Abstract
Background:
Short-chain fatty acids (SCFAs) are fermented dietary components that regulate immune responses, promote colonic health and suppress mast cell-mediated diseases. However, the effects of SCFAs on human mast cell function, including the underlying mechanisms, remain unclear. Here, we investigated the effects of the SCFAs acetate, propionate and butyrate on mast cell-mediated pathology and human mast cell activation, including the molecular mechanisms involved.
Method:
Precision-cut lung slices (PCLS) of allergen-exposed guinea pigs were used to assess the effects of butyrate on allergic airway contraction. Human and mouse mast cells were co-cultured with SCFAs and assessed for degranulation after IgE- or non-IgE-mediated stimulation. The underlying mechanisms involved were investigated using knockout mice, small molecule inhibitors/agonists, and genomics assays.
Results:
Butyrate treatment inhibited allergen-induced histamine release and airway contraction in guinea pig PCLS. Propionate and butyrate, but not acetate, inhibited IgE and non-IgE-mediated human or mouse mast cell degranulation in a concentration-dependent manner. Notably, these effects were independent of the stimulation of SCFA receptors GPR41, GPR43 or PPAR, but instead were associated with inhibition of histone deacetylases. Transcriptome analyses revealed butyrate-induced downregulation of the tyrosine kinases BTK, SYK and LAT, critical transducers of FcεRI-mediated signals that are essential for mast cell activation. Epigenome analyses indicated that butyrate redistributed global histone acetylation in human mast cells, including significantly decreased acetylation at the BTK, SYK and LAT promoter regions.
Conclusion:
Known health benefits of SCFAs in allergic disease can, at least in part, be explained by epigenetic suppression of human mast cell activation.
Keywords: Mast cells, Short-chain fatty acids, Butyrate, Histone deacetylase, FcεRI signaling
Introduction
Short-chain fatty acids (SCFAs) are positive regulators of immune responses and colonic health(1,2). The three most prevalent SCFAs, acetate, propionate and butyrate, produced by fermentation of non-digestible dietary fiber in the gut, are known to regulate gut integrity, colonic mobility, mucus production, and gastrointestinal pH(1,2). SCFAs are important regulators of immune responses in human studies of IgE-mediated(3) and non-IgE-mediated(4) food allergy and mouse models of colitis, arthritis and allergic airway disease(5). Although mainly produced in the gut, human and mouse studies indicate significant immunoregulatory effects of SCFAs in other tissues including the lungs(6-8), skin(9-11) and bones(12).
Mast cells play a central role in initiating and maintaining inflammation, particularly in allergies and asthma, in which allergen re-exposure induces IgE-mediated FcεRI aggregation on the plasma membrane, rapidly triggering mast cell degranulation(15-17). Degranulation initiates the release of numerous inflammatory mediators(13) and subsequent downstream signaling responses initiate the production of inflammatory cytokines, including tumor necrosis factor-alpha (TNFα)(14,15) and interleukin 6 (IL-6)(16). FcɛRI aggregation induces phosphorylation of the Linker for Activation of T cells (LAT) adaptor molecule by the tyrosine kinases Lck/Yes-related Novel tyrosine kinase (LYN) and spleen tyrosine kinase (SYK)(17). This signaling cascade subsequently triggers Bruton’s tyrosine kinase (BTK) phosphorylation and activation of phospholipase (PLC)γ and protein kinase C, which increase the mobilization of calcium (Ca2+) to initiate mast cell degranulation(17).
Mast cells are present in tissues where the body contacts the outside world(18), including the gastrointestinal tract, the upper and lower airways and the skin. Due to their location in the gut and vascularized tissues, mast cells are exposed to high SCFA concentrations, reaching up to 140 mM in the colon (23, 24). Notably, diffusion and active transport significantly reduce SCFA concentrations in human stool samples (3,19). Importantly, SCFAs are also detectable in the blood(20,21) and previously implicated in protection against allergic airway inflammation, although the role of mast cells was not investigated(21).
Previous studies have suggested possible links between fiber intake, SCFA concentrations and mast cell-mediated pathology. Butyrate has been reported to decrease proliferation and increase histamine content in a mouse mast cell line(22). Butyrate also inhibits mouse mast cell degranulation and cytokine production(23) as well as mast cell degranulation and inflammatory mediator content in the gut mucosa of piglets(24). However, the underlying mechanistic basis for these effects of butyrate remains unclear(25). Moreover, studies in primary human mast cells are lacking, which is critical since significant functional and phenotypical differences exist between rodent and human mast cells(26).
Functional effects of SCFAs are often attributed to activation of membrane receptors GPR41 (or ‘FFAR3’) and GPR43 (or ‘FFAR2’)(5,27), the latter also expressed by mast cells(28). Nuclear peroxisome proliferator-activated receptors (PPARs) can also be stimulated by SCFAs, in particular butyrate(29). PPARs are expressed in mast cells (30) and PPARγ stimulation attenuated atopic and contact dermatitis, possibly inhibiting mast cell maturation(31). Finally, butyrate is a known inhibitor of histone deacetylases (HDACs)(32), a class of chromatin modifying enzymes that plays key roles in transcriptional regulation(33,34). Butyrate inhibits class I and II HDACs, but not class III enzymes (including the Sirtuins)(35). Dietary components, including SCFAs, were shown to promote gut homeostasis and immunity via control of histone acetylation and subsequent gene transcription(36,37). Furthermore, the therapeutic use of butyrate to modulate gene expression has been employed for various diseases, including cancer(38-41) and inflammatory disease(42-45). Notably, It was recently shown in mouse mast cells that butyrate suppressed FcεRI-dependent cytokine release, likely via inhibition of HDAC activity, without affecting β-Hexosaminidase(46).
Here, we investigated the effects of SCFAs in a clinically relevant ex vivo model of mast cell-mediated pathology. Importantly, using primary mouse and human mast cells for functional assays as well as transcriptome and epigenome profiling, we have uncovered a critical mechanism of action by which butyrate suppresses mast cell activation.
