Abstract
The levels and subcellular localizations of proteins regulate critical aspects of many cellular processes and can become targets of therapeutic intervention. However, high-throughput methods for the discovery of proteins that change localization either by shuttling between compartments, by binding larger complexes, or by localizing to distinct membraneless organelles are not available. Here we describe a scalable strategy to characterize effects on protein localizations and levels in response to different perturbations. We use CRISPR-Cas9-based intron tagging to generate cell pools expressing hundreds of GFP-fusion proteins from their endogenous promoters and monitor localization changes by time-lapse microscopy followed by clone identification using in situ sequencing. We show that this strategy can characterize cellular responses to drug treatment and thus identify nonclassical effects such as modulation of protein–protein interactions, condensate formation, and chemical degradation.
Currently available mass-spectrometry methods (Rix and Superti-Furga 2009; Martinez Molina et al. 2013; Savitski et al. 2014; Huber et al. 2015; Drewes and Knapp 2018) for monitoring the effects of cellular perturbations on proteomes cannot be scaled efficiently to monitor time-dependent effects in high throughput. A different approach to study drug action is live-cell imaging of protein dynamics in cells expressing a protein of interest fused to a fluorescent tag. Traditionally, such reporter cells are generated either by overexpression to nonphysiologic levels, by oligonucleotide-directed homologous recombination in yeast, or by using CRISPR-Cas9 and homology-directed repair (HDR) to endogenously tag proteins in human cells (Ghaemmaghami et al. 2003; Huh et al. 2003; Chong et al. 2015; Leonetti et al. 2016). In addition to those targeted approaches, “gene trapping” or “CD-tagging” strategies, which rely on the random, viral integration of fluorescent tags as synthetic exons, have been used for analyzing dynamic changes in response to drugs (Jarvik et al. 1996; Morin et al. 2001; Cohen et al. 2008; Kang et al. 2016), but they are limited by integration site biases and require the isolation and characterization of clones before using them in an arrayed format. Recently, a strategy combining genome engineering and gene trapping using homology-independent CRISPR-Cas9 editing to place a fluorescent tag as a synthetic exon into introns of individual target genes has been described (Serebrenik et al. 2019). The strategy relies on a generic sgRNA excising a fluorescent tag flanked by splice acceptor and donor sites from a generic donor plasmid, which is coexpressed with a gene-specific intron-targeting sgRNA specifying the integration site. Here we show the scalability of that strategy to enable pooled protein tagging of more than 900 metabolic enzymes and epigenetic modifiers. Exposing the GFP-tagged cells to compounds allows us to monitor drug effects on the localization and levels of hundreds of proteins in real time in a pooled format, followed by identification of responding clones by in situ sequencing of the expressed intron-targeting sgRNA that corresponds to the tagged protein (Fig. 1A).
Figure 1.
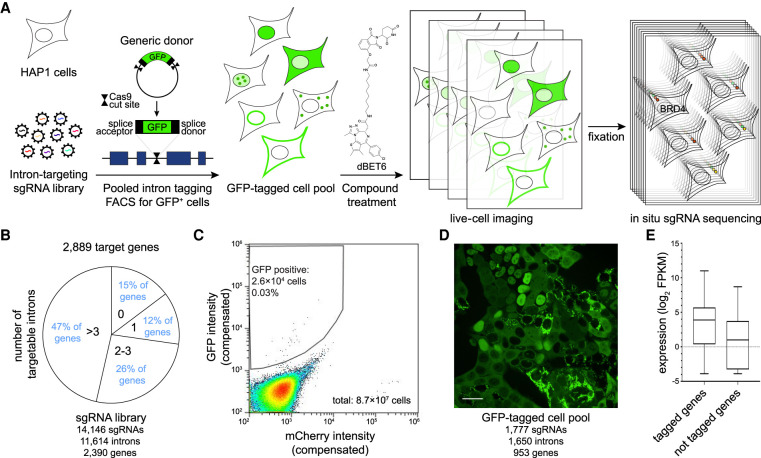
Pooled GFP intron-tagging of metabolic enzymes. (A) Schematic outline of the approach. (B) Identification of targetable introns within metabolic genes. (C) FACS sorting of clones with successful GFP-tagging by signal enrichment over background mCherry intensity used as control for autofluorescence. (D) Representative image of sorted GFP-tagged cell pool. Scale bar, 25 µm. (E) Comparison of RNA-seq expression in HAP1 cells between genes for which GFP-tagged cells could be isolated and genes that were targeted in the sgRNA library but did not result in successful clone isolation.
Results
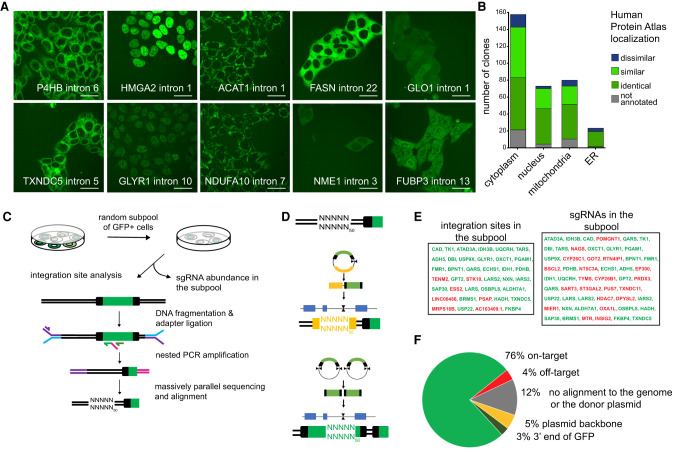
We selected to target 2889 genes comprising all classic metabolic enzymes (Birsoy et al. 2015; Corcoran et al. 2017) and epigenetic modifiers. For the 2387 genes from this set that harbor targetable introns in the selected reading frame, we designed a library comprising 14,049 sgRNAs targeting 11,614 introns (Fig. 1B; Supplemental Table S1). To generate a pool of GFP-tagged cells, we transduced HAP1 cells with that sgRNA library followed by cotransfection with a GFP donor plasmid and a plasmid expressing Cas9 and the donor-targeting sgRNA. We enriched for transfected cells using blasticidin for 24 h and sorted GFP-positive cells 6 d after transfection (Fig. 1C). Massively parallel sgRNA amplicon sequencing of the pool of GFP-positive cells identified 1777 sgRNAs targeting 1650 introns of 953 genes as highly enriched in the GFP-positive cell pool (Fig. 1D, Supplemental Table S2). Compared with genes for which intron targeting sgRNAs did not result in isolation of GFP-positive cells, successfully targeted genes have higher average expression in HAP1 cells (Fig. 1E; Schick et al. 2019). By single-cell dilution, we then isolated 335 clonal cell lines for which a massively parallel multiplex sgRNA amplicon sequencing strategy unambiguously identified the integrated sgRNAs indicating a single tagged protein (Supplemental Note S1; Supplemental Table S3). In all these clones, we mapped GFP localization (Fig. 2A; Supplemental Note S1), which in the majority of our cell lines was either cytoplasmic, nuclear, or mitochondrial, with some proteins showing a typical ER localization pattern (Fig. 2B). For 299 of the clonal cell lines, antibody-based annotations of the