Abstract
Retinoic acid receptor-related orphan receptor-alpha (RORα) is a transcription factor from the nuclear receptor family expressed by immune cells and involved in the development of obesity, insulin resistance (IR) and non-alcoholic steatohepatitis (NASH). It was recently reported that mice deficient for RORα in macrophages develop more severe NASH upon high fat diet (HFD) feeding due to altered Kupffer cell function. To better understand the role of RORα in obesity and IR, we independently generated a macrophage RORα-deficient mouse line. We report that RORα deletion in macrophages does not impact on HFD-induced obesity and IR. Surprisingly, we did not confirm an effect on NASH development upon HFD feeding nor in the more severe and obesity-independent choline-deficient, L-amino acid-defined diet model. Our results therefore show that RORα deletion in macrophages does not alter the development of obesity and IR and question its role in NASH.
Subject terms: Monocytes and macrophages, Kupffer cells, Non-alcoholic fatty liver disease, Non-alcoholic steatohepatitis, Obesity, Type 2 diabetes
Introduction
During the last few decades, the prevalence of obesity has dramatically increased worldwide, arising from excessive dietary intake and a sedentary lifestyle1. Obesity is a strong risk factor for type 2 diabetes (T2D) and non-alcoholic fatty liver disease (NAFLD). T2D is characterized by hyperglycemia resulting from insulin resistance (IR) and relative insulin deficiency2. The chronic and low-grade systemic inflammation developing during obesity is considered as one critical step in the pathogenesis of IR and represents a valuable therapeutic target3. In obesity, enlarged adipocytes promote the recruitment and activation of immune cells leading to the accumulation of monocytes, pro-inflammatory macrophages, neutrophils, T cells and B cells in adipose tissue (AT)4. AT macrophages (ATM) are the most abundant leukocytes in obese AT and among the most important immune cell types mediating inflammation and IR5,6. ATM secrete several pro-inflammatory cytokines such as IL-1β, TNF-α and IL-6, all interfering with the insulin signaling pathway7. Expansion of ATM mainly results from tissue recruitment of circulating monocytes followed by their differentiation into pro-inflammatory macrophages8.
NAFLD comprises a liver phenotype spectrum ranging from simple steatosis, also called non-alcoholic fatty liver (NAFL), to non-alcoholic steatohepatitis (NASH). NAFLD diagnosis is based on histology with steatosis, characterized by triglyceride accumulation in hepatocytes, being the first stage9. NAFL progresses into NASH with the appearance of lobular inflammation and hepatocyte ballooning. NASH predisposes to fibrosis and may ultimately evolve into cirrhosis and hepatocellular carcinoma10. NAFLD is strongly associated with obesity, IR and the capacity of AT to store lipids11. The AT-liver crosstalk is essential in NAFLD pathophysiology. Indeed, development of IR in AT increases free fatty acids (FFAs) release, leading to FFAs uptake and esterification into triglycerides in the liver. Excessive lipid accumulation within hepatocytes induces lipotoxicity and the release of damage-associated molecular patterns (DAMPs) that ultimately activate immune cells and favor leukocyte recruitment. Immune cells, and particularly the resident liver macrophages (Kupffer cells, KCs), play key roles in NAFLD by regulating hepatocyte metabolism, inflammation and fibrosis12,13.
These metabolic and inflammatory processes are tightly regulated at the transcriptional level by a complex network of interactions. Among them, transcription factors such as nuclear receptors play prominent roles. The retinoic acid receptor-related orphan receptors (RORs) family consists of RORα (NR1F1), RORβ (NR1F2), and RORγ (NR1F3). RORs have the common nuclear receptor structure with a variable N-terminal A/B domain, a two zinc fingers-containing DNA-binding domain (DBD), a hinge region and a C-terminal ligand-binding domain (LBD). The DBD is highly conserved between RORs and bind the same ROR response element (RORE). RORα and RORγ are widely expressed, whereas RORβ is more restricted to the central nervous system14. The RORα gene (RORA) produces four different isoforms in humans (RORα1-4) and two in mice (RORα1 and RORα4). RORα1 and RORα4 are generated by alternative promoter usage and diverge only in the N-terminal A/B domain. RORα regulates the expression of genes involved in lipid and glucose metabolism, circadian rhythm and immunity15. It has been shown that RORα regulates the development and function of several immune cell types, such as group 2 innate lymphoid cells16, Th17 cells17, IgA-producing B cells18 and macrophages19.
A growing body of evidence also suggests a role for RORα in obesity, T2D and NAFLD. In humans, mutations in the RORA gene is associated with obesity20, T2D21 and liver diseases22. The staggerer mice (Rorasg/sg), which are carrying a natural mutation in the RORα LBD, are protected against diet-induced obesity, IR and NAFLD23,24. However, due to the high expression of RORα in the cerebellum and its critical role in the development of Purkinje cells, Rorasg/sg mice exhibit a severe cerebellar ataxia, motor defect and tremor. Rorasg/sg also display increased energy expenditure which likely contributes to the protection against obesity23. Concomitantly, the development of new synthetic ligands also provided evidences for a role of RORα in IR and NAFLD25,26. Recently, hepatocyte-specific RORα deletion was reported to exacerbate NASH development27,28. However, in an independent study, no effect of hepatocyte-specific RORα deletion on NASH was observed29, leaving open the question about how RORα influences NASH. Since immune cells, and particularly macrophages, are important in the development of obesity-related diseases including T2D and NASH, we hypothesized that the role of RORα in these pathologies might in part result from its expression in macrophages. In the present work, we therefore generated a mouse line harboring RORα deletion in macrophages and evaluated its impact in models of obesity, IR and NASH.
Results
Characterization of RORα MKO mice
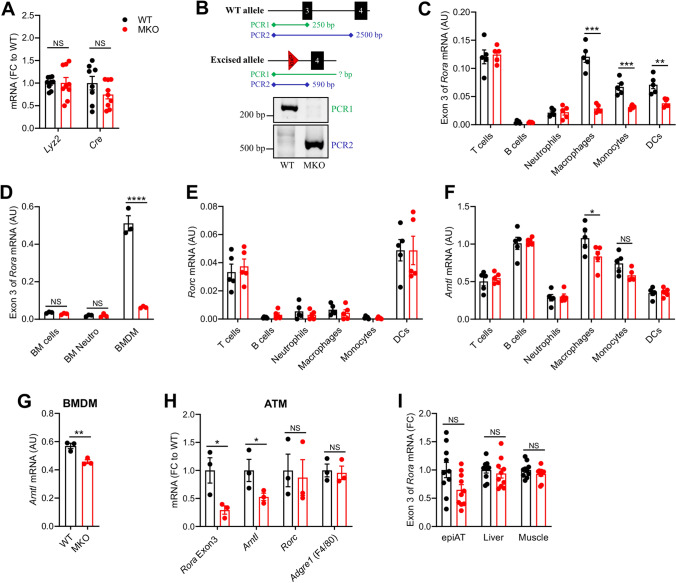
We generated Rora floxed mice by introducing loxP sites flanking exon 3 (Fig. S1A). Exon 3, the first common exon of isoforms 1 and 4, encodes for the DBD and includes the first zinc finger motif. Deletion of exon 3 leads to a frameshift resulting in a premature stop codon and the absence of functional domains (Fig. S1B,C). To achieve RORα deletion in macrophages, these mice were subsequently crossed with the LysM-Cre mice. Rorafl/fl Lyz2Cre/+ (MKO) mice and their littermate controls Rora+/+ Lyz2Cre/+ (WT), exhibiting comparable expression of Lyz2 and Cre (Fig. 1A), were further studied.
Figure 1.
