ABSTRACT
Nutritional and lifestyle changes remain at the core of healthy aging and disease prevention. Accumulating evidence underscores the impact of genetic, metabolic, and host gut microbial factors on individual responses to nutrients, paving the way for the stratification of nutritional guidelines. However, technological advances that incorporate biological, nutritional, lifestyle, and health data at an unprecedented scale and depth conceptualize a future where preventative dietary interventions will exceed stratification and will be highly individualized. We herein discuss how genetic information combined with longitudinal metabolomic, immune, behavioral, and gut microbial parameters, and bioclinical variables could define a digital replica of oneself, a “virtual digital twin,” which could serve to guide nutrition in a personalized manner. Such a model may revolutionize the management of obesity and its comorbidities, and provide a pillar for healthy aging.
Keywords: nutrition, obesity, nutrigenetics, immunome, microbiome, epigenome, digital twin, digital medicine
Introduction
Precision medicine is transforming clinical practice. A record-high number of new precision drugs, pharmacogenetic, and cancer risk-related genetic tests have been approved by the US FDA during the last 2 y, testimony to the practical benefits of extensive research on genetic and lifestyle factors impacting human health. The FDA approvals address the need to reshape health care by redefining drug target populations and provide patients with the most clinically effective, safe, and cost-effective personalized treatment options. The new era of data-driven medicine, vis-à-vis technological advances, provide the foundation for “the assessment and management of human health at an unprecedented level of resolution” (1), referred to as high-definition medicine. In a forward-thinking review article, Torkamani et al. (1), envisioned the future of high-definition technologies towards an “actual digital twin” health model in which deep phenotypic and molecular characteristics of a diseased person are most closely matched to thousands or millions of patients with the same disease and similar molecular profile, allowing better prediction of individualized treatment and health outcomes. Various computational, in vitro and in vivo “humanized” models align to this concept, particularly in oncology (2–4).
Several lines of evidence call for a major conceptual and practical shift in dietetics towards high-definition nutrition that would parallel progress in precision medicine. We herein discuss this evidence, emphasizing the need to combine genetic, immune, metabolic, behavioral, bioclinical, and meta-genomic data to guide nutrition in a truly personalized manner. Extending the “actual digital twin” health model (1), we conceptualize a future in which these health and biomolecular parameters will endow the definition of a digital replica of oneself, a “virtual digital twin” (Figure 1), that could provide the most acceptable, adaptable, efficient, and cost-effective dietary and lifestyle recommendations.
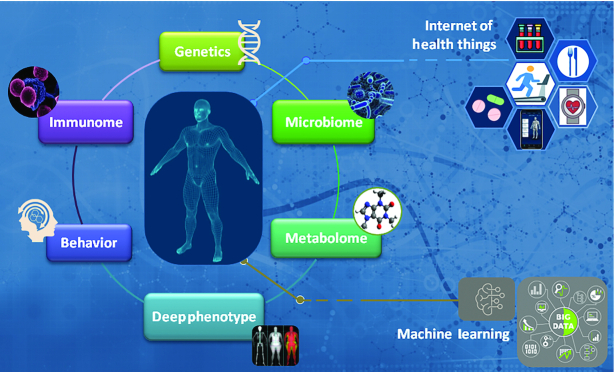
FIGURE 1.
Genetic, epigenetic, immune, metabolic, microbial, and behavioral paths to nutrition interact, highlighting the need for a “holistic” approach to precision nutrition. A “virtual digital twin” could be constructed as a digital replica of oneself by combining longitudinal high-definition -omics data, bioclinical and phenotypic variables, and behavioral aspects (dietary habits, physical activity, sleeping patterns, etc.) of the same individual, collected through internet of things health care platforms and managed by machine learning algorithms to simulate specific dietary and lifestyle strategies in silico. Such a highly individualized platform may revolutionize the management of obesity and its comorbidities, and provide a pillar for healthy aging.
Current Status of Knowledge
Genetic and epigenetic paths to precision nutrition
A plethora of twin and genome-wide association studies (GWAS) underscore a strong genetic influence on both human thinness (5) and obesity (6–8). Twin studies indicate that ∼40–70% of interindividual variability in BMI could be attributed to genetic factors (9), and >800 single nucleotide polymorphisms (SNPs) and copy number variations (CNVs) have been associated with obesity (6–8). Many of these genetic variants impinge on key hypothalamic circuits that regulate appetite, emotional eating, and food intake (6), indicating that eating behavior—influenced by genetics—may significantly affect body mass. Other SNPs relate to energy metabolism and expenditure, insulin secretion and action, adipogenesis, and other key biological processes (6). As individual variants may have small or intermediate effects on BMI, genetic risk scores (GRSs) have been proposed as the most relevant source of identifying high-risk individuals who would benefit most from early lifestyle changes or preventative treatments (10). It should be noted, however, that an obesogenic environment is likely to preferentially uncover inherited susceptibility among those with highest genetic risk, in accord to the “missing heritability” reported in several obesity GWAS. Polygenic risk scores are emerging as the new frontier, by evaluating a large number of genetic loci, not only those significantly associated with a defined phenotype or response (11).
There has been a long debate whether the macronutrient composition of dietary interventions impacts weight loss in individuals with obesity. In a seminal randomized clinical trial entailing 4 diets that differed in the percentages of essential macronutrients, the patterns of weight loss in overweight and obese were found similar for each diet (12). Notably, however, a considerable variation of weight loss was noted within each of the 4 dietary groups (12), indicating genetic influences in the response to dietary interventions. Beyond macronutrients, genetics may interplay with responses to micronutrients. For example, the association between vitamin D concentrations and cardiovascular disease risk varies among ethnic groups and likely reflects polymorphisms in genes involved in the biological activity of vitamin D (13).
