Abstract
Dysregulation of cellular ribose uptake can be indicative of metabolic abnormalities or tumorigenesis. However, analytical methods are currently limited for quantifying ribose concentration in complex biological samples. Here, we utilize the highly-specific recognition of ribose by ribose binding protein (RBP) to develop a single-protein ribose sensor detectable via a sensitive NMR technique known as hyperpolarized (hp) 129Xe chemical exchange saturation transfer (hyper-CEST). We demonstrate that RBP, with a tunable ribose binding site and further engineered to bind xenon, enables the quantitation of ribose over a wide concentration range (nM-mM). Ribose binding induces the RBP ‘closed’ conformation, which slows Xe exchange to a rate detectable by hyper-CEST. Such detection is remarkably specific for ribose, with minimal background signal from endogenous sugars of similar size and structure, e.g., glucose or ribose-6-phosphate. Ribose concentration was measured for mammalian cell lysate and serum, which led to estimates of low-mM ribose in a HeLa cell line. This highlights the potential for using genetically encoded periplasmic binding proteins such as RBP to measure metabolites in different biological fluids, tissues, and physiologic states.
Analytical methods for quantifying metabolites in vivo are critical to the diagnosis and assessment of human disease. Data from the Human Metabolome Database (HMDB) provide a lower limit of 150 000 human metabolites, of which only 1–2% can be identified via current profiling methods.1 Metabolomics research is being driven by advances in nuclear magnetic resonance spectroscopy (MRS) and mass spectrometry (MS).2,3 Notably, MRS analysis of metabolites is rapid, reproducible, automatable, nondestructive, and quantifiable.4 MR-based metabolomics methods have been applied in the analysis of amino acids, nucleotides and nucleosides, carbohydrates, peptides, and vitamins; and have been used to study metabolic profiles of Alzheimer’s disease, multiple sclerosis, prostate cancer, and colorectal cancer.1,5,6 However, the accurate and sensitive detection of numerous medically relevant small-molecule metabolites, both in vitro and in vivo, remains challenging.
Ribose (specifically d-ribose) is an abundant metabolite (~100 μM in human fasting serum),7 yet surprisingly little is known regarding its biodistribution and role in human disease.8 Ribose is a structural component of myriad biomolecules such as DNA, RNA and ATP, and ribose-5-phosphate is a key intermediate in the pentose phosphate pathway. Mammalian cells uptake extracellular ribose through a process known as “ribose salvage”, whereby ribose is imported via transporters such as GLUT2 and phosphorylated by ribokinase (RBKS) to produce ribose-5-phosphate.9–11 While the administration of exogenous ribose has been recognized as a potential treatment of ischemic cardiovascular disease,12 the underlying process of ribose salvage and metabolism is poorly understood. It is suspected that increased rates of ribose salvage are associated with higher rates of glucose and lipid metabolism, and it has been proposed that different cell types, including cancer cells, utilize ribose salvage to provide precursors for distinct cellular pathways.11,13 A recent PET study conducted by Clark and coworkers characterized the metabolism of exogenous ribose in vivo and reported greatly elevated levels of ribose salvage in the liver.11 Moreover, it was discovered that the dysregulation of ribose salvage in hepatic cells is associated with metabolic syndrome. Whereas baseline glucose levels have been well established, and glucose dysregulation is a known hallmark of several human metabolic disorders, relatively little is known regarding the range of ribose levels in healthy and disease states. Thus, there is a clear need for reliable MRS methods for quantifying free ribose at nM to mM concentrations. Here, we utilize the highly-specific ribose binding protein (RBP) to develop a single-protein ribose sensor that exploits the sensitivity of hyperpolarized (hp) 129Xe NMR spectroscopy.
Contrast agents detectable by 1H MRS and magnetic resonance imaging (1H MRI) enable the detection of metabolites in specific tissues at unlimited depth, without the use of ionizing radiation.14–17 To date, only fluorescent protein sensors18 and positron-emission tomography (PET) probes11 have been developed for in situ detection of ribose. The use of hp 129Xe MR overcomes many of the limitations in 1H MR detection sensitivity and chemical shift dispersion. Hp 129Xe is produced via spin-exchange optical pumping,19 which increases the overall magnetization of the 129Xe population by several orders of magnitude and greatly enhances the sensitivity of the MRS/MRI measurement.20–23 Atomic 129Xe has a large electron cloud (r = 2.15 Å), and its perturbation produces a well-dispersed chemical shift window for the spin-½ nucleus. The Xe atom is soluble in water (~5 mM/atm at rt), but it binds preferentially via dispersion forces to hydrophobic cavities of comparable size. A small number of proteins have been shown by X-ray crystallography and 129Xe NMR spectroscopy to bind xenon at interior sites.24–28 These low-affinity sites can be studied via a technique known as hp 129Xe chemical exchange saturation transfer (hyper-CEST).29 Using a radiofrequency (rf) saturation pulse specific to host-bound hyperpolarized 129Xe, magnetization is transferred to the 129Xe population in bulk solution, where loss of polarization can be readily observed, allowing for the indirect detection of the host-bound 129Xe pool. This technique has been used by our laboratory and others to analyze such systems as bacterial spores,30 tumor cells,31 and small molecule hosts.32–38 Protein-based hyper-CEST contrast agents have previously been developed for the sensitive detection of biological structures.39–41 The monomeric proteins TEM-1 β-lactamase (Bla)42–44 and maltose-binding protein (MBP)45 were previously identified by our laboratory as contrast agents for 129Xe hyper-CEST.
RBP and MBP belong to a family of bacterial proteins known as periplasmic binding proteins (PBPs), which undergo a significant conformational change upon binding to their respective ligands.46–49 The diverse ligand profile of the PBP family has allowed for the development of PBP-based sensors for targets such as amino acids,50–53 sugars,18,50,54–57 metal ions,50,58 anions,59–61 and phosphonates.62,63 PBPs offer opportunities to study metabolomics in vivo,64 and have been used to analyze the dynamic cellular metabolism of maltose,65 glucose,66 and ribose18 by fluorescence microscopy. Strategies for expanding the repertoire of analytes detectable by PBP-based biosensors include in silico design,67–69 metabolome panning,70 and directed evolution.71 However, the reliance of many PBP biosensors on optical detection methods limits their use in many biological fluids as well as in larger, opaque organisms.
Previously, we showed that the ‘closed’ conformation that MBP adopts upon maltose binding is required for generating hyper-CEST contrast, making MBP an ultrasensitive “smart” contrast agent for maltose detection.45 We sought to extend these studies to other PBPs as potential 129Xe MR contrast agents. Here, our laboratory investigated ribose-binding protein (RBP) as a hyper-CEST contrast agent for the detection and quantitation of ribose in biological fluids (Scheme 1). Prior work by Lowery and colleagues reported that a leucine-to-alanine mutation at residue 19 of RBP created a binding site for Xe (Figure 1), which was verified via 15N HSQC spectroscopy.72 We hypothesized that RBP(L19A) should generate 129Xe hyper-CEST signal that varies with ribose concentration.
Scheme 1.
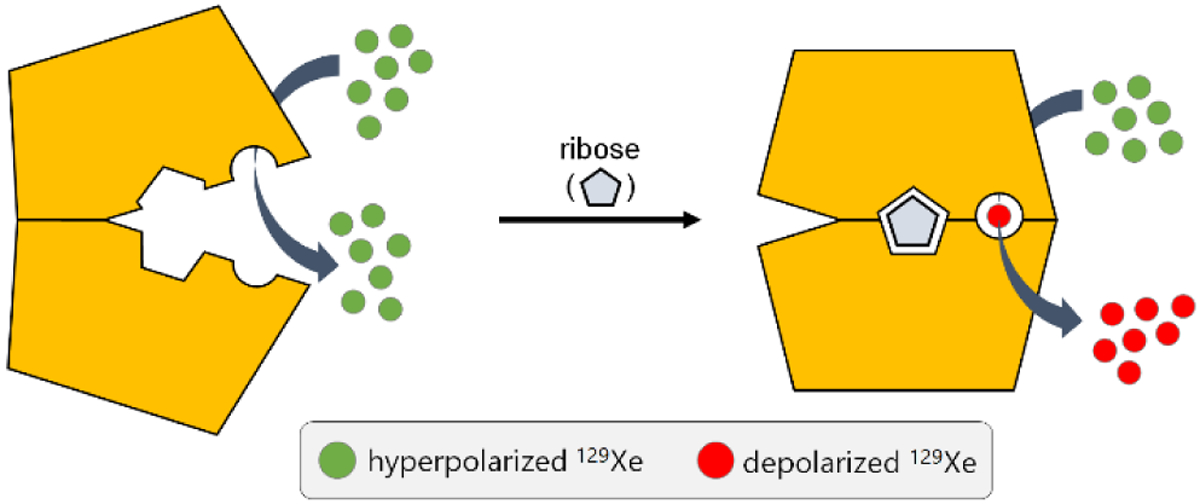
RBP (yellow) transitions from its “open” to “closed” conformation upon binding ribose. This conformational change allows for the binding and radiofrequency-selective depolarization of hp 129Xe, generating MR contrast.
