Abstract
Spinal muscular atrophy (SMA) is 1 of the leading causes of infant mortality. SMA is mostly caused by low levels of Survival Motor Neuron (SMN) protein due to deletion of or mutation in the SMN1 gene. Its nearly identical copy, SMN2, fails to compensate for the loss of SMN1 due to predominant skipping of exon 7. Correction of SMN2 exon 7 splicing by an antisense oligonucleotide (ASO), nusinersen (Spinraza™), that targets the intronic splicing silencer N1 (ISS-N1) became the first approved therapy for SMA. Restoration of SMN levels using gene therapy was the next. Very recently, an orally deliverable small molecule, risdiplam (Evrysdi™), became the third approved therapy for SMA. Here we discuss how these therapies are positioned to meet the needs of the broad phenotypic spectrum of SMA patients.
Keywords: SMA, splicing, SMN, ISS-N1, antisense oligonucleotide, risdiplam, Spinraza, nusinersen, Branaplam, Evrysdi, Zolgensma
Introduction
Spinal muscular atrophy (SMA) is the leading genetic cause of infant mortality affecting 1 in every ~10,000 live births.1,2 Low levels of the Survival Motor Neuron (SMN) protein due to deletion of or mutation in the SMN1 gene is the primary cause of SMA.3 A nearly identical copy of SMN1 universally present in humans, called SMN2, cannot compensate for the loss of SMN1 since SMN2 exon 7 is predominantly skipped. Skipping of exon 7 leads to production of a truncated unstable protein, SMNΔ7.4 SMA has a broad disease spectrum that is categorized into 5 types: 0, 1, 2, 3, and 4.5 Type 0 is the most severe, in which patients die before birth.6 Patients suffering from type 1 SMA (also called Werdnig-Hoffmann disease) need ventilatory support, cannot sit or walk and succumb to death before their 2nd birthday.7 The onset of type 2 SMA (also called Dubowitz disease) occurs before 18 months of age; here patients cannot walk but can sit.7 Type 3 SMA (also called Kugelberg-Welander disease) is manifested after 18 months of age; and patients can sit, walk, and survive into adulthood.7,8 Type 4 SMA is characterized by mild symptoms, it manifests during early adulthood, and patients are expected to have a normal lifespan.6,9 SMN is a multifunctional protein involved in RNA metabolism, DNA repair, cytoskeletal dynamics, and macromolecular trafficking.10-13 Low levels of SMN were shown to affect multiple signaling cascades, including STAT5, RhoA/ROCK, AKT/CREB and JNK pathways.14-18 SMN can bind RNA, preferentially interacting with GA-rich motifs.19,20 A recently published study indicated that the protein might play a critical role in the translation of a specific subset of mRNAs linked to SMA pathogenesis by “priming” ribosomes in a tissue-specific manner.21 A much broader role of the SMN genes could be envisioned based on a vast repertoire of transcripts generated from the SMN loci. These transcripts include alternatively spliced mRNA isoforms, non-coding antisense RNAs, and circular RNAs.22-31 Loss of SMN1 affects all tissues, including skeletal muscle, central, peripheral and autonomic nervous system, heart, liver, lung, kidney, pancreas, spleen, ovary, and testis.32-52 Hence, an ideal therapy for SMA must “target/remedy” the body-wide defects caused by the loss of SMN1. The severity of SMA correlates inversely with the SMN2 copy number: the higher the copy number, the lower the severity.53-56 Several factors, including Plastin (PLS3), Neuritin 1 (NRN1), Neurocalcin delta (NCALD), TIA1 cytotoxic granule associated RNA binding protein (TIA1), Ubiquitin specific peptidase 9 X-linked (USP9X), Ubiquitin like modifier activating enzyme 1 (UBA1), Stathmin-1 (STMN1), Myostatin (MSTN), ZPR1 zinc finger protein (ZPR1), and Senataxin (SETX), have been suggested to modify SMA severity.57-69 Due to broad differences in the age of the SMA onset and the diversity of SMA phenotypes, developing an ideal therapy for the disease remains a challenging task.
Considering SMN2 is universally present in SMA patients, correction of SMN2 exon 7 splicing remains one of the most promising avenues for the treatment of the disease.70 A critical C-to-T mutation at the 6th position (C6U substitution in RNA) of exon 7 is associated with the skipping of SMN2 exon 7.71,72 In general, skipping of exons is triggered by suboptimal splice sites defined by a combinatorial control of splicing cis-elements and transacting factors that recognize them.73 Being close to the 3′ splice site (3′ss), C6U substitution is proposed to weaken the 3′ss of SMN2 exon 7.74 Various other mechanisms including abrogation of an enhancer, creation of a silencer and strengthening of an extended inhibitory context (Exinct) have been put forward to explain the C6U substitution-induced skipping of exon 7.75-78 A breakthrough in our understanding of exon 7 splicing regulation came from a study performing in vivo selection of the entire exon that, among other important observations, confirmed that the 5′ss of exon 7 was suboptimal.79 Of note, in vivo selection of the entire exon 7 was the first experiment of its kind: here the relative significance of every exonic nucleotide was functionally analyzed in a single experiment.80 In addition, the unbiased method of in vivo selection revealed that not only the linear sequence(s) but also putative structural motifs were critical for inclusion of SMN exon 7.81,82 Significantly, findings of in vivo selection of the entire exon 7 of SMN1 turned out to be useful for validating the outcome of a machine learning program that analyzed the pathogenicity of all known point mutations within the human genome.83 As discussed below, two splicing modulating compounds currently approved for SMA therapy as well as an additional small molecule are currently in clinical trials are linked to the strengthening of the 5′ss of SMN2 exon 7.
Context of the Suboptimal 5′ss of SMN2 Exon 7
Most human introns, including all introns in the SMN genes, belong to the U2-type. The 5′ss of U2-type introns is defined by a total of eleven nucleotides, specifically the last 3 exonic residues and the first 8 intronic residues.84 An RNA:RNA duplex (U1:5′ss duplex) formed between these 11 nucleotides and the 5′-end of U1 snRNA, a component of U1 snRNP, sets the stage for exon definition and intron removal.84 With very few exceptions, a GU dinucleotide at the first 2 positions of the U2-type introns is required for the exon definition.85 Additional splicing cis-elements come into play when the size of the U1:5′ss duplex is less than 6 base pairs and/or the 5′ss is sequestered in a RNA structure.86,87 The finding that the 5′ss of exon 7 is suboptimal as revealed by the in vivo selection paved the way for the discovery of a number of inhibitory cis-elements located in the 5′ss vicinity.88 These include the intronic splicing silencer N1 (ISS-N1), the terminal stem-loop 2 (TSL2), the GC-rich sequence (GCRS) overlapping ISS-N1, and the internal stem formed by a long-distance interaction 1 (ISTL1) (Figure 1).89-94 Of note, as per recent estimates, more than 30 cis-elements and an even higher number of transacting factors have been implicated in the regulation of SMN exon 7 splicing.95,96 However, subsequent studies showed that some of these splicing factors were dispensable. For instance, SF2/ASF was initially thought to be critical for exon 7 inclusion.75 Yet, in cells lacking SF2/ASF no effect on exon 7 splicing was observed.76 Similarly, Tra2-β1, a positive regulator of exon 7 splicing, turned out to be dispensable for inclusion of this exon in a Tra2-β1-deficient mouse model.97,98 These observations are not entirely surprising given the redundancy and cross-regulation of splicing factors as, for example, observed for PTB, CELF2, and hnRNP C.99-101
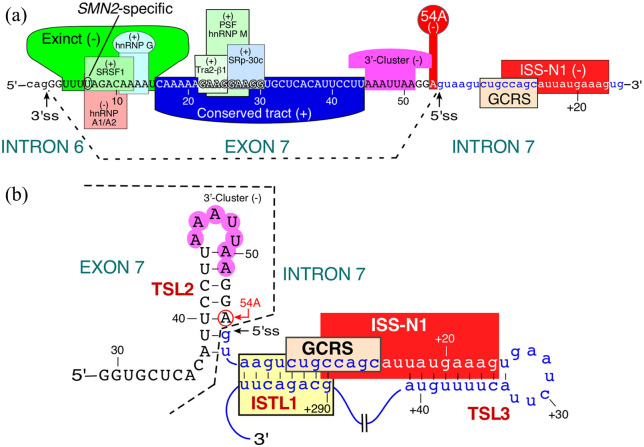
Figure 1.
