Abstract
3’, 5’-cyclic adenosine monophosphate (cAMP) mediates the effects of sympathetic stimulation on the rate and strength of cardiac contraction. Beyond this pivotal role, in cardiac myocytes cAMP also orchestrates a diverse array of reactions to various stimuli. To ensure specificity of response, the cAMP signaling pathway is intricately organized into multiple, spatially confined, subcellular domains, each governing a distinct cellular function. In this review, we describe the molecular components of the cAMP signalling pathway with a specific focus on adenylyl cyclases, A kinase anchoring proteins and phosphodiesterases. We discuss how they are organized are inside the intracellular space and how they achieve exquisite regulation of signalling within nanometer-size domains. We delineate the key experimental findings that lead to the current model of compartmentalised cAMP signaling and we offer an overview of our present understanding of how cAMP nanodomains are structured and regulated within cardiac myocytes. Furthermore, we discuss how compartmentalized cAMP signaling is affected in cardiac disease and consider the potential therapeutic opportunities arising from understanding such organization. By exploiting the nuances of compartmentalized cAMP signaling, novel and more effective therapeutic strategies for managing cardiac conditions may emerge. Finally, we highlight the unresolved questions and hurdles that must be addressed to translate these insights into interventions that may benefit patients.
Keywords: 3’, 5’-cyclic adenosine monophosphate; protein kinase A; G protein coupled receptors; signalling compartmentalisation; targeted therapy
1. Introduction
3’, 5’-cyclic adenosine monophosphate (cAMP), the first identified second messenger (1), is a small, hydrophilic molecule found across a wide range of living organisms, from bacteria to humans. It operates within virtually all organs and systems, playing a crucial role in the regulation of bodily functions. Intracellular cAMP levels are regulated by ligand binding to G-protein coupled receptors (GPCRs) that either activate or inhibit synthesis of cAMP by adenylyl cyclases (AC) via coupling to Gαs- or Gαi-proteins, respectively (2). GPCRs, the largest family of cell-surface receptors, modulate a plethora of physiological processes in response to various chemical ligands, including hormones, neurotransmitters, lipids, odorants, and even non-chemical stimuli such as light and mechanical stress (3). They are implicated in the pathophysiology of numerous diseases (4) and are the primary target of approved therapeutics (5). cAMP exerts its influence by initiating a cascade of events that impact cellular function through interactions with various protein effectors, including protein kinase A (PKA), exchange proteins activated by cAMP (EPACs), cyclic nucleotide-gated ion (CNG) channels, hyperpolarization-activated cyclic nucleotide-gated (HCN) channels, and Popeye domain-containing (POPDC) proteins. The levels of cAMP are continuously modulated through degradation to adenosine monophosphate (AMP) by cAMP-hydrolyzing phosphodiesterases (PDEs). Metabolism (6), gene regulation (7), secretion (8), immune function (9), cardiac contraction and relaxation (10), cognition and memory (11) are only a few examples of processes where cAMP plays a role. Exploitation of the cAMP pathway forms the basis for treatments across a wide array of human diseases, including cancer, diabetes, heart failure, inflammation, neurological disorders, and mood disorders (12).
The role of cAMP as a ubiquitous second messenger generated in response to a multitude of extracellular stimuli raises the question of how it achieves specificity of response. Over the past 25 years, evidence has accumulated that demonstrates that specificity is achieved via compartmentalisation of cAMP signalling in subcellular nanodomains. Within these domains, PKA anchoring proteins (AKAPs) organize multiprotein complexes, or signalosomes, where PKA is brought in close proximity to a specific phosphorylation target and where multiple other signalling components can be assembled to locally regulate the amplitude, duration and functional effects of the cAMP signal.
Here we review our current understanding of the mechanisms responsible for generating the intricate cAMP signaling landscape within cardiac myocytes. We explore the remodelling of this spatial regulation that arise in the context of disease and the potential that cAMP signalling compartmentalisation offers for the development of targeted therapeutic interventions. Finally, we discuss the ongoing challenges and gaps in our knowledge that must be addressed to translate these insights into potential clinical advancements.
2. Components of the cAMP signalling pathway in cardiac myocytes
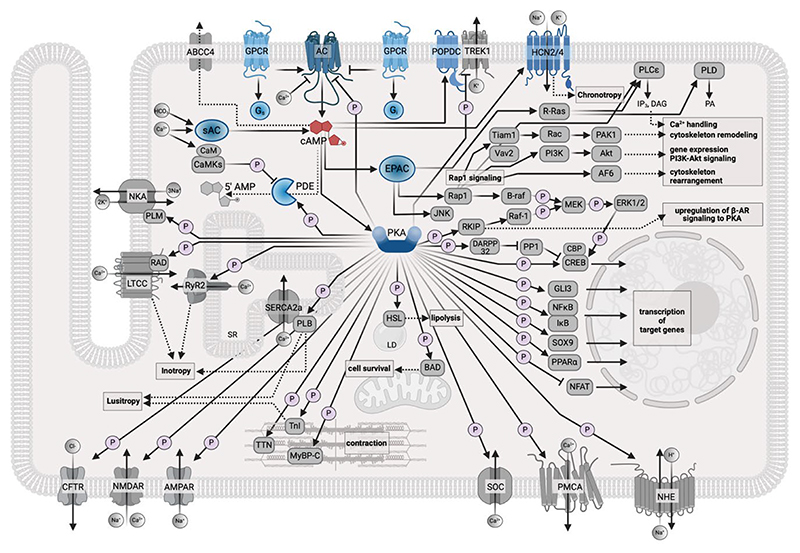
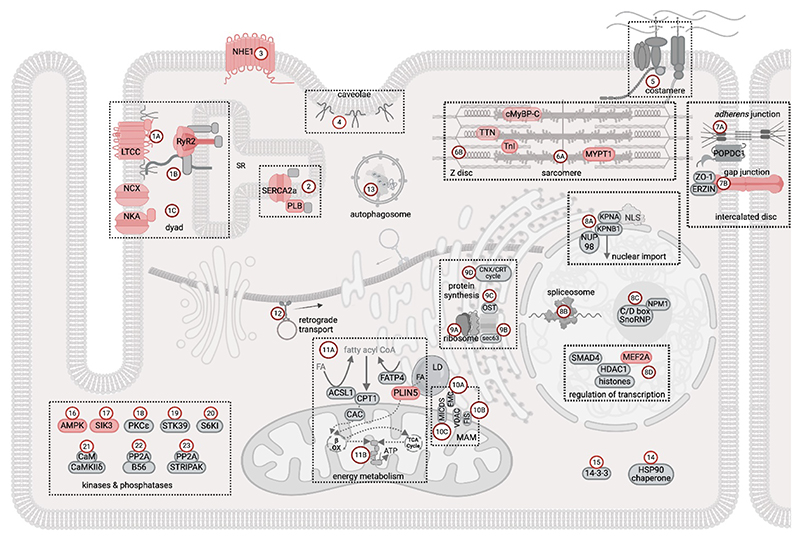
A representation of the cAMP signalling pathway map, based on the information currently available in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, is presented in Fig 1. As is evident from the ensuing discussion, while this annotation and similar ones provide a helpful overview of the cAMP pathway components, these canonical descriptions fall short in delineating the intricacies of the system and the significance of the subcellular spatial organization of this signalling network.
Figure 1. Map of the cAMP signalling pathway in cardiac myocytes.
The schematic shows the cAMP signalling map, as currently reported in the KEGG database, and adapted for a cardiac myocyte. The components of the pathway that are involved in the generation and degradation of cAMP and the cAMP effectors are shown in blue. Targets affected by cAMP signalling are shown in grey. The P symbol indicates phosphorylation. ABCC4, ATP binding cassette subfamily C member 4; AC, adenylyl cyclase; AF6, afadin; Akt, RAC serine/threonine-protein kinase; AMPAR, glutamatergicα-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor; B-raf, serine/threonine-protein kinase B-Raf; BAD, Bcl-2-associated death promoter; CaM, calmodulin; CaMKs, calcium/calmodulin-dependent protein kinases; CBP, E1A/CREB-binding protein; CFTR, cystic fibrosis transmembrane conductance regulator; CREB, cAMP-responsive element-binding protein 1; DAG, diacylglycerol; DARPP32, protein phosphatase 1 regulatory subunit 1B; EPAC, exchange protein directly activated by cAMP; ERK1/2, extracellular signaling-related kinase 1/2; Gi, inhibitory G protein; GLI3, zinc finger protein GLI3; GPCR, G protein-coupled receptor; Gs, stimulatory G protein; HCN2/4, hyperpolarization-activated cyclic nucleotide-gated potassium channel 2/4; HSL, hormone-sensitive lipase; I B, NF-kappa-B inhibitor alpha; IP3, inositol 1,4,5-trisphosphate; JNK, c-Jun N-terminal kinase; LTCC, L-type calcium channel; MEK, mitogen-activated protein kinase kinase 1; MyBPC, myosin binding protein C; NFAT, nuclear factor of activated T-cells, cytoplasmic 1; NFk B, nuclear factor NF-kappa-B p105 subunit; NHE, sodium/hydrogen exchanger; NKA, sodium/potassium-transporting ATPase; NMDAR, N-methyl-D-aspartate receptor; PA, phosphatidic acid; PAK1, p21-activated kinase 1; PDE, cyclic nucleotide phosphodiesterase ;PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; PKA, protein kinase A; PLB, phospholamban; PLCε, phospholipase C epsilon; PLD, phospholipase D; PLM, phospholemman; PMCA, plasma membrane calcium-ATPase; POPDC, popeye domain containing protein; PP1, serine/threonine-protein phosphatase PP1; PPARα, peroxisome proliferator-activated receptor alpha; R-Ras, Ras-related protein R-Ras; Rac, Ras-related C3 botulinum toxin substrate 1; RAD, Ras associated with diabetes; Raf-1, RAF proto-oncogene serine/threonine-protein kinase; Rap1, Ras-associated protein 1; RKIP; Raf kinase inhibitor protein; RyR2, ryanodine receptor 2; SERCA2a, sarco(endo)plasmic reticulum calcium-ATPase; SOC, store-operated calcium channel; SOX9, transcription factor SOX9; SR, Sarcoplasmic Reticulum; Tiam1, T-lymphoma invasion and metastasis-inducing protein 1; TnI, Troponin I; TREK1, TWIK-related potassium channel 1; TTN, Titin; Vav2, guanine nucleotide exchange factor VAV.
This figure was generated using Biorender.
Below, we delineate the main molecular components that are typically described as being associated with the cAMP signalling pathway in cardiac myocytes.
2.1. cAMP synthesis and degradation
2.1.1. GPCRs
GPCRs are seven-transmembrane receptors. They activate heterotrimeric G proteins, which consist of three subunits: α, β, and γ. Ligand binding to a GPCR stabilizes a conformation of the receptor that promotes the recruitment of G proteins to the plasma membrane. G proteins function as molecular switches in the transduction of intracellular signaling. When G proteins bind to GTP, they become active, and when bound to GDP, they assume an inactive state. When recruited to an activated GPCR, the exchange of GDP for GTP on the Gα subunit results in its activation. The GDP-GTP exchange causes the heterotrimeric G protein to dissociate into two units: the GTP-bound Gαsubunit and the dimeric Gβγ complex. On activation, Gαs subunits stimulate ACs to catalyse the synthesis of cAMP from ATP. In contrast, Gαi has an inhibitory effect on ACs, therefore dampening intracellular cAMP levels (13).
At least 25 different GPCR that signal via modulation of cAMP are expressed in cardiac myocytes (14). Among the Gαs-coupled receptors that stimulate cAMP production, the β-adrenergic receptors (β-ARs) are perhaps the most extensively studied because they regulate the cardiac response to sympathetic stimulation by catecholamines and are the target of drugs used for the treatment of heart failure (β-blockers). Other Gαs-coupled receptors expressed in the heart include the glucagon receptor, the glucagon-like peptide 1 receptors (GLP-1R), the prostaglandin EP4 receptors, the adenosine A2a receptor (A2R), each inducing distinct functional responses. The actions of these receptors are counteracted by Gαi-coupled receptors, with the muscarinic M2 receptor and the adenosine A1 receptor playing prominent roles in cardiac myocytes.
2.1.2. Adenylyl cyclases
ACs catalyse the synthesis of cAMP from ATP. Mammalian cells express 9 transmembrane ACs isoforms (AC1-AC9) (15), and one soluble AC (AC10 or sAC). The latter enzyme is not sensitive to Gαs/i modulation and is regulated by bicarbonate and Ca2+(16, 17).
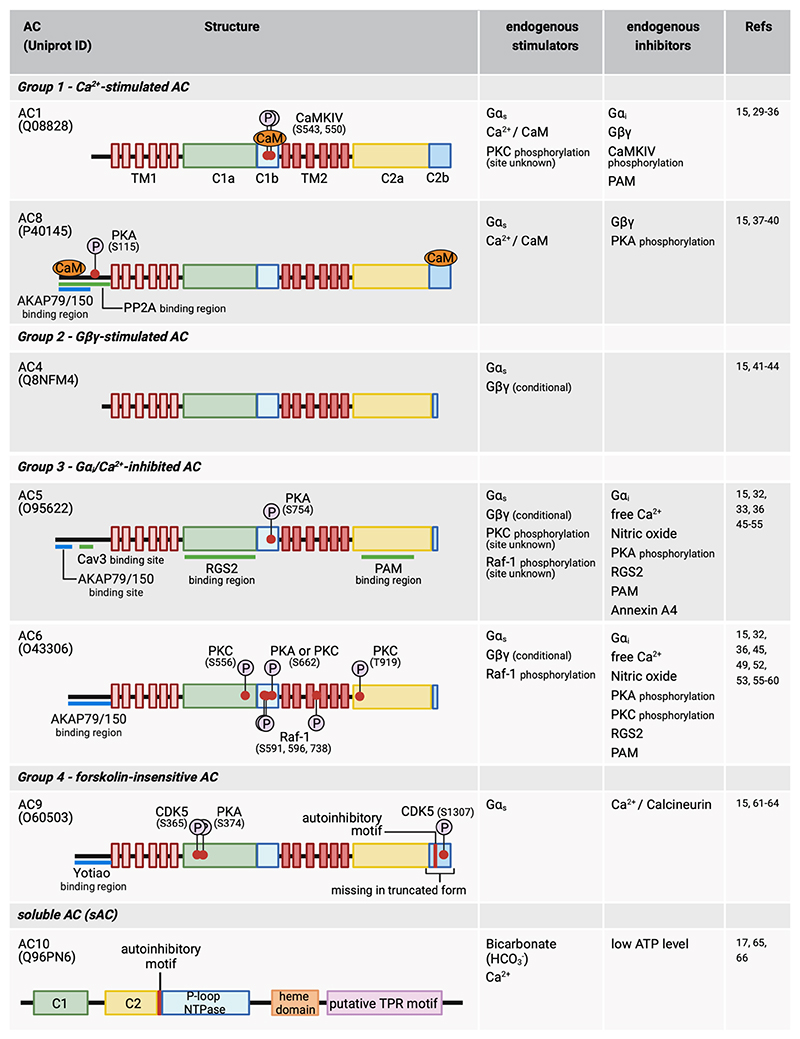
Cardiac myocytes express AC1, AC4, AC5, AC6, AC8, AC9 and AC10 isoforms (18) (Fig 2). AC5 and AC6 are the most abundant enzymes expressed in ventricular myocytes while AC1 and AC8 are expressed in sinoatrial node (SAN) cells where they have been suggested to drive heart rate and regulate cardiac rhythm (19–22). It should be noted though, that the role of AC8 in sinoatrial node was inferred from studies using AC8-overexpression systems (19, 20). AC9 activity has also been detected in SAN cells and independent studies support its role in the regulation of heart rate (23, 24). Although AC4 has been reported to be expressed in the heart (18), no information on its physiological role in this organ is currently available. In addition to Gαs/i-dependent regulation, the activity of transmembrane ACs is modulated in complex ways (15, 25). AC1 and AC8 are stimulated by Ca2+/calmodulin (CaM). High Ca2+ concentrations (mid μM) inhibit all ACs, while low Ca2+ inhibits selectively AC5 and AC6. Gβγ, Gαi, and Gαs subunits stimulate and/or inhibit specific AC in different ways. Gαs stimulates all AC isoforms, Gαi inhibits AC1, AC5 and AC6, whereas Gβγ inhibits AC1 and AC8 but conditionally stimulates AC4 AC5, and AC6 when another activator is present (15, 25). Protein kinase C (PKC) phosphorylation activates AC1and AC5 but inhibits AC6, while PKA phosphorylation inhibits AC5, AC6 and AC8 (15, 25). The natural diterpene forskolin activates all transmembrane AC, except AC9 (26).
Figure 2. Adenylyl cyclases expressed in cardiac myocytes.
The position of the phosphorylation sites (P symbols) refers to the human protein (Uniprot ID as indicated). The figure attempts to list all known post-translational modification modifications reported so far, although some may have not been confirmed in cardiac myocytes. AKAP, A-kinase anchoring protein; C1, cytoplasmic domain 1; C2, cytoplasmic domain 2; CaM, calmodulin; CaMKIV, calcium/calmodulin-dependent protein kinase IV; Cav3, Caveolin 3; CDK5, cyclin-dependent kinase 5; G i, inhibitory G protein alpha subunit; Gαs, stimulatory G protein alpha-subunit; Gβγ, G protein beta-gamma complex; P-loop NTPase, P-loop containing nucleoside triphosphate hydrolase; PAM, protein associated with Myc; PKA, Protein kinase A; PKC, protein kinase C; PP2A, protein phosphatase 2A; Raf-1, RAF proto-oncogene serine/threonine-protein kinase; RGS2, regulator of G-protein signaling 2; TM, transmembrane domain; TPR, Tetratricopeptide repeat.
This figure was generated using Biorender.
AC10 localises to multiple intracellular sites and has been proposed as a local source for compartmentalized cAMP signalling (27). In cardiac myocytes it has been associated with the regulation of mitochondrial function (17, 28).
2.2. cAMP degradation by phosphodiesterases
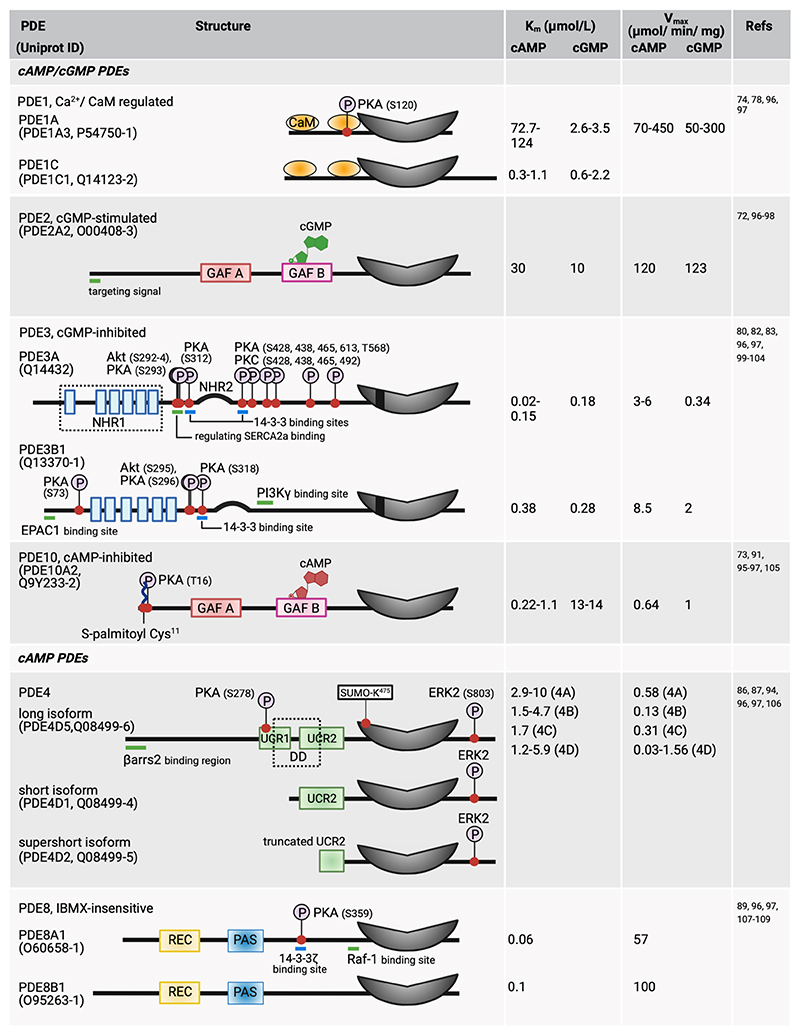
Although efflux of cAMP into the extracellular space via the transporter multidrug-resistance protein 4 (MRP4, gene name ABCC4) has been described as a process that can reduce intracellular levels of the second messenger in cardiac myocytes (67), hydrolysis of cAMP to adenosine monophosphate (AMP) by cyclic nucleotide-hydrolysing phosphodiesterases (PDEs) is the main mechanism that terminates cAMP signalling. PDEs are a superfamily of enzymes that comprises 11 families encoded by 21 genes. As a result of alternative splicing and different translation initiation sites, more than 100 different PDE isoforms are predicted (68). Six PDE families that can degrade cAMP have been characterised in cardiac myocytes, namely the dual-specificity enzymes PDE1, PDE2, PDE3, and PDE10, which degrade both cAMP and cGMP, and the cAMP selective PDE4 and PDE8 (Fig 3). An extensive review of PDEs structure, regulation, physiological roles, pharmacology, and expression in the heart of different species and involvement in cardiovascular disease can be found elsewhere (69–71).
Figure 3. cAMP-hydrolysing phosphodiesterases expressed in cardiac myocytes.
The Figure attempts to list all known post-translational modifications reported so far, although some may have not been confirmed in cardiac myocytes. The position of phosphorylation sites (P symbols) refers to the human sequence (Uniprot ID as indicated). The catalytic domain is shown in grey. Length of the protein sequence is drawn to scale, except for the catalytic domain. Only one isoform of each PDE is used as an example to illustrate regulatory domains, post-translational modifications, and protein-binding regions. Akt, protein kinase B; CaM, calmodulin; EPAC1, exchange protein directly activated by cAMP isoform 1; ERK2, extracellular signaling-related kinase 2; DD, dimerization domain, GAF, cGMP-dependent phosphodiesterase, Anabaena adenylyl cyclase, Escherichia coli FhlA; NHR, N-terminal hydrophobic region; PAS, Per-Arnt-Sim; PI3Kγ, phosphoinositide 3-kinase gamma; PKA, protein kinase A; PKC, protein kinase C; Raf-1, RAF proto-oncogene serine/threonine-protein kinase; REC, response regulator receiver ; SERCA2a, sarco(endo)plasmic reticulum calcium-ATPase; UCR, upstream conserved region; βarrs2, β-arrestin-2.
This figure was generated using Biorender.
The N-terminal domain of these enzymes contains a variety of elements engaged in enzyme dimerization, small molecule binding, phosphorylation, protein-protein interactions, and localization. GAF domains (named after mammalian cGMP-dependent phosphodiesterase, Anabaena adenylyl cyclase, Escherichia coli FhlA) are present in PDE2 and PDE10 and enable allosteric regulation by cGMP. In PDE2 isoforms, cGMP binding to the GAF-B domains enhances the affinity of the enzyme for cAMP by 10-folds (72). cAMP binds to the GAF domain in PDE10 and, depending on the cAMP concentration, cGMP hydrolysis is either activated (low cAMP) or inhibited (high cAMP) (73). Other PDE families feature distinctive modulatory domains. For example, PDE1 contains a CaM-binding domain that, upon interaction with Ca2+/CaM upregulates the activity of the enzyme by 10-folds (74). Upstream conserved regions (UCRs) in PDE4 isoforms are involved in the dimerization of the enzyme and in the regulation of its activity (75, 76). The response regulator receiver (REC) and Per−Arnt−Sim (PAS) domains in PDE8 are involved in protein-protein interactions (77).
PDEs are also regulated by phosphorylation. PKA and CaMKII phosphorylate on S120 and S124 of PDE1A and PDE1B (numbering according to the canonical sequence of human PDE1A and PDE1B with UniProt ID P54750 and Q01064), respectively, resulting in decreased sensitivity to Ca2+/CaM and reduced activation (78, 79). PKA phosphorylation of PDE3A1 on S292/293 not only enhances the enzyme activity but also results in its recruitment to a multiprotein complex localised to the SR that includes SERCA2a, PLB, the A Kinase Anchoring Protein 18 (AKAP18), PKA-RII, and protein phosphatase 2A (80). The hydrolytic activity of PDE3A1 at that location regulates local cAMP levels and basal contractility via modulation of PLB phosphorylation, SERCA2a activity and SR Ca2+ reuptake (81). PKA phosphorylation of PDE3A1 on S312 (numbering according to the canonical sequence of human PDE3A with UniProt ID Q14432), a 14-3-3 interaction site, also enhances enzyme activity and affects its ability to interact with other proteins, although the identity of the proteins involved in this complex is not clear (82). PKB/AKT also phosphorylates and activates PDE3 isoforms (83). PKA, together with CaMKII, phosphorylates the UCR1 region at the N terminus of long forms of the PDE4 enzymes, enhancing their catalytic activity (84–86), while ERK2 phosphorylates all PDE4 isoforms at their C terminus, inhibiting these enzymes (87). PKA phosphorylation of PDE4D3 also increases the binding affinity of the enzyme for mAKAP, promoting faster hydrolysis of cAMP at this signalosome (88). Finally, PDE8A and PDE10 were also shown to be activated by PKA phosphorylation on S359 (89) and T16 (90) (numbering according to the canonical sequence of human PDE8A and human PDE10A2 with UniProt ID O60658 and Q9Y233-2), respectively. Phosphorylation of PDE8 enhances the enzyme activity (89), whereas phosphorylation of PDE10 affects its subcellular localization (91).
Several other post-translational modifications have also been reported to modulate PDEs activity or localization. For example, ubiquitylation by the E3 ubiquitin ligase Smurf2 affects PDE4B stability by controlling its degradation (92), while hydroxylation of proline residues on PDE4D increases its recognition by E3 ligase complexes in cardiac myocytes (93). SUMOylation can enhance the activity of PDE4A and PDE4D isoforms by augmenting the positive effects of PKA phosphorylation and by repressing the inhibitory effects induced by ERK phosphorylation (94). Palmitoylation of PDE10A2 in its N-terminal region promotes translocation of the enzyme to the plasma membrane, an effect that can be blocked by PKA phosphorylation (95).
2.3. cAMP effectors
2.3.1. PKA
PKA is a tetrameric serine/threonine kinase. In the holoenzyme conformation PKA consists of a dimer of cAMP-binding regulatory (R) subunits, each including two cAMP-binding domains, and two catalytic (C) subunits. In the PKA holoenzyme, R subunits autoinhibit the catalytic subunits via a sequence of amino acids that occupy the substrate-binding interface of the kinase. cAMP binding to the two cAMP-binding domains in the R subunits results in a conformational change that frees the C catalytic pocket resulting in PKA activation. Five different human C subunit genes have been identified: of these, PRKACA and PRKACB, encoding for proteins Cα and Cβ, respectively, are the two principal genes expressed in humans (110) and are considered to be redundant. The R subunits, instead, diversify PKA function. Four separate genes code for four functionally non-redundant R subunits (RIα, RIβ, RIIα, RIIβ). While PKA holoenzymes containing RII (PKAII) are predominantly anchored to subcellular scaffolding proteins known as AKAPs (111, 112), RI containing enzymes (PKAI) also exist free in the cytosol (113). R subunits also confer different sensitivity to cAMP, with PKAI being more readily activated than PKAII (114, 115). A more rigid conformation has also been observed in RIIβ compared to RIIα holoenzymes (116), potentially impacting the range of action for the associated C subunit and the level of PKA phosphorylation of proximal substrates (117). R subunits are also differently affected by interaction with myristoylated C subunits, resulting in increased rigidity for RI holoenzymes and enhanced flexibility and ability to associate with membranes for RII holoenzymes (118, 119). Via phosphorylation, PKA modulates the activity of hundreds of substrates, including, but not limited to, transcription factors, ion channels, transporters, exchangers, intracellular Ca2+-handling proteins, and the contractile machinery in muscle cells (Figure 1).
In cardiac myocytes PKA mediates the fight-or-flight response triggered by catecholamine-induced stimulation of β-adrenergic receptors (β-AR) and controls the activity of key proteins involved in excitation-contraction coupling (ECC). PKA increases ICa,L by modulating the activity of voltage gated, L-type calcium channels (LTCC). As recently established, rather than a consequence of the direct phosphorylation of the channel by PKA (120), this regulation requires PKA-dependent phosphorylation of the regulatory GTP-binding protein RAD, which relieves a constitutive inhibition that RAD exerts on the channel subunit CaV1.2. RAD phosphorylation results in an increase in channel open probability (121, 122). PKA also phosphorylates the ryanodine receptor 2 (RyR2) (123, 124), a Ca2+-release channel on the sarcoplasmic reticulum (SR) membrane responsible for the bulk cytosolic Ca2+ increase that triggers cardiac myocyte contraction. However, the functional significance of this phosphorylation remains controversial (125). PKA also enhances Ca2+ re-uptake in the SR by the sarco/endoplasmic reticulum calcium ATPase (SERCA2) pump. Phosphorylation of the SERCA2-associated protein phospholamban (PLB) releases the inhibitory effect that PLB exerts on SERCA. PKA phosphorylation of phospholemman (PLM), a regulatory protein associated with the Na+/K+-ATPase (NKA), relieves its inhibitory effect of PLM on NKA, driving Na+ efflux (126, 127). This, in turn, activates the forward mode of the Na+/Ca2+ exchanger (NCX) to promote Ca2+ extrusion during relaxation, allowing for greater calcium influx via LTCC during excitation (128). PKA-dependent phosphorylation of the myofilament proteins troponin I (TnI) and myosin binding protein C (MyBPC) impacts myofilament contraction and relaxation efficiency (129). TnI phosphorylation results in accelerated troponin C-Ca2+ off-rate and facilitates quicker force development and shortening during systole, as well as faster force relaxation and re-lengthening during diastole (130). MyBPC phosphorylation by PKA regulates cycling of myosin heads, reduces myofilament Ca2+ sensitivity and promotes cross-bridge formation (131). PKA also phosphorylates the sarcomeric protein titin (TTN), reducing its stiffness (132). In addition to phosphorylation of targets involved in the control of ECC, cardiac PKA also activates glycogenolytic (133) and other metabolic enzymes (134) and regulates multiple other proteins involved in a variety of cellular processes, including modulation of gene transcription (2, 135, 136).
2.3.2. EPACs
EPAC proteins function as guanine nucleotide exchange factors (GEFs) for both Rap1 and Rap2 (137). Various effector proteins, including adaptor proteins implicated in modulation of the actin cytoskeleton, regulators of G proteins of the Rho family, and phospholipases, relay signaling downstream from Rap (138).
The cAMP effector EPAC1 is involved in the regulation of ECC as part of an EPAC-Rap axis that intersects with Ca2+ signalling through EPAC-dependent activation of phospholipase Cε (PLCε) and CaMKII (139). In response to β-AR stimulation, EPAC−Rap2b induces both Rap GEF activity and hydrolytic activity of PLCε. This leads to sustained Rap activation and the induction of PKCε and CaMKII phosphorylation downstream from PIP2 breakdown. These signaling events lead to enhanced SR Ca2+-induced Ca2+ release (CICR) (140, 141), contributing to modulation of contraction.
It is interesting to note that EPAC proteins can have subcellular localisation distinct from PKA and mediate functional effects of cAMP that are different, and in some cases opposite, to the effects mediated by PKA (142, 143).
2.3.3. HCN channels
Another type of cAMP effector expressed in the heart is the hyperpolarization-activated cyclic nucleotide gated (HCN) channels. These channels are expressed in cells of the cardiac conduction system and direct binding of cAMP to a conserved cyclic-nucleotide binding domain in the distal C-terminus of the channel potentiates the channel by shifting the voltage- dependence of activation to more depolarized membrane potentials, resulting in the modulation of heart rate associated with the fight-or-flight response (144). HCN4 have been shown to interact with caveolin (145), an association that has been suggested to promote localisation of the channel in proximity to β-ARs and facilitate enhanced firing of action potential in SAN cells in response to catecholamine stimulation (146).
2.3.4. POPDC proteins
POPDC proteins are a more recent addition to the list of cAMP effectors. Of the three known isoforms, POPDC1 (gene name BVES) and POPDC2 proteins are expressed in the heart, with particularly high expression levels in the conduction system. They interact directly with cAMP and contribute to cardiac automatism and cardiac adaptation to stress, although the precise role of these proteins remains poorly understood (147).
3. Compartmentalisation of cAMP signalling
While β-ARs are critically involved in regulating the cAMP response to stress, they are not the sole receptors utilizing this pathway in cardiac myocytes. Analysis of mRNA obtained from cardiac myocytes reveals the expression of approximately 200 different GPCRs in cardiac cells (148), several of which signal via modulation of cAMP levels. Consequently, within the multitude of chemical messages influencing the heart, several lead to activation or inhibition of cAMP generation, prompting the question of how myocytes interpret these signals, prevent inappropriate phosphorylation of unsuitable targets, and achieve the required response to each specific stimulus. The regulation of strength and rate of cardiac contraction by catecholamines enables the critical adaptations necessary to face a situation of danger or stress. Such regulation is achieved in milliseconds, only lasts for the required time, and is precisely directed to the relevant targets only. Critically, these adaptations are not triggered by the continuous fluctuation of other hormones that also signal via cAMP, implying mechanisms that, depending on the stimulus, can direct the effector machinery to the appropriate targets while shielding other destinations. How is this achieved?
In the late ‘70s, early ‘80s, the initial indication that not all cAMP signals evoke identical downstream responses emerged from studies conducted on isolated perfused hearts. These studies revealed that isoproterenol (Iso) and prostaglandin E1 (PGE1), despite raising intracellular cAMP to comparable levels and similarly influencing the PKA activity ratio, exerted markedly different effects on PKA substrates. Specifically, Iso induced the phosphorylation of phosphorylase kinase, TnI, and various other PKA phosphorylation substrates while PGE1 stimulation showed no increase in the phosphorylation of these targets (133, 149, 150). Notably, prior research had established that cAMP can bind to both soluble and particulate sites, with up to 50% of PKA activity being associated with the particulate fraction of heart homogenates (151, 152). This indicated that PKA is directed to specific locations within the cell. Building on these findings, the hypothesis emerged that distinct and spatially segregated cAMP pathways exist (152, 153). The discovery that perfusion with Iso activates the particulate fraction of PKA, while PGE1 increases the activity ratio of soluble PKA, supported this hypothesis (153). It suggested that a selective activation of PKA subsets confined to specific intracellular compartments occurs, leading to distinct functional responses (154).
The hypothesis of compartmentalized cAMP signalling was initially received with some skepticism, partly due to concerns that, while it had not been possible at that point to correlate ‘soluble’ and ‘particulate’ fraction with any specific subcellular structure, the high-speed centrifugations in high salt concentrations used to separate these fractions could result in artifactual redistribution of PKA. However, investigations into the negative regulation of the adrenergic response, triggered by the activation of the muscarinic M2 receptor, also indicated compartmentalisation of cAMP. While muscarinic M2 receptor is a Gαi-coupled GPCR, and its activation is anticipated to result in AC inhibition and a decrease in cAMP, several studies found that the adverse impact of acetylcholine (ACh) on inotropy and Ca2+-dependent action potentials can manifest even in the absence of a measurable overall decline in cAMP levels (155–157). These observations prompted a realization that ‘the question of compartmentalisation of cAMP and whether whole-cell cAMP measurements reflect important changes in subpools of cAMP, while distasteful subject to some people, must not be ignored’ (158).
In the following decade, further data supporting the functional compartmentalisation of cAMP in cardiac myocytes emerged. Studies showed that low levels of forskolin, while elevating cAMP to higher levels than Iso, induced less protein phosphorylation and a smaller effect on contractility (159). Experiments utilizing various cAMP-elevating agents demonstrated that the magnitude of myocyte shortening and Ca2+ transients exhibit a stronger correlation with the augmentation of particulate cAMP concentration than with total cAMP levels (160). Another illustration of the compartmentalised action of cAMP came from the observation that stimulation of distinct β-AR subtype results in different functional outcomes. While activation of β1-AR led to the phosphorylation of LTCC, PLB, RyR, TnI and MyBPC, resulting in the typical positive inotropic and lusitropic effects associated with catecholamines, β2-AR activation showed more limited effects, selectively inducing the phosphorylation of LTCC, causing a less pronounced positive inotropic effect and no lusitropic effect (161). Once again, the observed functional disparities appeared to align with the differential activation of particulate versus soluble fractions of PKA (162). In further support of compartmentalised signalling, the β-AR agonist Iso and the gut hormone GLP-1 elevated cAMP to a similar amount in adult rat cardiac myocytes; however, GLP-1 failed to produce a robust positive inotropic effect and, in fact, mildly reduced myocyte contraction (163).
