Abstract
Abstract
Helicobacter pylori, a member of Epsilonproteobacteria, is a Gram-negative microaerophilic bacterium that colonizes gastric mucosa of about 50% of the human population. Although most infections caused by H. pylori are asymptomatic, the microorganism is strongly associated with serious diseases of the upper gastrointestinal tract such as chronic gastritis, peptic ulcer, duodenal ulcer, and gastric cancer, and it is classified as a group I carcinogen. The prevalence of H. pylori infections varies worldwide. The H. pylori genotype, host gene polymorphisms, and environmental factors determine the type of induced disease. Currently, the most common therapy to treat H. pylori is the first line clarithromycin–based triple therapy or a quadruple therapy replacing clarithromycin with new antibiotics. Despite the enormous recent effort to introduce new therapeutic regimens to combat this pathogen, treatment for H. pylori still fails in more than 20% of patients, mainly due to the increased prevalence of antibiotic resistant strains. In this review we present recent progress aimed at designing new anti-H. pylori strategies to combat this pathogen. Some novel therapeutic regimens will potentially be used as an extra constituent of antibiotic therapy, and others may replace current antibiotic treatments.
Key points
• Attempts to improve eradication rate of H. pylori infection.
• Searching for new drug targets in anti-Helicobacter therapies.
Keywords: Helicobacter pylori, Antibiotic resistance, Biofilm, AMP, Probiotics, Therapy
Introduction
Helicobacter pylori, a Gram-negative spiral-shaped bacterium, is a member of Epsilonproteobacteria that specifically colonizes gastric epithelium of humans causing one of the most common infections worldwide. Infection with H. pylori affects about half of the world’s population, yet its prevalence varies throughout the world. Currently, incidence rates of H. pylori infection remains high in most developing countries, and it is generally related to socioeconomic status and levels of hygiene. Colonization of the stomach mucosa results in various diseases of the upper part of the gastrointestinal tract; however, the disease affects only a subset of infected individuals. H. pylori infection elicits gastric mucosal inflammatory responses and causes peptic ulcers (in about 10% of those infected). H. pylori is also associated with a high risk for development of mucosa-associated lymphoid tissue lymphoma and adenocarcinoma of the stomach (about 1% those of infected) (De Falco et al. 2015; Cover 2016). In 1994, The World Health Organization classified H. pylori infections as class I carcinogens (IARC 1994).
This bacterium was isolated for the first time from human stomach biopsies in 1983, and the first H. pylori genome from the 26695 strain was sequenced in 1997 (Marshall and Warren 1984; Tomb et al. 1997). Two years later, H. pylori became the first bacterium for which genomes of two strains were compared (Alm et al. 1999). Since then, the number of publicly available H. pylori genomes has increased rapidly to more than 1500. There are now (as of 18.05.2020) 1850 H. pylori genomes available on PATRIC (Pathosystem Resource Integration Center) (Wattam et al. 2014). These were isolated from patients with various disease symptoms, as well as from a 5300-year-old European natural mummy, named Iceman (Maixner et al. 2016). The H. pylori 26695, J99, G27, P12, and N6 are the most commonly used strains in both basic as well as applied research (Ferrero et al. 1992; Tomb et al. 1997; Alm et al. 1999; Belogolova et al. 2013). Comparative genomics experiments have revealed a high level of genomic diversity that results from intra-genomic diversification (for example, point mutations, recombination, and slipped-strand mispairing), as well as inter-genomic recombination, the ability of H. pylori to take up exogenous DNA and incorporate it into its genome (Suerbaum and Josenhans 2007; Morelli et al. 2010). While, the average genome size of H. pylori is relatively small (~ 1.6 Mbp) and compact (~ 1500 ORF), the high level of diversity can complicate comparative genomics analyses between many strains, and new methods of analysis have to be developed, such as a feature frequency profiling (FFP), an alignment-free analysis approach (Van Vliet and Kusters 2015).
Several international guidelines (Maastricht V Florence Consensus, the Kyoto Global Consensus and the Toronto Consensus Report) elaborated by experts in various fields of microbiology have recommended different drug regimens for treatment of H. pylori infections (Sugano et al. 2015; Fallone et al. 2016; Malfertheiner et al. 2017). However, treatment for H. pylori still fails in more than 20% of patients, and one would hope to diminish that failure rate to a more acceptable level. The two main obstacles to effective therapy are (1) H. pylori resistance to different antibiotics, which correlates with use of antibiotics in the general population, and (2) a lack of patient compliance. Clarithromycin-resistant strains of H. pylori have recently been recognized by WHO as one of the 12 priority pathogens for which novel antibiotics are urgently needed (WHO 2017).
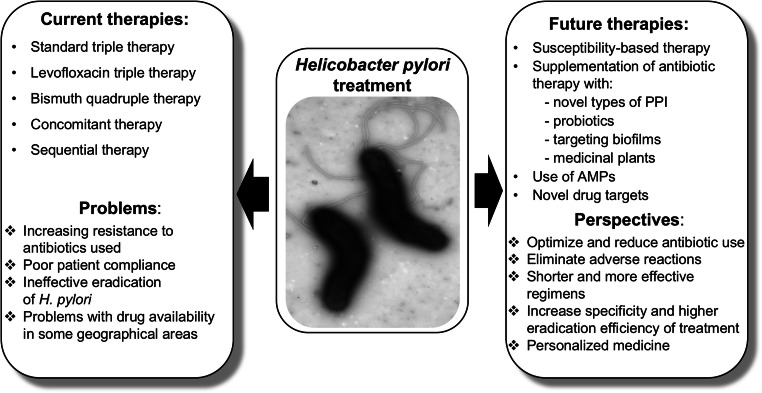
In this review, we present various novel therapeutic regiments against H. pylori that can serve either as a supplement or a replacement for current therapies. Some, such as probiotics, plant extracts, or inhibitors of biofilm formation, should improve eradication rate achieved by standard therapies and should restrict the increase of antibiotic-resistant bacteria. Others have a potential to replace antibiotics. Global screening approaches that combine genomic, proteomic, and metabolic analyses conducted on multiple H. pylori strains have led to identification of many possible new drug targets that potentially could eliminate H. pylori without interfering with the physiological microbiota. In silico screening that compared H. pylori metabolic pathways with those of Homo sapiens have indicated proteins unique to the pathogen. Another way to establish new classes of antibacterial agents is targeting virulence rather than the viability of bacteria. In case of H. pylori, an understanding of the molecular mechanisms of pathogenesis, in combination with analysis of huge numbers of genomes of strains, responsible for serious disease outcomes, permits selection of a new relevant drug targets. This research concentrates on virulence factors involved in the colonization process and those implicated in gastric cancer development. Solving the structures of many H. pylori proteins allows design of effective inhibitors by in silico docking studies. Figure 1 presents principal issues discussed in this review.
Fig. 1.
Schematic representation of currently used treatment of H. pylori infection and promising future approaches
Pathogenesis of H. pylori infections
Considerable experimental evidence indicates that the H. pylori genotype is a substantial factor determining the type of induced disease (Bridge and Scott Merrell 2013; McLean and El-Omar 2014). However, there are also some host factors, such as polymorphism of the IL-1β, which influences gastric acid secretion, or polymorphism of the CYP2C19, which is the main enzyme metabolizing PPI (proton pump inhibitor), that are associated with the risk of gastric cancer development (Uotani et al. 2015). Some studies indicate that environmental factors, such as nutrition, should additionally be taken into account (Noto et al. 2013; Gaddy et al. 2013).
Analyses of the H. pylori pan-genome (defined as the entire gene repertoire in a given species) revealed that this species possesses an open pan-genome. The estimated size of its core genome (the number of genes present in all isolates) varies depending on the number of analyzed strains, their geographical origin, and the methods used; the core genome ranges between more than 1000 to only 244 genes (Salama et al. 2000; Gressmann et al. 2005; van Tonder et al. 2014). Most of the virulence factors are classified as part of the dispensable genome.
H. pylori is one of the most common microorganisms able to colonize human gastric mucosa. The bacterium develops many factors or strategies to adapt to this harsh environment and establish persistent infection. Urease, which neutralizes the acidic environment of the stomach, flagella, and a helical shape, in combination with an effective chemotaxis system, aid H. pylori to reach the protective mucus layer of gastric mucosa (Sachs et al. 2005; Lertsethtakarn et al. 2011; Sycuro et al. 2012).
Apart from factors involved in the adaptation to acidic stomach conditions, many virulence factors produced by some H. pylori clinical isolates engage in the development of disease symptoms. The most extensively studied are those affecting the pathogen’s interaction with host cells, both epithelial and immune system cells, with a consequence of inducing gastric cancer development. This set of virulence factors includes several outer-membrane proteins (OMP) playing a role in bacterial adhesion to gastric epithelial cells (Javaheri et al. 2016; Königer et al. 2016; Moonens et al. 2018), the cytotoxin-associated protein A (CagA) encoded within the cagA pathogenicity island (CagPAI) and translocated to eukaryotic cells by type four secretion system (T4SS), and the vacuolating cytotoxin A (VacA). For more details of VacA and CagA biology, we recommend several excellent review papers by Foegeding et al., McClain et al., Tegtmeyer et al., and Backert et al. (Foegeding et al. 2016; Tegtmeyer et al. 2017; Backert and Tegtmeyer 2017; McClain et al. 2017).
Although the majority of gene polymorphisms present in H. pylori genomes are distributed randomly, many strains isolated from gastric cancer or peptic ulcer patients possess a specific set of certain gene alleles, e.g., a majority of cagA PAI positive strains contain s1 type VacA and type I hopQ and produce BabA adhesin (Cover 2016).
Currently used and recommended antibiotic therapies against H. pylori
There are various recommended regimens for H. pylori eradication. One of the most commonly used therapies in the world is a standard triple therapy, which consists of a proton pump inhibitor (PPI), amoxicillin (AMX), and clarithromycin (CLR) or metronidazole (MTZ). This regimen should last for 10–14 days to get the best result in H. pylori eradication, and it is acceptable only in areas of low CLR resistance (< 15% efficacy). The reason for worldwide increasing resistance to CLR is the high use of macrolides in treatment (Kamboj et al. 2017; Matsumoto et al. 2019). Another regimen due to increasing resistance to CLR is the use of levofloxacin (LVF) triple therapy (LVF, AMX, PPI) because LVF is a broad-spectrum quinolone. However, LVF triple therapy is not recommended as a first-line choice for most patients because resistance to LVF is increasing worldwide, but in areas of low LVF resistance and high CLR and MTZ resistance or unavailability of bismuth, this therapy can be used (O’Morain et al. 2018; Matsumoto et al. 2019; Fallone et al. 2019).
