Abstract
Bacteriocins are potent antimicrobial peptides that are produced by bacteria. Since their discovery almost a century ago, diverse peptides have been discovered and described, and some are currently used as commercial food preservatives. Many bacteriocins exhibit extensively post-translationally modified structures encoded on complex gene clusters, whereas others have simple linear structures. The molecular structures, mechanisms of action and resistance have been determined for a number of bacteriocins, but most remain incompletely characterized. These gene-encoded peptides are amenable to bioengineering strategies and heterologous expression, enabling metagenomic mining and modification of novel antimicrobials. The ongoing global antimicrobial resistance crisis demands that novel therapeutics be developed to combat infectious pathogens. New compounds that are target-specific and compatible with the resident microbiota would be valuable alternatives to current antimicrobials. As bacteriocins can be broad or narrow spectrum in nature, they are promising tools for this purpose. However, few bacteriocins have gone beyond preclinical trials and none is currently used therapeutically in humans. In this Review, we explore the broad diversity in bacteriocin structure and function, describe identification and optimization methods and discuss the reasons behind the lack of translation beyond the laboratory of these potentially valuable antimicrobials.
Introduction
Antimicrobial peptides are produced ubiquitously across the kingdoms of life as an ancient form of host defence1. Bacteriocins are a subset of bacterially produced, ribosomally synthesized antimicrobial peptides that have been studied for almost a century2. In that time, bacteriocins have been the subjects of thousands of publications, describing diverse peptide families, many of which have reported the ability of the peptides and/or their producers to control the outgrowth of food spoilage organisms and to preserve foods3. Many bacteriocins are the products of safe and food grade fermenting species such as lactic acid bacteria, and some, exemplified by nisin, have transitioned in the past century from unknown antimicrobial by-products2 to characterized4, regulated and approved natural food additives5 in widely consumed commercial products. Bacteriocins have found success as food preservatives in the present but are not limited to it for the future, as they have also been investigated for various applications such as growth promotion in production animals6, plant growth promotion7 and clinical therapeutics for the treatment of fungal8, protozoal9, viral10 and, foremostly, bacterial infections11.
The increasing rates of antimicrobial resistance threatens the use of current antibiotics, and it is predicted that antibiotic-resistant microorganisms will cause millions of deaths worldwide every year by 2050, severely affecting what are now considered routine medical treatments12.
The WHO has generated a priority pathogen list highlighting an urgent need for novel treatments against antibiotic-resistant pathogens, with special mention of Mycobacterium tuberculosis and carbapenem-resistant Gram-negative bacteria for which only a few treatment options are available, followed by the Gram-positive vancomycin-resistant Enterococcus faecium and other high-priority pathogens13. Bacteriocins represent a potential solution as current antibiotic alternatives or adjuncts14–17. They offer several pharmacological advantages such as activity against pathogens in the nanomolar and picomolar ranges, narrow spectra, general non-toxicity and distinct mechanisms of action18. The diversity in bacteriocin structures and mechanisms also represents a range of ribosomally produced antimicrobials that are amenable to genetic engineering strategies enabling the production of bioengineered variants with superior efficacy against target microorganisms19.
As natural agents of microbial competition, bacteriocins offer an advantage to bacteria over sensitive neighbouring species, enabling invasion, colonization and protection of a niche in complex environments such as the gut microbiome20. By exploiting these natural mechanisms, they can be used as a tool for humans to deliver antimicrobial activity as pure peptides and composite nanoparticles or can be codified within live biotherapeutics for in situ expression in microbiomes. Advances in microbiome metagenomics and other ‘omics’ technologies over the past two decades have revealed catalogues of genome-mined and metabolome-mined bacteriocins, and new families are continually being discovered, but currently discovery is outpacing characterization and application. Narrow-target spectrum bacteriocins are also increasingly desirable as we advance our understanding of host– microbiota interactions (response and regulation of host processes to microbiota composition and products21) and are realizing that the impact of antibiotics on microbiome diversity should be considered22. Despite lack of investment, bacteriocins are currently undergoing clinical studies to treat infections caused by antimicrobial-resistant pathogens; the outlook is bright for diverse bacteriocin applications in the next century.
In this Review, we summarize recent advances in bacteriocin research, including the identification of novel peptides and mechanisms of action. We discuss bioengineering as a means to optimize bacteriocin activity and expression in addition to novel methods of bacteriocin discovery in the omics era. Finally, we outline the potential applications of bacteriocins producers, including live biotherapeutics for microbiome modulation, and highlight those peptides that are currently under investigation for clinical applications.
Bacteriocin diversity
Structural diversity
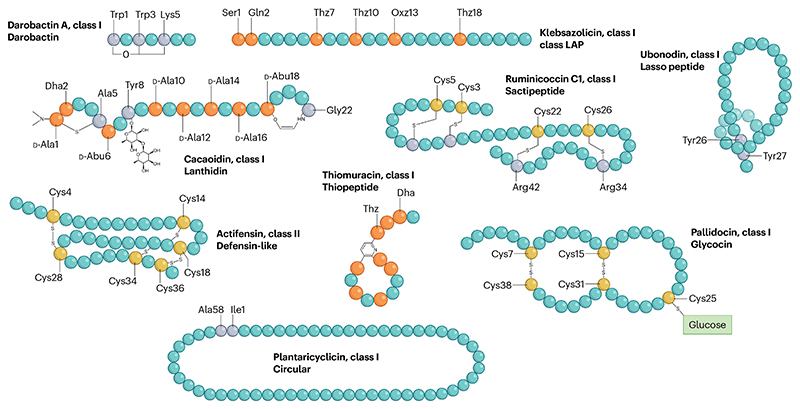
Bacteriocins are diverse and widespread across the phyla that produce them, with enormous heterogeneity in gene cluster organization, bio-synthetic machinery and peptide structures (Table 1 and Fig. 1). Despite such diversity, bacteriocins are usually classified into two major groups: those with post-translational modifications (class I) and unmodified peptides (class II) (Box 1). In recent years, there have been increased reports of novel ribosomally produced and post-translationally modified peptides (RiPPs) and characterization of their biosynthetic machinery (extensively reviewed elsewhere23). However, not all RiPPs are bacteriocins and nor are all bacteriocins RiPPs as some do not undergo substantial post-translational modification. Bacteriocins are generally considered large molecule antimicrobials relative to small-molecule antibiotics but can range from bicyclic peptides, such as the recently described darobactin (965 Da)16 that is smaller than the clinically used colistin (~1,200 Da)24, to larger circular peptides such as pumilarin (7,083 Da)25 (Fig. 1).
Table 1. Suggested updated bacteriocin classification scheme with examples.
| Bacteriocin classa | Subgroup | Defining features and conserved enzyme (if known or present) | Examples |
|---|---|---|---|
| I | Bottromycin | Macrolactamidine, YcaO | Bottromycin A2 (ref. 173) |
| I | Cyanobactin | N-terminal proteolysis enzyme, PatA | Kawaguchipeptin B174 |
| I | Darobactin | Unusual double ring structure from tryptophan-to-tryptophan, lysine or arginine linkages, rSAM | Darobactin16 |
| I | Epipeptide | d-amino acids, rSAM | EpeX175 |
| I | Glycocin | S-glycosylation and O-glycosylation of serine/threonine | Pallidocin121 |
| I | Lanthipeptide, type I | Methyllanthionine and/or lanthionine residues, LanBC | Kunkecin A176 |
| I | Lanthipeptide, type II | Methyllanthionine and/or lanthionine residues, LanM | Roseocin177 |
| I | Lanthipeptide, type V (lanthidin) | Methyllanthionine and/or lanthionine residues, LanKY | Cacaoidin178 |
| I | Lasso peptide | Macrolactam with threaded C-terminal tail | Ubonodin179 |
| I | Linaridin | Dehydrobutyrine, no lanthionine | Corynaridin27 |
| I | Linear azole-containing or azoline-containing peptide (LAP) | Azol(ine)s, YcaO | Spongiicolazolicin A/B180 |
| I | Pantocin | Glu–Glu crosslink, PaaA | Pantocin A181 |
| I | Pyritide (including thiopeptides) | Six-membered nitrogenous heterocycle | Thiomuracin182 |
| I | Sactipeptide | Intramolecular sulfur-to-α-carbon thioether (sactionine) crosslink | Ruminococcin C183 |
| I | Circular | Covalently linked N-terminal and C-terminal residues resulting in circular peptide backbone | Pumilarin25 |
| I | Microcins with non-ribosomal siderophore | Serine-rich C terminus with a non-ribosomal siderophore-type modification | MccH47184 |
| II | Pediocin-like | Contains YGNGVXC motif | Maltaricin CPN31 |
| II | Linear, two-component | Two peptides, both required for activity | Plantaricin EF29 |
| II | Linear, non-pediocin, non-two component | Lacks defining features of other groups | Bactofencin185 |
| II | Defensin-like | Conserved disulfide pattern of eukaryotic defensins | Actifensin32 |
| II | Leaderless | Leaderless peptides with or without four-helix motif | BacSp222, Enterocin DD14 (refs. 30,186) |
rSAM, radical S-adenosylmethionine.
Bacteriocins can be generally classified into two major groups: post-translationally modified (class I) and non-significantly post-translationally modified peptides (class II). Both groups are further subdivided on the basis of conserved features unique to the subgroup. Within class I bacteriocins, groups can be defined according to a specific modification that is installed by one or more modification enzymes.
Fig. 1. The structural diversity of bacteriocins.
Bacteriocins are typically classified into classes and subgroups based on their mechanism of production and secondary structures (Table 1). Lanthipeptides and other class I bacteriocins may contain complex intrapeptide bonds that are installed through enzymatic modifications of specific residues in the peptide backbone. These bonds may form small ring structures such as the C–O–C Trp–Trp ether bond in darobactin A (key residues coloured in grey) and the cacaoidin AviMeCys ring (modified residues coloured in orange). Multiple intrapeptide crosslinks may be formed following modification within one peptide such as the sacthioine bonds in ruminicoccin C1 (involved cysteine residues coloured in yellow). Class I bacteriocins may also feature post-translational installation of other moieties, shown here in glycosylated pallidocin at cysteine 25 and tyrosine 8 of cacaoidin. Modified residues are not always part of larger ring structures, such as the thiazole and oxazole residues present in the linear azoline-containing peptide, klebsazolicin. Large ring structures, however, are prominent features of different subgroups of class I bacteriocins and take different forms such as the loop structure present in the lasso peptide ubonodin that is sterically locked by bulky tyrosine side chains or the macrocycle of the heavily modified thiomuracin A. The largest rings are present within the circular bacteriocins wherein N-terminal and C-terminal residues are covalently linked resulting in a closed loop structure. Class II bacteriocins do not undergo enzymatic modification and are therefore less enzymatically decorated but may feature disulfide bonds that can also result in intercalated structures of such as those in defensin-like bacteriocins, such as actifensin. LAP, linear azole-containing or azoline-containing peptide.
Class I bacteriocins undergo extensive modification with enzymes that may dehydrate residues and form inter-amino acid bonds, leading to their intricate and stable structures. Such modifications can result in peptides that are extensively cyclized and dehydrated, such as polyheterocyclic klebsazolicin26 and corynaridin, the latter of which contains eight dehydrated residues27. Lantibiotics (lanthionine-containing antibiotics) are well-established class I bacteriocins, but novel structurally related peptide families continue to be identified; such as the type V lanthipeptides (lanthidins), which are glycosylated and N-terminally dimethylated, as evident in the founding member cacaoidin28. Class II bacteriocin structures include simple linear α-helical peptides such as plantaricin EF29, peptides with conserved complex secondary structures such as the four-helix peptide BacSp222 (ref. 30) or linear peptides with specific conserved disulfides such as pediocin-like maltaricin CPN and actifensin31,32. Class II bacteriocins do not feature the substantial modifications, such as dehydration and formation of unusual amino acids or linkages of class I, and previously included circular bacteriocins that are subject to N-terminal to C-terminal covalent linkage resulting in their simple circular shape33 and the microcins that are post-translationally conjugated to the siderophore (iron binding) moiety34. For consistency within the two major classification schemes (Box 1), we suggest that circular and siderophore-modified peptides are henceforth referred to as class I bacteriocins, as they undergo modification despite relatively simple mature structures.
For most bacteriocins, the structural peptide-encoding gene also encodes a leader sequence, which comprises N-terminal residues removed from the mature peptide, and can fulfil various functions in peptide export, cleavage and modification23. These roles are linked to the specific type of bacteriocin they accompany, shaping peptide production and post-translational alterations. For example, in the case of lantibiotics, conserved motifs such as the FNLD motif are observed within leader sequences, whereas circular bacteriocins exhibit considerable diversity in their leader sequences, ranging from minimal lengths as seen in lactocyclin Q (two residues35) to more extensive sequences such as in amylocyclicin (47 residues36). The flexibility of circular bacteriocin leader sequences was highlighted by modification of circularin A, which demonstrated functional expression with a leader of only one residue (Met), suggesting a limited role for the leader in active circularin A biosynthesis37. Additionally, specific motifs within the nisin leader sequence have been shown to guide peptide export, modification38 and cleavage39, whereas in lanthipeptides, such as mersacidin, the leader undergoes a two-step cleavage process involving initial cleavage upon exiting the cell by the transporter-peptidase MrsT40 and subsequent cleavage outside the cell by specific extracellular proteases such as AprE41. Although many bacteriocins with leader sequences possess a dedicated leader sequence essential for their biosynthesis and activity, with exceptions such as circularin A described earlier, leaderless bacteriocins lack this and instead are directly translated as active peptides33 and the mechanisms of their production and export are poorly understood. Two pleckstrin homology domain-containing proteins DdE and DdF have been linked to the export of enterocin DD14 peptides in Enterococcus faecalis42, but peptide export mechanisms are yet to be characterized.