3.1. AKAPs and the organization of signalosomes
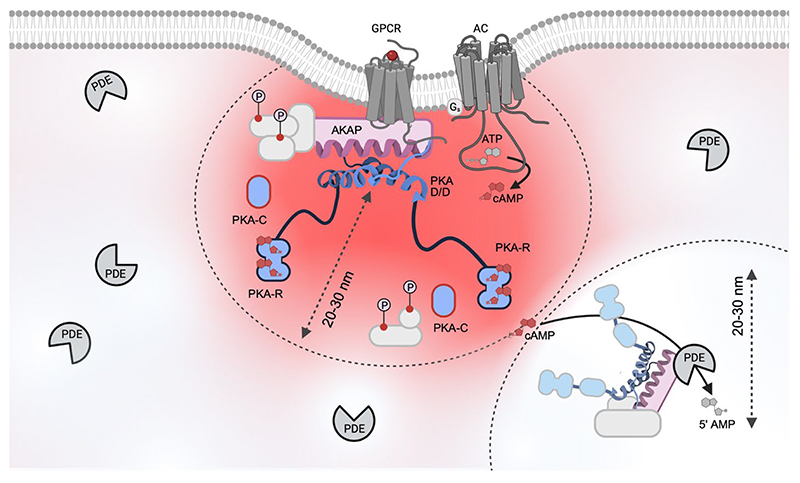
The observation that, depending on the GPCR activated, different subset of PKA targets are phosphorylated, led to the hypothesis that not all PKA enzymes expressed in the cell have equal access to all PKA targets present in that cell (150). It later became clear that the specific activation of PKA and its ability to deliver the signal to the appropriate target, relies on PKA immobilization at particular subcellular sites through interactions with AKAPs (111) (FIG 4). AKAPs contain targeting motifs that mediate their localization to organelles or other cellular structures, thus bringing PKA near one or more of its phosphorylation targets. AKAPs nucleate signaling hubs, or signalosomes, where they assemble, via protein-protein interaction, multi-protein complexes comprising various combinations of signaling elements, including GPCRs (164, 165), ACs (51), phosphatases (PPs) (166, 167), and cAMP-hydrolyzing PDEs (168) (Fig 4). Each signalosome localises at a specific subcellular site including, among others, the plasma membrane (e.g., AKAP18α, AKAP18β, AKAP79, and Yotiao, the smallest splice variant of AKAP9 (166, 169, 170)), the SR, (e.g., AKAP18δ (171)), the cytosol (e.g., GSKIP (172)), the cytoskeleton (e.g., gravin and ezrin (173)), the mitochondria (e.g., D-AKAP1 (174) and SKIP (175)) and the centrosome (e.g., pericentrin (176) and AKAP350, a long isoform of AKAP9 (177)). As individual signalosomes result from the assembly of a distinct assortment of protein components, each signalling hub enables local regulation of a specific cellular function.
Figure 4. AKAP-dependent signalosome activated by a cAMP nanodomain.
The schematic illustrates a generic AKAP-dependent signalosome localized in proximity of a GPCR and AC at the plasma membrane. Activated PKA is shown in blue. The interaction between the AKAP amphipathic helix and the R D/D domain is modelled on the AKAP18:PKA-RII crystal structure (PDB 4ZP3) (186). Activation of the receptor generates a pool of high [cAMP] that remains confined and, depending on the nature and amplitude of the stimulus, can be limited to a radius of about 20-30 nm. This distance also approximately equates to the range within which the AKAP-anchored PKA R subunits can extend. PKA targets (light grey), are phosphorylated only if they reside within the active cAMP nanodomain. PDEs hydrolyse cAMP in their immediate neighborhood (20-30 nm), creating region within which [cAMP] remains below the activation threshold of PKA. Within this domain PKA targets are not phosphorylated. Thus, PDEs contribute to compartmentalise cAMP and protect targets from inappropriate phosphorylation. AC, adenylyl cyclase; AKAP, A-kinase anchoring protein; D/D, dimerization/docking domain; GPCR, G protein coupled receptor; PKA, protein kinase A; PDE, phosphodiesterase.
This figure was generated using Biorender.
Over 60 AKAP have been identified in the human genome (178). Anchoring of PKA to AKAPs occurs via a structurally conserved amphipathic helix of 14−18 amino acids in the AKAP protein into which a hydrophobic groove formed by the N-terminal dimerization/docking (D/D) domains of a PKA R subunits dimer docks (Fig 4) (179–181). Although the amphipathic helix does not contain a specific conserved amino acids motif, it contains alternating pairs of polar and hydrophobic residues, that allows identification of members of this functionally related scaffolding proteins (182, 183). Some of the hydrophilic residues appear to be essential for strong binding (181, 184–186) and polymorphisms in the helix that disrupt this pattern cause functional deficits (187). While most known AKAPs bind to RII subunits, some dual-specific AKAPs anchor both RI and RII (174) and some AKAPs are RI specific (175, 188, 189). Electron microscopy and 3D reconstruction studies show that the AKAP:PKA macromolecular assembly is not rigid but dynamic. Data show that the intrinsically disordered regions located between the D/D domain and the autoinhibitory sites of each R subunit provide flexibility to the holoenzyme enabling movement of the C subunit within a 20−40-nm radius (Fig 4) (117, 190). To ensure that downstream phosphorylation is restricted to nearby targets, the assumption is that, on cAMP binding, either the catalytic subunit dissociates and rapidly re-associates to the free R subunits resident within the domain (191, 192), or that substrate phosphorylation can occur without complete dissociation of C subunits (190).
Multiple AKAPs are expressed in cardiac myocyte and for a number of these the functional role has been studied in some detail (193). Several AKAPs have been implicated in the regulation of Ca2+ handling and myocyte contractility, including AKAP79, AKAP18α, AKAP18γ, AKAP18δ and mAKAP. The AKAP Yotiao has been linked to cardiac repolarization through PKA-dependent modulation of the KCNQ1 subunit of the slowly activating delayed rectifier K+ current (IKs) potassium channel and several have been involved in the development of cardiac disease, as detailed below.
3.2. Imaging tools and direct visualization of cAMP compartments
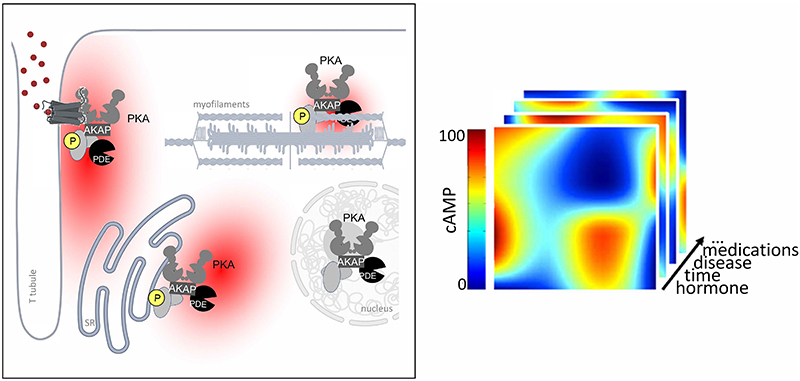
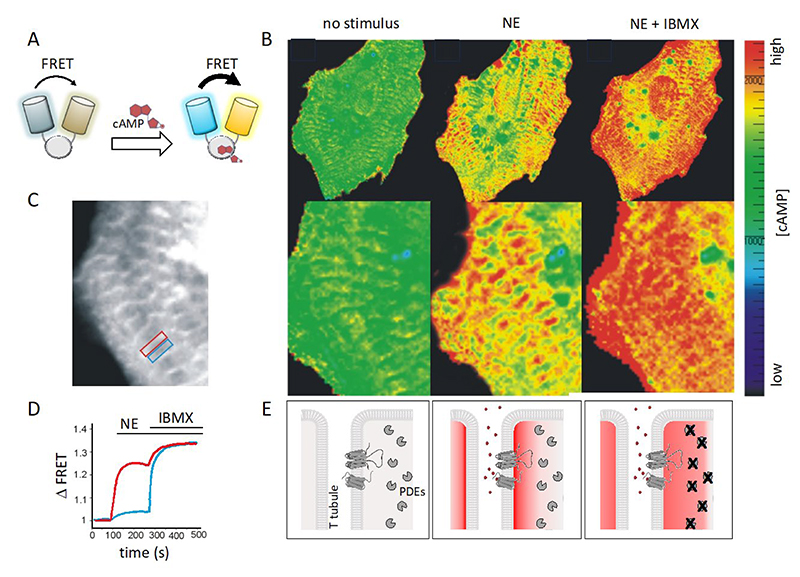
For AKAP-anchored PKA to be able to modulate specific functional effects through spatially restricted phosphorylation of targets and avoid generalized activation of all PKA signalosomes, mechanisms must be in place to ensure that [cAMP] above the PKA activation threshold is achieved only at selected locations. Indeed, elevation of cAMP in response to stimuli has been demonstrated to be spatially restricted. Significant advancement in elucidating the compartmentalised nature of cAMP signaling was powered by the introduction of molecular tools designed for real-time visualization and monitoring of intracellular cAMP in intact, living cells (194, 195) (Fig 5). Compared to previous methods based on cAMP detection in cell homogenates which measure bulk cAMP at a given time point, with the new tools it was possible to monitor the intracellular dynamics of free cAMP with sub-micrometer resolution. With this approach, direct evidence of cAMP compartmentalisation in cardiac myocytes was provided (196) (Fig 5).
Figure 5. Visualisation of compartmentalized cAMP signals in cardiac myocytes.
A. Schematic representation of a Forster Resonance Energy transfer (FRET) based reporter for cAMP. These sensors are genetically encoded and are typically constituted by a cAMP binding domain sandwiched between the cyan (CFP) and yellow (YFP) variant of the green fluorescent protein. Binding of cAMP to the cAMP binding domain results in a conformational change that affects the distance between the two fluorescent proteins modifying their fluorescent emission. Several variants of these sensors with slightly different designs are now available (217). B. Image of a cardiac myocyte showing, in pseudocolor, different concentrations of cAMP in response to norepinephrine (NE) and subsequent inhibition of PDEs with IBMX. C. Same myocyte as in B. The image highlights the subcellular localization of the FRET-based cAMP sensor. As the FRET reporter used in this experiment is based on the full PKAII holoenzyme and includes the RII D/D domain, the probe localises heavily at AKAPs, with an enrichment in correspondence of the T-tubules. D. Kinetics of FRET change, which report the changes in cAMP concentration, recorded within the red box (region where the reporter is AKAP-bound) and blue box (where the reporter is not bound to an AKAP) shown in C. E. Left panel: in the absence of agonist, [cAMP] is low. Middle panel: application of norepinephrine (NE) generates a local cAMP response at signalosomes organized along T tubules. Right panel: inhibition of PDEs disrupts the local cAMP gradients and the cAMP signal equilibrates across the cell. Modified from (196).
Part of this figure was generated using Biorender.
The method relies on Fluorescence Resonance Energy Transfer (FRET) between a donor and an acceptor fluorophore genetically fused to one or two cAMP-binding domains. The sensor design is such that the fluorophores come in proximity of each other or move further away of each other on cAMP binding. The change in distance between the two fluorophores affects energy transfer, resulting in a change in emitted fluorescence that can be measured as Δ FRET (Fig 5). The approach enables detection of signalling events with high temporal and spatial resolution in intact, living cells, where the complexity of the intracellular environment is preserved. The fluorescent signal generated by the sensor is captured by a sensitive camera and transferred to a computer that generates a curve representing the changes in cAMP levels over time (197–200) (Fig 5). The subcellular precision provided by this technique, coupled with the capability to quantitatively evaluate changes in real-time, enabled the first direct demonstration that, on activation of β-AR with norepinephrine, cAMP does not uniformly equilibrate within cardiac myocytes but instead forms intracellular gradients, with higher concentrations at specific subcellular sites (196) (Fig 5). In the original study, the changes in cAMP concentration were tracked by monitoring FRET changes between the PKA RII subunit linked to a cyan fluorophore (CFP) and the PKA C subunit linked to a yellow fluorophore (YFP) (197). With this sensor, it became evident that the cAMP response induced by norepinephrine was confined to subcellular compartments where PKA is tethered to AKAPs via its D/D domain. When employing a probe variant where the RII component lacked the domain essential for anchoring, the sensor failed to detect any substantial cAMP signal in response to norepinephrine, confirming the highly localized nature of the cAMP elevation (196, 201).
A variety of cAMP sensors based on FRET and with different design have been subsequently developed (198–200, 202) and multiple studies have confirmed cAMP compartmentalisation, both in cardiac myocytes (203–209) and various other cell types (210–214). cAMP FRET reporters targeted to defined subcellular compartments, clearly show that not only activation of different Gαs-coupled receptors generates distinct pools of cAMP (113) but also that the cAMP response generated by a specific stimulus is compartmentalised (202, 215, 216).
3.3. Role of phosphodiesterases in cAMP compartmentalisation
A crucial role in the establishment of confined cAMP pools is attributed to cAMP-hydrolysing PDEs, as substantiated by extensive evidence (70, 96, 209, 218–221). This is underscored by consistent observations that inhibiting PDEs results in the dissipation of intracellular cAMP gradients, as detected by live-cell imaging of cAMP reporters (196, 202, 222, 223).
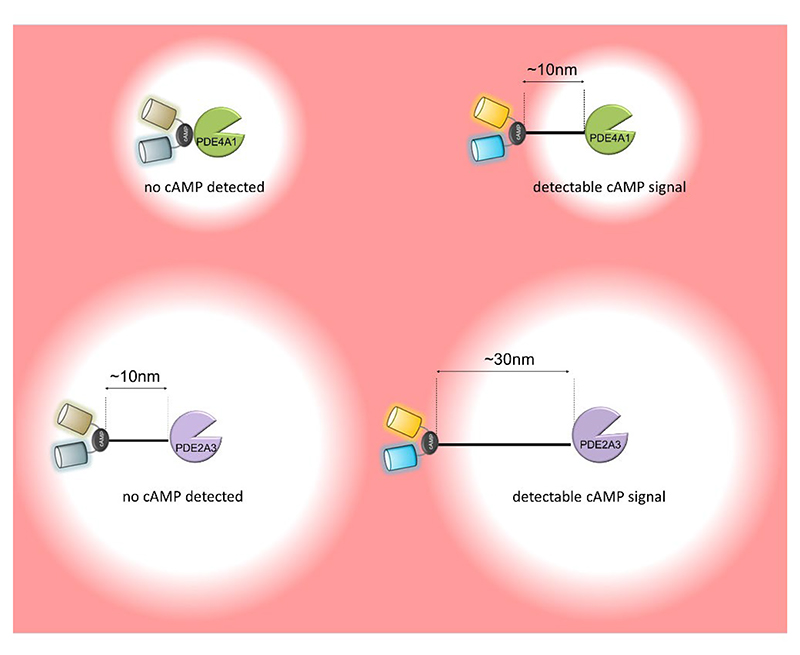
Experiments involving the direct fusion of FRET-based cAMP sensors to a PDE clearly illustrate that these enzymes can establish a region of low cAMP concentration in their immediate surroundings (Fig 6). Interestingly, evidence suggests that the range of action of a PDE depends on the enzymatic properties of the specific PDE isoform involved. For instance, PDE4A1, a relatively high-affinity (low μM) but low-turnover (1-5 cAMP molecules/s) enzyme (97), can effectively reduce cAMP levels within a 10 nm radius. In contrast, PDE2A3, a lower-affinity but faster enzyme (97), can deplete cAMP within a radius exceeding 30 nm (224). When a PKA target is located within this domain, then the activity of the PDE can block the target phosphorylation, as demonstrated by experiments where the PDE was fused to FRET reporters that measure PKA activity (224). It should be noted that these experiments were conducted using cell lysates, where potential physical barriers to cAMP diffusion are minimized, indicating that the PDEs alone can completely quench the cAMP signal, at least in the space immediately surrounding the enzyme.
Figure 6. Phosphodiesterases effectively degrade cAMP in their immediate surroundings.
Using a cAMP FRET reporter genetically fused to PDE enzymes it was possible to establish that the PDEs can effectively degrade cAMP and create a nanodomain devoid of second messenger in the immediate enzyme neighborhood. The radius of the nanodomain cleared of the second messenger depends on the enzymatic property of the specific PDE involved.
This figure was generated using Biorender.
The ability of PDEs to maintain cAMP levels below the PKA activation threshold helps explain the puzzling finding that the estimated intracellular concentration of cAMP in resting, unstimulated cells, calculated to be around 1μM (151, 225, 226), is significantly above the EC50 of cAMP binding to the regulatory subunit of PKA (estimated in vitro to be in the 100−300 nM range) (194) and matches the EC50 measured in intact cells (~5 μM for PKA type II) (227). Based on these values, one would expect to find PKA to be already robustly activated in unstimulated cells. A relatively high basal cAMP concentration in the bulk cytosol can be reconciled with inactive PKA if PDEs are anchored in proximity of the kinase and its substrates.
One interesting feature that has been demonstrated for some members of the PDE superfamily is that their subcellular localization is dynamic and can be regulated by signalling events. For example, the PDE3A1 isoform has been shown to be recruited to the SERCA/AKAP18 signalosome at the SR upon PKA phosphorylation of its unique N-terminus (80). Another example is the recruitment of PDE4 family members to the plasma membrane by β-arrestins, which results in augmented cAMP degradation near an active GPCR (228). A β-arrestin-dependent process has been described also for PDE4D5 recruitment to GPCRs located in endosomes, resulting in PDE4D5 depletion from the nucleus, increased nuclear cAMP and enhanced cAMP-dependent transcription (229).
The diversity in the enzymatic properties and the regulatory mechanisms that the PDE system displays (Table 2) allows for highly sophisticated regulation of the second messenger levels and the creation of complex and dynamic patterns of cAMP pools. Local control of cAMP levels at individual signalosomes can be executed by a single PDE isoform or by multiple PDEs acting synergistically (230, 231). Each PDE can be distinctly regulated by Ca2+, cGMP, and various post-translational modifications, including PKA-dependent phosphorylation, facilitating feedback regulatory loops (219). The observation that a specific PDE isoform can uniquely control the cAMP level in its proximal environment holds important implications. Firstly, it signifies that, even in the absence of a physical barrier to cAMP diffusion, the presence of a PDE is sufficient to generate a steep cAMP concentration gradient (Fig 4). Secondly the cAMP level within a specific cellular domain, and consequently, the ensuing cellular response, is determined by the identity and activation state of the resident PDE(s) and by the conditions that may promote PDE recruitment or release from that location. This dynamic landscape of cAMP gradients, shaped by the identity of the different PDE isoforms expressed in a specific cell type, their localization, and their coordinated regulation, determines the ensuing cellular response (232).
Table 2. AKAP-dependent signalosomes characterised in cardiac myocytes.
| Gene name |
Protein names |
Known Signalosome components |
Reported localisation |
Reported function |
Refs. |
|---|---|---|---|---|---|
| AKAP1 | D-AKAP1 AKAP121 AKAP149 S-AKAP84 |
PKAI, PKAII, CaN, RSK1, PP2A, Drp1, NDUFS1 |
mitochondria, nuclear envelop, sarcoplasmic reticulum | Cardiac stress response, Anti-hypertrophic |
(193, 313–321) |
| AKAP5 | AKAP79 AKAP150 |
PKAII, PKC, CaN, β-AR, AC5, AC6, SAP97, caveolin 3, LTCC, Orai1, Kir2.1 (KCNJ2), Kir2.2 (KCNJ12), |
plasma membrane, T tubule |
ECC, β -AR desensitization/resensitization cycle, arrhythmias, anti-hypertrophic |
(165, 193, 322–329) |
| AKAP6 | AKAP100 mAKAP |
PKAII, PDE4D3, RyR2, AC5, CaNAβ, PP2A, NFATc, MEF2 ERK5, MEK5, Epac1, Rap1, Siah2, RSK3, NCX1, nesprin-1α, PHD, pVHL, HIF-1α, PLCε, PKCε, PKD |
nuclear envelope, sarcoplasmic reticulum | EEC, pro-hypertrophic | (124, 193, 330–342) |
| AKAP7 | AKAP15 AKAP18 | PKAII, LTCC, PLB, SERCA2, PDE3A1, PP1, inhibitor 1 (I-1), CaMKII, RyR2, Kir2.1, Kir2.2 | plasma membrane, endoplasmic reticulum | ECC, Anti-hypertrophic | (80, 171, 193, 332, 343–347) |
| AKAP9 | Yotiao, AKAP350 AKAP450 CG-NAP |
PKAII, PP1, PP2A, PKC, PKN1, kinase 1, AC9, PDE4D3, Kv7.1 (KCNQ1), KCNE1, CLIC | plasma membrane, Golgi, centrosome | cardiac repolarization, arrhythmias | (193, 290, 310, 322, 328, 348–350) |
| AKAP10 | D-AKAP2 | PKAI, PKAII, Rab4, Rab11, PDZK1, NHERF-1, GIRK | Plasma membrane, endocytic vesicles, mitochondrial membrane | cardiac repolarization, arrhythmias | (351–357) |
| AKAP12 | Gravin AKAP250 |
PKAII, PDE4D, PKC, β-AR, Src | plasma membrane |
ECC, β -AR desensitization/resensitization cycle |
(193, 322, 358–361) |
| AKAP13 | AKAP-Lbc Ht31 Brx-1 Proto-Lbc |
PKAII, PDE4, PKC, PKD, PKNα, MLTKβ, MKK3β, p38α, RhoA, IKKβ, HSP20, Shp2, actin | cytoskeleton | cardiac stress response, pro-hypertrophic | (193, 322, 346, 362–374) |
| SPHKAP | SKIP | PKAI | Inner mitochondrial membrane | Cardiac stress response | (175, 188, 375) |
| PDE4DIP | myomegalin | PKAI, PKAII, PDE4D, desmin, MyBPC, cTnI | Golgi/ centrosome |
Hh signalling, atrial fibrillation MyBPC and TnI phosphorylation |
(376–379) |
| TNNT2 | troponin T (TnT) | PKAI, PKAII | sarcomere thin filament | MyBPC and TnI phosphorylation | (380, 381) |
| SYNM | synemin | PKAII, PP2A, α-actinin, titin, talin, vinculin, α-dystrophin, α-dystrobrevin |
sarcomere Z line, M band, costamere | anti-hypertrophic, arrhythmia | (382–384) |
| ARFGEF2 | BIG2 | PKAI, PKAII, PDE3A, PP1γ, ARFs |
plasma membrane | vesicular trafficking | (385–387) |
| LDB3 | Zasp cypher |
PKAII, CaN, β-catenin, LTCC, calsarcin-1, myotilin, PGM1 |
sarcomere Z line, intercalated disk | ECC, cardiac remodelling |
(388–392) |
| TLN1 | talin 1 | PKAII | costamere, focal adhesions | Mechano transduction |
(393) |
3.4. cAMP nanodomains
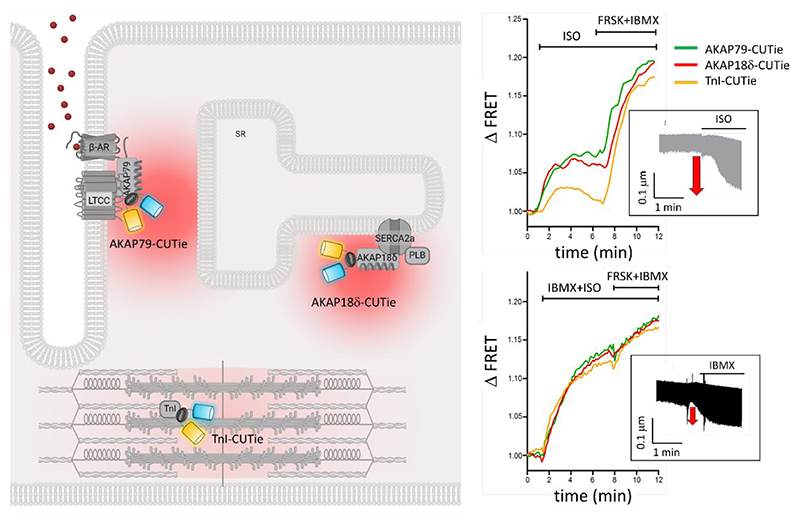
The organization of signalosomes, wherein PKA is in proximity of specific phosphorylation targets, along with the capacity of cells to create steep cAMP gradients (Fig 4), is an effective strategy for achieving hormonal specificity. As discussed above, experimental evidence supports the notion that the compartmentalized increase in cAMP levels on ligand binding to a specific GPCR enables the activation of a limited subset of PKA effectors, ensuring the initiation of the relevant cellular response only. For example, in cardiac myocytes the activation of β-AR leads to increased PKA-dependent phosphorylation of PLB and Tn while the activation of the prostaglandin receptor does not yield a similar effect (233, 234). This distinction persists despite both receptors generating a similar overall amount of cAMP upon activation. Imaging experiments utilizing targeted FRET reporters have indicated that the phosphorylation of distinct PKA targets in response to activation of a specific GPCR relies on cAMP pools being generated where specific AKAP-PKA signalosomes are positioned (113). This elucidates how catecholamines, via β-adrenergic receptors, and prostaglandin E1, via EP receptors, induce a comparable elevation of cAMP in the heart, yet only isoproterenol enhances cardiac contractile force and activates glycogen metabolism (234). The utilization of targeted cAMP reporters also demonstrated that the cAMP response triggered by β-AR results in a fast, larger, and sustained cAMP increase at the plasmalemma and SR compared to a slower, smaller, and transient cAMP response at the myofilament. PDEs inhibition abolishes this difference but also results in an attenuated inotropic response (202), indicating that the subcellular heterogeneity of the cAMP pools generated in response to catecholamines is required to optimize the contractile response (Figure 7).
Figure 7. The heterogeneous cAMP signal generated by β-AR stimulation is required to achieve optimal inotropic response.
Detection of local cAMP signals at specific locations is possible by using targeted FRET reporters generated by fusing the sensor to a protein component of a specific signalosome. In the example illustrated here, the FRET reporter CUTie was fused to AKAP79 for targeting to the plasmalemma, to AKAP18δ for targeting to the sarcoplasmic reticulum (SR), or to the myofilament protein troponin I (TnI) for targeting to the sarcomere. Activation of the β-AR with isoproterenol generated distinct responses at these locations. As illustrated in the top right panel, the cAMP level detected at the plasma membrane and SR were comparable and showed sustained kinetics, while the cAMP response at the sarcomere was delayed and significantly smaller than at the other sites and transient in nature. The compartmentalisation of the cAMP signal was dependent on the presence of active PDEs as, on application of the PDE inhibitor IBMX, the signal equilibrated at the three locations (bottom right panel). The insets show a measure of cardiac myocyte contractility, as determined by quantification of sarcomere fractional shortening (μm) and illustrate that compartmentalized cAMP elevation achieves significantly larger inotropic effect compared to homogeneous cAMP increase. Modified from (202).
Part of this figure was generated using Biorender.
The differences in amplitude and kinetics of the cAMP response detected at the plasmalemma, SR, and myofilaments also indicated that neighboring cAMP domains are demarcated by extremely narrow boundaries as the subcellular structures where these three sensors localize are in close proximity to each other and only tens of nanometer apart from each other (235). The nanometer-size of cAMP domains has been directly confirmed in HEK293 cells, a cell type characterized by a simpler subcellular architecture compared to cardiac myocytes. In this study, the cAMP response to different agonists was measured in proximity of the GLP-1R by fusing a FRET reporter directly to the receptor or by introducing a spacer of known length between the receptor and the probe. The analysis showed that, at low agonist concentrations, cAMP could be detected at up to 60 nm away from the receptor but not at a larger distance (215). The nanodomain generated by activation of the GLP-1R was also shown to be shielded from the influx of cAMP generated by activation of the β-AR (215) confirming very tight regulation of the spatial propagation of cAMP.
The nanometer size of the cAMP pools generated in proximity of GPCRs in HEK293 cells aligns well with the estimated 15-25 nm size of a single AKAP:PKA complex (190) (Fig 4) and is in the same range of the domain within which PDEs can effectively regulate cAMP levels (Fig 6). In addition, the cAMP nanodomain size is consistent with the dimensions of AKAP:PKA clusters observed at the plasma membrane of CHO cells using super-resolution microscopy, showing an average diameter of these clusters of approximately 100 nm (236). Thus, it appears that the cAMP nanodomains can extend as needed to facilitate the activation of individual PKA/AKAP signalosomes.
3.4.1. cAMP buffering
Despite the clear role of PDEs, a full understanding of the mechanisms enabling cAMP nanodomains remains elusive. Multiple measurements of cAMP mobility in cells yielded diffusion coefficients exceeding 100 μm2/s, and up to nearly 800 μm2/s, indicative of free diffusion, as it would occur in aqueous medium (195, 198, 237, 238). In addition, mathematical modeling identified a “mismatch” of four orders of magnitude between the apparent rapid cAMP diffusion rates and the rate of cAMP degradation by PDEs, suggesting that cAMP concentrations should rapidly equalize throughout a cell, preventing the formation of any significant spatial confinement (239). One way of reconciling fast diffusion of cAMP and slow PDE hydrolytic activity with the formation of cAMP gradients would be to introduce cAMP buffering capacity in the equation, and recent research indicates that a combination of various mechanisms may effectively contribute cAMP buffering.
For a long time, intracellular buffering of cAMP was considered negligible. However, measurements of cAMP diffusion have typically been carried out at high levels of cAMP, raising the possibility that, in those conditions, cAMP binding site may be saturated. Recent experiments utilizing a fluorescent analog of cAMP revealed that at low, physiological concentrations, cAMP appears to be effectively entirely bound and, consequently, immobile (224). This finding is consistent with quantitative immunoblotting studies showing that, in a large panel of cell types, including cardiac myocytes, PKA R subunits are in large molar excess of C subunits (on average ~17-fold) (191). Excess free R subunits can bind and buffer cAMP. Other cAMP binding sites may also exist as their abundance was estimated to exceed 10 μM (224), enough to completely bind cAMP at basal and even moderately elevated concentrations. However, robust elevation of cAMP, such as through intense receptor stimulation, or treatment with a combination of PDE inhibitors and the AC stimulator forskolin, overcame the cAMP binding capacity. If cAMP can be significantly buffered by binding to proteins, the concentration of free cAMP in the cell may be much lower than initially thought, allowing PDEs to establish substantial concentration gradients, at least over a short distance and at physiological agonist concentrations (224).
3.4.2. Liquid-liquid phase separation
Liquid-liquid phase separation (LLPS) has been proposed as an additional mechanism that could contribute to maintaining low levels of free cytosolic cAMP (240). LLPS occurs when, surpassing a specific concentration threshold, a molecule in a solution undergoes de-mixing into two distinct phases − a concentrated, condensed phase and a diluted phase (241). Experimental evidence suggests that PKA RIα is capable of phase separation, dynamically sequestering cAMP within RIα liquid condensates. Interestingly, the excess expression of R subunits discussed above predominantly involves RI isoforms (191). The creation of phase-separated RIα bodies appears to result from multivalent interactions involving RIα D/D domain and linker region and seems to be stimulated by cAMP. Using targeted FRET sensors, disruption of RIαcondensates using the compound 1,6-hexanediol was shown to lead to enhanced cAMP levels detected in the immediate vicinity of a PDE, which was interpreted as the result of cAMP being released from the RIα condensates and being able to ‘flood’ the domain with low cAMP established by the PDE hydrolytic activity (240). To what extent RIα LLPS occurs in cardiac myocytes and whether this phenomenon affects compartmentalized cAMP signalling and cardiac myocyte function remains to be established.
3.5. Signalling from internal GPCRs
While the long-established mechanism for GPCRs signalling involves receptors that span the plasma membrane where they can be activated by extracellular stimuli, more recently an alternative modality has emerged that involves receptors signalling from internal membranes (201). At these internal locations, GPCRs can provide an alternative local source of cAMP. While in cardiac myocytes β1-AR have been reported to be activated and generate cAMP from the plasmalemma, the Golgi apparatus (242) and nuclear membranes (243), β2-AR have been reported to generate cAMP from the plasma membrane and endosomes (244). Consistent with the notion of compartmentalized cAMP signalling, activation of β-ARs localized at internal membranes appears to regulate distinct functions. Data support a model where β1-AR located on the nuclear membrane regulate gene transcription (243), while Golgi localized β1-AR mediate cardiac hypertrophy via a mechanism that involves phosphoinositide hydrolysis, an effect that is not induced by stimulation of β-AR at the cell surface (245). Notably, the internal receptors appear to be activated by norepinephrine entering the cell via an OCT3 organic cation transporter, as blockade of OCT3, or specific blockade of Golgi resident β1-ARs, prevented the norepinephrine-induced hypertrophy (245). A recent study where local generation of cAMP was induced using an optogenetic approach, confirmed that β1-AR signalling at the Golgi and plasma membrane serves distinct functions. When cAMP generated at the plasmalemma increased force of contraction, cAMP generated at the Golgi promoted cardiac myocyte relaxation rate (246). β1-ARs have also been reported to exist at the SR where their activation leads to PKA-mediated phosphorylation of PLB and enhanced cardiac contractility upon catecholamine stimulation. Again, this effect appears to require catecholamine transport into the cell via OCT3 (247).
4. Disruption of cAMP compartmentalisation in cardiac disease
The use of targeted reporters for cAMP has clearly demonstrated that not only different GPCRs generate spatially distinct cAMP signals, but also that the cAMP response induced by the activation of a specific receptor is spatially heterogeneous. For instance, as discussed above, the activation of cardiac β-AR results in a larger cAMP response at the plasmalemma and sarcoplasmic reticulum compared to the cAMP signal generated at the myofilament. This heterogeneity holds functional significance, as imposing a homogeneous cAMP increase through pharmacological inhibition of PDEs leads to a diminished inotropic response to catecholamine stimulation (202) (Fig 7). Given the pervasive involvement of cAMP signaling in cellular functions, it is unsurprising to observe remodeling of the cAMP signalling landscape in pathological conditions (248–251).
In a system where the functional outcome is highly dependent on the specific subcellular location of the molecular components involved, a change in the level of expression of these components is expected to have a profound impact. For example, overexpression of a certain AKAP may potentiate the PKA response at a certain location. Similarly, downregulation or upregulation of a certain PDE isoform will impact cAMP levels in a specific nanodomain which may then trigger inappropriate target phosphorylation.
Genetic polymorphisms in the sequence encoding for several AKAPs have been reported. The nucleotides involved affect the interaction of the AKAP with components of the signalosome they assemble, and these polymorphisms have been associated with increased risk of developing various cardiovascular disease conditions (252).
Polymorphisms in the AKAP helix that anchors PKA have been shown to reduce the RII-binding affinity and determine relocalisation of some polymorphic variants (187). Similarly, polymorphisms in genes encoding for PDE have been associated with atrial fibrillation (253), endothelial dysfunction and stroke (254) and cardiometabolic traits (255).
Multiple transcriptomics studies comparing healthy controls with diseased hearts, both from human and animal models, from a variety of etiologies including ischemic cardiomyopathy (256, 257), heart failure (258), genetic hypertrophic cardiomyopathy (HCM) (259) and rhythm disturbances (260) show up- or down-regulation of multiple components of the cAMP signalling pathway, including ACs, AKAPs, PKA subunits and PDEs, indicating that cardiac disease is associated with a profound rearrangement of local cAMP signalling. These findings are confirmed at the protein level both in rat (261) and human (262) tissue obtained from failing hearts. For example, a chemical proteomics study where a cAMP-bound resin was used to enrich for pathway components, found severe alteration in the composition of AKAP-PKA signalosomes in human failing hearts compared to healthy controls (251).
Disruption of PKA-dependent phosphorylation involving selected PKA targets has been reported in samples from both patients affected by HCM (263) and in a HCM mouse model that carries a truncating mutation with haploinsufficiency of MyBPC (263, 264). In both instances, TnI phosphorylation in response to β-AR stimulation was found to be significantly attenuated while phosphorylation of PLB was unaffected, indicating local disruption of cAMP signalling. Similar results were reported in a rat model of HF induced by chronic exposure to catecholamines, where the cAMP signal generated by sympathetic stimulation was found to be dramatically reduced at the myofilament and, to a lesser extent, at the SR, while the cAMP signal was unaffected at the plasmalemma (202).
Below, we give an overview of the information available on cAMP pathway remodeling in cardiac disease.
4.1. GPCR signalling in cardiac pathophysiology
β-AR compartmentalized signalling has been extensively studied in the context of cardiac disease. β1-AR accounts for about 80% of the β-ARs in the heart, 15%−18% are β2-AR, and the remaining 2%-5% are β3-AR (265). All β-AR subtypes can couple with Gαs and stimulate cAMP synthesis, while β2-AR and β3-AR can also couple with Gαi to inhibit cAMP generation. A negative feed-back loop protects from excessive catecholamine stimulation. This involves phosphorylation of the agonist-bound receptor by G-protein coupled kinases (GRKs), leading to recruitment of β-arrestin to the receptor. This interaction blocks G protein binding and promotes receptor internalization. The recruited β-arrestin can also provide a scaffold for PDE recruitment, further contributing to local quenching of cAMP (228). Data indicate that different β-AR are differently distributed on the plasmalemma and are coupled with distinct signalosomes. While β1-AR are present across the entire membrane and generate a cAMP signal that propagates a long distance from the membrane, β2-AR are localised to caveolin-rich domains and predominantly in T-tubules and the cAMP they generate remains confined to a smaller domain proximal to the T-tubular membrane (223). The distinct compartmentalisation of the cAMP signal has been attributed to the action of PDEs, rather than coupling of β2-AR with Gαi, as inhibition of PDE3 and PDE4 enzymes equalised the cAMP response generated by activation of the two receptors (238). β1-ARs contribute the majority of the inotropic response to catecholamines (266), are coupled to both AC5 and AC6 and their chronic stimulation leads to cardiac myocyte apoptosis, hypertrophy and heart failure (50, 266, 267). By contrast, β2-ARs are predominantly coupled to AC5, their activation results in a smaller inotropic response and have been associated with cardioprotective effects (268, 269) (but see section 4.2.2 below regarding the role of AC5 in stress-induced responses).
