPCR was performed using 20 μl SYBR Green reaction mixture using a real-time PCR system (Agilent AriaMx Real-time PCR System, Agilent, CA, USA). The ChIP experiments and qPCR were performed in triplicate. The enrichment/input values were calculated as: ΔCT=CT(ChIP)-[CT(Input)-LogE (Input dilution factor)] where E is the specific primer efficiency value; % Enrichment/Input=E–ΔCT. All the primers that were used in this study are listed in the Table S2.
Immunoprecipitation
The different grades of oral and stomach tissues were suspended in RIPA buffer to carry out lysis (20 mMTris-HCl, pH 8.0, 137 mM NaCl, 1% NP-40, 2 mM EDTA) along with protease inhibitor cocktail (Roche) and manually homogenized on ice. The mixture was spun down at 4°C at ~7000 g for 20 min. The obtained supernatant was incubated with the anti-p53 antibody (SP5, Invitrogen) overnight at 4°C under rotation. On the next day, 100 μl of Sepharose G beads (Thermo Fisher Scientific) were added to the solution and incubated at 4°C under constant agitation for 4hrs. The solution was then centrifuged at ~1000 g for 1 min to precipitate the beads. The supernatant was discarded, and the elution was performed by mixing the equal amount of SDS loading dye to the beads and the samples were heated in a dry bath for 10 min at 95°C. The eluted fractions were loaded in the SDS gel and western blot was performed with anti-mouse p63 and p73 primary antibodies (Santa Cruz Biotechnology; 1:500) as mentioned above. All the original uncropped western blot images are shown as a separate file (Fig. S8F–I).
Database analysis
The TCGA data and its specific grade information were retrieved from the UCSC Xena site (https://xena.ucsc.edu/). We previously established the p53 amyloid-specific alteration of gene expressions in cells (Navalkar et al., 2021, 2022). Utilizing those unique gene-sets, we examined the single-sample gene set enrichment analysis (ssGSEA) (Subramanian et al., 2005) using the python package gseapy (https://github.com/zqfang/gseapy). This quantifies the amyloid-specific uniquely differential expressed genes with the grades of various cancer types in the TCGA database. R version 4.2.0 was used for all statistical analysis and the plots were generated using ggplot2 function.
Statistical analysis
The statistical significances for this study were calculated using one-way ANOVA followed by the Tukey’s multiple comparison test. The P-values for the significance are marked as *P<0.05, **P<0.01, ***P<0.001; non-significant (NS, P≥0.05). GraphPad prism was used to calculate the statistical significance.
Supplementary Material
Acknowledgements
We acknowledge the National Tumor Tissue Repository (NTTR), Indian Council for Medical Research (ICMR) at the Tata Memorial Hospital (TMH), Mumbai, India, for providing human cancer tissue; Dr Omshree Shetty, Tata Memorial Hospital, Dr Sushil Kumar, IIT Bombay for providing valuable inputs, Mr. Pradeep Kadu for making the schematic and Ms. Manisha Poudyal for her inputs in figure rearrangements. We also acknowledge the Center for Research in Nano Technology and Science (CRNTS) and Industrial Research and Consultancy Centre (IRCC), IIT Bombay, for the FTIR, electron microscopy and confocal facility.
Funding
This work was support by the Department of Biotechnology (DBT) (BT/PR9797/NNT/28/774/2014) Government of India, the Wadhwani Research Center for Bioengineering (WRCB), the Department of Science and Technology, Ministry of Science and Technology, India (EMR/2014/001233 and CRG/2019/001133) for financial support. The authors also acknowledge an The Wellcome Trust DBT India Alliance fellowship (IA/E/17/1/503663) awarded to S.S. for financial support. A.P.l acknowledges the Council of Scientific and Industrial Research (CSIR), Government of India, for the Shyama Prasad Mukherjee fellowship (SPMF).
Footnotes
Author contributions
Conceptualization: S.S., S.K.M.; Methodology: S.S., N.S., M.J.; Validation: S.S., D.C., S.K.M.; Formal analysis: S.S., N.S., A.P., D.D., D.C., S. Mukherjee, L.G., J.D., Y.M., M.J., S.K.M.; Investigation: S.S., N.S., S.K.M.; Resources: S.S.; Data curation: N.S., A.P., D.D., D.C., S. Mukherjee, L.G., Y.M., M.J., S.K.M.; Writing - original draft: S.S., S.K.M.; Visualization: S.S., N.S., A.P., D.C., M.J., S.K.M.; Supervision: S.K.M.; Funding acquisition: S.S., S.K.M.
