Abstract
Background
Circumsporozoite surface protein (CSP) of malaria parasites has been recognized as one of the leading vaccine candidates. Clinical trials of vaccines for vivax malaria incorporating Plasmodium vivax CSP (PvCSP) have demonstrated their effectiveness in preventing malaria, at least in part. However, genetic diversity of pvcsp in the natural population remains a major concern.
Methods
A total of 171 blood samples collected from patients infected with Plasmodium vivax in Myanmar were analysed in this study. The pvcsp was amplified by polymerase chain reaction, followed by cloning and sequencing. Polymorphic characteristics and natural selection of pvcsp population in Myanmar were analysed using DNASTAR, MEGA6 and DnaSP programs. The polymorphic pattern and natural selection of publicly accessible global pvcsp sequences were also comparatively analysed.
Results
Myanmar pvcsp sequences were divided into two subtypes VK210 and VK247 comprising 143 and 28 sequences, respectively. The VK210 subtypes showed higher levels of genetic diversity and polymorphism than the VK247 subtypes. The N-terminal non-repeat region of pvcsp displayed limited genetic variations in the global population. Different patterns of octapeptide insertion (ANKKAEDA in VK210 and ANKKAGDA in VK247) and tetrapeptide repeat motif (GGNA) were identified in the C-terminal region of global pvcsp population. Meanwhile, the central repeat region (CRR) of Myanmar and global pvcsp, both in VK210 and VK247 variants, was highly polymorphic. The high level of genetic diversity in the CRR has been attributed to the different numbers, types and combinations of peptide repeat motifs (PRMs). Interestingly, 27 and 5 novel PRMs were found in Myanmar VK210 and VK247 variants, respectively.
Conclusion
Comparative analysis of the global pvcsp population suggests a complex genetic profile of pvcsp in the global population. These results widen understanding of the genetic make-up of pvcsp in the global P. vivax population and provide valuable information for the development of a vaccine based on PvCSP.
Keywords: Plasmodium vivax, Circumsporozoite protein, Genetic polymorphism, Natural selection, Myanmar
Background
Malaria caused by Plasmodium species has threatened human health since ancient time [1]. The mortality and morbidity of malaria has greatly decreased in recent years; however, an estimated 219 million cases and 435,000 deaths due to malaria were reported globally in 2017 [2]. Among five human malaria parasites, Plasmodium vivax is the most prevalent species outside of Africa. More than 7.5 million cases of malaria were caused by the parasite, which accounted for 56% of total malaria cases in South East Asia [2]. Plasmodium vivax has been linked to benign malarial infection due to its mild clinical manifestations compared with Plasmodium falciparum; however, concerns that P. vivax also causes serious clinical illnesses and even death have increased [3, 4]. The emergence and spread of drug-resistant strains also increases the burden of the parasite [5, 6]. Furthermore, the liver stages in the life cycle of P. vivax, known as hypnozoites, can remain dormant and survive for weeks to months in liver cells, and be reactivated [7]. This unique biological characteristic is a challenge in the control and elimination of this parasite. Therefore, the development of an effective vaccine is imperative for effective control and elimination of the parasite.
Circumsporozoite protein (CSP) is the most abundantly expressed protein on the surface of sporozoites. CSP plays multiple and crucial roles in the development, migration and invasion of sporozoites into hepatocytes [8]. Thus, this protein has been studied extensively as one of the most promising vaccine candidates. The gene encoding CSP consists of three distinct regions: a conserved non-repeat N-terminal region, a highly polymorphic central repeat region (CRR), and a conserved non-repeat C-terminal region (Additional file 1: Figure S1). The CRR of P. vivax CSP (PvCSP) consists of peptide repeat motifs (PRMs). Based on the composition of one of the three major PRMs in the CRR, pvcsp is classified into three allelic variants: VK210, VK247 and P. vivax-like. The two most abundant alleles include VK210 and VK247, which carry nonapeptide repeat motifs GDRA(D/A)GQPA and ANGAG(N/D)QPG, respectively [9]. Meanwhile, the P. vivax-like variant contains APGANQ(E/G)GAA motifs in the CRR [10].
Although no licensed vaccine against vivax malaria is currently available, a few notable advances in the development of a potential vaccine have been reported. A vivax malaria subunit vaccine named VMP001 has been clinically tested. This vaccine uses a chimeric recombinant protein containing repeat sequences of two major alleles of pvcsp, including VK210 and VK247 [11]. The phase 1 trial with VMP001 showed a significant delay in parasitaemia, even though vaccination did not induce fully sterilizing protective immunity [12]. However, the geographical variation in the genetic make-up of pvcsp population may influence the effectiveness of PvCSP-based vaccine, which may be a barrier for the development of a universal vaccine. Therefore, a comprehensive analysis of genetic diversity and structure of the global pvcsp population is necessary. In this study, genetic polymorphism and natural selection of Myanmar pvcsp has been examined. A comparative analysis of global pvcsp was also conducted to obtain a deeper insight into the genetic nature of pvcsp in the global P. vivax population.
Methods
Blood samples
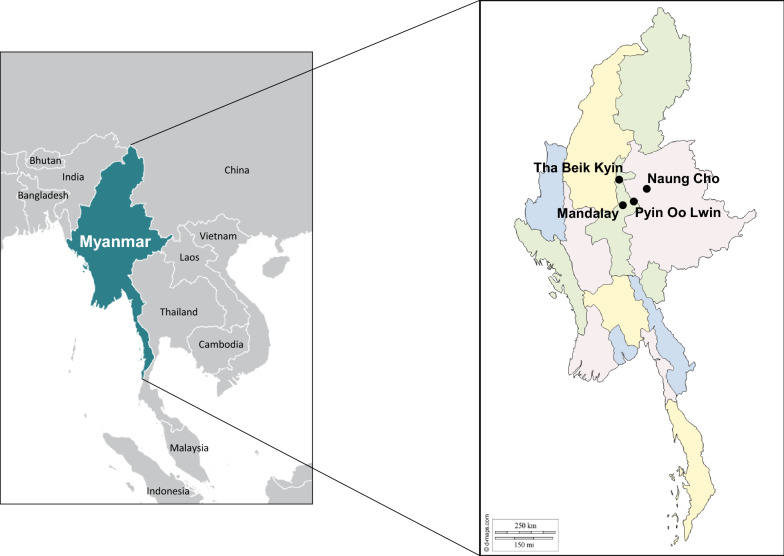
One hundred seventy-one blood samples from P. vivax-infected Myanmar patients were used in this study. The blood samples were collected from the patients in field surveys in towns and villages in Naung Cho, Pyin Oo Lwin, and Tha Beik Kyin in Upper Myanmar during 2013 to 2015 (Fig. 1). The age range of patients was 13–62 years, with median age of 28.4 years. Initial screening for malaria infection was done by microscopic examination of thin and thick blood smears. Finger-prick blood samples were taken from the P. vivax-infected patients before drug treatment and spotted on Whatman 3 MM filter paper (GE Healthcare, Maidstone, UK) for confirmation by species-specific polymerase chain reaction (PCR) targeting 18S ribosomal RNA (rRNA) gene [13, 14]. Informed consent was obtained from all of the patients before blood collection. The study protocol was reviewed and approved by either the Ethics committee of the Ministry of Health, Myanmar (97/Ethics 2015) and the Biomedical Research Ethics Review Board of Inha University School of Medicine, Republic of Korea (INHA 15-013).
Fig. 1.
Map of the study sites. Blood samples were collected from P. vivax infected patients, who resided in villages or townships in Naung Cho, Pyin Oo Lwin and Tha Beik Kyin, Myanmar and used in this study
Genomic DNA extraction and amplification of pvcsp
Genomic DNA was extracted from dried blood spots using QIAamp DNA Blood Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Amplification of pvcsp gene was performed with primers by using nested PCR. The primers for first round PCR were 5ʹ-ATGTAGATCTGTCCAAGGCCATAAA-3ʹ and 5ʹ-AATTGAATAATGCTAGGACTAACAATATG-3ʹ. The thermal cycling parameters for primary PCR were as follows: one cycle of initial denaturation at 95 °C for 5 min, 25 cycles of 94 °C for 1 min, annealing at 58 °C for 2 min and extension at 72 °C for 2 min, followed by a final extension at 72 °C for 5 min. The nested PCR was performed with primers, 5ʹ-GCAGAACCAAAAAATCCACGTGAAAATAAG-3ʹ and 5ʹ-CCAACGGTAGCTCTAACTTTATCTAGGTAT-3ʹ, and similar amplification condition except the annealing temperature was 68 °C. Ex Taq DNA polymerase (Takara, Otsu, Japan) with proof-reading activity was used in all PCR steps to minimize the nucleotide mismatching during the amplification. Each PCR product was analysed by electrophoresis on 2% agarose gel. The resulting PCR product was extracted from the gel and was cloned into T&A cloning vector (Real Biotech Corporation, Banqiao City, Taiwan). Each ligation mixture was transformed into Escherichia coli DH5α competent cells. To identify positive clones with appropriate insert, colony PCR with nested PCR primers was performed. The nucleotide sequences of the cloned pvcsp were analysed by the Sanger method with M13 forward and reverse primers. Plasmids from at least two independent clones from each transformation mixture were analysed to confirm the sequence accuracy. These nucleotide sequences analysed in this study were deposited at GenBank under the accession numbers MN821829–MN821999.