Results
Butyrate reduces histamine release and inhibits OVA-induced airway narrowing
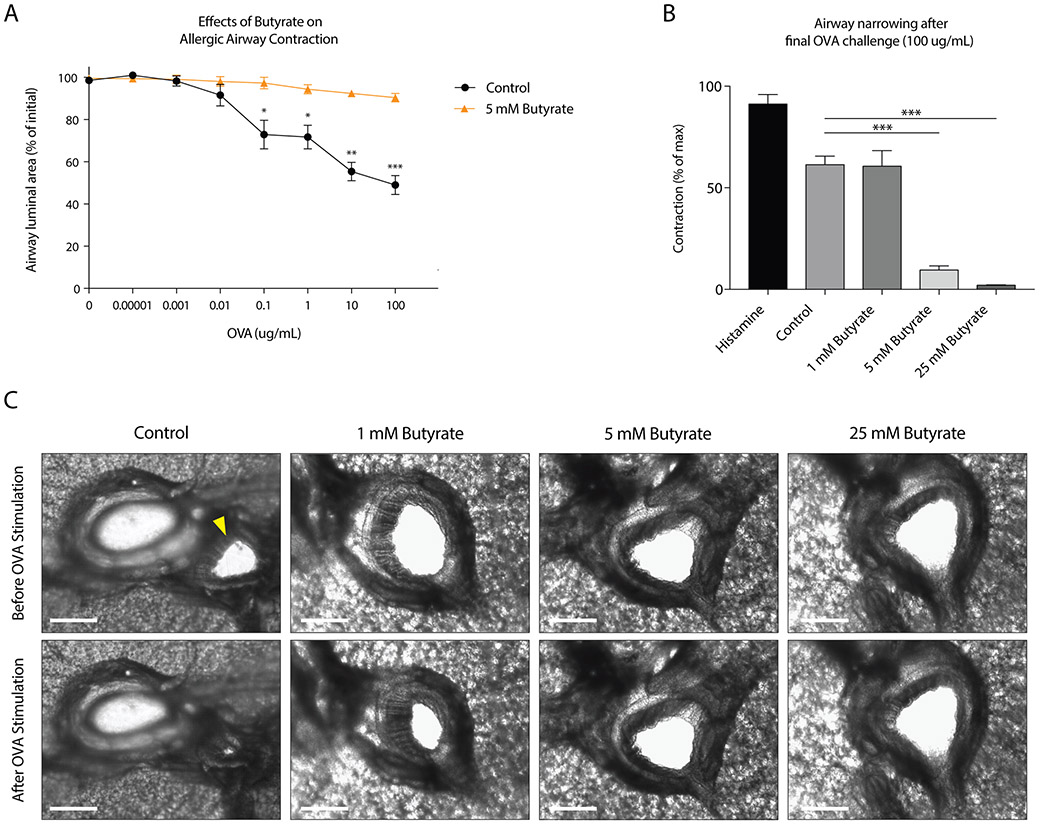
To mimic the mast cell-driven airway narrowing seen in asthma patients, we prepared precision cut lung slices (PCLS) from the lower airways of guinea pigs sensitized to the model allergen ovalbumin (OVA). Subsequent OVA challenges induced airway narrowing in a concentration-dependent manner (Fig.1A). To assess the functional effects of butyrate on mast cell-mediated airway contraction, we treated PCLS with different concentrations of butyrate for 24 hours. Butyrate inhibited IgE- and allergen-induced airway contraction in a concentration-dependent manner (Fig.1A-B). Normal tissue viability and responsiveness were confirmed via a histamine challenge following the final OVA challenge (Appendix S1A).
Figure 1. Butyrate inhibits OVA-induced airway contraction in an ex vivo model of bronchoconstriction.
A, OVA-induced reduction of airway luminal area in precision cut lung slices (PCLS) of OVA-sensitized guinea pigs either untreated or treated with butyrate for 24 h. B, Effect of different concentrations of butyrate on the airway luminal area after the final OVA challenge. As a control, slices were stimulated with histamine to induce strong contraction (1.84 mg/ml). C, Video stills depicting the effects of butyrate on OVA-induced airway contraction in PCLS. The white scale bar indicates 500 μm. OVA stimulation in vehicle treated PCLS induced strong airway contraction (‘control’). Note that the airway, indicated by a yellow arrow, is located close to a blood vessel. Pre-treatment of 5 and 25 mM butyrate inhibits OVA-induced airway contraction. Data represent mean ± SEM, statistical significance was tested using a (A) two-way ANOVA test or (B) one-way ANOVA test: Results in A and B are pooled from 2-3 independent experiments performed with PCLS from different animals (n=2-3).
Histamine release by mast cells, and subsequent stimulation of the H1-receptor on airway smooth muscle cells, is a common inducer of airway contraction(47,48). We measured histamine release in our ex vivo assay and found that OVA-challenged PCLS showed increased histamine release (Appendix S1B). Butyrate treatment of tissue slices abolished histamine release upon OVA stimulation (Appendix S1B). The concentration-dependent inhibition of airway narrowing by butyrate is visualized by representative photographs in Fig.1C and in supplementary videos 1-4. Together, these data demonstrate that butyrate inhibits IgE-dependent mast cell activation in a clinically relevant model of allergen-induced airway narrowing.
Short-chain fatty acids propionate and butyrate inhibit primary mast cell activation
Given the continuous exposure of mast cell populations to high concentrations of acetate, propionate and butyrate in vivo, we investigated the direct effects of these SCFA on the activation of cultured mast cells. Murine primary bone marrow-derived mast cells and human peripheral blood mononuclear cell-derived mast cells were incubated with increasing concentrations of the SCFAs acetate, propionate and butyrate for 24 hours. Propionate and butyrate, but not acetate, inhibited IgE/antigen-induced mast cell degranulation in a concentration-dependent manner (Fig.2A). Propionate and butyrate – but not acetate - , at millimolar concentrations, induced up to 90% inhibition of both mouse and human mast cell degranulation after IgE/antigen-induced stimulation. Mast cell activation induced by ionomycin was also markedly inhibited by butyrate and, to a lesser extent, by propionate treatment (Appendix S2A), as was degranulation triggered by compound 48/80 or substance P (Fig.2B). In addition, mast cell secretion of IL-6, a typical inflammatory mediator produced by activated mast cells, was inhibited by both propionate and butyrate (Fig.2C). SCFAs did not affect total beta-hexosaminidase levels (data not shown), did not induce spontaneous mast cell degranulation (Appendix S2A) and did not induce cellular toxicity as measured by LDH leakage (Appendix S2B). Together, these data show that butyrate and propionate, but not acetate, potently inhibit both IgE and non-IgE-mediated mast cell activation.
Figure 2. Butyrate and propionate, but not acetate, inhibit IgE and non-IgE mediated mast cell activation.
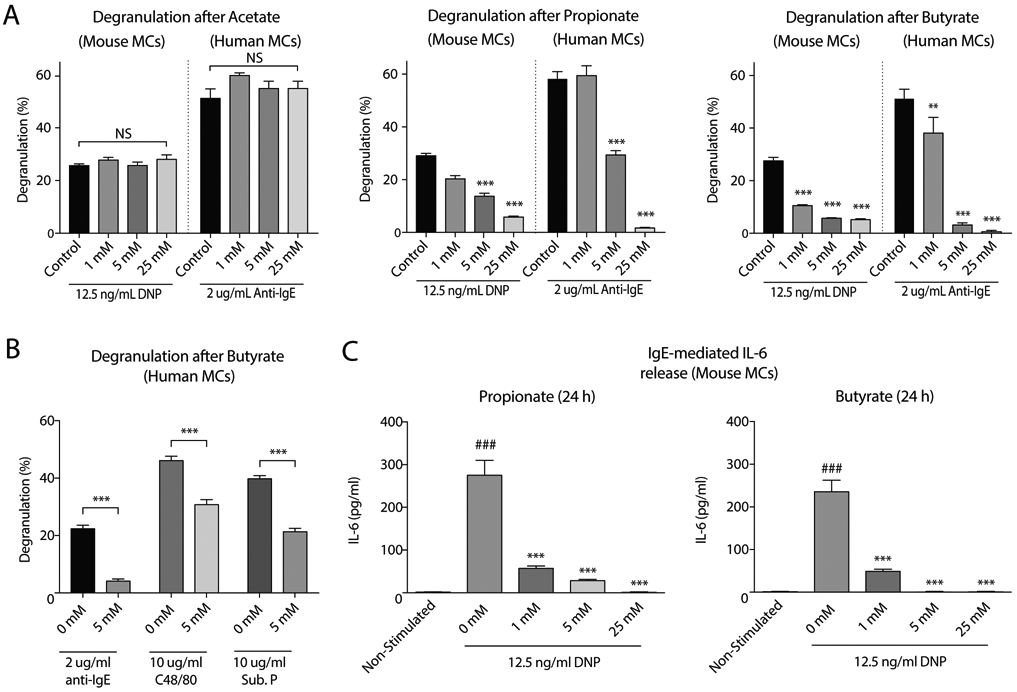
A, Percentage of mast cell degranulation (as measured by beta-hexosaminidase release) after IgE-mediated activation using either DNP-HSA or anti-IgE stimulation in mouse and human mast cells treated with increasing concentrations of acetate (left), propionate (middle) and butyrate (right). B, Percentage of degranulation after IgE- and non-IgE (C48/80 or Substance P) mediated mast cell activation in untreated or treated (5 mM butyrate, 24 h) human mast cells. C, Effects of butyrate and propionate on IL-6 cytokine production in mouse mast cells after IgE-mediated activation. Results are pooled from 3 independent experiments performed with mouse or human mast cells from 3 different donors (n=3/group). Data represent mean ± SEM, statistical significance was tested using a one-way ANOVA test: #, significantly increased compared to non-stimulated; *, Significantly decreased compared to control (P < 0.05). * P < 0.05; ** P < 0.01; ***, ### P < 0.001. NS = not significant, DNP-HSA = Dinitrophenyl - Human Serum Albumins, Sub. P. = Substance P.