subcellular localization of the tagged protein are available on The Human Protein Atlas (Thul et al. 2017), and in 90% of those clones, the protein localization is either identical (all main and additional localizations are the same) or similar (additional localizations in either the clonal cell line or The Human Protein Atlas). For 36 clonal cell lines, there is no previous localization data available for the tagged protein, showing how pooled protein tagging can be used to characterize those proteins. Furthermore, 35 proteins were represented by multiple cell lines harboring the GFP tag at different introns. We observed that for 29 of those 35 proteins, the subcellular protein localization is the same when targeted at different introns, again illustrating that in the majority of editing events GFP tagging does not interfere with protein localization. However, some independent clones for the same tagged protein showed differences in fluorescence intensities. Therefore, we implemented a sequencing approach to directly analyze genomic GFP integration sites in a subset of cells isolated from the pool and compared them to the desired tagging sites based on the sgRNA sequence (Fig. 2C). In particular, we wanted to analyze whether off-target integrations, or aberrant integrations of the entire plasmid or of multiple GFP tags, can be observed (Fig. 2D). We observed that the vast majority of GFP integration sites identified by massively parallel sequencing in the isolated cell pool have a corresponding sgRNA sequence in that cell pool (Fig. 2E). Overall, 76% of sequencing reads are indicative of on-target GFP integrations, 5% map to integration of the plasmid backbone and 3% indicate multiple GFP insertions (Fig. 2F). Four percent of reads of integrations sites identified in that subpool did not have a corresponding sgRNA and are most likely off-target integrations of the donor plasmid. In additional subpools that were analyzed, we observed similar rates of plasmid backbone and multiple GFP integrations, but lower on-target rates and an increased number of reads that cannot be mapped to the genome or plasmid, indicating that there are editing events that cannot be mapped using this strategy (Supplemental Table S4). It should be noted that also in cases in which the plasmid backbone or additional GFP sequences are integrated, the insertion site cannot be mapped with our approach. Such clones likely reflect those sgRNAs in the pool for which no corresponding integration site has been found (Fig. 2E). They will still harbor the GFP tag on the desired protein, but the larger insertions may alter protein levels and the additional GFP will increase fluorescence intensities.
Figure 2.
Subcellular protein localizations and GFP integration sites in GFP-tagged clones isolated from the pool of tagged cells. (A) Representative images of individual clones isolated by single-cell dilution and identified by massively parallel sgRNA sequencing. Scale bars, 25 µm. (B) Comparison of localizations of 335 individually isolated clones to localization annotations in The Human Protein Atlas. (C) Outline of integration site analysis in a subpool of approximately 50 different GFP-tagged cells. (D) The 50-bp region upstream of the integration site aligns either to an on- or off-target genomic region or to the plasmid backbone in case the donor was only cut once, leading to integration of the whole donor plasmid, or to the 3′ end of an additional GFP fragment in case of a double integration event. (E) Integration sites and gene names of sgRNAs identified in the subpool by massively parallel sequencing (ranked by abundance). (F) Alignment of the identified integration sites in the subpool.
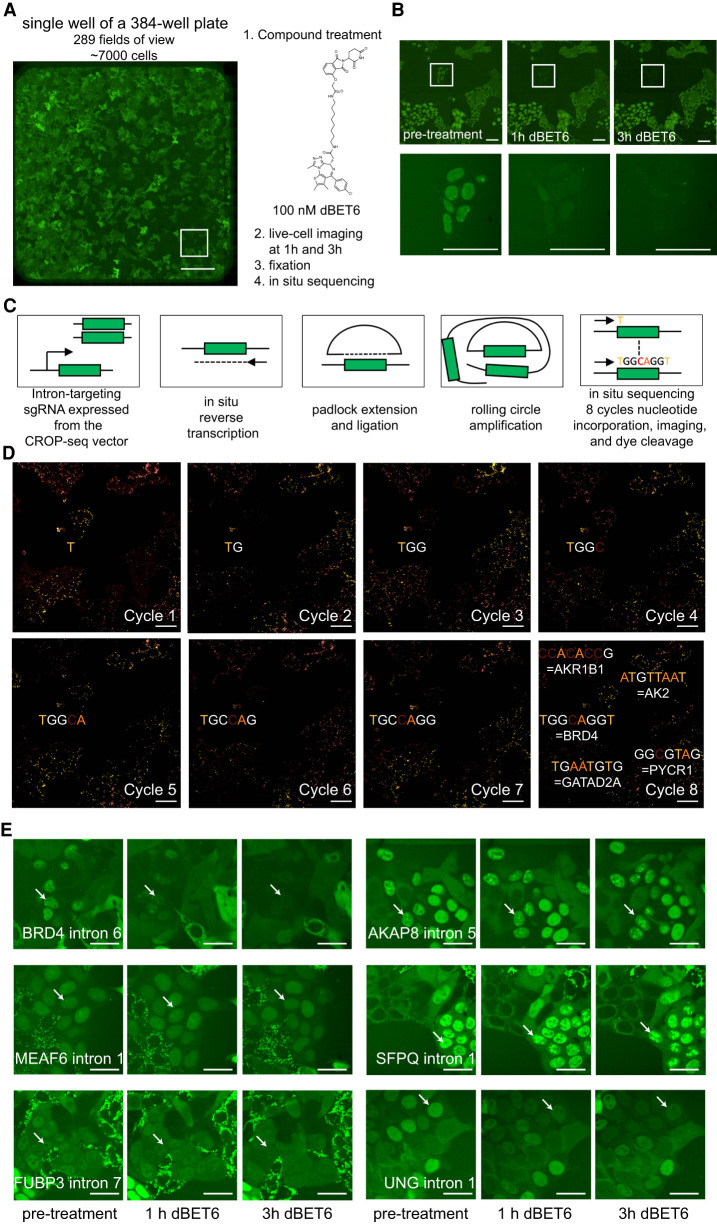
We reasoned that the highly diverse pool of cells expressing GFP-tagged proteins can be used to identify compounds that change protein levels or localization of any of the tagged proteins. Therefore, we treated the cell pool with the BRD4-targeting PROTAC dBET6 (Winter et al. 2017) and used high-content live-cell imaging to track protein dynamics of GFP-tagged proteins over 3 h in approximately 7000 cells in a single well on a 384-well plate (Fig. 3A). We observed a drastic loss of GFP signal in selected clones already 1 h after compound treatment. These clones had a nuclear GFP localization pattern with few selected foci, compatible with the known phase separation behavior of BRD4 (Fig. 3B; Supplemental Fig. S1; Sabari et al. 2018). The application of the CROP-seq vector (Datlinger et al. 2017) that expresses the sgRNA sequence in a polyadenylated mRNA transcript enabled us to cell-specifically identify the targeted intron by situ sequencing (Larsson et al. 2010; Ke et al. 2013; Feldman et al. 2019). To identify the sgRNA sequence integrated into individual cells, we fixed the cell pool and developed a two-color in situ sequencing protocol compatible with the presence of the GFP tag (Fig. 3C; Supplemental Movie S1). Based on the library diversity, eight cycles of nucleotide incorporation and imaging were sufficient to unambiguously assign sgRNA sequences. Application of this protocol to the cell pool confirmed that in clones with drastic loss of signal, GFP was indeed targeted to BRD4 (Fig. 3D). Analysis of the entire cell pool suggested several other effects of the compound, including the loss of subnuclear localization patterns of MEAF6 and FUBP3, gain of nuclear foci of AKAP8 and SFPQ, and loss of nuclear intensity for UNG (Fig. 3E), none of which are identifiable by global proteomics profiling (Winter et al. 2017).