Characterization of RORα MKO mice. (A) mRNA expression levels measured by RT-qPCR for Lyz2 and Cre genes in spleen from WT (Rora+/+ Lyz2Cre/+) and MKO (Rorafl/fl Lyz2Cre/+) mice. (B) PCR analysis of genomic DNA extracted from sorted splenic macrophages of WT and MKO mice. Locations of primer hybridization are represented with filled diamond. Agarose gels were acquired with a Gel Doc XR system (Bio-Rad) and the Image Lab software verion 2.0 build 8 for PC (Bio-Rad, https://www.bio-rad.com/fr-fr/product/image-lab-software?ID=KRE6P5E8Z). Images were cropped for sake of clarity and full-length gels are presented in Supplementary Fig. 6. (C) mRNA expression levels measured by RT-qPCR for the exon 3 of Rora in B cells, T cells, neutrophils, macrophages, monocytes, and dendritic cells (DCs) sorted from the spleen of WT and MKO mice. (D) mRNA expression levels measured by RT-qPCR for the exon 3 of Rora in total bone marrow (BM) cells, in neutrophils purified from BM and in BM-derived macrophages (BMDM) from WT and MKO mice. (E,F) mRNA expression levels measured by RT-qPCR for Rorc (D) and Arntl (E) genes in B cells, T cells, neutrophils, macrophages, monocytes, and DCs sorted from the spleen of WT and MKO mice. (G) mRNA expression levels measured by RT-qPCR for Arntl in BMDM. (H) mRNA expression levels measured by RT-qPCR for the exon 3 of Rora, Arntl, Rorc and Adgre1 in adipose tissue macrophages (ATM) sorted from epididymal adipose tissue of WT and MKO mice. (I) mRNA expression levels measured by RT-qPCR for the exon 3 of Rora in liver, epididymal adipose tissue (epiAT) and skeletal muscle from WT and MKO mice. Data are shown as mean ± SEM. * p < 0.05; ** p < 0.01; *** p < 0.001 by 2-way ANOVA followed by Sidak’s multiple comparisons test or unpaired t test. All statistical analyses were carried out using GraphPad Prism 8 for Windows (GraphPad Software). n = 8–10 mice per genotype for A & I. n = 5 mice per genotype for C,E & F. n = 3 mice per genotype for D & G. For H, each dot represents a pool of 3–4 mice. NS: Not significant; AU: Arbitrary unit; FC: Fold change. See also Fig. S1.
To validate and characterize the RORα deletion in MKO mice, we demonstrated the excision of exon 3 in DNA from sorted splenic macrophages (Fig. 1B). Since the LysM-Cre mouse model mainly targets macrophages and neutrophils, but also a fraction of monocytes and DCs30, we sorted these populations from spleen and quantified the deletion of exon 3 in Rora mRNA by RT-qPCR (Fig. 1C). MKO mice displayed an important deletion of exon 3 in macrophages and to a lesser extent in monocytes and DCs, but surprisingly not in neutrophils. No deletion of exon 3 in Rora mRNA was observed in T cells and B cells. To confirm the absence of deletion in neutrophils, we also analyzed the bone marrow (BM) cells which mainly contain neutrophils. Neither total BM cells nor purified neutrophils from BM showed a deletion of exon 3, while bone marrow-derived macrophages (BMDM) did (Fig. 1D). Collectively, our results show that macrophages, monocytes and DCs, but not neutrophils, exhibit a LyzM-Cre-induced deletion of Rora exon 3.
Rorc expression was not affected by Rora deletion (Fig. 1E) and Rorb was undetectable in both WT and MKO immune cells (data not shown), suggesting no compensatory regulation of other ROR genes. The expression of a key ROR target gene Arntl, encoding for BMAL1, was significantly lower in RORα-deficient splenic macrophages and BMDM (Fig. 1F,G). A tendency to a lower Arntl expression was also observed in monocytes, but not in DCs, probably due to the high expression of Rorc (Fig. 1E). Thus, despite the significant LysM-Cre-induced deletion of exon 3 in monocytes and DCs, MKO mice foremost displayed a phenotype in macrophages. As macrophages display some tissue-specific characteristics, we sorted ATM (CD45+ CD3ε− TCRβ− CD19− CD20− F4/80+ CD64+) from epididymal AT to evaluate the efficiency of Rora deletion in these cells. Similar to the splenic macrophages and BMDM, MKO mice showed a significant deletion of exon 3 and a significant decrease of the ROR target gene Arntl in ATM (Fig. 1H). No significant deletion of exon 3 occurred in total epididymal AT, liver nor skeletal muscle (Fig. 1I), suggesting the absence of an unwanted recombination in parenchymal cells of metabolic tissues.
RORα deletion in macrophages does not affect obesity, IR nor steatosis
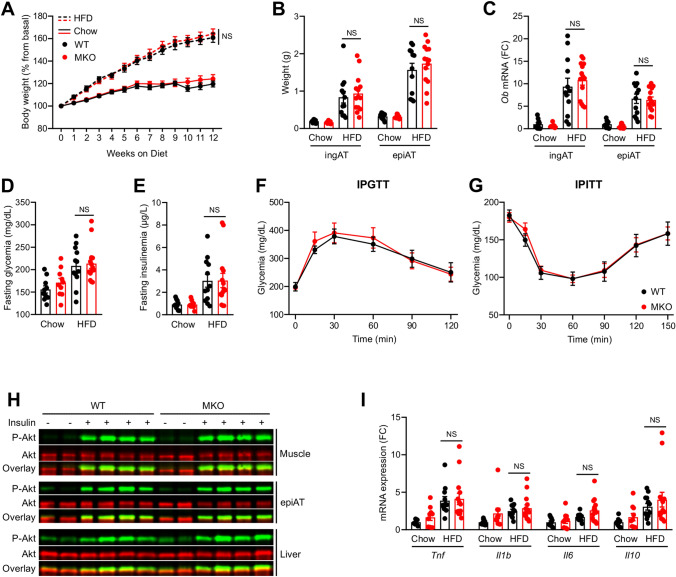
To investigate the role of macrophages RORα in metabolic functions, in particular obesity and IR, we fed WT and MKO mice with a 60% high fat diet (HFD) for 12 weeks. WT and MKO mice under HFD significantly gained weight, AT mass and increased leptin expression similarly in both genotypes (Fig. 2A–C). Fasting serum glucose and insulin concentrations increased significantly upon HFD feeding although no difference between genotypes was observed (Fig. 2D,E). Furthermore, we observed no difference in glucose levels between WT and MKO mice during IPGTT and IPITT (Fig. 2F,G). Moreover, insulin signaling in skeletal muscle, epididymal AT and liver was similar in both genotypes (Fig. 2H and S2A). AT inflammation is a central driver for obesity-induced IR4. No difference in Tnf, Il1b, Il6, and Il10 expression was observed in epididymal AT between WT and MKO mice (Fig. 2I). Likewise, plasma cholesterol and triglyceride levels were not different between WT and MKO mice (Fig. S2B,C). Taken together, our results show that RORα deletion in macrophages has no impact on HFD-induced obesity and IR.
Figure 2.
Effect of RORα deletion in macrophages on HFD-induced obesity and IR. Ten weeks old WT and MKO mice were fed with either a chow or a HFD for 12 weeks. (A) Body weight gain. (B) Weight of inguinal (ingAT) and epidydimal (epiAT) adipose tissues. (C) mRNA expression levels measured by RT-qPCR for the Ob gene in both ingAT and epiAT. (D,F) After 10 weeks of diet, mice were fasted for 5 h and glycemia (D) and insulinemia (E) were measured, then intraperitonealy injected with 1 g/kg glucose for a glucose tolerance test (IPGTT) (F). (G) Intraperitoneal insulin tolerance test (IPITT) was performed by injecting 1 IU/kg insulin after 11 weeks of diet and 5 h of fasting. (H) Western blot of phospho-AKTSer473 in skeletal muscle, epiAT and liver. Mice were injected with either PBS or 1 IU of insulin 15 min before sacrifice. Western blots were scanned with an Odyssey CLx Imaging System (LI-COR) and with the Image studio Version 4.0.21 software (LI-COR, https://www.licor.com/bio/image-studio/). Images were cropped for sake of clarity and full-length blots are presented in Supplementary Figs. 7 to 9. (I) mRNA expression levels measured by RT-qPCR for Tnf, Il1b, Il6, and Il10 genes in epiAT. Data are shown as mean ± SEM. 2-way ANOVA followed by Sidak’s multiple comparisons test was performed. All statistical analyses were carried out using GraphPad Prism 8 for Windows (GraphPad Software). n = 10–15 mice per group. NS: Not significant; FC: Fold Change. See also Fig.S2.