Analyses of genetic diversity and natural selection in Myanmar pvcsp
The nucleotide and deduced amino acid sequences of Myanmar pvcsp were analysed using Editseq and Seqman in the DNASTAR package (DNASTAR, Madison, WI, USA). Two major variants of pvcsp sequences, Salvador I (Sal I; GU339059) and Papua New Guinea (PNG; M69059), were used as reference sequences to analyse Myanmar pvcsp sequences. The values of number of segregating sites (S), haplotypes (H), haplotype diversity (Hd), nucleotide diversity (π), and average number of pair-wise nucleotide differences within a population (K) were calculated with DnaSP ver. 5.10.00 [15]. The rate of synonymous (dS) and nonsynonymous (dN) substitutions were calculated and compared using the Z-test (P < 0.05) with MEGA6 program [16] using Nei and Gojobori’s method [17] with the Juke and Cantor (JC) correction of 1000 bootstrap replications. Based on the acquired vales, the dN–dS value was calculated. Positive value for dN–dS imply to positive natural selection whereas negative value correspond to negative or purifying natural selection [18]. Tajima’s D test, Fu and Li’s D and F statistics analysis was performed using DnaSP ver. 5.10.00 [15] to evaluate the neutral theory of natural selection. An excess of high-frequency variation is consistent with balancing selection and is indicated by a positive Tajima’s D and/or Fu and Li’s D [19, 20]. A negative value of Tajima’s D and/or Fu and Li’s D indicates an excess of rare alleles, which may result from a recent selective sweep or purifying selection.
Genetic diversity and natural of pvcsp among the global P. vivax population
The genetic diversity of pvcsp among the global P. vivax population was also analysed. The pvcsp sequences deposited in public database were used in this study; Cambodia (n = 41), India (n = 79), Iran (n = 50), South Korea (n = 39), Brazil (n = 41), Mexico (n = 19), Colombia (n = 25), Sudan (n = 30), and Vanuatu (n = 21) (Additional file 2: Table S1). Genetic polymorphism and test of neutrality were examined for each pvcsp population with DnaSP ver 5.10.00 [15] and MEGA6 [16] as described above. To analyse the polymorphic patterns of the N- and C-terminal non-repeat regions in global pvcsp, a logo plot was constructed for each population using the WebLogo program (https://weblogo.berkeley.edu/logo.cgi).
Results
Amplification and sequence analysis of Myanmar pvcsp
A total of 171 Myanmar pvcsp genes were successfully amplified form the genomic DNA samples used in this study. The size of the amplified pvcsp genes ranged from 0.5 to 1.3 kb. Sequence analysis of the amplified pvcsp genes revealed only two variants of pvcsp, VK210 and VK247, in Myanmar pvcsp, but not the P. vivax-like variant. The VK210 variants were prevalent (n = 143, 83.6%) and the frequency of VK247 variants was 16.4% (n = 28). No mixed infection with the two different variants was detected.
Genetic diversity of the N-terminal non-repeat region of Myanmar pvcsp
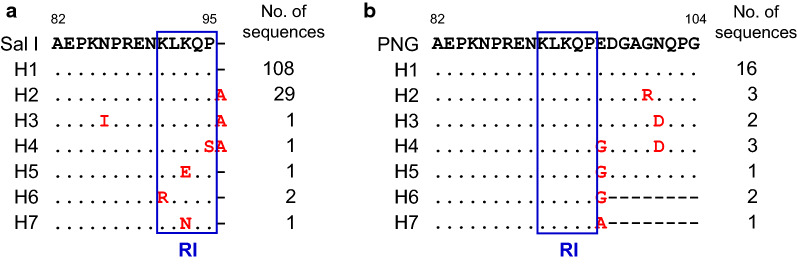
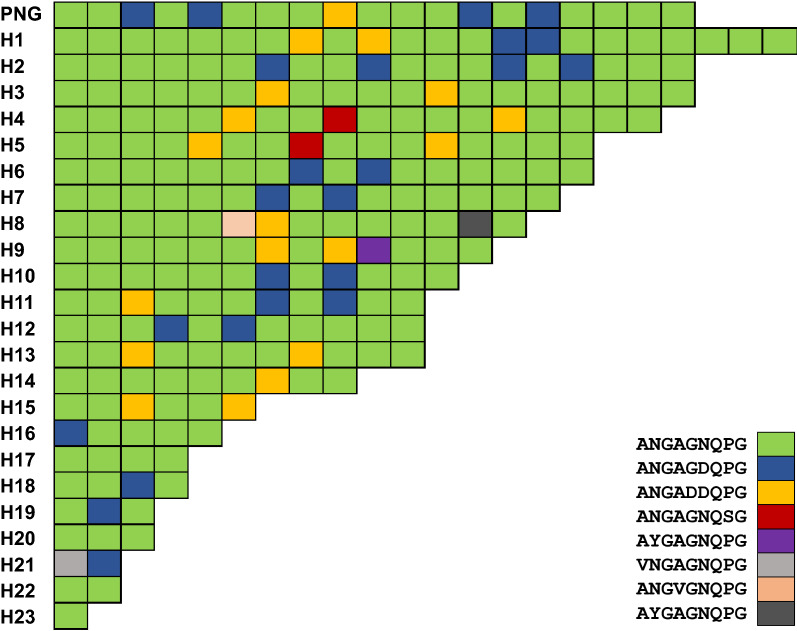
The N-terminal non-repeat region of Myanmar pvcsp showed a limited range of genetic diversity. Alignment of the deduced amino acid sequences of Myanmar pvcsp revealed 7 distinct haplotypes of VK210 variants and 7 haplotypes of VK247 variants (Fig. 2). The N-terminal non-repeat region of Myanmar VK210 variants was highly conserved. Compared with Sal I (GU339059) sequence, only few amino acid substitutions was found in the latter portion of the N-terminal non-repeat region. Haplotype 1, which was identical to Sal I sequence, was predominant (n = 108, 75.5%). An alanine insertion at the end of the RI conserved motif (KLKQP) was identified in haplotypes 2, 3 and 4. The N86I was found in haplotype 3. Two dimorphic (K91R and P95S) and one trimorphic (K93E/N) amino acid changes were observed in the RI motif of haplotypes 4, 5, 6, and 7 (Fig. 2a). Changes in the amino acid sequences were also identified in VK247 variants of Myanmar pvcsp. Haplotype 1, which shared the same sequence with the PNG (M69059) sequence, was the most prevalent, accounting for 57.1% of 28 Myanmar VK247 variants. All amino acid changes identified in Myanmar VK247 variants were all dimorphic (E96G/A, G99R, and N100D). Furthermore, eight amino acids (97DGAGNQPG104) were not detected in haplotypes 6 and 7 (Fig. 2b).
Fig. 2.
