Abstract
Objective
Normalizing clinical mentions to concepts in standardized medical terminologies, in general, is challenging due to the complexity and variety of the terms in narrative medical records. In this article, we introduce our work on a clinical natural language processing (NLP) system to automatically normalize clinical mentions to concept unique identifier in the Unified Medical Language System. This work was part of the 2019 n2c2 (National NLP Clinical Challenges) Shared-Task and Workshop on Clinical Concept Normalization.
Materials and Methods
We developed a hybrid clinical NLP system that combines a generic multilevel matching framework, customizable matching components, and machine learning ranking systems. We explored 2 machine leaning ranking systems based on either ensemble of various similarity features extracted from pretrained encoders or a Siamese attention network, targeting at efficient and fast semantic searching/ranking. Besides, we also evaluated the performance of a general-purpose clinical NLP system based on Unstructured Information Management Architecture.
Results
The systems were evaluated as part of the 2019 n2c2 challenge, and our original best system in the challenge obtained an accuracy of 0.8101, ranked fifth in the challenge. The improved system with newly designed machine learning ranking based on Siamese attention network improved the accuracy to 0.8209.
Conclusions
We demonstrate the successful practice of combining multilevel matching and machine learning ranking for clinical concept normalization. Our results indicate the capability and interpretability of our proposed approach, as well as the limitation, suggesting the opportunities of achieving better performance by combining general clinical NLP systems.
Keywords: clinical natural language processing, concept normalization, attention, CUI, UMLS
INTRODUCTION
Electronic health records (EHRs), which detailly document patient’s medical history and clinical activities, contain a lot of useful information such as diseases, symptoms, medications, treatments, and so on.1 This information holds great value for various applications in both the healthcare industry and academia, including clinical decision support, risk evaluation, disease modeling, healthcare quality measurements, etc.1–4 However, much of the information is embedded in unstructured data of EHRs such as progress notes, discharge summaries, procedure notes, and so on, which typically are difficult for direct use or query compared with structured data.
Clinical natural language processing (NLP), which serves to unlock the information by processing those free-text clinical notes in EHRs, has gained great interest recently from the healthcare industry and medical informatics community. In clinical NLP, one of the critical tasks is information extraction which typically consists of 2 steps: (1) named entity recognition, which locates or extracts the syntaxial mentions from the text, and (2) named entity normalization, which maps the extracted mentions to concept identifiers in a standardized terminology. The concept unique identifier (CUI) in the Unified Medical Language System (UMLS)5 is one of the most widely used medical terminologies. The clinical concept normalization task is critical as it generalized the extracted medical information crossing different contexts, notes, and patients, leading to more exchangeable and effective medical data usage. However, it is also challenging because of the variation and ambiguity of the terms used in clinical notes. Therefore, clinical NLP systems that can accurately normalize the medical mentions to corresponding concepts are highly desirable.
The 2019 National NLP Clinical Challenges (n2c2) Shared-Task and Workshop on Clinical Concept Normalization6 was organized for this topic. The challenge provided annotated medical mentions/entities as well as the corresponding clinical notes, and asked for NLP systems that can automatically process the mentions and normalize them to CUI in UMLS.
In this article, we describe a hybrid clinical NLP system for clinical concept normalization, which employs a generic multilevel matching framework combining customizable matching components and machine learning (ML) ranking systems. In addition, we also investigated a general clinical NLP (GCNLP) system that is built with Unstructured Information Management Architecture (UIMA).7 Evaluation and analysis were conducted upon different aspects between these systems with the n2c2 challenge data. Our best system submitted to n2c2 was ranked fifth in the challenge. We demonstrate the advantages of our approaches for feasible and reliable concept normalization as well as their limitations.
Clinical concept normalization links free-text mentions into standardized clinical concepts. It increases the interoperability of medical data and removes ambiguities associated with text, which is critical for a wide range of applications in research and industry such as cohort selection and clinical coding. For instance, “cardiomegaly,” “enlarged heart,” and “increased heart size” are semantically identical and all refer to the symptom of heart enlargement. If we want to select the patients with the heart enlargement condition for a clinical trial based on their medical records, it could be very difficult to recognize the similarity among patients without clinical concept normalization, as different words and expressions are used. For another instance, “ASA,” “Aspirin,” “Acetylsalicylic Acid,” “Durlaza,” and “Ecotrin” are commonly used terms in clinical notes referring to the same generic medication of aspirin. Variation in the expression of medical concepts is very common, which is also the primary challenge a clinical concept normalization system aiming to tackle.
Many previous studies, NLP systems, as well as competitions, have focused on addressing this issue. Among them, several general clinical NLP systems have been developed, such as MedLEE,8 MetaMap,9 cTAKES,10 CLAMP,11 etc. Those systems provide end-to-end solutions for clinical concept extraction, including both mention extraction and concept normalization. Though those general clinical NLP systems have been widely used in the community, the normalization is typically based on lexical or syntactic patterns and morphological variations. Though those systems can serve as decent baselines, they are hard to be competitive against later on more sophisticated concept normalization systems due to the intrinsic limitation of lacking semantic information.12,13 Besides, a series of challenges have been organized and played as significant roles to push forward state of the art, including ShARe/CLEF eHealth 2013 Task 1,14SemEval-2014 Task 7,15 and SemEval-2015 Task 14.16 Among the top-performing systems in those challenges, Leaman et al17 introduced a normalization system with term frequency–inverse document frequency (TF-IDF) representations of mentions and concept descriptions and a scoring function trained with similarity matrix and pairwise learning-to-rank technique. Zhang et al18 proposed a vector space model (VSM)–based approach in which the cosine similarity over TF-IDF representations was adopted to rank the candidates. Ghiasvand and Kate19 presented a pattern-based system that combined exact match and edit distance patterns learned from UMLS and training data. D’Souza and Ng20 introduced a sieve system employing rule-based matching and lexical variations.
