Abstract
Biological sex profoundly conditions organismal development and physiology, imposing wide-ranging effects on cell signaling, metabolism, and immune response. These effects arise from sex-specified differences in hormonal exposure, and from intrinsic genetic and epigenetic differences associated with the presence of an XX versus XY chromosomal complement. In addition, biological sex is now recognized to be a determinant of the incidence, presentation, and therapeutic response of multiple forms of cancer, including cancers not specifically associated with male or female anatomy. While multiple factors contribute to sex-based differences in cancer, a growing body of research emphasizes a role for differential activity of X- and Y- linked tumor suppressor genes in males and females. Among these, the X-linked KDM6A/UTX and KDM5C/JARID1C/SMCX, and their Y-linked paralogs UTY/KDM6C and KDM5D/JARID1D/SMCY encode lysine demethylases. These epigenetic modulators profoundly influence gene expression, based on enzymatic activity in demethylating H3K27me3 and H3K4me3, and non-enzymatic scaffolding roles for large complexes that open and close chromatin for transcription. In a growing number of cases, mutations affecting these proteins have been recognized to strongly influence cancer risk, prognosis, and response to specific therapies. However, sex-specific patterns of mutation, expression, and activity of these genes, coupled with tissue-specific requirement for their function as tumor suppressors, together exemplify the complex relationship between sex and cancer vulnerabilities. In this review, we summarize and discuss the current state of the literature on the roles of these proteins in contributing to sex bias in cancer, and the status of clinical agents relevant to their function.
Introduction.
In studies of cancer risk, prognosis, and therapeutic response, sex is often underexplored as a relevant variable (1,2), even though a recent comprehensive study of 30 types of human cancer found significant sexual dimorphism of cancer incidence and presentation (3). These trends remain after controlling for epidemiologic risk factors, geographical origin and ethnicity, and excluding sex-specific cancers affecting the ovary, testis or prostate (2,3). Mechanistically, sex-based differences relevant to cancer include metabolism, immune function, exposure to mutagens, the pattern and frequency of mutations, gene dosage and expression of clinically actionable genes, and the prognostic impact of individual mutations or gene expression signatures (4–9) (Figure 1). Based on this growing recognition of the impact of sex, the National Institutes of Health and health advisory groups in the European Union have mandated sex-balanced representation of cells, biological samples and experimental animals in preclinical studies (10,11).
Figure 1: Sources of sex-based bias in disease.

A. Male versus female differences relevant to cancer include mutational spectrum, gene expression profile, metabolism, immune cell interactions with tumors, microbiome composition, pharmacokinetics and tissue distribution of drugs, and other factors; all now recognized as important contributors to tumor phenotypes (2,111–113) . B. Both X and Y evolved from an ancestral autosomal chromosome pairs; except for two small pseudo-autosomal regions (PARs) common to both chromosomes, X and Y encode distinct complements of genes. The X chromosome is a large and gene-rich, encoding over 1,000 genes and many non-coding RNAs; by contrast, the much smaller Y chromosome encodes 568 genes, with only 71 having protein-coding potential. While it has long been considered as a “genetic wasteland” undergoing rapid evolutionary deterioration except for the sex-determining region, the Y chromosome is now recognized as having biological functions beyond its role in male sex determination by bearing a small but stable group of essential genes critically important for the health and survival of males. Among the 17 surviving ancestral genes that have survived as moderately divergent paralogs on both X and Y, 8 pairs (KDM6A, UTY; KDM5C, KDM5D; EIF1AX, EIF1AY; ZFX, ZFY; RPS4X, RPS4Y1; DDX3X, DDX3Y; USP9X, USP9Y; TBL1X and TBL1Y) encode X- and Y-specific isoforms of global regulators of gene and protein expression (26).
Typically, studies of sex-specified physiological differences focus on the roles of gonadal steroid hormones, including estrogens, androgens, and progestogens, which have profound genomic and non-genomic effects that condition cell identity and signaling (12–14). Complementing this work, recent studies have emphasized the role of genetic architecture in causing sexual dimorphism of cancer presentation (2,11), and in particular focused on contributions of the allosomes, X and Y (12,15,16). Multiple allosomally-encoded tumor suppressor genes (TSGs) have been characterized as epigenetic regulators, which function as components of large chromatin-modifying complexes that broadly influence gene expression, and modulate the efficiency of other cancer-relevant processes such as DNA repair, in a sex-biased manner (5,16). The literature defining the biological roles of allosomal TSGs in sex-specified differences in cancer is expanding rapidly; we here focus on two exemplar allosomal TSG pairs, KDM6A/UTX and UTY/KDM6C, and KDM5C/JARID1C and KDM5D/JARID1D.
Tumor suppressive chromatin modifiers on the X and Y chromosomes.
Epigenetic factors mediate transcriptional responses to oncogenic stimuli, and influence the propensity of DNA to become mutated, affecting cancer risk and response to therapy (17). Typically, epigenetic regulators directly modify DNA, or modify protein components of chromatin (Figure 2). Modifications such as methylation, acetylation, and mono-ubiquitination specify chromatin that is “open/permissive” or “closed/restrictive” for access by the machineries governing recombination, gene transcription, replication, and repair. Aberrant permissive and restrictive chromatin states promote cancer (17).
Figure 2. Epigenetic modulation of chromatin.
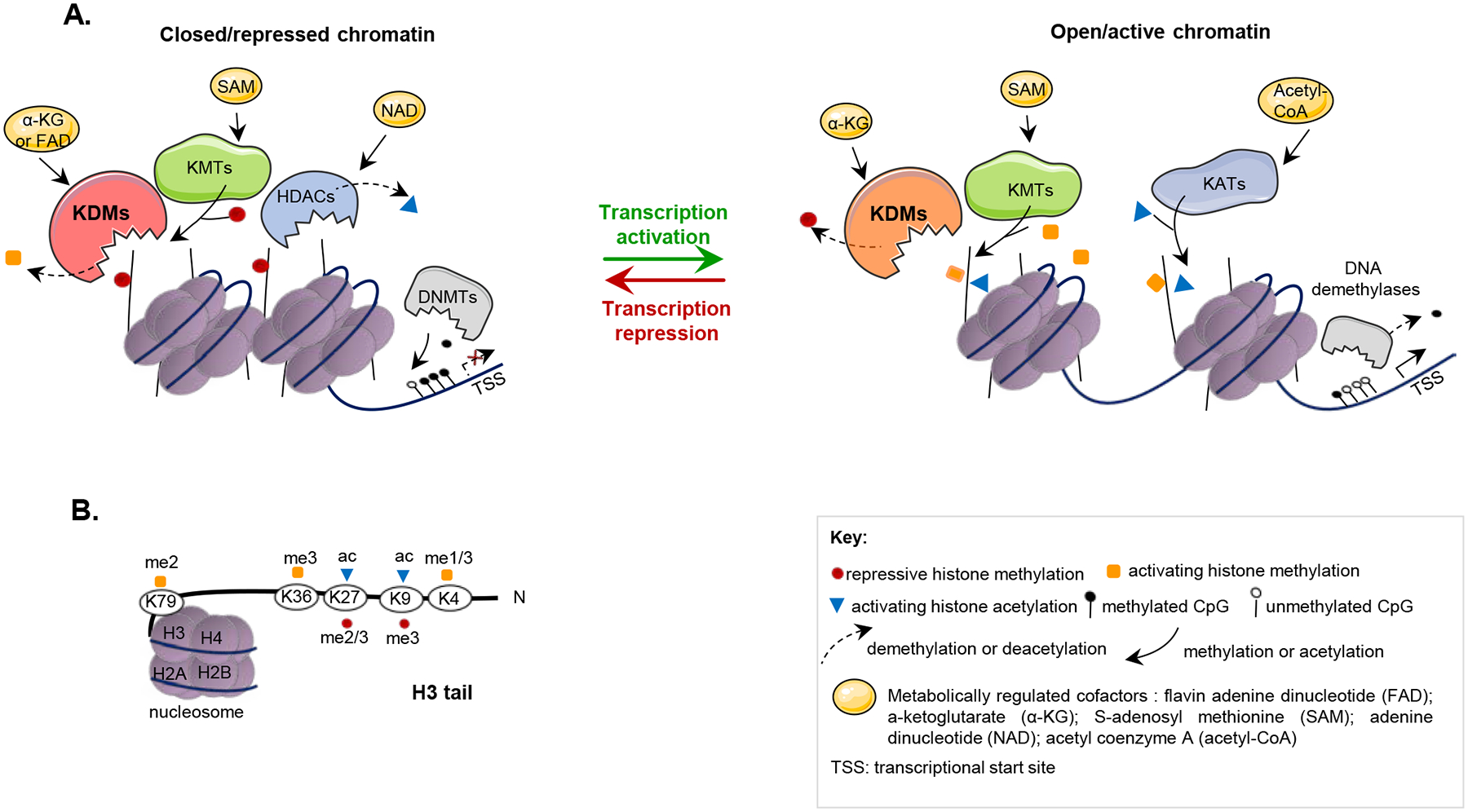
A. Transcription, replication, and repair of DNA are regulated by chromatin accessibility. DNA is wrapped around nucleosomes, composed of histone octamers (two molecules each of histones H2A, H2B, H3, and H4). Selective histone modification in the promoter and enhancer regions of highly regulated genes specifies whether transcription initiates. Amino-terminal sequences (or “tails”) of histones H3 and H4 are subject to extensive post-translational modifications that govern “open” versus “closed” chromosomes, with acetylation and methylation on specific lysines in the tails promoting transcription or repression. Lysine methyltransferases (KMTs) and lysine acetyltransferases (KATs) (often called “writers”) place marks on the tails; lysine demethylases (KDMs) and histone deacetylases (HDACs) (often called “erasers”) remove marks. Specific marks recruit “readers”. Repressive marks recruit DNA-modifying enzymes such as DNA methyltransferase (DNMT), which methylate CpG sites on DNA, further contributing to gene silencing. Both activating and repressive marks recruit protein complexes that can either reinforce the open or closed state of chromatin, or act to reverse these states (see also Figure 4). Pioneer transcription factors include members of the GATA, FOXA, and other protein families; these factors can target histone-modifying enzymes to specific genes, enabling switching between repressed and active states; some pioneer factors interact with transcription factors responsive to hormones including estrogen and androgens, contributing to sex-specific targeting of histone modifying complexes. In addition to having silenced gene expression, repressed chromatin is less accessible to enzyme complexes regulating DNA repair. B. Marks typically associated with open chromatin typically include H3K4me1/3, H3K36me3, H3K79me3, H3K27ac and H3K9ac; marks associated with closed chromatin included H3K9m3, H3K27me2/3 (114). TSS: transcriptional start site.