Figure 3.
Compound screening on cell pools followed by in situ sequencing enables the detection of protein-specific compound effects. (A) Stitched image of 289 fields of view representing an entire well on a 384-well plate containing approximately 7000 individual cells. Scale bar, 500 µm. (B) Identification of a clone with rapid loss of GFP signal following treatment with 100 nM dBET6, whereas neighboring clones are unaffected. Scale bars, 50 µm. (C) Outline of the in situ sequencing approach. (D) Images from eight cycles in situ sequencing of the area shown in panel B. Scale bar, 25 µm. (E) Selected images for cells within the pool showing localization changes following dBET6 treatment. Scale bars, 25 µm.
We then tested whether the cell pool also reveals complex cellular responses to compounds that act by conventional mechanisms. We therefore treated the cell pool with methotrexate (MTX), an antimetabolite impairing DNA and RNA synthesis and causing DNA damage by inhibiting tetrahydrofolate metabolism. We observed changes to the localizations of several proteins in the cell pool (Supplemental Fig. S5). Some of our findings are consistent with the known effects of the drug. For example, in cell lines expressing either GFP-tagged RPA1 or RPA2, which are part of a heterotrimeric DNA single-strand binding complex, we observed the formation of nuclear foci in response to treatment, presumably by the recruitment of the proteins to sites of DNA damage (Raderschall et al. 1999).
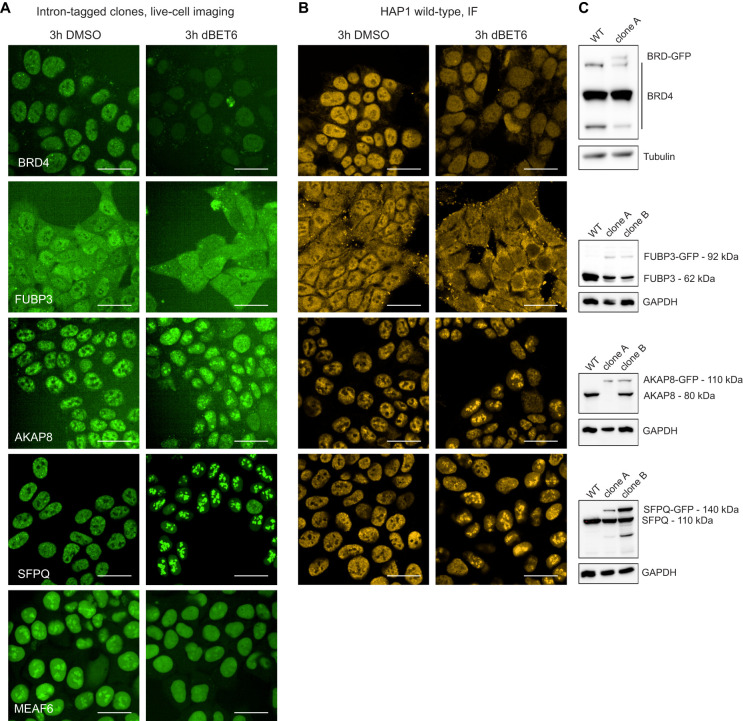
To validate the observations we made on cell pools, we first generated novel individual clones for these candidate factors using the same intron tagging strategy in an arrayed format with individual intron-targeting sgRNAs (Fig. 4A; Supplemental Fig. S2). Although changes in UNG and ACLY localization appear to be caused by cell cycle effects, dBET6 treatment confirmed changes in nuclear signal of MEAF6 and FUBP3 and gain of foci of AKAP8 and SFPQ. For FUBP3, AKAP8, and SPFQ, for which high-quality antibodies were available, we could also validate these findings on endogenous protein in untagged wild-type cells (Fig. 4B). Validation of the integration in GFP-tagged clones by western blot (Fig. 4C), PCR, and Sanger sequencing (Supplemental Figs. S3 and S4) confirmed the on-target integration but also highlighted differences in abundance of the tagged protein compared with wild-type levels. Such effects might be technical owing to reduced antibody affinity but more likely reflect reduced levels owing to splicing defects or changes in protein folding or stability. However, we observe that the relative cell-specific changes caused by compound treatment are nevertheless highly relevant and, in the majority of cases, can be validated on endogenous wild-type proteins.
Figure 4.
Validation of the compound effects in GFP-tagged clonal cell lines and wild-type cells. (A) Effects of treatment with 100 nM dBET6 for 3 h in newly generated GFP-tagged clonal cell lines. (B) Effects of treatment with 100 nM dBET6 for 3 h in HAP1 wild-type cells shown by immunofluorescence staining. (C) Western blot of wild-type HAP1 cells and GFP-tagged clonal cell lines.