Figure 1.
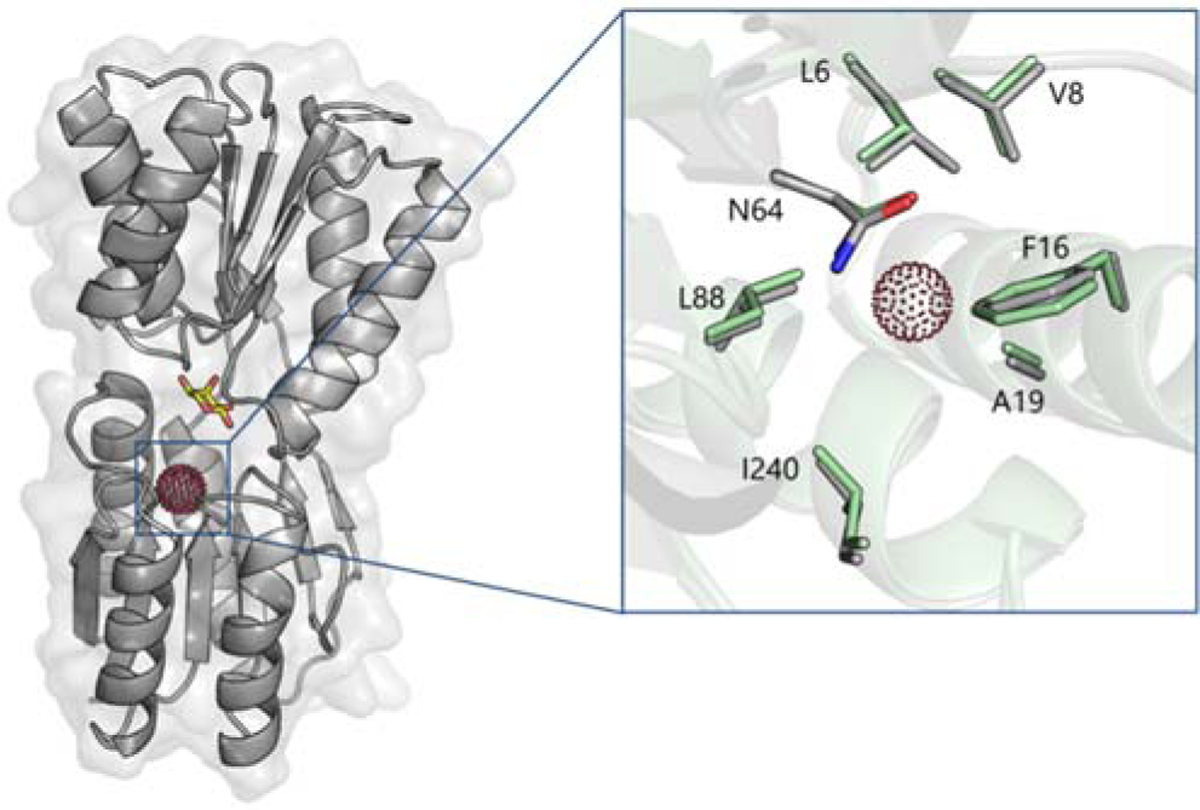
Proposed Xe binding site in RBP(L19A). The protein model is based on the crystal structure of ribose-bound RBP in its closed conformation (PDB ID 2DRI). Xe (red dots) was modeled at the center of the cavity created by the L19A mutation. Bound ribose shown as yellow sticks. (Inset) Close-up view of the Xe binding site of RBP(L19A) in its closed (gray) and open (green; PDB ID 1URP) conformations.
EXPERIMENTAL SECTION
Plasmid preparation.
The codon-optimized gene for ribose binding protein (RBP) from Escherichia coli (UniProt accession no. P02925) incorporating a L19A mutation was synthesized and cloned into a pJ411 vector by ATUM. The RBP(L19A) gene was amplified using the primers listed in Table S1 and cloned into a pET-His-GFP-TEV LIC cloning vector, a gift from Scott Gradia acquired via Addgene (plasmid #29663), via ligation-independent cloning. The resulting GFP-RBP(L19A) gene was sequenced at the University of Pennsylvania DNA Sequencing Facility to verify the integrity of the fusion construct.
Site-directed mutagenesis.
Mutations were introduced to the GFP-RBP(L19A) gene via site-directed mutagenesis using the forward and reverse primers listed in Table S2. The mutated plasmids were amplified in NEB-5α competent E. coli cells and purified using a miniprep kit (New England Biolabs). The mutated plasmids were sequenced to verify the incorporation of the desired mutation and integrity of the gene.
Protein expression and purification.
The GFP-RBP(L19A) plasmid was transformed into BL21(DE3) competent E. coli cells (New England Biolabs). The cells were grown in 4 × 1 L of LB Miller broth supplemented with 50 μg/mL kanamycin to a final OD600 of roughly 0.8, at which point the cells were induced with 1 mM IPTG. The cells were incubated overnight at 25 °C, pelleted by centrifugation, and frozen at −80 °C.
Frozen cells were resuspended in 20 mM sodium phosphate (pH 7.4) to a total volume of 80 mL, lysed with lysozyme (Sigma), and treated with benzonase nuclease (Sigma) to reduce the viscosity of the lysate. After stirring the lysate at rt for 30 min, NaCl (0.5 M) and imidazole (20 mM) were added. The lysate was clarified by centrifugation, and supernatant was loaded onto a 5-mL HisTrap nickel affinity column (GE Life Sciences) pre-equilibrated with 20 mM sodium phosphate (pH 7.4), 0.5 M NaCl, 20 mM imidazole. GFP-RBP(L19A) bound to the column was unfolded with 20 column volumes (100 mL) of 20 mM sodium phosphate (pH 7.4), 0.5 M NaCl, 20 mM imidazole, 8 M urea to remove any endogenous ribose bound to the protein. GFP-RBP(L19A) was refolded on-column via a 12-column volume (60 mL) gradient to 20 mM sodium phosphate (pH 7.4), 0.5 M NaCl, 20 mM imidazole, and then eluted with 20 mM sodium phosphate (pH 7.4), 0.5 M NaCl, 500 mM imidazole. The eluate was concentrated and further purified by size-exclusion chromatography in PBS (HyClone) using a HiLoad 16/600 Superdex column (GE Life Sciences). Fractions containing pure protein (over 95% as indicated by SDS-PAGE, Figure S1) were pooled and concentrated to ~1 mL. Protein concentration was determined from the absorbance at 280 nm (ε280 = 27 850 M−1 cm−1). Protein structure and ribose binding were confirmed using CD and ITC, respectively. The same procedure was carried out for the expression and purification of RBP mutants.
Isothermal titration calorimetry (ITC).
ITC experiments were performed at 298 K on a GE Healthcare MicroCal™ iTC200 instrument. GFP-RBP(L19A) was prepared at 30 μM in PBS. Ribose was prepared at 10x the protein concentration in PBS. The sample cell was filled with 300 μL of protein solution, and reference cell contained deionized water. Calorimetric data were analyzed by performing nonlinear regression fitting to the binding isotherms using ORIGIN software. Enthalpograms are shown in Figure S2.