Polymorphic patterns of the N-terminal non-repeat region of Myanmar pvcsp sequences. a VK210 variants. Seven distinct haplotypes of the N-terminal non-repeat region was identified in 143 Myanmar VK210 sequences. The dots represent residues identical to the reference sequence of Sal I (GU339059). Amino acid changes at particular amino acid positions are indicated as red. The dashes represent gaps to maximize the alignment. b VK247 variants. Seven distinct haplotypes of the N-terminal non-repeat region was identified in 28 Myanmar VK247 sequences. The dots represent residues identical to the reference sequence of PNG (M69059). Amino acid changes at particular amino acid positions are indicated as red. The dashes represent gaps to maximize the alignment. RI means KLKQP motif that is involved in the sporozoite invasion of mosquito salivary gland and in binding to hepatocytes prior to invasion. The total number of sequences for each hapoltype is listed in the right panel
Genetic polymorphisms of the N-terminal non-repeat region in global pvcsp
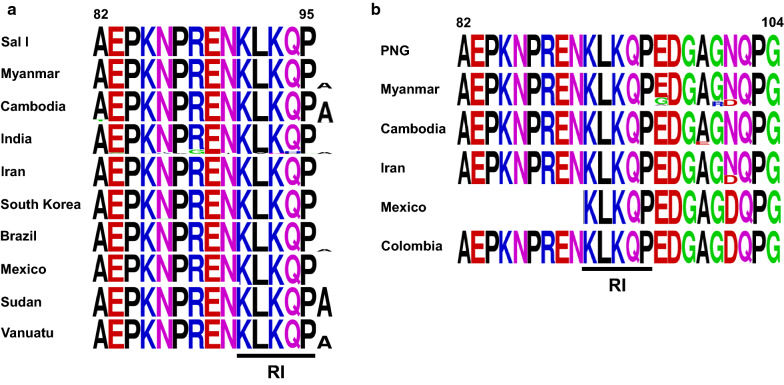
Overall genetic polymorphisms of the N-terminal non-repeat region in the global pvcsp population were analysed. A comparative analysis of the region revealed that the region is relatively well-conserved in the global pvcsp. Alanine insertion at the end of the RI in the VK210 variants was the major variation identified in the global pvcsp, but the prevalence of the insertion differed by geographically (Fig. 3a). The pvcsp from Sudan showed 100% alanine insertion followed by pvcsp from Cambodia (67.7%), Vanuatu (47.6%), Myanmar (21.7%), Brazil (9.8%), and India (7.6%). No alanine insertion was found in the VK210 sequences identified in Iran, South Korea, and Mexico. Indian VK210 variants showed the highest genetic diversity with amino acid substitutions at 10 positions including E83K/G, K85T/Q, N86K/Y/I, P87A, R88G, N90I, L92V, K93N, Q94H/P and P95G, even though their frequencies were low, less than 15%. Meanwhile, A82T, N86I, N86S, K91R, P95S, and K93E/N showed uneven geographic distribution in the global VK210 variants with very low frequencies. The N-terminal non-repeat region of global VK247 variants were also well-conserved, although low frequencies of uneven amino acid changes were identified (Fig. 3b). The most remarkable variation involved N100D, which was observed in VK247 variants from Colombia (100%), Mexico (100%), Iran (27.3%), and Myanmar (17.9%). The amino acid changes such as E96G/A, A99E, G100R, and N101D showed uneven geographic distribution with low frequencies.
Fig. 3.
Comparative analysis of polymorphic patterns of N-terminal non-repeat region in global pvcsp. a Global VK210 variants. b Global VK247 variants. The pattern of amino acid changes differed by country or geographic continent. A logo plot was constructed for each pvcsp population using the WebLogo program. Sal I, Salvador I (GU339059); PNG, Papua New Guinea (M69059). RI region was marked as an underline
Polymorphic pattern of the CRR in Myanmar pvcsp
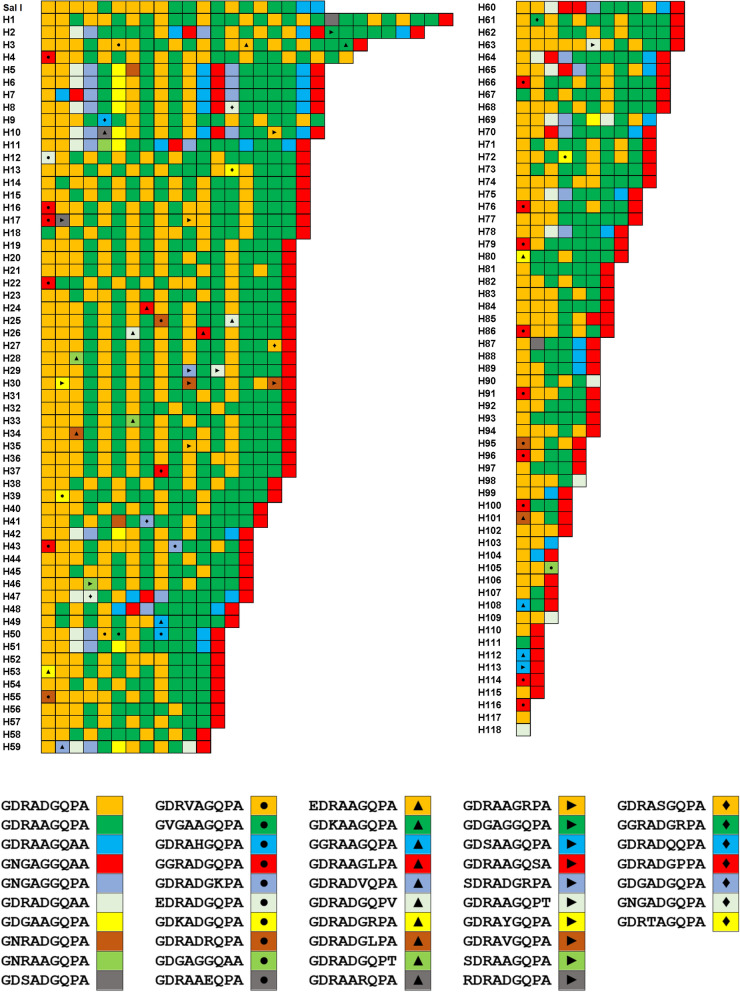
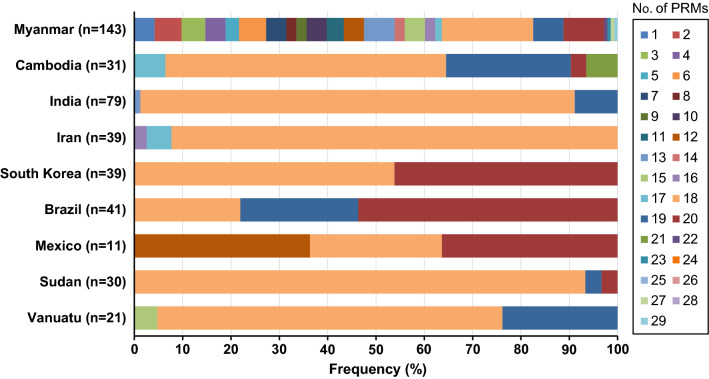
The CRR of Myanmar pvcsp showed extreme diversity in both VK210 and VK247 variants. A total of 118 and 23 haplotypes were identified in VK210 and VK247 variants, respectively. As expected, the greatest diversity of Myanmar pvcsp CRR was mainly attributed to differences in numbers, types, and arrangements of PRMs in each haplotype. A total of 47 different types of PRMs have been identified in the CRR of Myanmar VK210 variants (Fig. 4). Among these PRMs, two major types GDRADGQPA and GDRAAGQPA were the dominant ones for VK210 variants. Twenty-seven novel PRMs including GDRVAGQPA, GDRAHGQPA, GDRADGKPA, GDRADRQPA, GDGAGGQAA, EDRAAGQPA, GDKAAGQPA, GDRAAGLPA, GDRADVQPA, GDRADGQPV, GDRADGRPA, GDRADGLPA, GDRADGQPT, GDRAARQPA, GDRAAGRPA, GDGAGGQPA, GDRAAGQSA, SDRADGRPA, GDRAAGQPT, GDRAYGQPA, SDRAAGQPA, RDRADGQPA, GDRASGQPA, GGRADGRPA, GDRADQQPA, GDRADGPPA and GNGADGQPA, which were not previously reported, were found in Myanmar VK210 variants. Interestingly, 109 haplotypes out of 118 VK210 variants were terminated with GNGAGGQAA motif. The number of PRMs consisting CRR of Myanmar VK210 haplotypes varied from 1 to 29. Sequences with 18 PRMs were the most prevalent accounting for 18.9% of 143 Myanmar VK210 sequences (Fig. 5).
Fig. 4.
Genetic polymorphism in the central repeat region (CRR) of Myanmar VK210 variants. Sequence alignment revealed that the CRR of Myanmar VK210 variants showed polymorphic characters with 118 distinct haplotypes. A total of 47 different types of peptide repeat motifs (PRMs) were identified in CRR of Myanmar VK210 variants. Differences in numbers, types, and combinations of PRMs produced the high levels of genetic diversity of the CRR in Myanmar VK210 variants
Fig. 5.