Biosynthetic gene clusters
Bacteriocin-encoding gene clusters encode essential factors involved in their production and protection, typically incorporating genes encoding a core peptide, modifying enzymes for modified peptides43, export machinery, self-immunity proteins that protect the producer44 and many clusters also contain genes encoding regulatory proteins45. In the case of the unmodified bacteriocin bactofencin, a ~4 kb gene cluster encodes the peptide and an ABC transporter with a protease domain that exports and removes the leader sequence. These two genes alone (bfnA and DLSL_0052) have been demonstrated to produce active peptide when expressed heterologously in Escherichia coli46. By contrast, the native producer of bactofencin A, Ligilactobacillus salivarius DPC6502, produces an immunity protein that provides self-protection and an additional protein containing a thioredoxin family domain that is hypothesized to assist in disulfide bond formation46. Other small gene clusters result in more complicated peptide structures such as a cluster of 6.2 kb that contains genes for darobactin A production: a core peptide (darA), an ABC transporter (darBCD) and a single-radical S-adenosylmethionine enzyme (encoded by darE) that catalyse the formation of a Trp–Lys C–C bond and a C–O–C Trp–Trp ether bond via an unknown mechanism16.
Radical S-adenosylmethionine enzymes also catalyse the formation of α-carbon to thioether (sactionine) bonds in sactibiotics (sactionine-containing antibiotics), such as the ruminococcins, in which the gene cluster in Ruminococcus gnavus E1 consists of 17 open reading frames over 15 kb that can result in five distinct peptides47. In the case of the lanthidin cacaoidin, the gene cluster spans 30 kb and contains 27 genes that encode the necessary enzymes for multiple modifications, including rhamnose synthesis and glycosyl transferases, dehydration and C-terminal S-[(Z)-2-aminovinyl-3-methyl]-d-cysteine (AviMeCys) formation28. Many bacteriocin gene clusters are not constrained to specific phyla, such as glycocins and lasso peptides, which are encoded within the genomes of both Gram-positive and Gram-negative bacteria48,49. Bacteriocin gene clusters may be present on plasmids capable of conjugation or transposable elements, thus facilitating transfer of the bacteriocin production phenotype in a competitive environment50. Recently, it was found that up to 3% of prophage encode biosynthetic gene clusters, the majority of which are for bacteriocin production and can demonstrably transfer production phenotype to nascent strains51.
Functional diversity
Although this Review mainly focuses on the antibacterial nature of bacteriocins, they are known to have a diverse set of other biological functions. A wide range of peptides possess antiviral activities52 such as subtilosin from Bacillus amyloliquefaciens that displays antiviral activity against herpes simplex viruses53. Peptides have been demonstrated to have cytotoxic effects towards cancer cell lines such as Laterosporulin10 (ref. 54) or possess in vivo antitumour activity as was shown for microcin E492 against human colorectal cancer cells using a xenografted zebrafish model55. Bacteriocins can modulate immune systems such as sublancin that can enhance phagocytic activity of macrophages and increase T cell frequency in mesenteric lymph nodes in mouse models of methicillin-resistant Staphylococcus aureus infection56,57. Thuricin-17, which is produced by Bacillus thuringiensis, has been shown to promote the growth of canola plants (Brassica napus L.) at high temperatures through an unknown mechanism, displaying a potential role for bacteriocins in supporting plant growth in changing climates7. Finally, human streptococcal pathogens co-regulate complex bacteriocin production and competence pathways linking fratricide and natural transformation, implicating a role for bacteriocins in gene acquisition58.
Narrow-to-broad spectrum peptides
Bacteriocins, such as antimicrobial peptides across all kingdoms of life, are typically cationic hydrophobic peptides, many of which are active at negatively charged bacterial cell surfaces59–61. The activities of bacteriocins, once they reach the cell surface, are diverse, including specific interactions with cell-wall or cell-membrane components, general membrane interaction and pore formation, or receptor-mediated cell penetration and interference with intracellular processes (for extensive mechanistic reviews, see refs. 62–64) (Fig. 2).
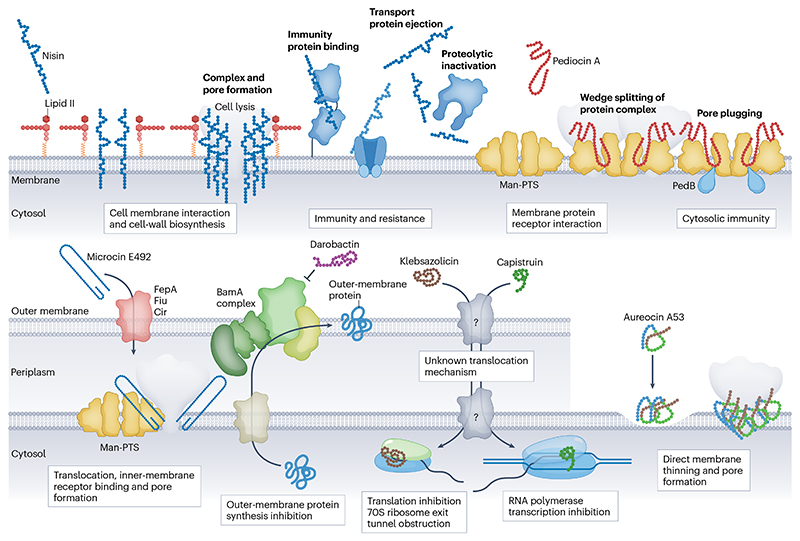
Fig. 2. Bacteriocins mechanisms of action and resistance mechanisms.
Bacteriocins are heterogeneous in function and exhibit various mechanisms of action. Bacteriocins are typically cationic hydrophobic peptides that are drawn towards anionic bacterial cell surfaces. At the surface, they can interact with specific receptors, such as nisin that complexes specifically to the cell-wall precursor molecule lipid II and subsequently forms pores across the membrane, which leads to cell death. Mechanisms of immunity and resistance protect strains from nisin by binding, ejecting or proteolytically cleaving the peptides. Pediocin-like bacteriocins (class II) attach to the extracellular surface of the mannose-phosphotransferase system (Man-PTS) core domain, penetrate the membrane and shift the core domain away from its V-motif, thereby forming a pore in the membrane. PedB proteins in the targeted bacterium can physically plug the pore formed by pediocin PA-1 and the Man-PTS, thereby providing cytosolic immunity. Other peptides target the membrane directly, such as aureocin 53 that initially causes Gram-positive membrane thinning by accumulating in the upper leaflet of the phospholipid bilayer, which is mediated by specific α-helices of the peptide, and then causes poration favoured by other α-helices within the peptide. Bacteriocins can also interrupt essential cellular processes such as outer-membrane protein integration of BamA by darobactin or inhibition of protein and nucleic acid synthesis of klebsazolicin and capistruin, respectively. Microcin E492 translocates across the outer membrane using a ‘Trojan horse’ strategy, wherein the siderophore moiety is recognized by outer-membrane transporters that facilitate uptake of the peptide into the periplasm, followed by binding to the mannose-PTS at the inner membrane and subsequent lethal disruption of membrane potential. Immunity to microcin E492 is provided by the action of the immunity protein MceB, via an unknown mechanism.
Extracellular targets
Nisin is well established to have a dual mechanism of action, binding the cell-wall precursor molecule lipid II, thereby inhibiting peptidoglycan biosynthesis and subsequent pore formation leading to cell death65. Initially, it was believed to form a pore complex of eight nisin and four lipid II molecules, although more recent evidence has shown that the pore complexes continue to grow causing massive cell membrane damage66 (Fig. 2). Numerous post-translationally modified bacteriocins share a conserved nisin-like N-terminal lipid II binding domain (rings A and B), but do not possess the same C-terminal domain, and do not form pores as a result67. This has been recently been demonstrated for the nisin-like peptides rombocin A68 and cesin69. Many bacteriocins do not target lipid II and instead use a different receptor or lipid component as a binding site70,71.
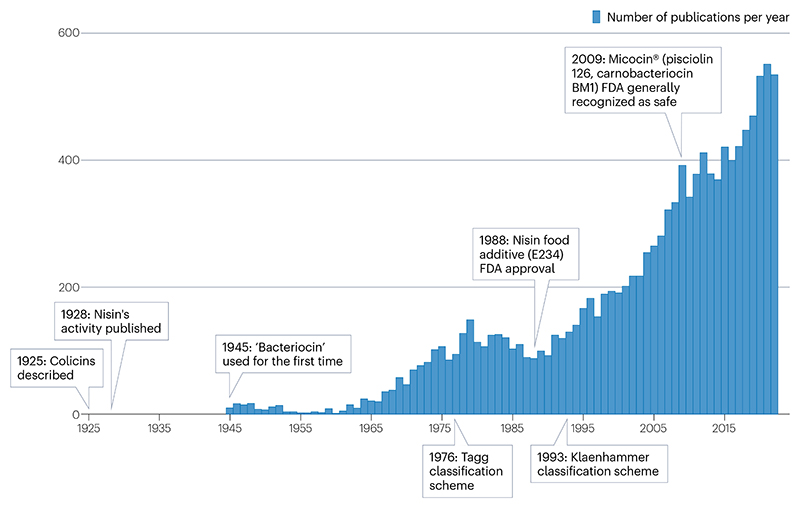
Box 1. History of bacteriocin nomenclature.
Searching the literature with the term bacteriocin returns more than 11,000 publications, describing a heterogenous group of compounds that mostly share two general features: bacterial ribosomally synthesized peptides/proteins that are antagonistic towards other bacteria. These compounds can range from multiprotein contractile nanomachines191 to antimicrobial secreted peptides192. The lack of a precise definition after a century of bacteriocin research results in a field that can often be contradictory and confusing.
Jacob et al.193 first coined ‘proteins of the colicin type’ as bacteriocins following the initial work of Gratia194 in 1925 describing colicins as the source of strain-specific antagonistic activity between Escherichia coli (see the figure). Subsequent publications used bacteriocin to describe lytic proteins from Gram-positive bacteria, such as megacin from Bacillus megaterium195. The discovery of nisin was published in the same decade as colicin2 but Gram-positive bacteria-derived antimicrobials were not as widely described as bacteriocins for decades196. In 1976, Tagg et al.196 acknowledged that no universally accepted definition existed and criticized the use of the term bacteriocin for poorly characterized compounds. It was proposed that bacteriocin should describe bactericidal compounds containing a biologically active protein moiety. The 1993 Klaenhammer scheme expanded this definition, which is the basis for modern bacteriocin classification. Peptides were grouped into class I (post-translationally modified) and class II (unmodified), with larger proteins grouped as class III (ref. 197). Research on bacteriocins from Gram-positive bacteria grew considerably, and in 2005 Cotter et al.198 suggested that bacteriocin be reserved for peptides rather than heat-labile proteins. The term has not ceased to be used for these larger proteins. Other schemes have been proposed including up to four classes, and many bacteriocins are now popularly referred to as RiPPs (ribosomally produced and post-translationally modified peptides)3,23,199. We suggest consistent usage of the two major class schemes for bacteriocins to provide clarity for future researchers navigating the literature.
Pediocin-like bacteriocins (class II) interact with the mannose phosphotransferase system (Man-PTS) forcing the channel into an open state, which was recently confirmed by cryo-electron microscopy72. The Man-PTS transporter is a receptor for several class II bacteriocins, such as lactococcins A and B and garvicins A, B and C, as was recently confirmed by deletion mutants, protein modelling and cryo-electron microscopy73,74, and is the target for microcin E492 after ‘Trojan horse’ translocation across the outer membrane75. The circular bacteriocin garvicin ML relies on the maltose ABC transporter to target Lactococcus lactis, whereas AS-48 is believed to interact based solely on electrostatic and hydrophobic interactions to insert directly into the lipid bilayer, leading to destabilization76,77. Direct membrane interaction is also the mechanism for the four helix-bundle bacteriocins epidermicin NI01 and aureocin A53, which have been found to have distinct multimodal ‘flowering’ pore-forming activity61.