Discussion
We here described the first large pool of GFP-tagged human cells generated using intron tagging. Compared with other large-scale approaches based on CD tagging that have been used to generate collections of fluorescently tagged cells for analyzing dynamic changes in response to drug treatments (Cohen et al. 2008; Kang et al. 2016), our approach enables specifying both the tagged genes and introns for targeted generation of cell pools. Additionally, by combining intron tagging with in situ sequencing, the drug treatment can be performed in a pooled format as opposed to the arrayed screening that first requires the isolation and characterization of individual tagged clones.
As every tagging event, GFP insertion by intron tagging bears the risk of altering protein function. Although all functions cannot be comprehensively assessed for the large number proteins represented in the cell pool, at least regarding localization, we observe that for the majority of proteins the GFP tag does not cause alterations. For those genes for which GFP integrations at sites accessible by intron tagging do affect function and for genes that do not contain targetable introns, tagging at exonic sites (Lackner et al. 2015), tagging using homologous recombination (Leonetti et al. 2016), or targeted integrations using prime editing (Anzalone et al. 2019) are alternatives compatible with our overall strategy.
We here showed that the generation of targeted GFP-tagged cell pools enables the identification of cellular responses to perturbations by time-lapse microscopy. In contrast to indirect approaches that measure transcription changes (Lamb et al. 2006; Subramanian et al. 2017), this method directly follows proteins as primary targets of most drugs. Its low cost and fast timescales enable applications both in large-scale screening and in the deep phenotypic characterization of dose-dependence and response kinetics. This approach is especially useful for the discovery and development of PROTACs and molecular glue degraders, for which activity can easily be determined by the disappearance of the tagged protein. However, also for classical drugs like MTX, the method not only confirmed known phenotypes but also uncovered novel previously undescribed protein localization changes. More broadly, intron tagging can easily be applied for other sets of genes beyond metabolic enzymes and potentially in a genome-wide manner to study protein dynamics at scale not only in response to drug treatment or other physiological perturbations.
Methods
Generation of an intron-targeting sgRNA library
To design an intron-targeting sgRNA library for metabolic enzymes and epigenetic modifiers, we started with a list of 2889 genes by combining a published list of all classic metabolic enzymes (Corcoran et al. 2017), most genes in a human CRISPR metabolic gene knockout library (Birsoy et al. 2015), as well as genes annotated with the Gene Ontology (GO) terms “histone modification,” “DNA methylation,” or “DNA demethylation.” We then used the Ensembl BioMart data mining tool to obtain chromosomal coordinates of introns of the primary transcripts of those genes and selected only those introns in which integration of our donor plasmid does not lead to frameshift mutations after splicing, because our donor plasmid starts with a full codon and is not compatible to all exon–exon junctions. By using Ensembl BioMart, this filtering was performed by only selecting introns that are preceded by an exon with the attribute “end phase = 0.” We then used GuideScan (Perez et al. 2017) to obtain the top 20 guides for each selected intronic region based on the GuideScan cutting efficiency score. Those 20 guides we then ranked based on a combined on- and off-target score using the scores provided by GuideScan. For genes that have only one intron that can be targeted, we selected up to three sgRNAs per intron; for genes with two or three introns that can be targeted, we selected up to two sgRNAs per intron; and for genes that have more than three introns that can be targeted, we selected the top ranked sgRNA of each intron. By using that strategy, we selected 14,049 sgRNAs targeting 11,614 introns of 2387 genes. We also added 75 nontargeting sgRNAs to our library that we obtained from the human Brunello CRISPR KO library (Doench et al. 2016). For cloning of our library into the CROPseq-Guide-Puro vector (Addgene 86708) (Datlinger et al. 2017) using Gibson Assembly, we added adapter sequences to our sgRNA sequences and ordered the 74 nucleotide oligos as an oligo pool (Twist Biosciences). Additional adapters were added to the pooled oligos by PCR (eight cycles, NEB Q5) to generate fragments with a size of 140 nt that were purified (Qiagen MinElute PCR purification) before being used for Gibson Assembly. The vector was digested with BsmBI (NEB), size-selected using agarose gel electrophoresis, and gel-purified (Qiagen QIAquick gel extraction kit) followed by an additional column purification (Qiagen QIAquick PCR purification kit). Four Gibson Assembly reactions (10 µL NEBuilder HiFi DNA assembly, 60-ng vector, 10-ng insert) were prepared and incubated for 45 min at 50°C. Reactions were pooled and purified (Qiagen MinElute PCR purification) before being used for transformation in Lucigen Endura electrocompetent bacteria (four reactions, 25 µL each). Bacteria were plated on four 245 × 245 × 25 mm bioassay dishes and dilution plates (1:10,000) and incubated for 16 h at 32°C. Cells were scraped off the plates, and plasmid DNA was extracted using multiple Qiagen plasmid plus midi kits. Library coverage was 211× and was estimated based on the number of colonies on the dilution plates.
Cloning
The GFP-donor plasmid with the coding sequence of EGFP flanked by generic sgRNA targeting sites, splice acceptor and splice donor sites, and 20-amino-acid linkers was assembled from four fragments using Gibson Assembly to generate a donor plasmid that is similar in design to a previously published donor plasmid that can be used for intron tagging (Serebrenik et al. 2019). The DNA fragment with a 25-nt overlap to the pUC19 vector and 32-nt overlap to the N terminus of EGFP was generated from overlapping oligos (Sigma-Aldrich) and comprises a generic sgRNA targeting site that is not present in the human genome (He et al. 2016) followed by a splice acceptor site (Guzzardo et al. 2017) and a flexible 20-amino-acid glycine-serine linker. This fragment is followed by a fragment with the coding sequence of EGFP without a start or stop codon that was generated by PCR. The third fragment has a 27-nt overlap to the C terminus of EGFP and a 25-nt overlap to the pUC19 vector and was generated from overlapping oligos (Sigma-Aldrich) and comprises a flexible 20-amino-acid glycine-serine linker followed by a splice donor site (Guzzardo et al. 2017) and the generic sgRNA targeting site. The pUC19 vector was linearized by PCR for Gibson Assembly (NEBuilder HiFi DNA assembly) with the other three fragments.
