ABSTRACT
Sirtuin 6 (SIRT6) exerts a protective effect on health and extends the lives of model organisms. We, therefore, aimed to clarify whether age-related epigenetic drift is responsible for differences in SIRT6 expression in peripheral blood mononuclear cells (PBMCs) of healthy young (n = 55, mean age 27.5 ± 4.4 years), middle-aged (n = 51, 65.4 ± 3.3 years), and long-lived (n = 51, 93.9 ± 3.6 years) humans. In silico analysis was performed using the STRING network. No age-related differences were observed in the percentage of SIRT6 CpG island methylation. However, the age affected the expression of miR-34a-5p, miR-125a-5p, miR-186-5p, miR-342-5p and miR-766-3p (all p < 0.0001), miR-181-2-3p and Let-7c (both p = 0.0003), and miR-103a-3p (p = 0.0069). A negative association was observed between SIRT6 mRNA and miR-186-5p (rs = −0.25, p = 0.026), and a positive association was observed with miR-34a-5p (rs = 0.31, p = 0.0055) and miR-181a-2-3p (rs = 0.39, p = 0.0002). SIRT6 mRNA also negatively correlated with the expression of TP53 (rs = −0.41, p = 0.0126) and MYC (rs = −0.35, p = 0.0448). Notably, the expression of several miRNAs and genes was similar in young and long-lived groups but different from the middle-aged group. We conclude that age-related epigenetic changes can affect the expression of SIRT6 in PBMCs and, in this way, possibly influence immunosenescence. Moreover, molecular events could differentiate ‘normal’ ageing from that of long-lived individuals.
KEYWORDS: SIRT6, interaction network, miRNA, methylation, peripheral blood mononuclear cells (PBMCs), aging, longevity
Introduction
Sirtuins form a conserved family of nicotinamide adenine dinucleotide (NAD+)-dependent deacetylases. Invertebrates have a single sirtuin isoform, whereas mammals have seven sirtuins (SIRT1-7) that are expressed in different cell compartments [1]. SIRT6, which is present mainly in the nucleus, plays an essential role in the post-translational epigenetic modification of histone proteins, chromatin formation, and maintenance of genome integrity [2,3]. It decreases oxidative stress, increases resistance to DNA damage [4,5], and takes part in DNA repair [6] and metabolic homoeostasis [7]. SIRT6 also plays a role in acute and chronic inflammatory responses, where it exhibits both pro- and anti-inflammatory activities [8,9].
Overexpression or increased activity of SIRT6 orthologs have beneficial effects on the lifespans of model organisms. For example, overexpression of sir2 (silent information regulator 2) in S. cerevisiae extended its life by 30 to 50% [10–12], the presence of additional copies of dSir2 extended life of D. melanogaster by 57% [13], while SIRT6 overexpression in male mice resulted in a 15% lifespan elongation [14]. Notably, SIRT6 appears also to affect the length of life in humans. The C allele of the SIRT6 rs350846 polymorphism is positively associated [15], while the T allele of rs117385980 is negatively associated with longevity [16]. Furthermore, individuals with the CC or CT genotype of rs107251 had a greater 5-year mean survival time compared to carriers of the TT genotype [17]. In addition to the ageing-delaying functions of SIRT6 related to DNA repair and telomere homoeostasis, SIRT6 exerts protective effects on human health through various molecular mechanisms (e.g., by regulation of stem cell biology, cancerogenesis, and metabolic homoeostasis), which may be the underlying causes for the extended lifespan [18,19].
Epigenetic modifications, such as DNA methylation, post-translational modifications of histone proteins, and the action of small non-coding RNA molecules, undergo ageing-related changes [20]. In the context of DNA methylation, this phenomenon was called an ‘epigenetic drift’ [21]. However, this term is currently extended to the other epigenetic changes (e.g., miRNA interference) that occur with age, since all of them affect age-related changes in the transcriptome [22–24].
In our previous work, we showed that SIRT6 expression in human peripheral blood mononuclear cells (PBMCs) significantly decreases with age; however, such a decrease was not observed in PBMCs of long-lived, healthy study subjects (Supplementary Figure S1) [23]. In the current study, using unique biological material obtained from healthy middle-aged and long-lived individuals, we aimed to clarify whether two epigenetic regulatory mechanisms (i.e., DNA methylation and the action of miRNAs) could be responsible for the age-related differences in SIRT6 expression in PBMCs and whether these SIRT6 alterations correlated with the expression of interacting genes associated with vital cellular processes, including apoptosis and immunity.
Results
Analysis of the methylation status of the SIRT6 promoter region
In silico analysis identified a 564-bp fragment within the SIRT6 gene, located between −292 bp and +272 bp relative to the transcription start site (TSS). For methylation analysis, this CpG island was divided into two parts, and each fragment contained additional surrounding sequences. The first fragment was located between −412 and −10 bp (32 potential methylation sites), and the second fragment was located between −3 to +302 bp (29 potential methylation sites) relative to the TSS. Our method of methylation analysis revealed that the first fragment had a higher percentage of the methylated CG pairs than the second; however, no age-related differences in the percentage of methylation in both analysed regions were observed (Supplementary Figure S2). There was also no correlation between the percentage of methylation and SIRT6 mRNA levels, suggesting that methylation does not play an essential role in ageing-related SIRT6 expression changes in PBMCs.
Expression of in silico-selected SIRT6 mRNA-interacting miRNAs in PBMCs
miRNAs that could potentially interact with SIRT6 mRNA, were selected using the MirWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/), miRanda (http://www.microrna.org/microrna/home.do), and TargetScan (http://www.targetscan.org/) programs, as well as literature data [25]. Results of the in silico analyses are summarized in Supplementary Table S1. Two out of eight studied miRNAs, let-7c and miR-342-5p, appeared to interact with the coding sequence (CDS), while miR-181a-2-3p with the 5´UTR. Taking into account an increasing amount of data indicating the role of 5´UTR and CDS regions in the regulation of gene expression, we have decided to include these miRNAs into our experiments as they could have additional interaction sites in the 3'UTR [26,27].
miRNA expression analysis was performed using real-time PCR. No significant gender-related differences in miRNA levels were observed, and further analyses were performed for all study subjects combined. To normalize the data, we intended to use U6 small nuclear RNA (U6 snRNA) expression as a reference, but in our data set, its levels significantly differed between the studied groups. However, the age did not affect the mean value of the miR-125b-5p cross point (Y: 32.7, E: 32.8, LL: 32.2); therefore, its expression was used as a reference for other analyses.