Bacteriocin activities are typically restricted within the Gram status of the producer; that is, peptides derived from Gram-positive bacteria are typically far less active against Gram-negative bacteria without assistance of an outer-membrane permeabilizing agent78,79. Contradictory to this, a semipurified supernatant containing bacteriocins from Lacticaseibacillus paracasei CNCM I-5369 was found to be active against a panel of Gram-negative bacteria, which was confirmed by heterologous expression80. Bioengineering and rational design have been utilized in the absence of an outer-membrane permeabilizing compound (which would typically be required for Gram-positive-derived bacteriocins to inhibit Gram-negative bacteria) to produce variants of nisin with up to 12-fold greater activity including bactericidal activity against E. coli and Acinetobacter baumanii81. A hybrid peptide consisting of enterocin CRL35 fused with microcin V (Ent35–MccV) was shown to be active against the targets of both individual peptides, including Listeria monocytogenes and E. coli82.
Intracellular targets
Some bacteriocins interfere with essential intracellular processes, in particular those that target Gram-negative bacteria, which lack exposed peptidoglycan and inner cell membrane components. The TOMM klebsazolicin inhibits protein synthesis by blocking the ribosome exit tunnel, but its extracellular uptake mechanism is unknown26. The crystal structures of the lasso peptides microcin J25 and capistruin have revealed their specific sites of transcription inhibition within the RNA polymerase secondary channel following uptake mediated by a host of transport proteins83. Sactibiotics are a class known for their narrow spectra of inhibition. Recently, it was suggested that the R. gnavus- produced and Clostridioides- targeting sactibiotic ruminicoccin C could act through nucleic acid synthesis inhibition in a manner similar to metronidazole, but this has yet to be confirmed47.
Unknown targets
Despite ongoing characterization of the mechanisms of action of bacteriocins, many peptides of interest remain uncharacterized. Similarly, the recently described bacteroidetocins, a family of anti-Bacteroidales pediocin-like bacteriocins, have an unknown mechanism of action as Bacteroides species lack the mannose-PTS84. Although most research to date has focused on mechanisms of bacteriocins as free agents in liquid culture, a two-component Gram-negative bacteria-derived bacteriocin CdzCD was recently identified on the basis of cell-to-cell contact-dependent killing, for which the outer-membrane protein PerA was required for sensitivity, and has been suggested as the target receptor for CdzCD binding85. With vast recent improvements in protein structural modelling and prediction, we may begin to see more in-depth bacteriocin–target complex structures and gain insights into yet unknown mechanisms.
Immunity and resistance
The risk of target strains developing resistance against bacteriocins because of continuous use is a key concern, given the current situation with antimicrobial resistance and the antimicrobial arms race. Bacteriocin producers are self-protected by the action of immunity proteins, which may bind or export the peptides44, or block the action of peptides on protein complexes72. Resistance to bacteriocin activity can occur through general mechanisms, such as cell-wall thickening causing resistance to the lipid II binding lasso peptide, siamycin I59, or by truncation of a protein target similar to the transporter of the mannose-PTS86 but high-level resistance occurs through acquisition of specific resistance genes. Bacteriocins can be inactivated owing to the activity of specific enzymes such as the nisin resistance protein, which degrades nisin87. Worryingly, some pathogenic species express generalized transporter proteins such as Streptococcus agalactiae bacitracin type efflux protein (SaNsrFP) that protects against numerous cell-wall targeting compounds including lantibiotics88. As bacteriocins have not been extensively used in the clinical setting, the extent or risk for resistance in clinical use is unknown and to our knowledge for many classes of bacteriocins, no resistance mechanisms have been characterized yet.
Production and bioengineering
One of the most appealing attributes of bacteriocins is their gene-encoded nature, which enables the alteration of peptides using simple techniques and the generation of new-to-nature structures with unknown characteristics (Fig. 3). Wild-type bacteriocin producers often generate low bacteriocin yields because production is a high-energy process for the cell and is thus tightly controlled. This can result in excessive production costs that render bacteriocins non-viable commercially89. Production optimization has therefore been examined by altering growth parameters in addition to exploiting their gene-encoded nature by manipulating the native producers90 and/or peptide expression in more efficient or peptide-insensitive heterologous hosts91,92.
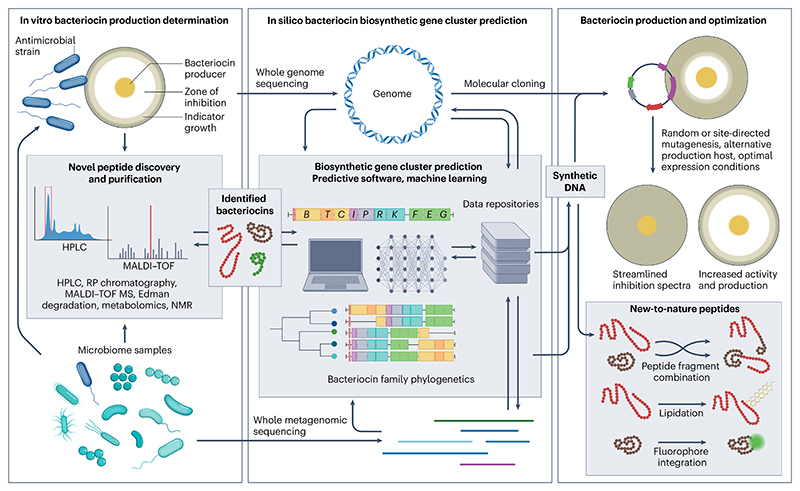
Fig. 3. Methods of in silico bacteriocin identification and translation to in vitro production, optimization and bioengineering.
Until the recent proliferation of large-scale whole metagenomics, bacteriocins were mainly identified in vitro through screening with agar-based assays. Specific antimicrobial peptides could be identified and purified through a number of techniques including mass spectrometry and HPLC (left panel) and corresponded to a biosynthetic gene cluster identified from a sequenced genome (middle panel, top). Now large data sets can be more easily generated or sourced from data repositories and mined for bacteriocin gene clusters with improved predictive software and corresponded with metabolomic data to identify peptides (left panel, bottom and middle panel). In silico mined peptides and known peptides can be heterologously expressed in desirable hosts to optimize production and/or modified to generate mutant peptides with desirable characteristics such as improved activity or streamlined inhibition spectra (right panel, top). Novel techniques have also been developed to further modify expressed peptides through recombination of peptide fragments generating new-to-nature peptides or modification with compounds of specific properties, such as lipids or fluorophores (right panel, bottom). HPLC, high-performance liquid chromatography; MALDI–TOF MS, matrix-assisted laser desorption ionization–time-of-flight mass spectrometry; RP, reverse phase.
Heterologous expression optimization
Bacteriocins of all classes have been subject to heterologous expression to express novel silent genes93, understand their mechanism of biosynthesis94,95 or to improve their production for commercial viability92. Although post-translationally modified (class I) bacteriocins require essential modification enzymes to produce active mature peptides, class II bacteriocins are particularly suited to heterologous expression as many are ‘simple’ linear peptides. A range of Gram-positive bacteria, Gram-negative bacteria, eukaryotic hosts and diverse plasmids have been exploited for this such as E. coli for expression of pediocin PA-1 (syn. pediocin AcH), bactofencin A and lactocin AB46,96, L. lactis for expression of plantaricin JK and enterocin NKR-5-3B97,98 and the yeasts Pichia pastoris (syn. Komagataella phaffii) and Kluyveromyces lactis for the expression of enterocins99,100.
Beyond heterologous expression, simple optimization of culture conditions can increase bacteriocin yield, as demonstrated by researchers by improving garvicin KS production from Lactococcus garvieae as high as 60-fold when cultured in pasteurized milk and tryptone, compared with GM17 media90. Combining the improved media with increased gene dosage of the core peptide of the native producer further increased production to the high concentration of 1.2 g of peptide per litre90. Increasing gene copy number has also improved production of the sactibiotic thurincin H, in which an additional three copies of the structural gene, thnA, resulted in double the yield from broth culture101. Synthesis of plantaricin E/F in Lactobacillus plantarum strains is induced by both an auto-inducing peptide PlnA1 and acetate via a distinct interaction with the PlnB1 response regulator102. Exploiting this mechanistic understanding enabled an increase in the yield of plantaricin in culture by addition of sodium acetate at a concentration of 1 g l−1 (ref. 102). Limited studies report extensive optimization or comparative purification yields, as peptide purification is typically a means of characterizing peptides at small scale in vitro rather than for potential commercial application. However, to scale up to in vivo experiments with pure peptide and for bacteriocin production to become more commercially viable, optimization must be explored in greater detail.
Expanding the functional diversity of bacteriocins
Nisin is perhaps the most genetically manipulated bacteriocin with thousands of unique variants having been generated using random and site-direct PCR mutagenesis techniques, in addition to an entire expression system developed based on the tight regulation of nisin production103. Mutagenesis methods have produced nisin variants with increased activity against biofilms and variants with greater activity against Gram-negative bacteria such as Cronobacter sakazakii, Salmonella typhimurium and E. coli104. Rombocin K, a bioengineered variant of nisin-like rombocin A, was found to have a twofold lower minimum inhibitory concentration than the original peptide when tested against Listeria monocytogenes68. The inhibition spectrum of nisin has also been refined in the derivatives M17Q, T2L and HTK, which possess increased anti-Staphylococcus activity and reduced anti-Lactococcus activity19,105. Similarly, saturation mutagenesis of mutacin 1140 generated 418 single amino acid variants of which Phe1Ile ultimately emerged as the most promising in vivo candidate for Clostridioides difficile infection and associated diarrhoea, which in turn underwent a second round of mutagenesis optimization resulting in a more potent peptide106,107. Such methods may improve a promising bacteriocin for application or be used to overcome a limitation, such as the oxidation-resistant M31L variant of pediocin PA-1, in which oxidation-associated activity reduction challenges the wider application of wild-type peptide108. Bioengineering can be used as a means to overcome proteolytic degradation, such as nisin S29P that is immune to the action of the nisin resistance serine protease109 and cesin R15G that has improved resistance to trypsin69.
Recently developed methods have utilized incorporation of non-canonical amino acids into the peptide backbone, which can then be conjugated to other compounds via ‘click chemistry’. Using the non-canonical and more-hydrophobic amino acid ethionine in the place of methionine, heterodimeric and fluorescently labelled nisin variants have been generated, each of which retained antimicrobial activity110. A similar method lipidated nisin variants, which had improved bio-activity and specificity against pathogenic strains111. Conjugation of bacteriocin peptides to molecules such as fatty acids can be exploited to produce bacteriocins that form colloidal stable assemblies, a feature which could be utilized to overcome problems with solubility of hydrophobic peptides112. Novel methods such as this can be used to further expand, edit and hone already diverse peptide functionality and future downstream applications.
Discovery in the meta-omics era
In recent years, bacteriocin discovery has been catapulted into the meta-omics era with the availability of vast genomic data sets and dedicated bioinformatic software enabling in silico screening of bacterial genomes and large-scale metagenomic mining for the presence of novel bacteriocin gene clusters (Fig. 3). Despite convincing evidence that bacteriocin-encoding genes are widely distributed across many genera, confirming bacteriocin production from individual strains isolated during screening studies has proven difficult. Before the increased accessibility of next-generation sequencing technologies, the majority of novel bacteriocins were identified in vitro through laborious screening of hundreds or thousands of bacterial isolates113. These culture-dependent screening methods face logistical microbiological limitations such as the choice of media on which samples are plated, the choice of indicator species and the growth conditions (pH, atmosphere composition and temperature), which can all lower the likelihood of bacteriocin detection. Failure to detect bacteriocin production does not mean lack of bacteriocin presence, but could be due to the inadvertent choice of an insensitive indicator species or a limited or lack of induction and production of an active bacteriocin. Genomic approaches that are agnostic of active expression and rely on bioinformatic predictions of the presence of biosynthetic gene clusters can overcome these issues, but are not without their own limitations. For example, in silico approaches alone do not confirm biological activity nor do they enable the purification and characterization of a bacteriocin. It is likely that a combination of in silico and culture-dependent methods will prove most effective in terms of identifying and isolating novel functional bacteriocins.
Bacteriocin gene cluster prediction
As sequencing technology has improved and sequence databases have grown, a range of tools have been developed for mining bacteriocin genes encoded in bacterial DNA. Tools have been developed that span from ‘point-and-click’ interfaces suitable for non-bioinformaticians to command-line controlled packages to detect biosynthetic gene clusters in genomic and metagenomic data sets at a range of assembly levels. BAGEL and antiSMASH are well-tested and frequently updated tools that are designed for the identification of bacteriocin (and secondary metabolite) biosynthetic gene clusters in assembled bacterial genomes114,115. Both tools are useful for the characterization of an unknown antimicrobial-producing isolate but are limited by predictions based on enzyme homology and predicted core peptide characteristics, which may miss truly novel compounds. In addition, they can be low to medium throughput based on the skill level of the user. Despite this, they have enabled the discovery of bacteriocins in several hundreds to thousands of genomes, such as the identification of 101 lanthipeptide gene clusters from 223 Bacillus cereus strains116, the characterization of the biotechnological potential of 213 diverse lactic acid bacteria117 and the genome-guided expansion of a bacteriocin family from an initial 11 members to widespread detection of homologues in hundreds of strains25. More recently, a simple tool for mining specific RiPP classes based on BAGEL has been developed to interrogate metagenomic data sets for bacteriocin biosynthetic gene clusters118.