The pX330 plasmid expressing Cas9 and the generic sgRNA targeting the donor plasmid was generated by digesting pU6-(BbsI)_CBh-Cas9-T2A-mCherry (Addgene 64324) (Chu et al. 2015) with BbsI followed by ligation with an annealed oligo duplex as described previously (Ran et al. 2013). mCherry was replaced with a blasticidin resistance (BSD) using Gibson Assembly. Intron-Tagging-EGFP-Donor (Addgene plasmid 159740), Intron-Tagging-pX330-Cas9-Blast (Addgene plasmid 159741), and Intron-Tagging-pX330-Cas9-mCherry (Addgene plasmid 159742) will be made available via Addgene.
Pooled protein tagging
To generate lentiviral particles, HEK293T cells were transiently transfected with the intron-targeting library and packaging plasmids psPAX2, pMD2.G using PEI transfection. After 12 h, the media were replaced with IMDM supplemented with 10% FBS and penicillin-streptomycin. Viral supernatant was collected 48 h after transfection and stored at −80°C. HAP1 cells were transduced with virus and selected with puromycin for 3 d. Multiplicity of infection (MOI) was 0.2, and transduction was performed at a coverage of 500×. After puromycin selection, cells were grown for 1 d in media without puromycin before being seeded for transfection (8 million cells per 15-cm dish, 48 million cells in total). One day after seeding, each dish was cotransfected with 20 µg pX330 expressing Cas9-BSD and the generic sgRNA and 10 µg EGFP donor plasmid with 90 µL TurboFectin in 2.5 mL Opti-MEM as described by the manufacturer. Transfection efficiency was ∼10% as determined by a transfection performed in parallel with pX330 Cas9-mCherry and the EGFP donor plasmid using the same ratio. The next day, cells were subjected to a transient selection using blasticidin (10 µg/mL) for 24 h. After selection, cells were maintained in full media without blasticidin and sorted 5 d after transfection by flow cytometry using a Sony cell sorter SH800ZD; 0.03% cells were selected as GFP-positive relative to mCherry used as autofluorescence control. In total, 24,300 of those GFP-positive cells were sorted, and the cell population was expanded for 7 d before DNA was isolated to determine sgRNA abundance in the cell population.
Massively parallel sequencing
To generate a sequencing library, genomic DNA from 1 million cells of the GFP-positive cell population was isolated, and the sgRNA region was amplified by PCR (two reactions using 500 ng genomic DNA, NEB Q5 high-fidelity Polymerase). Illumina adapter ligation and sequencing were performed by a commercial sequencing service. To determine sgRNA abundance, sgRNA sequences were extracted from sequencing reads using cutadapt, and sgRNA read counts were determined using the MAGeCK count function to match the extracted reads to the sgRNA library. Of the 14,049 sgRNAs in the library, we considered 1777 as highly enriched as these sgRNAs accounted for 90% of the obtained sequencing reads, whereas the majority of sgRNAs was not detectable anymore. The remaining 10% of sequencing reads comprise an additional 1622 sgRNAs that we do not consider as enriched, as each of them is only supported by a few sequencing reads that might be the result of cells being transduced with two sgRNAs or the result of off-target integration and expression of the GFP-tag. Our library also includes 75 nontargeting sgRNAs, making up 0.53% of the sgRNAs in our library. As expected, they are depleted in the pool of GFP-positive, making up 0.15% of the sequencing reads with only three nontargeting sgRNAs among the 1777 sgRNAs we consider enriched.
Isolation, imaging, and sequencing of clonal cell lines
To obtain clonal cell lines, cells were seeded at a concentration of 0.7 cells per well in 96-well cell culture plates. After 9 d of clonal expansion, 720 colonies were harvested using trypsin, and cell suspensions were transferred in equal amounts to eight 96-well imaging plates (PerkinElmer CellCarrier Ultra) and eight corresponding 96-well cell culture plates. After 24 h, cells on the imaging plates were imaged on a PerkinElmer Opera Phenix high content screening system (five fields of view per well, 63× water-immersion objective, confocal mode, excitation laser: 488 nm, emission filter: 500–550 nm, 700-msec exposure time). TIFF images of all 720 wells can be found in Supplemental Data S1. Images were processed using CellProfiler. To identify the intron-targeting sgRNAs expressed in imaged cells, we performed multiplexed amplicon sequencing of the sgRNA regions in the corresponding clones on the eight 96-well cell culture plates. Cells were lysed, and cell lysates were used for PCR to amplify the sgRNA region in each clone using barcoded primers flanking the sgRNA region (36 different 5-mers added to the 5′ end of the forward primer and 24 different 5-mers added to the 5′ end of the reverse primer; 720 of all possible 864 combinations were used). PCR reactions were pooled and column-purified before being sent for sequencing by a commercial sequencing service (Genewiz). Sequencing reads were demultiplexed using cutadapt (Martin 2011), and sgRNA read counts for each individual well were obtained using MAGeCK (Li et al. 2014). For further analysis, we excluded clones for which we either had no cells in any of the five fields of view that were imaged or no sequencing reads for the corresponding well or for which we observed polyclonal cell populations as determined by imaging or detection of multiple sgRNAs per well. By using that strategy, we obtained images of 335 clones for which we could identify the expressed intron-targeting sgRNA corresponding to the tagged protein.
Comparison of subcellular localization to The Human Protein Atlas
Comparison of subcellular protein localizations of GFP-tagged protein in 335 clones to the localization patterns as annotated on The Human Protein Atlas was performed as described previously for the comparison of N- or C-terminally GFP-tagged proteins to IF-based annotations on The Human Protein Atlas (Stadler et al. 2013). Briefly, the overlap was defined as “identical” if one or multiple main and additional localizations were the same in the intron-tagged clone compared with The Human Protein Atlas, “similar” if one localization is the same in the clone compared with The Human Protein Atlas with additional localization(s) observed either in the clone or on The Human Protein atlas, or “dissimilar” if there were no common subcellular localization patterns. We did not take into account extended localization annotations such as nucleoplasm, nuclear speckles, or nucleoli that we all considered nuclear.