Remodelling of the β-AR signalling domains has been described in human and animal models of HF, characterised by altered levels of receptor expression, disrupted coupling with ACs and anomalous cAMP signal propagation (270). Associated with derangement of the T-tubular system and a significant loss of T-tubules associated with HF (271), a broader distribution of β2-ARs to non-tubular membrane has been observed (223). This was accompanied with a more diffuse cAMP signal, similar to that generated by β1-AR (223), which is consistent with earlier observations that β2-AR stimulation in failing human and mouse hearts induces functional effects that are typical β1-AR signalling (272). Interestingly, studies in rats show that restoration of the T-tubular network in conditions that reverse HF also rectifies the distribution of β2-AR and the associated pattern of cAMP propagation (273).
Very sparse information is available on the involvement of other cardiac GPCRs on local modulation of cAMP signals and how this impacts cardiac disease. As sympathetic control of cardiac function occurs concomitantly with fluctuations of multiple other hormones and stimuli, how signalling from other GPCRs integrates and is coordinated with sympathetic regulation in healthy and diseased conditions is an obvious area of investigations where efforts should be focused in the future. Some evidence that GPCRs other than adrenergic receptors also generate compartmentalised pools of cAMP is available. For example, early studies had demonstrated that activation of the Gαs-coupled glucagon-like peptide-1 (GLP-1) receptor leads to elevation of cAMP but has no positive inotropic effect (163). Imaging studies have linked the response to GLP-1 to subcellular compartments distinct from those engaged by catecholamines and to the predominant activation of PKAI rather than PKAII, the latter being the main isoform activated by adrenergic stimulation (113). GLP-1 has been shown to be cardioprotective both in pre-clinical and clinical studies (274, 275) and, although the mechanisms responsible remain to be fully understood, a direct effect of GLP-1 on the heart and involving cAMP-dependent pro-survival signalling has been suggested (276, 277).
A recent study (278) further supports the notion that, by participating in different signalosomes, different GPCRs can mediate deleterious or protective effects on the heart. The data indicate that histamine H2 receptors, together with β1-AR, couple to AC5 to generate cAMP that leads to PKA-dependent phosphorylation of pannexin-1. This promotes ATP release in the extracellular space and activation of a pro-death pathway. Conversely, activation of adenosine A2 receptors, calcitonin-gene-related-peptide-receptor and relaxin-family peptide-1 receptor couple to AC6 to generate cAMP that is extruded from the cell via a proximally located MRP4, and triggers pro-survival signalling (278).
4.2. Adenylyl cyclases in cardiac pathophysiology
While all transmembrane ACs are activated by Gαs proteins, not all ACs are activated by all Gαs-coupled receptors, making the various AC isoforms non-redundant (Table 1). By localizing to defined domains within the plasma membrane, at least in part through partitioning within or outside cholesterol-rich membrane regions (15), ACs can be activated in a receptor-selective manner. In this way they contribute to compartmentalisation of cAMP signalling and specificity of response. For example, the engagement of distinct ACs and the generation of cAMP in distinct compartments has been implicated in the ability of muscarinic M2 receptors to generated cAMP-dependent effects that are both inhibitory and stimulatory (279, 280). Evidence suggests that these opposing effects are achieved via the involvement of both AC5/AC6, which are Gαi inhibited, and AC4, which is not modulated by Gαi but is instead activated by free Gβγ subunits released on activation of Gαi (204, 279). The localization of AC5/AC6 to caveolar domains and of AC4 to extra-caveolar domains (281) would provide compartmentation of the excitatory and inhibitory effects. The inclusion of ACs in multiprotein complexes organized by AKAPs provides further opportunity for isoform-specific regulation.
Table 1. Adenylyl cyclases characterised in cardiac myocytes.
| Gene | Protein name |
Reported selective coupling |
Reported association with cardiac disease |
Refs |
|---|---|---|---|---|
| ADCY1 | AC1 | HCN4 (SAN cells) | SAN dysfunction | (21, 282) |
| ADCY5 | AC5 | AKAP79/150 (T tubules) | Cardiac hypertrophy | (50, 283, 284) |
| ADCY6 | AC6 | AKAP79/150 (outside T tubules) PHLPP2 |
Cardioprotective | (50, 285–287) |
| ADCY8 | AC8 | IP3R (SAN cells) | Cardiac rhythm variability, Hypertrophy fibrosis |
(19, 20, 288) |
| ADCY9 | AC9 | POPDC1, TREK-1 Yotiao, KCNQ1 Hsp20 endosomes |
Long QT | (23, 24, 289–291) |
| ADCY10 | AC10 | ? | Redox damage, Ischemic injury |
(292, 293) |
4.2.1. AC1
AC1 is expressed in SAN cells (21, 22) and genetic deletion of this isoform has been associated with SAN dysfunction. Mechanistic studies suggest that HCN4 channels form functional domains where they specifically couple with AC1 which, via generation of a local pool of cAMP, plays a critical role in mediating the sustained rise in SAN automaticity in response to β-AR (21, 282). In addition, pharmacological studies also suggest that AC1 is involved in the regulation of the pacemaker activity in response to α-AR stimulation, via a mechanism that involves IP3-mediated Ca2+ release (288, 294).
4.2.2. AC5 and AC6
AC5 and AC6 are the main isoforms expressed in ventricular myocytes (50). They both interact with AKAP79/150 (49, 51) but AC5 is enriched in T-tubules where it couples preferentially with β2-ARs while AC6 localises outside T-tubules (50). The PKA anchored at these signalosomes can phosphorylate and inhibit both AC5 and AC6, providing a negative feed-back mechanism that rapidly and locally controls cAMP synthesis (51), while phosphorylation by the AKAP79/150-anchored PKC activates AC5 but inhibits AC6 (15), again enabling differentiation of the downstream response. Although AC5 and AC6 are structurally very similar (25), genetic manipulation studies clearly illustrate the non-redundant properties of these ACs and the importance of their subcellular location. Genetic ablation of AC5 was reported to affect both sympathetic and parasympathetic regulation of ECC (295) and has been associated with cardioprotective effects in conditions of chronic pressure overload, chronic β-AR stimulation and aging (283, 296, 297). AC5 ablation did not show beneficial effects when the pathogenic stressor was delivered via Gαq signalling (298). By contrast, AC6 overexpression, rather than ablation, was shown to be cardioprotective (285, 286). The beneficial effect of cAMP generated by AC6 has been suggested to rely on AC6 inhibition by Gαi, which would limit the cAMP response downstream of β1-ARs activation (299). cAMP-dependent activation of the AC6-associated phosphatase PHLPP2 and enhanced dephosphorylation of Akt/PKB has also been hypothesized (287), although cAMP-independent mechanisms have also been suggested (300).
4.2.3. AC8
AC8 is Ca2+ activated and is expressed in atrial myocytes and SAN cells, where modulation of its activity by IP3R-released Ca2+ has been proposed as a possible trigger for atrial fibrillation (288). Overexpression of AC8 appears to be detrimental to the heart, leading to increased heart rate and decreased heart rate variability, enhanced age-related heart dysfunction, early cardiac myocyte hypertrophy and interstitial fibrosis (19, 20).
4.2.4. AC9
AC9 is also expressed in the SAN and genetic ablation studies support a role for this isoform in the control of heart rate (23, 289). AC9 interacts with the AKAP Yotiao (290, 301), forming a signalosome that controls PKA-dependent phosphorylation of KCNQ1 and modulation of IKs currents, as confirmed by AC9 knock out studies (301). AC9 is recruited to the two-pore domain potassium (K2P) channel TREK-1 (KCNK2)-POPDC1 complex, where local cAMP production by AC9 facilitates the POPDC1 regulation of TREK-1 K+ currents. ADCY9 deletion results in bradycardia and diastolic dysfunction with preserved ejection fraction (23). AC9 may also be involved in the regulation of basal cardiac stress responses, via PKA phosphorylation of heat shock protein 20 (Hsp20), which may be directly associated with the cyclase (23). AC9 has also been reported to localize to internal membranes upon internalization of ligand-bound β1-AR, and to be able to generate cAMP from endosomes (291).
4.2.5. AC10
AC10 has been found to localize to multiple intracellular locations, including the mitochondria, the centrioles and the nucleus (27, 65, 302). As discussed for internal GPCRs, the possibility of producing cAMP from a source distal to the plasma membrane provides an additional modality to supply local PKA- or EPAC-dependent signalosomes, aiding in spatial control of signal propagation (303). AC10 is activated by bicarbonate and, as such, is considered to be responsive to changes in cellular energy metabolism. In the cell, bicarbonate is in dynamic equilibrium with CO2, a waste product of the mitochondrial Krebs cycle that fuels ATP synthesis. Activation of mitochondrial AC10 has been linked to increased mitochondrial ATP production (16). The exact location of AC10 and the pathway triggered by AC10-generated cAMP remains, however, controversial. While earlier studies suggested that AC10 is present in the mitochondrial matrix (28, 65, 304) where it activates local PKA in response to nutrient availability and regulates the electron transport chain (305), other studies challenge this model and suggest that in cardiac myocytes AC10 is located in the mitochondrial intermembrane space where the cAMP it generates activates EPAC and Rap1, leading to increased ATP production in response to nutrient consumption (66). The lack of evidence that the mitochondrial matrix hosts a local subset of PKA (304, 306, 307) supports the latter model and other data show activation of EPAC downstream of AC10, for example for the regulation of fatty acid uptake and oxidation (308) and for activation of glycogenolysis (142), although the latter effect has not been established in cardiac myocytes. Additional work is required to identify the target(s) downstream of EPAC/Rap1 and to clarify the molecular mechanisms involved in the cAMP-dependent modulation of energy metabolism, particularly in view of the fact that mitochondrial AC10 has been implicated in protection from ischemic cardiac injury (292, 293) and damage induced by reactive oxygen species (ROS) (309).
4.3. AKAP-PKA signalosomes in cardiac pathophysiology
The arrangement of signalosomes, where PKA is closely situated in proximity to specific phosphorylation targets, coupled with the ability of the cell to create steep local gradients of cAMP, represents an effective strategy for achieving hormonal specificity of signalling (Fig 4). Compartmentalised cAMP enables the activation of a limited subset of PKA targets, thereby triggering only the relevant cellular functions. The critical importance of PKA anchoring to AKAPs for appropriate information processing and correct functional outcome is clearly illustrated by pathological conditions that are associated with altered signalosome composition. For example, a S1570L point mutation in AKAP9 that alters its association with the KCNQ1 subunit of the IKs potassium channel, leads to defective phosphorylation of the channel and long-QT syndrome (310). In another example, mutations in the gene encoding for the PKA C subunit that affect the interaction of the mutant protein with the R subunits, result in defective C recruitment to AKAP signalosomes. The dispersal of the C subunits away from the anchoring complex (250, 311) or its subcellular redistribution with preferential association with AKAP-anchored RI subunits (312) and the ensuing phosphorylation of inappropriate targets is thought to be responsible for the Cushing’s syndrome phenotype associated with these mutations.
A number of AKAP-dependent signalosomes have been described in cardiac myocytes (Table 2). The list is likely not exhaustive, and future discoveries may reveal additional complexes. While limited information exists for some cardiac signalosomes regarding their role in disease, several have undergone thorough characterization and have been shown to be linked to arrhythmia, cardiac remodeling, and HF.
4.3.1. AKAP1
AKAP1 is a dual-specificity AKAP that localises to the outer mitochondrial membrane and regulates mitochondrial morphology via PKA-dependent phosphorylation of Drp1 (320). The signalosome organized by AKAP1 has been linked to the control of cellular respiration, ROS production, and cell survival (394, 395) and to cardioprotective effects (313, 314). The mechanism appears to involve the inhibition of nuclear translocation of the nuclear factor of activated T-cells (NFATc3), that would otherwise enable hypertrophic gene expression (320). In support of its cardioprotective role, AKAP1 overexpression leads to reduction of cardiac myocyte size and inhibition of isoproterenol-induced hypertrophy (313), while its expression is downregulated in conditions of pressure overload, leading to mitochondrial dysfunction, oxidative stress, and cell death (314). AKAP1 expression has also been reported to result in reduced infarct size and mortality associated with myocardial infarction (396).
4.3.2. AKAP5
The signalosome organized by AKAP79 (in humans) or AKAP150 (in mouse) anchors PKAII to LTCC at the plasma membrane and controls phosphorylation-dependent modulation of the channel ion conductance (397), at least in part through association of AKAP79/150 with the channel subunit Cav1.2 (398, 399). The multiprotein complex organized by this AKAP also includes β-AR, PKA, AC5/AC6, PKC, protein phosphatase calcineurin (PP2B) and caveolin 3 (CAV3) (325). The signalosome is found on T-tubules and disruption of the complex by deletion of the AKAP5 gene alters CAV3 and AC6 localization (325). The interaction with AC5/AC6 has been shown to regulate the cyclase activity by enabling a negative feed-back loop where cAMP production is inhibited by PKA-dependent phosphorylation of the AC (51). Inhibition of PKA in cardiac myocytes reduces calcium currents downstream of β-AR stimulation (397) and cardiac myocytes from AKAP5 knock out mice do not respond to β-adrenergic stimulation (325). Deletion of AKAP5 significantly reduces stimulated Ca2+ transients and Ca2+ sparks as well as phosphorylation of RyR and PLB in response to isoproterenol. Multiple studies have analysed the impact of genetic ablation of AKAP5. AKAP5-/- myocytes show enhanced contraction rate in response to isoproterenol and increased cell size, suggesting a cardioprotective role for this signalosome (323), supported by studies where AKAP5-/- mice were subjected to pressure overload or myocardial infarction (326, 400). By contrast, ablation of AKAP150 ameliorates glucose toxicity and diastolic disfunction in cardiac myocytes from diabetic rats. In this model, overexpression of AKAP150 and enhanced PKC activity at the plasma membrane promote activation of NFκB and NADPH oxidase and generation of ROS, resulting in diabetes-induced cardiovascular injury (401, 402). Genetic deletion of AKAP5 also impacts trafficking of β-AR, suggesting a role for this AKAP in receptor re-sensitisation (323). In atrial myocytes AKAP150-bound PKC has also been associated with regulation of Ca2+ sparklets (403).
4.3.3. AKAP6
The protein encoded by AKAP6, originally named AKAP100, exists in two main isoforms (mAKAPα and mAKAPβ) (404). mAKAPβ is the predominant isoform in the heart. mAKAPβ localizes to the nuclear envelope through interaction with the integral membrane protein nesprin1-α (338). It also interacts with AC5 localised to T-tubules, where the plasma membrane invaginations come in proximity to the nuclear membrane (330), to facilitate cardiac signaling (405, 406). As described for AKAP5, PKA anchoring to the mAKAP signalosome enables a negative feedback loop leading to phosphorylation and inhibition of AC5 (406). mAKAP assembles a number of proteins that are implicated in cardiac myocyte hypertrophy, including protein phosphatases 2A and 2B (PP2A/2B), PDE4D3, hypoxia-inducible factor 1 (HIF1), phospholipase Cε (PLCε), myocyte enhancer factor-2 (MEF2) and p90 ribosomal S6 kinase 3 (RSK3) (168, 333, 336, 340–342). The signalosome also associates indirectly with EPAC1 and the ERK5 and MEK5 mitogen-activated protein kinases via interactions with PDE4D3 (334). Extensive investigations indicate that the main role of the signalosome organized by mAKAP is to control gene expression involved in cardiac remodeling. The mAKAP associated phosphatase calcineurin dephosphorylates the transcription factors NFATc and MEF2, triggering their translocation to the nucleus and leading to expression of pro-hypertrophic genes (331, 341, 407). mAKAP also interacts with PKD, leading to phosphorylation of HDAC4 and activation of hypertrophic genes (331, 336). Deletion of the AKAP6 gene has been shown to be cardioprotective in animal models subjected to pressure overload to induce hypertrophy (331) while anchoring of the E3 ubiquitin ligases to the signalosome with hypoxia-inducible factor 1 (HIF-1α) stabilises HIF-1α, promoting its translocation to the nucleus and inducting pro-survival gene expression (340). A shorter isoform of mAKAP (mAKAPβ) has been reported to associate with the ryanodine receptor (RyR) at the SR, where PKA phosphorylation would enhance opening of RyR and Ca2+ release in the cytosol (124), whereas association with the Na+/Ca2+ exchanger (NCX) at the plasmalemma would result in increased NCX activity and Ca2+ efflux (337, 408).
4.3.4. AKAP7
AKAP7 encodes for AKAP18 and is expressed as at least five isoforms (AKAP18α, AKAP18β, AKAP18γ, AKAP18δ, and AKAP18ε) that vary in length, localization, tissue-specific distribution, and function (170). All isoforms are expressed in the heart. Initial studies found an association of AKAP18 (also called AKAP15) with cardiac LTCC (169, 409). AKAP18α localises to plasma membranes via myristylation and palmitoylation of its N-terminus (169) and interacts with a conserved leucine zipper motif in the LTCC C-terminus (344, 410). Although PKA phosphorylation of Cav1.2 on multiple sites (S1700, T1704, S1928, and S1518, numbering according to the rabbit Cav1.2 sequence with UniProt ID P15381) has been shown to increase channel conductance and contractile force (411–414), as mentioned above the channel activity has recently been shown to relay on PKA-dependent phosphorylation of Rad (121).
AKAP18δ contains a unique targeting sequence for localization to the SR. This signalosome anchors PKA is proximity to SERCA2 and PLN (171) enabling PKA positioning in proximity to PLN. PLN phosphorylation facilitates SERCA2 activity, with impact on cardiac myocyte relaxation and contraction (171). Studies also demonstrate that AKAP18δ scaffolds CAMKII to ryanodine receptors (RyR). Within this signalosome, the N-terminal region of AKAP18δ interacts with and inhibits CaMKII, while binding at the C-terminal region of AKAP18δ enhances CAMKII activity and promotes PLN phosphorylation. The net effect is faster Ca2+ release by RyRs and reuptake into the SR (347).
4.3.5. AKAP9
Yotiao is a short splice variant (250 kDa) encoded by AKAP9 expressed in the heart. It is localised to the plasma membrane where it interacts selectively with AC9 (290, 415) to modulate cardiac repolarization. Yotiao interacts with the KCNQ1 subunit of IKs channel, a critical component for the late phase repolarization of the cardiac action potential in humans (348). This interaction is critical for cardiac repolarization as it involves a leucine zipper motif in the KCNQ1 subunit which is disrupted by a mutation (G589D) associated with long-QT syndrome (348). PKA phosphorylation of KCNQ1 on S27 following sympathetic stimulation accounts for most of the cAMP-dependent modulation of IKs, as evidenced by the observation that a S27D mutation of this residue reconstitutes most of the effects of PKA activation (416, 417). In addition, on adrenergic stimulation PKA also phosphorylates Yotiao on S43. While this phosphorylation does not affect the association of Yotiao with KCNQ1 or channel phosphorylation, it impacts channel activity, indicating that the AKAP functions not only to anchor and recruit PKA but also as an allosteric modulator of the ion channel, through a mechanism that is distinct from PKA (349, 418). In addition to PKA and KCNQ1, Yotiao also scaffolds negative regulators of KCNQ1 phosphorylation, including protein phosphatase 1 (PP1) and PDE4D3 (348, 419–421).
4.3.6. AKAP10
D-AKAP2 is a multi-domain protein that anchors both PKAI and PKAII and is involved in the late stages of endocytosis (357). In addition to the PKA binding domain, D-AKAP2 also includes two tandem regulators of G protein signalling (RGS)−like homology domains that interact with two small GTPases, Rab4, and Rab11 (357) and a PSD-95/DlgA/ZO-1(PDZ)-binding motif, through which the AKAP interacts with the multi-PDZ domain proteins PDZK1 and NHERF-1 (354, 422). Mutations in mouse models (352) or single nucleotide polymorphism (351, 353) in the PKA binding domain of D-AKAP2 result in cardiac rhythm anomalies and sudden cardiac death and may be associated with cardiac dysfunction in the ageing population (423).
4.3.7. AKAP12
AKAP12 encodes for the protein Gravin which organizes a signalosome that includes PKA, PKC, G-protein-receptor kinase 2 (GRK2), β-arrestin and PP2B (358, 424). Gravin anchors PKA in proximity to β2-ARs by interacting to the receptor C-terminal tail (425). Mice lacking Gravin show increased contractility both in the absence and in the presence of β-AR activation (360, 361). Consistently, cardiac overexpression of Gravin is associated with defective contraction and cardiac dilatation and patients with end-stage HF have upregulated AKAP12 both at the RNA and protein level (426). Interestingly, genetic deletion of Gravin has no impact on PLB phosphorylation or SERCA activity and cardiac overexpression does not affect TnI phosphorylation, suggesting a local effect of the signalosome organized by Gravin, possibly in the regulation of the desensitization/resensitization cycle of β2-AR (360, 361).
4.3.8. AKAP13
AKAP-Lbc is encoded by the AKAP13 gene and is highly expressed in the heart (363). It scaffolds multiple signalling enzymes in addition to exerting its activity as a guanine nucleotide exchange factor (GEF) that selectively activates the small molecular weight GTPases RhoA and RhoC (363, 427). Extensive characterisation of this anchoring protein uncovered a complex role in the coordination of multiple distinct signalling pathways that contribute to cardiac myocyte adaptation to stress stimuli and to cardioprotection (362). AKAP-Lbc forms a p38-activating transduction unit that includes p38α and its upstream activators protein kinase Nα (PKN α), mixed lineage kinase-like mitogen-activated protein triple kinase (MLTK), and mitogen-activated protein kinase kinase 3 (MKK3) (374). This pathway leads to induction of mammalian target of rapamycin (mTOR) and increase in protein synthesis (374). AKAP-Lbc also interacts with PKD1 and PKCη. This pathway responds to α1-AR and ET1-R activation, PKC activation and phosphorylation of anchored PKD1. Activated PKD1 is then released from the signalosome upon phosphorylation by local PKA. Once released, PKD1 goes on to phosphorylate targets that are involved in regulation of gene transcription. These include HDAC5, leading to HDAC5 nuclear export, de-repression of MEF2 and activation of pro-hypertrophic gene transcription (365), and CREB and cofilin2-phosphatase slingshot-1L (SSH1L), leading to protection from apoptosis and resistance to doxorubicin toxicity (428). AKAP-Lbc also facilitates PKA-dependent phosphorylation of Hsp20, another component of the signalosome (429). This phosphorylation appears to be required for the anti-apoptotic effects of Hsp20 (430), which involves inhibition of apoptosis signal-regulating kinase 1 (Ask1) and Bax, and the preservation of the pro-survival activity of Akt1/PKB. Hsp20 phosphorylation by PKA is regulated by recruitment to the signalosome of PDE4 isoforms which reduce cAMP levels locally (431). Upon chronic β-adrenergic stimulation, cAMP levels rise in cardiac myocytes and overcome the hydrolyzing capacity of the PDE, leading to Hsp20 phosphorylation (432).
4.3.9. SPHKAP
SKIP is a PKAI specific AKAP (188) that interacts with and inhibits the activity of sphingosine kinase 1 (SPHK1) (375), an enzyme activated by growth factors and leading to the phosphorylation of sphingosine to generate sphingosine 1 phosphate (S1P). S1P is released at sites of tissue injury affecting cellular responses through activation of GPCRs that promote cell survival following in vivo myocardial ischemia-reperfusion (433), suggesting a role for this signalosome in the regulation of the cardiac stress response. SKIP localises to the mitochondrial intermembrane space where it associates with the protein ChChd3 (175), a target of PKA phosphorylation and a component of the mitochondrial contact site and organizing system (MICOS) complex, which has also been shown to interact with PDE2A2 (434).
4.4. Phosphodiesterases in cardiac pathophysiology
As discussed above, PDEs play a key role in cAMP compartmentalisation. Some PDEs are integrated in signalling complexes organized by AKAPs in combination with ACs, enabling a local feed-back loop regulation of cAMP levels (15, 435). In multiple cases, PDEs have been shown to be functionally coupled with specific targets or localized at certain subcellular locations via AKAP-independent interactions (Table 3). Not always information on the specific PDE isoform involved is available. The identification of the PDE isoform associated with a certain signalosome can be challenging. Functional studies rely on genetic ablation that often affect multiple PDE isoforms transcribed from the same gene and pharmacological inhibitors discriminate between PDE families but not between isoforms within a family. In addition, antibodies available for immunolocalization studies and biochemical characterisation not always are very specific or able to unequivocally discriminate PDE isoforms with small differences in molecular weight.
Table 3. Phosphodiesterases characterised in cardiac myocytes.
| PDE family |
PDE genes |
Reported cardiac splice variants |
Reported subcellular localisation |
Reported association with cardiac disease |
Refs |
|---|---|---|---|---|---|
| PDE1 | PDE1A PDE1B PDE1C |
PDE1A PDE1C1 PDE1C3 |
Soluble fractions Nucleus Z- line and M- line |
HF, cardiac hypertrophy, cardiac fibrosis | (436–441) |
| PDE2 | PDE2A | PDE2A1 PDE2A2 |
Soluble fraction Mitochondria (MOM, MIM, matrix) Plasma membrane Intercalated disk Nuclear envelope SR |
HF, hypertrophy, arrhythmia | (28, 98, 442–448) |
| PDE3 | PDE3A PDE3B |
PDE3A1 PDE3A2 PDE3B |
Microsomal fraction, SR, Nucleus Cytoplasm |
HF, hypertension, ischemia/reperfusion, apoptosis, cardioprotection |
(80, 81, 101, 232, 449–458) |
| PDE4 | PDE4A PDE4B PDE4C PDE4D |
PDE4D3 PDE4D5 PDE4D8 PDE4D9 |
Plasma membrane SR nucleus |
HF, cardiac hypertrophy, arrhythmia | (216, 452, 459, 460) |
| PDE8 | PDE8A PDE8B |
PDE8A1 PDE8A2 PDE8B |
Plasma membrane, Cytosol Mitochondria |
Atrial fibrillation | (461, 462) |
| PDE10 | PDE10A | PDE10A2 | Golgi Cytosol |
HF, cardiac hypertrophy, Dox-induced cardiotoxicity | (91, 463, 464) |
4.4.1. PDE1
PDE1 is a dual cAMP/cGMP esterase and binding of the two cyclic nucleotides is mutually competitive. PDE1 is expressed as three different isoforms: PDE1A and 1B have a higher affinity for cGMP than cAMP, whereas PDE1C has same affinity for the two nucleotides (97). While PDE1A is the dominant cardiac isoform in mice, PDE1C predominates in humans (437).
Multiple studies show that enhanced expression of PDE1 is detrimental to the heart whereas its inhibition is beneficial (465–467). PDE1A expression was found to be upregulated in human failing hearts and animal models of HF (439). A role for PDE1 in the regulation of a localised pool of cAMP has been described in cardiac myocytes from multiple species. In studies using the PDE1 selective inhibitor ITI-214, a positive inotropic and lusitropic effect was observed in the absence of an increase in the Ca2+ transient or myofilament protein phosphorylation. The data suggest that PDE1 may operate in a compartment that is independent of β-AR signalling and is coupled instead to adenosine A2B receptor signalling (440, 468). Genetic ablation of PDE1C was found to be cardioprotective in a model of pressure overload, where lack of this isoform was associated with reduced hypertrophy and apoptosis, an effect that was linked to cAMP/PKA and PI3K/AKT pathways (439). PDE1 inhibition or PDE1A gene silencing was also found to reduce cardiac myocyte hypertrophy induced by phenylephrine. These findings suggest PDE1A upregulation may contribute to the hypertrophy induced by α1-AR agonists via suppression of cAMP/PKA signalling (438). PDE1A is upregulated in cardiac fibroblasts when exposed to pro-fibrotic stimuli (469). The data suggests that this specific isoform plays a pivotal role in regulating localized pools of cAMP that trigger the activation of the EPAC1-Rap1 pathway. Furthermore, PDE1A is implicated in the modulation of cGMP, resulting in the activation of protein kinase G (PKG), with effects on collagen synthesis within the cardiac fibroblasts (469).
4.4.2. PDE2
PDE2A is a dual-substrate enzyme expressed as three splice variants (PDE2A1, PDE2A2 and PDE2A3). These have distinct subcellular localisation conferred by a unique sequence at their N terminus (98, 470). PDE2A has been shown to regulate cAMP pools generated by β1-AR and β2-AR, but also to control cGMP resulting from activation of β3-AR and NO synthesis (471). In addition to modulation of ECC, PDE2A2 also plays a role in the regulation morphology and function of cardiac mitochondria. By regulating a local pool of cAMP, this isoform controls PKA-dependent phosphorylation of the pro-fission protein Drp1, promoting mitochondria fragmentation (443). In addition, PDE2A2 localises to the mitochondria intermembrane space where it is integrated in the MICOS complex. At this location, PDE2A2 controls the PKA-dependent phosphorylation of MIC60 with impact on Parkin recruitment and mitophagy (434). PDE2A has also been implicated in the regulation of a cAMP pool in the mitochondrial matrix, with effects on ATP production (28).
PDE2A expression is increased in failing human hearts and in animal models of HF (251, 445, 446). Work from different groups suggests that high levels of PDE2A may contribute to the pathology. Inhibition of PDE2A was shown to reduce cardiac remodeling in a rat models of pressure overload and chronic catecholamine stimulation (472, 473). In addition, pharmacological inhibition or expression of a catalytically inactive PDE2A2 enzyme was found to normalize cardiac release of noradrenaline from stellate ganglia neurons from rat and humans with sympathetic hyperactivity, an effect that could contribute to cardioprotection in HF (474). In contrast, it has been proposed that elevated PDE2A expression may be cardioprotective based on the observation that transgenic mice overexpressing this enzyme show blunted response to β-AR stimulation, reduced hypertrophy and attenuated propensity to arrhythmic events (447, 448).
4.4.3. PDE3
PDE3 constitutes the majority of the cAMP-hydrolysing activity in human myocardium (475) and PDE3 inhibitors are approved for the treatment of acute, refractory HF (476). Two genes encode for PDE3, PDE3A and PDE3B, both of which are expressed in the heart, although PDE3A is more abundant. PDE3 binds with high affinity and hydrolyses both cAMP and cGMP; however, as the Vmax for cGMP hydrolysis is about 10-fold slower than for cAMP, cGMP acts effectively as a competitive inhibitor of cAMP degradation (477, 478). PDE3 isoforms have been implicated in the regulation of ECC via modulation of LTCC activity (479), regulation of phospholemman phosphorylation and control the N+/K+-ATPase (480) and modulation of Ca2+ reuptake in the SR (80, 81).
PDE3B has been reported to localize to T tubules in CAV3-rich areas and in proximity to mitochondria (456). Genetic ablation of PDE3B has been associated with protection from ischemic damage but had no impact on cardiac myocyte contractility (81, 456).
Two of the three PDE3A isoforms (PDE3A1 and PDE3A2) are expressed in cardiac myocytes and gene deletion studies show that PDE3A is responsible for the positive inotropic effects of PDE3 inhibitors (81). PDE3A isoforms differ in their N terminus, which determines their subcellular localization, which includes microsomal fraction, SR, nucleus and cytosol (101, 232, 450–452). PDE3A1 is known to be part of a signalosome organized by AKAP18 at the SR membrane (80).
Several studies report PDE3A downregulation in failing hearts, both in humans and animal models (454, 481), while other studies find no change (453, 482). In animal studies, genetic ablation or inhibition of PDE3 also leads to contradictory results. For example, infarct size was shown to be reduced by both PDE3 pharmacological inhibition and PDE3A gene ablation (483) and ventricular remodelling was found to be reduced in a model of pressure overload (484). By contrast, other studies found that PDE3 is cardioprotective. PDE3 inhibition was reported to worsen the cardiac remodelling induced chronic by β-AR stimulation (485) and to inhibit the hypertrophy induced by EPAC-mediated activation of PLCβ (486). In addition, evidence suggests that cardiac-specific overexpression of PDE3A1 protects from ischemia reperfusion (487), and downregulation of PDE3A promotes the transcription of ICER, promoting apoptosis (455). A cardioprotective role for PDE3A is also supported by the finding that patients carrying a gain of function mutation in the PDE3A gene responsible for a condition known as hypertension with brachydactyly (HTNB) do not develop cardiac hypertrophy or HF, despite the severe chronic hypertension, a phenotype that is recapitulated in a rat models carrying the same mutation (458, 488). The mechanism responsible for the cardioprotective effect of these mutations remains to be established. However, a possible modality is suggested by recent studies demonstrating that PDE3A2 is part of a multiprotein complex localized to the nucleus and that includes the regulators of transcription SMAD4 and HDAC1. At this location, the PDE3A2 hydrolytic activity maintains the local cAMP concentration below the activation threshold of a local PKA. As a result, PKA-dependent phosphorylation of HDAC1 is minimized, resulting in reduced histone deacetylation and pro-hypertrophic genes transcription is silenced (232). A hyperactive PDE3A2 at this signalosome may be able to shield the local PKA targets from inappropriate phosphorylation resulting from excessive adrenergic stimulation associated with the chronic hypertensive state, whereas a PDE3A2 with normal hydrolytic activity may be overwhelmed in those conditions.
Further studies will be necessary to work out the exact and apparently varied roles of PDE3A isoforms in cardiac pathophysiology. The contrasting results reported in the literature may reflect the fact that the different PDE3A isoforms localize to distinct subcellular sites where they are involved in different interactions and mediate different functional effects; the dominant outcome may then depend on other concurrent conditions that contribute to the resulting phenotype. In vitro studies where catalytically inactive mutants of PDE3A1 or PDE3A2 were overexpressed in cardiac myocytes chronically treated with β-AR agonists support this notion. These mutants act as dominant negative constructs and displace the endogenous, active enzyme from its subcellular localization sites (232, 472). Expression of these mutants demonstrated that while overexpression of inactive PDE3A1 induces hypertrophy, overexpression of PDE3A2 inhibits the hypertrophy resulting from chronic catecholamine stimulation, suggesting that the two isoforms control cAMP levels in different domains with opposing effects on cardiac myocyte hypertrophic growth (232).
4.4.4. PDE4
PDE4 selectively hydrolyses cAMP and is expressed as more than 20 different isoforms from four genes (PDE4A-PDE4D). In the heart, isoforms belonging to the PDE4A, PDE4B and PDE4D subfamilies are expressed (459) and localise at various sites, including the plasma membrane, where they contribute to regulate the phosphorylation of β-ARs and LTCC; at the SR, where they regulate RyR2, PLB and SERCA2a, and at the myofilaments, where their exact involvement in the regulation of specific targets remains to be established.
Multiple PDE4D variants interact at the plasma membrane with β1-AR and β2-AR subtypes, either directly or via β-arrestin, modulating receptor desensitization (489–491). PDE4D5 recruitment to β2-AR by β-arrestin was shown to prevent the hypertrophy induced by β2-AR activation through a pathway that involves inhibition of EPAC1 and CaMKII (492). At the SR, PDE4D3 has been reported to form a complex with the RYR2. This association was found to be reduced in human failing hearts and in mouse model of pressure overload (493, 494). The consequent enhanced phosphorylation of the RYR was proposed to lead to Ca2+ leak from the SR and susceptibility to arrhythmia, specifically in response to β2-AR stimulation (493, 494). At the SR PDE4D is also in a complex with SERCA2:PLB (495), an association that is affected in cardiac hypertrophy (216). PDE4D isoforms have also been implicated in determining heterogeneities in cAMP breakdown in different regions of the mouse heart, with faster hydrolytic activity at the apex compared to the base. These heterogeneities drive nonuniform changes in the duration of the action potential and repolarization kinetics. Interestingly, these differences were more evident in female hearts, suggesting a possible link with sex-dependent differences in arrhythmogenic activity (496).
PDE4B isoforms have been suggested to be involved in the regulation of β-AR-dependent stimulation of LTCC activity, supported by the observation that PDE4B KO mice show increased susceptibility to arrhythmia (460) and a recent study using a mouse model of type 2 diabetes and HF with preserved ejection fraction (HFpEF), reported a loss of PDE4 activity selectively at the SERCA/PLB complex, leading to exaggerated PLB phosphorylation in response to β2-AR stimulation (497).
Localization of PDE4 at the nuclear envelop occurs via interaction with mAKAP and data indicate that it plays a role in the control of gene expression, hypertrophy and apoptosis through regulation of both PKA and EPAC activation (168, 498, 499).
4.4.5. PDE8
PDE8 is a high-affinity, cAMP-specific PDE encoded by two genes, PDE8A and PDE8B (500) and transcribed as at least nine splice variants. Both genes are expressed in the human heart (462). PDE8A has been reported to have a predominant cytosolic distribution in human atrial cells (462) and to be localized predominantly at mitochondria in other cell types (501, 502). A prominent PDE8 activity has also been reported at the outer membrane of mitochondria in mouse ventricular myocytes, an activity that was significantly enhanced in cardiac myocytes isolated from a Duchenne Muscular Dystrophy mouse model (503). By contrast, PDE8B localises to the plasma membrane in human atrial cells (462). Genetic ablation of PDE8A in mice affects ECC, with increased basal Ca2+ sparks frequency, higher Ca2+ transients and increased LTCC current in response to β-AR stimulation (461), which however does not result in a cardiac phenotype in vivo. PDE8A expression has been shown to be elevated in the atrium of patients affected by atrial fibrillation (260) and a recent study reported upregulation of PDE8B isoforms in patients affected by persistent, chronic atrial fibrillation (462). Mechanistic studies support direct interaction of PDE8B with the Cav1.2α1c subunit of the LTCC. The enhanced PDE8B hydrolytic activity would result in enhanced local degradation of cAMP and reduced calcium current and abbreviated action potential duration, leading to persistent atrial fibrillation (462).