With continuous growth in computing power and bioinformatic tool development, artificial intelligence has also been introduced into bacteriocin research, with machine learning being utilized to predict biosynthetic gene clusters and predict chemical structures following post-translational modification. RODEO (Rapid ORF Description and Evaluation Online) was the first tool available to use machine learning and was able to expand the number of known lasso peptides 10-fold upon its initial publication and subsequently increased the number of known glycocins from 6 to 10 distinct characterized members using only a single enzyme as a starting point49,119.
From in silico to in vitro
Bioinformatics can have an important role in identifying potential bacteriocins, but this should be supported by functional characterization by isolation of a producing strain, chemical synthesis or heterologous expression from genomic or synthetic DNA in a suitable host. When candidate bacteriocin-producing strains are identified in silico but production cannot be demonstrated, or when putative producers are not available, E. coli has been used as a heterologous bacteriocin expression ‘workhorse’. Expression in E. coli strains has enabled the characterization of structural peptides and biosynthetic machinery for glycocins and lantibiotics40,120,121. Bioinformatically mined class II bacteriocins have also been expressed in E. coli, including pediocin PA-1 and lactobacilli- derived pediocin-like peptides, enabling the identification of genes essential for bacteriocin production46,93. Gram-positive hosts have also been extensively utilized for the expression of bacteriocins, including the reliable nisin-inducible expression system in L. lactis, which has recently been used for the functional characterization of the circular bacteriocin circularin A mined from a clostridial genome95. Genome mining paired with heterologous expression is a powerful combination to explore novel compounds but can be low throughput and failure-prone depending on the choice of the host and vector. Researchers recently developed a powerful platform combining high-throughput bioinformatics and an integrated robotic system to express 96 RiPP gene clusters from diverse bacterial phyla involving 383 bio-synthetic genes, with a success rate of 86% (ref. 122). High-throughput methods such as this could be used to explore the Integrated Microbial Genome Atlas of biosynthetic Gene Clusters (IMG-ABC) database that consists of 20,000 gene clusters, of which a small number has been experimentally validated123.
Integrating omics technologies
Whole metabolomic methods are of growing interest in bacteriocin and RiPPs research (for an extensive recent review, see ref. 124). Peptidogenomics, which is mass spectrometry-guided genome mining for peptides, have successfully identified a novel lanthipeptide and production gene cluster from the cell extract and genome of a single strain125. MetaMiner has been developed to search for novel compounds from billions of mass spectra and compare with diverse metagenomic data sets such as human microbiome samples126. ‘Omic-mining’ can successfully identify novel peptides and biosynthetic gene clusters in an untargeted manner, but without ‘wet-laboratory’ demonstration of compound-specific antimicrobial activity, all such compounds should not be classed as bacteriocins. As the number of known biosynthetic gene clusters continues to grow, it is inevitable that biological characterization will lag behind and will have to be upscaled to explore applications of novel peptides.
Translation from food to clinic
Food preservatives
Fermentation as a means of food preservation, and bacteriocins unknowingly as by-products of thereof, has been utilized by humanity for millennia. Much of early bacteriocin research was performed in the context of food-fermenting organisms, such as the description of nisin in the context of delayed dairy fermentation owing to producer inhibition, and continues to be studied to this day2,127. Vast amounts of historical and ongoing research continue to describe the efficacy of bacteriocin producers (as fermentation starters or adjuncts) and their products to inhibit spoilage organisms in different products. However, despite decades of bacteriocin use in the food industry (nisin was designated as generally recognized as safe by the FDA in 1988 (ref. 128)) and the array of bacteriocins that have been identified to date, only a small number are currently commercially available as food preservatives (mainly nisin A, nisin Z and pediocin PA-1). Most bacteriocin-preserving agents are available as partially purified fermentates, some of which also contain the producing organism. Nisin is available as a powder with degrees of purity under the product names Nisaplin® (Danisco, Dupont), Chrisin® (Chr. Hansen) and vegetal or white NisinA/Z® and NisinA/Z® P (Handary). Pediocin PA-1 is used as an antilisterial additive in food production in a number of powdered fermentate forms such as ALTA 2351/2341 (Kerry Group) and Fargo 23® (formerly in production by now defunct Quest International) approved by EFSA’s Qualified Presumption of Safety status129. A formulation containing Carnobacterium maltaromaticum CB1 and derived bacteriocins labelled Micocin® (Griffith Foods) has been approved for use in North America and Canada for the preservation of meats130.
Live strains and microbiome modulators
Microbiome modulation
Microbiome research continues to grow exponentially with considerable efforts directed towards associating particular microorganisms or microbial compositions with human diseases or conditions131–133. Identifying such correlations will naturally lead to attempts to develop tools for modulating the microbiome to enhance specific health-associated taxa or to reduce disease-associated taxa. Nutrition and lifestyle are obvious microbiome modulating strategies but these are essentially ‘blunt tools’, generating largely nonspecific and potentially widespread changes to gut microbiota composition and functionality. Antibiotics are also blunt tools, and despite being necessary for treatment and prevention of infectious agents, their lack of precision causes collateral damage to beneficial gut microorganisms, risking adverse health events such as C. difficile over-growth or the expansion of antibiotic-resistant strains22,134,135. This effect is not restricted to the gut, with antibiotic treatment also restructuring microbiomes of the skin, vagina and oral cavity136–138. Bacteriocins may prove to be an exciting alternative as potent antimicrobials, which may have narrow target spectra to target specific genera and species, and can therefore be utilized to more precisely inhibit pathogens or pathobionts, leaving benign commensals unharmed. Several class II bacteriocins have been found not to alter the overall gut microbiota composition but to reduce specific taxa such as Enterococcaceae and Clostridium spp. when consumed by mice139. Thuricin CD, produced by B. thuringiensis, has also been shown to specifically reduce clostridia in a faecal fermentation model, whereas vancomycin, a commonly used treatment in clinical settings, has a broad impact on the bacterial community140. Even broad-spectrum bacteriocins can induce only subtle changes in microbiome compositions when compared with traditional antibiotics, as was demonstrated with microcin J25 relative to rifampicin in a continuous culture porcine model141. Targeted microbiome modulation should also consider potential ecosystem effects as removal of a targeted species or genus could lead to unintended increases or decreases in others142. Simplified model ecosystems such as the OMM12 model have shown that bacteriocin production can have broad effects on neighbouring strains, and removal of a single species, in this case, a bacteriocin-producing strain, leads to the increased abundance of numerous others143.
Delivery to the gut
Therapeutics for human or animal consumption must be carefully formulated to be delivered in an active form to the correct anatomical site for function. For example, a small-molecule antibiotic must survive harsh environments during gastrointestinal transit to be absorbed and function systemically to prevent or treat infection. Bacteriocins have long been thought to be proteolytically degraded during gastrointestinal transit144 and therefore must be protected by encapsulation for applications directed at the large intestine. Recent evidence has challenged this dogma and shows that some bacteriocins, such as nisin, may be more resistant than previously thought, with detectable intact nisin surviving in vivo gastrointestinal transit in pigs and microcin J25 resisting degradation during simulated human digestion37,38. Despite this, many bacteriocins can be readily degraded by proteases in gastric transit145,146, and peptide delivery to the gut faces challenges of producing sufficient quantities to administer to large animals and the degree of purity required for in vivo studies. Fortunately, bacteriocins can also be deployed in the form of a probiotic or live-biotherapeutic agent that arrives at the desired destination and delivers a bacteriocin dose in situ. Researchers recently showed that genetically modified E. coli Nissle 1917 expressing microcin l47 in a preclinical mouse model significantly reduced carbapenem-resistant Klebsiella pneumoniae after 1 week oral administration of 108 cells per day without alteration of the resident microbiota34. E. coli expressing the unmodified enterocins A/B and hiracin JM79 have also been shown to reduce vancomycin-resistant Enterococcus when provided in drinking water ad libitum (5 × 108 cfu ml−1) in a vancomycin-resistant Enterococcus-colonized mouse model14. Live bacteriocin producers can also be used to treat or modulate microbiomes outside the gut, such as the use of an L. lactis strain producing lacticin 3147 in a liquid paraffin-based emulsion that was used to treat bovine mastitis (109 cfu 5 ml−1 dose) as successfully as the standard antibiotic treatment147.
Bacteriocin producers that are closely related to the intended target species can have the added benefit of potentially being able to occupy the same niche, potentially providing future colonization resistance against pathogens. Bacteriocin-21-producing E. faecalis were found to evict their drug-resistant relatives in the mouse intestine when provided in drinking water ad libitum (5 × 108 cfu ml−1) without perturbation of the overall community50. However, the plasmid-encoded nature of this bacteriocin resulted in conjugation to native enterococci providing immunity and the capacity to produce the bacteriocin. These data facilitate a scenario in which a multidrug-resistant strain could gain another tool to aid colonization and therefore must be cautiously considered when developing bacteriocin producers as targeted live biotherapeutics.
Other methods for ensuring unscathed bacteriocin delivery to site of action include through formulation with other compounds that increase peptide stability under biologically relevant conditions148. Silica mesoporous matrices have been utilized as protective delivery vehicles for bactofencin A and nisin A, resulting in active peptides that are resistant to trypsin149 and pepsin150 degradation, respectively. Hydrogels have been developed to ensure sustained release of bacteriocins over time, including an injectable polysaccharide gel containing nisin A active for up to 10 days151, and a two-phase release gel of subtilosin developed for inhibition of Gardnerella vaginalis, which causes bacterial vaginosis152. Formulation of bacteriocins into nanoparticles to increase the antimicrobial activity or widen the spectrum of the resulting nanoconjugate is a growing trend for food preservation and clinical application (reviewed elsewhere153). Recently, a mix of five bacteriocins adsorbed on alginate nanoparticles was found to be 500-fold more active than the peptides against clinically relevant E. coli154, and microcin J25-chitosan nanoparticles were active against enterotoxigenic E. coli in addition to ameliorating lipopolysaccharide-induced cytotoxicity of macrophages and downregulating lipopolysaccharide-stimulated pro-inflammatory cytokines155. Emerging technologies such as these should continue to improve and develop bacteriocin candidates for application in food preservation and in clinical settings.
Clinical application
In addition to few commercial bacteriocin products on the market, none is currently approved for human therapeutic use. Research and development for clinical application of bacteriocins has been chronically underfunded, with 70 novel antimicrobials under development in 2020, of which none were bacteriocins156. Despite this lack of considerable investment, many preclinical and some clinical studies have been performed demonstrating that bacteriocins, and particularly lantibiotics, exert antimicrobial activity in vivo in tolerable quantities (Table 2). Several bacteriocins and their producers have been investigated for the oral treatment of C. difficile infection and associated diarrhoea. Preclinical trials have assessed OG-716 (mutacin derivative) for the treatment of C. difficile infection and associated diarrhoea when administered via oral gavage (20 mg kg−1 bodyweight) in golden hamsters, finding the peptide to be safe and effective at preventing relapse107,157. A bacteriocin-producing Bacillus velezensis strain as a live biotherapeutic for the treatment of C. difficile infection has been proven effective preclinically in mice (single oral dose of 5 × 108 cfu) and in miniature swine (single oral dose of 5 × 108 cfu and 28 days administration of 5 × 109 cfu in capsules) and recently concluded a phase I clinical trial158. The semi-synthetic thiopeptide LFF571 (derived from GE2270A) has undergone phase I and II clinical trials (oral administration of 200 mg four times a day for 10 days) for C. difficile infections and was determined to be comparable to vancomycin treatment159,160.
Table 2. Bacteriocins and producers undergoing investigation for therapeutic application.
| Form | Bacteriocin | Class | Producer | Clinical trial phase (completed) | Treatment and animal model (if applicable) | Administration | Result | Company |
|---|---|---|---|---|---|---|---|---|
| Peptide | NVB333 (ref. 187) | Lantibiotic | Actinoplanes liguriae NCIMB41362 | Preclinical | Mouse thigh infection model, MRSA disseminated | Intravenous | Equal efficacy to vancomycin | Novacta |
| Peptide | NVB302 (ref. 188) | Lantibiotic | A. liguriae NCIMB41362 | Phase I | Clostridioides difficile infection | Oral | Not reported189 | Novacta |
| Peptide | Mutacin 1140 and analogues163 | Lantibiotic | Streptococcus mutans JH1000 | Preclinical | Mouse MRSA skin and systemic infection | Intravenous or topical | Effective intravenous treatment | Sano Chemicals |
| Peptide | OG716 (mutacin 1140 derivative)107 | Lantibiotic | S. mutans | Preclinical | Hamster model of Clostridioides difficile infection and associated diarrhoea | Oral | 100% survival and no relapse | – |
| Peptide | NAI-107 (microbisporicin)190 | Lantibiotic | Microbispora corallina | Preclinical | Acute severe infections in mouse and rat models | Intravenous | Lower effective dose than best alternative, rapid bactericidal activity | Naicons SRL, Sentinella Pharmaceuticals |
| Peptide | Moli1901 (duramycin/lancuvotide)169,170 | Lantibiotic | Streptomyces cinnamoneum | Phases I and II | Cystic fibrosis symptoms | Intranasal, inhalation | Well tolerated, no difference from placebo | Lantibio/AOP Orphan Pharmaceuticals |
| LBT | Sh-lantibiotic-α/β (ref. 162) | Lantibiotic | Staphylococcus hominis ShA9 | Phase I | Atopic dermatitis | Topical | Achieved primary safety end point and decreased Staphylococcus aureus | – |
| LBT | Abp-118 (ref. 164) | Pediocin-like | Ligilactobacillus salivarius PS7 | Phase I | Recurrent acute otitis media | Oral consumption | Significant decrease (84%) in AOM episodes | ProbiSearch |
| LBT | Multiple158 | Multiple | Bacillus velezensis ADS024 | Preclinical | C. difficile infection | Oral | Inhibited C. difficile infection in mice and did not colonize mice or swine after 28-day repeat dose | Adiso Therapeutics |
AOM, acute otitis media; LBT, live biotherapeutic; MRSA, methicillin-resistant Staphylococcus aureus.
