Abstract
Human induced pluripotent stem cell (hiPSC)‐derived blood‐brain barrier (BBB) models established to date lack expression of key adhesion molecules involved in immune cell migration across the BBB in vivo. Here, we introduce the extended endothelial cell culture method (EECM), which differentiates hiPSC‐derived endothelial progenitor cells to brain microvascular endothelial cell (BMEC)‐like cells with good barrier properties and mature tight junctions. Importantly, EECM‐BMEC‐like cells exhibited constitutive cell surface expression of ICAM‐1, ICAM‐2, and E‐selectin. Pro‐inflammatory cytokine stimulation increased the cell surface expression of ICAM‐1 and induced cell surface expression of P‐selectin and VCAM‐1. Co‐culture of EECM‐BMEC‐like cells with hiPSC‐derived smooth muscle‐like cells or their conditioned medium further increased the induction of VCAM‐1. Functional expression of endothelial ICAM‐1 and VCAM‐1 was confirmed by T‐cell interaction with EECM‐BMEC‐like cells. Taken together, we introduce the first hiPSC‐derived BBB model that displays an adhesion molecule phenotype that is suitable for the study of immune cell interactions.
Keywords: blood‐brain barrier, human induced pluripotent stem cells, T‐cell migration, adhesion molecules, VCAM‐1
Abbreviations
- BBB
blood‐brain barrier
- BLECs
brain‐like endothelial cells
- BMECs
brain microvascular endothelial cells
- CNS
central nervous system
- CM
conditioned medium
- DMM
defined medium method
- EECM
Extended endothelial cell culture method
- EPC
endothelial progenitor cell
- FBS
fetal bovine serum
- hiPSC
human induced pluripotent stem cell
- ICAM‐1
intercellular adhesion molecule‐1
- LFA‐1
lymphocyte function‐associated antigen 1
- NaFl
sodium fluorescein
- NS
non‐stimulated
- Pe
permeability coefficient
- PECAM‐1
platelet endothelial cell adhesion molecule
- RT
room temperature
- SMLC
smooth muscle‐like cell
- TEER
transendothelial electrical resistance
- Th cells
T helper cells
- UMM
unconditioned medium method
- VCAM‐1
vascular cell adhesion molecule‐1
- vWF
von Willebrand factor
1. INTRODUCTION
Under physiological conditions, the blood‐brain barrier (BBB) maintains central nervous system (CNS) homeostasis by protecting the CNS from the constantly changing milieu in the bloodstream. The BBB is established by brain microvascular endothelial cells (BMECs), which inhibit free paracellular diffusion of water‐soluble molecules by complex tight junctions that connect the endothelial cells. 1 Combined with their characteristically low pinocytotic activity and lack of fenestrations, which inhibit transcellular passage of molecules across the BBB, these features establish the physical barrier of the BBB. 2 At the same time, the BBB establishes a functional barrier, in which the expression of specific transporters and enzymes in BMECs ensures that nutrients pass into the CNS and toxic metabolites are removed from the CNS. In the absence of neuroinflammation, the BBB also limits immune cell trafficking to specific immune cell subsets that ensure CNS immune surveillance. 3 Importantly, the segments of the microvasculature mediating immune cell trafficking vs regulating transport of solutes are not identical; while solute transport is predominantly localized to CNS capillaries, immune cell trafficking occurs at the level of CNS postcapillary venules. 4 , 5
Current knowledge about the cellular and molecular mechanisms mediating immune cell migration across the BBB during CNS immune surveillance and neuroinflammation have to a large degree been derived from animal models of neuroinflammatory diseases. These studies showed that the migration of immune cells across the BBB follows a multistep process that is regulated by the sequential interaction of different signaling and adhesion molecules on the BBB endothelium and the immune cells. 4 Because of the unique tightness of this vascular bed, immune cell migration across the BBB is characterized by unique adaptations. These range from a predominant role of α4β1‐integrin in mediating interaction of T cells with endothelial VCAM‐1 6 to the extended crawling of T cells mediated by endothelial ICAM‐1 and ICAM‐2 against the direction of the blood flow in search of rare sites permissive for diapedesis across the BBB. 7 , 8
There are some limitations when applying knowledge from animal models to humans. It has been observed that levels of some adhesion molecules (eg, activated leukocyte cell adhesion molecule (ALCAM) or junctional adhesion molecules (JAMs)) are different between rodents and human. 9 As a result, there is a significant need for human BBB models that are suitable for the study of immune cell trafficking across the BBB. To this end, human brain endothelial cell lines (eg, hCMEC/D3 10 ) have been established allowing the study of barrier properties and immune cell trafficking under inflammatory conditions. Unfortunately, these BBB models often fail to establish complex tight junctions and barrier properties characterized by high transendothelial electrical resistance (TEER) and low permeability of soluble tracers. 11 Appropriate barrier properties are prerequisite for appropriate modeling of the unique mechanisms involved in T‐cell diapedesis across the BBB. 12 Primary human brain microvascular endothelial cells (hBMECs) have proven useful to study T‐cell/BBB interactions; however, these cells are often not readily available to researchers 12 , 13 and they come from non‐autologous sources. Recent advances in stem cell technology have allowed derivation of hBMEC‐like cells from various stem cell sources including human cord blood‐derived endothelial progenitors and human pluripotent stem cells (hPSCs). 14 , 15 In particular, patient‐sourced human induced pluripotent stem cell (hiPSC)‐derived BMEC‐like cells uniquely enable the study of BBB dysfunction by providing a scalable and renewable source of BMEC‐like cells. For immune cell studies, one could also envision combining hiPSC and autologous immune cells sourced from the same patient cohort. Human iPSC‐derived in vitro models of the BBB have been established 14 , 16 , 17 , 18 and proven useful for modeling BBB dysfunction in inheritable neurological disorders in vitro. 19 , 20 , 21
Presently available hiPSC‐derived in vitro BBB models are well characterized with respect to their barrier properties and expression of BBB‐specific transporters and efflux pumps 14 , 16 , 17 and have proven useful for the study of barrier regulation, molecular transport, and brain drug delivery. However, much less is known about the immune phenotypes such as the expression and cytokine‐induced upregulation of adhesion molecules in hiPSC‐derived in vitro BBB models. These properties are extremely important when aiming to apply hiPSC‐derived in vitro BBB models to study cerebrovascular pathologies involved in multiple sclerosis, stroke, or CNS infections, where immune cell trafficking across the BBB critically contributes to disease pathogenesis. Previous reports have suggested that inflammatory stimuli could induce the expression of ICAM‐1 17 , 22 and VCAM‐1 22 in hiPSC‐derived BMEC‐like cells, but detailed characterization of the full panel of adhesion molecules shown to mediate the multistep immune cell migration across the BBB has not been reported. Here, we show that two different well‐established protocols developed in our laboratory (co‐differentiation unconditioned medium method (UMM) 14 , 16 , 23 and chemically defined medium method (DMM) 17 ) for differentiating hiPSCs to BMEC‐like cell models yield BMEC‐like cells lacking expression of several key adhesion molecules known to be involved in immune cell migration across the BBB in vivo. To address these specific shortcomings in current hiPSC‐derived BBB models, we developed a new protocol to differentiate hiPSC‐derived endothelial progenitor cells (EPCs) to BMEC‐like cells that display an improved immune phenotype, while also displaying barrier properties similar to those observed for primary human brain endothelial cells. 24 , 25 , 26 , 27 , 28 Overall, these cells faithfully reproduce the required molecular repertoire needed for the study of immune cell trafficking across the BBB.
2. MATERIALS AND METHODS
2.1. Human induced pluripotent stem cells
Human induced pluripotent stem cells (hiPSCs) were reprogrammed, expanded, and characterized for pluripotency and differentiation capacity as described before. 29 In brief, erythroblasts from three donors (age/sex: donor 1: 27/F, donor 2: 50/M, and donor 3: 49/F) were reprogrammed by nucleofection of plasmids encoding for OCT4, shRNA‐p53, SOX2, KLF4, L‐Myc, and Lin28. Human iPSCs were cultured using ReproTeSR medium (STEMCELL Technologies) and expanded using StemMACS iPSC‐Brew XF medium (Miltenyi Biotec, Bergisch Gladbach). Five hiPSC clones from three donors (donor 1: 2 clones, donor 2: 2 clones, and donor 3: 1 clone) were used in this study. For some experiments, we also used the IMR90‐4 iPSC line. 30 Human iPSCs were maintained on Matrigel (Corning)‐coated plates in mTeSR1 medium (STEMCELL Technologies).
2.2. Brain‐like endothelial cells (BLECs) and primary BMECs
Brain‐like endothelial cells (BLECs) were obtained exactly as described before. 15 , 31 In brief, CD34+ cells were isolated from human umbilical cord blood and differentiated to endothelial cells in ECM basal medium (ScienCell) supplemented with 20% (v/v) fetal bovine serum (FBS; Life Technologies) and 50 ng/mL of VEGF165 (PeproTech Inc.). To induce a BBB phenotype, CD34+ cell‐derived endothelial cells were cultured on Transwell filters (0.4 μm pore size, polycarbonate membrane, Corning 3401) and start co‐culture with bovine pericytes at the same day for 6 days using ECM‐5 medium (ECM basal medium (ScienCell) supplemented with 5% FBS, 1% endothelial cell growth supplement (ECGS; ScienCell), and 50 mg/mL gentamycin (Biochrom AG)). Primary human microvascular endothelial cells (pHBMECs) derived from autopsy tissue were purchased from PELOBiotech (Planegg/Martinsried). HBMECs were grown in endothelial cell medium supplemented with FBS, endothelial cell growth supplement, and penicillin/streptomycin solution (ScienCell Research Laboratories).
2.3. Differentiation of hiPSCs into brain microvascular endothelial‐like cells (BMEC‐like cells)
Unconditioned medium method (UMM) 14 , 16 and defined medium method (DMM) 17 were slightly modified and used to differentiate BMEC‐like cells. In brief, hiPSCs were seeded onto a Matrigel‐coated plate in mTeSR1 supplemented with 10 μmol/L ROCK inhibitor Y‐27632 (Selleckchem, Houston, TX, USA) for 24 hours (day ‐3 or day ‐4). Seeding densities at day ‐3 or day ‐4 were optimized at 8,000/cm2 to 21,000/cm2 for UMM, and between 35,000/cm2 to 150,000/cm2 for DMM depending on donor and passage number (Table S1), in order to obtain transendothelial electrical resistance (TEER) of more than 2,000 Ω × cm2 and permeability to sodium fluorescein (NaFl, 376.3 Da) less than 0.6 × 10−4 cm/min. hiPSCs were then expanded in mTeSR1 medium for 2‐3 days depending on hiPSC clone. At day 0, cells were induced to differentiate using specific medium for each method: For UMM, medium was switched to unconditioned medium (DMEM/F12 with 20% Knockout serum replacement, 1× nonessential amino acids, 0.5× Glutamax, and 0.1 mmol/L β‐mercaptoethanol) and changed every day for 6 days. For DMM, medium was switched to DeSR1 (DMEM/F12 with 1× nonessential amino acids, 0.5× Glutamax, and 0.1 mmol/L β‐mercaptoethanol) supplemented with 6 μmol/L CHIR99021 (Selleckchem) and cultured for 24 hours. Then, the medium was switched to DeSR2 (DeSR1 plus 1× B27 supplement) every day for another 5 days. At day 6, for both UMM and DMM, medium was switched to hECSR (human endothelial serum free medium (hESFM, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 1× B27 and 20 ng/mL bFGF) and cultured for 2 days. For retinoic acid (RA) treatment, 10 μmol/L RA was added to hECSR medium. At day 8, cells were replated onto collagen IV (400 μg/mL)/fibronectin (100 μg/mL)‐ or Matrigel‐coated, respectively, for UMM and DMM, Transwell filters (0.4 μm pore size, polycarbonate membrane, Corning 3401) or culture plates at 1,000,000/cm2 in hECSR medium. At day 9, the medium was switched to hECSR2 (hECSR lacking bFGF or RA) and cells were used for assays at day 10.
2.4. Differentiation of hiPSCs into endothelial progenitor cells (EPCs)
CD34+ CD31+ endothelial progenitor cells (EPCs) were differentiated from hiPSCs as described. 32 , 33 In brief, hiPSCs were seeded onto a Matrigel‐coated plate in mTeSR1 supplemented with 5 μmol/L ROCK inhibitor Y‐27632 for 24 hours (day ‐3). Seeding densities at day ‐3 were optimized between 16,000/cm2 and 132, 000/cm2 depending on donor and passage in order to obtain high number of CD34+ CD31+ EPCs. At day 0 and day 1, medium was changed to LaSR basal medium (Advanced DMEM/F12, 2.5 mmol/L GlutaMAX, and 60 μg/mL ascorbic acid) supplemented with 8 μmol/L CHIR99021. At day 2, medium was switched to LaSR basal medium and changed every day for another 3 days. At day 5, CD31+ EPCs were purified using FITC‐conjugated human CD31 antibody (Miltenyi Biotec, clone AC128) and EasySep Human FITC Positive Selection Kit II (STEMCELL Technologies) with an EasySep Magnet kit (STEMCELL Technologies). Purified EPCs were seeded onto collagen IV/fibronectin‐coated Transwell filters at a density of 100,000/cm2 or collagen IV (10 μg/mL)‐coated culture plates at a density of 10,000‐20,000/cm2 in hECSR medium and used for assay or further extended EC culture.