Indeed, various clinical studies, including the POUNDS (Preventing Overweight Using Novel Dietary Strategies) Lost (12), the DioGenes (Diet, Obesity, and Genes) trial (14), PREDIMED (Prevención con Dieta Mediterránea) (15), Food4Me, and others, have uncovered genetic variants that may predict weight loss mostly in relation to specific macronutrients (16, 17). For example, obese individuals carrying the AA genotype of FTO rs1558902 respond to a high-protein diet with a significant reduction in BMI compared to those with a TT genotype (18). The AA genotype of HNF1A rs7957197 is associated with greater weight loss among individuals assigned to a low-fat diet compared to those with TT alleles (19). Eating behavior has also been linked to specific variants. Thus, a high-protein diet significantly reduces the appetite of carriers of the A allele of rs9939609 at the FTO locus, whereas individuals with an A allele at MC4R rs7227255 display increased appetite when assigned a low-fat diet (20, 21). Variants at the AMY1-AMY2 (amylase) locus (rs11185098) have been associated with weight loss irrespective of diet (22). Although association does not connote causation, several gene-diet associations have been experimentally validated in model systems [see, for example, (17, 23, 24)].
GRSs have also been explored in relation to responses to dietary components. For example, high coffee consumption has beneficial effects on weight loss only among those individuals with a high obesity GRS (25). A 32 SNP-based GRS can predict those individuals who are more susceptible to the adverse effects of sugar-sweetened beverages on obesity (26, 27) and a 13 risk allele GRS predicts reduction in blood triglycerides in individuals fed a diet supplemented with n–3 PUFAs (28). These are only a few examples of gene-diet interactions which, collectively, underscore the potential of genetically guided dietary recommendations for the management of obesity and for personalized healthy lifestyles (29). This area is rigorously explored by both the academic (30) and industrial sectors (31), including our teams.
Unlike the wealth of available information regarding the impact of genetic variants on BMI and nutritional responses, data on epigenetic effects is lagging behind. Whole genome DNA methylation profiles of adipose tissue and blood sampled from obese versus never obese individuals have identified a relatively small number of methylated genetic loci associated with BMI (32). Differential CpG methylation was predicted to affect genes involved in lipid and lipoprotein metabolism, substrate transport, and inflammatory pathways but appears to be predominantly the consequence rather than the cause of adiposity (32).
Nevertheless, there are several observations that underscore the need to continue exploring epigenetics in the context of precision nutrition. First, nutrition is a major environmental factor that may affect the epigenome, including DNA methylation. Second, the association of a subset of DNA methylation markers with BMI is stronger in certain populations (32). Third, the epigenetic heterogeneity observed at some of the differentially methylated loci among individuals, coupled with the graded relation between methylation and BMI, are indicative of epigenetic reprogramming, likely in adipocytes (32), which could be addressed by dietary and lifestyle interventions. This hypothesis aligns with the “epigenetic clock” model proposed by Steve Horvath in 2013 (33), which stipulates that “epigenetic age” is not only significantly related to biological age but is also both an indicator of overall health and subject to a healthy lifestyle (34). Along these lines, epigenetic reprogramming can ameliorate age-related hallmarks in vivo (35).
The immunome path to precision nutrition
Obesity is characterized by low-grade, subclinical systemic inflammation which affects several tissues and organs and impacts insulin sensitivity and secretion, and glucose homeostasis. As a result, obesity-associated inflammation is regarded as a driver of type 2 diabetes mellitus and a predisposing factor for the development of nonalcoholic fatty liver disease (NAFLD), nonalcoholic steatohepatitis (NASH), and several types of human malignancies (36).
Several studies have addressed changes in immune cell content in the peripheral blood in relation to BMI (37). The consensus drawn from the majority of these studies is that the immunophenotype of individuals with obesity differs from that of normal weight people, having increased numbers of white blood cells, neutrophils, intermediate CD14+CD16+ monocytes, and total CD3+ and CD4+ T cells, but reduced numbers of CD8+ T cells and of regulatory T (Treg) lymphocytes, a CD4+ T cell subset which functions to counterbalance exaggerated inflammatory responses and maintain self-tolerance. Individuals with very low Treg cell counts (<1.06%) have a 9.6-fold higher risk of developing an inflammatory obese phenotype (38). Elevated total lymphocyte counts, monocytes, and CD4+ T cells are also found in the peripheral blood of overweight (39, 40). In contrast, effector memory CD4+ T cells, which produce IFN-γ and TNF, are increased in obese but not in overweight individuals (40). We have recently reported experimental obesity-related changes in monocyte-macrophage responses to insulin characterized by increased glycolysis and a unique phenotype which may contribute to insulin resistance-associated inflammation (41). Several proinflammatory factors are also elevated in the plasma of individuals with obesity, including leptin, TNF, IL-6, and C-reactive protein (CRP) (42). CRP concentrations increase progressively with BMI (40) and decrease following weight loss (43). The aforementioned observations rationalized the development and use of inflammatory scores, which have uncovered associations with insulin resistance, BMI, waist circumference, and blood pressure (44).
Notably, systemic low-level inflammation is also a hallmark of aging. Dubbed “InflammAging” by Franceschi and colleagues (45), this aging-related inflammatory phenotype is typified by elevated serum concentrations of proinflammatory factors that include TNF, IL-6, and CRP, and are associated with frailty and increased morbidity and mortality in older adults (46). Elevated mortality risk in older people is also influenced by BMI (47). Effector memory CD4+ T cells also increase with age (48), similar to the obese immunophenotype (40). Therefore, monitoring inflammatory markers could serve to mobilize dietary and lifestyle changes towards both healthy aging and obesity prevention.
Along these lines, dietary supplementation with n–3 PUFAs in healthy adults, and overweight and obese individuals has been correlated with reduced serum CRP, TNF, and IL-6 concentrations in several studies (49, 50). Notably, genetic variants have been found to interact with dietary fatty acids to influence inflammatory responses (28, 51, 52). Micronutrients may also have significant effects on immune function; vitamin D3, for example, influences innate and adaptive immunity and confers anti-inflammatory properties, including positive effects on Treg cell numbers (53). Several polymorphisms have been identified in vitamin D receptor and vitamin D metabolism-associated genes that affect organismal vitamin D status and health outcomes (54), further underscoring the need to combine genotypic, bioclinical, and nutritional data to achieve improved immunological outcomes.
The advent of novel technologies, such as single-cell profiling with mass cytometry (CyTOF), which enables >50 parameters to be assayed on single cells, is anticipated to provide high-definition immunophenotypic information and revolutionize our understanding of the interplay between nutrition and immunity. The “10,000 Immunomes Project” (55), which aspires to define all the genes and proteins that constitute the immune system, will also accelerate research towards this goal.