Competing interests
The authors declare no competing or financial interests.
Data Availability
RNA-seq data from this study has been deposited in the Sequence Read Archive (SRA) under Bioproject ID PRJNA1008865.
References
- Aguzzi A, Heppner FL. Pathogenesis of prion diseases: a progress report. Cell Death Differ. 2000;7:889–902. doi: 10.1038/sj.cdd.4400737. [DOI] [PubMed] [Google Scholar]
- Arsic N, Slatter T, Gadea G, Villain E, Fournet A, Kazantseva M, Allemand F, Sibille N, Seveno M, de Rossi S, et al. Δ133p53β isoform pro-invasive activity is regulated through an aggregation-dependent mechanism in cancer cells. Nat Commun. 2021;12:5463. doi: 10.1038/s41467-021-25550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018;25:104–113. doi: 10.1038/cdd.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer MR, Krämer A, Settanni G, Jones RN, Ni X, Khan Tareque R, Fersht AR, Spencer J, Joerger AC. Targeting cavity-creating p53 cancer mutations with small-molecule stabilizers: the Y220X paradigm. ACS Chem Biol. 2020;15:657–668. doi: 10.1021/acschembio.9b00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandino G, Dobbelstein M. p73 and p63: why do we still need them? Cell Cycle. 2004;3:886–894. doi: 10.4161/cc.3.7.996. [DOI] [PubMed] [Google Scholar]
- Bosari S, Viale G, Bossi P, Maggioni M, Coggi G, Murray JJ, Lee AK. Cytoplasmic accumulation of p53 protein: an independent prognostic indicator in colorectal adenocarcinomas. J Natl Cancer Inst. 1994;86:681–687. doi: 10.1093/jnci/86.9.681. [DOI] [PubMed] [Google Scholar]
- Bosari S, Viale G, Roncalli M, Graziani D, Borsani G, Lee AK, Coggi G. p53 gene mutations, p53 protein accumulation and compartmentalization in colorectal adenocarcinoma. Am J Pathol. 1995;147:790–798. [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;99(Suppl.4):16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JS, Loh SN. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 2003;42:2396–2403. doi: 10.1021/bi026635n. [DOI] [PubMed] [Google Scholar]
- Chatterjee D, Jacob RS, Ray S, Navalkar A, Singh N, Sengupta S, Gadhe L, Kadu P, Datta D, Paul A, et al. Co-aggregation and secondary nucleation in the life cycle of human prolactin/galanin functional amyloids. Elife. 2022;11:e73835. doi: 10.7554/eLife.73835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- Crews L, Masliah E. Molecular mechanisms of neurodegeneration in Alzheimer’s disease. Hum Mol Genet. 2010;19:R12–R20. doi: 10.1093/hmg/ddq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet F, Saiz Rubio M, Hompes D, Naus E, De Baets G, Langenberg T, Hipp MS, Houben B, Claes F, Charbonneau S. Nuclear inclusion bodies of mutant and wild-type p53 in cancer: a hallmark of p53 inactivation and proteostasis remodelling by p53 aggregation. J Pathol. 2017;242:24–38. doi: 10.1002/path.4872. [DOI] [PubMed] [Google Scholar]
- Dobson CM. The structural basis of protein folding and its links with human disease. Philos Trans R Soc Lond Ser B Biol Sci. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dötsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73 the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes HK, Khamar HJ, Winchester C-L, Greenler AJ, Zhou A, McGlinchey RP, Gorkovskiy A, Wickner RB. Sporadic distribution of prion-forming ability of Sup35p from yeasts and fungi. Genetics. 2014;198:605–616. doi: 10.1534/genetics.114.166538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti GDS, Quarti J, Dos Santos G, Rangel LP, Silva JL. Anticancer therapeutic strategies targeting p53 aggregation. Int J Mol Sci. 2022;23:11023. doi: 10.3390/ijms231911023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget KJ, Tremblay G, Roucou X. p53 Aggregates penetrate cells and induce the co-aggregation of intracellular p53. PLoS One. 2013;8:e69242. doi: 10.1371/journal.pone.0069242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Kelly JW. Functional amyloidogenesis and cytotoxicity —insights into biology and pathology. PLoS Biol. 2012;10:e1001459. doi: 10.1371/journal.pbio.1001459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional Amyloid Formation within Mammalian Tissue. PLoS Biol. 2005;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, Bellefroid E, Marine J-C. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci USA. 2006;103:3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Salot S, Sengupta S, Navalkar A, Ghosh D, Jacob R, Das S, Kumar R, Jha NN, Sahay S, et al. p53 amyloid formation leading to its loss of function: implications in cancer pathogenesis. Cell Death Differ. 2017;24:1784–1798. doi: 10.1038/cdd.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltia M, Prelli F, Ghiso J, Kiuru S, Somer H, Palo J, Frangione B. Amyloid protein in familial amyloidosis (Finnish type) is homologous to gelsolin, an actin-binding protein. Biochem Biophys Res Commun. 1990;167:927–932. doi: 10.1016/0006-291X(90)90612-Q. [DOI] [PubMed] [Google Scholar]