2.5. Extended endothelial cell culture method (EECM) and culture of smooth muscle‐like cells (SMLCs)
EPCs purified as described above were plated on collagen IV (10 μg/mL)‐coated plates in hECSR medium in the presence of 5 μmol/L ROCK inhibitor Y‐27632 of 10,000‐20,000/cm2 as described before. 33 Twenty‐four hours later, medium was changed to hECSR without ROCK inhibitor and then, medium was changed every 2 days. Once the cells reached 100 % confluency (Figure 3B (a)), the cells were passaged using Accutase (Innovative Cell Technologies). In detail, medium was removed and 1 mL of Accutase was added to the 6‐well plate and cells were carefully observed under an inverted microscope (Zeiss Axiovert 25). Once endothelial cells started to become round (Figure 3B (b)), the endothelial cells were physically detached by tapping the 6‐well plate (Figure 3B (c)). Since endothelial cells detached earlier than non‐endothelial cells, preferentially detached endothelial cells were collected in a 15 mL conical tube containing 4 mL of hECSR and centrifuged, resuspended, and seeded onto collagen IV (400 μg/mL)/fibronectin (100 μg/mL)‐coated Transwell filters at a density of 100,000/cm2 or collagen IV (10 μg/mL)‐coated culture plates at a density of 10,000‐20,000/cm2 in hECSR medium. These cell densities were found optimal for achieving closed monolayers. Coating with collagen IV/fibronectin for Transwell filters was chosen as it was previously shown to be optimal for growing primary BMECs and for UMM‐BMEC‐like cells. Here, we found that the combination of collagen IV and fibronectin also enhanced adhesion of both EECM‐BMEC‐like cells and SMLCs. Therefore, we used only collagen IV (10 μg/mL)‐coated culture plates during the selective passaging process to aid in SMLC depletion (Figure 3A). For clarity, we refer to D5 CD34+CD31+ cells as EPCs, and refer to cells that transitioned from EPCs to ECs after 6 days of culture in the EC culture medium as naïve ECs, and refer to “EECM‐BMEC‐like cells” starting at passage 2 (Figure 3A). In some experiments, EECM‐BMEC‐like cells were cultured using ECM‐5 medium. Human iPSC‐derived SMLCs were obtained after harvesting preferentially detached endothelial cells (Figure 3B (c)) by adding 2 mL of hECSR medium to 6‐well plate and culturing ~6‐10 days until confluence. Culture medium was collected every 2 days for SMLC‐derived conditioned medium (CM). List of materials used in the EECM‐BMEC‐like cell differentiation protocol is shown in Table S2.
FIGURE 3.
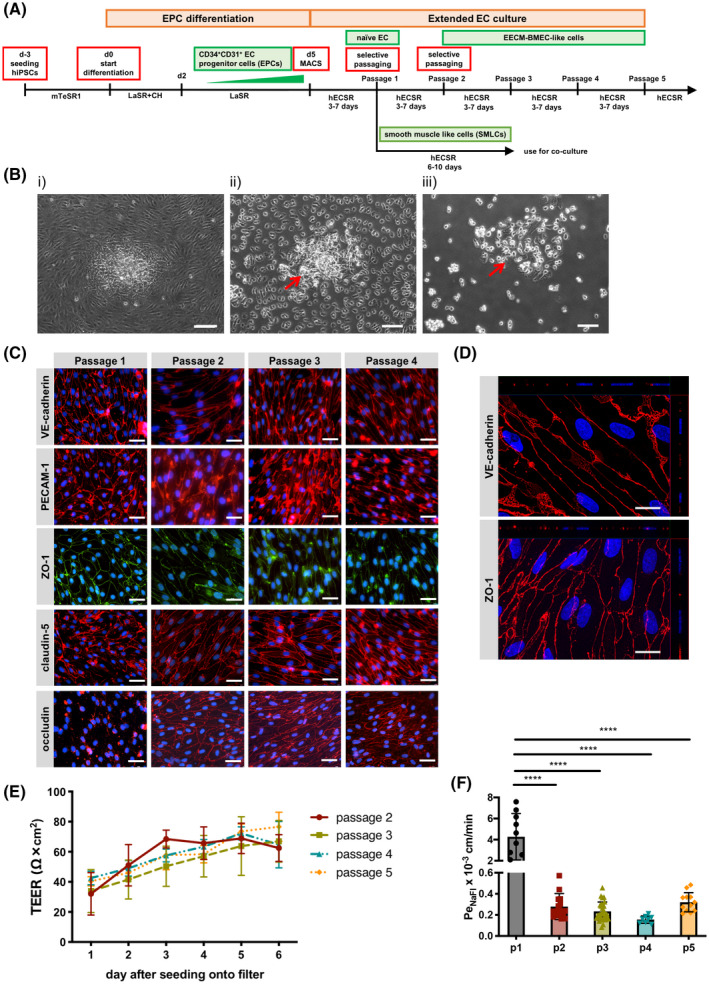
Extended endothelial cell culture method (EECM). A, Schematic representation of the protocols used for extended endothelial cell culture method (EECM). B, Phase contrast images of hiPSC‐derived naïve ECs before (i) and after (ii) incubation with Accutase, and after detachment of ECs by tapping the plate (iii). Red arrow shows non‐ECs remaining attached to the plate. Scale bars = 100 μm. C, Immunofluorescence staining of naïve ECs (passage 1) or EECM‐BMEC‐like cells (passages 2‐4) grown on chamber slides. Junctions were stained for VE‐cadherin (red), PECAM‐1 (red), ZO‐1 (green), claudin‐5 (red), or occludin (red), and nuclei were stained with DAPI (blue). Scale bars = 50 μm. D, Immunofluorescence images of EECM‐BMEC‐like cells (passage 5) grown on 0.4 µm pore Transwell filters. Cell junctions were stained for VE‐cadherin (red) or zonula occludens 1 (ZO‐1, red), and nuclei were stained with DAPI (blue). Each panel shows a xy en face view of a maximum‐intensity projection through the z‐axis. The top (xz) and right (yz) side panels indicate the flat morphology of the EECM‐BMEC‐like cells with staining for cell nuclei and ZO‐1 at the same level in the z‐axis. Scale bars = 20 µm. (C, D) Representative images from donor 2 are shown. Five hiPSCs clones from three donors (donor 1, 2, and 3) were used in this assay and at least three independent differentiations were performed for each clone producing comparable results. E, Time‐dependent progression of transendothelial electrical resistance (TEER) of EECM‐BMEC‐like cell monolayers derived from donor 2. EECM‐BMEC‐like cells (passage 2: red, passage 3: green, passage 4: blue, passage 5: orange) were grown to confluency on 0.4 μm pore size Transwell filters for 6 days. Passage 1 naïve ECs show no measurable TEER. Plotted data are mean TEER values ± SD. Data are combined from at least three independent differentiations for each passage, each measurement performed in triplicates. F, Permeability of 0.37 kDa sodium fluorescein (NaFl): naïve ECs (passage 1: black) or EECM‐BMEC‐like cells (passage 2: red, passage 3: green, passage 4: blue, passage 5: orange) derived from donor 2 were cultured to confluency on 0.4 μm pore size Transwell filters for 6 days and permeability was measured at day 6. Bars show the mean permeability coefficients (Pe) ± SD. Data are combined from at least three independent differentiations each performed in triplicate per condition. Statistical analysis: one‐way ANOVA followed by Tukey’s multiple comparison test. (****P < .0001).
2.6. Co‐culture of BMEC‐like cells with pericytes, astrocytes, or SMLCs
Bovine pericytes, 15 the human brain pericyte cell line (HBPCT), 34 the human astrocyte cell line (hAST) 35 , and hiPSC‐derived astrocytes (iPS_AC) 29 were obtained and maintained as previously described. Since HBPCT and hAST cell lines were immortalized with a temperature‐dependent SV40‐antigen, these cells were expanded at 33°C and switched to 37°C when seeded for co‐culture with EECM‐BEMC‐like cells. HBPCT and hAST cells were shown before to stop proliferating 2 days after the temperature switch. 36 Generally, pericytes and astrocytes were seeded at a density of 26,000/cm2 for 2 days before starting the co‐culture with EECM‐BMEC‐like cells using hECSR and incubated at 37°C (5% CO2) to become confluent. SMLCs were seeded at a density of 13, 000/cm2 1 day before starting the co‐culture using hECSR. EECM‐BMEC‐like cells were seeded on collagen IV (400 μg/mL)/fibronectin (100 μg/mL)‐coated Transwell inserts at a density of 100,000/cm2 and the co‐culture was initiated on the same day. Co‐cultures were maintained for 6 days using hECSR for both the apical and basolateral compartment, with medium changes every other day. Transendothelial electrical resistance (TEER) was measured using a Volt‐Ohm‐Meter (Millicell ERS‐2, MERSSTX01‐electrode). In order to calculate the net resistance in Ω × cm2 of the cell monolayers, TEER value of an empty filter was subtracted from each measurement and TEER values in Ω were multiplied by the surface area of the filters (1.12 cm2) as follows: TEER (Ω × cm2) = (cell monolayer resistance (Ω) − empty Transwell filter resistance (Ω)) × surface area (cm2).
2.7. Treatment of EPCs with RA or pericyte conditioned medium (PCM)
EPCs purified as described above were plated on collagen IV (10 μg/mL)‐coated plates in respective medium. For RA treatment, 10 μmol/L of RA was added to hECSR for 3 days and then, switched to hECSR medium without RA. For PCM treatment, bovine pericytes were cultured in hECSR medium at a density of 26,000/cm2 and culture medium was collected 24 hours later and used as PCM. EPCs were treated with PCM for 6 days. After 6 days of culture, EPCs were detached with Accutase and seeded onto collagen IV/fibronectin‐coated Transwell filters at a density of 100,000/cm2 in hECSR medium. TEER values were measured over 6 days and permeability assay was done at day 6 after seeding onto filters.
2.8. Investigation of cell surface expression of adhesion molecules by flow cytometry
BMEC‐like cells differentiated by UMM, DMM, or EECM and naïve ECs were cultured on collagen IV (10 μg/mL)‐coated plates at a density of 10,000/cm2 or collagen IV (400 μg/mL)/fibronectin (100 μg/mL)‐coated Transwell inserts at a density of 100,000/cm2 in respective media. Some wells were stimulated with 10 ng/mL of recombinant human TNF‐α (R&D Systems, 210TA) and 200 IU/mL recombinant human IFN‐γ (R&D systems, 285IF) for UMM‐ or DMM‐differentiated BMEC‐like cells, and 1 ng/mL of recombinant human TNF‐α and 20 IU/mL recombinant human IFN‐γ, or 1 ng/mL of recombinant human IL‐1β (R&D systems, 201‐LB/CF) for EECM‐BMEC‐like cells or naïve ECs for 16 hours at 37°C (5% CO2). Stimulated and non‐stimulated control cells were gently detached with Accutase, washed, and resuspended in FACS‐buffer (DPBS (1×), 2.5% FBS, 0.1% NaN3). Then, 1 × 105 cells per well were transferred to a 96‐well microtiter plate and incubated 20 minutes on ice with the fluorochrome‐conjugated antibodies or respective isotype controls (Table S3). After staining, cells were washed twice with DPBS and measured with an Attune NxT Flow Cytometer (Thermo Fisher Scientific). Data were analyzed using FlowJo 10 software (Tree Star).
2.9. Western blotting
Naïve ECs (passage 1) and EECM‐BMEC‐like cells (passages 2 and 3) were lysed with RIPA buffer supplemented with Halt protease inhibitor cocktail (Thermo Scientific, 78430). The BCA assay (Pierce BCA Protein Assay Kit: Thermo Scientific, 23227) was used to determine the protein concentration and 23 μg of protein resolved on 4%‐12% tris‐glycine gels. Proteins were transferred to nitrocellulose membranes and blocked for 1 hour at room temperature in tris‐buffered saline supplemented with 0.1% Tween 20 (TBST) and 5% nonfat dry milk. Membranes were incubated with primary antibodies (Table S4) diluted in TBST + 5% nonfat dry milk overnight at 4°C. Membranes were washed 5× with TBST and incubated with infrared‐labeled secondary antibodies (Table S4) diluted in TBST + 5% nonfat dry milk for 1 hour at room temperature. Membranes were washed 5× with TBST and imaged using a LI‐COR Odyssey Classic (9120). Quantification of band intensity was performed using LI‐COR Image Studio software.
2.10. Immunofluorescence staining
BMEC‐like cells differentiated by UMM, DMM, or EECM, naïve ECs, and SMLCs were cultured on Transwell inserts as described above or collagen IV (10 μg/mL)‐coated chamber slides (Thermo Fisher Scientific). Same amounts of pro‐inflammatory cytokine (1 ng/mL TNF‐α + 20 IU/mL IFN‐γ or 1 ng/mL IL‐1β) for Transwell inserts disrupted the EECM‐BMEC‐like cell monolayers grown on chamber slides; therefore, we titrated and used 0.1 ng/mL TNF‐α + 2 IU/mL IFN‐γ or 0.1 ng/ml IL1‐β for stimulation of EECM‐BMEC‐like cells cultured on chamber slides. To stain for claudin‐5, occludin, VE‐cadherin, PECAM‐1, ZO‐1, and N‐cadherin cells were fixed with −20°C precooled methanol for 20 seconds and rehydrated during subsequent washing steps in PBS. Cells were blocked and permeabilized with 5% skimmed milk containing 0.1% Triton X‐100 for 10 minutes at RT, and then, stained with primary antibodies diluted in 5% skimmed milk in PBS for 1 hour at RT as described. 16 , 17 , 37 For staining of the adhesion molecules ICAM‐1, ICAM‐2, VCAM‐1, ALCAM, and MCAM, primary antibodies were diluted in hECSR medium and added to live cells and incubated at 37°C (5% CO2) for 15 minutes. This allowed for selective detection of cell surface‐expressed adhesion molecules, which are functionally available for immune cell interaction. After washing with PBS, cells were fixed with 1% (w/v) formaldehyde in PBS for 10 minutes at RT. Cells were washed with PBS and blocked with 5% skimmed milk in PBS for 10 minutes at RT and then, incubated with secondary antibodies for 1 hour at RT as described. 37 For the staining of P‐selectin, E‐selectin, CD99, ⍺‐smooth muscle actin, NG2, and vimentin, cells were first fixed with 1% (w/v) formaldehyde in PBS for 10 minutes at RT and blocked and permeabilized with 5% skimmed milk in PBS containing 0.1% Triton X‐100 for 10 minutes at RT. Cells were then incubated with primary antibodies for 1 hour at RT diluted in 5% skimmed milk in PBS. To stain for calponin and SM22⍺, cells were fixed with 4% (w/v) formaldehyde in PBS, blocked and permeabilized with 3% BSA in PBS containing 0.1% Triton X‐100 for 1 hour at RT, and then, stained with primary antibodies at 4°C overnight as described. 38 After three washes, cells were incubated with respective secondary antibodies for 1 hour at RT. Nuclei were stained with DAPI at 1 µg/mL or Hoechst 33342 at 4 µmol/L. After washing with DPBS, cell monolayers on filters or chamber slides were mounted with Mowiol (Sigma‐Aldrich). Images were acquired using a Nikon Eclipse E600 microscope or Nikon Eclipse Ti2‐E microscope using the Nikon NIS‐Elements BR3.10 software (Nikon) or inverted research microscope Axio observer Z1 (63x immersion oil, Apotome mode, Zeiss) equipped with a camera (Zeiss Axiocam MRm) and the ZEN‐blue software. List of antibodies used in this study is shown in Table S5. Quantification of EC shape was performed using VE‐cadherin immunofluorescence images by an observer blind to the experimental groups. For each image, FIJI (ImageJ) software was used to trace ten cell outlines, and the Shape Descriptors tool under the Set Measurements function of FIJI was used to calculate circularity values (4 × π × area/perimeter2); average cell circularity for each image is reported.