Microbiota: a novel path to precision nutrition
Advances in genomic technologies have dramatically expanded our appreciation of the extent of microbial colonization in different organs and tissues and its impact on both human physiology and disease (56, 57). These analyses so far suggest that the microbiota is an essential partner of various biological functions of the host, including development and training of the immune system, defense against pathogenic microorganisms, digestion of food components, and synthesis of certain micronutrients and bioactive compounds such as SCFAs and other metabolites [(56); see also next section].
In general, a low abundance of the phylum Proteobacteria and a high abundance of the genera Bacteroides, Prevotella, and Ruminococcus have been associated with a healthy adult intestinal microbiota (58). Compared with lean people, obese individuals have a different microbiota profile that is largely typified by a higher Firmicutes-to-Bacteroidetes ratio, reduced bacterial diversity, and altered representation of bacterial genes and metabolic pathways (59). Such microbial shifts are likely to pose a functional impact on obesity. Indeed, a 20% increase in Firmicutes phylum abundance translates to elevated energy harvest from food in humans (60). Similarly, the microbiota of leptin-deficient ob/ob mice enables higher energy extraction from nutrients in the absence of an increase in food consumption (61). These seminal observations raise important questions pertinent to the exploitation of microbiota composition in precision nutrition.
Currently, microbiome signatures are explored as prognostic markers. Thus, a low bacterial diversity is predictive of a propensity towards developing obesity (62) and cardiovascular diseases in relation to the consumption of certain nutrients (63, 64). Individuals with obesity bearing a low gene-count microbiome were also more refractory to improvement of inflammatory variables during weight loss interventions (65). Conversely, stratification according to the Prevotella-to-Bacteroidesratio improves the outcomes of nutritional interventions on weight loss (66). Dysbiosis and reduced abundance of SCFA-producing bacterial genera are observed in children with a high risk of developing type 1 diabetes mellitus even before the overt manifestations of the disease (67). Another example entails an algorithm that integrates microbiota and can accurately predict postprandial glycemic responses despite variations among individuals (68).
Due to the complexity and individuality of each person's microbiota, the extent to which modulation of microbiota composition can be realized in the context of precision nutrition and the management of obesity is currently unknown (69). Ideally, personalized intervention strategies could be developed to “normalize” a deregulated microbiota or further improve the response to a specific diet. Probiotics, prebiotics, synthetic stools, and stool transplantation have demonstrated efficacy towards weight reduction in experimental models of obesity which warrants further studies in humans. Along these lines, the obesity-associated reduction in Treg cell numbers is reversed in mice administered probiotics (70), pertinent to the inverse correlation between Treg and the inflammatory obese phenotype in humans (38).
However, the key determinant of human gut microbiota is diet (71, 72) which is also more amenable to intervention. Adherence to the Mediterranean diet, for example, leads to increases in the genera Bacteroides, Prevotella, and Bifidobacteria (73), and long-term consumption of fiber results in a high abundance of Prevotella genus (74), both of which are associated with a healthy intestinal microbiota. Although further clinical studies are required, the available evidence indicates that normalization of gut microbiota has the potential to contribute to metabolic health and weight loss (57). In the context of precision nutrition, it would be important to proceed beyond the compositional analysis of microbiota towards understanding its interaction with the host, including host genetics (57), and microbial metabolites that may serve as key mediators of the host-microbiota connection, as discussed in the next section.
Advances in metabolomics of nutrition
The metabolome entails the end-products of metabolic processes and it has thus attracted significant attention in nutrition sciences for the identification of biomarkers of dietary intake and obesity. Several conventional metabolites, such as nonesterified free fatty acids, triglycerides, and products of carbohydrate catabolism (e.g. pyruvate and lactate) are higher in the serum of obese subjects (42, 75). Additional metabolic alterations identified in obese compared with lean subjects included several branched-chain essential amino acids, the presence of which correlated with obesity-associated gut microbiota (42, 75). Functional studies showed that rats fed a high-fat diet supplemented with branched-chain amino acids rapidly developed insulin resistance (42), linking microbiota with circulating amino acids and obesity-related pathologies. This association was further corroborated by the transcriptomic analysis of microflora from human twins discordant for obesity which uncovered induction of the de novo biosynthetic pathway for branched-chain amino acids in the microbiota of obese compared with lean twins (76). Remarkably, gut bacteria also produce metabolites capable of stimulating the central nervous system control of appetite, brain reward signaling, and emotional eating [reviewed in (77)]. These observations highlight the interplay between host genetic, metabolic, and microbial factors and further underscore the need for a holistic approach to nutritional interventions by combining several -omics outcomes.
Metabolomics has also been employed to identify biomarkers for differential responses to dietary interventions. Thus, weight loss in metabolically healthy obese women is accompanied by changes in plasma metabolites that include microbial products, lipids, and amino acids (78). For example, trimethylamine, an intermediate metabolite from the microbial metabolism of dietary carnitine and choline, is decreased after weight loss (78). Metabolomic studies in blood and urine from postmenopausal women with low bone density who received calcium and vitamin D supplements revealed different metabolic profiles which segregated with genotype and could partly predict nonresponders to treatment (79). Another study identified a metabolomic profile predictive of liver dysfunction in individuals fed a choline-depleted diet (80). Increased blood concentrations of acylcarnitines—intermediates of fatty acid and amino acid oxidation—are associated with cardiovascular diseases. Mediterranean diet interventions were found to diminish the risk of stroke associated with higher plasma concentrations of acylcarnitines before the intervention (81). These examples underscore the potential of exploring the human metabolome towards the identification of biomarkers that could predict responses to food components and dietary interventions in conjunction with additional -omics outcomes. Beyond obesity, metabolomics could be further explored to determine how food metabolites may influence food allergies or intolerances.