Circular dichroism (CD) spectroscopy.
CD spectra of all RBP variants were measured on a Jasco J-1500 CD spectrometer equipped with a Peltier temperature controller (Figure S3). Spectra were acquired from 5 μM protein in 10 mM sodium phosphate (pH 8.0) buffer inside a quartz cuvette with a 1 mm path length. CD spectra were taken at 20 °C with a wavelength step of 1 nm.
Cell growth in serum-free medium.
HeLa cell stocks (ATCC; passage #8) frozen in liquid nitrogen were thawed and grown in suspension at 37 °C in serum-free medium (Sigma 14591C) supplemented with L-glutamine (Sigma 59202C) to a final concentration of 6 mM in a T-25 culture flask in a humidified incubator with 5% CO2. Media was changed every 3 days. Cell growth was monitored by hemocytometer. After cell density reached 1.8 × 106 cells/mL, the cells were split into two T-75 flasks. After cell density reached 0.9 × 106 cells/mL, the cells were split into three T-75 flasks. After 3 additional days, the cells were harvested, resuspended in PBS, counted using a hemocytometer, and lysed using five freeze-thaw cycles from −80 °C to 4 °C. Total cell lysis was confirmed via hemocytometer.
Cell growth in FBS-supplemented medium.
HeLa cell stocks (ATCC; passage #6) frozen in liquid nitrogen were thawed and grown at 37 °C in DMEM media supplemented with fetal bovine serum (FBS) and penicillin/streptomycin in a T-75 culture flask in a humidified incubator with 5% CO2. After reaching about 80–90% confluency, the cells were split into 6 flasks and regrown until once again reaching 80–90% confluency. At this point, the cells were harvested, resuspended in PBS, counted using a hemocytometer, and lysed using five freeze-thaw cycles from −80 °C to 4 °C. Total cell lysis was confirmed via hemocytometer. Immediately after thawing, the lysate was refrozen at −80 °C until ready for use in order to prevent enzymatic activity from affecting the free ribose concentration.
129Xe NMR hyper-CEST frequency scans.
Hyper-CEST samples were prepared in PBS using RBP and lysate at indicated concentrations. Hyperpolarized (hp) 129Xe was generated using the spin-exchange optical pumping (SEOP) method19 with a home-built 129Xe polarizer based on the IGI.Xe.2000 commercial model by GE. A Shark 65 W tunable ultra-narrow band diode laser (OptiGrate) set to 795 nm was used for optical pumping of Rb vapor. A gas mixture of 88% helium, 10% nitrogen, and 2% natural abundance xenon (Linde Group, NJ) was used as the hyperpolarizer input. 129Xe hyperpolarization level was roughly 10–15%. To determine the magnitude and frequency of the CEST effect for a given sample, shaped saturation pulses were scanned across a specific chemical shift range, and the normalized integral of the resulting 129Xe(aq) signal was plotted as a function of saturation frequency, generating what is known as a z-spectrum. For each data point in the hyper-CEST z-spectra, hp 129Xe was bubbled into a 10-mm NMR tube containing 2.5 mL of sample through capillaries for 20 s, followed by a 3 s delay allowing bubbles to collapse. After this, a d-SNOB saturation pulse with 690 Hz bandwidth was used for FID acquisition. Pulse length tpulse = 3.80 ms, field strength B1, max = 77 μT, number of pulses npulse = 600, total saturation time Tsat = 2.29 s. The gas pressure downfield of the inlet valve to the NMR tube was ca. 63 psi and the gas flow was controlled at a rate of ca. 0.70 standard liters per minute. NMR experiments were performed using a Bruker BioDRX 500 MHz NMR spectrometer and a 10-mm PABBO probe at 300 K. A 90° hard pulse of this probe has a pulse length of 40.6 μs. For all experiments, 0.1% (v/v) Pluronic L81 (Aldrich) was added to mitigate foaming.
129Xe NMR hyper-CEST depolarization curves.
To assess the detection sensitivity of GFP-RBP, time-dependent saturation transfer experiments were performed by measuring 129Xe(aq) polarization as a function of saturation time. Shaped saturation pulses were applied at the chemical shift of 129Xe@RBPclosed, and the residual aqueous 129Xe signal after saturation transfer was measured as an on-resonance CEST response. Saturation frequencies of Dsnob-shaped pulses were positioned at +42.5 and −42.5 ppm, referenced to the 129Xe(aq) peak, for on- and off-resonance, respectively. The pulse length was 1.727 ms, and the field strength was 170 μT. The normalized difference between on- and off-resonance signals was represented by the saturation contrast.
Quantitation of ribose in HeLa cell lysate by LC-MS.
HeLa cell lysate was prepared as described in the Cell growth in serum-free medium section, with the only exception being that cells were resuspended in ddH2O instead of PBS buffer. To remove proteins, cell lysate was extracted with methanol following a previously published procedure.73 The extracted metabolites were resuspended in a solution of 1:2 ddH2O:MeOH v/v to a final concentration of 2 million/mL lysed cells. Uniformly labeled 13C d-ribose ([U-13C5] ribose, Cambridge Isotope Labs) was spiked into cell lysate solutions pre- and post-extraction as an internal standard to a final concentration of 20 μM. A standard series was generated over a concentration range of 3.13 to 100 μM ribose with 20 μM [U-13C5] ribose internal standard.
LC-MS analysis was performed in positive and negative ion modes using a Thermo Scientific Vanquish Horizon UHPLC system with an Imtakt Unison UK-Amino column and a Thermo Scientific Q Exactive HF-X mass spectrometer. LC solvents were 10 mM ammonium acetate (solvent A) and acetonitrile (solvent B), and separation used a gradient of 91% solvent B for 7 min, 91 to 79% solvent B over 23 min, 20% solvent B for 5 min, and 91% solvent B for 10 min. Full MS scans were acquired at 120k resolution with a scan range of 65–975 m/z. All samples were injected twice.
MS data were analyzed using TraceFinder 4.1 from Thermo Scientific. Ribose and [U-13C5] ribose were quantified as acetate adducts at 209.06665 and 214.08345 m/z, respectively. A calibration curve was generated from the standard concentration series with linear-fit and 1/x2 weighting.
RESULTS AND DISCUSSION
129Xe hyper-CEST NMR with GFP-RBP(L19A) in PBS.
The GFP-RBP(L19A) construct was expressed in BL21(DE3) E. coli, purified by column chromatography, and characterized by SDS-PAGE (Figure S1), ITC (Figure S2) and CD spectroscopy (Figure S3). These methods confirmed the identity and purity of RBP, as well as its ribose-binding function.
A series of 129Xe hyper-CEST z-spectra was acquired for GFP-RBP(L19A) (Figure S4). Saturation pulses were scanned over the chemical shift range of 93 to 358 ppm in 5 ppm steps, and the 129Xe(aq) signal was measured as a function of saturation pulse offset. GFP-RBP(L19A) in its apo form showed only a single saturation response corresponding to free 129Xe in aqueous solution centered at 193 ppm. To assess the magnitude and specificity of contrast response of GFP-RBP(L19A) to metabolite binding, 1 mM ribose, 1 mM glucose, and 1 mM ribose-5-phosphate were individually added to the protein. Notably, the intracellular concentration of phosphorylated pentose is ca. 1.3 mM in E. coli.74 Upon adding 1 mM ribose, the z-spectrum of GFP-RBP(L19A) showed a second pronounced response at 233 ppm, determined via Lorentzian line fitting, 40 ppm downfield of the 129Xe(aq) peak. Notably, the addition of 1 mM glucose or ribose-5-phosphate to GFP-RBP(L19A) produced only weak contrast at 233 ppm (Figure S4). This corroborated the findings of Lager and coworkers using an RBP-based FRET sensor, which showed poor affinity for hexoses, and only appreciable conformational change induced by physiologic ribose, of several common pentoses tested.18
To quantify the relationship between ribose concentration and the 129Xe(aq) post-saturation signal at 233 ppm, a series of z-spectra of GFP-RBP(L19A) was acquired in PBS at ribose concentrations corresponding to specific percentages of GFP-RBP(L19A) in the closed conformation (Figure 2). The percentage of protein in the closed conformation was calculated using the dissociation constant for ribose binding to GFP-RBP(L19A) experimentally measured by isothermal titration calorimetry (ITC), Kd = 0.3 ± 0.1 μM (Figure S2). The magnitude of the CEST effect at 233 ppm was then plotted against the percentage of ‘closed’ GFP-RBP(L19A) (Figure 2 inset). The strong linear relationship indicates that the magnitude of the GFP-RBP(L19A) CEST effect can be used to determine the percentage of protein in the closed conformation, and consequently the ribose concentration.