Expression of the remaining miRNAs was affected by age: p < 0.0001 for miR-34a-5p, miR-125a-5p, miR-186-5p, miR-342-5p and miR-766-3p; p = 0.0003 for miR-181-2-3p and Let-7c; p = 0.0069 for miR-103a-3p. The multiple comparisons tests (post-hoc) showed significant differences between the age groups (Figure 1). Age-related alterations in miR-34a-5p expression have been previously described [23] and confirmed in the current study.
Figure 1.
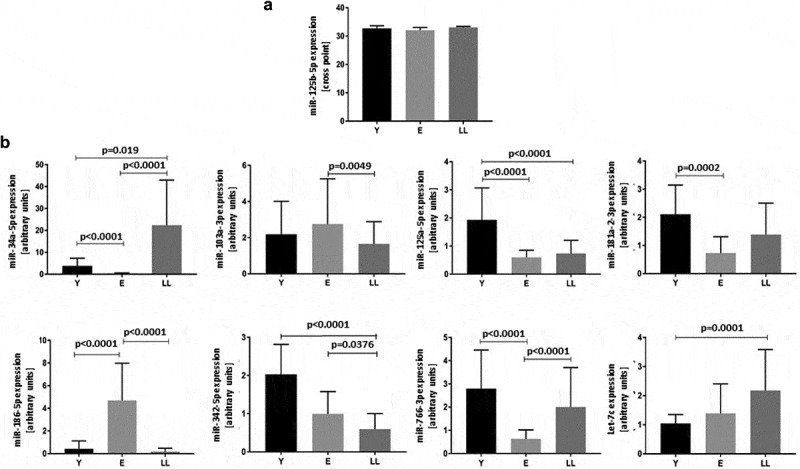
Age-related expression of miRNAs indicated in silico as interacting with SIRT6 mRNA in PBMCs. (a) Levels of miR-125b-5p are not affected by age. (b) Levels of miR-34a-5p, miR-103-3p, miR-125a-5p, miR-181a-2-3p, miR-186-5p, miR-342-5p, miR-776-3p and Let-7c are significantly affected by age. Results are presented as median with interquartile range. Statistical analysis was performed with the Kruskal-Wallis test and Dunn’s multiple comparisons test. Y: young (mean age 27.5 ± 4.4 years), E: middle-aged (65.4 ± 3.3 years), LL: long-lived (93.9 ± 3.6 years)
Analysis of the interaction between selected miRNAs and SIRT6 mRNA
Interactions of miR-34a-5p and miR-766-3p with the SIRT6 mRNA have been previously described [28,29]. Therefore, only miR-125a-5p, miR-181a-2-3p, , miR-186-5p, and Let-7c were subjected to functional analysis. We performed co-transfection of HEK293 with synthetic miRNA precursors and pmirGLO_SIRT6 reporter vector containing the entire 3´UTR of SIRT6 mRNA. There was no significant reduction in luminescence following co-transfection of the pmirGLO_SIRT6 vector with miR-125a-5p, pre-miR-181a-2-3p, , or pre-let-7c. However, co-transfection of pmirGLO_SIRT6 and pre-miR-186-5p caused a 24% decrease (p = 0.0006) in luminescence (Figure 2(a)). Moreover, overexpression of miR-186-5p (80% transfection efficiency) resulted in a 54% decrease in endogenous SIRT6 protein levels (Figure 2(b)). The Western blot analysis was performed once, and the results of densitometric scanning are provided in Supplementary Table S2.
Figure 2.
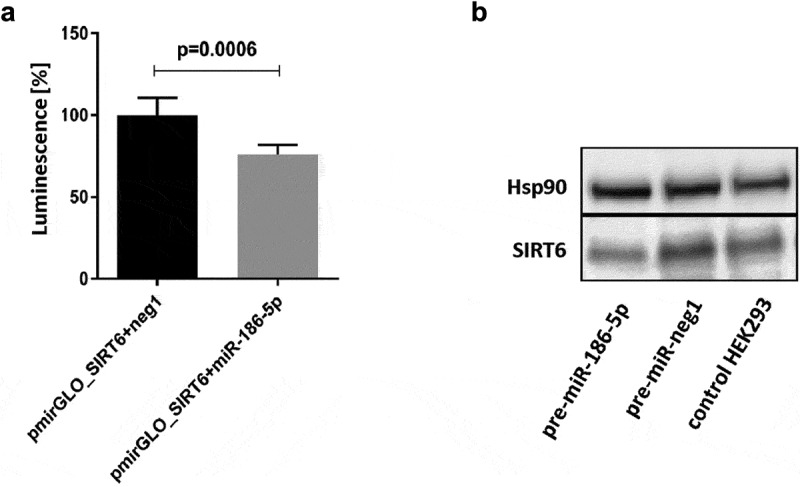
Effects of interaction of the 3´UTR of SIRT6 mRNA with miR-186-5p. (a) Co-transfection of the pmirGLO_SIRT6 vector containing the full-length 3´UTR of SIRT6 mRNA and the firefly luciferase reporter gene with pre-miR-186-5p results in a significant decrease in luminescence as compared to co-transfection with the pre-neg-miR negative control (neg1). Results are presented as mean with standard deviation. Statistical analysis was performed with a two-tailed Student t-test. (b) Transfection of HEK293 cells with pre-miR-186-5p results in a decrease in the expression of endogenous SIRT6 protein. Hsp90 – heat shock protein 90, control protein expression of which is not affected by miR-186-5p. The Western blot analysis was performed once, and the results of densitometric scanning are provided in Supplementary Table S2
Correlation of miRNAs expression with the expression of SIRT6
A lack of evidence confirming a direct interaction of a given miRNA with SIRT6 mRNA does not mean that the miRNA does not affect SIRT6 expression because such an effect can be the result of indirect interactions. Therefore, we performed correlation analyses between all miRNAs affected by age and SIRT6 mRNA expression in all studied age groups. Detailed data regarding the number of individuals included in each correlation analysis are presented in Supplementary Table S3.