The 2016 Maastricht V/Florence Consensus Report (Malfertheiner et al. 2017) and the 2016 Toronto Consensus (Fallone et al. 2016) recommended the first-line 14-day bismuth quadruple therapy (PPI, bismuth, MTZ, tetracycline (TC)) when there is above 15% CLR resistance or a concomitant therapy (PPI, AMX, MTZ, CLR), if bismuth is not available. The bismuth quadruple therapy is the best choice (especially recommended in cases of penicillin allergy) because it shows effectiveness even against in vitro MTZ resistant strains, and it is well tolerated by patients (Kamboj et al. 2017; Matsumoto et al. 2019; Suzuki et al. 2019; Fallone et al. 2019). There are also new types of bismuth quadruple therapy in which TC or MTZ can be replaced by AMX in areas where TC is difficult to obtain or resistance to MTZ has increased. Sequential therapy is also possible; it consists of PPI and AMX for the first half of therapy (5 days) and PPI, MTZ, and CLR for the second half (Kamboj et al. 2017).
If the first-line therapies fail, different types of second-line rescue treatments for H. pylori infection can be used, such as a bismuth quadruple therapy which consist of PPI, bismuth, MTZ, and TC. It is recommended after failure of standard triple therapy (Malfertheiner et al. 2017; Lin and Hsu 2018; Matsumoto et al. 2019). A 10-day TC-LVF quadruple therapy provides the best regimen for a second-line treatment. It consists of an esomeprazole (a type of PPI) and bismuth salts in addition to antibiotics (Lin and Hsu 2018). Furthermore, the 2016 Maastricht V/Florence Consensus Report (Malfertheiner et al. 2017) also recommends fluoroquinolone-AMX triple/quadruple therapy as a second-line treatment. They have comparable effectiveness and consist of LVF, AMX, PPI, and bismuth (only in quadruple therapy).
Facing resistance of H. pylori: novel therapies
Antibiotic resistance of H. pylori has reached alarming levels worldwide, prompting an urgent search for more efficient treatments. A recent systematic review and meta-analysis of 178 studies demonstrated an increased rate of primary and secondary H. pylori resistance in six World Health Organization Regions (Savoldi et al. 2018; Matsumoto et al. 2019). Success in all antibacterial therapies depends on susceptibility to given drugs, doses, formulations, use of adjuvants (agents that demonstrate additive and/or synergistic effect with antibiotic therapy), treatment duration, and reinfection rates. Antimicrobial susceptibility testing seems to be the best solution to optimize and reduce antibiotics use in H. pylori treatment. A meta-analysis conducted by Chen et al., consisting of 13 controlled clinical trials with 3512 participants, showed better efficacy of tailored therapies compared with empirically chosen regimens (Chen et al. 2016). Moreover, establishing H. pylori resistance surveillance programs worldwide to provide local resistance reports may be beneficial for preparation of guidelines to treat infection caused by H. pylori in particular regions. Matsumoto et al. propose incorporating susceptibility-based therapy against H. pylori into practice (Matsumoto et al. 2019). Nonetheless, antibiotic susceptibility testing may also be a challenge; it requires usually biopsy to start bacterial culture. H. pylori culture is expensive, intricate, and time-consuming. However, empirical therapies are less effective and face challenges due to the changing profile of H. pylori antibiotic resistance across the world. Unnecessary use of antibiotics in H. pylori treatment should be avoided because it can influence not only regimen efficacy but also increase the possibility of creating new multi-drug-resistant strains.
Use of novel components in classical antibiotic therapies of H. pylori
One of the new strategies to combat H. pylori infections is dual therapy with AMX and PPI. This has demonstrated promising results in eradication of H. pylori as well as decreasing the amount of antibiotics used (Suzuki et al. 2019). In the past this approach did not achieve satisfactory outcomes (Bayerdörffer et al. 1995; Koizumi et al. 1998). The successful turning point for this dual therapy seems to be the substitution of conventional PPIs with a novel type of PPI, the potassium-competitive acid blocker, vonoprazan, that has a strong and durable inhibitory effect on acid secretion and generates a neutral environment in the stomach, suitable for H. pylori growth. The neutral pH of the colonized niche causes dormant H. pylori to enter the replication phase, which makes the bacteria sensitive to antibiotics such as AMX and CLR (Graham and Fischbach 2010). The study of Furuta et al. showed that a 7-day regimen consisting of AMX-vonoprazan provided an eradication rate of 93.8% for H. pylori infections (Furuta et al. 2019). The possible limitations of this dual therapy are unavailability of vonoprazan (which is only approved in some countries) and penicillin allergy reactions in some patients (Suzuki et al. 2019).
Rifabutin, a derivative of rifamycin, structurally resembles rifampicin, and it is characterized by a broad spectrum of antibacterial activity. Rifabutin is routinely used as an anti-tuberculosis drug against Mycobacterium tuberculosis strains that are resistant to rifampicin, and it is also used against M. avium for HIV patients (Gisbert and Calvet 2012). Because this antibiotic exhibits high in vitro activity against H. pylori and is not degraded by acid in the stomach, and because rifabutin-resistant H. pylori strains have rarely been isolated from patients, rifabutin-based treatment has been considered as a potential option to combat H. pylori for more than 20 years. The antibiotic inhibits the ß-subunit of RNA polymerase, which is encoded by the rpoB gene, and H. pylori rpoB mutants that are resistant to rifabutin have been created in the lab (Heep et al. 1999). To date, rifabutin-based triple treatment has mainly been a rescue therapy for difficult-to-treat patients, after multiple failures to eradicate H. pylori (Gisbert and Calvet 2012; Ribaldone et al. 2019). A review paper by Gisbert and Calvet used meta-analysis of data published through 2010 to evaluate the role of rifabutin in H. pylori treatment. The mean H. pylori eradication rate was 73%, but the data in various studies were contradictory and difficult to compare. The observed eradication rates ranged from higher than 80% to 40%, depending on patient status, rifabutin dosage, and duration of treatment (Gisbert and Calvet 2012). A recent study by Graham et al. showed that enrichment of a dual regimen (AMX and PPI) with rifabutin significantly increased treatment efficacy (Graham et al. 2020). Also, the addition of bismuth to a standard rifabutin triple therapy (AMX, PPI, rifabutin) resulted in higher treatment efficacy (Ciccaglione et al. 2016). These results indicate that rifabutin-based therapies have potential as a first- or second-line empirical treatment for H. pylori. There are, however, concerns with respect to rifabutin treatment of H. pylori: the drug is extremely expensive, severe adverse effects (myelotoxicity, leucopenia) have been observed, and widespread use of rifabutin could cause a surge of resistant H. pylori strains, especially because this drug is also commonly used against mycobacteria.
Antimicrobial peptides in the treatment of H. pylori
Antimicrobial peptides (AMPs) have been recently widely verified as an efficient replacement for antibiotics to combat pathogenic microorganism (Sierra et al. 2017; Xu et al. 2020). These compounds are produced by the cells of various organisms as part of their innate immunity, and they provide protection against various pathogens (bacteria, viruses, and fungi), as well as acting as mediators of immune responses (Lei et al. 2019). AMPs usually consist of fewer than 100 amino acids, and they possess an amphipathic structure that allows cationic AMPs to act precisely on anionic bacterial membranes to increase membrane permeability and cause cell death without interacting with the surface of the neutral eukaryotic cell membranes. Apart from acting on cell membranes some AMPs interfere with diverse intracellular processes, such as transcription, translation, or with biogenesis of the cell wall (Mishra et al. 2017).
Neshani et al. searched PubMed, Scopus, and Web of Science databases and found 22 antimicrobial peptides with anti-H. pylori activity. The 3 peptides with the strongest anti-H. pylori activity were pexiganan, tilapia piscidin 4 (TP4), and PGLa-AM1. These 5-kDa peptides with α-helical structures are cationic and have high positive charge and isoelectric point (Neshani et al. 2019). Most anti-H. pylori peptides have the activity against some antibiotic-resistant strains. However, there are some disadvantages to use of AMPs to treat people. Many of natural AMPs can be degraded by both host and bacterial proteases (Pero et al. 2017), and some can cause toxic side effects on human cells. However, the structure of some AMPs has been solved which may allow a synthesis of their analogs lacking cytotoxicity. Additionally, bacteria can become resistant to AMPs after prolonged use (Lei et al. 2019). To avoid AMP degradation engineered probiotic bacteria (lactic acid bacteria-LAB) that colonize the human intestinal tract were tested as a delivery vehicle. However, prolonged release of AMPs by engineered probiotics may kill commensal bacteria and thus may reduce their beneficial effects (Mishra et al. 2017). Despite disadvantages, AMP have synergistic effects in combination with known antibiotics; however, in some cases this may increase their cytotoxic activity. Further in vivo testing is required (Zharkova et al. 2019).
Probiotics in the treatment of H. pylori
Probiotics are defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (Hill et al. 2014). They have been used in the prevention and treatment of some gastrointestinal disorders, such as diarrhea, irritable bowel syndrome, and inflammatory bowel disease (Sarowska et al. 2013). Administration of probiotics brings beneficial effects to host microbiota, enhances the immune system, and shows inhibitory effects on pathogens.
Probiotic supplementation is an emerging field in H. pylori treatment (Chey et al. 2017). In vitro studies report inhibitory effects of probiotics on the expression level of genes encoding virulence factors, including ureB, vacA, flaA, and sabA (Lesbros-Pantoflickova et al. 2007; de Klerk et al. 2016; Urrutia-Baca et al. 2018). Some probiotics, such as Lactococcus lactis, Lactobacillus reuteri, and Lactobacillus bulgaricus produce peptide and nonpeptide antipathogen substances that can inhibit the growth, as well as the adhesion process, of H. pylori (Kim et al. 2003; Boyanova et al. 2017; Urrutia-Baca et al. 2018). Moreover, Emara et al. found that using probiotics can improve H. pylori-induced histological features by lowering the density of the pathogen on the luminal side of the epithelium, which results in improving the histological inflammatory response that persists even after termination of probiotic supplementation (Emara et al. 2016).
Many clinical studies explored the potential of probiotics in H. pylori treatment. They include the effect of a single treatment of probiotics as well as the effect of combining probiotics with standard H. pylori therapy (Boonyaritichaikij et al. 2009). However, a recent systematic review by Losurdo et al. based on 11 independent studies suggests that using probiotics alone is not sufficient for H. pylori clearance (Losurdo et al. 2018). The use of probiotics in combination with antibiotic therapy appears to be more effective, in both children and adults, compared with a standard therapy alone; however, the increases in eradication rates differ among studies (McFarland et al. 2016; Feng et al. 2017; Wang et al. 2017). Moreover, probiotics can provide a beneficial effect on adverse reactions, such as nausea, vomiting, diarrhea, and taste disturbance, that may occur during eradication therapy for H. pylori infections (Tong et al. 2007; Zou et al. 2009; Szajewska et al. 2015; McFarland et al. 2016; Zhang et al. 2017), though the decrease in side effects varied among the studies. The variation may result from different kinds of bacteria and yeasts used as probiotics. Ji et al. suggested that only carefully chosen and examined probiotic strains applied in an appropriate manner have a chance to be effective in H. pylori treatment (Ji and Yang 2020). There are still many questions regarding the use of probiotics that need to be explored further, including dosages, duration of probiotic treatment, effectiveness of specific probiotic strains, and the interaction between selected probiotics and antibiotics (Kamiya et al. 2019).