Frequency comparison of peptide repeat motifs (PRMs) in the central repeat region (CRR) of global VK210 variants. The numbers of PRMs differed in the global VK210 varaints
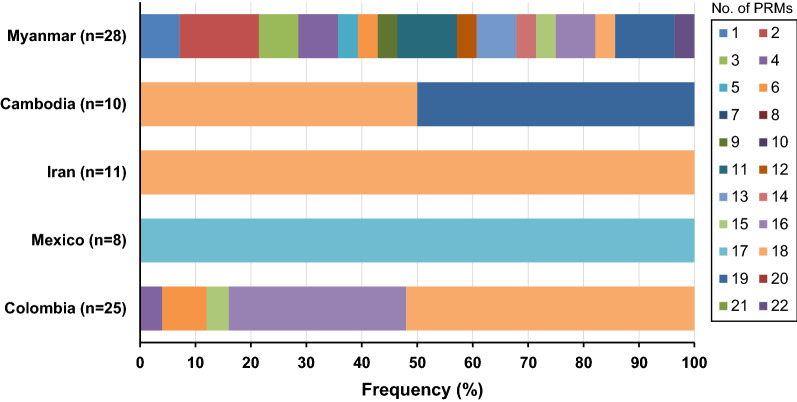
Compared with global VK210 variants, the CRR of Myanmar VK210 variants showed a high level of length polymorphisms. The VK210 variants from other countries analysed in this study showed only a few (2 to 5) variations in length of polymorphisms in CRR; however, the CRR of Myanmar VK210 variants had 24 different length polymorphisms consistent with various types and different compositions of PRMs. The overall genetic diversity in CRR of Myanmar VK247 variants was much lower than that of Myanmar VK210 variants. A total of 23 VK247 haplotypes, each CRR comprising different numbers and combinations of 8 types of PRMs, were identified (Fig. 6). Five of 8 PRMs, ANGAGNQSG, AYGAGNQPG, VNGAGNQPG, ANGVGNQPG and AYGAGNQPG, identified in Myanmar VK247 variants were novel ones that have not reported previously. The CRR of Myanmar VK247 variants carried different numbers of PRMs ranging from 1 to 22, and the CRR with 2 PRMs was the most prevalent (14.3%) (Fig. 7). Similar to VK210 variants, the Myanmar VK247 variants also displayed higher levels of diversity than the VK247 variants identified from other geographical origins. Sixteen different size polymorphisms of CRR were identified in Myanmar VK247 variants, whereas less size variations (1 to 5) were found in CRR of all other countries analysed in this study.
Fig. 6.
Genetic polymorphism in the central repeat rsegion (CRR) of Myanmar VK247 varaints. Sequence alignment revealed that the CRR of Myanmar VK247 variants showed polymorphic characters with 23 distinct haplotypes. A total of 8 different types of peptide repeat motifs (PRMs) were identified in CRR of Myanmar VK247 variants. Differences in numbers, types, and combinations of PRMs produced the high levels of genetic diversity of the CRR in Myanmar VK247 variants
Fig. 7.
Frequency comparison of peptide repeat motifs (PRMs) in the central repeat region of global VK247 variants. The numbers of PRMs differed in the global VK210 varaints
Genetic diversity in the C-terminal non-repeat region of Myanmar and global pvcsp
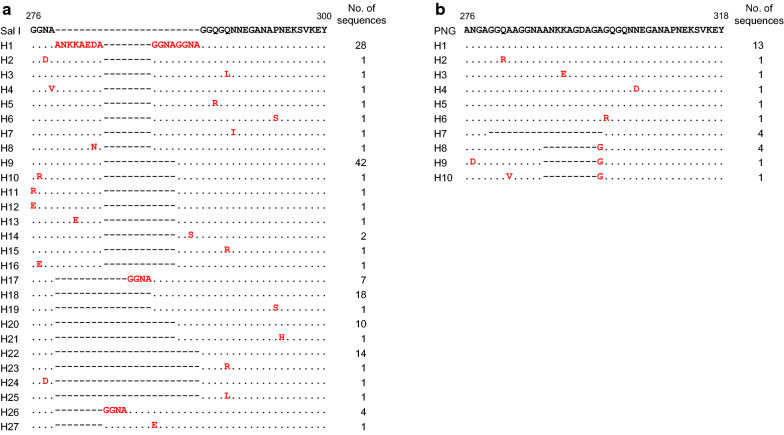
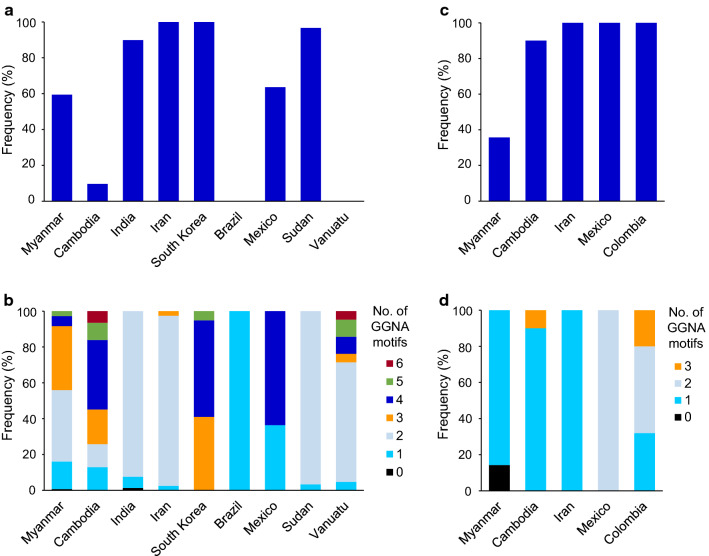
Sequence analysis of the C-terminal non-repeat region of Myanmar VK210 variants revealed 27 distinct haplotypes (Fig. 8a). These sequence diversities were attributed to differences in the arrangement of ANKKAEDA octapeptide insertion and GGNA tetrapeptide repeat motifs with different amino acid substitutions throughout the region. The sequence of haplotype 22 was identical with Sal I (GU339059), and accounted for 9.8% of all the VK210 sequences. The octapeptide insertions were observed in haplotypes 1 to 16. All the inserted sequences in these 16 haplotypes were ANKKAEDA except for haplotypes 8 and 13, which contained ANKKAENA and ANKEAENA, respectively. The C-terminal non-repeat region of Myanmar VK247 variants showed a lower level of genetic diversity than that of Myanmar VK210 variants (Fig. 8b). A total of 10 haplotypes were identified, and haplotype 1, which carried a sequence identical to that of the reference sequence of PNG (M69059), was the most prevalent haplotype with a frequency of 46.4%. The most noteworthy polymorphic characteristics identified in the C-terminal non-repeat region of Myanmar VK247 variants were the deletions of GGQAAGGNAANKKAGDAG in haplotype 7 and ANKKAGDAG in haplotypes 8, 9 and 10. Analysis of sequence polymorphisms in the C-terminal non-repeat region of the global pvcsp suggested a high level of genetic diversity. Among VK210 variants, the frequency of ANKKAEDA insertion differed by country (Fig. 9a). All sequences from Iran and South Korea VK210 variants contained an octapeptide insertion, but the frequencies in VK210 variants from Sudan, India, Mexico and Cambodia were 96.7, 89.9, 63.6 and 9.7%, respectively. Interestingly, no insertion of the octapeptide was identified in VK210 variants from Brazil and Vanuatu. The numbers of GGNA motifs in the C-terminal non-repeat region of global VK210 variants also differed by country (Fig. 9b). The number of repeated GGNA motifs found in global VK210 variants ranged from 0 to 6. Similar to VK210 variants, the presence of ANKKAGDAG octapeptide insertion and the number of GGNA motifs also differed in the C-terminal non-repeat region of global VK247 variants. The frequency of ANKKAGDAG octapeptide insertion was high in VK247 variants isolated from Cambodia, Iran, Mexico, and Colombia, but was low in Myanmar VK247 variants (Fig. 9c). The number of GGNA motifs found in the C-terminal non-repeat region of global VK247 variants ranged from 0 to 3 (Fig. 9d). The number of GGNA motifs in global VK247 also differed by country.
Fig. 8.