The Spearman’s rank correlation showed a relationship between SIRT6 expression and three miRNAs. A negative association was observed for miR-186-5p (rs = −0.25, p = 0.026), and positive correlations were found for miR-34a-5p (rs = 0.31, p = 0.0055) and miR-181a-2-3p (rs = 0.39, p = 0.0002) (Table 1). Considering the values of the rs coefficient, these correlations were weak; however, such a situation is common if only selected elements of complicated networks and multiple interactions are analysed.
Table 1.
Correlation of miRNAs expression with the expression of SIRT6 in human PBMCs
| miRNA | rs | p value* |
|---|---|---|
| miR-34a-5p | 0.3097 | 0.0055 |
| miR-103a-3p | −0.1336 | 0.2094 |
| miR-125a-5p | 0.1212 | 0.2553 |
| miR-181a-2-3p | 0.3933 | 0.0002 |
| miR-186-5p | −0.2525 | 0.0257 |
| miR-342-5p | 0.0097 | 0.9293 |
| miR-766-3p | 0.2054 | 0.0521 |
| Let-7c | −0.1343 | 0.2068 |
*The Spearman’s rank correlation was used for the analysis of the association between the expression of SIRT6 and miRNAs.
mRNA expression of SIRT6-interacting genes and proteins
Because we were interested in the ageing of immune cells, we evaluated genes and their protein products that are involved in the regulation of three cellular processes: immunity, apoptosis, and ageing, which are also direct or indirect SIRT6 interactors. To study the connection network between SIRT6 and proteins involved in the processes mentioned above, the STRING network version 10.5 (https://string-db.org/) was used with a minimum required interaction score of 0.4 (medium confidence).
Based on an extensive search of the literature, we selected 15 genes (ICAM1, CD48, STAT2, RDH11, DISC1, TP53, BCL2, IGF1 R, MYC, BAP1, BAX, BAK, CASP8, CASP9, FAS), whose products may be involved in the processes mentioned above. Next, using STRING, we verified whether these proteins form an interaction network with SIRT6, and allowing up to 5 interactors in the first shell to fill the ‘gaps’ in the resulting network. In our search, we marked only text mining, database, and experiments as data sources. We rejected proteins that were not SIRT6 interactors (DISC1, RDH11, and BAP1) and added MDM2, which was indicated by STRING as a direct interactor of this sirtuin. The results of the analysis are presented in Figure 3 [30–47].
Figure 3.
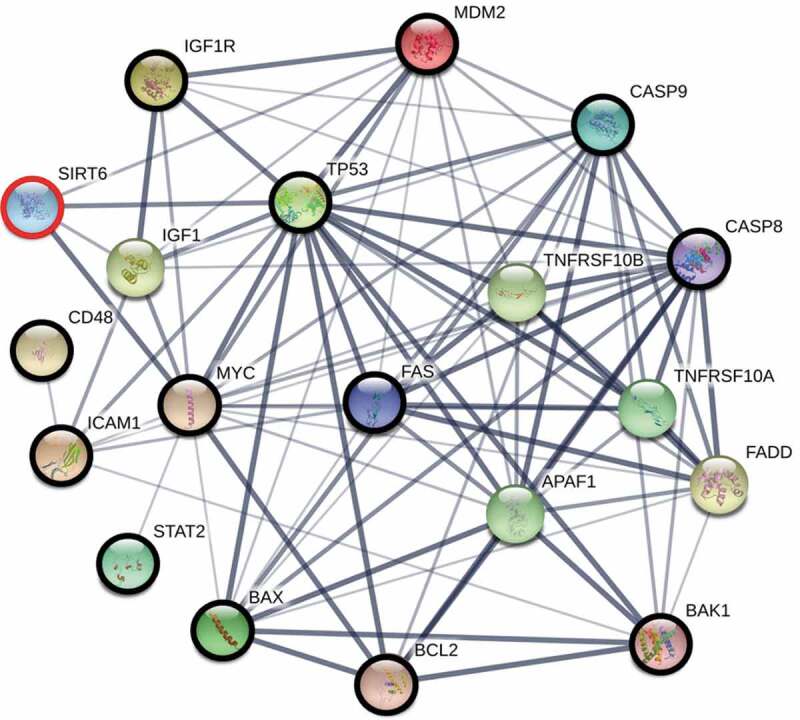
The interaction network between proteins, each of which is involved in the regulation of immunity, apoptosis, and ageing (bold circles), which are also direct or indirect SIRT6 interactors. The network was constructed using the STRING functional protein association network version 10.5 (https://string-db.org/) with the minimum required interaction score of 0.4 (medium confidence). To fill ‘gaps’ between SIRT6 and these proteins, the program was allowed to include up to 5 additional interactors in the first shell
We also tested whether the mRNA expression of direct SIRT6 interactors (TP53, MYC, and MDM2), as well as indirect SIRT6 interactors (BCL2, CD48, and ICAM1), changed in PBMCs in an age-related manner (Figure 4(a,b), respectively). We confirmed previous findings that ageing affected the expression of multiple genes, but its effect was much less pronounced in long-lived, healthy individuals compared to average-ageing people, represented here by the E group [23,48–50].
Figure 4.
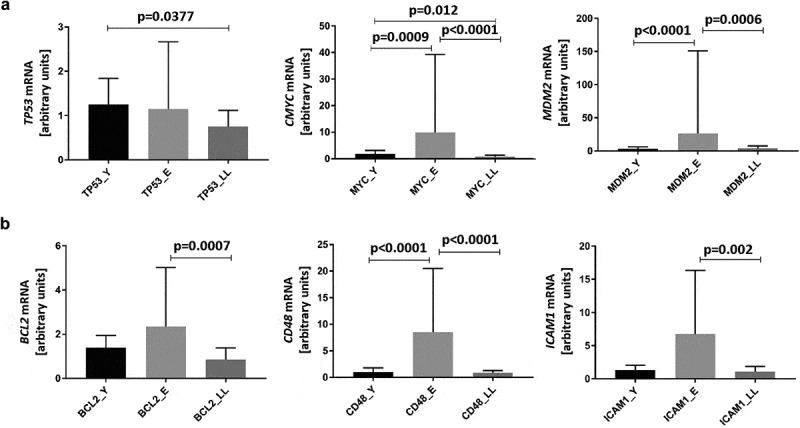
Age-related mRNA expression of proteins interacting with SIRT6 in PBMCs. Results, normalized against the expression of the β-actin gene (ACTB), are presented in arbitrary units as median mRNA levels with interquartile range. Statistical analysis was performed with the Kruskal-Wallis test and Dunn’s multiple comparisons test. (a) Direct SIRT6 interactors TP53, MYC, and MDM2. (b) Indirect SIRT6 interactors BCL2, CD48, and ICAM1. Y: young (mean age 27.5 ± 4.4 years), E: middle-aged (65.4 ± 3.3 years), LL: long-lived (93.9 ± 3.6 years)
Notably, SIRT6 expression was negatively correlated with both TP53 (r = −0.41, p = 0.0126) and MYC (r = −0.35, p = 0.0448) mRNA levels. However, there was no correlation between SIRT6 and MDM2 mRNA levels (r = −0.28, p = 0.1115). We did not find any significant correlations between the expression of SIRT6 and its indirect interactors. Detailed data regarding the number of individuals included in each correlation analysis has been presented in Supplementary Table S4.