Figure 2.
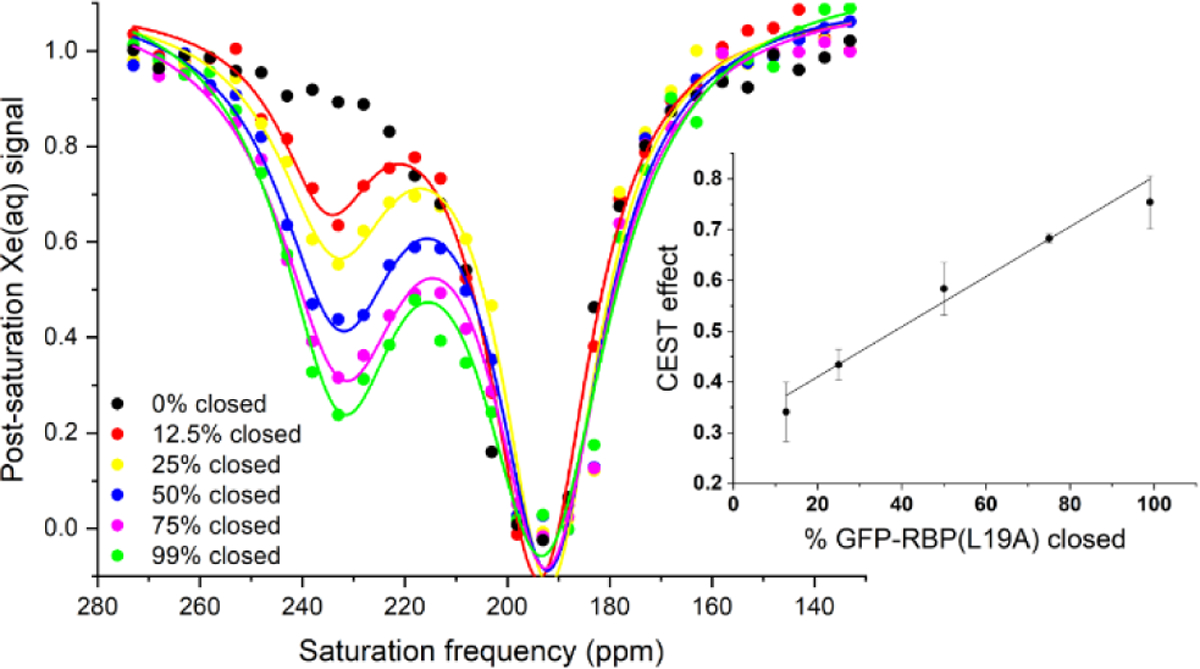
Hyper-CEST z-spectra of GFP-RBP(L19A) in pH 7.2 PBS buffer (circles). Spectra were acquired with [ribose] = 0, 2.5, 5.1, 10.3, 15.9, 49.5 μM to achieve the corresponding % RBP closed, according to the measured Kd. All z-spectra were obtained with 20 μM protein at 300 K. Pulse length, τpulse = 3.80 ms; field strength, B1,max = 77 μT. Data shown as an average of 3 trials. Inset: Magnitude of the CEST effect from each z-spectrum plotted against the percentage of closed RBP. R2 = 0.982.
129Xe hyper-CEST NMR with GFP-RBP (L19A) in HeLa cell lysate.
We then investigated whether GFP-RBP(L19A) is capable of quantifying ribose concentration in a cellular environment. In cellulo detection of ribose in HeLa cells expressing GFP-RBP(L19A) was unfeasible due to the partial and inconsistent lysis of cells caused by Xe bubbling during hyper-CEST. For more consistent results, HeLa cells were grown in a serum-free medium, washed and resuspended in PBS, counted by hemocytometer, and then lysed prior to the hyper-CEST measurement. Cell growth in serum-free conditions was important to verify that no exogenous ribose was complicating the CEST measurement. FBS, which is normally added to cell growth media, showed a significant CEST peak at 233 ppm in the presence of GFP-RBP(L19A) corresponding to 30 μM ribose, whereas the serum-free medium revealed no such peak (Figure S5). To increase the robustness of the RBP-CEST assay, we treated cell lysate with methanol to precipitate macromolecules (e.g., endogenous proteins) that could potentially contribute to a CEST response.73 To estimate the ribose extraction efficiency, we prepared two test samples of cell lysate: one spiked with 20 μM [U-13C5] ribose before extraction, and one spiked with 20 μM [U-13C5] ribose after extraction. LC-MS analysis revealed that the peak area of the former was ca. 0.46-fold that of the latter (Figure S6), corresponding to a ribose extraction efficiency of 46%.
Extracted HeLa cell lysate in the absence of GFP-RBP(L19A) showed a CEST effect comparable to that of apo-protein when rf pulses were applied at 233 ppm. Upon addition of 20 μM GFP-RBP(L19A), z-spectra showed that increasing concentrations of cell lysate gave increased hyper-CEST contrast (Figure 3). At 1 million cells/mL, the CEST effect was measured to be 0.56 ± 0.04, while at 2 million cells/mL, the CEST effect was measured to be 0.71 ± 0.05. Plotted against the standard curve shown in the inset of Figure 2, these values correspond to 50 ± 7% and 82 ± 10% GFP-RBP(L19A) in its closed conformation, with lysate-ribose concentrations of 22 ± 3 μM and 40 ± 8 μM, respectively, corrected for extraction efficiency. Within error, these ribose concentrations have a ratio of 1:2, consistent with the ratio of the cell lysate concentrations.
Figure 3.
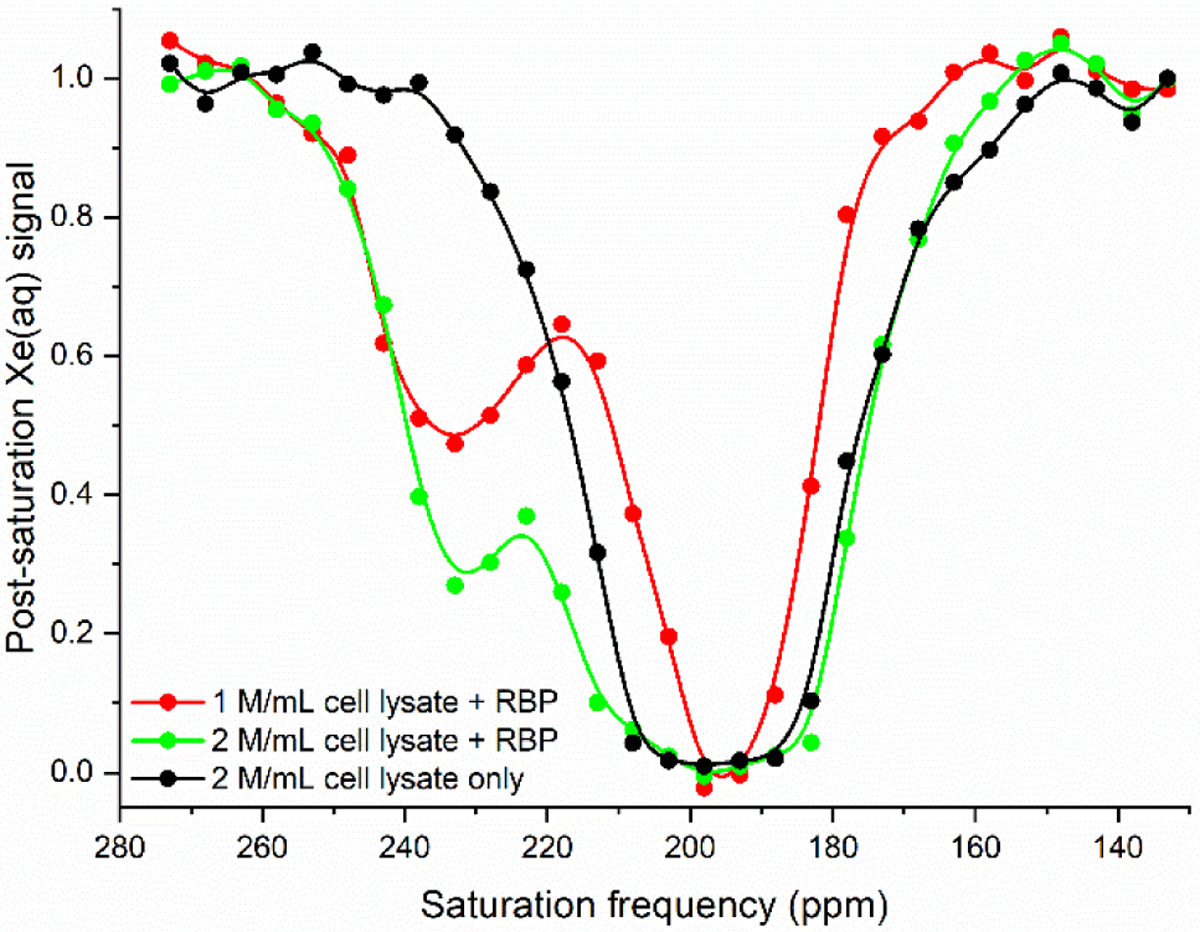
Hyper-CEST z-spectra of 20 μM GFP-RBP(L19A) with extracted lysate from HeLa cells grown in serum-free conditions. The z-spectrum of 2 million/mL cell lysate without RBP is shown for reference. All z-spectra were obtained at 300 K. Pulse length, τpulse = 3.80 ms; field strength, B1,max = 77 μT. Data shown as an average of 3 trials.
