Abstract
Activation of protein phosphatase 2A (PP2A) is a promising anti-cancer therapeutic strategy, as this tumor suppressor has the ability to coordinately downregulate multiple pathways involved in the regulation of cellular growth and proliferation. In order to understand the systems-level perturbations mediated by PP2A activation, we carried out mass spectrometry-based phosphoproteomic analysis of two KRAS mutated non-small cell lung cancer (NSCLC) cell lines (A549 and H358) treated with a novel Small Molecule Activator of PP2A (SMAP). Overall, this permitted quantification of differential signaling across over 1,600 phosphoproteins and 3,000 phosphosites. Kinase activity assessment and pathway enrichment implicated collective downregulation of RAS and cell cycle kinases in the case of both cell lines upon PP2A activation. However, the effects on RAS-related signaling was attenuated for A549 compared to H358, while the effects on cell cycle related kinases were noticeably more prominent in A549. Network-based analyses and validation experiments confirmed these detailed differences in signaling. These studies reveal the power of phosphoproteomics studies, coupled to computational systems biology, to elucidate global patterns of phosphatase activation and understand the variations in response to PP2A activation across genetically similar NSCLC cell lines.
Keywords: Label-free, non-small cell lung cancer, phosphatase, phosphoproteomics, signaling
1. Introduction
As a serine/threonine phosphatase that antagonizes the effects of multiple kinases, PP2A is a promising anti-cancer target whose activation may disrupt many cancer-promoting pathways simultaneously. PP2A is a well-known tumor suppressor that in many cancer models dephosphorylates prominent downstream effectors of KRAS, such as AKT, MEK, ERK, and MYC, among others [1–3]. Furthermore, PP2A inactivation is a common feature of cancer development, as it is frequently disabled in multiple cancer types [4–7]. Overall, activated PP2A has the potential to reverse the general responses mediated by oncogenic kinases often required for cancer development and maintenance [1,8–10].
PP2A activation has been shown to be a promising anti-cancer strategy in numerous models including chronic myeloid leukemia cell lines [7] and T cell acute lymphoblastic leukemia [11]. More recently, the pro-apoptotic and anti-tumor properties of a class of anti-psychotic drugs have been described [12], and their anti-cancer activity has been attributed to their ability to activate PP2A [11]. Reverse engineering of these drugs resulted in a first-in-class series of SMAPs that activates this family of phosphatases but lacks the main dose-limiting toxicities associated with the parent molecules [13]. Follow-up assays in NSCLC models have confirmed that these molecules bind to the PP2A trimer, and they demonstrated pro-apoptotic, anti-proliferative properties that are blunted upon PP2A inhibition [14].
A thorough understanding of these compounds’ biological effects and clinical utility requires comprehensive identification of their net signaling effects. Notably, the detailed characterization of SMAPs’ cellular effects at the protein level, where phosphorylation-mediated signaling is directly regulated, will enable us to identify key pathways and targets of SMAP regulation. Due to the ever-expanding catalog of known PP2A targets, SMAP molecules may induce a complex signaling response that would be difficult to fully characterize through targeted approaches. High throughput, mass spectrometry-based phosphoproteomics can identify and quantitate thousands of peptides that may be differentially phosphorylated in the context of disease and/or drug treatment [15], and these data, in turn, help identify cellular signaling patterns that reflect the protein-level pathway and network response phenotype.
Ultimately, the experimental and computational pipelines presented here intend to accomplish the following: 1) generate phosphoproteomic datasets of two KRAS mutant NSCLC cell lines treated with our PP2A activator; and 2) characterize the major systems-level signaling perturbations induced by SMAP treatment. Furthermore, the bioinformatic findings would reveal unique response signatures between these cell lines, which share mutations in a major oncogene. Ultimately, these findings can contribute to a better understanding of SMAP’s mechanism of action and reflect the potential variations in patient-specific responses to drug at the protein level.
2. Materials and methods
2.1. Cell Culture
All cell lines were acquired from the American Type Culture Collection (ATCC) and maintained in media (RPMI-1640, 10% FBS, 0.5% Pen/Strep) at 37°C and 5% CO2. The TRC-794 compound was dissolved in DMSO solvent and stored at room temperature.
2.2. Cell Viability Assay
Cell viability was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) kit (Sigma-Aldrich). All measurements were performed 24 hours after treatment with the increasing concentrations of TRC-794. Values in the plot are relative to the DMSO control samples.
2.3. Annexin V
Cells were treated with DMSO, TRC-794 (20μM), Z-VAD (100μM), and a combination of TRC-794 + Z-VAD for 24 hours. Annexin V staining was performed using Annexin V conjugate Alexa Fluor 488 from Invitrogen (Life Technologies), Annexin binding buffer (no. V13246) from Invitrogen (Life Technologies), and Z-VAD (OMe)-FMK (no. sc-311561A) from Santa Cruz Biotechnology according to the manufacturer’s protocol. Flow cytometric analysis was performed on the FACSCalibur.
2.4. Cell Cycle Analysis
Cells were treated with either TRC-794 or DMSO for 24 hours and subsequently fixed in 100% ethanol at −20°C overnight. Cells were then resuspended in propidium iodide (Sigma-Aldrich) and RNAseA (Roche) and allowed to stain for 20 minutes prior to flow cytometry analysis, performed on the FACSCalibur.