In recent years, deep learning-based approaches have been proposed on this topic. Among them, convolutional neural networks,21 bidirectional long short-term memory networks,12 and more recently, bidirectional encoder representations from transformers (BERT)22 have been investigated for concept normalization in biomedical domain. With pretrained embeddings or language models, those deep learning systems have intrinsic advantages in capturing semantic information. And they typically outperform the traditional systems, achieving state-of-the-art performance. However, typical deep learning models require domain-specific pretrained models and sufficient task-specific labeled data for fine-tuning. Besides, they typically require longer running time, are more sensitive to data quality issues such as annotation errors or bias, and have a lack of interpretability and transparency. Thus, traditional approaches at some point still hold their own value for better efficiency, robustness, interpretability, and transferability, and are still the mainstream methods in commercial products.23
In this article, we propose a hybrid method that combines and utilizes the traditional morphology-based dictionary lookup, semantics-based text representations, and ML methods in an efficient way. More specifically, we designed a generic multilevel matching framework that can apply cascading matching and choose the appropriate normalization methods for a given mention. The rationale is that there is no need to apply heavy deep learning models or pretrained representations if the mention-concept linking can be found by low-cost morphology-based dictionary matching. For those cases cannot be solved by morphological matching, semantic modeling, and ML ranking system are required.
For the ML ranking system design, we adapted the idea from previous works12–22 that concept normalization benefits from either task-specific similarity scoring functions or task-specific text representations. We explored 2 lightweight ML ranking systems under these 2 directions: (1) a system with trainable ensemble-based similarity score but various predefined text representations (e.g. word embedding, BERT) and (2) a system with predefined similarity score (Cosine similarity) but trainable text representation with Siamese attention networks. The first system aimed to leverage the advantages of various pretrained representations and to be fine-tuned with ensemble weights in the scoring function, while the second system aimed to use a simple scoring function but with representation fine-tuned with attention.
The multilevel matching framework not only combines traditional morphological matching and ML ranking, but also provides the flexibility to define the usage and priority of each method, ensuring both accuracy and efficiency. Our hybrid systems were evaluated using the 2019 n2c2 track3 corpus and achieved good performance in the challenge. We also investigated an in-house general clinical NLP system on this task for comparison.
MATERIALS AND METHODS
Task and data
In 2019 n2c2 challenge on clinical concept normalization, participants were provided with annotated clinical-related mentions as well as the original notes and were asked to map those mentions to CUIs in the UMLS 2017AB version. For example, “heart attack” and “myocardial infarction” should be mapped to the same CUI of C0027051. Furthermore, only 1 CUI was expected as the output per mention. If there was no proper CUI to be assigned to the given mention, a “CUI-less” label should be outputted. The n2c2 challenge used the Medical Concept Normalization corpus.24 The Medical Concept Normalization corpus contains 100 discharge summaries from Partners HealthCare, which provides the normalization for a total of 10 919 concept mentions, with 3792 unique concepts from 2 vocabularies, SNOMED-CT (Systematized Nomenclature of Medicine Clinical Terms)25 and RxNorm,26 in UMLS. It covers diverse categories of clinical concepts such as diseases, disorders, symptoms, medications, exams, procedures, etc. Table 1 shows more details of the data.
Table 1.