Diagrammatic representation of cis-elements and transacting factors that regulate SMN2 exon 7 splicing. (a) Relative positioning of cis-elements within exon 7 and downstream intron 7 of SMN. Cis-elements and transacting factors that bind them are highlighted in different colors. Note, the diagram presented here is not inclusive of all reported exon 7 splicing regulators. Please refer to recent reviews for more comprehensive information.95,96 Positive and negative regulators of exon 7 splicing are indicated by (+) and (−), respectively. Neutral numbering of nucleotides starts from the first position of exon 7. Positive numbering of nucleotides starts from the first position of intron 7. Exonic and intronic sequences are shown in upper- and lower-case letters, respectively. SMN2-specific C6U substitution is marked. Exinct, the Conserved tract and the 3′-Cluster were identified by in vivo selection of the entire exon 7. In vivo selection of the entire exon also revealed the strong negative effect of an “A” residue at the 54th position (54A) of exon 7 and (b) structural context of the 5′ss of SMN exon 7. Numbering is the same as described in panel A. Only a portion of exon 7 and intron 7 is shown. Cis-elements that promote exon 7 skipping are highlighted in colors. Abbreviations: 3′ss, 3′ splice site; 5′ss, 3′ splice site; Exinct, extended inhibitory context; GCRS, GC-rich sequence; ISS-N1, intronic splicing silencer; ISTL1, an internal stem (inhibitory RNA structure) formed by long-distance interaction; TSL2, terminal stem-loop structure 2; TSL3, terminal stem-loop structure 3.
The discovery of the 15-nucleotide long ISS-N1 propelled the development of an antisense oligonucleotide (ASO)-directed therapy for SMA.89,102,103 Based on its strong inhibitory effect, ISS-N1 was dubbed as a master regulator of both splicing checkpoint and exon definition.104 Nusinersen (Spinraza™), the first FDA-approved drug for SMA, is an ISS-N1-targeting ASO that is intrathecally delivered for the treatment of the disease.105,106 Methods and mechanisms associated with the splicing correction by an ISS-N1-targeting ASO are reviewed elsewhere.107-109 Collaborative studies conducted in the Krainer lab at Cold Spring Harbor Laboratories, New York and by Ionis Pharmaceuticals, Carlsbad, California played a pivotal role in the therapeutic development of nusinersen.110 Several recent reports describe the efficacy of nusinersen in SMA patients.111-113 Of note, similar to ISS-N1, GCRS and ISS-N2 are additional targets that could be potentially exploited for correction of SMN2 exon 7 splicing by abrogating the inhibitory context at the 5′ss of exon 7.91,93,114,115 Indeed, in vivo studies employing ASOs targeting GCRS and ISS-N2 have shown therapeutic benefits in mouse models of SMA.116,117 Zolgensma®, an adeno-associated virus 9 (AAV9) based gene delivery, became the second FDA-approved therapy for SMA.118 The success of gene therapy was enabled by pre-clinical and clinical studies conducted by Kaspar and colleagues at Nationwide Children’s Hospital, Columbus, Ohio.119 Unlike nusinersen that relies on the endogenous SMN2 transcripts for the production of SMN, gene therapy produces SMN from exogenously delivered DNA coding for SMN1. Hence, risks of the generation of autoantibodies against SMN due to overexpression of this protein as a consequence of gene therapy could not be ruled out. Of note, a recent study has found a correlation between autoantibodies against SMN and systemic sclerosis.120 Both nusinersen and gene therapy have the limitations of an invasive administration process and having poor body-wide delivery/distribution.121 The recent approval of risdiplam (Evrysdi™), an orally deliverable small molecule, addresses these concerns.122-124 Here we review the mechanism of action of risdiplam, its target specificity, and potential off-target effects. We also discuss how available SMA drugs would potentially complement each other for a better treatment of the disease. Other SMA therapies currently in preclinical and clinical studies/trials have been described elsewhere.5,125,126
Discovery of Risdiplam as a Therapeutic Candidate
A joint endeavor by PTC-Roche (PTC Therapeutics, South Plainfield, New Jersey and Hoffmann-La Roche, Basel, Switzerland) to identify an orally available molecule for the treatment of SMA began about a decade ago. Investigators at these companies screened a library of small molecules and reported three orally deliverable compounds, namely SMN-C1 (isocoumarin), SMN-C2 (coumarin), and SMN-C3 (pyrido-pyrimidinone derivative); each promoted exon 7 inclusion from SMN2 minigene expressed in HEK293H human embryonic kidney cell line127 (Figure 2). These compounds also promoted exon 7 inclusion in mRNAs generated from the endogenous SMN2 and increased SMN levels in SMA patient fibroblasts and patient-derived induced pluripotent stem cells (iPSCs). The results of RNA-seq conducted using type 1 SMA fibroblasts treated with 500 nM of SMN-C3 showed high target specificity of the compound for SMN2 exon 7 splicing correction, although limited off-target effects were also captured. For instance, expression of important genes including DNA polymerase N (POLN) and PAP-associated domain containing 4 protein (PAPD4) were significantly altered.127 Also, SMN-C3 promoted inclusion of several exons in mRNAs generated from Pyridoxal-dependent Decarboxylase Domain Containing 1 (PDXDC1), a gene associated with an increased risk for brain cancer.127,128 In vivo studies employing a mild SMA mouse model (allele C model) as well as a severe SMA mouse model (SMA Δ7 model) confirmed splicing correction of SMN2 exon 7 and upregulation of SMN upon oral or intraperitoneal (IP) administration of SMN-C3.127 IP administration of SMN-C3 conferred substantial gain of lifespan and improvement of neuromuscular junction (NMJ) phenotype of Δ7 mice. However, based on their potential genotoxicity, phototoxicity, and/or chemical instability in plasma or aqueous buffers, none of the above-mentioned molecules advanced on to the human clinical trials.
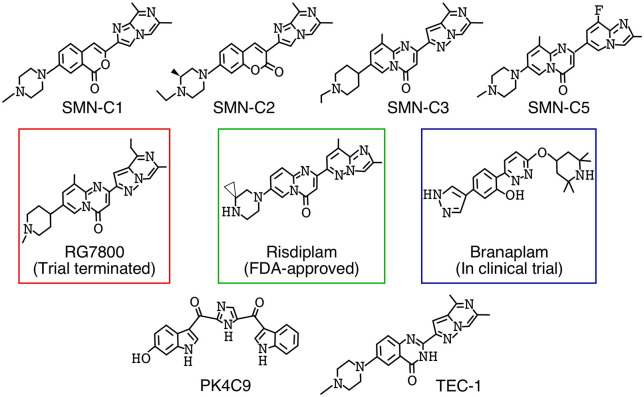
Figure 2.
Structure of orally available small molecules used in pre-clinical and clinical studies for the treatment of SMA. SMN-C1, SMNC-2, and SMN-C3 were the first set of compounds reported by PTC-Roche (PTC Therapeutics, South Plainfield, New Jersey and Hoffmann-La Roche, Basel, Switzerland) to correct SMN2 exon 7 splicing with high specificity. Most mechanistic studies have been done using SMN-C3 and SMN-C5. Clinical trial of RG-7800 by PTC-Roche was terminated due to its toxicity in cynomolgus monkeys. Risdiplam has gone through multiple clinical trials by PTC-Roche and has recently been approved by FDA. Branaplam is in clinical trial by Novartis Pharmaceuticals. PK4C9 and TEC-1 are the newly reported compounds to show specific splicing correction of SMN2 exon 7.
With the realization that the compounds present in the existing library have a high potential for direct clinical applications, PTC-Roche began the process of designing improved versions of their active small molecules. As a result, three novel pyrido-pyrimidinone derivatives were “created,” namely, compounds 3, 4, and 5; all displayed negative genotoxicity in the universally used Ames assay and showed very high therapeutic efficacy in the SMAΔ7 mouse model.129 Compound 3, also known as RG7800, was selected for the subsequent human clinical trial that began in 2014. In parallel to the human clinical trial, RG7800 was also evaluated in cynomolgus monkeys for chronic toxicity. Due to nonreversible adverse effects on monkey retina, RG7800 clinical trial was put on hold.129 Uncertain about the success of RG7800, PTC-Roche chose another small molecule, risdiplam (also known as RG7916), an improved version of RG7800, for clinical trials.123 Risdiplam selection was based on its superior in vivo efficacy in the SMAΔ7 mouse model as well as its reduced off-target effects tested in SMA patient fibroblasts as compared to RG7800.99 Risdiplam went through a relatively rapid clinical development, from the phase 1 clinical trial (January of 2016) to its FDA approval (August of 2020).130 The fast approval of risdiplam was possible in part due to the well-defined parameters of therapeutic efficacy established during the preclinical and clinical studies of nusinersen and gene therapy.