2.5. Phosphoproteomics Experimental Design
Two cell lines, A549 and H358, were tested, with each one having 3 independent replicates of DMSO (control) treatment and 3 independent replicates of SMAP treatment, all incubated for 12hrs. After protein lysis, each sample was further divided into 2 technical replicates to assess the consistency of the LC-MS/MS runs. In total, there were 6 Controls + 6 Treatments = 12 runs per cell line. Note that one technical replicate in the treated A549 group was ultimately excluded due to poor detection of phospho-enriched peptides. Based on optimization studies (unpublished), a 12hr time-point allowed for reproducible dephosphorylation of ERK1/2, a target previously shown to be differentially phosphorylated with SMAP [13,14], without induction of apoptosis. This strategy was intended to limit the secondary signaling that may accumulate from cell death. Additionally, a priori power analysis required n = 3 per group in order to achieve a power = 0.8 and alpha = 0.05 based on the effect size of phospho-ERK2.
Peptide/protein data from the featured phosphoproteomics experiment were compiled into Supporting Information Table 1. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD005698 and 10.6019/PXD005698. Supporting Information Table 3 matches the uploaded file names to the specific treatment conditions.
2.6. Label-Free Phosphoproteomics Sample Preparation and Phosphopeptide Enrichment
Cells were treated with fresh media containing either 0.025% (v/v%) DMSO (control) or 20μM of TRC-794 for 12hrs. Samples were harvested and triple-washed with ice-cold PBS prior to storing cell pellets in −80°C for subsequent phosphoproteomic analysis. Cell pellets from the previous step were pulse sonicated in 2% SDS, 50mM Tris buffer, pH 8 over ice and in the presence of protease and phosphatase inhibitors (Sigma Aldrich). The samples were centrifuged, and the supernatants were reduced (25mM DTT for 1 hour at 37C), alkylated (25mM iodoacetamide for 30min at room temperature in the dark), and cleared of SDS using the previously-published FASP protocol [16]. The protein concentration in the cleaned supernatant was then measured using a protein assay kit (Bio-Rad). 800 micrograms of protein per sample were then digested with Lys-C for 1 hour at 37°C, followed by Trypsin overnight at 37°C (1:20 enzyme (g) to protein (g) ratio for each enzyme; final mixture concentrations were adjusted to 2M Urea and 50mM Tris buffer, pH 8). Digested peptides were desalted using C18 cartridges (Oasis 1cc HLB. Waters), eluted in 0.1% formic acid and 70% acetonitrile, and lyophilized for 1 hour. Samples were reconstituted with buffer containing 0.3% trifluoroacetic acid (TFA), 60% acetonitrile, and 13% lactic acid. Subsequent phosphopeptide enrichment was performed using a titanium dioxide spin tips (Thermo Fisher, IL). Samples were eluted using 1.5% ammonium hydroxide and concentrated before combined with 0.1% formic acid (FA) in HPLC grade water.
2.7. LC-MS/MS
400 nanograms of each phospho-enriched peptide mixture were analyzed by LC-MS/MS using a LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, CA) equipped with a nanoAcquity Ultra-high pressure liquid chromatography system (Waters, MA). Separation and detection of peptides were performed as previously described [17,18]. LC-MS/MS raw data were acquired in a data-dependent mode using Xcalibur (Thermo Scientific, 2.2 SP1). The injection order was randomized, and the technician was blinded to the sample labels.
2.8. Data Processing
The LC-MS/MS raw files were imported into Rosetta Elucidator™ (Rosetta, 3.3.0.1.SP.25) and processed as previously described [18]. The aligned and normalized peaks were annotated at the feature level by generating database search files (*.dta). The files were searched by Mascot (version 2.3.01) against the human Universal Protein Resources (UniProt) (20,233 sequences) database using the following parameters: trypsin enzyme specificity; mass accuracy window for precursor ion, 10 ppm; mass accuracy window for fragment ions, 0.8 Da; carbamidomethylation of cysteines as fixed modifications; oxidation of methionine, phosphorylation of serine, threonine and tyrosine as variable modification; and 1 missed cleavage. Comparison of the runs against decoy searches yielded an average false discovery rate (FDR) of 1.3% ± 0.3% across all samples.
Relative quantitation was based on area under the curve. Subsequently, each peptide ion intensity was normalized by the median intensity within its replicate sample. The phosphopeptide intensity values for every technical replicate were then averaged, and the resulting datasets were analyzed for differential phosphorylation between DMSO vs. TRC-794 using Welch’s t-test, 2-tailed (Supporting Information Table 1). The resulting p-values were adjusted for multiple-hypothesis testing using the Benjamini-Hochberg method and reported in the supplementary table. However, since at this step we were performing a discovery-based study, we refrained from any filtering based on the corrected p-values in order to maximize hypothesis generation. Unless noted, all subsequent discussion of the t-test results involved the raw p-values.
2.9. Kinase-Substrate Enrichment Analysis (KSEA)
KSEA was used to infer kinases’ relative activities upon SMAP treatment using the global phosphoproteomics data. This method relies on a fundamental assumption that a kinase’s differential activity is positively correlated with the collective phosphorylation changes of its substrates. Calculations were performed using R version 3.3.0. Please refer to the original publication [19] for details on KSEA and the statistical scoring. Our analysis was based on the third formula variation described in their Materials and Methods. We assigned kinase-substrate (K-S) links based on the Kinase Substrate Dataset from PhosphoSitePlus (July 2016 release) [20], search restricted to human proteins. To maximize the coverage of usable phosphosites from our experiment, this K-S dataset was supplemented with predicted K-S links using the NetworKIN method [21]. We pulled these additional relationships from pre-computed data, downloaded from the KinomeXplorer-DB website [22], for all available kinase predictors against ENSEMBL version 59. Only predictions with scores 5 and above were considered. Each site’s fold change was derived by taking the ratio of the mean peptide ion intensity of SMAP over that of DMSO. Normalized scores (calculated from the weighted z-scores, as described in original paper) were reported. Fold change values were averaged for identical phosphosites detected across multiple peptides.