2.11. Permeability (Pe) assay
Permeability of EC monolayers was assessed by measuring the clearance of sodium fluorescein (NaFl, 376.3 Da, Sigma‐Aldrich) as previously described. 16 Briefly, NaFl was added to the upper compartment of the Transwell inserts at a concentration of 10 μmol/L. Medium samples containing fluorescent tracer that had diffused across the monolayers were collected from the bottom well every 15 minutes for a total of 60 minutes, and fluorescence intensity was measured in a Tecan Infinite M1000 multi‐well reader (Tecan Trading AG). The clearance principle was used to calculate the permeability coefficient (Pe) and to obtain a concentration‐independent transport parameter as previously described in detail. 16 This method includes blank filters without cells as a control to measure clearance across the filter membrane for appropriate calculation of the Pe across the endothelium (Pe). The experiments were done in triplicates for each condition.
2.12. Human Th1* cells
Human CD4+ T cells were isolated and sorted as previously described. 39 , 40 In brief, human Th1* cells were isolated by fluorescence‐activated cell sorting according to their specific expression pattern of chemokine receptors (CXCR3+CCR4−CCR6+) from the peripheral blood of healthy donors. T cells were expanded for 20 days with periodic re‐stimulation with 1 μg/mL phytohemagglutinin, irradiated allogeneic peripheral blood mononuclear cells, and human interleukin 2 (IL‐2, 500 IU/mL) as previously described. 39 , 40 After 20 days of expansion, T cells were frozen and stored in liquid nitrogen until employed in the experiments. T cells were thawed 1 day prior to the respective experiment and labeled with 1 μmol/L CellTracker Green (CMFDA Dye, Life technologies) at 37°C (5% CO2) for 30 minutes on the day of the experiment. After labeling, T cells were washed and dead cells were removed by Ficoll‐Hypaque gradient (780g, 20 minutes, 20°C). T cells were washed twice and resuspended in migration assay medium (DMEM supplemented with 5% FBS, 4 mmol/L L‐Glutamine and 25 mmol/L HEPES) at concentrations as described below.
2.13. Adhesion assay under static condition
EECM‐BMEC‐like cells were cultured on collagen IV (10 μg/mL)‐coated 16‐well chamber slides (Thermo Fisher Scientific) at a density of 100,000/cm2 in hECSR medium or CM derived from SMLCs. EECM‐BMEC‐like cells were stimulated with 0.1 ng/mL recombinant human IL‐1β or 0.1 ng/mL TNF‐α + 2 IU/mL IFN‐γ for 16 hours at 37°C (5% CO2), or maintained as non‐stimulated controls. EECM‐BMEC‐like cell monolayers were washed twice with migration assay medium and 20,000 Th1* cells were added on top of EECM‐BMEC‐like cell monolayers and incubated at RT for 30 minutes using a rocking platform. Chamber slides were gently washed twice and fixed with 2.5% glutaraldehyde for 2 hours on ice. Chamber slides were then washed with DPBS and adherent fluorescencently labeled Th1* cells per pre‐defined field of view (FOV) were analyzed by fluorescence microscopy (Nikon Eclipse E600) and FIJI software (Version 2.0.0, Image J). The average number of adherent cells/FOV was determined counting two fields per well. Assays were performed in at least triplicates for each condition.
2.14. T‐cell arrest to EECM‐BMEC‐like cells under flow
EECM‐BMEC‐like cells were seeded in cloning rings placed on collagen IV (10 μg/mL)‐coated Ibidi μ‐dishes (Ibidi) at a density of 100,000/cm2. EECM‐BMEC‐like cells were stimulated with 0.1 ng/mL recombinant human IL‐1β for 16 hours at 37°C (5% CO2). Live cell imaging was done as described. 41 In brief, Th1* cells were perfused over EECM‐BMEC‐like cell monolayers and allowed to accumulate for 4 minutes using low shear stress (0.1 dynes/cm2). Thereafter, flow was increased to physiological shear stress (1.5 dynes/cm2). Thirty seconds after increasing shear stress, the number of arrested Th1* cells was counted.
2.15. In vitro live cell imaging
EECM‐BMEC‐like cells were cultured in cloning rings placed on collagen IV (10 μg/mL)‐coated Ibidi μ‐dishes (Ibidi) at a density of 75,000/cm2. EECM‐BMEC‐like cells were stimulated with 0.1 ng/mL recombinant human TNF‐α + 2 IU/mL IFN‐γ for 16 hours at 37°C (5% CO2) diluted in CM from SMLC. Fluorescencently labeled Th1* cells were allowed to accumulate on the EECM‐BMEC‐like cell monolayer at a low flow rate of 0.1 dynes/cm2 for 4 minutes from the first frame after the first Th1* cells appeared in the FOV (accumulation phase until 3 minutes 55 seconds). After the accumulation phase of precisely 3 minutes 55 seconds, the flow rate was set to the physiological level of 1.5 dynes/cm2 for 16 minutes (shear phase). The dynamic T‐cell interactions with the EECM‐BMEC‐like cell monolayers under the physiological flow were recorded at a 10x magnification with a Zeiss Axiocam MRm camera. During the recording, 1 image was acquired every 5 seconds, then the video was exported with a frame rate of 30 images/second. T‐cell behavior on the EECM‐BMEC‐like cell monolayer was categorized as described previously. 37 In brief, T cells found to polarize upon arrest and to migrate across the EECM‐BMEC‐like cells monolayer with or without prior crawling or probing on the EECM‐BMEC‐like cells were categorized as “diapedesis.” T cells that crawled on the surface of the EECM‐BMEC‐like cells for the entire observation time were categorized as “crawling.” T cells that remained stationary without displacing beyond a distance exceeding their own diameter and presenting dynamic cellular protrusions were categorized as “probing.”
2.16. Statistical analysis
Statistical analyses comprising calculation of degrees of freedom were done using GraphPad Prism 7 software (Graphpad software, La Jolla, CA, USA). Data are shown as the mean ± SD. To compare two groups, statistical significance was assessed by paired or unpaired t test, while more groups were analyzed by one‐way ANOVA followed by Tukey’s multiple comparison test (*P < .05, **P < .01, ***P < .001, ****P < .0001). The respective statistical methodology used for each assay is specified in corresponding figure legends.
3. RESULTS
3.1. BMEC‐like cells differentiated by UMM or DMM are not suitable for studying immune cell interactions
We first explored if BMEC‐like cells differentiated by the UMM or DMM express and display cytokine‐induced upregulation of adhesion molecules described for the BBB in vivo. 4 To this end, we performed flow cytometry analysis of non‐stimulated (NS) or pro‐inflammatory cytokine‐stimulated (10 ng/mL TNF‐α + 200 IU/mL IFN‐γ) BMEC‐like cells differentiated by UMM or DMM for the BBB adhesion molecules ICAM‐1, ICAM‐2, VCAM‐1, P‐selectin, E‐selectin, and the junctional molecules CD99 and PECAM‐1. UMM‐ and DMM‐differentiated BMEC‐like cells expressed CD99, PECAM‐1, and ICAM‐1, the latter only being further increased after stimulation with pro‐inflammatory cytokines (Figure 1A,B,E). At the same time, UMM‐ and DMM‐differentiated BMEC‐like cells did not stain positive for ICAM‐2, VCAM‐1, E‐selectin, or P‐selectin. Since RA has been reported to inhibit upregulation of endothelial VCAM‐1 in vitro, 42 we asked if omitting RA treatment influences the adhesion molecule profile of UMM‐ and DMM‐differentiated BMEC‐like cells. Completely omitting RA treatment during the whole differentiation process allowed for enhanced cytokine‐induced upregulation of ICAM‐1 on UMM‐ or DMM‐differentiated BMEC‐like cells, but did not affect lack of detection of P‐selectin, E‐selectin, ICAM‐2, and VCAM‐1 (Figure 1C‐E; Figure S1). UMM‐ and DMM‐differentiated BMEC‐like cells thus lack expression of the full array of vascular cell adhesion molecules necessary to support the sequential multistep immune cell adhesion cascade characterized by rolling, arrest, polarization, crawling of the immune cell on, and diapedesis across the BBB under physiological flow. Thus, the well‐established methods for differentiating hiPSC‐derived BMEC‐like cells are not well suited for modeling immune cell interactions at the BBB in the configurations studied here.
FIGURE 1.
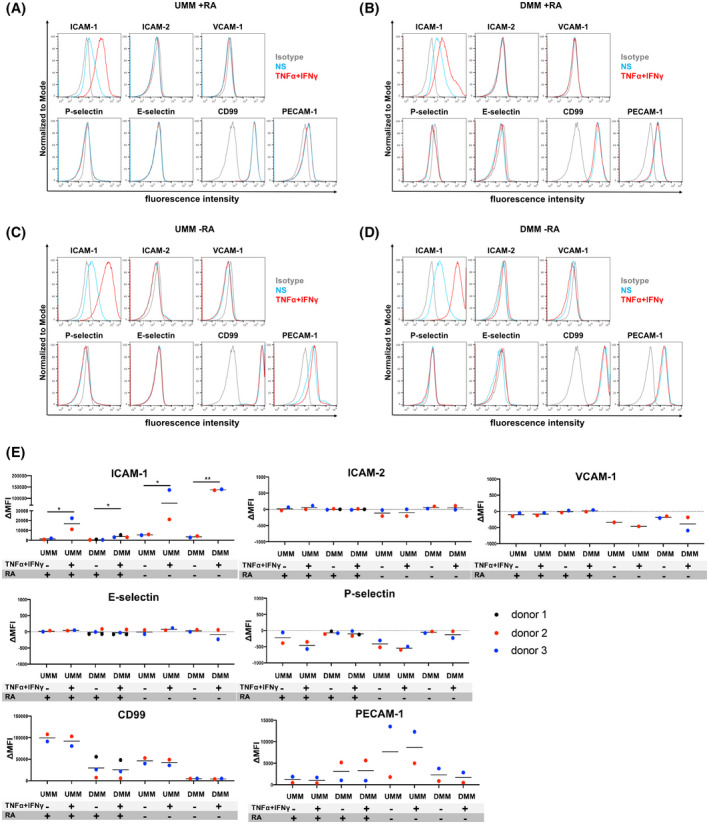
Adhesion molecule phenotype of BMEC‐like cells differentiated by the unconditioned medium method (UMM) or the defined medium method (DMM). Cell surface staining of BMEC‐like cells differentiated by UMM (A, C) or DMM (B, D) in the presence or absence of RA for the adhesion molecules ICAM‐1, ICAM‐2, VCAM‐1, P‐selectin, E‐selectin, CD99, and PECAM‐1 was analyzed by flow cytometry. Isotype control, non‐stimulated (NS), and 16 h pro‐inflammatory cytokine‐stimulated condition (10 ng/mL TNF‐α + 200 IU/mL IFN‐γ) are represented in gray, blue, and red, respectively, in a histogram overlay. Representative data from donor 2 are shown. At least three independent differentiations were performed in each condition using two different hiPSC clones derived from two different donors (donor 2 and 3) for UMM‐differentiated BMEC‐like cells and three hiPSC clones from three donors (donor 1, 2, and 3) for DMM‐differentiated BMEC‐like cells with comparable data observed (eg, Figure S1). E, The Δ geometric mean (MFI staining–MFI isotype) of cell surface adhesion molecules of UMM‐ or DMM‐BMEC‐like cells were analyzed by flow cytometry. Displayed are the mean ∆MFI for each donor (donor 1: black, donor 2: red, and donor 3: blue). Mean ± S.D. from triplicate differentiations were used in a paired students t test to determine statistically significant changes upon stimulation (*P < .05, **P < .01).
3.2. hiPSC‐derived ECs display a BBB immune phenotype but lack barrier properties
We have previously shown that human CD34+ cord blood stem cells can be differentiated into CD34+ EPCs and finally by co‐culture with pericytes into BLECs that display endothelial adhesion molecule expression similar to that observed in vivo. 15 , 31 Therefore, we first asked if hiPSC‐derived naïve ECs could exhibit a mature immune phenotype with respect to expression of endothelial adhesion molecules. To this end, we generated CD34+CD31+ EPCs from hiPSCs using an established protocol (Figure 3A) 32 , 33 and transitioned them to naïve ECs by 6 days of culture in the EC culture medium (hECSR medium: human endothelial serum‐free medium + 1× B‐27 supplement + 20 ng/mL bFGF) and investigated the cell surface expression of adhesion molecules. Under NS conditions, hiPSC‐derived ECs expressed ICAM‐1, ICAM‐2, E‐selectin, CD99, and PECAM‐1, while under pro‐inflammatory cytokine (1 ng/mL TNF‐α + 20 IU/mL IFN‐γ)‐stimulated conditions, ICAM‐1 was upregulated, as expected (Figure 2A,B, Figure S2A). In addition, we found that hiPSC‐derived ECs expressed VCAM‐1 and P‐selectin upon pro‐inflammatory cytokine stimulation (Figure 2A,B, Figure S2A). Thus, these naïve ECs may be suitable for modeling aspects of the BBB immune phenotype. However, since these ECs were not specified to a BMEC‐like fate, barrier characteristics of hiPSC‐derived naïve ECs as measured by TEER and permeability to the small molecule tracer sodium fluorescein with an average molecular weight of 0.37 kDa (PeNaFl), were minimal. We failed to detect a measurable TEER across the naïve EC monolayer and detected a high permeability to sodium fluorescein (Figure 3F, P1; PeNaFl: 2‐10 × 10−3 cm/min). Thus, while hiPSC‐derived ECs exhibit improved adhesion molecule expression compared to UMM‐ and DMM‐differentiated BMEC‐like cells, they fail to establish barrier properties characteristic of the BBB.