In vitro studies have also implicated PDE8 in the formation of a transient signal termination complex that involves the interaction of PDE8 with RIα. The RIα:PDE8 complex shows higher PDE catalysis than PDE8 alone and facilitates cAMP hydrolysis through translocation of cAMP from RIα directly to the PDE8 hydrolysis site through a channel that is formed across the R:PDE interface (504, 505). This mechanism would facilitate processive hydrolysis of bursts of cAMP, thereby leading to rapid PKA signalosome reset to the inactive holoenzyme state. Whether this mechanism also takes place in intact cells remains to be established.
4.4.6. PDE10
PDE10 hydrolyses both cAMP and cGMP. While PDE10 is present at low levels in healthy hearts recently its expression has been reported to be upregulated in HF, both in humans and mouse models (463). Consistently, PDE10 inhibition was shown to reduce cardiac hypertrophy and remodelling both in vitro, in myocyte treated with angiotensin II, phenylephrine or isoprenaline, and in vivo, in models of cardiac dysfunction triggered by pressure overload (463). In another study, PDE10 was found to contribute to doxorubicin-induced cardiotoxicity by modulating both cAMP/PKA and cGMP/PKG pathways and inhibition of PDE10 was cardioprotective in a nude mice model with implanted ovarian cancer xenografts and treated with doxorubicin (464).
4.5. EPAC in cardiac pathophysiology
EPAC signalling has been involved in cardiac disease of various aetiologies through activation of pathways that are largely independent of PKA (506). As PKA, EPAC1 can localise to multiple subcellular compartments, including the nucleus, the plasmalemma and mitochondria, and can mediate cellular responses in a spatially regulated fashion. For example, interaction of EPAC1 with β-arrestin was shown to be required for CaMKII-dependent phosphorylation of HDAC4, HDAC4 extrusion from the nucleus and consequent activation of the pro-hypertrophic transcription factor MEF2, resulting in hypertrophy (492, 507). This signalling pathway could be activated by β-ARs localized to the Golgi and involves an EPAC1/PLC signalosome organised by mAKAP at the nuclear envelope (245, 486). Conversely, EPAC2 localised to T-tubules has been implicated in ventricular arrhythmia through modulation of Ca2+ release from RyR (508). Both activation (509) and inhibition (510) of mitochondrial EPAC1 has been reported to be protective against ischemia/reperfusion injury. Evidence suggests that mitochondrial EPAC1 responds to cAMP generated by a local AC10 and that the signalling of this signalosome is involved in the regulation of ATP production in response to increased functional demands (66). In ischemic conditions, the same signalosome appears to be involved in mitochondrial Ca2+ overload and ROS production (510).
4.6. cAMP signalling landscape in cardiac disease
Apart from specific mutations that directly impact protein-protein interactions within a signalosome, it is currently uncertain whether the profound remodelling of cAMP nanodomain signalling observed in diseased hearts is a driver in the pathogenesis of the cardiac condition or whether it is an adaptive or maladaptive response, or simply an epiphenomenon. As discussed above, there are many contradictory reports on how individual pathway components are affected and currently we don’t have a clear picture of the cAMP signalling landscape associated with different cardiac pathologies. While the lack of consensus in the published data can be ascribed to differences in the models and methodologies used, in the stage of the disease investigated and, for studies using human tissue, in the heterogeneity inherent to those samples, it is nevertheless clear that reorganisation of compartmentalised cAMP signalling pinpoints heart disease. The remodeling of the T-tubular system observed in failing cardiac myocytes and the consequent redistribution of β-AR from tubular to non-tubular plasmalemma is expected to dramatically affect the receptor molecular neighborhood with significant impact on signal propagation. The rearrangement of the cytoskeleton (511, 512) and of other intracellular structures (513, 514) observed in cardiac disease is bound to similarly affect the spatial distribution and appropriate coupling of pathway components downstream to the receptors and the localization of signalosomes. Irrespective of the extent to which cAMP nanodomain remodeling contributes to the development of heart disease, given the role of cAMP in regulating cardiac force generation, cardiac reserve and exercise tolerance— all factors directly influencing patients’ quality of life—it becomes paramount to meticulously examine this remodeling process, as it may unveil novel therapeutic avenues for intervention.
5. Harnessing cAMP nanodomain signalling for targeted therapeutic intervention
The cAMP signaling pathway has long been at the center of efforts to develop drugs to treat cardiac disease. The introduction of β-blockers in the management of HF has been transformative, significantly enhancing clinical outcomes and firmly establishing these medications as indispensable elements in treatment protocols. Despite this progress, the 5-year mortality rate post-diagnosis remains alarmingly high, exceeding 50% for all types of HF patients (515, 516). Notably, while β-blockers are effective in the treatment of HF with reduce ejection fraction (HFrEF), their benefit in patients suffering from HFpEF remains uncertain (517). Clearly, there is a pressing need for identification and implementation of novel and improved treatment strategies. While drugs targeting GPCRs affect cells throughout the body and the entire signaling cascade downstream of the receptor, directing interventions toward specific cAMP nanodomains offers the potential for cell-type specificity and subcellular precision. This approach holds promise for heightened efficacy and minimized side effects (69, 96, 178, 221).
The use of PDE inhibitors has been investigated as a treatment for HF. Milrinone is a PDE3 inhibitor approved for the treatment of acute congestive HF. In addition to increasing cardiac contractility and lusitropy, it is also a potent vasodilator and inhibitor of platelet aggregation and it enhances cardiac output in the short term (476). However, chronic administration to patients with HF has been linked to adverse effects including cardiac remodeling, increased incidence of arrhythmias, and sudden death, (518), ultimately outweighing its short-term benefits (519). A polymorphism in the human PDE3A promoter that affects PDE3A gene transcription in a cAMP−PKA dependent fashion may explain the variable tolerance to milrinone observed in patients with HFrEF (520). However, the detrimental effects of milrinone may also be attributed to its lack of selectivity among different PDE3 isoforms and the consequent elevation of cAMP levels in various subcellular compartments where the different PDE3 isoforms are localized, contributing to the diverse range of responses observed in patients. To mitigate the undesired effects of PDE3 inhibition, an international multicenter trial explored combined therapy using low doses of another PDE3 inhibitor, enoximone, in conjunction with β-blockers. Despite showing no adverse effects on survival or other clinical outcomes, the trial failed to demonstrate a significant advantage of the combination therapy (521).
PDE1 is also under investigation as a potential therapeutic target for patients with HFrEF, drawing from promising results observed in animal models. In these models, the PDE1 inhibitor ITI-214 showed positive effects including increased contractility, improved relaxation, and arterial vasodilation (440). In a phase I/II randomized single rising dose study, ITI-214 demonstrated overall good tolerance and acute reductions in arterial vascular resistance. Additionally, it enhanced cardiac contractility, cardiac output, and heart rate (522).
The structural similarities among PDE isoforms within a family present a challenge in designing traditional small-molecule inhibitors that target a specific isoform and, as multiple isoforms from the same PDE family are typically expressed in cardiac myocytes and are embedded in a variety of signalosomes, pharmacological inhibition is unlikely to provide the desired specificity. As an alternative, overexpression of a PDE isoform has been suggested as an approach that aims for more selective and efficient modulation of cAMP levels (447, 523), although these studies remain at the preclinical stage. A similar approach has been used to selectively manipulate cAMP synthesis. Based on investigations in animal models showing benefits of cardiac AC6 overexpression and the observation that AC6 is reduced in human failing hearts (524), a randomized, double blind clinical trial including 59 patients evaluated the benefit of delivering the AC6 gene via intracoronary adenoviral injection and found significantly improved left ventricular function without increased adverse events (525). Although gene manipulation of signalosome components can attain a level of selectivity, particularly when employing cell type-specific promoters and viral vectors with cardiac-specific tropism - e.g. adeno-associated virus serotype 9, AAV9-based vectors (526) - regulating transgene expression levels may be challenging, and mislocalisation of the overexpressed protein could compromise or nullify the intended benefits.
A more sophisticated approach to fully capitalize on the advantages offered by cAMP signaling compartmentalization involves selectively displacing an individual protein component from a specific signalosome. This method offers the potential for exceptional precision while minimizing disruption to the broader intracellular signaling machinery. Its implementation requires peptides or small molecules capable of selectively disrupting protein-protein interactions that anchor specific components to the targeted signalosome. Research into this approach has been conducted using an animal model of cardiac hypertrophy induced by pressure overload. In this study, intraperitoneal injections of a peptide disrupting the interaction between PDE4 and HSP20 demonstrated mitigation of hypertrophy-induced cardiac remodeling (432, 527). A similar strategy was effective also in the context of cancer cell spreading (528, 529).
A significant hurdle in leveraging compartmentalised cAMP signaling for the development of novel therapeutics to treat cardiac disease lies in the limited number of nanodomains that have been characterized so far. A recent study conducted using rat cardiac myocytes employed PDE isoform-affinity purification mass spectrometry alongside the analysis of PDE family-dependent phosphoproteomes to attempt a non-biased, at scale, identification of cAMP nanodomains under the control of specific PDEs. This study revealed that the involvement of individual PDE isoforms in protein complexes localized across various subcellular structures is much more extensive than previously anticipated (232). To exemplify this complexity, Fig 8 shows the subcellular localization of potential PDE3A1-regulated cAMP nanodomains as define in (232). The data were obtained by concurrent analysis of the PDE3A1-specific interactome and of the phosphopeptides selectively upregulated on inhibition of PDE3 isoforms in cardiac myocytes stimulated with a β-AR agonist. The two independent datasets were combined using protein network analysis using STRING database, which generates multiple subnetworks each comprising both PDE3A1 interactors and PDE3 inhibitor-dependent phosphotargets. As the two datasets are generated using independent approaches, the identified subnetworks represent, with high-confidence, potential novel cAMP nanodomains under the control of PDE3A1 (232). While some of the signalosome emerging from this analysis include well established PKA targets, many others have not been associated before with cAMP/PKA signalling. Notably, and in line with prior research (530), only approximately 40% of the phosphopeptides upregulated upon PDE3 inhibition contained a PKA motif. This finding is consistent with the observation that PDE3A1 participates in multiple signalosomes that involve kinases other than PKA (group 16-21 in Fig 8), establishing a framework for extensive cross-talk and integration with other signaling pathways.
Figure 8. Map of the PDE3A1-dependent cAMP nanodomains in rat cardiac myocytes.
The cartoon shows the PDE3A1-dependent cAMP nanodomains distributed across various subcellular structures. This depiction is based on data obtained by the integrated STRING network analysis of two orthogonal datasets − the PDE3A1 interactome and the PDE3-dependent phosphoproteome − generated by mass spectrometry analysis of rat cardiac myocyte samples stimulated with isoproterenol (232). The output of this analysis is multiple protein interaction subnetworks, or modulomes, each including one or more PDE3A1 interactors and one or more phosphotargets. Each subnetwork is given a number and a letter following the number denotes subnetworks that can be grouped within a certain location or are involved in the same functional activity. Previously known PKA targets identified in this study are highlighted in red. The protein components for each group, identified by their gene name, are listed below. Interactors are in bold, phosphotargets are underscored.
1: dyad − includes the L-type calcium channel (LTCC) modulome (1A; CACNA1C, CACNB2, AHNAK), the RyR2 modulome (1B; ASPH, CASQ2, JPH2, HRC, RYR2, SPEG) and the Na+, K+-ATPase (NKA) and Na+, Ca2+ exchanger or NCX modulome (1C; ATP1A1, ATP1B1, and SLC8A1).
2: SERCA2a (sarco(endo)plasmic reticulum calcium-ATPase) modulome, (ATP2A2, HAX1, PLN, SRL). Involvement of PDE3A in this complex had been previously reported (80, 81).
3: Na+, H+ exchanger NHE1 modulome (SLC9A1), also previously reported (531).
4: caveolae modulome (CAV1, CAV2, CAV3, CAVIN 1).
5: costamere modulome (ITGB1, LAMB1, LAMB2, LAMC1, DMD, SGCA, SNTB1).
6: sarcomere - comprises myofibril modulome (6A; MYH6, MYH7, MYL2, MYL3, TTN, LMOD2, MYBPC3, PPP1R12A, TNNI3) and Z disc modulome (6B; ACTN1, ACTN2, ACTN4, DES, FLNC, MYOZ2, OBSCN).
7: intercalated disc, comprises adherens junction modulome (7A; CTNNB1, MYZAP, XIRP1, BVES (POPDC1), CTNNA1, CTNNA3, CXADR, TJP1, XIRP2) and gap junction modulome (7B; EZR (erzin), GJA1 (Cx43), TJP1 (ZO-1)).
8: nucleus, comprises nuclear transport machinery modulome (8A; KPNA1, KPNA2, KPNB1, KPNA3, NUP98), spliceosome modulome (8B; DHX9, EFTUD2, HNRNPA0, HNRNPA2B1, HNRNPA1, HNRNPAB, HNRNPC, HNRNPM, HNRNPU, LUC7L2, MATR3, PRPF8, RBMX, SNRPF, U2AF2, SRSF3, SRSF7, YBX1, PRPF31, RBM20, SERBP1, SF1, SRPK1, SRPK3, SRRM1, THRAP3, ZRANB2), nucleolus modulome (8C; FBL and NOP58 of C/D box SnoRNP and NPM1), and a modulome including proteins that regulate gene expression (8C; SMAD4, HDAC1, MEF2A, together with histones H1, H2A, H2B, H3, H4).
9: protein synthesis machineries at the ER, including ribosomal proteins modulome (9A; RPL3-6, 7a, 9, 12-15, 18, 19, 22, 23a, 32, 35a, RPLP0, RPLP2, RPS14, 18, 26, 27l, EEF1B2, EIF3A, EIF3C, EIF3G, EIF3S6IP, ABCF1, EEF1D, EIF4EBP1, EIF4G1, EIF5B, NACA), transport into the ER modulome (9B; SEC63, HSPA5), N-linked glycosylation oligosaccharyl-transferase complex, or OST, modulome (9C; DDOST, RPN1, RPN2, STT3A, STT3B), and ER chaperone system for glycoproteins or calnexin-calreticulin (CNX-CRT) cycle modulome (9D; CANX, CAL).
10: mitochondria-associated membrane (MAM), including ER membrane complex (EMC) modulome (10A; EMC1, EMC2, EMC3, EMC10) and (10B; FIS1, VDAC1, VDAC2) and MICOS-MIB (mitochondrial contact site - mitochondrial intermembrane space bridging) modulome (10C; APOOL, CHCHD3, IMMT, RHOT2, SAMM50).
11: energy metabolism regulation, including fatty acid mitochondria uptake modulome (11A; CPT1A, CPT1B, SLC27A4 (FATP4), SLC25A20 (CAC), ACSL1, and lipid droplet (LD) protein, PLIN5) and electron transport chain (11B, ATP5F1A, ATP5F1B-D, ATP5PB, ATP5PD, ATP5MG, ATP5PO, ATP5PF, COX15, COX5B, MT-CO2, COX10, NDUFA13, NDUFA9, NDUFS7-8, UQCRQ, UQCRC1, UQCRH).
12: retrograde transport (DCTN2 and DYNC1LI1).
13 autophagosome, comprising the chaperone-assisted selective autophagy (CASA) complex (BAG3, HSPA8, HSPB8) and cargo protein SQSTM1.
14: HSP90 chaperone (CDC37, HSP90AA1, HSPA1A)
15: 14-3-3 scaffold proteins (YWHAB, YWHAG, YWHAH, YWHAQ), which are known interactors of PDE3A (82).
16 -21: kinases, including AMPK (16; PRKAA2, PRKAB1), SIK3 (17), PKC (18; PRKCE), STK39 (19), S6KI (20; RPS6KB1), Ca2+/calmodulin (CaM)-dependent protein kinase (CaMK) II delta (21; CALM1, CAMK2D).
22-23: protein phosphatase 2 (PP2) complexes, (22; PPP2R1A and PPP2R5A) (previously reported (532) and PP2A-STRIPAK (23; PPP2R1A, SLMAP, STRN, STRN3, STRN4).
This figure was generated using Biorender.
This type of analysis offers a glimpse into the extraordinary complexity of the intracellular cAMP signaling network. While the data collected in this study provide a snapshot of the cAMP nanodomains regulated by PDE3A1 in a healthy myocyte responding to β-AR stimulation, the global cAMP signaling landscape is influenced by multiple other PDEs, is anticipated to be dynamic, stimulus-dependent, and subject to remodeling in diseased conditions. These findings are in sharp contrast with the typical description of the cAMP signaling pathway as it is represented, for example, in the KEGG database. While this canonical representation may appear complex enough (Fig 1), it fails to acknowledge the presence of multiple isoforms for many of the pathway protein components and the fact that each individual protein is expressed in multiple copies, each of which has the potential to interact with distinct partners, forming multiple signalosomes with unique localization, composition, and function. While determining the full map of the cAMP nanodomains may seem a daunting task, tackling this complexity will provide a rich framework for mechanistic studies and potentially identify actionable targets for disease treatment.
6. Open questions and outlook
As highlighted above, acknowledging the compartmentalization of cAMP signaling holds significant implications for understanding disease mechanisms and exploring avenues for more effective therapeutic strategies. Nevertheless, numerous unresolved questions require additional investigation to fully capitalize on the potential that this intricate system offers. For example, it will be important to understand how different GPCRs precisely couple with the intracellular molecular machinery to ensure cAMP levels rise only in the appropriate compartment, especially when the signalosomes activated by cAMP are distant from the cell surface where the receptors are located. Similarly, how the regulation of PDEs is orchestrated to achieve specific cAMP signals only in the appropriate nano-space, and how the multiple signalosomes coordinate their signalling activity, remains to be established. Subcellular spatial proteomic approaches, such as proximity tagging (533) or biochemical fractionation followed by quantitative mass spectrometry, combined with machine learning applications (534) will help dissect the complexity of the system and define how cAMP nanodomain signalling is impacted by the dynamic molecular context within which each signalosome operates (535).
Alongside the current approach of assessing effects on overall cellular cAMP levels, it will be important to develop innovative tools and assays for monitoring local cAMP signalling in a high-throughput manner, so that they can be integrated into drug discovery pipelines. This will enable early evaluation of whether drug candidates influence cAMP dynamics within nanodomains—an effect that is overlooked by tests solely measuring global changes in the second messenger. Implementing such methods may help mitigate current attrition rates in drug development.
To date, only a scant number of cAMP nanodomains have been characterized in cardiac myocytes, especially in human cells. Progress will require substantial efforts to delineate a comprehensive map of the subcellular cAMP nanodomain network and to gain a detailed understanding of how this signaling landscape changes in response to various stressors. Defining how individual nanodomains link to specific cellular functions and define how signal transmission is impacted by the local molecular milieu will help identify potential actionable points for therapeutic intervention. In summary, unraveling the intricacies of cAMP nanodomain organization presents a formidable challenge, yet it is essential for understanding how cardiac myocytes work and how cardiac disease develops. This endeavor will greatly enhance our capacity to infer disease mechanisms from genome-wide association studies and omics data and will lay the groundwork for developing therapeutic approaches that precisely target cAMP signaling with nanoscale subcellular precision.
Clinical Highlights.
The pursuit of effective cardiac disease treatments has long revolved around the cAMP signaling pathway. The advent of β-blockers has revolutionised heart failure management, significantly improving clinical outcomes. However, despite these strides, mortality rates post-diagnosis remain disturbingly high. Thus, there is an urgent need to identify and implement novel, enhanced therapeutic strategies. While targeting β-adrenergic receptors impacts cells systemically and affects the entire downstream intracellular signaling cascades, an alternative approach is to target individual cAMP nanodomains. This approach holds potential for cell-specificity and the option to intervene with subcellular precision, offering prospects for improved efficacy and minimized side effects.
Acknowledgements
M.Z acknowledges funding by the British Heart Foundation (RG/17/6/32944, RE/18/3/34214) and the Oxford NIHR Biomedical Research Centre. D.K. is supported by the Thai Ministry of Higher Education, Science, Research and Innovation under the PMU-B program 1 (RGNS63-182).
Footnotes
Contributions
MZ and DK reviewed the literature, wrote the manuscript, and generated the figures.
References
- 1.Sutherland EW, Rall TW. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J Biol Chem. 1958;232:1077–1091. [PubMed] [Google Scholar]
- 2.Fimia GM, Sassone-Corsi P. Cyclic AMP signalling. J Cell Sci. 2001;114:1971–1972. doi: 10.1242/jcs.114.11.1971. [DOI] [PubMed] [Google Scholar]
- 3.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 4.Heng BC, Aubel D, Fussenegger M. An overview of the diverse roles of G-protein coupled receptors (GPCRs) in the pathophysiology of various human diseases. Biotechnol Adv. 2013;31:1676–1694. doi: 10.1016/j.biotechadv.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 5.Sriram K, Insel PA. G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol Pharmacol. 2018;93:251–258. doi: 10.1124/mol.117.111062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.London E, Stratakis CA. The regulation of PKA signaling in obesity and in the maintenance of metabolic health. Pharmacol Ther. 2022;237:108113. doi: 10.1016/j.pharmthera.2022.108113. [DOI] [PubMed] [Google Scholar]
- 7.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahuja M, Jha A, Maleth J, Park S, Muallem S. cAMP and Ca(2)(+) signaling in secretory epithelia: crosstalk and synergism. Cell Calcium. 2014;55:385–393. doi: 10.1016/j.ceca.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serezani CH, Ballinger MN, Aronoff DM, Peters-Golden M. Cyclic AMP: master regulator of innate immune cell function. Am J Respir Cell Mol Biol. 2008;39:127–132. doi: 10.1165/rcmb.2008-0091TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Triposkiadis F, Karayannis G, Giamouzis G, Skoularigis J, Louridas G, Butler J. The sympathetic nervous system in heart failure physiology, pathophysiology, and clinical implications. J Am Coll Cardiol. 2009;54:1747–1762. doi: 10.1016/j.jacc.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci U S A. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan K, Gao LN, Cui YL, Zhang Y, Zhou X. The cyclic AMP signaling pathway: Exploring targets for successful drug discovery (Review. Mol Med Rep. 2016;13:3715–3723. doi: 10.3892/mmr.2016.5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilman AG. G proteins and dual control of adenylate cyclase. Cell. 1984;36:577–579. doi: 10.1016/0092-8674(84)90336-2. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Li B, Chen WD, Wang YD. Role of G-protein coupled receptors in cardiovascular diseases. Front Cardiovasc Med. 2023;10:1130312. doi: 10.3389/fcvm.2023.1130312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrom KF, LaVigne JE, Brust TF, Seifert R, Dessauer CW, Watts VJ, Ostrom RS. Physiological roles of mammalian transmembrane adenylyl cyclase isoforms. Physiol Rev. 2022;102:815–857. doi: 10.1152/physrev.00013.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buck J, Sinclair ML, Schapal L, Cann MJ, Levin LR. Cytosolic adenylyl cyclase defines a unique signaling molecule in mammals. Proc Natl Acad Sci U S A. 1999;96:79–84. doi: 10.1073/pnas.96.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Liu D, Varin A, Nicolas V, Courilleau D, Mateo P, Caubere C, Rouet P, Gomez AM, Vandecasteele G, Fischmeister R, et al. A cardiac mitochondrial cAMP signaling pathway regulates calcium accumulation, permeability transition and cell death. Cell Death Dis. 2016;7:e2198. doi: 10.1038/cddis.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T, Brown MJ. Differential expression of adenylyl cyclase subtypes in human cardiovascular system. Mol Cell Endocrinol. 2004;223:55–62. doi: 10.1016/j.mce.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Moen JM, Matt MG, Ramirez C, Tarasov KV, Chakir K, Tarasova YS, Lukyanenko Y, Tsutsui K, Monfredi O, Morrell CH, Tagirova S, et al. Overexpression of a Neuronal Type Adenylyl Cyclase (Type 8) in Sinoatrial Node Markedly Impacts Heart Rate and Rhythm. Front Neurosci. 2019;13:615. doi: 10.3389/fnins.2019.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mougenot N, Mika D, Czibik G, Marcos E, Abid S, Houssaini A, Vallin B, Guellich A, Mehel H, Sawaki D, Vandecasteele G, et al. Cardiac adenylyl cyclase overexpression precipitates and aggravates age-related myocardial dysfunction. Cardiovasc Res. 2019;115:1778–1790. doi: 10.1093/cvr/cvy306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattick P, Parrington J, Odia E, Simpson A, Collins T, Terrar D. Ca2+-stimulated adenylyl cyclase isoform AC1 is preferentially expressed in guinea-pig sino-atrial node cells and modulates the I(f) pacemaker current. J Physiol. 2007;582:1195–1203. doi: 10.1113/jphysiol.2007.133439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Younes A, Lyashkov AE, Graham D, Sheydina A, Volkova MV, Mitsak M, Vinogradova TM, Lukyanenko YO, Li Y, Ruknudin AM, Boheler KR, et al. Ca(2+)-stimulated basal adenylyl cyclase activity localization in membrane lipid microdomains of cardiac sinoatrial nodal pacemaker cells. J Biol Chem. 2008;283:14461–14468. doi: 10.1074/jbc.M707540200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Baldwin TA, Wang Y, Subramaniam J, Carbajal AG, Brand CS, Cunha SR, Dessauer CW. Loss of type 9 adenylyl cyclase triggers reduced phosphorylation of Hsp20 and diastolic dysfunction. Sci Rep. 2017;7:5522. doi: 10.1038/s41598-017-05816-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rautureau Y, Deschambault V, Higgins ME, Rivas D, Mecteau M, Geoffroy P, Miquel G, Uy K, Sanchez R, Lavoie V, Brand G, et al. ADCY9 (Adenylate Cyclase Type 9) Inactivation Protects From Atherosclerosis Only in the Absence of CETP (Cholesteryl Ester Transfer Protein. Circulation. 2018;138:1677–1692. doi: 10.1161/CIRCULATIONAHA.117.031134. [DOI] [PubMed] [Google Scholar]
- 25.Dessauer CW, Watts VJ, Ostrom RS, Conti M, Dove S, Seifert R. International Union of Basic and Clinical Pharmacology. CI. Structures and Small Molecule Modulators of Mammalian Adenylyl Cyclases. Pharmacol Rev. 2017;69:93–139. doi: 10.1124/pr.116.013078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hacker BM, Tomlinson JE, Wayman GA, Sultana R, Chan G, Villacres E, Disteche C, Storm DR. Cloning, chromosomal mapping, and regulatory properties of the human type 9 adenylyl cyclase (ADCY9. Genomics. 1998;50:97–104. doi: 10.1006/geno.1998.5293. [DOI] [PubMed] [Google Scholar]
- 27.Zippin JH, Chen Y, Nahirney P, Kamenetsky M, Wuttke MS, Fischman DA, Levin LR, Buck J. Compartmentalization of bicarbonate-sensitive adenylyl cyclase in distinct signaling microdomains. Faseb J. 2003;17:82–84. doi: 10.1096/fj.02-0598fje. [DOI] [PubMed] [Google Scholar]
- 28.Di Benedetto G, Scalzotto E, Mongillo M, Pozzan T. Mitochondrial Ca(2)(+) uptake induces cyclic AMP generation in the matrix and modulates organelle ATP levels. Cell Metab. 2013;17:965–975. doi: 10.1016/j.cmet.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Taussig R, Iniguez-Lluhi JA, Gilman AG. Inhibition of adenylyl cyclase by Gi alpha. Science. 1993;261:218–221. doi: 10.1126/science.8327893. [DOI] [PubMed] [Google Scholar]
- 30.Taussig R, Quarmby LM, Gilman AG. Regulation of purified type I and type II adenylylcyclases by G protein beta gamma subunits. J Biol Chem. 1993;268:9–12. [PubMed] [Google Scholar]
- 31.Tang WJ, Krupinski J, Gilman AG. Expression and characterization of calmodulin-activated (type I) adenylylcyclase. J Biol Chem. 1991;266:8595–8603. [PubMed] [Google Scholar]
- 32.Taussig R, Tang WJ, Hepler JR, Gilman AG. Distinct patterns of bidirectional regulation of mammalian adenylyl cyclases. J Biol Chem. 1994;269:6093–6100. [PubMed] [Google Scholar]
- 33.Yoshimura M, Cooper DM. Cloning and expression of a Ca(2+)-inhibitable adenylyl cyclase from NCB-20 cells. Proc Natl Acad Sci U S A. 1992;89:6716–6720. doi: 10.1073/pnas.89.15.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masada N, Schaks S, Jackson SE, Sinz A, Cooper DM. Distinct mechanisms of calmodulin binding and regulation of adenylyl cyclases 1 and 8. Biochemistry. 2012;51:7917–7929. doi: 10.1021/bi300646y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wayman GA, Wei J, Wong S, Storm DR. Regulation of type I adenylyl cyclase by calmodulin kinase IV in vivo. Mol Cell Biol. 1996;16:6075–6082. doi: 10.1128/mcb.16.11.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scholich K, Pierre S, Patel TB. Protein associated with Myc (PAM) is a potent inhibitor of adenylyl cyclases. J Biol Chem. 2001;276:47583–47589. doi: 10.1074/jbc.M107816200. [DOI] [PubMed] [Google Scholar]
- 37.Willoughby D, Halls ML, Everett KL, Ciruela A, Skroblin P, Klussmann E, Cooper DM. A key phosphorylation site in AC8 mediates regulation of Ca(2+)-dependent cAMP dynamics by an AC8-AKAP79-PKA signalling complex. J Cell Sci. 2012;125:5850–5859. doi: 10.1242/jcs.111427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steiner D, Saya D, Schallmach E, Simonds WF, Vogel Z. Adenylyl cyclase type-VIII activity is regulated by G(betagamma) subunits. Cell Signal. 2006;18:62–68. doi: 10.1016/j.cellsig.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 39.Crossthwaite AJ, Ciruela A, Rayner TF, Cooper DM. A direct interaction between the N terminus of adenylyl cyclase AC8 and the catalytic subunit of protein phosphatase 2A. Mol Pharmacol. 2006;69:608–617. doi: 10.1124/mol.105.018275. [DOI] [PubMed] [Google Scholar]
- 40.Gu C, Cooper DM. Calmodulin-binding sites on adenylyl cyclase type VIII. J Biol Chem. 1999;274:8012–8021. doi: 10.1074/jbc.274.12.8012. [DOI] [PubMed] [Google Scholar]
- 41.Tang WJ, Gilman AG. Type-specific regulation of adenylyl cyclase by G protein beta gamma subunits. Science. 1991;254:1500–1503. doi: 10.1126/science.1962211. [DOI] [PubMed] [Google Scholar]
- 42.Zimmermann G, Taussig R. Protein kinase C alters the responsiveness of adenylyl cyclases to G protein alpha and betagamma subunits. J Biol Chem. 1996;271:27161–27166. doi: 10.1074/jbc.271.43.27161. [DOI] [PubMed] [Google Scholar]
- 43.Gao BN, Gilman AG. Cloning and expression of a widely distributed (type IV) adenylyl cyclase. Proc Natl Acad Sci U S A. 1991;88:10178–10182. doi: 10.1073/pnas.88.22.10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Federman AD, Conklin BR, Schrader KA, Reed RR, Bourne HR. Hormonal stimulation of adenylyl cyclase through Gi-protein beta gamma subunits. Nature. 1992;356:159–161. doi: 10.1038/356159a0. [DOI] [PubMed] [Google Scholar]
- 45.Gao X, Sadana R, Dessauer CW, Patel TB. Conditional stimulation of type V and VI adenylyl cyclases by G protein betagamma subunits. J Biol Chem. 2007;282:294–302. doi: 10.1074/jbc.M607522200. [DOI] [PubMed] [Google Scholar]
- 46.Brand CS, Sadana R, Malik S, Smrcka AV, Dessauer CW. Adenylyl Cyclase 5 Regulation by Gbetagamma Involves Isoform-Specific Use of Multiple Interaction Sites. Mol Pharmacol. 2015;88:758–767. doi: 10.1124/mol.115.099556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwami G, Kawabe J, Ebina T, Cannon PJ, Homcy CJ, Ishikawa Y. Regulation of adenylyl cyclase by protein kinase A. J Biol Chem. 1995;270:12481–12484. doi: 10.1074/jbc.270.21.12481. [DOI] [PubMed] [Google Scholar]
- 48.Heinick A, Husser X, Himmler K, Kirchhefer U, Nunes F, Schulte JS, Seidl MD, Rolfes C, Dedman JR, Kaetzel MA, Gerke V, et al. Annexin A4 is a novel direct regulator of adenylyl cyclase type 5. FASEB J. 2015;29:3773–3787. doi: 10.1096/fj.14-269837. [DOI] [PubMed] [Google Scholar]
- 49.Efendiev R, Samelson BK, Nguyen BT, Phatarpekar PV, Baameur F, Scott JD, Dessauer CW. AKAP79 interacts with multiple adenylyl cyclase (AC) isoforms and scaffolds AC5 and-6 to alpha-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) receptors. J Biol Chem. 285:14450–14458. doi: 10.1074/jbc.M110.109769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Timofeyev V, Myers RE, Kim HJ, Woltz RL, Sirish P, Heiserman JP, Li N, Singapuri A, Tang T, Yarov-Yarovoy V, Yamoah EN, et al. Adenylyl cyclase subtype-specific compartmentalization: differential regulation of L-type Ca2+ current in ventricular myocytes. Circ Res. 2013;112:1567–1576. doi: 10.1161/CIRCRESAHA.112.300370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bauman AL, Soughayer J, Nguyen BT, Willoughby D, Carnegie GK, Wong W, Hoshi N, Langeberg LK, Cooper DM, Dessauer CW, Scott JD. Dynamic regulation of cAMP synthesis through anchored PKA-adenylyl cyclase V/VI complexes. Mol Cell. 2006;23:925–931. doi: 10.1016/j.molcel.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salim S, Sinnarajah S, Kehrl JH, Dessauer CW. Identification of RGS2 and type V adenylyl cyclase interaction sites. J Biol Chem. 2003;278:15842–15849. doi: 10.1074/jbc.M210663200. [DOI] [PubMed] [Google Scholar]