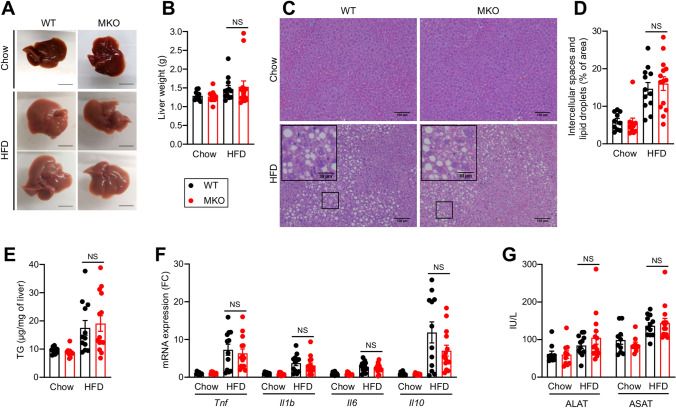
In interconnection with obesity and IR, HFD feeding also induces liver steatosis. Liver weight, steatosis level determined by histology and hepatic triglyceride content were increased upon HFD feeding to the same extent in WT and MKO mice (Fig. 3A–E). Histology analysis did not reveal cell infiltrates upon HFD feeding despite the significantly increased expression of inflammatory genes such as Tnf, Il1b, Il6 and Il10 (Fig. 3F). Likewise, we did not observe any significant differences in hepatic cytokine expression between WT and MKO mice besides a tendency to a lower Il10 expression in MKO mice (p = 0.07). HFD feeding induced a similar mild increase of plasma transaminase activity in both WT and MKO mice (Fig. 3G). Liver fibrosis and expression of collagen genes slightly increased upon HFD feeding for 12 weeks at comparable level between WT and MKO mice (Fig. S3A–C).
Figure 3.
Effect of RORα deletion in macrophages on liver upon HFD feeding. Ten weeks old WT and MKO mice were fed with either a chow or a HFD for 12 weeks. (A) Representative liver gross morphology. (B) Liver weight. (C, D) Hematoxylin & Eosin staining of liver sections (C) and quantification of lipid droplets (D). Images were acquired on an Eclipse Ti-U microscope (Nikon) and quantified with Image J version 1.51j8 software (NIH, https://imagej.nih.gov/ij/). (E) Hepatic triglycerides (TG) content. (F) mRNA expression levels measured by RT-qPCR for Tnf, Il1b, Il6, and Il10 genes in liver. (G) Plasma transaminases activity. Data are shown as mean ± SEM. 2-way ANOVA followed by Sidak’s multiple comparisons test was performed. All statistical analyses were carried out using GraphPad Prism 8 for Windows (GraphPad Software). n = 10–15 mice per group. NS: Not significant; FC: Fold Change. See also Fig. S3.
RORα deletion in macrophages does not affect NASH
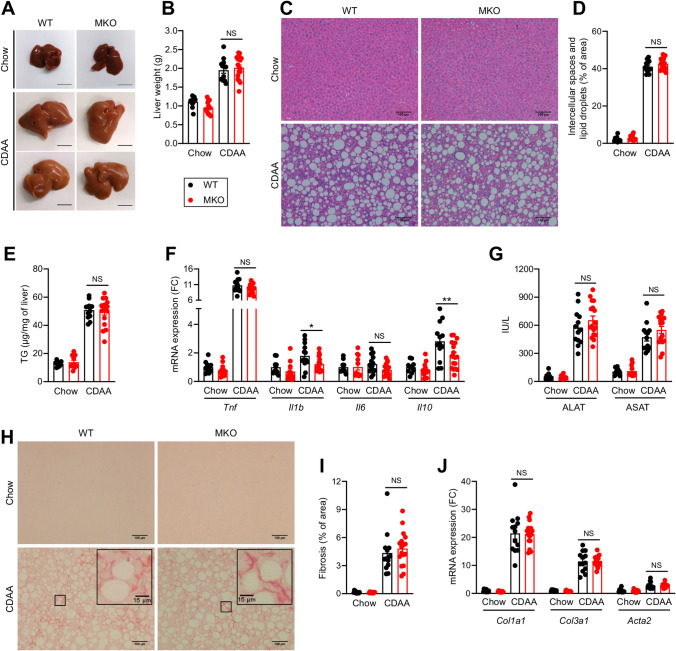
While HFD feeding efficiently promotes obesity and IR, the liver pathology is mainly characterized by triglyceride accumulation corresponding to the NAFL stage9. To investigate whether the deletion of RORα in macrophages might play a role in advanced NAFLD, WT and MKO mice were fed with the choline-deficient, L-amino acid-defined (CDAA) diet supplemented with sucrose and 2% cholesterol for 8 weeks, leading to a pronounced hepatic steatosis, inflammation and fibrosis, but dissociated from obesity and IR allowing the analysis of liver-centric responses9. As expected and unlike HFD, the CDAA diet did not induce obesity, hyperglycemia nor AT inflammation, but led to a 10% weight loss and a drop of glycemia of 30% in both WT and MKO (Fig. S4A–C). The CDAA diet produced a massive hepatic steatosis, but RORα deletion in macrophages did not impact on liver weight nor steatosis level (Fig. 4A–E). Histological analysis showed evidences of cell infiltrates upon CDAA diet feeding but no manifest difference between WT and MKO mice was observed (Fig. 4C & S4D). Further analysis of the expression of inflammatory genes revealed no difference in Tnf and Il6 expression whereas Il1b and Il10 mRNA significantly decreased in livers of MKO mice (Fig. 4F). Despite this modest decrease in cytokine expression, we did not observe any significant differences in plasma transaminase activity nor fibrosis, while a strong effect of the CDAA diet was found on these parameters (Fig. 4G–J & S4E,F). These results indicate that deletion of RORα in macrophages does not affect CDAA-induced NASH.
Figure 4.
Effect of RORα deletion in macrophages on liver upon CDAA diet feeding. Ten weeks old WT and MKO mice were fed with either a chow or a CDAA diet for 8 weeks. (A) Representative liver gross morphology. (B) Liver weight. (C,D) Hematoxylin & Eosin staining of liver sections (C) and quantification of lipid droplets (D). Images were acquired on an Eclipse Ti-U microscope (Nikon) and quantified with Image J version 1.51j8 software (NIH, https://imagej.nih.gov/ij/). (E) Hepatic triglycerides (TG) content. (F) mRNA expression levels measured by RT-qPCR for Tnf, Il1b, Il6, and Il10 genes in liver. (G) Plasma transaminases activity. (H,I) Sirius red staining of liver sections (H) and quantification of fibrosis (I). Images were acquired on an Eclipse Ti-U microscope (Nikon) and quantified with Image J version 1.51j8 software (NIH, https://imagej.nih.gov/ij/). (J) mRNA expression levels measured by RT-qPCR for Col1a1, Col3a1, and Acta2 genes in the liver. Data are shown as mean ± SEM. *p < 0.05 by 2-way ANOVA followed by Sidak’s multiple comparisons test. All statistical analyses were carried out using GraphPad Prism 8 for Windows (GraphPad Software). n = 10–17 mice per group. NS: Not significant; FC: Fold Change. See also Fig. S4.
