Abstract
The tumor microenvironment (TME) is programmed by cancer cells and critically influences antitumor immune responses1,2. Within the TME, CD8+ T cells undergo full effector differentiation and acquire cytotoxic antitumor functions in specialized niches3–7. While interactions with type-1 conventional dendritic cells (cDC1s) have been implicated in this process3–5,8–10, underlying cellular players and molecular mechanisms remain incompletely understood. Here, we show that inflammatory monocytes can adopt a critical role in intratumoral T cell stimulation. They express Cxcl9, Cxcl10 and Il15, but as opposed to cDC1s that cross-present antigens, inflammatory monocytes obtain and present peptide-major histocompatibility complex class I (pMHCI) complexes from tumor cells through “cross-dressing”. Hyperactivation of MAPK signaling in cancer cells hampers this process by coordinately blunting the production of type I interferon (IFN-I) and inducing the secretion of prostaglandin E2 (PGE2), impairing the inflammatory monocyte state and intratumoral T cell stimulation. Enhancing IFN-I production and blocking PGE2 secretion restores this process and thereby re-sensitizes tumors to T cell-mediated immunity. Together, our work uncovers a central role of inflammatory monocytes in intratumoral T cell stimulation, elucidates how oncogenic signaling disrupts T cell responses via counter-regulation of PGE2 and IFN-I, and proposes rational combination therapies to enhance immunotherapies.
Although T cell responses are often suppressed in the TME by inhibitory signals, emerging insights suggest that the TME also plays a pivotal role in supporting T cell function1,11. The activation and differentiation of CD8+ T cells was thought to occur primarily in the lymph node. However, recent findings indicate that upon reaching the tumor, antigen-committed memory or effector T cells require further restimulation to expand, differentiate, and effectively control tumor growth3,4,8,11,12. This process is thought to take place in discrete niches within the TME, where T cells spatially organize with myeloid cells3–7, in particular activated CCR7- cDC1s and cDCs in a stimulatory CCR7+ state3,8–10,13,14. Such multicellular hubs have increasingly been linked to positive outcomes in immunotherapy, underscoring their therapeutic relevance4,5. The growing recognition of T cell restimulation within the TME has prompted the addition of “the TME subcycle” as a new step in the cancer immunity cycle11. However, the processes that facilitate restimulation of primed CD8+ T cells within the TME remain incompletely understood.
To identify mechanisms of T cell restimulation in the tumor, we capitalized on matched pairs of tumor models that were derived by exposing targeted therapy (TT)-naive (NTT) BrafV600E-driven melanoma models to MAPK pathway inhibitors (RAFi, RAFi/MEKi) until they acquired resistance (RTT). We previously demonstrated that while NTT tumors are susceptible to eradication via immune checkpoint blockade (ICB) and adoptive T cell transfer (ACT), RTT tumors harbor an immune-evasive TME rendering them cross-resistant to immunotherapies15. Prospective clinical studies16–18 have confirmed that resistance to targeted therapy jeopardizes a subsequent response to immunotherapy. Here, using these models we discovered that inflammatory monocytes facilitate intratumoral expansion of primed T cells even in absence of cDC1s. We gained mechanistic insights into their mode of action, identified cancer cell-derived cues that disrupt this process, and put forward mechanism-based therapies that can reinstate antitumor immunity.
Results
Myeloid polarization underlies immune escape
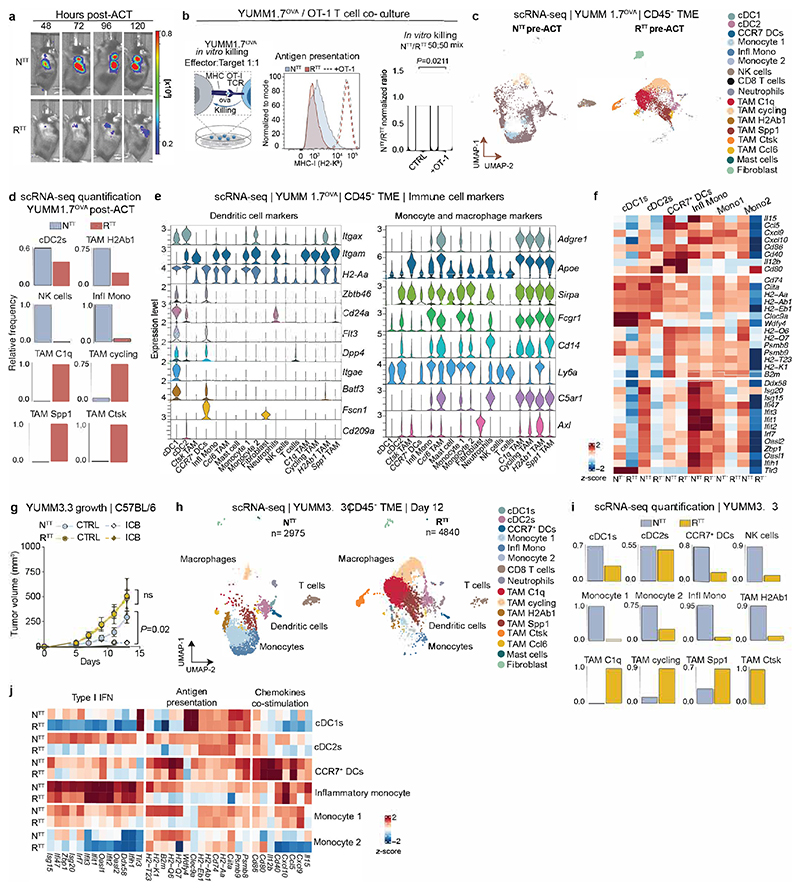
We established NTT and RTT tumors of the YUMM1.7 murine melanoma model (BrafV600E, Pten-/- Cdkn2a-/-) expressing the model antigen ovalbumin (OVA) in Rag2-/- mice and performed ACT by intravenously injecting activated tumor antigen-specific CD8+ T cells (OT-1Luc) (Fig. 1a). As we previously showed15, in NTT tumors, T cells infiltrated and controlled tumor growth, whereas RTT tumors were resistant (Fig. 1b and Extended Data Fig. 1a). Notably, OT-1Luc T cells effectively killed NTT and RTT tumor cells in vitro (Extended Data Fig. 1b), suggesting a role for the TME in mediating resistance to T cell killing in vivo. To define the composition and transcriptional states of the immune cells within the TME of NTT and RTT tumors, we performed single-cell RNA sequencing (scRNA-seq) of CD45+ immune cells before ACT and 72 hours post-ACT. We found that the immune landscape was drastically different between NTT and RTT YUMM1.7OVA tumors, with a reduction of the total CD45+ abundance in RTT tumors and prominent differences within the myeloid cell compartment (Fig. 1c, d and Extended Data Fig. 1c-e). Monocytes, the most abundant immune population in the TME of NTT tumors, were strongly reduced in RTT tumors (Fig. 1c, d and Extended Data Fig. 1d). This population expressed typical monocyte markers (Ly6a, Ly6c2, C5ar1, Fcgr1), low levels of macrophage markers (Adgre, Apoe), and lacked classical cDC markers (Cd24a, Flt3, Dpp4, Zbtb46) (Extended Data Fig. 1e and Supplementary Table 1). Moreover, in RTT tumors, conventional dendritic cells (cDCs) (Zbtb46+, Flt3+, Cd24+), including cDC1s and activated CCR7+ DCs (often referred to as mregDCs)19–22 were severely reduced and in a dysfunctional state (Fig. 1c, d and Extended Data Fig. 1d-f). Furthermore, immunosuppressive macrophages21,23,24 (e.g. Spp1+, Ctsk+) and cycling macrophages were increased in RTT compared to NTT tumors. We observed a similar repolarization of the myeloid compartment in the YUMM3.3 (BrafV600E, Cdkn2a-/-) model in which RTT tumors failed to respond to immune checkpoint blockade (ICB; i.e. anti-PD-1/CTLA-4)15 (Extended Data Fig. 1g-j). This suggests a conserved regulation and functional importance of the myeloid TME across models and immunotherapies.
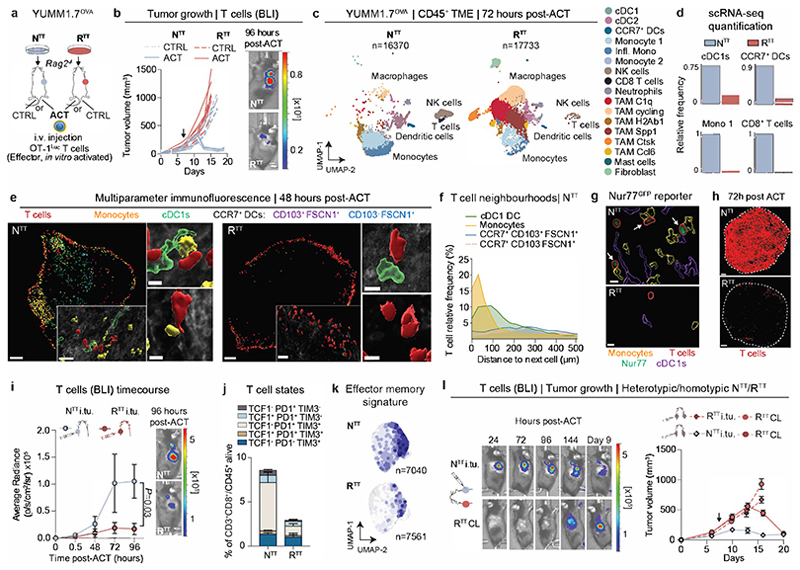
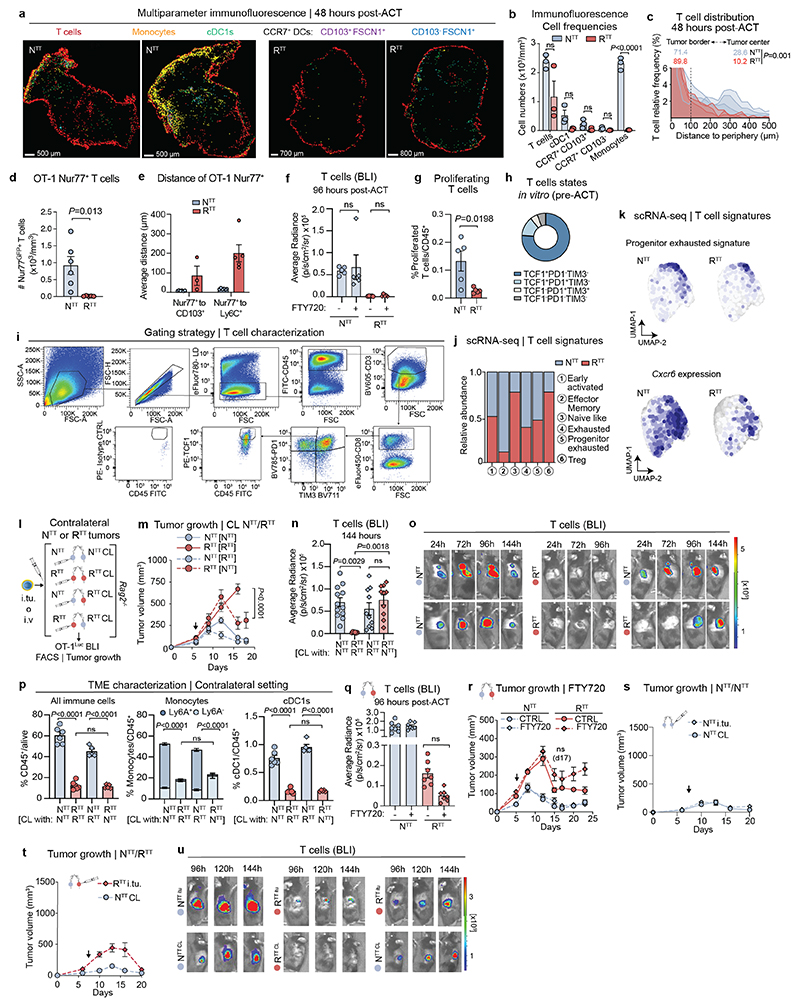
Fig. 1. Tumor infiltrating T cells are restimulated in permissive TMEs.
a, Subcutaneous injection of YUMM1.7OVA NTT and RTT cells in Rag2-/- mice and OT-1Luc adoptive T cell transfer (ACT). b, Left, ACT response of NTT and RTT tumors (n=5 mice/group). Right, representative bioluminescence images (BLI) of T cells 96 hours post-ACT. c, UMAP of scRNA-seq of CD45+ cells in NTT and RTT tumors 72 hours post-ACT (n=4 tumors pooled/group). d, Relative cell frequencies from scRNA-seq. e, Representative immunofluorescence (IF) of YUMM1.7OVA NTT and RTT tumors 48 hours post-ACT, scale bar=400 μm, zoom-ins=50 μm and 10 μm (n=3 tumors/group). f, Relative T cell frequency and distance to next immune cell in NTT tumors (n=3 tumors/group). g, Representative IF of Nur77GFP OT-1 cells in YUMM1.7OVA NTT and RTT tumors 48 hours post-ACT (n=3 tumors/group), scale bar=10μm. Arrows indicate Nur77+ OT-1. h, Representative IF of YUMM1.7OVA NTT and RTT tumors 72 hours post-ACT, dashed-lines depict tumor border, scale bar=500 μm, (n=2 tumors/group). i, Left, quantification of T cells by BLI over time after intratumoral (i.tu.) ACT (mean ± s.e.m, n=4 mice/group). Two-way ANOVA with Sidak’s multiple comparisons test. Right, representative BLI 96 hours post-ACT. j, T cell states by flow cytometry 120 hours post-i.tu. ACT (mean ± s.e.m, n=5 NTT, n=4 RTT tumors). k, Effector memory T cell signature29 on scRNA-seq of T cells (n=14 NTT, n=18 RTT pooled tumors). l, Left, representative BLI of T cells in NTT and contralateral (CL) RTT tumors post-i.tu ACT. Right, growth curves (mean ± s.e.m, n=6 mice/group). Arrows in b and l indicate day of ACT.
T cells are restimulated in permissive TMEs
Recent studies suggest that activated CD8+ T cells require additional stimuli from intratumoral myeloid cells to acquire full effector functions and sustain a T cell response3,6,10,12. To examine this further, we used multiparameter immunofluorescence (IF) microscopy for discriminatory myeloid cell markers to determine the main interaction partner(s) of tumor-infiltrating T cells (Fig. 1e and Extended Data Fig. 2a-c). In NTT tumors, tumor specific CD8+ T cells were in close proximity to cDC1s and monocytes, and often organized in multicellular clusters at the tumor margin (Fig. 1f and Extended Data Fig. 2a-c). Intriguingly, tumor-infiltrating T cells interacted with monocytes more commonly than with cDC1s, possibly due to the high abundance of monocytes in the TME (Fig. 1f and Extended Data Fig. 2a,b). Within these hubs, T cells stained positive for Nur77, a marker indicative of recent TCR-signaling, indicating not just proximity but also direct antigen-specific stimulation by the interacting myeloid cells (Fig. 1g and Extended Data Fig. 2d,e). Within 72 hours post-ACT, T cells in NTT tumors permeated the entire tumor parenchyma, which was not abolished by blocking T cell egress from the lymph node with FTY720, suggesting local expansion (Fig. 1h and Extended Data Fig. 2f). In contrast, RTT tumors did not contain such hubs, T cells remained confined to the periphery, rarely interacted with the few cDCs and monocytes and Nur77+ T cells were strongly reduced (Fig. 1g and Extended Data Fig. 2d,e).
To investigate T cell functionality within TMEs, we injected the same amount of activated OT-1Luc T cells intratumorally into NTT and RTT tumors (Fig. 1i). We found that in contrast to RTT, in NTT tumors, CD8+ T cells quickly underwent a proliferative burst and expanded (Fig. 1i, Extended Data Fig. 2g). Five days post intratumoral injection, we observed a significant reduction in differentiated CD8+ T cells (TCF1- PD1+ TIM3+) in RTT compared to NTT tumors, while in both conditions a small fraction (∼5%) of all T cells remained in a stem cell-like state (TCF1+PD1+TIM3-), required for the expansion of the T cell effector pool12,25–28 (Fig. 1j and Extended Data Fig. 2h, i). In NTT tumors, but not in RTT tumors, T cells acquired features of effector memory T cells29, and the majority displayed upregulated expression of the T cell effector marker CXCR630 (Fig. 1k, Extended Data Fig. 2j, k and Supplementary Table 2). Altogether, these findings suggest that multicellular hubs containing cDCs and, unexpectedly, a substantial fraction of monocytes, are associated with T cell expansion and that T cell proliferation and effector differentiation is facilitated only within the TME of NTT, but not of RTT tumors.
Local licensing of systemic immunity
To examine whether an immune-permissive TME can act as a reservoir for T cell restimulation, we established NTT and RTT tumors in opposite flanks within the same mouse and performed intravenous ACT. T cells expanded in NTT tumors, and, at later time points also infiltrated contralateral RTT tumors and transiently controlled their growth (Extended Data Fig. 2l-o). Of note, we did not observe differences in the myeloid composition of RTT tumors in the presence of a contralateral NTT tumor (Extended Data Fig. 2p). Treatment with FTY720 only marginally reduced T cell infiltration into contralateral RTT tumors, still resulting in tumor control, suggesting that T cells may traffic partly through the lymph node but also directly through the circulation (Extended Data Fig. 2q, r). To further confirm that restimulated T cells traffic between tumors, we directly injected activated OT-1Luc T cells into one tumor and evaluated T cell infiltration in contralateral tumors. T cells introduced into an NTT tumor demonstrated the capacity to expand locally and infiltrate contralateral tumors, irrespective of whether these were NTT or RTT tumors. Contralateral NTT tumors fully regressed and even contralateral RTT tumors were temporarily controlled (Fig. 1l and Extended Data Fig. 2s-u). Interestingly, T cells directly injected into RTT tumors initially failed to expand but trafficked to contralateral NTT tumors where they expanded and controlled tumor growth and, eventually, re-infiltrated RTT tumors (Extended Data Fig. 2t, u). Collectively, our data indicate that after initial priming of T cells, restimulation and subsequent effector functions are strongly dictated by the characteristics of the TME. We conclude that T cells are capable of trafficking between tumors and that T cell restimulation in an immune-permissive TME can facilitate the control of distant, resistant tumors.
Immune-stimulatory role of monocytes
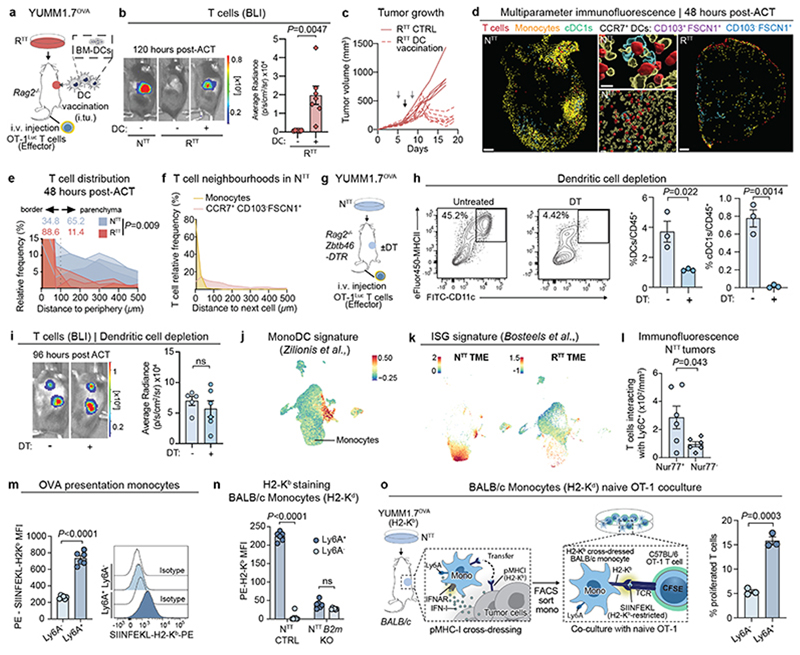
Activated cDC1s and CCR7+ cDCs have long been implicated in antitumor immunity13,31–35 and recently in intratumoral CD8+ T cell restimulation3,4,8,9. Consistently, cDC1 vaccination in RTT tumors restored T cell infiltration and led to transient tumor control (Extended Data Fig. 3a-c). Interestingly, when we injected NTT cells in Rag2-/- Batf3-/- mice that lack functional cDC1s and performed ACT, T cells still infiltrated, expanded, and controlled NTT tumors (Fig. 2a). In absence of functional cDC1s, multiparameter IF showed retained immune hubs with monocytes clustering together with T cells in NTT, but not in RTT tumors (Extended Data Fig. 3d-f). Furthermore, in NTT tumors grown in Zbtb46-DTR bone marrow chimeras, where all cDC subsets were depleted, T cells were effectively restimulated and expanded (Extended Data Fig. 3g-i).