Mechanism of Action of Risdiplam
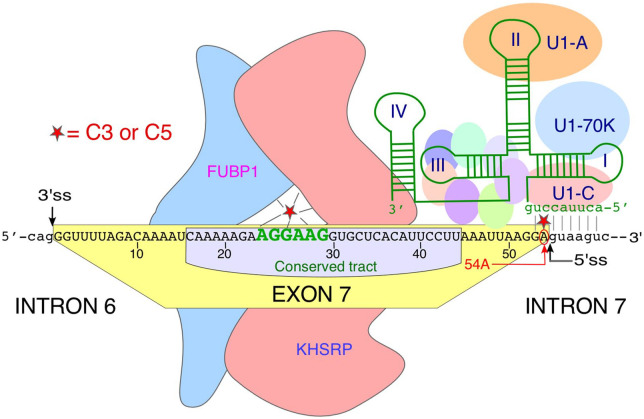
Currently, there is no consensus on the mechanism by which risdiplam (molecular mass 401.46 Da) promotes SMN2 exon 7 inclusion with high specificity. A study led by investigators at California Institute for Biomedical Research (CIBR) showed that SMN-C3, one of the analogs of risdiplam, interacts with an AG-rich motif, AGGAAG, located in the middle of exon 7 (Figure 3).131 Authors employed a series of in vitro and in vivo techniques to demonstrate a high specificity of direct interactions between SMN-C3 and this AG-rich motif. Further, binding of SMN-C3 to this AG-rich motif was proposed to recruit stimulatory splicing factors Far Upstream Element Binding Protein 1 (FUBP1) and its homolog KH-type Splicing Regulatory Protein (KHSRP) (Figure 3).131 Supporting this hypothesis, depletion of FUBP1/KHSRP diminished the effect of SMN-C3 on SMN2 exon 7 splicing, particularly at low nanomolar concentrations of SMN-C3.131 A different study led by investigators at Hoffmann-La Roche suggested that the interaction of SMN-C class of compounds with the AG-rich motif displaces hnRNP G.132 Previous studies have implicated the role of hnRNP G in promoting of SMN2 exon 7 inclusion, although it has been also argued that the stimulatory effect of hnRNP G on exon 7 splicing is mediated through Tra2-β1, which in turn interacts with the purine-rich motif located in the middle of exon 7.133,134 It is likely that the displacement of hnRNP G is accompanied by the recruitment of stimulatory factors, including FUBP1/ KHSRP as proposed by investigators at CIBR.
Figure 3.
Potential mechanism of action of risdiplam. Risdiplam analogs SMN-C3 and -C5 are depicted as red stars. SMN-C3 has been shown to interact with an AG-rich motif (shown in green letters) located in the middle of exon 7.131 SMN-C3 has been proposed to recruit splicing factors FUBP1 and KHSRP.107 SMN-C5 has been proposed to promote recruitment of U1 snRNP by directly binding to 54A at the 5′ss of exon 7.135 Interaction of U1 snRNP with the 5′ss of exon 7 has been depicted. Drawing is not to scale.
In addition to the interaction with the AG-rich motif, SMN-C class of compounds have been shown to interact with the 5′ss of exon 7, particularly with the adenosine residue at the last exonic position (54A) (Figure 3).132 Of note, the inhibitory effect of 54A was first uncovered by in vivo selection of the entire exon 7.79 Consistently, replacement of 54A with 54G (A54G substitution) fully restores SMN2 exon 7 inclusion even when the Tra2-β1-binding site in exon 7 is destroyed.79 Importantly, 54A also strengthens a stem-loop structure (TSL2) that sequesters the 5′ss of exon 7 (Figure 1). When it comes to the 5′ss recognition, 54A creates a bulge (a mismatch base pair) in the duplex formed between the U1 snRNA and the 5′ss of exon 7.90 Strengthening of the U1:5′ss duplex by a compensatory mutation within U1 snRNA has been shown to have the similar stimulatory effect on SMN2 exon 7 inclusion as the one observed with the A54G substitution.90
A recent study by Allain and colleagues employing NMR confirmed the interaction between SMN-C5 and 54A in the context of the U1:5′ss duplex (Figure 3).135 The authors proposed that SMN-C5 stabilizes the U1:5′ss duplex by “5′ss bulge repair,” restoring the accessibility of the U1-C zinc finger for the interaction with the minor groove of the duplex.135 While stabilization of the U1:5′ss duplex by SMN-C5 seems to be sufficient to promote SMN2 exon 7 inclusion, the authors did not rule out the role of additional factors recruited by SMN-C5 to the AG-rich motif. A caveat in the SMN-C5-induced U1:5′ss duplex model is its inability to explain why lower concentrations of SMN-C series of compounds were ineffective in the promotion of SMN2 exon 7 inclusion upon depletion of splicing factors FUBP1/KHSRP.131
All mechanistic studies proposed thus far have been performed employing risdiplam analogs but not risdiplam itself. Some of the disparities in the proposed mechanisms of action could lie in the methods employed and risdiplam analogs used. Given the structural differences between risdiplam and its analogs (Figure 2), it is not a matter of fact that the mechanism proposed for a risdiplam analog will also hold true for risdiplam itself. Additional studies including analysis of exons associated with the off-target effects of risdiplam would be needed to fully understand its mechanism of action.
Off-Target Effects of Risdiplam
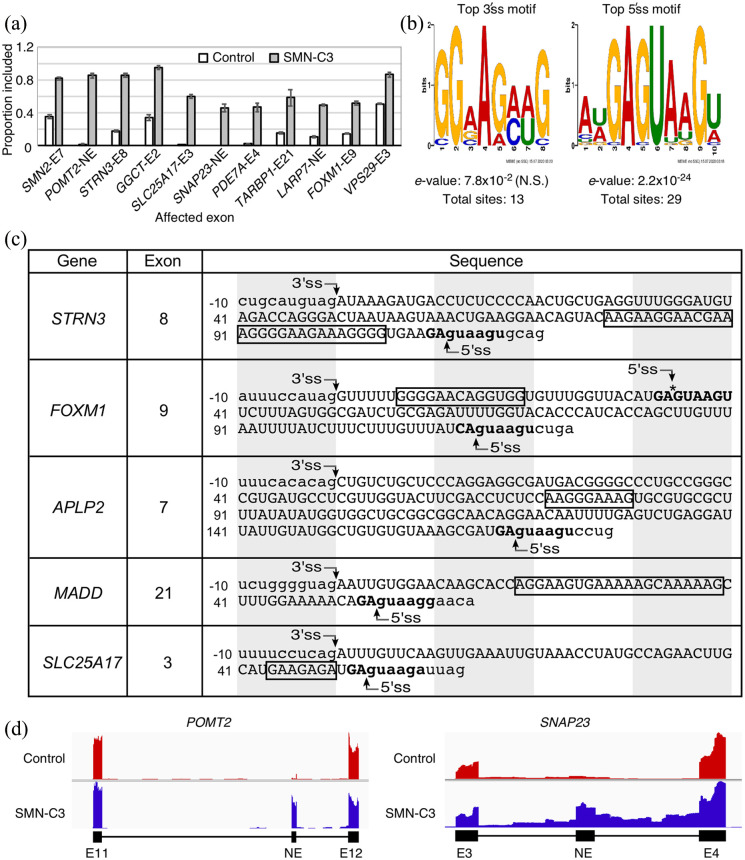
The first RNA-seq performed on transcripts isolated from SMN-C3-treated SMA type I fibroblasts provides insight into the nature of off-target effects of C-series of small molecules that are analogs of risdiplam.127 Analysis of this RNA-seq revealed that SMN-C3 treatment altered splicing of 42 exons, 6 of which underwent a change of greater than 40%. The effect on splicing of the top 10 candidate exons including SMN2 exon 7 is shown in Figure 4a. Analysis of the sequences surrounding the 3′ss of the affected exons revealed a slight, but not significant enrichment in the AG-rich motif (Figure 4b). However, similar motifs were not enriched in total exonic sequences or in the vicinity of the 5′ss (Figure 4b). SMN-C3-affected “off-target” exons had a strong enrichment for a GA dinucleotide at their two last positions followed by a consensus GUAAGU 5′ss sequence (Figure 4b and c). Interestingly, SMN-C3 also triggered the inclusion of previously unannotated exons and in at least one case promoted intron retention (Figure 4d). For instance, inclusion of an unannotated exon positioned between exons 3 and 4 of the SNAP23 gene was accompanied by a significant retention of the downstream intron (Figure 4d). As compared to its precursors, low concentrations of risdiplam showed similar off-target effects on pre-mRNA splicing in a cell culture model.123 However, it is predicted to exhibit superior target specificity and stability in vivo due to novel modifications that were introduced to prevent its conversion into potentially harmful active metabolite(s).123 It should be noted that at high concentrations, risdiplam did produce off-target effect on splicing of several genes, including STRN3, FOXM1, APLP2, MADD, and SLC25A17 (Figure 4).123 The off-target effect of risdiplam on splicing of exons of several genes could be attributed to the similarity of the 5′ss context and sequence motifs present within the affected “off-target” exons (Figure 4).