Beyond C. difficile infection, the lantibiotic NAI-107 (microbisporicin) has undergone a series of preclinical rodent studies for severe antimicrobial-resistant infections with promising results161. In animal studies, nisin and lacticin-3147 have shown promise in the treatment of mastitis147, and lantibiotic-producing Staphylococcus hominis ShA9 decreased S. aureus, improving atopic dermatitis when delivered to the arms in a lotion (106 cfu cm−2 applied twice daily for 1 week) and meeting primary and secondary endpoints in a phase I clinical trial162. Mutacin 1140-derived topical treatments (5 or 10 mg kg−1 per day for 7 days) have been proven safe and more effective than vancomycin for methicillin-resistant S. aureus infection in mice and have also been effective when administered intravenously (10 mg kg−1 per day for 6 days)163. Fewer trials have been performed with class II bacteriocins, but recently the effect of an Abp-118-producing L. salivarius live biotherapeutic on recurrent acute otitis media (middle ear infection) in children was examined164. The treatment was proven highly effective (84% reduction in episodes over 6 months of daily consumption of 109 cfu) and as one of the most common infections in children, a commercial bacteriocin therapy could greatly reduce antibiotic prescription (Table 2).
Beyond antimicrobials
Thiostrepton, derived from Streptomyces spp., has long been used in veterinary medicine for the topical treatment of acute otitis media, but has never been approved for human use nor have any other thiopeptides165. However, it is one of several bacteriocins gaining interest beyond antimicrobial application. Thiostrepton selectively and potently represses the cancer-associated transcriptional activator and therapeutic target, FoxM1 (ref. 166). Diverse bacteriocins have been studied in vitro and in vivo for chemotherapeutic treatment of different cancer types including nisin167, microcin E492 (ref. 55) and enterocin LNS18 (ref. 168), with none having yet progressed to phase I clinical trials. Moli1901 (duramycin/lancuvotide) has undergone phase I and II clinical trials as a therapeutic for cystic fibrosis as a host chloride channel activator, although with limited efficacy169,170. Bacteriocins are not immune to the pitfalls of drug development and can be abandoned in early clinical trials owing to the poor performance or difficulties in production. Ultimately, the pharmacokinetic and pharmacodynamic attributes of bacteriocins under investigation are critical limiting factors in enabling them to become therapeutic agents. Vastly increased funding of discovery, characterization, preclinical and human trials is required to adequately advance suitable bacteriocin candidates to the frontline of clinics, which currently continues to be sorely lacking.
Conclusions
Bacteriocins are a diverse group of antimicrobial peptides produced across a number of bacterial phyla, and ongoing discovery efforts continue to reveal biological insights. These discoveries include not only novel peptide families but also instances of structurally similar peptides and biosynthetic machinery present in taxonomically unrelated species. This phenomenon may shed light on aspects of horizontal gene transfer and the evolution of antimicrobial peptides, akin to the convergent evolution observed in Eukaryotic defensins171.
Bacteriocin discovery has been transformed in recent years from the labour-intensive process of identifying producer isolates in vitro to the in silico genome mining that can yield hundreds of related peptide sequences within seconds. Machine learning has become an invaluable tool in detecting bacteriocin sequences within vast genomic data sets, and with continuous advancements in computing power and artificial intelligence training, we can expect the identification of even more novel peptides and perhaps even novel bacteriocin classes in the future. Despite the growing number of known peptides, our understanding of their mechanisms of action lags behind. Investigating the cellular targets of bacteriocins is crucial for choosing the correct antimicrobial for specific pathogenic species and for understanding resistance mechanisms. This will also enable the rational design of bacteriocins through peptide bioengineering, potentially leading to the development of highly targeted and potent therapeutic agents.
Bacteriocins hold great promise in complex microbial communities such as the gut microbiome with their ability to selectively target specific pathogens or pathobionts without disrupting the overall community structure and function. However, the potential unintended consequences of such interventions must be carefully considered to avoid disruptions to microbial ecosystems that could lead to undesirable health effects. One substantial challenge is translating laboratory discoveries into clinical applications. Bacteriocins offer a viable alternative or adjunct therapy in the treatment of infections, especially against multidrug-resistant pathogens. Realizing these opportunities will require substantial funding and rigorous clinical studies. In addition to their antimicrobial properties, structurally related peptides have been identified that lack antimicrobial activity but possess other valuable bioactivities, such as pain relief or anti-inflammatory effects172. These findings expand the horizon of bacteriocins beyond antimicrobials, conceivably offering multipurpose therapeutic agents.
In conclusion, bacteriocins have the potential to act as precise and versatile tools in an ongoing battle against antimicrobial-resistant microorganisms. As they continue to be explored, a path is paved whereby these remarkable molecules could have a pivotal role in safe-guarding human health and well-being. Although challenges lie ahead, the potential for bacteriocins as tools to eliminate clinically relevant microorganisms and precisely shape microbial communities is an exciting and promising avenue for research.
Footnotes
Author contributions
I.S. researched data for article. C.H., I.S. and R.P.R. substantially contributed to discussion of the content, wrote the article and reviewed and edited the manuscript before submission.
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Reviews Microbiology thanks Djamel Drider, Michael Chikindas and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
References
- 1.Koehbach J, Craik DJ. The vast structural diversity of antimicrobial peptides. Trends Pharmacol Sci. 2019;40:517–528. doi: 10.1016/j.tips.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Rogers LA. The inhibiting effect of Streptococcus lactis on Lactobacillus bulgaricus. J Bacteriol. 1928;16:321. doi: 10.1128/jb.16.5.321-325.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson EM, et al. Bacteriocins as food preservatives: challenges and emerging horizons. Crit Rev Food Sci. 2018;58:2743–2767. doi: 10.1080/10408398.2017.1340870. [DOI] [PubMed] [Google Scholar]
- 4.Gross E, Morell JL. Structure of nisin. J Am Chem Soc. 1971;93:4634–4635. doi: 10.1021/ja00747a073. [DOI] [PubMed] [Google Scholar]
- 5.European Food Safety Authority. Opinion of the Scientific Panel on food additives, flavourings, processing aids and materials in contact with food (AFC) related to the use of nisin (E234) as a food additive. EFSA J. 2006;4:314. [Google Scholar]
- 6.Vieco-Saiz N, et al. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front Microbiol. 2019;10:57. doi: 10.3389/fmicb.2019.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nazari M, Yaghoubian I, Smith DL. The stimulatory effect of Thuricin 17, a PGPR-produced bacteriocin, on canola (Brassica napus L.) germination and vegetative growth under stressful temperatures. Front Plant Sci. 2022;13:1079180. doi: 10.3389/fpls.2022.1079180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz MR, et al. Structural and functional analysis of EntV reveals a 12 amino acid fragment protective against fungal infections. Nat Commun. 2022;13:6047. doi: 10.1038/s41467-022-33613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martínez-García M, et al. Autophagic-related cell death of Trypanosoma brucei induced by bacteriocin AS-48. Int J Parasitol Drug. 2018;8:203–212. doi: 10.1016/j.ijpddr.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quintana VM, et al. Antiherpes simplex virus type 2 activity of the antimicrobial peptide subtilosin. J Appl Microbiol. 2014;117:1253–1259. doi: 10.1111/jam.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meade E, Slattery MA, Garvey M. Bacteriocins, potent antimicrobial peptides and the fight against multidrug resistant species: resistance is futile? Antibiotics. 2020;9:32. doi: 10.3390/antibiotics9010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. IACG, Interagency Coordination Group on Antimicrobial Resistance. Report to the Secretary General of the United Nations. No Time to Wait: Securing the Future from Drug-Resistant Infections. 2019 https://www.who.int/publications/i/item/no-time-to-wait-securing-the-future-from-drug-resistant-infections .
- 13.World Health Organization. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis. Technical document. 2017. https://www.who.int/publications/i/item/WHO-EMP-IAU-2017.12 .
- 14.Geldart KG, et al. Engineered E. coli Nissle 1917 for the reduction of vancomycin-resistant Enterococcus in the intestinal tract. Bioeng Transl Med. 2018;3:197–208. doi: 10.1002/btm2.10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telhig S, et al. Evaluating the potential and synergetic effects of microcins against multidrug-resistant Enterobacteriaceae. Microbiol Spectr. 2022;10:e02752–21. doi: 10.1128/spectrum.02752-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai Y, et al. A new antibiotic selectively kills Gram-negative pathogens. Nature. 2019;576:459–464. doi: 10.1038/s41586-019-1791-1. [This study provides a detailed discovery and characterization of a novel bacteriocin class that inhibits Gram-negative pathogens] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng F, et al. Plantaricin A reverses resistance to ciprofloxacin of multidrug-resistant Staphylococcus aureus by inhibiting efflux pumps. Environ Microbiol. 2022;24:4818–4833. doi: 10.1111/1462-2920.16158. [DOI] [PubMed] [Google Scholar]
- 18.Soltani S, et al. Bacteriocins as a new generation of antimicrobials: toxicity aspects and regulations. FEMS Microbiol Rev. 2021;45:fuaa039. doi: 10.1093/femsre/fuaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Field D, et al. Bio-engineered nisin with increased anti-Staphylococcus and selectively reduced anti-Lactococcus activity for treatment of bovine mastitis. Int J Mol Sci. 2021;22:3480. doi: 10.3390/ijms22073480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heilbronner S, Krismer B, Brötz-Oesterhelt H, Peschel A. The microbiome-shaping roles of bacteriocins. Nat Rev Microbiol. 2021;19:726–739. doi: 10.1038/s41579-021-00569-w. [DOI] [PubMed] [Google Scholar]
- 21.Cryan JF, et al. The microbiota–gut–brain axis. Physiol Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 22.Becattini S, Taur Y, Pamer EG. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med. 2016;22:458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montalbán-López M, et al. New developments in RiPP discovery, enzymology and engineering. Nat Prod Rep. 2021;38:130–239. doi: 10.1039/d0np00027b. [This article provides an intricately detailed review of ribosomally produced and post-translationally modified peptide groups, biosynthesis and biochemistry, of which many are bacteriocins] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trimble MJ, Mlynárčik P, Kolář M, Hancock REW. Polymyxin: alternative mechanisms of action and resistance. CSH Perspect Med. 2016;6:a025288. doi: 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Heel AJ, Montalban-Lopez M, Oliveau Q, Kuipers OP. Genome-guided identification of novel head-to-tail cyclized antimicrobial peptides, exemplified by the discovery of pumilarin. Microb Genom. 2017;3:e000134. doi: 10.1099/mgen.0.000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metelev M, et al. Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel. Nat Chem Biol. 2017;13:1129–1136. doi: 10.1038/nchembio.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pashou E, et al. Identification and characterization of corynaridin, a novel linaridin from Corynebacterium lactis. Microbiol Spectr. 2023;11:e01756–22. doi: 10.1128/spectrum.01756-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deisinger JP, et al. Dual targeting of the class V lanthipeptide antibiotic cacaoidin. iScience. 2023;26:106394. doi: 10.1016/j.isci.2023.106394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ekblad B, et al. Structure–function analysis of the two-peptide bacteriocin plantaricin EF. Biochemistry. 2016;55:5106–5116. doi: 10.1021/acs.biochem.6b00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowakowski M, et al. Spatial attributes of the four-helix bundle group of bacteriocins — the high-resolution structure of BacSp222 in solution. Int J Biol Macromol. 2018;107:2715–2724. doi: 10.1016/j.ijbiomac.2017.10.158. [DOI] [PubMed] [Google Scholar]
- 31.Hammi I, et al. Maltaricin CPN, a new class IIa bacteriocin produced by Carnobacterium maltaromaticum CPN isolated from mould-ripened cheese. J Appl Microbiol. 2016;121:1268–1274. doi: 10.1111/jam.13248. [DOI] [PubMed] [Google Scholar]
- 32.Sugrue I, O’Connor PM, Hill C, Stanton C, Ross RP. Actinomyces produces defensin-like bacteriocins (actifensins) with a highly degenerate structure and broad antimicrobial activity. J Bacteriol. 2020;202:e00529–e00619. doi: 10.1128/JB.00529-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perez RH, Zendo T, Sonomoto K. Circular and leaderless bacteriocins: biosynthesis, mode of action, applications, and prospects. Front Microbiol. 2018;9:2085. doi: 10.3389/fmicb.2018.02085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortzfeld BM, et al. Microcin MccI47 selectively inhibits enteric bacteria and reduces carbapenem-resistant Klebsiella pneumoniae colonization in vivo when administered via an engineered live biotherapeutic. Gut Microb. 2022;14:2127633. doi: 10.1080/19490976.2022.2127633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawa N, et al. Identification and characterization of lactocyclicin Q, a novel cyclic bacteriocin produced by Lactococcus sp. strain QU 12. Appl Environ Microbiol. 2009;75:1552–1558. doi: 10.1128/AEM.02299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scholz R, et al. Amylocyclicin, a novel circular bacteriocin produced by bacillus amyloliquefaciens FZB42. J Bacteriol. 2014;196:1842–1852. doi: 10.1128/JB.01474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu F, Van Heel AJ, Kuipers OP. Leader- and terminal residue requirements for circularin A biosynthesis probed by systematic mutational analyses. ACS Synth Biol. 2023;12:852–862. doi: 10.1021/acssynbio.2c00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lagedroste M, Reiners J, Smits SHJ, Schmitt L. Impact of the nisin modification machinery on the transport kinetics of NisT. Sci Rep. 2020;10:12295. doi: 10.1038/s41598-020-69225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagedroste M, Smits SHJ, Schmitt L. Substrate specificity of the secreted nisin leader peptidase NisP. Biochemistry. 2017;56:4005–4014. doi: 10.1021/acs.biochem.7b00524. [DOI] [PubMed] [Google Scholar]