- 53.Hill J, Howlett A, Klein C. Nitric oxide selectively inhibits adenylyl cyclase isoforms 5 and 6. Cell Signal. 2000;12:233–237. doi: 10.1016/s0898-6568(99)00082-0. [DOI] [PubMed] [Google Scholar]
- 54.Kawabe J, Iwami G, Ebina T, Ohno S, Katada T, Ueda Y, Homcy CJ, Ishikawa Y. Differential activation of adenylyl cyclase by protein kinase C isoenzymes. J Biol Chem. 1994;269:16554–16558. [PubMed] [Google Scholar]
- 55.Ding Q, Gros R, Gray ID, Taussig R, Ferguson SS, Feldman RD. Raf kinase activation of adenylyl cyclases: isoform-selective regulation. Mol Pharmacol. 2004;66:921–928. [PubMed] [Google Scholar]
- 56.Katsushika S, Chen L, Kawabe J, Nilakantan R, Halnon NJ, Homcy CJ, Ishikawa Y. Cloning and characterization of a sixth adenylyl cyclase isoform: types V and VI constitute a subgroup within the mammalian adenylyl cyclase family. Proc Natl Acad Sci U S A. 1992;89:8774–8778. doi: 10.1073/pnas.89.18.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas JM, Hoffman BB. Isoform-specific sensitization of adenylyl cyclase activity by prior activation of inhibitory receptors: role of beta gamma subunits in transducing enhanced activity of the type VI isoform. Mol Pharmacol. 1996;49:907–914. [PubMed] [Google Scholar]
- 58.Lai HL, Yang TH, Messing RO, Ching YH, Lin SC, Chern Y. Protein kinase C inhibits adenylyl cyclase type VI activity during desensitization of the A2a-adenosine receptor-mediated cAMP response. J Biol Chem. 1997;272:4970–4977. doi: 10.1074/jbc.272.8.4970. [DOI] [PubMed] [Google Scholar]
- 59.Chen Y, Harry A, Li J, Smit MJ, Bai X, Magnusson R, Pieroni JP, Weng G, Iyengar R. Adenylyl cyclase 6 is selectively regulated by protein kinase A phosphorylation in a region involved in Galphas stimulation. Proc Natl Acad Sci U S A. 1997;94:14100–14104. doi: 10.1073/pnas.94.25.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin TH, Lai HL, Kao YY, Sun CN, Hwang MJ, Chern Y. Protein kinase C inhibits type VI adenylyl cyclase by phosphorylating the regulatory N domain and two catalytic C1 and C2 domains. J Biol Chem. 2002;277:15721–15728. doi: 10.1074/jbc.M111537200. [DOI] [PubMed] [Google Scholar]
- 61.Antoni FA. The chilling of adenylyl cyclase 9 and its translational potential. Cell Signal. 2020;70:109589. doi: 10.1016/j.cellsig.2020.109589. [DOI] [PubMed] [Google Scholar]
- 62.Palvolgyi A, Simpson J, Bodnar I, Biro J, Palkovits M, Radovits T, Skehel P, Antoni FA. Auto-inhibition of adenylyl cyclase 9 (AC9) by an isoform-specific motif in the carboxyl-terminal region. Cell Signal. 2018;51:266–275. doi: 10.1016/j.cellsig.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 63.Baldwin TA, Li Y, Brand CS, Watts VJ, Dessauer CW. Insights into the Regulatory Properties of Human Adenylyl Cyclase Type 9. Mol Pharmacol. 2019;95:349–360. doi: 10.1124/mol.118.114595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antoni FA, Palkovits M, Simpson J, Smith SM, Leitch AL, Rosie R, Fink G, Paterson JM. Ca2+/calcineurin-inhibited adenylyl cyclase, highly abundant in forebrain regions, is important for learning and memory. J Neurosci. 1998;18:9650–9661. doi: 10.1523/JNEUROSCI.18-23-09650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Acin-Perez R, Salazar E, Kamenetsky M, Buck J, Levin LR, Manfredi G. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell Metab. 2009;9:265–276. doi: 10.1016/j.cmet.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greiser M, Karbowski M, Kaplan AD, Coleman AK, Verhoeven N, Mannella CA, Lederer WJ, Boyman L. Calcium and bicarbonate signaling pathways have pivotal, resonating roles in matching ATP production to demand. Elife. 2023;12 doi: 10.7554/eLife.84204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sassi Y, Abi-Gerges A, Fauconnier J, Mougenot N, Reiken S, Haghighi K, Kranias EG, Marks AR, Lacampagne A, Engelhardt S, Hatem SN, et al. Regulation of cAMP homeostasis by the efflux protein MRP4 in cardiac myocytes. FASEB J. 2012;26:1009–1017. doi: 10.1096/fj.11-194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bolger GB. The PDE-Opathies: Diverse Phenotypes Produced by a Functionally Related Multigene Family. Trends Genet. 2021;37:669–681. doi: 10.1016/j.tig.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 69.Baillie GS, Tejeda GS, Kelly MP. Therapeutic targeting of 3’,5’-cyclic nucleotide phosphodiesterases: inhibition and beyond. Nat Rev Drug Discov. 2019;18:770–796. doi: 10.1038/s41573-019-0033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kamel R, Leroy J, Vandecasteele G, Fischmeister R. Cyclic nucleotide phosphodiesterases as therapeutic targets in cardiac hypertrophy and heart failure. Nat Rev Cardiol. 2023;20:90–108. doi: 10.1038/s41569-022-00756-z. [DOI] [PubMed] [Google Scholar]
- 71.Keravis T, Lugnier C. Cyclic nucleotide phosphodiesterase (PDE) isozymes as targets of the intracellular signalling network: benefits of PDE inhibitors in various diseases and perspectives for future therapeutic developments. Br J Pharmacol. 2012;165:1288–1305. doi: 10.1111/j.1476-5381.2011.01729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martinez SE, Beavo JA, Hol WG. GAF Domains: Two-Billion-Year-Old Molecular Switches that Bind Cyclic Nucleotides. Mol Intervent. 2002;2:317–323. doi: 10.1124/mi.2.5.317. [DOI] [PubMed] [Google Scholar]
- 73.Jager R, Russwurm C, Schwede F, Genieser HG, Koesling D, Russwurm M. Activation of PDE10 and PDE11 phosphodiesterases. J Biol Chem. 2012;287:1210–1219. doi: 10.1074/jbc.M111.263806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sonnenburg WK, Seger D, Kwak KS, Huang J, Charbonneau H, Beavo JA. Identification of inhibitory and calmodulin-binding domains of the PDE1A1 and PDE1A2 calmodulin-stimulated cyclic nucleotide phosphodiesterases. J Biol Chem. 1995;270:30989–31000. doi: 10.1074/jbc.270.52.30989. [DOI] [PubMed] [Google Scholar]
- 75.Richter W, Conti M. Dimerization of the type 4 cAMP-specific phosphodiesterases is mediated by the upstream conserved regions (UCRs. J Biol Chem. 2002;277:40212–40221. doi: 10.1074/jbc.M203585200. [DOI] [PubMed] [Google Scholar]
- 76.Cedervall P, Aulabaugh A, Geoghegan KF, McLellan TJ, Pandit J. Engineered stabilization and structural analysis of the autoinhibited conformation of PDE4. Proc Natl Acad Sci U S A. 2015;112:E1414–1422. doi: 10.1073/pnas.1419906112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kobayashi T, Gamanuma M, Sasaki T, Yamashita Y, Yuasa K, Kotera J, Omori K. Molecular comparison of rat cyclic nucleotide phosphodiesterase 8 family: unique expression of PDE8B in rat brain. Gene. 2003;319:21–31. doi: 10.1016/s0378-1119(03)00809-6. [DOI] [PubMed] [Google Scholar]
- 78.Florio VA, Sonnenburg WK, Johnson R, Kwak KS, Jensen GS, Walsh KA, Beavo JA. Phosphorylation of the 61-kDa calmodulin-stimulated cyclic nucleotide phosphodiesterase at serine 120 reduces its affinity for calmodulin. Biochemistry. 1994;33:8948–8954. doi: 10.1021/bi00196a012. [DOI] [PubMed] [Google Scholar]
- 79.Sharma RK, Wang JH. Calmodulin and Ca2+-dependent phosphorylation and dephosphorylation of 63-kDa subunit-containing bovine brain calmodulin-stimulated cyclic nucleotide phosphodiesterase isozyme. J Biol Chem. 1986;261:1322–1328. [PubMed] [Google Scholar]
- 80.Ahmad F, Shen W, Vandeput F, Szabo-Fresnais N, Krall J, Degerman E, Goetz F, Klussmann E, Movsesian M, Manganiello V. Regulation of sarcoplasmic reticulum Ca2+ ATPase 2 (SERCA2) activity by phosphodiesterase 3A (PDE3A) in human myocardium: phosphorylation-dependent interaction of PDE3A1 with SERCA2. J Biol Chem. 2015;290:6763–6776. doi: 10.1074/jbc.M115.638585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Beca S, Ahmad F, Shen W, Liu J, Makary S, Polidovitch N, Sun J, Hockman S, Chung YW, Movsesian M, Murphy E, et al. Phosphodiesterase type 3A regulates basal myocardial contractility through interacting with sarcoplasmic reticulum calcium ATPase type 2a signaling complexes in mouse heart. Circ Res. 2013;112:289–297. doi: 10.1161/CIRCRESAHA.111.300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vandeput F, Szabo-Fresnais N, Ahmad F, Kho C, Lee A, Krall J, Dunlop A, Hazel MW, Wohlschlegel JA, Hajjar RJ, Houslay MD, et al. Selective regulation of cyclic nucleotide phosphodiesterase PDE3A isoforms. Proc Natl Acad Sci U S A. 2013;110:19778–19783. doi: 10.1073/pnas.1305427110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Han SJ, Vaccari S, Nedachi T, Andersen CB, Kovacina KS, Roth RA, Conti M. Protein kinase B/Akt phosphorylation of PDE3A and its role in mammalian oocyte maturation. EMBO J. 2006;25:5716–5725. doi: 10.1038/sj.emboj.7601431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beard MB, Olsen AE, Jones RE, Erdogan S, Houslay MD, Bolger GB. UCR1 and UCR2 domains unique to the cAMP-specific phosphodiesterase family form a discrete module via electrostatic interactions. The Journal of biological chemistry. 2000;275:10349–10358. doi: 10.1074/jbc.275.14.10349. [DOI] [PubMed] [Google Scholar]
- 85.Mika D, Richter W, Conti M. A CaMKII/PDE4D negative feedback regulates cAMP signaling. Proc Natl Acad Sci U S A. 2015;112:2023–2028. doi: 10.1073/pnas.1419992112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.MacKenzie SJ, Baillie GS, McPhee I, MacKenzie C, Seamons R, McSorley T, Millen J, Beard MB, van Heeke G, Houslay MD. Long PDE4 cAMP specific phosphodiesterases are activated by protein kinase A-mediated phosphorylation of a single serine residue in Upstream Conserved Region 1 (UCR1. Br J Pharmacol. 2002;136:421–433. doi: 10.1038/sj.bjp.0704743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hoffmann R, Baillie GS, MacKenzie SJ, Yarwood SJ, Houslay MD. The MAP kinase ERK2 inhibits the cyclic AMP-specific phosphodiesterase HSPDE4D3 by phosphorylating it at Ser579. EMBO J. 1999;18:893–903. doi: 10.1093/emboj/18.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carlisle Michel JJ, Dodge KL, Wong W, Mayer NC, Langeberg LK, Scott JD. PKA-phosphorylation of PDE4D3 facilitates recruitment of the mAKAP signalling complex. Biochem J. 2004;381:587–592. doi: 10.1042/BJ20040846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brown KM, Lee LC, Findlay JE, Day JP, Baillie GS. Cyclic AMP-specific phosphodiesterase, PDE8A1, is activated by protein kinase A-mediated phosphorylation. FEBS Lett. 2012;586:1631–1637. doi: 10.1016/j.febslet.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 90.Kotera J, Fujishige K, Yuasa K, Omori K. Characterization and phosphorylation of PDE10A2, a novel alternative splice variant of human phosphodiesterase that hydrolyzes cAMP and cGMP. Biochemical and biophysical research communications. 1999;261:551–557. doi: 10.1006/bbrc.1999.1013. [DOI] [PubMed] [Google Scholar]
- 91.Kotera J, Sasaki T, Kobayashi T, Fujishige K, Yamashita Y, Omori K. Subcellular localization of cyclic nucleotide phosphodiesterase type 10A variants, and alteration of the localization by cAMP-dependent protein kinase-dependent phosphorylation. J Biol Chem. 2004;279:4366–4375. doi: 10.1074/jbc.M308471200. [DOI] [PubMed] [Google Scholar]
- 92.Cai Y, Huang G, Ma L, Dong L, Chen S, Shen X, Zhang S, Xue R, Sun D, Zhang S. Smurf2, an E3 ubiquitin ligase, interacts with PDE4B and attenuates liver fibrosis through miR-132 mediated CTGF inhibition. Biochim Biophys Acta Mol Cell Res. 2018;1865:297–308. doi: 10.1016/j.bbamcr.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 93.Huo Z, Ye JC, Chen J, Lin X, Zhou ZN, Xu XR, Li CM, Qi M, Liang D, Liu Y, Li J. Prolyl hydroxylase domain protein 2 regulates the intracellular cyclic AMP level in cardiomyocytes through its interaction with phosphodiesterase 4D. Biochemical and biophysical research communications. 2012;427:73–79. doi: 10.1016/j.bbrc.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 94.Li X, Vadrevu S, Dunlop A, Day J, Advant N, Troeger J, Klussmann E, Jaffrey E, Hay RT, Adams DR, Houslay MD, et al. Selective SUMO modification of cAMP-specific phosphodiesterase-4D5 (PDE4D5) regulates the functional consequences of phosphorylation by PKA and ERK. Biochem J. 2010;428:55–65. doi: 10.1042/BJ20091672. [DOI] [PubMed] [Google Scholar]
- 95.Charych EI, Jiang LX, Lo F, Sullivan K, Brandon NJ. Interplay of palmitoylation and phosphorylation in the trafficking and localization of phosphodiesterase 10A: implications for the treatment of schizophrenia. J Neurosci. 2010;30:9027–9037. doi: 10.1523/JNEUROSCI.1635-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fu Q, Wang Y, Yan C, Xiang YK. Phosphodiesterase in heart and vessels: from physiology to diseases. Physiol Rev. 2024;104:765–834. doi: 10.1152/physrev.00015.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev. 2006;58:488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- 98.Acin-Perez R, Russwurm M, Gunnewig K, Gertz M, Zoidl G, Ramos L, Buck J, Levin LR, Rassow J, Manfredi G, Steegborn C. A phosphodiesterase 2A isoform localized to mitochondria regulates respiration. J Biol Chem. 2011;286:30423–30432. doi: 10.1074/jbc.M111.266379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Palmer D, Jimmo SL, Raymond DR, Wilson LS, Carter RL, Maurice DH. Protein kinase A phosphorylation of human phosphodiesterase 3B promotes 14-3-3 protein binding and inhibits phosphatase-catalyzed inactivation. J Biol Chem. 2007;282:9411–9419. doi: 10.1074/jbc.M606936200. [DOI] [PubMed] [Google Scholar]
- 100.Wilson LS, Baillie GS, Pritchard LM, Umana B, Terrin A, Zaccolo M, Houslay MD, Maurice DH. A phosphodiesterase 3B-based signaling complex integrates exchange protein activated by cAMP 1 and phosphatidylinositol 3-kinase signals in human arterial endothelial cells. J Biol Chem. 2011;286:16285–16296. doi: 10.1074/jbc.M110.217026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kenan Y, Murata T, Shakur Y, Degerman E, Manganiello VC. Functions of the N-terminal region of cyclic nucleotide phosphodiesterase 3 (PDE 3) isoforms. J Biol Chem. 2000;275:12331–12338. doi: 10.1074/jbc.275.16.12331. [DOI] [PubMed] [Google Scholar]
- 102.Hunter RW, Mackintosh C, Hers I. Protein kinase C-mediated phosphorylation and activation of PDE3A regulate cAMP levels in human platelets. J Biol Chem. 2009;284:12339–12348. doi: 10.1074/jbc.M807536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maass PG, Aydin A, Luft FC, Schachterle C, Weise A, Stricker S, Lindschau C, Vaegler M, Qadri F, Toka HR, Schulz H, et al. PDE3A mutations cause autosomal dominant hypertension with brachydactyly. Nat Genet. 2015;47:647–653. doi: 10.1038/ng.3302. [DOI] [PubMed] [Google Scholar]
- 104.Khalil JS, Law R, Raslan Z, Cheah LT, Hindle MS, Aburima AA, Kearney MT, Naseem KM. Protein Kinase A Regulates Platelet Phosphodiesterase 3A through an A-Kinase Anchoring Protein Dependent Manner. Cells. 2024;13 doi: 10.3390/cells13131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rybalkin SD, Hinds TR, Beavo JA. Enzyme assays for cGMP hydrolyzing phosphodiesterases. Methods Mol Biol. 2013;1020:51–62. doi: 10.1007/978-1-62703-459-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bolger GB, McCahill A, Huston E, Cheung YF, McSorley T, Baillie GS, Houslay MD. The unique amino-terminal region of the PDE4D5 cAMP phosphodiesterase isoform confers preferential interaction with beta-arrestins. J Biol Chem. 2003;278:49230–49238. doi: 10.1074/jbc.M303772200. [DOI] [PubMed] [Google Scholar]
- 107.Brown KM, Day JP, Huston E, Zimmermann B, Hampel K, Christian F, Romano D, Terhzaz S, Lee LC, Willis MJ, Morton DB, et al. Phosphodiesterase-8A binds to and regulates Raf-1 kinase. Proc Natl Acad Sci U S A. 2013;110:E1533–1542. doi: 10.1073/pnas.1303004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mukherjee S, Roy S, Mukherjee S, Harikishore A, Bhunia A, Mandal AK. 14-3-3 interaction with phosphodiesterase 8A sustains PKA signaling and downregulates the MAPK pathway. J Biol Chem. 2024;300:105725. doi: 10.1016/j.jbc.2024.105725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gamanuma M, Yuasa K, Sasaki T, Sakurai N, Kotera J, Omori K. Comparison of enzymatic characterization and gene organization of cyclic nucleotide phosphodiesterase 8 family in humans. Cell Signal. 2003;15:565–574. doi: 10.1016/s0898-6568(02)00146-8. [DOI] [PubMed] [Google Scholar]
- 110.Taylor SS, Zhang P, Steichen JM, Keshwani MM, Kornev AP. PKA: lessons learned after twenty years. Biochim Biophys Acta. 2013;1834:1271–1278. doi: 10.1016/j.bbapap.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Scott JD, Dessauer CW, Tasken K. Creating order from chaos: cellular regulation by kinase anchoring. Annu Rev Pharmacol Toxicol. 2013;53:187–210. doi: 10.1146/annurev-pharmtox-011112-140204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Theurkauf WE, Vallee RB. Molecular characterization of the cAMP-dependent protein kinase bound to microtubule-associated protein 2. J Biol Chem. 1982;257:3284–3290. [PubMed] [Google Scholar]
- 113.Di Benedetto G, Zoccarato A, Lissandron V, Terrin A, Li X, Houslay MD, Baillie GS, Zaccolo M. Protein kinase A type I and type II define distinct intracellular signaling compartments. Circ Res. 2008;103:836–844. doi: 10.1161/CIRCRESAHA.108.174813. [DOI] [PubMed] [Google Scholar]
- 114.Ueland PM, Doskeland SO. Adenosine 3’:5’-cyclic monophosphate-dependence of protein kinase isoenzymes from mouse liver. Biochem J. 1976;157:117–126. doi: 10.1042/bj1570117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dostmann WR, Taylor SS. Identifying the molecular switches that determine whether (Rp)-cAMPS functions as an antagonist or an agonist in the activation of cAMP-dependent protein kinase I. Biochemistry. 1991;30:8710–8716. doi: 10.1021/bi00099a032. [DOI] [PubMed] [Google Scholar]
- 116.Zhang P, Smith-Nguyen EV, Keshwani MM, Deal MS, Kornev AP, Taylor SS. Structure and allostery of the PKA RIIbeta tetrameric holoenzyme. Science. 2012;335:712–716. doi: 10.1126/science.1213979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Smith FD, Reichow SL, Esseltine JL, Shi D, Langeberg LK, Scott JD, Gonen T. Intrinsic disorder within an AKAP-protein kinase A complex guides local substrate phosphorylation. Elife. 2013;2:e01319. doi: 10.7554/eLife.01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang P, Ye F, Bastidas AC, Kornev AP, Wu J, Ginsberg MH, Taylor SS. An Isoform-Specific Myristylation Switch Targets Type II PKA Holoenzymes to Membranes. Structure. 2015;23:1563–1572. doi: 10.1016/j.str.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tillo SE, Xiong WH, Takahashi M, Miao S, Andrade AL, Fortin DA, Yang G, Qin M, Smoody BF, Stork PJS, Zhong H. Liberated PKA Catalytic Subunits Associate with the Membrane via Myristoylation to Preferentially Phosphorylate Membrane Substrates. Cell Rep. 2017;19:617–629. doi: 10.1016/j.celrep.2017.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Man KNM, Bartels P, Horne MC, Hell JW. Tissue-specific adrenergic regulation of the L-type Ca(2+) channel Ca(V)1.2. Sci Signal. 2020;13 doi: 10.1126/scisignal.abc6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liu G, Papa A, Katchman AN, Zakharov SI, Roybal D, Hennessey JA, Kushner J, Yang L, Chen BX, Kushnir A, Dangas K, et al. Mechanism of adrenergic Ca(V)1.2 stimulation revealed by proximity proteomics. Nature. 2020;577:695–700. doi: 10.1038/s41586-020-1947-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Papa A, Del Rivero Morfin PJ, Chen BX, Yang L, Katchman AN, Zakharov SI, Liu G, Bohnen MS, Zheng V, Katz M, Subramaniam S, et al. A membrane-associated phosphoswitch in Rad controls adrenergic regulation of cardiac calcium channels. J Clin Invest. 2024 doi: 10.1172/JCI176943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Witcher DR, Kovacs RJ, Schulman H, Cefali DC, Jones LR. Unique phosphorylation site on the cardiac ryanodine receptor regulates calcium channel activity. J Biol Chem. 1991;266:11144–11152. [PubMed] [Google Scholar]
- 124.Marx SO, Reiken S, Hisamatsu Y, Jayaraman T, Burkhoff D, Rosemblit N, Marks AR. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell. 2000;101:365–376. doi: 10.1016/s0092-8674(00)80847-8. [DOI] [PubMed] [Google Scholar]
- 125.Dobrev D, Wehrens XH. Role of RyR2 phosphorylation in heart failure and arrhythmias: Controversies around ryanodine receptor phosphorylation in cardiac disease. Circ Res. 2014;114:1311–1319. doi: 10.1161/CIRCRESAHA.114.300568. discussion 1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Despa S, Bossuyt J, Han F, Ginsburg KS, Jia LG, Kutchai H, Tucker AL, Bers DM. Phospholemman-phosphorylation mediates the beta-adrenergic effects on Na/K pump function in cardiac myocytes. Circ Res. 2005;97:252–259. doi: 10.1161/01.RES.0000176532.97731.e5. [DOI] [PubMed] [Google Scholar]
- 127.Han F, Bossuyt J, Martin JL, Despa S, Bers DM. Role of phospholemman phosphorylation sites in mediating kinase-dependent regulation of the Na+-K+-ATPase. Am J Physiol Cell Physiol. 2010;299:C1363–1369. doi: 10.1152/ajpcell.00027.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Aronsen JM, Swift F, Sejersted OM. Cardiac sodium transport and excitation-contraction coupling. J Mol Cell Cardiol. 2013;61:11–19. doi: 10.1016/j.yjmcc.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 129.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 130.Bers DM. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- 131.Song T, Landim-Vieira M, Ozdemir M, Gott C, Kanisicak O, Pinto JR, Sadayappan S. Etiology of genetic muscle disorders induced by mutations in fast and slow skeletal MyBP-C paralogs. Exp Mol Med. 2023;55:502–509. doi: 10.1038/s12276-023-00953-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kotter S, Gout L, Von Frieling-Salewsky M, Muller AE, Helling S, Marcus K, Dos Remedios C, Linke WA, Kruger M. Differential changes in titin domain phosphorylation increase myofilament stiffness in failing human hearts. Cardiovasc Res. 2013;99:648–656. doi: 10.1093/cvr/cvt144. [DOI] [PubMed] [Google Scholar]
- 133.Hayes JS, Brunton LL, Brown JH, Reese JB, Mayer SE. Hormonally specific expression of cardiac protein kinase activity. Proc Natl Acad Sci U S A. 1979;76:1570–1574. doi: 10.1073/pnas.76.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Aslam M, Ladilov Y. Emerging Role of cAMP/AMPK Signaling. Cells. 2022;11 doi: 10.3390/cells11020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sands WA, Palmer TM. Regulating gene transcription in response to cyclic AMP elevation. Cell Signal. 2008;20:460–466. doi: 10.1016/j.cellsig.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 136.Muller FU, Boknik P, Knapp J, Linck B, Luss H, Neumann J, Schmitz W. Activation and inactivation of cAMP-response element-mediated gene transcription in cardiac myocytes. Cardiovasc Res. 2001;52:95–102. doi: 10.1016/s0008-6363(01)00361-3. [DOI] [PubMed] [Google Scholar]
- 137.de Rooij J, Zwartkruis FJ, Verheijen MH, Cool RH, Nijman SM, Wittinghofer A, Bos JL. Epac is a Rap1 guanine-nucleotide-exchange-factor directly activated by cAMP. Nature. 2003;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 138.Kawasaki H, Springett GM, Mochizuki N, Toki S, Nakaya M, Matsuda M, Housman DE, Graybiel AM. A family of cAMP-binding proteins that directly activate Rap1. Science. 1998;282:2275–2279. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
- 139.Ruiz-Hurtado G, Morel E, Dominguez-Rodriguez A, Llach A, Lezoualc’h F, Benitah JP, Gomez AM. Epac in cardiac calcium signaling. J Mol Cell Cardiol. 2013;58:162–171. doi: 10.1016/j.yjmcc.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 140.Oestreich EA, Wang H, Malik S, Kaproth-Joslin KA, Blaxall BC, Kelley GG, Dirksen RT, Smrcka AV. Epac-mediated activation of phospholipase C(epsilon) plays a critical role in beta-adrenergic receptor-dependent enhancement of Ca2+ mobilization in cardiac myocytes. J Biol Chem. 2007;282:5488–5495. doi: 10.1074/jbc.M608495200. [DOI] [PubMed] [Google Scholar]
- 141.Oestreich EA, Malik S, Goonasekera SA, Blaxall BC, Kelley GG, Dirksen RT, Smrcka AV. Epac and phospholipase Cepsilon regulate Ca2+ release in the heart by activation of protein kinase Cepsilon and calcium-calmodulin kinase II. J Biol Chem. 2009;284:1514–1522. doi: 10.1074/jbc.M806994200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bizerra PFV, Gilglioni EH, Li HL, Go S, Oude Elferink RPJ, Verhoeven AJ, Chang JC. Opposite regulation of glycogen metabolism by cAMP produced in the cytosol and at the plasma membrane. Biochim Biophys Acta Mol Cell Res. 2024;1871:119585. doi: 10.1016/j.bbamcr.2023.119585. [DOI] [PubMed] [Google Scholar]
- 143.Mei FC, Qiao J, Tsygankova OM, Meinkoth JL, Quilliam LA, Cheng X. Differential signaling of cyclic AMP: opposing effects of exchange protein directly activated by cyclic AMP and cAMP-dependent protein kinase on protein kinase B activation. J Biol Chem. 2002;277:11497–11504. doi: 10.1074/jbc.M110856200. [DOI] [PubMed] [Google Scholar]
- 144.DiFrancesco D. The role of the funny current in pacemaker activity. Circ Res. 2010;106:434–446. doi: 10.1161/CIRCRESAHA.109.208041. [DOI] [PubMed] [Google Scholar]
- 145.Barbuti A, Scavone A, Mazzocchi N, Terragni B, Baruscotti M, Difrancesco D. A caveolin-binding domain in the HCN4 channels mediates functional interaction with caveolin proteins. J Mol Cell Cardiol. 2012;53:187–195. doi: 10.1016/j.yjmcc.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 146.Barbuti A, Terragni B, Brioschi C, DiFrancesco D. Localization of f-channels to caveolae mediates specific beta2-adrenergic receptor modulation of rate in sinoatrial myocytes. J Mol Cell Cardiol. 2007;42:71–78. doi: 10.1016/j.yjmcc.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 147.Gruscheski L, Brand T. The Role of POPDC Proteins in Cardiac Pacemaking and Conduction. J Cardiovasc Dev Dis. 2021;8 doi: 10.3390/jcdd8120160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tang CM, Insel PA. GPCR expression in the heart; “new” receptors in myocytes and fibroblasts. Trends Cardiovasc Med. 2004;14:94–99. doi: 10.1016/j.tcm.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 149.Keely SL. Activation of cAMP-dependent protein kinase without a corresponding increase in phosphorylase activity. Res Commun Chem Pathol Pharmacol. 1977;18:283–290. [PubMed] [Google Scholar]
- 150.Brunton LL, Hayes JS, Mayer SE. Hormonally specific phosphorylation of cardiac troponin I and activation of glycogen phosphorylase. Nature. 1979;280:78–80. doi: 10.1038/280078a0. [DOI] [PubMed] [Google Scholar]
- 151.Terasaki WL, Brooker G. Cardiac adenosine 3’:5’-monophosphate. Free and bound forms in the isolated rat atrium. J Biol Chem. 1977;252:1041–1050. [PubMed] [Google Scholar]
- 152.Corbin JD, Sugden PH, Lincoln TM, Keely SL. Compartmentalization of adenosine 3’:5’-monophosphate and adenosine 3’:5’-monophosphate-dependent protein kinase in heart tissue. J Biol Chem. 1977;252:3854–3861. [PubMed] [Google Scholar]
- 153.Hayes JS, Brunton LL, Mayer SE. Selective activation of particulate cAMP-dependent protein kinase by isoproterenol and prostaglandin E1. J Biol Chem. 1980;255:5113–5119. [PubMed] [Google Scholar]
- 154.Hayes JS, Brunton LL. Functional compartments in cyclic nucleotide action. J Cyclic Nucleotide Res. 1982;8:1–16. [PubMed] [Google Scholar]
- 155.Watanabe AM, Besch HR., Jr Interaction between cyclic adenosine monophosphate and cyclic gunaosine monophosphate in guinea pig ventricular myocardium. Circ Res. 1975;37:309–317. doi: 10.1161/01.res.37.3.309. [DOI] [PubMed] [Google Scholar]
- 156.Keely SL, Jr, Lincoln TM, Corbin JD. Interaction of acetylcholine and epinephrine on heart cyclic AMP-dependent protein kinase. Am J Physiol. 1978;234:H432–438. doi: 10.1152/ajpheart.1978.234.4.H432. [DOI] [PubMed] [Google Scholar]
- 157.Biegon RL, Pappano AJ. Dual mechanism for inhibition of calcium-dependent action potentials by acetylcholine in avian ventricular muscle. Relationship to cyclic AMP. Circ Res. 1980;46:353–362. doi: 10.1161/01.res.46.3.353. [DOI] [PubMed] [Google Scholar]
- 158.Hartzell HC. Regulation of cardiac ion channels by catecholamines, acetylcholine and second messenger systems. Prog Biophys Mol Biol. 1988;52:165–247. doi: 10.1016/0079-6107(88)90014-4. [DOI] [PubMed] [Google Scholar]
- 159.England PJ, Shahid M. Effects of forskolin on contractile responses and protein phosphorylation in the isolated perfused rat heart. Biochem J. 1987;246:687–695. doi: 10.1042/bj2460687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hohl CM, Li QA. Compartmentation of cAMP in adult canine ventricular myocytes. Relation to single-cell free Ca2+ transients. Circ Res. 1991;69:1369–1379. doi: 10.1161/01.res.69.5.1369. [DOI] [PubMed] [Google Scholar]
- 161.Kuschel M, Zhou YY, Cheng H, Zhang SJ, Chen Y, Lakatta EG, Xiao RP. G(i) protein-mediated functional compartmentalization of cardiac beta(2)-adrenergic signaling. J Biol Chem. 1999;274:22048–22052. doi: 10.1074/jbc.274.31.22048. [DOI] [PubMed] [Google Scholar]
- 162.Xiao RP, Hohl C, Altschuld R, Jones L, Livingston B, Ziman B, Tantini B, Lakatta EG. Beta 2-adrenergic receptor-stimulated increase in cAMP in rat heart cells is not coupled to changes in Ca2+ dynamics, contractility, or phospholamban phosphorylation. J Biol Chem. 1994;269:19151–19156. [PubMed] [Google Scholar]
- 163.Vila Petroff MG, Egan JM, Wang X, Sollott SJ. Glucagon-like peptide-1 increases cAMP but fails to augment contraction in adult rat cardiac myocytes. Circ Res. 2001;89:445–452. doi: 10.1161/hh1701.095716. [DOI] [PubMed] [Google Scholar]
- 164.Valentine CD, Haggie PM. Confinement of beta(1)- and beta(2)-adrenergic receptors in the plasma membrane of cardiomyocyte-like H9c2 cells is mediated by selective interactions with PDZ domain and A-kinase anchoring proteins but not caveolae. Mol Biol Cell. 2011;22:2970–2982. doi: 10.1091/mbc.E11-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gardner LA, Tavalin SJ, Goehring AS, Scott JD, Bahouth SW. AKAP79-mediated targeting of the cyclic AMP-dependent protein kinase to the beta1-adrenergic receptor promotes recycling and functional resensitization of the receptor. J Biol Chem. 2006;281:33537–33553. doi: 10.1074/jbc.M601809200. [DOI] [PubMed] [Google Scholar]
- 166.Dell’Acqua ML, Faux MC, Thorburn J, Thorburn A, Scott JD. Membrane-targeting sequences on AKAP79 bind phosphatidylinositol-4, 5-bisphosphate. Embo J. 1998;17:2246–2260. doi: 10.1093/emboj/17.8.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Coghlan VM, Perrino BA, Howard M, Langeberg LK, Hicks JB, Gallatin WM, Scott JD. Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science. 1995;267:108–111. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 168.Dodge KL, Khouangsathiene S, Kapiloff MS, Mouton R, Hill EV, Houslay MD, Langeberg LK, Scott JD. mAKAP assembles a protein kinase A/PDE4 phosphodiesterase cAMP signaling module. EMBO J. 2001;20:1921–1930. doi: 10.1093/emboj/20.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fraser ID, Tavalin SJ, Lester LB, Langeberg LK, Westphal AM, Dean RA, Marrion NV, Scott JD. A novel lipid-anchored A-kinase Anchoring Protein facilitates cAMP-responsive membrane events. EMBO J. 1998;17:2261–2272. doi: 10.1093/emboj/17.8.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Trotter KW, Fraser ID, Scott GK, Stutts MJ, Scott JD, Milgram SL. Alternative splicing regulates the subcellular localization of A-kinase anchoring protein 18 isoforms. The Journal of cell biology. 1999;147:1481–1492. doi: 10.1083/jcb.147.7.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lygren B, Carlson CR, Santamaria K, Lissandron V, McSorley T, Litzenberg J, Lorenz D, Wiesner B, Rosenthal W, Zaccolo M, Tasken K, et al. AKAP complex regulates Ca2+ re-uptake into heart sarcoplasmic reticulum. EMBO Rep. 2007;8:1061–1067. doi: 10.1038/sj.embor.7401081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hundsrucker C, Skroblin P, Christian F, Zenn HM, Popara V, Joshi M, Eichhorst J, Wiesner B, Herberg FW, Reif B, Rosenthal W, et al. Glycogen synthase kinase 3beta interaction protein functions as an A-kinase anchoring protein. J Biol Chem. 2010;285:5507–5521. doi: 10.1074/jbc.M109.047944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tasken K, Aandahl EM. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- 174.Huang LJ, Durick K, Weiner JA, Chun J, Taylor SS. Identification of a novel protein kinase A anchoring protein that binds both type I and type II regulatory subunits. J Biol Chem. 1997;272:8057–8064. doi: 10.1074/jbc.272.12.8057. [DOI] [PubMed] [Google Scholar]
- 175.Means CK, Lygren B, Langeberg LK, Jain A, Dixon RE, Vega AL, Gold MG, Petrosyan S, Taylor SS, Murphy AN, Ha T, et al. An entirely specific type I A-kinase anchoring protein that can sequester two molecules of protein kinase A at mitochondria. Proc Natl Acad Sci U S A. 2011;108:E1227–1235. doi: 10.1073/pnas.1107182108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Diviani D, Langeberg LK, Doxsey SJ, Scott JD. Pericentrin anchors protein kinase A at the centrosome through a newly identified RII-binding domain. Curr Biol. 2000;10:417–420. doi: 10.1016/s0960-9822(00)00422-x. [DOI] [PubMed] [Google Scholar]
- 177.Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Omar MH, Scott JD. AKAP Signaling Islands: Venues for Precision Pharmacology. Trends Pharmacol Sci. 2020;41:933–946. doi: 10.1016/j.tips.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Carr DW, Hausken ZE, Fraser ID, Stofko-Hahn RE, Scott JD. Association of the type II cAMP-dependent protein kinase with a human thyroid RII-anchoring protein. Cloning and characterization of the RII-binding domain. J Biol Chem. 1992;267:13376–13382. [PubMed] [Google Scholar]
- 180.Scott JD, Stofko RE, McDonald JR, Comer JD, Vitalis EA, Mangili JA. Type II regulatory subunit dimerization determines the subcellular localization of the cAMP-dependent protein kinase. J Biol Chem. 1990;265:21561–21566. [PubMed] [Google Scholar]
- 181.Gold MG, Lygren B, Dokurno P, Hoshi N, McConnachie G, Tasken K, Carlson CR, Scott JD, Barford D. Molecular basis of AKAP specificity for PKA regulatory subunits. Mol Cell. 2006;24:383–395. doi: 10.1016/j.molcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 182.Carr DW, Stofko-Hahn RE, Fraser ID, Bishop SM, Acott TS, Brennan RG, Scott JD. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J Biol Chem. 1991;266:14188–14192. [PubMed] [Google Scholar]