Figure 4.
Off-target effects of risdiplam or its analog SMN-C3. (a) Splicing pattern of SMN2 exon 7 and ten other splicing events affected by SMN-C3 treatment as reported by Naryshkin and coworkers.127 Y axis indicates the proportion of total spliced transcript that has the exon in question included. X axis labels indicate the host gene and exon number of the target exons. NE: novel (unannotated) exon, (b) top enriched sequence motifs near the 3′ and 5′ss of the exons, splicing of which was changed by SMN-C3. Letter height in each motif corresponds to nucleotide enrichment at that position, (c) the sequences of 5“off-target” exons, splicing of which was affected by risdiplam, as reported by Ratni and coworkers.123 Numbering is given relative to the first position of each exon. Uppercase letters represent exonic sequences, lowercase letters represent intronic sequences. The longest AG-rich motif in each exon is boxed. The last two exonic nucleotides and the first six intronic nucleotides of the 5′ss are shown in bold. Each shaded/clear area “cover” ten consecutive nucleotides. An additional 5′ss within exon 9 of FOXM1 is indicated with an asterisk, and (d) genomic overview of two examples of splicing events induced by SMN-C3 treatment. POMT2 (top panel) contains a novel, unannotated exon located in the region between exons 11 and 12. Inclusion of this unannotated exon is caused by SMN-C3 treatment, as shown by the increased read depth. SNAP23 (bottom panel) has a novel exon (between exons 3 and 4) that undergoes inclusion. This is coupled with intron retention, as indicated by increased read depth in the flanking introns.
In Vivo Efficacy of Risdiplam in Mouse Models
Risdiplam showed enhancement in expression of SMN in brain and quadriceps muscle upon oral administration in a mild SMA mouse model (allele C model).123 Intraperitoneal (IP) mode of delivery was used to monitor the efficacy of risdiplam in severe (SMAΔ7) mouse model. IP administration of risdiplam at a concentration as low as 1 mg/kg of body weight produced a robust enhancement in SMN levels in brain and quadriceps muscle of these mice.123 Also, risdiplam-treated SMAΔ7 mice showed a dose-dependent improvement of NMJ phenotype and an increase in the number of motor neurons and the size of the extensor digitorum longus (EDL) muscles. Higher IP doses of risdiplam (10 mg/kg of body weight) provided one of the best life expectancy gains reported in the literature.123 For instance, more than 70% of SMAΔ7 mice survived beyond seven months upon treatment with risdiplam at 10 mg/kg of body weight. In further studies in several SMA mouse models as well as in rats and non-human primates, risdiplam displayed excellent pharmacokinetic and pharmacodynamic properties, such as body-wide distribution and stable plasma levels over extended dosing periods.124 These results were sufficient to launch clinical trials of risdiplam for its evaluation in SMA patients.
Clinical Trials and FDA Approval of Risdiplam
Several clinical trials of risdiplam have been performed to evaluate the safety, tolerability, and efficacy of the drug in both healthy and SMA patients. Two of these clinical trials, first in the infantile-onset (NCT02913482) and second in the later-onset SMA patients (NCT02908685) were significant for the approval of risdiplam.130 The clinical trial for infantile-onset SMA was an open-label study in which 21 patients, whose average age was ~6.7 months, participated.130 About 41% patients showed ability to independently sit after 12-month treatment. Also, the patients showed more than 81% survival without permanent ventilation after 23 or more months of treatment. These results were considered as a significant improvement over the untreated patients in a similar category. The clinical trial with the later-onset SMA patients was randomized and placebo-controlled in which 180 SMA patients aged from 2 to 25 years participated.130 Risdiplam-treated patients performed significantly better in motor function tests than untreated patients. On August 7, 2020 FDA granted approval of risdiplam (Evrysdi™) under the fast-track designation and rare pediatric disease priority review process.130
Side Effects of Risdiplam
The most common side effects in clinical trials of risdiplam were fever, rash, ulcers of the mouth area, joint pain (arthralgia), diarrhea, and urinary tract infections.130 The infantile-onset population receiving risdiplam had additional side effects including upper respiratory tract infection, pneumonia, vomiting, and constipation.130 Currently, it is not known if the side effects are directly linked to the off-target effects of risdiplam.
Conclusion
Recent approval of risdiplam, an orally deliverable small molecule, is a major advancement for the treatment of SMA. The noninvasive mode of administration coupled with body-wide distribution provide risdiplam with clear advantages over other approved therapies. Risdiplam availability is particularly great news for a group of SMA patients that might have tolerability and/or immune response concerns when it comes to nusinersen and gene therapy. Storage and shipping at ambient temperatures as well as its comparatively low cost are added benefits of risdiplam for its worldwide availability/distribution. In comparison to its “parent analogs,” risdiplam is predicted to exhibit reduced off-target effect in vivo, particularly at lower concentrations. Similar to other approved drugs for SMA, side effects encountered during the clinical trials of risdiplam remain a cause of concern. Future studies will reveal if the side effects associated with the frequent administration of risdiplam would pose a hurdle for its acceptance for the long-term treatment. In addition, risdiplam may not be useful for SMA patients that carry pathogenic mutations at the 5′ss of SMN exon 7.136 Activation of a cryptic 5′ss downstream of exon 7 by an engineered U1 snRNP could be an alternative therapeutic approach in this case.136,137 In fact, in vivo efficacy of the engineered U1 snRNP has been validated in a mouse model of SMA.138 Future studies aimed at the activation of the cryptic 5′ss downstream of exon 7 of the SMN genes by a small molecule will cater to the needs of a broader patient population.
One of the exciting aspects of risdiplam’s approval is the validation of the utility of a small molecule for targeted splicing correction as a promising therapy. Another orally available small molecule, branaplam, that modulates SMN2 exon 7 splicing with high specificity is about to conclude the phase 2 clinical trial (NCT02268552) conducted by Novartis Pharmaceuticals (Figure 2).139,140 Branaplam (synonyms: NVS-SM1 and LMI070) was identified by high-throughput screening of the Novartis compound library, followed by chemistry optimization.139,141 It was shown to modulate splicing, elevate levels of the full-length SMN protein and increase the survival of a severe SMA mouse model.141 Despite structural differences between branaplam and risdiplam, the proposed mode of action of branaplam appears to be similar to that of risdiplam.141 Both drugs stabilize the U1:5′ss duplex at the 5′ss of SMN2 exon 7.135,141 Two more small molecules, PK4C9 and TEC-1, have been recently reported to enhance SMN2 exon 7 inclusion with high specificity (Figure 2).142,143 TEC-1 has been found to be permeable to the central nervous system and confer therapeutic efficacy in a mouse model of SMA.142 While the mechanism of TEC-1 action has not yet been examined, PK4C9 has been shown to interact with a structural element, namely the tri-loop of TSL2.142,144 Incidentally, sequences encompassing the tri-loop of TSL2 has been found to overlap the “3′-cluster,” a negative element identified by in vivo selection of the entire exon 7 (Figure 1).79,81 These findings expand the number of potential targets that could be exploited for developing small molecules therapies for SMA. In addition, several orally available small molecules that work downstream of SMN or independent of SMN are currently undergoing pre-clinical and clinical studies.5,140,145 Diverse treatment options currently being exploited for SMA are commensurate with the varied needs of the broad spectrum of SMA patients.
To harness the full potential of available treatment options, it is likely that the combined therapies would become the desired approach for the treatment of SMA. Recent studies of the combined therapies (in mouse models of SMA) in which one of the components was an ISS-N1 targeting ASO have shown promising results.25,146-148 Now that risdiplam is approved, future studies will reveal if it could be combined with other drugs for a better therapeutic outcome. For example, risdiplam could be used together with an “SMN-independent” treatment(s) targeting muscle or neurological functional deficits observed in SMA to further alleviate symptoms of the disease. Using risdiplam together with other splicing-modulating drugs that work by complementary mechanisms, such as nusinersen, holds the promise to enhance the expression of full-length SMN, while maintaining minimum off-target effects on other splicing events due to lowering the treatment dose. With the precedence-setting success of risdiplam coupled with the discovery of additional small molecules capable of modulation of SMN2 exon 7 splicing with high specificity, prospects of small molecule therapeutics for the treatment of SMA appears to be on the fast track. In addition, these advancements should serve as a catalyst for the development of novel therapeutics for other genetic diseases amenable by splicing modulation.