Some differences in chromatin regulation pertinent to sex-bias stem from the fundamental biological distinction between the mammalian sexes: the presence of two X chromosomes in females, versus one X and one Y chromosome in males (Figure 1A) (18,19). Among a limited number of X- and Y-linked genes with paralogous function (Figure 1B), KDM6A, UTY and KDM5C, KDM5D encode lysine demethylases (KDMs), a broad class of enzymes that modify histones, opposing the function of lysine methyltransferases (KMTs) (20). In addition, these KDMs have critical non-catalytic roles, including the recruitment of other epigenetic modifiers and transcription factors to specific sites on chromatin (21). KDM6A, UTY, KDM5C and KDM5D have significant tumor suppressor activity in several types of cancer ((22–24), and Table 1). Importantly, factors governing the expression of allosomal genes can lead to mutations in these KDMs having distinct penetrance in males and females.
Table 1.
KDM6A, UTY, KDM5C and KDM5D in cancer therapy.
| Context | Mechanistic Observation | Therapeutic opportunity | Ref. |
|---|---|---|---|
| Targeting KDM6A | |||
| T-ALL driven by oncogenic transcription factor TAL-1 | TAL-1 recruits KDM6A to aberrantly activate transcription of its target genes through H3K27me3 demethylation. | The H3K27 demethylase inhibitor GSK-J4 represses TAL-1-KDM6A target genes and kills TAL-1-positive T-ALL cells. The treatment is efficient in vivo in PDX models of TAL-1 positive T-ALL. | (107) |
| Chemotherapy for CRC | Low H3K27me3 correlates with poor prognosis and oxaliplatin resistance. | GSK-J4 combined with oxaliplatin inhibits growth of oxaliplatin-resistant PDX. | (108) |
| Drug resistance in ESR1/ERα-positive breast cancer | KDM1A/LSD1 and KDM6A are co-expressed and colocalized with ESR1/ERα. | The dual KDM1A-KDM6A inhibitor MC3324 down-regulates ESR1/ERα and attenuates hormone signaling. | (77) |
| Chemoinformatics approach to computationally predict biomolecular targets of metformin with experimental validation | Metformin inhibits the demethylation activity of purified KDM6A. Pharmacological dosage of metformin augments global H3K27me3 in vivo. | Low levels of H3K27me3 are a predictor of cancer aggressiveness. Metformin promotes a more resilient H3K27me3 enriched epigenome. It is currently in several clinical trials for cancer, including a phase 2 in bladder cancer NCT03379909. | (87) |
| Exploiting KDM6A deficiency | |||
| Loss or inactivation of KDM6A in MM | KDM6A loss leads to abnormal PRC2-mediated repression. | Rebalancing KDM6A-PRC2 activities with EZH2 inhibitors (GSK343, GSK126) causes death of KDM6A-mutated MM cells. | (79) |
| Loss or inactivation of KDM6A in bladder cancer | KDM6A loss leads to abnormal PRC2-mediated repression. | The EZH2 inhibitors GSK503 and EPZ6438* significantly attenuate the growth of KDM6A-null but not KDM6A-wt cells and engrafted tumors. | (78) |
| Loss of KDM6A in poorly differentiated and squamous-like pancreatic cancer | KDM6A functions in pancreatic cancer are largely non-catalytic. Kdm6a loss deregulates the COMPASS-like complex, activating oncogenic super-enhancers that drive squamous differentiation and metastasis. | KDM6A deficient pancreatic cell lines are sensitive to BET inhibitors (e.g. JQ1) that disrupt long range interactions between promoters and super-enhancers. JQ1 reverses the squamous differentiation of KDM6A-deficient cancers in vivo. | (54) |
| KDM6A loss blocks HSPC differentiation in mouse model for MDS and AML | H3K4 methylation is crucially involved in the differentiation block caused by KDM6A deficiency | Inhibition of the H3K4 KDM KDM1A by SP2509 (HCl2509) induces differentiation in Kdm6a-null cells. | (109) |
| Mouse model for Kdm6a-deficient bladder cancer | Kdm6a deficiency induces growth-promoting cytokine and chemokine signaling. | Combined inhibition of IL6 and CCL2 effectively suppresses cell growth. | (98) |
| Recurrent KDM6A mutation in AML patients relapsing after chemotherapy | Loss of KDM6A decreases expression of the drug influx transporter ENT1, providing a selective advantage after Cytarabine (AraC)-based chemotherapy. | Re-expression of KDM6A in KDM6A-null cells suppresses cell growth and resensitizes cells to AraC therapy. | (110) |
| Targeting KDM5C | |||
| KDM5C is upregulated in CRPC | BRD4 transcriptionally induces KDM5C, which suppresses PTEN transcription to promote tumorigenesis. | Knockdown of KDM5C sensitizes the responses of CRPC cells to treatment with a BET inhibitor. | (80) |
| Exploiting KDM5D deficiency | |||
| Loss of KDM5D causes DNA replication stress and poor prognosis in prostate cancer | Loss of KDM5D activates ATR signaling and alters histone methylation of promoter regions to increase expression of G2/M checkpoint mediators. | In KDM5D-deficient cells, blocking ATR activity with the ATR inhibitor VX-970* enhances DNA damage and causes apoptosis. | (70) |
| KDM pan inhibitor | |||
| In silico molecular docking to identify with drugs targeting KDMs, to support repurposing | Deferiprone (DFP, Ferriprox), approved for thalassemia, chelates the Fe2+ ion at the active sites of multiple KDMs; KDM6A is the highest affinity target. | In breast cancer cell lines DFP causes a dose-dependent increase in H3K4me3 and H3K27me3 levels and cytotoxicity, with potentially greater activity in triple negative breast cancer. May be useful as a combination sensitizing agent. | (84) |
EPZ6438/Tazemetostat is in phase I/II clinical trials (NCT03854474) as is VX-970/Berzosertib (e.g. NCT03517969). CRC, colorectal cancer; ESR1/ERα, estrogen-receptor-α; MM, multiple myeloma; HSPC, hematopoietic stem and progenitor cells; MDS, myelodysplastic syndrome; AML, acute myeloid leukemia; CRPC, castration-resistant prostate cancer; T-ALL, T-cell acute lymphoblastic leukemia.
Briefly (Figure 3), the most important mechanism mammals use to adjust X chromosome gene dosage between males and females is X chromosome inactivation (XCI). XCI makes females functionally haploid mosaics with respect to most X-linked genes, conferring both positive and negative features associated with genetic heterogeneity (25). In contrast, in males, mutations in X-linked genes subject to XCI have a dominant effect. However, KDM6A and KDM5C typically escape X inactivation (16,26), making them an example of EXITS (Escape from X-inactivation of tumor suppressor) genes, in which the ability to express both copies of X-linked TSGs can buffer the effect of single inherited or somatic gene-inactivating mutations in females, and increase overall gene dosage relative to males (16). Conversely, in male aging, somatic loss of the Y chromosome (LOY) (27), and the more recently defined extreme down-regulation of chromosome Y gene expression (EDY; inclusive both of LOY, and abnormal methylation of a retained Y-chromosome associated with gene silencing) can limit expression of Y-linked TSGs. LOY and EDY have emerged as signatures for cancer risk in men (15,27). Together, these mechanisms governing TSG activity contribute to sex-biased phenotypes in cancer.
Figure 3: Mechanisms regulating the dosage of allosomal genes influence cancer risk.