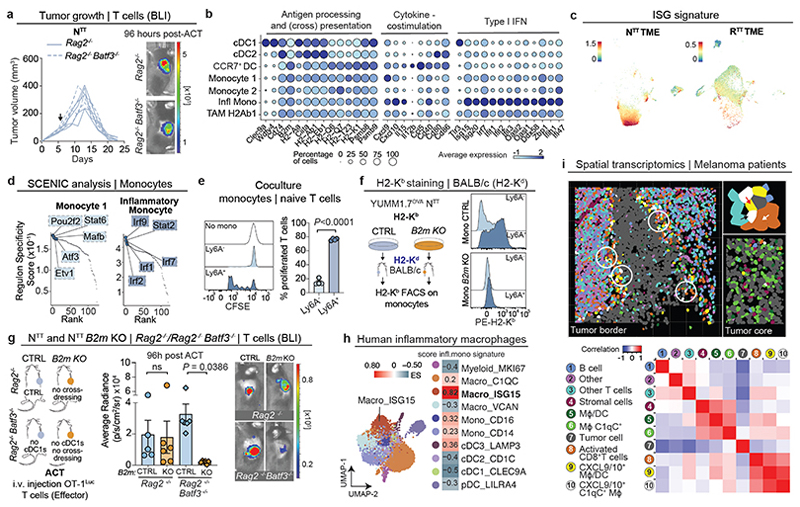
Fig. 2. Inflammatory monocytes restimulate T cells within the TME.
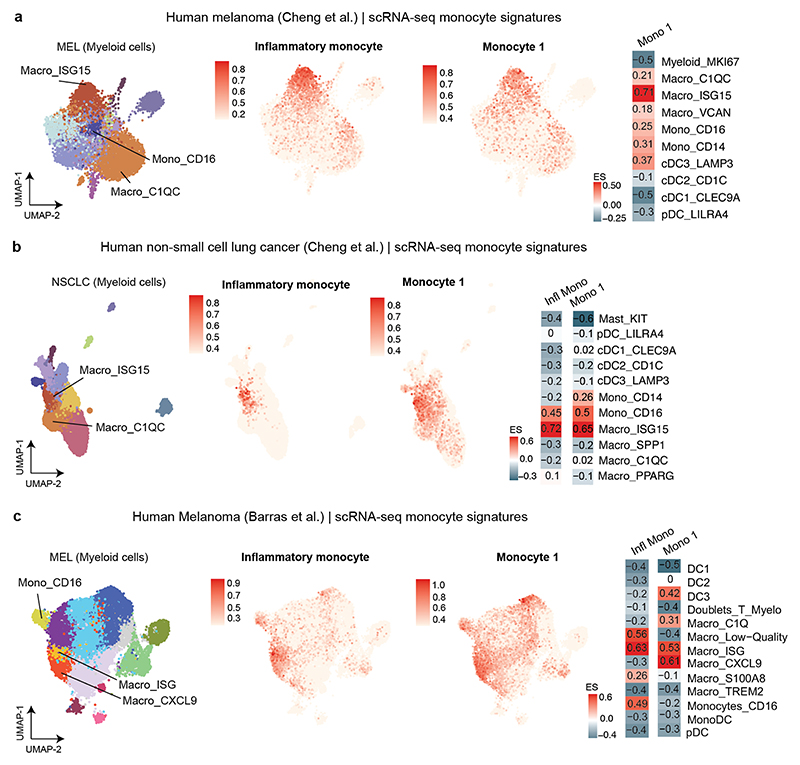
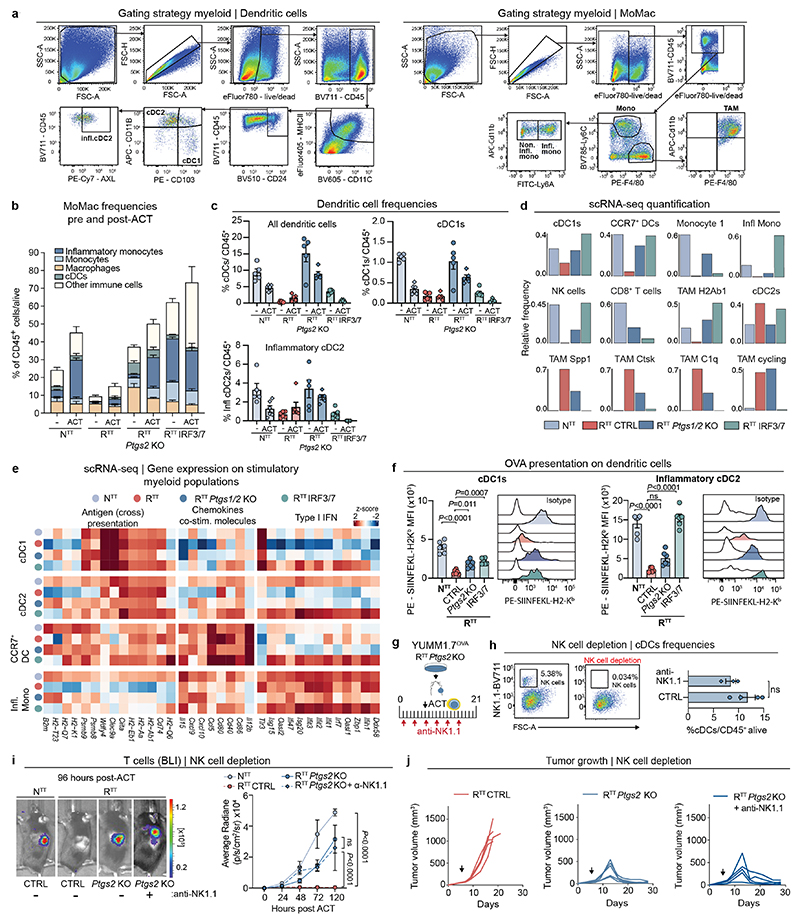
a, Left, ACT response of YUMM1.7OVA NTT tumors in Rag2-/- (n=4 mice) and Batf3-/- Rag2-/- (n=5 mice). Right, representative BLI of T cells 96 hours post-ACT. b, Gene expression in individual clusters from scRNA-seq from Fig. 1c. c, Scoring of an interferon-stimulated gene (ISG) signature37 on our scRNA-seq data from Fig. 1c. d, Regulon Specificity Score on monocytes calculated with SCENIC in NTT tumors. e, Left, representative histograms depicting CFSE intensity. Right, quantification of T cell proliferation after 72 hours of co-culture of naive CFSE-labeled T cells and inflammatory (Ly6A+) or non-inflammatory monocytes (Ly6A-) isolated from NTT tumors in Rag2-/- mice (n=4 tumors). Two-tailed unpaired student’s t-test. f, Left, injection of YUMM1.7OVA NTT CTRL or B2m KO cells in BALB/c mice. Right, representative histograms depicting H2-Kb levels. g, Left, injection of NTT CTRL or NTT B2m KO tumors in Rag2-/- or Batf3-/- Rag2-/-. Middle, quantification of T cell infiltration by BLI. Right, representative BLI of T cell infiltration 96 hours post-ACT (n=5 NTT in Rag2-/- and n=6 mice for other groups). One-way ANOVA with Tukey’s multiple comparisons test. h, Left, UMAP of human melanoma scRNA-seq myeloid data-set23. Right, enrichment scores (ES) of the inflammatory monocyte gene signature for each cell cluster. i, Top, representative field of view (FOV) of human metastatic melanoma (n=2 of 72 FOVs) analyzed by CosMx spatial transcriptomic profiling. Bottom, Pearson correlation values between cell types across FOVs (n=72) were determined and displayed as a heatmap (n=34 melanoma samples). White arrow depicts activated CD8+ T cells, black arrow CXCL9+/10+ C1qC+ MΦ and red arrow CXCL9+/10+ MΦ/DCs. MΦ=Macrophages. Bar graphs depict the mean ± s.e.m.
Next, we probed monocytes for expression of genes linked to T cell stimulation. We found that monocytes expressed elevated levels of the antigen presentation machinery (Psmb8/9, MHCI and MHCII), Il15, which mediates T cell survival and effector differentiation3, as well as Cxcl9 and Cxcl10, essential for T cell recruitment and linked to positive immunotherapy responses33 (Fig. 2b). These monocytes did not score positively for an established monocyte-derived DC (moDC) signature21, and interestingly, a large subset of these monocytes also expressed high levels of interferon-stimulated genes (ISGs), identifying them as inflammatory monocytes (Fig. 1c, 2b, and Extended Data Fig. 3j). These inflammatory monocytes expressed the IFN-induced surface marker Ly6A (Extended Data Fig. 1e), consistent with analyses of human and murine tumors and viral infection models, showing that myeloid cells transition to an inflammatory state upon IFN-I signaling23,36–40. As expected, previously defined ISG signatures (See Supplementary Table 3) scored the highest in the monocyte and inflammatory monocyte cluster in NTT tumors (Fig. 2c and Extended Data Fig 3k). Single-cell regulatory network inference and clustering (SCENIC), predicted transcriptional activity of Irf9, Irf7, Irf2 and Stat2, major effectors of IFN signaling, specifically in inflammatory monocytes (Fig. 2d). Altogether, these data suggest that even in the absence of cDCs, monocytes are capable of mediating expansion of tumor-specific CD8+ T cells and promoting antitumor immunity.
MHCI-dressed monocytes stimulate T cells
Given that T cells close to monocytes within immune hubs expressed Nur77, indicative of TCR stimulation (Fig. 1g and Extended Data Fig. 3l), we assessed whether monocytes are able to present tumor antigen. H2-Kb-SIINFEKL staining revealed that inflammatory monocytes, but not their non-inflammatory counterparts, display tumor-derived antigens on MHCI (Extended Data Fig. 3m), and could induce naive T cell activation and proliferation ex vivo, measured by CFSE dilution (Fig. 2e). However, inflammatory monocytes do not express genes involved in cross-presentation (Clec9a, Wdfy4)14 (Fig. 2b), prompting us to investigate if inflammatory monocytes can acquire and display antigens through the direct transfer of intact peptide-MHCI (pMHCI) complexes from adjacent cells - a process called “MHCI cross-dressing”37,41,42.
To investigate if monocytes are able to cross-dress, we established NTT tumors, which are derived from C57BL/6 mice and therefore express H2-Kb, into MHCI haplotype-mismatched (H2-Kd) BALB/c mice and examined cancer cell-derived H2-Kb expression on myeloid subsets. Only inflammatory monocytes (Ly6A+), harbored cancer cell-derived H2-Kb and were capable of activating naive T cells ex vivo (Fig. 2f and Extended Data Fig. 3n, o). Notably, in NTT tumors harboring a knockout in β2-microglobulin (NTT B2m KO), no H2-Kb signal was detected on BALB/c inflammatory monocytes, demonstrating that pMHCI complexes on inflammatory monocytes are sourced from cancer cells (Fig. 2f and Extended Data Fig. 3n). To test the relative contribution of cross-presenting cDC1s and cross-dressed inflammatory monocytes to T cell restimulation, we injected NTT and NTT B2m KO tumors, which abolishes the ability of inflammatory monocytes to present tumor antigens through MHCI cross-dressing, into Rag2-/- or Rag2-/- Batf3-/- mice and performed ACT (Fig. 2g). When pMHCI cross-dressing on monocytes was intact, T cells expanded in NTT tumors, even in absence of cDC1s. Conversely, when pMHCI cross-dressing on inflammatory monocytes was abolished (B2m KO tumors) but cDC1s were present, T cells were also able to expand, reflecting the established capacity of cDC1s to re-stimulate CD8+ T cells through classical cross-presentation. Only when both, cDC1s and cross-dressed inflammatory monocytes, were absent (B2m KO tumors engrafted in Rag2-/- Batf3-/-) T cells failed to become restimulated (Fig. 2g). Collectively, our data highlight that pMHCI cross-dressing by inflammatory monocytes, together with stimulatory cytokine expression, underlies their ability to promote restimulation of primed CD8+ T cells in the TME.
ISG+ macrophages in human melanoma
We scored our murine inflammatory monocyte signature in human myeloid scRNA-seq datasets of melanoma and non-small cell lung cancer (NSCLC)23,43 and found ISG+ (CXCL9+/10+) macrophages and CD16+ monocytes as the analogous inflammatory populations across human cancers (Fig. 2h and Extended Data Fig. 4a-c, Supplementary Table 2). Importantly, analyzing the spatial distribution of T cells with immune cells in human melanoma samples, revealed that activated CD8+ T cells preferentially co-localized with CXCL9+/10+ inflammatory macrophages and cDCs in immune hubs, whereas regions that lacked CXCL9+/10+ macrophages were devoid of T cells (Fig. 2i).
Cancer cells produce PGE2 and dampen IFN-I
To identify factors derived from NTT and RTT cells determining the intratumoral immune landscape pivotal for T cell restimulation, we isolated cancer cells from tumors and performed RNA-seq. Pathway enrichment analysis of differentially regulated genes revealed upregulation of the prostaglandin synthesis pathway and downregulation of IFN-I signaling in RTT cells as the top differential pathways compared to NTT cells (Fig. 3a and Supplementary Table 3). Metabolomic analysis identified prostaglandin E2 (PGE2) as the most enriched eicosanoid in RTT tumors (Extended Data Fig. 5a). PGE2 has recently been shown to limit cDC1-mediated support of CD8+ T cells9,34,44, but its impact on inflammatory monocytes and on their ability to promote T cell restimulation is unknown.
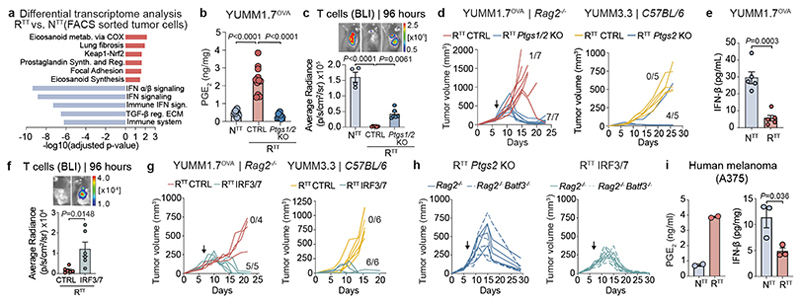
Fig. 3. Cancer cells produce PGE2 and downregulate IFN-I conferring immunotherapy resistance.
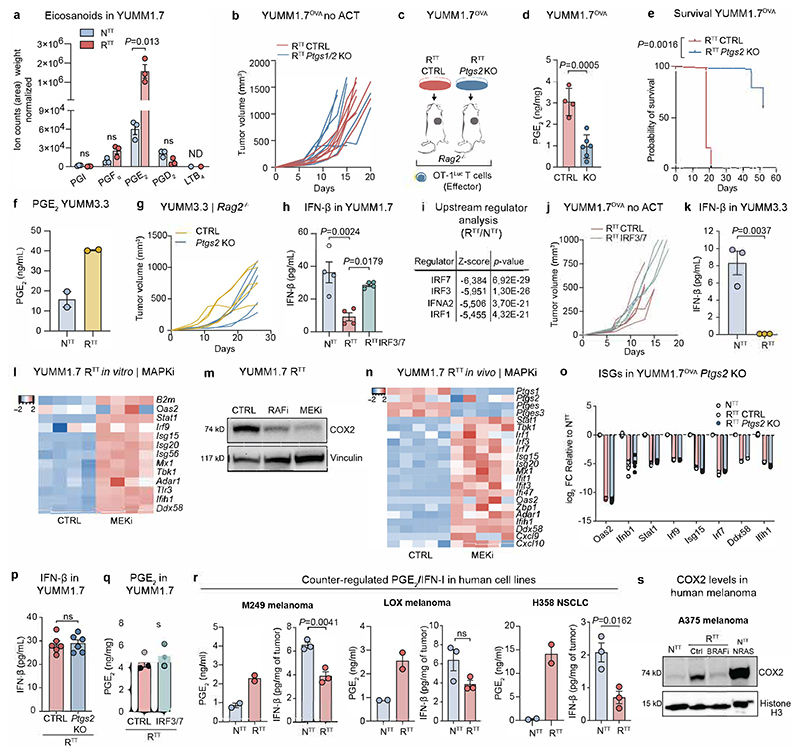
a, Pathway enrichment analysis of differential gene expression in cancer cells isolated from YUMM1.7OVA NTT and RTT tumors (n=3 tumors/group, See Supplementary Table 3). Adjusted p-value computed using the Benjamini-Hochberg correction. b, PGE2 ELISA of YUMM1.7OVA tumors in Rag2-/- mice (n=10 NTT, n=11 RTT CTRL, n=7 RTT Ptgs1/2 KO over 2 independent experiments). c, Top, representative BLI of T cells. Bottom, BLI quantification (n=4 mice NTT, n=6 mice RTT CTRL and RTTPtgs1/2 KO). d, Left, response to ACT (n=7 mice/group). Right, YUMM3.3 in C57BL/6 mice (n=5 mice/group). e, IFN-β ELISA of supernatants from YUMM1.7OVA NTT and RTT cells (n=5 replicates per group over 2 independent experiments). f, Top, representative BLI of T cells in YUMM1.7OVA RTT CTRL and RTT IRF3/7 tumors in Rag2-/- mice. Bottom, BLI quantification (n=6 mice/group). g, Left, response to ACT (n=4 mice RTT CTRL, n=5 mice RTT IRF3/7). Right, YUMM3.3 in C57BL/6 mice (n=6 mice/group). h, Response to ACT of YUMM1.7OVA RTT Ptgs2 KO and RTT IRF3/7 tumors in Rag2-/- (n=4 mice/group) and Batf3-/- Rag2-/- mice (n=5 mice/group). i, PGE2 and IFN-β ELISAs of NTT and RTT A375 human melanoma (n=2 replicates/group for PGE2, n=3 tumors/group for IFN-β). Arrows in d, g and h indicate day of ACT. Bar graphs depict the mean ± s.e.m. Statistical analysis was performed with a two-tailed unpaired student’s t-test in e, f and i, a one-way ANOVA with Tukey’s multiple comparisons test in b and c.
To understand the role of cancer cell-derived PGE2 in immune-evasion we ablated PGE2 production through cyclooxygenase-1 (COX1, Ptgs1) and COX2 (Ptgs2) KO in RTT cells and engrafted them into Rag2-/- mice. Ptgs1/2-deficient RTT tumors displayed diminished PGE2 levels comparable to those of NTT tumors (Fig. 3b) and increased T cell infiltration (Fig. 3c). Remarkably, Ptgs1/2 deletion fully re-sensitized RTT tumors to ACT and grew unperturbed without ACT (Fig. 3d and Extended Data Fig. 5b). Genetic ablation of only Ptgs2 also led to re-sensitization of RTT tumors to ACT (Extended Data Fig. 5c-e). Similarly, Ptgs2 deletion in the YUMM3.3 RTT model, resulted in tumor rejection in immunocompetent C57BL/6 mice in a T cell dependent manner, even without ICB treatment (Fig. 3d and Extended Data Fig. 5f, g). Similar effects of Ptgs2 inactivation have been reported in other mouse models34,44–46, emphasizing the role of PGE2 in the TME as a strong modulator of T cell responses.
Given the effects of PGE2, we wondered whether the transcriptional downregulation of the IFN-I program in RTT cells (Fig. 3a) was merely a reflection of low IFN levels in a PGE2-induced immune suppressive TME or an independent driver of immune evasion. IFN-β was significantly reduced in RTTcompared to NTT tumor lysates (Extended Data Fig. 5h), with RTT cells producing less IFN-β in vitro, indicating cancer cell-intrinsic regulation of IFN-I without microenvironmental cues (Fig. 3e). Upstream regulator analysis (Ingenuity) predicted that these transcriptional changes stemmed from decreased activity of the transcription factors IRF3 and IRF7, important regulators of interferon production (Extended Data Fig. 5i).
To test whether re-establishment of a functional IFN-I pathway in RTT cancer cells was sufficient to restore response to ACT, we overexpressed IRF3 and IRF7 in RTT cells and established tumors in Rag2-/- mice. This restored IFN-I levels in the TME (Extended Data Fig. 5h) and increased T cell infiltration leading to full tumor control upon ACT (Fig. 3f, g and Extended Data Fig. 5j). YUMM3.3 RTT tumors also harbored lower IFN-β levels than their NTT counterparts (Extended Data Fig. 5k) and upon re-establishment of an interferon response and injection in C57BL/6 mice they were controlled (Fig. 3g). Moreover, similar to NTT tumors, YUMM1.7OVA RTT tumors with Ptgs2 KO or IRF3/7 overexpression were controlled in Rag2-/- Batf3-/- mice that lack cDC1s (Fig. 3h). Collectively, these findings suggest that RTT cancer cells establish an immune-evasive TME by increasing PGE2 and simultaneously reducing IFN-I production. Reverting either of these events re-sensitizes tumors to killing by activated T cells, even in the absence of cDC1s (Fig. 3h).
MAPK signaling regulates PGE2 and IFN-I
We recently showed that cross-resistance between targeted therapy (BRAFi/MEKi) and immunotherapy is driven by reactivated oncogenic RAF-MEK-ERK signaling15. To explore whether the hyperactivated MAPK pathway in RTT tumors is the common regulator of both immune evasive programs, we inhibited the MAPK pathway in cancer cells, in vitro and in vivo. This induced ISGs, and reduced COX2 levels (Extended Data Fig. 5l-n), consistent with previous reports44,47. We asked whether PGE2 could be the cause of the dampened IFN-I program in RTT cancer cells, but Ptgs2 deletion did not restore ISG expression, and did not increase IFN-I levels in the TME (Extended Data Fig. 5o, p). Similarly, IRF3/7 overexpression in RTT tumors, did not attenuate PGE2 production (Extended Data Fig. 5q).