Genetic polymorphism of the C-terminal non-repeat region of Myanmar pvcsp sequences. a VK210 variants. A total of 27 distinct haplotypes of the C-terminal non-repeat region was identified in 143 Myanmar VK210 sequences. The dots represent residues identical to the reference sequence of Sal I (GU339059). Amino acid changes at particular amino acid positions are indicated as red. The dashes represent gaps to maximize the alignment. b VK247 variants. Ten distinct haplotypes of the C-terminal non-repeat region was identified in 28 Myanmar VK247 sequences. The dots represent residues identical to the reference sequence of PNG (M69059). Amino acid changes at particular amino acid positions are indicated as red. The dashes represent gaps to maximize the alignment. The total number of sequences for each hapoltype is listed in the right panel
Fig. 9.
Frequency of octa-peptide insertion and GGNA motif in the C-terminal non-repeat region of global pvcsp. a Frequency of ANKKAEDA octa-peptide insertion in the C-terminal non-repeat region of global VK210 variants. b Frequency of GGNA repeat motifs in the C-terminal non-repeat region of global VK210 variants. c Frequency of ANKKAGDA insertion in the C- terminal region of global VK247 variants. d) Frequency of GGNA repeat motifs in the C- terminal region of global VK247 variants
Nucleotide diversity and natural selection in the N- and C-terminal non-repeat regions of Myanmar pvcsp
Since the CRR of Myanmar pvcsp sequences showed a high degree of length polymorphisms, the nucleotide diversity and genetic differentiation of the N- and C-terminal non-repeat regions were analysed separately by omitting the CRR. In the N-terminal non-repeat region of Myanmar VK210 variants, the average number of nucleotide differences (K), overall haplotype diversity (Hd), and nucleotide diversity (π) were 0.098, 0.096 ± 0.034, and 0.0024 ± 0.0009, respectively (Table 1). The estimated dN–dS value in the N-terminal non-repeat regions was 0.0008. These results suggested that the N-terminal non-repeat region of Myanmar VK210 variants was under positive natural selection. Meanwhile, the average number of nucleotide differences (K), the overall haplotype diversity (Hd), and the nucleotide diversity (π) for the C-terminal non-repeat region of Myanmar VK210 variants were 0.209, 0.186 ± 0.044, and 0.0035 ± 0.0009, respectively (Table 1). The dN–dS value was –0.0035. These findings indicated that the C-terminal region was influenced by negative natural selection. The Tajima’s D test was also performed to further elucidate the effect of natural selection on the N- and C-terminal non-repeat regions in Myanmar VK210 variants. Tajima’s D values for the N- and C-terminal non-repeat regions were –1.9359 (P < 0.05) and –2.2251 (P < 0.01), respectively (Table 1). The Fu and Li’s D and F values of these regions also showed negative values. In the Myanmar VK247 variants, the average number of nucleotide differences (K), overall haplotype diversity (Hd) and nucleotide diversity (π) of the N-terminal non-repeat region were 0.405, 0.405 ± 0.094 and 0.0090 ± 0.0021, respectively (Table 1). These values for the C-terminal non-repeat region were 0.495, 0.331 ± 0.114 and 0.0067 ± 0.0027, respectively. The dN–dS values of the N- and C-terminal non-repeat regions of Myanmar VK247 variants were 0.0117 and –0.0150, respectively (Table 1). The Tajima’s D values were –0.4445 (P > 0.1) and –1.9719 (P < 0.05) for the N- and C-terminal non-repeat regions, respectively (Table 1). The Fu and Li’s D and F values for both regions were all negative.
Table 1.
Genetic polymorphism and tests of neutrality in the N-terminal and C-terminal regions of Myanmar pvcsp
| Variant | Region | n | K | S | Eta | H | Hd ± SD | π ± SD | dN-dS | Tajima’s DP value | Fu and Li’s DP value | Fu and Li’s FP value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VK210 | N-terminal | 143 | 0.098 | 5 | 6 | 7 | 0.096 ± 0.034 | 0.0024 ± 0.0009 | 0.0008 | − 1.9359c | − 3.9086b | − 3.8409b |
| C-terminal | 143 | 0.209 | 10 | 11 | 11 | 0.186 ± 0.044 | 0.0035 ± 0.0009 | −0.0035 | − 2.2251d | − 3.5089b | − 3.6329b | |
| VK247 | N-terminal | 28 | 0.405 | 1 | 2 | 3 | 0.405 ± 0.094 | 0.0090 ± 0.0021 | 0.0117 | − 0.4445a | − 0.7114a | − 0.7369a |
| C-terminal | 28 | 0.495 | 6 | 6 | 6 | 0.331 ± 0.114 | 0.0067 ± 0.0027 | − 0.0150 | − 1.9719c | − 2.5946c | − 2.8039c |
n number of analysed sequences, K average number of nucleotide differences, S number of segregating sites, Eta total number of mutations, H number of haplotypes, Hd haplotype diversity, π observed average pairwise nucleotide diversity, SD standard deviation, dN rate of non-synonymous mutations, dS rate of synonymous mutations
aP > 0.1
bP < 0.02
cP < 0.05
dP < 0.01
Nucleotide diversity and natural selection in the N- and C-terminal non-repeat regions of global VK210 variants
To further examine the nucleotide diversity and natural selection in the global pvcsp population, the nucleotide diversity of the N- and C-terminal non-repeat regions of global pvcsp was analysed. For the N-terminal non-repeat region of VK210 variants, nucleotide diversity and pattern of natural selection were differed by country (Table 2). VK210 variants from India showed the highest nucleotide diversity; the values of K, Hd, and π were 1.972, 0.661 ± 0.063, and 0.0470 ± 0.0074, respectively. The N-terminal non-repeat region of Cambodia VK210 variants also showed relatively high nucleotide diversity comparable to Myanmar VK210 variants. Substantial nucleotide diversity was found in the VK210 variants from South Korea and Brazil. However, VK210 variants from Iran, Mexico, Sudan, and Vanuatu were genetically well-conserved. The N-terminal non-repeat region of VK210 variants showed different patterns of natural selection by country. The dN–dS value was positive for VK210 variants from Myanmar, Cambodia, and South Korea, which indicated positive natural selection may occur in the region. Meanwhile, the values for VK210 variants from India and Brazil were negative, suggesting negative selection. The values of Tajima’s D for all VK210 variants derived from Myanmar, Cambodia, India, South Korea, and Brazil were negative, indicating that they were under purifying selection. The C-terminal non-repeat region of global VK210 variants also revealed nucleotide diversity and pattern of natural selection (Table 2). Indian VK210 variants showed the greatest nucleotide diversity with K, Hd, and π values of were 0.276, 0.123 ± 0.051, and 0.0043 ± 0.0020, respectively. Meanwhile, no nucleotide diversity was detected in VK210 variants from Cambodia, Iran, Brazil, Mexico, and Vanuatu. Similar to the N-terminal non-repeat region, the C-terminal non-repeat region of global VK210 was also under the effect of purifying selection based on the negative values of Tajima’s D. The values of Fu and Li’s D and Fu and Li’s F were also negative for the C-terminal region of VK210 variants from Myanmar, India, South Korea, and Sudan.
Table 2.