Summary of the mentions and CUIs in each semantic type group
| Full |
Train |
Test |
||||||
|---|---|---|---|---|---|---|---|---|
| Semantic type | Type description | Mention | CUI | Mention | CUI | Mention | CUI | Mention examples |
| DiseaseDisorder | Disease or Syndrome, Acquired Abnormality, etc. | 2460 | 799 | 1242 | 471 | 1218 | 535 | “congestive heart failure,” “coronary artery disease,” “diabetes mellitus” |
| Exam | Laboratory or Test Result, Laboratory Procedure, etc. | 2263 | 463 | 1142 | 308 | 1121 | 318 | “creatinine,” “hematocrit,” “platelet count” |
| Medication | Clinical Drug, Pharmacologic Substance, etc. | 1816 | 522 | 959 | 353 | 857 | 345 | “Coumadin,” “Lasix,” “Colace,” “Aspirin,” “Percocet” |
| Concept | Qualitative Concept, Spatial Concept, Quantitative Concept, etc. | 1749 | 396 | 798 | 263 | 951 | 300 | “some,” “left,” “mild,” “nontender,” “elevated,” “multiple” |
| Finding | Finding, Phenomenon or Process, etc. | 1376 | 504 | 630 | 300 | 746 | 334 | “afebrile,” “mass,” “No known drug allergies,” “lesion,” “weight loss” |
| Procedure | Therapeutic or Preventive Procedure | 1138 | 385 | 573 | 226 | 565 | 253 | “the procedure,” “chemotherapy,” “resection,” “radiation therapy” |
| SignSymptom | Sign or Symptom | 992 | 183 | 535 | 128 | 457 | 127 | “pain,” “nausea,” “chest pain,” “shortness of breath,” “fever” |
| Activity | Health Care Activity, Social Behavior, etc. | 554 | 72 | 249 | 38 | 305 | 51 | “blood pressure,” “heart rate,” “cardiac catheterization,” “pulse,” “evaluation” |
| Anatomy | Body Part, Organ, or Organ Component, Body Location or Region, etc. | 530 | 294 | 248 | 160 | 282 | 192 | “left lower lobe,” “bibasilar,” “LAD,” “lower extremity,” “transverse mesocolon” |
| Others | Bacterium, Body Substance, Medical Device, etc. | 363 | 173 | 157 | 83 | 206 | 123 | “drains,” “bacteria,” “yeast,” “protocol,” “somomabodies” |
| CUI-less | No corresponding CUI in UMLS | 368 | 1 | 151 | 1 | 217 | 1 | “no acute distress,” “further,” “apparent,” “atraumatic,” “workup” |
| Total | 13 609 | 3791 | 6684 | 2330 | 6925 | 2578 | ||
CUI: concept unique identifier; UMLS: Unified Medical Language System.
During the challenge, these 100 discharge summaries were split into training and testing datasets with a 50/50 split. All the mentions extracted from those notes were annotated with span locations and corresponding CUI or “CUI-less” label. Only the training dataset was released during the development phase, and the final evaluation was performed by the organizer against the held-out test dataset.
Systems overview
We developed 3 NLP systems for this task: (1) a hybrid multilevel matching system embedded with an ensemble-based ML ranking subsystem with similarity features extracted from pretrained encoders; (2) a hybrid multilevel matching system embedded with an attention-based ML ranking subsystem trained with Siamese attention networks; and (3) an in-house general clinical NLP (GCNLP) system based on UIMA framework and Lucene27 lookup to serve as the baseline.
As a comparison, the 2 hybrid systems shared the same set of methods in each matching level and only differed in the ML ranking method. The cascading design of the multilevel matching system ensures both effectiveness and efficiency of applying normalization methods. As mentioned above, the rationale of exploring these 2 ML ranking methods is to benefit from either a trainable task-specific similarity scoring function or text representation. In contrast to these hybrid systems, the GCNLP system was not conducted with fine-tuning with n2c2 data, and it searched among all the CUIs in UMLS without considering the annotation preference from the training data. Besides, the GCNLP system used a more sophisticated disambiguation module pretrained with a much larger dataset. These facts indicate its potential of being a good complement of the hybrid systems and draw our interest to test it on this task.
Multilevel matching system
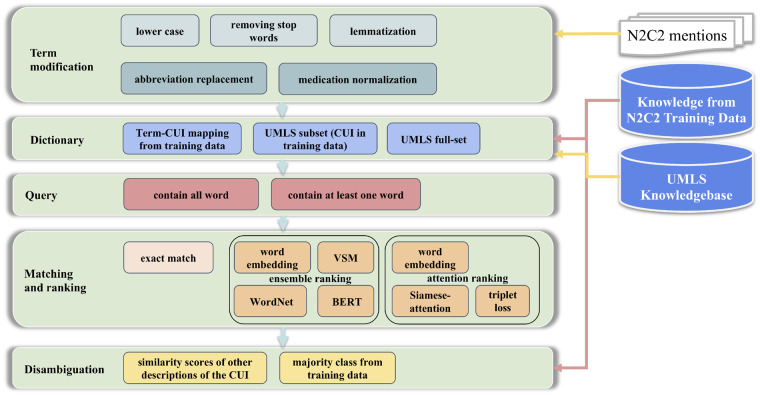
This matching system employs a generic multilevel matching framework. The basic operation in this system is “matching,” which consists of 5 components: term modification, dictionary, query, matching and ranking, and disambiguation. Figure 1 shows the high-level architecture of this system. With this framework, one can customize the criteria of the components in each matching level and define the sequence of matching to be performed. For the n2c2 task, we specifically implemented the following components and 11 matching levels according to error analysis during the development process. More details of the development process and the matching levels can be found in the Supplementary Appendix. The components are:
Figure 1.
Architecture of the hybrid system combining the multilevel matching with machine learning ranking systems. BERT: bidirectional encoder representations from transformers; CUI: concept unique identifier; UMLS: Unified Medical Language System; VSM: vector space model.
Term modification: This component defines the form of the given mention to be used for searching. 5 modification methods were employed, including lower case, removing stop words, lemmatization, abbreviation replacement, and medication normalization. In this study, we used the default English stop words from NLTK28 package and list of medical abbreviations from Wikipedia29 for abbreviation replacement. A medication brand name to generic name mapping table extracted from UMLS was used for medication normalization.
Dictionary: Dictionary contains the mapping between CUIs and their synonyms/descriptions. This component defines the range of CUI and synonyms to be used for matching. Three levels of dictionaries were used, including annotated Term-CUI mapping from the training data, UMLS subset (with CUI included in training dataset), and the UMLS full set.