Discussion
It is estimated that in the majority of people, genetic factors determine their lifespan in only approximately 25% [48,49]. A small percentage of the population can achieve longevity due to the favourable combination of polymorphisms in genes regulating the rate of ageing, which predisposes to an efficient metabolism or an efficient response to different stresses. Those who are not genetically predisposed to longevity can modify their lifespan by targeting the same pathways with a proper lifestyle or interventions. Epigenetic factors mediate this beneficial effect of lifestyle on genetic predisposition [50–52].
We previously reported that SIRT6 expression significantly decreases with age, being the lowest in the PBMCs of middle-aged subjects and rising again in long-lived individuals, where it reaches a level similar to that in cells of young people [23]. Having in mind that ageing is associated with an epigenetic drift, we decided to investigate whether epigenetic mechanisms stand behind the age-related fluctuations in the SIRT6 level in PBMCs. As hypermethylation is one of the most common mechanisms responsible for ageing-associated decreases in gene expression, we first analysed SIRT6 promoter methylation. However, we found neither significant differences in the percentage of methylation between the different age groups nor significant correlations between methylation and SIRT6 expression levels. These results are contrary to previously published data that showed higher methylation levels in the SIRT6 promoter in 20 to 79-year-old study subjects compared to those aged 9 to 19 years. These inconsistent results are likely due to the many methodological differences between the two studies. In our study, groups were approximately 3-fold larger, and all participants were healthy. The age groups were defined differently, and the methods of analysis were also different [53]. Nevertheless, the age-related expression pattern of SIRT6 in our study subjects seemed to match the methylation trend described by Sahin et al. [53]. Thus, the role of methylation in the age-related SIRT6 expression changes remains to be clarified.
Another epigenetic mechanism that can affect gene expression is the activity of miRNAs. Analysis of the SIRT6 mRNA using the TargetScan, miRWalk, and miRanda programs indicated that several miRNAs could potentially interact with this mRNA. Direct interactions between SIRT6 mRNA and some of the candidates were previously demonstrated [28,29]. In this study, we showed that miR-186-5p bound to the 3´UTR of SIRT6 mRNA and negatively affected endogenous SIRT6 expression in HEK293 cells, which is consistent with data obtained in non-small cell lung carcinoma [54] and suggests that this interaction can be responsible for the negative correlation between SIRT6 mRNA and miR-186-5p also in PBMCs. However, an in vitro study with a reporter vector with cloned SIRT6 3´UTR sequence bearing mutations in the miR-186-5p binding sites would further support the notion that miR-186-5p interaction with SIRT6 mRNA is of functional significance.
In contrary, under our experimental conditions, miR-125a-5p, miR-181a-2-3p, and Let-7c did not interact with SIRT6 mRNA. The fact that not all in silico-selected miRNAs interacted with the target mRNA could reflect the lack of full compatibility between mathematical predictions and biological events. It should be taken into account, however, that our experimental model might not be optimal for the detection of weaker interactions.
For the SIRT6 mRNA-interacting miRNAs, miR-34a-5p correlated positively, miR-186-5p negatively, and miR-766-3p did not correlate with SIRT6 mRNA expression. A negative correlation between miR-186-5p and SIRT6 expression is compatible with the assumption that while miRNA expression increases, its target gene expression decreases. However, we detected a positive correlation between SIRT6 mRNA and miR-34a-5p. It has been previously indicated that gene regulatory networks often include feedback motifs (i.e., ‘feed-forward loops’ overrepresented in pure transcription regulatory networks or less abundant ‘feedback loops’). As part of complex regulatory networks, miRNAs coordinate transcriptional regulation in signalling networks. Positive correlations could be explained by downstream effects, such as inhibited repression or disturbances in feedback loops. Moreover, miRNA-mRNA positive correlations have been previously reported [55–57]. An important limitation of our results is the fact that the observed correlations, measured by the rs value, should be interpreted as weak. Nevertheless, ageing-associated changes in the expression of miRNAs, especially miR-186-5p, are most likely an important mechanism responsible for age-related differences in SIRT6 expression.
Maintenance of SIRT6 expression might be one of the mechanisms promoting stress resistance and longevity. Indeed, Sirt6-knockout mice have a significantly shortened lifespan accompanied by degenerative and metabolic defects reminiscent of premature ageing [58]. SIRT6-deleted embryonic stem cells and mouse embryonic fibroblasts demonstrate impaired proliferation and increased sensitivity to DNA-damaging agents, resulting in chromosomal abnormalities [4]. The depletion of SIRT6 leads to telomere dysfunction with end-to-end chromosomal fusions and premature cellular senescence [59–61]. Therefore, the high expression of SIRT6 that we previously described in PBMCs of long-lived individuals could suggest a function for SIRT6 in the immune system to delay immunosenescence, which promotes longevity. Indeed, SIRT6 overexpression results in a significant decrease in TNF-α, IL-1β, sRANKL, IL-6, and IL-17 levels in rheumatoid arthritis mouse serum and delays the replicative senescence of human fibroblast-like foetal lung cells by attenuating NF-κB signalling [62,63]. In addition, old transgenic mice overexpressing SIRT6 have a lower level of adipose inflammation compared to wild-type mice [64]. In line with this hypothesis, we observed low SIRT6 expression in PBMCs of middle-aged study participants representing a population that is not predisposed to longevity.