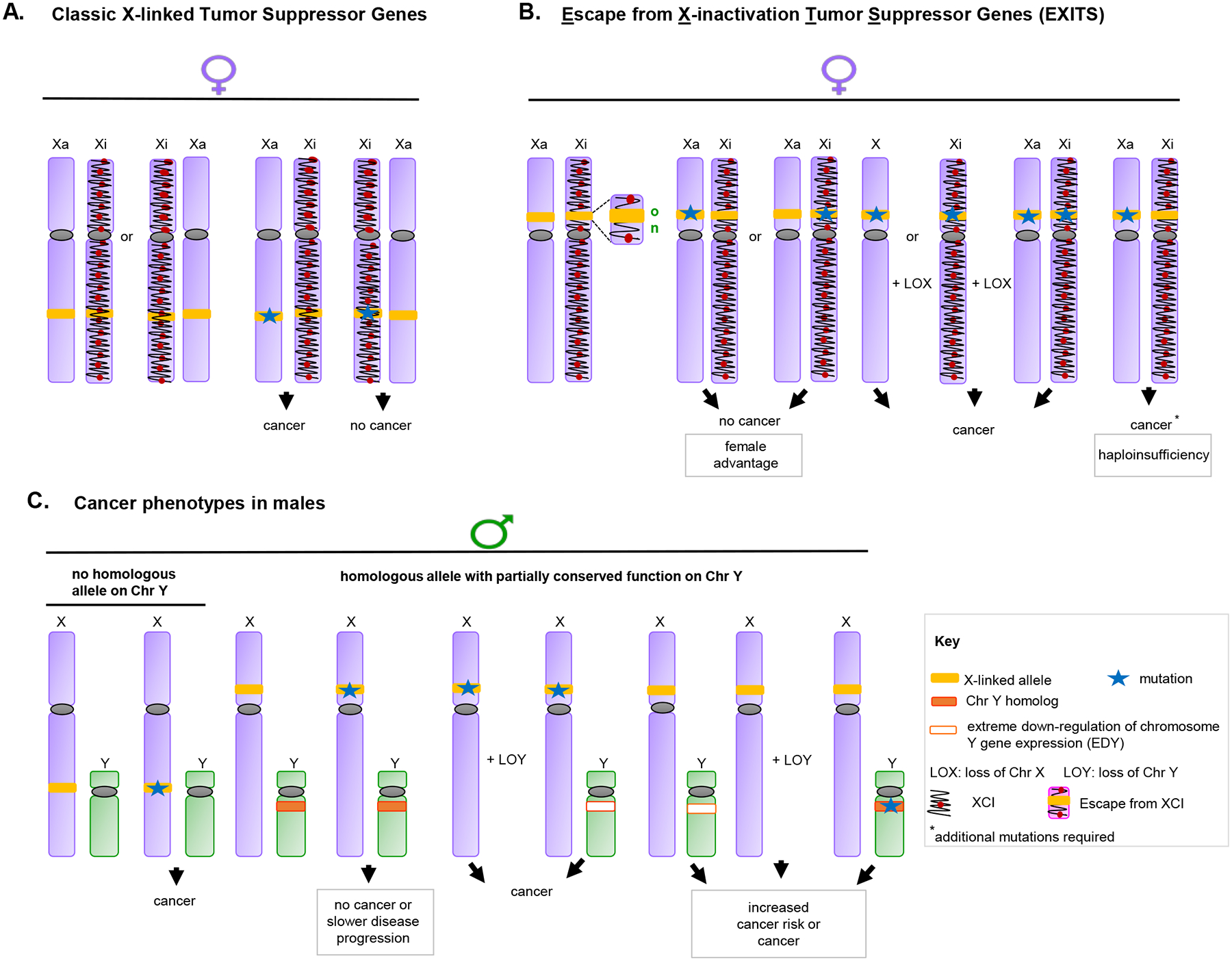
A-C. In X chromosome inactivation (XCI), one of the two X chromosomes is epigenetically modified during embryogenesis to silence transcription. Chromatin features distinguishing the inactive X (Xi) from the active X (Xa) include specific histone post-translational modifications, incorporation of variant histones into nucleosomes, association with the long non-coding RNA (lncRNA) XIST, and extensive CpG DNA methylation (115,116). However, up to 23% of X-linked genes escape inactivation. X-linked genes with Y-linked homologs are more likely to escape X inactivation. These Escape from X-inactivation tumor suppressor (EXITS) genes include the examples discussed in this article. Shown, different scenarios for cancer risk in females (A, B) and males (C). A represents cancer risk in females with wild type or mutated tumor suppressor genes (TSGs) subject to XCI. B represents cancer risk in females with wild type or mutated EXITS. The two active alleles of an EXITS TSG protect females from developing cancer after mutation of a single allele. Mutations on both alleles or on a single allele with loss of the other X are required to develop cancer. In addition, concomitant mutations in KMTs (e.g. KMT2C and KMT2D) might contribute to the cancer phenotype in EXITS genes (e.g. KDM6A) haploinsufficient tissues in females. C represents cancer risks in males with or without Y-linked TSGs that moderately to significantly conserve functions with X-linked homologs. Mutation of a single allele of an EXITS gene with no Y chromosome homolog is required to develop cancer in males. Alternatively, LOY or EDY enhances cancer development in males with mutation in an EXITS gene with a Y-chromosome homolog with partially conserved function. For some allosomal TSGs, this elevated risk occurs in a tissue-specific manner.
In addition (not shown), sporadic reactivation of X-linked genes can occur during aging, and in transformed cells. In parallel, male versus female differences in gene imprinting can cause differences in the expression of specific alleles dependent on their maternal versus paternal origin (117). In some cases, XCI occurs with skewing (the preferential inactivation of one of the two X chromosomes), amplifying or reducing the effect of inherited mutations. These factors can influence degree of sex-bias for phenotypes arising from mutations in allosomal genes, sometimes in a tissue-specific manner.
The KDM5 and KDM6 protein families
KDM5 and KDM6 lysine demethylases each contain a signature catalytic motif, the Jumonji C (JmjC) domain (Figure 4A, B). These enzymes require ferrous iron Fe(II) as a cofactor, and use the TCA cycle intermediate α-ketoglutarate (α-KG, also known as 2-oxoglutarate (2OG)) and oxygen as co-factors (28,29). Because of these dependencies, their activity is influenced by cancer-associated mutations in the tricarboxylic acid (TCA) cycle enzymes isocitrate dehydrogenase 1 and 2 (IDH1 and 2), succinate dehydrogenase (SDH), and fumarase hydratase (FH), which regulate α-KG availability (30); and by tumor hypoxia, which limits oxygen levels (31).
Figure 4. KDM6A, UTY, KDM5C and KDM5D: structure and function.
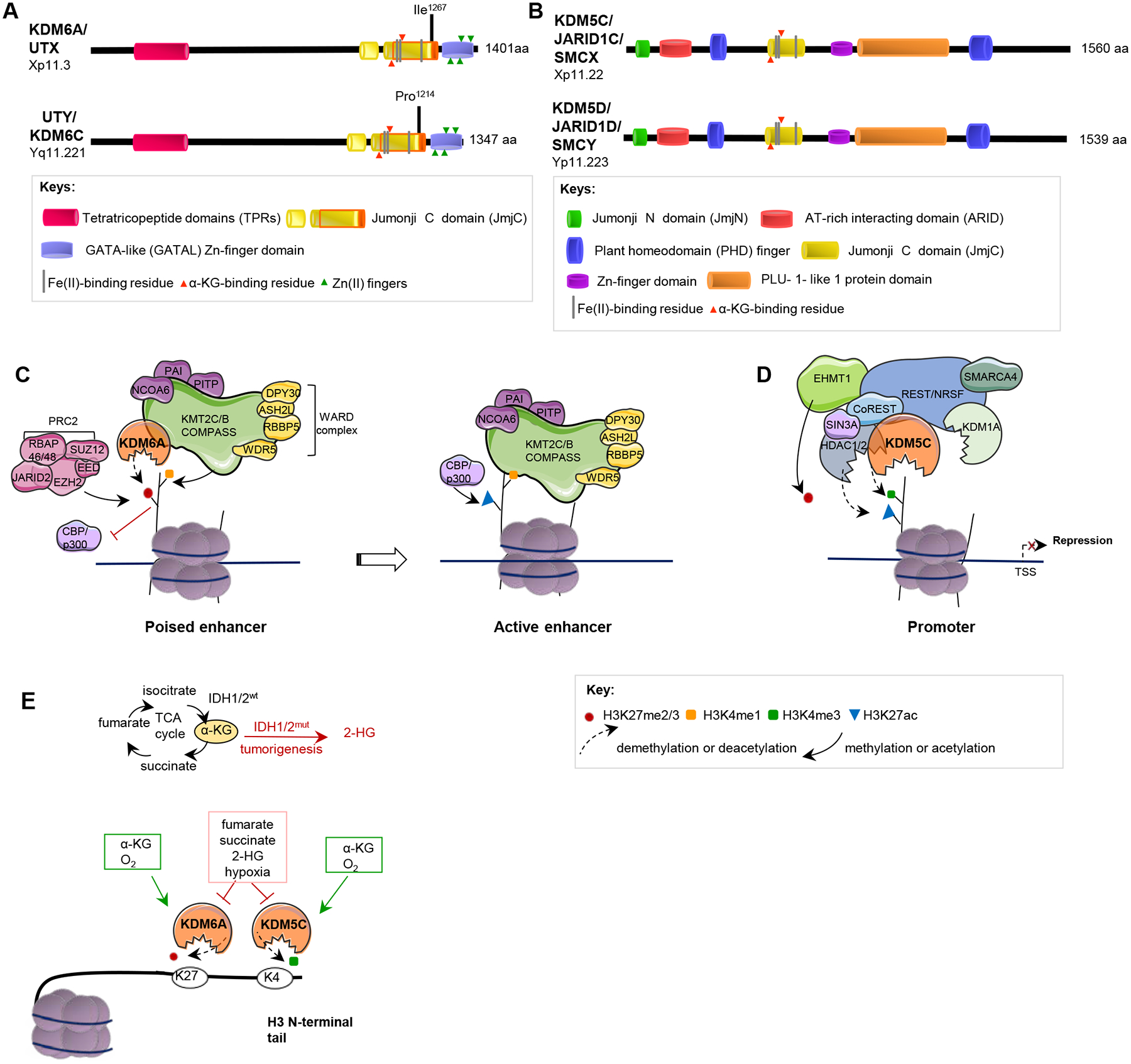
A, B. Domain structures of KDM6A and UTY (A), and for KDM5C and KDM5D (B) are indicated. The core catalytic domain, JmjC, demethylates histones by an oxidative mechanism requiring Fe(II) and alpha-ketoglutarate (α−KG) as cofactors; JmjN, in JARID proteins, interacts with JmjC, inducing a conformational change that promotes enzymatic activity. ARID and PLU-1 are DNA binding domains. TPR domains mediate interactions with other proteins including the KMT2C-KMT2B complex (49). The Zn-fingers mediate histone tail recognition; the PHD domain binds H3K9me3, coordinating H3K4 demethylation. Note, each protein family contains additional paralogous members (KDM5A (Chr 12), KDM5B (Chr 1) and KDM6B (Chr 17)), which have related but not equivalent function. See legend to Supp Fig 1 for extended comment on placement of motif boundaries. C. Through its activity as a histone H3K27me2/3 demethylase, KDM6A opposes activity of the repressive PRC2 complex, converting poised enhancers to transcriptionally active enhancers. KDM6A is also a component of the KMT2C-KMT2B COMPASS-like complex. As part of this complex, KDM6A contributes to protein recruitment at enhancers and cooperates with other complex components, including the lysine methyltransferases (KMTs) KMT2C and KMT2B, which monomethylate H3K4 (H3K4me1) and the histone acetyltransferases (HATs) p300 and CBP, which acetylate H3K27. These actions result in enhancer activation (118). Loss of KDM6A activates PRC2-regulated transcription repression, similar to EZH2 gain-of-function mutations, and sensitizes tumors to EZH2 inhibitors (78). KDM6A mutations damaging the TPR domain disrupt interaction with KMT2C-KMT2B complexes (49). D. KDM5C associates with Co-REST, Sin3a and other proteins to demethylate H3K4me3 at specific promoters, causing gene repression. E. Metabolic factors influencing the activity of KDM6A, UTY, KDM5C and KDM5D. Mutations in fumarate hydratase (FH), succinate dehydrogenase (SDH) or isocitrate dehydrogenase (IDH1, IDH2) affecting pools of the metabolites fumarate, succinate, or 2-hydroxyglutarate (2-HG), or hypoxia, repress KDM6A and KDM5C activity (119,120).