Inhibitory effects of SCFAs on mast cell activation are independent of GPR41, GPR43 and PPAR receptors
Next, we investigated whether the inhibitory effects of SCFAs depend on signaling through their known GPR41/43 and PPAR receptors. We found that FcεRI-mediated degranulation was highly similar in mast cells from GPR41−/− and GPR43−/− mice as compared to wildtype (WT) mast cells (Fig.3A). Importantly, GPR41 and GPR43 were not required for the inhibitory effects of butyrate (Fig.3A) or propionate (Appendix S3A-B) on mast cell degranulation. In line with these observations, direct agonist stimulation of the GPR43 receptor did not suppress mast cell degranulation (Fig.3A, right panel). Potential signal transduction of SCFAs via GPR109a was not further pursued due to very low expression detected in our gene expression analysis of primary human mast cells (see below; data not shown). Exposure of mast cells to PPARα, PPARβ or PPARγ agonists also did not result in reduced mast cell degranulation (Fig.3B). Moreover, a specific PPARγ antagonist did not prevent the inhibition mast cell degranulation by butyrate (Appendix S3C). These results indicate that the effects of SCFAs on mast cell activity are not mediated via GPR41, GPR43 or PPAR receptor stimulation.
Figure 3. The effects of SCFAs are independent of GPR41, GPR43 and PPAR stimulation, but dependent on HDAC activity.
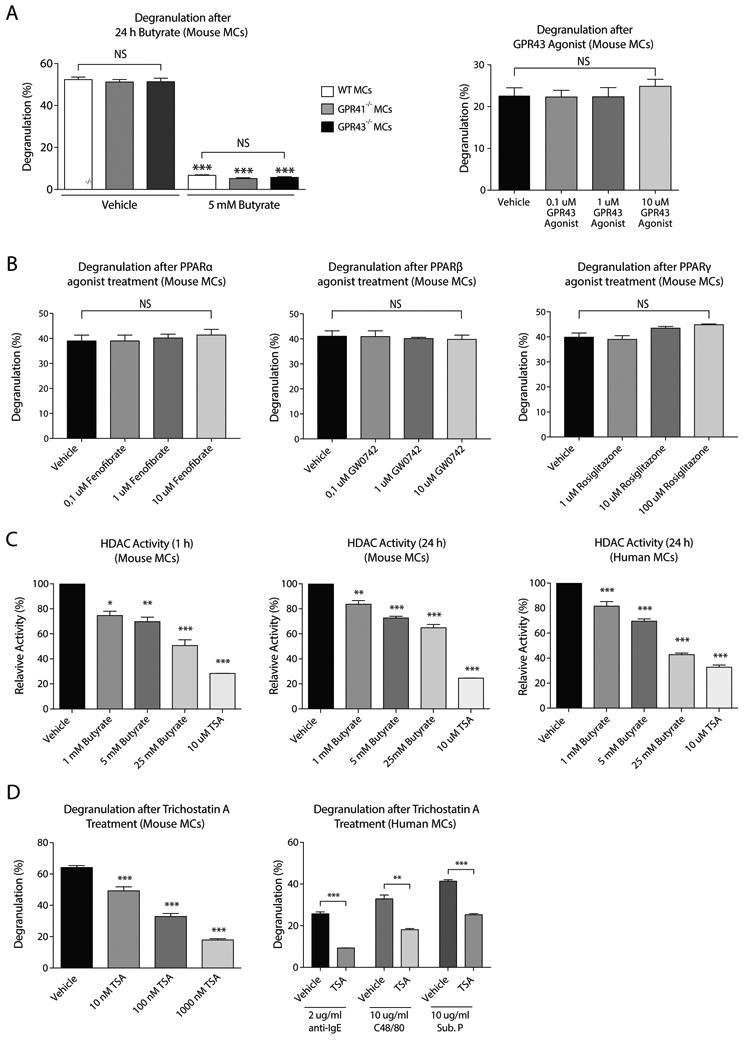
A, Percentage of mast cell degranulation after IgE-mediated activation in mouse mast cells from WT, GPR41−/− or GPR43−/− mice (left panel). Effects of a GPR43 agonist on mast cell degranulation (right panel). B, Effect of different PPAR agonists on the percentage of degranulation via IgE-mediated activation in mouse mast cells. C, The effect of butyrate or TSA (10 μM) on HDAC activity in mouse and human mast cells. HDAC activity was measured by adding cell permeable HDAC substrate containing an acetylated lysine side chain; subsequent deacetylation by HDAC releases a detectable fluorophore. D, Percentage of degranulation after TSA treatment (left panel: 10 – 1000 nM, right panel: 1000 nM) in mouse and human mast cells. Results are pooled from 3 independent experiments performed with mouse and human mast cells from 3 different donors (n=3/group). Data represent mean ± SEM, statistical significance was tested using a one-way ANOVA test or an unpaired Student’s t test: *P < 0.05; **P < 0.01; ***P < 0.001. TSA = Trichostatin A, Sub. P. = Substance P.
HDAC activity is regulated by propionate and butyrate and modulates mast cell degranulation
SCFAs, and butyrate in particular, are known to act as inhibitors of HDAC activity. Indeed, administration of butyrate led to attenuated HDAC activity in both human and mouse mast cells (Fig.3C). Propionate also inhibited HDAC activity in mouse mast cells, albeit less potently than butyrate (Appendix S3D). In mouse mast cells, inhibition of HDAC activity was strongest after 1 hour of incubation (Fig.3C). Trichostatin A (or TSA, a potent pan-HDAC inhibitor) treatment also caused a significant decrease in human and mouse mast cell degranulation (Fig.3D), with a quantitatively similar impact as SCFAs (i.e. compared to Fig.2A). Therefore, SCFAs may regulate mast cell degranulation via modulation of HDAC activity and, as a consequence, gene expression.