Acknowledgments
Authors acknowledge members of the Singh lab for critical reading of the manuscript. While authors have attempted to include relevant research contributions to the field of SMA therapeutic targets and risdiplam, they regret of not being able to incorporate several related references due to the lack of space.
Footnotes
Author Contributions: RNS and NNS reviewed the literature and wrote and edited the manuscript. EWO reviewed the literature, performed the data analysis for Figure 4, and edited the manuscript. All authors reviewed and gave approval of the intellectual content of the final manuscript.
Declaration of conflicting interests:The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: ISS-N1 target (US patent # 7,838,657) mentioned in this review was discovered in the Singh lab at UMASS Medical School (Worcester, MA, USA). Inventors, including NNS, RNS, and UMASS Medical School, are currently benefiting from licensing of ISS-N1 target to IONIS Pharmaceuticals/Biogen, which is marketing Spinraza™ (Nusinersen), the FDA-approved drug, based on ISS-N1 target. RNS is co-founder of RNACorrect, Inc., an Iowa-based small business engaged in research and development.
Funding:The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a grant from the National Institutes of Health (R01 NS055925).
ORCID iD: Ravindra N Singh  https://orcid.org/0000-0001-5399-2662
https://orcid.org/0000-0001-5399-2662
References
- 1. Lally C, Jones C, Farwell W, Reyna SP, Cook SF, Flanders WD. Indirect estimation of the prevalence of spinal muscular atrophy Type I, II, and III in the United States. Orphanet J Rare Dis. 2017;12:175. doi: 10.1186/s13023-017-0724-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jha NN, Kim JK, Monani UR. Motor neuron biology and disease: a current perspective on infantile-onset spinal muscular atrophy. Future Neurol. 2018;13:161-172. doi: 10.2217/fnl-2018-0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lefebvre S, Bürglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155-165. [DOI] [PubMed] [Google Scholar]
- 4. Cho SC, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24:438-442. doi: 10.1101/gad.1884910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wirth B, Karakaya M, Kye MJ, Mendoza-Ferreira N. Twenty-five years of spinal muscular atrophy research: from phenotype to genotype to therapy, and what comes next. Annu Rev Genomics Hum Genet. 2020;21:231-261. doi: 10.1146/annurev-genom-102319-103602 [DOI] [PubMed] [Google Scholar]
- 6. Dubowitz V. Very severe spinal muscular atrophy (SMA type 0): an expanding clinical phenotype. Eur J Paediatr Neurol. 1999;3:49–51. [DOI] [PubMed] [Google Scholar]
- 7. Zerres K, Rudnik-Schöneborn S. Natural history in proximal spinal muscular atrophy. Clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch Neurol. 1995;52:518-523. doi: 10.1001/archneur.1995.00540290108025 [DOI] [PubMed] [Google Scholar]
- 8. Wijngaarde CA, Stam M, Otto LAM, et al. Population-based analysis of survival in spinal muscular atrophy. Neurology. 2020;94:e1634-e1644. doi: 10.1212/WNL.0000000000009248 [DOI] [PubMed] [Google Scholar]
- 9. Zerres K, Rudnik-Schoneborn S, Forkert R, Wirth B. Genetic basis of adult-onset spinal muscular atrophy. Lancet. 1995;346:1162. [DOI] [PubMed] [Google Scholar]
- 10. Singh RN, Howell MD, Ottesen EW, Singh NN. Diverse role of survival motor neuron protein. Biochim Biophys Acta. 2017;1860:299-315. doi: 10.1016/j.bbagrm.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang ET, Taliaferro JM, Lee JA, et al. Dysregulation of mRNA localization and translation in genetic disease. J Neurosci. 2016;36:11418-11426. doi: 10.1523/JNEUROSCI.2352-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gruss OJ, Meduri R, Schilling M, Fischer U. UsnRNP biogenesis: mechanisms and regulation. Chromosoma. 2017;126:577-593. doi: 10.1007/s00412-017-0637-6 [DOI] [PubMed] [Google Scholar]
- 13. Price PL, Morderer D, Rossoll W. RNP assembly defects in spinal muscular atrophy. Adv Neurobiol. 2018;20:143-171. doi: 10.1007/978-3-319-89689-2_6 [DOI] [PubMed] [Google Scholar]
- 14. Farooq F, Molina FA, Hadwen J, et al. Prolactin increases SMN expression and survival in a mouse model of severe spinal muscular atrophy via the STAT5 pathway. J Clin Invest. 2011;121:3042-3050. doi: 10.1172/JCI46276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Biondi O, Branchu J, Ben Salah A, et al. IGF-1R reduction triggers neuroprotective signaling pathways in spinal muscular atrophy mice. J Neurosci. 2015;35:12063-12079. doi: 10.1523/JNEUROSCI.0608-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahmad S, Bhatia K, Kannan A, Gangwani L. Molecular mechanisms of neurodegeneration in spinal muscular atrophy. J Exp Neurosci. 2016;10:39-49. doi: 10.4137/JEN.S33122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Genabai NK, Ahmad S, Zhang Z, Jiang X, Gabaldon CA, Gangwani L. Genetic inhibition of JNK3 ameliorates spinal muscular atrophy. Hum Mol Genet. 2015;24:6986-7004. doi: 10.1093/hmg/ddv401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang X, Kannan A, Gangwani L. ZPR1-dependent neurodegeneration is mediated by the JNK signaling pathway. J Exp Neurosci. 2019;13. doi: 10.1177/1179069519867915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bertrandy S, Burlet P, Clermont O, et al. The RNA-binding properties of SMN: deletion analysis of the zebrafish orthologue defines domains conserved in evolution. Hum Mol Genet. 1999;8:775-782. doi: 10.1093/hmg/8.5.775 [DOI] [PubMed] [Google Scholar]
- 20. Ottesen EW, Singh NN, Luo D, Singh RN. High-affinity RNA targets of the survival motor neuron protein reveal diverse preferences for sequence and structural motifs. Nucleic Acids Res. 2018;46:10983-11001. doi: 10.1093/nar/gky770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lauria F, Bernabò P, Tebaldi T, et al. SMN-primed ribosomes modulate the translation of transcripts related to spinal muscular atrophy. Nat Cell Biol. 2020;22:1239-1251. doi: 10.1038/s41556-020-00577-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singh NN, Seo J, Rahn SJ, Singh RN. A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes. PLoS One. 2012;7:e49595. doi: 10.1371/journal.pone.0049595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seo J, Singh NN, Ottesen EW, Lee BM, Singh RN. A novel human-specific splice isoform alters the critical C-terminus of survival motor neuron protein. Sci Rep. 2016;6:30778. doi: 10.1038/srep30778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seo J, Singh NN, Ottesen EW, Sivanesan S, Shishimorova M, Singh RN. Oxidative stress triggers body-wide skipping of multiple exons of the spinal muscular atrophy gene. PLoS One. 2016;11:e0154390. doi: 10.1371/journal.pone.0154390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. d’Ydewalle C, Ramos DM, Pyles NJ, et al. The antisense transcript SMN-AS1 regulates SMN expression and is a novel therapeutic target for spinal muscular atrophy. Neuron. 2017;93:66-79. doi: 10.1016/j.neuron.2016.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Woo CJ, Maier VK, Davey R, et al. Gene activation of SMN by selective disruption of lncRNA-mediated recruitment of PRC2 for the treatment of spinal muscular atrophy. Proc Natl Acad Sci U S A. 2017;114:E1509-E1518. doi: 10.1073/pnas.1616521114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ottesen EW, Seo J, Singh NN, Singh RN. A multilayered control of the human. Front Microbiol. 2017;8:2252. doi: 10.3389/fmicb.2017.02252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ottesen EW, Luo D, Seo J, Singh NN, Singh RN. Human survival motor neuron genes generate a vast repertoire of circular RNAs. Nucleic Acids Res. 2019;47:2884-2905. doi: 10.1093/nar/gkz034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pagliarini V, Jolly A, Bielli P, Di Rosa V, De la Grange P, Sette C. Sam68 binds Alu-rich introns in SMN and promotes pre-mRNA circularization. Nucleic Acids Res. 2020;48:633-645. doi: 10.1093/nar/gkz1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ottesen EW, Singh RN. Characteristics of circular RNAs generated by human survival motor neuron genes. Cell Signal. 2020;73:109696. doi: 10.1016/j.cellsig.2020.109696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh NN, Ottesen EW, Singh RN. A survey of transcripts generated by spinal muscular atrophy genes. Biochim Biophys Acta Gene Regul Mech. 2020;1863:194562. doi: 10.1016/j.bbagrm.2020.194562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mendonça RH, Rocha AJ, Lozano-Arango A, et al. Severe brain involvement in 5q spinal muscular atrophy type 0. Ann Neurol. 2019;86:458-462. doi: 10.1002/ana.25549 [DOI] [PubMed] [Google Scholar]