The STRING analysis revealed that SIRT6, along with other proteins, is involved in many cellular processes, including immunity and apoptosis. Subsequently, we found that expression levels of the majority of the analysed downstream genes encoding proteins belonging to the SIRT6 network were age-dependent. Among them were MYC and p53 being not only direct SIRT6 targets but also transcription factors playing critical roles in many cellular events. Although the criteria we set may have affected the result of the STRING analysis, the mRNA levels of both MYC and p53 negatively correlated with SIRT6 expression, strongly suggesting that age-related differences in their expression in PBMCs, at least to some extent, depended on ageing-related changes in SIRT6 expression.
The mechanistic studies of Zhang et al. established that p53 directly activates the expression of SIRT6 [38], while Li and co-workers pointed out physical interactions between p53 and SIRT6 in vitro and in vivo, making the last one a co-activator for downstream processes [65]. TP53 induces cell cycle arrest, DNA repair, apoptosis, and senescence, and also affects other, non-canonical pathways [66]. It inhibits inflammation by antagonizing the action of NF-ĸB, suppressing inflammatory cytokines, and controlling T-cell differentiation [67,68]. Therefore, its decreased expression in PBMCs of long-lived individuals could contribute to inflammaging, the process of low-grade, chronic inflammation that accompanies ageing, which is a risk factor for the development of many diseases [69]. Moreover, in some individuals, it can be a risk factor for the development of lymphomas and chronic lymphocytic leukaemia, blood diseases most commonly diagnosed in people of advanced age.
In contrast, decreased expression of MYC could have beneficial effects. MYC accelerates proliferation, promotes cell growth and survival, enhances cellular metabolism, and triggers or sensitizes cells to apoptosis [70]. Its overexpression plays a crucial role in multiple cancers, including leukaemias and lymphomas [71]. It upregulates IL-8, IL-10, TNF-α, and TGF-β [72]. Therefore, its low expression in PBMCs of long-lived individuals lowers the risk of neogenesis and inflammaging, which can be one of the mechanisms underlying longevity. Our findings regarding the possible SIRT6 – MYC interactions are confirmed in functional studies where SIRT6 by deacetylation of acH3K56 residues at MYC target genes contributed to the inhibition of MYC transcriptional output [72,73].
Of note in this study, we showed that not only the expression of various miRNAs differed between middle-aged and long-lived individuals, but also SIRT6 expression was lower, and the expression levels of other genes (e.g., MYC, MDM2, BCL2, CD48, and ICAM1) were higher in middle-aged study subjects. These and other molecular differences could lead to different susceptibilities to age-related diseases and underlie differences in the rate of ageing and life expectancy.
In conclusion, this study demonstrated age-associated alterations in miRNA control that could lead to alterations in the expression of SIRT6 and some SIRT6 interactors. These effects, in turn, could affect the function of PBMCs. In addition, the differences in the expression of various miRNAs and genes in middle-aged and long-lived individuals and similarities between young and long-lived individuals described here emphasize that average ageing differs from that of individuals predisposed to longevity, who have already reached a very advanced age.
Materials and methods
Materials
PBMCs were obtained from non-obese individuals (BMI < 30 kg/m2) belonging to three age groups: young (Y, n = 55, 29 women and 26 men, age range 19 to 42 years, mean age 27.5 ± 4.4 years), middle-aged (E, n = 51, 28 women and 23 men, age range 60 to 73 years, mean age 65.4 ± 3.3 years), and long-lived (LL, n = 51, 31 women and 20 men, age range 90 to 102 years, mean age 93.9 ± 3.6 years). Members of the E group had only a 1:97 chance to reach the mean age of the LL group (Statistics Poland, https://stat.gov.pl/obszary-tematyczne/ludnosc/ludnosc/ludnosc-piramida/). Therefore, in this study, the E group represented people who were not predisposed to longevity and aged in a ‘standard’ way.
The study participants underwent a comprehensive medical examination, and those from the middle-aged (E) and long-lived (LL) groups were evaluated using the Activities of Daily Living (ADL) and Mini-Mental State Examination (MMSE) scales [74]. All study participants were non-obese, without a history of myocardial infarction, stroke, type 2 diabetes mellitus, cancer, or neurodegeneration. However, in the E and LL groups, moderate hypertension was allowed, and some study participants belonging to the LL group had a mild degree physical or cognitive disability (ADL score 5–6 and/or MMSE score 20–23) [75,76].
The study protocol was approved by the Bioethics Committee of the Medical University of Warsaw. All participants gave informed written consent for participation in the study. The anonymity of patients was preserved at all stages of this investigation.
Isolation of PBMCs and RNA
PBMCs and RNA isolation were performed as previously described [77].
DNA methylation
A 3-kb fragment of SIRT6 located from 1.5 kb upstream to 1.5 kb downstream of the primary TSS (transcript ENST00000337491.6, 94 release of the ENSEMBL database) was analysed using the MethPrimer 2.0 online tool (http://www.urogene.org/methprimer2/) with the following settings: CpG island longer than 200 kb with a minimum of 50% GC content. Methylation status was assessed using the OneStep qMethyl™-Lite Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocol. The sequences of the primers and probes used for the 5´ fragment of the CpG island were forward 5´GTAACTCTGCGTGGCATTCA3´, reverse 5´CCCCACGGGAACAATAAAGT3´, and UPL probe #56 (Roche Diagnostics, Mannheim, Germany). For the 3´ end of the CpG island, the primers and probe were forward 5´AGGATGTCGGTGAATTACGC3´, reverse 5´CCTGGAACTACATCCCCTCTC3´, and UPL probe #81 (Roche Diagnostics). Methylation-sensitive digestion was carried out for 2 h at 37°C. Digestion was followed by an initial denaturation for 10 min at 95°C, then 45 cycles of 30 s at 95°C, 30 s at 59°C for the 5´ fragment or 60°C for the 3´ fragment, 30 s at 72°C, and one cooling cycle. Reactions were performed in the LightCycler 480 II (Roche Diagnostics).
Reverse transcription and RT-PCR
The Universal cDNA Synthesis Kit and the miRCURY LNA™ Universal RT microRNA PCR system (EXIQON, Vedbaek, Denmark) were used for reverse transcription and evaluation of the expression of miRNAs, respectively. Reverse transcription was performed with 20 µg of total RNA as a template. miRNA expression analysis was performed with cDNA corresponding to 0.02 ng of input RNA in the Light Cycler 480 II (Roche Diagnostics). Primers were designed using miRprimer software [78] and are listed in Supplementary Table S5. The reaction conditions were as follows: 10 min at 95°C, 45 cycles of 10 s at 95°C, 1 min at 60°C, and a melting curve cycle.
