Abstract
Background
Although recent advances in immunotherapy have transformed the treatment landscape for many anatomically defined cancers, these therapies are currently not approved for patients diagnosed with cancer of unknown primary (CUP). Molecular cancer classification using gene expression profiling (GEP) assays has the potential to identify tumor type and putative primary cancers and thereby may allow consideration of immune checkpoint inhibitor (ICI) therapy options for a subset of patients with CUP. Herein, we evaluated and characterized the ability of a 92‐gene assay (CancerTYPE ID) to provide a molecular diagnosis and identify putative tumor types that are known to be sensitive to ICI therapies in patients with CUP or uncertain diagnosis.
Findings
A total of 24,426 cases from a large‐scale research database of 92‐gene assay clinical cases were classified, of which 9,350 (38%) were predicted to have an ICI‐eligible tumor type. All ICIs with approved indications as of March 2020 were included in the analysis. Non‐small cell lung cancer (NSCLC) was the most frequent molecular diagnosis and accounted for 33% of the ICI‐eligible tumor types identified and 13% of the overall reportable results. In addition to NSCLC, the assay also frequently identified urothelial carcinomas, gastric cancer, and head and neck squamous cell carcinoma. The distributions of identified tumor types with indications for ICI therapy were similar across age and gender.
Conclusions
Results suggest that molecular profiling with the 92‐gene assay identifies a subset of ICI‐eligible putative primary cancers in patients with CUP. We propose a treatment strategy based on available tests, including clinicopathologic features, GEP, and ICI biomarkers of response.
Short abstract
Regulatory approval of immune checkpoint inhibitors (ICI) is restricted to anatomically defined cancers with a known primary. This article reports cases submitted for 92‐gene assay testing with an unknown or uncertain diagnosis for which the subsequent post‐test report included a tumor type linked to an FDA‐approved ICI therapy, with the goal of identifying characteristics of cancers of unknown primary tumors that might benefit from immunotherapy.
Introduction
Immune checkpoint inhibitors (ICIs) have provided dramatic improvements in survival outcomes across diverse tumor types [1, 2]. However, despite its proven clinical utility in metastatic and in some cases early stage disease [3], the regulatory approval of ICI is currently restricted to anatomically defined cancer or cancers with a known primary.
Cancer of unknown primary (CUP) represents a heterogeneous group of cancers for which the anatomic site of origin or tumor type has not been identified. Uncertain cancers include those in which radiographic and pathologic determination suggests it is not CUP and further characterization is difficult. Unlike for patients with known cancers, immunotherapy is not approved for on‐label use in patients with CUP [4, 5, 6]. Instead, contemporary management of CUP most often remains limited to conventional cytotoxic chemotherapies that are selected according to the putative primary site as inferred by clinicopathologic data [7, 8]. With this approach, prognosis remains poor, with limited options beyond front line platinum‐based doublet therapy [9].
Innovative technologies such as gene expression profiling (GEP) can be used to augment clinicopathologic evaluations in cases of unknown, uncertain, or difficult to diagnose tumors, as well as poorly differentiated tumors and/or specimens with limited tissue [4]. The 92‐gene assay is a validated gene expression‐based assay that classifies 50 tumor types and subtypes using an algorithmic‐based comparison of a tumor's gene expression profile with a reference database of known tumor types encompassing 96% of cancers based on incidence [10, 11, 12]. Gene characteristics in the 92‐gene assay biomarker panel do not overlap with standard immunohistochemical biomarkers [13]. Unlike immunohistochemistry (IHC), the 92‐gene assay algorithm is based on examination of collective expression of 87 tumor‐related genes and 5 reference genes versus using a tissue‐specific approach. The 92‐gene assay has demonstrated improved diagnostic accuracy compared with IHC, particularly in poorly differentiated tumors [12], and identified the tumor type and primary site in 85%–90% of metastases [11]. Prospective outcome trials have shown mixed results, with one study reporting encouraging overall survival for site‐directed therapy for “treatment‐sensitive” tumors compared with empiric CUP treatment for these tumors [10, 14]. The increasing number of tumor types now known to be sensitive to ICI has expanded this treatment‐sensitive group and suggests that a significant number of patients with CUP might benefit from ICI treatment. Herein, we sought to identify the percentage and characteristics of CUP tumors that might benefit from immunotherapy by investigating cases submitted for 92‐gene assay testing with an unknown or uncertain diagnosis in which the subsequent post‐test report to the physician included a tumor type linked to a U.S. Food and Drug Administration (FDA)‐approved ICI therapy.