To address the human relevance of these findings, we assessed PGE2 and IFN-I production in matched pairs of the human RAFi-sensitive (NTT) and RAFi-resistant (RTT) melanoma cell-lines A375, M249 and LOX48. We consistently found an increased production of PGE2 in TT-resistant cells, together with a decrease in IFN-I. This was also confirmed in the KRAS-driven NSCLC cell-line NCI-H358 after it acquired resistance to a targeted KRASi (Fig. 3i and Extended Data Fig. 5r). COX2 protein levels were regulated by the MAPK pathway in the A375 cell-line, upon BRAF inhibition or upon NRAS overexpression (Extended Data Fig. 5s). Collectively, these studies indicate a common regulatory module driven by oncogenic MAPK signaling that upregulates PGE2 and downregulates IFN-I in cancer cells to drive immune evasion.
PGE2/IFN-I instruct myeloid polarization
To understand how genetic ablation of PGE2 synthesis or restoration of IFN-I in RTT cancer cells reinstates an immune-permissive TME rich in inflammatory monocytes, we performed scRNA-seq and flow cytometry analysis of CD45+ cells from YUMM1.7OVA NTT, RTT CTRL, RTT Ptgs1/2 KO and RTT IRF3/7 overexpressing tumors 72 hours post-ACT (Fig. 4a and Extended Data Fig. 6a-e). The most pronounced TME changes upon deletion of Ptgs1/2 or overexpression of IRF3/7 were indeed in the monocyte (Mo) and macrophage (Mac) compartment (MoMac). In RTT IRF3/7 tumors, inflammatory monocytes were the predominant population together with a TAM cluster with enhanced stimulatory functions (TAM H2Ab1) that was absent in RTT tumors (Fig. 4a and Extended Data Fig. 6d). In RTT Ptgs1/2 KO tumors we observed an increase of both monocytes and inflammatory monocytes, with a reduction of immune-suppressive TAMs (Spp1+, C1q+, Ctsk+) and an increase in H2Ab1 TAMs. Upon PGE2 reduction or IRF3/7 overexpression, cDC1 abundance and functionality, as well as CCR7+ DCs were significantly increased and antigen presentation capacity enhanced (Fig. 4a and Extended Data Fig. 6c-f). Lastly, NK cells were also rescued in RTT Ptgs1/2 KO and IRF3/7 overexpressing tumors, but NK cell depletion in Ptgs2 KO tumors did not significantly change the infiltration of cDCs and T cells or overall tumor control in our models (Extended Data Fig. 6g-j).
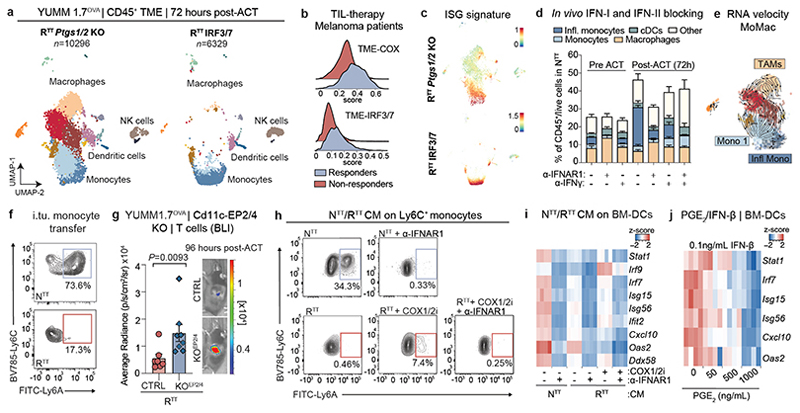
Fig. 4. PGE2 and IFN-I determine myeloid cell abundance and their functional inflammatory state in the TME.
a, UMAP of scRNA-seq of CD45+ cells in YUMM1.7OVA RTT Ptgs1/2 KO and RTT IRF3/7 tumors 72 hours post-ACT (n=3 tumors pooled/group). See Fig. 1c for cell cluster annotation. b, Scoring of the TME-COX and TME-IRF3/7 signatures (Supplementary Table 4) in myeloid fractions of responder (n=6) and non-responder (n=7) patients pre-TIL infusion43. c, Scoring of an interferon-stimulated gene (ISG) signature37 in the scRNA-seq from a. d, Flow cytometry quantification of myeloid populations normalized to CD45+ fraction from YUMM1.7OVA NTT tumors in Rag2-/- mice treated with α-IFNAR1/α-IFN-γ (n=5 tumors/group, except n=4 in α-IFNAR1+ACT and α-IFN-γ+ACT). e, RNA-velocity of MoMac from RTT Ptgs1/2 KO tumors. f, Representative contour plots of Ly6C+ monocytes depicting Ly6A expression 72 hours post intratumoral transfer into NTT and RTT tumors in Rag2-/- mice (n=5 tumors/group). g, Left, BLI quantification of T cells 96 hours post-ACT of YUMM1.7OVA RTT cells into CD11cCre-Ptger2-/-Ptger4fl/fl mice (KOEP2/EP4) or CD11cCre (CTRL) (n=7 mice/CTRL group and n=8 mice/KOEP2/EP4 group), two-tailed Mann-Whitney U test. Right, representative BLI. h, Representative contour plots of Ly6C+ monocytes depicting expression of Ly6A 48 hours after treatment with conditioned media (CM) from cancer cells ± COX1/2i (indomethacin) ± α-ΙFNAR1 or isotype (n=3 biological replicates). i, Heatmap of scaled ISG expression in BM-DCs exposed to CM from NTTor RTT ± COX1/2i in the presence of α-ΙFNAR1 or isotype (n=4 technical replicates) measured by RT-qPCR. j, Heatmap of scaled ISG expression in BM-DCs treated with IFN-β and PGE2 measured by RT-qPCR (n=4 technical replicates). Bar graphs depict the mean ± s.e.m.
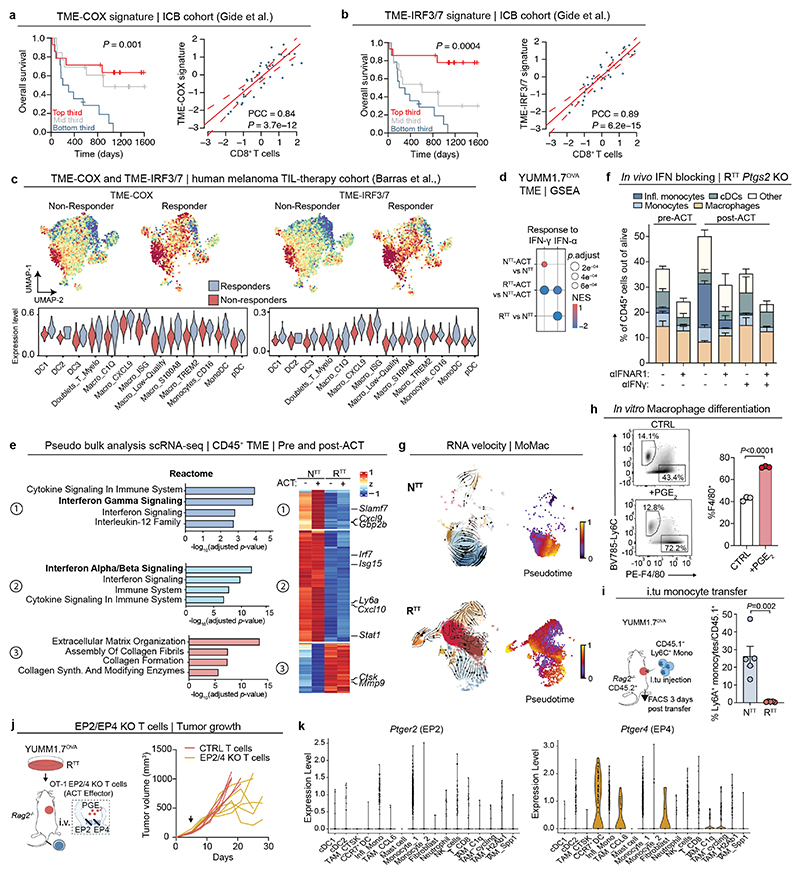
To investigate the distinctive features of PGE2 depletion or IFN-I reinstatement in cancer patients, we established a signature of the top upregulated genes in immune cells upon Ptgs1/2 KO (TME-COX signature, n=26) and the top upregulated genes upon IRF3/7 overexpression (TME-IRF3/7 signature, n=40, see Supplementary Table 4). Both signatures strongly correlated with a gene expression signature of CD8+ T cell infiltration and were highly predictive of survival in melanoma patients treated with ICB49 (Extended Data Fig. 7a, b). Furthermore, both signatures were also enriched in a cohort of patients that responded to subsequent tumor-infiltrating lymphocyte (TIL) therapy43 (Fig. 4b and Extended Data Fig. 7c). From these studies, we conclude that PGE2 depletion and IFN-I increase is associated with an immune-permissive state of the TME and better response to therapy in preclinical models and patients.
IFN-I/IFN-γ drive the inflammatory state
The TME of NTT, RTT Ptgs1/2 KO and RTT IRF3/7 tumors share an inflammatory state, with monocytes being the most abundant cell type in this transcriptional state (Fig. 4c). We found that upon ACT, inflammatory monocytes strongly expanded, especially in NTT and RTT Ptgs2 KO tumors, suggesting that both cancer cell-derived IFN-I and T cell-derived IFN-γ play a role at inducing the inflammatory state (Extended Data Fig. 6b). Indeed, looking at the most variable genes in the immune compartment of the TME before and after ACT, we found that a core IFN-γ response gene set (e.g., Cxcl9, Gbp2 and Slamf7) was strongly increased in NTT tumors. Independently of the effects of T cell transfer, a set of genes characteristic of an IFN-α response (e.g., Irf7, Isg15, Cxcl10) was higher in NTT compared to RTT TMEs suggesting that both type I and type II IFNs modulate an inflammatory TME in NTT tumors (Extended Data Fig. 7d, e).
To assess how interferons modulate the inflammatory TME and monocytes, we selectively blocked IFN-I and IFN-II, pre- and post-ACT. Blocking IFN-I before ACT, prevented the formation of inflammatory monocytes, with low levels of IFN-I produced by RTT Ptgs2 KO cells being sufficient for inducing an inflammatory state. Post-ACT, IFN-γ depletion substantially reduced (NTT) or completely abolished (RTT Ptgs2 KO) inflammatory monocytes (Fig. 4d and Extended Data Fig. 7f). In NTT tumors that produce high levels of IFN-I, the combinatorial depletion of type I and type II IFNs was necessary to fully abolish inflammatory monocytes after ACT (Fig. 4d). These data indicate that monocytes initially rely on tumor-derived IFN-I to transition to an inflammatory state, which shifts towards IFN-γ when T cells infiltrate the TME.
The expansion of inflammatory monocytes upon T cell transfer, is coupled with a reduction in macrophages, suggesting a shift in myeloid differentiation towards inflammatory monocytes at the expense of TAM maturation (Extended Data Fig. 6b). To further understand myeloid differentiation trajectories, we performed RNA-velocity analysis of the MoMac compartment in NTT, RTT and RTT Ptgs1/2 KO tumors, predicting that a common monocyte precursor population (Monocyte 1 cluster) can give rise to both TAMs and inflammatory monocytes (Fig. 4e and Extended Data Fig. 7g). This suggests that in RTT tumors, high PGE2 and low IFN-I drive differentiation of monocytes into suppressive macrophages, whereas in NTT monocytes are maintained, with a subset acquiring the inflammatory state. Indeed, treatment of Ly6C+ monocytes with PGE2 in vitro promoted their differentiation towards F4/80+ macrophages (Extended Data Fig. 7h). In absence of PGE2 (RTT Ptgs1/2 KO), TAMs are reduced, and monocytes increased with a fraction becoming inflammatory (Fig. 4a). In RTT IRF3/7 tumors, despite high levels of PGE2, the elevated levels of IFN-I can mediate inflammatory monocyte formation (Fig. 4a). To further probe monocyte differentiation trajectories in vivo, we intratumorally injected bone marrow-derived Ly6C+ monocytes. In NTT tumors, 70-80% of these became inflammatory, compared to only 10-20% in RTT tumors (Fig. 4f and Extended Data Fig. 7i). Thus, both cancer cells and T cells shape the immune status of the TME, by driving differentiation of monocytes and their inflammatory state in therapy-responsive tumors.
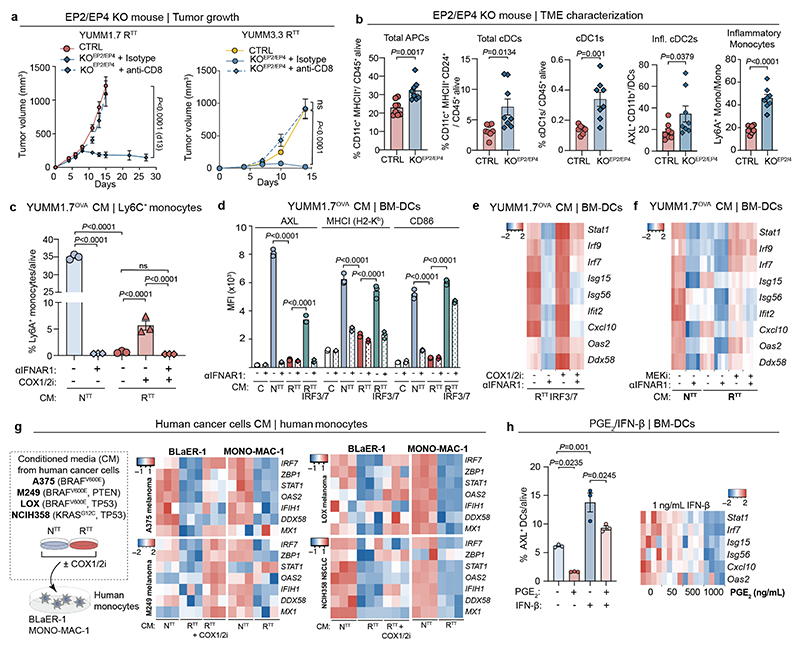
PGE2 impairs the inflammatory state
To rule-out that high levels of PGE2 in the TME directly impairs T cell function30,50, we knocked out the PGE2 receptors, EP2 (encoded by Ptger2) and EP4 (encoded by Ptger4) in OT-1 T cells, followed by their intravenous injection into RTT tumor-bearing mice. This had modest effects and did not achieve tumor control in this model (Extended Data Fig. 7j). We then examined effects of PGE2 on the myeloid compartment using CD11cCre(Itgax-Cre)/Ptger2-/-/Ptger4fl/fl mice in which EP2 and EP4 are selectively ablated in CD11c+ cells. We observed improved T cell infiltration and RTT tumors were controlled in a CD8-dependent manner, in both the YUMM1.7 and YUMM3.3 models (Fig. 4g, Extended Data Fig. 7k and Extended Data Fig. 8a). In line with these data, inflammatory monocytes and cDC2s, along with cDC1s, were increased in CD11cCre(Itgax-Cre)/Ptger2-/-/Ptger4fl/fl mice, compared to control mice (Extended Data Fig. 8b). These data suggest that disruption of PGE2 signaling, either by inhibiting PGE2 production in cancer cells or by blocking downstream signaling in myeloid cells, is sufficient to restore the inflammatory state in myeloid cells and subsequent T cell function.
Given that IFN-I and PGE2 represent distinct biological classes of mediators, we investigated their individual and combined effects on the inflammatory state of myeloid cells. We exposed murine Ly6C+ monocytes and bone marrow-derived DCs (BM-DCs) to conditioned media (CM) from NTT, RTT, and RTT IRF3/7 cells. CM from NTT or RTT IRF3/7, but not RTT cells, induced an inflammatory state in BM-DCs and monocytes (Fig. 4h, i and Extended Data Fig. 8c-e). This inflammatory response was blocked by α-IFNAR1 treatment. Using CM of RTT cells treated with a COX1/2 inhibitor (COX1/2i) also increased the inflammatory state, which was fully dependent on the marginal remaining IFN-I produced by RTT cells (Fig. 4h, i and Extended Data Fig. 8c-e). Furthermore, inhibiting the MAPK pathway with a MEKi in RTT cells during media conditioning also resulted in an increase in ISG expression in BM-DCs, highlighting the role of oncogenic MAPK signaling in regulating PGE2 and IFN-I (Extended Data Fig. 8f). Exposure of human monocytes to CM from NTT or RTT cells of various human melanoma and NSCLC cell-lines, with and without COX1/2i, demonstrated effects similar to those in murine monocytes, supporting a role for counter-regulated PGE2 and IFN-I in myeloid dysfunction in human cancers (Extended Data Fig. 8g). Finally, low IFN-β levels were sufficient to induce ISGs and the inflammatory marker AXL in BM-DCs, and adding PGE2 significantly impaired their response to IFN-I, thereby hampering the acquisition of an inflammatory state (Fig. 4j and Extended Data Fig. 8h).
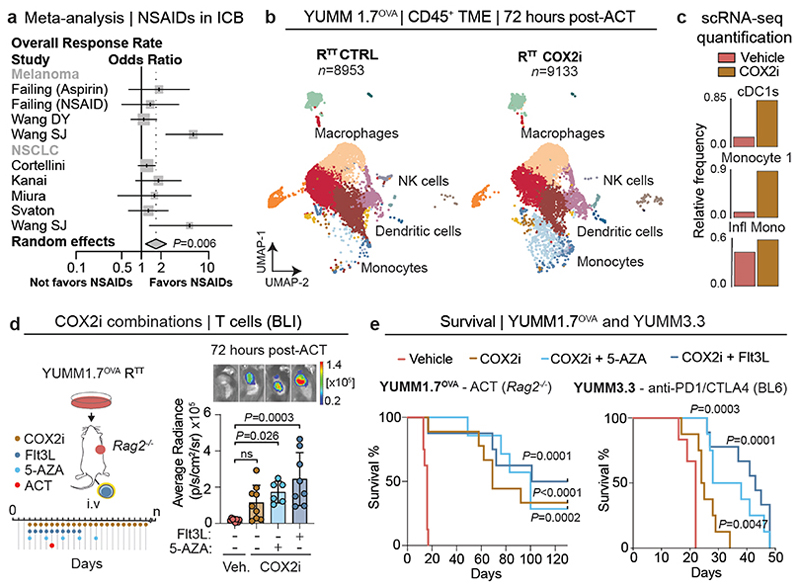
Pharmacological targeting of PGE2/IFN-I
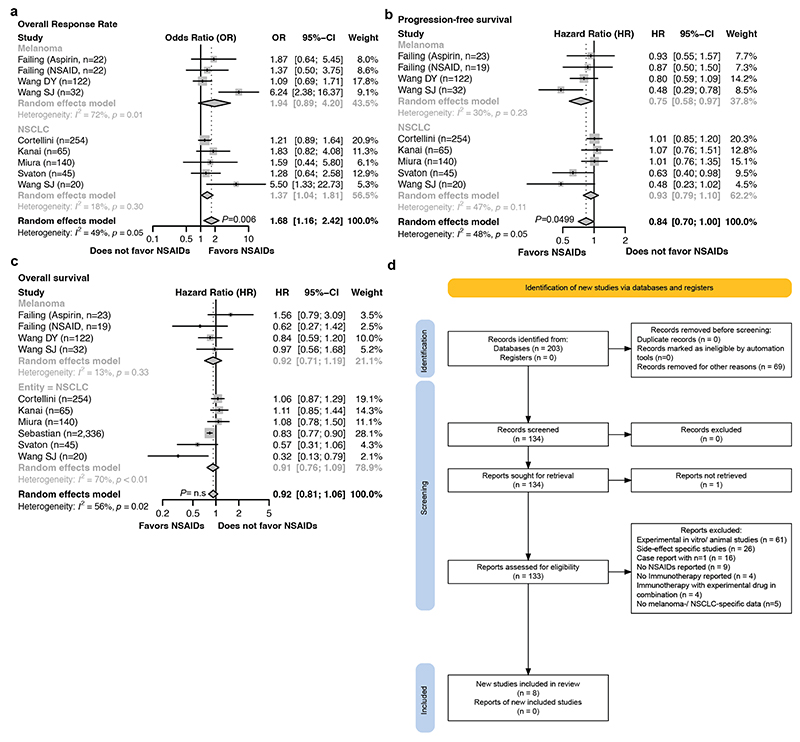
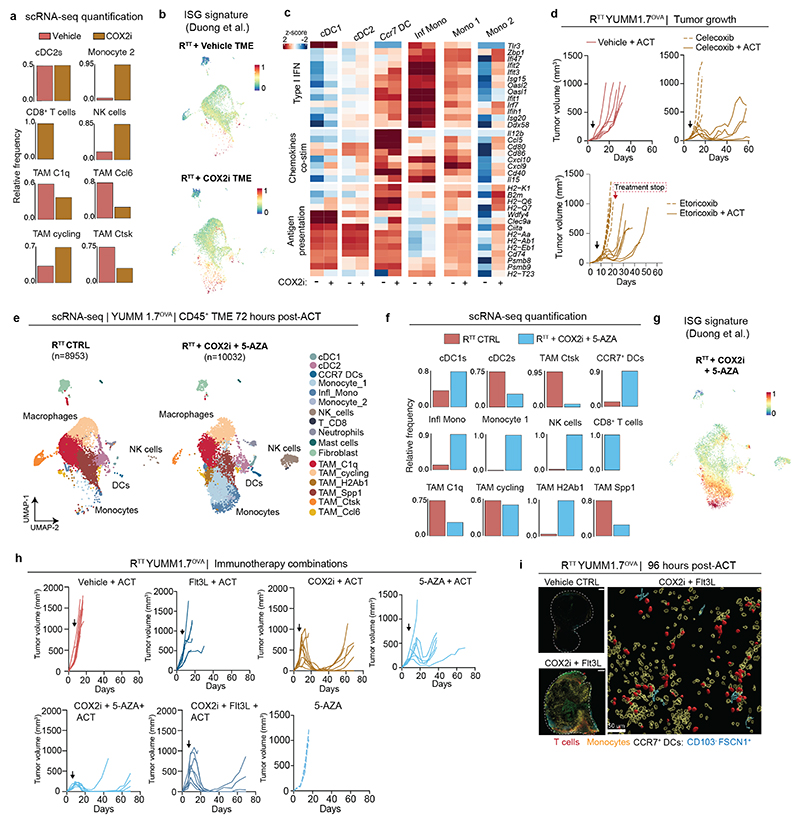
Non-steroidal anti-inflammatory drugs (NSAIDs) inhibit PGE2 production by COX enzymes and are commonly used for pain management in cancer patients. Retrospective studies51,52 and our own meta-analysis revealed that NSAID co-medication significantly improves overall response rate to ICB in melanoma and NSCLC patients (odds ratio 1.68, 95% CI 1.16 – 2.42, p=0.006), as well as progression-free survival, compared to patients receiving only ICB (Fig. 5a, Extended Data Fig. 9a-d and Supplementary Tables 5, 6). Nevertheless, these benefits did not translate into durable response and long-term survival (Extended Data Fig. 9c). To examine how COX2 inhibitors (COX2i) affect the immune TME, we treated RTT tumor-bearing Rag2-/- mice with celecoxib, adoptively transferred T cells intravenously and performed scRNA-seq (Fig. 5b). In line with findings in other models53, COX2 inhibition significantly increased the total amount of cDCs, including cDC1s and CCR7+ DCs, monocytes and their inflammatory state, and immunostimulatory TAMs, while reducing suppressive TAMs (Fig. 5b, c and Extended Data Fig. 10a-c). We observed both an increase in the abundance and functionality of cDCs and monocytes (Extended Data Fig. 10c). Continuous COX2 inhibition with celecoxib and etoricoxib, increased CD8+ T cell infiltration and led to shrinkage of RTT tumors in combination with ACT (Extended Data Fig. 10d). Notably, when COX2i treatment was stopped, tumors rapidly relapsed, suggesting that a continuous TME remodeling is required for T cell restimulation (Extended Data Fig. 10d).