- 33. Michaud M, Arnoux T, Bielli S, et al. Neuromuscular defects and breathing disorders in a new mouse model of spinal muscular atrophy. Neurobiol Dis. 2010;38:125-135. doi: 10.1016/j.nbd.2010.01.006 [DOI] [PubMed] [Google Scholar]
- 34. Wijngaarde CA, Veldhoen ES, van Eijk RPA, et al. Natural history of lung function in spinal muscular atrophy. Orphanet J Rare Dis. 2020. a;15:88. doi: 10.1186/s13023-020-01367-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Genabai NK, Kannan A, Ahmad S, Jiang X, Bhatia K, Gangwani L. Deregulation of ZPR1 causes respiratory failure in spinal muscular atrophy. Sci Rep. 2017;7:8295. doi: 10.1038/s41598-017-07603-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vitte JM, Davoult B, Roblot N, et al. Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am J Pathol. 2004;165:1731-1741. doi: 10.1016/S0002-9440(10)63428-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szunyogova E, Zhou H, Maxwell GK, et al. Survival motor neuron (SMN) protein is required for normal mouse liver development. Sci Rep. 2016;6:34635. doi: 10.1038/srep34635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sintusek P, Catapano F, Angkathunkayul N, et al. Histopathological defects in intestine in severe spinal muscular atrophy mice are improved by systemic antisense oligonucleotide treatment. PLoS One. 2016;11:e0155032. doi: 10.1371/journal.pone.0155032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deguise MO, Baranello G, Mastella C, et al. Abnormal fatty acid metabolism is a core component of spinal muscular atrophy. Ann Clin Transl Neurol. 2019;6:1519-1532. doi: 10.1002/acn3.50855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shanmugarajan S, Tsuruga E, Swoboda KJ, Maria BL, Ries WL, Reddy SV. Bone loss in survival motor neuron (Smn(-/-) SMN2) genetic mouse model of spinal muscular atrophy. J Pathol. 2009;219:52-60. doi: 10.1002/path.2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shababi M, Habibi J, Yang HT, Vale SM, Sewell WA, Lorson CL. Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum Mol Genet. 2010;19:4059-4071. doi: 10.1093/hmg/ddq329 [DOI] [PubMed] [Google Scholar]
- 42. Heier CR, Satta R, Lutz C, DiDonato CJ. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum Mol Genet. 2010;19:3906-3918. doi: 10.1093/hmg/ddq330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Šoltić D, Shorrock HK, Allardyce H, et al. Lamin A/C dysregulation contributes to cardiac pathology in a mouse model of severe spinal muscular atrophy. Hum Mol Genet. 2019;28:3515-3527. doi: 10.1093/hmg/ddz195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bowerman M, Swoboda KJ, Michalski JP, et al. Glucose metabolism and pancreatic defects in spinal muscular atrophy. Ann Neurol. 2012;72:256-268. doi: 10.1002/ana.23582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gombash SE, Cowley CJ, Fitzgerald JA, et al. SMN deficiency disrupts gastrointestinal and enteric nervous system function in mice. Hum Mol Genet. 2015;24:5665. doi: 10.1093/hmg/ddv292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomson AK, Somers E, Powis RA, et al. Survival of motor neurone protein is required for normal postnatal development of the spleen. J Anat. 2017;230:337-346. doi: 10.1111/joa.12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deguise MO, Beauvais A, Schneider BL, Kothary R. Blood flow to the spleen is altered in a mouse model of spinal muscular atrophy. J Neuromuscul Dis. 2020;7:315-322. doi: 10.3233/JND-200493 [DOI] [PubMed] [Google Scholar]
- 48. Kim JK, Jha NN, Feng Z, et al. Muscle-specific SMN reduction reveals motor neuron-independent disease in spinal muscular atrophy models. J Clin Invest. 2020;130:1271-1287. doi: 10.1172/JCI131989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nery FC, Siranosian JJ, Rosales I, et al. Impaired kidney structure and function in spinal muscular atrophy. Neurol Genet. 2019;5:e353. doi: 10.1212/NXG.0000000000000353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Motyl AAL, Faller KME, Groen EJN, et al. Pre-natal manifestation of systemic developmental abnormalities in spinal muscular atrophy. Hum Mol Genet. 2020;29: 2674-2683. doi: 10.1093/hmg/ddaa146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ottesen EW, Howell MD, Singh NN, Seo J, Whitley EM, Singh RN. Severe impairment of male reproductive organ development in a low SMN expressing mouse model of spinal muscular atrophy. Sci Rep. 2016;6:17. doi: 10.1038/srep20193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chang WF, Xu J, Lin TY, et al. Survival motor neuron protein participates in mouse germ cell development and spermatogonium maintenance. Int J Mol Sci. 2020;21:794. doi: 10.3390/ijms21030794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Monani UR, Sendtner M, Coovert DD, et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(-/-) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet. 2000;9:333-339. doi: 10.1093/hmg/9.3.333 [DOI] [PubMed] [Google Scholar]
- 54. Swoboda KJ, Prior TW, Scott CB, et al. Natural history of denervation in SMA: relation to age, SMN2 copy number, and function. Ann Neurol. 2005;57:704-712. doi: 10.1002/ana.20473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wirth B, Brichta L, Schrank B, et al. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum Genet. 2006;119:422-428. doi: 10.1007/s00439-006-0156-7 [DOI] [PubMed] [Google Scholar]
- 56. Prior TW, Krainer AR, Hua Y, et al. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet. 2009;85:408-413. doi: 10.1016/j.ajhg.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Oprea GE, Kröber S, McWhorter ML, et al. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524-527. doi: 10.1126/science.1155085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yener İH, Topaloglu H, Erdem-Özdamar S, Dayangac-Erden D. Transcript levels of plastin 3 and neuritin 1 modifier genes in spinal muscular atrophy siblings. Pediatr Int. 2017;59:53-56. doi: 10.1111/ped.13052. [DOI] [PubMed] [Google Scholar]
- 59. Riessland M, Kaczmarek A, Schneider S, et al. Neurocalcin delta suppression protects against spinal muscular atrophy in humans and across species by restoring impaired endocytosis. Am J Hum Genet. 2017;100:297-315. doi: 10.1016/j.ajhg.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Singh NN, Seo J, Ottesen EW, Shishimorova M, Bhattacharya D, Singh RN. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol Cell Biol. 2011;31:935-954. doi: 10.1128/MCB.00945-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Howell MD, Ottesen EW, Singh NN, et al. TIA1 is a gender-specific disease modifier of a mild mouse model of spinal muscular atrophy. Sci Rep. 2017;7:18. doi: 10.1038/s41598-017-07468-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Han KJ, Foster DG, Zhang NY, et al. Ubiquitin-specific protease 9x deubiquitinates and stabilizes the spinal muscular atrophy protein-survival motor neuron. J Biol Chem. 2012;287:43741-43752. doi: 10.1074/jbc.M112.372318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Powis RA, Karyka E, Boyd P, et al. Systemic restoration of UBA1 ameliorates disease in spinal muscular atrophy. JCI Insight. 2016;1:e87908. doi: 10.1172/jci.insight.87908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Villalón E, Kline RA, Smith CE, et al. AAV9-stathmin1 gene delivery improves disease phenotype in an intermediate mouse model of spinal muscular atrophy. Hum Mol Genet. 2019;28:3742-3754. doi: 10.1093/hmg/ddz188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou H, Meng J, Malerba A, et al. Myostatin inhibition in combination with antisense oligonucleotide therapy improves outcomes in spinal muscular atrophy. J Cachexia Sarcopenia Muscle. 2020;11:768-782. doi: 10.1002/jcsm.12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ahmad S, Wang Y, Shaik GM, Burghes AH, Gangwani L. The zinc finger protein ZPR1 is a potential modifier of spinal muscular atrophy. Hum Mol Genet. 2012;21:2745-2758. doi: 10.1093/hmg/dds102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kannan A, Jiang X, He L, Ahmad S, Gangwani L. ZPR1 prevents R-loop accumulation, upregulates SMN2 expression and rescues spinal muscular atrophy. Brain. 2020;143:69-93. doi: 10.1093/brain/awz373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhao DY, Gish G, Braunschweig U, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2016;529:48-53. doi: 10.1038/nature16469 [DOI] [PubMed] [Google Scholar]