Behavioral aspects of nutrition
High-resolution monitoring of dietary behavior is an integral part of precision nutrition. A precise quantification of the type, amount, and frequency of food consumed by a person should ideally be accomplished by real-time monitoring rather than 24-h dietary recalls or short-term measurement of food consumption. Innovative technologies are being developed and will be increasingly utilized for the recording of dietary parameters. Table-embedded scales (82), automatic ingestion monitors that incorporate sensors for jaw motion with hand gestures and an accelerometer (83), smart-phone camera applications that calculate contents and calories through deep learning algorithms (84), and tooth-mounted sensors capable of recording a wide range of nutrients (85) are some examples of devices that aim to provide a more accurate means to log and adjust food consumption than dietary recalls. In addition to food quantity and quality, monitoring of food timing is also important as it affects weight loss effectiveness in conjunction with circadian cycle-related gene variants (86, 87). Such technologies are often combined with wearable sensors that offer continuous or frequent recording of several health parameters, including heart rate, oxygen saturation, respiratory rate, and blood pressure.
Activity tracking devices may provide additional insight into physiological measurements, a convenient and indirect estimate of energy consumption, and an objective reference for the motivation of specific health-promoting behaviors. Activity tracking includes monitoring of sleep duration. Short sleeping behaviors have been associated with irregular eating, higher total energy intake, and lower quality diets (88). Therefore, improving sleeping patterns has been proposed as an integral part of health promotion strategies that extend to weight management (88).
New technological advances may also assist in the assessment of basic emotions which are relevant to eating behavior, including measurement of hunger and food reward (89). For example, voice and image analysis has been used to detect mood disturbances relevant to eating disorders (90). As emotional functions are influenced by social interactions (91), environmental monitoring at high resolution may further contribute to the modulation of physiological, cognitive, and emotional states in the context of precision nutrition.
Furthermore, recent evidence suggests that dietary habits and behavioral eating have a strong genetic influence (92, 93). For example, of 85 curated habits analyzed, 83 were found to be significantly heritable and to be associated with several SNPs (93). Collectively, current evidence suggests that behavioral aspects of nutrition should be considered through a combination of genetics and real-time monitoring of food intake, physical activity, and emotional states.
Future directions and challenges: towards a “virtual digital twin” model for human nutrition
The adoption of high-definition technologies is leading to the accumulation of a wealth of data regarding basic health units (e.g. whole genome SNP profiles, immunophenotypes, bioclinical data, etc.), behavioral information (e.g. physical activity tracking), and environmental monitoring (e.g. food consumption). The UK Biobank represents a prime example of these efforts that have also led to major discoveries in the context of gene-diet interactions. Entries from such sources could aid the identification of “actual digital twins,” a set of people with similar characteristics at their most basic health units (1). The health outcomes of “actual digital twins” could be utilized to formulate basic prognostic models and actions that are predicted to be more relevant to the person in question and lead to improved outcomes compared with current practices.
A “virtual digital twin” could be constructed as a digital replica of oneself by high-definition data of the same individual, thereby extending the “actual digital twin” health model. To achieve a truly personalized platform in nutrition, the “virtual digital twin” should include a personal health baseline and enable the incorporation of health and lifestyle data in young (disease-free) versus old (disease-prone) age, along the lines of the longitudinal holo'ome monitoring we have previously proposed for colon cancer (94). For example, the obesity GRS could serve as an early-life, albeit incomplete, virtual representation of a baseline risk of weight gain which could progressively be enriched by the incorporation of genomic, metagenomic, immune, behavioral, and deep phenotypic data to build a “virtual digital twin” (Figure 1). Integrative -omics technologies, wearable sensors, internet of things (IoT), and consumer internet behavior are available tools and potential sources of such data with increasing capabilities that could spearhead early preventive actions for health maintenance.
Deviations from health states, such as an increase in BMI, a reduction in gut bacterial diversity, changes in bioclinical parameters (e.g. CRP, serum lipid profile), or an elevated inflammatory score, should enable health professionals to simulate further specific dietary and lifestyle strategies in silico by using the patient's “virtual digital twin.” In this manner, unwanted, unnecessary, or even harmful dietary or lifestyle choices could be eliminated and only the most promising interventions could be applied and monitored. A “virtual digital twin” may also serve as a powerful motivational tool that reinforces specific health care choices. Indeed, various public surveys have indicated that individuals are more likely to make nutritional choices and adhere to dietary recommendations if these are based on their molecular profile (95, 96).
The applicability of this model is underscored by a study by Price et al. (97) in which several metabolic, bioclinical, microbiome, and physical activity data were collected from 108 people 3 times over a 9-mo period and coupled to their genotype. Polygenic risk scores (PRS) and correlation networks were generated by integrating these data and identified clusters of analytes associated with physiology and disease which assisted in the implementation of behavioral coaching that helped individuals to improve health biomarkers. The results of this study represent an elegant example of high-definition health monitoring comprising a set of health and behavioral parameters that could serve as a pillar for the “digital twin” model of precision nutrition and healthy lifestyle.
Despite technological progress, several conceptual and practical barriers to the development and implementation of “digital twin” models need to be overcome. Personalized dietary advice is strongly determined by the way molecular, phenotypic, and bioclinical data are appreciated and utilized, calling for a change in the training that health professionals receive and a stronger interaction among relevant scientific disciplines. The former is highlighted by a recently published survey reporting that whereas 93.5% of dieticians perceived personalized nutrition as of high importance, only 9.5% of them had received some training on nutrigenetics (98). Although the cost of -omics technologies rapidly decreases and their performance increases, stronger computational tools and capabilities are likely to be required to accommodate and interrogate the large amount of data generated. Mathematical models are being developed for both polygenic prediction of weight/obesity trajectories (7, 26) and in the context of genetically guided nutrition (99) but further development of machine learning algorithms will be required to fully realize the potential of big data exploration in precision nutrition (100). IoT health care platforms are rapidly emerging to support the networking of mobiles, wearables, and other physical devices but need to be extended to facilitate the assembly and integration of longitudinal biological, nutritional, lifestyle, and health data. The implementation of “digital twin” models will also require carefully defined technical means and safeguards to protect sensitive personal information along with a specialized, detailed legal framework. Important questions such as who processes these data, where and how it is stored, who/under what conditions has access to these data, how can maximum data access protection be achieved, as well as how safe and effective database interoperability is ensured, will need to be carefully addressed.