Fig. 5. Pharmacological modulation of PGE2 and IFN-I reinstates an immune-permissive TME and immunotherapy response.
a, Forest plot of pooled odds ratios and 95% confidence intervals across clinical studies for overall response rates in patients receiving immune-checkpoint blockade (ICB) ± non-steroidal anti-inflammatory drug (NSAIDs) co-medication (n=722 patients over 8 independent cohorts, see also Extended Data Fig. 9a). Statistical analysis was performed with a random effects model, data are presented as mean values ± 95% confidence interval (CI). b, UMAP of scRNA-seq of CD45+ cells of RTT CTRL and COX2 inhibitor (COX2i)-treated YUMM1.7OVA RTT tumors 72 hours post-ACT (n=3 tumors pooled/condition). See Fig. 1c for cell cluster annotation. c, Relative frequency of cell types across conditions. d, Left, treatment schedule of YUMM1.7OVA RTT tumors in Rag2-/- mice with celecoxib (COX2i) in combination with Flt3L or 5-AZA. Right, BLI quantification. Top, representative BLI for vehicle (n=8 mice), COX2i (n=9 mice), COX2i+5-AZA (n=6 mice) and COX2i+Flt3L (n=8 mice) groups. Bar graphs depict the mean ± s.e.m. One-way ANOVA with Tukey’s multiple comparisons test. e, Left, Survival of Rag2-/- mice bearing YUMM1.7OVA RTT tumors treated with ACT and vehicle (n=8 mice), COX2i (n=9 mice), COX2i+5-AZA (n=7 mice) and COX2i+Flt3L (n=8 mice). Right, Survival of C57BL/6 mice bearing YUMM3.3 RTT tumors treated with anti-PD1/CTLA-4 with vehicle (n=6 mice), COX2i (n=8 mice), COX2i+Flt3L (n=9 mice) and COX2i+5-AZA (n=9 mice). Log-rank Mantel Cox test.
To further enhance the effects of COX2i, we explored different mechanism-based drug combinations aimed at inducing IFN-I or expanding antigen-presenting cells. To investigate whether the induction of IFN-I in RTT tumors could synergize with COX2i, we used 5-Azacitidine (5-AZA), a clinically approved DNA methyltransferase inhibitor that induces IFN-I54. Treatment with 5-AZA and its combination with COX2i led to a robust repolarization of the immune TME increasing inflammatory monocytes and T cell infiltration, leading to tumor regression in all Rag2-/- mice receiving ACT (Fig. 5d and Extended Data Fig. 10e-h). Similarly to COX2i, short-term treatment with 5-AZA only led to transient tumor control, further underlining the need for a continuous stimulatory TME (Extended Data Fig. 10h).
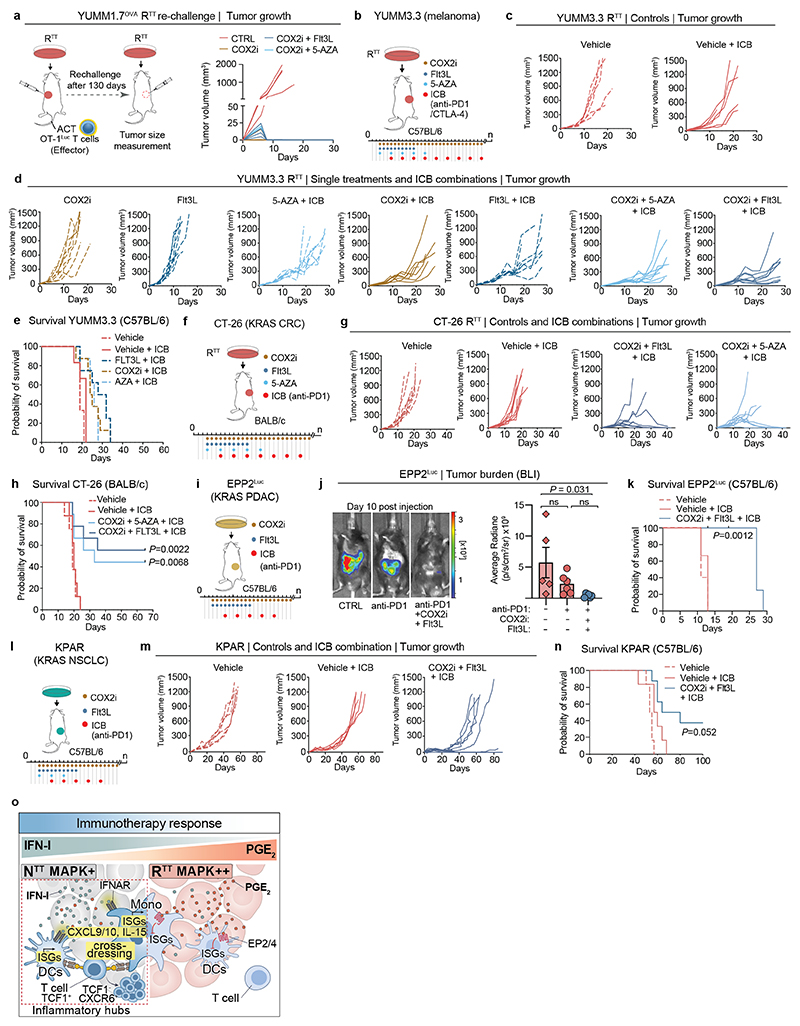
Next, we combined ACT with COX2i and Flt3L, a cytokine that promotes the expansion of cDCs32,55, which increased inflammatory hubs (Extended Data Fig. 10i), improved T cell expansion and led to more durable responses compared to COX2i alone (Fig. 5d, e and Extended Data Fig. 10h). Of note, Flt3L treatment alone did not lead to significant tumor control (Extended Data Fig. 10h). After 130 days, 3/9 mice remained alive in the COX2i group, 4/8 mice in the COX2i+Flt3L and 2/7 in the COX2i+5-AZA groups (Fig. 5e). Reinjection of YUMM1.7OVA RTT cells in long term tumor-free mice did not form tumors, indicating immune memory and T cell recall (Extended Data Fig. 11a). The benefit of combining COX2i with Flt3L or 5-AZA was even more pronounced in the YUMM3.3 RTT model, where COX2i+ICB had modest effects, but the addition of Flt3L or 5-AZA induced tumor control and significantly improved survival (Fig. 5e and Extended Data Fig. 11b-e). Similar benefits of these combination therapies were observed in several KRAS-driven models, including the colorectal cancer model CT-26, the NSCLC model KPAR and the PDAC model EPP2 (Extended Data Fig. 11f-n). Altogether, our results demonstrate that an immune evasive TME orchestrated by cancer cell-derived PGE2 and low IFN-I can be pharmacologically targeted using rational therapy combinations (Extended Data Fig. 11o).
Discussion
While the lymph node has long been recognized as a critical environment for determining CD8+ T cell function, findings by us and others assign a complementary role to the TME3–6,24. In this study, we show that inflammatory monocytes, which in human tumors correspond to CXCL9/10+ macrophages, drive T cell restimulation in the TME. We find that PGE2 and IFN-I, controlled by oncogenic MAPK signaling in cancer cells, disrupt this process. These findings provide mechanistic insights into the recent discoveries that CXCL10+ macrophages56, often present in immune hubs together with CD8+ T cells in patients5 and CXCL9:SPP1 macrophage polarity24 are predictive of response to ICB.
We find that inflammatory monocytes exhibit immune stimulatory capacities, evidenced by their expression of CXCR3 ligands (Cxcl9, Cxcl10) which recruit and position T cells, and IL-15, which promotes expansion and survival of the effector pool3. Unlike cDC1s that cross-present antigens, inflammatory monocytes obtain intact pMHCI complexes from tumor cells through cross-dressing. Both inflammatory monocytes and cDC1s facilitated intratumoral T cell restimulation in our models. In future studies it will be important to dissect these seemingly redundant functions of cDC1s and inflammatory monocytes/macrophages, and the contribution of other cell types, such as cDC2s37 and CD4+ T cells4,57,58. This will help determine whether the signals provided by these cells, including different modes of TCR engagement (cross-presentation versus cross-dressing), enhance the robustness of intratumoral CD8+ T cell restimulation or differentially impact T cell function. The plasticity, high abundance and short half-life of monocytes/macrophages render them promising therapeutic targets for boosting the efficacy of immunotherapies, in particular in tumors where functional intratumoral cDC1s are limited, a common feature of immune evasive tumors14,34. Ultimately it will be pivotal to understand if T cell restimulation within the TME is strictly required across tumor entities and/or modes of immunotherapy.
We show that patients with NSCLC and metastatic melanoma receiving ICB and NSAIDs concomitantly, display improved therapy response and progression-free survival51,52. However, use of NSAIDs did not translate into long-term benefit possibly due to incomplete inhibition of PGE2 production or discontinuation of treatment, which, in our preclinical models, led to rapid tumor regrowth. We propose mechanism-based interventions that combine immunotherapy with suppression of PGE2 levels via COX2i and increase in IFN-I or cDC function via 5-AZA or Flt3L administration, respectively. Given that combining these clinically-approved agents could cause toxicity and/or chronic IFN-I signaling resulting in T cell exhaustion59, further optimization in terms of drug timing and sequence will be required prior to patient use. Previous studies have shown that PGE2 limits inflammatory gene expression in infection models60. A thorough understanding of the pathways that underlie this process in intratumoral myeloid cells has the potential to reveal novel therapeutic targets for counteracting immune evasion.
Methods
Cell lines
YUMM1.7 and YUMM3.3 mouse melanoma61 cell lines (obtained from Marcus Bosenberg, Yale University, New Haven) were cultured in Dulbecco’s modified Eagle’s medium (DMEM)-F12 produced in-house. A375, M24962 (obtained from Joan Massague, MSKCC, New York), KPAR63 (obtained from Julian Downward, Francis Crick Institute, London) and EPP264 (obtained from Johannes Zuber, IMP, Vienna) cell lines were cultured in DMEM (Gibco). LOX48 (obtained from Joan Massague, MSKCC, New York), CT-2665 and NCI-H358 cell lines were purchased from ATCC and cultured in RPMI-1640 (Gibco). The NCI-H358 RTT derivative was generated by culturing NCI-H358 parental cells in the presence of 1 μM KRAS inhibitor (Amgen) for 90 days until cells became resistant. YUMM1.7OVA clones and all NTT and RTT derivatives were generated before15. RTT BRAFi-resistant cancer cells (YUMM1.7 and YUMM3.3 model) and all genetically engineered derivatives were cultured continuously in 100 nM of dabrafenib (Selleckchem). MEKi-resistant cancer cells were cultured continuously in 10 nM of trametinib (Selleckchem). Human NTT and RTT melanoma cell line derivatives (A375, M249 and LOX) were generated as described before48 and RTT maintained in culture on 1 μM of vemurafenib (LC-Labs). HEK-293T cells were purchased from Takara (Lenti-X 293T, 632180) and cultured in DMEM high glucose produced in-house. Bone marrow-derived dendritic cells (BM-DCs) were cultured according to an adapted version of a previously described protocol66. In brief, for the first 6-7 days, cells were cultured at a density of 1×106 cells/mL. On day 4, fresh medium was added to minimize cell death. After that, cells were either seeded for assays or counted and re-seeded at a density of 300,000 cells/mL. BM-DCs were cultured in full T cell medium supplemented with 200 ng/mL Flt-3L-Ig (BioXcell) and 5 ng/mL of GM-CSF (in-house produced). Bone marrow-derived Ly6C+ monocytes were cultured in DMEM media (Gibco). Human MONO-MAC-1 (obtained from Johannes Zuber, IMP, Vienna) and BLaER-167 (obtained from Moritz Gaidt, IMP, Vienna) cell lines were cultured in RPMI-1640 (Gibco). All media for cell lines were supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine (Gibco) and 100 IU/mL of penicillin-streptomycin (ThermoFisher). BLaER-1 and NCI-H358 cells were additionally supplemented with 1X sodium pyruvate. CD8+ T cells were cultured in full T cell media containing RPMI-1640 supplemented with 10% FBS, 2 mM L-glutamine, and 100 IU/mL of penicillin-streptomycin, 1X sodium pyruvate (Gibco), 1X non-essential amino acids (NEAA) (Gibco), 20 mM HEPES (produced in-house) and 0.05 mM β-mercaptoethanol (Millipore). All cells were cultured at 37°C and 5% CO2. Cells were routinely tested negative for Mycoplasma contamination. STR Profiling was performed in-house for the YUMM1.7, YUMM3.3, EPP2 and KPAR cell lines. Moreover, sensitivity to MAPK inhibitors was confirmed for A375, M249 and LOX (BRAFi), CT-26 (MEKi) and for NCI-H358 (KRASi).
Animal experiments and ethics
All mice were bred and housed in pathogen-free conditions with a housing temperature of 22±1°C, 55 ± 5% humidity, and a photoperiod of 14 hours light and 10 hours dark. Within each experiment, age and sex-matched groups were used. B6.129S(C)-Batf3tm1Kmm/J (Batf3-/-) mice, B6(Cg)-Zbtb46tm1(HBEGF)Mnz/J (zDC-DTR) mice, B6.Cg-Tg(Itgax-cre)1-1Reiz/J (CD11c-Cre) mice and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from Jackson Laboratories. B6.Cg-Rag2tm1.1Cgn/J Ly5.2 (Rag2-/-) mice, BALB/c and C57BL/6J were obtained from the Vienna Biocenter in-house breeding facility. ItgaxCrePtger2–/–Ptger4fl/fl mice were kindly provided by Dr. Jan Boettcher (TUM, Munich). For Rag2-/- Batf3-/- strain generation, Batf3-/- were crossed to Rag2-/- mice and homozygous offspring (Rag2-/- x Batf3-/-) were confirmed by genotyping and used in subsequent experiments to evaluate the lack of cDC1s in the context of adoptive T cell transfer. For Rag2-/- zDC-DTR strain generation, zDC-DTR mice were crossed to Rag2-/- mice and homozygous offspring were confirmed by genotyping and used in subsequent experiments to evaluate the effects of DC depletion. For adoptive T cell transfer experiments and injection of YUMM1.7OVA cell lines, Rag2-/- mice were used. For the injection of YUMM3.3, KPAR and EPP2 cell lines, C57BL/6 mice were used. For the injection of the CT-26 cell line, BALB/c mice were used. For the generation of bone marrow-derived dendritic cells (BM-DCs) and Ly6C+ monocytes, bones (femurs and tibias) were collected from in-house-bred C57BL/6 mice. For all above strains mice were used between 6-12 weeks old. For OT-1Luc CD8+ T cell isolations, 6-24 week-old OT-1Luc Thy1.1 mice68 were used. All mouse experiments were performed according to our license approved by the Austrian Ministry (GZ: MA58-2260492-2022-22, GZ: 340118/2017/25, BMBWF-66.015/0009-V/3b/2019, GZ: 801161/2018/17 and GZ: 2021-0.524.218 and their amendments). Mice were euthanized when the humane end point was reached (e.g weight loss>20%, signs of distress and pain), when tumors displayed signs of continuous necrosis or when tumors reached the maximum allowed tumor volume of 1500 mm3.
Tumor cell injections
For subcutaneous injections, mice were anesthetized with 2-4% isoflurane. For the YUMM1.7OVA model and all its derivatives 0.5-1 × 106 YUMM1.7OVA cancer cells were injected subcutaneously into the flank of each mouse in a volume of 50 μL. For contralateral experiments, alternating flanks were used for the injection of NTT and RTT cells to avoid preferential growth biases. For the YUMM3.3 model, 0.3-1 × 106 cells were injected subcutaneously in a volume of 50 μL. For the CT-26 model, 0.25 × 106 cells were injected subcutaneously in 50 μL. For the KPAR model 0.35 × 106 cells were injected subcutaneously in 50 μL. For the EPP2Luc cell line derivative, orthotopic injections were performed as previously described64. Briefly, surgeries were performed under isoflurane (2-4%) anesthesia on a heated plate. A small incision on the upper left quadrant of the shaved abdomen was made and the spleen identified, followed by externalization of the pancreas, 1 × 106 cells were injected intra-pancreatically. Organs were re-situated, and the peritoneum closed with a resorbable 6-0 Vicryl suture, followed by skin closure with sterile wound clips. Animals received intraperitoneal injections of 5 mg/kg carprofen preemptively and every 12 to 48 hours after surgery. Health status of mice was monitored daily and tumor burden was assessed by BLI. All cell lines were resuspended in PBS mixed 1:1 with Matrigel (Corning) in the final injection volume. Subcutaneous tumors were monitored by caliper measurements every 2-4 days, and tumor volume was calculated according to the following formula: Volume = (D x d2)/2, in which D and d are the long and short tumor diameters, respectively.
Isolation and activation of naive OT-1Luc CD8+ T cells
Spleen and lymph nodes were isolated from OT-1Luc mice, and red blood-cell lysis was performed with Ammonium-chloride-potassium (ACK) lysis buffer (Thermo Fisher) according to the manufacturer’s protocol. T cell isolation was performed with the Magnisort mouse CD8+ naïve T cell enrichment kit (ThermoFisher) according to the manufacturer’s protocol. T cells were activated for the first 24 hours by seeding them on a plate coated with 2 μg/mL anti-CD3 (145-2C11, eBioscience) overnight, and adding 1 μg/mL anti-CD28 (37.51, eBioscience), and 20 ng/mL carrier-free IL-2 (BioLegend). T cells were expanded for approximately 6-7 days in the presence of IL-2 and maintained daily at a concentration of 1 × 106 cells/mL in fresh T cell medium.
Adoptive T cell transfer (ACT), intratumoral injection and bioluminescence imaging (BLI)
Unless otherwise specified, when tumors reached a volume of 100-150 mm3, 4 × 106 in vitro activated OT-1Luc CD8+ T cells were injected intravenously (i.v) into mice in a volume of 100 μL of PBS. For intratumoral (i.tu) injections, 4 × 106 in vitro activated OT-1Luc CD8+ T cells were injected in a volume of 50 μL of PBS. For measuring T cell infiltration by BLI, D-luciferin (150 mg/kg, Goldbio) was injected retro-orbitally or via tail vein injection into anesthetized mice and mice were imaged with an IVIS machine (Caliper Life Sciences) and analyzed using Living Image Software v4.4 (Caliper Life Sciences). In NTT tumors, T cell recruitment to the tumor is detectable by BLI within 24 to 48 hours. This initial recruitment is followed by a phase of T cell expansion, with peak BLI signal between 96 to 120 hours. Hence, we depict 96 hours post-ACT images (unless otherwise specified in figure legends), as a suitable time point to assess T cell expansion in immune permissive TMEs.