- Higashimoto Y, Asanomi Y, Takakusagi S, Lewis MS, Uosaki K, Durell SR, Anderson CW, Appella E, Sakaguchi K. Unfolding, aggregation, and amyloid formation by the tetramerization domain from mutant p53 associated with lung cancer. Biochemistry. 2006;45:1608–1619. doi: 10.1021/bi051192j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Fry EA. Alterations of p63 and p73 in human cancers. Subcell Biochem. 2014;85:17–40. doi: 10.1007/978-94-017-9211-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru D, Andrade LR, Teixeira LSP, Quesado PA, Maiolino LM, Lopez PM, Cordeiro Y, Costa LT, Heckl WM, Weissmuller G, et al. Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry. 2003;42:9022–9027. doi: 10.1021/bi034218k. [DOI] [PubMed] [Google Scholar]
- Iwahashi N, Ikezaki M, Komohara Y, Fujiwara Y, Noguchi T, Nishioka K, Sakai K, Nishio K, Ueda M, Ihara Y, et al. Cytoplasmic p53 aggregates accumulated in p53-mutated cancer correlate with poor prognosis. PNAS Nexus. 2022;1:pgac128. doi: 10.1093/pnasnexus/pgac128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Mantsch HH. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit Rev Biochem Mol Biol. 1995;30:95–120. doi: 10.3109/10409239509085140. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Ang HC, Fersht AR. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc Natl Acad Sci U S A. 2006;103:15056–15061. doi: 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- Kayed R, Head E, Sarsoza F, Saing T, Cotman CW, Necula M, Margol L, Wu J, Breydo L, Thompson JL, et al. Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Giese A, Deppert W. Wild-type p53 in cancer cells: when a guardian turns into a blackguard. Biochem Pharmacol. 2009;77:11–20. doi: 10.1016/j.bcp.2008.08.030. [DOI] [PubMed] [Google Scholar]
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh Y-A, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. Gain of function of a p53 hot spot mutation in a mouse model of li-fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Lee AS, Galea C, DiGiammarino EL, Jun B, Murti G, Ribeiro RC, Zambetti G, Schultz CP, Kriwacki RW. Reversible amyloid formation by the p53 tetramerization domain and a cancer-associated mutant. J Mol Biol. 2003;327:699–709. doi: 10.1016/S0022-2836(03)00175-X. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53 the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/S0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Levy CB, Stumbo AC, Ano Bom APD, Portari EA, Cordeiro Y, Silva JL, De Moura-Gallo CV. Co-localization of mutant p53 and amyloid-like protein aggregates in breast tumors. Int J Biochem Cell Biol. 2011;43:60–64. doi: 10.1016/j.biocel.2010.10.017. [DOI] [PubMed] [Google Scholar]
- Liang SH, Clarke MF. Regulation of p53 localization. Eur J Biochem. 2001;268:2779–2783. doi: 10.1046/j.1432-1327.2001.02227.x. [DOI] [PubMed] [Google Scholar]
- Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191:1041–1072. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DP, Song H, Xu Y. A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene. 2010;29:949–956. doi: 10.1038/onc.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Pochampally R, Chen L, Traidej M, Wang Y, Chen J. Nuclear exclusion of p53 in a subset of tumors requires MDM2 function. Oncogene. 2000;19:232–240. doi: 10.1038/sj.onc.1203262. [DOI] [PubMed] [Google Scholar]
- Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KPR, Simon R, Schubert D, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manterola L, Aguirre P, Larrea E, Arestìn M, Gaafar A, Elorriaga K, Goicoechea I, Armesto M, Fernández-Mercado M, Zabalza I, et al. Mutational profiling can identify laryngeal dysplasia at risk of progression to invasive carcinoma. Sci Rep. 2018;8:6613. doi: 10.1038/s41598-018-24780-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019;26:199–212. doi: 10.1038/s41418-018-0246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine J-C, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- Maritschnegg E, Heinzl N, Wilson S, Deycmar S, Niebuhr M, Klameth L, Holzer B, Koziel K, Concin N, Zeillinger R. Polymer-ligandbased ELISA for robust, high-throughput, quantitative detection of p53 aggregates. Anal Chem. 2018;90:13273–13279. doi: 10.1021/acs.analchem.8b02373. [DOI] [PubMed] [Google Scholar]
- Marques MA, de Andrade GC, Silva JL, de Oliveira GAP. Protein of a thousand faces: The tumor-suppressive and oncogenic responses of p53. Front Mol Biosci. 2022;9:944–955. doi: 10.3389/fmolb.2022.944955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon FD. p63 and p73 in tumor suppression and promotion. Cancer Res Treat. 2004;36:6–12. doi: 10.4143/crt.2004.36.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello SS, Attardi LD. Not all p53 gain-of-function mutants are created equal. Cell Death Differ. 2013;20:855–857. doi: 10.1038/cdd.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello SS, Attardi LD. Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol. 2018;51:65–72. doi: 10.1016/j.ceb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo Dos Santos N, de Oliveira GAP, Ramos Rocha M, Pedrote MMD, da Silva Ferretti G, Pereira Rangel L, Morgado-Diaz JA, Silva JL, Rodrigues Pereira Gimba E. Loss of the p53 transactivation domain results in high amyloid aggregation of the Δ40p53 isoform in endometrial carcinoma cells. J Biol Chem. 2019;294:9430–9439. doi: 10.1074/jbc.RA119.007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LM, Bourassa MW, Smith RJ. FTIR spectroscopic imaging of protein aggregation in living cells. Biochim Biophys Acta Biomembr. 2013;1828:2339–2346. doi: 10.1016/j.bbamem.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Riou G, Levine AJ. Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc Natl Acad Sci U S A. 1992;89:7262–7266. doi: 10.1073/pnas.89.15.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, LaQuaglia M, Benard J, Riou G. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc Natl Acad Sci U S A. 1995;92:4407–4411. doi: 10.1073/pnas.92.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Ostermeyer AG, Haladay R, Winkfield B, Frazier M, Zambetti G. Cytoplasmic sequestration of wild-type p53 protein impairs the G1 checkpoint after DNA damage. Mol Cell Biol. 1996;16:1126–1137. doi: 10.1128/MCB.16.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti P, Menichini P, Speciale A, Cutrona G, Fais F, Taiana E, Neri A, Bomben R, Gentile M, Gattei V, et al. Heterogeneity of TP53 mutations and P53 protein residual function in cancer: does it matter? Front Oncol. 2020;10:593383. doi: 10.3389/fonc.2020.593383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navalkar A, Ghosh S, Pandey S, Paul A, Datta D, Maji SK. Prion-like p53 amyloids in cancer. Biochemistry. 2020;59:146–155. doi: 10.1021/acs.biochem.9b00796. [DOI] [PubMed] [Google Scholar]
- Navalkar A, Pandey S, Singh N, Patel K, Datta D, Mohanty B, Jadhav S, Chaudhari P, Maji SK. Direct evidence of cellular transformation by prion-like p53 amyloid infection. J Cell Sci. 2021;134:jcs258316. doi: 10.1242/jcs.258316. [DOI] [PubMed] [Google Scholar]
- Navalkar A, Paul A, Sakunthala A, Pandey S, Dey AK, Saha S, Sahoo S, Jolly MK, Maiti TK, Maji SK. Oncogenic gain of function due to p53 amyloids occurs through aberrant alteration of cell cycle and proliferation. J Cell Sci. 2022;135:jcs259500. doi: 10.1242/jcs.259500. [DOI] [PubMed] [Google Scholar]
- Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol Cell Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J, Song B-D, Rogel JK, Kujawara D, Altobel L, III, Izharrudin A, Boldt GE, Grover RK, Wentworth AD, Wentworth PJ. Cholesterol secosterol aldehydes induce amyloidogenesis and dysfunction of wild-type tumor protein p53. Chem Biol. 2011;18:920–927. doi: 10.1016/j.chembiol.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeyer AG, Runko E, Winkfield B, Ahn B, Moll UM. Cytoplasmically sequestered wild-type p53 protein in neuroblastoma is relocated to the nucleus by a C-terminal peptide. Proc Natl Acad Sci U S A. 1996;93:15190–15194. doi: 10.1073/pnas.93.26.15190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otzen D. Functional amyloid: turning swords into plowshares. Prion. 2010;4:256–264. doi: 10.4161/pri.4.4.13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanikumar L, Karpauskaite L, Al-Sayegh M, Chehade I, Alam M, Hassan S, Maity D, Ali L, Kalmouni M, Hunashal Y, et al. Protein mimetic amyloid inhibitor potently abrogates cancer-associated mutant p53 aggregation and restores tumor suppressor function. Nat Commun. 2021;12:3962. doi: 10.1038/s41467-021-23985-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel LP, Costa DCF, Vieira TCRG, Silva JL. The aggregation of mutant p53 produces prion-like properties in cancer. Prion. 2014;8:75–84. doi: 10.4161/pri.27776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigacci S, Bucciantini M, Relini A, Pesce A, Gliozzi A, Berti A, Stefani M. The (1-63) region of the p53 transactivation domain aggregates in vitro into cytotoxic amyloid assemblies. Biophys J. 2008;94:3635–3646. doi: 10.1529/biophysj.107.122283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2:466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammons MA, Nguyen T-AT, McDade SS, Fischer M. Tumor suppressor p53: from engaging DNA to target gene regulation. Nucleic Acids Res. 2020;48:8848–8869. doi: 10.1093/nar/gkaa666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Ghufran SM, Khan A, Biswas S, Roychoudhury S. Transition of amyloid/mutant p53 from tumor suppressor to an oncogene and therapeutic approaches to ameliorate metastasis and cancer stemness. Cancer Cell Int. 2022;22:416. doi: 10.1186/s12935-022-02831-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JL, Cino EA, Soares IN, Ferreira VF, de Oliveira GAP. Targeting the prion-like aggregation of mutant p53 to combat cancer. Acc Chem Res. 2018;51:181–190. doi: 10.1021/acs.accounts.7b00473. [DOI] [PubMed] [Google Scholar]
- Skinner HD, Sandulache VC, Ow TJ, Meyn RE, Yordy JS, Beadle BM, Fitzgerald AL, Giri U, Ang KK, Myers JN. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin Cancer Res. 2012;18:290–300. doi: 10.1158/1078-0432.CCR-11-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soragni A, Janzen DM, Johnson LM, Lindgren AG, Thai-Quynh Nguyen A, Tiourin E, Soriaga AB, Lu J, Jiang L, Faull KF, et al. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell. 2016;29:90–103. doi: 10.1016/j.ccell.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T, Ishioka C, Claustres M, Béroud C. Locus-specific mutation databases: pitfalls and good practice based on the p53 experience. Nat Rev Cancer. 2006;6:83–90. doi: 10.1038/nrc1783. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinidad AG, Muller PAJ, Cuellar J, Klejnot M, Nobis M, Valpuesta JM, Vousden KH. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol Cell. 2013;50:805–817. doi: 10.1016/j.molcel.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Wang G, Fersht AR. First-order rate-determining aggregation mechanism of p53 and its implications. Proc Natl Acad Sci USA. 2012;109:13590–13595. doi: 10.1073/pnas.1211557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Fersht AR. Mechanism of initiation of aggregation of p53 revealed by Φ-value analysis. Proc Natl Acad Sci U S A. 2015a;112:2437–2442. doi: 10.1073/pnas.1500243112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Fersht AR. Propagation of aggregated p53: Crossreaction and coaggregation vs. seeding. Proc Natl Acad Sci USA. 2015b;112:2443–2448. doi: 10.1073/pnas.1500262112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcken R, Wang G, Boeckler FM, Fersht AR. Kinetic mechanism of p53 oncogenic mutant aggregation and its inhibition. Proc Natl Acad Sci U S A. 2012;109:13584–13589. doi: 10.1073/pnas.1211550109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KJ, Cyr DM. Amyloid in neurodegenerative diseases: friend or foe? Semin Cell Dev Biol. 2011;22:476–481. doi: 10.1016/j.semcdb.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, Cornelis A, Rozenski J, Zwolinska A, Marine J-C, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data from this study has been deposited in the Sequence Read Archive (SRA) under Bioproject ID PRJNA1008865.