Phytomedicine in the treatment of H. pylori
The use of natural compounds in H. pylori treatment is gaining popularity due to their low side effects and low toxicity. Folk medicine, especially traditional Chinese medicine (TCM), constitutes a valuable guide for using plants to combat various diseases. Numerous studies have been performed to evaluate the usefulness of diverse plant extracts in anti-H. pylori therapy. Based on in vitro studies (evaluation of MIC—minimal inhibitory concentration), plant extracts were assigned to four categories. The majority were classified as possessing weak-to-moderate activity. An extract of Impatiens balsamina L. (Balsaminaceae), a Taiwanese folk medicinal plant, showed the strongest activity (Wang et al. 2009).
One patented Chinese drug with two ingredients, Chenopodium ambrosioides L. (CAL) and Adina pilulifera (AP), is commonly used as a therapy for H. pylori infection gastritis and peptic ulcers. Recently, Ye et al. have reported that Ch. ambrosioides alone also showed effective antimicrobial activity against H. pylori, both in vitro and in vivo. The active ingredients of this plant and mechanism of action have not been elucidated yet, but its inhibitory effect on growth of H. pylori is promising for further research (Ye et al. 2015).
Encouraging results from in vitro and in vivo studies were also reported by Kouticheu Mabeku et al., who analyzed the anti-H. pylori activity of Bryophyllum pinnatum, a plant known for its antiulcer properties. The B. pinnatum activity is probably due to active compounds present in the methanol extract, such as phenolics and flavonoids that react with hydroxyl radicals, superoxide anion radicals, and lipid peroxy radicals. They can protect gastric mucosa against reactive oxygen species produced during infection by H. pylori (Kouitcheu Mabeku et al. 2017).
Apart from crude plant extracts, purified plant compounds also were tested for their anti-Helicobacter activities (Bonifácio et al. 2014). Although the mechanisms of their action require clarification some were recognized as inhibitors of H. pylori urease (Xiao et al. 2012; Yu et al. 2015). Structures of the H. pylori urease and the urease complex with various ligands have been solved, which allowed a wide range in silico structure-based analysis (molecular docking) of the interactions between this enzyme and plant bioactive components (Xiao et al. 2012; Yu et al. 2015; Hassan and Žemlička 2016). Additionally, several synthetic molecules were also tested. Gains in knowledge may accelerate the design and synthesis of effective inhibitors (for a profound review paper see Kafarski and Talma (2018)).
Combining of phytomedicine with standard medicine may offer effective H. pylori treatment. Judaki et al. reported that addition of a turmeric extract (curcumin) to the standard triple therapy significantly increased the eradication rate of H. pylori and decreased oxidative damage to DNA (Judaki et al. 2017). Treatment with curcumin reduced the number of bacteria colonizing gastric mucosa and reduced the activity of lipid peroxidases and urease in H. pylori infected mice (Kwiecien et al. 2019).
Although many studies of plant extracts or purified plant compounds have been reported, reliable randomized and controlled clinical trials that treat H. pylori infections with plant extracts and compare their efficacy with recommended triple therapies are lacking. The only systematic review of clinical trials with TCM conducted between 1982 and 2008 showed an average eradication rate of about 72%. This indicates that plant extract therapy cannot be used as monotherapy, although it has a great potential to assist treatment (Lin and Huang 2009).
Searching for new drug targets to combat H. pylori infection
Discovery of new therapies against H. pylori requires identification and validation of novel drug targets. Therapeutic agents are usually designed against basic cellular processes, so the proteins encoded by the core genome are specific and promising candidates as drug targets. Thus, research into H. pylori genomics could lead to new therapies. In principle, ideal targets for anti-bacterial agents are H. pylori proteins whose homologs are absent in corresponding host and resident microbiota. Various combinations of in silico analyses, together with genomic, proteomic, and metabolomic studies, have been used to identify possible new drug-specific targets. Sarkar et al. used subtractive analysis of metabolic pathways of H. pylori HPAG1 and humans and found 25 candidate target proteins that are likely to be essential for viability of the pathogen but have no homologs in human. The largest contributors to the list of potential drug targets in this study were proteins (9 out of 25) involved in the LPS biosynthesis pathway (Sarkar et al. 2012). In a study by Uddin et al. metabolic sets of 61 H. pylori strains were extracted and analyzed by pan-genomics and subtractive genomics to reveal 38 non-homologs to human proteins that are potential drug targets. Only one of these was essential for bacterial survival, namely, the saccharopine dehydrogenase that is involved in lysine degradation (Uddin and Khalil 2020). Nammi et al. compared 23 genomes of H. pylori with genomes of Homo sapiens and 3 others Helicobacter species to identify 29 unique proteins that may be potential drug targets; all were essential for H. pylori growth and 15 were important for survival of pathogen in the host (Nammi et al. 2016). Dutta et al. used subtractive genomics to identify 10 surface-exposed proteins that may be potential drug targets (Dutta et al. 2006). Screening of 23 genomes of H. pylori strains by Nammi et al., in 5-step in silico analyses, revealed 31 genes of interested that are localized within pathogenic islands of H. pylori and may be potential drug targets in novel therapies (Nammi et al. 2017). Moreover, pan-genome analysis of global representative of 39 genomes led to characterization of 28 non-host homolog proteins, as universal therapeutic targets (Ali et al. 2015). In summary, some newly identified in silico targets already have been validated and tested by using molecular docking, interactome studies, and structural studies to discover inhibitors that next need to be validated (Sarkar et al. 2012; Nammi et al. 2016; Nammi et al. 2017; Uddin and Khalil 2020). There are also various attempts to find a strategy to develop effective drug delivery systems to eradicate H. pylori (El-Zahaby et al. 2014; Verma et al. 2016).
Targeting biofilms of H. pylori
One of the reasons standard therapies are not highly effective in eradication of H. pylori may be H. pylori’s ability to form biofilms. Since the discovery of this pathogen most studies have focused on the planktonic form (free-living) of H. pylori. However, some recent studies indicate that H. pylori can develop biofilm structures, both in vitro and in vivo. The presence of biofilms has been observed in gastric mucosa, within glands and gastrointestinal tracts in mice or patients infected with H. pylori (Sigal et al. 2015; Keilberg et al. 2016). Most strains of H. pylori studied could form biofilms, yet they showed variations in morphology, and some clinical isolates had higher levels of biofilm formation than other strains (Yonezawa et al. 2010). Multiple approaches, including genomic (Wong et al. 2016), transcriptomic (Anderson et al. 2015), proteomic (Shao et al. 2013), and target-specific studies (Cole et al. 2004; Anderson et al. 2015), have been used to identify factors associated with development of biofilms by H. pylori; still, the molecular processes need to be elucidated. Bacteria in biofilms exhibit different characteristics when compared with planktonic forms; one of these is an increase resistance to antimicrobial agents (Mah and O’Toole 2001). Recently, an in vitro study by Yonezawa et al. showed that resistance to CLR, MTZ, and AMX increased after H. pylori biofilm formation (Yonezawa et al. 2019). Alternative treatment approaches that target H. pylori biofilms already have shown some promising results. The use of N-acetylcysteine (NAC), a mucolytic and thiol-containing antioxidant agent, was effective in disrupting biofilm development and prevented formation of new biofilm in vitro and in vivo. Additionally, administration of NAC before traditional antibiotic treatment of patients with a history of at least 4 anti-H. pylori therapy failures resulted in a more than 3 fold better eradication rate of H. pylori infection (65%) comparing with non-NAC group (20%) (Cammarota et al. 2010).
Targeting proteins of the primary metabolism of H. pylori
Drug targets in anti-H. pylori therapy often are proteins involved in the primary metabolism, and their inhibition is bactericidal. There are long lists of targets in this category (see previous subchapter of this article or the paper by Salillas and Sancho). Some of the targets identified in silico have been already tested, and they reveal various level of effectiveness (Salillas and Sancho 2020). The majority of studies described above paid attention to homology of the identified gene products with human proteins but ignored the potential impacts of the proposed therapy on the human microbiota. Recent studies indicate that the interaction of H. pylori with the natural microbiota is extremely complex and must be kept in mind while designing new anti-Helicobacter drugs (Pero et al. 2019). Here, we present some examples of proteins belonging in the primary metabolism that are promising candidates as novel drug targets.
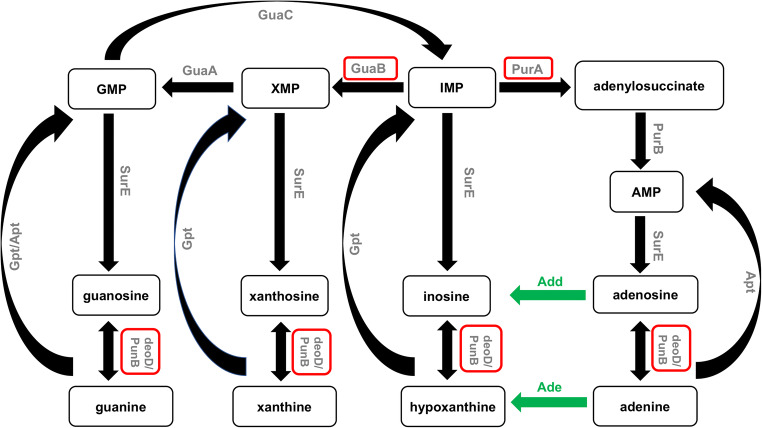
Purine metabolism is the growth-limiting step for both prokaryotic and eukaryotic cells. H. pylori, similar to other pathogens that have evolved in close association with their hosts, lacks a de novo purine nucleoside synthesis pathway. Therefore, it obtains purine nucleosides only via the purine salvage route (Liechti and Goldberg 2012; Štefanić et al. 2017). Figure 2 presents the purine nucleotide biosynthesis pathway of H. pylori. Purine nucleoside phosphorylase (PNP) and adenylosuccinate synthetase (AdSS) are crucial enzymes of the purine salvage pathway. In recent years, the biophysical and biochemical properties of both enzymes have been extensively studied. Purine nucleoside phosphorylase (PNP) catalyzes, in the presence of orthophosphate as a second substrate, the reversible phosphorolytic cleavage of the glycosidic bond of purine nucleosides (Bzowska et al. 2000). Štefanić et al. reported for the first time the unique structure of this enzyme (Štefanić et al. 2017) that influences its interactions with formycin (Narczyk et al. 2018). Formycin A (an analogue of adenosine) is a well-known inhibitor of hexameric PNPs (Bzowska et al. 2000). The formycin A inhibition constant for PNP from H. pylori is about threefold higher than reported for E. coli PNP (Narczyk et al. 2018). This compound appears to be a good inhibitor of H. pylori PNP, and a good lead compound to design more potent inhibitors, but its effect should still be tested in vivo. Adenylosuccinate synthetase (AdSS) is an enzyme that catalyzes the condensation of IMP (inosine-5′-monophosphate) with L-aspartate to adenylosuccinate. Bubić et al. described the biochemical properties of AdSS from H. pylori. They showed, inter alia, that AdSS from H. pylori is active in vivo as a dimer even at low concentrations and in the absence of ligands (IMP, GTP, GDP). Monomers and dimers of this enzyme seem not to be in a dynamic equilibrium in contrast to E. coli AdSS (Bubić et al. 2018). One of the known AdSS inhibitors is hadacidin, which are a natural antibiotic (the fermentation product of Penicillium frequentans) and an analogue of L-aspartate (Tibrewal and Elliott 2011). Hadacidin inhibits the enzyme of H. pylori strain 26695 in vitro (Bubić et al. 2018). Unfortunately, hadacidin does not significantly affect the growth of H. pylori in vivo. The reason could be that the inhibitor does not penetrate into the cells of these Gram-negative bacteria. Solving the structure of this protein alone or with its substrate will help in designing better inhibitors of this enzyme.