Genetic polymorphism and tests of neutrality in the N-terminal and C-terminal non-repeat regions of global VK210 variants
| Region | Country | n | K | S | Eta | H | Hd ± SD | π ± SD | dN-dS | Tajima’s DP value | Fu and Li’s DP value | Fu and Li’s FP value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N-terminal | Myanmar | 143 | 0.098 | 5 | 6 | 7 | 0.096 ± 0.034 | 0.0024 ± 0.0009 | 0.0008 | − 1.9359c | − 3.9086b | − 3.8409b |
| Cambodia | 31 | 0.125 | 1 | 1 | 2 | 0.125 ± 0.077 | 0.0030 ± 0.0018 | 0.0039 | − 0.7737a | 0.5907a | 0.2450a | |
| India | 79 | 1.972 | 23 | 28 | 28 | 0.661 ± 0.063 | 0.0470 ± 0.0074 | − 0.0909 | − 2.0261c | − 1.3952a | − 1.9514e | |
| Iran | 39 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| South Korea | 39 | 0.051 | 1 | 1 | 2 | 0.051 ± 0.048 | 0.0012 ± 0.0011 | 0.0016 | − 1.1264a | − 1.7662a | − 1.8293a | |
| Brazil | 41 | 0.049 | 1 | 1 | 2 | 0.049 ± 0.046 | 0.0012 ± 0.0011 | − 0.0059 | − 1.1219a | − 1.7816a | − 1.8406a | |
| Mexico | 11 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Sudan | 30 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Vanuatu | 21 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| C-terminal | Myanmar | 143 | 0.209 | 10 | 11 | 11 | 0.186 ± 0.044 | 0.0035 ± 0.0009 | − 0.0035 | − 2.2251d | − 3.5089b | − 3.6329b |
| Cambodia | 31 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| India | 79 | 0.276 | 8 | 9 | 6 | 0.123 ± 0.051 | 0.0043 ± 0.0020 | − 0.0149 | − 2.1953d | − 4.4419b | − 4.3524b | |
| Iran | 39 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| South Korea | 39 | 0.103 | 2 | 2 | 3 | 0.101 ± 0.065 | 0.0008 ± 0.0005 | − 0.0037 | − 1.4889a | − 2.4148e | − 2.4864e | |
| Brazil | 41 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Mexico | 11 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Sudan | 30 | 0.067 | 1 | 1 | 2 | 0.067 ± 0.061 | 0.0009 ± 0.0008 | − 0.0041 | − 1.1470a | − 1.6821a | − 1.7655a | |
| Vanuatu | 21 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
n number of analysed sequences, K avarage number of nucleotide differences, S number of segregating sites, Eta total number of mutations, H number of haplotypes, Hd haplotype diversity, π observed average pairwise nucleotide diversity, SD standard deviation, dN rate of non-synonymous mutations, dS rate of synonymous mutations
aP > 0.1
bP < 0.02
cP < 0.05
dP < 0.01
e0.05 < P < 0.1
Nucleotide diversity and natural selection in the N-terminal and C-terminal non-repeat regions of global VK247 variants
The nucleotide diversity and natural selection in the global VK247 variants were analysed (Table 3). Analysis of the N-terminal non-repeat region of VK247 variants revealed the greatest nucleotide diversity in VK247 variants from Iran, with the values of K, Hd, and π of 1.309, 0.436 ± 0.133, and 0.0190 ± 0.0058, respectively. VK247 variants from Cambodia also revealed nucleotide diversity in the region. However, no nucleotide diversity was found in VK247 variants from Mexico and Colombia. The N-terminal non-repeat region of VK247 variants from Iran showed negative dN–dS (–0.0543) and positive Tajima’s D (0.9518), Fu and Li’s D (1.1271), and Fu and Li’s F (1.2185). However, Myanmar and Cambodia VK247 variants revealed positive values for dN–dS and negative values for Tajima’s D, Fu and Li’s D, and Fu and Li’s F. The C-terminal non-repeat region of VK267 variants from Cambodia and Colombia also showed nucleotide diversity, which was lower than that of Myanmar VK247 variants. Similar to Myanmar VK247 variants, the C-terminal non-repeat region of Cambodia VK247 variants showed a negative values of dN–dS (–0.0172), Tajima’s D (–1.4009), Fu and Li’s D (–1.5866), and Fu and Li’s F (–1.7190). However, the dN–dS of Colombia variants was estimated to be positive (0.0017), although the values of Tajima’s D, Fu and Li’s D, and Fu and Li’s F were negative.
Table 3.
Genetic polymorphism and tests of neutrality in N-terminal and C-terminal regions of global pvcsp VK247 variants
| Region | Country | n | K | S | Eta | H | Hd ± SD | π ± SD | dN-dS | Tajima’s DP value | Fu andLi’s DP value | Fu and Li’s FP value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N-terminal | Myanmar | 28 | 0.405 | 1 | 2 | 3 | 0.405 ± 0.094 | 0.0090 ± 0.0021 | 0.0117 | − 0.4445a | − 0.7114a | − 0.7369a |
| Cambodia | 10 | 0.2 | 1 | 1 | 2 | 0.200 ± 0.154 | 0.0029 ± 0.0022 | 0.0038 | − 1.1117a | − 1.2434a | − 1.3467a | |
| Iran | 11 | 1.309 | 3 | 3 | 2 | 0.436 ± 0.133 | 0.0190 ± 0.0058 | − 0.0543 | 0.9518a | 1.1271a | 1.2185a | |
| Mexico | 8 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Colombia | 25 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| C-terminal | Myanmar | 28 | 0.495 | 6 | 6 | 6 | 0.331 ± 0.114 | 0.0067 ± 0.0027 | − 0.0150 | − 1.9719c | − 2.5946c | − 2.8039c |
| Cambodia | 10 | 0.400 | 2 | 2 | 2 | 0.200 ± 0.154 | 0.0039 ± 0.0030 | − 0.0172 | − 1.4009a | − 1.5866a | − 1.7190a | |
| Iran | 11 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Mexico | 8 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Colombia | 25 | 0.500 | 2 | 2 | 3 | 0.44 ± 0.095 | 0.0039 ± 0.0010 | 0.0017 | − 0.1215a | − 0.6754a | − 0.6012a |
n number of analysed sequences, K average number of nucleotide differences, S number of segregating sites, Eta total number of mutations, H number of haplotypes, Hd haplotype diversity, π observed average pairwise nucleotide diversity, SD standard deviation, dN rate of non-synonymous mutations, dS rate of synonymous mutations
aP > 0.1
bP < 0.02
cP < 0.05
dP < 0.01
e0.05 < P < 0.1
Discussion
PvCSP is one of the leading candidates for vivax malaria vaccine. Several recent PvCSP-derived vaccines were designed as multivalent formulations or chimeric molecules in an attempt to induce protective immunity against P. vivax [11, 21, 22]. However, the impact of natural genetic variations in the global pvcsp population on vaccine efficacy remains unclear. In this study, genetic polymorphism and natural selection in Myanmar pvcsp and global pvcsp populations were comprehensively analysed. Among the three pvcsp allelic variants, only two allelic variants, VK210 and VK247, were identified in P. vivax isolates from Myanmar analysed in this study. The VK210 was the dominant allele occurring in 83.6% of Myanmar pvcsp population, which is consistent with previous studies reporting that VK210 is the most common allelic variant in the pvcsp populations from Iran, India, China, Brazil, Thailand, Bangladesh, and Azerbaijan [23–31]. Meanwhile, VK247 was more prevalent in certain regions of Colombia [32]. It has been suggested that the distribution of Anopheles mosquito species may affect the prevalence of VK210 and VK247 variants in endemic areas [32, 33]. In Myanmar, diverse species of Anopheles are distributed throughout the country, and at least 10 species including Anopheles dirus, Anopheles minimus, and Anopheles aconitus may transmit malaria [34]. Differences in infectivity of VK210 and VK247 variants in certain Anopheles species is not clear; however, the differences in adaptability of each allelic variant to mosquito vectors affects the prevalence of VK210 in Myanmar. The predominance of VK210 may also be associated with its genetic diversity. VK247 showed a lower level of genetic diversity than VK210 in the Myanmar pvcsp population, suggesting different polymorphisms may increase the adaptability of VK210 to different mosquito vectors.
The N-terminal non-repeat region of Myanmar pvcsp was relatively well-conserved. Only a few amino acid changes were identified in the latter portion of the N-terminal non-repeat region of Myanmar pvcsp. The RI motif (KLKQP), a cell adhesive motif exposed by proteolytic cleavage after the interaction between sporozoite and hepatocyte [8], was well conserved in both Myanmar VK210 and VK247 variants. Global VK210 variants also displayed limited polymorphism in the N-terminal non-repeat region. An alanine insertion at the end of the RI was the major variation identified in global pvcsp, although the prevalence of the insertion differed by geographically. Amino acid changes were also identified in global VK210 variants, but they showed uneven geographic distribution with low frequencies. The N-terminal non-repeat region of global VK247 variants was also relatively well-conserved, although uneven and minor amino acid changes were also identified. It has been shown that the invasion of malaria parasite is inhibited by antibodies directed against RI motif [35]. The highly conserved genetic characteristic of the N-terminal region of global pvcsp suggest that this region including RI motif may represent an attractive candidate for formulation of a PvCSP-based vaccine.
As expected, high level of genetic diversity was identified in the CRR of Myanmar pvcsp. Both VK210 and VK247 variants showed great genetic diversity in CRR resulting in 118 and 23 different haplotypes for VK210 and VK247, respectively. Insertions and deletions of nonapeptide sequences probably resulted from either sexual recombination during meiosis or intrahelical strand slippage during mitotic DNA replication [24], which may generate novel haplotypes in Myanmar pvcsp. Interestingly, 47 different types of PRMs including 27 novel types were identified in the CRR of Myanmar VK210 variants. In the case of VK247 variants, a total of 8 distinct PRMs were identified, in which 5 were novel ones. These findings also suggest point mutations as one of the major factors increasing the genetic complexity of Myanmar pvcsp. Similar patterns of high genetic polymorphisms were also identified in the CRR of global pvcsp population; however, the overall diversity due to different types and repeats of PRMs was greater in the pvcsp from Myanmar compared with other countries. Especially, the repeated numbers of PRMs varied widely in both Myanmar VK210 and VK247 variants compared with those from other countries.