Reverse transcription for the analysis of gene expression was performed as previously described [23]. The PCR reaction was carried out as follows: 5 min at 95°C, 45 cycles of 10 s at 95°C, 10 s at 60°C, 10 s at 72°C. The primer sequences for the expression analysis are presented in Supplementary Table S6. The CD48 primers were designed by our laboratory. All other primer sequences were based on previous reports [77,79–83]. Results, normalized against the expression of the β-actin gene (ACTB) were presented in arbitrary units (AU) as mean mRNA levels.
Reporter vector for mRNA-miRNA interaction analysis
DNA corresponding to the full-length 3´UTR of SIRT6 mRNA (514 bp) was amplified using Dream Taq polymerase (Thermo Scientific, Vilnius, Lithuania) from 100 ng of cDNA prepared from the RNA isolated from PBMCs. The forward (5´ACTAGAGCTCCAGCTGACCAGGGTGCTT3´, STOP codon is underlined) and reverse (5´CTAACTCGAGGCAAGAAAGAAATTGTTTTTATTG3´) primers contained SacI or XhoI restriction sites (bolded sequences), respectively. The PCR conditions consisted of 4 min at 94°C, 39 cycles of 30 s at 94°C, 30 s at 59°C, 30 s at 72°C, and a final extension of 4 min at 72°C. The PCR product was cloned into the pmirGLO reporter vector (Promega, Madison, WI, USA) and sequenced (pmirGLO_SIRT6).
Cell culture and transfection
Human embryonic kidney cells 293 (HEK293; Sigma-Aldrich, St. Louis, MO, USA) were grown in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated foetal bovine serum at 37°C in 5% CO2. Twenty-four hours before transfection, cells were seeded in a 96-well plate for evaluation of miRNA interaction with the 3´UTR of SIRT6 mRNA (3 x 104 per well) or 12-well plate for the assessment of the effect of miRNA on endogenous SIRT6 expression (2.2 x 105 per well). Cells were transfected at 80% confluency using Lipofectamine 2000 according to the manufacturer’s protocol (Life Technologies, Carlsbad, CA, USA).
For miRNA interaction with the 3´UTR of SIRT6 mRNA, 40 ng of the reporter plasmid and 5 pmol (30 nM final concentration) of the synthetic pre-miRNA (Life Technologies) were used. Pre-miR miRNA Precursor Negative Control #1 (Ambion, Life Technologies) served as a negative control. Cells were cultured for 24 h after transfection, washed with phosphate-buffered saline, and lysed for 15 min with 30 μl Passive Lysis Buffer (Promega) on a rocking platform. The luminescence was measured using the Centro XS3 LB 960 luminometer (Berthold Technologies, Bad Wilbad, Germany). The luminescence of the firefly luciferase substrate was normalized against that of the Renilla luciferase substrate. The mean luciferase activity in extracts from cells co-transfected with the pmirGLO_SIRT6 vector and synthetic pre-neg-miRNA was normalized to 100%. All experiments were repeated six times.
For the assessment of the miRNA effect on endogenous SIRT6 expression, 40 pmol (30 nM final concentration) of synthetic pre-miR-186-5p or pre-miR miRNA Precursor Negative Control #1 were used .
Western blot analysis
For the evaluation of endogenous SIRT6, cells were harvested 24 h after transfection using radioimmunoprecipitation assay (RIPA) buffer. Total protein (30 µg) was resolved on a 4 to 15% Mini-PROTEAN® TGX™ gel (Bio-Rad, Hercules, CA, USA) and transferred to a nitrocellulose membrane. The membrane was incubated with mouse anti-SIRT6 primary antibody (1:200, sc-517196, Santa Cruz Biotechnology, Dallas, TX, USA) and anti-mouse IgG secondary antibody (1:2000, sc-516102, Santa Cruz Biotechnology). As a control, blots were incubated with rabbit anti-Hsp90 primary antibody (1:1000, sc-7947, Santa Cruz Biotechnology) and anti-rabbit IgG secondary antibody (1:10000, AP132P, Millipore, Darmstadt, Germany). Proteins were detected using the Clarity™ ECL Western Blotting Substrate (Bio-Rad) and visualized with the GeneGnome XRQ bioimaging system (Syngene, Cambridge, MA, USA). Densitometric measurement was performed with Image Studio Lite software (LI-COR Biosciences, Lincoln, NE, USA).
Statistical analysis
The Shapiro-Wilk test was used to assess the normality of distributions. Whenever three age groups were analysed, and the distribution was not normal, the Kruskal-Wallis test and Dunn’s multiple comparisons test were used. The two-tailed Student t-test was used for the analysis of the interaction of miRNAs with SIRT6 mRNA. The U Mann-Whitney test was used to examine the expression of genes in two age cohorts. The Spearman’s rank correlation was used for the analysis of the association between the expression of SIRT6 and other factors and the analysis of other associations. The strength of the correlation was interpreted as ‘weak’ when Spearman’s correlation coefficient value (rs, a statistical measure of the strength of a monotonic relationship between paired data) was between 0.2 and 0.39, ‘moderate’ when 0.4 to 0.59, ‘strong’ when 0.6 to 0.79 and ‘very strong’ when rs was between 0.8 and 1.0 [84]. All calculations were performed with GraphPad Prism 7.0. Significance was indicated by p < 0.05.
Supplementary Material
Funding Statement
This work was supported by the National Science Centre grant 2013/09/N/NZ3/01363.
Disclosure statement
The authors declare no conflict of interest.