In vivo treatments
For treatment with immune checkpoint blockade (ICB), anti-PD1 (clone RMP1-14, BioXcell) and anti-CTLA4 (clone 9D9, BioXcell) were injected intraperitoneally (i.p) in 100 μL of PBS when tumors reached a volume of 150-200 mm3 (usually between 6-8 days post-injection). The YUMM3.3 model was treated with 200 μg of anti-PD1/anti-CTLA4, the CT-26 model with 100 μg of anti-PD1 and the EPP2 model with 100 μg of anti-PD1. ICB treatment was administered every 3 days and continued at least for 3 weeks, as indicated in the figure legends. Control mice were treated with an isotype control antibody (rat IgG2a anti-trinitrophenol, clone 2A3, BioXcell, and mouse IgG2b, clone MPC-11, BioXcell). For COX2i treatment, celecoxib (LC Laboratories) was reconstituted in a 60:40 (DMSO:PEG400, dH2O) mixture as previously described53. Etoricoxib (Selleckem) was dissolved first in a small volume of DMSO and then in 1% sodium carboxymethyl cellulose (CMC-Na). COX2i were given by oral gavage every day (30 mg/kg) in a volume of 200 μL. For both COX2i regiments (celecoxib and etoricoxib), the treatment was started at day 3 post-injection (p.i), when tumors were palpable, and continued every day until the termination of the experiment. 5-Azacytidine (5-AZA) (Sigma-Aldrich) was reconstituted in DMSO to a stock concentration of 10 mg/mL, and further diluted in PBS for in vivo treatments and administered i.p (1 mg/kg) in 100-250 μL every 3 days, as described before54. For NK cell depletion, 200 μg of anti-NK1.1 (clone PK136, BioXcell) was administered every 3 days i.p, starting at day 1 p.i. NK cell depletion was confirmed by flow cytometry. For blocking T cell egress from the lymph node, mice were injected i.p with 20 μg per mouse of FTY720 (Sigma) in 100 μL of saline. Treatment was started on the day of T cell transfer and administered for 5-7 consecutive days. Control mice received saline injection. Flt3L (recombinant Flt-3l-Ig, hum/hum, BioXCell) treatment (30 μg/mouse in 100 μL PBS i.p) was started at day 3 p.i and administered every day for 9 consecutive days. In vivo IFNAR blockade was performed with InVivoMab anti-mouse IFNAR-1 (clone MAR1-5A3, BioXcell) and was administered i.p (200 μg/mouse) in 100uL. For IFN-γ, the neutralizing anti-mouse IFN-γ mAb was used (clone XMG1.2, BioXcell). Treatment was started on the day of tumor engraftment and administered every 3 days. InVivoMab IgG1 isotype control (BioXCell) was used as control. For experiments were CD8 depletion was performed, mice were treated with 50 μg of anti-CD8 (clone 2.43, in-house produced), while control mice were treated with isotype control (rat IgG2b anti-keyhole limpet hemocyanin, clone LTF-2) starting the day before tumor engraftment and then every 3 days.
Dendritic cell (DC) vaccination with bone marrow-(BM)-derived DCs
BM-DCs were cultured with Flt3L and GM-CSF as described above. At day 10-12 post isolation, DCs were activated overnight with PolyI:C (5 μg/mL, Invitrogen), pulsed with recombinant SIINFEKL peptide (5 μg/mL, Genscript) and sorted with fluorescence-activated cell sorting (FACS) on the basis of alive MHCII+CD103+CD11c+ cells. 1 × 106 cells in a volume of 50 μL of PBS were injected intratumorally. Control mice received 50 μL of PBS. For DC vaccinations, 2 doses of intratumoral injections were administered on day 4 and day 6 post tumor engraftment.
In vivo depletion of DCs with diphtheria toxin
For generation of bone marrow chimeras, Rag2-/- Ly5.1 mice were preconditioned (2x 5 Gy), before transferring back 10 × 106 bone marrow cells via iv injection. As donor mice, Rag2-/- Ly5.2 zDC-DTR mice were used. After 8 weeks of reconstitution, mice were used for experiments. NTT cells were injected and DCs were depleted by injecting 25 μg/kg of body weight of diphtheria toxin (DT) (Sigma-Aldrich) i.p in PBS, starting on the day of tumor engraftment and then every 3 days for 3-4 doses. Reconstitution efficiency and depletion of intratumoral DCs was confirmed by flow-cytometry.
Lentivirus generation and cell transduction
Lenti-X (HEK-293T) cells were transfected with 4,000 ng of the plasmid of interest, 2,000 ng of VSV-G plasmid, and 1,000 ng of PAX2 plasmid using polyethylenimine (PEI) (Avantor). Virus-containing supernatant was collected 24 hours and 48 hours post-transfection and subsequently filtered through a 0.45 μm filter. The cell lines of interest were transduced with the collected virus mixed with 8 μg/mL polybrene (Merck).
Generation of CRISPR-Cas9 knockout and overexpression cell lines
Doxycycline-inducible Cas9 (iCas9) clones from parental cell lines were generated to allow for inducible expression of Cas9. sgRNAs were chosen based on the best VBC score69 (see Supplementary Table 7) and were cloned into a vector containing a puromycin selection marker and mCherry or eGFP (hU6-sgRNA–PuroR–mCherry/eGFP). sgRNAs targeting the ROSA26 locus were used as controls for knockout cell lines. After transduction, cells were selected with puromycin (5-8 μg/mL) for 5 days. All sgRNA sequences can be found in Supplementary Table 7. For the generation of single cell-derived clonal cell lines, cells were FACS-sorted based on the fluorescent marker on the sgRNA backbone, at one cell per well into 96-well plates. To avoid immunogenicity caused by antibiotic selection markers or fluorophores in the YUMM3.3 model, we transiently transfected the cell lines with an all-in-one vector containing Cas9, the sgRNA of interest and eGFP (U6-IT-EF1As-Cas9-P2A-eGFP). For transient transfection, 7,000 ng of the plasmid with PEI was used and single cell clones were established. For IRF3 and IRF7 overexpression, synthesized cDNA sequences were ordered from Twist Biosciences and cloned into two different expression vectors with distinctive selection/fluorescent markers (SFFV-IRF3-mCherry and SFFV-IRF7-PuroR). After transduction, cells were selected with puromycin (5-8 μg/mL for 5 days) and bulk FACS-sorted on the basis of mCherry expression. The same cell line engineered with an empty vector containing an mCherry and a puromycin resistance cassette was used as a control. Knockout and overexpression of the target proteins was confirmed by genotyping, Western Blot or quantitative real time PCR (RT-qPCR). For the YUMM1.7 and YUMM3.3 Ptgs2 KO cell lines, single cell-derived clonal cell lines were generated and several were tested in vivo for growth kinetics.
EP2/4 KO in T cells
sgRNAs targeting the Ptger2 and Ptger4 mouse genes were designed according to the VBC score69 and cloned into the dual hU6-sgRNA-mU6-sgRNA-EF1α-mCherry-PuroR backbone (See also supplementary Table 7). As a control we used an sgRNA targeting a gene desert in Chromosome 1. Lentiviral vector was produced as described above. T cells were isolated from Cas9-OT-1 mice, which were a kind gift from Dr. Johannes Zuber (IMP, Vienna), as described above. Twelve hours after CD3/CD28 activation, T cells were spin-infected with the lentiviral vector containing the sgRNAs in a 1:1 ratio for 1 hour at 32°C and 800g. 12 hours post infection, T cells were removed from the activation plate, washed with PBS and cultured in the presence of 20 ng/mL of IL-2. Selection with puromycin was performed 30 hours after viral transduction. Prior to ACT, mCherry levels were assessed and knockout was confirmed by functional in vitro assays.
Flow cytometry and cell sorting
For flow cytometry-based characterization of the TME, tumors were isolated between day 7-11 p.i, cut into pieces, and digested for 1.5 hours at 37°C with collagenase A (1 mg/mL, Roche) and DNAse (20 μg/mL, Worthington) in unsupplemented RPMI-1640 medium. Digested tumors were strained through a 70 μm filter and resuspended in FACS buffer (0.5% BSA, 2 mM EDTA in PBS). Fc-block was performed with anti-CD16/32 (clone 2.4G2, Pharmingen) for 10 minutes at 4°C to avoid Fc-specific antibody capture, and staining for cell-surface markers was performed for 30 minutes at 4°C. For intracellular staining, the Foxp3 Transcription Factor staining kit was used (eBioscience). Live/dead exclusion was performed by staining with the fixable viability dye eFluor780 (1:1,000, eBioscience). DCs were defined in most experiments as MHCII+ CD11c+ CD24+ out of alive CD45+ cells. cDC1s were identified as CD103+ CD11b- out of the total DCs, cDC2s as CD103- CD11b+ and inflammatory cDC2 as CD103- CD11b+ AXL+. AXL was previously described to identify inflammatory cDC2s37. Monocytes were defined as Ly6C+ CD11b+ F4/80-, and inflammatory monocytes were identified as monocytes that were Ly6A+. Ly6A was previously described to identify monocytes expressing high levels of interferon-stimulated genes (ISGs)38. Macrophages were defined as Ly6C- F4/80+ Cd11b+. Acquisition of the samples was performed using a BD LSR Fortessa machine (BD Biosciences) with FACS Diva software v9.0.1, and analysis was conducted with the FlowJo software v10.8 or newer. For cell sorting, a BD Aria cell sorter (BD Biosciences) with FACS Diva software v9.0.1 was used.
Antibodies for flow-cytometry
The following antibodies (all anti-mouse) were used for flow cytometry stainings (target (clone, CAT#, manufacturer, dilution)): AXL PE-Cy7 (MAXL8DS, 25-1084-82, eBioscience, 1:200), CD103 PerCP/Cyanine5.5 (2E7, 121415, BioLegend, 1:100), CD103 PE (2E7, BioLegend, 121405, 1:100), CD11b APC (M1/70,17-0112-81, eBioscience, 1:200), CD11b PerCP/Cyanine5.5 (M1/70, 101229, BioLegend, 1:200), CD11c BV605 (HL3, 563057, BD Pharmigen, 1:100), CD11c FITC (N418, 117305, BioLegend, 1:100), CD24 BV510 (M1/69, 101831, BioLegend, 1:100), CD24 FITC (M1/69, 11-0242-82, eBioscience, 1:100), CD279/PD-1 BV785 (29F.1A12, 135225, BioLegend, 1:200), CD279/PD-1 FITC (29F.1A12, 135213, BioLegend, 1:200), CD40 APC (3/23, 124611, BioLegend, 1:200), CD45 BV711 (30-F11, 103147, BioLegend, 1:500), CD45 FITC (30-F11, 103107, BioLegend, 1:500), CD86 BV510 (GL-1, 105039, BioLegend, 1:100), CD3 BV605 (17A2, 564009, BD Horizon, 1:100), CD3 AF647 (17A2, 100209, BD Horizon, 1:100), CD3 AF488 (17A2, 100212, BD Horizon, 1:100), CD8a eFluor 450 (53-6.7, 48-0081-80, eBioscience, 1:100), CD8a AF647 (53-6.7, 128041, BioLegend, 1:100), MHCI (H-2Kb) APC (AF6-88.5.5.3, 17-5958-82, Bioscience, 1:200), MHCI (H-2Kb) PE (AF6-88.5.5.3, 17-5958-80, Bioscience, 1:200), MHCII (I-A/I-E) eFluor450 (M5/114.15.2, 48-5321-80, eBioscience, 1:200), MHCII (I-A/I-E) APC (M5/114.15.2, 107613, BioLegend, 1:200), NK-1.1 BV711 (PK136, 108745, BioLegend, 1:100), TCF1 PE (S33-966, 564217, BD Pharmigen, 1:50), TIM3 BV711 (RMT3-23, 119727, BioLegend, 1:100), CD88 PE (20/70, 135805, BioLegend, 1;100), Ly-6A/E (Sca-1) FITC (D7, 108105, BioLegend, 1:100), SIINFEKL-HK2B PE (25-D1.16, 12-5743-81, Invitrogen, 1:100), F4/80 PE (BM8, B123110, BioLegend, 1:200), Rat IgG1, K Isotype control PE (R3-34, 5546, BD Pharmigen). Further information can be found in Supplementary Table 8.
RNA extraction of cancer cells sorted from tumors, in vitro cell lines, and myeloid cells
Tumors were surgically removed between days 10-12 p.i. The tissue was processed as described above, and cancer cells were isolated via flow cytometry on the basis of alive, CD45-. For cancer cell lines and in vitro assays with myeloid cells, cells were washed with PBS and snap-frozen in liquid nitrogen and kept at -70°C until further processing. RNA was extracted using a magnetic bead-based RNA extraction protocol (in-house produced). In brief, cells were lysed and incubated with beads together with DNase I (NEB) followed by magnetic isolation. RNA was purified by further elution with nuclease-free water.
Reverse transcription and quantitative real-time PCR (RT-qPCR)
Reverse transcription was performed for complementary DNA (cDNA) formation with 1 μg of RNA per sample utilizing the LunaScript® RT SuperMix Kit (NEB) according to the manufacturer’s instructions. Quantitative real-time PCR (RT-qPCR) was performed with 10 ng of cDNA per sample either with the Luna® Universal qPCR Master Mix (NEB) or the in-house produced MTD qPCR Dye 2x HS Master Mix according to the manufacturer’s protocol. Each sample included 4 technical replicates. The RT-qPCR reaction was carried out in a Bio-RadCFX384 Real-Time Cycler and contained 1 min of initial denaturation (95°C) and 45 annealing cycles lasting 15 seconds at 95°C and 30 seconds at 60°C. The analysis of gene expression levels was determined by the quantification cycles (Cq). Internal controls and the housekeeping gene GAPDH were used to correct for differences in sample quality and to normalize expression values. qPCR primer pair sequences are listed in Supplementary table 7.
In vitro assays with BM-DCs
For cancer cell conditioned media experiments, supernatants (in full T cell media) from confluent cancer cells were collected after 48 hours, filtered through a 45 μm filter, and frozen at -70°C until further use. Full T cell medium was supplemented with 20 μM of the COX1/2 inhibitor indomethacin (Selleckchem) or 5 nM of the MEK inhibitor trametinib (Selleckchem) for the evaluation of MAPK and COX1/2 activity before media conditioning. BM-DCs were differentiated as described above, and harvested at day 6. 0.5-1 × 106 cells were seeded in triplicates in a 12-well plate in conditioned medium and treated with 10 μg/mL of InVivoMab anti-mouse IFNAR-1 antibody or InVivoMab IgG1 isotype control (BioXCell). Cells were cultured for 24 hours, collected, and processed for flow cytometry analysis or RNA extraction. For treatment with PGE2 and IFN-β, cells were collected at day 6 and seeded at a concentration of 0.5-1 × 106 cells/mL. Cells were treated for 24-48 hours with recombinant PGE2 (100 ng/mL, Sigma-Aldrich) and recombinant murine/human IFN-β (R&D Systems) at the concentrations indicated in the corresponding figures. Same volumes of acetone and PBS were used as a control for PGE2 and IFN-β, respectively.
Isolation of bone marrow-derived Ly6C+ monocytes for intratumoral injection and in vitro assays
Ly6C+ monocytes were directly isolated from the bone marrow of CD45.1+ C57BL/6 mice using the monocyte isolation kit (Miltenyi Biotec) following the manufacturer’s instructions. For intratumoral monocyte transfer, 1 × 106 monocytes were injected intratumorally into NTT and RTT tumors established in CD45.2+ Rag2-/- mice. Tumors were isolated for FACS analysis 72 hours post intratumoral transfer. For in vitro assays to assess effects of PGE2, Ly6C+ monocytes were seeded at a density of 1 × 106 cells/mL and cultured in recombinant IL-4 and GM-CSF (both produced in-house), and exposed to 200 ng/mL of PGE2 or vehicle for 3 or 5 days. For conditioned media experiments, monocytes were seeded at a density of 1 × 106 cells/mL in conditioned media obtained from from NTT, RTT or RTT IRF3/7 cells ± 20 μM COX1/2i (Indomethacin) during media conditioning and subsequently supplemented ± 10 μg/mL InVivoMab anti-mouse IFNAR1 anti-mouse (BioXCell) or isotype IgG1 control (BioXCell).
In vitro monocyte co-culture assay
Ly6C+ Ly6A+ or Ly6C+ Ly6A- monocytes were FACS sorted from NTT tumors grown in Rag2-/- mice or BALB/c mice and co-cultured for 72 hours with naive OT-1 T cells (1:3 ratio: 100,000 monocytes for 300,000 naive OT-1 cells) previously labeled with 0.25 μM CFSE for 30 minutes at 37°C.
In vitro human monocyte assays
BLaER-1 cells were trans-differentiated into monocytes as previously described67. Briefly, BlaER-1 trans-differentiation media was freshly prepared by adding 10 ng/mL human recombinant (hr-)IL-3 (PeproTech), 10 ng/mL hr-M-CSF (PeproTech) and 100 nM β-Estradiol (Sigma-Aldrich) to complete RPMI medium. Cells were resuspended in trans-differentiation media and plated in a 12-well plate at 0.7 × 106 cells/mL. Cells were incubated at 37°C for 5-6 days until mature monocytes were differentiated. For conditioned media experiments, BLaER-1 or MONO-MAC-1 human monocytes were seeded at a density of 0.7 × 106 cells/mL in conditioned media obtained from from NTT or RTT cells from the human melanoma cell lines A375, M249, LOX or the human NSCLC cell line NCI-H358 ± 20 μM COX1/2i (Indomethacin) during media conditioning. Cells were cultured in CM for 24 hours and collected for RNA extraction, as described above.
Evaluation of pMHC-I cross-dressing on monocytes
For mismatched MHC-I haplotype experiments, 1×106 YUMM1.7OVA NTT cells from C57BL/6 origin (H-2Kb) were injected in the flank of BALB/c (H2-Kd) mice. BALB/c mice were treated with anti-CD8 (50 μg in 100 μl, in-house produced), while control mice were treated with isotype control (rat IgG2b anti-keyhole limpet hemocyanin, clone LTF-2) starting the day before tumor engraftment and then every 3 days, to avoid T cell-mediated mismatched MHC-I rejection of YUMM1.7 cells. On day 10, tumors were collected and processed for flow cytometry staining of H2-Kb or FACS-sorted on the basis of Ly6A expression for in vitro assays.