Butyrate induces gene expression changes associated with cytokine signaling and activation of human mast cells
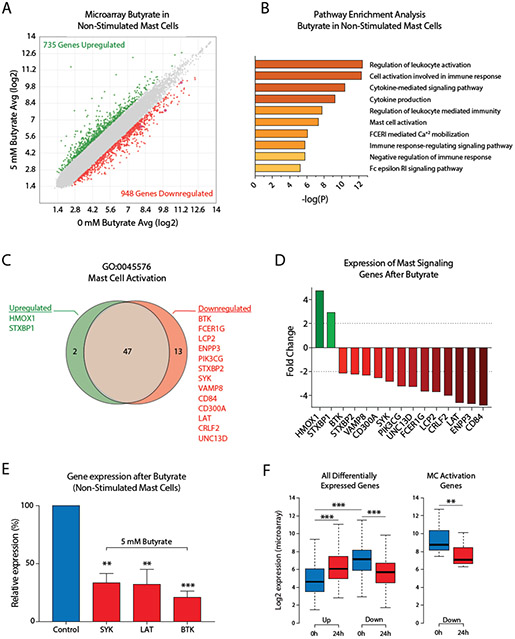
To assess whether butyrate treatment induced gene expression changes in primary human mast cells, we employed microarray gene expression profiling. The effect of butyrate on the human mast cell transcriptome was studied in non-stimulated mast cells and anti-IgE stimulated mast cells. In non-stimulated mast cells, 1683 genes were differentially expressed (>2-fold up- or downregulated) following butyrate treatment. Of these genes, 735 were upregulated (43.7%) and 948 downregulated (56.3%) (Fig.4A).
Figure 4. Butyrate induces human mast cell gene expression changes associated with cytokine signaling and mast cell activation.
A, Scatterplot comparison of gene expression levels measured by microarray analysis in unstimulated human mast cells either untreated or exposed to 5 mM butyrate for 24 h. Upregulated genes (>1.0 log2 fold change) are indicated in green, downregulated genes (<−1.0 log2 fold change) in red. B, Selected pathways that were strongly affected by butyrate treatment in unstimulated human mast cells. Y-axis denotes P-values on a −log10 scale. C, D, Butyrate-induced upregulation or downregulation of genes associated with the GO:0045576 – ‘Mast Cell Activation’ pathway represented as a Venn diagram (C) or bar graph (D) in unstimulated human mast cells treated with or without butyrate for 24 h. E, Butyrate-induced downregulation of BTK, SYK and LAT was validated using qPCR in human mast cells from 3 additional independent donors. F, Genes upregulated in response to 24h butyrate treatment (compared to vehicle treated cells, indicated as 0 h) showed overall low expression levels in vehicle treated mast cells, while downregulated genes showed significantly higher expression levels prior to treatment. Downregulated mast cell activation genes in particular are highly expressed before butyrate treatment (right boxplot). A-D, F, Results are from a single culture of human mast cells. E, Results are pooled from 3 independent experiments performed in human mast cells from 3 different donors. Data represent mean ± SEM, statistical significance was tested using a one-way ANOVA test: *P < 0.05; **P < 0.01; ***P < 0.001. GO = Gene ontology.
To investigate the cellular functions of butyrate-regulated genes and to link such functions to the regulatory effects of butyrate on mast cell activation, we performed a gene ontology (GO) pathway enrichment analysis. Human mast cells treated with butyrate for 24 h predominantly showed expression changes in genes associated with the regulation of immune cell activation and (cytokine) signaling processes (Fig.4B). Butyrate treatment strongly affected transcription of genes associated with mast cell activation and FcεRI signaling. 15 of 62 genes linked to the mast cell activation pathway (GO:0045576) were found to be differently expressed, including reduced expression of the signaling genes BTK (−2,12 fold), SYK (−2,84 fold) and LAT (−4,6 fold), each of which is essential for full mast cell activation(49-52) (Fig.4C-D). Downregulation of these essential genes following 24 h of butyrate treatment was validated using qPCR in human mast cells from 3 additional donors (Fig.4E). Accordingly, 24 h of butyrate exposure reduced BTK and SYK protein levels in mouse mast cells (Appendix S4A-B). The butyrate-induced transcriptional changes in FcεRI signaling pathway genes and how they explain the impaired mast cell response is further visualized in Appendix S4C. To further define the roles of butyrate-targeted genes in human mast cell activation, we examined the basal expression levels of these genes. Interestingly, genes upregulated in response to butyrate treatment showed overall low expression levels in the vehicle-treated mast cells, while downregulated genes showed significantly higher expression levels prior to treatment (Fig.4F). The 13 downregulated mast cell activation genes in particular were highly expressed before butyrate treatment (Fig.4F, right boxplot).
In human mast cells stimulated via IgE-crosslinking, butyrate treatment also induced a robust transcriptional response, with 1767 differentially expressed genes following butyrate treatment (1092 upregulated and 675 downregulated, see Appendix S5A). Similar to non-stimulated mast cells, gene expression changes were associated with mast cell activation and cytokine production (Appendix S5B). Although butyrate primarily induced transcriptional upregulation in stimulated mast cells, genes in pathways associated with the activation of immune responses (e.g. mast cell activation, FcεRI-mediated signaling and cytokine production) were strongly downregulated (Appendix S5C-D). Butyrate-induced downregulation of key mast cell activation genes BTK, SYK and LAT also occurred in stimulated mast cells (Appendix S5E).
Finally, our microarray analysis showed that butyrate treatment regulated expression of genes associated with asthma and bronchoconstriction (ALOX5, LTC4S and IFNGR2)(53-57), JAK/STAT signal transduction (JAK3, STAT6), cytokine receptors associated with different immune responses (IL2RG, IL1RL1, IL18R1, IL18RAP, CRLF2) and the negative regulator of NFkB signaling, TNFAIP3 (Supporting Information1 - MicroArray_Genelist). Furthermore, expression of TET2, a gene associated with epigenetic regulation of mast cell proliferation and activation, was downregulated(58-61). Together, these data indicate that butyrate regulates the expression of genes associated with mast cell activation, inflammatory responses and cytokine signaling.