FIGURE 2.
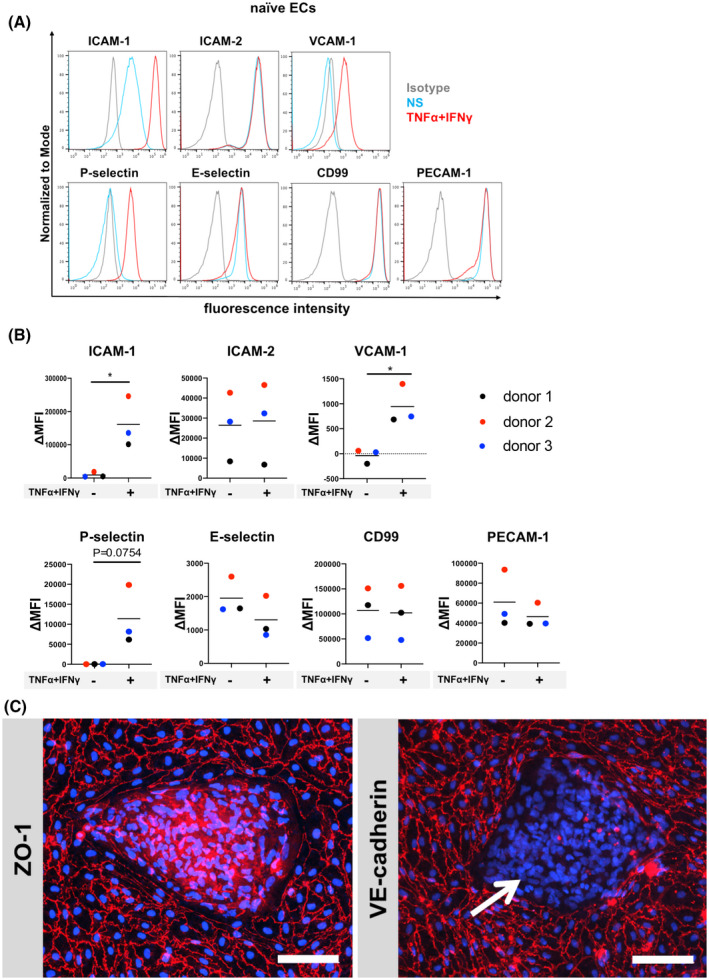
Adhesion molecule phenotype and morphology of hiPSC‐derived naïve ECs. A, Cell surface staining of hiPSC‐derived naïve ECs for the adhesion molecules ICAM‐1, ICAM‐2, VCAM‐1, P‐selectin, E‐selectin, CD99, and PECAM‐1 was analyzed by flow cytometry. Isotype control, non‐stimulated (NS), and 16 h pro‐inflammatory cytokine‐stimulated condition (1 ng/mL TNF‐α + 20 IU/mL IFN‐γ) are represented in gray, blue, and red, respectively, in a histogram overlay. Representative data from donor 2 are shown. Three independent differentiations were performed using three hiPSC clones from three donors (donor 1, 2, and 3) showing comparable data (eg, Figure S2). B, The Δ geometric mean (MFI staining–MFI isotype) of cell surface adhesion molecules of naïve ECs were analyzed by flow cytometry. Displayed are the mean ∆MFI for each donor (donor 1: black, donor 2: red, and donor 3: blue). Mean ± S.D. from triplicate differentiations were used in a paired students t test to determine statistically significant changes upon stimulation (*P < .05). C, Immunofluorescence staining of hiPSC‐derived naïve ECs grown on 0.4 µm pore Transwell filter inserts for ZO‐1 (red) or VE‐cadherin (red), and nuclei (DAPI, blue) is shown. Arrow depicts VE‐cadherin negative cells. Representative data from donor 2 are shown. Each staining is representative of at least three independent differentiations. Three hiPSC clones from three donors (donor 1, 2, and 3) were used in this assay and at least three independent differentiations were performed in each donor. Scale bars = 100 μm.
3.3. Extended passaging drives naïve hiPSC‐derived ECs to BMEC‐like cells
Since studying immune cell interactions at the BBB requires both adhesion molecule phenotypes and tight junction phenotypes critical to the process of T‐cell diapedesis, we next aimed to identify strategies to drive the development of barrier properties in hiPSC‐derived EC monolayers. When EPCs were transitioned to ECs, a mixed population of ZO‐1+/VE‐cadherin+ ECs and VE‐cadherinneg cells resulted (Figure 2C). Thus, despite the presence of junction‐associated ZO‐1‐ and adherens junction‐associated VE‐cadherin‐expressing cells, even a minimal barrier could not be detected as a complete EC monolayer could not form. Since EPCs have been reported to have the capacity to differentiate to both ECs and smooth muscle cells, depending on the medium used for differentiation, 32 PECAM‐1neg cells were tested to determine if they expressed ⍺‐smooth muscle actin (⍺‐SMA). Indeed, the majority of PECAM‐1neg cells stained positive for ⍺‐SMA (Figure S2B,C), underscoring that a fraction of the EPCs differentiated to SMLCs despite being cultured in EC‐inductive medium.
We, therefore, aimed to generate pure naïve EC monolayers in order to test the hypothesis that reducing SMLC contamination in the EC monolayers would improve their basal barrier properties. Careful microscopic observation of the EC cultures during the detachment process when performing standard culture passaging showed that ECs detached earlier compared to non‐ECs (Figure 3B). Selective passaging of the preferentially detached ECs yielded EC monolayers with enriched purity that by passage 2, developed elevated TEER (at day 6 after passaging: 62.4 ± 9.0 Ω × cm2) and ~10‐fold lower permeability (PeNaFl = (0.279 ± 0.124) × 10‐3 cm/min) when compared to naïve ECs (Figure 3E,F). These ECs formed by the EECM, also maintain barrier characteristics over several passages. EECM‐differentiated ECs showed increased purity of VE‐cadherin+ cells (Figure S2D) and stable barrier characteristics with TEER above 60 Ω × cm2 and low permeability to sodium fluorescein (PeNaFl < (0.32 ± 0.1) × 10−3 cm/min) until at least passage 5 (Figure 3E,F). The barrier properties of EECM‐differentiated ECs are comparable to or even tighter than those measured in primary human brain microvascular endothelial cells, 13 , 28 and similar to those observed in BLECs differentiated from primary CD34+ progenitors. 15 Correlating with the enhanced barrier properties upon passage, the passage 2‐4 EECM‐differentiated ECs exhibited increased expression of claudin‐5 and improved junctional localization of both occludin and claudin‐5 when compared to passage 1 naïve ECs (Figure 3C). Western blotting confirmed expression of adherens and tight junction‐associated proteins (VE‐cadherin, occludin, claudin‐5, ZO‐1), Von Willebrand factor (vWF), and caveolin‐1, with claudin‐5 abundance increasing after extended passage culture (Figure S3A,B). Thus, improved barrier properties as measured by TEER and PeNaFl are likely a result of improved EC purity and improved expression and localization of important BBB tight junction proteins occludin and claudin‐5. We further confirmed that the EECM differentiation protocol works independently of hiPSC origin using the fibroblast‐reprogrammed IMR90‐4 line (Figure S3C‐E). In addition, immunofluorescence staining of passage 5 ECs showed that these ECs develop a flat morphology with flat nuclei which overall resembles the typical morphology of brain endothelial cells (Figure 3C,D, Figure S1C). Since EECM‐differentiated ECs formed a diffusion barrier as marked by a low PeNaFl value, and showed cellular and junctional morphology reminiscent of that observed for BBB endothelium in vivo, we term the cells generated by this new protocol EECM‐BMEC‐like cells.
3.4. Astrocytes and pericytes affect morphology but not diffusion barrier characteristics of EECM‐BMEC‐like cells
Barrier properties of brain endothelial cells are not intrinsic and rather rely on developmental cues and continuous cross‐talk with cells from the neurovascular unit such as pericytes and astrocytes. Indeed, previous observations from us and others have shown that astrocyte or pericyte co‐culture can induce barrier characteristics in BLECs and hiPSC‐derived BMEC‐like cells. 15 , 38 , 43 To determine if barrier properties of EECM‐BMEC‐like cells (passages 3‐5) could be improved by co‐culture with astrocytes or pericytes, we grew EECM‐BMEC‐like cells (passages 3‐5) on Transwell filters and co‐cultured them for 6 days with bovine pericytes (bovine‐PC 15 ), a human brain pericyte cell line (HBPCT 34 ), a human astrocyte cell line (hAST 35 ), or hiPSC‐derived astrocytes (iPSC_AC 29 ) in the lower chamber (Figure 4A). We also tested RA treatment for the first 2 days of passage 3 since RA has been reported to improve barrier properties of hiPSC‐derived BMEC‐like cells. 23 Barrier characteristics of monocultured EECM‐BMEC‐like cells were not enhanced by co‐culture or RA treatment as indicated by indistinguishable TEER and NaFl permeability measurements (Figure 4B,C; Figure S4A‐F). Immunofluorescent detection of VE‐cadherin, claudin‐5, and occludin did not reveal any obvious differences in staining levels or junctional localization upon co‐culture with hiPSC‐derived astrocytes or the HBPCT. However, EECM‐BMEC‐like cells exhibited a higher density cell monolayer and a more elongated morphology compared to the monoculture condition (Figure 4D‐F). Thus, EECM‐BMEC‐like cells respond to factors secreted by astrocytes and pericytes, but these factors did not enhance diffusion barrier properties.
FIGURE 4.
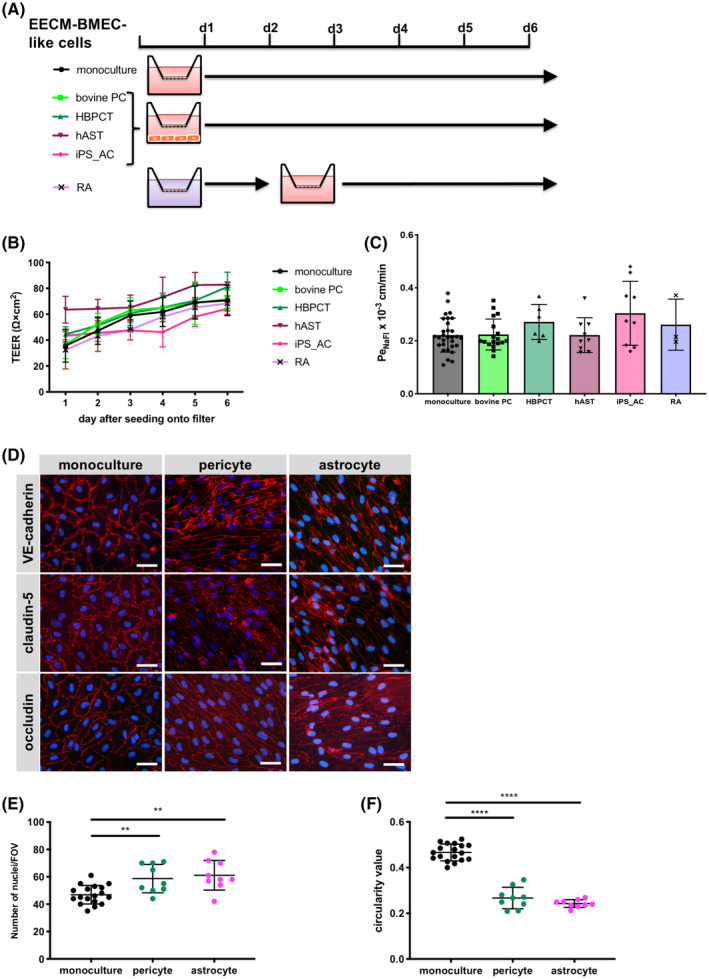
Effect of co‐culture with astrocytes or pericytes on EECM‐BMEC‐like cell monolayers. A, Schematic representation of the protocols for EECM‐BMEC‐like cells co‐cultured with astrocytes or pericytes, or RA treatment is shown. B, TEER and (C) permeability of sodium fluorescein of EECM‐BMEC‐like cell monolayers are shown. EECM‐BMEC‐like cells (from passages 3‐5) were seeded onto 0.4 μm pore size Transwell filters and co‐cultured for 6 days with bovine pericytes ((bovine PC): light green), a human brain pericyte cell line ((HBPCT): green), a human astrocyte cell line ((hAST): dark red), hiPSC‐derived astrocytes ((iPS_AC): pink), or treated with RA for 2 days (purple). Monoculture was used as a control (black). B, Plotted data are mean TEER values ± SD. C, Bars show the mean permeability coefficients (Pe) ± SD. (B, C) Data are from at least three independent differentiations for co‐cultures each performed in triplicate using the same hiPSC clone. Representative data from donor 2 are shown. The following clones were tested for each condition: three hiPSC clones from two donors (donor 1 and 2) for bovine PC, two hiPSC clones from two donors (donor 1 and 2) for HBPCT, two hiPSC clones from two donors (donor 1 and 2) for hAST, and three hiPSC clones from two donors (donor 1, and 2) for iPS_AC and yielded comparable results (eg, Figure S4). D, Immunofluorescence staining of EECM‐BMEC‐like cells cultured on 0.4 μm pore size Transwell filters. EECM‐BMEC‐like cells were seeded onto Transwell filters and either co‐cultured with HBPCT (pericyte) or hiPSC‐derived astrocytes over 6 days. Monoculture was used as a control. Junctions were stained for VE‐cadherin (red), claudin‐5 (red), or occludin (red), and nuclei were stained with DAPI (blue). Representative data from donor 2 are shown. Each staining is representative of at least three independent differentiations performed on three distinct filters. Scale bars = 50 μm. E, The number of nuclei per pre‐defined field of view (FOV) of EECM‐BMEC‐like cells grown on 0.4 μm pore size Transwell filters is shown. ImageJ software was used to automatically count the nuclei. F, EC shape was analyzed using circularity values (4 × π × area/perimeter2) calculated from VE‐cadherin immunofluorescence images using ImageJ software. A value of 1.0 indicates a perfect circle. As the value approaches 0, it indicates an increasingly elongated shape of the cell. Each dot represents average circularity values of 10 randomly chosen cells/FOV. Data are shown as mean ± SD. Statistical analysis: one‐way ANOVA followed by Tukey’s multiple comparison test. (**P < .01, ****P < .0001).