Effect of RORα deletion in KC is offset by high RORγ expression
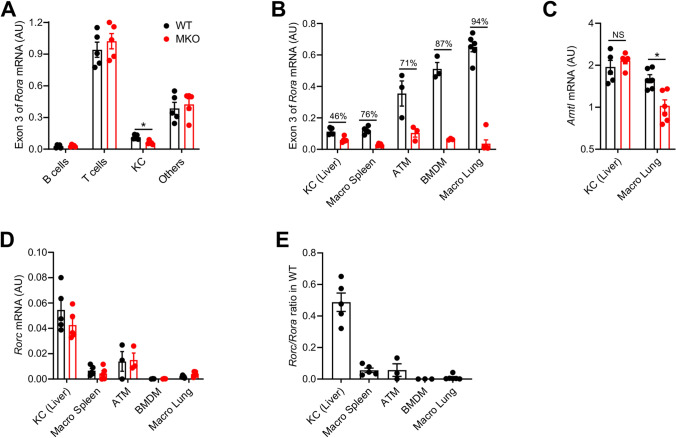
Contrasting with these results in 2 independent and complementary models of NAFLD, it was reported that RORα deletion in macrophages, achieved by using a similar strategy (Rorafl/fl Lyz2Cre), predisposes mice to NASH after 12 weeks of HFD feeding31. NASH exacerbation was attributed to a key role of RORα in Kupffer Cell (KC) function31. To determine a possible cause for this discrepancy, we first assessed the extent of Rora exon 3 deletion in KC from chow diet fed mice. KC and other liver immune cell populations, sorted to a high level of purity (Fig. S5A–G), revealed efficient and specific deletion of Rora exon 3 in KC (Fig. 5A). Interestingly, and similarly to publicly available transcriptomic data (Fig. S5H), we observed that KC expressed lower levels of Rora than other macrophage populations, with lung macrophages expressing the highest level (Fig. 5B).
Figure 5.
Impact of RORα deletion on Kupffer Cells (KCs). (A,B) mRNA expression levels measured by RT-qPCR for the exon 3 of Rora gene in B cells, T cells, KCs, and CD45+ non T, B or KC (others) sorted from liver (A) and in different macrophage populations (B) from WT and MKO mice fed with chow diet. (C,D) mRNA expression levels measured by RT-qPCR for Arntl (C) and Rorc (D) genes in different macrophage populations. (E) Rorc/Rora ratio in different macrophage populations from WT mice. Data are shown as mean ± SEM. *p < 0.05 by 2-way ANOVA followed by Sidak’s multiple comparisons test. All statistical analyses were carried out using GraphPad Prism 8 for Windows (GraphPad Software). n = 3–6 mice per genotype except for ATM where each dot represents a pool of 3–4 mice. NS not significant, AU arbitrary unit. See also Fig. S5.
Despite a significant Rora exon 3 deletion, expression of the ROR target gene Arntl did not decrease in KC, unlike lung macrophages (Fig. 5C), splenic macrophages (Fig. 1F), BMDM (Fig. 1G) and ATM (Fig. 1H). The lack of effect of RORα deletion on Arntl expression in KC suggests a compensatory mechanism by another ROR protein. Rorb was not expressed in KC, similarly to other macrophage populations tested (data not shown), but Rorc expression was detected (Fig. 5D). No compensation of Rorc expression was found in any macrophage subsets from MKO mice compared to littermate controls. However, basal Rorc expression was between 5- and tenfold higher in KC compared to other macrophage subsets. The low Rora expression and the high Rorc expression in KC led to a Rorc/Rora ratio 10- to 50-fold higher than in other macrophage populations (Fig. 5E). Collectively, these results suggest that RORα deficiency is unlikely to induce major transcriptional effect in KC due to a concomitant high RORγ expression.
Discussion
Over the past years, a considerable number of studies contributed to our understanding about the role of nuclear receptors, including RORs, in the regulation of lipid and glucose metabolism, circadian rhythm as well as the development and function of the immune system. Due to the pleiotropic effects and wide cell distribution of RORα and considering the key role of macrophages in metabolic diseases, we investigated the impact of RORα deletion in these cells on obesity, IR and NASH by using the LysM-Cre mice. Evaluation of deletion efficiency and decreased expression of a major RORα target gene in several myeloid subsets showed that macrophages were mostly impacted by RORα deletion in basal conditions although we cannot formally exclude that other myeloid subsets are also affected upon metabolic challenge.
Yet, importantly, we found no impact of LysM-Cre-mediated RORα deletion neither on HFD-induced obesity, IR and steatosis nor on CDAA diet-induced NASH. These findings contrast with an earlier report showing that LysM-Cre-mediated RORα deletion increases the susceptibility to HFD-induced NASH31, while both studies used the same HFD reference and duration of treatment. As feeding a HFD for 12 weeks is not per se leading to NASH, but rather to NAFL9, only a more pronounced steatosis (higher liver weight and triglyceride content) was reported in MKO mice in the earlier study31. Indicators of increased hepatic lipotoxicity such as higher plasma transaminase activity, increased expression of pro-inflammatory cytokines (Tnf, Il1b and Il6) and decreased expression of Il10 in liver were also observed31. It was proposed that RORα directly regulates M2 polarization of KCs leading to increased IL-10 production which would protect hepatocytes against lipid accumulation31. While we also observed a tendency to lower Il10 expression in livers from MKO mice upon HFD feeding (Fig. 3F) and a significant decrease upon CDAA diet feeding (Fig. 4F), we did not observe any exacerbation of steatosis or NASH. As the putative protective role for IL-10 was only based on in vitro findings, we believe that the reduced Il10 expression in liver of MKO mice upon HFD is unlikely to contribute to aggravated steatosis. Further supporting this finding, IL-10-deficient mice fed a HFD for 12 weeks developed increased liver inflammation, but decreased steatosis and transaminase activity32, suggesting that beyond its well described anti-inflammatory function, endogenous IL-10 is not protective against HFD-induced steatosis.
Several factors might account for the discrepancy between our results and these earlier findings31. The microbiological status of the animal facility and housing conditions are two underestimated factors that might account for major differences in experimental outcome and it is not uncommon for this information to be missing. Specific pathogen free (SPF) status indicates that mice are free of defined pathogens including the mouse hepatitis virus and Helicobacter hepaticus which may interfere with liver function. Gut microbiota also impacts on NAFLD development33. The microbiota composition is highly variable between animal facilities, mouse lines and even between cages, especially when two genotypes are bred separately. Generation of littermate animals is critical to insure that both deficient and control mice harbor identical gut microbiota while post-weaning cohousing is ineffective34. Our mice were housed in SPF facility and bred to obtain WT and MKO littermate animals. No information was provided in the earlier study about the animal facility and littermate status.
Two additional factors that may also account for the observed discrepancies between studies are the floxed mice and the breeding strategy used. We generated mice with floxed Rora exon 3 while exon 4 was targeted in the earlier study31. Deletion of exon 4 may still result in the translation of a truncated protein containing a portion of the DBD that includes the exon 3-encoded zinc finger motif (Fig. S1C). This difference in deletion strategy is however unlikely to account for the observed phenotypic differences. The selected control animals represent a second major difference between the studies. The LysM-Cre mice carry an insertion of Cre recombinase into the Lyz2 gene, leading to Cre expression under the control of the Lyz2 promoter and enhancers, but abolishing endogenous Lyz2 expression. While we intentionally maintained similar Cre and Lyz2 expression between WT and MKO by using only hemizygous animals for this locus (comparing Rora+/+ Lyz2Cre/+ with Rorafl/fl Lyz2Cre/+), floxed mice (Rorafl/fl Lyz2+/+) were used as WT control and compared with MKO mice missing information about the Lyz2 locus (Rorafl/fl Lyz2Cre/?) in the earlier study31. This latter strategy results in differential Cre and Lyz2 expression between the WT, expressing no Cre, and the MKO mice expressing less or no Lyz2. Even though mammalian genomes possess no loxP sites, active Cre recombinase recognition sites, called pseudo loxP sites, have been reported35 and Cre expression mediates DNA damage, cell toxicity and apoptosis even in the absence of floxed alleles36. In addition to RORα deletion and exogenous Cre expression in macrophages, MKO mice generated with this strategy possess also at least one inactivated Lyz2 allele and possibly a homozygous (whole body) Lyz2 deletion. Indeed, based on the described genotyping strategy using a single PCR reaction to only analyze the Cre gene31, it is highly likely that MKO mice were actually also Lyz2 deficient.