Conclusions
The rapid progress in the field of precision medicine paves the way for a major conceptual and practical shift in dietetics towards high-definition nutrition. It is now evident that the wealth of information extracted from the human genome, combined with longitudinal metabolomic, immune, behavioral, and gut microbial parameters and bioclinical variables could be explored towards improved dietary choices. A guide for the future will be to develop user-friendly, artificial intelligence-based “virtual digital twin” platforms (Figure 1) that will endow health professionals with fully informed decision-making capabilities, surpassing stratification and providing nutritional and lifestyle recommendations that are highly individualized. We propose that such a platform may revolutionize the management of obesity and its comorbidities, and provide a pillar for healthy aging.
ACKNOWLEDGEMENTS
The authors’ contributions were as follows—KG, JV, DS, and AGE: contributed to the conception of the article; and all authors contributed to the preparation, and read and approved the final manuscript.
Notes
DS is supported by CURE-PLaN, a grant from the Leducq Foundation for Cardiovascular Research.
Author disclosures: The authors report no conflicts of interest.
Perspective articles allow authors to take a position on a topic of current major importance or controversy in the field of nutrition. As such, these articles could include statements based on author opinions or point of view. Opinions expressed in Perspective articles are those of the author and are not attributable to the funder(s) or the sponsor(s) or the publisher, Editor, or Editorial Board of Advances in Nutrition. Individuals with different positions on the topic of a Perspective are invited to submit their comments in the form of a Perspectives article or in a Letter to the Editor.
Abbreviations used: CRP, C-reactive protein; GRS, genetic risk score; GWAS, genome-wide association studies; IoT, internet of things; SNP, single nucleotide polymorphism.
Contributor Information
Kalliopi Gkouskou, Department of Biology, School of Medicine, National and Kapodistrian University of Athens, Athens, Greece; Embiodiagnostics, Biology Research Company, Heraklion, Crete, Greece.
Ioannis Vlastos, Department of Biology, School of Medicine, National and Kapodistrian University of Athens, Athens, Greece.
Petros Karkalousos, Department of Biomedical Sciences, University of West Attica, Athens, Greece.
Dimitrios Chaniotis, Department of Biomedical Sciences, University of West Attica, Athens, Greece.
Despina Sanoudou, Clinical Genomics and Pharmacogenomics Unit, 4th Department of Internal Medicine, School of Medicine, National and Kapodistrian University of Athens, Athens, Greece; Center for New Biotechnologies and Precision Medicine, School of Medicine, National and Kapodistrian University of Athens, Athens, Greece; Center of Basic Research, Biomedical Research Foundation of the Academy of Athens, Athens, Greece.
Aristides G Eliopoulos, Department of Biology, School of Medicine, National and Kapodistrian University of Athens, Athens, Greece; Center for New Biotechnologies and Precision Medicine, School of Medicine, National and Kapodistrian University of Athens, Athens, Greece; Center of Basic Research, Biomedical Research Foundation of the Academy of Athens, Athens, Greece.
References
- 1. Torkamani A, Andersen KG, Steinhubl SR, Topol EJ. High-definition medicine. Cell. 2017;170(5):828–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Filippo MD, Damiani C, Vanoni M, Maspero D, Mauri G, Alberghina L, Pescini D. Single-cell digital twins for cancer preclinical investigation. Methods Mol Biol. 2020;2088:331–43. [DOI] [PubMed] [Google Scholar]
- 3. Sontheimer-Phelps A, Hassell BA, Ingber DE. Modelling cancer in microfluidic human organs-on-chips. Nat Rev Cancer. 2019;19(2):65–81. [DOI] [PubMed] [Google Scholar]
- 4. De La Rochere P, Guil-Luna S, Decaudin D, Azar G, Sidhu SS, Piaggio E. Humanized mice for the study of immuno-oncology. Trends Immunol. 2018;39(9):748–63. [DOI] [PubMed] [Google Scholar]
- 5. Riveros-McKay F, Mistry V, Bounds R, Hendricks A, Keogh JM, Thomas H, Henning E, Corbin LJ, Understanding Society Scientific Group, O'Rahilly S et al. Genetic architecture of human thinness compared to severe obesity. PLoS Genet. 2019;15(1):e1007603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khera AV, Chaffin M, Wade KH, Zahid S, Brancale J, Xia R, Distefano M, Senol-Cosar O, Haas ME, Bick A et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell. 2019;177(3):587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stunkard AJ, Harris JR, Pedersen NL, McClearn GE. The body-mass index of twins who have been reared apart. N Engl J Med. 1990;322(21):1483–7. [DOI] [PubMed] [Google Scholar]
- 10. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19(9):581–90. [DOI] [PubMed] [Google Scholar]
- 12. Sacks FM, Bray GA, Carey VJ, Smith SR, Ryan DH, Anton SD, McManus K, Champagne CM, Bishop LM, Laranjo N et al. Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N Engl J Med. 2009;360(9):859–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pilz S, Verheyen N, Grubler MR, Tomaschitz A, Marz W. Vitamin D and cardiovascular disease prevention. Nat Rev Cardiol. 2016;13(7):404–17. [DOI] [PubMed] [Google Scholar]
- 14. Larsen TM, Dalskov SM, van Baak M, Jebb SA, Papadaki A, Pfeiffer AF, Martinez JA, Handjieva-Darlenska T, Kunesova M, Pihlsgard M et al. Diets with high or low protein content and glycemic index for weight-loss maintenance. N Engl J Med. 2010;363(22):2102–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fito M, Melander O, Martinez JA, Toledo E, Carpene C, Corella D. Advances in integrating traditional and omic biomarkers when analyzing the effects of the Mediterranean diet intervention in cardiovascular prevention. IJMS. 2016;17(9)1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bray GA, Krauss RM, Sacks FM, Qi L. Lessons learned from the POUNDS Lost Study: genetic, metabolic, and behavioral factors affecting changes in body weight, body composition, and cardiometabolic risk. Curr Obes Rep. 2019;8(3):262–83. [DOI] [PubMed] [Google Scholar]