Sample preparation for single-cell RNA sequencing (scRNA-seq)
For scRNA-seq experiments involving the TME characterization, tumors were isolated at day 10 p.i (72 hours post-ACT) and were processed as described above. The CD45+ live fraction was isolated by FACS-sorting and approximately 1 × 105 cells were collected. For scRNA-seq of OT–1 T cells, tumors were isolated 5 days after intratumoral injection of 4 × 106 T cells. Alive T cells were isolated from tumors by FACS-sorting for CD45+CD3+CD8+ markers. Dissociated cell concentrations were measured using NucleoCounter NC250 (Chemometec) following the manufacturer’s instructions. For scRNA-seq samples from exp3-4 (see below), the Chromium Next GEM Single Cell Fixed RNA Sample preparation kit was used according to the manufacturer’s protocol. In brief, 1 × 106 cells were fixed for 22 hours at 4°C, quenched, and long-term stored at -80°C according to 10X genomic Fixation of Cells & Nuclei for Chromium Fixed RNA profiling (CG000478) using Chromium Next GEM Single Cell Fixed RNA Sample preparation kit (PN-1000414, 10X Genomics). 250,000 cells per sample were used for probe hybridization using the Chromium Fixed RNA Kit, Mouse Transcriptome, 4rxn x 4BC (PN-1000496, 10X Genomics), pooled equally and washed following the Pooled Wash Workflow as described in the Chromium Fixed RNA Profiling Reagent kit protocol (CG000527, 10X Genomics). For all the other scRNA-seq samples, the Chromium Next GEM Single cell 3’ kit with Dual Index was used according to the manufacturer’s instructions. GEMs were generated on Chromium X (10X Genomics) with a target of 10,000 cells recovered and libraries prepared according to the manufacturer’s instructions (CG000527, 10X Genomics). Sequencing was performed on NovaSeq S4 lane PE150 (Illumina) with a target of 15,000 reads per cell.
scRNA-seq analysis of CD45+ TME
CD45+ immune cells were collected in 4 different 10x Genomics sequencing experiments: Exp1, Chromium Single Cell 3’ scRNA-seq samples pre-processed using cellranger count v6.1.1 (YUMM3.3 samples: NTT/108155 and RTT/108157). Exp2, 3’ CellPlex multiplex experiment with 4 samples pre-processed using cellranger multi v6.1.1 (YUMM1.7OVA samples: NTT+ACT, RTT+ACT, RTT Ptgs1/2 KO+ACT, RTT CTRL ROSA26+ACT). Exp3 and Exp4, Chromium Flex multiplex experiments with 4 samples each pre-processed using cellranger multi v7.1.0 and the built in Probe Set v1.0.1 mm10-2020-A. Exp3, YUMM1.7OVA samples: RTT mCherry CTRL, RTT IRF3/7, RTT COX2i and RTT COX2i + 5-AZA, all ACT treated. Exp4, YUMM1.7OVA contained biological replicates of Exp2 samples and untreated YUMM1.7OVA samples (noA): NTTnoA/271221, RTTnoA/271222, NTT/271223 ACT, RTT/271224 ACT). The prebuilt 10X mm10 reference refdata-gex-mm10-2020-A was used. Further processing was performed in R v4.2.2 with Seurat v4.3.0. For generating a CD45+ immune reference map we integrated cells from the first three experiments as follows. The cellranger filtered feature-barcode matrices were used, retaining cells with more than 1000 detected genes and less than 15% of mitochondrial and less than 40% of ribosomal RNA reads. An integrated feature-barcode matrix from the 3 experimental batches was generated accounting for the inclusion of a probe-based assay by keeping genes found in at least 5 cells in each experiment, and excluding ribosomal and mitochondrial genes. Data was log-normalized, scaled (regressing out the difference between the G2M and S phase signature scores), dimensionality reduction was performed using PCA on the top 3000 most variable genes, batch correction across batches was performed using Harmony70 v0.1.1, the 40 harmony embeddings were used for UMAP visualizations. The first 40 harmony dimensions were used to identify immune cell subclusters with a resolution of 0.5, that were further assigned to cell types using known markers and publicly available myeloid reference data-sets21,71. Cells were scored for the expression of published signatures using the AddModuleScore function72. Wilcoxon rank sum test implemented in Presto v1.0.0 was used to identify differentially expressed genes. Seurat’s reference-based mapping was used to predict celltype identity and map cells of the biological replicate experiment to our annotated reference set using the FindTransferAnchors and MapQuery functions, after a quality control process retaining cells between 1000 and 4500 detected genes for 271222 and 271224, and 1300 and 8000 detected genes for 271221 and 271223, and limiting count tables to the gene universe of the reference. Depth normalized counts for pseudobulk and GSEA functional analyses of this experiment were generated using cellranger aggr. Differences between ACT and untreated conditions (no ACT) from the replicate experiment (Exp4) were explored on a pseudo-bulk level in an unsupervised clustering analysis with heatmap visualization. The fibroblast cluster was removed prior to further processing. Sum aggregation on the depth-normalized UMI counts, was followed by variance stabilizing transformation, selection of 300 most variable genes, standardization, kmeans clustering (k=3) and enrichr analysis against the Reactome_2022 using enrichR. The relative frequency bar plots depict the changes in the relative abundance of a cell type across different experimental conditions. For each condition, we calculated the normalized abundance of a specific cell type by comparing the absolute number of the cell type to the absolute number of all cells in the same condition. This normalization accounts for differences in total number of cells captured between conditions. We then calculated the relative cell abundance of the cell type in all conditions of the experiment. This was done by comparing the normalized abundance of the cell type to the sum of normalized abundances of the same cell type across conditions of the experiment. This step produces values between 0 and 1 for each condition for each cell type, with the sum of these values across all conditions of the experiment equaling 1 for each cell type.
scRNA-seq analysis intratumoral CD8+ OT-1Luc T cells
Single-cell gene-expression of isolated NTT and RTT T cells was assayed in a Chromium Flex experiment, read processing was performed using cellranger multi v7.1.0 using probeset v1.0.1 mm10-2020-A. Cellranger filtered feature-barcode matrices were used on, further filtered to retain cells with more than 800 detected genes, less than 10 percent of mitochondrial and less than 10 percent of ribosomal RNAs reads and remove cells of contaminant clusters identified using SingleR and ImmGen reference (fibroblasts, MoMac populations). Data was log-normalized, scaled, dimensionality reduction was performed using PCA on the top 2000 most variable genes, harmony was used for the integration of cells from different samples, 15 harmony embeddings were used for UMAP visualizations, and published tumor single cell data for signature scoring29. Gene lists are deposited in Supplementary Table 2.
RNA velocity analysis
To understand differentiation trajectories of myeloid cells within the TME, we performed RNA-velocity analysis73 of the MoMac compartment. Loom files containing the splicing annotation were created for each sample using the velocyto run command from the package velocyto (0.17,17), with default parameters, with no masked intervals. Then, the loom files were combined with the scRNA-seq object that had been filtered to keep the data for monocyte and macrophage populations (Monocyte_1, Monocyte_2, Infl_Mono, TAM_CCL6, TAM_Ctsk, TAM_C1q, TAM_H2Ab1, TAM_Spp1, TAM_cycling), for each condition (NTT, RTT, RTT Ptgs1/2 KO). First and second order moments were computed using scvelo (0.2.5) pp.moments (n_pcs = 30, n_neighbors = 30) and the dynamical model was run with default parameters. Python version 3.8.12 was used.
SCENIC analysis
Gene regulatory networks for each cell population in each condition were calculated using SCENIC74. The motif database used was mm9-tss-centered-10kb-7species.mc9nr.feather. Co-expression network was calculated using GENIE3. The gene regulatory network was built using SCENIC wrapper functions.
Analysis of publicly available myeloid datasets and inflammatory signatures
For the MEL and LUNG samples from the Cheng et al23. dataset (GSE154763), the raw counts were pre-processed as described in the publication, and clustering was calculated using a resolution of 0.8. The monocyte and inflammatory monocyte gene set was derived by using the wilcoxauc() function from presto and by selecting the genes with a logFC > 0.6 (See Supplementary Table 2). Then, the gene symbols were converted to human symbols. The human inflammatory monocyte gene set was used to calculate an enrichment score per cluster. Briefly, the gene average expression was calculated for each cluster in the LUNG, and MEL datasets, on normalized data. Then, an enrichment score was calculated using GSVA with the following parameters: minSize = 5, maxSize = 500, kcdf = “Gaussian”. The projection of the signature on the UMAP embedding was done using the function AddModuleScore() and then plotting the resulting score using FeaturePlot() with min.cutoff = 0.3 for the Inflammatory monocyte score and min.cutoff = 0.4 for the Monocyte_1 score. For the Barras et al.43 dataset, the annotated seurat-object for myeloid populations corresponding to the original Fig.4a was obtained and gene sets were analyzed as described above. For querying previously published inflammatory gene signatures, the Duong et al. ISG+ DC signature was previously generated by taking the top differentially expressed genes (DEGs) in the cDC2 cluster37. The Bosteels Inf-cDC2 DC signature was generated previously by Duong et al. and was obtained by reanalyzing the scRNAseq data set (GSM4505993), where the top 20 DEGs were taken in the identified inflammatory cDC2 cluster. All of them were subsequently scored in our dataset using the AddModuleScore72 and plotted the resulting score using FeaturePlot(). Gene lists are deposited in Supplementary Table 2.
Single cell spatial transcriptomics of human melanoma samples
Single cell spatial transcript profiling was performed using the CosMx technology (Nanostring, Seattle, WA). Biopsies were obtained from patients with an age at diagnosis that ranged from 24 to 85 with a median of 66. 34% were female patients and 66% were male patients. We obtained cell-segmented data for 74 FOVs (Field of View, an area of 500×500 μm) from TMA cores of 34 melanoma metastases, in total consisting of 980 genes × 171,536 cells. Tumor tissues were obtained from 21 lymph nodes, 7 subcutaneous metastases, 1 lung metastasis and 1 brain metastasis (4 NAs), from 31 patients containing 72 FOVs. 2 FOVs were from tonsils as control. Most tumor tissue were from patients that were treatment naïve at the time of surgery. Tissue collection was approved by the Regional Ethics committee at Lund University (Dnr. 191/2007 and 101/2013). Patients signed an informed consent. The majority of TMA cores contained tertiary lymphoid structures (TLS) and FOVs were preferentially directed to these regions. Low-quality FOVs, cells with < 20 counts and potential multiplets of cells (area exceeding the sample geometric mean + 5 standard deviation) were discarded. Using Seurat, genes whose mean expression was below 3 x median of negative probe mean expression, and genes with the highest 99% quantile expression, MALAT1 and IGKC (due to potential spill-over to neighboring cells), were removed, which retained 641 genes. The data were normalized using SCTransform75, counts that were zero before SCTransform were restored, and counts were log-transformed as log2 (counts+1). The top 30 principal components were used for UMAP reduction and clustering (k.param=15, resolution=0.5, Louvain algorithm). Resulting clusters were assigned to biological annotations using known marker genes, and annotations were mapped back to FOV coordinates. Expression of C1QC, CXCL9 or CXCL10 > 0 was considered as “Positive”. Cell type fractions were derived for each FOV. Pearson correlation values between cell type fractions across FOVs were determined and displayed. In Fig. 2i, CXCL9+/10+ MΦ/DCs (#9 and #10) were either CXCL9+ or CXCL10+. MΦ/DCs (#5) were negative for CXCL9, CXCL10, and C1qC.
Generation of the TME-COX and TME-IRF3/7 signature
For the TME-COX signature, the FindMarkers function was used in Seurat, with tresh.use=0.25 and min.pct=0.1, to compare RTT CTRL (ROSA26) and RTT Ptgs1/2 KO scRNA-seq samples. The top DEGs (log2FC<-1.5, adj-p-value< 0.05) were used and converted to human orthologs using DIOPT76. For the TME-IRF3/7 signature, the FindMarkers function was used in Seurat, comparing RTT CTRL (mCherry) and RTT IRF3/7 and taking the top 40 DEGs.
TME signatures in immunotherapy-treated human samples
Gene expression data for patients receiving checkpoint blockade were obtained from Gide et al.49 (accession no. PRJEB23709). The TME-COX, TME-IRF3/7 and CD8+ T cell scores for each tumor sample were defined as the geometric mean of the expression values of each of the gene sets, respectively (Supplementary Table 4). Univariate Cox proportional hazards models, in which the TME-COX and TME-IRF3/7 scores were included as continuous variables, were used for testing the statistical association between gene signature expression and patient survival, separately for both signatures. The tumor samples were then divided into three groups based on the signature score (bottom third, mid third, top third) and Kaplan-Meier plots were generated for visualization. The association between signature expression and CD8+ T cell abundance was evaluated by calculating the Person’s correlation coefficient between the signature score and a CD8+ score for each signature separately. For this all scores were normalized to a median of zero and standard deviation of one. The two overlapping genes were removed from the CD8+ signature before comparing it to TME-IRF3/7 signature expression. For evaluating the enrichment of TME-COX and TME-IRF37 gene signatures in responder and non-responder patients to TIL therapy (baseline) from Barras et al.43 mouse gene ids were first converted to human orthologs (with DIOPT v9; Best Score = Yes, Best Score Reverse = Yes, DIOPT Score > 7) and single cell level signature enrichment scores for the “humanized” gene sets were calculated using AddModuleScore_UCell77.
Analysis of transcriptomic data
For plots shown in Fig. 3a a cut-off of adjusted p-value<0.05 and log2FC>2 and <-2 was used on DEGs expressed in YUMM1.7OVA NTT and RTT GFP+ cancer cells FACS-sorted out of tumors (Supplementary Table 3). Pathway enrichment analysis was performed with Enrichr78,79. For plot in Extended Data Fig. 5i Upstream regulator analysis (Ingenuity)80 was used to identify upstream regulators using DEGs with an adj p-value<0.05.
Quantification of PGE2 and IFN-β by ELISA
For in vitro analysis of PGE2 production, 2 × 106 cells were seeded in 10 mL of media and supernatants were collected after 48 hours and kept at -70°C, until analysis. For IFN-β, 0.3 × 106 cells were seeded and 1 mL of supernatant was collected from confluent cells in a 6-well plate after 48 hours of culture and kept at -70°C until analysis. For analysis of PGE2 and IFN-β from mouse tumors, whole tumors were isolated between days 4-10 post engraftment, accurately weighted, and immediately snap-frozen in liquid nitrogen. They were stored at -70°C until further processing. For PGE2 analysis, tumors were subsequently digested using the MACS dissociator according to the manufacturer’s protocol in PBS supplemented with 1 mM EDTA and 10 μM indomethacin. Lysate was further diluted in dissociation buffer depending on the tumor condition and weight (100 μL per mg of tumor) and further quantified by PGE2 ELISA (Cayman) or by Mouse IFN-beta Quantikine ELISA Kit (Biotechne) according to the manufacturer’s protocol. Values were normalized taking into account dilution factors and tumor weight. For human IFN-β analysis from human cells, 1 × 106 A375, M249, LOX or NCI-H358 cells were injected into NSG mice and harvested on day 21. Tumors were processed as described above and quantified by Human IFN-beta Quantikine ELISA (Biotechne) according to the manufacturer’s protocol.
Eicosanoids analysis from tumors by HPLC-MS
YUMM1.7OVA NTT and RTT tumors were isolated at day 10 p.i, weighed, and a solution of isopropanol:methanol (1:1, v/v) was added to the tissue for metabolite extraction. The material was subsequently homogenized and incubated for 1 hour at -20°C. The samples were then centrifuged at 14,000 g for 3 minutes. A second extraction round was performed by adding 80% methanol:H2O (vol/vol) to the pellet, centrifuged, and both supernatants were combined. Finally, the samples were incubated for another 2 hours at -20°C, and after final centrifugation, the supernatants were stored at -70°C until further analysis. Samples were subsequently measured on a ZIC-pHILIC column or a RP column. Metabolites were annotated using the compound discoverer 3.0 software (Thermo Fisher), using an internal database or the mzCloud database (at least 75% match based on measured molecular weight and MS2 spectra). For filtering, an RSD of corrected QC areas was used, being less than or equal to 25%. Group CV of at least 1 group is less than or equal to 40%.
Western blotting
Cells were lysed with RIPA buffer (Cell Signaling Technology) supplemented with complete Protease Inhibitor Cocktail (Sigma Aldrich) and HALT phosphatase inhibitor (Thermo Fisher Scientific). Lysates were sonicated and cleared by centrifugation at 14,000g for 10 minutes at 4°C. Protein concentrations were quantified according to the manufacturer’s instructions using a BCA Protein Assay kit (Pierce, Thermo Fisher Scientific). Immunoblotting was conducted according to standard protocols. The primary antibodies used for immunoblotting were as follows: anti-Vinculin (Sigma-Aldrich, 1:1,000), anti-COX2 (CST,1:1,000), anti-H3 (acetyl K27) (abcam, 1:5000). The secondary antibodies used were as follows: anti-rabbit IgG HRP-linked (Cell Signaling Technology, 1:10,000) and anti-mouse IgG HRP-linked (Cell Signaling Technology, 1:10,000).
Volumetric immunofluorescence microscopy and image analysis
Volumetric microscopy of mouse tumors was performed as described previously9. In brief, tumors were fixed in Antigenfix solution (Diapath) for 6-8 hours, dehydrated in 30% sucrose overnight, embedded in TissueTek OCT freezing medium (Sakura Finetek) and stored at -80°C. Using a Leica CM3050 S cryostat, consecutive sections of 50 μm thickness were generated, subsequently permeabilized, blocked, and stained in 0.1 M Tris (Carl Roth) supplemented with 1% BSA, 0.3% Triton X-100 (Merck), normal mouse serum (Merck) and donkey serum (Merck). Stained sections were mounted in Mowiol (Merck) and imaged on an inverted TCS SP8 confocal microscope (Leica) using a HC PL APO CS2 20x/0.75 NA objective. Images were acquired as tiled image stacks, covering whole tumor sections in the XY plane, with 2 μm Z-spacing to provide three-dimensional (3D) image volumes of at least 20 μm depth. For further analyses, images were adaptively deconvoluted using the Leica TCS SP8 LIGHTNING tool v3.5.7.23225 and analyzed using Imaris 9.9 software (Oxford Instruments). The Imaris surface generation tool was used to reconstruct and visualize 3D objects for individual cells. Where indicated, signals outside rendered cells were masked to visualize intracellular proteins. For analysis of immune cell infiltration by histo-cytometry, statistics for object localizations were exported into Excel v16.88 (Microsoft) and analyzed by GraphPad Prism software (GraphPad). Quantification of the number of cells was performed relative to the volume of the imaged section. Interacting cells were described as being less than <5 μm apart from each other.
Antibodies for immunofluorescence microscopy
The following antibodies were used for staining of mouse tissues: anti-CD3 (Biolegend, clone 17A2), anti-CD103 (R&D Systems, goat polyclonal), anti-FSCN1 (Santa Cruz Biotechnology, clone 55-k2), anti-LY6C (Biolegend, clone HK1.4), anti-MHCII I-A/I-E (Biolegend, clone M5/114.15.2). All antibodies were either validated by the manufacturer or were previously reported for immunofluorescence microscopy. The populations were defined as follows: T cells (CD3+), Monocytes (Ly6C+ CD103- MHCII+), cDC1s (FSCN1- CD103+ MHCII+), CCR7+ cDC1 (FSCN1+ CD103+ MHCII+) and CCR7+ cDC2 (FSCN1+ CD103- MHCII+).
Nur77-GFP was directly assessed by transferring Nur77-GFP reporter OT-1 T cells.
Meta-analysis of NSAID immunotherapy cohorts
The meta-analysis was performed in accordance with the updated Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) reporting guidelines81. The literature search was conducted using the PubMed (MEDLINE) database and last updated on 31 December 2023. The full search strategy is available in the Supplementary Table 6. The literature review considered to include studies on a) adult patients with b) melanoma or non-small cell lung cancer c) undergoing FDA approved immunotherapy including anti-PD1, anti-PD-L1 or anti-CTLA4, d) comedication with NSAIDs and e) available sufficient patients’ outcome data to calculate odds ratios for overall response rates or hazard ratios for progression-free and overall survival. Patients were not excluded when receiving concomitant chemo- and/or radiotherapy. Included studies report time of overall survival (OS), time of progressions-free survival (PFS) and overall response rates (ORR; defined as complete responses and partial responses divided by patient population). All studies published since 1 January 2011 (FDA-approval of first immunotherapy, i.e. ipilimumab) were included. Survival data was reported as univariate or multivariate hazard ratio, if both available, multivariate analysis was prioritized. Odds ratios and hazard ratios with 95% confidence interval for overall response rates, progression-free and overall survival from included studies were utilized to calculate the pooled odds and hazard ratios. The heterogeneity of the pooled results was evaluated using Q test to assess between-study heterogeneity and quantified by the Higgins I2 test. If p was < 0.10 for the Q-test or I2 was >50%, significant heterogeneity was assumed, and the random-effects model was employed to summarize the data. Statistical analysis was performed using R software (version 4.3.2) with “meta” (General Package for Meta-Analysis, version 7.0-0).
Statistical analysis and reproducibility
Statistical analyses were performed using Graphpad Prism v9.1.2 or newer and Microsoft Excel v16.88. Normality of the data distribution was calculated using a D’agostino & Pearson test or Shapiro-Wilk test. The number of samples (n) used per experiment and the statistical test used are indicated in the figure legends. All in vitro and in vivo experiments were repeated at least twice and always with multiple replicates, except for the following experiments which were performed only once: scRNA-seq involving pharmacological treatment of the YUMM1.7 RTT model, intratumoral injection of T cells and the YUMM3.3 model. IF stainings for which representative images are shown were repeated at least twice, except for the NTT in Batf3-/- and Nur77 reporter experiment which was performed once but with n=3 tumors and was also confirmed with flow cytometry. Pharmacological combination treatments of the KPAR model were performed once. No statistical methods were used to determine sample size for in vivo experiments, numbers were chosen based on previous preliminary experiments. Scientists were not blinded to experimental groups, and experiments were repeated by different investigators. Mice were randomly assigned to treatment groups based on tumor size at the day of treatment start or randomly allocated across separate cages when treatment had to be started at day 3. p-values < 0.05 were considered significant.
Extended Data
Extended Data Fig. 1. Characterization of immune permissive versus suppressive TMEs in mouse melanoma models.
a, Representative images of bioluminescent live imaging (BLI) of T cell infiltration after adoptive CD8+ OT-1Luc T cell transfer (ACT) in YUMM1.7OVA NTT and RTT tumors. b, Scheme outlining the in vitro killing of NTT and RTT cells with activated CD8+ OT-1 T cells (1:1 ratio) (left), histograms (flow cytometry) displaying MHCI levels of NTT and RTT cells (middle) and normalized ratio of 50:50 NTT/RTT cells mixed cultures in an in vitro killing assay with CD8+ OT-1 T cells. Normalization was performed with NTT and RTT percentages from the same 50:50 mixed cultures without CD8+ OT-1 T cells to account for proliferation differences (n=3 biological replicates) (right). Two-tailed unpaired Student’s t-test. c, UMAP of scRNA-seq of CD45+ cells of YUMM1.7OVA NTT and RTT tumors in Rag2-/- before ACT (n=3 tumors pooled/group). d, Relative frequency of cell types across conditions in YUMM1.7OVA tumors post-ACT (n=4 tumors pooled/group). e, Expression of myeloid cell markers in each cluster (scRNA-seq). f, Heatmap of scaled gene expression (scRNA-seq) across cell clusters between NTT and RTT YUMM1.7OVA tumors post-ACT. g, Response to immune checkpoint blockade (ICB, anti-PD1/CTLA-4) of YUMM3.3 NTT and RTT tumors (n=6 NTT, n=7 NTT + ICB, n=8 RTT, n=9 RTT+ ICB). Two-way ANOVA with Tukey’s multiple comparison. h, UMAP of scRNA-seq of CD45+ cells of YUMM3.3 NTT and RTT (n=1 tumor/group). i, Relative frequency of cell types across conditions. j, Heatmap of scaled gene expression (scRNA-seq) across cell clusters between YUMM3.3 NTT and RTT. Bar graphs and growth curves depict the mean ± s.e.m. ns=not significant.