To understand the relevance of culture medium composition on barrier properties of EECM‐BMEC‐like cells, we also employed ECM‐5 medium, which we have previously used in the human CD34+ cord blood stem cell‐derived BLECs model. 15 Culturing EECM‐BMEC‐like cells (passage 3‐5) in the presence of bovine pericytes in ECM‐5 medium improved barrier properties as shown by the significant reduction of their permeability to NaFl when compared to monoculture conditions (Figure S4G). However, irrespective of monoculture or co‐culture, PeNaFl was extremely high when compared to EECM‐BMEC‐like cells cultured in hECSR medium (Figure S4G vs Figure S4C; ECM‐5 monoculture 2.875 ± 0.641 × 10−3 cm/min, ECM‐5 bovine pericyte co‐culture 1.180 ± 0.424 × 10−3 cm/min, hECSR monoculture 0.221 ± 0.064 × 10−3 cm/min, hECSR bovine pericyte co‐culture 0.223 ± 0.058 × 10−3 cm/min). Thus, hESCR medium plays an important role in driving barrier formation in EECM‐BMEC‐like cells.
We next tested if differentiating cells in the potentially more plastic, early stages of EPC‐to‐EC transition (ie, first several days of culture after EPC purification by MACS, Figure 3A) were more responsive to RA treatment or secreted factors present in PCM. After EPC purification, EPCs were cultured with either RA for 3 days or PCM for 6 days and then, upon reaching passage 2, the resulting ECs were seeded onto Transwell filters. Comparing the TEER and permeability for NaFl, we found that neither RA nor PCM pre‐treatment improved barrier properties. Instead, PCM pre‐treatment increased the PeNaFl of the EECM‐BMEC‐like cell monolayer (Figure S4H,I). Given the minimal impact of co‐cultured astrocytes, pericytes and RA treatment on the barrier properties in EECM‐BMEC‐like cells, these conditions were not further investigated.
3.5. Adhesion molecule phenotype of EECM‐BMEC‐like cells
We next asked if the extended passaging of naïve ECs to EECM‐BMEC‐like cells impacted the adhesion molecule repertoire observed for naïve ECs in Figure 2. Much like naïve ECs, EECM‐BMEC‐like cells expressed cell surface ICAM‐1, ICAM‐2, CD99, PECAM‐1, E‐selectin, and P‐selectin under NS conditions (Figure 5A‐C; Figure S5A,B). Pro‐inflammatory cytokine (1 ng/mL TNF‐α + 20 IU/mL IFN‐γ) stimulation‐induced upregulated expression of ICAM‐1 and P‐selectin, but did not change the levels of ICAM‐2 and CD99 (Figure 5A, B). With this adhesion molecule phenotype, EECM‐BMEC‐like cells differ substantially from UMM‐ or DMM‐differentiated BMEC‐like cells and resemble brain like endothelial cells (BLECs 15 ) and primary human brain endothelial cells previously shown to express BBB adhesion molecules 44 (Figure S6). We also confirmed ALCAM and MCAM expression by EECM‐BMEC‐like cells using immunostaining (Figure S5C). In contrast to the findings with naïve ECs, we found only subtle upregulation of VCAM‐1 on TNF‐α/ IFN‐γ‐stimulated EECM‐BMEC‐like cells (Figure 5A‐C and Figure S5A) or 1 ng/mL IL‐1β‐stimulated EECM‐BMEC‐like cells (Figure S5B). TNF‐α/ IFN‐γ‐ or IL‐1β‐stimulated EECM‐BMEC‐like cells showed similar levels of cell surface ICAM‐1 and VCAM‐1 (Figure S5D). Moreover, increasing the concentration of pro‐inflammatory cytokines or using different combinations thereof resulted in disruption of EECM‐BMEC‐like cell monolayers (Figure S5E) rather than increasing the endothelial surface staining for ICAM‐1 and VCAM‐1.
FIGURE 5.
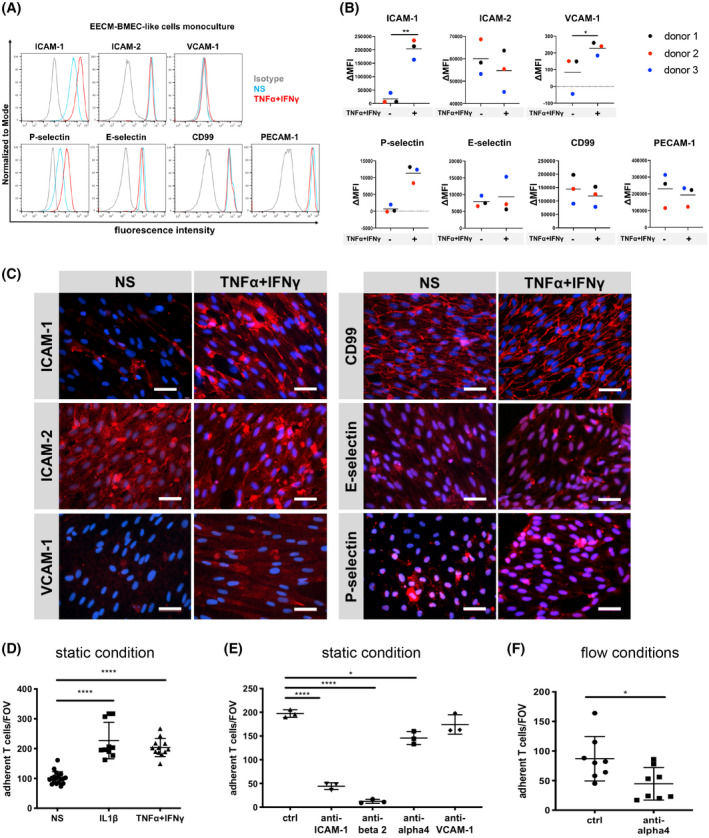
Adhesion molecule phenotype of EECM‐BMEC‐like cells. A, Cell surface staining of EECM‐BMEC‐like cells for the adhesion molecules ICAM‐1, ICAM‐2, VCAM‐1, P‐selectin, E‐selectin, CD99, and PECAM‐1 was analyzed by flow cytometry. Isotype control, non‐stimulated (NS), and 16 h pro‐inflammatory cytokine‐stimulated condition (1 ng/mL TNF‐α + 20 IU/mL IFN‐γ) are represented in gray, blue, and red lines, respectively, in a histogram overlay. Representative data from donor 3 are shown. Three hiPSC clones from three donors (donor 1, 2, and 3) were used in this assay. B, The Δ geometric mean (MFI staining–MFI isotype) of cell surface adhesion molecules of EECM‐BMEC‐like cells were analyzed by flow cytometry. Displayed are the mean ∆MFI for each donor (donor 1: black, donor 2: red, and donor 3: blue). Mean ± S.D. from triplicate differentiations were used in a paired students t test to determine statistically significant changes upon stimulation (*P < .05, **P < .01). C, Immunofluorescence staining of EECM‐BMEC‐like cell monolayers grown to confluency in chamber slides for ICAM‐1 (red), ICAM‐2 (red), VCAM‐1 (red), P‐selectin (red), E‐selectin (red), or CD99 (red) are shown. Nuclei were stained with DAPI (blue). Representative data from donor 2 are shown. NS and 0.1 ng/mL TNF‐α + 2 IU/mL IFN‐γ stimulated conditions are shown. Each staining is representative of at least three independent differentiations performed on three distinct chamber slides. Scale bars = 50 μm. D, The number of cell tracker‐labeled Th1* cells adherent to non‐stimulated (NS), 0.1 ng/mL IL‐1β‐, or 0.1 ng/mL TNF‐α + 2 IU/mL IFN‐γ stimulated EECM‐BMEC‐like cell monolayers derived from donor 2 were counted after 30 minutes of adhesion under static conditions. Adherent cells/FOV is the mean number of cells from two fields per well automatically counted using ImageJ. Data are shown as the mean ± SD of at least three individual differentiations each performed in triplicate. Statistical analysis: one‐way ANOVA followed by Tukey’s multiple comparison test. (****P < .0001). E, The number of cell tracker‐labeled Th1* cells adherent to 0.1 ng/mL IL‐1β stimulated EECM‐BMEC‐like cell monolayers were measured after 30 minutes of adhesion under static conditions. Adherent cells/FOV is the mean number of cells from two fields per well automatically counted using ImageJ. EECM‐BMEC‐like cells derived from donor 2 were preincubated with either anti‐human ICAM‐1 (10 μg/mL, clone R6.5), anti‐human VCAM‐1 (10 μg/mL, polyclonal), or isotype controls for 30 minutes at 37°C. Th1* cells were pretreated with either mouse anti‐human beta2 integrin (10 μg/mL, clone TS1/18), humanized anti‐human α4 integrin IgG4 (10 μg/mL, Natalizumab), or isotype controls for 30 minutes at 37°C. Data are shown as the mean ± SD of a representative experiment of three individual differentiations each performed in at least triplicate. Statistical analysis: one‐way ANOVA followed by Tukey’s multiple comparison test. (*P < .05, ****P < .0001). F, The numbers of Th1* cells adherent to 0.1 ng/mL IL‐1β stimulated EECM‐BMEC‐like cell monolayers derived from donor 2 under flow conditions were measured. Th1* cells were pretreated with either 10 μg/mL Natalizumab or isotype control for 30 minutes at 37°C. Data are shown as the mean ± SD of eight experiments in each condition. Three independently differentiated BMEC‐like cell populations derived from donor 2 were used in this study. Statistical analysis: unpaired t test (*P < .05)
3.6. EECM‐BMEC‐like cells express VCAM‐1 and functional ICAM‐1
Since EECM‐BMEC‐like cells possess both barrier and immune phenotypes that would potentially be valuable for investigating immune cell interactions with the BBB, we wished to determine the functional relevance of endothelial ICAM‐1 and VCAM‐1 on EECM‐BMEC‐like cells. To this end, we investigated the adhesion of Th1* cells on NS and TNF‐α/ IFN‐γ‐stimulated and IL‐1β‐stimulated EECM‐BMEC‐like cells (passages 3‐5) under static conditions. Cytokine stimulation significantly increased the numbers of Th1* cells adhering to EECM‐BMEC‐like cell monolayers (Figure 5D). Antibody‐mediated blocking of endothelial ICAM‐1 and its respective β2‐integrin ligand on Th1* cells significantly reduced adhesion of Th1* cells to cytokine stimulated EECM‐BMEC‐like cells (Figure 5E). Antibody‐mediated blocking of α4β1‐integrins on Th1* cells also reduced adhesion of Th1* cells to cytokine stimulated EECM‐BMEC‐like cells, but blocking of the endothelial α4β1‐integrin ligand VCAM‐1 did not significantly reduce Th1* adhesion (Figure 5E). α4β1‐integrin‐mediated arrest of Th1* cells was also confirmed on cytokine‐stimulated EECM‐BMEC‐like cells under physiological flow (Figure 5F). Taken together, these data demonstrate that EECM‐BMEC‐like cells express functional ICAM‐1 supporting β2‐integrin‐dependent adhesion of Th1* cells. Furthermore, EECM‐BMEC‐like cells also support α4β1‐integrin‐mediated arrest of Th1* cells, where VCAM‐1 does not visibly contribute to this process.
3.7. Smooth muscle‐like cells (SMLCs) induce robust functional VCAM‐1 expression on EECM‐BMEC‐like cells
As the α4‐integrin/VCAM‐1 interaction, in addition to LFA‐1/ICAM‐1, has been reported to mediate CNS entry of T and B cells in CNS autoimmunity, 6 , 45 we explored further strategies to enhance expression of VCAM‐1 on EECM‐BMEC‐like cells. In peripheral endothelial cell cultures, co‐culture with smooth muscle cells was shown to enhance endothelial VCAM‐1 expression. 46 Therefore, we asked if hiPSC‐derived SMLCs could increase the expression of VCAM‐1 in EECM‐BMEC‐like cells. Human iPSC‐derived SMLCs were obtained by continued culture of adherent cells after selective detachment of endothelial cells for derivation of EECM‐BMEC‐like cells (Figure 3A,B). These SMLCs expressed proteins characteristic of smooth muscle cells, including ⍺‐SMA, calponin, and smooth muscle protein‐22⍺ (Figure S7). Next, the SMLCs were co‐cultured with EECM‐BMEC‐like cells (passage 3‐5) on Transwell filters for 6 days and VCAM‐1 staining on EECM‐BMEC‐like cells was compared before and after cytokine stimulation. We found that co‐culture of EECM‐BMEC‐like cells with SMLCs significantly increased cell surface expression of VCAM‐1 after TNF‐α/ IFN‐γ‐stimulation, while there was no overt change to cell surface levels of ICAM‐1, ICAM‐2, PECAM‐1, CD99, E‐selectin, or P‐selectin under either NS or stimulated conditions compared with monocultured EECM‐BMEC‐like cells (Figure 6A vs Figure 5A and Figure 6C; Figure S8). IL‐1β‐stimulation also showed similar effects on cell surface ICAM‐1 and VCAM‐1 staining of SMLC co‐cultured EECM‐BMEC‐like cells (Figure S9A). Interestingly, the SMLC co‐culture could be replaced by providing CM from SMLCs to the abluminal side of EECM‐BMEC‐like cells monolayers (Figure 6B; Figure S8). Immunofluorescence staining of EECM‐BMEC‐like cells cultured in the presence of SMLC co‐culture or SMLC‐derived CM confirmed increased induction of VCAM‐1 after cytokine stimulation (Figure 6D; Figure S9B).