The C-terminal regions of Myanmar and global pvcsp also showed genetic polymorphisms. Differences in ANKKAEDA octapeptide insertion and GGNA tetrapeptide repeat motifs and amino acid substitutions throughout the region contributed to genetic diversity in Myanmar and global VK210 variants. Despite the unclear biological functions of ANKKAEDA octapeptide and GGNA tetrapeptide in VK210 variants, they may also be generated via intrahelical recombination, and parasites carrying the ANKKAEDA octapeptide show a high degree of delayed infections [11]. Similar to VK210 variants, ANKKAGDAG octapeptide and GGNA motifs were important factors contributing to genetic diversity in Myanmar and global VK247 variants. Although the frequency of octapeptide insertion and number of GGNA motifs in global pvcsp, both in VK210 and VK247 variants, were differed by country, no clear geographic clustering was identified. Considering the limited genetic information of the C-terminal region in global pvcsp in current, a further study employing larger numbers of pvcsp sequences in diverse geographical origins is necessary to understand the genetic nature and evolutionary aspects of the C-terminal region in global pvcsp population.
Analysis of the two non-repeat regions in global pvcsp suggests that these regions are likely to be under natural selection, which may maintain or generate genetic diversity in the global pvcsp population. The dN–dS values for Myanmar and Cambodia VK210 and VK247 variants were positive, implying that balancing selection might act in these regions. However, the values were 0 or negative for both VK210 and VK247 variants from other countries except the N-terminal region of South Korea VK210 and the C-terminal region of Colombia VK247. Tajima’s D and Fu and Li’s D and F values also revealed complex patterns of natural selection that were unique to pvcsp from each country. While pvcsp populations from a few countries showed no genetic diversity involving the N- and C-terminal regions, a few pvcsp populations showed negative Tajima’s D values, suggesting purifying natural selection. The overall trend indicated that the global pvcsp population decreased the genetic diversity in the two terminal non-repeat regions. Due to size polymorphisms induced by different numbers and arrangements of PRMs in CRR, a direct analysis of natural selection in CRR was not possible in this study. However, evidence supporting natural selection in the CRR have been reported [36–38]. These findings collectively suggest that complex natural selection phenomena may act on the global pvcsp.
Despite the remarkable reduction in malaria transmission in Myanmar during the last decades [2, 38], the high levels of genetic diversity among malaria parasites appears to persist in the country. As demonstrated in this study, the overall genetic polymorphisms of Myanmar pvcsp were greater than those reported from other countries analysed. Similar patterns of high genetic diversity in several polymorphic genetic markers including apical membrane antigen-1, merozoite surface protein-1 and -2, and CSP of Myanmar P. falciparum population have also been reported [39–42]. Despite the lack of clear insight into the high genetic diversity underlying the population decline, asymptomatic carriers may act as fundamental reservoirs contributing to malaria transmission, which provides adequate population size to maintain or generate genetic diversity of malaria parasites in Myanmar [42]. This study also had a limitation. The Myanmar P. vivax isolates analysed in this study were collected in restricted areas of Myanmar and, therefore, not fully representative of the nation-wide genetic diversity and population structure of Myanmar pvcsp. A further comprehensive study involving a larger number of P. vivax isolates collected from different regions in Myanmar is necessary.
Conclusions
PvCSP is a leading candidate for vivax vaccine; however, the impact of natural genetic variations in the global pvcsp population on the efficacy of PvCSP-based vaccine remains unclear. This study highlights an additional level of complexity in the application of this antigen for the development of a vivax vaccine. Global pvcsp population revealed polymorphic characters, especially in the CRR. The size polymorphisms and the appearance of novel PRMs as well as new CRR arrays primarily contribute to the genetic diversity of global pvcsp. Natural selection may also be a major force affecting the genetic diversity of global pvcsp. The results of this study provide not only an insight into the genetic nature of global pvcsp but also valuable information for the development of a universal vaccine based on PvCSP. Continuous monitoring of genetic diversity of global pvcsp population is necessary to elucidate the polymorphic nature and evolutionary aspect of pvcsp in global P. vivax population.
Supplementary information
Additional file 1: Figure S1. Schematic structure of pvcsp. The gene is separated into three regions; an N-terminal non-repeat region, a central repeat region (CRR), and a C-terminal non-repeat region. The CRR consists of two major repeat peptide motifs (PRMs), termed VK210 and VK247.
Additional file 2: Table S1. Global pvcsp sequences analysed in this study.
Acknowledgements
We thank the staffs in the Department of Medical Research Pyin Oo Lwin Branch and the health professionals in Naung Cho and Pyin Oo Lwin townships for their contribution and technical support during blood collection.
Abbreviations
- CRR
Central repeat region
- CSP
Circumsporozoite surface protein
- dN
Rate of non-synonymous mutations
- dS
Rate of synonymous mutations
- H
Number of haplotypes
- Hd
Haplotype diversity
- K
Average number of nucleotide differences
- PCR
Polymerase chain reaction
- PRM
Peptide repeat motif
- PvCSP
Plasmodium vivax circumsporozoite surface protein
- rRNA
18S ribosomal RNA
- S
Number of segregating sites
- π
Observed average pairwise nucleotide diversity
Authors’ contributions
TCV, HGL and JMK performed the genetic analysis of pvcsp. MM, HN, MKM, JL and KL contributed to blood sample collection. TCV, GHL, JMK, JL, WMS and BKN analysed and interpreted the data. BKN designed and supervised the experiments. TCV and BKN drafted the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the National Research Foundation of Korea (NRF) Grants funded by the Korean Government (NRF-2019K1A3A9A01000005).
Availability of data and materials
The data supporting the conclusions of this article are provided within the article and its additional files. The original datasets analysed in this current study are available from the corresponding author upon request. The nucleotide sequences reported in this study have been deposited in the GenBank database under the Accession Numbers MN821829–MN821999.
Ethics approval and consent to participate
This study was approved by the Ethics Review Committee, Department of Medical Research, Myanmar (97/Ethics 2015) and by the Ethical Review Committee of Inha University School of Medicine, Korea (INHA 15-013). Informed written consent and permission were obtained from each individual.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12936-020-03366-7.
References
- 1.Cox FE. History of human parasitology. Clin Microbiol Rev. 2002;15:595–612. doi: 10.1128/CMR.15.4.595-612.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. World Malaria report 2018. Geneva, World Health Organization, 2018.