- 17. Valsesia A, Wang QP, Gheldof N, Carayol J, Ruffieux H, Clark T, Shenton V, Oyston LJ, Lefebvre G, Metairon S et al. Genome-wide gene-based analyses of weight loss interventions identify a potential role for NKX6.3 in metabolism. Nat Commun. 2019;10(1):540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang X, Qi Q, Zhang C, Smith SR, Hu FB, Sacks FM, Bray GA, Qi L. FTO genotype and 2-year change in body composition and fat distribution in response to weight-loss diets: the POUNDS LOST Trial. Diabetes. 2012;61(11):3005–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang T, Wang T, Heianza Y, Sun D, Ivey K, Durst R, Schwarzfuchs D, Stampfer MJ, Bray GA, Sacks FM et al. HNF1A variant, energy-reduced diets and insulin resistance improvement during weight loss: The POUNDS Lost Trial and DIRECT. Diabetes Obes Metab. 2018;20(6):1445–52. [DOI] [PubMed] [Google Scholar]
- 20. Huang T, Zheng Y, Hruby A, Williamson DA, Bray GA, Shen Y, Sacks FM, Qi L. Dietary protein modifies the effect of the MC4R genotype on 2-year changes in appetite and food craving: The POUNDS Lost Trial. J Nutr. 2017;147(3):439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang T, Qi Q, Li Y, Hu FB, Bray GA, Sacks FM, Williamson DA, Qi L. FTO genotype, dietary protein, and change in appetite: the Preventing Overweight Using Novel Dietary Strategies trial. Am J Clin Nutr. 2014;99(5):1126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heianza Y, Sun D, Wang T, Huang T, Bray GA, Sacks FM, Qi L. Starch digestion-related amylase genetic variant affects 2-year changes in adiposity in response to weight-loss diets: The POUNDS Lost Trial. Diabetes. 2017;66(9):2416–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Larder R, Sim MFM, Gulati P, Antrobus R, Tung YCL, Rimmington D, Ayuso E, Polex-Wolf J, Lam BYH, Dias C et al. Obesity-associated gene TMEM18 has a role in the central control of appetite and body weight regulation. Proc Natl Acad Sci USA. 2017;114(35):9421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang T, Huang T, Kang JH, Zheng Y, Jensen MK, Wiggs JL, Pasquale LR, Fuchs CS, Campos H, Rimm EB et al. Habitual coffee consumption and genetic predisposition to obesity: gene-diet interaction analyses in three US prospective studies. BMC Med. 2017;15(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ebbeling CB, Feldman HA, Chomitz VR, Antonelli TA, Gortmaker SL, Osganian SK, Ludwig DS. A randomized trial of sugar-sweetened beverages and adolescent body weight. N Engl J Med. 2012;367(15):1407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brunkwall L, Chen Y, Hindy G, Rukh G, Ericson U, Barroso I, Johansson I, Franks PW, Orho-Melander M, Renstrom F. Sugar-sweetened beverage consumption and genetic predisposition to obesity in 2 Swedish cohorts. Am J Clin Nutr. 2016;104(3):809–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rudkowska I, Guenard F, Julien P, Couture P, Lemieux S, Barbier O, Calder PC, Minihane AM, Vohl MC. Genome-wide association study of the plasma triglyceride response to an n-3 polyunsaturated fatty acid supplementation. J Lipid Res. 2014;55(7):1245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ordovas JM, Ferguson LR, Tai ES, Mathers JC. Personalised nutrition and health. BMJ. 2018;361:bmj.k2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arkadianos I, Valdes AM, Marinos E, Florou A, Gill RD, Grimaldi KA. Improved weight management using genetic information to personalize a calorie controlled diet. Nutr J. 2007;6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Floris M, Cano A, Porru L, Addis R, Cambedda A, Idda ML, Steri M, Ventura C, Maioli M. Direct-to-consumer nutrigenetics testing: an overview. Nutrients. 2020;12(2):566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, Tsai PC, Ried JS, Zhang W, Yang Y et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541(7635):81–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, Ritz B, Bandinelli S, Neuhouser ML, Beasley JM et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging. 2017;9(2):419–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ocampo A, Reddy P, Martinez-Redondo P, Platero-Luengo A, Hatanaka F, Hishida T, Li M, Lam D, Kurita M, Beyret E et al. In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell. 2016;167(7):1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee YS, Wollam J, Olefsky JM. An integrated view of immunometabolism. Cell. 2018;172(1–2):22–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pecht T, Gutman-Tirosh A, Bashan N, Rudich A. Peripheral blood leucocyte subclasses as potential biomarkers of adipose tissue inflammation and obesity subphenotypes in humans. Obes Rev. 2014;15(4):322–37. [DOI] [PubMed] [Google Scholar]
- 38. Wagner NM, Brandhorst G, Czepluch F, Lankeit M, Eberle C, Herzberg S, Faustin V, Riggert J, Oellerich M, Hasenfuss G et al. Circulating regulatory T cells are reduced in obesity and may identify subjects at increased metabolic and cardiovascular risk. Obesity. 2013;21(3):461–8. [DOI] [PubMed] [Google Scholar]
- 39. Womack J, Tien PC, Feldman J, Shin JH, Fennie K, Anastos K, Cohen MH, Bacon MC, Minkoff H. Obesity and immune cell counts in women. Metabolism. 2007;56(7):998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mauro C, Smith J, Cucchi D, Coe D, Fu H, Bonacina F, Baragetti A, Cermenati G, Caruso D, Mitro N et al. Obesity-induced metabolic stress leads to biased effector memory CD4(+) T cell differentiation via PI3K p110delta-Akt-mediated signals. Cell Metab. 2017;25(3):593–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ieronymaki E, Theodorakis EM, Lyroni K, Vergadi E, Lagoudaki E, Al-Qahtani A, Aznaourova M, Neofotistou-Themeli E, Eliopoulos AG, Vaporidi K et al. Insulin resistance in macrophages alters their metabolism and promotes an M2-like phenotype. J Immunol. 2019;202(6):1786–97. [DOI] [PubMed] [Google Scholar]