The lysate-ribose measurements allowed for estimation of the initial intracellular ribose concentration. Due to the spherical shape of HeLa cells in suspension, it is possible to estimate an average cell volume of 4000 μm3 from the average radius of the cell, 10 μm.75 This results in an average intracellular ribose concentration of 5 ± 1 mM, which is markedly higher than a previous unpublished estimate of < 10 μM.76 To our knowledge, this is the first report of a mammalian intracellular ribose concentration.
This method was also used to quantify ribose concentration in HeLa cells grown in a medium supplemented with FBS (Figure S7), which, as noted above, contains exogenous ribose. Using the standard curve shown in the Figure 2 inset, it was determined that a solution of 0.5 million cells/mL contained 10 ± 1 μM ribose, and a solution of 2 million cells/mL contained 42 ± 7 μM ribose, corrected for extraction efficiency. While these values are comparable to those measured with cells grown in a serum-free medium, the cells in the two cultures have different morphologies. HeLa cells grown in a FBS-supplemented medium have a half-sphere shape, and an average volume of 2600 μm3.77 Given this reduced cellular volume, the initial intracellular ribose concentration for FBS-supplemented cells was estimated to be 8 ± 1 mM. The relevant values determined for solutions of GFP-RBP(L19A) in the presence of cell lysate via hyper-CEST NMR are summarized in Table 1.
Table 1.
Summary of HeLa cell lysate measurements
| Serum-free | FBS-supplemented | |||
|---|---|---|---|---|
| [cell lysate] | 1 million/mL | 2 million/mL | 0.5 million/mL | 2 million/mL |
| CEST effect | 0.56 ± 0.04 | 0.71 ± 0.05 | 0.43 ± 0.01 | 0.73 ± 0.05 |
| % RBP closed | 50 ± 7 | 82 ± 10 | 24 ± 1 | 85 ± 9 |
| [ribose in solution] (μM) | 22 ± 3 | 40 ± 8 | 10 ± 1 | 42 ± 7 |
| [intracellular ribose] (mM) | 6 ± 1 | 5 ± 1 | 8 ± 1 | 8 ± 1 |
Quantitation of ribose in HeLa cell lysate by LC-MS.
To support our results obtained from the 129Xe NMR method, we analyzed ribose levels in HeLa cell lysate by LC-MS. A calibration curve was first generated for a concentration range of 3.13 to 100 μM ribose, using 20 μM [U-13C5] ribose as an internal standard (R2 = 0.995, Figure S8 and Table S3). By plotting the ratio of the standard ribose peak area to the [U-13C5] ribose internal standard peak area as a function of standard ribose concentration, the ribose concentration of solutions spiked with known amounts of [U-13C5] ribose could be determined. Using this calibration curve, a sample of 2 million/mL lysed HeLa cells spiked pre-methanol extraction with [U-13C5] ribose to a final concentration of 20 μM was found to contain 39 ± 3 μM ribose (Table S3). Importantly, this value is nearly identical to that obtained via the 129Xe NMR method (40 ± 8 μM, Table 1) and supports our estimate of ca. 5 mM intracellular ribose concentration.
This study provides new insights into the biodistribution of ribose in mammalian systems. The unexpected finding that the intracellular ribose concentration is nearly three orders of magnitude higher than a previous estimate76 requires a reevaluation of our current knowledge regarding the biodistribution and biosynthesis of ribose. It is well understood that cells can synthesize ribose-5-phosphate from glucose via the pentose phosphate pathway. Notably, the DMEM cell growth medium contains approximately 25 mM glucose. Much less is known about the subsequent conversion of ribose-5-phosphate into ribose. A phosphatase enzyme with the ability to perform this biochemistry was recently discovered in bacteria.78 Similar phosphatases are likely to exist in mammalian cells and may contribute to a high intracellular ribose concentration. Finally, because these studies were performed with the HeLa cancer cell line, it is likely that transport, biosynthesis, and metabolism of these sugars differ from healthy cell types. Hepatic cells, for example, have been shown to display elevated rates of ribose salvage, and would thus concentrate ribose to a greater extent than other cell types.11 Additional studies will be undertaken to determine how ribose concentration differs as a function of cell type.
Time-dependent saturation transfer experiments with GFP-RBP(L19A) and mutants.
Finally, we set out to assess the detection sensitivity of GFP-RBP(L19A). Time-dependent saturation transfer experiments were performed by measuring 129Xe(aq) polarization as a function of saturation time and quantifying the normalized difference between on- and off-resonance saturation transfer as on-resonance hyper-CEST contrast. By this method, 100 nM ribose-bound GFP-RBP(L19A) reported a maximum of 0.30 ± 0.01 saturation contrast (Figure S9), which improves upon the values previously reported for 100 nM maltose-bound MBP (0.26 ± 0.01)45 and 100 nM TEM-1 β-lactamase (0.23 ± 0.01)42 using the same total saturation time and 129Xe concentration. Measuring saturation contrast as a function of percent GFP-RBP(L19A) in its closed conformation showed a linear relationship (R2 > 0.99, Figure S10), providing additional evidence that GFP-RBP(L19A) can serve as a ribose sensor in the nM to μM range.
To allow for linearly responsive CEST measurements at higher concentrations of ribose, and thereby expand the potential for in cellulo measurements, we tested the saturation contrast response of RBP mutants with lower ribose affinity. These mutations were based on those first reported by Lager and coworkers18 and any alterations to side chains lining the Xe binding site were avoided. Adding the Q235A mutation to GFP-RBP(L19A) increased the Kd for ribose to 10 μM, as measured by ITC (Figure S2). Further introducing the T135A mutation increased the Kd for ribose to 130 μM. Measuring saturation transfer for 100 nM GFP-RBP(L19A/T135A/Q235A) as a function of percent protein in the closed conformation showed a linear relationship for ribose in the μM to mM range (R2 > 0.99, Figure S10). Z-spectra of the two new RBP variants showed a slight decrease in CEST effect at 233 ppm relative to GFP-RBP(L19A) in the presence of 1 mM ribose, but the 129Xe chemical shift remained unchanged (Figure S11). The combination of the L19A and L19A/T135A/Q235A variants of RBP covers a large (nM-to-mM) range of ribose concentrations measurable by 129Xe hyper-CEST NMR.