Lysozyme is an antimicrobial protein that catalyzes the hydrolysis of peptidoglycan between N-acetylglucosamine and N-acetylmuramic acid, contributing to the degradation of the Gram-positive bacterial cell wall. In humans, lysozyme is encoded by a single gene LYZ, whereas two genes exist in mouse, Lyz1 and Lyz2 encoding for lysozyme P and lysozyme M respectively. Lyz2 is the most expressed lysozyme gene in mice with a high expression in the myeloid lineage, similar to the human LYZ gene. Expectedly, Lyz2-deficient mice develop more severe bacterial infections37–39. In addition to its antimicrobial properties, lysozyme is also an anti-inflammatory factor. Lysozyme inhibits serum complement activation40 and possesses a LPS-binding ability that reduces LPS-related inflammation41. Peptides derived from lysozyme cleavage inhibit production of pro-inflammatory cytokines, including TNF-α and IL-1β, by macrophages42. Moreover, lysozyme improves the antioxidant capacity of hepatocytes in vitro and in vivo, leading to a protection against oxidative stress in liver43. Lysozyme expression and activity is affected in various liver diseases44–46, but, to our knowledge, the role of Lyz2 in NASH was never investigated. These roles in infection, inflammation and probably in liver homeostasis, suggest that Lyz2 may play a role in NASH development. Thus, caution should be exerted when comparing mouse lines with different Lyz2 copy numbers.
By using a 12-week HFD that induces liver steatosis (NAFL) or an 8-week CDAA diet that leads to NASH, we observed no differences between WT and MKO mice for any tested hepatic parameters including liver weight, triglyceride content, inflammatory and fibrosis gene expression, histology and transaminase activity. We also found no impact on obesity and glucose homeostasis. Investigations on KC showed that, despite an effective deletion of RORα, KCs were the only macrophage population without transcriptional effect on the ROR target gene Arntl. We found no compensatory expression of other ROR genes in MKO, but we evidenced a high level of Rorc expression specifically in KC that might offset the RORα deletion. The redundancy between ROR proteins is well established17,47–49. Hepatocytes express high levels of RORγ in addition to RORα48. It was shown that a single deletion of RORα or RORγ in hepatocytes affects only 2 and 6 transcripts respectively in whole liver transcriptome, while RORα/γ deletion leads to a broader effect with 299 genes affected, unambiguously demonstrating the redundancy between RORα and RORγ in hepatocytes49. Such a redundancy might be at play in KCs and further investigation with a double RORα/γ deficient mouse model might address this question. Moreover, usage of the new KC-specific Clec4f-Cre mouse line would improve the specificity towards KC and may result in a better deletion efficiency than the one reached with the LysM-cre mice50.
In conclusion RORα deletion in macrophages using the LyzM-Cre system has no impact on the development of obesity, IR and NASH. We suggest that the previously reported impact of RORα deletion in macrophages on NASH31 likely does not result from a specific effect of RORα deletion, but rather from a different Cre or Lyz2 copy number between WT and MKO.
Methods
Generation of RORα MKO Mice
We generated mice harboring a floxed allele of Rora (Rorafl/fl) by flanking exon 3 with two loxP sequences. The strategy and outcome on RORα1 and RORα4 are summarized in figures S1A and S1B respectively. Briefly, a targeting vector containing loxP sites, a FRT-floxed neomycin cassette and homologous regions surrounding exon 3 was constructed and transfected in embryonic stem (ES) cells derived from 129/Sv. After screening for homologous recombination by southern blot and PCR, ES cells were injected into C57BL/6 blastocysts. Neomycin cassette was removed in vivo by using FLP deleter mice in C57BL/6 background. Finally, mice were backcrossed with C57BL/6 J mice for at least six generations.
To achieve the deletion of RORα in macrophages, Rorafl/fl mice were crossed with LysM-Cre transgenic mice (Jackson laboratory) in which Cre recombinase is expressed under the control of endogenous Lyz2 promoter. Littermates RORα-deficient (MKO, Rorafl/fl Lyz2Cre/+) and WT (Rora+/+ Lyz2Cre/+) mice were generated by crossing Rorafl/+ Lyz2Cre/Cre with Rorafl/+ Lyz2+/+ mice. All mice were genotyped twice.
Mouse studies
Mice were kept on a 12-h light/dark cycle in the SPF animal facility from the Institut Pasteur de Lille with ad libitum access to food and water. Littermate WT and MKO mice were maintained all along the experiment procedures in 904 cm2 cages (Green line GR900, Tecniplast) with 6–12 mice per cage and a ratio WT:MKO tending to 1:1. Only male mice were used for the experiments. All animal procedures were approved by the ethical committee for animal experimentation of the Nord-Pas-de-Calais Region (CEEA75) (APAFIS#7160-2017040313471173) in accordance with European guidelines on the protection of animals used for scientific purposes (2010/63/UE).
Ten-week-old RORα WT and their littermates RORα MKO mice were fed with a 60% high-fat diet (HFD, Research Diet, D12492) for 12 weeks or maintained under chow diet (SAFE, #A04). To induce a NASH-like disease, ten-week-old RORα WT and MKO mice were fed with a choline-deficient, L-amino acid-defined (CDAA) diet with 35% sucrose, 21% fat and 2% cholesterol (Ssniff, custom diet) for 8 weeks. In addition to CDAA diet, mice also received monosaccharides in the drinking water (42 g/L, fructose:glucose ratio of 55:45). Weight of mice was measured weekly. Before sacrifice, mice were fasted for 5 h. Mice were sacrificed at ZT3 (10 am) for HFD and at ZT7 (2 pm) for CDAA experiment.
Mouse genotyping
DNA was extracted from tail with REDExtract-N-Amp Tissue PCR Kit (Sigma, #XNAT-1000RXN). Floxed Rora was detected by PCR with the Rora genotyping primers (Supplementary Table 1) and the following cycling conditions: 1 cycle at 94 °C for 3 min; 35 cycles at 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min; and 1 cycle at 72 °C for 10 min; hold at 4 °C. Samples were separated by gel electrophoresis on a 1.5% agarose gel. Rora+/+ gave a single band at 250 bp, Rorafl/fl at 340 bp and Rorafl/+ had both bands. Endogenous Lyz2 and Cre were detected with the Lyz2 genotyping and Cre genotyping primers respectively (Supplementary Table 1) and the following cycling conditions: 1 cycle at 94 °C for 3 min; 35 cycles at 94 °C for 1 min, 63 °C for 1 min, 72 °C for 90 s; and 1 cycle at 72 °C for 10 min; hold at 4 °C. Samples were separated by gel electrophoresis on a 1.5% agarose gel. Lyz2+/+ gave a single band at 350 bp for Lyz2 PCR while Lyz2Cre/Cre gave a single band at 700 bp for Cre PCR. Lyz2Cre/+ gave bands for both Lyz2 and Cre PCR. Agarose gels were acquired with a Gel Doc XR system (Bio-Rad) and the Image Lab software verion 2.0 build 8 for PC (Bio-Rad, https://www.bio-rad.com/fr-fr/product/image-lab-software?ID=KRE6P5E8Z). The following acquisition settings were chosen: application: SYBR Safe; Image exposure: automatically optimized.