Supplementary materials
Supplemental data for this article can be accessed here
References
- [1].Kupis W, Pałyga J, Tomal E, et al. The role of sirtuins in cellular homeostasis. J Physiol Biochem. 2016;72:371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tennen RI, Berber E, Chua KF.. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech Ageing Dev. 2010;131:185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gao Y, Tan J, Jin J, et al. SIRT6 facilitates directional telomere movement upon oxidative damage. Sci Rep. 2018;8:5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mostoslavsky R, Chua KF, Lombard DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. [DOI] [PubMed] [Google Scholar]
- [5].Pan H, Guan D, Liu X, et al. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 2016;26:190–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vazquez BN, Thackray JK, Serrano L.. Sirtuins and DNA damage repair: SIRT7 comes to play. Nucleus. 2017;8:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Giblin W, Skinner ME, Lombard DB. Sirtuins: guardians of mammalian healthspan. Trends Genet. 2014;30:271–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li X, Kazgan N. Mammalian sirtuins and energy metabolism. Int J Biol Sci. 2011;7:575–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li P, Jin Y, Qi F, et al. SIRT6 acts as a negative regulator in Dengue virus-induced inflammatory response by targeting the DNA binding domain of NF-κB p65. Front Cell Infect Microbiol. 2018;8:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. [DOI] [PubMed] [Google Scholar]
- [12].Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:48–56. [DOI] [PubMed] [Google Scholar]
- [13].Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kanfi Y, Naiman S, Amir G, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. [DOI] [PubMed] [Google Scholar]
- [15].Li Y, Qin J, Wei X, et al. Association of SIRT6 gene polymorphisms with human longevity. Iran J Public Health. 2016;45:1420–1426. [PMC free article] [PubMed] [Google Scholar]
- [16].Hirvonen K, Laivuori H, Lahti J, et al. SIRT6 polymorphism rs117385980 is associated with longevity and healthy aging in Finnish men. BMC Med Genet. 2017;18:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].TenNapel MJ, Lynch CF, Burns TL, et al. SIRT6 minor allele genotype is associated with >5-year decrease in lifespan in an aged cohort. PLoS ONE. 2014;9:e115616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Vitiello M, Zullo A, Servillo L, et al. Multiple pathways of SIRT6 at the crossroads in the control of longevity, cancer, and cardiovascular diseases. Ageing Res Rev. 2017;35:301–311. [DOI] [PubMed] [Google Scholar]
- [19].Khan RI, Nirzhor SSR, Akter R. A review of the recent advances made with SIRT6 and its implications on aging related processes, major human diseases, and possible therapeutic targets. Biomolecules. 2018;8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lara E, Calvanese V, Fraga MF. Epigenetic drift and aging. In: Tollefsbol TO, editor. Epigenetics of aging. New York: Springer New York; 2010. p. 257–273. ISBN 978-1-4419-0639-7. [Google Scholar]
- [21].Teschendorff AE, West J, Beck S. Age-associated epigenetic drift: implications, and a case of epigenetic thrift? Hum Mol Genet. 2013;22:R7–R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wallace RG, Twomey LC, Custaud MA, et al. Potential diagnostic and prognostic biomarkers of epigenetic drift within the cardiovascular compartment. Biomed Res Int. 2016;2016:2465763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Owczarz M, Budzinska M, Domaszewska-Szostek A, et al. miR-34a and miR-9 are overexpressed and SIRT genes are downregulated in peripheral blood mononuclear cells of aging humans. Exp Biol Med (Maywood). 2017;242:1453–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Budzinska M, Owczarz M, Pawlik-Pachucka E, et al. miR-96, miR-145 and miR-9 expression increases, and IGF-1R and FOXO1 expression decreases in peripheral blood mononuclear cells of aging humans. BMC Geriatr. 2016;16:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gu W, Xu Y, Xie X, et al. The role of RNA structure at 5´ untranslated region in microRNA-mediated gene regulation. RNA. 2014;20:1369–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liu B, Li J, Cairns MJ. Identifying miRNAs, targets, and functions. Brief Bioinform. 2014;15:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sharma A, Diecke S, Zhang WY, et al. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. J Biol Chem. 2013;288:18439–18447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lefort K, Brooks Y, Ostano P, et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. Embo J. 2013;32:2248–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gazin C, Wajapeyee N, Gobeil S, et al. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Munitz A, Bachelet I, Finkelman FD, et al. CD48 is critically involved in allergic eosinophilic airway inflammation. Am J Respir Crit Care Med. 2007;175:911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. [DOI] [PubMed] [Google Scholar]
- [34].Lawson C, Wolf S. ICAM-1 signaling in endothelial cells. Pharmacol Rep. 2009;61:22–32. [DOI] [PubMed] [Google Scholar]
- [35].Lappas M. Anti-inflammatory properties of sirtuin 6 in human umbilical vein endothelial cells. Mediators Inflamm. 2012;2012:597514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sundaresan NR, Vasudevan P, Zhong L, et al. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med. 2012;18:1643–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu Y, He Y, Jin A, et al. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc Natl Acad Sci USA. 2014;111:E2414–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang P, Tu B, Wang H, et al. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc Natl Acad Sci USA. 2014;111:10684–10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Thirumurthi U, Shen J, Xia W, et al. MDM2-mediated degradation of SIRT6 phosphorylated by AKT1 promotes tumorigenesis and trastuzumab resistance in breast cancer. Sci Signal. 2014;7:ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zwaans BMM, Lombard DB. Interplay between sirtuins, MYC and hypoxia-inducible factor in cancer-associated metabolic reprogramming. Dis Model Mech. 2014;7:1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shahni R, Cale CM, Anderson G, et al. Signal transducer and activator of transcription 2 deficiency is a novel disorder of mitochondrial fission. Brain. 2015;138:2834–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ran L-K, Chen Y, Zhang -Z-Z, et al. SIRT6 overexpression potentiates apoptosis evasion in hepatocellular carcinoma via BCL2-associated X protein-dependent apoptotic pathway. Clin Cancer Res. 2016;22:3372–3382. [DOI] [PubMed] [Google Scholar]
- [43].Sipos F, Firneisz G, Műzes G. Therapeutic aspects of c-MYC signaling in inflammatory and cancerous colonic diseases. World J Gastroenterol. 2016;22:7938–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Riscal R, Schrepfer E, Arena G, et al. Chromatin-bound MDM2 regulates serine metabolism and redox homeostasis independently of p53. Mol Cell. 2016;62:890–902. [DOI] [PubMed] [Google Scholar]