- 40.Viel JH, Jaarsma AH, Kuipers OP. Heterologous expression of mersacidin in Escherichia coli elucidates the mode of leader processing. ACS Synth Biol. 2021;10:600–608. doi: 10.1021/acssynbio.0c00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viel JH, Kuipers OP. Mutational studies of the mersacidin leader reveal the function of its unique two-step leader processing mechanism. ACS Synth Biol. 2022;11:1949–1957. doi: 10.1021/acssynbio.2c00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pérez-Ramos A, Ladjouzi R, Benachour A, Drider D. Evidence for the involvement of pleckstrin homology domain-containing proteins in the transport of enterocin DD14 (EntDD14); a leaderless two-peptide bacteriocin. Int J Mol Sci. 2021;22:12877. doi: 10.3390/ijms222312877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Der Donk WA, Nair SK. Structure and mechanism of lanthipeptide biosynthetic enzymes. Curr Opin Struct Biol. 2014;29:58–66. doi: 10.1016/j.sbi.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khosa S, Lagedroste M, Smits SH. J. Protein defense systems against the lantibiotic nisin: function of the immunity protein NisI and the resistance protein NSR. Front Microbiol. 2016;7:504. doi: 10.3389/fmicb.2016.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spieß T, Korn SM, Kötter P, Entian K-D. Autoinduction specificities of the lantibiotics subtilin and nisin. Appl Environ Microbiol. 2015;81:7914–7923. doi: 10.1128/AEM.02392-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mesa-Pereira B, et al. Controlled functional expression of the bacteriocins pediocin PA-1 and bactofencin A in Escherichia coli. Sci Rep. 2017;7:3069. doi: 10.1038/s41598-017-02868-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiumento S, et al. Ruminococcin C, a promising antibiotic produced by a human gut symbiont. Sci Adv. 2019;5:eaaw9969. doi: 10.1126/sciadv.aaw9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh V, Rao A. Distribution and diversity of glycocin biosynthesis gene clusters beyond Firmicutes. Glycobiology. 2021;31:89–102. doi: 10.1093/glycob/cwaa061. [DOI] [PubMed] [Google Scholar]
- 49.Tietz JI, et al. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat Chem Biol. 2017;13:470–478. doi: 10.1038/nchembio.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kommineni S, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–722. doi: 10.1038/nature15524. [This detailed paper illustrates not only the efficacy of using related species to inhibit pathogens in vivo but also the risks of bacteriocin gene transfer to host microorganisms] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dragoš A, et al. Phages carry interbacterial weapons encoded by biosynthetic gene clusters. Curr Biol. 2021;31:3479–3489.:e5. doi: 10.1016/j.cub.2021.05.046. [This study is an interesting description of bacteriocin gene clusters present in bacteriophage and demonstration of bacteriocin gene transfer via phage infection] [DOI] [PubMed] [Google Scholar]
- 52.Fu Y, Jaarsma AH, Kuipers OP. Antiviral activities and applications of ribosomally synthesized and post-translationally modified peptides (RiPPs) Cell Mol Life Sci. 2021;78:3921–3940. doi: 10.1007/s00018-021-03759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torres NI, et al. Safety, formulation, and in vitro antiviral activity of the antimicrobial peptide subtilosin against herpes simplex virus type 1. Probiot Antimicrob Prot. 2013;5:26–35. doi: 10.1007/s12602-012-9123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baindara P, Gautam A, Raghava GPS, Korpole S. Anticancer properties of a defensin like class IId bacteriocin Laterosporulin10. Sci Rep. 2017;7:46541. doi: 10.1038/srep46541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varas MA, et al. Exploiting zebrafish xenografts for testing the in vivo antitumorigenic activity of microcin E492 against human colorectal cancer cells. Front Microbiol. 2020;11:405. doi: 10.3389/fmicb.2020.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S, et al. Enhancement of macrophage function by the antimicrobial peptide sublancin protects mice from methicillin-resistant Staphylococcus aureus. J Immunol Res. 2019;2019:1–13. doi: 10.1155/2019/3979352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J, Chen J, Yang G, Tao L. Sublancin protects against methicillin-resistant Staphylococcus aureus infection by the combined modulation of innate immune response and microbiota. Peptides. 2021;141:170533. doi: 10.1016/j.peptides.2021.170533. [DOI] [PubMed] [Google Scholar]
- 58.Shanker E, Federle M. Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes. 2017;8:15. doi: 10.3390/genes8010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan S, Ludwig KC, Müller A, Schneider T, Nodwell JR. The lasso peptide siamycin-I targets lipid II at the Gram-positive cell surface. ACS Chem Biol. 2019;14:966–974. doi: 10.1021/acschembio.9b00157. [DOI] [PubMed] [Google Scholar]
- 60.Zhu L, Zeng J, Wang C, Wang J. Structural basis of pore formation in the mannose phosphotransferase system by pediocin PA-1. Appl Environ Microbiol. 2022;88:e01992–21. doi: 10.1128/AEM.01992-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammond K, et al. Flowering poration — a synergistic multi-mode antibacterial mechanism by a bacteriocin fold. iScience. 2020;23:101423. doi: 10.1016/j.isci.2020.101423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ongpipattanakul C, et al. Mechanism of action of ribosomally synthesized and post-translationally modified peptides. Chem Rev. 2022;122:14722–14814. doi: 10.1021/acs.chemrev.2c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pérez-Ramos A, Madi-Moussa D, Coucheney F, Drider D. Current knowledge of the mode of action and immunity mechanisms of LAB-bacteriocins. Microorganisms. 2021;9:2107. doi: 10.3390/microorganisms9102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antoshina DV, Balandin SV, Ovchinnikova TV. Structural features, mechanisms of action, and prospects for practical application of class II bacteriocins. Biochemistry. 2022;87:1387–1403. doi: 10.1134/S0006297922110165. [DOI] [PubMed] [Google Scholar]
- 65.Field D, Fernandez de Ullivarri M, Ross RP, Hill C. After a century of nisin research — where are we now? FEMS Microbiol Rev. 2023;47:fuad023. doi: 10.1093/femsre/fuad023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scherer KM, Spille J-H, Sahl H-G, Grein F, Kubitscheck U. The lantibiotic nisin induces lipid II aggregation, causing membrane instability and vesicle budding. Biophys J. 2015;108:1114–1124. doi: 10.1016/j.bpj.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dickman R, et al. A chemical biology approach to understanding molecular recognition of lipid II by nisin(1–12): synthesis and NMR ensemble analysis of nisin(1–12) and analogues. Chem Eur J. 2019;25:14572–14582. doi: 10.1002/chem.201902814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo L, et al. Rombocin, a short stable natural nisin variant, displays selective antimicrobial activity against Listeria monocytogenes and employs a dual mode of action to kill target bacterial strains. ACS Synth Biol. 2024;13:370–383. doi: 10.1021/acssynbio.3c00612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo L, et al. Cesin, a short natural variant of nisin, displays potent antimicrobial activity against major pathogens despite lacking two C-terminal macrocycles. Microbiol Spectr. 2023;11:e05319–e05322. doi: 10.1128/spectrum.05319-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heeney DD, Yarov-Yarovoy V, Marco ML. Sensitivity to the two peptide bacteriocin plantaricin EF is dependent on CorC, a membrane-bound, magnesium/cobalt efflux protein. MicrobiologyOpen. 2019;8:e827. doi: 10.1002/mbo3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kjos M, et al. Sensitivity to the two-peptide bacteriocin lactococcin G is dependent on Uppp, an enzyme involved in cell-wall synthesis. Mol Microbiol. 2014;92:1177–1187. doi: 10.1111/mmi.12632. [DOI] [PubMed] [Google Scholar]
- 72.Zhu L, Zeng J, Wang J. Structural basis of the immunity mechanisms of pediocin-like bacteriocins. Appl Environ Microbiol. 2022;88:e00481–22. doi: 10.1128/aem.00481-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tymoszewska A, Diep DB, Aleksandrzak-Piekarczyk T. The extracellular loop of Man-PTS subunit IID is responsible for the sensitivity of Lactococcus garvieae to garvicins A, B and C. Sci Rep. 2018;8:15790. doi: 10.1038/s41598-018-34087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li R, Duan J, Zhou Y, Wang J. Structural basis of the mechanisms of action and immunity of lactococcin A, a class IId bacteriocin. Appl Environ Microbiol. 2023;89:e00066–23. doi: 10.1128/aem.00066-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang K, Zeng J, Liu X, Jiang T, Wang J. Structure of the mannose phosphotransferase system (Man-PTS) complexed with microcin E492, a pore-forming bacteriocin. Cell Discov. 2021;7:20. doi: 10.1038/s41421-021-00253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gabrielsen C, Brede DA, Hernández PE, Nes IF, Diep DB. The maltose ABC transporter in Lactococcus lactis facilitates high-level sensitivity to the circular bacteriocin garvicin ML. Antimicrob Agents Chemother. 2012;56:2908–2915. doi: 10.1128/AAC.00314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cebrián R, et al. The bacteriocin AS-48 requires dimer dissociation followed by hydrophobic interactions with the membrane for antibacterial activity. J Struct Biol. 2015;190:162–172. doi: 10.1016/j.jsb.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 78.Li Q, et al. Outer-membrane-acting peptides and lipid II-targeting antibiotics cooperatively kill Gram-negative pathogens. Commun Biol. 2021;4:31. doi: 10.1038/s42003-020-01511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Helander IM, Mattila-Sandholm T. Permeability barrier of the Gram-negative bacterial outer membrane with special reference to nisin. Int J Food Microbiol. 2000;60:153–161. doi: 10.1016/s0168-1605(00)00307-x. [DOI] [PubMed] [Google Scholar]
- 80.Belguesmia Y, Bendjeddou K, Kempf I, Boukherroub R, Drider D. Heterologous biosynthesis of five new class II bacteriocins from Lactobacillus paracasei CNCM I-5369 with antagonistic activity against pathogenic Escherichia coli strains. Front Microbiol. 2020;11:1198. doi: 10.3389/fmicb.2020.01198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Q, Montalban-Lopez M, Kuipers OP. Increasing the antimicrobial activity of nisin-based lantibiotics against Gram-negative pathogens. Appl Environ Microbiol. 2018;84:e00052–18. doi: 10.1128/AEM.00052-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Acuña L, Picariello G, Sesma F, Morero RD, Bellomio A. A new hybrid bacteriocin, Ent35–MccV, displays antimicrobial activity against pathogenic Gram-positive and Gram-negative bacteria. FEBS Open Biol. 2012;2:12–19. doi: 10.1016/j.fob.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Braffman NR, et al. Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin. Proc Natl Acad Sci USA. 2019;116:1273–1278. doi: 10.1073/pnas.1817352116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coyne MJ, et al. A family of anti-Bacteroidales peptide toxins wide-spread in the human gut microbiota. Nat Commun. 2019;10:3460. doi: 10.1038/s41467-019-11494-1. [Description of a novel family of bacteriocins widespread in Bacteroidetes that are related to Gram-positive bacteriocins] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.García-Bayona L, Gozzi K, Laub MT. Mechanisms of resistance to the contact-dependent bacteriocin CdzC/D in Caulobacter crescentus. J Bacteriol. 2019;201:e00538–e00618. doi: 10.1128/JB.00538-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Geldart K, Kaznessis YN. Characterization of class IIa bacteriocin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2017;61:e02033–16. doi: 10.1128/AAC.02033-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun Z, et al. Novel mechanism for nisin resistance via proteolytic degradation of nisin by the nisin resistance protein NSR. Antimicrob Agents Chemother. 2009;53:1964–1973. doi: 10.1128/AAC.01382-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gottstein J, et al. New insights into the resistance mechanism for the BceAB-type transporter SaNsrFP. Sci Rep. 2022;12:4232. doi: 10.1038/s41598-022-08095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ongey EL, Neubauer P. Lanthipeptides: chemical synthesis versus in vivo biosynthesis as tools for pharmaceutical production. Microb Cell Fact. 2016;15:97. doi: 10.1186/s12934-016-0502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Telke AA, et al. Over 2000-fold increased production of the leaderless bacteriocin garvicin KS by increasing gene dose and optimization of culture conditions. Front Microbiol. 2019;10:389. doi: 10.3389/fmicb.2019.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cui Y, et al. Mining, heterologous expression, purification, antibactericidal mechanism, and application of bacteriocins: a review. Comp Rev Food Sci Food Safe. 2021;20:863–899. doi: 10.1111/1541-4337.12658. [DOI] [PubMed] [Google Scholar]
- 92.Mesa-Pereira B, Rea MC, Cotter PD, Hill C, Ross RP. Heterologous expression of biopreservative bacteriocins with a view to low cost production. Front Microbiol. 2018;9:1654. doi: 10.3389/fmicb.2018.01654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Collins FWJ, et al. Reincarnation of bacteriocins from the lactobacillus pangenomic graveyard. Front Microbiol. 2018;9:1298. doi: 10.3389/fmicb.2018.01298. [This publication describes mining of peptides from functionally ‘dead’ biosynthetic gene clusters and expressing them using characterized machinery] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Himes PM, Allen SE, Hwang S, Bowers AA. Production of sactipeptides in Escherichia coli: probing the substrate promiscuity of subtilosin A biosynthesis. ACS Chem Biol. 2016;11:1737–1744. doi: 10.1021/acschembio.6b00042. [DOI] [PubMed] [Google Scholar]