Butyrate triggers elevated global H3K27 acetylation but decreased acetylation at the transcription start site of human mast cell activation genes
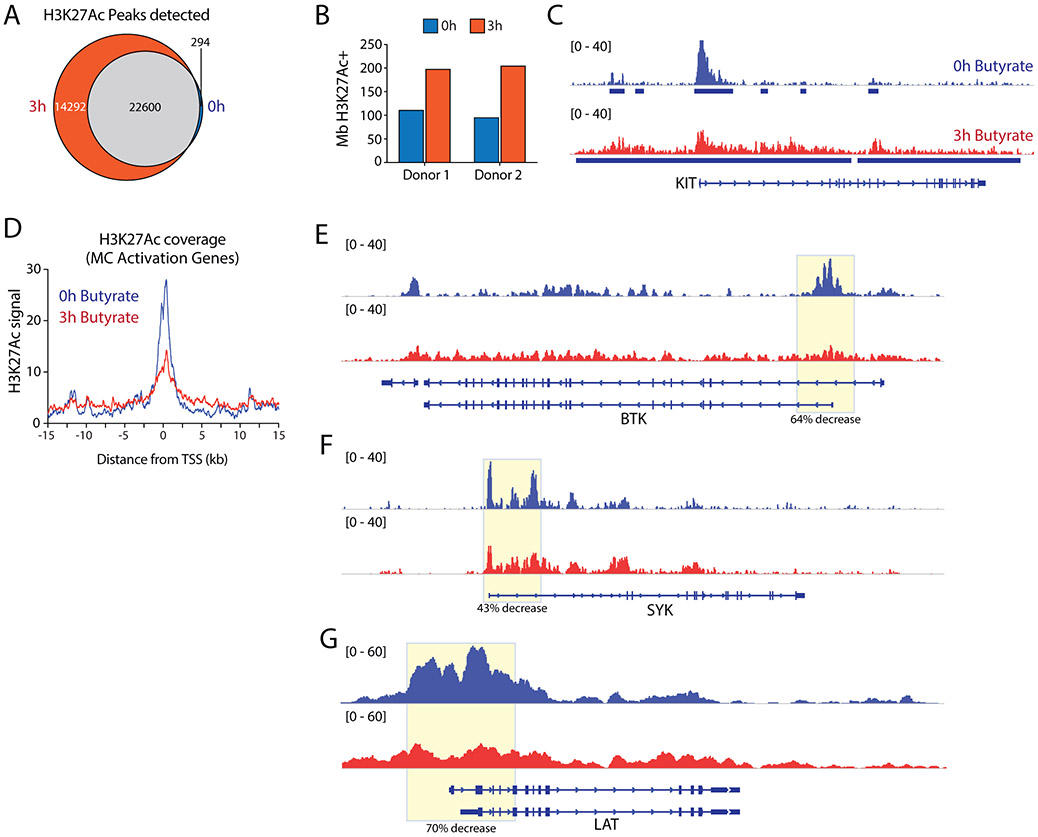
Acetylated histones are associated with active gene transcription and represent a major substrate for HDACs. To assess whether butyrate-induced changes in HDAC activity result in an altered histone acetylation landscape in mast cells, we performed ChIP-Seq analysis of H3 lysine 27 acetylation (H3K27Ac) - a well-characterized epigenetic modification that recognizes active promoters and enhancers(62) - on two independent butyrate (or vehicle) treated primary human mast cell cultures. To capture the immediate early effects of butyrate treatment on the mast cell chromatin landscape, we analyzed H3K27Ac levels after 3 hours of treatment. In line with its inhibitory effect on HDAC activity, butyrate treatment rapidly triggered increased global H3K27Ac levels, both in terms of genome-wide acetylation coverage and enrichment peaks detected (Fig.5A-B, shown for the KIT gene, as an example in Figure 5C). These observations were largely independent of analysis parameter settings (Appendix S6A-B). Notably, 3 hours of butyrate treatment predominantly triggered de novo acetylation events, with only a small minority (~1%) of regions that completely lost H3K27Ac (Fig.5A). De novo acquisition of H3K27Ac induced relatively modest acetylation mostly outside of or between regions that were highly acetylated in vehicle-treated mast cells (Fig.5C, Appendix S6C-E). Thus, butyrate exposure has a rapid and substantial impact on global mast cell histone acetylation.
Figure 5. Butyrate induces a loss of histone acetylation at the transcription start site of key genes involved in FcεRI-mediated signaling.
A, Venn diagram depicting the overlap in H3K27Ac enrichment peaks (reproducibly detected in both donors) detected at 0h and 3h of butyrate treatment. Only a minority (~1%) of regions lose all H3K27Ac signal, in contrast to de novo enrichment of 14292 unique peaks following butyrate treatment. B, Butyrate treatment induces elevated megabases (Mb) of genome covered by H3K27Ac. C, Genome browser view of histone 3 lysine 27 acetylation (H3K27Ac) levels at the KIT locus, as measured by ChIP-Seq before and after 3h of 5 mM butyrate exposure. D, Average H3K27Ac levels around the TSS of mast cell activation genes downregulated by 24 hours of butyrate exposure (see Fig.4D). E-G, Genome browser views depicting H3K27ac levels across loci encoding key signaling molecules involved in FcεRI-mediated signaling. TSS regions show reduced acetylation levels for BTK (64% decrease), SYK (43% decrease) and LAT (70% decrease), correlating with their loss of expression upon butyrate treatment. Similar results were obtained using mast cells from 2 independent cultures of 2 different donors; data obtained from donor 2 is shown. TSS = transcription start site.
We next integrated H3K27Ac ChIP-Seq data with butyrate-induced transcriptional changes in non-stimulated human mast cells (see Fig.4). H3K27Ac levels around the transcription start site (TSS) of the 735 genes upregulated by butyrate treatment remained largely unchanged (Appendix S6D), although there was a minor reduction in peak height. In contrast, genes that were downregulated after 24 hours of butyrate exposure showed substantial deacetylation already at 3 hours of treatment (Appendix S6E). These included the set of 62 GO mast cell activation genes, which showed high H3K27Ac levels in the absence of butyrate, but were rapidly deacetylated around their TSS upon butyrate treatment (Fig.5D). Specifically, H3K27ac covering the TSS of BTK (64% decrease), SYK (43% decrease) and LAT (70% decrease) was substantially reduced (Fig.5E-G), correlating with their loss of expression upon butyrate treatment.
Together, these data show that exposure of mast cells to butyrate has a profound impact on their chromatin landscape. Butyrate evokes a low-level global histone hyper-acetylation, but also induces a specific loss of acetylated transcription-competent chromatin around highly expressed genes critical for FcεRI-mediated mast cell activation.
Discussion
The immunomodulatory effects of SCFAs in mast cell-mediated disease, such as allergies and asthma, have been extensively studied. While SCFAs such as butyrate appear to suppress mast cell activity, the underlying mechanisms remain unclear and the direct effects of SCFAs on human mast cells have not yet been explored. Here, we validate that butyrate strongly reduces mast cell-driven airway narrowing in an ex vivo model of mast cell-mediated bronchoconstriction. These findings were further substantiated in a primary human mast cell culture system, as SCFAs propionate and especially butyrate inhibited both IgE and non-IgE mediated degranulation. Importantly, our studies in primary mast cells indicate that these effects are not dependent on the membrane receptors GRP41 and GPR43, nor on the nuclear PPAR receptors, which all have been implicated in mediating the biological effects of SCFAs. Instead, we show that HDAC activity in both mouse and human mast cells can be suppressed by propionate and butyrate, evoking a redistribution of global histone acetylation. Altered histone acetylation included a loss of acetylation and expression at genes encoding key signaling molecules mediating FcεRI-mediated degranulation, providing a plausible mechanism for how SCFAs can provide health benefits in the context of mast cell-mediated allergic disease.