- 3.Rogerson SJ, Carter R. Severe vivax malaria: newly recognised or rediscovered. PLoS Med. 2008;5:e136. doi: 10.1371/journal.pmed.0050136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anstey NM, Russell B, Yeo TW, Price RN. The pathophysiology of vivax malaria. Trends Parasitol. 2009;25:220–227. doi: 10.1016/j.pt.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Baird JK. Chloroquine resistance in Plasmodium vivax. Antimicrob Agents Chemother. 2004;48:4075–4083. doi: 10.1128/AAC.48.11.4075-4083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM. Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007;77:79–87. doi: 10.4269/ajtmh.2007.77.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 8.Coppi A, Natarajan R, Pradel G, Bennett BL, James ER, Roggero MA, et al. The malaria circumsporozoite protein has two functional domains, each with distinct roles as sporozoites journey from mosquito to mammalian host. J Exp Med. 2011;208:341–356. doi: 10.1084/jem.20101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg R, Wirtz RA, Lanar DE, Sattabongkot J, Hall T, Waters AP, et al. Circumsporozoite protein heterogeneity in the human malaria parasite Plasmodium vivax. Science. 1989;245:973–976. doi: 10.1126/science.2672336. [DOI] [PubMed] [Google Scholar]
- 10.Qari SH, Shi YP, Goldman IF, Udhayakumar V, Alpers MP, Collins WE, et al. Identification of Plasmodium vivax-like human malaria parasite. Lancet. 1993;341:780–783. doi: 10.1016/0140-6736(93)90559-Y. [DOI] [PubMed] [Google Scholar]
- 11.Yadava A, Sattabongkot J, Washington MA, Ware LA, Majam V, Zheng H, et al. A novel chimeric Plasmodium vivax circumsporozoite protein induces biologically functional antibodies that recognize both VK210 and VK247 sporozoites. Infect Immun. 2007;75:1177–1185. doi: 10.1128/IAI.01667-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett JW, Yadava A, Tosh D, Sattabongkot J, Komisar J, Ware LA, et al. Phase 1/2a trial of Plasmodium vivax malaria vaccine candidate VMP001/AS01B in malaria-naive adults: safety, immunogenicity, and efficacy. PLoS Negl Trop Dis. 2016;10:e0004423. doi: 10.1371/journal.pntd.0004423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993;58:283–292. doi: 10.1016/0166-6851(93)90050-8. [DOI] [PubMed] [Google Scholar]
- 14.Kang JM, Cho PY, Moe M, Lee J, Jun H, Lee HW, et al. Comparison of the diagnostic performance of microscopic examination with nested polymerase chain reaction for optimum malaria diagnosis in Upper Myanmar. Malar J. 2017;16:119. doi: 10.1186/s12936-017-1765-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 16.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 18.Escalante AA, Cornejo OE, Rojas A, Udhayakumar V, Lal AA. Assessing the effect of natural selection in malaria parasites. Trends Parasitol. 2004;20:388–395. doi: 10.1016/j.pt.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gimenez AM, Lima LC, Francoso KS, Denapoli PMA, Panatieri R, Bargieri DY, et al. Vaccine containing the three allelic variants of the Plasmodium vivax circumsporozoite antigen induces protection in mice after challenge with a transgenic rodent malaria parasite. Front Immunol. 2017;8:1275. doi: 10.3389/fimmu.2017.01275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teixeira LH, Tararam CA, Lasaro MO, Camacho AG, Ersching J, Leal MT, et al. Immunogenicity of a prime-boost vaccine containing the circumsporozoite proteins of Plasmodium vivax in rodents. Infect Immun. 2014;82:793–807. doi: 10.1128/IAI.01410-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shabani SH, Zakeri S, Mehrizi AA, Mortazavi Y, Djadid ND. Population genetics structure of Plasmodium vivax circumsporozoite protein during the elimination process in low and unstable malaria transmission areas, southeast of Iran. Acta Trop. 2016;160:23–34. doi: 10.1016/j.actatropica.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Kim JR, Imwong M, Nandy A, Chotivanich K, Nontprasert A, Tonomsing N, et al. Genetic diversity of Plasmodium vivax in Kolkata, India. Malar J. 2006;5:71. doi: 10.1186/1475-2875-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machado RL, Povoa MM. Distribution of Plasmodium vivax variants (VK210, VK247 and P. vivax-like) in three endemic areas of the Amazon region of Brazil and their correlation with chloroquine treatment. Trans R Soc Trop Med Hyg. 2000;94:377–381. doi: 10.1016/S0035-9203(00)90110-X. [DOI] [PubMed] [Google Scholar]
- 26.Cui L, Mascorro CN, Fan Q, Rzomp KA, Khuntirat B, Zhou G, et al. Genetic diversity and multiple infections of Plasmodium vivax malaria in Western Thailand. Am J Trop Med Hyg. 2003;68:613–619. doi: 10.4269/ajtmh.2003.68.613. [DOI] [PubMed] [Google Scholar]
- 27.Leclerc MC, Menegon M, Cligny A, Noyer JL, Mammadov S, Aliyev N, et al. Genetic diversity of Plasmodium vivax isolates from Azerbaijan. Malar J. 2004;3:40. doi: 10.1186/1475-2875-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pratt-Riccio LR, Baptista BO, Torres VR, Bianco-Junior C, Perce-Da-Silva DS, Riccio EKP, et al. Chloroquine and mefloquine resistance profiles are not related to the circumsporozoite protein (CSP) VK210 subtypes in field isolates of Plasmodium vivax from Manaus. Brazilian Amazon. Mem Inst Oswaldo Cruz. 2019;114:e190054. doi: 10.1590/0074-02760190054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kibria MG, Elahi R, Mohon AN, Khan WA, Haque R, Alam MS. Genetic diversity of Plasmodium vivax in clinical isolates from Bangladesh. Malar J. 2015;14:267. doi: 10.1186/s12936-015-0790-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li YC, Wang GZ, Meng F, Zeng W, He CH, Hu XM, et al. Genetic diversity of Plasmodium vivax population before elimination of malaria in Hainan Province China. Malar J. 2015;14:78. doi: 10.1186/s12936-015-0545-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaur H, Sehgal R, Kumar A, Sehgal A, Bharti PK, Bansal D, et al. Exploration of genetic diversity of Plasmodium vivax circumsporozoite protein (Pvcsp) and Plasmodium vivax sexual stage antigen (Pvs25) among North Indian isolates. Malar J. 2019;18:308. doi: 10.1186/s12936-019-2939-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez JM, Hurtado S, Arevalo-Herrera M, Herrera S. Variants of the Plasmodium vivax circumsporozoite protein (VK210 and VK247) in Colombian isolates. Mem Inst Oswaldo Cruz. 2001;96:709–712. doi: 10.1590/S0074-02762001000500023. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez MH, Gonzalez-Ceron L, Hernandez JE, Nettel JA, Villarreal C, Kain KC, et al. Different prevalences of Plasmodium vivax phenotypes VK210 and VK247 associated with the distribution of Anopheles albimanus and Anopheles pseudopunctipennis in Mexico. Am J Trop Med Hyg. 2000;62:122–127. doi: 10.4269/ajtmh.2000.62.122. [DOI] [PubMed] [Google Scholar]
- 34.Ool TT, Storch V, Becker N. Review of the anopheline mosquitoes of Myanmar. J Vector Ecol. 2004;29:21–40. [PubMed] [Google Scholar]
- 35.Ying P, Shakibaei M, Patankar MS, Clavijo P, Beavis RC, Clark GF, et al. The malaria circumsporozoite protein: interaction of the conserved regions I and II-plus with heparin-like oligosaccharides in heparan sulfate. Exp Parasitol. 1997;85:168–182. doi: 10.1006/expr.1996.4134. [DOI] [PubMed] [Google Scholar]
- 36.Hughes AL. The evolution of amino acid repeat arrays in Plasmodium and other organisms. J Mol Evol. 2004;59:528–535. doi: 10.1007/s00239-004-2645-4. [DOI] [PubMed] [Google Scholar]
- 37.Patil A, Orjuela-Sanchez P, da Silva-Nunes M, Ferreira MU. Evolutionary dynamics of the immunodominant repeats of the Plasmodium vivax malaria-vaccine candidate circumsporozoite protein (CSP) Infect Genet Evol. 2010;10:298–303. doi: 10.1016/j.meegid.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parobek CM, Bailey JA, Hathaway NJ, Socheat D, Rogers WO, Juliano JJ. Differing patterns of selection and geospatial genetic diversity within two leading Plasmodium vivax candidate vaccine antigens. PLoS Negl Trop Dis. 2014;8:e2796. doi: 10.1371/journal.pntd.0002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WHO. Malaria in the Greater Mekong Subregion: regional and country profiles. Geneva, World Health Organization, 2010.
- 40.Kang JM, Lee J, Moe M, Jun H, Le HG, Kim TI, et al. Population genetic structure and natural selection of Plasmodium falciparum apical membrane antigen-1 in Myanmar isolates. Malar J. 2018;17:71. doi: 10.1186/s12936-018-2215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le HG, Kang JM, Moe M, Jun H, Thai TL, Lee J, et al. Genetic polymorphism and natural selection of circumsporozoite surface protein in Plasmodium falciparum field isolates from Myanmar. Malar J. 2018;17:361. doi: 10.1186/s12936-018-2513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le HG, Kang JM, Jun H, Lee J, Thai TL, Myint MK, et al. Changing pattern of the genetic diversities of Plasmodium falciparum merozoite surface protein-1 and merozoite surface protein-2 in Myanmar isolates. Malar J. 2019;18:241. doi: 10.1186/s12936-019-2879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Schematic structure of pvcsp. The gene is separated into three regions; an N-terminal non-repeat region, a central repeat region (CRR), and a C-terminal non-repeat region. The CRR consists of two major repeat peptide motifs (PRMs), termed VK210 and VK247.
Additional file 2: Table S1. Global pvcsp sequences analysed in this study.
Data Availability Statement
The data supporting the conclusions of this article are provided within the article and its additional files. The original datasets analysed in this current study are available from the corresponding author upon request. The nucleotide sequences reported in this study have been deposited in the GenBank database under the Accession Numbers MN821829–MN821999.