- 42. Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tchernof A, Nolan A, Sites CK, Ades PA, Poehlman ET. Weight loss reduces C-reactive protein levels in obese postmenopausal women. Circulation. 2002;105(5):564–9. [DOI] [PubMed] [Google Scholar]
- 44. Recasens M, Lopez-Bermejo A, Ricart W, Vendrell J, Casamitjana R, Fernandez-Real JM. An inflammation score is better associated with basal than stimulated surrogate indexes of insulin resistance. J Clin Endocrinol Metab. 2005;90(1):112–6. [DOI] [PubMed] [Google Scholar]
- 45. Franceschi C, Bonafe M, Valensin S, Olivieri F, De Luca M, Ottaviani E, De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2006;908:244–54. [DOI] [PubMed] [Google Scholar]
- 46. Pansarasa O, Pistono C, Davin A, Bordoni M, Mimmi MC, Guaita A, Cereda C. Altered immune system in frailty: genetics and diet may influence inflammation. Ageing Res Rev. 2019;54:100935. [DOI] [PubMed] [Google Scholar]
- 47. de Hollander EL, Van Zutphen M, Bogers RP, Bemelmans WJ, De Groot LC. The impact of body mass index in old age on cause-specific mortality. J Nutr Health Aging. 2012;16(1):100–6. [DOI] [PubMed] [Google Scholar]
- 48. Saule P, Trauet J, Dutriez V, Lekeux V, Dessaint JP, Labalette M. Accumulation of memory T cells from childhood to old age: central and effector memory cells in CD4(+) versus effector memory and terminally differentiated memory cells in CD8(+) compartment. Mech Ageing Dev. 2006;127(3):274–81. [DOI] [PubMed] [Google Scholar]
- 49. He K, Liu K, Daviglus ML, Jenny NS, Mayer-Davis E, Jiang R, Steffen L, Siscovick D, Tsai M, Herrington D. Associations of dietary long-chain n-3 polyunsaturated fatty acids and fish with biomarkers of inflammation and endothelial activation (from the Multi-Ethnic Study of Atherosclerosis [MESA]). Am J Cardiol. 2009;103(9):1238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Connaughton RM, McMorrow AM, McGillicuddy FC, Lithander FE, Roche HM. Impact of anti-inflammatory nutrients on obesity-associated metabolic-inflammation from childhood through to adulthood. Proc Nutr Soc. 2016;75(2):115–24. [DOI] [PubMed] [Google Scholar]
- 51. Lankinen MA, Fauland A, Shimizu BI, Agren J, Wheelock CE, Laakso M, Schwab U, Pihlajamaki J. Inflammatory response to dietary linoleic acid depends on FADS1 genotype. Am J Clin Nutr. 2019;109(1):165–75. [DOI] [PubMed] [Google Scholar]
- 52. Keramat L, Sadrzadeh-Yeganeh H, Sotoudeh G, Zamani E, Eshraghian M, Mansoori A, Koohdani F. Apolipoprotein A2 -265 T>C polymorphism interacts with dietary fatty acids intake to modulate inflammation in type 2 diabetes mellitus patients. Nutrition. 2017;37:86–91. [DOI] [PubMed] [Google Scholar]
- 53. Wei R, Christakos S. Mechanisms underlying the regulation of innate and adaptive immunity by vitamin D. Nutrients. 2015;7(10):8251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jolliffe DA, Walton RT, Griffiths CJ, Martineau AR. Single nucleotide polymorphisms in the vitamin D pathway associating with circulating concentrations of vitamin D metabolites and non-skeletal health outcomes: review of genetic association studies. J Steroid Biochem Mol Biol. 2016;164:18–29. [DOI] [PubMed] [Google Scholar]
- 55. Zalocusky KA, Kan MJ, Hu Z, Dunn P, Thomson E, Wiser J, Bhattacharya S, Butte AJ. The 10,000 Immunomes Project: building a resource for human immunology. Cell Rep. 2018;25(2):513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Panagi M, Georgila K, Eliopoulos AG, Apidianakis Y. Constructing personalized longitudinal holo'omes of colon cancer-prone humans and their modeling in flies and mice. Oncotarget. 2019;10(41):4224–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cuevas-Sierra A, Ramos-Lopez O, Riezu-Boj JI, Milagro FI, Martinez JA. Diet, gut microbiota, and obesity: links with host genetics and epigenetics and potential applications. Adv Nutr. 2019;10(suppl_1):S17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hollister EB, Gao C, Versalovic J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology. 2014;146(6):1449–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94(1):58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. [DOI] [PubMed] [Google Scholar]
- 62. Zmora N, Zeevi D, Korem T, Segal E, Elinav E. Taking it personally: personalized utilization of the human microbiome in health and disease. Cell Host Microbe. 2016;19(1):12–20. [DOI] [PubMed] [Google Scholar]
- 63. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500(7464):585–8. [DOI] [PubMed] [Google Scholar]
- 66. Hjorth MF, Blaedel T, Bendtsen LQ, Lorenzen JK, Holm JB, Kiilerich P, Roager HM, Kristiansen K, Larsen LH, Astrup A. Prevotella-to-Bacteroides ratio predicts body weight and fat loss success on 24-week diets varying in macronutrient composition and dietary fiber: results from a post-hoc analysis. Int J Obes. 2019;43(1):149–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, Lernmark A, Hagopian WA, Rewers MJ, She JX et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562(7728):589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–94. [DOI] [PubMed] [Google Scholar]
- 69. Kolodziejczyk AA, Zheng D, Elinav E. Diet-microbiota interactions and personalized nutrition. Nat Rev Microbiol. 2019;17(12):742–53. [DOI] [PubMed] [Google Scholar]
- 70. Poutahidis T, Kleinewietfeld M, Smillie C, Levkovich T, Perrotta A, Bhela S, Varian BJ, Ibrahim YM, Lakritz JR, Kearney SM et al. Microbial reprogramming inhibits Western diet-associated obesity. PLoS One. 2013;8(7):e68596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yang Q, Liang Q, Balakrishnan B, Belobrajdic DP, Feng QJ, Zhang W. Role of dietary nutrients in the modulation of gut microbiota: a narrative review. Nutrients. 2020;12(2):381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci. 2010;107(33):14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, Shi J, Zhao S, Liu W, Wang X et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23(7):859–68. [DOI] [PubMed] [Google Scholar]