- 95.Liu F, Van Heel AJ, Chen J, Kuipers OP. Functional production of clostridial circularin A in Lactococcus lactis NZ9000 and mutational analysis of its aromatic and cationic residues. Front Microbiol. 2022;13:1026290. doi: 10.3389/fmicb.2022.1026290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu W, et al. Expression and purification of recombinant Lactobacillus casei bacteriocin and analysis of its antibacterial activity. CyTA J Food. 2020;18:301–308. [Google Scholar]
- 97.Xu Y, Yang L, Li P, Gu Q. Heterologous expression of class IIb bacteriocin plantaricin JK in Lactococcus lactis. Protein Expr Purif. 2019;159:10–16. doi: 10.1016/j.pep.2019.02.013. [DOI] [PubMed] [Google Scholar]
- 98.Perez RH, et al. Functional analysis of genes involved in the biosynthesis of enterocin NKR-5-3B, a novel circular bacteriocin. J Bacteriol. 2016;198:291–300. doi: 10.1128/JB.00692-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arbulu S, et al. Cloning and expression of synthetic genes encoding native, hybrid- and bacteriocin-derived chimeras from mature class IIa bacteriocins, by Pichia pastoris (syn. Komagataella spp) Food Res Int. 2019;121:888–899. doi: 10.1016/j.foodres.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 100.Jiménez JJ, et al. Use of synthetic genes for cloning, production and functional expression of the bacteriocins enterocin A and bacteriocin E 50-52 by Pichia pastoris and Kluyveromyces lactis. Mol Biotechnol. 2014;56:571–583. doi: 10.1007/s12033-014-9731-7. [DOI] [PubMed] [Google Scholar]
- 101.Gaona-Mendoza AS, Barboza-Corona JE, Casados-Vázquez LE. Improving the yields of thurincin H in a native producer strain. Anton Leeuw. 2020;113:1061–1066. doi: 10.1007/s10482-020-01408-3. [DOI] [PubMed] [Google Scholar]
- 102.Meng F, et al. Expression of a novel bacteriocin — the plantaricin Pln1-in Escherichia coli and its functional analysis. Protein Expr Purif. 2016;119:85–93. doi: 10.1016/j.pep.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 103.Mierau I, Kleerebezem M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl Microbiol Biotechnol. 2005;68:705–717. doi: 10.1007/s00253-005-0107-6. [DOI] [PubMed] [Google Scholar]
- 104.Field D, et al. A bioengineered nisin derivative to control biofilms of Staphylococcus pseudintermedius. PLoS ONE. 2015;10:e0119684. doi: 10.1371/journal.pone.0119684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Twomey E, Hill C, Field D, Begley M. Bioengineered nisin derivative M17Q has enhanced activity against Staphylococcus epidermidis. Antibiotics. 2020;9:305. doi: 10.3390/antibiotics9060305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kers JA, et al. Blueprints for the rational design of therapeutic mutacin 1140 variants. Chem Biol Drug Des. 2018;92:1940–1953. doi: 10.1111/cbdd.13365. [DOI] [PubMed] [Google Scholar]
- 107.Kers JA, et al. Mutacin 1140 lantibiotic variants are efficacious against Clostridium difficile infection. Front Microbiol. 2018;9:415. doi: 10.3389/fmicb.2018.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kuniyoshi TM, et al. An oxidation resistant pediocin PA-1 derivative and penocin A display effective anti-Listeria activity in a model human gut environment. Gut Microb. 2022;14:2004071. doi: 10.1080/19490976.2021.2004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Field D, et al. Bioengineering nisin to overcome the nisin resistance protein. Mol Microbiol. 2019;111:717–731. doi: 10.1111/mmi.14183. [DOI] [PubMed] [Google Scholar]
- 110.Deng J, Viel JH, Chen J, Kuipers OP. Synthesis and characterization of heterodimers and fluorescent nisin species by incorporation of methionine analogues and subsequent click chemistry. ACS Synth Biol. 2020;9:2525–2536. doi: 10.1021/acssynbio.0c00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo L, Wang C, Broos J, Kuipers OP. Lipidated variants of the antimicrobial peptide nisin produced via incorporation of methionine analogs for click chemistry show improved bioactivity. J Biol Chem. 2023;299:104845. doi: 10.1016/j.jbc.2023.104845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wiman E, et al. Development of novel broad-spectrum antimicrobial lipopeptides derived from plantaricin NC8 β. Sci Rep. 2023;13:4104. doi: 10.1038/s41598-023-31185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rea MC, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci USA. 2010;107:9352–9357. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van Heel AJ, et al. BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018;46:W278–W281. doi: 10.1093/nar/gky383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Blin K, et al. AntiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023;51:W46–W50. doi: 10.1093/nar/gkad344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xin B, et al. The Bacillus cereus group is an excellent reservoir of novel lanthipeptides. Appl Environ Microbiol. 2015;81:1765–1774. doi: 10.1128/AEM.03758-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sun Z, et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun. 2015;6:8322. doi: 10.1038/ncomms9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Costa SS, Da Silva Moia G, Silva A, Baraúna RA, De Oliveira Veras AA. BADASS: bacteriocin-diversity assessment software. BMC Bioinf. 2023;24:24. doi: 10.1186/s12859-022-05106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ren H, Biswas S, Ho S, Van Der Donk WA, Zhao H. Rapid discovery of glycocins through pathway refactoring in Escherichia coli. ACS Chem Biol. 2018;13:2966–2972. doi: 10.1021/acschembio.8b00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Biswas S, Garcia De Gonzalo CV, Repka LM, Van Der Donk WA. Structure–activity relationships of the S-linked glycocin sublancin. ACS Chem Biol. 2017;12:2965–2969. doi: 10.1021/acschembio.7b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kaunietis A, Buivydas A, Čitavičius DJ, Kuipers OP. Heterologous biosynthesis and characterization of a glycocin from a thermophilic bacterium. Nat Commun. 2019;10:1115. doi: 10.1038/s41467-019-09065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ayikpoe RS, et al. A scalable platform to discover antimicrobials of ribosomal origin. Nat Commun. 2022;13:6135. doi: 10.1038/s41467-022-33890-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hadjithomas M, et al. IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. mBio. 2015;6:e00932–15. doi: 10.1128/mBio.00932-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zdouc MM, Van Der Hooft JJJ, Medema MH. Metabolome-guided genome mining of RiPP natural products. Trends Pharmacol Sci. 2023;44:532–541. doi: 10.1016/j.tips.2023.06.004. [DOI] [PubMed] [Google Scholar]
- 125.Mohimani H, et al. Automated genome mining of ribosomal peptide natural products. ACS Chem Biol. 2014;9:1545–1551. doi: 10.1021/cb500199h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cao L, et al. MetaMiner: a scalable peptidogenomics approach for discovery of ribosomal peptide natural products with blind modifications from microbial communities. Cell Syst. 2019;9:600–608.:e4. doi: 10.1016/j.cels.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.van Gijtenbeek LA, et al. Gene-trait matching and prevalence of nisin tolerance systems in Lactococus lactis. Front Bioeng Biotechnol. 2021;9:622835. doi: 10.3389/fbioe.2021.622835. [This systematic study teases apart the relationship between nisin genes and production and tolerance phenotypes of lactococci in dairy fermentation] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.GovInfo. US Food and Drug Administration (FDA). ‘Nisin preparation’. Code of Federal Regulations, Title 21. 2011. https://www.govinfo.gov/app/details/CFR-2011-title21-vol3/CFR-2011-title21-vol3-sec184-1538 .
- 129.European Food Safety Authority (EFSA) Introduction of a qualified presumption of safety (QPS) approach for assessment of selected microorganisms referred to EFSA — Opinion of the Scientific Committee. EFSA J. 2007;587:1–16. [Google Scholar]
- 130.US Food and Drug Administration. GRAS Notice No 305 Carnobacterium maltaromaticum Strain CB1 (Viable and Heat-treated) 2010. https://www.cfsanappsexternal.fda.gov/scripts/fdcc/index.cfm?set=GRASNotices&id=305 .
- 131.Glassner KL, Abraham BP, Quigley EMM. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol. 2020;145:16–27. doi: 10.1016/j.jaci.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 132.Cryan JF, O’Riordan KJ, Sandhu K, Peterson V, Dinan TG. The gut microbiome in neurological disorders. Lancet Neurol. 2020;19:179–194. doi: 10.1016/S1474-4422(19)30356-4. [DOI] [PubMed] [Google Scholar]
- 133.Dabke K, Hendrick G, Devkota S. The gut microbiome and metabolic syndrome. J Clin Invest. 2019;129:4050–4057. doi: 10.1172/JCI129194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yip AYG, et al. Antibiotics promote intestinal growth of carbapenem-resistant Enterobacteriaceae by enriching nutrients and depleting microbial metabolites. Nat Commun. 2023;14:5094. doi: 10.1038/s41467-023-40872-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Maier L, et al. Unravelling the collateral damage of antibiotics on gut bacteria. Nature. 2021;599:120–124. doi: 10.1038/s41586-021-03986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dessinioti C, Katsambas A. Propionibacterium acnes and antimicrobial resistance in acne. Clin Dermatol. 2017;35:163–167. doi: 10.1016/j.clindermatol.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 137.Jo J-H, et al. Alterations of human skin microbiome and expansion of antimicrobial resistance after systemic antibiotics. Sci Transl Med. 2021;13:eabd8077. doi: 10.1126/scitranslmed.abd8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dunlop AL, et al. Stability of the vaginal, oral, and gut microbiota across pregnancy among African American women: the effect of socioeconomic status and antibiotic exposure. PeerJ. 2019;7:e8004. doi: 10.7717/peerj.8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Umu ÖCO, et al. The potential of class II bacteriocins to modify gut microbiota to improve host health. PLoS ONE. 2016;11:e0164036. doi: 10.1371/journal.pone.0164036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rea MC, et al. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci USA. 2011;108:4639–4644. doi: 10.1073/pnas.1001224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Naimi S, et al. Impact of microcin J25 on the porcine microbiome in a continuous culture model. Front Microbiol. 2022;13:930392. doi: 10.3389/fmicb.2022.930392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ríos Colombo NS, et al. Impact of bacteriocin-producing strains on bacterial community composition in a simplified human intestinal microbiota. Front Microbiol. 2023;14:1290697. doi: 10.3389/fmicb.2023.1290697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Weiss AS, et al. Nutritional and host environments determine community ecology and keystone species in a synthetic gut bacterial community. Nat Commun. 2023;14:4780. doi: 10.1038/s41467-023-40372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gough R, et al. Simulated gastrointestinal digestion of nisin and interaction between nisin and bile. LWT. 2017;86:530–537. [Google Scholar]
- 145.Rea MC, et al. Bioavailability of the anti-clostridial bacteriocin thuricin CD in gastrointestinal tract. Microbiology. 2014;160:439–445. doi: 10.1099/mic.0.068767-0. [DOI] [PubMed] [Google Scholar]
- 146.Soltani S, et al. Gastrointestinal stability and cytotoxicity of bacteriocins from Gram-positive and Gram-negative bacteria: a comparative in vitro study. Front Microbiol. 2022;12:780355. doi: 10.3389/fmicb.2021.780355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kitching M, et al. A live bio-therapeutic for mastitis, containing Lactococcus lactis DPC3147 with comparable efficacy to antibiotic treatment. Front Microbiol. 2019;10:2220. doi: 10.3389/fmicb.2019.02220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Flynn J, Ryan A, Hudson SP. Pre-formulation and delivery strategies for the development of bacteriocins as next generation antibiotics. Eur J Pharm Biopharm. 2021;165:149–163. doi: 10.1016/j.ejpb.2021.05.015. [DOI] [PubMed] [Google Scholar]
- 149.Durack E, et al. Protecting bactofencin A to enable its antimicrobial activity using mesoporous matrices. Int J Pharm. 2019;558:9–17. doi: 10.1016/j.ijpharm.2018.12.035. [DOI] [PubMed] [Google Scholar]
- 150.Flynn J, Mallen S, Durack E, O’Connor PM, Hudson SP. Mesoporous matrices for the delivery of the broad spectrum bacteriocin, nisin A. J Colloid Interface Sci. 2019;537:396–406. doi: 10.1016/j.jcis.2018.11.037. [DOI] [PubMed] [Google Scholar]
- 151.Flynn J, Durack E, Collins MN, Hudson SP. Tuning the strength and swelling of an injectable polysaccharide hydrogel and the subsequent release of a broad spectrum bacteriocin, nisin A. J Mater Chem B. 2020;8:4029–4038. doi: 10.1039/d0tb00169d. [DOI] [PubMed] [Google Scholar]
- 152.Sundara Rajan S, et al. Polyethylene glycol-based hydrogels for controlled release of the antimicrobial subtilosin for prophylaxis of bacterial vaginosis. Antimicrob Agents Chemother. 2014;58:2747–2753. doi: 10.1128/AAC.02446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sulthana R, Archer AC. Bacteriocin nanoconjugates: boon to medical and food industry. J Appl Microbiol. 2021;131:1056–1071. doi: 10.1111/jam.14982. [DOI] [PubMed] [Google Scholar]
- 154.Belguesmia Y, Hazime N, Kempf I, Boukherroub R, Drider D. New bacteriocins from Lacticaseibacillus paracasei CNCM I-5369 adsorbed on alginate nanoparticles are very active against Escherichia coli. Int J Mol Sci. 2020;21:8654. doi: 10.3390/ijms21228654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Haitao Y, Yifan C, Mingchao S, Shuaijuan H. A novel polymeric nanohybrid antimicrobial engineered by antimicrobial peptide MccJ25 and chitosan nanoparticles exerts strong antibacterial and anti-inflammatory activities. Front Immunol. 2022;12:811381. doi: 10.3389/fimmu.2021.811381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.World Health Organisation. 2020 Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis. 2021. https://www.who.int/publications/i/item/9789240021303 .