2.10. Reactome Pathway Enrichment Analysis
We performed Fisher’s exact test to identify the processes that are enriched in proteins differentially phosphorylated between the two cell lines. Analysis was performed at the protein level, in which the p-value and phosphorylation fold change values for each protein were selected from the peptide with the lowest p-value from Welch’s t-test. The Reactome database [23] (version 60) served as the reference pathway annotation. The “background” for which enrichment is calculated against was not the list of all universally-known proteins; rather, it was restricted to all the proteins that were quantified from the phosphoproteomics experiment. Per pathway, a 2X2 contingency table was constructed to assess counts of proteins included/excluded from the pathway and meeting/lacking the specified inclusion criteria, described as follows: For enrichment of dephosphorylated proteins, the criteria were p < 0.05 from Welch’s t-test and log2FC < 0 (FC = fold change determined from the peptide ion intensity ratio of (mean TRC-794) / (mean DMSO)). For enrichment of hyperphosphorylated proteins, the criteria were p < 0.05 from Welch’s t-test and log2FC > 0. The raw one-tailed p-value from Fisher’s exact test on each contingency table was reported. All calculations were performed in R version 3.3.0.
2.11. MoBaS Analysis
Using the STRING database of protein interactions [24], we first built a PPI network, one per cell line, with all identified proteins from the unfiltered dataset. We scored the nodes of the networks by taking the minimum of the p-values from Welch’s t-test across identified peptides for each protein and taking −log of this p-value. We assigned a null score to proteins for which phosphorylation data was not available. From that, we then applied the Steiner Tree algorithm to recruit additional nodes that would optimize the connectivity amongst the experimentally-identified nodes. Subsequently, we applied MoBaS [25] to identify subnetworks that are enriched with proteins that are tightly interconnected and differentially phosphorylated (having significant p-value scores). Additional details on this method are available in “Supporting Information (Materials and Methods cont’d)”.
2.12. Additional Materials and Methods
Procedures exclusively featured in the Supporting Information figures are described in the document “Supporting Information (Materials and Methods cont’d)”.
3. Results and discussion
3.1. SMAP reduces cell viability and induces apoptosis
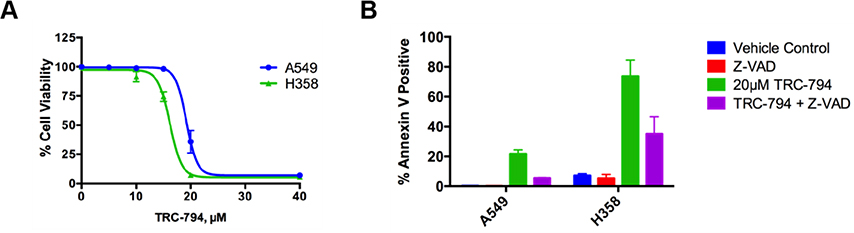
Previous work from our group and collaborators have 1) described the derivatization and structures of our series of small molecules [13], 2) studied its mechanism of action as a PP2A activator [14], and 3) demonstrated its anti-tumor properties in murine models [14]. We have selected TRC-794 (aliases RT-30, DT-794, and SMAP3 in other publications) as the featured molecule for our global phosphoproteomics studies; Supporting Information Fig. 1 illustrates its chemical structure. In line with other members of the SMAP family of compounds, TRC-794 exhibits consistent dose-dependent inhibition of cell viability between two KRAS mutated NSCLC cell lines, A549 and H358 (Figure 1A). Increased Annexin V staining in treated cells indicates induction of cell death, which is partly reversed with co-treatment of Z-VAD, a pan-caspase inhibitor (Figure 1B). This suggests that TRC-794 promotes cell death that is partially caspase-dependent.
Figure 1.
TRC-794 decreases cell viability and induces apoptosis.
(A) MTT cell viability assay against increasing concentrations of the SMAP TRC-794 after 24hr incubation. All measurements are normalized to DMSO control. (B) Annexin V staining of cells treated with DMSO, 100uM Z-VAD (caspase inhibitor), TRC-794, or a combination of Z-VAD + TRC-794 after 24hrs. Annexin V staining in the A549 control and Z-VAD groups was barely detectable. The means ± SD across 3 independent replicates are reported.