CONCLUSIONS
In summary, we demonstrated that RBP(L19A) and mutants thereof generate quantifiable 129Xe hyper-CEST contrast in response to ribose. Specifically, we showed a linear relationship between the magnitude of MR saturation contrast and the percentage of protein in its closed, ribose-bound form. This relationship was used to calculate the ribose concentration of lysate solutions from HeLa cells grown in either serum-free or serum-supplemented conditions; the ribose concentration of FBS was also determined. From these assays, it was possible to estimate the intracellular ribose concentration in a mammalian cell line. This objective has been difficult to achieve using conventional analytical chemistry approaches and is important for understanding the distribution of free ribose among various cell types and physiologic states. Prior work has demonstrated efficient cellular uptake of ribose from extracellular milieu,79 and RBP-CEST provides a method for quantifying ribose uptake in different cell media.
RBP gave minimal CEST response in the presence of the structurally similar sugars glucose and ribose-5-phosphate, consistent with a prior study showing strong preference of RBP for ribose over several common pentoses and hexoses.18 Experiments confirmed that CEST signal was RBP-dependent and originated predominantly from the methanol-extracted, ribose-containing lysate fraction. More in-depth studies will be needed to confirm that interfering CEST signal cannot arise from additional small molecule metabolites in the cell. Nonetheless, the low-mM HeLa cell ribose concentration determined via RBP-CEST was strongly validated using an LC-MS method. It is striking that the intracellular ribose concentration is nearly three orders of magnitude higher than had been previously estimated. Using methods described herein, it will be possible to estimate intracellular ribose concentrations in a wide variety of cell lines.
Overall, these experiments show that Xe-binding PBPs can be used as quantitative tools for analyzing the presence of small-molecule ligands via 129Xe NMR. We demonstrated RBP mutants that tune RBP-ribose CEST response over the nM-to-mM ribose concentration range, expanding the versatility of RBP for in vitro and in vivo applications. We note that the strong hyper-CEST response observed for ribose-bound RBP(L19A) at 233 ppm is spectrally well resolved from our previous measurement of 129Xe-MBP at 288 ppm in the presence of maltose,45 which facilitates multiplexing applications. Combining the versatility and adaptability of the PBP scaffold with the sensitivity of the hyper-CEST technique offers exciting potential for developing PBPs as hyper-CEST contrast agents for detection and simultaneous quantitation of metabolites, ions, and small molecules.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to the National Institutes of Health (R01-GM-097478 and R35-GM-131907) and the University of Pennsylvania, Department of Chemistry for supporting this work. K.L.F. received financial support from the Vagelos Molecular Life Sciences program. The LC-MS analysis was performed at The Wistar Institute Proteomics and Metabolomics Shared Resource on a Thermo Q-Exactive HF-X mass spectrometer purchased with NIH grant S10 OD023586. We thank Hsin-Yao Tang for advice regarding LC-MS methodology.
REFERENCES
- (1).Markley JL; Brüschweiler R; Edison AS; Eghbalnia HR; Powers R; Raftery D; Wishart DS The Future of NMR-Based Metabolomics. Curr. Opin. Biotechnol 2017, 43, 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Dunn WB; Broadhurst DI; Atherton HJ; Goodacre R; Griffin JL Systems Level Studies of Mammalian Metabolomes: The Roles of Mass Spectrometry and Nuclear Magnetic Resonance Spectroscopy. Chem. Soc. Rev 2011, 40, 387–426. [DOI] [PubMed] [Google Scholar]
- (3).Minuto MN; Shintu L; Caldarelli S Proteomics, and Metabolomics: Magnetic Resonance Spectroscopy for the Presurgical Screening of Thyroid Nodules. Curr. Genomics 2014, 15, 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Emwas A-H; Roy R; McKay RT; Tenori L; Saccenti E; Gowda GAN; Raftery D; Alahmari F; Jaremko L; Jaremko M; Wishart DS NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9 (7), 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zhang A; Sun H; Wang P; Han Y; Wang X Modern Analytical Techniques in Metabolomics Analysis. Analyst 2012, 137 (2), 293–300. [DOI] [PubMed] [Google Scholar]
- (6).Kim ER; Kwon HN; Nam H; Kim JJ; Park S; Kim YH Urine-NMR Metabolomics for Screening of Advanced Colorectal Adenoma and Early Stage Colorectal Cancer. Sci. Rep 2019, 9 (1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Gross M; Zöllner N Serum Levels of Glucose, Insulin, and C-Peptide during Long-Term D-Ribose Administration in Man. Klin. Wochenschr 1991, 69 (1), 31–36. [DOI] [PubMed] [Google Scholar]
- (8).Patra KC; Hay N The Pentose Phosphate Pathway and Cancer. Trends Biochem. Sci 2014, 39 (8), 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mahoney DE; Hiebert JB; Thimmesch A; Pierce JT; Vacek JL; Clancy RL; Sauer AJ; Pierce JD Understanding D-Ribose and Mitochondrial Function. Adv. Biosci. Clin. Med 2018, 6 (1), 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Park J; van Koeverden P; Singh B; Gupta RS Identification and Characterization of Human Ribokinase and Comparison of Its Properties with E. Coli Ribokinase and Human Adenosine Kinase. FEBS Lett. 2007, 581 (17), 3211–3216. [DOI] [PubMed] [Google Scholar]
- (11).Clark PM; Flores G; Evdokimov NM; McCracken MN; Chai T; Nair-Gill E; O’Mahony F; Beaven SW; Faull KF; Phelps ME; Jung ME; Witte ON Positron Emission Tomography Probe Demonstrates a Striking Concentration of Ribose Salvage in the Liver. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (28), E2866–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Pauly DF; Johnson C; St.Cyr JA The Benefits of Ribose in Cardiovascular Disease. Med. Hypotheses 2003, 60 (2), 149–151. [DOI] [PubMed] [Google Scholar]
- (13).Reitzer LJ; Wice BM; Kennell D The Pentose Cycle. Control and Essential Function in HeLa Cell Nucleic Acid Synthesis. J. Biol. Chem 1980, 255 (12), 5616–5626. [PubMed] [Google Scholar]
- (14).Shapiro MG; Westmeyer GG; Romero PA; Szablowski JO; Küster B; Shah A; Otey CR; Langer R; Arnold FH; Jasanoff A Directed Evolution of a Magnetic Resonance Imaging Contrast Agent for Noninvasive Imaging of Dopamine. Nat. Biotechnol 2010, 28 (3), 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Minn I; Bar-Shir A; Yarlagadda K; Bulte JWM; Fisher PB; Wang H; Gilad AA; Pomper MG Tumor-Specific Expression and Detection of a CEST Reporter Gene. Magn. Reson. Med 2015, 74 (2), 544–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bar-Shir A; Bulte JWM; Gilad AA Molecular Engineering of Nonmetallic Biosensors for CEST MRI. ACS Chem. Biol 2015, 10 (5), 1160–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Harney AS; Meade TJ Molecular Imaging of In Vivo Gene Expression. Futur. Med Chem 2010, 2 (3), 503–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lager I; Fehr M; Frommer WB; Lalonde S Development of a Fluorescent Nanosensor for Ribose. FEBS Lett. 2003, 553 (1–2), 85–89. [DOI] [PubMed] [Google Scholar]