Query: Considering the computing cost and speed, not all the synonyms in the dictionary should be considered for matching with the given mention. Thus, this component defines the criteria for selecting synonym or CUI candidates. Two types of queries were considered: (1) the synonyms should contain all words from the mention and (2) the synonyms should contain at least 1 word from the mention.
Matching and ranking: This component defines the method to be used for concept lookup. In this study, exact match of the mention with modifications against CUI synonyms was used as the baseline. However, for cases without any exact match, 2 ML ranking systems considering semantic information were developed. More details regarding these 2 ranking systems can be found in the following section.
Disambiguation: Sometimes, a given mention can trigger multiple CUIs with the matched synonyms. This component defines the method to select the preferred CUI among them. Two methods were considered in this component: (1) the majority class from training data (ie, the preferred CUI from the training set) and (2) similarity scores of other synonyms instead of the best-matching one.
The process performed in each matching level is (1) by given a “term” or modification of the term, generate “query” to fetch the CUI candidates from the “dictionary”; (2) then use one method from “matching and ranking” to select the matched CUIs; and (3) use “disambiguation” system to select only one best CUI if the matching triggered multiple CUIs. This operation is performed recursively with different combinations of the methods until an appropriate CUI has been found.
ML ranking systems
Two ML ranking systems based on semantic similarity were developed as part of the multilevel matching system: (1) an ensemble system based on similarity features extracted from word embedding, TF-IDF vectors, WordNet,30 and BERT/BioBERT31,32 and (2) a simple similarity system merely based on Siamese attention networks and word embedding.
Instead of classifying whether yes or no for each candidate to be selected as the preferred CUI, we approached this task as a ranking problem which focused more on the relative similarity scores among the candidates. Both of the ML systems employed the same processes of candidate generation, training sample preparation and loss function. For candidate generation, we used the average-pooling of the word embedding as the default representations of the mentions and CUI synonyms. The word embedding was trained with word2vec algorithm and MIMIC III (Medical Information Mart for Intensive Care-III)33 data, and with a dimension of 200. Cosine similarities between the mention and CUI synonyms were calculated as the default ranking scores to select the candidates. Then top 30 CUIs with the highest scores (maximum score among all synonyms of each CUI) were selected as the candidates.
The triplet loss algorithm is a widely used algorithm for ranking problems originally introduced in face recognition.34 Here, we used it as our loss function and prepared samples accordingly. The triplet loss is defined as:
| (1) |
Where a, p, n represents anchor input, positive input, and negative input, respectively. D is the distance function and margin is a positive hyperparameter. The basic idea of triplet loss is to guide the ML model to minimize the anchor-positive distance and maximize the anchor-negative distance. In the training dataset, the given mention which was asked for a matched CUI was regarded as the anchor. The synonyms of the annotated CUI (ground truth) were regarded as positive inputs. And the synonyms of other candidate CUIs were regarded as negative inputs. The distance was defined as negative value of the final similarity score outputted from the models. During training, samples were prepared in form of triplets and were fed into the ML ranking models.
For the ensemble model, we directly trained a single layer dense neural network with the similarity features extracted from pretrained encoding representations of the mentions and CUI synonyms. The similarity features are:
Word embedding–based: An average-pooling or max-pooling of the word embeddings were used as the representations. For each pooling method, we calculated the cosine similarity and Manhattan similarity as features.
VSM-based: The mention was treated as query and CUI synonyms were treated as documents in VSM model. The cosine similarity and Manhattan similarity over TF-IDF representations were calculated as features.
WordNet-based: Similarity score based on WordNet and corpus statistics30 was generated as a feature. This similarity score was implemented as a linear combination of semantic similarity and word order similarity as introduced by Li et al,30 where the semantic similarity was calculated based on the hierarchical semantic distance between words in WordNet.
BERT-based: Sentence-level vectors generated by pretrained encoders, BERT31 and BioBERT,32 were used as the representations. Here, we only encoded the given mention or CUI synonyms themselves without considering the surrounding context. Then cosine similarity and Manhattan similarity for each encoded mention-synonym pair were calculated as features.
In order to obtain a better mention and synonym representation for fast and low-cost pairing or searching, we developed another simple ML ranking system based on Siamese attention networks. The system contains 4 parts, as shown in Figure 2:
Figure 2.