- 183.Newlon MG, Roy M, Morikis D, Carr DW, Westphal R, Scott JD, Jennings PA. A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. EMBO J. 2001;20:1651–1662. doi: 10.1093/emboj/20.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Newlon MG, Roy M, Morikis D, Hausken ZE, Coghlan V, Scott JD, Jennings PA. The molecular basis for protein kinase A anchoring revealed by solution NMR. Nat Struct Biol. 1999;6:222–227. doi: 10.1038/6663. [DOI] [PubMed] [Google Scholar]
- 185.Alto NM, Soderling SH, Hoshi N, Langeberg LK, Fayos R, Jennings PA, Scott JD. Bioinformatic design of A-kinase anchoring protein-in silico: a potent and selective peptide antagonist of type II protein kinase A anchoring. Proc Natl Acad Sci U S A. 2003;100:4445–4450. doi: 10.1073/pnas.0330734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gotz F, Roske Y, Schulz MS, Autenrieth K, Bertinetti D, Faelber K, Zuhlke K, Kreuchwig A, Kennedy EJ, Krause G, Daumke O, et al. AKAP18:PKA-RIIalpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA. Biochem J. 2016;473:1881–1894. doi: 10.1042/BCJ20160242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Smith FD, Omar MH, Nygren PJ, Soughayer J, Hoshi N, Lau HT, Snyder CG, Branon TC, Ghosh D, Langeberg LK, Ting AY, et al. Single nucleotide polymorphisms alter kinase anchoring and the subcellular targeting of A-kinase anchoring proteins. Proc Natl Acad Sci U S A. 2018;115:E11465–E11474. doi: 10.1073/pnas.1816614115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kovanich D, van der Heyden MA, Aye TT, van Veen TA, Heck AJ, Scholten A. Sphingosine kinase interacting protein is an A-kinase anchoring protein specific for type I cAMP-dependent protein kinase. Chembiochem. 2010;11:963–971. doi: 10.1002/cbic.201000058. [DOI] [PubMed] [Google Scholar]
- 189.Burgers PP, Ma Y, Margarucci L, Mackey M, van der Heyden MA, Ellisman M, Scholten A, Taylor SS, Heck AJ. A small novel A-kinase anchoring protein (AKAP) that localizes specifically protein kinase A-regulatory subunit I (PKA-RI) to the plasma membrane. J Biol Chem. 2012;287:43789–43797. doi: 10.1074/jbc.M112.395970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Smith FD, Esseltine JL, Nygren PJ, Veesler D, Byrne DP, Vonderach M, Strashnov I, Eyers CE, Eyers PA, Langeberg LK, Scott JD. Local protein kinase A action proceeds through intact holoenzymes. Science. 2017;356:1288–1293. doi: 10.1126/science.aaj1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Walker-Gray R, Stengel F, Gold MG. Mechanisms for restraining cAMP-dependent protein kinase revealed by subunit quantitation and cross-linking approaches. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1701782114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chao YC, Surdo NC, Pantano S, Zaccolo M. Imaging cAMP nanodomains in the heart. Biochemical Society transactions. 2019;47:1383–1392. doi: 10.1042/BST20190245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ercu M, Klussmann E. Roles of A-Kinase Anchoring Proteins and Phosphodiesterases in the Cardiovascular System. J Cardiovasc Dev Dis. 2018;5 doi: 10.3390/jcdd5010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Adams SR, Harootunian AT, Buechler YJ, Taylor SS, Tsien RY. Fluorescence ratio imaging of cyclic AMP in single cells. Nature. 1991;349:694–697. doi: 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- 195.Bacskai BJ, Hochner B, Mahaut-Smith M, Adams SR, Kaang BK, Kandel ER, Tsien RY. Spatially resolved dynamics of cAMP and protein kinase A subunits in Aplysia sensory neurons. Science. 1993;260:222–226. doi: 10.1126/science.7682336. [DOI] [PubMed] [Google Scholar]
- 196.Zaccolo M, Pozzan T. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science. 2002;295:1711–1715. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
- 197.Zaccolo M, De Giorgi F, Cho CY, Feng L, Knapp T, Negulescu PA, Taylor SS, Tsien RY, Pozzan T. A genetically encoded, fluorescent indicator for cyclic AMP in living cells. Nat Cell Biol. 2000;2:25–29. doi: 10.1038/71345. [DOI] [PubMed] [Google Scholar]
- 198.Nikolaev VO, Bunemann M, Hein L, Hannawacker A, Lohse MJ. Novel single chain cAMP sensors for receptor-induced signal propagation. J Biol Chem. 2004;279:37215–37218. doi: 10.1074/jbc.C400302200. [DOI] [PubMed] [Google Scholar]
- 199.DiPilato LM, Cheng X, Zhang J. Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signalling within discrete subcellular compartments. Proc Natl Acad Sci. 2004;101:16513–16518. doi: 10.1073/pnas.0405973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ponsioen B, Zhao J, Riedl J, Zwartkruis FJ, van der Krogt G, Zaccolo M, Moolenaar WH, Bos JL, Jalink K. Detecting cAMP-induced activation by fluorescence resonance energy transfer: Epac as a novel cAMP indicator. EMBO Rep. 2004;5:1–5. doi: 10.1038/sj.embor.7400290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lohse MJ, Bock A, Zaccolo M. G Protein-Coupled Receptor Signaling: New Insights Define Cellular Nanodomains. Annu Rev Pharmacol Toxicol. 2024;64:387–415. doi: 10.1146/annurev-pharmtox-040623-115054. [DOI] [PubMed] [Google Scholar]
- 202.Surdo NC, Berrera M, Koschinski A, Brescia M, Machado MR, Carr C, Wright P, Gorelik J, Morotti S, Grandi E, Bers DM, et al. FRET biosensor uncovers cAMP nano-domains at beta-adrenergic targets that dictate precise tuning of cardiac contractility. Nat Commun. 2017;8:15031. doi: 10.1038/ncomms15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Terrin A, Di Benedetto G, Pertegato V, Cheung YF, Baillie G, Lynch MJ, Elvassore N, Prinz A, Herberg FW, Houslay MD, Zaccolo M. PGE(1) stimulation of HEK293 cells generates multiple contiguous domains with different [cAMP]: role of compartmentalized phosphodiesterases. The Journal of cell biology. 2006;175:441–451. doi: 10.1083/jcb.200605050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Iancu RV, Jones SW, Harvey RD. Compartmentation of cAMP signaling in cardiac myocytes: a computational study. Biophys J. 2007;92:3317–3331. doi: 10.1529/biophysj.106.095356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Shcherbakova OG, Hurt CM, Xiang Y, Dell’Acqua ML, Zhang Q, Tsien RW, Kobilka BK. Organization of beta-adrenoceptor signaling compartments by sympathetic innervation of cardiac myocytes. J Cell Biol. 2007;176:521–533. doi: 10.1083/jcb.200604167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen W, Levine H, Rappel WJ. A mathematical analysis of second messenger compartmentalization. Phys Biol. 2008;5:046006. doi: 10.1088/1478-3975/5/4/046006. [DOI] [PubMed] [Google Scholar]
- 207.Leroy J, Abi-Gerges A, Nikolaev VO, Richter W, Lechene P, Mazet JL, Conti M, Fischmeister R, Vandecasteele G. Spatiotemporal dynamics of beta-adrenergic cAMP signals and L-type Ca2+ channel regulation in adult rat ventricular myocytes: role of phosphodiesterases. Circ Res. 2008;102:1091–1100. doi: 10.1161/CIRCRESAHA.107.167817. [DOI] [PubMed] [Google Scholar]
- 208.Feinstein WP, Zhu B, Leavesley SJ, Sayner SL, Rich TC. Assessment of cellular mechanisms contributing to cAMP compartmentalization in pulmonary microvascular endothelial cells. Am J Physiol Cell Physiol. 2012;302:C839–852. doi: 10.1152/ajpcell.00361.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mika D, Leroy J, Vandecasteele G, Fischmeister R. PDEs create local domains of cAMP signaling. Journal of molecular and cellular cardiology. 2012;52:323–329. doi: 10.1016/j.yjmcc.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 210.Annamdevula NS, Sweat R, Griswold JR, Trinh K, Hoffman C, West S, Deal J, Britain AL, Jalink K, Rich TC, Leavesley SJ. Spectral imaging of FRET-based sensors reveals sustained cAMP gradients in three spatial dimensions. Cytometry A. 2018;93:1029–1038. doi: 10.1002/cyto.a.23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Maiellaro I, Lohse MJ, Kittel RJ, Calebiro D. cAMP Signals in Drosophila Motor Neurons Are Confined to Single Synaptic Boutons. Cell Rep. 2016;17:1238–1246. doi: 10.1016/j.celrep.2016.09.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Agarwal SR, Miyashiro K, Latt H, Ostrom RS, Harvey RD. Compartmentalized cAMP responses to prostaglandin EP(2) receptor activation in human airway smooth muscle cells. British journal of pharmacology. 2017;174:2784–2796. doi: 10.1111/bph.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hansen JN, Kaiser F, Leyendecker P, Stuven B, Krause JH, Derakhshandeh F, Irfan J, Sroka TJ, Preval KM, Desai PB, Kraut M, et al. A cAMP signalosome in primary cilia drives gene expression and kidney cyst formation. EMBO Rep. 2022;23:e54315. doi: 10.15252/embr.202154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Monterisi S, Favia M, Guerra L, Cardone RA, Marzulli D, Reshkin SJ, Casavola V, Zaccolo M. CFTR regulation in human airway epithelial cells requires integrity of the actin cytoskeleton and compartmentalized cAMP and PKA activity. J Cell Sci. 2012;125:1106–1117. doi: 10.1242/jcs.089086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Anton SE, Kayser C, Maiellaro I, Nemec K, Moller J, Koschinski A, Zaccolo M, Annibale P, Falcke M, Lohse MJ, Bock A. Receptor-associated independent cAMP nanodomains mediate spatiotemporal specificity of GPCR signaling. Cell. 2022 doi: 10.1016/j.cell.2022.02.011. [DOI] [PubMed] [Google Scholar]
- 216.Sprenger JU, Perera RK, Steinbrecher JH, Lehnart SE, Maier LS, Hasenfuss G, Nikolaev VO. In vivo model with targeted cAMP biosensor reveals changes in receptor-microdomain communication in cardiac disease. Nat Commun. 2015;6:6965. doi: 10.1038/ncomms7965. [DOI] [PubMed] [Google Scholar]
- 217.Demby A, Zaccolo M. Investigating G-protein coupled receptor signalling with light-emitting biosensors. Front Physiol. 2023;14:1310197. doi: 10.3389/fphys.2023.1310197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Mongillo M, Zaccolo M. A complex phosphodiesterase system controls beta-adrenoceptor signalling in cardiomyocytes. Biochem Soc Trans. 2006;34:510–511. doi: 10.1042/BST0340510. [DOI] [PubMed] [Google Scholar]
- 219.Houslay MD. Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem Sci. 2010;35:91–100. doi: 10.1016/j.tibs.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 220.Zaccolo M. Phosphodiesterases and compartmentalized cAMP signalling in the heart. Eur J Cell Biol. 2006;85:693–697. doi: 10.1016/j.ejcb.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 221.Kokkonen K, Kass DA. Nanodomain Regulation of Cardiac Cyclic Nucleotide Signaling by Phosphodiesterases. Annual review of pharmacology and toxicology. 2017;57:455–479. doi: 10.1146/annurev-pharmtox-010716-104756. [DOI] [PubMed] [Google Scholar]
- 222.Jurevicius J, Fischmeister R. cAMP compartmentation is responsible for a local activation of cardiac Ca2+ channels by beta-adrenergic agonists. Proc Natl Acad Sci U S A. 1996;93:295–299. doi: 10.1073/pnas.93.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Nikolaev VO, Moshkov A, Lyon AR, Miragoli M, Novak P, Paur H, Lohse MJ, Korchev YE, Harding SE, Gorelik J. Beta2-adrenergic receptor redistribution in heart failure changes cAMP compartmentation. Science. 2010;327:1653–1657. doi: 10.1126/science.1185988. [DOI] [PubMed] [Google Scholar]
- 224.Bock A, Annibale P, Konrad C, Hannawacker A, Anton SE, Maiellaro I, Zabel U, Sivaramakrishnan S, Falcke M, Lohse MJ. Optical Mapping of cAMP Signaling at the Nanometer Scale. Cell. 2020;182:1519–1530.:e1517. doi: 10.1016/j.cell.2020.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Beavo JA, Bechtel PJ, Krebs EG. Activation of protein kinase by physiological concentrations of cyclic AMP. Proc Natl Acad Sci U S A. 1974;71:3580–3583. doi: 10.1073/pnas.71.9.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Borner S, Schwede F, Schlipp A, Berisha F, Calebiro D, Lohse MJ, Nikolaev VO. FRET measurements of intracellular cAMP concentrations and cAMP analog permeability in intact cells. Nat Protoc. 2011;6:427–438. doi: 10.1038/nprot.2010.198. [DOI] [PubMed] [Google Scholar]
- 227.Koschinski A, Zaccolo M. Activation of PKA in cell requires higher concentration of cAMP than in vitro: implications for compartmentalization of cAMP signalling. Sci Rep. 2017;7:14090. doi: 10.1038/s41598-017-13021-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Willis MJ, Baillie GS. Arrestin-dependent localization of phosphodiesterases. Handb Exp Pharmacol. 2014;219:293–307. doi: 10.1007/978-3-642-41199-1_15. [DOI] [PubMed] [Google Scholar]
- 229.Martinez JM, Shen A, Xu B, Jovanovic A, de Chabot J, Zhang J, Xiang YK. Arrestin-dependent nuclear export of phosphodiesterase 4D promotes GPCR-induced nuclear cAMP signaling required for learning and memory. Sci Signal. 2023;16:eade3380. doi: 10.1126/scisignal.ade3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Beltejar MG, Lau HT, Golkowski MG, Ong SE, Beavo JA. Analyses of PDE-regulated phosphoproteomes reveal unique and specific cAMP-signaling modules in T cells. Proc Natl Acad Sci U S A. 2017;114:E6240–E6249. doi: 10.1073/pnas.1703939114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Mika D, Bobin P, Lindner M, Boet A, Hodzic A, Lefebvre F, Lechene P, Sadoune M, Samuel JL, Algalarrondo V, Rucker-Martin C, et al. Synergic PDE3 and PDE4 control intracellular cAMP and cardiac excitation-contraction coupling in a porcine model. Journal of molecular and cellular cardiology. 2019;133:57–66. doi: 10.1016/j.yjmcc.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 232.Subramaniam G, Schleicher K, Kovanich D, Zerio A, Folkmanaite M, Chao YC, Surdo NC, Koschinski A, Hu J, Scholten A, Heck AJR, et al. Integrated Proteomics Unveils Nuclear PDE3A2 as a Regulator of Cardiac Myocyte Hypertrophy. Circ Res. 2023;132:828–848. doi: 10.1161/CIRCRESAHA.122.321448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Brunton LL, Hayes JS, Mayer SE. Functional compartmentation of cyclic AMP and protein kinase in heart. Adv Cyclic Nucleotide Res. 1981;14:391–397. [PubMed] [Google Scholar]
- 234.Buxton IL, Brunton LL. Compartments of cyclic AMP and protein kinase in mammalian cardiomyocytes. J Biol Chem. 1983;258:10233–10239. [PubMed] [Google Scholar]
- 235.Schleicher K, Zaccolo M. Using cAMP Sensors to Study Cardiac Nanodomains. J Cardiovasc Dev Dis. 2018;5 doi: 10.3390/jcdd5010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mo GC, Ross B, Hertel F, Manna P, Yang X, Greenwald E, Booth C, Plummer AM, Tenner B, Chen Z, Wang Y, et al. Genetically encoded biosensors for visualizing live-cell biochemical activity at super-resolution. Nat Methods. 2017;14:427–434. doi: 10.1038/nmeth.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Chen C, Nakamura T, Koutalos Y. Cyclic AMP diffusion coefficient in frog olfactory cilia. Biophys J. 1999;76:2861–2867. doi: 10.1016/S0006-3495(99)77440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nikolaev VO, Bunemann M, Schmitteckert E, Lohse MJ, Engelhardt S. Cyclic AMP imaging in adult cardiac myocytes reveals far-reaching beta1-adrenergic but locally confined beta2-adrenergic receptor-mediated signaling. Circ Res. 2006;99:1084–1091. doi: 10.1161/01.RES.0000250046.69918.d5. [DOI] [PubMed] [Google Scholar]
- 239.Lohse C, Bock A, Maiellaro I, Hannawacker A, Schad LR, Lohse MJ, Bauer WR. Experimental and mathematical analysis of cAMP nanodomains. PLoS One. 2017;12:e0174856. doi: 10.1371/journal.pone.0174856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang JZ, Lu TW, Stolerman LM, Tenner B, Yang JR, Zhang JF, Falcke M, Rangamani P, Taylor SS, Mehta S, Zhang J. Phase Separation of a PKA Regulatory Subunit Controls cAMP Compartmentation and Oncogenic Signaling. Cell. 2020;182:1531–1544.:e1515. doi: 10.1016/j.cell.2020.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 242.Irannejad R, Pessino V, Mika D, Huang B, Wedegaertner PB, Conti M, von Zastrow M. Functional selectivity of GPCR-directed drug action through location bias. Nat Chem Biol. 2017;13:799–806. doi: 10.1038/nchembio.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Boivin B, Lavoie C, Vaniotis G, Baragli A, Villeneuve LR, Ethier N, Trieu P, Allen BG, Hebert TE. Functional beta-adrenergic receptor signalling on nuclear membranes in adult rat and mouse ventricular cardiomyocytes. Cardiovasc Res. 2006;71:69–78. doi: 10.1016/j.cardiores.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 244.Irannejad R, Tomshine JC, Tomshine JR, Chevalier M, Mahoney JP, Steyaert J, Rasmussen SG, Sunahara RK, El-Samad H, Huang B, von Zastrow M. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495:534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Nash CA, Wei W, Irannejad R, Smrcka AV. Golgi localized beta1-adrenergic receptors stimulate Golgi PI4P hydrolysis by PLCepsilon to regulate cardiac hypertrophy. Elife. 2019;8 doi: 10.7554/eLife.48167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lin TY, Mai QN, Zhang H, Wilson E, Chien HC, Yee SW, Giacomini KM, Olgin JE, Irannejad R. Cardiac contraction and relaxation are regulated by distinct subcellular cAMP pools. Nat Chem Biol. 2024;20:62–73. doi: 10.1038/s41589-023-01381-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wang Y, Shi Q, Li M, Zhao M, Reddy Gopireddy R, Teoh JP, Xu B, Zhu C, Ireton KE, Srinivasan S, Chen S, et al. Intracellular beta(1)-Adrenergic Receptors and Organic Cation Transporter 3 Mediate Phospholamban Phosphorylation to Enhance Cardiac Contractility. Circ Res. 2021;128:246–261. doi: 10.1161/CIRCRESAHA.120.317452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Fields LA, Koschinski A, Zaccolo M. Sustained exposure to catecholamines affects cAMP/PKA compartmentalised signalling in adult rat ventricular myocytes. Cell Signal. 2016;28:725–732. doi: 10.1016/j.cellsig.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Abi-Gerges A, Richter W, Lefebvre F, Mateo P, Varin A, Heymes C, Samuel JL, Lugnier C, Conti M, Fischmeister R, Vandecasteele G. Decreased expression and activity of cAMP phosphodiesterases in cardiac hypertrophy and its impact on beta-adrenergic cAMP signals. Circ Res. 2009;105:784–792. doi: 10.1161/CIRCRESAHA.109.197947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Omar MH, Byrne DP, Jones KN, Lakey TM, Collins KB, Lee KS, Daly LA, Forbush KA, Lau HT, Golkowski M, McKnight GS, et al. Mislocalization of protein kinase A drives pathology in Cushing’s syndrome. Cell Rep. 2022;40:111073. doi: 10.1016/j.celrep.2022.111073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Aye TT, Soni S, van Veen TA, van der Heyden MA, Cappadona S, Varro A, de Weger RA, de Jonge N, Vos MA, Heck AJ, Scholten A. Reorganized PKA-AKAP associations in the failing human heart. J Mol Cell Cardiol. 2011 doi: 10.1016/j.yjmcc.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 252.Suryavanshi SV, Jadhav SM, Anderson KL, Katsonis P, Lichtarge O, McConnell BK. Human muscle-specific A-kinase anchoring protein polymorphisms modulate the susceptibility to cardiovascular diseases by altering cAMP/PKA signaling. American journal of physiology Heart and circulatory physiology. 2018;315:H109–H121. doi: 10.1152/ajpheart.00034.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zhou Z, Wang Y, Li X, Zhang Y, Yuan L, Chen D, Wang X. PDE3A and GSK3B as Atrial Fibrillation Susceptibility Genes in the Chinese Population via Bioinformatics and Genome-Wide Association Analysis. Biomedicines. 2023;11 doi: 10.3390/biomedicines11030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Traylor M, Amin Al Olama A, Lyytikainen LP, Marini S, Chung J, Malik R, Dichgans M, Kahonen M, Lehtimaki T, Anderson CD, Raitakari OT, et al. Influence of Genetic Variation in PDE3A on Endothelial Function and Stroke. Hypertension. 2020;75:365–371. doi: 10.1161/HYPERTENSIONAHA.119.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Brotman SM, El-Sayed Moustafa JS, Guan L, Broadaway KA, Wang D, Jackson AU, Welch R, Currin KW, Tomlinson M, Vadlamudi S, Stringham HM, et al. Adipose tissue eQTL meta-analysis reveals the contribution of allelic heterogeneity to gene expression regulation and cardiometabolic traits. bioRxiv. 2023 doi: 10.1038/s41588-024-01982-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wang C, Taskinen JH, Segersvard H, Immonen K, Kosonen R, Tolva JM, Mayranpaa MI, Kovanen PT, Olkkonen VM, Sinisalo J, Laine M, et al. Alterations of Cardiac Protein Kinases in Cyclic Nucleotide-Dependent Signaling Pathways in Human Ischemic Heart Failure. Front Cardiovasc Med. 2022;9:919355. doi: 10.3389/fcvm.2022.919355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Gladka MM, Molenaar B, de Ruiter H, van der Elst S, Tsui H, Versteeg D, Lacraz GPA, Huibers MMH, van Oudenaarden A, van Rooij E. Single-Cell Sequencing of the Healthy and Diseased Heart Reveals Cytoskeleton-Associated Protein 4 as a New Modulator of Fibroblasts Activation. Circulation. 2018;138:166–180. doi: 10.1161/CIRCULATIONAHA.117.030742. [DOI] [PubMed] [Google Scholar]
- 258.Das S, Frisk C, Eriksson MJ, Walentinsson A, Corbascio M, Hage C, Kumar C, Asp M, Lundeberg J, Maret E, Persson H, et al. Transcriptomics of cardiac biopsies reveals differences in patients with or without diagnostic parameters for heart failure with preserved ejection fraction. Sci Rep. 2019;9:3179. doi: 10.1038/s41598-019-39445-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Chen S, Hu J, Xu Y, Yan J, Li S, Chen L, Zhang J. Transcriptome analysis of human hypertrophic cardiomyopathy reveals inhibited cardiac development pathways in children. iScience. 2024;27:108642. doi: 10.1016/j.isci.2023.108642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Garnier A, Bork NI, Jacquet E, Zipfel S, Munoz-Guijosa C, Baczko I, Reichenspurner H, Donzeau-Gouge P, Maier LS, Dobrev D, Girdauskas E, et al. Mapping genetic changes in the cAMP-signaling cascade in human atria. Journal of molecular and cellular cardiology. 2021;155:10–20. doi: 10.1016/j.yjmcc.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 261.Bai H, Sun K, Wu JH, Zhong ZH, Xu SL, Zhang HR, Gu YH, Lu SF. Proteomic and metabolomic characterization of cardiac tissue in acute myocardial ischemia injury rats. PloS one. 2020;15:e0231797. doi: 10.1371/journal.pone.0231797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Sebastiao MJ, Almeida HV, Serra M, Hamdani N, Saraiva F, Lourenco AP, Barros AS, Vasques-Novoa F, Leite-Moreira A, Alves PM, Falcao-Pires I, et al. Unveiling Human Proteome Signatures of Heart Failure with Preserved Ejection Fraction. Biomedicines. 2022;10 doi: 10.3390/biomedicines10112943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Najafi A, Schuldt M, Pham TV, Michels M, Schlossarek S, Carrier L, Jimenez C, Zaccolo M, van der Velden J, Kuster DWD. PKA’s favorite son: prioritizing phosphorylation of phospholamban over cardiac troponin I contributes to diastolic dysfunction in hypertrophic cardiomyopathy. Journal of Molecular and Cellular Cardiology. 2018;120:39–40. [Google Scholar]
- 264.Najafi A, Sequeira V, Helmes M, Bollen IA, Goebel M, Regan JA, Carrier L, Kuster DW, Van Der Velden J. Selective phosphorylation of PKA targets after beta-adrenergic receptor stimulation impairs myofilament function in Mybpc3-targeted HCM mouse model. Cardiovasc Res. 2016;110:200–214. doi: 10.1093/cvr/cvw026. [DOI] [PubMed] [Google Scholar]
- 265.Capote LA, Mendez Perez R, Lymperopoulos A. GPCR signaling and cardiac function. Eur J Pharmacol. 2015;763:143–148. doi: 10.1016/j.ejphar.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 266.Engelhardt S, Hein L, Wiesmann F, Lohse MJ. Progressive hypertrophy and heart failure in beta1-adrenergic receptor transgenic mice. Proc Natl Acad Sci U S A. 1999;96:7059–7064. doi: 10.1073/pnas.96.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Schafer M, Frischkopf K, Taimor G, Piper HM, Schluter KD. Hypertrophic effect of selective beta(1)-adrenoceptor stimulation on ventricular cardiomyocytes from adult rat. Am J Physiol Cell Physiol. 2000;279:C495–503. doi: 10.1152/ajpcell.2000.279.2.C495. [DOI] [PubMed] [Google Scholar]
- 268.Kuschel M, Zhou YY, Spurgeon HA, Bartel S, Karczewski P, Zhang SJ, Krause EG, Lakatta EG, Xiao RP. beta2-adrenergic cAMP signaling is uncoupled from phosphorylation of cytoplasmic proteins in canine heart. Circulation. 1999;99:2458–2465. doi: 10.1161/01.cir.99.18.2458. [DOI] [PubMed] [Google Scholar]
- 269.Milano CA, Allen LF, Rockman HA, Dolber PC, McMinn TR, Chien KR, Johnson TD, Bond RA, Lefkowitz RJ. Enhanced myocardial function in transgenic mice overexpressing the beta 2-adrenergic receptor. Science. 1994;264:582–586. doi: 10.1126/science.8160017. [DOI] [PubMed] [Google Scholar]
- 270.De Jong KA, Nikolaev VO. Multifaceted remodelling of cAMP microdomains driven by different aetiologies of heart failure. FEBS J. 2021;288:6603–6622. doi: 10.1111/febs.15706. [DOI] [PubMed] [Google Scholar]
- 271.Lyon AR, MacLeod KT, Zhang Y, Garcia E, Kanda GK, Lab MJ, Korchev YE, Harding SE, Gorelik J. Loss of T-tubules and other changes to surface topography in ventricular myocytes from failing human and rat heart. Proc Natl Acad Sci U S A. 2009;106:6854–6859. doi: 10.1073/pnas.0809777106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Dorn GW, 2nd, Tepe NM, Lorenz JN, Koch WJ, Liggett SB. Low-and high-level transgenic expression of beta2-adrenergic receptors differentially affect cardiac hypertrophy and function in Galphaq-overexpressing mice. Proc Natl Acad Sci U S A. 1999;96:6400–6405. doi: 10.1073/pnas.96.11.6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lyon AR, Nikolaev VO, Miragoli M, Sikkel MB, Paur H, Benard L, Hulot JS, Kohlbrenner E, Hajjar RJ, Peters NS, Korchev YE, et al. Plasticity of surface structures and beta(2)-adrenergic receptor localization in failing ventricular cardiomyocytes during recovery from heart failure. Circ Heart Fail. 2012;5:357–365. doi: 10.1161/CIRCHEARTFAILURE.111.964692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Bao W, Aravindhan K, Alsaid H, Chendrimada T, Szapacs M, Citerone DR, Harpel MR, Willette RN, Lepore JJ, Jucker BM. Albiglutide, a long lasting glucagon-like peptide-1 analog, protects the rat heart against ischemia/reperfusion injury: evidence for improving cardiac metabolic efficiency. PloS one. 2011;6:e23570. doi: 10.1371/journal.pone.0023570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Marso SP, Daniels GH, Brown-Frandsen K, Kristensen P, Mann JF, Nauck MA, Nissen SE, Pocock S, Poulter NR, Ravn LS, Steinberg WM, et al. Liraglutide and Cardiovascular Outcomes in Type 2 Diabetes. The New England journal of medicine. 2016;375:311–322. doi: 10.1056/NEJMoa1603827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Noyan-Ashraf MH, Momen MA, Ban K, Sadi AM, Zhou YQ, Riazi AM, Baggio LL, Henkelman RM, Husain M, Drucker DJ. GLP-1R agonist liraglutide activates cytoprotective pathways and improves outcomes after experimental myocardial infarction in mice. Diabetes. 2009;58:975–983. doi: 10.2337/db08-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Bose AK, Mocanu MM, Carr RD, Brand CL, Yellon DM. Glucagon-like peptide 1 can directly protect the heart against ischemia/reperfusion injury. Diabetes. 2005;54:146–151. doi: 10.2337/diabetes.54.1.146. [DOI] [PubMed] [Google Scholar]
- 278.Zhang Y, Chen S, Luo L, Greenly S, Shi H, Xu JJ, Yan C. Role of cAMP in Cardiomyocyte Viability: Beneficial or Detrimental? Circ Res. 2023;133:902–923. doi: 10.1161/CIRCRESAHA.123.322652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Belevych AE, Sims C, Harvey RD. ACh-induced rebound stimulation of L-type Ca(2+) current in guinea-pig ventricular myocytes, mediated by Gbetagamma-dependent activation of adenylyl cyclase. J Physiol. 2001;536:677–692. doi: 10.1111/j.1469-7793.2001.00677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Belevych AE, Harvey RD. Muscarinic inhibitory and stimulatory regulation of the L-type Ca2+ current is not altered in cardiac ventricular myocytes from mice lacking endothelial nitric oxide synthase. J Physiol. 2000;528(Pt 2):279–289. doi: 10.1111/j.1469-7793.2000.00279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Cooper DM. Compartmentalization of adenylate cyclase and cAMP signalling. Biochem Soc Trans. 2005;33:1319–1322. doi: 10.1042/BST0331319. [DOI] [PubMed] [Google Scholar]
- 282.Ren L, Thai PN, Gopireddy RR, Timofeyev V, Ledford HA, Woltz RL, Park S, Puglisi JL, Moreno CM, Santana LF, Conti AC, et al. Adenylyl cyclase isoform 1 contributes to sinoatrial node automaticity via functional microdomains. JCI Insight. 2022;7 doi: 10.1172/jci.insight.162602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Yan L, Vatner DE, O’Connor JP, Ivessa A, Ge H, Chen W, Hirotani S, Ishikawa Y, Sadoshima J, Vatner SF. Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell. 2007;130:247–258. doi: 10.1016/j.cell.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 284.Vatner SF, Park M, Yan L, Lee GJ, Lai L, Iwatsubo K, Ishikawa Y, Pessin J, Vatner DE. Adenylyl cyclase type 5 in cardiac disease, metabolism, and aging. Am J Physiol Heart Circ Physiol. 2013;305:H1–8. doi: 10.1152/ajpheart.00080.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Roth DM, Bayat H, Drumm JD, Gao MH, Swaney JS, Ander A, Hammond HK. Adenylyl cyclase increases survival in cardiomyopathy. Circulation. 2002;105:1989–1994. doi: 10.1161/01.cir.0000014968.54967.d3. [DOI] [PubMed] [Google Scholar]
- 286.Lai NC, Roth DM, Gao MH, Tang T, Dalton N, Lai YY, Spellman M, Clopton P, Hammond HK. Intracoronary adenovirus encoding adenylyl cyclase VI increases left ventricular function in heart failure. Circulation. 2004;110:330–336. doi: 10.1161/01.CIR.0000136033.21777.4D. [DOI] [PubMed] [Google Scholar]
- 287.Gao MH, Miyanohara A, Feramisco JR, Tang T. Activation of PH-domain leucine-rich protein phosphatase 2 (PHLPP2) by agonist stimulation in cardiac myocytes expressing adenylyl cyclase type 6. Biochem Biophys Res Commun. 2009;384:193–198. doi: 10.1016/j.bbrc.2009.04.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Capel RA, Bose SJ, Collins TP, Rajasundaram S, Ayagama T, Zaccolo M, Burton RB, Terrar DA. IP3-mediated Ca(2+) release regulates atrial Ca(2+) transients and pacemaker function by stimulation of adenylyl cyclases. Am J Physiol Heart Circ Physiol. 2021;320:H95–H107. doi: 10.1152/ajpheart.00380.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Baldwin TA, Li Y, Marsden AN, Rinne S, Garza-Carbajal A, Schindler RFR, Zhang M, Garcia MA, Venna VR, Decher N, Brand T, et al. POPDC1 scaffolds a complex of adenylyl cyclase 9 and the potassium channel TREK-1 in heart. EMBO Rep. 2022;23:e55208. doi: 10.15252/embr.202255208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Li Y, Chen L, Kass RS, Dessauer CW. The A-kinase anchoring protein Yotiao facilitates complex formation between adenylyl cyclase type 9 and the IKs potassium channel in heart. J Biol Chem. 2012;287:29815–29824. doi: 10.1074/jbc.M112.380568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Lazar AM, Irannejad R, Baldwin TA, Sundaram AB, Gutkind JS, Inoue A, Dessauer CW, Von Zastrow M. G protein-regulated endocytic trafficking of adenylyl cyclase type 9. Elife. 2020;9 doi: 10.7554/eLife.58039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Rinaldi L, Pozdniakova S, Jayarajan V, Troidl C, Abdallah Y, Aslam M, Ladilov Y. Protective role of soluble adenylyl cyclase against reperfusion-induced injury of cardiac cells. Biochim Biophys Acta Mol Basis Dis. 2019;1865:252–260. doi: 10.1016/j.bbadis.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 293.Siraj MA, Mundil D, Beca S, Momen A, Shikatani EA, Afroze T, Sun X, Liu Y, Ghaffari S, Lee W, Wheeler MB, et al. Cardioprotective GLP-1 metabolite prevents ischemic cardiac injury by inhibiting mitochondrial trifunctional protein-alpha. J Clin Invest. 2020;130:1392–1404. doi: 10.1172/JCI99934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Bose SJ, Read MJ, Akerman E, Capel RA, Ayagama T, Russell A, Terrar DA, Zaccolo M, Burton RAB. Inhibition of adenylyl cyclase 1 by ST034307 inhibits IP(3)-evoked changes in sino-atrial node beat rate. Front Pharmacol. 2022;13:951897. doi: 10.3389/fphar.2022.951897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Okumura S, Kawabe J, Yatani A, Takagi G, Lee MC, Hong C, Liu J, Takagi I, Sadoshima J, Vatner DE, Vatner SF, et al. Type 5 adenylyl cyclase disruption alters not only sympathetic but also parasympathetic and calcium-mediated cardiac regulation. Circ Res. 2003;93:364–371. doi: 10.1161/01.RES.0000086986.35568.63. [DOI] [PubMed] [Google Scholar]
- 296.Okumura S, Takagi G, Kawabe J, Yang G, Lee MC, Hong C, Liu J, Vatner DE, Sadoshima J, Vatner SF, Ishikawa Y. Disruption of type 5 adenylyl cyclase gene preserves cardiac function against pressure overload. Proc Natl Acad Sci U S A. 2003;100:9986–9990. doi: 10.1073/pnas.1733772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Okumura S, Vatner DE, Kurotani R, Bai Y, Gao S, Yuan Z, Iwatsubo K, Ulucan C, Kawabe J, Ghosh K, Vatner SF, et al. Disruption of type 5 adenylyl cyclase enhances desensitization of cyclic adenosine monophosphate signal and increases Akt signal with chronic catecholamine stress. Circulation. 2007;116:1776–1783. doi: 10.1161/CIRCULATIONAHA.107.698662. [DOI] [PubMed] [Google Scholar]