Fig. 2.
The purine nucleotide biosynthesis pathway in H. pylori. Enzymes that have been studied in H. pylori by mutant analysis and/or biochemical analysis are shown in gray. Enzymes described in this work are shown in red frames. Enzymes with likely functions, whose genes have not yet been identified, are shown in green (figure adapted from Liechti and Goldberg (2012) and Miller et al. (2012)). Abbreviations: GuaB, IMP dehydrogenase; GuaA, GMP synthetase; GuaC, GMP reductase; PurA, adenylosuccinate synthetase; PurB, adenylosuccinate lyase; Gpt, hypoxanthine-guanine phosphoribosyl-transferase; Apt, adenine phosphoribosyltransferase; SurE, 5′-nucleotidase; deoD, gene encoding purine nucleoside phosphorylase; PunB, purine nucleoside phosphorylase; Ade, adenine deaminase; Add, adenosine deaminase; IMP, inosine monophosphate; XMP, xanthosine monophosphate; GMP, guanosine monophosphate; AMP, adenosine monophosphate
Recently, the bacterial IMPDH, GuaB (inosine-5′-monophosphate dehydrogenase) enzyme, a compound of the purine nucleotide biosynthetic pathway, has become popular as a potential target for eradication multi-drug resistant pathogens such as Cryptosporidium parvum or Mycobacterium tuberculosis. This enzyme is responsible for the oxidation of IMP to xanthosine 5′-monophosphate (XMP). In subsequent enzymatic reactions, GMP is formed and continues to generate nucleotides for the synthesis of DNA and RNA. Juvale et al. performed kinetic studies on H. pylori GuaB and compared the activity of a previously tested benzimidazole-based inhibitor with newly synthetized indole-based small molecules. They found that the indole-based molecules have superiority over previously tested inhibitors. They do not affect activity of the human enzyme IMPDH-II and show a non-competitive inhibition against both Hp-IMPDH substrates, IMP and NAD+. The results of these studies are stimulating the development of better bacterial IMPDH inhibitors to overcome infections (Juvale et al. 2019).
Flavodoxin, which is a small soluble electron transfer protein, has also been recognized as a new therapeutic target against H. pylori because it is a constituent of the pyruvate-oxidoreductase complex that catalyzes the oxidative decarboxylation of pyruvate. Blocking this metabolic pathway causes bacterial death. Flavodoxins are present in bacterial cells of various species but are absent in animals and humans. Therefore, potential inhibitors will not cause side effects in patients. Solving the H. pylori flavodoxin (Hp-Fld) structure, as compared with other flavodoxin, revealed that it possesses a specific cofactor (FMN) binding site, in which a tryptophan residue is replaced by alanine. As a result, a specific cavity on the Hp-Fld surface is created, and this facilitates a search for molecules that bind there and block enzyme action (Cremades et al. 2005). A high-throughput screening method of a library of 10,000 small organic molecules identified 29 compounds that bind to Hp-Fld and stabilize its structure. Detailed experiments to improve the therapeutic and pharmacokinetic properties of selected compounds resulted in designed molecules that inhibit the in vitro electron transfer to flavodoxin physiological partners and also inhibit growth of H. pylori strains, including those resistant to commonly used antibiotics (Cremades et al. 2009). Some of the synthetized molecules also significantly reduced H. pylori gastric colonization in a mouse model of infection (Cremades et al. 2009; Salillas and Sancho 2020).
Encouraging results arose from a series of experiments which explored the possibility of using MTAN (5′-methylthioadenosine/ S-adenosylhomocysteine nucleosidase) as a target for novel anti-Helicobacter drug. The enzyme involved in quorum sensing, menaquinone synthesis, and 5′-methylthioadenosine recycling to S-adenosylmethionine in most bacteria is not essential. However, in members of Epsilonproteobacteria—H. pylori and Campylobacter jejuni, MTAN plays a critical role in their metabolism. These bacteria utilize the unusual futalosine pathway for the biosynthesis of menaquinone, an essential electron transfer agent. It was shown that the inhibition of this pathway is lethal for H. pylori (Wang et al. 2012). As this pathway is absent in humans and in human commensal bacteria MTAN is a promising candidate as a target for new drugs. Several the transition state analogues of MTAN have been designed and tested as potential antibiotics in in vitro and in vivo experiments. Solving the crystal structures of MTAN co-crystallized with several inhibitors provided significant information about inhibitor-target interactions, and this is an essential step in exploring the potential of using the transition-state enzyme analogues as antibacterial drugs. Some of the tested inhibitors were able to penetrate the H. pylori cell wall as they inhibited the pathogen growth at low concentration (Wang et al. 2012; Wang et al. 2015; Harijan et al. 2019). Aminofutalosine synthase (MqnE) is another enzyme of the futalosine pathway of menaquinone biosynthesis indicated as a potential target for new antibiotics (Joshi et al. 2019).
The essential global gene expression regulator, HsrA, has been shown recently to be a promising target for drug development (González et al. 2019). HsrA belongs to the OmpR-like response regulator that acts as a transcription activator for itself and many others genes, including those involve in redox homeostasis, nitrogen metabolism, energy metabolism, and protein synthesis processes (Olekhnovich et al. 2014). González et al. carried out affinity binding high-throughput screening (HTS) to test in vitro, 1120 FDA-approved small molecules for their ability to a specifically bind to HsrA and inhibit its function. Eventually, four natural flavonoids, apigenin, chrysin, kaempferol, and hesperetin, were demonstrated to exhibit potent bactericidal activity against three different H. pylori strains, including those resistant to antibiotics (CLR and MTZ). Subsequent biophysical and molecular docking analyses were used to investigate the mechanism of inhibition of selected potential therapeutics (González et al. 2019).
Targeting virulence factors of H. pylori
Another way to combat infectious disease is to inhibit virulence rather than bacterial viability. This strategy has some advantages over currently used antibiotic therapies. First, it does not induce pressure to select antibiotic-resistant strains and does not disturb the host microbiota. Second, most of the virulence factors are extracytoplasmic molecules that should be easily accessible to potential inhibitors. Additionally, the structures of many bacterial virulence factors have been solved, so the design of new inhibitors and testing their mechanisms of action can be investigate in silico using molecular docking. These types of inhibitors, with respect of their mode of actions, can be divided into two categories: those blocking a single virulence factor and those blocking global bacterial processes involved in pathogen virulence, such as the two-component system, the quorum sensing system, secretion systems, or post-translational protein modifications (Duncan et al. 2012; Krachler and Orth 2013; Brackman and Coenye 2015; Bocian-Ostrzycka et al. 2017). As H. pylori has accompanied humankind for more than 100,000 years, its interdependence with the host is extremely complicated (Moodley et al. 2012). There is an evidence that host colonization by H. pylori influences the composition of the microbiota not only in the stomach but also in distantly located organs (Kienesberger et al. 2016). Additionally, some inverse association between H. pylori infection and some diseases, such as Barrett’s esophagus and its consequences, or allergic asthma, were also reported (Chen and Blaser 2008; Blaser 2011; Rubenstein et al. 2014). In light of this, therapy that targets virulence factors has great potential, as it will not remove H. pylori but only disturb its virulence properties. For H. pylori, research in this field is still in its infancy, though both types of inhibitors have been recently tested or indicated as potentially effective.
H. pylori has a specific helical shape that enhances microorganism motility through the viscous mucus layer of the stomach. Several proteases, acting on the peptide chain of peptidoglycan, play an important role in cell shape determination (Sycuro et al. 2010; Sycuro et al. 2012). One of them, Csd4, was tested as a target for a new drug created through structure-based design, followed by analysis of the Csd4-inhibitor complex. The inhibitor successfully altered on the shape of H. pylori in vitro (Liu et al. 2016).
T4SS and its effector molecule CagA are recognized as key virulence factors responsible for gastric cancer development. Blocking this system in patients infected with cagA positive strains should be an efficient treatment. Hilleringmann et al. used HTS strategy to identify a small molecule that blocks CagA (a part of the T4SS apparatus, containing an NTP binding motif), and the molecule prevented CagA from translocation into AGS cells. Moreover, the researchers showed that pretreatment of H. pylori with this molecule impaired gastric colonization in mice (Hilleringmann et al. 2006). Also, Shaffer et al. identified two compounds that block CagA and peptidoglycan translocation. One interfered with pilus biogenesis, and the second impaired its activity (Shaffer et al. 2016).
Proteins of the Dsb (disulfide bond) system are present in all bacterial cells and are responsible for generation of disulfide bonds between cysteine residues of proteins. As a consequence, they determine protein structure and function. Dsb proteins represent possible targets for antibacterial molecules because virulence factors of several bacteria require this post-translational modification to achieve their proper structures. Inactivation of dsb genes attenuates many pathogens (Smith et al. 2016; Bocian-Ostrzycka et al. 2017; Landeta et al. 2019). The H. pylori Dsb system is different from Dsb systems operating in cells of Gram-negative bacteria, so its potential inhibitors should not interfere with the physiological human microbiota (Roszczenko et al. 2012; Bocian-Ostrzycka et al. 2015). Several OMPs that play a role in H. pylori adhesion with host cells contain cysteine residues, and their proper conformation depends on the action of Dsb proteins (Pang et al. 2014; Javaheri et al. 2016; Moonens et al. 2016). An H. pylori strain lacking the major Dsb oxidoreductase, encoded by hp0231, is deficient in CagA translocation into host epithelial cells as Hp0231, among other activities, influences the structure of HopQ, an outer membrane protein that mediates pathogen adhesion to epithelial cells by interaction with CECAM receptors (Javaheri et al. 2016; Königer et al. 2016). Lack of Hp0231 abolishes the H. pylori-induced IL-8 production and the development of the hummingbird phenotype in AGS cells (Zhong et al. 2016; Grzeszczuk et al. 2018). Moreover, an H. pylori hp0231-deficient strain was not able to colonize the gastric mucosa of mice. Additionally, DsbI, encoded by hp0595, another component of the H. pylori Dsb system, affects the colonization process (Raczko et al. 2005; Godlewska et al. 2006). Therefore, H. pylori proteins of the Dsb system have potential to be used as targets for new drugs.