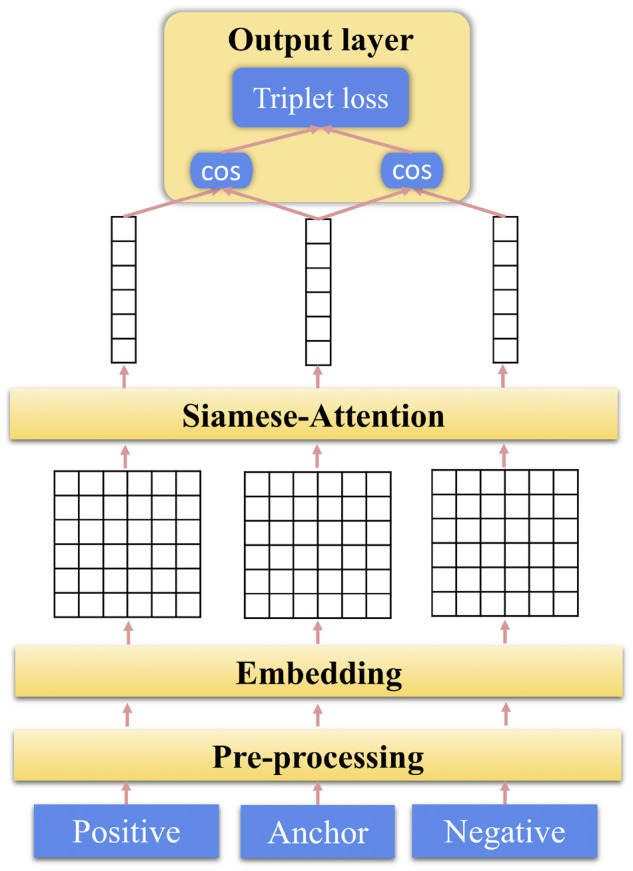
Architecture of the machine learning ranking system based on the Siamese attention network.
Preprocessing: This module takes in the triplet inputs consisted of anchor, positive, and negative phrases. Stop words and special characters are removed or replaced during this process.
Embedding layer: In this layer, the input phrases are tokenized. And each word in a phrase is transferred into a real-valued vector. Then the phrase initially as a sequence of words is represented as a sequence of word embedding vectors.
Siamese attention layer: Attention provides a trainable weight vector that guides the system to focus on task-specific semantic information. Here, weighted-pooling is applied among word embeddings to generate phrase-level vectors. More details can be found in Supplementary Appendix.
Output layer: After gathering the vector representations of the anchor, positive, and negative phrases, cosine similarities of anchor-positive and anchor-negative pairs are calculated and fed into the triplet loss.
In contrast to the arbitrary average-pooling or max-pooling, the attention layer provides a more sophisticated way of pooling the word-level encodings into a phrase-level representation. The attention layer can help the system to focus more on the words with higher task-specific semantic importance, leading to better performance for other NLP tasks, as reported elsewhere.35–37 In addition, compared with the ensemble system that relies on various pretrained encoders, this system only uses word embedding and cosine similarly. This simple design leads to a faster, lighter, and more interpretable solution for semantic searching or matching.
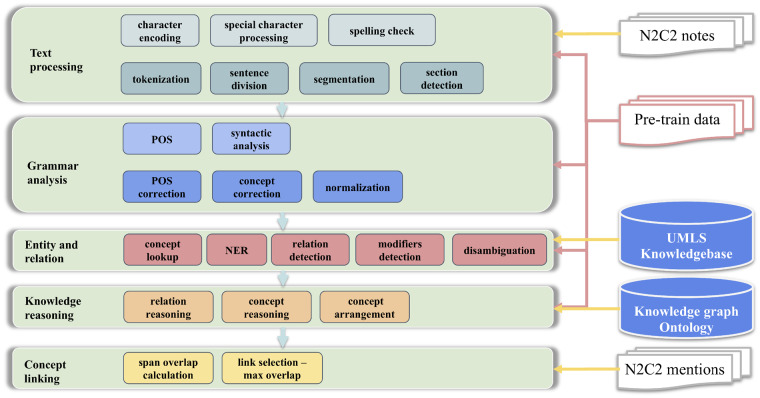
GCNLP system
The GCNLP system was modified from an in-house general medical information extraction system that was initially designed for computer-assistant coding.38 This system provides an end-to-end solution for clinical concept extraction, covering both mention and entity extraction and concept normalization. This UIMA-based system contains 5 modules, as shown in Figure 3. For medical entity extraction, it employs Lucene27 lookup of CUI in UMLS. A disambiguation submodule based on PageRank algorithm39 and VSM40 is also included considering context information, semantic type as well as the co-occurrence among concepts. More details about this system can be found in our previous works37,41 and the Supplementary Appendix.
Figure 3.
Architecture of the general clinical natural language processing system. This system is based on the Unstructured Information Management Architecture framework and Lucene lookup. NER: named entity recognition; POS: part of speech.
RESULTS
The systems were evaluated as part of the 2019 n2c2 challenge. System-generated CUIs are compared against the ground-truth CUIs provided by the organizers, and the overall accuracy was considered as the evaluation metric. Table 2 shows the results (accuracy) of our systems: (1) GCNLP: the general clinical NLP system; (2) Hybrid1: the hybrid system combing multilevel matching with the ensemble-based ML ranking; (3) Hybrid2: the hybrid system combing multilevel matching with the attention-based ML ranking; (4) Meta1: a meta system used the Hybrid1 as the base but replaced some of the predictions (predictions with low confident scores or trigging disambiguation) with the GCNLP outputs; and (5) Meta2: a meta system similar to Meta1 by replacing some Hybrid2 outputs with the GCNLP outputs.
Table 2.
Overall systems’ performance
| Our Systems | Score |
|---|---|
| GCNLP (General Clinical NLP)a | 0.6974 |
| Hybrid1 (Matching with Ensemble-Ranking)a | 0.8009 |
| Hybrid2 (Matching with Attention-Ranking)b | 0.8116 |
| Meta1a | 0.8101 |
| Meta2b | 0.8209 |
GCNLP: general clinical natural language processing.