Histone 3 mono-methylated on lysine 4 (H3K4me1) is associated with closed/poised, primed, and active enhancers; H3K4me2/3 is associated with active promoters. These methylations are introduced by the SETD1A/SET1- and KMT2C/MLL3-KMT2B/MLL4-containing COMPASS complexes. Conversely, H3K27me3 is a repressive mark found in promoters and enhancers, introduced by EZH2/KMT6A, a component of Polycomb Repressive Complex 2 (PRC2) (17). By demethylating H3K27 at enhancers, KDM6 proteins such as KDM6A and UTY contribute to gene activation. In contrast, by demethylating H3K4 at promoters, KDM5 family proteins such as KDM5C and KDM5D tip the balance toward gene repression (Figure 4C, D). In addition, these KDMs also have non-catalytic functions that promote transcription in a context-specific manner (32), pertinent to their tissue-specific actions in cancer, discussed below. For example, through interaction with other chromatin-modifying complexes (e.g. p300/CBP and the KMT2B complex), these proteins also influence other histone modifications (33).
KDM6A/UTX and UTY/KDM6C
KDM6A, formerly known as UTX (Ubiquitously transcribed Tetratricopeptide repeat (TPR) X chromosome) (Figure 4A) is located at Xp11.3; UTY (also known under the alias KDM6C), located at Yq11.221, encodes a protein that has ~84% homology with KDM6A (34). However, UTY has markedly lower demethylase activity than KDM6A, in part due to a single amino acid difference (I1267 in KDM6A, P1214 at the comparable position in UTY) in the catalytic JmjC domain, which reduces substrate binding (35). Of note, the Km of KDM6A for α-KG is twice as high as that of UTY (35), and KDM6A is more oxygen-dependent (31,36). Based on these differences, KDM6A is a more active enzyme, and more responsive to environmental cues, than is UTY.
Studies in conditional knock-out mice indicate a sex-specific requirement for Kdm6a during development, and inform assessment of functional differences between KDM6A and UTY in cancer. In two different Kdm6a conditional mouse models, homozygous loss of Kdm6a is embryonic lethal in females, while heterozygous loss causes minor developmental defects. Simultaneous loss of Kdm6a and Uty is embryonic lethal in males, but in contrast to females, most Kdm6a-null, Uty-wild type male embryos also die at midgestation; the ~25% of males surviving are smaller and with reduced lifespan (37,38). Mechanistically, studies in cell lines derived from Kdm6a-null mice indicate Kdm6a demethylation of H3K27me3 is important for proper activation of developmental genes, and not complemented by Uty, which lacks efficient H3K27me3 demethylase activity (37,38). These data imply an important non-enzymatic function of Uty sufficient for some, but not all, aspects of development (37).
In considering KDM6A and UTY mutations in cancer, an emerging concept is that while some of their roles depend on enzymatic activities, which differ between males and females, others depend on their adaptor roles, which do not (21,33). Hence, the impact of mutations in KDM6A on sex-specific transcription in cancer will depend on the domain of the protein they affect; e.g., mutations in the JmjC domain, affecting enzymatic activity, would likely be associated with distinct penetrance in males and females. In addition, as KDM6A is an EXITS gene, the encoded protein is expressed from both alleles in females, blunting the effect of heterozygous mutations.
Multiple cancer types bear frequent somatic mutations inactivating KDM6A; these include multiple myeloma, esophageal squamous cell carcinoma, non-muscle-invasive bladder cancer (NMIBC) (the most common form of urothelial bladder carcinomas), renal clear cell carcinoma and T-cell acute lymphoblastic leukemia (T-ALL) (16,39,40) (Supp Figure 1). In most of these cancers, somatic mutations in KDM6A tend to be more common in males than in females ((16); see also an extended analysis, based on data in cBioPortal (41), in Supp Table 1). In females, when such mutations occur, they are commonly biallelic. In some male cancer cell lines, derived from acute myeloid leukemia (AML), esophageal squamous cell carcinoma and others, inactivating KDM6A mutations are often accompanied by the loss of its paralog UTY (81%) (40,42), suggesting selection pressure; in the absence of mutation of KDM6A, UTY is less frequently lost (49%) (40).
KDM6A and UTY are expressed in many tissues; notably, because their gene targeting and functionality is modulated by interaction with additional transcription factors, their activity is also tissue specific (e.g. (43)). Although somatic mutations of KDM6A have been associated with sex-biased incidence of multiple forms of cancer, how this sex bias manifests is variable between different cancer types. For instance, incidence of T-ALL occurs at a male to female ratio of 3:1 (44). Loss of KDM6A tumor suppressor activity is important in T-ALL pathogenesis, and depends on intact demethylase activity, reflected in the high frequency of somatic mutations in KDM6A, most of which inactivate the JmjC enzymatic domain. Such mutations predominate in males, and likely reflect the fact that the less active UTY enzyme is unable to replace the catalytic activity of KDM6A.
In contrast, bladder cancer also has a male:female incidence ratio of >3:1, and the highest overall KDM6A mutation frequency among cancers, at 29%−41% ((45,46), Supp Table 1). However, in this case, there is an increased frequency of KDM6A mutations in females relative to males in NMIBC (47–49). KDM6A mutations have variously been reported as homozygous (48) or heterozygous in females (50), with one study suggesting that KDM6A may be haploinsufficient in the female urothelium (50). In this cancer type, mutations are not concentrated in the JmjC domain, but dispersed, implying dependence on both catalytic and non-catalytic roles for tumor suppression (50). In bladder and some additional cancers, the scaffold function of KDM6A may be more important than the enzymatic activity for tumor suppression, influencing sex-specific manifestation of KDM6A mutations. This interpretation is further supported by the fact that in some cancer settings, pro-oncogenic changes in transcription induced by KDM6A loss can be reversed by re-expression of enzymatically inactive KDM6A, or of UTY (51). Interestingly, in females with bladder cancer, heterozygous mutations in KDM6A often co-occur with mutations in other COMPASS components, such as KMT2C and KMT2D. In males, KDM6A and/or UTY alterations co-occur with KMT2C and KMT2D only in a small fraction of bladder cancer (50) (Figure 4C). Reflecting the cooperative activity of histone-modifying enzymes, mutations in KMT2C and KMT2D significantly influence KDM6A function, resulting in complex disruptions in gene expression in multiple mutated tumors (52). In addition, germline mutations in either KMT2D (type 1, MIM #147920) or KDM6A (type 2, MIM #300867) result in the developmental disorder Kabuki Syndrome, causing similar phenotypes (53), further supporting the idea of closely linked function.
In most tumor types, KDM6A mutations are dispersed throughout the coding exons, suggesting a predominant role in scaffolding. In these cases, KDM6A loss is associated with extensive changes in transcription that include both gene repression and activation mediated through inappropriate gene-targeting of larger chromatin modifier complexes. In human pancreatic cancer, such KDM6A mutations are common, and typically result in protein loss. In males, loss of KDM6A is frequently accompanied by EDY (whether by silencing of UTY, loss of the UTY locus at Yq11, or complete loss of the Y chromosome), and associated with a squamous phenotype with poor prognosis. In a mouse model of Kras-driven pancreatic cancer, total Kdm6a deficiency in females greatly accelerates pancreatic intraepithelial neoplasia (PanIN) and induces a squamous phenotype, while heterozygous loss of Kdm6a in females, or Kdm6a loss in males with intact Uty, causes less tumor acceleration (54). Notably, homozygous loss of Kdm6a in female mice causes premalignant changes, even in the absence of Kras driver mutations. In both human and murine pancreatic cancer, Kdm6a loss selectively activates a cluster of super-enhancers regulating oncogenes including Tp63, Myc and Runx3, increasing H3K4me1 modifications at these loci; while Kdm6a loss inactivates a suite of other genes in a non-sex biased manner (54). These findings suggest the critical sex-specific role of Kdm6a may be to influence lineage selection in tumor precursor cells (43). As another example, in AML, KDM6A reduces tumor-suppressive GATA-dependent transcription programs while upregulatiing oncogenic ETS-dependent programs (51). These extensive transcriptional changes, accompanied by minimal changes in H3K27me3 levels, have been suggested to reflect changes in the enhancer targeting of the COMPASS complex in the absence of KDM6A and UTY scaffolding.
Although the chromosomal complement of KDM6A and UTY is clearly the major driver of their sex-biased tumor suppressive function, it is notable that sex hormones also play a role in modulating their activity (55). For example, KDM6A enhances the expression of hormone-dependent nuclear receptors such as estrogen receptor α (ERα), but is also itself transactivated by ERα, forming a feed-forward regulatory loop of hormone response (55). In addition, KDM6A is regulated by cellular metabolites (Figure 4E), which are produced in sexually dimorphic abundance (56). As a result, the expression of the KDM6A and UTY cofactor α-KG, is 2.3 fold higher in males than females (57), which would be expected to elevate the enzymatic activity of the more metabolite-responsive KDM6A protein in males. These interactions between gene complement and metabolic landscape can exacerbate or quench the effect of specific mutations.
KDM5C/JARID1C and KDM5D/JARID1D
KDM5C/JARID1C/SMCX localizes to Xp11.22; its paralog KDM5D/JARID1D/SMCY, on Yq11.223, encodes a protein with ~84% similarity (Figure 4B). KDM5C binds to the activating mark H3K9me3, present in heterochromatin, demethylates mono-, di- or tri-methylated H3K4, and associates with protein complexes regulating heterochromatin assembly; depending on the genomic element bound, KDM5C can act as context-dependent transcriptional repressor or activator. For example, KDM5C interacts with histone deacetylases (HDACs) and KMTs to restrain transcription at promoters (Figure 4D), but may also interact with distal elements to stimulate the activity of enhancers (58). Mechanistically, loss of KDM5C has been proposed to support tumor growth in several ways, with critical targets varying in different tumor types. For example, in VHL-deficient renal cancers, loss of the hypoxia-induced transcription factor HIF2α, PBRM1 (the nucleosome-targeting subunit of the SWI/SNF complex), or KDM5C reduces an ISGF3-dependent interferon signature that is an important negative feedback mechanism for tumor growth (59). Loss of KDM5C in renal cancer also disrupts heterochromatin stability, causing anomalous transcription of non-coding RNAs, and triggering genomic instability (24).