Materials and Methods
Patient Information and Analysis Plan
A database was created under an institutional review board‐approved protocol that integrated deidentified patient information (age at diagnosis and testing, gender, date of biopsy and assay) and 92‐gene assay (CancerTYPE ID, Biotheranostics, Inc., San Diego, CA) results where the assay had been ordered during routine care. At the time of analysis, the database contained 24,486 clinical cases with sufficient tissue for analysis and adequate RNA quality that had undergone 92‐gene assay testing between March 2010 and December 2016 (supplemental online Fig. 1). Biopsy sites were separated into 9 anatomic categories (abdomen, bone, brain, head and neck, liver, lung, lymph node, renal, and thoracic cavity). All ICIs with FDA‐approved indications of use as of 2020 were included in the analysis, with the exception of specific subsets such as triple‐negative breast cancer, microsatellite instability‐high (MSI‐H) colon cancer, and advanced endometrial carcinoma. Analyses included the proportional distributions of biopsy sites across tumor types identified by the 92‐gene assay with an ICI indication and distribution by patient gender and age at diagnosis. χ2 tests were used to determine the statistical significance of observed differences in the analyses of proportional distributions.
92‐Gene Assay
Gene expression analysis was performed on formalin‐fixed paraffin‐embedded tumor tissue as previously described [11]. Tumor cells were enriched by macrodissection or laser microdissection (LMD 6000, Leica Microsystems) and subsequently analyzed by real‐time reverse transcription polymerase chain reaction (RT‐PCR) using isolated total RNA. Tumor classification is based on the collective expression of 87 tumor‐related and 5 reference genes using a prespecified computational algorithm that generates probabilities for candidate tumor types based on the degree of similarity of the queried sample to a reference database of more than 2,000 tumor samples covering 50 tumor types and subtypes. Testing is performed in a Clinical Laboratory Improvement Amendments‐certified College of American Pathologists‐accredited laboratory and analyzed with a proprietary software that generates an automated test report of the percentage probability match of tumor type and subtype.
Results
Baseline characteristics of cases included in the analysis (n = 24,426) are provided in supplemental online Table 1. Among patients for whom the 92‐gene assay identified a tumor type, 38% (n = 9,350) were found to have a tumor type associated with an FDA‐approved ICI (Fig. 1). Non‐small cell lung cancer (lung adenocarcinoma and lung squamous; supplemental online Table 2) accounted for 33% of molecular diagnoses with an FDA‐approved ICI and 13% of the overall reportable results (Fig. 1A). Additionally, the assay frequently identified urothelial carcinomas (14% of ICI‐eligible tumors; 5.4% of the overall results), gastric cancer (14% of ICI‐eligible; 5.3% of the overall results), and head and neck squamous cell carcinoma (13% of ICI‐eligible, 4.8% of the overall results) in potential metastatic locations such as abdomen, bone, brain, liver, and lymph node (Fig. 1B). The distributions of identified tumor types with indications for ICI therapy were similar across age (<40 years, 30%; ≥40 years, 39%) and gender (female, 33%; male, 45%; supplemental online Table 1). Current FDA‐approved ICI and indications are displayed in online supplemental Table 3. A full list of tumor types and subtypes classified by the 92‐gene assay is provided in supplemental online Table 4.
Figure 1.