FIGURE 6.
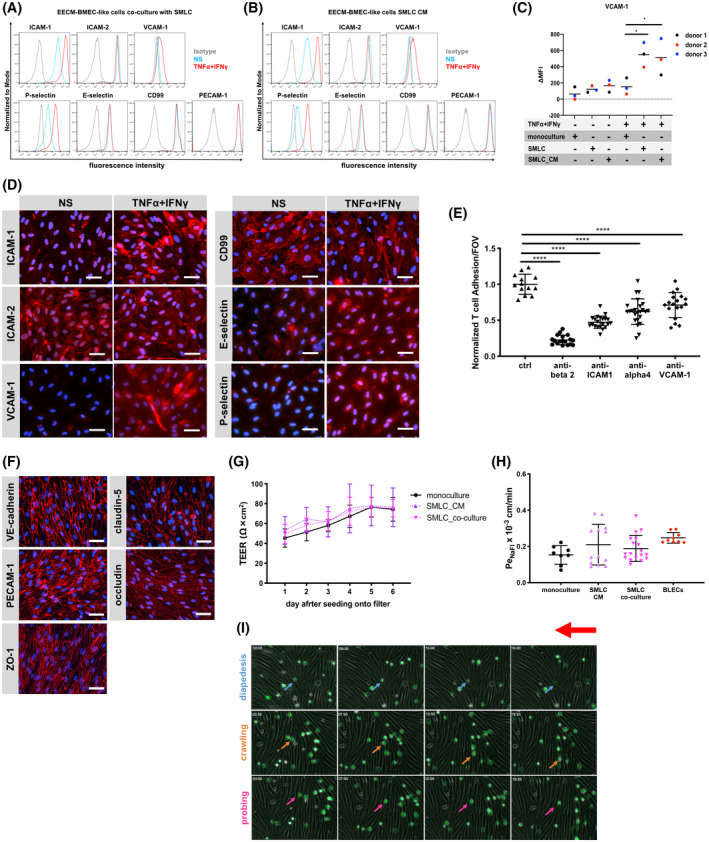
Human iPSC‐derived smooth muscle‐like cells (SMLCs) or conditioned medium enhances VCAM‐1 expression on EECM‐BMEC‐like cells without impairing barrier properties. Cell surface staining of EECM‐BMEC‐like cells co‐cultured with hiPSC‐derived SMLCs (A) or conditioned medium from hiPSC‐derived SMLC (B) for the adhesion molecules ICAM‐1, ICAM‐2, VCAM‐1, P‐selectin, E‐selectin, CD99, and PECAM‐1 was analyzed by flow cytometry. Isotype control, non‐stimulated (NS), and 16 h pro‐inflammatory cytokine‐stimulated condition (1 ng/mL TNF‐α + 20 IU/mL IFN‐γ) are represented in gray, blue, and red lines, respectively, in a histogram overlay. Representative data from donor 3 are shown. Three hiPSC clones from three donors (donor 1, 2, and 3) were used in this assay (eg, Figure S8). C, The Δ geometric mean (MFI staining–MFI isotype) of cell surface VCAM‐1 of EECM‐BMEC‐like cells were analyzed by flow cytometry. Monocultured EECM‐BMEC‐like cells, EECM‐BMEC‐like cells co‐cultured with hiPSC‐derived SMLCs, or cultivated with conditioned medium from hiPSC‐derived SMLC are shown. Each symbol (donor 1: black, donor 2: red, and donor 3: blue) shows the mean of at least three independent differentiations (co‐culture condition, donor 1: n=3, donor 2: n= 4, and donor 3: n= 7). Statistical analysis: one‐way ANOVA followed by Tukey’s multiple comparison test. (*P < .05). D, Immunofluorescence staining of EECM‐BMEC‐like cells derived from donor 3 grown to confluency in chamber slides using conditioned medium from hiPSC‐derived SMLCs for ICAM‐1 (red), ICAM‐2 (red), VCAM‐1 (red), P‐selectin (red), E‐selectin (red), or CD99 (red) are shown. Nuclei were stained with DAPI (blue). NS and 16 h pro‐inflammatory cytokine‐stimulated condition (0.1 ng/mL TNF‐α + 2 IU/mL IFN‐γ) are shown. Each staining is representative of at least three independent differentiations performed on three distinct chamber slides. Scale bars = 50 μm. E, The number of Th1* cells adherent to 16 h pro‐inflammatory cytokine‐stimulated (0.1 ng/mL TNF‐α + 2 IU/mL IFN‐γ) EECM‐BMEC‐like cell monolayers derived from donor 2 cultured in the presence of SMLC‐derived conditioned medium, normalized to control condition was measured after 30 minutes of adhesion under static conditions. Adherent cells/FOV is the mean number of cells from two fields per well automatically counted using ImageJ. EECM‐BMEC‐like cells were preincubated with either anti‐human ICAM‐1 (10 μg/mL, clone R6.5), anti‐human VCAM‐1 (10 μg/mL, polyclonal), or isotype control for 30 minutes at 37°C. Th1* cells were pretreated with either anti‐human beta2 integrin (10 μg/mL, clone TS1/18), humanized anti‐human α4 integrin IgG4 (10 μg/mL, Natalizumab), or isotype controls for 30 minutes at 37°C. Data are shown as the mean ± SD of three individual differentiations each performed in at least triplicates. Statistical analysis: one‐way ANOVA followed by Tukey’s multiple comparison test (****P < .0001). F, Immunofluorescence staining on EECM‐BMEC‐like cells grown on 0.4 μm pore size Transwell filters co‐cultured with SMLCs. Junctions were stained for VE‐cadherin (red), PECAM‐1 (red), ZO‐1 (red), claudin‐5 (red), or occludin (red), and nuclei were stained with DAPI (blue). Representative data from donor 2 is shown. Each staining is representative of at least three independent differentiations performed on three distinct filters. Five hiPSC clones from three donors (donor 1, 2, and 3) were used in this assay. Scale bars = 50 μm. G, TEER and (H) permeability of sodium fluorescein of EECM‐BMEC‐like cell monolayers derived from donor 2 co‐cultured with SMLCs or cultured in the presence of SMLC conditioned medium. EECM‐BMEC‐like cells were grown on 0.4 μm pore size Transwell filters in the presence of either SMLCs (pink) or conditioned medium from SMLCs in the lower chamber (abluminal side) (purple) for 6 days. G, Plotted data are mean TEER values ± SD. H, Bars show the mean permeability coefficients (Pe) ± SD. (G, H) Data are representative of at least three independent differentiations with three filters per conditions. Three hiPSC clones from three donors (donor 1, 2, and 3) were used in this assay yielding comparable results. Human CD34+ cord blood stem cell‐derived brain‐like endothelial cells (BLECs, red) were used for a comparison to a previously developed human in vitro BBB model. I, In vitro live cell imaging of the Th1* cell/EECM‐BMEC‐like cells interactions under flow. A temporal snapshot illustrating that the different Th1* cell‐EECM‐BMEC‐like cells interactions have been made with several frames taken from a video recorded with a 10× magnification. EECM‐BMEC‐like cells from donor 2 were cultured in cloning rings placed on collagen IV (10 μg/mL)‐coated Ibidi μ‐dishes at a density of 75,000/cm2. EECM‐BMEC‐like cells were stimulated with 0.1 ng/mL recombinant human TNF‐α + 2 IU/mL IFN‐γ for 16 h at 37°C (5% CO2) diluted in conditioned medium from SMLC. Fluorescencently labeled Th1* cells were allowed to accumulate on the EECM‐BMEC‐like cell monolayer at a low flow rate of 0.1 dyne/cm2 for 4 min from the first frame after the first Th1* cells appear in the field of view (accumulation phase until 3 min 55 s). After the accumulation phase of precisely 3 min 55 sec, the flow rate was set to 1.5 dyne/cm2 for 16 min (shear phase). Each row of images shows a different behavior (diapedesis (blue), crawling (orange), probing (pink)) of the Th1* cells with the EECM‐BMEC‐like cells. The red arrow shows the direction of flow and the time is displayed on the top left of each image (min:sec format). Time‐lapse video showing Th1* cell interaction on EECM‐BMEC‐like cells is provided as Video S1. During the recording, 1 picture is acquired every 5 second then the video is exported with a framerate of 30 images/second. The video is repeated four times, the first run shows the full‐scale video, the second run is a zoom in of area highlighting T cells diapedesis (blue circles) and the third run is a zoom in crawling (orange circle) and diapedesis (blue circle), and the last run is a zoom in of one area highlighting crawling (orange circles) and probing (pink circle) events. The red arrow shows the direction of flow and the time is displayed on the top left of the video (min:sec format).
Considering the SMLC‐enhanced cell surface expression of VCAM‐1 on EECM‐BMEC‐like cells, we next asked if VCAM‐1 would now play a role in Th1* cell adhesion to EECM‐BMEC‐like cells. As observed in the adhesion assays with monocultured EECM‐BMEC‐like cells, antibody‐mediated blocking of endothelial ICAM‐1 or its Th1* ligand, β2‐integrin, significantly reduced adhesion to SMLC‐derived CM enhanced EECM‐BMEC‐like cells (Figure 6E). However, in key contrast to monocultured EECM‐BMEC‐like cells, antibody‐mediated blocking of endothelial VCAM‐1 in addition to its ligand, α4‐integrin, on Th1* cells significantly decreased T‐cell adhesion to EECM‐BMEC‐like cells indicating that SMLC‐derived CM enhanced VCAM‐1 expression on EECM‐BMEC‐like cells to levels of functional relevance.
Since smooth muscle cells can increase endothelial permeability in the periphery, 46 we also examined if SMLC‐derived factors would impair barrier properties of the EECM‐BMEC‐like cells. Although both co‐culture of EECM‐BMEC‐like cells with SMLC‐ or SMLC‐derived CM enhanced variability of permeability measures, there was no significant effect on barrier characteristics of EECM‐BMEC‐like cells as determined by TEER values or permeability to NaFl (Figure 6G,H). Furthermore, co‐culture of EECM‐BMEC‐like cells with SMLC did not change the junctional localization of VE‐cadherin, PECAM‐1, ZO‐1, claudin‐5, or occludin (Figure 6F). Taken together, SMLC‐enhanced EECM‐BMEC‐like cells retain the barrier properties while displaying an optimized cell adhesion molecule phenotype, thus making them a highly suitable model to study immune cell interactions with the human BBB in vitro. To verify their capability to fully support immune cell migration, we investigated the interaction of human Th1* cells superfused over TNF‐α/ IFN‐γ stimulated EECM‐BMEC‐like cell monolayers under physiological flow. In vitro live cell imaging demonstrated that Th1* cells arrest and polarize on EECM‐BMEC‐like cell monolayers. This was followed by Th1* crawling and probing prior to crossing the EECM‐BMEC‐like cell monolayers (Figure 6I, Video S1), as observed by us previously on CD34+‐derived BLECs. 37 The EECM‐BMEC‐like cells thus allow for the observation of the individual steps of the multistep T‐cell extravasation cascade across the BBB.
4. DISCUSSION
We set out to identify a human in vitro BBB model that is suitable for the study of BBB‐immune cell interactions, which are important in CNS immune surveillance and neuroinflammatory diseases. 3 , 4 , 47 , 48 Human BBB models have been generated from diverse sources, including primary or immortalized brain ECs 10 , 49 , 50 and cord blood‐derived endothelial progenitors. 15 , 51 We focused, however, on models generated from hiPSCs, as they offer benefits in scalability and the potential for studying immune cell interactions in an autologous fashion. We first evaluated well‐established protocols for generating hiPSC‐derived BMEC‐like cells via the UMM and DMM. 17 , 23 These and similar methods generate cells that achieve very high TEER, express known BBB solute and efflux transporters, and yield molecular permeabilities that correlate well with in vivo observations and these BMEC‐like cells are, therefore, well suited to study small and large molecule permeability. 14 , 17 , 21 , 23 , 52 , 53 , 54 , 55 However, we found that BMEC‐like cells differentiated by the UMM and DMM do not express all adhesion molecules shown to mediate the interaction of immune cells with the BBB, 4 , 8 , 48 , 56 , 57 , 58 and thus an alternative approach is required to examine such interactions. We, therefore, evaluated the immune cell adhesion molecule profile of naïve ECs differentiated from hiPSC‐derived EPCs, 32 and observed basal or pro‐inflammatory cytokine‐inducible expression of ICAM‐1, ICAM‐2, VCAM‐1, E‐selectin, P‐selectin, CD99, and PECAM‐1. However, these cells did not develop a passive barrier characteristic of the BBB. In contrast, after extended culture, the resulting cells displayed well‐developed tight junctions, moderate TEER, and a flat cellular morphology and junctional architecture characteristic of primary BMECs. Importantly, after extended culture, the resulting cells retained an immune cell adhesion molecule profile similar to primary BMECs, 26 , 27 , 59 and expressed ICAM‐1, ICAM‐2, E‐selectin, P‐selectin, CD99, and PECAM‐1. Pro‐inflammatory cytokine stimulation led to upregulation of ICAM‐1 and P‐selectin and induction of VCAM‐1 that could be further enhanced by SMLC co‐culture or SMLC‐derived CM treatment. Non‐inflammatory expression of E‐selectin on EECM‐BMEC‐like cells may be due to mechanisms involved in growth of EECM‐BMEC‐like cells as previously reported 60 and must be considered when using EECM‐BMEC‐like cells to study immune cell trafficking across the non‐inflamed BBB, which does not show constitutive E‐selectin expression in vivo.