As with KDM6A and UTY, genetic studies suggest similar but non-equivalent function between KDM5C and KDM5D, and both enzymatic and scaffolding activity. Specifically, inherited mutations of KDM5C are one of the most common sources of X-linked intellectual disability in males (Mental Retardation, X-linked, Syndromic, Claes-Jensen type (MRXSCJ, MIM #300534); inherited mutations in KDM5D, however, do not cause mental retardation. While most MRXSCJ-associated pathogenic variants impair the enzymatic activity of KDM5C, at least some do not, even though they significantly influence the gene expression profile (60). KDM5D, in turn, is specifically required in testicular germ cells, where it acts as part of a complex that promotes chromatin condensation prior to meiosis (61). Part of the difference in activity between KDM5C and KDM5D may also reflect differences in expression. KDM5C is expressed in almost all tissues in adults, and at higher levels than KDM5D, which is predominantly detectable in testis, prostate, and small intestine (62).
Mutation or gene silencing of KDM5C is common and removes tumor suppressive activity in clear cell renal carcinomas (ccRCCs), gastric cancer, follicular thyroid carcinoma, salivary duct carcinoma, human papillomavirus (HPV)-associated cancers, and mantle cell lymphoma (63–67). Many of these cancers are more prevalent in males than females, except thyroid carcinoma and human papillomavirus (HPV)-associated cancers ((68), and Supp Table 1). Based on public data in cBioPortal (41), mutations in KDM5C are dispersed throughout the coding sequence for many of these cancers, emphasizing the importance of a non-catalytic (or scaffolding) role for the protein (Supp Figure 2). In addition, for some of these cancers, there is evidence of sex-biased mutation ((64) and Supp Table 1).
KDM5D also contributes to tumor suppression, with evidence for a more important TSG role than KDM5C in some cancers. Downregulation, loss or inactivating mutations of KDM5D occur in ~40% of male ccRCCs (16), while KDM5C is mutated in only a small fraction of tumors in males (6.2%) and females (0.5%) (64). In gastric cancer, KDM5D overexpression in cancer cells significantly reduces viability, suggesting a direct growth suppressive role (23). Gastric cancer occurs with an imbalanced male-to-female ratio of ~2:1, with mortality also higher in males (55). Mechanistically, gene knockdown of KDM5D in male gastric cancer cells inhibits the demethylation of CUL4A, reducing the expression of CUL4A target genes such as the tumor suppressor CDKN1A/p21 and TP53, and promoting metastasis (23). Loss of KDM5D has been shown to cause atypical patterns of H3K4me3 at promoters, targeting gene programs controlling proliferation, apoptosis, and invasion (69). In prostate cancer, loss of KDM5D by gene silencing or mutation is common during progression, causing defects in control of genomic stability, including DNA replication stress and ATR activation (70), and is associated with shorter overall survival (71). Interestingly, indicating non-equivalent function with KDM5D, KDM5C is overexpressed in prostate cancer, promotes cellular proliferation and has emerged as a predictive marker for therapy failure in patients after prostatectomy (72). Additionally, a pro-oncogenic role for KDM5C upregulation has been proposed in breast and hepatocellular cancers (73), implying tissue specificity in tumor-suppressive versus tumor-promoting action. The reasons for this difference are not yet well understood.
Targeting histone modifications
Aberrant chromatin states are increasingly being targeted in cancer (74–76). Drugs developed to modify chromatin include two major classes. Broad reprogramming agents significantly alter gene expression based on inhibition of widely expressed targets that include histone deacetylase inhibitors (HDACis), DNA methyltransferase inhibitors (DNMTis), and bromodomain and extra-terminal (BET)-targeting agents. Targeted agents focus more exactly on individual components of the epigenetic machinery that are mutated in specific cancers, including the H3K27 histone N-methyltransferase EZH2, and the TCA cycle components IDH1 and IDH2, which regulate α-KG availability (e.g. (77)).
Because KDM6A, UTY, KDM5C and KDM5D are components of large multimeric complexes (Figure 4), their mutation and expression can affect cellular response to clinically advanced drugs targeting complex components (Table 1). For example, in urothelial bladder cancer (78), and in multiple myeloma (79), KDM6A loss confers sensitivity to inhibitors of EZH2 (78), while KDM6A loss in pancreatic cancers (54) or knockdown in prostate cancer models (80) sensitizes to BET inhibitors. However, in aggressive prostate cancer, loss of KDM5D increases DNA replication stress and accelerates mitotic entry; this causes resistance to docetaxel, but sensitization to ATR inhibition, emphasizing the complex action of the KDMs (70,71). It is likely that many drugs targeting the epigenome indirectly influence KDM6A and KDM5C activity. For example, dual treatment of cells with low levels of an EZH2 inhibitor (GSK126) and a histone deacetylase inhibitor (SAHA, also known as vorinostat) caused striking defects in XCI (a process maintained by extensive H3K27me3 modification on the X chromosome). In this study, dual drug treatment resulted in loss of H3K27 methylation on the Xi chromosome, causing reprogramming of transcription. Interestingly, the effects on transcription were uneven, with these drugs selectively increasing expression of genes near the center of XCI, such as TSPAN7 at Xp11.4, and FOXP3 at Xp11.2; KDM6A (Xp11.3) and KDM5C (Xp11.22) are also located near this center. Such reprogramming could lead to undesirable side effects specifically in females treated with such drugs. Moreover, as noted above, EZH2 is a component of the PRC2 complex; intact KDM6A opposes PRC2 activity, and hence would function similarly as an EZH2 inhibitor; hence, administration of SAHA or other epigenetic drugs targeting PRC2 function in tumors bearing mutations inactivating KDM6A might lead to defects in maintenance of XCI, leading to greater disruption of gene expression patterns in females than in males (81).
Although KDM6A, UTY, KDM5C and KDM5D are typically tumor-suppressive, as a class, KDMs have attracted significant interest as targets for enzymatic inhibition (82). This interest was driven by the recognition that some KDMs, including KDM1A/LSD1, members of the JMJD2 subfamily, and other JmjC proteins KDM5B/JARID1B and KDM2B/FBXL10 are often overexpressed in cancers, and are thought to be pro-oncogenic. Most drugs developed for Jumonji-family KDMs target the active site of the Jumonji domain (Table 1). These agents typically bind competitively with α-KG and chelate the active site Fe(II) residue. Because the active site is similar across 17 histone demethylases in humans, this strategy has resulted in limited drug selectivity, with most compounds targeting multiple members of the KDM4, KDM5, and KDM6 families. Perhaps for this reason, use of these compounds results in significant cytotoxicity (83). Some more recently developed compounds inhibit a more restrictive subset of KDMs, with KDM6A being among the targets (84). In addition, KDM-inhibiting activity has been identified in some drugs originally developed for alternative targets. For example, metformin is currently widely used for treatment of Type 2 diabetes, and has also shown promising activity as a cancer therapeutic (85,86) through downregulation of mitochondrial complex 1 and reduced expression of enzymes mediating gluconeogenesis. Unexpectedly, metformin has also been recognized as a catalytic inhibitor of KDM6A, with activity at biologically relevant concentrations (87).
Given that mutations in KDM6A are sensitizing for some drug treatments, these agents may be useful in combination approaches. However, a fundamental safety issue with the use of these compounds in patients is concern that broadly specific agents will inhibit catalysis-dependent tumor-suppressive activity of KDM6A, UTY, KDM5C and KDM5D, particularly in cell types where tumor suppression depends on the catalytic activity of the protein (e.g. KDM6A in T-ALL). As most Jumonji KDMs achieve in vivo target specificity by residing in larger multimeric complexes that regulate gene localization and histone targeting, developing drugs that disrupt the interaction of specific oncogenic KDMs with protein partners may be one approach to increase specificity. Alternatively, use of degradation-inducing agents such as PROTACs (88), targeted to specific oncogenic KDM family members, may be a more effective approach.
Overall, there has been surprisingly little evaluation to date of sex-based differences in response to agents targeting the cellular machinery regulating the epigenome, nor of the specific roles of KDM6A, UTY, KDM5C, or KDM5D mutation or expression in mediating sex-biased response. Given the extensive evidence for sex-specific differences in their activity, such investigations would be timely. Interestingly, two recent studies evaluating metformin in colorectal cancer (CRC) in patients with concurrent diabetes identified a significantly greater reduction of CRC-specific mortality in females (89,90). Whether these results in part reflect inhibition of KDM6A, and sex-specific effects on the epigenome, is currently unknown.
Future prospects
While much of the above discussion addresses the impact of somatic mutation or silencing of KDM6A, UTY, KDM5C, or KDM5D on histone methylation, emerging topics may suggest a broader role for these proteins in influencing sexual differences in cancer incidence or treatment. Of note, a new enzymatic activity as methylarginine demethylases (RDMs) has been recently demonstrated for a subset of JmjC KDMs, including KDM5C (91). The biological significance of this class of modification is not well-understood and may reflect an important biological activity for this X-linked enzyme.
Immune checkpoint inhibitors (ICIs) have attracted considerable interest as a promising new class of anti-cancer agents. Part of the anti-tumoral activity of epigenetic inhibitors has been determined to depends in part on enhancement of innate and acquired immune responses (92), and a number of trials combining epigenetic inhibitors with ICIs are in progress (e.g., NCT02453620; see also (93)). Notably, KDM6A regulates multiple immune response genes (94–97). Further, KDM6A mutation status has recently been shown to influence the activity of drugs targeting the immune system in bladder cancer cells, where loss of KDM6A activated cytokine and chemokine pathways; these cells were more responsive to two clinically approved agents, the IL6 receptor inhibitor tocilizumab, and the CCR2 inhibitor propagermanium (98). Given the role of KDM6A and KDM5C in immune response and their ability to escape X inactivation, it is reasonable to speculate that their different gene dosage or inactivation in males and females may contribute to differential responses to immunotherapy observed in the two sexes. Moreover, these data also suggest that epigenetic therapies targeting KDM6A and KDM5C might “boost” antitumor immune response in a sex-dependent manner. Potential sex-specific differences in the interaction of epigenetic therapies and ICIs has not been extensively investigated, but merit study.