- 69. Kannan A, Bhatia K, Branzei D, Gangwani L. Combined deficiency of Senataxin and DNA-PKcs causes DNA damage accumulation and neurodegeneration in spinal muscular atrophy. Nucleic Acids Res. 2018;46:8326-8346. doi: 10.1093/nar/gky641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Singh RN, Seo J, Singh NN. RNA in spinal muscular atrophy: therapeutic implications of targeting. Expert Opin Ther Targets. 2020;24:731-743. doi: 10.1080/14728222.2020.1783241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96:6307-6311. doi: 10.1073/pnas.96.11.6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Monani UR, Lorson CL, Parsons DW, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177-1183. doi: 10.1093/hmg/8.7.1177 [DOI] [PubMed] [Google Scholar]
- 73. Shenasa H, Hertel KJ. Combinatorial regulation of alternative splicing. Biochim Biophys Acta Gene Regul Mech. 2019;1862:194392. doi: 10.1016/j.bbagrm.2019.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lim SR, Hertel KJ. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3' splice site pairing. J Biol Chem. 2001;276:45476-45483. doi: 10.1074/jbc.M107632200 [DOI] [PubMed] [Google Scholar]
- 75. Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377-384. doi: 10.1038/ng854 [DOI] [PubMed] [Google Scholar]
- 76. Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34:460-463. doi: 10.1038/ng1207 [DOI] [PubMed] [Google Scholar]
- 77. Singh NN, Androphy EJ, Singh RN. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem Biophys Res Commun. 2004;315:381-388. doi: 10.1016/j.bbrc.2004.01.067 [DOI] [PubMed] [Google Scholar]
- 78. Singh NN, Androphy EJ, Singh RN. The regulation and regulatory activities of alternative splicing of the SMN gene. Crit Rev Eukaryot Gene Expr. 2004;14:271-285. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.30 [DOI] [PubMed] [Google Scholar]
- 79. Singh NN, Androphy EJ, Singh RN. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291-1305. doi: 10.1261/rna.7580704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Buratti E, Baralle FE. Another step forward for SELEXive splicing. Trends Mol Med. 2005;11:5-9. doi: 10.1016/j.molmed.2004.11.007 [DOI] [PubMed] [Google Scholar]
- 81. Singh RN. Unfolding the mystery of alternative splicing through a unique method of in vivo selection. Front Biosci. 2007;12:3263-3272. doi: 10.2741/2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Singh RN. Evolving concepts on human SMN pre-mRNA splicing. RNA Biol. 2007;4:7-10. doi: 10.4161/rna.4.1.4535 [DOI] [PubMed] [Google Scholar]
- 83. Xiong HY, Alipanahi B, Lee LJ, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lund M, Kjems J. Defining a 5' splice site by functional selection in the presence and absence of U1 snRNA 5' end. RNA. 2002;8:166-179. doi: 10.1017/s1355838202010786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Parada GE, Munita R, Cerda CA, Gysling K. A comprehensive survey of non-canonical splice sites in the human transcriptome. Nucleic Acids Res. 2014;42:10564-10578. doi: 10.1093/nar/gku744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Baralle FE, Singh RN, Stamm S. RNA structure and splicing regulation. Biochim Biophys Acta Gene Regul Mech. 2019;1862:194448. doi: 10.1016/j.bbagrm.2019.194448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ptok J, Müller L, Theiss S, Schaal H. Context matters: regulation of splice donor usage. Biochim Biophys Acta Gene Regul Mech. 2019;1862:194391. doi: 10.1016/j.bbagrm.2019.06.002 [DOI] [PubMed] [Google Scholar]
- 88. Singh NN, Singh RN. Alternative splicing in spinal muscular atrophy underscores the role of an intron definition model. RNA Biol. 2011;8:600-606. doi: 10.4161/rna.8.4.16224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26:1333-1346. doi: 10.1128/MCB.26.4.1333-1346.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35:371-389. doi: 10.1093/nar/gkl1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341-350. doi: 10.4161/rna.6.3.8723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Singh NN, Hollinger K, Bhattacharya D, Singh RN. An antisense microwalk reveals critical role of an intronic position linked to a unique long-distance interaction in pre-mRNA splicing. RNA. 2010;16:1167-1181. doi: 10.1261/rna.2154310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Singh NN, Lawler MN, Ottesen EW, Upreti D, Kaczynski JR, Singh RN. An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res. 2013;41:8144-8165. doi: 10.1093/nar/gkt609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Singh NN, Lee BM, Singh RN. Splicing regulation in spinal muscular atrophy by an RNA structure formed by long-distance interactions. Ann N Y Acad Sci. 2015;1341:176-187. doi: 10.1111/nyas.12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Singh NN, Howell MD, Singh RN. Transcription and splicing regulation of spinal muscular atrophy genes. In: Sumner CJ, Paushkin S, Ko C-P, eds. Spinal Muscular Atrophy: Disease Mechanisms and Therapy. Academic Press; 2017: 75–97. doi: 10.1016/B978-0-12-803685-3.00005-7 [DOI] [Google Scholar]
- 96. Singh RN, Singh NN. Mechanism of splicing regulation of spinal muscular atrophy genes. Adv Neurobiol. 2018;20:31-61. doi: 10.1007/978-3-319-89689-2_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc Natl Acad Sci U S A. 2000;97:9618-9623. doi: 10.1073/pnas.160181697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Mende Y, Jakubik M, Riessland M, et al. Deficiency of the splicing factor Sfrs10 results in early embryonic lethality in mice and has no impact on full-length SMN/Smn splicing. Hum Mol Genet. 2010;19:2154-2167. doi: 10.1093/hmg/ddq094 [DOI] [PubMed] [Google Scholar]
- 99. Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420-434. doi: 10.1016/j.molcel.2007.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Schultz AS, Preussner M, Bunse M, Karni R, Heyd F. Activation-dependent TRAF3 exon 8 alternative splicing is controlled by CELF2 and hnRNP C binding to an upstream intronic element. Mol Cell Biol. 2017;37:e00488-16. doi: 10.1128/MCB.00488-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mallory MJ, McClory SP, Chatrikhi R, Gazzara MR, Ontiveros RJ, Lynch KW. Reciprocal regulation of hnRNP C and CELF2 through translation and transcription tunes splicing activity in T cells. Nucleic Acids Res. 2020;48:5710-5719. doi: 10.1093/nar/gkaa295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Singh RN, Singh NN, Singh NK, Androphy EJ. Spinal muscular atrophy (SMA) treatment via targeting of SMN2 splice site inhibitory sequences. US Patent 7,838,657. November 1, 2010. [Google Scholar]
- 103. Sivanesan S, Howell MD, Didonato CJ, Singh RN. Antisense oligonucleotide mediated therapy of spinal muscular atrophy. Transl Neurosci. 2013;4. doi: 10.2478/s13380-013-0109-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494-3510. doi: 10.1093/nar/gkl498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ottesen EW. ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Translat Neurosci. 2017;8:1–6. doi: 10.1515/tnsci-2017-0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Singh NN, Howell MD, Androphy EJ, Singh RN. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther. 2017;24:520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Seo J, Ottesen EW, Singh RN. Antisense methods to modulate pre-mRNA splicing. Methods Mol Biol. 2014;1126:271-283. doi: 10.1007/978-1-62703-980-2_20 [DOI] [PubMed] [Google Scholar]
- 108. Singh NN, Luo D, Singh RN. Pre-mRNA splicing modulation by antisense oligonucleotides. Methods Mol Biol. 2018;1828:415-437. doi: 10.1007/978-1-4939-8651-4_26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Singh NN, Lee BM, DiDonato CJ, Singh RN. Mechanistic principles of antisense targets for the treatment of spinal muscular atrophy. Future Med Chem. 2015;7:1793-1808. doi: 10.4155/fmc.15.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hua Y, Sahashi K, Rigo F, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123-126. doi: 10.1038/nature10485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Finkel RS, Chiriboga CA, Vajsar J, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017-3026. doi: 10.1016/S0140-6736(16)31408-8 [DOI] [PubMed] [Google Scholar]
- 112. Finkel RS, Mercuri E, Darras BT, et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med. 2017;377:1723-1732. doi: 10.1056/NEJMoa1702752 [DOI] [PubMed] [Google Scholar]
- 113. Mercuri E, Darras BT, Chiriboga CA, et al. Nusinersen versus sham control in later-onset spinal muscular atrophy. N Engl J Med. 2018;378:625-635. doi: 10.1056/NEJMoa1710504 [DOI] [PubMed] [Google Scholar]