We confirmed the functionality of ICAM‐1 and VCAM‐1 in our model by employing blocking antibodies and observed reduced Th1* cell adhesion. While EECM‐BMEC‐like cells have weaker barrier properties than UMM‐ or DMM‐differentiated BMEC‐like cells, they are similar to other human stem cell‐derived models 15 , 18 , 51 and primary mouse BMECs, 61 , 62 and have stronger barrier properties than immortalized human or rodent cell lines. 10 , 50 , 63 EECM‐BMEC‐like cells are similar to recently reported hiPSC‐derived brain capillary‐like endothelial cells (BCLECs) 18 as both proceed through an EPC intermediate and develop a moderate TEER of ~60‐80 Ω × cm2. However, in contrast to the work of Praça et al. 18 we found that EECM‐BMEC‐like cells demonstrated pro‐inflammatory cytokine‐inducible expression of VCAM‐1, a prerequisite for studying immune cell interactions with the BBB. 4 , 6 Taken together, while other hiPSC‐derived BBB models are likely better suited to evaluate drug permeability, drug efflux, and nutrient transport, EECM‐BMEC‐like cells have a robust adhesion molecule profile, functionally interact with Th1* cells, and have good paracellular barrier properties, making this model well suited to study immune cell‐BBB interactions.
The EECM‐BMEC‐like cell differentiation protocol was adapted from an existing method to generate CD34+CD31+ EPCs from hiPSCs via small molecule activation of canonical Wnt signaling. 32 Prior work demonstrated that culturing these endothelial progenitors for 10 days in EGM‐2, a widely used endothelial cell medium that contains serum and several growth factors (EGF, IGF, bFGF, and VEGF), yielded ECs with TNF⍺‐inducible expression of ICAM‐1 and TEER of ~40 Ω × cm2. 32 We, therefore, asked whether hECSR, a serum‐free medium containing B‐27 supplement and bFGF that has been previously used to culture BMEC‐like cells, 17 would enhance the barrier phenotype of the resulting ECs without compromising the robust immune cell adhesion molecule profile we observed. We found that after extended culture, the resulting ECs developed increased TEER of 60‐80 Ω × cm2 and reduced NaFl permeability, phenotypes attributable both to improved EC monolayer purity and improvements to claudin‐5 and occludin expression and junctional localization. Importantly, the presence of continuous localization of occludin to tight junctions, a hallmark of BBB endothelium, 64 suggests specification of these naïve ECs to a barrier forming, BMEC‐like phenotype. RA treatment or co‐culture of the resulting EECM‐BMEC‐like cells with pericytes or astrocytes did not lead to further improvements in barrier properties, and conversely, culturing these cells in serum‐ and mitogen‐rich ECM‐5 medium yielded markedly higher sodium fluorescein permeability. In contrast, Praça et al. 18 generated BCLECs from similar hiPSC‐derived mesodermal endothelial progenitors using EGM‐2 medium, and found that VEGF, Wnt3a, and RA supplementation increased TEER from ~20 to 60 Ω × cm2, decreased Lucifer yellow permeability, increased claudin‐5 abundance, and increased efflux transporter and solute carrier transcript expression. 18 Together, these observations suggest a strong dependence of endothelial barrier properties on basal cell culture medium, and that extended culture in minimal hECSR medium is sufficient to generate cells with a robust paracellular barrier in the absence of co‐culture or additional exogenous factors. We note, however, that co‐culture or supplementation of Wnts, RA, or other factors with known roles in BBB development 65 , 66 could potentially improve aspects of BBB phenotype in EECM‐BMEC‐like cells, but the barrier properties, junctional architecture, and immune cell adhesion molecule profile of the present model is well suited for the intended application. In fact, a major advantage of the EECM‐BMEC‐like model is the presence of functional VCAM‐1, which plays a major role in the capture of T cells on endothelium via ⍺4β1 integrin binding. 4 , 6 While recent work demonstrated TNF⍺‐inducible expression of VCAM‐1 in BMEC‐like cells differentiated via the UMM, maintained under flow in collagen channels, and assayed after fixation and permeabilization, 22 we did not observe cell surface VCAM‐1 expression in UMM or DMM‐differentiated BMEC‐like cells. Furthermore, cytokine‐inducible expression of VCAM‐1 in ECs differentiated from hiPSC‐derived endothelial progenitors has been variable, with some reports of positive 67 and negative 18 , 68 results, and our monocultured EECM‐BMEC‐like cells only exhibited a modest response to inflammatory stimulation. Given the importance of mural cells in endothelial function and previous observations that smooth muscle cell‐EC interactions induce VCAM‐1 in peripheral ECs, 46 we asked whether hiPSC‐derived SMLCs could modulate VCAM‐1 expression. Indeed, we found that co‐culturing EECM‐BMEC‐like cells with hiPSC‐derived SMLCs or SMLC‐derived CM improved VCAM‐1 inducibility on EECM‐BMEC‐like cells without significantly influencing barrier properties. Furthermore, VCAM‐1 functionally contributed to Th1* cell adhesion to SMLC‐enhanced EECM‐BMEC‐like cells as assessed using a VCAM‐1 blocking antibody. Importantly, the SMLCs are a byproduct generated during the EECM‐BMEC‐like cell differentiation and as such, do not require independent differentiation of multiple cell types in parallel and are derived from the same hiPSC line as the EECM‐BMEC‐like cells, which should make this system readily applied to isogenic disease modeling. Taken together, EECM‐BMEC‐like cells represent a novel hiPSC‐derived in vitro model of the BBB possessing key molecular and functional attributes required to evaluate the interactions of immune cells with the BBB.
Cellular and molecular mechanisms mediating immune cell trafficking across the BBB have largely been studied in mouse models. 4 Human in vitro BBB models have however been essential to confirm the translational importance of these mechanisms and verify novel therapeutic targets for inhibiting immune cell trafficking into the CNS in neuroinflammatory diseases in humans. 12 , 31 , 62 Primary hBMECs have proven especially useful to study immune cell/BBB interactions and have allowed the identification of additional molecules expressed on the BBB like ALCAM, 12 , 26 MCAM, 69 and ninjurin‐1, 70 , 71 which seem to contribute to the recruitment of pathogenic T cells and myeloid cells into the CNS in the context of neuroinflammation. However, primary human brain microvascular endothelial cells mainly originate from surgical specimens, limiting the ability to establish patient‐derived in vitro BBB model from diseases where biopsy or surgery is not common. The contribution of BBB alterations mediating immune cell trafficking has been largely studied in autopsy brain samples. Indeed adhesion molecules like ICAM‐1, VCAM‐1, ALCAM, MCAM, and ninjurin‐1 were reported to be upregulated on brain endothelial cells in neuroinflammatory diseases such as multiple sclerosis. 12 , 26 , 59 , 69 , 70 , 71 , 72 However, direct contribution of these endothelial adhesion molecules to disease pathogenesis remains unknown since autopsy brain samples mainly reflect advanced stages of the disease and robust and reproducible functional assays using autopsy samples is quite difficult. Our novel EECM‐BMEC‐like cells, which possess proper adhesion molecules, open the field to study functional contributions of patient‐derived brain endothelium in mediating altered immune cell migration into the CNS. Furthermore, isolating immune cells from the very same donors will facilitate the study immune cell migration across the BBB in an entirely autologous fashion. This will eventually also allow for novel observations on how antigen‐specific processes may contribute to immune cell/BBB interactions including BBB disruption or antigen presentation by brain endothelium in human neurological disorders. 73
In conclusion, EECM‐BMEC‐like cells establish an in vitro BBB model that resembles, in morphology, molecular junctional architecture, diffusion barrier characteristics, and adhesion molecule profile, primary BMECs. Thus, this is the first model specifically adapted to study the role of the BBB in CNS immunity including immune cell migration across the BBB during CNS immune surveillance or inflammation. Additional molecular and cellular characterization of EECM‐BMEC‐like cells under non‐stimulated and stimulated conditions will expand our understanding of how closely this novel model mimics neuroimmune conditions at the BBB ranging from CNS immune surveillance to different neuroinflammatory disorders. Employing patient‐derived or genetically modified hiPSCs in concert with the EECM‐BMEC‐like cell differentiation protocol will facilitate study of the contribution of BBB impairment to CNS disorders ranging from multiple sclerosis to Alzheimer’s disease. Employing immune cells from the very same donors, will enable the unique opportunity to study immune cell interactions with the BBB in an autologous fashion.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
For the hiPSC work, all subjects gave their written informed consent according to institutional review board guidelines (Consent form 107/13 for Lausanne subjects and OFSEP consent form for Nantes subjects). For the BLEC model, The French Ministry of Higher Education and Research approved the protocol regarding the use of human tissues and cells (CODECOH Number DC2011‐1321).
CONFLICT OF INTERESTS
BE received a grant from Biogen to study extended natalizumab dosing on T‐cell migration across the blood‐brain barrier. HN, BDG, SPP, EVS, and BE are inventors on a provisional US patent application related to this work.
AUTHORS' CONTRIBUTIONS
BE: Conceptualization and supervision of study, funding acquisition; project administration, writing and editing of the manuscript. HN: performing and analyzing experiments, developing methodology, funding acquisition, and writing of the original manuscript. BDG: performing and analyzing experiments, developing methodology, and manuscript writing and editing. EVS: Conceptualization and supervision of study, funding acquisition, project administration, and writing and editing of the manuscript. SPP: providing reagents, cells and methodology, and manuscript editing. SS: performing and analyzing experiments. SP: providing reagents, cells and methodology, and manuscript editing. AM: providing reagents, cells and methodology, and manuscript editing. RDP: providing reagents, cells and methodology, and manuscript editing. YS: providing reagents, cells and methodology, and manuscript editing. FS: providing reagents, cells and methodology, and manuscript editing. TK: providing reagents, cells and methodology, and manuscript editing. FG: providing reagents and cells, methodology, and contribution to manuscript editing.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8
Fig S9
Table S1 and S2
Table S3 and S4
Table S5
Video S1
ACKNOWLEDGMENTS
The authors thank William A. Muller for providing anti‐CD99 antibody and Andrew Chan and Robert Hoepner for providing Natalizumab.
Nishihara H, Gastfriend BD, Soldati S, et al. Advancing human induced pluripotent stem cell‐derived blood‐brain barrier models for studying immune cell interactions. The FASEB Journal. 2020;34:16693–16715. 10.1096/fj.202001507RR
Funding informationThis study was funded by the Bangerter‐Rhyner Foundation and the Bern Center for Precision Medicine to BE, the Swiss MS Society to BE and RDP, an ECTRIMS Postdoctoral Research Exchange Fellowship, the Uehara Memorial Foundation, and JSPS Overseas Research Fellowships to HN, and National Institutes of Health Grant NS103844 to EVS and SPP. RDP was also supported by a SNF 320030‐179531. BDG was supported by the National Science Foundation Graduate Research Fellowship program under grant number 1747503 and the National Institutes of Health Biotechnology Training Program grant T32 GM008349.
Contributor Information
Eric V. Shusta, Email: eshusta@wisc.edu.
Britta Engelhardt, Email: bengel@tki.unibe.ch, Email: eshusta@wisc.edu.