It has long been known that many lifestyle factors, including diet, physical activity, tobacco smoking, and alcohol consumption, distinguish males and females (99). Some of these factors may plausibly influence the activity of KDM6A, UTY, KDM5C, and KDM5D (as well as other epigenetic regulators). For example, tobacco smoking is nearly five times more common in men than in women. Tobacco smoke causes overexpression of TCA cycle enzymes including fumarate hydratase (FH) and isocitrate dehydrogenase (IDH1/3) (100). In some tissues, tobacco smoke increases driver mutations in genes including IDH1 (101). By modulating FH and IDH1/3 activity, tobacco smoke can affect levels of α-KG, or of α-KG competitive antagonists such as fumarate or the oncometabolite 2HG; this in turn would affect the activity of KDM6A and KDM5C, influencing H3K27me2/3 or H3K4me1/3 levels. As another example, caloric restriction, high fat diets, low protein diets, and other diets affect metabolic pathways and can cause significantly global epigenetic changes, affecting levels of H3K27me2/3 or H3K4me1/3 histone (102). Sex-specific differences have been reported in dietary preferences and nutrient intakes (103). Based on an extensive literature, it is reasonable to speculate that sex differences in nutrient metabolism might influence the availability of metabolic co-factors such as Fe(II) and α-KG, contributing to the selective modulation of the enzymatic activities of KDM6A, UTY, KDM5C, and KDM5D in men and women. These possibilities have received limited if any scrutiny.
Finally, in provocative work in mouse models, mutations in KDM6A were found to cause sex-specific intergenerational epigenetic inheritance and cancer susceptibility (104). In humans, germline variance in the lysine demethylase KDM1A/LSD1 impact cancer risk (105). Similarly, drugs targeting EZH1/2 significantly disrupt the female germline epigenome, causing irreversible changes in H3K27 modification accompanied by growth restriction and other anomalies in offspring that resemble those observed in Ezh2-mutant mice (106). Little is currently known about the degree to which X- or Y- linked KDM-targeting drugs, or inherited alleles impacting activity of these proteins, might provide sex bias in hereditary cancer risk. Of note, although the topic has not attracted significant study, in at least one case the defective allele of KDM6A identified in a ccRCC tumor was identified as germline (40). There is much scope for future work.
Supplementary Material
Funding:
The authors are supported by NIH DK108195 and CA228187 (to EAG), by NCI Core Grant CA006927 (to Fox Chase Cancer Center), and by a Marie Curie Individual Fellowship from the Horizon 2020 EU Program (to RT).
Footnotes
Conflict Statement: The authors declare no conflict of interest.
References.
- 1.Ozdemir BC, Csajka C, Dotto GP, Wagner AD. Sex Differences in Efficacy and Toxicity of Systemic Treatments: An Undervalued Issue in the Era of Precision Oncology. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2018;36(26):2680–3 doi 10.1200/jco.2018.78.3290. [DOI] [PubMed] [Google Scholar]
- 2.Wilson MA, Buetow KH. Novel Mechanisms of Cancer Emerge When Accounting for Sex as a Biological Variable. Cancer research 2020;80(1):27–9 doi 10.1158/0008-5472.can-19-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng D, Trynda J, Williams C, Vold JA, Nguyen JH, Harnois DM, et al. Sexual dimorphism in the incidence of human cancers. BMC cancer 2019;19(1):684 doi 10.1186/s12885-019-5902-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agarwal I, Przeworski M. Signatures of replication timing, recombination, and sex in the spectrum of rare variants on the human X chromosome and autosomes. Proceedings of the National Academy of Sciences of the United States of America 2019;116(36):17916–24 doi 10.1073/pnas.1900714116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haupt S, Caramia F, Herschtal A, Soussi T, Lozano G, Chen H, et al. Identification of cancer sex-disparity in the functional integrity of p53 and its X chromosome network. Nature communications 2019;10(1):5385 doi 10.1038/s41467-019-13266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan Y, Liu L, Chen H, Wang Y, Xu Y, Mao H, et al. Comprehensive Characterization of Molecular Differences in Cancer between Male and Female Patients. Cancer cell 2016;29(5):711–22 doi 10.1016/j.ccell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conforti F, Pala L, Bagnardi V, Viale G, De Pas T, Pagan E, et al. Sex-based heterogeneity in response to lung cancer immunotherapy: a systematic review and meta-analysis. Journal of the National Cancer Institute 2019. doi 10.1093/jnci/djz094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W, Warrington NM, Taylor SJ, Whitmire P, Carrasco E, Singleton KW, et al. Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Science translational medicine 2019;11(473) doi 10.1126/scitranslmed.aao5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ippolito JE, Yim AK, Luo J, Chinnaiyan P, Rubin JB. Sexual dimorphism in glioma glycolysis underlies sex differences in survival. JCI insight 2017;2(15) doi 10.1172/jci.insight.92142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karp NA, Reavey N. Sex bias in preclinical research and an exploration of how to change the status quo. British journal of pharmacology 2018. doi 10.1111/bph.14539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner AD, Oertelt-Prigione S, Adjei A, Buclin T, Cristina V, Csajka C, et al. Gender Medicine and Oncology: Report and consensus of an ESMO Workshop. Annals of oncology : official journal of the European Society for Medical Oncology 2019. doi 10.1093/annonc/mdz414. [DOI] [PubMed] [Google Scholar]
- 12.Clocchiatti A, Cora E, Zhang Y, Dotto GP. Sexual dimorphism in cancer. Nature reviews Cancer 2016;16(5):330–9 doi 10.1038/nrc.2016.30. [DOI] [PubMed] [Google Scholar]
- 13.Groner AC, Brown M. Role of steroid receptor and coregulator mutations in hormone-dependent cancers. The Journal of clinical investigation 2017;127(4):1126–35 doi 10.1172/JCI88885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng D, Williams C, Vold JA, Nguyen JH, Harnois DM, Bagaria SP, et al. Regulation of sex hormone receptors in sexual dimorphism of human cancers. Cancer letters 2018;438:24–31 doi 10.1016/j.canlet.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caceres A, Jene A, Esko T, Perez-Jurado LA, Gonzalez JR. Extreme down-regulation of chromosome Y and cancer risk in men. Journal of the National Cancer Institute 2020. doi 10.1093/jnci/djz232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunford A, Weinstock DM, Savova V, Schumacher SE, Cleary JP, Yoda A, et al. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nature genetics 2017;49(1):10–6 doi 10.1038/ng.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science (New York, NY) 2017;357(6348) doi 10.1126/science.aal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC. Genomic Imprinting and Physiological Processes in Mammals. Cell 2019;176(5):952–65 doi 10.1016/j.cell.2019.01.043. [DOI] [PubMed] [Google Scholar]
- 19.Amos-Landgraf JM, Cottle A, Plenge RM, Friez M, Schwartz CE, Longshore J, et al. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. American journal of human genetics 2006;79(3):493–9 doi 10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Molecular cell 2012;48(4):491–507 doi 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aubert Y, Egolf S, Capell BC. The Unexpected Noncatalytic Roles of Histone Modifiers in Development and Disease. Trends in genetics : TIG 2019;35(9):645–57 doi 10.1016/j.tig.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 2014;514(7523):513–7 doi 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen X, Hu K, Cheng G, Xu L, Chen Z, Du P, et al. KDM5D inhibit epithelial-mesenchymal transition of gastric cancer through demethylation in the promoter of Cul4A in male. Journal of cellular biochemistry 2019;120(8):12247–58 doi 10.1002/jcb.27308. [DOI] [PubMed] [Google Scholar]
- 24.Rondinelli B, Rosano D, Antonini E, Frenquelli M, Montanini L, Huang D, et al. Histone demethylase JARID1C inactivation triggers genomic instability in sporadic renal cancer. The Journal of clinical investigation 2015;125(12):4625–37 doi 10.1172/jci81040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 2005;434(7031):400–4 doi 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 26.Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 2014;508(7497):494–9 doi 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forsberg LA, Rasi C, Malmqvist N, Davies H, Pasupulati S, Pakalapati G, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nature genetics 2014;46(6):624–8 doi 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature reviews Genetics 2006;7(9):715–27 doi 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 29.Pilka ES, James T, Lisztwan JH. Structural definitions of Jumonji family demethylase selectivity. Drug Discov Today 2015;20(6):743–9 doi 10.1016/j.drudis.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 30.Tran TQ, Lowman XH, Kong M. Molecular Pathways: Metabolic Control of Histone Methylation and Gene Expression in Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2017;23(15):4004–9 doi 10.1158/1078-0432.ccr-16-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batie M, Rocha S. JmjC histone demethylases act as chromatin oxygen sensors. Molecular & cellular oncology 2019;6(4):1608501 doi 10.1080/23723556.2019.1608501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harmeyer KM, Facompre ND, Herlyn M, Basu D. JARID1 Histone Demethylases: Emerging Targets in Cancer. Trends in cancer 2017;3(10):713–25 doi 10.1016/j.trecan.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang SP, Tang Z, Chen CW, Shimada M, Koche RP, Wang LH, et al. A UTX-MLL4-p300 Transcriptional Regulatory Network Coordinately Shapes Active Enhancer Landscapes for Eliciting Transcription. Molecular cell 2017;67(2):308–21.e6 doi 10.1016/j.molcel.