Extended Data Fig. 2. Permissive TMEs act as local reservoirs for T cell expansion.
a, Representative immunofluorescence (IF) of YUMM1.7OVA NTT and RTT tumors 48 hours post-ACT(n=3 tumors/group). Scale bars depicted in image. b, Quantification of immune populations based on IF staining (n=3 tumors/group). c, Quantification of relative T cell frequency and distance to the tumor periphery in NTT and RTT tumors. Percentages of T cells found in the tumor border or tumor center are depicted (n=3 tumors per condition). d, Quantification of Nur77+ OT-1 cells in YUMM1.7OVA NTT and RTT tumors 48 hours post-ACT (n=6 tumors for NTT, n=5 for RTT). e, Average distances between Nur77+ OT-1 cells and cDC1s or monocytes based on IF stainings (n=6 tumors for NTT, n=5 for RTT). f, Quantification of CD8+ OT-1Luc T cell infiltration by BLI post-ACT in YUMM1.7OVA NTT and RTT in Rag2-/- mice treated with FTY720 (n=5 mice/group). g, Quantification of T cell proliferation by CFSE dilution via flow cytometry 72 hours post intratumoral (i.tu.) T cell injection into NTT and RTT tumors in Rag2-/- (n=5 mice/group). h, Relative frequencies of T cell phenotypes (flow cytometry) after activation in vitro prior to ACT (n=1, experiment repeated 3 times). i, Gating strategy for the phenotypic characterization of T cells. j, Relative frequencies of CD8+ T cell populations based on defined T cell signatures29 of i.tu. injected T cells isolated and subjected to scRNA-seq 5 days post-transfer (n=14 NTT, n=18 RTT pooled tumors). k, Projection of a progenitor exhausted T cell signature score29 and Cxcr6 gene expression on scRNA-seq from intratumoral T cells. l, Scheme outlining the contralateral (CL) injection of tumors in opposite flanks of the same mouse followed by intravenous (i.v) or i.tu. ACT in one of the tumors. m, Growth curves depicting the response to i.v ACT of primary or CL tumor in the same mouse (n=4 mice NTT/NTT, n=3 mice RTT/RTT and n=6 mice NTT/RTT). n, Quantification of T cell infiltration by BLI in mice harboring CL NTT/NTT tumors (n=6 mice), RTT/RTT tumors (n=3 mice) or an NTT and RTT tumor (n=10 mice) after iv ACT. o, Representative BLI images from Extended Data Fig. 2m, n. p, Flow cytometry characterization of the TME of tumors in the CL setting (n=3 mice NTT/NTT, n=3 mice RTT/RTT, n=5 mice NTT/RTT tumors). q, Quantification of T cell infiltration by BLI post-ACT in NTT and RTT in Rag2-/- mice treated with FTY720 (n=7 mice/group). r, Response to ACT of mice bearing CL tumors (NTT/RTT) and treated with FTY720 or untreated (CTRL), (n=7 mice/group). s, Response to ACT of Rag2-/- mice bearing CL tumors (NTT/NTT) with one of the NTT tumors injected i.tu (n=6 mice). t, Response to ACT of Rag2-/- mice bearing CL tumors (NTT/RTT) with RTT tumors injected i.tu (n=6 mice). u, Representative BLI images of T cell infiltration in mice harboring NTT/NTT, RTT/ RTT or RTT/ NTT tumors, with one of the tumors receiving i.tu. T cell transfer (n=6 mice). Bar graphs and growth curves depict the mean ± s.e.m. Error bars in s and t not visible due to synchronous tumor regression. Statistical analysis was performed with a two-tailed unpaired student’s t-test in b, c, d, and g. A one-way ANOVA with Tukey’s multiple comparisons test in f, n, p and q. Two-way ANOVA with Tukey’s multiple comparisons test in m and r on day 17 pi. ns=not significant. Arrow indicates day of ACT.
Extended Data Fig. 3. Inflammatory monocytes cross-dress with cancer-derived pMHCI complexes.
a, Scheme outlining the dendritic cell (DC) vaccination of YUMM1.7OVA RTT tumors with bone marrow-derived cDC1s (BM-DCs) injected i.tu. and treated with ACT. b, Left, representative BLI images and right, quantification of T cell infiltration by BLI (n=6 mice CTRL and n=7 mice for DC vaccination). c, Response to ACT of RTT tumors vaccinated with BM-DCs followed by ACT. Black arrow indicates the day of ACT and gray arrows the days of cDC1 vaccination (n=6 mice CTRL and n=7 mice for DC vaccination). d, Representative IF staining of YUMM1.7OVA NTT and RTT tumors in Batf3-/- Rag2-/- mice, 48 hours post ACT, scale bar in NTT=500 μm, zoom-ins=20 μm and 10μm, scale bar in RTT=1000 μm, (n=3 tumors/group). e, Quantification of relative T cell frequency and distance to the tumor periphery in NTT and RTT tumors injected in Rag2-/- Batf3-/- mice. Percentages of T cells found in the tumor border or tumor center are depicted (n=3 tumors/group). f, Quantification of relative T cell frequency and distance to the next immune cell in NTT tumors in Rag2-/- Batf3-/- mice (n=3 tumors). g, Scheme outlining the depletion of dendritic cells with diphtheria toxin (DT) in NTT tumors in Rag2-/- mice reconstituted with bone marrow from Rag2-/- zDC-DTR mice. DT was administered every 3 days starting on the day of tumor injection for the duration of the experiment. h, Contour plots depicting the frequency of dendritic cells (left) and flow quantification (flow cytometry) (n=3 tumors/group). i, Representative BLI images (right) and quantification of T cell infiltration (n=5 mice for untreated, n=6 DT treated) (left). j, Scoring of a monocyte derived dendritic cell (MonoDC) signature21 in the YUMM1.7OVA scRNA-seq. k, Scoring of the Bosteels et al.40 ISG signature (Supplementary Table 2) in the YUMM1.7OVA scRNA-seq. l, Quantification of Nur77+ or Nur77- OT-1 T cells interacting with Ly6c+ monocytes based on IF staining of NTT tumors 48 hours post-ACT (n=6 tumors). m, Median fluorescence intensity (MFI) quantification of SIINFEKL-H2Kb staining (n=6 mice/group) (left) and histograms (right). n, MFI quantification of NTT-derived H2-Kb on BALB/c pMHC-I cross-dressed monocytes (n=6 mice/group) in NTT and NTT B2m KO. o, Scheme of the process of cross-dressing and in vitro T cell activation (left) and quantification of OT-1 T cell proliferation after 72 hours of co-culture with monocytes FACS-sorted from YUMM1.7OVA NTT tumors in BALB/c mice (n=3 technical replicates). Bar graphs depict the mean ± s.e.m. Statistical analysis was performed with a two-tailed unpaired student’s t-test in b, e, h, i, l, m, n and o. ns=not significant.
Extended Data Fig. 4. Inflammatory monocytes correspond to ISG+ and CXCL9+ macrophages in human data-sets.
a, Inflammatory monocyte and Monocyte 1 gene signature projection on a UMAP, and enrichment scores (ES) calculated with Gene Set Variance Analysis (GSVA) for different myeloid populations in a human melanoma scRNA-seq myeloid data-set (MEL) (Cheng et al.23). b, Inflammatory monocyte and Monocyte 1 gene signature projection on a UMAP, and ES calculated with GSVA for different myeloid populations, in human non-small cell lung cancer (NSCLC) scRNA-seq myeloid data-set (Cheng et al.23). c, Inflammatory monocyte and Monocyte 1 gene signature projection on a UMAP, and ES calculated with GSVA for different myeloid populations, in human melanoma scRNA-seq myeloid data-set (Barras et al.43). See also Supplementary Table 2.
Extended Data Fig. 5. Oncogenic MAPK signaling counter-regulates the production of PGE2 and IFN-I.
a, Targeted metabolomic analysis of eicosanoids in YUMM1.7OVA NTT and RTT tumors isolated at day 10 post-injection from Rag2-/- mice (n=3 tumors/group). b, Tumor growth of non-ACT RTT CTRL and RTT Ptgs1/2 KO (n=7 mice/group) in Rag2-/- mice. c, Scheme outlining the injection of YUMM1.7OVA RTT Ptgs2 KO into Rag2-/- mice followed by ACT. d, Quantification of PGE2 by ELISA from YUMM1.7OVA RTT Ptgs2 KO tumors isolated at day 10 post-injection (n=4 for RTT CTRL and n=6 for RTT Ptgs2 KO, over 2 independent experiments). e, Survival of Rag2-/- mice harboringYUMM1.7OVA RTT CTRL or RTT Ptgs2 KO tumors upon ACT treatment (n=5 mice/group). f, Quantification of PGE2 by ELISA from YUMM3.3 NTT and RTT tumors (n=2 biological replicates). g, Tumor growth of YUMM3.3 RTT CTRL and RTT Ptgs2 KO (n=5 mice/group) in Rag2-/- mice. h, Quantification of IFN-β by ELISA from YUMM1.7OVA NTT, RTT and RTT IRF3/7 tumors grown in Rag2-/- mice normalized to tumor weight (n=4 tumors/group, pooled from 2 independent experiments). i, Upstream regulator analysis (Ingenuity) of differentially expressed genes in cancer cells sorted from RTT vs. NTT YUMM1.7OVA tumors grown in Rag2-/- mice. Benjamini-Hochberg correction for multiple testing. j, Tumor growth of non-ACT treated YUMM1.7OVA RTT CTRL (n=5 mice) and RTT IRF3/7 (n=4 mice) in Rag2-/- mice. k, Quantification of IFN-β by ELISA from YUMM3.3 NTT and RTT tumors grown in C57BL/6 mice normalized to tumor weight (n=3 tumors/group). l, Heatmap of scaled ISG expression in YUMM1.7OVA RTT upon treatment with MEK inhibitor (MEKi) for 48 hours. m, Western blot of RTT YUMM1.7OVA depicting COX2 protein levels upon treatment with RAF inhibitor (RAFi) or MEKi for 48 hours. For gel source data see Supplementary Fig. 1. Experiment repeated 2 independent times. n, Heatmap of scaled gene expression in YUMM1.7OVA RTT cancer cells upon treatment with MEKi in vivo (n=5 tumors per condition). o, ISG expression in YUMM1.7OVA RTT Ptgs2 KO cell line compared to NTT and RTT CTRL by RT-qPCR (n=4 technical replicates). All expression values are depicted as the log2FC of the expression of each gene compared to NTT. p, Quantification of IFN-β by ELISA from YUMM1.7OVA RTT CTRL and RTT Ptgs2 KO tumors grown in Rag2-/- mice normalized to tumor weight (n=6 tumors/group). q, Quantification of PGE2 by ELISA from YUMM1.7OVA RTT CTRL and RTT IRF3/7 tumors grown in Rag2-/- mice (n=3 tumors/group). r, Quantification of IFN-β and PGE2 by ELISA from NTT and RTT variants of M249, LOX and H358 cell lines. For IFN-β analysis, tumors were grown NSG mice and normalized to tumor tumor weight (n=3 tumors/group). For PGE2, supernatants from in vitro cell lines were analyzed (n=2 biological replicates/group). s, Western blot of RTT, NTT and NTT NRAS variants of A375 human melanoma cell lines depicting COX2 protein levels upon treatment with RAFi for 48 hours. For gel source data see Supplementary Fig. 1. Experiment performed once. Bar graphs depict the mean ± s.e.m. Statistical analysis was performed with a two-tailed student’s t-test in a, d, k, p, q and r and one-way ANOVA with Tukey’s multiple comparison in h. p-value in e was calculated using a Log-rank Mantel Cox test. ns=not significant. ND=Not detected.
Extended Data Fig. 6. High PGE2 and low IFN-I instruct an immune evasive TME.
a, Gating strategy for the identification of intratumoral myeloid populations. b, Flow cytometry myeloid characterization pre and 72 hours post-ACT (n=6 for RTT ± ACT, IRF37 RTT ± ACT, n=5 for NTT ± ACT, RTT Ptgs2 KO + ACT, n=4 RTT Ptgs2 KO). c, Flow cytometry characterization of dendritic cell populations pre and 72 hours post-ACT, (n=5 for NTT, RTT Ptgs2 KO ± ACT, IRF37 RTT + ACT, n=6 for NTT + ACT, RTT ± ACT, IRF37 RTT). d, Relative frequency of cell types across conditions in YUMM1.7OVA (scRNA-seq), for RTT CTRL the RTT mCherry sample was used. e, Heatmap of scaled gene expression (scRNA-seq) between NTT, RTT CTRL, RTT Ptgs1/2 KO and RTT IRF3/7 for individual cell clusters. f, Flow cytometry characterization of SIINFEKL peptide on MHCI of dendritic cells isolated from tumors and pulsed ex vivo with SIINFEKL peptide (n=6 tumors/group) and MFI quantification. g, Scheme outlining in vivo NK cell depletion in Rag2-/- mice harboring YUMM1.7OVA RTT Ptgs2 KO tumors. h, Representative plot depicting NK1.1+ cells in NK cell-depleted vs CTRL RTT Ptgs2 KO tumors measured by flow cytometry (left) and quantification of cDCs (n=4 CTRL and n=3 anti-NK1.1 treated tumors) (right). i, Representative BLI images (left) and quantification of CD8+ OT-1Luc T cell infiltration by BLI (right), (n=3 mice for NTT, n=5 mice for all other groups). j, Tumor response to ACT (n=5 mice/group). Arrow indicates day of ACT. Bar graphs and growth curves depict the mean ± s.e.m. Data in i depicts the mean ± s.e.m. Statistical analysis was performed with a two-tailed unpaired student’s t-test in h. A one-way ANOVA with Tukey’s multiple comparison in f. Two-way ANOVA with Tukey’s multiple comparison in i. ns=not significant.
Extended Data Fig. 7. Type I and type II IFNs determine the inflammatory state.
a, b, Overall survival of melanoma patients stratified according to the TME-COX and TME-IRF3/7 signatures (Supplementary Table 4) (left) and correlation of intratumoral CD8+ T cell signature (right) in the Gide et al. RNA-seq data-set49. PCC=Pearson correlation coefficient. c, TME-COX and TME-IRF3/7 signatures projected on the myeloid UMAP from baseline tumors (split for responder and non-responders to TIL therapy) from the Barras et al.43 patient cohort (above) and individual score across different myeloid populations (below). d, Gene set enrichment analysis (GSEA) of NTT and RTT tumors pre and post-ACT based on scRNA-seq analysis. Normalized enrichment score (NES). Multiple correction testing with false discovery rate. e, Pathway enrichment analysis of the top variable genes in the immune tumor microenvironment (TME) from NTT and RTT tumors pre and post-ACT analyzed by scRNA-seq. 1, 2 and 3 depict each gene cluster identified. Benjamini-Hochberg correction for multiple testing. f, TME characterization of YUMM1.7OVA RTT Ptgs2 KO tumors pre and 72 hours post-ACT upon αIFN-γ and αIFNAR1 treatment (n=5 tumors/group, except n=4 for isotype and αIFNAR + ACT). g, Trajectory analysis and pseudotime of the MoMac compartment of YUMM1.7OVA NTT and RTT tumors. h, Density plots of F4/80+ macrophages after exposure of BM-derived monocytes to PGE2 for 48 hours (left) and quantification (right) (n=3 replicates per condition). i, Scheme outlining the intratumoral (i.tu.) transfer of CD45.1+ BM-derived monocytes into NTT and RTT tumors injected into CD45.2+ Rag2-/- mice (left) and inflammatory monocytes frequencies 3 days after transfer (right) (n=5 tumors/group). j, Scheme outlining the knockout of EP2 (Ptger2) and EP4 (Ptger4) in OT-1 T cells and i.v. injection into YUMM1.7OVA RTT tumor-bearing Rag2-/- mice (left) and tumor growth (n=6 mice/group) (right). Arrow indicates the day of ACT. k, Expression of Ptger2 and Ptger4 across populations from the YUMM1.7OVA scRNA-seq. Bar graphs depict the mean ± s.e.m. Statistical analysis was performed with a two-tailed unpaired student’s t-test in h and i. p-value in a and b was calculated using a Cox’s proportional hazards model (survival) and with a two-sided Pearson’s correlation (correlations).
Extended Data Fig. 8. PGE2 suppresses myeloid responsiveness to IFN-I.
a, Tumor growth of YUMM1.7 (left) and YUMM3.3 (right) RTT in Itgax-Cre (CTRL) (n=7 mice for YUMM3.3 and n=8 for YUMM1.7) or CD11cCre(Itgax-Cre)/Ptger2-/-/Ptger4fl/fl treated with anti-CD8 to deplete CD8+ T cells (n=5 mice for YUMM3.3 and n=6 for YUMM1.7) or isotype control (n=7 mice for YUMM3.3 and n=10 for YUMM1.7). b, TME characterization (flow cytometry) of RTT tumors in CD11cCre-Ptger2-/-Ptger4fl/fl mice (KOEP2/EP4) or CD11cCre control mice (CTRL) (n=8 tumors/group). c, Quantification of Ly6A+ cells after exposing BM-derived Ly6C+ monocytes to conditioned media (CM) from cancer cell lines ± COX1/2 inhibitor (COX1/2i, indomethacin) during media conditioning ± αΙFNAR1 during 48 hours of culture (n=3 biological replicates per condition). d, MFI quantification of AXL, MHC-I and CD86 (flow cytometry) of BM-DCs treated with fresh media (C) or CM from NTT, RTT and RTT IRF3/7 cells for 24h ± αΙFNAR1 or isotype control (n=3 biological replicates per condition). e, Heatmap of scaled expression of ISGs in BM-DCs exposed to CM from RTT IRF3/7 treated with COX1/2i (indomethacin) or untreated, in the presence of αΙFNAR1 or isotype control measured by RT-qPCR (n=4 technical replicates). f, Heatmap of scaled ISG expression in BM-DCs exposed to CM from NTT and RTT treated with MEKi or untreated, in the presence of αΙFNAR1 or isotype control (n=4 technical replicates) measured by RT-qPCR. g, Scheme outlining the culture of human monocyte models to CM from cancer cell lines ± COX1/2i during media conditioning (left) and heatmaps of scaled ISG expression (right). h, Quantification of AXL+ BM-DCs treated with IFN-β and PGE2 (flow cytometry) (left) (n=3 biological replicates) and heatmap of scaled ISG expression measured by RT-qPCR (right) (n=4 technical replicates). Bar graphs and growth curves depict the mean ± s.e.m. Statistical analysis were performed with a two-way ANOVA with Tukey’s multiple comparisons test in a, a two-tailed unpaired student’s t-test (APCs, cDC1 and infl.monocytes) and two-tailed Mann Whitney U (total cDCs and infl.cDC2s) in b, one-way ANOVA with Tukey’s multiple comparison in c, d and h. ns=not significant.
Extended Data Figure 9. Meta-analysis of retrospective clinical studies.
a-c, Forest plot of pooled odds ratios (ORs) and hazard ratios (HRs) with 95% confidence intervals (CI) across included studies of patients receiving ICB with and without concomitant non-steroidal anti-inflammatory drugs (NSAIDs) medication. Number of participants are included in each figure panel. a, Odds ratios comparing overall response rates. b, Hazard ratios comparing progression-free survival. c, Hazard ratios comparing overall survival. I2: I-squared; P: probability. d, PRISMA flow diagram of literature search and study selection process for meta-analysis. Statistical analysis was performed with a random effects model, data are presented as mean values ± 95% confidence interval. See also Supplementary Tables 5 and 6.
Extended Data Figure 10. Pharmacological inhibition of PGE2 and type I IFN modulators resensitize cross-resistant tumors to immunotherapy.
a, Relative frequency of each cell type across conditions (scRNA-seq). b, Scoring of the Duong et al. inflammatory signature37 in RTT COX2i-treated tumors. c, Heatmap of scaled gene expression (scRNA-seq) in YUMM1.7 OVA RTT vehicle-treated tumors (CTRL) and RTT celecoxib (COX2i)-treated tumors (n=3 tumors were pooled/group). d, Response of YUMM1.7OVA RTT tumors to vehicle + adoptive T cell transfer (ACT) (n=7 mice), celecoxib (n=4 mice), celecoxib + ACT (n=6 mice) and etoricoxib ± ACT (n=7 mice/group). Red arrow and box indicate COX2i treatment stop. e, UMAP of scRNA-seq of CD45+ cells from YUMM1.7OVA RTT mice treated with vehicle or COX2i+5-AZA isolated 72 hours post-ACT (n=3 tumors pooled/group). f, Relative frequency of each cell type across conditions in e (scRNA-seq). g, Scoring of the Duong et al. inflammatory signature37 in COX2i+5-AZA treated tumors. h, Response of YUMM1.7OVA RTT tumors to 5-azacytidine (5-AZA) (n=4 mice), 5-AZA + ACT (n=7 mice), vehicle+ACT (n=8 mice), Flt3L+ACT (n=7 mice), COX2i+ACT (n=9 mice), COX2i+5-AZA+ACT (n=7 mice) and COX2i+Flt3L+ACT (n=8 mice). i, Representative image of IF staining of vehicle-treated and COX2i+Flt3L RTT YUMM1.7OVA tumors 96 hours post-ACT (n=2 tumors per condition). Scale bar=1000μm, zoom-in=50μm. Black arrows indicates day of ACT.