- 298.Timofeyev V, Porter CA, Tuteja D, Qiu H, Li N, Tang T, Singapuri A, Han PL, Lopez JE, Hammond HK, Chiamvimonvat N. Disruption of adenylyl cyclase type V does not rescue the phenotype of cardiac-specific overexpression of Galphaq protein-induced cardiomyopathy. Am J Physiol Heart Circ Physiol. 2010;299:H1459–1467. doi: 10.1152/ajpheart.01208.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Bull Melsom C, Cosson MV, Orstavik O, Lai NC, Hammond HK, Osnes JB, Skomedal T, Nikolaev V, Levy FO, Krobert KA. Constitutive inhibitory G protein activity upon adenylyl cyclase-dependent cardiac contractility is limited to adenylyl cyclase type 6. PLoS One. 2019;14:e0218110. doi: 10.1371/journal.pone.0218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Gao MH, Lai NC, Giamouridis D, Kim YC, Guo T, Hammond HK. Cardiac-directed expression of a catalytically inactive adenylyl cyclase 6 protects the heart from sustained beta-adrenergic stimulation. PLoS One. 2017;12:e0181282. doi: 10.1371/journal.pone.0181282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Li Y, Hof T, Baldwin TA, Chen L, Kass RS, Dessauer CW. Regulation of I(Ks) Potassium Current by Isoproterenol in Adult Cardiomyocytes Requires Type 9 Adenylyl Cyclase. Cells. 2019;8 doi: 10.3390/cells8090981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Feng Q, Zhang Y, Li Y, Liu Z, Zuo J, Fang F. Two domains are critical for the nuclear localization of soluble adenylyl cyclase. Biochimie. 2006;88:319–328. doi: 10.1016/j.biochi.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 303.Chen J, Levin LR, Buck J. Role of soluble adenylyl cyclase in the heart. American journal of physiology Heart and circulatory physiology. 2012;302:H538–543. doi: 10.1152/ajpheart.00701.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Lefkimmiatis K, Leronni D, Hofer AM. The inner and outer compartments of mitochondria are sites of distinct cAMP/PKA signaling dynamics. The Journal of cell biology. 2013;202:453–462. doi: 10.1083/jcb.201303159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Acin-Perez R, Gatti DL, Bai Y, Manfredi G. Protein phosphorylation and prevention of cytochrome oxidase inhibition by ATP: coupled mechanisms of energy metabolism regulation. Cell Metab. 2011;13:712–719. doi: 10.1016/j.cmet.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Covian R, French S, Kusnetz H, Balaban RS. Stimulation of oxidative phosphorylation by calcium in cardiac mitochondria is not influenced by cAMP and PKA activity. Biochim Biophys Acta. 2014;1837:1913–1921. doi: 10.1016/j.bbabio.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Burdyga A, Surdo NC, Monterisi S, Di Benedetto G, Grisan F, Penna E, Pellegrini L, Zaccolo M, Bortolozzi M, Swietach P, Pozzan T, et al. Phosphatases control PKA-dependent functional microdomains at the outer mitochondrial membrane. Proc Natl Acad Sci U S A. 2018;115:E6497–E6506. doi: 10.1073/pnas.1806318115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Laudette M, Sainte-Marie Y, Cousin G, Bergonnier D, Belhabib I, Brun S, Formoso K, Laib L, Tortosa F, Bergoglio C, Marcheix B, et al. Cyclic AMP-binding protein Epac1 acts as a metabolic sensor to promote cardiomyocyte lipotoxicity. Cell Death Dis. 2021;12:824. doi: 10.1038/s41419-021-04113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Jayarajan V, Appukuttan A, Aslam M, Reusch P, Regitz-Zagrosek V, Ladilov Y. Regulation of AMPK activity by type 10 adenylyl cyclase: contribution to the mitochondrial biology, cellular redox and energy homeostasis. Cell Mol Life Sci. 2019;76:4945–4959. doi: 10.1007/s00018-019-03152-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Chen L, Marquardt ML, Tester DJ, Sampson KJ, Ackerman MJ, Kass RS. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc Natl Acad Sci U S A. 2007;104:20990–20995. doi: 10.1073/pnas.0710527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Omar MH, Kihiu M, Byrne DP, Lee KS, Lakey TM, Butcher E, Eyers PA, Scott JD. Classification of Cushing’s syndrome PKAc mutants based upon their ability to bind PKI. Biochem J. 2023;480:875–890. doi: 10.1042/BCJ20230183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Omar MH, Byrne DP, Shrestha S, Lakey TM, Lee KS, Lauer SM, Collins KB, Daly LA, Eyers CE, Baird GS, Ong SE, et al. Discovery of a Cushing’s syndrome protein kinase A mutant that biases signaling through type I AKAPs. Sci Adv. 2024;10:eadl1258. doi: 10.1126/sciadv.adl1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Abrenica B, AlShaaban M, Czubryt MP. The A-kinase anchor protein AKAP121 is a negative regulator of cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2009;46:674–681. doi: 10.1016/j.yjmcc.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 314.Perrino C, Feliciello A, Schiattarella GG, Esposito G, Guerriero R, Zaccaro L, Del Gatto A, Saviano M, Garbi C, Carangi R, Di Lorenzo E, et al. AKAP121 downregulation impairs protective cAMP signals, promotes mitochondrial dysfunction, and increases oxidative stress. Cardiovasc Res. 2010;88:101–110. doi: 10.1093/cvr/cvq155. [DOI] [PubMed] [Google Scholar]
- 315.Jun YW, Park H, Lee YK, Kaang BK, Lee JA, Jang DJ. D-AKAP1a is a signal-anchored protein in the mitochondrial outer membrane. FEBS Lett. 2016;590:954–961. doi: 10.1002/1873-3468.12123. [DOI] [PubMed] [Google Scholar]
- 316.Ma Y, Taylor SS. A molecular switch for targeting between endoplasmic reticulum (ER) and mitochondria: conversion of a mitochondria-targeting element into an ER-targeting signal in DAKAP1. J Biol Chem. 2008;283:11743–11751. doi: 10.1074/jbc.M710494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Marin W. A-kinase anchoring protein 1 (AKAP1) and its role in some cardiovascular diseases. J Mol Cell Cardiol. 2020;138:99–109. doi: 10.1016/j.yjmcc.2019.11.154. [DOI] [PubMed] [Google Scholar]
- 318.Chaturvedi D, Cohen MS, Taunton J, Patel TB. The PKARIalpha subunit of protein kinase A modulates the activation of p90RSK1 and its function. J Biol Chem. 2009;284:23670–23681. doi: 10.1074/jbc.M109.032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Gao X, Lin B, Sadayappan S, Patel TB. Interactions between the regulatory subunit of type I protein kinase A and p90 ribosomal S6 kinase1 regulate cardiomyocyte apoptosis. Mol Pharmacol. 2014;85:357–367. doi: 10.1124/mol.113.090613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Kim H, Scimia MC, Wilkinson D, Trelles RD, Wood MR, Bowtell D, Dillin A, Mercola M, Ronai ZA. Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Mol Cell. 2011;44:532–544. doi: 10.1016/j.molcel.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Qi B, He L, Zhao Y, Zhang L, He Y, Li J, Li C, Zhang B, Huang Q, Xing J, Li F, et al. Akap1 deficiency exacerbates diabetic cardiomyopathy in mice by NDUFS1-mediated mitochondrial dysfunction and apoptosis. Diabetologia. 2020;63:1072–1087. doi: 10.1007/s00125-020-05103-w. [DOI] [PubMed] [Google Scholar]
- 322.Welch EJ, Jones BW, Scott JD. Networking with AKAPs: context-dependent regulation of anchored enzymes. Mol Interv. 2010;10:86–97. doi: 10.1124/mi.10.2.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Li X, Nooh MM, Bahouth SW. Role of AKAP79/150 protein in beta1-adrenergic receptor trafficking and signaling in mammalian cells. J Biol Chem. 2013;288:33797–33812. doi: 10.1074/jbc.M113.470559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Cheng EP, Yuan C, Navedo MF, Dixon RE, Nieves-Cintron M, Scott JD, Santana LF. Restoration of normal L-type Ca2+ channel function during Timothy syndrome by ablation of an anchoring protein. Circ Res. 2011;109:255–261. doi: 10.1161/CIRCRESAHA.111.248252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Nichols CB, Rossow CF, Navedo MF, Westenbroek RE, Catterall WA, Santana LF, McKnight GS. Sympathetic stimulation of adult cardiomyocytes requires association of AKAP5 with a subpopulation of L-type calcium channels. Circ Res. 2010;107:747–756. doi: 10.1161/CIRCRESAHA.109.216127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Nieves-Cintron M, Hirenallur-Shanthappa D, Nygren PJ, Hinke SA, Dell’Acqua ML, Langeberg LK, Navedo M, Santana LF, Scott JD. AKAP150 participates in calcineurin/NFAT activation during the down-regulation of voltage-gated K(+) currents in ventricular myocytes following myocardial infarction. Cell Signal. 2016;28:733–740. doi: 10.1016/j.cellsig.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Bahouth SW, Nooh MM, Mancarella S. Involvement of SAP97 anchored multiprotein complexes in regulating cardiorenal signaling and trafficking networks. Biochem Pharmacol. 2023;208:115406. doi: 10.1016/j.bcp.2022.115406. [DOI] [PubMed] [Google Scholar]
- 328.Seyler C, Scherer D, Kopple C, Kulzer M, Korkmaz S, Xynogalos P, Thomas D, Kaya Z, Scholz E, Backs J, Karle C, et al. Role of plasma membrane-associated AKAPs for the regulation of cardiac I(K1) current by protein kinase A. Naunyn Schmiedebergs Arch Pharmacol. 2017;390:493–503. doi: 10.1007/s00210-017-1344-9. [DOI] [PubMed] [Google Scholar]
- 329.Kar P, Lin YP, Bhardwaj R, Tucker CJ, Bird GS, Hediger MA, Monico C, Amin N, Parekh AB. The N terminus of Orai1 couples to the AKAP79 signaling complex to drive NFAT1 activation by local Ca(2+) entry. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2012908118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Passariello CL, Li J, Dodge-Kafka K, Kapiloff MS. mAKAP-a master scaffold for cardiac remodeling. J Cardiovasc Pharmacol. 2015;65:218–225. doi: 10.1097/FJC.0000000000000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Kritzer MD, Li J, Passariello CL, Gayanilo M, Thakur H, Dayan J, Dodge-Kafka K, Kapiloff MS. The scaffold protein muscle A-kinase anchoring protein beta orchestrates cardiac myocyte hypertrophic signaling required for the development of heart failure. Circ Heart Fail. 2014;7:663–672. doi: 10.1161/CIRCHEARTFAILURE.114.001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Redden JM, Dodge-Kafka KL. AKAP phosphatase complexes in the heart. J Cardiovasc Pharmacol. 2011;58:354–362. doi: 10.1097/FJC.0b013e31821e5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Dodge-Kafka KL, Bauman A, Mayer N, Henson E, Heredia L, Ahn J, McAvoy T, Nairn AC, Kapiloff MS. cAMP-stimulated protein phosphatase 2A activity associated with muscle A kinase-anchoring protein (mAKAP) signaling complexes inhibits the phosphorylation and activity of the cAMP-specific phosphodiesterase PDE4D3. J Biol Chem. 2010;285:11078–11086. doi: 10.1074/jbc.M109.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Dodge-Kafka KL, Soughayer J, Pare GC, Carlisle Michel JJ, Langeberg LK, Kapiloff MS, Scott JD. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 2005;437:574–578. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Zhang L, Malik S, Kelley GG, Kapiloff MS, Smrcka AV. Phospholipase C epsilon scaffolds to muscle-specific A kinase anchoring protein (mAKAPbeta) and integrates multiple hypertrophic stimuli in cardiac myocytes. J Biol Chem. 2011;286:23012–23021. doi: 10.1074/jbc.M111.231993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Zhang L, Malik S, Pang J, Wang H, Park KM, Yule DI, Blaxall BC, Smrcka AV. Phospholipase Cepsilon hydrolyzes perinuclear phosphatidylinositol 4-phosphate to regulate cardiac hypertrophy. Cell. 2013;153:216–227. doi: 10.1016/j.cell.2013.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Schulze DH, Muqhal M, Lederer WJ, Ruknudin AM. Sodium/calcium exchanger (NCX1) macromolecular complex. J Biol Chem. 2003;278:28849–28855. doi: 10.1074/jbc.M300754200. [DOI] [PubMed] [Google Scholar]
- 338.Pare GC, Easlick JL, Mislow JM, McNally EM, Kapiloff MS. Nesprin-1alpha contributes to the targeting of mAKAP to the cardiac myocyte nuclear envelope. Exp Cell Res. 2005;303:388–399. doi: 10.1016/j.yexcr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 339.Martinez EC, Passariello CL, Li J, Matheson CJ, Dodge-Kafka K, Reigan P, Kapiloff MS. RSK3: A regulator of pathological cardiac remodeling. IUBMB Life. 2015;67:331–337. doi: 10.1002/iub.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Wong W, Goehring AS, Kapiloff MS, Langeberg LK, Scott JD. mAKAP compartmentalizes oxygen-dependent control of HIF-1alpha. Sci Signal. 2008;1:ra18. doi: 10.1126/scisignal.2000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Li J, Vargas MA, Kapiloff MS, Dodge-Kafka KL. Regulation of MEF2 transcriptional activity by calcineurin/mAKAP complexes. Exp Cell Res. 2013;319:447–454. doi: 10.1016/j.yexcr.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Li J, Kritzer MD, Michel JJ, Le A, Thakur H, Gayanilo M, Passariello CL, Negro A, Danial JB, Oskouei B, Sanders M, et al. Anchored p90 ribosomal S6 kinase 3 is required for cardiac myocyte hypertrophy. Circ Res. 2013;112:128–139. doi: 10.1161/CIRCRESAHA.112.276162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Singh A, Redden JM, Kapiloff MS, Dodge-Kafka KL. The large isoforms of A-kinase anchoring protein 18 mediate the phosphorylation of inhibitor-1 by protein kinase A and the inhibition of protein phosphatase 1 activity. Mol Pharmacol. 2011;79:533–540. doi: 10.1124/mol.110.065425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Hulme JT, Lin TW, Westenbroek RE, Scheuer T, Catterall WA. Beta-adrenergic regulation requires direct anchoring of PKA to cardiac CaV1.2 channels via a leucine zipper interaction with A kinase-anchoring protein 15. Proc Natl Acad Sci U S A. 2003;100:13093–13098. doi: 10.1073/pnas.2135335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Bunemann M, Gerhardstein BL, Gao T, Hosey MM. Functional regulation of L-type calcium channels via protein kinase A-mediated phosphorylation of the beta(2) subunit. J Biol Chem. 1999;274:33851–33854. doi: 10.1074/jbc.274.48.33851. [DOI] [PubMed] [Google Scholar]
- 346.Johnson KR, Nicodemus-Johnson J, Carnegie GK, Danziger RS. Molecular evolution of A-kinase anchoring protein (AKAP)-7: implications in comparative PKA compartmentalization. BMC Evol Biol. 2012;12:125. doi: 10.1186/1471-2148-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Carlson CR, Aronsen JM, Bergan-Dahl A, Moutty MC, Lunde M, Lunde PK, Jarstadmarken H, Wanichawan P, Pereira L, Kolstad TRS, Dalhus B, et al. AKAP18delta Anchors and Regulates CaMKII Activity at Phospholamban-SERCA2 and RYR. Circ Res. 2022;130:27–44. doi: 10.1161/CIRCRESAHA.120.317976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Marx SO, Kurokawa J, Reiken S, Motoike H, D’Armiento J, Marks AR, Kass RS. Requirement of a macromolecular signaling complex for beta adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science. 2002;295:496–499. doi: 10.1126/science.1066843. [DOI] [PubMed] [Google Scholar]
- 349.Kurokawa J, Motoike HK, Rao J, Kass RS. Regulatory actions of the A-kinase anchoring protein Yotiao on a heart potassium channel downstream of PKA phosphorylation. Proc Natl Acad Sci U S A. 2004;101:16374–16378. doi: 10.1073/pnas.0405583101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Berryman MA, Goldenring JR. CLIC4 is enriched at cell-cell junctions and colocalizes with AKAP350 at the centrosome and midbody of cultured mammalian cells. Cell Motil Cytoskeleton. 2003;56:159–172. doi: 10.1002/cm.10141. [DOI] [PubMed] [Google Scholar]
- 351.Kammerer S, Burns-Hamuro LL, Ma Y, Hamon SC, Canaves JM, Shi MM, Nelson MR, Sing CF, Cantor CR, Taylor SS, Braun A. Amino acid variant in the kinase binding domain of dual-specific A kinase-anchoring protein 2: a disease susceptibility polymorphism. Proc Natl Acad Sci U S A. 2003;100:4066–4071. doi: 10.1073/pnas.2628028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Tingley WG, Pawlikowska L, Zaroff JG, Kim T, Nguyen T, Young SG, Vranizan K, Kwok PY, Whooley MA, Conklin BR. Gene-trapped mouse embryonic stem cell-derived cardiac myocytes and human genetics implicate AKAP10 in heart rhythm regulation. Proc Natl Acad Sci U S A. 2007;104:8461–8466. doi: 10.1073/pnas.0610393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Loniewska B, Kaczmarczyk M, Clark JS, Goracy I, Horodnicka-Jozwa A, Ciechanowicz A. Association of functional genetic variants of A-kinase anchoring protein 10 with QT interval length in full-term Polish newborns. Arch Med Sci. 2015;11:149–154. doi: 10.5114/aoms.2013.34172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Huang LJ, Durick K, Weiner JA, Chun J, Taylor SS. D-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain. Proc Natl Acad Sci U S A. 1997;94:11184–11189. doi: 10.1073/pnas.94.21.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Wang L, Sunahara RK, Krumins A, Perkins G, Crochiere ML, Mackey M, Bell S, Ellisman MH, Taylor SS. Cloning and mitochondrial localization of full-length D-AKAP2, a protein kinase A anchoring protein. Proc Natl Acad Sci U S A. 2001;98:3220–3225. doi: 10.1073/pnas.051633398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Gisler SM, Madjdpour C, Bacic D, Pribanic S, Taylor SS, Biber J, Murer H. PDZK1: II. an anchoring site for the PKA-binding protein D-AKAP2 in renal proximal tubular cells. Kidney Int. 2003;64:1746–1754. doi: 10.1046/j.1523-1755.2003.00267.x. [DOI] [PubMed] [Google Scholar]
- 357.Eggers CT, Schafer JC, Goldenring JR, Taylor SS. D-AKAP2 interacts with Rab4 and Rab11 through its RGS domains and regulates transferrin receptor recycling. J Biol Chem. 2009;284:32869–32880. doi: 10.1074/jbc.M109.022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Nauert JB, Klauck TM, Langeberg LK, Scott JD. Gravin, an autoantigen recognized by serum from myasthenia gravis patients, is a kinase scaffold protein. Curr Biol. 1997;7:52–62. doi: 10.1016/s0960-9822(06)00027-3. [DOI] [PubMed] [Google Scholar]
- 359.Lin F, Wang H, Malbon CC. Gravin-mediated formation of signaling complexes in beta 2-adrenergic receptor desensitization and resensitization. J Biol Chem. 2000;275:19025–19034. doi: 10.1074/jbc.275.25.19025. [DOI] [PubMed] [Google Scholar]
- 360.Guillory AN, Yin X, Wijaya CS, Diaz Diaz AC, Rababa’h A, Singh S, Atrooz F, Sadayappan S, McConnell BK. Enhanced cardiac function in Gravin mutant mice involves alterations in the beta-adrenergic receptor signaling cascade. PLoS One. 2013;8:e74784. doi: 10.1371/journal.pone.0074784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Li Z, Singh S, Suryavanshi SV, Ding W, Shen X, Wijaya CS, Gao WD, McConnell BK. Force development and intracellular Ca(2+) in intact cardiac muscles from gravin mutant mice. Eur J Pharmacol. 2017;807:117–126. doi: 10.1016/j.ejphar.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Diviani D, Osman H, Reggi E. A-Kinase Anchoring Protein-Lbc: A Molecular Scaffold Involved in Cardiac Protection. J Cardiovasc Dev Dis. 2018;5 doi: 10.3390/jcdd5010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Diviani D, Soderling J, Scott JD. AKAP-Lbc anchors protein kinase A and nucleates Galpha 12-selective Rho-mediated stress fiber formation. J Biol Chem. 2001;276:44247–44257. doi: 10.1074/jbc.M106629200. [DOI] [PubMed] [Google Scholar]
- 364.Appert-Collin A, Cotecchia S, Nenniger-Tosato M, Pedrazzini T, Diviani D. The A-kinase anchoring protein (AKAP)-Lbc-signaling complex mediates alpha1 adrenergic receptor-induced cardiomyocyte hypertrophy. Proc Natl Acad Sci U S A. 2007;104:10140–10145. doi: 10.1073/pnas.0701099104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Carnegie GK, Soughayer J, Smith FD, Pedroja BS, Zhang F, Diviani D, Bristow MR, Kunkel MT, Newton AC, Langeberg LK, Scott JD. AKAP-Lbc mobilizes a cardiac hypertrophy signaling pathway. Mol Cell. 2008;32:169–179. doi: 10.1016/j.molcel.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Taglieri DM, Johnson KR, Burmeister BT, Monasky MM, Spindler MJ, DeSantiago J, Banach K, Conklin BR, Carnegie GK. The C-terminus of the long AKAP13 isoform (AKAP-Lbc) is critical for development of compensatory cardiac hypertrophy. Journal of molecular and cellular cardiology. 2014;66:27–40. doi: 10.1016/j.yjmcc.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Klussmann E, Edemir B, Pepperle B, Tamma G, Henn V, Klauschenz E, Hundsrucker C, Maric K, Rosenthal W. Ht31: the first protein kinase A anchoring protein to integrate protein kinase A and Rho signaling. FEBS Lett. 2001;507:264–268. doi: 10.1016/s0014-5793(01)02995-7. [DOI] [PubMed] [Google Scholar]
- 368.Carnegie GK, Smith FD, McConnachie G, Langeberg LK, Scott JD. AKAP-Lbc nucleates a protein kinase D activation scaffold. Mol Cell. 2004;15:889–899. doi: 10.1016/j.molcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 369.Burmeister BT, Taglieri DM, Wang L, Carnegie GK. Src homology 2 domain-containing phosphatase 2 (Shp2) is a component of the A-kinase-anchoring protein (AKAP)-Lbc complex and is inhibited by protein kinase A (PKA) under pathological hypertrophic conditions in the heart. J Biol Chem. 2012;287:40535–40546. doi: 10.1074/jbc.M112.385641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Mayers CM, Wadell J, McLean K, Venere M, Malik M, Shibata T, Driggers PH, Kino T, Guo XC, Koide H, Gorivodsky M, et al. The Rho guanine nucleotide exchange factor AKAP13 (BRX) is essential for cardiac development in mice. J Biol Chem. 2010;285:12344–12354. doi: 10.1074/jbc.M110.106856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Smith FD, Langeberg LK, Cellurale C, Pawson T, Morrison DK, Davis RJ, Scott JD. AKAP-Lbc enhances cyclic AMP control of the ERK1/2 cascade. Nat Cell Biol. 2010;12:1242–1249. doi: 10.1038/ncb2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Cariolato L, Cavin S, Diviani D. A-kinase anchoring protein (AKAP)-Lbc anchors a PKN-based signaling complex involved in alpha1-adrenergic receptor-induced p38 activation. J Biol Chem. 2011;286:7925–7937. doi: 10.1074/jbc.M110.185645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.del Vescovo CD, Cotecchia S, Diviani D. A-kinase-anchoring protein-Lbc anchors IkappaB kinase beta to support interleukin-6-mediated cardiomyocyte hypertrophy. Mol Cell Biol. 2013;33:14–27. doi: 10.1128/MCB.00887-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Perez Lopez I, Cariolato L, Maric D, Gillet L, Abriel H, Diviani D. A-kinase anchoring protein Lbc coordinates a p38 activating signaling complex controlling compensatory cardiac hypertrophy. Mol Cell Biol. 2013;33:2903–2917. doi: 10.1128/MCB.00031-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Lacana E, Maceyka M, Milstien S, Spiegel S. Cloning and characterization of a protein kinase A anchoring protein (AKAP)-related protein that interacts with and regulates sphingosine kinase 1 activity. J Biol Chem. 2002;277:32947–32953. doi: 10.1074/jbc.M202841200. [DOI] [PubMed] [Google Scholar]
- 376.Peng H, Zhang J, Ya A, Ma W, Villa S, Sukenik S, Ge X. Myomegalin regulates Hedgehog pathway by controlling PDE4D at the centrosome. Mol Biol Cell. 2021;32:1807–1817. doi: 10.1091/mbc.E21-02-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Verde I, Pahlke G, Salanova M, Zhang G, Wang S, Coletti D, Onuffer J, Jin SL, Conti M. Myomegalin is a novel protein of the golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J Biol Chem. 2001;276:11189–11198. doi: 10.1074/jbc.M006546200. [DOI] [PubMed] [Google Scholar]
- 378.Uys GM, Ramburan A, Loos B, Kinnear CJ, Korkie LJ, Mouton J, Riedemann J, Moolman-Smook JC. Myomegalin is a novel A-kinase anchoring protein involved in the phosphorylation of cardiac myosin binding protein C. BMC Cell Biol. 2011;12:18. doi: 10.1186/1471-2121-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Abou Ziki MD, Bhat N, Neogi A, Driscoll TP, Ugwu N, Liu Y, Smith E, Abboud JM, Chouairi S, Schwartz MA, Akar JG, et al. Epistatic interaction of PDE4DIP and DES mutations in familial atrial fibrillation with slow conduction. Hum Mutat. 2021;42:1279–1293. doi: 10.1002/humu.24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Sumandea CA, Garcia-Cazarin ML, Bozio CH, Sievert GA, Balke CW, Sumandea MP. Cardiac troponin T, a sarcomeric AKAP, tethers protein kinase A at the myofilaments. J Biol Chem. 2011;286:530–541. doi: 10.1074/jbc.M110.148684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Sfichi-Duke L, Garcia-Cazarin ML, Sumandea CA, Sievert GA, Balke CW, Zhan DY, Morimoto S, Sumandea MP. Cardiomyopathy-causing deletion K210 in cardiac troponin T alters phosphorylation propensity of sarcomeric proteins. J Mol Cell Cardiol. 2010;48:934–942. doi: 10.1016/j.yjmcc.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Russell MA, Lund LM, Haber R, McKeegan K, Cianciola N, Bond M. The intermediate filament protein, synemin, is an AKAP in the heart. Arch Biochem Biophys. 2006;456:204–215. doi: 10.1016/j.abb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 383.Garcia-Pelagio KP, Chen L, Joca HC, Ward C, Jonathan Lederer W, Bloch RJ. Absence of synemin in mice causes structural and functional abnormalities in heart. J Mol Cell Cardiol. 2018;114:354–363. doi: 10.1016/j.yjmcc.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Russell MA. Synemin Redefined: Multiple Binding Partners Results in Multifunctionality. Front Cell Dev Biol. 2020;8:159. doi: 10.3389/fcell.2020.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Li H, Adamik R, Pacheco-Rodriguez G, Moss J, Vaughan M. Protein kinase A-anchoring (AKAP) domains in brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2. Proc Natl Acad Sci U S A. 2003;100:1627–1632. doi: 10.1073/pnas.0337678100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Kuroda F, Moss J, Vaughan M. Regulation of brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1) and BIG2 activity via PKA and protein phosphatase 1gamma. Proc Natl Acad Sci U S A. 2007;104:3201–3206. doi: 10.1073/pnas.0611696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Puxeddu E, Uhart M, Li CC, Ahmad F, Pacheco-Rodriguez G, Manganiello VC, Moss J, Vaughan M. Interaction of phosphodiesterase 3A with brefeldin A-inhibited guanine nucleotide-exchange proteins BIG1 and BIG2 and effect on ARF1 activity. Proc Natl Acad Sci U S A. 2009;106:6158–6163. doi: 10.1073/pnas.0901558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Lin C, Guo X, Lange S, Liu J, Ouyang K, Yin X, Jiang L, Cai Y, Mu Y, Sheikh F, Ye S, et al. Cypher/ZASP is a novel A-kinase anchoring protein. J Biol Chem. 2013;288:29403–29413. doi: 10.1074/jbc.M113.470708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Arimura T, Inagaki N, Hayashi T, Shichi D, Sato A, Hinohara K, Vatta M, Towbin JA, Chikamori T, Yamashina A, Kimura A. Impaired binding of ZASP/Cypher with phosphoglucomutase 1 is associated with dilated cardiomyopathy. Cardiovasc Res. 2009;83:80–88. doi: 10.1093/cvr/cvp119. [DOI] [PubMed] [Google Scholar]
- 390.Zheng M, Cheng H, Li X, Zhang J, Cui L, Ouyang K, Han L, Zhao T, Gu Y, Dalton ND, Bang ML, et al. Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum Mol Genet. 2009;18:701–713. doi: 10.1093/hmg/ddn400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Yu H, Yuan C, Westenbroek RE, Catterall WA. The AKAP Cypher/Zasp contributes to beta-adrenergic/PKA stimulation of cardiac Ca(V)1.2 calcium channels. J Gen Physiol. 2018;150:883–889. doi: 10.1085/jgp.201711818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Lv J, Pan Z, Chen J, Xu R, Wang D, Huang J, Dong Y, Jiang J, Yin X, Cheng H, Guo X. Phosphoproteomic Analysis Reveals Downstream PKA Effectors of AKAP Cypher/ZASP in the Pathogenesis of Dilated Cardiomyopathy. Front Cardiovasc Med. 2021;8:753072. doi: 10.3389/fcvm.2021.753072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Kang M, Otani Y, Guo Y, Yan J, Goult BT, Howe AK. The focal adhesion protein talin is a mechanically gated A-kinase anchoring protein. Proc Natl Acad Sci U S A. 2024;121:e2314947121. doi: 10.1073/pnas.2314947121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Czachor A, Failla A, Lockey R, Kolliputi N. Pivotal role of AKAP121 in mitochondrial physiology. Am J Physiol Cell Physiol. 2016;310:C625–628. doi: 10.1152/ajpcell.00292.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Gabrovsek L, Collins KB, Aggarwal S, Saunders LM, Lau HT, Suh D, Sancak Y, Trapnell C, Ong SE, Smith FD, Scott JD. A-kinase-anchoring protein 1 (dAKAP1)-based signaling complexes coordinate local protein synthesis at the mitochondrial surface. J Biol Chem. 2020;295:10749–10765. doi: 10.1074/jbc.RA120.013454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Schiattarella GG, Cattaneo F, Pironti G, Magliulo F, Carotenuto G, Pirozzi M, Polishchuk R, Borzacchiello D, Paolillo R, Oliveti M, Boccella N, et al. Akap1 Deficiency Promotes Mitochondrial Aberrations and Exacerbates Cardiac Injury Following Permanent Coronary Ligation via Enhanced Mitophagy and Apoptosis. PLoS One. 2016;11:e0154076. doi: 10.1371/journal.pone.0154076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Gao T, Yatani A, Dell’Acqua ML, Sako H, Green SA, Dascal N, Scott JD, Hosey MM. cAMP-dependent regulation of cardiac L-type Ca2+ channels requires membrane targeting of PKA and phosphorylation of channel subunits. Neuron. 1997;19:185–196. doi: 10.1016/s0896-6273(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 398.Catterall WA. Structure and regulation of voltage-gated Ca2+ channels. Annu Rev Cell Dev Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 399.Navedo MF, Santana LF. CaV1.2 sparklets in heart and vascular smooth muscle. J Mol Cell Cardiol. 2013;58:67–76. doi: 10.1016/j.yjmcc.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Li L, Li J, Drum BM, Chen Y, Yin H, Guo X, Luckey SW, Gilbert ML, McKnight GS, Scott JD, Santana LF, et al. Loss of AKAP150 promotes pathological remodelling and heart failure propensity by disrupting calcium cycling and contractile reserve. Cardiovasc Res. 2017;113:147–159. doi: 10.1093/cvr/cvw221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Matheus AS, Tannus LR, Cobas RA, Palma CC, Negrato CA, Gomes MB. Impact of diabetes on cardiovascular disease: an update. Int J Hypertens. 2013;2013:653789. doi: 10.1155/2013/653789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Zeng C, Wang J, Li N, Shen M, Wang D, Yu Q, Wang H. AKAP150 mobilizes cPKC-dependent cardiac glucotoxicity. Am J Physiol Endocrinol Metab. 2014;307:E384–397. doi: 10.1152/ajpendo.00175.2014. [DOI] [PubMed] [Google Scholar]
- 403.Navedo MF, Nieves-Cintron M, Amberg GC, Yuan C, Votaw VS, Lederer WJ, McKnight GS, Santana LF. AKAP150 is required for stuttering persistent Ca2+ sparklets and angiotensin II-induced hypertension. Circ Res. 2008;102:e1–e11. doi: 10.1161/CIRCRESAHA.107.167809. [DOI] [PubMed] [Google Scholar]
- 404.McCartney S, Little BM, Langeberg LK, Scott JD. Cloning and characterization of A-kinase anchor protein 100 (AKAP100). A protein that targets A-kinase to the sarcoplasmic reticulum. J Biol Chem. 1995;270:9327–9333. doi: 10.1074/jbc.270.16.9327. [DOI] [PubMed] [Google Scholar]
- 405.Kapiloff MS, Schillace RV, Westphal AM, Scott JD. mAKAP: an A-kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. J Cell Sci. 1999;112(Pt 16):2725–2736. doi: 10.1242/jcs.112.16.2725. [DOI] [PubMed] [Google Scholar]
- 406.Kapiloff MS, Piggott LA, Sadana R, Li J, Heredia LA, Henson E, Efendiev R, Dessauer CW. An adenylyl cyclase-mAKAPbeta signaling complex regulates cAMP levels in cardiac myocytes. J Biol Chem. 2009;284:23540–23546. doi: 10.1074/jbc.M109.030072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Li J, Negro A, Lopez J, Bauman AL, Henson E, Dodge-Kafka K, Kapiloff MS. The mAKAPbeta scaffold regulates cardiac myocyte hypertrophy via recruitment of activated calcineurin. J Mol Cell Cardiol. 2010;48:387–394. doi: 10.1016/j.yjmcc.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Ruknudin A, He S, Lederer WJ, Schulze DH. Functional differences between cardiac and renal isoforms of the rat Na+-Ca2+ exchanger NCX1 expressed in Xenopus oocytes. J Physiol. 2000;529(Pt 3):599–610. doi: 10.1111/j.1469-7793.2000.00599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Gray PC, Tibbs VC, Catterall WA, Murphy BJ. Identification of a 15-kDa cAMP-dependent protein kinase-anchoring protein associated with skeletal muscle L-type calcium channels. J Biol Chem. 1997;272:6297–6302. doi: 10.1074/jbc.272.10.6297. [DOI] [PubMed] [Google Scholar]
- 410.Hulme JT, Ahn M, Hauschka SD, Scheuer T, Catterall WA. A novel leucine zipper targets AKAP15 and cyclic AMP-dependent protein kinase to the C terminus of the skeletal muscle Ca2+ channel and modulates its function. J Biol Chem. 2002;277:4079–4087. doi: 10.1074/jbc.M109814200. [DOI] [PubMed] [Google Scholar]
- 411.Hulme JT, Westenbroek RE, Scheuer T, Catterall WA. Phosphorylation of serine 1928 in the distal C-terminal domain of cardiac CaV1.2 channels during beta1-adrenergic regulation. Proc Natl Acad Sci U S A. 2006;103:16574–16579. doi: 10.1073/pnas.0607294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Fu Y, Westenbroek RE, Scheuer T, Catterall WA. Phosphorylation sites required for regulation of cardiac calcium channels in the fight-or-flight response. Proc Natl Acad Sci U S A. 2013;110:19621–19626. doi: 10.1073/pnas.1319421110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Fu Y, Westenbroek RE, Scheuer T, Catterall WA. Basal and beta-adrenergic regulation of the cardiac calcium channel CaV1.2 requires phosphorylation of serine 1700. Proc Natl Acad Sci U S A. 2014;111:16598–16603. doi: 10.1073/pnas.1419129111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Pallien T, Klussmann E. New aspects in cardiac L-type Ca2+ channel regulation. Biochem Soc Trans. 2020;48:39–49. doi: 10.1042/BST20190229. [DOI] [PubMed] [Google Scholar]
- 415.Piggott LA, Bauman AL, Scott JD, Dessauer CW. The A-kinase anchoring protein Yotiao binds and regulates adenylyl cyclase in brain. Proc Natl Acad Sci U S A. 2008;105:13835–13840. doi: 10.1073/pnas.0712100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Kurokawa J, Chen L, Kass RS. Requirement of subunit expression for cAMP-mediated regulation of a heart potassium channel. Proc Natl Acad Sci U S A. 2003;100:2122–2127. doi: 10.1073/pnas.0434935100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Yang T, Kanki H, Roden DM. Phosphorylation of the IKs channel complex inhibits drug block: novel mechanism underlying variable antiarrhythmic drug actions. Circulation. 2003;108:132–134. doi: 10.1161/01.CIR.0000082708.86266.B8. [DOI] [PubMed] [Google Scholar]
- 418.Chen L, Kurokawa J, Kass RS. Phosphorylation of the A-kinase-anchoring protein Yotiao contributes to protein kinase A regulation of a heart potassium channel. J Biol Chem. 2005;280:31347–31352. doi: 10.1074/jbc.M505191200. [DOI] [PubMed] [Google Scholar]
- 419.Westphal RS, Tavalin SJ, Lin JW, Alto NM, Fraser ID, Langeberg LK, Sheng M, Scott JD. Regulation of NMDA receptors by an associated phosphatase-kinase signaling complex. Science. 1999;285:93–96. doi: 10.1126/science.285.5424.93. [DOI] [PubMed] [Google Scholar]
- 420.Lin JW, Wyszynski M, Madhavan R, Sealock R, Kim JU, Sheng M. Yotiao, a novel protein of neuromuscular junction and brain that interacts with specific splice variants of NMDA receptor subunit NR1. J Neurosci. 1998;18:2017–2027. doi: 10.1523/JNEUROSCI.18-06-02017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Terrenoire C, Houslay MD, Baillie GS, Kass RS. The cardiac IKs potassium channel macromolecular complex includes the phosphodiesterase PDE4D3. J Biol Chem. 2009;284:9140–9146. doi: 10.1074/jbc.M805366200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Sarma GN, Moody IS, Ilouz R, Phan RH, Sankaran B, Hall RA, Taylor SS. D-AKAP2:PKA RII:PDZK1 ternary complex structure: insights from the nucleation of a polyvalent scaffold. Protein Sci. 2015;24:105–116. doi: 10.1002/pro.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Neumann SA, Tingley WG, Conklin BR, Shrader CJ, Peet E, Muldoon MF, Jennings JR, Ferrell RE, Manuck SB. AKAP10 (I646V) functional polymorphism predicts heart rate and heart rate variability in apparently healthy, middle-aged European-Americans. Psychophysiology. 2009;46:466–472. doi: 10.1111/j.1469-8986.2009.00802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Qasim H, McConnell BK. AKAP12 Signaling Complex: Impacts of Compartmentalizing cAMP-Dependent Signaling Pathways in the Heart and Various Signaling Systems. J Am Heart Assoc. 2020;9:e016615. doi: 10.1161/JAHA.120.016615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Fan G, Shumay E, Wang H, Malbon CC. The scaffold protein gravin (cAMP-dependent protein kinase-anchoring protein 250) binds the beta 2-adrenergic receptor via the receptor cytoplasmic Arg-329 to Leu-413 domain and provides a mobile scaffold during desensitization. J Biol Chem. 2001;276:24005–24014. doi: 10.1074/jbc.M011199200. [DOI] [PubMed] [Google Scholar]