Systems submitted to n2c2 2019 for official evaluation.
Systems developed after n2c2 challenge.
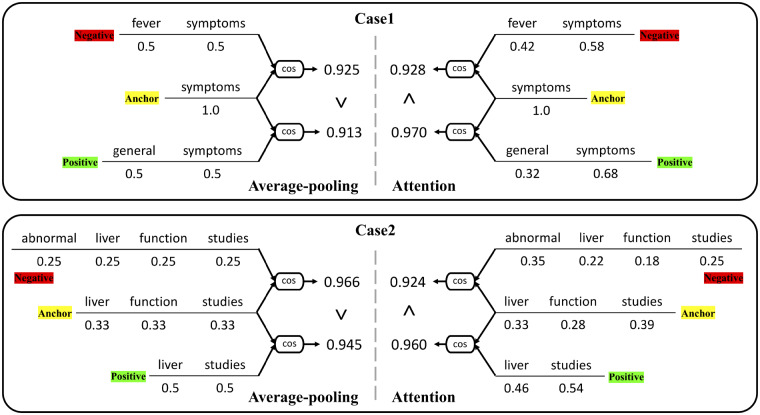
Among these systems, the GCNLP system, the Hybrid1 system, and the Meta1 system were developed during the 2019 n2c2 challenge time frame and submitted for official evaluation. The Hybrid2 and the Meta2 system were developed after the challenge. As shown in Table 2, our Hybrid1 and Meta1 system achieved high accuracies of 0.8009 and 0.8101, respectively, and both were within the scope of the top 10 teams’ best systems. And the Meta1 system won the fifth place among 33 teams in the challenge. Moreover, the Hybrid2 and Meta2 system, which replaced with the new ML ranking system based on Siamese attention, obtained even higher accuracy, of 0.8116 and 0.8209, respectively, indicating the importance of using attention in this task. Figure 4 provides 2 examples to demonstrate the effect of attention. For each case, a triplet of anchor, negative, and positive phrases are given with the weight of each word under either average-pooling or attention-based pooling algorithm. And then cosine similarity scores of anchor-negative, anchor-positive pairs are calculated for ranking. For average-pooling, every word is equally weighted. However, with attention, each word is weighted based on its task-specific semantic importance. In Case1 the attention layer helps the system to recognize that “fever” carries more important information than “general.” As a result, the system prefers the positive phrase with a higher semantic similarity. Similarly in Case2, “abnormal” is assigned with the highest weight by attention in the negative phrase, which plays a key role for the system to distinguish the negative and anchor phrases semantically.
Figure 4.
Examples to demonstrate the effect of attention on semantic similarity ranking.
The system performances regarding mention length (number of words) are established in Table 3. As shown in Table 3, in general, all the systems experienced performance drop with the increase of mention length. In addition, the performance of Hybrid1 and Hybrid2 systems were initially pretty close when the mention length equaled 1. However, with the increase of mention length, the Hybrid2 system with attention-based ML ranking remarkably outperformed the Hybrid1 system, with accuracy difference as high as 1.9%-5.5% when mention length equaled to 2-4. This result provides another aspect supporting our explanation of the advantages of using attention for semantic ranking. Table 4 also provides the system performances on each semantic type. Here, the “SignSymptom,” “Concept,” and “Medication” groups form the high-performance tier with accuracy over 85% for all the Hybrid or Meta systems, which may due to the high population of short mentions in these groups. Besides, almost all systems didn’t perform well on “Anatomy,” “Others,” and “CUI-less” groups with accuracy below 70%. Especially for “CUI-less,” the most of the correct predictions were contributed from the already labeled mention in training data. For an unknown mention, the systems were trying to assign a CUI in UMLS instead of “CUI-less.”
Table 3.
Systems’ performance on mention length with test dataset
| Number of words | Mention | GCNLP | Hybrid1 | Hybrid2 | Meta1 | Meta2 |
|---|---|---|---|---|---|---|
| 1 | 3840 | 0.7698 | 0.8586 | 0.8607 | 0.8711 | 0.8753 |
| 2 | 1778 | 0.6755 | 0.7570 | 0.7762 | 0.7559 | 0.7733 |
| 3 | 858 | 0.5175 | 0.7389 | 0.7576 | 0.7448 | 0.7634 |
| 4 | 272 | 0.5588 | 0.6581 | 0.7132 | 0.6875 | 0.7279 |
| 5 | 87 | 0.3793 | 0.4828 | 0.5172 | 0.4828 | 0.5172 |
| ≥6 | 90 | 0.4889 | 0.5333 | 0.5111 | 0.5889 | 0.5667 |
CUI: concept unique identifier; GCNLP: general clinical natural language processing.
Table 4.