Bone marrow–derived macrophages
Bone marrow was isolated from tibia and femur of mice. Bone marrow cells were culture in RPMI 1640 with Hepes and L-glutamine supplemented with 10% fetal bovine serum, 20% L929-conditioned medium and 25 µg/mL gentamycin for one week. After three days of culture, fresh medium was added. After one week of culture, supernatant was discarded and adherent bone marrow–derived macrophages (BMDMs) were washed two times with PBS. BMDMs were collected by using a cell scraper, counted and plated at a concentration of 106 cells/ml. After 24 h, BMDMs were treated with 100 nM dexamethasone (Sigma, #D1756) for 2 h to synchronize cells, washed and maintained in complete medium for 32 h to reach the pic of RORα activity corresponding to ZT0 in vivo.
Insulin and glucose tolerance tests
After 10 weeks of HFD, a Glucose Tolerance Test (GTT) was performed by intraperitoneal injection of glucose (1 g/kg). Tail blood sample was collected before glucose injection to measure fasting insulin by ELISA (Mercodia #10-1247-10) according to manufacturer instruction. After 11 weeks of HFD, an Insulin Tolerance Test (ITT) was performed by intraperitoneal injection of human insulin (1 IU/kg) (Actrapid, Novo Nordisk). Glycemia was measured from tail before and 15, 30, 60, 90 and 120 min after glucose or insulin injection by using a glucose meter (Accu-Check performa, Roche). Before GTT and ITT, mice were fasted for 5 h at ZT2 (9 am) and the tests were performed at ZT7 (2 pm).
Cell sorting
Chow diet fed mice were sacrificed at ZT0 (7 am) and tissue kept in PBS on ice until processing. For epididymal adipose tissue, between 3 and 4 mice were pooled. Spleen was gently pressed on a 70 µm cell strainer by using a 1 mL syringe plunger. Adipose tissue, liver and lung were minced with scissors and digested in RPMI containing 1 mg/mL collagenase D (Roche # 11088882001) at 37 °C under agitation for 30, 45 and 60 min respectively and then carefully passed several times through an 18 or 19 G needle to obtain a single cell suspension. Red blood cells were lysed with ammonium chloride-based buffer and cells were blocked by using a combination of anti-CD16/CD32 (clone 2.4G2) (BD Biosciences, #553142) and anti-FcγRIV (clone 9E9) (BioLegend, #149502). Antibodies used for staining are provided in Supplementary Table 2. Cell suspension was directly run into an Influx sorter (Becton Dickinson) (Plateau d’Immunophenotypage Metabolique, Inserm U1011) equipped with an 86 µm nozzle and tuned at a pressure of 24.7 psi and a frequency of 48.25 kHz. Sample fluid pressure was adjusted to reach an event rate of maximum 10 000 events/sec. In spleen, cells of interest were selected in the following order: T cells (CD45+ CD3ε+ TCRβ+ MHC-II−), B cells (CD45+ CD19+ MHC-II+), Neutrophils (CD45+ CD3ε− TCRβ− CD19− CD11b+ Ly6G+), Macrophages (CD45+ CD3ε− TCRβ− CD19− Ly6G− F4/80+), monocytes (CD45+ CD3ε− TCRβ− CD19− Ly6G− F4/80− CD11b+ CD115+) and DCs (CD45+ CD3ε− TCRβ− CD19− Ly6G− F4/80− CD115− CD11c+ MHC-II+). In liver, cells of interest were selected in the following order: T cells (CD45+ CD3ε+ TCRβ+ CD19− CD20−), B cells (CD45+ CD19+ CD20+ MHC-II+), Kupffer cells (CD45+ CD3ε− TCRβ− CD19− CD20− F4/80+ Clec4F+) and other (CD45+ CD3ε− TCRβ− CD19− CD20− F4/80− Clec4F−). Adipose tissue macrophages were selected as CD45+ CD3ε− TCRβ− CD19− CD20− F4/80+ CD64+ and lung macrophages as CD45+ CD3ε− TCRβ− CD19− F4/80+ CD64+ CD11blow SiglecF+.
For bone marrow (BM) neutrophils, BM was first isolated from tibia and femur of mice. Red blood cells were lysed with ammonium chloride-based buffer and BM cells were blocked by using a combination of anti-CD16/CD32 (clone 2.4G2) (BD Biosciences, #553142) and anti-FcγRIV (clone 9E9) (BioLegend, #149502). Cells were incubated with PE rat anti-mouse Ly-6G (Clone 1A8) (BD Biosciences, #551461) for 30 min and purified by magnetic separation with anti-PE MicroBeads (Miltenyi Biotec, #130-048-801) according to the manufacturer’s instructions.
Histology
The median lobes of liver were collected and fixed in 4% paraformaldehyde for 48 to 72 h. Tissues were embedded in paraffin by using a STP 120 Spin Tissue Processor (Microm Microtech) and an EG1160 Tissue Embedding Station (Leica). Paraffin-embedded samples were cut at a thickness of 3 μm and sections were transferred on a gelatin-coated slide for hematoxylin and eosin (H&E) staining and on a Superfrost Plus slides (Thermo Scientific, # J1800AMNZ) for Sirius Red staining. Staining was performed with a Leica autostainer XL and the following steps for H&E staining: xylene (2 min), xylene (2 min), 100% ethanol (2 min), tap water (2 min), hematoxylin (Sigma, #HHS128) (3 min), tap water (2 min), 70% ethanol 0.25% HCl (6 s), tap water (2 min), 90% ethanol (2 min), eosin (Sigma, #HT1101128) (2 min), 90% ethanol (6 s), 100% ethanol (1 min) and finished in xylene before mounting with Mercoglas (Merck, #103973). For Sirius red staining, the process was the following: xylene (2 min), xylene (2 min), 100% ethanol (2 min), tap water (2 min), 0.1% Direct Red 80 (Sigma, #365548) in saturated picric aqueous solution (60 min), tap water (8 min), 100% ethanol (1 min) and finished in xylene before mounting with Mercoglas. Images were acquired on an Eclipse Ti-U microscope (Nikon) and quantified with Image J version 1.51j8 software (NIH, https://imagej.nih.gov/ij/).
Metabolic parameters
Before sacrifice and after 5 h of fasting, blood samples were collected from the retro orbital sinus of mice. Plasma alanine aminotransferase (ALAT), aspartate aminotransferase (ASAT), total cholesterol and triglycerides were measured on a Konelab 20 (Thermo Fisher) with reagents from Thermo Scientific for ALAT (#981769) and ASAT (#981771) and reagents from DiaSys for cholesterol (#113009910026) and triglycerides (#157109910026). The plasma ALAT and ASAT measurement under CDAA diet was performed without fasting for the time point T0, 2, 4 and 6 weeks.
Measurement of liver triglycerides
Lipids were extracted from the liver caudate lobe. A weighted piece of tissue was homogenized with T10 Ultra-Turrax (Ika) in PBS 1% Triton. Samples were transferred into glass tubes and mixed with a 2:1 chlorofrom:methanol mixture. After centrifugation, upper- and inter-phase were discarded. The lower organic phase was evaporated under nitrogen flow and reconstituted in 1% Triton X100. Triglyceride content was measured with Triglycerides FS kit (DiaSys, #157109910026).
RT-qPCR
Total RNA was extracted from the snap-frozen tissues or sorted cell pellets by using TRIzol reagent (Ambion, #15596018) according to the manufacturer’s instructions. DNase treatment was performed (Thermo Scientific, #EN0521) and RNA was reverse-transcribed to complementary DNA (cDNA) by using the high capacity cDNA reverse transcription kit (Applied Biosystems, #4368813). qPCR was performed by using Brilliant II SYBR Green QPCR Master Mix (Agilent, #600828) and ran on a Mx3000P qPCR system (Agilent). Relative changes in mRNA expression (Fold change, FC) were calculated by the 2−ΔΔCT method. Absolute changes in mRNA expression were calculated with the formula 2−ΔCT × 100 and expressed as arbitrary unit (AU). The mean of four housekeeping genes (Rplp0, Ppia, Rpl4 and Rps29) was used to normalize mRNA expression. The primers used for qRT-PCR are listed in Supplementary Table 1.