The SCFA levels that we found to inhibit mast cell activation in vitro or ex vivo were non-toxic and comparable to physiological SCFA concentrations in the gut and blood of humans(20,63) or rodents(64). Although several mechanisms have been proposed to underlie the immunomodulatory effects of SCFAs(5,27,29,30,65), our data indicate that mast cell suppression by SCFAs is relayed via modulation of HDAC activity, rather than by the stimulation of membrane GPR41/43 or PPAR nuclear receptors. The non-instantaneous (>12 h) effects of SCFAs on mast cell degranulation indicate that they are more likely to originate from alterations in gene expression, rather than by direct receptor stimulation. Indeed, we observed maximum HDAC inhibition directly after butyrate incubation (1 h), followed by rapid changes in histone acetylation (3 h) and a subsequent transcriptional silencing of FcεRI signaling genes for optimal suppression of mast cell degranulation (18-24 h). Zhang et al. recently reported that butyrate did not affect β-Hexosaminidase release in mouse mast cells. We believe that this discrepancy arises from the shorter exposure time (i.e. 12 h) used in this study, which is likely not enough to allow for the abovementioned sequence of molecular events to sufficiently deplete the levels of FcεRI signaling components or other, possibly FcεRI-independent, mediators of degranulation.
Although butyrate-induced local histone deacetylation and subsequent transcriptional downregulation at highly expressed genes appears counterintuitive at first glance, earlier studies have also reported gene repression induced by HDAC inhibitors(66), particularly for highly expressed genes heavily occupied by HDACs(67-69). Of note, HDACs were reported to boost transcription by restricting histone acetylation specifically to the TSS(68). We postulate that HDAC inhibition by SCFAs triggers global ‘aspecific’ low level histone hyper-acetylation, especially near highly active gene loci that are preferentially targeted by HDACs. This redistribution lowers acetylation levels at transcription start sites, resulting in reduced mRNA expression and histone acetylation at highly expressed genes (e.g. for BTK, Fig.5). Regulation of mast cell function by HDAC activity is further supported by a recent study demonstrating that TSA can diminish FcεRI-mediated cytokine production and degranulation in mouse mast cells(70).
Our analyses suggest that a strong transcriptional silencing of critical molecules for IgE receptor-induced signal transduction, including BTK, SYK and LAT, represents the mechanism that underlies SCFA-mediated inhibition of human mast cell activation. Mast cells deficient in these signaling genes display reduced FcεRI-mediated degranulation(49-52). Importantly, BTK, SYK and LAT are directly upstream of JNK and NFAT activation, potentially explaining the results of studies showing reduced JNK and NFAT phosphorylation or binding upon butyrate treatment (23,24). Butyrate might additionally modulate AP-1 and NF-AT DNA binding through altered histone acetylation to suppress the late phase of mast cell activation. Whether butyrate exposure also modulates other (unknown) regulators of mast cell activity via histone acetylation levels is an important topic for future investigations.
The observation that butyrate prevented allergen-induced histamine release in PCLS of OVA-sensitized guinea pigs and markedly attenuated OVA-induced airway contraction indicates that SCFA-mediated HDAC inhibition of mast cell activation could potentially be of therapeutic interest in allergic diseases. Comparable effects were reported in a similar PCLS model using other HDAC inhibitors(71,72). Notably, short-term butyrate treatment (2 hours) showed little effect on contraction of guinea pig PCLS(71), which parallels our findings that longer treatment with butyrate (24 hours) is needed for demonstration of inhibitory responses.
In summary, our findings indicate that SCFAs suppress human mast cell degranulation, cytokine production and allergen-induced airway contraction via HDAC inhibition and the subsequent transcriptional downregulation of critical mast cell signal transducers via an epigenetic mechanism. Hence, the acknowledged health benefits of SCFAs for allergic disease can be, in part, attributed to inhibition of mast cell activation via histone deacetylation. Additional insight into the inhibitory mechanisms of SCFAs may be of clinical importance and could reveal new approaches to inhibit pathogenic mast cell activity in allergic diseases.
Methods
Animals
All protocols described in this study were approved by the University of Groningen Committee for Animal Experimentation, Groningen, The Netherlands. Guinea pigs were housed under a 12-hour light/dark cycle in a temperature- and humidity-controlled room with food and tap water ad libitum. Animals were actively IgE-sensitized to ovalbumin as described previously(72). The animals were used for experiments 4 weeks after sensitization.
Peripheral blood mononuclear cell-derived human mast cells
Human peripheral blood mononuclear cell-derived mast cells were generated as previously described by Gaudenzio et al.(73). Briefly, peripheral blood mononuclear cells were obtained from buffy coats of healthy blood donors and CD34+ precursor cells were isolated using the EasySep Human CD34 Positive Selection Kit (STEMCELL Technologies). CD34+ cells were maintained for 4 weeks under serum-free conditions using StemSpan medium (STEMCELL Technologies) supplemented with recombinant human IL-6 (50 ng/ml; Peprotech), human IL-3 (10 ng/ml; Peprotech) and human Stem Cell Factor (100 ng/mL Peprotech, Rocky Hill, NJ). Thereafter, the cells were maintained in IMDM Glutamax I, sodium pyruvate, 2-mercaptoethanol, 0.5% BSA, insulin- 175 transferrin selenium (all from Invitrogen), ciprofloxacin (10 ug/ml; Sigma-Aldrich), IL-6 (50 ng/ml) and human Stem Cell Factor (100 ng/mL Peprotech, Rocky Hill, NJ). After 8-12 weeks, PBCMCs were tested for maturity by Giemsa or toluidine blue staining and beta-hexosaminidase release assays (see below).
Statistical analysis
Statistical tests were performed with Graphpad Prism 7 (GraphPad Software, Inc). Two-tailed Student’s t-tests (unpaired or paired) and one-way ANOVA tests were performed as described in the respective figure legends. A p-value of less than 0.05 was considered statistically significant.
A full description of all methods is available in the Supplementary Methods.
Supplementary Material
Acknowledgements
We thank Chen Liu for technical assistance, Reinoud Gosens and colleagues (Department of Molecular Pharmacology, University of Groningen) for their advice and technical support in performing the PCLS measurements. J.F. is supported by a Fulbright Fellowship (financed by the Netherlands-America Foundation). R.S. is supported by an NWO Veni Fellowship (grant no. 91617114) and an Erasmus MC Fellowship. This work was partly supported by the Lung Foundation Netherlands Grant 4.1.18.226 to R.W.H. SJG and SYT are supported by NIH/NIAID U19AI104209, NIH/NIAMS R01 AR067145, NIH/NIAID R01 AI132494, and United States-Israel Binational Science Foundation, Grant 2017182 (S. J. Galli).