3.2. Phosphoproteomics enabled system-wide characterization of differential phosphorylation changes
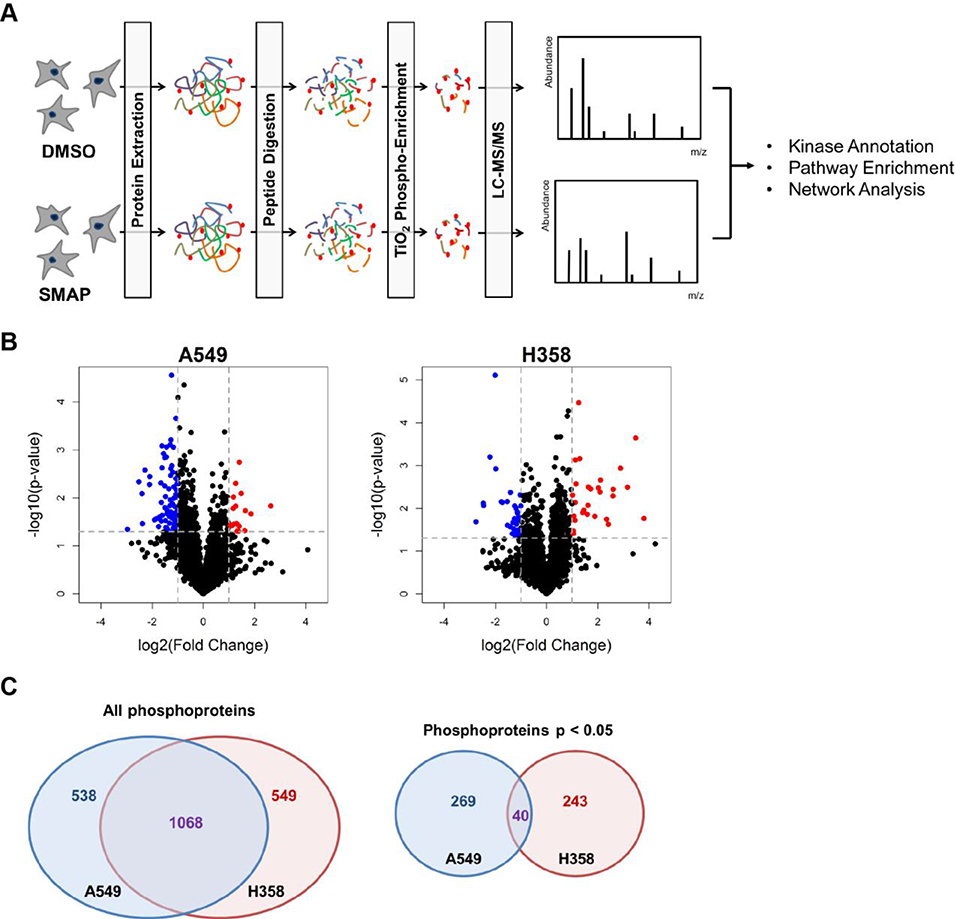
Label-free shotgun phosphoproteomics was used to interrogate the differential phosphorylation changes with drug treatment in the A549 and H358 cell lines. TRC-794 and DMSO-treated samples were treated for 12hrs and processed in parallel as illustrated in the workflow (Figure 2A). Principle component analysis (PCA) and Spearman correlation assessment of the two datasets yielded good separation between DMSO and TRC-794-treated samples and consistency among the technical and independent replicates (Supporting Information Fig. 2).
Figure 2.
Phosphoproteomics identified numerous differentially phosphorylated peptides between the two treatment groups.
(A) The phosphoproteomics workflow that was applied: two NSCLC cell lines were treated separately with DMSO control and the PP2A activator TRC-794 for 12hrs. Three independent replicates per condition were analyzed. (B) Volcano plots of A549 and H358 at the phosphosite level. The horizontal dotted line represents Welch’s t-test p = 0.05; vertical lines represent log2(fold change) ± 1, corresponding to 2-fold change in magnitude. Blue dots = hits with p < 0.05 and mean log2(FC) ≤ −1 in the TRC-794 group. Red dots = hits with p < 0.05 and mean log2(FC) ≥ 1 in the TRC-794 group. (C) Venn diagram depicting phosphoprotein-level overlap between A549 and H358 between all proteins (left diagram) and significant proteins (right diagram).
We identified 3,396 unique phosphosites mapping to 2,999 unique phosphopeptides mapping to 1,605 unique phosphoproteins in the A549 cells. Of this total, 400 phosphosites (mapping to 363 phosphopeptides and 308 phosphoproteins) passed a p < 0.05 threshold of being differentially phosphorylated between DMSO and TRC-794 treatment. As for H358, we identified 3,135 unique phosphosites mapping to 2,895 unique phosphopeptides mapping to 1,617 unique proteins; 372 of these phosphosites (337 peptides and 283 proteins) met a p < 0.05 criteria for significance. Additional summary statistics are outlined in Supporting Information Fig. 3.
Although PP2A activation is expected to lead to reduced phosphorylation for direct targets of PP2A, we expected a wide-ranging response of both increases and decreases in phosphorylation. In fact, phosphosite-level volcano plots (Figure 2B) illustrate significantly larger numbers of dephosphorylated phosphosites meeting p < 0.05 and −log2(fold change) ≤ −1.0 for the A549 cell lines compared to H358. In contrast, the H358 dataset included a greater number of significantly hyper-phosphorylated sites upon drug treatment. Although these cell lines have ~70% of their identified proteins in common (58% of phosphopeptides and 39% of phosphosites), there are striking differences in their responses to the PP2A activator: out of the phosphoproteins meeting the p < 0.05 criterion, no more than 25% of these proteins (16% of phosphopeptides and 13% of phosphosites) are shared between the two (Figure 2C, Supporting Information Fig. 4). Thus, a detailed examination of the pathways and network differences was carried out to understand the molecular basis for the sharply differing cellular responses.
3.3. Kinase-Substrate Enrichment Analysis (KSEA) implicated downregulation of MAPK and cell cycle signaling
Since phosphorylation-based signaling is heavily dependent on the actions of kinases, we first sought to characterize changes in treatment-induced signaling by estimating the relative activities of kinases. Consequently, we employed Kinase-Substrate Enrichment Analysis (KSEA) [19] to compute scores that reflect the directional shifts in each kinase’s signaling output. Ultimately, a negative score corresponds to a kinase with substrates that are generally dephosphorylated with TRC-794, thereby suggesting that the overall kinase activity is decreased with treatment relative to DMSO control. Inversely, a positive score implies upregulated activity with treatment.
Altogether, 149 unique kinases were scored in both cell lines using data from 632 and 535 phosphosites with known kinase-substrate (K-S) relationships in A549 and H358, respectively (Supporting Information Table 2). Both cell lines exhibited robust SMAP-induced downregulation of RAF1, MAP2K1/MAP2K2, and RPS6KA1—all of which annotate to the canonical MAPK pathway downstream of (K)RAS (Figure 3). This pattern is consistent with early published observations of diminished MAPK signaling with SMAP treatment [13,14]. Interestingly, though, H358-specific downregulation of AKT1, RPS6KB1, and MAPK3 (all downstream of RAS) suggests stronger suppression of the RAS family cascade in this cell line. Additionally, while some cell cycle kinases (PLK1 and AURKA) experienced negative scores in both cells, many other related kinases (CDK1, CDK2, CDK4, AURKB, and TTK) showed downregulation unique to A549. Targeted Western blots and quantitative PCR validation confirmed downregulation of select MAPK and cell cycle pathway members (Supporting Information Fig. 5). These KSEA findings altogether suggest that SMAP treatment induced a stronger suppression of cell cycle members in A549 and more robust inhibition of RAS effectors in H358.