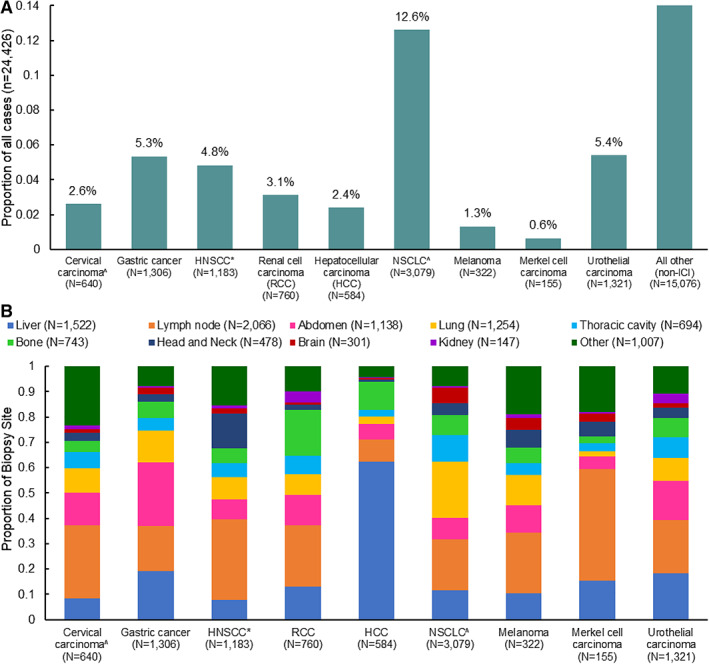
Tumor types identified by the 92‐gene assay. (A): Proportion of tumor types identified by the 92‐gene assay with U.S. Food and Drug Administration (FDA)‐approved ICIs. (B): Distribution of tumor types identified by the 92‐gene assay with an FDA‐approved ICI, across anatomic biopsy site categories. ^, Cervical carcinoma and NSCLC include squamous and adenocarcinoma histologies. *, This 92‐gene subtype includes skin squamous cell carcinoma. Abbreviations: HCC, hepatocellular carcinoma; ICI, immune checkpoint inhibitor; HNSCC, head and neck squamous cell carcinoma; NSCLC, non‐small cell lung cancer; RCC, renal cell carcinoma.
Discussion
Although they represent an attractive treatment option, ICIs are not currently indicated for CUP, except for rare occurrences [15]: patients with MSI‐H tumors; more recently, tumor mutational burden‐high (TMB‐H) tumors [16]; and off‐label use. Therefore, identification of ICI‐eligible CUP patients represents an important step toward improving treatment options and outcomes.
Efforts have been made to identify biomarkers of tumor responsiveness to ICI [17], and such biomarkers have the potential to inform the use of ICI in CUP [15, 18, 19]. The 92‐gene assay is based on collective GEP and is therefore distinct from DNA‐ and protein‐based methods such as next‐generation sequencing (NGS) and measurement of ICI‐related biomarkers such as MSI‐H, TMB‐H, and PD‐L1. The assay is supported for reimbursement to date primarily through Medicare and Medicare Advantage coverage [20]. The data presented here indicate that the 92‐gene assay identifies nearly 40% of patients with a CUP diagnosis who would be eligible for treatment with an FDA‐approved ICI, highlighting the potential utility of molecular cancer classification for such patients. The likelihood of benefit in this ICI‐eligible subset would depend on the cellular context plus additional biomarkers of response.
Figure 2 delineates the possible impact of available tests in CUP, including clinicopathologic data, GEP, and known biomarkers of ICI responsiveness (e.g., PD‐L1, TMB, and MSI‐H). Some of these overlap as well, and this influences the likelihood of benefit with ICI therapy. Extrapolating from known cancer data, we propose a treatment strategy that separates CUP patients into three categories based on the test results: 0%–5% ICI response, 10%–20% ICI response, and >30% ICI response. The MSI‐H plus TMB‐H category has the highest predicted response to ICI and is tissue agnostic, although only representing approximately 3% of cases, whereas most of the other test categories require cellular or tissue context for determining likelihood of response. Additionally, because our results indicate that a large subset (nearly 60%) of patients with a CUP diagnosis belong to a likely immune‐resistant phenotype, clinical trials of ICI alone in a nonstratified CUP sample are unlikely to yield promising results without the prior enrichment with a cancer classifier tool.
Figure 2.