- (19).Walker TG; Happer W Spin-Exchange Optical Pumping of Noble-Gas Nuclei. Rev. Mod. Phys 1997, 69 (2), 629–642. [Google Scholar]
- (20).Albert MS; Cates GD; Driehuys B; Happer W; Saam B; Springer CS; Wishnia A Biological Magnetic Resonance Imaging Using Laser-Polarized 129Xe. Nature 1994, 370 (6486), 199–201. [DOI] [PubMed] [Google Scholar]
- (21).Mortuza MG; Anala S; Pavlovskaya GE; Dieken TJ; Meersmann T Spin-Exchange Optical Pumping of High-Density Xenon-129. J. Chem. Phys 2003, 118 (4), 1581–1584. [Google Scholar]
- (22).Berthault P; Huber G; Desvaux H Biosensing Using Laser-Polarized Xenon NMR/MRI. Prog. Nucl. Magn. Reson. Spectrosc 2009, 55 (1), 35–60. [Google Scholar]
- (23).Barskiy DA; Coffey AM; Nikolaou P; Mikhaylov DM; Goodson BM; Branca RT; Lu GJ; Shapiro MG; Telkki V; Zhivonitko VV; Koptyug IV; Salnikov OG; Kovtunov KV; Bukhtiyarov VI; Rosen MS; Barlow MJ; Safavi S; Hall IP; Schröder L; Chekmenev EY NMR Hyperpolarization Techniques of Gases. Chemistry 2017, 23 (4), 725–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Horn JR; Shoichet BK Allosteric Inhibition through Core Disruption. J. Mol. Biol 2004, 336 (5), 1283–1291. [DOI] [PubMed] [Google Scholar]
- (25).Desvaux H; Dubois L; Huber G; Quillin ML; Berthault P; Matthews BW Dynamics of Xenon Binding inside the Hydrophobic Cavity of Pseudo-Wild-Type Bacteriophage T4 Lysozyme Explored through Xenon-Based NMR Spectroscopy. J. Am. Chem. Soc 2005, 127 (33), 11676–11683. [DOI] [PubMed] [Google Scholar]
- (26).Rubin SM; Lee S-Y; Ruiz EJ; Pines A; Wemmer DE Detection and Characterization of Xenon-Binding Sites in Proteins by 129Xe NMR Spectroscopy. J. Mol. Biol 2002, 322 (2), 425–440. [DOI] [PubMed] [Google Scholar]
- (27).Gröger C; Möglich A; Pons M; Koch B; Hengstenberg W; Kalbitzer HR; Brunner E NMR-Spectroscopic Mapping of an Engineered Cavity in the I14A Mutant of HPr from Staphylococcus Carnosus Using Xenon. J. Am. Chem. Soc 2003, 125 (29), 8726–8727. [DOI] [PubMed] [Google Scholar]
- (28).Tilton RF; Kuntz ID; Petsko GA Cavities in Proteins: Structure of a Metmyoglobin-Xenon Complex Solved to 1.9 A. Biochemistry 1984, 23 (13), 2849–2857. [DOI] [PubMed] [Google Scholar]
- (29).Schröder L; Lowery TJ; Hilty C; Wemmer DE; Pines A Molecular Imaging Using a Targeted Magnetic Resonance Hyperpolarized Biosensor. Science 2006, 314 (5798), 446–449. [DOI] [PubMed] [Google Scholar]
- (30).Bai Y; Wang Y; Goulian M; Driks A; Dmochowski IJ Bacterial Spore Detection and Analysis Using Hyperpolarized 129 Xe Chemical Exchange Saturation Transfer (Hyper-CEST) NMR. Chem. Sci 2014, 5 (8), 3197–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhang H; Chen S; Yuan Y; Li Y; Jiang Z; Zhou X 129 Xe Hyper-CEST/19F MRI Multimodal Imaging System for Sensitive and Selective Tumor Cells Detection. ACS Appl. Bio Mater 2019, 2 (1), 27–32. [DOI] [PubMed] [Google Scholar]
- (32).Bai Y; Hill PA; Dmochowski IJ Utilizing a Water-Soluble Cryptophane with Fast Xenon Exchange Rates for Picomolar Sensitivity NMR Measurements. Anal. Chem 2012, 84 (22), 9935–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wang Y; Dmochowski IJ Cucurbit[6]Uril Is an Ultrasensitive 129 Xe NMR Contrast Agent. Chem. Commun 2015, 51 (43), 8982–8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Riggle BA; Wang Y; Dmochowski IJA “Smart” 129Xe NMR Biosensor for pH-Dependent Cell Labeling. J. Am. Chem. Soc 2015, 137 (16), 5542–5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kunth M; Döpfert J; Witte C; Rossella F; Schröder L Optimized Use of Reversible Binding for Fast and Selective NMR Localization of Caged Xenon. Angew. Chem., Int. Ed 2012, 51 (33), 8217–8220. [DOI] [PubMed] [Google Scholar]
- (36).Hane FT; Li T; Smylie P; Pellizzari RM; Plata JA; DeBoef B; Albert MS In Vivo Detection of Cucurbit[6]uril, a Hyperpolarized Xenon Contrast Agent for a Xenon Magnetic Resonance Imaging Biosensor. Sci. Rep 2017, 7, 41027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hane FT; Fernando A; Prete BRJ; Peloquin B; Karas S; Chaudhuri S; Chahal S; Shepelytskyi Y; Wade A; Li T; Deboef B; Albert MS Cyclodextrin-Based Pseudorotaxanes: Easily Conjugatable Scaffolds for Synthesizing Hyperpolarized Xenon-129 Magnetic Resonance Imaging Agents. ACS Omega 2018, 3 (1), 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Schnurr M; Sloniec-Myszk J; Döpfert J; Schröder L; Hennig A Supramolecular Assays for Mapping Enzyme Activity by Displacement-Triggered Change in Hyperpolarized 129Xe Magnetization Transfer NMR Spectroscopy. Angew. Chem., Int. Ed 2015, 54 (45), 13444–13447. [DOI] [PubMed] [Google Scholar]
- (39).Shapiro MG; Ramirez RM; Sperling LJ; Sun G; Sun J; Pines A; Schaffer DV; Bajaj VS Genetically Encoded Reporters for Hyperpolarized Xenon Magnetic Resonance Imaging. Nat. Chem 2014, 6 (7), 629–634. [DOI] [PubMed] [Google Scholar]
- (40).Kunth M; Lu GJ; Witte C; Shapiro MG; Schröder L Protein Nanostructures Produce Self-Adjusting Hyperpolarized Magnetic Resonance Imaging Contrast through Physical Gas Partitioning. ACS Nano 2018, 12 (11), 10939–10948. [DOI] [PubMed] [Google Scholar]
- (41).Farhadi A; Ho G; Kunth M; Ling B; Lakshmanan A; Lu GJ; Bourdeau RW; Schröder L; Shapiro MG Recombinantly Expressed Gas Vesicles as Nanoscale Contrast Agents for Ultrasound and Hyperpolarized MRI. AIChE J. 2018, 64 (8), 2927–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Wang Y; Roose BW; Palovcak EJ; Carnevale V; Dmochowski IJ A Genetically Encoded β-Lactamase Reporter for Ultrasensitive 129Xe NMR in Mammalian Cells. Angew. Chem., Int. Ed 2016, 55 (31), 8984–8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Roose BW; Zemerov SD; Wang Y; Kasimova M; Carnevale V; Dmochowski IJ A Structural Basis for 129Xe Hyper-CEST Signal in TEM-1 β-Lactamase. ChemPhysChem 2019, 20 (2), 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhao Z; Roose BW; Zemerov SD; Stringer MA; Dmochowski IJ Detecting Protein–Protein Interactions by Xe-129 NMR. Chem. Commun 2020. DOI: 10.1039/D0CC02988B [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Roose BW; Zemerov SD; Dmochowski IJ Nanomolar Small-Molecule Detection Using a Genetically Encoded 129Xe NMR Contrast Agent. Chem. Sci 2017, 8 (11), 7631–7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Dwyer MA; Hellinga HW Periplasmic Binding Proteins: A Versatile Superfamily for Protein Engineering. Curr. Opin. Struct. Biol 2004, 14 (4), 495–504. [DOI] [PubMed] [Google Scholar]
- (47).Ribeiro LF; Amarelle V; Ribeiro LFC; Guazzaroni ME Converting a Periplasmic Binding Protein into a Synthetic Biosensing Switch through Domain Insertion. Biomed Res. Int 2019, 2019 (i), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ha J-H; Loh SN Protein Conformational Switches: From Nature to Design. Chem. - Eur. J 2012, 18 (26), 7984–7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Li L; Ghimire-Rijal S; Lucas SL; Stanley CB; Wright E; Agarwal PK; Myles DA; Cuneo MJ Periplasmic Binding Protein Dimer Has a Second Allosteric Event Tied to Ligand Binding. Biochemistry 2017, 56 (40), 5328–5337. [DOI] [PubMed] [Google Scholar]