Figure 3.
KSEA calculations identified common downregulation of select MAPK proteins but a high proportion of cell cycle kinases in A549.
Heat map reporting KSEA results according to the normalized scores. Only kinases that are shared between the two datasets and that have 5+ substrates are included. Asterisks indicate the scores meeting the p < 0.05 statistical cutoff. Blue color represents negative kinase scores, and red represents positive.
3.4. Reactome pathway enrichment identified dephosphorylation of numerous cell cycle-related proteins
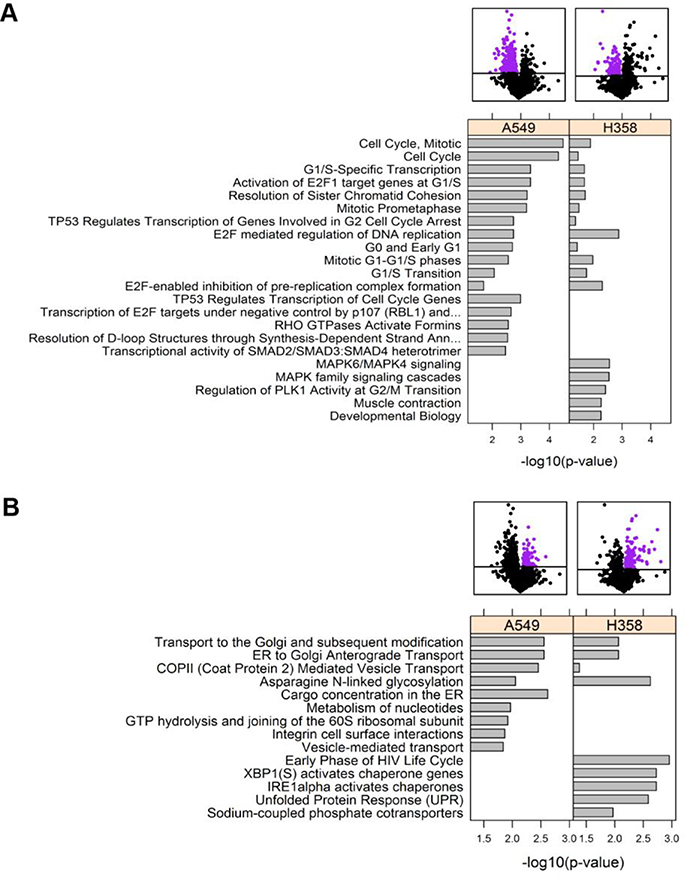
This initial observation provided very interesting insight on differential signaling changes from a kinase perspective. However, since only a small subset of the experimental phosphosites had K-S annotations and were thus used for KSEA calculations, we wanted to expand our analysis to general pathway enrichment, so we could potentially characterize processes that have too few identified substrates to generate robust KSEA signatures. Consequently, we performed Reactome pathway enrichment scored by Fisher’s exact test against a background of all the proteins from the phosphoproteomics dataset. Interestingly, when focusing on the dephosphorylated proteins, cell cycle pathways were significant for both cell types (Figure 4A); however, cell cycle annotations (notably, those linked with transcriptional regulation) were more numerous for the A549 cell line. Furthermore, H358 displayed notable dephosphorylation of MAPK-related signaling, while these pathways were not significant for A549. Analysis of hyperphosphorylated pathways revealed common hits on intracellular transport (Figure 4B). Network-based pathway enrichment using the YourCrosstalker™ software also revealed similar trends as with Figure 4A: there was a stronger cell cycle effect in A549 and preferential enrichment of IGF1R, mTOR, EGFR, and SOS-mediated signaling pathways—all of which overlap with RAS and/or MAPK signaling—in H358 (Supporting Information Fig. 6; http://proteomics.case.edu/ct/nsclc/dwiredja/).
Figure 4.
Reactome pathway enrichment revealed dephosphorylation of cell cycle pathways.
Analysis was performed at the protein level, in which the p-value and phosphorylation fold change values for each protein was selected from the peptide with the lowest p-value from the Welch’s t-test. All the pathways with visible bars met the Fisher’s exact p < 0.05 cutoff in the designated cell line(s). All shared pathways are listed, but only the top 5 unique pathways to each cell line are included. The purple dots in the volcano plots indicate the proteins that enrichment was calculated for. (A) Pathways enriched with dephosphorylated proteins. (B) Pathways enriched with hyper-phosphorylated proteins.
3.5. MoBaS network-based scoring identified a protein cluster that reflected differential SMAP response between A549 and H358 cells
Since canonical pathways are pre-defined and rigid, the earlier Reactome enrichment results may miss protein groupings that are not classically established but may better capture the differential signaling response in our model. Motivated by this challenge, we decided to take a protein-protein interaction (PPI) network approach and employed Modularity-Based Scoring (MoBaS) [25]. Overall, this method aims to identify densely-connected subnetworks of proteins that 1) are functionally related and 2) exhibit robust differential phosphorylation with treatment. Statistical scoring of the subnetworks is illustrated in Supporting Information Fig. 7, and the top ten readouts are listed in Supporting Information Fig. 8 for both cell lines.