Conclusions
Many strategies to develop anti-H. pylori therapies have been designed and tested at various levels since this bacterium was first recognized as a human pathogen. Nevertheless, we still do not have efficacious and universal drugs to combat this bacterium. There are still important issues concerning diseases induced by H. pylori waiting to be resolved. First, which H. pylori-infected people should be treated, a decision that will vary according to geographic regions? Anti-H. pylori therapy is recommended for cancer and ulcer patients and for patients with some precancer symptoms. However, the treatment of asymptomatic patients is still controversial. The interaction of H. pylori with the human microbiota is complex and should be taken into account when new drugs are introduced into the market. As the outcomes of infection are dependent on the H. pylori genotype and on host gene polymorphisms, personalized treatment may be needed. The increase in H. pylori strains resistant to antibiotics, mainly clarithromycin, is a substantial medical problem. Recent progress in genome sequencing and comparative genomics indicates several housekeeping gene products, that likely are essential for the pathogen viability but have no homologs in humans; these can serve as targets for small-molecule inhibitors. However, this application requires detailed studies of the genomes of the human microbiota. Another approach to fight H. pylori infection is to target its virulence factors. Their inhibition attenuates infection and can serve as an individual therapy or in combination with already used antibiotics. This may increase specificity and produce a higher eradication efficacy of treatment. In another approach, probiotics and plant extracts may be administered as a cocktail with antibiotics. It must be stressed that all novel strategies should be tested in animal models and clinical trials, before they are put into practice.
Acknowledgments
We thank Dr. Jeff Hansen and Prof. Agnieszka Bzowska for the critical reading of the manuscript.
Authors’ contributions
KJK conceived the general idea of the article; all authors were engaged in writing the manuscript. PRJ and MW prepared figures. PRJ prepared the reference part of the manuscript; KJK and PRJ gather all parts of the manuscript and formed its final version.
Funding
This study was funded by the National Science Centre, Poland (grant No 2018/29/B/NZ1/00140).
Data availability
Not applicable.
Compliance with ethical standards
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
All authors approved the final version of the manuscript.
Code availability
Not applicable.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ali A, Naz A, Soares SC, Bakhtiar M, Tiwari S, Hassan SS, Hanan F, Ramos R, Pereira U, Barh D, Figueiredo HCP, Ussery DW, Miyoshi A, Silva A, Azevedo V (2015) Pan-genome analysis of human gastric pathogen H. pylori: comparative genomics and pathogenomics approaches to identify regions associated with pathogenicity and prediction of potential core therapeutic targets. Biomed Res Int:139580. 10.1155/2015/139580 [DOI] [PMC free article] [PubMed]
- Alm RA, Ling LSL, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, DeJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, Guillemin K. Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. MBio. 2015;6:e00379. doi: 10.1128/MBIO.00379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Tegtmeyer N. Type IV secretion and signal transduction of Helicobacter pylori CagA through interactions with host cell receptors. Toxins (Basel) 2017;9:115. doi: 10.3390/toxins9040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayerdörffer E, Miehlke S, Mannes GA, Sommer A, Höchter W, Weingart J, Heldwein W, Klann H, Simon T, Schmitt W. Double-blind trial of omeprazole and amoxicillin to cure Helicobacter pylori infection in patients with duodenal ulcers. Gastroenterology. 1995;108:1412–1417. doi: 10.1016/0016-5085(95)90689-4. [DOI] [PubMed] [Google Scholar]
- Belogolova E, Bauer B, Pompaiah M, Asakura H, Brinkman V, Ertl C, Bartfeld S, Nechitaylo TY, Haas R, Machuy N, Salama N, Churin Y, Meyer TF. Helicobacter pylori outer membrane protein HopQ identified as a novel T4SS-associated virulence factor. Cell Microbiol. 2013;15:1896–1912. doi: 10.1111/cmi.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser M. Stop the killing of beneficial bacteria. Nature. 2011;476:393–394. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- Bocian-Ostrzycka KM, Łasica AM, Dunin-Horkawicz S, Grzeszczuk MJ, Drabik K, Dobosz AM, Godlewska R, Nowak E, Collet J-F, Jagusztyn-Krynicka EK. Functional and evolutionary analyses of Helicobacter pylori HP0231 (DsbK) protein with strong oxidative and chaperone activity characterized by a highly diverged dimerization domain. Front Microbiol. 2015;6:1065. doi: 10.3389/fmicb.2015.01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocian-Ostrzycka KM, Grzeszczuk MJ, Banaś AM, Jagusztyn-Krynicka EK. Bacterial thiol oxidoreductases—from basic research to new antibacterial strategies. Appl Microbiol Biotechnol. 2017;101:3977–3989. doi: 10.1007/s00253-017-8291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifácio BV, dos Santos Ramos MA, da Silva PB, Bauab TM. Antimicrobial activity of natural products against Helicobacter pylori: a review. Ann Clin Microbiol Antimicrob. 2014;13:54. doi: 10.1186/s12941-014-0054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaritichaikij S, Kuwabara K, Nagano J, Kobayashi K, Koga Y. Long-term administration of probiotics to asymptomatic pre-school children for either the eradication or the prevention of Helicobacter pylori infection. Helicobacter. 2009;14:202–207. doi: 10.1111/j.1523-5378.2009.00675.x. [DOI] [PubMed] [Google Scholar]
- Boyanova L, Gergova G, Markovska R, Yordanov D, Mitov I. Bacteriocin-like inhibitory activities of seven Lactobacillus delbrueckii subsp. bulgaricus strains against antibiotic susceptible and resistant Helicobacter pylori strains. Lett Appl Microbiol. 2017;65:469–474. doi: 10.1111/lam.12807. [DOI] [PubMed] [Google Scholar]
- Brackman G, Coenye T. Quorum sensing inhibitors as anti-biofilm agents. Curr Pharm Des. 2015;21:5–11. doi: 10.2174/1381612820666140905114627. [DOI] [PubMed] [Google Scholar]
- Bridge DR, Scott Merrell D. Polymorphism in the Helicobacter pylori CagA and VacA toxins and disease. Gut Microbes. 2013;4:101–117. doi: 10.4161/gmic.23797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubić A, Mrnjavac N, Stuparević I, Łyczek M, Wielgus-Kutrowska B, Bzowska A, Luić M, Leščić Ašler I. In the quest for new targets for pathogen eradication: the adenylosuccinate synthetase from the bacterium Helicobacter pylori. J Enzyme Inhib Med Chem. 2018;33:1405–1414. doi: 10.1080/14756366.2018.1506773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzowska A, Kulikowska E, Shugar D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol Ther. 2000;88:349–425. doi: 10.1016/S0163-7258(00)00097-8. [DOI] [PubMed] [Google Scholar]
- Cammarota G, Branca G, Ardito F, Sanguinetti M, Ianiro G, Cianci R, Torelli R, Masala G, Gasbarrini A, Fadda G, Landolfi R, Gasbarrini G. Biofilm demolition and antibiotic treatment to eradicate resistant Helicobacter pylori: a clinical trial. Clin Gastroenterol Hepatol. 2010;8:817–820.e3. doi: 10.1016/j.cgh.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Chen Y, Blaser MJ. Helicobacter pylori colonization is inversely associated with childhood asthma. J Infect Dis. 2008;198:553–560. doi: 10.1086/590158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Dang Y, Zhou X, Liu B, Liu S, Zhang G. Tailored therapy versus empiric chosen treatment for Helicobacter pylori eradication a meta-analysis. Med (United States) 2016;95:e2750. doi: 10.1097/MD.0000000000002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chey WD, Leontiadis GI, Howden CW, Moss SF. ACG clinical guideline: treatment of Helicobacter pylori infection. Am J Gastroenterol. 2017;112:212–238. doi: 10.1038/ajg.2016.563. [DOI] [PubMed] [Google Scholar]
- Ciccaglione AF, Tavani R, Grossi L, Cellini L, Manzoli L, Marzio L. Rifabutin containing triple therapy and rifabutin with bismuth containing quadruple therapy for third-line treatment of Helicobacter pylori infection: two pilot studies. Helicobacter. 2016;21:375–381. doi: 10.1111/hel.12296. [DOI] [PubMed] [Google Scholar]
- Cole SP, Harwood J, Lee R, She R, Guiney DG. Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol. 2004;186:3124–3132. doi: 10.1128/JB.186.10.3124-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL. Helicobacter pylori diversity and gastric cancer risk. MBio. 2016;7:e01869–e01815. doi: 10.1128/MBIO.01869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremades N, Bueno M, Toja M, Sancho J. Towards a new therapeutic target: Helicobacter pylori flavodoxin. Biophys Chem. 2005;115:267–276. doi: 10.1016/j.bpc.2004.12.045. [DOI] [PubMed] [Google Scholar]
- Cremades N, Velázquez-Campoy A, Martínez-Júlvez M, Neira JL, Pérez-Dorado I, Hermoso J, Jiménez P, Lanas A, Hoffman PS, Sancho J. Discovery of specific flavodoxin inhibitors as potential therapeutic agents against Helicobacter pylori infection. ACS Chem Biol. 2009;4:928–938. doi: 10.1021/cb900166q. [DOI] [PubMed] [Google Scholar]
- De Falco M, Lucariello A, Iaquinto S, Esposito V, Guerra G, De Luca A. Molecular mechanisms of Helicobacter pylori pathogenesis. J Cell Physiol. 2015;230:1702–1707. doi: 10.1002/jcp.24933. [DOI] [PubMed] [Google Scholar]
- de Klerk N, Maudsdotter L, Gebreegziabher H, Saroj SD, Eriksson B, Eriksson OS, Roos S, Lindén S, Sjölinder H, Jonsson A-B. Lactobacilli reduce Helicobacter pylori attachment to host gastric epithelial cells byinhibiting adhesion gene expression. Infect Immun. 2016;84:1526–1535. doi: 10.1128/IAI.00163-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MC, Linington RG, Auerbuch V. Chemical inhibitors of the type three secretion system: disarming bacterial pathogens. Antimicrob Agents Chemother. 2012;56:5433–5441. doi: 10.1128/AAC.00975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Singh SK, Ghosh P, Mukherjee R, Mitter S, Bandyopadhyay D. In silico identification of potential therapeutic targets in the human pathogen Helicobacter pylori. In Silico Biol. 2006;6:43–47. [PubMed] [Google Scholar]