REFERENCES
- 1. Tietz S, Engelhardt B. Brain barriers: Crosstalk between complex tight junctions and adherens junctions. J Cell Biol. 2015;209:493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Engelhardt B, Sorokin L. The blood‐brain and the blood‐cerebrospinal fluid barriers: function and dysfunction. Semin Immunopathol. 2009;31:497–511. [DOI] [PubMed] [Google Scholar]
- 3. Engelhardt B, Vajkoczy P, Weller RO. The movers and shapers in immune privilege of the CNS. Nat Immunol. 2017;18:123–131. [DOI] [PubMed] [Google Scholar]
- 4. Engelhardt B, Ransohoff RM. Capture, crawl, cross: the T cell code to breach the blood‐brain barriers. Trends Immunol. 2012;33:579–589. [DOI] [PubMed] [Google Scholar]
- 5. Sweeney MD, Zhao Z, Montagne A, Nelson AR, Zlokovic BV. Blood‐brain barrier: from physiology to disease and back. Physiol Rev. 2019;99:21–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vajkoczy P, Laschinger M, Engelhardt B. Alpha4‐integrin‐VCAM‐1 binding mediates G protein‐independent capture of encephalitogenic T cell blasts to CNS white matter microvessels. J Clin Investig. 2001;108:557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abadier M, Haghayegh Jahromi N, Cardoso Alves L, et al. Cell surface levels of endothelial ICAM‐1 influence the transcellular or paracellular T‐cell diapedesis across the blood‐brain barrier. Eur J Immunol. 2015;45:1043–1058. [DOI] [PubMed] [Google Scholar]
- 8. Bartholomaus I, Kawakami N, Odoardi F, et al. Effector T cell interactions with meningeal vascular structures in nascent autoimmune CNS lesions. Nature. 2009;462:94–98. [DOI] [PubMed] [Google Scholar]
- 9. Lecuyer MA, Saint‐Laurent O, Bourbonniere L, et al. Dual role of ALCAM in neuroinflammation and blood–brain barrier homeostasis. Proc Natl Acad Sci USA. 2017;114(4):E524–E533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weksler BB, Subileau EA, Perriere N, et al. Blood‐brain barrier‐specific properties of a human adult brain endothelial cell line. FASEB J. 2005;19:1872–1874. [DOI] [PubMed] [Google Scholar]
- 11. Helms HC, Abbott NJ, Burek M, et al. In vitro models of the blood‐brain barrier: An overview of commonly used brain endothelial cell culture models and guidelines for their use. J Cereb Blood Flow Metab. 2016;36:862–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lyck R, Lecuyer MA, Abadier M, et al. ALCAM (CD166) is involved in ALCAM (CD166) is involved in extravasation of monocytes rather than T cells across the blood‐brain barrier. J Cereb Blood Flow Metab. 37(8):2894‐2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alvarez JI, Dodelet‐Devillers A, Kebir H, et al. The Hedgehog pathway promotes blood‐brain barrier integrity and CNS immune quiescence. Science (New York. 2011;N.Y.) 334:1727–1731. [DOI] [PubMed] [Google Scholar]
- 14. Lippmann ES, Azarin SM, Kay JE, et al. Derivation of blood‐brain barrier endothelial cells from human pluripotent stem cells. Nat Biotechnol. 2012;30:783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cecchelli R, Aday S, Sevin E, et al. A stable and reproducible human blood‐brain barrier model derived from hematopoietic stem cells. PLoS One. 2014;9:e99733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stebbins MJ, Wilson HK, Canfield SG, Qian T, Palecek SP, Shusta EV. Differentiation and characterization of human pluripotent stem cell‐derived brain microvascular endothelial cells. Methods. 2016;101:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qian T, Maguire SE, Canfield SG, et al. Directed differentiation of human pluripotent stem cells to blood‐brain barrier endothelial cells. Science advances. 2017;3:e1701679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Praca C, Rosa SC, Sevin E, Cecchelli R, Dehouck MP, Ferreira LS. Derivation of brain capillary‐like endothelial cells from human pluripotent stem cell‐derived endothelial progenitor cells. Stem Cell Rep. 2019;13:599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lim RG, Quan C, Reyes‐Ortiz AM, et al. Huntington's disease iPSC‐derived brain microvascular endothelial cells reveal WNT‐mediated angiogenic and blood‐brain barrier deficits. Cell Rep. 2017;19:1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vatine GD, Al‐Ahmad A, Barriga BK, et al. Modeling psychomotor retardation using iPSCs from MCT8‐deficient patients indicates a prominent role for the blood‐brain barrier. Cell Stem Cell. 2017;20:831–843.e835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vatine GD, Barrile R, Workman MJ, et al. Human iPSC‐derived blood‐brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell. 2019;24:995–1005.e1006. [DOI] [PubMed] [Google Scholar]
- 22. Linville RM, DeStefano JG, Sklar MB, et al. Human iPSC‐derived blood‐brain barrier microvessels: validation of barrier function and endothelial cell behavior. Biomaterials. 2019;190–191:24–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lippmann ES, Al‐Ahmad A, Azarin SM, Palecek SP, Shusta EV. A retinoic acid‐enhanced, multicellular human blood‐brain barrier model derived from stem cell sources. Sci Rep. 2014;4:4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Urich E, Lazic SE, Molnos J, Wells I, Freskgård PO. Transcriptional profiling of human brain endothelial cells reveals key properties crucial for predictive in vitro blood‐brain barrier models. PLoS One. 2012;7:e38149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malin D, Strekalova E, Petrovic V, et al. αB‐crystallin: a novel regulator of breast cancer metastasis to the brain. Clin Cancer Res. 2014;20:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cayrol R, Wosik K, Berard JL, et al. Activated leukocyte cell adhesion molecule promotes leukocyte trafficking into the central nervous system. Nat Immunol. 2008;9:137–145. [DOI] [PubMed] [Google Scholar]
- 27. Cayrol R, Haqqani AS, Ifergan I, Dodelet‐Devillers A, Prat A. Isolation of human brain endothelial cells and characterization of lipid raft‐associated proteins by mass spectroscopy. Methods Mol Biol. 2011;686:275–295. [DOI] [PubMed] [Google Scholar]
- 28. Podjaski C, Alvarez JI, Bourbonniere L, et al. Netrin 1 regulates blood‐brain barrier function and neuroinflammation. Brain. 2015;138:1598–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perriot S, Mathias A, Perriard G, et al. Human induced pluripotent stem cell‐derived astrocytes are differentially activated by multiple sclerosis‐associated cytokines. Stem Cell Rep. 2018;11:1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu J, Vodyanik MA, Smuga‐Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 318, 1917‐1920. [DOI] [PubMed] [Google Scholar]
- 31. Nishihara H, Soldati S, Mossu A, et al. Human CD4(+) T cell subsets differ in their abilities to cross endothelial and epithelial brain barriers in vitro. Fluids Barriers CNS. 2020;17:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lian X, Bao X, Al‐Ahmad A, et al. Efficient differentiation of human pluripotent stem cells to endothelial progenitors via small‐molecule activation of WNT signaling. Stem Cell Rep. 2014;3:804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bao X, Lian X, Palecek SP. Directed endothelial progenitor differentiation from human pluripotent stem cells via Wnt activation under defined conditions. Methods Mol Biol. 2016;1481:183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shimizu F, Sano Y, Abe MA, et al. Peripheral nerve pericytes modify the blood‐nerve barrier function and tight junctional molecules through the secretion of various soluble factors. J Cell Physiol. 2011;226:255–266. [DOI] [PubMed] [Google Scholar]
- 35. Haruki H, Sano Y, Shimizu F, et al. NMO sera down‐regulate AQP4 in human astrocyte and induce cytotoxicity independent of complement. J Neurol Sci. 2013;331:136–144. [DOI] [PubMed] [Google Scholar]
- 36. Sano Y, Shimizu F, Abe M, et al. Establishment of a new conditionally immortalized human brain microvascular endothelial cell line retaining an in vivo blood‐brain barrier function. J Cell Physiol. 2010;225:519–528. [DOI] [PubMed] [Google Scholar]
- 37. Mossu A, Rosito M, Khire T, et al. A silicon nanomembrane platform for the visualization of immune cell trafficking across the human blood‐brain barrier under flow. J Cereb Blood Flow Metab. 39(3):395‐410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stebbins MJ, Gastfriend BD, Canfield SG, et al. Human pluripotent stem cell‐derived brain pericyte‐like cells induce blood‐brain barrier properties. Science advances. 2019;5:eaau7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Engen SA, Valen Rukke H, Becattini S, et al. The oral commensal Streptococcus mitis shows a mixed memory Th cell signature that is similar to and cross‐reactive with Streptococcus pneumoniae. PLoS One. 2014;9:e104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sallusto F, Schaerli P, Loetscher P, et al. Rapid and coordinated switch in chemokine receptor expression during dendritic cell maturation. Eur J Immunol. 1998;28:2760–2769. [DOI] [PubMed] [Google Scholar]
- 41. Coisne C, Lyck R, Engelhardt B. Live cell imaging techniques to study T cell trafficking across the blood‐brain barrier in vitro and in vivo. Fluids Barriers CNS. 2013;10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mizee MR, Nijland PG, van der Pol SM, et al. Astrocyte‐derived retinoic acid: a novel regulator of blood‐brain barrier function in multiple sclerosis. Acta Neuropathol. 2014;128:691–703. [DOI] [PubMed] [Google Scholar]
- 43. Canfield SG, Stebbins MJ, Morales BS, et al. An isogenic blood‐brain barrier model comprising brain endothelial cells, astrocytes, and neurons derived from human induced pluripotent stem cells. J Neurochem. 2017;140:874–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Herich S, Schneider‐Hohendorf T, Rohlmann A, et al. Human CCR5high effector memory cells perform CNS parenchymal immune surveillance via GZMK‐mediated transendothelial diapedesis. Brain. 2019;142(11):3411–3427. [DOI] [PubMed] [Google Scholar]
- 45. Alter A, Duddy M, Hebert S, et al. Determinants of human B cell migration across brain endothelial cells. J Immunol. 2003;170:4497–4505. [DOI] [PubMed] [Google Scholar]
- 46. Chang SF, Chen LJ, Lee PL, Lee DY, Chien S, Chiu JJ. Different modes of endothelial‐smooth muscle cell interaction elicit differential beta‐catenin phosphorylations and endothelial functions. Proc Natl Acad Sci USA. 2014;111:1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lopes Pinheiro MA, Kooij G, Mizee MR, et al. Immune cell trafficking across the barriers of the central nervous system in multiple sclerosis and stroke. Biochem Biophys Acta. 2016;1862:461–471. [DOI] [PubMed] [Google Scholar]
- 48. Engelhardt B, Carare RO, Bechmann I, Flugel A, Laman JD, Weller RO. Vascular, glial, and lymphatic immune gateways of the central nervous system. Acta Neuropathol. 2016;132:317–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bernas MJ, Cardoso FL, Daley SK, et al. Establishment of primary cultures of human brain microvascular endothelial cells to provide an in vitro cellular model of the blood‐brain barrier. Nat Protoc. 2010;5:1265–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eigenmann DE, Xue G, Kim KS, Moses AV, Hamburger M, Oufir M. Comparative study of four immortalized human brain capillary endothelial cell lines, hCMEC/D3, hBMEC, TY10, and BB19, and optimization of culture conditions, for an in vitro blood‐brain barrier model for drug permeability studies. Fluids Barriers CNS. 2013;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boyer‐Di Ponio J, El‐Ayoubi F, Glacial F, et al. Instruction of circulating endothelial progenitors in vitro towards specialized blood‐brain barrier and arterial phenotypes. PLoS One. 2014;9:e84179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Clark PA, Al‐Ahmad AJ, Qian T, et al. Analysis of Cancer‐Targeting Alkylphosphocholine Analogue Permeability Characteristics Using a Human Induced Pluripotent Stem Cell Blood‐Brain Barrier Model. Mol Pharm. 2016;13:3341–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hollmann EK, Bailey AK, Potharazu AV, Neely MD, Bowman AB, Lippmann ES. Accelerated differentiation of human induced pluripotent stem cells to blood‐brain barrier endothelial cells. Fluids Barriers CNS. 2017;14:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ribecco‐Lutkiewicz M, Sodja C, Haukenfrers J, et al. A novel human induced pluripotent stem cell blood‐brain barrier model: Applicability to study antibody‐triggered receptor‐mediated transcytosis. Sci Rep. 2018;8:1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Neal EH, Marinelli NA, Shi Y, et al. A Simplified, Fully Defined Differentiation Scheme for Producing Blood‐Brain Barrier Endothelial Cells from Human iPSCs. Stem cell reports. 2019;12:1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kerfoot SM, Norman MU, Lapointe BM, Bonder CS, Zbytnuik L, Kubes P. Reevaluation of P‐selectin and alpha 4 integrin as targets for the treatment of experimental autoimmune encephalomyelitis. J Immunol. 2006;176:6225–6234. [DOI] [PubMed] [Google Scholar]
- 57. Schlager C, Korner H, Krueger M, et al. Effector T‐cell trafficking between the leptomeninges and the cerebrospinal fluid. Nature. 2016;530:349–353. [DOI] [PubMed] [Google Scholar]
- 58. Steiner O, Coisne C, Cecchelli R, et al. Differential roles for endothelial ICAM‐1, ICAM‐2, and VCAM‐1 in shear‐resistant T cell arrest, polarization, and directed crawling on blood‐brain barrier endothelium. J Immunol. 2010;185:4846–4855. [DOI] [PubMed] [Google Scholar]
- 59. Alvarez JI, Cayrol R, Prat A. Disruption of central nervous system barriers in multiple sclerosis. Biochem Biophys Acta. 2011;1812:252–264. [DOI] [PubMed] [Google Scholar]
- 60. Luo J, Paranya G, Bischoff J. Noninflammatory expression of E‐selectin is regulated by cell growth. Blood. 1999;93:3785–3791. [PubMed] [Google Scholar]
- 61. Castro Dias M, Coisne C, Lazarevic I, et al. Claudin‐3‐deficient C57BL/6J mice display intact brain barriers. Sci Rep. 2019;9:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wimmer I, Tietz S, Nishihara H, et al. PECAM‐1 stabilizes blood‐brain barrier integrity and favors paracellular T‐cell diapedesis across the blood‐brain barrier during neuroinflammation. Front Immunol. 2019;10:711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Steiner O, Coisne C, Engelhardt B, Lyck R. Comparison of immortalized bEnd5 and primary mouse brain microvascular endothelial cells as in vitro blood‐brain barrier models for the study of T cell extravasation. J Cereb Blood Flow Metab. 2011;31:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hirase T, Staddon JM, Saitou M, et al. Occludin as a possible determinant of tight junction permeability in endothelial cells. J Cell Sci. 1997;110(Pt 14):1603–1613. [DOI] [PubMed] [Google Scholar]
- 65. Liebner S, Corada M, Bangsow T, et al. Wnt/beta‐catenin signaling controls development of the blood‐brain barrier. J Cell Biol. 2008;183:409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bonney S, Harrison‐Uy S, Mishra S, et al. Diverse Functions of Retinoic Acid in Brain Vascular Development. J Neurosci. 2016;36:7786–7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vazao H, Rosa S, Barata T, et al. High‐throughput identification of small molecules that affect human embryonic vascular development. Proc Natl Acad Sci USA. 2017;114:E3022–E3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Halaidych OV, Freund C, van den Hil F, et al. Inflammatory responses and barrier function of endothelial cells derived from human induced pluripotent stem cells. Stem Cell Rep. 2018;10:1642–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Larochelle C, Cayrol R, Kebir H, et al. Melanoma cell adhesion molecule identifies encephalitogenic T lymphocytes and promotes their recruitment to the central nervous system. Brain. 2012;135:2906–2924. [DOI] [PubMed] [Google Scholar]
- 70. Ifergan I, Kebir H, Terouz S, et al. Role of Ninjurin‐1 in the migration of myeloid cells to central nervous system inflammatory lesions. Ann Neurol. 2011;70:751–763. [DOI] [PubMed] [Google Scholar]
- 71. Odoardi F, Sie C, Streyl K, et al. T cells become licensed in the lung to enter the central nervous system. Nature. 2012;488:675–679. [DOI] [PubMed] [Google Scholar]
- 72. Alvarez JI, Saint‐Laurent O, Godschalk A, et al. Focal disturbances in the blood‐brain barrier are associated with formation of neuroinflammatory lesions. Neurobiol Dis. 2015;74:14–24. [DOI] [PubMed] [Google Scholar]
- 73. Lopes Pinheiro MA, Kamermans A, Garcia‐Vallejo JJ, et al. Internalization and presentation of myelin antigens by the brain endothelium guides antigen‐specific T cell migration. eLife. 2016;5:e13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8
Fig S9
Table S1 and S2
Table S3 and S4
Table S5
Video S1