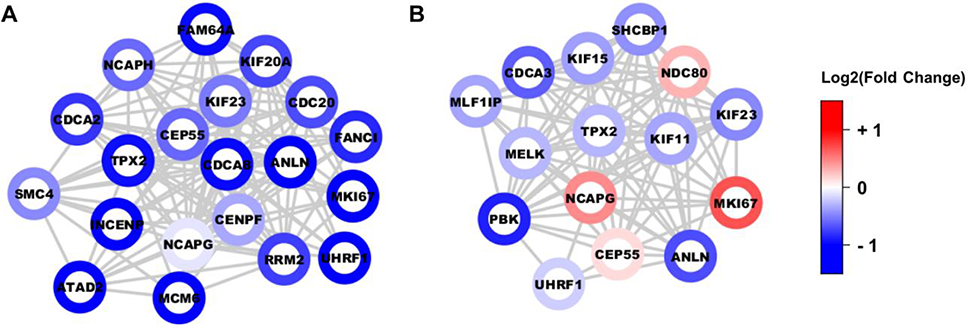
Not only did the resulting top module for A549 comprise entirely of proteins that participate in cell cycle processes, but all the protein nodes were also uniformly dephosphorylated in drug treatment (Figure 5A). Analysis of the H358 dataset yielded a similar module comprising of cell cycle players; however, these proteins displayed no coherent directional phosphorylation change (Figure 5B). To verify that MoBaS offers relevant cluster identification, we performed a parallel analysis using the popular MCODE method [26]. Indeed, for both A549 and H358, MCODE also extracted high-scoring modules populated with cell cycle proteins (Supporting Information Fig. 9), and these constituents significantly overlapped with the proteins from their MoBaS cluster counterparts (p-value < 2.2e−16 for both cell lines, Fisher’s exact test). Along with previous kinase and pathway findings, MoBaS analysis confirms that A549 has greater cell cycle sensitivity to drug treatment.
Figure 5.
MoBaS identified a tightly-interconnected subnetwork involved in cell cycle progression.
(A) The top-scoring protein-protein interaction (PPI) subnetwork in A549, identified from the MoBaS approach. (B) Second top-scoring subnetwork from MoBaS analysis on H358 cells, which enriched for cell cycle proteins. Each node is a phosphoprotein represented by its most significant peptide. Node color is based on log2(fold change) relative to DMSO control.
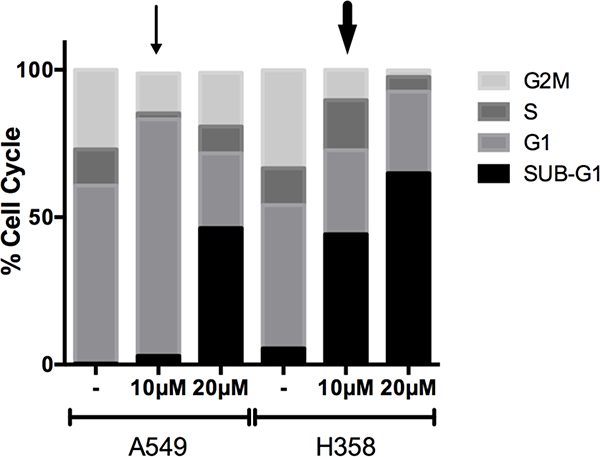
3.6. TRC-794 induces A549-specific cell cycle disruption
Thus far, although our SMAP compound frequently induced dephosphorylation of many cell cycle and RAS/MAPK-related proteins, bioinformatic analyses have consistently uncovered a more vigorous downregulation of cell cycle processes in A549. We sought to confirm this collective observation by testing if these system-level patterns translate into specific phenotypic differences between A549 and H358’s response to TRC-794. Cells were differentially treated with DMSO or two varying doses of our compound for 24hrs and subsequently incubated with propidium iodide (PI) to stain the DNA content. Flow cytometric analysis against the PI staining would then determine the distribution of cells across the different phases of the cell cycle. Interestingly, A549 treatment with 10μM SMAP induced a striking increase in the G1 population and shrinkage of the G2M phase (Figure 6, thin arrow), whereas H358 experienced a more apoptotic/cell death reaction at the same dose (Figure 6, thick arrow), as indicated by the rise in cells that had undergone DNA fragmentation (Sub-G1). Ultimately, however, the higher concentration of TRC-794 had marked increase in the Sub-G1 population in both cell lines. Collectively, these observations reaffirm our computational findings and suggest that our SMAP promotes diverging intermediate cellular responses within KRAS mutant lines at the given time points, which ultimately converge on cell death.
Figure 6.
SMAP resulted in A549-specific cell cycle disruption.
Cell cycle analysis measured by propidium iodide staining of DNA in the samples. Cells were treated with DMSO or two different concentrations of TRC-794 for 24 hours prior to analysis. The thin and thick arrows mark specific conditions featured in the results and discussion text.
4. Concluding remarks
Our study applied quantitative phosphoproteomics to explore the global signaling effects of a novel phosphatase activator in KRAS mutant NSCLC, and subsequent bioinformatics analyses revealed stronger disruption of cell cycle proteins in A549, while H358 had a more prominent dysregulation of RAS/MAPK-related pathways. These observations not only corroborate previous studies that suggest diverging pathway dependencies between the two [27–29], but they also exemplify how SMAP therapy may target unique combinations of pathways that eventually promote cell death. Although our current experimental study did not explore the temporal aspect of the signaling response and cannot distinguish whether or not a given dephosphorylation event is due to direct PP2A-substrate interaction, our work highlights the intricacies of the primary and secondary signaling patterns that accumulate from small molecule phosphatase activation. Ultimately, the observed variations in response of similar cell lines depict potential complexities in drug response for individual patients harboring identical mutations. Phosphoproteomics, coupled to advanced bioinformatics, is a valuable tool to dissect the phospho-signaling basis of these differences.
Supplementary Material
Significance of the study.