Systems’ performance on semantic type with test dataset
| Semantic Type | Mention | GCNLP | Hybrid1 | Hybrid2 | Meta1 | Meta2 |
|---|---|---|---|---|---|---|
| DiseaseDisorder | 1218 | 0.7118 | 0.8177 | 0.8268 | 0.8333 | 0.8489 |
| Exam | 1121 | 0.7743 | 0.8359 | 0.8385 | 0.8475 | 0.8519 |
| Concept | 951 | 0.7466 | 0.8738 | 0.8707 | 0.8864 | 0.8864 |
| Medication | 857 | 0.8588 | 0.8670 | 0.8775 | 0.8646 | 0.8798 |
| Finding | 746 | 0.6005 | 0.7359 | 0.7507 | 0.7386 | 0.7547 |
| Procedure | 565 | 0.5504 | 0.6973 | 0.7133 | 0.7115 | 0.7274 |
| SignSymptom | 457 | 0.8468 | 0.9081 | 0.9190 | 0.9234 | 0.9234 |
| Activity | 305 | 0.7574 | 0.7902 | 0.8197 | 0.8098 | 0.8295 |
| Anatomy | 282 | 0.4468 | 0.6489 | 0.6844 | 0.6596 | 0.6950 |
| Others | 206 | 0.4951 | 0.6893 | 0.6748 | 0.6990 | 0.6845 |
| CUI-less | 217 | 0.2028 | 0.5300 | 0.5899 | 0.5023 | 0.5207 |
CUI: concept unique identifier; GCNLP: general clinical natural language processing.
Without any fine-tuning using the training data, the GCNLP system obtained the lowest performances on the overall test data as well as on almost all the subcategories as shown in Tables 2, 3, and 4. However, the GCNLP system has its own value and served as a good complement to our Hybrid systems. As shown in Table 2, both the Meta1 and Meta2 systems gained ∼1% accuracy enhancement comparing to Hybrid1 and Hybrid2 systems, respectively. The GCNLP system assisted the Hybrid systems in the following aspects. First, for the CUI candidates, the queries used in Hybrid systems required the synonyms of CUI candidates to contain either one or all word from the mention. Thus, mentions with spelling errors or lexical variants could end up with no CUI candidates or CUIs with low confident scores. Thus, the GCNLP with spelling check and greedy searching for all the CUI in UMLS served as an alternative solution. Second, in disambiguation, our Hybrid systems employed a simple disambiguation module without considering the context around the mention. In contrast, the GCNLP system used a more sophisticated disambiguation module considering context information, semantic type of CUIs and pretrained with a much larger dataset. These facts indicate the opportunities of achieving better performance by either combining the general clinical NLP and the hybrid systems or developing more sophisticated algorithms for CUI candidate generation and disambiguation.
CONCLUSION
In this study, we have described a hybrid clinical NLP system that can automatically normalize clinical mentions into concepts in standardized medical ontologies such as CUIs in UMLS. This hybrid system is based on a generic multilevel matching framework that integrates customizable matching components and ML ranking systems. We also demonstrated 2 simple ML ranking systems based on either ensemble of various similarity features extracted from pretrained encoders or a Siamese attention network, targeting at efficient and fast semantic searching and ranking. The evaluations of our systems with the data from 2019 n2c2 challenge on Clinical Concept Normalization task fully demonstrated the capability of our approaches. In addition, we also discussed the limitation of our current systems and the opportunities for achieving better performance by combining general clinical NLP systems. Besides, we believe our concept normalization approaches are designed in a generic manner with good transferability and interpretability, which can be applied to applications in other terminologies or domains such as International Classification of Diseases–Tenth Revision mapping, or entity linking in general NLP domain. Moreover, the ML ranking systems can be used independently for applications related to semantic searching and ranking.
AUTHOR CONTRIBUTIONS
LC devised the main idea for the work. LC designed the study, carried out the data collection, developed the systems, analyzed the results, and wrote the article. WF and YG assisted with study design, module development, and analysis. ZS, HL, EL, and LJ contributed to the NLP system design and implementation. YH and YG contributed significant edits. YH supervised this study. All the authors discussed the results and contributed to the final manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the 2019 n2c2 challenge organizers for organizing the challenge and providing the data used in training and testing. The authors also thank the participants of the 2019 n2c2 workshop for helpful discussions.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform 2009; 42 (5): 760–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Casey JA, Schwartz BS, Stewart WF, et al. Using electronic health records for population health research: a review of methods and applications. Annu Rev Public Health 2016; 37 (1): 61–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: a literature review. J Biomed Inform 2018; 77: 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review. J Biomed Inform 2017; 73: 14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Unified Medical Language System (UMLS). https://www.nlm.nih.gov/research/umls/ Accessed January 15, 2019.
- 6. N2C2: National NLP Clinical Challenges. https://n2c2.dbmi.hms.harvard.edu/ Accessed February 10, 2020.
- 7. Apache UIMA. https://uima.apache.org/ Accessed January 15, 2019.
- 8. Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004; 11 (5): 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010; 17 (3): 229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Soysal E, Wang J, Jiang M, et al. CLAMP—a toolkit for efficiently building customized clinical natural language processing pipelines. J Am Med Inform Assoc 2018; 25 (3): 331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luo Y-F, Sun W, Rumshisky A. A hybrid normalization method for medical concepts in clinical narrative using semantic matching. AMIA Jt Summits Transl Sci Proc 2019; 2019: 732–40. [PMC free article] [PubMed] [Google Scholar]
- 13. Kate RJ. Normalizing clinical terms using learned edit distance patterns. J Am Med Inform Assoc 2016; 23 (2): 380–6. [DOI] [PubMed] [Google Scholar]
- 14. Suominen H, Salanterä S, Velupillai S, et al. Overview of the ShARe/CLEF eHealth evaluation lab 2013 In: International Conference of the Cross-Language Evaluation Forum for European Languages. Berlin: Germany: Springer; 2013: 212–31. [Google Scholar]
- 15. Pradhan S, Elhadad N, Chapman W, et al. SemEval-2014 Task 7: Analysis of Clinical Text. In: proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014). Association for Computational Linguistics (ACL); 2015: 54–62.