Integration site analysis
Analysis of genomic GFP integration sites was performed on small subpools of clones isolated from the pool of GFP-positive cells. To obtain random subpools, cells were seeded at a concentration of 50 cells per well and expanded for 2 wk. Genomic DNA was isolated from 5 × 105 cells to determine the sgRNA abundance in the subpools as described above and to perform an integration site analysis. To prepare DNA libraries for sequencing of integration sites, genomic DNA was first fragmented by sonication using a Bioruptor Pico (Diagenode) to obtain DNA fragments of an average size of 500 bp (500 ng in 50 µL 1 × TE buffer, 15 sec on, 30 sec off, two cycles). The NEBNext Ultra II DNA library prep kit for Illumina was used for end repair of DNA fragments, adapter ligation, and cleavage of the hairpin adapter as described by the manufacturer, but with a modified adapter instead of the provided NEBNext adapter. The modified adapter has the same hairpin structure as the NEBNext adapter, including a cleavable uracil, but does not have a binding site for the Illumina p5 indexing primers. To enrich for fragments containing GFP and to add the binding site for the Illumina p5 index primers to the fragments, a nested PCR was performed with adapter-ligated and column-purified DNA (Qiagen MinElute PCR purification). In the first PCR reaction, a forward primer binding the p7 part of the adapter and a reverse primer binding GFP was used (NEB OneTaq HotStart 2× master mix, 18 cycles), and 10% of the reaction was used as a template in the second PCR using the same forward primer binding the p7 part of the adapter and a nested reverse primer binding GFP and an overhang providing a binding site for Illumina p5 index primers (NEB OneTaq HotStart 2× master mix, 18 cycles). PCR products were purified and library amplification was analyzed by gel electrophoresis before being sent for sequencing by a commercial sequencing service (2 × 250-bp paired-end sequencing).
To map genomic integration sites to the human genome, the forward sequencing reads starting within the GFP fragment, extending beyond the splice acceptor and the 5′ junction into the genomic DNA of the integration site, were analyzed. First, cutadapt was used to filter for reads with a minimum length of 210 bp to exclude shorter reads that do not extend at least 68 bp beyond the 5′ junction into the genomic DNA. These reads were then shortened to a length of 50 bp so that remaining trimmed sequencing reads that were used for aligning to the genome start 68 bp upstream of the integrated fragment (assuming no insertions or deletions) and end 18 bp upstream of the integrated fragment. Bowtie 2 (Langmead and Salzberg 2012) was used for aligning these 50-bp reads to the hg38 version of the human genome, and featureCounts and the hg38 Ensembl GTF file were used to annotate the obtained alignments with gene names and count the identified integration site per gene or nonannotated regions. Additionally, Bowtie 2 was used to align reads to plasmid backbone, specifically the 210-bp region upstream of the sgRNA cut site on the donor plasmid and to the 3′ end of the donor fragment, where alignment would indicate two integrated GFP fragments.
Live-cell imaging
Live-cell imaging was performed on a PerkinElmer Opera Phenix microscope with excitation laser 488 nm, and emission filter 500–550 nm, 700-msec exposure time.
In situ sequencing
Identification of the expressed sgRNAs by in situ sequencing was performed by following and modifying published protocols (Larsson et al. 2010; Ke et al. 2013; Feldman et al. 2019). After live-cell imaging after treatment with MTX or dBET6, cells were fixed with 4% paraformaldehyde for 30 min, washed with PBS, permeabilized with 70% ethanol for 30 min, and washed with PBS-T (PBS + 0.05% Tween-20) twice. Reverse transcription mix (1× RevertAid RT buffer, 250 µM dNTPs, 0.2 mg/mL BSA, 1 µM RT primer, 0.8 U/mL Ribolock RNase inhibitor, and 4.8 U/mL RevertAid H minus reverse transcriptase) was added to the sample and incubated for 16 h at 37°C. Following reverse transcription, cells were washed five times with PBS-T and postfixed with 3% paraformaldehyde and 0.1% glutaraldehyde for 30 min at room temperature and washed five times with PBS-T. Cells were incubated in a padlock probe and extension-ligation reaction mix (1× Ampligase buffer, 0.4 U/mL RNase H, 0.2 mg/mL BSA, 100 nM padlock probe, 0.02 U/mL KlenTaq polymerase, 0.5 U/mL Ampligase, and 50 nM dNTPs) for 5 min at 37°C and 90 min at 45°C and then washed 2 times with PBS-T. Circularized padlocks were amplified with rolling circle amplification mix (1× Phi29 buffer, 250 µM dNTPs, 0.2 mg/mL BSA, 5% glycerol, and 1 U/mL Phi29 DNA polymerase) for 4 h at 30°C. Rolling circle amplicons were prepared for sequencing by hybridizing a mix containing sequencing primer oSBS_CROP-seq (1 µM primer in 2× SSC + 10% formamide) for 30 min at room temperature. Barcodes were read out using sequencing-by-synthesis reagents from the Illumina NextSeq 500/550 kit v2 (Illumina 15057934). First, samples were washed with incorporation buffer (NextSeq 500/550 buffer cartridge, position 35) and incubated for 4 min in incorporation mix (NextSeq 500/550 reagent cartridge, position 31) at 60°C. Samples were then washed with incorporation buffer (four washes, for 4 min at 60°C at the last wash) and placed in scan mix (NextSeq 500/550 reagent cartridge, position 30) for imaging. Imaging was performed on a PerkinElmer Opera Phenix microscope using a 63× water immersion objective in confocal mode on the yellow channel (excitation laser: 561 nm, emission filter: 570-630, 500-msec exposure time) and the red channel (excitation laser: 640 nm, emission filter: 650–760 nm, 500-msec exposure). Bases were detected as follows: base T, signal in the 561 nm channel; base C, signal in the 640 nm channel; base A, signal in both channels; base G, no signal. Following each imaging cycle, samples were washed with the cleavage mix (NextSeq 500/550 reagent cartridge, position 29) once followed by incubation with cleavage mix for 4 min at 60°C to remove dye terminators. Samples were washed five times with incorporation buffer before starting the next cycle.