PP2A is a documented phosphatase and tumor suppressor that downregulates numerous oncogenic targets. Its activation thus provides a promising anti-cancer therapeutic strategy that can potentially mirror the signaling effects of kinase inhibitor combinations. This study features high-throughput phosphoproteomic characterization of KRAS mutated NSCLC cell lines treated with a novel class of small molecules previously shown to bind to PP2A and potentiate its activity. The results would 1) provide one of the earliest glimpses of the differential phosphorylation changes upon phosphatase activation on a global scale; 2) resolve the compounds’ mechanism of action at the signaling level; and 3) demonstrate how similar cell lines may exhibit unique response signatures that ultimately converge and manifest as cell death.
Acknowledgements
Funding sources
This work has been supported by the National Institutes of Health: P30-CA-043703, UL1TR000439, and TL1 TR000441.
ABBREVIATIONS
- NSCLC
Non-Small Cell Lung Cancer
- PP2A
Protein Phosphatase 2A
- SMAP
Small Molecule Activator of PP2A
- KSEA
Kinase-Substrate Enrichment Analysis
- K-S
Kinase-Substrate
- PPI
Protein-Protein Interaction
- MoBaS
Modularity-Based Scoring
- PI
Propidium Iodide
Footnotes
Conflict of interest statement
M.R.C. has an ownership interest in, receives compensation from, and serves as officer to NeoProteomics, Inc., the parent company that developed YourCrosstalker software. G.N. has filed patents covering composition of matter on the SMAP molecules for the treatment of human cancer and other diseases.
5 References
- [1].Perrotti D, Neviani P Protein Phosphatase 2A: A Target for Anticancer Therapy. Lancet Oncol, 2013, 14, e229–e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kuo Y, Huang K, Chiang C-W, Yang C-H, Yang Y-S, Lee W-Y Regulation of Phosphorylation of Thr-308 of Akt, Cell Proliferation, and Survival by the B55alpha Regulatory Subunit Targeting of the Protein Phosphatase 2A Holoenzyme to Akt. J. Biol. Chem, 2008, 283, 1882–1892. [DOI] [PubMed] [Google Scholar]
- [3].Zhou B, Wang Z, Zhao Y, Brautigan DL, Zhang Z-Y, Erk PPA The Specificity of Extracellular Signal-Regulated Kinase 2 Dephosphorylation by Protein Phosphatases. J. Biol. Chem, 2002, 277, 31818–31825. [DOI] [PubMed] [Google Scholar]
- [4].Ruvolo PP, Qui YH, Coombes KR, Zhang N, Ruvolo VR, Borthakur G, Konopleva M, Andreeff M, Kornblau SM Low Expression of PP2A Regulatory Subunit B55α Is Associated with T308 Phosphorylation of AKT and Shorter Complete Remission Duration in Acute Myeloid Leukemia Patients. Leukemia, 2011, 25, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cristobal I, Garcia-Orti L, Cirauqui C, Cortes-Lavaud X, Garcia-Sanchez MA, Calasanz MJ, Odero MD Overexpression of SET Is a Recurrent Event Associated with Poor Outcome and Contributes to Protein Phosphatase 2A Inhibition in Acute Myeloid Leukemia. Haematologica, 2012, 97, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Xu P, Xu X-L, Huang Q, Zhang Z-H, Zhang Y-B CIP2A with Survivin Protein Expressions in Human Non-Small-Cell Lung Cancer Correlates with Prognosis. Med. Oncol, 2011. [DOI] [PubMed] [Google Scholar]
- [7].Neviani P, Harb JG, Oaks JJ, Santhanam R, Walker CJ, Ellis JJ, Ferenchak G, Dorrance AM, Paisie CA, Eiring AM, Ma Y, Mao HC, Zhang B, Wunderlich M, May PC, Sun C, Saddoughi SA, Bielawski J, Blum W, Klisovic RB, Solt JA, Byrd JC, Volinia S, Cortes J, Huettner CS, Koschmieder S, Holyoake TL, Devine S, Caligiuri MA, Croce CM, Garzon R, Ogretmen B, Arlinghaus RB, Chen C-S, Bittman R, Hokland P, Roy D-C, Milojkovic D, Apperley J, Goldman JM, Reid A, Mulloy JC, Bhatia R, Marcucci G, Perrotti D PP2A-Activating Drugs Selectively Eradicate TKI-Resistant Chronic Myeloid Leukemic Stem Cells. J. Clin. Invest, 2013, 123, 4144–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Oaks JJ, Santhanam R, Walker CJ, Roof S, Harb JG, Ferenchak G, Eisfeld AK, Van Brocklyn JR, Briesewitz R, Saddoughi S. a., Nagata K, Bittman R, Caligiuri M. a., Abdel-Wahab O, Levine R, Arlinghaus RB, Quintas-Cardama A, Goldman JM, Apperley J, Reid A, Milojkovic D, Ziolo MT, Marcucci G, Ogretmen B, Neviani P, Perrotti D Antagonistic Activities of the Immunomodulator and PP2A-Activating Drug FTY720 (Fingolimod, Gilenya) in Jak2-Driven Hematologic Malignancies. Blood, 2013, 122, 1923–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Neviani P, Santhanam R, Trotta R, Notari M, Blaser BW, Liu S, Mao H, Chang JS, Galietta A, Uttam A, Roy DC, Valtieri M, Bruner-Klisovic R, Caligiuri MA, Bloomfield CD, Marcucci G, Perrotti D The Tumor Suppressor PP2A Is Functionally Inactivated in Blast Crisis CML through the Inhibitory Activity of the BCR/ABL-Regulated SET Protein. Cancer Cell, 2005, 8, 355–368. [DOI] [PubMed] [Google Scholar]