Western blotting
Tissues were weighted and homogenized with a T10 Ultra-Turrax (Ika) in protect buffer (water with cOmplete protease inhibitor cocktail (Roche, #11836145001) and PhosSTOP (Roche, #04906837001)) (3.75 µL/mg for adipose tissue and 7.5 µL/mg for muscle and liver). Next, total protein lysates were mixed with 4X Laemmli buffer (Tris–HCl 250 mM pH 6.8, 40% glycerol, 8% SDS, 12% β-mercaptoethanol and bromophenol blue) and heated at 100 °C for 5 min. Total protein extracts were subjected to SDS-PAGE (10% polyacrylamide gel) and transferred on a 0.22 µm nitrocellulose membrane (Li-Cor, #P/N 926-31092). After blocking in 5% BSA, membranes were probed with mouse anti-Akt (Cell Signaling, #2920) and rabbit anti-phospho-Akt (Ser473) (Cell Signaling, #4058) monoclonal antibodies. The secondary antibodies used for the revelation were Alexa Fluor 680 anti-mouse IgG (Jackson ImmunoResearch, #715-625-150) and Alexa Fluor 790 anti-rabbit IgG (Jackson ImmunoResearch, #711-655-152). Immunoblots were scanned with an Odyssey CLx Imaging System (LI-COR) and with the Image studio Version 4.0.21 software (LI-COR, https://www.licor.com/bio/image-studio/). The following acquisition settings were chosen: setup: Western; acquisition channels: auto; scan resolution: 169 µm; scan quality: low; focus offset: 0.0 mm. Images were quantified with the Image studio Lite Version 4.0.21 software (LI-COR, https://www.licor.com/bio/image-studio-lite/).
Statistical analyses
All statistical analyses were carried out using GraphPad Prism 8 for Windows (GraphPad Software) and presented as means ± SEM. The study was done blinded for genotype. Only one mouse (WT) was excluded from the analysis because it did not reach an appropriate weight gain under HFD feeding (Body weight: 31.2 g; ingAT: 0.165 g; epiAT: 0.348 g). No mice were excluded in CDAA diet group. Data were analyzed with 2-way ANOVA and Sidak’s multiple comparisons post-hoc test or Student's t-test. Values with P < 0.05 were considered as significant. All statistical details including statistical test used and exact value of n are described in each figure legend. Each data point represents genuine replication, also called true replicate, and were obtained from a single measurement or multiple measurements illustrated by the mean, such as gene expression by RT-qPCR that was assessed in duplicate or histology quantification made on five random fields per mice. For sake of clarity, results of statistical comparisons between WT and MKO mice upon Chow feeding were not illustrated on the figures but were not significant for all the parameters. In Figs. 2, 3, and 4, the diet effect was analyzed by 2-way ANOVA and was significant for all the parameters with the exception of liver weight (Fig. 3B) that was not statistically significant between chow and HFD (p = 0.06). The datasets used in this study are available from GEO under the accession number GSE10434251 and GSE5668252 and from data assembled by the ImmGen consortium (http://www.immgen.org/)53.
Supplementary information
Acknowledgements
This work was supported in part by grants from the ANR and the European Union: EGID ANR-10-LABX-46 (to DD & BS), OSEO Anvar (to BS), European Foundation for the Study of Diabetes (EFSD) (to DD), Fondation Francophone pour la Recherche sur le Diabète (FFRD) (to DD), Fondation pour la Recherche Médicale (FRM, grant number DPC20171138951) (to DD) and by the University of Liege and the European Union in the context of the FP7-PEOPLE-COFUND-BeIPD project (to LL). BS is a recipient of an Advanced European Research Council (ERC) grant (number 694717). We thank Dr S. Ivanov (Inserm U1065, Nice, France) for his critical reading of the manuscript.
Author contributions
Conceptualization: L.L., B.P.S., O.M.C., B.S., D.D.; Formal Analysis: L.L., B.P.S.; Funding acquisition: L.L., B.S., D.D.; Investigation: L.L., B.P.S., O.M.C., S.F., S.Q., M.L.M., M.L.J., L.P., C.D., B.G., E.V.; Methodology: L.L., B.P.S., O.M.C., B.S., D.D.; Project administration: B.S., D.D.; Resources: L.L., B.P.S., O.M.C., S.Q., C.D., B.G., E.V., A.M.T.; Supervision: B.S., D.D.; Visualization: L.L., B.P.S.; Writing—original draft: L.L., B.P.S., D.D.; Writing—review & editing: L.L., B.P.S., O.M.C., S.F., S.Q., M.L.M., M.L.J., L.P., C.D., B.G., E.V., A.M.T., B.S., D.D.
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by David Dombrowicz (david.dombrowicz@inserm.fr).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Laurent L’homme, Benan Pelin Sermikli and Olivier Molendi-Coste.
These authors jointly supervised this work: Bart Staels and David Dombrowicz.
Supplementary information
is available for this paper at 10.1038/s41598-020-77858-6.
References
- 1.Ng M, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2014;37(Suppl 1):S81–90. doi: 10.2337/dc14-S081. [DOI] [PubMed] [Google Scholar]
- 3.Esser N, Paquot N, Scheen AJ. Anti-inflammatory agents to treat or prevent type 2 diabetes, metabolic syndrome and cardiovascular disease. Expert Opin. Investig. Drugs. 2015;24:283–307. doi: 10.1517/13543784.2015.974804. [DOI] [PubMed] [Google Scholar]
- 4.Grant RW, Dixit VD. Adipose tissue as an immunological organ. Obes. Silver Spring MD. 2015;23:512–518. doi: 10.1002/oby.21003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng B, et al. Clodronate liposomes improve metabolic profile and reduce visceral adipose macrophage content in diet-induced obese mice. PLoS ONE. 2011;6:e24358. doi: 10.1371/journal.pone.0024358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patsouris D, et al. Ablation of CD11c-positive cells normalizes insulin sensitivity in obese insulin resistant animals. Cell Metab. 2008;8:301–309. doi: 10.1016/j.cmet.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNelis JC, Olefsky JM. Macrophages, immunity, and metabolic disease. Immunity. 2014;41:36–48. doi: 10.1016/j.immuni.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Bai Y, Sun Q. Macrophage recruitment in obese adipose tissue. Obes. Rev. Off. J. Int. Assoc. Study Obes. 2015;16:127–136. doi: 10.1111/obr.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrell G, et al. Mouse models of nonalcoholic steatohepatitis: Toward optimization of their relevance to human nonalcoholic steatohepatitis. Hepatol. Baltim. Md. 2019;69:2241–2257. doi: 10.1002/hep.30333. [DOI] [PubMed] [Google Scholar]
- 10.Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M. From NASH to HCC: Current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 2019;16:411–428. doi: 10.1038/s41575-019-0145-7. [DOI] [PubMed] [Google Scholar]
- 11.Azzu V, Vacca M, Virtue S, Allison M, Vidal-Puig A. Adipose tissue-liver cross talk in the control of whole-body metabolism: Implications in nonalcoholic fatty liver disease. Gastroenterology. 2020;158:1899–1912. doi: 10.1053/j.gastro.2019.12.054. [DOI] [PubMed] [Google Scholar]