- (50).de Lorimier RM; Smith JJ; Dwyer MA; Looger LL; Sali KM; Paavola CD; Rizk SS; Sadigov S; Conrad DW; Loew L; Hellinga HW Construction of a Fluorescent Biosensor Family. Protein Sci. 2002, 11 (11), 2655–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Okumoto S; Looger LL; Micheva KD; Reimer RJ; Smith SJ; Frommer WB Detection of Glutamate Release from Neurons by Genetically Encoded Surface-Displayed FRET Nanosensors. Proc. Natl. Acad. Sci. U. S. A 2005, 102 (24), 8740–8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Bogner M; Ludewig U Visualization of Arginine Influx into Plant Cells Using a Specific FRET-Sensor. J. Fluoresc 2007, 17 (4), 350–360. [DOI] [PubMed] [Google Scholar]
- (53).Ko W; Kim S; Lee HS Engineering a Periplasmic Binding Protein for Amino Acid Sensors with Improved Binding Properties. Org. Biomol. Chem 2017, 15 (41), 8761–8769. [DOI] [PubMed] [Google Scholar]
- (54).Marvin JS; Schreiter ER; Echevarría IM; Looger LL A Genetically Encoded, High-Signal-to-Noise Maltose Sensor. Proteins 2011, 79 (11), 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Marvin JS; Hellinga HW Engineering Biosensors by Introducing Fluorescent Allosteric Signal Transducers: Construction of a Novel Glucose Sensor. J. Am. Chem. Soc 1998, 120 (1), 7–11. [Google Scholar]
- (56).Ribeiro LF; Bressan F; Furtado GP; Meireles F; Ward RJ D-Xylose Detection in Escherichia Coli by a Xylose Binding Protein-Dependent Response. J. Biotechnol 2013, 168 (4), 440–445. [DOI] [PubMed] [Google Scholar]
- (57).Tian Y; Cuneo MJ; Changela A; Höcker B; Beese LS; Hellinga HW Structure-Based Design of Robust Glucose Biosensors Using a Thermotoga Maritima Periplasmic Glucose-Binding Protein. Protein Sci. 2007, 16 (10), 2240–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Salins LLE; Goldsmith ES; Ensor CM; Daunert S A Fluorescence-Based Sensing System for the Environmental Monitoring of Nickel Using the Nickel Binding Protein from Escherichia Coli. Fresenius. J. Anal. Chem 2002, 372 (1), 174–180. [DOI] [PubMed] [Google Scholar]
- (59).Gu H; Lalonde S; Okumoto S; Looger LL; Scharff-Poulsen AM; Grossman AR; Kossmann J; Jakobsen I; Frommer WB A Novel Analytical Method for in Vivo Phosphate Tracking. FEBS Lett. 2006, 580 (25), 5885–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Shrestha S; Salins LLE; Ensor CM; Daunert S Rationally Designed Fluorescently Labeled Sulfate-Binding Protein Mutants: Evaluation in the Development of a Sensing System for Sulfate. Biotechnol. Bioeng 2002, 78 (5), 517–526. [DOI] [PubMed] [Google Scholar]
- (61).Solscheid C; Kunzelmann S; Davis CT; Hunter JL; Nofer A; Webb MR Development of a Reagentless Biosensor for Inorganic Phosphate, Applicable over a Wide Concentration Range. Biochemistry 2015, 54 (32), 5054–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Rizk SS; Cuneo MJ; Hellinga HW Identification of Cognate Ligands for the Escherichia Coli PhnD Protein Product and Engineering of a Reagentless Fluorescent Biosensor for Phosphonates. Protein Sci. 2006, 15 (7), 1745–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Alicea I; Marvin JS; Miklos AE; Ellington AD; Looger LL; Schreiter ER Structure of the Escherichia Coli Phosphonate Binding Protein PhnD and Rationally Optimized Phosphonate Biosensors. J. Mol. Biol 2011, 414 (3), 356–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Liu D; Evans T; Zhang F Applications and Advances of Metabolite Biosensors for Metabolic Engineering. Metab. Eng 2015, 31, 35–43. [DOI] [PubMed] [Google Scholar]
- (65).Fehr M; Frommer WB; Lalonde S Visualization of Maltose Uptake in Living Yeast Cells by Fluorescent Nanosensors. Proc. Natl. Acad. Sci. U. S. A 2002, 99 (15), 9846–9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Fehr M; Lalonde S; Lager I; Wolff MW; Frommer WB In Vivo Imaging of the Dynamics of Glucose Uptake in the Cytosol of COS-7 Cells by Fluorescent Nanosensors. J. Biol. Chem 2003, 278 (21), 19127–19133. [DOI] [PubMed] [Google Scholar]
- (67).Marvin JS; Hellinga HW Conversion of a Maltose Receptor into a Zinc Biosensor by Computational Design. Proc. Natl. Acad. Sci. U. S. A 2001, 98 (9), 4955–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Dwyer M. a; Looger LL; Hellinga HW Computational Design of a Zn2+ Receptor That Controls Bacterial Gene Expression. Proc. Natl. Acad. Sci. U. S. A 2003, 100 (20), 11255–11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Looger LL; Dwyer MA; Smith JJ; Hellinga HW Computational Design of Proteins with Novel Structure and Functions. Nature 2003, 423, 185–190. [DOI] [PubMed] [Google Scholar]
- (70).Vetting MW; Al-Obaidi N; Zhao S; San Francisco B; Kim J; Wichelecki DJ; Bouvier JT; Solbiati JO; Vu H; Zhang X; Rodionov DA; Love JD; Hillerich BS; Seidel RD; Quinn RJ; Osterman AL; Cronan JE; Jacobson MP; Gerlt JA; Almo SC Experimental Strategies for Functional Annotation and Metabolism Discovery: Targeted Screening of Solute Binding Proteins and Unbiased Panning of Metabolomes. Biochemistry 2015, 54 (3), 909–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Guntas G; Mansell TJ; Kim JR; Ostermeier M Directed Evolution of Protein Switches and Their Application to the Creation of Ligand-Binding Proteins. Proc. Natl. Acad. Sci. U. S. A 2005, 102 (32), 11224–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Lowery TJ; Rubin SM; Ruiz EJ; Pines A; Wemmer DE Design of a Conformation-Sensitive Xenon-Binding Cavity in the Ribose-Binding Protein. Angew. Chem., Int. Ed 2004, 43 (46), 6320–6322. [DOI] [PubMed] [Google Scholar]
- (73).Nagana Gowda GA; Raftery D Quantitating Metabolites in Protein Precipitated Serum Using NMR Spectroscopy. Anal. Chem 2014, 86 (11), 5433–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Bennett BD; Kimball EH; Gao M; Osterhout R; Van Dien SJ; Rabinowitz JD Absolute Metabolite Concentrations and Implied Enzyme Active Site Occupancy in Escherichia Coli. Nat. Chem. Biol 2009, 5 (8), 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Das D; Kamil FA; Biswas K; Das S Evaluation of Single Cell Electrical Parameters from Bioimpedance of a Cell Suspension. RSC Adv. 2014, 4 (35), 18178–18185. [Google Scholar]
- (76).Fortpied J; Gemayel R; Vertommen D; Van Schaftingen E Identification of Protein-Ribulosamine-5-Phosphatase as Human Low-Molecular-Mass Protein Tyrosine Phosphatase-A. Biochem. J 2007, 406, 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Zhao L; Kroenke CD; Song J; Piwnica-Worms D; Ackerman JJH; Neil JJ Intracellular Water Specific MR of Microbead-Adherent Cells: The HeLa Cell Intracellular Water Exchange Lifetime. NMR Biomed. 2008, 21 (2), 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Maleki S; Hrudikova R; Zotchev SB; Ertesvåg H Identification of a New Phosphatase Enzyme Potentially Involved in the Sugar Phosphate Stress Response in Pseudomonas Fluorescens. Appl. Environ. Microbiol 2017, 83 (2), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Horowitz SB; Pearson TW Intracellular Monosaccharide and Amino Acid Concentrations and Activities and the Mechanisms of Insulin Action. Mol. Cell. Biol 1981, 1 (9), 769–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