- 157.Pulse ME, et al. Pharmacological, toxicological, and dose range assessment of OG716, a novel lantibiotic for the treatment of Clostridium difficile-associated infection. Antimicrob Agents Chemother. 2019;63:e01904–e01918. doi: 10.1128/AAC.01904-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Murphy CK, et al. Novel, non-colonizing, single-strain live biotherapeutic product ADS024 protects against Clostridioides difficile infection challenge in vivo. World J Gastrointest Pathophysiol. 2023;14:71–85. doi: 10.4291/wjgp.v14.i4.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bhansali SG, et al. Pharmacokinetics of LFF571 and vancomycin in patients with moderate Clostridium difficile infections. Antimicrob Agents Chemother. 2015;59:1441–1445. doi: 10.1128/AAC.04252-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mullane K, et al. Multicenter, randomized clinical trial to compare the safety and efficacy of LFF571 and vancomycin for Clostridium difficile infections. Antimicrob Agents Chemother. 2015;59:1435–1440. doi: 10.1128/AAC.04251-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lepak AJ, Marchillo K, Craig WA, Andes DR. In vivo pharmacokinetics and pharmacodynamics of the lantibiotic NAI-107 in a neutropenic murine thigh infection model. Antimicrob Agents Chemother. 2015;59:1258–1264. doi: 10.1128/AAC.04444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nakatsuji T, et al. Development of a human skin commensal microbe for bacteriotherapy of atopic dermatitis and use in a phase 1 randomized clinical trial. Nat Med. 2021;27:700–709. doi: 10.1038/s41591-021-01256-2. [A phase I clinical trial demonstrates the effect of a bacteriocin producer as a treatment for human illness] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ju M, et al. Evaluation of analogs of mutacin 1140 in systemic and cutaneous methicillin-resistant Staphylococcus aureus infection models in mice. Front Microbiol. 2022;13:1067410. doi: 10.3389/fmicb.2022.1067410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cárdenas N, et al. Prevention of recurrent acute otitis media in children through the use of Lactobacillus salivarius PS7, a target-specific probiotic strain. Nutrients. 2019;11:376. doi: 10.3390/nu11020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Studdert V, Hughes K. A clinical trial of a topical preparation of miconazole, polymyxin and prednisolone in the treatment of otitis externa in dogs. Aust Vet J. 1991;68:193–195. doi: 10.1111/j.1751-0813.1991.tb03188.x. [DOI] [PubMed] [Google Scholar]
- 166.Bailly C. The bacterial thiopeptide thiostrepton. An update of its mode of action, pharmacological properties and applications. Eur J Pharmacol. 2022;914:174661. doi: 10.1016/j.ejphar.2021.174661. [DOI] [PubMed] [Google Scholar]
- 167.Norouzi Z, Salimi A, Halabian R, Fahimi H. Nisin, a potent bacteriocin and anti-bacterial peptide, attenuates expression of metastatic genes in colorectal cancer cell lines. Microb Pathog. 2018;123:183–189. doi: 10.1016/j.micpath.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 168.Al-Madboly LA, et al. Purification, characterization, identification, and anticancer activity of a circular bacteriocin from Enterococcus thailandicus. Front Bioeng Biotechnol. 2020;8:450. doi: 10.3389/fbioe.2020.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Grasemann H, et al. Inhalation of Moli1901 in patients with cystic fibrosis. Chest. 2007;131:1461–1466. doi: 10.1378/chest.06-2085. [DOI] [PubMed] [Google Scholar]
- 170.Oliynyk I, Varelogianni G, Roomans GM, Johannesson M. Effect of duramycin on chloride transport and intracellular calcium concentration in cystic fibrosis and non-cystic fibrosis epithelia. APMIS. 2010;118:982–990. doi: 10.1111/j.1600-0463.2010.02680.x. [DOI] [PubMed] [Google Scholar]
- 171.Shafee TM, Lay FT. Convergent evolution of defensin sequence, structure and function. Cell Mol Life Sci. 2017;74:663–682. doi: 10.1007/s00018-016-2344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Iorio M, et al. A glycosylated, labionin-containing lanthipeptide with marked antinociceptive activity. ACS Chem Biol. 2014;9:398–404. doi: 10.1021/cb400692w. [DOI] [PubMed] [Google Scholar]
- 173.Gouda H, et al. Three-dimensional solution structure of bottromycin A2: a potent antibiotic active against methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococci. Chem Pharm Bull. 2012;60:169–171. doi: 10.1248/cpb.60.169. [DOI] [PubMed] [Google Scholar]
- 174.Ishida K, Matsuda H, Murakami M, Yamaguchi K. Kawaguchipeptin B, an antibacterial cyclic undecapeptide from the cyanobacterium Microcystis aeruginosa. J Nat Prod. 1997;60:724–726. doi: 10.1021/np970146k. [DOI] [PubMed] [Google Scholar]
- 175.Kalamara M, Abbott J, Sukhodub T, MacPhee C, Stanley-Wall NR. The putative role of the epipeptide EpeX in Bacillus subtilis intra-species competition. Microbiology. 2023;169:001344. doi: 10.1099/mic.0.001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zendo T, et al. Kunkecin A, a new nisin variant bacteriocin produced by the fructophilic lactic acid bacterium, Apilactobacillus kunkeei FF30-6 isolated from honey bees. Front Microbiol. 2020;11:571903. doi: 10.3389/fmicb.2020.571903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Singh M, Chaudhary S, Sareen D. Roseocin, a novel two-component lantibiotic from an actinomycete. Mol Microbiol. 2020;113:326–337. doi: 10.1111/mmi.14419. [DOI] [PubMed] [Google Scholar]
- 178.Ortiz-López FJ, et al. Cacaoidin, first member of the new lanthidin RiPP family. Angew Chem Int Ed Engl. 2020;59:12654–12658. doi: 10.1002/anie.202005187. [DOI] [PubMed] [Google Scholar]
- 179.Cheung-Lee WL, et al. Discovery of ubonodin, an antimicrobial lasso peptide active against members of the Burkholderia cepacia complex. ChemBioChem. 2020;21:1335–1340. doi: 10.1002/cbic.201900707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Suzuki M, et al. Isolation and structure determination of new linear azole-containing peptides spongiicolazolicins A and B from Streptomyces sp. CWH03. Appl Microbiol Biotechnol. 2021;105:93–104. doi: 10.1007/s00253-020-11016-w. [DOI] [PubMed] [Google Scholar]
- 181.Jin M, Liu L, Wright SAI, Beer SV, Clardy J. Structural and functional analysis of pantocin A: an antibiotic from Pantoea agglomerans discovered by heterologous expression of cloned genes. Angew Chem Int Ed Engl. 2003;42:2898–2901. doi: 10.1002/anie.200351053. [DOI] [PubMed] [Google Scholar]
- 182.Hudson GA, Zhang Z, Tietz JI, Mitchell DA, Van Der Donk WA. In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin. J Am Chem Soc. 2015;137:16012–16015. doi: 10.1021/jacs.5b10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Balty C, et al. Ruminococcin C, an anti-clostridial sactipeptide produced by a prominent member of the human microbiota Ruminococcus gnavus. J Biol Chem. 2019;294:14512–14525. doi: 10.1074/jbc.RA119.009416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Palmer JD, et al. Microcin H47: a class IIb microcin with potent activity against multidrug resistant Enterobacteriaceae. ACS Infect Dis. 2020;6:672–679. doi: 10.1021/acsinfecdis.9b00302. [DOI] [PubMed] [Google Scholar]
- 185.O’Shea EF, et al. Bactofencin A, a new type of cationic bacteriocin with unusual immunity. mBio. 2013;4:e00498–13. doi: 10.1128/mBio.00498-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ladjouzi R, Lucau-Danila A, Benachour A, Drider D. A leaderless two-peptide bacteriocin, Enterocin DD14, is involved in its own self-immunity: evidence and insights. Front Bioeng Biotechnol. 2020;8:644. doi: 10.3389/fbioe.2020.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Boakes S, Weiss WJ, Vinson M, Wadman S, Dawson MJ. Antibacterial activity of the novel semisynthetic lantibiotic NVB333 in vitro and in experimental infection models. J Antibiot. 2016;69:850–857. doi: 10.1038/ja.2016.47. [DOI] [PubMed] [Google Scholar]
- 188.Boakes S, Dawson MJ. In: Natural Products. Osbourn A, Goss RJ, Carter GT, editors. Wiley; 2014. Discovery and development of NVB302, a semisynthetic antibiotic for treatment of Clostridium difficile infection; pp. 455–468. [Google Scholar]
- 189.Febbraro S. Assessment of the safety and distribution of NVB302 in healthy volunteers. ISRCTN. 2016 doi: 10.1186/ISRCTN40071144. [DOI] [Google Scholar]
- 190.Jabés D, et al. Efficacy of the new lantibiotic NAI-107 in experimental infections induced by multidrug-resistant Gram-positive pathogens. Antimicrob Agents Chemother. 2011;55:1671–1676. doi: 10.1128/AAC.01288-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ge P, et al. Action of a minimal contractile bactericidal nanomachine. Nature. 2020;580:658–662. doi: 10.1038/s41586-020-2186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hols P, Ledesma-García L, Gabant P, Mignolet J. Mobilization of microbiota commensals and their bacteriocins for therapeutics. Trends Microbiol. 2019;27:690–702. doi: 10.1016/j.tim.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 193.Jacob F, Lwoff A, Siminovitch A, Wollman E. [Definition of some terms relative to lysogeny] Ann Inst Pasteur. 1953;84:222–224. [PubMed] [Google Scholar]
- 194.Gratia A. Sur un remarquable exemple d’antagonisme entre deux souches de coilbacille. CR Seances Soc Biol Fil. 1925;93:1040–1041. [Google Scholar]
- 195.Ivanovics G, Alfoldi L, Nagy E. Mode of action of megacin. J Gen Microbiol. 1959;21:51–60. doi: 10.1099/00221287-21-1-51. [DOI] [PubMed] [Google Scholar]
- 196.Tagg JR, Dajani AS, Wannamaker LW. Bacteriocins of Gram-positive bacteria. Bacteriol Rev. 1976;40:722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Klaenhammer T. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–85. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 198.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3:777. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 199.Ahmad V, et al. Antimicrobial potential of bacteriocins: in therapy, agriculture and food preservation. Int J Antimicrob Agents. 2017;49:1–11. doi: 10.1016/j.ijantimicag.2016.08.016. [DOI] [PubMed] [Google Scholar]