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Shilatifard A. UTX Mutations in Human Cancer. Cancer cell 2019;35(2):168–76 doi 10.1016/j.ccell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walport LJ, Hopkinson RJ, Vollmar M, Madden SK, Gileadi C, Oppermann U, et al. Human UTY(KDM6C) is a male-specific N-methyl lysyl demethylase. The Journal of biological chemistry 2014;289(26):18302–13 doi 10.1074/jbc.M114.555052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakraborty AA, Laukka T, Myllykoski M, Ringel AE, Booker MA, Tolstorukov MY, et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science (New York, NY) 2019;363(6432):1217–22 doi 10.1126/science.aaw1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shpargel KB, Starmer J, Wang C, Ge K, Magnuson T. UTX-guided neural crest function underlies craniofacial features of Kabuki syndrome. Proceedings of the National Academy of Sciences of the United States of America 2017;114(43):E9046–e55 doi 10.1073/pnas.1705011114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, et al. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proceedings of the National Academy of Sciences of the United States of America 2012;109(32):13004–9 doi 10.1073/pnas.1210787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawlyn C, Kaiser MF, Heuck C, Melchor L, Wardell CP, Murison A, et al. The Spectrum and Clinical Impact of Epigenetic Modifier Mutations in Myeloma. Clinical cancer research : an official journal of the American Association for Cancer Research 2016;22(23):5783–94 doi 10.1158/1078-0432.Ccr-15-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature genetics 2009;41(5):521–3 doi 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2012;2(5):401–4 doi 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Zhang Y, Zheng L, Liu M, Chen CD, Jiang H. UTX is an escape from X-inactivation tumor-suppressor in B cell lymphoma. Nature communications 2018;9(1):2720 doi 10.1038/s41467-018-05084-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalisz M, Bernardo E, Beucher A, Maestro MA, Del Pozo N, Millan I, et al. HNF1A recruits KDM6A to activate differentiated acinar cell programs that suppress pancreatic cancer. The EMBO journal 2020:e102808 doi 10.15252/embj.2019102808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood 2015;125(1):13–21 doi 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim PH, Cha EK, Sfakianos JP, Iyer G, Zabor EC, Scott SN, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. European urology 2015;67(2):198–201 doi 10.1016/j.eururo.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017;171(3):540–56.e25 doi 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanli O, Dobruch J, Knowles MA, Burger M, Alemozaffar M, Nielsen ME, et al. Bladder cancer. Nature reviews Disease primers 2017;3:17022 doi 10.1038/nrdp.2017.22. [DOI] [PubMed] [Google Scholar]
- 48.Nassar AH, Umeton R, Kim J, Lundgren K, Harshman L, Van Allen EM, et al. Mutational Analysis of 472 Urothelial Carcinoma Across Grades and Anatomic Sites. Clinical cancer research : an official journal of the American Association for Cancer Research 2019;25(8):2458–70 doi 10.1158/1078-0432.CCR-18-3147. [DOI] [PubMed] [Google Scholar]
- 49.Kato H, Asamitsu K, Sun W, Kitajima S, Yoshizawa-Sugata N, Okamoto T, et al. Cancer-derived UTX TPR mutations G137V and D336G impair interaction with MLL3/4 complexes and affect UTX subcellular localization. Oncogene 2020;39(16):3322–35 doi 10.1038/s41388-020-1218-3. [DOI] [PubMed] [Google Scholar]
- 50.Hurst CD, Alder O, Platt FM, Droop A, Stead LF, Burns JE, et al. Genomic Subtypes of Non-invasive Bladder Cancer with Distinct Metabolic Profile and Female Gender Bias in KDM6A Mutation Frequency. Cancer cell 2017;32(5):701–15.e7 doi 10.1016/j.ccell.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gozdecka M, Meduri E, Mazan M, Tzelepis K, Dudek M, Knights AJ, et al. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nature genetics 2018;50(6):883–94 doi 10.1038/s41588-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lang A, Yilmaz M, Hader C, Murday S, Kunz X, Wagner N, et al. Contingencies of UTX/KDM6A Action in Urothelial Carcinoma. Cancers 2019;11(4) doi 10.3390/cancers11040481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adam MP, Banka S, Bjornsson HT, Bodamer O, Chudley AE, Harris J, et al. Kabuki syndrome: international consensus diagnostic criteria. Journal of medical genetics 2019;56(2):89–95 doi 10.1136/jmedgenet-2018-105625. [DOI] [PubMed] [Google Scholar]
- 54.Andricovich J, Perkail S, Kai Y, Casasanta N, Peng W, Tzatsos A. Loss of KDM6A Activates Super-Enhancers to Induce Gender-Specific Squamous-like Pancreatic Cancer and Confers Sensitivity to BET Inhibitors. Cancer cell 2018;33(3):512–26.e8 doi 10.1016/j.ccell.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie G, Liu X, Zhang Y, Li W, Liu S, Chen Z, et al. UTX promotes hormonally responsive breast carcinogenesis through feed-forward transcription regulation with estrogen receptor. Oncogene 2017;36(39):5497–511 doi 10.1038/onc.2017.157. [DOI] [PubMed] [Google Scholar]
- 56.Hedrington MS, Davis SN. Sexual Dimorphism in Glucose and Lipid Metabolism during Fasting, Hypoglycemia, and Exercise. Frontiers in endocrinology 2015;6:61 doi 10.3389/fendo.2015.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan S, Yeon A, Shahid M, Anger JT, Eilber KS, Fiehn O, et al. Sex-associated differences in baseline urinary metabolites of healthy adults. Scientific reports 2018;8(1):11883 doi 10.1038/s41598-018-29592-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Outchkourov NS, Muino JM, Kaufmann K, van Ijcken WF, Groot Koerkamp MJ, van Leenen D, et al. Balancing of histone H3K4 methylation states by the Kdm5c/SMCX histone demethylase modulates promoter and enhancer function. Cell reports 2013;3(4):1071–9 doi 10.1016/j.celrep.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 59.Liao L, Liu ZZ, Langbein L, Cai W, Cho EA, Na J, et al. Multiple tumor suppressors regulate a HIF-dependent negative feedback loop via ISGF3 in human clear cell renal cancer. eLife 2018;7 doi 10.7554/eLife.37925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vallianatos CN, Farrehi C, Friez MJ, Burmeister M, Keegan CE, Iwase S. Altered Gene-Regulatory Function of KDM5C by a Novel Mutation Associated With Autism and Intellectual Disability. Frontiers in molecular neuroscience 2018;11:104 doi 10.3389/fnmol.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Akimoto C, Kitagawa H, Matsumoto T, Kato S. Spermatogenesis-specific association of SMCY and MSH5. Genes Cells 2008;13(6):623–33 doi 10.1111/j.1365-2443.2008.01193.x. [DOI] [PubMed] [Google Scholar]
- 62.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 2014;13(2):397–406 doi 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang S, Yim S, Park H. The cancer driver genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism. Experimental & molecular medicine 2019;51(6):66 doi 10.1038/s12276-019-0230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ricketts CJ, Linehan WM. Gender Specific Mutation Incidence and Survival Associations in Clear Cell Renal Cell Carcinoma (CCRCC). PloS one 2015;10(10):e0140257 doi 10.1371/journal.pone.0140257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicolson NG, Murtha TD, Dong W, Paulsson JO, Choi J, Barbieri AL, et al. Comprehensive Genetic Analysis of Follicular Thyroid Carcinoma Predicts Prognosis Independent of Histology. The Journal of clinical endocrinology and metabolism 2018;103(7):2640–50 doi 10.1210/jc.2018-00277. [DOI] [PubMed] [Google Scholar]
- 66.Gargano SM, Senarathne W, Feldman R, Florento E, Stafford P, Swensen J, et al. Novel therapeutic targets in salivary duct carcinoma uncovered by comprehensive molecular profiling. Cancer Med 2019;8(17):7322–9 doi 10.1002/cam4.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Q, Wang HY, Liu X, Roth MH, Shestov AA, Lee SC, et al. Dynamic Changes in Gene Mutational Landscape With Preservation of Core Mutations in Mantle Cell Lymphoma Cells. Front Oncol 2019;9:568 doi 10.3389/fonc.2019.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69(1):7–34 doi 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 69.Li N, Dhar SS, Chen TY, Kan PY, Wei Y, Kim JH, et al. JARID1D Is a Suppressor and Prognostic Marker of Prostate Cancer Invasion and Metastasis. Cancer research 2016;76(4):831–43 doi 10.1158/0008-5472.can-15-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Komura K, Yoshikawa Y, Shimamura T, Chakraborty G, Gerke TA, Hinohara K, et al. ATR inhibition controls aggressive prostate tumors deficient in Y-linked histone demethylase KDM5D. The Journal of clinical investigation 2018;128(7):2979–95 doi 10.1172/jci96769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Komura K, Jeong SH, Hinohara K, Qu F, Wang X, Hiraki M, et al. Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression. Proceedings of the National Academy of Sciences of the United States of America 2016;113(22):6259–64 doi 10.1073/pnas.1600420113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stein J, Majores M, Rohde M, Lim S, Schneider S, Krappe E, et al. KDM5C is overexpressed in prostate cancer and is a prognostic marker for prostate-specific antigen-relapse following radical prostatectomy. Am J Pathol 2014;184(9):2430–7 doi 10.1016/j.ajpath.2014.05.022. [DOI] [PubMed] [Google Scholar]
- 73.Ji X, Jin S, Qu X, Li K, Wang H, He H, et al. Lysine-specific demethylase 5C promotes hepatocellular carcinoma cell invasion through inhibition BMP7 expression. BMC cancer 2015;15:801 doi 10.1186/s12885-015-1798-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kelly AD, Issa JJ. The promise of epigenetic therapy: reprogramming the cancer epigenome. Current opinion in genetics & development 2017;42:68–77 doi 10.1016/j.gde.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 75.Loo Yau H, Ettayebi I, De Carvalho DD. The Cancer Epigenome: Exploiting Its Vulnerabilities for Immunotherapy. Trends in cell biology 2018. doi 10.1016/j.tcb.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 76.Miranda Furtado CL, Dos Santos Luciano MC, Silva Santos RD, Furtado GP, Moraes MO, Pessoa C. Epidrugs: targeting epigenetic marks in cancer treatment. Epigenetics 2019:1–13 doi 10.1080/15592294.2019.1640546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benedetti R, Dell’Aversana C, De Marchi T, Rotili D, Liu NQ, Novakovic B, et al. Inhibition of Histone Demethylases LSD1 and UTX Regulates ERalpha Signaling in Breast Cancer. Cancers 2019;11(12) doi 10.3390/cancers11122027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ler LD, Ghosh S, Chai X, Thike AA, Heng HL, Siew EY, et al. Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Science translational medicine 2017;9(378) doi 10.1126/scitranslmed.aai8312. [DOI] [PubMed] [Google Scholar]