- 16. Elhadad N, Pradhan S, Gorman S, et al. SemEval-2015 Task 14: Analysis of Clinical Text. In: proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015). Association for Computational Linguistics (ACL); 2015: 303–10.
- 17. Leaman R, Islamaj Dogan R, Lu Z. DNorm: Disease name normalization with pairwise learning to rank. Bioinformatics 2013; 29 (22): 2909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Wang J, Tang B. UTH_CCB: A report for SemEval 2014—Task 7 Analysis of Clinical Text. In: proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014). Association for Computational Linguistics (ACL); 2015: 802–6.
- 19. Ghiasvand O, Kate R. UWM: Disorder Mention Extraction from Clinical Text Using CRFs and Normalization Using Learned Edit Distance Patterns. In: proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015). Association for Computational Linguistics (ACL); 2015: 828–32.
- 20.D’Souza J, Ng V. Sieve-based entity linking for the biomedical domain. In: proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing; 2015: 297–302.
- 21. Li H, Chen Q, Tang B, et al. CNN-based ranking for biomedical entity normalization. BMC Bioinformatics 2017; 18 (S11): 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ji Z, Wei Q, Xu H. BERT-based Ranking for Biomedical Entity Normalization. arXiv: 1908.03548; 2019. [PMC free article] [PubMed]
- 23. Chiticariu L, Li Y, Reiss FR. Rule-based information extraction is dead! long live rule-based information extraction systems! In: proceedings of the 2013 Conference on Empirical Methods in Natural Language Processing; 2013: 827–32.
- 24. Luo YF, Sun W, Rumshisky A. MCN: a comprehensive corpus for medical concept normalization. J Biomed Inform 2019; 92: 103132. [DOI] [PubMed] [Google Scholar]
- 25. Spackman KA, Campbell KE, Côté RA. SNOMED RT: a reference terminology for health care. In: proceedings: a conference of the American Medical Informatics Association AMIA Fall Symposium; 1997: 640–4. [PMC free article] [PubMed]
- 26. Liu S, Ma W, Moore R, et al. RxNorm: Prescription for electronic drug information exchange. IT Prof 2005; 7: 17–23. [Google Scholar]
- 27. Apache Lucene. http://lucene.apache.org/ Accessed January 16, 2019.
- 28. Natural Language Toolkit—NLTK. https://www.nltk.org/ Accessed January 30, 2019.
- 29. List of medical abbreviations—Wikipedia. https://en.wikipedia.org/wiki/List_of_medical_abbreviations Accessed February 10, 2020.
- 30. Li Y, McLean D, Bandar ZA, et al. Sentence similarity based on semantic nets and corpus statistics. IEEE Trans Knowl Data Eng 2006; 18 (8): 1138–50. [Google Scholar]
- 31. Devlin J, Chang M-W, Lee K, et al. BERT: pretraining of deep bidirectional transformers for language understanding. arXiv: 1810.04805; 2019.
- 32. Lee J, Yoon W, Kim S, et al. BioBERT: a pretrained biomedical language representation model for biomedical text mining. Bioinformatics 2020; 36(4): 1234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schroff F, Kalenichenko D, Philbin J. FaceNet: A unified embedding for face recognition and clustering. In: proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. IEEE Computer Society; 2015: 815–23.
- 35. Zhou P, Shi W, Tian J, et al. Attention-based bidirectional long short-term memory networks for relation classification. In: proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers); 2016.
- 36. Chen L. Assertion detection in clinical natural language processing: a knowledge-poor machine learning approach. In: 2019 IEEE 2nd International Conference on Information and Computer Technologies, ICICT 2019. Institute of Electrical and Electronics Engineers; 2019: 37–40.
- 37. Chen L, Gu Y, Ji X, et al. Extracting medications and associated adverse drug events using a natural language processing system combining knowledge base and deep learning. J Am Med Inform Assoc 2020; 27 (1): 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Crawford M. Truth about computer-assisted coding: a consultant, him professional, and vendor weigh in on the real CAC impact. J AHIMA 2013; 84: 24–7. [PubMed] [Google Scholar]
- 39. Agirre E, Soroa A, Stevenson M. Graph-based word sense disambiguation of biomedical documents. Bioinformatics 2010; 26 (22): 2889–96. [DOI] [PubMed] [Google Scholar]
- 40. Melamud O, Levy O, Dagan I. A simple word embedding model for lexical substitution. In: proceedings of the 1st Workshop on Vector Space Modeling for Natural Language Processing. Stroudsburg, PA: Association for Computational Linguistics; 2015: 1–7.
- 41. Chen L, Gu Y, Ji X, et al. Clinical trial cohort selection based on multilevel rule-based natural language processing system. J Am Med Inform Assoc 2019; 26 (11): 1218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.