Abbreviations
- BMCMCs
Bone marrow–derived cultured mast cells
- GO
Gene ontology
- GPR
G protein-coupled receptor
- HDAC
Histone deacetylase
- OVA
Ovalbumin
- PBCMC
Peripheral blood mononuclear cell-derived human mast cell
- PCLS
Precision-cut lung slices
- PPARs
Peroxisome proliferator-activated receptors
- SCFA
Short-chain fatty acids
- TSA
Trichostatin A
- TSS
Transcription Start Site
- WT
Wild-type
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol 2014;121:91–119. [DOI] [PubMed] [Google Scholar]
- 2.Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016;165:1332–1345. [DOI] [PubMed] [Google Scholar]
- 3.Roduit C, Frei R, Ferstl R, Loeliger S, Westermann P, Rhyner C et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 2018;40:833. [DOI] [PubMed] [Google Scholar]
- 4.Berni Canani R, De Filippis F, Nocerino R, Paparo L, Di Scala C, Cosenza L et al. Gut microbiota composition and butyrate production in children affected by non-IgE-mediated cow's milk allergy. Sci Rep 2018;8:12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009;461:1282–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun 2015;6:7320. [DOI] [PubMed] [Google Scholar]
- 7.Thio CLP, Chi PY, Lai ACY, Chang YJ. Regulation of type 2 innate lymphoid cell–dependent airway hyperreactivity by butyrate. J Allergy Clin Immunol 2018;142:1867–1883.e12. [DOI] [PubMed] [Google Scholar]
- 8.Cait A, Hughes MR, Antignano F, Cait J, Dimitriu PA, Maas KR et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol 2018;11:785–795. [DOI] [PubMed] [Google Scholar]
- 9.Sanford JA, Zhang L-J, Williams MR, Gangoiti JA, Huang C-M, Gallo RL. Inhibition of HDAC8 and HDAC9 by microbial short-chain fatty acids breaks immune tolerance of the epidermis to TLR ligands. Sci Immunol 2016;1:eaah4609–eaah4609. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz A, Bruhs A, Schwarz T. The Short-Chain Fatty Acid Sodium Butyrate Functions as a Regulator of the Skin Immune System. J Invest Dermatol 2017;137:855–864. [DOI] [PubMed] [Google Scholar]
- 11.Krejner A, Bruhs A, Mrowietz U, Wehkamp U, Schwarz T, Schwarz A. Decreased expression of G-protein-coupled receptors GPR43 and GPR109a in psoriatic skin can be restored by topical application of sodium butyrate. Arch Dermatol Res 2018;310:751–758. [DOI] [PubMed] [Google Scholar]
- 12.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell 2010;38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siebenhaar F, Redegeld FA, Bischoff SC, Gibbs BF, Maurer M. Mast Cells as Drivers of Disease and Therapeutic Targets. Trends Immunol 2018;39:151–162. [DOI] [PubMed] [Google Scholar]
- 14.Gordon JR, Galli SJ. Mast cells as a source of both preformed and immunologically inducible TNF-alpha/cachectin. Nature 1990;346:274–276. [DOI] [PubMed] [Google Scholar]
- 15.Bischoff SC, Lorentz A, Schwengberg S, Weier G, Raab R, Manns MP. Mast cells are an important cellular source of tumour necrosis factor alpha in human intestinal tissue. Gut 1999;44:643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krüger-Krasagakes S, Möller A, Kolde G, Lippert U, Weber M, Henz BM. Production of interleukin-6 by human mast cells and basophilic cells. J Invest Dermatol 1996;106:75–79. [DOI] [PubMed] [Google Scholar]
- 17.Gilfillan AM, Tkaczyk C. Integrated signalling pathways for mast-cell activation. Nature Reviews Immunology 2006;6:218–230. [DOI] [PubMed] [Google Scholar]
- 18.Krystel-Whittemore M, Dileepan KN, Wood JG. Mast Cell: A Multi-Functional Master Cell. Front Immunol 2015;6:620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 2010;107:14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987;28:1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014;20:159–166. [DOI] [PubMed] [Google Scholar]
- 22.Galli SJ, Dvorak AM, Marcum JA, Ishizaka T, Nabel G, Simonian HD et al. Mast cell clones: A model for the analysis of cellular maturation. J Cell Biol 1982;95:435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diakos C, Prieschl EE, S emann MD, B hmig GA, Csonga R, Sobanov Y et al. n-Butyrate inhibits Jun NH(2)-terminal kinase activation and cytokine transcription in mast cells. Biochem Biophys Res Commun 2006;349:863–868. [DOI] [PubMed] [Google Scholar]
- 24.Wang CC, Wu H, Lin FH, Gong R, Xie F, Peng Y et al. Sodium butyrate enhances intestinal integrity, inhibits mast cell activation, inflammatory mediator production and JNK signaling pathway in weaned pigs. Innate Immun 2018;24:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folkerts J, Stadhouders R, Redegeld FA, Tam S-Y, Hendriks RW, Galli SJ et al. Effect of Dietary Fiber and Metabolites on Mast Cell Activation and Mast Cell-Associated Diseases. Front Immunol 2018;9:1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bischoff SC. Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data. Nature Reviews Immunology 2007;7:93–104. [DOI] [PubMed] [Google Scholar]
- 27.Sina C, Gavrilova O, Förster M, Till A, Derer S, Hildebrand F et al. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J Immunol 2009;183:7514–7522. [DOI] [PubMed] [Google Scholar]
- 28.Karaki S-I, Mitsui R, Hayashi H, Kato I, Sugiya H, Iwanaga T et al. Short-chain fatty acid receptor, GPR43, is expressed by enteroendocrine cells and mucosal mast cells in rat intestine. Cell Tissue Res 2006;324:353–360. [DOI] [PubMed] [Google Scholar]
- 29.Alex S, Lange K, Amolo T, Grinstead JS, Haakonsson AK, Szalowska E et al. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor γ. Mol Cell Biol 2013;33:1303–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugiyama H, Nonaka T, Kishimoto T, Komoriya K, Tsuji K, Nakahata T. Peroxisome proliferator-activated receptors are expressed in mouse bone marrow-derived mast cells. FEBS Lett 2000;467:259–262. [DOI] [PubMed] [Google Scholar]
- 31.Tachibana M, Wada K, Katayama K, Kamisaki Y, Maeyama K, Kadowaki T et al. Activation of peroxisome proliferator-activated receptor gamma suppresses mast cell maturation involved in allergic diseases. Allergy 2008;63:1136–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr 2003;133:2485S–2493S. [DOI] [PubMed] [Google Scholar]
- 33.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 2009;10:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seto E, Yoshida M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol 2014;6:a018713–a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ververis K, Hiong A, Karagiannis TC, Licciardi PV. Histone deacetylase inhibitors (HDACIS): Multitargeted anticancer agents. Biologics 2013;7:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schilderink R, Verseijden C, de Jonge WJ. Dietary inhibitors of histone deacetylases in intestinal immunity and homeostasis. Front Immunol 2013;4:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A 2014;111:2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q, Cao L, Tian Y, Zhang P, Ding C, Lu W et al. Butyrate Suppresses the Proliferation of Colorectal Cancer Cells via Targeting Pyruvate Kinase M2 and Metabolic Reprogramming. Mol Cell Proteomics 2018;17:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han A, Bennett N, Ahmed B, Whelan J, Donohoe DR. Butyrate decreases its own oxidation in colorectal cancer cells through inhibition of histone deacetylases. Oncotarget 2018;9:27280–27292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Z, Tao J, Chen P, Chen L, Sharma S, Wang G et al. Sodium Butyrate Inhibits Colorectal Cancer Cell Migration by Downregulating Bmi-1 Through Enhanced miR-200c Expression. Mol Nutr Food Res 2018;62:e1700844. [DOI] [PubMed] [Google Scholar]