Image analysis of in situ sequencing
Base calling and sgRNA identification in cells in the GFP pool responding to dBet6 or MTX treatment was performed manually by analyzing in situ sequencing spots only in the respective cells of interest (eight cycles of in situ sequencing analyzed in one field view). For identification of sgRNAs in cells in a complete well of a 384-well plate, spot detection and base calling were partially automated using ImageJ and CellProfiler (McQuin et al. 2018). First, all 289 fields of view per in situ sequencing cycle were merged using ImageJ for alignment of all cycles using the DAPI image. For further analysis, the merged images covering the whole well were split into nine tiles to create smaller images that can be processed using CellProfiler. The CellProfiler pipeline can be found in the Supplemental Data S2 together with the output and calculation table and example images. In brief, for images of each cycle, the “EnhanceOrSupressFeatures” module in CellProfiler was used to enhance foci speckles in the red and yellow channel. Then, the “IdentifyPrimaryObjects” module was used to detect foci based on object size and based on an automatically calculated threshold that adapts to differences in background intensities in the different cycles. For all foci detected in either of the two channels, the maximum intensity values in both the red and yellow channel were determined using the “MeasureObjectIntensity” module. Then, the “RelateObjects” module was used to determine which foci belongs to which cell. Cells were identified by first detecting nuclei using the DAPI image followed by identification of the cell by using the “IdentifySecondaryObject” module and the background staining in the yellow channel to determine cell boundaries. Identification of cell objects was performed with the images of the last imaging cycles only and used to relate foci in all cycles to the correct cell. The output of the CellProfiler analysis was a table of all detected foci with maximum intensity values and the identity and spatial location of the overlapping cell. By using that table, the first seven bases of the sgRNA sequence present in each cell was determined by determining a base for each cell and cycle. Because we used a two-color sequencing chemistry in which the base “A” is characterized by a signal in both channels, we first used the maximum intensity values in the red and yellow channel of foci detected in the yellow channel to determine which foci are considered as “A” and “T.” Specifically, if the maximum intensity value in the yellow channel divided by the maximum intensity in the red channel was greater than 2, that foci was considered as “T.” If the calculated value was between 0.9 and 2, the foci was considered as “A.”
Foci in the red channel were considered as “C” if their maximum intensity value in the red channel divided by the maximum intensity value in the yellow channel was greater than 1.5. Finally, for each cell the foci considered as A, T, or C were counted, and a base for each cell and cycle was determined. If foci considered as different bases were detected in a cell in the same cycle, the cell was only considered as A, T, or C if the respective base accounted for ≥60% of all foci in that cell and cycle. Cells were considered as “G” (characterized by no signal in the two-color sequencing chemistry) for a cycle if in that cycle ≤20% foci were detected compared with the cycle of that cell with most reads. If none of that applied in any of the seven cycles or if a cell had no more than five foci in any of the seven cycles, that cell was excluded for further analysis and no sgRNA was determined. For the remaining cells, the identified sequence was compared to the first seven bases of the sgRNAs present in the pool of GFP cells to determine the tagged protein. ImageJ was used to annotate the live-cell image of the entire well with the names of the tagged proteins for better visualization of the automated in situ sequencing results.
Generation of GFP-tagged clonal cell lines
Clonal cell lines to validate hits identified with in situ sequencing were generated in an arrayed format. First, individual intron-targeting sgRNAs were cloned into the CROPseq plasmid as described previously (Datlinger et al. 2017). HAP1 cells seeded in a 12-well plate were cotransfected with 400 ng of the CROPseq plasmid with the intron-targeting sgRNA, 400 ng of the pX330 plasmid expressing Cas9 and the donor-targeting sgRNA, and 200 ng of the GFP donor plasmid using TurboFectin as described by the manufacturer. GFP-positive cells were sorted 48 h after transfection and expanded for 1 wk before single cells were sorted and expanded for further experiments.
Western blot
Cell pellets were lysed rotating for 30 min at 4°C in RIPA buffer containing 1× completeTM, EDTA-free protease inhibitor cocktail (Sigma-Aldrich 4693132001). After centrifugation for 10 min at 4°C at 13,000 rpm, the supernatant was collected and protein content was measured using the Bradford assay (AppliChem A6932). Equal amounts of protein were mixed with 4× SDS loading buffer (250 mM Tris at pH 6.8, 40% glycerol, 8% SDS, 0.08% bromophenol blue, 20% β-mercaptoethanol) and incubated for 5 min at 95°C. Samples were loaded on acrylamide gels together with a protein ladder (precision plus protein dual color standards, Bio-Rad 1610394). After gel electrophoresis, proteins were transferred to 0.45-µm nitrocellulose membranes (Amersham Protran western blotting membranes, GE10600002). After blocking in TBST + 5% nonfat dry milk, the membranes were incubated overnight at 4°C with primary antibody in 5% milk in TBST. On the next day, the membranes were washed three times with TBST and then incubated for 1 h at room temperature with secondary antibodies in 5% milk in TBST. After washing three times with TBST, membranes were developed using Clarity western ECL substrate (Bio-Rad 170-5060) and imaged on a Bio-Rad ChemiDoc MP.
Immunofluorescence staining
Cells in 96-well plates were fixed with 3.7% formaldehyde (Merck 1.04002) for 10 min at room temperature and washed three times with DPBS. For blocking and permeabilization, cells were then incubated with DPBS + 3% bovine serum albumin + 0.1% Triton X-100 for 45 min at room temperature. The blocking/permeabilization solution was removed and the primary antibodies diluted in DPBS + 3% bovine serum albumin + 0.1% Triton X-100 were added and incubated for 1 h at room temperature. Cells were washed twice with PBS and incubated with the secondary antibodies + DAPI diluted in DPBS + 3% bovine serum albumin + 0.1% Triton X-100 overnight at 4°C in the dark. On the next day, cells were washed twice with PBS and then imaged on an Opera Phenix high-content screening system.
PCR validation of GFP integration in clonal cell lines
For PCR analysis of GFP integration sites, clonal cell lines were lysed and cell lysate was used for PCR using primers producing 200–300 bp amplicons at the 5′ and 3′ junctions. To amplify the 5′ junction, an intronic primer binding upstream of the integration was used together with a primer binding to GFP and to amplify the 3′ junction, an intronic primer binding downstream from the GFP integration together with a primer binding GFP was used. PCR products were analyzed by gel electrophoresis and Sanger sequencing.
Data access
Sequence information of the donor plasmid, a list of primers and oligos, and a list of antibodies used in this study can be found in Supplemental Table S5. TIFF images of all single-clones are available in Supplemental Data S1. The CellProfiler pipeline, a calculation table, and example images for image analysis of the in situ sequencing can be found in Supplemental Data S2. Supplemental Data files have been published at CyVerse Data Commons (https://datacommons.cyverse.org/browse/iplant/home/shared/commons_repo/curated/Reicher_PooledProteinTagging_2020). Image data from this study have also been submitted to the Image Data Resource (https://idr.openmicroscopy.org) under accession number idr0097.
Competing interest statement
A.R. and S.K. have filed a European patent application EP19211077.3 based on the findings described in this paper.