- El-Zahaby S, Kassem A, El-Kamel A. Helicobacter pylori: an overview on antimicrobials and drug delivery systems for its eradication. Curr Drug Deliv. 2014;11:306–312. doi: 10.2174/1567201811666140327145049. [DOI] [PubMed] [Google Scholar]
- Emara MH, Elhawari SA, Yousef S, Radwan MI, Abdel-Aziz HR. Emerging role of probiotics in the management of Helicobacter pylori infection: histopathologic perspectives. Helicobacter. 2016;21:3–10. doi: 10.1111/hel.12237. [DOI] [PubMed] [Google Scholar]
- Fallone CA, Chiba N, van Zanten SV, Fischbach L, Gisbert JP, Hunt RH, Jones NL, Render C, Leontiadis GI, Moayyedi P, Marshall JK. The Toronto consensus for the treatment of Helicobacter pylori infection in adults. Gastroenterology. 2016;151:51–69.e14. doi: 10.1053/j.gastro.2016.04.006. [DOI] [PubMed] [Google Scholar]
- Fallone CA, Moss SF, Malfertheiner P. Reconciliation of recent Helicobacter pylori treatment guidelines in a time of increasing resistance to antibiotics. Gastroenterology. 2019;157:44–53. doi: 10.1053/j.gastro.2019.04.011. [DOI] [PubMed] [Google Scholar]
- Feng JR, Wang F, Qiu X, McFarland LV, Chen PF, Zhou R, Liu J, Zhao Q, Li J. Efficacy and safety of probiotic-supplemented triple therapy for eradication of Helicobacter pylori in children: a systematic review and network meta-analysis. Eur J Clin Pharmacol. 2017;73:1199–1208. doi: 10.1007/s00228-017-2291-6. [DOI] [PubMed] [Google Scholar]
- Ferrero RL, Cussac V, Courcoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol. 1992;174:4212–4217. doi: 10.1128/jb.174.13.4217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foegeding N, Caston R, McClain M, Ohi M, Cover T. An overview of Helicobacter pylori VacA toxin biology. Toxins (Basel) 2016;8:173. doi: 10.3390/toxins8060173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta T, Yamade M, Kagami T, Uotani T, Suzuki T, Higuchi T, Tani S, Hamaya Y, Iwaizumi M, Miyajima H, Umemura K, Osawa S, Sugimoto K. Dual therapy with vonoprazan and amoxicillin is as effective as triple therapy with vonoprazan, amoxicillin and clarithromycin for eradication of Helicobacter pylori. Digestion. 2019;3192:1–9. doi: 10.1159/000502287. [DOI] [PubMed] [Google Scholar]
- Gaddy JA, Radin JN, Loh JT, Zhang F, Washington MK, Peek RM, Algood HMS, Cover TL. High dietary salt intake exacerbates Helicobacter pylori- induced gastric carcinogenesis. Infect Immun. 2013;81:2258–2267. doi: 10.1128/IAI.01271-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisbert JP, Calvet X. Review article: Rifabutin in the treatment of refractory Helicobacter pylori infection. Aliment Pharmacol Ther. 2012;35:209–221. doi: 10.1111/j.1365-2036.2011.04937.x. [DOI] [PubMed] [Google Scholar]
- Godlewska R, Dzwonek A, Mikuła M, Ostrowski J, Pawłowski M, Bujnicki JM, Jagusztyn-Krynicka EK. Helicobacter pylori protein oxidation influences the colonization process. Int J Med Microbiol. 2006;296:321–324. doi: 10.1016/j.ijmm.2005.11.010. [DOI] [PubMed] [Google Scholar]
- González A, Salillas S, Velázquez-Campoy A, Espinosa Angarica V, Fillat MF, Sancho J, Lanas Á. Identifying potential novel drugs against Helicobacter pylori by targeting the essential response regulator HsrA. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-019-47746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DY, Fischbach L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut. 2010;59:1143–1153. doi: 10.1136/GUT.2009.192757. [DOI] [PubMed] [Google Scholar]
- Graham DY, Canaan Y, Maher J, Wiener G, Hulten KG, Kalfus IN. Rifabutin-based triple therapy (RHB-105) for Helicobacter pylori eradication: a double-blind, randomized, controlled trial. Ann Intern Med. 2020;172:795–802. doi: 10.7326/M19-3734. [DOI] [PubMed] [Google Scholar]
- Gressmann H, Linz B, Ghai R, Pleissner K-P, Schlapbach R, Yamaoka Y, Kraft C, Suerbaum S, Meyer TF, Achtman M. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1:e43. doi: 10.1371/journal.pgen.0010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeszczuk MJ, Bocian-Ostrzycka KM, Banaś AM, Roszczenko-Jasinska P, Malinowska A, Stralova H, Haas R, Meyer TF, Jagusztyn-Krynicka EK. Thioloxidoreductase HP0231 of Helicobacter pylori impacts HopQ-dependent CagA translocation. Int J Med Microbiol. 2018;308:977–985. doi: 10.1016/j.ijmm.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Harijan RK, Hoff O, Ducati RG, Firestone RS, Hirsch BM, Evans GB, Schramm VL, Tyler PC. Selective inhibitors of Helicobacter pylori methylthioadenosine nucleosidase and human methylthioadenosine phosphorylase. J Med Chem. 2019;62:3286–3296. doi: 10.1021/acs.jmedchem.8b01642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan STS, Žemlička M. Plant-derived urease inhibitors as alternative chemotherapeutic agents. Arch Pharm (Weinheim) 2016;349:507–522. doi: 10.1002/ardp.201500019. [DOI] [PubMed] [Google Scholar]
- Heep M, Beck D, Bayerdörffer E, Lehn N. Rifampin and rifabutin resistance mechanism in Helicobacter pylori. Antimicrob Agents Chemother. 1999;43:1497–1499. doi: 10.1128/aac.43.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. Expert consensus document: the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- Hilleringmann M, Pansegrau W, Doyle M, Kaufman S, MacKichan ML, Gianfaldoni C, Ruggiero P, Covacci A. Inhibitors of Helicobacter pylori ATPase Cagα block CagA transport and cag virulence. Microbiology. 2006;152:2919–2930. doi: 10.1099/mic.0.28984-0. [DOI] [PubMed] [Google Scholar]
- IARC (1994) Schistosomes, liver flukes and Helicobacter pylori. Iarc Monogr Eval Carcinog Risks To Humans 61:1–241 [PMC free article] [PubMed]
- Javaheri A, Kruse T, Moonens K, Mejías-Luque R, Debraekeleer A, Asche CI, Tegtmeyer N, Kalali B, Bach NC, Sieber SA, Hill DJ, Königer V, Hauck CR, Moskalenko R, Haas R, Busch DH, Klaile E, Slevogt H, Schmidt A, Backert S, Remaut H, Singer BB, Gerhard M. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat Microbiol. 2016;2:161189. doi: 10.1038/nmicrobiol.2016.189. [DOI] [PubMed] [Google Scholar]
- Ji J, Yang H. Using probiotics as supplementation for Helicobacter pylori antibiotic therapy. Int J Mol Sci. 2020;21:1136. doi: 10.3390/ijms21031136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Fedoseyenko D, Mahanta N, Ducati RG, Feng M, Schramm VL, Begley TP. Antibacterial strategy against H. pylori: inhibition of the radical SAM enzyme MqnE in menaquinone biosynthesis. ACS Med Chem Lett. 2019;10:363–366. doi: 10.1021/acsmedchemlett.8b00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judaki A, Rahmani A, Feizi J, Asadollahi K, Hafezi Ahmadi MR. Curcumin in combination with triple therapy regimes ameliorates oxidative stress and histopathologic changes in chronic gastritis-associated Helicobacter pylori infection. Arq Gastroenterol. 2017;54:177–182. doi: 10.1590/s0004-2803.201700000-18. [DOI] [PubMed] [Google Scholar]
- Juvale K, Shaik A, Kirubakaran S. Inhibitors of inosine 5′-monophosphate dehydrogenase as emerging new generation antimicrobial agents. Medchemcomm. 2019;10:1290–1301. doi: 10.1039/c9md00179d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafarski P, Talma M. Recent advances in design of new urease inhibitors: a review. J Adv Res. 2018;13:101–112. doi: 10.1016/j.jare.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj AK, Cotter TG, Oxentenko AS. Helicobacter pylori: the past, present, and future in management. Mayo Clin Proc. 2017;92:599–604. doi: 10.1016/j.mayocp.2016.11.017. [DOI] [PubMed] [Google Scholar]
- Kamiya S, Yonezawa H, Osaki T. Role of probiotics in eradication therapy for Helicobacter pylori infection. Adv Exp Med Biol. 2019;1149:243–255. doi: 10.1007/5584_2019_369. [DOI] [PubMed] [Google Scholar]
- Keilberg D, Zavros Y, Shepherd B, Salama NR, Ottemann KM. Spatial and temporal shifts in bacterial biogeography and gland occupation during the development of a chronic infection. MBio. 2016;7:e01705–e01716. doi: 10.1128/mBio.01705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienesberger S, Cox LM, Livanos A, Zhang XS, Chung J, Perez-Perez GI, Gorkiewicz G, Zechner EL, Blaser MJ. Gastric Helicobacter pylori infection affects local and distant microbial populations and host responses. Cell Rep. 2016;14:1395–1407. doi: 10.1016/j.celrep.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Hur JW, Yu MA, Cheigh CI, Kim KN, Hwang JK, Pyun YR. Antagonism of Helicobacter pylori by bacteriocins of lactic acid bacteria. J Food Prot. 2003;66:3–12. doi: 10.4315/0362-028X-66.1.3. [DOI] [PubMed] [Google Scholar]
- Koizumi W, Tanabe S, Hibi K, Imaizumi H, Ohida M, Okabe H, Saigenji K, Okayasu I. A prospective randomized study of amoxycillin and omeprazole with and without metronidazole in the eradication treatment of Helicobacter pylori. J Gastroenterol Hepatol. 1998;13:301–304. doi: 10.1111/j.1440-1746.1998.01559.x. [DOI] [PubMed] [Google Scholar]
- Königer V, Holsten L, Harrison U, Busch B, Loell E, Zhao Q, Bonsor DA, Roth A, Kengmo-Tchoupa A, Smith SI, Mueller S, Sundberg EJ, Zimmermann W, Fischer W, Hauck CR, Haas R. Helicobacter pylori exploits human CEACAMs via HopQ for adherence and translocation of CagA. Nat Microbiol. 2016;2:16188. doi: 10.1038/nmicrobiol.2016.188. [DOI] [PubMed] [Google Scholar]
- Kouitcheu Mabeku LB, Eyoum Bille B, Tchouangueu TF, Nguepi E, Leundji H. Treatment of Helicobacter pylori infected mice with Bryophyllum pinnatum, a medicinal plant with antioxidant and antimicrobial properties, reduces bacterial load. Pharm Biol. 2017;55:603–610. doi: 10.1080/13880209.2016.1266668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachler AM, Orth K. Targeting the bacteria-host interface strategies in anti-adhesion therapy. Virulence. 2013;4:284–294. doi: 10.4161/viru.24606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiecien S, Magierowski M, Majka J, Ptak-Belowska A, Wojcik D, Sliwowski Z, Magierowska K, Brzozowski T. Curcumin: a potent protectant against esophageal and gastric disorders. Int J Mol Sci. 2019;20:1477. doi: 10.3390/ijms20061477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeta C, McPartland L, Tran NQ, Meehan BM, Zhang Y, Tanweer Z, Wakabayashi S, Rock J, Kim T, Balasubramanian D, Audette R, Toosky M, Pinkham J, Rubin EJ, Lory S, Pier G, Boyd D, Beckwith J. Inhibition of Pseudomonas aeruginosa and Mycobacterium tuberculosis disulfide bond forming enzymes. Mol Microbiol. 2019;111:918–937. doi: 10.1111/mmi.14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Sun LC, Huang S, Zhu C, Li P, He J, Mackey V, Coy DH, He QY. The antimicrobial peptides and their potential clinical applications. Am J Transl Res. 2019;11:3919–3931. [PMC free article] [PubMed] [Google Scholar]
- Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol. 2011;65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesbros-Pantoflickova D, Corthésy-Theulaz I, Blum AL. Helicobacter pylori and probiotics. J Nutr. 2007;137:812S–818S. doi: 10.1093/jn/137.3.812S. [DOI] [PubMed] [Google Scholar]
- Liechti G, Goldberg JB. Helicobacter pylori relies primarily on the purine salvage pathway for purine nucleotide biosynthesis. J Bacteriol. 2012;194:839–854. doi: 10.1128/JB.05757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T-F, Hsu P-I. Second-line rescue treatment of Helicobacter pylori infection: where are we now? World J Gastroenterol. 2018;24:4548–4553. doi: 10.3748/wjg.v24.i40.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Huang WW. A systematic review of treating Helicobacter pylori infection with traditional Chinese medicine. World J Gastroenterol. 2009;15:4715–4719. doi: 10.3748/wjg.15.4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Frirdich E, Taylor JA, Chan ACK, Blair KM, Vermeulen J, Ha R, Murphy MEP, Salama NR, Gaynor EC, Tanner ME. A bacterial cell shape-determining inhibitor. ACS Chem Biol. 2016;11:981–991. doi: 10.1021/acschembio.5b01039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losurdo G, Cubisino R, Barone M, Principi M, Leandro G, Ierardi E, Di Leo A. Probiotic monotherapy and Helicobacter pylori eradication: a systematic review with pooled-data analysis. World J Gastroenterol. 2018;24:139–149. doi: 10.3748/wjg.v24.i1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah TFC, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Maixner F, Krause-Kyora B, Turaev D, Herbig A, Hoopmann MR, Hallows JL, Kusebauch U, Egarter Vigl E, Malfertheiner P, Megraud F, Cipollini G, Coia V, Samadelli M, Engstrand L, Linz B, Moritz RL, Grimm R, Krause J, Nebel A, Moodley Y, Rattei T, Zink A. The 5,300-year-old Helicobacter pylori genome of the iceman HHS public access. Science. 2016;351:162–165. doi: 10.1126/science.aad2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfertheiner P, Megraud F, O’Morain C, Gisbert JP, Kuipers EJ, Axon A, Bazzoli F, Gasbarrini A, Atherton J, Graham DY, Hunt R, Moayyedi P, Rokkas T, Rugge M, Selgrad M, Suerbaum S, Sugano K, El-Omar E, Agreus L, Andersen LP, Coelho L, Delchier JC, Di Mario F, Dinis-Ribeiro M, Fischbach W, Flahou B, Fock KM, Gasbarrini G, Gensini G, Goh KL, Herrero R, Kupcinskas L, Lanas A, Leja M, Machado JC, Mahachai V, Milosavljevic T, Niv Y, Ristimaki A, Tepes B, Vaira D, Vieth M, You W. Management of Helicobacter pylori infection-the Maastricht V/Florence consensus report. Gut. 2017;66:6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- Marshall B, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;323:1311–1315. doi: 10.1016/S0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Shiotani A, Graham DY. Current and future treatment of Helicobacter pylori infections. Adv Exp Med Biol. 2019;1149:211–225. doi: 10.1007/5584_2019_367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain M, Beckett A, Cover T. Helicobacter pylori vacuolating toxin and gastric cancer. Toxins (Basel) 2017;9:316. doi: 10.3390/toxins9100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland LV, Huang Y, Wang L, Malfertheiner P. Systematic review and meta-analysis: multi-strain probiotics as adjunct therapy for Helicobacter pylori eradication and prevention of adverse events. United Eur Gastroenterol J. 2016;4:546–561. doi: 10.1177/2050640615617358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11:664–674. doi: 10.1038/nrgastro.2014.143. [DOI] [PubMed] [Google Scholar]
- Miller EF, Vaish S, Maier RJ. Efficiency of purine utilization by Helicobacter pylori: roles for adenosine deaminase and a NupC homolog. PLoS One. 2012;7:e38727. doi: 10.1371/journal.pone.0038727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B, Reiling S, Zarena D, Wang G. Host defense antimicrobial peptides as antibiotics: design and application strategies. Curr Opin Chem Biol. 2017;38:87–96. doi: 10.1016/j.cbpa.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley Y, Linz B, Bond RP, Nieuwoudt M, Soodyall H, Schlebusch CM, Bernhöft S, Hale J, Suerbaum S, Mugisha L, van der Merwe SW, Achtman M. Age of the association between Helicobacter pylori and man. PLoS Pathog. 2012;8:e1002693. doi: 10.1371/journal.ppat.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonens K, Gideonsson P, Subedi S, Bugaytsova J, Romaõ E, Mendez M, Nordén J, Fallah M, Rakhimova L, Shevtsova A, Lahmann M, Castaldo G, Brännström K, Coppens F, Lo AW, Ny T, Solnick JV, Vandenbussche G, Oscarson S, Hammarström L, Arnqvist A, Berg DE, Muyldermans S, Borén T, Remaut H. Structural insights into polymorphic ABO glycan binding by Helicobacter pylori. Cell Host Microbe. 2016;19:55–66. doi: 10.1016/j.chom.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonens K, Hamway Y, Neddermann M, Reschke M, Tegtmeyer N, Kruse T, Kammerer R, Mejías-Luque R, Singer BB, Backert S, Gerhard M, Remaut H. Helicobacter pylori adhesin HopQ disrupts trans dimerization in human CEACAMs. EMBO J. 2018;37:e98665. doi: 10.15252/embj.201798665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli G, Didelot X, Kusecek B, Schwarz S, Bahlawane C, Falush D, Suerbaum S, Achtman M. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 2010;6:e1001036. doi: 10.1371/journal.pgen.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nammi D, Srimath-Tirumala-Peddinti RCPK, Neelapu NRR. Identification of drug targets in Helicobacter pylori by in silico analysis: possible therapeutic implications for gastric cancer. Curr Cancer Drug Targets. 2016;16:79–98. doi: 10.2174/1568009615666150602143239. [DOI] [PubMed] [Google Scholar]
- Nammi D, Yarla NS, Chubarev VN, Tarasov VV, Barreto GE, Corolina Pasupulati AM, Aliev G, Reddy Neelapu NR. A systematic in-silico analysis of Helicobacter pylori pathogenic islands for identification of novel drug target candidates. Curr Genomics. 2017;18:450–465. doi: 10.2174/1389202918666170705160615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narczyk M, Bertoša B, Papa L, Vuković V, Leščić Ašler I, Wielgus-Kutrowska B, Bzowska A, Luić M, Štefanić Z. Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features. FEBS J. 2018;285:1305–1325. doi: 10.1111/febs.14403. [DOI] [PubMed] [Google Scholar]
- Neshani A, Zare H, Akbari Eidgahi MR, Hooshyar Chichaklu A, Movaqar A, Ghazvini K. Review of antimicrobial peptides with anti-Helicobacter pylori activity. Helicobacter. 2019;24:e12555. doi: 10.1111/hel.12555. [DOI] [PubMed] [Google Scholar]
- Noto JM, Gaddy JA, Lee JY, Piazuelo MB, Friedman DB, Colvin DC, Romero-Gallo J, Suarez G, Loh J, Slaughter JC, Tan S, Morgan DR, Wilson KT, Bravo LE, Correa P, Cover TL, Amieva MR, Peek RM. Iron deficiency accelerates Helicobacter pylori–induced carcinogenesis in rodents and humans. J Clin Invest. 2013;123:479–492. doi: 10.1172/JCI64373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Morain NR, Dore MP, O’Connor AJP, Gisbert JP, O’Morain CA. Treatment of Helicobacter pylori infection in 2018. Helicobacter. 2018;23:1–9. doi: 10.1111/hel.12519. [DOI] [PubMed] [Google Scholar]
- Olekhnovich IN, Vitko S, Valliere M, Hoffman PS. Response to metronidazole and oxidative stress is mediated through homeostatic regulator HsrA (HP1043) in Helicobacter pylori. J Bacteriol. 2014;196:729–739. doi: 10.1128/JB.01047-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang SS, Nguyen STS, Perry AJ, Day CJ, Panjikar S, Tiralongo J, Whisstock JC, Kwok T. The three-dimensional structure of the extracellular adhesion domain of the sialic acid-binding adhesin SabA from Helicobacter pylori. J Biol Chem. 2014;289:6332–6340. doi: 10.1074/jbc.M113.513135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pero R, Coretti L, Nigro E, Lembo F, Laneri S, Lombardo B, Daniele A, Scudiero O. β-defensins in the fight against Helicobacter pylori. Molecules. 2017;22:1–17. doi: 10.3390/molecules22030424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pero R, Brancaccio M, Laneri S, De Biasi M-G, Lombardo B, Scudiero O. A novel view of human Helicobacter pylori infections: interplay between microbiota and beta-Defensins. Biomolecules. 2019;9:237. doi: 10.3390/biom9060237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczko AM, Bujnicki JM, Pawłowski M, Godlewska R, Lewandowska M, Jagusztyn-Krynicka EK. Characterization of new DsbB-like thiol-oxidoreductases of Campylobacter jejuni and Helicobacter pylori and classification of the DsbB family based on phylogenomic, structural and functional criteria. Microbiology. 2005;151:219–231. doi: 10.1099/mic.0.27483-0. [DOI] [PubMed] [Google Scholar]
- Ribaldone D, Fagoonee S, Astegiano M, Durazzo M, Morgando A, Sprujevnik T, Giordanino C, Baronio M, De Angelis C, Saracco G, Pellicano R. Rifabutin-based rescue therapy for Helicobacter pylori eradication: a long-term prospective study in a large cohort of difficult-to-treat patients. J Clin Med. 2019;8:199. doi: 10.3390/jcm8020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszczenko P, Radomska KA, Wywial E, Collet J-F, Jagusztyn-Krynicka EK. A novel insight into the oxidoreductase activity of Helicobacter pylori HP0231 protein. PLoS One. 2012;7:e46563. doi: 10.1371/journal.pone.0046563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JH, Inadomi JM, Scheiman J, Schoenfeld P, Appelman H, Zhang M, Metko V, Kao JY. Association between Helicobacter pylori and Barrett's esophagus, erosive esophagitis, and gastroesophageal reflux symptoms. Clin Gastroenterol Hepatol. 2014;12:239–245. doi: 10.1016/j.cgh.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs G, Weeks DL, Wen Y, Marcus EA, Scott DR, Melchers K. Acid acclimation by Helicobacter pylori. Physiology. 2005;20:429–438. doi: 10.1152/physiol.00032.2005. [DOI] [PubMed] [Google Scholar]
- Salama N, Guillemin K, McDaniel TK, Sherlock G, Tompkins L, Falkow S. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc Natl Acad Sci. 2000;97:14668–14673. doi: 10.1073/PNAS.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salillas S, Sancho J. Flavodoxins as novel therapeutic targets against Helicobacter pylori and other gastric pathogens. Int J Mol Sci. 2020;21:1881. doi: 10.3390/ijms21051881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M, Maganti L, Ghoshal N, Dutta C. In silico quest for putative drug targets in Helicobacter pylori HPAG1: molecular modeling of candidate enzymes from lipopolysaccharide biosynthesis pathway. J Mol Model. 2012;18:1855–1865. doi: 10.1007/s00894-011-1204-3. [DOI] [PubMed] [Google Scholar]