- 41.Pant K, Yadav AK, Gupta P, Islam R, Saraya A, Venugopal SK. Butyrate induces ROS-mediated apoptosis by modulating miR-22/SIRT-1 pathway in hepatic cancer cells. Redox Biol 2017;12:340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berni Canani R, Di Costanzo M, Leone L. The epigenetic effects of butyrate: potential therapeutic implications for clinical practice. Clin Epigenetics 2012;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva LG, Ferguson BS, Avila AS, Faciola AP. Sodium propionate and sodium butyrate effects on histone deacetylase (HDAC) activity, histone acetylation, and inflammatory gene expression in bovine mammary epithelial cells. J Anim Sci 2018;96:5244–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chriett S, Dąbek A, Wojtala M, Vidal H, Balcerczyk A, Pirola L Prominent action of butyrate over β-hydroxybutyrate as histone deacetylase inhibitor, transcriptional modulator and anti-inflammatory molecule. Sci Rep 2019;9:742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cleophas MCP, Ratter JM, Bekkering S, Quintin J, Schraa K, Stroes ES et al. Effects of oral butyrate supplementation on inflammatory potential of circulating peripheral blood mononuclear cells in healthy and obese males. Sci Rep 2019;9:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Du M, Yang Q, Zhu M-J. Butyrate suppresses murine mast cell proliferation and cytokine production through inhibiting histone deacetylase. J Nutr Biochem 2015;27:299–306. [DOI] [PubMed] [Google Scholar]
- 47.Bryce PJ, Mathias CB, Harrison KL, Watanabe T, Geha RS, Oettgen HC. The H1 histamine receptor regulates allergic lung responses. J Clin Invest 2006;116:1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margulis A, Nocka KH, Brennan AM, Deng B, Fleming M, Goldman SJ et al. Mast cell-dependent contraction of human airway smooth muscle cell-containing collagen gels: influence of cytokines, matrix metalloproteases, and serine proteases. J Immunol 2009;183:1739–1750. [DOI] [PubMed] [Google Scholar]
- 49.Hata D, Kawakami Y, Inagaki N, Lantz CS, Kitamura T, Khan WN et al. Involvement of Bruton's tyrosine kinase in FcepsilonRI-dependent mast cell degranulation and cytokine production. J Exp Med 1998;187:1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwaki S, Tkaczyk C, Satterthwaite AB, Halcomb K, Beaven MA, Metcalfe DD et al. Btk plays a crucial role in the amplification of Fc epsilonRI-mediated mast cell activation by kit. J Biol Chem 2005;280:40261–40270. [DOI] [PubMed] [Google Scholar]
- 51.Siraganian RP, de Castro RO, Barbu EA, Zhang J. Mast cell signaling: The role of protein tyrosine kinase Syk, its activation and screening methods for new pathway participants. FEBS Lett 2010;584:4933–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saitoh S, Arudchandran R, Manetz TS, Zhang W, Sommers CL, Love PE et al. LAT is essential for Fc(epsilon)RI-mediated mast cell activation. Immunity 2000;12:525–535. [DOI] [PubMed] [Google Scholar]
- 53.Kalayci O, Birben E, Sackesen C, Keskin O, Tahan F, Wechsler ME et al. ALOX5 promoter genotype, asthma severity and LTC production by eosinophils. Allergy 2006;61:97–103. [DOI] [PubMed] [Google Scholar]
- 54.Mougey E, Lang JE, Allayee H, Teague WG, Dozor AJ, Wise RA et al. ALOX5 polymorphism associates with increased leukotriene production and reduced lung function and asthma control in children with poorly controlled asthma. Clin Exp Allergy 2013;43:512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson MD, Capra V, Clunes MT, Rovati GE, Stankova J, Maj MC et al. Cysteinyl Leukotrienes Pathway Genes, Atopic Asthma and Drug Response: From Population Isolates to Large Genome-Wide Association Studies. Front Pharmacol 2016;7:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song Y, Schwager MJ, Backer V, Guo J, Porsbjerg C, Khoo S-K et al. Environment Changes Genetic Effects on Respiratory Conditions and Allergic Phenotypes. Sci Rep 2017;7:6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar A, Das S, Agrawal A, Mukhopadhyay I, Ghosh B. Genetic association of key Th1/Th2 pathway candidate genes, IRF2, IL6, IFNGR2, STAT4 and IL4RA, with atopic asthma in the Indian population. J Hum Genet 2015;60:443–448. [DOI] [PubMed] [Google Scholar]
- 58.Montagner S, Leoni C, Emming S, Chiara Della G, Balestrieri C, Barozzi I et al. TET2 Regulates Mast Cell Differentiation and Proliferation through Catalytic and Non-catalytic Activities. Cell Rep 2017;20:1744. [DOI] [PubMed] [Google Scholar]
- 59.Monticelli S, Leoni C. Epigenetic and transcriptional control of mast cell responses. F1000Res 2017;6:2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soucie E, Hanssens K, Mercher T, Georgin-Lavialle S, Damaj G, Livideanu C et al. In aggressive forms of mastocytosis, TET2 loss cooperates with c-KITD816V to transform mast cells. Blood 2012;120:4846–4849. [DOI] [PubMed] [Google Scholar]
- 61.Traina F, Visconte V, Jankowska AM, Makishima H, O'Keefe CL, Elson P et al. Single nucleotide polymorphism array lesions, TET2, DNMT3A, ASXL1 and CBL mutations are present in systemic mastocytosis. PLoS ONE 2012;7:e43090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 2015;16:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 2006;40:235–243. [DOI] [PubMed] [Google Scholar]
- 64.Høverstad T, Midtvedt T. Short-chain fatty acids in germfree mice and rats. J Nutr 1986;116:1772–1776. [DOI] [PubMed] [Google Scholar]
- 65.Vinolo MAR, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients 2011;3:858–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther 2003;2:151–163. [PubMed] [Google Scholar]
- 67.Kim YJ, Greer CB, Cecchini KR, Harris LN, Tuck DP, Kim TH. HDAC inhibitors induce transcriptional repression of high copy number genes in breast cancer through elongation blockade. Oncogene 2013;32:2828–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greer CB, Tanaka Y, Kim YJ, Xie P, Zhang MQ, Park I-H et al. Histone Deacetylases Positively Regulate Transcription through the Elongation Machinery. Cell Rep 2015;13:1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009;138:1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krajewski D, Kaczenski E, Rovatti J, Polukort S, Thompson C, Dollard C et al. Epigenetic Regulation via Altered Histone Acetylation Results in Suppression of Mast Cell Function and Mast Cell-Mediated Food Allergic Responses. Front Immunol 2018;9:2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Assem E-SK, Peh KH, Wan BYC, Middleton BJ, Dines J, Marson CM. Effects of a selection of histone deacetylase inhibitors on mast cell activation and airway and colonic smooth muscle contraction. Int Immunopharmacol 2008;8:1793–1801. [DOI] [PubMed] [Google Scholar]
- 72.Bos IST, Gosens R, Zuidhof AB, Schaafsma D, Halayko AJ, Meurs H et al. Inhibition of allergen-induced airway remodelling by tiotropium and budesonide: a comparison. Eur Respir J 2007;30:653–661. [DOI] [PubMed] [Google Scholar]
- 73.Gaudenzio N, Sibilano R, Marichal T, Starkl P, Reber LL, Cenac N et al. Different activation signals induce distinct mast cell degranulation strategies. J Clin Invest 2016;126:3981–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.