- [10].Arnold HK, Sears RC A Tumor Suppressor Role for PP2A-B56alpha through Negative Regulation of c-Myc and Other Key Oncoproteins. Cancer Metastasis Rev, 2008, 27, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gutierrez A, Pan L, Groen RWJ, Baleydier F, Kentsis A, Marineau J, Grebliunaite R, Kozakewich E, Reed C, Pflumio F, Poglio S, Uzan B, Clemons P, VerPlank L, An F, Burbank J, Norton S, Tolliday N, Steen H, Weng AP, Yuan H, Bradner JE, Mitsiades C, Look a. T., Aster JC. Phenothiazines Induce PP2A-Mediated Apoptosis in T Cell Acute Lymphoblastic Leukemia. J. Clin. Invest, 2014, 124, 644–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mortensen PB Neuroleptic Treatment and Other Factors Modifying Cancer Risk in Schizophrenic Patients. Acta Psychiatr. Scand, 1987, 75, 585–590. [DOI] [PubMed] [Google Scholar]
- [13].Kastrinsky DB, Sangodkar J, Zaware N, Izadmehr S, Dhawan NS, Narla G, Ohlmeyer M Reengineered Tricyclic Anti-Cancer Agents. Bioorg. Med. Chem, 2015, 23, 6528–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sangodkar J, Perl A, Tohme R, Kiselar J, Kastrinsky DB, Zaware N, Izadmehr S, Mazhar S, Wiredja DD, O’Connor CM, Hoon D, Dhawan NS, Schlatzer D, Yao S, Leonard D, Borczuk AC, Gokulrangan G, Wang L, Svenson E, Farrington CC, Yuan E, Avelar RA, Stachnik A, Smith B, Gidwani V, Giannini HM, McQuaid D, McClinch K, Wang Z, Levine AC, Sears RC, Chen EY, Duan Q, Datt M, Haider S, Ma’ayan A, DiFeo A, Sharma N, Galsky MD, Brautigan DL, Ioannou YA, Xu W, Chance MR, Ohlmeyer M, Narla G Activation of Tumor Suppressor Protein PP2A Inhibits KRAS-Driven Tumor Growth. J. Clin. Invest, 2017, 109, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Olsen JV, Mann M Status of Large-Scale Analysis of Post-Translational Modifications by Mass Spectrometry. Mol. Cell. Proteomics, 2013, 12, 3444–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wiśniewski JR, Zougman A, Nagaraj N, Mann M Universal Sample Preparation Method for Proteome Analysis. Nat. Methods, 2009, 6, 359–362. [DOI] [PubMed] [Google Scholar]
- [17].Schlatzer DM, Sugalski J, Dazard J-E, Chance MR, Anthony DD A Quantitative Proteomic Approach for Detecting Protein Profiles of Activated Human Myeloid Dendritic Cells. J. Immunol. Methods, 2012, 375, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tomechko SE, Liu G, Tao M, Schlatzer D, Powell CT, Gupta S, Chance MR, Daneshgari F Tissue Specific Dysregulated Protein Subnetworks in Type 2 Diabetic Bladder Urothelium and Detrusor Muscle. Mol. Cell. Proteomics, 2015, 14, 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Casado P, Rodriguez-Prados J-C, Cosulich SC, Guichard S, Vanhaesebroeck B, Joel S, Cutillas PR Kinase-Substrate Enrichment Analysis Provides Insights into the Heterogeneity of Signaling Pathway Activation in Leukemia Cells. Sci. Signal, 2013, 6, rs6–rs6. [DOI] [PubMed] [Google Scholar]
- [20].Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M PhosphoSitePlus: A Comprehensive Resource for Investigating the Structure and Function of Experimentally Determined Post-Translational Modifications in Man and Mouse. Nucleic Acids Res, 2012, 40, D261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Linding R, Jensen LJ, Ostheimer GJ, van Vugt MATM, Jørgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T Systematic Discovery of in Vivo Phosphorylation Networks. Cell, 2007, 129, 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Horn H, Schoof EM, Kim J, Robin X, Miller ML, Diella F, Palma A, Cesareni G, Jensen LJ, Linding R KinomeXplorer: An Integrated Platform for Kinome Biology Studies. Nat. Methods, 2014, 11, 603–604. [DOI] [PubMed] [Google Scholar]
- [23].Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S, Matthews L, May B, Milacic M, Rothfels K, Shamovsky V, Webber M, Weiser J, Williams M, Wu G, Stein L, Hermjakob H, D’Eustachio P The Reactome Pathway Knowledgebase. Nucleic Acids Res, 2016, 44, D481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C STRING v10: Protein-Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res, 2015, 43, D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ayati M, Erten S, Chance MR, Koyutürk M MOBAS: Identification of Disease-Associated Protein Subnetworks Using Modularity-Based Scoring. EURASIP J. Bioinforma. Syst. Biol, 2015, 2015, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bader GD, Hogue CWV An Automated Method for Finding Molecular Complexes in Large Protein Interaction Networks. BMC Bioinformatics, 2003, 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J A Gene Expression Signature Associated with “K-Ras Addiction” Reveals Regulators of EMT and Tumor Cell Survival. Cancer Cell, 2009, 15, 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yoon YK, Kim HP, Han SW, Oh DY, Im S. a., Bang YJ, Kim TY KRAS Mutant Lung Cancer Cells Are Differentially Responsive to MEK Inhibitor due to AKT or STAT3 Activation: Implication for Combinatorial Approach. Mol. Carcinog, 2010, 49, 353–362. [DOI] [PubMed] [Google Scholar]
- [29].Wang C-Y, Chao T-T, Chang F-Y, Chen Y-L, Tsai Y-T, Lin H-I, Huang Y-CT, Shiau C-W, Yu C-J, Chen K-F CIP2A Mediates Erlotinib-Induced Apoptosis in Non-Small Cell Lung Cancer Cells without EGFR Mutation. Lung Cancer, 2014, 85, 152–160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.