- 114. Singh RN, Singh NN. Spinal muscular atrophy treatment via targeting smn2 catalytic core. US Patent 9,217,147. November 11, 2015. [Google Scholar]
- 115. Singh RN, Singh NN. Deep intronic target for splicing correction on spinal muscular atrophy gene. US Patent 9,856,474. January 2, 2018. [Google Scholar]
- 116. Keil JM, Seo J, Howell MD, Hsu WH, Singh RN, DiDonato CJ. A short antisense oligonucleotide ameliorates symptoms of severe mouse models of spinal muscular atrophy. Mol Ther Nucleic Acids. 2014;3:e174. doi: 10.1038/mtna.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Howell MD, Ottesen EW, Singh NN, Anderson RL, Singh RN. Gender-specific amelioration of SMA phenotype upon disruption of a deep intronic structure by an oligonucleotide. Mol Ther. 2017;25:1328-1341. doi: 10.1016/j.ymthe.2017.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Al-Zaidy SA, Mendell JR. From clinical trials to clinical practice: practical considerations for gene replacement therapy in SMA type 1. Pediatr Neurol. 2019;100:3-11. doi: 10.1016/j.pediatrneurol.2019.06.007 [DOI] [PubMed] [Google Scholar]
- 119. Meyer K, Ferraiuolo L, Schmelzer L, et al. Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA: a dose-response study in mice and nonhuman primates. Mol Ther. 2015;23:477-487. doi: 10.1038/mt.2014.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Landon-Cardinal O, Baril-Dionne A, Hoa S, et al. Recognising the spectrum of scleromyositis: HEp-2 ANA patterns allow identification of a novel clinical subset with anti-SMN autoantibodies. RMD Open. 2020;6:e001357. doi: 10.1136/rmdopen-2020-001357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Singh RN. More is needed to complement the available therapies of spinal muscular atrophy. Future Med Chem. 2019;11:2873-2876. doi: 10.4155/fmc-2019-0239 [DOI] [PubMed] [Google Scholar]
- 122. Dhillon S. Risdiplam: first approval. Drugs. Published online October 12, 2020. doi: 10.1007/s40265-020-01410-z [DOI] [PubMed] [Google Scholar]
- 123. Ratni H, Ebeling M, Baird J, et al. Discovery of risdiplam, a selective survival of motor neuron-2 (SMN2) gene splicing modifier for the treatment of spinal muscular atrophy (SMA). J Med Chem. 2018;61:6501-6517. doi: 10.1021/acs.jmedchem.8b00741 [DOI] [PubMed] [Google Scholar]
- 124. Poirier A, Weetall M, Heinig K, et al. Risdiplam distributes and increases SMN protein in both the central nervous system and peripheral organs. Pharmacol Res Perspect. 2018;6:e00447. doi: 10.1002/prp2.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Seo J, Howell MD, Singh NN, Singh RN. Spinal muscular atrophy: an update on therapeutic progress. Biochim Biophys Acta. 2013;1832:2180-2190. doi: 10.1016/j.bbadis.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Howell MD, Singh NN, Singh RN. Advances in therapeutic development for spinal muscular atrophy. Future Med Chem. 2014;6:1081-1099. doi: 10.4155/fmc.14.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Naryshkin NA, Weetall M, Dakka A, et al. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345:688-693. doi: 10.1126/science.1250127 [DOI] [PubMed] [Google Scholar]
- 128. Albright LAC, Farnham JM, Stevens J, et al. Genome-wide analysis of high-risk primary brain cancer pedigrees identifies PDXDC1 as a candidate brain cancer predisposition gene. Neuro Oncol. Published online July 9, 2020. doi: 10.1093/neuonc/noaa161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Ratni H, Karp GM, Weetall M, et al. Specific correction of alternative survival motor neuron 2 splicing by small molecules: discovery of a potential novel medicine to treat spinal muscular atrophy. J Med Chem. 2016;59:6086-6100. doi: 10.1021/acs.jmedchem.6b00459 [DOI] [PubMed] [Google Scholar]
- 130. O’Keefe L. FDA approves oral treatment for spinal muscular atrophy. FDA News Release, August 7, 2020. https://www.fda.gov/news-events/press-announcements/fda-approves-oral-treatment-spinal-muscular-atrophy
- 131. Wang J, Schultz PG, Johnson KA. Mechanistic studies of a small-molecule modulator of SMN2 splicing. Proc Natl Acad Sci U S A. 2018;115:E4604-E4612. doi: 10.1073/pnas.1800260115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sivaramakrishnan M, McCarthy KD, Campagne S, et al. Binding to SMN2 pre-mRNA-protein complex elicits specificity for small molecule splicing modifiers. Nat Commun. 2017;8:1476. doi: 10.1038/s41467-017-01559-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Hofmann Y, Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum Mol Genet. 2002;11:2037-2049. doi: 10.1093/hmg/11.17.2037 [DOI] [PubMed] [Google Scholar]
- 134. Moursy A, Allain FH, Cléry A. Characterization of the RNA recognition mode of hnRNP G extends its role in SMN2 splicing regulation. Nucleic Acids Res. 2014;42:6659-6672. doi: 10.1093/nar/gku244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Campagne S, Boigner S, Rüdisser S, et al. Structural basis of a small molecule targeting RNA for a specific splicing correction. Nat Chem Biol. 2019;15:1191-1198. doi: 10.1038/s41589-019-0384-5 [DOI] [PubMed] [Google Scholar]
- 136. Singh NN, Del Rio-Malewski JB, Luo D, Ottesen EW, Howell MD, Singh RN. Activation of a cryptic 5' splice site reverses the impact of pathogenic splice site mutations in the spinal muscular atrophy gene. Nucleic Acids Res. 2017;45:12214-12240. doi: 10.1093/nar/gkx824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Singh RN, Singh NN. A novel role of U1 snRNP: splice site selection from a distance. Biochim Biophys Acta Gene Regul Mech. 2019;1862:634-642. doi: 10.1016/j.bbagrm.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Donadon I, Bussani E, Riccardi F, et al. Rescue of spinal muscular atrophy mouse models with AAV9-exon-specific U1 snRNA. Nucleic Acids Res. 2019;47:7618-7632. doi: 10.1093/nar/gkz469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Cheung AK, Hurley B, Kerrigan R, et al. Discovery of small molecule splicing modulators of survival motor neuron-2 (SMN2) for the treatment of spinal muscular atrophy (SMA). J Med Chem. 2018;61:11021-11036. doi: 10.1021/acs.jmedchem.8b01291 [DOI] [PubMed] [Google Scholar]
- 140. Chen TH. New and developing therapies in spinal muscular atrophy: from genotype to phenotype to treatment and where do we stand? Int J Mol Sci. 2020;21:3297. doi: 10.3390/ijms21093297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Palacino J, Swalley SE, Song C, et al. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat Chem Biol. 2015;11:511-517. doi: 10.1038/nchembio.1837 [DOI] [PubMed] [Google Scholar]
- 142. Garcia-Lopez A, Tessaro F, Jonker HRA, et al. Targeting RNA structure in SMN2 reverses spinal muscular atrophy molecular phenotypes. Nat Commun. 2018;9:2032. doi: 10.1038/s41467-018-04110-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Ando S, Suzuki S, Okubo S, et al. Discovery of a CNS penetrant small molecule SMN2 splicing modulator with improved tolerability for spinal muscular atrophy. Sci Rep. 2020;10:17472. doi: 10.1038/s41598-020-74346-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Singh NN, Singh RN. How RNA structure dictates the usage of a critical exon of spinal muscular atrophy gene. Biochim Biophys Acta Gene Regul Mech. 2019;1862:194403. doi: 10.1016/j.bbagrm.2019.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Andrews JA, Miller TM, Vijayakumar V, et al. CK-2127107 amplifies skeletal muscle response to nerve activation in humans. Muscle Nerve. 2018;57:729-734. doi: 10.1002/mus.26017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Zhou H, Meng J, Malerba A, et al. Myostatin inhibition in combination with antisense oligonucleotide therapyimproves outcomes in spinal muscular atrophy. J Cachexia Sarcopenia Muscle. 2020;11:768-782. doi: 10.1002/jcsm.12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Pagliarini V, Guerra M, Di Rosa V, Compagnucci C, Sette C. Combined treatment with the histone deacetylase inhibitor LBH589 and a splice-switch antisense oligonucleotide enhances SMN2 splicing and SMN expression in spinal muscular atrophy cells. J Neurochem. 2020;153:264-275. doi: 10.1111/jnc.14935 [DOI] [PubMed] [Google Scholar]
- 148. Farrelly-Rosch A, Lau CL, Patil N, Turner BJ, Shabanpoor F. Combination of valproic acid and morpholino splice-switching oligonucleotide produces improved outcomes in spinal muscular atrophy patient-derived fibroblasts. Neurochem Int. 2017;108:213-221. doi: 10.1016/j.neuint.2017.02.016 [DOI] [PubMed] [Google Scholar]