- 12.Huang W, et al. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes. 2010;59:347–357. doi: 10.2337/db09-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koyama Y, Brenner DA. Liver inflammation and fibrosis. J. Clin. Invest. 2017;127:55–64. doi: 10.1172/JCI88881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jetten AM, Joo JH. Retinoid-related orphan receptors (RORs): Roles in cellular differentiation and development. Adv. Dev. Biol. Amst. Neth. 2006;16:313–355. doi: 10.1016/S1574-3349(06)16010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jetten AM, Kang HS, Takeda Y. Retinoic acid-related orphan receptors α and γ: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Front. Endocrinol. 2013;4:1. doi: 10.3389/fendo.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong SH, et al. Transcription factor RORα is critical for nuocyte development. Nat. Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang XO, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang NS, et al. Divergent transcriptional programming of class-specific B cell memory by T-bet and RORα. Nat. Immunol. 2012;13:604–611. doi: 10.1038/ni.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao L, et al. Retinoid acid receptor-related orphan receptor alpha (RORα) regulates macrophage M2 polarization via activation of AMPKα. Mol. Immunol. 2016;80:17–23. doi: 10.1016/j.molimm.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Klar J, et al. RAR-related orphan receptor A isoform 1 (RORa1) is disrupted by a balanced translocation t(4;15)(q22.3;q21.3) associated with severe obesity. Eur. J. Hum. Genet. 2005;13:928–934. doi: 10.1038/sj.ejhg.5201433. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Liu Y, Liu Y, Zhang Y, Su Z. Genetic variants of retinoic acid receptor-related orphan receptor alpha determine susceptibility to type 2 diabetes mellitus in Han Chinese. Genes. 2016;7:66. doi: 10.3390/genes7090066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chambers JC, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat. Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang HS, et al. Transcriptional profiling reveals a role for RORalpha in regulating gene expression in obesity-associated inflammation and hepatic steatosis. Physiol. Genomics. 2011;43:818–828. doi: 10.1152/physiolgenomics.00206.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau P, et al. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: Staggerer (SG/SG) mice are resistant to diet-induced obesity. J. Biol. Chem. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 25.Han Y-H, et al. RORα decreases oxidative stress through the induction of SOD2 and GPx1 expression and thereby protects against nonalcoholic steatohepatitis in mice. Antioxid. Redox Signal. 2014;21:2083–2094. doi: 10.1089/ars.2013.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Retinoic acid receptor-related orphan receptor α stimulates adipose tissue inflammation by modulating endoplasmic reticulum stress. J. Biol. Chem. 2017;292:13959–13969. doi: 10.1074/jbc.M117.782391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H-J, et al. Liver-specific deletion of RORα aggravates diet-induced nonalcoholic steatohepatitis by inducing mitochondrial dysfunction. Sci. Rep. 2017;7:16041. doi: 10.1038/s41598-017-16077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim K, et al. RORα controls hepatic lipid homeostasis via negative regulation of PPARγ transcriptional network. Nat. Commun. 2017;8:162. doi: 10.1038/s41467-017-00215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molinaro A, et al. Liver-specific RORα deletion does not affect the metabolic susceptibility to western style diet feeding. Mol. Metab. 2019;23:82–87. doi: 10.1016/j.molmet.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abram CL, Roberge GL, Hu Y, Lowell CA. Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J. Immunol. Methods. 2014;408:89–100. doi: 10.1016/j.jim.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han Y-H, et al. RORα induces KLF4-mediated M2 polarization in the liver macrophages that protect against nonalcoholic steatohepatitis. Cell Rep. 2017;20:124–135. doi: 10.1016/j.celrep.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 32.Miller AM, et al. Inflammation-associated interleukin-6/signal transducer and activator of transcription 3 activation ameliorates alcoholic and nonalcoholic fatty liver diseases in interleukin-10-deficient mice. Hepatol. Baltim. Md. 2011;54:846–856. doi: 10.1002/hep.24517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolodziejczyk AA, Zheng D, Shibolet O, Elinav E. The role of the microbiome in NAFLD and NASH. EMBO Mol. Med. 2019;11:66. doi: 10.15252/emmm.201809302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson SJ, et al. Comparison of co-housing and littermate methods for microbiota standardization in mouse models. Cell Rep. 2019;27:1910–1919.e2. doi: 10.1016/j.celrep.2019.04.023. [DOI] [PubMed] [Google Scholar]
- 35.Thyagarajan B, Guimarães MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/S0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 36.Janbandhu VC, Moik D, Fässler R. Cre recombinase induces DNA damage and tetraploidy in the absence of loxP sites. Cell Cycle Georget. Tex. 2014;13:462–470. doi: 10.4161/cc.27271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganz T, et al. Increased inflammation in lysozyme M-deficient mice in response to Micrococcus luteus and its peptidoglycan. Blood. 2003;101:2388–2392. doi: 10.1182/blood-2002-07-2319. [DOI] [PubMed] [Google Scholar]
- 38.Markart P, Korfhagen TR, Weaver TE, Akinbi HT. Mouse lysozyme M is important in pulmonary host defense against Klebsiella pneumoniae infection. Am. J. Respir. Crit. Care Med. 2004;169:454–458. doi: 10.1164/rccm.200305-669OC. [DOI] [PubMed] [Google Scholar]
- 39.Shimada J, et al. Lysozyme M deficiency leads to an increased susceptibility to Streptococcus pneumoniae-induced otitis media. BMC Infect. Dis. 2008;8:134. doi: 10.1186/1471-2334-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogundele MO. A novel anti-inflammatory activity of lysozyme: Modulation of serum complement activation. Mediators Inflamm. 1998;7:363–365. doi: 10.1080/09629359890893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurasawa T, Takada K, Ohno N, Yadomae T. Effects of murine lysozyme on lipopolysaccharide-induced biological activities. FEMS Immunol. Med. Microbiol. 1996;13:293–301. doi: 10.1111/j.1574-695X.1996.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 42.Ibrahim HR, Hamasaki K, Miyata T. Novel peptide motifs from lysozyme suppress pro-inflammatory cytokines in macrophages by antagonizing toll-like receptor and LPS-scavenging action. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2017;107:240–248. doi: 10.1016/j.ejps.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 43.Liu H, et al. Amelioration of oxidant stress by the defensin lysozyme. Am. J. Physiol. Endocrinol. Metab. 2006;290:E824–832. doi: 10.1152/ajpendo.00349.2005. [DOI] [PubMed] [Google Scholar]
- 44.Kelly PM, Heryet AR, McGee JO. Kupffer cell number is normal, but their lysozyme content is reduced in alcoholic liver disease. J. Hepatol. 1989;8:173–180. doi: 10.1016/0168-8278(89)90005-6. [DOI] [PubMed] [Google Scholar]
- 45.Manifold IH, Bishop FM, Cloke P, Triger DR, Underwood JC. Lysozyme in chronic liver disease: A biochemical and histological study. J. Clin. Pathol. 1982;35:815–819. doi: 10.1136/jcp.35.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.vanVliet AC, Bakker WH, Lindemans J, Wilson JH, vanZanten RA. Plasma lysozyme level and reticuloendothelial system function in human liver disease. Clin. Chim. Acta Int. J. Clin. Chem. 1981;113:193–199. doi: 10.1016/0009-8981(81)90153-4. [DOI] [PubMed] [Google Scholar]
- 47.Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 48.Kang HS, et al. Gene expression profiling reveals a regulatory role for ROR alpha and ROR gamma in phase I and phase II metabolism. Physiol. Genomics. 2007;31:281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, et al. The hepatic circadian clock fine-tunes the lipogenic response to feeding through RORα/γ. Genes Dev. 2017;31:1202–1211. doi: 10.1101/gad.302323.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott CL, et al. The transcription factor ZEB2 is required to maintain the tissue-specific identities of macrophages. Immunity. 2018;49:312–325.e5. doi: 10.1016/j.immuni.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoyer FF, et al. Tissue-specific macrophage responses to remote injury impact the outcome of subsequent local immune challenge. Immunity. 2019;51:899–914.e7. doi: 10.1016/j.immuni.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heng TSP, Painter MW. Immunological Genome Project Consortium. The immunological genome project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further information and requests for resources and reagents should be directed to and will be fulfilled by David Dombrowicz (david.dombrowicz@inserm.fr).