- 79.Ezponda T, Dupere-Richer D, Will CM, Small EC, Varghese N, Patel T, et al. UTX/KDM6A Loss Enhances the Malignant Phenotype of Multiple Myeloma and Sensitizes Cells to EZH2 inhibition. Cell reports 2017;21(3):628–40 doi 10.1016/j.celrep.2017.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hong Z, Wu G, Xiang ZD, Xu CD, Huang SS, Li C, et al. KDM5C is transcriptionally regulated by BRD4 and promotes castration-resistance prostate cancer cell proliferation by repressing PTEN. Biomed Pharmacother 2019;114:108793 doi 10.1016/j.biopha.2019.108793. [DOI] [PubMed] [Google Scholar]
- 81.Laskowski AI, Fanslow DA, Smith ED, Kosak ST. Clinical Epigenetic Therapies Disrupt Sex Chromosome Dosage Compensation in Human Female Cells. Gender and the genome 2018;2(1):2–7 doi 10.1177/2470289718787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov 2013;12(12):917–30 doi 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 83.Hatch SB, Yapp C, Montenegro RC, Savitsky P, Gamble V, Tumber A, et al. Assessing histone demethylase inhibitors in cells: lessons learned. Epigenetics & chromatin 2017;10:9 doi 10.1186/s13072-017-0116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khodaverdian V, Tapadar S, MacDonald IA, Xu Y, Ho PY, Bridges A, et al. Deferiprone: Pan-selective Histone Lysine Demethylase Inhibition Activity and Structure Activity Relationship Study. Scientific reports 2019;9(1):4802 doi 10.1038/s41598-019-39214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morales DR, Morris AD. Metformin in cancer treatment and prevention. Annu Rev Med 2015;66:17–29 doi 10.1146/annurev-med-062613-093128. [DOI] [PubMed] [Google Scholar]
- 86.Arrieta O, Barron F, Padilla MS, Aviles-Salas A, Ramirez-Tirado LA, Arguelles Jimenez MJ, et al. Effect of Metformin Plus Tyrosine Kinase Inhibitors Compared With Tyrosine Kinase Inhibitors Alone in Patients With Epidermal Growth Factor Receptor-Mutated Lung Adenocarcinoma: A Phase 2 Randomized Clinical Trial. JAMA Oncol 2019:e192553 doi 10.1001/jamaoncol.2019.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cuyas E, Verdura S, Llorach-Pares L, Fernandez-Arroyo S, Luciano-Mateo F, Cabre N, et al. Metformin directly targets the H3K27me3 demethylase KDM6A/UTX. Aging cell 2018;17(4):e12772 doi 10.1111/acel.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cermakova K, Hodges HC. Next-Generation Drugs and Probes for Chromatin Biology: From Targeted Protein Degradation to Phase Separation. Molecules 2018;23(8) doi 10.3390/molecules23081958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Y, Xiao J, Zhao Y, Du S, Du J. Effect of metformin on the mortality of colorectal cancer patients with T2DM: meta-analysis of sex differences. Int J Colorectal Dis 2020;35(5):827–35 doi 10.1007/s00384-020-03539-5. [DOI] [PubMed] [Google Scholar]
- 90.Park JW, Lee JH, Park YH, Park SJ, Cheon JH, Kim WH, et al. Sex-dependent difference in the effect of metformin on colorectal cancer-specific mortality of diabetic colorectal cancer patients. World journal of gastroenterology 2017;23(28):5196–205 doi 10.3748/wjg.v23.i28.5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Walport LJ, Hopkinson RJ, Chowdhury R, Schiller R, Ge W, Kawamura A, et al. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nature communications 2016;7:11974 doi 10.1038/ncomms11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jones PA, Ohtani H, Chakravarthy A, De Carvalho DD. Epigenetic therapy in immune-oncology. Nature reviews Cancer 2019;19(3):151–61 doi 10.1038/s41568-019-0109-9. [DOI] [PubMed] [Google Scholar]
- 93.Mazzone R, Zwergel C, Mai A, Valente S. Epi-drugs in combination with immunotherapy: a new avenue to improve anticancer efficacy. Clinical epigenetics 2017;9:59 doi 10.1186/s13148-017-0358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cribbs AP, Terlecki-Zaniewicz S, Philpott M, Baardman J, Ahern D, Lindow M, et al. Histone H3K27me3 demethylases regulate human Th17 cell development and effector functions by impacting on metabolism. Proceedings of the National Academy of Sciences of the United States of America 2020;117(11):6056–66 doi 10.1073/pnas.1919893117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cribbs A, Hookway ES, Wells G, Lindow M, Obad S, Oerum H, et al. Inhibition of histone H3K27 demethylases selectively modulates inflammatory phenotypes of natural killer cells. The Journal of biological chemistry 2018;293(7):2422–37 doi 10.1074/jbc.RA117.000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Itoh Y, Golden LC, Itoh N, Matsukawa MA, Ren E, Tse V, et al. The X-linked histone demethylase Kdm6a in CD4+ T lymphocytes modulates autoimmunity. The Journal of clinical investigation 2019;129(9):3852–63 doi 10.1172/JCI126250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu L, Cao J, Cai WL, Lang SM, Horton JR, Jansen DJ, et al. KDM5 histone demethylases repress immune response via suppression of STING. PLoS Biol 2018;16(8):e2006134 doi 10.1371/journal.pbio.2006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kobatake K, Ikeda KI, Nakata Y, Yamasaki N, Ueda T, Kanai A, et al. Kdm6a Deficiency Activates Inflammatory Pathways, Promotes M2 Macrophage Polarization, and Causes Bladder Cancer in Cooperation with p53 Dysfunction. Clinical cancer research : an official journal of the American Association for Cancer Research 2020;26(8):2065–79 doi 10.1158/1078-0432.CCR-19-2230. [DOI] [PubMed] [Google Scholar]
- 99.Alegria-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics 2011;3(3):267–77 doi 10.2217/epi.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Solanki HS, Babu N, Jain AP, Bhat MY, Puttamallesh VN, Advani J, et al. Cigarette smoke induces mitochondrial metabolic reprogramming in lung cells. Mitochondrion 2018;40:58–70 doi 10.1016/j.mito.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 101.Yoshida K, Gowers KHC, Lee-Six H, Chandrasekharan DP, Coorens T, Maughan EF, et al. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature 2020;578(7794):266–72 doi 10.1038/s41586-020-1961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Molina-Serrano D, Kyriakou D, Kirmizis A. Histone Modifications as an Intersection Between Diet and Longevity. Frontiers in genetics 2019;10:192 doi 10.3389/fgene.2019.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li KK, Concepcion RY, Lee H, Cardinal BJ, Ebbeck V, Woekel E, et al. An examination of sex differences in relation to the eating habits and nutrient intakes of university students. J Nutr Educ Behav 2012;44(3):246–50 doi 10.1016/j.jneb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 104.Lesch BJ, Tothova Z, Morgan EA, Liao Z, Bronson RT, Ebert BL, et al. Intergenerational epigenetic inheritance of cancer susceptibility in mammals. eLife 2019;8 doi 10.7554/eLife.39380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wei X, Calvo-Vidal MN, Chen S, Wu G, Revuelta MV, Sun J, et al. Germline Lysine-Specific Demethylase 1 (LSD1/KDM1A) Mutations Confer Susceptibility to Multiple Myeloma. Cancer research 2018;78(10):2747–59 doi 10.1158/0008-5472.CAN-17-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Prokopuk L, Hogg K, Western PS. Pharmacological inhibition of EZH2 disrupts the female germline epigenome. Clinical epigenetics 2018;10:33 doi 10.1186/s13148-018-0465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Benyoucef A, Palii CG, Wang C, Porter CJ, Chu A, Dai F, et al. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes & development 2016;30(5):508–21 doi 10.1101/gad.276790.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang Q, Chen X, Jiang Y, Liu S, Liu H, Sun X, et al. Elevating H3K27me3 level sensitizes colorectal cancer to oxaliplatin. J Mol Cell Biol 2020;12(2):125–37 doi 10.1093/jmcb/mjz032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu B, Pan X, Chen X, Chen M, Shi K, Xu J, et al. Epigenetic drug library screening identified an LSD1 inhibitor to target UTX-deficient cells for differentiation therapy. Signal Transduct Target Ther 2019;4:11 doi 10.1038/s41392-019-0040-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stief SM, Hanneforth AL, Weser S, Mattes R, Carlet M, Liu WH, et al. Loss of KDM6A confers drug resistance in acute myeloid leukemia. Leukemia 2020;34(1):50–62 doi 10.1038/s41375-019-0497-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McNairn AJ, Chuang CH, Bloom JC, Wallace MD, Schimenti JC. Female-biased embryonic death from inflammation induced by genomic instability. Nature 2019;567(7746):105–8 doi 10.1038/s41586-019-0936-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gal-Oz ST, Maier B, Yoshida H, Seddu K, Elbaz N, Czysz C, et al. ImmGen report: sexual dimorphism in the immune system transcriptome. Nature communications 2019;10(1):4295 doi 10.1038/s41467-019-12348-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pala L, Nezi L, De Pas T, Pennacchioli E, Cocorocchio E, Ferrucci P, et al. Sex Differences in Efficacy and Toxicity of Systemic Cancer Treatments: Role of the Microbiome. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2019;37(5):439 doi 10.1200/jco.18.01270. [DOI] [PubMed] [Google Scholar]
- 114.Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harbor perspectives in biology 2016;8(4):a019521 doi 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Galupa R, Heard E. X-Chromosome Inactivation: A Crossroads Between Chromosome Architecture and Gene Regulation. Annual review of genetics 2018. doi 10.1146/annurev-genet-120116-024611. [DOI] [PubMed] [Google Scholar]
- 116.Migeon BR. Choosing the Active X: The Human Version of X Inactivation. Trends in genetics : TIG 2017;33(12):899–909 doi 10.1016/j.tig.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 117.Leseva M, Knowles BB, Messerschmidt DM, Solter D. Erase-Maintain-Establish: Natural Reprogramming of the Mammalian Epigenome. Cold Spring Harb Symp Quant Biol 2015;80:155–63 doi 10.1101/sqb.2015.80.027441. [DOI] [PubMed] [Google Scholar]
- 118.Fagan RJ, Dingwall AK. COMPASS Ascending: Emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT2D proteins in cancer. Cancer letters 2019;458:56–65 doi 10.1016/j.canlet.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Berger SL, Sassone-Corsi P. Metabolic Signaling to Chromatin. Cold Spring Harbor perspectives in biology 2016;8(11) doi 10.1101/cshperspect.a019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Laukka T, Myllykoski M, Looper RE, Koivunen P. Cancer-associated 2-oxoglutarate analogues modify histone methylation by inhibiting histone lysine demethylases. Journal of molecular biology 2018;430(18 Pt B):3081–92 doi 10.1016/j.jmb.2018.06.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