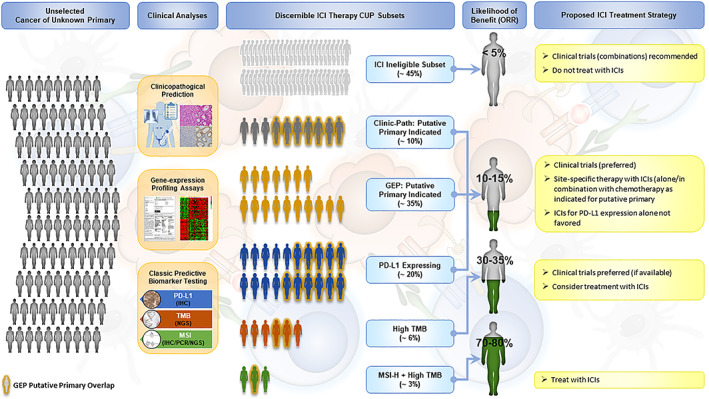
Clinical analysis and treatment of patients with CUP. The represented treatment strategy separates patients with CUP into three categories based on the test results: those with 0%–5% ICI response, those with 10%–20% ICI response, and those with >30% ICI response. GEP overlap with Clinic‐Path, PD‐L1 expressing, high TMB, and MSI‐H plus High TMB is indicated. Abbreviations: CUP, cancer of unknown primary; GEP, gene expression profiling; ICI, immune checkpoint inhibitor; MSI‐H, microsatellite instability‐high; ORR, overall response rate; PD‐L1, programmed death‐ligand 1; TMB, tumor mutational burden.
It is important to note that ICIs have a unique side‐effect profile of immune‐related adverse events compared with chemotherapeutic agents or targeted therapies. However, they are easier to tolerate because they lack the acute hematologic and gastrointestinal toxicity of many doublet chemotherapies used for CUP cancers and, therefore, have the potential to impact drug‐related quality of life.
Our study limitations include the retrospective nature of the data, the inability to acquire patient follow‐up to determine if tumor type identification allowed patients access to ICI therapy or a clinical trial with ICI, and difficulty incorporating comprehensive corroborative IHC data.
Conclusion
Development of a predictive model of ICI responsiveness that considers various tumor‐host interactions and tumor‐specific immunoregulation is under way, with cellular context and the identification of tumor type by GEP continuing to play important roles. In this study of real‐world patients with CUP who underwent GEP, the 92‐gene assay identified nearly 40% of cases from a wide array of anatomic biopsy sites as having a tumor type for which an FDA‐approved ICI wis available. Future prospective trials are needed to define the role of immunotherapy in selected CUP subsets. Additionally, we need to develop integrative and tissue‐sensitive algorithms to stratify patients with current testing (i.e., IHC + GEP + NGS) to push the therapeutic envelope for patients with CUP.
Disclosures
Graham M. Poage: Biotheranostics, Inc. (E, OI); Harris S. Soifer: Biotheranostics, Inc., BridgeBio, QED Therapeutics, Inc. (OI), Biotheranostics, Inc. (E); Catherine A. Schnabel: Biotheranostics Inc (E, OI). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Supplementary Figure 1 Methodology and case selection
Supplemental Table 1 Patient baseline characteristics.
Supplemental Table 2. 92‐gene assay tumor type and subtype(s) and corresponding ICI indications.
Supplementary Table 3. US FDA‐approved ICIs, biomarkers (PDL‐1, TMB and MSI) and indications.
Supplementary Table 4. Scope of 92‐gene assay tumor classification.
Acknowledgments
Data are available upon reasonable request. The data that support these findings are available upon request to Catherine A. Schnabel, Ph.D., at cathy.schnabel@biotheranostics.com.
Disclosures of potential conflicts of interest may be found at the end of this article.