- Sarowska J, Choroszy-Król I, Regulska-Ilow B, Frej-Mądrzak M, Jama-Kmiecik A. The therapeutic effect of probiotic bacteria. Adv Clin Exp Med. 2013;22:759–766. [PubMed] [Google Scholar]
- Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization regions. Gastroenterology. 2018;155:1372–1382.e17. doi: 10.1053/J.GASTRO.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer CL, Good JAD, Kumar S, Krishnan KS, Gaddy JA, Loh JT, Chappell J, Almqvist F, Cover TL, Hadjifrangiskou M. Peptidomimetic small molecules disrupt type IV secretion system activity in diverse bacterial pathogens. MBio. 2016;7:e00221–e00216. doi: 10.1128/MBIO.00221-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Sun Y, Wang N, Yu H, Zhou Y, Chen C, Jia J. Changes of proteome components of Helicobacter pylori biofilms induced by serum starvation. Mol Med Rep. 2013;8:1761–1766. doi: 10.3892/mmr.2013.1712. [DOI] [PubMed] [Google Scholar]
- Sierra JM, Fusté E, Rabanal F, Vinuesa T, Viñas M. An overview of antimicrobial peptides and the latest advances in their development. Expert Opin Biol Ther. 2017;17:663–676. doi: 10.1080/14712598.2017.1315402. [DOI] [PubMed] [Google Scholar]
- Sigal M, Rothenberg ME, Logan CY, Lee JY, Honaker RW, Cooper RL, Passarelli B, Camorlinga M, Bouley DM, Alvarez G, Nusse R, Torres J, Amieva MR. Helicobacter pylori activates and expands Lgr5+ stem cells through direct colonization of the gastric glands. Gastroenterology. 2015;148:1392–1404.e21. doi: 10.1053/j.gastro.2015.02.049. [DOI] [PubMed] [Google Scholar]
- Smith R, Paxman J, Scanlon M, Heras B. Targeting bacterial Dsb proteins for the development of anti-virulence agents. Molecules. 2016;21:811. doi: 10.3390/molecules21070811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Štefanić Z, Mikleušević G, Luić M, Bzowska A, Leščić Ašler I. Structural characterization of purine nucleoside phosphorylase from human pathogen Helicobacter pylori. Int J Biol Macromol. 2017;101:518–526. doi: 10.1016/j.ijbiomac.2017.03.101. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5:441–452. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P. Kyoto global consensus report on Helicobacter pylori gastritis. Gut. 2015;64:1353–1367. doi: 10.1136/GUTJNL-2015-309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Esaki M, Kusano C, Ikehara H, Gotoda T. Development of Helicobacter pylori treatment: how do we manage antimicrobial resistance? World J Gastroenterol. 2019;25:1907–1912. doi: 10.3748/wjg.v25.i16.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, Salama NR. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, Salama NR. Multiple peptidoglycan modification networks modulate Helicobacter pylori's cell shape, motility, and colonization potential. PLoS Pathog. 2012;8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajewska H, Horvath A, Kołodziej M. Systematic review with meta-analysis: Saccharomyces boulardii supplementation and eradication of Helicobacter pylori infection. Aliment Pharmacol Ther. 2015;41:1237–1245. doi: 10.1111/apt.13214. [DOI] [PubMed] [Google Scholar]
- Tegtmeyer N, Neddermann M, Asche CI, Backert S. Subversion of host kinases: a key network in cellular signaling hijacked by Helicobacter pylori CagA. Mol Microbiol. 2017;105:358–372. doi: 10.1111/mmi.13707. [DOI] [PubMed] [Google Scholar]
- Tibrewal N, Elliott GI. Evaluation of hadacidin analogues. Bioorganic Med Chem Lett. 2011;21:517–519. doi: 10.1016/j.bmcl.2010.10.088. [DOI] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Craig Venter J. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Tong JL, Ran ZH, Shen J, Zhang CX, Xiao SD. Meta-analysis: the effect of supplementation with probiotics on eradication rates and adverse events during Helicobacter pylori eradication therapy. Aliment Pharmacol Ther. 2007;25:155–168. doi: 10.1111/j.1365-2036.2006.03179.x. [DOI] [PubMed] [Google Scholar]
- Uddin R, Khalil W. A comparative proteomic approach using metabolic pathways for the identification of potential drug targets against Helicobacter pylori. Genes Genomics. 2020;42:519–541. doi: 10.1007/s13258-020-00921-z. [DOI] [PubMed] [Google Scholar]
- Uotani T, Miftahussurur M, Yamaoka Y. Effect of bacterial and host factors on Helicobacter pylori eradication therapy. Expert Opin Ther Targets. 2015;19:1637–1650. doi: 10.1517/14728222.2015.1073261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia-Baca VH, Escamilla-García E, de la Garza-Ramos MA, Tamez-Guerra P, Gomez-Flores R, Urbina-Ríos CS. In vitro antimicrobial activity and downregulation of virulence gene expression on Helicobacter pylori by reuterin. Probiotics Antimicrob Proteins. 2018;10:168–175. doi: 10.1007/s12602-017-9342-2. [DOI] [PubMed] [Google Scholar]
- van Tonder AJ, Mistry S, Bray JE, Hill DMC, Cody AJ, Farmer CL, Klugman KP, von Gottberg A, Bentley SD, Parkhill J, Jolley KA, Maiden MCJ, Brueggemann AB. Defining the estimated core genome of bacterial populations using a Bayesian decision model. PLoS Comput Biol. 2014;10:e1003788. doi: 10.1371/journal.pcbi.1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vliet AHM, Kusters JG. Use of alignment-free phylogenetics for rapid genome sequence-based typing of Helicobacter pylori virulence markers and antibiotic susceptibility. J Clin Microbiol. 2015;53:2877–2888. doi: 10.1128/JCM.01357-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A, Dubey J, Hegde RR, Rastogi V, Pandit JK. Helicobacter pylori: past, current and future treatment strategies with gastroretentive drug delivery systems. J Drug Target. 2016;24:897–915. doi: 10.3109/1061186X.2016.1171326. [DOI] [PubMed] [Google Scholar]
- Wang Y-C, Wu D-C, Liao J-J, Wu C-H, Li W-Y, Weng B-C. In vitro activity of Impatiens balsamina L. against multiple antibiotic-resistant Helicobacter pylori. Am J Chin Med. 2009;37:713–722. doi: 10.1142/S0192415X09007181. [DOI] [PubMed] [Google Scholar]
- Wang S, Haapalainen AM, Yan F, Du Q, Tyler PC, Evans GB, Rinaldo-Matthis A, Brown RL, Norris GE, Almo SC, Schramm VL. A picomolar transition state analogue inhibitor of MTAN as a specific antibiotic for Helicobacter pylori. Biochemistry. 2012;51:6892–6894. doi: 10.1021/bi3009664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Cameron SA, Clinch K, Evans GB, Wu Z, Schramm VL, Tyler PC. New antibiotic candidates against Helicobacter pylori. J Am Chem Soc. 2015;137:14275–14280. doi: 10.1021/jacs.5b06110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Feng J, Chen P, Liu X, Ma M, Zhou R, Chang Y, Liu J, Li J, Zhao Q. Probiotics in Helicobacter pylori eradication therapy: systematic review and network meta-analysis. Clin Res Hepatol Gastroenterol. 2017;41:466–475. doi: 10.1016/j.clinre.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Wattam AR, Gabbard JL, Shukla M, Sobral BW. Comparative genomic analysis at the PATRIC, a bioinformatic resource center. Methods Mol Biol. 2014;1197:287–308. doi: 10.1007/978-1-4939-1261-2_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2017) Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics
- Wong EHJ, Ng CG, Chua EG, Tay ACY, Peters F, Marshall BJ, Ho B, Goh KL, Vadivelu J, Loke MF. Comparative genomics revealed multiple Helicobacter pylori genes associated with biofilm formation in vitro. PLoS One. 2016;11:1–16. doi: 10.1371/journal.pone.0166835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao ZP, Wang XD, Peng ZY, Huang S, Yang P, Li QS, Zhou LH, Hu XJ, Wu LJ, Zhou Y, Zhu HL. Molecular docking, kinetics study, and structure-activity relationship analysis of quercetin and its analogous as Helicobacter pylori urease inhibitors. J Agric Food Chem. 2012;60:10572–10577. doi: 10.1021/jf303393n. [DOI] [PubMed] [Google Scholar]
- Xu L, Shao C, Li G, Shan A, Chou S, Wang J, Ma Q, Dong N. Conversion of broad-spectrum antimicrobial peptides into species-specific antimicrobials capable of precisely targeting pathogenic bacteria. Sci Rep. 2020;10:1–9. doi: 10.1038/s41598-020-58014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Liu Y, Li N, Yu J, Cheng H, Li J, Zhang XZ. Anti-Helicobacter pylori activities of Chenopodium ambrosioides L. in vitro and in vivo. World J Gastroenterol. 2015;21:4178–4183. doi: 10.3748/wjg.v21.i14.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa H, Osaki T, Kurata S, Zaman C, Hanawa T, Kamiya S. Assessment of in vitro biofilm formation by Helicobacter pylori. J Gastroenterol Hepatol. 2010;25:S90–S940. doi: 10.1111/j.1440-1746.2009.06213.x. [DOI] [PubMed] [Google Scholar]
- Yonezawa H, Osaki T, Hojo F, Kamiya S. Effect of Helicobacter pylori biofilm formation on susceptibility to amoxicillin, metronidazole and clarithromycin. Microb Pathog. 2019;132:100–108. doi: 10.1016/j.micpath.2019.04.030. [DOI] [PubMed] [Google Scholar]
- Yu XD, Zheng RB, Xie JH, Su JY, Huang XQ, Wang YH, Zheng YF, Mo ZZ, Wu XL, Wu DW, Liang YE, Zeng HF, Su ZR, Huang P. Biological evaluation and molecular docking of baicalin and scutellarin as Helicobacter pylori urease inhibitors. J Ethnopharmacol. 2015;162:69–78. doi: 10.1016/j.jep.2014.12.041. [DOI] [PubMed] [Google Scholar]
- Zhang X, Jiang A, Wang G, Yu H, Qi B, Xiong Y, Zhou G, Qin M, Dou J, Wang J. Fusion expression of the PGLa-AM1 with native structure and evaluation of its anti-Helicobacter pylori activity. Appl Microbiol Biotechnol. 2017;101:5667–5675. doi: 10.1007/s00253-017-8302-9. [DOI] [PubMed] [Google Scholar]
- Zharkova MS, Orlov DS, Golubeva OY, Chakchir OB, Eliseev IE, Grinchuk TM, Shamova OV. Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics-a novel way to combat antibiotic resistance? Front Cell Infect Microbiol. 2019;9:128. doi: 10.3389/fcimb.2019.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Anderl F, Kruse T, Schindele F, Jagusztyn-Krynicka EK, Fischer W, Gerhard M, Mejías-Luque R. Helicobacter pylori HP0231 influences bacterial virulence and is essential for gastric colonization. PLoS One. 2016;11:e0154643. doi: 10.1371/journal.pone.0154643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Dong J, Yu X. Meta-analysis: Lactobacillus containing quadruple therapy versus standard triple first-line therapy for Helicobacter pylori eradication. Helicobacter. 2009;14:97–107. doi: 10.1111/j.1523-5378.2009.00716.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.