- 76. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Torres-Fuentes C, Schellekens H, Dinan TG, Cryan JF. The microbiota-gut-brain axis in obesity. Lancet Gastroenterol Hepatol. 2017;2(10):747–56. [DOI] [PubMed] [Google Scholar]
- 78. Almanza-Aguilera E, Brunius C, Bernal-Lopez MR, Garcia-Aloy M, Madrid-Gambin F, Tinahones FJ, Gomez-Huelgas R, Landberg R, Andres-Lacueva C. Impact in plasma metabolome as effect of lifestyle intervention for weight-loss reveals metabolic benefits in metabolically healthy obese women. J Proteome Res. 2018;17(8):2600–10. [DOI] [PubMed] [Google Scholar]
- 79. Elnenaei MO, Chandra R, Mangion T, Moniz C. Genomic and metabolomic patterns segregate with responses to calcium and vitamin D supplementation. Br J Nutr. 2011;105(1):71–9. [DOI] [PubMed] [Google Scholar]
- 80. Sha W, da Costa KA, Fischer LM, Milburn MV, Lawton KA, Berger A, Jia W, Zeisel SH. Metabolomic profiling can predict which humans will develop liver dysfunction when deprived of dietary choline. FASEB J. 2010;24(8):2962–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Guasch-Ferre M, Zheng Y, Ruiz-Canela M, Hruby A, Martinez-Gonzalez MA, Clish CB, Corella D, Estruch R, Ros E, Fito M et al. Plasma acylcarnitines and risk of cardiovascular disease: effect of Mediterranean diet interventions. Am J Clin Nutr. 2016;103(6):1408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mattfeld RS, Muth ER, Hoover A. Measuring the consumption of individual solid and liquid bites using a table-embedded scale during unrestricted eating. IEEE J Biomed Health Inform. 2017;21(6):1711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fontana JM, Farooq M, Sazonov E. Automatic ingestion monitor: a novel wearable device for monitoring of ingestive behavior. IEEE Trans Biomed Eng. 2014;61(6):1772–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mandracchia F, Llaurado E, Tarro L, Del Bas JM, Valls RM, Pedret A, Radeva P, Arola L, Sola R, Boque N. Potential use of mobile phone applications for self-monitoring and increasing daily fruit and vegetable consumption: a systematized review. Nutrients. 2019;11(3):686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tseng P, Napier B, Garbarini L, Kaplan DL, Omenetto FG. Functional, RF-Trilayer sensors for tooth-mounted, wireless monitoring of the oral cavity and food consumption. Adv Mater. 2018;30(18):1703257. [DOI] [PubMed] [Google Scholar]
- 86. Garaulet M, Gomez-Abellan P, Alburquerque-Bejar JJ, Lee YC, Ordovas JM, Scheer FA. Timing of food intake predicts weight loss effectiveness. Int J Obes. 2013;37(4):604–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Garaulet M, Vera B, Bonnet-Rubio G, Gomez-Abellan P, Lee YC, Ordovas JM. Lunch eating predicts weight-loss effectiveness in carriers of the common allele at PERILIPIN1: the ONTIME (Obesity, Nutrigenetics, Timing, Mediterranean) study. Am J Clin Nutr. 2016;104(4):1160–6. [DOI] [PubMed] [Google Scholar]
- 88. Dashti HS, Scheer FA, Jacques PF, Lamon-Fava S, Ordovas JM. Short sleep duration and dietary intake: epidemiologic evidence, mechanisms, and health implications. Adv Nutr. 2015;6(6):648–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gibbons C, Hopkins M, Beaulieu K, Oustric P, Blundell JE. Issues in measuring and interpreting human appetite (satiety/satiation) and its contribution to obesity. Curr Obes Rep. 2019;8(2):77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Leppanen J, Dapelo MM, Davies H, Lang K, Treasure J, Tchanturia K. Computerised analysis of facial emotion expression in eating disorders. PLoS One. 2017;12(6):e0178972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Fowler JH, Christakis NA. Dynamic spread of happiness in a large social network: longitudinal analysis over 20 years in the Framingham Heart Study. BMJ. 2008;337:a2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Merino J, Dashti HS, Li SX, Sarnowski C, Justice AE, Graff M, Papoutsakis C, Smith CE, Dedoussis GV, Lemaitre RN et al. Genome-wide meta-analysis of macronutrient intake of 91,114 European ancestry participants from the cohorts for heart and aging research in genomic epidemiology consortium. Mol Psychiatry. 2019;24(12):1920–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cole JB, Florez JC, Hirschhorn JN. Comprehensive genomic analysis of dietary habits in UK Biobank identifies hundreds of genetic associations. Nat Commun. 2020;11(1):1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Apidianakis Y, Eliopoulos AG. A holo'ome approach in colon cancer: we change as we age. EMBO Rep. 2015;16(10):1239–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Nielsen DE, Shih S, El-Sohemy A. Perceptions of genetic testing for personalized nutrition: a randomized trial of DNA-based dietary advice. J Nutrigenet Nutrigenomics. 2014;7(2):94–104. [DOI] [PubMed] [Google Scholar]
- 96. Vallee Marcotte B, Cormier H, Garneau V, Robitaille J, Desroches S, Vohl MC. Current knowledge and interest of French Canadians regarding nutrigenetics. Genes Nutr. 2019;14:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Price ND, Magis AT, Earls JC, Glusman G, Levy R, Lausted C, McDonald DT, Kusebauch U, Moss CL, Zhou Y et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat Biotechnol. 2017;35(8):747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kaufman-Shriqui V, Salem H, Boaz M, Birk R. Knowledge and attitudes towards nutrigenetics: findings from the 2018 Unified Forces Preventive Nutrition Conference (UFPN). Nutrients. 2020;12(2):335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ramos-Lopez O, Cuervo M, Goni L, Milagro FI, Riezu-Boj JI, Martinez JA. Modeling of an integrative prototype based on genetic, phenotypic, and environmental information for personalized prescription of energy-restricted diets in overweight/obese subjects. Am J Clin Nutr. 2020;111(2):459–70. [DOI] [PubMed] [Google Scholar]
- 100. Perakakis N, Yazdani A, Karniadakis GE, Mantzoros C. Omics, big data and machine learning as tools to propel understanding of biological mechanisms and to discover novel diagnostics and therapeutics. Metabolism. 2018;87:A1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]