References
- 1. Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015;27:450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vansteenkiste J, Wauters E, Reymen B et al. Current status of immune checkpoint inhibition in early‐stage NSCLC. Ann Oncol 2019;30:1244–1253. [DOI] [PubMed] [Google Scholar]
- 4. Economopoulou P, Mountzios G, Pavlidis N et al. Cancer of unknown primary origin in the genomic era: Elucidating the dark box of cancer. Cancer Treat Rev 2015;41:598–604. [DOI] [PubMed] [Google Scholar]
- 5. Hirsch FR, Scagliotti GV, Mulshine JL et al. Lung cancer: Current therapies and new targeted treatments. Lancet 2017;389:299–311. [DOI] [PubMed] [Google Scholar]
- 6. Schapira DV, Jarrett AR. The need to consider survival, outcome, and expense when evaluating and treating patients with unknown primary carcinoma. Arch Intern Med 1995;155:2050–2054. [PubMed] [Google Scholar]
- 7. Pavlidis N, Pentheroudakis G. Cancer of unknown primary site. Lancet 2012;379:1428–1435. [DOI] [PubMed] [Google Scholar]
- 8. Varadhachary GR, Raber MN. Cancer of unknown primary site. N Engl J Med 2014;371:757–765. [DOI] [PubMed] [Google Scholar]
- 9. Hemminki K, Riihimäki M, Sundquist K et al.Site‐specific survival rates for cancer of unknown primary according to location of metastases. Int J Cancer 2013;133:182–189. [DOI] [PubMed] [Google Scholar]
- 10. Hainsworth JD, Rubin MS, Spigel DR et al. Molecular gene expression profiling to predict the tissue of origin and direct site‐specific therapy in patients with carcinoma of unknown primary site: A prospective trial of the Sarah cannon research institute. J Clin Oncol 2013;31:217–223. [DOI] [PubMed] [Google Scholar]
- 11. Kerr SE, Schnabel CA, Sullivan PS et al. Multisite validation study to determine performance characteristics of a 92‐gene molecular cancer classifier. Clin Cancer Res 2012;18:3952–3960. [DOI] [PubMed] [Google Scholar]
- 12. Weiss LM, Chu P, Schroeder BE et al. Blinded comparator study of immunohistochemical analysis versus a 92‐gene cancer classifier in the diagnosis of the primary site in metastatic tumors. J Mol Diagn 2013;15:263–269. [DOI] [PubMed] [Google Scholar]
- 13. Ma XJ, Patel R, Wang X et al. Molecular classification of human cancers using a 92‐gene real‐time quantitative polymerase chain reaction assay. Arch Pathol Lab Med 2006;130:465–473. [DOI] [PubMed] [Google Scholar]
- 14. Fizazi K, Maillard A, Penel N et al. A phase III trial of empiric chemotherapy with cisplatin and gemcitabine or systemic treatment tailored by molecular gene expression analysis in patients with carcinomas of an unknown primary (CUP) site (GEFCAPI 04). Ann Oncol 2019;30(suppl 5):v851. [Google Scholar]
- 15. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merino DM, McShane LM, Fabrizio D et al; TMB Harmonization Consortium. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: Phase I of the Friends of Cancer Research TMB Harmonization Project. J Immunother Cancer 2020;8:e000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arora S, Velichinskii R, Lesh RW et al. Existing and emerging biomarkers for immune checkpoint immunotherapy in solid tumors. Adv Ther 2019;36:2638–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gatalica Z, Xiu J, Swensen J et al. Comprehensive analysis of cancers of unknown primary for the biomarkers of response to immune checkpoint blockade therapy. Eur J Cancer 2018;94:179–186 [DOI] [PubMed] [Google Scholar]
- 19. Gröschel S, Bommer M, Hutter B et al. Integration of genomics and histology revises diagnosis and enables effective therapy of refractory cancer of unknown primary with PDL1 amplification. Cold Spring Harb Mol Case Stud 2016;2:a001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Local coverage determination for MolDX: Breast cancer indexTM (BCI) gene expression test (L37822). CMS.gov. https://www.cms.gov/medicare‐coverage‐database/details/lcd‐details.aspx?LCDId=37822&ContrId=360&ver=7&ContrVer=1&DocID=DL37822&bc=gAAAAAgAgAAA&. Accessed August 4, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Supplementary Figure 1 Methodology and case selection
Supplemental Table 1 Patient baseline characteristics.
Supplemental Table 2. 92‐gene assay tumor type and subtype(s) and corresponding ICI indications.
Supplementary Table 3. US FDA‐approved ICIs, biomarkers (PDL‐1, TMB and MSI) and indications.
Supplementary Table 4. Scope of 92‐gene assay tumor classification.