- 426.Qasim H, Rajaei M, Xu Y, Reyes-Alcaraz A, Abdelnasser HY, Stewart MD, Lahiri SK, Wehrens XHT, McConnell BK. AKAP12 Upregulation Associates With PDE8A to Accelerate Cardiac Dysfunction. Circ Res. 2024 doi: 10.1161/CIRCRESAHA.123.323655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Diviani D, Reggi E, Arambasic M, Caso S, Maric D. Emerging roles of A-kinase anchoring proteins in cardiovascular pathophysiology. Biochim Biophys Acta. 2016;1863:1926–1936. doi: 10.1016/j.bbamcr.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 428.Caso S, Maric D, Arambasic M, Cotecchia S, Diviani D. AKAP-Lbc mediates protection against doxorubicin-induced cardiomyocyte toxicity. Biochim Biophys Acta Mol Cell Res. 2017;1864:2336–2346. doi: 10.1016/j.bbamcr.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 429.Edwards HV, Scott JD, Baillie GS. The A-kinase-anchoring protein AKAP-Lbc facilitates cardioprotective PKA phosphorylation of Hsp20 on Ser(16. Biochem J. 2012;446:437–443. doi: 10.1042/BJ20120570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Edwards HV, Scott JD, Baillie GS. PKA phosphorylation of the small heat-shock protein Hsp20 enhances its cardioprotective effects. Biochem Soc Trans. 2012;40:210–214. doi: 10.1042/BST20110673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Wang L, Burmeister BT, Johnson KR, Baillie GS, Karginov AV, Skidgel RA, O’Bryan JP, Carnegie GK. UCR1C is a novel activator of phosphodiesterase 4 (PDE4) long isoforms and attenuates cardiomyocyte hypertrophy. Cell Signal. 2015;27:908–922. doi: 10.1016/j.cellsig.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Sin YY, Edwards HV, Li X, Day JP, Christian F, Dunlop AJ, Adams DR, Zaccolo M, Houslay MD, Baillie GS. Disruption of the cyclic AMP phosphodiesterase-4 (PDE4)-HSP20 complex attenuates the beta-agonist induced hypertrophic response in cardiac myocytes. J Mol Cell Cardiol. 2011;50:872–883. doi: 10.1016/j.yjmcc.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 433.Means CK, Xiao CY, Li Z, Zhang T, Omens JH, Ishii I, Chun J, Brown JH. Sphingosine 1-phosphate S1P2 and S1P3 receptor-mediated Akt activation protects against in vivo myocardial ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2007;292:H2944–2951. doi: 10.1152/ajpheart.01331.2006. [DOI] [PubMed] [Google Scholar]
- 434.Lobo MJ, Reverte-Salisa L, Chao YC, Koschinski A, Gesellchen F, Subramaniam G, Jiang H, Pace S, Larcom N, Paolocci E, Pfeifer A, et al. Phosphodiesterase 2A2 regulates mitochondria clearance through Parkin-dependent mitophagy. Communications Biology. 2020;3 doi: 10.1038/s42003-020-01311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Halls ML, Cooper DM. Adenylyl cyclase signalling complexes - Pharmacological challenges and opportunities. Pharmacol Ther. 2017;172:171–180. doi: 10.1016/j.pharmthera.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 436.Lukyanenko YO, Younes A, Lyashkov AE, Tarasov KV, Riordon DR, Lee J, Sirenko SG, Kobrinsky E, Ziman B, Tarasova YS, Juhaszova M, et al. Ca(2+)/calmodulin-activated phosphodiesterase 1A is highly expressed in rabbit cardiac sinoatrial nodal cells and regulates pacemaker function. J Mol Cell Cardiol. 2016;98:73–82. doi: 10.1016/j.yjmcc.2016.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Vandeput F, Wolda SL, Krall J, Hambleton R, Uher L, McCaw KN, Radwanski PB, Florio V, Movsesian MA. Cyclic nucleotide phosphodiesterase PDE1C1 in human cardiac myocytes. The Journal of biological chemistry. 2007;282:32749–32757. doi: 10.1074/jbc.M703173200. [DOI] [PubMed] [Google Scholar]
- 438.Miller CL, Oikawa M, Cai Y, Wojtovich AP, Nagel DJ, Xu X, Xu H, Florio V, Rybalkin SD, Beavo JA, Chen YF, et al. Role of Ca2+/calmodulin-stimulated cyclic nucleotide phosphodiesterase 1 in mediating cardiomyocyte hypertrophy. Circ Res. 2009;105:956–964. doi: 10.1161/CIRCRESAHA.109.198515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Knight WE, Chen S, Zhang Y, Oikawa M, Wu M, Zhou Q, Miller CL, Cai Y, Mickelsen DM, Moravec C, Small EM, et al. PDE1C deficiency antagonizes pathological cardiac remodeling and dysfunction. Proc Natl Acad Sci U S A. 2016;113:E7116–E7125. doi: 10.1073/pnas.1607728113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Hashimoto T, Kim GE, Tunin RS, Adesiyun T, Hsu S, Nakagawa R, Zhu G, O’Brien JJ, Hendrick JP, Davis RE, Yao W, et al. Acute Enhancement of Cardiac Function by Phosphodiesterase Type 1 Inhibition. Circulation. 2018;138:1974–1987. doi: 10.1161/CIRCULATIONAHA.117.030490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Chen S, Knight WE, Yan C. Roles of PDE1 in Pathological Cardiac Remodeling and Dysfunction. J Cardiovasc Dev Dis. 2018;5 doi: 10.3390/jcdd5020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Lugnier C, Keravis T, Le Bec A, Pauvert O, Proteau S, Rousseau E. Characterization of cyclic nucleotide phosphodiesterase isoforms associated to isolated cardiac nuclei. Biochim Biophys Acta. 1999;1472:431–446. doi: 10.1016/s0304-4165(99)00145-2. [DOI] [PubMed] [Google Scholar]
- 443.Monterisi S, Lobo MJ, Livie C, Castle JC, Weinberger M, Baillie G, Surdo NC, Musheshe N, Stangherlin A, Gottlieb E, Maizels R, et al. PDE2A2 regulates mitochondria morphology and apoptotic cell death via local modulation of cAMP/PKA signalling. Elife. 2017;6 doi: 10.7554/eLife.21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Lobo MJ, Reverte-Salisa L, Chao YC, Koschinski A, Gesellchen F, Subramaniam G, Jiang H, Pace S, Larcom N, Paolocci E, Pfeifer A, et al. Phosphodiesterase 2A2 regulates mitochondria clearance through Parkin-dependent mitophagy. Commun Biol. 2020;3:596. doi: 10.1038/s42003-020-01311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Mehel H, Emons J, Vettel C, Wittkopper K, Seppelt D, Dewenter M, Lutz S, Sossalla S, Maier LS, Lechene P, Leroy J, et al. Phosphodiesterase-2 is up-regulated in human failing hearts and blunts beta-adrenergic responses in cardiomyocytes. J Am Coll Cardiol. 2013;62:1596–1606. doi: 10.1016/j.jacc.2013.05.057. [DOI] [PubMed] [Google Scholar]
- 446.Galindo-Tovar A, Vargas ML, Kaumann AJ. Phosphodiesterase PDE2 activity, increased by isoprenaline, does not reduce beta-adrenoceptor-mediated chronotropic and inotropic effects in rat heart. Naunyn Schmiedebergs Arch Pharmacol. 2018;391:571–585. doi: 10.1007/s00210-018-1480-x. [DOI] [PubMed] [Google Scholar]
- 447.Vettel C, Lindner M, Dewenter M, Lorenz K, Schanbacher C, Riedel M, Lammle S, Meinecke S, Mason FE, Sossalla S, Geerts A, et al. Phosphodiesterase 2 Protects Against Catecholamine-Induced Arrhythmia and Preserves Contractile Function After Myocardial Infarction. Circ Res. 2017;120:120–132. doi: 10.1161/CIRCRESAHA.116.310069. [DOI] [PubMed] [Google Scholar]
- 448.Wagner M, Sadek MS, Dybkova N, Mason FE, Klehr J, Firneburg R, Cachorro E, Richter K, Klapproth E, Kuenzel SR, Lorenz K, et al. Cellular Mechanisms of the Anti-Arrhythmic Effect of Cardiac PDE2 Overexpression. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22094816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Wechsler J, Choi YH, Krall J, Ahmad F, Manganiello VC, Movsesian MA. Isoforms of cyclic nucleotide phosphodiesterase PDE3A in cardiac myocytes. J Biol Chem. 2002;277:38072–38078. doi: 10.1074/jbc.M203647200. [DOI] [PubMed] [Google Scholar]
- 450.Liu H, Maurice DH. Expression of cyclic GMP-inhibited phosphodiesterases 3A and 3B (PDE3A and PDE3B) in rat tissues: differential subcellular localization and regulated expression by cyclic AMP. British journal of pharmacology. 1998;125:1501–1510. doi: 10.1038/sj.bjp.0702227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Shakur Y, Takeda K, Kenan Y, Yu ZX, Rena G, Brandt D, Houslay MD, Degerman E, Ferrans VJ, Manganiello VC. Membrane localization of cyclic nucleotide phosphodiesterase 3 (PDE3). Two N-terminal domains are required for the efficient targeting to, and association of, PDE3 with endoplasmic reticulum. J Biol Chem. 2000;275:38749–38761. doi: 10.1074/jbc.M001734200. [DOI] [PubMed] [Google Scholar]
- 452.Mongillo M, McSorley T, Evellin S, Sood A, Lissandron V, Terrin A, Huston E, Hannawacker A, Lohse MJ, Pozzan T, Houslay MD, et al. Fluorescence resonance energy transfer-based analysis of cAMP dynamics in live neonatal rat cardiac myocytes reveals distinct functions of compartmentalized phosphodiesterases. Circ Res. 2004;95:67–75. doi: 10.1161/01.RES.0000134629.84732.11. [DOI] [PubMed] [Google Scholar]
- 453.Movsesian MA, Smith CJ, Krall J, Bristow MR, Manganiello VC. Sarcoplasmic reticulum-associated cyclic adenosine 5’-monophosphate phosphodiesterase activity in normal and failing human hearts. J Clin Invest. 1991;88:15–19. doi: 10.1172/JCI115272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Smith CJ, Huang R, Sun D, Ricketts S, Hoegler C, Ding JZ, Moggio RA, Hintze TH. Development of decompensated dilated cardiomyopathy is associated with decreased gene expression and activity of the milrinone-sensitive cAMP phosphodiesterase PDE3A. Circulation. 1997;96:3116–3123. doi: 10.1161/01.cir.96.9.3116. [DOI] [PubMed] [Google Scholar]
- 455.Yan C, Miller CL, Abe J. Regulation of phosphodiesterase 3 and inducible cAMP early repressor in the heart. Circ Res. 2007;100:489–501. doi: 10.1161/01.RES.0000258451.44949.d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Chung YW, Lagranha C, Chen Y, Sun J, Tong G, Hockman SC, Ahmad F, Esfahani SG, Bae DH, Polidovitch N, Wu J, et al. Targeted disruption of PDE3B, but not PDE3A, protects murine heart from ischemia/reperfusion injury. Proc Natl Acad Sci U S A. 2015;112:E2253–2262. doi: 10.1073/pnas.1416230112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Movsesian M, Ahmad F, Hirsch E. Functions of PDE3 Isoforms in Cardiac Muscle. J Cardiovasc Dev Dis. 2018;5 doi: 10.3390/jcdd5010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Ercu M, Mucke MB, Pallien T, Marko L, Sholokh A, Schachterle C, Aydin A, Kidd A, Walter S, Esmati Y, McMurray BJ, et al. Mutant Phosphodiesterase 3A Protects From Hypertension-Induced Cardiac Damage. Circulation. 2022;146:1758–1778. doi: 10.1161/CIRCULATIONAHA.122.060210. [DOI] [PubMed] [Google Scholar]
- 459.Richter W, Xie M, Scheitrum C, Krall J, Movsesian MA, Conti M. Conserved expression and functions of PDE4 in rodent and human heart. Basic Res Cardiol. 2011;106:249–262. doi: 10.1007/s00395-010-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Leroy J, Richter W, Mika D, Castro LR, Abi-Gerges A, Xie M, Scheitrum C, Lefebvre F, Schittl J, Mateo P, Westenbroek R, et al. Phosphodiesterase 4B in the cardiac L-type Ca(2)(+) channel complex regulates Ca(2)(+) current and protects against ventricular arrhythmias in mice. J Clin Invest. 2011;121:2651–2661. doi: 10.1172/JCI44747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Patrucco E, Albergine MS, Santana LF, Beavo JA. Phosphodiesterase 8A (PDE8A) regulates excitation-contraction coupling in ventricular myocytes. Journal of molecular and cellular cardiology. 2010;49:330–333. doi: 10.1016/j.yjmcc.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Grammatika Pavlidou N, Dobrev S, Beneke K, Reinhardt F, Pecha S, Jacquet E, Abu-Taha IH, Schmidt C, Voigt N, Kamler M, Schnabel RB, et al. Phosphodiesterase 8 governs cAMP/PKA-dependent reduction of L-type calcium current in human atrial fibrillation: a novel arrhythmogenic mechanism. Eur Heart J. 2023;44:2483–2494. doi: 10.1093/eurheartj/ehad086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Chen S, Zhang Y, Lighthouse JK, Mickelsen DM, Wu J, Yao P, Small EM, Yan C. A Novel Role of Cyclic Nucleotide Phosphodiesterase 10A in Pathological Cardiac Remodeling and Dysfunction. Circulation. 2020;141:217–233. doi: 10.1161/CIRCULATIONAHA.119.042178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Chen S, Chen J, Du W, Mickelsen DM, Shi H, Yu H, Kumar S, Yan C. PDE10A Inactivation Prevents Doxorubicin-Induced Cardiotoxicity and Tumor Growth. Circ Res. 2023;133:138–157. doi: 10.1161/CIRCRESAHA.122.322264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Wu MP, Zhang YS, Xu X, Zhou Q, Li JD, Yan C. Vinpocetine Attenuates Pathological Cardiac Remodeling by Inhibiting Cardiac Hypertrophy and Fibrosis. Cardiovasc Drugs Ther. 2017;31:157–166. doi: 10.1007/s10557-017-6719-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Zhang Y, Knight W, Chen S, Mohan A, Yan C. Multiprotein Complex With TRPC (Transient Receptor Potential-Canonical) Channel, PDE1C (Phosphodiesterase 1C), and A2R (Adenosine A2 Receptor) Plays a Critical Role in Regulating Cardiomyocyte cAMP and Survival. Circulation. 2018;138:1988–2002. doi: 10.1161/CIRCULATIONAHA.118.034189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Zhang H, Pan B, Wu P, Parajuli N, Rekhter MD, Goldberg AL, Wang X. PDE1 inhibition facilitates proteasomal degradation of misfolded proteins and protects against cardiac proteinopathy. Sci Adv. 2019;5:eaaw5870. doi: 10.1126/sciadv.aaw5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Muller GK, Song J, Jani V, Wu Y, Liu T, Jeffreys WPD, O’Rourke B, Anderson ME, Kass DA. PDE1 Inhibition Modulates Ca(v)1.2 Channel to Stimulate Cardiomyocyte Contraction. Circ Res. 2021;129:872–886. doi: 10.1161/CIRCRESAHA.121.319828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Miller CL, Cai Y, Oikawa M, Thomas T, Dostmann WR, Zaccolo M, Fujiwara K, Yan C. Cyclic nucleotide phosphodiesterase 1A: a key regulator of cardiac fibroblast activation and extracellular matrix remodeling in the heart. Basic Res Cardiol. 2011;106:1023–1039. doi: 10.1007/s00395-011-0228-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Russwurm C, Zoidl G, Koesling D, Russwurm M. Dual acylation of PDE2A splice variant 3: targeting to synaptic membranes. J Biol Chem. 2009;284:25782–25790. doi: 10.1074/jbc.M109.017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Mongillo M, Tocchetti CG, Terrin A, Lissandron V, Cheung YF, Dostmann WR, Pozzan T, Kass DA, Paolocci N, Houslay MD, Zaccolo M. Compartmentalized phosphodiesterase-2 activity blunts beta-adrenergic cardiac inotropy via an NO/cGMP-dependent pathway. Circ Res. 2006;98:226–234. doi: 10.1161/01.RES.0000200178.34179.93. [DOI] [PubMed] [Google Scholar]
- 472.Zoccarato A, Surdo NC, Aronsen JM, Fields LA, Mancuso L, Dodoni G, Stangherlin A, Livie C, Jiang H, Sin YY, Gesellchen F, et al. Cardiac Hypertrophy Is Inhibited by a Local Pool of cAMP Regulated by Phosphodiesterase 2. Circ Res. 2015;117:707–719. doi: 10.1161/CIRCRESAHA.114.305892. [DOI] [PubMed] [Google Scholar]
- 473.Baliga RS, Preedy MEJ, Dukinfield MS, Chu SM, Aubdool AA, Bubb KJ, Moyes AJ, Tones MA, Hobbs AJ. Phosphodiesterase 2 inhibition preferentially promotes NO/guanylyl cyclase/cGMP signaling to reverse the development of heart failure. Proc Natl Acad Sci U S A. 2018;115:E7428–E7437. doi: 10.1073/pnas.1800996115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Liu K, Li D, Hao G, McCaffary D, Neely O, Woodward L, Ioannides D, Lu CJ, Brescia M, Zaccolo M, Tandri H, et al. Phosphodiesterase 2A as a therapeutic target to restore cardiac neurotransmission during sympathetic hyperactivity. JCI Insight. 2018;3 doi: 10.1172/jci.insight.98694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Hambleton R, Krall J, Tikishvili E, Honeggar M, Ahmad F, Manganiello VC, Movsesian MA. Isoforms of cyclic nucleotide phosphodiesterase PDE3 and their contribution to cAMP hydrolytic activity in subcellular fractions of human myocardium. J Biol Chem. 2005;280:39168–39174. doi: 10.1074/jbc.M506760200. [DOI] [PubMed] [Google Scholar]
- 476.Jaski BE, Fifer MA, Wright RF, Braunwald E, Colucci WS. Positive inotropic and vasodilator actions of milrinone in patients with severe congestive heart failure. Dose-response relationships and comparison to nitroprusside. J Clin Invest. 1985;75:643–649. doi: 10.1172/JCI111742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Shakur Y, Holst LS, Landstrom TR, Movsesian M, Degerman E, Manganiello V. Regulation and function of the cyclic nucleotide phosphodiesterase (PDE3) gene family. Prog Nucleic Acid Res Mol Biol. 2001;66:241–277. doi: 10.1016/s0079-6603(00)66031-2. [DOI] [PubMed] [Google Scholar]
- 478.Grant PG, Colman RW. Purification and characterization of a human platelet cyclic nucleotide phosphodiesterase. Biochemistry. 1984;23:1801–1807. doi: 10.1021/bi00303a034. [DOI] [PubMed] [Google Scholar]
- 479.Fischmeister R, Hartzell HC. Regulation of calcium current by low-Km cyclic AMP phosphodiesterases in cardiac cells. Mol Pharmacol. 1990;38:426–433. [PubMed] [Google Scholar]
- 480.Bastug-Ozel Z, Wright PT, Kraft AE, Pavlovic D, Howie J, Froese A, Fuller W, Gorelik J, Shattock MJ, Nikolaev VO. Heart failure leads to altered beta2-adrenoceptor/cyclic adenosine monophosphate dynamics in the sarcolemmal phospholemman/Na,K ATPase microdomain. Cardiovasc Res. 2019;115:546–555. doi: 10.1093/cvr/cvy221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Ding B, Abe JI, Wei H, Huang Q, Walsh RA, Molina CA, Zhao A, Sadoshima J, Blaxall BC, Berk BC, Yan C. Functional role of phosphodiesterase 3 in cardiomyocyte apoptosis: implication in heart failure. Circulation. 2005;111:2469–2476. doi: 10.1161/01.CIR.0000165128.39715.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.von der Leyen H, Mende U, Meyer W, Neumann J, Nose M, Schmitz W, Scholz H, Starbatty J, Stein B, Wenzlaff H, et al. Mechanism underlying the reduced positive inotropic effects of the phosphodiesterase III inhibitors pimobendan, adibendan and saterinone in failing as compared to nonfailing human cardiac muscle preparations. Naunyn Schmiedebergs Arch Pharmacol. 1991;344:90–100. doi: 10.1007/BF00167387. [DOI] [PubMed] [Google Scholar]
- 483.Sanada S, Kitakaze M, Papst PJ, Asanuma H, Node K, Takashima S, Asakura M, Ogita H, Liao Y, Sakata Y, Ogai A, et al. Cardioprotective effect afforded by transient exposure to phosphodiesterase III inhibitors: the role of protein kinase A and p38 mitogen-activated protein kinase. Circulation. 2001;104:705–710. doi: 10.1161/hc3201.092216. [DOI] [PubMed] [Google Scholar]
- 484.Polidovitch N, Yang S, Sun H, Lakin R, Ahmad F, Gao X, Turnbull PC, Chiarello C, Perry CGR, Manganiello V, Yang P, et al. Phosphodiesterase type 3A (PDE3A), but not type 3B (PDE3B), contributes to the adverse cardiac remodeling induced by pressure overload. Journal of molecular and cellular cardiology. 2019;132:60–70. doi: 10.1016/j.yjmcc.2019.04.028. [DOI] [PubMed] [Google Scholar]
- 485.Nakata TM, Suzuki K, Uemura A, Shimada K, Tanaka R. Contrasting Effects of Inhibition of Phosphodiesterase 3 and 5 on Cardiac Function and Interstitial Fibrosis in Rats With Isoproterenol-Induced Cardiac Dysfunction. J Cardiovasc Pharmacol. 2019;73:195–205. doi: 10.1097/FJC.0000000000000652. [DOI] [PubMed] [Google Scholar]
- 486.Nash CA, Brown LM, Malik S, Cheng X, Smrcka AV. Compartmentalized cyclic nucleotides have opposing effects on regulation of hypertrophic phospholipase Cepsilon signaling in cardiac myocytes. Journal of molecular and cellular cardiology. 2018;121:51–59. doi: 10.1016/j.yjmcc.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Oikawa M, Wu M, Lim S, Knight WE, Miller CL, Cai Y, Lu Y, Blaxall BC, Takeishi Y, Abe J, Yan C. Cyclic nucleotide phosphodiesterase 3A1 protects the heart against ischemia-reperfusion injury. J Mol Cell Cardiol. 2013;64:11–19. doi: 10.1016/j.yjmcc.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Ercu M, Marko L, Schachterle C, Tsvetkov D, Cui Y, Maghsodi S, Bartolomaeus TUP, Maass PG, Zuhlke K, Gregersen N, Hubner N, et al. Phosphodiesterase 3A and Arterial Hypertension. Circulation. 2020;142:133–149. doi: 10.1161/CIRCULATIONAHA.119.043061. [DOI] [PubMed] [Google Scholar]
- 489.Shi Q, Li M, Mika D, Fu Q, Kim S, Phan J, Shen A, Vandecasteele G, Xiang YK. Heterologous desensitization of cardiac beta-adrenergic signal via hormone-induced betaAR/arrestin/PDE4 complexes. Cardiovasc Res. 2017;113:656–670. doi: 10.1093/cvr/cvx036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.De Arcangelis V, Liu R, Soto D, Xiang Y. Differential association of phosphodiesterase 4D isoforms with beta2-adrenoceptor in cardiac myocytes. The Journal of biological chemistry. 2009;284:33824–33832. doi: 10.1074/jbc.M109.020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Richter W, Day P, Agrawal R, Bruss MD, Granier S, Wang YL, Rasmussen SG, Horner K, Wang P, Lei T, Patterson AJ, et al. Signaling from beta1-and beta2-adrenergic receptors is defined by differential interactions with PDE4. EMBO J. 2008;27:384–393. doi: 10.1038/sj.emboj.7601968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Berthouze-Duquesnes M, Lucas A, Sauliere A, Sin YY, Laurent AC, Gales C, Baillie G, Lezoualc’h F. Specific interactions between Epac1, beta-arrestin2 and PDE4D5 regulate beta-adrenergic receptor subtype differential effects on cardiac hypertrophic signaling. Cellular signalling. 2013;25:970–980. doi: 10.1016/j.cellsig.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 493.Lehnart SE, Wehrens XH, Reiken S, Warrier S, Belevych AE, Harvey RD, Richter W, Jin SL, Conti M, Marks AR. Phosphodiesterase 4D deficiency in the ryanodine-receptor complex promotes heart failure and arrhythmias. Cell. 2005;123:25–35. doi: 10.1016/j.cell.2005.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Berisha F, Gotz KR, Wegener JW, Brandenburg S, Subramanian H, Molina CE, Ruffer A, Petersen J, Bernhardt A, Girdauskas E, Jungen C, et al. cAMP Imaging at Ryanodine Receptors Reveals beta(2)-Adrenoceptor Driven Arrhythmias. Circ Res. 2021;129:81–94. doi: 10.1161/CIRCRESAHA.120.318234. [DOI] [PubMed] [Google Scholar]
- 495.Beca S, Helli PB, Simpson JA, Zhao D, Farman GP, Jones P, Tian X, Wilson LS, Ahmad F, Chen SRW, Movsesian MA, et al. Phosphodiesterase 4D regulates baseline sarcoplasmic reticulum Ca2+ release and cardiac contractility, independently of L-type Ca2+ current. Circ Res. 2011;109:1024–1030. doi: 10.1161/CIRCRESAHA.111.250464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Caldwell JL, Lee IJ, Ngo L, Wang L, Bahriz S, Xu B, Bers DM, Navedo MF, Bossuyt J, Xiang YK, Ripplinger CM. Whole-heart multiparametric optical imaging reveals sex-dependent heterogeneity in cAMP signaling and repolarization kinetics. Sci Adv. 2023;9:eadd5799. doi: 10.1126/sciadv.add5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Lai P, Hille SS, Subramanian H, Wiegmann R, Roser P, Muller OJ, Nikolaev VO, De Jong KA. Remodelling of cAMP dynamics within the SERCA2a microdomain in heart failure with preserved ejection fraction caused by obesity and type 2 diabetes. Cardiovasc Res. 2024;120:273–285. doi: 10.1093/cvr/cvad178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Bedioune I, Lefebvre F, Lechene P, Varin A, Domergue V, Kapiloff MS, Fischmeister R, Vandecasteele G. PDE4 and mAKAPbeta are nodal organizers of beta2-ARs nuclear PKA signalling in cardiac myocytes. Cardiovasc Res. 2018;114:1499–1511. doi: 10.1093/cvr/cvy110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Haj Slimane Z, Bedioune I, Lechene P, Varin A, Lefebvre F, Mateo P, Domergue-Dupont V, Dewenter M, Richter W, Conti M, El-Armouche A, et al. Control of cytoplasmic and nuclear protein kinase A by phosphodiesterases and phosphatases in cardiac myocytes. Cardiovasc Res. 2014;102:97–106. doi: 10.1093/cvr/cvu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Soderling SH, Bayuga SJ, Beavo JA. Cloning and characterization of a cAMP-specific cyclic nucleotide phosphodiesterase. Proc Natl Acad Sci U S A. 1998;95:8991–8996. doi: 10.1073/pnas.95.15.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Lounas A, Vernoux N, Germain M, Tremblay ME, Richard FJ. Mitochondrial sub-cellular localization of cAMP-specific phosphodiesterase 8A in ovarian follicular cells. Sci Rep. 2019;9:12493. doi: 10.1038/s41598-019-48886-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Shimizu-Albergine M, Tsai LC, Patrucco E, Beavo JA. cAMP-specific phosphodiesterases 8A and 8B, essential regulators of Leydig cell steroidogenesis. Molecular pharmacology. 2012;81:556–566. doi: 10.1124/mol.111.076125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Brescia M, Chao YC, Koschinski A, Tomek J, Zaccolo M. Multi-Compartment, Early Disruption of cGMP and cAMP Signalling in Cardiac Myocytes from the mdx Model of Duchenne Muscular Dystrophy. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21197056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Tulsian NK, Krishnamurthy S, Anand GS. Channeling of cAMP in PDE-PKA Complexes Promotes Signal Adaptation. Biophys J. 2017;112:2552–2566. doi: 10.1016/j.bpj.2017.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Tulsian NK, Ghode A, Anand GS. Adenylate control in cAMP signaling: implications for adaptation in signalosomes. Biochem J. 2020;477:2981–2998. doi: 10.1042/BCJ20200435. [DOI] [PubMed] [Google Scholar]
- 506.Bouvet M, Blondeau JP, Lezoualc’h F. The Epac1 Protein: Pharmacological Modulators, Cardiac Signalosome and Pathophysiology. Cells. 2019;8 doi: 10.3390/cells8121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Mangmool S, Shukla AK, Rockman HA. beta-Arrestin-dependent activation of Ca(2+)/calmodulin kinase II after beta(1)-adrenergic receptor stimulation. The Journal of cell biology. 2010;189:573–587. doi: 10.1083/jcb.200911047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Pereira L, Cheng H, Lao DH, Na L, van Oort RJ, Brown JH, Wehrens XH, Chen J, Bers DM. Epac2 mediates cardiac beta1-adrenergic-dependent sarcoplasmic reticulum Ca2+ leak and arrhythmia. Circulation. 2013;127:913–922. doi: 10.1161/CIRCULATIONAHA.12.148619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Khaliulin I, Bond M, James AF, Dyar Z, Amini R, Johnson JL, Suleiman MS. Functional and cardioprotective effects of simultaneous and individual activation of protein kinase A and Epac. British journal of pharmacology. 2017;174:438–453. doi: 10.1111/bph.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Fazal L, Laudette M, Paula-Gomes S, Pons S, Conte C, Tortosa F, Sicard P, Sainte-Marie Y, Bisserier M, Lairez O, Lucas A, et al. Multifunctional Mitochondrial Epac1 Controls Myocardial Cell Death. Circ Res. 2017;120:645–657. doi: 10.1161/CIRCRESAHA.116.309859. [DOI] [PubMed] [Google Scholar]
- 511.Wang X, Li F, Campbell SE, Gerdes AM. Chronic pressure overload cardiac hypertrophy and failure in guinea pigs: II. Cytoskeletal remodeling. Journal of molecular and cellular cardiology. 1999;31:319–331. doi: 10.1006/jmcc.1998.0885. [DOI] [PubMed] [Google Scholar]
- 512.Tagawa H, Koide M, Sato H, Zile MR, Carabello BA, Cooper GT. Cytoskeletal role in the transition from compensated to decompensated hypertrophy during adult canine left ventricular pressure overloading. Circ Res. 1998;82:751–761. doi: 10.1161/01.res.82.7.751. [DOI] [PubMed] [Google Scholar]
- 513.Pinali C, Bennett H, Davenport JB, Trafford AW, Kitmitto A. Three-dimensional reconstruction of cardiac sarcoplasmic reticulum reveals a continuous network linking transverse-tubules: this organization is perturbed in heart failure. Circ Res. 2013;113:1219–1230. doi: 10.1161/CIRCRESAHA.113.301348. [DOI] [PubMed] [Google Scholar]
- 514.Rakusan K, Tomanek RJ. Distribution of mitochondria in normal and hypertrophic myocytes from the rat heart. Journal of molecular and cellular cardiology. 1986;18:299–305. doi: 10.1016/s0022-2828(86)80412-6. [DOI] [PubMed] [Google Scholar]
- 515.Gerber Y, Weston SA, Redfield MM, Chamberlain AM, Manemann SM, Jiang R, Killian JM, Roger VL. A contemporary appraisal of the heart failure epidemic in Olmsted County, Minnesota, 2000 to 2010. JAMA Intern Med. 2015;175:996–1004. doi: 10.1001/jamainternmed.2015.0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Tsao CW, Lyass A, Enserro D, Larson MG, Ho JE, Kizer JR, Gottdiener JS, Psaty BM, Vasan RS. Temporal Trends in the Incidence of and Mortality Associated With Heart Failure With Preserved and Reduced Ejection Fraction. JACC Heart Fail. 2018;6:678–685. doi: 10.1016/j.jchf.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Kaddoura R, Madurasinghe V, Chapra A, Abushanab D, Al-Badriyeh D, Patel A. Beta-blocker therapy in heart failure with preserved ejection fraction (B-HFpEF): A systematic review and meta-analysis. Curr Probl Cardiol. 2024;49:102376. doi: 10.1016/j.cpcardiol.2024.102376. [DOI] [PubMed] [Google Scholar]
- 518.Amsallem E, Kasparian C, Haddour G, Boissel JP, Nony P. Phosphodiesterase III inhibitors for heart failure. Cochrane Database Syst Rev. 2005;2005:CD002230. doi: 10.1002/14651858.CD002230.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Packer M, Carver JR, Rodeheffer RJ, Ivanhoe RJ, DiBianco R, Zeldis SM, Hendrix GH, Bommer WJ, Elkayam U, Kukin ML, et al. Effect of oral milrinone on mortality in severe chronic heart failure. The PROMISE Study Research Group. N Engl J Med. 1991;325:1468–1475. doi: 10.1056/NEJM199111213252103. [DOI] [PubMed] [Google Scholar]
- 520.Sucharov CC, Nakano SJ, Slavov D, Schwisow JA, Rodriguez E, Nunley K, Medway A, Stafford N, Nelson P, McKinsey TA, Movsesian M, et al. A PDE3A Promoter Polymorphism Regulates cAMP-Induced Transcriptional Activity in Failing Human Myocardium. J Am Coll Cardiol. 2019;73:1173–1184. doi: 10.1016/j.jacc.2018.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Metra M, Eichhorn E, Abraham WT, Linseman J, Bohm M, Corbalan R, DeMets D, De Marco T, Elkayam U, Gerber M, Komajda M, et al. Effects of low-dose oral enoximone administration on mortality, morbidity, and exercise capacity in patients with advanced heart failure: the randomized, double-blind, placebo-controlled, parallel group ESSENTIAL trials. Eur Heart J. 2009;30:3015–3026. doi: 10.1093/eurheartj/ehp338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Gilotra NA, DeVore AD, Povsic TJ, Hays AG, Hahn VS, Agunbiade TA, DeLong A, Satlin A, Chen R, Davis R, Kass DA. Acute Hemodynamic Effects and Tolerability of Phosphodiesterase-1 Inhibition With ITI-214 in Human Systolic Heart Failure. Circ Heart Fail. 2021;14:e008236. doi: 10.1161/CIRCHEARTFAILURE.120.008236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Karam S, Margaria JP, Bourcier A, Mika D, Varin A, Bedioune I, Lindner M, Bouadjel K, Dessillons M, Gaudin F, Lefebvre F, et al. Cardiac Overexpression of PDE4B Blunts beta-Adrenergic Response and Maladaptive Remodeling in Heart Failure. Circulation. 2020;142:161–174. doi: 10.1161/CIRCULATIONAHA.119.042573. [DOI] [PubMed] [Google Scholar]
- 524.Bristow MR, Ginsburg R, Minobe W, Cubicciotti RS, Sageman WS, Lurie K, Billingham ME, Harrison DC, Stinson EB. Decreased catecholamine sensitivity and beta-adrenergic-receptor density in failing human hearts. N Engl J Med. 1982;307:205–211. doi: 10.1056/NEJM198207223070401. [DOI] [PubMed] [Google Scholar]
- 525.Hammond HK, Penny WF, Traverse JH, Henry TD, Watkins MW, Yancy CW, Sweis RN, Adler ED, Patel AN, Murray DR, Ross RS, et al. Intracoronary Gene Transfer of Adenylyl Cyclase 6 in Patients With Heart Failure: A Randomized Clinical Trial. JAMA Cardiol. 2016;1:163–171. doi: 10.1001/jamacardio.2016.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 527.Martin TP, Hortigon-Vinagre MP, Findlay JE, Elliott C, Currie S, Baillie GS. Targeted disruption of the heat shock protein 20-phosphodiesterase 4D (PDE4D) interaction protects against pathological cardiac remodelling in a mouse model of hypertrophy. FEBS Open Bio. 2014;4:923–927. doi: 10.1016/j.fob.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Delyon J, Servy A, Laugier F, Andre J, Ortonne N, Battistella M, Mourah S, Bensussan A, Lebbe C, Dumaz N. PDE4D promotes FAK-mediated cell invasion in BRAF-mutated melanoma. Oncogene. 2017;36:3252–3262. doi: 10.1038/onc.2016.469. [DOI] [PubMed] [Google Scholar]
- 529.Serrels B, Sandilands E, Serrels A, Baillie G, Houslay MD, Brunton VG, Canel M, Machesky LM, Anderson KI, Frame MC. A complex between FAK, RACK1, and PDE4D5 controls spreading initiation and cancer cell polarity. Curr Biol. 2010;20:1086–1092. doi: 10.1016/j.cub.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 530.Lundby A, Andersen MN, Steffensen AB, Horn H, Kelstrup CD, Francavilla C, Jensen LJ, Schmitt N, Thomsen MB, Olsen JV. In vivo phosphoproteomics analysis reveals the cardiac targets of beta-adrenergic receptor signaling. Sci Signal. 2013;6:rs11. doi: 10.1126/scisignal.2003506. [DOI] [PubMed] [Google Scholar]
- 531.Richards MA, Simon JN, Ma R, Loonat AA, Crabtree MJ, Paterson DJ, Fahlman RP, Casadei B, Fliegel L, Swietach P. Nitric oxide modulates cardiomyocyte pH control through a biphasic effect on sodium/hydrogen exchanger-1. Cardiovasc Res. 2020;116:1958–1971. doi: 10.1093/cvr/cvz311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Corradini E, Klaasse G, Leurs U, Heck AJ, Martin NI, Scholten A. Charting the interactome of PDE3A in human cells using an IBMX based chemical proteomics approach. Mol Biosyst. 2015;11:2786–2797. doi: 10.1039/c5mb00142k. [DOI] [PubMed] [Google Scholar]
- 533.Go CD, Knight JDR, Rajasekharan A, Rathod B, Hesketh GG, Abe KT, Youn JY, Samavarchi-Tehrani P, Zhang H, Zhu LY, Popiel E, et al. A proximity-dependent biotinylation map of a human cell. Nature. 2021;595:120–124. doi: 10.1038/s41586-021-03592-2. [DOI] [PubMed] [Google Scholar]
- 534.Christopher JA, Stadler C, Martin CE, Morgenstern M, Pan Y, Betsinger CN, Rattray DG, Mahdessian D, Gingras AC, Warscheid B, Lehtio J, et al. Subcellular proteomics. Nat Rev Methods Primers. 2021;1 doi: 10.1038/s43586-021-00029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Kovanich D, Low TY, Zaccolo M. Using the Proteomics Toolbox to Resolve Topology and Dynamics of Compartmentalized cAMP Signaling. Int J Mol Sci. 2023;24 doi: 10.3390/ijms24054667. [DOI] [PMC free article] [PubMed] [Google Scholar]