- [45].Huang N, Liu Z, Zhu J, et al. Sirtuin 6 plays an oncogenic role and induces cell autophagy in esophageal cancer cells. Tumour Biol. 2017;39:1010428317708532. [DOI] [PubMed] [Google Scholar]
- [46].Garcia-Peterson LM, Ndiaye MA, Singh CK, et al. SIRT6 histone deacetylase functions as a potential oncogene in human melanoma. Genes Cancer. 2017;8:701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wu D, Prives C. Relevance of the p53-MDM2 axis to aging. Cell Death Differ. 2018;25:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Herskind AM, McGue M, Holm NV, et al. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97:319–323. [DOI] [PubMed] [Google Scholar]
- [49].van Hjelmborg J, Iachine I, Skytthe A, et al. Genetic influence on human lifespan and longevity. Hum Genet. 2006;119:312–321. [DOI] [PubMed] [Google Scholar]
- [50].Sebastiani P, Solovieff N, Dewan AT, et al. Genetic signatures of exceptional longevity in humans. PLoS ONE. 2012;7:e29848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Beekman M, Nederstigt C, Suchiman HE, et al. Genome-wide association study (GWAS)-identified disease risk alleles do not compromise human longevity. Proc Natl Acad Sci USA. 2010;107:18046–18049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Passarino G, De Rango F, Montesanto A. Human longevity: genetics or lifestyle? It takes two to tango. Immun Ageing. 2016;13:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sahin K, Yilmaz S, Gozukirmizi N. Changes in human sirtuin 6 gene promoter methylation during aging. Biomed Rep. 2014;2:574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ruan L, Chen J, Ruan L, et al. MicroRNA-186 suppresses lung cancer progression by targeting SIRT6. Cancer Biomark. 2018;21:415–423. [DOI] [PubMed] [Google Scholar]
- [55].Laxman N, Rubin C-J, Mallmin H, et al. Global miRNA expression and correlation with mRNA levels in primary human bone cells. RNA. 2015;21:1433–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. [DOI] [PubMed] [Google Scholar]
- [57].Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. [DOI] [PubMed] [Google Scholar]
- [58].Peshti V, Obolensky A, Nahum L, et al. Characterization of physiological defects in adult SIRT6-/- mice. PLoS ONE. 2017;12:e0176371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Michishita E, McCord RA, Berber E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cardus A, Uryga AK, Walters G, et al. SIRT6 protects human endothelial cells from DNA damage, telomere dysfunction, and senescence. Cardiovasc Res. 2013;97:571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Nagai K, Matsushita T, Matsuzaki T, et al. Depletion of SIRT6 causes cellular senescence, DNA damage, and telomere dysfunction in human chondrocytes. Osteoarthr Cartil. 2015;23:1412–1420. [DOI] [PubMed] [Google Scholar]
- [62].Lee H-S, Ka S-O, Lee S-M, et al. Overexpression of sirtuin 6 suppresses inflammatory responses and bone destruction in mice with collagen-induced arthritis. Arthritis Rheum. 2013;65:1776–1785. [DOI] [PubMed] [Google Scholar]
- [63].Zhang N, Li Z, Mu W, et al. Calorie restriction-induced SIRT6 activation delays aging by suppressing NF-κB signaling. Cell Cycle. 2016;15:1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Roichman A, Kanfi Y, Glazz R, et al. SIRT6 overexpression improves various aspects of mouse healthspan. J Gerontol A Biol Sci Med Sci. 2017;72:603–615. [DOI] [PubMed] [Google Scholar]
- [65].Li M, Hou T, Gao T, et al. . p53 cooperates with SIRT6 to regulate cardiolipin de novo biosynthesis. Cell Death Dis. 2018;9:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Komarova EA, Krivokrysenko V, Wang K, et al. p53 is a suppressor of inflammatory response in mice. FASEB J. 2005;19:1030–1032. [DOI] [PubMed] [Google Scholar]
- [68].Gudkov AV, Komarova EA. p53 and the carcinogenicity of chronic inflammation. Cold Spring Harb Perspect Med. 2016;6:pii: a026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S4–S9. [DOI] [PubMed] [Google Scholar]
- [70].Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013;3. DOI: 10.1101/cshperspect.a014217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Trop-Steinberg S, Azar Y. Is Myc an important biomarker? Myc expression in immune disorders and cancer. Am J Med Sci. 2018;355:67–75. [DOI] [PubMed] [Google Scholar]
- [72].Liu T, Zhou Y, Ko KS, et al. Interactions between Myc and mediators of inflammation in chronic liver diseases. Mediators Inflamm. 2015;2015:276850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sebastián C, Zwaans BM, Silberman DM, et al. . The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185‐1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mossakowska M, Broczek K, Wieczorowska-Tobis K, et al. Cognitive performance and functional status are the major factors predicting survival of centenarians in Poland. J Gerontol A Biol Sci Med Sci. 2014;69:1269–1275. [DOI] [PubMed] [Google Scholar]
- [75].Katz S, Ford AB, Moskowitz RW, et al. Studies of illness in the aged: the index of ADL: A standardized measure of biological and psychosocial function. JAMA. 1963;185:94–99. [DOI] [PubMed] [Google Scholar]
- [76].Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. [DOI] [PubMed] [Google Scholar]
- [77].Polosak J, Roszkowska-Gancarz M, Kurylowicz A, et al. Decreased expression and the Lys751Gln polymorphism of the XPD gene are associated with extreme longevity. Biogerontology. 2010;11:287–297. [DOI] [PubMed] [Google Scholar]
- [78].Busk PK. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinformatics. 2014;15:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Takaoka A, Hayakawa S, Yanai H, et al. Integration of interferon-α/β signalling to p53 responses in tumour suppression and antiviral defence. Nature. 2003;424:516–523. [DOI] [PubMed] [Google Scholar]
- [80].Hellebrekers DMEI, Melotte V, Viré E, et al. Identification of epigenetically silenced genes in tumor endothelial cells. Cancer Res. 2007;67:4138–4148. [DOI] [PubMed] [Google Scholar]
- [81].Huang H-P, Chen P-H, Yu C-Y, et al. Epithelial cell adhesion molecule (EpCAM) complex proteins promote transcription factor-mediated pluripotency reprogramming. J Biol Chem. 2011;286:33520–33532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Huang Q, Li L, Li L, et al. MDM2 knockdown mediated by a triazine-modified dendrimer in the treatment of non-small cell lung cancer. Oncotarget. 2016;7:44013–44022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kim JA, Kim Y, Kwon B-M, et al. The natural compound cantharidin induces cancer cell death through inhibition of heat shock protein 70 (HSP70) and Bcl-2-associated athanogene domain 3 (BAG3) expression by blocking heat shock factor 1 (HSF1) binding to promoters. J Biol Chem. 2013;288:28713–28726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Mukaka MM. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med J. 2012;24:69–71. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