Supplementary Material
Acknowledgments
The plasmid pU6-(BbsI)_CBh-Cas9-T2A-mCherry was a gift from Ralf Kuehn (Addgene plasmid 64324; http://n2t.net/addgene:64324; RRID:Addgene_64324). CROPseq-Guide-Puro was a gift from Christoph Bock (Addgene plasmid 86708; http://n2t.net/addgene:86708; RRID:Addgene_86708). Research in the Kubicek laboratory is supported by the Austrian Academy of Sciences, the Austrian Federal Ministry for Digital and Economic Affairs, and the National Foundation for Research, Technology, and Development, the Austrian Science Fund (FWF) F4701-614 B20 and the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (ERC-CoG-772437). A.R. is supported by a Boehringer Ingelheim Fonds PhD fellowship.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.261503.120.
Freely available online through the Genome Research Open Access option.
References
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, et al. 2019. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576: 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. 2015. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162: 540–551. 10.1016/j.cell.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong YT, Koh JL, Friesen H, Duffy SK, Cox MJ, Moses A, Moffat J, Boone C, Andrews BJ. 2015. Yeast proteome dynamics from single cell imaging and automated analysis. Cell 161: 1413–1424. 10.1016/j.cell.2015.04.051 [DOI] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. 2015. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33: 543–548. 10.1038/nbt.3198 [DOI] [PubMed] [Google Scholar]
- Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, et al. 2008. Dynamic proteomics of individual cancer cells in response to a drug. Science 322: 1511–1516. 10.1126/science.1160165 [DOI] [PubMed] [Google Scholar]
- Corcoran CC, Grady CR, Pisitkun T, Parulekar J, Knepper MA. 2017. From 20th century metabolic wall charts to 21st century systems biology: database of mammalian metabolic enzymes. Am J Physiol Renal Physiol 312: F533–F542. 10.1152/ajprenal.00601.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C. 2017. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods 14: 297–301. 10.1038/nmeth.4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34: 184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G, Knapp S. 2018. Chemoproteomics and chemical probes for target discovery. Trends Biotechnol 36: 1275–1286. 10.1016/j.tibtech.2018.06.008 [DOI] [PubMed] [Google Scholar]
- Feldman D, Singh A, Schmid-Burgk JL, Carlson RJ, Mezger A, Garrity AJ, Zhang F, Blainey PC. 2019. Optical pooled screens in human cells. Cell 179: 787–799.e17. 10.1016/j.cell.2019.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Guzzardo PM, Rashkova C, Dos Santos RL, Tehrani R, Collin P, Bürckstümmer T. 2017. A small cassette enables conditional gene inactivation by CRISPR/Cas9. Sci Rep 7: 16770 10.1038/s41598-017-16931-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Tan C, Wang F, Wang Y, Zhou R, Cui D, You W, Zhao H, Ren J, Feng B. 2016. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res 44: e85 10.1093/nar/gkw064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KV, Olek KM, Müller AC, Tan CS, Bennett KL, Colinge J, Superti-Furga G. 2015. Proteome-wide drug and metabolite interaction mapping by thermal-stability profiling. Nat Methods 12: 1055–1057. 10.1038/nmeth.3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Jarvik JW, Adler SA, Telmer CA, Subramaniam V, Lopez AJ. 1996. CD-tagging: a new approach to gene and protein discovery and analysis. BioTechniques 20: 896–904. 10.2144/96205rr03 [DOI] [PubMed] [Google Scholar]
- Kang J, Hsu CH, Wu Q, Liu S, Coster AD, Posner BA, Altschuler SJ, Wu LF. 2016. Improving drug discovery with high-content phenotypic screens by systematic selection of reporter cell lines. Nat Biotechnol 34: 70–77. 10.1038/nbt.3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, Nilsson M. 2013. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods 10: 857–860. 10.1038/nmeth.2563 [DOI] [PubMed] [Google Scholar]
- Lackner DH, Carré A, Guzzardo PM, Banning C, Mangena R, Henley T, Oberndorfer S, Gapp BV, Nijman SMB, Brummelkamp TR, et al. 2015. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun 6: 10237 10.1038/ncomms10237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. 2006. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313: 1929–1935. 10.1126/science.1132939 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Grundberg I, Söderberg O, Nilsson M. 2010. In situ detection and genotyping of individual mRNA molecules. Nat Methods 7: 395–397. 10.1038/nmeth.1448 [DOI] [PubMed] [Google Scholar]
- Leonetti MD, Sekine S, Kamiyama D, Weissman JS, Huang B. 2016. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc Natl Acad Sci 113: E3501–E3508. 10.1073/pnas.1606731113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15: 554 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P. 2013. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341: 84–87. 10.1126/science.1233606 [DOI] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, et al. 2018. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol 16: e2005970 10.1371/journal.pbio.2005970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci 98: 15050–15055. 10.1073/pnas.261408198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez AR, Pritykin Y, Vidigal JA, Chhangawala S, Zamparo L, Leslie CS, Ventura A. 2017. GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol 35: 347–349. 10.1038/nbt.3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T. 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci 96: 1921–1926. 10.1073/pnas.96.5.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rix U, Superti-Furga G. 2009. Target profiling of small molecules by chemical proteomics. Nat Chem Biol 5: 616–624. 10.1038/nchembio.216 [DOI] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361: eaar3958 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski MM, Reinhard FB, Franken H, Werner T, Savitski MF, Eberhard D, Martinez Molina D, Jafari R, Dovega RB, Klaeger S, et al. 2014. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346: 1255784 10.1126/science.1255784 [DOI] [PubMed] [Google Scholar]
- Schick S, Rendeiro AF, Runggatscher K, Ringler A, Boidol B, Hinkel M, Májek P, Vulliard L, Penz T, Parapatics K, et al. 2019. Systematic characterization of BAF mutations provides insights into intracomplex synthetic lethalities in human cancers. Nat Genet 51: 1399–1410. 10.1038/s41588-019-0477-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebrenik YV, Sansbury SE, Kumar SS, Henao-Mejia J, Shalem O. 2019. Efficient and flexible tagging of endogenous genes by homology-independent intron targeting. Genome Res 29: 1322–1328. 10.1101/gr.246413.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler C, Rexhepaj E, Singan VR, Murphy RF, Pepperkok R, Uhlén M, Simpson JC, Lundberg E. 2013. Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat Methods 10: 315–323. 10.1038/nmeth.2377 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. 2017. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171: 1437–1452.e17. 10.1016/j.cell.2017.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, et al. 2017. A subcellular map of the human proteome. Science 356: eaal3321 10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, Reyes JM, di Iulio J, Souza A, Ott CJ, et al. 2017. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol Cell 67: 5–18.e19. 10.1016/j.molcel.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.