Extended Data Figure 11. Combination therapies in melanoma, lung, pancreatic and colorectal murine cancer models.
a, Scheme outlining the re-injection of YUMM1.7OVA RTT cells into tumor-free responder mice from Fig. 5e (left) and tumor growth of re-challenged mice (n=4 mice for COX2i + Ftl3L, n=3 COX2i and n=2 COX2i+ 5-AZA) or naive control mice (n=5 mice). b, Scheme outlining the injection of YUMM3.3 RTT tumors into C57BL/6 mice and the treatment regimen with COX2i, Flt3L, 5-AZA and anti-PD1/CTLA-4 (ICB) or isotype control. c, Response to control vehicle treatment or vehicle + ICB treatment of YUMM3.3 RTT tumors (n=6 mice). d, Response to single treatments and ICB combination treatments of YUMM3.3 RTT tumors. COX2i, Flt3L, 5-AZA+ICB, COX2i+ICB and Flt3L+ICB (n=8 mice), COX2i+5-AZA+ICB and COX2i+Flt3L+ICB (n=10 mice). e, Survival plot. f, Scheme outlining the injection of CT-26 RTT tumors into BALB/c mice and the treatment regimen with COX2i, Flt3L, 5-AZA and anti-PD1 (ICB) or isotype control. g, Response to treatment of CT26 RTT tumors. Vehicle and Vehicle+ICB (n=8 mice), COX2i+5-AZA+ICB and COX2i+Flt3L+ICB (n=9 mice). h, Survival plot. i, Scheme outlining the intrapancreatic injection of EPP2Luc cells into C57BL/6 mice and the treatment regimen with COX2i, Flt3L and anti-PD1 (ICB) or isotype control. j, Left, representative BLI images of EPP2Luc tumor burden and right, quantification of control (n=5 mice), anti-PD1 (n=6 mice), anti-PD1 + COX2i + Ftl3L (n=6 mice). k, Survival plot of j. l, Scheme outlining the injection of KPAR tumors into C57BL/6 mice and the treatment regimen with COX2i, Flt3L and anti-PD1 (ICB) or isotype control. m, Response to treatment of KPAR tumors, vehicle ± ICB (n=6 mice) and ICB + COX2i + Ftl3L (n=8 mice). n, Survival plot of m. o, Scheme summarizing the role of inflammatory monocytes in T cell restimulation and the functional convergence of PGE2 and IFN-I to determine an inflammatory TME, T cell restimulation and immunotherapy response. Bar graphs depict the mean ± s.e.m. Statistical analysis was performed with a one-way ANOVA with Tukey’s multiple comparisons test in j. Log-rank Mantel Cox test in h, k, n. ns=not significant.
Supplementary Material
Acknowledgements
We thank the members of A.C.O.’s laboratory and J.Z.’s laboratory (Research Institute of Molecular Pathology) for discussions and proof-reading the manuscript. P. S. Jung for the help with the summary scheme, S. Balay, E. Mancheño, and J. van der Veeken for critical reading of the manuscript, G. Jonsson and J. Penninger for providing Itgax-Cre mice, J. Moon (University of Michigan) for providing OT-1Luc Thy1.1 mice, V. Buchholz (TUM) for providing Nur77-GFP-OT-1 T cells and M. Bosenberg (Yale School of Medicine) for providing the parental YUMM1.7 and YUMM3.3 cell lines. Z. Zhang and S. Cheng for sharing the raw count matrix for their scRNA-seq analysis (Cheng et al.). M. Gaidt for sharing the BLaER-1 cell line. The BioOptics facility at the Vienna Biocenter for cell-sorting. We thank T. Grentzinger and the Next Generation Sequencing Facility at the Vienna Biocenter for the single-cell RNA sequencing. We also thank T. Köcher from the Metabolomics Facility at the Vienna Biocenter for the metabolomic analysis of eicosanoids. Finally, we want to thank M. Kostic for editing the manuscript. Research in the laboratory of A.C.O. is supported by Boehringer Ingelheim, the Austrian Research Promotion Agency (Headquarter grant FFG-852936), European Research Council (‘CombaTCancer’, grant no. 759590 and ‘UnlockIT’ grant no. 101125797) and the Vienna Science and Technology Fund (LS-16-063). M.V. is supported by the SNF postdoc mobility grant (grant no. 214359), the Marie Skłodowska–Curie postdoctoral fellowship program (grant no. 101108907) and the VIP2 postdoc program, which has received funding from the European Union’s Framework Programme for Research and Innovation Horizon 2020 (2014-2020) (Marie Curie Skłodowska Grant Agreement Nr. 847548). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska–Curie grant agreement No 955951. S.V. is supported by the Sigrid Jusélius Foundation. For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Footnotes
Author contributions
A.E., G.E., and A.C.O. conceived the study, designed the experiments and interpreted the results. A.C.O. supervised the study. A.E. and G.E. developed experimental tools, performed in vitro experiments, in vivo treatment studies, flow-cytometry analysis, gene expression profiling and parts of the computational analysis, and analyzed the data. F.B. and A.P. performed immunofluorescence stainings. L.H.H established parental cell lines and phenotypes, experimental design and expression profiling. M.N. and L.C. analyzed gene expression data, single-cell RNAseq results including the SCENIC and RNA velocity analyses. E.P., F.A., T.N., M.M. and N.F. contributed to the generation of experimental tools and in vivo and in vitro studies. M.S. performed the meta-analysis of patient data. I.K. performed western blotting and helped with mouse colony maintenance and genotyping. F.B., M.V., S.M.C., S.R., F.H. J.Z., and T.W. contributed to experimental design, generation of experimental tools, or in vivo experiments. G.C., D.D.L. and D.B. provided the TIL-therapy melanoma patient scRNA-seq data-set. G.J. and M.L., provided spatial CosMx data and analysis of human melanoma samples. J.P.B., provided EP2/EP4 KO mice and supervised immunofluorescence stainings and analysis. C.Q. and X.B. analyzed patient data. S.V. analyzed RNA-seq data and probed signatures derived from the scRNA-seq in melanoma patient datasets. A.E., G.E. and A.C.O. wrote the manuscript, with input from S.V., J.P.B., J.Z., and all authors read and approved it.
Competing interests declaration
A.C.O. and J.Z. labs received research support and funding from Boehringer Ingelheim. J.Z. is a founder and scientific advisor of Quantro Therapeutics. G.C. has received grants from Celgene, Boehringer-Ingelheim, Roche, BMS, Iovance Therapeutics, and Kite Pharma. The institution G.C. is affiliated with has received fees for his participation on an advisory board or for presentation at a company-sponsored symposium from Genentech, Roche, BMS, AstraZeneca, NextCure, Geneos Tx, and Sanofi/Avensis. G.C. has patents in the domain of antibodies and vaccines targeting the tumor vasculature as well as technologies related to T cell expansion and engineering for T cell therapy. G.C. has received royalties from the University of Pennsylvania regarding CAR T cell technology. G.C. is inventor on patent applications filed by the Ludwig Institute for Cancer Research Ltd pertaining to the subject matter disclosed herein. D.D.L. has received a grant from Hoffmann-La Roche AG. All other authors declare no conflicts of interest.
Data availability
Gene expression data from YUMM1.7 NTT and RTT cancer cells sorted from tumors was previously deposited under the Gene Expression Omnibus accession number GSE132443. Specifically samples from NTT tumors (GSM3864154,GSM3864155, GSM3864157) and RTT tumors (GSM3864170, GSM3864172,GSM3864173) were used to generate plots in Fig. 3a and Extended Data Fig. 5i. Raw and processed files of the single-cell RNA sequencing generated in this study have been deposited under the GEO accession number GSE241750. Expression data from the Gide et al. study is publicly available (accession no. PRJEB23709). Single cell RNA data from the Barras et al. study, including the Seurat object for the myeloid compartment was obtained upon direct request. Expression data from the Cheng et al. study was publicly available (GSE154763) and the raw count matrix was obtained upon request. Source data is provided for all graphs that are shown in the manuscript. All other formats of raw data are available upon request.
References
- 1.Binnewies M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer. 2018;18:139–147. doi: 10.1038/nrc.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Pilato M, et al. CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell. 2021;184:4512–4530.:e22. doi: 10.1016/j.cell.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magen A, et al. Intratumoral dendritic cell–CD4+ T helper cell niches enable CD8+ T cell differentiation following PD-1 blockade in hepatocellular carcinoma. Nat Med. 2023:1–11. doi: 10.1038/s41591-023-02345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen JH, et al. Human lung cancer harbors spatially organized stem-immunity hubs associated with response to immunotherapy. Nat Immunol. 2024;25:644–658. doi: 10.1038/s41590-024-01792-2. [DOI] [PubMed] [Google Scholar]
- 6.Duraiswamy J, et al. Myeloid antigen-presenting cell niches sustain antitumor T cells and license PD-1 blockade via CD28 costimulation. Cancer Cell. 2021 doi: 10.1016/j.ccell.2021.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jansen CS, et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature. 2019;576:465–470. doi: 10.1038/s41586-019-1836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meiser P, et al. A distinct stimulatory cDC1 subpopulation amplifies CD8+ T cell responses in tumors for protective anti-cancer immunity. Cancer Cell. 2023 doi: 10.1016/j.ccell.2023.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Bayerl F, et al. Tumor-derived prostaglandin E2 programs cDC1 dysfunction to impair intratumoral orchestration of anti-cancer T cell responses. Immunity. 2023;56:1341–1358.:e11. doi: 10.1016/j.immuni.2023.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Garris CS, et al. Successful Anti-PD-1 Cancer Immunotherapy Requires T Cell-Dendritic Cell Crosstalk Involving the Cytokines IFN-γ and IL-12. Immunity. 2018;49:1148–1161.:e7. doi: 10.1016/j.immuni.2018.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellman I, Chen DS, Powles T, Turley SJ. The cancer-immunity cycle: Indication, genotype, and immunotype. Immunity. 2023;56:2188–2205. doi: 10.1016/j.immuni.2023.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Prokhnevska N, et al. CD8+ T cell activation in cancer comprises an initial activation phase in lymph nodes followed by effector differentiation within the tumor. Immunity. 2023;56:107–124.:e5. doi: 10.1016/j.immuni.2022.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broz ML, et al. Dissecting the Tumor Myeloid Compartment Reveals Rare Activating Antigen-Presenting Cells Critical for T Cell Immunity. Cancer Cell. 2014;26:638–652. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wculek SK, et al. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol. 2020;20:7–24. doi: 10.1038/s41577-019-0210-z. [DOI] [PubMed] [Google Scholar]
- 15.Haas L, et al. Acquired resistance to anti-MAPK targeted therapy confers an immune-evasive tumor microenvironment and cross-resistance to immunotherapy in melanoma. Nature Cancer. 2021;2(7):693–708. doi: 10.1038/s43018-021-00221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seitter SJ, et al. Impact of Prior Treatment on the Efficacy of Adoptive Transfer of Tumor-Infiltrating Lymphocytes in Patients with Metastatic Melanoma. Clin Cancer Res. 2021;27:5289–5298. doi: 10.1158/1078-0432.CCR-21-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ascierto PA, et al. Sequencing of Ipilimumab Plus Nivolumab and Encorafenib Plus Binimetinib for Untreated BRAF-Mutated Metastatic Melanoma (SECOMBIT): A Randomized, Three-Arm, Open-Label Phase II Trial. J Clin Oncol. 2023;41:212–221. doi: 10.1200/JCO.21.02961. [DOI] [PubMed] [Google Scholar]
- 18.Atkins MB, et al. Combination Dabrafenib and Trametinib Versus Combination Nivolumab and Ipilimumab for Patients With Advanced BRAF-Mutant Melanoma: The DREAMseq Trial-ECOG-ACRIN EA6134. J Clin Oncol. 2022:JCO2201763. doi: 10.1200/JCO.22.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, et al. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell. 2019;179:829–845.:e20. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Maier B, et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature. 2020;580:257–262. doi: 10.1038/s41586-020-2134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zilionis R, et al. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity. 2019;50:1317–1334.:e10. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nirschl CJ, et al. IFNγ-Dependent Tissue-Immune Homeostasis Is Co-opted in the Tumor Microenvironment. Cell. 2017;170:127–141.:e15. doi: 10.1016/j.cell.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng S, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184:792–809.:e23. doi: 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Bill R, et al. CXCL9:SPP1 macrophage polarity identifies a network of cellular programs that control human cancers. Science. 2023;381:515–524. doi: 10.1126/science.ade2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siddiqui I, et al. Intratumoral Tcf1+PD-1+CD8+ T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity. 2019;50:195–211.:e10. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 26.Krishna S, et al. Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science. 2020;370:1328–1334. doi: 10.1126/science.abb9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurtulus S, et al. Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1-CD8+ Tumor-Infiltrating T Cells. Immunity. 2019;50:181–194.:e6. doi: 10.1016/j.immuni.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller BC, et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20:326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreatta M, et al. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat Commun. 2021;12:2965. doi: 10.1038/s41467-021-23324-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lacher SB, et al. PGE2 limits effector expansion of tumour-infiltrating stem-like CD8+ T cells. Nature. 2024:1–9. doi: 10.1038/s41586-024-07254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salmon H, et al. Expansion and Activation of CD103(+) Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity. 2016;44:924–938. doi: 10.1016/j.immuni.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spranger S, Dai D, Horton B, Gajewski TF. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell. 2017;31:711. doi: 10.1016/j.ccell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Böttcher JP, et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 2018;172:1022–1037.:e14. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammerich L, et al. Systemic clinical tumor regressions and potentiation of PD1 blockade with in situ vaccination. Nat Med. 2019;25:814–824. doi: 10.1038/s41591-019-0410-x. [DOI] [PubMed] [Google Scholar]
- 36.Gungabeesoon J, et al. A neutrophil response linked to tumor control in immunotherapy. Cell. 2023;186:1448–1464.:e20. doi: 10.1016/j.cell.2023.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duong E, et al. Type I interferon activates MHC class I-dressed CD11b+ conventional dendritic cells to promote protective anti-tumor CD8+ T cell immunity. Immunity. 2021 doi: 10.1016/j.immuni.2021.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwart D, et al. Cancer cell-derived type I interferons instruct tumor monocyte polarization. Cell Rep. 2022;41:111769. doi: 10.1016/j.celrep.2022.111769. [DOI] [PubMed] [Google Scholar]
- 39.Tadepalli S, et al. Rapid recruitment and IFN-I-mediated activation of monocytes dictate focal radiotherapy efficacy. Sci Immunol. 2023;8:eadd7446. doi: 10.1126/sciimmunol.add7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosteels C, et al. Inflammatory Type 2 cDCs Acquire Features of cDC1s and Macrophages to Orchestrate Immunity to Respiratory Virus Infection. Immunity. 2020;52:1039–1056.:e9. doi: 10.1016/j.immuni.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakim LM, Bevan MJ. Cross-dressed dendritic cells drive memory CD8+ T-cell activation after viral infection. Nature. 2011;471:629–632. doi: 10.1038/nature09863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacNabb BW, et al. Dendritic cells can prime anti-tumor CD8+ T cell responses through major histocompatibility complex cross-dressing. Immunity. 2022;55:982–997.:e8. doi: 10.1016/j.immuni.2022.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barras D, et al. Response to tumor-infiltrating lymphocyte adoptive therapy is associated with preexisting CD8+ T-myeloid cell networks in melanoma. Sci Immunol. 2024;9:eadg7995. doi: 10.1126/sciimmunol.adg7995. [DOI] [PubMed] [Google Scholar]
- 44.Zelenay S, et al. Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell. 2015;162:1257–1270. doi: 10.1016/j.cell.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caronni N, et al. IL-1β+ macrophages fuel pathogenic inflammation in pancreatic cancer. Nature. 2023;623:415–422. doi: 10.1038/s41586-023-06685-2. [DOI] [PubMed] [Google Scholar]
- 46.Bonavita E, et al. Antagonistic Inflammatory Phenotypes Dictate Tumor Fate and Response to Immune Checkpoint Blockade. Immunity. 2020;53:1215–1229.:e8. doi: 10.1016/j.immuni.2020.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muthalagu N, et al. Repression of the Type I Interferon Pathway Underlies MYC- and KRAS-Dependent Evasion of NK and B Cells in Pancreatic Ductal Adenocarcinoma. Cancer Discov. 2020;10:872–887. doi: 10.1158/2159-8290.CD-19-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obenauf AC, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520:368–372. doi: 10.1038/nature14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gide TN, et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell. 2019;35:238–255.:e6. doi: 10.1016/j.ccell.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Morotti M, et al. PGE2 inhibits TIL expansion by disrupting IL-2 signalling and mitochondrial function. Nature. 2024:1–9. doi: 10.1038/s41586-024-07352-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S-J, et al. Effect of cyclo-oxygenase inhibitor use during checkpoint blockade immunotherapy in patients with metastatic melanoma and non-small cell lung cancer. J Immunother Cancer. 2020;8 doi: 10.1136/jitc-2020-000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang DY, et al. The Impact of Nonsteroidal Anti-Inflammatory Drugs, Beta Blockers, and Metformin on the Efficacy of Anti-PD-1 Therapy in Advanced Melanoma. Oncologist. 2020;25:e602–e605. doi: 10.1634/theoncologist.2019-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pelly VS, et al. Anti-Inflammatory Drugs Remodel the Tumor Immune Environment to Enhance Immune Checkpoint Blockade Efficacy. Cancer Discov. 2021;11:2602–2619. doi: 10.1158/2159-8290.CD-20-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiappinelli KB, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barry KC, et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med. 2018;24:1178–1191. doi: 10.1038/s41591-018-0085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Elsas MJ, et al. Immunotherapy-activated T cells recruit and skew late-stage activated M1-like macrophages that are critical for therapeutic efficacy. Cancer Cell. 2024;42:1032–1050.:e10. doi: 10.1016/j.ccell.2024.04.011. [DOI] [PubMed] [Google Scholar]
- 57.Espinosa-Carrasco G, et al. Intratumoral immune triads are required for immunotherapy-mediated elimination of solid tumors. Cancer Cell. 2024;42:1202–1216.:e8. doi: 10.1016/j.ccell.2024.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kruse B, et al. CD4+ T cell-induced inflammatory cell death controls immune-evasive tumours. Nature. 2023;618:1033–1040. doi: 10.1038/s41586-023-06199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacquelot N, et al. Sustained Type I interferon signaling as a mechanism of resistance to PD-1 blockade. Cell Res. 2019;29:846–861. doi: 10.1038/s41422-019-0224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cilenti F, et al. A PGE2-MEF2A axis enables context-dependent control of inflammatory gene expression. Immunity. 2021;54:1665–1682.:e14. doi: 10.1016/j.immuni.2021.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meeth K, Wang JX, Micevic G, Damsky W, Bosenberg MW. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res. 2016;29:590–597. doi: 10.1111/pcmr.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boumelha J, et al. An Immunogenic Model of KRAS-Mutant Lung Cancer Enables Evaluation of Targeted Therapy and Immunotherapy Combinations. Cancer Res. 2022;82:3435–3448. doi: 10.1158/0008-5472.CAN-22-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rathert P, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Castle JC, et al. Immunomic, genomic and transcriptomic characterization of CT26 colorectal carcinoma. BMC Genomics. 2014;15:190. doi: 10.1186/1471-2164-15-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayer CT, et al. Selective and efficient generation of functional Batf3-dependent CD103+ dendritic cells from mouse bone marrow. Blood. 2014;124:3081–3091. doi: 10.1182/blood-2013-12-545772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gaidt MM, Rapino F, Graf T, Hornung V. Modeling Primary Human Monocytes with the Trans-Differentiation Cell Line BLaER1. Methods Mol Biol. 2018;1714:57–66. doi: 10.1007/978-1-4939-7519-8_4. [DOI] [PubMed] [Google Scholar]
- 68.Ochyl LJ, Moon JJ. Whole-animal imaging and flow cytometric techniques for analysis of antigen-specific CD8+ T cell responses after nanoparticle vaccination. J Vis Exp. 2015:e52771. doi: 10.3791/52771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Michlits G, et al. Multilayered VBC score predicts sgRNAs that efficiently generate loss-of-function alleles. Nat Methods. 2020;17:708–716. doi: 10.1038/s41592-020-0850-8. [DOI] [PubMed] [Google Scholar]
- 70.Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerhard GM, Bill R, Messemaker M, Klein AM, Pittet MJ. Tumor-infiltrating dendritic cell states are conserved across solid human cancers. J Exp Med. 2021;218 doi: 10.1084/jem.20200264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu Y, et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andreatta M, Carmona SJ. UCell: Robust and scalable single-cell gene signature scoring. Comput Struct Biotechnol J. 2021;19:3796–3798. doi: 10.1016/j.csbj.2021.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krämer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Page MJ, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data from YUMM1.7 NTT and RTT cancer cells sorted from tumors was previously deposited under the Gene Expression Omnibus accession number GSE132443. Specifically samples from NTT tumors (GSM3864154,GSM3864155, GSM3864157) and RTT tumors (GSM3864170, GSM3864172,GSM3864173) were used to generate plots in Fig. 3a and Extended Data Fig. 5i. Raw and processed files of the single-cell RNA sequencing generated in this study have been deposited under the GEO accession number GSE241750. Expression data from the Gide et al. study is publicly available (accession no. PRJEB23709). Single cell RNA data from the Barras et al. study, including the Seurat object for the myeloid compartment was obtained upon direct request. Expression data from the Cheng et al. study was publicly available (GSE154763) and the raw count matrix was obtained upon request. Source data is provided for all graphs that are shown in the manuscript. All other formats of raw data are available upon request.